Abstract
RNA helicases catalyze the ATP-dependent destabilization of RNA duplexes. DEAD-box helicases share a helicase core that mediates ATP binding and hydrolysis, RNA binding and unwinding. Most members of this family contain domains flanking the core that can confer RNA substrate specificity and guide the helicase to a specific RNA. However, the in vivo RNA substrates of most helicases are currently not defined. The DEAD-box helicase Hera from Thermus thermophilus contains a helicase core, followed by a dimerization domain and an RNA binding domain that folds into an RNA recognition motif (RRM). The RRM mediates high affinity binding to an RNA hairpin, and an adjacent duplex is then unwound by the helicase core. Hera is a cold-shock protein, and has been suggested to act as an RNA chaperone under cold-shock conditions. Using crosslinking immunoprecipitation of Hera/RNA complexes and sequencing, we show that Hera binds to a large fraction of T. thermophilus RNAs under normal-growth and cold-shock conditions without a strong sequence preference, in agreement with a structure-specific recognition of RNAs and a general function in RNA metabolism. Under cold-shock conditions, Hera is recruited to RNAs with high propensities to form stable secondary structures. We show that selected RNAs identified, including a set of tRNAs, bind to Hera in vitro, and activate the Hera helicase core. Gene ontology analysis reveals an enrichment of genes related to translation, including mRNAs of ribosomal proteins, tRNAs, tRNA ligases, and tRNA-modifying enzymes, consistent with a key role of Hera in ribosome and tRNA metabolism.
Keywords: Hera, helicase, RNA binding, chaperone, CLIP, cold-shock response
INTRODUCTION
RNA helicases catalyze the ATP-dependent destabilization of RNA duplexes. DEAD-box helicases are the largest family of RNA helicases (for reviews, see Hilbert et al. 2009; Jankowsky 2011; Jarmoskaite and Russell 2011; Linder and Jankowsky 2011; Henn et al. 2012; Putnam and Jankowsky 2013). Members of this family share a helicase core that consists of two RecA domains connected by a short, flexible linker. The helicase core mediates ATP binding and hydrolysis, RNA binding, and RNA unwinding (Rogers et al. 2002). Many DEAD-box helicases comprise additional domains flanking the core region that affect nucleotide binding and hydrolysis, and contribute to RNA binding and specificity, to binding of protein partners or to duplex destabilization (for review, see Rudolph and Klostermeier 2015). Dedicated RNA binding domains (RBDs) confer RNA substrate specificity and guide the helicase core to a unique RNA target. For example, the Escherichia coli DEAD-box helicase DbpA and its Bacillus subtilis homolog YxiN contain an RNA binding domain at the carboxyl terminus which forms an RNA recognition motif (RRM) and mediates specific binding of DbpA/YxiN to hairpin 92 of the 23S ribosomal RNA (Pugh et al. 1999; Kossen et al. 2002; Karginov and Uhlenbeck 2004; Karginov et al. 2005; Wang et al. 2006). Other helicases bind RNAs nonspecifically through the helicase core, or via ancillary domains (Tijerina et al. 2006; Grohman et al. 2007; Mallam et al. 2011; Busa et al. 2017). DEAD-box proteins are involved in all aspects of RNA metabolism from transcription to degradation, and most organisms have a set of helicases with different functions: The human genome contains more than 40 DEAD-box helicase genes, E. coli and B. subtilis comprise five and four DEAD-box proteins, respectively. Genetic studies have provided insight into the processes in which these helicases are involved (Jagessar and Jain 2010; Lehnik-Habrink et al. 2013). However, the physiological targets of individual RNA helicases in the cell are often unknown. With the advent of next-generation sequencing, it has become possible to identify RNAs bound to helicases under in vivo conditions by isolation of helicase/RNA complexes, generation of cDNA libraries from the RNAs bound, library sequencing, and mapping of the sequences against the genome or transcriptome (for reviews, see Van Nostrand et al. 2016; Lee and Ule 2018; Wheeler et al. 2018). Such studies have been performed for a number of DEAD-box proteins, including eIF4A and Ded1p from S. cerevisiae, involved in translation initiation (Sen et al. 2015; Guenther et al. 2018), the exon junction complex helicase eIF4A-III (Sauliere et al. 2012), and the DEAD-box protein Dbp2p (Tedeschi et al. 2018).
Hera (Heat-resistant RNA-dependent ATPase) is the only DEAD-box protein of the thermophilic bacterium Thermus thermophilus (Morlang et al. 1999; for review, see Klostermeier 2013). Hera consists of a helicase core, followed by a bipartite carboxy-terminal extension that contains a dimerization domain (DD) and an RBD (Fig. 1; Rudolph et al. 2006, 2009; Klostermeier and Rudolph 2009; Rudolph and Klostermeier 2009). Both RecA domains of the helicase core contribute to binding of duplex RNA (Samatanga and Klostermeier 2014). The DD mediates the formation of a stable dimer (Klostermeier and Rudolph 2009), even at picomolar concentrations (Linden et al. 2008), in which the two helicase cores are facing each other (Fig. 1A; Klostermeier and Rudolph 2009). It is conceivable that both cores functionally cooperate in the structural remodeling of large RNA substrates (Klostermeier 2013). The Hera RBD is attached to the DD by a short double-β-hairpin structure (Rudolph and Klostermeier 2009). It forms an RNA recognition motif (RRM), consisting of a central four-stranded β-sheet, flanked by an α-helix (Rudolph and Klostermeier 2009). A carboxy-terminal tail of ten amino acids is disordered. The RBD interacts with single-stranded RNA through the RRM, and with double-stranded RNA via the carboxy-terminal tail (Steimer et al. 2013). The RBD mediates binding of Hera to 23S rRNA fragments comprising hairpin 92 and to ribonuclease (RNase) P RNA (Linden et al. 2008; Steimer et al. 2013). Binding of these RNAs to Hera induces a conformational change of the helicase core to a compact, closed state (Linden et al. 2008), leading to the formation of the catalytic site for ATP hydrolysis and the bipartite RNA binding site extending over both RecA domains (Sengoku et al. 2006). Hera unwinds a minimal RNA substrate comprising hairpin 92 and the adjacent helix 91 of the 23S rRNA in an ATP-dependent reaction (Linden et al. 2008). However, the cellular RNA substrates of Hera are unknown, and the in vivo role of Hera is currently unclear (see Klostermeier 2013). The expression of the hera gene is increased within 30 sec after a temperature shift to 45°C (Mega et al. 2010), implying that Hera may be a cold-shock protein that remodels RNA structures stabilized at lower temperatures (Klostermeier 2013). To test this hypothesis, and to identify physiologically relevant RNAs that could serve as model substrates for Hera in future mechanistic studies, we set out to identify the RNA substrates of Hera in vivo. To this end, we isolated Hera/RNA complexes from T. thermophilus by enhanced cross-linking and immunoprecipitation (eCLIP Fig. 1B; Van Nostrand et al. 2016), and performed RNA sequencing of the cDNA libraries obtained. We observe that Hera binds to a broad spectrum of cellular RNAs, consistent with a general function in RNA metabolism. This central function of Hera is mirrored in a reduced growth rate of T. thermophilus hera deletion mutants. After cold shock, Hera preferentially associates with RNAs that have a high tendency to form stable secondary structures. Gene ontology analysis of RNAs identified under normal-growth and cold-shock conditions reveals an enrichment of RNAs associated with ribosomes and translation, in agreement with key functions of Hera in ribosome and tRNA metabolism.
FIGURE 1.
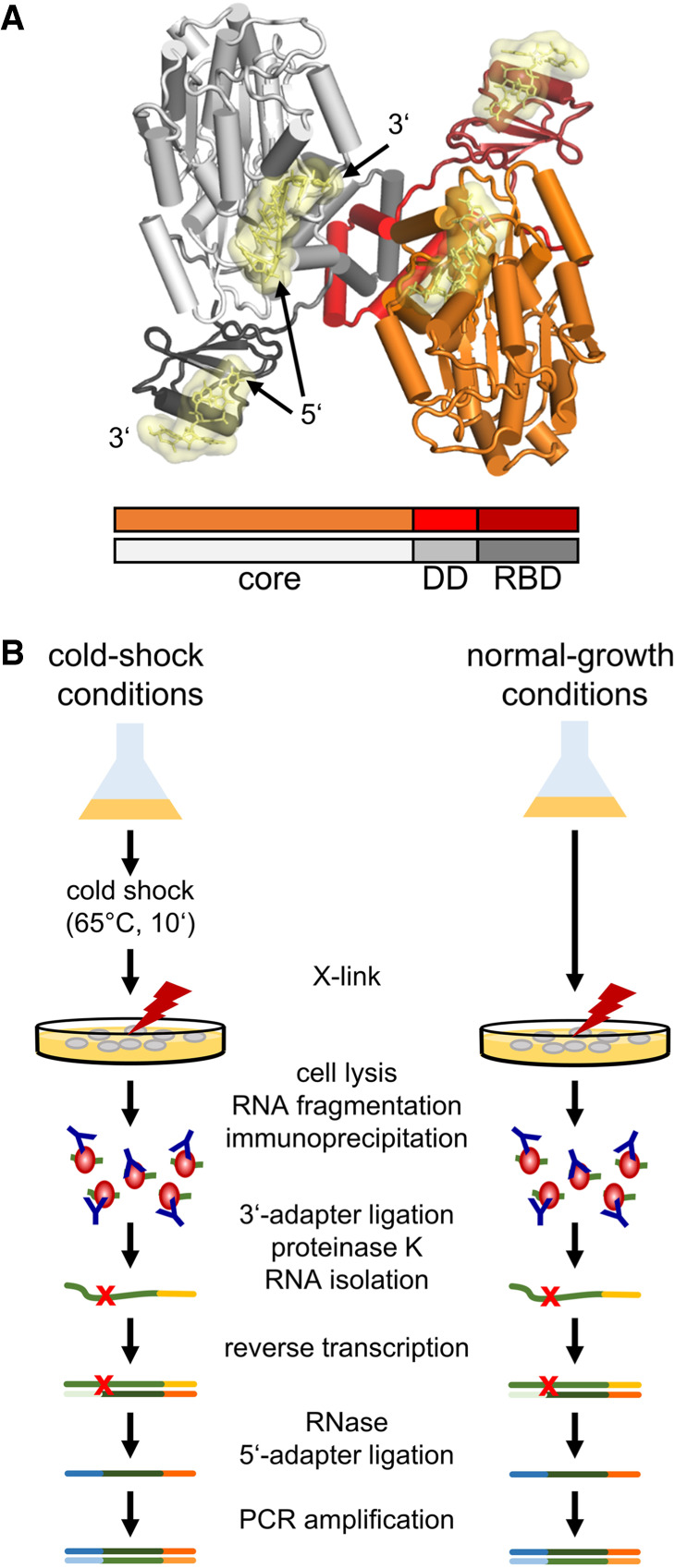
Hera, RNA binding and workflow. (A) Structural model for the Hera dimer. The model was generated by superposition of the structure of a Hera deletion variant comprising the second RecA-domain of the core, the dimerization domain (DD), and the RNA-binding domain (Rudolph and Klostermeier 2009) (RBD; PDB-ID 3i32) and a homology model of the Hera helicase core in the closed, RNA- and nucleotide-bound state, based on the structure of the DEAD-box helicase Vasa (Sengoku et al. 2006) (PBB-ID 2db3), onto the carboxy-terminal RecA-domain (Klostermeier 2013). One Hera protomer is depicted in red hues, the second protomer is shown in gray. RNA bound to the RBDs (Steimer et al. 2013; PDB-ID 4i67) and to the cores (according to the Vasa structure) is shown in yellow. (B) Workflow of the eCLIP experiments (Van Nostrand et al. 2016). T. thermophilus cultures were grown under normal-growth conditions (Hera1, Hera2), or subjected to a cold shock (Hera3, Hera 4). After UV cross-linking, cell lysates were treated with RNase, Hera/RNA complexes were isolated by immunoprecipitation using an anti-Hera antibody (blue Y), followed by ligation of an adapter (yellow) to the 3′-end, protein removal with proteinase K, RNA isolation and reverse transcription. The RNA was removed, a second adapter (blue) was ligated to the 5′-end of the cDNA, and DNA libraries for sequencing were generated by PCR amplification. Experiments were performed in duplicate with paired size-matched input control libraries generated from a portion of the corresponding cell lysates. The red x marks the site of the cross-link.
RESULTS
The role of RNA helicases as cold-shock proteins that remove RNA secondary structures stabilized under cold-shock conditions has been discussed increasingly (Cavicchioli et al. 2000; Jarmoskaite and Russell 2011; Klostermeier 2013; Nagaoka et al. 2013; Virtanen et al. 2018), and several DEAD-box helicases have been associated with cold adaptation and the cold-shock response (Jones et al. 1996; Chamot et al. 1999; Yu and Owttrim 2000; Charollais et al. 2004; Hunger et al. 2006; Cartier et al. 2010; Kuhn 2012; Nagaoka et al. 2013; Virtanen et al. 2018). A previous study determined mRNA expression profiles of T. thermophilus (Mega et al. 2010) and identified the gene TTHA0109 as a cold-shock gene with a 10-fold increase of mRNA levels within 10 min after applying a cold shock. TTHA0109 encodes the DEAD-box helicase Hera (Morlang et al. 1999), whose physiological function is unknown. To identify cellular RNA targets of Hera under normal-growth and cold-shock conditions, we cultivated T. thermophilus under normal-growth conditions (70°C–75°C; Oshima and Imahori 1971, 1974) up to an OD600 of 0.8–1.0 (Supplemental Fig. 1). The cultures were split, one half was subjected to a cold shock to 65°C, while the other half was kept at 75°C. After 10 min, both cultures were irradiated with UV light to cross-link protein/RNA complexes present, and cells were harvested by centrifugation (see Materials and Methods), Hera/RNA complexes were isolated by immunoprecipitation with an anti-Hera antibody, and RNAs were converted into cDNA libraries (Van Nostrand et al. 2016; see Materials and Methods) and sequenced. Experiments were performed in duplicate (Hera1, Hera2: normal-growth conditions, Hera3, Hera4: after cold shock). For each immunoprecipitation sample, a size-matched input library was prepared from a portion of the corresponding cell lysate and used as a reference to identify binding sites with significant read enrichment over input (significant peaks) (Van Nostrand et al. 2016, 2017). Next-generation sequencing and data analysis recovered 7129 significant peaks in Hera1, 5939 significant peaks in Hera2, 5259 significant peaks in Hera3, and 4940 significant peaks in Hera4 (Supplemental Table 1). The overlap between the biological replicates, Hera1/Hera2 or Hera3/Hera4, was similar to the overlap between nonrelated data sets Hera1/Hera3, Hera1/Hera4, Hera2/Hera3, and Hera2/Hera4 (Supplemental Fig. 2a–c).
RNAs bound to Hera under normal-growth and cold-shock conditions
Hera1 contained significant peaks in 1956 genes, Hera2 significant peaks in 1763 genes; 1701 genes were present in both data sets (87% or 96%, respectively). Hera3 contained significant peaks in 1740 genes, Hera4 significant peaks in 1698 genes; 1535 genes were present in both data sets (88% or 90%, respectively; Supplemental Fig. 2d). Notably, a similarly high overlap was detected with genes containing peaks in unrelated samples (Hera1–Hera3, Hera1–Hera4, Hera2–Hera3, Hera2–Hera4; Supplemental Fig. 2e), indicating that the spectrum of bound RNAs does not change drastically under cold-shock conditions. The entire T. thermophilus genome contains 2263 annotated genes, 2210 protein genes and 53 RNA genes (Henne et al. 2004). Hera thus binds to ∼75% of the RNAs in the T. thermophilus transcriptome, consistent with a broad spectrum of functions and a global role in RNA metabolism.
For further analysis of the RNAs bound, a consolidated list of peaks was generated from the overlapping peaks in Hera1 and Hera2, containing 2797 peaks in 1370 genes (no cold shock; Supplemental Table 2), and from the overlapping peaks in Hera3 and Hera4, containing 1654 peaks in 1025 genes (cold shock; Supplemental Table 3). These two lists of potential RNA substrates for Hera have 893 genes in common, one of which is the gene coding for Hera (TT_RS09640; see below). 477 genes (corresponding to 21% of the transcriptome) were only identified under normal-growth conditions, 132 (6%) only under cold-shock conditions.
Hera binds to multiple sites within genes without a positional preference
Many genes identified had several peaks (Supplemental Fig. 3a,b), indicating that Hera binds to multiple sites within the corresponding RNAs. The genes identified under normal-growth and cold-shock conditions were enriched significantly in genes that were longer than average (Fig. 2A). The gene length did not differ significantly between the genes identified in the absence or presence of a cold shock. This most likely reflects the higher probability of Hera binding to a longer RNA. In agreement with this hypothesis, the number of peaks identified per gene scaled roughly linearly with gene length (Supplemental Fig. 3c,d). Peaks were distributed over the entire reading frame (Fig. 2B,C). The peak frequency was highest close to the 5′-end, and decreased linearly toward the 3′-end, but the overall change was small (Fig. 2C), indicating that Hera exhibits little positional binding preference within RNAs.
FIGURE 2.
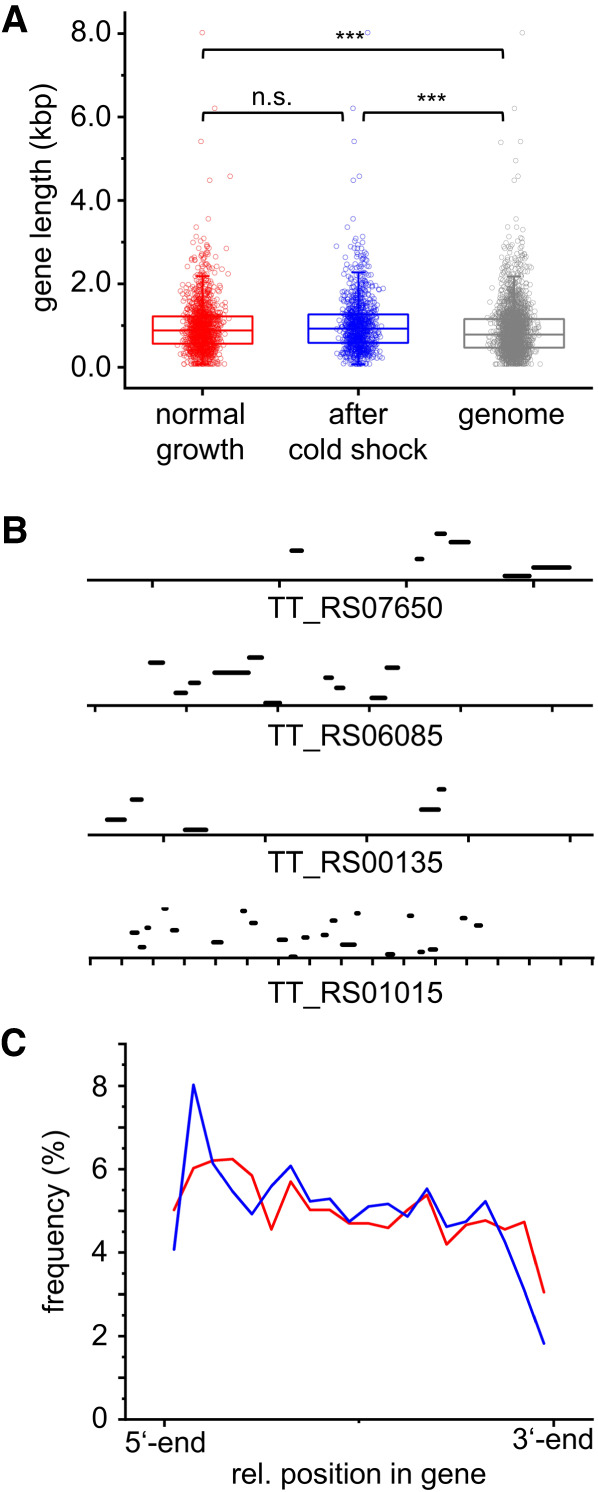
Hera binds to multiple sites within RNAs without a positional preference. (A) Gene length of genes identified as Hera substrates under normal-growth (red) and cold-shock conditions (blue). Longer genes are enriched significantly in the pools compared to the gene length distribution in the genome (gray). n.s.: not significant, (***) P < 0.001. (B) Position of peaks in selected genes. Each tick mark corresponds to 500 bp. (C) Global distribution of peaks along genes identified under normal-growth conditions (red) and under cold-shock conditions (blue).
Given that Hera contains two helicase cores and two RBDs, we hypothesized that adjacent peaks might reflect binding of a Hera dimer to two sites on the same RNA. An analysis of the distance between the centers of adjacent peaks in the genome, either globally or within genes with only two peaks, revealed a predominant separation of the peak centers by ∼150 bases (up to 50 nm in distance), which was more prominent if the analysis was restricted to genes with only two peaks (P < 0.001; Supplemental Fig. 3e,f). However, the Hera dimer is too small to span a distance of 50 nm. The distance of 150 bases might merely reflect the minimum possible distance between peaks in our experiment.
Hera binds RNAs with a high propensity to form secondary structures
A global analysis of the nucleotide distribution in all T. thermophilus genes, in the sequences identified under normal-growth and cold-shock conditions, and in the top 25 sequences with the highest log2-fold enrichment did not reveal a prominent nucleotide bias in the RNAs bound to Hera (Supplemental Fig. 4a–d). The only change observed is a small increase in the fraction of G and a slight decrease in the fraction of U compared to the entire genome for the 25 highest-enriched sequences under cold-shock conditions, which leads to an increased fraction of purines and a concomitant decrease in the fraction of pyrimidines. These differences are within 1.2-fold, and probably not significant.
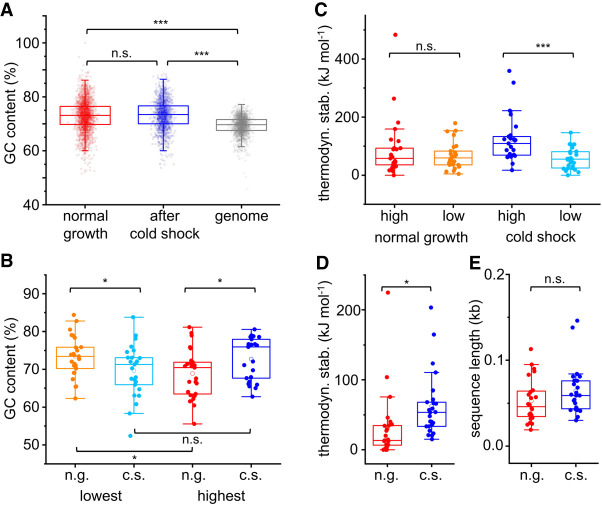
To test whether Hera acts predominantly on RNAs that form stable secondary structures, we analyzed the GC-content of the peak sequences identified under normal-growth and cold-shock conditions. The GC-content of RNAs bound to Hera is higher than the average GC-content of the T. thermophilus genome, both in the absence and presence of a cold shock (Fig. 3A). A subanalysis of the 25 sequences with the highest log2-fold enrichment over input for GC-content and thermodynamic stability was performed. The GC-content of the 25 top sequences identified under cold-shock conditions is (slightly) higher than that of the top 25 identified under normal-growth conditions (P < 0.05; Fig. 3B). Interestingly, a comparison of the 25 least enriched sequences revealed a (slightly) lower GC-content of the sequences identified under cold-shock conditions (P < 0.05). When we compared the GC-content of the top 25 sequences with the 25 sequences with the lowest log2-fold change, we observed a lower GC-content in the top 25 under normal-growth conditions (P < 0.01), but no significant difference under cold-shock conditions (P > 0.05; Fig. 3B). Altogether, we thus observe a preference of Hera for binding to GC-rich RNAs, which becomes slightly more pronounced under cold-shock conditions.
FIGURE 3.
Hera binds RNAs with high GC content and a high propensity to form secondary structures. (A) GC content of sequences identified as bound to Hera under normal-growth (red) and cold-shock conditions (blue), and GC content of all genes in the genome (gray). Hera-bound sequences have a significantly higher-than-average GC content. (B) GC content of the 25 sequences with the lowest (orange, light blue) and highest (red, blue) log2-fold enrichment identified under normal-growth (red hues) and cold-shock conditions (blue hues). (C) Thermodynamic stability at 37°C of the 25 sequences with the highest and lowest log2-fold enrichment under normal-growth conditions (red, orange) and cold-shock conditions (blue, light blue). (D) Thermodynamic stability of the 25 sequences with the highest log2-fold enrichment under normal-growth (red; calculated at 75°C) and cold-shock conditions (blue; calculated at 65°C). (E) Length of peak sequences analyzed in panel D. n.s.: not significant, (*) P < 0.05, (***) P < 0.001, n.g.: normal-growth conditions, c.s.: cold-shock conditions.
We next analyzed the propensity of the RNAs to form secondary structures using RNAstructure (see Materials and Methods). The thermodynamic stability of RNA structures in the 25 top sequences identified under cold-shock and normal-growth conditions was similar at 25°C, 65°C, and 75°C (Supplemental Fig. 4e,f). When we compared the thermodynamic stabilities of the 25 RNAs with the highest and the 25 RNAs with the lowest log2-fold change, no difference was observed under normal-growth conditions (Fig. 3C). In contrast, under cold-shock conditions, the thermodynamic stability of the top 25 enriched RNAs was significantly higher than the stability of the 25 with the lowest enrichment (P < 0.001; Fig. 3C). When the difference in temperatures between the experiments is taken into account (65°C for cold-shock conditions, 75°C for normal-growth conditions), the thermodynamic stability of the 25 top RNAs identified under cold-shock conditions is significantly higher than the stability of the top 25 RNAs identified under normal-growth conditions (P < 0.05; Fig. 3D; Supplemental Fig. 4f), despite the similar lengths of these RNAs (Fig. 3E). Altogether, our data support the conclusion that Hera tends to bind to GC-rich RNAs with higher thermodynamic stability, and interacts preferentially with RNAs that form stable secondary structures under cold-shock conditions.
Sequence motifs enriched
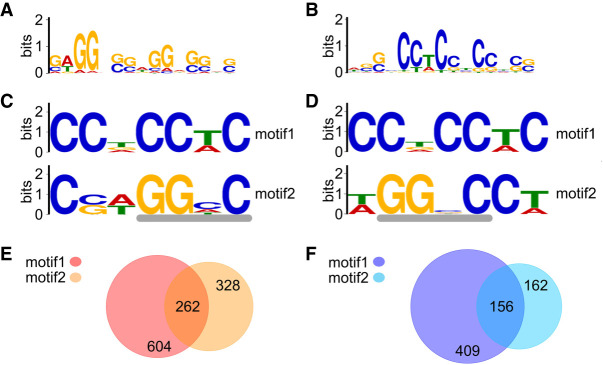
Analysis of sequences using MEME (Multiple Em for Motif, Bailey et al. 2009) discovered a 15-nucleotide GWGGVSSWGGRSSHS motif (H = A/C/U, S = C/G, V = A/C/G, W = A/U) in 2690 sites (P < 0.0001) within the RNA sequences identified under normal-growth conditions, and a 15-nucleotide WSSBCCTCCYCCHSS motif (B = C/G/T, Y = C/U) in 1613 sequences (P < 0.0001) under cold-shock conditions (Fig. 4A,B). A FIMO (Find Individual Motif Occurrence, Grant et al. 2011) search with these motifs identified 5628 occurrences of the GWGGVSSWGGRSSHS motif within the RNAs bound to Hera under normal-growth conditions, and 3326 occurrences of the WSSBCCTCCYCCHSS motif within the RNAs bound to Hera under cold-shock conditions. Cross-searches with the GWGGVSSWGGRSSHS motif among RNAs bound under cold-shock conditions and with the WSSBCCTCCYCCHSS motif among RNAs bound under normal-growth conditions gave a comparable number of occurrences, 3487 and 5104 occurrences, respectively.
FIGURE 4.
Sequence motifs in Hera-bound RNAs. (A) Fifteen nucleotide motif identified with MEME (normal-growth conditions). (B) Fifteen nucleotide motif identified with MEME (cold-shock conditions). (C) Seven nucleotide motifs identified with DREME (normal-growth conditions). The gray line marks a GGNY motif compatible with binding to the Hera RBD. (D) Seven nucleotide motifs identified with DREME (cold-shock conditions). The gray line marks a GGNY motif compatible with binding to the Hera RBD. (E) Occurrence according to FIMO of motifs 1 and 2 discovered with DREME in all peak sequences identified under normal-growth conditions. (F) Occurrence according to FIMO of motifs 1 and 2 discovered with DREME in all peak sequences identified under cold-shock conditions.
A motif search with DREME (Discriminative Regular Expression Motif Elicitation, Bailey 2011) identified shorter motifs (Fig. 4C,D). A CCDCCWC motif (D = A/G/U) was identified with highest significance both among the sequences bound to Hera under normal-growth and under cold-shock conditions. The second-most significant motif discovered was a CSWGGHC motif. This motif contains the GGNY motif (N = A/C/G/U; underlined) previously deduced from a crystal structure of the Hera RBD bound to single-stranded RNA (Steimer et al. 2013) as a recognition element for RNA binding by the Hera RBD. A GGNY sequence is also part of the second-most significant motif identified under cold-shock conditions, a WGGNCCW motif. It is conceivable that these motifs represent RNAs bound to the RBD, while the CCDCCWC motif may reflect binding of RNA to the RBD or the helicase core of Hera. An analysis of the occurrence of these motifs in all sequences using FIMO revealed 1273 occurrences of motif 1 and 770 occurrences of motif 2 (normal-growth conditions), or 846 occurrences of motif 1 and motif 2 (cold-shock conditions). Under normal-growth conditions, 604 sequences contained only motif 1, 388 only motif 2, and 262 contained both motifs (Fig. 4E). Motif 1 occurred between one and nine times in these sequences, motif 2 between one and five times. From the sequences identified after cold shock, 409 sequences contained only motif 1 (one to seven times), 162 only motif 2 (one to four times), and 156 contained both motives (Fig. 4F). We also searched all sequences for the GGNY motif (Supplemental Fig. 5). FIMO identified this motif in 12883 instances in the 2797 sequences under normal-growth conditions, and in 7933 instances under cold-shock conditions (P = 0.01). Both under normal-growth and under cold-shock conditions, 85% of the sequences identified contained at least one GGNY motif. 37% of the sequences contained one type of motif, 33% contained two of the motifs, 13% contained three of the motifs, and 2% contained all four motifs (Supplemental Fig. 5b,c). However, the average log2-fold enrichment of the sequences with one or more GGNY motifs is not higher than the enrichment of sequences lacking the motif (Supplemental Fig. 5d).
In vitro binding of RNAs identified by eCLIP to Hera
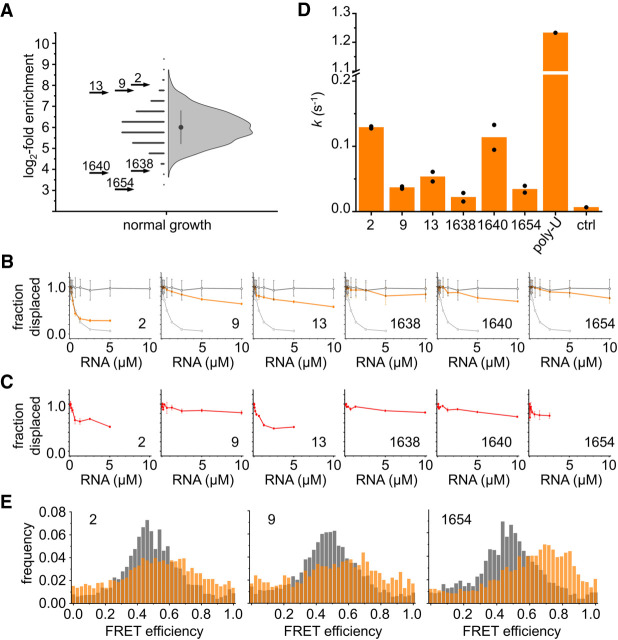
To test if RNAs identified as potential Hera binding partners by eCLIP also bind to Hera in vitro, we selected three RNA sequences from the top of the consolidated list Hera3/Hera4, showing a high log2-fold enrichment of 7–8 (RNAs 2, 9, and 13), and three RNAs from the bottom of the list with a low log2-fold enrichment of 3–4 (1638, 1640, 1654; Fig. 5A). While the former RNAs can fold into stem–loop structures with high thermodynamic stability (∼66 kJ × mol−1 at 37°C; Zuker 1989, 2003), the less enriched RNAs form less stable structures (∼24 kJ × mol−1). RNA 1640 contains both motifs 1 and 2 discovered by DREME, the remaining sequences contain neither of the two motifs according to FIMO. We measured binding of these RNAs to Hera in displacement titrations of a Hera/32mer RNA or an RBD/32mer RNA complex, using the decrease in anisotropy of fluorescein attached to the 5′-end of the 32mer as a spectroscopic probe for binding of the unlabeled RNAs. As a control, we performed a displacement with unlabeled 32mer (Supplemental Fig. 6). The homopurine 50A does not displace the 32mer and serves as a negative control, whereas the homopyrimidine 50U displaces the 32mer more efficiently than the unlabeled 32mer (positive control; Supplemental Fig. 6). In vitro experiments with peak sequences most likely underestimate the affinities of Hera for the respective RNAs, as the isolated sequences will lack structure or fold into different structures in the absence of their cellular context. Nevertheless, RNAs 2, 9, and 13 displace the 32mer from Hera, although with different efficiencies (Fig. 5B). Thus, Hera binds these RNAs in vitro in the absence of other cellular factors. RNAs 1638, 1640, and 1654 do not significantly displace the 32mer RNA in the concentration range tested. A lower affinity of Hera for these RNAs is consistent with their low log2-fold enrichment. It is also possible that binding of Hera to these RNAs requires an additional cellular factor. The titration data are thus consistent with a higher-affinity binding of Hera to RNAs with a higher log2-fold enrichment, and with a higher thermodynamic stability of structures formed. Only RNAs 2 and 13 displace the 32mer efficiently from an RBD/32mer complex (Fig. 5C). RNA 2 displace the 32mer from Hera and the RBD with similar efficiencies, and possibly interacts only with the RBD, not with the core. Overall, the affinity of Hera and of the RBD for the RNAs tested is thus higher for those RNAs with a high log2-fold enrichment.
FIGURE 5.
Binding of selected RNAs to Hera in vitro. (A) Half-Violin plot showing the range of log2-fold enrichment for all RNA sequences identified under normal-growth conditions. On the left side, the log2-fold change for each RNA is indicated by a symbol, the right-hand side shows the distribution. The filled circle marks the mean log2-fold enrichment, the line indicates one standard deviation. The log2-fold enrichment of the selected RNAs 2, 9, and 13 (log2-fold enrichment 8.0, 7.7, and 7.6) and 1638, 1640, and 1654 (log2-fold enrichment 3.9, 3.9, and 3.0) is indicated by arrows. (B) Displacement titrations of a Hera/32mer complex with RNAs 2, 9, 13, 1638, 1640, and 1654. The fluorescence anisotropy of a fluorescein attached to the 5′-end of the 32mer was used as a probe for 32mer dissociation due to binding of unlabeled RNAs. The open black circles depict values for a control experiment with a nonbinding 50A-RNA (negative control), the open gray circles show the control experiment with a binding 50U-RNA (positive control; see Supplemental Fig. 6). The average values from two replicate experiments are shown; error bars denote the standard error of the mean (SEM). Values are normalized: A value of one corresponds to no displacement of the fluorescein-labeled 32mer, a value of zero corresponds to complete displacement. (C) Displacement titrations of an RBD/32mer complex with RNAs 2, 9, 13, 1638, 1640, and 1654. The average of two replicate experiments is shown. Values are normalized: A value of one corresponds to no displacement of the fluorescein-labeled 32mer, a value of zero corresponds to complete displacement. (D) Rate constants k for ATP hydrolysis by Hera in the presence of different RNAs (10 µM) and saturating concentrations of poly(U) RNA (500 µM bases, see Linden et al. 2008) as an internal control. Experiments were performed in duplicate; individual values are depicted as circles, the bars show the mean. Ctrl: background in the presence of poly(U) and absence of enzyme. (E) Normalized single-molecule FRET histograms of Hera without RNA (gray), and in the presence of RNA and nucleotide (orange). RNA 2 and to a lesser extent also RNA 9 causes a shift in FRET efficiency to higher values, indicating that the Hera core closes in part of the Hera molecules. RNA 1654 induces a clear shift in FRET efficiency to higher values, consistent with closure and activation of the helicase core (see Supplemental Fig. 9 for control experiments).
Activation of the Hera core by RNAs identified by eCLIP
To test if the RNAs that bind to Hera in vitro interact with the Hera helicase core, we investigated the activation of the helicase core by these RNAs in steady-state ATPase assays (Fig. 5D) and in single-molecule FRET experiments with Hera carrying donor and acceptor fluorophores in the two RecA domains (Fig. 5E; Linden et al. 2008; Steimer et al. 2013). The RNAs stimulate the intrinsic ATPase activity of Hera to different extents; the maximum stimulation by RNA 2 is ∼10% of the stimulation achieved by saturating concentrations of poly(U) RNA (Linden et al. 2008). Except for RNA 2 that binds most tightly to Hera and also elicits the highest ATPase rate, there is no clear correlation between log2-fold enrichment, binding, and ATPase stimulation. For single-molecule FRET experiments, we selected RNAs 2, 9, and 1654, which cover the range from little to highest ATPase stimulation, and are bound by Hera and the RBD with different affinities. All RNAs caused an increase in FRET efficiency in the presence of the nonhydrolyzable ATP analog ADPNP, and thus induced closing of the Hera helicase core on binding, although the effect is most pronounced in the presence of RNA 1654. Two of these RNAs do not contain a motif identified by DREME, indicating that core activation is independent of the presence of these motifs. The (partial) activation of the helicase core by RNAs 2, 9, and 1654 suggests that all of them can interact with the Hera core in the presence of nucleotide, but stimulate the ATPase activity to different extents.
Genes identified and gene ontology
A visual inspection of genes with peaks identified under normal-growth and cold-shock conditions revealed that 23S rRNA and RNase P, two RNAs identified previously as in vitro substrates for Hera (Morlang et al. 1999; Linden et al. 2008) were absent from the consolidated lists of genes, although both genes showed coverage in the sequencing reads. 23S rRNA was also not found in the individual replicates, whereas the rnpB gene coding for RNase P RNA was identified in samples Hera2 and Hera3. Hairpin 92 and the adjacent helix 91 in the peptidyl-transferase center, which form the Hera binding site in 23S rRNA, are accessible in ribosome assembly intermediates, but not in fully assembled ribosomes (Tsu and Uhlenbeck 1998; Sharpe Elles et al. 2009), which may rationalize why this RNA is not recovered by eCLIP. The Hera binding site(s) in RNase P RNA might also not be exposed in the fully assembled ribonucleoprotein complex. Alternatively, 23S rRNA and RNase P RNA might not be physiological substrates of Hera (Steimer et al. 2013).
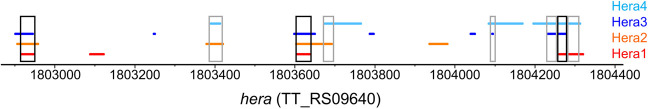
Interestingly, the hera gene itself (TT_RS09640) was among the peak-containing genes both under normal-growth conditions and after cold shock, indicating that Hera binds to its own mRNA. The peak sequences are more GC-rich than the Thermus genome, but show no increased GC content compared to the entire hera gene (Supplemental Fig. 7). The log2-fold enrichment was 7.00 and 6.62 under normal-growth- and 6.56 and 5.97 under cold-shock conditions, which is above the average (6.00 under normal-growth conditions, 5.75 under cold-shock conditions). However, the peaks were in different positions within the gene in the four individual experiments performed (Fig. 6), and no specific binding site within the mRNA could be identified from the data.
FIGURE 6.
Position of peaks in the hera gene. Position of peaks in the hera gene in Hera1 (red), Hera2 (orange), Hera3 (blue), and Hera4 (light blue). The black boxes highlight regions identified in three of the four experiments, the gray boxes highlight regions identified in two experiments.
Among the genes identified were numerous genes related to protein biosynthesis (tRNAs, tRNA-aminoacyl synthetases, tRNA-modifying enzymes, ribosomal proteins). As an example, genes for ribosomal proteins were highly represented with 18 out of the 22 genes (82%) for small ribosomal proteins, (12 after cold shock, 55%), and 31 out of 34 genes (91%) for large ribosomal subunit proteins (22 after cold shock, 65%). Other protein families represented were chaperones (e.g., dnaK, dnaJ, grpE, but not dafA, groES, groEL, clpB), cold-shock and general stress proteins (TT_RS08235, _09210; hsp22, hsp30; uspA), topoisomerase genes (gyrA, gyrB, topA), genes for transporters and their subunits (often both the gene for the substrate-binding protein and for the permease, for example, branched-chain amino acid ABC transporter, TT_RS01115 and _01120), and genes related to sugar- and cell wall metabolism and cell division, as well as CRISPR-related genes. In many cases, genes for all subunits of protein complexes (e.g., genes for cytochrome c oxidase subunits I and II; clpA, clpX coding for the ClpAX protease) were present.
A systematic gene ontology analysis using the DAVID server 6.7 (https://david-d.ncifcrf.gov/home.jsp) (Huang da et al. 2009a,b) with the consolidated gene lists for Hera1/Hera2, and Hera3/Hera4, translated into UniProt accession numbers, revealed three annotation clusters with significant enrichment (Supplemental Table 4a,b). Under normal-growth conditions, the most enriched cluster (enrichment score 11.4) comprised genes related to ribonucleoproteins, ribosomes, ribosomal proteins, and translation. The second-most enriched cluster (enrichment score 3.8) comprised genes belonging to protein biosynthesis and tRNA metabolism, including aminoacyl-tRNA synthetases. A third cluster (enrichment score 2.2) comprised genes related to nucleotide-binding proteins. Among the genes with peaks under cold-shock conditions, the same three annotation clusters were identified, with enrichment scores 6.5, 4.1, and 2.5, respectively.
We also analyzed the genes identified under both normal-growth and cold-shock conditions, only under normal-growth conditions, or only under cold-shock conditions. (Supplemental Table 4c–e). The same three annotation clusters (ribosome and translation, tRNA metabolism, and nucleotide binding, enrichment scores 6.4, 3.8, and 2.7, respectively) were reported for genes that contained peaks under both normal-growth and cold-shock conditions. Genes with peaks only under normal-growth conditions were enriched for genes related to ribosomes and translation, genes with peaks only under cold-shock conditions showed no specific enrichment. Genes that neither contained a peak under normal-growth nor under cold-shock conditions also showed no significant enrichment of a particular family of proteins (Supplemental Table 4f).
As an example for genes related to annotation cluster 1, 18 out of the 22 genes (82%) for proteins of the 30S ribosomal subunit were identified under normal-growth conditions, 12 (55%) after cold shock (11 common genes, three without peaks). For the proteins of the 50S ribosomal subunit, the picture was similar: Out of 34 genes, 31 (91%) were identified under normal-growth, 22 (65%) under cold-shock conditions (20 common genes, one without peaks).
For annotation cluster 2, we specifically examined the genes for aminoacyl-tRNA synthetases. T. thermophilus contains 21 aminoacyl-tRNA synthetase genes (Supplemental Table 5a). All 21 genes contained peaks under normal-growth conditions, 18 (86%) under cold-shock conditions. Thus, the gene ontology analysis supports an important role of Hera in protein biosynthesis, particularly in ribosome and tRNA metabolism.
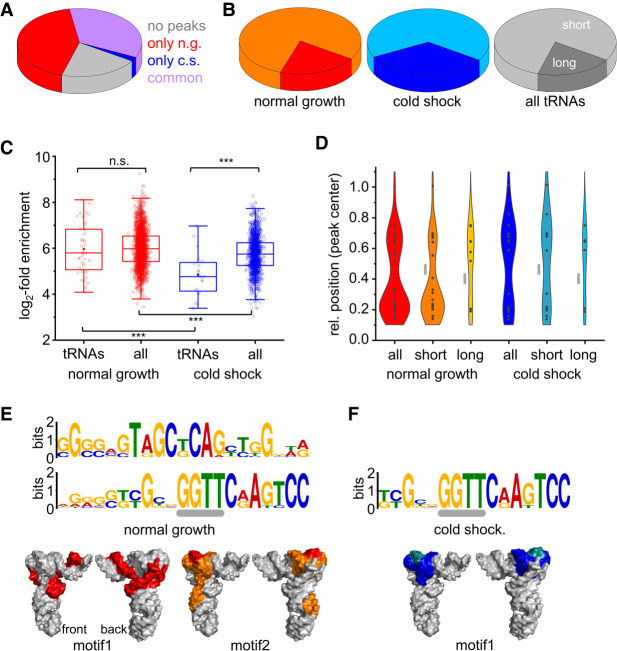
The gene ontology analysis focused on protein-coding genes. We therefore looked at the RNA genes separately. The T. thermophilus genome contains 53 annotated RNA genes: two genes each for 5S rRNA (TT_RS08185, _10125), 16S rRNA (TT_RS06975, _09465), and 23S rRNA (TT_RS08180, _10130), one tmRNA gene (TT_RS11335), RNase A RNA (TT_RS11270), and SRP RNA (TT_RS11430), and 49 genes for tRNAs. The only RNA genes that were enriched, both under normal-growth and cold-shock conditions, were those for tRNAs (Supplemental Table 5b), suggesting that Hera might perform an important role in tRNA metabolism. Under normal-growth conditions, Hera bound to 38 of the 49 tRNAs (78%; Fig. 7A), which is a similar fraction as the ∼75% of the RNA transcriptome bound by Hera overall. However, tRNA genes are very short (72–94 nt), and all tRNA genes of T. thermophilus comprise 3802 nt, less than 0.2% of the genome. After cold shock, 18 tRNAs out of 49 contain peaks (Fig. 7A), corresponding to 37%. Although tRNAs are abundant, they are often underrepresented in RNA-seq data (Jacob et al. 2019). Binding to this type of RNA is thus a strong indication for an involvement of the respective protein in tRNA metabolism (Tuck and Tollervey 2013; Gerstberger et al. 2014). A potential role of Hera in tRNA metabolism is also supported by an overrepresentation of sequences of genes coding for aminoacyl-tRNA synthetases and tRNA-modifying enzymes (Supplemental Table 5a,c): Peaks are present in 21 or 19 out of the 22 T. thermophilus amino-acyl-tRNA synthetase genes under normal-growth and cold-shock conditions, respectively (Supplemental Table 5a).
FIGURE 7.
The interaction of Hera with tRNAs. (A) Distribution of peaks in tRNA genes. n.g.: normal-growth conditions, c.s.: cold-shock conditions. Common: peaks under both normal-growth and cold-shock conditions. (B) Fraction of genes for short and long tRNAs among all tRNAs (light gray: short tRNAs, dark gray: long tRNAs), and distribution in genes with peaks under normal-growth (orange: short tRNAs, red: long tRNAs) and cold-shock conditions (cyan: short tRNAs, blue: long tRNAs). (C) Log2-fold enrichment of peaks in tRNA genes and in all genes under normal-growth (red) and under cold-shock conditions. n.s.: not significant, (***) P < 0.001. (D) Violin plot of the distribution of peaks in tRNAs identified under normal-growth (red: all tRNAs, orange: short tRNAs, yellow: long tRNAs) and cold-shock conditions (blue: all tRNAs, dark cyan: short tRNAs, light blue: long tRNAs). The gray bar marks the position of the anticodon. (E) Sequence motifs discovered with MEME (normal-growth conditions). (Top) Motif. The gray line marks a GGNY stretch compatible with binding to the Hera RBD. (Bottom) Position of peak in the cloverleaf structure of tRNAs, mapped onto the structure of T. thermophilus tRNAPhe (Klipcan et al. 2012; PDB-ID 3tup). The GGNY stretch in motif 2 is highlighted in red. (F) Sequence motifs discovered with MEME (cold-shock conditions). (Top) Motif. The gray line marks a GGNY motif compatible with binding to the Hera RBD. (Bottom) Position of peak in tRNA structure. The GGNY stretch is highlighted in cyan.
T. thermophilus contains short tRNAs (72–77 nt) and long tRNAs (85–94 nt). The long tRNAs contain insertions of various lengths between the anticodon and the TψC arm (Supplemental Fig. 8). Under normal-growth conditions, the fraction of short and long tRNAs bound to Hera reflected the distribution in the genome (Fig. 7B). In contrast, long tRNAs were overrepresented under cold-shock conditions. (Fig. 7B). The log2-fold enrichment of peak sequences in tRNAs was not significantly different from all peak sequences identified both under normal-growth conditions, and lower for cold-shock conditions (Fig. 7C), pointing to a preferred interaction with tRNAs under normal-growth conditions. Peaks in the tRNA genes were distributed bimodally, with maxima in the centers of the 5′- and 3′-half, and a minimum at the position of the anticodon (Fig. 7D). Under normal-growth conditions, Hera showed a preference for binding to the 5′-half (Fig. 7D), although for the long tRNA genes a slight preference for the 3′-half was observed. Under cold-shock conditions, the preference was shifted to the 3′-half for all tRNAs (Fig. 7D).
A motif search with MEME retrieved two 21-nt consensus motifs (motifs 1 and 2; Fig. 7E) under normal-growth conditions, and one 16-nt motif under cold-shock conditions (Fig. 7F). Motif 1 reflects peak sequences located in the 5′-half of tRNAs, motif 2 corresponds to peak sequences located in the 3′-half. The motif identified under cold-shock conditions is a subset of motif 2 identified under normal-growth conditions, and also maps to the 3′-half of tRNAs. Structurally, the peak sequences map to the elbow of the tRNAs, formed by the dihydrouridine and TψC-loops (Fig. 7F), which is a binding platform for interacting proteins and RNAs (Zhang and Ferre-D'Amare 2016).
In vitro binding of Hera to tRNAs and activation of the helicase core
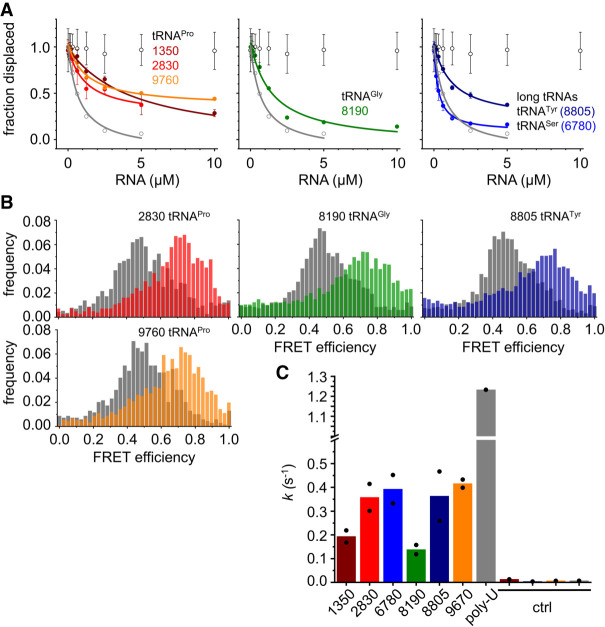
To validate if tRNAs are indeed substrates for Hera, we tested binding of three different tRNAPro (TT_RS02830: short tRNA, peak at 5′-end; TT_RS01350: short tRNA, peak at 3′-end; TT_RS09670: short tRNA, two peaks), of tRNAGly (TT_RS08190: short tRNA), and of the two long tRNAs tRNATyr (TT_RS08805) and tRNASer (TT_RS06780) to Hera in vitro in anisotropy displacement titrations (Fig. 8A). The tRNAs bound to Hera with Kd values in the range of 0.06–1.4 µM, confirming that no additional factors are needed for the interaction of Hera with these RNAs. Within the error of the experiment, the affinity of Hera for tRNAPro (TT_RS09670) that gave rise to two peaks in the eCLIP experiments was similar to the other tRNAs that gave only one peak.
FIGURE 8.
Interaction of Hera with tRNAs in vitro and activation of the helicase core. (A) Displacement titrations of a Hera/32mer complex with tRNAs. Data for 50A-RNA (open black circles) and 50U-RNA (open gray circles) are shown as negative and positive controls, respectively (see Supplemental Fig. 6). Data points are average values from two replicate experiments; error bars denote the standard error of the mean (SEM). Lines depict curves obtained from analyses with Equation (1); Kd values are 1.4 µM (1350), 0.6 µM (2830), 0.4 µM (9760), 0.4 µM (8190), 0.06 µM (6780), and 0.4 µM (8805), and 0.24 µM (50U). (B) Normalized single-molecule FRET histograms of Hera without RNA (gray), and in the presence of tRNA and nucleotide (colored as in A). All tRNAs cause an increase in FRET efficiency, indicative of closing of the Hera helicase core and core activation. See Supplemental Figure 9 for positive and negative control experiments. (C) Rate constants k for ATP hydrolysis by Hera in the presence of different tRNAs (5 µM) and poly(U) RNA (500 µM bases) as an internal control. Experiments were performed in duplicate; individual values are depicted as circles, the bar show the mean. Ctrl: Background in the presence of tRNAs 1350, 8805, 9670, and poly(U) RNA in the absence of enzyme. Numbers refer to the gene name (1350 = TT_RS01350, etc.).
Finally, we examined whether tRNA binding is coupled to the activation of the Hera helicase core in single-molecule FRET experiments (Fig. 8B; Supplemental Fig. 9) and steady-state ATPase assays (Fig. 8C). All tRNAs stimulated the intrinsic ATPase activity of Hera substantially. tRNASer, tRNATyr, and two of the tRNAPro (TT_RS02830 and _09670) elicited the highest rate constant for ATP hydrolysis, corresponding to ∼30%–40% of the stimulation achieved by saturating concentrations of poly(U) RNA (Linden et al. 2008). tRNAPro (TT_RS01350) and tRNAGly (TT_RS08190) achieved ∼10%–15% of the stimulation by poly(U). Thus the stimulation of ATP hydrolysis does not correlate with the affinity of Hera for the respective RNA. We also tested whether tRNAs can elicit closing of the Hera helicase core in single-molecule FRET experiments (Fig. 8B). Poly(U) RNA served as a positive control that elicits a shift to the closed, high-FRET state of the closed Hera conformer, whereas poly(G) RNA does not induce closing of the helicase core to the same extent (Supplemental Fig. 9). Binding of tRNAGly, tRNAPro (TT_RS02830, TT_RS09760), and tRNATyr caused a large increase in FRET efficiency in the presence of ADPNP, confirming that short and long tRNAs can bind to Hera and activate the helicase core.
Effect of a Hera deletion in vivo
The association of Hera with a large fraction of the T. thermophilus mRNAs both under normal-growth and cold-shock conditions suggest a key function for Hera in T. thermophilus in protein synthesis and tRNA metabolism, and in RNA metabolism in general. We therefore generated hera deletion strains (Supplemental Fig. 10a) and tested the effect of the hera deletion on T. thermophilus growth both at physiological and at low temperatures (72°C and 55°C, respectively; Fig. 9; Supplemental Fig. 10b,c). Cells lacking Hera are viable, demonstrating that the Hera gene is not essential. However, growth curves reveal a longer lag phase for deletion strains after inoculation of cultures compared to wild-type cells. The doubling time of mutants lacking Hera is increased (to >60 min) compared to about 45 min for T. thermophilus wild-type, in line with a general function of Hera in RNA metabolism. After changing the growth temperature to 55°C, wild-type and mutants are viable (Fig. 9; Supplemental Fig. 10c), but the growth rates are reduced tremendously. However, no difference in growth between wild-type and mutants is detected (Supplemental Fig. 10c). The appearance of wild-type and mutant cells is similar in the exponential phase, but during the stationary phase minor shape changes, aberrant morphologies, and a tendency toward elongated cells are visible (Supplemental Fig. 10d), suggesting a defective adaptation to the stationary phase. Altogether, the reduced growth of hera deletion mutants at physiological temperatures and their altered appearance are consistent with a key function of Hera in RNA metabolism in T. thermophilus.
FIGURE 9.
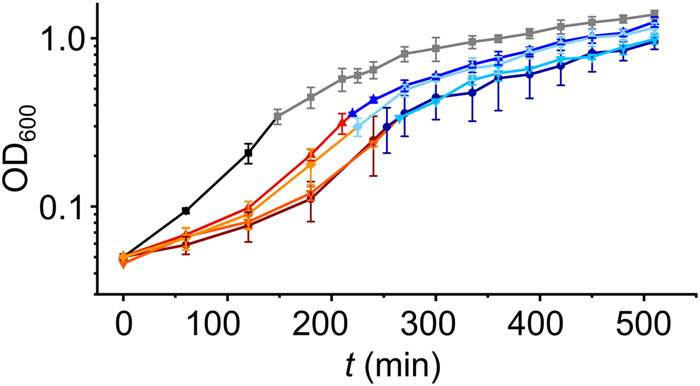
Growth of T. thermophilus wild-type and hera deletion mutants at high and low temperatures. Growth curves of wild-type T. thermophilus HB27 (black, gray) and of 4 hera deletion mutants (red/blue). Cultures were cultivated at 72°C (black, red hues for mutants) and rapidly transferred to 55°C during exponential growth (OD600 0.3–0.4), cultivation was then continued at 55°C (gray, blue hues for mutants). Data are the mean from two biological replicates, the error bars depict the standard error of the mean. See Supplemental Figure 10b,c for a linear plot of growth in the two different growth phases.
DISCUSSION
Hera: a house-keeping helicase with a broad spectrum of functions in RNA metabolism
Hera is the only DEAD-box protein present in T. thermophilus. The large number of eCLIP peaks in more than 75% of the T. thermophilus genes is in agreement with a wide range of functions of this helicase in RNA metabolism. The broad distribution of sequences bound by Hera along genes, with a very moderate preference for the 5′-proximal region, is consistent with a role of Hera as a general RNA chaperone, facilitating RNA folding and preventing misfolding as soon as RNAs emerge from the RNA polymerase during transcription. A key function of Hera in RNA metabolism under normal-growth conditions would also explain the observed lag phase, reduced growth rate and altered appearance of T. thermophilus hera deletion strains.
Hera shows a binding preference for RNAs with higher-than-average thermodynamic stability, and is recruited to regions that form stable secondary structures under cold-shock conditions, in agreement with a central function in the cold-shock response and cold adaptation (Mega et al. 2010). The sequences identified in the eCLIP experiments represent RNAs bound to Hera by the RBD without contributions from the helicase core, or by interactions with both the RBD and the helicase core. At this point, we cannot exclude that interaction partners affect the range of RNAs bound to Hera in vivo. However, the RNAs for which we validated binding in in vitro experiments are bound to Hera with high affinity, and activate the helicase core of Hera in the presence of nucleotide, indicating that they interact with the RBD and with the helicase core of Hera, even in the absence of other factors. Although sequence motifs were identified, their presence in only a subset of the peak sequences suggests that RNA recognition by Hera is structure- rather than sequence-specific. We have shown previously that Hera can unwind RNA duplexes flanked by hairpins with stems of four to six nucleotides, and loops of four to six nucleotides (Steimer et al. 2013). Consistent with a key role of hairpins in RNA binding to Hera, the six peak sequences for which we confirmed in vitro binding all form stem–loop structures. tRNAs, which consist of four stem–loop elements folded into the characteristic three-dimensional L-shape, also bind to Hera in vivo and in vitro. Binding to structured regions by Hera is reminiscent of the RNA-binding mode of other helicases such as the splicing helicases and general RNA chaperones Cyt-19 (Tijerina et al. 2006; Grohman et al. 2007) and Mss116 (Mallam et al. 2011).
Hera as a cold-shock protein
Hera has been identified as a cold-shock protein previously (Mega et al. 2010). We show here that Hera binds to its own mRNA. Although the binding sites are scattered over the entire Hera mRNA, this observation may point to a possible auto-regulatory mechanism. Hera is associated with a number of RNAs related to the general stress response, in agreement with a regulatory role in the response to stress conditions. Low temperatures reduce membrane fluidity, slow down mRNA translation and protein folding, and stabilize nucleic acid structures. The high enrichment of RNAs related to ribosomes and translation is consistent with a function of Hera as an RNA chaperone and thus a general regulator of ribosomal protein levels to maintain protein biosynthesis. Under cold-shock conditions, Hera interacts with RNAs that have a high propensity to form stable secondary structures, exemplified by the enrichment of longer, GC-rich, structured RNAs such as the group of long tRNAs identified in eCLIP experiments under cold-shock conditions. Although we cannot exclude that the changes in RNAs bound under cold-shock conditions partially reflect changes in the transcriptome, our data suggest that Hera might be directed to “severe cases” after a cold shock, which require remodeling to maintain the flexibility and structure required for the physiological function of these RNAs. The bacterial cold-shock DEAD-box protein CsdA is essential for growth at low temperatures in E. coli, Yersinia pseudotuberculosis and Psychrobacter arcticus (Jones et al. 1996; Bergholz et al. 2009; Palonen et al. 2012). CsdA is involved in translation (Lu et al. 1999), ribosome biogenesis (Charollais et al. 2004; Peil et al. 2008), mRNA stability and degradation (Rocak and Linder 2004; Awano et al. 2007). The high association of Hera with mRNAs of ribosomal proteins and tRNAs, as well as its interaction with ribosomal RNAs in vitro, suggest Hera has similar functions to CsdA. In agreement with similar functions, Hera and CsdA share structural similarity: Both enzymes form a stable dimer, mediated by dimerization domains with similar folds, and contain a carboxy-terminal RBD (Rudolph et al. 2006; Klostermeier and Rudolph 2009; Rudolph and Klostermeier 2009; Xu et al. 2017). Despite the large body of evidence for a function of Hera in the cold-shock response, hera deletion strains do not show significant impairment in growth at low temperatures. While Hera might play an important role in the immediate cold-shock response, the wild-type-like growth of the hera mutants at low temperatures suggests that alternative mechanisms exist that enable T. thermophilus to cope with increased RNA stability at reduced temperatures and to achieve cold adaptation.
Hera binding to tRNAs points to a key role in tRNA metabolism
Among the genes identified by eCLIP, tRNAs, tRNA synthetases, and tRNA modification enzymes are overrepresented. Hera binds to the majority of the tRNAs in T. thermophilus, again highlighting its preference for binding to stem–loop structures (Steimer et al. 2013). Formation of the tertiary structure of tRNAs relies on stable secondary structure elements, which can serve as kinetic traps and frequently lead to misfolding (Treiber and Williamson 1999; Bhaskaran et al. 2012). According to structure predictions, Thermus tRNAs show a strong tendency to form aberrant structures deviating from the four-hairpin consensus that forms into a cloverleaf shape. In addition, Thermus contains a number of long tRNAs with an expanded variable loop which can fold into an extra hairpin at lower temperatures, rendering these tRNAs particularly prone to misfolding. These longer tRNAs are preferentially bound to Hera under cold-shock conditions. It is conceivable that the helicase function of Hera is needed to remodel such off-pathway folding intermediates of tRNAs, particularly under cold-shock conditions. The increased lag phase in growth observed for the hera mutants would be consistent with a chaperone function of Hera for tRNAs.
Binding sites in tRNAs map to the elbow of the L-shaped structure, formed by the dihydrouridine- and TψC-loops. The elbow is recognized by numerous tRNA-binding proteins, including tRNA modification enzymes, and by noncoding RNAs such as 23S rRNA, RNase P, and T-box riboswitches (for review, see Zhang and Ferre-D'Amare 2016). The interaction mode of RNAs with the elbow is conserved, while protein binding is more diverse. Notably, a number of enzymes recognizing the elbow region act on distant parts of the tRNAs. The end-processing enzymes RNase P (Reiter et al. 2010) and RNase Z (Pellegrini et al. 2012), as well as CCA-adding or CC- and A- adding enzymes establish interactions with the elbow (Xiong and Steitz 2004; Yamashita et al. 2014, 2015), but act on the 5′- or 3′-ends at the end of the acceptor stem. tRNA modification enzymes either contact the elbow and disrupt its structure to modify a nucleotide within this region (Hoang and Ferre-D'Amare 2001; Ishitani et al. 2003; Byrne et al. 2015), or they mediate nucleotide modification elsewhere in the tRNA. Some tRNA synthetases also recognize tRNAs through the elbow region, which may provide an additional identity element for correct aminoacylation at the 3′-end (Fukai et al. 2000; Oshikane et al. 2006; Rock et al. 2007). Although the region of the anticodon is underrepresented in tRNA sequences bound to Hera, it is conceivable that Hera preferentially interacts with the elbow region, possibly through the RBD, but the helicase core can act on any region of the tRNA within reach, including the anticodon loop.
Nucleotides in tRNAs are highly modified in all kingdoms of life. It is thought that these modifications increase the thermal stability of tRNAs, particularly in thermophilic genera such as Thermus (Tomikawa et al. 2010; for review, see Hori 2019). On the other hand, tRNA modifications are also frequent in psychrophilic bacteria (Dalluge et al. 1997), and have been linked to cold adaptation of T. thermophilus (Shigi et al. 2006; Ishida et al. 2011). Our eCLIP results strongly support a role for Hera in tRNA modification under normal-growth and cold-shock conditions. A prominent role of helicases in RNA modification is not unprecedented: The yeast helicases Prp43 and Rok1 promote release of snoRNAs which hybridize to modification sites of rRNA in preribosomal particles and mark them for RNA modification enzymes (Bohnsack et al. 2008, 2009). The acetyltransferase TmcA catalyzes the acetylation of a cytosine in the anticodon loop of elongator-tRNAMet (Ikeuchi et al. 2008; Chimnaronk et al. 2009), a modification that enhances codon–anticodon interactions and ribosome fidelity (Kawai et al. 1989). TmcA consists of a modified helicase domain, fused to an acetylase domain. Possibly, Hera as the only DEAD-box protein in Thermus provides the general unwinding function to aid tRNA modification at high and low temperatures.
Implications for the mechanism of RNA unwinding by the Hera dimer
Previous studies indicate that Hera is a stable dimer at lower temperatures, and might remain a dimer under physiological conditions at high temperatures (Klostermeier and Rudolph 2009), suggesting that the two helicase cores could functionally cooperate in remodeling structures of large RNA substrates. Several genes identified from eCLIP sequences contain multiple peaks, which could be caused by binding of these RNAs through the helicase core and the RBD, by multiple copies of Hera binding to these RNAs, or by binding of the two helicase cores to adjacent sites, enabling a concerted action of one dimer on different regions within the same RNA molecule. The RNAs identified here will provide a starting point to characterize the interaction of Hera with physiological RNA substrates, as well as the mechanism of a possible functional cooperation between the two helicase cores in RNA and ribonucleoprotein remodeling.
MATERIALS AND METHODS
Protein production and purification
All proteins were produced in E. coli Rosetta(DE3) grown in autoinducing medium (Studier 2005). Hera, Hera_E115C/E260C, and the GST-Hera_RBD fusion protein were purified as described (Linden et al. 2008; Steimer et al. 2013).
eCLIP
Thermus thermophilus HB27 (DSM 7039, DSMZ; ATCC BAA-163) was grown in liquid media (8 g × L−1 peptone, 4 g × L−1 yeast extract, 2 g × L−1 NaCl, pH 7.5) at 75°C. At an OD600 of 0.8 (Hera2, Hera3) or 1.0 (Hera1, Hera4), cells from cultures Hera1 and Hera2 were spread in a thin layer in a heated Petri dish, and RNA–protein complexes were cross-linked by illumination with UV light (254 nm, 400 J cm−2). Cultures Hera3 and Hera4 were subjected to a cold shock by rapidly reducing the growth temperature to 65°C for 10 min, followed by cross-linking. UV-irradiated cells were pelleted by centrifugation (4000 rcf, 10 min), the pellets were shock-frozen in liquid nitrogen and stored at −80°C until library preparation was performed.
cDNA library preparation and sequencing
The isolation of RNA–protein complexes, trimming of the RNA, and preparation of the cDNA library was performed as described (Fig. 1B; Van Nostrand et al. 2016). Briefly, cells were thawed in lysis buffer (50 mM Tris/HCl, pH 7.4, 100 mM NaCl, 1% [w/v] NP-40, 0.1% [w/v] SDS, 0.5% [w/v] sodium deoxycholate, and Protein Inhibitor Cocktail III [1:200, EMD Millipore]) and disrupted by sonication. The crude extract was treated with Turbo DNase (ThermoFisher) and RNase I (ThermoFisher) to remove DNA and to trim the protein-bound RNA. Hera/RNA complexes were immunoprecipitated using an anti-Hera antibody from rabbit (Eurogentec, Supplemental Fig. 11), and isolated using sheep anti-rabbit Dynabeads (ThermoFisher). Subsequent steps were performed with RNA bound to the beads. After dephosphorylation with FastAP (ThermoFisher) and T4 PNK (NEB), a barcoded RNA adapter was ligated to the 3′ end (T4 RNA Ligase, NEB). RNA–protein complexes were separated by SDS-PAGE and transferred to nitrocellulose membranes. The region from 55–130 kDa (75 kDa above the molecular mass of Hera) was excised, the membrane was suspended in buffer and treated with proteinase K (NEB). RNA was isolated by phenol/chloroform extraction, purified on spin columns (Zymo Research), and reverse-transcribed with AffinityScript (Agilent). Excess oligonucleotides were removed by treatment with ExoSAP-IT (Affymetrix). The second DNA adapter (containing a random-mer of 5 [N5] or 10 [N10] random bases at the 5′-end) was ligated to the 5′-end of the cDNA (T4 RNA Ligase, NEB). After cleanup on Dynabeads MyOne Silane (ThermoFisher), the required number of PCR cycles was determined from qPCR experiments, the DNA was amplified by PCR, purified with the PippinPrep system (Sage Science), and sequenced on the Illumina HiSeq 4000 platform. Paired size-matched input libraries were generated in parallel from supernatants of lysates subjected to immunoprecipitation with the anti-Hera antibody. Normalization of the eCLIP data to the size-matched input eliminates biases for structured RNAs that may arise from the preference of RNase I for cleaving single-stranded RNA. Libraries were analyzed for fragment size distribution on a D1000 Screentape (Agilent; Supplemental Figs. 12, 13). The sequencing data have been deposited in the Gene Expression Omnibus (GEO) database (accession number GSE135435).
eCLIP data analysis
Reads were processed as described for mapping to mammalian genomes (Van Nostrand et al. 2016), with the indicated modifications. Briefly, reads were demultiplexed and trimmed to remove adapters and to account for double ligation events (Cutadapt 1.14). Repeat element sequences aligning to RepBase (18.05) with STAR (2.4.0i) were removed and remaining reads mapped to the T. thermophilus HB27 genome assembly (GCF_000008125.1) (Henne et al. 2004), allowing for mapping to up to 10 genomic loci per read. PCR duplicate analysis on uniquely mapped reads using the unique molecular identifiers (UMIs) in the 5′-adapter showed an average 85% unique library molecules. Peaks were called in the immunoprecipitation (IP) samples with CLIPper as described (Lovci et al. 2013). Each peak was normalized to the corresponding size-matched input (SMInput) by calculating the fraction of the number of reads from the IP sample to the number of reads from the corresponding SMInput sample. To a first approximation, this normalization also accounts for changes in transcript levels between different samples. Peaks were deemed significant at greater than or equal to eightfold enrichment (log2-fold change ≥ 3) and P ≤ 10−3 (χ2 test). Sequencing reads were mapped to the T. thermophilus HB27 genome (Henne et al. 2004). Peak sequences were retrieved using the Galaxy getfastabed tool (www.usegalaxy.org; Blankenberg et al. 2007). Analysis for nucleotide bias and GC content was performed with the Genomics %G∼C Content Calculator (www.sciencebuddies.org) and with EMBOSS Infoseq (www.usegalaxy.org; Rice et al. 2000; Blankenberg et al. 2007). Sequence motifs were identified with MEME (Multiple Em for Motif; Bailey et al. 2009), DREME (Discriminative Regular Expression Motif Elicitation; Bailey 2011), and FIMO (Find Individual Motif Occurrence; Grant et al. 2011); all available at www.meme-suite.org; version 5.0.5. RNA structures were predicted and their thermodynamic stabilities were calculated using the program RNAstructure (www.usegalaxy.org; http://rna.urmc.rochester.edu/RNAstructure.html).
RNA substrates
RNA oligonucleotides were purchased PAGE-purified from Sigma. Sequences were
5′-UAGGGGCGUG UGGGCCCACC CGGCGUGGCG CUGGCUCCAC C-3′ (RNA 2: TT_RS08430, 1577621-1577662), 5′-CUGGCUUGCC CGGGAGAUCC UGGGCUACCC CGA-3′ (RNA 9: TT_RS01805, 342075-342108), 5′-UUUGGGGCUG CGGGGCUGGA CCCCACGGGC GCCGCCACCC-3′ (RNA 13: TT_RS07980, 1497551-1497591), 5′-GCCAAGCCGG GCCUGGAC-3′ (RNA 1638: TT_ RS03440, 660264-660282), 5′-CUCGUGGCCCUGGCGAGGAG GCUCGUGGAG GAGUAC-3′ (RNA 1640: TT_RS09625, 1800084-1800120), and 5′-CCUGCACGUU CCCACCGCCU GGGU-3′ (RNA 1654: TT_RS05260, 1015666-1015690). The numbers are the rank of these sequences in a list of peaks sorted after the log2-fold enrichment over input (consolidated list from Hera3/Hera4, after cold shock, see Results, Supplemental Table 3).
tRNAs were ordered HPLC-purified from Dharmacon. Sequences were
5′-CGGGGAGUAG CGCAGCCCGG UAGCGCACCU CGUUCGGGAC GAGGGGGUCG CUGGUUCAAA UCCAGUCUCC CCGACCA-3′ (tRNAPro, TT_RS02830), 5′-CGGGGCGUAG CGCAGGCCGG UAGCGCACCU GCUUUGGGAG CAGGGGGUCG CCCGUUCAAA UCGGGCCGCC CCGACCA-3′ (tRNAPro, TT_RS01350), 5′-CGGGGAGUAG CGCAGCCUGG UAGCGCACAC GCUUGGGGUG CGUGGGGUCG UCGGUUCAAA UCCGGCCUCC CCGACCA-3′ (tRNAPro, TT_RS09670), 5′-GCGGGAGUAG CUCAGUCGGU AGAGCACGAC CUUGCCAAGG UCGGGGUCGC GGGUUCAAGU CCCGUCUCCC GCUCCA-3′ (tRNAGly, TT_RS08190), 5′-GGGCAGGUGC CCGAGCGGCC AAAGGGGACG GUCUGUAAAA CCGUUGGCGU AUGCCUUCGC UGGUUCGAAU CCAGCCCUGC CCACCA-3′ (tRNATyr, TT_RS08805), and 5′-GGAGAGGUGG CCGAGUGGUC GAAGGCGGCA CCCUGCUAAG GUGUUGCACC GGGAAAACCG GUGCCGCGGG UUCGAAUCCC GCCCUCUCCG CCA-3′ (tRNASer, TT_RS06780).
Poly(U) and poly(G) RNA were from Sigma. The 9mer DNA (5′-AGGTCCCAA-3′) was purchased PAGE-purified from Sigma.
Determination of Kd values in fluorescence equilibrium titrations
Kd values for Hera/RNA complexes were determined in displacement titrations of 0.3 µM 32mer RNA labeled with fluorescein at the 5′-end and 0.3 µM Hera or 0.5 µM GST-Hera_RBD with unlabeled RNA in 50 mM Tris/HCl, pH 7.5, 150 mM NaCl, 5 mM MgCl2 at 25°C with Hera or GST-Hera_RBD (Linden et al. 2008), using a Jobin Yvon FluoroMax4 fluorimeter. Fluorescence was excited at 496 nm (2 nm bandwidth), and detected at 520 nm (4 nm bandwidth). To ensure equilibrium conditions, individual samples were prepared for each data point, and preequilibrated for at least 45 min before the measurement. Measurements were performed in duplicate; data points were averaged. For titrations with tRNAs, data were analyzed with error-weighted fits using Equation 1 and the program Origin:
The parameter r is the measured anisotropy, rb and rf are the anisotropies of bound and free RNA1 (fluorescein-labeled 32mer), respectively, [RNA1]tot and [RNA2]tot are the total concentrations for RNA1 and RNA2 (unlabeled RNA), and Kd,1 and Kd,2 are the dissociation constants of the Hera/RNA1 and Hera/RNA2 complexes. Kd,1 was determined independently in a control measurement prior to the displacement titration and kept constant during the fit. Note that the Kd,2 values reflect the dissociation constants of the complex of Hera with the unlabeled RNA, and are independent of the identity of the RNA displaced. See Supplemental Figure 6 for control experiments, and the Supplemental Methods for the derivation of Equation 1.
Steady-state ATPase activity
Steady-state ATPase activity was measured in a coupled enzymatic assay as described (Linden et al. 2008) with 0.3 µM Hera at 37°C in 50 mM Tris/HCl pH 7.5, 150 mM NaCl, 5 mM MgCl2, and 5 mM ATP at 37°C. RNAs 2, 9, 13, 1638, 1640, or 1654 were added at a concentration of 10 µM, tRNAs at a concentration of 5 µM; poly(U) RNA was used at saturating concentrations (500 µM bases; see Linden et al. 2008). Reaction velocities v for ATP hydrolysis were obtained from the change in absorption with time, and converted to rate constants k by division by the enzyme concentration. Measurements were performed in duplicate; errors for k are standard errors of the mean (SEM).
Single-molecule FRET experiments
Single-molecule FRET experiment were performed on a Microtime 200 (Picoquant) with Hera_E115C/R260C, labeled with Alexa488 (donor) and Alexa546 (acceptor) as described (Linden et al. 2008). Unlabeled Hera was added in a fivefold molar excess to labeled Hera_E115C/R260C and incubated for 1 h at 65°C to allow for subunit exchange and the formation of heterodimers in which only one of the protomers in the Hera dimer is labeled. Concentrations were 200 pM Hera (donor concentration, corresponding to 2.5 nM total Hera concentration), 8 mM ADPNP, 2.4 µM RNA in 50 mM Tris/HCl, pH 7.5, 150 mM NaCl, 5 mM MgCl2. Poly(U) and poly(G) RNA were used at a concentration of 1 mM (bases). Measurements were performed at 25°C.
Strains and growth conditions for the analysis of hera deletion variants
T. thermophilus strains were grown on TB medium (8 g × L−1 of Trypticase [BBL], 4 g × L−1 of yeast extract [Oxoid], and 3 g × L−1 of NaCl in carbonate rich mineral water, adjusted to pH 7.5) at the indicated temperatures with mild shaking (180 rpm). Escherichia coli DH5α [supE44 Δ(lacZYA-argF)U169 Φ80lacZΔM15 hsdR17 recA1 endA1 gyrA96 thi-1 relA1] was used for plasmid construction. E. coli was routinely grown in LB medium at 37°C. Agar (1.5% [w/v]) was added to the TB or LB media for growth on plates. Ampicillin (100 μg × mL−1), or Hygromycin B (100 μg × mL−1) were used for selection when needed.
Isolation of Δhera::hph mutants
The gene TT_RS09640 (TTC-1895) encoding Hera was deleted following a double recombination insertion procedure. For this, an upstream recombination arm of 1126 bp, flanked by restriction sites for EcoRI and XbaI, and a downstream recombination arm of 1012 bp, flanked by restriction sites for XbaI and SalI, were amplified with the proofreading Phusion High-Fidelity DNA Polymerase (NEB Biolabs; see Supplemental Table 6 for primer sequences), and cloned in an oriented form into plasmid pUC19 (Vieira and Messing 1982) using the indicated restriction sites. Further, the hph17 gene encoding a thermostable form of the Hygromycin phosphotransferase evolved from the E. coli gene (S Bosch, J Quesada, J Berenguer, and A Hidalgo, unpubl.) was cloned into the single XbaI site located between the recombination arms. The resulting plasmid pUC-Δhera::hph was linearized with EcoRI and used to transform wild-type cells of T. thermophilus HB27 by natural competence (Alvarez et al. 2019). Insertion mutants were selected on TB plates containing 100 µg mL−1 of Hygromycin B after incubation at 65°C for 48 h. Colonies were restreaked twice, grown in TB medium with Hygromycin B and conserved as frozen pellets for further analysis. The absence of the hera gene in the mutants was confirmed by PCR amplification with appropriate primers (Supplemental Table 6) and further sequencing of the amplification products. Four strains (hera1–hera4) were selected for further studies.
Growth analysis
The wild-type and the four hera mutants were grown to stationary phase on TB at 65°C, with Hygromycin B (100 mg L−1) for the mutants, and diluted in duplicate samples to OD600 = 0.05 into preheated TB medium without antibiotics. The cultures were incubated at 72°C under stirring, and growth was followed at regular time intervals until the cultures reached approximately OD600 = 0.3–0.4. At this point, the cultures were shifted to 55°C by transferring them to a water bath for two minutes to allow rapid decrease of temperature, and the growth was further followed in an air shaking bath at 55°C.
DATA DEPOSITION
The sequencing data have been deposited in the Gene Expression Omnibus (GEO) database (accession number GSE135435). An annotated UCSC Genome Browser session with the tracks is available under http://genome.ucsc.edu/s/brianfbb/hub_1497567_hb27_ann.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
COMPETING INTEREST STATEMENT
G.W.Y. is cofounder, member of the Board of Directors, on the SAB, equity holder, and paid consultant for Locanabio and Eclipse BioInnovations. G.W.Y. is a visiting professor at the National University of Singapore. G.W.Y.’s interests have been reviewed and approved by the University of California San Diego in accordance with its conflict of interest policies. The authors declare no other competing financial interests.
Supplementary Material
ACKNOWLEDGMENTS
We thank Jessica Guddorf for excellent technical assistance, Angel Angelov (Technical University of Munich, Germany) for helpful advice on gene ontology analyses, James R. Williamson (The Scripps Research Institute, La Jolla, USA) for support, and members of the Klostermeier, Yeo, and Williamson laboratories for help, advice, and discussions. This work was supported by the DFG (KL1153/7-1 and 7-2 [D.K.]), the Spanish Ministry of Economy and Competitiveness (BIO2016-77031-R [J.B.]), and the U.S. National Institutes of Health (HG004659 and HG009889 [G.W.Y.])
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.075580.120.
REFERENCES
- Alvarez L, Sanchez-Hevia D, Sanchez M, Berenguer J. 2019. A new family of nitrate/nitrite transporters involved in denitrification. Int Microbiol 22: 19–28. 10.1007/s10123-018-0023-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awano N, Xu C, Ke H, Inoue K, Inouye M, Phadtare S. 2007. Complementation analysis of the cold-sensitive phenotype of the Escherichia coli csdA deletion strain. J Bacteriol 189: 5808–5815. 10.1128/JB.00655-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL. 2011. DREME: motif discovery in transcription factor ChIP-seq data. Bioinformatics 27: 1653–1659. 10.1093/bioinformatics/btr261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. 2009. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res 37: W202–W208. 10.1093/nar/gkp335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergholz PW, Bakermans C, Tiedje JM. 2009. Psychrobacter arcticus 273-4 uses resource efficiency and molecular motion adaptations for subzero temperature growth. J Bacteriol 191: 2340–2352. 10.1128/JB.01377-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaskaran H, Rodriguez-Hernandez A, Perona JJ. 2012. Kinetics of tRNA folding monitored by aminoacylation. RNA 18: 569–580. 10.1261/rna.030080.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blankenberg D, Taylor J, Schenck I, He J, Zhang Y, Ghent M, Veeraraghavan N, Albert I, Miller W, Makova KD, et al. 2007. A framework for collaborative analysis of ENCODE data: making large-scale analyses biologist-friendly. Genome Res 17: 960–964. 10.1101/gr.5578007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohnsack MT, Kos M, Tollervey D. 2008. Quantitative analysis of snoRNA association with pre-ribosomes and release of snR30 by Rok1 helicase. EMBO Rep 9: 1230–1236. 10.1038/embor.2008.184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohnsack MT, Martin R, Granneman S, Ruprecht M, Schleiff E, Tollervey D. 2009. Prp43 bound at different sites on the pre-rRNA performs distinct functions in ribosome synthesis. Mol Cell 36: 583–592. 10.1016/j.molcel.2009.09.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busa VF, Rector MJ, Russell R. 2017. The DEAD-Box protein CYT-19 uses arginine residues in its C-tail to tether RNA substrates. Biochemistry 56: 3571–3578. 10.1021/acs.biochem.7b00362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne RT, Jenkins HT, Peters DT, Whelan F, Stowell J, Aziz N, Kasatsky P, Rodnina MV, Koonin EV, Konevega AL, et al. 2015. Major reorientation of tRNA substrates defines specificity of dihydrouridine synthases. Proc Natl Acad Sci 112: 6033–6037. 10.1073/pnas.1500161112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartier G, Lorieux F, Allemand F, Dreyfus M, Bizebard T. 2010. Cold adaptation in DEAD-box proteins. Biochemistry 49: 2636–2646. 10.1021/bi902082d [DOI] [PubMed] [Google Scholar]
- Cavicchioli R, Thomas T, Curmi PM. 2000. Cold stress response in archaea. Extremophiles 4: 321–331. 10.1007/s007920070001 [DOI] [PubMed] [Google Scholar]
- Chamot D, Magee WC, Yu E, Owttrim GW. 1999. A cold shock-induced cyanobacterial RNA helicase. J Bacteriol 181: 1728–1732. 10.1128/JB.181.6.1728-1732.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charollais J, Dreyfus M, Iost I. 2004. CsdA, a cold-shock RNA helicase from Escherichia coli, is involved in the biogenesis of 50S ribosomal subunit. Nucleic Acids Res 32: 2751–2759. 10.1093/nar/gkh603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chimnaronk S, Suzuki T, Manita T, Ikeuchi Y, Yao M, Suzuki T, Tanaka I. 2009. RNA helicase module in an acetyltransferase that modifies a specific tRNA anticodon. EMBO J 28: 1362–1373. 10.1038/emboj.2009.69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalluge JJ, Hamamoto T, Horikoshi K, Morita RY, Stetter KO, McCloskey JA. 1997. Posttranscriptional modification of tRNA in psychrophilic bacteria. J Bacteriol 179: 1918–1923. 10.1128/JB.179.6.1918-1923.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukai S, Nureki O, Sekine S, Shimada A, Tao J, Vassylyev DG, Yokoyama S. 2000. Structural basis for double-sieve discrimination of L-valine from L-isoleucine and L-threonine by the complex of tRNAVal and valyl-tRNA synthetase. Cell 103: 793–803. 10.1016/S0092-8674(00)00182-3 [DOI] [PubMed] [Google Scholar]
- Gerstberger S, Hafner M, Tuschl T. 2014. A census of human RNA-binding proteins. Nat Rev Genet 15: 829–845. 10.1038/nrg3813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant CE, Bailey TL, Noble WS. 2011. FIMO: scanning for occurrences of a given motif. Bioinformatics 27: 1017–1018. 10.1093/bioinformatics/btr064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grohman JK, Campo MD, Bhaskaran H, Tijerina P, Lambowitz AM, Russell R. 2007. Probing the mechanisms of DEAD-Box proteins as general RNA chaperones: the C-terminal domain of CYT-19 mediates general recognition of RNA. Biochemistry 46: 3013–3022. 10.1021/bi0619472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther UP, Weinberg DE, Zubradt MM, Tedeschi FA, Stawicki BN, Zagore LL, Brar GA, Licatalosi DD, Bartel DP, Weissman JS, et al. 2018. The helicase Ded1p controls use of near-cognate translation initiation codons in 5′ UTRs. Nature 502: 385–388. 10.1038/nature12543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henn A, Bradley MJ, De La Cruz EM. 2012. ATP utilization and RNA conformational rearrangement by DEAD-Box proteins. Annu Rev Biophys 41: 247–267. 10.1146/annurev-biophys-050511-102243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henne A, Bruggemann H, Raasch C, Wiezer A, Hartsch T, Liesegang H, Johann A, Lienard T, Gohl O, Martinez-Arias R, et al. 2004. The genome sequence of the extreme thermophile Thermus thermophilus. Nat Biotechnol 22: 547–553. 10.1038/nbt956 [DOI] [PubMed] [Google Scholar]
- Hilbert M, Karow AR, Klostermeier D. 2009. The mechanism of ATP-dependent RNA unwinding by DEAD box proteins. Biol Chem 390: 1237–1250. 10.1515/BC.2009.135 [DOI] [PubMed] [Google Scholar]
- Hoang C, Ferre-D'Amare AR. 2001. Cocrystal structure of a tRNA Psi55 pseudouridine synthase: nucleotide flipping by an RNA-modifying enzyme. Cell 107: 929–939. 10.1016/S0092-8674(01)00618-3 [DOI] [PubMed] [Google Scholar]
- Hori H. 2019. Regulatory factors for tRNA modifications in extreme-Thermophilic Bacterium Thermus thermophilus. Front Genet 10: 204 10.3389/fgene.2019.00204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. 2009a. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 37: 1–13. 10.1093/nar/gkn923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. 2009b. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44–57. 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- Hunger K, Beckering CL, Wiegeshoff F, Graumann PL, Marahiel MA. 2006. Cold-induced putative DEAD box RNA helicases CshA and CshB are essential for cold adaptation and interact with cold shock protein B in Bacillus subtilis. J Bacteriol 188: 240–248. 10.1128/JB.188.1.240-248.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeuchi Y, Kitahara K, Suzuki T. 2008. The RNA acetyltransferase driven by ATP hydrolysis synthesizes N4-acetylcytidine of tRNA anticodon. EMBO J 27: 2194–2203. 10.1038/emboj.2008.154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida K, Kunibayashi T, Tomikawa C, Ochi A, Kanai T, Hirata A, Iwashita C, Hori H. 2011. Pseudouridine at position 55 in tRNA controls the contents of other modified nucleotides for low-temperature adaptation in the extreme-thermophilic eubacterium Thermus thermophilus. Nucleic Acids Res 39: 2304–2318. 10.1093/nar/gkq1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishitani R, Nureki O, Nameki N, Okada N, Nishimura S, Yokoyama S. 2003. Alternative tertiary structure of tRNA for recognition by a posttranscriptional modification enzyme. Cell 113: 383–394. 10.1016/S0092-8674(03)00280-0 [DOI] [PubMed] [Google Scholar]
- Jacob D, Thuring K, Galliot A, Marchand V, Galvanin A, Ciftci A, Scharmann K, Stock M, Roignant JY, Leidel SA, et al. 2019. Absolute quantification of noncoding RNA by microscale thermophoresis. Angew Chem Int Ed Engl 58: 9565–9569. 10.1002/anie.201814377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagessar KL, Jain C. 2010. Functional and molecular analysis of Eschericia coli strains lacking multiple DEAD-box helicases. RNA 16: 1386–1392. 10.1261/rna.2015610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankowsky E. 2011. RNA helicases at work: binding and rearranging. Trends Biochem Sci 36: 19–29. 10.1016/j.tibs.2010.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarmoskaite I, Russell R. 2011. DEAD-box proteins as RNA helicases and chaperones. WIREs RNA 2: 135–152. 10.1002/wrna.50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PG, Mitta M, Kim Y, Jiang W, Inouye M. 1996. Cold shock induces a major ribosomal-associated protein that unwinds double-stranded RNA in Escherichia coli. Proc Natl Acad Sci 93: 76–80. 10.1073/pnas.93.1.76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karginov FV, Uhlenbeck OC. 2004. Interaction of Escherichia coli DbpA with 23S rRNA in different functional states of the enzyme. Nucleic Acids Res 32: 3028–3032. 10.1093/nar/gkh640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karginov FV, Caruthers JM, Hu Y, McKay DB, Uhlenbeck OC. 2005. YxiN is a modular protein combining a DExD/H core and a specific RNA-binding domain. J Biol Chem 280: 35499–35505. 10.1074/jbc.M506815200 [DOI] [PubMed] [Google Scholar]
- Kawai G, Hashizume T, Miyazawa T, McCloskey JA, Yokoyama S. 1989. Conformational characteristics of 4-acetylcytidine found in tRNA. Nucleic Acids Symp Ser 1989: 61–62. [PubMed] [Google Scholar]
- Klipcan L, Moor N, Finarov I, Kessler N, Sukhanova M, Safro MG. 2012. Crystal structure of human mitochondrial PheRS complexed with tRNAPhe in the active “open” state. J Mol Biol 415: 527–537. 10.1016/j.jmb.2011.11.029 [DOI] [PubMed] [Google Scholar]
- Klostermeier D. 2013. Rearranging RNA structures at 75°C? Towards the molecular mechanism and physiological function of the Thermus thermophilus DEAD-box helicase Hera. Biopolymers 99: 1137–1146. [DOI] [PubMed] [Google Scholar]
- Klostermeier D, Rudolph MG. 2009. A novel dimerization motif in the C-terminal domain of the Thermus thermophilus DEAD box helicase Hera confers substantial flexibility. Nucleic Acids Res 37: 421–430. 10.1093/nar/gkn947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kossen K, Karginov FV, Uhlenbeck OC. 2002. The carboxy-terminal domain of the DExDH protein YxiN is sufficient to confer specificity for 23S rRNA. J Mol Biol 324: 625–636. 10.1016/S0022-2836(02)01140-3 [DOI] [PubMed] [Google Scholar]
- Kuhn E. 2012. Toward understanding life under subzero conditions: the significance of exploring psychrophilic “cold-shock” proteins. Astrobiology 12: 1078–1086. 10.1089/ast.2012.0858 [DOI] [PubMed] [Google Scholar]
- Lee FCY, Ule J. 2018. Advances in CLIP technologies for studies of protein-RNA interactions. Mol Cell 69: 354–369. 10.1016/j.molcel.2018.01.005 [DOI] [PubMed] [Google Scholar]
- Lehnik-Habrink M, Rempeters L, Kovacs AT, Wrede C, Baierlein C, Krebber H, Kuipers OP, Stulke J. 2013. DEAD-Box RNA helicases in Bacillus subtilis have multiple functions and act independently from each other. J Bacteriol 195: 534–544. 10.1128/JB.01475-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linden MH, Hartmann RK, Klostermeier D. 2008. The putative RNase P motif in the DEAD box helicase Hera is dispensable for efficient interaction with RNA and helicase activity. Nucleic Acids Res 36: 5800–5811. 10.1093/nar/gkn581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder P, Jankowsky E. 2011. From unwinding to clamping—the DEAD box RNA helicase family. Nat Rev Mol Cell Biol 12: 505–516. 10.1038/nrm3154 [DOI] [PubMed] [Google Scholar]
- Lovci MT, Ghanem D, Marr H, Arnold J, Gee S, Parra M, Liang TY, Stark TJ, Gehman LT, Hoon S, et al. 2013. Rbfox proteins regulate alternative mRNA splicing through evolutionarily conserved RNA bridges. Nat Struct Mol Biol 20: 1434–1442. 10.1038/nsmb.2699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Aoki H, Ganoza MC. 1999. Molecular characterization of a prokaryotic translation factor homologous to the eukaryotic initiation factor eIF4A. Int J Biochem Cell Biol 31: 215–229. 10.1016/S1357-2725(98)00142-3 [DOI] [PubMed] [Google Scholar]
- Mallam AL, Jarmoskaite I, Tijerina P, Del Campo M, Seifert S, Guo L, Russell R, Lambowitz AM. 2011. Solution structures of DEAD-box RNA chaperones reveal conformational changes and nucleic acid tethering by a basic tail. Proc Natl Acad Sci 108: 12254–12259. 10.1073/pnas.1109566108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mega R, Manzoku M, Shinkai A, Nakagawa N, Kuramitsu S, Masui R. 2010. Very rapid induction of a cold shock protein by temperature downshift in Thermus thermophilus. Biochem Biophys Res Commun 399: 336–340. 10.1016/j.bbrc.2010.07.065 [DOI] [PubMed] [Google Scholar]
- Morlang S, Weglohner W, Franceschi F. 1999. Hera from Thermus thermophilus: the first thermostable DEAD-box helicase with an RNase P protein motif. J Mol Biol 294: 795–805. 10.1006/jmbi.1999.3282 [DOI] [PubMed] [Google Scholar]
- Nagaoka E, Hidese R, Imanaka T, Fujiwara S. 2013. Importance and determinants of induction of cold-induced DEAD RNA helicase in the hyperthermophilic archaeon Thermococcus kodakarensis. J Bacteriol 195: 3442–3450. 10.1128/JB.00332-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshikane H, Sheppard K, Fukai S, Nakamura Y, Ishitani R, Numata T, Sherrer RL, Feng L, Schmitt E, Panvert M, et al. 2006. Structural basis of RNA-dependent recruitment of glutamine to the genetic code. Science 312: 1950–1954. 10.1126/science.1128470 [DOI] [PubMed] [Google Scholar]
- Oshima T, Imahori K. 1971. Isolation of an extreme thermophile and thermostability of its transfer ribonucleic acid and ribosomes. J Gen Appl Microbiol 17: 513–517. 10.2323/jgam.17.513 [DOI] [Google Scholar]
- Oshima T, Imahori K. 1974. Description of Thermus thermophilus (Yoshida and Oshima) comb. nov., a nonsporulating thermophilic bacterium from a Japanese thermal spa. Int J Syst Bacteriol 24: 102–112. 10.1099/00207713-24-1-102 [DOI] [Google Scholar]
- Palonen E, Lindstrom M, Somervuo P, Johansson P, Bjorkroth J, Korkeala H. 2012. Requirement for RNA helicase CsdA for growth of Yersinia pseudotuberculosis IP32953 at low temperatures. Appl Environ Microbiol 78: 1298–1301. 10.1128/AEM.07278-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peil L, Virumae K, Remme J. 2008. Ribosome assembly in Escherichia coli strains lacking the RNA helicase DeaD/CsdA or DbpA. FEBS J 275: 3772–3782. 10.1111/j.1742-4658.2008.06523.x [DOI] [PubMed] [Google Scholar]
- Pellegrini O, de la Sierra-Gallay I L, Piton J, Gilet L, Condon C. 2012. Activation of tRNA maturation by downstream uracil residues in B. subtilis. Structure 20: 1769–1777. 10.1016/j.str.2012.08.002 [DOI] [PubMed] [Google Scholar]
- Pugh GE, Nicol SM, Fuller-Pace FV. 1999. Interaction of the Escherichia coli DEAD box protein DbpA with 23 S ribosomal RNA. J Mol Biol 292: 771–778. 10.1006/jmbi.1999.3112 [DOI] [PubMed] [Google Scholar]
- Putnam AA, Jankowsky E. 2013. DEAD-box helicases as integrators of RNA, nucleotide and protein binding. Biochim Biophys Acta 1829: 884–893. 10.1016/j.bbagrm.2013.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter NJ, Osterman A, Torres-Larios A, Swinger KK, Pan T, Mondragon A. 2010. Structure of a bacterial ribonuclease P holoenzyme in complex with tRNA. Nature 468: 784–789. 10.1038/nature09516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice P, Longden I, Bleasby A. 2000. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet 16: 276–277. 10.1016/S0168-9525(00)02024-2 [DOI] [PubMed] [Google Scholar]
- Rocak S, Linder P. 2004. DEAD-box proteins: the driving forces behind RNA metabolism. Nat Rev Mol Cell Biol 5: 232–241. 10.1038/nrm1335 [DOI] [PubMed] [Google Scholar]
- Rock FL, Mao W, Yaremchuk A, Tukalo M, Crepin T, Zhou H, Zhang YK, Hernandez V, Akama T, Baker SJ, et al. 2007. An antifungal agent inhibits an aminoacyl-tRNA synthetase by trapping tRNA in the editing site. Science 316: 1759–1761. 10.1126/science.1142189 [DOI] [PubMed] [Google Scholar]
- Rogers GWJ, Komar AA, Merrick WC. 2002. eIF4A: the godfather of the DEAD box helicases. Prog Nucleic Acid Res Mol Biol 72: 307–331. 10.1016/S0079-6603(02)72073-4 [DOI] [PubMed] [Google Scholar]
- Rudolph MG, Klostermeier D. 2009. The Thermus thermophilus DEAD box helicase Hera contains a modified RNA recognition motif domain loosely connected to the helicase core. RNA 15: 1993–2001. 10.1261/rna.1820009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolph MG, Klostermeier D. 2015. When core competence is not enough: functional interplay of the DEAD-box helicase core with ancillary domains and auxiliary factors in RNA binding and unwinding. Biol Chem 396: 849–865. 10.1515/hsz-2014-0277 [DOI] [PubMed] [Google Scholar]
- Rudolph MG, Heissmann R, Wittmann JG, Klostermeier D. 2006. Crystal structure and nucleotide binding of the Thermus thermophilus RNA helicase hera N-terminal domain. J Mol Biol 361: 731–743. 10.1016/j.jmb.2006.06.065 [DOI] [PubMed] [Google Scholar]
- Rudolph MG, Wittmann JG, Klostermeier D. 2009. Crystallization and preliminary characterization of the Thermus thermophilus RNA helicase Hera C-terminal domain. Acta Crystallogr Sect F 65: 248–252. 10.1107/S1744309108043145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samatanga B, Klostermeier D. 2014. DEAD-box RNA helicase domains exhibit a continuum between complete functional independence and high thermodynamic coupling in nucleotide and RNA duplex recognition. Nucleic Acids Res 42: 10644–10654. 10.1093/nar/gku747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauliere J, Murigneux V, Wang Z, Marquenet E, Barbosa I, Le Tonqueze O, Audic Y, Paillard L, Crollius HR, Le Hir H. 2012. CLIP-seq of eIF4AIII reveals transcriptome-wide mapping of the human exon junction complex. Nat Struct Mol Biol 19: 1124–1131. 10.1038/nsmb.2420 [DOI] [PubMed] [Google Scholar]
- Sen ND, Zhou F, Ingolia NT, Hinnebusch AG. 2015. Genome-wide analysis of translational efficiency reveals distinct but overlapping functions of yeast DEAD-box RNA helicases Ded1 and eIF4A. Genome Res 25: 1196–1205. 10.1101/gr.191601.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengoku T, Nureki O, Nakamura A, Kobayashi S, Yokoyama S. 2006. Structural basis for RNA unwinding by the DEAD-Box protein Drosophila Vasa. Cell 125: 287–300. 10.1016/j.cell.2006.01.054 [DOI] [PubMed] [Google Scholar]
- Sharpe Elles LM, Sykes MT, Williamson JR, Uhlenbeck OC. 2009. A dominant negative mutant of the E. coli RNA helicase DbpA blocks assembly of the 50S ribosomal subunit. Nucleic Acids Res 37: 6503–6514. 10.1093/nar/gkp711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigi N, Suzuki T, Terada T, Shirouzu M, Yokoyama S, Watanabe K. 2006. Temperature-dependent biosynthesis of 2-thioribothymidine of Thermus thermophilus tRNA. J Biol Chem 281: 2104–2113. 10.1074/jbc.M510771200 [DOI] [PubMed] [Google Scholar]
- Steimer L, Wurm JP, Linden MH, Rudolph MG, Wohnert J, Klostermeier D. 2013. Recognition of two distinct elements in the RNA substrate by the RNA binding domain of the T. thermophilus DEAD box helicase Hera. Nucleic Acids Res 41: 6259–6272. 10.1093/nar/gkt323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier FW. 2005. Protein production by auto-induction in high density shaking cultures. Protein Expr Purif 41: 207–234. 10.1016/j.pep.2005.01.016 [DOI] [PubMed] [Google Scholar]
- Tedeschi FA, Cloutier SC, Tran EJ, Jankowsky E. 2018. The DEAD-box protein Dbp2p is linked to noncoding RNAs, the helicase Sen1p, and R-loops. RNA 24: 1693–1705. 10.1261/rna.067249.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tijerina P, Bhaskaran H, Russell R. 2006. Nonspecific binding to structured RNA and preferential unwinding of an exposed helix by the CYT-19 protein, a DEAD-box RNA chaperone. Proc Natl Acad Sci 103: 16698–16703. 10.1073/pnas.0603127103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomikawa C, Yokogawa T, Kanai T, Hori H. 2010. N7-Methylguanine at position 46 (m7G46) in tRNA from Thermus thermophilus is required for cell viability at high temperatures through a tRNA modification network. Nucleic Acids Res 38: 942–957. 10.1093/nar/gkp1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treiber DK, Williamson JR. 1999. Exposing the kinetic traps in RNA folding. Curr Opin Struct Biol 9: 339–345. 10.1016/S0959-440X(99)80045-1 [DOI] [PubMed] [Google Scholar]
- Tsu CA, Uhlenbeck OC. 1998. Kinetic analysis of the RNA-dependent adenosinetriphosphatase activity of DbpA, an Escherichia coli DEAD protein specific for 23S ribosomal RNA. Biochemistry 37: 16989–16996. 10.1021/bi981837y [DOI] [PubMed] [Google Scholar]
- Tuck AC, Tollervey D. 2013. A transcriptome-wide atlas of RNP composition reveals diverse classes of mRNAs and lncRNAs. Cell 154: 996–1009. 10.1016/j.cell.2013.07.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Nostrand EL, Pratt GA, Shishkin AA, Gelboin-Burkhart C, Fang MY, Sundararaman B, Blue SM, Nguyen TB, Surka C, Elkins K, et al. 2016. Robust transcriptome-wide discovery of RNA-binding protein binding sites with enhanced CLIP (eCLIP). Nat Methods 13: 508–514. 10.1038/nmeth.3810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Nostrand EL, Nguyen TB, Gelboin-Burkhart C, Wang R, Blue SM, Pratt GA, Louie AL, Yeo GW. 2017. Robust, cost-effective profiling of RNA binding protein targets with single-end enhanced crosslinking and immunoprecipitation (eCLIP). Methods Mol Biol 1648: 177–200. 10.1007/978-1-4939-7204-3_14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira J, Messing J. 1982. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene 19: 259–268. 10.1016/0378-1119(82)90015-4 [DOI] [PubMed] [Google Scholar]
- Virtanen JP, Keto-Timonen R, Jaakkola K, Salin N, Korkeala H. 2018. Changes in transcriptome of Yersinia pseudotuberculosis IP32953 grown at 3 and 28°C detected by RNA sequencing shed light on cold adaptation. Front Cell Infect Microbiol 8: 416 10.3389/fcimb.2018.00416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Hu Y, Overgaard MT, Karginov FV, Uhlenbeck OC, McKay DB. 2006. The domain of the Bacillus subtilis DEAD-box helicase YxiN that is responsible for specific binding of 23S rRNA has an RNA recognition motif fold. RNA 12: 959–967. 10.1261/rna.5906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler EC, Van Nostrand EL, Yeo GW. 2018. Advances and challenges in the detection of transcriptome-wide protein-RNA interactions. Wiley Interdiscip Rev RNA 9: e1436 10.1002/wrna.1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y, Steitz TA. 2004. Mechanism of transfer RNA maturation by CCA-adding enzyme without using an oligonucleotide template. Nature 430: 640–645. 10.1038/nature02711 [DOI] [PubMed] [Google Scholar]
- Xu L, Wang L, Peng J, Li F, Wu L, Zhang B, Lv M, Zhang J, Gong Q, Zhang R, et al. 2017. Insights into the structure of dimeric RNA helicase CsdA and indispensable role of its C-terminal regions. Structure 25: 1795–1808 e1795. 10.1016/j.str.2017.09.013 [DOI] [PubMed] [Google Scholar]
- Yamashita S, Takeshita D, Tomita K. 2014. Translocation and rotation of tRNA during template-independent RNA polymerization by tRNA nucleotidyltransferase. Structure 22: 315–325. 10.1016/j.str.2013.12.002 [DOI] [PubMed] [Google Scholar]
- Yamashita S, Martinez A, Tomita K. 2015. Measurement of acceptor-TPsiC helix length of tRNA for terminal A76-addition by A-adding enzyme. Structure 23: 830–842. 10.1016/j.str.2015.03.013 [DOI] [PubMed] [Google Scholar]
- Yu E, Owttrim GW. 2000. Characterization of the cold stress-induced cyanobacterial DEAD-box protein CrhC as an RNA helicase. Nucleic Acids Res 28: 3926–3934. 10.1093/nar/28.20.3926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Ferre-D'Amare AR. 2016. The tRNA elbow in structure, recognition and evolution. Life (Basel) 6: E3 10.3390/life6010003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M. 1989. Computer prediction of RNA structure. Methods Enzymol 180: 262–288. 10.1016/0076-6879(89)80106-5 [DOI] [PubMed] [Google Scholar]
- Zuker M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31: 3406–3415. 10.1093/nar/gkg595 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.