Abstract
Rearrangements of the mixed lineage leukemia gene MLL are associated with aggressive lymphoid and myeloid leukemias. The resulting MLL fusion proteins enforce high-level expression of HOX genes and the HOX cofactor MEIS1, which is pivotal for leukemogenesis. Both wild-type MLL and MLL fusion proteins interact with the tumor suppressor menin and with the Hoxa9 locus in vivo. Here, we show that MLL sequences between amino acids 5 and 44 are required for interaction with menin and for the transformation of hematopoietic progenitors. Blocking the MLL-menin interaction by the expression of a dominant negative inhibitor composed of amino terminal MLL sequences down-regulates Meis1 expression and inhibits cell proliferation, suggesting that targeting this interaction may be an effective therapeutic strategy for leukemias with MLL rearrangements.
Introduction
Translocations involving the mixed lineage leukemia (MLL) gene, the human homologue of Drosophila trithorax, are a common cause of acute lymphoid and myeloid leukemias. A pivotal feature of the more than 40 different MLL translocations identified is that their expression is associated with high-level expression of the clustered HOX genes, including HOXA7, HOXA9, and the HOX cofactor MEIS1 (1–3). MLL directly binds to HOX loci and other targets and methylates histone H3 at lysine 4 via the intrinsic methyltransferase activity of the COOH-terminal SET domain (4, 5). MLL has been reported to associate with a “core” complex composed of homologues of proteins present in the yeast Set1 methyltransferase complex including Ash2, Rbbp5, and WDR5 (6, 7). In addition, the mammalian MLL complex includes the tumor suppressor menin. Mutations in menin, which is encoded by the MEN1 gene, are associated with the multiple endocrine neoplasia syndrome type 1 (MEN1; ref. 8). This tumor predisposition syndrome is characterized by parathyroid hyperplasia and tumors of the pituitary gland and pancreatic islets (9). Importantly, menin is required for normal transcriptional regulation of MLL target genes (7, 10). Menin knockout mice show defects in Hox gene expression and severe defects in Hox gene expression in cell lines derived from these animals, which can be completely corrected by menin reexpression (10). Furthermore, MLL and menin bind directly to the p18Ink4c and p27Kip1 loci (6), suggesting that menin’s tumor suppressor activity is the result of its cooperative regulation of cyclin-dependent kinase inhibitors.
Although the mechanism by which menin acts as a transcriptional coactivator is unclear, the preponderance of the data suggest that menin is involved in recruiting MLL to transcriptional targets (6). Menin interacts either directly or indirectly with MLL as well as with RNA polymerase II (6, 10). Importantly, the point mutations in menin which occur in patients that have been tested thus far markedly reduce its binding to target genes (6).
Here, we show that this MLL-menin interaction occurs via a localized region of MLL and is crucial for continual immortalization. These findings extend previous observations on the role of menin in transformation by MLL fusion proteins (11, 12) and suggest that the menin-MLL interaction is a promising therapeutic target for leukemias with MLL rearrangements.
Materials and Methods
Cell lines
HEK 293 cells were grown in DMEM containing 10% FCS. MLL-AF9 cells were grown in Iscove’s modified Dulbecco’s medium (IMDM) containing 15% fetal bovine serum (FBS; Stem Cell Technologies) and 10 ng/mL of interleukin (IL)-3. MLL-ENL-ER cells were grown in RPMI 1640 containing IL-3, IL-6, granulocyte macrophage colony-stimulating factor (GM-CSF), stem cell factor (SCF; all from R&D Systems) as previously described (3), with the addition of 4-hydroxy-tamoxifen (4-OHT) at a final concentration of 100 nmol/L to MLL-ENL-ER cells.
Immunoprecipitations
A DNA fragment corresponding to amino acids 2 to 167 of human MLL was amplified by PCR and cloned into a MIGR1 vector (13) containing two FLAG epitopes, three nuclear localization signals, and two Myc epitopes, collectively adding ~10 kDa to the expressed MLL2–167 fragment. Further deletions were generated either by PCR or restriction enzyme digestion followed by religation (details available upon request). HEK 293 cells (1–1.5 × 106) were transfected with 10 μg of DNA using FuGene 6 (Roche Diagnostics Corporation). Two days after transfection, the cells were rinsed with PBS and lysed in 50 mmol/L of Tris-HCl (pH 8.0), 150 mmol/L of NaCl, 5 mmol/L of EDTA, and 0.5% NP40. Protein extracts were incubated with anti–FLAG M2-agarose (Sigma) or MLL612 anti-MLL antibody (14) plus Protein A-Sepharose (Amersham Biosciences), overnight and washed with 50 mmol/L of Tris-HCl (pH 8.0), 100 mmol/L of NaCl, 1 mmol/L of EDTA, and 0.2% NP40 thrice. The bound proteins were resolved on 10% or 4% to 12% NuPAGE gels (Invitrogen). The primary antibodies used for Western blot included MLL612 anti-MLL antibody, anti-menin antibody from Bethyl Laboratories, anti-FLAG antibodies from Sigma, and anti-hemagglutinin antibody from Roche. Horseradish peroxidase–conjugated secondary antibodies were obtained from Amersham Biosciences. The protein bands were visualized with SuperSignal West Dura Extended Duration Substrate (Pierce).
Chromatin immunoprecipitation assays
Chromatin immunoprecipitations (ChIP) were done as described (15) except that mouse antibodies were incubated overnight, then incubated with 2 μg of anti-mouse IgG for 7 h, then incubated with agarose A for 4 h, all at 4°C. Anti–estrogen receptor (ER) antibodies Ab10 and Ab3 were from Lab Vision-Neomarkers. The Ab1 antibody, which unlike Ab10 and Ab3, recognizes endogenous ER but not the ER in the fusion protein, was used as a negative control. ChIP was quantified as the percentage of inputs using TaqMan Real-Time PCR (Applied Biosystems) as previously described (15). TaqMan primer and probe sequences have been reported (16).
Retroviral transformation assays
MSCV retrovirus was produced by FuGene 6 transfection of Plat-E packaging cells (17). Virus was harvested 48 to 72 h after transfection, filtered and stored at −80°C. Titering on NIH 3T3 cells (18) yielded viral titers in the range of 106 to 107 colony-forming units (CFU)/mL. Retroviral transduction of primary bone marrow (BM) cells with MSCV-MLL-AF9, various deletion mutants of MSCV-MLL-AF9, or MSCV-Neo retrovirus was done as previously described (19). Briefly, 6-week-old C57Bl/6 mice (Taconic Farms) were primed with i.p. injections of 150 mg/kg of 5-fluorouracil (5-FU). Four days later, BM cells enriched in noncycling progenitors were harvested from the femurs and tibiae of mice and activated overnight in IMDM containing 15% FBS, IL-3, IL-6, and SCF. BM cells were transduced by spin-infection (SI) on 2 consecutive days with retroviral supernatants in the presence of Polybrene (Sigma). Cells were cultured in triplicate (104 cells/mL) in MethoCult M3234 methylcellulose medium (Stem Cell Technologies) in IMDM, 15% FBS, IL-3, IL-6, and GM-CSF, all at 10 ng/mL, and SCF at 100 ng/mL. G418 (Life Technologies) was added to a final concentration of 1 mg/mL. Cell growth and colony morphology and numbers were assessed during the course of three rounds of serial replating of methylcellulose cultures. After the third round of plating in methylcellulose, transduced cells were harvested and propagated in suspension cultures containing IL-3 at 10 ng/mL. For dominant negative experiments, the G418-resistant, MLL-AF9–transformed BM cells obtained from third-round passage in methylcellulose, or primary BM cells obtained from 6-week-old C57Bl/6 mice primed with 5-FU were transduced by spin-infection with MSCV retroviruses expressing FLAG-tagged MLL NH2-terminal fragments that interact with menin, or MSCV-MLL2–35, MSCV-MLL15–167, or the empty MSCV expression vector, which do not interact with menin. All of the MSCV constructs also express green fluorescent protein (GFP) from an internal ribosome entry site. Following transduction, the cells were sorted into GFP-positive cells on a FACSVantage SE (BD) flow cytometer to >95% purity and tested for methylcellulose colony–forming assay. Liquid cultures were analyzed by quantitating cell number by trypan blue exclusion, and reanalyzed by flow cytometric analysis at 4 and 11 days after sorting for GFP fluorescence.
Quantitative real-time PCR analysis of gene expression
Real-time PCR was used to quantify mRNA levels following reverse transcription. To determine gene expression, contaminating DNA from the RNA preparations was removed using TURBO DNA-free (Ambion). RNA was quantified and then reverse-transcribed (RT) using SuperScript First-strand Synthesis System for RT-PCR (Invitrogen). Random decamers and anchored oligo-dT were used for priming. For each sample, the reverse transcription step was done with 400 ng of total RNA. TaqMan sets for Hoxa9, Meis1, and Gapdh gene expression analysis were purchased from Applied Biosystems. The TaqMan set for MLL-AF9 fusion gene expression has been previously described (16). The TaqMan universal master mix (Applied Biosystems) was used for PCR. Relative quantification of each gene transcript in the different samples was done using the comparative CT method (Applied Biosystems; User Bulletin no. 2).
Results
Menin interacts with evolutionarily conserved sequences in the NH2 terminus of MLL
Preliminary immunoprecipitations using FLAG epitope–tagged MLL-AF9 showed that menin was immunoprecipitated with the fusion protein, and deletion of sequences spanning the amino terminal and AT hook region of MLL resulted in a loss of interaction. Conversely, amino terminal sequences of MLL spanning the AT hooks interacted with similar or better avidity than MLL-AF9 itself. Additional immunoprecipitations were done to further map the site of interaction. MSCV-based vectors expressing FLAG epitope–tagged deletion mutants of MLL, along with the GFP protein (Fig. 1; Supplementary Fig. 1), were transfected into 293 cells and immunoprecipitations were done with an M2 anti-FLAG antibody, followed by Western blot detection using an anti-menin antibody.
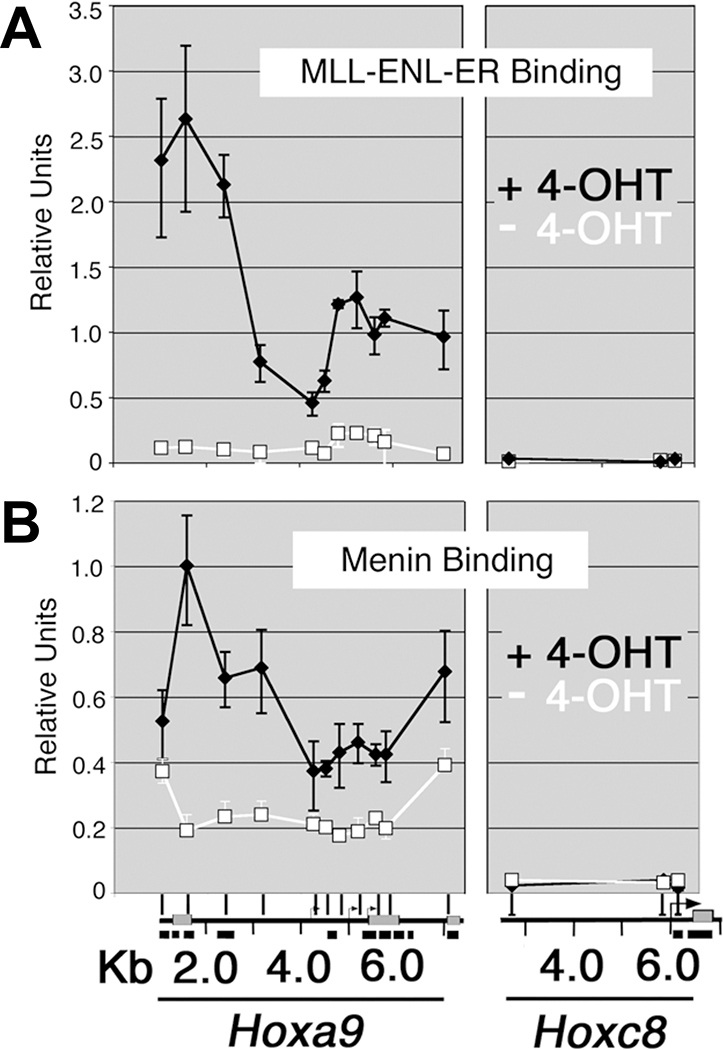
Figure 1.
Menin interacts with evolutionarily conserved sequences in the NH2 terminus of MLL. A, sequence alignment of the first 167 amino acids of human MLL with Fugu MLL. Dashes were introduced to achieve better alignment. “+”, similarity. B, schematic of the MLL NH2-terminal deletions and whether they coimmunoprecipitate with menin. C, 293 cells were transfected with individual FLAG-MLL deletion fragments, immunoprecipitated with M2 anti-FLAG antibody, and probed with anti-menin antibody. The extract was also probed by anti-FLAG antibody to show the expression and expected size of the FLAG-MLL fusion proteins (bottom).
These experiments show that MLL sequences between 5 and 44 are required for maximal menin binding. This spans the sequence CRWRFPARPG, which is 100% conserved with Fugu, Xenopus, and Zebrafish MLL (20), and includes the sequence SCRWR, which is conserved with the chromatin-associated enhancer of Polycomb-1 protein (Epc-1; ref. 21). Of note, sequences between 35 and 44 were necessary, but not sufficient, for high-affinity binding (Fig. 1C), which extends considerably beyond the five amino acid MLL-menin interaction domain RXRFP previously reported (11). The amino acids ALL between 37 and 39 reconstituted in the internal deletion mutant MLL Δ35–103 seem to be important as this reconstitution restores high affinity binding.
MLL fusion protein activation results in increased localization of menin at the Hoxa9 locus in vivo
To localize binding of menin and MLL fusion proteins across the Hoxa9 locus in vivo, we used a myeloblastic cell line transformed with the MLL-ENL-ER fusion protein (3), the activity of which is conditionally induced by 4-OHT. Hoxa9 expression in these cells is exquisitely dependent on the presence of 4-OHT (3). ChIP was done with antibodies specific for either menin or the MLL-ENL-ER fusion protein coupled with quantitative PCR detection (Fig. 2). In the absence of 4-OHT, no MLL fusion protein is bound to the Hoxa9 locus (Fig. 2A, white line), but low levels of menin are bound in both coding and noncoding regions in the locus (Fig. 2B, white line). In the presence of 4-OHT, the fusion protein associates with the locus and menin binding is increased in a similar distribution (Fig. 2A and B, black line) that overlaps the region bound by wild-type MLL (16). Hoxc8 is not expressed in these cells and is included as a negative control. Importantly, the close correlation of menin and MLL fusion protein binding strongly suggests that these proteins not only interact in solution, but also at target promoters.
Figure 2.
MLL fusion protein activation results in increased localization of menin at the Hoxa9 locus in MLL-ENL-ER–transformed cells. A, ChIP was done using an antibody directed against the estrogen receptor in MLL-ENL-ER and a series of PCR primers representing the Hoxa9 and Hoxc8 loci as described in Materials and Methods. The plot in (A), which is included for comparison with (B), is based on previously published data (16). Bottom, a schematic of the two Hox loci. In the presence of 4-OHT, the MLL fusion protein binds across a broad region of the Hoxa9 locus (black line). In the absence of 4-OHT (white line), MLL-ENL-ER binding is abolished and Hoxa9 expression is reduced (3). The Hoxc8 locus is not expressed in these cells and represents background levels of binding. B, ChIP using antibodies directed against menin shows that menin binding is highly correlated with binding of the MLL fusion protein. Menin binds across a broad region of the Hoxa9 locus, but not the Hoxc8 locus, in the presence of 4-OHT (black line). In the absence of 4-OHT, menin binding drops dramatically, although some residual binding remains (white line).
MLL sequences that interact with menin are required for transformation of hematopoietic progenitors
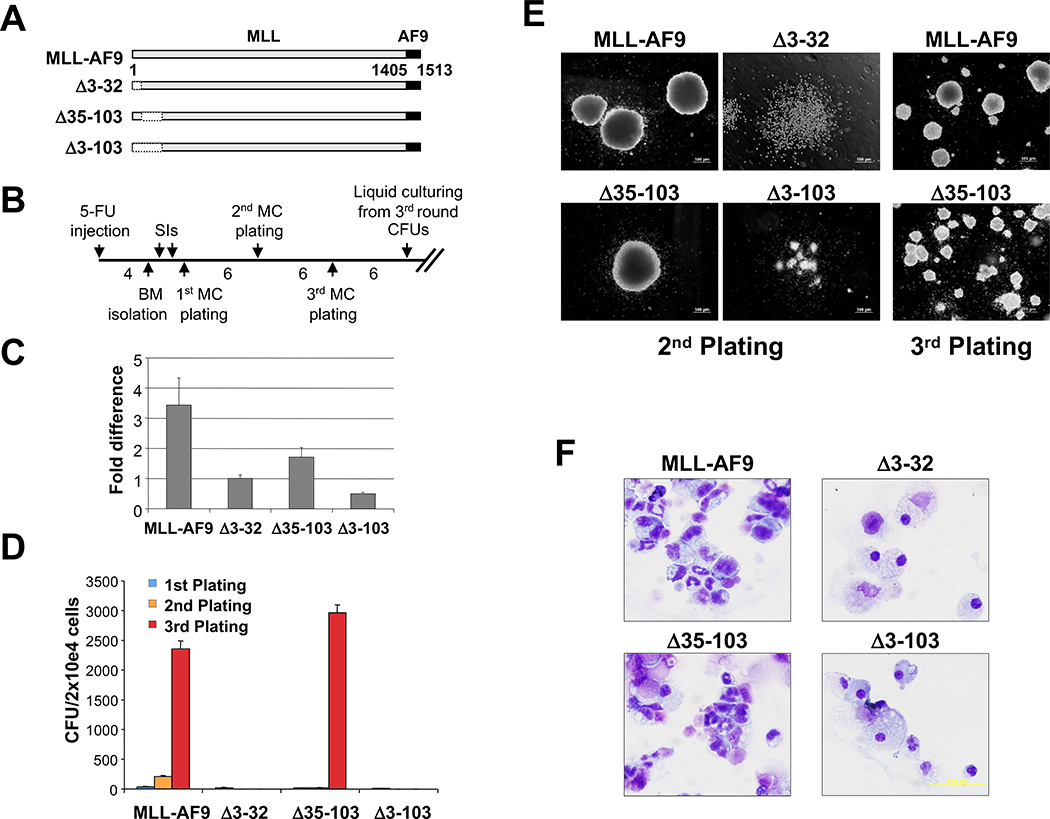
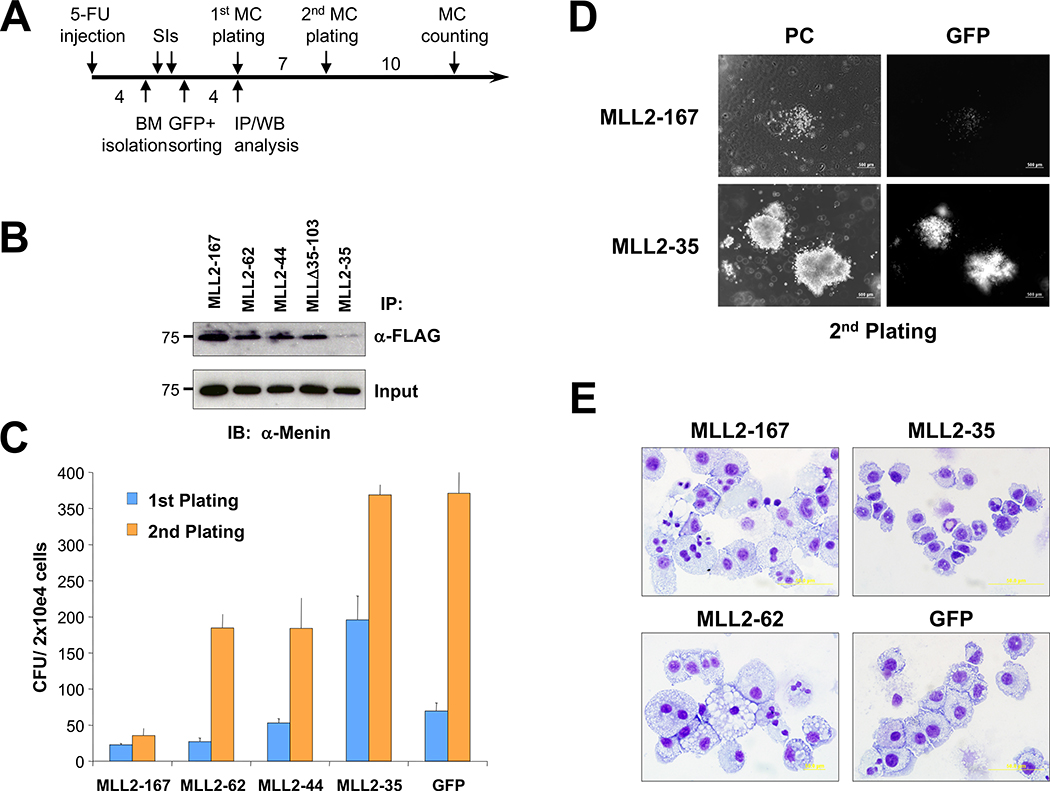
To test whether the MLL sequences involved with menin interaction, as well as the serine-rich sequences adjacent to this domain were required for transformation by MLL fusion proteins, MSCV retroviruses were produced that express MLL-AF9, or the internal deletion mutants MLL-AF9 Δ3–32, MLL-AF9 Δ35–103, or MLL-AF9 Δ3–103, along with the G418 resistance gene (Fig. 3A). These were transduced into BM cells from 5-FU primed mice (Fig. 3B) and their relative expression levels compared by quantitative real-time RT-PCR on total RNA extracted from G418-resistant BM cells at 9 days after transduction (Fig. 3C). Cells were replated in methylcellulose at ~6-day intervals, and the colony number (Fig. 3D) and morphology (Fig. 3E) were assessed as previously described (15). A similar number of colonies were present in the first plating with the different constructs; however, by the second plating, compact colonies characteristic of MLL fusion protein or Hoxa9/Meis1 transformation (19) were seen only in progenitors transduced with either the MLL-AF9 or MLL-AF9 Δ35–103 retroviruses. Wright-Giemsa–stained cytospins of MLL-AF9 or MLL-AF9 Δ35–103–transduced cell cultures obtained from colonies of third-round plating in methylcellulose showed a homogeneous population composed predominantly of blasts with occasional monocytes and apoptotic cells (Fig. 3F). BM transduced with MLL-AF9 Δ3–103, MLL-AF9 Δ3–32 (Fig. 3F), or MSCV-Neo (data not shown) showed a heterogeneous mixture of macrophages and mast cells with a smaller number of other differentiated myeloid cells and rare blasts. Continuously proliferating myeloblastic cell lines that grow in liquid medium containing IL-3 could only be established with cells transduced with MLL-AF9 or MLL-AF9 Δ35–103 retroviruses (Supplementary Fig. S3). Relative protein expression levels were also analyzed after transfection into 293 cells. Importantly, the nontransforming MLL-AF9Δ3–32 showed even higher levels of expression than the transforming MLL-AF9 or MLL-AF9Δ35–103 (Supplementary Fig. S3). These experiments indicate that the amino terminal MLL sequences that interact with menin, but not the serine-rich sequences between amino acids 35 and 103, are required for transformation by MLL-AF9.
Figure 3.
MLL sequences that interact with menin are required for transformation of hematopoietic progenitors. A, schematic of MLL deletion mutants made in the context of MLL-AF9. Left, the amino acid numbers (deletion mutants are not drawn to scale). B, schematic of the experimental procedure (see Materials and Methods). C, relative expression levels of transduced MLL-AF9 and MLL-AF9 deletion alleles analyzed by real-time quantitative RT-PCR normalized to Gapdh at 7 d of culturing in the presence of G418. D, the results of myeloid progenitor immortalization assays are summarized as the average of CFU per 2 × 104 cells plated in triplicate for each round of plating in methylcellulose. Columns, means of three independent experiments; bars, SD. E, morphology of the colonies formed after the second (left) and third (right) rounds of plating in methylcellulose of BM-transduced cells. F, Wright-Giemsa–stained cytospins of liquid cultures obtained from colonies of third-round plating in methylcellulose.
MLL peptides that disrupt MLL-menin interaction inhibit the growth of MLL-AF9 transformed cells
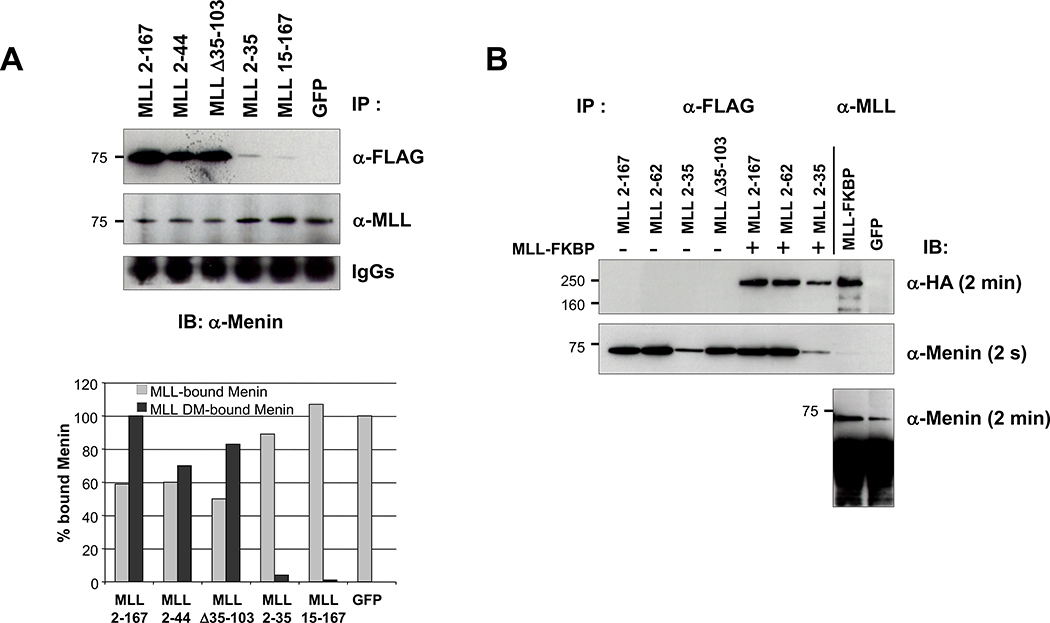
We then examined if the expression of a peptide derived from amino terminal MLL sequences would function as a dominant negative inhibitor of the MLL-menin interaction. We transfected 293 cells with plasmids expressing MLL deletion mutants and isolated GFP-positive cells by flow cytometric sorting. Cell lysates were immunoprecipitated using an M2 anti-FLAG antibody or an affinity-purified MLL612 anti-MLL antibody (14), followed by Western blot detection using an anti-menin antibody (Fig. 4A). The MLL612 antibody recognizes an epitope that is carboxyl terminal to the first 167 amino acids spanned by the deletion mutants and therefore reacts only with the endogenous MLL protein. Transfections of MLL2–167, MLL2–44, or MLL Δ35–103 deletion mutants, which bind menin with high affinity, resulted in ~50% reduction of menin binding with endogenous MLL, whereas transfection of MLL2–35 and MLL15–167 deletion mutants with low or no affinity did not alter the amount of menin bound to endogenous MLL (Fig. 4A, compare lanes 1–3 with lanes 4–6). Additional immunoprecipitations were done with a hemagglutinin epitope-tagged MLL fusion protein (MLL-FKBP; ref. 15) and the FLAG-tagged MLL dominant negative expression vectors. These experiments also show that amino terminal MLL sequences are apparently associated in vivo, because immunoprecipitation with anti-FLAG antibodies also immunoprecipitate the MLL-FKBP (Fig. 4B). In aggregate, these experiments show that MLL dominant negative mutants successfully inhibit menin interaction with MLL amino terminal sequences.
Figure 4.
Competition of MLL deletion fragments for menin binding with endogenous MLL. A, equal amounts of transfected cell extracts from GFP-positive–sorted cells were immunoprecipitated with M2 (anti-FLAG) or MLL612 (anti-MLL) antibodies, and then probed with anti-menin antibody. The representative Western blot was quantitatively determined by the ImageJ program (bottom), and the results presented as a percentage of endogenous MLL-bound menin relative to MSCV-GFP (gray columns) and as a percentage of MLL deletion mutant–bound menin relative to MSCV-MLL2–167 (black columns). B, MLL deletion mutants binding with cotransfected hemagglutinin-tagged MLL-FKBP into 293 cells. After transfections, cell extracts were immunoprecipitated with anti-FLAG and anti-MLL antibodies and Western blot analyzed with anti-hemagglutinin and anti-menin. MLL deletion mutants and MLL-FKBP coimmunoprecipitate through their association with menin. Immunoprecipitations for each MLL fragment were done at least twice (some immunoprecipitations were shown twice for figure consistency).
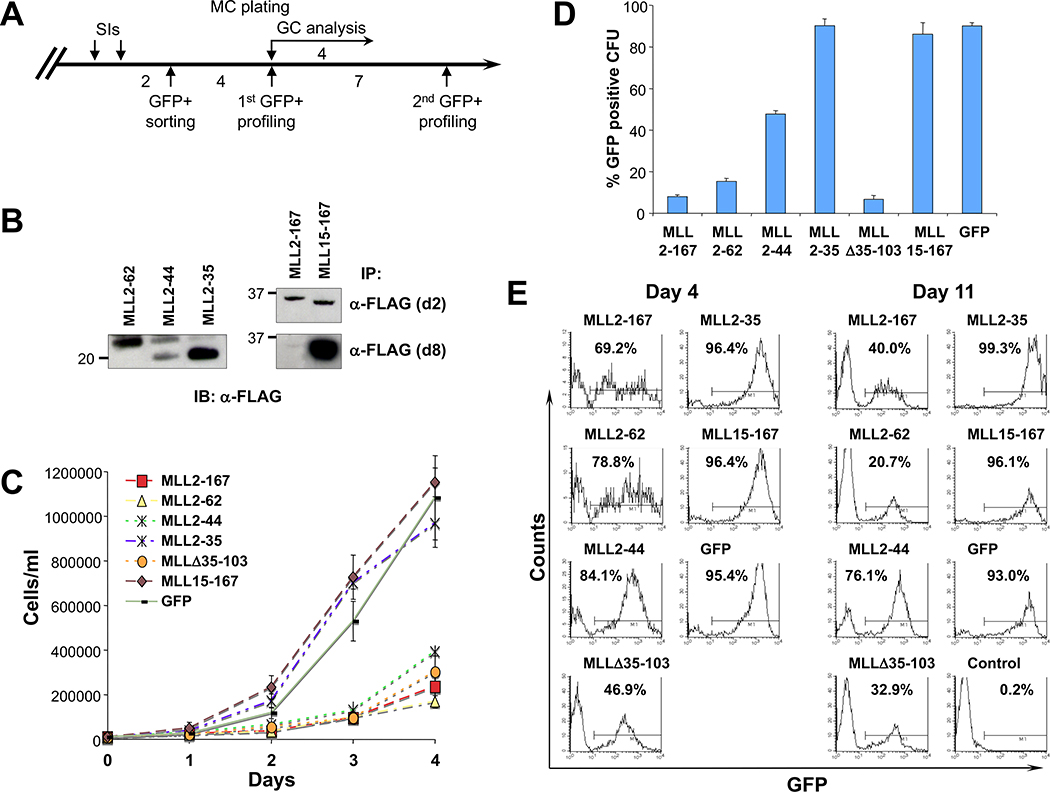
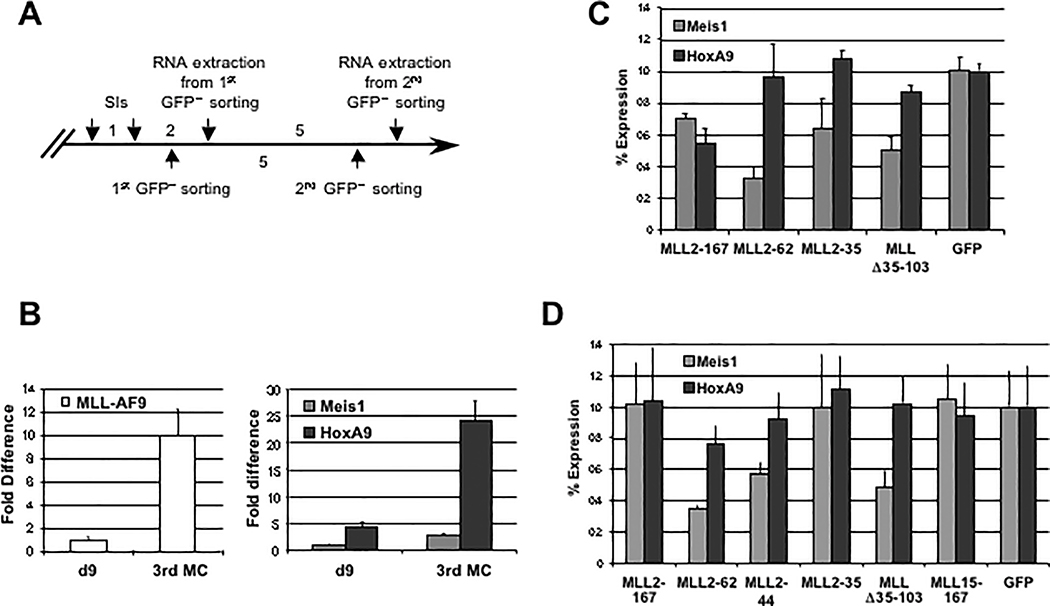
To determine if blocking the MLL-menin interaction inhibits growth, MLL-AF9–transformed cells derived from colonies at third-round methylcellulose plating were transduced with viruses encoding dominant negative amino terminal MLL sequences and flow sorted to obtain nearly 100% GFP-positive cells (Fig. 5A), and the level of ectopically expressed proteins was analyzed by Western blot at 2 and 8 days after cell sorting (Fig. 5B). When compared with GFP-positive cells transduced with MLL2–35, MLL15–167, or MSCV-GFP empty vector, cells expressing the MLL dominant negatives MLL2–167, MLL2–62, MLL2–44, and MLL Δ35–103 showed markedly impaired growth in liquid cultures (Fig. 5C).
Figure 5.
Transduction of MLL fragments with high menin-binding avidity inhibits cell growth. A, schematic of the experimental procedure (see Materials and Methods). B, immunoprecipitation analysis of MLL deletion mutants at 2 and 8 d after GFP sorting. Equal amounts of cell extracts from GFP-positive–sorted BM cells were immunoprecipitated with M2 anti-FLAG monoclonal antibody, and then immunoblotted with anti-FLAG polyclonal antibody. At 8 d postsorting, the expression levels of MLL2–167 were significantly reduced compared with other MLL dominant negative mutants. C, GC analysis on GFP-sorted cells shows that all MLL dominant negative cells profoundly inhibited cell growth, whereas all the other transduced cells proliferated rapidly. Points, means; bars, SD. D, results of methylcellulose colony-forming assay summarized as a percentage of GFP-positive CFUs obtained from 2 × 104 transduced cells 4 d after plating. MLL-AF9–transformed cells transduced with MLL2–167, MLL2–62, or MLL Δ35–103 fragments with high menin-binding avidity show a highly reduced percentage of GFP-positive colonies, whereas for MLL2–44 dominant negative cells, the reduction was ~50% compared with MLL fragments with weak or no interaction with menin. A representative of three independent experiments. Bars, SD. E, flow cytometric analysis of GFP signal of MLL-AF9–transformed cells after MLL fragment transduction and GFP sorting. The percentage of GFP-positive cells at 4 and 11 d after cell sorting. Expression of GFP in MLL2–167, MLL2–62, or MLL Δ35–103 is rapidly down-regulated or outcompeted by GFP-negative cells. In contrast, equally high and stable percentages of GFP-positive cells are seen within all the remaining transductions.
These findings were further supported by methylcellulose colony assays. When GFP-sorted cells were plated in methylcellulose, cells transduced with MLL2–167, MLL2–62, or MLL Δ35–103 generated <20% GFP-positive colonies, suggesting that these MLL dominant negative mutants strongly inhibited the growth of MLL-AF9–transformed BM cells (Fig. 5D; Supplementary Fig. S4). Of the colonies that formed in transduced cells, high colony numbers and percentages of GFP-positive colonies were seen only with the MLL2–35 and MLL15–167–transduced cells corresponding with peptides that did not interact with menin, or in MSCV-GFP control cells. The MLL2–44 transduced cells showed >40% GFP-positive colonies, indicating less growth inhibition than the larger dominant negative peptides. This might be the result of lower levels of MLL2–44 expression, as observed at 8 days after cell sorting (Fig. 5B). Alternatively, it is possible that the inclusion of additional MLL sequences disrupts interactions with menin more effectively or blocks interaction with other yet to be identified cofactors.
The growth-inhibitory effect of the MLL dominant negative mutants was also supported by long-term culture experiments (Fig. 5E; Supplementary Fig. S5). Flow cytometric analysis of GFP-sorted cells transduced with MLL dominant negative mutants showed the appearance of GFP-negative cells at 4 days after cell sorting and, with the exception of cells transduced with MLL2–44, a marked diminution in the percentage of GFP-positive cells at 11 days. Conversely, GFP-positive cells transduced with MLL2–35, MLL15–167, or MSCV-GFP empty vector maintained uniformly equal percentages, respectively, at 4 and 11 days after sorting, indicating that these cells did not inhibit growth. Together, these data show that MLL dominant negative inhibitors which strongly interact with menin inhibit the growth and colony-forming ability of MLL-AF9–transformed cells.
MLL dominant negative mutants down-regulate Meis 1 gene expression in MLL-AF9 transformed cells
We then tested whether the dominant negative inhibitors of the MLL-menin interaction blocked the expression of target genes important for maintaining transformation. Menin is required for the maintenance of HOX gene expression in HeLa and MLL-transformed cells (7, 11). The effect of retroviral transduction of MLL dominant negative mutants on Hoxa9 and Meis1 gene expression was tested on MLL-AF9 transformed myeloid progenitors obtained from colonies at third passage in methylcellulose (Figs. 3B and 6A). These cells showed a 10-fold increase of MLL-AF9 expression compared with MLL-AF9 immortalized BM cells at 9 days posttransduction, and a 5.4- and 2.7-fold increase of Hoxa9 and Meis1 expression levels, respectively (Fig. 6B). The effect of the dominant negative inhibitors on target gene transcription was then analyzed. Total RNA was extracted 24 h after GFP-positive flow sorting at 2 and 7 days after transduction, and quantitated by real-time PCR for the expression of Hoxa9 and Meis1 relative to that of Gapdh. The expression of Hoxa9 was reduced by ~50% of the dominant negative inhibitor in MLL2–167 GFP-sorted cells at 3 days posttransduction, but not in the other transduced cells (Fig. 6C). However, full Hoxa9 expression was reconstituted at 8 days posttransduction, possibly because of a significant decrease in MLL2–167 expression levels (Figs. 5B and 6D). In contrast, Meis1 expression was significantly decreased in MLL2–62 and MLL Δ35–103 transduced cells when compared with MSCV-GFP empty vector, and this down-regulation persisted at 8 days posttransduction (Fig. 6C and D).
Figure 6.
MLL dominant negative mutants interfere with Meis1 gene expression in MLL-AF9–transformed BM cells. A, schematic of the experimental procedure. MLL-AF9–transformed BM cells obtained from the third round of passage in methylcellulose were spin-infected (SI) with MSCV-GFP empty vector or encoding for the MLL deletion mutants indicated. The GFP-positive cells were flow-sorted at 2 and 7 d following transductions and total RNA was extracted 24 h later. B, quantitative real-time PCR comparison of MLL-AF9, Meis1, and HoxA9 expression among MLL-AF9 immortalized BM cells at 9 d posttransduction and MLL-AF9–transformed cells at the third round of passage in methylcellulose. C, quantitative real-time PCR for Meis1 and Hoxa9 expression levels normalized to Gapdh at 3 d posttransduction. D, quantitative real-time PCR for Meis1 and Hoxa9 expression levels normalized to Gapdh at 8 d posttransduction. Columns, percentages relative to MLL-AF9–transformed/MSCV-GFP vector–transduced BM cells; bars, SD.
MLL2–44 showed a >40% reduction on Meis1 expression, whereas MLL2–35 and MLL15–167 retroviral transductions, whose expressed peptides did not interact with menin (Fig. 1), had no significant effect on both Hoxa9 and Meis1 expression in MLL-AF9–transformed myeloid progenitors. These findings suggest that one mechanism of growth inhibition by the dominant negative inhibitors of the MLL-menin interaction is by inhibiting the MLL-menin interaction resulting in down-regulation of Meis1 expression.
MLL dominant negative mutants inhibit the colony-forming ability of wild-type myeloid progenitors
To determine whether the expression of MLL dominant negative mutants inhibits the growth of nontransformed hematopoietic progenitors, BM cells extracted from 5-FU–primed mice were transduced with viruses encoding MLL dominant negative mutants and then flow-sorted for their GFP expression (Fig. 7A). In GFP-sorted BM cells, a strong interaction of transduced MLL dominant negative mutants with endogenous menin was confirmed by immunoprecipitation with anti-FLAG antibody followed by immunoblot detection using anti-menin antibody (Fig. 7B).
Figure 7.
MLL dominant negative mutants inhibit the colony-forming ability of wild-type myeloid progenitors. A, experimental procedure (see Materials and Methods). B, interaction of transduced MLL dominant negative mutants with endogenous menin in GFP-sorted BM cells. Equal amounts of cell extracts from GFP-positive–sorted BM cells were immunoprecipitated with M2 anti-FLAG monoclonal antibody and then immunoblotted with anti-menin polyclonal antibodies. A 5% aliquot of each cell lysate was analyzed by Western blot as control for endogenous levels of menin (Input). C, results of methylcellulose colony-forming assay summarized as a percentage of GFP-positive CFU obtained from 2 × 104 transduced cells at 7 to 10 d from plating. Wild-type myeloid progenitor cells transduced with MLL dominant negative fragments with high menin-binding avidity show a reduced percentage of GFP-positive colonies compared with cells transduced with MLL fragments with weak or no interaction with menin (MLL2–35) or GFP alone. Bars, SD. D, morphology and density of GFP-positive colonies at 14 d from second methylcellulose plating. E, Wright-Giemsa–stained cytospins of cells obtained from colonies at the second round of plating in methylcellulose.
GFP-positive BM cells transduced with any of the MLL deletion mutants or MSCV-GFP empty vector do not proliferate when cultured in liquid medium in the presence of SCF, GM-CSF, IL-6 and IL-3, or IL-3 alone (Supplementary Fig. S3). When plated in methylcellulose at the second passage, elevated numbers of GFP-positive colonies, with morphology typical of CFU-M and CFU-GM, were seen only with MLL2–35 or MSCV-GFP empty vectors. Meanwhile, a lower number of GFP-positive colonies with reduced size and GFP intensity were counted with MLL dominant negative mutants (Fig. 7C and D).
Wright-Giemsa–stained cytospins of MLL dominant negative mutants or MSCV-GFP empty vector–transduced cells from colonies of second-round plating in methylcellulose showed a heterogeneous mixture of differentiated myeloid cells including granulocytes, macrophages, and mast cells (Fig. 7E). Collectively, these findings reveal that MLL peptides that interact with menin show inhibitory effects not only in leukemic cells transformed by MLL-AF9 but also in normal hematopoietic progenitors.
Discussion
The mechanism by which menin regulates transcription and the factors that regulate the targeting of menin to specific promoters are not well understood. Menin binds DNA nonspecifically, although the protein may also recognize specific DNA sequences (22). In addition, protein-protein interactions may play a role in menin recruitment to promoters. Menin interacts with a variety of transcription factors including activator protein-1 family members Jun D and c-Jun, nuclear factor κB, and others (8). In addition, menin interacts with phosphorylated serine 5 of the COOH-terminal domain in RNA polymerase II, and has a distribution at target loci nearly identical to RNA polymerase II (9). Our previous studies suggest that MLL association with transcriptionally active loci is highly menin-dependent (6). In aggregate, these data suggest that one role of menin is to recruit MLL histone methyltransferase complexes that promote the transcriptional elongation of target genes through interactions with progressive RNA polymerase II, which would result not only in methyltransferase activity, but potentially also the recruitment of the histone acetyltransferases CBP and/or MOF, both of which interact with MLL (23). However, our ChIP results suggest that alternatively, MLL fusion proteins may recruit additional menin molecules to target promoters.
Our data show that menin interacts with a limited amino terminal region of MLL. This domain is larger than the RXRFP interaction domain previously reported by Yokoyama et al. (11). In MLL deletion mutants, we found that sequences between 35 and 44 are required along with sequences between 5 and 15 (including amino acids RWRFP) for high-affinity binding with menin, inhibition of growth, and down-regulation of Meis1 gene expression in MLL-AF9–transformed cells, although additional MLL downstream sequences seem to be required for effective dominant negative interference with MLL-AF9 transformation. This has important implications for designing inhibitors of the MLL-menin interaction. Importantly, our data show that this interaction domain, but not the adjacent serine-rich regions of MLL, are required for transformation by MLL-AF9. Previous studies by our lab and collaborators, and work by Yokoyama et al. (11), show that continual menin expression is required for maintaining transformation by MLL fusion proteins, but not other leukemogenic fusion oncoproteins. The studies reported here extend these findings by providing proof of principle that inhibition of the MLL-menin interaction could be an effective therapeutic strategy in leukemias with MLL rearrangements and, possibly, in those lacking such rearrangements. Structural studies leading to a detailed view of the MLL-menin interaction will be an important next step in the development of small molecule inhibitors that disrupt the menin-MLL interaction. One complication we anticipate is that inhibitors of the MLL-menin interaction may have a low therapeutic index, as these inhibitors also impair normal hematopoiesis as judged by colony-forming assays.
The findings also raise the possibility that normal down-regulation of Hox gene expression might involve the abrogation of the MLL-menin interaction, by either interacting proteins or posttranslational modification. This would help explain how transcriptional down-regulation would occur in the face of continual MLL expression.
Supplementary Material
Acknowledgments
Grant support: NIH grants CA92251 (J. Hess), CA78815 (J. Hess), and by a Specialized Center of Research grant from the Leukemia and Lymphoma Society of America (J. Hess).
Footnotes
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
References
- 1.Ayton PM, Cleary ML. Transformation of myeloid progenitors by MLL oncoproteins is dependent on Hoxa7 and Hoxa9. Genes Dev 2003;17:2298–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hess JL. MLL: a histone methyltransferase disrupted in leukemia. Trends Mol Med 2004;10:500–7. [DOI] [PubMed] [Google Scholar]
- 3.Zeisig BB, Milne T, Garcia-Cuellar MP, et al. Hoxa9 and Meis1 are key targets for MLL-ENL-mediated cellular immortalization. Mol Cell Biol 2004;24:617–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Milne TA, Briggs SD, Brock HW, et al. MLL targets SET domain methyltransferase activity to Hox gene promoters. Mol Cell 2002;10:1107–17. [DOI] [PubMed] [Google Scholar]
- 5.Nakamura T, Mori T, Tada S, et al. ALL-1 is a histone methyltransferase that assembles a supercomplex of proteins involved in transcriptional regulation. Mol Cell 2002;10:1119–28. [DOI] [PubMed] [Google Scholar]
- 6.Milne TA, Hughes CM, Lloyd R, et al. Menin and MLL cooperatively regulate expression of cyclin-dependent kinase inhibitors. Proc Natl Acad Sci U S A 2005;102: 749–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yokoyama A, Wang Z, Wysocka J, et al. Leukemia proto-oncoprotein MLL forms a SET1-like histone methyltransferase complex with menin to regulate Hox gene expression. Mol Cell Biol 2004;24:5639–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chandrasekharappa SC, Guru SC, Manickam P, et al. Positional cloning of the gene for multiple endocrine neoplasia-type 1. Science 1997;276:404–7. [DOI] [PubMed] [Google Scholar]
- 9.Agarwal SK, Lee Burns A, Sukhodolets KE, et al. Molecular pathology of the MEN1 gene. Ann N Y Acad Sci 2004;1014:189–98. [DOI] [PubMed] [Google Scholar]
- 10.Hughes CM, Rozenblatt-Rosen O, Milne TA, et al. Menin associates with a trithorax family histone methyltransferase complex and with the hoxc8 locus. Mol Cell 2004;13:587–97. [DOI] [PubMed] [Google Scholar]
- 11.Yokoyama A, Somervaille TC, Smith KS, Rozenblatt-Rosen O, Meyerson M, Cleary ML. The menin tumor suppressor protein is an essential oncogenic cofactor for MLL-associated leukemogenesis. Cell 2005;123:207–18. [DOI] [PubMed] [Google Scholar]
- 12.Chen YX, Yan J, Keeshan K, et al. The tumor suppressor menin regulates hematopoiesis and myeloid transformation by influencing Hox gene expression. Proc Natl Acad Sci U S A 2006;103:1018–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pear WS, Miller JP, Xu L, et al. Efficient and rapid induction of a chronic myelogenous leukemia-like myeloproliferative disease in mice receiving P210 bcr/abl-transduced bone marrow. Blood 1998;92:3780–92. [PubMed] [Google Scholar]
- 14.Caslini C, Serna Alarcon A, Hess JL, Tanaka R, Murti KG, Biondi A. The amino terminus targets the mixed lineage leukemia (MLL) protein to the nucleolus, nuclear matrix and mitotic chromosomal scaffolds. Leukemia 2000;14:1898–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martin ME, Milne TA, Bloyer S, et al. Dimerization of MLL fusion proteins immortalizes hematopoietic cells. Cancer Cell 2003;4:197–207. [DOI] [PubMed] [Google Scholar]
- 16.Milne TA, Martin ME, Brock HW, Slany RK, Hess JL. Leukemogenic MLL fusion proteins bind across a broad region of the Hox a9 locus, promoting transcription and multiple histone modifications. Cancer Res 2005;65:11367–74. [DOI] [PubMed] [Google Scholar]
- 17.Morita S, Kojima T, Kitamura T. Plat-E: an efficient and stable system for transient packaging of retroviruses. Gene Ther 2000;7:1063–6. [DOI] [PubMed] [Google Scholar]
- 18.Pear WS, Nolan GP, Scott ML, Baltimore D. Production of high-titer helper-free retroviruses by transient transfection. Proc Natl Acad Sci U S A 1993; 90:8392–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lavau C, Szilvassy SJ, Slany R, Cleary ML. Immortalization and leukemic transformation of a myelomonocytic precursor by retrovirally transduced HRX-ENL. EMBO J 1997;16:4226–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Caldas C, Kim MH, MacGregor A, Cain D, Aparicio S, Wiedemann LM. Isolation and characterization of a pufferfish MLL (mixed lineage leukemia)-like gene (fMll) reveals evolutionary conservation in vertebrate genes related to Drosophila trithorax. Oncogene 1998; 16:3233–41. [DOI] [PubMed] [Google Scholar]
- 21.Stankunas K, Berger J, Ruse C, Sinclair DA, Randazzo F, Brock HW. The enhancer of polycomb gene of Drosophila encodes a chromatin protein conserved in yeast and mammals. Development 1998;125:4055–66. [DOI] [PubMed] [Google Scholar]
- 22.La P, Silva AC, Hou Z, et al. Direct binding of DNA by tumor suppressor menin. J Biol Chem2004;279:49045–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dou Y, Milne TA, Tackett AJ, et al. Physical association and coordinate function of the H3 K4 methyltransferase MLL1 and the H4 K16 acetyltransferase MOF. Cell 2005;121:873–85. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.