Abstract
Cellular quiescence and cell cycle reentry regulate vital biological processes such as cellular development and tissue homeostasis and are controlled by precise regulation of gene expression. The roles of long noncoding RNAs (lncRNAs) during these processes remain to be elucidated. By performing genome-wide transcriptome analyses, we identify differential expression of several hundreds of lncRNAs, including a significant number of the less-characterized class of microRNA-host-gene (MIRHG) lncRNAs or lnc-MIRHGs, during cellular quiescence and cell cycle reentry in human diploid fibroblasts. We observe that MIR222HG lncRNA displays serum-stimulated RNA processing due to enhanced splicing of the host nascent pri-MIR222HG transcript. The pre-mRNA splicing factor SRSF1 negatively regulates the microprocessor-catalyzed cleavage of pri-miR-222, thereby increasing the cellular pool of the mature MIR222HG. Association of SRSF1 to pri-MIR222HG, including to a mini-exon, which partially overlaps with the primary miR-222 precursor, promotes serum-stimulated splicing over microRNA processing of MIR222HG. Further, we observe that the increased levels of spliced MIR222HG in serum-stimulated cells promote the cell cycle reentry post quiescence in a microRNA-independent manner. MIR222HG interacts with DNM3OS, another lncRNA whose expression is elevated upon serum-stimulation, and promotes cell cycle reentry. The double-stranded RNA binding protein ILF3/2 complex facilitates MIR222HG:DNM3OS RNP complex assembly, thereby promoting DNM3OS RNA stability. Our study identifies a novel mechanism whereby competition between the splicing and microprocessor machinery modulates the serum-induced RNA processing of MIR222HG, which dictates cell cycle reentry.
Keywords: quiescence, cell cycle, lncRNAs, microRNA-host genes
INTRODUCTION
Cell proliferation is a complex biological process that is controlled by differential gene and protein expression. A typical eukaryotic cell cycle consists of four consecutive stages: G1, S (synthesis), G2, and M (mitotic) phases (Harashima et al. 2013). Additionally, there exist three types of nondividing cell states: terminally differentiated, senescent, and quiescent. Among them, quiescence/G0 is unique due to its reversibility. Cells in quiescence temporarily exit the cell cycle and can resume cell division capacity by entering back to G1 upon environmental cues (Coller 2007; So and Cheung 2018). In higher multicellular organisms, undergoing cellular quiescence is essential for the regulation of tissue regeneration and homeostasis, immune responses, and aging (Yusuf and Fruman 2003; Liu et al. 2007; Lemons et al. 2010; Coller 2011; Bainbridge 2013; Yao 2014). In the context of disease, cancer stem cells remain in quiescent state and resume proliferation during tumor reoccurrence (Cheung and Rando 2013). Dysregulation of cellular quiescence is correlated with proliferation-related diseases such as fibrosis, immune disorders, and cancer (Liu et al. 2007; Lemons et al. 2010; Coller 2011; Yao 2014). A deep understanding of quiescence induction and maintenance, and further exit out of quiescence or cell cycle reentry, will allow the discovery of therapeutic targets for treating quiescence-related diseases.
Earlier studies have established the roles of signaling pathways/protein factors/microRNAs (miRNAs) in quiescence maintenance and cell cycle reentry. For instance, certain metabolic processes are found to be reprogrammed in quiescent cells (Lemons et al. 2010; Laporte et al. 2011; Valcourt et al. 2012). Likewise, various signaling pathways are highly regulated during quiescence (Coller et al. 2006; Lemons et al. 2010; Valcourt et al. 2012; So and Cheung 2018). Several groups have also reported aberrant levels and/or functions of protein-coding genes and miRNAs during quiescence establishment or cell cycle reentry (Gos et al. 2005; Coller et al. 2006; Coller 2007, 2011; Liu et al. 2007; Cheung et al. 2012; Suh et al. 2012; Cheung and Rando 2013; Polioudakis et al. 2013; Yao 2014; Martinez et al. 2017). Long noncoding RNAs (lncRNAs) have been shown, by us and others, to play important roles in cell cycle progression and therefore have the potential to regulate quiescence and cell cycle reentry. However, only few studies so far have reported the role of lncRNAs in these processes (Venkatraman et al. 2013; Bierhoff et al. 2014).
LncRNAs are noncoding RNAs that are longer than 200 nt, most of which do not code for any proteins or peptides. They are ubiquitously found in many species from prokaryotes to eukaryotes and can play diverse roles in multiple biological events including differentiation, cell cycle progression, immune response, apoptosis, and diseases (Kitagawa et al. 2013; Flynn and Chang 2014; Fang and Fullwood 2016; Li et al. 2016; Schmitt and Chang 2016; Chen et al. 2017). LncRNAs function in both the nucleus and the cytoplasm via various mechanisms such as regulating nuclear structure, chromatin, gene transcription, RNA stability, or translation, often by interacting with other protein/RNA/DNA elements (Wang and Chang 2011; Marchese et al. 2017; Noh et al. 2018; Sun et al. 2018a). Several lncRNAs can also produce short and functional peptides (Choi et al. 2019).
Some lncRNA genes display intriguing genomic structures wherein they harbor miRNA genes in their gene loci and are therefore categorized as “microRNA-host-gene lncRNAs (lnc-MIRHGs).” MiRNAs are ∼22 nt noncoding RNAs that play crucial roles in posttranscriptional regulation of gene expression (Ameres and Zamore 2013). Most miRNAs are encoded within protein-coding or long noncoding genes (Berezikov et al. 2007a; Kahl 2009; Monteys et al. 2010). A significant fraction of miRNAs (17.5%) in humans are encoded within lnc-MIRHGs (Dhir et al. 2015b). Earlier studies have reported that lnc-MIRHGs can serve as diagnosis/prognosis markers and also play regulatory roles in various cellular processes (Augoff et al. 2012; Morenos et al. 2014; Montes et al. 2015; Kato et al. 2016; Molinari et al. 2016; Su et al. 2018). These studies have brought crucial insights into the significance of lnc-MIRHGs. However, most of these studies have focused on their role merely as a source of miRNA synthesis. On the other hand, several recent studies, including work from our laboratory, have provided compelling evidence demonstrating miRNA-independent roles of the mature and processed lnc-MIRHGs (Ng et al. 2013; Tseng et al. 2014; Colombo et al. 2015; Raveh et al. 2015; Shih et al. 2017; Sun et al. 2018b; Du et al. 2019; Profumo et al. 2019). For instance, we have demonstrated that lncRNA MIR100HG, whose gene locus hosts miR-125-b1 and miR-100, regulates cell cycle progression by modulating the activity of RNA-binding protein HuR in a miRNA-independent manner (Sun et al. 2018b). Two other recent studies described MIR205HG’s role in regulating basal-luminal differentiation as well as the production of growth hormone and prolactin (Du et al. 2019; Profumo et al. 2019).
At present, very little is known about the function of lnc-MIRHGs during quiescence and cell cycle reentry. The major “road-block” to such studies is the lack of genome-wide analysis of lncRNA expression, including lnc-MIRHG, during these processes. Keeping this in mind, we performed a systematic transcriptome profiling to identify differentially expressed protein-coding and lncRNA genes during cellular quiescence and subsequent reentry into the cell cycle in normal human diploid fibroblasts. From this, we identified several dynamically regulated lnc-MIRHGs, and further focused on mechanistic studies of MIR222HG, which showed enhanced expression during cell cycle reentry. We found that the spliced (and mature) MIR222HG levels were significantly elevated in serum-stimulated cells due to the enhanced processing of pri-MIR222HG transcripts, mediated by the pre-mRNA splicing factor SRSF1. We further demonstrated that the spliced MIR222HG facilitated cell cycle reentry in a miRNA-independent manner. Mechanistic studies revealed that MIR222HG forms an RNP (Ribonucleoprotein) complex along with another lncRNA DNM3OS and ILF3/ILF2 complex to stabilize DNM3OS, which in turn is instrumental in modulating serum-stimulated cell cycle reentry.
RESULTS
Transcriptome profiling of quiescent and serum-stimulated cells reveals differential expression of both protein- and noncoding genes
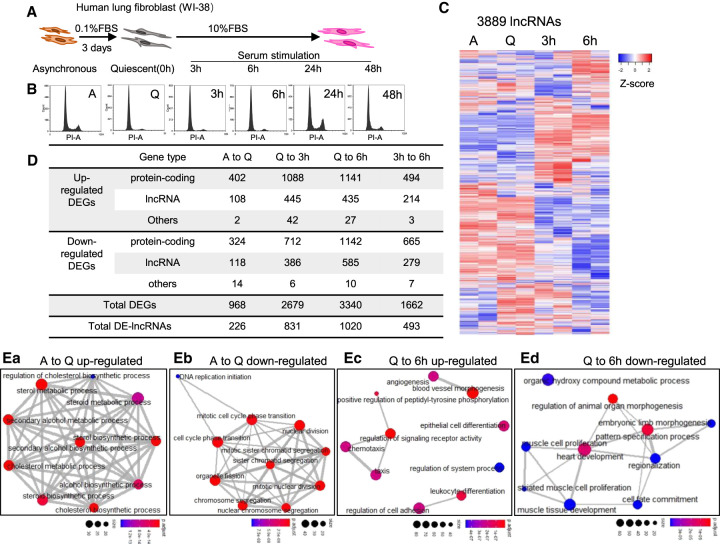
Cellular quiescence can be induced experimentally via serum deprivation, loss of adhesion, or cell contact inhibition (Coller et al. 2006). In the present study, we triggered cellular quiescence in normal human diploid lung fetal fibroblast WI-38 cells by serum starvation (Fig. 1A). WI-38 cells become quiescent after 72-h of serum starvation, reflected by the almost undetectable populations of S-phase or G2/M phase cells (Fig. 1B). Quiescent WI-38 cells reenter the cell cycle upon serum stimulation that results in increase in S and G2/M phase populations (Fig. 1B).
FIGURE 1.
Transcriptome landscape of human lung fibroblasts during quiescence and cell cycle reentry. (A) Schematic of inducing WI-38 cellular quiescence and cell cycle reentry using serum starvation and serum stimulation method. (B) PI-flow cytometry analyses in WI-38 during cellular quiescence and cell cycle reentry. A: asynchronous; Q: quiescent; 3 h/6 h/24 h/48 h: corresponding serum stimulation time postserum-starvation. (C) Heatmaps showing the relative expression levels of 3889 lncRNAs detected from RNA-seq. Biological duplicates from four time points are represented. Genes (rows of heatmap) are hierarchically clustered using average-linkage clustering method. (D) Table representing the number of differentially expressed genes (DEGs) detected from multiple comparisons between RNA-seq samples. Detailed DEG information is available in Supplemental Table S2. (E) Gene ontology (GO) analyses showing the top enriched biological processes of (a) up-regulated DEGs and (b) down-regulated DEGs from asynchronous to quiescence, as well as (c) up-regulated DEGs and (d) down-regulated DEGs from quiescence to 6 h serum stimulation. Nodes represent the gene size in each biological process, edges represent the overlapping of genes in different processes. Full GO analyses results are listed in Supplemental Table S3.
To understand the roles of lncRNAs in cellular quiescence and cell cycle reentry, we performed RNA-seq from asynchronous, quiescent, and serum-stimulated (3 and 6 h of serum-stimulation post quiescence) WI-38 cells. We reasoned that genes that play vital roles in cell cycle reentry would show differential expression during the early time points of serum stimulation. Hence, we chose 3 and 6 h of serum stimulation as time points for RNA isolation. The polyA+ paired-end RNA-seq was performed in biological duplicates with high-quality, high-depth, and high reproducibility between replicates (Supplemental Fig. S1A,B). We detected the expression of 17,640 genes including 13,574 protein-coding and 4066 noncoding genes, in which 3889 genes were annotated as lncRNAs (Supplemental Fig. S1C; Supplemental Table S1). We observed dramatic changes in both complete transcriptome profile (17,640 genes) and lncRNA transcriptome profile (3889 genes) of quiescent and serum-stimulated fibroblasts (Fig. 1C; Supplemental Fig. S1D).
We then performed statistical analyses to identify differentially expressed genes (DEGs; |log2 (fold change)| > 1 and FDR < 0.05) by comparing different RNA-seq samples, including asynchronous (A) to quiescence (Q) (A to Q), quiescence to serum stimulation (SS) (Q to 3 h SS, Q to 6 h SS), and 3 h SS to 6 h SS. We identified the differential expression of ∼1000 to ∼3000 genes during these cell cycle transitions (968 DEGs from A to Q, 2679 from Q to 3 h SS, 3340 from Q to 6 h SS, 1662 from 3 h to 6 h SS), in which ∼20%–30% of them were lncRNAs (226 between A to Q, 831 between Q to 3 h SS, 1020 between Q to 6 h SS, 493 between 3 h to 6 h SS) (Fig. 1D; Supplemental Table S2). The differential expression of a significant number of lncRNAs during quiescence or cell cycle reentry implies that lncRNAs play vital roles during such cell cycle transitions.
To understand the biological processes that are involved in cellular quiescence and subsequent cell cycle reentry, we performed gene set enrichment analysis (GSEA) and gene ontology (GO) analyses. GSEA showed that multiple quiescence-associated processes were regulated between asynchronous (A) to quiescent (Q) states, exemplified by reduced rRNA processing (Supplemental Fig. S1Ea; Supplemental Table S3). We then performed GO analysis by stratifying DEGs into up-regulated and down-regulated groups. During A to Q transition, several key metabolic processes were found to be activated (Fig. 1Ea; Supplemental Table S3). On the contrary, genes controlling cell cycle-/cell division-related events showed consistent down-regulation in quiescent cells (Fig. 1Eb; Supplemental Table S3). Pathway analyses also revealed key pathways that were affected during quiescence establishment, including the cell cycle (Supplemental Fig. S1Fa; Supplemental Table S3). Similarly, serum-stimulated cells exhibited enhanced levels of processes such as rRNA processing, MAPK cascade, and extracellular matrix organization (Supplemental Fig. S1Eb–d; Supplemental Table S3). In addition, up-regulated DEGs in serum-stimulated cells (Q to 6 h SS) participate in biological processes such as chemotaxis and angiogenesis, whereas genes controlling morphogenesis and development seem to be down-regulated during serum stimulation (Fig. 1Ec,d; Supplemental Table S3). We also observed that pathways controlling cell growth, metabolism, and development (MAPK/Ras/Hippo/PI3K-Akt pathways) showed significant changes during early serum response (Supplemental Fig. S1Fb,c; Supplemental Table S3).
The genome-wide transcriptome landscape analyses revealed that cells undergo complex, precise, and rapid changes in gene expression during both quiescence induction and exit out of quiescence. However, the role of lncRNAs during such processes is least understood. The large number of differentially expressed lncRNAs identified in this study hence serves as a useful resource for further mechanistic studies.
Genome-wide analysis of miRNA host genes identifies potential lnc-MIRHGs as regulators for cell cycle reentry
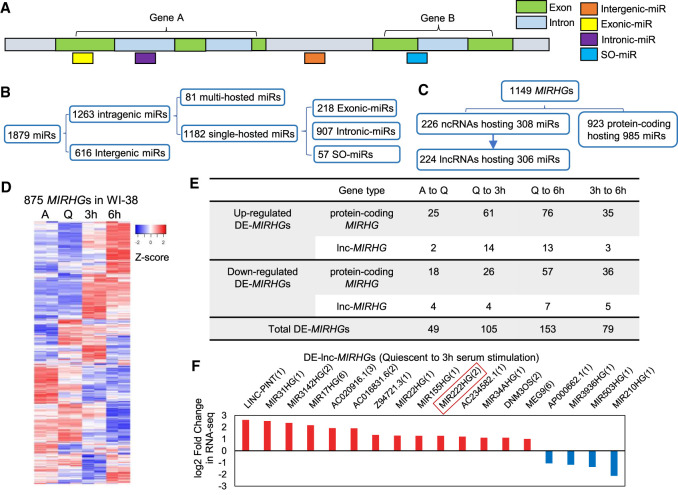
Earlier studies have uncovered the role of miRNAs in promoting/repressing quiescence (Cheung et al. 2012; Suh et al. 2012; Cheung and Rando 2013; Polioudakis et al. 2013). However, very little is known about the function of lncRNAs that are processed from miRNA-host gene loci (termed as lnc-MIRHGs), during quiescence and serum stimulation. To identify the lnc-MIRHG repertoire in the human genome, we performed a genome-wide screen to categorize the miRNAs and MIRHGs in the recent version of the human genome assembly GRCh38 (p12.v94). We classified miRNAs based on their relative location to the host genes (Fig. 2A). Our genomic analyses revealed that ∼2/3 of human miRNAs (1263 out of 1879) were “intragenic miRs” and are encoded within 1149 host genes (MIRHGs), out of which 224 are composed of lncRNAs (lnc-MIRHGs). Furthermore, we found that most of the intragenic miRNAs (72%, 907 out of 1263) reside within the introns of MIRHGs (“intronic-miRs”), and 17% were located in the exonic regions (“exonic-miRs”). Interestingly, ∼5% (57 out of 1263) of miRNAs were located at exon–intron junctions of protein-coding or lncRNA genes (Fig. 2B,C; Supplemental Tables S4, S5). Such miRNAs were previously identified as “SO-miRs” (splice site overlapping miRNAs), so we kept the same nomenclature (Mattioli et al. 2013, 2014; Pianigiani et al. 2018). Finally, the majority of MIRHGs (1028 out of 1149) host one miRNA and a small fraction of them contains multiple miRNAs (Supplemental Fig. S2A; Supplemental Table S5).
FIGURE 2.
MIRHGs show dynamic expression during cellular quiescence and upon cell cycle reentry. (A) Schematic showing different types of miRNAs (miRs) categorized in this study. Detailed list is available in Supplemental Table S4. (B) Categorization of miRNAs (miRs) in human genome GRCh38 version in terms of their relative locations to host genes (MIRHGs). Detailed list is shown in Supplemental Table S4. (C) Categorization of all identified miRNA host genes (MIRHGs) in human genome GRCh38 version. Detailed list is shown in Supplemental Table S5. (D) Heatmap showing the relative expression levels of 875 MIRHGs that are expressed in WI-38 RNA-seq. Duplicates are represented. Genes (rows of heatmap) are hierarchically clustered using average-linkage clustering method. (E) Table representing the number of differentially expressed MIRHGs (DE-MIRHGs) detected from multiple comparisons between RNA-seq samples. Detailed DE-MIRHG information is available in Supplemental Table S5. (F) Fold change (log2 scale) of 18 DE-lnc-MIRHGs from quiescent to 3-h serum stimulation. Numbers in the parentheses refer to the number of miRNAs encoded within each lnc-MIRHG. See Supplemental Table S5 for detailed DE-lnc-MIRHGs information.
Eight hundred and seventy-five MIRHGs were expressed in WI-38 cells, wherein 91 belonged to the lnc-MIRHG repertoire (Fig. 2D; Supplemental Fig. S2B). We examined the differential expression (DE) of MIRHGs during quiescence and serum stimulation and further focused on the differentially expressed lnc-MIRHGs (DE-lnc-MIRHGs) (Fig. 2E; Supplemental Table S5). By this, we identified six DE-lnc-MIRHGs from asynchronous to quiescent transition, 18 DE-lnc-MIRHGs from quiescence to 3 h serum stimulation, 20 DE-lnc-MIRHGs from quiescence to 6 h serum stimulation (SS), and 8 DE-lnc-MIRHGs from 3 h SS to 6 h SS (Fig. 2E,F; Supplemental Fig. S2C).
LncRNA MIR222HG is induced during cell cycle reentry post cellular quiescence
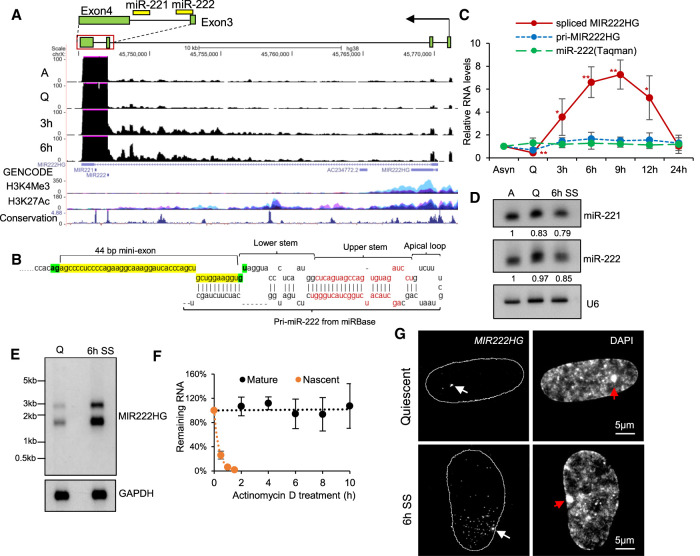
Based on the existing knowledge in the case of protein-coding genes, we hypothesized that early serum-induced lncRNA genes play crucial roles in regulating cell cycle reentry. Among the several up-regulated DE-lnc-MIRHGs from quiescence to 3 h serum stimulation, we focused our investigations on MIR222HG for several reasons. MIR222HG (ENSG00000270069 from Ensembl, lnc-CXorf36-46:7 from LNCipedia) is transcribed from the X-chromosome and it is up-regulated upon 3 h and 6 h of serum stimulation (Fig. 2F; Supplemental Fig. S2Cb; Supplemental Table S5). The MIR222HG gene locus encodes two miRNAs, miR-221 and miR-222 (Fig. 3A), both of which were previously reported to play roles in cell proliferation and cancer progression (Felli et al. 2005; Medina et al. 2008; Cardinali et al. 2009; Chun-Zhi et al. 2010; Garofalo et al. 2012; Li et al. 2014). In addition, lncRNA MIR222HG was also reported to play a pro-tumorigenic role (Sun et al. 2018c), further implying that its induction during serum stimulation could promote cell proliferation. The existing Ensembl gene annotation indicates that MIR222HG contains two exons and a long intron, within which both miR-221 and 222 reside (Fig. 3A, ENSG00000270069, GENCODE track). RNA-seq revealed significant number of reads from the 3′ end of the last intron, spanning miR-221 and miR-222 (Supplemental Fig. S3A), implying intron retention. Our high-depth RNA-seq (∼100 million paired reads/sample) analyses revealed that the spliced (and mature) MIR222HG expressed in WI-38 cells contained an unannotated 44 bp mini-exon, which partially overlapped with the 5′ end of the primary precursor of miR-222 (Fig. 3A; Supplemental Fig. S3Ba,b). The 10 bp at the 3′ end of the 44 bp exon is located just upstream of the lower stem of the pri-miR-222 hairpin structure and formed imperfect complementarity with the 3′ end of the pri-miR-222 sequence (Fig. 3B).
FIGURE 3.
MIR222HG is induced during the cell cycle reentry post cellular quiescence. (A) UCSC genomic browser view of MIR222HG genomic locus and RNA-seq coverage tracks in WI-38 cells. Y-axis displays the reverse strand of BigWig (bw) files. Max value of y-axis is set at 100 for a better visualization of exons 1 – 3 of MIR222HG. Full display of the last exon's signal is shown in Supplemental Figure S3A. H3K4Me3 and H3K27Ac mark tracks contain all of the seven ENCODE cell lines. Conservation track presents Vertebrate Multiz Alignment & Conservation (100 Species). Schematic of the ∼1.3 kb MIR222HG isoform expressed in WI-38 is drawn on the top to represent its genomic location; regions spanning the last two exons and last intron are presented in detail. The genomic positions of miRs are shown as yellow rectangles. (B) Schematic of the 44 bp mini-exon genomic position with respect to the hairpin structure of primary miR-222. Nucleotide sequence of the 44 bp mini-exon is highlighted in yellow. The 5′GU and 3′AG spliced site on either side of the mini-exon are highlighted in green. Sequences in red represent the miRNA duplex of miR-222. Primary miR-222 structure is obtained from miRbase database (http://www.mirbase.org/cgi-bin/mirna_entry.pl?acc=MI0000299). (C) Relative levels of MIR222HG (pri- and mature forms) and miR-222 (TaqMan assay) in WI-38s during quiescence and various time points (h/hours) postserum stimulation. (D) Northern blot to detect human miR-221 and miR-222 from asynchronous, quiescent, and 6-h postserum-stimulated WI-38 cells. U6 is used as loading control. Relative levels of miR-221 and miR-222 are included below the blots, by normalization to U6 levels and comparison to asynchronous sample. Quantification is performed using ImageJ. (E) Northern blot to detect MIR222HG using polyA+ RNA from quiescent and 6-h serum-stimulated WI-38 cells. GAPDH is used as loading control. MIR222HG construct used for probe preparation contains sequence from exons 2 – 4; please see cloning primer position in Supplemental Figure S3D. (F) RNA stability analyses of mature and pri-MIR222HG. RNA is extracted from WI-38s treated by actinomycin D for different time points. The levels of MIR222HG RNA (pri-MIR222HG as well as mature MIR222HG) are quantified via RT-qPCR and compared to 0 h time point to calculate the remaining levels of RNA. (G) Single molecule RNA-FISH (smRNA-FISH) to detect MIR222HG in quiescent and serum stimulated WI-38 cells. DNA is counterstained with DAPI. Contour represents the nuclei location. The magnification bar represents 5 µm. White arrows indicate transcription site and red arrows indicate Barr body. (*) P ≤ 0.05, (**) P ≤ 0.01, (***) P ≤ 0.001, (****) P ≤ 0.0001 by two-tailed Student's t-test, n = 3 for all figures. Error bars represent standard deviation.
This observation indicates that in WI-38 cells, miR-222 is expressed as a SO-miRNA instead of an intronic miRNA. In support of this, an earlier study had categorized miR-222 as a SO-miRNA in the Bos tauros genome (Melamed et al. 2013). In addition, we have identified the transcription start site of MIR222HG isoform in WI-38 cells. The 5′ end of MIR222HG (based on RNA-seq reads and 5′ RACE), which is different from the current GENCODE/Ensembl annotation, is located ∼24 kb upstream of miR-222 and the promoter of MIR222HG is decorated with chromatin marks (H3K4Me3 and H3K27Ac) associated with active transcription (Fig. 3A). We confirmed the 5′ and 3′ ends of MIR222HG by RACE experiment, and the full-length of the annotated spliced/mature MIR222HG was ∼1.3 kb (please see the full sequence in Supplemental File). The PhyloCSF score and ribosome footprint from ribo-seq signals indicated that MIR222HG did not code for any protein (Supplemental Fig. S3A). Further, ribosome fractionation followed by RT-qPCR analyses showed that MIR222HG was majorly associated with the monosome fraction, whereas the protein-coding GAPDH mRNA showed a strong association with heavy polysome fractions (Supplemental Fig. S3C). In summary, MIR222HG is a multiexonic lnc-MIRHG containing a 44 bp mini-exon partially overlapping with the primary precursor of miR-222.
We next performed RT-qPCR to quantitate the relative levels of both nascent and mature MIR222HG using primers from exonic or inronic regions or exon–intron or exon–exon junctions of MIR222HG (Supplemental Fig. S3D). The pri-MIR222HG transcript, detected by the exon–intron junction primer, showed moderate up-regulation (approximately twofold) from quiescence to 3-h serum stimulation (Fig. 3C, blue line) and the levels remained stable afterward. Primers detecting either the intronic or exonic regions of MIR222HG also displayed similar patterns (Supplemental Fig. S3D,E). However, the spliced, mature MIR222HG was strongly up-regulated in serum-stimulated cells (eightfold from Q to 3 h SS, 15-fold from Q to 6 h SS), as detected by two distinct sets of exon–exon junction primers (Fig. 3C, red line, Supplemental Fig. S3D,E). This data implies that the addition of serum only moderately enhanced the transcription of MIR222HG but dramatically elicited the posttranscriptional processing of pri-MIR222HG to produce spliced MIR222HG. Since both miR-221 and miR-222, encoded in the MIR222HG, are known to play broad functions in multiple cellular processes including cell proliferation and quiescence (Felli et al. 2005; Medina et al. 2008; Cardinali et al. 2009; Chun-Zhi et al. 2010; Mayoral et al. 2011; Garofalo et al. 2012; Li et al. 2014), we examined if they show altered expression during serum stimulation. Both quantitative TaqMan assay and Northern blot analyses showed that miR-221 and miR-222 levels remained unchanged among asynchronous, quiescent, and serum-stimulated samples, suggesting that their levels were not coregulated with the mature MIR222HG (Fig. 3C green line, Fig. 3D). Northern blot using poly(A)+ RNA detected two discrete bands at ∼1.5 kb and <3 kb of MIR222HG in both quiescent and 6 h SS WI-38 cells (Fig. 3E). We reasoned that the 1.5 kb RNA coded for the mature poly-adenylated MIR222HG, whereas the <3 kb band most likely corresponded to the miRNA-bearing intron 3-retained isoform of MIR222HG. Both isoforms displayed enhanced expression in serum-stimulated cells (Fig. 3E). Northern blot data further supported the RT-qPCR analyses that showed increased levels of spliced/mature and not the pri-MIR222HG transcripts upon serum stimulation.
Earlier studies have reported Drosha-mediated transcription termination of noncoding MIRHGs in human cells (Dhir et al. 2015a; Pianigiani et al. 2018). In such a scenario, cleavage of pri-MIRHG by Drosha results in the generation of poly(A)− and unstable MIRHG transcripts of no apparent function. Unlike the other lnc-MIRHG transcripts that were identified in earlier studies, MIR222HG contains a discrete poly(A)+ tail. We further determined the stability of MIR222HG by transcription inhibition followed by a chase experiment. We observed that spliced MIR222HG constituted a very stable pool compared to the nascent form of MIR222HG, pri-MIR222HG (Fig. 3F). Pri-MIR222HG is relatively abundant, present at 300–500 copies/cell and predominantly localized in the nucleus, whereas the low-abundant spliced MIR222HG was localized in both nuclear and cytoplasmic compartments (Supplemental Fig. S3F,G). We next performed single-molecule fluorescence RNA in situ hybridization (smRNA-FISH) to determine the cellular localization of MIR222HG in quiescent and serum-stimulated cells. smRNA-FISH revealed that MIR222HG was expressed only from the active X-chromosome, as observed by a single prominent locus, reminiscent of the transcription site (Fig. 3G; white arrow). No smRNA-FISH signal was observed on the DAPI-rich inactive-X-chromosome (Barr body) (Fig. 3G, red arrows). In the quiescent cell nucleus, most of MIR222HG RNA decorated at the transcription site, and this fraction of RNA could represent the nascent transcripts, pri-MIR222HG. In serum-stimulated cells, we continued to see enrichment of MIR222HG RNA at its transcription site. In addition, we observed 20–30 bright RNA spots in the nucleoplasm, located in proximity to the MIR222HG transcription site (Fig. 3G; Supplemental Fig. S3H). Since RNA splicing is predominantly a cotranscriptional event, we concluded that the RNA spots that were dispersed in the nucleoplasm represent spliced and mature MIR222HG. Thus, the smRNA-FISH results further confirmed our molecular analyses (RNA-seq and RT-qPCR) that serum stimulation enhanced the RNA splicing of MIR222HG. Both smRNA-FISH (Supplemental Fig. S3H) and RT-qPCR (Fig. 3C) results showed reduced expression of mature MIR222HG post 12 h serum stimulation (compared to 6 h). This result implies that serum stimulation enhances the splicing of pri-MIR222HG that are concentrated at its site of transcription, whereas the spliced, mature MIR222HG diffuse into the nucleoplasm.
SRSF1 antagonizes Drosha/DGCR8 for spliced MIR222HG biogenesis upon serum stimulation
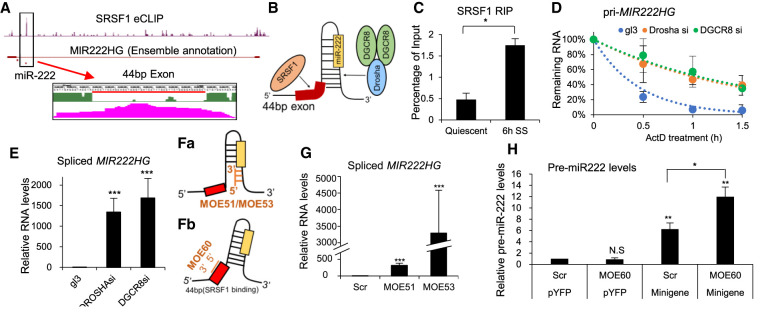
Increased levels of spliced MIR222HG in serum-stimulated cells indicate serum-responsive posttranscriptional processing of pri-MIR222HG. An earlier study reported that pre-mRNA splicing factor SRSF1 (Serine/arginine-rich splicing factors) promotes the Drosha-mediated cleavage of miR-222 in HeLa cells (Wu et al. 2010). SRSFs constitute a conserved family of essential pre-mRNA splicing factors, which bind to the exonic regions on pre-mRNAs by recognizing exonic splicing enhancers (ESEs) (Busch and Hertel 2012). RBPmap prediction analyses identified a consensus SRSF1 binding site (ugrwgvh/ugcugga) within the 44 bp mini-exon (Paz et al. 2014). The publicly available eCLIP data set from human HepG2 cells (ENCODE project ENCSR989VIY) displayed significant binding of SRSF1 at the 44 bp mini-exon (Fig. 4A). We therefore hypothesized that the increased binding of SRSF1 on the mini-exon in serum-stimulated cells could facilitate the recruitment of the microprocessor complex (Fig. 4B). In support of this, SRSF1 RNA-IP- (RIP) followed by RT-qPCR using primers that amplify the 44 bp mini-exon region displayed enhanced association of SRSF1 to MIR222HG in serum-stimulated cells (Fig. 4C; Supplemental Figs. S3D, S4A). Serum-stimulated increase in the association of SRSF1 was not observed at the intronic region of MIR222HG (Supplemental Fig. S4Ba), nor other known SRSF1 target RNAs such as MALAT1 lncRNA (Supplemental Fig. S4Bb), implying the specificity of the interaction. Serum-stimulated cells did not show any significant change in the cellular levels of SRSF1, indicating that differential binding of SRSF1 to MIR222HG mini-exon is a regulated event (Supplemental Fig. S4C). Finally, stable depletion of SRSF1 in WI-38 cells led to the skipping of the 44 bp mini-exon (Supplemental Fig. S4D,E). Based on this, we concluded that SRSF1 showed enhanced association to an exonic region within MIR222HG, which partially overlapped with the pri-miR-222, during serum stimulation, resulting in increased pri-MIR222HG splicing.
FIGURE 4.
Mature spliced MIR222HG lncRNA is processed via the competition between SRSF1 and microprocessor complex. (A) SRSF1 eCLIP signal from HepG2 cells at the 44 bp mini-exon position. The peak at the 44 bp mini-exon is zoomed in and displayed at the bottom. A red line is drawn under the nucleotide sequence to represent the position of the 44 bp mini-exon. Pink peak represents the SRSF1 eCLIP signal. (B) Schematic of hairpin structure of miR-221 primary precursor and the 44 bp mini-exon from MIR222HG. Yellow box represents the mature 21 nt miR-222 position. Arrows indicate the binding sites of splicing factor SRSF1 and microprocessor (Drosha/DGCR8). (C) SRSF1 RIP in quiescent and 6-h serum-stimulated (SS) WI-38 cells followed by RT-qPCR to quantify the levels of MIR222HG, using primers detecting the 44 bp mini-exon region. (D) RNA stability analysis of pri-MIR222HG in control (gl3), Drosha-depleted, and DRCR8-depleted WI-38 cells treated by actinomycin D for indicated time points. Remaining levels of RNA are measured via RT-qPCR. (E) RT-qPCR to quantify the levels of spliced MIR222HG in control (gl3), Drosha-depleted, and DRCR8-depleted WI-38 cells. (F) Schematic figures showing the position of MOEs used in this study: (a) MOE51 and MOE53, block hairpin formation within primary miR-222, and (b) MOE60 blocks SRSF1 binding to the 44 bp mini-exon of MIR222HG. (G) RT-qPCR to quantify the levels of spliced MIR222HG in scramble and MOE-treated WI-38 cells. (H) RT-qPCR to quantify the levels of pre-miR-222 levels in scrambled and MOE-treated WI-38 cells, under both control (pYFP transfected) and MIR222HG minigene overexpressed backgrounds. (*) P ≤ 0.05, (**) P ≤ 0.01, (***) P ≤ 0.001, (****) P ≤ 0.0001 by two-tailed Student's t-test, n = 3 for all figures. Error bars represent standard deviation.
To test whether SRSF1 regulates the Drosha-mediated processing of pri-miR-222 during serum stimulation, we examined the levels of miR-222 in control and SRSF1 overexpressed or depleted WI-38 cells. To our surprise, unlike what had been reported previously in HeLa cells, both overexpression and stable depletion of SRSF1 in serum-stimulated WI-38 cells did not alter the levels of miR-222 (Supplemental Fig. S4F,G). These results argue against the earlier model that SRSF1 facilitates Drosha-mediated miR-222 synthesis (Wu et al. 2010). The disparate expression patterns of miR-221/miR-222 and spliced MIR222HG during serum stimulation indicate that miRNA biogenesis and splicing of MIR222HG, both using pri-MIR222HG as substrate, are regulated independently. Earlier studies have reported that, during the processing of SO-miRNA-host transcripts, differential splicing versus Drosha-mediated cleavage dictates the levels of spliced host gene transcripts versus miRNAs (Mattioli et al. 2013; Melamed et al. 2013; Agranat-Tamir et al. 2014; Pianigiani et al. 2018). In such a situation, we hypothesized that SRSF1-mediated splicing of the mini-exon in serum-stimulated cells negatively regulates the Drosha-mediated cleavage. Depletion of members of the microprocessor complex (Drosha or DRCR8) stabilized pri-MIR222HG and significantly increased the cellular level of spliced mature MIR222HG (Fig. 4D,E; Supplemental Fig. S4H,I), implying that Drosha-mediated cleavage negatively regulates the levels of mature MIR222HG. To further investigate the crosstalk between miR-221/222 miRNA biogenesis and MIR222HG splicing, we blocked the binding of the microprocessor or SRSF1 to the pri-miR-222 hairpin or to the mini-exon, respectively, by using independent 2′-O-methoxyethyl-modified antisense oligonucleotides (MOE) (see schematic in Fig. 4F). Transfection of MOEs (MOE51 and 53) that disrupt the binding of the microprocessor robustly increased the levels of spliced MIR222HG (Fig. 4G). Next, we designed another MOE (MOE60) that binds to the 44 bp mini-exon of MIR222HG and inhibits the exon inclusion by preventing the binding of the splicing factor at the mini-exon. MOE60 transfection inhibited the inclusion of the 44 bp mini-exon of MIR222HG, confirming that it successfully blocked splicing factor activity (Supplemental Fig. S4J). Next, we tested whether MOE60 treatment influences miR-222 synthesis. We performed the splicing blockage experiment using MOE60 together with a MIR222HG minigene. Specifically, the minigene construct contained key elements of the MIR222HG gene (see schematic in Supplemental Fig. S4K) and could be efficiently spliced upon overexpression (data not shown). We observed that MOE60-transfected cells, which also expressed the minigene, showed a significantly increased level of pre-miR-222 (Scr vs MOE60 under minigene background, Fig. 4H), implying that reduced binding of the splicing factor at the mini-exon facilitated miR-222 synthesis. Based on these, we speculate that Drosha/DGCR8 competes with SRSF1 on the pri-MIR222HG, which hosts SO-miRNA miR-222. Upon serum stimulation, enhanced SRSF1 binding on pri-MIR222HG leads to the increased synthesis of the mature, spliced MIR222HG.
Spliced mature MIR222HG facilitates the cell cycle reentry post quiescence in a microRNA-independent manner
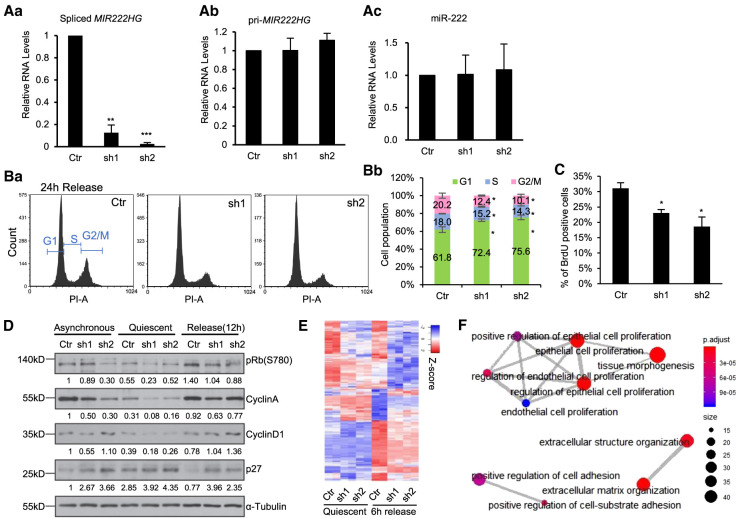
Increased levels of spliced MIR222HG in serum-stimulated cells prompted us to investigate whether it plays any role in cell cycle reentry. By transiently transducing WI-38 cells with two independent lentiviral shRNAs, we preferentially depleted the nuclear and cytoplasmic pool of only the mature spliced MIR222HG (Fig. 5Aa; Supplemental Fig. S5A), but not pri-MIR222HG (Fig. 5Ab), nor miR-222 (Fig. 5Ac). We then analyzed the ability for control or mature MIR222HG-depleted quiescent cells to reenter the cell cycle upon serum stimulation. PI (Propidium Iodide)-flow cytometry analyses revealed that mature MIR222HG-depleted cells showed a defect in cell cycle reentry, reflected by the pronounced G1 arrest and a concomitant reduction of S-phase and G2/M-phase populations (Fig. 5B). BrdU incorporation assays (45 min pulse; 24-h postserum stimulation) also revealed a reduced percentage of S-phase cells upon mature MIR222HG-depletion (Fig. 5C). Reduced levels of phosphorylated Retinoblastoma (pRb; S780) and Cyclin A in mature MIR222HG-depleted serum-stimulated cells further supported the defects in cell cycle reentry (Fig. 5D). Finally, we observed increased levels of Cyclin D1 and p27 in serum-stimulated MIR222HG-depleted WI-38 cells, implying that mature MIR222HG-depleted cells failed to enter into S-phase of the cell cycle, consistent with the reduced Cyclin A and BrdU incorporation (Fig. 5D).
FIGURE 5.
Spliced mature MIR222HG facilitates cell cycle reentry post quiescence in a micro-RNA independent manner. (A) RT-qPCR to quantify the levels of (a) spliced MIR222HG, (b) pri-MIR222HG, and (c) mature miR-222 (TaqMan assay), in control and MIR222HG shRNA treated WI-38 cells. (B) (a) PI-flow cytometry analyses in control and MIR222HG-depleted WI-38 cells at 24 h postserum stimulation. (b) Percentage of cells at a specific cell cycle stage in control and MIR222HG-depleted cells. Data obtained from biological triplicates. Quantification is performed using FCS Express. (C) Percentage of BrdU-incorporated cells in control and MIR222HG-depleted WI-38s at 24 h postserum stimulation. (D) Immunoblot to detect the levels of several cell cycle marker proteins in control and MIR222HG-depleted asynchronous, quiescent, and serum-stimulated (12 h) WI-38 cells. α-Tubulin is used as loading control. Quantification was performed with Image J. Relative protein levels were calculated by normalizing with GAPDH, followed by comparing to the asynchronous control sample. Values are labeled on the bottom of each blot. (E) Heatmaps showing the relative expression of 398 differentially expressed genes in control and MIR222HG-depleted cells during quiescence and 6-h serum stimulation, obtained by RNA-seq. Biological duplicates are represented. Genes (rows of heatmap) are hierarchically clustered using average-linkage clustering method. Detailed DEG information is available in Supplemental Table S6. (F) Gene ontology (GO) analyses showing the top enriched biological processes that show changes in MIR222HG-depleted cells. (*) P ≤ 0.05, (**) P ≤ 0.01, (***) P ≤ 0.001, (****) P ≤ 0.0001 by two-tailed Student's t-test, n = 3 for all figures. Error bars represent standard deviation.
To capture the changes of the transcriptome upon MIR222HG knockdown, we performed RNA-seq using control and mature MIR222HG-depleted WI-38 cells in both quiescence and 6-h serum stimulation conditions. Our analyses identified 398 genes whose expression levels were significantly altered upon knockdown by both MIR222HG shRNAs (Fig. 5E; Supplemental Table S6). Gene ontology analysis showed that the top biological processes, represented by those 398 genes, formed three clusters: cell proliferation, extracellular matrix organization, and cell adhesion (Fig. 5F). Pathway analysis showed that these genes were enriched in pathways including adherens, focal adhesion, Ras signaling pathway, and MAPK pathway (Supplemental Fig. S5B). All these events/pathways play crucial roles for quiescent cells to resume proliferation upon serum stimulation. Based on all these data, we concluded that the increased levels of spliced mature MIR222HG during serum stimulation promotes cell cycle reentry post quiescence.
Spliced MIR222HG depletion did not alter the levels of miR-222 (Fig. 5Ac). In addition, the levels of known canonical miR-221/222 target genes, including CDKN1B (p27), KIT, and PTEN (Felli et al. 2005; Cardinali et al. 2009; Chun-Zhi et al. 2010; Li et al. 2014), remained unchanged in mature MIR222HG-depleted cells (Supplemental Table S6). Finally, we examined the activity of miR-221/222 by performing a luciferase reporter assay using a reporter plasmid containing the 3′-UTR of the CDKN1B gene. We observed that the luciferase signals remained unchanged upon MIR222HG knockdown (Supplemental Fig. S5C), suggesting that the activity of both miRNAs were not affected. These results indicate that the cell cycle phenotype observed in MIR222HG-depleted cells is not due to altered levels or activity of MIR222HG-encoded miRNAs, implying that spliced MIR222HG and miR-221/222 have independent functions.
MIR222HG forms an RNP complex with ILF3/2 and other RNAs
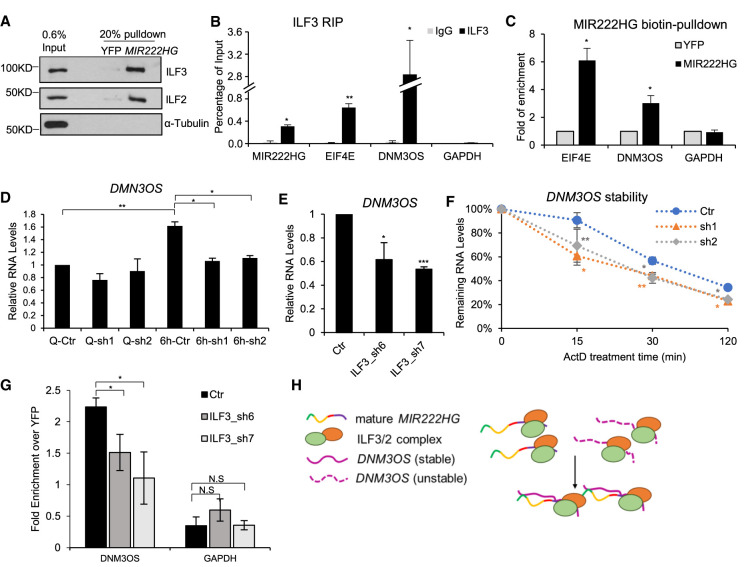
Several lncRNAs, by interacting with proteins/RNA/DNA elements, act as a “scaffold” to modulate RNA–protein, RNA–RNA, or RNA–DNA interactions (Wang and Chang 2011; Kopp and Mendell 2018). In order to determine the molecular function of MIR222HG during cell cycle reentry, we investigated the MIR222HG-interacting proteins and RNAs. We performed streptavidin pulldown in WI-38 cell lysate using in vitro transcribed biotinylated full-length spliced MIR222HG or biotinylated YFP (as negative control), followed by mass spectrometry. From the protein interactome of MIR222HG, we identified two proteins, ILF3 (Interleukin Enhancer Binding Factor 3, or NF90) and ILF2 (Interleukin Enhancer Binding Factor 2, or NF45), as the top hits in all the three biological replicates (Supplemental Table S7). We validated the MIR222HG–ILF3/ILF2 interaction by immunoblotting in the biotin RNA pulldown samples (Fig. 6A). Furthermore, ILF3 RIP confirmed the interaction between endogenous ILF3 and MIR222HG (Fig. 6B; Supplemental Fig. S6A).
FIGURE 6.
MIR222HG forms RNP complex containing ILF3/2 protein and other RNAs. (A) In vitro biotin-RNA pull-down followed by immunoblotting to detect the interaction between MIR222HG and ILF3/ILF2. YFP RNA is used as negative RNA control. α-Tubulin is used as negative protein control. (B) ILF3 RIP followed by RT-qPCR to quantify the interaction between ILF3 and MIR222HG or EIF4E (3′-UTR region) or DNM3OS lncRNA in WI-38 cells. GAPDH is used as negative control. (C) In vitro biotin-RNA pull-down followed by RT-qPCR to determine the interaction between MIR222HG and ILF3/2-target RNAs in WI-38 cells. Relative fold enrichment of EIF4E mRNA and DNM3OS lncRNA are calculated by comparing to biotin-YFP. GAPDH is used as negative control. (D) RT-qPCR to quantify the levels of DNM3OS lncRNA in control and MIR222HG-depleted WI-38 cells in quiescent and 6-h post-serum-stimulated WI-38 cells. (E) RT-qPCR to quantify the levels of DNM3OS in control and ILF3-depleted WI-38 cells post 6-h serum-stimulation. (F) RNA stability analyses of DNM3OS in control and MIR222HG-depleted WI-38 cells. RNA is extracted from cells incubated with actinomycin D for indicated time points (min). Relative levels of RNA are measured via RT-qPCR. (G) In vitro biotin-RNA pull-down followed by RT-qPCR to determine the interaction between MIR222HG and DNM3OS in control and ILF3-depleted serum-stimulated WI-38 cells. Relative fold enrichment of DNM3OS lncRNA is calculated by comparing to biotin-YFP. GAPDH is used as negative control. (H) Schematic of ILF3/2-MIR222HG-DNM3OS RNP complex. (*) P ≤ 0.05, (**) P ≤ 0.01, (***) P ≤ 0.001, (****) P ≤ 0.0001 by two-tailed Student's t-test, n = 3 for all figures. Error bars represent standard deviation.
ILF3/ILF2 is an RNA-binding-protein (RBP) complex that specifically binds to double-stranded RNAs (dsRNA) and regulates RNA stabilization or translation or even ADAR-mediated A-to-I editing (Kuwano et al. 2010; Castella et al. 2015; Jiang et al. 2017; Freund et al. 2020). We examined whether MIR222HG forms intermolecular double stranded RNA structure with other cellular RNAs, and if such RNA–RNA structure is stabilized by the ILF3/2 complex. To test this, we performed a genome-wide bioinformatic prediction to identify RNAs that display sequence complementarity with MIR222HG. RNA complementarity analysis revealed that EIF4E mRNA and DNM3OS lncRNA contained multiple stretches of sequence elements that shared sequence complementarity with different regions of MIR222HG (Supplemental Table S8). Therefore, we performed RNA pull-down using in vitro transcribed biotinylated full-length spliced MIR222HG in total cell extracts and successfully validated its interaction with endogenous EIF4E mRNA and DNM3OS (Fig. 6C). Interestingly, both EIF4E and DNM3OS RNAs were previously identified as ILF3 interacting RNAs (Kuwano et al. 2010; Das et al. 2018). We further confirmed the interaction between ILF3 and EIF4E and DNM3OS in WI-38 cells by RIP experiment (Fig. 6B). Our results indicate that MIR222HG along with ILF3/2 form a ribonucleoprotein (RNP) complex and interacts with several of the known ILF3/2-interacting mRNAs and lncRNAs.
We then sought to investigate whether MIR222HG has a role in regulating the levels or complex assembly of ILF3 and ILF2. Spliced MIR222HG-depleted cells did not show changes in the total cellular levels of ILF2 or both isoforms of ILF3 (NF90 and NF110) (Supplemental Fig. S6B). By performing Proximity Ligation Assay (PLA), we found that the loss of MIR222HG did not cause a biologically significant change in the interaction between IL3 and ILF2 (Supplemental Fig. S6C). We then investigated whether MIR222HG regulates the expression or stability of RNAs that interact with both MIR222HG and IL3/2 complex. DNM3OS (Dynamin-3 opposite strand) is one such candidate lncRNA that is transcribed from the intron of the DNM3 (Dynamin-3) gene and is conserved across mammals. DNM3OS is also a lnc-MIRHG, which encodes miR-199a-2 and miR-214. We observed that DNM3OS interacts with both MIR222HG and ILF3 (Fig. 6B,C). Our genome-wide transcriptome analyses revealed that, similar to MIR222HG, the level of DNM3OS was also significantly elevated during Q to SS transition (Fig. 2F; Supplemental Table S2). RT-qPCR results also confirmed serum-stimulated induction of DNM3OS in WI-38 cells (Fig. 6D). Furthermore, RNA-seq as well as RT-qPCR results revealed that MIR222HG-depleted cells, upon serum stimulation, failed to induce DNM3OS levels (Fig. 6D; Supplemental Table S6). These results imply that both MIR222HG and DNM3OS are induced upon serum stimulation, and MIR222HG positively regulates the levels of DNM3OS. On the other hand, EIF4E mRNA and protein levels were not changed in MIR222HG-depleted cells (Supplemental Fig. S6B,D).
Several recent studies have reported the role of DNM3OS in epithelial-mesenchymal transition (EMT), cell proliferation, and inflammation, and identified DNM3OS as a positive regulator of cell proliferation (Watanabe et al. 2008; Mitra et al. 2017; Das et al. 2018; Ai and Yu 2019). DNM3OS-depleted WI-38 cells showed defects in cell cycle reentry postserum stimulation (Supplemental Fig. S6E,F), supporting its role as a mediator of pro-proliferation. Finally, MIR222HG depletion not only decreased DNM3OS levels but also reduced the level of miR-214, an exonic miRNA generated from the last exon of DNM3OS (Supplemental Fig. S6G). In summary, our data suggested a possibility that MIR222HG-DNM3OS axis could promote the cell cycle reentry of quiescent cells upon serum stimulation.
We focused our initial efforts on understanding the involvement of MIR222HG in regulating DNM3OS levels. We found that ILF3-depleted cells showed defects in the serum-stimulated induction of DNM3OS (Fig. 6E; Supplemental Fig. S6H). Reduced levels of DNM3OS in both MIR222HG- (Fig. 6D) and ILF3-depleted cells (Fig. 6E) suggest that ILF3/2-MIR222HG RNP complex could synergistically regulate the levels of DNM3OS. RNA stability assay showed that DNM3OS stability was significantly reduced upon MIR222HG knockdown (Fig. 6F), suggesting that MIR222HG facilitates the stability of DNM3OS RNA. We examined whether MIR222HG regulates DNM3OS stability by modulating the interaction between ILF3 and DNM3OS. Control and MIR222HG-depleted cells did not show any significant difference in the interaction between ILF3 and DNM3OS, as observed by ILF3 RIP followed by RT-qPCR (Supplemental Fig. S6I). Next, we asked whether the ILF3/2 complex facilitates the interaction between MIR222HG and DNM3OS. MIR222HG biotin-RNA pull-down followed by RT-qPCR in control and ILF3-depleted serum-stimulated cell extracts revealed reduced interactions between MIR222HG and DNM3OS upon ILF3 depletion (Fig. 6G). Based on our results, we speculate that during serum stimulation, MIR222HG along with IL3/2 form an RNP complex to stabilize DNM3OS (Fig. 6H), thereby promoting cell cycle reentry post cellular quiescence.
DISCUSSION
In the present study, we focus on understanding the role of lncRNAs during cell cycle reentry. Within the lncRNA repertoire, lnc-MIRHGs (microRNA-host-gene lncRNAs) constitute an understudied family because they are often considered as primary miRNA transcripts, without additional functions. In support of this argument, studies have reported that several pri-MIRHGs are cleaved by Drosha to make unproductive transcripts (Dhir et al. 2015a; Pianigiani et al. 2018). However, earlier independent studies from our laboratory as well as other groups have documented miRNA-independent roles of lnc-MIRHGs (Ng et al. 2013; Tseng et al. 2014; Colombo et al. 2015; Raveh et al. 2015; Shih et al. 2017; Sun et al. 2018b; Du et al. 2019; Profumo et al. 2019). In the present study, we systematically annotated the role of human MIR222HG in cell cycle reentry. These examples strongly support the idea that it is not an uncommon but a rather general genomic mechanism for a single gene locus to generate two classes of independently functional noncoding RNAs.
From our RNA-seq analyses, we identified several differentially expressed lnc-MIRHGs (DE-lnc-MIRHGs) during the course of quiescence onset and cell cycle reentry. The up-regulated DE-lnc-MIRHGs during serum stimulation have the potential to facilitate the quiescence exit/cell cycle reentry, which is an event strongly associated with tumor reoccurrence. Hence, it is not surprising that some of the up-regulated DE-lnc-MIRHGs that we identified in our study (MIR31HG, MIR22HG, and MIR17HG) were previously recognized as cancer biomarkers or regulators of cancer progression (Montes et al. 2015; Shih et al. 2017; Su et al. 2018; Xu et al. 2019; Yuan et al. 2019). For instance, MIR17HG can promote tumorigenesis and metastasis of colorectal cancer and gastric cancer (Xu et al. 2019; Yuan et al. 2019). MIR31HG has been shown to play roles in senescence modulation (Montes et al. 2015), MIR31HG has also been shown to play miR-31-independent functions in regulating the HIF-1 transcriptional network to drive oral cancer progression (Shih et al. 2017). These studies have substantiated the biological significance of the DE-lnc-MIRHGs identified in our study.
Previous studies have reported coregulated or independent expression patterns of miRNAs and their host genes (Baskerville and Bartel 2005; Lutter et al. 2010; Budach et al. 2016; Liu et al. 2018; Steiman-Shimony et al. 2018). For example, DLEU2, which is down-regulated in pediatric acute myeloid leukemia, was shown to express independently from miR-15a/16-1, which are encoded within DLEU2 (Morenos et al. 2014). Also, we previously demonstrated that MIR100HG was up-regulated in G1 phase of the cell cycle, independently of its encoded miRNAs, such as miR-100 (Sun et al. 2018b). Similarly, in the present study, we showed that spliced MIR222HG, but not miR-221 or miR-222, was dramatically up-regulated during serum stimulation, implying that the mature MIR222HG and miR-221/222 display independent expression patterns. We also demonstrated that the major inciting factor for the induction of mature MIR222HG upon serum stimulation was the increased splicing efficiency of pri-MIR222HG, due to enhanced SRSF1 binding. Our findings suggest that transcriptional and posttranscriptional regulations cooperate to fine-tune the levels of mature lnc-MIRHGs and the miRNAs that are encoded within the MIRHGs. It is not yet known how the association of SRSF1 to MIR222HG is being regulated. We speculate that SRSF1 binding could be modulated by serum-induced posttranslational modifications of SRSF1 (i.e., phosphorylation). Alternatively, serum induction probably could promote the recruitment of SRSF1-interacting factors to MIR222HG, which stabilizes the association of SRSF1 to the MIR222HG transcript.
During the processing of SO-miRNAs, the crosstalk between the splicing factors and microprocessor proteins dictates the relationship between miRNA biogenesis and host gene RNA splicing. This falls under two general models: “synergic” and “competitive,” as described by several previous studies (Wu et al. 2010; Janas et al. 2011; Westholm and Lai 2011; Mattioli et al. 2013, 2014; Melamed et al. 2013; Agranat-Tamir et al. 2014; Pianigiani et al. 2018). Synergic cases are exemplified by a type of intronic miRNAs “mirtrons” whose production depends on the splicing of their host gene (Berezikov et al. 2007b; Westholm and Lai 2011). On the other hand, MCM7 and MIR202HG, which host miR-106b-25 and miR-202, respectively, identify examples of the competitive model (Melamed et al. 2013; Agranat-Tamir et al. 2014). Notably, both miR-106b-25 and miR-202 are splice site overlapping miRNAs (SO-miRNAs) (Mattioli et al. 2014). We defined miR-222 as a SO-miRNA in light of the newly identified 44 bp mini-exon that partially overlaps with pri-miR-222. We concluded that the splicing and microprocessor machinery compete to process the pri-MIR222HG transcripts during serum stimulation, reflected by the observation that Drosha/DGCR8 complex antagonizes the access of SRSF1 to pri-MIR222HG. In this context, an earlier study reported that SRSF1, instead of playing its canonical role as a splicing factor, facilitated Drosha-mediated processing of pre-miRNAs (Wu et al. 2010). The authors reported that the overexpression of SRSF1 in HeLa cells caused increased levels of several miRNAs, including miR-7, miR-221, and miR-222. In addition to the mature miRNA levels, SRSF1 overexpression also increased the levels of pre-miRNAs when cells expressed minigenes containing miRNA precursors. Based on this, the authors claimed that SRSF1 promotes the Drosha cleavage step of pri-miRNA processing, resulting in enhanced syntheses of multiple miRNAs, including miR-222/221. However, in WI-38 diploid fibroblasts, overexpression or knockdown of SRSF1 did not affect the levels of miR-222. One possible explanation for the discrepancy in results could be the fact that the two studies utilized different cell lines and physiological conditions (serum-stimulated WI-38 diploid fibroblasts versus asynchronous HeLa cancer cell line). LncRNA genes are shown to generate a variety of isoforms in different cell lines/tissues/species/biological scenarios (Ziegler and Kretz 2017). We speculate that Hela cells might not express the serum-responsive MIR222HG isoform that contains the 44 bp mini-exon. In such a scenario, miR-222 would behave as an intronic miRNA and could rely on splicing for its biogenesis. In summary, these observations indicate that MIR222HG gene expression is regulated by differential splicing and intragenic miRNA biogenesis in a cell type-specific manner.
LncRNAs can modulate transcription, pre-mRNA splicing, RNA stability, and protein translation by regulating the recruitment of proteins to chromatin and/or RNA (Wang and Chang 2011; Noh et al. 2018; Sun et al. 2018a; He et al. 2019). We found that spliced MIR222HG associated with the ILF3/2 complex. ILF3/2 heterodimeric protein complex regulates both transcription and posttranscriptional events, such as RNA stability and translation (Kuwano et al. 2010; Hu et al. 2013; Castella et al. 2015; Nakadai et al. 2015; Jiang et al. 2017; Wu et al. 2018). Earlier studies demonstrated the involvement of the ILF3/2 complex in modulating lncRNA functions. For example, lncRNA LincIN-ILF3 complex regulates breast cancer metastasis by inhibiting p21 translation (Jiang et al. 2017), and ILF3-SALNR lncRNA complex modulates miRNA biogenesis (Wu et al. 2015). Our data indicated that MIR222HG, DNM3OS, and ILF3/2 form RNP complex, and MIR222HG/ILF3 complex positively regulate the stability of DNM3OS lncRNA. Furthermore, we demonstrated that ILF3/2-facilitates the interaction between MIR222HG and DNM3OS. Hence, we hypothesize that ILF3/2, with their inherent ability to bind to double-stranded RNAs, act as a “scaffold” to promote MIR222HG-DNM3OS duplex formation, and thereby stabilize DNM3OS. A similar mechanism has been previously identified in the case of HOTAIR-hnRNP B1 complex (Meredith et al. 2016; Nguyen et al. 2018). HnRNP B1, by forming an RNP-complex with HOTAIR facilitates RNA–RNA interactions between HOTAIR and several target RNAs to initiate the heterochromatinization. We therefore speculate that, ILF3, like hnRNP B1, functions as an “RNA matchmaker protein.”
We found that DNM3OS-depleted WI-38 cells displayed defects in cell proliferation post quiescence, which suggests that DNM3OS could be one of the downstream effectors of MIR222HG–ILF3/2 axis. DNM3OS has been previously shown to be involved in cell growth/proliferation (Watanabe et al. 2008; Mitra et al. 2017; Ai and Yu 2019). In human chondrocyte cells, DNM3OS enhanced cell proliferation (Ai and Yu 2019). In mice, DNM3OS was found to be essential for normal growth, based on a study that showed its depletion caused defects in birth and/or development (Watanabe et al. 2008). DNM3OS is also known to facilitate the mesenchymal-to-epithelial transition, cancer cell migration, and invasion; and elevated levels of DNM3OS in patients were associated with poor survival (Mitra et al. 2017). In this particular study, the authors have reported that depletion of DNM3OS altered multiple pathways including cytoskeleton regulation, adherens junction, focal adhesion, and MAPK signaling pathways. Interestingly, MIR222HG-depleted cells also displayed defects in similar events/pathways (Fig. 5F; Supplemental Fig. S5B), implying the involvement of MIR222HG-DNM3OS axis in influencing crucial pathways that regulate cell cycle reentry. Based on this, we speculate that MIR222HG depletion caused cell cycle defects by reducing the cellular level of another pro-proliferative lnc-MIRHG, DNM3OS. Future studies will focus on the molecular mechanism by which MIR222HG controls DNM3OS activity.
Although, in the current study, we identified the involvement of MIR222HG–ILF3 RNP complex in regulating the RNA stability of DNM3OS, a cell cycle regulator, we cannot rule out the possibility that MIR222HG–ILF3 could in addition engage in other molecular processes. For example, ILF3 has been shown, by multiple studies, to have pro-proliferative roles by serving as transcription factor or cofactor to facilitate the expression of early serum-responsive genes, including EGR1, MYC, JUN, and FOS (Nakadai et al. 2015; Wu et al. 2018, 2019). Hence, it might also be possible that MIR222HG plays additional roles in regulating transcription via modulating ILF3/ILF2 transcription factor/cofactor activity.
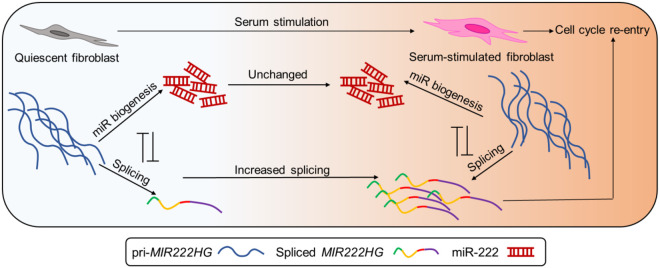
We have proposed a working model to summarize our findings about how MIR222HG promotes cell cycle reentry post quiescence (Fig. 7). We showed that the abundant and unstable pri-MIR222HG could undergo two mutually independent processes. The dominant role of pri-MIR222HG is to act as a substrate for miR-221/222 biogenesis. However, upon serum stimulation, a small but significant fraction of pri-MIR222HG undergoes splicing, which is mediated by enhanced SRSF1 binding, resulting in elevated levels of the spliced mature MIR222HG lncRNA. The mature lncRNA MIR222HG exerts miRNA-independent functions to facilitate cell cycle reentry. Our preliminary studies support a model that MIR222HG–ILF3/2 axis assures the success of cell cycle reentry in WI-38 cells.
FIGURE 7.
Schematic of MIR222HG biogenesis and potential mode of action.
MATERIALS AND METHODS
Cell culture
WI-38 cells were cultured in 10% FBS + MEM with nonessential amino acids (NEAA) and sodium pyruvate. To induce cellular quiescence, cells were cultured in 0.1% serum for 3 d. Then, serum reactivation was performed by adding back the regular 10% serum growth medium. For lentivirus packaging, HEK293T cells were cultured using DMEM containing 10%FBS (heat inactivated).
RNA-seq and bioinformatics analyses
Total RNA was extracted using RNeasy Mini Kit and DNA was removed using DNase I (Qiagen, Cat# 74104). The RNA-seq libraries were prepared with Illumina's TruSeq Stranded mRNAseq Sample Prep kit and polyadenylated mRNAs were captured with oligo(dT) beads. Paired-end, poly(A)+ RNA-sequencing was performed on the Illumina platform (Novaseq 6000, SP flowcell) at the Roy J. Carver Biotechnology Center at UIUC. Quality of RNA-seq reads was examined by FASTQC. RNA-seq reads were aligned to the human reference genome GRCh38 assembly using HISAT2 (Kim et al. 2015). Transcript assembly and expression assessment was performed using Stringtie (Pertea et al. 2015) to get the TPM (Transcripts Per Million) values for each gene. Read coverage BigWig (bw) files were generated with RPKM normalization using bamCoverage in deepTools (Ramirez et al. 2016). Bw files from two biological replicates were further merged using bigWigMerge and bedGraghToBigWig. For statistical analyses, raw gene counts were first analyzed by HTseq-Count (Anders et al. 2015), then analyzed using edgeR (Robinson et al. 2010). Qualifiable expression was defined by CPM ≥ 0.1 in at least two samples out of total eight samples. Normalization of library size was performed by calcNormFactors in edgeR with the default TMM method. For visualization of the transcriptome profile, heatmaps were plotted using coolmap function from limma package (Ritchie et al. 2015), with row centering and scaling. Hierarchical clustering of genes (rows) was performed with average-linkage method. Differential expression analyses were performed using exactTest between every two adjacent cell cycle phases. Differentially expressed genes (DEGs) were defined by |log2(fold change)| > 1 and FDR < 0.05.
Gene ontology analyses and GSEA (gene set enrichment analysis) were performed using clusterProfiler of Bioconductor (Yu et al. 2012). Specifically, gene ontology for biological process was performed using enrichGO function, Kegg pathway analyses were performed using enrichKEGG. All enrichment analyses include using a background gene list containing all 17,640 genes that showed qualifiable expression in the RNA-seq. GSEA analysis was performed using gseGO function and gene lists were ranked using logFC values.
MIRHGs, in this study, were defined when the coordinates of a gene include the coordinates of a miRNA gene. The coordinate information of the gene/exons was extracted from the GRCh38 assembly GTF file (v94 of Ensembl). For intragenic miRNA categorization, miRNAs that completely locate within any exon of a MIRHG were defined as exonic-miRs. MiRs were defined as intronic miRs when they do not overlap with any exonic regions of any genes. The rest of intragenic miRNAs were then defined as spliced site overlapping miRNAs (SO-miRs).
MicroRNA quantification
TaqMan assays were performed for measuring the mature miRNAs as per manufacturer's instruction. hsa-miR-222 was measured and normalized to U6 (TaqMan, Cat# 4427975 with AssayID 002276 and 001973, respectively). The method of measuring precursor miRNA (pre-miRs) was adapted from a previous study (Kawahara 2012). Small RNAs were isolated using miRVANA (Ambion, Cat# AM1560). 1ug small RNA was then used for 3′ Polyadenylation addition using Poly(A) Polymerase Tailing Kit (Epicentre, Cat# PAP5104H), followed by DNase I treatment (Sigma, Cat# AMPD1) and reverse transcription (Applied Biosystems, Cat# 4368814) with a customized RT primer (Customized Oligo-dT RT primer, see primer table in Supplemental Materials and Methods). Quantification of pre-miRNAs was performed by RT-qPCR with customized primers (222-3tail-qPCR-F, U6-3tail-qPCR-F, 3tail-qpcr-R, see primer table in Supplemental Materials and Methods). U6 served as loading control.
Single-molecule fluorescence RNA in situ hybridization (smRNA-FISH)
The MIR222HG smRNA FISH probe set was designed using Stellaris Probe Designer, and consisted of 29 20-mer DNA oligonucleotides. Full-length (1.3 kb) MIR222HG sequence was used as input for probe design. 3′amino group modified oligonucleotides (LGC Biosearch Technologies) were pooled and coupled with Cy3 Mono NHS Ester (GE Healthcare). WI-38 cells were seeded on coverslips for serum starvation and release. At harvest, cells were fixed with 10% neutral buffered formalin (Sigma, HT501128) for 10 min at room temperature and permeabilized with 70% ethanol for overnight at −20°C. The coverslips were then washed with Buffer A (20% Stellaris RNA FISH Wash Buffer A [Biosearch Technologies, SMF-WA1-60] and 10% formamide in nuclease-free water) for 5 min at room temperature and incubated with hybridization buffer (90% Stellaris RNA FISH Hybridization Buffer [Biosearch Technologies, SMF-HB1-10] and 10% formamide) containing ∼125 nM of the Cy3-labeled MIR222HG smFISH probe in a humidified chamber in the dark at 37°C for 16 h. After hybridization, the coverslips were washed twice with Buffer A, 30 min for each of the washes, in the dark at 37°C. DNA were counterstained by DAPI during the second wash. The coverslips were then washed with Stellaris RNA FISH Wash Buffer B (Biosearch Technologies, SMF-WB1-20) for 5 min in the dark at room temperature and mounted onto microscope slides with VectaShield Antifade Mounting Medium (Vector Laboratories, H-1000). Z-stack images were taken using DeltaVision microscope (GE) equipped with 60×/1.42 NA oil immersion objective (Olympus) and CoolSNAP-HQ2 camera. Images were then processed through deconvolution and maximum intensity projection.
DATA DEPOSITION
RNA-seq files were submitted to GEO. The asynchronous, quiescent, and serum stimulated WI-38 RNA-seq files are in GSE144710. The RNA-seq data of MIR222HG-depletion experiment are in GSE145212. Mass spectrometric data has been submitted to ProteomeXchange via the PRIDE database (PXD017585).
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
COMPETING INTEREST STATEMENT
S.M.F. is an employee of Ionis Inc. and receives a salary from Ionis Inc.
Supplementary Material
ACKNOWLEDGMENTS
We would like to thank K.V. Prasanth and S.G. Prasanth laboratory members for their comments and suggestions. We thank Dr. Brian Freeman (UIUC) for sharing equipment and Dr. Alvaro Hernandez (J. Carver Biotechnology Center Directory, UIUC) for technical support for RNA-seq. We also thank the CCR, National Institutes of Health (NIH) sequencing core for help with RNA-seq. Work in the K.V.P. laboratory was supported by grants from the National Institute on Aging/NIH (R21AG065748), Cancer Center at Illinois seed grant, Prairie Dragon Paddlers, and the National Science Foundation (NSF) (EAGER; MCB1723008). Work in the S.G.P. laboratory is supported by the NSF (1243372 and 1818286) and the NIH (GM125196). Work in the A.K. laboratory is supported by the NIH (R01HL126845, R01AA010154), Muscular Dystrophy Association (MDA514335), Planning Grant Award from the Cancer Center at Illinois, and a Beckman Fellowship from the Center for Advanced Study at the University of Illinois Urbana-Champaign. S.B. is supported by the NIH Tissue Microenvironment Training Program (T32-EB019944). J.-H.C. and J.-H.Y were funded by the Medical University of South Carolina.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.075309.120.
REFERENCES
- Agranat-Tamir L, Shomron N, Sperling J, Sperling R. 2014. Interplay between pre-mRNA splicing and microRNA biogenesis within the supraspliceosome. Nucleic Acids Res 42: 4640–4651. 10.1093/nar/gkt1413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ai D, Yu F. 2019. LncRNA DNM3OS promotes proliferation and inhibits apoptosis through modulating IGF1 expression by sponging MiR-126 in CHON-001 cells. Diagn Pathol 14: 106 10.1186/s13000-019-0877-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ameres SL, Zamore PD. 2013. Diversifying microRNA sequence and function. Nat Rev Mol Cell Biol 14: 475–488. 10.1038/nrm3611 [DOI] [PubMed] [Google Scholar]
- Anders S, Pyl PT, Huber W. 2015. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics 31: 166–169. 10.1093/bioinformatics/btu638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augoff K, McCue B, Plow EF, Sossey-Alaoui K. 2012. miR-31 and its host gene lncRNA LOC554202 are regulated by promoter hypermethylation in triple-negative breast cancer. Mol Cancer 11: 5 10.1186/1476-4598-11-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bainbridge P. 2013. Wound healing and the role of fibroblasts. J Wound Care 22: 407–408. 10.12968/jowc.2013.22.8.407 [DOI] [PubMed] [Google Scholar]
- Baskerville S, Bartel DP. 2005. Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes. RNA 11: 241–247. 10.1261/rna.7240905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berezikov E, Chung WJ, Willis J, Cuppen E, Lai EC. 2007a. Mammalian mirtron genes. Mol Cell 28: 328–336. 10.1016/j.molcel.2007.09.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berezikov E, Chung WJ, Willis J, Cuppen E, Lai EC. 2007b. Mammalian mirtron genes. Mol Cell 28: 328–336. 10.1016/j.molcel.2007.09.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierhoff H, Dammert MA, Brocks D, Dambacher S, Schotta G, Grummt I. 2014. Quiescence-induced lncRNAs trigger H4K20 trimethylation and transcriptional silencing. Mol Cell 54: 675–682. 10.1016/j.molcel.2014.03.032 [DOI] [PubMed] [Google Scholar]
- Budach S, Heinig M, Marsico A. 2016. Principles of microRNA regulation revealed through modeling microRNA expression quantitative trait loci. Genetics 203: 1629–1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch A, Hertel KJ. 2012. Evolution of SR protein and hnRNP splicing regulatory factors. Wiley Interdiscip Rev RNA 3: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardinali B, Castellani L, Fasanaro P, Basso A, Alema S, Martelli F, Falcone G. 2009. MicroRNA-221 and microRNA-222 modulate differentiation and maturation of skeletal muscle cells. PLoS ONE 4: e7607 10.1371/journal.pone.0007607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castella S, Bernard R, Corno M, Fradin A, Larcher JC. 2015. Ilf3 and NF90 functions in RNA biology. Wiley Interdiscip Rev RNA 6: 243–256. 10.1002/wrna.1270 [DOI] [PubMed] [Google Scholar]
- Chen YG, Satpathy AT, Chang HY. 2017. Gene regulation in the immune system by long noncoding RNAs. Nat Immunol 18: 962–972. 10.1038/ni.3771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung TH, Rando TA. 2013. Molecular regulation of stem cell quiescence. Nat Rev Mol Cell Biol 14: 329–340. 10.1038/nrm3591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung TH, Quach NL, Charville GW, Liu L, Park L, Edalati A, Yoo B, Hoang P, Rando TA. 2012. Maintenance of muscle stem-cell quiescence by microRNA-489. Nature 482: 524–528. 10.1038/nature10834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi SW, Kim HW, Nam JW. 2019. The small peptide world in long noncoding RNAs. Brief Bioinformatics 20: 1853–1864. 10.1093/bib/bby055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun-Zhi Z, Lei H, An-Ling Z, Yan-Chao F, Xiao Y, Guang-Xiu W, Zhi-Fan J, Pei-Yu P, Qing-Yu Z, Chun-Sheng K. 2010. MicroRNA-221 and microRNA-222 regulate gastric carcinoma cell proliferation and radioresistance by targeting PTEN. BMC Cancer 10: 367 10.1186/1471-2407-10-367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coller HA. 2007. What's taking so long? S-phase entry from quiescence versus proliferation. Nat Rev Mol Cell Biol 8: 667–670. 10.1038/nrm2223 [DOI] [PubMed] [Google Scholar]
- Coller HA. 2011. Cell biology. The essence of quiescence. Science 334: 1074–1075. 10.1126/science.1216242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coller HA, Sang L, Roberts JM. 2006. A new description of cellular quiescence. PLoS Biol 4: e83 10.1371/journal.pbio.0040083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo T, Farina L, Macino G, Paci P. 2015. PVT1: a rising star among oncogenic long noncoding RNAs. Biomed Res Int 2015: 304208 10.1155/2015/304208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S, Reddy MA, Senapati P, Stapleton K, Lanting L, Wang M, Amaram V, Ganguly R, Zhang L, Devaraj S, et al. 2018. Diabetes mellitus-induced long noncoding RNA Dnm3os regulates macrophage functions and inflammation via nuclear mechanisms. Arterioscler Thromb Vasc Biol 38: 1806–1820. 10.1161/ATVBAHA.117.310663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhir A, Dhir S, Proudfoot NJ, Jopling CL. 2015a. Microprocessor mediates transcriptional termination of long noncoding RNA transcripts hosting microRNAs. Nat Struct Mol Biol 22: 319–327. 10.1038/nsmb.2982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhir A, Dhir S, Proudfoot NJ, Jopling CL. 2015b. Microprocessor mediates transcriptional termination of long noncoding RNA transcripts hosting microRNAs. Nat Struct Mol Biol 22: 319–327. 10.1038/nsmb.2982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Q, Hoover AR, Dozmorov I, Raj P, Khan S, Molina E, Chang TC, de la Morena MT, Cleaver OB, Mendell JT, et al. 2019. MIR205HG is a long noncoding RNA that regulates growth hormone and prolactin production in the anterior pituitary. Dev Cell 49: 618–631 e615. 10.1016/j.devcel.2019.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y, Fullwood MJ. 2016. Roles, functions, and mechanisms of long non-coding RNAs in cancer. Genomics Proteomics Bioinformatics 14: 42–54. 10.1016/j.gpb.2015.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felli N, Fontana L, Pelosi E, Botta R, Bonci D, Facchiano F, Liuzzi F, Lulli V, Morsilli O, Santoro S, et al. 2005. MicroRNAs 221 and 222 inhibit normal erythropoiesis and erythroleukemic cell growth via kit receptor down-modulation. Proc Natl Acad Sci 102: 18081–18086. 10.1073/pnas.0506216102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn RA, Chang HY. 2014. Long noncoding RNAs in cell-fate programming and reprogramming. Cell Stem Cell 14: 752–761. 10.1016/j.stem.2014.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freund EC, Sapiro AL, Li Q, Linder S, Moresco JJ, Yates JR III, Li JB. 2020. Unbiased identification of trans regulators of ADAR and A-to-I RNA editing. Cell Rep 31: 107656 10.1016/j.celrep.2020.107656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garofalo M, Quintavalle C, Romano G, Croce CM, Condorelli G. 2012. miR221/222 in cancer: their role in tumor progression and response to therapy. Curr Mol Med 12: 27–33. 10.2174/156652412798376170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gos M, Miloszewska J, Swoboda P, Trembacz H, Skierski J, Janik P. 2005. Cellular quiescence induced by contact inhibition or serum withdrawal in C3H10T1/2 cells. Cell Prolif 38: 107–116. 10.1111/j.1365-2184.2005.00334.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harashima H, Dissmeyer N, Schnittger A. 2013. Cell cycle control across the eukaryotic kingdom. Trends Cell Biol 23: 345–356. 10.1016/j.tcb.2013.03.002 [DOI] [PubMed] [Google Scholar]
- He RZ, Luo DX, Mo YY. 2019. Emerging roles of lncRNAs in the post-transcriptional regulation in cancer. Genes Dis 6: 6–15. 10.1016/j.gendis.2019.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Q, Lu YY, Noh H, Hong S, Dong Z, Ding HF, Su SB, Huang S. 2013. Interleukin enhancer-binding factor 3 promotes breast tumor progression by regulating sustained urokinase-type plasminogen activator expression. Oncogene 32: 3933–3943. 10.1038/onc.2012.414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janas MM, Khaled M, Schubert S, Bernstein JG, Golan D, Veguilla RA, Fisher DE, Shomron N, Levy C, Novina CD. 2011. Feed-forward microprocessing and splicing activities at a microRNA-containing intron. PLoS Genet 7: e1002330 10.1371/journal.pgen.1002330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Z, Slater CM, Zhou Y, Devarajan K, Ruth KJ, Li Y, Cai KQ, Daly M, Chen X. 2017. LincIN, a novel NF90-binding long non-coding RNA, is overexpressed in advanced breast tumors and involved in metastasis. Breast Cancer Res 19: 62 10.1186/s13058-017-0853-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahl G. 2009. The dictionary of genomics, transcriptomics and proteomics. Wiley-Blackwell, Hoboken, NJ. [Google Scholar]
- Kato M, Wang M, Chen Z, Bhatt K, Oh HJ, Lanting L, Deshpande S, Jia Y, Lai JY, O'Connor CL, et al. 2016. An endoplasmic reticulum stress-regulated lncRNA hosting a microRNA megacluster induces early features of diabetic nephropathy. Nat Commun 7: 12864 10.1038/ncomms12864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara Y. 2012. Quantification of adenosine-to-inosine editing of microRNAs using a conventional method. Nat Protoc 7: 1426–1437. 10.1038/nprot.2012.073 [DOI] [PubMed] [Google Scholar]
- Kim D, Langmead B, Salzberg SL. 2015. HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12: 357–360. 10.1038/nmeth.3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa M, Kitagawa K, Kotake Y, Niida H, Ohhata T. 2013. Cell cycle regulation by long non-coding RNAs. Cell Mol Life Sci 70: 4785–4794. 10.1007/s00018-013-1423-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopp F, Mendell JT. 2018. Functional classification and experimental dissection of long noncoding RNAs. Cell 172: 393–407. 10.1016/j.cell.2018.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwano Y, Pullmann R Jr, Marasa BS, Abdelmohsen K, Lee EK, Yang X, Martindale JL, Zhan M, Gorospe M. 2010. NF90 selectively represses the translation of target mRNAs bearing an AU-rich signature motif. Nucleic Acids Res 38: 225–238. 10.1093/nar/gkp861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laporte D, Lebaudy A, Sahin A, Pinson B, Ceschin J, Daignan-Fornier B, Sagot I. 2011. Metabolic status rather than cell cycle signals control quiescence entry and exit. J Cell Biol 192: 949–957. 10.1083/jcb.201009028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemons JM, Feng XJ, Bennett BD, Legesse-Miller A, Johnson EL, Raitman I, Pollina EA, Rabitz HA, Rabinowitz JD, Coller HA. 2010. Quiescent fibroblasts exhibit high metabolic activity. PLoS Biol 8: e1000514 10.1371/journal.pbio.1000514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Liang C, Ma H, Zhao Q, Lu Y, Xiang Z, Li L, Qin J, Chen Y, Cho WC, et al. 2014. miR-221/222 promotes S-phase entry and cellular migration in control of basal-like breast cancer. Molecules 19: 7122–7137. 10.3390/molecules19067122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Tian H, Yang J, Gong Z. 2016. Long noncoding RNAs regulate cell growth, proliferation, and apoptosis. DNA Cell Biol 35: 459–470. 10.1089/dna.2015.3187 [DOI] [PubMed] [Google Scholar]
- Liu H, Adler AS, Segal E, Chang HY. 2007. A transcriptional program mediating entry into cellular quiescence. PLoS Genet 3: e91 10.1371/journal.pgen.0030091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Shyr Y, Cai J, Liu Q. 2018. Interplay between miRNAs and host genes and their role in cancer. Brief Funct Genomics 18: 255–266. 10.1093/bfgp/elz002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutter D, Marr C, Krumsiek J, Lang EW, Theis FJ. 2010. Intronic microRNAs support their host genes by mediating synergistic and antagonistic regulatory effects. BMC Genomics 11: 224 10.1186/1471-2164-11-224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchese FP, Raimondi I, Huarte M. 2017. The multidimensional mechanisms of long noncoding RNA function. Genome Biol 18: 206 10.1186/s13059-017-1348-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez I, Hayes KE, Barr JA, Harold AD, Xie M, Bukhari SIA, Vasudevan S, Steitz JA, DiMaio D. 2017. An Exportin-1-dependent microRNA biogenesis pathway during human cell quiescence. Proc Natl Acad Sci 114: E4961–E4970. 10.1073/pnas.1618732114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattioli C, Pianigiani G, Pagani F. 2013. A competitive regulatory mechanism discriminates between juxtaposed splice sites and pri-miRNA structures. Nucleic Acids Res 41: 8680–8691. 10.1093/nar/gkt614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattioli C, Pianigiani G, Pagani F. 2014. Cross talk between spliceosome and microprocessor defines the fate of pre-mRNA. Wiley Interdiscip Rev RNA 5: 647–658. 10.1002/wrna.1236 [DOI] [PubMed] [Google Scholar]
- Mayoral RJ, Deho L, Rusca N, Bartonicek N, Saini HK, Enright AJ, Monticelli S. 2011. MiR-221 influences effector functions and actin cytoskeleton in mast cells. PLoS ONE 6: e26133 10.1371/journal.pone.0026133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina R, Zaidi SK, Liu CG, Stein JL, van Wijnen AJ, Croce CM, Stein GS. 2008. MicroRNAs 221 and 222 bypass quiescence and compromise cell survival. Cancer Res 68: 2773–2780. 10.1158/0008-5472.CAN-07-6754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melamed Z, Levy A, Ashwal-Fluss R, Lev-Maor G, Mekahel K, Atias N, Gilad S, Sharan R, Levy C, Kadener S, et al. 2013. Alternative splicing regulates biogenesis of miRNAs located across exon-intron junctions. Mol Cell 50: 869–881. 10.1016/j.molcel.2013.05.007 [DOI] [PubMed] [Google Scholar]
- Meredith EK, Balas MM, Sindy K, Haislop K, Johnson AM. 2016. An RNA matchmaker protein regulates the activity of the long noncoding RNA HOTAIR. RNA 22: 995–1010. 10.1261/rna.055830.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra R, Chen X, Greenawalt EJ, Maulik U, Jiang W, Zhao Z, Eischen CM. 2017. Decoding critical long non-coding RNA in ovarian cancer epithelial-to-mesenchymal transition. Nat Commun 8: 1604 10.1038/s41467-017-01781-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molinari C, Salvi S, Foca F, Teodorani N, Saragoni L, Puccetti M, Passardi A, Tamberi S, Avanzolini A, Lucci E, et al. 2016. miR-17-92a-1 cluster host gene (MIR17HG) evaluation and response to neoadjuvant chemoradiotherapy in rectal cancer. Onco Targets Ther 9: 2735–2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montes M, Nielsen MM, Maglieri G, Jacobsen A, Hojfeldt J, Agrawal-Singh S, Hansen K, Helin K, van de Werken HJG, Pedersen JS, et al. 2015. The lncRNA MIR31HG regulates p16INK4A expression to modulate senescence. Nat Commun 6: 6967 10.1038/ncomms7967 [DOI] [PubMed] [Google Scholar]
- Monteys AM, Spengler RM, Wan J, Tecedor L, Lennox KA, Xing Y, Davidson BL. 2010. Structure and activity of putative intronic miRNA promoters. RNA 16: 495–505. 10.1261/rna.1731910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morenos L, Chatterton Z, Ng JL, Halemba MS, Parkinson-Bates M, Mechinaud F, Elwood N, Saffery R, Wong NC. 2014. Hypermethylation and down-regulation of DLEU2 in paediatric acute myeloid leukaemia independent of embedded tumour suppressor miR-15a/16-1. Mol Cancer 13: 123 10.1186/1476-4598-13-123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakadai T, Fukuda A, Shimada M, Nishimura K, Hisatake K. 2015. The RNA binding complexes NF45-NF90 and NF45-NF110 associate dynamically with the c-fos gene and function as transcriptional coactivators. J Biol Chem 290: 26832–26845. 10.1074/jbc.M115.688317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng SY, Bogu GK, Soh BS, Stanton LW. 2013. The long noncoding RNA RMST interacts with SOX2 to regulate neurogenesis. Mol Cell 51: 349–359. 10.1016/j.molcel.2013.07.017 [DOI] [PubMed] [Google Scholar]
- Nguyen ED, Balas MM, Griffin AM, Roberts JT, Johnson AM. 2018. Global profiling of hnRNP A2/B1-RNA binding on chromatin highlights lncRNA interactions. RNA Biol 15: 901–913. 10.1080/15476286.2018.1474072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noh JH, Kim KM, McClusky WG, Abdelmohsen K, Gorospe M. 2018. Cytoplasmic functions of long noncoding RNAs. Wiley Interdiscip Rev RNA 9: e1471 10.1002/wrna.1471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paz I, Kosti I, Ares M Jr, Cline M, Mandel-Gutfreund Y. 2014. RBPmap: a web server for mapping binding sites of RNA-binding proteins. Nucleic Acids Res 42: W361–W367. 10.1093/nar/gku406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea M, Pertea GM, Antonescu CM, Chang TC, Mendell JT, Salzberg SL. 2015. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol 33: 290–295. 10.1038/nbt.3122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pianigiani G, Licastro D, Fortugno P, Castiglia D, Petrovic I, Pagani F. 2018. Microprocessor-dependent processing of splice site overlapping microRNA exons does not result in changes in alternative splicing. RNA 24: 1158–1171. 10.1261/rna.063438.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polioudakis D, Bhinge AA, Killion PJ, Lee BK, Abell NS, Iyer VR. 2013. A Myc-microRNA network promotes exit from quiescence by suppressing the interferon response and cell-cycle arrest genes. Nucleic Acids Res 41: 2239–2254. 10.1093/nar/gks1452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Profumo V, Forte B, Percio S, Rotundo F, Doldi V, Ferrari E, Fenderico N, Dugo M, Romagnoli D, Benelli M, et al. 2019. LEADeR role of miR-205 host gene as long noncoding RNA in prostate basal cell differentiation. Nat Commun 10: 307 10.1038/s41467-018-08153-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez F, Ryan DP, Gruning B, Bhardwaj V, Kilpert F, Richter AS, Heyne S, Dundar F, Manke T. 2016. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res 44: W160–W165. 10.1093/nar/gkw257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raveh E, Matouk IJ, Gilon M, Hochberg A. 2015. The H19 long non-coding RNA in cancer initiation, progression and metastasis—a proposed unifying theory. Mol Cancer 14: 184 10.1186/s12943-015-0458-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. 2015. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43: e47 10.1093/nar/gkv007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt AM, Chang HY. 2016. Long noncoding RNAs in cancer pathways. Cancer Cell 29: 452–463. 10.1016/j.ccell.2016.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih JW, Chiang WF, Wu ATH, Wu MH, Wang LY, Yu YL, Hung YW, Wang WC, Chu CY, Hung CL, et al. 2017. Long noncoding RNA LncHIFCAR/MIR31HG is a HIF-1α co-activator driving oral cancer progression. Nat Commun 8: 15874 10.1038/ncomms15874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- So WK, Cheung TH. 2018. Molecular regulation of cellular quiescence: a perspective from adult stem cells and its niches. Methods Mol Biol 1686: 1–25. 10.1007/978-1-4939-7371-2_1 [DOI] [PubMed] [Google Scholar]
- Steiman-Shimony A, Shtrikman O, Margalit H. 2018. Assessing the functional association of intronic miRNAs with their host genes. RNA 24: 991–1004. 10.1261/rna.064386.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su W, Feng S, Chen X, Yang X, Mao R, Guo C, Wang Z, Thomas DG, Lin J, Reddy RM, et al. 2018. Silencing of long noncoding RNA MIR22HG triggers cell survival/death signaling via oncogenes YBX1, MET, and p21 in lung cancer. Cancer Res 78: 3207–3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh EJ, Remillard MY, Legesse-Miller A, Johnson EL, Lemons JM, Chapman TR, Forman JJ, Kojima M, Silberman ES, Coller HA. 2012. A microRNA network regulates proliferative timing and extracellular matrix synthesis during cellular quiescence in fibroblasts. Genome Biol 13: R121 10.1186/gb-2012-13-12-r121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q, Hao Q, Prasanth KV. 2018a. Nuclear long noncoding RNAs: key regulators of gene expression. Trends Genet 34: 142–157. 10.1016/j.tig.2017.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q, Tripathi V, Yoon JH, Singh DK, Hao Q, Min KW, Davila S, Zealy RW, Li XL, Polycarpou-Schwarz M, et al. 2018b. MIR100 host gene-encoded lncRNAs regulate cell cycle by modulating the interaction between HuR and its target mRNAs. Nucleic Acids Res 46: 10405–10416. 10.1093/nar/gky696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun T, Du SY, Armenia J, Qu F, Fan J, Wang X, Fei T, Komura K, Liu SX, Lee GM, et al. 2018c. Expression of lncRNA MIR222HG co-transcribed from the miR-221/222 gene promoter facilitates the development of castration-resistant prostate cancer. Oncogenesis 7: 30 10.1038/s41389-018-0039-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng YY, Moriarity BS, Gong W, Akiyama R, Tiwari A, Kawakami H, Ronning P, Reuland B, Guenther K, Beadnell TC, et al. 2014. PVT1 dependence in cancer with MYC copy-number increase. Nature 512: 82–86. 10.1038/nature13311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valcourt JR, Lemons JM, Haley EM, Kojima M, Demuren OO, Coller HA. 2012. Staying alive: metabolic adaptations to quiescence. Cell Cycle 11: 1680–1696. 10.4161/cc.19879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatraman A, He XC, Thorvaldsen JL, Sugimura R, Perry JM, Tao F, Zhao M, Christenson MK, Sanchez R, Yu JY, et al. 2013. Maternal imprinting at the H19-Igf2 locus maintains adult haematopoietic stem cell quiescence. Nature 500: 345–349. 10.1038/nature12303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KC, Chang HY. 2011. Molecular mechanisms of long noncoding RNAs. Mol Cell 43: 904–914. 10.1016/j.molcel.2011.08.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Sato T, Amano T, Kawamura Y, Kawamura N, Kawaguchi H, Yamashita N, Kurihara H, Nakaoka T. 2008. Dnm3os, a non-coding RNA, is required for normal growth and skeletal development in mice. Dev Dyn 237: 3738–3748. 10.1002/dvdy.21787 [DOI] [PubMed] [Google Scholar]
- Westholm JO, Lai EC. 2011. Mirtrons: microRNA biogenesis via splicing. Biochimie 93: 1897–1904. 10.1016/j.biochi.2011.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Sun S, Tu K, Gao Y, Xie B, Krainer AR, Zhu J. 2010. A splicing-independent function of SF2/ASF in microRNA processing. Mol Cell 38: 67–77. 10.1016/j.molcel.2010.02.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CL, Wang Y, Jin B, Chen H, Xie BS, Mao ZB. 2015. Senescence-associated long non-coding RNA (SALNR) delays oncogene-induced senescence through NF90 regulation. J Biol Chem 290: 30175–30192. 10.1074/jbc.M115.661785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu TH, Shi L, Adrian J, Shi M, Nair RV, Snyder MP, Kao PN. 2018. NF90/ILF3 is a transcription factor that promotes proliferation over differentiation by hierarchical regulation in K562 erythroleukemia cells. PLoS ONE 13: e0193126 10.1371/journal.pone.0193126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu TH, Shi L, Lowe AW, Nicolls MR, Kao PN. 2019. Inducible expression of immediate early genes is regulated through dynamic chromatin association by NF45/ILF2 and NF90/NF110/ILF3. PLoS ONE 14: e0216042 10.1371/journal.pone.0216042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Meng Q, Li X, Yang H, Xu J, Gao N, Sun H, Wu S, Familiari G, Relucenti M, et al. 2019. Long noncoding RNA MIR17HG promotes colorectal cancer progression via miR-17-5p. Cancer Res 79: 4882–4895. 10.1158/0008-5472.CAN-18-3880 [DOI] [PubMed] [Google Scholar]
- Yao G. 2014. Modelling mammalian cellular quiescence. Interface Focus 4: 20130074 10.1098/rsfs.2013.0074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G, Wang LG, Han Y, He QY. 2012. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16: 284–287. 10.1089/omi.2011.0118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J, Tan L, Yin Z, Zhu W, Tao K, Wang G, Shi W, Gao J. 2019. MIR17HG-miR-18a/19a axis, regulated by interferon regulatory factor-1, promotes gastric cancer metastasis via Wnt/β-catenin signalling. Cell Death Dis 10: 454 10.1038/s41419-019-1685-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusuf I, Fruman DA. 2003. Regulation of quiescence in lymphocytes. Trends Immunol 24: 380–386. 10.1016/S1471-4906(03)00141-8 [DOI] [PubMed] [Google Scholar]
- Ziegler C, Kretz M. 2017. The more the Merrier—complexity in long non-coding RNA loci. Front Endocrinol (Lausanne) 8: 90 10.3389/fendo.2017.00090 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.