Abstract
Background
Differentiating treatment-induced injury from recurrent high-grade glioma is an ongoing challenge in neuro-oncology, in part due to lesion heterogeneity. This study aimed to determine whether different MR features were relevant for distinguishing recurrent tumor from the effects of treatment in contrast-enhancing lesions (CEL) and non-enhancing lesions (NEL).
Methods
This prospective study analyzed 291 tissue samples (222 recurrent tumor, 69 treatment-effect) with known coordinates on imaging from 139 patients who underwent preoperative 3T MRI and surgery for a suspected recurrence. 8 MR parameter values were tested from perfusion-weighted, diffusion-weighted, and MR spectroscopic imaging at each tissue sample location for association with histopathological outcome using generalized estimating equation models for CEL and NEL tissue samples. Individual cutoff values were evaluated using receiver operating characteristic curve analysis with 5-fold cross-validation.
Results
In tissue samples obtained from CEL, elevated relative cerebral blood volume (rCBV) was associated with the presence of recurrent tumor pathology (P < 0.03), while increases in normalized choline (nCho) and choline-to-NAA index (CNI) were associated with the presence of recurrent tumor pathology in NEL tissue samples (P < 0.008). A mean CNI cutoff value of 2.7 had the highest performance, resulting in mean sensitivity and specificity of 0.61 and 0.81 for distinguishing treatment-effect from recurrent tumor within the NEL.
Conclusion
Although our results support prior work that underscores the utility of rCBV in distinguishing the effects of treatment from recurrent tumor within the contrast enhancing lesion, we found that metabolic parameters may be better at differentiating recurrent tumor from treatment-related changes in the NEL of high-grade gliomas.
Keywords: diffusion-weighted imaging, DSC perfusion imaging, glioblastoma, high-grade glioma, image-guided tissue acquisition, MRI, MRSI, spectroscopic imaging, treatment effect
Key Points.
1. MR signatures that distinguish treatment effect and high-grade tumor vary spatially.
2. MR spectroscopic metrics within nonenhancing regions are the most predictive.
Importance of the Study.
It is estimated that 25‒35% of patients experience treatment-induced effects that can mimic recurrent high-grade gliomas, which poses a great diagnostic challenge. If recurrence is mistakenly diagnosed, a patient may be removed from an effective therapy, which may invalidate the results of a clinical trial and/or expose that patient to unnecessary surgical intervention. Alternatively, mistaking tumor progression for treatment effect would allow an aggressive tumor to exist without further treatment. The coexistence of treatment effects and recurrent tumor within the same patient is thought to impact the classification of lesions as treatment effect or tumor using summarized MRI characteristics. This study aims to determine an optimal combination of physiologic and metabolic imaging metrics that can distinguish pathological features of recurrent tumor from the effects of treatment by leveraging a unique dataset of image-guided tissue samples that directly maps pathology to MR characteristics.
Tumor recurrence in patients with high-grade glioma (HGG) is difficult to diagnose because treatment-induced injury often appears identical on conventional anatomic MRI. It is estimated that 25‒35% of patients who undergo standard-of-care radiation and chemotherapy in the form of temozolomide for HGG experience treatment-related injury,1–3 and its appearance is even more common with the recent advent of immuno- and other targeted therapies in clinical trials. If recurrence is incorrectly diagnosed, a patient may be removed from an effective therapy, which could invalidate the results of a clinical trial or expose a patient to unnecessary surgical intervention. To mitigate these risks, identifying the exact location and extent of treatment-related changes within newly enlarging lesions is critical.
Despite the known pitfalls of using anatomic imaging, the current Response Assessment in Neuro-Oncology criteria for HGG relies solely on standard T1- and T2-weighted MR imaging.4 These techniques allow for visualization of anatomic abnormalities but are limited in their ability to capture the underlying biology that differentiates true recurrent glioma from treatment effects. Emerging data suggest that incorporating more advanced MRI techniques may be useful for probing underlying biological differences: diffusion-weighted imaging for capturing the restricted water movement from the density of proliferating tumor cells5–10; perfusion-weighted imaging for evaluating the increased vasculature recruited to support a growing mass11–15; and spectroscopic imaging for elucidating the metabolic differences between inflammation and proliferating tumor cells.6,14,16 Although several groups have investigated the potential for distinguishing between treatment effects and recurrent tumor using these techniques, the majority of prior studies typically involve calculating the mean diffusion-, perfusion-, or spectroscopic-derived parameter value from an anatomic region of interest (ROI) and normalizing that value against that obtained from contralateral normal-appearing white matter in order to obtain a threshold that can distinguish treatment effects from true tumor recurrence. These ROI-based methods suffer from widely varying cutoff values for parameters due to interobserver dependence, intratumoral heterogeneity, and the coexistence of treatment effect and tumor within the same lesion. Such studies also typically use radiographic observations or a single tissue sample for outcome determination, ascribing a single diagnosis to a mixture of tissue types that could mask the heterogeneity of the lesion and limit the accuracy of the cutoff value concluded from the study and overall clinical diagnosis.
To overcome the complications introduced by tissue heterogeneity in ROI-based studies, one strategy is to use image-guided tissue samples of known coordinates to directly map MRI characteristics to histopathology. In 2002, Rock et al pioneered this technique in distinguishing radiation necrosis from recurrent disease using metabolite ratios derived from 1H MR spectroscopic imaging (MRSI) at the location of sampled tissue.17 Their findings suggested that the ratios of choline and lactate/lipid to creatine could differentiate samples with pure necrosis and tumor, but not those with mixed pathology. In 2009, Hu et al utilized this technique in conjunction with dynamic susceptibility contrast (DSC) perfusion-weighted imaging to distinguish post-treatment radiation effect from recurrent tumor with high sensitivity and specificity using relative cerebral blood volume (rCBV) values from 13 patients.
The goal of this study was to determine whether the relevance of different MR characteristics for distinguishing pathological features of recurrent tumor from the effects of treatment in the contrast enhancing and non-enhancing lesions of recurrent high-grade gliomas by leveraging a unique dataset of image-guided tissue samples of known coordinates to avoid complications of tissue heterogeneity that confound most lesion-level analyses. Based on prior literature, we expect that samples from recurrent tumor will have increased blood volume and abnormal metabolism, with decreased diffusion compared with samples containing treatment effect. We also hypothesize that: (i) this difference would be more pronounced in diffusion and perfusion metrics for samples within the contrast enhancing lesion (CEL), while metabolic measures would be equally effective at differentiating recurrent tumor from treatment effect in both the contrast enhancing and non-enhancing lesions (NEL); and (ii) the addition of multiparametric physiologic and metabolic MRI in conjunction with tissue sample level analyses will provide increased sensitivity and specificity in distinguishing recurrent tumor from the effects of treatment in both types of lesions compared with anatomic imaging.
Materials and Methods
Patient Recruitment
Institutional Review Board approval was obtained at our institution to prospectively enroll consecutive patients with an initial pathological diagnosis of a World Health Organization grade III or IV glioma who were suspected of recurrence between 2007 and 2017. A total of 173 patients (median age, 52; range, 21–84) were enrolled who provided written informed consent to participate and underwent MR imaging between 1 and 3 days prior to surgical resection and tissue sample collection. Demographics were typical for this population as shown in Table 1. The majority of patients received an initial surgical resection followed by the standard-of-care radiotherapy (119 patients) plus temozolomide (114 patients). Forty-one patients also received 1 or more of 19 different additional therapies (18 bevacizumab, 8 immunotherapy). Prior treatment history was unknown for 12 patients.
Table 1.
Clinical and demographic characteristics of the patient population included in this study
| Clinical/Demographic Characteristics | Patients, n | Patients, % | |
|---|---|---|---|
| Totals | 139 | 100 | |
| Sex | Female | 56 | 40.3 |
| Race | White | 112 | 80.6 |
| American Indian | 1 | 0.7 | |
| Asian | 6 | 4.3 | |
| Pacific Islander | 2 | 1.4 | |
| Other | 18 | 12.9 | |
| Clinical diagnosis | Grade III astrocytoma | 11 | 7.2 |
| Grade III oligodendroglioma | 15 | 10.8 | |
| Grade IV glioblastoma | 87 | 62.6 | |
| Grade IV gliosarcoma | 3 | 2.2 | |
| Treatment effect | 23 | 16.5 | |
| Age at recurrent surgery | Median, range | 53 | 21–84 |
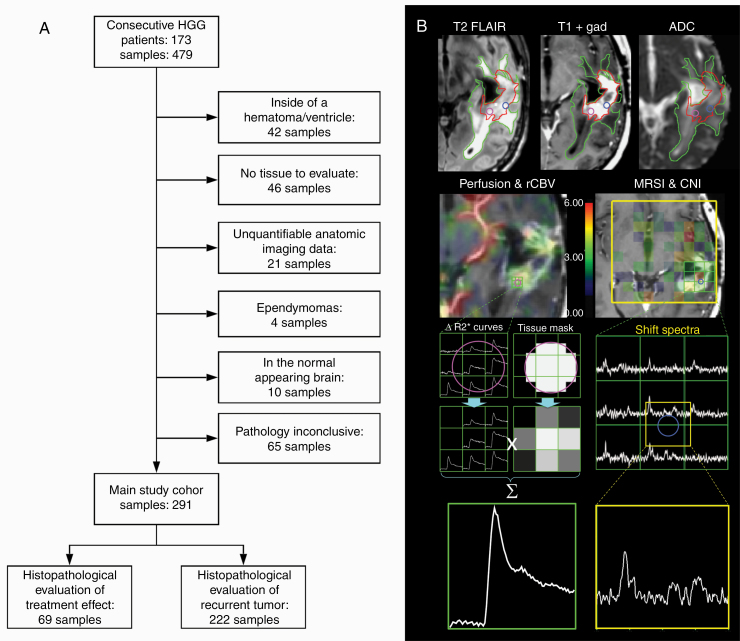
From each patient, 1–4 tissue samples were obtained (mean samples/patient was 2.3, with 1.18 samples/patient within the CEL and 0.91 samples/patient within the NEL). Of the initial 479 samples from 173 patients, a sample was only included in the analysis if it: (i) had a conclusive histopathological outcome; (ii) came from a patient whose initial diagnosis was as high-grade glioma; (iii) did not come from a region of hematoma or extensive necrosis; (iv) had quantifiable anatomic imaging data; and (v) was located within either the CEL or NEL (Fig. 1A). This resulted in a total of 291 samples from 139 patients, 26 of whom had a diagnosis at surgery of grade III and 90 of grade IV, and 23 were exclusively treatment effect.
Fig. 1.
(A) Flowchart depicting sample inclusion criteria and final sample outcome distribution in the main study cohort. (B) Anatomic imaging derived ROIs (top row) and processing steps for the generation of perfusion and metabolic metrics at the location of the tissue samples.
MR Acquisition
MR examinations were performed on a 3T scanner (GE Healthcare Technologies) using an 8-channel phased-array head coil. Standard anatomic imaging included T2-weighted fluid attenuated inversion recovery (FLAIR) and fast spin echo images, along with 3D T1-weighted inversion recovery spoiled gradient imaging pre- and post-injection of a gadolinium-based contrast agent. Diffusion-tensor images (DTIs) were obtained in the axial plane with b = 1000 s/mm2 and either 6 gradient directions and 4 excitations or 24 gradient directions and 1 excitation or b = 2000 s/mm2 and 55 gradient directions (repetition time [TR]/echo time [TE] = 1000/108 ms, voxel size = 1.7–2.0 × 1.7–2.0 × 2.0–3.0 mm). DSC perfusion-weighted images were obtained following a 3 mL/s bolus injection of 0.1 mmol/kg body weight gadolinium diethyltriamine pentaacetic acid using a series of T2*-weighted echo-planar images (TR/TE/flip angle = 1250–1500/35–54 ms/30°–35°, 128 × 128 matrix, slice thickness = 3–5 mm, 7–24 slices with 60–80 time points) before, during, and after the arrival of the contrast agent bolus. The temporal resolution was between 1 and 1.5 seconds, with total acquisition time ranging 1–2 min. The 3D 1H MRSI was acquired using point-resolved spectroscopic selection for volume localization and very selective saturation pulses for lipid signal suppression (excited volume = 80 × 80 × 40 mm, TR = 1100–1250 ms, TE = 144 ms, overpress factor = 1.5 if lactate edited, otherwise 1.2, field of view = 16 × 16 × 16 or 18 × 18 × 16 cm, nominal voxel size = 1 × 1 × 1 cm), flyback echo-planar readout gradient in the superior–inferior direction, 988 Hz sweep width and 712 dwell points. A dual-cycle lactate-edited sequence18 was used for 42 patients (83 samples, 11 min), while a standard single-cycle sequence was used for the remaining 38 patients (68 samples, 6 min).
MR Data Processing
Anatomic, diffusion, and perfusion data were aligned to the T1 post-contrast image using either FMRIB’s FSL Linear Image Registration Tool19,20 or Slicer’s BRAINSFit tool with B-spline warping.21 Spherical 5-mm-diameter ROIs were generated at the location of the spatial coordinates recorded during surgery in order to balance the potential error introduced by tissue shift and the need to restrict the ROI to the immediate vicinity of the sampled tissue.22 All locations were then visually verified for accuracy on anatomic imaging using screenshots taken during the surgery, and excluded if there was a mismatch between the coordinate locations and visualized location on imaging.
A pipeline that utilized components of FMRIB’s Diffusion Toolkit was applied to estimate relevant diffusion parameters from the DWI and DTI data as previously described.19 In order to account for differences in acquisition parameters over the 10-year study duration, voxel values for the apparent diffusion coefficient (ADC) and fractional anisotropy (FA) maps were normalized to the mode of intensities in normal-appearing brain tissue (resulting in normalized [n]ADC and nFA maps). Normalized ADC increases as average diffusivity of water within a voxel increases and therefore its decrease should act as a marker for glial proliferation, while nFA is an index for the amount of directional movement of water resulting from the parallel orientation of axonal fibers in white matter.
From the DSC perfusion data, rCBV maps were first calculated on a voxel-by-voxel basis utilizing a modified gamma-variate function that takes into account leakage of the contrast agent.23 To generate a single concentration-time curve per sample, unquantifiable voxels of noise were automatically excluded and the percentage of the tissue sample ROI within each perfusion voxel was determined before taking a weighted average of the remaining dynamic curves based on their percentage overlap with the ROI (Fig. 1B).24 This method helped mitigate inaccuracies in quantification of perfusion parameters from tissue sample ROIs due to the relatively small size of the ROI compared with the low resolution of the perfusion scan and the presence of susceptibility artifacts and necrosis. Relative CBV for each tissue sample was calculated as the area under the final gamma-variate fitted single concentration-time curve after leakage correction. An increase in rCBV reflects an increased volume of blood vessels in a given amount of brain tissue, and is therefore expected to be elevated in regions of recurrent tumor as it recruits blood vessels to supply oxygen to an enlarging mass.
Spectroscopic data were reconstructed and postprocessed using in-house software, as previously described.25–27 To generate a single spectrum centered at the location of each tissue sample, 3D spectral arrays were first shifted in k-space to reconstruct a spectral voxel on the center coordinates of each tissue sample location (Fig. 1B). Peak heights and areas were determined from baseline-subtracted and frequency- and phase-corrected spectra on a voxel-by-voxel basis.28 The choline-to-NAA index (CNI) and the choline-to-creatine index (CCrI) were obtained as previously described, using the entire 3D array of spectra in the iterative regression.29 Normalized total choline (nCho), creatine (nCre), and N-acetylaspartate (nNAA) intensities were calculated using their median value in voxels that had been identified during the CNI calculation as being from normal brain. Normalized Cho increases with increased cellular turnover, nNAA is a neuronal marker, and nCre is thought to be relatively constant regardless of tissue makeup. Using indices such as CNI and CCrI are appealing because they can capture more complex information (eg, areas of high cellular turnover with low neuron density [CNI]), which can be indicative of tumor tissue.
Together, these post-processing steps resulted in 8 different MRI parameters: nADC and nFA from DTI; rCBV from DSC perfusion; and CNI, CCrI, nCho, nCre, and nNAA from MRSI.
Tissue Sampling and Histopathological Assessment
A minimum of 4 tissue samples at least 1 cm apart were preoperatively planned to maximize heterogeneity within the hyperintense region on the T2 FLAIR image. Edges of cavities or necrotic regions were avoided, and the accessibility of the tissue target to the surgeon was considered during planning. During surgery, an intraoperative navigation system (BrainLab or Surgical Stealth) guided the neurosurgeon to the targeted locations and was used to record target coordinates for excised tissue samples. Samples were immediately formalin fixed and paraffin embedded.30
Hematoxylin and eosin (H&E) stained slides from tissue samples were evaluated by a board-certified pathologist (J.J.P.). Slides were assessed for the presence of tumor cells, necrosis, and treatment-related abnormal vasculature. Samples that had both signs of treatment-related changes and zero tumor cells were considered treatment effect. The presence of tumor cells was scored based upon review of H&E-stained sections by a neuropathologist as 0 = no tumor present, 1 = infiltrating tumor with rare cells, 2 = infiltrating cellular tumor, and 3 = highly cellular infiltrating tumor involving >75% of the tissue. Only samples with more than rare infiltrating tumor cells (tumor scores of 2 or greater) were considered recurrent tumor.
Statistical Analysis
Diffusion, perfusion, and spectroscopy parameters summarized from a 5-mm ROI centered on the tissue sample coordinates were tested for association with pathological outcome denoting treatment effect or recurrent HGG. Samples were analyzed both together and separately based on their location in the contrast enhancing or non-enhancing lesion. To account for the potential correlation among multiple samples derived from the same patient, univariate generalized estimating equations (GEE) were fit to the data for each MR parameter to estimate population-average coefficients, conditioning only on the fixed design matrix. The cutoff for determining significance was a P-value <0.05 after a Benjamini–Hochberg (B-H) correction for multiple testing was applied. Samples that did not have a value for a particular parameter were excluded from the analysis of that parameter. To complement these analyses, univariate GEE was also performed to evaluate the association of parameters from all samples combined irrespective of the presence of contrast enhancement. Finally, beginning with all MR parameters in a model, a backward stepwise GEE with elimination by least statistical significance was performed on all samples, while taking into account the presence or absence of contrast enhancement with an interaction term to evaluate whether individual parameter significance was upheld in a multiparametric setting.
To evaluate whether significant parameters from the previous analysis were able to separate samples into treatment effect and recurrent tumor categories, we used a cross-validation thresholding approach, where the samples were first divided into enhancing and non-enhancing groups based on their location. To create 5 folds for cross-validation within each group, samples were assigned to a fold randomly while stratifying by outcome in each fold. All samples from a single patient were included in the same fold in order to ensure the independence of each fold. To evaluate cutoff values for these imaging metrics, 4 folds were used to calculate the area under the receiver operator characteristic curve (AUC ROC) for all cutoff values and the threshold that yielded the highest AUC was chosen. This value was then applied to separate the fifth fold by outcome, and the sensitivity, specificity and accuracy of this classification were calculated to gain insight into the performance of the cutoff value. This process was repeated 5 times, providing 5 different cutoff value estimates and performance metrics. The mean and standard deviation of all thresholds and metrics derived from all 5 cross-validation experiments were calculated and reported.
In order to evaluate whether a combination of parameters could better predict outcome than thresholds alone, a logistic regression (LR) model was fit using independent significant parameters and all available samples (CEL + NEL) with that parametric information. To ensure that the validation (or fifth) fold remained independent for each cross-validation experiment, standardization was performed on the 4 folds used for training and these normalization parameters were saved for application to the validation fold. This normalization was necessary to be able to compare coefficients among regression models. The accuracy, sensitivity, and specificity of the LR models from training on the 4 folds and subsequent application of the trained model on the fifth fold were recorded. All modeling analyses were performed in R using the caret and pROC packages.31,32 To verify this procedure we also performed a complementary bootstrapping analysis that randomly selected only one sample per patient 1000 times (200/fold), with priority given to treatment-effect samples if present.
Results
Contrast-Enhancing Samples
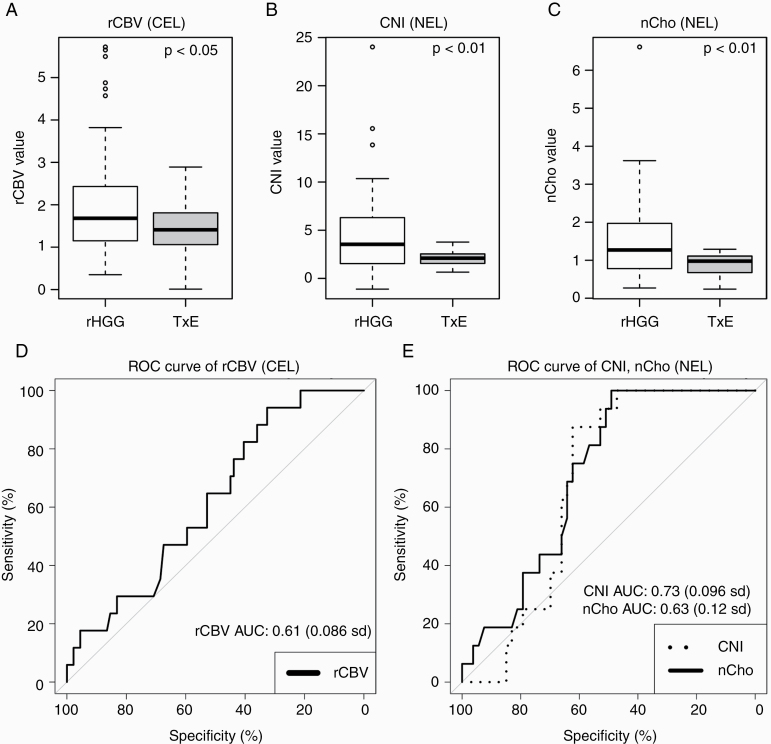
Of the 8 MRI/MRSI parameters evaluated for samples within the CEL, the perfusion parameter rCBV was significantly associated with the binary outcome of treatment effect or recurrent tumor (P < 0.03). Figures 2C, D and 3A demonstrate the elevated levels of rCBV values among tumor samples compared with treatment-effect samples. Supplementary Table 1 reports the number of samples included for each test. The model estimations, standard errors, Wald test statistics, and P-values for all tested parameters are reported in Table 2.
Fig. 2.
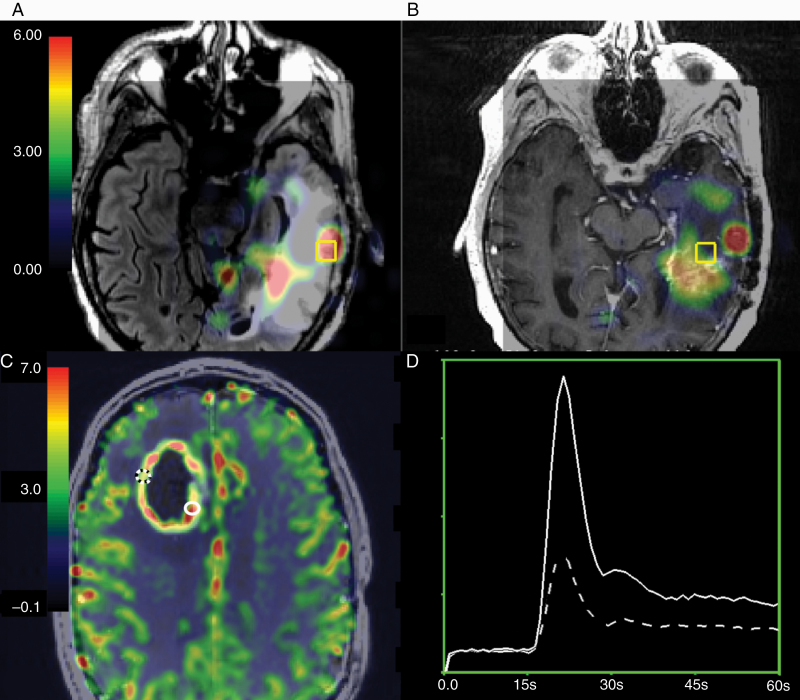
Within-patient imaging differences between treatment-induced injury and recurrent HGG. (A) Non-enhancing CNI parameter maps overlaid on the T2 FLAIR image are elevated in HGG samples. (B) Reduced CNI in a sample with treatment-induced injury within the same patient (overlaid on the post-contrast T1-weighted image). (C) Enhancing rCBV parameter maps are elevated in HGG (solid white location) versus treatment-induced injury (dashed location). (D) The corresponding ΔR2* curves.
Fig. 3.
Boxplots representing distributions of values in recurrent HGG samples and treatment-induced injury samples. Visualization of (A) rCBV values from samples in the CEL, (B) CNI, and (C) nCho values from samples in the NEL. In (D) the average ROC curve of rCBV in the CEL and (E) the average ROC curve of CNI and nCho in the NEL samples. Relative CBV was significantly associated with treatment effect versus recurrent tumor (P < 0.03) in the CEL, while spectroscopic parameters nCho and CNI were significant in the non-enhancing lesion (P = 0.008).
Table 2.
Generalized estimating equation (GEE) results from imaging values associated with pathology
| Parameter | Anatomic Region | Coeff Estimate | Std Error | Wald Value | B-H adjusted P-value |
|---|---|---|---|---|---|
| nADC | CEL | 0.04 | 0.39 | 0.01 | 0.918 |
| NEL | 0.29 | 0.36 | 0.66 | 0.56 | |
| CEL+NEL | 0.33 | 0.3 | 1.18 | 0.448 | |
| FA | CEL | 0.11 | 0.45 | 0.07 | 0.913 |
| NEL | −0.25 | 0.57 | 0.19 | 0.66 | |
| CEL+NEL | −0.25 | 0.37 | 0.45 | 0.571 | |
| rCBV | CEL | 0.31 | 0.11 | 8.72 | 0.024* |
| NEL | 0.29 | 0.29 | 0.99 | 0.512 | |
| CEL+NEL | 0.23 | 0.15 | 2.49 | 0.22 | |
| nNAA | CEL | 0.06 | 0.17 | 0.12 | 0.913 |
| NEL | 0.47 | 0.82 | 0.33 | 0.977 | |
| CEL+NEL | −0.37 | 0.40 | 0.85 | 0.48 | |
| nCho | CEL | 0 | 0 | 2.04 | 0.411 |
| NEL | 1.4 | 0.41 | 11.48 | 0.008** | |
| CEL+NEL | 0.19 | 0.07 | 6.76 | 0.024* | |
| nCre | CEL | 0 | 0 | 2.04 | 1.029 |
| NEL | 0.93 | 0.46 | 4.16 | 0.08 | |
| CEL+NEL | −0.06 | 0.15 | 0.19 | 0.66 | |
| CNI | CEL | 0 | 0.17 | 0.12 | 0.211 |
| NEL | 0.29 | 0.09 | 9.85 | 0.008** | |
| CEL+NEL | 0.05 | 0.01 | 15.08 | 0.0008** | |
| CCrI | CEL | 0 | 0 | 2.62 | 0.211 |
| NEL | 0.16 | 0.08 | 4.41 | 0.096 | |
| CEL+NEL | 0.06 | 0.02 | 8.98 | 0.012* |
*P < 0.05, **P < 0.001.
Non-Enhancing Samples
In the NEL samples, however, spectroscopic parameters nCho (P = 0.008) and CNI (P = 0.008) were significantly associated with the presence of recurrent tumor versus treatment effect. Figures 2A, B and 3B, C demonstrate the elevated levels of total choline and CNI values among tumor samples compared with samples containing purely treatment effect. Supplementary Table 1 reports the number of samples included for each test. The model estimations, standard errors, Wald test statistics, and P-values for all tested parameters are reported in Table 2.
All Samples Combined
When combining the CEL and NEL regions, elevated nCho (P = 0.024), CNI (P = 0.0008), and CCrI (P = 0.012) values were significantly associated with the presence of recurrent tumor. When parameters were combined in a backward stepwise GEE that included the anatomic lesion as an interaction term, rCBV (P = 0.036) and CNI (P = 0.003) remained significant in the final model.
Cutoff Analyses
After dividing the data into 5 folds for cutoff determination, the proportion of treatment-effect samples in each fold ranged from 18% to 29%. The mean rCBV cutoff value for distinguishing treatment effect from recurrent tumor within the CEL was 1.62 (0.21 SD). However, when this cutoff value was applied to the fifth validation fold, the mean accuracy was only 50%, with a sensitivity of 0.50 and specificity of 0.59. For samples in the NEL, the best cutoff values to separate tumor and treatment effect were 2.71 for CNI and 1.10 for nCho. This resulted in mean AUC ROC values of 0.73 (0.10 SD) and 0.63 (0.12 SD), for CNI and nCho respectively. When applied to the validation fold, the mean accuracy for CNI was 68%, while sensitivity and specificity were 0.61 and 0.85, respectively. The cutoff threshold for nCho resulted in an accuracy of 66%, while the mean sensitivity was 0.62 and specificity was 0.65. Table 3 reports these values and the standard deviations across all 5-fold tests and the corresponding ROC curves are shown in Figures 3D, E.
Table 3.
Threshold and logistic regression analysis results
| Modality | DSC | MRSI | DSC + MRSI | |
|---|---|---|---|---|
| Analysis Type | Cutoff | Cutoff | Logistic Regression | |
| Parameter | rCBV | CNI | nCho | rCBV, CNI |
| Anatomic Region | CEL | NEL | NEL | CEL+NEL |
| Threshold | 1.62 (0.21) | 2.71 (0.06) | 1.09 (0.03) | n/a |
| AUC ROC | 0.61 (0.09) | 0.73 (0.10) | 0.63 (0.12) | 0.69 (0.09) |
| Accuracy % | 50% (7%) | 68% (9%) | 66% (13%) | 64% (16%) |
| Sensitivity | 0.50 (0.10) | 0.61 (0.07) | 0.62 (0.11) | 0.65 (0.16) |
| Specificity | 0.59 (0.25) | 0.85 (0.22) | 0.65 (0.22) | 0.62 (0.21) |
Values in parentheses are means and standard deviations.
Multivariate Model
Parameter combination into an LR using all significant MR parameters resulted in a mean AUC of 0.69 (0.09 SD). When tested on the fifth fold, the LR predicted with an average accuracy of 64% and sensitivity and specificity of 0.65 and 0.62, respectively (Table 3). The single-sample bootstrapping analysis confirmed these results with a median AUC of 0.70 (95% CI: 0.69, 0.72) and an mean accuracy, sensitivity, and specificity of 68%, 0.64, and 0.79, respectively, providing confidence in our predictions despite the use of multiple samples per patient.
Discussion
While distinguishing recurrent HGG from treatment-induced effects is an important clinical objective, it has remained an immense challenge in part due to the coexistence of the 2 phenomena in the same lesion. This study attempts to overcome the problem of lesion heterogeneity by using surgical tissue samples with known coordinates on imaging acquired with neuronavigation tools. To our knowledge, this study includes the greatest number of patients with spatially mapped tissue samples to distinguish recurrent HGG from treatment-induced effects to date and takes advantage of incorporating metabolic and physiologic derived metrics from DSC perfusion weighted imaging, DTI, and MRSI. Through our statistical and predictive approaches, we demonstrated the importance of MRSI in distinguishing recurrent tumor from the effects of treatment in the non-enhancing lesion and examined whether the combination of vascular and metabolic metrics could lead to the generation of more accurate predictions.
Our results support the use of 1H-MRSI for identifying regions of abnormal metabolism in the non-enhancing region that are indicative of infiltrative recurrent tumor cells rather than the effects of treatment. Tumor samples demonstrated higher levels of choline-containing metabolites (nCho) and elevated CNI compared with treatment-effect samples, consistent with our current understanding of the biological underpinnings of choline and NAA metabolite flux.16 After employing cross-validation to gain insight into how a given cutoff threshold would perform when applied to external data that were not used in determining its value, our cutoffs for nCho (1.1) and CNI (2.7) separated NEL samples into treatment-effect or recurrent tumor categories with mean sensitivity of 0.62, 0.61 and specificity of 0.85, 0.65, for nCho and CNI respectively. While many prior studies have used single-voxel spectroscopy in the enhancing lesion,33–35 our results indicate that spectroscopic coverage of the non-enhancing lesion area would benefit in the accurate diagnosis of recurrence. In turn, a multivoxel spectroscopic approach would provide greater utility in assessing the metabolic lesion than single-voxel sequences, as they are often centered on the contrast enhancing region. These spectroscopic findings are similar to metabolic differences observed between vasogenic edema and enhancing metastatic disease in patients with brain metastases.34–37 Although recent studies apply machine learning techniques to dynamic contrast enhanced (DCE) perfusion, DTI, and anatomic imaging,38,39 the innate biological differences between metastatic brain tumors and gliomas prohibit their generalizability. Though more rigorous validation is necessary before incorporation into a clinical workflow, this study lays the groundwork for future investigation into the utility of these parameters in a prospective, independent cohort with image-guided tissue samples using more sophisticated machine learning algorithms. This has potential to direct surgeons to which part of the non-enhancing lesion contains infiltrating tumor.
Although the recent meta-analysis of ROI-based studies by Van Dijken et al found that the greatest sensitivity and specificity for distinguishing recurrent tumor from treatment effects lies in the spectroscopic parameters derived from the CEL, we were not able to replicate these results with our analyses. This could be because ROI-based studies do not account for the spatial heterogeneity that exists in metabolism within the lesion. Despite our voxel shifting methods to reconstruct spectra at the center of the tissue sample location to avoid errors from interpolation, it was still possible that pathological heterogeneity existed within the 1 cc spectral voxel. Additionally, spectral voxels at the location of CEL tissue samples often overlapped with necrotic regions and non-enhancing regions because of their larger size, potentially affecting the quantification of CNI.
Our findings in tissue samples obtained from within the contrast enhancing lesion suggest that elevated rCBV is significantly associated with recurrent tumor compared with treatment effects. The GEE and cutoff analysis suggest that rCBV is useful for differentiating recurrent tumor from treatment effects in the CEL. These findings are consistent with ROI-based studies that report as high as 87% sensitivity and 86% specificity when differentiating recurrent tumor from treatment-induced effects from the contrast enhancing ROI. Although these individual smaller studies report higher sensitivity and specificity, their cutoff values were highly variable, ranging from 0.71 to 3.7 for rCBV, reflecting the difficulty in recommending a universal cutoff. It was our hope that our analysis would provide clarity to this body of work by mapping local MRI characteristics directly to pathology. Although the cutoff for rCBV in our study was 1.59 (0.21 SD), our sensitivity and specificity were not significantly better than random chance. These results may be in part attributed to the very large range of rCBV values observed in high-grade tumor tissue samples (min: 0.10, max: 5.72, med: 1.61). This large range, taken in combination with previous rCBV reports, supports the notion that a “signature” rCBV value that can distinguish high-grade tumor from treatment-related injury remains difficult to define and that multiparametric analyses with more advanced machine learning methodologies in larger datasets may be necessary to adequately address this problem.
After analyzing the association of singular MR parameters to outcome, we assessed whether the combination of parameters improves classification of samples into treatment effects or recurrent HGG. Although it does not model the potential correlation among samples derived from the same patients, logistic regression was chosen for its interpretability and reluctance to overfitting, and was further validated by comparing with the result attained from randomly selecting 1 sample per patient and bootstrapping the data. For this analysis, 5-fold cross-validation was used where each fold was separated by patient and stratified by outcome to control for information leakage and optimistic prediction. Only parameters from modalities that were determined to be useful for differentiating treatment effects from recurrent HGG in the univariate analyses were retained in the multivariate models. Combining the results into a logistic regression resulted in a model that, compared with cutoff analyses, had similar sensitivity (0.65) and specificity (0.63) when tested on the fifth fold, suggesting that modeling parameters together may not improve the classification of tissue samples by pathology. Though these results seem counterintuitive at first, it is likely that combining the anatomic regions of CEL and NEL averages out the signal that was present in each separate anatomic region, further substantiating our hypothesis that recurrent tumor in these regions have distinct metabolic and physiologic characteristics. For example, because the appearance of the CEL is driven by the extravasation of contrast by leaky blood vessels, rCBV values in the CEL have a significantly different value distribution from those in the NEL; therefore, the signal driving the difference in rCBV in the CEL is lost when combined with samples in the NEL.
The low sensitivity in predicting pathology from MR parameters and reliably classify subregions of a lesion in our dataset can be attributed to 3 main causes. First, the parameters that were determined as being the most important for prediction of treatment effects and tumor were obtained in only 52% (perfusion) or 67% (MRSI) of patients (Supplementary Table 1) because they were not part of routine clinical evaluation. This, along with imbalanced classes, limited our ability to build a predictive model that could be tested on an unseen dataset with these parameters. Still, using a 5-fold cross-validation approach allowed us to instead estimate the predictive value of our MR metrics, whereby we could iterate over all available treatment-effect samples and observe the stability of the prediction. Although we removed samples that largely consisted of necrosis, our modest results could also be explained by the possibility of some necrosis coexisting within tissue samples made up of mostly tumor or treatment effect. Although a recent review article summarizing 25 studies of brain shift reported maximum shifts between 4 and 31 mm during the course of resection,39 the vast majority of our tissue samples were acquired with a biopsy needle before opening the dura and resecting the tumor tissue, where most reported shifts have been 2–5 mm, with maximum shifts of <10 mm. These results informed our rationale for using a 5 mm ROI around the center of the biopsied sample, even though the diameter of the excised tissue was 2 mm. This is less of an issue for MRSI than other imaging metrics, because the voxel size is 1 mm3 and we shift the reconstruction of the spectra voxel in k-space so that it is centered on the location of the tissue sample coordinates. Despite our efforts to further correct for errors due to tissue shift by performing extensive quality control through manual visualization of each tissue sample location and exclusion of samples that were structurally inaccurate, it is highly likely that this shift is the main contributor to reducing the accuracy of our results, especially when lesion heterogeneity is pronounced.
In conclusion, this study attempts to overcome the heterogeneity inherent in treated HGG lesions by mapping pathological findings directly to MR parameters. Our results suggest the need for separate MR markers of recurrent tumor for enhancing and non-enhancing lesions, highlight the potential utility of using 3D MRSI to obtain CNI maps that include the non-enhancing region in the recurrent high-grade setting, and support previous studies that suggest rCBV should be used to differentiate treatment effects from recurrent tumor within the contrast enhancing region. These findings lay the foundation for a larger, multi-institutional investigation that includes MRSI of the non-enhancing region and multiparametric MRI, along with machine learning for differentiation of treatment-induced injury from true recurrent tumor.
Supplementary Material
Acknowledgments
We would like to acknowledge the patients and their families for their participation in this study and the numerous prior lab members, clinical coordinators, and technologists for their help with scanning, scheduling, planning and collecting tissue, and compiling data.
Funding
This work was supported by the National Institutes of Health National Cancer Institute Programmatic Project grant P01 CA118816 and a T32 Ruth L. Kirschstein Institutional National Research Service Award (JC).
Conflict of interest statement.
The authors have no conflicts of interest to report with respect to the content of this manuscript.
Authorship statement.
Study design: JML, SJN. Data collection: MF, DN, MSB. Image processing and analysis: JC, QW, SJN, JML, PAL, MPO. Pathological analysis: JJP, AS. Statistical analysis: JC, AMM, PC. Data interpretation: JEVM, SC, SMC. Manuscript preparation: JC, JML, AMM, JJP, SC, MSB, JEVM
References
- 1. Ellingson BM, Wen PY, Cloughesy TF. modified criteria for radiographic response assessment in glioblastoma clinical trials. Neurotherapeutics. 2017;14(2):307–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Abbasi AW, Westerlaan HE, Holtman GA, Aden KM, van Laar PJ, van der Hoorn A. Incidence of tumour progression and pseudoprogression in high-grade gliomas: a systematic review and meta-analysis. Clin Neuroradiol. 2018;28(3):401–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ellingson BM, Chung C, Pope WB, Boxerman JL, Kaufmann TJ. Pseudoprogression, radionecrosis, inflammation or true tumor progression? Challenges associated with glioblastoma response assessment in an evolving therapeutic landscape. J Neurooncol. 2017;134(3):495–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wen PY, Macdonald DR, Reardon DA, et al. Updated response assessment criteria for high-grade gliomas: Response Assessment in Neuro-Oncology working group. J Clin Oncol. 2010;28(11):1963–1972. [DOI] [PubMed] [Google Scholar]
- 5. Hein PA, Eskey CJ, Dunn JF, Hug EB. Diffusion-weighted imaging in the follow-up of treated high-grade gliomas: tumor recurrence versus radiation injury. AJNR Am J Neuroradiol. 2004;25(2):201–209. [PMC free article] [PubMed] [Google Scholar]
- 6. Jena A, Taneja S, Jha A, et al. Multiparametric evaluation in differentiating glioma recurrence from treatment-induced necrosis using simultaneous 18F-FDG-PET/MRI: a single-institution retrospective study. AJNR Am J Neuroradiol. 2017;38(5):899–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kazda T, Bulik M, Pospisil P, et al. Advanced MRI increases the diagnostic accuracy of recurrent glioblastoma: single institution thresholds and validation of MR spectroscopy and diffusion weighted MR imaging. Neuroimage Clin. 2016;11:316–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Alexiou GA, Zikou A, Tsiouris S, et al. Comparison of diffusion tensor, dynamic susceptibility contrast MRI and (99m)Tc-Tetrofosmin brain SPECT for the detection of recurrent high-grade glioma. Magn Reson Imaging. 2014;32(7):854–859. [DOI] [PubMed] [Google Scholar]
- 9. Narang J, Jain R, Arbab AS, et al. Differentiating treatment-induced necrosis from recurrent/progressive brain tumor using nonmodel-based semiquantitative indices derived from dynamic contrast-enhanced T1-weighted MR perfusion. Neuro Oncol. 2011;13(9):1037–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jain R, Narang J, Schultz L, et al. Permeability estimates in histopathology-proved treatment-induced necrosis using perfusion CT: can these add to other perfusion parameters in differentiating from recurrent/progressive tumors? AJNR Am J Neuroradiol. 2011;32(4):658–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Boxerman JL, Ellingson BM, Jeyapalan S, et al. Longitudinal DSC-MRI for distinguishing tumor recurrence from pseudoprogression in patients with a high-grade glioma. Am J Clin Oncol. 2017;40(3):228–234. [DOI] [PubMed] [Google Scholar]
- 12. Kim TH, Yun TJ, Park CK, et al. Combined use of susceptibility weighted magnetic resonance imaging sequences and dynamic susceptibility contrast perfusion weighted imaging to improve the accuracy of the differential diagnosis of recurrence and radionecrosis in high-grade glioma patients. Oncotarget. 2017;8(12):20340–20353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Prager AJ, Martinez N, Beal K, Omuro A, Zhang Z, Young RJ. Diffusion and perfusion MRI to differentiate treatment-related changes including pseudoprogression from recurrent tumors in high-grade gliomas with histopathologic evidence. AJNR Am J Neuroradiol. 2015;36(5):877–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Seeger A, Braun C, Skardelly M, et al. Comparison of three different MR perfusion techniques and MR spectroscopy for multiparametric assessment in distinguishing recurrent high-grade gliomas from stable disease. Acad Radiol. 2013;20(12):1557–1565. [DOI] [PubMed] [Google Scholar]
- 15. Hu LS, Baxter LC, Smith KA, et al. Relative cerebral blood volume values to differentiate high-grade glioma recurrence from posttreatment radiation effect: direct correlation between image-guided tissue histopathology and localized dynamic susceptibility-weighted contrast-enhanced perfusion MR imaging measurements. AJNR Am J Neuroradiol. 2009;30(3):552–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Elias AE, Carlos RC, Smith EA, et al. MR spectroscopy using normalized and non-normalized metabolite ratios for differentiating recurrent brain tumor from radiation injury. Acad Radiol. 2011;18(9):1101–1108. [DOI] [PubMed] [Google Scholar]
- 17. Rock JP, Hearshen D, Scarpace L, et al. Correlations between magnetic resonance spectroscopy and image-guided histopathology, with special attention to radiation necrosis. Neurosurgery. 2002;51(4):912–919; discussion 919. [DOI] [PubMed] [Google Scholar]
- 18. Park I, Chen AP, Zierhut ML, Ozturk-Isik E, Vigneron DB, Nelson SJ. Implementation of 3 T lactate-edited 3D 1H MR spectroscopic imaging with flyback echo-planar readout for gliomas patients. Ann Biomed Eng. 2011;39(1):193–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Duarte-Carvajalino JM, Sapiro G, Harel N, Lenglet C. A framework for linear and non-linear registration of diffusion-weighted MRIs using angular interpolation. Front Neurosci. 2013;7:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jenkinson M, Bannister P, Brady M, Smith S. Improved optimization for the robust and accurate linear registration and motion correction of brain images. Neuroimage. 2002;17(2):825–841. [DOI] [PubMed] [Google Scholar]
- 21. Johnson HJ, Harris G, Williams K. BRAINSFit: mutual information registrations of whole-brain 3d images, using the insight toolkit.http://hdl.handle.net/1926/1291. Published 2007. Accessed May 1, 2019.
- 22. Kelly PJ, Kall BA, Goerss S, Earnest F 4th. Computer-assisted stereotaxic laser resection of intra-axial brain neoplasms. J Neurosurg. 1986;64(3):427–439. [DOI] [PubMed] [Google Scholar]
- 23. Weisskoff RM, Boxerman JL, Sorensen AG, et al. Simultaneous blood volume and permeability mapping using a single Gd‐based contrast injection. In: Proc. Intl. Soc. Mag. Reson. Med. 2. San Francisco, CA: International Society for Magnetic Resonance in Medicine; 1994:279. [Google Scholar]
- 24. Lupo JM, Wen Q, Chang SM, et al. Weighted-average model curve preprocessing strategy for quantification of DSC perfusion imaging metrics from image-guided tissue samples in patients with brain tumors. In: Proc. Intl. Soc. Mag. Reson. Med. 23. Toronto, Ontario, Canada: International Society for Magnetic Resonance in Medicine; 2015:4377. [Google Scholar]
- 25. Li Y, Osorio JA, Ozturk-Isik E, et al. Considerations in applying 3D PRESS H-1 brain MRSI with an eight-channel phased-array coil at 3 T. Magn Reson Imaging. 2006;24(10):1295–1302. [DOI] [PubMed] [Google Scholar]
- 26. Crane JC, Olson MP, Nelson SJ. SIVIC: open-source, standards-based software for DICOM MR spectroscopy workflows. Int J Biomed Imaging. 2013;2013:169526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nelson SJ, Kadambi AK, Park I, et al. Association of early changes in 1H MRSI parameters with survival for patients with newly diagnosed glioblastoma receiving a multimodality treatment regimen. Neuro Oncol. 2017;19(3):430–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Li Y, Lupo JM, Parvataneni R, et al. Survival analysis in patients with newly diagnosed glioblastoma using pre- and postradiotherapy MR spectroscopic imaging. Neuro Oncol. 2013;15(5):607–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. McKnight TR, Noworolski SM, Vigneron DB, Nelson SJ. An automated technique for the quantitative assessment of 3D-MRSI data from patients with glioma. J Magn Reson Imaging. 2001;13(2):167–177. [DOI] [PubMed] [Google Scholar]
- 30. Barajas RF Jr, Phillips JJ, Parvataneni R, et al. Regional variation in histopathologic features of tumor specimens from treatment-naive glioblastoma correlates with anatomic and physiologic MR Imaging. Neuro Oncol. 2012;14(7):942–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Robin X, Turck N, Hainar A, et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics. 2011;12:77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kuhn M. Building predictive models in R using the caret package. J Stat. Softw 2008;28(5):1–26.27774042 [Google Scholar]
- 33. Nakajima T, Kumabe T, Kanamori M, et al. Differential diagnosis between radiation necrosis and glioma progression using sequential proton magnetic resonance spectroscopy and methionine positron emission tomography. Neurol Med Chir (Tokyo). 2009;49(9): 394–401. [DOI] [PubMed] [Google Scholar]
- 34. Peca C, Pacelli R, Elefante A, et al. Early clinical and neuroradiological worsening after radiotherapy and concomitant temozolomide in patients with glioblastoma: tumour progression or radionecrosis? Clin Neurol Neurosurg. 2009;111(4):331–334. [DOI] [PubMed] [Google Scholar]
- 35. D’Souza MM, Sharma R, Jaimini A, et al. 11C-MET PET/CT and advanced MRI in the evaluation of tumor recurrence in high-grade gliomas. Clin Nucl Med. 2014;39(9):791–798. [DOI] [PubMed] [Google Scholar]
- 36. Stummer W. Mechanisms of tumor-related brain edema. Neurosurg Focus. 2007;22(5):E8. [DOI] [PubMed] [Google Scholar]
- 37. Server A, Josefsen R, Kulle B, et al. Proton magnetic resonance spectroscopy in the distinction of high-grade cerebral gliomas from single metastatic brain tumors. Acta Radiol. 2010;51(3):316–325. [DOI] [PubMed] [Google Scholar]
- 38. van Dijken BRJ, van Laar PJ, Holtman GA, van der Hoorn A. Diagnostic accuracy of magnetic resonance imaging techniques for treatment response evaluation in patients with high-grade glioma, a systematic review and meta-analysis. Eur Radiol. 2017;27(10): 4129–4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gerard IJ, Kersten-Oertel M, Petrecca K, Sirhan D, Hall JA, Collins DL. Brain shift in neuronavigation of brain tumors: a review. Med Image Anal. 2017;35:403–420. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.