Abstract
We hypothesized that a joint analysis of cancer risk-associated single-nucleotide polymorphism (SNP) and somatic mutations in tumor samples can predict functional and potentially causal SNPs from GWASs. We used mutations reported in the Catalog of Somatic Mutations in Cancer (COSMIC). Confirmed somatic mutations were subdivided into two groups: (1) mutations reported as SNPs, which we call mutational/SNPs and (2) somatic mutations that are not reported as SNPs, which we call mutational/noSNPs. It is generally accepted that the number of times a somatic mutation is reported in COSMIC correlates with its selective advantage to tumors, with more frequently reported mutations being more functional and providing a stronger selective advantage to the tumor cell. We found that mutations reported ≥10 times in COSMIC—frequent mutational/SNPs (fmSNPs) are likely to be functional. We identified 12 cancer risk-associated SNPs reported in the Catalog of published GWASs at least 10 times as confirmed somatic mutations and therefore deemed to be functional. Additionally, we have identified 42 SNPs that are tightly linked (R2 ≥ 0.8) to SNPs reported in the Catalog of published GWASs as cancer risk associated and that are also reported as fmSNPs. As a result, 54 candidate functional/potentially causal cancer risk associated SNPs were identified. We found that fmSNPs are more likely to be located in evolutionarily conserved regions compared with cancer risk associated SNPs that are not fmSNPs. We also found that fmSNPs also underwent positive selection, which can explain why they exist as population polymorphisms.
The study identified candidates for causal SNPs among GWAS-detected cancer risk SNPs and SNPs that are tightly linked to cancer GWAS-reported ones. The results of the analysis can be used to guide functional studies of cancer risk-associated SNPs.
Introduction
Two major cancer genetics research activities are as follows: (1) identification of cancer risk-associated single-nucleotide polymorphisms (SNPs) by genome-wide association studies (GWASs) and (2) identification of somatic mutations in tumor samples. These disciplines barely talk to each other even though cross talk between these two areas is likely to be beneficial. Here we jointly consider cancer risk-associated SNPs and somatic mutations detected in tumor samples. Our hypothesis was that considering SNPs and somatic mutations together will help to identify cancer risk-associated SNPs that are functional.
GWASs have identified a very large number of SNPs associated with cancer risk (1–3). It is generally accepted that the majority of GWAS-detected SNPs are not functional/causal SNPs, but are rather proxies linked to unknown causal variants (4,5). Detection of causal/functional variants among GWAS-detected SNPs is challenging. Several bioinformatics tools have been developed to predict functional/potentially causal SNPs. These tools use SNP characteristics including the level of evolutionary conservation of the site (6), projected effect of the SNP on protein structure (7) and other SNP features (8) for assessing the SNP’s functionality. To our best knowledge, these tools never used somatic mutation data to predict SNP functionality.
To identify tumor somatic mutations that also exist as SNPs, we have overlapped SNPs reported in the dbSNP database (9,10), with somatic mutations from the Catalog Of Somatic Mutations In Cancer (COSMIC) (11,12). Several millions of unique somatic mutations are reported in COSMIC. Mutations differ by the number of times they are reported in COSMIC, with the majority of them being singletons. To a great extent, cancer development is driven by an acquisition of driver mutations providing selective advantage (e.g. a higher proliferation rate or better survival) to the mutant clone. Clone-carrying driver mutations survive better and propagate faster (13). A higher propagation rate of cell clones with driver mutations results in an excess of driver mutations when compared with selectively neutral passenger mutations (14,15). Therefore, mutations providing a selective advantage (driver mutations) are detected more frequently in tumor samples compared with selectively neutral passenger mutations (15). Therefore, the number of times a somatic mutation is reported can be used as an indicator of functionality (16,17). If a frequent and therefore potentially functional somatic mutation also exists as a SNP, one can expect that it will be functional as an SNP also. In this study, we used somatic mutations detected in tumor samples jointly with SNP to identify candidate functional/potentially causal variants among GWAS-detected cancer risk-associated SNPs.
Materials and methods
Identification of somatic mutations that also exist as SNPs
Our first step was to identify somatic mutations that also exist as SNPs, regardless of whether SNPs were reported as cancer risk-associated or not. Hereafter we call them mutational/SNPs (mSNPs). To identify mSNPs we overlapped confirmed somatic mutations, detected by whole genome screens and reported in COSMIC (Build 88) with SNPs reported in dbSNP database. Confirmed somatic mutations are mutations established to be somatic rather than germline polymorphisms based on sequencing of paired normal tissue or in some cases comparison of the detected variant with the SNP database. Matching somatic mutations to SNPs was done based on the condition that the following three characteristics are the same for the somatic mutation and the SNP: (i) chromosome number, (ii) nucleotide position on the chromosome and (iii) the type of nucleotide substitution, e.g. C>T. We used human genome Build 38 for both SNPs and somatic mutations.
The number of times a somatic mutation is reported in COSMIC was used as a measure of its functionality. Before counting individual mutations, we excluded duplicates related to using different reference transcripts for annotation. Exactly the same mutation can be reported in COSMIC several times depending on the transcript used as a reference. To remove reference transcript-related duplicates, we identified mutations with the same chromosomal position, same type of nucleotide substitution and the same sample ID and removed all duplicates. A total of 1 719 388 annotation duplicates were detected and removed. After removing annotation duplicates, 2 955 675 mSNPs were identified. Those mSNPs were detected in 40 550 tumor samples across 42 cancer types.
Minor allele frequency and number of mutational counts
mSNPs are genetic variants that exist as both somatic mutations and germline polymorphisms. As a result of their dual nature, mSNPs have two key characteristics: the number of counts in COSMIC and minor allele frequency (MAF). Mutational counts were estimated for confirmed somatic mutations detected by whole genome sequencing. MAFs were estimated based on the data from Trans-Omics for Precision Medicine (TOPMed) project (18). We found that the majority of mSNPs are rare: >95% of them have MAFs < 0.001. mSNPs with MAF < 0.001 were excluded from the analysis because they may be false positives and also because their practical/clinical significance is questionable. After removing rare mSNPs, the total number of mSNPs was 599 630. Among these, we studied mSNPs reported in COSMIC at least 10 times as somatic mutations. The goal of implementing this threshold was to exclude mSNPs that are likely not functional. The justification for using 10 counts as a threshold is given below. Hereafter we refer to the mSNPs reported at least 10 times in COSMIC as frequent mutational/SNPs (fmSNPs). In total, 8533 fmSNPs were identified (Supplementary Table S1, available at Carcinogenesis Online). The table includes mutation ID, SNP ID, number of times the mutation is reported in COSMIC, MAF from TOPMed database and prediction of functionality by functional analysis through hidden Markov model (FATHMM) (19).
Selecting threshold for mSNPs that are likely to be functional
We found that the majority of COSMIC mutations are singletons. Singletons are likely to be selectively neutral and their presence in tumor samples reflects the randomness of the mutational process (20). On the other hand, somatic mutations frequently detected in tumor samples are likely to be functional: they are positively selected because tumor cells need them to proliferate and survive (21–23). Even though we know that singletons are likely to be selectively neutral and frequent somatic mutations are likely to be functional it is difficult, however, to decide where to put a threshold between neutral and functional mutations.
We used two approaches to decide on the boundary between functional mSNPs and mSNPs that are likely to be noise. In the first approach, we categorized mSNPs based on the number of times they are reported in COSMIC. In each category, we estimated the proportion of mSNPs predicted to be deleterious by FATHMM method (19). Our assumption was that the proportion of mSNPs predicted to be deleterious by FATHMM reflects a proportion of functional SNPs in the group.
The second approach was gene based. We first identified genes linked to mSNPs and then checked if those genes cluster in cancer pathways. For this analysis, all mSNPs were divided into five categories based on the number of counts in COSMIC: singletons, mSNPs reported 2–4 times, SNPs reported 5–9 times, mSNPs reported 9–19 times and mSNPs reported ≥20 times in COSMIC. The grouping strategy was chosen to ensure that the numbers of mSNPs across categories are comparable to each other. For each category of genes, we conducted the pathway enrichment analysis and counted the number of cancer-related pathways among the 20 top pathways. The pathways were defined by Kyoto Encyclopedia of Genes and Genomes (KEGG). KEGG pathways were designed using published data on proteins interactions as well as experimental evidence (24). Therefore, circular reasoning (selecting cancer-relevant genes based on how frequently they are mutated in cancer) is unlikely to be an issue in this analysis. However, it would be an issue, if we used the proportions of COSMIC-defined cancer census genes as a measure of cancer relevance.
Cancer risk-associated SNPs
Cancer risk-associated SNPs were retrieved from the Catalog of the published GWASs (25). The database was accessed 14 October 2019, and findings from 194 cancer GWASs were available. Table 1 shows the numbers of SNPs, genes and numbers of published GWASs for different cancer types. Cancer types are defined in the table exactly how they defined in the catalog. Supplementary Table S2 (available at Carcinogenesis Online) shows the complete list of cancer risk-associated SNPs used in this study. A total of 1013 unique SNPs were reported in the catalog as cancer risk associated at the GWA significance level—P ≤ 5 × 10−8. These SNPs are linked to 1011 genes. Because some SNPs are associated with risk in multiple cancers, the number of reports/lines in Supplementary Table S1 (available at Carcinogenesis Online)is larger than the number of unique SNPs.
Table 1.
Number of SNPs and linked genes associated with risk of different cancer types based on the data from the Catalog of the published GWASs
| Cancer type | Number of SNPs | Number of unique genes | Number of studies |
|---|---|---|---|
| Prostate cancer | 161 | 136 | 30 |
| Breast cancer | 134 | 147 | 37 |
| Lung cancer | 75 | 95 | 23 |
| Colorectal cancer | 74 | 84 | 30 |
| Testicular germ cell tumor | 59 | 68 | 8 |
| Basal cell carcinoma | 45 | 53 | 8 |
| Squamous cell lung carcinoma | 43 | 59 | 4 |
| Breast cancer (estrogen-receptor negative) | 43 | 45 | 3 |
| Non-melanoma skin cancer | 41 | 56 | 2 |
| Lung cancer in ever smokers | 33 | 47 | 18 |
| Pancreatic cancer | 32 | 36 | 9 |
| Lung adenocarcinoma | 30 | 37 | 8 |
| Multiple myeloma | 26 | 27 | 4 |
| Glioma | 22 | 15 | 6 |
| Breast cancer (early onset) | 18 | 13 | 2 |
| Epithelial ovarian cancer | 18 | 22 | 2 |
| Thyroid cancer | 18 | 17 | 5 |
| Bladder cancer | 15 | 22 | 7 |
| Esophageal adenocarcinoma | 14 | 22 | 2 |
| Squamous cell carcinoma | 14 | 17 | 3 |
| Melanoma | 14 | 17 | 9 |
| Nasopharyngeal carcinoma | 13 | 16 | 5 |
| Endometrial cancer | 12 | 14 | 4 |
| Renal cell carcinoma | 11 | 10 | 6 |
| Cutaneous squamous cell carcinoma | 11 | 15 | 1 |
| Esophageal cancer | 11 | 12 | 2 |
| Ovarian cancer | 10 | 13 | 5 |
| Cervical cancer | 9 | 12 | 5 |
| Endometrial endometrioid carcinoma | 6 | 10 | 1 |
fmSNPs linked to the cancer risk-associated SNPs
GWASs typically report a single most significant risk-associated SNP in the region. The reported most significant SNPs are not necessarily causal/functional variants. Functional and potentially causal variants can be linked to the reported SNPs but not reported because they may have happened to be less significant. Therefore, one needs to look for functional (potentially causal) SNPs among the SNPs linked to the reported most significant variant. We identified fmSNPs among SNPs linked to those reported in the catalog of published GWASs. As a first step, we identified SNPs located in the adjacent ±50 kb regions. We used a 50 kb region because it is about the size of an average linkage disequilibrium (LD) block in the human genome (26). For SNPs located in the human leukocyte antigen region we used ±100 kb adjacent region because LD blocks in the human leukocyte antigen region are larger (27). We obtained LDs between the GWAS-reported SNP and the SNPs from the adjacent region from the LDLink database. Pairwise LDs were assessed separately for five major ethnic groups: Africans, Mixed Americans, East Asians, Europeans and South Asians (28). SNPs with R2 ≥ 0.8 in at least one group were considered to be proxy for the reported cancer risk-associated SNP. Among those proxies we have identified fmSNPs as candidate functional SNPs.
Estimates of selection pressure on mSNPs
Evolutionary conservation of the site is often used as a measure of functionality of the genetic variant (29). Genetic polymorphisms, e.g. SNP, located in a site with a signature of negative or positive selection are likely to be functional, whereas SNPs located in sites with no evidence of selection are likely to be neutral. We compared mSNPs and mutations that are not reported as SNPs (not-mSNP) by evolutionary conservation. We used PhyloP method to estimate strength and direction of natural selection on a given site (30). The PhyloP analyzes the distribution of nucleotide substitutions in an evolutionary tree of 44 vertebrate species. The method estimates the expected number of substitutions per site under the assumption of neutral evolution and compares them with the number of substitutions that have actually occurred in the site on the tree to generate likelihood score. Positive scores indicate slower-than-neutral evolution and negative ones—faster-than-neutral evolution of the site. We categorized not-mSNPs and mSNPs by the number of counts in COSMIC and estimated PhyloP scores for each count category.
In a separate analysis, we used Phylogenetic Analysis with Space/Time models (PHAST) to identify SNPs located in evolutionary conserved regions (31). PHAST uses multiple alignments of sequences from 100 vertebrate species to identify evolutionarily conserved regions. We estimated proportions of SNPs located in evolutionary conserved regions for mSNPs stratified by number of COSMIC counts and GWAS-detected associations with cancer risk.
Results
Majority of mSNPs are singletons
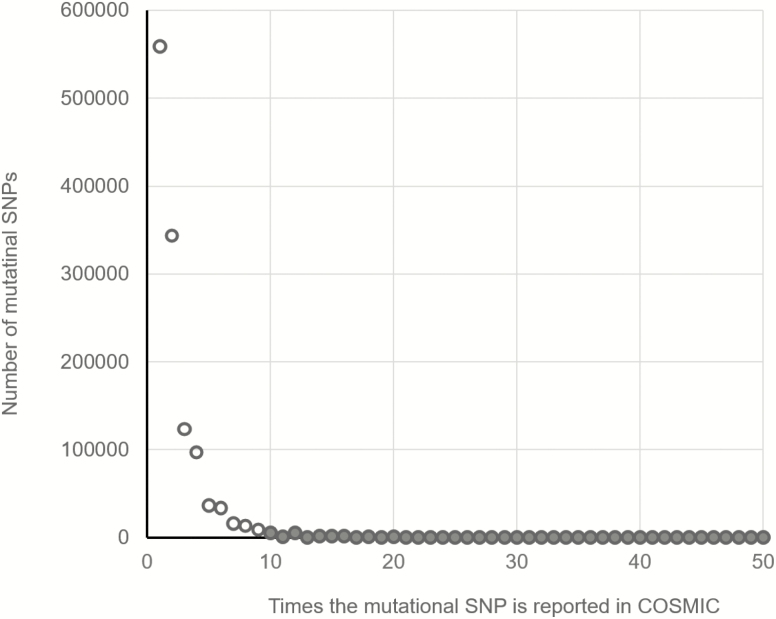
Figure 1 shows the distribution of mSNPs by the number of counts in COSMIC. One can see that the majority of mSNPs are singletons. There were >3 million singletons that comprise >45% of all mSNPs. The proportion of the mSNPs with two counts was 27%, and the proportion of the mSNPs with three counts was 10%. The proportions of SNPs with at least 10 COSMIC counts was <0.5%. The complete list of mSNPs categorized by the number of times they are reported in COSMIC with corresponding counts (number of cases) and their percentages can be found in Supplementary Table S1 (available at Carcinogenesis Online).
Figure 1.
The number of singletons and other number of counts categories among mutational SNPs. The absolute majority of mutational SNPs in COSMIC are singletons. mSNPs with at least 10 counts are shown as filled circles and with <10 counts as open circles.
Proportions of mSNPs predicted to be pathogenic by FATHMM in count categories
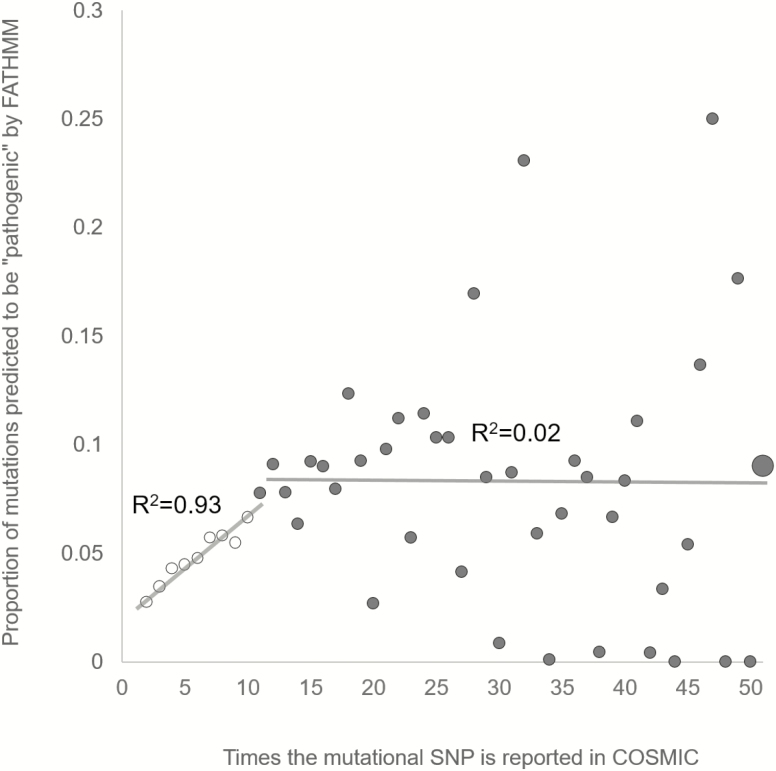
Figure 2 shows proportions of mutational SNPs classified as ‘pathogenic’ by FATHMM. mSNPs were categorized in 51 groups based on the number of counts in COSMIC with mSNPs with >50 counts combined in one group. One can see that the association between the number of counts and the proportion of mSNPs predicted to be pathogenic by FATHMM is not uniform across count categories. mSNPs with one to nine counts show positive linear association between the number of counts and the proportion of pathogenic mSNPs (orange dots in Figure 2). However, among mSNPs with at least 10 COSMIC counts there is no association between the number of counts and the proportion of mutations predicted to be pathogenic (blue dots in Figure 2).
Figure 2.
Proportions of the mSNPs predicted to be pathogenic by FATHMM. There is a positive linear association between the number of counts and the proportion of mutations predicted to be pathogenic in first nine categories (small open circles) (Pearson r = 0.96, n = 8, P = 0.00004). Categories with 10–50 counts (small filled circles) or more (a large filled circle) show no association between the number of counts and the proportion of pathogenic mutations (Pearson r = −0.06, n = 39, P = 0.72). Increasing variance after nine COSMIC counts is likely due to small sample sizes in those groups (see Figure 1).
Genes identified by linkage to the frequent mSNPs cluster in cancer-related pathways
For this analysis, we used mutation-linked genes provided by COSMIC annotation somatic mutations. We identified genes linked to the mSNPs from 5 count categories: singletons, mSNPs with 2–4 counts, mSNPs with 5–9 counts, mSNPs with 10–19 counts, and the mSNPs with >19 counts in COSMIC and then ran a pathway enrichment analysis. Table 2 reports 30 most significant pathways for each count category. Among 30 most significant pathways, we were interested to identify pathways directly related to cancer (shown in red in Table 2). No cancer pathways were identified in the singleton category or mSNPs with two to four counts in COSMIC. There was one cancer-related pathway in the group with five to nine counts. Four cancer-related pathways were identified in the group with 10–19 counts and 7 in the group with ≥20 counts. We also considered cancer-related pathways (shown in orange in Table 2) that do not mention cancer directly. The results of the analysis of the cancer-related pathways are consistent with the results of the analysis of pathways directly associated with cancer. Therefore, pathway enrichment analysis shows that genes linked to the mSNPs reported ≥10 times in COSMIC tend to cluster in cancer-related pathways.
Table 2.
Top pathways for the genes linked to mSNPs categorized by the number of counts in COSMIC
| COSMIC singletons (7290) | 2–4 COSMIC counts (8430) | 5–9 COSMIC counts (5340) | 10–19 COSMIC counts (2747) | >19 COSMIC counts (968) | |||||
|---|---|---|---|---|---|---|---|---|---|
| KEGG pathway | P-value | KEGG pathway | P-value | KEGG pathway | P-value | KEGG pathway | P-value | KEGG pathway | P-value |
| Pancreatic secretion | 0.00000058 | Endocytosis | 0.00011 | Ascorbate and aldarate metabolism | 0.00078 | Adherens junction | 0.00015 | Drug metabolism—cytochrome P450 | 0.0000083 |
| Endocytosis | 0.000087 | Pancreatic secretion | 0.00033 | Pancreatic secretion | 0.00085 | Fc gamma R-mediated phagocytosis | 0.00035 | Metabolism of xenobiotics by cytochrome P450 | 0.000023 |
| Calcium signaling pathway | 0.00057 | Hematopoietic cell lineage | 0.0027 | Endocrine and other factor-regulated calcium reabsorption | 0.0028 | Pentose and glucuronate interconversions | 0.00051 | Pentose and glucuronate interconversions | 0.000034 |
| Gastric acid secretion | 0.0015 | Epstein-Barr virus infection | 0.0036 | Fc gamma R-mediated phagocytosis | 0.0031 | Pathways in cancer | 0.0023 | Ascorbate and aldarate metabolism | 0.000048 |
| Neuroactive ligand-receptor interaction | 0.0022 | Renin secretion | 0.0068 | Protein digestion and absorption | 0.0041 | Cell adhesion molecules | 0.0026 | Chemical carcinogenesis | 0.000058 |
| Endocrine and other factor-regulated calcium reabsorption | 0.0037 | Leukocyte transendothelial migration | 0.0082 | Pentose and glucuronate interconversions | 0.0042 | Endometrial cancer | 0.0027 | Endometrial cancer | 0.00006 |
| Leukocyte transendothelial migration | 0.004 | TNF signaling pathway | 0.0086 | Long-term depression | 0.0044 | Long-term depression | 0.0052 | Prostate cancer | 0.00017 |
| Carbohydrate digestion and absorption | 0.0064 | Synaptic vesicle cycle | 0.0096 | Cell adhesion molecules | 0.0079 | Serotonergic synapse | 0.0055 | Porphyrin and chlorophyl metabolism | 0.00025 |
| Thyroid hormone synthesis | 0.0098 | Proximal tubule bicarbonate reclamation | 0.0099 | Metabolism of xenobiotics by cytochrome P450 | 0.0087 | Axon guidance | 0.0056 | Adherens junction | 0.00027 |
| Fc gamma R-mediated phagocytosis | 0.01 | Calcium signaling pathway | 0.017 | Inflammatory mediator regulation of TRP channels | 0.0097 | Proteoglycans in cancer | 0.0068 | Pathways in cancer | 0.00032 |
| Starch and sucrose metabolism | 0.013 | Endocrine and other factor-regulated calcium reabsorption | 0.018 | Endocytosis | 0.016 | Focal adhesion | 0.007 | Drug metabolism—other enzymes | 0.00052 |
| Protein digestion and absorption | 0.015 | Biosynthesis of antibiotics | 0.02 | Chemical carcinogenesis | 0.017 | ECM-receptor interaction | 0.0072 | Central carbon metabolism in cancer | 0.0016 |
| Glycerolipid metabolism | 0.016 | Gastric acid secretion | 0.031 | cAMP signaling pathway | 0.019 | Prostate cancer | 0.0083 | Retinol metabolism | 0.0016 |
| Synaptic vesicle cycle | 0.018 | Fc gamma R-mediated phagocytosis | 0.036 | Axon guidance | 0.021 | Endocrine and other factor-regulated calcium reabsorption | 0.012 | Proteoglycans in cancer | 0.0017 |
| TGF-beta signaling pathway | 0.018 | Circadian entrainment | 0.041 | Thyroid hormone signaling pathway | 0.023 | HIF-1 signaling pathway | 0.012 | Arrhythmogenic right ventricular cardiomyopathy | 0.0023 |
| cAMP signaling pathway | 0.019 | Cell adhesion molecules | 0.044 | Thyroid hormone synthesis | 0.024 | Morphine addiction | 0.012 | Thyroid hormone signaling pathway | 0.0026 |
| Cell adhesion molecules | 0.02 | Long-term depression | 0.045 | Insulin secretion | 0.024 | GnRH signaling pathway | 0.012 | Thyroid cancer | 0.0034 |
| Epstein-Barr virus infection | 0.028 | Glycerolipid metabolism | 0.045 | Bladder cancer | 0.024 | Ascorbate and aldarate metabolism | 0.013 | Colorectal cancer | 0.0046 |
| Phosphatidylinositol signaling system | 0.028 | Mineral absorption | 0.049 | Epstein-Barr virus infection | 0.025 | Arrhythmogenic right ventricular cardiomyopathy | 0.016 | Cell adhesion molecules | 0.0071 |
| Mucin type O-Glycan biosynthesis | 0.033 | Inflammatory mediator regulation of TRP channels | 0.049 | Histidine metabolism | 0.026 | Leukocyte transendothelial migration | 0.017 | Steroid hormone biosynthesis | 0.01 |
| Renin secretion | 0.041 | Adherens junction | 0.053 | TNF signaling pathway | 0.027 | Vibrio cholera infection | 0.017 | Melanoma | 0.011 |
| Aldosterone synthesis and secretion | 0.044 | Complement and coagulation cascades | 0.054 | Calcium signaling pathway | 0.027 | Biosynthesis of antibiotics | 0.019 | Calcium signaling pathway | 0.012 |
| Salivary secretion | 0.046 | Epithelial cell signaling in Helicobacter pylori infection | 0.055 | Purine metabolism | 0.028 | Fc epsilon RI signaling pathway | 0.019 | Bladder cancer | 0.019 |
| Signaling pathways regulating pluripotency of stem cells | 0.047 | p53 signaling pathway | 0.055 | Circadian entrainment | 0.028 | Cholinergic synapse | 0.02 | Hepatitis B | 0.019 |
| AMPK signaling pathway | 0.049 | Arrhythmogenic right ventricular cardiomyopathy | 0.055 | Salivary secretion | 0.029 | Type II diabetes mellitus | 0.02 | MAPK signaling pathway | 0.024 |
| Aldosterone-regulated sodium reabsorption | 0.049 | Protein digestion and absorption | 0.055 | Drug metabolism—cytochrome P450 | 0.029 | Circadian entrainment | 0.02 | Oxytocin signaling pathway | 0.025 |
| Olfactory transduction | 0.05 | Salivary secretion | 0.057 | Adherens junction | 0.029 | Calcium signaling pathway | 0.021 | B cell receptor signaling pathway | 0.027 |
| Hippo signaling pathway | 0.055 | NF-kappa B signaling pathway | 0.069 | Inositol phosphate metabolism | 0.039 | Thyroid cancer | 0.041 | Cholinergic synapse | 0.04 |
Number of genes used in pathway analysis is shown in parenthesis.
About 1% of cancer risk-associated SNPs are fmSNPs
Based on the results described in the two previous sections, mSNPs reported ≥10 times in COSMIC (fmSNPs) are considered to be functional. One can expect that fmSNPs are functional not only as mutations but also as germline polymorphisms SNPs. Therefore, we checked if fmSNPs are represented among GWAS-detected cancer risk-associated SNPs reported in the Catalog of published GWASs. A total of 12 fmSNPs were identified among cancer risk SNPs (Table 3). Taking into account that there are 1013 unique cancer risk-associated SNPs reported in the Catalog of published GWASs, ~1% of SNPs from there are potentially functional fmSNPs.
Table 3.
Frequent mutational SNPs reported in the Catalog of published GWASs as cancer risk associated
| rs ID | COSMIC ID | position | Ref | Alt | Gene | SNP | Count | MAF | GWAS cancer type | PubMed ID | P-value | Odds ratio |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs10934853 | 87135561 | 3:128319530 | C | A | EEFSEC | intronic | 24 | 0.43 | Prostate cancer | 19767754 | 3.1E-10 | 1.12 |
| rs10936599 | 89751629 | 3:169774313 | C | T | MYNN | syn | 12 | 0.22 | Colorectal cancer | 20972440 | 3E-8 | 1.04 |
| rs1801516 | 88127386 | 11:108304735 | G | A | ATM | nonsyn | 15 | 0.09 | Melanoma | 21983787 | 3.3E-09 | 1.19 |
| rs2274223 | 94964513 | 10:94306584 | A | G | PLCE1 | nonsyn | 26 | 0.31 | Esophageal cancer | 21642993 | 4E-20 | 1.35; 1.34 |
| rs2292884 | 88401131 | 2:237534583 | A | G | MLPH | nonsyn | 72 | 0.35 | Prostate cancer | 21743057 | 4E-8 | 1.14 |
| rs3765524 | 94964492 | 10:94298541 | C | T | PLCE1 | nonsyn | 24 | 0.32 | Esophageal and gastric cancer | 20729852 | 2E-9 | 1.35 |
| rs3781264 | 94964531 | 10:94310618 | A | G | PLCE1 | intronic | 18 | 0.25 | Esophageal and gastric cancer | 20729852 | 4E-9 | 1.36 |
| rs5768709 | 103312035 | 22:48533757 | A | G | FAM19A5 | intronic | 15 | 0.35 | Pancreatic cancer | 22158540 | 1E-10 | 1.25 |
| rs6983267 | 151207513 | 8:127401060 | G | T | CASC8; CCAT2 | intronic; non-coding | 16 | 0.37 | Colorectal and prostate cancer | 28960316; 26034056 | 2E-21; 3E-27 | 1.18; 1.25 |
| rs8034191 | 106432881 | 15:78513681 | T | C | HYKK | intronic | 16 | 0.26 | Lung cancer | 19654303 | 3E-26 | 1.29 |
| rs8100241 | 99852108 | 19:17282085 | G | A | ANKLE1; USHBP1 | nonsyn; intronic | 11 | 0.46 | Breast cancer | 22976474 | 4E-8 | 1.14 |
| rs9364554 | 86706928 | 6:160412632 | C | T | SLC22A3 | intronic | 10 | 0.21 | Prostate cancer | 26034056 | 6E-12 | 1.14 |
SNPs linked to the reported cancer risk-associated SNPs
We have identified 54 fmSNPs linked to the SNPs reported to be cancer risk associated by the Catalog of published GWASs (Supplementary Table 3, available at Carcinogenesis Online). Together with 12 cancer risk SNPs identified earlier (Table 3), the total number of candidate functional SNPs is 66. Those candidate SNPs are mapped to 47 unique genes (see Supplementary Table 3, available at Carcinogenesis Online, for gene information).
Comparison of selective pressure on not-mSNPs and mSNPs
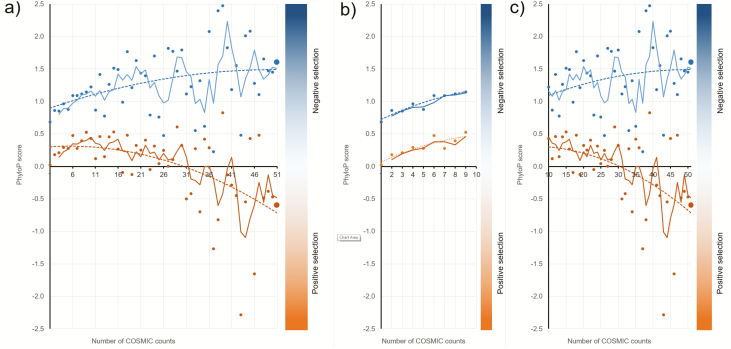
Positive PhyloP score for a given nucleotide position (site) indicates negative selection, meaning that the substitution rate for the site is lower compared with the substitution rate expected under neutral evolution. Negative PhyloP score is indicative of positive selection for a given site—the substitution rate for the site is higher compared with the substitution rate expected under neutral evolution. Figure 3 shows PhyloP scores for the somatic mutations categorized based on the number of COSMIC counts. mSNPs and not-mSNPs were analyzed separately. Mean PhyloP score for not-mSNPs are shown as blue dots and PhyloP scores for mSNPs are shown as orange dots. Overall PhyloP scores for not-mSNPs are higher compared with mSNPs, indicating that mutations that do not exist as polymorphisms are under stronger negative selection compared with the mutations that also exist as germline polymorphisms.
Figure 3.
PhyloP scores for somatic mutations stratified by the number of times they are reported in COSMIC. Somatic mutations not reported as SNPs (blue dots) and somatic mutations reported as SNPs (orange dots) were analyzed separately. Large dots show mean PhyloP scores for the somatic mutations reported >50 times in COSMIC. Dotted lines show polynomial regression and solid lines moving averages. (a) All count categories; (b) somatic mutations with 1–9 COSMIC counts; and (c) somatic mutations reported ten or more times in COSMIC.
Because the level of evolutionary conservation of the site reflects the strength of purifying selection one can expect that mutations frequently detected in COSMIC and, therefore, expected to be functional, will be preferentially located in evolutionary conserved sites. This is exactly what we observed for not-mSNPs (blue trend line in Figure 3). However for mSNPs (orange dots in Figure 3) the picture is more complicated. At the beginning—COSMIC counts from 1 to 9 we observed a positive correlation between number of counts and PhyloP score (Figure 3b). In this range mSNPS curve parallels the curve for not-mSNPs. Starting from the count 10, however, the curve for not-mSNPs continues to rise, whereas the curve for mSNPs becomes down-bound (Figure 3c). The downward trend for mSNPs indicates that functional somatic mutations that also exist as germline polymorphisms tend to be under positive selection.
fmSNPs are more likely to be located in evolutionary conserved regions than not-fmSNPs
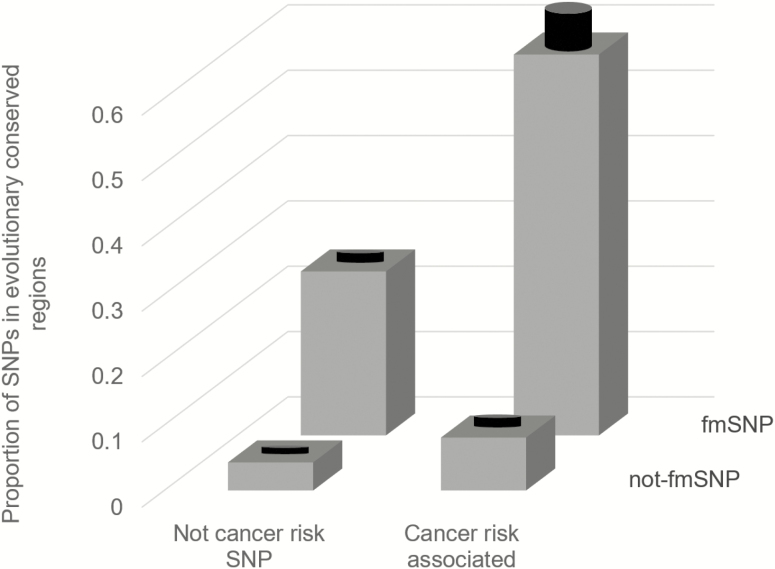
We estimated the proportion of SNPs located in evolutionary conserved regions. We subdivided all SNPs in those reported to be cancer risk-associated by the Catalog of published GWASs and SNPs that are located in physical proximity to the cancer risk-associated SNP (±1000 nucleotides) but not reported as cancer risk-associated. We selected physically linked SNPs for comparison because they are expected to be similar in terms of nucleotide and gene content. Each SNP category was further stratified into fmSNPs and not-fmSNPs. SNPs not reported to be cancer risk associated and not reported as fmSNPs were used as a reference. The results of the analysis are shown in Figure 4. The proportion of SNPs located in evolutionary conserved regions in the reference group was 0.043 ± 0.001. Proportions of SNPs in evolutionary conserved regions were significantly higher for all other three categories. The highest proportion of SNPs in conserved regions (58.3 ± 7.1%) was observed among fmSNPs reported to be cancer risk associated.
Figure 4.
Proportions of SNPs located in evolutionary conserved regions among SNPs categorized based on being cancer risk associated and frequently reported as somatic mutation (fmSNP). Black cylinders indicate standard error of the proportion.
Discussion
Our analysis was based on two assumptions: (1) somatic mutations frequently detected in tumor samples are functional and (2) if a functional mutation exists as a SNP, it is also functional as SNP. Our data indicate that fmSNPs, that is, somatic mutations with at least 10 counts in COSMIC, are likely to be functional. A total of 8536 unique fmSNPs have been identified in the analysis. Twelve fmSNPs have been reported to be associated with cancer risk by GWASs. An additional 54 fmSNPs tightly linked to the cancer risk SNPs have been identified making the total number of potentially functional cancer risk-related SNPs equal to 66. Those SNPs represent only a small fraction of all identified fmSNPs, as there are 8536–66 = 8470 fmSNPs that are not reported as cancer risk-associated. We think that many of the remaining 8470 fmSNPs may be cancer relevant: they may be associated with cancer progression, survival and/or response to treatment. Below we provide our pilot analysis supporting this hypothesis. Our analysis identified 23 fmSNPs reported at least 100 times in COSMIC. These SNPs are linked to 17 genes (listed here according to the number of COSMIC counts): CACNA1C, CACNB2, SMIM4, FCRLA, IRF5, MDM4, PCDHGA11, YAP1, MADCAM1, CACNA1G, PDE9A, SPATA3, PCDHAC2, PCDHA10, HSD17B4, ERBB2 and MYBPC1. Ten of them have published evidence of an association with cancer progression, survival and/or response to treatment. For example, it has been demonstrated that somatic mutations in CACNA1C are associated with adverse prognosis of endometrial cancer (32). Loss of IRF5 expression in ductal carcinoma contributes to metastasis (33). It has been demonstrated that MDMF influences immune response in breast cancer (34). Dysregulation of PCDHGA11 is associated with progression of various cancers (35). Overexpression of YAP1 is associated with poor prognosis in breast cancer (36). MADCAM1 plays an important role in response to oxorubicin treatment (37). Inactivation of CACNA1G in colorectal cancer increases cell proliferation and suppresses apoptosis (38). It has been demonstrated that PDE9A suppression induces apoptosis of breast cancer cells (39). HSD17B4 has been shown to increase liver cancer progression (40). ERBB2 plays a critical role in the development and progression of various cancer types, especially breast cancer (41).
Our pilot analysis, therefore, demonstrates that fmSNPs are enriched by functional cancer-related SNPs. Frequent mutational SNPs can be used to identify causal variants among SNPs detected by GWASs as well as for targeted association analysis of cancer-related phenotypes. The hypothesis that fmSNPs are enriched by functional polymorphisms is further supported by the observation that fmSNPs more frequently located in evolutionary conserved regions than not-fmSNPs (Figure 4).
Comparative analysis of the frequency of fmSNPs and known driver mutations shows that fmSNPs are not as frequently detected in tumor samples as known driver mutations. The maximal count of mSNPs was 286 for mutation COSM3931613 or SNP ID rs201777030. Considering that these counts were detected among 40 550 tumor samples, the frequency of the most frequent mSNP is 0.7%. The median count of fmSNPs is 13, which transforms into the median frequency of fmSNPs of 0.03%. Known driver mutations are typically detected at the frequency of 2%, with some them being much more frequent (found in 50% of samples) (42). Therefore, the estimated frequencies of fmSNPs by two orders of magnitude are lower compared with the frequencies of known driver mutations. The difference in mutation frequencies between known drivers and fmSNPs may be related to the fact that mSNPs exist also as polymorphisms. It is known that genetic polymorphisms are mostly neutral or slightly deleterious (43). If a mutation is only slightly functional, genetic drift and other random factors may increase its population frequency because the pressure of negative selection is too weak to completely eliminate them from the population genetic pool.
Strong driver mutations do not exist as germline polymorphisms since they occur in the genes with important cellular functions (cell cycle, apoptosis) (44,45). To be able to exist as germline polymorphisms, genetic variants have to be only marginally functional or positively selected. We found that mSNPs reported at least 10 times as somatic mutations tend to be positively selected (Figure 3c). This observation suggests that genetic variants with relatively strong functional effects can exist as polymorphism only if they are positively selected. One can expect that positive selection will finally lead to fixation of the advantageous mutations but it will take some time during which the variant will exist as a polymorphism.
One of the advantages of using fmSNPs to identify cancer-related functional SNPs is that counts of fmSNPs are linkage independent. If, for example, we have several SNPs in the region that are in perfect LD and only one of them exists as a frequent somatic mutation it is an indication that this specific SNP is functional but not the others in LD with it. The drawback of fmSNP approach to identify functional SNPs relates to the fact that we are using the number of mutational counts as a proxy for functionality. Thus functionality in our analysis is narrowly defined: we are talking about functionality related to the selective advantage of tumor cells, e.g. a higher proliferation rate, or better survival. Other relevant functional variants that do not affect the behavior of tumor cells directly (e.g. influencing smoking behavior) cannot be identified using fmSNP approach.
The positive association between the number of times a somatic mutation is reported in COSMIC and the probability that the somatic mutation is functional is likely to be continuous: the more frequent the mutation, the more likely it is to be functional. Therefore, using a threshold for the number of times a mutation is reported in COSMIC to define a functional SNPs is a simplification. There is no doubt that there are functional SNPs among mutational SNPs that do not meet the frequency criterion to be considered fmSNPs. As the number of studies deposited in COSMIC increases (the repository is updated quarterly), the total number of mutations reported in COSMIC is expected to increase, which will require to modify (elevate) the threshold for functional significance. Therefore, the researchers wishing to use somatic mutations data as a guidance for the identification of potentially functional SNPs will need to consider the frequency of a given count category in the current COSMIC version.
In conclusion, by overlapping somatic mutations reported in COSMIC and SNPs reported in dbSNP, we have identified genetic variants of a dual nature: existing as somatic mutations and as population polymorphisms, termed mSNPs. We used the number of mutational counts in COSMIC as a proxy for cancer-relevant functionality: mutations reported more often were assumed to be more functional. We identified >8000 frequent mSNPs—those reported in COSMIC ≥10 times are considered to be likely functional polymorphisms based on the result of this study. Twelve of fmSNPs are reported as cancer risk associated by a GWAS. Additionally, we have identified 54 fmSNPs linked to the GWAS-detected SNPs. These SNPs are candidates for functional/potentially causal cancer risk-associated SNPs. fmSNPs can be used to identify causal SNPs associated with cancer risk, survival, progression and response to treatment.
Supplementary Material
Acknowledgements
We would like to thank the members of the Institute for Clinical and Translational Research (Baylor College of Medicine) for helpful discussion of the study.
Glossary
Abbreviations
- COSMIC
Catalog of Somatic Mutations in Cancer
- FATHMM
functional analysis through hidden Markov model
- fmSNPs
frequent mutational/SNPs
- GWASs
genome-wide association studies
- LD
linkage disequilibrium
- MAF
minor allele frequency
- mSNPs
mutational/SNPs
- SNPs
single-nucleotide polymorphisms
Funding
This work was supported in part by the National Institutes of Health U19 CA148127 and P01 CA206980-01A1. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Conflict of Interest Statement: None declared.
References
- 1. Benafif S. et al. ; PRACTICAL Consortium (2018) A review of prostate cancer genome-wide association studies (GWAS). Cancer Epidemiol. Biomarkers Prev., 27, 845–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bossé Y. et al. (2018) A decade of GWAS results in lung cancer. Cancer Epidemiol. Biomarkers Prev., 27, 363–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Farashi S. et al. (2019) Post-GWAS in prostate cancer: from genetic association to biological contribution. Nat. Rev. Cancer, 19, 46–59. [DOI] [PubMed] [Google Scholar]
- 4. Han B. et al. (2010) A Markov blanket-based method for detecting causal SNPs in GWAS. BMC Bioinformatics, 11(Suppl 3), S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Schmitt A.O. et al. (2010) CandiSNPer: a web tool for the identification of candidate SNPs for causal variants. Bioinformatics, 26, 969–970. [DOI] [PubMed] [Google Scholar]
- 6. Johansen M.B. et al. (2013) Prediction of disease causing non-synonymous SNPs by the Artificial Neural Network Predictor NetDiseaseSNP. PLoS One, 8, e68370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mueller S.C. et al. (2015) BALL-SNP: combining genetic and structural information to identify candidate non-synonymous single nucleotide polymorphisms. Genome Med., 7, 65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yang Y. et al. (2019) AWESOME: a database of SNPs that affect protein post-translational modifications. Nucleic Acids Res., 47(D1), D874–D880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Day I.N. (2010) dbSNP in the detail and copy number complexities. Hum. Mutat., 31, 2–4. [DOI] [PubMed] [Google Scholar]
- 10. Saccone S.F. et al. (2011) New tools and methods for direct programmatic access to the dbSNP relational database. Nucleic Acids Res., 39(database issue), D901–D907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Forbes S.A. et al. (2016) COSMIC: high-resolution cancer genetics using the Catalogue of Somatic Mutations in Cancer. Curr. Protoc. Hum. Genet., 91, 10.11.1–10.11.37. [DOI] [PubMed] [Google Scholar]
- 12. Tate J.G. et al. (2019) COSMIC: the Catalogue Of Somatic Mutations In Cancer. Nucleic Acids Res., 47(D1), D941–D947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bailey M.H. et al. ; MC3 Working Group; Cancer Genome Atlas Research Network (2018) Comprehensive characterization of cancer driver genes and mutations. Cell, 173, 371–385.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Merid S.K. et al. (2014) Distinguishing between driver and passenger mutations in individual cancer genomes by network enrichment analysis. BMC Bioinformatics, 15, 308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pon J.R. et al. (2015) Driver and passenger mutations in cancer. Annu. Rev. Pathol., 10, 25–50. [DOI] [PubMed] [Google Scholar]
- 16. Gorlov I.P. et al. (2018) Gene characteristics predicting missense, nonsense and frameshift mutations in tumor samples. BMC Bioinformatics, 19, 430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lawrence M.S. et al. (2013) Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature, 499, 214–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kowalski M.H. et al. ; NHLBI Trans-Omics for Precision Medicine (TOPMed) Consortium; TOPMed Hematology & Hemostasis Working Group (2019) Use of >100,000 NHLBI Trans-Omics for Precision Medicine (TOPMed) Consortium whole genome sequences improves imputation quality and detection of rare variant associations in admixed African and Hispanic/Latino populations. PLoS Genet., 15, e1008500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rogers M.F. et al. (2018) FATHMM-XF: accurate prediction of pathogenic point mutations via extended features. Bioinformatics, 34, 511–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Harris K. (2018) The randomness that shapes our DNA. Elife, 7, 1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gopal P. et al. (2019) Clonal selection confers distinct evolutionary trajectories in BRAF-driven cancers. Nat. Commun., 10, 5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hodgkin P.D. (2018) Modifying clonal selection theory with a probabilistic cell. Immunol. Rev., 285, 249–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Steele E.J. (2017) Reverse transcriptase mechanism of somatic hypermutation: 60 years of clonal selection theory. Front. Immunol., 8, 1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kanehisa M. (2002) The KEGG database. Novartis Found. Symp., 247, 91–101; discussion 101. [PubMed] [Google Scholar]
- 25. Buniello A. et al. (2019) The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res., 47(D1), D1005–D1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Olivier M. (2003) A haplotype map of the human genome. Physiol. Genomics, 13, 3–9. [DOI] [PubMed] [Google Scholar]
- 27. Osoegawa K. et al. (2019) Tools for building, analyzing and evaluating HLA haplotypes from families. Hum. Immunol., 80, 633–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Machiela M.J. et al. (2015) LDlink: a web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants. Bioinformatics, 31, 3555–3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cooper G.M. et al. (2008) Qualifying the relationship between sequence conservation and molecular function. Genome Res., 18, 201–205. [DOI] [PubMed] [Google Scholar]
- 30. Pollard K.S. et al. (2010) Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res., 20, 110–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ramani R. et al. (2019) PhastWeb: a web interface for evolutionary conservation scoring of multiple sequence alignments using phastCons and phyloP. Bioinformatics, 35, 2320–2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Qiao Z. et al. (2019) Mutations in KIAA1109, CACNA1C, BSN, AKAP13, CELSR2, and HELZ2 are associated with the prognosis in endometrial cancer. Front. Genet., 10, 909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bi X. et al. (2011) Loss of interferon regulatory factor 5 (IRF5) expression in human ductal carcinoma correlates with disease stage and contributes to metastasis. Breast Cancer Res., 13, R111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Haupt S. et al. (2017) The role of MDM2 and MDM4 in breast cancer development and prevention. J. Mol. Cell Biol., 9, 53–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Berx G. et al. (2009) Involvement of members of the cadherin superfamily in cancer. Cold Spring Harb. Perspect. Biol., 1, a003129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Guo L. et al. (2019) YAP1 overexpression is associated with poor prognosis of breast cancer patients and induces breast cancer cell growth by inhibiting PTEN. FEBS Open Bio, 9, 437–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wang J. et al. (2015) Doxorubicin induces apoptosis by targeting Madcam1 and AKT and inhibiting protein translation initiation in hepatocellular carcinoma cells. Oncotarget, 6, 24075–24091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Toyota M. et al. (1999) Inactivation of CACNA1G, a T-type calcium channel gene, by aberrant methylation of its 5′ CpG island in human tumors. Cancer Res., 59, 4535–4541. [PubMed] [Google Scholar]
- 39. Saravani R. et al. (2012) Inhibition of phosphodiestrase 9 induces cGMP accumulation and apoptosis in human breast cancer cell lines, MCF-7 and MDA-MB-468. Cell Prolif., 45, 199–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lu X. et al. (2019) 17β-Hydroxysteroid dehydrogenase 4 induces liver cancer proliferation-associated genes via STAT3 activation. Oncol. Rep., 41, 2009–2019. [DOI] [PubMed] [Google Scholar]
- 41. Pegram M.D. (2013) Treating the HER2 pathway in early and advanced breast cancer. Hematol. Oncol. Clin. North Am., 27, 751–765, viii. [DOI] [PubMed] [Google Scholar]
- 42. Iranzo J. et al. (2018) Cancer-mutation network and the number and specificity of driver mutations. Proc. Natl Acad. Sci. USA, 115, E6010–E6019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ohta T. (1973) Slightly deleterious mutant substitutions in evolution. Nature, 246, 96–98. [DOI] [PubMed] [Google Scholar]
- 44. Tokheim C.J. et al. (2016) Evaluating the evaluation of cancer driver genes. Proc. Natl Acad. Sci. USA, 113, 14330–14335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Vogelstein B. et al. (2013) Cancer genome landscapes. Science, 339, 1546–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.