Abstract
Background
Mesenchymal stemlike cells (MSLCs) have been detected in many types of cancer including brain tumors and have received attention as stromal cells in the tumor microenvironment. However, the cellular mechanisms underlying their participation in cancer progression remain largely unexplored. The aim of this study was to determine whether MSLCs have a tumorigenic role in brain tumors.
Methods
To figure out molecular and cellular mechanisms in glioma invasion, we have cultured glioma with MSLCs in a co-culture system.
Results
Here, we show that MSLCs in human glioblastoma (GBM) secrete complement component C5a, which is known for its role as a complement factor. MSLC-secreted C5a increases expression of zinc finger E-box-binding homeobox 1 (ZEB1) via activation of p38 mitogen-activated protein kinase (MAPK) in GBM cells, thereby enhancing the invasion of GBM cells into parenchymal brain tissue.
Conclusion
Our results reveal a mechanism by which MSLCs undergo crosstalk with GBM cells through the C5a/p38 MAPK/ZEB1 signaling loop and act as a booster in GBM progression.
Key Points
1. MSLCs activate p38 MAPK-ZEB1 signaling in GBM cells through C5a in a paracrine manner, thereby boosting the invasiveness of GBM cells in the tumor microenvironment.
2. Neutralizing of C5a could be a potential therapeutic target for GBM by inhibition of mesenchymal phenotype.
Keywords: C5a, glioblastoma, invasiveness, mesenchymal stem-like cells, tumor microenvironment
Importance of the Study.
Cancer treatments have been commonly focused on the tumor itself. However, Tumor tissue contained not only proliferating cancer cells but also normal cells, such as fibroblasts, macrophages, pericytes and endothelial cells are recruit and crosstalk with the cancer cells. MSCs has also been identified in several types of tumors, including GBM. Herein, we defined MSLC mediated GBM infiltration mechanism that C5a secreted from MSLCs activates the p38 signaling pathway in GBM cells, thereby increasing the expression of ZEB1 and changing to a mesenchymal phenotype. Eventually, phenotype of GBM cells was changed by MSLC become malignant and contribute to poor prognosis of the patient survival. Therefore, targeting C5a in MSLC isolatable GBM patients can be potential therapeutic approach through suppressed the infiltration of GBM cells, thereby increasing patient survival.
Glioblastoma (GBM) is one of the most lethal human tumor types. Despite treatment with a standard regimen of surgery and concomitant chemoradiation therapy followed by adjuvant chemotherapy, the median survival of GBM patients is 14.6 months.1 Although there have been many efforts to develop targeted molecular agents,2 these agents have failed to improve the overall survival of primary GBM patients.3 Accordingly, a new conceptual approach is urgently needed to overcome treatment failure.
Current treatment strategies reflect a long-standing focus on the intrinsic properties of cancer cells. However, a tumor is not a solitary mass of only proliferating cancer cells. Instead, the tumor microenvironment includes a repertoire of recruited normal cells, such as fibroblasts, macrophages, pericytes, and endothelial cells.4 Emerging evidence suggests that these tumor-associated stromal cells behave as active participants in tumorigenesis rather than passive bystanders in the tumor microenvironment.5 Among the stromal cells, mesenchymal stemlike cells (MSLCs) have been recently identified as stromal components in many types of cancers, including GBM.6,7 Mesenchymal stem cells (MSCs), which were originally isolated from the bone marrow, are non-hematopoietic multipotent precursors characterized in vitro by their adherence to plastic, multipotency for mesenchymal trilineage differentiation, and expression of distinguishing surface markers.8 Although bone marrow–derived MSCs (BM-MSCs) are the prototypical MSCs, this cell type has also been detected in almost all tissues, including brain tissue.9 MSCs display wound tropism and have the potential to repair damaged tissues and regenerate native tissues owing to their differentiation potential.10 Given the innate tropism of MSCs for injury sites under pathological conditions such as inflammation, it would be reasonable to assume that MSCs are recruited to tumor sites and become tumor-associated stromal cells. However, whether MSCs or MSLCs in the tumor microenvironment play a role in cancer promotion or suppression has remained obscure, presumably reflecting differences in their origin and the type of tumor studied.11,12 Bioinformatics analysis show that the majority of GBM samples have a mesenchymal profile rather than epithelial. If epithelial-mesenchymal transition (EMT) is induced, this phenotype can be shifted toward an even more mesenchymal type phenotype in glioma cells. Increased EMT related markers can induce mesenchymal transitions particularly invasion and migration of glioma cells.13
Here, we used MSLCs isolated from primary GBM patients to investigate heterotypic tumor-stroma signaling in the GBM microenvironment. Collectively, our findings reveal the molecular and cellular mechanisms underlying the pathological role of MSLCs as active contributors to the invasiveness of GBM cells.
Materials and Methods
Details of this section can be found in the Supplementary Material.
Culture of GBM Cells and MSLCs
X01 GBM cells established previously from an acutely resected human GBM biopsy were kindly provided by Dr Akio Soeda (University of Virginia).14 TS11-16 (referred to as gCSC0329) and TS09-03 (referred to as gCSC0504) GBM cells were established from GBM biopsies.15 Patient-derived GSC11 GBM cells were kindly provided by Frederick F. Lang’s laboratory (The University of Texas MD Anderson Cancer Center).16 Human BM-MSCs were obtained from Yonsei Severance Hospital, Seoul, Korea.
3D Invasion Assays
The GBM cells were loaded in the upper chamber and incubated for 48 h. To visualize infiltration, GSC11 cells were transduced with green fluorescent protein (GFP). Infiltration was quantified as a percentage of the spheroid area at the initial time according to the formula (AT–A0)/A0 × 100, where AT is the area covered by invaded cells at time T and A0 is the area of the spheroid at the initial time.
Transfection
Cells were transfected with small interfering (si)RNA using a Microporator-mini (Digital Bio Technology) according to the procedure recommended by the manufacturer. The sequences of the siRNAs are given in Supplementary Table 2.
Transduction
For retrovirus production, H29D packaging cells were transfected with pSuper-sh-control or pSuper-sh-C5. Forty-eight hours after transfection, the viral supernatant was collected, passed through a 0.45-μm filter, and then used for transduction.
Western Blot Analysis
Proteins in cell lysates were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis and transferred to a nitrocellulose membrane (Amersham). These proteins were visualized by enhanced chemiluminescence procedures (Amersham).
Real-Time Quantitative PCR
Total RNA was isolated using Trizol (Invitrogen). Quantitative (q)PCR was performed using a KAPA SYBR FAST qPCR kit (KAPA Biosystems) in Rotor Gene Q (Qiagen). Fold changes were calculated by the 2-ΔΔCt method using ACTB as the internal normalization control.
Co-culture of MSLCs and GBM Cells
MSLCs were seeded in the upper chamber, and GBM cells were seeded in the lower chamber of a Transwell system with a pore size of 0.4 μm. Phenotypic changes were analyzed after 3 days of co-culture.
Cytokine Array
A human cytokine array (Proteome Profiler Array Human Cytokine, R&D Systems, #ARY005) was performed according to the manufacturer’s instructions.
Immunocytochemistry
Cells were fixed with 4% paraformaldehyde and permeabilized with 0.1% Triton X-100 in phosphate buffered saline. The cells were visualized using an anti-mouse Alexa Fluor 488–conjugated secondary antibody (Molecular Probes). The stained cells were visualized with a fluorescence microscope (Olympus).
Animal Studies
Male athymic nude mice (5‒8 wk old) (Central Lab, Animal Inc) were used to assess tumorigenesis by GBM cells and MSLCs. GBM cells (5 × 105) alone or combined with MSLCs or BM-MSCs (1:1 ratio) were injected at a speed of 0.5 μL/min into the right frontal lobe of the mouse brain via a Hamilton syringe (Dongwoo Science) using a guide-screw system as described previously.9,17,18
Immunohistochemistry
Paraffin-embedded tissue sections were deparaffinized in xylene and stained with hematoxylin. Observation and imaging were conducted using an IX71 microscope (Olympus).
Fluorescence Activated Cell Sorting Analysis
For FACS, GBM cells or MSLCs were gently dissociated and filtered through cell strainers (70 μm; BD Falcon) followed by incubation with conjugated primary antibodies. Cell surface markers were analyzed with a FACS Vantage SE flow cytometer (BD Biosciences) equipped with FlowJo software (TreeStar), and 10 000 events were recorded for each sample.
Study Ethics Approval
Animal experimental procedures were approved by the Institutional Animal Care and Use Committee, Severance Hospital, Yonsei University College of Medicine. Human studies were approved by the institutional review boards of Severance Hospital, Yonsei University College of Medicine (4-2012-0212). Informed consent was obtained from patients according to the Declaration of Helsinki. Neuropathologists diagnosed each surgical specimen according to World Health Organization classifications.19
Statistical Analysis
Comparisons between values were performed using an unpaired two-tailed Student’s t-test, and ANOVA was used for multivariate analysis. Survival curves were plotted using the Kaplan–Meier method, and P-values were determined by the log-rank test. All statistical analyses were performed using GraphPad Prism 7.0 software. P-values <0.05 were considered significant.
Results
Tumor-Associated MSLCs Correlate with the Invasion of GBM Cells
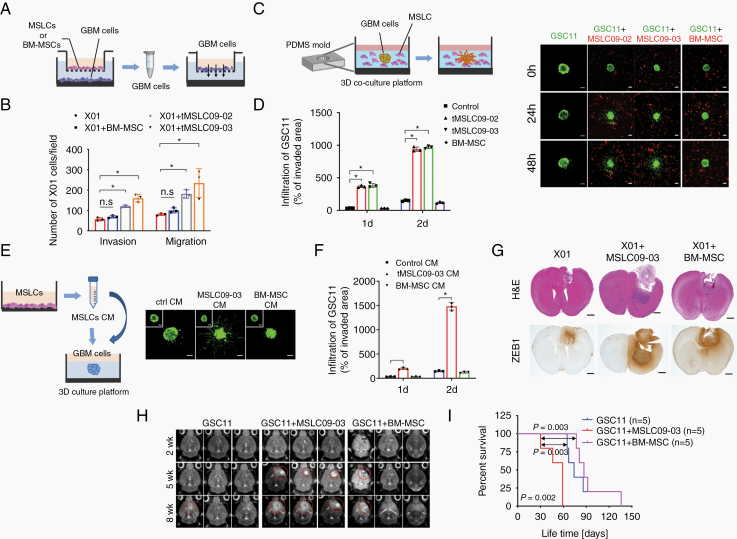
Earlier, we isolated human MSLCs from glioma specimens (Supplementary Table 1).18 To investigate their functional role in the GBM microenvironment, we studied 3 MSLC lines (MSLC09-03, MSLC11-31, MSLC09-02). Importantly, these cells are similar to MSCs20 based on their spindle-shaped morphology (Supplementary Figure 1A) and surface antigens (CD105, CD73, CD90) but not leukocyte (CD45), endothelial cell (CD31), or pericyte (NG2) markers (Supplementary Figure 1B). We next tested mesenchymal differentiation potential as described previously.18 Of note, these cells were able to differentiate into trilineage mesenchymal cells, including osteogenic, adipogenic, and chondrogenic cells (Supplementary Figure 1C). Critically, orthotopic injection of these MSLCs into the mouse brain failed to form tumors, even at 6 months, indicating that MSLCs lack an intrinsic tumorigenic ability (Supplementary Figure 1D). To determine whether MSLCs contribute to phenotypic changes in GBM cells, we co-cultured patient-derived X01, TS11-16, and TS09-03 GBM cells with MSLCs or BM-MSCs in transwells. In this co-culture system, the GBM cells were separated from the MSLCs or BM-MSCs but could communicate with each other through a porous membrane (Fig. 1A). After co-culture, the migration and invasion of the GBM cells were analyzed in a Transwell system. Importantly, the GBM cells co-cultured with the MSLCs (09-02, 09-03) were more migratory and invasive than the cells cultured alone or co-cultured with the BM-MSCs (Fig. 1B, Supplementary Figure 2A–C). To confirm this finding, we labeled GSC11 GBM cells with GFP and then visualized the infiltration of the GBM cells into a collagen-based matrix. Consistently, the invasiveness of the GBM cells was markedly increased by co-culture with MSLCs but not by co-culture with BM-MSCs (Fig. 2C, D). Since phenotypic changes were caused by soluble factors secreted by MSLCs, we next sought to determine whether the invasiveness of GBM cells could be enhanced by treatment with conditioned media (CM) from MSLCs. To this end, we treated GSC11 spheroids with MSLC CM or BM-MSC CM for 48 hours and then examined the infiltration of the GBM cells into the collagen-based matrix. As expected, compared with treatment with control CM or the BM-MSC CM, treatment with the MSLC CM enhanced the invasiveness of the GSC11 and X01 GBM cells (Fig. 1E, F, Supplementary Figure 3A, Movie S1A, B).
Fig. 1.
Phenotypic change observed in GBM cells co-cultured with MSLCs but not with BM-MSCs. (A) Schematic illustration of the invasion/migration assays used to analyze GBM cells after co-culturing with MSLCs or BM-MSCs in transwells. (B) Quantification of the invasion/migration assays with X01 GBM cells shown in (A) (n = 3). (C) Infiltration of spheroid GSC11 cells (green) in co-culture with MSLCs or BM-MSCs (red) into a 3D collagen matrix. Scale bars: 100 μm. (D) Quantification of GSC11 cell infiltration into a 3D collagen matrix (n = 3). (E) Infiltration of GSC11 spheroid cells into a 3D collagen matrix filled with MSLC09-03 CM or BM-MSC CM. The inset shows spheroids at the initial time. Scale bars: 100 μm. (F) Quantification of GSC11 cell infiltration into a 3D collagen matrix (n = 3). (G) Hematoxylin and eosin (H&E) for ZEB1 in coronally sectioned mouse brain samples (n = 5 mice/group). Scale bar: 1 mm. (H) MRI of mouse brain samples harvested at 2, 5, 8 weeks after implantation with GSC11 alone or combined with either MSLC09-03 or BM-MSCs. (I) Kaplan–Meier survival of mice implanted with GSC11 cells alone or combined with either MSLC09-03 or BM-MSCs. *P < 0.05, **P < 0.01, ***P < 0.001.
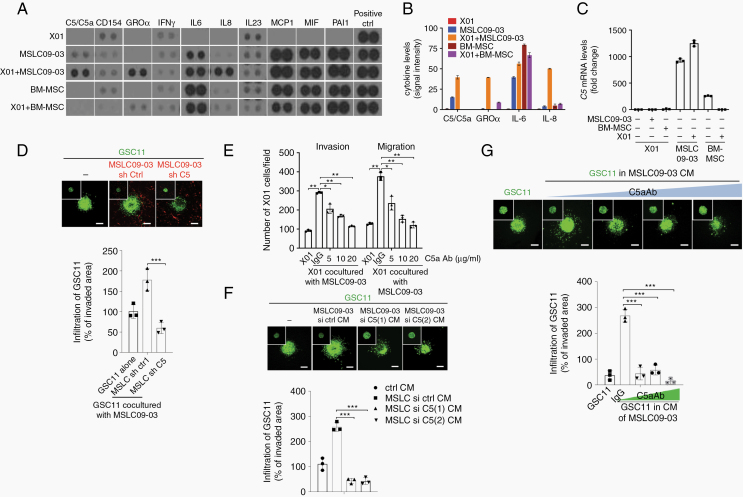
Fig. 2.
MSLCs promote the invasion of GBM cells through C5a. (A) Cytokine array of CM from each cell type alone or co-culture as indicated (n = 2). (B) Densitometry analysis of the cytokine array shown in (A). (C) RT-qPCR for the C5 expression (n = 3). (D) Infiltration of GSC11 spheroid cells (green) into a 3D collagen matrix premixed with MSLC09-03 (red) transduced with the C5-shRNA. The inset shows spheroids at the initial time. Scale bars: 100 μm (n = 3). (E) Invasion/migration with GBM cells co-cultured with MSLC09-03 with anti-C5a antibody (n = 3). (F, G) Infiltration of GSC11 spheroid cells into a 3D collagen matrix filled with the CM of MSLCs pretreated with C5-siRNAs (F) or CM of MSLCs supplemented with the anti-C5a antibody (G) (n = 3). The insets show spheroids at the initial time. Scale bar: 100 μm. *P < 0.05, **P < 0.01, ***P < 0.001.
Among molecularly defined GBM subtypes, the mesenchymal subtype exhibits more invasiveness and a worse prognosis than the other subtypes.21,22 Notably, the non-mesenchymal subtypes tend to acquire mesenchymal features upon relapse after aggressive anticancer treatment, which is reminiscent of epithelial-mesenchymal transitions.23,24 In addition, EMT regulators (SNAIL, SLUG, TWIST, ZEB) have been shown to play critical roles in the malignant phenotypes of GBM.13 Among EMT transcription factors, zinc finger E-box-binding homeobox 1 (ZEB1) was exclusively induced along with loss of epithelial markers E-cadherin and mucin 1 in the GBM cells after co-culture with MSLCs (Supplementary Figure 2D, Supplementary Figure 3B).
To validate these cellular events in an in vivo system, we orthotopically implanted X01 cells alone or with MSLCs into the mouse brain. Consistent with the in vitro data, the GBM cells co-injected with the MSLCs showed greater infiltration into adjacent regions following tumor formation than the X01 cells injected alone (Fig. 1GSupplementary Figure 3C). To a lesser extent, co-inoculation with BM-MSCs slightly enhanced the infiltration of GBM cells; however, this effect was not statistically significant. In agreement with the in vitro data, immunohistochemistry (IHC) analysis revealed that ZEB1 levels were higher in the xenograft tumors formed by the mixture of X01 cells and MSLC09-03 than in the tumor margin formed by the GBM cells alone or combined with BM-MSCs. In addition, brain tumor formation was observed at an earlier time point, and the survival rate was poorer in the mice co-implanted with GSC11 cells and MSLCs than in the counterpart mice (Fig. 1H, I). Also, there was no significant changes in tumor growth observed in mice co-implanted with GBM cells and MSLCs compared with only GBM groups (Supplementary Figure 3D). These results suggest that MSLCs contribute to a shift in GBM cells toward the mesenchymal state, presumably via paracrine factors released by the MSLCs.
MSLCs Contribute to the Invasiveness of GBM Cells Through C5a
Since the above data indicate that MSLCs promote a malignant GBM phenotype in a paracrine manner, we sought to define the soluble factor secreted by MSLCs that is responsible for the shift toward the more invasive type of GBM cells. Using a cytokine array, we found that the levels of the soluble factors C5a, growth-regulated oncogene alpha (GROα), interleukin (IL)-6, and IL-8 were drastically increased in a co-culture of GBM cells and MSLCs (Fig. 2A, B, Supplementary Figure 4A). Among those soluble factors, C5, the precursor of C5a, was highly expressed in MSLCs compared with GBM cells and BM-MSCs (Fig. 2C, Supplementary Figure 4B). The levels were elevated to a relatively high extent in the MSLCs co-cultured with GBM cells, as were the levels of GROα, IL-6, and IL-8 (Supplementary Figure 4C–E), indicating crosstalk between the GBM cells and MSLCs. To test the possibility that these soluble factors mediate the MSLC-induced invasiveness of GBM cells, we analyzed the migration and invasion of GBM cells after co-culturing with MSLCs transfected with siRNA against C5, GROα, IL-6, or IL-8. Notably, the effect of MSLCs on the migration and invasion of GBM cells was abolished by pretreatment with the C5 siRNA but not with the siRNA against GROα, IL-6, or IL-8 (Supplementary Figure 4F, G, Supplementary Figure 5A), indicating that C5a is responsible for the MSLC-induced invasiveness of GBM cells. To further confirm the specificity of this effect, we depleted C5a in MSLCs (09-02, 09-03) using 2 siRNAs targeting different sites of the C5 transcript or a C5 short hairpin (sh)RNA. Consistently, treatment with either the siRNAs or shRNA inhibited the enhancing effect of MSLCs (09-02, 09-03) on the invasion and migration of GBM cells (Supplementary Figure 5B‒D). In agreement with these results, C5 depletion also blocked the effect of MSLC09-03 cells on the infiltration of GSC11 cells in a collagen-based matrix (Fig. 2D). To validate the critical role of C5a in the invasiveness of GBM cells, we next sought to block C5a functionally using an anti-C5a antibody. Consistently, treatment with the anti-C5a antibody attenuated the MSLC-enhanced migration and invasion of GBM cells in a dose-dependent manner (Fig. 2E, Supplementary Figure 6A). GBM cells also infiltrated less in a collagen-based matrix filled with C5-depleted MSLC CM than in a matrix filled with control MSLC CM (Fig. 2F, Supplementary Figure 6B). Likewise, supplementation with the anti-C5a antibody diminished the effect of MSLC CM on the infiltration of GBM cells into a collagen-based matrix in a manner proportional to the concentration of the anti-C5a antibody (Fig. 2G). To further confirm the effect of C5a on invasiveness, we treated X01 cells with a human recombinant (rh) C5a protein. Importantly, rhC5a enhanced the migration and invasion of GBM cells in a Transwell system in a concentration-dependent manner (Supplementary Figure 6C). A similar observation was also made in a collagen-based matrix (Supplementary Figure 6D). In agreement with the in vitro data, IHC analysis revealed that C5a levels were higher in orthotopic xenograft tumors formed by a mixture of X01 cells and MSLC09-03 than in tumors formed by X01 alone or combined with BM-MSCs (Supplementary Figure 6E).
MSLC-Secreted C5a Promotes ZEB1-Driven Phenotypic Changes in GBM Cells
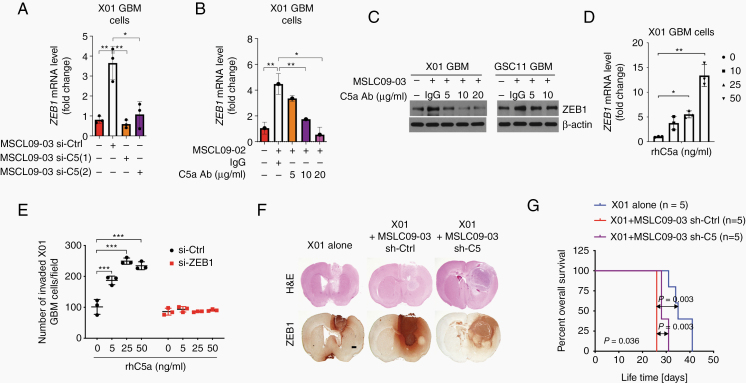
Given that ZEB1 was highly induced in GBM cells co-cultured with MSLCs, we next examined whether MSLC-secreted C5a is responsible for the increase in the ZEB1 level in GBM cells. Real-time (RT)-qPCR showed that the ZEB1 level was increased in X01 and GSC11 cells by co-culture with MSLCs; however, this effect was not observed after co-culture with C5-depleted MSLCs (Fig. 3A, Supplementary Figure 7A). Treatment with a C5a function-blocking antibody also attenuated the effect of MSLCs on ZEB1 induction in a concentration-dependent manner (Fig. 3B, Supplementary Figure 7B). A similar observation was made by western blot (Fig. 3C). In accordance with these data, we further confirmed the role of C5a in the induction of ZEB1 expression in GBM cells by treatment with rhC5a. As anticipated, treatment with rhC5a effectively increased the levels of ZEB1 in both GBM cells treated with increasing concentrations of rhC5a (Fig. 3D, Supplementary Fig. 7B).
Fig. 3.
C5a promotes the invasion of GBM cells via the upregulation of ZEB1 expression. (A) RT-qPCR for ZEB1 in X01 GBM cells after co-culture with MSLC09-03 transfected with a C5-siRNA (n = 3). (B, C) RT-qPCR and western blot of ZEB1 in X01 co-cultured with MSLC09-02 or MSLC09-03 with anti-C5a antibody (n = 3). (D) RT-qPCR for ZEB1 in X01 after treatment with rhC5a (n = 3). (E) Effect of rhC5a on the invasion of X01 in transwells after transfection with a ZEB1-siRNA (n = 3). (F) H&E of ZEB1 in coronally sectioned mouse brain tumors (n = 5 mice/group). Scale bar: 1 mm. (G) Kaplan–Meier survival of mice bearing brain tumors formed by X01 cells (n = 5 mice/group). *P < 0.05, **P < 0.01, ***P < 0.001.
Since ZEB1 expression in GBM cells was increased by C5a, we next examined whether the effect of C5a on the invasiveness of GBM cells occurs via ZEB1. To this end, we tested whether ZEB1 depletion can attenuate the effect of C5a on the invasiveness of GBM cells. Consistently, treatment with rhC5a enhanced X01 cell invasion in a concentration-dependent manner; however, pretreatment with ZEB1 siRNA blocked the effect of rhC5a on X01 GBM invasiveness (Fig. 3E).
To validate our in vitro observation, we transduced MSLC09-03 cells with a C5 shRNA or scrambled control shRNA prior to orthotopic co-inoculation with X01 cells into the mouse brain. Upon tumor formation, the GBM cells co-implanted with MSLCs invaded more into adjacent regions of the brain than the GBM cells implanted alone; however, C5 depletion in the MSLCs blocked the effect on GBM cell invasion (Fig. 3F, Supplementary Figure 7D). Additionally, we observed that growth of glioma cells was not affected after either co-culture with MSLC or rC5a treatment, indicating our mechanism shows motility independent cell growth pathways (Supplementary Figure 7E). The co-inoculation of MSLCs with X01 cells produced a higher number of ZEB1-positive cells in tumors than the injection of GBM cells alone; however, the number of ZEB1-positive cells was not increased by the co-inoculation including C5-depleted MSLCs. In parallel, the mice co-inoculated with GBM and C5a-depleted MSLCs showed longer survival time than the mice inoculated with GBM cells combined with MSLCs transduced with the control shRNA (Fig. 3G).
C5a Increases ZEB1 Expression in GBM Cells Through C5aR1-Mediated p38 Mitogen-Activated Protein Kinase
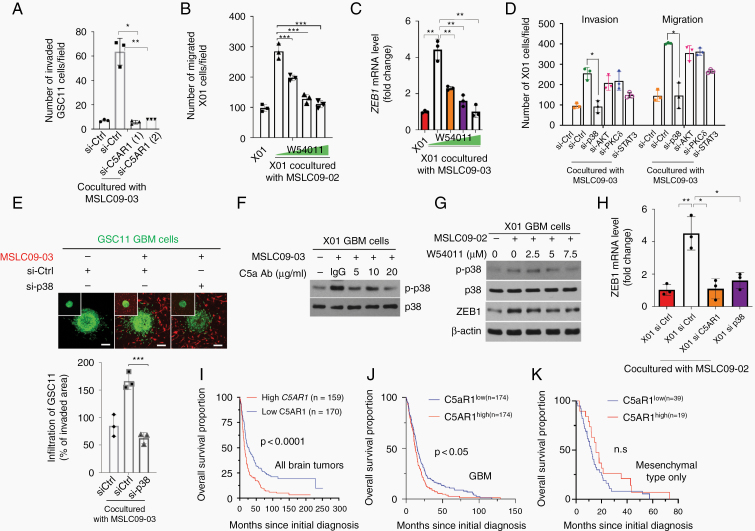
To investigate the regulatory mechanisms of C5a, we examined whether the effect of C5a occurs through its cognate receptor in GBM cells. To this end, we treated GBM cells with siRNA against C5a receptor 1 (C5aR1) (Supplementary Figure 8A) and examined invasion in a co-culture with MSLC09-03. Consistently, invasion by the GBM cells was enhanced by co-culturing with the MSLCs; however, this was not the case when C5aR1 was depleted in the GBM cells (Fig. 4A, Supplementary Figure 8B). Notably, treatment with the C5aR1 antagonist W54011 also diminished the effect of MSLC co-culture on the invasiveness of GBM cells and ZEB1 induction (Fig. 4B, C, Supplementary Figure 8C, D). In accordance, we further examined the signaling mechanism underlying C5a/C5aR1-mediated phenotypic changes in GBM cells. To this end, we examined the main downstream effectors of C5aR1—mitogen-activated protein kinases (MAPKs) (p38, JNK, ERK), JAK/STAT, SRC, PKCδ—in GBM cells after co-culture with MSLCs. We found that the phosphorylation of p38, AKT, PKCδ, and STAT3 was increased in the GBM cells co-cultured with the MSLCs (Supplementary Figure 8E, F). In accordance with these findings, we knocked down the expression of each signaling component by siRNA in GBM cells and analyzed GBM cell motility. The downregulation of p38 expression effectively attenuated the MSLC-enhanced migration and invasion of GBM cells, whereas the downregulation of the expression of the other signaling factors had no such effect (Fig. 4D, Supplementary Figures 8G, 9A, B). To confirm the contribution of p38 to MSLC-induced infiltration, we treated GBM spheroids with MSLC09-03 CM supplemented with the p38 inhibitor SB203580. Interestingly, treatment with the MSLC CM enhanced the invasion of GBM cells; however, this effect was abrogated by SB203580 (Supplementary Figure 9C). When p38 was depleted in GBM cells by siRNA, co-culture with MSLCs no longer enhanced the infiltration of GBM cells into a collagen-based matrix (Fig. 4E). Given that p38 was activated by MSLCs and contributed to the invasion of GBM cells, we tested whether p38 activity depends on C5a-mediated C5aR1 activation. Treatment with the anti-C5a antibody or C5aR1 antagonist W54011 attenuated the effect of MSLCs (09-03, 09-02 cells) on p38 activation in a concentration-dependent manner (Fig. 4F, G, Supplementary Figure 9D). Similar effect was observed in presence of shC5a (Supplementary Figure 9E). Because ZEB1 expression is increased by C5a, we next validated whether ZEB1 expression can be upregulated by C5aR1 and p38. Consistently, ZEB1 expression was greatly increased in GBM cells by co-culture with MSLCs; however, it was not increased in GBM cells in which C5aR1 or p38 was depleted prior to co-culturing with MSLCs (Fig. 4H, Supplementary Figure 9F).
Fig. 4.
C5a increases ZEB1 expression via C5aR1-mediated p38 MAPK activation. (A) Invasion of GSC11 GBM cells co-cultured with MSLC09-03 cells in transwells after transfection with a C5aR1-siRNA (A) or treatment with the C5aR1 antagonist W54011 (B) as indicated (n = 3). (C) RT-qPCR for ZEB1 levels in X01 cells treated with the C5aR1 antagonist W54011 in co-culture with MSLC09-03 (n = 3). (D) Invasion/migration with GBM cells transfected with siRNA as indicated and co-cultured with MSLC09-03. (E) Infiltration of GBM cells into a 3D collagen matrix premixed with MSLC09-03 after transfection with a p38-siRNA. The inset shows spheroids at the initial time. (F) Western blot for p-p38 in X01 co-cultured with MSLC09-03 in the presence of an anti-C5a antibody (n = 3). (G) Western blot for p-p38 and ZEB1 in X01 treated with the C5aR1 antagonist W54011 in co-culture with MSLC09-02 (n = 3). (H) RT-qPCR for ZEB1 in X01 co-cultured with MSLC09-02 after siRNA transfection (n = 3). (I–K) Kaplan–Meier survival of human all brain tumors from REMBRANDT (I), TCGA GBM (J), and mesenchymal type of TCGA GBM (K) patients with high or low C5aR1 median expression. *P < 0.05, **P < 0.01, ***P < 0.001.
Since MSLC-induced C5a increases ZEB1 expression in GBM cells through C5aR1-mediated p38 activation, we also evaluated whether C5aR1 levels in human brain tumors including mesenchymal or only GBM cases correlate with patient survival. An analysis using the Repository of Molecular Brain Neoplasia Data (REMBRANDT) and The Cancer Genome Atlas (TCGA) database showed that C5aR1 levels were well correlated with the survival of glioma and GBM patients (Fig. 4I, J), while there is no correlation in the mesenchymal type of GBM cases (Fig. 4K).
MSLCs Residing in GBM Tumors Correlate with Clinical Outcome
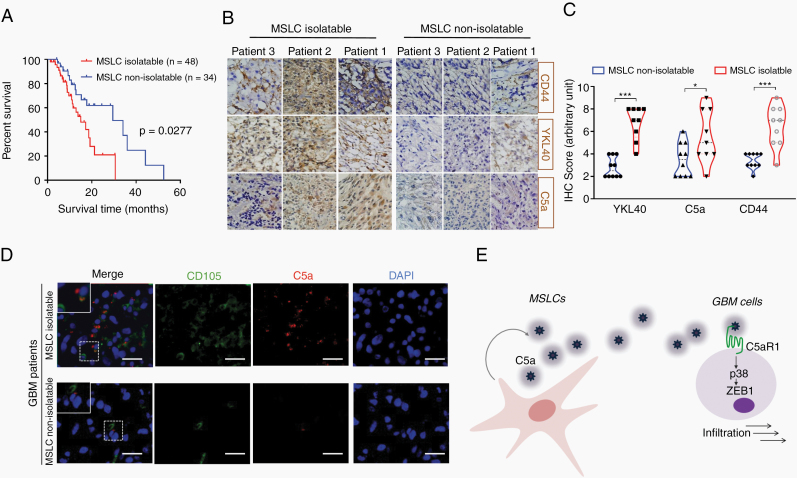
MSLCs have previously been obtained from primary GBM patient biopsies. However, intriguingly, MSLCs could not be isolated from all GBM patients. Notably, in our previous work tracking the overall survival of GBM patients (n = 82; MSLC isolatable and non-isolatable), we observed that the survival time was significantly shorter in the MSLC-isolatable GBM patients than in the non-isolatable patients, presumably owing to the absence or rarity of these cells in GBM (Fig. 5A).25 Out of the 82 patients in our trial, 48 patients were successfully isolated and 34 patients were not isolated. There are 4 steps to define MSLC isolatable. The case of failure to isolate MSLCs is well documented in our previous report.18 If it is determined that the 4 steps are not suitable for the MSLCs defined in each step, they are defined as non-isolatable MSLCs. In addition, IHC analysis using randomly selected patient samples revealed that the expression of YKL40 and CD44, two well-known mesenchymal markers, was higher in the MSLC-isolatable samples than in the non-isolatable GBM patient specimens (Fig. 5B, C). Because our data showed that MSLCs promoted mesenchymal features and the invasion of GBM cells through C5a secretion, we also examined the level of C5a in these patient samples. As anticipated, C5a-positive cells were observed more frequently in MSLC-isolatable GBM than in non-isolatable GBM. Immunofluorescence analysis revealed that C5a-expressing cells were mainly colocalized with MSLCs (CD105+) in the MSLC-isolatable GBM patient specimens, whereas MSLCs (CD105+) and C5a-expressing cells were rarely detected in the MSLC-non-isolatable specimens (Fig. 5D, Supplementary Figure 10A). Since C5a can be expressed by macrophages, we also examined the colocalization of C5a with macrophages (ionized calcium binding adaptor molecule 1 [Iba1] positive). Noticeably, most C5a overlapped with Iba1-positive cells in MSLC non-isolatable GBM; however, we easily found C5a-expressing cells that were negative for Iba1 in the MSLC-isolatable GBM patient specimens (Supplementary Figure 10B). These findings suggest that MSLCs residing in human GBM participate in the mesenchymal phenotypic change in GBM cells, at least though C5a secretion, and are associated with the prognosis of GBM patients.
Fig. 5.
MSLCs residing in GBM correlate with patient clinical outcomes and mesenchymal transformation in GBM cells. (A) Kaplan–Meier survival of GBM patients25 from whom MSLCs could be isolated (n = 48) or could not be isolated (n = 34). (B) IHC of YKL40 and CD44 in MSLC-isolatable (n = 9) and MSLC-non-isolatable (n = 10) GBM patient specimens. (C) Quantification of YKL40, C5a, CD44 levels in IHC. (D) Immunofluorescence of MSLCs (CD105+) and C5a in MSLC-isolatable and MSLC-non-isolatable GBM patient. Scale bar: 50 µm. (E) Schematic summarizing the phenotypic change in GBM cells in the MSLC-supported tumor microenvironment. The n values in A and C indicate the number of GBM patients. *P < 0.05, ***P < 0.001.
Discussion
During the invasion, cancer cells loosen their contact adhesion to neighboring cells and the extracellular matrix (ECM), degrade the adjacent cell tissues, and enhance their motility. Also, invasion of metastatic cancer cells throughout ECM has been believed to be facilitated by protease-mediated matrix degradation that makes a movement track for cancer cells migration.26 The detection of MSLCs in many invasive types of cancer has raised interest in their role in the tumor microenvironment.6,7,27 However, whether MSLCs positively or negatively regulate cancer progression has remained obscure.28,29 Using co-culture systems, we found that MSLCs increased ZEB1 levels in GBM cells through the C5a/C5aR1/p38 MAPK signaling and thereby increased the invasiveness of GBM (Fig. 5E).
In accordance with our findings, several lines of evidence have also suggested that MSLCs in the tumor microenvironment contribute to cancer progression. Xu et al reported that intravenous injection of human BM-MSCs into human osteosarcoma tumor-bearing mice stimulated tumor growth and further showed that osteosarcoma cell proliferation was increased by MSC CM.30 Moreover, subcutaneous administration of cancer cells mixed with human BM-MSCs was shown to promote the growth of human colon carcinoma in mice.31 Bourkoula et al reported that non-tumorigenic multipotent stem cells were present as stromal cells in the tumor microenvironment and increased the aggressiveness of GBM.32 Likewise, brain tumor–derived MSLCs have been shown to be involved in glioma progression in mice.11 However, other studies have suggested the opposite effect, reporting antitumor activity by MSCs. Otsu et al observed that direct inoculation of rat BM-MSCs into subcutaneous melanoma tumors induced apoptosis and abrogated tumor growth in mice.12 Using a model of Kaposi’s sarcoma, Khakoo et al found that intravenously injected human BM-MSCs homed to sites of tumorigenesis and inhibited tumor growth, implying anti-tumorigenic effects mediated by MSCs.33 Although the reasons for these contrasting observations remain largely unknown, the role of MSLCs in the tumor microenvironment might depend on their origin, their degree of differentiation, and the type of tumor cells they interact with. To precisely define the biological role of MSLCs in cancer progression, their site of origin must be considered. Hence, we used MSLCs that were isolated from GBM patient biopsies18 and examined the effect of these MSLCs on GBM progression. Notably, survival time was significantly shorter in patients from whom MSLCs were isolated than in those from whom MSLCs could not be isolated. Moreover, orthotopic co-inoculation of GBM cells with MSLCs enhanced tumorigenesis in mice and shortened the survival time of the mice compared with that of mice inoculated with GBM cells alone or GBM cells combined with human BM-MSCs, suggesting that endogenous MSLCs in tumor sites are intrinsically different from BM-MSCs despite their similarities in regard to MSC marker expression and multipotency for mesenchymal trilineage differentiation.
In this study, C5a was secreted by MSLCs but not by BM-MSCs. Importantly, the secretion of C5a was markedly increased in MSLCs co-cultured with GBM cells, indicating crosstalk between the GBM cells and MSLCs. C5a, a complement component, triggers the degranulation of mast cells and neutrophils, enhancing the phagocytosis of pathogens.34 Our findings suggest that in addition to performing this well-known role, C5a acts as a cue that promotes the invasion of GBM cells in the tumor microenvironment. MSLC-secreted C5a increased the expression of ZEB1, one of the EMT transcription factors, via p38 activation in GBM cells. In agreement, ZEB1 levels were higher in xenograft tumors formed by GBM cells co-injected with MSLCs than in tumors formed by GBM cells alone. Although the term “EMT” in cancer biology is more suitable for carcinoma cancers that are developed from epithelial cells, the EMT-like process and EMT activating transcription factors have also begun to gain attention in non-epithelial-origin cancer cells. In line with this notion, our findings show that the EMT transcription factor ZEB1 is induced in GBM and promotes the invasion of GBM cells. Importantly, treatment with rhC5a enhanced the invasion of GBM cells; however, rhC5a had no such effect on ZEB1-depleted GBM cells, indicating that ZEB1 acts as the downstream effector of C5a in the invasion of GBM cells. In agreement with our findings, previous studies have also suggested that the ZEB1 pathway correlates with the initiation, invasion, and stemness of GBM, consequently affecting the survival of GBM patients.35 Besides, we showed that there was no reliable relationship between C5a and glioma growth in vitro (Supplementary Figure 7E). To define this correlation more clearly, further studies including in vivo experiments would be needed. In summary, our findings show that non-tumorigenic MSLCs promote the invasion of GBM cells through the secretion of C5a into the tumor microenvironment, which further increases ZEB1 expression in the GBM cells through the C5aR1/p38 MAPK signaling pathway.
Funding
This study was supported by the Bio & Medical Technology Development Program of the National Research Foundation (NRF) funded by the Korean government (MSIT) (No.2019M3E5D1A01069361), National Research Foundation of Korea (NRF) grant funded by the Korea government (MEST) (NRF-2019R1A2C3004155), and a grant of the Korea Health Technology, R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea (HI14C1324).
Conflict of interest statement
The authors declare no conflict of interest.
Authorship statement
Concept design: EJL, SGK, SJL, Designed and performed most of the experiments: EJL, RKK, SK, HJL, 3D invasion experimental and data analysis: YJO, JC, MSLC characterization, animal experiments data collection, and analysis: JHL, Pathological analysis, clinical insight in animal experiments: SHK, JC, Animal experiments: JKS, Clinical insight, data analysis and interpretation: JHC, YKH, YMH, Chromosome Spreading Analysis and relative data interpretation: EJL, Intellectual support and data analysis: MJK, MJP, All data reviewed and wrote the manuscript: YS, NK, SGK, SJL, Conceived and supervised the project.: SJL, SGK, PK
Supplementary Material
Acknowledgments
We thank Dr Lang (Department of Neurosurgery, The University of Texas, MD Anderson Cancer Center) for providing patient-derived GSC11 GBM cells. We also thank Dr Akio Soeda (Department of Neurological Surgery, University of Virginia) for providing patient-derived X01 GBM cells.
References
- 1. Stupp R, Mason WP, van den Bent MJ, et al. ; European Organisation for Research and Treatment of Cancer Brain Tumor and Radiotherapy Groups; National Cancer Institute of Canada Clinical Trials Group Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352(10):987–996. [DOI] [PubMed] [Google Scholar]
- 2. Thomas AA, Brennan CW, DeAngelis LM, Omuro AM. Emerging therapies for glioblastoma. JAMA Neurol. 2014;71(11):1437–1444. [DOI] [PubMed] [Google Scholar]
- 3. Chinot OL, Wick W, Mason W, et al. Bevacizumab plus radiotherapy-temozolomide for newly diagnosed glioblastoma. N Engl J Med. 2014;370(8):709–722. [DOI] [PubMed] [Google Scholar]
- 4. Junttila MR, de Sauvage FJ. Influence of tumour micro-environment heterogeneity on therapeutic response. Nature. 2013;501(7467):346–354. [DOI] [PubMed] [Google Scholar]
- 5. Malanchi I, Santamaria-Martínez A, Susanto E, et al. Interactions between cancer stem cells and their niche govern metastatic colonization. Nature. 2011;481(7379):85–89. [DOI] [PubMed] [Google Scholar]
- 6. Kim SM, Kang SG, Park NR, et al. Presence of glioma stroma mesenchymal stem cells in a murine orthotopic glioma model. Childs Nerv Syst. 2011;27(6):911–922. [DOI] [PubMed] [Google Scholar]
- 7. Ho CM, Chang SF, Hsiao CC, Chien TY, Shih DT. Isolation and characterization of stromal progenitor cells from ascites of patients with epithelial ovarian adenocarcinoma. J Biomed Sci. 2012;19:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bianco P, Robey PG, Simmons PJ. Mesenchymal stem cells: revisiting history, concepts, and assays. Cell Stem Cell. 2008;2(4):313–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kang SG, Shinojima N, Hossain A, et al. Isolation and perivascular localization of mesenchymal stem cells from mouse brain. Neurosurgery. 2010;67(3):711–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Phinney DG, Prockop DJ. Concise review: mesenchymal stem/multipotent stromal cells: the state of transdifferentiation and modes of tissue repair—current views. Stem Cells. 2007;25(11):2896–2902. [DOI] [PubMed] [Google Scholar]
- 11. Behnan J, Isakson P, Joel M, et al. Recruited brain tumor-derived mesenchymal stem cells contribute to brain tumor progression. Stem Cells. 2014;32(5):1110–1123. [DOI] [PubMed] [Google Scholar]
- 12. Otsu K, Das S, Houser SD, Quadri SK, Bhattacharya S, Bhattacharya J. Concentration-dependent inhibition of angiogenesis by mesenchymal stem cells. Blood. 2009;113(18):4197–4205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Iser IC, Pereira MB, Lenz G, Wink MR. The epithelial-to-mesenchymal transition-like process in glioblastoma: an updated systematic review and in silico investigation. Med Res Rev. 2017;37(2):271–313. [DOI] [PubMed] [Google Scholar]
- 14. Soeda A, Park M, Lee D, et al. Hypoxia promotes expansion of the CD133-positive glioma stem cells through activation of HIF-1alpha. Oncogene. 2009;28(45):3949–3959. [DOI] [PubMed] [Google Scholar]
- 15. Kong BH, Park NR, Shim JK, et al. Isolation of glioma cancer stem cells in relation to histological grades in glioma specimens. Childs Nerv Syst. 2013;29(2):217–229. [DOI] [PubMed] [Google Scholar]
- 16. Bhat KPL, Balasubramaniyan V, Vaillant B, et al. Mesenchymal differentiation mediated by NF-κB promotes radiation resistance in glioblastoma. Cancer Cell. 2013;24(3):331–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lal S, Lacroix M, Tofilon P, Fuller GN, Sawaya R, Lang FF. An implantable guide-screw system for brain tumor studies in small animals. J Neurosurg. 2000;92(2):326–333. [DOI] [PubMed] [Google Scholar]
- 18. Kim YG, Jeon S, Sin GY, et al. Existence of glioma stroma mesenchymal stemlike cells in Korean glioma specimens. Childs Nerv Syst. 2013;29(4):549–563. [DOI] [PubMed] [Google Scholar]
- 19. Louis DN, Ohgaki H, Wiestler OD, et al. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114(2):97–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dominici M, Le Blanc K, Mueller I, et al. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy. 2006;8(4): 315–317. [DOI] [PubMed] [Google Scholar]
- 21. Verhaak RG, Hoadley KA, Purdom E, et al. ; Cancer Genome Atlas Research Network Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17(1):98–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Carro MS, Lim WK, Alvarez MJ, et al. The transcriptional network for mesenchymal transformation of brain tumours. Nature. 2010;463(7279):318–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Halliday J, Helmy K, Pattwell SS, et al. In vivo radiation response of proneural glioma characterized by protective p53 transcriptional program and proneural-mesenchymal shift. Proc Natl Acad Sci U S A. 2014;111(14):5248–5253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Phillips HS, Kharbanda S, Chen R, et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9(3):157–173. [DOI] [PubMed] [Google Scholar]
- 25. Yoon SJ, Shim JK, Chang JH, et al. Tumor mesenchymal stem-like cell as a prognostic marker in primary glioblastoma. Stem Cells Int. 2016;2016:6756983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lim EJ, Suh Y, Kim S, Kang SG, Lee SJ. Force-mediated proinvasive matrix remodeling driven by tumor-associated mesenchymal stem-like cells in glioblastoma. BMB Rep. 2018;51(4):182–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kwak J, Shin HJ, Kim SH, et al. Isolation of tumor spheres and mesenchymal stem-like cells from a single primitive neuroectodermal tumor specimen. Childs Nerv Syst. 2013;29(12):2229–2239. [DOI] [PubMed] [Google Scholar]
- 28. Karnoub AE, Dash AB, Vo AP, et al. Mesenchymal stem cells within tumour stroma promote breast cancer metastasis. Nature. 2007;449(7162):557–563. [DOI] [PubMed] [Google Scholar]
- 29. Shinagawa K, Kitadai Y, Tanaka M, et al. Mesenchymal stem cells enhance growth and metastasis of colon cancer. Int J Cancer. 2010;127(10):2323–2333. [DOI] [PubMed] [Google Scholar]
- 30. Xu WT, Bian ZY, Fan QM, Li G, Tang TT. Human mesenchymal stem cells (hMSCs) target osteosarcoma and promote its growth and pulmonary metastasis. Cancer Lett. 2009;281(1):32–41. [DOI] [PubMed] [Google Scholar]
- 31. Zhu W, Xu W, Jiang R, et al. Mesenchymal stem cells derived from bone marrow favor tumor cell growth in vivo. Exp Mol Pathol. 2006;80(3):267–274. [DOI] [PubMed] [Google Scholar]
- 32. Bourkoula E, Mangoni D, Ius T, et al. Glioma-associated stem cells: a novel class of tumor-supporting cells able to predict prognosis of human low-grade gliomas. Stem Cells. 2014;32(5):1239–1253. [DOI] [PubMed] [Google Scholar]
- 33. Khakoo AY, Pati S, Anderson SA, et al. Human mesenchymal stem cells exert potent antitumorigenic effects in a model of Kaposi’s sarcoma. J Exp Med. 2006;203(5):1235–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Peng Q, Li K, Sacks SH, Zhou W. The role of anaphylatoxins C3a and C5a in regulating innate and adaptive immune responses. Inflamm Allergy Drug Targets. 2009;8(3):236–246. [DOI] [PubMed] [Google Scholar]
- 35. Siebzehnrubl FA, Silver DJ, Tugertimur B, et al. The ZEB1 pathway links glioblastoma initiation, invasion and chemoresistance. EMBO Mol Med. 2013;5(8):1196–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.