Abstract
Regulation of actin cytoskeleton dynamics in dendritic spines is crucial for learning and memory formation. Hence, defects in the actin cytoskeleton pathways are a biological trait of several brain diseases, including Alzheimer's disease. Here, we describe a novel synaptic mechanism governed by the cyclase-associated protein 2, which is required for structural plasticity phenomena and completely disrupted in Alzheimer's disease. We report that the formation of cyclase-associated protein 2 dimers through its Cys32 is important for cyclase-associated protein 2 binding to cofilin and for actin turnover. The Cys32-dependent cyclase-associated protein 2 homodimerization and association to cofilin are triggered by long-term potentiation and are required for long-term potentiation-induced cofilin translocation into spines, spine remodelling and the potentiation of synaptic transmission. This mechanism is specifically affected in the hippocampus, but not in the superior frontal gyrus, of both Alzheimer's disease patients and APP/PS1 mice, where cyclase-associated protein 2 is down-regulated and cyclase-associated protein 2 dimer synaptic levels are reduced. Notably, cyclase-associated protein 2 levels in the cerebrospinal fluid are significantly increased in Alzheimer's disease patients but not in subjects affected by frontotemporal dementia. In Alzheimer's disease hippocampi, cofilin association to cyclase-associated protein 2 dimer/monomer is altered and cofilin is aberrantly localized in spines. Taken together, these results provide novel insights into structural plasticity mechanisms that are defective in Alzheimer's disease.
Keywords: dementia, synapse, actin, cytoskeleton
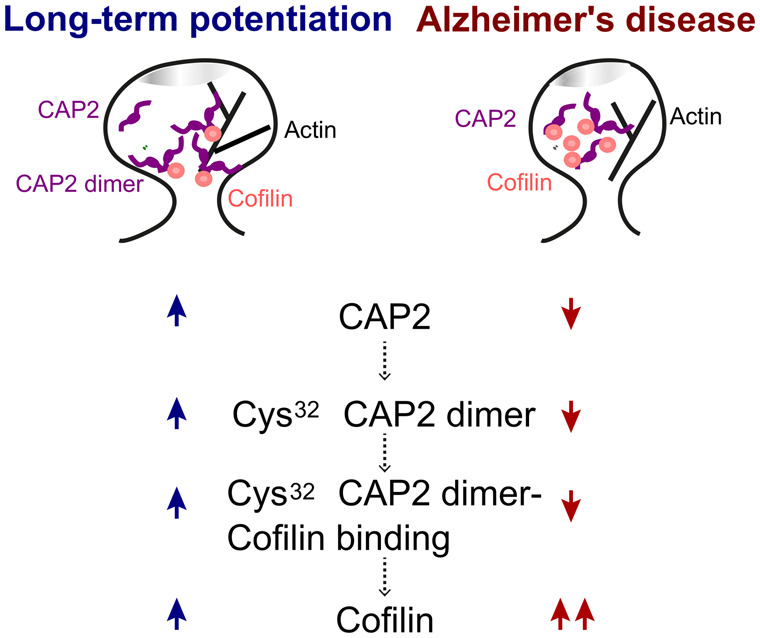
Long-term potentiation triggers cyclase-associated protein 2 translocation to spines, the formation of Cys32-dependent CAP2 dimers and binding to cofilin, thus leading to an increase in cofilin synaptic localization. In Alzheimer's disease, patients’ synapses this pathway is altered: cyclase-associated protein 2 dimer is reduced and aberrantly associated to cofilin, while cofilin synaptic localization is increased.
Graphical Abstract
Graphical Abstract.
Introduction
Dendritic spines are small dendritic protrusions containing excitatory postsynaptic machinery (Bourne and Harris, 2008). Since changes in spine morphology account for functional differences at the synaptic level (Yuste and Bonhoeffer, 2001), spine remodelling and modifications in spine density are believed to be important for learning and memory (Holtmaat and Svoboda, 2009) and are thereby associated with brain diseases characterized by cognitive decline, including Alzheimer’s Disease (Penzes et al., 2011). Indeed, it is widely accepted that spines constitute the anatomical locus of plasticity, where short-term alterations in synaptic strength are converted into long-lasting changes that are embedded in stable structural modifications (Sala and Segal, 2014). Hence, spine structural plasticity is tightly coordinated with synaptic function and plasticity: spine enlargement parallels modification of the number, types and properties of surface glutamate α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptors during long-term potentiation (LTP), whereas during long-term depression (LTD) the decrease in synaptic strength is associated with spine shrinkage (Kasai et al., 2010).
In this frame, the actin cytoskeleton is important for postsynaptic structure, function and plasticity as it confers spine plasticity and stability (Cingolani and Goda, 2008; Hotulainen and Hoogenraad, 2010; Pelucchi et al., 2020). Actin is highly enriched in spines and filamentous actin (F-actin) is the major structural backbone of spines since it forms organized bundles in spine necks (Star et al., 2002). Only a relatively small fraction of actin in spines is stable (Kasai et al., 2010), while the most abundant dynamic fraction of the actin cytoskeleton provides the driving force behind structural remodelling of spines and contributes to synaptic plasticity (Matus, 2005).
Regulation of actin dynamics is also relevant in Alzheimer's disease pathogenesis, since the signalling pathways influencing actin cytoskeleton remodelling have been shown to be impaired in Alzheimer's Disease (Penzes and Vanleeuwen, 2011; Pelucchi et al., 2020). In particular, among the actin-binding proteins implicated in Alzheimer's disease pathology, cofilin plays a critical role. Indeed, abnormalities of cofilin have been reported in Alzheimer's disease patients (Bamburg and Bernstein, 2016) and β-amyloid oligomers affect cofilin activation (Henriques et al., 2015). Cofilin is a key bidirectional regulator of spine structural plasticity, as it is implicated in both spine enlargement (Bosch et al., 2014) and spine shrinkage (Zhou et al., 2004; Hotulainen et al., 2009; Rust et al., 2010; Pontrello et al., 2012). Cofilin controls F-actin assembly and disassembly in a complex, concentration-dependent manner (Hild et al., 2014; Rust, 2015b). At low concentrations, cofilin promotes F-actin disassembly by accelerating the dissociation of monomeric actin (G-actin) from the filaments’ minus ends and by severing F-actin (Blanchoin and Pollard, 1999). Conversely, at high concentrations, cofilin can promote F-actin assembly by nucleating new and by stabilizing pre-existing filaments (Andrianantoandro and Pollard, 2006). Indeed, during LTP cofilin is transported to the spine where it promotes the F-actin assembly that is required for spine expansion (Bosch et al., 2014).
The actin dynamizing activity of cofilin is enhanced by binding partners as the cyclase-associated proteins (CAPs) (Normoyle and Brieher, 2012). CAPs are evolutionary highly conserved multi-domain actin binding proteins capable of regulating actin dynamics at multiple levels (Ono, 2013). Indeed, CAP and cofilin synergize to accelerate the depolymerization of the pointed end of actin filaments (Kotila et al., 2019; Shekhar et al., 2019). CAP deficiency results in defects in vesicle trafficking, endocytosis and in an altered cell morphology and cell growth (Noegel et al., 1999). Two closely related homologs of CAP have been described in mammals. CAP1 is expressed in nearly all cells, whereas CAP2 expression is restricted to a limited number of tissues, including the brain (Bertling et al., 2004; Peche et al., 2007), suggesting that CAP2 may have unique roles, particularly in neuronal cells. CAP2 gene deletion has been described in a rare developmental disorder, named 6p22 syndrome, which is characterized by developmental delays and autism spectrum disorders (Field et al., 2015). In addition, alterations in spine morphology and dendrite architecture have been reported in CAP2 knock-out neurons (Kumar et al., 2016).
Here, we introduce a novel mechanism, altered in both Alzheimer's disease patients and APP/PS1 mice hippocampi, through which the dimerization of CAP2, dependent on Cys32, is relevant for actin dynamics and is necessary to target cofilin to spines upon LTP induction. The mutation of CAP2 Cys32 is sufficient to prevent the LTP-triggered changes in spine morphology and function. These findings may open new ways in our understanding and targeting synaptic dysfunction and spine dysmorphogenesis in Alzheimer's disease.
Materials and methods
Human brain tissue
The hippocampus and superior frontal gyrus (SFG) samples of Alzheimer's disease patients and of age-matched healthy controls (HC) were obtained from the Netherlands Brain Bank (NBB). Established Braak and Braak criteria were used to categorize Alzheimer's disease tissues (Braak and Braak, 1991). Alzheimer's disease patients fulfilled Braak 4 and 5 stages. Accordingly, in Alzheimer's disease cases, there were tangles and neuritic plaques in hippocampus. HC had no history of psychiatric or neurological disease. Detailed information is reported in Tables 1 and 2.
Table1.
Demographic and neuropathological characteristics of Alzheimer's disease and HC cases that were selected for the analysis of hippocampal specimens
| Subjects | Gender | Age at death (years) | PMD (hours) | pH CSF | Brain weight (g) | Braak level |
|---|---|---|---|---|---|---|
| 1 (AD) | M | 82 | 5.15 | 6.34 | 1182 | 5 |
| 2 (AD) | F | 84 | 5.55 | 6.42 | 1017 | 5 |
| 3 (AD) | F | 88 | 6.45 | 6.5 | 1148 | 5 |
| 4 (AD) | F | 91 | 6.25 | 6.05 | 1026 | 4 |
| 5 (AD) | M | 90 | 5.55 | 6.37 | 1080 | 4 |
| 6 (AD) | M | 91 | 4.1 | 6.28 | 1117 | 4 |
| 7 (AD) | F | 86 | 4.10 | 6.34 | 1083 | 4 |
| 8 (AD) | M | 81 | 4.05 | 6.42 | 1253 | 4 |
| 9 (AD) | F | 86 | 5.5 | 6.85 | 950 | 4 |
| Mean ± STD | 86.56 ± 3.75 | 5.19 ± 0.92 | 6.40 ± 0.21 | 1095 ± 92.24 | ||
| 1 (HC) | F | 85 | 6.25 | 6.6 | 1080 | 3 |
| 2 (HC) | F | 89 | 6.35 | 6.73 | 1139 | 3 |
| 3 (HC) | F | 91 | 4.1 | 6.58 | 1052 | 3 |
| 4 (HC) | M | 83 | 5.15 | 6.6 | 1372 | 1 |
| 5 (HC) | M | 80 | 4.25 | 6.59 | 1429 | 2 |
| 6 (HC) | M | 89 | 6.5 | 6.23 | 1185 | 2 |
| 7 (HC) | F | 81 | 6.40 | 7.16 | 1164 | 1 |
| 8 (HC) | M | 80 | 7.15 | 5.8 | 1376 | 0 |
| 9 (HC) | F | 85 | 4.40 | 6.71 | 1165 | 2 |
| Mean ± STD | 84.78 ± 4.15 | 5.62 ± 1.15 | 6.56 ± 0.37 | 1218± 138.23 |
AD = Alzheimer’s disease; CSF = cerebrospinal fluid; F = female; HC = healthy controls; M = male; PMD = postmortem delay; STD = standard deviation.
Table 2.
Demographic and neuropathological characteristics of Alzheimer's disease and HC cases that were selected for the analysis of SFG specimens
| Subjects | Gender | Age at death (years) | PMD (hours) | pH CSF | Brain weight (g) | Braak level |
|---|---|---|---|---|---|---|
| 1 (AD) | F | 91 | 3.45 | 6.36 | 1011 | 4 |
| 2 (AD) | F | 91 | 4.15 | 6.27 | 1202 | 4 |
| 3 (AD) | F | 86 | 4.10 | 6.34 | 1083 | 4 |
| 4 (AD) | F | 86 | 5.05 | 6.62 | 998 | 4 |
| 5 (AD) | M | 81 | 4.05 | 6.42 | 1253 | 4 |
| 6 (AD) | F | 86 | 5.5 | 6.85 | 950 | 4 |
| mean ± STD | 86.83 ± 3.76 | 4.38 ± 0.75 | 6.48 ± 0.22 | 1082 ± 120 | ||
| 1 (HC) | F | 84 | 4.45 | 6.26 | 1179 | 1 |
| 2 (HC) | F | 81 | 6.40 | 7.16 | 1164 | 1 |
| 3 (HC) | M | 80 | 7.15 | 5.8 | 1376 | 0 |
| 4 (HC) | M | 84 | 7.05 | 5.9 | 1385 | 1 |
| 5 (HC) | F | 85 | 5.00 | 6.72 | 1257 | 1 |
| 6 (HC) | F | 85 | 4.40 | 6.71 | 1165 | 2 |
| mean ± STD | 83.17 ± 2.14 | 5.74 ± 1.28 | 6.43 ± 0.53 | 1254± 103 |
AD = Alzheimer’s disease; CSF = cerebrospinal fluid; F = female; HC = healthy controls; M = male; PMD = postmortem delay; STD = standard deviation.
Triton-insoluble fraction synaptic membrane preparation
Triton insoluble fraction (TIF), a fraction highly enriched in all categories of postsynaptic density proteins (i.e. receptor, signalling, scaffolding and cytoskeletal elements) absent of presynaptic markers (Gardoni et al., 2001), was obtained from human hippocampus and SFG specimens and APP/PS1 and wild-type (WT) hippocampal samples. In order to avoid protein degradation, Alzheimer's disease samples were paired to HC samples and processed at the same time. The procedure was performed at least twice to have two experimental replicates.
Samples of human hippocampal and SFG specimens were homogenized at 4°C in an ice-cold buffer with Roche cOmplete™ Protease Inhibitor Cocktail, Ser/Thr and Tyr phosphatase inhibitors (Sigma-Aldrich), 0.32 M Sucrose, 1 mM Hepes, 1 mM NaF, 0.1 mM PMSF, 1 mM MgCl2 using a glass-teflon homogenizer. An aliquot of homogenate (Homo) was kept for western blot (WB) analysis. Homo were then centrifuged at 1000 g for 5 min at 4°C, to remove nuclear contamination and white matter. The supernatant was collected and centrifuged at 13 000 g for 15 min at 4°C. The resulting pellet (crude membrane fraction) was resuspended in resuspension buffer [1 mM Hepes with protease inhibitors (Roche cOmplete™)] and then centrifuged at 100 000 g for 1 h at 4°C. Triton-X extraction of the resulting pellet was carried out at 4°C for 15 min in an extraction buffer [1% Triton-X, 75 mM KCl and protease inhibitors (Roche cOmplete™ Protease Inhibitor Cocktail)]. After extraction, the samples were centrifuged at 100 000 g for 1 h at 4 °C and the TIFs obtained were resuspended in 20 mM HEPES with protease inhibitors (Roche cOmplete™ Protease Inhibitor Cocktail).
To obtain the TIF fractions from mouse hippocampi and hippocampal cultures, we used a slightly modified protocol according to the lower amount of tissue. The samples were homogenized at 4°C in a the ice-cold buffer described above using a glass-teflon homogenizer for tissues and a glass–glass homogenizer for the cultures samples. An aliquot of Homo was kept for WB analysis. Homo samples were centrifuged at 13 000 g for 15 min at 4°C. Triton-X extraction of the resulting pellet was carried out at 4°C for 15 min in an extraction buffer [0.5% Triton-X, 75 mM KCl and protease inhibitors (Roche cOmplete™ Protease Inhibitor Cocktail)]. After extraction, the samples were centrifuged at 100 000 g for 1 h at 4°C and the TIFs obtained were resuspended in 20 mM HEPES with protease inhibitors (Roche cOmplete™ Protease Inhibitor Cocktail).
Treatments of neuronal cultures
To induce chemical LTP (cLTP), hippocampal neuronal cultures were first incubated in artificial cerebrospinal fluid (ACSF) for 30 min: 125 mM NaCl, 2.5 mM KCl, 1 mM MgCl2, 2 mM CaCl2, 33 mM D-glucose and 25 mM HEPES (pH 7.3; 320 mosM final), followed by stimulation with 50 μM forskolin, 0.1 μM rolipram and 100 μM picrotoxin (Tocris) in ACSF without MgCl2. After 16 min of stimulation, neurons were replaced in regular ACSF for 15 min and, after treatment, samples were processed (Marcello et al., 2013). To induce chemical LTD (cLTD), neuronal cultures were incubated in ACSF for 30 min, followed by stimulation with 50 μM NMDA (Sigma-Aldrich) in ACSF. After 10 min of stimulation, neurons were replaced in regular ACSF for 20 min and then subjected to the biochemical and imaging studies (Marcello et al., 2013). Stimulation of synaptic NMDA receptors (synaptic stimulation) was obtained by treating hippocampal neurons with 50 mM Bicuculline (Tocris), 2.5 mM 4-AP and 5 mM ifenprodil in Neurobasal medium supplemented with B27 (Dinamarca et al., 2016).
Co-immunoprecipitation assays
Aliquots of 20 μg of TIF obtained from human hippocampus were incubated with an antibody against cofilin overnight at 4°C in a final volume of 150 μL of RIA buffer [200 mM NaCl, 10 mM ethylene-diaminetetra-acetic acid (EDTA), 10 mM Na2HPO4, 0.5% NP-40, 0.1% sodium dodecyl sulfate (SDS)]. SureBeads Protein A/G Magnetic Beads (Bio-Rad) were used to precipitate the immunocomplex. After 3 washes with RIA buffer, the beads were resuspended in sample buffer without β-mercaptoethanol and heated for 3 min before the loading onto SDS-PAGE to detect CAP2 dimer and monomer. Beads were collected by centrifugation and applied onto SDS-PAGE; the precipitated immunocomplex was revealed by anti-cofilin and anti-CAP2 antibody. Heterologous co-immunoprecipitation experiments were carried out from lysates of either COS-7 or HEK293 cells transfected with different combinations of Myc-CAP2, EGFP-CAP2 or EGFP tagged truncated constructs of CAP2. Cells were harvested and proteins extracted as previously described (Marcello et al., 2010). The same immunoprecipitation protocol was used, incubating aliquots of 100 μg of HEK293/COS-7 lysates with an anti-Myc antibody. Protein A/G Agarose beads (Pierce) were added, and incubation was continued for 2 h at room temperature on the rotator mixer. Beads were collected and washed with RIA buffer for three times. Sample buffer for SDS-PAGE was added and the mixture was heated for 10 min. The precipitated immunocomplex was revealed with either anti-GFP or anti-Myc or anti-actin antibodies.
In situ proximity ligation assay
Proximity ligation assay (PLA) was performed in primary neuronal cultures as previously described (Dinamarca et al., 2016). Primary hippocampal neurons were fixed with 4% paraformaldehyde (PFA)—4% sucrose in PBS for 5 min at 4°C and washed several times with PBS. Neurons were permeabilized with 0.1% Triton X-100 in PBS for 15 min at RT. After incubation with the blocking solution of the PLA kit (Duolink® PLA Technology), cells were incubated overnight with the primary antibodies at 4°C. According to the manufacturer’s instructions, after several washing with solution A, secondary probes attached to oligonucleotides were added and the oligonucleotides of the bound probes were ligated and amplified by a fluorescent polymerase that visualizes the PLA signal. To stain MAP2 or the transfected GFP, cells were washed with PBS for 30 min and then the immunocytochemistry protocol was used. Cells were mounted on slides in Fluoromount™ Aqueous Mounting Medium (Sigma-Aldrich).
Cell culture electrophysiology
Whole cell voltage-clamp recordings were performed on rat hippocampal neurons transfected at day in vitro (DIV) 10 and maintained in culture for 15–16 DIV. Recording pipettes were fabricated from borosilicate glass capillary with a tip resistance of 3–5 MΩ and filled with an intracellular solution of the following composition (in mM): 130 potassium gluconate, 10 KCl, 1 EGTA, 10 Hepes, 2 MgCl2, 4 MgATP and 0.3 Tris-GTP. During recordings of miniature excitatory postsynaptic currents (mEPSCs), cells were bathed in a standard external solution containing (in mM): 125 NaCl, 5 KCl, 1.2 MgSO4, 1.2 KH2PO4, 2 CaCl2, 6 glucose and 25 HEPES-NaOH, pH 7.4, with also tetrodotoxin (TTX-1μM), bicuculline (20 μM) and strychnine (1 μM). For chemical LTP experiments, recordings of mEPSCs were performed applying glycine (100 μM) for 3 min at room temperature in Mg2+-free KRH, also containing TTX, bicuculline and strychnine. Recordings were performed at room temperature in voltage clamp mode using a Multiclamp 700B amplifier (Molecular Devices) and pClamp-10 software (Axon Instruments). Series resistance ranged from 10 MΩ to 20 MΩ and was monitored for consistency during recordings. Cells in culture with leak currents >200 pA were excluded from the analysis. Signals were amplified, sampled at 10 kHz, filtered to 2 or 3 kHz and analyzed using pClamp 10 data acquisition and analysis program.
Ethical approvals
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional research committee (Brescia Ethics Committee, protocol #NP2806) and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
All procedures performed in studies involving animals were in accordance with the ethical standards of the Institutional Animal Care and Use Committee of University of Milan (Italian Ministry of Health permit #326/2015, #5247B.N.YCK/2018, #295/2012-A, #497/2015-PR) and of the internal animal welfare authorities of the University of Marburg (references: AK-6-2014-Rust), at which the studies were conducted. Animals were maintained on a 12-h light/dark cycle in a temperature-controlled room (22°C) in cages with free access to food and water. Housing in the animal facility is performed in conformity with local and European Community regulations under the control of veterinarians with the assistance of trained personnel.
Experimental design and quantification of data
To minimize the possibility of bias in experimental results, randomization and blinding were used in the experimental design, imaging acquisitions and analyses. Acquisition and quantification of western blotting was performed by means of computer-assisted imaging (ChemiDoc system and Image lab 4.0 software; Bio-Rad). Data obtained by coimmunoprecipitation assays were normalized on the amount of immunoprecipitated protein. As far as concern imaging analysis, cells were randomly chosen from at least two cultures. Morphological analysis of dendritic spines was performed on the total length of the dendrites using ImageJ software to measure spine length, head and neck width (Malinverno et al., 2010). Sholl analysis was performed using Fiji free software as described in Mauceri et al. (2011).
Cofilin spine localization analysis was carried out assuming that the spine volume and the amount of fusion protein were proportional to the integrated intensity of the RFP signal (Bosch et al., 2014). Z-stack images were processed and analysed using Nikon Elements software and ImageJ. To analyse the distribution of PSD-95 and Bassoon within the spine, a line was drawn across the spine head semi-perpendicular to the dendrite, and the proteins intensity profiles were evaluated (Smith et al., 2014). The analysis of PLA experiments was performed with ImageJ software and the number of clusters was normalized on the total dendritic length. Total internal reflection fluorescence image analysis was done using Fiji. For spontaneous depolymerization, kymographs of single filaments (30–50 filaments per condition) were generated and depolymerization rates were calculated from the slope of the graphs. To assess cofilin cutting rates, at least 30 filaments in the frame were selected.
Statistical analysis
Linear mixed-effects (LMMs) models were used in most of the imaging experiments (Tables 3 and 4) as outcome variables were mostly continuous. To account for the hierarchical structure of dendritic spine morphology designs, we specified one mixed-effects model (Pinheiro and Bates, 2006) for each experiment under investigation and outcome variable of interest within the experiment (Paternoster et al., 2018). Generalized linear mixed-effects models with a logit link function and binomial family were fitted for the Type outcome variable in the experiments assessing spine morphology; in these analyses, the outcome variable was restricted to assume categories Mushroom (reference category) and Stubby only. For each mixed-effects model, one or two random intercepts were specified depending on the available levels of the hierarchy. The culture random intercept was present in any model, whereas the neuron level of the hierarchy was present when the experimental unit of analysis was spine and it was not estimable when neuron itself was the experimental unit. Correlations between random effects were allowed to be present and fitted within the model to better capture the hierarchical structure of the experiment. We reported information on which dependent and independent variables were used within each mixed-effects model in Table 3 (columns on the left). For model fitting, restricted maximum likelihood (REML) estimates were used, as they provided better estimates of variance components. To test model assumptions, we plotted residuals versus fitted values for each model; when outliers were detected, they were checked at the experimental level. When model convergence issues persisted after changing the estimation method, we fitted an equivalent model showing a fixed-effects term instead of the corresponding mixed-effect one. If two hierarchical levels were available in the experiment, the fitted model was still a mixed-effects model splitting the hierarchical levels into one random- and one fixed-effect term; when neuron was the experimental unit, a fixed-effects model was estimated using ordinary least squares method. Hypothesis testing on the significance of single parameters was performed within the linear mixed-effects or generalized linear mixed-effects models with a Student t distribution or a Gaussian distribution, respectively. The F statistics from the ANOVA analysis provided information on the overall significance of the independent variables entered into each model. When necessary, contrasts of interest were extracted from the linear mixed-effects to calculate estimated mean differences and Standard errors (SEs) between any pair of categories of the effect of interest. The corresponding P-values were adjusted for multiple comparisons according to the False Discovery Rate (FDR) or Benjamini–Hochberg method (Table 4) (Benjamini and Hochberg, 1995). Finally, the chi-squared test of independence was used to assess the presence of an association between spine type and genetic modification.
Table 3.
Estimates of the mean and standard error from mixed-effects models fitted to the available datasets of the corresponding figures
| Figure | Y | Random effects at different hierarchical levels |
Variable | Estimate | Standard error | P value | |
|---|---|---|---|---|---|---|---|
| Culture | Neuron | ||||||
| 3 B, C | Spine length |
|
|
|
0.1657 | 0.0181 | <0.0001 |
| Spine width |
|
|
|
0.0312 | 0.0087 | 0.00035 | |
| Spine type |
|
|
Genetic modification CAP2-shRNA versus SCR | −0.3122 | 0.0784 | <0.0001 | |
| 3 E, F | Dendritic length |
|
N/Eb |
|
−299.67 | 136.12 | 0.4995 |
| Culture, ANOVA, P = 0.7505 | 44.85 | 137.53 | 0.75 | ||||
| Intersection |
|
|
Genetic modification CAP2-shRNA, ANOVA, P = 0.4457 | −0.5065 | 0.6634 | 0.4457 | |
| Length, ANOVA, P < 0.0001 | 0.0627 | 0.0075 | <0.0001 | ||||
| Culture, ANOVA, P = 0.7804 | 0.0892 | 0.3199 | 0.7803 | ||||
| Interaction between genetic modification and length, ANOVA, P = 0.0394 | −0.0219 | 0.0106 | 0.0394 | ||||
| 5 F | PLA clusters/μm |
|
N/Eb |
|
−0.7997 | 0.1486 | <0.0001 |
| 6 G | PLA clusters/μm |
|
N/Eb |
|
−0.0176 | 0.0064 | 0.0126 |
| Culture 2, ANOVA, P = 0.2579 | −0.0070 | 0.0075 | 0.3670 | ||||
| Culture 3, ANOVA, P = 0.2579 | 0.0074 | 0.0078 | 0.3556 | ||||
| 7 F | PLA clusters/μm |
|
N/Eb | MycCAP2 and cLTP | 0.5791 | 0.1640 | 0.00086 |
| Myc-(C32G) CAP2 and CTRL | −0.5914 | 0.177 | 0.0016 | ||||
| Myc-(C32G) CAP2 and cLTP | −0.6388 | 0.177 | 0.00070 | ||||
| ANOVA, P < 0.0001 | |||||||
| 7 H | PLA clusters/μm |
|
N/Eb | Myc-CAP2 and cLTP | 0.0668 | 0.0096 | <0.0001 |
| Myc-(C32G) CAP2 and CTRL | −0.0178 | 0.0104 | 0.0960 | ||||
| Myc-(C32G) CAP2 and cLTP | −0.0001 | 0.0096 | 0.9901 | ||||
| ANOVA, P < 0.0001 | |||||||
| 8 B | IntDen Cofilin/IntDen RFP |
|
|
Genetic modification and treatment, CAP2-shRNA and cLTP | 0.0051 | 0.0243 | 0.833 |
| Genetic modification and treatment, SCR and cLTP | 0.0992 | 0.0218 | <0.0001 | ||||
| Genetic modification and treatment, shRNA + shr-C32G-CAP2 and cLTP | 0.0244 | 0.0240 | 0.309 | ||||
| Genetic modification and treatment, shRNA + shr-wt-CAP2 and cLTP | 0.1386 | 0.0239 | <0.0001 | ||||
| ANOVA P < 0.0001 | |||||||
| 8 D, E | Spine length |
|
|
Genetic modification and treatment, CAP2-shRNA and cLTP | 0.3468 | 0.0173 | <0.0001 |
| Genetic modification and treatment, SCR and cLTP | 0.2984 | 0.0177 | <0.0001 | ||||
| Genetic modification and treatment, shRNA + shr-C32G-CAP2 and cLTP | 0.3675 | 0.0165 | <0.0001 | ||||
| Genetic modification and treatment, shRNA + shr-wt-CAP2 and cLTP | 0.1881 | 0.0184 | <0.0001 | ||||
| ANOVA P < 0.0001 | |||||||
| Spine width |
|
|
Genetic modification and treatment, CAP2-shRNA and cLTP | 0.0116 | 0.0094 | 0.219 | |
| Genetic modification and treatment, SCR and cLTP | 0.2100 | 0.0096 | <0.0001 | ||||
| Genetic modification and treatment, shRNA + shr-C32G-CAP2 and cLTP | −0.0472 | 0.0090 | <0.0001 | ||||
| Genetic modification and treatment, shRNA + shr-wt-CAP2 and cLTP | 0.2821 | 0.0100 | <0.0001 | ||||
| ANOVA P < 0.0001 | |||||||
| Spine type |
|
|
Genetic modification and treatment, CAP2-shRNA and cLTP | −0.7528 | 0.1077 | <0.0001 | |
| Genetic modification and treatment, SCR and cLTP | −1.5111 | 0.1019 | <0.0001 | ||||
| Genetic modification and treatment, shRNA + shr-C32G-CAP2 and cLTP | −1.1777 | 0.0988 | <0.0001 | ||||
| Genetic modification and treatment, shRNA + shr-wt-CAP2 and cLTP | −2.0932 | 0.1037 | <0.0001 | ||||
| Culture | 0.0364 | 0.0627 | 0.562 | ||||
The hierarchical structure of the experiments was introduced in the left part of the table, including the standard deviation (STD) of the random-effects terms when available.
Due to convergence issues, fixed-effects terms were introduced instead of the corresponding mixed-effects ones for some hierarchical levels.
N/E = not estimable due to the presence of one hierarchical level only (i.e. neuron is the experimental unit).
Table 4.
Pairwise contrasts of interest derived from mixed-effects models introduced in Figs 7 and 8
| Figure | Y | Contrast | Estimate | Standard error | P value |
|---|---|---|---|---|---|
| 7 F | PLA clusters/μm | EGFP-CAP2/Myc-(C32G)CAP2-CTRL versus EGFP-CAP2/Myc-CAP2-CTRL | −0.5914 | 0.1777 | <0.0001 |
| EGFP-CAP2/Myc-(C32G)CAP2-cLTP versus EGFP-CAP2/Myc-CAP2-CTRL | −0.6388 | 0.1777 | 0.00129 | ||
| EGFP-CAP2/Myc-CAP2-cLTP versus EGFP-CAP2/Myc-CAP2-CTRL | 0.5791 | 0.1640 | 0.00129 | ||
| EGFP-CAP2/Myc-(C32G)CAP2-cLTP versus EGFP-CAP2/Myc-(C32G)CAP2-CTRL | −0.0475 | 0.1894 | 0.8030 | ||
| EGFP-CAP2/Myc-CAP2-cLTP versus EGFP-CAP2/Myc-(C32G)CAP2-CTRL | 1.1705 | 0.1777 | <0.0001 | ||
| EGFP-CAP2/Myc-CAP2(C32G)-cLTP versus EGFP-CAP2/Myc-CAP2-cLTP | −1.2180 | 0.1777 | <0.0001 | ||
| 7 H | PLA clusters/μm | Myc-(C32G)CAP2/Cofilin-CTRL versus Myc-CAP2/Cofilin-CTRL | −0.0178 | 0.0104 | 0.115 |
| Myc-(C32G)CAP2/Cofilin-cLTP versus Myc-CAP2/Cofilin-CTRL | −0.0001 | 0.0096 | 0.9901 | ||
| Myc-CAP2/Cofilin-cLTP versus Myc-CAP2/Cofilin -CTRL | 0.0668 | 0.0096 | <0.0001 | ||
| Myc-(C32G)CAP2/Cofilin -cLTP versus Myc-(C32G)CAP2/Cofilin-CTRL | 0.0177 | 0.0102 | 0.115 | ||
| Myc-CAP2/Cofilin-cLTP versus Myc-(C32G)CAP2/Cofilin-CTRL | 0.0846 | 0.0102 | <0.0001 | ||
| Myc-(C32G)CAP2/Cofilin-cLTP versus Myc-CAP2/Cofilin-cLTP | −0.0669 | 0.0094 | <0.0001 | ||
| 8 B | IntDen Cofilin/IntDen RFP | CAP2-shRNA-cLTP versus SCR-CTRL | −0.0051 | 0.0243 | 0.834 |
| SCR-cLTP versus SCR-CTRL | 0.0992 | 0.0218651 | <0.0001 | ||
| shRNA + shr-C32G-CAP2-cLTP versus SCR-CTRL | 0.0245 | 0.0240298 | 0.386 | ||
| shRNA + shr-wt-CAP2-cLTP versus SCR-CTRL | 0.1386 | 0.0239044 | <0.0001 | ||
| CAP2-shRNA-cLTP versus SCR-cLTP | −0.09412 | 0.0236968 | 0.00016 | ||
| CAP2-shRNA-cLTP versus shRNA + shr-C32G-CAP2-cLTP | −0.01937 | 0.0250532 | 0.489 | ||
| CAP2-shRNA-cLTP versus shRNA + shr-wt-CAP2-cLTP | −0.1335 | 0.0238909 | <0.0001 | ||
| shRNA + shr-C32G-CAP2-cLTP versus SCR-cLTP | −0.07475 | 0.0233884 | 0.00246 | ||
| shRNA + shr-wt-CAP2-cLTP versus SCR-cLTP | 0.0394 | 0.0231644 | 0.128 | ||
| shRNA + shr-C32G-CAP2-cLTP versus shRNA + shr-wt-CAP2-cLTP | −0.1142 | 0.0240324 | <0.0001 | ||
| 8 D | Spine length | CAP2-shRNA-cLTP versus SCR-CTRL | 0.3468 | 0.0173 | <0.0001 |
| SCR-cLTP versus SCR-CTRL | 0.2984 | 0.0177 | <0.0001 | ||
| shRNA + shr-C32G-CAP2-cLTP versus SCR-CTRL | 0.3676 | 0.0165 | <0.0001 | ||
| shRNA + shr-wt-CAP2-cLTP versus SCR-CTRL | 0.1881 | 0.0184 | <0.0001 | ||
| CAP2-shRNA-cLTP versus SCR-cLTP | 0.0484 | 0.0180 | 0.00795 | ||
| shRNA + shr-C32G-CAP2-cLTP versus CAP2-shRNA-cLTP | 0.0207 | 0.0171 | 0.2255 | ||
| shRNA + shr-wt-CAP2-cLTP versus CAP2-shRNA-cLTP | −0.1588 | 0.0187 | <0.0001 | ||
| shRNA + shr-C32G-CAP2-cLTP versus SCR-cLTP | 0.0691 | 0.0171 | <0.0001 | ||
| shRNA + shr-wt-CAP2-cLTP versus SCR-cLTP | −0.1104 | 0.0187 | <0.0001 | ||
| shRNA + shr-C32G-CAP2-cLTP versus shRNA + shr-wt-CAP2-cLTP | 0.1795 | 0.0184 | <0.0001 | ||
| Spine width | CAP2-shRNA-cLTP versus SCR-CTRL | 0.0116 | 0.0094 | 0.2185 | |
| SCR-cLTP versus SCR-CTRL | 0.2100 | 0.0096 | <0.0001 | ||
| shRNA + shr-C32G-CAP2-cLTP versus SCR-CTRL | −0.0473 | 0.0090 | <0.0001 | ||
| shRNA + shr-wt-CAP2-cLTP versus SCR-CTRL | 0.2821 | 0.0100 | <0.0001 | ||
| CAP2-shRNA-cLTP versus SCR-cLTP | −0.1984 | 0.0098 | <0.0001 | ||
| shRNA + shr-C32G-CAP2-cLTP versus CAP2-shRNA-cLTP | −0.0589 | 0.0093 | <0.0001 | ||
| shRNA + shr-wt-CAP2-cLTP versus CAP2-shRNA-cLTP | 0.2705 | 0.0102 | <0.0001 | ||
| shRNA + shr-C32G-CAP2-cLTP versus SCR-cLTP | −0.2573 | 0.0093 | <0.0001 | ||
| shRNA + shr-wt-CAP2-cLTP versus SCR-cLTP | 0.0721 | 0.0102 | <0.0001 | ||
| shRNA + shr-C32G-CAP2-cLTP versus shRNA + shr-wt-CAP2-cLTP | −0.3294 | 0.0100 | <0.0001 |
P values were derived after adjustment for multiple comparisons according to the false discovery rate (FDR) approach.
For all the other experiments, as data followed a normal distribution, either Student t-test or one-way ANOVA, followed by a post hoc adjustment, were carried out.
Throughout the manuscript, when continuous variables were considered, values were reported as mean ± SE, and when qualitative variables were analysed, absolute frequencies (raw numbers) or percentages were indicated. The type of parametric test used for each experiment and the corresponding P-values, as well the type of adjustment for multiple comparisons (if any), were provided in figure legends. All statistical tests were two-sided and significance was assumed if P < 0.05. Calculations were carried out using the open-source statistical computing environment R (R Core Team, 2019) with its libraries lme4 (Bates et al., 2015), lmerTest (Kuznetsova et al., 2017) and emmeans (Lenth, 2020), for the imaging experiments and with Prism 6 (GraphPad, La Jolla, CA, USA) for all the other quantitative evaluations.
Data availability
Data are available on direct request to the corresponding author.
Results
Synaptic localization of CAP2 and cofilin is impaired in Alzheimer’s disease hippocampi
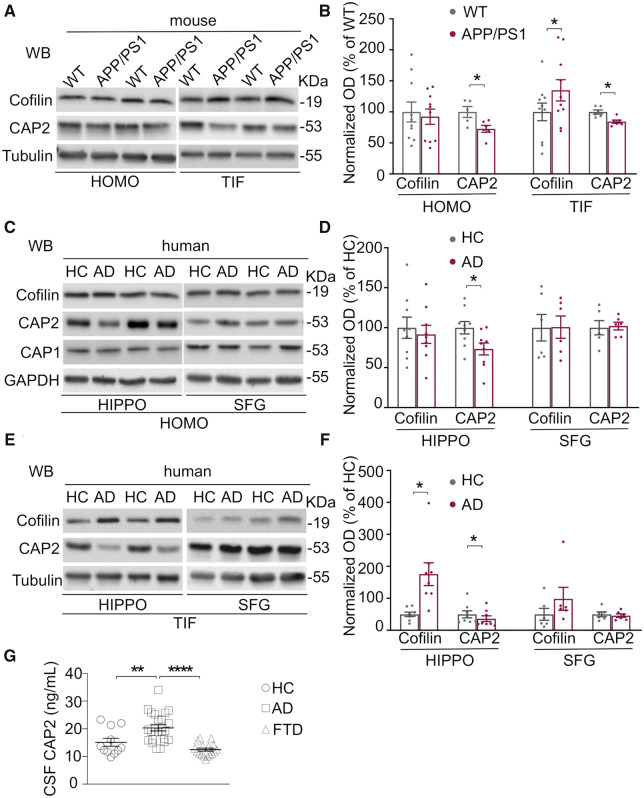
The actin-binding protein CAP2, that is expressed in several brain areas including the hippocampus (Supplementary Fig. 1A), has been described as a cofilin binding partner (Kumar et al., 2016). An impaired activation of cofilin has been reported in the hippocampus of Alzheimer's disease patients (Minamide et al., 2000; Barone et al., 2014). Cofilin postsynaptic localization, that plays a crucial role in synaptic plasticity phenomena (Bosch et al., 2014; Mikhaylova et al., 2018), is not altered in the frontal cortex of Alzheimer's disease patients and in the brain of animal models (Rush et al., 2018). Considering that the hippocampus is one of the first brain areas affected in Alzheimer's disease patients (Frisoni et al., 2010), we first assessed total and synaptic levels of cofilin and CAP2 in the hippocampus of APP/PS1 mice at 6 months of age, when they begin to develop amyloid deposits (Jankowsky et al., 2004). We purified the triton-insoluble fraction (TIF), which is highly enriched in postsynaptic proteins (Gardoni et al., 2001). WB analysis performed on the total homogenate and on the TIF samples shows a significant increase in cofilin synaptic levels in APP/PS1 mice hippocampi compared to WT mice, while no changes in the total protein levels were detected (Fig. 1A and B). Interestingly, the CAP2 protein levels in the total homogenate and, consistently, in the postsynaptic fraction of APP/PS1 mice were significantly reduced compared to WT mice (Fig. 1A and B).
Figure 1.
The synaptic localization of cofilin and CAP2 is specifically altered in the hippocampus of Alzheimer's disease patients and APP/PS1 mice. (A) Representative WB showing the expression of cofilin, CAP2 and Tubulin in homogenate (HOMO) and TIF hippocampal samples of APP/PS1 mice at 6 months of age compared to wild type (WT) animals. Non-cropped blots are shown in Supplementary Fig. 1C. (B) Quantification of optical density (OD) and normalization on loading control OD (Tubulin) shows that CAP2 but not cofilin total levels are reduced in APP/PS1 mice (HOMO, CAP2, APP/PS1 versus WT, unpaired t-test: *P = 0.023, n WT = 5 mice, n APP/PS1 = 6 mice; cofilin, APP/PS1 versus WT, unpaired t-test: P > 0.05, n WT = 10 mice, n APP/PS1 = 10 mice). In the TIF fraction, the quantitative analysis reveals that cofilin levels increase in transgenic mice, while CAP2 levels decrease (TIF, cofilin, APP/PS1 versus WT, paired t-test: *P = 0.015, n WT = 10 mice, n APP/PS1 = 10 mice; CAP2, APP/PS1 versus WT, paired t-test: *P = 0.026, n WT = 6 mice, n APP/PS1 = 6 mice). (C) Representative WB showing the expression of cofilin, CAP2, CAP1 and GAPDH in homogenate samples (HOMO) of Alzheimer's Disease (AD) and HC hippocampi (HIPPO) and SFG. Non-cropped blots are shown in Supplementary Fig. 1D. (D) Quantification of optical density (OD) and normalization on loading control OD (GAPDH) shows that CAP2 but not cofilin total levels are reduced in AD patients (CAP2, AD versus HC, unpaired t-test: *P = 0.027, n HC = 9, n AD = 9). In SFG HOMO samples cofilin and CAP2 levels were not affected in AD patients compared to HC (AD versus HC, unpaired t-test: *P > 0.05; n HC = 6, n AD = 6). No changes in CAP1 levels in the HOMO of both HIPPO and SFG were observed (CAP1, HIPPO, HC = 100 ± 8.72%, AD = 89.83 ± 9.65% AD versus HC, unpaired t-test: P > 0.05, n HC = 8, n AD = 9; SFG, HC = 100 ± 5.35%, AD = 110.8 ± 12.3%, AD versus HC, unpaired t-test: P > 0.05, n HC = 6, n AD = 6). (E) Representative WB showing cofilin, CAP2 and Tubulin levels in postsynaptic TIF samples of the HIPPO and SFG of AD and HC. Non-cropped blots are shown in Supplementary Fig. 1E. (F) The quantitative analysis reveals that cofilin levels increase in the TIF of AD patients while CAP2 levels decrease (cofilin, AD versus HC, paired t-test: *P = 0.0109, n HC = 8, n AD = 8; CAP2, AD versus HC, paired t-test: *P = 0.0172, n HC = 8, n AD = 8). No significant modifications of cofilin and CAP2 synaptic localization were detected in the SFG (AD versus HC, paired t-test: *P > 0.05; n HC = 6, n AD = 6). (G) CSF concentration of CAP2 in healthy controls (HC), Alzheimer's Disease patients (AD) and frontotemporal dementia patients (FTD). CAP2 levels are higher in AD patients, but not in FTD patients, compared to HC (HC: CAP2 = 15.13 ± 1.5 ng/ml, n = 11; AD: CAP2 = 20.34 ± 1.12 ng/ml, n = 23; FTD: CAP2 = 12.45 ± 0.56 ng/ml, n = 17; one-way ANOVA, Bonferroni multiple comparison test: **P = 0.0075; ****P < 0.0001).
These data suggest an altered synaptic availability of cofilin and of its binding partner CAP2 in the hippocampal synapses of APP/PS1 mice at the early stages of the pathology.
To strengthen these results, and verify the relevance in human pathology, we took advantage of autoptic specimens obtained from sporadic Alzheimer's Disease patients, fulfilling criteria for Braak 4 and 5 stage, and age-matched control subjects (HC) (Tables 1 and 2). In particular, we examined hippocampus and SFG, two areas differentially affected by the pathology.
The analysis of the total homogenate revealed no changes in cofilin total protein levels and a significant down-regulation of CAP2 in Alzheimer's disease patients hippocampi (Fig. 1C and D), as previously reported in microarray studies (Blalock et al., 2004). This Alzheimer's disease-associated alteration of CAP2 is specific since no modifications in the total levels of the homolog CAP1 have been detected in Alzheimer's disease patients' hippocampi and SFG when compared to HC (Fig. 1C and D). After the validation of the postsynaptic fraction purification protocol from human specimens (Supplementary Fig. 1B), we performed WB analysis of the TIF samples. A significant increase in cofilin synaptic levels and a concomitant decrease in CAP2 synaptic localization have been detected (Fig. 1E and F), as observed in APP/PS1 hippocampi (Fig. 1A and B). Interestingly, these alterations are specific for the hippocampus because no modifications of CAP2 and cofilin total levels and synaptic localization were found in SFG (Fig. 1 C–F), consistent with previous data showing no changes of cofilin levels in the postsynaptic densities (PSD) fraction of the frontal cortex of Alzheimer's disease patients (Rush et al., 2018).
Furthermore, we challenged cerebrospinal fluid (CSF) for the presence of CAP2. A preliminary screening experiment of CAP2 levels in the CSF has been performed on 23 Alzheimer's disease patients, 11 HC, 17 frontotemporal dementia patients as non-Alzheimer's disease neurodegenerative controls. CAP2 levels in Alzheimer's disease patients (20.34 ± 1.12 ng/ml) were significantly higher than in HC (15.13 ± 1.45 ng/ml) and in frontotemporal dementia patients (12.45 ± 0.56 ng/ml) (Fig. 1G), suggesting that the changes in CAP2 are specifically associated to Alzheimer's disease.
CAP2 is a synaptic protein relevant for synaptic function and neuronal structure
Since the Alzheimer's disease-related alterations of CAP2 and cofilin have been detected in the postsynaptic compartment, we decided to further investigate CAP2 role in dendritic spines. We employed stimulated emission depletion nanoscopy to analyse primary hippocampal neurons stained for endogenous CAP2, pre- and postsynaptic markers and F-actin, labelled by phalloidin. Endogenous CAP2 has a distribution pattern similar to phalloidin, is localized throughout the dendrites and is detectable in dendritic spines, as previously reported (Kumar et al., 2016) (Supplementary Fig. 2A). A linescan through the spine length shows that peaks of fluorescence for CAP2 and the presynaptic marker Bassoon do not overlap (Fig. 2A), while CAP2 fluorescence profile shows a partial overlap with the postsynaptic protein PSD-95 profile (Fig. 2B) (Smith et al., 2014).
Figure 2.
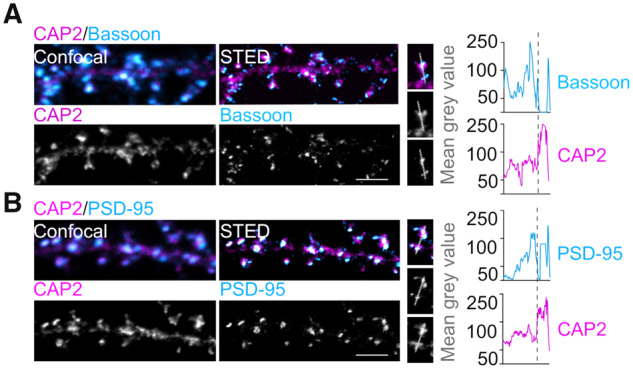
CAP2 is a postsynaptic protein. Confocal and stimulated emission depletion images of hippocampal neurons fixed and stained for CAP2 (magenta) and synaptic markers, either Bassoon (A) or PSD-95 (B) (cyan). Linescan through spine head was used to evaluate the mean grey value of the stained proteins. Scale bar = 2 μm.
To further confirm the localization of CAP2 in the postsynaptic compartment, we performed biochemical fractionation of rat hippocampi to isolate the PSD. WB analysis demonstrated that CAP2 is present in the synaptic fractions but not enriched in highly detergent-insoluble fractions PSD2, which corresponds to the ‘core’ of the PSD (Supplementary Fig. 2B).
To gain insights into the specific role of CAP2 in hippocampal neuronal cells and considering the reduction of CAP2 protein levels in the hippocampi of Alzheimer's disease patients and APP/PS1 mice (Fig. 1A–D), we took advantage of small hairpin RNA (shRNA) to down-regulate CAP2 expression. We first identified a shRNA that reduced by about 80% the levels of co-transfected Myc-CAP2 in heterologous cells (CAP2-shRNA, Supplementary Fig. 2C). The same shRNA reduced by about 80% endogenous CAP2 mRNA levels when packaged into recombinant adeno-associated virus (rAAV) particles and used to infect primary hippocampal neurons (Supplementary Fig. 2D).
Since CAP2 is a postsynaptic actin binding protein, we investigated whether it’s down-regulation affects spine morphology and synaptic transmission in hippocampal neurons.
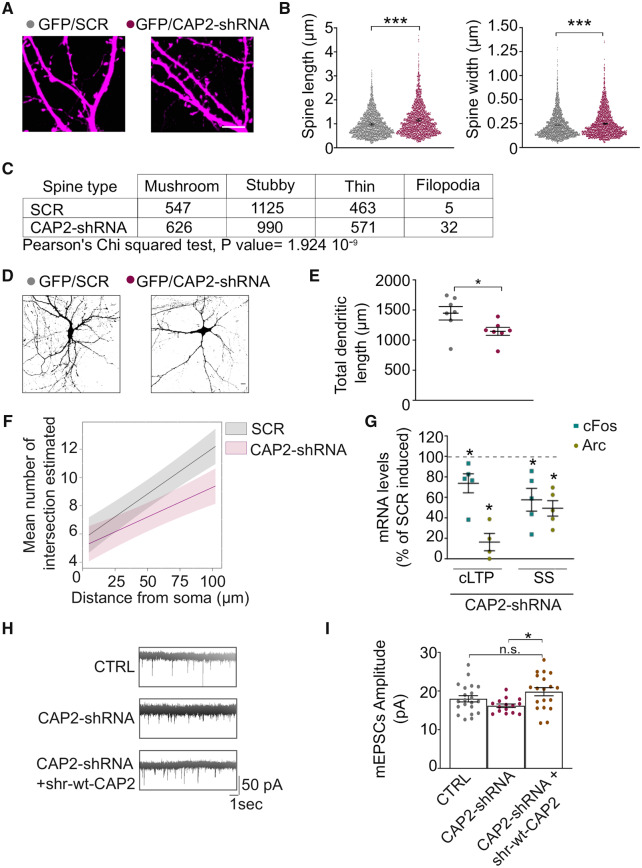
The analysis of the spine shape of hippocampal neurons transfected with CAP2-shRNA and GFP revealed a significant increase in spine length and head width, while no changes in density of spines were detected (Fig. 3A and B, SCR: 2.61 ± 0.30 spines/10 μm; CAP2-shRNA: 2.65 ± 0.36 spines/10 μm). The examination of the different spines categories showed that the different spines populations were associated with CAP2-down-regulation (Fig. 3C), highlighting altered neuronal spine morphology in CAP2-shRNA transfected neurons. Notably, in primary hippocampal neurons transfected with CAP2-shRNA and GFP, to visualize dendrite architecture, the Sholl analysis revealed a reduction in the complexity of the dendritic tree (Fig. 3D and F). This effect was accompanied by a significant reduction in total dendritic length (Fig. 3E). Interestingly, the down-regulation of CAP2 levels also reduces the mRNA levels of BDNF, a neurotrophin that controls dendrite morphology (Horch and Katz, 2002) (Supplementary Fig. 2D). These data suggest that the down-regulation of CAP2 affects neuronal architecture and spine morphology. Therefore, we asked whether the decreased CAP2 expression could influence synaptic function. Firstly, we observed that the knockdown of CAP2 impairs the expected increase in the transcription of immediate early genes (cFos and Arc) after the induction of both chemical LTP (cLTP) and synaptic stimulation (Fig. 3G). This suggests the relevance of CAP2 in synaptic plasticity phenomena and is consistent with an alteration of neuronal architecture leading to impaired integration of signal-regulated transcription. Secondly, we recorded the miniature excitatory postsynaptic currents (mEPSCs) in neurons that revealed impairments in the post-synaptic AMPA receptors sensitivity as suggested by the lower mEPSCs amplitude in CAP2-shRNA neurons respect to control conditions (Fig. 3H and I). To assess whether this effect could be specifically ascribed to CAP2, we generated and validated a shRNA-resistant (shr) ‘rescue’ form of Myc-CAP2 (shr-wt-CAP2) that was insensitive to the CAP2-shRNA (Supplementary Fig. 2E). As shown in Fig. 3I, the mEPSCs amplitude was restored to control levels by the shr-wt-CAP2, demonstrating the specificity of the effect on synaptic transmission. These results indicate that the down-regulation of CAP2 leads to a less responsive/functional postsynaptic compartment.
Figure 3.
CAP2 is critical for neuronal structure and function. (A) Representative confocal images of primary hippocampal neurons transfected with GFP and either CAP2-shRNA or scramble sequence (SCR); scale bar = 5 μm. (B) Graphs show the quantification of dendritic spine length and width (length, SCR = 0.956 ± 0.061 μm, CAP2-shRNA = 1.121 ± 0.066 μm; width, SCR = 0.4686 ± 0.0097 μm, CAP2-shRNA = 0.4998 ± 0.0099 μm, t-test from mixed-effects model: ***P < 0.001, SCR n = 2140 spines, CAP2-shRNA n = 2219 spines). (C) The joint distribution of dendritic spine type and genetic modification is represented in the table together with the P value of the Chi-squared test of independence. (D) Representative micrographs of hippocampal neurons transfected with GFP and either CAP2-shRNA or SCR. Scale bar = 10 μm. (E) Graph show the quantification of the total dendritic length (SCR = 1446.78 ± 113.3 μm, CAP2-shRNA = 1147.12 ± 65.67 μm, unpaired t-test from fixed-effects ordinary least squares regression model: *P = 0.04995, n SCR = 7 neurons, n CAP2-shRNA = 7 neurons). (F) The graph of Sholl analysis displays predicted values across available values of distance from soma (X axis) and corresponding 95% confidence bands (for details on how P values were calculated see Table 3, n SCR = 8 neurons, n CAP2-shRNA = 8 neurons). (G) QRT-PCR analysis of cFos and Arc expression in hippocampal neurons infected with either rAAV-CAP2-shRNA or rAAV-SCR and with or without treatment to induce either cLTP or synaptic stimulation (SS). The treated samples are normalized on either the cLTP-SCR or the SS-SCR condition. The down-regulation of CAP2 prevents the induction of Arc and cFos upon cLTP and SS (cLTP CAP2-shRNA versus cLTP-SCR, paired t-test: cFos *P = 0.0485, Arc *P = 0.0137; SS-CAP2-shRNA versus SS-SCR: cFos *P = 0.0188, Arc *P = 0.003, n = 5 independent experiments). (H) Representative traces of excitatory miniature postsynaptic currents (mEPSCs) recorded from 15 DIV hippocampal neurons obtained from CTRL, CAP2-shRNA and CAP2-shRNA+shr-wt-CAP2 transfected cultures. (I) Electrophysiological analysis of mEPSCs amplitude (pA): CTRL n = 21; CAP2 shRNA n = 14; CAP2 shRNA + shr-wt-CAP2 n = 19; one-Way ANOVA followed by Tukey's multiple comparison test: *P = 0.013, n.s. non-significant.
Collectively these data suggest that CAP2 is a postsynaptic protein relevant for shaping dendritic and spine morphology as well as for the regulation of synaptic transmission and plasticity, which are all deeply linked to the actin polymerization through cofilin activity (Hotulainen et al., 2009; Gu et al., 2010).
CAP2 forms disulfide-crosslinked dimers pivotal for association to cofilin and for actin turnover
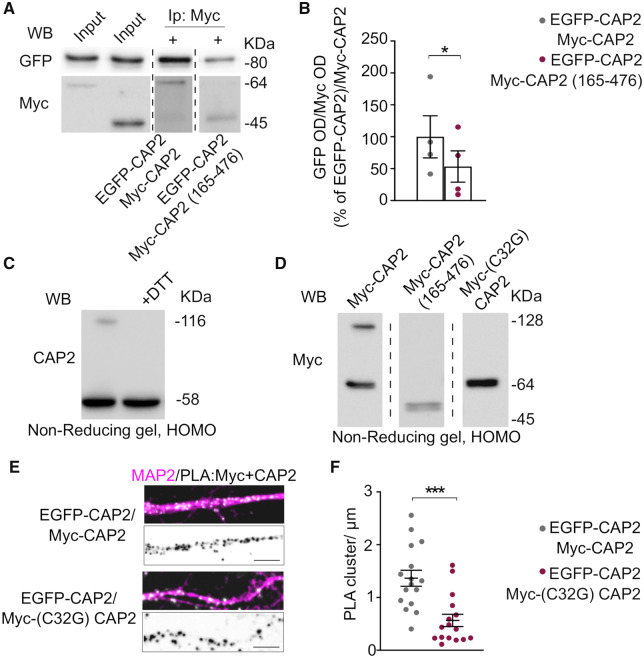
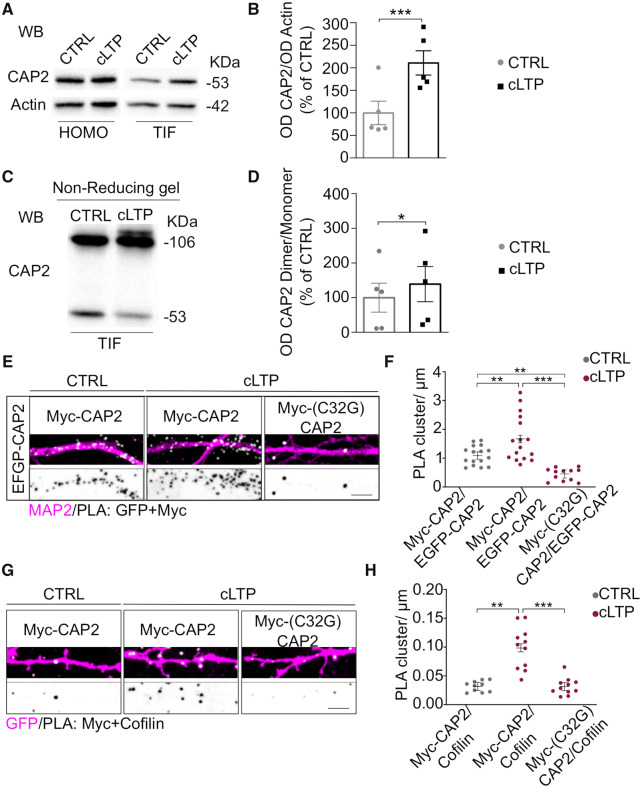
The importance of protein self-association has been previously demonstrated for Srv2/CAP, the yeast homologous form of CAP2 (Hubberstey and Mottillo, 2002; Ono, 2013). Indeed, Srv2/CAP complex catalytically accelerates cofilin-dependent actin turnover (Balcer et al., 2003). The hexameric CAP molecules interact with the pointed ends of cofilin-decorated filaments promoting the disassembly (Chaudhry et al., 2013; Kotila et al., 2019; Shekhar et al., 2019). In vitro experiments have shown that the N-terminal domain of CAP2 forms tetramers have a high affinity for F-actin (Purde et al., 2019). Therefore, we wondered whether the mammalian CAP2 is also able to dimerize and oligomerize in a cellular system. Different experimental approaches were used to test this hypothesis. Either Myc-CAP2 or EGFP-CAP2 were transfected in COS-7 cells and we observed that both constructs are able to form aggregates in the cytoplasm, independently of the tag (Fig. 4A, lower panels). The transfection of both CAP2 constructs in COS-7 cells demonstrates that Myc-CAP2 and EGFP-CAP2 perfectly colocalize in cytoplasmic clusters, suggesting that CAP2 can self-associate (Fig. 4A). Coimmunoprecipitation assays performed from lysates of COS-7 cells cotransfected with Myc-CAP2 and EGFP-CAP2 confirmed that Myc-CAP2 interacts with EGFP-CAP2 (Fig. 4B).
Figure 4.
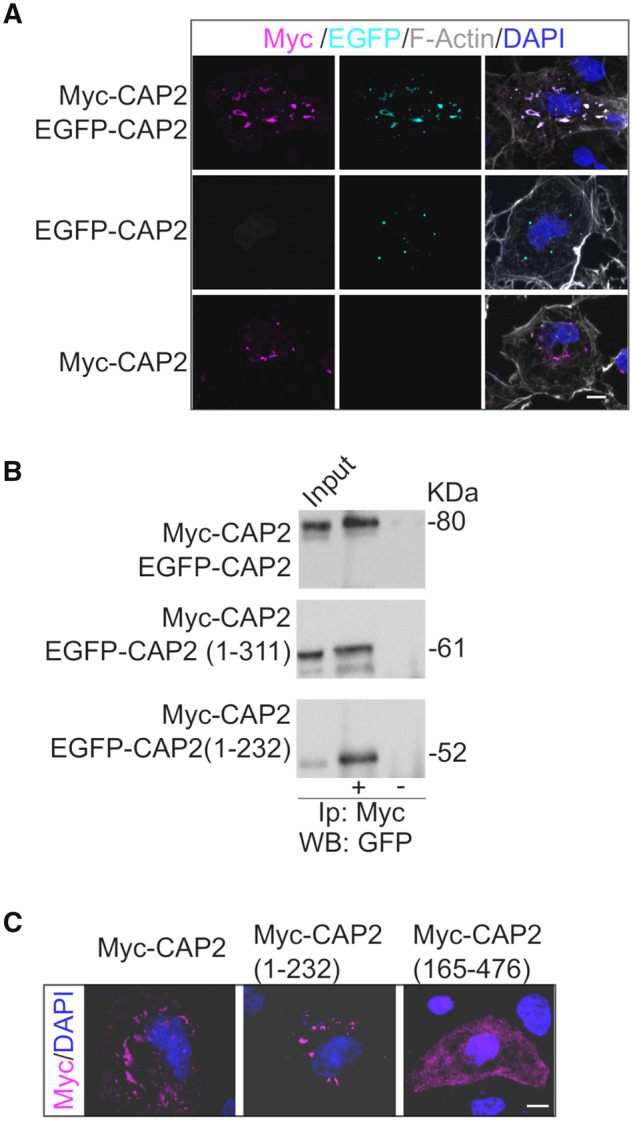
CAP2 self-associates. (A) Representative confocal images of COS-7 cells co-transfected with either Myc-CAP2 (magenta) or EGFP-CAP2 (cyan) or both constructs show the capability of CAP2 to form clusters. Scale bar = 10 μm. (B) Homogenate of HEK293 cells transfected with Myc-CAP2 and either EGFP-CAP2 or EGFP-CAP2(1-232) or EGFP-CAP2(1-311) deletion mutants were immunoprecipitated (Ip) with anti-Myc antibody and the WB analysis carried out with anti-GFP antibody. The sequence 1-232 of CAP2 is sufficient for CAP2 self-association. Full uncropped blots are available in the Supplementary material. (C) Representative confocal images of COS-7 cells transfected with Myc-CAP2, Myc-CAP2(1-232) and Myc-CAP2(165-476) reveal that the region 1-232 is sufficient for clusters formation while the deletion of the region 1-164 completely changes the protein intracellular pattern. Scale bar = 5 μm.
The full three-dimensional (3D) structure of the human CAP2 is not available, but several studies have underlined the multi-domains organization of the protein (Ono, 2013). The only structural information that can be useful to propose a possible 3D organization of the CAP2 dimer comes from the X-ray structure of the Helical Folded Domain (HFD) of the CAP homolog from Dictyostelium discoideum (Yusof et al., 2004). In details, this region is composed of an initial Coiled-coil region followed by a stable bundle of six antiparallel α-helices (HFD domain) (Supplementary Fig. 3A). This region (i.e. Coiled-coil and HFD) is not only conserved in CAP1 and CAP2 from human and mouse but also in the homologous CAP protein from Dictyostelium discoideum (Supplementary Fig. 3B). This evidence strengthens the idea that the dimerization/oligomerization of CAP2 is crucial for its cellular functions, such as the interaction with other protein partners (i.e. actin and cofilin).
To better characterize the region of CAP2 responsible for the self-association, we transfected COS-7 cells with Myc-CAP2 and either EGFP-CAP2 or the deletion mutant EGFP-CAP2(1-311) deleted of the CARP domain or the EGFP-CAP2(1-232) mutant lacking of the CARP domain, the proline rich sequences and the WH2 region (Peche et al., 2013) (Supplementary Fig. 3A). Coimmunoprecipitation experiments revealed that CAP2 self-association depends on its N-terminal region, similarly to what previously described for Srv2/CAP (Quintero-Monzon et al., 2009) (Fig. 4B). To investigate the involvement of N-terminus in CAP2-self-association, we performed imaging analysis in COS-7. The deletion mutant lacking the coiled-coil region and HFD, Myc-CAP2(165-476), displayed a diffuse staining throughout the cell, while Myc-CAP2 and Myc-CAP2(1-232) showed a clustered cytoplasmic distribution (Fig. 4C). In addition, coimmunoprecipitation assays confirmed that the deletion of the region 1–164 significantly reduces the self-association (Fig. 5A and B).
Figure 5.
CAP2 forms dimers through Cys32. (A) Co-immunoprecipitation assays carried out from homogenate of HEK293 cells transfected with EGFP-CAP2 and either Myc-CAP2 or Myc-CAP2(165-476). The samples derive from the same experiment and gels/blots were processed in parallel. Full uncropped blots are available in the Supplementary material. (B) The analysis of the optical density (OD) reveals that the lack of the sequence 1-164 of CAP2 significantly reduces CAP2 self-association [Myc-CAP2(165-476)/EGFP-CAP2 versus Myc-CAP2/EGFP-CAP2, paired t-test P = 0.036, n = 4 independent experiments]. (C) Representative WB of homogenates of rat brain hippocampus loaded onto a non-denaturating gel, without (left side) or with dithiothreitol (DTT) treatment (right side), as indicated. The band corresponding to the dimeric form of CAP2 is eliminated by DTT treatment. Full uncropped blots are available in the supplementary material. (D) Representative WB of homogenates of HEK293 cells expressing Myc-CAP2, Myc-CAP2(165-476) or Myc-(C32G)CAP2. The deletion of the 1-164 region or the single point mutation Cys32 to Gly avoids the capability to form dimers. The samples derive from the same experiment and gels/blots were processed in parallel. Full uncropped blots are available in the supplementary material. (E) Representative confocal images of PLA experiments showing proximity between EGFP-CAP2 and either Myc-CAP2 or Myc-(C32G)CAP2 (white) along MAP2 positive dendrites (magenta); lower panels, inverted images of PLA signal (black), scale bar = 5 μm. (F) Graphs show the quantification of the PLA clusters/μm (EGFP-CAP2/Myc-CAP2 = 1.366 ± 0.150, EGFP-CAP2/Myc-(C32G)CAP2 = 0.566 ± 0.117, t-test from mixed-effects model: ***P < 0.0001; n EGFP-CAP2/Myc-CAP2 = 16 neurons, n EGFP-CAP2/Myc-(C32G)CAP2 = 16 neurons).
It has been recently demonstrated that CAP1 homodimerization depends on the formation of a disulfide bond that requires the Cys29 of CAP1 (Liu et al., 2018). Therefore, we verified the presence of a disulfide cross-linked CAP2 dimer by loading an aliquot of hippocampal homogenate onto a non-reducing SDS-PAGE. As shown in Fig. 5C, both a band corresponding to CAP2 monomer at 58 kDa and a species of CAP2 with an apparent molecular weight of approximately double the CAP2 monomer (116 kDa) were detected. Importantly, the band corresponding to CAP2 dimer was eliminated by DTT, consistent with a disulfide cross-linked CAP2 dimer (Fig. 5C). To exclude the possibility that intra- and inter-molecular disulfide bonds were non-specifically formed during the lysis process of the tissue, we carried-out the homogenization of rat hippocampus using a buffer containing iodoacetamide to protect free sulfhydryl groups. The samples were loaded onto a non-reducing SDS-PAGE and the WB analysis revealed the presence of both the CAP2 monomer and the CAP2 DTT-sensitive dimer (Supplementary Fig. 3C), thus confirming the existence of specific CAP2 disulfide cross-linked dimers in the hippocampus.
To identify the region of CAP2 responsible for dimerization, we transfected COS-7 cells with either Myc-CAP2 or a deletion mutant lacking of the N-terminal region [Myc-CAP2(165-476)] and the lysates were loaded onto a non-reducing SDS-PAGE. As shown in Fig.5D, CAP2 monomer and dimer were detectable when Myc-CAP2 was expressed, while the deletion of the N-terminal region [Myc-CAP2(165-476)] abolishes the CAP2 capability to form dimers, demonstrating that this is the domain involved in the dimerization. Interestingly, only one Cys residue is present in the CAP2 region spanning from amino acid 1 to 164. Notably, the Myc-CAP2 mutant carrying the mutation of the Cys32 to Gly [Myc-(C32G)CAP2] is not able to dimerize in non-reducing SDS-PAGE (Fig. 5D). To strengthen these data, we performed a PLA in hippocampal neurons transfected with EGFP-CAP2 and either Myc-CAP2 or Myc-(C32G)CAP2. In neurons expressing EGFP-CAP2 and Myc-CAP2, a large number of PLA signals were detected when the two antibodies anti-GFP and anti-Myc were used, indicating that these two CAP2 constructs are in close proximity to each other along MAP2-positive dendrites (Fig. 5E). In neurons transfected with EGFP-CAP2 and the mutant Myc-(C32G)CAP2, the density of PLA signals significantly decreased when compared to cells overexpressing EGFP-CAP2 and Myc-CAP2 (Fig. 5F). For control experiments, only the anti-Myc primary antibody was used and no PLA signal was generated (Supplementary Fig. 3D). Overall, these findings suggest that the Cys32 is fundamental for the formation of CAP2 dimers and for CAP2 clustering in neurons.
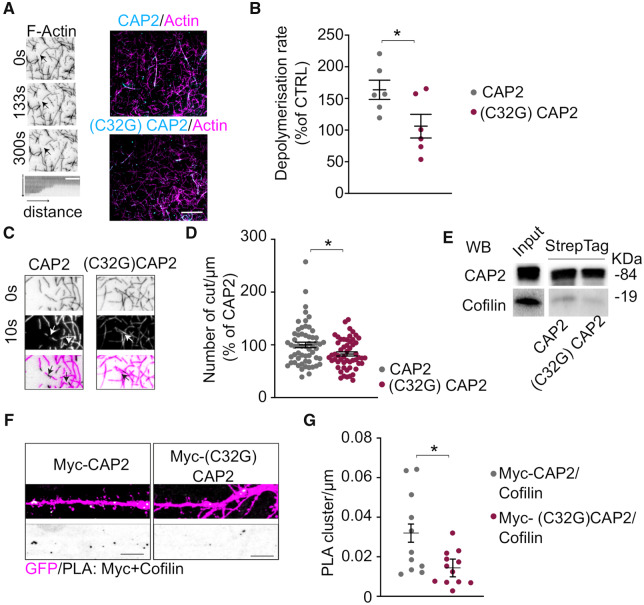
To investigate the direct effect of monomeric and dimeric CAP2 on the assembly and stability of individual actin filaments, we employed in vitro dual-colour total internal reflection fluorescence microscopy. In vitro polymerization of a mixture of unconjugated and Alexa-Fluor-561-conjugated actin occurs spontaneously at certain actin concentrations until it reaches equilibrium of G- and F-actin. We then added the purified recombinant CAP2 and (C32G)CAP2 proteins and determined the rate of F-actin depolymerization caused by G-actin washout, using kymograph analysis. The data showed a clear increase of actin depolymerization in the presence of CAP2 that was prevented by the mutation of Cys32 to glycine (Fig. 6A and B). In addition, the analysis of the filaments showed that the CAP2-dependent increase in actin depolymerization rate was related to an increase in the severing activity (Fig. 6C and D). Non-reducing SDS-PAGE assays of the recombinant proteins confirmed that CAP2, but not the mutant (C32G)CAP2, forms dimers (Supplementary Fig. 4A). Remarkably, WB analysis revealed that cofilin was co-purified with CAP2, but in a smaller extent with the mutant (C32G)CAP2 (Fig. 6E).
Figure 6.
Cys32-dependent CAP2 dimerization is relevant for association with cofilin and for controlling actin depolymerization. (A) Spontaneous in vitro depolymerization of F-actin-Alexa Fluor 568 visualized by total internal reflection fluorescence microscopy. Left panels, actin filament undergoing depolymerization upon G-actin wash-out, arrowheads indicate the barbed ends of the filament. Right panels, overlay images show the actin filaments (magenta) and either CAP2 or (C32G)CAP2 mutant (cyan). Scale bar = 5 μm. (B) Kymographs analysis of F-actin depolymerization rate indicates that CAP2 increases the depolymerization rate of F-actin while the mutant (C32G)CAP2 abolished this effect [(C32G)CAP2 versus CAP2, unpaired t-test: *P = 0.038, n = 6 independent experiments]. (C) Representative images of the analysis of the severing activity show an actin filament undergoing depolymerization in presence of either CAP2 or (C32G)CAP2 and arrowheads indicate the presence of cutting sites. (D) The number of cuts/μm significantly decreases in presence of (C32G)CAP2 compared to CAP2 [(C32G)CAP2 versus CAP2, unpaired t-test: *P = 0.01, n (C32G)CAP2 = 54 filaments, n CAP2 = 58 filaments]. (E) WB of the Streptag CAP2 and (C32G)CAP2 fusion proteins shows that the mutation of C32 to G reduces CAP2 capability to bind cofilin. Full uncropped blots are available in the supplementary material. (F) Images showing PLA signal between cofilin and either Myc-CAP2 or Myc-(C32G)CAP2 (white) along MAP2 positive dendrites (magenta). Scale bar = 5 μm. Lower panels, inverted images of PLA signal (black). (G) The mutation of Cys32 significantly reduces association with cofilin, as shown in the graphs reporting the quantification of the PLA clusters/μm (Myc-CAP2 = 0.0320 ± 0.005; Myc-(C32G)CAP2 = 0.0144 ± 0.005, unpaired t-test from fixed-effects ordinary least squares regression model: *P = 0.0146, Myc-CAP2 n = 11 neurons, Myc-(C32G)CAP2 n = 12 neurons).
To confirm that the lack of Cys32 affects CAP2 association with cofilin, PLA assays were performed in primary hippocampal neurons transfected with either Myc-CAP2 or Myc-(C32G)CAP2. The analysis of the PLA signals confirms a significant decrease in the association between Myc-(C32G)CAP2 and cofilin when compared to the binding of Myc-CAP2 to cofilin (Fig. 6F and G). For control experiments, when anti-Myc primary antibody or anti-cofilin antibody alone was used, no PLA signal was generated (Supplementary Fig. 4B). As control, we verified that the association of CAP2 with actin was not affected by the mutation of the Cys32 to Gly (Supplementary Fig. 4C). Therefore, the lack of the disulfide bond involving CAP2 Cys32 abolishes the CAP2 dimerization, reduces the cofilin binding and, thereby, prevents the increase in actin depolymerization rate in total internal reflection fluorescence microscopy experiments.
Long-term potentiation triggers CAP2 dimer formation as a crucial step for structural plasticity
Given the key role of cofilin-mediated actin dynamics in the structural remodelling of spines during activity-dependent synaptic plasticity phenomena (Bosch et al., 2014), we tested whether either LTP or LTD could modulate CAP2 dimerization, and consequently its association with cofilin.
First, we investigated CAP2 homodimer localization in the postsynaptic compartment. WB analyses showed that CAP2 dimer levels are higher in the postsynaptic TIF compared to the total homogenate (Supplementary Fig. 5A) in both rat and human hippocampus, thus confirming the existence of CAP2 dimer in human and revealing the enrichment of the CAP2 dimer in this subcellular compartment.
Second, we asked whether LTP regulates CAP2 synaptic availability in the dendritic spines. To this, we induced cLTP in hippocampal cultures that results in prolonged NMDA receptor-dependent LTP (Marcello et al., 2013). Thirty minutes after cLTP induction, we purified TIF from control and cLTP-treated hippocampal neurons. WB analysis revealed that cLTP elicited an increase in the CAP2 levels in the TIF fraction compared to control neurons, without affecting the total levels of the protein (Fig. 7A and B). Notably, cLTP stimulation caused also a significant increase in CAP2 dimer levels in TIF (Fig. 7C and D). Secondly, to evaluate whether these effects could be ascribed to the CAP2 Cys32-dependent dimerization, we analysed the PLA signal upon cLTP induction in hippocampal neurons transfected with EGFP-CAP2 and either Myc-CAP2 or Myc-(C32G)CAP2. The cLTP induction significantly increased the PLA signal in neurons transfected with EGFP-CAP2 and Myc-CAP2, confirming the LTP-triggered augment in CAP2 self-association. On the other hand, no modifications in PLA signals upon LTP induction were detected when EGFP-CAP2 and Myc-(C32G)CAP2 were expressed (Fig. 7E and F). This effect is specifically related to LTP, since both PLA and WB analysis of the synaptic fraction revealed no variations in CAP2 synaptic levels and in dimer formation after cLTD induction (Supplementary Fig. 5E–G).
Figure 7.
LTP promotes CAP2 synaptic localization, dimer formation and association with cofilin. (A) Representative WB of CAP2 and actin in Homo and TIF samples obtained from control and cLTP-treated neurons, 30 min after the cLTP induction. Non-cropped blots are shown in Supplementary Fig. 5B and C. (B) cLTP promotes CAP2 enrichment in the TIF (cLTP versus CTRL, paired t-test: ***P = 0.0008, n = 5 independent experiments). (C) Representative WB of CAP2 dimer and monomer in TIF samples obtained from control and cLTP-treated neurons, 30 min after the cLTP induction Non-cropped blots are shown in Supplementary Fig. 5D. (D) cLTP increases CAP2 dimer/monomer ratio in the TIF fraction of cLTP-treated neurons (cLTP versus CTRL, paired t-test: *P = 0.042, n = 5 independent experiments). (E) PLA representative images showing the proximity between EGFP-CAP2 and either Myc-CAP2 or Myc-(C32G)CAP2 (white) along MAP2 positive dendrites (magenta) 30 min after cLTP induction. Lower panels, inverted images of PLA signal (black), scale bar = 5 μm. (F) Graphs show the quantification of the PLA clusters/μm (EGFP-CAP2/Myc-CAP2-CTRL = 1.100 ± 0.128, EGFP-CAP2/Myc-CAP2-cLTP = 1.679 ± 0.128, EGFP-CAP2/Myc-(C32G)CAP2-cLTP = 0.461 ± 0.148, linear mixed-effects model, false discovery rate adjustment for multiple comparisons for the contrasts of interest: **P = 0.00129, ***P < 0.0001, P values for all the multiple comparisons are reported in Table 4; EGFP-CAP2/Myc-CAP2-CTRL n = 16 neurons, EGFP-CAP2/Myc-CAP2-cLTP n = 16 neurons, EGFP-CAP2/Myc-(C32G)CAP2-cLTP n = 12 neurons). (G) PLA images revealing the closeness between cofilin and either Myc-CAP2 or Myc-(C32G)CAP2 (white) along dendrites of GFP transfected hippocampal neurons (magenta) after cLTP induction. Lower panels, inverted images of PLA signal (black), scale bar = 5 μm. (H) Graphs show the quantification of the PLA clusters/μm (MycCAP2/cofilin-CTRL = 0.0317 ± 0.0073, MycCAP2/cofilin-cLTP = 0.0984 ± 0.0070, Myc-(C32G)-CAP2/cofilin-cLTP = 0.0315 ± 0.0070, linear mixed-effects model, false discovery rate adjustment for multiple comparisons for the contrasts of interest: ***P < 0.0001, P values for all the multiple comparisons are reported in Table 4; MycCAP2/cofilin-CTRL n = 10 neurons, MycCAP2/cofilin-cLTP n = 11 neurons, Myc-(C32G)CAP2/cofilin-cLTP n = 11 neurons).
To determine if the increase in LTP-driven CAP2 dimer formation affects the association with cofilin, we performed PLA assays after cLTP induction in neurons transfected with either Myc-CAP2 or Myc-(C32G)CAP2. As shown in Fig. 7G and H, LTP fosters the association of cofilin with CAP2. Likewise, we detected a significant increase in the association between endogenous CAP2 and cofilin upon LTP induction, indicating that this effect was not related to the overexpression of Myc-CAP2 construct (Supplementary Fig. 5H). The Cys32-dependent self-association of CAP2 is required because the Myc-(C32G)CAP2 mutant prevents the LTP-induced increase in CAP2/cofilin interaction (Fig. 7G and H).
Overall, these findings suggest that LTP triggers CAP2 translocation to the spine and the formation of Cys32-dependent dimers, thus promoting the association of CAP2 with cofilin.
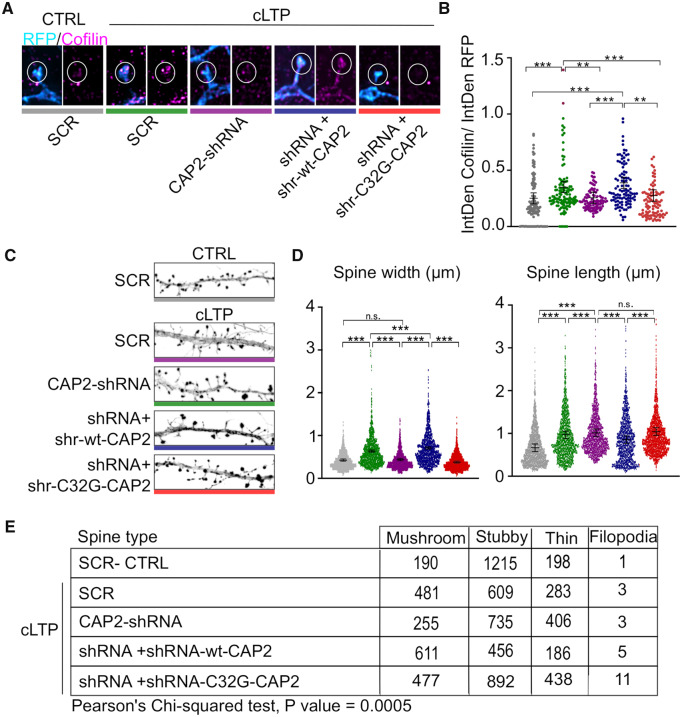
In the initial phase of LTP, the actin cytoskeleton is rapidly remodelled while active cofilin is transported to the spine (Bosch et al., 2014). In light of this observation, we attempted to determine whether the hampering CAP2 dimerization impairs LTP-driven enrichment of cofilin in the spine. To this, we induced cLTP in primary hippocampal neurons transfected with either CAP2-shRNA or the corresponding control scramble sequence. These constructs express the fluorescent protein mCherry under the control of the neuron-specific Ca2+/calmodulin-dependent protein kinase II promoter. To assess the magnitude of accumulation, we took advantage of structured illumination microscopy to calculate the relative concentration of cofilin in the spine by dividing cofilin intensity by mCherry intensity (Bosch et al., 2014). As previously demonstrated (Bosch et al., 2014), after 30 min from cLTP induction, cofilin is highly enriched in the spines of neurons transfected with the scramble sequence, while the down-regulation of CAP2 prevents the LTP-triggered translocation of cofilin in the spine. To assess whether this effect could be specifically ascribed to CAP2 and to determine the role of the Cys32, we used shr-wt-CAP2 as a rescue and we generated and validated the mutant shr-C32G-CAP2 that was insensitive to the CAP2-shRNA (Supplementary Fig. 5I). Importantly, the transfection of the shRNA-resistant CAP2 construct (shr-wt-CAP2), but not the expression of shr-C32G-CAP2, restores the LTP-driven translocation of cofilin to the spines (Fig. 8A and B). These data demonstrate that CAP2 and the Cys32-dependent dimerization of the protein are essential for the LTP-triggered cofilin trafficking to spines.
Figure 8.
Cys32-dependent CAP2 dimer is required for cofilin translocation and for structural plasticity changes. (A) Structured illumination microscopy images of cofilin (magenta) and RFP (cyan) staining of primary hippocampal neurons transfected with vectors expressing RFP and either scramble sequence (SCR) or CAP2-shRNA or CAP2-shRNA + shr-wt-CAP2 or CAP2-shRNA + shr-C32G-CAP2, 30 min after cLTP induction. Scale bar = 2 μm. (B) Graphs show the quantification of the ratio between the integrated density (IntDen) of cofilin and RFP staining in dendritic spines (SCR-CTRL = 0.257 ± 0.049, SCR-cLTP = 0.357 ± 0.049, CAP2-shRNA-cLTP = 0.263 ± 0.050, CAP2-shRNA + shr-wt-CAP2-cLTP = 0.396 ± 0.050, CAP2-shRNA + shr-C32G-CAP2-cLTP = 0.282 ± 0.050, linear mixed-effects model, false discovery rate adjustment for multiple comparisons for the contrasts of interest: *** P < 0.0001; ** P < 0.001, SCR-CTRL n = 107 spines, SCR-cLTP n = 120 spines, CAP2-shRNA-cLTP n = 94 spines, CAP2-shRNA + shr-wt-CAP2-cLTP n = 104 spines, CAP2-shRNA + shr-C32G-CAP2-cLTP n = 96 spines). (C) Confocal images of RFP staining of primary hippocampal neurons transfected with vectors expressing RFP and either SCR or CAP2-shRNA or CAP2-shRNA + shr-wt-CAP2 or CAP2-shRNA + shr-C32G-CAP2, 30 min after cLTP induction; scale bar = 5 μm. (D) Graphs show the quantification of dendritic spine head length and width (length, SCR-CTRL = 0.672 ± 0.089 μm, SCR-cLTP = 0.971 ± 0.089 μm, CAP2-shRNA-cLTP = 1.019 ± 0.089 μm, CAP2-shRNA + shr-wt-CAP2-cLTP = 0.860 ± 0.089 μm, CAP2-shRNA + shr-C32G-CAP2-cLTP = 1.039 ± 0.089 μm; width, SCR-CTRL = 0.428 ± 0.026 μm, SCR-cLTP = 0.638 ± 0.026 μm, CAP2-shRNA-cLTP = 0.440 ± 0.026 μm, CAP2-shRNA + shr-wt-CAP2-cLTP = 0.711 ± 0.026 μm, CAP2-shRNA + shr-C32G-CAP2-cLTP = 0.381 ± 0.026 μm; linear mixed-effects model, false discovery rate adjustment for multiple comparisons for the contrasts of interest: *** P < 0.0001, n.s. non significant, P values for all the multiple comparisons are reported in Table 4; SCR-CTRL n = 1604 spines, SCR-cLTP n = 1376 spines, CAP2-shRNA-cLTP n = 1399 spines, CAP2-shRNA + shr-wt-CAP2-cLTP n = 1258 spines, CAP2-shRNA + shr-(C32G)-CAP2-cLTP n = 1818 spines). (E) The joint distribution of dendritic spine type and the combination of genetic modification/treatment is represented in the table together with the P value of the Chi-squared test of independence.
Since cofilin translocation to spines during LTP is required for actin cytoskeleton dynamics and spine enlargement, we verified whether the loss of Cys32 and, thereby, of CAP2 dimerization impairs LTP-induced spine remodelling. An accurate analysis of dendritic spine morphology demonstrated that cLTP induces the expected spine head enlargement in control conditions, but not in neurons transfected with CAP2-shRNA (Fig. 8C and D). cLTP triggers a significant increase in spine length that is not restored to control levels by CAP2 down-regulation (Fig. 8C and D). The expression of the shr-wt-CAP2, but not of the mutant shr-C32G-CAP2, slightly potentiates the LTP-induced increase in spine head width (Fig. 8C and D). Regarding the spine length, the mutant shr-C32G-CAP2 does not rescue the effect of CAP2 down-regulation in cLTP-treated neurons, while shr-wt-CAP2 expression reduces the cLTP-triggered increase in spine length (Fig. 8C and D). The analysis of the different spines categories revealed the presence of a significant association between spines populations and the combination of cLTP treatment and manipulation of CAP2 expression (Fig. 8E), showing the importance of CAP2 in LTP-triggered modifications in spine morphology.
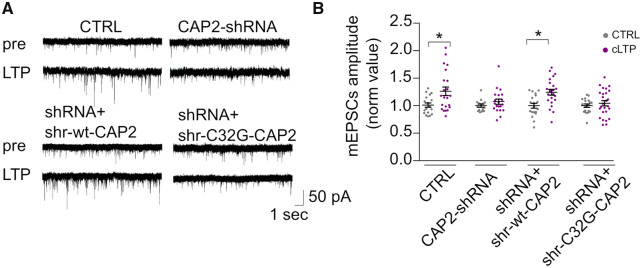
Furthermore, taking advantage of electrophysiological recordings of mEPSCs, we addressed the functional impact of CAP2 down-regulation on hippocampal plasticity by induction of cLTP. As indicated in Fig. 9A and B, cLTP mediates a significant increment in mEPSCs amplitude of excitatory synaptic events only in control cultures whereas no potentiation occurs in CAP2-shRNA neurons. Similarly, cultures transfected with the shr-C32G-CAP2 mutants do not display any potentiation that is rescued only in the shRNA+shr-wt-CAP2 control group (Fig. 9A and B). Taken together, these results suggest that synaptic plasticity is affected by CAP2 and that mutation in the residue Cys32 is sufficient for defective CAP2 activity and LTP induction.
Figure 9.
Cys32-dependent CAP2 dimer is relevant for synaptic transmission potentiation upon LTP induction. (A) Electrophysiological recordings of mEPSCs before and after chemical LTP (cLTP) induction in 15 DIV hippocampal neurons. Representative mEPSC traces. (B) Differently from CTRL and shr-wt-CAP2 cells, CAP2-shRNA and CAP2-shRNA+shr-C32G-CAP2 neurons are unable to undergo cLTP. mEPSCs amplitude (normalized values): CTRL = 1 ± 0.03 (n = 18) versus cLTP: 1.26 ± 0.07 (n = 22); CAP2-shRNA: 1 ± 0.02 (n = 15) versus cLTP: 1 ± 0.02 (n = 20); shRNA+shr-wt-CAP2: 1 ± 0.04 (n = 19) versus cLTP: 1.25 ± 0.05 (n = 23); shRNA+shr-C32G-CAP2: 1 ± 0.031 (n = 18) versus cLTP: 1 ± 0.04 (n = 26). One-way ANOVA, Holm-Sidak's multiple comparisons test: P < 0.0001.
CAP2 dimer association with cofilin is altered in Alzheimer's disease
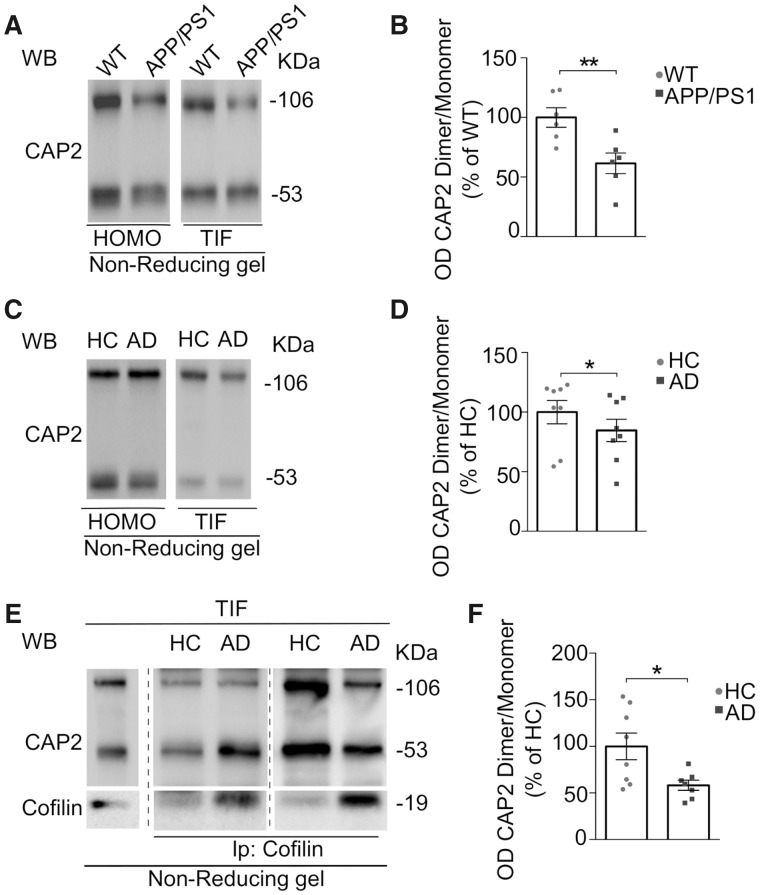
Since Cys32-dependent CAP2 homodimerization is crucial for the regulation of dendritic spine morphology and synaptic plasticity via cofilin binding, we wondered whether alterations of this mechanism were detectable in Alzheimer's disease synapses, where we found an aberrant increased level of cofilin along with a significant reduction in CAP2 (Fig. 1).
The analysis of the CAP2 dimer/monomer ratio in APP/PS1 and WT mice hippocampi revealed a significant decrease of dimer levels in the postsynaptic fraction, but not in the total homogenate, of APP/PS1 mice when compared to WT mice (Fig. 10A and B, Supplementary Fig. 6A). We examined the CAP2 homodimer formation in Alzheimer's disease patients' hippocampi and the quantitative results showed a significant reduction in CAP2 dimer/monomer ratio specifically in the synapse of Alzheimer's disease patients compared to HC, while no changes were detected in the total homogenate (Fig. 10C and D, Supplementary Fig. 6B).
Figure 10.
Cys32-dependent CAP2 dimer levels and CAP2 association to cofilin are affected in Alzheimer's disease synapses. (A) Representative WB of CAP2 dimer level in the HOMO and TIF hippocampal fraction of APP/PS1 and WT animals. Full uncropped blots are available in the supplementary material. (B) Quantitative analysis shows the decreased ratio of CAP2 dimer OD on the CAP2 monomer OD of APP/PS1 mice compared to WT (TIF, APP/PS1 versus WT, paired t-test: **P = 0.003, n WT = 6 mice, n APP/PS1 = 6 mice). (C) Representative WB showing the levels of CAP2 monomer and dimer in the HOMO and TIF samples of Alzheimer's disease (AD) and HC hippocampal samples. Full uncropped blots are available in the Supplementary material. (D) Quantitative analysis shows that CAP2 dimer/monomer ratio is reduced in AD patients TIF but not in HOMO (TIF, AD versus HC paired t-test: *P = 0.014, n HC = 8, n AD = 8). (E) TIF from hippocampi of HC and AD patients were immunoprecipitated (Ip) with anti-cofilin antibody and CAP2 coprecipitation evaluated in non-denaturating condition. Samples derive from the same experiment and gels/blots were processed in parallel. Full uncropped blots are available in the Supplementary material. (F) Cofilin binding to CAP2 is altered in TIF fraction of AD compared with HC (CAP2 dimer/monomer, AD versus HC, unpaired t-test: *P = 0.023, n HC = 8, n AD = 7).
Since CAP2 dimer formation is relevant for CAP2 association with cofilin and for cofilin localization in the postsynaptic compartment, we verified whether cofilin binding to CAP2 monomer and dimer is affected in Alzheimer's disease. Given that cofilin is aberrantly increased in the Alzheimer's disease hippocampi postsynapse, we focused on TIF samples obtained from HC and Alzheimer's disease patients. TIF samples were immunoprecipitated with cofilin antibody and loaded onto a non-reducing gel. WB analysis carried out with a CAP2 antibody revealed that cofilin specifically precipitates both CAP2 monomer and dimer in human postsynaptic compartment (Supplementary Fig. 6C), confirming CAP2 dimer and monomer as binding partners of cofilin in human. The quantitative analysis of coimmunoprecipitation experiments revealed that the ratio of CAP2 dimer/monomer bound to cofilin is significantly reduced in Alzheimer's Disease patients when compared to HC (Fig. 10E and F; Supplementary Fig. 6D), that is in line with the availability of the CAP2 dimer/monomer in the postsynaptic compartment.
In light of the above, we tested whether the association between single nucleotide polymorphisms of CAP2 Cys32 (rs750613178 variant) and Alzheimer's disease onset might occur. To this aim, a preliminary screening experiment of genetic polymorphism has been performed on 70 Alzheimer's disease patients (age = 68.2 ± 16.3; female% = 76.8). No genetic variations of CAP2 Cys32 were detected in Alzheimer's disease patients, suggesting that other cellular mechanisms might be responsible for the decreased synaptic dimer levels in Alzheimer's disease.
Discussion
A major question in Alzheimer's disease pathogenesis is to define the mechanism guiding alteration of synaptic plasticity and spine dysmorphogenesis. Here, we show that CAP2 is able to control cofilin synaptic availability during activity-dependent plasticity phenomena. Remarkably, this mechanism is specifically altered in the hippocampus of Alzheimer's disease patients and of an Alzheimer's disease mouse model.
CAP2 has been reported as a multi-domain actin binding protein, expressed in specific tissues, including the brain (Peche et al., 2007; Kumar et al., 2016). Our study provides a detailed characterization of CAP2 in neurons bringing into focus conclusive evidences for both CAP2 localization in the postsynaptic compartment and its pivotal role in synaptic plasticity phenomena. This has been demonstrated with different approaches. First, stimulated emission depletion nanoscopy and biochemical fractionation approaches showed that CAP2 is localized in the postsynaptic compartment, in the region just underneath the PSD where F-actin is enriched (Korobova and Svitkina, 2010). Secondly, we observed that the down-regulation of CAP2 impairs neuronal architecture and spine shape, together with a decrease in synaptic excitatory transmission. Such alterations are translated into an impairment of plasticity phenomena.
Furthermore, the down-regulation of CAP2 decreases the levels of BDNF, a neurotrophin important for neuronal survival and synaptic plasticity (Lynch et al., 2008) and reduced early in the course of the disease (Kaminari et al., 2017). Its delivery could rescue memory deficits dendrite outgrowth and spine loss (de Pins et al., 2019). Therefore, we can hypothesize a role for CAP2 in the alterations of BDNF in Alzheimer's disease and in the consequent impairment of plasticity phenomena.
It has been shown that CAPs and cofilin synergize to depolymerize actin filaments (Chaudhry et al., 2013; Kotila et al., 2019; Shekhar et al., 2019), highlighting the critical role of this protein family in actin-dependent processes. In line with these results, total internal reflection fluorescence experiments showed that CAP2 increases the actin depolymerization rate. Considering that an efficient actin turnover in spines is required for functional LTP (Okamoto et al., 2004), we observed that the down-regulation of CAP2 leads to the lack of both potentiation and spine head enlargement expected upon cLTP induction.
Why is CAP2 critical for plasticity phenomena? CAP2 has been reported as a binding partner of cofilin (Kumar et al., 2016), a master regulator of postsynaptic actin dynamics and structural plasticity (Rust, 2015a). Upon LTP induction, cofilin activity is first increased and subsequently decreased, allowing first for actin remodelling and then for an increase in F-actin and associated spine enlargement (Fukazawa et al., 2003; Chen et al., 2007; Gu et al., 2010). Even though cofilin phosphorylation is considered one of the major mechanisms controlling its activity (Van Troys et al., 2008; Bernstein and Bamburg, 2010; Rust, 2015a), the regulation of its availability and retention in spines could also contribute to cofilin-dependent sculpturing of the cytoskeleton. Cofilin indeed accumulates in regions of the spine that contain a dynamic F-actin network responsible for spine morphological changes during synaptic plasticity (Racz and Weinberg, 2006). Importantly, cofilin is translocated to the spine upon LTP induction (Bosch et al., 2014) and our findings show that CAP2 down-regulation blocks the LTP-triggered enrichment of cofilin in spines. The impaired cofilin localization in spines and, thereby, the defects in structural and functional synaptic plasticity could be rescued by cotransfection of the cells with an shRNA-resistant CAP2 construct, confirming that the described CAP2-knockdown phenotypes indeed resulted from diminished CAP2 expression.
Furthermore, we also addressed the key question regarding the molecular determinants relevant for CAP2/cofilin association. A careful mapping of CAP2 structure was carried out and, by using different approaches, we showed that the N-terminal domain of CAP2 is responsible for its self-association, as previously demonstrated for Svr2/CAP protein (Quintero-Monzon et al., 2009). In particular, in this region we identified the Cys32 as the amino acid involved in the formation of disulfide cross-linked CAP2 dimers. The mutation of Cys32 to Gly dramatically reduces, but not completely abolishes, CAP2 self-association and the binding to cofilin, suggesting that CAP2 can still form aggregates capable of interacting with cofilin. However, the lack of Cys32-dependent CAP2 homodimer results in a loss of function of CAP2/cofilin complex on actin depolymerization, highlighting the importance of such disulfide bond for the operational role of CAP2. In fact, the CAP2 construct carrying the mutation Cys32 to Gly significantly attenuates the capability of CAP2 of promoting actin filaments depolymerization in an in vitro assay. A similar molecular pathway has been shown for CAP1, since Cys29-dependent homodimerization of CAP1 affects binding to cofilin and F-actin stability (Liu et al., 2018). Notably, the Cys29 residue of CAP1 is not conserved in human, thus limiting the relevance of CAP1-dependent mechanism and highlighting the role of CAP2 as main regulator of cofilin in human neuronal cells.
To strengthen the critical role of CAP2 homodimer in the spine, we show that it is localized in the postsynaptic compartment and it undergoes a dynamic regulation of its synaptic levels by activity-dependent synaptic plasticity. The LTP-induced increase in CAP2 localization in spines promotes the formation of CAP2 dimers through Cys32, and thereby fosters CAP2 association with cofilin. This event is required for LTP-induced translocation of cofilin to the spine and for structural changes and the potentiation of synaptic transmission underlying LTP expression. Indeed, the WT CAP2 construct, but not the CAP2 mutant carrying the Cys32 mutated to Gly, restores and slightly increases the LTP-triggered enlargement of spine head in CAP2-knockdown neurons. In addition, the structured illumination microscopy analysis of cofilin localization in spines revealed that the CAP2 mutant failed to rescue the expected enrichment of cofilin in spines upon LTP. These data suggest that the increased synaptic availability of CAP2 monomers in the synapse triggers the dimer formation and the downstream events, even though we cannot exclude that post-translational modifications or interacting proteins could affect this pathway. Of note, the induction of LTD does not affect CAP2 localization in the spine and dimer formation, highlighting the specificity of this mechanism for LTP. Further experiments are required in ex vivo systems stimulating specific synaptic afferents, in order to assess input specificity of LTP induction that cannot be evaluated using a global chemical stimulation of the synapses.
Cofilin has been implicated in the pathophysiology of Alzheimer's disease, since cofilin accumulates in neurites close to senile plaques in Alzheimer's disease tissue and Alzheimer's disease mouse models (Bamburg and Bernstein, 2016). However, the mechanisms by which cofilin contributes to Alzheimer's disease pathogenesis remain quite controversial, as either an excessive activation (Kim et al., 2013) or inactivation (Barone et al., 2014; Rush et al., 2018) have been reported in Alzheimer's disease patients. Here, we changed perspective, and bring into focus cofilin localization in spines in Alzheimer's disease, because it is directly regulated by LTP, as well as its association with CAP2 (Bosch et al., 2014). The CAP2 dimer-dependent mechanism that regulates cofilin availability at the synapse is specifically affected in the hippocampus, but not in the SFG, of both Alzheimer's disease patients and APP/PS1 mice. Remarkably, we detected a significant increase in the CSF CAP2 levels in Alzheimer's disease patients but not in frontotemporal dementia patients, indicating the specificity of the alteration for this form of dementia. Accurate early and differential diagnosis across different forms of dementia represents a clinical challenge due to the complexity of neurodegenerative processes and is crucial for establishing prognosis and accessing adequate treatment (Palop et al., 2006; Oxtoby et al., 2017). Our preliminary study could set the stage for further studies aimed at confirming CAP2 as a potential biomarker of hippocampal synaptic dysfunction in Alzheimer's disease.
In the hippocampal synapses of Alzheimer's disease patients and Alzheimer's disease mouse model, we measured a dramatic increase in cofilin levels, along with a reduction in CAP2 synaptic availability, and accordingly a decrease in CAP2 dimer formation at the synapse. Furthermore, we noticed that in Alzheimer's disease patients' hippocampi cofilin precipitates a different pattern of CAP2 monomeric and dimeric forms, suggesting the presence of an ineffective CAP2/cofilin complex in Alzheimer's disease hippocampal synapses that could contribute to the loss of structural plasticity of spines in Alzheimer's disease.
In the current paper, we explored the use of mixed-effects models to fully account for the complex design of multi-level hierarchically structured imaging data (Paternoster et al., 2018); this approach improved our ability to control variability at culture and neuron levels and provided us more precise mean estimates (Lazic, 2010).
Together, our findings describe a novel CAP2-dependent mechanism controlling cofilin synaptic availability and actin turnover, which is closely interdependent to LTP. Furthermore, we show that the main players of this pathway are altered in Alzheimer's disease, adding new pieces to the puzzle in the understanding of the complex and coordinated events leading to early synaptic dysfunction and plasticity alterations in Alzheimer's disease pathogenesis.
Supplementary material
Supplementary material is available at Brain Communications online.
Supplementary Material
Acknowledgements
We thank A. Longhi and E. Zianni for technical assistance and Alessia Mariani, Filippo La Greca, Ramona Stringhi, and Michael Wolf for excellent practical work, Chiara Macchi and Massimiliano Ruscica for advice on ELISA assays. We thank UMIF (UKE, Hamburg) for access to their stimulated emission depletion imaging system. Part of this work was carried out at NOLIMITS, an advanced imaging facility established by the Università degli Studi di Milano. The brain tissues were obtained from the Netherlands Brain Bank, Netherlands Institute for Neuroscience, Amsterdam.
Funding
This project has received funding from the European Union’s Horizon 2020 research and innovation program under the Marie Skłodowska-Curie grant agreement No 676144 (Synaptic Dysfunction in Alzheimer Disease, SyDAD) to M.D.L., from the Italian Ministry of Education, University and Research (MIUR) (PRIN 2015N4FKJ4 to M.D.L., PRIN 2017B9NCSX to E.M., Fondo per il Finanziamento delle Attività Base di Ricerca FFABR18_10 to E.M., MIUR Progetto Eccellenza), from Fondazione Cariplo to E.M. (Grant no. 2018—0511), from AIRAlzh Onlus-COOP Italia (fellowship to S.P.), from Boehringer Ingelheim Fonds (travel grant to S.P.), from an intramural grant of University of Milan to E.M. (Fondo di sviluppo unimi- linea2—PSR2015-1716GRACA02_05_M and PSR2017_DIP_022_03 and PSR2019_EMARC) and to V.E. (PSR2015-1719FBRAV), from the Deutsche Forschungsgemeinschaft (DFG) [Emmy-Noether Programm (MI 1923/1-1) and FOR2419 (MI 1923/2-1 and MI 1923/2-2)] to M.M. and from DFG (SFB1158, project A08) to D.M. This work was supported by MIUR - PON ‘Ricerca e Innovazione’ PerMedNet project (ARS01_01226).
Competing interests
The authors report no competing interests.
Glossary
- AMPA =
α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid
- CAP2 =
cyclase-associated protein 2
- CSF =
cerebrospinal fluid
- F-actin =
filamentous actin
- G-actin =
globular actin
- HC =
healthy controls
- LTD =
long-term depression
- LTP =
long-term potentiation
- PLA =
proximity ligation assay
- PSD-95 =
postsynaptic density protein 95
- PSD =
postsynaptic densities
- rAAV =
recombinant adeno-associated virus
- SFG =
superior frontal gyrus
- shr =
shRNA-resistant
- shRNA =
small hairpin RNA
- SIM =
structured illumination microscopy
- SS =
synaptic stimulation
- TIF =
triton-insoluble fraction
- WB =
western blot
- WT =
wild-type
References
- Andrianantoandro E, Pollard TD. Mechanism of actin filament turnover by severing and nucleation at different concentrations of ADF/cofilin. Mol Cell 2006; 24: 13–23. [DOI] [PubMed] [Google Scholar]
- Balcer HI, Goodman AL, Rodal AA, Smith E, Kugler J, Heuser JE, et al. Coordinated regulation of actin filament turnover by a high-molecular-weight Srv2/CAP complex, cofilin, profilin, and Aip1. Curr Biol 2003; 13: 2159–69. [DOI] [PubMed] [Google Scholar]
- Bamburg JR, Bernstein BW. Actin dynamics and cofilin-actin rods in Alzheimer disease. Cytoskeleton (Hoboken) 2016; 73: 477–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barone E, Mosser S, Fraering PC. Inactivation of brain cofilin-1 by age, Alzheimer's disease and γ-secretase. Biochim Biophys Acta 2014; 1842: 2500–9. [DOI] [PubMed] [Google Scholar]
- Bates D, Mächler M, Bolker B, Walker S. Fitting linear mixed-effects models using lme4. J Stat Soft 2015; 67: 1–48. [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B 1995; 57: 289–300. [Google Scholar]
- Bernstein BW, Bamburg JR. ADF/cofilin: a functional node in cell biology. Trends Cell Biol 2010; 20: 187–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertling E, Hotulainen P, Mattila PK, Matilainen T, Salminen M, Lappalainen P. Cyclase-associated protein 1 (CAP1) promotes cofilin-induced actin dynamics in mammalian nonmuscle cells. Mol Biol Cell 2004; 15: 2324–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blalock EM, Geddes JW, Chen KC, Porter NM, Markesbery WR, Landfield PW. Incipient Alzheimer's disease: microarray correlation analyses reveal major transcriptional and tumor suppressor responses. Proc Natl Acad Sci USA 2004; 101: 2173–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchoin L, Pollard TD. Mechanism of interaction of Acanthamoeba actophorin (ADF/Cofilin) with actin filaments. J Biol Chem 1999; 274: 15538–46. [DOI] [PubMed] [Google Scholar]
- Bosch M, Castro J, Saneyoshi T, Matsuno H, Sur M, Hayashi Y. Structural and molecular remodeling of dendritic spine substructures during long-term potentiation. Neuron 2014; 82: 444–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourne JN, Harris KM. Balancing structure and function at hippocampal dendritic spines. Annu Rev Neurosci 2008; 31: 47–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braak H, Braak E. Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol 1991; 82: 239–59. [DOI] [PubMed] [Google Scholar]
- Chaudhry F, Breitsprecher D, Little K, Sharov G, Sokolova O, Goode BL. Srv2/cyclase-associated protein forms hexameric shurikens that directly catalyze actin filament severing by cofilin. Mol Biol Cell 2013; 24: 31–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LY, Rex CS, Casale MS, Gall CM, Lynch G. Changes in synaptic morphology accompany actin signaling during LTP. J. Neurosci 2007; 27: 5363–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cingolani LA, Goda Y. Actin in action: the interplay between the actin cytoskeleton and synaptic efficacy. Nat Rev Neurosci 2008; 9: 344–56. [DOI] [PubMed] [Google Scholar]
- de Pins B, Cifuentes-Díaz C, Farah AT, López-Molina L, Montalban E, Sancho-Balsells A, et al. Conditional BDNF delivery from astrocytes rescues memory deficits, spine density, and synaptic properties in the 5xFAD mouse model of Alzheimer disease. J Neurosci 2019; 39: 2441–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinamarca MC, Guzzetti F, Karpova A, Lim D, Mitro N, Musardo S, et al. Ring finger protein 10 is a novel synaptonuclear messenger encoding activation of NMDA receptors in hippocampus. Elife 2016; 5: e12430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field J, Ye DZ, Shinde M, Liu F, Schillinger KJ, Lu M, et al. CAP2 in cardiac conduction, sudden cardiac death and eye development. Sci Rep 2015; 5: 17256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisoni GB, Fox NC, Jack CR, Scheltens P, Thompson PM. The clinical use of structural MRI in Alzheimer disease. Nat Rev Neurol 2010; 6: 67–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukazawa Y, Saitoh Y, Ozawa F, Ohta Y, Mizuno K, Inokuchi K. Hippocampal LTP is accompanied by enhanced F-actin content within the dendritic spine that is essential for late LTP maintenance in vivo. Neuron 2003; 38: 447–60. [DOI] [PubMed] [Google Scholar]
- Gardoni F, Bellone C, Cattabeni F, Di Luca M. Protein kinase C activation modulates alpha-calmodulin kinase II binding to NR2A subunit of N-methyl-D-aspartate receptor complex. J Biol Chem 2001; 276: 7609–13. [DOI] [PubMed] [Google Scholar]
- Gu J, Lee CW, Fan Y, Komlos D, Tang X, Sun C, et al. ADF/cofilin-mediated actin dynamics regulate AMPA receptor trafficking during synaptic plasticity. Nat Neurosci 2010; 13: 1208–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriques AG, Oliveira JM, Carvalho LP, da CE, Silva O. Aβ influences cytoskeletal signaling cascades with consequences to Alzheimer's disease. Mol Neurobiol 2015; 52: 1391–407. [DOI] [PubMed] [Google Scholar]
- Hild G, Kalmár L, Kardos R, Nyitrai M, Bugyi B. The other side of the coin: functional and structural versatility of ADF/cofilins. Eur J Cell Biol 2014; 93: 238–51. [DOI] [PubMed] [Google Scholar]
- Holtmaat A, Svoboda K. Experience-dependent structural synaptic plasticity in the mammalian brain. Nat Rev Neurosci 2009; 10: 647–58. [DOI] [PubMed] [Google Scholar]
- Horch HW, Katz LC. BDNF release from single cells elicits local dendritic growth in nearby neurons. Nat Neurosci 2002; 5: 1177–84. [DOI] [PubMed] [Google Scholar]
- Hotulainen P, Hoogenraad CC. Actin in dendritic spines: connecting dynamics to function. J Cell Biol 2010; 189: 619–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotulainen P, Llano O, Smirnov S, Tanhuanpää K, Faix J, Rivera C, et al. Defining mechanisms of actin polymerization and depolymerization during dendritic spine morphogenesis. J Cell Biol 2009; 185: 323–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubberstey AV, Mottillo EP. Cyclase-associated proteins: CAPacity for linking signal transduction and actin polymerization. FASEB J 2002; 16: 487–99. [DOI] [PubMed] [Google Scholar]
- Jankowsky JL, Fadale DJ, Anderson J, Xu GM, Gonzales V, Jenkins NA, et al. Mutant presenilins specifically elevate the levels of the 42 residue beta-amyloid peptide in vivo: evidence for augmentation of a 42-specific gamma secretase. Hum Mol Genet 2004; 13: 159–70. [DOI] [PubMed] [Google Scholar]
- Kaminari A, Giannakas N, Tzinia A, Tsilibary EC. Overexpression of matrix metalloproteinase-9 (MMP-9) rescues insulin-mediated impairment in the 5XFAD model of Alzheimer's disease. Sci Rep 2017; 7: 683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasai H, Fukuda M, Watanabe S, Hayashi-Takagi A, Noguchi J. Structural dynamics of dendritic spines in memory and cognition. Trends Neurosci 2010; 33: 121–9. [DOI] [PubMed] [Google Scholar]
- Kim T, Vidal GS, Djurisic M, William CM, Birnbaum ME, Garcia KC, et al. Human LilrB2 is a β-amyloid receptor and its murine homolog PirB regulates synaptic plasticity in an Alzheimer's model. Science 2013; 341: 1399–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korobova F, Svitkina T. Molecular architecture of synaptic actin cytoskeleton in hippocampal neurons reveals a mechanism of dendritic spine morphogenesis. Mol Biol Cell 2010; 21: 165–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotila T, Wioland H, Enkavi G, Kogan K, Vattulainen I, Jégou A, et al. Mechanism of synergistic actin filament pointed end depolymerization by cyclase-associated protein and cofilin. Nat Commun 2019; 10: 5320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Paeger L, Kosmas K, Kloppenburg P, Noegel AA, Peche VS. Neuronal actin dynamics, spine density and neuronal dendritic complexity are regulated by CAP2. Front Cell Neurosci 2016; 10: 180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuznetsova A, Brockhoff PB, Christensen R. lmerTest package: tests in linear mixed effects models. J Stat Soft 2017; 82: 1–26. [Google Scholar]
- Lazic SE. The problem of pseudoreplication in neuroscientific studies: is it affecting your analysis?. BMC Neurosci 2010; 11: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenth R. emmeans: Estimated Marginal Means, aka Least-Squares Means. R package version 1.4.5, 2020. Available from: https://CRAN.R-project.org/package=emmeans (20 April 2020, date last accessed).
- Liu Y, Xiao W, Shinde M, Field J, Templeton DM. Cadmium favors F-actin depolymerization in rat renal mesangial cells by site-specific, disulfide-based dimerization of the CAP1 protein. Arch Toxicol 2018; 92: 1049–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch G, Rex CS, Chen LY, Gall CM. The substrates of memory: defects, treatments, and enhancement. Eur J Pharmacol 2008; 585: 2–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinverno M, Carta M, Epis R, Marcello E, Verpelli C, Cattabeni F, et al. Synaptic localization and activity of ADAM10 regulate excitatory synapses through N-cadherin cleavage. J. Neurosci 2010; 30: 16343–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcello E, Gardoni F, Luca Pérez-Otaño DM. I. An arginine stretch limits ADAM10 exit from the endoplasmic reticulum. J Biol Chem 2010; 285: 10376–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcello E, Saraceno C, Musardo S, Vara H, la Fuente de AG, Pelucchi S, et al. Endocytosis of synaptic ADAM10 in neuronal plasticity and Alzheimer's disease. J Clin Invest 2013; 123: 2523–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matus A. Growth of dendritic spines: a continuing story. Curr Opin Neurobiol 2005; 15: 67–72. [DOI] [PubMed] [Google Scholar]
- Mauceri D, Freitag HE, Oliveira AMM, Bengtson CP, Bading H. Nuclear calcium-VEGFD signaling controls maintenance of dendrite arborization necessary for memory formation. Neuron 2011; 71: 117–30. [DOI] [PubMed] [Google Scholar]
- Mikhaylova M, Bär J, van Bommel B, Schätzle P, Yuanxiang P, Raman R, et al. Caldendrin directly couples postsynaptic calcium signals to actin remodeling in dendritic spines. Neuron 2018; 97: 1110–25.e14. [DOI] [PubMed] [Google Scholar]
- Minamide LS, Striegl AM, Boyle JA, Meberg PJ, Bamburg JR. Neurodegenerative stimuli induce persistent ADF/cofilin-actin rods that disrupt distal neurite function. Nat Cell Biol 2000; 2: 628–36. [DOI] [PubMed] [Google Scholar]
- Noegel AA, Rivero F, Albrecht R, Janssen KP, Köhler J, Parent CA, et al. Assessing the role of the ASP56/CAP homologue of Dictyostelium discoideum and the requirements for subcellular localization. J Cell Sci 1999; 112: 3195–203. [DOI] [PubMed] [Google Scholar]
- Normoyle KPM, Brieher WM. Cyclase-associated protein (CAP) acts directly on F-actin to accelerate cofilin-mediated actin severing across the range of physiological pH. J Biol Chem 2012; 287: 35722–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto K-I, Nagai T, Miyawaki A, Hayashi Y. Rapid and persistent modulation of actin dynamics regulates postsynaptic reorganization underlying bidirectional plasticity. Nat Neurosci 2004; 7: 1104–12. [DOI] [PubMed] [Google Scholar]
- Ono S. The role of cyclase-associated protein in regulating actin filament dynamics - more than a monomer-sequestration factor. J Cell Sci 2013; 126: 3249–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oxtoby NP, Alexander DC, Consortium E. Imaging plus X: multimodal models of neurodegenerative disease. Curr Opin Neurol 2017; 30: 371–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palop JJ, Chin J, Mucke L. A network dysfunction perspective on neurodegenerative diseases. Nature 2006; 443: 768–73. [DOI] [PubMed] [Google Scholar]
- Paternoster V, Rajkumar AP, Nyengaard JR, Børglum AD, Grove J, Christensen JH. The importance of data structure in statistical analysis of dendritic spine morphology. J Neurosci Methods 2018; 296: 93–8. [DOI] [PubMed] [Google Scholar]
- Peche V, Shekar S, Leichter M, Korte H, Schröder R, Schleicher M, et al. CAP2, cyclase-associated protein 2, is a dual compartment protein. Cell Mol Life Sci 2007; 64: 2702–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peche VS, Holak TA, Burgute BD, Kosmas K, Kale SP, Wunderlich FT, et al. Ablation of cyclase-associated protein 2 (CAP2) leads to cardiomyopathy. Cell Mol Life Sci 2013; 70: 527–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelucchi S, Stringhi R, Marcello E. Dendritic spines in Alzheimer's disease: how the actin cytoskeleton contributes to synaptic failure. Int J Mol Sci 2020; 21: 908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penzes P, Cahill ME, Jones KA, Vanleeuwen J-E, Woolfrey KM. Dendritic spine pathology in neuropsychiatric disorders. Nat Neurosci 2011; 14: 285–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penzes P, Vanleeuwen J-E. Impaired regulation of synaptic actin cytoskeleton in Alzheimer's disease. Brain Res Rev 2011; 67: 184–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinheiro J, Bates D. Mixed-effects models in S and S-PLUS. New York: Springer-Verlag; 2006. [Google Scholar]
- Pontrello CG, Sun M-Y, Lin A, Fiacco TA, DeFea KA, Ethell IM. Cofilin under control of β-arrestin-2 in NMDA-dependent dendritic spine plasticity, long-term depression (LTD), and learning. Proc Natl Acad Sci USA 2012; 109: E442–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purde V, Busch F, Kudryashova E, Wysocki VH, Kudryashov DS. Oligomerization affects the ability of human cyclase-associated proteins 1 and 2 to promote actin severing by cofilins. Int J Mol Sci 2019; 20: 5647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintero-Monzon O, Jonasson EM, Bertling E, Talarico L, Chaudhry F, Sihvo M, et al. Reconstitution and dissection of the 600-kDa Srv2/CAP complex: roles for oligomerization and cofilin-actin binding in driving actin turnover. J Biol Chem 2009; 284: 10923–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. R: a language and environment for statistical computing. Vienna, Austria: R Foundation For Statistical Computing; 2019.
- Racz B, Weinberg RJ. Spatial organization of cofilin in dendritic spines. Neuroscience 2006; 138: 447–56. [DOI] [PubMed] [Google Scholar]
- Rush T, Martinez-Hernandez J, Dollmeyer M, Frandemiche ML, Borel E, Boisseau S, et al. Synaptotoxicity in Alzheimer's disease involved a dysregulation of actin cytoskeleton dynamics through cofilin 1 phosphorylation. J Neurosci 2018; 38: 10349–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rust MB, Gurniak CB, Renner M, Vara H, Morando L, Görlich A, et al. Learning, AMPA receptor mobility and synaptic plasticity depend on n-cofilin-mediated actin dynamics. Embo J 2010; 29: 1889–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rust MB. ADF/cofilin: a crucial regulator of synapse physiology and behavior. Cell Mol Life Sci 2015. a; 72: 3521–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rust MB. Novel functions for ADF/cofilin in excitatory synapses - lessons from gene-targeted mice. Commun Integr Biol 2015. b; 8: e1114194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sala C, Segal M. Dendritic spines: the locus of structural and functional plasticity. Physiol Rev 2014; 94: 141–88. [DOI] [PubMed] [Google Scholar]
- Shekhar S, Chung J, Kondev J, Gelles J, Goode BL. Synergy between cyclase-associated protein and Cofilin accelerates actin filament depolymerization by two orders of magnitude. Nat Commun 2019; 10: 5319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith KR, Kopeikina KJ, Fawcett-Patel JM, Leaderbrand K, Gao R, Schürmann B, et al. Psychiatric risk factor ANK3/ankyrin-G nanodomains regulate the structure and function of glutamatergic synapses. Neuron 2014; 84: 399–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Star EN, Kwiatkowski DJ, Murthy VN. Rapid turnover of actin in dendritic spines and its regulation by activity. Nat Neurosci 2002; 5: 239–46. [DOI] [PubMed] [Google Scholar]
- Van Troys M, Huyck L, Leyman S, Dhaese S, Vandekerkhove J, Ampe C. Ins and outs of ADF/cofilin activity and regulation. Eur J Cell Biol 2008; 87: 649–67. [DOI] [PubMed] [Google Scholar]
- Yusof AM, Hu N-J, Wlodawer A, Hofmann A. Structural evidence for variable oligomerization of the N-terminal domain of cyclase-associated protein (CAP). Proteins 2004; 58: 255–62. [DOI] [PubMed] [Google Scholar]
- Yuste R, Bonhoeffer T. Morphological changes in dendritic spines associated with long-term synaptic plasticity. Annu Rev Neurosci 2001; 24: 1071–89. [DOI] [PubMed] [Google Scholar]
- Zhou Q, Homma KJ, Poo M-M. Shrinkage of dendritic spines associated with long-term depression of hippocampal synapses. Neuron 2004; 44: 749–57. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are available on direct request to the corresponding author.