Abstract
Background
Fosfomycin is an antibiotic that has seen a revival in use due to its unique mechanism of action and efficacy against isolates resistant to many other antibiotics. In Escherichia coli, fosfomycin often selects for loss-of-function mutations within the genes encoding the sugar importers, GlpT and UhpT. There has, however, not been a genome-wide analysis of the basis for fosfomycin susceptibility reported to date.
Methods
Here we used TraDIS-Xpress, a high-density transposon mutagenesis approach, to assay the role of all genes in E. coli involved in fosfomycin susceptibility.
Results
The data confirmed known fosfomycin susceptibility mechanisms and identified new ones. The assay was able to identify domains within proteins of importance and revealed essential genes with roles in fosfomycin susceptibility based on expression changes. Novel mechanisms of fosfomycin susceptibility that were identified included those involved in glucose metabolism and phosphonate catabolism (phnC-M), and the phosphate importer, PstSACB. The impact of these genes on fosfomycin susceptibility was validated by measuring the susceptibility of defined inactivation mutants.
Conclusions
This work reveals a wider set of genes that contribute to fosfomycin susceptibility, including core sugar metabolism genes and two systems involved in phosphate uptake and metabolism previously unrecognized as having a role in fosfomycin susceptibility.
Introduction
The increasing prevalence of bacteria that are resistant to clinically important antibiotics has led to searches for alternative options to treat problematic infections.1 There has been limited progress in the development of new antibiotics and one strategy has been to revive older drugs that may be effective but are not commonly used in clinical practice.2 One example is fosfomycin, which has seen an increase in clinical use in recent years. Fosfomycin has a unique mode of action where it targets the initial stages of peptidoglycan biosynthesis by acting as a phosphoenolpyruvate analogue and inhibiting MurA.3 Consequently, fosfomycin retains activity against bacterial strains expressing β-lactamases that inhibit later peptidoglycan biosynthetic reactions. This is useful given the prevalence of different β-lactamase enzymes in important pathogens. Fosfomycin is a phosphonic acid molecule produced in nature by Streptomyces species and is commonly used for treatment of complicated urinary tract infections and increasingly for more serious systemic infections. In Enterobacteriaceae, fosfomycin enters the cell by acting as a mimic for two nutrient importer systems, GlpT and UhpT.4
Resistance to fosfomycin has been shown to be relatively easy to select in vitro and resistant mutants often show loss of function of GlpT or UhpT, or have mutations in adenylcyclase (cyaA) or phosphotransferase (ptsI) genes, both of which control intracellular levels of cyclic AMP, which in turn regulates expression of glpT and uhpT.5–9 In addition, alterations within the drug target MurA (particularly those altering a Cys115 residue near the active site) can decrease susceptibility by reducing its affinity for fosfomycin.10–12 Over-expression of murA has also been observed in fosfomycin-resistant isolates and is thought to act by saturating the drug.13,14 Resistance to fosfomycin may also be acquired by genetic transfer of fosA and fosB coding for functions that disrupt the fosfomycin oxirane ring.15–17
Whilst it has been easy to select for fosfomycin-resistant isolates in vitro, there is evidence that selection of resistance carries a major fitness cost, and that fosfomycin-resistant isolates may be compromised in virulence.18–20 Various studies have looked for the prevalence of fosfomycin resistance in different settings, and resistance rates in general have remained relatively low, even in high-use settings.21,22
Given the recent increase in the use of fosfomycin, we used a genome-wide transposon mutagenesis approach in Escherichia coli to identify loci involved in fosfomycin susceptibility.23 We identified new loci as being involved in fosfomycin susceptibility, including the phosphonate uptake and utilization system and phosphate import system, as well as identifying sub-domains within some proteins involved in fosfomycin susceptibility.
Materials and methods
TraDIS-Xpress library
We recently described the construction of a high-density (insert approximately every 6 bp) TraDIS-Xpress library in E. coli BW25113, which was used in this work.24 BW25113 is a commonly used reference strain, a full and well-annotated genome sequence is available, and it was the parent strain for the KEIO collection of defined insertion mutants, thus enabling easier validation of the roles of candidate genes.24 The transposon used {a mini-Tn5 transposon coding for kanamycin resistance [aph(3′)-Ia]} incorporates an outward-transcribing tac promoter 3′ of the kanamycin cassette that is inducible by IPTG, allowing over-expression or repression of genes adjacent to insertion sites (depending on insert orientation) as well as gene inactivation. This allows the roles of essential genes in response to a stress to be analysed based on expression changes; traditionally these loci have been cryptic in TraDIS experiments as insertions within them are lethal.
Fosfomycin exposure conditions and TraDIS-Xpress sequencing
The MIC of fosfomycin for BW25113 was determined using microbroth dilution in Mueller–Hinton (MH) and LB broth (the same growth medium that was used for TraDIS-Xpress experiments). For TraDIS-Xpress experiments, approximately 107 mutants were inoculated into 1.7 mL LB broth in deep 96-well plates containing doubling concentrations of fosfomycin ranging from 0.25× to 2× MIC (and a drug-free control). Replicate experiments were completed with no induction, or with the addition of 0.2 or 1 mM IPTG to induce transcription from the outward-transcribing promoter. Mutants were grown for 24 h at 37°C. All experiments were performed in duplicate to give a total of 30 independent TraDIS-Xpress experiments (Figure S1, available as Supplementary data at JAC Online).
After growth under experimental conditions, DNA was extracted from pools of mutants using a Quick-DNA™ Fungal/Bacterial 96 Kit (Zymo Research). DNA was then fragmented using a Nextera DNA library preparation kit (Illumina) except that Tnp-i5 oligonucleotides were used instead of i5 index primers, and 28 PCR cycles. The resulting DNA was sequenced on a NEXTSeq 500 sequencing machine using a NextSeq 500/550 High Output v2 kit (75 cycles). All sequence data have been deposited with EBI under project accession number PRJEB29311.
Bioinformatics
Results were analysed using BioTraDIS (version 1.4.1) and AlbaTraDIS (version 0.0.5), developed for TraDIS-Xpress analysis and recently described.25,26 Briefly, BioTraDIS was used to map sequence reads against the BW25113 reference genome (CP009273) using SMALT and to create insertion plots.
The patterns of inserts were compared between fosfomycin-exposed and control conditions, AlbaTraDIS then calculated the number of inserts within each gene as well as assessing the number of forward and reverse insertions per gene and within a window of 198 bp upstream and downstream of each gene. This length for a window was chosen as likely to include regions within which inserts were likely to influence expression of the relevant gene. The number of sequence reads was modelled on a per-gene basis using a negative binomial distribution and an adapted exact test as implemented in edgeR followed by multiple testing correction to identify significant differences between conditions (each test condition compared with insert patterns from drug-free controls).27,28 A set of default cut-offs for significance and number of reads were applied (q-value ≤0.05, logFC ≥1, logCPM >8). This resulted in a list of candidate genes involved in fosfomycin susceptibility as well as a prediction as to whether a change in expression of a gene (either down- or up-regulated) influences survival. The insertion patterns at candidate loci were visually inspected using Artemis, which was also used to capture images for figures.29
Validation experiments
A total of 18 mutants were selected from the KEIO library to validate predictions about susceptibility to fosfomycin made by TraDIS-Xpress.30 These included genes in the phosphonate uptake and metabolism and phosphorus import systems identified as major contributors to fosfomycin susceptibility as well as a set of randomly selected control genes not expected to have any impact on fosfomycin susceptibility. Both replicate mutants present in the KEIO collection (the collection contains two independent insertion mutants for each gene in BW25113) were analysed. Mutants were tested for their susceptibility to fosfomycin by determination of fosfomycin MIC. All experiments were duplicated (giving at least four datasets for each gene: two repeats from each of the two mutant alleles of each gene). BW25113 was included in all experiments as a control.
The potential for the bis-phosphonate etidronate to antagonize fosfomycin was evaluated using chequerboard assays where dilutions of fosfomycin (from 16 to 0.125 mg/L) and dilutions of etidronate (from 400 to 0.4 mg/L) were combined. Plates were incubated for 24 h at 37°C in a FLUOstar Omega plate reader (BMG Labtech) with OD measurements being taken every 15 min to capture growth kinetics.
Results
Susceptibility of BW25113 to fosfomycin
The MIC of fosfomycin for BW25113 was determined to be 4 mg/L in both MH and LB broth, and cultures were prepared in LB broth without or with fosfomycin at 0.25×, 0.5×, 1× and 2× MIC, combined with IPTG at two concentrations to induce transcription from the outward-transcribing transposon promoter, or without IPTG to allow promoter repression. Each condition was performed in duplicate to give a total of 30 independent experiments, and each was inoculated with ∼107 cfu from the transposon mutant library. After incubation overnight, DNA was extracted from all cultures, and transposon insertion sites were identified using TraDIS-Xpress. Figure S2 shows the concordance between independent repeats and the impacts of increasing drug concentrations on numbers of nucleotide sequence reads locating to each gene of the whole genome.
Genes involved in fosfomycin susceptibility
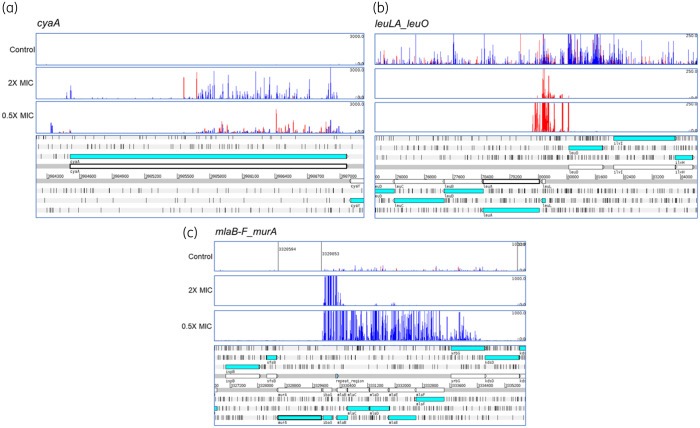
Whilst exposure to different concentrations of fosfomycin identified some concentration-dependent genes, 31 were involved in susceptibility under all fosfomycin conditions (Table 1, Figure S3). These included the majority of known, chromosomally encoded loci that determine fosfomycin susceptibility, with strong signals identified for murA, the target for fosfomycin, glpT and cyaA. This validates the specificity of TraDIS-Xpress in identifying genes involved in fosfomycin susceptibility. The TraDIS-Xpress method also proved able to assay essential genes. For example, murA is an essential gene but mutants with inserts that are positioned upstream and in the same orientation as murA were highly enriched in the presence of IPTG (Figure 1). These mutants will over-express murA, which will help the target saturate the activity of fosfomycin. The high density of the library also allows very high-resolution analysis; for example, cyaA was identified as a significant mechanism but enrichment of inserts was restricted to that part of the gene encoding the regulatory domain (Figure 1). Mutations within this domain (but not the remainder of the protein) have previously been reported as being important in determining fosfomycin susceptibility and TraDIS-Xpress identified that part of the gene encoding this domain. However, no signal was identified at the uhpT locus, which has also previously been implicated in fosfomycin import, although it is not expressed in the test conditions so knockout mutations would have had no selective advantage.
Table 1.
Loci significantly altered in all exposure conditions
| Locus | Predicted impacts of insertiona | Function and interpretation |
|---|---|---|
| ahr | Over-expression | Aldehyde reductase |
| alsR | Inactivation and up-regulation of alsBACEK | Allose binding transporter |
| cmk | Inactivated | Cytidylate kinase; nucleotide binding |
| cra | Inactivated | Catabolite repressor/activator. Global regulator of carbon metabolism |
| crp | Antisense knockdown | Global regulator and carbon catabolite repression |
| cyaA | Inactivation of regulatory domain | Adenylate cyclase; regulates cyclase activity by carbon source |
| dacB | Intragenic insert at one site | Peptidoglycan biosynthesis |
| galU | Inactivated | Synthesis of UDP-d-glucose |
| glpE | Inactivation and overexpression of glpD | Glycerol metabolism |
| glpG | Inactivated | Membrane protein protease |
| glpK | Inactivated | Glycerol kinase; key player in glucose control of glycerol metabolism |
| glpT | Inactivated | Glycerol uptake; known fosfomycin importer |
| glpX | Protected | Fructose bisphosphatase |
| guaA | Inactivation | GMP synthesis |
| ibaG | Inactivated, inserts upstream of murA | Increase in murA expression |
| leuA | Protected and over-expression of leuO | Regulation of stringent response, pleiotropic effects from leuO |
| leuL | Inactivated and over-expression of leuO | Pleiotropic effects from leuO |
| mlaB-F | Inactivated, inserts upstream of murA | Peptidoglycan recycling; fosfomycin target expression increased |
| murA | Upstream inserts | Peptidoglycan biosynthesis; fosfomycin target; expression increased |
| mutL | Protected | DNA repair |
| nagB | Inactivated with up-regulation of nagE | Over-expression of sugar importer (nagE) |
| phnC-M | Inactivated (all 14 members of operon in sense) | Phase-variable phosphonate transporter and degradation complex |
| phoU | Inactivation (downstream of pstB) | Repressor of pstABCS phosphate uptake system |
| pstABCS | Inactivated | Phosphate uptake system |
| ptsH | Inactivated | Decoration of imported sugars with phosphates |
| purA | Inactivated | Purine biosynthesis |
| rfaH | Inactivated (inserts antisense to tatD) | Transcription anti-terminator |
| tatD | Protected | DNA repair |
| treC | Inactivated | Hydrolyses trehalose to glucose |
| waaFGP | Inactivated | LPS branching |
| yhfA | Protected and up-regulated | Conserved hypothetical product, known to be Regulated by CRP |
Over-expression indicates selective advantage for insertions upstream. ‘Protected’ indicates a loss of insertion mutants in the selective condition.
Figure 1.
Differential selection of transposon mutants at cyaA (a), leuO (b) and murA (c). The bottom of each panel illustrates the genomic context and the panels above illustrate the mapped reads. Red bars indicate reads orientated left to right and blue bars the opposite. The height of each bar reflects the abundance of each insert. Data shown are from conditions with IPTG present as an inducer.
Identification of new mechanisms that reduce susceptibility to fosfomycin
As well as loci known to be involved in determining fosfomycin susceptibility, several new loci were identified by TraDIS-Xpress. These included the cra, crp, cyaA, galU, glpK, glpX and treC genes, which are involved in control of available glucose within the cell (Table 1) and will influence expression of the glucose importers that are known routes of entry for fosfomycin. Mutants that over-expressed leuO, a pleiotropic regulator with known roles in control of stress responses, were strongly selected by fosfomycin (Figure 1).
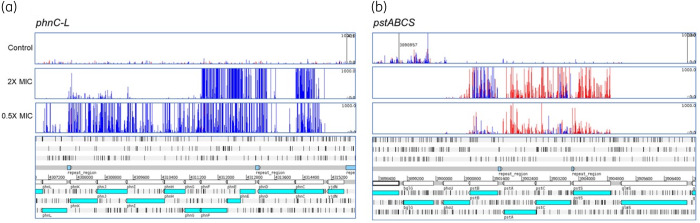
Mutants with transposon insertion mutations within the phn operon coding for phosphonate uptake and degradation were very strongly selected by fosfomycin (Figure 2). The phosphonate uptake and degradation operon comprises 14 genes (phnCDEFGHIJKLMNOP) coding for an ABC importer (PhnCDE), a regulator (PhnF), a multi-subunit methylphosphonate degradation complex (PhnGHIJKL), which includes the carbon–phosphorus bond cleavage activity (PhnGHIJ), and additional functions required to yield phosphate from phosphonate (PhnMNOP). The phn operon has been reported to be cryptic in E. coli K12 due to three, not two, copies of an 8 bp tandem repeat within phnE (encoding the integral membrane component of the importer) that cause a frameshift.31 Sequencing of the parent strain of the transposon library, and mapping of reads obtained from TraDIS experiments after fosfomycin exposure confirmed that the BW25113 parent strain, used to make the transposon mutant library, also has three copies of the tandem repeat within phnE, which should therefore not be expressed. However, following growth with fosfomycin at 2× MIC, transposon insertions in the phnC, phnE and phnF genes conferred a large selective advantage (Figure 2a). These insertions are all oriented such that the transposon promoter will transcribe the rest of the operon, most likely leading to its expression, and suggesting that the phosphonate degradation complex encoded by these genes can modify or degrade fosfomycin. At lower concentrations (0.5× MIC) of fosfomycin, insertion mutations in the same orientation were observed across most of the operon but not within phnMNOP, suggesting that at this lower concentration these gene products alone provide sufficient inactivation of fosfomycin to confer a significant selective advantage.
Figure 2.
Inactivation of the phn (a) and pst (b) operons is selected by fosfomycin. The bottom of each panel illustrates the genomic context and the panels above illustrate the mapped reads. Red bars indicate reads orientated left to right and blue bars the opposite. The height of each bar reflects the abundance of each insert.
Insertion mutations in the pstSACB operon coding for an ABC phosphate importer also conferred a selective advantage during growth with fosfomycin (Figure 2). Insertion mutations in this operon generally showed an antisense orientation bias for at least two of the genes, which probably reflects the relative selective advantage or disadvantage of inactivation and altered expression of the operon components resulting from the different insertions sites of the transposon and its outward-directed promoter.
In contrast, the DNA repair functions encoded by mutL and tatD were beneficial for growth with fosfomycin, as insertion mutants for these genes were lost at all fosfomycin concentrations.
Changes in the expression of some bacterial cell functions reduce susceptibility to several antibiotics simultaneously. These functions include efflux transporters, porins and proteins that regulate their expression directly or indirectly, such as AcrR, MarA and SoxR.32 The TraDIS-Xpress data indicate that none of these functions contributes to susceptibility at any of the fosfomycin concentrations tested, suggesting that selection of cross-resistance to other antibacterials by fosfomycin is limited.
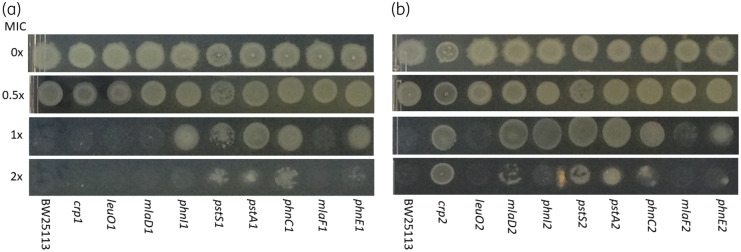
Validation of targets
To test the predictions made by TraDIS-Xpress we tested nine pairs of insertion mutants from the KEIO collection for susceptibility to fosfomycin by growth on LB agar containing different concentrations of the drug (Figure 3). As expected, BW25113 was inhibited by the MIC. Growth of both the leuO mutants was also inhibited at the MIC, and this is also expected as the TraDIS-Xpress data predicted that altered leuO expression, rather than inactivation, was important for reduced fosfomycin susceptibility. For all the other mutant pairs, one or both showed a 2- to 4-fold increase in MIC of fosfomycin compared with the BW25113 parent strain. These mutant pairs included three for the phn operon and both the pst mutants tested (Figure 3). Given the indicated role for the phosphonate uptake system, we tested whether addition of an exogenous phosphonate would impact fosfomycin susceptibility and found that 100 mg/L of the bis-phosphonate etidronate increased the MIC of fosfomycin against BW25113 by 2- to 4-fold. In addition, growth curve experiments indicated that the addition of 100 mg/L etidronate resulted in improvements in growth rate of BW25113 cultures supplemented with different concentrations of fosfomycin (Figure S3). Etidronate alone showed no antibacterial activity (Figure S3).
Figure 3.
Validation of specific mutants by inoculation onto agar containing different fosfomycin concentrations. Panels (a) and (b) represent analysis of both independent mutants for each gene present in the KEIO collection. For each strain, 5 μL spots representing ∼104 cfu were inoculated and incubated overnight at 37°C. Concentrations are indicated as fractions of the MIC (4 mg/L) for BW25113.
Discussion
The mechanisms of fosfomycin action and resistance known to date have largely been elucidated by the study of individual mutants and isolates that demonstrate fosfomycin resistance.18 Here, we used TraDIS-Xpress to assay the role of the whole genome of E. coli in fosfomycin susceptibility in a series of parallel experiments. The data showed the sensitivity of this approach, with many of the known chromosomal mechanisms of resistance being identified as important contributors to fosfomycin susceptibility (apart from uhpT, which is not expressed in the assay conditions used). Additionally, TraDIS-Xpress was also able to identify domains within targets that are important; for example, the regulatory domain of CyaA is known to be involved in fosfomycin susceptibility by controlling cAMP levels and therefore expression of glpT and uhpT.19 Inactivation of the CyaA domain responsible for this function results in lower expression of both GlpT and UhpT importers, and reduced fosfomycin susceptibility. TraDIS-Xpress clearly indicated the role for this regulatory domain but not the rest of cyaA (Figure 1). In line with the identification of cyaA as important, our data also identified inactivation of glpT as providing a selective advantage with fosfomycin as expected, although there was no signal for uhpT.
Traditional TraDIS experiments have not been able to assay many essential genes, which often reduce susceptibility to antibiotics. However, the current work demonstrates that TraDIS-Xpress did assay essential genes following growth in fosfomycin, as exemplified by the selection of many mutants with transposon insertions upstream of murA and oriented to promote transcription from the outward-transcribing transposon promoter. These insertions most likely increase expression, increasing the number of copies of MurA beyond that which can be inhibited by fosfomycin.
In addition to genes known to be involved in fosfomycin resistance, a wider set of new loci were also identified as contributing to susceptibility to the drug (Table 1). These included genes involved in glucose metabolism, which are likely to mediate expression of the glucose importers that facilitate transport of fosfomycin across the inner membrane. This extends our current understanding of how central metabolism can impact susceptibility to fosfomycin and shows how specific growth conditions and associated gene expression may influence susceptibility to this drug by altering gene regulation.
These experiments also found that knockout mutations in mutL and tatD were selectively disadvantageous for growth in fosfomycin. These genes encode DNA repair functions, suggesting that fosfomycin causes some DNA damage resulting from altered metabolism following inhibition of the primary target, MurA, and consequent bactericidal effect. This has been proposed as a common impact of many bactericidal drugs.33–35
The TraDIS-Xpress data indicated that transposon insertions into the phosphonate uptake and catabolism operon (phnCDEFGHILJLMNO) provided a considerable selective advantage for growth in fosfomycin. All these insertions were oriented such that the transposon outward-transcribing promoter would promote expression of downstream genes, suggesting that the products of this system can inactivate fosfomycin. At 0.5× MIC fosfomycin, insertions were found across most of the operon, but not within phnMNOP, suggesting that at lower fosfomycin concentrations one or more of these functions is sufficient for fosfomycin inactivation. At the highest fosfomycin concentration investigated, insertions were upstream of phnG, and orientated towards phnG-P, thereby over-expressing the lyase components. The lack of transposon insertions in the reverse orientation within phnCDE indicates the selective advantage is not likely to be due to inactivation of transport, which should remain inactive throughout the experiment due to the phnE frameshift mutation. There was, however, a phenotype for the phnC mutant, and phnCD will be expressed as they are upstream of the frameshift even though phnE is inactive. Therefore, these results indicate that it is most likely expression of the phosphonate metabolism system, phnGHIJKLMNOP, that provides a selective advantage during growth with fosfomycin.
Whilst UhpT and GlpT are known import systems for fosfomycin, mutation of either the PhnC-M or PstBCSA system was strongly selected by fosfomycin exposure (Figure 2).
The PstBCAS phosphate transporter was also identified as a likely mechanism of fosfomycin entry into the cell, as insertion mutations into the pstBCAS operon conferred a selective advantage during growth with fosfomycin (Figure 2). These data suggest that multiple importers, including novel systems, are involved in fosfomycin susceptibility. Testing of defined mutants in both the Phn and Pst systems confirmed a phenotype, with the mutants able to grow above the MIC of the drug (Figure 3).
In addition, growth in the presence of the bis-phosphonate etidronate resulted in limited but consistent (2-fold increase in the MIC) rescue of growth in the presence of fosfomycin. Figure S4 shows the impact of addition of 100 mg/L of etidronate on growth in the presence of a series of concentrations of fosfomycin, with improved growth evident at most concentrations (etidronate had no intrinsic antimicrobial activity). These data are supportive of specific phosphonate importers being important routes of entry into the cell for fosfomycin. The potential for phosphonate moieties to mediate import of molecules into the cell may have potential utility. A major challenge in therapy is getting drugs into cells and new routes to modify molecules and promote their uptake are likely to enhance efficacy of drugs. It has become clear in recent years that most drugs cross the inner membrane by active import rather than passive diffusion and identifying side groups that may improve uptake by changing importer specificity is important.36
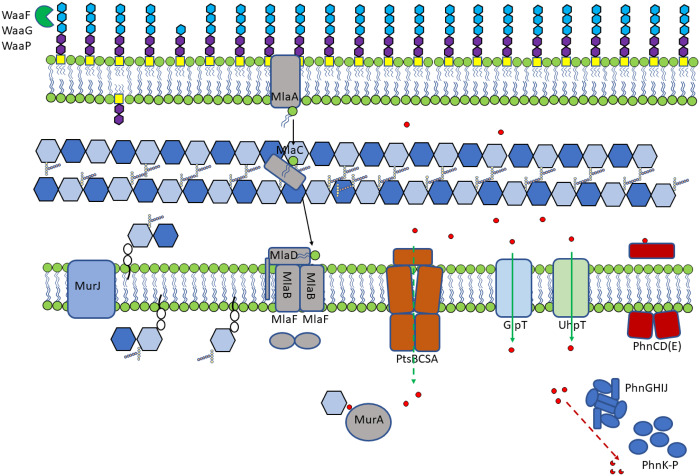
One major worry with some drugs is selection of cross-resistance to other agents, often mediated by generic mechanisms of resistance, including multidrug efflux and/or porins, where expression changes can influence accumulation of many drugs.32 There was not a strong signal for these pathways after fosfomycin exposure, which suggests that the major mechanisms of fosfomycin resistance (we propose a model in Figure 4) are likely to be relatively specific and not influence other classes of agent. This is potentially important as, whilst fosfomycin resistance is not hard to select, a key feature of this drug is its activity against strains resistant to other drugs, in particular various β-lactams.
Figure 4.
Summary of pathways implicated in fosfomycin entry into the cell. Proposed model of roles for genes identified as being involved in fosfomycin susceptibility. Fosfomycin is indicated by red circles and known and potentially novel mechanisms of uptake are indicated by solid and dashed green arrows, respectively.
Taken together, these data show a genome-wide analysis of genes not known previously to be involved in susceptibility to fosfomycin. These include a role for Pst as a transporter and the phosphonate catabolism pathway as a putative inactivator of fosfomycin.
Supplementary Material
Acknowledgements
The author(s) gratefully acknowledge the support of the Biotechnology and Biological Sciences Research Council (BBSRC).
Funding
S.B., A.K.T., M.Y., M.A.W. and I.G.C. were supported by the Biotechnology and Biological Sciences Research Council (BBSRC) Institute Strategic Programme Microbes in the Food Chain BB/R012504/1 and its constituent project BBS/E/F/000PR10349. Genomic analysis used the MRC ‘CLIMB’ cloud computing environment supported by grant MR/L015080/1. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Transparency declarations
None to declare.
Supplementary data
Figures S1 to S4 and an Excel file are available as Supplementary data at JAC Online.
References
- 1. Livermore DM. Has the era of untreatable infections arrived? J Antimicrob Chemother 2009; 64: 29–36. [DOI] [PubMed] [Google Scholar]
- 2. Falagas ME, Grammatikos AP, Michalopoulos A.. Potential of old-generation antibiotics to address current need for new antibiotics. Expert Rev Anti Infect Ther 2008; 6: 593–600. [DOI] [PubMed] [Google Scholar]
- 3. Michalopoulos AS, Livaditis IG, Gougoutas V.. The revival of fosfomycin. Int J Infect Dis 2011; 15: E732–9. [DOI] [PubMed] [Google Scholar]
- 4. Kahan FM, Kahan JS, Cassidy PJ. et al. Mechanism of action of fosfomycin (phosphonomycin). Ann N Y Acad Sci 1974; 235: 364–86. [DOI] [PubMed] [Google Scholar]
- 5. Alper MD, Ames BN.. Transport of antibiotics and metabolite analogs by systems under cyclic AMP control: positive selection of Salmonella Typhimurium cya and Crp mutants. J Bacteriol 1978; 133: 149–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Brown ED, Vivas EI, Walsh CT. et al. MurA (MurZ), the enzyme that catalyzes the first committed step in peptidoglycan biosynthesis, is essential in Escherichia coli. J Bacteriol 1995; 177: 4194–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Falagas ME, Giannopoulou KP, Kokolakis GN. et al. Fosfomycin: use beyond urinary tract and gastrointestinal infections. Clin Infect Dis 2008; 46: 1069–77. [DOI] [PubMed] [Google Scholar]
- 8. Larson TJ, Ye SH, Weissenborn DL. et al. Purification and characterization of the repressor for the sn-glycerol 3-phosphate regulon of Escherichia coli K12. J Biol Chem 1987; 262: 15869–74. [PubMed] [Google Scholar]
- 9. Sonna LA, Ambudkar SV, Maloney PC.. The mechanism of glucose-6-phosphate transport by Escherichia coli. J Biol Chem 1988; 263: 6625–30. [PubMed] [Google Scholar]
- 10. Castañeda-García A, Blázquez J, Rodríguez-Rojas A.. Molecular mechanisms and clinical impact of acquired and intrinsic fosfomycin resistance. Antibiotics 2013; 2: 217–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cordaro JC, Melton T, Stratis JP. et al. Fosfomycin resistance: selection method for internal and extended deletions of phosphoenolpyruvate:sugar phosphotransferase genes of Salmonella typhimurium. J Bacteriol 1976; 128: 785–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kim DH, Lees WJ, Kempsell KE. et al. Characterization of a Cys115 to Asp substitution in the Escherichia coli cell wall biosynthetic enzyme UDP-GlcNAc enolpyruvyl transferase (MurA) that confers resistance to inactivation by the antibiotic fosfomycin. Biochemistry 1996; 35: 4923–8. [DOI] [PubMed] [Google Scholar]
- 13. Takahata S, Ida T, Hiraishi T. et al. Molecular mechanisms of fosfomycin resistance in clinical isolates of Escherichia coli. Int J Antimicrob Agents 2010; 35: 333–7. [DOI] [PubMed] [Google Scholar]
- 14. Venkateswaran PS, Wu HC.. Isolation and characterization of a phosphonomycin resistant mutant of Escherichia coli K-12. J Bacteriol 1972; 110: 935–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Couce A, Briales A, Rodriguez-Rojas A. et al. Genomewide overexpression screen for fosfomycin resistance in Escherichia coli: MurA confers clinical resistance at low fitness cost. Antimicrob Agents Chemother 2012; 56: 2767–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Horii T, Kimura T, Sato K. et al. Emergence of fosfomycin-resistant isolates of Shiga-like toxin-producing Escherichia coli O26. Antimicrob Agents Chemother 1999; 43: 789–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mendes AC, Rodrigues C, Pires J. et al. Importation of fosfomycin resistance fosA3 gene to Europe. Emerg Infect Dis 2016; 22: 346–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fillgrove KL, Pakhomova S, Newcomer ME. et al. Mechanistic diversity of fosfomycin resistance in pathogenic microorganisms. J Am Chem Soc 2003; 125: 15730–1. [DOI] [PubMed] [Google Scholar]
- 19. Nilsson AI, Berg OG, Aspevall O. et al. Biological costs and mechanisms of fosfomycin resistance in Escherichia coli. Antimicrob Agents Chemother 2003; 47: 2850–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rigsby RE, Fillgrove KL, Beihoffer LA. et al. Fosfomycin resistance proteins: a nexus of glutathione transferases and epoxide hydrolases in a metalloenzyme superfamily In: Sies H, Packer L, eds. Glutathione Transferases and Gamma-Glutamyl Transpeptidases. Elsevier Academic Press, 2005; 367–79. [DOI] [PubMed] [Google Scholar]
- 21. Schito GC. Why fosfomycin trometamol as first line therapy for uncomplicated UTI? Int J Antimicrob Agents 2003; 22 Suppl 2: 79–83. [DOI] [PubMed] [Google Scholar]
- 22. Tsuruoka T, Yamada Y.. Characterization of spontaneous fosfomycin (phosphonomycin)-resistant cells of Escherichia coli B in vitro. J Antibiot 1975; 28: 906–11. [DOI] [PubMed] [Google Scholar]
- 23.Health and Social Care Information Centre. Prescription cost analysis England 2012/2013. V1.0 (2013/2014). 2014.
- 24. Yasir M, Turner AK, Bastkowski S. et al. TraDIS-Xpress: a high-resolution whole-genome assay identifies novel mechanisms of triclosan action and resistance. Genome Res 2020; 30: 239–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Barquist L, Mayho M, Cummins C. et al. The TraDIS toolkit: sequencing and analysis for dense transposon mutant libraries. Bioinformatics 2016; 32: 1109–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bastkowski S, Page AJ, Yasir M. et al. AlbaTraDIS: comparative analysis of large datasets from parallel transposon mutagenesis experiments. PLoS Comput Biol 2020; in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Benjamini Y, Hochberg Y.. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Royal Stat Soc 1995; 57: 289–300. [Google Scholar]
- 28. Robinson MD, McCarthy DJ, Smyth GK.. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010; 26: 139–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Carver T, Harris SR, Berriman M. et al. Artemis: an integrated platform for visualization and analysis of high-throughput sequence-based experimental data. Bioinformatics 2012; 28: 464–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Baba T, Ara T, Hasegawa M. et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2006; 2: 2006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Makino K, Kim SK, Shinagawa H. et al. Molecular analysis of the cryptic and functional phn operons for phosphonate use in Escherichia coli K-12. J Bacteriol 1991; 173: 2665–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Blair JM, Webber MA, Baylay AJ. et al. Molecular mechanisms of antibiotic resistance. Nat Rev Microbiol 2015; 13: 42–51. [DOI] [PubMed] [Google Scholar]
- 33. Belenky P, Ye JD, Porter CB. et al. Bactericidal antibiotics induce toxic metabolic perturbations that lead to cellular damage. Cell Rep 2015; 13: 968–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chen YC, Li CL, Hsiao YY. et al. Structure and function of TatD exonuclease in DNA repair. Nucleic Acids Res 2014; 42: 10776–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ordabayev YA, Nguyen B, Niedziela-Majka A. et al. Regulation of UvrD helicase activity by MutL. J Mol Biol 2018; 430: 4260–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kell DB, Oliver SG.. How drugs get into cells: tested and testable predictions to help discriminate between transporter-mediated uptake and lipoidal bilayer diffusion. Front Pharmacol 2014; 5: 231. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.