Abstract
Background
Although most observational studies identify viral or bacterial pathogens in 50% or less of patients hospitalized with community-acquired pneumonia (CAP), we previously demonstrated that a multi-test bundle (MTB) detected a potential pathogen in 73% of patients. This study compares detection rates for potential pathogens with the MTB versus the Biofire® Pneumonia FilmArray® panel (BPFA) multiplex PCR platform and presents an approach for integrating BPFA results as a foundation for subsequent antibiotic stewardship (AS) activities.
Methods
Between January 2017 to March 2018, all patients admitted for CAP were enrolled. Patients were considered evaluable if all elements of the MTB and the BPFA were completed, and they met other a priori inclusion criteria. The primary endpoint was the percentage of potential pathogens detected using the MTB (8 viral and 6 bacterial targets) versus the BPFA (8 viral and 18 bacterial targets). Blood and sputum cultures were performed on all patients. Two or more procalcitonin (PCT) levels assisted clinical assessments as to whether detected bacteria were invading or colonizing.
Results
Of 585 enrolled patients, 274 were evaluable. A potential viral pathogen was detected in 40.5% with MTB versus 60.9% of patients with BPFA with an odds ratio (95% CI) of 9.00 (4.12 to 23.30) p<0.01. A potential bacterial pathogen was identified in 66.4% with the MTB vs 75.5% with the BPFA odds ratio (95% CI) of 2.09 (1.24 to 3.59), p 0.003). Low PCT levels helped identify detected bacteria as colonizers.
Key words: Community-acquired pneumonia, diagnostics, pneumonia, filmarray, procalcitonin
SUMMARY.
Compared to a standard multi-test diagnostic bundle, the sputum Biofire\elsamp #x00AE; Pneumonia FilmArray\elsamp #x00AE; multiplex platform, significantly increased the detection of potential viral and bacterial pathogens in hospitalized adult patients with community-acquired pneumonia.
Alt-text: Unlabelled box
Summary.
For hospitalized patients with CAP, the Biofire\elsamp #x00AE; Pneumonia FilmArray\elsamp #x00AE; multiplex platform significantly increased the number of patients with detectable viral and bacterial potential pathogens and, thereby, provided key data for AS activities.
Alt-text: Unlabelled box
1. INTRODUCTION
Pneumonia is the leading cause of death due to infection in the United States with an estimated 1.2 million annual hospitalizations (Self et al., 2017). Management of community-acquired pneumonia (CAP) entails empiric antibiotic therapy that targets the most likely bacterial pathogens. The American Thoracic Society-Infectious Diseases Society of America CAP guidelines reserve microbiologic testing for patients with severe CAP (Metlay et al., 2019). Further, “rapid, cost-effective, sensitive, and specific diagnostic tests to identify organisms causing CAP have potential to improve routine care by supporting the use of targeted therapy . . .” (Metlay et al., 2019)
A benchmark study reported that microbiologic testing identified the microbiologic etiology in only 853 of 2259 (38%) hospitalized CAP patients (Jain et al., 2015). To improve this diagnostic yield, we reported that a multi-test bundle (MTB) consisting of sputum and blood bacterial cultures, polymerase chain reaction (PCR) testing of nasopharyngeal swabs (NP) samples by the multiplex Biofire® (Biofire® Co., Salt Lake City, Utah) Respiratory FilmArray® panel, anterior nasal swab PCR for Staphylococcus aureus, nasopharyngeal swab for Streptococcus pneumoniae, and urine antigen testing for S. pneumoniae and Legionella pneumophila detected a potential bacterial and/or viral pathogen in 73% of patients (Gilbert et al., 2016, Carvalho Mda et al., 2007). The feasibility of this approach was hampered by long turn-around times (TAT) for multiple MTB components, and the failure of the MTB to detect common potential bacterial pathogens, such as Moraxella and Haemophilus species.
The Biofire® Pneumonia FilmArray® (BPFA) panel probes sputum samples for 8 respiratory viruses and 18 bacteria with a 1 to 2-hour TAT. However, the performance of BPFA as compared with multi-element testing, such as MTB, has neither been evaluated in large studies, nor integrated into clinical practice to identify invasive vs colonizing pathogens. Herein, we compare the detection of potential viral and bacterial pathogens with MTB as compared with BPFA. We hypothesized that the BPFA would detect more potential viral and bacterial pathogens than MTB. We also present our approach to integrating BPFA results into clinical practice by eliminating unnecessary tests and serving as the basis of antibiotic stewardship (AS) activities.
2. MATERIALS AND METHODS
2.1. Study Patients
The study was registered with clinicaltrials.gov (NC02580384) as a prospective, non-blinded comparison of pathogen detection with patients as their own controls was conducted at Providence Portland Medical Center, a 480-bed community teaching hospital in Portland, Oregon between January 2017 and March 2018 and approved by the Privacy Board and the Institutional Board (IRB) that waived informed consent. Subjects were identified from adults (over the age of 18) presenting to the Emergency Department with respiratory symptoms diagnosed by emergency physicians as CAP based on their usual clinical practice. All patients were given a consecutive study number and their clinical data were entered into an online database (FileMaker 13®, Claris International Inc, Santa Clara, CA). Within two days, a study investigator (DNG or JEL) reviewed the database to ascertain if the patient was evaluable based on the presence of all inclusion criteria:
-
1.
Production of sputum, or sputum equivalent (e.g. endotracheal tube aspirate, bronchoalveolar lavage) for testing.
-
2.
Availability of all elements of the MTB and BPFA (Sputum or sputum equivalent) and blood samples for PCT levels.
-
3.
Absence of any extrapulmonary infection that required antibiotics (e.g., pyelonephritis).
-
4.
No transition to comfort care within 24 hours of admission.
-
5.
No history of a witnessed aspiration of gastric contents.
-
6.
Diagnosis not changed by treating physicians to a non-infectious disease (e.g., pulmonary edema) within 48 hours.
2.2. Specimen collection
Emergency Department (ED) nurses collected two blood culture samples and expectorated or hypertonic saline-induced sputum (or endotracheal aspirate) for BPFA; in addition, sputum was submitted for culture. For MTB, nasopharyngeal swab samples were submitted for the BioFire® Respiratory FilmArray® panel and lab-generated PCR for S. pneumoniae (Carvalho Mda et al., 2007); anterior nasal swab for S. aureus PCR (BD Max SR®), and urine for antigen detection (Binaxnow, Abbott Labs, Abbott ParK, IL) of S. pneumoniae and L. pneumophila (Table 1 ). Blood was collected for PCT testing (Used Vidas B.R.A.H.M.S instrument and PCT reagents from Biomerieux, Durham, NC) with PCT retesting 4-6 hours later. A rapid response laboratory adjacent to the ED received all samples.
Table 1.
Components of Multi-Test Bundle and BioFire® Pneumonia FilmArray® Testing
| Multi-Test Bundle | BioFire® Pneumonia FilmArray® |
|---|---|
| Blood cultures x 2 | Blood cultures x 2 |
| Sputum culture if sputum purulent | Sputum culture if sputum purulent |
| NP swab for Biofire® Respiratory FilmArray®⁎⁎ | Sputum* for Biofire® Pneumonia FilmArray®⁎⁎ |
| Nasal swab for Staphylococcus aureus PCR (BD Max SR®) | |
| NP swab for Streptococcus pneumoniae PCR | |
| Urine for Streptococcus pneumoniae and Legionella pneumophila antigens (Binaxnow®) |
NP = nasopharyngeal, PCR = polymerase chain reaction
sputum or sputum equivalent (ie, endotracheal aspirate, bronchoalveolar lavage)
Organism targets for BioFire® FilmArray® panels listed in Table 2.
2.3. Data collection
The laboratory entered MTB results into the electronic medical record (EMR), and study investigators re-entered EMR data in the FileMaker® database. Being investigational, the BPFA results were entered by investigators into the study database. Three authors (DNG, JEL, SF) verified the accuracy of all transcribed data.
2.4. Study objectives
The primary objective was to determine if BPFA improved detection rates for potential viral and bacterial pathogens as compared with MTB. We also present an approach to integrating BPFA results into clinical practice aided by PCT testing. Lastly, we suggest that use of BPFA may eliminate the need for selected diagnostic tests in the current MTB.
2.5. MTB and BPFA Test Components
Table 1 lists the components of the MTB and BPFA entered into comparative analyses. See Table 2 for components of the Biofire® Respiratory and Pneumonia FilmArray® panels used in MTB and BPFA.
Table 2.
Comparison of organism targets in the Biofire® Respiratory FilmArray and the Biofire Pneumonia FilmArray® Panels. The Respiratory Panel requires a nasopharyngeal swab and the Pneumonia Panel requires sputum*.
| Viruses | Respiratory Panel | Pneumonia Panel | Atypical Bacteria | Respiratory Panel | Pneumonia Panel | Bacteria | Respiratory Panel | Pneumonia Panel |
|---|---|---|---|---|---|---|---|---|
| Adenovirus | X | X | Bordetella pertussis | X | O | Acinetobacter calcoaceticus-baumannii complex | O | X |
| Coronavirus⁎⁎ | X | X | Chlamydophila pneumonia | X | X | Enterobacter cloacae complex | O | X |
| Human metapneumovirus | X | X | Legionella pneumophila | O | X | Escherichia coli | O | X |
| Human rhinovirus/enterovirus | X | X | Mycoplasma pneumoniae | X | X | Haemophilus influenzae | O | X |
| Influenza A | X | X | Klebsiella aerogenes | O | X | |||
| Influenza B | X | X | Klebsiella oxytoca | O | X | |||
| Parainfluenza virus | X | X | Klebsiella pneumoniae group | O | X | |||
| Respiratory syncytial virus | X | X | Moraxella catarrhalis | O | X | |||
| Proteus spp | O | X | ||||||
| Pseudomonas aeruginosa | O | X | ||||||
| Serratia marcescens | O | X | ||||||
| Staphylococcus aureus | O | X | ||||||
| Streptococcus agalactae | O | X | ||||||
| Streptococcus pneumoniae | O | X | ||||||
| Streptococcus pyogenes | O | X |
Sputum or sputum equivalent (ie, endotracheal aspirate or BAL)
Includes 4 strains of coronavirus, but not SARS-CoV-2
2.6. Clinical Categorization and Adjudication
The FileMaker® database calculated a pneumonia severity index (PSI) for each patient (Fine et al., 2020). To assess the clinical importance of detected pathogens, patients were divided into four categories (Table 3 ) based on their clinical and laboratory presentation, PCT test results, and likelihood based on clinical judgment (adjudication) as to whether detected pathogens were colonizing or invasive.
Table 3.
Clinical Classification of Patients Based on Integration of Test Results with Clinical Judgment
| Uninfected: No evidence of CAP |
| Post-admission clinical, laboratory, and imaging studies document an alternative non-infectious diagnosis: e.g., congestive heart failure. |
| Pure Bacterial Pneumonia |
| Proven: A compatible clinical syndrome (fever, cough, new pulmonary infiltrates, elevated white blood count) with detection of only bacterial pathogen(s) by one or more of: Biofire® Respiratory FilmArray®, NP S. pneumoniae PCR, nasal swab for S. aureus PCR, urine antigens, sputum and/or blood culture, or BPFA. |
| Presumptive: Clinical CAP syndrome with compatible chest radiographic abnormalities without detection of a potential bacterial pathogen by any of the same tests listed in the last paragraph. A PCT level ≥0.25 ng/ml with no other explanation was interpreted as presumptive evidence of invasion by an undetected bacterial pathogen. |
| Pure Viral Pneumonia |
| Presumptive: High probability of viral pneumonia based on compatible clinical presentation, detection of a virus by the BioFire® Respiratory FilmArray® or BPFA absent detected bacterial pathogen by FilmArray®, and serum PCT level <0.25 ng/ml. |
| Viral and Bacterial Co-Infection |
| Presumptive: Detection of a virus by either the BioFire® Respiratory FilmArray® or BPFA. In addition, detection of a bacterial pathogen in sputum and/or blood cultures, urine antigen tests, the Biofire® Respiratory FilmArray®, the BPFA, the nasal swab for S. aureus and/or the NP PCR for S. pneumoniae. |
| Pneumonia of Unclear Etiology |
| Clinical criteria for CAP but no bacterial or viral pathogen detected by any test and no non-infectious disease diagnosed. |
NP = nasopharyngeal; PCR = polymerase chain reaction; PCT = procalcitonin; BPFA = BioFire® Pneumonia FilmArray®
In adults with CAP, detection of a potential viral respiratory pathogen is accepted as compatible with the etiology of the patient's invasive infection as the frequency of asymptomatic colonization is very low. In a prospective controlled detection trial in adult CAP patients, detection of influenza, hMPV, and RSV “probably indicate an etiologic role.” Detection of other respiratory viruses, especially in children, requires further study (Self et al., 2016).
The biomarker procalcitonin (PCT) was used to assist in the clinical decision as to whether a detected virus and/or bacteria was a commensal. PCT serum levels do not increase with pure viral infection but do increase with invasive bacterial infection or the combination of viral and bacterial disease (Gilbert, 2020, Gilbert, 2017).
The BPFA panel includes genomic semi-quantitation of 15 of 18 bacterial targets with the premise that higher bacterial “loads” would be consistent with invasive infection. However, concordance between genomic semi-quantitation and sputum culture CFU/ml is reported between 40 and 54% and the degree of concordance varied by organism (Biofire 2020, Lee et al., 2019, Murphy et al., 2020). For these reasons, we elected to not use the BPFA semi-quantitation result in the adjudication of positive pathogen detection.
2.7. Cost Considerations
We determined the costs of BPFA versus all elements in the MTB based on 2017-2018 cost data.
2.8. Statistics
The performance between MTB and the BPFA was compared based on the number of pathogen species (bacterial and/or viral) detected in each patient by the two methods, using the paired t-test and Wilcoxon signed rank test. The same analysis was repeated for the number of bacterial species detected and for the number of viral species detected.
McNemar test assessed the difference in the proportions of patients detected with pathogen(s) (combined bacterial plus viral, bacterial, viral, and selected individual specific pathogen species) between MTB and BPFA.
The detection of individual specific pathogen species by the BPFA method was evaluated and presented as positive percent agreement (PPA) and negative percent agreement (NPA) against the MTB method; p-value <0.05 considered statistically significant. The R statistical program (www.r-project.org, R Foundation for Statistical Computing, Vienna, Austria) performed all analyses.
3. RESULTS
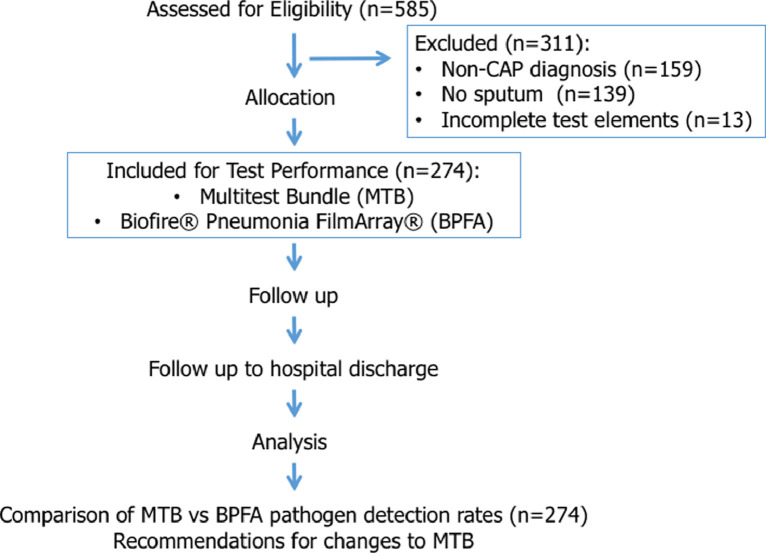
Of 585 enrolled patients, 274 (47.2%) were evaluable and 311 were excluded (Figure 1 ). Non-evaluability most commonly resulted from a final diagnosis other than CAP [159 of 311 (51.1%)], not obtaining a sputum or equivalent for BPFA [139 of 311 (44.7%)], and/or incomplete MTB results [13 of 311 (4.2%)]. The 274 (47.2%) evaluable patients were divided into four categories based on detected potential pathogens:
-
1.
Virus only: 31 (11.3%)
-
2.
Bacteria only: 87 (31.8%)
-
3.
Virus and bacteria detected: 143 (52.2%)
-
4.
No potential pathogen detected: 13 (4.7%)
Figure 1.
Flow diagram of 585 enrolled patients
This older patient population had many co-morbidities, and 64.2% had PSI scores >91, Table 4 . BioFire® results returned within 2 hours, and other PCR and urine antigen tests returned from 2 to 24 hours.
Table 4.
Demographics and Pneumonia Severity of 274 evaluable patients
| Mean (SD) | Median (IQR) | |
|---|---|---|
| Age | 66.3 (16.6) | 68 (56,79) |
| Weight (kg) | 77.2 (28.0) | 72.6 (59.4, 88.5) |
| Gender female, n (%) | 126 (46.0%) | |
| Comorbidity | n (%) | |
| Alcoholism | 21 (7.7%) | |
| Cerebrovascular disease | 33 (12.0%) | |
| Chronic liver disease | 22 (8.0%) | |
| Congestive heart failure | 65 (23.7%) | |
| COPD | 114 (41.6%) | |
| Diabetes mellitus | 84 (30.7%) | |
| Illicit drug use | 46 (16.8%) | |
| Malignancy | 55 (20.1%) | |
| Obesity (BMI>30) | 66 (24.1%) | |
| Renal insufficiency(CrCl<2) | 11 (4.0%) | |
| Pneumonia Severity Index | n (%) | |
| ____≤70 Risk classes I & II | 46 (16.8) | |
| ____71-90 Risk class III | 52 (19.0) | |
| ____91-130 Risk class IV | 94 (34.3) | |
| ____>130 Risk class V | 82 (29.9) |
3.1. MTB and BPFA Pathogen Detection Rates
The proportion of all patients detected by MTB or BPFA with potential bacterial pathogens (± a viral pathogen) or potential viral pathogens (± a bacterial pathogen) are shown in Table 5 along with the aggregate proportion of patients detected with any potential pathogen (bacterial and/or viral pathogens). The BPFA panel as compared with MTB detected more patients with bacterial pathogens (75.5% vs 66.4%, p=0.003) and viral pathogens (60.9% vs 40.5%, p=<0.001). Significantly more patients were detected by BPFA as having any potential pathogen (viral, bacterial or both) (90.6% vs 80.9%, p=0.001), Table 5.
Table 5.
Number of patients with bacterial, viral, or any (bacterial and/or viral) potential pathogens detected by MTB and BPFA
| Bacteria | MTB | BPFA | Effect Measure | p-value* |
|---|---|---|---|---|
| Any bacteria, patient No. (%) | 182 (66.4%) | 207 (75.5%) | OR (95% CI) = 2.09 (1.24,3.59) | 0.003 |
| Number of bacterial species per patient | ||||
| Mean (SD) | 0.92 (0.82) | 1.35 (1.19) | Diff (95% CI=0.43(0.30,0.56) | <0.001 |
| Median (IQR) | 1 (0,1) | 1 (1,2) | NA | <0.001 |
| Virus | ||||
| Any virus, patient No.(%) | 111 (40.5%) | 167 (60.9%) | OR (95%CI)=9.00(4.12,23.30) | <0.001 |
| Number of virus species per patient | ||||
| Mean (SD) | 0.43 (0.55) | 0.68(0.61) | Diff (95%CI)=0.25 (0.18,0.31) | <0.001 |
| Median IQR) | 0 (0,1) | 1 (0,1) | NA | <0.001 |
| Combined bacteria and virus species | ||||
| Any pathogen detected, patient No.(%) | 224 (80.9%) | 251 (90.6%) | OR (95%CI)=3.33 (1.71, 6.98) | <0.001 |
| Number of pathogen species per patient | ||||
| Mean (SD) | 1.35 (0.94) | 2.03 (1.27) | Diff (95%CI)=0.68 (0.53,0.83) | <0.001 |
| Median IQR) | 1 (1,2) | 2 (1,3) | NA | <0.001 |
MTB = Multi-test bundle; BPFA = BioFire® Pneumonia FilmArray®
p-value was based on McNemar test for odds ratio, paired t-test for mean difference, and Wilcoxon signed rank test for distribution
3.2. Comparison of Detection Rates for Specific Bacterial and Viral Pathogens
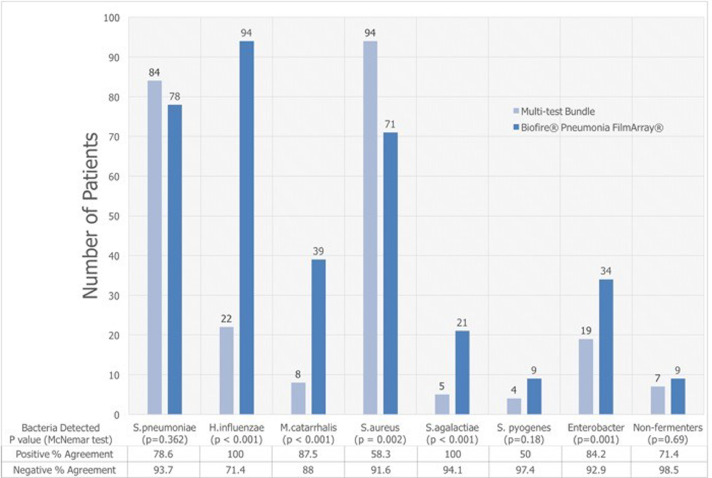
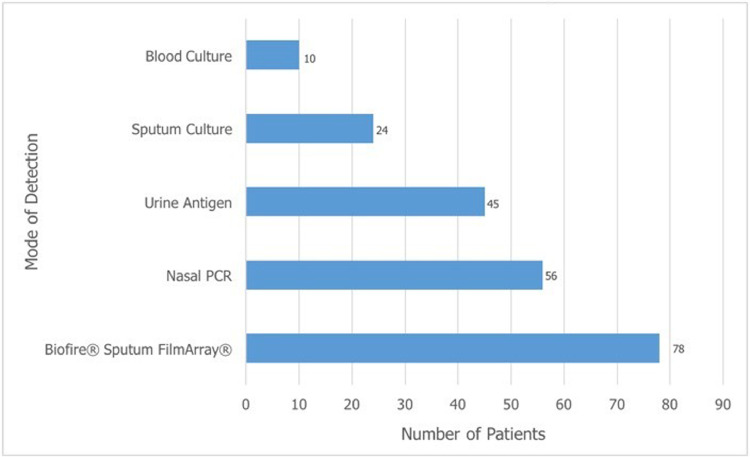
The BPFA outperformed the MTB in identifying all patients with Haemophilus influenzae, Moraxella catarrhalis, Streptococcus agalactiae, and Enterobacteriaceae, p ≤0.001 (Figure 2 ). Detection rates for S. pneumoniae varied widely in the 274 patients between individual elements of MTB (Figure 3 ). The BPFA detected S. pneumoniae more often (in 78 patients, 28.5%), than any individual component in MTB but not with the composite MTB (Figure 3).
Figure 2.
Comparison of MTB versus BPFA for detecting number of patients with potential bacterial pathogens, with or without detectable virus. Some patients were detected with more than one bacterial species.
Figure 3.
Comparison of number of patients detected with Streptococcus pneumoniae by individual elements of the multi-test bundle vs the BioFire® Pneumonia FilmArray®. Some patients had more than one test element positive for S. pneumoniae. PCR denotes polymerase chain reaction.
S. aureuswas detected by nasal swab PCR in 94 of 274 (34.3%) patients vs. 71 of 274 (25.9%) by BPFA, p=0.002. Six patients (all detected by BPFA probe of sputum) had S. aureus bacteremia, three methicillin-sensitive S. aureus (MSSA) and three methicillin-resistant S. aureus (MRSA).
Only one patient had a positive urine antigen test for Legionella pneumophila.
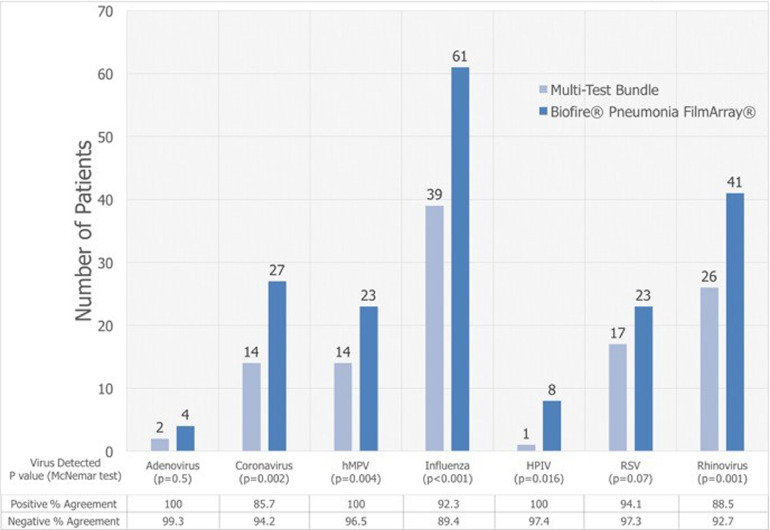
The BPFA detected significantly more patients with potential viral pathogens (coronavirus, hMPV, influenza, parainfluenza, and rhinovirus) than the BioFire® Respiratory FilmArray® component of MTB (Figure 4 ). Influenza virus was detected in 61 (22.3%) with BPFA as compared to 39 (14.2%) with MTB, p <0.001. The number of patients with one or more detectable viruses increased from 41% with MTB to 61% with BPFA, p<0.001.
Figure 4.
Comparison of MTB versus BPFA for detecting number of patients with potential viral pathogens, with or without detectable bacteria Some patients were detected with more than one viral species.
3.3. Positive Blood Cultures
Of 274 CAP patients, 22 (8%) had positive blood cultures deemed pathogens rather than contaminants. The same organism was detected in the sputum culture in 9 of the 22 (41%) patients. On the other hand, S. aureus by sputum BPFA in 17 of the 22 (77%) patients.
S. pneumoniae was identified in the blood of 10 patients, S. aureus in 6, S. pyogenes in 2, and Enterobacter cloacae, E. coli, S. agalactiae, and Aerococcus viridans each in one patient. A concomitant viral pathogen was detected by BPFA in 10 of the 22 patients with positive blood cultures.
3.4. Multiple Pathogen Detection
The superiority of the BPFA as compared with MTB in detecting co-infections is reported in Table 6 . In one extreme example, BPFA detected 1 virus (RSV) and 5 bacterial species (E.coli, P. aeruginosa, K. pneumoniae, S. marcescens, and MRSA) while the MTB detected no virus and 3 bacteria (P. aeruginosa, E. coli, and MSSA).
Table 6.
Comparative detection of multiple bacterial and viral species with BPFA versus MTB
| Type of Pathogen | Number of species | Number of patients with detected species |
|
|---|---|---|---|
| MTB* | BPFA* | ||
| Only virus | 1 | 24 | 25 |
| 2 | 1 | 5 | |
| Only bacteria | 1 | 43 | 37 |
| 2 | 17 | 21 | |
| ≥3 | 5 | 19 | |
| Virus (V) and Bacteria (B) | 1 (V) + 1 (B) | 54 | 62 |
| 1 (V) + 2 (B) | 10 | 30 | |
| 1 (V) + ≥3 (B) | 3 | 20 | |
| 2 (V) + 3 (B) | 5 | 12 | |
+Representative data. Less frequent combinations are not included
MTB: Multi-test bundle; BPFA: Biofire® Pneumonia FilmArray®
3.5. Potential Role of Serum PCT Levels
Detected pathogens are either colonizing or invading and stimulating host innate immune inflammatory response. All 274 evaluable patients had a clinical syndrome consistent with pneumonia; one or more potential pathogens were detected in 261. As the clinical syndromes lack discriminating features, we assessed whether the biomarker PCT could help adjudicate whether the detected potential virus and/or bacteria were colonizing or invading.
PCT levels are a marker of activation of the host innate immune inflammatory response by invading bacterial pathogens but not by pathogenic respiratory viruses (Gilbert, 2020, Gilbert, 2017). PCT levels increase in the presence of concomitant invasive disease by a virus and/or bacteria.
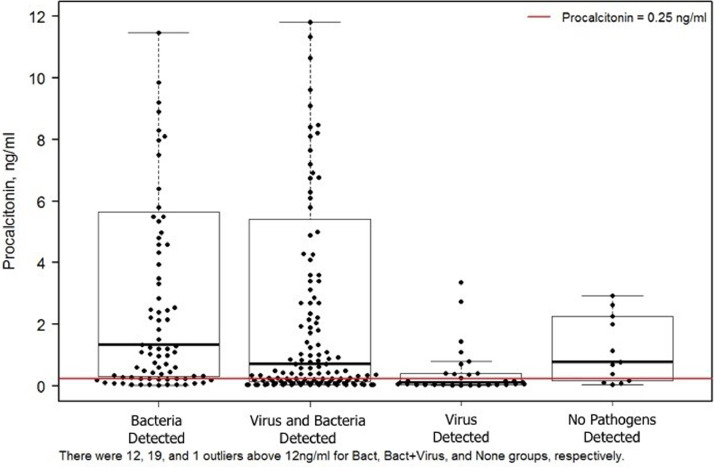
We divided our patients into four categories depending on the pathogens detected (Figure 5 ). By convention, a serum PCT elevation of ≥0.25 ng/ml is selected as a clinically meaningful increase.
Figure 5.
Box plot of distribution of serum procalcitonin levels in each of the 4 categories of pathogen detection. Each dot represents a single patient. The red line marks a serum procalcitonin level of 0.25 ng/ml
In 87 of 274 patients, only potential bacterial pathogens were detected by the MTB and/or BPFA of which 69 (79.3%) had PCT levels ≥0.25 ng/ml, a result consistent with invasive bacterial infection. In the remaining 18 (20.7%) patients, the PCT of <0.25 ng/ml was considered consistent with colonization by the detected bacteria.
For 143 patients, both viral and bacterial pathogens were detected by MTB and/or BPFA. Ninety-six (96) (67.1%) of these patients had PCT levels ≥0.25 ng/ml, which was interpreted as consistent with bacterial and viral co-infection. Forty-seven (47) patients (32.8%) had PCT values <0.25 ng/ml consistent with bacterial colonization and viral infection.
For 31 patients, only a potential viral pathogen was detected. In 11 (35.5%) the PCT level was ≥0.25 ng/ml consistent with viral infection and a superimposed, but undetected, bacterial pathogen.
The range of PCT levels in the 13 patients with no detected pathogens may reflect the presence of undetected viral and/or bacterial pathogens.
3.6. Comparative Cost of MTB vs Test Bundle with BPFA
Our laboratory estimated the costs for MTB as compared with the projected cost of a new test bundle anchored by the BPFA, Table 7 . Results indicate cost savings for the BPFA approach partially because three nasal swab PCRs and the two urine antigen tests would no longer be needed.
Table 7.
Comparative laboratory cost of MTB vs projected cost of BPFA bundle
| Test | MTB, $ | BPFA, $ |
|---|---|---|
| Urine antigen | No need for urine antigens | |
| Legionella | 30.00 | |
| S. pneumoniae | 19.00 | |
| Nasal swabs – PCRs | No need for nasal swabs | |
| S. aureus | 42.00 | |
| S. pneumoniae | 25.00 | |
| PCR “resp panel” | 91.00 | |
| Biofire Pneumonia FilmArray | 180.00 | |
| Sputum C&S | 8.23 | 8.23 |
| Blood culture x 2 | 9.80 | 9.80 |
| Tech x 1 hr @$36.50/hr | 36.50 | |
| Tech x ½ hr @ 36.50/hr | 18.00 | |
| PCT @ $23 ea x 2 | 46.00 | 46.00 |
| SUBTOTAL | 307.53 | 262.03 |
| Overhead @ 30% | 92.00 | 79.00 |
| TOTAL | 399.53 | 341.03 |
$ = Dollars: represents estimated cost as per 2018
4. DISCUSSION
Our results demonstrate, in adult patients hospitalized for CAP, that the Biofire® Pneumonia FilmArray® (BPFA) detected more patients with potential viral and bacterial pathogens (90.6%) than the MTB (80.9%), the latter having been our standard of practice (Gilbert et al., 2016).
BPFA sputum detection of 6 of the 7 respiratory viruses was statistically more frequent than testing for the same 7 viruses with the nasopharyngeal Biofire® respiratory FilmArray® panel which is part of the MTB.
The higher yield with the BPFA may reflect higher viral loads in the lower as compared with upper airways (Jeong et al., 2014, Wunderink, 2017). Alternatively, the higher yield may be due to differences in the viral primers used in the BPFA vs the primers in the Biofire respiratory FilmArray® (BioFire® FilmArray® User Instruction booklets).
For bacterial pathogens, BPFA detected more patients with 4 of the common pathogens that cause CAP. Notably, BPFA detected more patients with S. pneumoniae than individual urine antigen testing, nasal PCR, or sputum and blood cultures although statistically equivalent to the composite MTB results.
S. aureus was the only bacterium detected more frequently using the the nasal swab PCR in the MTB vs the sputum BPFA, p=0.001. The difference may result from differences in the PCR primers used for the nasal swab PCR in the MTB as compared with BPFA primers. In addition, replication of S. aureus is enhanced at 35°C (anterior nares) vs 37°C in the lower airway source of sputum sent for BPFA (Matoos-Mora et al., 1988, Milne et al., 1987).
An emerging body of evidence supports the use of PCR for the detection of etiologic pathogens for CAP. Previous studies combined cultures of sputum and blood, urine antigen detection, and NP multiplex platforms that primarily focused on viral pathogens and a few atypical bacteria. The reported detection rates for pathogens have varied between 38-53% of patients (Self et al., 2017, Jain et al., 2015, Stockmann et al., 2018, Musher et al., 2013, Musher et al., 2017).
First generation multiplex PCR respiratory panels only detected three atypical bacteria. Cultures and antigen detection are needed to detect common pneumonia pathogens: i.e., S. pneumoniae, H. influenzae, M. catarrhalis and others. We generated a MTB that included PCR probes for S. pneumoniae and S. aureus plus the Biofire® Respiratory FilmArray® panel which includes 7 viruses (Gilbert et al., 2016).
Traditional sputum cultures and antigen detection may miss bacterial pathogens obscured by in vitro overgrowth of normal oral flora and/or suppression of growth by empiric antibiotic therapy. Hence, we compared the present study to determine if the BPFA, with the 18-targeted bacterial targets, would increase detection as compared to our standard of care (SOC) MTB.
4.1. Study of BPFA vs SOC by Others
Other studies have compared pathogen detection by BPFA with standard of care (SOC) diagnostics. The SOC is not standardized from study to study but includes some combination of cultures of sputum and blood, urine antigens and nasopharyngeal swabs for multiplex PCR panels that most often focus on seasonal respiratory viruses.
In representative studies, the BPFA panel detected a viral and/or bacterial potential pathogen in 47.4 to 72% of the CAP patients (Lee et al., 2019, Murphy et al., 2020, Webber et al., 2020, Buchan et al., 2020). The number of patients varied from 595 to 1682. Patient population, comorbidities, severity of illness, season of the year, variations in PCR primers are a few of many pertinent variables.
Akin to our results, Webber et al detected potential bacterial pathogens in 60 patients by both SOC and BPFA plus an additional 92 detections of bacteria by the BPFA (Webber et al., 2020). Buchan et al reported similar results from 259 adult patients (Buchan et al., 2020).
Gadsby's research group has developed a PCR pneumonia panel with probes for 13 bacteria and 7 viruses. In 323 adult CAP patients, a potential bacterial pathogen was identified in 81.1%. H. influenzae and S. pneumoniae were the most frequently identified bacteria (Gadsby et al., 2016).
The BPFA is not the only commercial rapid syndromic molecular test panel for pneumonia pathogen identification (Noviello and Huang, 2019, Poole and Clark, 2020). The Curetis Unyvaro Hospitalized Pneumonia panel is FDA-approved and includes 17 bacterial, 3 atypical bacteria, 1 fungal, and 19 resistance genes but no viral targets. There are at least 4 other commercial entities developing pneumonia diagnostic panels (Poole and Clark, 2020).
4.2. Colonization versus Infection
Several studies report increased detection of potential viral and bacterial pathogens in lower respiratory secretions of CAP patients when SOC diagnostics are compared to use of the BPFA (Lee et al., 2019, Murphy et al., 2020, Webber et al., 2020, Poole and Clark, 2020). The positive and negative percent agreements concur with some variation as expected with variations in the SOC tests employed.
Rapid, sensitive, and specific identification of potential bacterial pathogens is a premise of improving the care of CAP patients. In theory, identification of a pathogen will allow switching from empiric antibiotic therapy to more focused specific therapy. Before switching, clinicians need to consider whether the detected bacteria are only colonizing and not a concern.
The BPFA platform provides a semi-quantitative number of bacterial genomes/ml present. Traditional indicators of bacterial invasion correlate with greater the numbers of bacteria, but patients with selected co-morbidities may be only colonized and yet have a high density of bacteria: e.g., cystic fibrosis, bronchiectasis, colonized tracheostomy. Growth characteristics of organisms are another variable. Antibiotic therapy can influence bacterial density. Hence, we elected to not utilize the results of bacterial quantitation in our adjudication of whether the detected bacteria are colonizing or invading.
We elected to use the serum biomarker procalcitonin (PCT) to assist in the assessment of whether detected bacteria were colonizing or invading. Elevations of serum PCT indicate activation of innate immunity (Gilbert, 2020, Gilbert, 2017). PCT gene activation does not occur with pure viral infection but promptly increases in response to invasion by bacteria to include dual infection with a virus (Branche et al., 2019, Schuetz et al., 2017, US Food and Drug Administration 2017). The other variables that influence PCT serum levels; e.g., impaired renal function, are known (Gilbert, 2020).
Jain and Pavia point out that the “holy grail” would apply molecular diagnostics to lung tissue (Jain and Pavia, 2016). In the absence of lung tissue, we integrated BPFA results with the clinical presentation and PCT test results to characterize with an a priori classification system the likelihood of CAP due to bacterial and/or viral pathogens (Table 3). This approach allows clinicians to focus the antimicrobial regimen and promote antibiotic stewardship.
Based on previous research, we selected PCT serum levels of ≥0.25 ng/ml as consistent with activation of an innate immune response by invasive bacteria (Branche et al., 2019, Schuetz et al., 2017). PCT levels of <0.25 ng/ml were interpreted as consistent with colonization (negative predictive value for invasive bacterial infection is >90%) (Self et al., 2017, Gilbert, 2017, Stockmann et al., 2018). The potential value of pathogen detection coupled to PCT levels is acknowledged in multiple publications (Branche et al., 2019, Schuetz et al., 2017, US Food and Drug Administration 2017, Jain and Pavia, 2016, Wunderink et al., 2020). A recent economic model supported use of PCT serum levels as a way to reduce hospital costs, decrease the risk of antibiotic resistance and Clostridium difficile infections, and enhance clinical decision-making beyond clinical judgment and heuristic approaches (Mewes et al., 2020).
4.3. Suggested Revised Multi-Test Bundle
The diagnostic yield of BPFA creates an opportunity to simplify diagnostic testing for CAP. The BPFA has PCR primers for all Legionella species causing disease in humans suggesting elimination of the urine antigen test for L. pneumophila. Also, the urine antigen test and NP PCR for S. pneumoniae may be eliminated because BPFA outperformed both. The low yield of blood cultures compared to BPFA suggests they can be reserved for ICU patients (Metlay et al., 2019). Of note, nasal swabs for S. aureus PCR may remain an optional test added to BPFA as nasal samples were 9% more sensitive than sputum samples submitted to BPFA. Sputum cultures remain necessary to provide antibiotic susceptibility testing of detected bacteria.
A new test bundle would include BPFA, sputum culture and sensitivity, blood cultures for the critically ill, and perhaps nasal S. aureus PCR. Two (2) PCT levels 4-6 hours apart for their high negative predictive value and to help discriminate between infection and colonization. This bundle replaces traditional testing, which may take 1-2 days to complete vs a bundle anchored by BPFA that takes less than 2 hours. Moreover, this proposed bundle decreases the cost of testing (Table 7) and, in theory, can reduce antibiotic consumption, and shorten length of hospitalization (Buchan et al., 2020).
4.4. Limitations
Patients inability to provide sputum and the difficulty of ED physicians in this single center study to make an accurate diagnosis of CAP excluded a majority of patients from the study. To avoid excluding patients from BPFA testing, studies are needed to assess the potential value of oropharyngeal swab specimens in lieu of sputum for testing, as has been investigated for children (Wang et al., 2019). Only 13 patients were excluded because of incomplete test results beyond failure to collect sputum. Second, despite the large panel size, a failure of the PCR panels to detect bacteria or virus does not exclude the presence of an undetected virus and/or bacteria. Third, without systematic antibody seroconversion testing, we could not provide definite differentiation between invasive viral disease vs viral colonization. However, protracted viral colonization of adults is rare. Lastly, we did not document the influence of rapid pathogen detection on antibiotic therapy, antibiotic adverse events to include C. difficle infection, antibiotic resistance, or need for patient isolation.
5. Conclusion
For hospitalized adult patients with CAP, the BPFA multiplex platform detected significantly more common viral and bacterial potential pathogens as compared to a MTB. Serum PCT levels can help in the judgement as to whether detected bacteria are colonizing or causing infection.
Future studies are needed to critically assess the influence of enhanced pathogen detection on antibiotic usage, prevalence of antibiotic adverse effects and antibiotic resistance, and other facets of antimicrobial stewardship
Funding
This study was funded by Biofire® Diagnostics, Salt Lake City, Utah
Acknowledgement
Thanks to Justin Jin, Brian Kendall, Robert Lusk, Veronica Schims, Ronald Dworkin, Ben
Pedroja, Jesse Powell, Lindsey McKinley, Stephanie Munoz, Chris Siegenthaler, Cami
Hilsendager, and the ED physicians and nursing staff of Providence Portland Medical
Center.
REFERENCES
- Self WH, Balk RA, Grijalva CG. Procalcitonin as a marker of etiology in adults hospitalized with community-acquired pneumonia. Clin Infect Dis. 2017;65:183–190. doi: 10.1093/cid/cix317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metlay JP, Waterer GW, Long AC. Diagnosis and treatment of adults with community-acquired pneumonia. Am J Respir Crit Care Med. 2019;200:e45–e67. doi: 10.1164/rccm.201908-1581ST. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain S, Self WH, Wunderkink RG. Community-acquired pneumonia requiring,hospitalization among U.S. adults. N Engl J Med. 2015;373:415–427. doi: 10.1056/NEJMoa1500245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert D, Gelfer G, Wang L. The potential of molecular diagnostics and serum procalcitonin levels to change the antibiotic management of community-acquired pneumonia. Diag Microbiol Infect Dis. 2016;86:102–107. doi: 10.1016/j.diagmicrobio.2016.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho Mda G, Tondella ML, McCaustland K. Evaluation and improvement of real-time PCR assays targeting lyta, ply and psaA genes for detection of pneumococcal DNA. J Clin Microbiol. 2007;45:2460–2466. doi: 10.1128/JCM.02498-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fine MJ, Auble TE, Yealy DM et al. A prediction rule to identify low-risk patients with community-required pneumonia. N Engl J Med 1997;336:243-250. [DOI] [PubMed]
- Self WH, Williams DJ, Zhu Y. Respiratory viral detection in children and adults: Comparing asymptomatic controls and patients with community-acquired pneumonia. J Infect Dis. 2016;213:584–591. doi: 10.1093/infdis/jiv323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert DN. Neglected variables in the interpretation of serum procalcitonin levels in patients with septic shock. J Infect Dis. 2020;222(S):S96–102. doi: 10.1093/infdis/jiaa204. [DOI] [PubMed] [Google Scholar]
- Gilbert DN. Role of procalcitonin in the management of infected patients in the intensive care unit. Infect Dis Clin N Amer. 2017;31:435–453. doi: 10.1016/j.idc.2017.05.003. [DOI] [PubMed] [Google Scholar]
- Biofire diagnostics – FilmArray Pneumonia panel. Available at https://biofiredx.com/products/the-filmarray-panels/filmarray-pneumonia/. Accessed September 11, 2020.
- Lee SH, Ruan S-Y, Pan S-C, Lee T-F, Chien J-Y, Hsueh P-R. Performance of a multiplex PCR pneumonia panel for the identification of respiratory pathogens and the main determinants of resistance from the lower respiratory tract specimens of adult patients in intensive care units. J Microbiol Immunol Infect. 2019;52:920–928. doi: 10.1016/j.jmii.2019.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy CN, Fowler R, Baloda-Llasat JM et al. Multicenter evaluation of the Biofire® FilmArray® pneumonia/pneumonia plus panel for the detection and quantification of agents of lower respiratory tract infection. J Clin Microbiol. 2020;58:e00128-20. [DOI] [PMC free article] [PubMed]
- Jeong JH, Kim KH, Jeong SH, Park JW, Lee SM, Seo YH. Comparison of sputum and nasopharyngeal swabs for detection of respiratory viruses. J Med Virol. 2014;86:2122–2127. doi: 10.1002/jmv.23937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wunderink RG. Other community respiratory viruses. Clin Chest Med. 2017;38:37–43. doi: 10.1016/j.ccm.2016.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matoos-Mora M, Knapp CC, Washington JA. Characterization of resistance phenotype and cephalosporin activity in oxacillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 1988;32:170–174. doi: 10.1128/aac.32.2.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milne LM, Curtis GDW, Crow M, Kraak WAG, Selkon JB. Comparison of culture media for detecting methicillin resistance in Staphylococcus aureus and coagulase-negative staphylococci. J Clin Path. 1987;40:1178–1181. doi: 10.1136/jcp.40.10.1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockmann C, Ampofo K, Killpack J. Procalcitonin accurately identifies hospitalized children with low risk of bacterial community-acquired pneumonia. J Ped Infect Dis Soc. 2018;7:46–53. doi: 10.1093/jpids/piw091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musher DM, Roig IL, Stazer CG, Logan N, Safar H. Can an etiologic agent be identified in adults who are hospitalized for community-acquired pneumonia: results of a one year study. J Infect. 2013;67:11–18. doi: 10.1016/j.jinf.2013.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musher DM, Aben MS, Bartlett JG. Evolving understanding of the causes of pneumonia in adults, with special attention to the role of pneumococcus. Clin Infect Dis. 2017;65:1736–1744. doi: 10.1093/cid/cix549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webber DM, Wallace MA, Burnham C-AD, Anderson NW. Evaluation of the Biofire® FilmArray® pneumonia panel for detection of viral and bacterial pathogens in lower respiratory tract specimens in the setting of a tertiary care academic medical center. J Clin Microbiol 2020; doi:10.1128/JCM 00343-20. [DOI] [PMC free article] [PubMed]
- Buchan BW, Windham S, Balada-Llasat J-M. Practical comparison of the Biofire ElmArray Pneumonia panel to routine diagnostic methods and potential impact on antimicrobial stewardship in adult hospitalized patient with lower respiratory tract infections. J Clin Microbiol. 2020;58 doi: 10.1128/JCM.00135-20. e00135-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadsby NJ, Russell CD, McHugh MP. Comprehensive molecular testing for respiratory pathogens in community-acquired pneumonia. Clin Infect Dis. 2016;62:817–823. doi: 10.1093/cid/civ1214. 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noviello S, Huang DM. The basics and advancements in diagnosis of bacterial lower respiratory tract infection. Diagnostics. 2019;9:37. doi: 10.3390/diagnostics9020037. Doi co.3390/diagnostics 9020037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole S, Clark TW. Rapid syndromic molecular testing in pneumonia. The current landscape and future potential. J Infect. 2020;80:1–7. doi: 10.1016/j.jinf.2019.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branche A, Nesser O, Mueller B, Schuetz P. Procalcitonin to guide antibiotic decision-making. Curr Opin Infect Dis. 2019;32:130–135. doi: 10.1097/QCO.0000000000000522. [DOI] [PubMed] [Google Scholar]
- Schuetz P, Wirz Y, Sager R. Procalcitonin to initiate or discontinue antibiotics in acute respiratory tract infections. Cochrane Database Syst Rev. 2017;10 doi: 10.1002/14651858.CD007498.pub3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- US Food and Drug Administration. FDA press release. FDA clears test to help manage antibiotic treatment for flower respiratory tract infections and sepsis. Feb 23, 2017.https://www.fda.gov/NewsEvents/Newsroom/PressAssnouncements/ucm543160.htm (accessed Oct 2, 2017).
- Jain S, Pavia AT. The modern quest for the “Holy Grail” of pneumonia. Clin Infect Dis. 2016;62:826–828. doi: 10.1093/cid/civ1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wunderink RG, Srinavasan A, Barie PS. Antibiotic stewardship in the Intensive Care unit. Ann Am Thorac Soc. 2020;17:531–540. doi: 10.1513/AnnalsATS.202003-188ST. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mewes JC, Pulia MS, Mansour MK, Broles MR, Nguyen HB, Steuten LM. (2019). The cost impact of PCT-guided antibiotic stewardship versus usual care for hospitalized patients with suspected sepsis or lower respiratory tract infections in the US: A health economic model analysis. PLoS One 14: e0214222 [DOI] [PMC free article] [PubMed]
- Wang L, Yang S, Yan X. Comparing the yield of oropharyngeal swabs and sputum for detection of 11 common pathogens in hospitalized children with lower respiratory tract infection. Virol J. 2019;16:84. doi: 10.1186/s12985-019-1177-x. Doi.org/10.1186/s12955-019-1177. [DOI] [PMC free article] [PubMed] [Google Scholar]