Abstract
The de novo synthesis of fatty acids has emerged as a therapeutic target for various diseases including cancer. Since cancer cells are intrinsically buffered to combat metabolic stress, it is important to understand how cells may adapt to loss of de novo fatty acid biosynthesis. Here we use pooled genome-wide CRISPR screens to systematically map genetic interactions (GIs) in human HAP1 cells carrying a loss-of-function mutation in FASN, whose product catalyzes the formation of long-chain fatty acids. FASN mutant cells show a strong dependence on lipid uptake that is reflected in negative GIs with genes involved in the LDL receptor pathway, vesicle trafficking, and protein glycosylation. Further support for these functional relationships is derived from additional GI screens in query cell lines deficient for other genes involved in lipid metabolism, including LDLR, SREBF1, SREBF2, ACACA. Our GI profiles also identify a potential role for the previously uncharacterized gene LUR1/C12orf49 in exogenous lipid uptake regulation through modulation of SREBF2 signalling in response to lipid starvation. Overall, our data highlight the genetic determinants underlying the cellular adaptation associated with loss of de novo fatty acid synthesis and demonstrate the power of systematic GI mapping for uncovering metabolic buffering mechanisms in human cells.
INTRODUCTION
It has long been recognized that cancer cells exploit lipid metabolism to fuel their proliferative demands and support oncogenic signalling. Notably, alterations in lipid metabolism, including the uptake of lipids and/or synthesis of fatty acids, are not only recognized hallmarks of cancer, but also occur commonly in diverse pathologic states such as fatty liver disease and metabolic syndrome, underscoring the importance of understanding this metabolic process1. De novo fatty acid synthesis in particular has gained significant traction as a targetable pathway following observations that overexpression of FASN, which encodes fatty acid synthase and catalyzes the formation of long chain fatty acids, and ACACA, which codes for Acetyl-CoA Carboxylase Alpha and acts directly upstream of FASN, are associated with decreased survival rates for numerous solid malignancies2–6. Efforts to develop and translate small molecule inhibitors of FASN (e.g. TVB-2640) have helped validate this enzyme as a targetable liability in cancer7,8, and have led to several clinical trials (e.g. NCT02223247, NCT02948569, NCT03179904, NCT02980029). Given that metabolic pathways are highly buffered to deal with environmental change, genetic screening approaches are a powerful strategy to reveal metabolic regulatory mechanisms that underscore metabolic redundancy, cross-talk and plasticity9,10. An understanding of how cells adapt to perturbation of de novo fatty acid synthesis could help identify new targetable vulnerabilities that may inform novel therapeutic strategies or biomarker approaches.
Mapping genetic interaction (GI) networks provides a powerful approach for identifying the functional relationships between genes and their corresponding pathways. The systematic exploration of pairwise GIs in model organisms revealed that GIs often occur among functionally related genes and that GI profiles organize a hierarchy of functional modules11,12. Thus, GI mapping has become an effective strategy for identifying functional modules and annotating the roles of previously uncharacterized genes. Model organism GI mapping has also provided insight into the mechanistic basis of cellular plasticity or phenotypic switching that occurs as cells evolve within their environments13,14. Accordingly, the insights gained through systematic interrogation of GIs have fuelled significant interest to leverage these approaches towards functionally annotating the human genome.
Recent technological advances using CRISPR-Cas enable the systematic mapping of GIs in human cells15,16. Here, we explore genome-wide GI screens within the context of human query mutant cells defective for de novo fatty acid synthesis. We systematically mapped genome-wide GI profiles for six genes involved in lipid metabolism, revealing cellular processes that pinpoint genetic vulnerabilities associated with defects in de novo fatty acid synthesis. In particular, negative GIs with known fatty acid synthesis genes tend to identify other genes that are associated with this process, including a previously uncharacterized gene C12orf49 (LUR1), which appears to function as a regulator of exogenous lipid uptake. Collectively, our data support the strategy of systematically mapping digenic interactions using knockout query cell lines for identifying buffering mechanisms for a given bioprocess (i.e. lipid metabolism).
RESULTS
Systematic identification of genetic interactions for de novo fatty acid synthesis
De novo fatty acid synthesis is a multi-step enzymatic process that converts cytosolic acetyl-CoA, malonyl-CoA, and NADPH to palmitate. Palmitate can be used directly or further elongated and/or undergo desaturation to form alternate lipid species. To systematically identify GIs associated with this metabolic process, we performed genome-wide CRISPR screens in co-isogenic cell lines either wild-type or deficient in FASN, a de novo fatty acid synthesis enzyme that is frequently overexpressed in malignancies6,17 (Fig. 1a). We chose the human near-haploid cell line HAP1 as a model system, given the relative ease for generating knockout (KO) mutations in this background18. We first validated our clonal FASN-KO cells by confirming loss of FASN protein levels by western blot (Extended Data Fig. 1a). We also performed targeted metabolite profiling of our parental HAP1 and FASN-KO cells, which revealed a significant increase in the FASN substrate malonyl-CoA in the FASN-KO cells, demonstrating their suitability as a model system for defective de novo fatty acid synthesis (Extended Data Fig. 1b).
Figure 1. Genome-scale identification of digenic interactions with FASN.
(a) Schematic outline for the identification of genetic interactions in co-isogenic HAP1 cell lines. FASN knockout (KO) and wildtype parental cells are infected with a lentiviral genome-wide CRISPR gene KO library (TKOv3) and gRNA abundance is determined using Illumina sequencing of guide RNA (gRNA) sequences amplified from extracted genomic DNA from the starting cell population (T0) and end time point (day 18, T18) of the screen.
(b) Schematic outline for scoring quantitative genetic interactions (qGI) across co-isogenic query cell lines. First, the log2 fold-change (LFC) for each gRNA comparing sequence abundance at the starting (T0) and end time point (T18) in a given query KO or wildtype (WT) cell population are computed. Differential LFC for each gRNA are then estimated by comparing its LFC in WT and query KO cells. A series of normalization steps and statistical tests are applied to these data to generate gene-level qGI scores and false discovery rates (see Methods). The LFC scatterplot (bottom left graph) visualizes differential fitness defects in a specific query KO and WT cells, whereas the volcano plot (bottom right graph) visualizes qGI scores for a specific query.
(c) Replicate analysis of gene loss of function fitness phenotypes in FASN screens. Scatter plots of LFC associated with perturbation of 17,804 individual genes derived from a FASN query KO mutant screen conducted in triplicate. Reproducibility of fitness effects were determined by measuring Pearson correlation coefficients (r) between all possible pairwise combinations of FASN-KO replicate screens.
(d) Evaluation of FASN quantitative genetic interactions (qGIs). qGI scores were measured by comparing the LFC for every gene represented in the TKOv3 gRNA library in a FASN-KO with those observed in a WT cell line, as described. Scatter plots show FASN genetic interactions (qGI scores) derived from all possible pairwise combinations of three biological replicate screens. The Pearson correlation coefficient (r), based on comparison of all qGI scores (r shown in grey, calculated on all the grey and purple data points in the scatter plots), or only genetic interactions that exceed a given significance threshold (|qGI| > 0.5, FDR < 0.5) in both screens (purple).
(e) Validation of FASN negative genetic interaction. Bar plots depict the ratio of WT and FASN-KO (2 independent clones, c1 and c2) cells carrying a gRNA targeting SLCO4A1, LDLR or C12orf49, which all showed a negative interaction with FASN, compared to a gRNA targeting AAVS1 (intergenic control). Experiments were performed with three independent gRNAs targeting each genetic interaction screen hit. All data are represented as means ± standard deviation; n = 3 (LDLR) or 4 (SLCO4A1, C12orf49) biologically independent experiments, one-way ANOVA (Kruskall-Wallis test).
(f) FASN negative and positive genetic interactions. A scatter plot illustrating the fitness effect (LFC) of 450 genes in a FASN-KO vs. WT parental HAP1 cell line, which exhibited a significant genetic interaction in at least 2 out of 3 FASN-KO replicate screens (|qGI| > 0.5, FDR < 0.5). Negative (blue) and positive (yellow) FASN genetic interactions are shown. Node size corresponds to the mean absolute qGI score derived from three biologically independent replicate screens. Genes with mean absolute qGI score > 1.5 as well as selected negative interactions involving genes with established roles in lipid metabolism are indicated. Inset displays scatter plot of the fitness effect (LFC) of all 17,804 genes targeted by the TKOv3 gRNA library in a FASN-KO vs. WT parental HAP1 cell line. The color indicates density of genes.
(g) Enrichment for Gene Ontology (GO) molecular function, GO bioprocesses and Reactome terms among genes that exhibited a significant negative genetic interaction with FASN (significant in at least 2 FASN replicates, |qGI| > 0.5, FDR < 0.5). The number of genes overlapping a particular term and term size are indicated next to each bar, respectively. The greyscale color legend for p-values is indicated on the right; p-values were calculated using gProfileR69.
(h) Schematic depicting the function of selected FASN negative interactions known to be involved in lipid uptake and homeostasis pathways (red), vesicle transport (black) and glycosylation (blue).
To map FASN GIs, we performed genome-wide CRISPR screens using the sequence optimized TKOv3 gRNA library19 in both the FASN-KO query cell line and control wild-type (WT) HAP1 cells, and we compared the relative abundance of individual gRNAs between the screen start (T0) and end (T18) time points (Fig. 1a–b). The relative abundance of gRNAs targeting each of ~18,000 genes in WT cells provides an estimate of single mutant fitness, whereas the relative abundance of gRNAs in a query mutant cell line provides an estimate of double mutant fitness. Since mutant phenotypes can strongly depend on culture conditions20 and most standard cell culture media contains supra-physiological nutrient levels that could mask phenotypic effects of perturbing certain metabolic pathways, we performed our screens utilizing media conditions containing the minimum amounts of glucose and glutamine required to sustain proliferation of HAP1 cells; termed limiting media (Extended Data Fig. 1c, see Methods).
We developed a quantitative genetic interaction (qGI) score that measures the strength and significance of genetic interactions by comparing the relative abundance of gRNAs in a given query mutant cell line to the relative abundance of gRNAs targeting the corresponding genes in an extensive panel of 21 genome-wide WT HAP1 screens (Fig. 1b, see Methods). In this context, negative interactions are identified as genes whose corresponding gRNAs exhibit significantly decreased abundance in a mutant KO background relative to the control WT HAP1 cell line, whereas positive interactions reflect genes with increased gRNA abundance in a mutant cell line relative to the parental line. The qGI score represents mean-summarized gRNA-level interactions after disagreeing reagents had been removed (Fig. 1b, see Methods).
We performed three independent genome-wide, GI screens using our FASN-KO query mutant cell line. Because GIs rely on accurate measurement of single and double mutant phenotypes, we first examined the reproducibility of our single and double mutant fitness measurements (see Methods). We observed a strong agreement of single gene fitness effects (LFC) among 21 replicate WT HAP1 (r > 0.87) (Extended Data Fig. 1d) and double mutant fitness effects derived from independent FASN-KO replicate screens (r > 0.89) (Fig. 1c). Moreover, all three FASN screens robustly discriminated a set of reference essential genes from non-essential genes (Extended Data Fig. 1e–f).
The identification of qGI scores depends on comparison of single mutant fitness measurements in a WT HAP1 cell screen and double mutant fitness measurements in a query mutant screen, both of which have inherent variability associated with them; therefore, the reproducibility of qGIs is expected to be more challenging than the measurement of either single or double mutant fitness phenotypes. Indeed, modest agreement was observed between qGI scores of the three FASN-KO replicate screens prior to filtering for significant interactions (pairwise r = 0.29 to 0.44) (Fig. 1d). The pairwise correlation between replicate screens increased substantially when we considered GIs found to be significant (|qGI| > 0.5, FDR < 0.5) in at least one (r = 0.52–0.69) or two (r = 0.86–0.94) FASN-KO replicate screens (Fig. 1d, Extended Data Fig. 1g–h, Supplementary Table 1).
Leveraging all 3 FASN-KO replicates, we developed a reproducibility score that measures each gene’s contribution to the covariance within two replicate screens and summarizes the resulting values across all available screen pairs (replicate 1–2, 1–3, 2–3) (Methods, Supplementary Table 1). This analysis confirms that both the strongest positive and negative qGI scores were highly reproducible across independent screens (Extended Data Fig. 1i). In particular, the most reproducible negative GIs with FASN were interactions with SLCO4A1, PGRMC2, LDLR, RABL3 and C12orf49 (Extended Data Fig. 1i, Supplementary Table 1). We tested three of these top five strongest negative GIs by independent validation assays and confirmed all three, examining WT and FASN-KO HAP1 cells expressing gRNAs against SLCO4A1, LDLR and C12orf49 (Fig. 1e, Extended Data Fig. 1j).
To generate an aggregate set of FASN GIs, we mean-summarised qGI scores across the three replicate screens (Fig. 1f, Supplementary Table 2). At a pathway level, significant negative GIs (qGI < −0.5, FDR < 0.5) with FASN were strongly enriched for genes annotated with roles in protein glycosylation, vesicle transport and cholesterol metabolism (FDR <0.05) (Fig. 1g, Supplementary Table 3). In the global yeast genetic network negative GIs often connect functionally related genes11,21, and we observed a similar general trend for the FASN negative GIs. For example, the FASN negative GIs included genes with established roles in the uptake, transport, and breakdown of low density lipoprotein (LDL), a major extracellular source of lipids, including the LDL receptor (LDLR) itself and its coreceptor adaptor protein (LDLRAP1). We also observed negative GIs between FASN and the transcription factor SREBF2, which controls expression of LDLR, as well as SCAP, MBTPS1 and MBTPS2, all of which are important for the activation and nuclear translocation of SREBF2 upon cholesterol depletion (Fig. 1h). Moreover, we observed negative GIs with additional lipid metabolic processes such as cholesterol biosynthesis (ACAT2), genes functioning in long chain fatty acid activation and β-oxidation (ACSL1, ACSL3), and vesicle trafficking genes (RAB18/10/1A, RABGEF1, RAB3GAP2/1) (Fig. 1h, Extended Data Fig. 1k), as well as a positive GI with the gene encoding stearoyl-CoA desaturase (SCD), the product of which catalyses the rate-limiting step in the biosynthesis of monounsaturated fatty acids. SCD acts downstream of FASN in the elongation and desaturation of fatty acids, and loss of SCD seems to be buffered by the upstream perturbation of fatty acid synthesis.
The FASN screen also highlighted an enrichment for genes functioning in protein N-linked glycosylation (e.g. ALG3/8/9/12, MOGS, DOLPP1, PRKCSH, MGAT2) (Fig. 1g–h, Extended Data Fig. 1k). Interestingly, the hexosamine biosynthetic and N-linked glycosylation pathways have been implicated in facilitating lipid accumulation from environmental sources through direct modulation of N-glycan branching on fatty acid transporters, possibly explaining the strong GIs we observe22. N-linked glycosylation is also known to play an important role in the activity of LDLR and activation of the SREBP transcriptional programs, providing a potential explanation for the interaction between loss of FASN and the glycosylation pathway23,24. Currently, we cannot exclude that glycosylation of additional targets may play a role in the adaptation to loss of de-novo fatty acid biosynthesis but nonetheless, our data highlights a role for N-linked glycosylation in fatty acid biosynthesis. Finally, we observed a significant negative GI between FASN and SLCO4A1 (Fig. 1f, Extended Data Fig. 1i). SLCO4A1 encodes a member of the organic anion-transporting polypeptides (OATPs), which can transport a wide range of structurally unrelated compounds including hormones, bile acids and lipid species (prostaglandins)25. Which and how these compounds may buffer loss of FASN, or whether SLCO4A1 may be involved in the transport of additional metabolites, awaits further investigation. To summarize, these results suggest that in the absence of cell autonomous de novo fatty acid synthesis, cells depend on uptake and breakdown of lipids from the environment or the synthesis of sterols, with our data illuminating the genetic determinants of how cells rewire to meet the demand for lipids in proliferating cells.
Expanding the genetic interaction landscape of de novo fatty acid synthesis
To better understand the GI landscape of de novo fatty acid synthesis, we next performed pooled genome-wide CRISPR screens using the TKOv3 library in five additional co-isogenic cell lines harbouring genetic KO of genes that exhibited significant negative GIs with our FASN-KO query, including LDLR, C12orf49 and SREBF2 (Supplementary Table 2), as well as two genes that did not show a negative GI with FASN, including SREBF1, which regulates the expression of FASN and other de novo fatty acid genes, and ACACA, which functions in the same pathway and immediately upstream of FASN (Fig. 2a)6,17,26. Each of these five query gene screens was performed in technical triplicate (i.e. parallel cultures from a common infection). Since these additional GI screens were performed under the same conditions as we used for the FASN-KO screens, we applied the same confidence threshold on the derived qGI scores (|qGI| > 0.5, FDR < 0.5; Methods) (Fig. 2b–f, Extended Data Fig. 2a–b, Supplementary Table 2). At this confidence threshold, we estimated a per-screen false discovery rate of ~0.3 and a false negative rate of ~0.6 (Methods; Extended Data Fig. 1l). Overall, 169 (36%, p < 2e-55) and 69 (49%, p < 2e-109) of the genes that interacted with FASN in two or three screens, respectively, also interacted with at least one of the five queries (Fig. 2g, Supplementary Table 2).
Figure 2. Querying five additional lipid metabolism genes for digenic interactions.
(a) Schematic diagram showing key steps in fatty acid metabolism. The genes encoding the proteins mediating these key steps, which are also query genes for genetic interaction screens described in the main text, are labelled in red.
(b-f) Volcano plots showing qGI scores and associated significance (-log10 p-value) for the 17,804 genes targeted by the TKOv3 gRNA library in the (b) LDLR-KO, (c) SREBF2-KO, (d) ACACA-KO, (e) SREBF1-KO and (f) C12orf49-KO screens. Colored dots indicate genes that meet the standard threshold of |qGI| > 0.5, FDR < 0.5, where positive GIs are indicated in yellow and negative GIs in blue. The dot size is proportional to both qGI and FDR, calculated as described in the Methods. Genes with |qGI| scores > 1.5 as well as selected top negative GI hits associated with lipid metabolism, citrate synthesis and transport are indicated.
(g) Heatmap showing overlapping genetic interactions across the six interrogated queries. Heatmap displaying genes (x-axis) significantly interacting with FASN across all three FASN replicate screens and at least one additional screened query genes (y-axis) (|qGI| > 0.5, FDR < 0.5). Positive and negative qGI scores are indicated in yellow and blue, respectively. The FASN qGI is represented as the mean qGI from the three FASN screens (same data as in Fig. 1f).
We next analyzed the functional enrichment across all GIs identified by our fatty acid synthesis-related query screens. While we did not detect strong functional enrichment amongst the positive GIs in our data set, we observed a clear 5-fold enrichment of negative GIs for genes annotated to functionally relevant pathways, which were defined by the metabolism-focused HumanCyc standard27 (Extended Data Fig. 2c). We further quantified enrichment for pathways annotated at different levels of the HumanCyc database hierarchy, including gene sets corresponding to general metabolic reaction categories, sub-categories, and finally specific metabolic pathways (Supplementary Table 4). At the most general level of the HumanCyc pathway hierarchy, negative GIs from all six genome-wide screens were most enriched for genes annotated to the biosynthesis and macromolecule modifications pathway categories (Fig. 3a). Further analysis of these terms at a more specific level of the HumanCyc hierarchy (i.e. sub-category level), we found that genes exhibiting negative GIs were associated with functions related to the roles of our six query genes, including fatty acid, lipid and carbohydrate biosynthesis (Fig. 3b, Extended Data Fig. 3a). At a more refined level of functional specificity within the fatty acid and lipid biosynthesis pathway, we found that each query gene was associated with a significant enrichment for negative GIs with functionally-related genes of distinct pathways. For example, the LDLR GI profile includes negative GIs with genes in the cholesterol/epoxysqualene biosynthesis pathway (i.e. HMGCS1, MSMO1, HMGCR, FDFT1, NSDHL, HSD17B7, SQLE, HSD17B7, ACT2, SQLE, LSS) and the ACACA, LDLR and SREBF2 GI profiles include negative GIs with fatty acid elongation and biosynthesis pathway genes (FASN, ACACA, OXSM) (Fig. 3c–d). Notably, the FASN GI profile, and to a lesser extent the ACACA and LDLR GI profiles, revealed negative GIs with pathways and genes involved in N-glycosylation initiation (ALG6, ALG13, ALG11, ALG1, ALG2, ALG8, ALG5, ALG3, ALG12, ALG9), processing (MOGS, PRKCSH), dolichol monophosphate mannose synthase activity (DPM2, DPM3, DPM1), and glycan transfer (STT3A, STT3B) (Fig. 3c, e, Supplementary Table 4).
Figure 3. Genetic interactions reveal multiple levels of functional enrichment.
(a) Dot plot of normalized pathway enrichment scores on the HumanCyc category level, calculated from qGIs across all six query genes (FASN, C12orf49, LDLR, SREBF2, ACACA, SREBF1). A GI is identified for a query-library pair if the |qGI| > 0.5 and FDR < 0.5. Enrichment for positive (yellow) and negative (blue) GIs is tested in each of the 10 HumanCyc main pathway categories using a hypergeometric test and considering all 17,804 genes targeted in the TKOv3 library as the universe. Enrichment with p-value < 0.05 are blue (negative GI) and yellow (positive GI). Dot size is proportional to the fold-enrichment in the indicated categories and specified in the legend. Categories indicated in bold are further expanded in part (b) and in Extended Data Fig. 3a.
(b) Dot plot of normalized pathway enrichment of GIs on a sub-category level, calculated as described in part (a), except that sub-categories were examined inside the Biosynthesis and Macromolecule Modification HumanCyc branches. Enrichment with p-value < 0.05 are blue (negative GI) and yellow (positive GI). Dot size is proportional to the fold-enrichment in the indicated categories and specified in the legend. Categories indicated in bold text are further expanded in part (c).
(c) Matrix dot plot of pathway enrichments of GIs for the fatty acid and lipid biosynthesis and protein modification sub-categories. Dots show positive (yellow) or negative (blue) z-transformed qGI scores summarized at a pathway-level. qGI scores were first z-score transformed at a gene-level for each genome-wide query screen separately. Then, a mean z-score was calculated for each pathway for a given query screen. Dot size corresponds to the absolute z-transformed mean qGI score, grey dots represent |z| < 0.5. Pathways marked with an asterisk are annotated to both protein modification and carbohydrate biosynthesis pathways. Bold pathways are shown in (d-e). Pathways were displayed if they shared an absolute z-score larger than 1.5 with any query gene.
(d-f) Gene-level heatmaps for genes involved in enriched pathways. qGI scores between query genes and all genes from the selected pathways. Positive and negative qGI scores are indicated in yellow and blue, respectively.
Our survey of GIs related to perturbation of de novo fatty acid synthesis or exogenous fatty acid uptake pathways provided unique insight into the genetic regulation of these processes. Specifically, for the SREBF2 screen, while we observed negative GIs with lipid uptake genes such as LDLR and LDLRAP1 (Fig. 3f, Supplementary Table 2), none were observed with the cholesterol biosynthesis pathway (Fig. 3d, 2d). This observation is consistent with SREBF2 being the predominant transcriptional regulator of cholesterol homeostasis26; its perturbation does not further reduce cellular fitness in cells deficient for cholesterol biosynthesis. In addition, we also detected a strong positive GI between SREBF2 and TFAP2C (Fig. 2d). Indeed, theTFAP2 transcription factor family has recently been proposed as a ‘master’ regulator of lipid droplet biogenesis28, with our data suggesting that reduced sequestration of lipids into lipid droplets may benefit SREBF2-KO cells to mitigate lipid starvation.
In contrast, SREBF1 did not show enrichment for GIs for either the cholesterol or fatty acid synthesis pathways (Fig. 3c, Supplementary Table 2). Instead, this query was found to show only a strong reciprocal negative GI with its paralog SREBF2, highlighting the functional redundancy between the paralog pair (Fig. 2e, Supplementary Table 2) and suggesting that SREBF2 may regulate some of the transcriptional targets of SREBF1 as previously described26,29. Furthermore, the imbalanced number of GIs between SREBF1 and SREBF2 may point towards asymmetric paralog evolution, whereby duplicated genes gain or lose functional roles at different rates while maintaining partially redundant functions, a process previously observed in yeast and human cells30–32.
A novel role for C12orf49 in lipid biosynthesis
One of the strongest negative GIs identified in both the FASN and the ACACA profiles involved the uncharacterized gene C12orf49, suggesting that this gene may have a role in lipid metabolism (Fig. 1f, 2c, Supplementary Table 2). C12orf49 is a 23.5 kDa protein that is part of the UPF0454 family of uncharacterized proteins, contains an N-terminal transmembrane sequence, a single uncharacterized DUF2054 domain of approximately 200 amino acid residues, 14 conserved cysteines (three of which are annotated to form CC-dimers), and a predicted glycosylation site33 (Extended Data Fig. 4a). In some plant proteins, the uncharacterized UPF0454 is found in juxtaposition to a glycosyltransferase domain and thus may be targeted into the lumen of the ER or Golgi34. By extension, the bulk of the C12orf49 protein may reside in the lumen of the ER or Golgi. In addition, C12orf49 is ubiquitously expressed across tissues and cell lines (http://www.proteinatlas.org )35. Notably, expression of C12orf49 is associated with differential prognoses on univariate analysis of TCGA data across multiple tumor types, including kidney, breast, liver and sarcoma36 (Extended Data Fig. 4b–e; p < 0.05), which further motivated us to study the functional role of this previously uncharacterized gene.
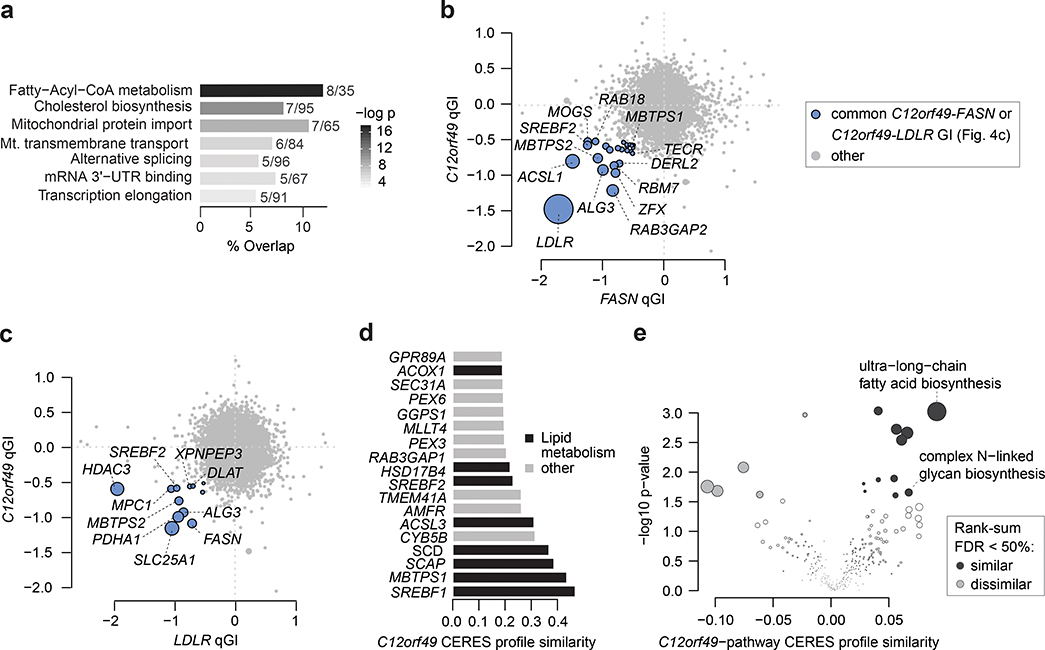
Genetic interactions derived from a genome-wide screen using a C12orf49-KO query cell line further supported a role for this gene in lipid biogenesis. Consistent with the results described above, C12orf49 showed a strong negative GI with both FASN and ACACA (Fig. 2f). C12orf49 also showed negative GIs with LDLR, ACSL1 (i.e. encoding acyl-CoA synthase), SLC25A1 (i.e. encoding mitochondrial citrate transporter), SCD and SREBF2, further supporting a role for this gene in fatty acid biosynthesis (Fig. 2f). Consistently, C12orf49 negative GIs were enriched for genes involved in fatty acid metabolism, cholesterol biosynthesis and additional metabolic pathways (FDR <0.05) (Fig. 4a, Supplementary Table 3). Moreover, as observed for the FASN GI profile, C12orf49 negative GIs involved genes functioning in vesicle-mediated trafficking and endocytosis, including RAB3GAP2, RABIF, RAB18, VPS18, VPS419 and VPS39 (Supplementary Table 2). Beyond vesicle trafficking, many of the genes that showed a negative GI with C12orf49 also displayed negative GIs with other query genes in our lipid metabolism panel (e.g. LDLR, ALG3, ASCL1, MBTPS2, SLC25A1, PDHA1), supporting the functional relatedness of these genes (Fig. 4b–c, Extended Data Fig. 4f–h). Thus, our lipid metabolism GI map strongly implicates C12orf49 as playing a functional role in lipid metabolism.
Figure 4. C12orf49 genetic interaction profile suggests a functional role in lipid metabolism.
(a) Bar plot depicting pathway enrichment of negative genetic interactions with C12orf49 (|qGI| > 0.5, FDR < 0.5) using GO molecular functions, GO bioprocesses and Reactome standards. Significantly enriched gene sets (p < 0.05, maximum term size 100). Bars depict mean percentage overlap with the indicated term, and the numbers next to each bar indicate the number of genes overlapping a particular term and term size, respectively. The greyscale color legend for p-values is indicated on the right. The greyscale color legend for p-values is indicated on the right; p-values were calculated using gProfileR69.
(b) Scatter plot of C12orf49 and FASN qGIs depicting GI overlap between C12orf49 and FASN qGI scores. FASN qGI scores are represented as the mean between three independent screens. A common negative GI is called if it is significant (qGI < −0.5, FDR < 0.5) in the C12orf49-KO screen and significant in 2 of 3 FASN-KO screens (indicated in blue). The top 10 strongest common GIs, lipid metabolism and vesicle trafficking genes are labelled.
(c) Scatter plot of C12orf49 and LDLR qGIs depicting GI overlap between C12orf49 and LDLR qGI scores. A common negative GI is called if it is significant (qGI < −0.5, FDR < 0.5) in both screens (indicated in blue). The top 10 strongest common GIs and lipid metabolism genes are labelled.
(d) Bar plot indicating the C12orf49 profile similarity across genome-wide DepMap CRISPR/Cas9 screens. Similarity (i.e. co-essentiality) was quantified by taking all pairwise gene-gene Pearson correlation coefficients of CERES score profiles across 563 screens (19Q2 DepMap data release). The top 18 out of 17,633 gene profiles most similar to C12orf49 are shown. Genes associated with lipid metabolism are indicated in black.
(e) Volcano plot of pathway enrichment for C12orf49 co-essential genes. C12orf49 co-essentiality profile scores for all 17,634 genes represented in the DepMap were mean-summarized by pathway as defined in the HumanCyc standard27. Tendencies towards pathway-level similarity (co-essentiality) and dissimilarity (exclusivity) with C12orf49 were tested using a two-sided Wilcoxon rank-sum test followed by multiple hypothesis correction with the Benjamini and Hochberg procedure.
To further confirm the predictions about C12orf49’s function based on our HAP1 GI data, we also examined publicly available data from the 19Q2 DepMap release and observed that C12orf49 is essential for fitness in 120 out of 563 cell lines with highest dependencies observed for lung, ovarian, pancreatic, colon and bile duct origins37,38. Other genes that shared similar cell line essentiality profiles to C12orf49 included SREBF1, SREBF2, MBTPS1, SCAP, SCD and ACSL3 (Fig. 4d, Extended Data Fig. 4i). The association of C12orf49 with lipid metabolism genes was corroborated by a pathway enrichment analysis of the co-essentiality profiles, which revealed strong enrichment for genes annotated to ultra-long-chain fatty acid biosynthesis (Fig. 4e, Extended Data Fig. 4j). Furthermore, we found that cell lines that depend on C12orf49 more frequently had missense mutations in FASN (FDR < 20%). Interestingly, germline variants in C12orf49 have also been reported to associate with serum lipid abnormalities in high-density lipoprotein (HDL) in a multi-ethnic cohort of the Million Veteran Program, further supporting a role for this gene in lipid metabolism39. Overall, these observations support a novel function for C12orf49 in lipid metabolism that is conserved across diverse cell types.
C12orf49 is a novel regulator of lipid uptake
Given the strong dependency on exogenous lipid uptake for FASN-deficient cells, we hypothesized a role for C12orf49 in this biological process. To explore this, we measured uptake of labelled LDL particles, which represent one of the major sources of extracellular lipids, upon serum or lipid starvation across several HAP1 KO lines. As expected, loss of LDLR resulted in abolishment of LDL-staining, while FASN-KO cells displayed increased uptake of exogenous lipid (Fig. 5a, Extended Data Fig. 5a). In contrast, loss of C12orf49 caused a significant reduction of LDL uptake, which was rescued by the exogenous expression of C12orf49 (Fig. 5a, Extended Data Fig. 5a). This phenotype was also observed in SREBF1- and SREBF2-deficient cells. Notably, this reduction of LDL was not secondary to a generalized impairment in receptor-mediated endocytosis, as loss of C12orf49 did not impair uptake of labelled transferrin (Extended Data Fig. 5b). We further confirmed a strong reduction in LDL uptake in C12orf49 KO cells upon lipoprotein starvation (Extended Data Fig. 5c). Overall, these results support the hypothesis that C12orf49 participates in lipid homeostasis through regulation of lipid uptake.
Figure 5. C12orf49 shuttles between ER and Golgi and regulates lipid uptake through modulation of SREBP2 processing.
(a) Bar plots showing the results of low density lipoprotein (LDL) uptake assays in the indicated cells using the Dil-LDL probe. All data are represented as means ± standard deviation; n = 4 (SREBF1, SREBF2), 6 (LDLR, WT + C12orf49-V5, C12orf49 + C12orf49-V5) or 8 (WT, FASN, C12orf49) independent biological replicates; one-way ANOVA.
(b) Schematic outlining proximal protein capture using BioID mass spectrometry analysis (upper panel) and analysis of subcellular localization of C12orf49 BioID preys (lower panel). Barplots depicting the fraction of proteins localizing to indicated cellular compartments for preys captured with N-terminal (grey) or C-terminal (black) miniTurbo-tagged C12orf49 in 293 cells. The inset shows a schematic representation of the predicted topology and orientation of C12orf49 with respect to the cytoplasm and ER.
(c) Immunofluorescence microscopy analysis of C-terminal V5-tagged C12orf49 in HAP1 cells under normal (left) or serum-starved (right) growth condition. C12orf49-V5 localization is shown in green, GOLGA2 is a marker of the Golgi apparatus and shown in red, and DAPI (blue) marks the nuclei. Scale bars correspond to 10 μm. Representative data from 2 independent biological replicates.
(d) Scatter plot displaying the specificity of detected preys captured with BioID. Average spectral counts of preys captured in proximity to N-terminal miniTurbo BirA-tagged C12orf49 exposed to serum starvation are plotted against their specificity across hundreds of baits listed in the Human Cell Map BioID data set (humancellmap.org)41 (left). The inset shows a comparison of the average spectral counts for preys involved in the regulation of SREBPs across the different miniTurbo-tagged constructs (i.e. N- and C-terminal) and growth conditions (i.e. normal and serum-starved) (right). BFDR was calculated using using SAINTexpress62 v3.6.1; n = 3 biologically independent experiments.
(e) Bar plots indicating FPKM expression values from RNA sequencing data for LDLR and LDLRAP1 in WT, C12orf49-KO, and SREBF2-KO cells under normal (+FBS) and serum-starved (-FBS) growth conditions as assessed by RNA sequencing. All data are represented as means ± standard deviation; n = 3 independent biological replicates.
(f) Bar plot of relative mRNA expression of LDLR across HAP1 WT, FASN-KO and C12orf49-KO cells. All data are represented as means ± standard deviation; n = 3 independent biological replicates; one-way ANOVA.
(g) Western blotting analysis of SREBP2, LDLR and β-Actin levels across the indicated HAP1 co-isogenic knockout cell lines cultured in presence of FBS or exposed to overnight serum (left panel) or lipoprotein withdrawal (middle panel) and a short refeeding period (−/+). Unprocessed full length (FL) and processed C-terminal SREBP2 products are indicated. The same analysis was repeated in HEK293T cells (right panel). Prior protein extraction, HEK293T cells were stably transduced with lentiviral Cas9 and gRNA expression cassettes, selected and cultured for 4 days. Representative data from 3 independent biological replicates.
To investigate why C12orf49 is required for optimal LDL uptake, we performed proximity-dependent biotinylation of proteins coupled to mass spectrometry (BioID) to reveal the physical neighbourhood in which C12orf49 resides. Because the C12orf49 single predicted N-terminal transmembrane domain may direct the C-terminal DUF2054 domain into the lumen of the secretory pathway, leaving the N-terminus facing the cytoplasm, BioID-MS was performed separately with both N- and C-terminal miniTurbo BirA*-tagged C12orf49 open reading frames (ORFs) expressed in HEK293 cells. Proximity-based labelling with the N-terminal construct captured proteins localizing to various cellular compartments including the ER, Golgi apparatus, plasma membrane and the cytosol, whereas the C-terminal miniTurbo construct revealed a strong enrichment of proteins localizing to the endoplasmic reticulum (ER) lumen (Fig. 5b, Supplementary Table 5). Furthermore, the BirA* ligase fused to the N-terminus captured proximal interactions that are enriched for proteins functioning in cholesterol biosynthesis and vesicle-mediated ER – Golgi transport, whereas the C-terminus labelled proteins enriched for functions related to protein glycosylation (Extended Data Fig. 5d, Supplementary Table 6).
To study the subcellular localization of C12orf49 we performed immunofluorescence analysis. This was performed under normal and starved conditions since SREBPs, the master regulators of lipid homeostasis, are known to travel between the ER and Golgi in response to lipid deprivation26,40. Under normal growth conditions (with serum), C12orf49 containing a C-terminal V5 tag (i.e. C12orf49-V5) was localized throughout the ER-Golgi network (Fig. 5c), consistent with our BioID results. Strikingly, C12orf49-V5 accumulated in the Golgi apparatus under serum starvation, as demonstrated by co-staining with GOLGA2, a Golgi membrane marker protein (Fig. 5c). These data thus suggest that localization of C12orf49 is regulated in a growth condition-dependent manner, involving the shuttling between the ER and the Golgi apparatus.
To gain insight into potential protein interactions of C12orf49 we further explored our BioID data performed under normal and starved conditions. While 1,688 proteins passed the high-confidence criteria (≤1% Bayesian FDR) against our negative controls across the four tested conditions (Supplementary Table 5), distinguishing between specific proximity partners versus the general footprint of the ER-Golgi neighbourhood is challenging. To do this, we therefore leveraged a recently-generated reference map of a human cell (humancellmap.org)41 containing 192 BioID experiments in HEK293 cells, and compared our quantitative profiles to that of these baits. This revealed high-specificity interactions with the master lipid homeostatic transcription factor, SREBF2 in addition to SREBF1, SCAP and MBTPS1 (Fig. 5d). Notably, GIs were also observed between C12orf49 and SREBF2, as well as the SREBF2 regulatory proteases, MBTPS1, MBTPS2, and major SREBF2 transcriptional target, LDLR. Further, similar to C12orf49, SREBF2 is also known to traffic to the Golgi upon serum starvation, whereby it is cleaved and subsequently translocates to the nucleus and activates the transcription of genes regulating lipid homeostasis26,40. Given this, we hypothesized that C12orf49 could have a role in the activation of the SREBF2.
To investigate a role for C12orf49 in the regulation of SREBF2, we performed RNA-sequencing experiments under normal and serum-starved conditions across HAP1 WT, C12orf49-KO and SREBF2-KO cells (Supplementary Table 7). As expected, serum-starvation resulted in the induction of a cholesterol biosynthetic transcriptomic signature in HAP1 WT cells but not in SREBF2-KO cells (Fig. 5e, Extended Data Fig. 6a–b). In C12orf49-KO cells, we observed a SREBF2-mediated transcriptional response similar to WT cells, suggesting that C12orf49 is not absolutely required for the activation of SREBF2 upon serum starvation (Fig. 5e, Extended Data Fig. 6a–b). However, we did notice a trend for lower expression of cholesterol biosynthesis and LDL uptake genes in C12orf49-KO cells, which was confirmed by analysis of LDLR levels by qRT-PCR and western blot (Fig. 5f–g). To more directly test the hypothesis that C12orf49 regulates SREBF2 processing, we performed western blots to assess cleavage of SREBF2 into its active form following serum and lipid starvation. This demonstrated a clear decrease in overall SREBF2 expression and its processing in C12orf49-deficient vs. WT HAP1 and HEK293T cells, a phenotype that was not rescued by inhibition of the proteasome (Fig. 5g, Extended Data Fig. 6c), confirming the requirement of this gene for optimal SREBF2 activation.
In summary, our unbiased GI screens, transcriptomic, proteomic and biochemical investigations reveal a novel role for the uncharacterized gene C12orf49 in the maintenance of lipid homeostasis. Our data indicate that C12orf49 localizes in a growth condition-dependent manner throughout the ER-Golgi network, in parallel with SREBF2, being required for full SREBF2 activation as a major lipid homeostasis transcription factor (Fig. 6). Based on our findings, we suggest that C12orf49 be named LUR1 for its role in Lipid Uptake Regulation.
Figure 6. LUR1/C12orf49 shuttles between ER and Golgi and regulates SREBF2 activation and lipid uptake.
Proposed model summarizing functions and locations of key players in lipid metabolism, including LUR1/C12orf49, and highlighting the processes induced upon lipid deprivation in presence (left) or upon loss of LUR1/C12orf49 (right).
Left panel: (1) upon lipid deprivation (e.g. -LDL) LUR1/C12orf49 and SREBF2 relocate from the ER to the Golgi; (2) SREBF2 gets activated in the Golgi through proteolytic cleavage; (3) the processed, transcriptionally active domain shuttles to the nucleus where it induces expression of target genes required for lipid homeostasis such as LDLR; (4) newly synthesized and recycled LDLR shuttle through the ER-Golgi network where they are post-translationally modified (incl. glycosylation) and traffic to the cell surface; (5) on the cell surface LDLR binds LDL particles; (6) LDL particles are taken up through receptor-mediated endocytosis; (7) LDL particles are degraded and lipoprotein becomes available for metabolic processes including the synthesis, modification or storage of fatty acids and cholesterol.
Right panel: (1) Loss of LUR1/C12orf49 results in (2) impaired SREBF2 processing (3–5) and subsequently reduced expression of LDLR; (6) LDL uptake levels are decreased as a consequence of reduced LDLR expression and uptake activity; (7) decreased availability of extracellular lipoprotein leads to increased dependence on de novo synthesis pathways such as de novo fatty acid and cholesterol synthesis, explaining the negative GI between LUR1/C12orf49 and FASN and other members of lipid metabolic pathways.
DISCUSSION
The systematic mapping of GIs in model organisms like yeast has provided a detailed view into the functional organisation of eukaryotic cells42. Recent advances in CRISPR-based genome engineering technologies provide a path for similar systematic GI studies in human cells43–48. Here, we apply genome-wide CRISPR-based fitness screens using query mutant HAP1 cell lines to systematically map GIs with a focus on lipid metabolism. Our data revealed a strong interaction between de novo fatty acid synthesis and lipid uptake processes, highlighting a system that balances synthesizing lipids intracellularly with their uptake from the extracellular environment. More generally, this analysis confirms that relatively strong negative GIs identify functionally related genes, mapping a functional wiring diagram for a particular cellular process.
We screened a FASN mutant query cell line multiple times and identified highly confident negative GIs, many of which were involved in lipid metabolism. Perturbation of de novo fatty acid synthesis has been suggested as a prominent cancer therapeutic approach and multiple compounds targeting FASN are currently being tested in clinical trials; for example, TVB-2640 is a FASN inhibitor that is being tested in solid tumors in phase 2 trials, while both Fatostatin and Betulin are inhibitors of the SREBP-SCAP interaction in pre-clinical development6,49. Since single agent therapies often lead to emergence of resistance and tumor relapse, it makes sense to pursue therapeutic targets that are synergistic with FASN inhibition. Thus, the strong GIs detected in our FASN screen may be informative towards future investigations of combinatorial targets or biomarkers to treat diseases that would benefit from disruption of de novo fatty acid biosynthesis.
Our focused GI landscape related to de novo fatty acid biosynthesis provides unique insight into the genetic dependencies required for response to perturbation of lipid metabolism. Several pathways emerge as being most commonly utilized to adapt to perturbations, including those involved in alternate fatty acid and cholesterol biosynthesis processes as well as lipid uptake. Interestingly, while our screens revealed strong negative GIs between de novo fatty acid synthesis and uptake of LDL, we failed to detect interactions with transporters of fatty acids. This may be a consequence of the genetic redundancy inherent amongst the SLC27A (FATP) fatty acid transporter family50. As previously shown in yeast31, functional redundancy between paralogs can mask genetic interactions associated with perturbation of a single gene of a duplicated pair and highlights an important need for multi-gene targeting systems to survey complex genetic interactions involving more than two genes. Nonetheless, our data suggest a strong functional relationship between de novo fatty acid synthesis and glycosylation, and may involve a mechanism wherein cells modify the FATP transporters through N-glycosylation, thereby enhancing lipid uptake as suggested by Ryczko et al.22. As such, this pathway serves as an obvious focal point not only for ongoing mechanistic investigation but also therapeutic development for anti-cancer strategies targeting de novo fatty acid synthesis.
Genome-wide GI profiling also revealed an important role for LUR1 (C12orf49) in lipid uptake. Interestingly, analysis of the DepMap data revealed that LUR1 is essential in the same set of cancer cell lines that also depend on other lipid biosynthesis-related genes for viability, including SREBF1, MBTPBS1, SCAP and SCD. Similarly, two recent studies identifying co-functional gene clusters, support a functional role of LUR1 in lipid metabolism across diverse genetic backgrounds51,52. Furthermore, genome-wide association studies with large patient cohorts have found LUR1 variants linked to abnormal HDL profiles39, neuroticism53–55 and body height54, all phenotypes that could have root causes in lipid metabolism defects.
In summary, we present an unbiased and genome-wide approach for uncovering genetic vulnerabilities related to lipid metabolism in human cells, which led us to identify a function for LUR1 in regulating SREBF2 activity (Fig. 6). Our GI profiles for de novo fatty acid synthesis and related lipid uptake genes provide a resource for studying metabolic rewiring and disease phenotypes linked to lipid metabolism. We also demonstrate the power of systematic GI profiling using query mutants in a co-isogenic cell line, an approach that can be applied to other bioprocesses and expanded to begin generating more comprehensive GI maps for human genes.
DATA AND CODE AVAILABILITY
The datasets generated and analysed in this study are included in the manuscript. The raw fastq files for the sequencing data are available upon request and have also been deposited to the Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/): RNA-sequencing data, GSE147770. All mass spectrometry data has been deposited to the MassIVE repository (https://massive.ucsd.edu/ProteoSAFe/static/massive.jsp) and assigned the accession number MSV000085005. The ProteomeXchange accession is PXD017719. Descriptions of the analyses, tools and algorithms are provided in the methods section and the Reporting Summary of this article. Custom code for generating gRNA counts from fastq files and code for generating qGI-scores will be made available on Github upon publication.
METHODS
Cell culture
Human HAP1 wild type cells were obtained from Horizon Genomics (clone C631, sex: male with lost Y chromosome, RRID: CVCL_Y019). The following HAP1 gene knockout cell lines were obtained from Horizon: FASN (HZGHC003700c006 – used for GI screening; HZGHC003700c011 – used for hit validation), ACACA (HZGHC004903c002), LDLR (HZGHC003978c007), SREBF1 (HZGHC001361c012), SREBF2 (HZGHC000683c004). All gene knockout cell lines were confirmed to carry the expected out-of-frame insertions or deletions by Sanger Sequencing of PCR products. HAP1 cells were maintained in low glucose (10 mM), low glutamine (1 mM) DMEM (Wisent, 319–162-CL) supplemented with 10% FBS (Life Technologies) and 1% Penicillin/Streptomycin (Life Technologies). This culture medium is referred to as “minimal medium”. Cells were dissociated using Trypsin (Life Technologies) and all cells were maintained at 37°C and 5% CO2. Cells were regularly monitored for mycoplasma infection.
HAP1 KO cell line generation
The HAP1 C12orf49 gene knockout cell line was constructed by first cloning a gRNA targeting C12orf49 (Supplementary Table 8) into the pX459v2 backbone (Addgene #62988), which was modified to carry the same restriction overhangs as the pLCKO vector (Addgene #73311). 350k HAP1 WT cells were seeded into a 6-well plate and 24 hours later cells were transfected with a mix of 2 μg pX459 plasmid (Addgene #62988) carrying a gRNA, 6 μl X-treme Gene transfection reagent (Roche), and 100 μl Opti-MEM media (Life Technologies). Twenty-four hours after transfection, cells were selected in medium containing 1 μg/ml puromycin for three days and single cells were sorted onto 96-well plates by manual seeding of a single cell suspension at 0.6 cells/well. Following amplification of cells from individual wells, genomic DNA was extracted with Extracta DNA Prep (Quanta Bio), Sanger sequencing was performed across the gRNA target sites following PCR amplification, and successful gene knockouts were identified following sequence analysis.
Library virus production and MOI determination
For CRISPR library virus production, 8 million HEK293T cells were seeded per 15 cm plate in DMEM medium containing high glucose, pyruvate and 10% FBS. Twenty-four hours after seeding, the cells were transfected with a mix of 8 μg lentiviral lentiCRISPRv2 vector containing the TKOv3 gRNA library19 (Addgene #90294), 4.8 μg packaging vector psPAX2, 3.2 μg envelope vector pMD2.G, 48 μl X-treme Gene transfection reagent (Roche) and 1.4 ml Opti-MEM media (Life Technologies). Twenty-four hours after transfection, the media was replaced with serum-free, high-BSA growth media (DMEM, 1.1g/100ml BSA, 1% Penicillin/Streptomycin). Virus-containing media was harvested 48 hours after transfection, centrifuged at 1,500 rpm for 5 minutes, aliquoted and frozen at −80°C.
For determination of viral titers, 3 million HAP1 cells seeded in 15 cm plates were transduced with different dilutions of the TKOv3 lentiviral gRNA library along with polybrene (8 μg/ml), in a total of 20 ml medium. After 24 hours, the virus-containing media was replaced with 25 ml of fresh media containing puromycin (1 μg/ml), and cells were incubated for an additional 48 hours. Multiplicity of infection (MOI) of the titrated virus was determined 72 hours post-infection by comparing percent survival of puromycin-selected cells to cells that were infected but not selected with puromycin (i.e. puro minus controls).
Pooled CRISPR dropout screens
For pooled CRISPR dropout screens, 3 million HAP1 cells were seeded in 15 cm plates in 20 ml of specified media. A total of 90 million cells were transduced with the lentiviral TKOv3 library at a MOI~0.3, such that each gRNA is represented in about 200–300 cells. Twenty-four hours after infection, transduced cells were selected with 25 ml medium containing 1 μg/ml puromycin for 48 hours. Cells were then harvested and pooled, and 30 million cells were collected for subsequent gDNA extraction and determination of the library representation at day 0 (i.e. T0 reference). The pooled cells were then seeded into three replicate plates, each containing 18 million cells (>200-fold library coverage), which were passaged every three days and maintained at >200-fold library coverage until T18. Genomic DNA pellets from each replicate were collected at each day of cell passage.
Preparation of sequencing libraries and Illumina sequencing
Genomic DNA was extracted using the Wizard Genomic DNA Purification Kit (Promega). The gDNA pellets were resuspended in TE buffer, and the concentration was estimated by Qubit using dsDNA Broad Range Assay reagents (Invitrogen). Sequencing libraries were prepared from 50 μg of the extracted gDNA in two PCR steps, the first to enrich guide-RNA regions from the genome, and the second to amplify guide-RNA and attach Illumina TruSeq adapters with i5 and i7 indices as described previously using staggered primers aligning in both orientations to the guide-RNA region (Supplementary Table 8)56. Barcoded libraries were gel purified and final concentrations were estimated by quantitative RT-PCR. Sequencing libraries were sequenced on an Illumina HiSeq2500 using single read sequencing and completed with standard primers for dual indexing with HiSeq SBS Kit v4 reagents. The first 21 cycles of sequencing were dark cycles, or base additions without imaging. The actual 36-bases read begins after the dark cycles and contains two index reads, reading the i7 first, followed by i5 sequences. The T0 and T18 time point samples were sequenced at 400- and 200-fold library coverage, respectively.
Construction of color-coded lentiCRISPRv2 vectors for co-culture assay
The color-coded lentiCRISPRv2 vectors were derived from the lentiCRISPRv2 vector (Addgene #52961) by inserting mCherry (Addgene #36084) or mClover3 (Addgene #74236) open reading frames between the Cas9 and PuroR expression cassette. To this end, the lentiCRISPRv2 vector was digested with BamHI, PCR products coding for the respective fluorescent protein flanked by T2A and P2A self-cleaving peptides were ligated into the vector using Gibson assembly. The two forward primers (Supplementary Table 8) were used at a 1:0.1:1 (P233:P234:P235) ratio in the same PCR reaction with the reverse primer (primers bind to both fluorescent proteins mCherry and mClover3).
Validation of genetic interactions using co-culture assays
For validation of genetic interactions, HAP1 parental and gene knockout clones were transduced with color-coded lentiCRISPRv2 vectors targeting either an intergenic site in the AAVS1 locus (i.e. negative control), or a specific target gene hit (e.g. LDLR). Each gene was targeted with three independent and unique gRNAs. Twenty-four hours after transduction, cells were selected with 1 μg/ml puromycin for 48 hours and seeded for co-culture proliferation assays as follow: 50k of green (e.g. lentiCRISPRv2-mClover3 AAVS1 gRNA) and red (e.g. lentiCRISPRv2-mCherry hit gene gRNA) cells were mixed (total 100k) in a 6-well plate in both color orientations for both parental and gene knockout cells, respectively. Cells were passaged every 4 days until day 12 (T12). Cells were trypsinized, washed and stained for dead cells using Zombie NIR (BioLegend). The relative proportion of red and green cells in the co-culture were assessed using an LSR Fortessa flow cytometer (BD Bioscience). The relative ratio of Hit:AAVS1 was calculated and averaged for the three gene-targeting guides and two color orientations.
Low-density lipoprotein and transferrin uptake assay
For uptake experiments with labelled probes 150k HAP1 cells were seeded in a 12-well plate. After 48 hours cells were serum-starved or deprived of lipoprotein overnight in minimal medium (described above) complemented with 0.3% BSA (BioShop) or lipoprotein-deprived FBS (MilliporeSigma, S5394) instead of standard FBS, respectively. After 16 hours cells were labelled with Dil-LDL (Invitrogen L3482), pHrodo Red LDL (Invitrogen L34356) or pHrodo Red Transferin (P35376) at 2 μg/ml (1:500) in minimal medium plus 0.3% BSA for 15 minutes at 37°C. Cells were washed in PBS, trypsinized and stained with 7-AAD (BioLegend 420404) or Zoombie NIR (BioLegend 423105) cell viability solution at 25 ng/ml (1:2,000) for 5 minutes at room temperature. Staining was measured using an LSR Fortessa flow cytometer (BD Bioscience). The gating strategy is outlined in Extended Data Fig. 7.
Proximity-based labelling of proteins capture to mass spectrometry (BioID-MS)
BioID-MS analysis was performed essentially as described previously57, with minor modifications. In brief, HEK293 Flp-In T-REx lines expressing inducible N- or C-terminal miniTurbo-FLAG-tagged C12ORF49 open reading frames were generated58. Five (for normal growth condition) and 12.5 (for serum-starvation) Mio. cells were seeded on 15 cm plates and after 24 hours, cells were treated with 1 μg/ml tetracycline to induce expression of baits. 24 hours later 50 μM biotin was added for labelling of proximal proteins for 3.5 hours. Cell pellets were collected and lysed in RIPA lysis buffer (50mM Tris-HCl pH 7.5, 150mM NaCl, 0.1% (w/v) SDS, 1% NP-40, 1mM EDTA, 1mM MgCl2; 0.5% Deoxycholate and Sigma protease inhibitors were added right before cell lysis.) at an 1:10 (g:ml) ratio, sonicated three times for 5 seconds with 2 seconds breaks. 1ul/sample TurboNuclease (BioVision) and 1ul/sample RNAse (Sigma) was added and samples were incubated at 4ºC for 30 minutes. 20% SDS was added to bring the sample’s final SDS concentration to 0.25%, samples were mixed well and centrifuged at 14,000 rpm (Microfuge) for 20 mins in 4ºC. The supernatant was added to Streptavidin resin (pre washed with lysis buffer) using 30μl bed volume and rotated at 4ºC for 3 hours. Beads were washed after binding as following: a) 1×1ml of 2% SDS buffer (2% SDS, 50mM Tris-Hcl pH7.5), b) 1×1ml of lysis buffer, c) 1×1ml of HEK293 lysis buffer (with 0.1% NP-40), d) 3×1ml of 50mM ammonium bicarbonate (made fresh). After purification of biotinylated preys using streptavidin sepharose, samples were digested on beads using trypsin. Samples were separated by liquid chromatography and analysed by tandem mass spectrometry on a AB SCIEX TripleTOF™ 5600 mass spectrometer.
The LC-MS/MS setup consisted of a TripleTOF™ 5600 (SCIEX, Concord, ON, Canada) equipped with a nanoelectrospray ion source connected in-line to an AS-2 Nano-HPLC system (Eksigent Technologies, Dublin, CA, USA). The fused silica column (10 cm x ID 75 μm, OD 360 μm) had an integrated emitter tip prepared in-house using a laser puller (Sutter Instrument Co., Novato, CA, USA). The column was packed with ~10 cm of C18 resin (Reprosil-Pur, 3.5 μm, Dr.Maisch HPLC GmbH, Germany). 5 μl of sample was loaded onto the column using the autosampler at 400nl/min, and the LC delivered the organic phase gradient at 200 nl/min over 90 min (2–35% acetonitrile with 0.1% formic acid). The MS instrument was operated in data-dependent acquisition mode with 1 MS scan (250 ms; mass range 400–1250 m/z) followed by up to 20 MS/MS scans (100 ms each). Only candidate ions between two and four charge states were considered, and ions were dynamically excluded for 15 s with a 50 mDa window. The isolation width was 0.7 m/z, and minimum threshold was set to 250. Between sample injections, 2 blank samples were injected (0.1% formic acid), each with 3 rapid gradient cycles at 300 nl/min over 60 min. Before another sample was injected, system performance was verified with a 30 min BSA quality control run and a 30 min BSA mass calibration run.
Raw files (.WIFF and .WIFF.SCAN) were converted to an MGF format and to an mzML format using ProteoWizard (v3.0.4468) and the AB SCIEX MS Data Converter (V1.3 beta), as implemented within ProHits59. For human samples, the database used for searches consisted of the human and adenovirus sequences in the RefSeq protein database (version 57). The database was supplemented with “common contaminants” from the Max Planck Institute (http://141.61.102.106:8080/share.cgi?ssid=0f2gfuB) and the Global Proteome Machine (GPM; http://www.thegpm.org/crap/index.html), and with commonly used epitope tags. The search databases consisted of forward and reverse sequences (labeled “gi 9999” or “DECOY”); in total, 72,481 entries were searched for the human database. Spectra were analyzed separately using Mascot (2.3.02; Matrix Science) and Comet [2018.01 rev.4] with trypsin specificity and up to two missed cleavages; deamidation (Asn or Gln) and oxidation (Met) were selected as variable modifications. The fragment mass tolerance was 0.15 Da, and the mass window for the precursor was ±35 ppm with charges of 2+ to 4+ (both monoisotopic mass). The resulting Comet and Mascot results were individually processed by PeptideProphet and combined into a final iProphet output using the Trans-Proteomic Pipeline (TPP; Linux version, v5.2.1-dev Flammagenitus, Build-201906251008-exported). TPP options were as follows: Peptide prophet were --minprob 0.05 --ppm --decoy DECOY --nonparam --accmass --expectscore --decoyprobs and iProphet options were --nonsp --nonrs --nonsi --nonsm --nonse. All proteins with a minimal iProphet probability of 0.95 and two unique peptides were used for analysis.
Data processing and analysis was performed within the ProHits LIMS60 searched against the RefSeq human and adenovirus data base, version 57; forward and reverse. Mascot and Comet search results were jointly analysed using the iProphet component of the Trans Proteomic Pipeline61.
High confidence interactions were determined by scoring bait samples against negative control samples (6 runs of miniTurbo-FLAG-EGFP) using the statistical tool SAINTexpress62 v3.6.1 with default parameters. Preys with a SAINT score (FDR) of less than 1% were considered as high confidence hits. To calculate specific enrichment of preys against a reference dataset, we leveraged the humancellmap.org, a compendium of 192 BioID baits profiled in HEK293s41. Each SAINTexpress-filtered list was uploaded to the “Analyze” module of the humancellmap, and processed using default options. Default specificity plots (calculating the specific enrichment of spectral counts for a prey with a bait against that across the entire database) were generated within the humancellmap site and visualized in ProHits-viz59.
Western blotting
HAP1 WT and FASN KO cells were lysed in buffer F (10 mM Tris pH 7.05, 50 mM NaCl, 30 mM Na pyrophosphate, 50 mM NaF, 10% Glycerol, 0.5% Triton X-100) and centrifuged at 14,000 rpm for 10 minutes. The supernatant was collected and protein concentration was determined using Bradford reagent (BioRad). 10–30 μg protein was resolved on 4–12% Bis-Tris gels (Life Technologies) and transferred to Immobilon-P nitrocellulose membrane (Millipore) at 66V for 90 minutes. Subsequently, proteins were detected using anti-FASN (1:2,000, Abcam ab128870), anti-SREBP2 (1:250, BD Biosciences 557037), anti-LDLR (1:250, ab52818), anti-GAPDH (1:10,000, Santa Cruz 166574) and anti-β-Actin (1:10,000, Abcam ab8226) antibodies and proteins were visualized on X-ray film using Super Signal chemiluminescence reagent (Thermo Scientific) and Western Lightning ECL Pro (PerkinElmer). Scans of uncropped western blots are provided in Source Data Figure 5 and Source Data Extended Figures 1, 6.
Immunofluorescence
Cells were seeded on cover slips and fixed with 4% paraformaldehyde in PBS for 10 minutes at room temperature. Cells were permeabilized with 1% NP-40 in antibody dilution solution (PBS, 0.2% BSA, 0.02% sodium azide) for 10 minutes and blocked with 1% goat serum for 45 minutes. Cells were incubated with anti-V5 (1:250, Abcam ab27671) and anti-GOLGA2 antibodies (1:250, Sigma HPA021799) for 1 hour at room temperature. Subsequently, cells were incubated with Alexa Fluor488 goat anti-mouse (1:500, Invitrogen A-11001) or Alexa Fluor647 anti-rabbit antibodies (1:500, Invitrogen A-21245) and counterstained with 1 μg/ml DAPI (Cell Signaling Technology, 4083S) for 45 minutes in the dark. Cells were visualized by confocal microscopy (Zeiss LSM 880).
RNA-sequencing
Sample preparation
HAP1 WT, C12orf49 and SREBF2 KO cells were cultured in minimal DMEM medium for 48h and either control treated or serum-starved for 4 hours as indicated. Each cell line was cultured and processed in three biological replicates. RNA was extracted using the RNeasy Kit (QIAGEN) according to manufacturer’s instructions. 18 total RNA samples were DNase treated using RNase-free DNase Set (Qiagen, 79254). Samples were submitted for mRNA-Seq at the Donnelly Sequencing Centre at the University of Toronto (http://ccbr.utoronto.ca/donnelly-sequencing-centre). RNA was quantified using Qubit RNA BR (Thermo Fisher Scientific, Q10211) fluorescent chemistry and 1 ng was used to obtain RNA Integrity Number (RIN) using the Bioanalyzer RNA 6000 Pico kit (Agilent Technologies, 5067–1513). Lowest RIN was 9.5; median RIN score was 9.8. 1000 ng per sample was then processed using the NEBNext Ultra II Directional RNA Library Prep Kit for Illumina (New England Biolabs, E7760L) and included polyA-enrichment using NEBNext Poly(A) mRNA Magnetic Isolation Module (New England Biolabs, E7490L), fragmentation for 15 minutes at 94°C prior to first strand synthesis, and 8 cycles of amplification after adapter ligation. 1μL top stock of each purified final library was run on an Agilent Bioanalyzer dsDNA High Sensitivity chip (Agilent Technologies, 5067–4626). The libraries were quantified using the Quant-iT dsDNA high-sensitivity (Thermo Fisher Scientific, Q33120) and were pooled at equimolar ratios after size-adjustment. The final pool was run on an Agilent Bioanalyzer dsDNA High Sensitivity chip and quantified using NEBNext Library Quant Kit for Illumina (New England Biolabs, E7630L). The quantified pool was hybridized at a final concentration of 400 pM and sequenced paired-end on the Illumina NovaSeq6000 platform using a S2 flowcell at 2×151 bp read lengths.
Data Processing
Samples were mixed to obtain an average of 35 million clusters that passed filtering. Reads shorter than 36bp on either read1 or read2 were removed prior to mapping. Reads were aligned to reference genome hg38 and Gencode V25 gene models using the STAR short-read aligner (v2.6.0a)63. Approximately 80% of the filtered reads mapped uniquely, and the read counts from each sample, computed by STAR, were merged into a single matrix using R. The raw and processed data will be deposited in the GEO database upon publication of manuscript.
Differential expression
Differentially expressed genes were identified using the Bioconductor packages limma (v3.32.10) and edgeR (v3.24.3). The read count matrix was filtered using the filterByExpr() function using default parameters. Principal Components Analysis was performed to examine the main treatment effects, and to exclude the presence of confounding batch effects, using the base R function prcomp(). Samples were normalized using calcNormFactors(method=”TMM”) from edgeR and transformed to log2 using voom(). Next, a design matrix was specified to fit coefficients for the CRISPR knockouts, presence or absence of FBS, and an interaction term to examined differences in the FBS effect in the mutant backgrounds. Differentially expressed genes were extracted using topTable() with log2( fold-change ) > 0.58 and adjusted P-value less than 0.05.
Quantitative real-time (qRT)-PCR analysis
HAP1 WT, FASN KO and C12orf49 KO cells were cultured in minimal DMEM medium for 48h and either control treated or serum-starved for 4 hours as indicated. RNA was extracted using the RNeasy Kit (QIAGEN) according to manufacturer’s instructions. RNA was converted into cDNA using the cVilo master mix (ThermoScientific) according to manufacturer’s instructions. The cDNA was amplified and quantified by quantitative PCR using a BioRad CFX96 real-time PCR detection system (BioRad) and using the Maxima SYBR Green PCR master mix (ThermoScientific) according to manufacturer’s instructions. Transcript levels were normalized to GAPDH (see Supplementary Table 8 for primer sequences).
Metabolite profiling
HAP1 WT and FASN-KO cells were cultured in minimal medium for 3 days. Cells were washed twice in warm PBS and subsequently flash frozen on liquid nitrogen. Cells were scraped in chilled extraction solvent (40% Acetonitrile: 40% Methanol: 20% water, all HPLC grade), transferred to clean tubes and shaken for one hour at 4°C and subsequently centrifuged at 4°C at max speed for 10 minutes. The supernatants were transferred to a clean tube and dried in a speedvac then stored at −80°C until mass spec analysis. Samples were reconstituted in water containing Internal Standards D7-Glucose and 13C15N-Tyrosine and injected twice through the HPLC (Dionex Corporation) for positive and negative mode analysis using a reverse phase column (Inertsil ODS-3, 4.6 mm internal diameter, 150 mm length, and 3 μM particle size). In positive mode analysis, the mobile phase gradient ramped from 5% to 90% acetonitrile in 16 minutes, remained for 1 minute at 90%, then returned to 5% acetonitrile in 0.1% acetic acid over two minutes. In negative mode, the acetonitrile composition ramped from 5 to 90% in 10 minutes, remained for 1 minute at 90%, then returned to 5% acetonitrile in mobile phase (0.1% tributylamine, 0.03% acetic acid, 10% methanol). The total runtime in both the positive and negative modes was 20 minutes, the samples were maintained at 4°C, and the injection volume was 10 μL. An automated washing procedure was included before and after each sample to avoid any sample carryover.
The eluted metabolites were analyzed at the optimum polarity in MRM mode on an electrospray ionization (ESI) triple-quadrupole mass spectrometer (ABSciex 5500 Qtrap). The mass spectrometric data acquisition time for each run was 20 minutes, and the dwell time for each MRM channel was 10 ms. Mass spectrometric parameters were as previously published64. Metabolite peak areas were determined using Multiquant software (SCIEX, Toronto, ON, Canada), normalized to internal standard in each mode yielding an area ratio and then further normalized to total cell number for each sample and Malonyl-CoA levels were further normalized to WT cells.
QUANTIFICATION AND STATISTICAL ANALYSIS
Guide mapping and quantification
FASTQ files from single read sequencing runs were first trimmed by locating constant sequence anchors and extracting the 20 bp gRNA sequence preceding the anchor sequence. Pre-processed paired reads were aligned to a FASTA file containing the TKOv3 library sequences using Bowtie (v0.12.8) allowing up to 2 mismatches and 1 exact alignment (specific parameters: -v2 -m1 -p4 --sam-nohead). Successfully aligned reads were counted, and merged along with annotations into a matrix.
Scoring of quantitative genetic interactions: the qGI score
To identify and quantify genetic interactions (GI), genome-wide CRISPR/Cas9 screens were performed using the TKOv3 gRNA library in HAP1 co-isogenic cell lines. Co-isogenic knockout (KO) “query” cell lines were obtained from Horizon Genomics (see above) or generated by introducing mutations in target genes of interest (see above) in the parental HAP1 cells, which we consider as wild-type (WT). The TKOv3 library contains 71,090 guide (g)RNAs that target ~18k human protein-coding genes, most of them with four sequence-independent gRNAs19. To quantify GIs, log2 fold-changes (LFC) between read-depth normalized gRNA abundance in the starting population (T0) and the endpoint (T18) were computed. Matched T0 measurement assured that differences between screens during library infection and Puromycin selection would not result in false positive GIs. Matched T0 were stabilized using the median across many T0 measurements (common T0), and those two estimates were combined in a weighted fashion to minimize correlation between GI scores and residual T0 (matched T0 – common T0).
gRNA-level residual scores were derived for a given genetic background by estimating a non-interacting model between LFC values in this background and 21 WT HAP1 backgrounds. To do so, for each WT-KO screen pair the population of LFC values were M-A-transformed, which contrast the per-gRNA LFC difference M with per-gRNA mean A. A Loess regression was fitted, which was additionally locally stabilized by binning the data along A and considered equal bin sizes and equal numbers of data points in every bin. For each gRNA, this resulted in 21 residual scores, which represent the contrasts of a given KO with the 21 WT HAP1 screen. Under the assumption that genetic interactions are sparse and that experimental artefacts such as batch effects would introduce additional signal into the population of residual values, we computed a weighted mean of its 21 residual scores by giving a higher weight to WT HAP1 screens with lower absolute residual mean of all 71k gRNAs. We refer to the resulting value for each gRNA as the “guide-level” GI score. Those guide-level GI scores were further normalized. First, locally-defined shifts towards negative or positive scores were identified and normalized, based on genome location of the target genes. Next, to remove unwanted effects that would arise from screen-to-screen variability, we quantified guide-level GI scores for each of the 21 WT HAP1 screens by contrasting a given WT screen to the remaining WT screens (as described for the KO-WT comparison above). Patterns that explain substantial variance among these WT guide-level GI scores are likely to correspond with unwanted experimental artefacts. To remove these artefacts from the GI data, we performed singular value decomposition (SVD) on guide-level GI scores of the HAP1 WT screens only. We then projected guide-level GI scores onto the left singular vectors, and subtracted the resulting signal from the GI scores.
Finally, we computed gene-level genetic interaction scores. First, gRNAs were excluded when their guide-level GI profile disagreed with those of the remaining gRNAs against the same gene. Specifically, the mean within-gene guide-level GI profile Pearson correlation coefficient was computed, which we call the gRNA quality score (QS) and a per-gene quality score (gQS) was computed using the median of the gRNA QS for a given gene. For each gene, we tested three criteria and excluded the gRNA with the lowest QS, if true. First, a gQS above a selected threshold th1 indicated that sufficient signal was present in the guide-level GI profiles. Second, a difference between the lowest QS and the gQS above a selected threshold th2 indicated sufficient within-gene disagreement. Third, a QS of the gRNA with the lowest QS for a given gene below a selected threshold th3 indicated lack of gene-specific signal. Based on these criteria, we excluded a gRNA for 648 out of the 17,804 genes. All remaining guide-level GI values per gene were mean-summarized and their significance was computed using limma’s moderated t-test followed by Benjamini-Hochberg multiple testing correction.
Screen reproducibility analysis
Reproducibility of the gRNA library screening data in FASN-KO cells was tested across three independent screens. The three screens were started from independent infections with lentivirus packaged gRNA library and performed as described above. To assess reproducibility of fitness effects, a log2 fold-change (LFC) quantifying the drop-out between T0 (after puromycin selection) and T18 (endpoint) was computed for each gene by mean-summarizing the respective four gRNA LFC values. The Pearson correlation coefficients (PCCs) were computed between LFC values of all three pairs of independent replicates.
Our experiments were designed to quantify fitness effect differences due to the introduction of a specific mutation into an otherwise isogenic background (i.e. GIs). To assess reproducibility of GIs, PCCs were computed between qGI values of all pairs of independent replicates. While we assessed screen reproducibility primarily at the gene-level, we also computed the gRNA-level GI PCC between the replicates as reference (Extended Data Fig. 1g).
To test reproducibility of genes, each gene’s contribution to the covariance between a pair of FASN-KO screens was computed and divided by the product of standard deviations of both given screens. The resulting three pairwise (for replicates A-B, A-C, B-C) gene-level scores were mean-summarized to a FASN qGI reproducibility score.
Reproducibility analysis of FASN interactions
We used an MCMC-based approach to measure the reproducibility of FASN GIs. Specifically, we first independently scored the three independent FASN replicate screens and applied an FDR threshold of FDR 50% to generate positive and negative GI profiles for each of the three screens. MCMC was then used to jointly infer false negative and false positive rates, as well as a binary consensus FASN GI profile (separately for positive/negative GI). Then, using this consensus profile as a standard for evaluation (assuming pairs with posterior probability of interaction of > 0.5 as positives), we measured precision and recall statistics (averaged across the three screens) at two different cut-offs: a “standard” cut-off (absolute qGI score > 0.5 and FDR 50%) and a “stringent” cut-off (absolute qGI score > 0.7 and FDR 20%).
Precision-recall analysis
To control quality of genome-wide gRNA screens, gene-level fitness effects were estimated by computing a log2 fold-change (LFC) quantifying the drop-out between T0 (after puromycin selection) and T18 (endpoint) for each gene and mean-summarizing the respective four gRNA LFC values. Gold-standard essential (reference) and non-essential (background) gene sets were taken from Hart et al., 201565 and Hart et al., 201719. For the identification of reference genes using LFC values of a given screen was assessed by computing precision over true positive statistics.
Functional evaluation of genetic interactions
To calculate the enrichment of metabolic GIs in different functional standards, we separated the metabolic GIs in two different sets: all (background) GI scores and high confidence (reference) GI (FDR < 0.5, |qGI| >= 0.5). Then we calculated the fold enrichment of the reference set against the background set in a particular functional standard. First, we computed the overlap of metabolic GI pairs as co-annotations in the standard. Then we divided the overlap density of the background set into the overlap density of the reference set to determine the fold enrichment. Once we got the fold enrichments, we calculated p-values on the actual overlap counts of the reference and background sets according to hypergeometric tests. We used four different functional standards: Human functional network66, GO biological processes67, Pathway (Canonical pathways from68), and HumanCyc27.
Gene ontology enrichment analysis
Gene ontology (GO) enrichment analysis for the FASN and C12orf49 GI screen and the BioID experiments were performed using the gProfileR R package using the GO-Bioprocesses, GO-Molecular Function and Reactome pathway standards69. For the GI screens, enrichment analysis was performed for significant negative GIs (qGI < −0.5, FDR < 0.5), enriched pathways (p<0.05, maximum term size 100) with a similarity of > 50% were collapsed using the Cytoscape Enrichment Map function and the mean percentage overlap of hits within the term were visualized on a bar plot. For the BioID experiments, GO-Cellular compartment and KEGG standards were also included in the analysis. Enrichment analysis was performed for significant hits (FDR < 0.01), enriched pathways (p<0.05) with a similarity of > 70% were collapsed using the Cytoscape Enrichment Map function and for pathways with at least 5 collapsed terms the mean percentage overlap of hits within the term were visualized on a bar plot.
Co-occurrence of missense mutations and gene essentiality
Starting from the DepMap (19Q2) dataset, we first filtered down to a subset of cell lines (523) that had no more than 1000 called missense mutations. Among this set, we identified the subset of cell lines (6 total) that exhibited dependency on C12orf49 (dependency scores threshold of < −1.0). Then, for each gene with at least one missense mutation with the candidate cell lines, we tested for association between the presence of a missense mutation in the corresponding gene and the dependency on C12orf49, using the hypergeometric distribution to assess significance and Benjamini and Hochberg multiple hypothesis correction. For FASN mutations, we observed 30 cell lines with at least one mutation with FASN, and 3 of this set were also dependent on C12orf49 (out of 6) (FDR=18.9%).
Statistical analysis
For all experiments the number of technical and/or biological replicates are listed in the figure legends or text. Unless otherwise indicated, statistical significance was assessed via one or two factor ANOVA with Fisher’s Least Significant Difference test. Statistical analyses were performed using GraphPad Prism 8 (GraphPad Software, La Jolla, California, USA) or the R language programming environment. For further information please also refer to the Reporting Summary.
Extended Data
Extended Data Fig. 1. Validation of FASN-KO cells and genetic interactions screens.
(a) Western blot depicting FASN and β-Actin levels in HAP1 parental wildtype (WT) and FASN-KO cells. Representative data from three biologically independent experiments.
(b) Bar plot depicting malonyl-CoA levels in HAP1 WT and FASN-KO cells as detected by mass spectrometry-based metabolite profiling, normalized to parent HAP1 WT cells. Data are represented as means ± standard deviation; n = four biologically independent experiments; two-tailed Mann Whitney U test.
(c) Growth curves of HAP1 WT cells depicting relative cell numbers over 3 days, plotted as a function of glucose concentration in mM, in either 0.5 mM (blue), 1 mM (red), 1.5 mM (yellow), or 2 mM (black) glutamine.
(d) Histogram showing a frequency distribution of all pairwise Pearson correlation coefficients for LFC values (T0/T18) of the 21 WT HAP1 screens.
(e) Precision-recall curves for the three CRISPR replicate screens in HAP1 FASN-KO cells using the reference core essential gene set (CEG2) defined in Hart et al., 201719.
(f) Fitness effect (log2 fold-change, LFC) distributions for reference core essential (CEG2) and non-essential gene sets defined in Hart et al., 201719 across the three FASN-KO query screens.
(g) Agreement of gRNA-level genetic interaction scores with FASN. Scatter plots show all possible pairwise combinations of three biological replicate screens. The Pearson correlation coefficient (r), based on comparison values for 70,152 gRNAs.
(h) Agreement of FASN quantitative genetic interactions (qGIs). Scatter plots show gene-level FASN genetic interactions (qGI scores) derived from all possible pairwise combinations of three biological replicate screens. The Pearson correlation coefficient (r), based on comparison of all qGI scores (r shown in grey, calculated on all the grey and purple data points in the scatter plots), or only genetic interactions that exceed a given significance threshold (|qGI| > 0.5, FDR < 0.5) in either one screen (logic OR; purple).
(i) Scatter plot showing reproducibility scores as a function of qGI scores for a single FASN-KO screen (replicate A). Pairwise reproducibility of a qGI score was calculated by computing the contribution of each of the 17,804 genes to the covariance between a pair of screens divided by the sum of standard deviations. The reproducibility score represents the sum of those values across the three pairwise comparisons. Five genes with highest reproducibility scores and the most negative qGI scores with the FASN-KO screen (replicate A) are labelled.
(j) Establishing the AAVS1 target locus as a good negative control site in HAP1 WT and FASN-KO cells. Schematic depicting co-culture validation assays (upper panel). Parental WT and FASN-KO cells were stably transduced with color-coded gRNA expression vectors carrying an intergenic control or screen hit-targeting gRNA. Color-coded cells are mixed at an equal ratio, cultured over two weeks and the relative proportion of green and red cells was quantified by flow cytometry. Control co-culture experiments performed in parallel to the validation of hit genes depicted in Fig. 1e as indicated above each barplot (lower panel). Bar plots are depicting the color ratio of cells carrying two colour-coded gRNAs targeting AAVS1 (intergenic control) across WT and two FASN-KO clones as indicated. Experiments were performed with three independent gRNA targeting AAVS1 and using both color orientations. All data are represented as means ± standard deviation; n = three (LDLR) or four (SLCO4A1, C12orf49) biologically independent experiments.
(k) Scatter plots reproducibility scores as a function of qGI scores for the negative genetic interaction hits depicted in Fig. 1h functioning in lipid uptake and homeostasis (red), vesicle transport genes (black) and glycosylation (blue).
(l) Precision and recall values for GIs with FASN measured at the standard (|qGI| > 0.5, FDR < 0.5) and stringent (|qGI| > 0.7, FDR < 0.2) thresholds. Precision and recall values were computed using an MCMC-based approach (see Methods).
Extended Data Fig. 2. Quality control of genetic interaction screens for fatty acid synthesis-related query genes.
(a) Precision-recall curves distinguishing the reference core essential gene set (CEG2) defined in Hart et al., 201719 and a non-essential gene set in CRISPR screens in five HAP1 knockout query cell lines (LDLR, C12orf49, SREBF2, ACACA, SREBF1-KO).
(b) Fitness effect (LFC) distributions for reference core essential (CEG2) and non-essential gene sets defined in Hart et al., 201719 across CRISPR screens in five HAP1 KO cell lines (LDLR, C12orf49, SREBF2, ACACA, SREBF1).
(c) Bar plot of enrichment of co-annotation as defined by the Human Functional Network, Gene Ontology Bioprocesses (BP), HumanCyc or and aggregation of pathway standards (including REACTOME, KEGG or BIOCARTA) for genetic interactions identified across all six query genome-wide screens (FASN, LDLR, C12orf49, SREBF2, ACACA, SREBF1). Enrichment was tested using a hypergeometric test. See methods for details of analysis.
Extended Data Fig. 3. Pathway enrichment analysis of genetic interactions for fatty acid synthesis-related query genes in additional HumanCyc sub-categories.
(a) Dot plot of normalized pathway enrichment values for aggregate GIs across the six query genes (FASN, C12orf49, LDLR, SREBF2, ACACA, SREBF1) with sub-categories from HumanCyc are indicated. A GI is identified for a query-library pair if the |qGI| > 0.5 and FDR < 0.5. Enrichment for positive (yellow) and negative (blue) GI is tested inside Glycan Pathways and Generation of precursor metabolite and energy HumanCyc branches using a hypergeometric test. Enrichment with p-value < 0.05 are blue (negative GI) and yellow (positive GI). Dot size is proportional to the fold-enrichment in the indicated categories and specified in the legend.
Extended Data Fig. 4. Overview of C12orf49, cancer associations, and functional correlates.
(a) Cartoon of C12orf49 protein sequence features and domains.
(b-e) Kaplan Meier survival plots displaying univariate analysis of TCGA data across multiple tumor types including kidney, breast, liver and sarcoma for C12orf49 high vs. low expressing tumor tissue (www.kmplot.com )36. Patient numbers at risk (n) are indicated below each plot; two-sided logrank test.
(f-h) GI overlap between the 17,804 C12orf49 and SREBF2, SREBF1 and ACACA qGI scores shown as pairwise scatter plots with C12orf49 as function of SREBF2 (f), SREBF1 (g) and ACACA (h). A common negative GI is called if it is significant (qGI < −0.5, FDR < 0.5) in both screens (indicated in blue). The top 10 strongest common GIs and lipid metabolism genes are labelled.
(i) Profile similarity of C12orf49 across genome-wide DepMap CRISPR/Cas9 screens. Similarity was quantified by taking all pairwise gene-gene Pearson correlation coefficients of CERES score profiles across 563 screens (19Q2 DepMap data release). The distribution of 17,633 CERES profile similarity is plotted as a quantile-quantile plot, and the top 18 most similar out of 17,633 genes are labelled. Genes associated with lipid metabolism are indicated in red.
(j) Pathway analysis of C12orf49 profile similarity. C12orf49 profile similarity scores for all 17,634 genes represented in the DepMap were mean-summarized by pathway as defined in the HumanCyc standard27. Tendencies towards pathway-level similarity (co-essentiality) and dissimilarity (exclusivity) with C12orf49 were tested using a two-sided Wilcoxon rank-sum test with multiple hypothesis correction using the Benjamini and Hochberg procedure.
Extended Data Fig. 5. Regulation of LDL uptake and LDLR expression by C12orf49.
(a) Bar plots showing the results of a low density lipoprotein (LDL) uptake assay across the indicated HAP1 cell lines using pHrodo-LDL probe. All data are represented as means ± standard deviation; n = two (SREBF1, SREBF2) or three (WT, FASN, LDLR, C12orf49, WT + C12orf49-V5, C12orf49 + C12orf49-V5) biologically independent experiments; one-way ANOVA.
(b) Bar plots showing the results of a transferin uptake assay across the indicated HAP1 cell lines using pHrodo-Transferin probe. All data are represented as means ± standard deviation; n = two (SREBF1, SREBF2) or three (WT, FASN, LDLR, C12orf49, WT + C12orf49-V5, C12orf49 + C12orf49-V5) biologically independent experiments; one-way ANOVA.
(c) Bar plots showing the results of a low density lipoprotein (LDL) uptake assay across the indicated in lipoprotein-deprived HAP1 cell lines using Dil-LDL probe. All data are represented as means ± standard deviation; n = two biologically independent experiments.
(d) Pathway enrichment analysis of BioID preys captured with N-terminal (top panel) or C-terminal (bottom panel) miniTurbo-tagged C12orf49 under normal growth conditions using the GO molecular function, biological process, cellular compartments, KEGG and Reactome standards. Terms for significantly enriched gene sets (p < 0.05, maximum term size 105) are indicated and bars depict mean percentage overlap with the indicated term. Numbers next to each bar indicate the mean overlap and term sizes, respectively. The greyscale color legend for p-values is indicated on the right. P-values were calculated using gProfileR69.
(e) Barplots depicting the number of proteins localizing to indicated cellular compartments for preys captured with N-terminal (grey) or C-terminal (black) miniTurbo-tagged C12orf49 in 293 cells under serum-starvation.
Extended Data Fig. 6. RNAseq and western blot analysis in response to serum or lipoprotein starvation and upon inhibition of the proteasome.
(a) Gene ontology enrichment analysis of genes upregulated under serum starvation in HAP1 wildtype (WT), C12orf49 or SREBF2-KO cells using GO molecular functions, GO bioprocesses and Reactome standards. Gene sets with overlapping members have been merged and bars depict mean percentage overlap with the indicated term. Numbers next to each bar indicate the mean overlap and term sizes, respectively. The greyscale color legend for p-values is indicated on the right; p-values were calculated using gProfileR69.
(b) Boxplots depicting mean expression and induction of genes assigned with the indicated term across HAP1 WT, C12orf49 and SREBF2-KO cells under normal (+FBS) and serum-starved (-FBS) conditions; n = three biologically independent experiments, two-tailed student’s t test. Boxes show IQR, 25th to 75th percentile, with the median indicated by a horizontal line. Whiskers extend to the quartile ± 1.5 x IQR.
(c) Western blotting analysis of SREBP2, LDLR and β-Actin levels across the indicated HAP1 co-isogenic knockout cell lines in response to overnight lipoprotein withdrawal (−) and a short refeeding period (−/+) in presence or absence of the proteasomal inhibitor MG132 (10μM MG132 for 5 hours) as indicated. Unprocessed full length and processed C-terminal SREBP2 products are indicated. Representative data from two biologically independent experiments.
Extended Data Fig. 7. Gating strategy for flow cytometry experiments.
Gating strategies for flow cytometry experiments for (a) co-culture assays and (b) LDL/Transferrin uptake assays. In all cases the following steps were taken: 1. Forward scatter area (FSC-A) vs. side scatter area (SSC-A) were used to separate cell events from debris. 2. Forward scatter height (FSC-H) vs. forward scatter width (FSC-W) was used separate single cells from aggregates. 3. Forward scatter area (FSC-A) vs. viability stain (7AAD/B695–40 or Zombie NIR/R780–60) was utilized to gate live cells. For co-culture assays, gating scheme for separating GFP/B530–30 vs RFP/YG610–20 positive cells including steps 1–3 are shown in panel (a). For Dil-LDL/YG585–15 quantification, marker-positive live cells were quantified relative to unstained controls following steps 1–3 as displayed panel (b).
Supplementary Material
ACKNOWLEDGMENTS
We thank members of the Moffat lab for helpful discussions. Queenie Huang, Michele Olivieri, Clare Sheene, Rummy Akthar and Shaan Sidhu are gratefully acknowledged for assistance with molecular biology experiments. Next-Generation Sequencing was performed at the Donnelly Sequencing Centre at the University of Toronto. Proteomics work was performed at the Network Biology Collaborative Centre at the Lunenfeld-Tanenbaum Research Institute, a facility supported by Canada Foundation for Innovation funding, by the Ontarian Government and by Genome Canada and Ontario Genomics (OGI-097, OGI-139). M.A. was supported by a Swiss National Science Foundation Postdoctoral Fellowship; K.A.L. was supported by a Vanier Canada Graduate Scholarship and Studentship award from the Kidney Cancer Research Network of Canada. M.B. was supported by a DFG Fellowship (Bi 2086/1–1). This research was funded by grants from the Canadian Institutes for Health Research (J.M., C.B, B.J.A. and A.-C.G.), Ontario Research Fund (B.J.A., C.B. and J.M) and Canada Research Chairs Program (J.M., C.B. and A.-C.G.). C.L.M., M.B., and M.R. are partially supported by grants from the National Science Foundation (MCB 1818293) and the National Institutes of Health (R01HG005084, R01HG005853).
Footnotes
COMPETING INTERESTS STATEMENT
J.M., B.A. and C.B are shareholders in Northern Biologics. J.M. is a shareholder in Pionyr Immunotherapeutics, is acting CSO and shareholder in Empirica Therapeutics, and is an SAB member and shareholder of Aelian Biotechnology. C.B. is an SAB member of Yumanity Therapeutics. The authors declare no competing interest.
REFERENCES
- 1.Chen M & Huang J The expanded role of fatty acid metabolism in cancer: new aspects and targets. Precis. Clin. Med. 2, 183–191 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen RR et al. Targeting of lipid metabolism with a metabolic inhibitor cocktail eradicates peritoneal metastases in ovarian cancer cells. Commun. Biol. 2, 281 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Imoto M Chemistry and biology for the small molecules targeting characteristics of cancer cells. Biosci. Biotechnol. Biochem. 83, 1–10 (2018). [DOI] [PubMed] [Google Scholar]
- 4.Menendez JA & Lupu R Fatty acid synthase (FASN) as a therapeutic target in breast cancer. Expert Opin. Ther. Targets 21, 1001–1016 (2017). [DOI] [PubMed] [Google Scholar]
- 5.Garber K Cancer anabolic metabolism inhibitors move into clinic. Nat. Biotechnol. 34, 794–795 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Röhrig F & Schulze A The multifaceted roles of fatty acid synthesis in cancer. Nat. Rev. Cancer 16, 732–749 (2016). [DOI] [PubMed] [Google Scholar]
- 7.Jones SF & Infante JR Molecular Pathways: Fatty Acid Synthase. Clin. Cancer Res. 21, 5434–5438 (2015). [DOI] [PubMed] [Google Scholar]
- 8.Benjamin DI et al. Diacylglycerol Metabolism and Signaling Is a Driving Force Underlying FASN Inhibitor Sensitivity in Cancer Cells. ACS Chem. Biol. 10, 1616–1623 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Birsoy K et al. Metabolic determinants of cancer cell sensitivity to glucose limitation and biguanides. Nature 508, 108–12 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Birsoy K et al. An Essential Role of the Mitochondrial Electron Transport Chain in Cell Proliferation Is to Enable Aspartate Synthesis. Cell 162, 540–551 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Costanzo M et al. A global genetic interaction network maps a wiring diagram of cellular function. Science 353, aaf1420–aaf1420 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fischer B et al. A map of directional genetic interactions in a metazoan cell. Elife 4, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harrison R, Papp B, Pál C, Oliver SG & Delneri D Plasticity of genetic interactions in metabolic networks of yeast. Proc. Natl. Acad. Sci. U. S. A. 104, 2307–12 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Szappanos B et al. An integrated approach to characterize genetic interaction networks in yeast metabolism. Nat. Genet. 43, 656–62 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wright AV, Nuñez JK & Doudna JA Biology and Applications of CRISPR Systems: Harnessing Nature’s Toolbox for Genome Engineering. Cell 164, 29–44 (2016). [DOI] [PubMed] [Google Scholar]
- 16.Doench JG Am i ready for CRISPR? A user’s guide to genetic screens. Nat. Rev. Genet. 19, 67–80 (2018). [DOI] [PubMed] [Google Scholar]
- 17.Currie E, Schulze A, Zechner R, Walther TC & Farese RV Cellular Fatty Acid Metabolism and Cancer. Cell Metab. 18, 153–161 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carette JE et al. Ebola virus entry requires the cholesterol transporter Niemann-Pick C1. Nature 477, 340–3 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hart T et al. Evaluation and Design of Genome-Wide CRISPR/SpCas9 Knockout Screens. G3 Genes, Genomes, Genet. 7, g3.117.041277 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Billmann M, Chaudhary V, ElMaghraby MF, Fischer B & Boutros M Widespread Rewiring of Genetic Networks upon Cancer Signaling Pathway Activation. Cell Syst. 6, 52–64.e4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Costanzo M et al. The genetic landscape of a cell. Science 327, 425–31 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ryczko MC et al. Metabolic Reprogramming by Hexosamine Biosynthetic and Golgi N-Glycan Branching Pathways. Sci. Rep. 6, 23043 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cheng C et al. Glucose-Mediated N-glycosylation of SCAP Is Essential for SREBP-1 Activation and Tumor Growth. Cancer Cell 28, 569–581 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang S et al. Site-specific O-glycosylation of members of the low-density lipoprotein receptor superfamily enhances ligand interactions. J. Biol. Chem. 293, 7408–7422 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Obaidat A, Roth M & Hagenbuch B The Expression and Function of Organic Anion Transporting Polypeptides in Normal Tissues and in Cancer. Annu. Rev. Pharmacol. Toxicol. 52, 135–151 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Horton JD, Goldstein JL & Brown MS SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J. Clin. Invest. 109, 1125–1131 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Romero P et al. Computational prediction of human metabolic pathways from the complete human genome. Genome Biol. 6, R2 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scott CC, Vossio S, Rougemont J & Gruenberg J TFAP2 transcription factors are regulators of lipid droplet biogenesis. Elife 7, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shimano H & Sato R SREBP-regulated lipid metabolism: convergent physiology — divergent pathophysiology. Nat. Rev. Endocrinol. 13, 710–730 (2017). [DOI] [PubMed] [Google Scholar]
- 30.Zhou Z, Zhou J, Su Z & Gu X Asymmetric Evolution of Human Transcription Factor Regulatory Networks. Mol. Biol. Evol. 31, 2149–2155 (2014). [DOI] [PubMed] [Google Scholar]
- 31.VanderSluis B et al. Genetic interactions reveal the evolutionary trajectories of duplicate genes. Mol. Syst. Biol. 6, 429 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ascencio D, Ochoa S, Delaye L & DeLuna A Increased rates of protein evolution and asymmetric deceleration after the whole-genome duplication in yeasts. BMC Evol. Biol. 17, 40 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.The UniProt Consortium. UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res. 47, D506–D515 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mitchell AL et al. InterPro in 2019: improving coverage, classification and access to protein sequence annotations. Nucleic Acids Res. 47, D351–D360 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Uhlen M et al. Tissue-based map of the human proteome. Science 347, 1260419–1260419 (2015). [DOI] [PubMed] [Google Scholar]
- 36.Nagy Á, Lánczky A, Menyhárt O & Győrffy B Validation of miRNA prognostic power in hepatocellular carcinoma using expression data of independent datasets. Sci. Rep. 8, 9227 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meyers RM et al. Computational correction of copy number effect improves specificity of CRISPR-Cas9 essentiality screens in cancer cells. Nat. Genet. 49, 1779–1784 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Behan FM et al. Prioritization of cancer therapeutic targets using CRISPR–Cas9 screens. Nature 568, 511–516 (2019). [DOI] [PubMed] [Google Scholar]
- 39.Klarin D et al. Genetics of blood lipids among ~300,000 multi-ethnic participants of the Million Veteran Program. Nat. Genet. 50, 1514–1523 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brown MS & Goldstein JL The SREBP pathway: Regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell 89, 331–340 (1997). [DOI] [PubMed] [Google Scholar]
- 41.Go CD et al. A proximity biotinylation map of a human cell. bioRxiv 796391 (2019). doi: 10.1101/796391 [DOI] [Google Scholar]
- 42.Costanzo M et al. Global Genetic Networks and the Genotype-to-Phenotype Relationship. Cell 177, 85–100 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Horlbeck MA et al. Mapping the Genetic Landscape of Human Cells. Cell 174, 953–967.e22 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Najm FJ et al. Orthologous CRISPR-Cas9 enzymes for combinatorial genetic screens. Nat. Biotechnol. 36, 179–189 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Han K et al. Synergistic drug combinations for cancer identified in a CRISPR screen for pairwise genetic interactions. Nat. Biotechnol. 35, 463–474 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Norman TM et al. Exploring genetic interaction manifolds constructed from rich single-cell phenotypes. Science 365, 786–793 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shen JP et al. Combinatorial CRISPR–Cas9 screens for de novo mapping of genetic interactions. Nat. Methods 14, 573–576 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gonatopoulos-Pournatzis T et al. Genetic interaction mapping and exon-resolution functional genomics with a hybrid Cas9-Cas12a platform. Nat. Biotechnol. 1–11 (2020). doi: 10.1038/s41587-020-0437-z [DOI] [PubMed] [Google Scholar]
- 49.Brenner A et al. Abstract P6–11-09: Heavily pre-treated breast cancer patients show promising responses in the first in human study of the first-In-class fatty acid synthase (FASN) inhibitor, TVB-2640 in combination with paclitaxel. in Cancer Research 77, P6–11-09–P6–11–09 (American Association for Cancer Research, 2017). [Google Scholar]
- 50.Gimeno RE Fatty acid transport proteins. Curr. Opin. Lipidol. 18, 271–6 (2007). [DOI] [PubMed] [Google Scholar]
- 51.Boyle EA, Pritchard JK & Greenleaf WJ High-resolution mapping of cancer cell networks using co-functional interactions. Mol. Syst. Biol. 14, e8594 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim E et al. A network of human functional gene interactions from knockout fitness screens in cancer cells. Life Sci. alliance 2, e201800278 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Luciano M et al. Association analysis in over 329,000 individuals identifies 116 independent variants influencing neuroticism. Nat. Genet. 50, 6–11 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kichaev G et al. Leveraging Polygenic Functional Enrichment to Improve GWAS Power. Am. J. Hum. Genet. 104, 65–75 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nagel M, Watanabe K, Stringer S, Posthuma D & van der Sluis S Item-level analyses reveal genetic heterogeneity in neuroticism. Nat. Commun. 9, 905 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Aregger M, Chandrashekhar M, Tong AHY, Chan K & Moffat J Pooled Lentiviral CRISPR-Cas9 Screens for Functional Genomics in Mammalian Cells. Methods Mol. Biol. 1869, 169–188 (2019). [DOI] [PubMed] [Google Scholar]
- 57.Hesketh GG, Youn JY, Samavarchi-Tehrani P, Raught B & Gingras AC Parallel exploration of interaction space by BioID and affinity purification coupled to mass spectrometry. Methods Mol. Biol. 1550, 115–136 (2017). [DOI] [PubMed] [Google Scholar]
- 58.Branon TC et al. Efficient proximity labeling in living cells and organisms with TurboID. Nat. Biotechnol. 36, 880–887 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Knight JDR et al. ProHits-viz: a suite of web tools for visualizing interaction proteomics data. Nat. Methods 14, 645–646 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu G et al. Data Independent Acquisition analysis in ProHits 4.0. J. Proteomics 149, 64–68 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Deutsch EW et al. Trans-Proteomic Pipeline, a standardized data processing pipeline for large-scale reproducible proteomics informatics. Proteomics. Clin. Appl. 9, 745–54 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Teo G et al. SAINTexpress: improvements and additional features in Significance Analysis of INTeractome software. J. Proteomics 100, 37–43 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dobin A et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Abdel Rahman AM, Ryczko M, Pawling J & Dennis JW Probing the Hexosamine Biosynthetic Pathway in Human Tumor Cells by Multitargeted Tandem Mass Spectrometry. ACS Chem. Biol. 8, 2053–2062 (2013). [DOI] [PubMed] [Google Scholar]
- 65.Hart T et al. High-Resolution CRISPR Screens Reveal Fitness Genes and Genotype-Specific Cancer Liabilities. Cell 163, 1515–26 (2015). [DOI] [PubMed] [Google Scholar]
- 66.Greene CS et al. Understanding multicellular function and disease with human tissue-specific networks. Nat. Genet. 47, 569–576 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ashburner M et al. Gene Ontology: tool for the unification of biology. Nat. Genet. 25, 25–29 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Liberzon A et al. Molecular signatures database (MSigDB) 3.0. Bioinformatics 27, 1739–1740 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Raudvere U et al. g:Profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res. 47, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and analysed in this study are included in the manuscript. The raw fastq files for the sequencing data are available upon request and have also been deposited to the Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/): RNA-sequencing data, GSE147770. All mass spectrometry data has been deposited to the MassIVE repository (https://massive.ucsd.edu/ProteoSAFe/static/massive.jsp) and assigned the accession number MSV000085005. The ProteomeXchange accession is PXD017719. Descriptions of the analyses, tools and algorithms are provided in the methods section and the Reporting Summary of this article. Custom code for generating gRNA counts from fastq files and code for generating qGI-scores will be made available on Github upon publication.