Abstract
Brain structural networks have been shown to consistently organize in functionally meaningful architectures covering the entire brain. However, to what extent brain structural architectures match the intrinsic functional networks in different functional domains remains under explored. In this study, based on independent component analysis, we revealed 45 pairs of structural-functional (S-F) component maps, distributing across nine functional domains, in both a discovery cohort (n = 6005) and a replication cohort (UK Biobank, n = 9214), providing a well-match multimodal spatial map template for public use. Further network module analysis suggested that unimodal cortical areas (e.g., somatomotor and visual networks) indicate higher S-F coherence, while heteromodal association cortices, especially the frontoparietal network (FPN), exhibit more S-F divergence. Collectively, these results suggest that the expanding and maturing brain association cortex demonstrates a higher degree of changes compared with unimodal cortex, which may lead to higher interindividual variability and lower S-F coherence.
Keywords: intrinsic brain networks, independent component analysis (ICA), structure-function coherence, unimodal cortex, heteromodal association cortex
Introduction
Human brain is a complex network of neurons that link physical neural structure to multiple human functions (Power et al. 2010). Multiple computational studies have suggested that the underlying anatomical architecture of cerebral cortex shapes resting state functional connectivity on multiple time scales (Misic et al. 2016). Recent evidences have suggested that functional architectures are topographically corresponds to equivalent structural patterns in particular networks (Greicius et al. 2009; Honey et al. 2009). Various multimodal fusion studies also revealed that impaired structure topography is correlated with functional damages in mental disease (Gao et al. 2018; Luo et al. 2019). However, when comparing the relationship between anatomic structure and functional connectivity, limited studies have used gray matter (GM) volume. For example, Seeley et al. (2009) demonstrated that GM architectures derived at group level partly resembled the intrinsic functional networks. The biological meaning of this structural covariance remains controversial, though it appears to reflect developmental coordination or synchronized maturation between areas of the brain (Alexander-Bloch et al. 2013). On the other hand, several studies suggested that the structural covariance networks reflect shared recruitment or common neurodevelopmental effects within functionally coactive regions (Zielinski et al. 2010; Clos et al. 2014). Moreover, Geng et al. (2017) revealed that the distributed structural covariance in adults may result from synchronized current and earlier maturation in regions that co-activate serving for some specific functional processing. Therefore, exploring where GM covariance corresponds to the functional network would aid in the understanding of human connectome. In addition, several recent studies have investigated how structure-function coupling is distributed across different domains, that is, unimodal cortex and heteromodal association cortex on hundreds of subjects from various perspectives (Paquola et al. 2019; Vázquez-Rodríguez et al. 2019; Baum et al. 2020). However, such work lacks a multimodal structure-function spatial template, which provided information of how structural spatial maps correspond to functional spatial maps in each network. Identification of patterns in a larger dataset (15 000+ participants) with more components can reveal a finer and more common degree of details, providing a stable structure-function correspondence template that may be of use to the broader neuroimaging community.
To this end, we have used a discovery dataset of 7104 functional magnetic resonance imaging (fMRI) scans (within 6005 structural MRI scans were matched for the same subjects) collected at the University of New Mexico (UNM) and the University of Colorado Boulder (UC Boulder), and a replication dataset of 9214 participants from UK Biobank. As shown in Figure 1, first, 100 “source-based morphometry networks” with spatially distinct regions were identified based on independent component analysis (ICA) on the structural data, providing information about localization of GM variation and their covariation among individuals (Xu et al. 2009). Similar job was done for the 7104 resting-state fMRI scans, which identified 100 intrinsic functional networks with common temporal features, providing robust measures of the intrinsic functional activity of the brain (Abrol et al. 2017; Xu et al. 2017). These GM and fMRI components were subsequently parcellated into nine brain network modules. Spatial coherences were measured between the effective GM networks and intrinsic functional connectivity networks by spatial correlation. Second, segmentation and spatial correlation on the GM of replication data were conducted, to verify the reproducibility of the identified structural-functional (S-F) paired components. Third, the replicated S-F component pairs were further compared across different domains. Interestingly, the unimodal cortical areas (e.g., somatomotor and visual networks) indicate higher S-F coherence in both discovery and replication data, while those made from heteromodal association cortices, for example, frontoparietal network (FPN), exhibit more S-F divergence. To the best of our knowledge, this is the first study to assess differences of structure-function coherence across different function domains on the currently largest dataset.
Figure 1.
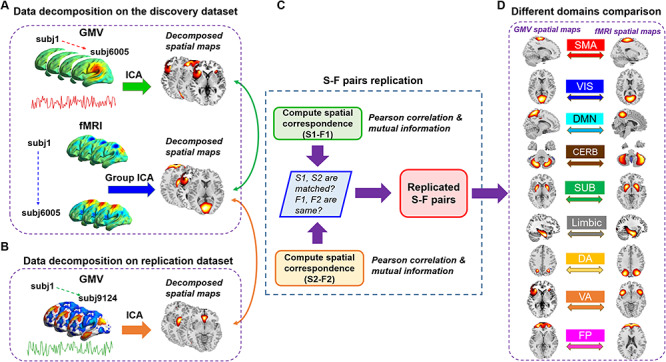
Schematic of preprocessing and analyses pipelines. (A) Structural data and functional data were preprocessed through an automated pipeline and then decomposed using spatial ICA. We then compared the correspondence between structural and functional components using PC and MI. (B) A replication dataset from UK Biobank, consisting of 9214 subjects, was preprocessed to validate the identified S-F pairs. We again applied ICA to decompose the structural replication data and measured the spatial correspondence between components in the discovery dataset and components in the replication dataset. (C) If structural components in one matched structural-structural pair between discovery and replication cohorts both show high correlation with the same functional component, then the S-F pair in the discovery dataset was regarded as replicated. (D) We further sorted the matched pairs into nine networks and compared S-F coherence across networks.
Materials and Methods
Data Acquisition and Preprocessing
Discovery Data
All 6101 structural scans and 7500 resting state functional scans were collected from anonymized subjects with informed consent at the UNM and the UC Boulder. Data from the UC Boulder site were collected using a 3T Siemens TIM Trio MRI scanner with 12 channel radio frequency coils, while data from the UNM site were acquired using the same type of 3T Siemens TIM Trio MRI scanner and a 1.5T Avanto MRI scanner. All the data were previously collected, anonymized, and had informed consent received from subjects including both healthy controls and patients. As it is a deidentified convenience dataset, we do not have access to the health and identifier information. We have confirmed that the brain images do not have any obvious pathology or atrophy. The fMRI data were used in a previous study that evaluated replicability in time-varying functional connectivity patterns (Abrol et al. 2017). The sMRI data were used in a previous study which measured age-related structural variations across the adult lifespan. The details of data acquisition and preprocessing are as bellows.
T 1-weighted structural images were acquired with a five-echo MPRAGE sequence with TE = 1.64, 3.5, 5.36, 7.22, and 9.08 ms, TI = 1.2 s, TR = 2.53 s, number of excitations = 1, flip angle = 7°, field of view = 256 mm, slice thickness = 1 mm, and resolution = 256 × 256. The structural images were then preprocessed using voxel-based morphometry (VBM) based on the SPM12 old segmentation, including: 1) spatial registration to a reference brain; 2) tissue classification into GM, white matter and cerebrospinal fluid (CSF); 3) bias correction of intensity nonuniformities; 4) spatial normalization to the standard Montreal Neurological Institute (MNI) space using nonlinear transformation; and 5) modulated by scaling with the amount of volume changes. The modulated GM data, representing the GM volumes, were resliced to 2 mm × 2 mm × 2 mm and smoothed with a 10-mm Gaussian model (Silver et al. 2011; Luo et al. 2020). Furthermore, we followed our previous paper (Chen et al. 2014) to investigate the correlations between individual GM images and the average image across all subjects, to exclude those exhibiting correlations 4 standard deviation (SD) less than the mean correlation. For our data, the mean correlation is 0.9166, with a SD of 0.0538. Thus, scans with a correlation less than 0.7 were removed, leaving behind a total number of 6005 scans for the subsequent correspondence analysis. The demographic information of the 6005 subjects was shown in Figure 2.
Figure 2.
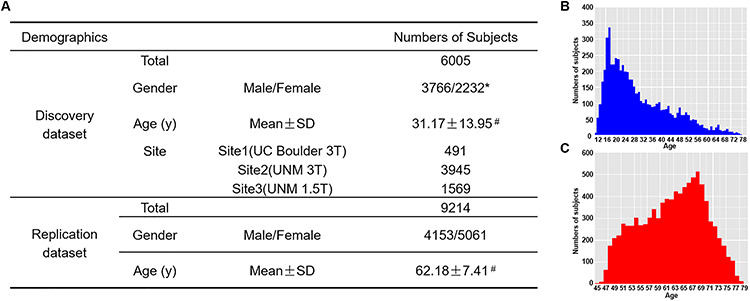
Demographic information of both datasets (A) and the distribution of age in the discovery data (B) and the replication data (C). Note: *the gender information of seven subjects is missing. #the distribution of age in the discovery data and replication data are presented in B and C.
T 2-weighted functional images were acquired using a gradient-echo EPI sequence with TE = 29 ms, TR = 2 or 1.3 s, slice thickness = 3.5 mm, flip angle = 75°, field of view = 240 mm, slice gap = 1.05 mm, voxel size = 3.75 mm × 3.75 mm × 4.55 mm, and matrix size = 64 × 64. The scans had variable length with the minimum scan length being 150 TRs; however, only the first 150 time-points of all scans were studied. The data preprocessing pipeline included discard of the first three images for the magnetization equilibrium, realignment using INRIalign, timing correction with the middle slice as reference, spatial normalization into the MNI space, reslicing to 3 × 3 × 3 mm, and smoothing with a 10 mm Gaussian model (Silver et al. 2011). To further evaluate the motion of the data, we computed the mean frame-wise displacements (FDs) for each subject (Power et al. 2012; Yan et al. 2013). The histogram of head motion was shown as Figure S1. Among all 6005 subjects, 88% scans were identified with mean FD less than 0.5. More details were provided in our previous study (Abrol et al. 2017). After preprocessing, 7104 functional scans were remained for the subsequent analysis, of which 6005 scans have corresponding structural images.
Replication Data
The UK Biobank is a large-scale prospective study of over 500 000 individuals from across the UK, with a major aim being to characterize subjects before disease onset. Participants were 40–69 years of age at baseline recruitments. Here, we used the sMRI data from the February 2017 release of ~10 000 participants (Alfaro-Almagro et al. 2018). VBM-related processing was performed with FMRIB Software Library v10.0. A study-specific template was created using an average T1-weighted image (provided by the UK Biobank) from 5000 subjects. To generate the template, brain extraction and tissue segmentation were performed on the average T1-weighted image. The GM image from the segmentation was then registered to the avg152T1_gray template available in FSL. Segmented GM images from each subject, available as part of the UK Biobank imaging data release, were nonlinearly registered to the study-specific template. Each registered GM image was also multiplied by the Jacobian of the warp field as a compensation (or “modulation”) for the contraction/enlargement due to the nonlinear component of the transformation. The resulting GM image was then smoothed with a 6-mm Gaussian kernel. The smoothed GM was then correlated with the mean of all scans to remove scans with a correlation less than 0.7, resulting in a total number of 9214 subjects for the analysis. The demographic information of the replication data was shown in Figure 2.
Group ICA on rsfMRI Data
ICA decomposition on the fMRI data was conducted in our previous study (Abrol et al. 2017) using Group ICA based on the GIFT toolbox (Calhoun et al. 2001; Cetin et al. 2016), with a model order of 100 components (Fig. 1A). The spatial maps and time courses of the components were examined to select physiologically nonartifactual and previously established functional networks, as reported in Allen et al. (2011)) and Du et al. (2015). Following this, 61 components were selected, which had local peak activations lying in GM, with time-courses dominated by low-frequency fluctuations, and exhibited high spatial overlap with the established rsfMRI networks.
Source-Based Morphometry
The segmented GM images were decomposed using spatial ICA through the GIFT toolbox (Xu et al. 2009; Cota Navin Gupta and Calhoun 2017), which linearly decomposed the GM matrix into a mixing matrix that represents the relative weight of each subject for every component, and the source matrix representing the maximally spatially independent GM regions. We chose a model order of 100 components to match the numbers of components used in the fMRI analysis (Fig. 1A). All 100 structural components were visually inspected by three experts. We excluded structural components that had significant spatial overlaps with ventricles, white matter, large vasculature, and the brainstem, or components located at the boundaries between these regions and GM. For the purpose of spatial correlation, the GM components were resliced to 3 mm × 3 mm × 3 mm to match the dimensions of the functional components.
We then defined nine domains/networks based on Yeo et al.’s seven-network template (Yeo et al. 2011), with two extended networks including the cerebellar and subcortical networks. The nine networks are: visual network (VIS), somatomotor network (SM), dorsal attention network (DA), ventral attention network (VA), subcortical network (SUB), limbic network (LIMBIC), FPN, default mode network (DMN), and cerebellar network (CB). All the effective GM and fMRI components were further grouped into the nine networks based on which network/domain the peak region belongs to.
Spatial Cross-Correlation between Structural and Functional Components
To assess both linear and nonlinear spatial correspondence, we calculated spatial correlation between the selected structural and functional spatial maps using Pearson correlation (PC) and mutual information (MI). Given two random variables 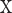 and
and 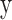 , their PC can be defined in terms of their covariance
, their PC can be defined in terms of their covariance 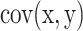 , SD of
, SD of 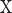 and
and 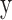 as equation (1), and their MI (Fig. 1C, computed using the mutualinfo package in MATLAB) is defined in terms of their probabilistic density functions
as equation (1), and their MI (Fig. 1C, computed using the mutualinfo package in MATLAB) is defined in terms of their probabilistic density functions 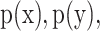 and
and 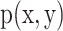 as equation (2)
as equation (2)
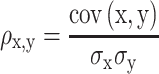 |
(1) |
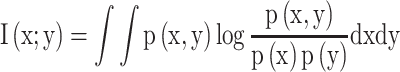 |
(2) |
Note that before computing the correspondence, the ICA-decomposed spatial maps have been converted to Z-scores and thresholded at |Z| > 2.
Replication using the UK Biobank Data
In order to validate the matched S-F component pairs, we used spatial ICA to decompose the replication data with the same model order of 100 components (Fig. 1B). The same inclusion criteria of component selection in the discovery dataset were applied to select good components in the replication dataset. We then computed the spatial correlation between GM components in the replication dataset and fMRI components in the discovery dataset, as well as GM components in the discovery dataset (Fig. 1C). If structural components in one matched structural–structural pair between discovery and replication cohorts both show high correlation with the same fMRI component, then the S-F pair in the discovery cohort was regarded as replicated.
Comparison between Different Network Modules
We subsequently counted the numbers of matched S-F pairs in each network module using the discovery dataset and the replicated percentage in each network module using the replication dataset. We then added up the values of PC and MI in both cohorts for each S-F pair and sorted them into a decreased order to explore which network module would present more S-F correspondence and which module indicate more S-F divergence (Fig. 1D). Moreover, we examined the S-F correspondence of different network modules using PC or MI separately.
Results
Structural Architectures Match Intrinsic Functional Networks
In the discovery dataset, 71 structural GM components (Fig. S2) and 61 fMRI components (Fig. S3) were retained for analysis after removing artifactual components through visual inspections by three professors. Out of the 71 GM versus 61 fMRI components comparisons, 44 (62%) structural components were matched with 47 (77.05%) functional components passing the predetermined PC coefficient threshold of 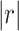 > 0.25 and an MI threshold of MI > 0.2 (Fig. S4). We followed Smith et al. (Smith et al. 2009) to select |r| > 0.25 as threshold for PC, which corresponds to a significance level of P < 1e−12, passing Bonferroni correlation P = 0.05/71/61). For MI, we computed the distribution of MI between structural and functional ICs as shown in Fig. S5 and then selected the top 2% (MI = 0.2) S-F pairs as significantly matched pairs. Moreover, there is a need for automated approaches to select the threshold in the future. As more than one functional component matched per structural component, as well as one functional component sometimes matched with several structural components, these matched components in discovery dataset together formed 70 S-F pairs. After sorting the matched S-F pairs into nine domains/networks, we computed the numbers of matched S-F pairs in each brain network of the discovery dataset (Fig. 3). The numbers of matched S-F pairs were higher in the VIS (15 pairs), DMN (13 pairs), and CB (11 pairs), but relatively lower in VA (5 pairs), DA (5 pairs), and LIMBIC (2 pairs).
> 0.25 and an MI threshold of MI > 0.2 (Fig. S4). We followed Smith et al. (Smith et al. 2009) to select |r| > 0.25 as threshold for PC, which corresponds to a significance level of P < 1e−12, passing Bonferroni correlation P = 0.05/71/61). For MI, we computed the distribution of MI between structural and functional ICs as shown in Fig. S5 and then selected the top 2% (MI = 0.2) S-F pairs as significantly matched pairs. Moreover, there is a need for automated approaches to select the threshold in the future. As more than one functional component matched per structural component, as well as one functional component sometimes matched with several structural components, these matched components in discovery dataset together formed 70 S-F pairs. After sorting the matched S-F pairs into nine domains/networks, we computed the numbers of matched S-F pairs in each brain network of the discovery dataset (Fig. 3). The numbers of matched S-F pairs were higher in the VIS (15 pairs), DMN (13 pairs), and CB (11 pairs), but relatively lower in VA (5 pairs), DA (5 pairs), and LIMBIC (2 pairs).
Figure 3.
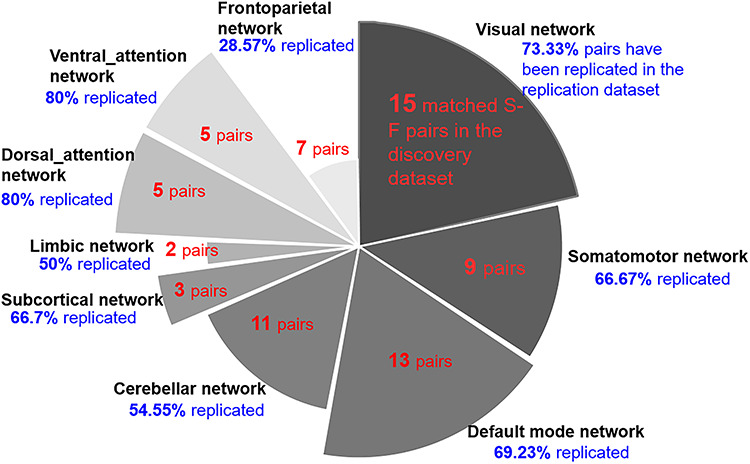
The numbers of matched pairs in the discovery data and the replicated percentages in the replication data. The S-F correspondence in visual (15 pairs), default model network (13 pairs), and cerebellar (11 pairs) networks is higher, whereas the correspondence in ventral_attention (5 pairs), dorsal_attention (5 pairs), and limbic networks (2 pairs) is relatively low. Moreover, replicated results indicate that the visual (73.33%) and DMN (69.23%) are highly replicated, while the FPN (28.57%) is not well replicated.
About 95 structural components in the replication data were selected as nonartifactual components after ICA decomposition. In the comparison of 95 structural components (replication dataset) and 61 functional components (discovery dataset), 66 (69.47%) structural components were matched with 49 (80.33%) functional components (Fig. S6). Meanwhile, 57 (60%) structural components in the replication dataset were matched with 50 (70.42%) structural components of discovery dataset (Fig. S7). If one matched structural–structural pair between discovery and replication cohorts showed high correlation with the same fMRI component, then the S-F pair in the discovery cohort was regarded as replicated. In total, 45 (64.28%) out of the 70 matched S-F pairs in the discovery dataset were replicated in the UK Biobank data. We set the same thresholds (|r| > 0.25 and MI > 0.2) as the discovery dataset to select the significant corresponding component pairs. The replicated percentages in each of networks are presented in Figure 3, which indicates that the VIS (73.33%) and DMN (69.23%) are highly replicated, while the FPN (28.57%) is not well replicated. The spatial maps of these matched 45 S-F pairs in discovery dataset and replication dataset are separately depicted in Figure 4A and 4B. The corresponding values are displayed in Figures S8–S11. To assess the significance of the spatial cross-correlations between the structural and functional independent components (ICs) among voxels, we randomly shuffled the structural IC across voxels and rerunning the correlation analysis with the functional IC 10 000 times as did in Sui et al. (2018) for each of the 45 identified S-F pairs. Then, we compared the strength of our observed correlations to this empirically generated null distribution. All the 45 S-F pairs quantified the probability P < 0.001 of obtaining the observed correlations between structural and functional ICs by chance. In addition, we also generated 1000 rotational permutations for the structural ICs using the framework proposed by Alexander-Bloch et al. (2018) to test the significance of spatial autocorrelation among voxels. As shown in Figure S12, none of the 45 S-F pairs presented a probability P > 0.05 of obtaining the observed correlations between structural and functional ICs by chance.
Figure 4.
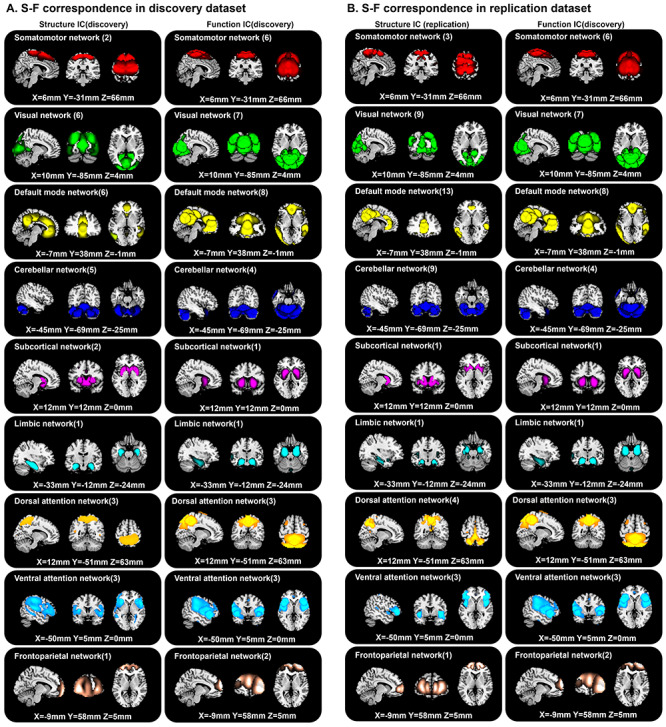
S-F correspondence in the discovery dataset (A) and replication dataset (B).
Furthermore, we computed the overlap between different structural ICs or functional ICs. About 94.05% of pair-wise structural ICs presented an overlap less than 0.1, and 92.29% of pair-wise function ICs exhibited an overlap less than 0.1 (Figure S13). Among all the 45 structure-function pairs, we separately computed the percentage of the matched structural or functional components of the whole brain. The results indicated that all the matched structural components covered 87.49% of the whole brain, while the functional components occupied 74.32%. For each network, we first added up all the matched structural ICs and all the corresponding functional ICs separately. Then, we counted overlapped dices between structural and functional ICs for each network and calculated the overlap percentages. As shown in Figure S14, the unimodal cortical areas (e.g., somatomotor and visual networks) still exhibited higher spatial dice-overlap (>70%) compared with heteromodal association cortices (<60%), except for the DMN.
Somatomotor Network
A large component, s-IC 12, spanning the supplementary motor areas, and bilateral pre- and post-central gyri are correlated with four rsfMRI components (Fig. S8A). The rs-IC 72, with peaks at the precentral gyri, presents the highest correspondence of PC and MI to the structural component for both discovery and replication dataset. The other three rsfMRI components are rs-IC49 and rs-IC52, centered at the paracentral lobule, and rs-IC99, which represents the bilateral postcentral gyri. A second structural component, s-IC19, which is also quite large and contains voxels spanning much of the supplementary motor area, is correlated to rs-IC36 and rs-IC13, with peaks at aspects of the supplementary motor area.
Visual Network
Notably, the visual network includes the largest numbers of S-F pairs, which is also observed in the replication dataset (Fig. 3). Structural component s-IC16, which largely centers at the calcarine gyrus, presents the second highest correspondence to functional component rs-IC25 in all matched S-F pairs and replicated in rep-sIC60 (Fig. S8B). The functional components rs-IC63 and rs-IC 96, centered at the calcarine gyrus, are also correlated with s-IC16. The other smaller structural component s-IC13 with peaks at calcarine gyrus and lingual gyrus is correlated with rs-IC25, as well as rs-IC17 and replicated in rep-sIC38. Components s-IC95 and s-IC96 comprised of a component pair, with peaks at right and left calcarine region, respectively, which are correlated with rs-56, rs-IC3, and rs-IC63. Another correlated region is the lingual gyrus, where component s-IC66 is associated with rs-IC 17 and replicated in rep-sIC66. The third region with a replicated S-F correspondence in the visual network is middle occipital gyrus. Structural components s-IC 79 is correlated with functional component rs-IC76, with peaks at the bilateral middle occipital gyrus.
Default Mode Network
The DMN also shows a high S-F correspondence (Fig. S9C). Component s-IC31, primarily comprised of the middle cingulate gyrus, is highly correlated with rs-IC 67 and rs-IC46. Component s-IC7, which contains voxels residing in the precuneus area, is correlated with three functional components. In order of correspondence magnitude, they are rs-IC34, rs-IC46, and rs-IC30, which presents aspects of precuneus and middle cingulate gyrus. Structural components, s-IC45 and s-IC52, are symmetrical components, which are respectively composed of right middle temporal gyrus and left middle temporal gyrus. They are correlated with a symmetrical pair rs-IC57 and rs-IC75, which are also replicated in a symmetrical pair rep-sIC25 and rep-sIC18. More interesting, structural components, s-IC40 and s-IC91, containing voxels in different subdivisions of anterior cingulate cortex, are correlated with two components rs-IC39 and rs-IC90 and replicated in rep-sIC 85 and rep-sIC 15, centered on the same subdivisions of anterior cingulate cortex.
Cerebellar Network
Components s-IC53 and s-IC46, respectively, represent left and right cerebellum, which are correlated with rs-IC26 and rs-IC40, peaking at left and right cerebellum, respectively, and replicated in nine components (Fig. S10D). Component s-IC36, primarily composed of vermis, is correlated with rsfMRI component rs-IC8 and replicated in rep-sIC1 and rep-sIC48, centered on vermis, a narrow midline zone in cerebellum. Structural components s-IC10 and s-IC11 are correlated with rs-IC26, replicated in rep-sIC53 and rep-sIC4, primarily comprised of bilateral cerebellum. Results from previous studies have found that higher scores on vocabulary, reading, working memory, and set-shifting were associated with increased GM in the posterior cerebellum (Moore et al. 2017), which is an example of how structural abnormalities directly relate to functional processing.
Subcortical Network
Subcortical structure s-IC3, comprising the putamen and parts of caudate, presents high S-F correspondence with functional component rs-IC33 in discovery dataset (Fig. S10E). The other subcortical component s-IC17, composed of bilateral caudate, is correlated with the same rsfMRI component, primarily comprising the bilateral putamen and caudate. The putamen and caudate contain the same types of neurons and circuits, which together form the dorsal striatum. The dorsal striatum plays an important role in the motor and reward systems, which receives inputs from cortical regions and then serves as the primary input to the rest of basal ganglia (Ferre et al. 2010). Thus, the putamen and caudate are likely to be decomposed in one functional component by ICA because of the similar brain function. However, as a white matter tract in the dorsal striatum structurally separates the caudate nucleus and the putamen (Ferre et al. 2010), the putamen and caudate are more likely to be decomposed into two components by ICA for the structural data.
Limbic Network
Only component s-IC23, primarily composed of the hippocampus, has been revealed to be correlated to rs-IC44 and replicated in rep-sIC 77 (Fig. S10F), primarily associated with memory function (van Strien et al. 2009). While S-F coherence of para-hippocampus was also observed in the discovery data as shown in Figure S4-1, the pair was not well replicated.
Dorsal_attention Network
Component s-IC67, which also largely covers the precuneus, is correlated to rs-IC30 and rs-IC35, replicated in five components (Fig. S11G). Two structural components s-IC 25 and s-IC27 are revealed to be correlated with the same functional component rs-IC76, which comprised of both parts of middle occipital gyrus and inferior parietal gyrus. Different from component s-IC79 and s-IC95, which also comprised of middle occipital but belongs to the visual network, s-IC25 and s-IC27 are more close to the inferior parietal gyrus, sorted to the dorsal attention network. Inferior parietal gyrus is associated with bottom-up attention (Igelstrom and Graziano 2017).
Ventral_attention Network
Structural component s-IC 30, centered at the supramarginal gyrus, is correlated with rs-IC 29 and replicated in rep-sIC73, which represents the aspects of the supramarginal gyrus (Fig. S11H). Several studies have reported the supramarginal gyrus in interoceptive attention/awareness tasks (Kashkouli Nejad et al. 2015). The other symmetrical components s-IC56 and s-IC76 are correlated with rs-IC31 and rs-IC94, respectively, and replicated in rep-sIC24 and rep-sIC68, primarily composed of the insula.
Frontoparietal Network
The FPN yielded the lowest degree of S-F correspondence, and the effects observed in the discovery sample were largely unreplicated (Fig. 3). Only component s-IC18, peaking in the superior frontal gyrus, is correlated to rs-IC 89 and rs-IC 85, and weakly replicates in rep-sIC 21, which represents activations over similar regions (Fig. S11I).
Unimodel Cortex Exhibited Better S-F Correspondence than Hetermodel Cortex Except DMN
Figure 5A depicts the values of PC and MI of each replicated S-F pair in both datasets. Results show that components from SM and VIS present the highest S-F correspondence when adding up values from both metrics (PC and MI) and both cohorts, followed by DMN, LIMBIC, and CB. In contrast, the values in VA, DA, and FPN are relatively lower than other networks, indicating more divergence between brain structure and function. Furthermore, we computed the S-F correspondence using PC or MI separately. As shown in Figure 5B and 5C, either using PC or MI, the correspondences consistently show that the SM, VIS, and DMN yield higher S-F correspondences, while components in the DA and FPNs show more divergence between structure and function.
Figure 5.
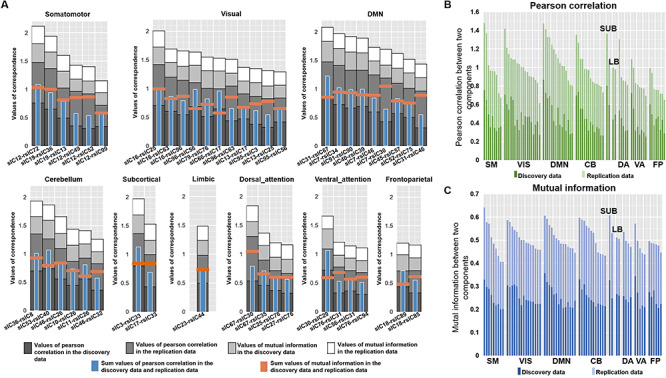
Comparison of S-F coherence across nine domains. (A) The sum of PC and MI in both discovery dataset and replication dataset of each component. The darker gray to white bars, respectively, represent PC values in the discovery dataset, PC values in the replication dataset, MI values in the discovery dataset, and MI values in the replication dataset. The blue bar indicates the sum of PC and MI in the discovery dataset, while the orange bar shows the sum of PC and MI in the replication dataset. (B) The S-F correspondence in the discovery and replication dataset measured only by PC. The dark green bar represents PC values in the discovery data, and the light green color indicates PC values in the replication data. (C) The S-F correspondence in the discovery and replication dataset measured only by MI. The dark blue bar represents MI values in the discovery data, and the light blue bar indicates MI values in the replication data.
Discussion
In this study, we demonstrate that human structural architectures match intrinsic functional networks across the entire brain. With an ICA decomposition on GM volumes and spontaneous fluctuations of discovery data (6000+ scans), the structural components found are largely correspond to functional networks (62% matched 77.05%). Thus, what we are seeing is the functional covariations in resting state BOLD signal, at the level of group networks across thousands of individuals, which correlates in some cases with the structural correlations. To aid the robustness of the identified pairs, we replicated the S-F coherence in another independent dataset (UK Biobank 9000+ subjects). While 64.28% pairs are validated, the replicated percentage in each domain network is not identical. For example, replicated percentage in the FPN was 28.57%, implying that individual difference was large in this area. This was consistent with previous studies, which states that the FPN has emerged as the most distinctive fingerprinting feature for identifying individuals (Finn et al. 2015). In total, 45 S-F component pairs with high spatial consistency in both discovery and replication cohorts were identified, which were divided into nine major networks, providing a stable S-F correspondence template that may be of use to the larger neuroimaing community.
The replicated S-F pairs and their correspondence values suggested that the unimodal cortical areas (especially the SM and VIS networks) show higher S-F correspondence, while heteromodal association cortical areas, especially the FPN, exhibit more divergence between intrinsic functional and structural networks. Consistent with this, Margulies et al. (2016) also reported that hetermodal regions standed at one extreme of a principal gradient hierarchy compared to primary sensory and motor regions, which allows them to process transmodal information that is unrelated to immediate sensory input. Unimodal cerebral cortex (Mendoza 2011) (somatomotor network and visual network), especially the precentral gyrus, supplementary motor area, and primary visual cortex (calcarine area, V1), exhibited higher S-F correpondence than other areas in both the discovery and replication datasets. The precentral gyrus and supplementary motor area are primarily associated with motor function (Graziano et al. 2002; Nachev et al. 2008). Visual area V1 receives sensory inputs from the thalamus and plays an important role in the extraction of early visual features (Mechelli et al. 2000). The somatomotor and visual regions have also been revealed to present the least individual differences in functional variability (Mueller et al. 2013). However, the heteromodal association cortex (Green 2004), except the DMN, shows relatively low S-F correspondence (DA, VA, and FPN). The FPN presents the least S-F correspondence compared with the other networks. It has been previously demonstrated that the regions presenting the most prominent intersubject variability are also the regions showing the most rapid expansion during human brain evolution (Mueller et al. 2013), as well as greatest postnatal enlargement and a low maturation rate (Hill et al. 2010). Wang et al. suggested that a greatly expanded and slowly maturing association cortex can provide a higher degree of freedom, both in physical space and time, for influences of environmental factors, potentially giving rise to interindividual variability (Wang and Liu 2014), leading to weak S-F correspondences in these areas. In contrast, the unimodal regions mature early in life with a low expansion rate and are more stable to the environmental factors. This may be why regions in this network are more highly structurally and functionally correlated, with more spatially contiguous components. Future studies might consider incorporating hypotheses about these differences or develop more systematic categorization approaches.
The spatial consistence in DMN is interesting; for example, the precuneus is split into two structural subregions (s-IC7 and s-IC67), which respectively correspond to two functional subregions (rs-IC34 and rs-IC30). The two subregions are consistent with previous subdivisions of the precuneus, which represent different functions: components s-IC7 and rs-IC34 are the posterior precuneus, which shows strong functional connectivity with visual-related areas, such as the cuneus (Margulies et al. 2009); components s-IC67 and rs-IC30 are the anterior precuneus, which exhibits strong functional connectivity with sensorimotor-related regions. Meanwhile, the other similar case is anterior cingulate cortex, where components s-IC40 and s-IC91 are correlated with two different anterior cingulate cortex in functional data (rs-IC39 and rs-IC90), consisting with previous subdivision of the anterior cingulate cortex (Bush et al. 2000). Component s-IC40 is the dorsal part of the anterior cingulate cortex, which is connected with the prefrontal cortex and parietal cortex, participating in cognitive control (Shenhav et al. 2016). In contrast, component s-IC91 is the ventral part of the anterior cingulate cortex, involving in generating emotional responses (Etkin et al. 2011). Besides, structural components, s-IC45 and s-IC52, are symmetrical components, which are respectively composed of right middle temporal gyrus and left middle temporal gyrus, correlated with symmetrical pair rs-IC57 and rs-IC75, and replicated in a symmetrical pair (rep-sIC25 and rep-sIC18). As shown in the Brainnetome Atlas (https://atlas.brainnetome.org/bnatlas.html), which yields functional characterization of subregions based on the BrainMap database using forward and reverse inferences (Fan et al. 2016), the left middle temporal gyrus is primarily related to cognition, language, and syntax, while the right middle temporal gyrus is involved in action and observation.
Despite the strengths of the current study, it still has some limitations. The first limitation is of course methodological: the structural images were generally collected at a much finer resolution (1-mm isotropic, generally) than the functional images (>3 mm on a side), and the functional imaging is susceptible to signal drop-out in inferior areas or near the tissue border. We attempted to address this by reslicing the structural images to match the functional and restricting the analysis to the areas of GM, where signal in both modalities was robust. The second limitation is the lack of careful assessment of health status for the individuals included in discovery dataset. However, our results were further replicated in a large independent dataset with only healthy subjects. We believe that the results may be more driven by the common characteristics on S-F correspondence. The third issue is that the MRI scanners and imaging protocols were not identical across the discovery and replication cohorts. However, since one important aim of this study is to identify a stable S-F template (i.e., a set of components with higher S-F correspondence), the replicated results in data with different scanners and imaging protocols provide a more generalizable result.
Author Contributions
V.D.C., J.S., and N.L. designed the experiments. N.L. conducted the analysis. V.D.C. and J.S. supervised and guided all aspects of the work. N.L., V.D.C., and J.S. wrote the paper. A.A. helped provide the information of the discovery data. Z.F. and E.D. helped select the components. D.C.G and A.L.R helped provide the information of the UK Biobank data. J.C., J.A.T, L.F., D.L., C.Z., Y.X., D.C.G, A.L.R, M.T.B., and G.D.P. helped revise the paper.
Funding
This work was supported by the National Institutes of Health (No. 2R01EB005846, P20GM103472, R01REB020407, R01MH117107), the National Science Foundation (No. 1539067), the Natural Science Foundation of China (No. 61773380, 81871052, 81801679, 81571319), the Strategic Priority Research Program of the Chinese Academy of Sciences (No. XDB32040100), Brain Science and Brain-Inspired Technology Plan of Beijing City (No. Z181100001518005), the Key Projects of the Natural Science Foundation of Tianjin, China (No. 17JCZDJC35700), and the National Key Research and Development Program of China (No. 2016YFC1307004). Conflict of Interest: The authors declare no competing interests.
Data and Materials Availability
The multimodal data used in the present study can be accessed upon request to the corresponding authors.
Supplementary Material
References
- Abrol A, Damaraju E, Miller RL, Stephen JM, Claus ED, Mayer AR, Calhoun VD. 2017. Replicability of time-varying connectivity patterns in large resting state fMRI samples. Neuroimage. 163:160–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander-Bloch A, Giedd JN, Bullmore E. 2013. Imaging structural co-variance between human brain regions. Nat Rev Neurosci. 14:322–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander-Bloch AF, Shou H, Liu S, Satterthwaite TD, Glahn DC, Shinohara RT, Vandekar SN, Raznahan A. 2018. On testing for spatial correspondence between maps of human brain structure and function. NeuroImage. 178:540–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfaro-Almagro F, Jenkinson M, Bangerter NK, Andersson JLR, Griffanti L, Douaud G, Sotiropoulos SN, Jbabdi S, Hernandez-Fernandez M, Vallee E, et al. 2018. Image processing and quality control for the first 10,000 brain imaging datasets from UK biobank. Neuroimage. 166:400–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen EA, Erhardt EB, Damaraju E, Gruner W, Segall JM, Silva RF, Havlicek M, Rachakonda S, Fries J, Kalyanam R, et al. 2011. A baseline for the multivariate comparison of resting-state networks. Front Syst Neurosci. 5(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum GL, Cui Z, Roalf DR, Ciric R, Betzel RF, Larsen B, Cieslak M, Cook PA, Xia CH, Moore TM, et al. 2020. Development of structure-function coupling in human brain networks during youth. Proc Natl Acad Sci USA. 117:771–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush G, Luu P, Posner MI. 2000. Cognitive and emotional influences in anterior cingulate cortex. Trends Cogn Sci. 4:215–222. [DOI] [PubMed] [Google Scholar]
- Calhoun VD, Adali T, Pearlson GD, Pekar JJ. 2001. A method for making group inferences from functional MRI data using independent component analysis. Hum Brain Mapp. 14:140–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cetin MS, Houck JM, Rashid B, Agacoglu O, Stephen JM, Sui J, Canive J, Mayer A, Aine C, Bustillo JR, et al. 2016. Multimodal classification of schizophrenia patients with MEG and fMRI data using static and dynamic connectivity measures. Front Neurosci. 10:466–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Liu J, Calhoun VD, Arias-Vasquez A, Zwiers MP, Gupta CN, Franke B, Turner JA. 2014. Exploration of scanning effects in multi-site structural MRI studies. J Neurosci Methods. 230:37–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clos M, Rottschy C, Laird AR, Fox PT, Eickhoff SB. 2014. Comparison of structural covariance with functional connectivity approaches exemplified by an investigation of the left anterior insula. Neuroimage. 99:269–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cota Navin Gupta JAT, Calhoun VD. 2017. Source Based Morphometry: Data-Driven Multivariate Analysis of Structural Brain Imaging Data, Brain Morphometry.
- Du YH, Pearlson GD, Liu JY, Sui J, Yu QB, He H, Castro E, Calhoun VD. 2015. A group ICA based framework for evaluating resting fMRI markers when disease categories are unclear: application to schizophrenia, bipolar, and schizoaffective disorders. Neuroimage. 122:272–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etkin A, Egner T, Kalisch R. 2011. Emotional processing in anterior cingulate and medial prefrontal cortex. Trends Cogn Sci. 15:85–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan LZ, Li H, Zhuo JJ, Zhang Y, Wang JJ, Chen LF, Yang ZY, Chu CY, Xie SM, Laird AR, et al. 2016. The human Brainnetome atlas: a new brain atlas based on connectional architecture. Cerebral Cortex. 26:3508–3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferre S, Lluis C, Justinova Z, Quiroz C, Orru M, Navarro G, Canela EI, Franco R, Goldberg SR. 2010. Adenosine-cannabinoid receptor interactions. Implications for striatal function. Brit J Pharmacol. 160:443–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn ES, Shen XL, Scheinost D, Rosenberg MD, Huang J, Chun MM, Papademetris X, Constable RT. 2015. Functional connectome fingerprinting: identifying individuals using patterns of brain connectivity. Nat Neurosci. 18:1664–1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao S, Calhoun VD, Sui J. 2018. Machine learning in major depression: from classification to treatment outcome prediction. CNS Neurosci Ther. 24:1037–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng X, Li G, Lu Z, Gao W, Wang L, Shen D, Zhu H, Gilmore JH. 2017. Structural and maturational covariance in early childhood brain development. Cereb Cortex. 27:1795–1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graziano MS, Taylor CS, Moore T. 2002. Complex movements evoked by microstimulation of precentral cortex. Neuron. 34:841–851. [DOI] [PubMed] [Google Scholar]
- Green R. 2004. Heteromodal association cortex in schizophrenia. Am J Psychiatry. 161:1723–1724author reply 1724. [DOI] [PubMed] [Google Scholar]
- Greicius MD, Supekar K, Menon V, Dougherty RF. 2009. Resting-state functional connectivity reflects structural connectivity in the default mode network. Cereb Cortex. 19:72–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill J, Inder T, Neil J, Dierker D, Harwell J, Van Essen D. 2010. Similar patterns of cortical expansion during human development and evolution. Proc Natl Acad Sci USA. 107:13135–13140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honey CJ, Sporns O, Cammoun L, Gigandet X, Thiran JP, Meuli R, Hagmann P. 2009. Predicting human resting-state functional connectivity from structural connectivity. Proc Natl Acad Sci USA. 106:2035–2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igelstrom KM, Graziano MSA. 2017. The inferior parietal lobule and temporoparietal junction: a network perspective. Neuropsychologia. 105:70–83. [DOI] [PubMed] [Google Scholar]
- Kashkouli Nejad K, Sugiura M, Nozawa T, Kotozaki Y, Furusawa Y, Nishino K, Nukiwa T, Kawashima R. 2015. Supramarginal activity in interoceptive attention tasks. Neurosci Lett. 589:42–46. [DOI] [PubMed] [Google Scholar]
- Luo N, Sui J, Abrol A, Lin D, Chen J, Vergara VM, Fu Z, Du Y, Damaraju E, Xu Y, et al. 2020. Age-related structural and functional variations in 5,967 individuals across the adult lifespan. Hum Brain Mapp. 41:1725–1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo N, Tian L, Calhoun VD, Chen J, Lin D, Vergara VM, Rao S, Yang J, Zhuo C, Xu Y, et al. 2019. Brain function, structure and genomic data are linked but show different sensitivity to duration of illness and disease stage in schizophrenia. NeuroImage Clinical. 23:101887–101887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulies DS, Ghosh SS, Goulas A, Falkiewicz M, Huntenburg JM, Langs G, Bezgin G, Eickhoff SB, Castellanos FX, Petrides M, et al. 2016. Situating the default-mode network along a principal gradient of macroscale cortical organization. Proc Natl Acad Sci USA. 113:12574–12579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulies DS, Vincent JL, Kelly C, Lohmann G, Uddin LQ, Biswal BB, Villringer A, Castellanos FX, Milham MP, Petrides M. 2009. Precuneus shares intrinsic functional architecture in humans and monkeys. Proc Natl Acad Sci USA. 106:20069–20074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mechelli A, Humphreys GW, Mayall K, Olson A, Price CJ. 2000. Differential effects of word length and visual contrast in the fusiform and lingual gyri during reading. Proc Biol Sci. 267:1909–1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza JE. 2011. Unimodal Cortex. In: Kreutzer JS, DeLuca J, Caplan B, editors. Encyclopedia of Clinical Neuropsychology New York. NY: Springer New York, pp. 2578–2578. [Google Scholar]
- Misic B, Betzel RF, Reus MA, Heuvel MP, Berman MG, McIntosh AR, Sporns O. 2016. Network-level structure-function relationships in human Neocortex. Cereb Cortex. 26:3285–3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore DM, D'Mello AM, McGrath LM, Stoodley CJ. 2017. The developmental relationship between specific cognitive domains and grey matter in the cerebellum. Dev Cognit Neurosci. 24:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller S, Wang D, Fox MD, Yeo BT, Sepulcre J, Sabuncu MR, Shafee R, Lu J, Liu H. 2013. Individual variability in functional connectivity architecture of the human brain. Neuron. 77:586–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachev P, Kennard C, Husain M. 2008. Functional role of the supplementary and pre-supplementary motor areas. Nat Rev Neurosci. 9:856–869. [DOI] [PubMed] [Google Scholar]
- Paquola C, Vos De Wael R, Wagstyl K, Bethlehem RAI, Hong SJ, Seidlitz J, Bullmore ET, Evans AC, Misic B, Margulies DS, et al. 2019. Microstructural and functional gradients are increasingly dissociated in transmodal cortices. PLoS Biol. 17:e3000284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power JD, Barnes KA, Snyder AZ, Schlaggar BL, Petersen SE. 2012. Spurious but systematic correlations in functional connectivity MRI networks arise from subject motion. Neuroimage. 59:2142–2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power JD, Fair DA, Schlaggar BL, Petersen SE. 2010. The development of human functional brain networks. Neuron. 67:735–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeley WW, Crawford RK, Zhou J, Miller BL, Greicius MD. 2009. Neurodegenerative diseases target large-scale human brain networks. Neuron. 62:42–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenhav A, Cohen JD, Botvinick MM. 2016. Dorsal anterior cingulate cortex and the value of control. Nat Neurosci. 19:1286–1291. [DOI] [PubMed] [Google Scholar]
- Silver M, Montana G, Nichols TE, Neuroimaging AD. 2011. False positives in neuroimaging genetics using voxel-based morphometry data. Neuroimage. 54:992–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SM, Fox PT, Miller KL, Glahn DC, Fox PM, Mackay CE, Filippini N, Watkins KE, Toro R, Laird AR, et al. 2009. Correspondence of the brain's functional architecture during activation and rest. Proc Natl Acad Sci USA. 106:13040–13045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui J, Qi S, Erp TGM, Bustillo J, Jiang R, Lin D, Turner JA, Damaraju E, Mayer AR, Cui Y, et al. 2018. Multimodal neuromarkers in schizophrenia via cognition-guided MRI fusion. Nat Commun. 9:3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strien NM, Cappaert NLM, Witter MP. 2009. The anatomy of memory: an interactive overview of the parahippocampal-hippocampal network. Nat Rev Neurosci. 10:272–282. [DOI] [PubMed] [Google Scholar]
- Vázquez-Rodríguez B, Suárez LE, Markello RD, Shafiei G, Paquola C, Hagmann P, Heuvel MP, Bernhardt BC, Spreng RN, Misic B. 2019. Gradients of structure-function tethering across neocortex. Proc Natl Acad Sci USA. 116:21219–21227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Liu H. 2014. Functional connectivity architecture of the human brain: not all the same. Neuroscientist. 20:432–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Groth KM, Pearlson G, Schretlen DJ, Calhoun VD. 2009. Source-based Morphometry: the use of independent component analysis to identify gray matter differences with application to schizophrenia. Hum Brain Mapp. 30:711–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P, Guo W, Jin T, Wang J, Fan D, Hao Z, Jing S, Han C, Du J, Jiang D, et al. 2017. TIMP-2 SNPs rs7342880 and rs4789936 are linked to risk of knee osteoarthritis in the Chinese Han population. Oncotarget. 8:1166–1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan CG, Cheung B, Kelly C, Colcombe S, Craddock RC, Di Martino A, Li Q, Zuo XN, Castellanos FX, Milham MP. 2013. A comprehensive assessment of regional variation in the impact of head micromovements on functional connectomics. Neuroimage. 76:183–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo BTT, Krienen FM, Sepulcre J, Sabuncu MR, Lashkari D, Hollinshead M, Roffman JL, Smoller JW, Zoller L, Polimeni JR, et al. 2011. The organization of the human cerebral cortex estimated by intrinsic functional connectivity. J Neurophysiol. 106:1125–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielinski BA, Gennatas ED, Zhou J, Seeley WW. 2010. Network-level structural covariance in the developing brain. Proc Natl Acad Sci USA. 107:18191–18196. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


