Abstract
tRNA molecules have well-defined sequence conservations that reflect the conserved tertiary pairs maintaining their architecture and functions during the translation processes. An analysis of aligned tRNA sequences present in the GtRNAdb database (the Lowe Laboratory, University of California, Santa Cruz) led to surprising conservations on some cytosolic tRNAs specific for alanine compared to other tRNA species, including tRNAs specific for glycine. First, besides the well-known G3oU70 base pair in the amino acid stem, there is the frequent occurrence of a second wobble pair at G30oU40, a pair generally observed as a Watson–Crick pair throughout phylogeny. Second, the tertiary pair R15/Y48 occurs as a purine–purine R15/A48 pair. Finally, the conserved T54/A58 pair maintaining the fold of the T-loop is observed as a purine–purine A54/A58 pair. The R15/A48 and A54/A58 pairs always occur together. The G30oU40 pair occurs alone or together with these other two pairs. The pairing variations are observed to a variable extent depending on phylogeny. Among eukaryotes, insects display all variations simultaneously, whereas mammals present either the G30oU40 pair or both R15/A48 and A54/A58. tRNAs with the anticodon 34A(I)GC36 are the most prone to display all those pair variations in mammals and insects. tRNAs with anticodon Y34GC36 have preferentially G30oU40 only. These unusual pairs are not observed in bacterial, nor archaeal, tRNAs, probably because of the avoidance of A34-containing anticodons in four-codon boxes. Among eukaryotes, these unusual pairing features were not observed in fungi and nematodes. These unusual structural features may affect, besides aminoacylation, transcription rates (e.g., 54/58) or ribosomal translocation (30/40).
Keywords: tRNA, anticodons, Ala, Gly, insects, mammals
INTRODUCTION
Transfer RNA specific for alanine has a long history. Fifty-five years ago, the sequence of yeast tRNAAla was published by Holley et al. (1965). The following year, the sequence of yeast tRNATyr led to the establishment of the cloverleaf secondary structure of transfer RNAs (Madison et al. 1966). Since the early days of the structure determination of tRNA molecules (Quigley and Rich 1976; Hingerty et al. 1978; Sussman et al. 1978), the structural roles of tertiary base pairs for the maintenance of the L-shape fold characteristic of tRNAs are well-appreciated. Further sequence and structure analyses led to additional conservation within the anticodon loop (Yarus 1982; Auffinger and Westhof 2001). Although we do not have a structure prototype for each of the basic 20 native tRNAs in any system (and by far), the sequence conservations overwhelmingly support the occurrence of most (if not all) identified tertiary pairs in all tRNA species (with variations for the long-arm tRNAs) (Biou et al. 1994; Westhof and Auffinger 2012). It has been remarked that tRNAs specific for a given amino acid are very similar throughout phylogeny, whereas, in contrast, within a given organism, the various tRNA species differ more between each other (Goodenbour and Pan 2006; Saks and Conery 2007; Ardell 2010). Clearly, tRNAs within a cell need to be distinct from each other in order to prevent misacylation by noncognate aminoacyl tRNA synthetases, and synthetases are known to possess characteristic determinants for specifically recognizing a single tRNA species (Saks et al. 1994; Giege et al. 1998). In other words, a tRNA sequence has stronger linkages to the amino acid it charges than to the organism in which it is encoded. Simultaneously, tRNAs need to maintain nucleotide conservations for folding, recognition by elongation factors like EF-Tu (Schrader and Uhlenbeck 2011), and, in eukaryotes, defined sequences are conserved in the internal promoters (A and B boxes) for transcription by polymerase III (Marck et al. 2006; Mitra et al. 2015). While searching and analyzing through the sequence alignments presented in the genomic tRNA database (version August 2019) (Chan and Lowe 2016), it came therefore as a surprise that some cytosolic tRNAAla presented different nucleotide conservations throughout phylogeny.
RESULTS
tRNA sequence alignments
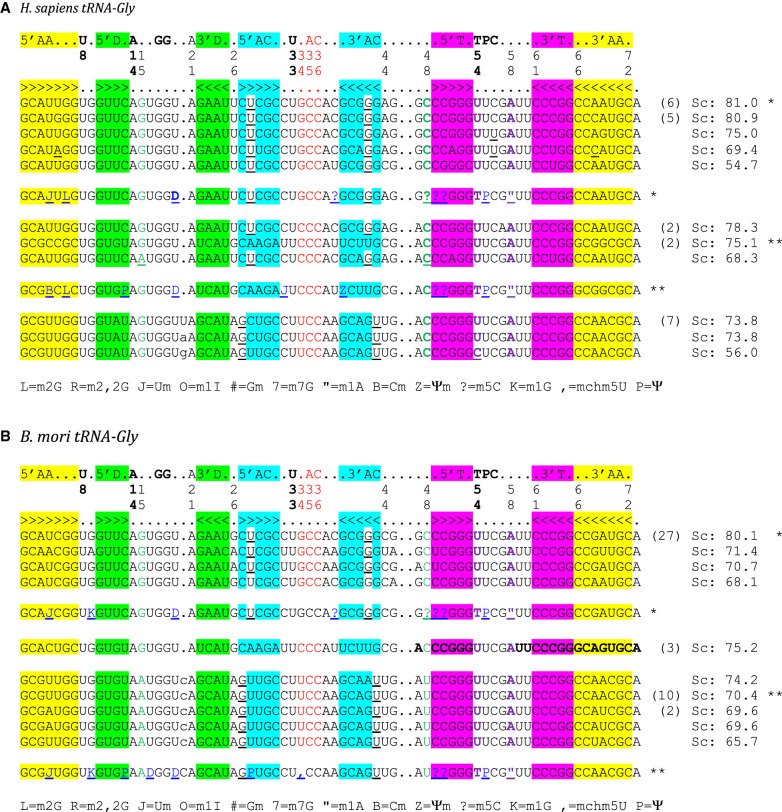
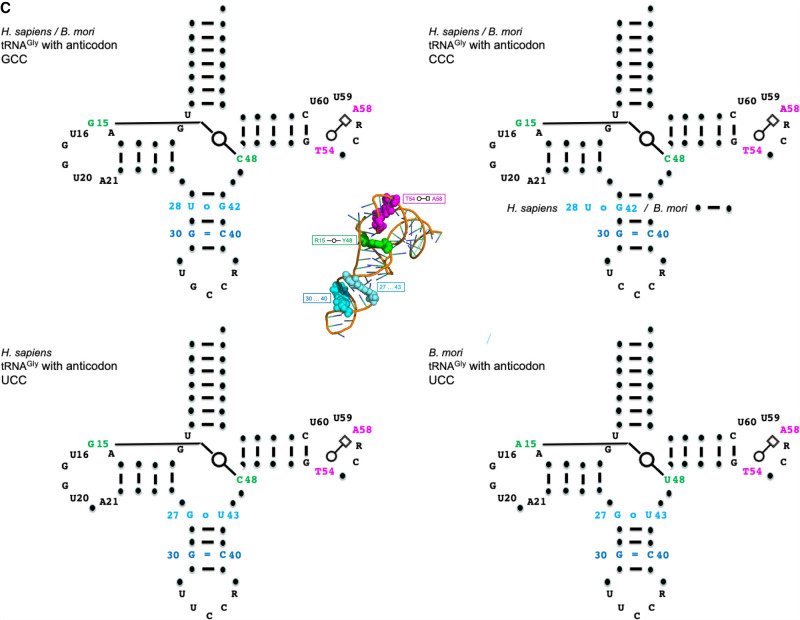
We focus our analysis here on mammals and insects especially for tRNAGly and tRNAAla, because for these two tRNAs the codon:anticodon triplets are G/C-rich and both belong to four-codon boxes. Besides, these two amino acids are the main components of silk-like fibers produced by several insects. Figure 1A,B shows the tRNA alignments, respectively, for Homo sapiens and Bombyx mori tRNAGly. In the text, we use the symbols “-” for A-U/U-A pair, “=” for G = C/C = G, “o” for wobble, and “/” for non-Watson–Crick pairs. The stems are colored, and the highly conserved residues are in bold. The numbering follows the usual tRNA-Phe nomenclature. Thus, the residues U8, A14, G18 and G19, U33, U54 (modified generally in T or thymine), U55 (modified in Ψ or pseudouridine), and C56 stand out. The residues in lowercase are weakly conserved or not at all in the Infernal covariance model (Nawrocki and Eddy 2013). It can be observed also that at the lower end of the scores, unexpected nucleotides appear (underlined) in those highly conserved positions. It is interesting to note the subtle variations in the base pairings and conservations as a function of the anticodon. For example, with the anticodon GCC, there is a U28oG42 pair (and C38 in the anticodon loop), whereas with the anticodon UCC, there is a G27oU43 (and A38 in the anticodon loop) in both H. sapiens and B. mori. Notice also the change from G15/C48 to A15/U48 in B. mori UCC-tRNAGly. The corresponding cloverleaf representations of the main nucleotide conservations are shown in Figure 1C, color-coded as in Figure 1A,B. Besides the presence of some GoU pairs within the AC-stem, all other features of tRNAGly follow the known nucleotide conservations, in marked contrast to what is observed for tRNAAla.
FIGURE 1.
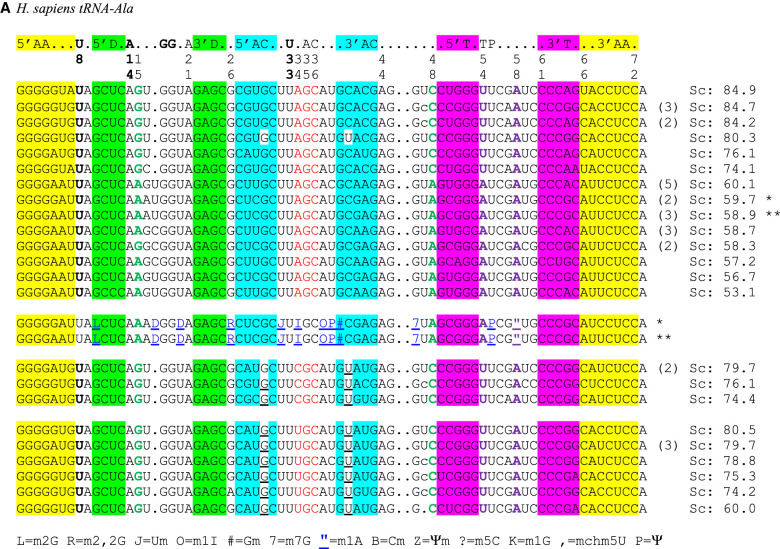
Structural alignments of tRNAGly from Homo sapiens (A) and Bombyx mori (B). The secondary structural elements of the tRNA cloverleaf are indicated above the alignments with key positions numbered following the standard nomenclature of yeast tRNAPhe. For clarity, the base-paired stems are colored (yellow for the amino acid stem; green for the dihydrouridine stem; cyan for the anticodon stem; purple for the thymine stem throughout the alignments), except for GoU pairs in the anticodon stem, which are white and underlined. The anticodon triplet is shown in red. At the right, the scores (Sc) from the tRNAscan-SE prediction algorithm (Lowe and Eddy 1997) as indicated in the database of transfer RNA genes GtRNAdb 2.0 (Chan and Lowe 2016) are shown. The number of potential tRNA gene copies is indicated next to the scores when greater than 1. Thus, for tRNAGly, there are 14 isoacceptors with anticodon GCC, five with CCC anticodons, and nine with UCC anticodons. The two tertiary pairs extensively discussed in the text are in bold and colored—green for 15/48 and magenta for 54/58. The same color code is used in the 2D and 3D representations. The sequences of the two isoacceptor tRNAGly present in the MODOMICS database (Boccaletto et al. 2018) are shown, and the code used is explained in the last line. D stands for dihydrouridine and I for inosine. The starred sequences are, respectively, the predicted and the experimentally determined sequences for a given isoacceptor. (C) Standard cloverleaf structures corresponding to the alignments with key contacts highlighted with the same color code. The Leontis–Westhof (2001) nomenclature is used for the non-Watson–Crick pairs. In the middle, a representative three-dimensional structure of a tRNA (PDB 1EHZ from Shi and Moore 2000) is shown together with base pairs discussed.
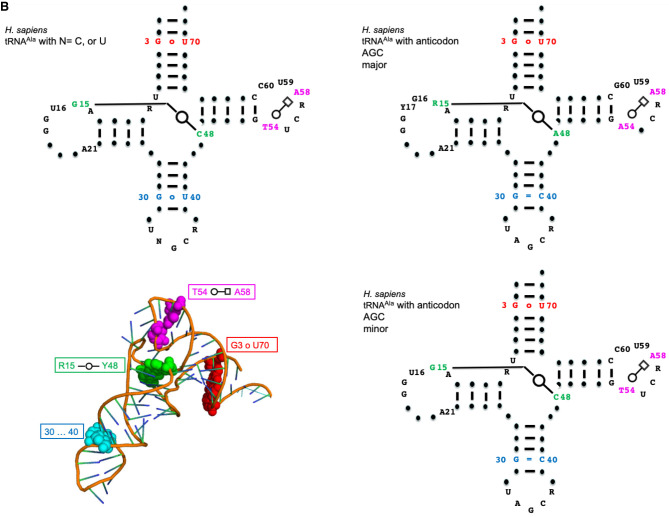
In tRNAAla, the key determinant for its cognate aminoacyl tRNA synthetase is a G3oU70 wobble pair in the amino acid stem (Hou and Schimmel 1988; McClain et al. 1988; Naganuma et al. 2014) and not the anticodon triplet (Giégé et al. 1998). This feature led to extensive studies mainly on the Escherichia coli system (Hou and Schimmel 1988, 1989; Hou et al. 1995). The base pair between residues 15 and 48 was especially studied (Hou et al. 1993, 1995). Nowadays, with so many more genomes available remarkably annotated (Chan and Lowe 2016), the analysis can be deepened. Figure 2A,C presents the alignments corresponding to tRNAAla in H. sapiens and B. mori. The tRNA sequences with the modifications of the two sets of tRNAs discussed (Ala and Gly) as extracted from MODOMICS (Boccaletto et al. 2018) are indicated in the figures. The corresponding cloverleaf representations of the main nucleotide conservations are shown in Figure 2B,D color-coded as in Figure 2A,C. We will now describe these new features.
FIGURE 2.
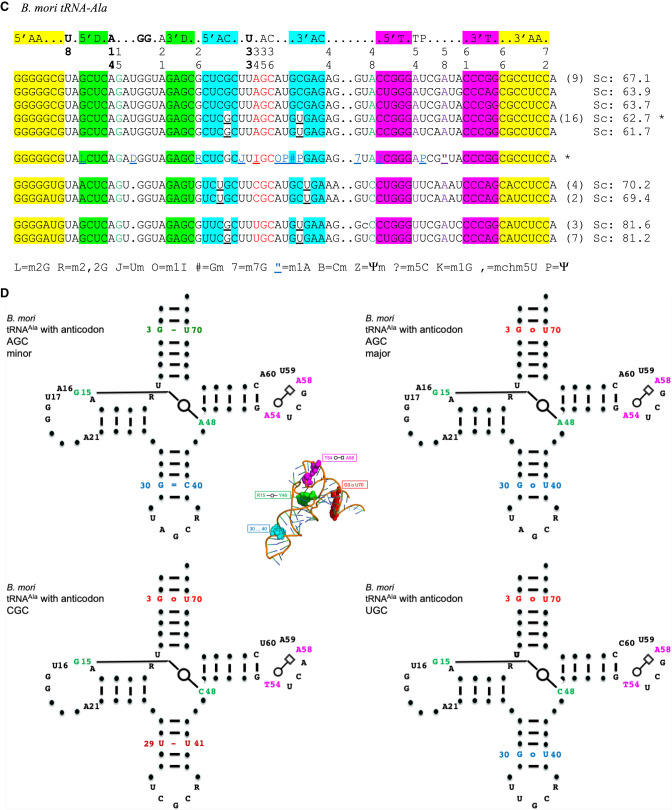
Structural alignments of tRNAAla from Homo sapiens (A) and Bombyx mori (C). The secondary structural elements of the tRNA cloverleaf are indicated above the alignments with key positions numbered following the standard nomenclature of yeast tRNAPhe. For clarity, the base-paired stems are colored (yellow for the amino acid stem; green for the dihydrouridine stem; cyan for the anticodon stem; purple for the thymine stem throughout the alignments), except for GoU pairs in the anticodon stem, which are white and underlined. The anticodon triplet is shown in red. At the right, the scores (Sc) from the tRNAscan-SE prediction algorithm (Lowe and Eddy 1997) as indicated in the database of transfer RNA genes GtRNAdb 2.0 (Chan and Lowe 2016) are shown. The number of potential tRNA gene copies is indicated next to the scores when greater than 1. The two tertiary pairs extensively discussed in the text are bold and colored—green for 15/48 and magenta for 54/58. The same color code is used in the 2D and 3D representations. The sequences of the two isoacceptor tRNAAla present in the MODOMICS database (Boccaletto et al. 2018) are shown, and the code used is explained in the last line. D stands for dihydrouridine and I for inosine. The starred sequences are, respectively, the predicted and the experimentally determined sequences for a given isoacceptor. (B,D) Standard cloverleaf structures corresponding to the alignments with key contacts highlighted with the same color code. The Leontis–Westhof (2001) nomenclature is used for the non-Watson–Crick pairs. A representative three-dimensional structure of a tRNA (PDB 1EHZ from Shi and Moore 2000) is shown together with base pairs discussed in each case.
Unusual structural features in eukaryotic tRNAAla
Two pairs in the secondary structure are unusual
The G3oU70 wobble pair in the amino acid stem
The first conserved unusual and well-documented feature in tRNAAla is the G3oU70 base pair as discussed above (Hou and Schimmel 1988, 1989; McClain et al. 1988; Hou et al. 1995; Naganuma et al. 2014). We did observe it in all alignments studied and we will not discuss it here further.
The G30oU40 wobble pair in the anticodon stem
The second unusual secondary pair is the presence of a GoU pair in the anticodon stem between nucleotides 30 and 40 in eukaryotic tRNAAla. This base pair is generally mainly a G = C pair, or, less frequently, a C = G pair, in the vast majority of tRNAs across phylogeny (Grosjean and Westhof 2016). In humans or insects, several tRNAAla isodecoders present a G30oU40 base pair. In B. mori, a U29oU41 pair occurs in tRNAs with anticodon CGC. Surprisingly, the only occurrence of a wobble pair at 30–40 occurs in eukaryotic tRNAIle but in the reverse order: between U30 and G40. Interestingly, in the same codon box, tRNAMet presents a U31–U39 in eukaryotes.
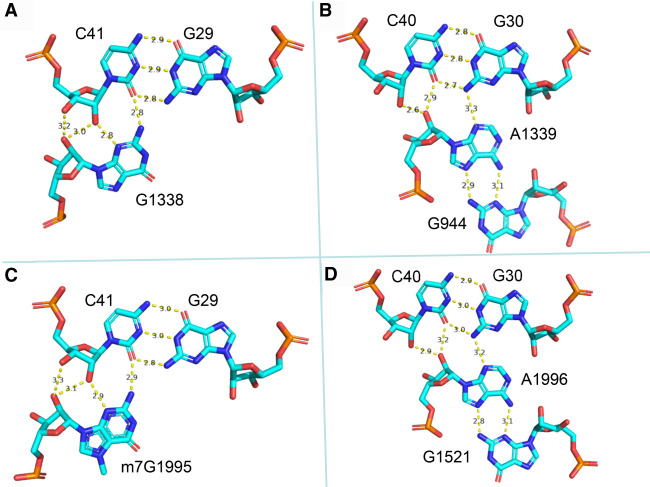
Residues 30 and 40 make contacts with the ribosome in the P-state
The base pair between nucleotides 30 and 40 makes important contacts with the ribosome during translation (Fig. 3). In the bacterial P-site, the tRNA residues 30 and 40 contact A1339 of the 16S rRNA (A-minor type interaction), with G1338 forming an A-minor contact with the preceding pair 29–41 (generally a Watson–Crick pair) (Selmer et al. 2006). Similar contacts are formed between the 30–40 pair and A1996 as well as between the 29–41 pair and G1995 in eukaryotic ribosomes in Leishmania (Shalev-Benami et al. 2017) or involving A1576/G1575 in Saccharomyces cerevisiae (Tesina et al. 2020). In a GoU wobble, the U is displaced in the major groove of the helix compared to the C in a G = C pair. Thus, in a G30oU40 wobble pair, U40 would be displaced into the major groove, away from the minor groove, thereby preventing the formation of H-bonding contacts with A1339(A1996). Alternatively, a movement of G30 into the minor groove with the maintenance of the H-bonding contacts between U40 and A1339(A1996) would require movements of many residues because A1339(A1996) forms a pair with G944(G1521). Besides, residue U40 is modified into pseudouridine (Sprague et al. 1977) and this modification would enforce the wobbling, because a pseudouridine does not tautomerize (Westhof et al. 2019).
FIGURE 3.
Interactions between the P-site tRNA and the large rRNA in bacterial ribosome (A,B) (from Watson et al. 2020) and in a eukaryotic ribosome (C,D) (from PDB 6AZ1, 6AZ3; Shalev-Benami et al. 2017). The contacts occur in the minor groove of base pairs 29–41 and 30–40 in the anticodon stem. Notice how the hydroxyl groups of 40 and 41 are forming H-bonds locking in those residues. Further, A1338 (A1996 in the eukaryotic ribosome) formed a trans-Hoogsteen/Watson–Crick pair (a “sheared” base pair) with G944 (G1521, respectively). Drawings made using PyMOL (PyMOL 1.7.7.6—Incentive Product © Schrodinger LLC).
On the other hand, with the reversed U30oG40 pair, the contacts between A1339(A1996) and G40 can form (with N3 of G40 H-bonding to N2 of A1339), as in C30 = G40 (a combination observed in tRNAAla of fungi). Pairs involving A and U at positions 30 and 40 would allow similar contacts with A1339. In eukaryotes, the only tRNAs with a C30 = G40 pair are found also in tRNAs for His and Leu (CAG). In the CGC-tRNAAla of B. mori, the U29oU41 pair should not disrupt the contact with G1338(G1995).
Two base pairs in the tertiary structure are unusual in tRNAAla
We first recall some observations about two structural pairs, essential for the maintenance of the tRNA L-shape fold, the non-Watson–Crick 15/48 and 54/58 pairs.
The 15–48 trans-Watson–Crick/Watson–Crick tertiary pair
Nucleotides at positions 15 and 48 form in tRNA structures a trans-Watson–Crick/Watson–Crick pair with, in most cases, position 15 a purine and position 48 a pyrimidine (Supplemental Fig. S1A). In bacteria and eukaryotes, both G15 = C48 and A15–U48 occur (see UCC-tRNAGly of B. mori in Fig. 1B). In archaea, only G15 = C48 pairs occur (with G15 modified into archaeosine, or 7-formamidino-7-deazaguanosine) (Watanabe et al. 1997). In E. coli tRNACys, a G15/G48 pair is present (Supplemental Fig. S1C; Hou et al. 1993). Such a G15/G48 pair is found in many other γ-proteobacteria (like Enterobacter, Klebsiella, Salmonella, or Shigella) but in neither vertebrates nor insects. Interestingly, in bacteria, there is a strong conservation of a G27oU43 wobble pair, whereas, in vertebrates and insects, that pair is reversed into U27oG43. There are other differential conservations between bacteria and eukaryotes. In bacteria, tRNACys occurs mainly with A9, A13oA23 (or G9, G13oA22), and Y21, irrespectively of G15oG48 or G15 = C48. In eukaryotes, tRNACys prefers A9, C13 = G22, and A21 always with C15 = G48. In both, there is a preference for Y60 and the possibility to form a pair between Y16 and residue 59 (that can be either R or Y).
The T-loop trans-Watson–Crick/Hoogsteen 54–58 tertiary pair
Nucleotides 54 and 58 in the T-loop stack on the last pair of the T-stem (a conserved G53 = C61) and form a trans-Watson–Crick/Hoogsteen pair between the highly conserved thymine at 54 and adenine at 58 (Supplemental Fig. S1B). The presence of the purine–purine A54oA58 pair is known in the T-loop of eukaryotic tRNAiMet (Basavappa and Sigler 1991). A similar type of pair can be formed between the Watson–Crick edge of A54 and the Hoogsteen edge of A58, but it is longer than the T54oA58 pair (12.5 Å vs. 9.8 Å) (Supplemental Fig. S1D; Leontis et al. 2002). The residue A58 is modified in m1A in eukaryotic initiator tRNAs (Boccaletto et al. 2018) and the effects of A58 hypomodification investigated (Saikia et al. 2010). Two characteristic features of the initiator tRNAiMet are three G = C pairs in the anticodon stem (Gs on the 5′ strand and Cs on the 3′ strand) before the anticodon loop and an A54oA58 pair in the T-loop.
The R15oA48 and the T-loop A54oA58 pairs occur together in AGC-tRNAAla
In tRNAAla, the trends are different, especially for the A(I)GC anticodons. In bacteria and archaea, there is no tRNAAla starting with A34 (Grosjean et al. 2010). However, A(I)34-containing tRNAAla do occur in eukaryotes. We did not observe these pairs in fungi or nematodes. Surprisingly, in such A(I)GC anticodons of other eukaryotes, the two purine–purine pairs, 15–48 and 54–58, occur simultaneously in a large proportion of sequences analyzed (with or without G30oU40). Residue R15 is mainly G15, but A15 does occur depending on the phyla. The residues R15, A48, and A54 are not modified, but the residue 58 is m1A58 (Fig. 2A,C).
These unusual pairs occur together with an additional residue in the D-loop, U17 (modified in dihydrouridine [D] D17), and the presence of a purine at positions 16 and 60 in the T-loop. In T-loops, a purine at 59 is more frequent than at position 60 (for steric constraints). In some crystal structures—for example, in tRNACys (Hauenstein et al. 2004)—residue Y16 has been observed paired trans-Watson–Crick/Watson–Crick with Y59. In tRNAAla, R16 could still form a trans-pair with Y59 and R60 stacking over. In which case, the additional U17(D17) residue would bulge out of the loop. Purine–purine pairs are longer than purine–pyrimidine pairs, and the way such pairs are accommodated in that key region of the tRNA core will depend on the immediate environment. Figure 4 illustrates the relative orientations and relationships between residues 48 and 54: If one moves in a direction, the other one should follow. The pair 54–58 has already been observed and discussed (see above). But what is the nature of the 15–48 bp? It can be either A15/A48 or G15/A48. Among non-Watson–Crick pairs, isosteric A/A and G/A pairs exist in the trans-Sugar-Edge/Hoogsteen family (as in GNRA tetraloops). But either a single H-bonded pair or a bifurcated pair (Supplemental Fig. S1C), like the one between G15/G48 in tRNACys (Nissen et al. 2009), is probable.
FIGURE 4.
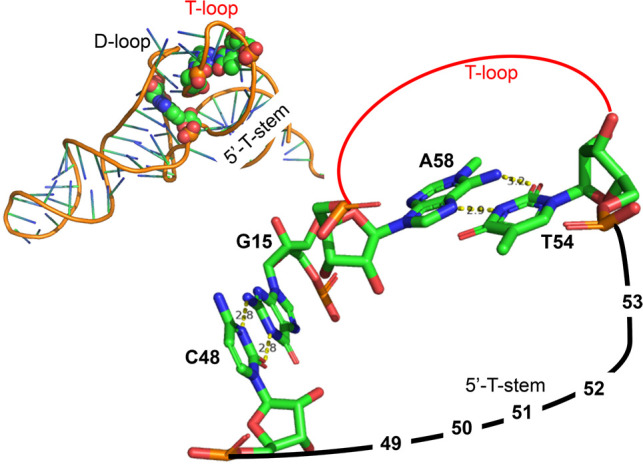
On the top left, a tRNA structure (PDB 1EHZ from Shi and Moore 2000) is shown with the two non-Watson–Crick pairs 15/48 (linking the beginning of the D-loop to the end of the variable loop) and 54/58 (closing the T-loop) highlighted with van der Waals spheres. On the right, a simplified diagram shows that both 48 and 54 are on the same strand so that a change from a pyrimidine to a purine would draw toward the reader the strand (with probably accompanying movement of the T-helix). Drawings made using PyMOL (PyMOL 1.7.7.6—Incentive Product © Schrodinger LLC).
DISCUSSION
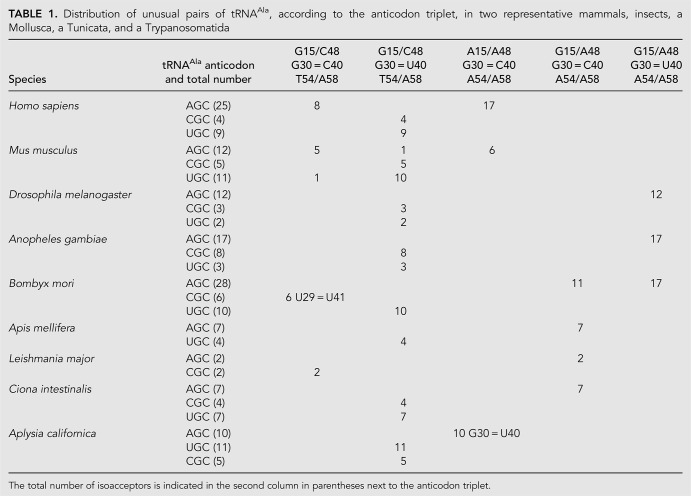
Several of these observations can be found scattered in the literature (Garel and Keith 1977; Sprague et al. 1977; Zuniga and Steitz 1977; Kawakami et al 1978; Fournier 1979). What the present comparisons (see Table 1) show is that (i) eukaryotic AGC-tRNAAla present unusual tertiary pairs: a GoU instead of a G = C pair in the anticodon, a pair recognized in the P-state, and two pairs that occur together, R15/A48 and A54/A58; (ii) in mammals, the YGC anticodons use the G30oU40 pair and the AGC ones the combination of both A15/A48 and A54/A58; (iii) in some insects (Drosophila species, Bombyx species, Anopheles gambiae), the AGC-tRNAAla present all unusual pairs, G30oU40 (G30oΨ40 in B. mori) and G15/A48 with A54/A58; in the Bombyx species, some AGC-tRNAAla present the G30 = C40 pair together with both G15/A48 and A54/A58; and (iv) the two purine–purine pairs occur with an additional residue in the D-loop and a purine at position 60 (instead of the usual pyrimidine). In Figure 5, the main trends are represented depending on some large divisions.
TABLE 1.
Distribution of unusual pairs of tRNAAla, according to the anticodon triplet, in two representative mammals, insects, a Mollusca, a Tunicata, and a Trypanosomatida
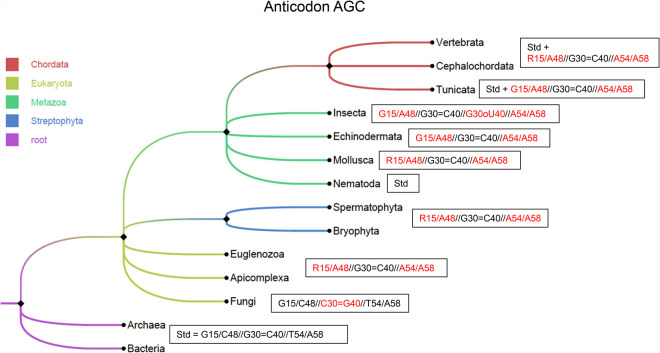
FIGURE 5.
Presence of unusual pairs in AGC-tRNAAla isodecoders in various branches of the phylogenetic tree. We checked the tRNA predictions present in GtRNAdb (Chan and Lowe 2016). As there were too many sequences, we used a threshold for the tRNAscan-SE score of 60. For the Spermatophyta, this represents 242 sequences and for the Vertebrata 199. The proportions of AGC-tRNAAla isodecoders vary strongly depending on species. Standard base pairs are in black and unusual pairs in red. The tree was built by the Taxonomy Common Tree tool (https://www.ncbi.nlm.nih.gov/Taxonomy/CommonTree/wwwcmt.cgi) and drawn using the software FigTree v1.4.4 (https://github.com/rambaut/figtree/).
Emphasis was made here on non-Watson–Crick pairs and especially GoU wobble pairs because such pairs have major structural impact on the three-dimensional fold of RNA molecules. In non-Watson–Crick pairs, various edges are used for H-bonding positioning the sugar-phosphate backbone so that the strands can be parallel or the anionic phosphate oxygens turned inside the fold and not to its exterior (Leontis et al. 2002). The GoU wobble pairs induce an over- or undertwisting in a helical stem depending on their 5′ or 3′ position so that a GoU pair can position differently in an apical loop (Masquida and Westhof 2000). However, the functional implications of those unusual features in AGC-tRNAAla are difficult to nail down on the basis of sequence alone. These features could participate in the recognition processes with the aminoacyl tRNA synthetases, with ribosomal states, or with both, and structures of complexes between such tRNAs and aminoacyl tRNA synthetases or the ribosomes would be required. The known structure of a complex between tRNAAla and aminoacyl tRNA synthetase is with the aminoacyl tRNA synthetase from the archaeon Archaeoglobus fulgidus (Naganuma et al. 2014). In that complex, the synthetase, besides contacting tightly the conserved G3oU70 pair, contacts also residues 12, 13, 20, and 47 and the conserved G19 = C56 pair at the extremity of the T-loop. Comparisons between yeast tRNAAsp free and in complex with its cognate aminoacyl synthetase show that deviations between the two tRNAs occur at a hinge point formed by the yeast-specific G30oU40 pair in the AC-stem (Ruff et al. 1991). In addition, residues 15, 54, and 58 involved in some of the unusual pairs belong to the A- and B-boxes for transcription by polymerase III (Marck et al. 2006; Mitra et al. 2015).
The presence of a G30oΨ40 wobble pair at a recognition point of the tRNA anticodon stem in the P-state could imply a role of that pair during translocation. It is known from past literature that, of the two isoacceptor tRNAAla species in B. mori, the one with the G30oΨ40 pair is twice as highly expressed in the posterior glands of the silkworm where silk translation occurs (Garel et al. 1974; Meza et al. 1977; Sprague et al. 1977). Interestingly, a similar phenomenon (Garel et al. 1974; Garel and Keith 1977; Zuniga and Steitz 1977) occurs for tRNAGly, where the most expressed isoacceptor in the posterior silk gland has the anticodon GCC in which there is a U28oG42 pair preceding the C29 = G41 pair. The C29 = G41 pair is recognized during translation in the P-state by an A-minor type contact with a G (Fig. 3; Selmer et al. 2006; Shalev-Benami et al. 2017). The presence of a 5′ U28oG42 pair will lead to a strong unstacked conformation (Masquida and Westhof 2000) with the following C29 = G41 so that G41 will lose its contact to the purine residue in the rRNA.
The two tRNA species discussed here (Ala and Gly) are major components of silk fibroin (∼30% and ∼43%, respectively [Fournier 1979]) and both display different peculiarities. For example, the aminoacyl tRNA synthetase specific for Ala does not recognize the anticodon of tRNAAla (Naganuma et al. 2014) and, in Eukarya, tRNAGly is the only tRNA species related to four-codon boxes with a G34 and not a A34(I) at the first base of the anticodon triplet (Grosjean et al. 2010). Also, both tRNA species lead to a high-GC content of the codon/anticodon triplet helix and require special conservations in the anticodon loop to guarantee smooth and uniform decoding in bacteria, especially at positions 32 and 38 (Ledoux et al. 2009; Murakami et al. 2009; Grosjean and Westhof 2016; Pernod et al. 2020). One can therefore wonder whether the observed GoU pairs in the anticodon helix of eukaryotic tRNAAla and tRNAGly do not contribute to smooth and uniform decoding.
MATERIALS AND METHODS
The analysis is based on the database of transfer RNA genes GtRNAdb 2.0 (Chan and Lowe 2016). The database contains alignments of tRNA genes based on the tRNAscan-SE prediction algorithm (Lowe and Eddy 1997). The sequences are organized as a function of an overall bit score. The score is composed of a primary sequence score and a secondary structure score based on the covariance model. A score below 55.0 may indicate the presence of a pseudogene. There are several isodecoders for the isoacceptor tRNAs (Goodenbour and Pan 2006). But, for most genomes, only a fraction of the predicted isodecoder tRNA genes have generally been experimentally observed and the tRNA modifications are known for still a smaller fraction of those on the basis of the MODOMICS database (Boccaletto et al. 2018). We extracted the tRNA alignment from the GtRNAdb 2.0 and realigned structurally by taking care of known tertiary structure conservations.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
E.W. wishes to thank G. and D. Crahay for their hospitality during the Coronavirus lockdown. Support from the National Natural Science Foundation of China (no. 31830094) is acknowledged.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.076299.120.
Freely available online through the RNA Open Access option.
REFERENCES
- Ardell DH. 2010. Computational analysis of tRNA identity. FEBS Lett 584: 325–333. 10.1016/j.febslet.2009.11.084 [DOI] [PubMed] [Google Scholar]
- Basavappa R, Sigler PB. 1991. The 3 Å crystal structure of yeast initiator tRNA: functional implications in initiator/elongator discrimination. EMBO J 10: 3105–3111. 10.1002/j.1460-2075.1991.tb07864.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biou V, Yaremchuk A, Tukalo M, Cusack S. 1994. The 2.9 Å crystal structure of T. thermophilus seryl-tRNA synthetase complexed with tRNASer. Science 263: 1404–1410. 10.1126/science.8128220 [DOI] [PubMed] [Google Scholar]
- Boccaletto P, Machnicka MA, Purta E, Piątkowski P, Bagiński B, Wirecki TK, de Crécy-Lagard V, Ross R, Limbach PA, Kotter A, et al. 2018. MODOMICS: a database of RNA modification pathways. 2017 update. Nucleic Acids Res 46: D303–D307. 10.1093/nar/gkx1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan PP, Lowe TM. 2016. GtRNAdb 2.0: an expanded database of transfer RNA genes identified in complete and draft genomes. Nucleic Acids Res 44: D184–D189. 10.1093/nar/gkv1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fournier A. 1979. Quantitative data on the Bombyx mori silkworm: a review. Biochimie 61: 283–320. 10.1016/S0300-9084(79)80073-5 [DOI] [PubMed] [Google Scholar]
- Garel JP, Keith G. 1977. Nucleotide sequence of Bombyx mori L tRNA1Gly. Nature 269: 350–352. 10.1038/269350a0 [DOI] [PubMed] [Google Scholar]
- Garel JP, Hentzen D, Daillie J. 1974. Codon responses of tRNAAla, tRNAGly and tRNASer from the posterior part of the silkgland of Bombyx mori L. FEBS Lett 39: 359–363. 10.1016/0014-5793(74)80149-3 [DOI] [PubMed] [Google Scholar]
- Giégé R, Sissler M, Florentz C. 1998. Universal rules and idiosyncratic features in tRNA identity. Nucleic Acids Res 26: 5017–5035. 10.1093/nar/26.22.5017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodenbour JM, Pan T. 2006. Diversity of tRNA genes in eukaryotes. Nucleic Acids Res 34: 6137–6146. 10.1093/nar/gkl725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosjean H, Westhof E. 2016. An integrated, structure- and energy-based view of the genetic code. Nucleic Acids Res 44: 8020–8040. 10.1093/nar/gkw608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosjean H, de Crécy-Lagard V, Marck C. 2010. Deciphering synonymous codons in the three domains of life: co-evolution with specific tRNA modification enzymes. FEBS Lett 584: 252–264. 10.1016/j.febslet.2009.11.052 [DOI] [PubMed] [Google Scholar]
- Hauenstein S, Zhang CM, Hou YM, Perona JJ. 2004. Shape-selective RNA recognition by cysteinyl-tRNA synthetase. Nat Struct Mol Biol 11: 1134–1141. 10.1038/nsmb849 [DOI] [PubMed] [Google Scholar]
- Hingerty B, Brown RS, Jack A. 1978. Further refinement of the structure of yeast tRNAPhe. J Mol Biol 124: 523–534. 10.1016/0022-2836(78)90185-7 [DOI] [PubMed] [Google Scholar]
- Holley RW, Apgar J, Everett GA, Madison JT, Marquisee M, Merrill SH, Penswick JR, Zamir A. 1965. Structure of a ribonucleic acid. Science 147: 1462–1465. 10.1126/science.147.3664.1462 [DOI] [PubMed] [Google Scholar]
- Hou YM, Schimmel P. 1988. A simple structural feature is a major determinant of the identity of a transfer RNA. Nature 333: 140–145. 10.1038/333140a0 [DOI] [PubMed] [Google Scholar]
- Hou YM, Schimmel P. 1989. Evidence that a major determinant for the identity of a transfer RNA is conserved in evolution. Biochemistry 28: 6800–6804. 10.1021/bi00443a003 [DOI] [PubMed] [Google Scholar]
- Hou YM, Westhof E, Giege R. 1993. An unusual RNA tertiary interaction has a role for the specific aminoacylation of a transfer RNA. Proc Natl Acad Sci 90: 6776–6780. 10.1073/pnas.90.14.6776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou YM, Sterner T, Jansen M. 1995. Permutation of a pair of tertiary nucleotides in a transfer RNA. Biochemistry 34: 2978–2984. 10.1021/bi00009a029 [DOI] [PubMed] [Google Scholar]
- Kawakami M, Nishio K, Takemura S. 1978. Nucleotide sequence of tRNAGly2 from the posterior silk glands of Bombyx mori. FEBS Lett 87: 288–290. 10.1016/0014-5793(78)80353-6 [DOI] [Google Scholar]
- Ledoux S, Olejniczak M, Uhlenbeck OC. 2009. A sequence element that tunes E. coli tRNAGGCAla to ensure accurate decoding. Nat Struct Mol Biol 16: 359–364. 10.1038/nsmb.1581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leontis NB, Westhof E. 2001. Geometric nomenclature and classification of RNA base pairs. RNA 7: 499–512. 10.1017/S1355838201002515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leontis NB, Stombaugh J, Westhof E. 2002. The non-Watson–Crick base pairs and their associated isostericity matrices. Nucleic Acids Res 30: 3497–3531. 10.1093/nar/gkf481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 25: 955–964. 10.1093/nar/25.5.955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madison JT, Everett GA, Kung H. 1966. Nucleotide sequence of a yeast tyrosine transfer RNA. Science 153: 531–534. 10.1126/science.153.3735.531 [DOI] [PubMed] [Google Scholar]
- Marck C, Kachouri-Lafond R, Lafontaine I, Westhof E, Dujon B, Grosjean H. 2006. The RNA polymerase III-dependent family of genes in hemiascomycetes: comparative RNomics, decoding strategies, transcription and evolutionary implications. Nucleic Acids Res 34: 1816–1835. 10.1093/nar/gkl085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masquida B, Westhof E. 2000. On the wobble GoU and related pairs. RNA 6: 9–15. 10.1017/S1355838200992082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClain WH, Chen YM, Foss K, Schneide J. 1988. Association of transfer RNA acceptor identity with a helical irregularity. Science 242: 1681–1684. 10.1126/science.2462282 [DOI] [PubMed] [Google Scholar]
- Meza L, Araya A, Leon G, Krauskopf M, Siddiqui MAQ, Garel JP. 1977. Specific alanine-tRNA species associated with fibroin biosynthesis in the posterior silk-gland of Bombyx mori. FEBS Lett 77: 255–260. 10.1016/0014-5793(77)80246-9 [DOI] [PubMed] [Google Scholar]
- Mitra S, Das P, Samadder A, Das S, Betai R, Chakrabarti J. 2015. Eukaryotic tRNAs fingerprint invertebrates vis-à-vis vertebrates. J Biomol Struct Dyn 33: 2104–2120. 10.1080/07391102.2014.990925. [DOI] [PubMed] [Google Scholar]
- Murakami H, Ohta A, Suga H. 2009. Bases in the anticodon loop of tRNAGGCAla prevent misreading. Nat Struct Mol Biol 16: 353–358. 10.1038/nsmb.1580 [DOI] [PubMed] [Google Scholar]
- Naganuma M, Sekine S, Chong YE, Guo M, Yang XL, Gamper H, Hou YM, Schimmel P, Yokoyama S. 2014. The selective tRNA aminoacylation mechanism based on a single G*U pair. Nature 510: 507–511. 10.1038/nature13440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrocki EP, Eddy SR. 2013. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics 29: 2933–2935. 10.1093/bioinformatics/btt509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissen P, Thirup S, Kjeldgaard M, Nyborg J. 2009. The crystal structure of Cys-tRNACys–EF-Tu–GDPNP reveals general and specific features in the ternary complex and in tRNA. Structure 7: 143–156. 10.1016/S0969-2126(99)80021-5 [DOI] [PubMed] [Google Scholar]
- Pernod K, Schaeffer L, Chicher J, Hok E, Rick C, Geslain R, Eriani G, Westhof E, Ryckelynck M, Martin F. 2020. The nature of the purine at position 34 in tRNAs of 4-codon boxes is correlated with nucleotides at positions 32 and 38 to maintain decoding fidelity. Nucleic Acids Res 48: 6170–6183. 10.1093/nar/gkaa221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quigley GJ, Rich A. 1976. Structural domains of transfer RNA molecules. Science 194: 796–806. 10.1126/science.790568 [DOI] [PubMed] [Google Scholar]
- Ruff M, Krishnaswamy S, Boeglin M, Poterszman A, Mitschler A, Podjarny A, Rees B, Thierry JC, Moras D. 1991. Class II aminoacyl transfer RNA synthetases: crystal structure of yeast aspartyl-tRNA synthetase complexed with tRNA(Asp). Science 252: 1682–1689. [DOI] [PubMed] [Google Scholar]
- Saikia M, Fu Y, Pavon-Eternod M, He C, Pan T. 2010. Genome-wide analysis of N1-methyl-adenosine modification in human tRNAs. RNA 16: 1317–1327. 10.1261/rna.2057810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saks ME, Conery JS. 2007. Anticodon-dependent conservation of bacterial tRNA gene sequences. RNA 13: 651–660. 10.1261/rna.345907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saks ME, Sampson JR, Abelson JN. 1994. The transfer RNA identity problem: a search for rules. Science 263: 191–197. 10.1126/science.7506844 [DOI] [PubMed] [Google Scholar]
- Schrader JM, Uhlenbeck OC. 2011. Is the sequence-specific binding of aminoacyl-tRNAs by EF-Tu universal among bacteria? Nucleic Acids Res 39: 9746–9758. 10.1093/nar/gkr641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selmer M, Dunham CM, Murphy FV, Weixlbaumer A, Petry S, Kelley AC, Weir JR, Ramakrishnan V. 2006. Structure of the 70S ribosome complexed with mRNA and tRNA. Science 313: 1935–1942. 10.1126/science.1131127 [DOI] [PubMed] [Google Scholar]
- Shalev-Benami M, Zhang Y, Rozenberg H, Nobe Y, Taoka M, Matzov D, Zimmerman E, Bashan A, Isobe T, Jaffe CL, et al. 2017. Atomic resolution snapshot of Leishmania ribosome inhibition by the aminoglycoside paromomycin. Nat Commun 8: 1587 10.1038/s41467-017-01664-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Moore PB. 2000. The crystal structure of yeast phenylalanine tRNA at 1.93 Å resolution: a classic structure revisited. RNA 6: 1091–1105. 10.1017/S1355838200000364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprague K, Hagenbüchle O, Zuniga MC. 1977. The nucleotide sequence of two silk gland alanine tRNAs: implications for fibroin synthesis and for initiator tRNA structure. Cell 11: 561–570. 10.1016/0092-8674(77)90074-5 [DOI] [PubMed] [Google Scholar]
- Sussman JL, Holbrook SR, Warrant RW, Church GM, Kim S-H. 1978. Crystal structure of yeast phenylalanine transfer RNA. I. Crystallographic refinement. J Mol Biol 123: 607–630. 10.1016/0022-2836(78)90209-7 [DOI] [PubMed] [Google Scholar]
- Tesina P, Lessen LN, Buschauer R, Cheng J, Wu CC, Berninghausen O, Buskirk AR, Becker T, Beckmann R, Green R. 2020. Molecular mechanism of translational stalling by inhibitory codon combinations and poly(A) tracts. EMBO J 39: e103365 10.15252/embj.2019103365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe M, Matsuo M, Tanaka S, Akimoto H, Asahi S, Nishimura S, Katze JR, Hashizume T, Crain PF, McCloskey JA, et al. 1997. Biosynthesis of archaeosine, a novel derivative of 7-deazaguanosine specific to archaeal tRNA, proceeds via a pathway involving base replacement on the tRNA polynucleotide chain. J Biol Chem 272: 20146–20151. 10.1074/jbc.272.32.20146 [DOI] [PubMed] [Google Scholar]
- Watson ZL, Ward FR, Méheust R, Ad O, Schepartz A, Banfield JF, Jamie HD, Cate JHD. 2020. Structure of the bacterial ribosome at 2 Å resolution. bioRxiv 10.1101/2020.06.26.174334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westhof E, Auffinger P. 2001. An extended structural signature for the tRNA anticodon loop. RNA 7: 334–341. 10.1017/S1355838201002382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westhof E, Auffinger P. 2012. Transfer RNA structure. In eLS. 10.1002/9780470015902.a0000527.pub2 [DOI] [Google Scholar]
- Yarus M. 1982. Translational efficiency of transfer RNA's: uses of an extended anticodon. Nature 218: 646–652. [DOI] [PubMed] [Google Scholar]
- Zuniga MC, Steitz JA. 1977. The nucleotide sequence of a major glycine transfer RNA from the posterior silk gland of Bombyx mori L. Nucleic Acids Res 4: 4175–4196. 10.1093/nar/4.12.4175 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.