Abstract
Background
HIV treatment guidelines have traditionally recommended that all HIV-positive individuals are tested for evidence of drug resistance prior to starting ART. Testing for resistance to reverse transcriptase inhibitors and PIs is well established in routine care. However, testing for integrase strand transfer inhibitor (InSTI) resistance is less consistent.
Objectives
To inform treatment guidelines by determining the prevalence of InSTI resistance in a national cohort of recently infected individuals.
Patients and methods
Recent (within 4 months) HIV-1 infections were identified using a Recent Infection Testing Algorithm of new HIV-1 diagnoses in the UK. Resistance-associated mutations (RAMs) in integrase, protease and reverse transcriptase were detected by ultradeep sequencing, which allows for the sensitive estimation of the frequency of each resistant variant in a sample.
Results
The analysis included 655 randomly selected individuals (median age = 33 years, 95% male, 83% MSM, 78% white) sampled in the period 2014 to 2016 and determined to have a recent infection. These comprised 320, 138 and 197 samples from 2014, 2015 and 2016, respectively. None of the samples had major InSTI RAMs occurring at high variant frequency (≥20%). A subset (25/640, 3.9%) had major InSTI RAMs occurring only as low-frequency variants (2%–20%). In contrast, 47/588 (8.0%) had major reverse transcriptase inhibitor and PI RAMs at high frequency.
Conclusions
Between 2014 and 2016, major InSTI RAMs were uncommon in adults with recent HIV-1 infection, only occurring as low-frequency variants of doubtful clinical significance. Continued surveillance of newly diagnosed patients for evidence of transmitted InSTI resistance is recommended to inform clinical practice.
Introduction
In 2007, raltegravir was the first integrase strand transfer inhibitor (InSTI) introduced into clinical practice, initially for treatment-experienced HIV-positive patients requiring rescue therapy1 and 2 years later for all patients, including treatment-naive patients. Raltegravir was followed by elvitegravir in 2012 as part of a fixed-dose, single-tablet combination (Stribild) that includes the booster cobicistat, emtricitabine and tenofovir disoproxil fumarate. Elvitegravir was subsequently reformulated in combination with tenofovir alafenamide instead of tenofovir disoproxil fumarate (Genvoya). Second-generation InSTIs comprise dolutegravir, which was approved in 2013, and more recently bictegravir coformulated with tenofovir alafenamide and emtricitabine (Biktarvy), which was approved in 2018.2 Large clinical trials have demonstrated that the second-generation InSTIs are potent suppressors of HIV replication and have good safety and high genetic barriers to the emergence of drug resistance.3–6 These features make them the preferred third agent for starting ART in combination with a backbone of two NRTIs.7,8 First-line regimens based on NNRTIs or boosted PIs (bPIs) are instead reserved for selected scenarios.
Mirroring European and American guidelines, the British HIV Association (BHIVA) guidelines for the treatment of HIV-1-positive adults recommend that resistance testing by viral partial genome sequencing be undertaken in all newly diagnosed patients prior to starting ART to allow the detection of transmitted drug resistance (TDR).7–10 Sequencing should be performed for reverse transcriptase and protease genes, based on studies showing that when the prevalence of TDR in the population exceeds a threshold of 1%–5% it is cost-effective to screen patients to guide treatment selection.11,12 In the UK, the prevalence of TDR affecting NRTIs, NNRTIs or PIs peaked at ∼14% in 2002 and has remained stable at 7%–9% since 2006.13,14 To date, there is no recommendation for baseline integrase sequencing as little evidence exists of the transmission of InSTI resistance-associated mutations (RAMs) in the UK and worldwide.15
Most routine resistance testing is performed using conventional Sanger sequencing technology, which has a variant frequency detection threshold of ∼20% and hence fails to detect variants that are present below this threshold in a patient’s viral population. Next-generation sequencing (NGS) technologies allow the detection of variants present in a sample at a frequency as low as 1%.16 The clinical significance of low-frequency resistant variants remains under debate. It has been shown that low-frequency variants with mutations affecting the NNRTIs, and to a lesser extent the NRTIs, significantly reduce responses to first-line therapy with two NRTIs plus one NNRTI, while showing no appreciable effect on bPI-based regimens.17–20 However, transmission is unlikely to be a source of the large majority of these low-frequency variants in individuals who have recently acquired HIV and consequently would have minimal to no impact on treatment outcome as they would not have been selected under drug pressure.21
Several studies have reported no evidence of InSTI major RAMs in treatment-naive or recently infected HIV-1-positive populations using Sanger sequencing.22–27 The few studies reporting apparent transmission of InSTI RAMs included mutations that are polymorphic among ART-naive patients (e.g. L74IM, T97A and E157Q).28,29 One notable exception is a study from Taiwan that observed InSTI major RAMs (e.g. Q148HKR and Y143R) in 1.2% of 1307 ART-naive individuals under a specific epidemiological circumstance where there was a large reservoir of InSTI resistance among the treated population.30
The UK national reference laboratory receives blood samples from half of all newly diagnosed cases of HIV-1 infection for incidence testing using a Recent Infection Testing Algorithm (RITA).31 Recently infected individuals are the most relevant population as they are treatment naive and detection of resistance is most likely due to transmission and prior to natural decay. Using this resource, we determined the national prevalence of TDR to InSTIs, NRTIs, NNRTIs and bPIs by performing NGS on samples from newly diagnosed patients identified as infected in the previous 4 months. The findings will inform the clinical utility of baseline resistance testing for integrase in the UK.
Patients and methods
Ethics
PHE has Section 251 approval, which is reviewed annually and provides the legal basis for the collection of HIV patient-level data for public health monitoring purposes. In addition, the HIV surveillance dataset is reviewed annually by the PHE Caldicott Panel to ensure compliance with information governance policies.
Study population
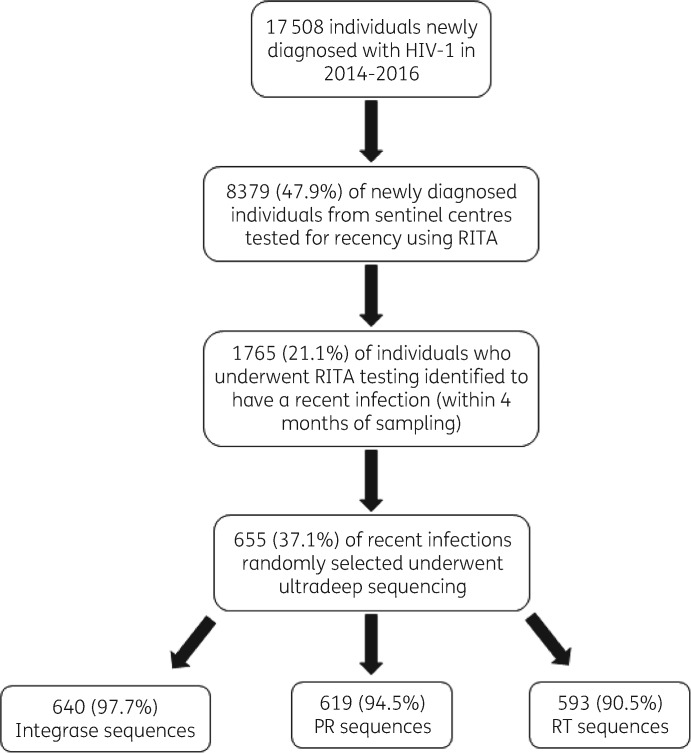
The UK national reference laboratory applies RITA to new HIV-1 diagnoses as a sentinel national surveillance programme. Blood samples from ART-naive individuals collected at HIV or Genitourinary Medicine Clinics in the UK are identified as likely recent infections (within 4 months of sample collection) using a limiting-antigen (LAg) avidity assay with an OD index <1.5. The assay differentiates likely recent from long-standing infection by the strength of HIV-specific antibody–antigen binding.32 The assay has a misclassification rate of long-standing HIV infections as recent of <1% when RITA is applied and samples close to the OD index cut-off values are more likely to be misclassified.33 The RITA algorithm also includes matching the sample to individual HIV records of the HIV and AIDS Reporting System (HARS). Individuals with an OD index <1.5 must also have a CD4+ cell count of >200 cells/mm3 and viral load of >1000 copies/mL to be assigned as ‘recent infections’. In the period 2014–16, RITA was applied to 8379 (47.9%) of new diagnoses and 1765 (21.1%) were identified as recent infections (Figure 1). We randomly selected 655 (37.1%) of these plasma specimens with residual volume for NGS analysis, comprising, by year, 320, 138 and 197 samples collected in 2014, 2015 and 2016, respectively. Linked demographic and clinical information was extracted from HARS. Prescription data were used to determine the use of InSTIs in clinical practice in England between 2010 and 2015.
Figure 1.
Flow chart showing study participant selection. Patients were eligible for inclusion if they were newly diagnosed and identified to be recently infected (within 4 months of sampling) using RITA. PR, protease; RT, reverse transcriptase.
NGS
Samples collected in 2014 were extracted using a QIAsymphony Virus/Pathogen DSP Mini Kit (QIAGEN) using 200 μL of plasma, eluted in a final volume of 60 μL and processed using a previously described PCR amplicon-based NGS assay for protease/reverse transcriptase34 and integrase was amplified in a nested PCR reaction. Briefly, cDNA was generated using 20 μL of RNA, a QIAGEN OneStep RT–PCR Kit (QIAGEN) and primers H10F2 (5′-GCACAYAARGGRATTGGAGGAAATGA-3′) and H10R3 (5′-CCTAGTGGRATGTGTACTTCTGA-3′), both at 1 μM, under the following cycling conditions: 50°C for 40 min; 95°C for 15 min; 35 cycles of 95°C for 30 s, 53°C for 30 s and 72°C for 1 min; and then a final elongation step at 72°C for 4 min. Two microlitres of cDNA was then used in a semi-nested PCR using a Platinum Taq DNA Polymerase Kit (Invitrogen) and 0.4 μM primer H10F2 and 1.6 μM primer H10R2 (5′-CATATGRTGYTTTACTAAACTHTTCCA-3′) under the following cycling conditions: 95°C for 5 min; 35 cycles of 94°C for 30 s, 53°C for 30 s and 72°C for 1 min; and then a final elongation step at 72°C for 2 min. Amplicons for protease/reverse transcriptase and integrase were pooled in equimolar concentration and then sequenced as previously described.34
Samples collected in 2015 and 2016 were processed using a sequence-capture WGS assay. Briefly, 350 μL of plasma was extracted using the NucliSENS system on the easyMag platform (bioMérieux) and eluted into a volume of 25 μL, all of which was subjected to DNAse digestion with 0.25 U of TURBO DNase (Thermo Fisher Scientific) in 30 μL reactions incubated for 30 min at 37°C. Digestion products were cleaned up using 2× AMPure XP Beads (Beckman Coulter) following the manufacturer’s instructions, with a final elution volume of 10 μL nuclease-free water. The 10 μL volume of DNAse-digested RNA extracts was used to generate DNA libraries, using the KAPA RNA HyperPrep Kit (Roche). DNA libraries were pooled in a total of 500 ng and hybridized using 120 nt HIV-specific biotinylated oligonucleotide probes and NimbleGen SeqCap target enrichment reagents (Roche) following the manufacturer’s specifications. Following hybridization, the HIV DNA libraries bound to the biotinylated probes were partitioned using magnetic streptavidin-coated beads and subjected to a further 14 cycles of PCR amplification. The concentration of the final pool was quantified using the KAPA SYBR FAST Universal qPCR Kit for Illumina libraries (KAPA Biosystems) on a 7500 Real-Time PCR System (Applied Biosystems) and analysed for fragment size distribution using the High Sensitivity DNA Kit (Agilent) on a 2100 Bioanalyser Instrument, following the specifications of both manufacturers. Sequencing was performed on an Illumina MiSeq instrument using the MiSeq Reagent Kit V2 (300 cycles) (Illumina) according to the manufacturer’s guidelines, with the following minor modifications. The final pool was diluted to 2 nM and denatured with 0.2 N sodium hydroxide for 2 min, incubated for 4 min at 95°C and diluted in kit reagent HT1 to produce 1 mL of a 20 pM solution. This was further diluted to make 700 μL of a 9 pM solution, of which 10% was substituted with 12.5 pM PhiX (Illumina). A total of 600 μL of this final solution was loaded onto the MiSeq cartridge.
Bioinformatic analysis
MiSeq paired-end FASTQs were trimmed for quality with Trimmomatic (v0.39, with LEADING and TRAILING set to 30 and MINLEN to 50)35 and human reads removed through BWA (v0.7.17) mapping to the human genome (GRCh37), retaining unmapped pairs. Contigs generated from dehumanized reads by de novo assembly using SPAdes (v3.13.1)36 were split into fragments of approximately 500 nt (depending on length) and BLASTed against 2427 reference genome sequences annotated and aligned by LANL (http://www.hiv.lanl.gov/). An in-house Python script was used to build a draft sequence from the contigs and their locations within the genome alignment, filling gaps from the reference sequence(s) informing the flanking termini (in a fashion somewhat analogous to LASTZ). Two rounds of BWA mapping and consensus derivation (using an in-house C++ script—QuasiBAM)37 were performed to obtain the final nucleotide frequency table, from which an in-house Perl script derived consensus genome sequences at 20% and 2% nucleotide frequencies at a minimum read depth of 30, the latter frequency previously established as the minimum threshold of the assay.34
The positive percentage agreement (PPA) between amplicon and sequence-capture methods was 99.8% (95% CI = 99.7%–99.9%) and 99.0% (95% CI = 98.8%–99.2%) at 20% and 2% variant frequency thresholds, respectively, at the nucleotide level (Tables S1 and S2 and Figure S1, available as Supplementary data at JAC Online).
Drug resistance analysis
The generated consensus sequences were analysed for surveillance drug resistance mutations (SDRMs) using the Calibrated Population Resistance (CPR) tool that uses the WHO 2009 list of SDRMs for PIs and reverse transcriptase inhibitors and the recently proposed 2019 list of SDRMs for InSTIs: T66AIK, E92GQ, G118R, F121Y, E138AKT, G140ACS, Y143CHRS, S147G, Q148HRK, N155H, S230R and R263K.38–40 The integrase sequences were also analysed for the presence of the following InSTI accessory mutations: H51Y, Q95K, T97A, A128T, V151A, S153FY, E157Q and G163KR. These mutations have minimal, if any, effect on InSTI susceptibility when present alone, but may contribute to reduced susceptibility in combination with InSTI major RAMs. For determination of mutational load, viral load and CD4+ cell count data were only used if performed within 30 days of the sample used for RITA and sequencing. Additional mutational load data for PIs, NRTIs and NNRTIs were obtained from previously reported reverse transcriptase and protease sequencing of recently infected individuals from 2011 to 2013.34
Statistical analyses
Descriptive statistics (medians and IQRs) are provided for continuous variables, whereas frequency distributions are provided for categorical variables. Mutational load datasets were compared using the Mann–Whitney U-test with the significance level set at P < 0.05.
Sequence data
Consensus HIV-1 pol sequences from this study have been submitted to GenBank and may be accessed by the following accession numbers: MT570368–MT571329.
Results
Study population characteristics
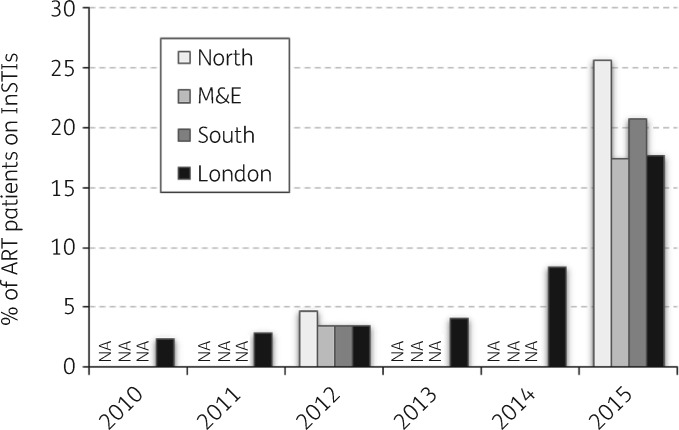
The proportion of adults using InSTIs increased significantly from a low of 2.4% in London in 2010 to between 17.4% and 25.7% in the Midlands & East of England and the North of England, respectively, in 2015 (Figure 2).
Figure 2.
Proportion of people on ART including InSTIs in England between 2010 and 2015. The data were estimated from ART prescribing data from NHS England and are stratified by the PHE regions of London, Midlands & East of England (M&E), North of England and South of England. Data for London in 2015 do not include figures for the month of December. Hospitals in the former North West Strategic Health Authority (SHA) preferred to use raltegravir over bPIs as the third agent for a significant portion of the survey period. Figures may include a source of overestimation as raltegravir was used as first-line post-exposure prophylaxis following sexual exposure (PEPSE). NA, data not available.
Sequencing was performed on plasma samples from 655 recently infected individuals collected between 2014 and 2016. The characteristics of the study population are summarized in Table 1. The majority were male (94.5%), of white ethnic background (77.9%) and with a risk factor for HIV infection being MSM (82.9%). Most of the recent infections were from the London region (57.1%) and the median age of the study population was 33 years (IQR = 26.5–41). Median viral load was 5.18 log10 copies/mL (IQR = 4.61–6.03) and median CD4+ cell count was 545 cells/mm3 (IQR = 408–723), in keeping with recency of infection. Subtyping using the pol gene showed most individuals were infected with subtype B (67.5%).
Table 1.
Characteristics of the study population
| Gender, n (%) | |
| male | 619 (94.5) |
| female | 36 (5.5) |
| Risk exposure, n (%) | |
| MSM | 543 (82.9) |
| heterosexual male | 48 (7.3) |
| heterosexual female | 33 (5.0) |
| IVDU | 3 (0.5) |
| other/unknown | 28 (4.3) |
| Ethnicity, n (%) | |
| white | 510 (77.9) |
| black (African/Caribbean/other) | 41 (6.3) |
| other/unknown | 104 (15.9) |
| Region, n (%) | |
| London | 374 (57.1) |
| North of England | 113 (17.3) |
| Midlands & East of England | 83 (12.7) |
| South of England | 78 (11.9) |
| Northern Ireland | 6 (0.9) |
| Wales | 1 (0.2) |
| Subtype, n (%) | |
| A | 24 (3.7) |
| B | 442 (67.5) |
| C | 38 (5.8) |
| D | 3 (0.5) |
| F | 27 (4.1) |
| G | 4 (0.6) |
| CRF01_AE | 28 (4.3) |
| CFR02_AG | 54 (8.2) |
| CRF06_cpx | 18 (2.7) |
| CRF07_BC | 1 (0.2) |
| CRF11_cpx | 3 (0.5) |
| CRF12_BF | 3 (0.5) |
| CRF24_BG | 1 (0.2) |
| complex recombinants | 9 (1.4) |
| Age (years), median (IQR) | 33 (26.5–41) |
| CD4+ cell count (cells/mm3), median (IQR) | 545 (408–723)a |
| Viral load (log10 copies/mL), median (IQR) | 5.18 (4.61–6.03)b |
n = 602, number of samples with CD4+ cell counts done within 30 days of date of collection of the sample used for RITA and sequencing.
n = 366, number of samples with viral load done within 30 days of date of collection of the sample used for RITA and sequencing.
Prevalence of integrase RAMs
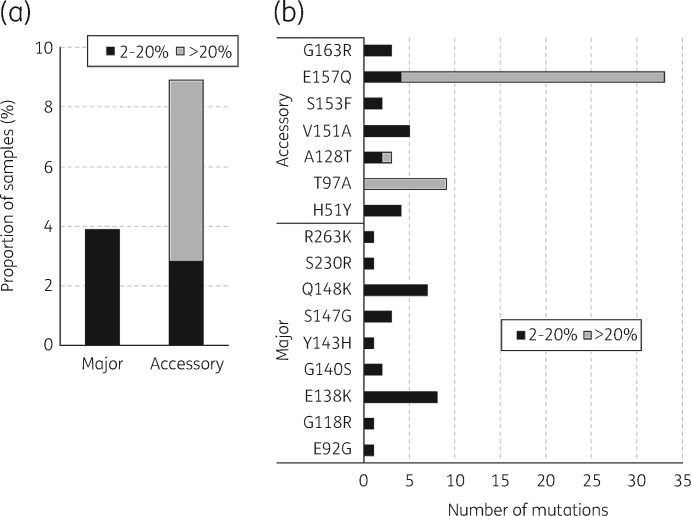
Of the 655 samples sequenced, 640 (97.7%) generated good-quality integrase gene sequence data (complete gene coverage at minimum read depth of 100): 316, 132 and 192 in 2014, 2015 and 2016, respectively. No InSTI major RAMs were detected in the 640 sequences as high-frequency variants (≥20%). A total of 25 (3.9%) sequences contained major InSTI RAMs as low-frequency variants occurring at a frequency between 2% and 20% (Figure 3a). By year, 18 (5.7%), 3 (2.3%) and 4 (2.1%) sequences contained InSTI major RAMs as low-frequency variants in 2014, 2015 and 2016, respectively. In contrast, 39 (6.1%) sequences contained InSTI accessory mutations as high-frequency variants and 18 (2.8%) as low-frequency variants (Figure 3a). By year, the numbers of sequences containing InSTI accessory mutations were 19 (6.0%), 11 (8.3%) and 9 (4.7%) as high-frequency variants and 8 (2.5%), 7 (5.3%) and 3 (1.6%) as low-frequency variants in 2014, 2015 and 2016, respectively.
Figure 3.
Prevalence of InSTI RAMs in recently infected individuals in the UK between 2014 and 2016. Stacked column graphs show the prevalence of InSTI major and accessory RAMs (a) and types of InSTI RAMs (b) at high (>20%) and low (2%–20%) variant frequency. This was determined from 640 integrase sequences from recently infected individuals consisting of 316, 132 and 192 sequences in 2014, 2015 and 2016, respectively. Major and accessory InSTI RAMs were determined using the Stanford HIV Drug Resistance Database 2019 InSTI SDRM list and InSTI accessory mutations as of October 2019.
The InSTI major RAMs and accessory mutations detected are shown in Figure 3(b). The most common InSTI major RAMs detected as low-frequency variants were E138K (8/25; 32.0%) and Q148K (7/25; 28.0%). On the other hand, the most common InSTI accessory mutation detected as a high-frequency variant was E157Q (29/39; 74.4%), whereas V151A was the most common InSTI accessory mutation detected as a low-frequency variant (5/18; 27.8%). Most of the InSTI accessory mutations present as high-frequency variants were associated with non-B subtypes (22/39; 56.4%) with the majority associated with the circulating recombinant forms CRF02_AG (n = 9) and CRF06_cpx (n = 8). The presence of low-frequency RAMs was confirmed by read-based RAM analysis using the variant frequency files produced by the QuasiBam software (Table S3).
Prevalence of reverse transcriptase and protease RAMs
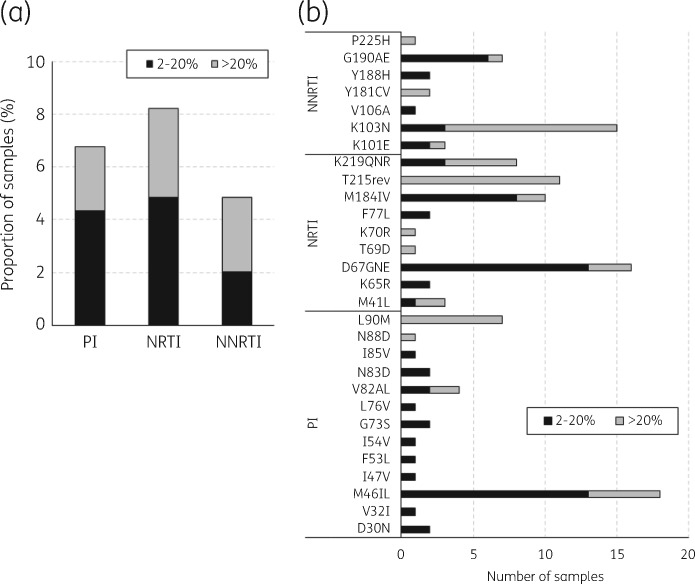
Of the 655 samples sequenced, 619, 593 and 588 generated good-quality sequence data for protease, reverse transcriptase and both gene regions, respectively. By year, 302, 129 and 188 protease and 295, 116 and 182 reverse transcriptase sequences were generated for 2014, 2015 and 2016, respectively. TDR prevalence for all drug classes for the period 2014–16 was 8.0% (47/588) for high-frequency variants and 10.9% (64/588) for low-frequency variants. TDR mutations were detected as high-frequency variants in 15 (2.4%), 20 (3.4%) and 17 (2.9%) sequences against PIs, NRTIs and NNRTIs, respectively (Figure 4a). The overall prevalence of TDR low-frequency variants against PIs, NRTIs and NNRTIs was 27 (4.4%), 29 (4.9%) and 12 (2.0%), respectively (Figure 4a and Table S3). The most common TDR mutations detected as high-frequency variants were L90M (7/15; 46.7%), T215rev (11/20; 55.0%) and K103N (12/17; 70.6%) against PIs, NRTIs and NNRTIs, respectively (Figure 4b). In contrast, the most common TDR mutations detected as low-frequency variants were M46IL (13/27; 48.1%), D67GNE (13/29; 44.8%) and G190E (6/12; 50.0%) against PIs, NRTIs and NNRTIs, respectively (Figure 4b).
Figure 4.
Prevalence of PI and reverse transcriptase inhibitor TDR mutations in recently infected individuals in the UK between 2014 and 2016. Stacked column graphs show the prevalence of PI and reverse transcriptase inhibitor TDR mutations (a) and types of PI, NRTI and NNRTI RAMs (b) at high (>20%) and low (2%–20%) variant frequency. This was determined from 619 protease and 593 reverse transcriptase sequences from recently infected individuals consisting of 302, 129 and 188 protease sequences and 295, 116 and 182 reverse transcriptase sequences in 2014, 2015 and 2016, respectively. PI, NRTI and NNRTI TDR mutations were determined using the WHO 2019 SDRM list.
Of the 655 sequences, 581 (88.7%) generated sufficient and good-quality data in all three polymerase gene regions. Two (0.3%) of the samples had a low-frequency InSTI major RAM and a high-frequency RAM in protease or reverse transcriptase: Q148K + G190A (NNRTI) and E92G + T215S (NRTI). Both were subtype B. Three samples (0.5%) had a low-frequency InSTI major RAM (E138K) and low-frequency RAM in protease and/or reverse transcriptase: D67N (NRTI) + M46I (PI), M46I (PI) and D67G (NRTI) (they were subtype F, B and CRF01_AE, respectively).
Mutational load of low-frequency RAMs
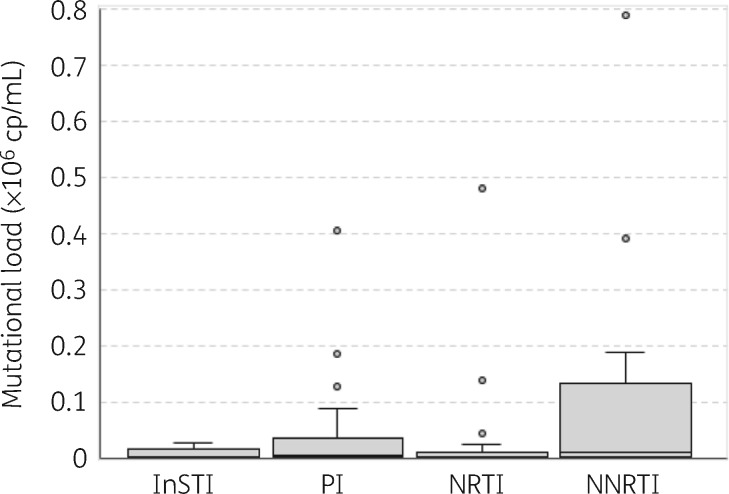
We determined the mutational load of low-frequency RAMs, as previously described.19 The median mutational load of low-frequency InSTI RAMs was 3833 copies/mL (IQR = 895–14 733) (Figure 5). The median mutational load of low-frequency InSTI RAMs was similar to that of low-frequency PI and NRTI RAMs at 5914 copies/mL (IQR = 1470–31 552) and 2706 copies/mL (IQR = 1387–11 726), respectively. In contrast, the median mutational load for low-frequency NNRTI RAMs was slightly higher and had a broad range at 10 188 copies/mL (IQR = 2170–95 313); however, the difference was not statistically significant (P > 0.05, Mann–Whitney U-test).
Figure 5.
Mutational load of low-frequency RAMs. Box-and-whisker plots of mutational load of low-frequency InSTI (n = 14), PI (n = 44), NRTI (n = 28) and NNRTI (n = 14) RAMs. Mutational load is defined as mutation frequency × viral load (copies/mL).
Discussion
Surveillance of transmitted InSTI resistance among 655 individuals with recent HIV infection in the UK who were sampled between 2014 and 2016 showed no evidence of major InSTI RAMs when considering mutants present at high frequency in the individuals’ samples. In contrast, at approximately 8%, the prevalence of TDR to PIs and reverse transcriptase inhibitors remains steady compared with last reported figures in 2014. This prevalence is still higher than the recommended threshold of 1%–5% where baseline resistance testing is considered of benefit at the population level.
On the other hand, accessory InSTI RAMs were detected as high-frequency variants in 6.1% of the study population. The most common accessory InSTI RAMs were T97A and E157Q; both are polymorphic and observed at a high prevalence (up to 7%) in InSTI-naive individuals infected with non-B subtypes, e.g. CRF02_AG. They are also selected in patients experiencing treatment failure with first-generation InSTIs, raltegravir and elvitegravir; however, they have little effect on InSTI susceptibility when present alone.41
Ultradeep sequencing allowed the detection of mutations below the Sanger sequencing variant frequency threshold of ∼20%. Major InSTI RAMs were detected as low-frequency variants in 3.9% of the study population, at a variant frequency between 2% and 20%. It has been argued that the mutational load of low-frequency RAMs, especially for NNRTIs, could play a role in treatment failure.42 The mutational load of low-frequency InSTI RAMs was comparable to that of NRTI and PI RAMs, but was lower and had a very narrow range compared with that of NNRTI RAMs. Compared with the data for low-frequency NNRTI RAMs, there is less compelling evidence that low-frequency PI and NRTI RAMs contribute to treatment failure.20 It is likely that the mutational load of low-frequency RAMs is associated with the impact on virus replication fitness, therefore InSTI, NRTI and PI RAMs that have a high impact on virus replication fitness are unlikely to accumulate to high absolute levels compared with NNRTI RAMs. In addition, we recently showed evidence that the majority of low-frequency RAMs to PIs and reverse transcriptase inhibitors in recently infected individuals are not a result of a transmission event and thus would not have been selected under drug pressure.21 Furthermore, a recent study showed no association between the presence of low-frequency InSTI RAMs prior to initiation of treatment and treatment outcomes.43 Taken together, these data suggest that the low-frequency InSTI RAMs in recently infected individuals are less likely to affect treatment outcome, especially as current second-generation InSTIs, dolutegravir and bictegravir, are highly effective and have very high barriers to resistance. However, low-frequency InSTI RAMs may still have an impact in individuals with poor adherence or those who harbour resistance to other components of their ART regimen.
The proportion of individuals on an ART regimen that included an InSTI was approximately 20% during the period covered by the study, as estimated using prescription data from NHS England. The use of InSTIs as part of first-line regimens is anticipated to continue to rise in the UK, reflecting national and international treatment guidelines.44 The use of InSTIs in the UK increased from less than 10% in 2014 to over 20% in 2015; thus, the effect of this and further projected increases in InSTI use may not be captured in this surveillance study. Nonetheless, the virological suppression rate for people on InSTI-based therapy in the UK is very high (>95%) and the likelihood of the emergence of drug resistance for those failing dolutegravir or bictegravir plus two NRTIs in first-line ART is negligible.3,4 However, raltegravir and elvitegravir have been used for longer than dolutegravir and bictegravir in ART-naive and ART-experienced patients and these drugs are more likely to result in treatment failure with resistance selection. Thus, these groups may have generated a pool of potential transmitters, which may later contribute to transmitted InSTI resistance. All these factors necessitate continued surveillance of InSTI TDR in the coming years.
Reflecting the focus on recent infection, another limitation of the study is that most of the sampled population was male of white ethnic background from England whose probable route of HIV exposure was sex between men and who were infected with subtype B virus. This is because gay and bisexual men are more likely to have recently acquired infection at HIV diagnosis. Thus, these findings may not be generalizable to the whole of the UK population living with HIV, particularly women and those infected with non-B subtypes. In addition, the frequency of InSTI use may be different in Scotland, Wales and Northern Ireland, data that were not captured in this study.
Two different sequencing methods were used in this study, an amplicon-based approach using gene-specific nested PCR followed by DNA library prep and a sequence-capture approach that is dependent on RNA library prep followed by enrichment using HIV-1-specific probe baits. Overall, the consensus sequence generated by both methods was highly concordant (>99%) using the 20% and 2% variant frequency at nucleotide level (see Table S1). Discordances were at mixed-base positions where one method detected only one of the mixed bases, with a tendency for low-frequency variants detected by the amplicon method being infrequently detected by the sequence-capture method, whereas the opposite was true (see Table S2 and Figure S1). This could either be due to an overcall of low-frequency variants by the amplicon method or a decreased sensitivity for detection of low-frequency variants by the sequence-capture method. This requires further investigation using standardized reference or control material with well-characterized low-frequency variants at specific frequencies. The validation of NGS methods to accurately reflect in vivo low-frequency variants is essential to determine their effect on clinical outcomes.
In conclusion, this study shows no evidence of transmitted InSTI resistance in the recently infected population in the UK. However, performing baseline integrase resistance testing is still important, especially for national reference laboratories in order to provide surveillance data and possibly in selected patients in routine clinical practice. One consideration is that the use of InSTIs as part of first-line combination ART (cART) is anticipated to continue to increase worldwide following WHO recommendations. The large-scale use of dolutegravir in resource-limited settings is likely to take place with limited viral load monitoring and thus could result in significant increases in transmitted InSTI resistance from a more global perspective. In parallel, prospective cohort studies to assess treatment outcomes in recently infected individuals harbouring low-frequency RAMs would best inform their clinical significance and diagnostic utility. The use of WGS adopted from 2015 onwards will also be useful in analysing other regions of the HIV-1 genome that have been postulated to be involved in development of resistance to second-generation InSTIs, e.g. envelope and 3′ polypurine tract (PPT).45,46
Supplementary Material
Acknowledgements
We thank the staff of the Antiviral Unit and Clinical Service Unit in the Virus Reference Department (PHE) for providing laboratory support. We also wish to thank the National Institute for Health Research (NIHR) Health Protection Research Unit in Blood Borne and Sexually Transmitted Infections Steering Committee [Caroline Sabin (Director), John Saunders (PHE Lead), Catherine Mercer, Gwenda Hughes, Jackie Cassell, Greta Rait, Samreen Ijaz, Tim Rhodes, Sema Mandal, Kholoud Porter and William Rosenberg] for reviewing the manuscript prior to submission.
Funding
This work was supported by ViiV Healthcare, Public Health England (PHE) and the National Institute for Health Research and undertaken by the National Institute for Health Research Health Protection Research Unit (NIHR HPRU) in Blood Borne and Sexually Transmitted Infections at University College London (UCL) in partnership with PHE, in collaboration with the London School of Hygiene and Tropical Medicine.
Transparency declarations
A.M.G. reports consultancy fees and research funding paid to the University of Liverpool from Gilead Sciences, Janssen and ViiV Healthcare, and personal fees from Roche Pharma Research & Early Development, outside of the presented work. All other authors: none to declare.
Author contributions
J.L.M. and J.L. contributed equally to this manuscript. J.L.M., A.M.G., V.D. and D.T.D. conceived the hypotheses and designed the study. J.L. and C.M. performed the sequencing experiments and all the laboratory work. P.K., A.S., G.M., A.B. and V.D. collected the metadata and coordinated RITA testing and the HARS database. J.L.M., J.L. and D.F.B. performed bioinformatic analysis. J.L.M. and A.M.G. drafted the manuscript. All authors provided critical reading that shaped the manuscript and approved the final version of the manuscript. The corresponding author attests that all listed authors meet authorship criteria and that no others meeting the criteria have been omitted and had final responsibility for the decision to submit for publication.
Disclaimer
The views expressed are those of the authors and not necessarily those of the NIHR, the Department of Health and Social Care or PHE.
Supplementary data
Tables S1 to S3 and Figure S1 are available as Supplementary data at JAC Online
References
- 1. Arribas JR, Eron J.. Advances in antiretroviral therapy. Curr Opin HIV AIDS 2013; 8: 341–9. [DOI] [PubMed] [Google Scholar]
- 2. Mesplede T, Wainberg MA.. Integrase strand transfer inhibitors in HIV therapy. Infect Dis Ther 2013; 2: 83–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Walmsley S, Baumgarten A, Berenguer J. et al. Brief report: dolutegravir plus abacavir/lamivudine for the treatment of HIV-1 infection in antiretroviral therapy-naive patients: week 96 and week 144 results from the SINGLE randomized clinical trial. J Acquir Immune Defic Syndr 2015; 70: 515–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sax PE, Pozniak A, Montes ML. et al. Coformulated bictegravir, emtricitabine, and tenofovir alafenamide versus dolutegravir with emtricitabine and tenofovir alafenamide, for initial treatment of HIV-1 infection (GS-US-380-1490): a randomised, double-blind, multicentre, phase 3, non-inferiority trial. Lancet 2017; 390: 2073–82. [DOI] [PubMed] [Google Scholar]
- 5. Gallant J, Lazzarin A, Mills A. et al. Bictegravir, emtricitabine, and tenofovir alafenamide versus dolutegravir, abacavir, and lamivudine for initial treatment of HIV-1 infection (GS-US-380-1489): a double-blind, multicentre, phase 3, randomised controlled non-inferiority trial. Lancet 2017; 390: 2063–72. [DOI] [PubMed] [Google Scholar]
- 6. Acosta RK, Willkom M, Martin R. et al. Resistance analysis of bictegravir-emtricitabine-tenofovir alafenamide in HIV-1 treatment-naive patients through 48 weeks. Antimicrob Agents Chemother 2019; 63: e02533–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Churchill D, Waters L, Ahmed N. et al. British HIV Association guidelines for the treatment of HIV-1-positive adults with antiretroviral therapy. HIV Med 2015; 17: s2–104. [DOI] [PubMed] [Google Scholar]
- 8.Panel on Antiretroviral Guidelines for Adults and Adolescents. Guidelines for the Use of Antiretroviral Agents in Adults and Adolescents with HIV. Department of Health and Human Services. http://www.aidsinfo.nih.gov/ContentFiles/AdultandAdolescentGL.pdf.
- 9.European AIDS Clinical Society. Guidelines Version 9.1. 2018. https://www.eacsociety.org/files/2018_guidelines-9.1-english.pdf.
- 10. Gunthard HF, Calvez V, Paredes R. et al. Human immunodeficiency virus drug resistance: 2018 recommendations of the International Antiviral Society–USA Panel. Clin Infect Dis 2019; 68: 177–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sax PE, Islam R, Walensky RP. et al. Should resistance testing be performed for treatment-naive HIV-infected patients? A cost-effectiveness analysis. Clin Infect Dis 2005; 41: 1316–23. [DOI] [PubMed] [Google Scholar]
- 12. Hirsch MS, Brun-Vezinet F, Clotet B. et al. Antiretroviral drug resistance testing in adults infected with human immunodeficiency virus type 1: 2003 recommendations of an International AIDS Society-USA Panel. Clin Infect Dis 2003; 37: 113–28. [DOI] [PubMed] [Google Scholar]
- 13.UK Collaborative Group on HIV Drug Resistance, UK Collaborative HIV Cohort Study, UK Register of HIV Seroconverters. Evidence of a decline in transmitted HIV-1 drug resistance in the United Kingdom. AIDS 2007; 21: 1035–9. [DOI] [PubMed] [Google Scholar]
- 14. Tostevin A, White E, Dunn D. et al. Recent trends and patterns in HIV-1 transmitted drug resistance in the United Kingdom. HIV Med 2017; 18: 204–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Geretti AM, Armenia D, Ceccherini-Silberstein F.. Emerging patterns and implications of HIV-1 integrase inhibitor resistance. Curr Opin Infect Dis 2012; 25: 677–86. [DOI] [PubMed] [Google Scholar]
- 16. Gega AK, Kozal MJ.. New technology to detect low-level drug-resistant HIV variants. Future Virol 2011; 6: 17–26. [Google Scholar]
- 17. Geretti AM, Paredes R, Kozal MJ.. Transmission of HIV drug resistance: lessons from sensitive screening assays. Curr Opin Infect Dis 2015; 28: 23–30. [DOI] [PubMed] [Google Scholar]
- 18. Li JZ, Paredes R, Ribaudo HJ. et al. Low-frequency HIV-1 drug resistance mutations and risk of NNRTI-based antiretroviral treatment failure: a systematic review and pooled analysis. JAMA 2011; 305: 1327–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cozzi-Lepri A, Noguera-Julian M, Di Giallonardo F. et al. Low-frequency drug-resistant HIV-1 and risk of virological failure to first-line NNRTI-based ART: a multicohort European case-control study using centralized ultrasensitive 454 pyrosequencing. J Antimicrob Chemother 2015; 70: 930–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Perrier M, Visseaux B, Landman R. et al. No impact of HIV-1 protease minority resistant variants on the virological response to a first-line PI-based regimen containing darunavir or atazanavir. J Antimicrob Chemother 2018; 73: 173–6. [DOI] [PubMed] [Google Scholar]
- 21. Mbisa JL, Kirwan P, Tostevin A. et al. Determining the origins of HIV-1 drug-resistant minority variants in people who are recently infected using phylogenetic reconstruction. Clin Infect Dis 2019; 69: 1136–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Casadella M, van Ham PM, Noguera-Julian M. et al. Primary resistance to integrase strand-transfer inhibitors in Europe. J Antimicrob Chemother 2015; 70: 2885–8. [DOI] [PubMed] [Google Scholar]
- 23. Doyle T, Dunn DT, Ceccherini-Silberstein F. et al. Integrase inhibitor (INI) genotypic resistance in treatment-naive and raltegravir-experienced patients infected with diverse HIV-1 clades. J Antimicrob Chemother 2015; 70: 3080–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Stekler JD, McKernan J, Milne R. et al. Lack of resistance to integrase inhibitors among antiretroviral-naive subjects with primary HIV-1 infection, 2007-2013. Antivir Ther 2015; 20: 77–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Alvarez M, Casas P, de Salazar A. et al. Surveillance of transmitted drug resistance to integrase inhibitors in Spain: implications for clinical practice. J Antimicrob Chemother 2019; 74: 1693–700. [DOI] [PubMed] [Google Scholar]
- 26. Karade S, Sen S, Sashindran VK.. Absence of integrase strand transfer inhibitor associated resistance in antiretroviral therapy naïve and experienced individuals from Western India. AIDS Res Hum Retroviruses 2019; 35: 567–71. [DOI] [PubMed] [Google Scholar]
- 27. Ji H, Patterson A, Taylor T. et al. Prevalence of primary drug resistance against HIV-1 integrase inhibitors in Canada. J Acquir Immune Defic Syndr 2018; 78: e1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Charpentier C, Lee GQ, Rodriguez C. et al. Highly frequent HIV-1 minority resistant variants at baseline of the ANRS 139 TRIO trial had a limited impact on virological response. J Antimicrob Chemother 2015; 70: 2090–6. [DOI] [PubMed] [Google Scholar]
- 29. Margot NA, Kitrinos KM, Fordyce M. et al. Rare emergence of drug resistance in HIV-1 treatment-naïve patients after 48 weeks of treatment with elvitegravir/cobicistat/emtricitabine/tenofovir alafenamide. HIV Clin Trials 2016; 17: 78–87. [DOI] [PubMed] [Google Scholar]
- 30. Chang SY, Lin PH, Cheng CL. et al. Prevalence of integrase strand transfer inhibitors (INSTI) resistance mutations in Taiwan. Sci Rep 2016; 6: 35779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Aghaizu A, Murphy G, Tosswill J. et al. Recent infection testing algorithm (RITA) applied to new HIV diagnoses in England, Wales and Northern Ireland, 2009 to 2011. Euro Surveill 2014; 19: 20673. [DOI] [PubMed] [Google Scholar]
- 32. Duong YT, Qiu M, De AK. et al. Detection of recent HIV-1 infection using a new limiting-antigen avidity assay: potential for HIV-1 incidence estimates and avidity maturation studies. PLoS One 2012; 7: e33328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kassanjee R, Pilcher CD, Busch MP. et al. Viral load criteria and threshold optimization to improve HIV incidence assay characteristics. AIDS 2016; 30: 2361–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cunningham E, Chan YT, Aghaizu A. et al. Enhanced surveillance of HIV-1 drug resistance in recently infected MSM in the UK. J Antimicrob Chemother 2017; 72: 227–34. [DOI] [PubMed] [Google Scholar]
- 35. Bolger AM, Lohse M, Usadel B.. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 2014; 30: 2114–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bankevich A, Nurk S, Antipov D. et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 2012; 19: 455–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Penedos AR, Myers R, Hadef B. et al. Assessment of the utility of whole genome sequencing of measles virus in the characterisation of outbreaks. PLoS One 2015; 10: e0143081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bennett DE, Camacho RJ, Otelea D. et al. Drug resistance mutations for surveillance of transmitted HIV-1 drug-resistance: 2009 update. PLoS One 2009; 4: e4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tzou PL, Rhee SY, Descamps D. et al. Integrase strand transfer inhibitor (INSTI)-resistance mutations for the surveillance of transmitted HIV-1 drug resistance. J Antimicrob Chemother 2020; 75: 170–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gifford RJ, Liu TF, Rhee SY. et al. The calibrated population resistance tool: standardized genotypic estimation of transmitted HIV-1 drug resistance. Bioinformatics 2009; 25: 1197–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Abram ME, Ram RR, Margot NA. et al. Lack of impact of pre-existing T97A HIV-1 integrase mutation on integrase strand transfer inhibitor resistance and treatment outcome. PLoS One 2017; 12: e0172206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gupta S, Lataillade M, Kyriakides TC. et al. Low-frequency NNRTI-resistant HIV-1 variants and relationship to mutational load in antiretroviral-naïve subjects. Viruses 2014; 6: 3428–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Nguyen T, Fofana DB, Le MP. et al. Prevalence and clinical impact of minority resistant variants in patients failing an integrase inhibitor-based regimen by ultra-deep sequencing. J Antimicrob Chemother 2018; 73: 2485–92. [DOI] [PubMed] [Google Scholar]
- 44.WHO. Update of Recommendations on First- and Second-Line Antiretroviral Regimens. 2019. https://apps.who.int/iris/bitstream/handle/10665/325892/WHO-CDS-HIV-19.15-eng.pdf?ua=1.
- 45. Van Duyne R, Kuo LS, Pham P. et al. Mutations in the HIV-1 envelope glycoprotein can broadly rescue blocks at multiple steps in the virus replication cycle. Proc Natl Acad Sci U S A 2019; 116: 9040–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Malet I, Subra F, Charpentier C. et al. Mutations located outside the integrase gene can confer resistance to HIV-1 integrase strand transfer inhibitors. mBio 2017; 8: e00922–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.