Abstract
Bioprinting holds great promise towards engineering functional cardiac tissue constructs for regenerative medicine and as drug test models. However, it is highly limited by the choice of inks that require maintaining a balance between the structure and functional properties associated with the cardiac tissue. In this regard, we have developed a novel and mechanically robust biomaterial-ink based on non-mulberry silk fibroin protein. The silk-based ink demonstrated suitable mechanical properties required in terms of elasticity and stiffness (~40 kPa) for developing clinically relevant cardiac tissue constructs. The ink allowed the fabrication of stable anisotropic scaffolds using a dual crosslinking method, which were able to support formation of aligned sarcomeres, high expression of gap junction proteins as connexin-43, and maintain synchronously beating of cardiomyocytes. The printed constructs were found to be non-immunogenic in vitro and in vivo. Furthermore, delving into an innovative method for fabricating a vascularized myocardial tissue-on-a-chip, the silk-based ink was used as supporting hydrogel for encapsulating human induced pluripotent stem cell derived cardiac spheroids (hiPSC-CSs) and creating perfusable vascularized channels via an embedded bioprinting technique. We confirmed the ability of silk-based supporting hydrogel towards maturation and viability of hiPSC-CSs and endothelial cells, and for applications in evaluating drug toxicity.
Keywords: Silk, GelMA, printing, human iPSC-derived cardiomyocytes, organ-on-a-chip
Graphical abstract
In this work, we have reported a novel non-mulberry silk based biomaterial-ink for developing mechanically robust and clinically relevant cardiac patches. Both anisotropic avascular constructs for mimicking the native tissue structure, as well as vascularized constructs using an innovative embedded gel technology have been fabricated using our designed ink. The vascularized constructs along with a microfluidic system offer great potential for drug sensing platforms.
1. Introduction
Engineering a cardiac tissue has become one of the greatest challenges in recent years due to the complexity associated with mimicking its native structure and the further translation from laboratory scale to clinical practices.[1] With the advent of three-dimensional (3D) bioprinting, fabricating biomimetic structures directly with cells over different length scales has resulted in greater fidelity and reproducibility over conventional fabrication methods.[2] The technique now stands as one of the most promising methods for fabricating native cardiac tissue-like architectures with anatomical precision in the spatio-temporal geometry.[1c, 3] Such fabricated biomimetic and spatially controlled constructs thus have immense potential to be used for tissue regeneration and as drug test models in vitro.[2, 4]
Over the years, 3D printing techniques have facilitated a large number of naturally derived polymers to be used alone or in combination as inks, enabling the printing of complex cardiac tissue constructs. Many of these natural polymers such as gelatin[5], collagen[6], alginate[7], gelatin methacryloyl (GelMA)[7a, 7b, 8] and fibrin[9] act as extracellular matrix (ECM) mimics which help support the growth and function of the desired cells.[10] For instance, cardiomyocyte laden hydrogels bioprinted using fibrin based bioink on a sacrificial poly-caprolactone (PCL) support, by Wang et al., showed dense and highly aligned cardiac tissues with synchronous contraction behaviors.[9] Subsequently, a co-axial extrusion based 3D bioprinting method was utilized by Zhang et al., for creating vascular structures using human umbilical vein endothelial cells (HUVEC) encapsulated in GelMA-alginate based bioink.[7a] Human induced pluripotent stem cells derived cardiomyocytes (hiPSC-CMs) seeded on these vascularized bioprinted scaffolds resulted in creating a mature and spontaneously contracting (75–80 beats min−1) endothelialized myocardial tissue. Providing a local elastic modulus and integrin based binding motifs similar to the native ECM of cardiac tissue, these materials help the cardiomyocytes to attach and mature.[11] The natural polymer based bioinks thus provide an ideal environment for the cardiomyocytes to be functionally active in an in vitro culture.[12] However, many of these natural polymer based bioinks lack the much required mechanical properties (elasticity or toughness) to enable the fabrication of mechanically robust and matured cardiac tissues in vitro to be used for clinical applications.[13] For instance, GelMA based bioprinted scaffolds display excellent biocompatibility and have high swelling behavior. However, their low mechanical properties and fast degradation capacity[14] offer low translation ability beyond their culture in an in vitro system.[15] At this juncture, the inclusion of synthetic polymers as reinforcing agents is highly debatable as they might provide the bioink with improved mechanical properties.[1a, 11a] However, the use of synthetic polymers might also lead to several drawbacks such as low degradability,[12] non-immunocompatibility of the degraded bi-products,[16] decreased porous structures, inhibition of biological activity of natural polymers,[17] and use of cytotoxic chemicals such as organic solvents to dissolve the synthetic polymers.[16a] Therefore, there is an urgent need to investigate polymeric materials that can act as strengthening components of the inks designed for cardiac tissue applications.
Amongst the naturally available biomaterials, silk fibroin (SF), merits its application in tissue engineering due to its several desirable qualities. SF is a mechanically robust, exceptionally resilient, biodegradable material and manifests minimal immunogenic response in comparison to synthetic polymers.[15, 18] Therefore, SF protein has become a popular choice as a biomaterial ink for printing several tissues such as cartilage, bone and skin.[15, 18a, 19] In addition, SF protein has also been to known to promote wound healing due to its excellent biomedical and mechanical properties.[18c, 20] Of the several silk varieties of silk protein, the non-mulberry SF (n-SF) derived from the silk glands of the endemic Indian silkworm variety Antheraea assamensis merits its potential as a propitious material for cardiac tissue engineering.[18b, 18d] Previous studies have reported the presence of intrinsic arginine-glycine-aspartate (RGD) binding sequences and other cell binding motifs (poly-arginine) which have been able to interact well with the integrin isoforms present in the cardiomyocytes, allowing for faster cardiomyocyte attachment and growth.[18b, 18d] Another remarkable feature of this n-SF protein is the ratio of the amorphous regions to β-crystallites in its secondary structure.[21] The presence of amorphous regions within the tightly packed β-nanocrystal arrangement in the n-SF protein has resulted in high elastic properties and tensile yield in comparison to its mulberry (Bombyx mori silk fibroin) counterpart.[21b, 22] Thus, n-SF protein can serve as a potential supporting material as a ink component in order to improve the mechanical strength of inks while maintaining their unique physical and biological properties, especially for cardiac tissue engineering.
In this work, we present a novel biomaterial ink comprising of n-SF as a reinforcing and biologically active component for engineering myocardial tissues. Completely characterizing the ink for its physico-chemical properties, we have delved into exploring the ability of the ink for fabricating both 3D non-vascular anisotropic cardiac constructs as well as a ink for vascularized myocardial tissues. Extrusion based 3D printing has been used to develop non-vascular anisotropic constructs to resemble the native tissue organization and mechanical properties of the heart and to evaluate the functional performance of the cardiomyocytes growing on the printed scaffolds. Furthermore, we have also investigated the translational prospects of such a printed construct by studying the in vivo response post their subcutaneous implantation in rat models.
Additionally, one of the merits of 3D printing is to develop miniature organ models in vitro such that they can be used as drug test models, further reducing the time and cost of production of several new pharma products. In this regard, miniature myocardial-on-a-chip tissues are a great boost to the pharma industries for testing several new drugs for their associated cardiotoxity. In this light, we have developed an endothelialized myocardial tissue as a close replica of native heart tissue by employing an innovative embedded gel bioprinting technique. This technique enables the printing of perfusable microvascular channels within a silk-based supporting hydrogel (using the biomaterial ink) encapsulating human induced pluripotent stem cell derived cardiac spheroids (hiPSC-CSs). Further, in order to explore the application attributes related to the fabricated endothelialized myocardial tissue, we combined the bioprinted vascularized myocardial constructs with a custom designed microfluidic bioreactor to develop an endothelialized myocardial tissue-on-a-chip platform. The combined platform was evaluated for its ability to screen pharmaceutical drugs for associated cardiotoxicity.
2. Results and Discussion
2.1. Biomaterial ink preparation and printability
The designed n-SF based biomaterial ink comprised of n-SF, polyethylene glycol dimethacrylate (PEGDMA), and GelMA (SPG) as its major components. The inability to obtain a high concentration (> 6% v/v) of n-SF in a solution form, partly due to its extraction method directly from the silkworm glands and also due to the formation of a silk gel at high concentrations, promoted the addition of both GelMA and PEGDMA as printable biomaterial ink components. The addition of GelMA supplemented the bioactive properties of the ink whereas the addition of a very small amount of PEGDMA (1% w/v) increased the stability of the printed constructs and the printing fidelity. The designed SPG ink featured a dual crosslinking mechanism that resulted in a stable fabrication of 3D microfibrous constructs. As indicated in Figure 1A, the biomaterial ink was initially crosslinked via temporary enzymatic crosslinking of n-SF in the presence of Horseradish Peroxidase (HRP) and H2O2. Incubation of n-SF at 37 °C with HRP-H2O2 resulted in the formation of di-tyrosinase bonds (n-SF gelation) via the oxidation of phenolic hydroxyl groups in the tyrosine residues present in n-SF.[23] During the preparation of the ink, this initial gelation helped to maintain the stable formation of microfibers in the printing process, ensuring structural integrity. Permanent chemical crosslinking of the printed microfibers was thereupon attained by introducing the 3D printed constructs under a UV-light in order to photo-crosslink the PEGDMA and GelMA components of the biomaterial ink. Through the dual crosslinking process, we successfully obtained the crosslinked SPG hydrogels with increased turbidity as shown in Figure S1A. An optimal ink composition was further determined to acquire a suitable viscosity that could provide a persistent extrusion of the microfibers without clogging the bioprinter needle. Multiple compositions of the biomaterial ink were gauged for their printability. Particularly, concentrations of the PEGDMA (1.0% w/v), photoinitiator (0.3% w/v), 10 mM HRP and 0.9% v/v H2O2 were maintained as constants, while the relative concentrations of GelMA and n-SF were varied in the range of 0–10% w/v and 0–4% v/v respectively (Figure 1B). The optimal composition of the ink was found to be 7.5% w/v GelMA and 3.0% v/v n-SF, in addition to 1.0% w/v PEGDMA (SPG7.5).
Figure 1.
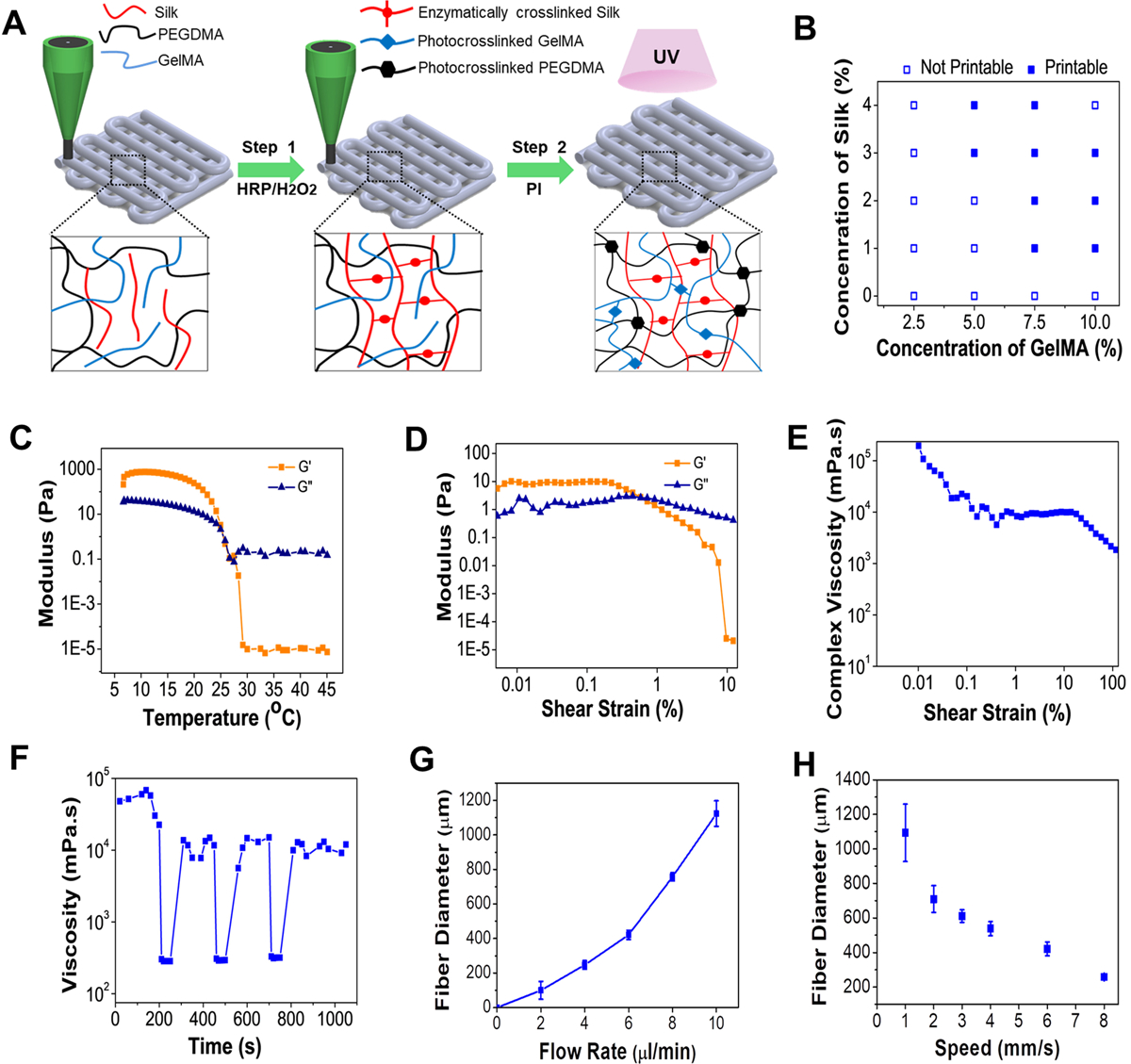
Biomaterial ink optimization and characterization (A) Schematic depicting the sequential crosslinking process of the biomaterial ink. (B) Printability of different concentrations of n-SF and GelMA (Printable: continuous cylindrical microfibers and Non-printable: non-continuous microfibers/liquid like dispensing of the ink). Rheological characterization of the biomaterial ink SPG7.5 with (C) temperature sweep and (D) amplitude sweep where G’ represents storage modulus and G” represents loss modulus (n=4). (E) Change in complex viscosity of the SPG7.5 ink with an amplitude sweep (n=4). (F) Thixotropic property analysis of the SPG7.5 ink at three alternate cycles of oscillation and rotation (n=3). The obtained fiber diameter using SPG7.5 ink at various (G) flow rates and (H) extrusion speeds (n=15).
The printability of the developed biomaterial ink was assessed through rheological characterizations where the storage modulus (G’) and loss modulus (G”) corresponded to the elastic and viscous behavior of the hydrogel ink respectively. The two major components of the biomaterial ink, GelMA and n-SF displayed temperature dependent gelation properties. Whereas, on one hand, GelMA displays gelation properties at lower temperatures, n-SF, on the other hand, resulted in gelation (self-crosslinking) above 37–40 °C. Therefore, in order to optimize the printing temperatures, rheological properties of the ink (SPG7.5) were studied at a temperature sweep ranging from 5–45 °C (Figure 1C). Optimal printing temperatures of the ink were obtained at 25–30 °C where a gel-sol transformation in the ink was observed. Furthermore, the elastic behavior of the SPG7.5 ink was assessed by performing an amplitude sweep at a constant angular frequency ω = 1 rad s−1 and by increasing the shear strain γ from 0.01% to 10%. At lower shear strains the elastic (G’ > G”) nature was evident but at higher shear strains the elastic nature was lost and a viscous nature was observed as G” > G’, assuring the shear thinning effect associated with the viscous behavior of the SPG7.5 ink (Figure 1D) similar to a pseudo-plastic. The non-Newtonian behavior (shear thinning nature) of the ink was also confirmed by analyzing the complex viscosity of the ink when subjected to an increase in shear strain γ from 0.01% to 10% (Figure 1E). Furthermore, the thixotropic nature of the SPG7.5 biomaterial ink was also evaluated for 3 alternate cycles of oscillation-rotation (Figure 1F). The SPG7.5 ink was found to regain its nature rapidly post deformation due to the shear thinning effect in the linear viscoelastic range (LVER) region, indicating the biomaterial ink would not tend to laterally diffuse post extrusion, thus resulting in obtaining stable bioprinted structures before crosslinking of PEGDMA and GelMA pre-polymers. Printing thin microfibers requires the optimization of flow rate and extrusion speed of the biomaterial ink (Figure S1B). By analyzing the fiber diameters obtained at different parameters (Figure 1G–H), a flow rate of 7 μL min−1 was adopted for the ink while the speed of the extruder was adjusted at 8 mm s−1. In these conditions, the smallest diameter of the printed fibers was 120–150 μm.
2.2. Characterization of the 3D printed constructs
In order to recapitulate the myocardium, the ink for fabricating the 3D printed constructs should be able to mimic the mechanical strength of the native tissue. The mechanical strength of the substrate not only serves as a cue for the regulation of cellular morphology,[24] attachment,[25] proliferation,[26] and differentiation[27] but also aids in translation when implanted in vivo after being cultured in an in vitro system. Several previous studies have revealed that cardiomyocytes are able to maintain their cytoskeletal tensions[28] and their maximum functional potential[27–29] by being cultured on substrates that offer stiffness (modulus in the range of 10–40 kPa) and elasticity similar to native cardiac tissue.[28] Using SPG7.5 biomaterial ink, we successfully obtained large scale and easy-to-handle 3D printed microfibrous constructs (4 cm (length) × 1 cm (width) × 0.8 cm (height)) within 4 mins which are difficult to achieve using pristine GelMA ink (due to difficulty in handling). (Figure 2A, Video S1). We then analyzed the tensile strength, elastic modulus and elongation at break of the 3D printed microfibrous constructs fabricated using SPG7.5 ink (Figure 2B–E). Similar constructs fabricated using 5% GelMA ink (G5; a well explored ink concentration) and 7.5% GelMA ink, printed while maintaining the bed temperature at 4 ⁰C, were used as controls. The 3D printed construct fabricated using SPG7.5 biomaterial ink exhibited an elastic modulus of 42.7 ± 8.4 kPa, more than three times greater (p ≤ 0.001) in comparison to the control G5 ink (13.1 ± 3.5 kPa) and almost 1.5 times greater (p ≤ 0.01) than the G7.5 (25.46 ± 1.6 kPa) ink based constructs as displayed in Figure 2D. Also, the 3D printed constructs fabricated using SPG7.5 ink displayed a higher elastic strength and could endure greater strains (up to 33.5 ± 3.3 % strain before complete breakage) as compared to those fabricated using G5 ink (break at 19.1 ± 5.9 % elongation; p ≤ 0.01) and G7.5 ink (break at 27.4 ± 0.78 % strain; p ≤ 0.05) as control samples, as seen in Figure 2E and Figure S2. One of the reasons behind this elastic behavior of the SPG7.5 fabricated constructs could be the due to the presence of β nanocrystals formed due to the enzymatic crosslinking of the n-SF component of the biomaterial ink. The presence of poly-alanine stretches in the secondary structure of the n-SF resulted in the formation of interspersed amorphous regions within the crosslinked β-sheet structures leading to an enhanced elastic nature.[8, 22] Furthermore, to understand the elastic behavior of the constructs in a dynamic condition similar to native cardiac tissue, constructs fabricated using SPG7.5, G5 and G7.5 inks were subjected to cyclic 5% strain for 40 cycles. The SPG7.5 3D printed constructs displayed better elastic strength and behavior in comparison to G5 and G7.5 under cyclic stress conditions (Figure 2F–G & Figure S3). During the first cycle of a dynamic tensile test the SPG7.5 printed constructs were seen to behave like a viscoelastic material. Eventually, the elastic property of the printed constructs was seen to improve through the 40 cycles. At the end of 40 cycles, the constructs behaved like a highly elastic material (rubber) thus justifying the application of the biomaterial ink for fabricating cardiac constructs. The elastic performance of the constructs fabricated using SPG7.5 ink were further analyzed under different cyclic strains (2.5%, 5%, 10%) for 40 cycles (Figure 2H). At higher strain values (10%), negative stress values were obtained at the same strain. This could be due to the re-orientation of silk β nanocrystals in the direction of stretch during the cyclic test as well due to the presence of less organized amorphous regions interspersed between these β-nanocrystallites.[21b, 30] Normal cardiac tissue undergoes a cyclic physiological strain between 5–10% during its function.[31] The ability of the 3D printed constructs to retain their elastic strength at these physiological strains at repeated cycles ensures the ability of the printed constructs to not only support the contractile properties of the cardiomyocytes but also the regular contraction cycles of the heart tissue in a dynamic system.
Figure 2.
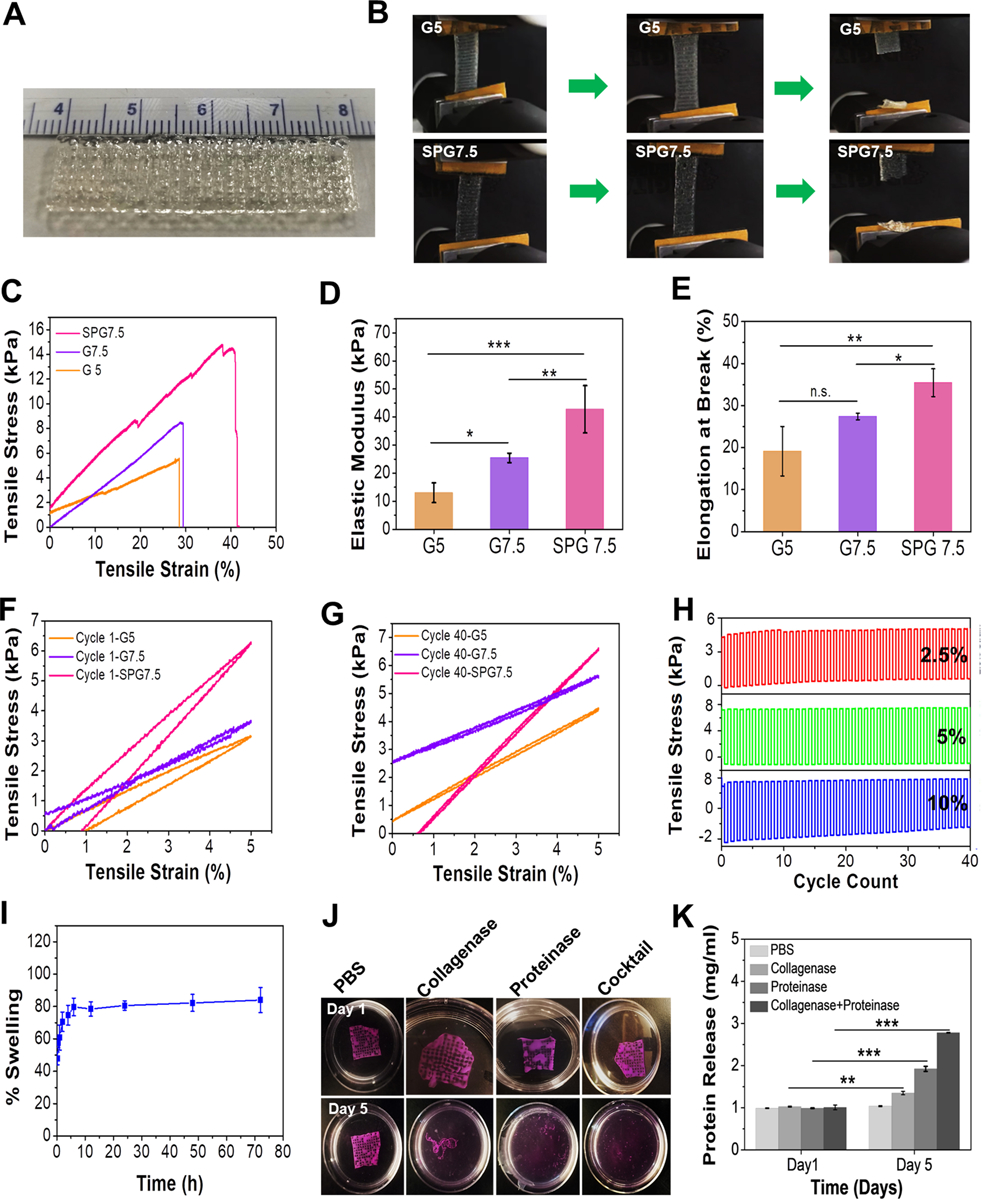
Characterization of 3D printed constructs using the optimized ink composition. (A) Large scale and easy-to-handle anisotropic 3D printed construct fabricated using SPG7.5 ink. (B) Images of the 3D printed construct with 5% GelMA inks as control (G5) and SPG7.5 ink under tensile tests. (C) Stress-strain curves, (D) evaluation of elastic moduli, and (E) percentage elongation at break of the bioprinted constructs fabricated using G5, G7.5 and SPG7.5 inks (n=4,*** p ≤ 0.001, **p ≤ 0.01 and *p ≤ 0.05). (F-G) Cyclic tensile tests of the 3D printed constructs using G5, G7.5 and SPG7.5 inks at 5% tensile strain for cycle 1 and cycle 40 (n=4). (H) Cyclic tensile tests of the 3D printed construct using SPG7.5 ink at 2.5% (red), 5% (green) and 10% (blue) strain for 40 cycles (n=4). (I) Swelling behavior of the 3D printed construct using SPG7.5 ink in DPBS (n=4). (J) Enzymatic degradation behavior of the 3D printed constructs in collagenase type II, proteinase K, and a cocktail (mixture of collagenase type II and proteinase K). Constructs kept in DPBS alone were used as controls. (K) Bradford assay for determing the amount of protein released as a result of degradation of the printed constructs in the presence of enzymes (n=3; *** p ≤ 0.001 and **p ≤ 0.01).
In order to evaluate the stability of the 3D printed constructs post printing, an assessment of swelling and degradation properties was performed. After 6 hours in DPBS, the 3D printed constructs using SPG7.5 ink had reached equilibrium and no significant changes in the degree of swelling were observed after this point with a maximum of 84.0 ± 7.8% (Figure 2I). Degradation analysis of the printed constructs was performed by subjecting them to three kinds of enzymatic treatments: 0.1U proteinase K for silk degradation, 0.1U collagenase type II for GelMA degradation and a combination of both (cocktail). 3D printed constructs placed in DPBS were used as controls. The enzymes collagenase II and Proteinase K attack specific sites of the GelMA and n-SF polypeptide chains, respectively, and break the polypeptide chains down into smaller units (peptides and amino-acids).[7b, 18b] The printed constructs when subjected to enzymatic treatments displayed complete disruption of their printed structures by day 5 as compared to the control set in which the constructs were stable as seen in Figure 2J. The amount of protein released on enzymatic degradation quantified using Bradford’s assay also confirmed a significant protein release in sets subjected to enzymatic degradation compared to control (Figure 2K). The highest degradation (p ≤ 0.001) was observed for constructs subjected to cocktail enzymatic treatment. Therefore, the SPG7.5 biomaterial ink displayed improved mechanical properties without inhibition of swelling and degradation behaviors.
2.3. Development and functional assessment of a 3D printed anisotropic cardiac tissue
In case of the myocardial tissue, the arrangement of cardiomyocytes was found to be dense and uni-directionally aligned in a highly organized quasi-lamellar structure, as revealed by previous investigations (Figure 3A).[32] Therefore, in order to mimic this anisotropy in the native tissue arrangement, 3D microfibrous scaffolds were 3D printed by varying the fiber distances in successive layers. Briefly, the layers comprising of fibers printed in the x direction had a spacing of 600 μm while the layers comprising of fibers printed in the y direction has a spacing of 300 μm to ensure anisotropy. Furthermore, to mimic the quasi-lamellar structure, the G-code was optimized to include an offset for the microfibers printed in the x direction as illustrated in Figure 3B. The printed fibers were then analyzed for their inter-fiber spacing which was found to be 543 ± 9 μm and 250 ± 15 μm in the x and y directions respectively. In order to develop a cardiac tissue, neonatal rat cardiomyocytes (NRCMs) were seeded onto these 3D printed microfibrous constructs and were analyzed for their proliferation activity. The proliferation activity of the seeded NRCMs was found to be consistent with previous reports for both GelMA and n-SF matrices alone, where each of these matrices were able to support the growth and maturation of NRCMs. [18b, 33] The presence of intrinsic RGD sequences in both n-SF and GelMA and the presence of long stretches of poly-arginine cell binding motifs in n-SF facilitated excellent cell attachment and growth of the seeded cardiomyocytes as observed in Figure 3C. Further, the printed microfibers acted as cues for the seeded cardiomyocytes in order to form anisotropic tissue. Arrangement of the NRCMs seeded on the 3D printed scaffolds analyzed by F-actin staining revealed that the NRCMs attached to the microfibers in all the successive layers leading to the 3D tissue formation (Figure 3D–F). The cardiomyocytes homogeneously adhered onto the microfibers of the 3D printed microfibrous scaffolds only leaving space at the intersection points formed between the microfibers in adjacent layers.
Figure 3.
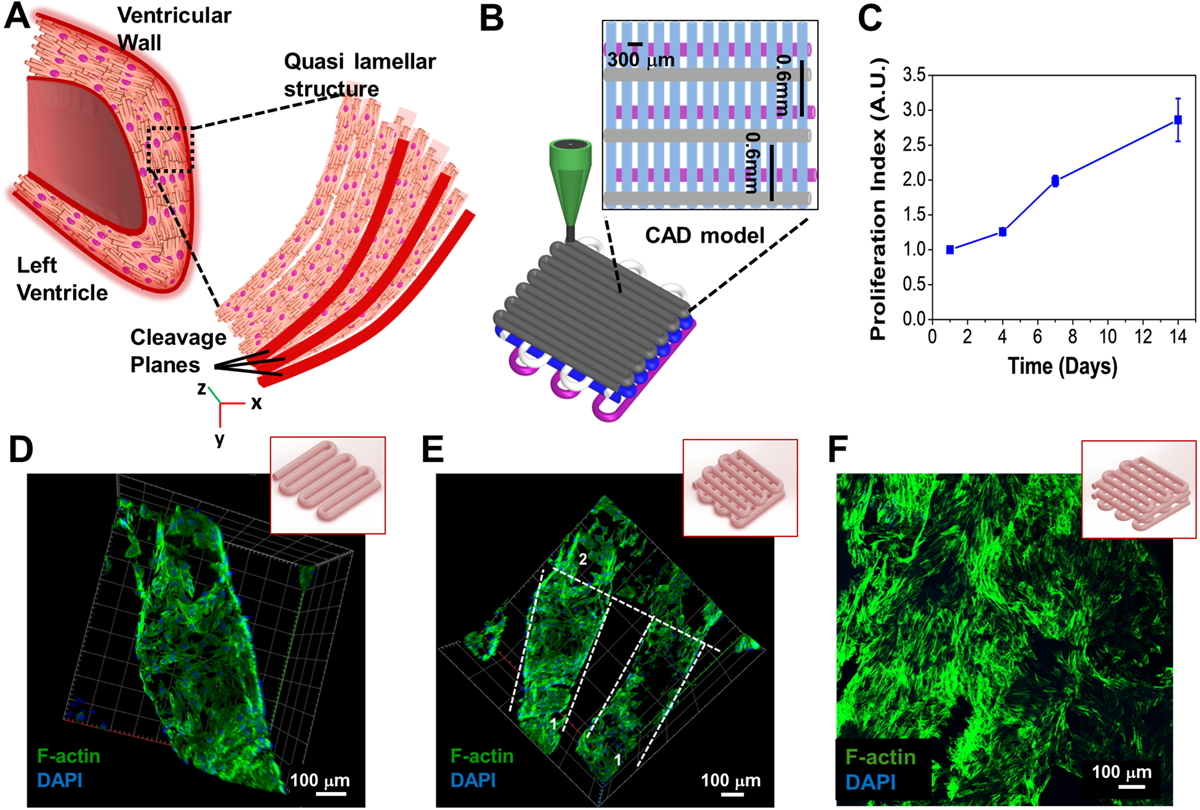
Designing of an anisotopic 3D printed cardiac like construct and its characterization. (A) Schematic of the native heart tissue with quasi-lamellar structure. (B) Schematic of the design of the 3D printed microfibrous scaffold to develop a quasi-lamellar 3D cardiac tissue construct similar to the native heart tissue. (C) Assessment of proliferation index of the seeded NRCMs on the printed 3D microfibrous scaffold using PrestoBlue assay (n=3). (D-F) Confocal fluorescence F-actin and DAPI staining exhibiting the cellular attachment and alignment along (D) a single printed microfiber, (E) 2 layers of the 3D printed construct (represented as 1 and 2), and (F) 4 layers of the 3D printed construct.
The seeded NRCMs on the 3D printed microfibrous constructs were further analyzed for functional performance. Immunostaining images from different regions of the 3D printed microfibrous construct demonstrated a high expression of proteins responsible for both contractile and conductive properties i.e. sarcomeric α-actinin and connexin 43 (Cx43) respectively (Figure 4A). The presence of aligned cardiomyocytes with highly organized sarcomeric bands on each 3D printed microfiber as well as the appearance of numerous conductive gap junctions (Cx43) suggested maturation of the growing NRCMs. High expression of these proteins exemplified the foundation for engendering synchronous beating of the 3D printed cardiac constructs. The NRCMs, when analyzed for beating frequencies overtime post being seeded onto the 3D printed microfibrous scaffolds, revealed ~70 ± 5 beats per minute by day 7 (Figure 4B and Video S2). The representative contraction plots of days 2, 4, and 7 for NRCMs seeded on the printed constructs, as shown in Figure 4C, illustrated robust and more uniform beating frequencies from day 4 to day 7. However, beating frequency was seen to decrease at day 10 (Figure 4B) due to the over proliferation of fibroblast present from the primary culture.[34] Maturation of NRCMs seeded onto the printed constructs was further analyzed by gene expression analysis of both cytoskeletal and gap junction genes (Figure 4D–F). High expression of Troponin T (Tnni3) and sarcomeric actinin (Actn1) observed on day 7 further confirmed cardiomyocyte maturation towards an adult phenotype. Similarly, high expression of Cx43 (a gap junction protein responsible for conducting electrical signals between cardiomyocytes) on day 7 confirmed the functional maturation of the NRCMs seeded on the 3D printed microfibrous constructs. After assessing the cardiac tissue maturation and organization on the SPG7.5 printed constructs using animal CMs, the printed scaffolds were then seeded with hiPSC-CMs and were analyzed for their functional performance. HiPSC-CMs have been widely studied for regenerative medicine applications including tissue engineering, because the unique nature of these cells lies in their capability, when cultured, for unlimited self–renewal and reproduction of all adult cell types.[35] Also, the use of autologous hiPSC-CMs minimizes the chances of adverse immune response after implantation of engineered cardiac tissue constructs.[36] The seeded hiPSC-CMs displayed a beating rate of 75–80 beats per minute on day 7 and were able to maintain beating for up to 28 days in culture, as shown in Figure 4G and Video S4. The beating behavior observed for the hiPSC-CMs was similar to that observed by other groups on seeding hiPSC-CMs on bioprinted scaffolds.[7a] Furthermore, the use of hiPSC-CMs as a better alternative to terminally differentiated primary cardiomyocytes (NRCMs) could be understood by the maintenance of beating behavior for long term culture.
Figure 4.
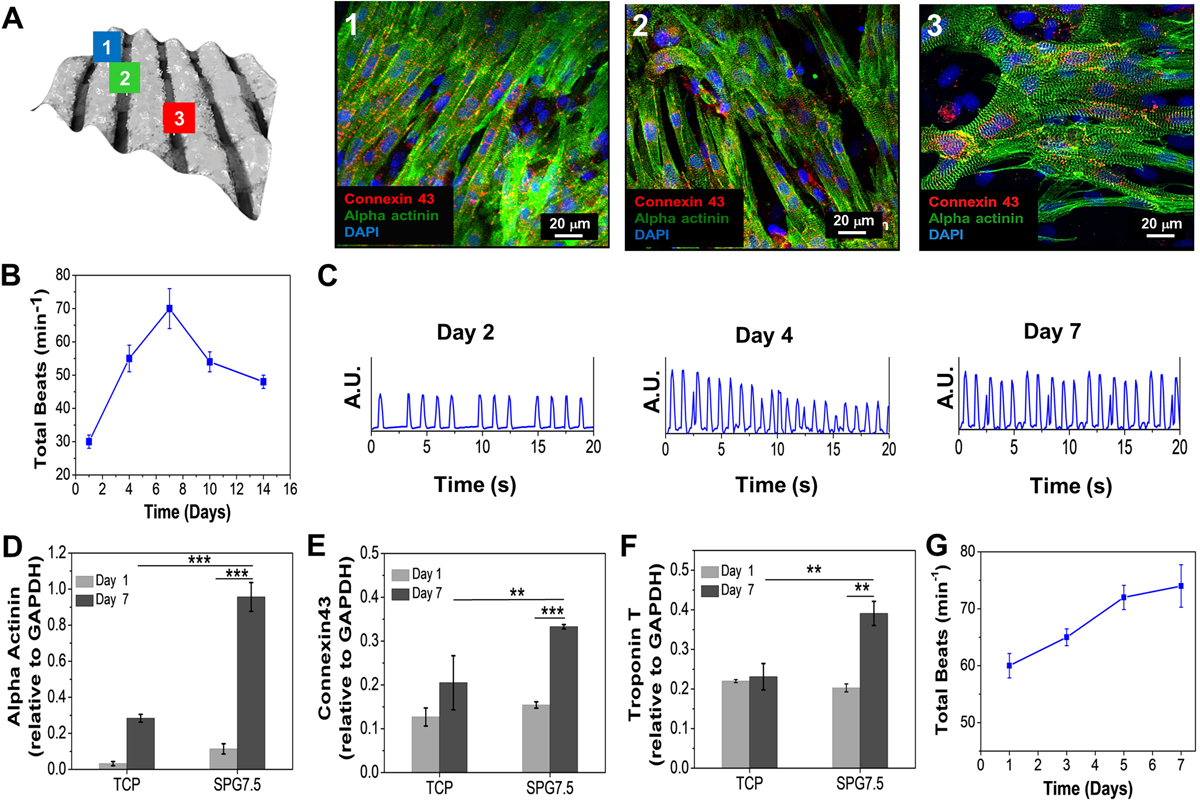
Functional assessment of cardiomyocytes seeded 3D printed constructs. (A) Computed bright field image of the 3D printed scaffold showing cell attachment on different microfibers; the vertical fibers as (1), horizontal fibers as (2) and the spaces in between them as (3). The seeded NRCMs on the three regions were analyzed for their orientation and maturation via immunostaining. Beating behavior of the seeded (B) NRCMs 3D printed micro-fibrous scaffold (n=5). (C) The beating signal of the seeded NRCMs on day 2, 4, and 7. (D-F) Gene expression analysis of cardiac specific biomarkers expressed by NRCMs seeded on 3D printed constructs (n=3; *** p ≤ 0.001 and **p ≤ 0.01). (G) Beating behavior of the seeded hiPSC-CMs on the 3D printed micro-fibrous scaffold (n=3).
2.4. Immunocompatibility assessment for successful implantability
In order to ensure successful implantation and tissue regeneration, the fabricated 3D printed constructs are required to be minimally immunogenic.[37] Macrophages are regarded as a prime source of pro-inflammatory chemokines, cytokines (interleukin-1β (IL-1β), tumor necrosis factor-α (TNF-α) and inflammation associated enzymes such as inducible nitric oxide synthase (iNOS), which are thereby known to regulate immune responses (inflammation).[38] An in vitro assessment for the release of pro-inflammatory biomarkers (IL-1β) and Nitric oxide (NO), synthesized by the action of released iNOS, by the macrophages in contact with the 3D printed constructs was done. The assays revealed a low immunogenic response to SPG7.5 which was comparable to tissue culture plates alone (TCP), used as negative control, as well as GelMA (G5) (Figure 5A–C). The in vitro immune response exhibited was much lower (p ≤ 0.001) than that exhibited in the presence of lipopolysaccharide (LPS; positive control) which is known to produce a high immune response by the secretion of such inflammatory molecules and cytokines (Figure 5A–C). Non-immunogenicity of both the biomaterial ink (SPG7.5; IB) and a completely crosslinked acellular 3D printed construct (IBC; cut into 0.5 cm (length) × 0.5 cm (width) × 0.5 cm (height)) was further analyzed by studying the in vivo response on days 7 and 14 via implantation in the subcutaneous pocket of Sprague Dawley (SD) rats (Figure 5D–F and Figure S5). CD68 protein is expressed in the cell membrane of all the macrophage phenotypes (M1 and M2 phenotypes). Assessment of the retrieved samples of 3D printed constructs (IBC) on day 7 and day 14 post implantation demonstrated infiltration of host tissue cells (mainly fibroblasts and few giant cells) into the constructs as revealed by hematoxylin and eosin (H&E) staining in Figure 5G. However, the absence of any macrophage infiltration from the host tissue was confirmed by immunostaining for the CD68 marker (Figure 5H). Furthermore, on CD68 immunostaining for IB, a moderate immune response was observed by the infiltration of a few giant cells (H&E) and macrophages (CD68 marker) near the implantation borders for the biomaterial ink retrieved on day 7 post implantation (Figure 5H). The presence of unperturbed fibroblast layers near the implanted materials was also observed. These results were coherent with the prior investigations which stated low immune response to pristine n-SF and GelMA hydrogels,[18b] thus showing the low immune response associated with the biomaterial ink. At this point, it is important to emphasize that the total amount of degraded construct could not be quantified post implantation. This is because the fabrication of the 3D printed constructs and the composition of the biomaterial ink itself includes GelMA, which is a derivative of collagen. Collagen is also found abundantly in host tissues. Hence, any kind of quantitative data for biochemical analysis (Hydroxyproline assay, total collagen content assay) of the retrieved samples in terms of degraded/remaining scaffold estimation would end in erroneous results.
Figure 5.
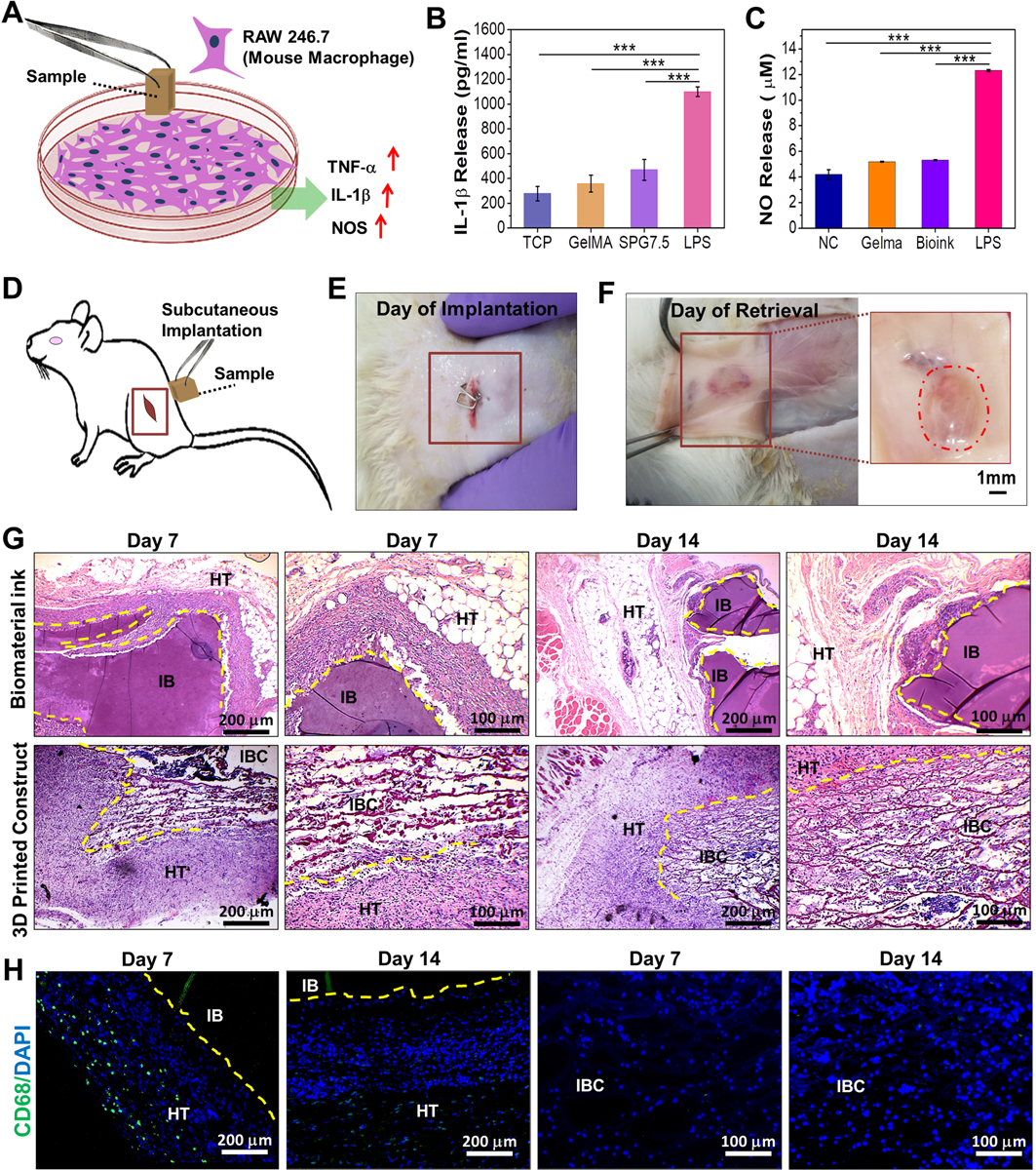
Immunocompatibility assessment of the biomaterial ink and 3D printed constructs. (A) Schematic depicting the release of pro-inflammatory molecules by mouse macrophage (RAW 246.7) when exposed to the 3D printed samples. Quantification of (B) IL-1β and (C) NO (nitric oxide) release by macrophages as determinants for in vitro immunocompatibility of the 3D printed SPG scaffolds compared to control samples (n=3; *** p ≤ 0.001) (D) Schematic depicting the implantation of samples in the left subcutaneous pocket of Sprague-Dawley rats. Images depicting the 3D printed scaffolds (E) after implantation and (F) on day 14 of retrieval (n=4). In vivo immunocompatibility assessment via (G) H&E staining of the implanted biomaterial ink and 3D printed scaffolds post retrieval at both low and high magnifications. Host cell infiltration was observed for IBC on subcutaneous implantation on day 7 and day 14. (H) CD68 immunostaining for determining the infiltration of macrophages post implantation of the bioprinted scaffolds on days 7 and 14 where HT stands for host tissue; IB stands for implanted biomaterial ink and IBC stands for implanted biomaterial ink based 3D printed construct.
2.5. Development and physical characterization of an endothelialized cardiac tissue construct
One of the major constraints of engineering a viable thick cardiac tissue (thickness: > 300 μm) is ensuring the exchange of nutrients and oxygen for strenuous cardiomyocytes in order to maintain their functionality. In the native tissue, this is taken care of by the presence of extensive microvascular networks. In order to mimic the native tissue organization, we have used an embedded gel bioprinting method for developing a vascularized construct. Briefly, a supporting bath comprising of an SPG hydrogel was used to encapsulate human induced pluripotent stem cell derived cardiac spheroids (hiPSC-CSs) (Diameter: 250–300 μm). Vascular channels were further embedded into the supporting hydrogel using a channel ink comprising of gelatin-GelMA biopolymers (GG) for encapsulating HUVECs (Figure 6A–B). The code was customized to create a spacing of 300 μm between channels in order to ensure efficient exchange of nutrients and oxygen to the encapsulated cardiomyocytes.
Figure 6.
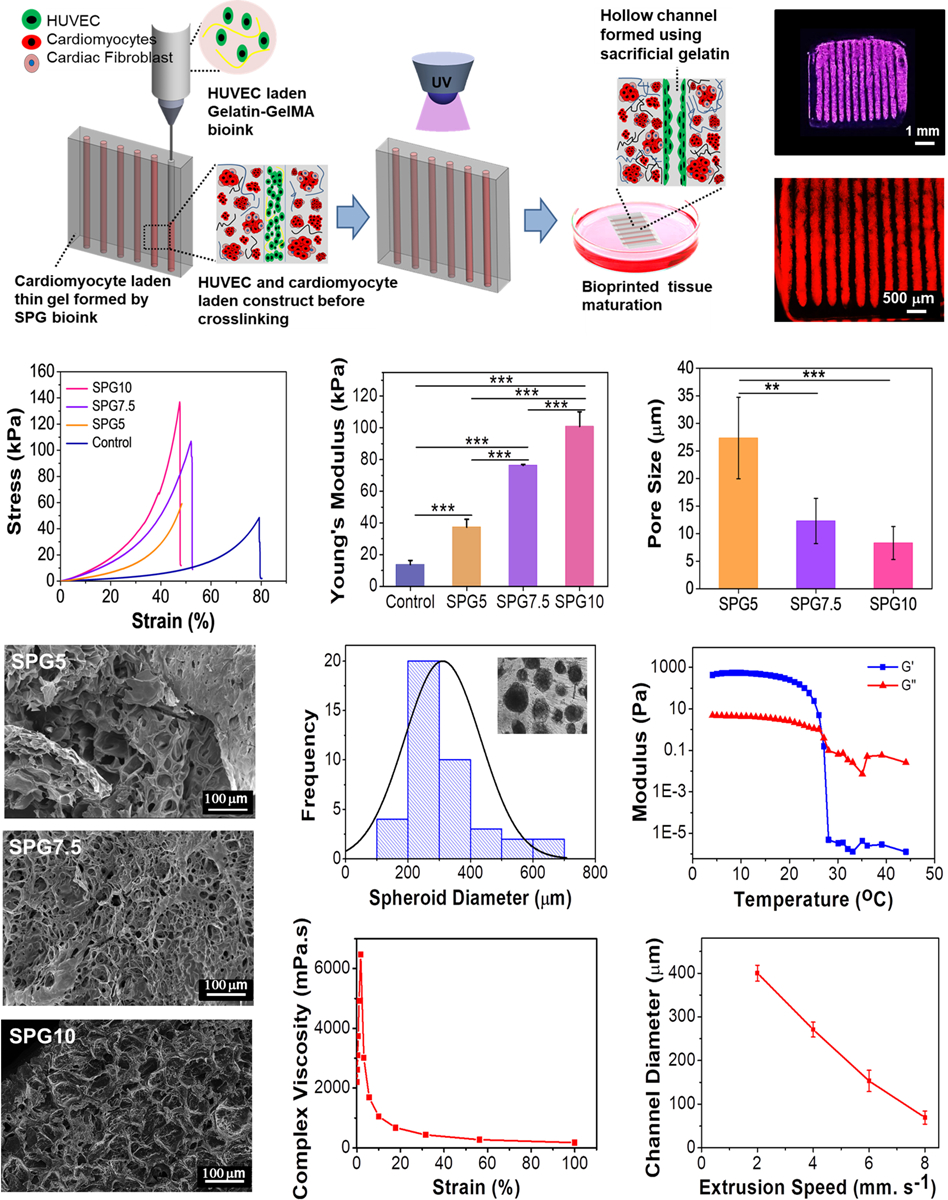
Embedded 3D bioprinting method for creating vascularized constructs and their physical characterization. (A) Schematic depicting 3D bioprinting of a vascularized cardiac tissue via embedded bioprinting method. The HUVEC-laden bioink was printed into a SPG bath (supporting) gel encapsulating hiPSC-CSs spheroids. (B) Representative image of the micro-channels printed into the bath (supporting) gel using the embedded bioprinting technique. (C) Strain-stress curves and (D) compressive modulus of different compositions of SPG5, SPG7.5 and SPG10 bath (supporting) gel. A supporting gel fabricated using 5% GelMA (G5) was used as control (n=4; *** p ≤ 0.001). (E) Pore size (n=3; *** p ≤ 0.001 and **p ≤ 0.01) and (F) SEM images of different compositions SPG5, SPG7.5 and SPG10 supporting gel post crosslinking. (G) Size distribution of hiPSC-CSs (Inset depicts the hiPSC-CSs encapsulated in the supporting bath gel). (H) Rheological characterization of the GG ink for determining optimal printing temperatures (n=3). (I) Complex viscosity of the GG ink at different strain rates (n=3). (J) Optimization of extrusion speed for printing small diameter micro-channels at a constant flow rate of 7mm s−1 (n=6).
Substrate stiffness has been known to alter the dynamics of cardiomyocytes.[15, 29b] Previous studies have shown that substrates encapsulating cardiomyocytes when mimicking the elasticity and strength of the developing myocardial environment were able to encourage actin-myosin cytoskeleton development and 1Hz beating in encapsulated cardiomyocytes.[25, 39] On the other hand, stiffer substrates have resulted in providing a fibrotic tough microenvironment similar to a scar tissue, thus attenuating cardiomyocyte beating behaviors and their cytoskeletal development.[25, 39b] In order to develop a supporting hydrogel for encapsulating the hiPSC-CSs, a local Young’s modulus in the range of 10 – 40 kPa similar to the native human cardiac tissue would be required.[40] In this respect, completely crosslinked hydrogels fabricated using three different compositions of biomaterial inks (by varying the amount of GelMA) were analyzed for their compressive strength in order to finalize the optimal concentration of the supporting hydrogel (Figure 6C–D). In particular, a Young’s modulus of 37.3 ± 4.9 kPa was obtained for ink comprising of 5% GelMA, 1% n-SF and 1% PEGDMA (SPG5) that was in the range of the local modulus of native ECM.[40] The ability to assure the viability of the encapsulated cells in polymer-based hydrogels requires them to be porous enough that an exchange of nutrients and oxygen can be facilitated. In this regard, supporting hydrogels fabricated using three different compositions of inks were analyzed for their porosity using SEM imaging (Figure 6E–F). The hydrogels with lower concentrations of GelMA were found to be more porous and could facilitate maintenance of viable cells post encapsulation. Hydrogels fabricated using SPG5, SPG7.5 and SPG10 inks displayed pore sizes of 31.1 ± 10.7μm, 18.2 ± 1.9μm and 7.4 ± 3.1μm respectively (p ≤ 0.001), suggesting a greater exchange of nutrients in the hydrogels fabricated using SPG5 biomaterial ink. Therefore, SPG5 biomaterial ink was chosen for encapsulating hiPSC-CSs in terms of its mechanical stiffness (20–40 kPa) similar to native cardiac ECM and high porosity.
For developing a vascular ink, sacrificial gelatin was used along with 1% w/v GelMA pre-polymer for encapsulating HUVECs. The channel ink (GG) exhibiting temperature dependent gelation was analyzed for its optimal printing temperatures (Figure 6H). Temperatures ranging from 25 – 30 °C were found to be optimal for the extrusion of the ink (GG). Moreover, the ink displayed a shear thinning behavior when subjected to increasing shear strain due to its viscous nature (Figure 6I). The optimal diameter of the vascular channel (~100 μm) embedded into the bath gel was obtained by setting the extrusion speed to 7 mm sec−1 (Figure 6J and Figure S4) and the ink flow rate to 4 μL min−1.
2.5. Generating an endothelialized myocardial tissue-on-a-chip as a potential drug screening platform for personalized medicine
The facile approach of developing vascularized myocardial constructs was primarily analyzed for cell viability post crosslinking. The bath (supporting) gel encapsulating the hiPSC-CS being viscous in nature facilitated the embedding of well-spaced vascular channels with ease. Post embedding, the construct was then crosslinked in the presence of UV light for 40 s for optimal cell viability. The crosslinked bioprinted vascularized construct displayed high cellular viability for both HUVECs and hiPSC-CS at day 1 and day 5 post printing (Figure 7A). For maturation of the fabricated vascularized myocardial construct, the bioprinted scaffolds were cultured for 14 days. The sacrificial nature of the gelatin comprising the channel bioink (leaches out slowly in culture at 37 °C) facilitated the proliferation and growth of HUVECs by allowing them to form a hollow tube like structure. F-actin staining of HUVEC channels at day 14 displayed the structure that HUVECs had coated on the hollow tube (Figure 7B). Furthermore, maturation of the construct comprising of both HUVECs and hiPSC-CSs was confirmed by immunostaining for CD31 and Troponin T (Figure 7C).
Figure 7.
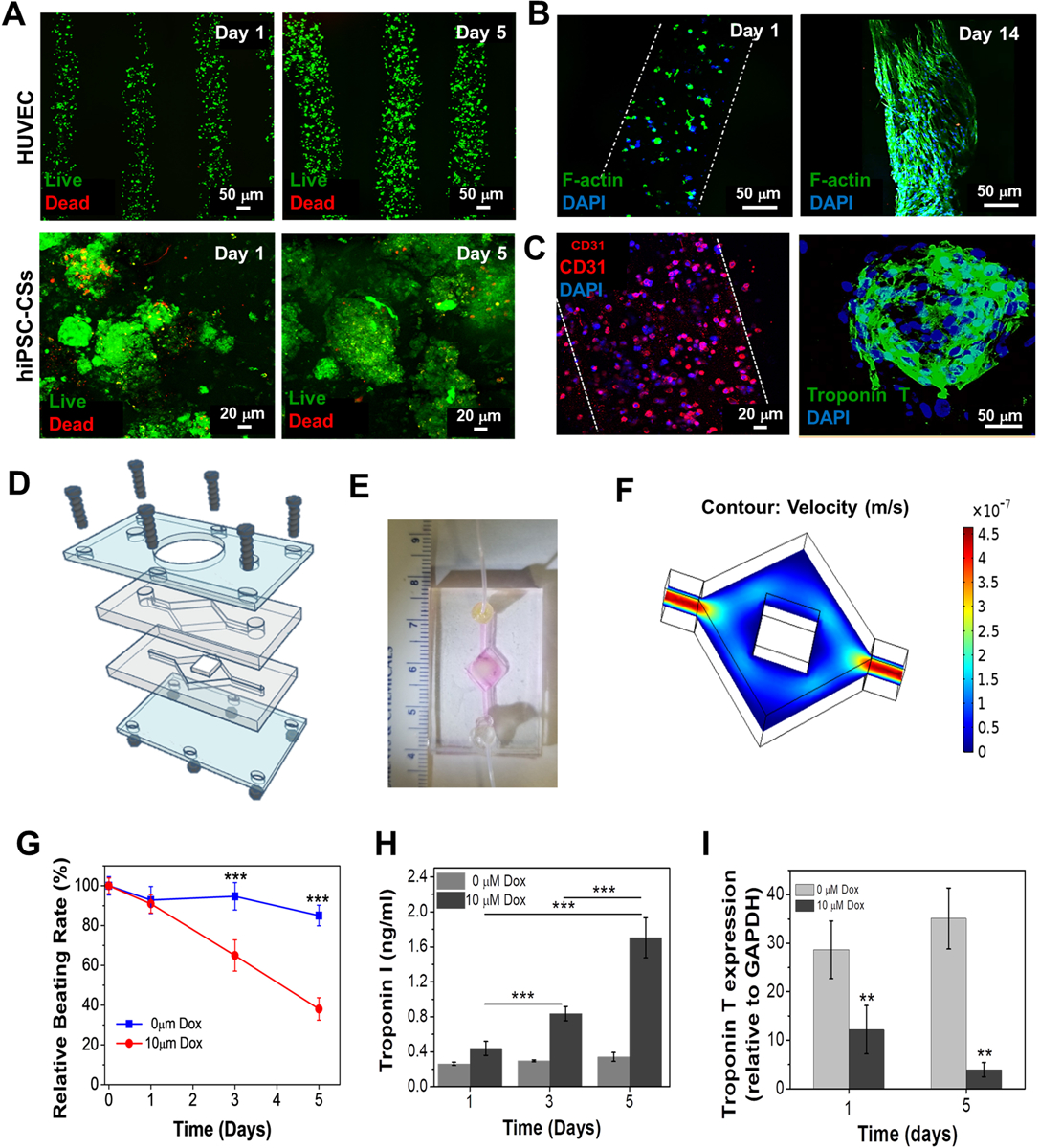
Biological characterization of a vascularized myocardium and its application as endothelialized myocardium-on-chip. (A) Live dead assay of bioprinted HUVECs and hiPSC-CSs in the bioprinted construct on days 1 and 5. (B) Confocal F-actin staining image showing the growth and distribution of the HUVECs in the bioprinted micro-channel on days 1 and 14. (C) Immunostaining for CD31 of bioprinted HUVECs in the micro-channel and for Troponin T of encapsulated hiPSC-CSs in the SPG gel bath. (D) Schematic of the microfluidic bioreactor construction. (E) Photograph of the bioreactor chamber harboring the bioprinted construct connected to inlet and outlet ports. (F) Simulation results for flow velocity in the bioreactor chamber. (G) Relative beating rate, (H) the levels of Troponin I biomarker expression (n=3; *** p ≤ 0.001) and (I) Troponin T gene expression by embedded hiPSC-CSs in the endothelialized myocardial tissue-on-a-chip to Dox (n=3; **p ≤ 0.01).
In order to sustain the long standing viability of the bioprinted vascularized myocardial constructs, a resealable bioreactor was fabricated for enabling perfusion-based culture as illustrated in Figure 7D. The design was similar to one of our previously published works.[29b] Briefly, the bioreactor was custom designed using a pair of micro featured PDMS gaskets which had hemi-chambers embedded in them. The hemi-chambers, upon being brought together, formed a small compartment in the center, in which the bioprinted vascularized myocardial constructs could be placed. The compartment was connected to inlet and outlet channels (1 mm) on either side (Figure 7E). In order to ensure hydraulic tightness, the PDMS hemi-chambers were packed in between two PMMA sheets for support on either side and were secured by being bolted together. The compartment for housing the bioprinted construct was a rhombic structure (0.7 cm2) and had a thickness of ~1mm post sealing. The edges of the compartment were slightly rounded in order to ensure the absence of any bubbles during the perfusion culture. One of the PMMA supports had a circular opening through which the beating of the encapsulated hiPSC-CS could be monitored. Furthermore, a computational model was developed using COMSOL Multiphysics (Finite Element Method) in order to simulate the flow velocity within the microfluidic bioreactor (Figure 7F). A flow rate of 70 μL min−1 was espoused, depending upon the size and thickness of the construct.
With several complications arising in drug development due to co-morbidities associated with a particular disease, personalized medicine has become the need of the hour. Several pharma based companies are now looking for in vitro alternatives which can greatly reduce the time and cost associated with animal testing and pre-clinical validation. In this respect, we have tried to create a physiological relevance to our 3D bioprinted vascularized myocardial construct cultured in a perfusion based system. We therefore expect that our facile approach of culturing a 3D bioprinted vascularized myocardial construct with a perfusion based method could help in studying the effects of several drugs on cardiac tissue. As a proof of concept, we exposed the 3D bioprinted vascularized myocardial construct to a pre-standardized dose of 10 μM doxorubicin (Dox).[41] Upon exposure to the Dox, the beating frequency/rate of the hiPSC-CS was reduced to 38% by day 5 (Figure 7G and Video S5–6). When quantified for cardiotoxicity biomarkers (Troponin I), an 8-fold increase of Troponin I (p ≤ 0.001) was revealed by ELISA on day 5 in comparison to non-treated controls. To further verify the effect of the drug on cardiomyocytes, gene expression analysis of myocardial cytoskeletal protein Troponin T was performed. Low expression of Troponin T on day 5 (p ≤ 0.01) of drug treatment could be due to the loss of cardiomyocytes (cardiotoxicity).
3. Conclusion
In summary, we have presented a novel silk-based biomaterial ink for engineering cardiac tissues. Partial enzymatic crosslinking of the ink resulted in a shear thinning nature, enabling in the fabrication of stable 3D constructs with ease. The biomaterial ink not only facilitated the fabrication of anisotropic cardiac constructs, which displayed an elastic behavior similar to the native heart tissue, but also promoted the functional attributes of the cardiomyocytes in terms of maturation, maintenance of cytoskeletal structure and beating potential. Furthermore, using an innovative gel embedding based bioprinting method, we have also tried to develop vascularized myocardial tissues using the silk-based ink. The silk-based ink both facilitated the encapsulation of beating hiPSC-CSs while maintaining their viability, and allowed for easy embedding of HUVEC channels due to its viscous nature. The endothelial cells, encapsulated inside the channel bioink, gradually migrated towards the margins of the channels, leading to the formation of endothelial based vasculature. The dual crosslinking step involved in the fabrication of such constructs enabled the fabrication of vascularized tissue without any cracks and abruptions. Upon maturation in culture, the vascularized myocardial tissue displayed maturation dependent expression of proteins for both cardiomyocytes and HUVECs. Essentially, in combination with the microfluidic perfusion-based bioreactor, the vascularized myocardial tissue-on-a-chip model could be utilized as a potential platform for screening several drugs. Also, a dose-dependent response of the cardiomyocytes to several drugs could be well evaluated using this proof-of-concept platform through further miniaturization and improvement via a throughput system in the future. It is now well understood that a combination of the appropriate inks and printing techniques is essential for developing viable next generation cardiac tissues in vitro. In this respect, we believe that our silk based ink will offer enormous potential towards fabricating such tissue while maintaining its functionality. More importantly, the ability of the bioprinted vascularized cardiac tissue fabricated using our ink to be used along with a microfluidic platform would offer numerous avenues for personalized drug screening towards cardiotoxicity. Future efforts using this ink will be directed towards engineering constructs which are able to display perfusable vascular networks serving as miniature hearts on a dish as well using such miniature models for elaborate on chip drug screening assays.
4. Experimental section
Materials:
Peroxidase from Horse Radish (HRP), 2-Hydroxy-4′-(2-hydroxyethoxy)-2-methylpropiophenone (Photoinitiator, Irgacure 2959), hydrogen peroxide (H2O2), 2-[4-(2-hydroxyethyl) piperazin-1-yl] ethane sulfonic acid (HEPES), gelatin (type A), methacrylic anhydride, lipopolysaccharide from Escherichia coli (LPS), CHIR99021, Wnt-C59, and paraformaldehyde ampules were purchased from Sigma-Aldrich (St. Louis, MO, USA). Polyethylene glycol dimethacrylate (PEGDMA, Mw = 1000 Da) was purchased from Polysciences Inc. (Warrington, PA, USA). Polydimethylsiloxane (PDMS) (Sylgard 184 Silicone Elastomer Kit) was purchased by Dow Corning (Midland, MI, EUA). Dulbecco’s phosphate-buffered saline (DPBS), fetal bovine serum (FBS), penicillin-streptomycin (P/S), dulbecco’s modified eagle medium (DMEM), LIVE/DEAD® viability/cytotoxicity Kit, prestoBlue™ kit, accutase, SYBR Green, mouse IL-1β kit and goat serum were purchased from Lifescience Technologies (Waltham, MA, USA). Endothelial growth medium (EBM-2 Basal Medium) and endothelial growth factor and supplements were obtained from Lonza (USA). Stem cell media was obtained from StemFit Basic 02 (Ajinomoto Co., Tokyo, Japan) and was supplemented with 100 ng/mL of recombinant human FGF-basic (Nacalai USA, San Diego, USA). Sodium dodecyl sulphate (SDS) was obtained from HiMedia (India). Alexa 488-phalloidin and Revita cell supplement was obtained from Thermo Fisher Scientific (Waltham, USA). A high capacity reverse transcription kit was purchased from Applied Biosystems (USA). All primary antibodies (mouse monoclonal anti-sarcomeric alpha actinin, rabbit polyclonal cardiac anti-Troponin T and rabbit polyclonal anti-connexin 43) and secondary antibodies (goat anti-mouse Alexa Flour 488, goat anti-rabbit Alexa Flour 594 and goat anti-rabbit Alexa Flour 488) were purchased from Abcam (USA).
Isolation of silk fibroin protein:
Silk fibroin protein was isolated from the glands of mature 5th instar larvae of the north-east Indian silk worm Antheraea assamensis by following a previously reported protocol. [18b, 42] The extracted gland fibroin protein was dissolved using a 1% w/v SDS solution, after which the silk fibroin solution was extensively dialyzed using a 12 kDa (MWCO) Sigma dialysis membrane against Milli-Q water at 4 ⁰C. The obtained regenerated silk solution was frozen and freeze dried for 48 h. The lyophilized protein was dissolved in deionized (DI) water in the desired concentrations.
Synthesis of GelMA:
GelMA was prepared by following a previously reported protocol. [7b] Briefly, a 10% w/v gelatin solution was prepared in DPBS and heated to 50°C for 1 h. Methacrylic anhydride (400 μL per g of gelatin) was added dropwise to this gelatin solution and was allowed to react for 2 h under constant stirring at 50°C. The reaction was stopped by further adding two times the volume of DPBS to the gelatin-methacrylate mixture. This solution was then extensively dialyzed using a 12 kDa (MWCO) Spectraphor dialysis membrane against deionized water for 5 days at 40°C, followed by freeze-drying. The freeze dried GelMA was dissolved in DPBS in the desired concentrations.
Cell culture:
Neonatal rat cardiomyocytes (NRCMs) were isolated from day old Wistar rat pups following our previously published protocol approved by the Institution of Animal Care and Use Committee at Brigham’s and Women’s Hospital through a collagenase based enzymatic digestion.[44] Briefly, hearts isolated from the neonatal rats were washed thoroughly using Hank’s balanced salt solution (HBSS) and incubated overnight with 0.05% trypsin at 4°C. This was followed by Collagenase Type II treatment at 37°C (~25–30 min) for heart tissue digestion. The isolated cardiomyocytes were enriched by pre-plating for 45–50 minutes. The isolated cardiomyocytes were cultured in DMEM, supplemented with 10% v/v FBS and 1% v/v penicillin-streptomycin at 37 ⁰C in a 95%O2/5%CO2 incubator. For printing vascular channels, human umbilical vein endothelial cells (HUVEC) were obtained from Lonza (U.S.A) and were cultured in EBM-2 supplemented with 1% v/v penicillin-streptomycin. The hiPSC cell line referred to as Cellartis® Human iPS Cell Line 12 (ChiPSC12) (Takara Bio, USA) was differentiated into cardiomyocytes (hiPSC-CMs) using a previously published protocol through the use of small molecules (CHIR99021 and Wnt-C59).[45] Human iPSC-derived cardiac spheroids (hiPSC-CSs) were shipped from the University of Hong Kong.
Biomaterial ink preparation and printability:
The biomaterial ink developed for 3D printing of cardiac constructs consisted of PEGDMA, GelMA, n-SF, 10U HRP, 0.09% H2O2 and 0.3% photoinitiator Irgacure 2959 dissolved in sterile DPBS supplemented with 10% v/v FBS. The optimized ink composition enabled a dual-step crosslinking procedure. At first, the ink was partially crosslinked at 37 °C via enzymatic crosslinking (~10 min) of SF by HRP-H2O2. Post printing, a stable and permanent gelation was attained by crosslinking GelMA and PEGDMA via exposure to UV light. The 3D printed construct was placed at a distance of 7 cm from a 800mW UV light source (Omnicure S2000, Excelitas Technologies, Salem, MA, USA)) and allowed to crosslink for 40 s. Continuous printing of microfibrous scaffolds was achieved by optimizing the composition of the ink by keeping the concentration of PEGDMA at 1% w/v while altering the relative concentrations of both GelMA and n-SF solutions.
Mechanical characterization:
Measurements for compressive stress under varied strain values were conducted to determine the Young’s modulus of the crosslinked hydrogels fabricated using different compositions of the biomaterial ink. The hydrogels were loaded onto an Instron 5944 having a 1N load cell. The compression was carried out at a 0.5 mm/min strain rate up to 50% deformation, at room temperature (RT; 25 ⁰C) in a DPBS bath. Young’s moduli were derived from the regression during the first 10% strain of the stress-strain curves. Each measurement was performed for four samples in each group. Single and cyclic tensile stress strain measurements were performed for the anisotropic 3D printed constructs (4 cm (length) × 1 cm (width) × 0.8 cm (height)). Elastic moduli were calculated from the tensile measurements carried out at a strain rate of 1 mm min−1 at room temperature. Cyclic tensile measurements were performed for 40 cycles at 2.5%, 5% and 10% strain for constructs fabricated using SPG7.5 and at 5% for those using G5 and G7.5 biomaterial inks.
Rheological measurements:
Rheological characterizations of the SPG7.5 ink were performed using a rheometer (MCR 302, Anton Paar, Austria) for characterizing (i) optimal printing temperatures using a temperature sweep analysis for a range of 5–45 °C at ω = 1 rad s−1, oscillatory strain; (ii) linear viscoelastic region (LVER) via amplitude sweep profiling performed at shear strain γ = 0.01 % to 10 % and a constant angular frequency ω = 1 rad s−1; (iii) the shear thinning nature of the ink through complex viscosity analysis via amplitude sweep performed at shear strain γ = 0.01 % to 100 % and a constant angular frequency ω = 1 rad s−1; (iv) the thixotropic nature of the ink using a three interval thixotropic test (at alternating low shear strain γ = 2 % and low angular frequency ω = 10 rad s−1, and high shear strain γ = 100 % and high angular frequency ω = 100 rad s−1). Rheological properties of the GG ink in terms of characterizing the shear thinning nature and complex viscosity were performed via amplitude sweep at a constant angular frequency ω = 5 rad s−1.
Printing of anisotropic cardiac tissue constructs:
For the printing of a construct, a commercial 3D bioprinter (Cellink Inkredible, Gothenburg, Sweden) was used in along with a customized extrusion system using 27G blunt syringe needles (OD: 410 mm; ID: 210 mm). The needles were connected to a syringe pump (New Era Pump Systems Inc., Suffolk County, NY) for the injection of the ink through PVC tubing (ColePalmer). All the junctions were sealed via epoxy glue. The customized extrusion system was attached to the commercial printer printhead using a custom-designed L-shaped holder made out of poly(methyl methacrylate) (PMMA) sheets. For the deposition of the ink into desired structures, a MATLAB was used to automatically generate the G-code for the bioprinter. Specifically, microfibrous scaffolds were printed through the deposition of one single continuous microfiber shaped in 3D for each scaffold.
Microscope imaging:
All confocal images were taken using a ZEISS LSM 880 with Airyscan Microscope (Carl Zeiss, Jena, Germany) and all bright field and fluorescent images were taken using a Nikon Eclipse Ti-S Microscope (Nikon, Tokyo, Japan). Scanning electron microscopy (SEM) images were taken using a LEO Electron Microscopy/Oxford (Cambridge, England).
Analysis of gene expression:
RNA extraction from each sample (n=3) was carried out using the Trizol method on days 1 and 7. The extracted RNA was quantified and converted into cDNA in a PCR thermal cycler (Applied Biosystems) using a high capacity reverse transcription kit pursuant to the provided the manufacturer’s protocol. The reaction volume was set to 20 μL and real time PCR (q-PCR) was performed using SYBR Green dye. The experiment was set to quantify the expression of cardiac specific genes such as connexin 43 (Gja1), sarcomeric alpha actinin (Actn1) and cardiac troponin T (Tnni3) through a comparative 2−ΔCt method where the expression of the above cardiac specific genes was normalized to the house keeping gene, glyceraldehyde −3-phosphate-dehydrogenase (GAPDH). For analyzing the drug treatment on vascularized myocardial-on-a-chip, gene expression analysis of human Cardiac Troponin T was also performed using a comparative 2−ΔCt method. Primer sequences for GAPDH, Gja1, Actn1, Tnni3 and human cardiac Troponin T are listed in Table S1 (Supporting information).
Live dead assay:
Cellular viability was assessed using calcein-AM-ethidium homodimer staining following manufacturer’s protocol. The cell laden constructs were washed once with 1X DPBS and incubated with 40 nM calcein-AM (green) and 20 nM ethidium homodimer (red), reconstituted in DPBS, for 30 min at 25 ⁰C. The dye was removed and the constructs were washed twice with 1X DPBS, subsequently visualized under hydrated conditions using a confocal microscope. Viable cells appeared green while dead cells appeared red.
Characterization of cellular proliferation and tissue formation:
Cell proliferation was analyzed using a PrestoBlue reagent following the manufacturer’s protocol. Briefly, a 1:10 ratio of PrestoBlue and media was added to the constructs for 3 h for analyzing cell proliferation. The absorbance was then taken at 570/600nm and used for calculation of reduction in PrestoBlue. The proliferation index was calculated by normalizing the PrestoBlue reduction values to day 1. Tissue formation was analyzed using F-actin staining. For all staining purposes, cells were fixed with 4% v/v paraformaldehyde in DPBS for 10 min and washed thrice with DPBS. For staining F-actin and the nucleus, Alexa Fluor 488 phalloidin and DAPI (1:500 dilution) were used. F-actin staining was performed by incubating the samples for 30 min at RT in a solution of Alexa 488-phalloidin at a 1:40 dilution ratio in 10% v/v goat serum and 0.1% v/v Triton X-100 in DPBS.
Analysis of beating rate:
Total beats per min for the NRCMs and hiPSC-CMs/-CS were derived from the videos. The video was recorded for both types of cardiomyocytes at different time points and was analyzed based on imaging using a customized MATLAB. The videos were analyzed for calculating beat frequency and amplitude.
Immunostaining:
NRCMs seeded on bioprinted constructs and cultured until maximum beating rate were obtained (day 7). This ensured that sarcomeres in the beating NRCMs were mature and fully developed. After gently washing with DPBS (pH 7.4), the samples were fixed using 4% v/v paraformaldehyde for 20 min at RT. The fixed cells were permeabilized with DPBS (pH 7.4) consisting of 0.1% v/v Triton X-100 for 20 min, washed thoroughly and then blocked using a blocking buffer (1% BSA, 0.2% Tween in DPBS) for 1 h. The samples were incubated at 4 °C overnight with primary antibodies such as rabbit polyclonal anti-connexin 43 and mouse monoclonal anti-sarcomeric α−actinin (1:100, Abcam). Subsequently, the samples were washed thrice with DPBS in 10 min intervals. For determining the maturation of vascularized myocardial tissue, the constructs were permeabilized and kept for blocking as described above. Furthermore, they were incubated at 4 °C overnight with primary antibodies such as rabbit polyclonal anti-troponin T (1:100, Abcam) and rabbit polyclonal CD31 (1:100, Abcam). Furthermore, the samples were treated by the corresponding secondary antibodies for 3 h at 1:200 dilutions. Nuclei were stained by incubating with 4’,6-diamidino-2-phenylindole (DAPI; 1:500) for 1.5 h. The stained samples were then visualized using a confocal microscope.
In vitro immunocompatibility assessment by determination of IL-1β and NOS release:
Murine macrophage cell line RAW 264.7 was grown in high glucose DMEM containing 10% FBS and 1.0% penicillin-streptomycin solution at 37 °C and 5% CO2. Cells were plated at a density of 2 × 104 cells per well in 24-well plates and incubated overnight at 37 °C. On the following day, conditioned hydrogels formed using the optimized biomaterial ink as well as n-SF and GelMA alone were transferred to the well plates pre-seeded with RAW 264.7. After 24 h of incubation at 37 °C, the media was collected from the wells and assayed for IL-1β and NO release. Media from the wells without any hydrogel samples, was used as a negative control, while LPS (500 ng/mL) was considered a positive control. The level of IL-1β released was measured by ELISA, following the manufacturer’s protocol. Similarly, estimation of NO release was measured by Griess reagent kit as indicated by the manufacturer’s protocol.
In vivo immunocompatibility assessment:
All in vivo implantation studies performed were approved by the Institutional Ethical Committee (IAEC) and the National Institute of Pharmaceutical Education and Research (NIPER-Guwahati). Sprague Dawley rats (body weight 200–300 g) were used to carry out the response studies of the biomaterial inks and the 3D printed constructs. UV sterilized ink and 3D printed constructs were subcutaneously implanted through a 0.7 cm incision on either lateral side of the thoraco-lumbar region of the rats and were secured in place using a surgical stapler. The site of incision was regularly inspected for the development of any infection as well as for monitoring the healing process. During the entire duration of the experiment, no loss of animals due to death was recorded. After 1 and 2 weeks of implantation, rats were sacrificed by cervical dislocation and the implantation regions were harvested and examined histologically using hematoxylin and eosin staining. The sections were also examined for macrophage infiltration to the implantation sites via CD68 immunostaining.
Bioprinting of endothelialized myocardial tissue constructs:
Endothelialized myocardium formation was carried out using embedded gel printing where vascular hollow channels were embedded into the un-crosslinked supporting bath gels (SPG5; held in a customized mold) using a gelatin based sacrificial ink (GG). A customized G-code was written to generate a 1 mm thick construct with hollow channels embedded with a gap of 300 μm. Briefly, Human iPSC derived cardiomyocytes were encapsulated in the optimized un-crosslinked SPG5 ink bathink. The supporting SPG5 ink encapsulating the cells was then subjected to the embedding of HUVEC encapsulated GG ink to develop vascular channels. Post printing, a stable gelation was attained by crosslinking GelMA and PEGDMA via UV exposure (40 s) and followed by enzymatic crosslinking of n-SF at 37 ⁰C. The constructs were then cultured in media containing 50% cardiomyocyte maintenance media and 50% endothelial media.
Computational Simulation of the Bioreactor:
Comsol Multiphysics (Version 5.2) was used to simulate the flow rates and velocity of media into the bioreactor using finite element method and laminar flow calculations. The simulations were based on uniform oxygen and nutrient supply to the fabricated endothelialized myocardial construct.
Generation of endothelialized-myocardium-on-a-chip model:
The bioprinted cell laden endothelialized myocardial construct was matured in a customized perfusion based bioreactor. The matured construct was then analyzed for the development of an organ-on a chip model by evaluating the effect of drug-doxorubicin (DOX). 10 μm of DOX was chosen as optimal from our previously reported studies to induce drug toxicity. The constructs without any exposure to DOX (0 μm) were used as suitable controls. This organ-on-a-chip model was further analyzed for its beating behavior in the presence (10 μm) and absence (0 μm) of DOX. Release of cardiomyocyte specific marker cardiac Troponin I on the action of DOX was analyzed using an ELISA kit, using the manufacturer’s protocol. Furthermore, gene expression studies using real time PCR were also performed for quantifying Troponin T gene to study the action of DOX.
Statistical analysis:
All data are expressed as mean ± standard deviation (SD). Results were statistically analyzed by one-way analysis of variance (ANOVA) via Turkey’s test and Holm-Sidak test to evaluate the level of significance among groups. For the experiments, * p ≤ 0.05 and ** p ≤ 0.01 were considered as significant whereas *** p ≤ 0.001 was considered as highly significant.
Supplementary Material
Figure S2. Analysis of the elastic nature of the 3D printed constructs post printing and conditioning in DPBS.
Figure S3. Understanding the behavior of 3D printed constructs using (A) SPG7.5 and (B) G5 (control) inks under cyclic tensile strain of 5% for 40 cycles (n=3).
Figure S1. (A) Dual crosslinking of the biomaterial ink confirmed by increase in turbidity. (B) Optimization of extrusion speed for SPG7.5 ink for fabrication of 3D printed cardiac construct (n=10).
Figure S4. Optimization of flowrate for the GG ink for printing vascular channels. The vascular channels were printed using the GG ink encapsulating fluorescent microparticles at flow rates of (A) 4 mm.s−1, (B) 6 mm.s−1, (C) 7 mm.s−1, and (D) 8mm.s−1 (n=10).
Figure S5. Assessment of host cell infiltration. Zoomed histological image of the implanted IBC in rat subcutaneous models where IBC stands for implanted 3D printed constructs, blue represents DAPI stain and green represents CD68, a macrophage marker (n=4).
Video S1: Printing of anisotropic scaffold using SPG7.5 ink
Video S2: Beating of NRCMs on 3D printed scaffold
Video S4: Beating of hiPSC-CMs on 3D printed scaffold
Video S5: HiPSC-CS encapsulated in silk based supporting hydrogel without DOX
Video S6: HiPSC-CS encapsulated in silk based supporting hydrogel with DOX
Video S3: Differentiation of human iPSC to beating cardiomyocytes
Acknowledgment
The authors declare no conflict of interests in this work. This paper was funded by the National Institutes of Health (R01AR074234, R21EB026824, and R01 AR073822-01), the Brigham Research Institute Stepping Strong Innovator Award, and AHA Innovative Project Award (19IPLOI34660079). B.B.M acknowledges the funding from Department of Biotechnology (DBT) and the Department of Science and Technology (DST), Govt. of India. S.M. acknowledges Fulbright Nehru Doctoral Research Program, IIE, USIEF and MHRD, India for her fellowship. S.M. and B.B.M. acknowledge the support from National Institute of Pharmaceutical Education and Research, Guwahati for the animal experiments. S.M. and B.B.M. also acknowledge (Centre of Instrument Facility) CIF, IIT-Guwahati for their facilities and support. S.M. and S.R.S acknowledge BWH-Neuroscience Department and BWH-HMS for their facilities.
Footnotes
Supporting Information
Supporting Information is available from the Wiley Online Library or the author.
Contributor Information
Shreya Mehrotra, Division of Engineering in Medicine, Department of Medicine, Harvard Medical School, Brigham and Women’s Hospital, Cambridge, MA 02139, USA.; Department of Biosciences and Bioengineering, Indian Institute of Technology Guwahati, Guwahati-781039, Assam, India.
Bruna A. G. de Melo, Division of Engineering in Medicine, Department of Medicine, Harvard Medical School, Brigham and Women’s Hospital, Cambridge, MA 02139, USA. Department of Engineering of Materials and Bioprocesses, School of Chemical Engineering, University of Campinas, Campinas, SP 13083-852, Brazil.
Minoru Hirano, Division of Engineering in Medicine, Department of Medicine, Harvard Medical School, Brigham and Women’s Hospital, Cambridge, MA 02139, USA.; Future Vehicle Research Department, Toyota Research Institute North America, Toyota Motor North America Inc., 1555 Woodridge Ave Ann Arbor, MI 48105, USA
Wendy Keung, Dr. Li Dak-Sum Research Centre, The University of Hong Kong, Hong Kong.; Ming Wai Lau Centre for Reparative Medicine, Karolinska Institutet, Hong Kong.
Ronald A. Li, Dr. Li Dak-Sum Research Centre, The University of Hong Kong, Hong Kong. Ming Wai Lau Centre for Reparative Medicine, Karolinska Institutet, Hong Kong.
Biman B. Mandal, Department of Biosciences and Bioengineering, Indian Institute of Technology Guwahati, Guwahati-781039, Assam, India.; Center for Nanotechnology, Indian Institute of Technology Guwahati, Guwahati-781039, Assam, India.
Su Ryon Shin, Division of Engineering in Medicine, Department of Medicine, Harvard Medical School, Brigham and Women’s Hospital, Cambridge, MA 02139, USA..
References
- [1].a) Vunjak-Novakovic G, Tandon N, Godier A, Maidhof R, Marsano A, Martens TP, Radisic M, Tissue Eng Part B Rev 2010, 16, 169; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Cheung DYC, Duan B, Butcher JT, in Essentials of 3D Biofabrication and Translation, DOI: 10.1016/B978-0-12-800972-7.00021-9 (Eds: Atala A, Yoo JJ), Academic Press, Boston: 2015, p. 351; [DOI] [Google Scholar]; c) Vukicevic M, Mosadegh B, Min JK, Little SH, JACC: Cardiovascular Imaging 2017, 10, 171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Mehrotra S, Moses JC, Bandyopadhyay A, Mandal BB, ACS Applied Bio Materials 2019, 2, 1385. [DOI] [PubMed] [Google Scholar]
- [3].a) Murphy SV, Atala A, Nature Biotechnology 2014, 32, 773; [DOI] [PubMed] [Google Scholar]; b) Derakhshanfar S, Mbeleck R, Xu K, Zhang X, Zhong W, Xing M, Bioactive Materials 2018, 3, 144; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Bishop ES, Mostafa S, Pakvasa M, Luu HH, Lee MJ, Wolf JM, Ameer GA, He T-C, Reid RR, Genes & Diseases 2017, 4, 185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].a) Ong CS, Nam L, Ong K, Krishnan A, Huang CY, Fukunishi T, Hibino N, BioMed research international 2018, 2018; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Zhang YS, Davoudi F, Walch P, Manbachi A, Luo X, Dell’Erba V, Miri AK, Albadawi H, Arneri A, Li X, Lab on a Chip 2016, 16, 4097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Gaetani R, Feyen DA, Verhage V, Slaats R, Messina E, Christman KL, Giacomello A, Doevendans PA, Sluijter JP, Biomaterials 2015, 61, 339. [DOI] [PubMed] [Google Scholar]
- [6].Jakab K, Norotte C, Damon B, Marga F, Neagu A, Besch-Williford CL, Kachurin A, Church KH, Park H, Mironov V, Tissue Engineering Part A 2008, 14, 413. [DOI] [PubMed] [Google Scholar]
- [7].a) Zhang YS, Arneri A, Bersini S, Shin S-R, Zhu K, Goli-Malekabadi Z, Aleman J, Colosi C, Busignani F, Dell’Erba V, Bishop C, Shupe T, Demarchi D, Moretti M, Rasponi M, Dokmeci MR, Atala A, Khademhosseini A, Biomaterials 2016, 110, 45; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Zhu K, Shin SR, van Kempen T, Li Y-C, Ponraj V, Nasajpour A, Mandla S, Hu N, Liu X, Leijten J, Lin Y-D, Hussain MA, Zhang YS, Tamayol A, Khademhosseini A, Advanced functional materials 2017, 27, 1605352; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Hinton TJ, Jallerat Q, Palchesko RN, Park JH, Grodzicki MS, Shue H-J, Ramadan MH, Hudson AR, Feinberg AW, Science Advances 2015, 1, e1500758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Koti P, Muselimyan N, Mirdamadi E, Asfour H, Sarvazyan NA, Journal of 3D Printing in Medicine 2019, 3, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Wang Z, Lee SJ, Cheng H-J, Yoo JJ, Atala A, Acta Biomaterialia 2018, 70, 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].a) Ikonen L, Kerkelä E, Kujala K, Haaparanta A, Ahola N, Ellä V, Poh T, Kellomäki M, Aalto-Setäläet K, J Clinic Experiment Cardiol S 2011, 4, 2; [Google Scholar]; b) Tang X, Thankappan SK, Lee P, Fard SE, Harmon MD, Tran K, Yu X, in Natural and Synthetic Biomedical Polymers, DOI: 10.1016/B978-0-12-396983-5.00022-3 (Eds: Kumbar SG, Laurencin CT, Deng M), Elsevier, Oxford: 2014, p. 351. [DOI] [Google Scholar]
- [11].a) Jang J, Bioengineering (Basel) 2017, 4, 71; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Rijal G, Li W, J Biol Eng 2018, 12, 20; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Hasan A, Waters R, Roula B, Dana R, Yara S, Alexandre T, Paul A, Macromol Biosci 2016, 16, 958; [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Arya AD, Hallur PM, Karkisaval AG, Gudipati A, Rajendiran S, Dhavale V, Ramachandran B, Jayaprakash A, Gundiah N, Chaubey A, ACS Applied Materials & Interfaces 2016, 8, 22005; [DOI] [PubMed] [Google Scholar]; e) Huyer LD, Montgomery M, Zhao Y, Xiao Y, Conant G, Korolj A, Radisic M, Biomed Mater 2015, 10, 034004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Reis LA, Chiu LLY, Feric N, Fu L, Radisic M, J Tissue Eng Regen Med 2016, 10, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].a) Chen Q-Z, Harding SE, Ali NN, Lyon AR, Boccaccini AR, Materials Science and Engineering: R: Reports 2008, 59, 1; [Google Scholar]; b) Rodrigues ICP, Kaasi A, Maciel Filho R, Jardini AL, Gabriel LP, Einstein (Sao Paulo) 2018, 16, eRB4538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Wang Y, Ma M, Wang J, Zhang W, Lu W, Gao Y, Zhang B, Guo Y, Materials 2018, 11, 1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Wang Q, Han G, Yan S, Zhang Q, Materials 2019, 12, 504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].a) Abbott RD, Kaplan DL, Current Stem Cell Reports 2016, 2, 140; [Google Scholar]; b) Sung H-J, Meredith C, Johnson C, Galis ZS, Biomaterials 2004, 25, 5735. [DOI] [PubMed] [Google Scholar]
- [17].Zhou DW, Lee TT, Weng S, Fu J, García AJ, Molecular Biology of the Cell 2017, 28, 1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].a) Chawla S, Midha S, Sharma A, Ghosh S, Advanced Healthcare Materials 2018, 7, 1701204; [DOI] [PubMed] [Google Scholar]; b) Mehrotra S, Nandi SK, Mandal BB, Journal of Materials Chemistry B 2017, 5, 6325; [DOI] [PubMed] [Google Scholar]; c) Chouhan D, Thatikonda N, Nilebäck L, Widhe M, Hedhammar M, Mandal BB, ACS Applied Materials & Interfaces 2018, 10, 23560; [DOI] [PubMed] [Google Scholar]; d) Patra C, Talukdar S, Novoyatleva T, Velagala SR, Mühlfeld C, Kundu B, Kundu SC, Engel FB, Biomaterials 2012, 33, 2673. [DOI] [PubMed] [Google Scholar]
- [19].a) Zheng Z, Wu J, Liu M, Wang H, Li C, Rodriguez MJ, Li G, Wang X, Kaplan DL, Advanced Healthcare Materials 2018, 7, 1701026; [DOI] [PubMed] [Google Scholar]; b) DeSimone E, Schacht K, Pellert A, Scheibel T, Biofabrication 2017, 9, 044104; [DOI] [PubMed] [Google Scholar]; c) Chawla S, Kumar A, Admane P, Bandyopadhyay A, Ghosh S, Bioprinting 2017, 7, 1; [Google Scholar]; d) Kim SH, Yeon YK, Lee JM, Chao JR, Lee YJ, Seo YB, Sultan MT, Lee OJ, Lee JS, Yoon S.-i., Hong I-S, Khang G, Lee SJ, Yoo JJ, Park CH, Nature Communications 2018, 9, 1620; [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Singh YP, Bandyopadhyay A, Mandal BB, ACS Applied Materials & Interfaces 2019, 11, 33684. [DOI] [PubMed] [Google Scholar]
- [20].Farokhi M, Mottaghitalab F, Fatahi Y, Khademhosseini A, Kaplan DL, Trends in biotechnology 2018, 36, 907. [DOI] [PubMed] [Google Scholar]
- [21].a) Mehrotra S, Chouhan D, Konwarh R, Kumar M, Jadi PK, Mandal BB, ACS Biomaterials Science & Engineering 2019, 5, 2054; [DOI] [PubMed] [Google Scholar]; b) Guo C, Zhang J, Jordan JS, Wang X, Henning RW, Yarger JL, Biomacromolecules 2018, 19, 906. [DOI] [PubMed] [Google Scholar]
- [22].Qi Y, Wang H, Wei K, Yang Y, Zheng R-Y, Kim IS, Zhang K-Q, International journal of molecular sciences 2017, 18, 237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Partlow BP, Hanna CW, Rnjak-Kovacina J, Moreau JE, Applegate MB, Burke KA, Marelli B, Mitropoulos AN, Omenetto FG, Kaplan DL, Advanced functional materials 2014, 24, 4615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Yeung T, Georges PC, Flanagan LA, Marg B, Ortiz M, Funaki M, Zahir N, Ming W, Weaver V, Janmey PA, Cell Motility 2005, 60, 24. [DOI] [PubMed] [Google Scholar]
- [25].Engler AJ, Carag-Krieger C, Johnson CP, Raab M, Tang H-Y, Speicher DW, Sanger JW, Sanger JM, Discher DE, Journal of Cell Science 2008, 121, 3794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Schrader J, Gordon-Walker TT, Aucott RL, van Deemter M, Quaas A, Walsh S, Benten D, Forbes SJ, Wells RG, Iredale JP, Hepatology 2011, 53, 1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].a) Bhana B, Iyer RK, Chen WLK, Zhao R, Sider KL, Likhitpanichkul M, Simmons CA, Radisic M, Biotechnology and bioengineering 2010, 105, 1148; [DOI] [PubMed] [Google Scholar]; b) Tallawi M, Rai R, Boccaccini AR, Aifantis KE, Tissue Eng Part B Rev 2015, 21, 157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Ribeiro AJS, Ang Y-S, Fu J-D, Rivas RN, Mohamed TMA, Higgs GC, Srivastava D, Pruitt BL, Proc Natl Acad Sci U S A 2015, 112, 12705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].a) Jacot JG, McCulloch AD, Omens JH, Biophysical Journal 2008, 95, 3479; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Boothe SD, Myers JD, Pok S, Sun J, Xi Y, Nieto RM, Cheng J, Jacot JG, Cell Biochem Biophys 2016, 74, 527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Krasnov I, Diddens I, Hauptmann N, Helms G, Ogurreck M, Seydel T, Funari SS, Müller M, Physical review letters 2008, 100, 048104. [DOI] [PubMed] [Google Scholar]
- [31].a) Kong M, Lee J, Yazdi IK, Miri AK, Lin Y-D, Seo J, Zhang YS, Khademhosseini A, Shin SR, Advanced Healthcare Materials 2019, 8, 1801146; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Ugolini GS, Pavesi A, Rasponi M, Fiore GB, Kamm R, Soncini M, Elife 2017, 6, e22847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].a) Zhang YS, Arneri A, Bersini S, Shin S-R, Zhu K, Goli-Malekabadi Z, Aleman J, Colosi C, Busignani F, Dell’Erba V, Biomaterials 2016, 110, 45; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Filipoiu FM, in Atlas of Heart Anatomy and Development, Springer; 2014, p. 151. [Google Scholar]
- [33].a) Chouhan D, Mehrotra S, Majumder O, Mandal BB, ACS Biomaterials Science & Engineering 2019, 5, 92; [DOI] [PubMed] [Google Scholar]; (b) Shin SR, Jung SM, Zalabany M, Kim K, Zorlutuna P, Kim SB, Nikkhah M, Khabiry M, Azize M, Kong J, ACS nano 2013, 7, 2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].a) Rother J, Richter C, Turco L, Knoch F, Mey I, Luther S, Janshoff A, Bodenschatz E, Tarantola M, Open Biology 5, 150038; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Vasquez C, Mohandas P, Louie KL, Benamer N, Bapat AC, Morley GE, Circulation Research 2010, 107, 1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Dakhore S, Nayer B, Hasegawa K, Stem Cells Int 2018, 2018, 7396905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].a) Shiba Y, Hauch KD, Laflamme MA, Curr Pharm Des 2009, 15, 2791; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Martins AM, Vunjak-Novakovic G, Reis RL, Stem Cell Rev Rep 2014, 10, 177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].a) Morais JM, Papadimitrakopoulos F, Burgess DJ, AAPS J 2010, 12, 188; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Mariani E, Lisignoli G, Borzì RM, Pulsatelli L, International journal of molecular sciences 2019, 20, 636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].a) Arango Duque G, Descoteaux A, Front Immunol 2014, 5, 491; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Kobayashi Y, Journal of leukocyte biology 2010, 88, 1157. [DOI] [PubMed] [Google Scholar]
- [39].a) Lee S, Serpooshan V, Tong X, Venkatraman S, Lee M, Lee J, Chirikian O, Wu JC, Wu SM, Yang F, Biomaterials 2017, 131, 111; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Jacot JG, Martin JC, Hunt DL, J Biomech 2010, 43, 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Pandey P, Hawkes W, Hu J, Megone WV, Gautrot J, Anilkumar N, Zhang M, Hirvonen L, Cox S, Ehler E, Developmental cell 2018, 44, 326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Shin SR, Zhang YS, Kim D-J, Manbohi A, Avci H, Silvestri A, Aleman J, Hu N, Kilic T, Keung W, Righi M, Assawes P, Alhadrami HA, Li RA, Dokmeci MR, Khademhosseini A, Anal Chem 2016, 88, 10019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Mandal BB, Kundu S, Biotechnology and Bioengineering 2008, 99, 1482. [DOI] [PubMed] [Google Scholar]
- [43].Nichol JW, Koshy ST, Bae H, Hwang CM, Yamanlar S, Khademhosseini A, Biomaterials 2010, 31, 5536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Tandon N, Cannizzaro C, Chao P-HG, Maidhof R, Marsano A, Au HTH, Radisic M, Vunjak-Novakovic G, Nat Protoc 2009, 4, 155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Wu KH, Wang SY, Xiao QR, Yang Y, Huang NP, Mo XM, Sun J, Journal of cellular biochemistry 2019, 120, 1318. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S2. Analysis of the elastic nature of the 3D printed constructs post printing and conditioning in DPBS.
Figure S3. Understanding the behavior of 3D printed constructs using (A) SPG7.5 and (B) G5 (control) inks under cyclic tensile strain of 5% for 40 cycles (n=3).
Figure S1. (A) Dual crosslinking of the biomaterial ink confirmed by increase in turbidity. (B) Optimization of extrusion speed for SPG7.5 ink for fabrication of 3D printed cardiac construct (n=10).
Figure S4. Optimization of flowrate for the GG ink for printing vascular channels. The vascular channels were printed using the GG ink encapsulating fluorescent microparticles at flow rates of (A) 4 mm.s−1, (B) 6 mm.s−1, (C) 7 mm.s−1, and (D) 8mm.s−1 (n=10).
Figure S5. Assessment of host cell infiltration. Zoomed histological image of the implanted IBC in rat subcutaneous models where IBC stands for implanted 3D printed constructs, blue represents DAPI stain and green represents CD68, a macrophage marker (n=4).
Video S1: Printing of anisotropic scaffold using SPG7.5 ink
Video S2: Beating of NRCMs on 3D printed scaffold
Video S4: Beating of hiPSC-CMs on 3D printed scaffold
Video S5: HiPSC-CS encapsulated in silk based supporting hydrogel without DOX
Video S6: HiPSC-CS encapsulated in silk based supporting hydrogel with DOX
Video S3: Differentiation of human iPSC to beating cardiomyocytes


