Abstract
Mice deficient in the IL-10 pathway are the most widely-used models of intestinal immunopathology.IL-17A is strongly implicated in gut disease in mice and humans, but conflicting evidence has drawn IL-17’s role in the gut into question.IL-22 regulates antimicrobial and repair activities of intestinal epithelial cells (IECs) and is closely associated with IL-17A responses but it’s role in chronic disease is uncertain. We report that IL-22, like IL-17A, is aberrantly expressed in colitic Il10−/− mice. While IL-22+Th17 cells were elevated in the colon, IL-22-producing ILC3s were highly enriched in the small intestines of Il10−/− mice. Remarkably, Il10−/−Il22−/− mice did not develop colitis despite retaining high levels of Th17 cells and remaining colonized with colitogenic Helicobacter spp.. Accordant with IL-22-induced IEC proliferation, the epithelia hyperplasia observed in Il10−/− animals was reversed in Il10−/−Il22−/− mice. Also, the high levels of antimicrobial IL-22-target genes, including Reg3g, were normalized in Il10−/−Il22−/− mice. Consistent with a heightened antimicrobial environment, Il10−/− mice had reduced diversity of the fecal microbiome that was reestablished in Il10−/−Il22−/− animals. These data suggest that spontaneous colitis in Il10−/− mice is driven by IL-22 and implicates an underappreciated IL-10-IL-22 axis in regulating intestinal homeostasis.
Introduction
Based on cues from the microenvironment, the mucosal immune system fine-tunes immune effector programs to maximize host defenses at barrier surfaces while preventing excess inflammation to avoid damage to host tissues. In the gut, dysregulation of this dynamic process can result in chronic inflammation and disease pathology. The etiology of inflammatory bowel disease (IBD), which encompasses Crohn’s disease and colitis, is still poorly understood despite being intensively studied. Our understanding of the immune and microbial factors that contribute to disease susceptibility have been complicated, at least in part, by difficulties in interpreting data generated under different experimental conditions, and with different animal models of IBD (1, 2).
IL-10 is an immunoregulatory cytokine that plays a central role in regulating intestinal inflammation in humans and mice (3).Mice deficient in IL-10 (4) or the IL-10 receptor (5) develop spontaneous colitis early in life and are one of the most widely used animal models for studying the pathogenesis of human IBD (6, 7).The development of colitis in IL-10-deficient mice is dependent on the intestinal microbiota (8). More specifically, co-colonization with “pathobionts” such as Helicobacter spp., which do not cause disease in immunocompetent mice (9), are required for the development of colitis in Il10−/− mice. Although it is clear that excessive immune reactivity to microbial antigens triggers colitis in Il10−/− mice (10, 11), remarkably, the host factors which drive intestinal pathology have been difficult to define.
Early work suggested that dysregulation of Th1 immunity (IL-12/IFN-γ) was responsible for colitis in Il10−/− mice (12). Prior to the discovery of the Th17 pathway (IL-23/IL-17), early work naturally focused on Th1-mediated responses (IL-12/IFN-γ).An important study by Yen and colleagues in 2006, specifically examined the contributions of IL-12-dependent Th1 and IL-23-dependent Th17 immunity to the development of colitis (13). They demonstrated that co-deletion of IL-23 (Il10−/−Il23p19−/−) but not IL-12 (Il10−/−Il12p35−/−), rescued Il10-deficient mice from spontaneous colitis (13). This study offered convincing evidence that excessive production of IL-17, driven by IL-23, was in fact primarily responsible for the development of colitis in Il10−/− animals (13). Thus, the prevailing model suggests that excessive Th17 development, driven by IL-23, is responsible for IBD pathology.
Additional studies have largely supported IL-23’s role in promoting intestinal inflammation (14–16), however, IL-17’s role has been somewhat less clear. A group of reports have accumulated suggesting that intestinal inflammation occurs independently of IL-17 and can be worsened when IL-17 is inhibited (15, 17–19).These data are consistent with the disappointing results from clinical trials using IL-17A- or IL-17RA-blocking antibodies to treat IBD. In these trials, Crohn’s disease patients receiving anti-IL-17A or -IL-17RA therapy had no clinical improvement and disease symptoms were exacerbated in some recipients (20, 21). These data highlight the need to reexamine existing models based on IL-17-mediated gut pathology and to reconsider other factors that may drive disease susceptibility.
IL-22 is closely associated with Th17 immunity, despite being a member of the IL-10 family, due to its complementary functions and overlapping expression with IL-17A (22). Although frequently co-expressed with IL-17A, IL-22-producing cells are far less abundant (23), which together confounds efforts to identify the individual contributions of IL-22 and IL-17 to host defense and disease pathogenesis (16, 24). Th17, Th22 and ILC3s are the primary sources of IL-22 in the gut and the cellular source of IL-22 appears to play an important part in determining it’s biological actions (23, 25). Recent evidence suggests that different IL-22-secreting subsets emerge during the course of immune responses depending on the nature of the insult (26) and may be selectively distributed in different anatomical locations within the GI tract (27). However, the factors that regulate IL-22 expression by these different cell types and mediate IL-22’s protective or pathogenic activities remain poorly understood.
Although it is clear that IL-10 and IL-22 contribute distinctly to intestinal health, it remains uncertain if or how the IL-10 and IL-22 pathways interconnect and to what extent, if any, that IL-22 contributes to pathogenesis in IL-10-deficient disease models. In this study, we provide compelling evidence indicating that IL-10 and IL-22 form a functional network which counterbalances antimicrobial and tissue repair defenses and anti-inflammatory responses to maintain both immune microbial homeostasis in the gut. We determined that IL-22-expressing ILC3s selectively accumulate in the small bowel of Il10−/− mice and IL-10/IL-22 double-deficient mice (Il10−/−Il22−/−) do not develop of colitis despite remaining colonized with colitogenic Helicobacter spp. and retaining high frequencies of Th17 cells in colonic tissues. Our data indicate that IL-22 is necessary for the development of spontaneous colitis in Il10−/− mice while excessive IL-17A is not sufficient to induce disease. Taken together, these data challenge current models of chronic intestinal inflammation and suggest that IL-10 and IL-22 form a regulatory axis which balances antimicrobial immunity and immune homeostasis.
Results
IL-22 is overexpressed in Il10−/− mice:
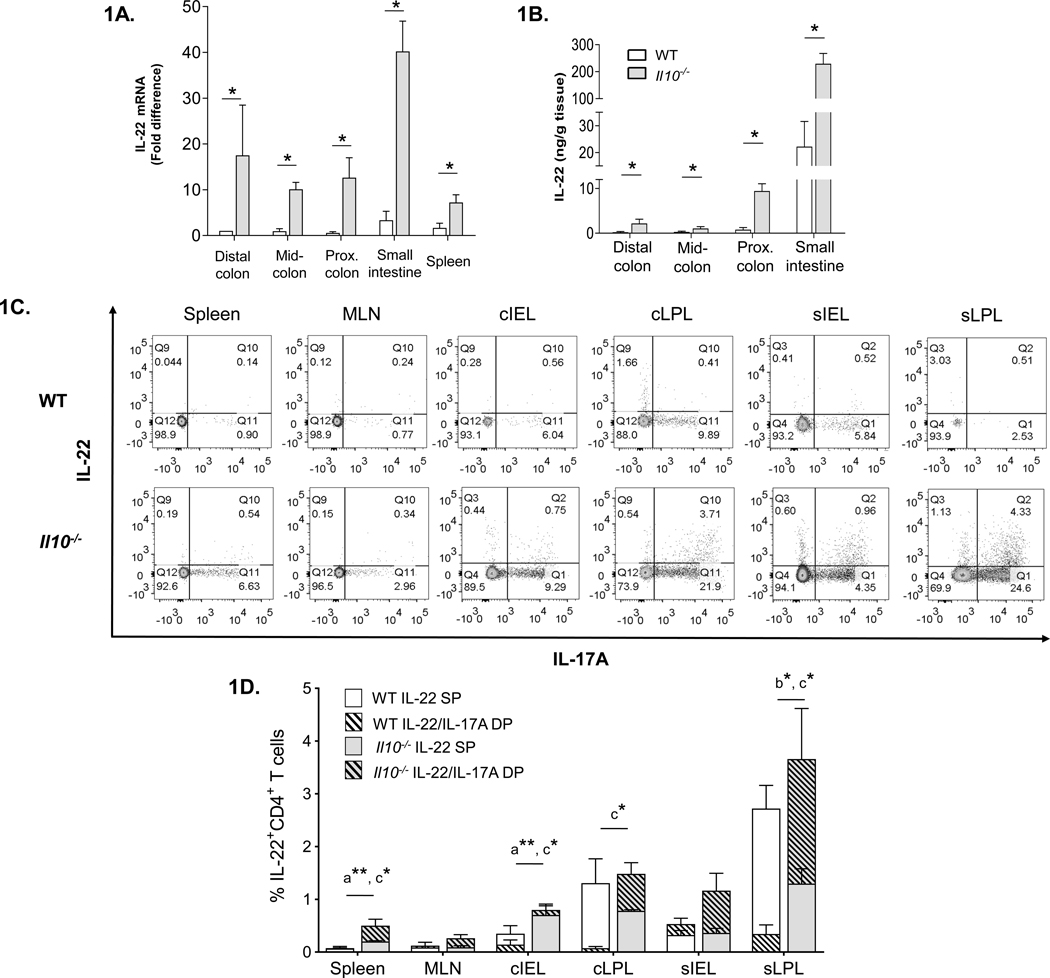
Helicobacter-colonized Il10−/− mice develop spontaneous colitis early in life (4, 8, 9). The progressive disease pathology is characterized by excessive epithelial hyperplasia, microbial dysbiosis and the accumulation of infiltrates in colonic tissues, including large numbers of Th17 cells that are thought to mediate immunopathology (11, 13, 28, 29). Though IL-22 is largely thought to be protective in the gut, it has also been linked to disease pathology (30, 31). Nevertheless, IL-22’s potential role in the development of colitis in Il10−/− mice has not been carefully-studied. Given the growing uncertainties about IL-17’s role in IBD, we began to explore IL-22’s potential role in the development of colitis in Il10−/− mice. Analysis of IL-22 mRNA expression in different regions of the colon as well as the small intestine (SI) and spleen indicated that IL-22 transcript levels were significantly elevated in all of the tissues tested from colitic Il10−/− compared to age-matched WT mice (Fig. 1A). Likewise, Il10−/− mice had significantly higher levels of IL-22 protein in tissue culture explants from the colon and SI (Fig. 1B). Interestingly, for both strains, the highest levels of IL-22 mRNA and protein were observed in the SI (Fig. 1A, B).
Figure 1. IL-22 is over-expressed and ILC3s accumulate in the small bowel of Il10−/− mice.
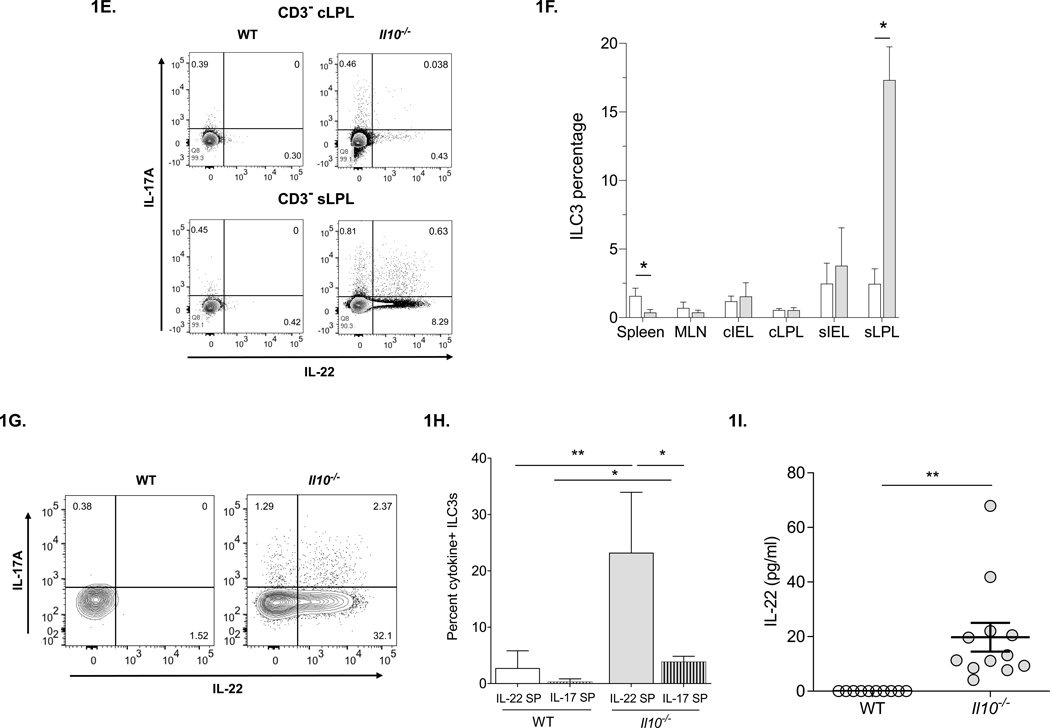
(A) IL-22 mRNA expression in distal, mid- and proximal colon and spleen of WT and Il10−/− mice (n=3–5). (B) IL-22 protein production in colon and small intestine explant supernatants from WT and Il10−/− mice cultured for 24 hours (n=4–7). (C) Representative flow cytometry plots of IL-22 and IL-17A expression in CD4+ T cells in various tissues of the GI tracts of WT and Il10−/− mice [MLN – mesenteric lymph nodes, cIEL – colonic intra-epithelial lymphocytes, cLPL – colonic lamina propria lymphocytes, sIEL – small intestine intra-epithelial lymphocytes, sLPL – small intestine lamina propria lymphocytes. (D) Summary of IL-22-expressing CD4+ T cells in “superimposed” graph showing percentages of single positive (IL-22 SP) and double positive (IL-22/IL-17A DP) IL-22-producing cells from Figure 1C. The total of all IL-22-producing CD4+ T cells is equal to IL-22 SP + IL-22/IL-17A DP, but not depicted on the graph. Data represent means from 3–5 individual experiments composed of cells obtained from 1–3 mice (males 20 weeks of age). Statistical differences in the percentages of IL-22+ T cells subsets between WT compared to Il10−/− mice for each tissue are indicated as follows: a = Total IL-22+; b = IL-22 SP and c = IL-22/IL-17A DP. Statistical analyses (*, p<0.05 **, p<0.01 using the Mann–Whitney U test - error bars represent SD). (E) Representative flow cytometry plots of IL-22 and IL-17A expression gated on live singlet, CD3− cells from the cLPL (top) and sLPL bottom of WT (left) and Il10−/− (right) mice. (F) Summary of total ILC3 percentage in the indicated tissues based on the gating strategy in Supplemental Fig 1. (G) Representative flow cytometry plots of IL-22 and IL-17A expression in ILC3s from the sLPL of WT (left) and Il10−/− (right) mice based on the gating strategy in Supplemental Fig 1. (H) Percentages of IL-22-expresssing (IL-22 SP) or IL-17A-expressing (IL-17A SP) ILC3s from the sLPL of WT (left) and Il10−/− (right) mice as represented in Fig 1G. (I) Serum IL-22 levels in WT and Il10−/− mice (n=10). Statistical analyses (*, p<0.05 **, p<0.01 using the Mann–Whitney U test - error bars represent SD).
To determine the cellular source(s) of IL-22 and patterns of IL-22 and IL-17A co-expression in mucosal and secondary lymphoid tissues of healthy WT and colitic Il10−/− mice, we performed flow cytometry analysis. First, we focused on characterizing IL-22 production in CD4+ T cells. In addition to the total percentage of CD4 T cells producing IL-22, we also examined IL-22-producing T cell subsets defined as IL-22+ IL-17A− single positive (SP) (i.e. Th22) and IL-22+ IL-17A+ double positive (DP) (i.e. Th17) as these subsets have been reported to be primary sources of IL-22 in the colon, mesenteric lymph nodes (MLN) and spleen during inflammatory challenge (32, 33). It is important to note that although the absolute numbers of cells isolated were invariably higher in Il10−/− compared to WT mice in all tissues tested, on average from 2–5 fold (data not shown), a relatively high degree inter-experiment variation in cell recovery from some tissues precluded a comprehensive flow cytometric analysis based on absolute cell numbers. Nonetheless, the relative percentages of the leukocyte subsets examined were quite consistent within groups and between experiments. In agreement with the mRNA and explant data (Fig. 1A–B), the highest percentages of IL-22-producing T cells were in the small intestine, specifically in the sLPL fraction, of both WT and Il10−/− mice (Fig. 1C–D). While we consistently observed higher levels of total IL-22-producing CD4+ T cells (IL-22 SP+ IL-22/IL-17A DP) in tissues from Il10−/− compared to WT mice, only the spleen and colonic intraepithelial lymphocytes (cIEL) reached statistical significance (Fig. 1D). The majority of IL-22-producing T cells from Il10−/− mice trended towards IL-17A co-expression in all tissues examined and were significantly higher in the spleen and cIEL compared to WT mice. This was also the case in the cLPL and sLPL fractions despite the fact that the total levels of IL-22-producing T cells were not significantly different between WT and Il10−/− mice (Fig. 1D). Interestingly, compared to Il10−/− mice, intestinal T cell-derived IL-22 in WT animals trended towards higher proportions of Th22 cells and was significantly higher in sLPLs.
Our data indicated that in tissues examined, the vast majority of IL-22+ cells in both WT and Il10−/− mice, were CD3+CD4+ Th17 and Th22 cells. However, we noted an exception exclusively in sLPLs from Il10−/− mice in which a sizeable population of IL-22+ CD45+CD3− lymphocytes were identified which secreted little IL-17A (Fig. 1E). Though usually found only in low abundance, we suspected that these cells could be ILC3s. ILC3s consist of at least 2 subpopulations in mice but their high degree of heterogeneity and plasticity has hindered efforts to classify ILC3 subsets phenotypically and functionally (27). Nevertheless, ILC3s are unified by RORγt expression and the preferential production of IL-22 and/or IL-17A (34). We examined the total levels of ILC3s (defined as CD45+CD3−RORγt+IL-7Rα+ (35) and refer to the gating strategy in Supplemental Fig 1) in mucosal and secondary lymphoid tissues from WT and colitic Il10−/− mice. Although Il10−/− mice had higher total numbers of leukocytes, including CD3− lymphocytes in all tissues tested compared to WT (data not shown and Supplemental Fig 1), the relative percentages of ILC3s were comparable in most tissues (Fig. 1F). As expected, a substantial population of ILC3s was observed specifically in sLPLs from Il10−/− mice (Fig. 1F). Interestingly, significantly higher levels of ILC3s were observed in spleens of WT compared to Il10−/− mice (Fig. 1F). We verified that a large fraction of the amassed ILC3s in Il10−/− sLPLs expressed only IL-22 (IL-22 SP) with only a small subset expressing IL-17A (Fig. 1G–H). As mentioned, ILC3s can be subdivided into smaller subsets based largely on the surface expression of NKp46 and CCR6 +/− CD4 (27). We found that the majority of total ILC3s as well as IL-22+ ILC3s in the sLPL of Il10−/− mice did not express NKp46 or CCR6, though the percentages of NKp46+ and CCR6+ ILC3s (total ILC3s) tended to be higher in Il10−/−compared to WT mice (Supplemental Fig. 1 and data not shown).
To determine if the elevated levels of IL-22-expressing cells and mRNA in Il10−/− mice were restricted to tissues, we examined IL-22 levels in the serum. While IL-22 was readily detectible in the serum of Il10−/− mice, no IL-22 was detected in the serum of matched WT animals (Fig. 1I). These data indicate that IL-22 is over-expressed both within mucosal tissues and the periphery and ILC3s accumulating in the small bowel are the primary source of IL-22 in Il10−/− mice.
IL-22-deficiency prevents spontaneous colitis in Il10−/− mice:
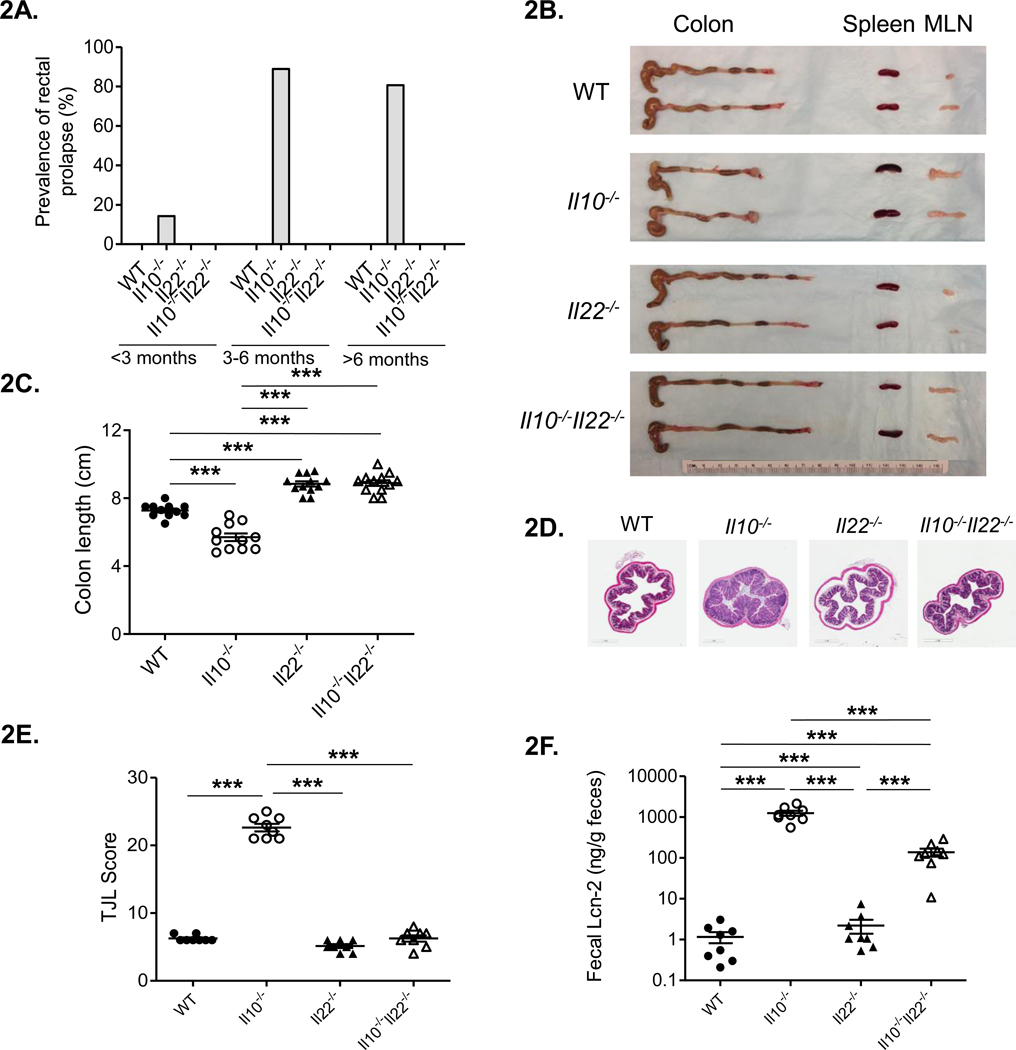
To determine the role of IL-22 in the development of spontaneous colitis, we crossed susceptible Il10−/− mice (colonized with Helicobacter typhlonius), with disease-free Il22−/− mice harboring both H. typhlonius and H. mastomyrinus. We have previously shown that H. typhlonius is associated with development of colitis in Il10−/−animals (28). Studies from other groups have demonstrated H. mastomyrinus to be even more pathogenic than H. typhlonius (36). We confirmed that while fully-backcrossed Il10−/−Il22−/− mice harbored both Helicobacter spp. (data not shown), Il10−/−Il22−/− mice did not develop chronic colitis (Fig. 2).
Figure 2. IL-22-deficiency prevents the development of spontaneous colitis in Il10−/− mice.
(A) Prevalence of rectal prolapse (n= >60 per group) in WT, Il10−/−, Il22−/−, and Il10−/−Il22−/− mice. (B) Representative gross images of organs of the gastrointestinal tracts from each strain. (C) Colon length for each strain (n=11 per group). (D) Representative photomicrographs of haemotoxylin and eosin stained colons (20×). (E) Histological disease scores (n=8/group). (F) Fecal Lcn-2 levels (n=8/group). Statistical analyses (***, p < 0.0001 using the Mann–Whitney U test (error bars represent SEM).
The development of colitis in Il10−/− mice colonized with Helicobacter spp. is highly correlated with rectal prolapse (37). We found that by 3 months of age, about 20% of Il10−/− mice had developed rectal prolapse and this number increased to 80% by 6 months of age. In contrast, but similar to WT animals, Il10−/−Il22−/− mice did not develop rectal prolapse (Fig. 2A). A subset of Il10−/−Il22−/− mice was observed for a period of more than 1.5 years during which they remained healthy and free of disease (data not shown). Macroscopically, Il10−/− mice exhibited thickening of the colonic wall, shortened colonic length, intussusception of the cecum and enlarged mesenteric lymph nodes (MLN) and spleens (Fig. 2B). Surprisingly, Il10−/−Il22−/− as well as Il22−/− exhibited significantly longer colon lengths than both WT and Il10−/− mice (Fig. 2B, C). The colonic thickening, cecal intussusception and splenic enlargement observed in Il10−/− mice was completely abrogated in Il10−/−Il22−/− mice. However, similar to Il10−/− mice, double-deficient mice had enlarged mesenteric lymph nodes compared to WT and Il22−/− mice (Fig. 2B).
Compared to WT controls, colons from Il10−/− mice had marked mucosal hyperplasia and thickening (Fig. 2D), increased cellular infiltrates, erosion and ulceration of the epithelium, and crypt abscesses as reported previously in Il10−/− mice (4, 28). Histology scores for colitis using the Jackson Laboratory (TJL) system reflected these findings and indicated that Il10−/−Il22−/− mice were protected from colitis (Fig. 2E). Lipocalin (Lcn-2) is strongly associated with intestinal inflammation (38, 39) and as expected, the high levels of fecal Lcn-2 in Il10−/− mice were significantly reduced in Il10−/−Il22−/− mice (Fig. 2F). Interestingly, the levels of Lcn-2 in Il10−/−Il22−/− mice were still significantly higher compared to fecal samples from both WT and Il22−/− mice. These data are in line with a recent report indicating that IL-17A and ILC3-derived IL-22 synergistically induce Lnc-2 expression from IECs (40) and suggest that that IL-17A levels remain high in disease-free Il10−/−Il22−/− mice. Taken together, these data indicate that IL-22 is necessary for the development of chronic colitis and plays a central, pathogenic role in the gut in the context of Il10-deficiency.
Overall, Il10−/− and Il10−/−Il22−/− mice have similar distributions of leukocyte subsets with some tissue-specific exceptions
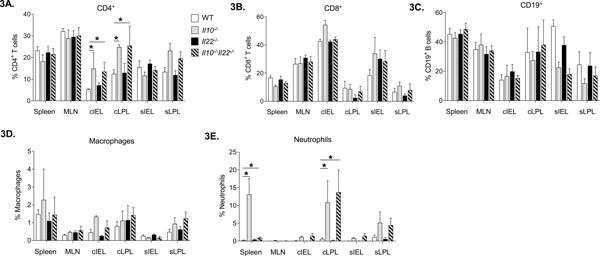
To characterize cell populations associated with absence of disease in Il10−/−Il22−/− mice we examined leukocyte distributions in the various mouse strains. As mentioned, Il10−/− mice have high numbers of leukocytes infiltrating secondary lymphoid and intestinal tissues (4, 28) and consistent with previous reports (12) we observed higher frequencies of CD4+ T cells in the cIEL and cLPL fractions compared to WT mice. Surprisingly, we also found higher frequencies of CD4+ T cells in the colons of Il10−/−Il22−/− mice compared to WT (Fig. 3A). No differences in the percentages of CD8+ cytotoxic T cells, CD3−CD19+ B cells or CD11b+Ly6C+Ly6G−F4/80+ side scatter low (SSClo) macrophages were observed between the groups (Fig. 3B–D respectively). However, higher frequencies of CD11b+Ly6C+Ly6G+F4/80− neutrophils in the spleens of Il10−/− mice, whereas the percentages of neutrophils from Il10−/−Il22−/− animals were comparable to levels observed in WT and Il22−/− spleens (Fig. 3E). Interestingly, the percentage of neutrophils was also elevated in the cIEL and cLPL fractions of both Il10−/− and Il10−/−Il22−/− mice compared to WT animals (Fig. 3E). Of note, the numbers and percentages of leukocyte subsets were comparable between WT and Il22−/− mice for all of the tissues examined (Fig 3).
Figure 3. Flow cytometric characterization of leukocyte populations in intestinal and secondary lymphoid tissues.
Leukocyte subsets as a percentage of live, CD45+ cells including (A) CD4+ T cells, (B) CD8+ T cells (C) CD19+ B cells, (D) macrophages (CD11b+ Ly6C+ Ly6G− F4/80+ side scatter low (SSClo)) and (E) neutrophils (CD11b+ Ly6C+ Ly6G+ F4/80-) in the indicated tissues of WT, Il10−/−, Il22−/−, and Il10−/− Il22−/− mice. (n=5 per group) Statistical analyses (*, p < 0.05 using the Mann-Whitney U test (error bars represent SD))].
The tissue-specific disparity in neutrophil frequencies prompted us to examine the distribution of leukocyte subsets in the periphery (Fig. S2). Although Il10−/− and Il10−/−Il22−/− mice trended towards higher numbers of leukocytes in the blood (Fig S2A), no significant differences between the groups were observed for white blood counts (WBC) or lymphocytes (Fig S2B). However, in agreement with findings in the spleen, complete blood counts (CBCs) revealed elevated numbers of neutrophils in the peripheral blood of Il10−/− mice compared to Il10−/−Il22−/− as well as WT and Il22−/− animals (Fig. S2C). In addition, Il10−/− mice harbored higher numbers of monocytes whereas Il10−/−Il22−/− mice had numbers comparable to WT (Fig. S1D). Similar to WBC, platelet counts trended higher in Il10−/− and Il10−/−Il22−/− mice but did not reach statistical significance (Fig. S2E). Thus, overall, Il10−/− and Il10−/−Il22−/− mice have higher levels of leukocytes in intestinal tissues and a selective enrichment of Th cells and neutrophils in the colon.
Il10−/− and Il10−/−Il22−/− mice have higher frequencies of Th17 cells in tissues
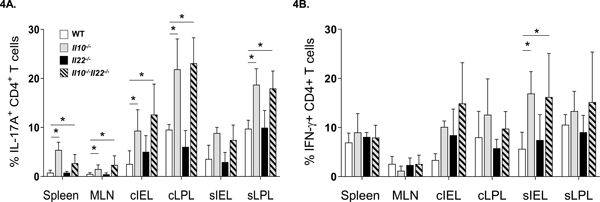
Dysregulation of Th1 and/or Th17 responses has been associated with intestinal inflammation (12, 28, 41, 42). In Il10−/− mice, the excessive accumulation of Th17 cells in the gut is thought to mediate intestinal disease (43) and we found that the levels of CD4+ T cells remained high in colonic tissues of Il10−/−Il22−/− mice (Fig 3A). Taken together with our finding that fecal Lcn-2 levels were reduced in Il10−/−Il22−/− compared to Il10−/− mice but elevated compared to WT and Il22−/− mice (Fig 2F) suggested that Th17 cells may remain high in disease-free Il10−/−Il22−/− mice. Thus, we examined the frequencies of CD4+ Th17 and Th1 subsets in intestinal and secondary lymphoid tissues based on the expression of IFN-γ and IL-17A. Consistent with elevated frequencies of CD4+ T cells in colonic tissues (Fig. 3A), Il10−/− mice had significantly higher percentages of IL-17A-expressing CD4+ Th17 cells in cIEL and cLPL fractions as expected, compared to WT and Il22−/− mice (Fig. 4A). Remarkably, disease-free Il10−/−Il22−/− mice had similarly-high levels of Th17 cells in colonic tissues. This suggests that IL-17A regulation is independent of IL-22. In addition, although we did not detect differences in the overall percentages of CD4+ T cells in other tissues (3A), the levels Th17 cells in the spleen, MLN and sLPL of both Il10−/− and Il10−/−Il22−/− were significantly elevated compared to WT (and Il22−/−) mice. Interestingly, though the percentages of IFN-γ+CD4+ Th1 cells tended to be higher in intestinal tissues of Il10−/− and Il10−/−Il22−/− mice, only the sIELs were significantly different (Fig. 4B). These data suggest that the well-known accumulation of Th17 cells in intestinal tissues of IL-10-deficient mice is not sufficient to induce colitis.
Figure 4. Flow cytometric characterization of Th17 and Th1 subsets in intestinal and secondary lymphoid tissues.
(A) percentage of IL-17A+ or (B) percentage of IFN-γ+ CD4+ T cells in various tissues of the GI tracts of WT, Il10−/−, Il22−/−, and Il10−/−Il22−/− mice. Data represent means from 4–5 individual experiments composed of cells obtained from 1–3 mice (male, between 8–12 weeks of age) per group (*, p < 0.05 using the Mann-Whitney U test (error bars represent SD)).
Il10/Il22 double-deficiency results in reduced fecal IgA and IgG
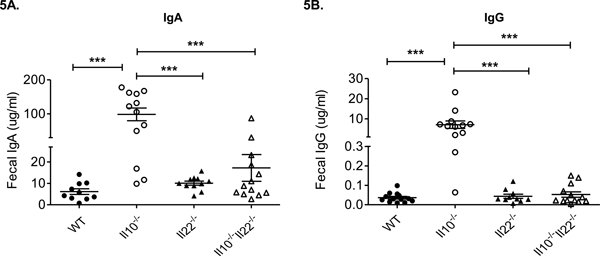
In addition to T cells, B cells and mucosal antibodies play important roles in maintaining immune-microbial homeostasis and barrier integrity in the gut (2). Studies in Il10−/− mice have shown that IgA and IgG1 levels are elevated in the serum (44) and serum IgG is reactive against enteric bacterial antigens (45). To determine the impact of double-deficiency in IL-10 and IL-22 on the levels of mucosal antibody we measured immunoglobulin levels in the feces in our mouse strains. While the levels of both IgA and IgG were significantly elevated in feces from Il10−/− mice compared to other strains, fecal antibody levels from Il10−/−Il22−/− mice were similar to those observed in WT (and Il22−/−) mice (Fig. 5A, B). Although the antigenic specificities and subclass of fecal immunoglobulins have not been determined, these data suggest that an imbalance in the IL-10/IL-22 axis results in dysregulation of mucosal antibody responses.
Figure 5. Fecal IgG and IgA levels are normalized in Il10−/−Il22−/− mice.
(A) Fecal IgA and (B) IgG levels in WT, Il10−/−, Il22−/−, and Il10−/−Il22−/− mice (male, 16–20 weeks of age, n = 10–12 mice per group) (***, p<0.001, using the Mann–Whitney U test (error bars represent SEM)).
Antimicrobial IL-22-target genes are upregulated in colonic tissues from Il10−/− mice but are normalized in disease-free Il10−/−Il22−/− mice
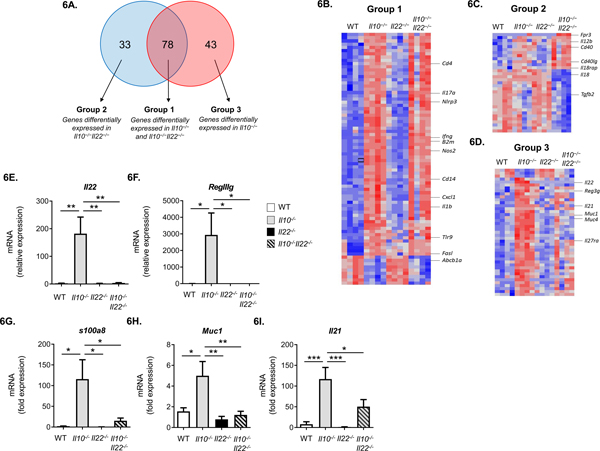
To identify genes that may underlie protection from colitis in Il10−/−Il22−/− mice, we examined mRNA expression profiles in tissue samples from the distal colons of age- and sex-matched WT, Il10−/−, Il22−/− and Il10−/−Il22−/− mice (n = 4 mice/group) using the The TaqMan® OpenArray® Mouse Inflammation Panel. For relative mRNA expression analyses, each experimental group (strain) was compared to WT to identify differentially expressed genes. Significantly different genes were defined based on q-values smaller than 0.05 and fold change greater than 2. Of 632 genes examined, 121 and 111 genes were differentially expressed in the distal colons from Il10−/− and Il10−/−Il22−/− mice, respectively, compared to WT (Fig. 6A). The gene expression profile of Il22−/− mice was remarkably similar to WT mice (Fig. 6B, C, D) and no significant differences were identified.
Figure 6. Distal colon tissues from Il10−/− and Il10−/−Il22−/− mice have similar gene expression patterns but IL-22 target genes associated with antimicrobial responses are normalized to WT levels in Il10−/−Il22−/− mice.
(A) Venn diagram from TaqMan OpenArray results depicting differential gene expression in distal colons of Il10−/− and/or Il10−/−Il22−/− mice compared to WT mice (n=4 mice per strain, 20 weeks of age) (q = 0.05, fold change boundary = 2). Heatmaps of (B) Group 1, (C) Group 2 and (D) Group 3 with select genes shown. Each line represents a single probe and each column a single mouse. Blue represents probes that were at least two-fold lower; red represents probes that were at least two-fold higher compared to WT mice. (E-I) Independent RT-PCR validation of mRNA expression for selected genes in the distal colons of WT, Il10−/−, Il22−/−, and Il10−/−Il22−/− mice (n=4–7) (***, p<0.001, **, p<0.01, *, p<0.05, using 1-way ANOVA and Bonferroni post-test - error bars represent SEM).
Genes differentially expressed in Il10−/− and Il10−/−Il22−/− mice were subdivided into 3 groups based on their patters of expression compared to WT (Fig. 6.A). Group 1 was comprised of genes differentially expressed in both Il10−/− and Il10−/−Il22−/− mice and likely represent genes affected by IL-10 independently of IL-22 (Fig. 6B; Supplementary Table 1). Like the similar distributions of inflammatory cells observed in Il10−/− and Il10−/−Il22−/− mice, there was a high degree of overlap in Group 1 genes (78 shared genes) (Fig. 6A). Group 1 genes included important mediators of inflammation such as Il17a, Cxcl1, Nos2, Il1b, and Cd4, as well as molecules important in antigen presentation such as Cd74 and B2m (Fig. 6B). Group 2 was comprised of genes differentially expressed only in Il10−/− Il22−/− mice compared to WT and likely represent genes affected by both IL-10 and IL-22 (Fig. 6C and Supplementary Table 2). Group 2 genes included Fpr3, Cd40 and Cd40L, Il18 and Il18rap.
Group 3 was comprised of genes differentially expressed only in Il10−/− mice compared to WT (Fig. 6D and Supplementary Table 3) and are likely to represent, at least in part, genes that are affected by overexpression of IL-22. Indeed, in agreement with our initial observations (Fig. 1A–B) mRNA for Il22 as well as several antimicrobial IL-22-target genes such as Reg3g and Muc1 were upregulated in Il10−/− mice but not in Il10−/−Il22−/− mice. Results from these experiments were validated by separate real-time qPCR analysis for Group 3 genes, which included Il22 (Fig. 6E), several IL-22-target genes (Reg3g (Fig. 6F), s100a8 (Fig. 6G), Muc1 (Fig. 6H)) and Il21 (Fig. 6I) which is known to regulate both IL-10 and IL-22 expression (46). In addition, several genes that were not differently expressed between the groups, including Il23a, Il23ra and Il22ra2, were independently confirmed by RT-qPCR (data not shown). Taken together, these data indicate that IL-10 is a negative regulator of IL-22 expression in the gut and in the absence of IL-10, aberrant IL-22 expression induces the overexpression of IL-22-target genes associated with antimicrobial immunity.
Microbial diversity is re-established in Il10−/−Il22−/− mice but with altered composition
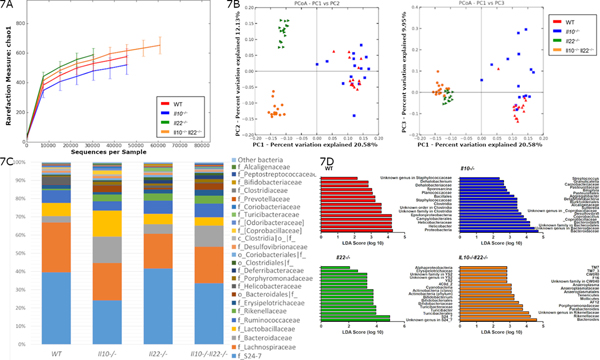
IL-22 promotes intestinal health in part, by regulating the microbiota (47). Chronic intestinal inflammation is associated with reduced microbial diversity in humans (48, 49) and Il10−/− mice (8, 50) but the mechanism remains unclear. Our data indicate that IL-22-dependent antimicrobial genes are highly upregulated in Il10−/− mice (Fig. 6). Thus, we explored the possibility that the IL-22 pathway drives microbial dysbiosis in Il10−/− mice by sequencing and analyzing fecal 16S rRNA isolated from each mouse strain. The number and relative abundance of bacterial phylotypes in the gastrointestinal tract are often decreased during states of chronic inflammation (51, 52). Accordingly, the Chao1 index (Fig. 7A) which measures the richness (total number) of phylotypes revealed that Il10−/− mice trended lower in diversity compared to WT mice (p=0.102, Supplementary Table 4). Lower diversity in Il10−/− mice was also evident when compared to Il10−/−Il22−/− mice (p=0.012) which indicates that loss of IL-22 significantly compensated for the negative impact of IL-10 deficiency on the number of phylotypes in the fecal microbiome. Interestingly, IL-22 deficiency was associated with increased diversity compared to both WT (p=0.03) and Il10−/− mice (p=0.006, Supplementary Table 4a). Similar results were obtained using the Shannon index, which measures richness and evenness of phylotypes (Supplementary Table 4b). These data support the histologic evidence that IL-22 deficiency appears to compensate for IL-10 deficiency in protecting Il10−/−Il22−/− mice against chronic colitis.
Figure 7. Reduced microbial diversity in Il10−/− mice is restored to WT levels in Il10−/−Il22−/− mice albeit a markedly altered microbial signature.
(A) Chao 1 index (B) Principal component analysis (unweighted, UniFrac analysis) based on 16S rRNA sequencing of DNA isolated from the feces of WT, Il10−/−, Il22−/−, and Il10−/−Il22−/− mice (10 male, 5 female per group, 12–16 weeks of age). Taxon-based analysis at (C) family level with (D) differentially expressed taxa by LEfSe analysis.
Principal component analysis of beta diversity using unweighted, UniFrac analysis, showed distinct clusters indicating distinct microbial community structures for each strain of mice (Fig. 7B). Analysis using permutational multivariate analysis of variance (PERMANOVA) demonstrated a significant effect on beta diversity by strain (p=0.001), and pairwise adonis comparisons revealed significant differences between every combination (p=0.006) with the exception of WT vs. Il10−/− mice (p=0.078). This suggests that together, IL-10 and IL-22 play an important role in regulating microbial diversity in the gut. .
Taxon-based analysis revealed that the microbiome of each group comprised mainly of Bacteroidetes, Firmicutes and Proteobacteria (Fig. 7C). At the family level, Il22−/− and Il10−/−Il22−/− mice had fewer Lactobacillaceae and Bacteroidacea compared to Il10−/− mice. Figure 7D and supplementary table 5 lists significant differences between the 4 strains of mice from the phylum down through genus level, when detectable. The pathogenic potential of most of these organisms is not clearly understood, but some detected differences may be relevant to the Il10−/− colitis model. Consistent with previous reports (8) and similarly to IBD in humans (48), species diversity was markedly reduced in the fecal microbiota of colitic Il10−/− mice compared to WT. Of note Il10−/− mice had increased abundance of the genus Sutterella which has been suspected to play a role in IBD pathogenesis (53). In addition, an unknown genus within the family Bacteroidaceae, was more common in Il10−/− mice than WT, Il22−/− and Il10−/−Il22−/− mice which is consistent with reports where select members of Bacteroidaceae have been associated with colitis in humans and mouse models (54). Interestingly, the genus Bacteroides was highest in the Il10−/−Il22−/− mice but was not associated with increased disease. Significantly, the Helicobacter genus was detected at the highest level in WT mice that did not develop colitis, as expected, and was detected at the lowest level in the feces of Il10−/− mice with severe colitis (Figure 7C) which is consistent with previous reports (55).
Taken together, these data suggest that without IL-10-mediated inhibition, IL-22 overexpression triggers excessive innate antimicrobial and tissue healing responses in the gut resulting in microbial dysbiosis, protracted tissue repair activity and a state of chronic inflammation (Fig. 8). Importantly, these findings may also help to explain the surprising and disappointing failure of IL-17A-blocking drugs in clinical trials (20, 21, 56) and shed new light on existing models of chronic intestinal inflammation.
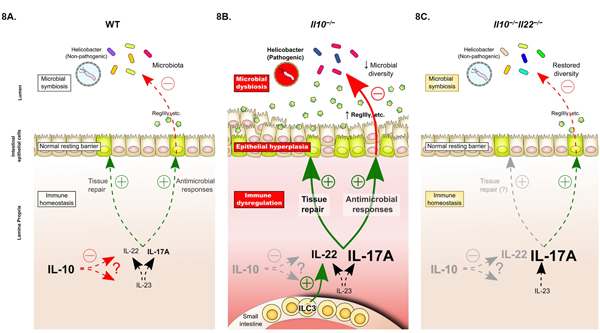
Figure 8. Proposed role of the IL-10/IL-22 axis in intestinal homeostasis and chronic inflammation.
(A) Under steady-state conditions in WT mice, Th17-axis cytokines (IL-23, IL-17A, IL-22) play a central role in regulating symbiosis between host and the intestinal microbiota by regulating the antimicrobial and tissue repair activities of IECs to control the growth of enteric bacteria, to maintain intestinal barrier integrity. Although the mechanism(s) are incompletely understood, IL-10 counterbalances these responses by restricting the expression of Th17-axis cytokines (including IL-22) to restore immune homeostasis. Helicobacter spp. are considered commensals and colonization does not trigger excessive inflammation or disease. (B) In Il10−/− mice, Helicobacter spp. triggers the development of spontaneous colitis within 2–3 months of birth. Dysregulation of IL-17A/Th17 cells have been strongly implicated in disease pathogenesis but IL-22 is also aberrantly expressed. Many of the pathological features of colitis in IL-10-deficient mice, such as epithelial hyperplasia, overexpression of pro-inflammatory and antimicrobial genes and dysbiosis of the intestinal microbiota, are consistent with unrestrained IL-22 production and sustained activation of IL-22-dependent tissue repair and antimicrobial pathways in the gut. (C) In Il10−/− Il22−/− double-deficient mice, animals are disease-free despite retaining high levels of Th17 cells and remaining colonized with Helicobacter spp.. Expression of antimicrobial IL-22-target genes are normalized, excessive proliferation of the colonic epithelium is reversed and microbial diversity is reestablished (albeit with an altered composition compared to WT mice).
Discussion
To maintain intestinal health, the immune system must balance between tolerance to commensal microorganisms and defense against invading pathogens and IL-17 and IL-22 play key roles in regulating host-protective responses at barrier surfaces. IL-22 facilitates the repair of damaged tissues by inducing regeneration of the intestinal epithelium (57, 58), while IL-22 and IL-17A regulate innate antimicrobial responses to defend against enteric pathogens (59). IL-10 acts in opposition by restricting host defense responses to prevent chronic inflammation and restore intestinal homeostasis (3). IL-10’s anti-inflammatory role in the gut is well-established (4, 8, 11, 60), however, it has been difficult to identify the immune effector mechanism(s) that mediate intestinal pathology in IL-10-deficient models. A landmark study by Yen, et al. in 2006 suggested that the IL-17/IL-23 (Th17) axis, not the previously implicated IL-12/IFN-γ (Th1) pathway, was responsible for intestinal disease (13).These data provided a compelling basis for the prevailing theory that pathogenic Th17 cells drive disease pathogenesis in IL-10-dependent models of intestinal inflammation. However, a growing number of conflicting studies report that intestinal inflammation is worsened by IL-17 depletion or occurs independently of IL-17 (15, 17–19). Taken together with the surprising failure of IL-17A or IL-17RA blocking antibodies to treat IBD in clinical trials (20, 21), these findings suggest that unlike IL-17’s pathogenic role in psoriasis (61, 62), its contributions to intestinal pathology may be less clear (63).
Here, we provide evidence that IL-22 overexpression accounts for much of the immunopathology associated with spontaneous colitis in Il10−/− mice and despite retaining high levels of Th17 cells in colonic tissues, Il10−/−Il22−/− double-deficient mice remained disease-free. Interestingly, while some groups have noted that IL-22, like IL-17, was markedly upregulated in IL-10/IL-10R-deficient mouse models of intestinal inflammation ironically, IL-22’s role in disease pathogenesis was not addressed and disease activity was correlated with IL-17 upregulation (64, 65). IL-10 receptor signaling was shown to play a role in controlling IL-22-dependent intestinal pathology in a T cell transfer model of colitis (66) and IL-10 has been shown to inhibit both IL-17 and IL-22 expression in vitro (67). However, to our knowledge, it has not been reported if IL-10/IL-17A-double-deficient mice develop colitis, which adds to the uncertainty of IL-17A’s role in gut pathology. Importantly, Morrison and colleagues used H.hepaticus-infected anti-IL-10 receptor-treated mice to compare the contributions of IL-17A and IL-22 to acute colonic and cecal inflammation (68). The authors found that IL-17A neutralization had no effect on colonic inflammation, but pathology in the cecum was exacerbated, while neutralization of IL-22 prevented inflammation in the colon, but not in the cecum. This study hinted at a pathogenic role for IL-22, and not IL-17, in IL-10-dependent acute colitis. Our data are in agreement with these findings and support a central role for an IL-10/IL-22-dependent axis of intestinal homeostasis.
Studies in Il22−/− mice suggest that IL-22’s role is largely protective in the gut (69) especially in the context of acute intestinal damage, such as during Citrobacter rodentium infection (31) or in the dextran sulfate sodium (DSS)-induced colitis model (33). Though ILC3s are an important early source of protective IL-22 (26), growing evidence in humans and mice suggests that chronic dysregulation of IL-22 and ILC3 responses in particular, results in intestinal pathology (14, 30, 70–75) and even tumorigenesis (76, 77). Accumulating data suggests that the distribution patterns of ILC3s within different intestinal tissues can dramatically impact host defense and tissue homeostasis (34, 71, 72). In the anti-CD40 model of acute colitis, the rapid redistribution of ILC3s from intestinal lymphoid structures precedes the development of inflammatory foci elsewhere in gut tissues (71) that is dependent on IL-22-, but not IL-17A-,expressing ILC3s (72, 78). In addition, a recent report found that IL-22-producing ILC3s accumulated in the small intestines of immunocompromised mice resulting in impaired host lipid metabolism (79). In our study, while the levels of IL-22+ Th17 cells were slightly elevated in colonic tissues, we observed a sizeable accumulation of IL-22+IL-17A− ILC3s selectively in the small bowel of Il10−/− compared to WT mice. Although it is tempting to speculate, it is important to note that our data do not indicate how, or if the expansion of IL-22-producing ILC3s in the small intestine relates to the development of chronic colitis in Il10−/−mice. Additional studies, will be required to parse out the potential role that IL-22+ILC3s, residing in the SI may play in chronic intestinal inflammation.
Indeed, the mechanisms by which ILC3s and IL-22 shape mucosal responses both locally and in distant tissues are poorly understood but, direct and indirect mechanisms have been described (34, 79, 80). For instance, ILC3s communicate bi-directionally with innate cells such as IECs, macrophages and dendritic cells (DCs) via cytokines (such as IL-23, IL-22, GM-CSF) to quickly shape innate responses in response to changes in the tissue microenvironment (71, 81, 82). In addition, MHCII+ ILC3s have been shown to present antigens and directly regulate antimicrobial CD4+ T cell activity and T cell-dependent antibody production (83, 84). Thus, despite being greatly outnumbered, ILC3s can employ various means to shape protective antimicrobial/tissue repair responses in the mucosa but more studies are needed to fully understand the underlying mechanism(s).
An important caveat, which has likely contributed to some confusion in the literature, is the inconsistent reporting of Helicobacter status in mouse colonies using IL-10-dependent models of colitis (85). Neither germfree nor conventionally-housed Il10−/− mice develop spontaneous colitis (8, 10), but rather, the development of intestinal disease is dependent on co-colonization with “pathobionts” such as Helicobacter spp. (9, 55). Helicobacter-colonized Il10−/− mice are easily identified clinically by the appearance of rectal prolapse (86), but in most other mouse strains infection is non-pathogenic or subclinical (8, 10, 37) and frequently, Helicobacter status is unknown (87). Pups acquire their intestinal microbiota through fecal-oral contact (7) and depending on the breeding strategy and status of the parental strains, the Helicobacter status of pups needs to be tested to confirm transmission, otherwise, the results may be confounded (37). Notably, Yen’s study did not report Helicobacter status of their mouse colonies, thus it is not clear if the resolution of colitis in Il10−/−p19−/− mice is due to the lack of IL-23 or Helicobacters (13). Similarly, Il10−/−Il17a−/− mice were previously generated but neither the Helicobacter status nor the incidence of intestinal disease was reported although these studies were focused on IL-17A’s role in lung pathology (88, 89). Here, we confirmed that Il10−/−Il22−/− mice remained colonized with Helicobacter spp. (Fig. 7) yet were disease free which strengthens our assertion that IL-22 is a critical effector cytokine driving the development of colitis in Il10−/− mice.
In both humans (90, 91) and IL-10-dependent models of colitis (8, 50) IBD is associated with reduced diversity of the intestinal microbiome, yet it remains unclear if microbial dysbiosis is a cause or result of intestinal inflammation (92). IL-22 has a vital role in shaping the commensal microbiota and protecting against enteric pathogens (80, 93, 94). In line with this, we observed that microbial diversity was restored in our Il10−/−Il22−/− mice suggesting that dysregulation of the ILC3/IL-22 axis drives microbial dysbiosis in Il10−/− mice. As mentioned previously, IL-22-producing ILC3s were found to gather in the small intestines of WT and Rag1−/− weanlings as they become colonized with commensals. In WT pups, the ILC3s were soon displaced by adaptive immune cell populations, but in Rag1−/− mice the levels of activated ILC3s remained elevated in resulting in altered homeostasis in gut tissues (79). The Helicobacter status of the mice in this study was not reported, but interestingly, work from others demonstrated that, like in Il10−/− mice, colonization of Rag-deficient mice with Helicobacter spp. results in spontaneous colitis (95, 96). Importantly, the development of colitis in Helicobacter-infected Rag−/− mice was shown to be dependent on IL-22 (97). It is important to note that our data do not indicate if the microbiota itself is sufficient to induce or protect against the development of spontaneous colitis. Additional studies will be needed to determine this. Together, these converging data along with our findings that microbial diversity is restored in Il10−/−Il22−/− mice, suggest that IL-10 plays an underappreciated role in establishing mutualism by regulating innate IL-22 responses to commensal bacteria and promoting the development of tolerogenic adaptive immune subsets (80).
This study highlights the mechanistic connection between IL-10 and IL-22 and offers important insights into the pathogenesis of chronic colitis in Il10−/− mice. Our data suggest a model in which IL-10 and IL-22 form a regulatory axis within the gut mucosa to maintain intestinal health. Thus, in the context of IL-10-deficiency, IL-22 is necessary while IL-17 is not sufficient to drive immune-mediated intestinal pathology. Importantly, the IL-10/IL-22 axis also provides a conceptual framework to link the development and resolution of intestinal inflammation with the mechanisms underlying epithelial repair. These findings may help to reconcile conflicting reports regarding IL-17’s role in intestinal disease and will be informative when considering new therapeutic approaches in IBD and other inflammatory disorders.
Materials and Methods
Mice
B6.129P2-IL10tm1Cgn (Il10−/−) as well as WT mice were originally obtained from Jackson Laboratories (Bar Harbor, ME) and have been maintained in separate breeding colonies within our animal facility for over 14 years. Il22−/−mice were obtained through an agreement with Genentech and have been maintained in separate breeding colonies within the same facility for over 7 years. The Il10−/−Il22−/− mice were generated in our facility through inter-crossing Il10−/− and Il22−/−mice. To examine IL-22’s role in regulating disease pathology and microbial diversity in Il10−/− mice, it was important to control for potential founder effects by generating mice genetically-deficient in Il10 and Il22 while maintaining the microbiota from Il10−/− mice. To do so, we used a breeding strategy which minimizes microbial cross-contamination (98). Briefly, we set up clean breeding cages with one female Il10−/− mouse in estrus and one male Il22−/− breeder and once impregnated, the female Il10−/− mouse was separated from the male and placed into a clean cage. For the F2 generation, a female Il10−/− mouse and a F1 male Il10+/−Il22+/− breeder were place in a clean cage and the impregnated female was separated from the male and placed into a clean cage. Il10−/−Il22−/− F2 mice were maintained then by interbreeding.
For experimental procedures male animals between 16–20 weeks of age were used unless stated otherwise. Mice on study were housed in individually ventilated cages (Allentown Caging Equipment, Allentown, PA), containing autoclaved corncob bedding (Harlan Teklad, Indianapolis, IN) with a cotton enrichment square. Mice were provided autoclaved rodent chow ad libitum (Harlan Teklad, Indianapolis, IN) and received reverse-osmosis-treated, hyperchlorinated water via automated in-cage watering system (Edstrom Industries, LLC, Waterford, WI). The room was maintained on a 14:10 hour light: dark cycle at 22.8±2°C, with relative humidity between 30 and 50%. All experimental procedures were approved by the Johns Hopkins University Animal Care and Use Committee, the program is accredited by AAALAC, International and procedures were consistent with the Guide for the Care and Use of Laboratory Animals.
mRNA isolation and quantitative RT-PCR analyses
Total RNA was isolated from spleen, mesenteric lymph node (MLN), terminal ileum and proximal and distal colon using TRIzol (Invitrogen, Carlsbad, CA) according to the manufacturer’s protocol. Reverse transcription was performed using a first strand cDNA synthesis kit (Roche, Indianapolis, IN). Quantitative PCR was performed using SYBR Green gene-specific primers on an ABI 7300 Real time PCR System. Results were normalized to Gapdh levels. For relative comparisons, samples were compared to the corresponding WT tissue that was assigned an arbitrary value of one.
Isolation of intraepithelial lymphocytes (IELs) and lamina propria lymphocytes (LPLs)
IELs and LPLs were isolated as described previously [1]. Briefly, colons and small intestines were harvested, cut open longitudinally and washed in cold 1xPBS to remove feces. Either two small intestines or three colons were cut into 1cm segments and incubated for 25 minutes at 37°C in RPMI containing 3% FBS, EDTA, DTT and Hepes, with shaking. Suspensions were strained and IELs were purified from the flow through using a percoll gradient. The remaining segments of small intestine and colon were further digested in RPMI containing DNAseI, Liberase TL and Hepes for 50 minutes at 37°C. The digested tissues were passed through a 70μm and a 40μm strainer respectively to obtain single cell suspensions of small intestinal and colonic LPLs. All procedures were carried out on ice.
Cell staining, antibodies and flow cytometry
For cellular phenotyping, cells were washed, incubated with anti-CD16 to block Fc receptors and stained with VIVID (Invitrogen, Carlsbad, CA) to exclude dead cells. Cells were surface stained in cocktails with different combinations of fluorescent dye-conjugated monoclonal antibodies (mAbs) depending on the panel that included antibodies raised against CD3ε, CD4, CD8, CD19, CD11b, Ly6C, Ly6G, F4/80, NKp46, CD127 (IL-7Rα), CCR6 and γδTCR (BD Biosciences, San Jose, CA). For intracellular cytokine staining (ICS) experiments, single cell suspensions of spleen, MLN, small intestinal IELs (sIEL) and sLPLs, colonic IELs (cIEL) and cLPLs were re-stimulated for 4 hours with 40ng/mL phorbol-12-myristate-13-acetate (PMA) and 2μg/mL ionomycin (Millipore, Billerica, MA, USA) in the presence of brefeldin A (BD Biosciences, San Jose, CA). Cells were surface stained as above then were fixed and permeabilized using the eBioscience Foxp3 buffer system and stained with antibodies against mouse IL-10, IFN-γ, IL-17, IL-22, RORγt and/or Foxp3 (BD Biosciences, San Jose, CA). Cells were acquired on a BD LSRII. Data were analyzed with FlowJo software (Tree Star, Ashland, OR).
Tissue explants
Colons and small intestines were harvested, cut open longitudinally and washed in cold 1xPBS to remove gut contents. For the colon, two 1 cm segments from the distal, mid and proximal regions as well as the cecum were cut, weighed and washed three additional times in sterile cold PBS. For the small intestine, two 1 cm segments from the midpoint were used. After washing, explants were cultured in a 24 well plate for 24 hours in 500 μL of complete IMDM media supplemented with 10% Fetal Bovine Serum (FBS, Atlanta biologicals, Inc. Flowery Branch, GA), 2 mM of L-glutamine (Cellgro®, Mediatech, Inc., Manassas, VA), 1X of Non-essential amino acids (Gibco®, life technologies, Grand Island, NY), 1mM of Sodium pyruvate (Gibco®, life technologies, Grand Island, NY), 10mM of 2-mercaptoethanol (Gibco®, Life Technologies, Grand Island, NY), 100 U/ml of penicillin (Cellgro®, Mediatech, Inc., Manassas, VA), and 100 mg/ml of streptomycin (CellgroR, Mediatech, Inc., Manassas, VA). Supernatants were collected, centrifuged to eliminate debris and stored at −80°C until analysis.
IL-22 ELISA
IL-22 in serum and colon explant supernatants was measured using the Mouse IL-22 ELISA MAX™ Deluxe (Biolegend, San Diego, CA) according to the manufacturer’s protocol.
Ig ELISA
Freshly collected or frozen fecal samples were suspended at a 100mg/mL in PBS containing 0.1% Tween 20 and Complete Mini (one tablet per 10mL, Roche, Mannheim, Germany). Samples were vortexed until a homogenous suspension was obtained and allowed to sit at room temperature for 20 minutes. Samples were centrifuged for 10 minutes at 12,000 rpm at 4°C. The clear supernatant was collected and stored at −20°C until analysis. Fecal IgA and IgG was measured by ELISA as described previously (99). ELISA plates were coated overnight at 4°C with goat anti-mouse Ig (Southern Biotech, Birmingham, AL), then washed and blocked with 1% BSA-PBS (30 minutes at room temperature). Serial dilutions of fecal supernatants were added to the 96-well plates, followed by HRP-conjugated, isotype-specific, secondary antibodies (1:1000 dilution in PBS-BSA; Southern Biotech, Birmingham, AL).
Pathology
Mice were monitored weekly for rectal prolapse. For histopathology, euthanasia was performed by CO2 inhalation, then exsanguination and blood collection by cardiocentesis. Mice were perfused with heparinized saline, followed by 10% neutral buffered formalin, via the left ventricle of the heart. Colons were collected, and either cut open longitudinally and prepared as a “Swiss roll”, or as cross sections. Fixed tissues were processed routinely to paraffin in graded alcohols, sectioned at 5u, and stained with Hematoxylin and Eosin (H&E). Inflammation was scored in a blinded manner according to The Jackson Laboratory (TJL) scoring system as described previously (100).
PCR array
Total RNA was isolated from the distal colons of WT, Il10−/−, Il22−/− and Il10−/−Il22−/− mice (n=4) using TRIzol (Invitrogen), according to the manufacturer’s protocol. RNA was reverse-transcribed using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). cDNA samples were then loaded onto the TaqMan™ OpenArray™ Mouse Inflammation Panel (Applied Biosystems) using the QuantStudio 12K Flex OpenArray AccuFill System (LTI). OpenArray Real-Time PCR results were analyzed using Partek genomic software. Gene expression was calculated using the comparative Cq method as fold change relative to WT using the reference gene Hprt1. Significantly different genes were calculated using ANOVA with cutoff at FDR< 0.05 and fold change of two. Data were visualized as heatmaps composed of Z-scores from standardized Cq values for each independent sample.
Bacterial 16S rRNA sequencing and qPCR.
The fecal microbiome was analyzed using feces from 10 male and 5 female mice of each genotype obtained between 12–16 weeks. DNA was extracted from fecal pellets using the PowerSoil DNA Kit (MO BIO Laboratories, Carlsbad, CA). Amplicons were generated using oligonucleotide primers that target approximately 300bp of the V4 variable region of the 16S rRNA gene (primers 515F and 806R) (101) and also were barcoded and pooled to construct the sequencing library, followed by sequencing with an Illumina MiSeq instrument to generate paired-end 150×150 reads. The software package QIIME 1.7.0 was used to analyze, display, and generate figures of microbiome data using a previously defined method (102).
Overlapping pair-end reads were aligned using SHE-RA (103) with subsequent analysis and normalization performed using QIIME 1.7.0 (104). Fecal communities were compared by using UniFrac, a phylogeny-based distance metric that measures the degree to which any two microbiota share branch length on a bacterial tree of life. As a first step, sequence data and metadata were combined to de-multiplex the barcoded reads followed by quality filtering using the default parameters in QIIME. Sequences were grouped into OTUs (Operational Taxonomic Units) at 97% sequence similarity using the UCLUST algorithm. Taxonomy was assigned using Ribosomal Database Project (RDP) classifier against the GreenGenes database, and sequences were aligned and a phylogenetic tree was built from reference sequences using FastTree. An OTU table showing counts of each OTU in each sample was produced. To control for differences in sequencing depth, OTU tables were rarified at a single sequencing depth (105, 106). Alpha diversity was determined using the Shannon and Chao1 indices, and statistical analysis was performed at a sampling depth of 30,450 sequences. Beta diversity was determined using unweighted and weighted UniFrac distance matrices (107) and the results presented as principal coordinate axis (PCoA) plots. Significant differences in the relative abundance of bacteria at different taxonomic levels were computed using Linear Discriminant Analysis Effect Size (LEfSe) comparing each mouse strain against the remaining samples with LDA scores > 2.0 and p-values <0.05 considered significant (108). Pairwise adonis comparisons were carried out using RStudio (version 3.4.3) with the pairwiseAdonis package (109).
Statistical measures of microbiota diversity
Data were analyzed at the highest sampling depth (30,450 reads) that was shared by all four genotypes of mice. All statistical analyses (α = 0.05) were performed using GraphPad Prism 5 (GraphPad Software, La Jolla, CA). If data were normally distributed based on the Kolmogorov-Smirnov test, groups were compared using one-way ANOVA and p-values generated using the Student-Neuman-Keuls multiple comparison procedure. If data sets were not normally distributed, due to either skew or non-continuous nature (histopathology metrics), the Mann–Whitney U test, Kruskal-Wallis test or Dunn’s multiple comparison procedure were used. Array findings were reported when the p value and the FDR were both less than 0.05.
Supplementary Material
Acknowledgements:
We thank Tricia Nilles and the Becton Dickinson Immune Function Lab in the Johns Hopkins School of Public Health (JHSPH) for assistance with flow cytometry, the Johns Hopkins Phenotyping Core and Nadine Forbes-McBean for assistance with pathology, and the Johns Hopkins Oncology Tissue Services (OTS) for histology processing. We thank Roxann Ashworth of the JHU Genetic Research Core facility for running the OpenArrays and Anne E. Jedlicka and Amanda Dziedzic of the JHSPH Genomic Analysis and Sequencing Core Facility for assistance with OpenArray data analysis. We thank Drs. Alan Scott, Fengyi Wan and Bruce Horwitz for careful review of this manuscript.
Funding: This work was supported in part by grants to J.H.B from the National Institutes of Health (NIH) (R01AI070594 and RO1AI113910) and to J.G.F. (T32OD 010978-28, R01OD011141, and P30ES002109).D.C.G. was supported in part by the Molecular and Cellular Basis of Infectious Diseases (MCBID) training grant from the NIH (T32AI007417-23). V.V. is supported by a scholarship from H.R.H. Maha Chakri Sirindhorn (Thailand).
Reference List
- 1.Whibley N, Gaffen SL. Gut-Busters: IL-17 Ain’t Afraid of No IL-23. Immunity. 2015;43(4):620–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Honda K, Littman DR. The microbiota in adaptive immune homeostasis and disease. Nature. 2016;535(7610):75–84. [DOI] [PubMed] [Google Scholar]
- 3.Shouval DS, Ouahed J, Biswas A, Goettel JA, Horwitz BH, Klein C, et al. Interleukin 10 receptor signaling: master regulator of intestinal mucosal homeostasis in mice and humans. Adv Immunol. 2014;122:177–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kühn R, Löhler J, Rennick D, Rajewsky K, Müller W. Interleukin-10-deficient mice develop chronic enterocolitis. Cell. 1993;75(2):263–74. [DOI] [PubMed] [Google Scholar]
- 5.Spencer SD, Di Marco F, Hooley J, Pitts-Meek S, Bauer M, Ryan AM, et al. The orphan receptor CRF2–4 is an essential subunit of the interleukin 10 receptor. J Exp Med. 1998;187(4):571–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kiesler P, Fuss IJ, Strober W. Experimental Models of Inflammatory Bowel Diseases. Cell Mol Gastroenterol Hepatol. 2015;1(2):154–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Whary MT, Fox JG. Natural and experimental Helicobacter infections. Comp Med. 2004;54(2):128–58. [PubMed] [Google Scholar]
- 8.Sellon RK, Tonkonogy S, Schultz M, Dieleman LA, Grenther W, Balish E, et al. Resident enteric bacteria are necessary for development of spontaneous colitis and immune system activation in interleukin-10-deficient mice. Infection and immunity. 1998;66(11):5224–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kullberg MC, Ward JM, Gorelick PL, Caspar P, Hieny S, Cheever A, et al. Helicobacter hepaticus triggers colitis in specific-pathogen-free interleukin-10 (IL-10)-deficient mice through an IL-12- and gamma interferon-dependent mechanism. Infect Immun. 1998;66(11):5157–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karrasch T, Kim J-S, Muhlbauer M, Magness ST, Jobin C. Gnotobiotic IL-10−/−; NF-κBEGFP mice reveal the critical role of TLR/NF-κB signaling in commensal bacteria-induced colitis. The Journal of Immunology. 2007;178(10):6522–32. [DOI] [PubMed] [Google Scholar]
- 11.Vieira P, O’Garra A. Regula’ten’the gut. Nature immunology. 2007;8(9):905–8. [DOI] [PubMed] [Google Scholar]
- 12.Berg DJ, Davidson N, Kuhn R, Muller W, Menon S, Holland G, et al. Enterocolitis and colon cancer in interleukin-10-deficient mice are associated with aberrant cytokine production and CD4(+) TH1-like responses. J Clin Invest. 1996;98(4):1010–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yen D, Cheung J, Scheerens H, Poulet F, McClanahan T, McKenzie B, et al. IL-23 is essential for T cell-mediated colitis and promotes inflammation via IL-17 and IL-6. J Clin Invest. 2006;116(5):1310–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Buonocore S, Ahern PP, Uhlig HH, Ivanov II, Littman DR, Maloy KJ, et al. Innate lymphoid cells drive interleukin-23-dependent innate intestinal pathology. Nature. 2010;464(7293):1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maxwell JR, Zhang Y, Brown WA, Smith CL, Byrne FR, Fiorino M, et al. Differential roles for interleukin-23 and interleukin-17 in intestinal immunoregulation. Immunity. 2015;43(4):739–50. [DOI] [PubMed] [Google Scholar]
- 16.Eyerich K, Dimartino V, Cavani A. IL-17 and IL-22 in immunity: Driving protection and pathology. Eur J Immunol. 2017;47(4):607–14. [DOI] [PubMed] [Google Scholar]
- 17.Ogawa A, Andoh A, Araki Y, Bamba T, Fujiyama Y. Neutralization of interleukin-17 aggravates dextran sulfate sodium-induced colitis in mice. Clinical immunology. 2004;110(1):55–62. [DOI] [PubMed] [Google Scholar]
- 18.Lee JS, Tato CM, Joyce-Shaikh B, Gulen MF, Cayatte C, Chen Y, et al. Interleukin-23-independent IL-17 production regulates intestinal epithelial permeability. Immunity. 2015;43(4):727–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.O’Connor W Jr, Kamanaka M, Booth CJ, Town T, Nakae S, Iwakura Y, et al. A protective function for interleukin 17A in T cell–mediated intestinal inflammation. Nature immunology. 2009;10(6):603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hueber W, Sands BE, Lewitzky S, Vandemeulebroecke M, Reinisch W, Higgins PD, et al. Secukinumab, a human anti-IL-17A monoclonal antibody, for moderate to severe Crohn’s disease: unexpected results of a randomised, double-blind placebo-controlled trial. Gut. 2012:gutjnl-2011–301668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Targan SR, Feagan B, Vermeire S, Panaccione R, Melmed GY, Landers C, et al. A Randomized, Double-Blind, Placebo-Controlled Phase 2 Study of Brodalumab in Patients With Moderate-to-Severe Crohn’s Disease. Am J Gastroenterol. 2016;111(11):1599–607. [DOI] [PubMed] [Google Scholar]
- 22.Dudakov JA, Hanash AM, van den Brink MR. Interleukin-22: immunobiology and pathology. Annu Rev Immunol. 2015;33:747–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sabat R, Ouyang W, Wolk K. Therapeutic opportunities of the IL-22-IL-22R1 system. Nature reviews Drug discovery. 2014;13(1):21–38. [DOI] [PubMed] [Google Scholar]
- 24.Akdis M, Palomares O, van de Veen W, van Splunter M, Akdis CA. TH17 and TH22 cells: a confusion of antimicrobial response with tissue inflammation versus protection. J Allergy Clin Immunol. 2012;129(6):1438–49; quiz50–1. [DOI] [PubMed] [Google Scholar]
- 25.Eberl G, Colonna M, Di Santo JP, McKenzie AN. Innate lymphoid cells. Innate lymphoid cells: a new paradigm in immunology. Science. 2015;348(6237):aaa6566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Basu R, O’Quinn DB, Silberger DJ, Schoeb TR, Fouser L, Ouyang W, et al. Th22 cells are an important source of IL-22 for host protection against enteropathogenic bacteria. Immunity. 2012;37(6):1061–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vivier E, Artis D, Colonna M, Diefenbach A, Di Santo JP, Eberl G, et al. Innate Lymphoid Cells: 10 Years On. Cell. 2018;174(5):1054–66. [DOI] [PubMed] [Google Scholar]
- 28.Ranatunga DC, Ramakrishnan A, Uprety P, Wang F, Zhang H, Margolick JB, et al. A protective role for human IL-10-expressing CD4+ T cells in colitis. J Immunol. 2012;189(3):1243–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang I, Eibach D, Kops F, Brenneke B, Woltemate S, Schulze J, et al. Intestinal microbiota composition of interleukin-10 deficient C57BL/6J mice and susceptibility to Helicobacter hepaticus-induced colitis. PLoS One. 2013;8(8):e70783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brand S, Beigel F, Olszak T, Zitzmann K, Eichhorst ST, Otte J-M, et al. IL-22 is increased in active Crohn’s disease and promotes proinflammatory gene expression and intestinal epithelial cell migration. American Journal of Physiology-Gastrointestinal and Liver Physiology. 2006;290(4):G827–G38. [DOI] [PubMed] [Google Scholar]
- 31.Zheng Y, Valdez PA, Danilenko DM, Hu Y, Sa SM, Gong Q, et al. Interleukin-22 mediates early host defense against attaching and effacing bacterial pathogens. Nature medicine. 2008;14(3):282. [DOI] [PubMed] [Google Scholar]
- 32.Sugimoto K, Ogawa A, Mizoguchi E, Shimomura Y, Andoh A, Bhan AK, et al. IL-22 ameliorates intestinal inflammation in a mouse model of ulcerative colitis. The Journal of clinical investigation. 2008;118(2):534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zenewicz LA, Yancopoulos GD, Valenzuela DM, Murphy AJ, Stevens S, Flavell RA. Innate and adaptive interleukin-22 protects mice from inflammatory bowel disease. Immunity. 2008;29(6):947–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Penny HA, Hodge SH, Hepworth MR. Orchestration of intestinal homeostasis and tolerance by group 3 innate lymphoid cells. Semin Immunopathol. 2018;40(4):357–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Villarino AV, Sciume G, Davis FP, Iwata S, Zitti B, Robinson GW, et al. Subset- and tissue-defined STAT5 thresholds control homeostasis and function of innate lymphoid cells. J Exp Med. 2017;214(10):2999–3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Eaton KA, Opp JS, Gray BM, Bergin IL, Young VB. Ulcerative typhlocolitis associated with Helicobacter mastomyrinus in telomerase-deficient mice. Veterinary pathology. 2011;48(3):713–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fox JG, Ge Z, Whary MT, Erdman SE, Horwitz BH. Helicobacter hepaticus infection in mice: models for understanding lower bowel inflammation and cancer. Mucosal Immunol. 2011;4(1):22–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peterson CG, Eklund E, Taha Y, Raab Y, Carlson M. A new method for the quantification of neutrophil and eosinophil cationic proteins in feces: establishment of normal levels and clinical application in patients with inflammatory bowel disease. The American journal of gastroenterology. 2002;97(7):1755–62. [DOI] [PubMed] [Google Scholar]
- 39.Chassaing B, Srinivasan G, Delgado MA, Young AN, Gewirtz AT, Vijay-Kumar M. Fecal lipocalin 2, a sensitive and broadly dynamic non-invasive biomarker for intestinal inflammation. PloS one. 2012;7(9):e44328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Coorens M, Rao A, Grafe SK, Unelius D, Lindforss U, Agerberth B, et al. Innate lymphoid cell type 3-derived interleukin-22 boosts lipocalin-2 production in intestinal epithelial cells via synergy between STAT3 and NF-kappaB. J Biol Chem. 2019;294(15):6027–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maloy KJ, Powrie F. Intestinal homeostasis and its breakdown in inflammatory bowel disease. Nature. 2011;474(7351):298. [DOI] [PubMed] [Google Scholar]
- 42.Feng T, Qin H, Wang L, Benveniste EN, Elson CO, Cong Y. Th17 cells induce colitis and promote Th1 cell responses through IL-17 induction of innate IL-12 and IL-23 production. The Journal of Immunology. 2011;186(11):6313–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu M, Pokrovskii M, Ding Y, Yi R, Au C, Harrison OJ, et al. c-MAF-dependent regulatory T cells mediate immunological tolerance to a gut pathobiont. Nature. 2018;554(7692):373–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gomes-Santos AC, Moreira TG, Castro-Junior AB, Horta BC, Lemos L, Cruz DN, et al. New insights into the immunological changes in IL-10-deficient mice during the course of spontaneous inflammation in the gut mucosa. Clinical and Developmental Immunology. 2012;2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Davidson NJ, Leach MW, Fort MM, Thompson-Snipes L, Kühn R, Müller W, et al. T helper cell 1-type CD4+ T cells, but not B cells, mediate colitis in interleukin 10-deficient mice. Journal of Experimental Medicine. 1996;184(1):241–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Leonard WJ, Wan C-K. IL-21 signaling in immunity. F1000Research. 2016;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hooper LV, Macpherson AJ. Immune adaptations that maintain homeostasis with the intestinal microbiota. Nat Rev Immunol. 2010;10(3):159–69. [DOI] [PubMed] [Google Scholar]
- 48.Ott S, Musfeldt M, Wenderoth D, Hampe J, Brant O, Fölsch U, et al. Reduction in diversity of the colonic mucosa associated bacterial microflora in patients with active inflammatory bowel disease. Gut. 2004;53(5):685–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dalal SR, Chang EB. The microbial basis of inflammatory bowel diseases. J Clin Invest. 2014;124(10):4190–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Whary MT, Taylor NS, Feng Y, Ge Z, Muthupalani S, Versalovic J, et al. Lactobacillus reuteri promotes Helicobacter hepaticus‐associated typhlocolitis in gnotobiotic B6. 129P2‐IL‐10tm1Cgn (IL‐10−/−) mice. Immunology. 2011;133(2):165–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Clemente JC, Ursell LK, Parfrey LW, Knight R. The impact of the gut microbiota on human health: an integrative view. Cell. 2012;148(6):1258–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim D, Zeng MY, Nunez G. The interplay between host immune cells and gut microbiota in chronic inflammatory diseases. Exp Mol Med. 2017;49(5):e339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hiippala K, Kainulainen V, Kalliomaki M, Arkkila P, Satokari R. Mucosal Prevalence and Interactions with the Epithelium Indicate Commensalism of Sutterella spp. Front Microbiol. 2016;7:1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Setoyama H, Imaoka A, Ishikawa H, Umesaki Y. Prevention of gut inflammation by Bifidobacterium in dextran sulfate-treated gnotobiotic mice associated with Bacteroides strains isolated from ulcerative colitis patients. Microbes and infection / Institut Pasteur. 2003;5(2):115–22. [DOI] [PubMed] [Google Scholar]
- 55.Whary MT, Cline J, King A, Ge Z, Shen Z, Sheppard B, et al. Long-term colonization levels of Helicobacter hepaticus in the cecum of hepatitis-prone A/JCr mice are significantly lower than those in hepatitis-resistant C57BL/6 mice. Comp Med. 2001;51(5):413–7. [PubMed] [Google Scholar]
- 56.Bilsborough J, Targan SR, Snapper SB. Therapeutic targets in inflammatory bowel disease: current and future. The American Journal of Gastroenterology Supplements. 2016;3(3):27. [Google Scholar]
- 57.Mitra A, Raychaudhuri SK, Raychaudhuri SP. IL-22 induced cell proliferation is regulated by PI3K/Akt/mTOR signaling cascade. Cytokine. 2012;60(1):38–42. [DOI] [PubMed] [Google Scholar]
- 58.Lindemans CA, Calafiore M, Mertelsmann AM, O’Connor MH, Dudakov JA, Jenq RR, et al. Interleukin-22 promotes intestinal-stem-cell-mediated epithelial regeneration. Nature. 2015;528(7583):560–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liang SC, Tan X-Y, Luxenberg DP, Karim R, Dunussi-Joannopoulos K, Collins M, et al. Interleukin (IL)-22 and IL-17 are coexpressed by Th17 cells and cooperatively enhance expression of antimicrobial peptides. Journal of Experimental Medicine. 2006;203(10):2271–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zigmond E, Bernshtein B, Friedlander G, Walker CR, Yona S, Kim K-W, et al. Macrophage-restricted interleukin-10 receptor deficiency, but not IL-10 deficiency, causes severe spontaneous colitis. Immunity. 2014;40(5):720–33. [DOI] [PubMed] [Google Scholar]
- 61.Rizzo HL, Kagami S, Phillips KG, Kurtz SE, Jacques SL, Blauvelt A. IL-23–Mediated Psoriasis-Like Epidermal Hyperplasia Is Dependent on IL-17A. The journal of immunology. 2011;186(3):1495–502. [DOI] [PubMed] [Google Scholar]
- 62.Martin DA, Towne JE, Kricorian G, Klekotka P, Gudjonsson JE, Krueger JG, et al. The emerging role of IL-17 in the pathogenesis of psoriasis: preclinical and clinical findings. Journal of Investigative Dermatology. 2013;133(1):17–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Whibley N, Gaffen SL. Brothers in arms: Th17 and Treg responses in Candida albicans immunity. PLoS pathogens. 2014;10(12):e1004456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chaudhry A, Samstein RM, Treuting P, Liang Y, Pils MC, Heinrich J-M, et al. Interleukin-10 signaling in regulatory T cells is required for suppression of Th17 cell-mediated inflammation. Immunity. 2011;34(4):566–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Krause P, Morris V, Greenbaum JA, Park Y, Bjoerheden U, Mikulski Z, et al. IL-10-producing intestinal macrophages prevent excessive antibacterial innate immunity by limiting IL-23 synthesis. Nature communications. 2015;6:7055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kamanaka M, Huber S, Zenewicz LA, Gagliani N, Rathinam C, O’Connor W, et al. Memory/effector (CD45RBlo) CD4 T cells are controlled directly by IL-10 and cause IL-22–dependent intestinal pathology. Journal of Experimental Medicine. 2011:jem. 20102149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gu Y, Yang J, Ouyang X, Liu W, Li H, Yang J, et al. Interleukin 10 suppresses Th17 cytokines secreted by macrophages and T cells. Eur J Immunol. 2008;38(7):1807–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Morrison PJ, Ballantyne SJ, Macdonald SJ, Moore JW, Jenkins D, Wright JF, et al. Differential requirements for IL-17A and IL-22 in cecal versus colonic inflammation induced by Helicobacter hepaticus. The American journal of pathology. 2015;185(12):3290–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schreiber F, Arasteh JM, Lawley TD. Pathogen Resistance Mediated by IL-22 Signaling at the Epithelial-Microbiota Interface. J Mol Biol. 2015;427(23):3676–82. [DOI] [PubMed] [Google Scholar]
- 70.Takayama T, Kamada N, Chinen H, Okamoto S, Kitazume MT, Chang J, et al. Imbalance of NKp44(+)NKp46(−) and NKp44(−)NKp46(+) natural killer cells in the intestinal mucosa of patients with Crohn’s disease. Gastroenterology. 2010;139(3):882–92, 92 e1–3. [DOI] [PubMed] [Google Scholar]
- 71.Pearson C, Thornton EE, McKenzie B, Schaupp AL, Huskens N, Griseri T, et al. ILC3 GM-CSF production and mobilisation orchestrate acute intestinal inflammation. Elife. 2016;5:e10066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bauche D, Joyce-Shaikh B, Jain R, Grein J, Ku KS, Blumenschein WM, et al. LAG3(+) Regulatory T Cells Restrain Interleukin-23-Producing CX3CR1(+) Gut-Resident Macrophages during Group 3 Innate Lymphoid Cell-Driven Colitis. Immunity. 2018;49(2):342–52 e5. [DOI] [PubMed] [Google Scholar]
- 73.Pantazi E, Powell N. Group 3 ILCs: Peacekeepers or Troublemakers? What’s Your Gut Telling You?! Front Immunol. 2019;10:676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Andoh A, Zhang Z, Inatomi O, Fujino S, Deguchi Y, Araki Y, et al. Interleukin-22, a member of the IL-10 subfamily, induces inflammatory responses in colonic subepithelial myofibroblasts. Gastroenterology. 2005;129(3):969–84. [DOI] [PubMed] [Google Scholar]
- 75.Shindo R, Ohmuraya M, Komazawa-Sakon S, Miyake S, Deguchi Y, Yamazaki S, et al. Necroptosis of Intestinal Epithelial Cells Induces Type 3 Innate Lymphoid Cell-Dependent Lethal Ileitis. iScience. 2019;15:536–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang C, Gong G, Sheh A, Muthupalani S, Bryant E, Puglisi D, et al. Interleukin-22 drives nitric oxide-dependent DNA damage and dysplasia in a murine model of colitis-associated cancer. Mucosal immunology. 2017;10(6):1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kirchberger S, Royston DJ, Boulard O, Thornton E, Franchini F, Szabady RL, et al. Innate lymphoid cells sustain colon cancer through production of interleukin-22 in a mouse model. Journal of Experimental Medicine. 2013;210(5):917–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Eken A, Singh A, Treuting P, Oukka M. IL-23R+ innate lymphoid cells induce colitis via interleukin-22-dependent mechanism. Mucosal immunology. 2014;7(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mao K, Baptista AP, Tamoutounour S, Zhuang L, Bouladoux N, Martins AJ, et al. Innate and adaptive lymphocytes sequentially shape the gut microbiota and lipid metabolism. Nature. 2018;554(7691):255–9. [DOI] [PubMed] [Google Scholar]
- 80.Fung TC, Bessman NJ, Hepworth MR, Kumar N, Shibata N, Kobuley D, et al. Lymphoid-Tissue-Resident Commensal Bacteria Promote Members of the IL-10 Cytokine Family to Establish Mutualism. Immunity. 2016;44(3):634–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kinnebrew MA, Buffie CG, Diehl GE, Zenewicz LA, Leiner I, Hohl TM, et al. Interleukin 23 production by intestinal CD103(+)CD11b(+) dendritic cells in response to bacterial flagellin enhances mucosal innate immune defense. Immunity. 2012;36(2):276–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mortha A, Chudnovskiy A, Hashimoto D, Bogunovic M, Spencer SP, Belkaid Y, et al. Microbiota-dependent crosstalk between macrophages and ILC3 promotes intestinal homeostasis. Science. 2014;343(6178):1249288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hepworth MR, Monticelli LA, Fung TC, Ziegler CG, Grunberg S, Sinha R, et al. Innate lymphoid cells regulate CD4+ T-cell responses to intestinal commensal bacteria. Nature. 2013;498(7452):113–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Melo-Gonzalez F, Kammoun H, Evren E, Dutton EE, Papadopoulou M, Bradford BM, et al. Antigen-presenting ILC3 regulate T cell-dependent IgA responses to colonic mucosal bacteria. J Exp Med. 2019;216(4):728–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Buchler G, Wos-Oxley ML, Smoczek A, Zschemisch NH, Neumann D, Pieper DH, et al. Strain-specific colitis susceptibility in IL10-deficient mice depends on complex gut microbiota-host interactions. Inflamm Bowel Dis. 2012;18(5):943–54. [DOI] [PubMed] [Google Scholar]
- 86.Whary MT, Fox JG. Detection, eradication, and research implications of Helicobacter infections in laboratory rodents. Lab Anim (NY). 2006;35(7):25–7, 30–6. [DOI] [PubMed] [Google Scholar]
- 87.Chichlowski M, Sharp JM, Vanderford DA, Myles MH, Hale LP. Helicobacter typhlonius and Helicobacter rodentium differentially affect the severity of colon inflammation and inflammation-associated neoplasia in IL10-deficient mice. Comp Med. 2008;58(6):534–41. [PMC free article] [PubMed] [Google Scholar]
- 88.Wilson MS, Madala SK, Ramalingam TR, Gochuico BR, Rosas IO, Cheever AW, et al. Bleomycin and IL-1beta-mediated pulmonary fibrosis is IL-17A dependent. J Exp Med. 2010;207(3):535–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Slight SR, Monin L, Gopal R, Avery L, Davis M, Cleveland H, et al. IL-10 restrains IL-17 to limit lung pathology characteristics following pulmonary infection with Francisella tularensis live vaccine strain. Am J Pathol. 2013;183(5):1397–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Manichanh C, Rigottier-Gois L, Bonnaud E, Gloux K, Pelletier E, Frangeul L, et al. Reduced diversity of faecal microbiota in Crohn’s disease revealed by a metagenomic approach. Gut. 2006;55(2):205–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lepage P, Hasler R, Spehlmann ME, Rehman A, Zvirbliene A, Begun A, et al. Twin study indicates loss of interaction between microbiota and mucosa of patients with ulcerative colitis. Gastroenterology. 2011;141(1):227–36. [DOI] [PubMed] [Google Scholar]
- 92.Ni J, Wu GD, Albenberg L, Tomov VT. Gut microbiota and IBD: causation or correlation? Nat Rev Gastroenterol Hepatol. 2017;14(10):573–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sonnenberg GF, Monticelli LA, Alenghat T, Fung TC, Hutnick NA, Kunisawa J, et al. Innate lymphoid cells promote anatomical containment of lymphoid-resident commensal bacteria. Science. 2012;336(6086):1321–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zenewicz LA, Yin X, Wang G, Elinav E, Hao L, Zhao L, et al. IL-22 deficiency alters colonic microbiota to be transmissible and colitogenic. The Journal of Immunology. 2013;190(10):5306–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bettelli E, Carrier Y, Gao W, Korn T, Strom TB, Oukka M, et al. Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature. 2006;441(7090):235–8. [DOI] [PubMed] [Google Scholar]
- 96.Mangan PR, Harrington LE, O’Quinn DB, Helms WS, Bullard DC, Elson CO, et al. Transforming growth factor-beta induces development of the T(H)17 lineage. Nature. 2006;441(7090):231–4. [DOI] [PubMed] [Google Scholar]
- 97.Wang C, Gong G, Sheh A, Muthupalani S, Bryant EM, Puglisi DA, et al. Interleukin-22 drives nitric oxide-dependent DNA damage and dysplasia in a murine model of colitis-associated cancer. Mucosal Immunol. 2017;10(6):1504–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bleich A, Fox JG. The Mammalian Microbiome and Its Importance in Laboratory Animal Research. ILAR J. 2015;56(2):153–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Peterson DA, McNulty NP, Guruge JL, Gordon JI. IgA response to symbiotic bacteria as a mediator of gut homeostasis. Cell host & microbe. 2007;2(5):328–39. [DOI] [PubMed] [Google Scholar]
- 100.Bleich A, Mähler M, Most C, Leiter EH, Liebler–Tenorio E, Elson CO, et al. Refined histopathologic scoring system improves power todetect colitis QTL in mice. Mammalian genome. 2004;15(11):865–71. [DOI] [PubMed] [Google Scholar]
- 101.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, et al. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proceedings of the National Academy of Sciences of the United States of America. 2011;108 Suppl 1:4516–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Swennes AG, Sheh A, Parry NM, Muthupalani S, Lertpiriyapong K, Garcia A, et al. Helicobacter hepaticus infection promotes hepatitis and preneoplastic foci in farnesoid X receptor (FXR) deficient mice. PLoS One. 2014;9(9):e106764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Rodrigue S, Materna AC, Timberlake SC, Blackburn MC, Malmstrom RR, Alm EJ, et al. Unlocking short read sequencing for metagenomics. PLoS One. 2010;5(7):e11840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nature methods. 2010;7(5):335–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hamady M, Knight R. Microbial community profiling for human microbiome projects: Tools, techniques, and challenges. Genome Res. 2009;19(7):1141–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kuczynski J, Costello EK, Nemergut DR, Zaneveld J, Lauber CL, Knights D, et al. Direct sequencing of the human microbiome readily reveals community differences. Genome Biol. 2010;11(5):210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lozupone C, Hamady M, Knight R. UniFrac--an online tool for comparing microbial community diversity in a phylogenetic context. BMC bioinformatics. 2006;7:371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, et al. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12(6):R60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Martinez AP. pairwiseAdonis: Pairwise multilevel comparison using adonis. R package version 0.0.1. 2017. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.