Summary
One in eight women develops invasive breast cancer in her lifetime. The frontline protection against this disease is mammography. While computer-assisted diagnosis algorithms have made great progress in generating reliable global predictions, few focus on simultaneously producing regions of interest (ROIs) for biopsy. Can we combine ROI-oriented algorithms with global classification of cancer status, which simultaneously highlight suspicious regions and optimize classification performance? Can the asymmetry of breasts be adopted in deep learning for finding lesions and classifying cancers? We answer the above questions by building deep-learning networks that identify masses and microcalcifications in paired mammograms, exclude false positives, and stepwisely improve performance of the model with asymmetric information regarding the breasts. This method achieved a co-leading place in the Digital Mammography DREAM Challenge for predicting breast cancer. We highlight here the importance of this dual-purpose process that simultaneously provides the locations of potential lesions in mammograms.
Keywords: breast cancer, digital mammography, asymmetry, deep learning, computer-assisted diagnosis
Highlights
-
•
A top-performing algorithm in the International DREAM Digital Mammography Challenge
-
•
Shows efficacy of asymmetric information from opposite breasts in identifying cancers
-
•
Integrated pixel-level localization and overall classification into the same software
The Bigger Picture
Breast cancer affects one out of eight women in their lifetime. Given the importance of the need, in this work we present a region-of-interest-oriented deep-learning pipeline for detecting and locating breast cancers based on digital mammograms. It is a leading algorithm in the well-received Digital Mammography DREAM Challenge, in which computational methods were evaluated on large-scale, held-out testing sets of digital mammograms. This algorithm connects two aims: (1) determining whether a breast has cancer and (2) determining cancer-associated regions of interest. Particularly, we addressed the challenge of variation of mammogram images across different patients by pairing up the two opposite breasts to examine asymmetry, which substantially improved global classification as well as local lesion detection. We have dockerized this code, envisioning that it will be widely used in practice and as a future reference for digital mammography analysis.
This paper reports a top-performing algorithm in the International DREAM Digital Mammography Challenge that leverages deep learning of the asymmetric information from opposite breasts in identifying cancers. The method integrates pixel-level localization and overall classification into the same software.
Introduction
Breast cancer is the most common solid cancer, affecting one in eight women. It is also the second leading cause of cancer deaths among women in the United States.1 Luckily, early detection of breast cancer can make a huge difference in prognosis. The most popular detection method today is mammography.2,3 Around the globe, women older than 40 or 45 years are recommended to start mammogram screening annually or biennially.4
Many computational approaches have been explored previously for the identification of breast calcification and masses, including traditional machine-learning classifiers,5, 6, 7 wavelet transformation,8, 9, 10 K-means clustering,11,12 and active contour models.13,14 In recent years, deep learning has surpassed traditional techniques in a large number of computer vision applications.15,16 For many tasks, deep-learning approaches have reached the accuracy of human experts.17,18 Many efforts have also been made to develop deep-learning algorithms for detecting breast cancers based on mammograms.19, 20, 21, 22, 23, 24
Revisiting and reflecting on the CAD (computer-assisted diagnosis) field of mammography, we see new opportunities to further improve the utility of the progress that has been made in this field. First, there is a need to synthesize region of interest (ROI)-based algorithms, which are informative to biopsy, and global classification algorithms, which are important for risk stratification. Additionally, prior to the deep-learning era, how to utilize the asymmetric information to identify lesions in mammography using traditional machine-learning or rule-based approaches has been explored.25, 26, 27, 28, 29 These methods typically rely on image registration of the left and right breasts and correct for the variance of the sizes and positions between two breasts. Subsequently, the texture differences between the aligned breasts are quantified, including features such as roughness, brightness, and directionality. The areas that differ significantly for these features are considered to be ROIs. These methods have reported significant performance improvement when referencing the target breast with the opposite breast. However, it remains unclear how to integrate the information of asymmetry into deep neural network models until today and whether or not it can improve the prediction performance.
We also see remaining challenges in mammogram interpretation: the high resolution of mammogram images and the heterogeneity of density and texture of human breasts.30,31 The high resolution of the image is necessary for the detection of tiny calcification dots that are smaller than a millimeter. On the other hand, human breasts are heterogeneous in their presentation: some are dense, others are fatty, and some naturally have calcifications spreading throughout the breast. It is therefore challenging to establish a model accounting for this heterogeneity.
Seeing the above opportunities and challenges, in this work we present an ROI-oriented deep-learning pipeline for detecting and locating breast cancers based on digital mammograms. It is a leading algorithm in the well-received Digital Mammography DREAM Challenge, in which computational methods were evaluated on large-scale, held-out testing sets of digital mammograms.32 This algorithm connects two aims: (1) determining whether a breast has cancer, and (2) determining cancer-associated ROIs. Particularly, we addressed the challenge of variation of mammogram images across different patients by pairing up the two opposite breasts to examine asymmetry, which substantially improved global classification as well as local lesion detection. We have dockerized this code, envisioning that it will be widely used in practice and as a future reference for digital mammography analysis.
Methods
Overview of the Workflow
The DREAM Digital Mammography Challenge was designed in a unique environment where the participants are not allowed to observe the training examples, but only the global label of whether a breast has cancers or not in three separate sets of data: training, validation, and final test. This design protects the privacy of the patients. In this study, we present the model performance based on experiments on the training set from the challenge.
In digital mammography images, cancer regions occupy a tiny proportion of the entire image. Thus, attempting to train a classification model solely based on global labels of the images is extremely challenging. We therefore sought public datasets that had locality information regarding breast cancers to initialize ROI-based algorithms for detecting and classifying breast cancer images. The INbreast data is a widely used, well-characterized, hand-labeled dataset that includes a total of 410 images.33 The image pixel size is 70 μm with 14-bit grayscale depth, and the image sizes are either 3,328 × 4,084 pixels or 2,560 × 3,328 pixels, depending on the compression plate used for the acquisition. Both INbreast and Digital Mammography DREAM Challenge data are digital mammograms, with the latter containing over 640,000 de-identified images ranging from 3,328 × 2,560 to 5,928 × 4,728 pixels.
We partitioned the DREAM data into three parts, denoted Part 1 (40%), Part 2 (40%), and Part 3 (20%), to serve the eventual purpose of identifying and locating malignancy (Figure 1A). This partition is done at the individual level. The reason for partitioning the data is to avoid contamination between the feature extraction, false-positive classification, and feature assembly step. Of note, the performance of individual models was estimated by data within their associated partitions.
Figure 1.
Overview of the Workflow of the Algorithm
(A) Data sources and training data partition.
(B) Training model 1 and model 2 with INbreast data, then using model 1 and model 2 to predict ROI regions in DREAM data Part 1.
(C) Training model 3 and model 4 using ROI-labeled DREAM Part 1, then using models 1, 2, 3, and 4 to extract features from DREAM Data Part 2.
(D) Training model 5 using features extracted from DREAM Part 2, then using DREAM Data Part 3 to evaluate the whole model.
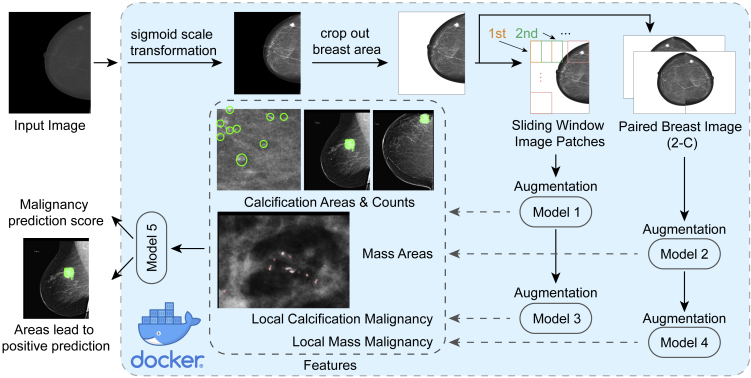
Globally, we built four models for feature extraction (Figures 1B and 1C). The INbreast dataset served to train the initial detection models to locate the regions of interest (model 1, the calcification detection patch model; model 2, the mass detection whole-image model) (Figure 1B). In building model 2 (mass detection model), the input images are paired breasts as the first channel and the horizontally flipped paired images as the second channel. We found this pairing substantially improved the performance of the model (see Results). When the patch model 1 was applied to the images, we used it as a moving window to identify all calcifications in the breasts. Using these detection models, we retrieved ROIs and their corresponding locations from the Part 1 data. These locations can either be cancerous or non-cancerous. Because we know the cancer labels of the breasts associated with these locations, this retrieval provided patches of true-positive examples and false-positive examples, and allowed us to train two additional models that classified a local patch (Figure 1C). Model 3 is the calcification false-positive area detection patch model, i.e., it detects large calcifications, or calcification in ducts, which are unlikely to be cancer. This model was used to lay on top of the regions identified in model 1 and mask out regions where there are supposed to be false positives. By laying model 3 on top of model 1, we obtained the maximal count of calcifications in a local patch, and the maximal size, across all patches. Model 4 is the mass true-positive patch model, which gave a score for whether an identified mass is likely to be true positive. It is applied to the largest mass area. Models 1–3 are for fundamentally identifying ROIs and we therefore used U-Net structures, which are designed for semantic segmentation. For Model 4, we used an end-to-end classification network.
Models 1–4 together can generate multiple quantitative features in Part 2 of the data including the size and count for calcification (after removing false-positive calcifications), size, and true-positive likelihood of the largest mass area identified. To predict the likelihood of a breast having cancer, we also included the corresponding features in the opposite breast and found that this step improves model performance (see Results). These quantitative features are then used to train a random forest model, i.e., model 5, based on Part 2 of the training data (Figure 1D). Performance estimation and model tuning and comparison were carried out by using models 1–4 for individual feature extraction and model 5 for feature integration on Part 3 of the images.
Color Profile Mapping between Linear and Sigmoid Scale to Homogenize INbreast Data to DREAM Data
The dicom file header in the INbreast database indicates that the original scale of the images was linear compared with the DREAM data, where the scale is sigmoid (i.e., has undergone contrast stretching). Thus we first mapped the INbreast data toward the sigmoid scale, which was necessary in accounting for the different modalities in equipment. As we do not know the parameters for contrast stretching, the idea is to create a percentile-wise match between the INbreast data and the DREAM data and fit a sigmoid curve between the two percentiles.
Specifically, we first separated the images into craniocaudal (CC) view and mediolateral oblique (MLO) views. As the images of the two views show very different pixel value distributions, the mapping was separately done for these two views. In creating a percentile profile, the first step is to identify the breast areas because breasts vary substantially by size, which will lead to biases in density estimation. We dilated each image using a 70 × 70 Gaussian kernel and then created a binary mask thresholding at 200 (original pixel values ranging from 0 to 4,095). Thereafter, we identified the largest connected component of each image, which represents the breast area. Using this mask, the breast can be cropped to make the surrounding area 0 (Figure 2A). We then generated the percentile distribution A of the pixel values within the breast areas for the INbreast data at 0.001 resolution, in the format of percentile: pixel value (e.g., 14.400%: 1,221; 14.500%: 1,222; 14.600: 1,223), going from 0% to 100%. Similarly, we generated the percentile distribution B from 500 randomly selected images from the DREAM data. We fit the percentile distribution A into percentile distribution B with the following sigmoid function:
where x is the pixel values in percentile distribution A, y is the corresponding pixel value in percentile distribution B, and a, b, and c are parameters to be fit in this sigmoid function (Figure 2B). We used Scipy's optimization function curve_fit to find the values of these parameters with an initiating search point of 1,800, 1,024, and 4,900, which correspond to the mean of the distribution B, the mean of the distribution A, and the potential largest range the pixel values can reach during the parameter fitting iterations, respectively. The fitted parameters are stable with small variations of these initiating search points. We then applied the above formula to the original images of INbreast, separately for CC and MLO images, and created a new set of INbreast images that match the pixel intensity distribution of the DREAM dataset (Figure S1).
Figure 2.
Image Processing for Deep Learning Models
(A) Calculating percentile distribution of breast areas.
(B) Color profile matching of INbreast images to DREAM images.
(C) Augmentation methods of training and test sets.
(D) Orientation detection.
(E) Image pairing.
(F) Performance evaluation of the three ways to integrate asymmetric information with AUC in detecting mass: single, single images; 1-C, paired left-right breasts; 2-C, paired left-right breasts with the horizontally flipped image as the second channel.
Training and Testing Augmentations and Model Ensembling to Avoid Overfitting and Improve Accuracy
As discussed above, to accurately predict breast cancer we developed four types of neural network models to capture different features of digital mammograms: model 1, calcification detection model; model 2, mass detection model; model 3, false-positive calcification detection; model 4, classification of the malignancy for mass patches. Overfitting is a shared problem in building deep-learning models. Thus, aggressive data augmentation was applied to each of the above models (Figure 2C). For model 1 and model 3, we used 0°–360° random rotation and left-right flipping for the input patches, because the orientation of the calcification does not affect calcification detection. However, scaling was not used because the size of the calcification is a strong indicator of whether it is malignant or not. For model 2, left-right flipping was applied, assuming the two breasts of a person are equivalent. We also applied scaling between 0.8 and 1.3, accounting for variations of breast size across individuals. No rotation was applied because this model is intended for the entire breast images, which is often well aligned and positioned in digital mammograms. For model 4, because it is a patch model and the direction of the mass does not affect its malignancy, we applied 0°–360° random rotation of the input patches.
We carried out test set augmentation to further improve prediction stability (Figure 2C). For model 1 (calcification detection model), model 3 (classification of the malignancy of the calcification patches), and model 4 (classification of the malignancy of mass patches), we rotated the testing patches by 90° four times and thus generated four patches. We then made predictions for each patch and used the average prediction values of each pixel across the rotations (model 1 and model 3), or for the patch (model 4) obtained through these four rotations of images, and uses these four images to generate four predictions and average as the final assembled value. Unlike patches, top-down flipping of a paired breast image does not generate equivalent images. Thus, for model 2 we left-right flipped the images to generate two sets of predictions and took the average as the final prediction.
For all of the above deep-learning models, we used a nested training strategy by subsampling and assembling in order to leverage the whole training partition and improve the robustness of models. This is a commonly used strategy in machine-learning challenges to increase the diversity of the model with the limitation of the set of training data. Specifically, for each of the models 1–4, we trained five submodels in parallel based on different training and validation data partitions (75% and 25% of the Part 1 data described above). The validation dataset was used to monitor the prediction loss, based on which the training process was monitored and the best-performing model was saved. Finally, the predictions from five submodels were averaged as the final prediction of the model.
We extracted the following features from the breast under investigation and its opposite breast: the maximal size of a mass detected in a breast, the likelihood of this mass to be malignant, the maximal calcification count, and total areas of a local patch.
Statistical Test of Significance of Performance Difference between Two Models
In many locations of this paper, we present and compare performance values of two methods, or two sets of feature input. To estimate the statistical significance of the differences in performance for each pair, we bootstrapped the examples for 10,000 times and computed the p values. Standard deviation and confidence intervals are calculated from the 10,000-time bootstrapping.
Results
Improving Mass Detection by Normalizing Density and Leveraging the Asymmetry of Breasts
Mammographic images are diverse in terms of the fraction of breast regions in the images and distribution of pixel intensity. Directly inputting the original images without normalization may result in dense breasts being classified as masses. We first segmented the breast areas by identifying the largest connected component in each image. We then calculated the average and standard deviation of the pixel values within the breast regions for normalizing the images for the input for mass detection model (model 1).
While most left breasts have their nipples pointing toward the left and right breasts have their nipples pointing toward the right, occasionally, there are left-right flipped images. In the DREAM dataset, 100% of the annotations of the “FieldOfViewHorizontalFlip” is ‘NO,’ but we still detected rare cases where the images are flipped. Thus we sought alternative approaches to detecting flipping. By calculating the average values of 10%–20% of the left and the right side of the original images, the side with higher average value was determined to be closer to the chest wall (Figure 2D). When we detected an image whose expected left-right orientation was inconsistent with the Dicom header, we horizontally flipped the image. This step was carried out in order to pair the two images together, with the chest wall side toward the middle (Figure 2D).
To detect an areas of interest, applications in other computer vision areas point us to semantic segmentation algorithms such as U-Net and fully convolutional networks.34,35 We used U-Net for mass detection (Figures 1B and S2–S6). The U-Net architecture consists of an encoder comprising convolution-max-pool blocks that extracts information from the images, and a decoder consisting of deconvolution blocks that generates prediction of the locality of the ROIs.
It has been reported that the asymmetric regions of paired left and right breast images contribute to the detection of breast cancer based on hand-crafted features from digital mammograms.25, 26, 27, 28, 29 Yet it remains unclear how to leverage this asymmetric information to improve prediction performance in neural network models. Here we investigated multiple approaches to integrate this information into our deep-learning models: (i) “single” refers to the model using a single breast image as the input; (ii) “1-channel” refers to the model using a pair of left and right breast images as one channel; and (iii) “2-channel” refers to the model combining two images of paired breasts as two channels, and the paired image is horizontally flipped in the second channel (Figure 2E). To avoid a confounding factor of performance comparison involving the number of samples, we maintained all models with the same amount of input. For example, suppose we have n individuals in the training set, then 2n images are used in training the model, and the only difference is how we combined the 2n images as the input. We found that although, as expected, the features derived from different methods are correlated (0.9188 between single and 1-channel, 0.8054 between 2-channel and single), there are cases where the extracted features are drastically different (Figure S7).
Steady improvement was achieved when we stepwisely added asymmetric information into the models. We estimated the prediction performance by area under the curve (AUC) for the mass areas identified by the above three approaches versus the global gold standard of the cancerous status of the breasts. Of note, later there will be many other features added into the final model, and thus the AUCs presented here are much lower than the eventual AUC. Nevertheless, they can serve as a fair comparison for the three mass segmentation models (i) to (iii). The performance of these three approaches is shown in Figure 2F. Compared with the “single” approach without the asymmetric information (AUC = 0.672), both the “1-channel” and “2-channel” approaches achieved higher AUCs of 0.692 (p = 0.0569 compared with “single”) and 0.708 (p < 0.0001 compared with “single”), respectively. The 2-channel approach has the highest AUC, since it directly contrasts and captures the differences between two breasts through stacking the two images as two channels. This result indicates that comparing and contrasting the asymmetric information from two breasts significantly improves the detection of masses in mammograms. We used this 2-channel approach in our final mass detection model. Such mass detection algorithm is carried out on resized and paired breast images, and included multiple rescaled sizes of 256 × 256, 384 × 384, and 512 × 512 to increase the diversity of the models and, thus, robustness (see Table 1 for complete list of all models), as we found that multiple resolutions stably improved overall performance of mass detection.
Table 1.
Summary of Feature Extraction Networks Used in the Model
| Input Size | Model Type | Input Type | Total Models | Training Set Augmentation | Testing Set Augmentation | |
|---|---|---|---|---|---|---|
| Model 1. Segmentation model for detecting calcifications | 256 × 256 full resolution | Seg-net v3 | patch | 4 | rotation, flip | rotation |
| Model 2. Segmentation model for detecting masses | pairs of 512 × 256 | Seg-net v6 with double input | whole paired images | 2 | scaling, left-right flip | left-right flip |
| pairs of 356 × 178 | Seg-net v3 with single input | 2 | scaling, left-right flip | left-right flip | ||
| pairs of 356 × 178 | Seg-net v6 with double input | 2 | scaling, left-right flip | left-right flip | ||
| pairs of 256 × 128 | Seg-net v6 with double input | 2 | scaling, left-right flip | left-right flip | ||
| Model 3. Segmentation model for false-positive calcification detection | 256 × 256 full resolution | Seg-net v3 | patch | 4 | rotation, flip | rotation |
| Model 4. End-to-end classification model for predicting malignancy score | 512 × 512 full resolution | end-to-end CNN | local patch centered at the detected mass | 8 | rotation, flip, scaling | rotation, flip |
| Ensemble model | random forest |
Full-Resolution Mass Malignancy Models Provide Additional Information to the Mass Segmentation Models in Detecting Breast Cancers
In the previous section, we described models to locate masses in paired mammograms. This segmentation network is unable to differentiate benign and malignant masses. Therefore, we built a secondary model for predicting the malignancy of the top-predicted mass area for each breast. To differentiate the false-positive masses against the true positives, we took advantage of full-resolution local patches and trained a classification convolutional neural network (CNN) to predict whether a locally cropped 3.6 cm × 3.6 cm (512 × 512 pixels) area centered at the detected mass is cancerous. Due to the lack of a patchwise training label, a gold standard was created based on the global label of the images. Predicted masses in negatively labeled breasts are identified as the negative examples (Figure 3A), and masses detected in positive breasts are identified as positive examples. For each breast, we predict the malignancy of the patch centered at the biggest mass. This classification model has an AUC of 0.725 when evaluated on the entire breast level, compared with 0.708 for the mass detection model, indicating that full-resolution local patches could provide additional information in classifying whether a detected mass is malignant (Figure 3B). This probability score of the malignancy of the local patches is used as an input feature in the final ensemble model.
Figure 3.
Generation and Performance of Patch Models
(A) Examples of false-positive mass.
(B) AUC of the mass local malignancy model.
(C) Using sliding windows to exhaustively find the patches with most calcification.
(D) AUC of the calcification segmentation model.
(E) Examples of false-positive calcifications.
(F) AUC of the detection of false-positive calcification patches model.
(G) Summary of stepwise improvement of the breast cancer prediction model.
Full-Resolution Calcification Models Give the Number of Microcalcifications in Local Patches
Given a screening digital mammogram, our end goal is to identify the maximal number of calcifications in a 1.8 cm × 1.8 cm (256 × 256 pixels) patch, because the presence of a cluster of microcalcifications is an important indicator of breast cancer. Specifically, from the top left corner of the image, 256 × 256 pixels were cropped out as the first patch, and the sliding window moved to the right for 128 pixels and captured another 256 × 256 patch until it reached the rightmost side (Figure 3C). If the rightmost patch could not have a clear cut, the window bounced back to make the right edge of the original image also to be the right edge of the patch (Figure 3C). The similar moving-window approach was applied vertically to generate multiple patches in a single mammogram.
Instead of using pixel counts, we counted the microcalcification dots. A pixel with all its adjacent pixels, as long as they are predicted by the U-Net to be positive, is counted as one dot. Therefore, one microcalcification may contain more than one pixel. The calcification model was trained using the sigmoid-transformed INbreast dataset. The INbreast dataset provided over 6,000 calcification annotations.33 Yet these annotations included large calcification areas and non-cancerous calcifications. Thus, we manually labeled all images for suspicious microcalcifications that are present in the breasts and are labeled to be positive of cancers. Furthermore, to exclude the microcalcifications that are unlikely to be cancers, we set up an upper limit as 35 pixels, with only the dots containing fewer pixels as valid counts. From the 410 INbreast images, we obtained 100,129 patches, including 98,808 negatives and 1,321 positives. During the training process, an oversampling operation was performed to increase the number of positive patches to match the amount of negatives. The training set was therefore balanced, containing approximately equal numbers of positive and negative patches.
We trained U-Nets to identify the microcalcifications, achieving an AUC of 0.95 on the testing dataset in classifying whether a patch contains microcalcification (Figure 3D). Of note, this AUC is a reflection of how accurately we can extract calcifications, rather than how accurate the global cancer detection method is, which will be discussed in later sections.
Microcalcifications exist in almost all breasts. However, large calcifications and calcifications regularly deposited in the milk ducts are unlikely to be cancers (Figure 3E). A separate segmentation model was thus trained to identify those false-positive calcification areas. In order to do so, we used the calcification models to make predictions on the DREAM training set Part 2, and separated the calcifications into true positives and false positives according to the global label of the breasts. False-positive calcification models, when examined by patches, had an AUC of 0.90 (Figure 3F). The calcification features in the eventual ensemble model were the predicted maximal calcification counts and areas in the patches within a breast (cut at probability of 0.5), masked by the false calcification predictions (if the pixel prediction value is bigger than 0.5). The calcification model performs at 0.75 AUC on the DREAM dataset when used by itself and evaluated with the global label of the breasts. This value is comparable with the AUC of 0.72 when we evaluated mass models alone, indicating that we detect cancers by each mechanism (calcification or mass) about an equal number of times.
The top two panels of Figure 3G summarize the performances of models and overall performances mentioned above: only using calcification models (model 1 and model 3) achieves an overall performance of 0.75, while only using mass models (model 2 and model 4) achieves an overall performance of 0.725.
Assembling Individual Features Reveals that Integrating Information from the Opposite Breast Improves Model Performance
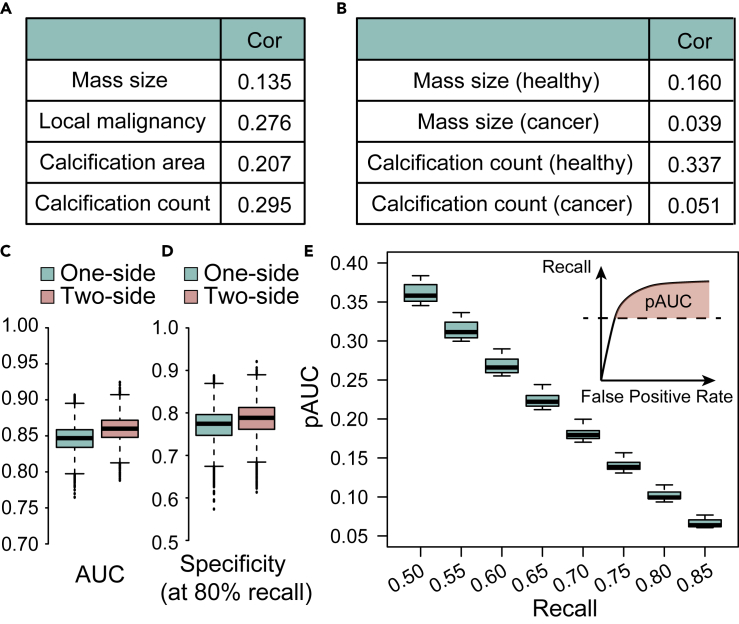
In clinical practice, a suspected abnormal region may be determined normal when a similar suspicious appearance occurs in the other breast of the patient. For example, scattered calcifications across both breasts are likely to be benign. Inspired by this phenomenon, we first examined whether correlation of various features exists between the two breasts across individuals. Intuitively, if two breasts are independent, correlations of 0 are expected. However, the correlation of predicted mass size between two breasts was 0.135, the correlation of local malignancy of the biggest mass 0.276, and the correlation of the maximal calcification number in a patch 0.295 (Figure 4A). These positive correlations indicate that if in one case both breasts have a lot of calcifications, this case is much less likely than the other case where a similar amount of calcifications only appeared in one of the breasts. Thus, taking into account the information from the other breast can be helpful.
Figure 4.
Evaluation of the Contribution of Symmetry Features
(A) Correlation of the features between left and right breasts extracted across different individuals.
(B) Correlation of the features between left and right breasts extracted across healthy individuals and cancer patients.
(C) AUC of 1-side and 2-side models.
(D) Specificity at 80% recall of 1-side and 2-side models.
(E) Partial AUC above different levels of recall of the 2-side model.
We next dissected whether such correlation changes on comparing the cancer population and the healthy controls. We found that for mass size, the correlation of healthy controls is 0.160, while that of cancer patients is 0.039; for maximal calcification numbers, the correlation of healthy controls is 0.337, while that of cancer patients is only 0.051. This result corroborates the hypothesis that asymmetry is more apparent in cancer patients (Figure 4B).
We thus constructed two models built with random forest, one using the features extracted from only the breast under study and the other one using both breasts, to predict the probability of cancer from breast mass area, local malignancy, calcification counts, and area. Taking in the features of both breasts can improve the overall performance measured by AUC from 0.844 to 0.856, although barely statistically significant (p = 0.0507, Figure 4C). The specificity at 80% recall improved from 0.768 to 0.780 (Figure 4D), which is lower than community-practice radiologists' specificity of 90.5% on this dataset.32 However, for both AUC and specificity this algorithm was a co-best performer, and an ensemble of this and other top algorithms with the radiologist's algorithm can further improve on the radiologist's performance.32 In Figure 4E, we plotted the partial AUC above different levels of recall. This result, together with the comparison results from the mass model (single breast versus paired breasts), supports the value of the information from the opposite breast in predicting breast cancers with deep learning.
The results of this part are also summarized in Figure 3G (bottom panel). When all the five models are combined together, using information from one breast leads to a performance of 0.844, while using information from two breasts leads to a performance of 0.856. As a whole, Figure 3G provides a summary of the stepwise improvement of the overall model performance.
Availability of Code and Docker Implementation
In order for the radiology community to use this algorithm in practice, we deployed the prediction part into a portable Docker (Figure 5). It is lightly weighted in terms of computations and can run on a computer without access to a graphics processing unit (GPU). The Docker consists of the following parts to address practical issues involved in mammogram analysis. First, the header file is read, and images with linear scale are transformed to sigmoid scale. Second, to eliminate the noise introduced by artifacts such as text labels, the program excludes the area outside of the breast by selecting only the largest connected component within the binary mask. Third, the deep-learning model takes in the processed image to extract features for mass areas, local malignancy, calcification counts, and calcification areas, which are then used to generate a highlighting map of the ROIs. Finally, the Docker ensembles all ROIs and generates a final prediction score, and visualizes the areas that lead to a positive prediction.
Figure 5.
Overview of Docker Deployment
We implemented and deployed the algorithms using the Docker system. Briefly, a Docker container is a lightweight virtual machine that contains not only the software but also the required libraries and environments to successfully run the software. This Docker container includes the image preprocessing steps, multiple neural network models, augmentation during model testing, feature extraction, and final prediction of breast cancer based on a mammogram.
Since INbreast data is already used in training and DREAM data do not allow distribution, we found another dataset, the breast cancer digital repository (BCDR).36 This dataset only has a very small number of cancer images, in .jpg format. Nonetheless, we used it to make several demonstration graphs shown in Figures 6 and S8. In the visualized image, calcification areas that are difficult to spot by the human eye can be identified by the algorithm, and false-positive areas can be identified and ignored by the model (Figures 6 and S8). Thus, the model will offer direct visualization of the suspicious areas as well as a prediction score. This could supplement the radiologist's diagnosis and help to identify subtle changes they may have missed.
Figure 6.
An Example of Model Output in the BCDR Database
Red regions in the visualized image indicate calcification areas. See also Figure S8.
Discussion
In this study, we present a machine-learning method that simultaneously detects and locates breast cancers on digital mammograms and gives a global classification score. In addition to image preprocessing and multiple customized deep CNN models, this model leverages the asymmetric information regarding a pair of breasts from the same subject to improve the detection accuracy.
In this work, we report a set of approaches to integrate two breast images into neural network models and demonstrate that pairing of the two images as two channels improves the model performance. Meanwhile, we report strong correlations between extracted features from two breasts across patients. Using features from both breasts achieved the highest predictive performance.
One important future work is to test this algorithm in a large-scale dataset that allows hand-labeling of the training data. The starting training set INbreast used in this study is relatively small, and was collected for known cancer patients rather than a screening population. INbreast was chosen as the starting point, as it was the only digital mammography database available to the public at the time, and DREAM's setup helps us to glean insight from a typical case where human subject-protected data do not allow direct annotation or observation. This is more challenging compared with the cases where the model developers are allowed to label the training images coming from the same population as the test. We envision that applying the same pipeline to a similar screening population can drastically improve the performance.
During the Digital Mammography DREAM Challenge, deep-learning methods were widely used by other leading teams around the globe, including single-shot multibox detector and Faster RCNN. We applaud the ingenuity of these methods in calculating losses on both bounding boxes of potential cancerous regions and the overall classification of ROIs. Similarly, in our analysis pipeline, we build both the segmentation and classification neural networks for masses and microcalcifications. A difference in our approach resides in the fact that pixel-level ROI maps are provided, whereas methods such as Faster RCNN focus on rectangular boxes of mass or calcification. Based on these pixel-level annotations, we further calculated features including the sizes of masses and numbers of calcifications, which could be useful for other clinical studies. This makes the model less like a black box and more useful when clinicians try to interpret the results for patients. However, the separation of ROI identification and classification was enforced mainly due to the limitation of GPU memory, which cannot fit two full-size mammographies. With the enlargement of GPU memory in the coming years, it is foreseeable that ROI identification and whole-breast classification can be seamlessly unified into a single network structure.
Experimental Procedures
Resource Availability
Lead Contact
Yuanfang Guan, gyuanfan@umich.edu.
Materials Availability
The study did not generate new unique reagents.
Data and Code Availability
INbreast data can be acquired from http://medicalresearch.inescporto.pt/breastresearch/index.php/Get_INbreast_Database. BCDR data can be acquired from https://bcdr.eu/information/about. Models used in this paper are implemented in Docker (https://www.synapse.org/#!Synapse:syn11313722).
Acknowledgments
This work was supported by NIH R35GM133346 and NSF#1452656.
Author Contributions
Y.G. conceived and implemented the algorithms. Y.G. and H.L. wrote the manuscript. Y.G. and Z.Z. carried out post-challenge analysis. X.W. and Y.G. prepared figures. All authors participated in manuscript editing.
Declaration of Interests
The authors declare no competing interests.
Published: September 21, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.patter.2020.100106.
Supplemental Information
References
- 1.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2019. Cancer J. Clin. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 2.Oeffinger K.C., Fontham E.T.H., Etzioni R., Herzig A., Michaelson J.S., Shih Y.-C.T., Walter L.C., Church T.R., Flowers C.R., LaMonte S.J. Breast cancer screening for women at average risk: 2015 guideline update from the American Cancer Society. J. Am. Med. Assoc. 2015;314:1599–1614. doi: 10.1001/jama.2015.12783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lehman C.D., Arao R.F., Sprague B.L., Lee J.M., Buist D.S.M., Kerlikowske K., Henderson L.M., Onega T., Tosteson A.N.A., Rauscher G.H. National performance benchmarks for modern screening digital mammography: update from the breast cancer surveillance consortium. Radiology. 2017;283:49–58. doi: 10.1148/radiol.2016161174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Siu A.L. Screening for breast cancer: U.S. preventive services task force recommendation statement. Ann. Intern. Med. 2016;164:279. doi: 10.7326/M15-2886. [DOI] [PubMed] [Google Scholar]
- 5.Chen Z., Strange H., Oliver A., Denton E.R.E., Boggis C., Zwiggelaar R. Topological modeling and classification of mammographic microcalcification clusters. IEEE Trans. Biomed. Eng. 2015;62:1203–1214. doi: 10.1109/TBME.2014.2385102. [DOI] [PubMed] [Google Scholar]
- 6.Chan H.-P., Wei J., Sahiner B., Rafferty E.A., Wu T., Roubidoux M.A., Moore R.H., Kopans D.B., Hadjiiski L.M., Helvie M.A. Computer-aided detection system for breast masses on digital tomosynthesis mammograms: preliminary experience. Radiology. 2005;237:1075–1080. doi: 10.1148/radiol.2373041657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wei J., Sahiner B., Hadjiiski L.M., Chan H.-P., Petrick N., Helvie M.A., Roubidoux M.A., Ge J., Zhou C. Computer-aided detection of breast masses on full field digital mammograms. Med. Phys. 2005;32:2827–2838. doi: 10.1118/1.1997327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Soltanian-Zadeh H., Rafiee-Rad F., Siamak Pourabdollah-Nejad D. Comparison of multiwavelet, wavelet, Haralick, and shape features for microcalcification classification in mammograms. Pattern Recognit. 2004;37:1973–1986. [Google Scholar]
- 9.Batchelder K.A., Tanenbaum A.B., Albert S., Guimond L., Kestener P., Arneodo A., Khalil A. Wavelet-based 3D reconstruction of microcalcification clusters from two mammographic views: new evidence that fractal tumors are malignant and Euclidean tumors are benign. PLoS One. 2014;9:e107580. doi: 10.1371/journal.pone.0107580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Görgel P., Sertbas A., Uçan O.N. Computer-aided classification of breast masses in mammogram images based on spherical wavelet transform and support vector machines. Expert Syst. 2015;32:155–164. [Google Scholar]
- 11.Singh N., Mohapatra A.G., Kanungo G. Breast cancer mass detection in mammograms using K-means and fuzzy C-means clustering. Int. J. Comput. Appl. Technol. 2011;22:15–21. [Google Scholar]
- 12.Patel B.C., Sinha G.R. An adaptive K-means clustering algorithm for breast image segmentation. Int. J. Comput. Appl. Technol. 2010;10:35–38. [Google Scholar]
- 13.Arikidis N.S., Karahaliou A., Skiadopoulos S., Korfiatis P., Likaki E., Panayiotakis G., Costaridou L. Size-adapted microcalcification segmentation in mammography utilizing scale-space signatures. Comput. Med. Imag. Graph. 2010;34:487–493. doi: 10.1016/j.compmedimag.2009.12.009. [DOI] [PubMed] [Google Scholar]
- 14.Duarte M.A., Alvarenga A.V., Azevedo C.M., Calas M.J.G., Infantosi A.F.C., Pereira W.C.A. Evaluating geodesic active contours in microcalcifications segmentation on mammograms. Comput. Methods Prog. Biomed. 2015;122:304–315. doi: 10.1016/j.cmpb.2015.08.016. [DOI] [PubMed] [Google Scholar]
- 15.LeCun Y., Bengio Y., Hinton G. Deep learning. Nature. 2015;521:436–444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 16.Litjens G., Kooi T., Bejnordi B.E., Setio A.A.A., Ciompi F., Ghafoorian M., van der Laak J.A.W.M., van Ginneken B., Sánchez C.I. A survey on deep learning in medical image analysis. Med. Image Anal. 2017;42:60–88. doi: 10.1016/j.media.2017.07.005. [DOI] [PubMed] [Google Scholar]
- 17.Taigman Y., Yang M., Ranzato M., Wolf L. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2014. Deepface: closing the gap to human-level performance in face verification; pp. 1701–1708.https://www.cv-foundation.org/openaccess/content_cvpr_2014/html/Taigman_DeepFace_Closing_the_2014_CVPR_paper.html [Google Scholar]
- 18.Szegedy C., Vanhoucke V., Ioffe S., Shlens J., Wojna Z. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2016. Rethinking the inception architecture for computer vision; pp. 2818–2826.https://www.cv-foundation.org/openaccess/content_cvpr_2016/html/Szegedy_Rethinking_the_Inception_CVPR_2016_paper.html [Google Scholar]
- 19.Ribli D., Horváth A., Unger Z., Pollner P., Csabai I. Detecting and classifying lesions in mammograms with Deep Learning. Sci. Rep. 2018;8:4165. doi: 10.1038/s41598-018-22437-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kooi T., Litjens G., van Ginneken B., Gubern-Mérida A., Sánchez C.I., Mann R., den Heeten A., Karssemeijer N. Large scale deep learning for computer aided detection of mammographic lesions. Med. Image Anal. 2017;35:303–312. doi: 10.1016/j.media.2016.07.007. [DOI] [PubMed] [Google Scholar]
- 21.Rodríguez-Ruiz A., Krupinski E., Mordang J.-J., Schilling K., Heywang-Köbrunner S.H., Sechopoulos I., Mann R.M. Detection of breast cancer with mammography: effect of an artificial intelligence support system. Radiology. 2019;290:305–314. doi: 10.1148/radiol.2018181371. [DOI] [PubMed] [Google Scholar]
- 22.Al-Masni M.A., Al-Antari M.A., Park J.-M., Gi G., Kim T.-Y., Rivera P., Valarezo E., Choi M.-T., Han S.-M., Kim T.-S. Simultaneous detection and classification of breast masses in digital mammograms via a deep learning YOLO-based CAD system. Comput. Methods Program. Biomed. 2018;157:85–94. doi: 10.1016/j.cmpb.2018.01.017. [DOI] [PubMed] [Google Scholar]
- 23.Shen L., Margolies L.R., Rothstein J.H., Fluder E., McBride R., Sieh W. Deep learning to improve breast cancer detection on screening mammography. Sci. Rep. 2019;9:12495. doi: 10.1038/s41598-019-48995-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yassin N.I.R., Omran S., El Houby E.M.F., Allam H. Machine learning techniques for breast cancer computer aided diagnosis using different image modalities: a systematic review. Comput. Methods Program. Biomed. 2018;156:25–45. doi: 10.1016/j.cmpb.2017.12.012. [DOI] [PubMed] [Google Scholar]
- 25.Lau T.K., Bischof W.F. Automated detection of breast tumors using the asymmetry approach. Comput. Biomed. Res. Int. J. 1991;24:273–295. doi: 10.1016/0010-4809(91)90049-3. [DOI] [PubMed] [Google Scholar]
- 26.Yang Q., Li L., Zhang J., Shao G., Zhang C., Zheng B. Computer-aided diagnosis of breast DCE-MRI images using bilateral asymmetry of contrast enhancement between two breasts. J. Digital Imag. 2014;27:152–160. doi: 10.1007/s10278-013-9617-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen B., Ma Z. 2008 International Workshop on Education Technology and Training 2008 International Workshop on Geoscience and Remote Sensing. Vol. 1. 2008. Automated image segmentation and asymmetry analysis for breast using infrared images; pp. 410–413. [Google Scholar]
- 28.Ericeira D.R., Silva A.C., de Paiva A.C., Gattass M. Detection of masses based on asymmetric regions of digital bilateral mammograms using spatial description with variogram and cross-variogram functions. Comput. Biol. Med. 2013;43:987–999. doi: 10.1016/j.compbiomed.2013.04.019. [DOI] [PubMed] [Google Scholar]
- 29.Zheng B., Sumkin J.H., Zuley M.L., Wang X., Klym A.H., Gur D. Bilateral mammographic density asymmetry and breast cancer risk: a preliminary assessment. Eur. J. Radiol. 2012;81:3222–3228. doi: 10.1016/j.ejrad.2012.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pisano E.D., Gatsonis C., Hendrick E., Yaffe M., Baum J.K., Acharyya S., Conant E.F., Fajardo L.L., Bassett L., D’Orsi C. Diagnostic performance of digital versus film mammography for breast-cancer screening. N. Engl. J. Med. 2005;353:1773–1783. doi: 10.1056/NEJMoa052911. [DOI] [PubMed] [Google Scholar]
- 31.Carney P.A., Miglioretti D.L., Yankaskas B.C., Kerlikowske K., Rosenberg R., Rutter C.M., Geller B.M., Abraham L.A., Taplin S.H., Dignan M. Individual and combined effects of age, breast density, and hormone replacement therapy use on the accuracy of screening mammography. Ann. Intern. Med. 2003;138:168–175. doi: 10.7326/0003-4819-138-3-200302040-00008. [DOI] [PubMed] [Google Scholar]
- 32.Schaffter T., Buist D.S.M., Lee C.I., Nikulin Y., Ribli D., Guan Y., Lotter W., Jie Z., Du H., Wang S. Evaluation of combined artificial intelligence and radiologist assessment to interpret screening mammograms. JAMA Netw. Open. 2020;3:e200265. doi: 10.1001/jamanetworkopen.2020.0265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moreira I.C., Amaral I., Domingues I., Cardoso A., Cardoso M.J., Cardoso J.S. INbreast: toward a full-field digital mammographic database. Acad. Radiol. 2012;19:236–248. doi: 10.1016/j.acra.2011.09.014. [DOI] [PubMed] [Google Scholar]
- 34.Ronneberger O., Fischer P., Brox T. U-Net: convolutional networks for biomedical image segmentation. Med. Image Comput. 2015;2015:234–241. [Google Scholar]
- 35.Çiçek Ö., Abdulkadir A., Lienkamp S.S., Brox T., Ronneberger O. 3D U-Net: learning dense volumetric segmentation from sparse annotation. In: Ourselin S., Joskowicz L., Sabuncu M.R., Unal G., Wells W., editors. Medical Image Computing and Computer-Assisted Intervention – MICCAI 2016 Proceedings Part II. Springer; 2016. pp. 424–432. [Google Scholar]
- 36.Moura D.C., Guevara López M.A. An evaluation of image descriptors combined with clinical data for breast cancer diagnosis. Int. J. Comput. Assist. Radiol. Surg. 2013;8:561–574. doi: 10.1007/s11548-013-0838-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
INbreast data can be acquired from http://medicalresearch.inescporto.pt/breastresearch/index.php/Get_INbreast_Database. BCDR data can be acquired from https://bcdr.eu/information/about. Models used in this paper are implemented in Docker (https://www.synapse.org/#!Synapse:syn11313722).