Abstract
The nuclear pore complex (NPC), comprised of individual nucleoporin (Nup) proteins, controls nucleo-cytoplasmic transport of RNA and protein, and is important for regulating plant growth and development. However, there are no reports on this complex in fruit tree species. In this study, we identified 38 apple Nups and named them based on the known Arabidopsis thaliana homologs. We also completed bioinformatics analyses of the intron and exon structural data for apple Nups. The proteins encoded by the apple Nups lacked a universally conserved domain. Moreover, a phylogenetic analysis separated the apple and A. thaliana Nups into three groups. The phylogenetic tree indicated that MdNup54 and MdNup62 are most closely related to genes in other Rosaceae species. To characterize the 38 candidate Malus domestica Nups, we measured their stage-specific expression levels. Our tests revealed these proteins were differentially expressed among diverse tissues. We analyzed the expression levels of seven apple Nups in response to an indole-3-acetic acid (IAA) treatment. The phytohormone treatment significantly inhibited apple flowering. A qRT-PCR analysis proved that an IAA treatment significantly inhibited the expression of these seven genes. A preliminary study regarding two members of the Nup62 subcomplex, MdNup54 and MdNup62, confirmed these two proteins can interact with each other. A yeast two-hybrid assay verified that MdNup54 can interact with MdKNAT4 and MdKNAT6. On the basis of the study results, we identified apple NPC and predicted its structure and function. The data generated in this investigation provide important reference material for follow-up research.
Subject terms: Developmental biology, Genetics, Plant sciences
Introduction
The nuclear pore complex (NPC), located within invaginations of the nuclear envelope, is a massive macromolecular conglomerate in cells1. It is composed of multiple copies of at least 30 diverse nucleoporins (Nups)2. Materials transported between the nucleus and cytoplasm have important effects on cell functions3, and the NPC is the only channel that controls nucleo-cytoplasmic transport of RNA and protein3,4. The initial research regarding the NPC involved analyses of vertebrates and yeast, and revealed that the Nups of yeast and metazoans are approximately 60 and 120 MDa, respectively1. An ultrastructural analysis indicated that the basic NPC framework is conserved in vertebrates, yeast, and plants2,5–7. The following three subcomplexes have been identified in vertebrate NPC: the Nup107 subcomplex (Nup37, Nup43, Nup85, Nup96, Nup107, Nup133,Nup160, Seh1, and Sec13); the Nup62 subcomplex (Nup45 , Nup54, Nup58, and Nup62); and the Nup93 subcomplex (Nup35, Nup93,Nup155, Nup188, and Nup205)8. The different subcomplexes have diverse functions. The remaining members Nup50, Nup88, Nup98, Nup136, NDC1, Tpr/NUA, CG1, RAE1a/b, ALADIN, GP210, HOS1, , and GLE1 do not form a subcomplex2, and they are also important parts of the NPC. Relatively little was known about plant NPC until recent studies involving electron microscopy7, proteomics2, and bioinformatics analyses confirmed that the NPCs of plants and other eukaryotes are structurally similar9,10. Tamura uncovered a greater sequence homology between plant and vertebrate NPCs than between plant and yeast NPCs2.
The research to date on plant NPC has revealed it influence plant immunity11–13, hormone signaling14–18, abiotic stress responses2,19–21, and flowering1–3,22,23. The mos3 Arabidopsis thaliana deletion mutant, which lacks a homolog of the Nup96 gene in animals, is more susceptible to pathogens than normal11. Additionally, MOS7, Seh1, and Nup160 are also involved in the plant immune pathway12,13,17. Nup160 and Nup96 mutations affect the nuclear output of mRNA and the subcellular localization of the auxin response transcriptional repressor IAA17 protein in A. thaliana, thereby partially restoring the auxin-resistant phenotype of axr-1 mutants14. Thus, Nup160 and Nup96 are likely involved in the auxin signal transduction pathway. The tpr/mlp1p/mlp2p mutations result in phenotypes that are similar to those due to mutations to Nup160 and Nup9615,16. In addition to altering auxin signal transduction, a mutation to Nup160 also increases the responsiveness of A. thaliana to ethylene, suggesting that it may help mediate the interaction between auxin and ethylene signals18. In terms of abiotic stress responses, both Nup160 and HOS1 participated in chilling stress by regulating CBF gene19–21. Furthermore, The HOS1 protein can specifically mediate the degradation of the ICE1 protein under cold conditions, thereby weakening A. thaliana responses to low temperatures21.And the Nup85 mutant reduced ABA and salt stress response in A. thaliana22. Additionally, Nup160, Nup96, and HOS1 also affect the flowering time of plants3. Specifically, HOS1 interacts with some nuclear genes to regulate its binding to FLC chromatin in flowering plants at low temperatures and weaken the transcriptional inhibition of FLC by HDA623. In A. thaliana, HOS1 interacts directly with CO. In the hos1 mutant, CO accumulates, which inhibits FLC expression and ultimately promotes flowering24. Mutations to Nup54, Nup58, Nup62, Nup136, and Nup160 result in an obvious early flowering phenotype in A. thaliana, whereas mutations to Nup62-2 and Nup160-4 lead to dwarfism1,2. An investigation of A. thaliana proved that Nup96 promotes the stability of HOS1, which binds to and degrades CO, resulting in delayed flowering. Moreover, HOS1 increases the stability of Nup96, thereby maintaining this regulatory pathway to control flowering time25.
A lot of research has been done on the model plants, which gives us a certain understanding of the plant NPC3,4,26. However, there is no report on NPC research of woody fruit trees. Considering the important function of NPC, it is necessary to carry out the related research on woody fruit trees. Apple is one of the most important fruit tree species worldwide. So we first identified candidate apple Nups based on A. thaliana Nups, after which we characterized the gene structure, protein structure, and tissue-specific expression patterns. We know that most apple species produce relatively few flowers or have stunted flower buds, which seriously affects the apple industry27,28. Although previous research has confirmed that some Nups in A. thaliana are involved in the flowering pathway, there have been no similar studies of apple Nups. So we made statistics on the effect of IAA treatment on apple flowering rate and detected the expression of some Nups to preliminarily reveal the effect of apple Nups on flowering. In addition, we conducted a preliminary study on the Nup62 subcomplex of apple. We studied the interaction between MdNup62 and MdNup54 and screened for proteins that interact with MdNup54. To the best of our knowledge, this study is the first comprehensive survey of the apple NPC, and the data presented herein will be useful for future analyses.
Materials and methods
Plant materials and treatments
The roots, stems, leaves, buds, flowers, and fruits of 6-year-old apple trees (Fu ji/T337/Malus robusta Rehd.) were collected for a tissue-specific gene expression analysis. We collected newly grown lateral roots (1–2 mm in diameter), new shoots (2–3 mm in diameter) near the tip, fully expanded leaves near buds, flower buds, blooming flowers, and young fruits, which were immediately frozen in liquid nitrogen and stored at − 80 °C for later use.
Regarding the hormone treatment, 40 apple trees (108° 04′ E, 34° 16′ N) growing in the experimental orchard of the Horticulture College of Northwest A&F University were randomly divided into two groups, which were treated with 300 mg/L IAA or water. During the study, apple leaves were dusted with a low-pressure manual duster, and samples were collected at 30, 50, and 70 days after flowering. The samples were immediately frozen in liquid nitrogen and stored at − 80 °C.
We also investigated the effects of an IAA treatment on the flowering rate of apple trees. Specifically, five similarly growing IAA- and water-treated apple trees were examined. The flowering rate was calculated as previously described29.
Identification of apple NPC
To identify apple NPC, we used the 30 identified NPC protein sequences of A. thaliana as queries to search the apple genome database (Malus domestica Genome GDDH13 V1.1, https://www.rosaceae.org/). The obtained sequences were then used as queries to search the conserved domain database (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi). The genes lacking the relevant Nup domain were eliminated. All non-redundant putative protein sequences were finally manually checked to confirm the presence of the Nup domain.
Analyses of phylogenetic relationships, gene structures, and tertiary protein structures
A phylogenetic tree comprising apple and A. thaliana Nups as well as Nup54 and Nup62 from 10 species (Arabidopsis thaliana, Malus domesica, Populus trichocarpa, Oryza sativa, Rosa chinensis, Pyrus communis, Ananas comosus, Vitis vinifera, Zea mays , and Prunus persica) were constructed with the MEGA-X program. The Gene Structure Display Server (https://gsds.cbi.pku.edu.cn/) was used to construct exon–intron structures. The gene structures were determined based on the coding sequences within the corresponding genomic sequences. The predicted Apple NPC tertiary structures were analyzed with the PHYRE server (version 2.0) (https://www.sbg.bio.ic.ac.uk/phyre2/html/page.cgi?id=index).
RNA extraction and qRT-PCR analysis
Total RNA was extracted from apple buds with the RNA Plant Plus Reagent Kit (TIANGEN, Beijing, China). The RNA was used as the template to synthesize cDNA with the PrimeScript RT Reagent Kit (TAKARA, Shiga, Japan). The expression levels of all identified Apple Nups were analyzed by qRT-PCR with primer pairs designed with Primer 6.0 (Table S1). The qRT-PCR analysis was conducted with the StepOnePlus Real-Time PCR System (THERMO FISHER SCIENTIFIC, USA). The reaction solution comprised 10 μL SYBR Green I Master Mix (CWBIO, Beijing, China), 0.5 μmol L−1 primers (SANGON BIOTECH, Shanghai, China), and 1 μL each template in a total volume of 20 μL.
The PCR program was as follows: 95 °C for 3 min; 40 cycles of 94 °C for 15 s, 62 °C for 20 s, and 72 °C for 20 s. The resulting fragments were immediately subjected to a melting-curve analysis to verify the amplification of gene-specific PCR products. The melting-curve analysis was completed with the following program: 94 °C for 15 s, followed by a constant increase from 60 to 95 °C at a 2% ramping rate. The apple actin gene (MD04G1127400) was used as an internal standard. All samples were analyzed with three biological replicates, each comprising three technical replicates. Relative gene expression levels were calculated according to the 2−ΔΔCt method30.
Yeast two-hybrid (Y2H) assay
The MdNup54175–339 and MdNup62508–613 truncated sequences were cloned into the pGBKT7 vector to generate the MdNup54175–339-pGBKT7 and MdNup62508–613-pGBKT7 recombinant plasmids. The MdNup54, MdKNAT4, and MdKNAT6 open reading frames were inserted into the pGADT7 vector to generate the MdNup54-pGADT7, MdKNAT4-pGADT7, and MdKNAT6-pGADT7 recombinant plasmids. The recombinant plasmids were inserted into Gold Yeast Two-Hybrid cells, which were then grown on selective medium. The primers used in the yeast experiment were designed with Primer 6.0 (Table S2). And these four gene sequences were submitted to NCBI (MdNup54: MT102239, MdNup62: MT102240, MdKNAT4: MT102238, and MdKNAT6: MT102237).
Split luciferase (LUC) complementation
The full-length MdNup54 coding sequence was cloned into CLUC vectors, whereas MdNup62 was cloned into NLUC vectors. The split-LUC complementation assay was performed with tobacco leaves. The LUC activity was quantified with the Dual-Luciferase Reporter Assay System. The primers used in the luciferase experiment were designed with Primer 6.0 (Table S2).
Statistical analysis
Data underwent an analysis of variance and the means were compared with a t-test at the 5% level using the SPSS 11.5 software package. Figures were prepared with Excel.
Results
Genome-wide identification of NPC in apple
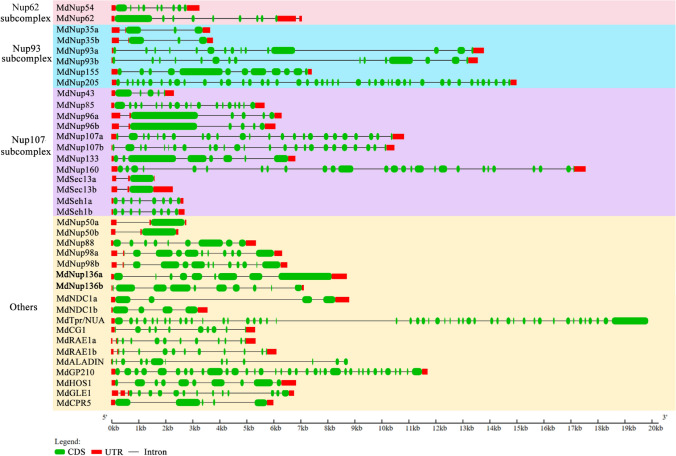
The apple Nups were detected and identified in the GDR database using BlastP. We obtained 38 candidate apple Nups after the genes with incomplete Nup-related domains and recurring genes were eliminated. The identified apple Nups are MdNup35a/b, MdNup43, MdNup50a/b, MdNup54, MdNup62, MdNup85, MdNup88, MdNup93a/b, MdNup96a/b, MdNup98a/b, MdNup107a/b, MdNup133, MdNup136a/b, MdNup155, MdNup160, MdNup205, MdSec13a/b, MdSeh1a/b, MdNDC1a/b, MdTpr/NUA, MdCG1, MdRAE1a/b, MdALADIN, MdGP210, MdHOS1, MdGLE1, and MdCPR5 (Table 1). Figure 1 also shows the gene locus, location, sequence length and other information of apple NUPs.
Table 1.
Information on the Nups in apple.
| Subcomplex | Gene | Gene locus | Location | CDS (bp) | Peptide (aa) | AtNPC | E value |
|---|---|---|---|---|---|---|---|
| Nup62 subcomplex | MdNup54 | MD16G1117500 | Chr16:8338184..8341499 | 1200 | 400 | AtNup54 | 1.5394E−161 |
| MdNup62 | MD07G1110700 | Chr07:12755345..12762516 | 2172 | 724 | AtNup62 | 1.69142E−135 | |
| Nup93 subcomplex | MdNup35a | MD09G1205800 | Chr09:19658131..19661853 | 993 | 331 | AtNup35 | 9.05037E−151 |
| MdNup35b | MD17G1186700 | Chr17:22318731..22322549 | 993 | 331 | AtNup35 | 3.02073E−147 | |
| MdNup93a | MD12G1080600 | Chr12:9823197..9837227 | 2592 | 864 | AtNup93 | 0 | |
| MdNup93b | MD14G1076700 | Chr14:8681861..8695666 | 2589 | 863 | AtNup93 | 0 | |
| MdNup155 | MD13G1020400 | Chr13:1280351..1287904 | 4407 | 1469 | AtNup155 | 0 | |
| MdNup205 | MD02G1032900 | Chr02:2574878..2590132 | 5643 | 1881 | AtNup205 | 0 | |
| Nup107 subcomplex | MdNup43 | MD10G1281400 | Chr10:37201121..37203464 | 1053 | 351 | AtNup43 | 1.53139E−127 |
| MdNup85 | MD15G1093200 | Chr15:6453444..6459227 | 2175 | 725 | AtNup85 | 0 | |
| MdNup96a | MD08G1215300 | Chr08:27827088..27833504 | 3096 | 1032 | AtNup96 | 0 | |
| MdNup96b | MD15G1399200 | Chr15:50011301..50017471 | 3348 | 1116 | AtNup96 | 0 | |
| MdNup107a | MD09G1178100 | Chr09:15235815..15246849 | 3246 | 1082 | AtNup107 | 0 | |
| MdNup107b | MD17G1148600 | Chr17:13573595..13584264 | 3219 | 1073 | AtNup107 | 0 | |
| MdNup133 | MD17G1113600 | Chr17:9738615..9745545 | 3915 | 1305 | AtNup133 | 0 | |
| MdNup160 | MD10G1009800 | Chr10:1350846..1368727 | 4521 | 1507 | AtNup160 | 0 | |
| MdSec13a | MD09G1041000 | Chr09:2640031..2641626 | 903 | 301 | AtSeh13 | 1.71982E−170 | |
| MdSec13b | MD17G1042300 | Chr17:3101074..3103384 | 903 | 301 | AtSeh13 | 4.377E−173 | |
| MdSeh1a | MD06G1103700 | Chr06:24176846..24179550 | 978 | 326 | AtSeh1 | 2.26035E−138 | |
| MdSeh1b | MD14G1122800 | Chr14:19731969..19734722 | 981 | 327 | AtSeh1 | 8.91422E−139 | |
| Others | MdNup50a | MD09G1214400 | Chr09:21079805..21082646 | 1272 | 424 | AtNup50a | 4.0775E−100 |
| MdNup50b | MD17G1196800 | Chr17:23575668..23578205 | 1293 | 431 | AtNup50a | 3.54614E−76 | |
| MdNup88 | MD01G1152200 | Chr01:26080908..26086372 | 2430 | 810 | AtNup88 | 0 | |
| MdNup98a | MD06G1126300 | Chr06:26827660..26834106 | 2973 | 991 | AtNup98a/b | 0 | |
| MdNup98b | MD14G1142000 | Chr14:23465594..23472220 | 3069 | 1023 | AtNup98a/b | 0 | |
| MdNup136a | MD02G1257800 | Chr02:31057936..31066675 | 3885 | 1294 | AtNup136 | 5.85327E−125 | |
| MdNup136b | MD07G1063100 | Chr07:5916974..5924113 | 2997 | 998 | AtNup136 | 2.51999E−117 | |
| MdNDC1a | MD05G1278400 | Chr05:41276844..41285832 | 1647 | 549 | AtNDC1 | 1.18565E−172 | |
| MdNDC1b | MD10G1257000 | Chr10:35129858..35133484 | 1653 | 551 | AtNDC1 | 2.80708E−159 | |
| MdTpr/NUA | MD05G1240600 | Chr05:37117000..37137248 | 6306 | 2102 | AtTpr/NUA | 0 | |
| MdCGI | MD04G1000600 | Chr04:62175..67597 | 1173 | 391 | AtCG1 | 3.48934E−63 | |
| MdRAE1a | MD08G1221600 | Chr08:28411242..28416688 | 1044 | 348 | AtRAE1 | 0 | |
| MdRAE1b | MD15G141250 | Chr15:51213459..51219697 | 1032 | 344 | AtRAE1 | 0 | |
| MdALADIN | MD12G1112000 | Chr12:17757407..17766342 | 1638 | 546 | AtALADIN | 1.53421E−105 | |
| MdGP210 | MD17G1026700 | Chr17:1907720..1919647 | 5913 | 1971 | AtGP210 | 0 | |
| MdHOS1 | MD04G1060900 | Chr04:8017657..8024612 | 2919 | 973 | AtHOS1 | 0 | |
| MdGLE1 | MD13G1104500 | Chr13:7472044..7478918 | 1935 | 645 | AtGLE1 | 1.86679E−127 | |
| MdCPR5 | MD01G1017700 | Chr01:7756340..7761421 | 1860 | 620 | AtCPR5 | 2.16518E−85 |
Figure 1.
Analysis of apple NPC gene structures. The Gene Structure Display Server (https://gsds.cbi.pku.edu.cn/) was used to construct exon–intron structures. Green boxes and black lines refer to exons and introns, respectively. Red boxes represent untranslated regions.
Gene structures in apple Nups
To structurally characterize the identified apple Nups, we generated exon–intron diagrams and revealed the coding sequences and untranslated regions (Fig. 1). An examination of all apple Nups indicated that MdTpr/NUA has the most exons, with 47, whereas MdSec13a/b and MdNup50a/b have the fewest exons, with only two. Of the Nup62 subcomplex genes, MdNup54 and MdNup62 have 9 and 10 exons, respectively. Regarding the Nup93 subcomplex, MdNup205 has the most exons, with 45, and MdNup35 has the fewest exons, with only four. The mean number of exons in the Nup93 subcomplex genes is 15.83. Among the Nup107–160 subcomplex genes, MdNup160 has the most exons, with 27, whereas MdSec13a/b has the fewest exons, with two. The mean number of exons is 11.58. Of the Other analyzed genes, MdTpr/NUA has the most exons, with 47, and MdNup50a/b has the fewest exons, with 2. The mean number of exons is 13.50. Additionally, we also predicted the tertiary structures of the Apple Nups, revealing α helices, β sheets, and random coils in all proteins (Fig. S1).
Conserved protein domains in apple Nups
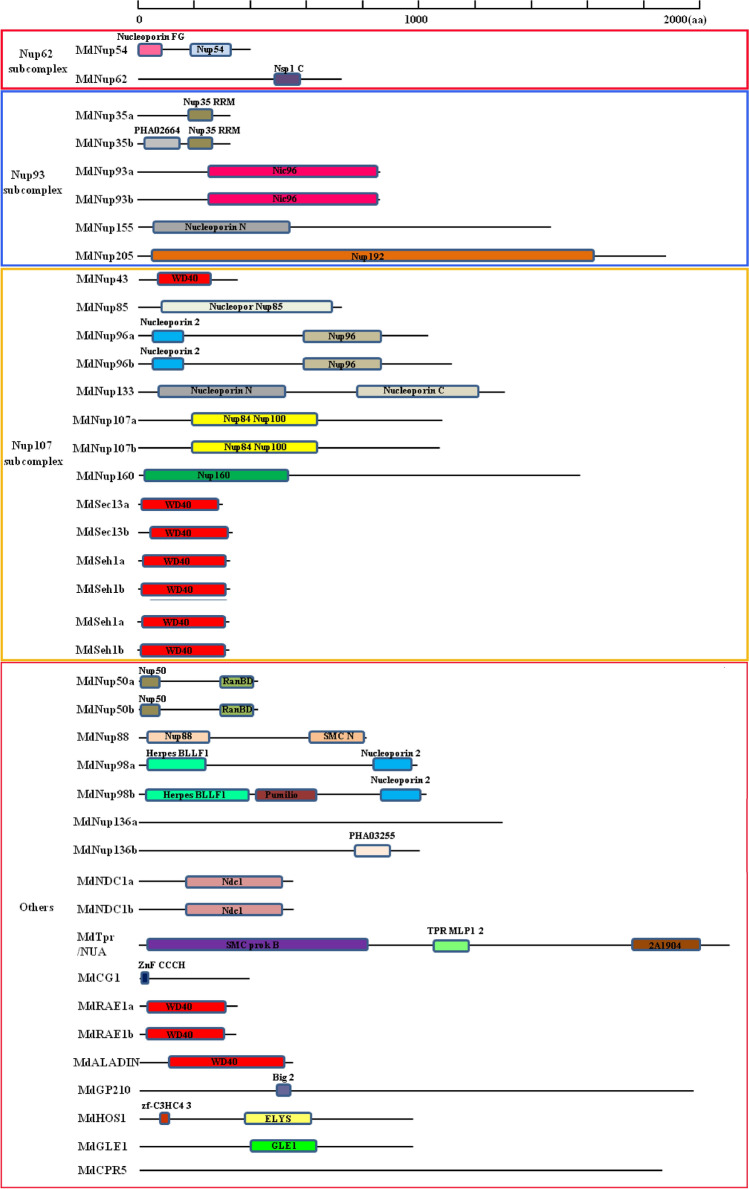
We used the NCBI BlastP tool to analyze the conserved protein domains of 38 apple Nups. As a class of complexes, Apple Nups have no common conserved domain. However, there are domains that are conserved among some members. For example, MdNup133 and MdNup155 have a conserved Nucleoporin N structural domain, whereas MdNup43, MdSec13a/b, MdSeh1a/b, MdRAE1a/b, and MdALADIN share a common conserved WD40 structural domain (Fig. 2). On the basis of previous studies, we divided the apple Nups into the following four subcomplex categories: Nup62 subcomplex, Nup93 subcomplex, Nup107 subcomplex and others. And there are 2 members (MdNup54 and MdNup62) in Nup62 subcomplex, 6 members (MdNup35a/b, MdNup93a/b, MdNup155, and MdNup205) in Nup93 subcomplex, 12 members (MdNup43, MdNup85, MdNup96a/b, MdNup107a/b, MdNup133, MdNup160, MdSec13a/b, and MdSeh1a/b) in Nup107 subcomplex, and 18 members (MdNup50a/b, MdNup88, MdNup98a/b, MdNup136a/b, MdNDC1a/b, MdTpr/NUA, MdCG1, MdRAE1a/b, MdALADIN, MdGP210, MdHOS1, MdGLE1, and MdCPR5) in Others.
Figure 2.
Schematic representation of the predicted domain features of apple NPCs. BlastP in NCBI (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) was used to get predicted domain information. Different squares represent different conserved domains of the protein, and the black line represents the number of amino acids.
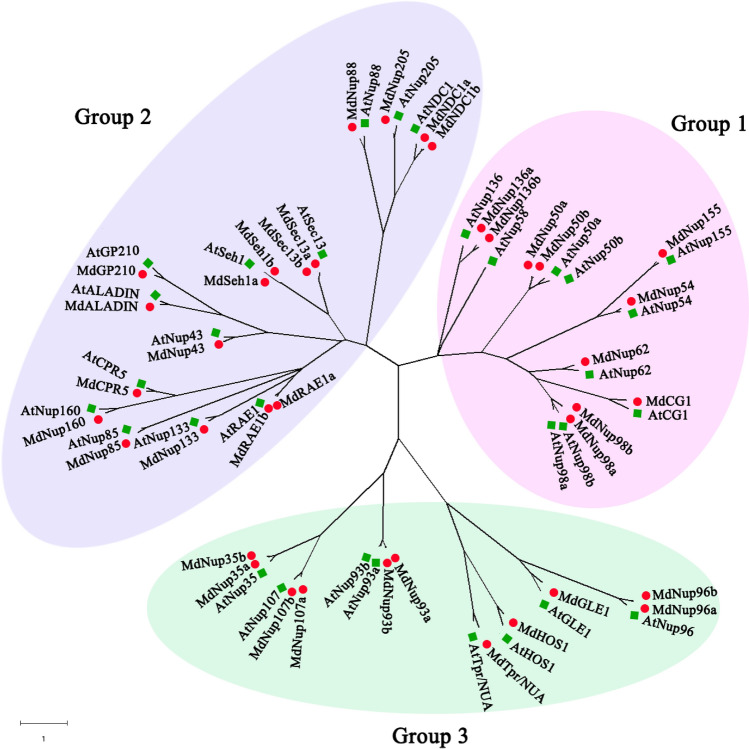
Analysis of evolutionary relationships among Nups
To elucidate the evolutionary relationships among Nups, we constructed a phylogenetic tree consisting of A. thaliana and apple Nups (Fig. 3). The Nups were divided into three groups, Groups 1, 2, and 3, which comprised 20, 30, and 19 members, respectively. Group 1 had 10 apple genes (MdNup50a/b, MdNup54, MdNup62, MdNup98a/b, MdNup136a/b, MdNup155, and MdCG1), while 17 apple genes (MdNup43, MdNup85, MdNup88, MdNup133, MdNup160, MdNup205, MdALADIN, MdNDC1a/b, MdGP210, MdRAE1a/b, MdSec13a/b, MdCPR5, and MdSeh1a/b) were clustered in Group 2 and 11 apple genes (MdNup35a/b, MdNup93a/b, MdNup96a/b, MdNup107a/b, MdGLE1, MdHOS1, and MdTpr/NUA) were clustered in Group 3.
Figure 3.
Phylogenetic analysis of apple and Arabidopsis thaliana NPCs. The phylogenetic trees were obtained through MEGA-X. Arabidopsis thaliana: At, green square; apple: Md, red circle.
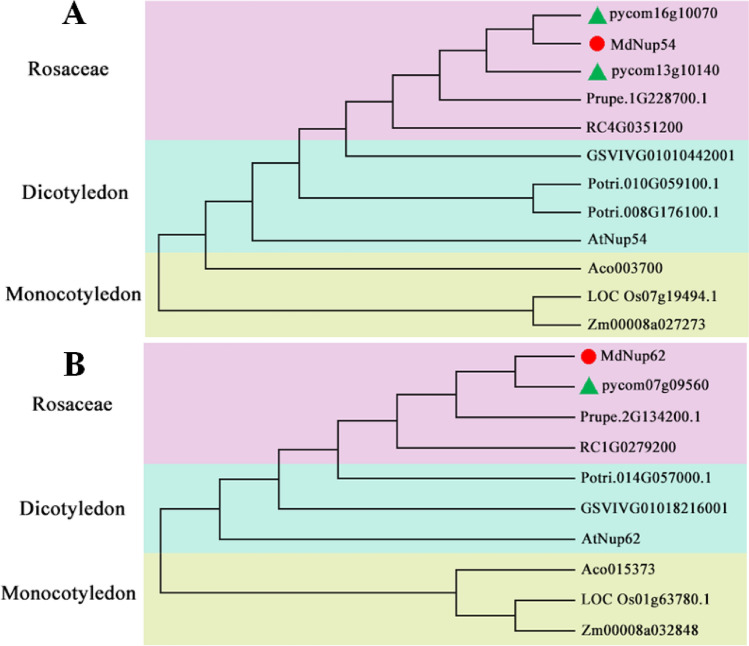
Evolutionary relationship between MdNup54 and MdNup62 among plant species
Nup62 subcomplex is located in the central part of the nuclear pore and plays an important role in the regulation of substances into and out of the nucleus1,2. And it has only two members in apple, MdNup54 and MdNup62. Given their importance, we analyzed the evolutionary relationship between Nup54 and Nup62 among the 10 species (Arabidopsis thaliana, Malus domestica, Populus trichocarpa, Oryza sativa, Rosa chinensis, Pyrus communis, Ananas comosus, Vitis vinifera, Zea mays , and Prunus persica) (Fig. 4). A phylogenetic analysis indicated that both MdNup54 and MdNup62 are closely related to genes in Rosa chinensis, Pyrus communis, and Prunus persica in the family Rosaceae, but are more distantly related to genes in monocotyledons (Oryza sativa, Ananas comosus, and Zea mays).
Figure 4.
Phylogenetic analysis of Nup54 (a) and Nup62 (b) in 10 species (Arabidopsis thaliana, apple, poplar, rice, rose, pear, pineapple, grape, corn, and peach). The phylogenetic trees were obtained through MEGA-X. The red circle represents apple gene, and the green triangle represents pear gene.
Expression levels of apple Nups in various tissues
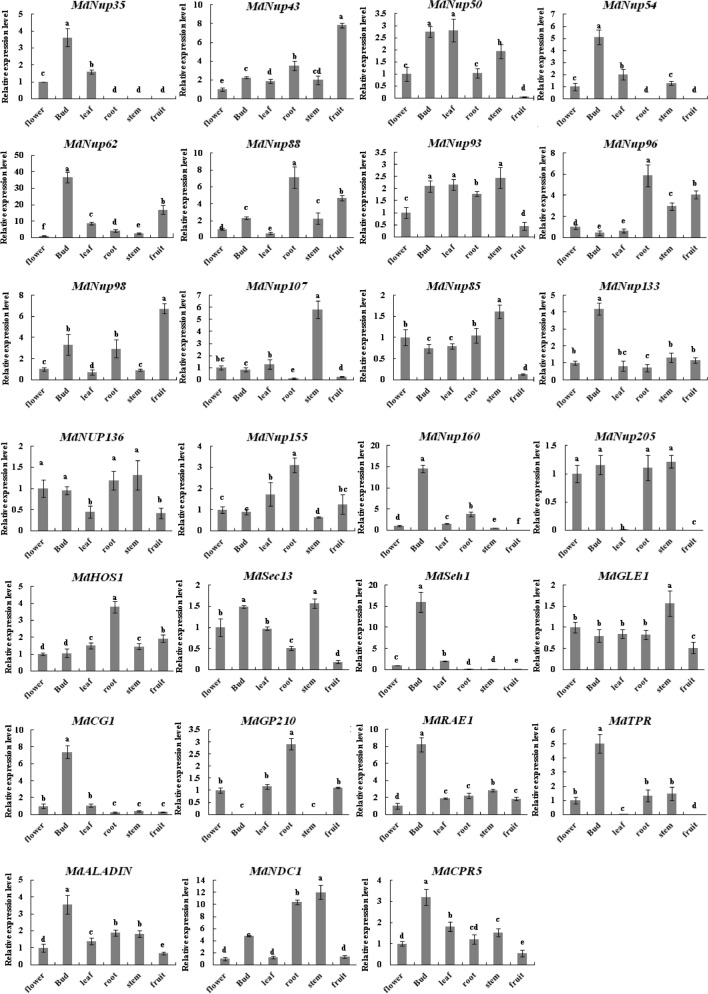
To functionally characterize apple Nups in apple, we completed a qRT-PCR assay to determine apple Nups expression levels in diverse tissues (flowers, buds, leaves, roots, stems and fruits) (Fig. 5). Because there are 11 pairs of highly similar homologous apple Nups (MdNup35a/b, MdNup93a/b, MdNup96a/b, MdNup107a/b, MdNup50a/b, MdNup98a/b, MdNup136a/b, MdSec13a/b, MdSeh1a/b, MdNDC1a/b, MdRAE1a/b), analyzing their expression levels separately was difficult. Therefore, we performed a combined analysis of the expression levels of each pair of homologous genes. The 38 candidate apple Nups produced varying expression patterns in different tissues. For example, MdNup35, MdNup54, MdNup62, MdNup133, MdNup160, MdSeh1, MdCG1, MdRAE1, MdTPR, MdALADIN, and MdCPR5 were most highly expressed in the buds, implying they may be involved in apple flower bud induction. In contrast, MdNup43 and MdNup98 were highly expressed in fruits, whereas MdNup107, MdGLE1, and MdNDC1 expression levels were high in the stems. Moreover, the highest MdNup88, MdNup96, MdNup155, MdHOS1, and MdGP210 expression levels were detected in the roots.
Figure 5.
Analysis of MdNPC expression levels in diverse ‘Nagafu No. 2’ tissues. Each sample was analyzed with three biological replicates, each comprising three technical replicates. The histograms were made by Excel 2007. Means followed by different lowercase letters are significantly different at the 0.05 level.
Apple Nups expression patterns in response to IAA treatments during the flower induction period
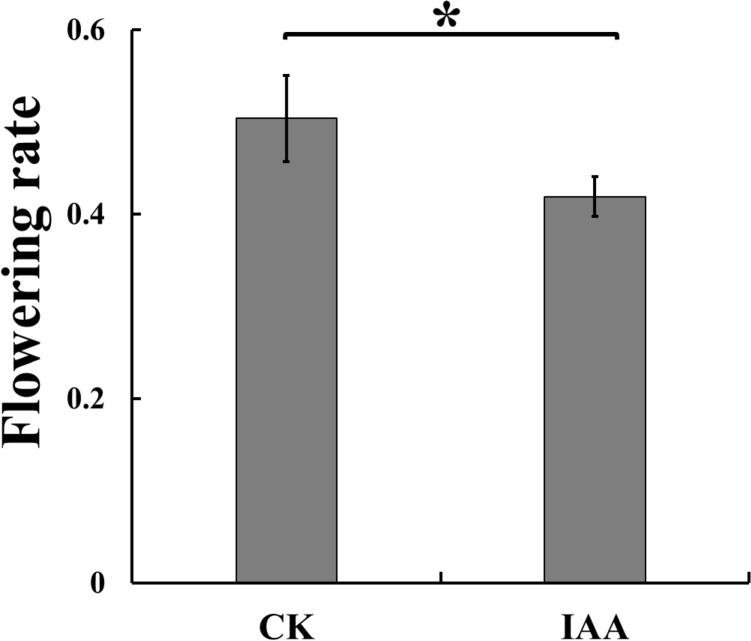
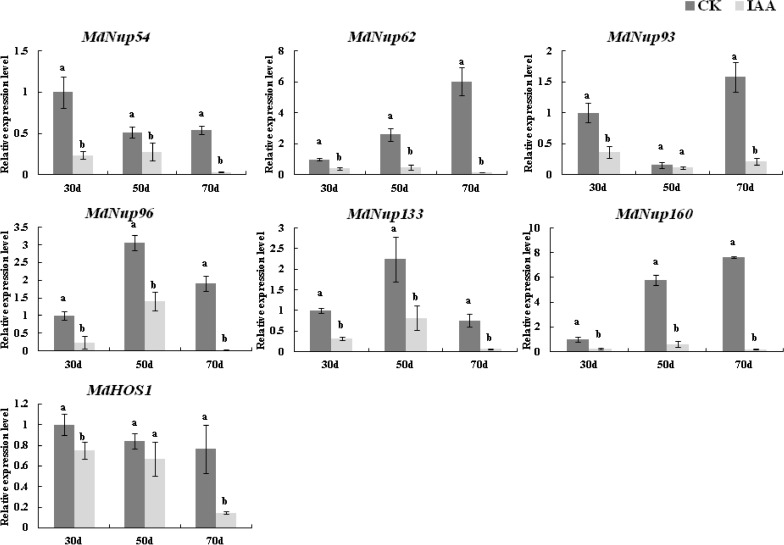
We investigated the effect of an IAA treatment on apple flower induction. The flowering rate following the IAA treatment was 41.9%, which was significantly lower than the 50.4% flowering rate after the water (control) treatment (Fig. 6). Previous studies have confirmed that some Nups (AtNup54, AtNup62, AtNup96, AtNup160, and AtHOS1) affect flowering time in Arabidopsis thaliana1,23–25. And the tissue-specificity analysis found that MdNup54, MdNup62, MdNup133, and MdNup160 were most highly expressed in buds, and MdNup93 was also highly expressed in buds, suggesting that these genes may be involved in the apple flowering pathway. So we analyzed the transcription of these seven candidate MdNups . And the expression of all seven candidate genes was significantly inhibited by the IAA treatment (Fig. 7). Moreover, the transcription of MdNup54, MdNup62, MdNup96, MdNup133, MdNup160 were significantly inhibited at 30, 50, and 70 days after flowering. In contrast, the MdNup93 and MdHOS1 expression levels were not significantly different following the IAA and water treatments at 50 days after flowering, but the expression levels were lower in the IAA-treated samples than in the control samples at 30 and 70 days after flowering. Accordingly, the IAA treatment can significantly inhibit the expression of these genes.
Figure 6.
Flowering rate of ‘Fuji’ apple trees following control (CK) and IAA treatments. The presented data are derived from five biological replicates. The histograms were made by Excel 2007. Asterisks denote a significant difference as determined by the t-test: *P < 0.05.
Figure 7.
Expression patterns of seven candidate MdNPCs in apple buds treated with IAA. Control buds were treated with water. Samples were collected at 30, 50, and 70 days after full bloom (DAFB). Each sample was analyzed with three biological replicates, each comprising three technical replicates. The histograms were made by Excel 2007. Means followed by different lowercase letters are significantly different at the 0.05 level.
MdNup62 interacts with MdNup54
Previous studies found that Nup54, Nup58, and Nup62 form a complex and function together in metazoan, and the interaction between the other two members in A. thaliana Nup62 subcomplex, AtNup58 and AtNup62, were also reported31. And MdNup62 and MdNup54 form the Nup62 subcomplex, we hypothesized that these two proteins interact with each other. To test this hypothesis, we performed a Y2H experiment. First, we observed that the truncated MdNup621–507 was self-activating, but MdNup62508–613 was not (Fig. S2). Therefore, MdNup62508–613-pGBKT7 was selected as the bait and was included in a co-transformation of yeast cells along with MdNup54-pGADT7. And they could grow normally on SD/ − Trp/ − Leu medium and SD/ − Trp/ − Leu/ − His/ − Ade/ + X-α-gal medium, and bacame significantly blue on SD/ − Trp/ − Leu/ − His/ − Ade/ + X-α-gal medium. But the co-transformation of MdNup62508–613-pGBKT7 and empty- pGADT7 could only grow on the SD/ − Trp/ − Leu medium, and could neither grow nor turn blue on the SD/ − Trp/ − Leu/ − His/ − Ade/ + X-α-gal medium. So the Y2H assay confirmed that MdNup62 can interact with MdNup54 (Fig. 8). This interaction was further verified in a split-LUC complementation assay. The co-expression of MdNup62-NLUC and MdNup54-CLUC resulted in higher LUC activity than the other combinations (Fig. 8). These results confirmed the interaction between MdNup62 and MdNup54.
Figure 8.
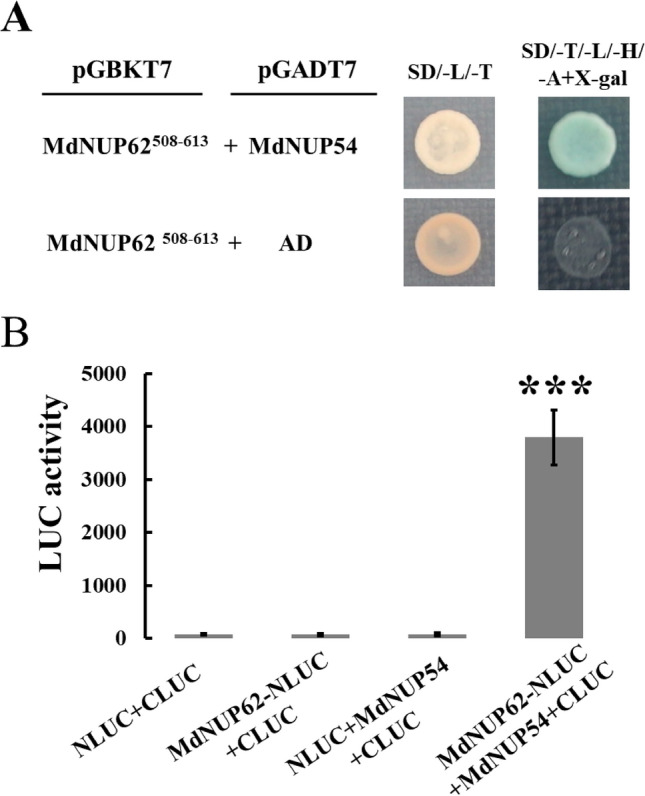
Interaction between MdNup62 and MdNup54. (A) MdNup62508–613 interacted with MdNup54 in Y2H assays. The MdNup62508–613 truncated sequence was cloned into pGBKT7, whereas MdNup54 was cloned into pGADT7. Empty pGADT7 plus MdNup62508–613-pGBKT7 was used as a control. Yeast cells grown in SD/ − Trp/ − Leu medium and SD/ − Trp/ − Leu/ − His/ − Ade/ + X-α-gal medium are presented. (B) The luciferase complementation experiment involving tobacco leaves revealed the interaction between MdNup62 and MdNup54. Empty NLUC and empty CLUC, MdNup62-NLUC and empty CLUC, and empty NLUC + MdNup54-CLUC were used as controls. The luciferase complementation experiment was repeated three times, with consistent results. Asterisks denote significant differences as determined by the t-test: *P < 0.01.
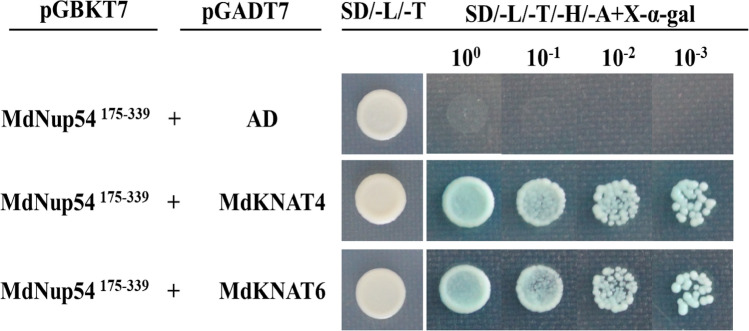
MdNup54 interacts with MdKNAT4 and MdKNAT6
Although MdNup54 is an important component of the Nup62 subcomplex and influences plant growth and development, there has been relatively little research on the MdNup54 gene in plants. Thus, we conducted a Y2H assay to explore the biological processes MdNup54 may contribute to. First, we observed that the truncated MdNup541–90 was self-activating, but MdNup54175–339 was not (Fig. S3). Accordingly, MdNup54175–339-pGBKT7 was selected as the bait and inserted into yeast cells, which were then transformed with plasmids from the apple bud plasmid library to screen for interacting proteins. Both MdKNAT4 and MdKNAT6 were detected as potential interacting proteins. The MdKNAT4 and MdKNAT6 sequences were cloned and ligated to separate pGADT7 vectors. To conduct a Y2H assay, we co-transformed yeast cells with MdKNAT4-pGADT7 or MdKNAT6-pGADT7 and MdNup54175–339-pGBKT7. And these two kinds co-transformed yeast cells could both grow normally on SD/ − Trp/ − Leu medium and SD/ − Trp/ − Leu/ − His/ − Ade/ + X-α-gal medium, and became significantly blue on SD/ − Trp/ − Leu/ − His/ − Ade/ + X-α-gal medium. But the control could only grow on SD/ − Trp/ − Leu medium. The assay results verified that both MdKNAT4 and MdKNAT6 can interact with MdNup54 (Fig. 9).
Figure 9.
Interactions between MdNup54175–339 and MdKNAT4 as well as MdKNAT6 in Y2H assays. The MdNup54175–339 truncated sequence was cloned into pGBKT7, whereas MdKNAT4 and MdKNAT6 were cloned into separate pGADT7 vectors. Empty pGADT7 plus MdNup54175–339-pGBKT7 was used as a control. Additionally, 100, 10−1, 10−2, and 10−3 represent the dilutions of the yeast solution. Yeast cells grown in SD/ − Trp/ − Leu medium and SD/ − Trp/ − Leu/ − His/ − Ade/ + X-α-gal medium are presented.
Discussion
The NPC controls the communication between the nucleus and cytoplasm, with consequences for diverse biological processes that influence plant growth and development. The relatively few studies that have examined plant NPC have been limited to model species, such as A. thaliana. Therefore, only the A. thaliana NPC has been systematically identified. We know very little about the corresponding apple genes. Thus, we identified and attempted to functionally characterize the apple NPC.
Genome-wide identification and characterization of Nups in apple
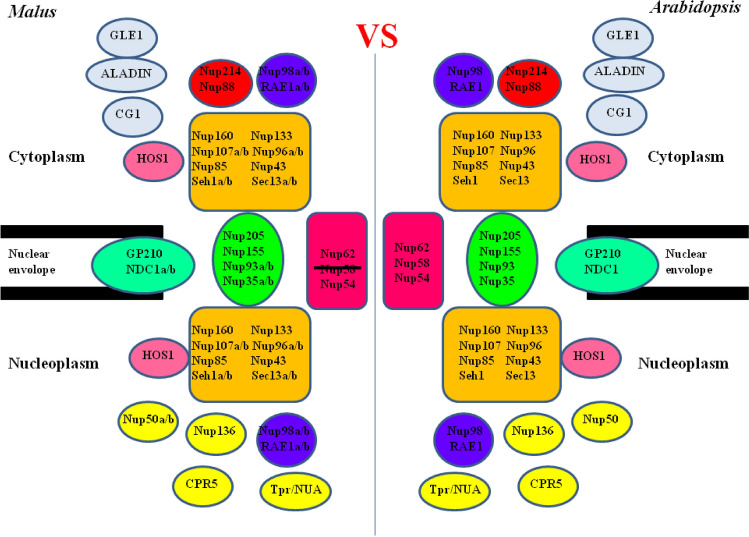
We identified 38 candidate apple Nups and more than 30 A. thaliana genes. Almost all of the A. thaliana Nups had a corresponding homologous sequence in apple, suggesting these are conserved plant genes. However, apple homologs of the A. thaliana Nup58 gene was not detected (Fig. 10). A. thaliana Nup58 belongs to Nup62 subcomplex, which means that lacking Nup58 of the apple Nup62 subcomplex must be functionally different from that of A. thaliana. The lack of Nup58 gene is bound to lead to differences in the function of the apple and A. thaliana. NPC. Additionally, because apple is a woody plant species, its genome is more complex than that of A. thaliana, with 11 genes (Nup35, Nup50, Nup93, Nup96, Nup98, Nup107,Nup136, Sec13, Seh1, NDC1, and RAE1) having two alleles each, whereas only Nup98 and Nup50 had two alleles in A. thaliana2. Although there are some differences between the apple and A. thaliana NPC, the basic structures of the encoded proteins are the same, including the Nup62, Nup93, and Nup107–160 subcomplexes as well as other Nups inside and outside the nuclear pore1,2. These findings imply the apple and A. thaliana NPC are functionally similar.
Figure 10.
Comparison between the apple and Arabidopsis thaliana NPC. The positions of each Nups refer to previous studies1,2,47.
The details regarding the introns and exons of the apple Nups suggested there is a lack of similar structures among the genes, including between the genes within the same subcomplex, further demonstrating the relative functional independence of the apple Nups. Apple Nups encode a class of compounds with no conserved domain among all members, unlike the members of other apple gene families (e.g., IDD, GRF, GASA and SBP-box genes)27,28,32,33. Only some of the apple Nups encode a conserved domain. Therefore, these proteins may have similar functions. Conserved domains were not detected among the other apple Nups, implying a lack of functional redundancy.
We also analyzed the phylogenetic relationships between Nups. First, we constructed a phylogenetic tree based on the A. thaliana and apple Nups and divided 69 Nups into three groups, consisting of 20, 30, and 19 members (Fig. 3). The apple Nups are most closely related to the corresponding A. thaliana Nups (e.g., MdNup96a/b and MdCG1 are most closely related to AtNup96 and AtCG1, respectively). These results suggest Nups are conserved and may have similar functions in diverse species.
The Nup62 subcomplex occupies an important position in the nuclear pore (i.e., central pore channel). An analysis of the evolutionary relationships involving MdNup54 and MdNup62 indicated that both genes are closely related to genes in other Rosaceae species, especially to genes in Pyrus communis, which is in the same subfamily as apple.
Apple Nups expression patterns
We performed a qRT-PCR assay to study the expression levels of apple Nups in six tissues of ‘Nagafu No. 2’. The MdNup35, MdNup54, MdNup62, MdNup133, MdNup160, MdSeh1, MdCG1, MdRAE1, MdTPR, MdALADIN, and MdCPR5 showed the highest expression in the buds, suggesting that they may be involved in the apple flowering pathway. NPC acts as a barrier and regulats the flow of RNA and proteins into and out of the nucleus. Therefore, the level of their expression should have an important effect on the growth and development of apple flower buds. Previous A. thaliana studies confirmed that the deletion of AtNup54, AtNup62, AtNup160, and AtTPR results in early flowering1. And this supports our speculation very well. Both MdNup43 and MdNup98 were highly expressed in fruits, implying they may contribute to apple fruit development. In contrast, MdNup107, MdGLE1, and MdNDC1 expression levels were highest in the stems, suggesting they are important for apple stem growth and development. Additionally, MdNup88, MdNup96, MdNup155, MdHOS1, and MdGP210 expression levels were highest in the roots, indicative of their potential roles in root development. However, Tissue specific expression only provides reference for the potential function of Nups, which need to be experimentally verified.
Arabidopsis thaliana studies have confirmed that some Nups (AtNup54, AtNup62, AtNup96, AtNup160, and AtHOS1) affect flowering time1,23–25. However, little is known about their potential roles in apple-induced flowering. Therefore, we investigated the expression patterns of MdNup54, MdNup62, MdNup93, MdNup96, MdNup133, MdNup160, and MdHOS1 to preliminarily explore whether they are associated with IAA-mediated flowering. We determined the flowering rates of ‘Nagafu No. 2’ treated with IAA. Our data indicated that the IAA treatment significantly inhibited flowering, which was consistent with the results of previous studies on apple and other species34,35. We subsequently performed a qRT-PCR assay to quantify the expression of these genes. The expression levels of the IAA-treated plants were significantly lower than those of the controls, suggesting these seven apple Nups are responsive to the application of exogenous IAA. Earlier investigations revealed that A. thaliana Nup62, Nup96, and Nup160 genes are involved in the auxin signaling pathway36, which is consistent with the results of this study. However, we cannot determine whether apple Nups are involved in the IAA treatment resulting in the reduction of flowering rates. And we only speculate that apple Nups are involved in the IAA regulation of apple flowering pathway. But it is certain that IAA treatment will reduce the expression of apple Nups and flower rates. And the relationship between apple Nups and flower rates needs to be verified by follow-up experiments.
Preliminary functional characterization of the Nup62 subcomplex in apple
The apple Nup62 subcomplex has only two members (MdNup54 and MdNup62), whereas the corresponding complex in A. thaliana has three members (AtNup54, AtNup58, and AtNup62)1,2. And we confirmed that MdNup54 and MdNup62 interact in apple. In other words, the complete biological functions of MdNup54 and MdNup62 may require the interaction between these two proteins. Additionally, because of a lack of Nup58, the apple Nup62 subcomplex and the corresponding A. thaliana subcomplex may be functionally diverse.
We conducted a Y2H assay to verify the interactions between MdNup54 and two members of the KNOX family (MdKNAT4 and MdKNAT6). To the best of our knowledge, this is the first study to reveal an interaction between an NPC and members of the KNOX family. The KNOX family members have crucial functions related to plant hormone signaling37–39, as well as leaf40,41 and flower development42. The apple MdKNAT4 and MdKNAT6 genes are homologs of the A. thaliana AtKNAT4 and AtKNAT6 genes, respectively. Earlier studies proved that AtKNAT4 influences seed dormancy43, while AtKNAT6 plays an important role in maintaining meristem integrity and flowering42. Apple MdNup54 may also affect similar pathways. Notably, the KNOX family is involved in cytokinin and gibberellin signaling pathways37–39. These two hormones are closely related to apple flowering27,44,45, suggesting that apple MdNup54 may indirectly affect cytokinin and gibberellin signaling pathways by controlling the transport of KNOX genes into the nucleus, thereby regulating apple flowering. But previous studies have shown that selective transport between the nucleus and cytoplasm depends on nuclear transport receptors (importin and exportin), which bind cargos and interact with the NPC selective barriers for cargo transport3,4,46. Recently, researchers have found that Nup85 and MED18 can interact directly with each other. And the two mutants had the same abiotic stress phenotype22. Therefore, the relationship between Nup54 and KNOX genes may provide a hypothesis for NPC studies that Nups may interact directly with transcription factors to control their nuclear transport.
Supplementary information
Acknowledgements
This work was financially supported by the National Natural Science Foundation of China (31801813); and the China Postdoctoral Science Foundation (2018M631207, 2017M623254).
Author contributions
M.H., N.A., L.X., C.Z., and X.R. conceived and designed the experiment. C.Z., P.J., W.Z., J.L., and X.Z. performed the experiment. C.Z., H.Z., and W.M. analyzed the data. L.X., and C.Z. wrote the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Chenguang Zhang, Na An and Peng Jia.
Contributor Information
Mingyu Han, Email: hanmy@nwsuaf.edu.cn.
Libo Xing, Email: libo_xing@nwsuaf.edu.cn.
Xiaolin Ren, Email: renxl@nwsuaf.edu.cn.
Supplementary information
is available for this paper at 10.1038/s41598-020-74171-0.
References
- 1.Parry G. Components of the Arabidopsis nuclear pore complex play multiple diverse roles in control of plant growth. J. Exp. Bot. 2014;65:6057–6067. doi: 10.1093/jxb/eru346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tamura K, Fukao Y, Iwamoto M, Haraguchi T, Hara-Nishimura I. Identification and characterization of nuclear pore complex components in Arabidopsis thaliana. Plant Cell. 2011;22:4084–4097. doi: 10.1105/tpc.110.079947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xu XM, Meier I. The nuclear pore comes to the fore. Trends Plant Sci. 2008;13:20–27. doi: 10.1016/j.tplants.2007.12.001. [DOI] [PubMed] [Google Scholar]
- 4.Meier I, Brkljacic J. The nuclear pore and plant development. Curr. Opin. Plant Biol. 2009;12:87–95. doi: 10.1016/j.pbi.2008.09.001. [DOI] [PubMed] [Google Scholar]
- 5.Roberts K, Northcote DH. Structure of the nuclear pore in higher plants. Nature. 1970;228:385–386. doi: 10.1038/228385a0. [DOI] [PubMed] [Google Scholar]
- 6.Goldberg MW, Allen TD. The nuclear pore complex and lamina: three-dimensional structures and interactions determined by field emission in-lens scanning electron microscopy. J. Mol. Biol. 1996;257:848–865. doi: 10.1006/jmbi.1996.0206. [DOI] [PubMed] [Google Scholar]
- 7.Fiserova J, Kiseleva E, Goldberg MW. Nuclear envelope and nuclear pore complex structure and organization in tobacco BY-2 cells. Plant J. 2009;59:243–255. doi: 10.1111/j.1365-313X.2009.03865.x. [DOI] [PubMed] [Google Scholar]
- 8.Tran EJ, Wente SR. Dynamic nuclear pore complexes: life on the edge. Cell. 2006;125:1041–1053. doi: 10.1016/j.cell.2006.05.027. [DOI] [PubMed] [Google Scholar]
- 9.Neumann N, Jeffares DC, Poole AM. Outsourcing the nucleus: nuclear pore complex genes are no longer encoded in nucleomorph genomes. Evol Bioinform Online. 2007;2:23–34. [PMC free article] [PubMed] [Google Scholar]
- 10.Neumann N, Lundin D, Poole AM. Comparative genomic evidence for a complete nuclear pore complex in the last eukaryotic common ancestor. PLoS ONE. 2010;5:e13241. doi: 10.1371/journal.pone.0013241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Y, Li X. A putative nucleoporin 96 is required for both basal defense and constitutive resistance responses mediated bysuppressor of Npr1-1, constitutive 1. Plant Cell. 2005;17:1306–1316. doi: 10.1105/tpc.104.029926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheng YT, et al. Nuclear pore complex component MOS7/Nup88 is required for innate immunity and nuclear accumulation of defense regulators in Arabidopsis. Plant Cell. 2009;21:2503–2516. doi: 10.1105/tpc.108.064519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roth C, Wiermer M. Nucleoporins Nup160 and Seh1 are required for disease resistance in Arabidopsis. Plant Signal Behav. 2012;7:1212–1214. doi: 10.4161/psb.21426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parry G, Ward S, Cernac A, Dharmasiri S, Estelle M. The Arabidopsis SUPPRESSOR of AUXIN RESISTANCE proteins are nucleoporins with an important role in hormone signaling and development. Plant Cell. 2006;18:1590–1603. doi: 10.1105/tpc.106.041566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jacob Y, Mongkolsiriwatana C, Veley KM, Kim SY, Michaels SD. The nuclear pore protein AtTPR is required for RNA homeostasis, flowering time, and auxin signaling. Plant Physiol. 2007;144:1383–1390. doi: 10.1104/pp.107.100735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu XM, et al. NUCLEAR PORE ANCHOR, the Arabidopsis homolog of Tpr/Mlp1/Mlp2/Megator, is involved in mRNA export and SUMO homeostasis and affects diverse aspects of plant development. Plant Cell. 2007;19:1537–1548. doi: 10.1105/tpc.106.049239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wiermer M, et al. Putative members of the Arabidopsis Nup107-160 nuclear pore sub-complex contribute to pathogen defense. Plant J. 2012;70:796–808. doi: 10.1111/j.1365-313X.2012.04928.x. [DOI] [PubMed] [Google Scholar]
- 18.Robles LM, Deslauriers SD, Alvarez AA, Larsen PB. A loss-of-function mutation in the nucleoporin AtNUP160 indicates that normal auxin signalling is required for a proper ethylene response in Arabidopsis. J. Exp. Bot. 2012;63:2231–2241. doi: 10.1093/jxb/err424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ishitani M, Xiong L, Lee H, Stevenson B, Zhu JK. HOS1, a genetic locus involved in cold-responsive gene expression in Arabidopsis. Plant Cell. 1998;10:1151–1161. doi: 10.1105/tpc.10.7.1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dong CH, et al. A putative arabidopsis nucleoporin, AtNUP160, is critical for RNA export and required for plant tolerance to cold stress. Mol. Cell. Biol. 2006;26:9533–9543. doi: 10.1128/MCB.01063-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dong C, Agarwal M, Zhang Y, Xie Q, Zhu J. The negative regulator of plant cold responses, HOS1, is a RING E3 ligase that mediates the ubiquitination and degradation of ICE1. Proc. Natl. Acad. Sci. USA. 2006;103:8281–8286. doi: 10.1073/pnas.0602874103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu Y, et al. An Arabidopsis nucleoporin NUP85 modulates plant responses to ABA and salt stress. PLoS Genet. 2017;13:e1007124. doi: 10.1371/journal.pgen.1007124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jung JH, et al. The cold signaling attenuator HIGH EXPRESSION of OSMOTICALLY RESPONSIVE GENE1 activates FLOWERING LOCUS C transcription via chromatin remodeling under short-term cold stress in Arabidopsis. Plant Cell. 2013;25:4378–4390. doi: 10.1105/tpc.113.118364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lazaro A, Mouriz A, Piñeiro M, Jarillo JA. Red light-mediated degradation of CONSTANS by the E3 ubiquitin ligase HOS1 regulates photoperiodic flowering in arabidopsis. Plant Cell. 2015;27:2437–2454. doi: 10.1105/tpc.15.00529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cheng, Z. et al. Nup96 and HOS1 are mutually stabilized and gate CONSTANS protein level, conferring long-day photoperiodic flowering regulation in arabidopsis. The Plant Cell. 661–2019 (2019). [DOI] [PMC free article] [PubMed]
- 26.Parry G. The plant nuclear envelope and regulation of gene expression. J. Exp. Bot. 2015;66:1673–1685. doi: 10.1093/jxb/erv023. [DOI] [PubMed] [Google Scholar]
- 27.Fan S, et al. Phylogenetic analysis of IDD gene family and characterization of its expression in response to flower induction in Malus. Mol. Genet. Genomics. 2017;292:755–771. doi: 10.1007/s00438-017-1306-4. [DOI] [PubMed] [Google Scholar]
- 28.Fan, S. et al. Comprehensive analysis of GASA family members in the Malus Domestica genome: identification, characterization, and their expressions in response to apple flower induction. BMC Genomics. 18, 1 (2017). [DOI] [PMC free article] [PubMed]
- 29.Xing L, et al. Shoot bending promotes flower bud formation by miRNA-mediated regulation in Apple (Malus Domestica Borkh.) Plant Biotechnol. J. 2016;14:749–770. doi: 10.1111/pbi.12425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 31.Ferrandez-Ayela A, et al. Arabidopsis TRANSCURVATA1 encodes NUP58, a component of the nucleopore central channel. PLoS ONE. 2013;8:e67661. doi: 10.1371/journal.pone.0067661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zheng, L. et al. Genome-wide identification and expression analysis of GRF genes regulating apple tree architecture. Tree Genet. Genomes. 14, (2018). [DOI] [PubMed]
- 33.Li, J. et al. Genome-wide identification and analysis of the SBP-box family genes in apple (Malus × Domestica Borkh.). Plant Physiol. Bioch.70, 100–114 (2013). [DOI] [PubMed]
- 34.Li Y, et al. Effect of iaa spraying on floral induction in ‘fuji’ apple (malus domestica borkh) Acta agriculturae boreali-occidentalis sinic. 2015;24:84–89. [Google Scholar]
- 35.Wilmowicz E, et al. The role of PnACO1 in light- and IAA-regulated flower inhibition in pharbitis nil. Acta Physiol. Plant. 2013;35:801–810. doi: 10.1007/s11738-012-1121-9. [DOI] [Google Scholar]
- 36.Boeglin M, et al. Reduced expression of AtNUP62 nucleoporin gene affects auxin response in arabidopsis. BMC Plant Biol. 2016;16:1. doi: 10.1186/s12870-015-0695-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sakamoto T, et al. Ectopic expression of KNOTTED1-like homeobox protein induces expression of cytokinin biosynthesis genes in rice. Plant Physiol. 2006;142:54–62. doi: 10.1104/pp.106.085811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bolduc N, Hake S. The maize transcription factor KNOTTED1 directly regulates the Gibberellin Catabolism Genega2ox1. Plant Cell. 2009;21:1647–1658. doi: 10.1105/tpc.109.068221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jasinski S, et al. KNOX action in arabidopsis is mediated by coordinate regulation of cytokinin and gibberellin activities. Curr. Biol. 2005;15:1560–1565. doi: 10.1016/j.cub.2005.07.023. [DOI] [PubMed] [Google Scholar]
- 40.Shani E, et al. Stage-specific regulation of solanum lycopersicum leaf maturation by class 1 KNOTTED1-LIKE HOMEOBOX proteins. Plant Cell. 2009;21:3078–3092. doi: 10.1105/tpc.109.068148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bharathan G, et al. Homologies in leaf form inferred from KNOXI gene expression during development. Science. 2002;296:1858–1860. doi: 10.1126/science.1070343. [DOI] [PubMed] [Google Scholar]
- 42.Khan M, et al. Repression of lateral organ boundary genes by PENNYWISE and POUND-FOOLISH is essential for meristem maintenance and flowering in Arabidopsis1[OPEN] Plant Physiol. 2015;169:2166–2186. doi: 10.1104/pp.15.00915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chai M, et al. A class II KNOX gene, KNOX4, controls seed physical dormancy. Proc. Natl. Acad. Sci. 2016;113:6997–7002. doi: 10.1073/pnas.1601256113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li Y, et al. Effect of exogenous 6-benzylaminopurine (6-BA) on branch type, floral induction and initiation, and related gene expression in ‘Fuji’ Apple (Malus Domestica Borkh) Plant Growth Regul. 2016;79:65–70. doi: 10.1007/s10725-015-0111-5. [DOI] [Google Scholar]
- 45.Zhang, S. et al. Effect of exogenous GA 3 and its inhibitor paclobutrazol on floral formation, endogenous hormones, and flowering-associated genes in ‘Fuji’ Apple (Malus Domestica Borkh.). Plant Physiol. Bioch.107, 178–186 (2016). [DOI] [PubMed]
- 46.Luo Y, et al. An Arabidopsis homolog of importin Β1 is required for ABA response and drought tolerance. Plant J. 2013;75:377–389. doi: 10.1111/tpj.12207. [DOI] [PubMed] [Google Scholar]
- 47.Gu Y, et al. Nuclear pore permeabilization is a convergent signaling event in effector-triggered immunity. Cell. 2016;166:1526–1538. doi: 10.1016/j.cell.2016.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.