Abstract
Staufen1 (STAU1) is an RNA-binding protein (RBP) that interacts with double-stranded RNA structures and has been implicated in regulating different aspects of mRNA metabolism. Previous studies have indicated that STAU1 interacts extensively with RNA structures in coding regions (CDSs) and 3′-untranslated regions (3′UTRs). In particular, duplex structures formed within 3′UTRs by inverted-repeat Alu elements (IRAlus) interact with STAU1 through its double-stranded RNA-binding domains (dsRBDs). Using 3′ region extraction and deep sequencing coupled to ribonucleoprotein immunoprecipitation (3′READS + RIP), together with reanalyzing previous STAU1 binding and RNA structure data, we delineate STAU1 interactions transcriptome-wide, including binding differences between alternative polyadenylation (APA) isoforms. Consistent with previous reports, RNA structures are dominant features for STAU1 binding to CDSs and 3′UTRs. Overall, relative to short 3′UTR counterparts, longer 3′UTR isoforms of genes have stronger STAU1 binding, most likely due to a higher frequency of RNA structures, including specific IRAlus sequences. Nevertheless, a sizable fraction of genes express transcripts showing the opposite trend, attributable to AU-rich sequences in their alternative 3′UTRs that may recruit antagonistic RBPs and/or destabilize RNA structures. Using STAU1-knockout cells, we show that strong STAU1 binding to mRNA 3′UTRs generally enhances polysome association. However, IRAlus generally have little impact on STAU1-mediated polysome association despite having strong interactions with the protein. Taken together, our work reveals complex interactions of STAU1 with its cognate RNA substrates. Our data also shed light on distinct post-transcriptional fates for the widespread APA isoforms in mammalian cells.
Keywords: 3′-untranslated regions, Alu elements, Staufen, mRNA-polysome association, alternative polyadenylation, mRNA isoforms
INTRODUCTION
The RNA-binding protein Staufen1 (STAU1) plays diverse roles in post-transcriptional gene regulation, including effects on mRNA localization, translation, and stability, some of which may link its function to cell-cycle progression (Park and Maquat 2013; Boulay et al. 2014). Many STAU1 properties are attributed to its (i) binding to double-stranded (ds)RNA structures through dsRNA-binding domain (dsRBD) 3 and dsRBD 4 and (ii) ability to self-assemble through intermolecular interactions between dsRBD 2 and dsRBD 5 and, separately, the STAU-swapping motif (Martel et al. 2010; Gleghorn et al. 2013; Lazzaretti et al. 2018). While dsRBDs 3 and 4 have appreciable affinities for RNA, the remaining dsRBDs have acquired other functions despite similar protein folding (Gleghorn and Maquat 2014). Recent structural studies have indicated that even though RNA binding by STAU1 is largely structure-based, some sequence specificity may be achieved through interactions with bases (Lazzaretti et al. 2018).
One class of dsRNA structures in the human transcriptome consists of inverted-repeat Alu elements (IRAlus), formed by intramolecular base-pairing between partially complementary Alus, a primate-specific, short interspersed element (SINE) that accounts for ∼11% of the human genome (DeCerbo and Carmichael 2005; Deininger 2011; Chen and Yang 2017). A full-length Alu is ∼300 nt and consists of 5′–part A–(A)5TAC(A)6–part B–poly(A)–3′, where parts A and B are similar nucleotide sequences connected by the denoted A-rich sequence (Häsler and Strub 2006; Elbarbary et al. 2016). While many Alus are transcribed by RNA polymerase III (Pol III) as independent transcriptional units, they are often embedded in protein-coding genes transcribed by Pol II. Alus in Pol II-transcribed genes have been reported to impact gene expression through a variety of mechanisms, including changes to chromatin structure, transcription, pre-mRNA processing, and other aspects of mRNA metabolism (Elbarbary et al. 2016; Chen and Yang 2017). Notably, a large fraction of Alus in the human genome resides within the 3′UTR of protein-coding genes. These 3′UTR Alus have the potential to contribute to polyadenylation site (PAS) evolution (Lee et al. 2008) and also regulate host mRNAs post-transcriptionally (Elbarbary et al. 2016; Chen and Yang 2017).
Most human genes produce 3′UTR isoforms through alternative polyadenylation (APA) (Tian and Manley 2017). Because 3′UTRs are enriched in regulatory motifs, APA that forms different 3′UTRs can play important roles in gene expression via changes in mRNA localization, translation, or decay (Mayr 2019). Here we use RNA immunoprecipitation (IP) coupled to 3′-end sequencing (3′READS + RIP) to examine STAU1 binding to protein-encoding transcripts, including alternative 3′UTR isoforms. We also analyze transcript abundance in cytoplasmic and polysomal fractions of STAU1-knockout (KO) cells, and correlate our findings with published STAU1-binding and RNA structure data. Analysis of 3′UTR isoforms enables specific interrogation of how 3′UTRs, especially alternative 3′UTRs, influence STAU1 binding and functions, which hitherto have been elusive. In these regards, we compare the contributions of IRAlus with other intramolecular RNA structures.
RESULTS
Global identification of poly(A)+ transcripts that bind STAU1
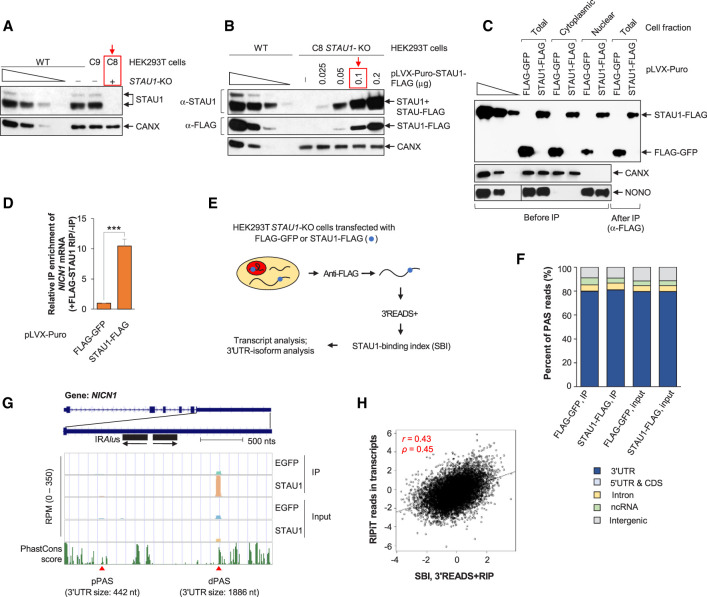
We aimed to define STAU1-bound transcripts genome-wide, with an ultimate interest in STAU1-binding to mRNA isoforms that differ in their 3′UTRs. To this end, we used CRISPR–Cas9n to generate STAU1 gene knockout (STAU1-KO) HEK293T cells (C8) and, in parallel, isogenic control cells (C9) in which the same guide RNAs failed to edit the STAU1 gene (Fig. 1A). We next established conditions in which carboxy-terminal FLAG-tagged STAU1 (STAU1-FLAG) expression in the STAU1-KO cells was ∼1.2-fold the level of STAU1 protein in untransfected wild-type (WT) cells (Fig. 1B). Like endogenous STAU1, STAU1-FLAG was expressed comparably in cytoplasmic and nuclear fractions (Fig. 1C; Elbarbary et al. 2013), indicating that FLAG-tagged STAU1 can serve as a reliable proxy for cellular STAU1. Note that the level of STAU2, the cellular paralog of STAU1, was not changed in STAU1-KO cells (Supplemental Fig. S1A).
FIGURE 1.
Transcriptome-wide analysis of STAU1 binding using 3′READS + RIP. (A) Western blots of lysates of wild-type (WT), STAU1-knockout (KO, C8, red box), or isogenic control (C9) HEK293T cell lines, the last two generated using CRISPR–Cas9n (i.e., Cas nickase). Calnexin (CANX) serves as a loading control. Here and elsewhere, lanes under the wedge are threefold dilutions of cell lysates and used for semiquantitative analyses. (B) Western blots of lysates of WT and STAU1-KO (C8) HEK293T cells transiently transfected with increasing amounts of STAU1-FLAG expression vector (µg per ∼3 × 106 cells in one 60-mm dish). The red box designates the amount of vector utilized in all subsequent experiments, where the level of STAU1-FLAG was ∼1.2-fold the level of endogenous STAU1 in WT HEK293T cells (the first lane of the anti-STAU1 titration corresponds to one cell equivalent). (C) Western blots of lysates from the specified fraction of STAU1-KO HEK293T cells transiently transfected with amounts of STAU1-FLAG expression vector equivalent to those shown in the red-boxed lane of panel B or, as a negative control, using the same amount of FLAG-GFP expression vector. Lysates were analyzed before or after immunoprecipitation (IP) using anti(α)-FLAG, where fivefold more cell equivalents were loaded after IP relative to before IP. CANX and NONO controls for the cytoplasmic and nuclear fraction, respectively, in before IP samples. (D) Histogram representation of RT-qPCR quantitations of NICN1 mRNA in the anti-FLAG IP relative to before IP using total-cell lysates of STAU1-KO HEK293T cells transiently expressing FLAG-GFP or STAU1-FLAG, where the value in FLAG-GFP-transfected cells is set to 1. Results are means ± standard deviations (SD). n = 3. (***) P < 0.001 (unpaired two-tailed t-test). (E) Schematic outlining the 3′READS + RIP procedure to analyze STAU1 binding to transcripts using samples prepared as in D. (F) Genomic distributions of poly(A) sites (PASs) detected in different input and RIP samples. (G) UCSC tracks for the NICN1 gene using 3′READS + RIP data. Reads per million (RPM) ranges are indicated on y-axis. Two APA isoforms were detected in HEK293T cells that derive from PASs different than those noted in the UCSC database. Their polyadenylation sites are depicted by red triangles, and their 3′UTR size is provided in nts. pPAS, proximal polyadenylation site; dPAS, distal polyadenylation site. Genomic sequence conservation, based on PhastCons scores derived from 100 vertebrate species, is shown in the bottom-most strip. (H) Correlation between the STAU1-Binding Index (SBI) derived from two biological replicates of 3′READS + RIP and STAU1 binding based on RIPiT data (Ricci et al. 2014) (accession number GSE52447). The SBI was calculated as the Log2(STAU1-FLAG IP RPM/FLAG-EGFP IP RPM), and STAU1 binding with RIPiT data was calculated by Log2(STAU1 WT IP/STAU1 MT IP RIPiT reads), where WT is wild-type STAU1 and MT is a STAU1 variant harboring a point mutation in dsRBD 3 and another in dsRBD 4, both known to disrupt RNA binding. For RIPiT, reads throughout transcripts were merged to calculate STAU1 binding in the whole transcript. Each point represents a transcript selected from one gene. For genes with multiple isoforms, the one with the highest expression level based on RPM in samples before IP was chosen. Pearson correlation r is indicated, as is the Spearman correlation coefficient ρ.
We next extracted RNA before and after ribonucleoprotein immunoprecipitation (RIP) using anti-FLAG and total-cell lysates of STAU1-KO HEK293T cells expressing either STAU1-FLAG or, as a negative control, FLAG-GFP (Fig. 1C). RIP efficiency was confirmed by using RT-qPCR to quantitate NICN1 mRNA, whose 3′UTR IRAlus are known to bind STAU1 (Elbarbary et al. 2013). As expected, NICN1 mRNA was enriched more than 10-fold in the RIP sample of cells expressing STAU1-FLAG compared to cells expressing FLAG-GFP (Fig. 1D).
Having validated the RIP method by RT-qPCR, we next subjected RNAs to 3′ Region Extraction And Deep Sequencing (3′READS), a method we previously developed to define and quantitate the abundance of poly(A)+ transcripts based on their 3′ ends (Zheng et al. 2016). This method, which we named 3′READS + RIP, readily addressed the issue of PCR overamplification (Supplemental Fig. S1B; Zheng et al. 2016) and was performed in biological duplicates (Supplemental Fig. S1C). As expected, most (∼80% of) polyadenylation site (PAS) reads in both input and IP samples were mapped to annotated 3′UTRs (Fig. 1F, blue). Approximately 5% of PAS reads in both STAU1-FLAG IP replicates were mapped to annotated introns (Fig. 1F, yellow) and corresponded to intronic polyadenylation isoforms. An additional 4%–6% and 9%–10% of reads were mapped to noncoding RNAs (ncRNAs) and intergenic regions, respectively (Fig. 1F, green and gray, respectively).
While 3′READS + RIP does not provide information on STAU1 binding sites per se, this method allows for the effective quantitation of STAU1-binding differences between 3′UTR isoforms produced from the same gene, which to date has been elusive. For example, 3′READS + RIP identified two NICN1 mRNA 3′UTR isoforms in HEK293T cells (Fig. 1G, red triangles). However, the identified long isoform (3′UTR size of 1886 nt), which contains one pair of IRAlus, was >100-fold more abundant than the shorter isoform (3′UTR size of 442 nt) based on analyses of input samples (Fig. 1G). Consistent with STAU1 binding to 3′UTR IRAlus (Elbarbary et al. 2013), only the long 3′UTR isoform was enriched in the STAU1-FLAG RIP relative to the FLAG-GFP RIP (Fig. 1G).
To quantify STAU1 binding to all transcripts, we calculated the log2 ratio of PAS read number in the STAU1-FLAG RIP sample relative to that in the FLAG-GFP RIP sample. For simplicity, this value was named the STAU1-binding index (SBI). Note that SBIs of two biological replicates were well correlated (r = 0.57, Pearson correlation, Supplemental Fig. S1C). In addition, we found that SBIs with or without normalization to input signals were highly similar (r = 0.86, Pearson correlation, Supplemental Fig. S1D). For simplicity, we therefore did not use input samples for normalization in subsequent SBI-based analyses. As an example, the SBI for the long 3′UTR isoform of NICN1 was 1.66, placing it at the third-percentile of all transcripts. In contrast, the SBI of the short 3′UTR isoform was −2.31 (99th-percentile), indicating that the short isoform is not enriched in the STAU1-FLAG RIP.
We next compared our 3′READS + RIP data with STAU1−RNA interaction data generated by other groups (Supplemental Fig. S2A). Ricci et al. (2014) used formaldehyde-based crosslinking and RNA immunoprecipitation in tandem (RIPiT) to identify transcript regions that interact in HEK293 cells with inducible FLAG-STAU1 but not with a FLAG-STAU1 variant containing point mutations in dsRBDs 3 and 4 that disrupt binding to dsRNA. We compared our 3′READS + RIP data with RIPiT reads mapped throughout the whole transcript region and found that the two data sets were generally correlated (r = 0.43, P < 2.2 × 10−16, Pearson correlation, Fig. 1H and Supplemental Fig. S2B), indicating that 3′READS + RIP effectively identifies STAU1-bound transcripts genome-wide. Note that the correlation was lower when RIPiT reads in 3′UTRs only were used (r = 0.37, Pearson correlation, Supplemental Fig. S1B), suggesting that the SBI value captures binding to not only the 3′UTR but also the CDS and/or 5′UTR.
We also compared our data to the FLAG-STAU1 iCLIP-seq data derived by Sugimoto et al. (2015) using HEK293 cells (Supplemental Fig. S2A). Interestingly, in contrast to our comparison with RIPiT, while STAU1 binding to full-length transcripts as calculated using the iCLIP-seq data showed a very modest correlation with our SBI (r = 0.15, Pearson correlation, Supplemental Fig. S2C), STAU1 binding to 3′UTRs showed a higher correlation (r = 0.26, Pearson correlation, Supplemental Fig. S1D). It is also noteworthy that correlation between the RIPiT and iCLIP-seq data is quite modest as well (r = 0.29 for 3′UTR and = 0.13 for full-length, Supplemental Fig. S2D), indicating that each method has its own intrinsic biases. Together, our analyses indicate that iCLIP may be more sensitive to STAU1 binding in 3′UTRs, that RIPiT preferentially maps STAU1 binding in CDSs and/or 5′UTRs, and that 3′READS + RIP efficiently detects STAU1 binding in both 3′UTRs and CDSs/5′UTRs.
STAU1 binds strongly to 3′UTR IRAlus
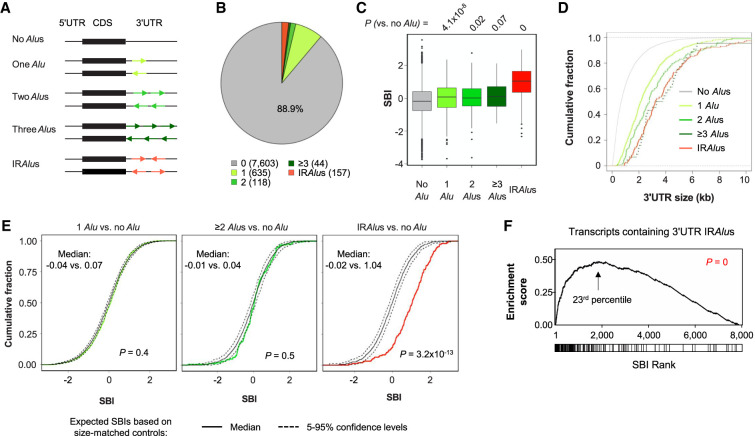
IRAlus in 3′UTRs have been shown to interact with STAU1 due to their dsRNA structures (Elbarbary et al. 2013; Sakurai et al. 2017). To examine IRAlus–STAU1 interactions in our data, we first identified all Alus in 3′UTRs using PolyA_DB3, a comprehensive PAS database which we recently created (Wang et al. 2018). We then grouped transcripts according to Alu configurations (Fig. 2A). Of the 8557 transcripts having a detectable PAS in STAU1 IP samples (more than two reads in all 3′READS + RIP samples), 954 transcripts had at least one 3′UTR Alu. Among these, 635 had one single Alu, 118 had two Alus in the same orientation, and 44 had three or more Alus in the same orientation (Fig. 2B). We also identified 157 transcripts which contained 3′UTR IRAlus, that is, at least one pair of Alus in opposite orientations (Fig. 2B). Note that both input and IP samples generated similar results (Supplemental Fig. S3).
FIGURE 2.
Alu-containing 3′UTRs and STAU1 binding. (A) Schematic showing different types of transcripts based on the number and orientation of Alus in their 3′UTR. Each group is specified by a different color. (B) Pie chart showing the percentage of transcripts harboring the different 3′UTR Alus configurations illustrated in panel A in all IP and input samples. The number of transcripts constituting each type is indicated in parentheses. (C) Boxplot of the SBIs for the different groups of transcripts in panel A. P-values (Wilcoxon test) comparing different groups with the “No Alu” group are indicated. (D) Cumulative fraction function curves of 3′UTR sizes for the different groups of transcripts in panel A. (E) Cumulative fraction function curves of SBIs of different groups with 3′UTR-size-matched control transcripts. Colored line in each plot corresponds to observed SBIs of a specific group of transcripts. Solid black line corresponds to expected SBIs based on bootstrapped transcripts with similar 3′UTR sizes. The bootstrapping process was repeated 100 times. Dotted black lines position the 0.05 or 0.95 percentile of bootstrapped SBIs. P-values (K–S test) comparing observed and expected SBIs, and their individual medians are shown. (F) Gene set enrichment analysis of 3′UTR IRAlus-containing transcripts based on SBI. Transcripts are ranked based on their SBIs. Transcripts containing 3′UTR IRAlus are marked by a vertical line in the box under the graph. The 23rd-percentile is the point at which the enrichment reaches the peak. P-value (K–S test comparing IRAlus-containing and other transcripts) indicating significance of enrichment toward high SBI values is indicated.
We found that the transcripts containing 3′UTR Alus, irrespective of the number of Alus, had higher SBIs than those without Alus (Fig. 2C). Transcripts containing 3′UTR IRAlus had the highest SBIs among all Alu-containing transcript groups (Fig. 2C). Because transcripts containing 3′UTR Alus also tend to have a longer 3′UTR than those without Alus (Fig. 2D), we next compared Alu-containing transcripts to randomly selected, 3′UTR Alu-less transcripts with similar 3′UTR sizes (see Materials and Methods for detail). Despite known examples of single 3′UTR Alus promoting STAU1-binding through intermolecular base-pairing (Gong and Maquat 2011; Gong et al. 2013), this size-corrected analysis indicated that, globally, neither one Alu nor multiple Alus in the same orientation impacted STAU1 binding (P = 0.4 and 0.5, respectively, compared to controls, K–S test, Fig. 2E, left and middle). In contrast, IRAlus-containing transcripts showed significantly higher STAU1 binding compared to size-matched controls (P = 3.2 × 10−13, K–S test, Fig. 2E, right).
Consistently, using gene set enrichment analysis (GSEA) (Subramanian et al. 2005), we found that transcripts containing 3′UTR IRAlus tended to be among the top transcripts based on SBI ranking, reaching a peak around the top 23rd-percentile based on enrichment score (Fig. 2F). We therefore defined the top 20% of transcripts based on their SBI values as high STAU1 binders in our subsequent analyses.
STAU1 binds secondary structures in both CDSs and 3′UTRs
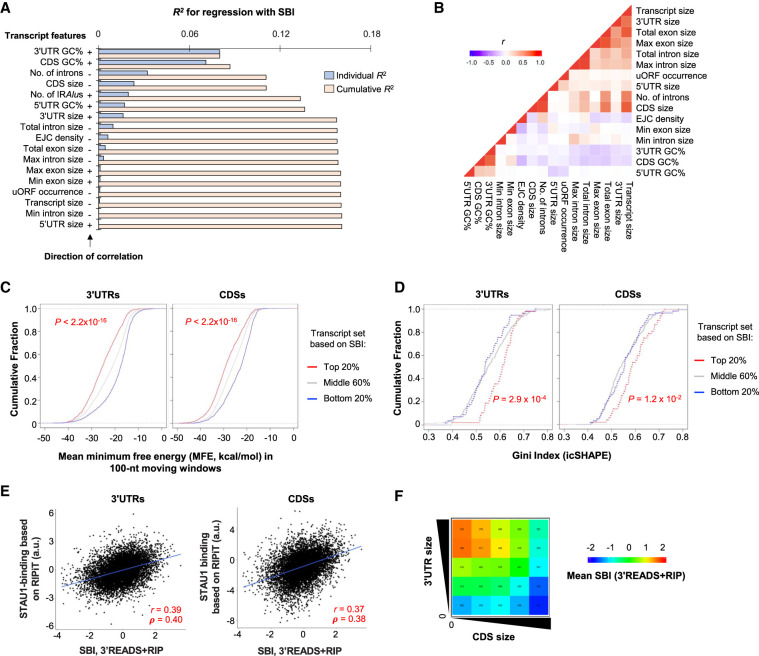
We next took an exploratory approach to systematically examine additional mRNA features that were correlated with SBIs. Based on a linear regression model (see Materials and Methods for detail), we found that the top five most significant features were GC content of the 3′UTR, GC content of the CDS, number of introns, CDS size, and number of IRAlus (Fig. 3A).
FIGURE 3.
Transcript features related to STAU1 binding. (A) Bar graph showing individual and cumulative coefficients of determination (R2) of transcript features for SBIs using a linear regression model. GC%, GC content; Max and Min refer to the largest and smallest exon or intron of a gene, respectively. +, positive correlation; −, negative correlation. Individual R2 is for a given transcript feature, and Cumulative R2 is for a combination of a given feature as well as all other features with a higher individual R2. Only the most abundant transcript per gene was used for this analysis and other analyses in this figure. (B) Correlation between individual features. Pearson correlation coefficient (r) is shown in a heatmap according to the indicated color scheme. (C) Cumulative fraction function curves of minimum free energy (MFE) by Mfold analysis, predicting the stability of RNA structures in the 3′UTR (left) and CDS (right) of genes grouped by SBI, including top 20%, middle 60%, and bottom 20%. (D) Cumulative fraction function curves of the Gini coefficient measuring the degree of RNA structures identified by icSHAPE analysis in the 3′UTR (left) and CDS (right) of genes grouped by SBI, as in panel C. (E) Correlation between SBI (3′READS + RIP) and a STAU1-binding score based on RIPiT data. The RIPiT score was individually calculated for 3′UTRs (left, 7907 genes) and CDSs (right, 8312 genes). Pearson correlation coefficient (r) is indicated in each comparison. a.u., arbitrary units. (F) Heatmap showing SBIs for genes grouped by 3′UTR size (y-axis) and CDS size (x-axis). Genes were divided into five equally sized bins based on each axis, resulting in 25 groups. The number of genes in each group is indicated, and the mean SBI for each group is represented by color based on the color scheme shown in the graph.
We found that the top two features, GC contents of 3′UTRs and CDSs, appeared very strongly correlated with one another among STAU1-bound transcripts (r = 0.75, Pearson correlation, Fig. 3B). Nonetheless, these two features were slightly additive, because the cumulative R2, which included both features, was greater than each individual alone (Fig. 3A), indicating that both features are, to some extent at least, independent determinants of STAU1 binding. Given that STAU1 binds dsRNAs, we reasoned that GC content might function to help the formation of stable RNA structures. We therefore used RNAfold (Hofacker and Stadler 2006) to predict RNA structures. Using a sliding window approach, involving RNA-folding prediction for every 100 nt with a 50-nt overlap between adjacent windows (Khaladkar et al. 2008), we found that transcripts with high SBIs (top 20%) had significantly lower minimum free energy (MFE) than those with low SBIs (bottom 20%) in both their 3′UTRs and CDSs (P < 2.2 × 10−16 for both, K–S test, Fig. 3C), suggesting that RNA secondary structures in both 3′UTRs and CDSs contribute to STAU1 binding. Note that, due to the use of 100-nt windows, this analysis is sensitive only to RNA structures shorter than IRAlus (each Alu arm is ∼300 nt) and thus reveals the contribution of short non-IRAlus RNA structures to STAU1 binding.
To examine RNA structures more directly, we analyzed RNA structure data generated by in vivo click-selective 2-hydroxyl acylation and profiling (icSHAPE) in HEK293 cells (Sun et al. 2019). When RNA structures were represented as Gini indices, we found that, compared to transcripts with low SBIs, transcripts with high SBIs tended to be more structured in both 3′UTRs (P = 2.9 × 10−4, K–S test, Fig. 3D, left) and CDSs (P = 1.2 × 10−2, K–S test, Fig. 3D, right). This result is consistent with the RNA structure prediction data (Fig. 3C), and it further confirms that both 3′UTR and CDS structures contribute to STAU1 binding.
Our findings are also supported by analysis of RIPiT reads in 3′UTRs and CDSs, which showed correlation between 3′UTR binding and CDS binding (r = 0.51, Pearson correlation, Supplemental Fig. S2E), both being similarly correlated with 3′READS + RIP SBI (r = 0.39 for 3′UTRs and = 0.37 for CDSs, Pearson correlation) (Fig. 3E).
Interestingly, the third and fourth top features, that is, number of introns and CDS size, contributed negatively to STAU1 binding (see their negative signs in Fig. 3A). These two features were correlated with each other (Fig. 3B). However, how they might cause inhibition of STAU1 binding is not immediately clear (see Discussion). In contrast, we found that 3′UTR size had a positive role in STAU1 binding (ranked seventh in our regression result), which is opposite to the role of CDS size. We thus wanted to further explore the relationship between CDS and 3′UTR sizes in STAU1 binding. We divided all transcripts into 25 groups based on CDS and 3′UTR sizes, independently. Supporting our conclusions, SBIs increased when 3′UTR size increased for a given CDS size group (Fig. 3F). This pattern was more obvious for transcripts with small CDS sizes as compared to those with large CDS sizes (the left two columns vs. the right two columns, Fig. 3F). Conversely, SBIs decreased as CDS size increased in all but the smallest 3′UTR size group (Fig. 3F). Taken together, our results indicate that while RNA structures in 3′UTRs and CDSs both contribute to STAU1 binding, 3′UTR size and CDS size have opposing roles in STAU1 binding.
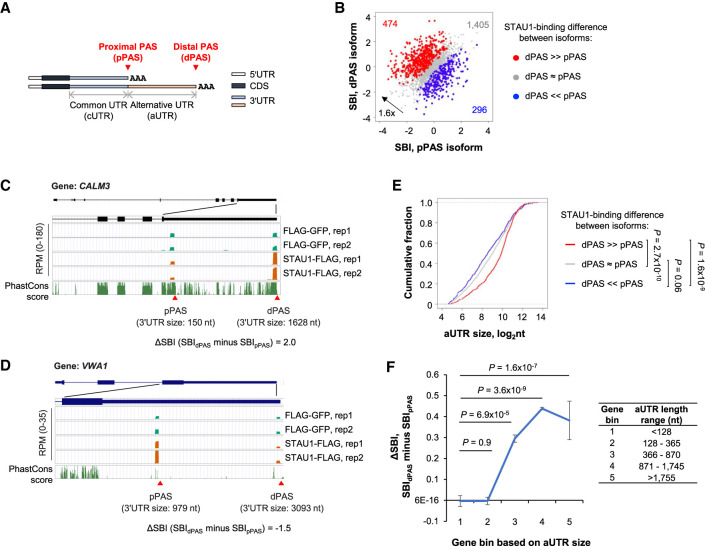
Longer 3′UTR isoforms have stronger STAU1 binding than short isoforms
Because 3′UTR isoforms from the same gene in general differ in their 3′UTR but not CDS or 5′UTR (illustrated in Fig. 4A), comparison of 3′UTR isoforms provides a means to interrogate the role of 3′UTRs without the influence of upstream RNA sequences. To this end, we compared the SBIs of the two most abundant 3′UTR isoforms produced from each gene, which we referred to as the proximal PAS (pPAS), and the distal PAS (dPAS) isoform (Fig. 4A). Note that pPAS demarcates the boundary between the common UTR (cUTR) and alternative UTR (aUTR), whereas dPAS defines the 3′ end of aUTR. We found that the number of genes whose longer dPAS isoform displayed greater STAU1 binding than the shorter pPAS isoform (474 red genes in Fig. 4B) outnumbered by 1.6-fold those showing the opposite trend (296 blue genes in Fig. 4B). This result was also confirmed by analysis of SBI deciles of short and long 3′UTR isoforms, which showed a depletion (blue colors) of genes whose short isoform had a larger SBI than the corresponding long isoform (Supplemental Fig. S4A).Two examples are shown in Figure 4C,D. Of the two 3′UTR isoforms deriving from the CALM3 gene, the longer isoform (3′UTR size = 1628 nt, SBI = 1.46) had stronger STAU1 binding than the shorter isoform (3′UTR size = 150 nt, SBI = −0.58) (Fig. 4C). In contrast, the shorter 3′UTR isoform of the VWA1 gene (3′UTR size = 979 nt, SBI = 1.4) had stronger STAU1 binding than its longer 3′UTR isoform (3′UTR size = 3093 nt, SBI = −0.05) (Fig. 4D).
FIGURE 4.
STAU1 binds APA isoforms with different affinities. (A) Schematics showing two alternative polyadenylation (APA) mRNA isoforms, that is, a proximal PAS (pPAS) isoform with a short 3′UTR, and a distal PAS (dPAS) isoform with a long 3′UTR. Common 3′UTR (cUTR) and alternative 3′UTR (aUTR) regions are indicated. (B) Scatter plot comparing the SBI of pPAS isoforms (x-axis) versus dPAS isoforms (y-axis). Red dots, genes whose dPAS isoform has a significantly higher SBI than its pPAS isoform. Blue dots, genes whose pPAS isoform has a significantly higher SBI than its dPAS isoform. Gray dots, genes whose dPAS and pPAS isoforms have similar SBIs. Significance of difference was based on P < 0.05 (DEXSeq) and fold change >1.2. (C) The CALM3 gene encodes a dPAS isoform with a higher SBI than its pPAS isoform. UCSC Genome Browser tracks are shown. 3′UTR sizes for the two isoforms are indicated. Genomic sequence conservation based on PhastCons scores derived from 100 vertebrate species is shown in the bottom-most strip. (D) As in panel C, except that the pPAS isoform encoded by the VWA1 gene has a higher SBI than the dPAS isoform. (E) Cumulative fraction function curves of aUTR sizes for three groups of genes (red, gray, and blue genes corresponding to those in panel B). P-values (K–S test) indicating significance of difference between gene groups are shown. (F) Mean ΔSBI between dPAS and pPAS for APA isoforms in different aUTR-size bins. Genes were evenly divided into five groups based on aUTR size (indicated in the table next to the graph).
In keeping with the overall trend that longer 3′UTRs had stronger STAU1 binding than shorter 3′UTRs (Fig. 2), we found that the regions present in longer 3′UTR isoforms but not in shorter 3′UTR isoforms (i.e., the aUTRs) of red genes were significantly longer than those of blue genes (P = 1.6 × 10−9, K–S test, Fig. 4E). To specifically explore the role of aUTRs, we divided genes into five bins based on aUTR size, again using the top two most abundant isoforms from each gene. Consistent with the notion that 3′UTR size plays a positive role in STAU1 binding, we found the longer the aUTR, the greater the SBI difference between the two isoforms, with longer isoforms being progressively stronger in STAU1 binding than shorter isoforms (P = 1.6 × 10−7 for gene bin 1 with the shortest aUTRs vs. gene bin 5 with the longest aUTRs, Wilcoxon test, Fig. 4F). Note that the difference in STAU1 binding between isoforms is not due to their differential abundance in the STAU1-FLAG- versus FLAG-EGFP-expressing cells because the blue and red genes in Figure 4B showed similar APA profiles in these cells (Supplemental Fig. S4B). Taken together, 3′UTR isoform analyses confirmed the positive role of 3′UTR size in STAU1 binding and indicate that APA isoforms can differ substantially in their interactions with STAU1.
aUTR IRAlus lead to 3′UTR isoform differences in STAU1 binding
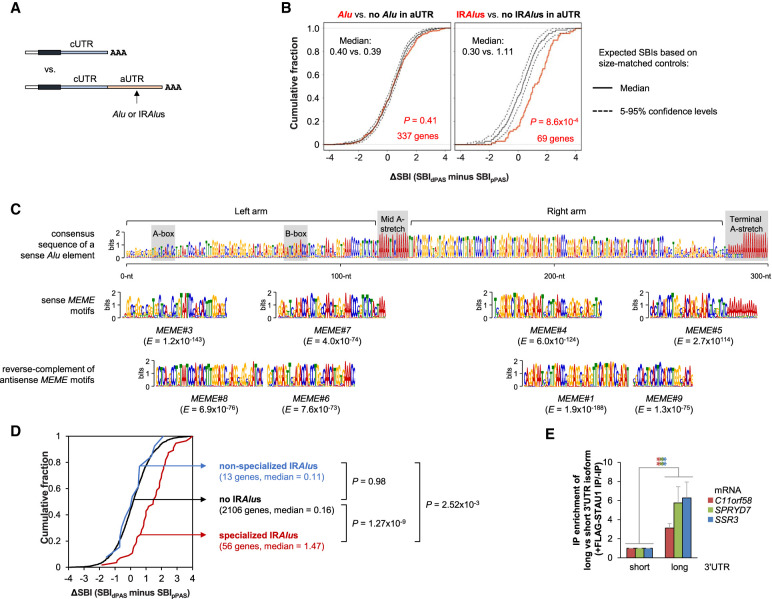
We next examined the transcript features that were responsible for STAU1 binding differences between 3′UTR isoforms. We first analyzed the contribution of Alus and IRAlus in alternative 3′UTRs to STAU1 mRNA binding (Fig. 5A). Consistent with whole transcript-based analyses (Fig. 2), using 3′UTR size-controlled sets, we found that the presence of single or multiple Alus in the same orientation in aUTRs did not differentiate 3′UTR isoforms in STAU1 binding (P = 0.41, K–S test) (Fig. 5B, left). In contrast, aUTR IRAlus significantly promoted STAU1 binding for the long isoform (P = 8.6 × 10−4, K–S test, Fig. 5B, right).
FIGURE 5.
Specialized IRAlus in alternative 3′UTRs lead to differential binding of STAU1 to different 3′UTR isoforms. (A) Schematic of SBI analysis using APA isoforms. The longer 3′UTR isoform with an Alu or IRAlus in its aUTR is compared to the shorter 3′UTR isoform. (B) Cumulative distribution function curves of ΔSBIs for gene transcripts having any number of Alus in the same orientation (left) or IRAlus (right) in their aUTRs, as illustrated in panel A. ΔSBI for each gene's transcripts is based on the SBI difference between the longer 3′UTR isoform and the shorter 3′UTR isoform. The two most abundant APA isoforms deriving from each gene were used for analysis. Observed values were compared to expected values calculated using aUTR size-matched controls. (C) Alignment of STAU1-binding MEME motifs with a consensus Alu element. Top, consensus sequence of a sense Alu element. Bottom, sense or antisense MEME motifs aligned to corresponding regions. Gray boxes indicate key features of an Alu element. The E-value (statistical value of a motif based on the log likelihood ratio, width, sites, the background letter frequencies, and the size of the training set) of each MEME motif is indicated. (D) Cumulative distribution function curves of the differential SBI of alternative APA mRNA isoforms classified based on their aUTR-bearing IRAlus and/or STAU1-binding MEME motifs. P-values (K–S test) indicating significance of difference between gene sets are indicated. (E) Histogram representation of RT-qPCR quantification of mRNAs whose aUTR promotes STAU1 binding and harbors a specialized IRAlus. RNA abundance after anti-FLAG immunoprecipitation (IP) of lysates of STAU1-KO HEK293T cells transiently expressing FLAG-STAU1, relative to abundance after anti-FLAG IP of lysates of STAU1-KO HEK293T cells expressing GFP-FLAG, were normalized to abundance before IP. Values depicting FLAG-STAU1 binding to the short isoforms are set to 1. Results are means ± SD. n = 3. (**) P < 0.01 by a two-tailed unpaired Student's t-test. Experimental design is detailed in Supplemental Figure S5D.
Using Multiple Em for Motif Elicitation (MEME) (Bailey and Elkan 1994) to identify motifs enriched in alternative 3′UTRs of transcripts with high SBIs (See Materials and Methods for detail), we found eight 29–50-nt motifs that corresponded to segments of Alu elements in either sense or antisense orientations (Fig. 5C). These motifs harbored notable differences as compared to the Alu consensus sequence (Supplemental Fig. S5A). Differences were largely conserved between sense and antisense MEME motifs, indicating that sense and antisense Alus harboring these motifs retain their ability to base-pair and form IRAlus specialized for efficient STAU1 binding. Consistent with this idea, we found that, in contrast to IRAlus-containing aUTRs lacking MEME motifs (i.e., nonspecialized IRAlus, blue line), IRAlus-containing aUTRs bearing sense and antisense MEME motifs (i.e., “specialized” IRAlus, red line) significantly increased STAU1 binding (Fig. 5D). Note that similar results were obtained when RIPiT or iCLIP-seq data were used (Supplemental Fig. S5B,C), corroborating 3′READS + RIP results.
We next developed an RT-qPCR assay to validate differential STAU1 binding to 3′UTR isoforms (illustrated in Supplemental Fig. S5D). In this assay, two sets of primers were used: One preferentially detected the short 3′UTR isoform, and the other exclusively detected the long 3′UTR isoform. Using this method and STAU1 RIP of lysates of STAU1-KO HEK293T cells transiently expressing FLAG-STAU1 or GFP-FLAG (as a control), we validated that STAU1 preferentially binds to the long 3′UTR isoform of three different transcripts, C11orf58, SPRYD7, and SSR3, all of which contain specialized IRAlus in their aUTR (Fig. 5E).
RNA structure and sequence motifs in aUTRs cause isoform differences in STAU1 binding
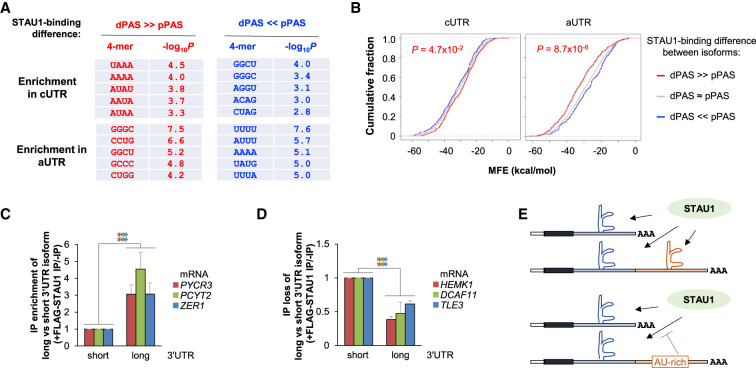
We next examined short sequence motifs in 3′UTR APA isoforms to confirm our finding that GC-rich sequences promote STAU1 binding (Fig. 3). Analyzing the enrichment of nucleotide tetramers, we found that the aUTRs deriving from genes whose longer isoform had greater binding (red genes in Fig. 4B) were enriched in GC-rich motifs (Fig. 6A, bottom left), whereas the aUTRs deriving from genes whose shorter isoform had greater binding (blue genes in Fig. 4B) were enriched in AU-rich motifs (Fig. 6A, bottom right). Interestingly, the cUTR of red genes was also enriched in AU-rich motifs, and the cUTR of blue genes was enriched in GC-rich motifs (Fig. 6A). Consistent with our previous findings, we found that RNAfold-predicted RNA structures in the aUTRs of red genes had significantly lower MFE values than those of blue genes (P = 8.7 × 10−6, K–S test, Fig. 6B, right). Conversely, predicted RNA structures in the cUTRs of blue genes had significantly lower MFE values than those of red genes (P = 4.7 × 10−2, K–S test, Fig. 6B, left).
FIGURE 6.
RNA sequence and structure features beyond IRAlus play roles in differential binding of 3′UTR isoforms to STAU1. (A) Top enriched tetramers (4-mers) in the cUTRs or aUTRs of different gene groups. Two groups of genes were analyzed for 4-mer enrichment in their cUTRs and aUTRs, that is, genes whose pPAS isoform has a higher SBI than its dPAS isoform (blue genes in Fig. 4B), and genes whose dPAS isoform has a higher SBI than its pPAS isoform (red genes in Fig. 4B). (B) Cumulative fraction function curves of minimum free energy (MFE) by Mfold analysis predicting the stability of RNA structures in the cUTRs or aUTRs of the three gene groups defined in Figure 4B. P-value (K–S test) indicating the significance of the difference between red genes (red line) and blue genes (blue line) is indicated. (C,D) RT-qPCR quantitation of mRNAs whose aUTR promotes (C) or inhibits (D) STAU1 binding. Experimental design is as in Figure 5E and detailed in Supplemental Figure S5D. mRNAs analyzed in C harbor predicted GC-rich secondary structures in their aUTR, whereas those in D contain sequences that reduce STAU1 binding, including U-rich (HEMK1), A-rich (DCAF11), or U-rich and A-rich (TLE3) motifs. (E) Schematic summarizing the sequence and structure analysis results.
Using RT-qPCR and STAU1 RIP of lysates of STAU1-KO HEK293T cells transiently expressing FLAG-STAU1, or as control GFP-FLAG, we validated for several genes differences in STAU1 binding between 3′UTR isoforms, including PYCR3, PCYT2, and ZER1 isoforms, whose aUTRs had high GC-content and low MFE values (Fig. 6C), as well as HEMK1, DCAF11, and TLE3 isoforms, whose aUTRs contained AU-rich stretches and were characterized by high MFE values (Fig. 6D). In summary, our 3′UTR isoform analyses validate the contributions of aUTR structures and sequences to STAU1 binding. Based on analyses of cUTRs and aUTRs, we conclude that AU-rich motifs that reside in cis to GC-rich secondary structures could mitigate STAU1-binding at GC-rich structures, possibly attributable to competition with other RBPs and/or remodeling of RNA secondary structures (illustrated in Fig. 6E).
STAU1 binding to 3′UTRs of IRAlus-less transcripts enhances polysome association
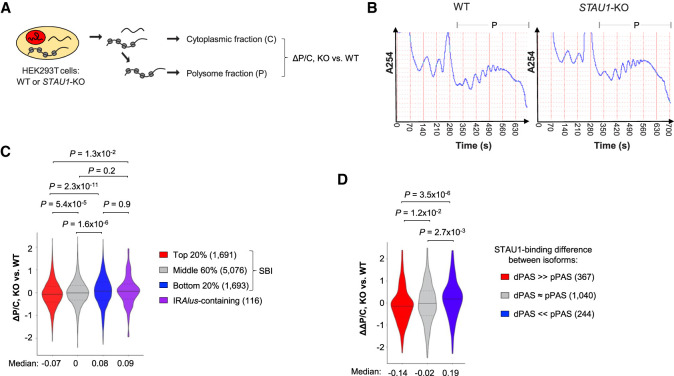
Given the extensive binding of STAU1 to translating transcripts (Ricci et al. 2014), we next wanted to understand how STAU1 binding impacts protein synthesis. To this end, we carried out polysome profiling using STAU1-KO and WT cells (Fig. 7A,B), extracted RNAs from polysomes as well as from total cytoplasm of each cell line, and subjected RNAs in each fraction to 3′READS+ analysis (Fig. 7A). For each transcript, we calculated the log2 ratio of its abundance (RPM) in the polysomal fraction relative to the total-cytoplasmic fraction as a measure of polysome association. This value is named P/C for simplicity. We then compared P/C values between STAU1-KO and WT cells, as represented by ΔP/C. As such, a low ΔP/C for a transcript indicates decreased polysome association in STAU1-KO cells as compared to WT cells.
FIGURE 7.
STAU1 binding promotes polysome association of IRAlus-less transcripts. (A) Schematic showing fractionation of WT or STAU1-KO (C8) HEK293T cells for polysome profiling and 3′READS analysis. Polysome association of a transcript is represented by P/C, which is the Log2(Ratio of RPM) for the transcript in the polysomal fraction relative to the cytoplasmic fraction. (B) Polysome profiles of WT (left) or STAU1-KO (right) HEK293T cells. The polysome fraction is indicated by “P.” (C) Violin plot showing P/C data in KO versus WT cells (ΔP/C) for different transcript groups based on SBI. Genes with 3′UTR IRAlus are analyzed as a separate group. Gene numbers are indicated in parentheses. P-values (Wilcoxon test) comparing different groups are shown. (D) Violin plot showing ΔΔP/C of genes, which represents the APA isoform difference in ΔP/C (KO vs. WT). Different groups of genes based on SBI difference between 3′UTR isoforms are shown. Gene numbers are indicated in parentheses. P-values (Wilcoxon test) comparing different groups are shown.
We found that transcripts with high SBIs (top 20% of IRAlus-less 3′UTRs) tended to have lower ΔP/C values as compared to transcripts with low SBIs (bottom 20%, median = −0.07 vs. 0.08, P = 2.3 × 10−11, Wilcoxon test, Fig. 7C). Interestingly, transcripts that contained 3′UTR IRAlus and a high SBI were characterized by much higher ΔP/C values than the IRAlus-less transcripts that had similar SBIs (P = 1.3 × 10−2, Wilcoxon test, Fig. 7C). In fact, no significant difference in ΔP/C could be discerned between the former and IRAlus-less transcripts that had low SBIs (P = 0.9, Wilcoxon test, Fig. 7C). Together, these results indicate that, in WT cells, STAU1 binding to its target transcripts enhances polysome association, unless binding is mediated by 3′UTR IRAlus.
We next compared the polysome association of 3′UTR isoforms (Fig. 7D). We used ΔΔP/C to reflect the difference in ΔP/C between two isoforms (long 3′UTR isoform/short 3′UTR isoform). As such, a low ΔΔP/C indicates that the long 3′UTR isoform has a greater decrease in polysome association than the short 3′UTR isoform in STAU1-KO cells as compared to WT cells. We found that genes whose long 3′UTR isoform had stronger STAU1 binding relative to their short isoform (red genes in Fig. 4B) displayed lower ΔΔP/C than those whose short isoform had stronger binding relative to their long isoform (blue genes in Fig. 4B) (median = −0.11 vs. 0.19, P = 3.5 × 10−6, Wilcoxon test, Fig. 7D). This result indicates that STAU1 binding to aUTRs selectively enhances polysome association to long 3′UTR isoforms but not to short 3′UTR isoforms. Note that we could not conduct reliable statistical tests using IRAlus-containing aUTRs because only a small number of genes (20) were available for analysis.
Taken together, our data indicate that STAU1 binding to 3′UTRs enhances polysome association, and 3′UTR isoforms can differ substantially in their polysome association due to selective STAU1 binding. However, IRAlus do not appear to enhance polysome association despite their strong STAU1 binding.
DISCUSSION
In this work, we examine STAU1 binding transcriptome-wide in HEK293T cells using 3′READS + RIP. Compared to other RIP methods, major advantages of our method are its level of sensitivity and quantitative analysis of APA isoforms. This allows us to specifically analyze 3′UTRs based on comparison of isoforms, without the confounding influences from the CDS or 5′UTR, which can be impacted by cotranscriptionally “imprinted” proteins at the nascent RNA stage, such as EJC components. As such, short isoforms are de facto internal controls for long isoforms. APA analysis would be difficult if RNA-seq or RNA fragment-based sequencing methods, such as RIPiT or iCLIP-seq, were used.
SBIs derived from 3′READS + RIP are generally correlated with RIPiT and iCLIP-seq data, instilling confidence in using our approach to unravel 3′UTR isoform-specific RNA–protein interactions. On the other hand, we note that our biological replicates were only modestly correlated. We think this may well be an intrinsic issue with STAU1 binding since STAU1 generally binds dsRNAs without a strong sequence preference. As such, changes in RNA structures during sample preparation can introduce data variability. In this vein, it is noteworthy that RIPiT and iCLIP data also showed a quite modest correlation, highlighting the complexity of studying STAU1 binding.
Consistent with the dsRNA-binding affinity of STAU1, transcripts with strong STAU1 binding tend to have either specialized IRAlus or a high GC content that is prone to form stable shorter secondary structures. This result is in line with previous studies by Ricci et al. (2014) and Sugimoto et al. (2015). Based on comparison of our data with RIPiT data (Ricci et al. 2014), the STAU1 binding we detect could be attributed to interactions with both 3′UTRs and CDSs. However, our 3′UTR isoform analysis allowed us to specifically pinpoint the contribution of 3′UTRs to STAU1 binding and also the consequence of 3′UTR STAU1 binding to polysome association. Not surprisingly, 3′UTR size also correlates with STAU1 binding, presumably because of the propensity of 3′UTRs to adopt additional RNA structures as they get longer. Using 3′UTR isoforms, we also found that AU-rich sequences appear to antagonize STAU1 binding to 3′UTR RNA structures.
One intriguing finding from our gene-feature analysis is the negative correlation between either CDS size or intron number and STAU1 binding. At this point, based on the level of the significance of the correlation, we cannot differentiate if CDS size or intron number is the primary cause. One could imagine that CDS size plays a more determinant role in STAU1 binding to translating mRNAs since remnants of introns should be gone from mRNAs once the post-splicing EJCs are removed by the translation of newly synthesized mRNA (Maquat et al. 2010). However, evidence of STAU1 binding to EJC constituents (Meyer and Gavis 2005) supports the hypothesis that intron number could influence STAU1 binding to nuclear mRNAs or to cytoplasmic mRNAs so as to promote their pioneer round(s) of translation, thereby predisposing subsequent steady-state rounds of translation (Maquat et al. 2010).
We detected significant changes in polysome association for STAU1 target transcripts in STAU1-KO cells as compared to WT cells, indicating that STAU1 generally promotes polysome association. This is consistent with previous reports implicating a role for STAU1 in translation (Ricci et al. 2014). However, 3′UTR IRAlus appear to have minimal impact on polysome association, indicating that 3′UTR IRAlus-containing transcripts are metabolized differently than other STAU1-bound transcripts. Whether this is due to structural or sequence differences and/or corecruitment or binding competition with other RNA-binding proteins is to be explored in the future.
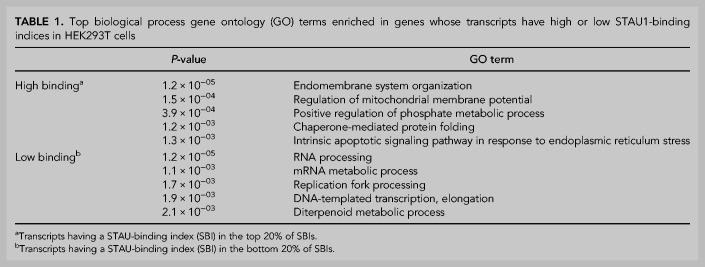
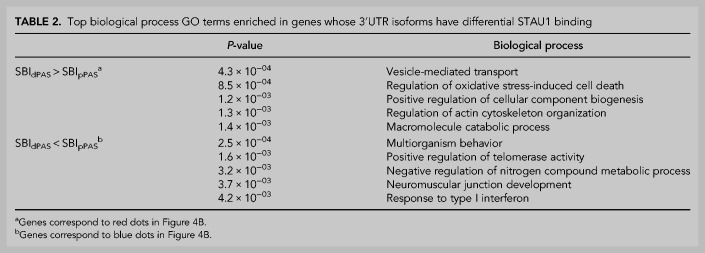
Using gene ontology (GO) analysis, we found that transcripts having a high SBI encode proteins involved in the organization of the endomembrane system and mitochondrial functions (Table 1). In addition, genes whose aUTR promotes STAU1 binding (red genes in Fig. 4B) tend to encode proteins that are associated with vesicle-mediated transport and cytoskeleton organization (Table 2). Together, these data indicate that STAU1 binding globally contributes to cellular and organelle membranes as well as cytoskeleton organization, function and regulation. In addition, we found that transcripts that are not preferentially bound by STAU1 or whose aUTR inhibits STAU1 binding (blue genes in Fig. 4B) are globally associated with the maintenance of genome integrity, the regulation of mRNA metabolism, and the inflammation response (Tables 1, 2). How APA alters those numerous aspects of cellular biology through perturbing STAU1 binding in different proliferation, differentiation, and stress conditions, under which APA is highly regulated (Tian and Manley 2017), is an open question.
TABLE 1.
Top biological process gene ontology (GO) terms enriched in genes whose transcripts have high or low STAU1-binding indices in HEK293T cells
TABLE 2.
Top biological process GO terms enriched in genes whose 3′UTR isoforms have differential STAU1 binding
MATERIALS AND METHODS
Cell lines and cell culture
Human embryonic kidney (HEK)293T cells were purchased from ATCC and propagated in Dulbecco's modified Eagle's medium (Gibco) supplemented with 10% fetal bovine serum. Where specified, cells were transfected with Lipofectamine 2000 (Invitrogen). STAU1-KO (STAU1 C8) and isogenic control (C9) HEK293T-cell lines were established using the CRISP–Cas9n double-nicking strategy as described previously (Elbarbary et al. 2017). Briefly, STAU1-KO C8 was generated using two small-guide (sg)RNAs synthesized from pSpCas9n(BB)-2A-Puro (Addgene) after individually inserting the following synthetic DNAs: STAU1-sgDNA5s (5′-CACCGCCATATTCTGGGCTTGTCT-3′ annealed to 3′-AAACAGACAAGCCCAGAATATGGC-3′), and STAU1-sgDNA6as (5′-CACCGGATCAATCCCATTAGCCGA-3′ annealed to 3′-AAACGTCGGCTAATCGGATTGATC-5′), where underlined residues represent the STAU1 gene-targeted sequences. Target-sites for the resulting sgRNAs were selected using CRISPR Design Tool (http://www.genome-engineering.org/crispr/). The isogenic control C9 line was selected among the isolated clones transfected with the plasmids encoding the CRISPR–Cas9n machinery and the guide RNAs, but in which STAU1 protein levels remained unchanged as compared to WT HEK293T.
Western blotting
Western blotting was performed as described (Elbarbary et al. 2013) using the following antibodies: anti-STAU1 (Lucas et al. 2018), anti-FLAG (Sigma), anti-CANX (Enzo Life Sciences), anti-NONO (Bethyl Labs), and anti-STAU2 (Sigma).
Anti-FLAG immunoprecipitations
Total-cell lysates from STAU1-KO HEK293T cells (C8) transiently expressing FLAG-GFP or STAU1-FLAG were prepared using Hypotonic Gentle Lysis Buffer (10 mM Tris [pH 7.4], 10 mM NaCl, 10 mM EDTA, 1% [w/w] Triton X-100, 1× Protease and Phosphatase Inhibitor [Pierce]) and two 30-sec rounds of sonication (Branson Sonifier 250, duty cycle 30%, output control = 3). The NaCl concentration was adjusted to 150 mM, and insoluble debris was pelleted by centrifugation at 16,000g for 10 min. Anti-FLAG immunoprecipitations (IPs) were then performed using supernatants as described previously (Cho et al. 2018).
RT-qPCR
RT was performed using total-cell RNA or coimmunoprecipitated RNA extracted using TRIzol reagent (Life Technologies) and then treated with DNase I (Promega Corp.). cDNA was synthesized for 2 h at 50°C using SuperScript III (Invitrogen), random hexamers (N6, 5 ng/µL), and a pool of anchored oligos (2.5 pmol/µL = 2.5 µM) that consisted of 5′-universal adapter-oligo(dT)-VN anchor-3′, where V is A,G, or C and N is any deoxynucleotide (5′-GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAG-TTTTTTTTTTTTTTTTTTTT-VN-3′). qPCR was undertaken using the StepOne Real-Time PCR System (Applied Biosystems), Fast SYBR-Green Master Mix (Applied Biosystems), and PCR primers specified in Supplemental Table S2. Briefly, cDNA deriving from short 3′UTR isoforms used a forward primer annealing to each cUTR and a reverse primer annealing to the universal adapter. qPCR of cDNA deriving from long (typically >1 kb) 3′UTRs will not be amplified by the cUTR-specific and universal primers due to short PCR-amplification times. Instead qPCR cDNA deriving from long 3′UTRs used forward and reverse primers specific to each aUTR.
Cell fractionation and polysome profiling
Separation of HEK293T-cell nuclear and cytoplasmic fractions (Fig. 1) was performed using NE-PER Nuclear and Cytoplasmic Extraction Reagents (Thermo Fisher Scientific) following manufacturer instructions. Polysome profiling (Fig. 7) was performed as described (Esposito et al. 2010) with minor modifications. HEK293T cells were grown to ∼80%–90% confluency. Ten minutes before harvesting, 100 μg/mL of cycloheximide (Sigma) was added to the cell-culture medium to freeze ribosomes on translationally active mRNAs. Cells were then washed once with PBS supplemented with 100 μg/mL of cycloheximide, exposed to trypsin-EDTA supplemented with 100 μg/mL of cycloheximide, washed twice with prechilled PBS supplemented with 100 μg/mL of cycloheximide, resuspended, and incubated in Lysis Buffer (20 mM HEPES-KOH pH 7.5, 15 mM MgCl2, 80 mM KCl, 1% Triton X-100, 2 mM DTT, 100 μg/mL cycloheximide, 1× SIGMAFAST Protease Inhibitor Cocktail [Sigma], and 200 U/mL SuperaseIn [Thermo Fisher Scientific]) for 10 min. During the incubation, cells were gently sheared three times through a 26-gauge needle. A percentage (10%) of each cell lysate was saved for total-cell RNA extraction. The rest of each cell lysate was centrifuged at 700g and 4°C for 5 min in a micro-centrifuge to pellet the nuclei. Cytoplasmic RNA was isolated from ∼20% of the supernatant using TRIzol (Thermo Fisher Scientific), while the rest of the supernatant was transferred to a new tube and centrifuged at 14,000g for 5 min to remove mitochondria. The resulting supernatant was layered onto a 10-mL 10%–50% linear sucrose gradient in a polyallomer tube (Beckman Coulter) containing 20 mM HEPES-KOH pH 7.5, 15 mM MgCl2, 80 mM KCl, 2mM DTT, and 100 μg/mL of cycloheximide, and centrifuged in an SW-41 Ti rotor at 39,000 rpm and 4°C for 2 h. The gradient was fractionated using a system comprised of a syringe pump (Harvard Apparatus model 11), a density-gradient fractionator (Brandel), and an ISCO UA-6 UV/VIS detector. The polysomal fraction was incubated at 65°C for 5 min with 25 mM EDTA pH 8.0, 10 mM Tris-HCl pH 7.0, and 1% SDS. RNA was extracted using phenol/chloroform and precipitated using ethanol. The quality of all RNA was checked using an Agilent RNA 6000 Pico Kit.
3′READS+ library construction and sequencing
The READS+ procedure has been previously described (Zheng et al. 2016). Briefly, poly(A)+ RNA in 1 µg of total-cell RNA was captured using 10 µL of oligo(dT)25 magnetic beads (NEB) in 100 µL of 1× Binding Buffer (10 mM Tris-Cl pH 7.5, 150 mM NaCl, 1 mM EDTA, and 0.05% TWEEN 20) and fragmented on the beads using 1.5 U of RNase III (NEB) in 30 µL of RNase III Buffer (10 mM Tris-Cl pH 8.3, 60 mM NaCl, 10 mM MgCl2, and 1 mM DTT) for 15 min at 37°C. After washing away unbound RNA fragments using Binding Buffer, poly(A)+ fragments were eluted from the beads using TE Buffer (10 mM Tris-Cl, 1 mM EDTA, pH 7.5) and precipitated using ethanol, followed by ligation to 3 pmol of heat-denatured 5′-adapter (5′-CCUUGGCACCCGAGAAUUCCANNNN-3′) in the presence of 1 mM ATP, 0.1 µL of SuperaseIn (ThermoFisher Scientific), and 0.25 µL of T4 RNA ligase 1 in a 5 µL reaction at 22°C for 1 h. Ligation products were captured using 10 pmol of biotin-T15-(+TT)5 attached to 12 µL of Dynabeads MyOne Streptavidin C1 (ThermoFisher Scientific). After washing with Washing Buffer (10 mM Tris-Cl pH 7.5, 1 mM NaCl, 1 mM EDTA, and 0.05% TWEEN 20), RNA fragments on the beads were incubated with 0.01 U/µL of RNase H (Epicentre) for 30 min at 37°C in 30 µL of RNase H Buffer (50 mM Tris-Cl pH 7.5, 5 mM NaCl, 10 mM MgCl2, and 10 mM DTT). After washing with RNase H Buffer, RNA fragments were eluted from the beads in Elution Buffer (1 mM NaCl, 1 mM EDTA pH7.5, and 0.05% TWEEN 20) at 50°C, precipitated using ethanol, and then ligated to 3 pmol of heat-denatured 5′-adenylated 3′-adapter (5′-rApp/NNNGATCGTCGGACTGTAGAACTCTGAAC/3ddC-3′) using 0.25 µL of T4 RNA ligase 2 (truncated KQ version) for 1 h at 22°C in a 5 µL reaction containing 15% PEG 8000 and 0.2 µL of SuperaseIn. Ligation products were precipitated and reverse-transcribed using M-MLV reverse transcriptase, and PCR-amplified using Phusion high-fidelity DNA polymerase (NEB) and bar-coded PCR primers (Sigma-Aldrich) for 15 cycles. Both RT primer (5′-GTTCAGAGTTCTACAGTCCGACGATC-3′) and PCR primers with indexes have been described previously (Hoque et al. 2013). PCR products were size-selected twice using 0.6 volume of AMPure XP beads (Beckman Coulter) relative to the PCR reaction volume to remove large DNA molecules, followed by an additional 0.4 volume of beads to remove small DNA molecules. The eluted DNA was selected again with 1 volume of AMPure XP beads to further remove small DNA molecules. The sizes of the libraries eluted from the AMPure beads were examined using a high-sensitivity DNA kit (Agilent) and an Agilent Bioanalyzer. Library concentrations were measured using the Qubit dsDNA HS Assay Kit (ThermoFisher Scientific). Equal amounts of libraries were mixed and sequenced using an Illumina NextSeq 500 (1 × 75 bases).
Analysis of 3′READS+ data
The sequence corresponding to the 5′-adapter was removed from raw 3′READS+ reads using Cutadapt (Martin 2011). Reads with short inserts (<23 nt) were discarded, and retained reads were mapped to the human (hg19) genome using STAR (Langmead and Salzberg 2012). The three or six 5′ random nts from the 3′-adapter were removed before mapping using the setting “−5 6” in bowtie2. Reads with a mapping quality score (MAPQ) of ≥10 were kept for further analysis. Reads with ≥2 nongenomic 5′-Ts after alignment were called PAS reads. Cleavage sites within 24 nt from one another were clustered as previously described (Hoque et al. 2013). Only PASs with ≥2 PAS reads were used for analysis.
Size-controlled analysis of Alu-containing transcripts
Transcripts were categorized into groups based on the number of 3′UTR Alus or the presence of 3′UTR IRAlus. For each group, a control set containing the same number of transcripts was generated. For each transcript in the Alu-containing group, an Alu-less transcript in the same 3′UTR size decile group was randomly selected to ensure control for size. The random selection was repeated 100 times. The fifth-percentile and 95th-percentile SBIs based on the 100 random sets were calculated.
Analysis of RiPIT, iCLIP, and icSHAPE data
STAU1 RIPiT bigwig data (Ricci et al. 2014) were downloaded from the NCBI GEO database. RIPiT read counts in different regions of each transcript were extracted from the bigwig data. Log2ratio of RPM (wild type STAU1 vs. mutant STAU1) was calculated to indicate STAU1 binding. STAU1 iCLIP (Sugimoto et al. 2015) raw data were downloaded from the NCBI GEO database. After computationally removing the Illumina universal adapter, sequences were mapped to the human genome (hg19) using STAR. Uniquely mapped reads were further processed using first-aligned positions. RPMs (reads per million mapped) were calculated for each sample. In vivo HEK293 icSHAPE scores (Sun et al. 2019) were extracted for different transcript regions. Gini indices were calculated using a sliding window of 20 nt over a given transcript region, and the median Gini index of each region was used for analysis.
Transcript feature analysis
Features were based on the RefSeq database. Linear model analysis was carried out by using the linear model (lm) function in R program. Pearson and Spearman correlation analyses were carried out to examine relationships between features.
MEME analysis and annotation of specialized IRAlus
MEME (Bailey and Elkan 1994) (v5.1.1) was used to discover medium-sized motifs (6−50 nt) that robustly and significantly promote STAU1 binding in each of the two 3′READS + RIP replicates (MEME discriminative mode, zero or one occurrence per sequence [zoops]). Transcripts whose aUTR robustly and significantly promotes STAU1 binding were defined as those for which the average SBI of the longest 3′UTR isoform was significantly (Fisher < 0.05) at least twofold higher (ΔSBI > 1) than the average SBI of the shortest 3′UTR isoform. Transcripts having a ΔSBI > 1 included those with a ΔSBI = + ∞, that is, genes whose long 3′UTR isoform was not detected in the FLAG-GFP IP, and also genes whose short 3′UTR isoform was not detected in the STAU1-FLAG IP. Controls utilized transcripts for which the average SBI of the longest 3′UTR isoform was significantly (Fisher < 0.05) at least twofold lower (ΔSBI < −1) than the average SBI of the shortest 3′UTR isoform. Transcripts having a ΔSBI < −1 included those with a ΔSBI = −∞, that is, genes whose short 3′UTR isoform is not detected in the FLAG-GFP IP, and also genes whose long 3′UTR isoform is not detected in the STAU1-FLAG IP. The consensus sense or reverse-complement sequence of eight of the ten MEME motifs with the highest significant enrichment score was aligned to a consensus Alu sequence using Clustal Omega (Madeira et al. 2019). The consensus Alu motif (and sequence) was derived from 342 Alu elements (Konkel et al. 2015) using MEME (classic mode, zoops). MEME motifs were then mapped onto aUTRs using FIMO (Find Individual Motif Occurrences, MEME suite v5.1.1) (Grant et al. 2011) and a corrected Q-value of 0.001. IRAlus in aUTRs having at least one sense and one antisense Alu MEME motif were annotated as specialized IRAlus.
Gene ontology analysis
Gene ontology (GO) analysis was carried out by using the GOstats Bioconductor package (Falcon and Gentleman 2007). Generic terms (associated with more than 1000 genes) were discarded. To reduce redundancy in reporting, any GO term with a >75% gene overlap with a more significant term was discarded.
DATA DEPOSITION
All custom-made code and scripts for processing of sequencing data and quantification analyses were written in Perl or R and will be provided upon request. Sequencing data sets generated in this study have been deposited in the NCBI GEO database under accession number GSE148771. All data sets used in this study are listed in Supplemental Table S1.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank Max Popp and members of the BT laboratory for comments on the manuscript. We thank Reyad Elbarbary, Jason R. Myers, Ashwin Kumar, and Bronwyn Lucas for participation at early stages of this work. Research on Alu elements in the Maquat laboratory is supported by National Institutes of Health (NIH) R37 GM074593 to L.E.M. H.C. and X.R. were partially supported by American Heart Association (AHA) postdoctoral fellowships 16POST27260273 and 18POST33960339, respectively. Work in the Tian laboratory is supported by NIH grants R01 GM129069 and GM084089.
Author contributions: B.T. and L.E.M. designed the research plan. H.C. evaluated the integrity of wild-type and STAU1-KO HEK293T cell lines with and without fractionation, and with and without immunoprecipitation, and confirmed RNA-seq and RIP-seq data using transcript-specific RT-qPCR. D.Z. fractionated wild-type and STAU1-KO cells, conducted polysome profiling, and generated all the 3′READS+ libraries. D.Z. and W.W. carried out computational data analysis. X.R. participated in discussions and provided guidance and some computational analyses. B.T. and L.E.M. wrote the manuscript with help from X.R.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.076133.120.
REFERENCES
- Bailey TL, Elkan C. 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol 2: 28–36. [PubMed] [Google Scholar]
- Boulay K, Ghram M, Viranaicken W, Trepanier V, Mollet S, Frechina C, DesGroseillers L. 2014. Cell cycle-dependent regulation of the RNA-binding protein Staufen1. Nucleic Acids Res 42: 7867–7883. 10.1093/nar/gku506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LL, Yang L. 2017. ALUternative regulation for gene expression. Trends Cell Biol 27: 480–490. 10.1016/j.tcb.2017.01.002 [DOI] [PubMed] [Google Scholar]
- Cho H, Rambout X, Gleghorn ML, Nguyen PQT, Phipps CR, Miyoshi K, Myers JR, Kataoka N, Fasan R, Maquat LE. 2018. Transcriptional coactivator PGC-1α contains a novel CBP80-binding motif that orchestrates efficient target gene expression. Genes Dev 32: 555–567. 10.1101/gad.309773.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeCerbo J, Carmichael GG. 2005. SINEs point to abundant editing in the human genome. Genome Biol 6: 216 10.1186/gb-2005-6-4-216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deininger P. 2011. Alu elements: know the SINEs. Genome Biol 12: 236 10.1186/gb-2011-12-12-236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbarbary RA, Li W, Tian B, Maquat LE. 2013. STAU1 binding 3′ UTR IRAlus complements nuclear retention to protect cells from PKR-mediated translational shutdown. Genes Dev 27: 1495–1510. 10.1101/gad.220962.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbarbary RA, Lucas BA, Maquat LE. 2016. Retrotransposons as regulators of gene expression. Science 351: aac7247 10.1126/science.aac7247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbarbary RA, Miyoshi K, Myers JR, Du P, Ashton JM, Tian B, Maquat LE. 2017. Tudor-SN-mediated endonucleolytic decay of human cell microRNAs promotes G1/S phase transition. Science 356: 859–862. 10.1126/science.aai9372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esposito AM, Mateyak M, He D, Lewis M, Sasikumar AN, Hutton J, Copeland PR, Kinzy TG. 2010. Eukaryotic polyribosome profile analysis. J Vis Exp 10.3791/1948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falcon S, Gentleman R. 2007. Using GOstats to test gene lists for GO term association. Bioinformatics 23: 257–258. 10.1093/bioinformatics/btl567 [DOI] [PubMed] [Google Scholar]
- Gleghorn ML, Maquat LE. 2014. ‘Black sheep’ that don't leave the double-stranded RNA-binding domain fold. Trends Biochem Sci 39: 328–340. 10.1016/j.tibs.2014.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleghorn ML, Gong C, Kielkopf CL, Maquat LE. 2013. Staufen1 dimerizes through a conserved motif and a degenerate dsRNA-binding domain to promote mRNA decay. Nat Struct Mol Biol 20: 515–524. 10.1038/nsmb.2528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong C, Maquat LE. 2011. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3′ UTRs via Alu elements. Nature 470: 284–288. 10.1038/nature09701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong C, Tang Y, Maquat LE. 2013. mRNA-mRNA duplexes that autoelicit Staufen1-mediated mRNA decay. Nat Struct Mol Biol 20: 1214–1220. 10.1038/nsmb.2664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant CE, Bailey TL, Noble WS. 2011. FIMO: scanning for occurrences of a given motif. Bioinformatics 27: 1017–1018. 10.1093/bioinformatics/btr064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Häsler J, Strub K. 2006. Alu elements as regulators of gene expression. Nucleic Acids Res 34: 5491–5497. 10.1093/nar/gkl706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofacker IL, Stadler PF. 2006. Memory efficient folding algorithms for circular RNA secondary structures. Bioinformatics 22: 1172–1176. 10.1093/bioinformatics/btl023 [DOI] [PubMed] [Google Scholar]
- Hoque M, Ji Z, Zheng D, Luo W, Li W, You B, Park JY, Yehia G, Tian B. 2013. Analysis of alternative cleavage and polyadenylation by 3′ region extraction and deep sequencing. Nat Methods 10: 133–139. 10.1038/nmeth.2288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaladkar M, Liu J, Wen D, Wang JT, Tian B. 2008. Mining small RNA structure elements in untranslated regions of human and mouse mRNAs using structure-based alignment. BMC Genomics 9: 189 10.1186/1471-2164-9-189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konkel MK, Walker JA, Hotard AB, Ranck MC, Fontenot CC, Storer J, Stewart C, Marth GT, Batzer MA. 2015. Sequence analysis and characterization of active human Alu subfamilies based on the 1000 Genomes Pilot Project. Genome Biol Evol 7: 2608–2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazzaretti D, Bandholz-Cajamarca L, Emmerich C, Schaaf K, Basquin C, Irion U, Bono F. 2018. The crystal structure of Staufen1 in complex with a physiological RNA sheds light on substrate selectivity. Life Sci Alliance 1: e201800187 10.26508/lsa.201800187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JY, Ji Z, Tian B. 2008. Phylogenetic analysis of mRNA polyadenylation sites reveals a role of transposable elements in evolution of the 3′-end of genes. Nucleic Acids Res 36: 5581–5590. 10.1093/nar/gkn540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas BA, Lavi E, Shiue L, Cho H, Katzman S, Miyoshi K, Siomi MC, Carmel L, Ares M Jr, Maquat LE. 2018. Evidence for convergent evolution of SINE-directed Staufen-mediated mRNA decay. Proc Natl Acad Sci 115: 968–973. 10.1073/pnas.1715531115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madeira F, Park YM, Lee J, Buso N, Gur T, Madhusoodanan N, Basutkar P, Tivey ARN, Potter SC, Finn RD, et al. 2019. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res 47: W636–W641. 10.1093/nar/gkz268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maquat LE, Tarn WY, Isken O. 2010. The pioneer round of translation: features and functions. Cell 142: 368–374. 10.1016/j.cell.2010.07.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martel C, Dugre-Brisson S, Boulay K, Breton B, Lapointe G, Armando S, Trepanier V, Duchaine T, Bouvier M, Desgroseillers L. 2010. Multimerization of Staufen1 in live cells. RNA 16: 585–597. 10.1261/rna.1664210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnetjournal 17: 3 10.14806/ej.17.1.200 [DOI] [Google Scholar]
- Mayr C. 2019. What are 3′ UTRs doing? Cold Spring Harb Perspect Biol 11 10.1101/cshperspect.a034728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer EL, Gavis ER. 2005. Staufen does double duty. Nat Struct Mol Biol 12: 291–292. 10.1038/nsmb0405-291 [DOI] [PubMed] [Google Scholar]
- Park E, Maquat LE. 2013. Staufen-mediated mRNA decay. Wiley Interdiscip Rev RNA 4: 423–435. 10.1002/wrna.1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricci EP, Kucukural A, Cenik C, Mercier BC, Singh G, Heyer EE, Ashar-Patel A, Peng L, Moore MJ. 2014. Staufen1 senses overall transcript secondary structure to regulate translation. Nat Struct Mol Biol 21: 26–35. 10.1038/nsmb.2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai M, Shiromoto Y, Ota H, Song C, Kossenkov AV, Wickramasinghe J, Showe LC, Skordalakes E, Tang HY, Speicher DW, et al. 2017. ADAR1 controls apoptosis of stressed cells by inhibiting Staufen1-mediated mRNA decay. Nat Struct Mol Biol 24: 534–543. 10.1038/nsmb.3403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci 102: 15545–15550. 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto Y, Vigilante A, Darbo E, Zirra A, Militti C, D'Ambrogio A, Luscombe NM, Ule J. 2015. hiCLIP reveals the in vivo atlas of mRNA secondary structures recognized by Staufen 1. Nature 519: 491–494. 10.1038/nature14280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Fazal FM, Li P, Broughton JP, Lee B, Tang L, Huang W, Kool ET, Chang HY, Zhang QC. 2019. RNA structure maps across mammalian cellular compartments. Nat Struct Mol Biol 26: 322–330. 10.1038/s41594-019-0200-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian B, Manley JL. 2017. Alternative polyadenylation of mRNA precursors. Nat Rev Mol Cell Biol 18: 18–30. 10.1038/nrm.2016.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Nambiar R, Zheng D, Tian B. 2018. PolyA_DB 3 catalogs cleavage and polyadenylation sites identified by deep sequencing in multiple genomes. Nucleic Acids Res 46: D315–D319. 10.1093/nar/gkx1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng D, Liu X, Tian B. 2016. 3′READS+, a sensitive and accurate method for 3′ end sequencing of polyadenylated RNA. RNA 22: 1631–1639. 10.1261/rna.057075.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.