Abstract
The pathological deposition of the transactive response DNA-binding protein of 43 kDa occurs in the majority (∼97%) of amyotrophic lateral sclerosis and in around 45% of frontotemporal lobar degeneration cases. Amyotrophic lateral sclerosis and frontotemporal lobar degeneration clinically overlap, presenting a continuum of phenotypes. Both amyotrophic lateral sclerosis and frontotemporal lobar degeneration lack treatments capable of interfering with the underlying pathological process and early detection of transactive response DNA-binding protein of 43 kDa pathology would facilitate the development of disease-modifying drugs. The real-time quaking-induced conversion reaction showed the ability to detect prions in several peripheral tissues of patients with different forms of prion and prion-like diseases. Despite transactive response DNA-binding protein of 43 kDa displays prion-like properties, to date the real-time quaking-induced conversion reaction technology has not yet been adapted to this protein. The aim of this study was to adapt the real-time quaking-induced conversion reaction technique for the transactive response DNA-binding protein of 43 kDa substrate and to exploit the intrinsic ability of this technology to amplify minute amount of mis-folded proteins for the detection of pathological transactive response DNA-binding protein of 43 kDa species in the cerebrospinal fluid of amyotrophic lateral sclerosis and frontotemporal lobar degeneration patients. We first optimized the technique with synthetic transactive response DNA-binding protein of 43 kDa–pre-formed aggregates and with autopsy-verified brain homogenate samples and subsequently analysed CSF samples from amyotrophic lateral sclerosis and frontotemporal lobar degeneration patients and controls. Transactive response DNA-binding protein of 43 kDa real-time quaking-induced conversion reaction was able to detect as little as 15 pg of transactive response DNA-binding protein of 43 kDa aggregates, discriminating between a cohort of patients affected by amyotrophic lateral sclerosis and frontotemporal lobar degeneration and age-matched controls with a total sensitivity of 94% and a specificity of 85%. Our data give a proof-of-concept that transactive response DNA-binding protein of 43 kDa is a suitable substrate for the real-time quaking-induced conversion reaction. Transactive response DNA-binding protein of 43 kDa real-time quaking-induced conversion reaction could be an innovative and useful tool for diagnosis and drug development in amyotrophic lateral sclerosis and frontotemporal lobar degeneration. The cerebrospinal fluid detection of transactive response DNA-binding protein of 43 kDa pathological aggregates may be exploited as a disease biomarker for amyotrophic lateral sclerosis and frontotemporal lobar degeneration patients.
Keywords: TDP-43, RT-QuIC, ALS, FTD, biomarker
We adapted the RT-QuIC (real-time quaking-induced conversion reaction) technology to the TDP-43 substrate. TDP-43 RT-QuIC detected as little as 15 pg of TDP-43 synthetic aggregates and reached 94% of sensitivity and 85% of specificity in the analysis of CSF from amyotrophic lateral sclerosis and frontotemporal lobar degeneration patients and age-matched controls.
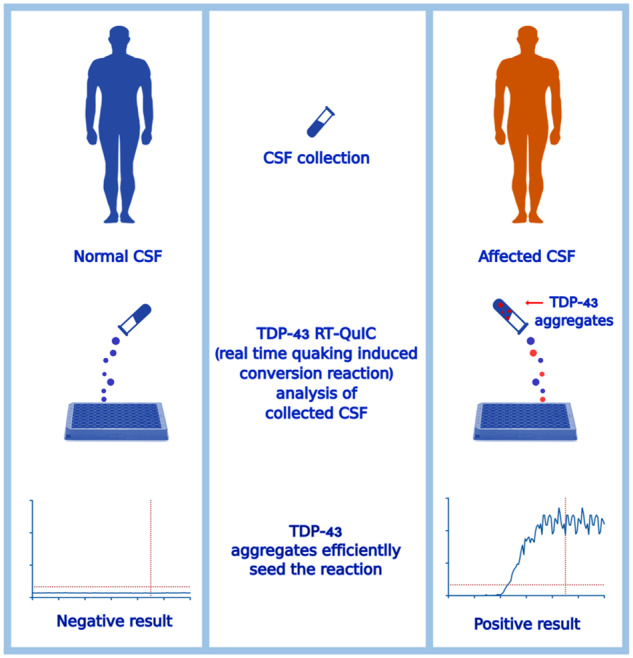
Graphical Abstract
Graphical Abstract.
Introduction
In 2006, two different groups identified the transactive response DNA-binding protein of 43 kDa (TDP-43) as the major component of aggregates deposited in the central nervous system (CNS) of patients affected by amyotrophic lateral sclerosis (ALS) and frontotemporal lobar degeneration (FTLD) (Arai et al., 2006; Neumann et al., 2006). TDP-43 pathology accounts for the majority of ALS (∼97%) and for around 45% of FTLD (Ling et al., 2013) cases. Subsequent studies showed abnormal accumulation of TDP-43 in neurodegenerative disorders other than FTLD and ALS, collectively named TDP-43 proteinopathies (Nonaka and Hasegawa, 2018). Amyotrophic lateral sclerosis and FTLD clinically overlap, presenting as a continuum phenotype, with ALS patients displaying cognitive impairment or signs of frontotemporal dementia (FTD) in approximately 50 and 15% of the cases, respectively (Tsermentseli et al., 2012). Both ALS and FTD display extensive clinical, pathological and genetic heterogeneity. Several genes showed to be linked to ALS and FTD pathology (Ng et al., 2015). More specifically, the presence of the C9orf72 expansion, mutations in TARDBP, the gene encoding for TDP-43, and in the Granulin gene, GRN, showed to be specifically related to CNS pathological TDP-43 deposition (Saberi et al., 2015; Mackenzie and Neumann, 2016; Turner et al., 2017). Increasing evidence suggests that TDP-43 pathology could progress following a prion-like mechanism (Maniecka and Polymenidou, 2015). According to the prion-like hypothesis, a mis-folded protein is able to interact with its native isoform and to convert it in its pathological aberrantly folded form. This process can amplify and propagate the aggregates, which can eventually be fragmented to form the so-called ‘seed’ (i.e. the smallest amount of the pathological protein able to replicate this process) (Maniecka and Polymenidou, 2015). In keeping with this view, TDP-43 aggregates appear to distribute progressively among axon-linked brain regions, suggesting the spreading of the prion-like agent (Ravits and La Spada, 2009; Braak et al., 2013; Brettschneider et al., 2014; Walker and Jucker, 2015; Ayers and Cashman, 2018) and several studies have shown the intrinsic propensity of TDP-43 to aggregate (Johnson et al., 2009; Furukawa et al., 2011; Jiang et al., 2013). The ability of TDP-43 aggregates to act as seeds in vitro (Furukawa et al., 2011; Nonaka et al., 2013; Smethurst et al., 2016) and recently, also in vivo, has been shown (Porta et al., 2018). Amyotrophic lateral sclerosis is a fatal neurodegenerative process usually diagnosed after 1 year from symptoms onset when disease is well established and the potential therapeutic window has passed (Feneberg et al., 2018). Frontotemporal dementia diagnosis is challenging and usually reached years after symptoms onset due to a close similarity between behavioural changes in patients with FTD and those seen in patients with psychiatric disorders (BaNg et al., 2015). Both ALS and FTD lack treatments capable of interfering with the underlying pathological process. Accurate detection of TDP-43 pathology is therefore of utmost importance to select and enroll patients in clinical trials in the earliest disease phase. In addition, pre-symptomatic screening would grant a larger therapeutic window for disease-modifying drugs. A consistent biochemical marker could also permit to monitor disease progression and to evaluate the effect of specific treatments. Recently, real-time quaking-induced conversion reaction (RT-QuIC) ante-mortem prion detection has proved as a robust technique for prion amplification (Orru et al., 2015; Fairfoul et al., 2016; Franceschini et al., 2017; Groveman et al., 2018). The RT-QuIC showed the ability to detect prions in several peripheral tissues of patients with different forms of prion diseases (Orru et al., 2014; Orru et al., 2015; Franceschini et al., 2017), misfolded amyloid beta (Aβ) oligomers in the cerebrospinal fluid (CSF) of patients with Alzheimer Disease (AD) (Salvadores et al., 2014), α-synuclein oligomers in the CSF of patients with Parkinson’s disease and Lewy body dementia (Fairfoul et al., 2016; Shahnawaz et al., 2017; Groveman et al., 2018; van Rumund et al., 2019; Shahnawaz et al., 2020) and tau in the CSF of patients with Pick’s disease (Saijo et al., 2017). Despite TDP-43 displayed prion-like properties (Johnson et al., 2009; Furukawa et al., 2011; Jiang et al., 2013; Nonaka et al., 2013; Smethurst et al., 2016; Porta et al., 2018), to date, the RT-QuIC technology has not yet been adapted to this protein. The aim of this study was to adapt the RT-QuIC technique to the TDP-43 substrate and to exploit this technology for the detection of pathological TDP-43 species in the CSF of ALS and FTD patients. As a first step, we characterized the kinetics of aggregation of full-length and truncated C-terminal TDP-43 and we observed that in vitro produced TDP-43 aggregates and patient-derived brain homogenates (BHs) efficiently seeded the reaction with both substrates. After this characterization, we selected the C-terminal fragment of TDP-43 as the most efficient substrate to perform the reaction. Finally, we collected and screened CSF samples from ALS/FTD patients and age-matched healthy controls. To increase the likelihood that the collected CSF really contained TDP-43 seeds, we focused our study on patients harbouring a pathological mutation in one of the genes known to be related with CNS TDP-43 pathology, TARDBP, GRN and C9orf72 (Saberi et al., 2015; Mackenzie and Neumann, 2016; Turner et al., 2017).
Materials and methods
Recombinant full-length and truncated TDP-43 production
The pET-11a plasmid containing human TDP-43 (HuTDP-43(1-414)) coding sequence was purchased from GenScript. The truncated construct (HuTDP-43(263-414)) was obtained by deleting the DNA region encoding for HuTDP-43 fragment 1-262 using mutagenesis. Polymerase chain reaction was carried out using site-directed mutagenesis kit (Agilent) with the following primers: 5′-ttaagaaggagatatacatatgaaacacaacagcaaccgtcagc-3′, 5′-gctgacggttgctgttgtgtttcatatgtatatctccttcttaa-3′. The protein production protocol was the same for both full-length and the truncated form of the protein. The construct was expressed in Escherichia coli BL21 (DE3) cells (Stratagene). Freshly transformed overnight culture was inoculated into Luria Bertani medium with 100 μg/mL ampicillin. At 0.8 OD600, protein expression was induced with isopropyl β-d-1-thiogalactopyranoside to a final concentration of 1 mM. After overnight incubation at 37°C, cells were lysed by a homogenizer (PandaPLUS 2000) and inclusion bodies were washed in a buffer containing 25 mM Tris-HCl, 5 mM ethylenediaminetetraacetic acid (EDTA), 0.8% TritonX100, (pH 8) and then in bi-distilled water (ddH2O) several times. Inclusion bodies containing HuTDP-43(1-414) or HuTDP-43(263-414) were dissolved in five volumes of 8 M guanidine hydrochloride (GdnHCl), loaded onto pre-equilibrated HiLoad 26/60 Superdex 200-pg column, and eluted in 25 mM Tris–HCl (pH 8), 5 mM EDTA and 5 M GdnHCl at a flow/rate of 1.5 mL/min. Protein refolding was performed by dialysis against different refolding buffers [10 mM PBS (pH = 8) for the full-length protein; 10 mM PBS (pH = 8) or 25 mM Tris-HCl (pH = 8) for the truncated fragment] using a Spectrapor membrane. Purified proteins were analysed by SDS-PAGE gel electrophoresis under reducing conditions and Western blot.
Electrophoretic mobility shift assay
A specific sequence of RNA oligonucleotides, termed AUG12 RNA, 5′-GUGUGAAUGAAU-3′, was purchased from Sigma-Aldrich. In brief, 12.6 μM of AUG12 RNA was mixed with 12.6 μM of HuTDP-43 or Bovine Serum Albumin (BSA), as a control. Binding reactions (20 μL) were incubated in water at room temperature (RT) for 20 min before loading onto an Agarose gel (1%) in 1× Tris–acetate–EDTA. Gels were run at 100–120 V for 45 min at 4 °C.
Brain homogenates collection e preparation
Post-mortem frozen frontal cortices samples of patients with a confirmed neuropathological diagnosis of FTLD-TDP associated with C9orf72 expansion (n = 1), FTLD-TDP with mutation in the GRN gene (n = 1), FTLD-TDP with unknown genetic background (n = 1) and of a normal control brain (n = 1) were collected. Brain samples (0.5 g) were homogenized as previously described with some modification (Nonaka et al., 2013). Briefly, samples were homogenized in 2.5 mL of homogenization buffer [HB: 10 mM Tris-HCl, 0.8 M NaCl, 1 mM EDTA, 1 mM dithiothreitol (DTT), 1× protease and phosphatase inhibitor cocktail tablets (Roche), pH 7.5]. Benzonase (Sigma) was added (0.25 μL) to aliquots of 500 μL for each of the lysates, which were then incubated in constant shaking (400 rpm) for 10 min at 37°C. After this, Sarkosyl was added to each aliquot (final concentration, 1%) and samples were then incubated for 20 min at 37°C under shaking (400 rpm). Ethanol was added to a final concentration of 20% and samples were incubated in constant shaking (400 rpm) for 10 min at 37°C. Samples were centrifuged at 150 000g for 60 min at RT. Supernatants were discarded, and pellets were suspended in 300 μL PBS by sonication and centrifuged at 150 000g for 60 min at RT. The resulting pellets were suspended in 50 μL of ddH2O by sonication and diluted at 10−3 v/v in ddH2O.
Cerebrospinal fluid sample collection and study population
Informed consent was given by all study participants or their next of kin. The CSF samples were collected by lumbar puncture after a standard procedure, centrifuged at 1000g for 10 min and stored in polypropylene tubes at −80°C, until analysis. Table 1 summarizes the characteristics of the study population. We retrospectively analysed CSF samples from a total of 36 ALS/FTD patients (21 males and 15 females) harbouring a pathological mutation in one of the genes known to be related to CNS TDP-43 deposition (i.e. C9orf72, TARDBP, GRN) (Supplementary Table 1). Samples were provided by several Italian institutions and specifically from: the Department of Neuroscience, Rehabilitation, Ophthalmology, Genetics, Maternal and Child Health, University of Genoa, IRCCS Ospedale Policlinico, San Martino, Genoa; the ‘Rita Levi Montalcini’ Department of Neuroscience, University of Turin, Turin; the Fondazione IRCCS Istituto Neurologico Carlo Besta, Unit of Neurology 5 and Neuropathology, Milan; the Department of Neurology and Laboratory of Neuroscience, IRCCS, Istituto Auxologico Italiano, Milan; the Molecular Markers Laboratory, IRCCS Istituto Centro San Giovanni di Dio Fatebenefratelli, Brescia; the Department of Clinical and Experimental Sciences, Neurology Unit, University of Brescia, Brescia and from the Clinical Unit of Neurology, Department of Medicine, Surgery and Health Sciences, Neurology Unit, University Hospital and Health Services of Trieste, University of Trieste, Trieste. We collected 19 CSF samples from patients with a C9orf72 expansion, three from patients with a TARDBP mutation and 14 GRN-mutated patients. The C9orf72 population was composed of 10 ALS patients (two of them with associated signs of cognitive decline, not reaching criteria for FTD diagnosis), eight FTD-affected patients and one patient with an AD-like phenotype (all of them without signs of motor neuron involvement). All TARDBP-mutated patients were clinically diagnosed as ALS. The diagnoses of ALS and FTD were made according to current diagnostic criteria (Brooks et al., 2000; Rascovsky et al., 2012). Among the GRN population, one sample was derived from a pre-symptomatic patient without any clinical sign. Except for this patient, all patients in the GRN population presented with cognitive impairment. In total, 10 GRN patients were classified as FTD and more specifically, 8 were diagnosed as behavioural variant FTD (bv-FTD) and 2 as agrammatic variant of primary progressive aphasia according to current re-defined clinical criteria (Gorno-Tempini et al., 2011; Rascovsky et al., 2012). Three GRN patients were not diagnosed as FTD. One had a clinical diagnosis of cortico-basal syndrome (CBS), one of mild cognitive impairment (MCI) and one was diagnosed as AD according to current clinical diagnostic criteria (McKhann et al., 2011; Armstrong et al., 2013). We also collected CSF from 27 age-matched control patients (14 males and 13 females), which underwent a lumbar puncture to exclude a neurological disease. In none of these cases was the suspected disease, ALS, FTD, or any other neurodegenerative disorder. Leucocyte count, glucose, total protein, blood pigments, lactate and oligoclonal IgG bands were all normal in the analysed CSF.
Table 1.
Characteristics of CSF study population
| Characteristic | C9orf72 | TARDBP | GRN | Totals patients | CTRLs |
|---|---|---|---|---|---|
| Men, n. (%) | 11 (58) | 2 (67) | 8 (57) | 21 (58) | 14 (52) |
| Age at CSF collection, y. (±SD) | 59 ± 10 | 58 ± 16 | 63 ± 8 | 60 ± 10 | 65 ± 13 |
| Age at onset, y. (±SD) | 58 ± 9 | 55 ± 15 | 59 ± 4 | 58 ± 9 | N.A. |
| Onset-CSF collection, m. (±SD) | 22 ± 17 | 32 ± 19 | 22 ± 9 | 23 ± 15 | N.A. |
| Pure ALS phenotype, n. | 8 | 3 | _ | 11 | N.A. |
| ALS plus cognitive decline n. | 2 | _ | _ | 2 | N.A. |
| Pure FTD, n. | 8 | _ | 10 | 18 | N.A. |
| CBS, n. | _ | _ | 1 | 1 | N.A. |
| MCI, n. | _ | _ | 1 | 1 | N.A. |
| AD, n. | 1 | _ | 1 | 2 | N.A. |
| Pre-symptomatic carrier, n. | _ | _ | 1 | 1 | N.A. |
| Total, n. | 19 | 3 | 14 | 36 | 27 |
| Positive in RT-QuIC, n. (%) | 18 (95) | 3 (100) | 13 (93) | 34 (94) | 4 (15) |
Data are presented as numbers (n.), (percentages) and means (±SD). The onset-CSF collection time was calculated in months and refers to the time passed between the onset of disease and the collection of the CSF sample. y.: years; m.: months; CTRLs: age-matched control patients; N.A.: not applicable. CSF: cerebrospinal fluid; ALS: amyotrophic lateral sclerosis; FTD: frontotemporal dementia; CBS: cortico-basal syndrome; MCI: mild cognitive impairment; AD: Alzheimer disease; RT-QuIC: real-time quacking-induced convertion.
In vitro generation of recombinant human TDP-43 aggregates
In vitro aggregation reactions of human TDP-43 were performed in 200 μL of reaction mix in black, clear-bottom, 96-well microplates. The reaction mix contained 10 mM PBS at pH 6.8, 5 mM DTT, 100 mM NaCl, 50 μM thioflavin T (ThT), 0.002% of sodium dodecyl sulfate (SDS) and 2 mg/mL of HuTDP-43. After sealing, the plate was incubated at 40°C, over a period of 50 h with intermittent cycles of shaking (60 s, 400 rpm, double-orbital) and rest (60 s). For HuTDP-43(263-414) aggregation, the reaction mix contained 10 mM PBS at pH 7.8, 100 mM NaCl, 10 μM ThT, 0.002% of SDS and 0.2 mg/mL of HuTDP-43(263-414). After sealing, the plate was incubated at 40°C, over a period of 24 h with intermittent cycles of shaking (60 s, 400 rpm, double-orbital) and rest (60 s). All reactions were performed in a FLUOstar OMEGA reader (BMG Labtech, Germany) and fluorescence intensity, expressed as relative fluorescence unit (RFU), was taken every 30 min using 450 ± 10 nm (excitation) and 480 ± 10 nm (emission) wavelengths and a gain of 1000. All solutions were filtered before use with 0.22 μm sterile filters. Recombinant proteins were subjected to 2 h of centrifugation at 186 000g at RT before their use in the reaction. The addition of a 3-mm glass bead (Sigma) was required to sustain protein aggregation. The final product of these reactions was collected by ultracentrifugation at 100 000g for 1 h and suspended by sonication in the same starting volume of ddH2O and used as synthetic seed. Protein aggregates were analysed by atomic force microscopy.
TDP-43 RT-QuIC optimization
All RT-QuIC reactions were performed in 200 μL of reaction mix in black, clear-bottom, 96-well microplates. To optimize the reaction for HuTDP-43 and its truncated fragment HuTDP-43(263-414), we used different experimental protocols. The RT-QuIC reaction mix contained 10 mM PBS at pH 6.8, 5 mM DTT, 100 mM NaCl, 25 μM ThT, 0.002% of SDS and 1 mg/mL of HuTDP-43. After sealing, the plate was incubated at 40 °C, over a period of 50 h with intermittent cycles of shaking (60 s, 400 rpm, double-orbital) and rest (60 s). For HuTDP-43(263-414) aggregation, the reaction mix contained 10 mM PBS at pH 7.8, 100 mM NaCl, 10 μM ThT, 0.002% of SDS and 0.2 mg/mL of HuTDP-43(263-414). After sealing, the plate was incubated at 40°C, over a period of 24 h with intermittent cycles of shaking (60 s, 400 rpm, double-orbital) and rest (60 s). We also optimized another reaction condition with HuTDP-43(263-414) in terms of reaction buffer and shaking protocol. The reaction buffer was changed as follows: 40 mM Tris (pH 8), 0.5 M GdnHCl (pH 8), 10 μM ThT, 0.002% of SDS and 0.05 mg/mL of HuTDP-43(263-414). After sealing, the plate was incubated at 40°C, over a period of 50 h exposed to 15 s of shaking every 30 min at 100 rpm (double-orbital). Reactions were performed in a FLUOstar OMEGA reader (BMG Labtech, Germany) and the fluorescence intensity, expressed as RFU, was taken every 30 min using 450 ± 10 nm (excitation) and 480 ± 10 nm (emission) wavelengths, with a bottom read and a gain of 1000. All solutions were filtered before use with 0.22-μm sterile filters. Recombinant proteins were subjected to 2 h of centrifugation at 186 000g at RT before their use in the reaction. The addition of a 3-mm glass bead (Sigma) was required to sustain protein aggregation. Reactions were seeded with 20 μL of 10−3 diluted BH samples.
CSF TDP-43 seed detection with RT-QuIC
For CSF TDP-43 seed detection, 6 μL of each CSF sample was added to 194 μL of reaction mix in black, clear-bottom, 96-well microplates. Samples were tested in triplicate. To test CSF samples, we used our optimized protocol for HuTDP-43(263-414). The RT-QuIC reaction mix contained 40 mM Tris at pH 8, 0.5 M GdnHCl (pH 8), 10 μM ThT, 0.002% of SDS and 0.05 mg/mL of HuTDP-43(263-414). All solutions were filtered before use with 0.22-μm sterile filters. Recombinant HuTDP-43(263-414) was centrifuged 2 h at 186 000g at RT to remove pre-formed aggregates before its use in the reaction. After sealing, the plate was incubated in a FLUOstar OMEGA reader (BMG Labtech, Germany) at 40°C, over a period of 72 h. Every 30 min, the reaction was exposed to 15 s of shaking at 100 rpm (double-orbital). The fluorescence intensity of the reaction was taken every 30 min using 450 ± 10 nm (excitation) and 480 ± 10 nm (emission) wavelengths, with a bottom read and a gain of 1000. The addition of a 3-mm glass bead (Sigma) was required to sustain protein aggregation. A CSF sample was considered positive in a single experiment if two out of three well reached a defined RFU threshold (five times their starting fluorescence value) before a time threshold of 56.6 h (see Lag-phase threshold definition section). To be considered as a definite positive, each sample had to test positive in two out of the three independent experiments. To investigate the detection power of our technique, we serially diluted (till 10−6) the in vitro obtained HuTDP-43(263-414) aggregates in a previously screened true-negative CSF sample which was analysed with the same protocol used to asses all other CSF samples.
Immune-depletion of TDP-43 species
For each sample, 50 μL of re-suspended PureProteome Protein A magnetic beads (MerkMillipore-LSKMAGA02) were used. After three washes with 500 μL of PBS 1× 0.1% Tween-20, beads were re-suspended in 100 μL of PBS 1× 0.1% Tween-20 and added to a cocktail composed of 10 μg of rabbit polyclonal anti-C-terminal (Sigma-Aldrich-T1580) and 10 μg of rabbit polyclonal anti-N-terminal (Sigma-Aldrich SAB4200006) TDP-43 antibodies. After 30 min of incubation at RT in constant agitation, the buffer was discarded and 60 μL of each sample was incubated with the antibody-coated beads for 24 h at 4°C with constant mixing. Depleted samples were subsequently analysed with the optimized TDP-43 RT-QuIC protocol.
In vitro generation of recombinant human α-synuclein (α-syn) and tau K18 aggregates
Recombinant full-length human α-syn and tau K18 fragment were provided from collaborators in our group. Expression and purification of recombinant tau K18 fragment and of full-length α-syn were performed as described previously (Barghorn et al., 2005; Huang et al., 2005). In vitro fibrillization reactions were performed at 37°C with intermittent cycles of shaking (50 s, 400 rpm, double-orbital) and rest (10 s) in a FLUOstar OMEGA reader (BMG Labtech, Germany). Newly formed aggregates were pelleted by ultracentrifugation (186 000 g for 1 h at 4°C) and re-suspended in sterile ddH2O. Before their use, aliquots were sonicated for 3 min in an ultrasonic bath (Branson 2510).
Atomic force microscopy analysis
TDP-43 in vitro-generated aggregates were dialysed against bi-distilled water at RT overnight and then subjected to atomic force microscopy (AFM) imaging. A volume of 10 μL was dropped onto freshly sliced muscovite mica (V-1) and incubated for 15 min for sample adhesion. The same volume of absolute ethanol was added, allowing the drop to evaporate. The sample was left to dry at RT and subjected to AFM imaging (NT-MDT Solver-Pro) in semi-contact mode, using commercial silicon cantilevers (NSG03, NT-MDT Spectrum Instruments, <10 nm radius of curvature, spring constant of 1.74 Nm−1).
Lag-phase threshold definition
We performed the RT-QuIC reaction with all control and patient samples in three separate experiments, each one in triplicate, and we collected for each well of each experiment the time needed to reach an arbitrary fluorescence threshold that we set at five times the initial fluorescence value of each well. We then calculated the median lag-phase time for each triplicate and, subsequently, the median of the medians resulting from the three independent experiments (that we will call ‘total median time’). This procedure provided a single and informative number for each patient (i.e. the total median time). After this, we plotted the distribution of total median times and from the observation of these distributions we derived the lag-phase threshold of 56.6 h (Supplementary Fig. 2A). In order to address the risk of overfitting, we cross-validated our data. For a 1000 times, we simulated to split the data set into a training set comprising 50% of the data, (i.e. 14 controls and 18 patients) and a testing set (i.e. the remaining 50%). For each simulation, we defined the threshold based on the training set and applied it to the testing set to compute the cross-validated accuracy. To illustrate how the choice of the lag-phase threshold would influence the true-positive rate and false-positive rate numbers, we built the receiver operating characteristic curve which highlights the points with the highest accuracies (Supplementary Fig. 2B).
Statistical analysis
For the correlation analysis between baseline patient characteristics and RT-QuIC positive results, we used different tests. For binary features, we performed Welch two-sample t-tests, for categorical baseline characteristics, we used ANOVAs, and for continuous features, both linear models and ANOVAs. All statistical analyses were conducted in R and figures were produced using the package ggplot2 (Wickham, 2009). All RT-QuIC graphs were produced with GraphPad (PRISM 7.0a) and the respective figures were generated with Inkscape 1.0.
Data availability
The data supporting the findings of this study are available from the corresponding author on request.
Results
Adaptation of RT-QuIC to TDP-43 amplification
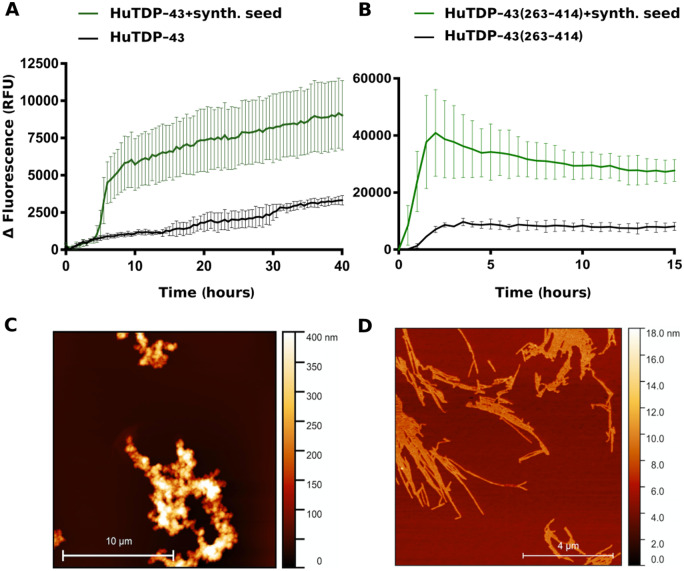
The first attempt to adapt the RT-QuIC methodology to TDP-43 was to obtain high amounts of soluble recombinant protein to be used as the substrate for the reaction. After several trials, we optimized an efficient protocol for full-length TDP-43 (HuTDP-43) production and refolding (Materials and methods section). After refolding, the recombinant protein product was highly pure, devoid of signs of degradation and functional in terms of its RNA-binding activity (Supplementary Fig. 1). The second critical step was to identify the best conditions for HuTDP-43 aggregation. In fact, several factors were modified to improve the efficiency and reproducibility of TDP-43 RT-QuIC, such as protein concentration, temperature, pH of the reaction and shaking protocol. Figure 1A shows the aggregation kinetics profile of HuTDP-43. The aggregation curve is characterized by a lag phase of about 15–20 h, followed by a slow and progressive increase in ThT fluorescence emission and a subsequent plateau reached after around 30–35 h (Fig. 1a, black line). We collected the final product of this reaction and employed it as a synthetic aggregate. When the synthetic seed was added to the reaction, we observed a clear anticipation of the lag phase associated with an increase in the amount of ThT fluorescence emission (Fig. 1A, green line). Interestingly, the aggregation profile observed in our experimental setting, substantially differed from those observed in the previous studies where the aggregation curve of HuTDP-43 completely lacked a lag phase (Johnson et al., 2009; Furukawa et al., 2011; Jiang et al., 2013). To characterize the aggregates used as synthetic seeds, we subjected them to AFM analysis, observing that they mainly displayed features of amorphous aggregates, lacking fibrillary structures (Fig. 1C). Since the C-terminal domain of TDP-43 is known to be more aggregation-prone (Saini and Chauhan, 2011; Budini et al., 2012), we produced a recombinant C-terminal fragment from amino acid 263 to the end of the protein (HuTDP-43(263-414)). Also in this case the final protein product was pure and devoid of signs of degradation (Supplementary Fig. 1). We analysed its aggregation properties and observed a typical prion-like aggregation kinetics (Fig. 1B, black line). Compared to HuTDP-43, HuTDP-43(263-414), aggregation curve showed a shorter lag phase and higher values of ThT fluorescence emission (Fig. 1). The final product of this reaction was also very efficient as synthetic seed (Fig. 1B, green line). The AFM analyses of HuTDP-43(263-414) aggregates differed substantially from those of HuTDP-43, presenting typical fibrillary features (Fig. 1D). We considered that the different aggregation profiles of HuTDP-43 and HuTDP-43(263-414) were strictly related to the different final conformation acquired by their aggregates (Fig. 1). Only a fraction of HuTDP-43 would acquire β-sheet conformation, thus binding ThT, whereas HuTDP-43(263-414) is readily converted in β-sheet-enriched fibrils.
Figure 1.
Aggregation kinetics of recombinant TDP-43 and AFM seeds characterization. Purified seed-free recombinant HuTDP-43 (2 mg/mL) (A) and HuTDP-43(263-414) (0.2 mg/mL) (B) were induced to aggregate by alternating cycles of 60 s (s) of shaking and 60 s of incubation at 40°C (black lines). The final product of their aggregation was collected and efficiently employed as synthetic seed (green lines). ThT fluorescence is plotted against time. Each graph has a specific range of ThT fluorescence values and duration of a given reaction. The experiment was performed in triplicate and each replica was performed three times. Curves represent means for all experimental replicates and bars indicate the standard deviations (SD). (C, D) AFM analysis of TDP-43 aggregates. HuTDP-43 aggregates showed features of amorphous aggregates (C), whereas HuTDP-43(263-414) (D) acquired a fibrillary structure after aggregation.
TDP-43 RT-QuIC optimization
To investigate which substrate (HuTDP-43 or HuTDP-43(263-414)) was more efficient to detect biological TDP-43 aggregates, we seeded the reaction with a small pool of pathological confirmed patient-derived BHs. Both HuTDP-43 and HuTDP-43(263-414) were efficiently seeded by three positive BH samples, whereas a negative-control BH at the same dilution (10−3) did not affect the aggregation kinetics of the reaction (Fig. 2A, B). This trial indicated that both proteins were suitable substrates for the amplification assay. However, since the reaction with HuTDP-43 required higher amounts of substrate, we selected HuTDP-43(263-414) as the most efficient product to perform further analyses. Another relevant point for the optimization of the TDP-43 RT-QuIC technology, as a tool to detect TDP-43 seeds, was to obtain a slower aggregation profile of the substrate, to provide the optimal time window for discrimination between positive and negative samples. For this purpose, we modified the reaction buffer with the addition of GdnHCl and the removal of NaCl. We also reduced the amount of the HuTDP-43(263-414) substrate from 0.2 to 0.05 mg/mL and modified the aggregation protocol (only 15 s of shaking every 30 min, at 100 rpm). After this optimization, all positive BH samples maintained their seeding activity but, as expected, with a slower aggregation kinetics (Fig. 2C).
Figure 2.
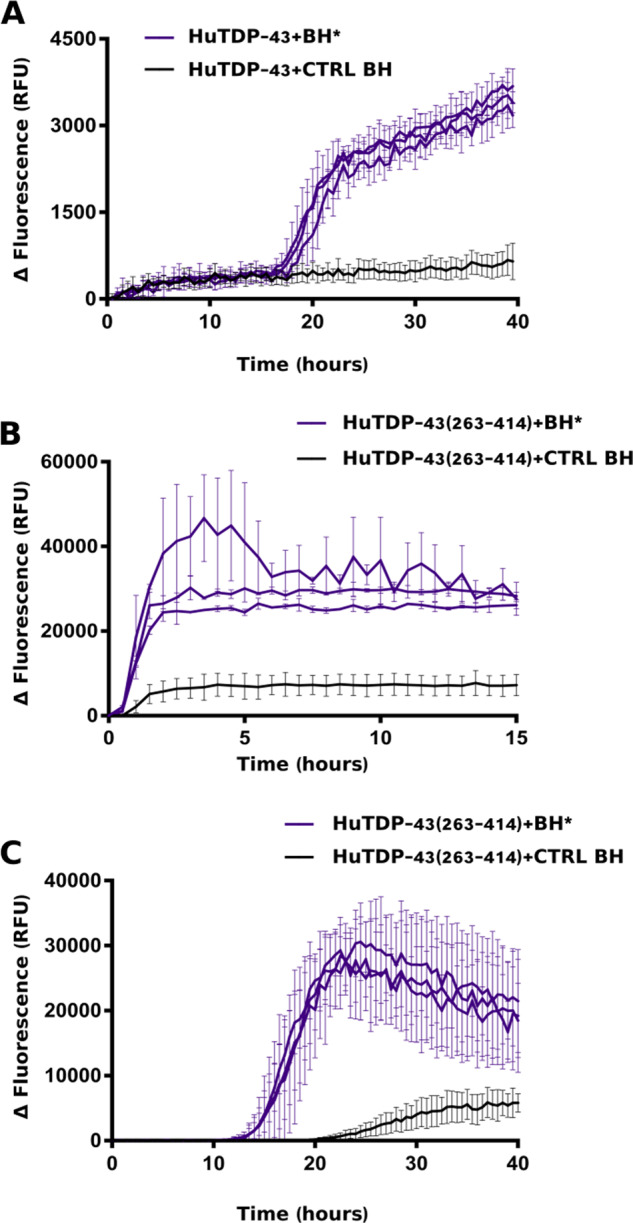
RT-QuIC analysis of BH samples. In brief, 20 μL of sonicated and diluted (10−3) BHs collected from three FTLD-TDP patients (BH*, purple lines) and one CTRL patient (CTRL BH, black line) were added to purified seed-free recombinant HuTDP-43 (1 mg/mL) (A) and HuTDP-43(263-414) (0.2 mg/mL) (B) and analysed by means of RT-QuIC. The reaction was exposed to 60 s of shaking and 60 s of rest. All positive BHs efficiently seeded the reaction, whereas the negative control BH at the same dilution (10−3) did not affect the aggregation kinetics of the reaction. (C) Optimized protocol for BH analysis using purified seed-free HuTDP-43(263-414) (0.05 mg/mL) as substrate with a different reaction buffer (no NaCl, Gdn-HCl added) and a modified protocol (shaking at 100 rpm for 15 s every 30 min at 40°C). ThT fluorescence intensity was plotted against time. Each graph has a specific range of ThT fluorescence values and duration of a given reaction. The experiment was performed in triplicate and each replica was performed three times. Curves represent means for all experimental replicates and bars indicate the standard deviations (SD).
CSF TDP-43 seed detection with RT-QuIC
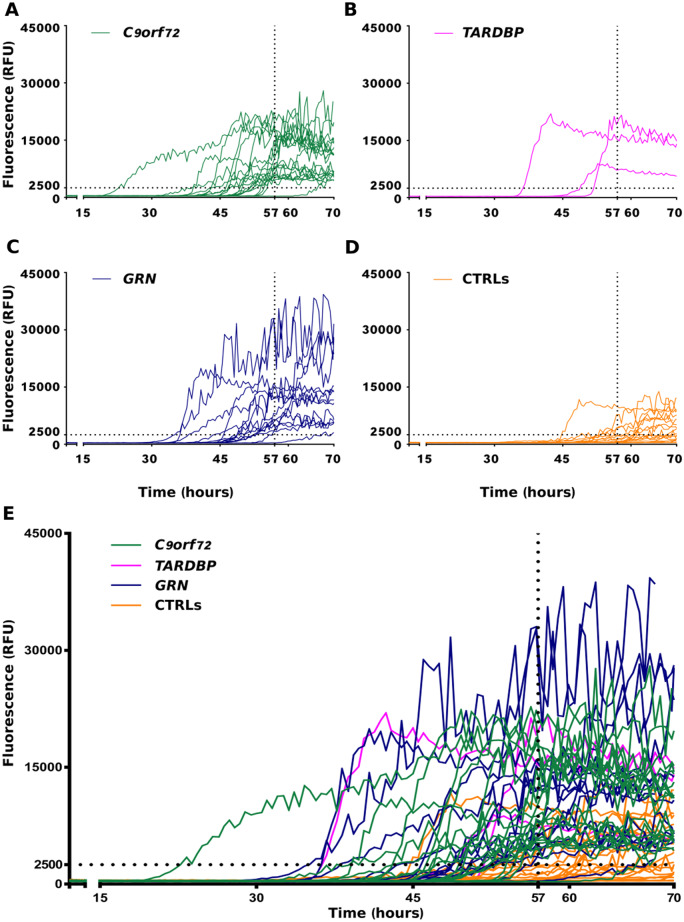
Since our final aim was to screen CSF samples, it was also important to take into account the effect of the CSF material to the reaction. When small aliquots of CSF (control or disease) were added to the reaction, we observed an inhibition of HuTDP-43(263-414) aggregation. This was expected, since this CSF inhibiting effect was also experienced by other groups (Shahnawaz et al., 2017). For this reason, the threshold that we set to discriminate between positive and negative CSF samples, derived from the comparison between all our CSF samples (disease and control) (Supplementary Fig. 2), since the spontaneous aggregation of the substrate (i.e. the reaction performed with the substrate alone without addition of CSF aliquots) was not a reliable parameter. CSF samples collected from C9orf72 expansion (n = 19), TARDBP (n = 3), GRN (n = 14) mutation carriers and age-matched controls (n = 27) (for clinical details, see Table 1) were analysed by means of RT-QuIC to investigate their effects on the kinetics of HuTDP-43(263-414) aggregation. We found that 18 out of 19 C9orf72, 3 out of 3 TARDBP and 13 out of 14 GRN samples efficiently seeded the RT-QuIC reaction (Fig. 3A, B, C, E). Among age-matched controls, we observed four positive results (Fig. 3D, E). These values correspond to a global sensitivity of 94% and specificity of 85%. In order to address the risk of overfitting, we cross-validated our data. The accuracy values predicted by the cross-validation model were 92 and 84% of sensitivity and specificity, respectively, confirming the observed experimental measurements. Among the GRN group, notably, one positive sample was derived from a healthy carrier of the mutation. There was no significant correlation between baseline characteristics (age at CSF collection, time between disease onset and CSF collection, ALS, FTD, ALS plus FTD or cognitive decline phenotypes) and RT-QuIC positive results. The one C9orf72 and the one GRN-negative patients did not display any difference compared to the other samples, possibly explaining their low seeding efficiency. More in detail, they presented clinical features (ALS without cognitive impairment and bv-FTD, respectively), age at CSF collection (45 and 58 years, respectively) and delay from onset to CSF collection (24 months for both patients) comparable to the rest of the patient population (Table 1). Regarding the four unexpected positive results among control patients (i.e. false-positive samples), we did not observe any specific feature implicated in their seeding ability. All four patients were age-matched against the study population and did not present any clinical condition related to neurodegeneration. Leucocyte count, glucose, total protein, blood pigments, lactate and oligoclonal IgG bands were all normal in their CSF. To investigate if the seeding activity of positive CSF samples was driven by the presence of TDP-43 aggregates, we used magnetic beads coated with a cocktail composed of two anti-TDP-43 antibodies to immune-deplete one true positive (i.e. a CSF sample from a patient which tested positive in the TDP-43 RT-QuIC assay) and two false-positive samples (i.e. samples from two controls which resulted positive in our analysis). After depletion, the true-positive sample had an almost abolished seeding activity and tested negative (Fig. 4A). This suggests that the procedure was able to deplete it from the majority of TDP-43 aggregates. Regarding the depleted false-positive samples, one of them became negative, whereas the other one remained positive, even if presenting a diminished intensity of ThT fluorescence emission (Figure 4B). This implies that some false-positive results may be associated with the presence of TDP-43 aggregates in the CSF of asymptomatic patients, whereas some others may be related to the presence/absence of other factors capable of interfering with the reaction in a non-specific way.
Figure 3.
RT-QuIC analysis of ALS and FTLD-TDP CSF samples. In brief, 6 μL of undiluted CSF collected from patients with a known pathological TDP-43-related mutation (C9orf72, green lines, TARDBP, magenta lines, GRN, blue lines) and from age-matched controls (CTRLs, orange lines) were added to purified seed-free recombinant HuTDP-43(263-414) (0.05 mg/mL) and analysed by means of RT-QuIC with the optimized protocol (shaking at 100 rpm for 15 s every 30 min at 40°C). ThT fluorescence intensity is plotted against time. Dotted black lines represent the ThT fluorescence and lag-phase thresholds. In the first four panels (A–D) the curves for each group are shown separately, whereas in (E) samples are shown all together. The experiment was performed in triplicate and each replica was performed three times. Each curve shows the median aggregation curve for each sample.
Figure 4.
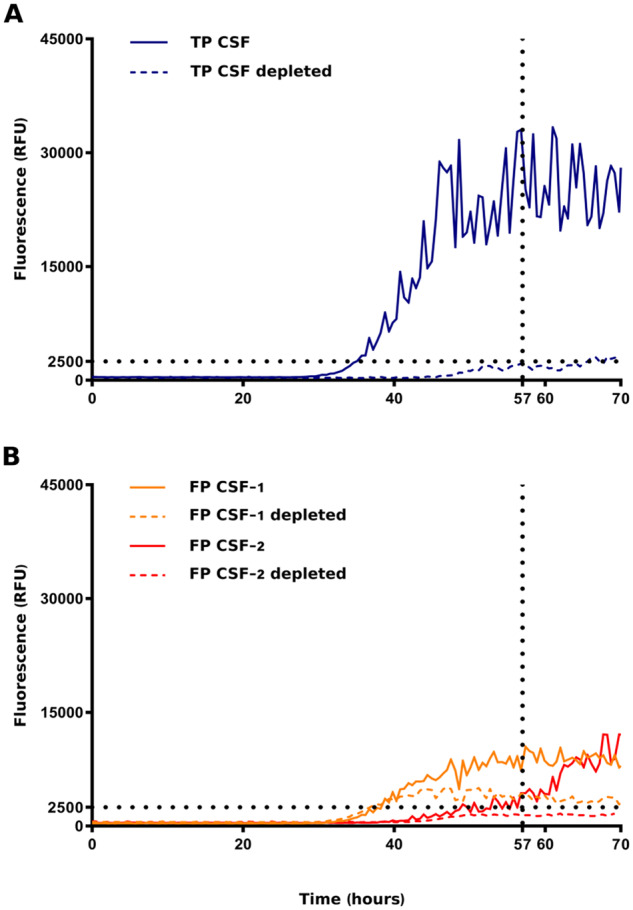
RT-QuIC analysis of immune-depleted CSF samples. One true-positive (TP) and two false-positive (FP) CSF samples were immune-depleted of TDP-43 aggregates using magnetic beads coated with a cocktail of two anti-TDP-43 antibodies directed against two different epitopes of the protein. Samples before and after immune-depletion were added to purified seed-free recombinant HuTDP-43(263-414) (0.05 mg/mL) and analysed by means of RT-QuIC (shaking at 100 rpm for 15 s every 30 min at 40°C). ThT fluorescence intensity is plotted against time. Dotted black lines represent the ThT fluorescence and lag-phase thresholds. (A) After depletion, the true-positive sample tested negative (blue dashed line). After depletion, one of the false-positive samples became negative (red dashed line), whereas the other one remained positive, even if presenting a diminished intensity of ThT fluorescence emission (yellow dashed line). The experiment was performed in triplicates and each replica was performed three times. The image represents the median aggregation curve for each sample.
Performance of TDP-43 RT-QuIC with tau and α-synuclein (α-syn) aggregates
To investigate the performance of TDP-43 RT-QuIC with aggregates composed of other pathological amyloids, we tested the seeding ability of pre-formed tau K18 and α-syn fibrils. Since we wanted to replicate the same experimental settings that we used for the CSF analysis, we diluted tau K18 or α-syn fibrils in a previously screened negative CSF from a control patients. The CSF sample spiked with 1.5 ng of tau K18 or α-syn fibrils presented no seeding activity in the TDP-43 RT-QuIC (Fig. 5). Notably, when the same amount of TDP-43 in vitro-obtained fibrils (1.5 ng) was diluted in the same negative CSF, the sample resulted positive (Fig. 5). Tau K18 and α-syn aggregates presented an efficient seeding activity when tested with their respective tau or α-syn RT-QuIC assays. These results suggest that, under these experimental conditions, TDP-43 RT-QuIC is not susceptible to the presence of tau or α-syn amyloids.
Figure 5.
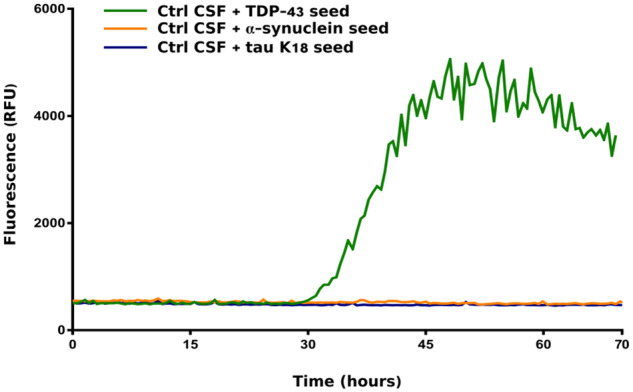
Performance of TDP-43 RT-QuIC with tau and α-synuclein aggregates. A previously screened negative CSF sample was spiked with 1.5 ng of tau K18, α-syn or TDP-43–pre-formed seeds. The CSF samples spiked with tau K18 (blue line) or α-syn (yellow line) fibrils presented no seeding activity in the optimized TDP-43 RT-QuIC. TDP-43 preformed aggregates diluted in the same negative CSF efficiently seeded the reaction (green line). Protocol details: 0.05 mg/mL of purified seed-free recombinant HuTDP-43(263-414) substrate; shaking at 100 rpm for 15 s every 30 min at 40°C. ThT fluorescence intensity is plotted against time. The experiment was performed in triplicates and each replica was performed three times. The image represents the median aggregation curve for each sample.
RT-QuIC detection of aggregated TDP-43
To measure the capabilities of RT-QuIC detection of aggregated TDP-43, we added different dilutions of the in vitro-obtained HuTDP-43(263-414) aggregates to a previously screened negative CSF sample and tested the seeding activity of these products. We observed that our assay was able to detect as little as 15 pg of TDP-43 pathological aggregates (Fig. 6). Since we knew the exact amount of the diluted TDP-43 aggregates, this was the most accurate method in our hands to characterize a dose–response correlation for the optimized RT-QuIC. Furthermore, standard detection techniques failed to quantify the content of pathological hyperphosphorylated TDP-43 in CSF samples from our cohort. For this reason, a dose–response analysis on patient-derived CSF was not feasible. Since our dose–response characterization was performed with the use of synthetic aggregates, which may or may not represent in vivo-circulating TDP-43 species, further experiments are needed to verify if this estimation also applies to patient-derived samples.
Figure 6.
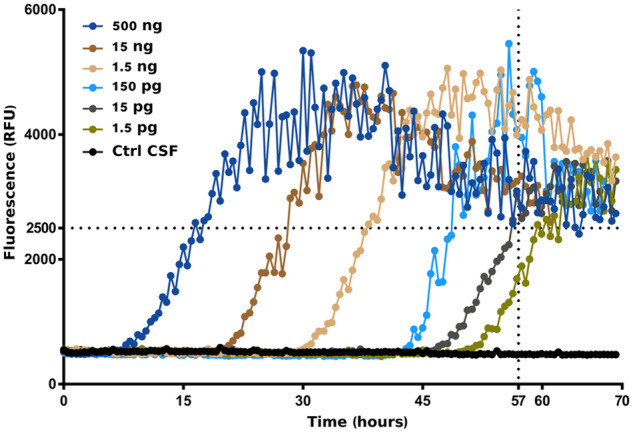
Performance of TDP-43 RT-QuIC with TDP-43 CSF. Different dilutions of in vitro-obtained HuTDP-43(263-414) aggregates were added to a previously screened negative CSF sample. TDP-43 RT-QuIC was able to detect as little as 15 pg of TDP-43 pathological aggregates. Protocol details: 0.05 mg/mL of purified seed-free recombinant HuTDP-43(263-414) substrate; shaking at 100 rpm for 15 s every 30 min at 40°C. ThT fluorescence intensity is plotted against time. Dotted black lines represent the ThT fluorescence and lag-phase thresholds. The experiment was performed in triplicate and each replica was performed three times. The image represents the median aggregation curve for each sample.
Discussion
Despite many efforts, ALS and FTD still lack treatments that are capable of interfering with the underlying pathological process. One reason is the absence of an early diagnostic biomarker able to detect the disease before irreversible neuronal damage has occurred. In fact, like amyloid β-deposition in AD (Bateman et al., 2012), it is possible that the formation and accumulation of pathological aggregates of mis-folded TDP-43 species could also begin decades before symptoms onset. As a consequence, patients are usually enrolled in trials in advanced clinical phases when neuronal damage and the neurodegenerative process have already started, thus decreasing the potential efficacy of tested compounds. The availability of a reliable biochemical marker could also help in monitoring disease progression in the evaluation of response to a specific treatment. Our approach to detect the presence of pathological TDP-43 species in the CSF of ALS and FTD patients was to exploit the RT-QuIC intrinsic ability to amplify minute amount of mis-folded proteins. Since this technology proved to efficiently perform with prions and several other prion-like proteins (Orru et al., 2014; Salvadores et al., 2014; Orru et al., 2015; Fairfoul et al., 2016; Franceschini et al., 2017; Saijo et al., 2017; Shahnawaz et al., 2017; Groveman et al., 2018; van Rumund et al., 2019) and since TDP-43 displayed prion-like features, we aimed at optimizing it for TDP-43. We report that recombinant human TDP-43 is a suitable substrate to perform the RT-QuIC reaction. We describe an efficient protocol for a large-scale production and purification of both full-length and truncated TDP-43, essential to set up and perform the RT-QuIC technique. As a first step in TDP-43 RT-QuIC adaptation, we used the final products of aggregation of both HuTDP-43 and HuTDP-43(263-414) proteins which efficiently seeded the reaction and we subsequently moved to the study of a small subset of BHs derived from autopsy-verified TDP-43-positive cases. We selected TDP-43 C-terminal fragment as the best substrate for a large-scale sample screening. We collected CSF samples from patients harbouring a pathological mutation in one of the genes known to cause TDP-43 deposition in the CNS. This approach implied at least two advantages: (i) even without a neuropathological confirmation, the presence of a known pathological TDP-43-related mutation increased the likelihood that those CSF contained toxic TDP-43 species; (ii) analysing CSF from patients with a TDP-43-related mutation reduced the pathological uncertainty related to heterogeneous clinical presentations. This second observation explains our decision to collect CSF also from patients with a clinical diagnosis different from classical ALS and FTD, as long as they harboured a TDP-43-related pathological mutation. Our data suggest that TDP-43 RT-QuIC is able to discriminate between a cohort of patients affected by ALS and FTD and age-matched controls with a total sensitivity of 94% and a specificity of 85%, detecting as little as 15 pg of TDP-43 seed. Among the GRN group, we also tested one pre-symptomatic carrier that resulted positive. This patient, to date, remains asymptomatic and is under clinical follow-up. Since the co-deposition of TDP-43 and other pathological proteins occurs frequently in many neurodegenerative disorders (Amador-Ortiz et al., 2007; Freeman et al., 2008; Lin and Dickson, 2008; Arai et al., 2009; Josephs et al., 2016; Berning and Walker, 2019), we considered inaccurate to use CSF samples derived from patients affected by other neurodegenerative diseases to assess the presence of cross-seeding. To overcome this potential limitation and to maintain the experimental settings of the CSF-optimized RT-QuIC technology, we diluted 1.5 ng of tau K18 or α-syn pre-formed aggregates in a negative CSF sample and we observed no cross-seeding activity. Our study is not without limitations. First, sensitivity and specificity of our protocol are lower than desired. According to the analysis on depleted false-positive samples, it seems that false positivity could be related to multiple factors. The fact that one false-positive sample became negative after the TDP-43 depletion protocol suggests that at least some of the observed false-positive results could be due to the presence of TDP-43 aggregates in the CSF of asymptomatic patients. Since we do not possess neuropathological confirmation for the examined samples, it is not possible to exclude that we have detected the presence of pathological TDP-43 seeds in healthy patients who would eventually develop a TDP-43-related disorder during their life span. In this regard, it should be mentioned that several reports observed the presence of age-dependent TDP-43 deposition in a discrete proportion of cognitively impaired but also healthy patients (Geser et al., 2010; Uchino et al., 2015). The fact that one false-positive sample remained positive after depletion also suggests that some of the observed false-positive results might be intrinsically related to CSF composition. The CSF samples, in fact, may differ in the presence or absence of specific molecules capable of modifying the kinetics of aggregation of the substrate in a non-specific way, independent of the presence of aggregates. A preparation method able to remove, or at least limit, possible TDP-43 interactors in the analysed samples needs to be implemented to overcome this potential limitation. We would also like to test other patient-derived tissues, to evaluate their characteristics and seeding efficiency. A second limitation of our study is the number of tested patients. On the one hand, the decision to focus on a selected population of patients (i.e. the ones with a known TDP-43-related pathogenic mutation) increased the likelihood that the suspected positive CSF really contained TDP-43 seeds. On the other hand, since these mutations are quite rare, it reduced our sample size. Even with this limitation, we considered this approach as the most efficient in the adaptation of this technology to TDP-43. In conclusion, we described a robust protocol for large-scale production of recombinant TDP-43 and for its use as a substrate in the RT-QuIC reaction. Our data on TDP-43 RT-QuIC represent a proof-of-concept of its potential for the detection of TDP-43 pathological aggregates. Together with prion, amyloid β, tau and α-syn RT-QuIC assays, a further optimization of the presented TDP-43 RT-QuIC protocol would increase the opportunity to perform the earliest and most accurate diagnosis at a single patient level.
Supplementary Material
Acknowledgements
The authors acknowledge F. Ansaloni, Scuola Internazionale Superiore di Studi Avanzati (SISSA), for his technical help in data analysis and E. De Cecco, SISSA, for providing human recombinant α-synuclein and tau proteins.
Funding
C.S. and G.L. received Scuola Internazionale Superiore di Studi Avanzati intramural funding for this study. This study was supported by the Italian Ministry of Health (Ricerca Corrente): L.B., R.G. and F.M. This study was also supported in part by the Italian Ministry of Health (GR-2013-02355724 to F.M. and GR-2013-02355764 to V.S.). The Association for Frontotemporal Degeneration Susan Marcus Memorial Fund Clinical Research Pilot Grant awarded to P.C.
Competing interests
The authors report no competing interests.
Glossary
- AD =
Alzheimer’s disease
- AFM =
atomic force microscopy
- ALS =
amyotrophic lateral sclerosis
- Aβ =
amyloid beta
- BH =
brain homogenate
- BSA =
bovine serum albumin
- bv-FTD =
behavioural variant of frontotemporal dementia
- CBS =
cortico-basal syndrome
- ddH2O =
bi-distilled water
- DTT =
dithiothreitol
- EDTA =
ethylenediaminetetraacetic acid
- EMSA =
electrophoretic mobility shift assay
- FTD =
frontotemporal dementia
- FTLD =
frontotemporal lobar degeneration
- GdnHCl =
guanidine hydrochloride
- MCI =
mild cognitive impairment
- RFU =
relative fluorescence unit
- RT =
room temperature
- RT-QuIC =
real time quaking induced conversion reaction
- SDS =
sodium dodecyl sulfate
- TDP-43 =
transactive response DNA-binding protein of 43 kDa
- ThT =
thioflavin T
- α-syn =
α-synuclein
Supplementary material
Supplementary material is available at Brain Communications online.
References
- Amador-Ortiz C, Lin WL, Ahmed Z, Personett D, Davies P, Duara R, et al. TDP-43 immunoreactivity in hippocampal sclerosis and Alzheimer’s disease. Ann Neurol 2007; 61: 435–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arai T, Hasegawa M, Akiyama H, Ikeda K, Nonaka T, Mori H, et al. TDP-43 is a component of ubiquitin-positive tau-negative inclusions in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Biochem Biophys Res Commun 2006; 351: 602–11. [DOI] [PubMed] [Google Scholar]
- Arai T, Mackenzie IR, Hasegawa M, Nonoka T, Niizato K, Tsuchiya K, et al. Phosphorylated TDP-43 in Alzheimer’s disease and dementia with Lewy bodies. Acta Neuropathol 2009; 117: 125–36. [DOI] [PubMed] [Google Scholar]
- Armstrong MJ, Litvan I, Lang AE, Bak TH, Bhatia KP, Borroni B, et al. Criteria for the diagnosis of corticobasal degeneration. Neurology 2013; 80: 496–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayers JI, Cashman NR. Prion-like mechanisms in amyotrophic lateral sclerosis. Handb Clin Neurol 2018; 153: 337–54. [DOI] [PubMed] [Google Scholar]
- Bang J, Spina S, Miller BL. Frontotemporal dementia. Lancet 2015; 386: 1672–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barghorn S, Biernat J, Mandelkow E. Purification of recombinant tau protein and preparation of Alzheimer-paired helical filaments in vitro. Methods Mol Biol 2005; 299: 35–51. [DOI] [PubMed] [Google Scholar]
- Bateman RJ, Xiong C, Benzinger TL, Fagan AM, Goate A, Fox NC, et al. Clinical and biomarker changes in dominantly inherited Alzheimer’s disease. N Engl J Med 2012; 367: 795–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berning BA, Walker AK. The pathobiology of TDP-43 C-terminal fragments in ALS and FTLD. Front Neurosci 2019; 13: 335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braak H, Brettschneider J, Ludolph AC, Lee VM, Trojanowski JQ, Tredici KD. Amyotrophic lateral sclerosis—a model of corticofugal axonal spread. Nat Rev Neurol 2013; 9: 708–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brettschneider J, Del Tredici K, Irwin DJ, Grossman M, Robinson JL, Toledo JB, et al. Sequential distribution of pTDP-43 pathology in behavioral variant frontotemporal dementia (bvFTD). Acta Neuropathol 2014; 127: 423–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks BR, Miller RG, Swash M, Munsat TL, World Federation of Neurology Research Group on Motor Neuron. El Escorial revisited: revised criteria for the diagnosis of amyotrophic lateral sclerosis. Amyotroph Lateral Scler Other Motor Neuron Disord 2000; 1: 293–9. [DOI] [PubMed] [Google Scholar]
- Budini M, Buratti E, Stuani C, Guarnaccia C, Romano V, Conti LD, et al. Cellular model of TAR DNA-binding protein 43 (TDP-43) aggregation based on its C-terminal Gln/Asn-rich region. J Biol Chem 2012; 287: 7512–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairfoul G, McGuire LI, Pal S, Ironside JW, Neumann J, Christie S, et al. Alpha-synuclein RT-QuIC in the CSF of patients with alpha-synucleinopathies. Ann Clin Transl Neurol 2016; 3: 812–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feneberg E, Gray E, Ansorge O, Talbot K, Turner MR. Towards a TDP-43-based biomarker for ALS and FTLD. Mol Neurobiol 2018; 55: 7789–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franceschini A, Baiardi S, Hughson AG, McKenzie N, Moda F, Rossi M, et al. High diagnostic value of second generation CSF RT-QuIC across the wide spectrum of CJD prions. Sci Rep 2017; 7: 10655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman SH, Spires-Jones T, Hyman BT, Growdon JH, Frosch MP. TAR-DNA binding protein 43 in Pick disease. J Neuropathol Exp Neurol 2008; 67: 62–7. [DOI] [PubMed] [Google Scholar]
- Furukawa Y, Kaneko K, Watanabe S, Yamanaka K, Nukina N. A seeding reaction recapitulates intracellular formation of Sarkosyl-insoluble transactivation response element (TAR) DNA-binding protein-43 inclusions. J Biol Chem 2011; 286: 18664–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geser F, Robinson JL, Malunda JA, Xie SX, Clark CM, Kwong LK, et al. Pathological 43-kDa transactivation response DNA-binding protein in older adults with and without severe mental illness. Arch Neurol 2010; 67: 1238–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorno-Tempini ML, Hillis AE, Weintraub S, Kertesz A, Mendez M, Cappa SF, et al. Classification of primary progressive aphasia and its variants. Neurology 2011; 76: 1006–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groveman BR, Orru CD, Hughson AG, Raymond LD, Zanusso G, Ghetti B, et al. Rapid and ultra-sensitive quantitation of disease-associated alpha-synuclein seeds in brain and cerebrospinal fluid by alphaSyn RT-QuIC. Acta Neuropathol Commun 2018; 6: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C, Ren G, Zhou H, Wang CC. A new method for purification of recombinant human alpha-synuclein in Escherichia coli. Protein Expr Purif 2005; 42: 173–7. [DOI] [PubMed] [Google Scholar]
- Jiang LL, Che MX, Zhao J, Zhou CJ, Xie MY, Li HY, et al. Structural transformation of the amyloidogenic core region of TDP-43 protein initiates its aggregation and cytoplasmic inclusion. J Biol Chem 2013; 288: 19614–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson BS, Snead D, Lee JJ, McCaffery JM, Shorter J, Gitler AD. TDP-43 is intrinsically aggregation-prone, and amyotrophic lateral sclerosis-linked mutations accelerate aggregation and increase toxicity. J Biol Chem 2009; 284: 20329–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josephs KA, Murray ME, Whitwell JL, Tosakulwong N, Weigand SD, Petrucelli L, et al. Updated TDP-43 in Alzheimer’s disease staging scheme. Acta Neuropathol 2016; 131: 571–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin WL, Dickson DW. Ultrastructural localization of TDP-43 in filamentous neuronal inclusions in various neurodegenerative diseases. Acta Neuropathol 2008; 116: 205–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling SC, Polymenidou M, Cleveland DW. Converging mechanisms in ALS and FTD: disrupted RNA and protein homeostasis. Neuron 2013; 79: 416–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie IR, Neumann M. Molecular neuropathology of frontotemporal dementia: insights into disease mechanisms from postmortem studies. J Neurochem 2016; 138: 54–70. [DOI] [PubMed] [Google Scholar]
- Maniecka Z, Polymenidou M. From nucleation to widespread propagation: a prion-like concept for ALS. Virus Res 2015; 207: 94–105. [DOI] [PubMed] [Google Scholar]
- McKhann GM, Knopman DS, Chertkow H, Hyman BT, Jack CR, Jr, Kawas CH, et al. The diagnosis of dementia due to Alzheimer’s disease: recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimers Dement 2011; 7: 263–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann M, Sampathu DM, Kwong LK, Truax AC, Micsenyi MC, Chou TT, et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science 2006; 314: 130–3. [DOI] [PubMed] [Google Scholar]
- Ng AS, Rademakers R, Miller BL. Frontotemporal dementia: a bridge between dementia and neuromuscular disease. Ann NY Acad Sci 2015; 1338: 71–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonaka T, Hasegawa M. TDP-43 Prions. Cold Spring Harb Perspect Med 2018; 8: a024463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonaka T, Masuda-Suzukake M, Arai T, Hasegawa Y, Akatsu H, Obi T, et al. Prion-like properties of pathological TDP-43 aggregates from diseased brains. Cell Rep 2013; 4: 124–34. [DOI] [PubMed] [Google Scholar]
- Orru CD, Bongianni M, Tonoli G, Ferrari S, Hughson AG, Groveman BR, et al. A test for Creutzfeldt-Jakob disease using nasal brushings. N Engl J Med 2014; 371: 519–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orru CD, Groveman BR, Hughson AG, Zanusso G, Coulthart MB, Caughey B. Rapid and sensitive RT-QuIC detection of human Creutzfeldt-Jakob disease using cerebrospinal fluid. MBio 2015; 6: e02451–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porta S, Xu Y, Restrepo CR, Kwong LK, Zhang B, Brown HJ, et al. Patient-derived frontotemporal lobar degeneration brain extracts induce formation and spreading of TDP-43 pathology in vivo. Nat Commun 2018; 9: 4220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rascovsky K, Hodges JR, Knopman D, Mendez MF, Kramer JH, Grossman M, on behalf of the International bvFTD Criteria Consortium (FTDC), et al. Sensitivity of revised diagnostic criteria for the behavioural variant of frontotemporal dementia. Brain 2012; 135: e214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravits JM, La Spada AR. ALS motor phenotype heterogeneity, focality, and spread: deconstructing motor neuron degeneration. Neurology 2009; 73: 805–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saberi S, Stauffer JE, Schulte DJ, Ravits J. Neuropathology of amyotrophic lateral sclerosis and its variants. Neurol Clin 2015; 33: 855–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saijo E, Ghetti B, Zanusso G, Oblak A, Furman JL, Diamond MI, et al. Ultrasensitive and selective detection of 3-repeat tau seeding activity in Pick disease brain and cerebrospinal fluid. Acta Neuropathol 2017; 133: 751–65. [DOI] [PubMed] [Google Scholar]
- Saini A, Chauhan VS. Delineation of the core aggregation sequences of TDP-43 C-terminal fragment. Chembiochem 2011; 12: 2495–501. [DOI] [PubMed] [Google Scholar]
- Salvadores N, Shahnawaz M, Scarpini E, Tagliavini F, Soto C. Detection of misfolded Abeta oligomers for sensitive biochemical diagnosis of Alzheimer’s disease. Cell Rep 2014; 7: 261–8. [DOI] [PubMed] [Google Scholar]
- Shahnawaz M, Mukherjee A, Pritzkow S, Mendez N, Rabadia P, Liu X, et al. Discriminating alpha-synuclein strains in Parkinson’s disease and multiple system atrophy. Nature 2020; 578: 273–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahnawaz M, Tokuda T, Waragai M, Mendez N, Ishii R, Trenkwalder C, et al. Development of a biochemical diagnosis of Parkinson disease by detection of alpha-synuclein misfolded aggregates in cerebrospinal fluid. JAMA Neurol 2017; 74: 163–72. [DOI] [PubMed] [Google Scholar]
- Smethurst P, Newcombe J, Troakes C, Simone R, Chen YR, Patani R, et al. In vitro prion-like behaviour of TDP-43 in ALS. Neurobiol Dis 2016; 96: 236–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsermentseli S, Leigh PN, Goldstein LH. The anatomy of cognitive impairment in amyotrophic lateral sclerosis: more than frontal lobe dysfunction. Cortex 2012; 48: 166–82. [DOI] [PubMed] [Google Scholar]
- Turner MR, Al-Chalabi A, Chio A, Hardiman O, Kiernan MC, Rohrer JD, et al. Genetic screening in sporadic ALS and FTD. J Neurol Neurosurg Psychiatry 2017; 88: 1042–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchino A, Takao M, Hatsuta H, Sumikura H, Nakano Y, Nogami A, et al. Incidence and extent of TDP-43 accumulation in aging human brain. Acta Neuropathol Commun 2015; 3: 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rumund A, Green AJE, Fairfoul G, Esselink RAJ, Bloem BR, Verbeek MM. alpha-Synuclein real-time quaking-induced conversion in the cerebrospinal fluid of uncertain cases of parkinsonism. Ann Neurol 2019; 85: 777–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker LC, Jucker M. Neurodegenerative diseases: expanding the prion concept. Annu Rev Neurosci 2015; 38: 87–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H. ggplot2: elegant graphics for data analysis. Berlin: Springer; 2009. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data supporting the findings of this study are available from the corresponding author on request.