Abstract
The human PUF-family proteins, PUM1 and PUM2, posttranscriptionally regulate gene expression by binding to a PUM recognition element (PRE) in the 3′-UTR of target mRNAs. Hundreds of PUM1/2 targets have been identified from changes in steady-state RNA levels; however, prior studies could not differentiate between the contributions of changes in transcription and RNA decay rates. We applied metabolic labeling to measure changes in RNA turnover in response to depletion of PUM1/2, showing that human PUM proteins regulate expression almost exclusively by changing RNA stability. We also applied an in vitro selection workflow to precisely identify the binding preferences of PUM1 and PUM2. By integrating our results with prior knowledge, we developed a “rulebook” of key contextual features that differentiate functional versus nonfunctional PREs, allowing us to train machine learning models that accurately predict the functional regulation of RNA targets by the human PUM proteins.
Keywords: RNA decay, Pumilio, machine learning
INTRODUCTION
The control of gene expression at the posttranscriptional level is critical for diverse biological processes including proper organismal development in multicellular organisms. Many regulators, including RNA-binding proteins (RBPs), control the stability of target mRNA transcripts through the recognition of key sequence elements in the 3′-UTRs of mRNAs (Wickens et al. 2002; Jonas and Izaurralde 2015). A recent survey of all known human RBPs indicated that a substantial fraction bind to mRNAs; however, for any given RBP, the binding specificity, set of mRNA targets, and functional role for the RBP at each target still remains poorly understood (Gerstberger et al. 2014).
The PUF (Pumilio and FBF [fem-3 binding factor]) family of proteins represent a well-studied class of RBPs (Wickens et al. 2002; Miller and Olivas 2011; Goldstrohm et al. 2018). PUF proteins possess a shared carboxy-terminal Pum homology domain (PUM-HD). Structurally, the human PUM-HD consists of eight helical repeats containing specific amino acids that both intercalate into and form hydrogen bonds and van der Waals contacts with the nucleobases of the target RNA, conferring specificity for a UGUANAUA consensus sequence, referred to as a PUM recognition element (PRE) (Wang et al. 2001, 2002). In general, RNA recognition by the PUM-HD is modular, predictable, and programmable (Wang et al. 2002; Dong et al. 2011; Campbell et al. 2014). Furthermore, interactions with protein partners can also alter sequence preference (Campbell et al. 2012; Weidmann et al. 2016; Qiu et al. 2019).
Functionally, the PUF family of proteins controls stem cell fate and developmental processes (Wickens et al. 2002). Mammalian PUM proteins have roles in regulating spermatogenesis and oogenesis (Fox et al. 2005; Chen et al. 2012), neuronal development and function (Vessey et al. 2010; Siemen et al. 2011; Gennarino et al. 2015; Zhang et al. 2017; Dong et al. 2018; Zahr et al. 2018), immune function (Narita et al. 2014; Brocard et al. 2018), and cancer (Kedde et al. 2010; Lee et al. 2016; Naudin et al. 2017; Tichon et al. 2018). PUM1 missense or deletion mutants lead to adult-onset ataxia (Pumilio1-related cerebellar ataxia [PRCA]), and loss of one copy leads to developmental delay and seizures (Pumilio1-associated developmental disability, ataxia, and seizure [PADDAS]) (Gennarino et al. 2018). Yet, the target mRNAs responsible for these biological outcomes are largely opaque. In humans, there are two members of the PUF family, PUM1 and PUM2, which share 75% overall sequence identity with 91% sequence identity in the PUM-HD. In addition, human PUM1 and PUM2 share 78% and 79% sequence identity in the PUM-HD to DmPum, respectively (Spassov and Jurecic 2002; Goldstrohm et al. 2018). Human PUM1 and PUM2 are expressed across tissues and their expression is highly overlapping (Spassov and Jurecic 2002; Goldstrohm et al. 2018). However, the presence of distinct phenotypes from PUM1 and PUM2 loss of function indicates likely functional differences, either in RNA binding and/or downstream effects of the human PUM proteins, which remain to be explored.
Targeted experiments have indicated that human PUM1 and PUM2 are capable of repressing expression through recognition of PREs in a reporter mRNA's 3′-UTR, likely through recruitment of the CCR4–NOT complex and subsequent degradation of the mRNA target (Van Etten et al. 2012). Additionally, similar assays have shown that repression by the human PUM2 PUM-HD alone—that is lacking the amino-terminal domains of PUM2—requires the poly(A) binding protein PABPC1, suggesting that the human PUMs could accelerate mRNA degradation by inhibiting translation (Weidmann et al. 2014). However, the importance of PUM-mediated decay relative to other potential contributing factors to regulation in vivo has still not been demonstrated. In addition, PUM-mediated repression is not the only type of gene regulation by human Pumilio proteins. Recently, expression of a key regulator of hematopoietic stem cell differentiation, FOXP1, was shown to be enhanced by human PUM1/2 binding to the 3′-UTR (Naudin et al. 2017). Similar increases in transcript abundance of PUM targets have also been observed transcriptome-wide (Bohn et al. 2018), yet the mechanism of PUM-mediated activation remains to be elucidated.
High-throughput measurements of PUM1 and PUM2 binding sites in vivo have confirmed specificity for a PRE and have identified a diverse set of PUM targets in human cell lines (Galgano et al. 2008; Morris et al. 2008; Hafner et al. 2010; Van Nostrand et al. 2016). Thus, sequence-specific recognition of the PRE is an important aspect of target recognition for the PUM proteins. However, key questions about PUM-mediated gene regulation remain. At the sequence level, there are on the order of 10,000 potential PRE sites across the full set of annotated human 3′-UTRs, but only ∼1000 genes exhibit changes in steady-state RNA levels in response to depletion of PUM1 and PUM2 (Bohn et al. 2018). Additionally, models using a simple count of PREs in the 3′-UTR of a transcript do not completely capture the complexity of PUM-mediated gene regulation (Bohn et al. 2018). The identification of additional sequence features that discriminate functional PREs from apparently nonfunctional PREs will improve the understanding of PUM-mediated gene regulation. Furthermore, the measurement of steady-state RNA levels does not allow for differentiation between the individual contributions of transcription and RNA stability, and thus the relative importance of PUM effects on stability versus transcription genome-wide remain unknown.
Through the use of genome-wide sequencing methodologies, we set out to answer two key questions in a consistent framework: First, what are the biochemical RNA binding activities of PUM1 and PUM2 and how do they differ; and second, what are the primary mechanisms through which PUM1/2 exert regulatory effects in vivo? We demonstrate that human PUM1/2 modulate the abundance of mRNA targets primarily through controlling mRNA stability and not transcription. We further show, through side-by-side high-throughput in vitro binding assays, that PUM1 and PUM2 PUM-HDs have highly similar preferences for the same sets of sequences, but that the proteins differ in their stringency of recognition for perfect vs. near-perfect target site matches. In addition, we identify a key set of contextual features around PREs that contribute meaningful information in predicting PUM-mediated regulation including proximity to the 3′ end of a transcript and the AU content around PRE sites. Taken together, our study illuminates key contributors to determining functional PRE sites and represents a rich resource for interrogating the control of mRNA stability by the PUM RBPs.
RESULTS
Global analysis of PUM-mediated regulation of mRNA stability
The effects of PUM1 and PUM2 on mRNA stability have not been measured on a transcriptome-wide scale; thus, we applied a bio-orthogonal labeling strategy coupled with RNAi depletion of the PUMs. Previously, we measured changes in steady-state RNA abundance after PUM depletion using RNA-seq (Bohn et al. 2018). A limitation of that approach is that it could not disentangle changes in transcription from changes in RNA stability. Given that targeted experiments have indicated that the human PUM proteins act to control gene expression through the modulation of RNA stability (Morris et al. 2008; Van Etten et al. 2012), we sought to determine whether this is the primary mode of PUM action at a transcriptome-wide scale using the Bru-seq and BruChase-seq methodology (Paulsen et al. 2014). In brief, Bru-seq and BruChase-seq involve the metabolic labeling of RNA using 5-bromouridine (BrU), which is readily taken up by the cells and incorporated into the nascent NTP pool (Ohtsu et al. 2008). After incubation with BrU over a short time period, newly synthesized and labeled RNAs are selectively purified from total RNA using an anti-BrdU antibody and sequenced (Bru-seq). Labeled RNA abundance is then tracked over time by chasing with high concentrations of uridine in the absence of BrU and isolating BrU-labeled RNA at additional time points (BruChase-seq).
To distinguish relative changes in transcription from changes in RNA stability between WT and PUM1/2 knockdown cells, we combined our Bru-seq and BruChase-seq results measured over two time points. A 0 h time point was taken at the transition to unlabeled media after 30 min of incubation in BrU-containing media, and a 6-h chase time point taken to coincide with the mean mRNA half-life in cultured mammalian cells (Yang et al. 2003; Sharova et al. 2009; Schwanhäusser et al. 2011; Lugowski et al. 2018). Post hoc analysis shows that our approach permitted the detection of PUM-mediated effects on mRNA stability over a wide range of mRNA half-lives (Supplemental Fig. S1A).
To determine the impact of PUM1/2 on relative RNA abundances, the experiment was performed in the presence of a mix of siRNAs targeting both PUM1 and PUM2 mRNAs (siPUM) or in the presence of scrambled nontargeting control siRNAs (NTC), as previously established (Fig. 1A; Van Etten et al. 2012; Bohn et al. 2018). Cells were treated with siRNAs for 48 h before BrU labeling, identical to the method used in Bohn et al. (2018). These previously validated siRNAs have high specificity for either PUM1 or PUM2 transcripts and do not appear to have off-target effects on nontarget transcripts (Supplemental Fig. S1B). Overall, four biological replicate samples were collected for each time point and RNAi condition resulting in a total of 16 samples and above the minimum recommendations for replicates suggested by the ENCODE consortium for RNA-seq and ChIP-seq experiments (ENCODE Project Consortium 2012; Landt et al. 2012). HEK293 cells were chosen for this study as they express both PUM1 and PUM2, have been previously used to analyze PUM activity (Van Etten et al. 2012; Bohn et al. 2018), support efficient BrU-labeling (Tani et al. 2012), and support robust RNA interference (Chang et al. 2012). As we have previously demonstrated (Van Etten et al. 2012; Bohn et al. 2018), knockdown of both PUM1 and PUM2 is necessary to alleviate PUM repression of PRE-containing mRNAs. The use of two time points permits measurements of relative changes in mRNA stability and relative changes in nascent mRNA abundance between the two conditions, but does not allow for determination of decay rate constants for each transcript. An in-depth discussion on the tradeoffs and interpretation of measuring global RNA decay with minimal time points is given in Wolfe et al. (2018).
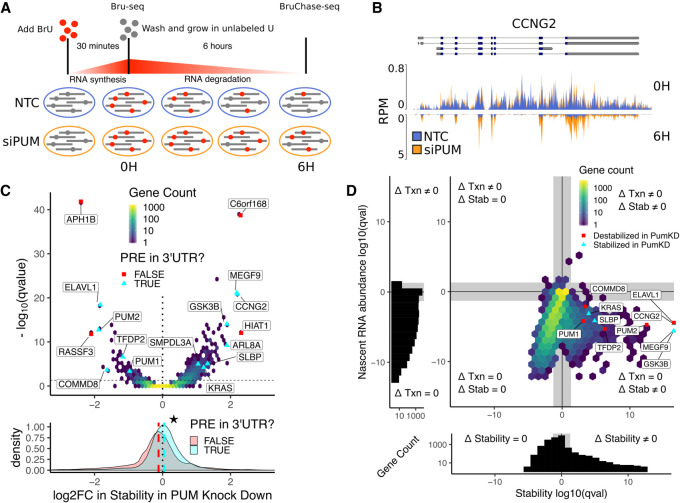
FIGURE 1.
Bru-seq and BruChase-seq allow for determination of PUM-mediated effects on RNA stability. (A) Experimental design for measuring PUM-mediated effects on RNA stability. HEK293 cells incubated for 30 min in the presence of 2 mM BrU prior to time 0. Cells were then washed and cultured in media containing 20 mM unlabeled uridine for 6 h. At 0 h (Bru-seq) and 6 h (BruChase-seq) timepoints, a portion of cells were harvested and BrU labeled RNA was isolated for sequencing. Changes in relative RNA abundance between the 0 and 6 h time points were compared between cells grown in the presence of silencing RNA targeting PUM1 and PUM2 (siPUM) and a nontargeting control siRNA (NTC). Cells were treated with siRNAs for 48 h prior to BrU labeling to allow for PUM depletion. (B) Read coverage traces for CCNG2 as measured in reads per million (RPM) at a resolution of 30 bp over the region shown (chr4:78077800–78092000, hg19). Traces are shown for siPUM (orange) and NTC (blue) conditions at both 0 h (top) and 6 h (inverted bottom) time points. Four replicates for each combination of siRNA and time point are overlaid. Known isoforms for CCNG2 are represented above. (C, top) Volcano hexbin plot displaying global changes in RNA stability under PUM knockdown conditions. The log2 fold change in stability under PUM knockdown conditions compared to the siNTC control after controlling for batch effects is displayed here, where positive values indicate stabilization upon PUM knockdown and negative values indicate destabilization upon PUM knockdown (see Materials and Methods for details). No change in stability is represented with a dotted line at 0. Statistical significance is displayed on the y-axis as the −log10 (FDR-corrected P-value) where larger values indicate a smaller P-value. An FDR-corrected P-value <0.05 is represented with a horizontal dashed line. A selection of genes known to be regulated by PUM (Morris et al. 2008; Bohn et al. 2018) and genes newly identified in this study are labeled. For selected genes only, cyan triangles indicate genes that have a PRE in any annotated 3′-UTR as determined by a match to the PUM1 motif we identified using SEQRS (Fig. 2A). Red squares indicate genes that did not have a PRE in their 3′-UTR. Unlabeled genes are binned into a two-dimensional histogram to avoid overplotting. (Bottom) Marginal distribution of log2FC in stability in PUM knockdown for genes with a PRE in their 3′-UTR (cyan) and genes without a PRE in their 3′-UTR (red). Median values for each distribution are plotted as a dashed line in the appropriate color. The star indicates a statistically significant difference in the median stability as measured by a two-sided permutation of shuffled labels (n = 1000, P < 0.001). (D) Analysis of changes in nascent RNA abundance versus changes in stability. Four separate statistical tests were calculated for each gene: (i) a test for statistically significant changes in RNA stability (Δ Stability ≠ 0), (ii) a test for statistically significant changes in nascent RNA abundance (Δ Txn ≠ 0), (iii) a test for no change in RNA stability (Δ Stability = 0), and (iv) a test for no change in nascent RNA abundance (Δ Txn = 0). Genes are plotted as an (x,y)-coordinate where each coordinate represents the ±log10 (FDR-corrected P-value) of the test with greater evidence (Δ ≠ 0, +log10; or Δ = 0, −log10) for each axis (see Materials and Methods for details). Representative genes displaying a range of stability effects are labeled. Red squares represent genes that were destabilized in PUM knockdown, whereas cyan triangles represent genes that were stabilized in PUM knockdown. All other genes were binned into a two-dimensional histogram. Gray rectangles represent a statistical significance cutoff of Q-value >0.05. (Left, below) Marginal histograms for each axis are plotted with matching gray rectangles to represent the same statistical significance cutoff of Q-value >0.05.
Clear changes in RNA abundance can be seen between time points and conditions at the gene level. Consider the Cyclin G2 (CCNG2) mRNA, which encodes a cyclin involved in the cell cycle, contains two PREs in its 3′-UTR, and was among the most dramatically affected mRNAs (Fig. 1B). At the 0 h time point, read coverage resulting from recent transcription for four distinct replicates in each condition can be seen with the trace for each replicate transparently overlaid (n.b. read coverage includes immature RNAs that still contain introns) (Fig. 1B, top, nonoverlaid tracks can be found in Supplemental Fig. S2A). At the 6 h time point, only mature RNA remains, with read coverage primarily observed at exons and no longer prevalent in the intronic regions (Fig. 1B, bottom). Here, silencing of both PUM1 and PUM2 clearly increases RNA abundance relative to the nontargeting control at the 6 h time point, but does not appear to impact transcription as seen at the 0 h time point.
To quantify the effect of silencing PUM1 and PUM2 on changes in relative labeled RNA abundance between the 0 and 6 h time points, we used DESeq2 (Love et al. 2014) to model the count of reads observed from each gene using a generalized linear model that allows us to separate the effects of PUM knockdown on RNA stability from those on nascent RNA abundance (transcription), and to account for any batch effects between replicates (Supplemental Fig. S2B) and changes in RNA stability not associated with the PUM knockdown treatment (see Materials and Methods for details). Using this methodology, we find that hundreds of genes show altered RNA stability under PUM knockdown conditions. Figure 1C displays an overview of PUM-mediated effects on stability as a volcano plot, with 10,132 genes represented in a two-dimensional histogram. Using an FDR-corrected P-value threshold of 0.05 and a fold-change cutoff of log2(1.75) (see Materials and Methods), we found 60 genes were statistically significantly destabilized (88 with no fold-change cutoff) and 248 genes were statistically significantly stabilized in the PUM knockdown condition (406 with no fold-change cutoff). Of these destabilized genes, 31 were also identified as having lower abundance under PUM knockdown in the Bohn et al. (2018) RNA-seq data set (46 with no fold-change cutoff). Likewise, of the stabilized genes, 104 were also identified as having higher abundance under PUM knockdown in the Bohn et al. (2018) RNA-seq data set (135 with no fold-change cutoff). Thus, here we identify 29 new mRNAs that are stabilized by PUM and 144 new mRNAs that are destabilized by PUM as compared to Bohn et al. (2018). As expected, both PUM1 and PUM2 were substantially destabilized in the PUM knockdown condition relative to the WT condition, indicating that the siRNAs were successful in disrupting PUM1/2 expression and that our methodology is capable of detecting known changes in RNA stability. Additionally, we found that genes with a PRE in their 3′-UTR were, on average, more stabilized in the PUM knockdown condition than those without a PRE in their 3′-UTR (Fig. 1C, bottom). Taken together, this data identifies hundreds of new PUM-regulated genes and provides broad evidence that PUM1/2 selectively modulate the stability of specific target transcripts.
To further dissect the effects of PUM knockdown on transcripts in our data set, we tested for statistically significant changes for both nascent RNA abundance and RNA stability for each gene under a null model centered around a log2 fold change of 0. In addition, we strove to identify genes for which we had robust evidence that no substantial changes were occurring under PUM knockdown conditions, at the level of nascent RNA abundance or RNA stability, by considering a null model centered around the boundary of a defined region of practical equivalence spanning from −log2(1.75) to log2(1.75) for each metric (see Materials and Methods for details); such a test is important because failure to reject the null hypothesis cannot, by itself, be taken as evidence favoring the alternative. Rejection of the null hypothesis in our practical equivalence test, however, permits us to confidently state that the abundance of a given transcript is not meaningfully changed. In total, four statistical tests were run for each gene: a test for change and a test for no change for both nascent RNA abundance and RNA stability. For each axis of Figure 1D, the smaller of the two FDR-corrected P-values (i.e., test for change vs. test for no change) was chosen as the coordinate for that term, which enabled classification of each gene into one of four quadrants: (i) genes that change in both stability and nascent RNA abundance (Fig. 1D, upper right quadrant), (ii) genes that change only in stability (Fig. 1D, lower right quadrant), (iii) genes that change only in nascent RNA abundance (Fig. 1D, upper left quadrant) and (iv) genes that change in neither stability nor nascent RNA abundance (Fig. 1D, lower left quadrant). Thus, using this methodology, we identified 308 genes with a statistically significant change in stability (Fig. 1D, upper and lower right quadrants). We were also able to identify a set of 5503 genes with evidence for no change in stability under our experimental conditions, that is, we are confident that PUM knockdown is having no effect on the stability of these genes (Fig. 1D, upper and lower left quadrants). Finally, we find that we have insufficient information to reliably classify 14,606 genes due to a failure to reject the null on either a test of change or test of no change in RNA stability under PUM knockdown conditions. At the level of nascent RNA abundance, we show only four genes (ETV1, C1S, ETV5, ANKRD30B), with a statistically significant change under PUM knockdown conditions (105 with no fold change cutoff) (Fig. 1D, right and left upper quadrants). Additionally, we find 12,245 genes with a statistically significant lack of change in nascent RNA abundance, that is, we are confident that PUM knockdown is having no effect on the nascent RNA abundance of these genes (Fig. 1D, right and left lower quadrants). Finally, we found that 10,542 genes have insufficient information to reliably classify due to a failure to reject the null on either a test of change or a test of no change in nascent RNA abundance under PUM knockdown conditions. Collectively, our results show that PUM meaningfully modulates RNA stability, with little or no effect on transcription. Throughout the remainder of the paper, we will use the words EFFECT and NOEFFECT to refer to genes for which PUM knockdown has a significant effect on RNA stability and those for which our testing indicated no practical change in RNA stability upon PUM knockdown, respectively.
High-definition specificity of PUM1 and PUM2
The sequence preferences of PUM proteins have been analyzed using a variety of approaches. RNA binding of full-length mammalian PUM1 and PUM2 to mRNAs has been previously probed in vivo (Galgano et al. 2008; Hafner et al. 2010; Van Nostrand et al. 2016; Sternburg et al. 2018), whereas the sequence preferences of their RNA-binding domains were probed in vitro (Campbell et al. 2012; Ray et al. 2013; Dominguez et al. 2018; Jarmoskaite et al. 2019). A general preference for the PRE consensus motif UGUANAUA has emerged, with subtle differences in the information content for the position weight matrices (PWMs) obtained from each technique, particularly at the 3′ end of the PWM. However, prior in vitro determination of human PUM sequence preferences have involved only one round of selection (Dominguez et al. 2018) or a selected subset of possible sequences (Jarmoskaite et al. 2019), yielding a limited and potentially biased view of PUM1 and PUM2 binding specificity. In vivo analyses of PUM1 and PUM2 RNA binding have reported only partially overlapping sets of bound RNAs, including many that have no identifiable consensus binding elements (Galgano et al. 2008; Hafner et al. 2010). These observations suggest the possibility that we do not fully know the RNA binding specificity of PUM1 and PUM2, indicating a need for unbiased approaches to profile the specificities of PUM1 and PUM2 for both high and low affinity sites. We applied in vitro selection and high-throughput sequencing of RNA and sequence specificity landscapes (SEQRS) to purified PUM-HDs of each protein (Lou et al. 2017). SEQRS allows for the determination of an RNA-binding protein's sequence specificity by selecting for RNAs that interact with the RBP out of a pool of randomized 20-mers generated by T7 transcription of a synthesized DNA library. The RNA pulled down from a previous round is reverse-transcribed into DNA to be used as the input for the next round of transcription and selection, allowing for exponential enrichment of preferred sequences that are then identified via sequencing. There are several key advantages to this approach over other in vitro methods including an unbiased starting pool, the use of 20-mers to probe a large sequence space, and multiple rounds of selection that allow for detection of specificity for a range of affinities. We performed five rounds of SEQRS to PUM1 or PUM2 and quantified the abundance of each of the 65,536 possible 8-mers for each round (including those that would overlap with the adjacent static adapter sequences; see Materials and Methods for details).
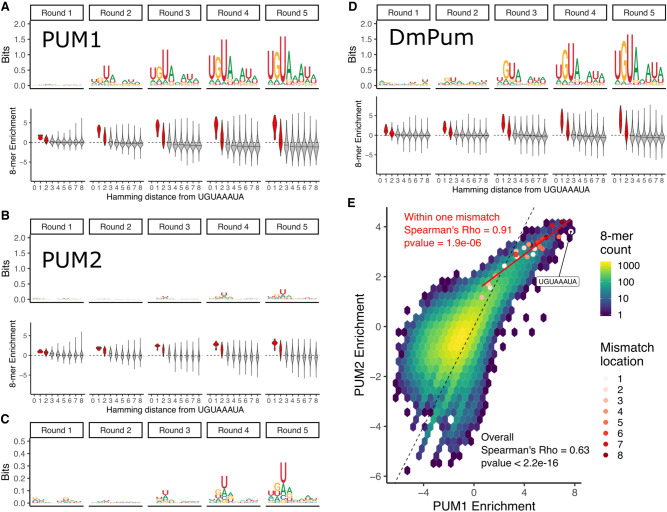
To obtain representative PWMs for each round of selection (Fig. 2A,B, top), we used the top enriched 8-mer, UGUAAAUA, as a seed sequence to create a multinomial model from the abundance of every possible single mismatched 8-mer to the seed sequence (see Materials and Methods for details). This data analysis approach has yielded similar results to that of expectation-maximization algorithms such as MEME (Bailey et al. 2006) and has been used successfully with SELEX experiments using DNA-binding proteins (Jolma et al. 2010, 2013). For comparison, we applied the same pipeline to previously published SEQRS data for the D. melanogaster Pumilio PUM-HD (Weidmann et al. 2016; Lou et al. 2017) and find that it readily captures the Pumilio sequence preference for the UGUANAUA PRE (Fig. 2D, top). Importantly, the PWMs defined here (Fig. 2A,B,D, top panels) are representative of only the most highly enriched sequences in each data set and round.
FIGURE 2.
SEQRS analysis of Human PUM1 and PUM2 PUM-HDs reveals preference for the canonical PUM recognition element. (A, top) Position weight matrices representing 8-mer sequence preferences for purified Human PUM1 PUM-HD, as determined for each SEQRS round. (Bottom) 8-mer enrichment, as measured by log2(Enrichment SEQRS round/Enrichment no protein) (see Materials and Methods for details) for each 8-mer as binned by Hamming distance from the canonical UGUAAAUA PUM recognition element. Enrichment scores for 8-mers within two mismatches are filled in red. (B) Same as in A, but for Human PUM2 PUM-HD. (C) Closer view of Human PUM2 PUM-HD PWMs. (D) Same as in A, but for Drosophila Pum PUM-HD. (E) Correlation of 8-mer enrichment between Human PUM1 and Human PUM2 PUM-HDs. Enrichment for all possible 8-mers are displayed in a two-dimensional histogram. The dashed black line represents one-to-one correspondence. All 8-mers within one mismatch to the UGUAAAUA sequence are plotted as red points with the color specifying the position within the motif where the mismatch occurs. The red line is a linear fit using only the UGUAAAUA 8-mer and all 8-mers within one mismatch.
To determine how representative the UGUANAUA consensus motif is for the entire data set of each protein, we grouped each 8-mer based on its distance from the UGUAAAUA seed sequence, and then considered the relative enrichment of a given 8-mer within each round. Scores above 0 indicate higher relative abundance relative to the input pool, and scores below 0 indicate lower relative abundance. We find that 8-mers within one to two mismatches of the UGUAAAUA seed sequence are highly enriched compared to 8-mers with more than two mismatches across each round for each protein (Fig. 2A,B,D, bottom).
The PWM obtained from SEQRS experiments for the PUM2 PUM-HD (Fig. 2B,C) suggests that PUM2 has weaker enrichment for the canonical PUM PRE compared to PUM1, whereas sequence preferences obtained from in vivo transcriptome-wide experiments were nearly indistinguishable (Galgano et al. 2008; Hafner et al. 2010). This may indicate differences between in vitro and in vivo conditions that specifically impact PUM2, that PUM2 PUM-HD does not bind as efficiently to RNA as the full-length PUM2 protein, or that additional protein partners mediate the downstream effects of PUM1 and PUM2 differently in vivo. However, comparing PWMs between these two proteins only considers the most highly enriched sequences in each data set. As seen in Figure 2C, the consensus motif emerging from the PUM2 SEQRS data strongly resembles those for other PUMs, albeit with less apparent stringency.
To compare the overall sequence preferences between PUM1 and PUM2, we plotted the enrichment scores for all possible 8-mers in each data set against each other (Fig. 2E). We find that the 8-mer enrichment scores between these two proteins are highly correlated (Spearman ρ = 0.63), which indicates that PUM1 and PUM2 PUM-HDs have overall similar sequence preferences when considering all possible sequences rather than only highly enriched sequences. PUM1 has an overall stronger enrichment for highly enriched sequences compared to PUM2, whereas PUM2 shows a larger dynamic range for nonideal targets, which may explain the differences in obtained PWMs for each protein. When considering only the 8-mers within one mismatch to the UGUAAAUA seed sequence used for creating the PWMs, we find that enrichment scores between PUM1 and PUM2 are nearly perfectly correlated (Spearman's ρ = 0.91). Furthermore, mismatches in the 3′ end of the motif appear to be less detrimental to enrichment by PUM1 and PUM2 compared to mismatches in the 5′ end of the motif, which is also represented by the lower information content at the 3′ end of the PWMs. Taken together, the SEQRS data and analysis provide a precise definition of the PUM1 and PUM2 PREs, demonstrating that PUM1 and PUM2 exhibit highly correlated specificities, but with differences in apparent binding affinities for nonideal targets. Below, we utilize this information to explore additional determinants of PUM regulation in target mRNAs and to develop predictive models. Due to the overall similarity in sequence preferences between these two proteins and the higher overall information content for PUM1, the SEQRS round 5 PWM for PUM1 will be used to define PREs throughout the text, unless otherwise indicated.
Contextual features around PREs are associated with PUM-mediated RNA stability effects
Determining what distinguishes a functional binding site from a nonfunctional binding site is a major question for any RBP. Taken as a whole, RBPs tend to bind similar low-sequence complexity motifs in vitro (Dominguez et al. 2018). Additionally, probing of RBP binding in vivo at a transcriptome-wide scale has indicated that the majority of predicted binding sites are not bound for some RBPs (Taliaferro et al. 2016). Global in vivo experiments with the Pumilio family of proteins have established that mammalian Pumilio proteins recognize the UGUANAUA PRE in the 3′-UTR of target genes (Galgano et al. 2008; Hafner et al. 2010; Van Etten et al. 2012; Zhang et al. 2017). However, predicting the PUM-mediated effect on gene expression from sequence information and/or PUM-binding measurements remains an elusive goal (Bohn et al. 2018).
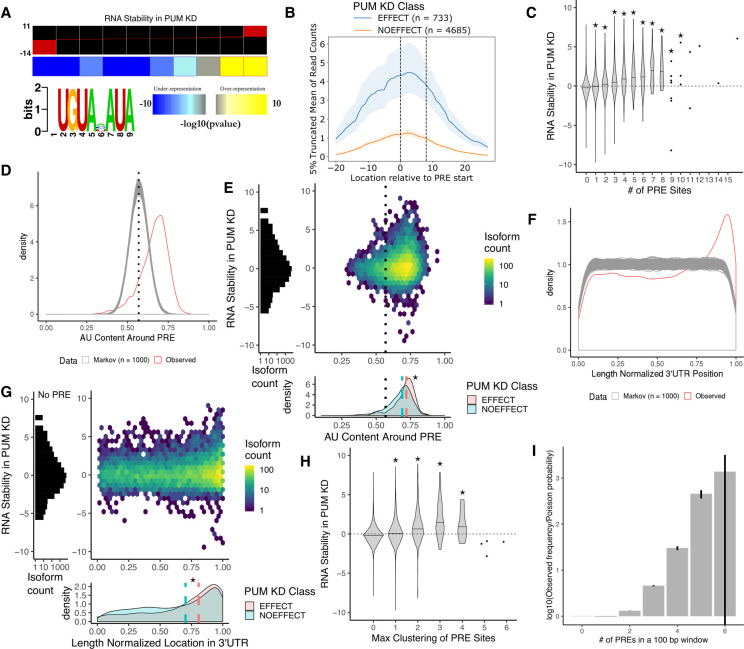
To determine sequence motifs de novo that have explanatory power for our RNA stability data set, we used FIRE (Elemento et al. 2007) to find motifs in the 3′-UTR of transcripts that share high mutual information with our RNA stability data set by taking the normalized interaction term (see Materials and Methods for details) and discretizing it into 10 bins, with an equal number of genes in each bin. Figure 3A shows that FIRE rediscovers the canonical UGUANAUA PRE using only the RNA stability data as input. Furthermore, the UGUANAUA PRE is enriched in transcripts that are highly stabilized under PUM knockdown conditions, indicating that these transcripts are directly regulated by PUM through recognition of a UGUANAUA PRE in their 3′-UTR.
FIGURE 3.
Features associated with a PUM recognition element (PRE) explain variability in PUM-mediated effect on decay. (A) Results of motif inference using FIRE (Elemento et al. 2007) on the stability in PUM knockdown data discretized into 10 equally populated bins. Red bars within each bin represent the spread of RNA stability values within each bin. Stability in PUM knockdown is represented by a normalized interaction term between time and condition throughout this figure, where positive values indicate stabilization upon PUM knockdown and negative values indicate destabilization upon PUM knockdown (see Materials and Methods for details). (B) Five percent truncated average of Pum2 PAR-CLIP read coverage (Hafner et al. 2010) over each PRE site in the 3′-UTRs of genes with a statistically significant change in RNA stability (blue) compared to genes in which there was a statistically significant lack of change in stability (orange; see Materials and Methods for details on NOEFFECT test). Shaded regions represent bootstrapping (n = 1000) within each group. Dashed lines indicate the PRE site. (C) Violin plots representing the distributions of RNA stability for genes with 0 to 15 PRE sites within their 3′-UTR. Stars represent statistical significance as measured by a Wilcoxon rank sum test using equality of pseudomedian with the 0 PRE case as the null hypothesis. (D) Distribution of AU content in a 100 bp window around all unique PRE sites in the 3′-UTRs of the human transcriptome. The observed distribution (red) is compared to the distribution of AU content around PRE sites in 1000 simulated sets of 3′-UTRs the same size as the true set of 3′-UTRs as simulated from a third-order Markov model trained on the true 3′-UTR sequences. The dotted line represents the average overall AU content of the entire set of 3′-UTRs in the human transcriptome. (E) Relationship of AU content in a 100 bp window around a PRE to RNA stability. (Left) Marginal histogram of RNA stability for genes with 0 PREs in their 3′-UTRs. (Right) 2D histogram of RNA stability and AU content around each PRE site for all genes with at least one PRE in the 3′-UTR. Dotted line represents the average AU content over the entire set of 3′-UTRs in the human transcriptome. (Bottom) Marginal kernel density plot of AU content around a PRE site split among genes with a statistically significant change in RNA stability (red) and genes with a statistically significant lack of change in stability (blue). Dotted black line represents the average AU content of 3′-UTRs. Dashed lines represent the median AU content around a PRE for the EFFECT (red) and NOEFFECT (blue) genes. The star represents a statistically significant difference in medians using a one-sided permutation test (n = 1000) of shuffled class labels. (F) Distribution of length-normalized locations of PRE sites in the 3′-UTRs of the human transcriptome. The observed distribution (red) is compared to that of PRE sites found in 1000 simulated sets of 3′-UTRs calculated as in D. (G) Relationship of normalized location of PRE site in 3′-UTR to RNA stability. Plots as in E. (H) Violin plots representing the distributions of RNA stability for genes with 0 to 6 full PRE sites clustered within a 100 bp window. Stars represent statistical significance as measured by a Wilcoxon rank sum test using the 0 PRE case as the null distribution. (I) Comparison of the observed frequencies of PRE site clustering over all possible 100 bp windows in the full set of human 3′-UTRs with at least one PRE in them to the probabilities expected from a Poisson null distribution. Error bars represent 95% confidence intervals based on 1000 bootstraps of the observed distribution.
To determine whether there was evidence for PUM binding at PREs associated with a change in RNA stability, we used publicly available in vivo binding data for human PUM2 obtained using photoactivatable ribonucleoside-enhanced cross-linking and immunoprecipitation (PAR-CLIP) (Hafner et al. 2010). The PAR-CLIP technique involves incorporation of 4-thiouracil (4sU) into the total cellular RNA pool allowing for efficient cross-linking of proteins that bind near an incorporated 4sU. Upon creation of sequencing libraries from PAR-CLIP samples, a T → C mutation is induced at the cross-linking site, which can be used as additional evidence for a protein binding. We used PAR-CLIP data from Hafner et al. (2010) to determine the amount of binding signal at PREs associated with transcripts that have a statistically significant change in RNA stability under PUM knockdown (EFFECT class, Fig. 1D) and compared it to transcripts with a statistically significant lack of change in RNA stability (NOEFFECT class, Fig. 1D). In Figure 3B, we report the average PAR-CLIP read coverage in a 40 bp window around PREs in the 3′-UTR of transcripts associated with the EFFECT and NOEFFECT classes. We use a 5% truncated mean to remove the impact of extreme outliers on the average coverage reported. To estimate a 95% confidence interval on the average coverage (shaded region), we performed bootstrapping (n = 1000) by sampling vectors of read coverage for individual PREs with replacement. Here, we clearly see that PREs in transcripts with a change in RNA stability have a higher binding signal than those with no change in RNA stability. This is consistent with higher overall PUM binding at PREs associated with changes in RNA stability but, as the PAR-CLIP signal is not normalized to RNA abundance, the possibility that these transcripts were simply more abundant under the PAR-CLIP conditions cannot be definitively ruled out.
Our analysis shows that PUM-mediated changes in RNA stability are associated with the presence of a 3′-UTR PRE and experimental evidence for in vivo PUM binding. However, knowledge of the presence or absence of a PRE in the 3′-UTR alone, or even the number of PREs, is not sufficient to predict the magnitude of PUM-mediated repression measured at the level of transcript abundance (Bohn et al. 2018). Here, we demonstrate that a similar level of variation can be seen in direct measurements of RNA stability. Figure 3C displays the overall distribution of RNA stability measurements for transcripts with increasing numbers of PREs in annotated 3′-UTRs. We find that an increase in the number of PREs is associated with an increase in RNA stability when PUMs are depleted. Even so, wide variations in RNA stability can be seen for each category. Thus, a simple count of PREs does not fully explain PUM-mediated action at a particular transcript, and other features likely play an important role in shaping the effect of PUM on each target.
The local sequence context around PREs is an important potential source for variations in their regulatory effects. We trained a third-order Markov model on the full set of unique annotated human (hg19) 3′-UTRs that were greater than 3 bp long (29,380 3′-UTRs). Using this Markov model, we simulated 1000 different sets of 29,380 3′-UTRs that were the same length and shared similar sequence composition to the set of true 3′-UTRs. We then searched for matching PREs in the simulated sets of 3′-UTRs and calculated the AU content in a 100 bp window around these PREs. On average, we discovered 12,200 matching PREs (standard deviation of 112) in simulated sets of 3′-UTRs compared to the 14,086 matching PREs in the annotated set of 3′-UTRs. We find that the true set of PREs have, on average, higher local AU content than PREs in simulated sets of 3′-UTRs (Fig. 3D). Additionally, in the simulated 3′-UTRs, the local AU content for PREs is centered around the average AU content for all 3′-UTRs, as would be expected if there was no selective pressure for PREs to occur in AU-rich areas of 3′-UTRs. This relationship is similar to the enrichment of PREs in AU-rich regions reported by Jiang et al. (2013). Importantly, we further show direct evidence that the local AU content surrounding a PRE is associated with a functional effect on PUM-mediated regulation.
To determine the relationship between local AU content and changes in RNA stability upon PUM knockdown, we plotted the AU content of a 100 bp window surrounding a PRE within a gene's 3′-UTR against the corresponding RNA stability measurement for that gene (Fig. 3E, top). For 3′-UTRs with more than one PRE, the PRE with the highest local AU content was considered. We find that large changes in RNA stability are associated with higher local AU content. Additionally, PREs in transcripts that had a statistically significant stability effect in PUM knockdown had higher local AU content compared to PREs in transcripts with no change in stability (P < 0.001, Fig. 3E, bottom). Together, these data indicate that local sequence context beyond the PREs plays a role in PUM regulatory function.
Previously proposed mechanisms of PUM-mediated control of RNA stability involve interaction with the CCR4–NOT complex and/or PABPs, both of which act at the 3′ end of mRNA transcripts to promote deadenylation or participate in translation initiation (Van Etten et al. 2012; Weidmann et al. 2014). Thus, the location of PUM binding sites within the 3′-UTR of target transcripts may play a role in determining PUM-mediated effects on stability by physically locating PUM near known coregulators. Using the Markov models described above, we determined the location of PREs within 3′-UTRs. As shown in Figure 3F, we see that the observed distribution of true PRE locations in length-normalized 3′-UTRs appear enriched toward the 3′ end of 3′-UTRs (red) as compared to PREs found within 1000 simulated sets of 3′-UTRs (gray). Again, this suggests a selective pressure for PRE sites to exist at the 3′ end of 3′-UTRs as compared to the uniform distribution of PREs found in simulated 3′-UTRs with similar sequence properties. This observation is consistent with Jiang et al. (2013) who reported enrichment toward the 3′ end for PREs in human 3′-UTRs compared to a shuffled PRE motif with preserved overall sequence content. While their analysis approach is complementary to ours, our statistical test allows for the exact identity of the PRE to remain intact, thereby maintaining a PRE-centric assessment rather than one based solely on the general sequence content within the motif. Uniquely, our analysis also incorporates mRNA stability measurements and shows that transcripts with a PRE toward the 3′ end of the 3′-UTR tend to have a larger RNA stability effect (Fig. 3G, center). Moreover, PREs in transcripts that had a statistically significant change in stability in PUM knockdown were, on average, closer to the 3′ end of the 3′-UTR than those with no change in RNA stability (P < 0.001, Fig. 3G, bottom), supporting a functional role for PRE location in the 3′-UTR of target transcripts.
Given that higher AU content around a PRE and the location of a PRE within the 3′-UTR are correlated both with PUM-mediated RNA decay and with each other, we assessed whether each was contributing independent information to predictions of PUM effects on RNA stability by fitting a logistic regression model to categorize PREs associated with EFFECT genes from PREs associated with NOEFFECT genes. We fitted three models, one using only AU content as a predictor, one using only the relative location of a PRE within the 3′-UTR as a predictor, and one using both AU content and the relative location of a PRE. We then compared the models using the Akaike information criterion (AIC) and the Bayesian information criterion (BIC), two measures used for selecting the most parsimonious model from a series of candidate models where the model with the lowest value is the most favored, with penalties applied for the inclusion of additional independent variables. Here, we find that including both variables is more favorable than a model using only the AU content as a predictor (ΔAIC = −6.26, ΔBIC = −0.16) or only the relative location as a predictor (ΔAIC = −30.55, ΔBIC = −24.45). These findings suggest that, despite the apparent correlation between AU content and PRE location, each feature still contributes meaningful and independent information to predicting PUM function.
High-throughput analysis of many human RBPs has indicated that some RBPs preferentially bind bipartite motifs, suggesting that clustering of RBP binding sites may contribute to binding specificity and subsequent function (Dominguez et al. 2018). To determine the relationship between PRE clustering and RNA stability in PUM knockdown, we discretized transcripts according to the maximum number of complete PREs that were within a sliding 100 bp window in the 3′-UTR of a transcript and plotted the distribution of RNA stability measurements for each cluster (Fig. 3H). Similar to the association with the number of PREs (Fig. 3C), we find that having more PREs clustered together is associated, on average, with a higher stabilization effect under PUM knockdown conditions. We also find that PREs tend to cluster together more than one would expect by chance by determining the divergence from a simple Poisson model (Fig. 3I, P < 0.001 for clusters 2–5; see Materials and Methods for details). Taken together, this analysis suggests that clustering of PREs occurs more often than expected by chance, and may facilitate PUM action on target transcripts. Taken together, our results connect multiple features (location, number, clustering, and the AU context of PREs) to PUM-mediated changes in RNA stability, providing new insights into the determinants of functional PUM output.
Pumilio proteins modulate the stability of genes involved in neural development, cell signaling, and gene regulation
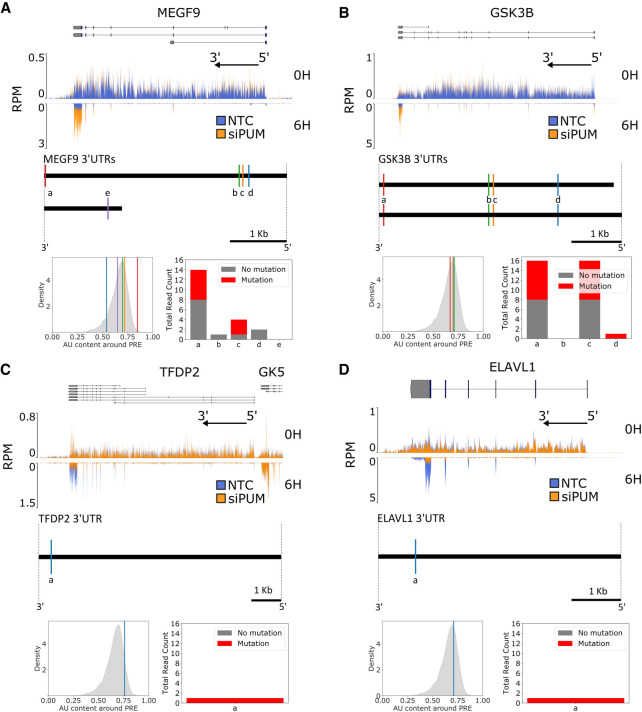
Mammalian Pumilio proteins have been shown to regulate the abundance of mRNAs from a diverse set of genes (Morris et al. 2008; Chen et al. 2012; Zhang et al. 2017; Bohn et al. 2018; Zahr et al. 2018). Given that our experiments were performed in HEK293 cells, which were derived from embryonic kidney and have neuronal and adrenal characteristics (Shaw et al. 2002), we would expect that many of the genes that are expressed in these cells would be associated with neuronal functions. Indeed, we found that PUM specifically modulates the RNA stability for genes involved in these functions. For example, we see strong stabilization of the MEGF9 transcript under PUM knockdown conditions (Fig. 4A, top). MEGF9 encodes a transmembrane protein that is highly expressed in the central and peripheral nervous system (Brandt-Bohne et al. 2007). Furthermore, of the five PREs we identify in two unique 3′-UTRs for MEGF9, we see the most PUM2 binding signal for the 3′-most PRE (Fig. 4A, bottom right), which also shows a high AU content compared to the overall distribution of PRE sites (Fig. 4A, bottom left). Taken together, these data implicate the PUM proteins as direct posttranscriptional regulators of MEGF9, acting through destabilization of the mRNA under native conditions.
FIGURE 4.
PUM-mediated effects on RNA stability under PUM knockdown include stabilization and destabilization. (A, top) Read coverage traces for MEGF9 and surrounding region (chr9:123348195–123491765, hg19) as measured in reads per million (RPM) at 100 bp resolution. Traces are shown for siPUM (orange) and NTC (blue) conditions at both 0H (upper track) and 6H (inverted lower track) time points. Four replicates for each combination of siRNA and time point are transparently overlaid. Known isoforms for MEGF9 are represented above. The black arrow indicates the direction of the 5′ and 3′ ends of the transcribed RNA molecule from the gene shown. Coverage plots were generated using pyGenomeTracks (Ramírez et al. 2018). N.b. in this and subsequent panels, the appearance of high density in the 3′-UTRs of the 6H samples is simply due to the presence of a small number of peaks that dominate the visualization when shown at the scale of the entire gene; the mean and median density in the 3′-UTR is in fact not substantively different from earlier exons. All sequencing data are available at GEO accession GSE145237. (Below) Diagram of unique MEGF9 3′-UTRs. Sites matching the PUM1 SEQRS motif are represented as vertical lines and labeled alphabetically from 3′ to 5′ for each UTR. (Below left) AU content of a 100 bp window around each PRE labeled above in the overall distribution of surrounding AU content for all PUM1 SEQRS motif matches in the entire set of 3′-UTRs. (Below right) PAR-CLIP read coverage (Hafner et al. 2010) of 40 bp around each indicated PRE. Number of reads with a T → C mutation are shown in red, whereas the number reads with no T → C mutation are shown in gray. (B) As in A, but for GSK3B and surrounding region (chr3:119509500–119848000). (C) As in A, but for TFDP2 and surrounding region (chr3:141630000–141900000). Annotations for the 3′ end of the GK5 gene are included due to their proximity to the TFDP2 5′ end. (D) As in A, but for ELAVL1 and surrounding region (chr19:8015000–8080000).
The GSK3B mRNA, which encodes a serine-threonine kinase that is associated with neurological disease, is strongly stabilized under PUM knockdown conditions (Fig. 4B, top; Jope and Johnson 2004; Jorge-Torres et al. 2018). GSK3B 3′-UTRs contain four PREs (Fig. 4B, below) with largely similar adjacent AU content (Fig. 4B, bottom left). The 3′ most distal PRE has evidence for PUM2 binding consistent with the global trends we describe in Figure 3. Thus, GSK3B provides another example of a direct target mRNA that is destabilized by PUMs.
Intriguingly, we also see examples of RNAs that are destabilized when PUM is knocked down, suggesting that PUM may normally act to stabilize these transcripts. As one example we highlight the TFDP2 mRNA, which encodes an E2F cofactor that is important in cell cycle progression and linked to cancer (Kent and Leone 2019). Our data show that TFDP2 is highly destabilized under PUM knockdown conditions (Fig. 4C, top). The TFDP2 mRNA has a single PRE site toward the 3′ end of the 3′-UTR and has high adjacent AU content (Fig. 4C, bottom and lower left), suggesting direct regulation by PUM, although the experimental evidence for PUM2 binding in PAR-CLIP data is limited (Fig. 4C, lower right).
Another example of a highly destabilized transcript under PUM knockdown conditions is the ELAVL1 mRNA, which encodes the HuR RNA-binding protein (Fig. 4D, top). The ELAVL1 RBP stabilizes RNA transcripts by binding to AU-rich elements in the 3′-UTR of transcripts (Lebedeva et al. 2011) and is dysregulated in several types of cancer (Wang et al. 2013). The 3′-UTR of ELAVL1 contains a PRE at the end of the 3′-UTR (Fig. 4D, bottom), consistent with regulatory potential. This data suggests that ELAV1 may be a direct target for PUM-mediated stabilization, though this conclusion is tempered by the average local AU enrichment of this PRE (Fig. 4D, lower left) and limited experimental evidence for PUM2 binding (Fig. 4D, lower right).
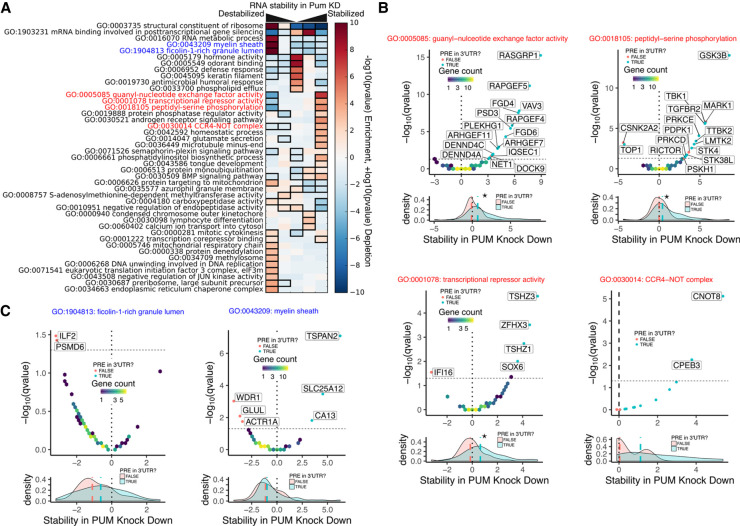
To discover categories of genes that are globally associated with RNA stability changes in PUM knockdown, we applied iPAGE—a computational tool that uses mutual information to find informative Gene Ontology (GO) terms associated with discretized gene expression data (Goodarzi et al. 2009)—to our stability data set as represented by the normalized interaction term discretized into five equally populated bins. This analysis will discover pathways regulated both indirectly and directly by PUM out of the full set of annotated GO terms. Figure 5A displays the iPAGE results with several GO terms that are either significantly overrepresented (red-filled box) or underrepresented (blue-filled box) within a discretized bin across the full range of stability data.
FIGURE 5.
Gene ontology terms associated with PUM-mediated changes in RNA stability. (A) Results of iPAGE analysis to find GO terms sharing mutual information with PUM-mediated effects on RNA stability, discretized into five equally populated bins. Red bins indicate overrepresentation of genes associated with the corresponding GO term. Blue bins indicate underrepresentation of genes associated with the corresponding GO term. A black box indicates a statistically significant over- or underrepresentation with a P-value <0.05 using a hypergeometric test (Goodarzi et al. 2009). Throughout this figure, stability in PUM knockdown is represented by a normalized interaction term between time and condition, where positive values indicate stabilization upon PUM knockdown and negative values indicate destabilization upon PUM knockdown (see Materials and Methods for details). (B) Selected GO terms whose members are overrepresented in the RNAs that are stabilized under PUM knockdown, as labeled in red in A. For each GO term, a volcano plot is shown for all genes within the GO term. Volcano plots are shown as two-dimensional histograms for genes below a statistical significance threshold (Q-value <0.05) and as individual points for genes above the statistical significance threshold. Individual points are blue if a PRE can be found within any annotated 3′-UTR for that gene and red otherwise. The dashed line represents the statistical significance threshold and the dotted line represents no change in RNA stability under PUM knockdown. Below each volcano plot is a marginal density plot for the RNA stability split into two categories within the specified GO term: Genes with a PRE in any annotated 3′-UTR (blue) and genes with no PRE in any annotated 3′-UTR (red). Medians for each distribution are shown as dashed lines in the appropriate color. The black dotted line represents no change in RNA stability, as in the volcano plot above. A star represents a statistically significant (P < 0.05) difference in the medians as tested by a two-sided permutation test of shuffled group labels (n = 1000). (C) As in B, but for selected GO terms whose members are overrepresented in the RNAs that are destabilized under PUM knockdown, as labeled in blue in A.
For a finer grain view of certain PUM-regulated pathways, we plotted the RNA stability results for each gene involved in selected GO terms that were enriched in genes destabilized by PUM KD (blue text, Fig. 5A) or enriched in genes stabilized by PUM KD (red text, Fig. 5A) as indicated by our iPAGE analysis.
In Figure 5B, we report specific GO terms that were enriched in genes that were stabilized under PUM knockdown and thus likely contain PUM-repressed targets. Highlighted GO terms include guanyl-nucleotide exchange factor activity (GO:0005085; Fig. 5B, far left), which includes guanine nucleotide exchange factors (GEFs) that regulate a diverse suite of cellular functions (Rossman et al. 2005); and genes involved in peptidyl-serine phosphorylation (GO:0018105; Fig. 5B, mid-left), representing a broad class of kinases including those involved in neurological disease and inflammation (Ahmad et al. 2016; Jorge-Torres et al. 2018); and genes involved in transcriptional repressor activity (GO:0001078, Fig. 5B, mid-right), including proteins involved in regulating hematopoiesis and controlling neurological development (Jankovic et al. 2008; Xu et al. 2010; Caubit et al. 2016). Supporting the idea that PUMs directly repress subsets of genes within these GO terms, we find that genes with a PRE in their 3′-UTR are significantly more stabilized under PUM knockdown than those lacking a PRE (Fig. 5B).
Nearly all the genes in the GO term for the CCR4–NOT deadenylase complex (GO:0030014; Fig. 5B, far right) were mildly stabilized under PUM knockdown. Several genes in this category have a PRE in their 3′-UTR, including both such genes with a statistically significant change in stability. These effects are particularly interesting because human Pumilio proteins have been shown to recruit the CCR4–NOT complex to repress target mRNAs (Goldstrohm et al. 2006; Van Etten et al. 2012; Weidmann et al. 2014; Arvola et al. 2020). These new observations suggest that PUM could also directly inhibit expression of CCR4–NOT and thus globally lower deadenylation rates, perhaps providing a feedback loop that modulates PUM activity.
In Figure 5C, we also highlight two GO terms enriched for genes that were destabilized by PUM knockdown: the ficolin-1-rich granule lumen (GO:1904813, left), which is involved in innate immunity, and myelin sheath (GO:0043209, right). Unlike the PUM-repressed GO terms, these categories show a limited association with PREs in their 3′-UTR. Moreover, among transcripts in these GO terms showing significant effects of PUM knockdown on stability, those that were destabilized by PUM knockdown do not have detectable PREs, whereas those with detectable PREs are stabilized by PUM knockdown. These findings indicate that PUM is indirectly regulating the putative PUM-activated transcripts in these GO terms, or that the PRE is not the primary determinant of such activity.
Overall, transcript-level analysis reveals two general trends: (i) Transcripts stabilized by PUM knockdown tend to have an identifiable bound PRE, providing high confidence that they represent direct PUM-repressed targets. (ii) Transcripts that are destabilized by PUM knockdown tend to not have an identifiable or bound PRE (although exceptions do exist), and thus either represent targets that PUM stabilizes through indirect mechanism(s) or for which the PRE is not the primary feature that PUM recognizes.
Conditional random forest models allow for prediction of PUM-mediated effects from sequence-specific features
A long-standing goal in the study of RBPs is to develop the ability to reliably predict their functional impact on any given RNA in the transcriptome. A previous model of PUM-mediated regulation exhibited modest performance by incorporating the number of PREs in various locations across the transcript including the 5′-UTR, CDS, and 3′-UTR (Bohn et al. 2018). Here, we use a different approach, which allows us to include a larger feature set of possible predictors for PUM-mediated regulation. Using conditional random forest models (Hothorn et al. 2006b), we divided genes into EFFECT and NOEFFECT classes, as shown in Figure 1D. Four different definitions for a PRE were incorporated into the analysis (Fig. 6A) including: (i) the PUM1 and (ii) the PUM2 SEQRS motifs (Fig. 2A,B); (iii) the PUM2 motif determined by Hafner et al. (2010) from PAR-CLIP data; and (iv) a direct match of the UGUANAUW consensus sequence (referred to here as regex, or regular expression, from computer terminology for a class of search patterns) defined in previous studies (Wang et al. 2002; Jiang et al. 2013) that emerged de novo from differential RNA expression changes induced by PUMs (Bohn et al. 2018).
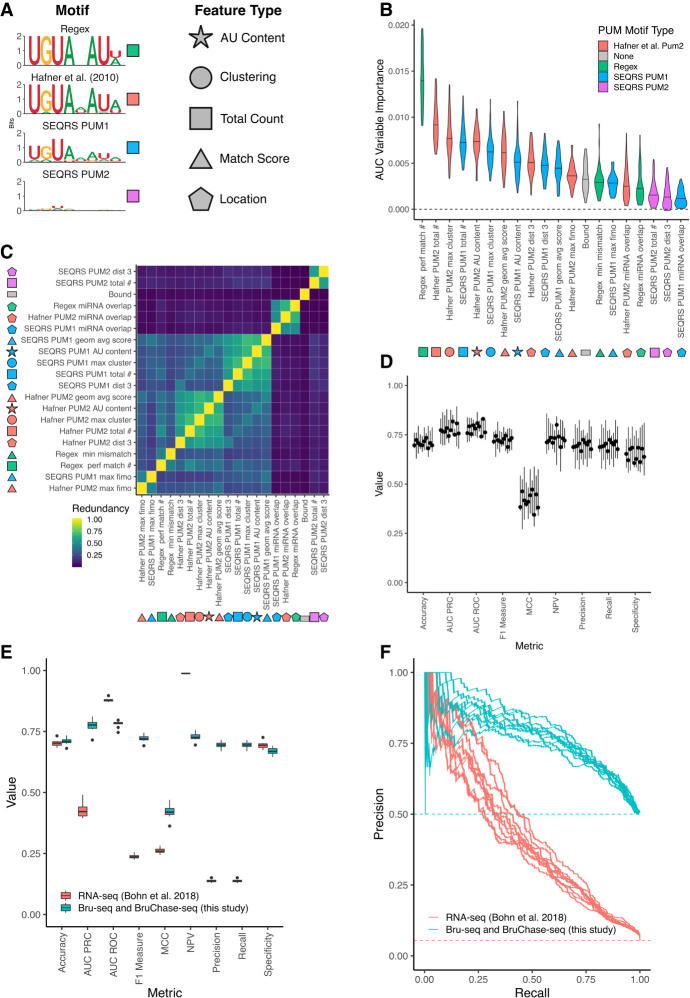
FIGURE 6.
Predicting PUM-mediated effect on decay using both sequence-based and experimental features. (A) Motifs used to calculate features for machine learning. Shapes indicate the type of feature calculated, whereas colors indicate the motif used to calculate those features. Total count is a simple count of motifs; match score refers to a numerical value indicating how well a sequence matches a motif; clustering indicates motif proximity to additional instances of the same motif; location indicates features associated with a single motif's location on the 3′-UTR. Shapes filled in with the appropriate color are used to label features throughout the rest of the figure. (B) Variable importance plot displaying the top 20 most important features, as determined by training a conditional random forest classifier on PUM decay data (see Materials and Methods for details including information on feature names). Violin plots represent density from 10 separate down-samplings of the majority class, each with fivefold cross-validation. An AUC-based variable importance measure is used as described in Janitza et al. (2013). (C) Calculation of the redundancy in information between the top 20 most important variables, as determined in A. Redundancy is calculated in the information-theoretic sense (see Materials and Methods for details) where 1 is completely redundant information and 0 is no redundancy in information between the two variables. (D) Cross-validation of conditional random forest classifier performance. Each boxplot represents a separate down-sample of the majority, no PUM-mediated effect class. Values for each boxplot represent the performance metric as calculated for each of fivefolds using a classification cutoff of 0.5. (E) Performance of conditional random forest models. Blue boxplots represent values from separate down-samplings of the majority, no PUM-mediated effect class used to train the model on the Bru-seq and BruChase-seq data set. Red boxplots indicate values from testing each model on the Bohn et al. (2018) steady-state RNA-seq data set. Metrics were calculated using a classification cutoff of 0.5. (F) Precision recall curves using the models in E. Each line represents one of 10 conditional random forest models trained on separate down-sampled sets of the entire Bru-seq and BruChase-seq data set and tested on the steady-state RNA-seq data set.
We focused on PREs found in the 3′-UTRs of target genes. For each definition of a PRE, we calculated several features based on our analysis in Figure 3, including AU content around a PRE, clustering of PREs, total count of PREs, a score for PRE match to the specific PRE definition, relative location of the PRE in the 3′-UTR, number of miRNA sites near a PRE, and predicted secondary structure around a PRE. In addition to these features, we included motif matches for additional human RBPs, in vivo PUM binding data, predictions of secondary structure, and the fraction of optimal codons for the CDS of target genes (see Materials and Methods for details). As our data is highly unbalanced (308 EFFECT genes versus 5503 NOEFFECT genes, after only including genes that have defined values for all features) we trained 10 different machine learning models where the NOEFFECT class was randomly down-sampled to match the number of EFFECT class genes in each model. Within each down-sampled data set, fivefold cross-validation was performed to assess performance.
To determine which features best predict EFFECT genes from NOEFFECT genes, we used an AUC-based permutation variable importance measure (Janitza et al. 2013), which indicates the average change in the area under the curve (AUC) of a receiver operator characteristic (ROC) plot across all trees with observations from both classes in the forest when the predictor of interest is permuted. By permuting the feature of interest and measuring the change in AUC of the ROC curve, one can measure the importance of that variable in predictive performance. Typically, values of the AUC of a ROC curve span from 0.5 to 1.0 where 1.0 indicates perfect classification performance and 0.5 indicates random guessing of class distinctions. Since the AUC-based variable importance measure is calculated using the change in AUC when the predictor is permuted, the expected values are much smaller and fall between 0.0 and 0.06 in simulated cases with 65 predictors and variable numbers of observations from n = 100 to n = 1000 (Janitza et al. 2013). Higher values indicate a larger drop in performance when that variable is permuted; thus, the variables can be ranked based on their unique contribution to the model, with higher values indicating a more important individual contribution.
Figure 6B displays the top 20 variables ranked according to their average AUC-based variable performance across all 50 models (10 sets of down-sampled models with fivefold cross-validation each). Count-based metrics enumerating the total number of PREs within the 3′-UTR appear to be the most important variable for predicting a PUM-mediated effect on stability in the Bru-seq and BruChase-seq data. In addition, local AU content and PRE clustering appear to be substantial contributors to the models. To a lesser extent, the number of miRNA sites around a PRE, the location of the PRE in the 3′-UTR, and the “Bound” status of the 3′-UTR also appear to contribute meaningfully to our models. It is possible that each of these variables contain largely the same information (i.e., whether or not the 3′-UTR has a PRE in it). To rule out the possibility that each feature was simply differentiating between genes with a PRE in their 3′-UTR from genes without a PRE, separate models were trained for each motif definition using only genes that have at least one PRE in their 3′-UTR. Each of these models also displayed substantial contributions for AU content, clustering, and total count in predicting PUM-mediated regulation of RNA stability (Supplemental Fig. S4A–D, left panel) suggesting that each of these features contributes meaningful information to the model.
Due to the high similarity of the PRE definitions, we explored how much redundant information is contained between each of the top 20 contributing features. To measure redundancy, we use an information theoretic definition based on discretization of each feature (see Materials and Methods for details). Figure 6C displays the redundancy between the top 20 features as a hierarchically clustered heatmap, where a value of 1.0 indicates that the features contain exactly the same information and a value of 0.0 indicates that the features share no information. As anticipated, features that are defined around the same motif definition or feature-type tend to share information; however, there are differences in information content between the motif definitions and feature types, indicating that there is information to be gained outside of a simple PRE count.
To assess the performance of our conditional random forest models we considered several performance measures including summary metrics (accuracy, F1 measure, Matthews correlation coefficient [MCC], area under the curve of a precision-recall curve [AUC PRC], and AUC ROC), and metrics more focused on performance for positive or negative cases (negative predictive value [NPV], precision, recall, specificity). We considered each of these metrics for all 50 models (10 down-sampled data sets with fivefold cross-validation each) at a classification probability cutoff of 0.5. The full range of values obtained are displayed in Figure 6D. It is evident that the models are robust to both down-sampling and cross-validation and the performance hovers ∼0.75 for each metric (and 0.5 for MCC), indicating balanced performance in predicting both positive and negative classes. These results are robust even in the case where we only use one PRE definition and only consider genes that contain a PRE in their 3′-UTR (Supplemental Fig. S4A–D).
To determine the predictive efficacy of our models, we tested their performance both on the training data using fivefold cross-validation, and on the RNA-seq-based differential expression analysis of PUM regulation by Bohn et al. (2018), which was not used to train the models (Fig. 6E). To observe the overall performance of the models, we display precision-recall curves on both the Bru-seq and BruChase-seq data on which the model was trained and the RNA-seq data for each of the 10 different down-sampled models (Fig. 6F). The baseline is defined separately for each data set as the overall class balance between the positive and negative class. A perfect model tends toward the upper right of the graph, and a poor model follows the dotted baseline for that data set. Despite the differences in technique and biological implications between RNA-seq (which measures equilibrium levels of transcripts) and Bru-seq and BruChase-seq (which track transcript stability) in determining PUM-mediated gene regulation, we find that the models trained on Bru-seq and BruChase-seq perform well in predicting PUM-mediated regulation in the RNA-seq data. Similar performance was achieved considering a single definition for a PRE and only considering genes that have a least one PRE in their 3′-UTR (Supplemental Fig. S4A–D). This analysis demonstrated a clear functional association and predictive utility for PUM motifs (i.e., match scores and count of PREs) as well as contextual features around PREs including the location, neighboring AU content, clustering of PREs, and overlap with predicted miRNA sites.
DISCUSSION
Through the combination of genome-wide measurements of RNA stability, RNA-binding specificity, and mining of sequence information, we established several general rules of PUM-mediated gene regulation in human cells. This knowledge will facilitate future efforts to discover PUM1 and PUM2 regulatory networks, biological functions, and roles in pathogenesis.
Human PUM proteins control gene expression by modulating RNA stability
Previous studies established that PUM1 and PUM2 control the levels of several PRE-containing transcripts (Morris et al. 2008; Van Etten et al. 2012; Bohn et al. 2018), but do not show whether the transcriptome-scale effects are due to changes in synthesis or degradation of those mRNAs. Through the use of metabolic labeling to track the effects of PUM1/2 knockdown on RNA stability and nascent RNA abundance, we found that reducing the expression of PUM1 and PUM2 has a widespread effect on the mRNA stability of many transcripts in HEK293 cells, but does not appear to perturb nascent RNA abundance in any significant way. As expected, the number of genes that are stabilized under PUM knockdown is much higher than the number of genes that were destabilized, providing strong, transcriptome-wide in vivo evidence for PUM's proposed role in reducing the expression levels of target genes through the recruitment of the CCR4–NOT deadenylase complex and subsequent destabilization of the transcript (Van Etten et al. 2012; Goldstrohm et al. 2018).
PUM1 and PUM2 have shared sequence preferences
Several prior in vitro studies explored the binding specificity of either PUM1 (Dominguez et al. 2018) or PUM2 (Jarmoskaite et al. 2019) separately using different approaches; we performed a direct comparison of the specificities and affinities of the RNA-binding domains of human PUM1 and PUM2, and of Drosophila Pumilio, using the SEQRS approach (Lou et al. 2017) in all cases. SEQRS is advantageous by virtue of the high diversity library of random 20-mers and multiple rounds of selection, and thus provides a comprehensive, unbiased, and directly comparable view of the binding affinity landscapes of the human PUM proteins. We find a strong preference for the UGUANAUA motif for PUM1 and, somewhat surprisingly, a weaker preference for this motif for PUM2. When considering the enrichment of all possible 8-mers, PUM1 and PUM2 preferences are highly correlated.
The PUM1 and PUM2 PRE motifs defined in our SEQRS analysis exhibit strong similarity to the sequences derived from RNAs that were associated with full-length PUMs immunoprecipitated from cells (Galgano et al. 2008; Morris et al. 2008; Hafner et al. 2010). Combining our results and those of prior studies, with past biochemical and structural analyses (Zamore et al. 1997, 1999; Wang et al. 2002; Cheong and Hall 2006; Lu and Hall 2011; Dominguez et al. 2018; Jarmoskaite et al. 2019), PUM specificity is now among the most precisely defined of any RNA-binding protein.
Human Pumilio proteins directly regulate genes involved in signaling pathways through large-scale transcript destabilization
Upon consideration of the classes of genes for which transcripts are stabilized under PUM knockdown, we find that many GO terms with evidence for direct repression by PUMs revolve around regulating signaling pathways mediated by proteins including kinases (GO:0018105) and GEFs (GO:0005085). The role of mammalian Pumilio proteins in modulating signaling through controlling mRNA levels has been established, particularly in developmental contexts (e.g., Wickens et al. 2002; Fox et al. 2005; Galgano et al. 2008; Morris et al. 2008; Chen et al. 2012; Zhang et al. 2017; Bohn et al. 2018; Goldstrohm et al. 2018). Our genome-wide profiling of the effects of PUM knockdown further showed that, for the vast majority of affected pathways, regulation occurs mainly through direct destabilization of target transcripts by PUM, whereas secondary effects on transcription are generally weak or nonexistent. We thus find that PUM tends to target many transcripts in parallel, and affects many components in a given pathway directly rather than relying on intermediate effects of a few regulatory hubs (as is often seen, for example, in transcriptional regulatory networks; Yu and Gerstein 2006; Song et al. 2016).
In many ways, posttranscriptional regulation of proteins involved in signaling cascades is an ideal way to rapidly modulate those pathways. In contrast to the delay in time between the control of mRNA synthesis and the resulting protein production involved in regulating a gene at the transcriptional level, posttranscriptional regulation allows for a rapid dampening of expression levels directly where protein synthesis is occurring (and subsequent ramp-up when PUM repression is removed) (Ross 1995). Furthermore, gene regulation in the cytosol allows for the possibility of localized control of expression (Hobert 2008). In fact, temporal and localized control of gene expression—important for proper development of the fly embryo—was exactly how Pumilio was initially discovered (Lehmann and Nüsslein-Volhard 1987). Given the emerging role for human PUM proteins in neuronal development and function (for review, see Goldstrohm et al. 2018) and the need for localized control of gene expression in neuronal tissue (Korsak et al. 2016), it is conceivable that PUM proteins could be heavily involved in RNA polarity within the neuron as has been observed in C. elegans olfactory neurons (Kaye et al. 2009), and in other contexts where spatially heterogeneous protein expression is required.
Mechanism of PUM-mediated activation remains elusive
The mechanism for the rarer case of PUM-mediated stabilization remains unclear. Previously reported measurements using reporter assays of PUM-activated transcripts showed a dependence on the presence of a PRE in the 3′-UTR (Bohn et al. 2018). Furthermore, direct binding of PUM1 or PUM2 to PREs present in the FOXP1 3′-UTR was reported to promote expression of the FOXP1 protein, an important regulator of the cell cycle in hematopoietic stem cells (Naudin et al. 2017). These observations supported a model wherein PUMs bind and stabilize a subset of PRE-containing mRNAs. In our analysis of transcript stability upon PUM knockdown, the evidence and number of examples for PUM binding and stabilizing of mRNAs was insufficient to draw general conclusions (Fig. 4C,D). An alternative hypothesis is that the destabilization of the transcripts in the absence of PUM is the result of indirect effects. Such effects could be mediated through another regulatory factor that PUM directly regulates or competes with for RNA binding. Consistent with this possibility, we observed multiple examples of pathways for which the majority of targets destabilized by PUM knockdown had no PREs (see Fig. 5). Collectively, it seems likely that the PUM-mediated activation of genes represents a combination of direct and indirect targets. General rules for predicting PUM-mediated activation remain elusive, and mechanistic insights into activation of key targets will require further study.
General principles for an ideal PUM target site
We identified an optimal set of determinants that allow prediction of PUM binding and transcript destabilization. A simple count of PREs in the 3′-UTR is the best single predictor for PUM activity, with varying performance for different motif representations. The sequence context of the PRE is also important for predicting PUM activity on any particular target. In particular, high AU content immediately surrounding the PRE appears to enhance its efficacy, as does a location near the 3′ end of the 3′-UTR. A similar correlative relationship was proposed by Jiang et al. (2013) based on sequence analysis. Here we demonstrate the importance of those properties for PUM-mediated target degradation. In our data, clustering of PREs becomes apparent as an additional contributor to PUM-mediated destabilization. In addition, we find that a count of predicted miRNA sites near PREs helps predict PUM effect, with a higher number of miRNA sites near a PRE indicating a larger stabilization under PUM knockdown (Supplemental Fig. S3A). This relationship is supported by previous studies that reported proximity of PREs to miRNA sites (Galgano et al. 2008; Miles et al. 2012; Jiang et al. 2013; Sternburg et al. 2018). Theoretically, PUM could act to block or enhance miRNA function through direct interactions with the miRNA machinery or through local rearrangements of RNA secondary structure, with examples reported for both scenarios (Miles et al. 2012; Sternburg et al. 2018).
Several other features hypothesized to play roles either in PUM-mediated regulation in particular, or mRNA decay in general, proved to be uninformative when applied to our data set. Secondary structure is predicted to affect many RBPs (Dominguez et al. 2018) and PUM is thought to change secondary structure upon binding (Kedde et al. 2010). We found that in silico predictions of RNA secondary structure around PREs were not predictive of PUM function (Supplemental Fig. S3C). In fact, regression models considering PRE count and structure performed worse when structural information was added (data not shown). Similarly, modifications of RNA nucleobases within PREs could limit recognition of mRNAs by PUM. However, using transcriptome-wide mapping data for m6A (Linder et al. 2015), we find limited to no overlap between m6A sites and PREs (data not shown). We also examined codon optimality, which is a determinant of mRNA decay in human cells (Forrest et al. 2018; Hanson and Coller 2018; Wu et al. 2019). We find that mRNAs undergoing PUM-mediated decay in our data set have a lower fraction of optimal codons on average than those that are not affected by PUMs (Supplemental Fig. S3B), suggesting that lower codon optimality might prime PUM target mRNAs for degradation. However, the fraction of optimal codons did not rank in the top 20 most important features in our machine learning models of PUM-mediated decay (Fig. 6). Thus, any interaction between PUM regulation and codon optimality appears to be minimal.
By combining high-throughput functional data with statistical modeling, we have identified several contextual features around PREs that have improved our understanding of PUM-mediated gene regulation and increased our ability to predict PUM targets. However, there is still room for improvement. Recent successes in Pumilio target prediction in Drosophila have come from characterizing RNA-binding partners of DmPum (Weidmann et al. 2016; Arvola et al. 2017). As summarized by Goldstrohm et al. (2018) multiple RBPs associate with mammalian PUMs and could modulate RNA-binding in vivo. Systematic incorporation of the effects of PUM binding partners will likely further improve our ability to predict targets of PUM-mediated decay. Likewise, recent studies have shown that RNA structural probing experiments used in tandem with in silico folding algorithms vastly improve biological predictions based on structural information (Mustoe et al. 2018). Incorporation of in vivo RNA structure data may enhance models of PUM-mediated regulation.
While we are able to draw the conclusions described above based on whole-transcriptome stability data, our inferences are necessarily correlative, and do not directly show a causative relationship between any particular feature and PUM-mediated decay. Importantly, the range of features identified here is informative across the entire transcriptome, and (as we have shown) constitute a set of independently informative features for determining the presence and magnitude of PUM-mediated transcript destabilization. Thus, the identified features must either be causally linked to target destabilization, or must somehow be tightly correlated across the transcriptome with some other underlying feature that in fact controls transcript stability. The biological plausibility, and consistency of our feature sets with prior targeted experiments, strongly argues that the factors that we have identified indeed constitute an important set of transcript features modulating PUM efficacy. Future experiments on artificial PRE contexts will permit testing of the extent to which PREs are tunable based on the features from our models.
MATERIALS AND METHODS
Experimental methodology
SEQRS protein purification
Methods are reproduced here from Weidmann et al. (2016). Recombinant Halo-tag PUM1 RBD (aa 828-1176) and Halo-tag PUM2 RBD (aa 705–1050) were expressed from plasmid pFN18A (Promega) in KRX E. coli cells (Promega) in 2×YT media with 25 µg/mL kanamycin and 2 mM MgSO4 at 37°C to OD600 of 0.7–0.9, at which point protein expression was induced with 0.1% (w/v) rhamnose for 3 h. These PUM RBD expression constructs were originally described in Van Etten et al. (2012). Cell pellets were washed with 50 mM Tris-HCl, pH 8.0, 10% (w/v) sucrose and pelleted again. Pellets were suspended in 25 mL of 50 mM Tris-HCl pH 8.0, 0.5 mM EDTA, 2 mM MgCl2, 150 mM NaCl, 1 mM DTT, 0.05% (v/v) Igepal CA-630, 1 mM PMSF, 10 µg/mL aprotinin, 10 µg/mL pepstatin, and 10 µg/mL leupeptin. To lyse cells, lysozyme was added to a final concentration of 0.5 mg/mL and cells were incubated at 4°C for 30 min with gentle rocking. MgCl2 was increased to 7 mM and DNase I (Roche) was added to 10 µg/mL, followed by incubation for 20 min. Lysates were cleared at 50,000g for 30 min at 4°C. Halotag-containing proteins were purified using Magnetic Halolink Resin (Promega) at 4°C. Beads were washed three times with 50 mM Tris-HCl (pH 8.0, 0.5 mM EDTA, 2 mM MgCl2, 1 M NaCl, 1 mM DTT, 0.5% [v/v] Igepal CA-630) and three times with elution buffer (50 mM Tris-HCl, pH 7.6, 150 mM NaCl, 1 mM DTT, 20% [v/v] glycerol).
To confirm protein expression, beads were resuspended in elution buffer with 30 U of AcTEV protease (Invitrogen), cleavage proceeded for 24 h at 4°C, and beads were removed by centrifugation through a micro-spin column (Bio-Rad). Concentration of eluted protein was measured by Bradford assay, followed by coomassie stained SDS-PAGE analysis.
SEQRS was conducted on PUM1 PUM-HD and PUM2 PUM-HD as described in Campbell et al. (2014) with minor modifications including the use of Magnetic Halolink beads (Promega). The PUM test proteins remained covalently bound via amino-terminal Halotag to the beads.
The initial RNA library was transcribed from 1 µg of input dsDNA using the AmpliScribe T7-Flash Transcription Kit (Epicentre). An amount of 200 ng of DNase treated RNA library was added to 100 nM of Halo-tagged proteins immobilized onto magnetic resin (Promega). The volume of each binding reaction was 100 µL in SEQRS buffer containing 200 ng yeast tRNA competitor and 0.1 units of RNase inhibitor (Promega). The samples were incubated for 30 min at 22°C prior to magnetic capture of the protein–RNA complex. The binding reaction was aspirated and the beads were washed four times with 200 µL of ice cold SEQRS buffer. After the final wash step, resin was suspended in elution buffer (1 mM Tris pH 8.0) containing 10 pmol of the reverse transcription primer. Samples were heated to 65°C for 10 min and then cooled on ice. A 5 µL aliquot of the sample was added to a 10 µL ImProm-II reverse transcription reaction (Promega). The ssDNA product was used as a template for 25 cycles of PCR using a 50 µL GoTaq reaction (Promega).
Bru-seq and BruChase-seq experimental procedure
Bru-seq and BruChase-seq were conducted as described in Paulsen et al. (2014) in HEK293 cells grown in the presence of siPUM1/2 or siNTC. RNAi conditions and siRNA sequences were previously described by Bohn et al. (2018) and include treatment with siRNAs for 48 h to allow for PUM depletion prior to BrU labeling. Four replicates were gathered for each time point and siRNA condition, resulting in 16 total samples. Resulting cDNA libraries were sequenced using an Illumina HiSeq 2000 via the University of Michigan Sequencing Core.
Bru-seq and BruChase-seq computational analysis
Rather than determine full decay rate constants for each transcript, which would have required the use of additional time points throughout the chase period of our experiment, we chose to determine relative changes in RNA stability using just two time points. The measurements obtained from these experiments cannot be interpreted on an absolute scale, but the rank order of stability measurements within the experiment is preserved, allowing us to determine the relative effects of PUM knockdown between any two genes (Wolfe et al. 2018) and require careful statistical analysis described below.
Modeling PUM-mediated RNA decay
Sequencing reads were aligned to the human genome (hg19) and processed according to Paulsen et al. (2014) up to obtaining read counts for exons and introns for each gene and sample. Our experimental design resulted in four different replicates of siNTC (WT) and siPUM1/2 (PUMKD) conditions with two different time points each: t0hr and t6hr. For the t0hr time points, read counts from both exons and introns were pooled for each gene. For the t6hr time points, only read counts from exons were used. Read abundance was modeled using DESeq2 (Love et al. 2014). As described in Love et al. (2014), DESeq2 models read count abundance K for gene i in sample j using the generalized linear model described below:
where αi is a gene-specific dispersion parameter for gene i and μij is defined by the following:
Here, sj is a sample-specific size factor used to put read count abundances on the same scale between samples. Finally, qi,j is defined according to our design matrix:
where c is an indicator variable that is 0 when the sample is in condition WT and 1 when the sample is in condition PUMKD. Likewise, t is an indicator variable that is 0 when sample is in the 0 h time point and 1 when the sample is in the 6 h time point. Finally, βrr is a series of three indicator variables and coefficients for each replicate to take into account batch effects; replicate 1 is taken as the baseline replicate. Since the interaction term captures changes in RNA abundance over time in the siPUM condition that differ from the siNTC condition, we interpret the βtc term to represent changes in RNA stability resulting specifically from the PUM KD condition. Similarly, since the condition term captures changes between the conditions at the 0 h time, we interpret the βc term to represent changes in nascent RNA abundance between the two conditions.Throughout the text, unless otherwise noted, we report βtc normalized by the reported standard error for the coefficient, which amounts to the Wald statistic computed for that term by DESeq2. Thus, the Wald statistic for the interaction term is denoted as “RNA stability in PUM KD” throughout the text and is a unitless quantity.
Analysis of transcriptional versus stability effects
To test for significant changes in transcription or stability, the Wald test statistic for the appropriate term—βc for transcription and βtc for stability—was calculated as described above. The Wald statistic was compared to a zero-centered normal distribution and a two-tailed P-value was calculated using statistical programming language R's pnorm function (n.b. this is virtually equivalent to the P-values calculated by the DEseq2 package for contrast [Love et al. 2014]). To test for a statistically significant lack of change in transcription or stability, the Wald statistic for the appropriate term was compared to a normal distribution centered at the nearest boundary of a region of practical equivalence (ROPE) and a two-tailed P-value was calculated using R's pnorm function. The ROPE was defined as log2(1/1.75) − log2(1.75) and was chosen to be within the range of fold expression change of a RnLuc reporter gene with between one and three PREs in its minimal 3′-UTR (Bohn et al. 2018). Each P-value was FDR-corrected using the Benjamini–Hochberg procedure (Benjamini and Hochberg 1995) and, for each term, the smaller of the two FDR-corrected P-values was reported. In order for a gene to be classified in the EFFECT class the following conditions had to be met: (i) its change in stability Q-value had to be smaller than its no change in stability Q-value; (ii) its change in stability Q-value had to pass a cutoff of 0.05 for statistical significance; and (iii) the original log2 fold-change value had to be outside the defined ROPE. In contrast, in order for a gene to be classified in the NOEFFECT class the following conditions had to be met: (i) it was not classified as an EFFECT gene; (ii) its no change in stability Q-value had to be smaller than its change in stability Q-value; (iii) its no change in stability Q-value had to pass a cutoff of 0.05 for statistical significance; and (iv) the original log2 fold-change value had to be within the defined ROPE. Genes not passing the criteria for either the EFFECT or NOEFFECT groups are those for which we lack sufficient information to make any strong statement on the effects of PUM knockdown.
SEQRS computational analysis
Each raw sequencing read from the SEQRS experiments has the following expected structure:
NNNNNN-CTGATCCTACCATCCGTGCT-NNNNNNNNNNNNNNNNNNNN-CACAGCTTCGTACCGAGCGG-GATCGGAAGA-XXXXXX-ATCTCGTA
where X represents a known barcode sequence used to split the reads from a multiplexed experiment and N represents a random variable base. The in vitro transcription reaction uses the above sequence as a template resulting in RNA with sequence starting from the 3′ end of the CACAGCTTCGTACCGAGCGG downstream from the 20-mer and going in the opposite direction. Thus, the RNA molecules in the SEQRS experiments are the reverse complement of the following:
CTGATCCTACCATCCGTGCT-NNNNNNNNNNNNNNNNNNNN-CACAGCTTCGTACCGAGCGG
Raw sequencing reads were split by barcode, allowing for up to two pairwise mismatches on both the upstream and downstream adapter sequences. The 20-mer variable regions and constant flanking adapter sequences of each read were reverse complemented and broken into all possible 8-mer sequences using a sliding window, and raw counts for all possible 8-mer abundances for each sequencing round for each protein were calculated using custom python scripts. For 8-mers that overlapped the constant flanking adapter sequences, only 8-mers that had at least one base in the variable region were considered.
To determine position-weight matrices that best represented selection by the protein of interest for that round, we followed the approach of Jolma et al. (2013) in the analysis of DNA-binding proteins using SELEX. Briefly, a seed sequence is determined from the most abundant N-mer within that round. From this seed sequence, the abundance of each base at a given position was tallied when all other positions match the seed sequence. The PWM frequencies were determined by dividing each column of the resulting count matrix by its column sum. For all PWMs determined by this method we used a UGUAAAUA seed sequence. Unlike Jolma et al. (2013), we do not include the correction for nonspecific carryover of nucleic acid from the previous cycle as the assumption that no more than 25% of 8-mers would be expected to be bound may not hold for RNA-binding proteins due to their promiscuous binding (Dominguez et al. 2018). Instead, we accounted for the bias of the initial sequencing pool by calculating a PWM for the initial pool using the UGUAAAUA seed sequence. We then divided the position frequency matrix of each PWM by the initial sequencing pool's position frequency matrix. Finally, we determined the bias-corrected frequency matrix by dividing each column of the matrix by its column sum.
In order to compare 8-mer selection between rounds or proteins, the enrichment of a particular 8-mer was calculated with the following equation:
where cs,i represents the count for 8-mer i in sample s, and cb,i represents the count for 8-mer i in the blank round where the input sequences were sampled. The DmPum data and corresponding blank sample were accessed from Weidmann et al. (2016) and only the first five rounds were considered.
GO term analysis and iPAGE
GO term analysis was performed using the integrative pathway analysis of the gene expression (iPAGE) software package (Goodarzi et al. 2009). Genes were discretized by the interaction term Wald test statistic into five equally populated bins and iPAGE was run with default settings.
Determination of matching PREs
The full set of 3′-UTRs for hg19 genome was downloaded using the TxDb.Hsapiens.UCSC.hg19.knownGene, BSgenome.Hsapiens.UCSC.hg19, and GenomicFeatures R packages. Matches to a given PWM across all 3′-UTRs were determined using the FIMO package with a uniform background using default cutoffs for reporting matches (Bailey et al. 2009). For PRE-centric figures, such as the heatmaps and violin plots in Figure 3 and Supplemental Figure S3, each unique 3′-UTR isoform is matched to its corresponding “RNA stability in PUM KD” value by gene name, and each feature's value is reported as the given summary statistic over a given 3′-UTR isoform for that feature, as described in the section below (i.e., for AU content, the value reported is the maximum AU content around any given PRE within that 3′-UTR isoform).
For de novo discovery of informative motifs in our Bru-seq and BruChase-seq data set, we applied the finding informative regulatory elements (FIRE) software (Elemento et al. 2007) with default settings to each unique 3′-UTR isoform matched to its “RNA stability in PUM KD” value and discretized into 10 equally populated bins.
To calculate the location and AU content of PREs in randomly generated sets of the 3′-UTRs, a third-order Markov model was trained on the annotated set of unique 3′-UTR isoforms from the hg19 genome. One thousand randomly simulated sets of 3′-UTRs—each with the same length as the annotated set of 3′-UTRs—was then generated using custom python scripts. For each of the thousand simulated sets of 3′-UTRs, the fifth round SEQRS PUM1 (Fig. 2A) was used to search for matches using FIMO as described above. Here each individual PRE was considered in the calculation of the kernel density plots shown in Figure 3.
To determine the PAR-CLIP read coverage at identified PRE sites in the set of known unique 3′-UTR isoforms, raw reads were downloaded from SRA with accession numbers SRR048967 and SRR048968. Raw fastq files were processed with trimmomatic (Bolger et al. 2014) and cutadapt (Martin 2011) to remove low quality reads and illumina adapters. Processed reads were aligned to the hg19 genome using the STAR aligner with default parameters (Dobin et al. 2013). Read coverage and T to C mutations were determined for reads within 20 bp of each PRE in each unique 3′-UTR isoform for both EFFECT and NOEFFECT genes, individually, using custom python scripts. Coverage over all PREs was aligned and the bottom and top 5% of read coverage at each position was removed from the average calculation. Error bars were determined by bootstrapping, with stratified sampling with replacement read coverage from individual PREs in each group separately.
Determination of PRE clustering
To determine whether the PREs cluster together more than would be expected by chance, we determined the ratio of the observed frequency of PUM sites within all possible 100 bp windows of 3′-UTRs with a least one PRE in them to a Poisson model with the rate parameter, λ, set to the average count of PREs within all 100 bp windows. 95% confidence intervals were determined by bootstrapping the observed distribution of PRE counts within all windows.
Predicting PUM-mediated regulation using conditional random forest models
In order to predict the PUM-mediated regulation on a given transcript, we used conditional random forest models as implemented by the cforest function from the party R package (Hothorn et al. 2006a; Strobl et al. 2007, 2008). Binary classification models were trained using default settings with no parameter tuning on the Bru-seq EFFECT and NOEFFECT classes and a permutation-based AUC variable importance metric was calculated for each individual model (Janitza et al. 2013). Due to the large class imbalance, 10 separate data sets were generated from the full data set, where the majority NOEFFECT class was randomly down-sampled to match the EFFECT class. Within each of the 10 data sets, fivefold cross-validation was performed to assess performance and detect overtraining. Final models were generated using the 10 down-sampled data sets without cross-validation and performance was tested on the RNA-seq data set from Bohn et al. (2018). Precision-recall plots were calculated using the PRROC package based on the methodology of Davis and Goadrich (2006).
Calculation of features associated with a PWM
For each of the features described, the values were first calculated individually for each unique 3′-UTR isoform. Values for each isoform were combined by taking the mean of the value for that feature and the isoform weighted by the number of isoforms that shared that unique 3′-UTR in the full set of annotated 3′-UTRs in the hg19 genome. For features ending in “fimo_best_bygene_max_fimo”, the maximum FIMO match score for each unique 3′-UTR isoform for that PWM was calculated by setting the P-value cutoff threshold in FIMO to 1.1, thereby allowing FIMO to consider every possible match for a given sequence. The maximum match score for each sequence was reported for each unique 3′-UTR isoform. For features ending in “fimo_best_bygene_total_num”, the total number of matching sites for a given unique 3′-UTR isoform was calculated as described above in the “Determination of matching PREs” section. For each sequence, the geometric average of FIMO scores for each matching PRE was calculated and reported in the “fimo_bygene_geom_avg_score”. The maximum match score, geometric average match score, and total match number were calculated for the SEQRS PUM1 round 5 PWM, SEQRS PUM2 round 5 PWM (Hafner et al. 2010) PUM2 PWM, and each of the PWMs for human RBPs found in the CISBP-RNA database (Ray et al. 2013).
For PREs, the shortest distance to the 3′-UTR for any given PRE is converted to normalized coordinates (i.e., 0.0 is the 5′ end and 1.0 is the 3′ end) and reported in the “fimo_best_bygene_dist_3”. For “fimo_bygene_at_content” the largest percentage AT content in a 100 bp window surrounding any PRE within a given sequence was reported. Similarly for “fimo_bygene_max_cluster”, the maximum number of full PRE sites within a sliding window of 100 bp was calculated. For both of these features, windows were truncated at the 3′ and 5′ ends of the sequence.
Predicted miRNA sites were determined using default predictions (conserved sites of conserved miRNA families) from TargetScan release 7.2 (Agarwal et al. 2015). Overlaps with PREs were calculated by counting miRNA sites within a 100 bp window surrounding each PRE. For 3′-UTRs with more than one PRE, the PRE with the maximum number of overlapping miRNA sites was considered.
Calculation of in silico base-pairing probabilities for PREs
For each identified PRE, the probability of the given PRE being base-paired within predicted secondary structure was calculated using RNAfold (Lorenz et al. 2011) by calculating the ensemble free energy of an unconstrained sequence Fu of 50 bp flanking each side of a given PRE and the ensemble free energy of a constrained sequence where no base within the PRE is allowed to form a base pair Fc. The probability of the PRE being constrained from base-pairing can be calculated using:
where T is the temperature (set to physiological temperature, 310.15 K), and R is the gas constant (set to 0.00198 kcal K−1 mol−1). Thus the probability of any given PRE being unpaired is Pc. We define two features associated with Pc for each PRE in a given 3′-UTR isoform. “_avgprob_unpaired” is the average Pc of all the PREs within a given 3′-UTR and “_maxprob_unpaired” is the maximum Pc of all the PREs within a given 3′-UTR. Values for each isoform were combined into gene level estimates, as described above.
Calculation of information redundancy between features
In order to calculate the information redundancy between features, each feature was discretized into 10 equally populated bins. The redundancy between feature 1 (F1) and feature 2 (F2) was calculated with the following equation:
where H is the entropy of a given vector X of discrete values, as defined below:
and the mutual information I(X;Y) of vectors X and Y of discrete values is defined as:
Determination of EFFECT and NOEFFECT classes for RNA-seq data
RNA-seq data was obtained from Bohn et al. (2018) and a gene was only considered if the FPKM for both the PUM1/2 knockdown condition and the siNTC condition were greater than five. Genes that passed this cutoff and that were considered to have statistically significant differential expression in the original analysis were considered EFFECT genes. Genes that passed the cutoff and were not considered to have statistically significant differential expression were considered NOEFFECT genes.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported in part by the National Institute of General Medical Sciences, National Institutes of Health grant R35 GM128637 to P.L.F. and grant R01 GM105707 to A.C.G. Additionally, this work was supported by National Institute of Neurological Disorders and Stroke, grant/award numbers R01NS100788, R01NS114018, and NIH grant/award number 1UG3TR003149 to Z.T.C., as well as NIH grant/award number UM1 HG009382 and NIH grant/award number R01 CA213214 01 to M.L. Work by M.B.W. was supported by the National Science Foundation Graduate Research Fellowship DGE1256260. Work by B.M. was supported by the NCI through the Rogel Cancer Center support grant P30CA046592.
Author contributions: Bioinformatics/computational analysis: Michael Wolfe (data analysis and primary manuscript author); Peter Freddolino (funding and concept, data analysis, writing); Bru-seq and BruChase-seq: Trista Schagat (RNAi, RNA labeling and purification); Aaron Goldstrohm (funding and concept, writing); Michelle Paulsen (BrU RNA Seq); Brian Magnuson (initial Bru-seq and BruChase-seq data analysis); Mats Ljungman (funding and concept); PUM Protein Purification for SEQRS: Daeyoon Park, Chi Zhang, and Zak Campbell (SEQRS and data analysis, funding).
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.077362.120.
REFERENCES
- Agarwal V, Bell GW, Nam J-W, Bartel DP. 2015. Predicting effective microRNA target sites in mammalian mRNAs. Elife 4: e05005 10.7554/eLife.05005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad L, Zhang S-Y, Casanova J-L, Sancho-Shimizu V. 2016. Human TBK1: a gatekeeper of neuroinflammation. Trends Mol Med 22: 511–527. 10.1016/j.molmed.2016.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arvola RM, Weidmann CA, Tanaka Hall TM, Goldstrohm AC. 2017. Combinatorial control of messenger RNAs by Pumilio, nanos and brain tumor proteins. RNA Biol 14: 1445–1456. 10.1080/15476286.2017.1306168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arvola RM, Chang C-T, Buytendorp JP, Levdansky Y, Valkov E, Freddolino PL, Goldstrohm AC. 2020. Unique repression domains of Pumilio utilize deadenylation and decapping factors to accelerate destruction of target mRNAs. Nucleic Acids Res 48: 1843–1871. 10.1093/nar/gkz1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Williams N, Misleh C, Li WW. 2006. MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res 34: W369–W373. 10.1093/nar/gkl198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. 2009. MEME suite: tools for motif discovery and searching. Nucleic Acids Res 37: W202–W208. 10.1093/nar/gkp335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B 57: 289–300. 10.1111/j.2517-6161.1995.tb02031.x [DOI] [Google Scholar]
- Bohn JA, Van Etten JL, Schagat TL, Bowman BM, McEachin RC, Freddolino PL, Goldstrohm AC. 2018. Identification of diverse target RNAs that are functionally regulated by human Pumilio proteins. Nucleic Acids Res 46: 362–386. 10.1093/nar/gkx1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt-Bohne U, Keene DR, White FA, Koch M. 2007. MEGF9: a novel transmembrane protein with a strong and developmentally regulated expression in the nervous system. Biochem J 401: 447–457. 10.1042/BJ20060691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brocard M, Khasnis S, David Wood C, Shannon-Lowe C, West MJ. 2018. Pumilio directs deadenylation-associated translational repression of the cyclin-dependent kinase 1 activator RGC-32. Nucleic Acids Res 46: 3707–3725. 10.1093/nar/gky038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell ZT, Bhimsaria D, Valley CT, Rodriguez-Martinez JA, Menichelli E, Williamson JR, Ansari AZ, Wickens M. 2012. Cooperativity in RNA-protein interactions: global analysis of RNA binding specificity. Cell Rep 1: 570–581. 10.1016/j.celrep.2012.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell ZT, Valley CT, Wickens M. 2014. A protein-RNA specificity code enables targeted activation of an endogenous human transcript. Nat Struct Mol Biol 21: 732–738. 10.1038/nsmb.2847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caubit X, Gubellini P, Andrieux J, Roubertoux PL, Metwaly M, Jacq B, Fatmi A, Had-Aissouni L, Kwan KY, Salin P, et al. 2016. TSHZ3 deletion causes an autism syndrome and defects in cortical projection neurons. Nat Genet 48: 1359–1369. 10.1038/ng.3681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang K, Marran K, Valentine A, Hannon GJ. 2012. RNAi in cultured mammalian cells using synthetic siRNAs. Cold Spring Harb Protoc 7: pdb.prot071076 10.1101/pdb.prot071076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Zheng W, Lin A, Uyhazi K, Zhao H, Lin H. 2012. Pumilio 1 suppresses multiple activators of P53 to safeguard spermatogenesis. Curr Biol 22: 420–425. 10.1016/j.cub.2012.01.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong C-G, Hall TM. 2006. Engineering RNA sequence specificity of Pumilio repeats. Proc Natl Acad Sci 103: 13635–13639. 10.1073/pnas.0606294103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis J, Goadrich M. 2006. The relationship between precision-recall and ROC curves. In Proceedings of the 23rd international conference on machine learning, pp. 233–240. ACM Press, New York. [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez D, Freese P, Alexis MS, Su A, Hochman M, Palden T, Bazile C, Lambert NJ, Van Nostrand EL, Pratt GA, et al. 2018. Sequence, structure, and context preferences of human RNA binding proteins. Mol Cell 70: 854–867.e9. 10.1016/j.molcel.2018.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong S, Wang Y, Cassidy-Amstutz C, Lu G, Bigler R, Jezyk MR, Li C, Tanaka Hall TM, Wang Z. 2011. Specific and modular binding code for cytosine recognition in Pumilio/FBF (PUF) RNA-binding domains. J Biol Chem 286: 26732–26742. 10.1074/jbc.M111.244889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong H, Zhu M, Meng L, Ding Y, Yang D, Zhang S, Qiang W, Fisher DW, Xu EY. 2018. Pumilio2 regulates synaptic plasticity via translational repression of synaptic receptors in mice. Oncotarget 9: 32134–32148. 10.18632/oncotarget.24345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elemento O, Slonim N, Tavazoie S. 2007. A universal framework for regulatory element discovery across all genomes and data types. Mol Cell 28: 337–350. 10.1016/j.molcel.2007.09.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ENCODE Project Consortium. 2012. An integrated encyclopedia of DNA elements in the human genome. Nature 489: 57–74. 10.1038/nature11247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forrest ME, Narula A, Sweet TJ, Arango D, Hanson G, Ellis J, Oberdoerffer S, Coller J, Rissland OS. 2018. Codon usage and amino acid identity are major determinants of mRNA stability in humans. bioRxiv 10.1101/488676 [DOI] [Google Scholar]
- Fox M, Urano J, Reijo Pera RA. 2005. Identification and characterization of RNA sequences to which human PUMILIO-2 (PUM2) and deleted in Azoospermia-like (DAZL) bind. Genomics 85: 92–105. 10.1016/j.ygeno.2004.10.003 [DOI] [PubMed] [Google Scholar]
- Galgano A, Forrer M, Jaskiewicz L, Kanitz A, Zavolan M, Gerber AP. 2008. Comparative analysis of mRNA targets for human PUF-family proteins suggests extensive interaction with the miRNA regulatory system. PLoS One 3: e3164 10.1371/journal.pone.0003164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gennarino VA, Singh RK, White JJ, De Maio A, Han K, Kim J-Y, Jafar-Nejad P, di Ronza A, Kang H, Sayegh LS, et al. 2015. Pumilio1 haploinsufficiency leads to SCA1-like neurodegeneration by increasing wild-type ataxin1 levels. Cell 160: 1087–1098. 10.1016/j.cell.2015.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gennarino VA, Palmer EE, McDonell LM, Wang L, Adamski CJ, Koire A, See L, Chen CA, Schaaf CP, Rosenfeld JA, et al. 2018. A mild PUM1 mutation is associated with adult-onset ataxia, whereas haploinsufficiency causes developmental delay and seizures. Cell 172: 924–936.e11. 10.1016/j.cell.2018.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstberger S, Hafner M, Tuschl T. 2014. A census of human RNA-binding proteins. Nat Rev Genet 15: 829–845. 10.1038/nrg3813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstrohm AC, Hook BA, Seay DJ, Wickens M. 2006. PUF proteins bind Pop2p to regulate messenger RNAs. Nat Struct Mol Biol 13: 533–539. 10.1038/nsmb1100 [DOI] [PubMed] [Google Scholar]
- Goldstrohm AC, Tanaka Hall TM, McKenney KM. 2018. Post-transcriptional regulatory functions of mammalian Pumilio proteins. Trends Genet 34: 972–990. 10.1016/j.tig.2018.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodarzi H, Elemento O, Tavazoie S. 2009. Revealing global regulatory perturbations across human cancers. Mol Cell 36: 900–911. 10.1016/j.molcel.2009.11.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M Jr, Jungkamp AC, Munschauer M, et al. 2010. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell 141: 129–141. 10.1016/j.cell.2010.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson G, Coller J. 2018. Codon optimality, bias and usage in translation and mRNA decay. Nat Rev Mol Cell Biol 19: 20–30. 10.1038/nrm.2017.91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobert O. 2008. Gene regulation by transcription factors and microRNAs. Science 319: 1785–1786. 10.1126/science.1151651 [DOI] [PubMed] [Google Scholar]
- Hothorn T, Bühlmann P, Dudoit S, Molinaro A, Van Der Laan MJ. 2006a. Survival ensembles. Biostatistics 7: 355–373. 10.1093/biostatistics/kxj011 [DOI] [PubMed] [Google Scholar]
- Hothorn T, Hornik K, Zeileis A. 2006b. Unbiased recursive partitioning: a conditional inference framework. J Comput Graph Stat 15: 651–674. 10.1198/106186006X133933 [DOI] [Google Scholar]
- Janitza S, Strobl C, Boulesteix A-L. 2013. An AUC-based permutation variable importance measure for random forests. BMC Bioinformatics 14: 119 10.1186/1471-2105-14-119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankovic D, Gorello P, Liu T, Ehret S, La Starza R, Desjobert C, Baty F, Brutsche M, Jayaraman PS, Santoro A, et al. 2008. Leukemogenic mechanisms and targets of a NUP98/HHEX fusion in acute myeloid leukemia. Blood 111: 5672–5682. 10.1182/blood-2007-09-108175 [DOI] [PubMed] [Google Scholar]
- Jarmoskaite I, Denny SK, Vaidyanathan PP, Becker WR, Andreasson JOL, Layton CJ, Kappel K, Shivashankar V, Sreenivasan R, Das R, et al. 2019. A quantitative and predictive model for RNA binding by human Pumilio proteins. Mol Cell 74: 966–981.e18. 10.1016/j.molcel.2019.04.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang P, Singh M, Coller HA. 2013. Computational assessment of the cooperativity between RNA binding proteins and microRNAs in transcript decay. PLoS Comput Biol 9: e1003075 10.1371/journal.pcbi.1003075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolma A, Kivioja T, Toivonen J, Cheng L, Wei G, Enge M, Taipale M, Vaquerizas JM, Yan J, Sillanpää MJ, et al. 2010. Multiplexed massively parallel SELEX for characterization of human transcription factor binding specificities. Genome Res 20: 861–873. 10.1101/gr.100552.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolma A, Yan J, Whitington T, Toivonen J, Nitta KR, Rastas P, Morgunova E, Enge M, Taipale M, Wei G, et al. 2013. DNA-binding specificities of human transcription factors. Cell 152: 327–339. 10.1016/j.cell.2012.12.009 [DOI] [PubMed] [Google Scholar]
- Jonas S, Izaurralde E. 2015. Towards a molecular understanding of microRNA-mediated gene silencing. Nat Rev Genet 16: 421–433. 10.1038/nrg3965 [DOI] [PubMed] [Google Scholar]
- Jope RS, Johnson GVW. 2004. The glamour and gloom of glycogen synthase kinase-3. Trends Biochem Sci 29: 95–102. 10.1016/j.tibs.2003.12.004 [DOI] [PubMed] [Google Scholar]
- Jorge-Torres OC, Szczesna K, Roa L, Casal C, Gonzalez-Somermeyer L, Soler M, Velasco CD, Segundo P Martínez-San, Petazzi P, Sáez MA, et al. 2018. Inhibition of Gsk3b reduces Nfkb1 signaling and rescues synaptic activity to improve the Rett syndrome phenotype in Mecp2-knockout mice. Cell Rep 23: 1665–1677. 10.1016/j.celrep.2018.04.010 [DOI] [PubMed] [Google Scholar]
- Kaye JA, Rose NC, Goldsworthy B, Goga A, L'Etoile ND. 2009. A 3′ UTR Pumilio-binding element directs translational activation in olfactory sensory neurons. Neuron 61: 57–70. 10.1016/j.neuron.2008.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedde M, van Kouwenhove M, Zwart W, Oude Vrielink JAF, Elkon R, Agami R. 2010. A Pumilio-induced RNA structure switch in P27-3′ UTR controls miR-221 and miR-222 accessibility. Nat Cell Biol 12: 1014–1020. 10.1038/ncb2105 [DOI] [PubMed] [Google Scholar]
- Kent LN, Leone G. 2019. The broken cycle: E2F dysfunction in cancer. Nat Rev Cancer 19: 326–338. 10.1038/s41568-019-0143-7 [DOI] [PubMed] [Google Scholar]
- Korsak LIT, Mitchell ME, Shepard KA, Akins MR. 2016. Regulation of neuronal gene expression by local axonal translation. Curr Genet Med Rep 4: 16–25. 10.1007/s40142-016-0085-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landt SG, Marinov GK, Kundaje A, Kheradpour P, Pauli F, Batzoglou S, Bernstein BE, Bickel P, Brown JB, Cayting P, et al. 2012. ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res 22: 1813–1831. 10.1101/gr.136184.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebedeva S, Jens M, Theil K, Schwanhäusser B, Selbach M, Landthaler M, Rajewsky N. 2011. Transcriptome-wide analysis of regulatory interactions of the RNA-binding protein HuR. Mol Cell 43: 340–352. 10.1016/j.molcel.2011.06.008 [DOI] [PubMed] [Google Scholar]
- Lee S, Kopp F, Chang T-C, Sataluri A, Chen B, Sivakumar S, Yu H, Xie Y, Mendell JT. 2016. Noncoding RNA NORAD regulates genomic stability by sequestering PUMILIO proteins. Cell 164: 69–80. 10.1016/j.cell.2015.12.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann R, Nüsslein-Volhard C. 1987. Involvement of the Pumilio gene in the transport of an abdominal signal in the Drosophila embryo. Nature 329: 167 10.1038/329167a0 [DOI] [Google Scholar]
- Linder B, Grozhik AV, Olarerin-George AO, Meydan C, Mason CE, Jaffrey SR. 2015. Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat Methods 12: 767–772. 10.1038/nmeth.3453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz R, Bernhart SH, Höner zu Siederdissen C, Tafer H, Flamm C, Stadler PF, Hofacker IL. 2011. ViennaRNA Package 2.0. Algorithms Mol Biol 6: 26 10.1186/1748-7188-6-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou T-F, Weidmann CA, Killingsworth J, Tanaka Hall TM, Goldstrohm AC, Campbell ZT. 2017. Integrated analysis of RNA-binding protein complexes using in vitro selection and high-throughput sequencing and sequence specificity landscapes (SEQRS). Methods 118–119: 171–181. 10.1016/j.ymeth.2016.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu G, Hall TM. 2011. Alternate modes of cognate RNA recognition by human PUMILIO proteins. Structure 19: 361–367. 10.1016/j.str.2010.12.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lugowski A, Nicholson B, Rissland OS. 2018. DRUID: a pipeline for transcriptome-wide measurements of mRNA stability. RNA 24: 623–632. 10.1261/rna.062877.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17: 10–12. 10.14806/ej.17.1.200 [DOI] [Google Scholar]
- Miles WO, Tschöp K, Herr A, Ji J-Y, Dyson NJ. 2012. Pumilio facilitates miRNA regulation of the E2F3 oncogene. Genes Dev 26: 356–368. 10.1101/gad.182568.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MA, Olivas WM. 2011. Roles of Puf proteins in mRNA degradation and translation. WIREs RNA 2: 471–492. 10.1002/wrna.69 [DOI] [PubMed] [Google Scholar]
- Morris AR, Mukherjee N, Keene JD. 2008. Ribonomic analysis of human Pum1 reveals cis-trans conservation across species despite evolution of diverse mRNA target sets. Mol Cell Biol 28: 4093–4103. 10.1128/MCB.00155-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustoe AM, Busan S, Rice GM, Hajdin CE, Peterson BK, Ruda VM, Kubica N, Nutiu R, Baryza JL, Weeks KM. 2018. Pervasive regulatory functions of mRNA structure revealed by high-resolution SHAPE probing. Cell 173: 181–195.e18. 10.1016/j.cell.2018.02.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narita R, Takahasi K, Murakami E, Hirano E, Yamamoto SP, Yoneyama M, Kato H, Fujita T. 2014. A novel function of human Pumilio proteins in cytoplasmic sensing of viral infection. PLoS Pathog 10: e1004417 10.1371/journal.ppat.1004417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naudin C, Hattabi A, Michelet F, Miri-Nezhad A, Benyoucef A, Pflumio F, Guillonneau F, Fichelson S, Vigon I, Dusanter-Fourt I, et al. 2017. PUMILIO/FOXP1 signaling drives expansion of hematopoietic stem/progenitor and leukemia cells. Blood 129: 2493–2506. 10.1182/blood-2016-10-747436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsu M, Kawate M, Fukuoka M, Gunji W, Hanaoka F, Utsugi T, Onoda F, Murakami Y. 2008. Novel DNA microarray system for analysis of nascent mRNAs. DNA Res 15: 241–251. 10.1093/dnares/dsn015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen MT, Veloso A, Prasad J, Bedi K, Ljungman EA, Magnuson B, Wilson TE, Ljungman M. 2014. Use of Bru-Seq and BruChase-Seq for genome-wide assessment of the synthesis and stability of RNA. Methods 67: 45–54. 10.1016/j.ymeth.2013.08.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu C, Bhat VD, Rajeev S, Zhang C, Lasley AE, Wine RN, Campbell ZT, Tanaka Hall TMT. 2019. A crystal structure of a collaborative RNA regulatory complex reveals mechanisms to refine target specificity. Elife 8: e48968 10.7554/eLife.48968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez F, Bhardwaj V, Arrigoni L, Lam KC, Grüning BA, Villaveces J, Habermann B, Akhtar A, Manke T. 2018. High-resolution TADs reveal DNA sequences underlying genome organization in flies. Nat Commun 9: 189 10.1038/s41467-017-02525-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray D, Kazan H, Cook KB, Weirauch MT, Najafabadi HS, Li X, Gueroussov S, Albu M, Zheng H, Yang A, et al. 2013. A compendium of RNA-binding motifs for decoding gene regulation. Nature 499: 172–177. 10.1038/nature12311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross J. 1995. mRNA stability in mammalian cells. Microbiol Rev 59: 423–450. 10.1128/MMBR.59.3.423-450.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossman KL, Der CJ, Sondek J. 2005. GEF means go: turning on RHO GTPases with guanine nucleotide-exchange factors. Nat Rev Mol Cell Biol 6: 167 10.1038/nrm1587 [DOI] [PubMed] [Google Scholar]
- Schwanhäusser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M. 2011. Global quantification of mammalian gene expression control. Nature 473: 337–342. 10.1038/nature10098 [DOI] [PubMed] [Google Scholar]
- Sharova LV, Sharov AA, Nedorezov T, Piao Y, Shaik N, Ko MSH. 2009. Database for mRNA half-life of 19,977 genes obtained by DNA microarray analysis of pluripotent and differentiating mouse embryonic stem cells. DNA Res 16: 45–58. 10.1093/dnares/dsn030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw G, Morse S, Ararat M, Graham FL. 2002. Preferential transformation of human neuronal cells by human adenoviruses and the origin of HEK 293 cells. FASEB J 16: 869–871. 10.1096/fj.01-0995fje [DOI] [PubMed] [Google Scholar]
- Siemen H, Colas D, Craig Heller H, Brüstle O, Reijo Pera RA. 2011. Pumilio-2 function in the mouse nervous system. PLoS ONE 6: e25932 10.1371/journal.pone.0025932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L, Huang SSC, Wise A, Castanon R, Nery JR, Chen H, Watanabe M, Thomas J, Bar-Joseph Z, Ecker JR. 2016. A transcription factor hierarchy defines an environmental stress response network. Science 354: aag1550 10.1126/science.aag1550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spassov DS, Jurecic R. 2002. Cloning and comparative sequence analysis of PUM1 and PUM2 genes, human members of the Pumilio family of RNA-binding proteins. Gene 299: 195–204. 10.1016/S0378-1119(02)01060-0 [DOI] [PubMed] [Google Scholar]
- Sternburg EL, Estep JA, Nguyen DK, Li Y, Karginov FV. 2018. Antagonistic and cooperative AGO2-PUM interactions in regulating mRNAs. Sci Rep 8: 15316 10.1038/s41598-018-33596-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strobl C, Boulesteix A-L, Zeileis A, Hothorn T. 2007. Bias in random forest variable importance measures: illustrations, sources and a solution. BMC Bioinformatics 8: 25 10.1186/1471-2105-8-25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strobl C, Boulesteix A-L, Kneib T, Augustin T, Zeileis A. 2008. Conditional variable importance for random forests. BMC Bioinformatics 9: 307 10.1186/1471-2105-9-307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taliaferro JM, Lambert NJ, Sudmant PH, Dominguez D, Merkin JJ, Alexis MS, Bazile CA, Burge CB. 2016. RNA sequence context effects measured in vitro predict in vivo protein binding and regulation. Mol Cell 64: 294–306. 10.1016/j.molcel.2016.08.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tani H, Mizutani R, Salam KA, Tano K, Ijiri K, Wakamatsu A, Isogai T, Suzuki Y, Akimitsu N. 2012. Genome-wide determination of RNA stability reveals hundreds of short-lived noncoding transcripts in mammals. Genome Res 22: 947–956. 10.1101/gr.130559.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tichon A, Perry RB-T, Stojic L, Ulitsky I. 2018. SAM68 is required for regulation of Pumilio by the NORAD long noncoding RNA. Genes Dev 32: 70–78. 10.1101/gad.309138.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Etten J, Schagat TL, Hrit J, Weidmann C, Brumbaugh J, Coon JJ, Goldstrohm AC. 2012. Human Pumilio proteins recruit multiple deadenylases to efficiently repress messenger RNAs. J Biol Chem 287: 36370–36383. 10.1074/jbc.M112.373522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Nostrand EL, Pratt GA, Shishkin AA, Gelboin-Burkhart C, Fang MY, Sundararaman B, Blue SM, Nguyen TB, Surka C, Elkins K, et al. 2016. Robust transcriptome-wide discovery of RNA-binding protein binding sites with enhanced CLIP (eCLIP). Nat Methods 13: 508–514. 10.1038/nmeth.3810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vessey JP, Schoderboeck L, Gingl E, Luzi E, Riefler J, Di Leva F, Karra D, Thomas S, Kiebler MA, Macchi P. 2010. Mammalian Pumilio 2 regulates dendrite morphogenesis and synaptic function. Proc Natl Acad Sci 107: 3222–3227. 10.1073/pnas.0907128107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Zamore PD, Tanaka Hall TM. 2001. Crystal structure of a Pumilio homology domain. Mol Cell 7: 855–865. 10.1016/S1097-2765(01)00229-5 [DOI] [PubMed] [Google Scholar]
- Wang X, McLachlan J, Zamore PD, Tanaka Hall TM. 2002. Modular recognition of RNA by a human Pumilio-homology domain. Cell 110: 501–512. 10.1016/S0092-8674(02)00873-5 [DOI] [PubMed] [Google Scholar]
- Wang J, Guo Y, Chu H, Guan Y, Bi J, Wang B. 2013. Multiple functions of the RNA-binding protein HuR in cancer progression, treatment responses and prognosis. Int J Mol Sci 14: 10015–10041. 10.3390/ijms140510015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidmann CA, Raynard NA, Blewett NH, Van Etten J, Goldstrohm AC. 2014. The RNA binding domain of Pumilio antagonizes poly-adenosine binding protein and accelerates deadenylation. RNA 20: 1298–1319. 10.1261/rna.046029.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidmann CA, Qiu C, Arvola RM, Lou T-F, Killingsworth J, Campbell ZT, Tanaka Hall TM, Goldstrohm AC. 2016. Drosophila Nanos acts as a molecular clamp that modulates the RNA-binding and repression activities of Pumilio. Elife 5: e17096 10.7554/eLife.17096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickens M, Bernstein DS, Kimble J, Parker R. 2002. A PUF family portrait: 3′UTR regulation as a way of life. Trends Genet 18: 150–157. 10.1016/S0168-9525(01)02616-6 [DOI] [PubMed] [Google Scholar]
- Wolfe MB, Goldstrohm AC, Freddolino PL. 2018. Global analysis of RNA metabolism using bio-orthogonal labeling coupled with next-generation RNA sequencing. Methods 155: 88–103. 10.1016/j.ymeth.2018.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q, Medina SG, Kushawah G, DeVore ML, Castellano LA, Hand JM, Wright M, Bazzini AA. 2019. Translation affects mRNA stability in a codon-dependent manner in human cells. Elife 8: e45396 10.7554/eLife.45396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Sankaran VG, Ni M, Menne TF, Puram RV, Kim W, Orkin SH. 2010. Transcriptional silencing of γ-globin by BCL11A involves long-range interactions and cooperation with SOX6. Genes Dev 24: 783–798. 10.1101/gad.1897310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang E, van Nimwegen E, Zavolan M, Rajewsky N, Schroeder M, Magnasco M, Darnell JE Jr. 2003. Decay rates of human mRNAs: correlation with functional characteristics and sequence attributes. Genome Res 13: 1863–1872. 10.1101/gr.997703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Gerstein M. 2006. Genomic analysis of the hierarchical structure of regulatory networks. Proc Natl Acad Sci 103: 14724–14731. 10.1073/pnas.0508637103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahr SK, Yang G, Kazan H, Borrett MJ, Yuzwa SA, Voronova A, Kaplan DR, Miller FD. 2018. A translational repression complex in developing mammalian neural stem cells that regulates neuronal specification. Neuron 97: 520–537.e6. 10.1016/j.neuron.2017.12.045 [DOI] [PubMed] [Google Scholar]
- Zamore PD, Williamson JR, Lehmann R. 1997. The Pumilio protein binds RNA through a conserved domain that defines a new class of RNA-binding proteins. RNA 3: 1421–1433. [PMC free article] [PubMed] [Google Scholar]
- Zamore PD, Bartel DP, Lehmann R, Williamson JR. 1999. The Pumilio-RNA interaction: a single RNA-binding domain monomer recognizes a bipartite target sequence. Biochemistry 38: 596–604. 10.1021/bi982264s [DOI] [PubMed] [Google Scholar]
- Zhang M, Chen D, Xia J, Han W, Cui X, Neuenkirchen N, Hermes G, Sestan N, Lin H. 2017. Post-transcriptional regulation of mouse neurogenesis by Pumilio proteins. Genes Dev 31: 1354–1369. 10.1101/gad.298752.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.