Abstract
In the last decade, carbohydrate microarrays have been core technologies for analyzing carbohydrate-mediated recognition events in a high-throughput fashion. A number of methods have been exploited for immobilizing glycans on the solid surface in a microarray format. This microarray-based technology has been widely employed for rapid analysis of the glycan binding properties of lectins and antibodies, the quantitative measurements of glycan–protein interactions, detection of cells and pathogens, identification of disease-related anti-glycan antibodies for diagnosis, and fast assessment of substrate specificities of glycosyltransferases. This review covers the construction of carbohydrate microarrays, detection methods of carbohydrate microarrays and their applications in biological and biomedical research.
1. Introduction
Carbohydrates are composed of a large group of biomolecules with highly diverse structures and exist largely in the form of glycoconjugates on the cell surface or inside cells. One major group of glycoconjugates are glycoproteins in which carbohydrates are conjugated to a serine/threonine (O-linked glycoproteins) or an asparagine residue (N-linked glycoproteins) (Fig. 1). Proteoglycans possess glycosaminoglycans that are linked to a serine/threonine side chain of the polypeptide backbone via a xylose moiety. Another important class of glycoconjugates is the glycosphingolipid in which mono or oligosaccharides are attached to ceramides.
Fig. 1.
Glycoconjugates in cells.
Glycans present in glycoconjugates are implicated in a variety of important cellular processes through interactions with glycan-binding proteins (GBPs).1–4 For example, cell-surface glycans mediate cell trafficking, adhesion and signaling by association with GBPs. In addition, pathogenic glycans are recognized by various receptors of the immune system, which leads to immune responses to many pathogens including yeast, bacteria and viruses.5–7 Importantly, glycan–protein interactions also play pivotal roles in various pathological events such as tumor metastasis,8 leukocyte recruitment to sites of inflammation,9 and infection of pathogens including toxins, bacteria and viruses.10,11 Therefore, the understanding of glycan–protein interactions at the molecular basis provides deep insights into glycan-mediated biological processes and enables the development of more efficacious drugs and diagnostic tools.
Various forms of ‘arrays’, in which glycoconjugates and glycans are attached to silica plates,12 beads13 or microplates,14 have been used to study glycan–protein interactions over the years. However, advances in high precision robotic arraying and high-resolution imaging enabled substantial miniaturization such that tens of thousands of glycans are immobilized on a standard size microscope slide and their binding to proteins is readily imaged. In efforts aimed at rapid analysis of glycan–protein interactions, carbohydrate microarrays, which are composed of diverse glycans densely and orderly attached to a solid surface, were first developed by several research groups in 2002.15–20 Since then, many elegant methods for the immobilization of glycans and the detection of binding events on the microarrays have been exploited.21–30 Nowadays, carbohydrate microarrays have become the leading edge tools for functional studies of glycans and GBPs because the microarray-based technology has the advantage of a simultaneous assessment of many glycan–protein interactions using small amounts of samples.31 Another important feature of carbohydrate microarrays is that glycans attached to the solid surface are displayed in a multivalent fashion and can form multivalent complexes with GPBs as a result of a cluster effect. Accordingly, proteins that weakly interact with monovalent glycans in solution can strongly bind to carbohydrates on the microarrays. These beneficial aspects make carbohydrate microarrays suitable for rapid analysis of glycan-mediated binding events.
This review article summarizes immobilization methods and library developments that have been used for the construction of carbohydrate microarrays. In addition, detection methods of binding events on carbohydrate microarrays are also included. Furthermore, various applications of carbohydrate microarrays in biological and biomedical research are presented. Because tens of thousands of small quantity samples can be analyzed simultaneously in large scale microarray systems unlike conventional microplate arrays which can be used to assess relatively small numbers of samples, the high-density carbohydrate microarrays are the major focus of this article.
2. Design and construction of carbohydrate microarrays
2.1. Preparation of glycan probes
One of the key aspects of successful glycan microarrays is the availability and strategy for synthesis of large glycan libraries. It is ideal that a single glycan microarray contains a broad repertoire of the representative glycome of an organism of interest to evaluate the binding property of GBPs. However, currently it is only realistically possible to display limited glycan libraries consisting of natural and synthetic glycans that can be practically obtained. The advantage of different glycan microarray platforms depends on the appropriate matching of the type of glycan structures and the specificity of the GBP to be analyzed.
Diverse glycans can be obtained by using glycosyltransferases, implicitly linking the glycome to the genome. Glycan diversity is enormous because the glycans produced within the same cells are highly heterogeneous. Owing to alternative branching patterns, incomplete glycosylation, and enzymatic sulfation and acetylation of glycans, the cellular glycome is estimated to encompass between 100 000 and 500 000 glycan structures.32 Furthermore, cells can assemble glycan structures independently of neighboring cells by regulating expression of the glycosyltransferases. However, the number of unique glycan determinants that comprise the terminal sequences of glycans present in glycoconjugates has been estimated to be only between 500 and 3500.33
An important concern when constructing glycan libraries is to cover the structural diversity of glycans. Classically, libraries designed to focus on a specific issue are small and convenient in size. The majority of GBPs have binding pockets for substructures of glycans composed of only a few monosaccharide residues. This fact is important to hold in mind when constructing a library. In addition, depending on the source of glycans (e.g. synthetic or naturally isolated), efficient functionalization and coupling reactions are particularly important factors. In many cases, glycans must contain a reactive group at the anomeric position that allows them to be immobilized on the surface in question (see Section 2.2).
When building the libraries from de novo synthesized approaches, such as chemical or chemoenzymatic, the glycans need to have a functional group chemically introduced from the start (Fig. 2). Functionalization of glycans is less of a problem when chemical synthesis of glycans is initiated, as the functionality is generally carried out by placing a linker at the reducing terminal sugar residue in a form suitable for flexible modifications. In contrast, constructing libraries with glycans isolated from natural sources, mostly available in tiny amounts, requires either a pre-existing aglycon, such as an amino group, peptide and lipid, or an efficient method to derivatize the reducing terminal sugar.34–36 Desired requirements for derivatization are to preserve as much as possible the natural structural integrity of the sugar linked to the aglycon. One conjugation method conserving the reducing end sugar in a ring-closed form is the reaction with an N-substituted N-hydroxylamine group37,38 or acylation of glycosylamines.39 These approaches were partially adopted to build the neoglycolipid microarray platform as well as for the immobilization of complex natural N-glycans and bacterial polysaccharides onto a hydrogel-functionalized glycan microarray platform by the Consortium for Functional Glycomics (www.functionalglycomics.org).31 Reduction-based methods generating open-chain structures have also been used to display various oligosaccharides on microarray surfaces.17
Fig. 2.
Functionalization of glycans via glycosidic linkages (I), N-hydroxylamines(II) and reductive amination (III).
Synthesis of the glycosidic linkages is the limiting factor when chemically generating glycan libraries. The complex linkages require extensive manipulation of the protecting group.40,41 Nevertheless, the established solution-phase and automated solid-phase synthesis as well as one-pot reactivity-based glycosylations allow for synthesis of diverse libraries comprising extended and branched glycans.42–44 An important complementary approach for the synthesis of glycans and other complex oligosaccharides is enzymatic glycosylation by using recombinant glycosyltransferases.45 They can be applied directly onto a simple monosaccharide acceptor, followed by a subsequent elongation to a more complex structure. Alternatively, they can be introduced after deprotection of chemically synthesized intermediates, or used to generate unnatural glycans.46 Introduction of sialic acid is particularly difficult to achieve chemically but easy to accomplish by sialyltransferases.47,48 Many mammalian glycosyltransferases have strict substrate and acceptor specificities, which limits their synthetic utility to only natural subsets of glycan structures.49 However, increased efforts in exploring alternative sources of glycosyltransferases such as bacteria have significantly expanded the enzymatic tool-box by obtaining transferases with more flexible donor and acceptor specificities and allowing easier production in bacterial expression systems such as E. coli.46
2.2. Immobilization methods
Efficient immobilization techniques of glycans on a solid surface are of great importance for construction of glycan microarrays. Although a variety of immobilization methods have been reported over the past decade, there are many practical issues to take into consideration when constructing glycan microarrays and one approach might not be superior to another. In this article, immobilization strategies are broadly classified into two categories – noncovalent and covalent immobilization – and each method is further divided into site-nonspecific and site-specific attachment of glycans on the surface (Fig. 3).
Fig. 3.
Immobilization strategy for construction of glycan microarrays. (a) Noncovalent, site-nonspecific attachment of glycans on the surface, (b) noncovalent, site-specific attachment of glycans on the surface, (c) covalent, sitenonspecific attachment of glycans on the surface, and (d) covalent, site-specific attachment of glycans on the surface.
2.2.1. NONCOVALENT IMMOBILIZATION
2.2.1.1. Noncovalent, site-nonspecific attachment of glycans.
Noncovalent immobilization of glycans relies on the adherence of free or derivatized glycans to modified or unmodified solid surfaces. The simplest procedure in this approach is site-nonspecific immobilization of free glycans on the surface. For instance, polysaccharide microarrays were constructed by spotting unmodified polysaccharides on nitrocellulose or oxidized black polystyrene slides (Fig. 4a).16,19 In this case, the glycans are noncovalently and site-nonspecifically adsorbed on the solid surface. Because of the nature of noncovalent attachment, glycans must be sufficiently large in order to provide a large enough contact area to adsorb efficiently on the surface. In contrast, small-sized glycans normally attach only weakly to the solid surface, which leads to the loss of glycans during washing steps.
Fig. 4.
Noncovalent and site-nonspecific immobilization. (a) Attachment of unmodified polysaccharides to the surface, (b) attachment of free heparin polysaccharides to the poly-L-lysine-coated surface, and (c) attachment of modified dextrans to the amine or semicarbazide-coated surface.
Polysaccharide microarrays were also prepared by using a different noncovalent immobilization approach. This includes noncovalent and site-nonspecific adsorption of unmodified heparin polysaccharides possessing sulfate groups on positively charged poly-L-lysine coated glass slides via electrostatic interactions (Fig. 4b).50,51 Alternatively, in order to search for glycans that interact with heparin-binding growth factors, chemically modified dextran polysaccharides were noncovalently and site-nonspecifically attached to the amine or semicarbazide-derivatized glass slide (Fig. 4c).52
2.2.1.2. Noncovalent, site-specific attachment of glycans.
Noncovalent, site-specific immobilization techniques of oligosaccharides on the surface were developed to prepare glycan microarrays containing simple carbohydrates and oligosaccharides. For example, lipid-conjugated glycans (termed neoglycolipids, NGLs) were attached to nitrocellulose or PVDF (polyvinylidenedifluoride) membranes via hydrophobic interactions (Fig. 5a).17,22,53 The required NGLs were prepared by reductive amination of sugars, which were generated by chemical or enzymatic methods, with an amino-conjugated lipid. Another example of this type of immobilization technique is the attachment of fluorous tag-conjugated glycans to the fluoroalkylated surface via fluorous–fluorous interactions (Fig. 5b).54–56 Fluorous tagged glycans have unique physical properties which allow for the easy purification by fluorous chromatography. The fluorous tag-based strategy was more recently applied to the construction of glycan microarrays by immobilizing fluorous tag-conjugated glycans on fluorous phosphonate derivatized on the aluminum oxide-coated surface (Fig. 5c).57,58 This immobilization was found to be useful for direct characterization of noncovalently attached glycans by mass spectrometry (MS) for label-free analysis. It should be noted that the lipid and fluorous tags, if long enough, are sufficient to retain glycans on the surface even after extensive washing.
Fig. 5.

Noncovalent and site-specific immobilization. (a) Attachment of lipid-conjugated glycans to nitrocellulose, (b) attachment of fluorous-tagged sugars to the fluoroalkylated surface, (c) attachment of fluorous-tagged glycans to fluorous phosphonate derivatized on the aluminum oxide-coated surface, (d) attachment of biotin-linked sugars to the streptavidin-coated surface, and (e) attachment of oligonucleotide-conjugated glycans to the complementary oligonucleotide-coated surface.
A strong biotin–streptavidin interaction (Kd ~ 10−15 M) was utilized to prepare glycan microarrays (Fig. 5d).59–62 In this case, biotin-conjugated glycans were immobilized on the streptavidin-coated surface. DNA hybridization was also employed to construct glycan microarrays, which relies on anchoring oligonucleotide-linked glycans onto the complementary oligonucleotide-derivatized surface (Fig. 5e).63 Oligonucleotides are conjugated to different carbohydrates and the glycoconjugates are immobilized on the complementary DNA sequence. To identify mediators of autoimmune brain inflammation, a microarray was prepared by noncovalent immobilization of intact glycosphigolipids to PVDF membranes.64 The ganglioside GM1 has also been incorporated into an immobilized fluid supported lipid bilayer approximating display in a cell membrane.65
2.2.2. COVALENT IMMOBILIZATION
2.2.2.1. Covalent, site-nonspecific attachment of glycans.
The simplest covalent immobilization includes attachment of unmodified glycans to the modified surface by a one-step procedure because modification of sugars is time-consuming and labor-intensive. As examples of this strategy, free carbohydrates were attached to the solid surface derivatized by photolabile groups, such as aryltrifluoromethyldiazirine,66 4-azido-2,3,5,6-tetrafluorophenyl67 and phthalimide groups,68,69 under light irradiation (Fig. 6a–c). The first two photolabile groups are converted after irradiation to highly reactive carbene and nitrene intermediates, which rapidly react with free glycans to form covalent bonds. In the case of the phthalimide group, the triple state carbonyl oxygen generated by UV irradiation abstracts a hydrogen atom from glycans and subsequently radical recombination takes place, which leads to covalent attachment of glycans to the surface. Recently, a different covalent and site-nonspecific immobilization method, which is based on recognition of 1,2- or 1,3-diols of sugars (e.g. glucose, fructose, galactose) by boronic acid, was developed (Fig. 6d).70 In this process, unmodified glycans are attached to the boronic acid-functionalized surface through complexation of the boronic acid with 1,2- or 1,3-diols of carbohydrates. Although the above approach has the advantage of easy construction of glycan microarrays, its major drawback is the site-nonspecific attachment of glycans to the surface.
Fig. 6.
Covalent and site-nonspecific immobilization. Attachment of free glycans on (a) aryltrifluoromethyldiazirine, (b) 4-azido-2,3,5,6-tetrafluorophenyl group, (c) phthalimide-derivatized surfaces by UV irradiation, and (d) phenylboronic acid-coated surfaces.
2.2.2.2. Covalent, site-specific attachment of glycans.
The most extensively developed method to prepare glycan microarrays is the covalent and site-specific attachment of modified glycans to the properly derivatized surface. In this approach, functional groups linked to glycans at the anomeric positions selectively react with those derivatized on the surface. Thus, most of these methods necessitate both chemically modified glycans and solid surfaces, and are appropriate for the creation of microarrays that contain simple carbohydrates and oligosaccharides. It has been shown that the nature of linkers between glycans and the solid surface affects protein binding to immobilized sugars as well as nonspecific adsorption of proteins. Glycans conjugated by oligo or poly(ethylene glycol)-based hydrophilic tethers on the microarrays have better binding properties for proteins than those conjugated by hydrophobic linkers. Furthermore, the lengths of tethers influence protein binding affinities to glycans on the surface because of different accessibility of proteins to the glycans depending on tether lengths.15,71
An early example of the application of this type of the immobilization method is based on the reaction between maleimide and thiol groups. In this process, maleimide-conjugated sugars were immobilized on the thiol-derivatized surface (Fig. 7a)15,71,72 or, reversely, thiol-linked sugars were attached to the maleimide-coated surface (Fig. 7b).73–78 Later, disulfide bond formation which has been widely used to prepare bioconjugates was employed to construct glycan microarrays. This immobilization methodology relies on attachment of respective thiosulfonate- and thiol-conjugated sugars to thiol and pyridyl disulfide-coated surfaces (Fig. 7c and d).79,80 It is worthwhile mentioning that because thiol-functionalized substances readily undergo air oxidation; great attention should be given when these substances are used for microarray construction.
Fig. 7.
Covalent and site-specific immobilization. (a) Attachment of maleimide-conjugated glycans to the thiol-coated surface, (b) attachment of thiol-linked glycans to the maleimide-coated surface, (c) attachment of thiosulfonate-conjugated glycans to thiol-coated surface, (d) attachment of thiol-conjugated sugars to the 2-pyridyl disulfide-coated surface, (e) attachment of cyclopentadiene-linked sugars to the benzoquinone-coated surface, (f) attachment of dienophile-conjugated sugars to the tetrazine-coated surface, (g) attachment of p-aminophenyl group-linked sugars to the cyanuric chloride-coated surface, (h) attachment of amine-linked sugars to the NHS ester-coated surface, (i) attachment of aminooxy-linked chondroitin oligosaccharides to the aldehyde-coated surface, and (j) attachment of heparin oligosaccharides, which are obtained from nitrous acid depolymerization of heparin, to the amine-coated surface.
A variety of other ligation reactions have been utilized to construct glycan microarrays. Diels–Alder reactions between dienes and dienophiles were applied to prepare glycan microarrays. For example, cyclopentadiene-linked sugars were covalently immobilized on the benzoquinone-coated surface (Fig. 7e).18 More recently, dienophile-conjugated sugars were immobilized on the tetrazine-coated surface via Diels–Alder reactions with inverse electron demand (Fig. 7f).81 Other widely used immobilization methods include attachment of aminelinked glycans to the cyanuric chloride-modified surface82,83 or N-hydroxysuccinimide (NHS) ester-derivatized surface (Fig. 7g and h).31,84 This immobilization method was later used to prepare glycan microarrays containing a variety of glycan probes.36,37,39,85–90 The ligation reactions between aldehydes and aminooxy or amino groups were used to fabricate glycosaminoglycan microarrays (Fig. 7i and j).91,92 In this method, synthetic chondroitin oligosaccharides conjugated by the aminooxy group or heparin oligosaccharides, which were prepared from nitrous acid depolymerization of heparin, were attached to aldehyde or amine-coated surface, respectively.
Selective ligation reaction between hydrazide and epoxide was developed to prepare glycan microarrays.93–98 In this process, hydrazide-linked glycans that were prepared on solid supports were selectively immobilized on the epoxide-coated surface even in the presence of other nucleophilic groups, such as thiol and amine, under weak acid conditions (Fig. 8a). Epoxide-coated slides were also utilized to construct glycan microarrays through immobilization of neoglycoproteins, such as glycan-conjugated bovine serum albumin (BSA) (Fig. 8b).99–101
Fig. 8.
Covalent and site-specific immobilization. (a) Attachment of hydrazide-linked sugars to the epoxide-coated surface, (b) attachment of glycan-linked BSA to the epoxide-coated surface, (c) attachment of azide-linked glycans to the alkyne-coated surface, (d) attachment of alkyne-linked glycans to the azidecoated surface, (e) attachment of azide-linked glycans to the phosphane-coated surface via Staudinger reaction, and (f) attachment of 4-azido-2,3,5,6tetrafluorophenyl group-conjugated glycans to the polymer monolayer by photochemistry.
Azide-based immobilization methods, which rely on the Staudinger ligation and click chemistry, have been exploited to create glycan microarrays. To date, click chemistry (or Cu(I)-catalyzed cycloaddition reaction between alkynes and azides) has been extensively employed to prepare conjugates of various substances, including polymers, nanomaterials and biomaterials.102–104 An advantage of this reaction is that it is highly compatible with a broad range of functional groups in various solvent systems. This ligation reaction was applied to prepare glycan microarrays by immobilizing azide-linked glycans on the alkyne-derivatized surface (Fig. 8c) or by an inverse strategy (Fig. 8d).105–109 The chemoselective Staudinger reaction between azides and phosphanes was used to prepare microarrays (Fig. 8e).110 Unlike click chemistry, the Staudinger process does not need metal ions. Azide group-conjugated glycans were also immobilized on the surface by different chemistry (Fig. 8f).67,111 In this process, 4-azido-2,3,5,6-tetrafluorophenyl group-conjugated glycans were attached to the polymer monolayer surface by photochemistry. Because photolabile groups are conjugated to glycans at the anomeric position, they are site-specifically attached to the surface unlike methods shown in Fig. 6b.
The covalent immobilization methods described above require modified sugars, which are typically prepared by multistep processes. To circumvent the labor-intensive and time-consuming nature of these synthetic routes, one-step procedures for the modification of free glycans with proper functional groups have been exploited. For example, simple carbohydrates and oligosaccharides, after reaction with 2,6-diaminopyridine in the presence of sodium cyanoborohydride or N-methylaminooxy-containing bifunctional linkers, yield acyclic or cyclic adducts, respectively (Fig. 9a).36,37,112,113 The derivatized glycans appended by amine groups are then printed on the NHS ester-coated surface for covalent immobilization. A more facile method for the preparation of glycan microarrays is the site-specific, covalent attachment of free glycans irrespective of their size to the properly modified surface. An early example of this approach involves immobilization of free glycans, including simple carbohydrates, oligosaccharides and polysaccharides, on the aminooxy- or hydrazide-derivatized surface (Fig. 9b).114,115 The hydrazide-based immobilization procedure was found to be more efficient for protein binding than the method using aminooxy surfaces. NMR studies showed that reducing sugar moieties reacted with hydrazide groups to predominantly generate cyclic structures with β-configurations at their anomeric positions, but reacted with aminooxy groups to mainly produce acyclic adducts. As a consequence, proteins may interact with their cognate glycans with cyclic structures on the hydrazide surface more strongly than those with acyclic structures on the aminooxy surface. Later, other research groups also applied this immobilization method to prepare glycan microarrays.116–118
Fig. 9.
(a) Immobilization of glycans obtained from one-step reactions on the NHS ester-derivatized surface and (b) immobilization of unmodified glycans on the hydrazide or aminooxy-derivatized surface.
2.3. Presentation issues
Glycan presentation is another important factor to consider when designing and constructing glycan microarrays. Monovalent interactions between a GBP and a glycan are typically quite weak, with Kd values in the high micromolar to millimolar range. To overcome this, many GBPs contain two or more glycan binding sites or assemble into oligomers with multiple binding sites, which allow them to simultaneously bind two or more glycans to produce a multivalent complex. These multivalent interactions have much higher functional affinity (avidity) and can have enhanced or altered selectivity relative to the monovalent interaction. To achieve a multivalent complex, however, the spacing and orientation of the carbohydrate ligands must match the spacing and orientation of the binding sites of proteins. Therefore, features of glycan presentation, such as glycan density, have a great influence on molecular recognition of carbohydrates.
For a given GBP, the optimal glycan structure and presentation are not easily predicted. The microarray format is an ideal platform for examining many potential combinations of glycans and displays of those glycans, and a number of groups have developed methods to vary presentation on the surface of a microarray. For example, several strategies for varying glycan density have been published.18,63,80,99,119–123 These strategies provide variations in the average spacing between glycan chains, but do not precisely control presentation at a molecular level. Nevertheless, glycan density on the surface of a microarray can have a profound effect on the affinity and specificity of lectins and antibodies, and variations in glycan density can be useful for distinguishing different subpopulations of serum antibodies that recognize the same glycan structure (Fig. 10).99 Other methods to regulate glycan presentation have also been examined. For example, glycan microarrays are typically produced by printing monovalent carbohydrates onto a surface. An alternative involves printing multivalent glycoconjugates onto surfaces. Some examples include immobilizing natural glycoproteins,124 neoglycoproteins/neoglycopeptides (proteins or peptides with glycans covalently attached via a non-native linkage),16,19,66,96,125–130 glycodendrimers,118,131–133 multivalent display on DNA,134–136 glycoclusters,137 and glycopolymers (Fig. 10c).109,138 By modulating the spacing and orientation of glycan attachment sites on the multivalent scaffold, as well as altering the physical properties and overall architecture of the scaffold itself, there are many modes of presentation that can be attained. In addition, immobilization of multivalent conjugates provides unique opportunities to vary scaffold presentation on the microarray surface.96,101,139 One can also vary the context of the glycans. For example, many glycans are naturally found attached to serine/threonine or asparagine residues of proteins. While the glycans are important, the peptide to which they are attached can also contribute to recognition. Several groups have immobilized glycopeptides on arrays and examined the effects of peptide sequence on glycan recognition.126,139–153 Peptide sequence can have a significant influence on anti-glycan antibody binding.152 Finally, several groups have examined microarrays containing mixtures of glycans at each spot and found that, in some cases, combinations of two glycans can provide better binding than homogeneous glycans.154,155
Fig. 10.
Glycan presentation on the solid surface. Multivalent binding is critical for strong glycan–protein recognition events. To form a high avidity multivalent complex, the spacing and orientation of glycans on the surface must match the spacing and orientation of binding sites on a GBP. (a) A lectin with short spacing between binding sites may bind strongly to the high density of glycans on the surface. (b) A lectin with short spacing between binding sites may not bind strongly to the low density of glycans on the surface. However, an antibody with longer spacing between binding sites can bind well to glycans at either high or low density. (c) Glycan microarrays can be constructed by immobilizing multivalent glycoconjugates such as glycodendrimers, neoglycoproteins/neoglycopeptides and glycopolymers. Use of multivalent glycoconjugates provides unique opportunities to modulate glycan presentation.
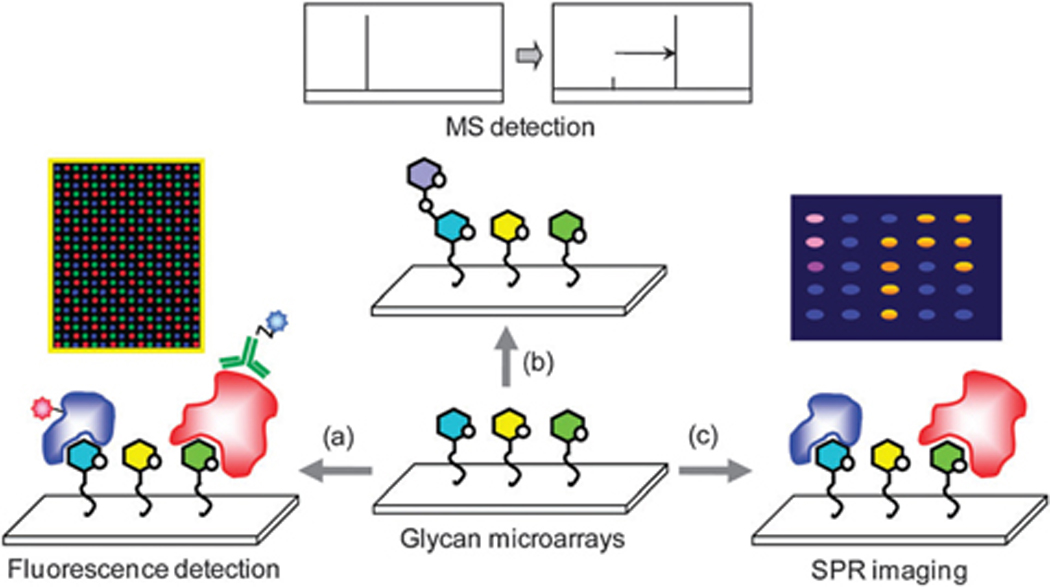
3. Detection methods
A variety of methods to detect binding events or enzymatic glycosylation taking place on glycan microarrays have been employed. The most common approach involves detection of fluorophore-labeled proteins directly or indirectly bound to glycans on the surface by using a fluorescence scanner (Fig. 11a). There are many variations of this approach; some examples include (1) direct fluorophore labeling of the GBP of interest, (2) use of a fluorophore-labeled secondary reagent that binds the GBP, and (3) use of a fluorophore-labeled secondary reagent that binds a tag (e.g. biotin, His tag) on the GBP. The advantage of this strategy is that there are many well-established systems and commercially available reagents for this approach. For example, biotinylated proteins can be readily monitored with fluorophore-labeled streptavidin, and anti-glycan antibodies can be readily detected with labeled secondary antibodies. In addition, microarray scanners with fluorescence detection are accessible in many laboratories due to their common use in DNA microarray experiments.
Fig. 11.
Detection methods of glycan microarrays. (a) Binding events of fluorophore-labeled proteins to glycans on microarrays can be monitored by using a fluorescence scanner. (b) Enzymatic reactions on microarrays can be detected using MS. (c) SPR imaging can be used as a label-free detection method for glycan microarrays.
While fluorescence detection is a versatile approach and has been used in the majority of glycan microarray studies, there are important limitations. First, modifications to GBPs, such as fluorophore-labeling, biotinylation and insertion of a His tag, can reduce activity or alter selectivity of binding.156 Second, fluorophore-labeled secondary reagents are not available for many proteins, especially newly discovered GBPs. Third, some secondary reagents can have glycan-binding properties of their own. For example, many polyclonal antibodies used to detect proteins contain carbohydrate-binding antibodies within the mixture. Therefore, one must ensure that proper controls are in place to correctly interpret results. Finally, most fluorophores are sensitive to light and prone to oxidative degradation. Therefore, signal intensity will decrease over time and can vary with experimental conditions. Due to these limitations, there has been considerable interest in developing alternative detection strategies, especially label-free methods.
One attractive alternative to fluorescence detection involves analysis by MS (Fig. 11b). This approach has primarily been used to monitor enzymatic reactions on slide surfaces that result in a change in mass (see Section 4.4).157–162 Surface plasmon resonance (SPR) imaging has also been used as a label-free detection method for glycan microarrays (Fig. 11c). SPR imaging offers real-time monitoring of binding events with the capacity to measure kinetics and thermodynamics of binding. A nice example comes from the work of de Boer and colleagues using a “natural glycan array”,130 an array containing glycans isolated directly from a natural source. The authors isolated N-linked glycans and glycolipids from the pathogen S. mansoni, constructed microarrays immobilized by these glycans, and then used an SPR imaging method to profile binding of human serum antibodies and serum lectins of infected and non-infected individuals. SPR imaging has also been used to profile plant lectins and identify potential inhibitors of the biological toxin ricin.163 While this approach has a number of advantages, the technology is somewhat limited in the total number of array components that can be accommodated.
In addition to these methods, several methods have been developed as alternative detection strategies. These include detection of radioactivity,164,165 oblique-incidence reflectivity difference (OI-RD) microscopy,166 electrochemoluminescence,167 complementary metal oxide semiconductor-based electric signal readout,168 and cantilever-based detection.169
4. Applications
4.1. Rapid analysis of glycan–protein interactions
Since their advent in 2002,12–16 applications of glycan microarrays to biological and biomedical research have been rapidly expanded. Most extensive use of this technology has been in the high-throughput analysis of the binding properties of proteins (Fig. 12). Basically, all alleged GBPs can be assessed by using glycan microarrays combined with any detection system, especially, a fluorescence detection method. In general, supplementary assays are required to obtain knowledge about key characteristic features of the samples before and after screening to assure sample activity and validation of the detected glycan binding.
Fig. 12.
Application of glycan microarrays for rapid analysis of glycan–protein interactions.
4.1.1. LECTINS.
Initially discovered in plants, lectins are now identified in most microbes and animals.4 Through well-defined carbohydrate-recognizing domains (CRD), lectins bind to carbohydrate structures without effectuating changes in glycan structures. Plant lectins are important tools for biological research and diagnosis and their binding specificities have long been extensively investigated by using conventional solution-based methods, in particular, the hemagglutination inhibition assay.170,171 Glycan microarrays probed with a wide range of plant lectins have provided important information about the detailed binding patterns and identified new glycan– lectin interactions (see collections of lectin binding specificity data obtained at website of CFG, www.functionalglycomics.org).18,31,71,125
Mammalian lectins, however, are more fascinating since they have important biological functions. Mammalian lectins interact with glycans in a wide range of essential biological processes and their specific binding to glycans can be useful in the development of innovative therapeutics. Carbohydrate microarrays have been used to estimate the binding patterns of several mammalian lectins, e.g. analysis of binding preferences of members of the C-type lectins (Ca2+-dependent lectins). Examples are DC-SIGN and DC-SIGNR.60,172 The first is a receptor primarily expressed on dendritic cells and encompasses innate immunity and pathogenesis of viruses. The latter is a receptor found on epithelial cells probably engaged in pathogen infection. Both receptors recognize high mannose oligosaccharides, but only DC-SIGN can interact with the fucose-containing sugars such as Lea, Leb, Lex and Ley, accompanying the idea that DC-SIGN can bind to pathogens containing high mannose glycans such as human immunodeficiency virus (HIV), hepatitis C virus (HCV), Ebola virus, M. tuberculosis and leishmania parasites. Other C-type lectins profiled by microarrays include langerin, a receptor on islets of Langerhans involved in innate and acquired immunity, the scavenger receptor C-type lectin (SRCL) and Dectin-1. The binding properties of langerin determined by glycan microarrays show binding to sulfated Lex sequences,172 however, only rare interactions with mannosylated glycans in contrast to DC-SIGN and DC-SIGNR. SRCL, an endothelial receptor which is also involved in innate immunity, binds to Lex-containing glycans lacking sialic acid and sulfate moieties.173 It has further been established that the primary binding site of SRCL is a fucose moiety. Dectin-1 and 2 are receptors expressed on leukocytes. Glycan microarrays have elucidated its binding to 1,3-linked glucose oligomers and interaction with high-mannose structures such as Man9GlcNAc2 > Man8GlcNAc2 and to a lesser degree Man7GlcNAc2.53,174
Another family of mammalian lectins, the Siglecs, is a subclass of the immunoglobulin gene superfamily. The binding preferences of Siglec-2, Siglec-F and Siglec-8 among others have been evaluated by glycan microarrays.31 Siglec-2 preferentially interacts with Neu5Acα2,6Galβ1,4GlcNAc epitope, mouse Siglec-F binds to the 6′-sulfo-sialyl Lex and human Siglec-8 interacts with 6′-sulfo-sialyl Lex.31,61,175
A third type of animal lectins is the β-galactoside-binding galectin (Gal). These lectins have also been evaluated by using microarrays and found to recognize the terminal and internal galactose moieties.31,176–178 Modifications of the basic LacNAc core on glycan recognition by Gal-1, Gal-2, and Gal-3 can either enhance or preclude recognition, suggesting that unique subsites exist with each CRD.86
4.1.2. MONOCLONAL ANTIBODIES.
Monoclonal antibodies have stronger binding affinities to glycans than lectins, and glycan microarrays have been applied to profiling of binding specificities of antibodies. The standard approach to screen mouse monoclonal antibodies is initiated by diluting the antibody to an appropriate working concentration (1–50 μg mL−1) and detecting with a fluorescently labeled anti-mouse antibody against the desired immunoglobulin isotype (e.g. IgG or IgM). The repertoire of glycans on the array will interact with the antibody if it contains a carbohydrate specific epitope. In 1979, Young et al. produced the first anti-carbohydrate mouse monoclonal antibody.179 Over the years multiple monoclonal antibodies to glycan determinants have been produced with well-defined validated specificities.180 The recent improvements of glycan microarray technology in the glycomic area provide new insight and have led to re-assessment of several monoclonal antibodies to re-confirm their specificities, fine-tuning of sub-specificities or to rule out cross-reactivity.181
Specificities of monoclonal antibodies to tumor-associated oligosaccharides, such as the Tn-, and STn-antigens, were found to have a more complex nature.126,152,181–183 For example, antibodies of IgM-type do react with corresponding spacered Tn displayed on the microarray, whereas antibodies of IgG-type frequently require parts of the underlying peptide backbone for complete epitope and/or correct glycan presentation.
4.1.3. OTHER PROTEINS.
A class of polysaccharides consisting of disaccharide repeating units is the glycoaminoglycans (GAG). Examples of GAGs are chondroitin sulfate, heparin/heparan sulfate, keratan sulfate, dermatan sulfate and hyaluronan. They are involved in homeostasis, cancer metastasis, cell growth, cell migration, development and other physiological processes through interactions with proteins such as growth factors, proteases, cytokines, chemokines, and cell adhesion molecules.184 It has been demonstrated, through glycan microarrays containing synthetic chondroitin sulfate and heparin oligosaccharides, that specific sulfation motifs in glycans are essential for their binding to proteins.91,185–187
4.1.4. IDENTIFICATION OF NEW GLYCAN-BINDING PROTEINS.
Glycan microarrays provide a powerful platform for identifying new GBPs and generating hypotheses regarding the function of those proteins. A nice illustration of this application comes from the work of Feizi and colleagues on a protein called malectin.188 Initial studies on this protein demonstrated that it was expressed in a wide variety of tissues and conserved across many species,189 suggesting that it plays a fundamental role in biology; however, its biological function was unknown. Glycan microarray analysis revealed strong and selective binding to a di-glucosyl-N-glycan (Glc2Man7). In addition to providing the key evidence that this protein has the glycan binding property, the results also provide crucial information for unraveling malectin’s biological function. The target glycan, Glc2Man7, is an intermediate in the biosynthesis of N-glycans, which suggested a role for malectin in the production and quality control of glycoproteins in the endoplasmic reticulum (ER). Based on this as well as additional studies, malectin is now thought to inhibit secretion of defective and/or misfolded proteins from the ER.
4.1.5. VIRAL AND BACTERIAL PROTEINS.
As glycans decorate most of the mammalian cell surface, many microbial pathogens do bind to cell surface glycans via their glycan receptors.190 Routinely, glycan microarrays are now also used to analyze the binding properties of viral and bacterial proteins. For example, human influenza viruses bind to the Neu5Acα2,6Gal residues on epithelial cells in the lungs and upper respiratory tract. Conversely, avian influenza viruses bind exclusively to Neu5Acα2,3Gal residues on intestinal epithelial cells. Microarrays have provided detailed information about the profiles of the glycan specificities of numerous kinds of influenza viruses.31,191–194 Glycan microarray analysis of two other viruses, adenovirus 37 and simian virus, shows that they bind to gangliosides, GM3 and GD1a, respectively.195,196 In addition, glycan microarrays were employed to evaluate binding properties of bacterial lectins. According to the results of glycan microarray experiments, the bacterial P. aeruginosa lectin I displays affinity for α-galactosylated glycans197,198 and B. cenocepacia lectin to high mannose glycans.199
4.2. Detection of viruses and whole cells
In addition to defined lectins and antibodies, glycan microarrays have been used to probe binding of viruses and whole cells (Fig. 13).200 While analysis can be more complicated due to the presence of more than one GBP, the GBPs are present in a natural context with native spacing and orientation of binding sites, which is not always true for lectins, antibodies and their subunits. Glycan microarrays have been used extensively to study binding properties of various influenza and parainfluenza strains.201–205 These studies have shed new light on the differences in binding specificities of various pandemic strains versus seasonal viruses and viruses that infect animals. In addition, glycan microarrays have been exploited to profile binding of adenoviruses,206,207 minute viruses,208 and simian virus 40.195
Fig. 13.
Application of glycan microarrays for detection of viruses and whole cells.
The ability to rapidly evaluate binding properties of whole cells has many potential applications and advantages. For example, one can probe binding properties without prior knowledge of the key GBP and one can monitor the cellular effects of binding. Although whole cells have been profiled on glycan microarrays previously, this approach is more challenging and there are relatively few examples in the literature, especially as compared to studies on GBPs. The majority of examples have focused on detecting pathogenic bacteria,84,114,115,128,166,209 but some examples of eukaryotic cells have also been published.83,210 Nevertheless, some optimization of experimental methods is needed for this approach to achieve more widespread use.
4.3. Determination of IC50 values and apparent dissociation constants
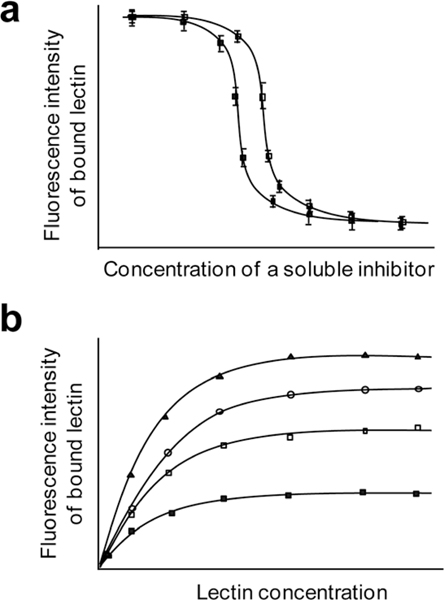
Results obtained from profiling of glycan–protein interactions using glycan microarrays normally provide information on qualitative binding properties of GBPs. This technology has been employed also to measure quantitative binding affinities of proteins to sugars. In initial studies in this area, IC50 values of soluble inhibitors for protein binding to glycans attached to the surface were determined. For this purpose, a series of pre-incubated mixtures of the fluorophore-labeled protein and an inhibitor are applied to microarrays containing specific glycans. The IC50 values of soluble inhibitors are then calculated by measuring fluorescence intensities of bound proteins on the microarrays after washing (Fig. 14a).18,71
Fig. 14.
Determination of (a) IC50 values of soluble inhibitors and (b) apparent dissociation constants (Kd values) for glycan–protein interactions using glycan microarrays.
More recently, glycan microarrays were applied to determine a number of Kd values (dissociation constants) between proteins and surface-immobilized glycans from a single experiment. In this procedure, microarrays containing a number of glycans are probed with various concentrations of labeled proteins.94–96,120,211 Apparent dissociation constants for surface-bound sugars with proteins are determined by measuring fluorescence intensities of bound proteins on the microarrays (Fig. 14b). Under equilibrium conditions, apparent Kd values are calculated by using the equation
where Flmax is the maximum fluorescence intensity, Fl is the mean fluorescence intensity and [P]o is the initial concentration of protein. It was found that the apparent Kd values obtained from these experiments were similar to those obtained in SPR experiments.94
4.4. Profiling of substrate specificity of enzymes
As described in Section 2.1, glycosyltransferases are useful biocatalysts to prepare oligosaccharides since extensive protection and deprotection steps are unnecessary. In order to efficiently use glycosyltransferases as synthetic catalysts, detailed knowledge about their acceptor specificities is important. Several methods for assessment of glycosyltransferase acceptor specificities have been established including radiochemical, spectrophotometric, immunological and chromatographic assays. Glycan microarrays can also be of great service to determine the extent of acceptor substrate utility of these enzymes. By using large glycan libraries displayed on microarray surfaces, enzyme specificities can be determined in more detail to confirm known specificities or identify new reactivities and thereby broaden their synthetic utilities. So far, only a few examples of enzymatic reactions taking place on glycan microarrays have been described in the literature. Enzymatic transfer can be measured by detection of the product either by lectin or antibody detection.71,94,95,212,213
Alternatively, substrate specificities of the 1,2-fucosyltransferases, which are involved in xyloglucan biosynthesis in the plant cell wall, were evaluated by monitoring [14C]fucose incorporated into xyloglucans attached to array surfaces.164 This method could detect as low as 45 cpm per spot (cpm = counts per minute). Blixt and coworkers used biotinylated cytidine-5′-monophospho-N-acetylneuraminic acid as a donor substrate for acceptor specificity screening of various recombinant sialyltransferases.214 After enzymatic reactions on the microarrays, biotinylated glycans were detected using fluorescein-conjugated streptavidin to generate a specificity profile for each enzyme. This study confirmed previously known specificities of enzymes and revealed additional specificity information to expand synthesis of glycan libraries.
Although there are tens of thousands of proteins tentatively assigned as glycosyltransferases based on sequence homology, only a small fraction has been characterized and validated. To identify new glycosyltransferases and characterize their substrate specificities, the Mrksich and Wang groups coupled glycan array technology to semi-automated SELDI-MS.157 Combinations of putative enzymes and glycosyl donors were incubated on arrays of glycosyl acceptors to profile nearly 60 000 potential glycosyltransferase reactions. Transfer of a monosaccharide residue to the surface bound acceptor results in a change in molecular weight, which was detected by MS. Using this approach, 4 previously unknown glycosyltransferases were identified and their substrate specificities were characterized. The polypeptide GalNAc transferases are key enzymes for initiation of O-glycosylation and thus knowledge of their substrate specificities is important.215 To address their substrate specificities, peptide and glycopeptide microarrays have been used.153,216,217 These enzymes have distinct but significantly overlapping peptide specificities.
4.5. Serodiagnosis and biomarkers
During a lifetime naturally high levels of immunoglobulins can remain practically unchanged with a conserved repertoire in serum and only limited data on human anti-glycan antibodies have been presented.218 Glycan microarrays have recently been explored as a serological tool.31,100,219,220 However, interpretation of autoantibody profiles from serum is complicated by the vast complexity of the serum composition.221 To date only a few methodological studies have been carried out with mammalian glycan microarrays and healthy blood donors, showing considerable binding reactivity to a wide range of carbohydrates (Fig. 15).31,100,219
Fig. 15.
Glycan binding properties of human intravenous immunoglobulin G (IgG). The antibody binds to a variety of glycans on the microarrays.
Aberrant glycosylation is one of the key features of cancer and, thus, identifying differences in cell surface glycan expression may be useful for diagnosing cancer.4 However, due to the complex serum reactivity profiles, diagnostic utility of glycan microarrays is limited.222–224 It is speculated that the broad reactivity may be caused by poly specific antibody reactivities, which make use of glycan microarrays complicated in serodiagnostics, and thus further sample processing before analysis is needed.221 In contrast, cancer-associated glycans displayed as glycopeptides on microarrays have shown some promise in detection of immunogenic glycopeptide epitopes (Fig. 16).150,153,225 In addition, the same strategy was applied to identify type-specific glycopeptide epitopes on virus envelope proteins expressing host-derived glycans.226
Fig. 16.
Detection of aberrant glycans displayed on proteins.
Microarrays have also been used for serodiagnosis of pathogen infections. For example, detection of the GalNAcβ1–4(Fucα1–3)-GlcNAc (LDNF) glycan antigen in trichinellosis infections,227 detection of IgA antibodies specific to synthetic PS-II hexasaccharide hapten in the stool of patients infected with C. difficile recognized228 and serum reactivities to type specific oligosaccharide antigens from Salmonella enterica sv., S. Paratyphi, S. Typhimurium,229 and S. mansoni-infected patients.130
4.6. Profiling of immune response
Glycan microarrays are advantageous tools for profiling immune responses induced by vaccines, immunotherapies, and organ or stem cell transplants.69,101,230–232 The diverse collection of antigens present on an array permits detection of a wide range of antibody populations and is particularly useful when evaluating responses to vaccines that contain a complex assortment of potential glycan antigens. By studying many possible responses in parallel, one increases the likelihood of identifying particular responses that contribute to efficacy, side effects or complications. Glycan microarrays have been exploited in a number of studies for the analysis of responses to pathogens. Some examples include monitoring anti-glycan immune responses to anthrax,69 SARS-coronavirus,233 P. falciparum sporozoites,228,234 and capsular polysaccharide from C. difficile.228 Glycan microarrays have also provided unique insight for the development and analysis of HIV vaccines. In addition to characterizing binding properties of broadly neutralizing monoclonal antibodies such as 2G12, glycan microarrays have been used to monitor immune responses induced by high mannose oligosaccharides101 and a mutant strain of S. cerevisiea to determine if antibodies with 2G12-like specificity are produced.235
Glycan microarrays have also been used to profile response to cancer vaccines. For example, Blixt and coworkers used glycan microarrays to evaluate antibody responses induced by a MUC1 glycopeptide vaccine.150,153 They demonstrated that antibodies to Tn-MUC1 were only generated in vaccinated patients and that these antibodies were specific for the glycopeptide. Additionally, the Gildersleeve group used glycan microarrays to profile immune responses induced by PROSTVAC-VF,101 a poxvirus-based vaccine currently in phase III clinical trials for the treatment of advanced prostate cancer.230–232 Increases in antibody levels to the Forssman antigen and the blood group A antigen were discovered in many patients.
Carbohydrates are also key antigens in organ transplantation and blood transfusions. The carbohydrates displayed on cells can vary from one person to another, and carbohydrates that are not expressed in an individual typically trigger a robust immune response. For example, the blood group antigens (A, B, and H) that determine the major blood types (A, B, and O, respectively) are glycans, and mismatching of blood group antigen expression between donor and recipient can cause serious complications in transplants and transfusions. Due to the shortage of human organs, there has been considerable interest in using animal organs (xenotransplantation), but immune responses to non-human glycans, such as the α-Gal antigen, present a major barrier. To more fully understand immune responses in xenotransplants and identify the antigen that can trigger host versus graft responses, Blixt and colleagues profiled anti-glycan antibody responses in patients transplanted with porcine fetal pig islet-like cell clusters. Using a glycan microarray, they observed a significant increase of antibodies to α-Gal after transplantation, as well as antibodies to Galα1, 3Lex and structures terminated with Neu5Gc. Glycan microarrays have also recently been used to determine cross-reactivities of a pathogen-specific maternal vaccine candidate elicited to Streptococal capsular polysaccharide group B type III (CPSIII), structurally similar to several mammalian cell surface glycans, and could be of potential importance for vaccine developments.236
4. Conclusions
Over the last decade, glycan microarray technology has emerged as a powerful and transformative tool for glycobiology. New glycan-binding proteins are now routinely screened for binding to hundreds of carbohydrates and specificity profiles for countless anti-glycan antibodies and lectins are now available. Glycan microarrays have also been used extensively to profile anti-glycan antibody populations in human sera, and these studies provide a much more comprehensive understanding of anti-glycan immunity as well as many new diagnostic and prognostic biomarkers for a range of diseases and conditions. Nonetheless, a number of barriers must be overcome to reach the full potential of this technology. First, existing microarrays contain only a small fraction of the glycan diversity found in nature. Expansion of glycan libraries through improved chemical and/or enzymatic synthesis as well as better isolation methods will be critical for enhancing diversity on glycan arrays. Second, new and better methods for varying glycan presentation are needed to improve the performance and capabilities of this technology. Third, better bioinformatics tools, especially those tailored to the unique attributes of glycans are needed to more fully extract valuable information from glycan array data. Finally, glycan microarray experiments are still primarily carried out in a handful of laboratories around the world. As this technology becomes more accessible directly to the broader community, new and unanticipated applications are likely to emerge.
Acknowledgements
This work was supported by grants from the National Creative Research Initiative (IS), WCU (IS), the intramural research program of the NIH (JG), NCI (JG), Benzon Foundation (OB), Danish Agency for Science (OB), EU FP7/2007–2013-EuroGlycoArrays 215536 (OB), EU FP7-GlycoBioM (OB), Danish National Research Foundation (OB) and University of Copenhagen Programme of Excellence (OB). Maya Bonde Haaland is acknowledged for linguistic help.
Biography

Sungjin Park received her BS degree in chemistry from the Hankuk University of Foreign Studies in 1998. Her MS and PhD degrees were awarded in chemistry in 2001 and 2009 at Yonsei University under the guidance of Professor Injae Shin. She had worked as a research fellow for 2 years in the Max Planck Institute of Colloids and Interfaces, Department of Biomolecular Systems (supervisor: Peter Seeberger). In 2012, she joined Center for Biofunctional Molecules (supervisor: Injae Shin) at Yonsei University as a research professor. Her research interests include functional studies of glycans using synthetic carbohydrates and glycan microarrays.

Jeffrey Gildersleeve received his BS degree in biology in 1993 from University of California at San Diego. He obtained his PhD degree in chemistry at Princeton University (supervisor: Professor Daniel Kahne), and completed postdoctoral training with Professor Peter Schultz at The Scripps Research Institute. In 2003, he began his independent career at the National Cancer Institute (NCI). He is currently a Senior Investigator and Head of Chemical Glycobiology Section in the Chemical Biology Laboratory of NCI. The Gildersleeve group uses chemical approaches and glycan microarrays to study the roles of anticarbohydrate immune responses in the development, progression, and treatment of cancer and HIV.

Ola Blixt received his BS degree in chemistry in 1994 from University of Umeå, Sweden. He obtained his PhD degree in organic chemistry at Swedish University of Agricultural Sciences (supervisor: Professor Thomas Norberg), and completed postdoctoral training with Professor James Paulson at The Scripps Research Institute. Between 2001 and 2007, he served as the Director of the Glycan microarray core facility at Consortium for Functional Glycomics. Currently, he is an Associate Professor at the Copenhagen Center for Glycomics at University of Copenhagen. The Blixt group develops glycan and glycopeptide microarrays and bead screening technologies to explore lectins, microbial interactions and immune responses to infections and human diseases.

Injae Shin received his BS degree in chemistry in 1985 and MS degree in 1987 from Seoul National University. His PhD studies were carried out at University of Minnesota with Professor Hung-wen Liu (1991–1995). After completing postdoctoral research at University of California at Berkeley with Professor Peter Schultz (1995–1998), he started his independent career as an Assistant Professor of Chemistry at Yonsei University in 1998 where he became an Associate Professor in 2001 and a Professor in 2006. His research interests include development of biofunctional molecules that can be used for biological and biomedical research and functional studies of glycans using chemical tools including glycan microarrays.
Footnotes
Part of the carbohydrate chemistry themed issue.
Notes and references
- 1.Varki A, Glycobiology, 1993, 3, 97–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bertozzi CR and Kiessling LL, Science, 2001, 291, 2357–2364. [DOI] [PubMed] [Google Scholar]
- 3.Park S, Lee M-R and Shin I, Chem. Soc. Rev, 2008, 37, 1579–1591. [DOI] [PubMed] [Google Scholar]
- 4.Essentials of Glycobiology, ed. Varki A, Cummings R, Esko J, Freeze H, Stanley P, Bertozzi CR, Hart G and Etzler ME, Cold Spring Harbor Laboratory Press, 2nd edn, 2009. [PubMed] [Google Scholar]
- 5.Crocker PR, Curr. Opin. Pharmacol, 2005, 5, 431–437. [DOI] [PubMed] [Google Scholar]
- 6.van Kooyk Y and Rabinovich GA, Nat. Immunol, 2008, 9, 593–601. [DOI] [PubMed] [Google Scholar]
- 7.Crocker PR, Paulson JC and Varki A, Nat. Rev. Immunol, 2007, 7, 255–266. [DOI] [PubMed] [Google Scholar]
- 8.Fuster MM and Esko JD, Nat. Rev. Cancer, 2005, 5, 526–542. [DOI] [PubMed] [Google Scholar]
- 9.Simanek EE, McGarvey GJ, Jablonowski JA and Wong C-H, Chem. Rev, 1998, 98, 833–862. [DOI] [PubMed] [Google Scholar]
- 10.Smith AE and Helenius A, Science, 2004, 304, 237–242. [DOI] [PubMed] [Google Scholar]
- 11.Imberty A and Varrot A, Curr. Opin. Struct. Biol, 2008, 18, 567–576. [DOI] [PubMed] [Google Scholar]
- 12.Tang PW, Gooi HC, Hardy M, Lee YC and Feizi T, Biochem. Biophys. Res. Commun, 1985, 132, 474–480. [DOI] [PubMed] [Google Scholar]
- 13.Liang R, Yan L, Loebach J, Ge M, Uozumi Y, Sekanina K, Horan N, Gildersleeve J, Thompson C, Smith A, Biswas K, Still WC and Kahne D, Science, 1996, 274, 1520–1522. [DOI] [PubMed] [Google Scholar]
- 14.Roy R, Curr. Opin. Struct. Biol, 1996, 6, 692–702. [DOI] [PubMed] [Google Scholar]
- 15.Park S and Shin I, Angew. Chem., Int. Ed, 2002, 41, 3180–3182. [DOI] [PubMed] [Google Scholar]
- 16.Wang D, Liu S, Trummer BJ, Deng C and Wang A, Nat. Biotechnol, 2002, 20, 275–281. [DOI] [PubMed] [Google Scholar]
- 17.Fukui S, Feizi T, Galustian C, Lawson AM and Chai W, Nat. Biotechnol, 2002, 20, 1011–1017. [DOI] [PubMed] [Google Scholar]
- 18.Houseman BT and Mrksich M, Chem. Biol, 2002, 9, 443–454. [DOI] [PubMed] [Google Scholar]
- 19.Willats WGT, Rasmussen SE, Kristensen T, Mikkelsen JD and Knox JP, Proteomics, 2002, 2, 1666–1671. [DOI] [PubMed] [Google Scholar]
- 20.Fazio F, Bryan MC, Blixt O, Paulson JC and Wong C-H, J. Am. Chem. Soc, 2002, 124, 14397–14402. [DOI] [PubMed] [Google Scholar]
- 21.Paulson JC, Blixt O and Collins BE, Nat. Chem. Biol, 2006, 2, 238–248. [DOI] [PubMed] [Google Scholar]
- 22.Feizi T and Chai W, Nat. Rev. Mol. Cell Biol, 2004, 5, 582–588. [DOI] [PubMed] [Google Scholar]
- 23.Park S, Lee M-R and Shin I, Chem. Commun, 2008, 4389–4399. [DOI] [PubMed] [Google Scholar]
- 24.Horlacher T and Seeberger PH, Chem. Soc. Rev, 2008, 37, 1414–1422. [DOI] [PubMed] [Google Scholar]
- 25.Oyelaran O and Gildersleeve JC, Curr. Opin. Chem. Biol, 2009, 13, 406–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu Y, Palma AS and Feizi T, Biol. Chem, 2009, 390, 647–656. [DOI] [PubMed] [Google Scholar]
- 27.Rillahan CD and Paulson JC, Annu. Rev. Biochem, 2011, 80, 797–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shin I, Park S and Lee M.-r., Chem.–Eur. J, 2005, 11, 2894–2901. [DOI] [PubMed] [Google Scholar]
- 29.Shin I, Cho JW and Boo DW, Comb. Chem. High Throughput Screening, 2004, 7, 565–574. [DOI] [PubMed] [Google Scholar]
- 30.Shin I, Tae J and Park S, Curr. Chem. Biol, 2007, 1, 187–199. [Google Scholar]
- 31.Blixt O, Head S, Mondala T, Scanlan C, Huflejt ME, Alvarez R, Bryan MC, Fazio F, Calarese D, Stevens J, Razi N, Stevens DJ, Skehel JJ, van Die I, Burton DR, Wilson IA, Cummings R, Bovin N, Wong C-H and Paulson JC, Proc. Natl. Acad. Sci. U. S. A, 2004, 101, 17033–17038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Freeze HH, Nat. Rev. Genet, 2006, 7, 537–551. [DOI] [PubMed] [Google Scholar]
- 33.Drickamer K and Taylor M, Genome Biol, 2002, 3, 1031–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Culf AS, Cuperlovic-Culf M and Ouellette RJ, OMICS, 2006, 10, 289–310. [DOI] [PubMed] [Google Scholar]
- 35.Larsen K, Thygesen MB, Guillaumie F, Willats WG and Jensen KJ, Carbohydr. Res, 2006, 341, 1209–1234. [DOI] [PubMed] [Google Scholar]
- 36.Seo JH, Kim CS, Hwang BH and Cha HJ, Nanotechnology, 2010, 21, 215101–215108. [DOI] [PubMed] [Google Scholar]
- 37.Bohorov O, Andersson-Sand H, Hoffmann J and Blixt O, Glycobiology, 2006, 16, 21C–27C. [DOI] [PubMed] [Google Scholar]
- 38.Liu Y, Chai W, Childs RA and Feizi T, Methods Enzymol, 2006, 415, 326–340. [DOI] [PubMed] [Google Scholar]
- 39.Song X, Lasanajak Y, Xia B, Smith DF and Cummings RD, ACS Chem. Biol, 2009, 4, 741–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nicolaou KC and Mitchell HJ, Angew. Chem., Int. Ed, 2001, 40, 1576–1624. [PubMed] [Google Scholar]
- 41.Garegg PJ, Adv. Carbohydr. Chem. Biochem, 2004, 59, 69–134. [DOI] [PubMed] [Google Scholar]
- 42.Seeberger PH, Chem. Soc. Rev, 2008, 37, 19–28. [DOI] [PubMed] [Google Scholar]
- 43.Tanaka H, Adachi M and Takahashi T, Chem.–Eur. J, 2005, 11, 849–862. [DOI] [PubMed] [Google Scholar]
- 44.Ye XS and Wong CH, J. Org. Chem, 2000, 65, 2410–2431. [DOI] [PubMed] [Google Scholar]
- 45.Palcic MM, Curr. Opin. Chem. Biol, 2011, 15, 226–233. [DOI] [PubMed] [Google Scholar]
- 46.Blixt O and Razi N, Methods Enzymol, 2006, 415, 137–153. [DOI] [PubMed] [Google Scholar]
- 47.Blixt O, Collins BE, van den Nieuwenhof IM, Crocker PR and Paulson JC, J. Biol. Chem, 2003, 278, 31007–31019. [DOI] [PubMed] [Google Scholar]
- 48.Blixt O, Vasiliu D, Allin K, Jacobsen N, Warnock D, Razi N, Paulson JC, Bernatchez S, Gilbert M and Wakarchuk W, Carbohydr. Res, 2005, 340, 1963–1972. [DOI] [PubMed] [Google Scholar]
- 49.Breton C, Mucha J and Jeanneau C, Biochimie, 2001, 83, 713–718. [DOI] [PubMed] [Google Scholar]
- 50.Shipp EL and Hsieh-Wilson LC, Chem. Biol, 2007, 14, 195–208. [DOI] [PubMed] [Google Scholar]
- 51.Rogers CJ, Clark PM, Tully SE, Abrol R, Garcia KC, Goddard WA 3rd. and Hsieh-Wilson C, Proc. Natl. Acad. Sci. U. S. A, 2011, 108, 9747–9752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Carion O, Lefebvre J, Dubreucq G, Dahri-Correia L, Correia J and Melnyk O, ChemBioChem, 2006, 7, 817–826. [DOI] [PubMed] [Google Scholar]
- 53.Palma AS, Feizi T, Zhang Y, Stoll MS, Lawson AM, DíazRodríguez E, Campanero-Rhodes MA, Costa J, Gordon S, Brown GD and Chai W, J. Biol. Chem, 2006, 281, 5771–5779. [DOI] [PubMed] [Google Scholar]
- 54.Ko K-S, Jaipuri FA and Pohl NL, J. Am. Chem. Soc, 2005, 127, 13162–13163. [DOI] [PubMed] [Google Scholar]
- 55.Mamidyala SK, Ko K-S, Jaipuri FA, Park G and Pohl NL, J. Fluorine Chem, 2006, 127, 571–579. [Google Scholar]
- 56.Chen G-S and Pohl NL, Org. Lett, 2008, 10, 785–788. [DOI] [PubMed] [Google Scholar]
- 57.Chang S-H, Han J-L, Tseng SY, Lee H-Y, Lin C-W, Lin Y-C, Jeng W-Y, Wang AHJ, Wu C-Y and Wong C-H, J. Am. Chem. Soc, 2010, 132, 13371–13380. [DOI] [PubMed] [Google Scholar]
- 58.Li Y, Arigi E, Eichert H and Levery SB, J. Mass Spectrom, 2010, 45, 504–519. [DOI] [PubMed] [Google Scholar]
- 59.Galanina OE, Mecklenburg M, Nifantiev NE, Pazynina GV and Bovin NV, Lab Chip, 2003, 3, 260–265. [DOI] [PubMed] [Google Scholar]
- 60.Guo Y, Feinberg H, Conroy E, Mitchell DA, Alvarez R, Blixt O, Taylor ME, Weis WI and Drickamer K, Nat. Struct. Mol. Biol, 2004, 11, 591–598. [DOI] [PubMed] [Google Scholar]
- 61.Bochner BS, Alvarez RA, Mehta P, Bovin NV, Blixt O, White JR and Schnaar RL, J. Biol. Chem, 2005, 280, 4307–4312. [DOI] [PubMed] [Google Scholar]
- 62.Godula K and Bertozzi CR, J. Am. Chem. Soc, 2010, 132, 9963–9965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chevolot Y, Bouillon C, Vidal S, Morvan F, Meyer A, Cloarec J-P, Jochum A, Praly J-P, Vasseur J-J and Souteyrand E, Angew. Chem., Int. Ed, 2007, 46, 2398–2402. [DOI] [PubMed] [Google Scholar]
- 64.Kanter JL, Narayana S, Ho PP, Catz I, Warren KG, Sobel RA, Steinman L and Robinson WH, Nat. Med, 2006, 12, 138–143. [DOI] [PubMed] [Google Scholar]
- 65.Yamazaki V, Sirenko O, Schafer RJ, Nguyen L, Gutsmann T, Brade L and Groves JT, BMC Biotechnol, 2005, 5, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Angeloni S, Ridet JL, Kusy N, Gao H, Crevoisier F, Guinchard S, Kochhar S, Sigrist H and Sprenger N, Glycobiology, 2004, 15, 31–41. [DOI] [PubMed] [Google Scholar]
- 67.Pei Z, Yu H, Theurer M, Waldén A, Nilsson P, Yan M and Ramström O, ChemBioChem, 2007, 8, 166–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Carroll GT, Wang D, Turro NJ and Koberstein JT, Langmuir, 2006, 22, 2899–2905. [DOI] [PubMed] [Google Scholar]
- 69.Wang D, Carroll GT, Turro NJ, Koberstein JT, Kováč P, Saksena R, Adamo R, Herzenberg LA, Herzenberg LA and Steinman L, Proteomics, 2007, 7, 180–184. [DOI] [PubMed] [Google Scholar]
- 70.Hsiao H-Y, Chen M-L, Wu H-T, Huang L-D, Chien W-T, Yu C-C, Jan F-D, Sahabuddin S, Chang T-C and Lin C-C, Chem. Commun, 2011, 47, 1187–1189. [DOI] [PubMed] [Google Scholar]
- 71.Park S, Lee M-R, Pyo S-J and Shin I, J. Am. Chem. Soc, 2004, 126, 4812–4819. [DOI] [PubMed] [Google Scholar]
- 72.Shin I, Zamfir AD and Ye B, Methods Mol. Biol, 2008, 441, 19–39. [DOI] [PubMed] [Google Scholar]
- 73.Houseman BT, Gawalt ES and Mrksich M, Langmuir, 2003, 19, 1522–1531. [Google Scholar]
- 74.Adams EW, Ratner DM, Bokesch HR, McMahon JB, O’Keefe BR and Seeberger PH, Chem. Biol, 2004, 11, 875–881. [DOI] [PubMed] [Google Scholar]
- 75.Ratner DM, Adams EW, Su J, O’Keefe BR, Mrksich M and Seeberger PH, ChemBioChem, 2004, 5, 379–382. [DOI] [PubMed] [Google Scholar]
- 76.Brun MA, Disney MD and Seeberger PH, ChemBioChem, 2006, 7, 421–424. [DOI] [PubMed] [Google Scholar]
- 77.Ratner DM and Seeberger PH, Curr. Pharm. Des, 2007, 13, 173–183. [DOI] [PubMed] [Google Scholar]
- 78.Seo JH, Adachi K, Lee BK, Kang DG, Kim YK, Kim KR, Lee HY, Kawai T and Cha HJ, Bioconjugate Chem, 2007, 18, 2197–2201. [DOI] [PubMed] [Google Scholar]
- 79.Harris LG, Schofield WCE, Doores KJ, Davis BG and Badyal JPS, J. Am. Chem. Soc, 2009, 131, 7755–7761. [DOI] [PubMed] [Google Scholar]
- 80.Smith EA, Thomas WD, Kiessling LL and Corn RM, J. Am. Chem. Soc, 2003, 125, 6140–6148. [DOI] [PubMed] [Google Scholar]
- 81.Beckmann HSG, Niederwieser A, Wiessler M and Wittmann V, Chem.–Eur. J, 2012, 18, 6548–6554. [DOI] [PubMed] [Google Scholar]
- 82.Schwarz M, Spector L, Gargir A, Shtevi A, Gortler M, Altstock RT, Dukler AA and Dotan N, Glycobiology, 2003, 13, 749–754. [DOI] [PubMed] [Google Scholar]
- 83.Nimrichter L, Gargir A, Gortler M, Altstock RT, Shtevi A, Weisshaus O, Fire E, Dotan N and Schnaar RL, Glycobiology, 2004, 14, 197–203. [DOI] [PubMed] [Google Scholar]
- 84.Disney MD and Seeberger PH, Chem. Biol, 2004, 11, 1701–1707. [DOI] [PubMed] [Google Scholar]
- 85.Noti C, de Paz JL, Polito L and Seeberger PH, Chem.–Eur. J, 2006, 12, 8664–8686. [DOI] [PubMed] [Google Scholar]
- 86.S. R. Stowell, C. M. Arthur, P. Mehta, K. A. Slanina, O. Blixt, H. Leffler, D. F. Smith and R. D. Cummings, J. Biol. Chem, 2008, 283, 10109–10123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Song X, Xia B, Stowell SR, Lasanajak Y, Smith DF and Cummings RD, Chem. Biol, 2009, 16, 36–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Serna S, Etxebarria J, Ruiz N, Martin-Lomas M and Reichardt N-C, Chem.–Eur. J, 2010, 16, 13163–13175. [DOI] [PubMed] [Google Scholar]
- 89.Park S, Pai J, Han E-H, Jun C-H and Shin I, Bioconjugate Chem, 2010, 21, 1246–1253. [DOI] [PubMed] [Google Scholar]
- 90.Arigi E, Blixt O, Buschard K, Clausen H and Levery SB, Glycoconjugate J, 2012, 29, 1–12. [DOI] [PubMed] [Google Scholar]
- 91.Tully SE, Rawat M and Hsieh-Wilson LC, J. Am. Chem. Soc, 2006, 128, 7740–7741. [DOI] [PubMed] [Google Scholar]
- 92.de PJL, Spillmann D and Seeberger PH, Chem. Commun, 2006, 3116–3118. [DOI] [PubMed] [Google Scholar]
- 93.M.-r. Lee and I. Shin, Angew. Chem., Int. Ed, 2005, 44, 2881–2884. [DOI] [PubMed] [Google Scholar]
- 94.Park S and Shin I, Org. Lett, 2007, 9, 1675–1678. [DOI] [PubMed] [Google Scholar]
- 95.Park S, Lee M-R and Shin I, Nat. Protocols, 2007, 2, 2747–2758. [DOI] [PubMed] [Google Scholar]
- 96.Tian X, Pai J and Shin I, Chem.–Asian J, 2012, 7, 2052–2060. [DOI] [PubMed] [Google Scholar]
- 97.Park S, Lee M-R and Shin I, Methods Mol. Biol, 2010, 669, 195–208. [DOI] [PubMed] [Google Scholar]
- 98.Lee M-R, Park S and Shin I, Methods Mol. Biol, 2012, 808, 103–116. [DOI] [PubMed] [Google Scholar]
- 99.Oyelaran O, Li Q, Farnsworth D and Gildersleeve JC, J. Proteome Res, 2009, 8, 3529–3538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Oyelaran O, McShane LM, Dodd L and Gildersleeve JC, J. Proteome Res, 2009, 8, 4301–4310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhang Y-L, Campbell C, Li Q and Gildersleeve JC, Mol. BioSyst, 2010, 6, 1583–1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Meldal M and Tornøe CW, Chem. Rev, 2008, 108, 2952–3015. [DOI] [PubMed] [Google Scholar]
- 103.Dondoni A, Chem.–Asian J, 2007, 2, 700–708. [DOI] [PubMed] [Google Scholar]
- 104.Tian X, Pai J, Baek K-H, Ko S-K and Shin I, Chem.–Asian J, 2011, 6, 2107–2113. [DOI] [PubMed] [Google Scholar]
- 105.Sun X-L, Stabler CL, Cazalis CS and Chaikof EL, Bioconjugate Chem, 2005, 17, 52–57. [DOI] [PubMed] [Google Scholar]
- 106.Huang C-Y, Thayer DA, Chang AY, Best MD, Hoffmann J, Head S and Wong C-H, Proc. Natl. Acad. Sci. U. S. A, 2006, 103, 15–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Michel O and Ravoo BJ, Langmuir, 2008, 24, 12116–12118. [DOI] [PubMed] [Google Scholar]
- 108.Barrett OJ, Pushechnikov A, Wu M and Disney MD, Carbohydr. Res, 2008, 343, 2924–2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Godula K, Rabuka D, Nam KT and Bertozzi CR, Angew. Chem., Int. Ed, 2009, 48, 4973–4976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Koehn M, Wacker R, Peters C, Schroeder H, Soulere L, Breinbauer R, Neimeyer CM and Waldmann H, Angew. Chem., Int. Ed, 2003, 42, 5830–5834. [DOI] [PubMed] [Google Scholar]
- 111.Tyagi A, Wang X, Deng L, Ramstroem O and Yan M, Biosens. Bioelectron, 2010, 26, 344–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Xia B, Kawar ZS, Ju T, Alvarez RA, Sachdev GP and Cummings RD, Nat. Methods, 2005, 2, 845–850. [DOI] [PubMed] [Google Scholar]
- 113.Song X, Lasanajak Y, Xia B, Heimburg-Molinaro J, Rhea JM, Ju H, Zhao C, Molinaro RJ, Cummings RD and Smith DF, Nat. Methods, 2011, 8, 85–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lee M.-r. and Shin I, Org. Lett, 2005, 7, 4269–4272. [DOI] [PubMed] [Google Scholar]
- 115.Park S, Lee M-R and Shin I, Bioconjugate Chem, 2009, 20, 155–162. [DOI] [PubMed] [Google Scholar]
- 116.Zhi Z-L, Powell AK and Turnbull JE, Anal. Chem, 2006, 78, 4786–4793. [DOI] [PubMed] [Google Scholar]
- 117.Zhou X and Zhou J, Biosens. Bioelectron, 2006, 21, 1451–1458. [DOI] [PubMed] [Google Scholar]
- 118.Zhou X, Turchi C and Wang D, J. Proteome Res, 2009, 8, 5031–5040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ngundi MM, Taitt CR, McMurry SA, Kahne D and Ligler FS, Biosens. Bioelectron, 2006, 21, 1195–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Liang P-H, Wang S-K and Wong C-H, J. Am. Chem. Soc, 2007, 129, 11177–11184. [DOI] [PubMed] [Google Scholar]
- 121.Mercey E, Sadir R, Maillart E, Roget A, Baleux F, Lortat-Jacob H and Livache T, Anal. Chem, 2008, 80, 3476–3482. [DOI] [PubMed] [Google Scholar]
- 122.Song X, Xia B, Lasanajak Y, Smith DF and Cummings RD, Glycoconjugate J, 2008, 25, 15–25. [DOI] [PubMed] [Google Scholar]
- 123.Grant CF, Kanda V, Yu H, Bundle DR and McDermott MT, Langmuir, 2008, 24, 14125–14132. [DOI] [PubMed] [Google Scholar]
- 124.Kilcoyne M, Gerlach JQ, Gough R, Gallagher ME, Kane M, Carrington SD and Joshi L, Anal. Chem, 2012, 84, 3330–3338. [DOI] [PubMed] [Google Scholar]
- 125.Manimala JC, Roach TA, Li Z and Gildersleeve JC, Angew. Chem., Int. Ed, 2006, 45, 3607–3610. [DOI] [PubMed] [Google Scholar]
- 126.Manimala JC, Li Z, Jain A, VedBrat S and Gildersleeve JC, ChemBioChem, 2005, 6, 2229–2241. [DOI] [PubMed] [Google Scholar]
- 127.Adams EW, Ueberfeld J, Ratner DM, O’Keefe BR, Walt DR and Seeberger PH, Angew. Chem., Int. Ed, 2003, 42, 5317–5320. [DOI] [PubMed] [Google Scholar]
- 128.Walz A, Odenbreit S, Mahdavi J, Borén T and Ruhl S, Glycobiology, 2005, 15, 700–708. [DOI] [PubMed] [Google Scholar]
- 129.Tateno H, Mori A, Uchiyama N, Yabe R, Iwaki J, Shikanai T, Angata T, Narimatsu H and Hirabayashi J, Glycobiology, 2008, 18, 789–798. [DOI] [PubMed] [Google Scholar]
- 130.de Boer AR, Hokke CH, Deelder AM and Wuhrer M, Glycoconjugate J, 2008, 25, 75–84. [DOI] [PubMed] [Google Scholar]
- 131.Fukuda T, Onogi S and Miura Y, Thin Solid Films, 2009, 518, 880–888. [Google Scholar]
- 132.H. M. Branderhorst, R. Ruijtenbeek, R. M. J. Liskamp and R. J. Pieters, ChemBioChem, 2008, 9, 1836–1844. [DOI] [PubMed] [Google Scholar]
- 133.Parera PN, Branderhorst HM, Kooij R, Maierhofer C, d. K. M. van, R. M. J. Liskamp, V. Wittmann, R. Ruijtenbeek and R. J. Pieters, ChemBioChem, 2010, 11, 1896–1904. [DOI] [PubMed] [Google Scholar]
- 134.Gorska K, Huang KT, Chaloin O and Winssinger N, Angew. Chem., Int. Ed, 2009, 48, 7695–7700. [DOI] [PubMed] [Google Scholar]
- 135.Huang KT, Gorska K, Alvarez S, Barluenga S and Winssinger N, ChemBioChem, 2011, 12, 56–60. [DOI] [PubMed] [Google Scholar]
- 136.Ciobanu M, Huang KT, Daguer JP, Barluenga S, Chaloin O, Schaeffer E, Mueller CG, Mitchell DA and Winssinger N, Chem. Commun, 2011, 47, 9321–9323. [DOI] [PubMed] [Google Scholar]
- 137.Moni L, Pourceau G, Zhang J, Meyer A, Vidal S, Souteyrand E, Dondoni A, Morvan F, Chevolot Y, Vasseur JJ and Marra A, ChemBioChem, 2009, 10, 1369–1378. [DOI] [PubMed] [Google Scholar]
- 138.Godula K and Bertozzi CR, J. Am. Chem. Soc, 2012, 134, 15732–15742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Zhang Y, Li Q, Rodriguez LG and Gildersleeve JC, J. Am. Chem. Soc, 2010, 132, 9653–9662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Li Q, Rodriguez LG, Farnsworth DF and Gildersleeve JC, ChemBioChem, 2010, 11, 1686–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Li Q, Anver MR, Li Z, Butcher DO and Gildersleeve JC, Int. J. Cancer, 2010, 126, 459–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Laurent N, Voglmeir J, Wright A, Blackburn J, Pham NT, Wong SCC, Gaskell SJ and Flitsch SL, ChemBioChem, 2008, 9, 883–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Westerlind U, Schröder H, Hobel A, Gaidzik N, Kaiser A, Niemeyer CM, Schmitt E, Waldmann H and Kunz H, Angew. Chem., Int. Ed, 2009, 48, 8263–8267. [DOI] [PubMed] [Google Scholar]
- 144.Li Q, Anver MR, Butcher DO and Gildersleeve JC, Mol. Cancer Ther, 2009, 8, 971–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Ohyabu N, Hinou H, Matsushita T, Izumi R, Shimizu H, Kawamoto K, Numata Y, Togame H, Takemoto H, Kondo H and Nishimura S-I, J. Am. Chem. Soc, 2009, 131, 17102–17109. [DOI] [PubMed] [Google Scholar]
- 146.Song X, Lasanajak Y, Rivera-Marrero C, Luyai A, Willard M, Smith DF and Cummings RD, Anal. Biochem, 2009, 395, 151–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Steentoft C, Schjoldager K, Cló E, Mandel U, Levery S, Pedersen J, Jensen K, Blixt O and Clausen H, Glycoconjugate J, 2010, 27, 571–582. [DOI] [PubMed] [Google Scholar]
- 148.Kračun SK, Cló E, Clausen H, Levery SB, Jensen KJ and Blixt O, J. Proteome Res, 2010, 9, 6705–6714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Pedersen JW, Blixt O, Bennett EP, Tarp MA, Dar I, Mandel U, Poulsen SS, Pedersen AE, Rasmussen S, Jess P, Clausen H and Wandall HH, Int. J. Cancer, 2011, 128, 1860–1871. [DOI] [PubMed] [Google Scholar]
- 150.Wandall HH, Blixt O, Tarp MA, Pedersen JW, Bennett EP, Mandel U, Ragupathi G, Livingston PO, Hollingsworth MA, Taylor-Papadimitriou J, Burchell J and Clausen H, Cancer Res, 2010, 70, 1306–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Weinrich D, Köhn M, Jonkheijm P, Westerlind U, Dehmelt L, Engelkamp H, Christianen PCM, Kuhlmann J, Maan JC,Nüsse D, Schröder H, Wacker R, Voges E, Breinbauer R, Kunz H, Niemeyer CM and Waldmann H, ChemBioChem, 2010, 11, 235–247. [DOI] [PubMed] [Google Scholar]
- 152.Borgert A, Heimburg-Molinaro J, Song X, Lasanajak Y, Ju T, Liu M, Thompson P, Ragupathi G, Barany G, Smith DF, Cummings RD and Live D, ACS Chem. Biol, 2012, 7, 1031–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Blixt O, Cló E, Nudelman AS, Sørensen KK, Clausen T, Wandall HH, Livingston PO, Clausen H and Jensen KJ, J. Proteome Res, 2010, 9, 5250–5261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Rinaldi S, Brennan KM, Goodyear CS, O’Leary C, Schiavo G, Crocker PR and Willison HJ, Glycobiology, 2009, 19, 789–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Liang C-H, Wang S-K, Lin C-W, Wang C-C, Wong C-H and Wu C-Y, Angew. Chem., Int. Ed, 2011, 50, 1608–1612. [DOI] [PubMed] [Google Scholar]
- 156.Fei Y, Sun YS, Li Y, Lau K, Yu H, Chokhawala HA, Huang S, Landry JP, Chen X and Zhu X, Mol. BioSyst, 2011, 7, 3343–3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Ban L, Pettit N, Li L, Stuparu AD, Cai L, Chen W, Guan W, Han W, Wang PG and Mrksich M, Nat. Chem. Biol, 2012, 8, 769–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Guan W, Ban L, Cai L, Li L, Chen W, Liu X, Mrksich M and Wang PG, Bioorg. Med. Chem. Lett, 2011, 21, 5025–5028. [DOI] [PubMed] [Google Scholar]
- 159.Ban L and Mrksich M, Angew. Chem., Int. Ed, 2008, 47, 3396–3399. [DOI] [PubMed] [Google Scholar]
- 160.Sanchez-Ruiz A, Serna S, Ruiz N, Martin-Lomas M and Reichardt N-C, Angew. Chem., Int. Ed, 2011, 50, 1801–1804. [DOI] [PubMed] [Google Scholar]
- 161.Beloqui A, Sanchez-Ruiz A, Martin-Lomas M and Reichardt N-C, Chem. Commun, 2012, 48, 1701–1703. [DOI] [PubMed] [Google Scholar]
- 162.Northen TR, Lee J-C, Hoang L, Raymond J, Hwang D-R, Yannone SM, Wong C-H and Siuzdak G, Proc. Natl. Acad. Sci. U. S. A, 2008, 105, 3678–3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Fais M, Karamanska R, Allman S, Fairhurst SA, Innocenti P, Fairbanks AJ, Donohoe TJ, Davis BG, Russell DA and Field RA, Chem. Sci, 2011, 2, 1952–1959. [Google Scholar]
- 164.Shipp M, Nadella R, Gao H, Farkas V, Sigrist H and Faik A, Glycoconjugate J, 2008, 25, 49–58. [DOI] [PubMed] [Google Scholar]
- 165.Pedersen HL, Fangel JU, McCleary B, Ruzanski C, Rydahl MG, Ralet M-C, Farkas V, von Schantz L, Marcos SE, Andersen MCF, Field R, Ohlin M, Knox JP, Clausen MH and Willats WGT, J. Biol. Chem, 2012, 287, 39429–39438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Fei YY, Schmidt A, Bylund G, Johansson DX, Henriksson S, Lebrilla C, Solnick JV, Borén T and Zhu XD, Anal. Chem, 2011, 83, 6336–6341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Han E, Ding L, Jin S and Ju H, Biosens. Bioelectron, 2011, 26, 2500–2505. [DOI] [PubMed] [Google Scholar]
- 168.Baader J, Klapproth H, Bednar S, Brandstetter T, Rühe J, Lehmann M and Freund I, Biosens. Bioelectron, 2011, 26, 1839–1846. [DOI] [PubMed] [Google Scholar]
- 169.Gruber K, Horlacher T, Castelli R, Mader A, Seeberger PH and Hermann BA, ACS Nano, 2011, 5, 3670–3678. [DOI] [PubMed] [Google Scholar]
- 170.Lectins The. Properties, Functions, and Applications in Biology and Medicine, ed. Liener IE, Sharon N and Goldstein IJ, Academic Press, Inc., 1986. [Google Scholar]
- 171.Van DEJM, Peumans WJ, Pusztai A and Bardocz S, Handbook of Plant Lectins: Properties and Biomedical Applications, Wiley, 1997. [Google Scholar]
- 172.Galustian C, Park CG, Chai W, Kiso M, Bruening SA, Kang YS, Steinman RM and Feizi T, Int. Immunol, 2004, 16, 853–866. [DOI] [PubMed] [Google Scholar]
- 173.Coombs PJ, Graham SA, Drickamer K and Taylor ME, J. Biol. Chem, 2005, 280, 22993–22999. [DOI] [PubMed] [Google Scholar]
- 174.McGreal EP, Rosas M, Brown GD, Zamze S, Wong SYC, Gordon S, Martinez-Pomares L and Taylor PR, Glycobiology, 2006, 16, 422–430. [DOI] [PubMed] [Google Scholar]
- 175.Tateno H, Crocker PR and Paulson JC, Glycobiology, 2005, 15, 1125–1135. [DOI] [PubMed] [Google Scholar]
- 176.Markova V, Smetana K Jr., Jenikova G, Lachova J, Krejcirikova V, Poplstein M, Fabry M, Brynda J, Alvarez RA, Cummings RD and Maly P, Int. J. Mol. Med, 2006, 18, 65–76. [PubMed] [Google Scholar]
- 177.Leppänen A, Stowell S, Blixt O and Cummings RD, J. Biol. Chem, 2005, 280, 5549–5562. [DOI] [PubMed] [Google Scholar]
- 178.Horlacher T, Oberli MA, Werz DB, Kröck L, Bufali S, Mishra R, Sobek J, Simons K, Hirashima M, Niki T and Seeberger PH, ChemBioChem, 2010, 11, 1563–1573. [DOI] [PubMed] [Google Scholar]
- 179.Young WW Jr., MacDonald EM, Nowinski RC and Hakomori SI, J. Exp. Med, 1979, 150, 1008–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Kannagi R and Hakomori S, Adv. Exp. Med. Biol, 2001, 491, 587–630. [DOI] [PubMed] [Google Scholar]
- 181.Blixt O, Boos I and Mandel U, Anticarbohydrate Antibodies, 2012, 283–306. [Google Scholar]
- 182.Blixt O, Lavrova OI, Mazurov DV, Clo E, Kracun SK, Bovin NV and Filatov AV, Glycobiology, 2012, 22, 529–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Manimala JC, Roach TA, Li Z and Gildersleeve JC, Glycobiology, 2007, 17, 17C–23C. [DOI] [PubMed] [Google Scholar]
- 184.Hacker U, Nybakken K and Perrimon N, Nat. Rev. Mol. Cell Biol, 2005, 6, 530–541. [DOI] [PubMed] [Google Scholar]
- 185.de Paz JL, Noti C and Seeberger PH, J. Am. Chem. Soc, 2006, 128, 2766–2767. [DOI] [PubMed] [Google Scholar]
- 186.Gama CI, Tully SE, Sotogaku N, Clark PM, Rawat M, Vaidehi N, Goddard WA, Nishi A and Hsieh-Wilson LC, Nat. Chem. Biol, 2006, 2, 467–473. [DOI] [PubMed] [Google Scholar]
- 187.de Paz JL, Moseman EA, Noti C, Polito L, von Andrian UH and Seeberger PH, ACS Chem. Biol, 2007, 2, 735–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Schallus T, Jaeckh C, Fehér K, Palma AS, Liu Y, Simpson JC, Mackeen M, Stier G, Gibson TJ, Feizi T, Pieler T and MuhleGoll C, Mol. Biol. Cell, 2008, 19, 3404–3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Takeda Y, Totani K, Matsuo I and Ito Y, Curr. Opin. Chem. Biol, 2009, 13, 582–591. [DOI] [PubMed] [Google Scholar]
- 190.Kulkarni AA, Weiss AA and Iyer SS, Med. Res. Rev, 2010, 30, 327–393. [DOI] [PubMed] [Google Scholar]
- 191.Lao WI, Wang YF, Kuo YD, Lin CH, Chang TC, Su IJ, Wang JR and Chang CF, Future Med. Chem, 2011, 3, 283–296. [DOI] [PubMed] [Google Scholar]
- 192.Liu Y, Childs R, Palma A, Campanero-Rhodes M, Stoll M, Chai W and Feizi T, Methods Mol. Biol, 2012, 808, 117–136. [DOI] [PubMed] [Google Scholar]
- 193.Stevens J, Blixt O, Glaser L, Taubenberger JK, Palese P, Paulson JC and Wilson IA, J. Mol. Biol, 2006, 355, 1143–1155. [DOI] [PubMed] [Google Scholar]
- 194.Stevens J, Blixt O, Tumpey TM, Taubenberger JK, Paulson JC and Wilson IA, Science, 2006, 312, 404–410. [DOI] [PubMed] [Google Scholar]
- 195.Campanero-Rhodes MA, Smith A, Chai W, Sonnino S, Mauri L, Childs RA, Zhang Y, Ewers H, Helenius A, Imberty A and Feizi T, J. Virol, 2007, 81, 12846–12858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Nilsson EC, Storm RJ, Bauer J, Johansson SMC, Lookene A, Angstrom J, Hedenstrom M, Eriksson TL, Frangsmyr L, Rinaldi S, Willison HJ, Domellof FP, Stehle T and Arnberg N, Nat. Med, 2011, 17, 105–109. [DOI] [PubMed] [Google Scholar]
- 197.Hsu K-L, Gildersleeve JC and Mahal LK, Mol. BioSyst, 2008, 4, 654–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Blanchard B, Nurisso A, Hollville E, Tetaud C, Wiels J, Pokorna M, Wimmerova M, Varrot A and Imberty A, J. Mol. Biol, 2008, 383, 837–853. [DOI] [PubMed] [Google Scholar]
- 199.Lameignere E, Malinovska L, Slavikova M, Duchaud E, Mitchell EP, Varrot A, Sedo O, Imberty A and Wimmerova M, Biochem. J, 2008, 411, 307–318. [DOI] [PubMed] [Google Scholar]
- 200.Heimburg-Molinaro J, Tappert M, Song X, Lasanajak Y, Air G, Smith D and Cummings R, Methods Mol. Biol, 2012, 808, 251–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Wang YF, Chang CF, Chi CY, Wang HC, Wang JR and Su IJ, J. Med. Virol, 2012, 84, 679–685. [DOI] [PubMed] [Google Scholar]
- 202.Chen LM, Rivailler P, Hossain J, Carney P, Balish A, Perry I, Davis CT, Garten R, Shu B, Xu X, Klimov A, Paulson JC, Cox NJ, Swenson S, Stevens J, Vincent A, Gramer M and Donis RO, Virology, 2011, 412, 401–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Bradley KC, Jones CA, Tompkins SM, Tripp RA, Russell RJ, Gramer MR, Heimburg-Molinaro J, Smith DF, Cummings RD and Steinhauer DA, Virology, 2011, 413, 169–182. [DOI] [PubMed] [Google Scholar]
- 204.Pearce MB, Jayaraman A, Pappas C, Belser JA, Zeng H, Gustin KM, Maines TR, Sun X, Raman R, Cox NJ, Sasisekharan R, Katz JM and Tumpey TM, Proc. Natl. Acad. Sci. U. S. A, 2012, 109, 3944–3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Yang H, Chen LM, Carney PJ, Donis RO and Stevens J, PLoS Pathog, 2010, 6, e1001081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Wu ZJ, Miller E, Agbandje-McKenna M and Samulski RJ, J. Virol, 2006, 80, 9093–9103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Bell CL, Vandenberghe LH, Bell P, Limberis MP, Gao GP, Van Vliet K, Agbandje-McKenna M and Wilson JM, J. Clin. Invest, 2011, 121, 2427–2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Nam H-J, Gurda-Whitaker B, Gan WY, Ilaria S, McKenna R, Mehta P, Alvarez RA and Agbandje-McKenna M, J. Biol. Chem, 2006, 281, 25670–25677. [DOI] [PubMed] [Google Scholar]
- 209.Day CJ, Tiralongo J, Hartnell RD, Logue C-A, Wilson JC, von Itzstein M and Korolik V, PLoS One, 2009, 4, e4927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Puvirajesinghe TM, Ahmed YA, Powell AK, Fernig DG, Guimond SE and Turnbull JE, Chem. Biol, 2012, 19, 553–558. [DOI] [PubMed] [Google Scholar]
- 211.Liang PH, Imamura M, Li X, Wu D, Fujio M, Guy RT, Wu BC, Tsuji M and Wong CH, J. Am. Chem. Soc, 2008, 130, 12348–12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Serna S, Yan S, Martin-Lomas M, Wilson IBH and Reichardt N-C, J. Am. Chem. Soc, 2011, 133, 16495–16502. [DOI] [PubMed] [Google Scholar]
- 213.Seibel J, Hellmuth H, Hofer B, Kicinska A-M and Schmalbruch B, ChemBioChem, 2006, 7, 310–320. [DOI] [PubMed] [Google Scholar]
- 214.Blixt O, Allin K, Bohorov O, Liu X, Andersson-Sand H, Hoffmann J and Razi N, Glycoconjugate J, 2008, 25, 59–68. [DOI] [PubMed] [Google Scholar]
- 215.Bennett EP, Mandel U, Clausen H, Gerken TA, Fritz TA and Tabak LA, Glycobiology, 2012, 22, 736–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Laurent N, Haddoub R and Flitsch SL, Trends Biotechnol, 2008, 26, 328–337. [DOI] [PubMed] [Google Scholar]
- 217.Pedersen JW, Bennett EP, Schjoldager KT-BG, Meldal M, Holmér AP, Blixt O, Cló E, Levery SB, Clausen H and Wandall HH, J. Biol. Chem, 2011, 286, 32684–32696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Lacroix-Desmazes S, Mouthon L, Coutinho A and Kazatchkine MD, Eur. J. Immunol, 1995, 25, 2598–2604. [DOI] [PubMed] [Google Scholar]
- 219.Huflejt ME, Vuskovic M, Vasiliu D, Xu H, Obukhova P, Shilova N, Tuzikov A, Galanina O, Arun B, Lu K and Bovin N, Mol. Immunol, 2009, 46, 3037–3049. [DOI] [PubMed] [Google Scholar]
- 220.von Gunten S, Smith DF, Cummings RD, Riedel S, Miescher S, Schaub A, Hamilton RG and Bochner BS, J. Allergy Clin. Immunol, 2009, 123, 1268–1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Obukhova P, Piskarev V, Severov V, Pazynina G, Tuzikov A, Navakouski M, Shilova N and Bovin N, Glycoconjugate J, 2011, 28, 501–505. [DOI] [PubMed] [Google Scholar]
- 222.Wu C-S, Yen C-J, Chou R-H, Li S-T, Huang W-C, Ren C-T, Wu C-Y and Yu Y-L, PLoS One, 2012, 7, e39466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Jacob F, Goldstein DR, Bovin NV, Pochechueva T, Spengler M, Caduff R, Fink D, Vuskovic MI, Huflejt ME and Heinzelmann-Schwarz V, Int. J. Cancer, 2012, 130, 138–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Bovin N, Obukhova P, Shilova N, Rapoport E, Popova I, Navakouski M, Unverzagt C, Vuskovic M and Huflejt M, Biochim. Biophys. Acta, Gen. Subj., 2012, 1820, 1373–1382. [DOI] [PubMed] [Google Scholar]
- 225.Blixt O, Bueti D, Burford B, Allen D, Julien S, Hollingsworth M, Gammerman A, Fentiman I, Taylor-Papadimitriou J and Burchell JM, Breast Cancer Res, 2011, 13, R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Clo E, Kracun SK, Nudelman AS, Jensen KJ, Liljeqvist JA, Olofsson S, Bergstrom T and Blixt O, J. Virol, 2012, 86, 6268–6278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Aranzamendi C, Tefsen B, Jansen M, Chiumiento L, Bruschi F, Kortbeek T, Smith DF, Cummings RD, Pinelli E and Van Die I,Exp. Parasitol, 2011, 129, 221–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Oberli MA, Hecht M-L, Bindschädler P, Adibekian A, Adam T and Seeberger PH, Chem. Biol, 2011, 18, 580–588. [DOI] [PubMed] [Google Scholar]
- 229.Blixt O, Hoffmann J, Svenson S and Norberg T, Glycoconjugate J, 2008, 25, 27–36. [DOI] [PubMed] [Google Scholar]
- 230.Gulley J, Arlen P, Madan R, Tsang K-Y, Pazdur M, Skarupa L, Jones J, Poole D, Higgins J, Hodge J, Cereda V, Vergati M, Steinberg S, Halabi S, Jones E, Chen C, Parnes H, Wright J, Dahut W and Schlom J, Cancer Immunol. Immunother, 2010, 59, 663–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Lechleider RJ, Arlen PM, Tsang K-Y, Steinberg SM, Yokokawa J, Cereda V, Camphausen K, Schlom J, Dahut WL and Gulley JL, Clin. Cancer Res, 2008, 14, 5284–5291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Madan RA, Gulley JL, Schlom J, Steinberg SM, Liewehr DJ, Dahut WL and Arlen PM, Clin. Cancer Res, 2008, 14, 4526–4531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Wang D and Lu J, Physiol. Genomics, 2004, 18, 245–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Kamena F, Tamborrini M, Liu X, Kwon Y-U, Thompson F, Pluschke G and Seeberger PH, Nat. Chem. Biol, 2008, 4, 238–240. [DOI] [PubMed] [Google Scholar]
- 235.Dunlop DC, Bonomelli C, Mansab F, Vasiljevic S, Doores KJ, Wormald MR, Palma AS, Feizi T, Harvey DJ, Dwek RA, Crispin M and Scanlan CN, Glycobiology, 2010, 20, 812–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Pincus SH, Moran E, Maresh G, Jennings HJ, Pritchard DG, Egan ML and Blixt O, Vaccine, 2012, 30, 4849–4858. [DOI] [PMC free article] [PubMed] [Google Scholar]