The outbreak of coronavirus disease 2019 (COVID-19) in December 2019 in Wuhan, China, introduced the third highly pathogenic coronavirus into humans in the 21st century. Scientific advance after the severe acute respiratory syndrome coronavirus (SARS-CoV) epidemic and Middle East respiratory syndrome coronavirus (MERS-CoV) emergence enabled clinicians to understand the epidemiology and pathophysiology of SARS-CoV-2. In this review, we summarize and discuss the epidemiology, clinical features, and virology of and host immune responses to SARS-CoV, MERS-CoV, and SARS-CoV-2 and the pathogenesis of coronavirus-induced acute respiratory distress syndrome (ARDS).
KEYWORDS: acute respiratory distress syndrome, hemophagocytic lymphohistiocytosis, human coronavirus
SUMMARY
The outbreak of coronavirus disease 2019 (COVID-19) in December 2019 in Wuhan, China, introduced the third highly pathogenic coronavirus into humans in the 21st century. Scientific advance after the severe acute respiratory syndrome coronavirus (SARS-CoV) epidemic and Middle East respiratory syndrome coronavirus (MERS-CoV) emergence enabled clinicians to understand the epidemiology and pathophysiology of SARS-CoV-2. In this review, we summarize and discuss the epidemiology, clinical features, and virology of and host immune responses to SARS-CoV, MERS-CoV, and SARS-CoV-2 and the pathogenesis of coronavirus-induced acute respiratory distress syndrome (ARDS). We especially highlight that highly pathogenic coronaviruses might cause infection-associated hemophagocytic lymphohistiocytosis, which is involved in the immunopathogenesis of human coronavirus-induced ARDS, and also discuss the potential implication of hemophagocytic lymphohistiocytosis therapeutics for combating severe coronavirus infection.
INTRODUCTION
Coronaviruses (CoVs) are classified into three groups: both group 1 (Alphacoronavirus) and group 2 (Betacoronavirus) include mammalian viruses, whereas group 3 (Gammacoronavirus) contains only avian viruses. Seven types of human CoVs have been identified to date (1, 2). Among them, HCoV-229E and HCoV-NL63 are part of the Alphacoronavirus genus; while the rest are part of the Betacoronavirus genus, including HCoV-OC43, HCoVHKU1, severe acute respiratory syndrome coronavirus (SARS-CoV), Middle East respiratory syndrome coronavirus (MERS-CoV), and SARS-CoV-2 (initially called 2019-nCoV). Coronaviruses did not attract worldwide attention until the SARS epidemic (3), followed by the MERS emergence (4) and, most recently, the coronavirus disease 2019 (COVID-19) pandemic outbreak (5, 6). These three highly pathogenic viruses (SARS-CoV, MERS-CoV, and SARS-CoV-2) derived from bats cause atypical pneumonia in humans that may evolve to acute lung injury (ALI) or acute respiratory distress syndrome (ARDS) leading to high morbidity and mortality, whereas infections with the other four low-pathogenic coronaviruses (HCoV-229E, HCoV-NL63, HCoV-OC43, and HCoVHKU1) lead to ailments such as mild upper respiratory illness in immunocompetent hosts, although they can also cause severe syndromes in those with weakened immunity (1, 6).
The identification of SARS-CoV in civets, MERS-CoV in domesticated camels, and SARS-CoV-2-like coronavirus in the intermediate horseshoe bat indicates that these viruses are able to leap the species barriers and may cause more outbreaks in the future. Although the intermediate host, transmissibility, and mortality among the three highly pathogenic viruses are distinct, the dysregulated host immune response to these viruses all resulted in ARDS, which is the primary cause of death among infected patients. The viral features of these highly pathogenic coronaviruses and advances in diagnosis and developing vaccines and therapeutics have been reviewed by Dhama et al. (7). This review was written to detail our current understanding of the epidemiology, clinical features, virology, dysregulated immune response of SARS, MERS, and COVID-19, and the pathogenesis of coronavirus-induced ARDS. We particularly highlight the potential role of the secondary hemophagocytic lymphohistiocytosis (HLH) on the immunopathogenesis of fatal coronavirus infection and discuss the future implication of using HLH therapeutics to combat severe coronavirus infection.
EPIDEMIOLOGY
The first SARS case probably occurred in live animal markets of Foshan, China, in November 2002 and increasingly spread around the globe in 2002 to 2003. The first major outbreak emerged and spread among medical workers and families through close contact in Guangzhou, China (3). Subsequently, a novel coronavirus was isolated from those atypical pneumonia patients and called SARS-CoV (8). After March 2003, the outbreak rapidly spread to other countries by air travel (9). Before the global outbreak was officially announced over in July 2003, it had caused 8,098 reported cases and 774 deaths (case-fatality rate, 9.6%) (10). After the 2003 epidemics, several patients with SARS were identified in 2005 because of contact with palm civets, which have been thought to be the source of infection (11).
In June 2012, a second Betacoronavirus called MERS-CoV was found in a patient who had died from a severe respiratory illness in Jeddah, Saudi Arabia (4). Following that, MERS broke out with 11 people affected, including eight health care workers, in a public hospital in Zarqa, Jordan. Subsequently, 30 MERS-infected cases were subsequently confirmed in Seoul, South Korea, in June 2015, which is the largest outbreak outside the Middle East (12). The nosocomial and travel-related transmission was reported to be increasing in the Middle East and other regions (12). The outbreaks of MERS-CoV were reported in 27 countries with 2,494 reported cases, including 722 deaths (case fatality rate, 34%) (13).
In December 2019, a number of atypical pneumonia patients associated with a seafood wholesale market were identified in Wuhan, China (14). A previously unknown coronavirus, later called SARS-CoV-2, was found in these patients (5). It infected men more than women, which is also a characteristic found in MERS-CoV, but not in SARS-CoV (15, 16). As of 21 June 2020, more than eight million confirmed cases with more than 461,000 deaths were reported globally (case fatality rate, 5.3%). The number is still increasing (Fig. 1) (17).
FIG 1.
The global spread of COVID-19. Location of COVID-19 cases and deaths, as of 10 June 2020. The countries colored in red are those where confirmed cases emerged. Darker colors indicate more cases. (Based on data from reference 213.)
Similar to the other two kinds of highly pathogenic coronaviruses, SARS-CoV-2 also spread mainly via large droplets and contact. There is a risk of fecal-oral and vertical transmission, but evidence of aerosol transmission is controversial (18). Since December 2019, a series of case reports confirmed human-to-human transmission of SARS-CoV-2 based on close patient contact with family members and health care providers (19). The superspreading events (SSEs) of SARS-CoV-2, which are associated with the rapidly increasing cases, tend to occur at close gatherings of households and large communities (20). Asymptomatic carriers may continue to transmit COVID-19 and lead to a possible epidemic rebound (21). Therefore, the World Health Organization (WHO) officially announced that COVID-19 is a pandemic, which means that the new coronavirus is a threat to humans around the world.
THE DISORDERS AND THE CORONAVIRUS
Manifestation of Disease
The main clinical features of highly pathogenic coronavirus are symptoms of acute pulmonary infection, ranging from asymptomatic or mild febrile illness to ALI and ARDS. ALI/ARD is a clinical syndrome characterized by decreased lung compliance, severe hypoxemia, and bilateral pulmonary infiltrates resulting from various diseases (sepsis, pneumonia, trauma) with extrapulmonary manifestations in some cases (Table 1) (22). The acute onset with fever, cough, myalgia, headache, and sore throat subsided in a few days in SARS patients (Table 1). The following clinical stage was characterized by high fever, shortness of breath, and hypoxemia, and two-thirds of the patients had atypical pneumonia (Table 1). After 2 weeks or so, approximately one-fifth developed ARDS with extrapulmonary manifestations, which is the primary cause of death in SARS-CoV infection (23). Compared with SARS-CoV, MERS-CoV infection progresses more rapidly to ARDS, septic shock, renal failure, and death, particularly in immunocompromised patients and those with comorbid conditions. Therefore, the time from onset to requiring ventilation in MERS-CoV-infected patients is shorter than that in SARS. The mortality rate of MERS (34%) is higher than that of SARS (10%), which may be associated with the prevalence of comorbidities (24, 25). Approximately three-fourths of patients who died of MERS were more likely to have at least one underlying comorbidity, including obesity, diabetes, systemic immunocompromising conditions, and chronic heart and pulmonary diseases (24). Similar to SARS, the gastrointestinal symptoms, such as vomiting and diarrhea, occurred in a third of patients with MERS (24). The main clinical features of COVID-19 vary from asymptomatic infection to severe atypical pneumonia with ARDS, which likely results in death. Compared to SARS and MERS, the symptoms of COVID-19 are more subtle, and many asymptomatic carriers or presymptomatic cases are less easily recognized, making SARS-CoV-2 more transmissible. At the onset of COVID-19, common manifestations were fever, dry cough, and fatigue, but only a minority of cases showed upper respiratory tract infection symptoms. Severe cases with dyspnea and hypoxemia usually occur 1 week after onset, and subacutely progress to ARDS and other multiple organ failures. Moreover, severe cases may manifest with low to moderate fever, even no fever, throughout the course of the disease, which may be because older patients are more likely to have severe disease and they may not have a good “fever response” (6, 26). The incidence of the probability of the progression to ARDS and mortality (approximately 5.7%) is lower than those with SARS and MERS. In addition, SARS-CoV-2 infection rarely causes diarrhea, which is more likely to occur in MERS or SARS (20 to 25%).
TABLE 1.
Comparison of clinical features among SARS, MERS, and COVID-19 patients
| Clinical feature | Value for disease (references) |
||
|---|---|---|---|
| SARS (24, 27) | MERS (24, 91) | COVID-19 (6, 26) | |
| Incubation period | |||
| Mean, days | 4.6 | 5.2 | 6.4 |
| 95% CI,a days | 3.8–5.8 | 1.9–14.7 | 2.1–11.1 |
| Serial interval, days | 8.4 | 7.6 | 7.5 |
| Basic reproduction no. | 2–3 | <1 | 2.2–3.6 |
| Patient characteristics | |||
| Age, median, yr | 50.0 | 39.9 | 55.5 |
| Sex (male:female), % | 43:57 | 64.5:35.5 | 68:32 |
| Disease progression (days) | |||
| Time from onset to ventilatory support | Mean 11 | Median 7 | Median 8 |
| Time from onset to death | Mean 23.7 | Median 11.5 | Mean 9.5 |
| Mortality, % | 9.6 | 34 | 5.3 |
| Presenting symptoms, % | |||
| Fever | 99–100 | 98 | 83 |
| Cough | 62–100 | 83 | 82 |
| Sputum production | 4–29 | 44 | 28 |
| Shortness of breath | 40–42 | 72 | 31 |
| Fatigue or malaise | 31–45 | 38 | 44 |
| Myalgia | 45–61 | 32 | 11 |
| Chills or rigors | 15–73 | 87 | NRb |
| Headache | 20–56 | 11 | 8 |
| Sore throat | 13–25 | 14 | 5 |
| Hemoptysis | 0–1 | 17 | 5 |
| Rhinorrhea | 2–24 | 6 | 4 |
| Diarrhea | 20–25 | 26 | 2–3 |
| Nausea and vomiting | 20–35 | 21 | 1 |
CI, confidence interval.
NR, not reported.
The laboratory characteristics of SARS and MERS were various degrees of pancytopenia, including lymphopenia and thrombocytopenia (Table 2) (8, 25). It was reported that the serum levels of lactate dehydrogenase (LDH), alanine aminotransferase (ALT), aspartate aminotransferase (AST), and creatine kinase (CK) are elevated in patients with fatal SARS and MERS (24, 25, 27, 28). Similar to SARS and MERS, the routine blood studies on admission showed lymphopenia in 35% of SARS-CoV-2-infected cases. ALT, AST, and LDH increased in 28% to 76% of patients. In severe SARS-CoV-2-infected cases, the D-dimer level was markedly elevated, and lymphocytes showed a progressive reduction. An estimated 63% of patients with COVID-19 had serum ferritin levels above the normal range (6, 26), whereas the data on ferritin concentrations in patients with SARS and MERS are unavailable. These laboratory features indicate that fatal coronavirus infections lead to multiple-organ damage, including the hematology, hepatic, and renal systems, among others.
TABLE 2.
Comparison of laboratory features among SARS, MERS, and COVID-19 patients
| Laboratory testa | % for disease (references) |
||
|---|---|---|---|
| SARS (27, 195, 196) | MERS (25, 91) | COVID-19 (6, 19, 26) | |
| WBC (<4.0 × 109/liter) | 25–35 | 14 | 25 |
| LYM (<1.5 × 109/liter) | 68–85 | 32 | 35 |
| PLT (<140 × 109/liter) | 40–45 | 36 | 12 |
| ALT (>50 U/liter) | 20–30 | 11 | 28 |
| AST (>40 U/liter) | 20–30 | 14 | 35 |
| LDH (>250 U/liter) | 50–71 | 48 | 76 |
Abbreviations: WBC, leukocytes; LYM, lymphocytes; PLT, platelets; ALT, alanine aminotransferase; AST, aspartate aminotransferase; LDH, lactate dehydrogenase.
Chest CT Findings in These Disorders
Pulmonary pathological types and imaging features among SARS, MERS, and COVID-19 patients share similarities (Table 3). The most common chest computed tomography (CT) imaging results for SARS are ground-glass opacification with or without consolidation. Overall, 24% of patients had ground-glass opacification only, 36% had consolidation only, and 39% had both (27). The pulmonary lesions were mainly located in the lower lobe and lateral belt of lungs with 30% being bilateral and 70% being unilateral (29). Multilobar involvement occurred in approximately half of the patients (27). The prominent chest imaging result of MERS was also ground-glass opacities, followed by consolidation. Moreover, MERS-CoV infection is more likely to lead to lesions in the lower lobe rather than upper lobes, and the lesions progressed more rapidly than those in SARS according to radiographic examination. The cardinal feature was peripheral distribution, followed by central distribution and combined distribution, and unifocal involvement was more common than multifocal involvement (30).
TABLE 3.
Comparison of pulmonary pathological types and imaging features among SARS, MERS, and COVID-19 patients
| Feature | Value for disease (references) |
||
|---|---|---|---|
| SARS (27, 29, 197–199) | MERS (30, 170, 200) | COVID-19 (31, 32, 120, 201) | |
| Pathologic types | DADa | DAD | DAD? |
| Bilateral pneumonia | 30% | 85.7% | 76% |
| Unilateral pneumonia | 70% | 14.3% | 24% |
| Ground-glass opacity | 63% | 65.5% | 86% |
| Peripheral distribution | 75% | 58% | 86% |
| Lower lung zone | 64.8% | 79.1% | 67% to ∼76% |
| Consolidations | 36% | 18.2% | 29% to ∼55% |
| Unifocal involvement | 54.6% | 69% | 29% |
| Multifocal involvement | 45.4% | 31% | 71% |
| Pneumothorax | 12% | 16.4% | 1% |
DAD, hyaline membrane formation was observed with exudate in the alveoli, and membranous organization was seen with the occlusion of alveoli, dilation of the alveolar ducts and sacs, and collapsing of the alveoli.
At the early stage of COVID-19, multiple small plaques were obvious in the subpleural, extraneous, posterior basal segment and lower lobe of the lungs. Furthermore, pulmonary multiple ground-glass shadows together with infiltration develop bilaterally; lung consolidation was seen in severe cases, while pleural effusion rarely occurred (6). Chest X-ray and CT findings show that 71% of patients had two or more lobes distributed, and the lower lobes were involved in 67% to 76% of patients. The lesions were distributed peripherally in 86% of patients, and 80% were located at the posterior part of the lungs (31). Approximately three-quarters of cases have bilateral pneumonia, and rest have unilateral pneumonia (32).
VIROLOGY
SARS-CoV, MERS-CoV, and SARS-CoV-2 belong to the Coronavirus genus in the Coronaviridae family. The possible origins of the three coronaviruses have been discussed (7), while further clarification is needed for genomic organization, protein domain composition, and cell entry receptors of these coronaviruses.
Genomic Organization and Protein Domain Composition of Coronavirus
Coronaviruses are the largest RNA viruses (100 to 160 nm in diameter) with enveloped and spherical particles. SARS-CoV and MERS-CoV contain a positive-sense, single-stranded RNA genome with approximately 30,000 bases comprising 11 potential open reading frames (ORFs). Among them, ORF1a and ORF1b encode nonstructural proteins (NSPs), which function in genome transcription and replication. The remaining ORFs encode major structural proteins spike (S), envelope (E), membrane (M), and nucleocapsid (N), which are related to cell invasion, virion formation, and release. These coronaviruses also encode many accessory proteins that are interspersed throughout the structural genes. Although these proteins are species specific, their functions are poorly understood since they are not necessary for replication (33).
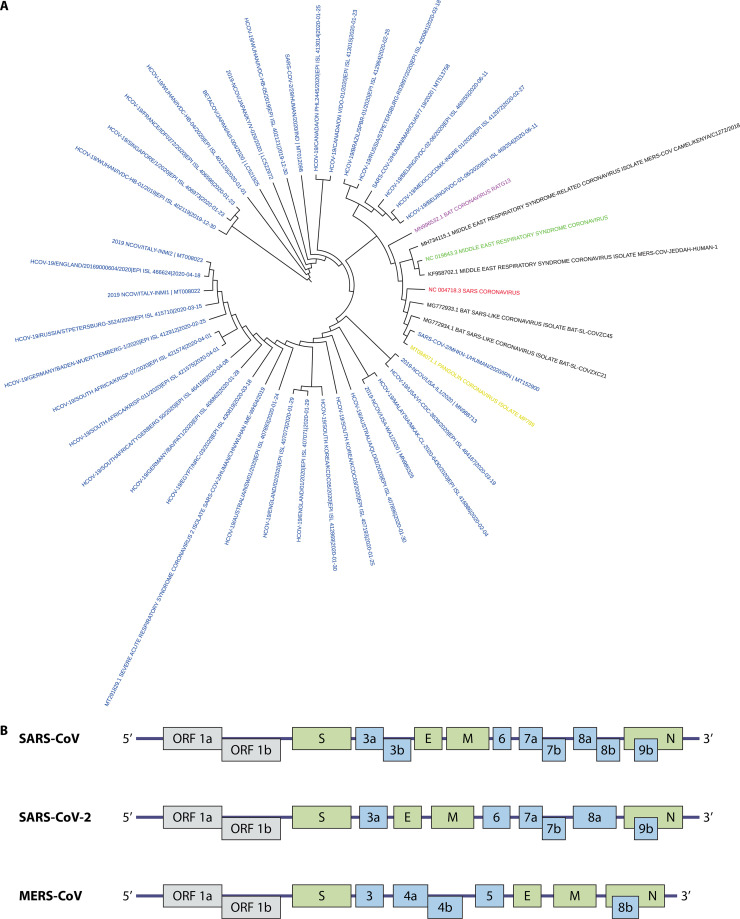
SARS-CoV-2, provisionally called 2019-nCoV, was identified in bronchoalveolar lavage fluid (BALF) samples from patients using next-generation sequencing (5). SARS-CoV-2 is classified into subgenus Sarbecovirus of the genus Betacoronavirus according to the phylogenetic analysis (Fig. 2A). SARS-CoV-2 possesses at least 12 coding regions, including 1ab, S, 3, E, M, 7, 8, 9, 10b, N, 13, and 14, three (10b, 13, and 14) of which are different from coding regions in SARS-CoV (Fig. 2B). As for the whole-genome sequence, SARS-CoV-2 is relatively closest to bat CoV RaTG13 and was distinct from SARS-CoV, indicating the transmission mode of SARS-CoV-2 from animals to humans.
FIG 2.
(A) The phylogenetic tree of representative betacoronavirus. Colors indicate different types of coronavirus: SARS-CoV (red), MERS-CoV (green), SARS-CoV-2 (blue), pangolin-CoV (yellow), bat CoV RaTG13 (purple). Whole-genome sequence was downloaded from NCBI and GISAID and underwent maximum-likelihood phylogenetic analyses. (B) Genome organization of three highly pathogenic coronaviruses (SARS-CoV, MERS-CoV, and SARS-CoV-2). The genes encoding structural proteins (spike [S], envelope [E], membrane [M], and nucleocapsid [N]) are in green. ORF 1a and ORF 1b, which encode nonstructural proteins, are in gray. The genes encoding accessory proteins are in blue.
The SARS-CoV-2 genome was reported to encode 27 proteins: four structural proteins (S, E, M, and N), eight accessory proteins (3a, 3b, p6, 7a, 7b, 8b, 9b, and orf14), and 15 nonstructural proteins (nsp1 to -10 and nsp12 to -16) (7, 34). Among four major structural proteins, the S protein of coronaviruses, a large class I fusion protein, participates in receptor binding and membrane fusion and plays a crucial role in host selection and transmissibility (7). Generally, S protein is divided into two parts: S1 domain, interacting with the host cell receptor, and S2 domain, mediating fusion with the cellular membrane. The receptor-binding domain (RBD) of Betacoronaviruses is located at the C-terminal end of the S1 domain consisting of one core surrounded by an external subdomain. The crystal structure analysis of SARS-CoV-2 RBD demonstrated the higher binding affinity to angiotensin-converting enzyme 2 (ACE2) receptor than that of SARS-CoV RBD, which provides a possible explanation for SARS-CoV-2 having strong infectivity (35, 36). The SARS-CoV-2 S protein has been extensively implicated in the development of diagnostic kits, vaccines, and therapeutic antibodies (7, 37, 38).
Cellular Entry of Coronavirus
The S protein of SARS-CoV binds to ACE2 on the cell membrane and predominantly infects ciliated bronchial epithelial cells and type I/type II pneumocytes (39–41). The ACE2 protein is expressed in human type I and type II pneumocytes, the luminal surface of ciliated bronchus cells of bronchus, enterocytes in the small intestine, the brush border of the proximal tubular cells, the endothelial cells, and arterial smooth muscle cells but is not expressed on T or B cells or macrophages in the spleen or lymphoid (42, 43). Although colonic enterocytes and liver cells lack the expression of ACE2 protein, viruses have been found in the colon and hepatocytes (42, 44). In contrast, ACE2 is present on the endothelial cells and the smooth muscle cells, but there is no evidence of viral particles and viral genome in these cells (42). The binding of spike protein to ACE2 resulted in the reduced expression of the receptor in the lungs and drove ALI during SARS because the downregulation of ACE2 leads to the excessive production of angiotensin II, which increases pulmonary vascular permeability (45, 46). The structural similarity between the receptor-binding domains of SARS-CoV-2 and SARS-CoV suggests that SARS-CoV-2 might use ACE2 as the receptor (2). SARS-CoV-2 was identified to use the cell entry receptor ACE2 in the ACE2-expressing HeLa cells (47). Besides, SARS-CoV was found to bind to dendritic cell (DC)-specific intercellular adhesion molecule 3 grabbing nonintegrin (DC-SIGN) on the surface of dendritic cells (DCs) and macrophages, which allows the cells to transfer infectious SARS-CoV to susceptible target cells, such as pneumocytes and monocytes, but does not facilitate viral infection of these cells (48). Liver/lymph node-specific ICAM-3-grabbing integrin (L-SIGN) can bind to SARS-CoV S protein, mediating viral entry and thus serving as an alternate receptor for SARS-CoV (49). MERS-CoV attaches to dipeptidyl peptidase 4 (DPP4; also known as CD26) receptor and infects unciliated bronchial epithelial cells and type II pneumocytes (50). The DPP4 highly expressed in the kidney accounts for common renal dysfunction or failure in patients. A recent study reported that the spike protein of SARS-CoV-2 bound to a novel receptor, CD147, and invaded the host cells, suggesting alternative receptors involving the invasion of SARS-CoV-2 (51). Whether the key residue variations in the receptor-binding region of SARS-CoV-2 affect ACE2 binding or change receptor tropism requires further study (2).
IMMUNE RESPONSE ASSOCIATED WITH SECONDARY HEMOPHAGOCYTIC LYMPHOHISTIOCYTOSIS IN FATAL CORONAVIRUS INFECTIONS
Immune response is essential to clear the coronavirus. After coronavirus invades the human body, the innate immune system is activated, which recognizes coronavirus and induces proinflammatory cytokines and chemokines. This process is followed by adaptive immune system activation, in which activated T cells directly kill virus-infected cells and B cells produce pathogen-specific antibodies. Immune response is essential for virus clearance, but it may also do harm to normal host tissues (52). Hyperinflammatory states have been confirmed to develop in the three highly pathogenic coronavirus-induced ARDS, and even death, which evoked considerable interest in cytokine-directed therapeutics to mitigate against such excessive immune responses (53, 54). The underlying mechanism of the exaggerated immune responses in fatal coronavirus infections is not understood. The observed severe lymphopenia and various degrees of pancytopenia, elevated ferritin, compromised liver function, abnormal clotting profiles, hypertriglyceridemia, and hypercytokinemia indicate that secondary hemophagocytic lymphohistiocytosis (sHLH) might play a crucial part in the pathogenesis of fatal COVID-19, SARS, and MERS (6, 26), although few clinical and laboratory indices in sHLH are distinct from fatal coronavirus infections (Table 4). The primary/familial or secondary hemophagocytic lymphohistiocytosis (fHLH or sHLH), a life-threatening syndrome related to severe hypercytokinemia, is thought to result from uncontrolled hyperactivation of gamma interferon (IFN-γ)-producing T cells and macrophages (55–57). The predominant causes of secondary HLH are the virus, neoplasms, and autoinflammatory and autoimmune diseases, whereas primary HLH is a typical autosomal recessive phenotype caused by mutations in the genes related to NK and CD8+ cytotoxic T cell functions (58). The secondary HLH associated with rheumatic diseases is also known as macrophage activation syndrome (MAS) (59). Infection-associated HLH has been reported to cause death in patients with Epstein-Barr virus (EBV), herpesviruses, HIV, influenza virus (H1N1 or H5N1), parvovirus, and hepatitis viruses (60). The cardinal clinical features of HLH include fever, hepatosplenomegaly, pancytopenia, fibrinolytic coagulopathy, hyperferritinemia, and hypohepatia, and the syndrome’s key immunological features are characterized by low cytotoxic lymphocyte activity (perforin and CD107a), increased T cell activation (soluble interleukin-2 receptor alpha chain, sCD25), increased macrophage activation (soluble CD163, soluble CD206, ferritin), and hemophagocytic activity. Although HLH is recognized more frequently, it is challenging to diagnose HLH due to strict criteria (61). In the following context, the role of NK, T lymphocytes, and macrophages in the dysregulated innate and adaptive immune responses associated with sHLH in severe coronavirus infection will be emphasized (Fig. 3).
TABLE 4.
Clinical and laboratory differences between COVID-19 and sHLH patients
| Finding | Value for disease (reference[s])a |
|||
|---|---|---|---|---|
| S-COVID-19 (202) | IAHS (203–205) | MAS (202) | MHLH (206) | |
| Fever | +++ | +++ | +++ | +++ |
| Hepatomegaly | + | +++ | ++ | +++ |
| Splenomegaly | − | +++ | ++ | +++ |
| Hemophagocytosis | +/− | +++ | +++ | +++ |
| Lymphopenia | ++ | +++ | + | +++ |
| Anemia | + | +++ | + | +++ |
| Low NK activity | + | + | + | NR |
| Elevated liver enzymes | ++ | ++ | ++ | +++ |
| Hypercytokinemia | +++ | +++ | +++ | NR |
| Hyperferritinemia | ++ | +++ | +++ | NR |
| Elevated sCD25 | + | NR | + | NR |
| Hypertriglyceridemia | +/− | +++ | ++ | NR |
| Hypofibrinogenemia | +/− | +++ | ++ | +++ |
| Coagulopathy | ++ | +++ | ++ | +++ |
| Multiorgan failure | +++ | +++ | + | +++ |
| ARDS | +++ | +++ | + | +++ |
Abbreviations: S-COVID-19, severe COVID-19; IAHS, infection-associated hemophagocytic syndrome; MAS, macrophage activation syndrome; MHLH, malignancy-associated HLH; NR, none reported; −, negative; +/−, not essential; +, slight; ++ moderate; +++, severe.
FIG 3.
A proposed model of the immunopathogenesis of human coronavirus-induced acute respiratory distress syndrome. In alveoli, coronavirus primarily infects pneumocytes through binding to specific receptors (ACE2 for SARS/SARS-CoV-2, DPP4 for MERS). These coronaviruses repress the induction of type I IFN through inhibiting the nuclear activation and translocation of IRF3, which allows coronavirus to replicate unrestrainedly. Meanwhile, pneumocytes produce proinflammatory cytokines and chemokines to mediate the recruitment of monocytes and lymphocytes. Usually, NK cells suppress viral replication by directly killing infected cells via granule or indirectly activating the macrophages via IFN-γ in the early phase of infection. However, unknown genetic or acquired factors cause NK cell cytotoxicity impairment in coronavirus infection. Specialized cross-presenting DCs ingest and process infected cells and present virus antigen to CD8+ T lymphocytes. Functional CD8+ CTLs then specifically destroy virus-infected cells through releasing granule and IFN-γ. Antigen or cytokine-induced T cell apoptosis might contribute to the lymphopenia observed in coronavirus infection. Alveolar macrophages and recruited monocytes accumulate in the lung microvasculature and are activated by persistent virus antigen stimulation, IFN-γ, or oxidized phospholipids. IFN-γ binds to IFNGR and subsequently induces the phosphorylation of STAT1 by JAK1/2 to promote the transcription of IFN-stimulated genes and proinflammatory cytokines/chemokines. Activated macrophages mainly contribute to the cytokine storm and hemophagocytosis, which might cause bone marrow hematopoietic inhibition and pancytopenia, and then lead to the inability of NK cells and cytolytic CD8+ T cells to lyse infected cells in the lung. Activated macrophages also release several toxic mediators inducing pneumocyte and lung endothelial cell apoptosis. Stimulated by virus particles or cytokines, endothelial cells express cell adhesion molecules and promote leukocyte extravasation.
Innate Immune Response to Coronavirus
The innate immune response forms the first line of host defense against coronavirus infection. It mainly consists of natural killer (NK) cells, macrophages, DCs, and molecules such as type I interferon (IFN), chemokines, and cytokines.
Defective type I IFN response.
Type I IFN, whose most important action is to inhibit viral replication in both infected and uninfected cells, is a key component of the innate immune response to combat the virus. Type I IFN is induced by several viral pathogen-associated molecular patterns (PAMPs) that are mediated by pattern recognition receptor (PRR), including endosomal Toll-like receptors (TLRs), cytoplasmic retinoic acid-inducible gene I protein (RIG-I), and melanoma differentiation-associated protein 5 (MDA5) (52). Type I IFN facilitates virus clearance through several parallel antiviral pathways (62). However, type I IFN was not detected in the lungs and serum of SARS or MERS patients, as well as in in vitro experiments (63–65). Moreover, SARS-CoV infection in vitro failed to activate nuclear transcriptional factor IFN regulatory factor 3 (IRF3) (66). In parallel, recent whole-blood transcriptome and serum cytokine profiles indicated that interferon-stimulated genes (ISGs) were significantly downregulated and type I IFN activity was low in severe COVID-19 patients, suggesting the impairment of type I IFN antiviral response (67). But it is crucial to investigate whether the production of IFN is delayed or decreased after a peak during the early onset of infection using longitudinal sera. Indeed, the delayed type I IFN response and unrestrained viral replication promote inflammatory responses and lung immunopathology in mice infected with SARS-CoV (68). Dynamically monitoring type I IFN production and interferon-stimulated gene expression in virus-susceptible cells or mice infected with SARS-CoV-2 will help clarify the role of type I IFN antiviral immunity in the pathogenesis of fatal cases.
The mechanism of the type I IFN defective response to SARS-CoV, MERS-CoV, and SARS-CoV-2 is not fully understood, but many viral components have been proved to participate in the process and help the virus escape the antiviral response. At least eight proteins in SARS-CoV antagonize IFN and interferon-stimulated gene (ISG) responses (69), and several proteins have been identified with similar functions in MERS-CoV (70). NSP14 and NSP10-16 complex can cap viral mRNAs, thus preventing the SARS-CoV mRNAs from being recognized by MDA5 and IFIT1 (70). The nucleocapsid protein of SARS-CoV also has an inhibitory effect on IFN induction (71). SARS-CoV ORF3b inhibits type I IFN by directly interfering in its production and indirectly preventing the phosphorylation of IRF3 (72). The membrane protein of SARS-CoV represses type I IFN production by preventing the formation of TRAF3 TANK/TBK1/IKKε complex, while MERS-CoV M protein is able to inhibit the nuclear translocation of IRF3 and the activation of type I IFN promoter (72). MERS-CoV ORF4a acts as an IFN suppressor by binding double-stranded RNA (dsRNA) and subsequently inhibiting MDA5 (73, 74). Moreover, MERS-CoV ORF4a, ORF4b, ORF5, and membrane protein all have an inhibitory effect on nuclear trafficking of IRF3 and activation of the IFNB promoter (75). Without adequate type I IFN, the highly pathogenic coronavirus replicates unrestrainedly to a high titer in the target cell, especially in pneumocytes, which further amplifies the aberrant inflammatory responses by increased viral PAMPs. The advanced understanding of the fundamentals of defective IFN production in fatal coronavirus infection will identify novel therapeutic targets.
NK cell cytotoxicity impairment in response to coronavirus.
NK cells can suppress viral replication by directly killing infected cells via releasing granules that contain perforin and granzymes or indirectly activating macrophages with phagocytosed microbes via IFN-γ release in the early course of infection (52). In addition, NK cells also possess an immunoregulatory function of restraining the overactivation and expansion of cytotoxic T lymphocytes to maintain immunological homeostasis. The role of NK cells in the highly pathogenic coronavirus clearance has not been fully elucidated. To date, only a few studies have shown a relationship between NK cell number reduction in the peripheral blood from SARS patients and the severity of disease (76, 77). Accordingly, recent data found that severe COVID-19 patients are characterized by depletion and functional exhaustion of NK cells, especially CD107a+ NK cells, perforin+ NK cells, and IFN-γ+ NK cells, which are possibly induced by the elevated inhibitory receptor NKG2A (78, 79). An interesting finding is the exhaustion of NK cells recovered during the convalescent stage after efficacious therapy in some patients.
The factors that trigger the exhaustion of NK cells in critical patients infected with virulent coronavirus might be classified as genetic or acquired. The genetic factors related to NK dysfunction in HLH mainly refer to the recessive hereditary defects in several genes encoding packaging and trafficking components of NK cytolytic granules, including perforin (PRF1), lysosomal-trafficking regulator (LYST), rab-27A (RAB27A), Munc 13-4 (UNC13D), syntaxin-binding protein 2 (STXBP2), syntaxin 11 (STX11), and adaptor-related protein complex 3 subunit beta 1 (AP3B1) in familial HLH. Thus, the absent granule-mediated cytotoxicity of NK cells and the inability of NK cells to lyse infected cells and eliminate pathogens do result in an uncontrolled but ineffective immune response including persistent antigenemia, constant IFN-γ-dependent stimulation, and prolonged innate cell and adaptive immune cell interactions. Such repeated antigen stimulation and presentation, in turn, lead to persistent proliferation and activation of T cells and excessive production of proinflammatory cytokines, especially IFN-γ, which can stimulate macrophages with a cytokine storm (80). Research has demonstrated that MAS with rheumatoid disease had the defective NK cell number and NK cell dysfunction, which are partially due to the rare biallelic protein-encoding or intronic mutations, even heterozygous variants or functional single nucleotide polymorphism (SNP) in fHLH-related genes. Moreover, several fHLH-associated gene variants were identified in fatal cases of H1N1 influenza virus infection. It cannot be excluded that some severe COVID-19 patients, especially those without comorbidity or who are not elderly, possess the genetic susceptibility in NK function-related genes, which remains to be elucidated, whereas NK functional impairment in most fatal coronavirus infections might be associated with acquired factors including age, comorbidities, viral pathogens, and excessive cytokines. Although a subset of severe coronavirus infection was reported to occur in youth without comorbidities, most deceased COVID-19 patients are characterized by aging and underlying diseases, which might indicate that disease- or age-related intrinsic NK cell impairments contribute to the uncontrolled immune response. The immunosenescence process is known to be related to a decrease in the function of innate and adaptive immunity in the elderly (81). The low proportion of perforin+ NK cells in elderly persons leads to the early defect in cytotoxic activity of NK cells in response to the virus (82). Therefore, aging and comorbidities are regarded as risk factors for poor outcome in patients with COVID-19. Evidence has shown that H1N1 influenza virions are capable of inhibiting the cytotoxicity of NK cells by directly infecting NK cells and inducing their apoptosis (83). It is unclear whether highly pathogenic coronavirus could enter into and replicate in NK cells, contributing to their exhaustion. For most MAS patients without causative mutations in cytotoxicity, elevated IL-6 under a hyperinflammatory milieu is thought to transiently impair NK cell cytotoxic function, which could reverse upon inflammation suppression. Other cytokines, such as IL-12 or IL-18, are also involved in NK cell overactivation, inducing cell death or exhaustion (84). These data suggested that the impairment of NK cell function might be attributed to hypercytokinemia in severe cases of coronavirus infection.
Two subsets of NK cells are usually described in the peripheral blood. The major subset is the CD56dim CD16bright population possessing cytolytic activity, whereas the minor subset is the CD56bright CD16−/dim population responding to inflammatory cytokines. During viral infection, NK cells express activating receptors for cytokines, antibody-coated cells, induced-self ligands, and virus-encoded ligands, including IFN-α receptors, interleukin-12 (IL-12) receptors, IL-18 receptors, CD16, NKG2D, DNAM-1 (CD226), and NKp46/NKp30, which are the surface markers for specific NK cell subsets (85). The unresolved issue is that the subsets of reduced NK cells in fatal coronavirus infection need further clarification since most studies assess the numbers and function of NK cells in COVID-19 patients using CD56 surface marker. The further investigation of the subsets, activation, and function of NK cells in the peripheral blood or postmortem lung and lymphoid organs might explain dysregulated NK function in fatal coronavirus infections.
Macrophage activation.
As a vital component of the innate immune response, macrophages engulf pathogens and infected cells and subsequently eliminate them through respiratory burst. In the meantime, they also secrete cytokines that facilitate the clearance of pathogens and promote tissue repair. Macrophages are commonly classified into two subsets: classically activated (M1) and alternatively activated (M2) macrophages. The former produce proinflammatory cytokines (e.g., IL-1β, IL-6, tumor necrosis factor alpha [TNF-α]), while the latter secrete anti-inflammatory cytokines (e.g., IL-10 and transforming growth factor beta [TGF-β]) (86). Usually, macrophages first exhibit the M1 phenotype to eliminate pathogens, and then there comes M2, which suppresses the inflammation and promotes tissue repair. Indeed, multinucleate giant macrophages are the prominent infiltrating leukocytes in pulmonary alveoli of severe SARS and COVID-19 cases (87–89). Also, RNA sequencing analysis found that mononuclear phagocyte (MNPs) consisted of 80% of total cells in the bronchoalveolar lavage fluid (BALF) from severe COVID-19 patients and most MNPs in BALF were monocyte-derived macrophages instead of alveolar macrophages (89), which indicates a possible macrophage activation in these patients. In line with the prominent infiltration of macrophages in the lung, dysregulated macrophage activation, which has been observed during coronavirus infection, is thought to participate in the pathogenesis of coronavirus-induced disease (90–92). High levels of ferritin and cytokine profiles in sera of deceased COVID-19 patients similar to those seen in HLH suggest that the hyperactivation of macrophages was highly involved in the disease progression, which has been linked to the pathogenesis of HLH. It will be noteworthy to detect the levels of soluble scavenger receptor (sCD163) and soluble mannose receptor (sCD206) in serum or the proportion of CD163+/CD206+ macrophages in the postmortem lung or lymphoid organs, which have been the indicators of macrophage activation observed in HLH.
Several mechanisms including PAMPs (e.g., virus components) and damage-associated molecular patterns (DAMPs) (e.g., cytokines, oxidative stress) may participate in the hyperactivation of macrophages in highly pathogenic coronavirus infection. Macrophages may not be the target cell for SARS-CoV, since monocyte-derived macrophages and purified monocyte macrophages are only abortively infected by SARS-CoV (93, 94). However, MERS-CoV replicates in monocyte-derived macrophages, stimulating the expression of tumor necrosis factor alpha (TNF-α), IL-6, IL-12, IFN-γ, and chemokines (CCL2, CCL3, CXCL8, and CXCL10) (95). However, whether macrophages are the target cell for SARS-CoV-2 remains elusive (92). Immunostaining analysis found that ACE2-expressing macrophages located in lymph node and spleen contained SARS-CoV-2 nucleoprotein (96), but this is more likely to be the result of other infected cells taken up by macrophages rather than direct viral infection, since ACE2 is undetectable on most tissue-resident macrophages from COVID-19 patients and viral gene expression had not been observed in peripheral blood mononuclear cells (PBMCs) from COVID-19 patients (96, 97). Although DC-SIGN and CD147 on the surface of macrophages have been suggested to bind to coronavirus and mediate the virus entry (48, 51), there is insufficient evidence to support the direct infection of SARS-CoV-2 into human macrophages.
Macrophages mainly recognize RNA viruses through PRRs such as Toll-like receptors (TLRs), retinoic acid-inducible gene I (RIG-I), and MDA5, which subsequently activate downstream signaling pathway NF-κB and IRF3/7 and promote the production of proinflammatory cytokines (98). It has been reported that coronavirus components such as viroporin A, E protein, ORF3 protein, and SARS-CoV ORF8b activate NLRP3 inflammasomes in macrophages (99–101). Stimulating murine macrophages with S protein of SARS-CoV in vitro induced an NF-κB-dependent production of proinflammatory cytokines (IL-6 and TNF-α) (102). Considering the similarity between SARS-CoV and SARS-CoV-2, genomic components and proteins of SARS-CoV-2 may also have a similar function. The hyperactivation of macrophages during coronavirus infection may also be due to the dysfunctional NK cell and cytotoxic T lymphocyte (CTL) response, which results in an impaired virus clearance and excessive IFN-γ production (103). Persistent coronavirus stimulation promotes the production of chemokines by pneumocytes and lung endothelial cells, which enhances the recruitment of monocytes in the lung; in turn, monocyte-derived macrophages are stimulated by unrestrained virus replication and secrete a large amount of cytokines. Recently, several studies have identified the expansion of inflammatory monocytes in the peripheral blood of severe COVID-19 cases (104–106). In addition, IFN-γ from dysfunctional NK cells and CTL cells binds the IFNGR on the surface of macrophages and subsequently facilitates STAT1 phosphorylation through JAK1/2, which promote the transcription of IFN-stimulated genes (CXCL10, CLXL9, and others) and proinflammatory cytokines. Macrophages engulf and degrade erythrocytes (RBC), leukocytes (WBC), and platelets through either CD163 on the surface or IFN-γ-induced STAT1 activation, which leads to hyperferritinemia and high soluble CD163 levels. This is called hemophagocytosis, which has been reported in the lung tissues from deceased SARS-CoV and COVID-19 patients (107, 108). Interestingly, ferritin’s H-chain also activates macrophage (109, 110). Moreover, the anti-S protein IgG and oxidized phospholipids (OxPLs) were also suggested to activate macrophages through Fcγ receptors (FcγRs) and Toll-like receptor 4 (TLR4)-TRIF signaling, respectively (92, 111). The exact pathways that trigger the hyperactivation of macrophages are not fully clarified, and further elucidation of other pathways involved in macrophage activation is necessary to develop potential therapeutics.
Dysregulated Cellular Immune Response to Coronavirus
Most previous studies focused on the innate immune response to SARS-CoV or MERS-CoV infection. Thus, the role that the adaptive cellular immune response plays in the host response to these highly pathogenic coronaviruses is largely unexplored. The cytotoxic T lymphocyte (CTL) is the key component of adaptive cellular immunity in combating viral infection. During this process, specialized cross-presenting DCs ingest infected cells in the lung and present the viral antigens to naive CD8+ T cells in the secondary lymphoid organs, resulting in the proliferation and differentiation of CD8+ T cells. Differentiated CD8+ CTLs then migrate into the infection site under the attraction of chemokine CXCL10 and specifically destroy virus-infected cells by releasing perforin and granzymes. Meanwhile, CTLs secrete IFN-γ at the site of infiltration, mediating the activation of macrophages. In addition, CTLs kill the antigen-processing DCs and limit sustained antigen presentation, thus suppressing the hyperactivation of T cells.
Clinically, the global lymphopenia is distinctly observed in many SARS, MERS, and COVID-19 patients, more prominently in fatal cases. However, it is controversial which kind of T lymphocyte, CD4+ or CD8+, has more pronouncedly reduced levels during coronavirus infection (26, 112–114). The mechanism of lymphopenia in highly pathogenic coronavirus infection is not known, but there are several possible explanations for this phenomenon including chemokine-mediated redistribution, virus-induced destruction, bone marrow suppression, and apoptosis. Considering the antiviral defense role of CTLs, it is reasonable to deduce that antigen-presenting cell (APC)-activating CTLs are recruited to the lung and clear the infected cells. Unexpectedly, the lymphocytic infiltration is scanty in the lung or bronchoalveolar lavage fluid in fatal SARS and COVID-19 autopsies (89, 107), although CD8+ T cells were abundant in the BALF from mild COVID-19 patients. Due to the lack of autopsy results, the cellular infiltration in the lungs of MERS patients is not available. These data might exclude the possibility that chemokine-mediated redistribution led to lymphopenia and also partly explain the unrestrained viral replication due to lack of CTL infiltration in the lung of severe coronavirus infection patients. Furthermore, it is unclear whether the direct invasion of T cells by coronavirus is linked to lymphopenia. In vitro experiments found that MERS-CoV infected T cells and induced T cell apoptosis through both extrinsic and intrinsic apoptosis pathways (115). In addition, SARS-CoV-2 was confirmed to abortively infect T cells by binding to the ACE2 receptor and might cause T cell apoptosis, although the level of ACE2 on the surface of T cells is low (116). Although there is no evidence indicating the productive infection of T cells by SARS-CoV, the obvious apoptosis of lymphocytes in peripheral lymphoid organs (spleen, lymph nodes, and lymphoid tissues of the gut) was found in deceased SARS or COVID-19 patients (96, 117). Antigen stimulation or cytokine storm might induce the apoptosis of T cells in secondary lymphoid organs. To date, excessive CXCL10 induced by viral infections has been found to rapidly recruit activated T lymphocytes, followed by cell apoptosis (117). Moreover, high levels of TNF-α and IFN-γ and a large amount of histiocytosis result in bone marrow hematopoietic inhibition and pancytopenia (80, 118). Myeloid-derived suppressor cells (MDSCs) have been recognized to promote the apoptosis of T lymphocytes (119). Recent data identified the expansion of MDSCs paralleled by the decrease of NK cell and T cells in COVID-19 patients, suggesting that the impairment of cytotoxic function in fatal SARS-CoV-2 infection might be related to MDSCs (78). Further exploration to ascertain the triggers of MDSC expansion in SARS-CoV-2 infection may be necessary.
In addition to the significant decrease in peripheral CD4+ and CD8+ T cell count, the cytotoxic activity of CTLs has also been impaired during SARS-CoV-2 infection, as represented by the decreased proportion of CD107a+ and granzyme B+ CD8+ cells (79). However, hyperactivation of T cells has been described in COVID-19 cases, characterized by the increased percentage of CCR4+ CCR6+ Th17 cells and pathogenic granulocyte-macrophage colony-stimulating factor-positive (GM-CSF+) IFN-γ+ Th1 cells in CD4 T cells (105, 120). In parallel, soluble IL-2 receptor (sCD25), a marker for T cell activation in HLH, was markedly increased in most severe COVID-19 patients (114). The possible explanation for T cell hyperactivation is likely related to the persistent antigen stimulation and presentation caused by the exhaustion of NK cells and CTLs. Regulatory T (Treg) cells might be another potential player in the T cell hyperactivation. Treg cells have a unique suppressive function in the immune system and prevent systemic inflammation. Recent data showed that COVID-19 patients have lower numbers of regulatory T cells, more pronounced in fatal infection (121). In an HLH mouse model mimicked by lymphocytic choriomeningitis virus-triggered perforin-deficient mice, increased IL-2 consumption by highly activated CD8+ T cells together with low IL-2 secretion by conventional CD4+ T cells caused a collapse of the Treg cell numbers and high sCD25 levels (122). Moreover, the patients experiencing HLH flares had low Treg cell numbers (122). The dysregulation of the IL-2/CD25/Treg cell axis provides a potential mechanistic explanation for uncontrolled immune response in HLH (123). The relationship between the overactivation and decline of number or cytolytic function of CTLs warrants further clarification. Dynamic monitoring and subsets of CD8+ T cells using single-cell RNA sequencing should clarify the critical role of CD8+ T cells in the dysregulated immune response in fatal coronavirus infection.
The role of cellular immune response in human coronavirus infection is also explored using animal models. Infecting young mice with MA15 virus (a SARS-CoV variant) induced respiratory disease accompanied by inefficiently activated DCs and a subsequently barely detectable antivirus T cell response (124). Depleting CD4+ T cells while preserving CD8+ T cells led to enhanced interstitial pneumonitis with delayed viral clearance, diminished virus-specific antibody, decreased cytokines, and reduced pulmonary recruitment of lymphocytes in SARS-CoV-infected mice (125). CD8+ T cells were proved to be necessary for MERS-CoV clearance in a mouse model (126, 127). Usually, elderly individuals present worse outcomes after coronavirus infection. Age-related increase in prostaglandin D (PGD) expression in the lungs impedes DCs migration, causing attenuated T cell responses and more severe clinical symptoms in older mice infected with respiratory viruses. PGD blockade accelerated DC migration, T cell responses, and survival (128). Details on the expansion, activation, and functions of CD4+ and CD8+ T cells would facilitate our understanding of coronavirus pathogenesis. The relationship between deficient NK cells and CTL functions and the proliferation and hyperactivation of macrophages and T cells is not fully understood. To date, accumulating evidence indicates that minor genetic predisposition and main acquired defect associated with the interplay between virus and host immune system converge to a hyperinflammatory state and death from coronavirus infection.
Cytokine Storm
Cytokine storm, a fatal systemic inflammatory response, refers to the rapid and massive production of various cytokines after infection or autoimmune disease (129). This excessive immune response promotes macrophage activation and aggregates the inflammation, leading to severe pneumonia, multiple organ failure, or even death (6). The activated macrophages secrete abundant proinflammatory cytokines (IL-1β, IL-6, IL-18, and TNF-α), CXCL10, ISG, immunoreceptor tyrosine-based activation motif (ITAM), and TRIF-related adaptor molecule (TRAM), which are thought to be the major sources accounting for the cytokine storm (68, 86, 89, 92). It has been reported that approximately 30 kinds of cytokines were significantly increased in COVID-19 patients on admission (130). The serum level of IL-1β, IL-1RA, IL-6, IL-7, IL-8, IL-9, IL-10, basic fibroblast growth factor, granulocyte colony-stimulating factor (G-CSF), granulocyte-macrophage CSF (GM-CSF), CXCL10, CCL2, CCL3, IFN-γ, TNF-α, and vascular endothelial growth factor (VEGF) is elevated in COVID-19 patients. Moreover, intensive care unit (ICU) patients with COVID-19 had higher serum levels of IL-2R, IL-6, IL-7, IL-10, interferon-inducible protein-10 (IP-10), monocyte chemotactic protein (MCP-1), macrophage inflammatory protein-1a (MIP-1A), TNF-α, CCL2, CCL3, and CXCL10 than did non-ICU patients (26, 114, 131). Various cytokines, including IL-6 and IL-10, IP-10, MCP-3, and MIP-1α, were implicated with disease severity (130, 132). A similar phenomenon was also observed in SARS-CoV and MERS-CoV infection (Table 5) (133, 134), indicating that cytokine storm may underlie the pathogenesis of the coronavirus-induced disease. It is essential to ascertain the role of cytokine storm associated with coronavirus infection to manage COVID-19 efficiently.
TABLE 5.
Comparison of serum levels of cytokines among SARS, MERS, and COVID-19 patientsa
| Cytokine | Value for disease (references) |
||
|---|---|---|---|
| SARS (134, 207–210) | MERS (133, 211) | COVID-19 (26, 130–132, 150, 212) | |
| IL-1β | E/U | U | E/U |
| IL-2 | E | N | E |
| IL-4 | E/L | N | E |
| IL-6 | E | E | E |
| IL-8 | E | N | E |
| IL-10 | E | N/E | E |
| IL-12 | E | N | E |
| IL-18 | E | NR | E |
| TNF-α | N/E | E | E |
| IFN-γ | E | E | E/N |
| IP-10 | N/E | E | E |
| MCP-1 | E | E | E |
| MIP-1A | NR | NR | E |
| M-CSF | NR | NR | E |
| G-CSF | NR | NR | E |
Abbreviations: NR, none reported; N, normal; E, elevated; L, lower; U, undetectable; IL, interleukin; TNF-α, tumor necrosis factor alpha; IFN-γ, interferon gamma; IP-10, interferon-inducible protein-10; MCP-1, monocyte chemotactic protein 1; MIP-1A, macrophage inflammatory protein-1a; M-CSF, macrophage colony-stimulating factor; G-CSF, granulocyte colony-stimulating factor.
Among proinflammatory cytokines, IL-6 usually released by macrophages was thought to be heavily involved in cytokine storm in COVID-19 patients (135–137), as well as in MERS and SARS cases (133, 134). Increased IL-6 performs various pathological functions that contribute to the development of COVID-19: (i) increasing vascular permeability directly or via inducing VEGF, resulting in interstitial edema and vascular leakage (138); (ii) weakening cytotoxicity of NK cells by downmodulating perforin and granzyme B expression (136, 139); and (iii) triggering monocytes recruitment by inducing the production of IL-8 and MCP-1 (140).
IFN-γ was increased in the sera and BALF of COVID-19 patients (26, 114, 141, 142), demonstrating the importance of IFN-γ in coronavirus infection. IFN-γ (type II IFN), mainly produced by T cells and NK cells, binds the IFNGR on the surface of macrophages and subsequently facilitates STAT1 phosphorylation through JAK1/2 promoting the transcription of IFN-stimulated genes (IP-10/CXCL10 and MIG/CXCL9, among others), which contributes to the cytokine storm and hemophagocytosis (143). Intriguingly, the level of IFN-γ is lower in severe COVID-19 patients than in mild or moderate cases (130, 132). A study showed that a high IL-6/IFN-γ ratio was related to disease severity (144). However, this could be the consequence of a decreased number of IFN-γ-expressing CD4+ T cells and CD8+ T cells in severe COVID-19 patients (114).
IL-1β and IL-18, members of the IL-1 family, are mainly cleaved and activated by caspase-1, which is activated by NLRP3 inflammasome upon stimulation of the danger signals (145). IL-1β is known for its proinflammatory property manifested as fever, pain, and vasodilatation, while IL-18 is able to induce IFN-γ either with IL-12 or with IL-15 on CD4+ and CD8+ T cells and NK cells, which further amplifies the activation of macrophages (145). Although a highly similar signal pathway is involved in the production of IL-1β and IL-18, the levels of IL-1β and IL-18 in the sera and BALF were completely different. IL-18 in the sera and BALF was significantly upregulated in COVID-19 patients in different stages, while IL-1β was barely detectable, especially in severe cases (26, 97, 141, 146). However, despite the low level of IL-1β, the transcription of IL-1β and IL-1R1 genes is elevated, indicating a strong response of IL-1β (67). Therefore, more studies are needed to ascertain whether IL-1β and IL-18 are increased in COVID-19 patients and to explore the role of the inflammasome in the immunopathogenesis of fatal COVID-19.
TNF-α, a potent inflammatory cytokine, is mainly produced by macrophages/monocytes leading to necrosis or apoptosis (147, 148). TNF-α is detectable in the blood from COVID-19 patients, and its levels are even higher in severe cases (26, 114, 121, 149). TNF-α has also been implicated in the severe immune-based pulmonary injury in SARS-CoV-infected patients (150). Moreover, the S protein of SARS-CoV induced TNF-α production by modulating TNF-α-converting enzyme (TACE or ADAM17) (151). Rather than resisting infection, highly expressed TNF-α leads to pathological complications (152). Most importantly, TNF-α can accentuate T cell apoptosis (153), which partially explains decreased T cell numbers.
In addition to proinflammatory cytokines, chemokines also play a crucial role in cytokine storm induced by coronavirus infection. COVID-19 patients have a high serum level of CXCL10, CCL2, and CCL3 (26, 97, 141). Neutrophil-recruiting attractants (CXCL1, CXCL2, CXCL8, CXCL10, CCL2, and CCL7) and monocyte attractants (CXCL6, CXCL11, CCL2, CCL3, CCL4, CCL7, CCL8, and CCL20) were also increased in the BALFs of COVID-19 patients (141), which is consistent with autopsy findings that monocytes and macrophages were the prominent infiltrating leukocytes in alveoli of COVID-19 patients (88, 154). Chemokines expression was also increased in SARS (CCL2, CXCL8/IL-8, CXCL9, and CXCL10) and MERS (CXCL10, CCL2, and CCL5) patients (65, 112, 155–158). Increased chemokines promote leukocyte infiltration and further aggravate disease severity, which is responsible for the development of pulmonary-centric disease in COVID-19 patients.
In summary, although these proinflammatory cytokines could be potential targets and have already prompted many clinical trials for COVID-19 using cytokine blockades, there is insufficient evidence for now demonstrating that using anticytokine therapies (IL-1β, IL-6, IL-18, and TNF-α) to treat COVID-19 patients is effective (159, 160). Blocking proinflammatory cytokines not only suppresses hyperinflammation but also hinders virus clearance. Therefore, when and how to use cytokine inhibitors affect patient outcomes.
Humoral Immune Response to Coronavirus
Adaptive humoral immunity acts through antibodies which can prevent the virus from binding and invading into cells. The most effective antibodies are high-affinity antibodies produced in T-dependent germinal center reactions. A limitation of antibodies is that they can work only before virus enters cells. Once viruses enter cells and begin to replicate intracellularly, they are inaccessible to antibodies (52). Neutralizing antibodies and immunoglobulin G (IgG) against S protein or N protein of SARS-CoV, MERS-CoV, and SARS-CoV-2 were present in infected patients during disease course (161, 162). Among four structural proteins of coronavirus, S protein was identified as the only active antigen which can induce the production of neutralizing antibodies (163). A number of virus-specific neutralizing monoclonal antibodies (MAbs) or related fragments targeted to S protein have been developed in SARS-CoV and MERS-CoV, but almost none of them have been assessed in clinical trials (164). So far, there are no available SARS-CoV-2-neutralizing MAbs for human use. It is urgent to figure out whether SARS-CoV-neutralizing MAbs show potential cross-neutralizing activity against S protein of SARS-CoV-2 and make it possible to prevent SARS-CoV-2 spread.
PATHOGENESIS OF ARDS
ARDS, a leading cause of respiratory failure, is theoretically characterized by severe impairment of gas exchange that may eventually lead to severe progressive hypoxemia, dyspnea, and impaired carbon dioxide excretion with a PaO2/FiO2 ratio (the ratio of arterial oxygen partial pressure to fractional inspired oxygen) less than 200 (165). The clinical features of SARS, MERS, and COVID-19 are markedly similar regardless of their subtle differences. These coronaviruses predominantly infect lower airways (terminal bronchus and pulmonary alveoli) and cause fatal pneumonia that may result in ALI and ARDS with high mortality. Since SARS-CoV-2 is insufficiently researched, the pathogenesis of the virus-induced ARDS remains unclear. However, these highly pathogenic coronaviruses may have a similar mechanism for inducing ARDS (Fig. 3).
Pathology of DAD
Diffuse alveolar damage (DAD), a characteristic pathological hallmark of the acute phase of ARDS, is roughly divided into two phases: the acute phase with hyaline membrane formation, acute interstitial inflammation, and edema and the organizing phase with loosely organized fibrosis and type II pneumocyte hyperplasia (166). DAD has been identified in SARS, MERS, and COVID-19 patients. Moreover, there is little difference in histopathological changes of DAD among these three different coronavirus infections.
The patients with SARS at the onset of illness generally showed DAD in the acute phase, including hyaline membrane formation, extensive edema, alveolus collapse, and pneumocyte desquamation (107, 167). Proliferative or organizing alteration with progression to fibrosis was present in the lung beyond day 10 (168, 169). Of note, macrophages are the primary cellular agent instead of neutrophils and fibroblasts in the alveoli of SARS patients with severe symptoms, even in the early stages of the disease (107). As for MERS-CoV infection, only one human autopsy is available, and the main histopathologic feature in the lung is DAD (alveolar fibrin deposits, type 2 pneumocyte hyperplasia, and thickened alveolar septa) accompanied by neutrophils and macrophage infiltration (170). In patients infected with COVID-19, similar histopathological changes of DAD with mononuclear inflammatory infiltrates were observed (171). In addition, alveolar septal vascular hyperemia, edema, and intravascular transparent thrombosis indicate the injury of lung endothelial cell (172).
Pneumocyte Injury
Both type I and II pneumocytes are crucial components of the epithelial-endothelial barrier that help to prevent pulmonary edema by limiting protein and ion transport (173). Pneumocytes not only participate in gas exchange but also are the target cells for coronavirus. After entering the alveolus, coronavirus first encounters pneumocytes which possess receptors for SARS-CoV/MERS-CoV/SARS-CoV-2 (47, 50, 174, 175), and then these viruses invade the pneumocytes mediated by receptors. The autopsies of SARS-CoV-infected patients identified the presence of the viral RNA and proteins in type II pneumocytes. In the aged macaque model of SARS, both types of pneumocytes were infected by coronaviruses (28, 39). However, SARS-CoV replication was observed only in primary human type II pneumocytes and not in type I-like cells in vitro (176). Similarly, MERS-CoV was predominantly located in pneumocytes and epithelial cells of terminal bronchioles in both patients and human lung tissue culture (95, 170). SARS-CoV-2 particles were also found in type II pneumocytes and macrophages (172). These results together suggest that pneumocytes, especially type II pneumocytes, play an essential role in mediating lung pathology and host susceptibility.
In coronavirus infection, injury to pneumocytes is an essential step in protein-rich alveolar edema via the destruction of the physical alveolar epithelial layer. The mechanisms responsible for pneumocytes injury in coronavirus infection are incompletely understood. However, rapid virus replication and macrophage hyperactivation were found to induce pneumocyte apoptosis during influenza virus infection. Lung-recruited exudate macrophages express and release IFN-β, which contributes to pneumocyte apoptosis by inducing TNF-related apoptosis inducing ligand (TRAIL) (177–179). Although type I IFN response is deficient in coronavirus infection patients, monocyte transepithelial migration and overactivated macrophages together with their products are likely to be the primary cause of pneumocyte injury in patients with ALI/ARDS induced by coronavirus infection. CCL2-mediated monocyte transepithelial migration has been shown to increase pneumocytes and endothelial permeability (180). Macrophage-producing reactive oxygen species (ROS) modify cellular proteins, lipids, and DNA in pneumocytes, impairing pneumocytes via either apoptotic or necrotic pathways (181). IFN-γ from hyperactivated T cells enhanced chemokine expression on pneumocytes, leading to the recruitment of inflammatory cells into alveoli and aggravating ALI following influenza virus infection (182, 183). In addition, the viral RNA genome can lead to cell death. After entering pneumocytes, SARS-CoV open reading frame 8b (ORF8b) forms insoluble intracellular aggregates that induce the death of epithelial cells (101). This could be due to the cytotoxicity of abnormal intracellular protein aggregation.
Since pneumocytes are indispensable in the gas-exchange barrier, the loss of epithelial cells has a considerable effect on lung architecture. After infection, pneumocytes produce proinflammatory cytokines and chemokines that can subsequently contribute to DAD (28, 39, 184). The infection of primary human type II pneumocytes with SARS-CoV in vitro induced markedly elevated levels of the mRNAs encoding type I and type III IFN (39). In addition, after SARS-CoV infection, type II pneumocytes generate a marked increase in chemokines (CCL5, CXCL8, CXCL10, and CXCL11) that can promote inflammatory cell recruitment (28). Infecting human airway epithelial cells with MERS-CoV induced a conspicuous but delayed proinflammatory cytokine response manifested as an increase of IL-1β, IL-6, and IL-8 (185). Among them, IL-1β decreases amiloride-sensitive epithelial sodium channel (ENaC) expression and activity via the p38 mitogen-activated protein kinase (MAPK)-dependent signaling pathway (186). Therefore, the osmotic gradient created by Na+ in the interstitium is insufficient to remove water from the alveolar lumen into alveolar epithelial cells, which may lead to alveolar edema in ALI (173, 187).
Macrophages in Lung
The macrophage is also important in the pathogenesis of ARDS in coronavirus patients (90–92). As previously mentioned, macrophages are the prominent infiltrating leukocytes in alveoli of severe SARS and COVID-19 patients, which indicates the importance of macrophages in the pathogenesis of ARDS during coronavirus infection. Macrophages in the patient’s lung tissue are potent producers of proinflammatory cytokines (68, 107). Mice infected with SARS-CoV showed delayed IFN-α/β response with pathogenic inflammatory monocyte-macrophage (IMM) influx. Signal transducer and activator of transcription 1 (STAT1) has been identified to regulate the perivascular infiltration of selectively activated macrophages in the lung and restrain fibrin deposition and alveolar collapse in SARS-CoV-challenged mice (28, 168, 188). These recruited macrophages produce and release proinflammatory cytokines (TNF-α, IL-6, and IL-1β) (68). Moreover, under the stimulation of IFN-α/β, accumulated IMMs produce monocyte chemokines such as CCL2, CCL7, and CCL12 (28). These monocyte chemoattractants promote macrophage infiltration, which further aggravates disease severity.
Lung Endothelial Cell Injury
In the alveoli, endothelial cells are closely apposed to epithelial cells on their basolateral side and directly contact circulating blood on their apical side (189). Due to this structure, lung endothelial cells are susceptible to cytokines in blood or alveoli that may cause lung endothelial cell injury and subsequently contribute to pulmonary edema and inflammatory cell infiltration.
Damage to lung endothelial cells, which may be the initial cause of DAD in patients with ALI/ARDS, can occur through several mechanisms. Neutrophils usually play a key role in endothelial cell injury by releasing several toxic mediators (190, 191). However, in coronavirus infection, it is macrophages instead of neutrophils that are the major cause of lung endothelial cell injury. Mechanistically, macrophages accumulated in the lung become activated and secrete several toxic agents, including proinflammatory cytokines (TNF, IL-6, and IL-1β) and ROS, which can promote lung endothelial cell apoptosis (28, 158, 192, 193). Coronavirus might also trigger endothelial cell death by direct infection, which damages the gas-exchange barrier. Receptors for SARS-CoV/MERS-CoV/SARS-CoV-2 are present on endothelial cells. Thus, lung endothelial cells could be the potential target for coronaviruses (95). Unfortunately, little evidence suggests the infection of endothelium with coronavirus.
Either directly or indirectly, coronaviruses induce lung endothelial cell injury, which leads to increased vascular permeability and alveolar edema, ultimately resulting in hypoxia. But it needs to be noted that lung endothelial injury is usually insufficient to cause pulmonary edema in the absence of pneumocyte injury (194), which indicates that pneumocyte injury is more important than endothelial injury in the development of ARDS. In addition, endothelial cells may promote leukocyte extravasation by expressing adhesion molecules, which facilitates infiltration of leukocytes, especially macrophages.
CONCLUSIONS
In the 21st century, three highly pathogenic coronaviruses have led to global emerging respiratory infectious diseases, indicating that these coronaviruses may cause additional outbreaks in the future. Although the epidemiology, transmission, pathogenesis, and treatment of these three coronavirus infections remain undiscovered, virus-induced ARDS is the most common cause of death and results in high mortality. Several studies provided evidence that DAD is a pathological feature characterized by the prominent infiltration of multinucleate giant macrophages in coronavirus-induced ARDS. The various degrees of cytopenia, hypohepatia, abnormal clotting profiles, hypercytokinemia, hyperferritinemia, and high predisposition of the elderly or those with comorbid conditions suggest the possible links between fatal coronavirus infection and secondary HLH. The pathophysiology of HLH remains elusive, but the cross talk among NK cells, lymphocytes, and macrophages is attracting increasing attention. The roles of the innate and adaptive cell-mediated immune response to these highly pathogenic coronaviruses are not entirely understood; however, the defect or delay of type I IFN response and exhaustion of NK cells and CTLs might lead to persistent antigenemia, enhanced antigen presentation, and prolonged innate-adaptive immune cell interactions, which induce the cytokine storms derived from uncontrolled hyperactivation of T cells and macrophages. Further studies on evaluating cytotoxic lymphocyte function, macrophage activation marker, and T cell activation marker and clarifying the related mechanism are needed to better explain exuberant immune responses in patients with coronavirus-induced ARDS. ROS, nitric oxide (NO), and cytokines mediated the damage of pneumocytes and lung endothelial cells, contributing to increased vascular permeability and alveolar edema. Therefore, interventions targeting specific cytokines to attenuate undesirable inflammatory responses in HLH might be useful strategies in patients with fatal coronavirus infection in the era of biologic therapy, but when and how to use cytokine-directed therapeutics remain challenging.
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (NSFC) (grant numbers 81771766, 81701621, 81871610, 81870071).
We thank Xianzhong Xiao (recipient of NSFC grant no. 81871610) and Sipin Tang (recipient of NSFC grant no. 81870071) for supervision and advice.
We declare no conflicts of interest.
Biographies

Chao Quan is an intern in the Department of Rheumatology at Xiangya Hospital and an 8-year medical student at Xiangya School of Medicine, Central South University, China. His research interest is in the field of sepsis pathogenesis, especially the role of myeloid-derived suppressor cells (MDSCs) in infectious diseases.

Caiyan Li, M.D., completed her M.D. degree (2010) in Harbin Medical University, China. She joined the Department of Rheumatology in Xianning Central Hospital as a physician in 2010. Currently, she is a Ph.D. student at the Department of Pathophysiology in Central South University, China. She is particularly interested in understanding the immunopathogenesis of myositis-related rapidly progressive interstitial lung disease, especially focusing on the NK cell dysfunction and macrophage activation.

Han Ma, B.M., obtained his B.M. degree (2019) in preventive medicine from Central South University, China, and then continued to pursue his Ph.D. degree at the Department of Pathophysiology, Central South University, in 2019. He is currently doing research in the Sepsis Translational Medicine Key Lab of Hunan Province, China. His research interests focus on the immunopathogenesis of sepsis-induced acute lung injury, especially the role of myeloid-derived suppressor cells (MDSCs) in acute lung injury.

Yisha Li, M.D., Ph.D., completed her M.D. (2003) degree from Xiangya School of Medicine and Ph.D. (2011) degree from Central South University, China. She joined the Department of Rheumatology in Xiangya Hospital of Central South University in 2003 and has served as chief physician since 2018. Currently, she is the vice director at the Department of Rheumatology in Xiangya Hospital and a young member of the Chinese Rheumatology Association. Dr. Li’s research interest focuses on the immunopathogenesis and management of interstitial lung disease in rheumatic diseases.

Huali Zhang, M.D., Ph.D., received her M.D. (1997) degree from Xiangya School of Medicine and Ph.D. (2001) degree in medical genetics from Central South University, China. She joined the Department of Pathophysiology in Central South University in 2002. She visited the University of Utah, Salt Lake City, Utah, USA, as a postdoc from 2007 to 2012. Since 2013, she has been working as a distinguished professor at the Department of Pathophysiology in Central South University and at the Department of Rheumatology in Xiangya Hospital. She is the chairman of the Department of Pathophysiology and the vice director of the Sepsis Translational Medicine Key Lab of Hunan Province. Dr. Zhang’s research interest is in the area of sepsis-associated multiple organ dysfunction and myositis-related rapidly progressive interstitial lung disease, with a focus on understanding the dysregulated immune response in sepsis and myositis.
REFERENCES
- 1.Su S, Wong G, Shi W, Liu J, Lai ACK, Zhou J, Liu W, Bi Y, Gao GF. 2016. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol 24:490–502. doi: 10.1016/j.tim.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lu R, Zhao X, Li J, Niu P, Yang B, Wu H, Wang W, Song H, Huang B, Zhu N, Bi Y, Ma X, Zhan F, Wang L, Hu T, Zhou H, Hu Z, Zhou W, Zhao L, Chen J, Meng Y, Wang J, Lin Y, Yuan J, Xie Z, Ma J, Liu WJ, Wang D, Xu W, Holmes EC, Gao GF, Wu G, Chen W, Shi W, Tan W. 2020. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet 395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhong NS, Zheng BJ, Li YM, Poon LL, Xie ZH, Chan KH, Li PH, Tan SY, Chang Q, Xie JP, Liu XQ, Xu J, Li DX, Yuen KY, Peiris JS, Guan Y. 2003. Epidemiology and cause of severe acute respiratory syndrome (SARS) in Guangdong, People’s Republic of China, in February, 2003. Lancet 362:1353–1358. doi: 10.1016/S0140-6736(03)14630-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus AD, Fouchier RA. 2012. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med 367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 5.Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, Niu P, Zhan F, Ma X, Wang D, Xu W, Wu G, Gao GF, Tan W, China Novel Coronavirus Investigating and Research Team . 2020. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med 382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen N, Zhou M, Dong X, Qu J, Gong F, Han Y, Qiu Y, Wang J, Liu Y, Wei Y, Xia J, Yu T, Zhang X, Zhang L. 2020. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet 395:507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dhama K, Khan S, Tiwari R, Sircar S, Bhat S, Malik YS, Singh KP, Chaicumpa W, Bonilla-Aldana DK, Rodriguez-Morales AJ. 2020. Coronavirus disease 2019-COVID-19. Clin Microbiol Rev 33:e00028-20. doi: 10.1128/CMR.00028-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peiris JS, Lai ST, Poon LL, Guan Y, Yam LY, Lim W, Nicholls J, Yee WK, Yan WW, Cheung MT, Cheng VC, Chan KH, Tsang DN, Yung RW, Ng TK, Yuen KY. 2003. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet 361:1319–1325. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Anonymous. . 2003. Centers for Disease Control and Prevention. Update: outbreak of severe acute respiratory syndrome–worldwide, 2003. JAMA 289:2059–2060. doi: 10.1001/jama.289.16.2059. [DOI] [PubMed] [Google Scholar]
- 10.WHO. 2003. Summary of probable SARS cases with onset of illness from 1 November 2002 to 31 July 2003. World Health Organization, Geneva, Switzerland. https://www.who.int/csr/sars/country/table2003_09_23/en/. Accessed 2 March 2020.
- 11.Wang M, Yan M, Xu H, Liang W, Kan B, Zheng B, Chen H, Zheng H, Xu Y, Zhang E, Wang H, Ye J, Li G, Li M, Cui Z, Liu YF, Guo RT, Liu XN, Zhan LH, Zhou DH, Zhao A, Hai R, Yu D, Guan Y, Xu J. 2005. SARS-CoV infection in a restaurant from palm civet. Emerg Infect Dis 11:1860–1865. doi: 10.3201/eid1112.041293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chan JF, Lau SK, To KK, Cheng VC, Woo PC, Yuen KY. 2015. Middle East respiratory syndrome coronavirus: another zoonotic betacoronavirus causing SARS-like disease. Clin Microbiol Rev 28:465–522. doi: 10.1128/CMR.00102-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.WHO. 2019. Middle East respiratory syndrome coronavirus (MERS-CoV). World Health Organization, Geneva, Switzerland. https://www.who.int/news-room/fact-sheets/detail/middle-east-respiratory-syndrome-coronavirus-(mers-cov). Accessed 2 March 2020.
- 14.Anonymous. . 2019. Report of clustering pneumonia of unknown etiology in Wuhan City. http://wjw.wuhan.gov.cn/xwzx_28/gsgg/202004/t20200430_1199588.shtml. Accessed 3 March 2020.
- 15.Badawi A, Ryoo SG. 2016. Prevalence of comorbidities in the Middle East respiratory syndrome coronavirus (MERS-CoV): a systematic review and meta-analysis. Int J Infect Dis 49:129–133. doi: 10.1016/j.ijid.2016.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Channappanavar R, Fett C, Mack M, Ten Eyck PP, Meyerholz DK, Perlman S. 2017. Sex-based differences in susceptibility to severe acute respiratory syndrome coronavirus infection. J Immunol 198:4046–4053. doi: 10.4049/jimmunol.1601896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.WHO. 2020. Novel coronavirus (2019-nCoV) situation report-153. World Health Organization, Geneva, Switzerland. https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports/. Accessed June 21 2020. [Google Scholar]
- 18.Special Expert Group for Control of the Epidemic of Novel Coronavirus Pneumonia of the Chinese Preventive Medicine Association. 2020. An update on the epidemiological characteristics of novel coronavirus pneumonia (COVID-19). Zhonghua Liu Xing Bing Xue Za Zhi 41:139–144. (In Chinese.) doi: 10.3760/cma.j.issn.0254-6450.2020.02.002. [DOI] [PubMed] [Google Scholar]
- 19.Chan JF, Yuan S, Kok KH, To KK, Chu H, Yang J, Xing F, Liu J, Yip CC, Poon RW, Tsoi HW, Lo SK, Chan KH, Poon VK, Chan WM, Ip JD, Cai JP, Cheng VC, Chen H, Hui CK, Yuen KY. 2020. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet 395:514–523. doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu Y, Eggo RM, Kucharski AJ. 2020. Secondary attack rate and superspreading events for SARS-CoV-2. Lancet 395:e47. doi: 10.1016/S0140-6736(20)30462-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu ZY. 2020. Asymptomatic and pre-symptomatic cases of COVID-19 contribution to spreading the epidemic and need for targeted control strategies. Zhonghua Liu Xing Bing Xue Za Zhi 41:E036. (In Chinese.) [DOI] [PubMed] [Google Scholar]
- 22.de Wit E, van Doremalen N, Falzarano D, Munster VJ. 2016. SARS and MERS: recent insights into emerging coronaviruses. Nat Rev Microbiol 14:523–534. doi: 10.1038/nrmicro.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peiris JS, Yuen KY, Osterhaus AD, Stohr K. 2003. The severe acute respiratory syndrome. N Engl J Med 349:2431–2441. doi: 10.1056/NEJMra032498. [DOI] [PubMed] [Google Scholar]
- 24.Zumla A, Hui DS, Perlman S. 2015. Middle East respiratory syndrome. Lancet 386:995–1007. doi: 10.1016/S0140-6736(15)60454-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Assiri A, Al-Tawfiq JA, Al-Rabeeah AA, Al-Rabiah FA, Al-Hajjar S, Al-Barrak A, Flemban H, Al-Nassir WN, Balkhy HH, Al-Hakeem RF, Makhdoom HQ, Zumla AI, Memish ZA. 2013. Epidemiological, demographic, and clinical characteristics of 47 cases of Middle East respiratory syndrome coronavirus disease from Saudi Arabia: a descriptive study. Lancet Infect Dis 13:752–761. doi: 10.1016/S1473-3099(13)70204-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, Zhang L, Fan G, Xu J, Gu X, Cheng Z, Yu T, Xia J, Wei Y, Wu W, Xie X, Yin W, Li H, Liu M, Xiao Y, Gao H, Guo L, Xie J, Wang G, Jiang R, Gao Z, Jin Q, Wang J, Cao B. 2020. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peiris JS, Chu CM, Cheng VC, Chan KS, Hung IF, Poon LL, Law KI, Tang BS, Hon TY, Chan CS, Chan KH, Ng JS, Zheng BJ, Ng WL, Lai RW, Guan Y, Yuen KY. 2003. Clinical progression and viral load in a community outbreak of coronavirus-associated SARS pneumonia: a prospective study. Lancet 361:1767–1772. doi: 10.1016/S0140-6736(03)13412-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Channappanavar R, Perlman S. 2017. Pathogenic human coronavirus infections: causes and consequences of cytokine storm and immunopathology. Semin Immunopathol 39:529–539. doi: 10.1007/s00281-017-0629-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leong HN, Chan KP, Oon LL, Koay E, Ng LC, Lee MA, Barkham T, Chen MI, Heng BH, Ling AE, Leo YS. 2006. Clinical and laboratory findings of SARS in Singapore. Ann Acad Med Singap 35:332–339. [PubMed] [Google Scholar]
- 30.Das KM, Lee EY, Al Jawder SE, Enani MA, Singh R, Skakni L, Al-Nakshabandi N, AlDossari K, Larsson SG. 2015. Acute Middle East respiratory syndrome coronavirus: temporal lung changes observed on the chest radiographs of 55 patients. AJR Am J Roentgenol 205:W267–W274. doi: 10.2214/AJR.15.14445. [DOI] [PubMed] [Google Scholar]
- 31.Song F, Shi N, Shan F, Zhang Z, Shen J, Lu H, Ling Y, Jiang Y, Shi Y. 2020. Emerging coronavirus 2019-nCoV pneumonia. Radiology 295:210–217. doi: 10.1148/radiol.2020200274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kanne JP. 2020. Chest CT findings in 2019 novel coronavirus (2019-nCoV) infections from Wuhan, China: key points for the radiologist. Radiology 295:16–17. doi: 10.1148/radiol.2020200241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cui J, Li F, Shi ZL. 2019. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol 17:181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu A, Peng Y, Huang B, Ding X, Wang X, Niu P, Meng J, Zhu Z, Zhang Z, Wang J, Sheng J, Quan L, Xia Z, Tan W, Cheng G, Jiang T. 2020. Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe 27:325–328. doi: 10.1016/j.chom.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shang J, Ye G, Shi K, Wan Y, Luo C, Aihara H, Geng Q, Auerbach A, Li F. 2020. Structural basis of receptor recognition by SARS-CoV-2. Nature 581:221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wrapp D, Wang N, Corbett KS, Goldsmith JA, Hsieh CL, Abiona O, Graham BS, McLellan JS. 2020. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu W, Liu L, Kou G, Zheng Y, Ding Y, Ni W, Wang Q, Tan L, Wu W, Tang S, Xiong Z, Zheng S. 2020. Evaluation of nucleocapsid and spike protein-based enzyme-linked immunosorbent assays for detecting antibodies against SARS-CoV-2. J Clin Microbiol 58:e00461-20. doi: 10.1128/JCM.00461-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhu FC, Li YH, Guan XH, Hou LH, Wang WJ, Li JX, Wu SP, Wang BS, Wang Z, Wang L, Jia SY, Jiang HD, Wang L, Jiang T, Hu Y, Gou JB, Xu SB, Xu JJ, Wang XW, Wang W, Chen W. 2020. Safety, tolerability, and immunogenicity of a recombinant adenovirus type-5 vectored COVID-19 vaccine: a dose-escalation, open-label, non-randomised, first-in-human trial. Lancet 395:1845–1854. doi: 10.1016/S0140-6736(20)31208-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Qian Z, Travanty EA, Oko L, Edeen K, Berglund A, Wang J, Ito Y, Holmes KV, Mason RJ. 2013. Innate immune response of human alveolar type II cells infected with severe acute respiratory syndrome-coronavirus. Am J Respir Cell Mol Biol 48:742–748. doi: 10.1165/rcmb.2012-0339OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li W, Moore MJ, Vasilieva N, Sui J, Wong SK, Berne MA, Somasundaran M, Sullivan JL, Luzuriaga K, Greenough TC, Choe H, Farzan M. 2003. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li F, Li W, Farzan M, Harrison SC. 2005. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science 309:1864–1868. doi: 10.1126/science.1116480. [DOI] [PubMed] [Google Scholar]
- 42.Hamming I, Timens W, Bulthuis ML, Lely AT, Navis G, van Goor H. 2004. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J Pathol 203:631–637. doi: 10.1002/path.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jia HP, Look DC, Shi L, Hickey M, Pewe L, Netland J, Farzan M, Wohlford-Lenane C, Perlman S, McCray PB, Jr. 2005. ACE2 receptor expression and severe acute respiratory syndrome coronavirus infection depend on differentiation of human airway epithelia. J Virol 79:14614–14621. doi: 10.1128/JVI.79.23.14614-14621.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Leung WK, To KF, Chan PK, Chan HL, Wu AK, Lee N, Yuen KY, Sung JJ. 2003. Enteric involvement of severe acute respiratory syndrome-associated coronavirus infection. Gastroenterology 125:1011–1017. doi: 10.1016/s0016-5085(03)01215-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Imai Y, Kuba K, Rao S, Huan Y, Guo F, Guan B, Yang P, Sarao R, Wada T, Leong-Poi H, Crackower MA, Fukamizu A, Hui CC, Hein L, Uhlig S, Slutsky AS, Jiang C, Penninger JM. 2005. Angiotensin-converting enzyme 2 protects from severe acute lung failure. Nature 436:112–116. doi: 10.1038/nature03712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kuba K, Imai Y, Rao S, Gao H, Guo F, Guan B, Huan Y, Yang P, Zhang Y, Deng W, Bao L, Zhang B, Liu G, Wang Z, Chappell M, Liu Y, Zheng D, Leibbrandt A, Wada T, Slutsky AS, Liu D, Qin C, Jiang C, Penninger JM. 2005. A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus-induced lung injury. Nat Med 11:875–879. doi: 10.1038/nm1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, Si HR, Zhu Y, Li B, Huang CL, Chen HD, Chen J, Luo Y, Guo H, Jiang RD, Liu MQ, Chen Y, Shen XR, Wang X, Zheng XS, Zhao K, Chen QJ, Deng F, Liu LL, Yan B, Zhan FX, Wang YY, Xiao GF, Shi ZL. 2020. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang ZY, Huang Y, Ganesh L, Leung K, Kong WP, Schwartz O, Subbarao K, Nabel GJ. 2004. pH-dependent entry of severe acute respiratory syndrome coronavirus is mediated by the spike glycoprotein and enhanced by dendritic cell transfer through DC-SIGN. J Virol 78:5642–5650. doi: 10.1128/JVI.78.11.5642-5650.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jeffers SA, Tusell SM, Gillim-Ross L, Hemmila EM, Achenbach JE, Babcock GJ, Thomas WD, Jr, Thackray LB, Young MD, Mason RJ, Ambrosino DM, Wentworth DE, Demartini JC, Holmes KV. 2004. CD209L (L-SIGN) is a receptor for severe acute respiratory syndrome coronavirus. Proc Natl Acad Sci U S A 101:15748–15753. doi: 10.1073/pnas.0403812101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Raj VS, Mou H, Smits SL, Dekkers DH, Muller MA, Dijkman R, Muth D, Demmers JA, Zaki A, Fouchier RA, Thiel V, Drosten C, Rottier PJ, Osterhaus AD, Bosch BJ, Haagmans BL. 2013. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature 495:251–254. doi: 10.1038/nature12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang K, Chen W, Zhou Y-S, Lian J-Q, Zhang Z, Du P, Gong L, Zhang Y, Cui H-Y, Geng J-J, Wang B, Sun X-X, Wang C-F, Yang X, Lin P, Deng Y-Q, Wei D, Yang X-M, Zhu Y-M, Zhang K, Zheng Z-H, Miao J-L, Guo T, Shi Y, Zhang J, Fu L, Wang Q-Y, Bian H, Zhu P, Chen Z-N. 2020. SARS-CoV-2 invades host cells via a novel route: CD147-spike protein. bioRxiv doi: 10.1101/2020.03.14.988345. [DOI] [PMC free article] [PubMed]
- 52.Abul K, Abbas AHL, Pillai S. 2014. Cellular and molecular immunology, 8th ed. Saunders, Elsevier, North York, Ontario, Canada. [Google Scholar]
- 53.Cao B, Wang Y, Wen D, Liu W, Wang J, Fan G, Ruan L, Song B, Cai Y, Wei M, Li X, Xia J, Chen N, Xiang J, Yu T, Bai T, Xie X, Zhang L, Li C, Yuan Y, Chen H, Li H, Huang H, Tu S, Gong F, Liu Y, Wei Y, Dong C, Zhou F, Gu X, Xu J, Liu Z, Zhang Y, Li H, Shang L, Wang K, Li K, Zhou X, Dong X, Qu Z, Lu S, Hu X, Ruan S, Luo S, Wu J, Peng L, Cheng F, Pan L, Zou J, Jia C, Wang J, Liu X, Wang S, Wu X, Ge Q, He J, Zhan H, Qiu F, Guo L, Huang C, Jaki T, Hayden FG, Horby PW, Zhang D, Wang C. 2020. A trial of lopinavir-ritonavir in adults hospitalized with severe Covid-19. N Engl J Med 382:1787–1799. doi: 10.1056/NEJMoa2001282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mehta P, McAuley DF, Brown M, Sanchez E, Tattersall RS, Manson JJ, HLH Across Speciality Collaboration, UK . 2020. COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet 395:1033–1034. doi: 10.1016/S0140-6736(20)30628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ramos-Casals M, Brito-Zerón P, López-Guillermo A, Khamashta MA, Bosch X. 2014. Adult haemophagocytic syndrome. Lancet 383:1503–1516. doi: 10.1016/S0140-6736(13)61048-X. [DOI] [PubMed] [Google Scholar]
- 56.Janka GE, Lehmberg K. 2014. Hemophagocytic syndromes–an update. Blood Rev 28:135–142. doi: 10.1016/j.blre.2014.03.002. [DOI] [PubMed] [Google Scholar]
- 57.Billiau AD, Roskams T, Van Damme-Lombaerts R, Matthys P, Wouters C. 2005. Macrophage activation syndrome: characteristic findings on liver biopsy illustrating the key role of activated, IFN-gamma-producing lymphocytes and IL-6- and TNF-alpha-producing macrophages. Blood 105:1648–1651. doi: 10.1182/blood-2004-08-2997. [DOI] [PubMed] [Google Scholar]
- 58.Griffin G, Shenoi S, Hughes GC. 2020. Hemophagocytic lymphohistiocytosis: an update on pathogenesis, diagnosis, and therapy. Best Pract Res Clin Rheumatol doi: 10.1016/j.berh.2020.101515. [DOI] [PubMed] [Google Scholar]
- 59.Carter SJ, Tattersall RS, Ramanan AV. 2019. Macrophage activation syndrome in adults: recent advances in pathophysiology, diagnosis and treatment. Rheumatology (Oxford) 58:5–17. doi: 10.1093/rheumatology/key006. [DOI] [PubMed] [Google Scholar]
- 60.Rouphael NG, Talati NJ, Vaughan C, Cunningham K, Moreira R, Gould C. 2007. Infections associated with haemophagocytic syndrome. Lancet Infect Dis 7:814–822. doi: 10.1016/S1473-3099(07)70290-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Henter JI, Horne A, Aricó M, Egeler RM, Filipovich AH, Imashuku S, Ladisch S, McClain K, Webb D, Winiarski J, Janka G. 2007. HLH-2004: diagnostic and therapeutic guidelines for hemophagocytic lymphohistiocytosis. Pediatr Blood Cancer 48:124–131. doi: 10.1002/pbc.21039. [DOI] [PubMed] [Google Scholar]
- 62.Cinatl J, Jr, Michaelis M, Scholz M, Doerr HW. 2004. Role of interferons in the treatment of severe acute respiratory syndrome. Expert Opin Biol Ther 4:827–836. doi: 10.1517/14712598.4.6.827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cheung CY, Poon LL, Ng IH, Luk W, Sia SF, Wu MH, Chan KH, Yuen KY, Gordon S, Guan Y, Peiris JS. 2005. Cytokine responses in severe acute respiratory syndrome coronavirus-infected macrophages in vitro: possible relevance to pathogenesis. J Virol 79:7819–7826. doi: 10.1128/JVI.79.12.7819-7826.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ziegler T, Matikainen S, Rönkkö E, Osterlund P, Sillanpää M, Sirén J, Fagerlund R, Immonen M, Melén K, Julkunen I. 2005. Severe acute respiratory syndrome coronavirus fails to activate cytokine-mediated innate immune responses in cultured human monocyte-derived dendritic cells. J Virol 79:13800–13805. doi: 10.1128/JVI.79.21.13800-13805.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cameron MJ, Ran L, Xu L, Danesh A, Bermejo-Martin JF, Cameron CM, Muller MP, Gold WL, Richardson SE, Poutanen SM, Willey BM, DeVries ME, Fang Y, Seneviratne C, Bosinger SE, Persad D, Wilkinson P, Greller LD, Somogyi R, Humar A, Keshavjee S, Louie M, Loeb MB, Brunton J, McGeer AJ, Kelvin DJCanadian SARS Research Network ,. 2007. Interferon-mediated immunopathological events are associated with atypical innate and adaptive immune responses in patients with severe acute respiratory syndrome. J Virol 81:8692–8706. doi: 10.1128/JVI.00527-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Spiegel M, Pichlmair A, Martínez-Sobrido L, Cros J, García-Sastre A, Haller O, Weber F. 2005. Inhibition of beta interferon induction by severe acute respiratory syndrome coronavirus suggests a two-step model for activation of interferon regulatory factor 3. J Virol 79:2079–2086. doi: 10.1128/JVI.79.4.2079-2086.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hadjadj J, Yatim N, Barnabei L, Corneau A, Boussier J, Pere H, Charbit B, Bondet V, Chenevier-Gobeaux C, Breillat P, Carlier N, Gauzit R, Morbieu C, Pene F, Marin N, Roche N, Szwebel T-A, Smith N, Merkling S, Treluyer J-M, Veyer D, Mouthon L, Blanc C, Tharaux P-L, Rozenberg F, Fischer A, Duffy D, Rieux-Laucat F, Kerneis S, Terrier B. 2020. Impaired type I interferon activity and exacerbated inflammatory responses in severe Covid-19 patients. medRxiv doi: 10.1101/2020.04.19.20068015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Channappanavar R, Fehr AR, Vijay R, Mack M, Zhao J, Meyerholz DK, Perlman S. 2016. Dysregulated type I interferon and inflammatory monocyte-macrophage responses cause lethal pneumonia in SARS-CoV-infected mice. Cell Host Microbe 19:181–193. doi: 10.1016/j.chom.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Totura AL, Baric RS. 2012. SARS coronavirus pathogenesis: host innate immune responses and viral antagonism of interferon. Curr Opin Virol 2:264–275. doi: 10.1016/j.coviro.2012.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shokri S, Mahmoudvand S, Taherkhani R, Farshadpour F. 2019. Modulation of the immune response by Middle East respiratory syndrome coronavirus. J Cell Physiol 234:2143–2151. doi: 10.1002/jcp.27155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lu X, Pan J, Tao J, Guo D. 2011. SARS-CoV nucleocapsid protein antagonizes IFN-β response by targeting initial step of IFN-β induction pathway, and its C-terminal region is critical for the antagonism. Virus Genes 42:37–45. doi: 10.1007/s11262-010-0544-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Freundt EC, Yu L, Park E, Lenardo MJ, Xu XN. 2009. Molecular determinants for subcellular localization of the severe acute respiratory syndrome coronavirus open reading frame 3b protein. J Virol 83:6631–6640. doi: 10.1128/JVI.00367-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Niemeyer D, Zillinger T, Muth D, Zielecki F, Horvath G, Suliman T, Barchet W, Weber F, Drosten C, Muller MA. 2013. Middle East respiratory syndrome coronavirus accessory protein 4a is a type I interferon antagonist. J Virol 87:12489–12495. doi: 10.1128/JVI.01845-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Siu KL, Yeung ML, Kok KH, Yuen KS, Kew C, Lui PY, Chan CP, Tse H, Woo PC, Yuen KY, Jin DY. 2014. Middle East respiratory syndrome coronavirus 4a protein is a double-stranded RNA-binding protein that suppresses PACT-induced activation of RIG-I and MDA5 in the innate antiviral response. J Virol 88:4866–4876. doi: 10.1128/JVI.03649-13. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 75.Yang Y, Zhang L, Geng H, Deng Y, Huang B, Guo Y, Zhao Z, Tan W. 2013. The structural and accessory proteins M, ORF 4a, ORF 4b, and ORF 5 of Middle East respiratory syndrome coronavirus (MERS-CoV) are potent interferon antagonists. Protein Cell 4:951–961. doi: 10.1007/s13238-013-3096-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cui W, Fan Y, Wu W, Zhang F, Wang JY, Ni AP. 2003. Expression of lymphocytes and lymphocyte subsets in patients with severe acute respiratory syndrome. Clin Infect Dis 37:857–859. doi: 10.1086/378587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.National Research Project for SARS, Beijing Group. 2004. The involvement of natural killer cells in the pathogenesis of severe acute respiratory syndrome. Am J Clin Pathol 121:507–511. doi: 10.1309/WPK7-Y2XK-NF4C-BF3R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bordoni V, Sacchi A, Cimini E, Notari S, Grassi G, Tartaglia E, Casetti R, Giancola L, Bevilacqua N, Maeurer M, Zumla A, Locatelli F, De Benedetti F, Palmieri F, Marchioni L, Capobianchi MR, D’Offizi G, Petrosillo N, Antinori A, Nicastri E, Ippolito G, Agrati C. 2020. An inflammatory profile correlates with decreased frequency of cytotoxic cells in COVID-19. Clin Infect Dis doi: 10.1093/cid/ciaa577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zheng M, Gao Y, Wang G, Song G, Liu S, Sun D, Xu Y, Tian Z. 2020. Functional exhaustion of antiviral lymphocytes in COVID-19 patients. Cell Mol Immunol 17:533–535. doi: 10.1038/s41423-020-0402-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Grom AA, Mellins ED. 2010. Macrophage activation syndrome: advances towards understanding pathogenesis. Curr Opin Rheumatol 22:561–566. doi: 10.1097/01.bor.0000381996.69261.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Salminen A, Kaarniranta K, Kauppinen A. 2019. Immunosenescence: the potential role of myeloid-derived suppressor cells (MDSC) in age-related immune deficiency. Cell Mol Life Sci 76:1901–1918. doi: 10.1007/s00018-019-03048-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rukavina D, Laskarin G, Rubesa G, Strbo N, Bedenicki I, Manestar D, Glavas M, Christmas SE, Podack ER. 1998. Age-related decline of perforin expression in human cytotoxic T lymphocytes and natural killer cells. Blood 92:2410–2420. doi: 10.1182/blood.V92.7.2410.2410_2410_2420. [DOI] [PubMed] [Google Scholar]
- 83.Mao H, Tu W, Liu Y, Qin G, Zheng J, Chan PL, Lam KT, Peiris JS, Lau YL. 2010. Inhibition of human natural killer cell activity by influenza virions and hemagglutinin. J Virol 84:4148–4157. doi: 10.1128/JVI.02340-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Vandenhaute J, Wouters CH, Matthys P. 2019. Natural killer cells in systemic autoinflammatory diseases: a focus on systemic juvenile idiopathic arthritis and macrophage activation syndrome. Front Immunol 10:3089. doi: 10.3389/fimmu.2019.03089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hammer Q, Rückert T, Romagnani C. 2018. Natural killer cell specificity for viral infections. Nat Immunol 19:800–808. doi: 10.1038/s41590-018-0163-6. [DOI] [PubMed] [Google Scholar]
- 86.Shapouri-Moghaddam A, Mohammadian S, Vazini H, Taghadosi M, Esmaeili SA, Mardani F, Seifi B, Mohammadi A, Afshari JT, Sahebkar A. 2018. Macrophage plasticity, polarization, and function in health and disease. J Cell Physiol 233:6425–6440. doi: 10.1002/jcp.26429. [DOI] [PubMed] [Google Scholar]
- 87.Wu JH, Li X, Huang B, Su H, Li Y, Luo DJ, Chen S, Ma L, Wang SH, Nie X, Peng L. 2020. Pathological changes of fatal coronavirus disease 2019 (COVID-19) in the lungs: report of 10 cases by postmortem needle autopsy. Zhonghua Bing Li Xue Za Zhi 49:568–575. (In Chinese.) doi: 10.3760/cma.j.cn112151-20200405-00291. [DOI] [PubMed] [Google Scholar]
- 88.Yao XH, Li TY, He ZC, Ping YF, Liu HW, Yu SC, Mou HM, Wang LH, Zhang HR, Fu WJ, Luo T, Liu F, Guo QN, Chen C, Xiao HL, Guo HT, Lin S, Xiang DF, Shi Y, Pan GQ, Li QR, Huang X, Cui Y, Liu XZ, Tang W, Pan PF, Huang XQ, Ding YQ, Bian XW. 2020. A pathological report of three COVID-19 cases by minimal invasive autopsies. Zhonghua Bing Li Xue Za Zhi 49:411–417. (In Chinese.) doi: 10.3760/cma.j.cn112151-20200312-00193. [DOI] [PubMed] [Google Scholar]
- 89.Liao M, Liu Y, Yuan J, Wen Y, Xu G, Zhao J, Chen L, Li J, Wang X, Wang F, Liu L, Zhang S, Zhang Z. 2020. The landscape of lung bronchoalveolar immune cells in COVID-19 revealed by single-cell RNA sequencing. medRxiv doi: 10.1101/2020.02.23.20026690. [DOI] [Google Scholar]
- 90.Lew TW, Kwek TK, Tai D, Earnest A, Loo S, Singh K, Kwan KM, Chan Y, Yim CF, Bek SL, Kor AC, Yap WS, Chelliah YR, Lai YC, Goh SK. 2003. Acute respiratory distress syndrome in critically ill patients with severe acute respiratory syndrome. JAMA 290:374–380. doi: 10.1001/jama.290.3.374. [DOI] [PubMed] [Google Scholar]
- 91.Drosten C, Seilmaier M, Corman VM, Hartmann W, Scheible G, Sack S, Guggemos W, Kallies R, Muth D, Junglen S, Müller MA, Haas W, Guberina H, Röhnisch T, Schmid-Wendtner M, Aldabbagh S, Dittmer U, Gold H, Graf P, Bonin F, Rambaut A, Wendtner CM. 2013. Clinical features and virological analysis of a case of Middle East respiratory syndrome coronavirus infection. Lancet Infect Dis 13:745–751. doi: 10.1016/S1473-3099(13)70154-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Merad M, Martin JC. 2020. Pathological inflammation in patients with COVID-19: a key role for monocytes and macrophages. Nat Rev Immunol 20:355–362. doi: 10.1038/s41577-020-0331-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yilla M, Harcourt BH, Hickman CJ, McGrew M, Tamin A, Goldsmith CS, Bellini WJ, Anderson LJ. 2005. SARS-coronavirus replication in human peripheral monocytes/macrophages. Virus Res 107:93–101. doi: 10.1016/j.virusres.2004.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Law HK, Cheung CY, Ng HY, Sia SF, Chan YO, Luk W, Nicholls JM, Peiris JS, Lau YL. 2005. Chemokine up-regulation in SARS-coronavirus-infected, monocyte-derived human dendritic cells. Blood 106:2366–2374. doi: 10.1182/blood-2004-10-4166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhou J, Chu H, Li C, Wong BH, Cheng ZS, Poon VK, Sun T, Lau CC, Wong KK, Chan JY, Chan JF, To KK, Chan KH, Zheng BJ, Yuen KY. 2014. Active replication of Middle East respiratory syndrome coronavirus and aberrant induction of inflammatory cytokines and chemokines in human macrophages: implications for pathogenesis. J Infect Dis 209:1331–1342. doi: 10.1093/infdis/jit504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chen Y, Feng Z, Diao B, Wang R, Wang G, Wang C, Tan Y, Liu L, Wang C, Liu Y, Liu Y, Yuan Z, Ren L, Wu Y. 2020. The novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) directly decimates human spleens and lymph nodes. medRxiv doi: 10.1101/2020.03.27.20045427. [DOI] [Google Scholar]
- 97.Xiong Y, Liu Y, Cao L, Wang D, Guo M, Jiang A, Guo D, Hu W, Yang J, Tang Z, Wu H, Lin Y, Zhang M, Zhang Q, Shi M, Liu Y, Zhou Y, Lan K, Chen Y. 2020. Transcriptomic characteristics of bronchoalveolar lavage fluid and peripheral blood mononuclear cells in COVID-19 patients. Emerg Microbes Infect 9:761–770. doi: 10.1080/22221751.2020.1747363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Jensen S, Thomsen AR. 2012. Sensing of RNA viruses: a review of innate immune receptors involved in recognizing RNA virus invasion. J Virol 86:2900–2910. doi: 10.1128/JVI.05738-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Chen IY, Moriyama M, Chang MF, Ichinohe T. 2019. Severe acute respiratory syndrome coronavirus viroporin 3a activates the NLRP3 inflammasome. Front Microbiol 10:50. doi: 10.3389/fmicb.2019.00050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Siu KL, Yuen KS, Castaño-Rodriguez C, Ye ZW, Yeung ML, Fung SY, Yuan S, Chan CP, Yuen KY, Enjuanes L, Jin DY. 2019. Severe acute respiratory syndrome coronavirus ORF3a protein activates the NLRP3 inflammasome by promoting TRAF3-dependent ubiquitination of ASC. FASEB J 33:8865–8877. doi: 10.1096/fj.201802418R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Shi CS, Nabar NR, Huang NN, Kehrl JH. 2019. SARS-coronavirus open reading frame-8b triggers intracellular stress pathways and activates NLRP3 inflammasomes. Cell Death Discov 5:101. doi: 10.1038/s41420-019-0181-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wang W, Ye L, Ye L, Li B, Gao B, Zeng Y, Kong L, Fang X, Zheng H, Wu Z, She Y. 2007. Up-regulation of IL-6 and TNF-alpha induced by SARS-coronavirus spike protein in murine macrophages via NF-kappaB pathway. Virus Res 128:1–8. doi: 10.1016/j.virusres.2007.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Crayne CB, Albeituni S, Nichols KE, Cron RQ. 2019. The immunology of macrophage activation syndrome. Front Immunol 10:119. doi: 10.3389/fimmu.2019.00119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zhang D, Guo R, Lei L, Liu H, Wang Y, Wang Y, Dai T, Zhang T, Lai Y, Wang J, Liu Z, He A, Dwyer M, Hu J. 2020. COVID-19 infection induces readily detectable morphological and inflammation-related phenotypic changes in peripheral blood monocytes, the severity of which correlate with patient outcome. medRxiv doi: 10.1101/2020.03.24.20042655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zhou Y, Fu B, Zheng X, Wang D, Zhao C, Qi Y, Sun R, Tian Z, Xu X, Wei H. 2020. Pathogenic T-cells and inflammatory monocytes incite inflammatory storms in severe COVID-19 patients. Natl Sci Rev 7:998–1002. doi: 10.1093/nsr/nwaa041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wen W, Su W, Tang H, Le W, Zhang X, Zheng Y, Liu X, Xie L, Li J, Ye J, Cui X, Miao Y, Wang D, Dong J, Xiao C-L, Chen W, Wang H. 2020. Immune cell profiling of COVID-19 patients in the recovery stage by single-cell sequencing. medRxiv doi: 10.1101/2020.03.23.20039362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Nicholls JM, Poon LL, Lee KC, Ng WF, Lai ST, Leung CY, Chu CM, Hui PK, Mak KL, Lim W, Yan KW, Chan KH, Tsang NC, Guan Y, Yuen KY, Peiris JS. 2003. Lung pathology of fatal severe acute respiratory syndrome. Lancet 361:1773–1778. doi: 10.1016/s0140-6736(03)13413-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Xu X, Chang XN, Pan HX, Su H, Huang B, Yang M, Luo DJ, Weng MX, Ma L, Nie X. 2020. Pathological changes of the spleen in ten patients with coronavirus disease 2019(COVID-19) by postmortem needle autopsy. Zhonghua Bing Li Xue Za Zhi 49:576–582. (In Chinese.) doi: 10.3760/cma.j.cn112151-20200401-00278. [DOI] [PubMed] [Google Scholar]
- 109.Rosário C, Zandman-Goddard G, Meyron-Holtz EG, D’Cruz DP, Shoenfeld Y. 2013. The hyperferritinemic syndrome: macrophage activation syndrome, Still’s disease, septic shock and catastrophic antiphospholipid syndrome. BMC Med 11:185. doi: 10.1186/1741-7015-11-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ruscitti P, Cipriani P, Di Benedetto P, Ciccia F, Liakouli V, Carubbi F, Berardicurti O, Rizzo A, Triolo G, Giacomelli R. 2015. Increased level of H-ferritin and its imbalance with L-ferritin, in bone marrow and liver of patients with adult onset Still’s disease, developing macrophage activation syndrome, correlate with the severity of the disease. Autoimmun Rev 14:429–437. doi: 10.1016/j.autrev.2015.01.004. [DOI] [PubMed] [Google Scholar]
- 111.Imai Y, Kuba K, Neely GG, Yaghubian-Malhami R, Perkmann T, van Loo G, Ermolaeva M, Veldhuizen R, Leung YH, Wang H, Liu H, Sun Y, Pasparakis M, Kopf M, Mech C, Bavari S, Peiris JS, Slutsky AS, Akira S, Hultqvist M, Holmdahl R, Nicholls J, Jiang C, Binder CJ, Penninger JM. 2008. Identification of oxidative stress and Toll-like receptor 4 signaling as a key pathway of acute lung injury. Cell 133:235–249. doi: 10.1016/j.cell.2008.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Shin HS, Kim Y, Kim G, Lee JY, Jeong I, Joh JS, Kim H, Chang E, Sim SY, Park JS, Lim DG. 2019. Immune responses to Middle East respiratory syndrome coronavirus during the acute and convalescent phases of human infection. Clin Infect Dis 68:984–992. doi: 10.1093/cid/ciy595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Li T, Qiu Z, Zhang L, Han Y, He W, Liu Z, Ma X, Fan H, Lu W, Xie J, Wang H, Deng G, Wang A. 2004. Significant changes of peripheral T lymphocyte subsets in patients with severe acute respiratory syndrome. J Infect Dis 189:648–651. doi: 10.1086/381535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Chen G, Wu D, Guo W, Cao Y, Huang D, Wang H, Wang T, Zhang X, Chen H, Yu H, Zhang X, Zhang M, Wu S, Song J, Chen T, Han M, Li S, Luo X, Zhao J, Ning Q. 2020. Clinical and immunological features of severe and moderate coronavirus disease 2019. J Clin Invest 130:2620–2629. doi: 10.1172/JCI137244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Chu H, Zhou J, Wong BH, Li C, Chan JF, Cheng ZS, Yang D, Wang D, Lee AC, Li C, Yeung ML, Cai JP, Chan IH, Ho WK, To KK, Zheng BJ, Yao Y, Qin C, Yuen KY. 2016. Middle East respiratory syndrome coronavirus efficiently infects human primary T lymphocytes and activates the extrinsic and intrinsic apoptosis pathways. J Infect Dis 213:904–914. doi: 10.1093/infdis/jiv380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Wang X, Xu W, Hu G, Xia S, Sun Z, Liu Z, Xie Y, Zhang R, Jiang S, Lu L. 2020. SARS-CoV-2 infects T lymphocytes through its spike protein-mediated membrane fusion. Cell Mol Immunol 17:894–894. doi: 10.1038/s41423-020-0498-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Gu J, Gong E, Zhang B, Zheng J, Gao Z, Zhong Y, Zou W, Zhan J, Wang S, Xie Z, Zhuang H, Wu B, Zhong H, Shao H, Fang W, Gao D, Pei F, Li X, He Z, Xu D, Shi X, Anderson VM, Leong AS. 2005. Multiple organ infection and the pathogenesis of SARS. J Exp Med 202:415–424. doi: 10.1084/jem.20050828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Schulert GS, Canna SW. 2018. Convergent pathways of the hyperferritinemic syndromes. Int Immunol 30:195–203. doi: 10.1093/intimm/dxy012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Mira JC, Gentile LF, Mathias BJ, Efron PA, Brakenridge SC, Mohr AM, Moore FA, Moldawer LL. 2017. Sepsis pathophysiology, chronic critical illness, and persistent inflammation-immunosuppression and catabolism syndrome. Crit Care Med 45:253–262. doi: 10.1097/CCM.0000000000002074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Xu Z, Shi L, Wang Y, Zhang J, Huang L, Zhang C, Liu S, Zhao P, Liu H, Zhu L, Tai Y, Bai C, Gao T, Song J, Xia P, Dong J, Zhao J, Wang FS. 2020. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir Med 8:420–422. doi: 10.1016/S2213-2600(20)30076-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Qin C, Zhou L, Hu Z, Zhang S, Yang S, Tao Y, Xie C, Ma K, Shang K, Wang W, Tian DS. 2020. Dysregulation of immune response in patients with COVID-19 in Wuhan, China. Clin Infect Dis 71:762–768. doi: 10.1093/cid/ciaa248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Humblet-Baron S, Franckaert D, Dooley J, Bornschein S, Cauwe B, Schönefeldt S, Bossuyt X, Matthys P, Baron F, Wouters C, Liston A. 2016. IL-2 consumption by highly activated CD8 T cells induces regulatory T-cell dysfunction in patients with hemophagocytic lymphohistiocytosis. J Allergy Clin Immunol 138:200–209.e8. doi: 10.1016/j.jaci.2015.12.1314. [DOI] [PubMed] [Google Scholar]
- 123.Humblet-Baron S, Franckaert D, Dooley J, Ailal F, Bousfiha A, Deswarte C, Oleaga-Quintas C, Casanova JL, Bustamante J, Liston A. 2019. IFN-γ and CD25 drive distinct pathologic features during hemophagocytic lymphohistiocytosis. J Allergy Clin Immunol 143:2215–2226.e7. doi: 10.1016/j.jaci.2018.10.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zhao J, Zhao J, Van Rooijen N, Perlman S. 2009. Evasion by stealth: inefficient immune activation underlies poor T cell response and severe disease in SARS-CoV-infected mice. PLoS Pathog 5:e1000636. doi: 10.1371/journal.ppat.1000636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Chen J, Lau YF, Lamirande EW, Paddock CD, Bartlett JH, Zaki SR, Subbarao K. 2010. Cellular immune responses to severe acute respiratory syndrome coronavirus (SARS-CoV) infection in senescent BALB/c mice: CD4+ T cells are important in control of SARS-CoV infection. J Virol 84:1289–1301. doi: 10.1128/JVI.01281-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Zhao J, Li K, Wohlford-Lenane C, Agnihothram SS, Fett C, Zhao J, Gale MJ, Jr, Baric RS, Enjuanes L, Gallagher T, McCray PB, Jr, Perlman S. 2014. Rapid generation of a mouse model for Middle East respiratory syndrome. Proc Natl Acad Sci U S A 111:4970–4975. doi: 10.1073/pnas.1323279111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Li K, Wohlford-Lenane CL, Channappanavar R, Park JE, Earnest JT, Bair TB, Bates AM, Brogden KA, Flaherty HA, Gallagher T, Meyerholz DK, Perlman S, McCray PB, Jr. 2017. Mouse-adapted MERS coronavirus causes lethal lung disease in human DPP4 knockin mice. Proc Natl Acad Sci U S A 114:E3119–E3128. doi: 10.1073/pnas.1619109114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Zhao J, Zhao J, Legge K, Perlman S. 2011. Age-related increases in PGD(2) expression impair respiratory DC migration, resulting in diminished T cell responses upon respiratory virus infection in mice. J Clin Invest 121:4921–4930. doi: 10.1172/JCI59777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Chousterman BG, Swirski FK, Weber GF. 2017. Cytokine storm and sepsis disease pathogenesis. Semin Immunopathol 39:517–528. doi: 10.1007/s00281-017-0639-8. [DOI] [PubMed] [Google Scholar]
- 130.Yang Y, Shen C, Li J, Yuan J, Wei J, Huang F, Wang F, Li G, Li Y, Xing L, Peng L, Yang M, Cao M, Zheng H, Wu W, Zou R, Li D, Xu Z, Wang H, Zhang M, Zhang Z, Gao GF, Jiang C, Liu L, Liu Y. 2020. Plasma IP-10 and MCP-3 levels are highly associated with disease severity and predict the progression of COVID-19. J Allergy Clin Immunol 146:119–127.e4. doi: 10.1016/j.jaci.2020.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Wu F, Zhao S, Yu B, Chen YM, Wang W, Song ZG, Hu Y, Tao ZW, Tian JH, Pei YY, Yuan ML, Zhang YL, Dai FH, Liu Y, Wang QM, Zheng JJ, Xu L, Holmes EC, Zhang YZ. 2020. A new coronavirus associated with human respiratory disease in China. Nature 579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Liu J, Li S, Liu J, Liang B, Wang X, Wang H, Li W, Tong Q, Yi J, Zhao L, Xiong L, Guo C, Tian J, Luo J, Yao J, Pang R, Shen H, Peng C, Liu T, Zhang Q, Wu J, Xu L, Lu S, Wang B, Weng Z, Han C, Zhu H, Zhou R, Zhou H, Chen X, Ye P, Zhu B, Wang L, Zhou W, He S, He Y, Jie S, Wei P, Zhang J, Lu Y, Wang W, Zhang L, Li L, Zhou F, Wang J, Dittmer U, Lu M, Hu Y, Yang D, Zheng X. 2020. Longitudinal characteristics of lymphocyte responses and cytokine profiles in the peripheral blood of SARS-CoV-2 infected patients. EBioMedicine 55:102763. doi: 10.1016/j.ebiom.2020.102763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Mahallawi WH, Khabour OF, Zhang Q, Makhdoum HM, Suliman BA. 2018. MERS-CoV infection in humans is associated with a pro-inflammatory Th1 and Th17 cytokine profile. Cytokine 104:8–13. doi: 10.1016/j.cyto.2018.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Wong CK, Lam CW, Wu AK, Ip WK, Lee NL, Chan IH, Lit LC, Hui DS, Chan MH, Chung SS, Sung JJ. 2004. Plasma inflammatory cytokines and chemokines in severe acute respiratory syndrome. Clin Exp Immunol 136:95–103. doi: 10.1111/j.1365-2249.2004.02415.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Park MD. 2020. Macrophages: a Trojan horse in COVID-19? Nat Rev Immunol 20:351. doi: 10.1038/s41577-020-0317-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Giamarellos-Bourboulis EJ, Netea MG, Rovina N, Akinosoglou K, Antoniadou A, Antonakos N, Damoraki G, Gkavogianni T, Adami ME, Katsaounou P, Ntaganou M, Kyriakopoulou M, Dimopoulos G, Koutsodimitropoulos I, Velissaris D, Koufargyris P, Karageorgos A, Katrini K, Lekakis V, Lupse M, Kotsaki A, Renieris G, Theodoulou D, Panou V, Koukaki E, Koulouris N, Gogos C, Koutsoukou A. 2020. Complex immune dysregulation in COVID-19 patients with severe respiratory failure. Cell Host Microbe 27:992–1000.e3. doi: 10.1016/j.chom.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.McGonagle D, Sharif K, O’Regan A, Bridgewood C. 2020. The role of cytokines including interleukin-6 in COVID-19 induced pneumonia and macrophage activation syndrome-like disease. Autoimmun Rev 19:102537. doi: 10.1016/j.autrev.2020.102537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Tanaka T, Narazaki M, Kishimoto T. 2016. Immunotherapeutic implications of IL-6 blockade for cytokine storm. Immunotherapy 8:959–970. doi: 10.2217/imt-2016-0020. [DOI] [PubMed] [Google Scholar]
- 139.Cifaldi L, Prencipe G, Caiello I, Bracaglia C, Locatelli F, De Benedetti F, Strippoli R. 2015. Inhibition of natural killer cell cytotoxicity by interleukin-6: implications for the pathogenesis of macrophage activation syndrome. Arthritis Rheumatol 67:3037–3046. doi: 10.1002/art.39295. [DOI] [PubMed] [Google Scholar]
- 140.Marin V, Montero-Julian FA, Grès S, Boulay V, Bongrand P, Farnarier C, Kaplanski G. 2001. The IL-6-soluble IL-6Ralpha autocrine loop of endothelial activation as an intermediate between acute and chronic inflammation: an experimental model involving thrombin. J Immunol 167:3435–3442. doi: 10.4049/jimmunol.167.6.3435. [DOI] [PubMed] [Google Scholar]
- 141.Yang Y, Shen C, Li J, Yuan J, Yang M, Wang F, Li G, Li Y, Xing L, Peng L, Wei J, Cao M, Zheng H, Wu W, Zou R, Li D, Xu Z, Wang H, Zhang M, Zhang Z, Liu L, Liu Y. 2020. Exuberant elevation of IP-10, MCP-3 and IL-1ra during SARS-CoV-2 infection is associated with disease severity and fatal outcome. medRxiv doi: 10.1101/2020.03.02.20029975. [DOI] [Google Scholar]
- 142.Song C-Y, Xu J, He J-Q, Lu Y-Q. 2020. COVID-19 early warning score: a multi-parameter screening tool to identify highly suspected patients. medRxiv doi: 10.1101/2020.03.05.20031906. [DOI] [Google Scholar]
- 143.Zoller EE, Lykens JE, Terrell CE, Aliberti J, Filipovich AH, Henson PM, Jordan MB. 2011. Hemophagocytosis causes a consumptive anemia of inflammation. J Exp Med 208:1203–1214. doi: 10.1084/jem.20102538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Lagunas-Rangel FA, Chávez-Valencia V. 2020. High IL-6/IFN-γ ratio could be associated with severe disease in COVID-19 patients. J Med Virol doi: 10.1002/jmv.25900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Dinarello CA. 2018. Overview of the IL-1 family in innate inflammation and acquired immunity. Immunol Rev 281:8–27. doi: 10.1111/imr.12621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Wan S, Yi Q, Fan S, Lv J, Zhang X, Guo L, Lang C, Xiao Q, Xiao K, Yi Z, Qiang M, Xiang J, Zhang B, Chen Y. 2020. Characteristics of lymphocyte subsets and cytokines in peripheral blood of 123 hospitalized patients with 2019 novel coronavirus pneumonia (NCP). medRxiv doi: 10.1101/2020.02.10.20021832. [DOI] [Google Scholar]
- 147.Idriss HT, Naismith JH. 2000. TNF alpha and the TNF receptor superfamily: structure-function relationship(s). Microsc Res Tech 50:184–195. doi:. [DOI] [PubMed] [Google Scholar]
- 148.Horiuchi T, Mitoma H, Harashima S, Tsukamoto H, Shimoda T. 2010. Transmembrane TNF-alpha: structure, function and interaction with anti-TNF agents. Rheumatology (Oxford) 49:1215–1228. doi: 10.1093/rheumatology/keq031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Gong J, Dong H, Xia SQ, Huang YZ, Wang D, Zhao Y, Liu W, Tu S, Zhang M, Wang Q, Lu F. 2020. Correlation analysis between disease severity and inflammation-related parameters in patients with COVID-19 pneumonia. medRxiv doi: 10.1101/2020.02.25.20025643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Tobinick E. 2004. TNF-alpha inhibition for potential therapeutic modulation of SARS coronavirus infection. Curr Med Res Opin 20:39–40. doi: 10.1185/030079903125002757. [DOI] [PubMed] [Google Scholar]
- 151.Haga S, Yamamoto N, Nakai-Murakami C, Osawa Y, Tokunaga K, Sata T, Yamamoto N, Sasazuki T, Ishizaka Y. 2008. Modulation of TNF-alpha-converting enzyme by the spike protein of SARS-CoV and ACE2 induces TNF-alpha production and facilitates viral entry. Proc Natl Acad Sci U S A 105:7809–7814. doi: 10.1073/pnas.0711241105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Ma X. 2001. TNF-alpha and IL-12: a balancing act in macrophage functioning. Microbes Infect 3:121–129. doi: 10.1016/s1286-4579(00)01359-9. [DOI] [PubMed] [Google Scholar]
- 153.Mehta AK, Gracias DT, Croft M. 2018. TNF activity and T cells. Cytokine 101:14–18. doi: 10.1016/j.cyto.2016.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Barton LM, Duval EJ, Stroberg E, Ghosh S, Mukhopadhyay S. 2020. COVID-19 autopsies, Oklahoma, USA. Am J Clin Pathol 153:725–733. doi: 10.1093/ajcp/aqaa062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Theron M, Huang KJ, Chen YW, Liu CC, Lei HY. 2005. A probable role for IFN-gamma in the development of a lung immunopathology in SARS. Cytokine 32:30–38. doi: 10.1016/j.cyto.2005.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Faure E, Poissy J, Goffard A, Fournier C, Kipnis E, Titecat M, Bortolotti P, Martinez L, Dubucquoi S, Dessein R, Gosset P, Mathieu D, Guery B. 2014. Distinct immune response in two MERS-CoV-infected patients: can we go from bench to bedside? PLoS One 9:e88716. doi: 10.1371/journal.pone.0088716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Min CK, Cheon S, Ha NY, Sohn KM, Kim Y, Aigerim A, Shin HM, Choi JY, Inn KS, Kim JH, Moon JY, Choi MS, Cho NH, Kim YS. 2016. Comparative and kinetic analysis of viral shedding and immunological responses in MERS patients representing a broad spectrum of disease severity. Sci Rep 6:25359. doi: 10.1038/srep25359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Cameron MJ, Bermejo-Martin JF, Danesh A, Muller MP, Kelvin DJ. 2008. Human immunopathogenesis of severe acute respiratory syndrome (SARS). Virus Res 133:13–19. doi: 10.1016/j.virusres.2007.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Russell B, Moss C, George G, Santaolalla A, Cope A, Papa S, Van Hemelrijck M. 2020. Associations between immune-suppressive and stimulating drugs and novel COVID-19-a systematic review of current evidence. Ecancermedicalscience 14:1022. doi: 10.3332/ecancer.2020.1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Jamilloux Y, Henry T, Belot A, Viel S, Fauter M, El Jammal T, Walzer T, François B, Sève P. 2020. Should we stimulate or suppress immune responses in COVID-19? Cytokine and anti-cytokine interventions. Autoimmun Rev 19:102567. doi: 10.1016/j.autrev.2020.102567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Nie Y, Wang G, Shi X, Zhang H, Qiu Y, He Z, Wang W, Lian G, Yin X, Du L, Ren L, Wang J, He X, Li T, Deng H, Ding M. 2004. Neutralizing antibodies in patients with severe acute respiratory syndrome-associated coronavirus infection. J Infect Dis 190:1119–1126. doi: 10.1086/423286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Liu W, Fontanet A, Zhang PH, Zhan L, Xin ZT, Baril L, Tang F, Lv H, Cao WC. 2006. Two-year prospective study of the humoral immune response of patients with severe acute respiratory syndrome. J Infect Dis 193:792–795. doi: 10.1086/500469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Buchholz UJ, Bukreyev A, Yang L, Lamirande EW, Murphy BR, Subbarao K, Collins PL. 2004. Contributions of the structural proteins of severe acute respiratory syndrome coronavirus to protective immunity. Proc Natl Acad Sci U S A 101:9804–9809. doi: 10.1073/pnas.0403492101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Jiang S, Hillyer C, Du L. 2020. Neutralizing antibodies against SARS-CoV-2 and other human coronaviruses. Trends Immunol 41:355–359. doi: 10.1016/j.it.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Radermacher P, Maggiore SM, Mercat A. 2017. Fifty years of research in ARDS. Gas exchange in acute respiratory distress syndrome. Am J Respir Crit Care Med 196:964–984. doi: 10.1164/rccm.201610-2156SO. [DOI] [PubMed] [Google Scholar]
- 166.Thille AW, Esteban A, Fernandez-Segoviano P, Rodriguez JM, Aramburu JA, Vargas-Errazuriz P, Martin-Pellicer A, Lorente JA, Frutos-Vivar F. 2013. Chronology of histological lesions in acute respiratory distress syndrome with diffuse alveolar damage: a prospective cohort study of clinical autopsies. Lancet Respir Med 1:395–401. doi: 10.1016/S2213-2600(13)70053-5. [DOI] [PubMed] [Google Scholar]
- 167.Gu J, Korteweg C. 2007. Pathology and pathogenesis of severe acute respiratory syndrome. Am J Pathol 170:1136–1147. doi: 10.2353/ajpath.2007.061088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Frieman MB, Chen J, Morrison TE, Whitmore A, Funkhouser W, Ward JM, Lamirande EW, Roberts A, Heise M, Subbarao K, Baric RS. 2010. SARS-CoV pathogenesis is regulated by a STAT1 dependent but a type I, II and III interferon receptor independent mechanism. PLoS Pathog 6:e1000849. doi: 10.1371/journal.ppat.1000849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Zornetzer GA, Frieman MB, Rosenzweig E, Korth MJ, Page C, Baric RS, Katze MG. 2010. Transcriptomic analysis reveals a mechanism for a prefibrotic phenotype in STAT1 knockout mice during severe acute respiratory syndrome coronavirus infection. J Virol 84:11297–11309. doi: 10.1128/JVI.01130-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Ng DL, Al Hosani F, Keating MK, Gerber SI, Jones TL, Metcalfe MG, Tong S, Tao Y, Alami NN, Haynes LM, Mutei MA, Abdel-Wareth L, Uyeki TM, Swerdlow DL, Barakat M, Zaki SR. 2016. Clinicopathologic, immunohistochemical, and ultrastructural findings of a fatal case of Middle East respiratory syndrome coronavirus infection in the United Arab Emirates, April 2014. Am J Pathol 186:652–658. doi: 10.1016/j.ajpath.2015.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Kan B, Wang M, Jing H, Xu H, Jiang X, Yan M, Liang W, Zheng H, Wan K, Liu Q, Cui B, Xu Y, Zhang E, Wang H, Ye J, Li G, Li M, Cui Z, Qi X, Chen K, Du L, Gao K, Zhao YT, Zou XZ, Feng YJ, Gao YF, Hai R, Yu D, Guan Y, Xu J. 2005. Molecular evolution analysis and geographic investigation of severe acute respiratory syndrome coronavirus-like virus in palm civets at an animal market and on farms. J Virol 79:11892–11900. doi: 10.1128/JVI.79.18.11892-11900.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.General Office of the National Health Commission. 2020. Diagnostic and treatment protocol for novel coronavirus pneumonia (trial version 7). National Health Commission, Beijing, China. http://www.nhc.gov.cn/yzygj/s7653p/202003/46c9294a7dfe4cef80dc7f5912eb1989.shtml?spm=C73544894212.P59511941341.0.0. Accessed 5 March 2020.
- 173.Folkesson HG, Matthay MA. 2006. Alveolar epithelial ion and fluid transport: recent progress. Am J Respir Cell Mol Biol 35:10–19. doi: 10.1165/rcmb.2006-0080SF. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Lu G, Hu Y, Wang Q, Qi J, Gao F, Li Y, Zhang Y, Zhang W, Yuan Y, Bao J, Zhang B, Shi Y, Yan J, Gao GF. 2013. Molecular basis of binding between novel human coronavirus MERS-CoV and its receptor CD26. Nature 500:227–231. doi: 10.1038/nature12328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Scobey T, Yount BL, Sims AC, Donaldson EF, Agnihothram SS, Menachery VD, Graham RL, Swanstrom J, Bove PF, Kim JD, Grego S, Randell SH, Baric RS. 2013. Reverse genetics with a full-length infectious cDNA of the Middle East respiratory syndrome coronavirus. Proc Natl Acad Sci U S A 110:16157–16162. doi: 10.1073/pnas.1311542110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Mossel EC, Wang J, Jeffers S, Edeen KE, Wang S, Cosgrove GP, Funk CJ, Manzer R, Miura TA, Pearson LD, Holmes KV, Mason RJ. 2008. SARS-CoV replicates in primary human alveolar type II cell cultures but not in type I-like cells. Virology 372:127–135. doi: 10.1016/j.virol.2007.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Herold S, Steinmueller M, von Wulffen W, Cakarova L, Pinto R, Pleschka S, Mack M, Kuziel WA, Corazza N, Brunner T, Seeger W, Lohmeyer J. 2008. Lung epithelial apoptosis in influenza virus pneumonia: the role of macrophage-expressed TNF-related apoptosis-inducing ligand. J Exp Med 205:3065–3077. doi: 10.1084/jem.20080201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Hogner K, Wolff T, Pleschka S, Plog S, Gruber AD, Kalinke U, Walmrath HD, Bodner J, Gattenlohner S, Lewe-Schlosser P, Matrosovich M, Seeger W, Lohmeyer J, Herold S. 2013. Macrophage-expressed IFN-beta contributes to apoptotic alveolar epithelial cell injury in severe influenza virus pneumonia. PLoS Pathog 9:e1003188. doi: 10.1371/journal.ppat.1003188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Rodrigue-Gervais IG, Labbe K, Dagenais M, Dupaul-Chicoine J, Champagne C, Morizot A, Skeldon A, Brincks EL, Vidal SM, Griffith TS, Saleh M. 2014. Cellular inhibitor of apoptosis protein cIAP2 protects against pulmonary tissue necrosis during influenza virus infection to promote host survival. Cell Host Microbe 15:23–35. doi: 10.1016/j.chom.2013.12.003. [DOI] [PubMed] [Google Scholar]
- 180.Herold S, von Wulffen W, Steinmueller M, Pleschka S, Kuziel WA, Mack M, Srivastava M, Seeger W, Maus UA, Lohmeyer J. 2006. Alveolar epithelial cells direct monocyte transepithelial migration upon influenza virus infection: impact of chemokines and adhesion molecules. J Immunol 177:1817–1824. doi: 10.4049/jimmunol.177.3.1817. [DOI] [PubMed] [Google Scholar]
- 181.Tasaka S, Amaya F, Hashimoto S, Ishizaka A. 2008. Roles of oxidants and redox signaling in the pathogenesis of acute respiratory distress syndrome. Antioxid Redox Signal 10:739–753. doi: 10.1089/ars.2007.1940. [DOI] [PubMed] [Google Scholar]
- 182.Ramana CV, DeBerge MP, Kumar A, Alia CS, Durbin JE, Enelow RI. 2015. Inflammatory impact of IFN-gamma in CD8+ T cell-mediated lung injury is mediated by both Stat1-dependent and -independent pathways. Am J Physiol Lung Cell Mol Physiol 308:L650–L657. doi: 10.1152/ajplung.00360.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Zhao MQ, Stoler MH, Liu AN, Wei B, Soguero C, Hahn YS, Enelow RI. 2000. Alveolar epithelial cell chemokine expression triggered by antigen-specific cytolytic CD8(+) T cell recognition. J Clin Invest 106:R49–R58. doi: 10.1172/JCI9786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.He L, Ding Y, Zhang Q, Che X, He Y, Shen H, Wang H, Li Z, Zhao L, Geng J, Deng Y, Yang L, Li J, Cai J, Qiu L, Wen K, Xu X, Jiang S. 2006. Expression of elevated levels of pro-inflammatory cytokines in SARS-CoV-infected ACE2+ cells in SARS patients: relation to the acute lung injury and pathogenesis of SARS. J Pathol 210:288–297. doi: 10.1002/path.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Lau SKP, Lau CCY, Chan KH, Li CPY, Chen H, Jin DY, Chan JFW, Woo PCY, Yuen KY. 2013. Delayed induction of proinflammatory cytokines and suppression of innate antiviral response by the novel Middle East respiratory syndrome coronavirus: implications for pathogenesis and treatment. J Gen Virol 94:2679–2690. doi: 10.1099/vir.0.055533-0. [DOI] [PubMed] [Google Scholar]
- 186.Roux J, Kawakatsu H, Gartland B, Pespeni M, Sheppard D, Matthay MA, Canessa CM, Pittet JF. 2005. Interleukin-1beta decreases expression of the epithelial sodium channel alpha-subunit in alveolar epithelial cells via a p38 MAPK-dependent signaling pathway. J Biol Chem 280:18579–18589. doi: 10.1074/jbc.M410561200. [DOI] [PubMed] [Google Scholar]
- 187.Berthiaume Y, Matthay MA. 2007. Alveolar edema fluid clearance and acute lung injury. Respir Physiol Neurobiol 159:350–359. doi: 10.1016/j.resp.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Page C, Goicochea L, Matthews K, Zhang Y, Klover P, Holtzman MJ, Hennighausen L, Frieman M. 2012. Induction of alternatively activated macrophages enhances pathogenesis during severe acute respiratory syndrome coronavirus infection. J Virol 86:13334–13349. doi: 10.1128/JVI.01689-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Husain AK. 2010. Robbins & Cotran pathologic basis of disease, 8th ed. Elsevier, Philadelphia, PA. [Google Scholar]
- 190.Matthay MA, Zimmerman GA. 2005. Acute lung injury and the acute respiratory distress syndrome: four decades of inquiry into pathogenesis and rational management. Am J Respir Cell Mol Biol 33:319–327. doi: 10.1165/rcmb.F305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Looney MR, Su X, Van Ziffle JA, Lowell CA, Matthay MA. 2006. Neutrophils and their Fc gamma receptors are essential in a mouse model of transfusion-related acute lung injury. J Clin Invest 116:1615–1623. doi: 10.1172/JCI27238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Jiang Y, Xu J, Zhou C, Wu Z, Zhong S, Liu J, Luo W, Chen T, Qin Q, Deng P. 2005. Characterization of cytokine/chemokine profiles of severe acute respiratory syndrome. Am J Respir Crit Care Med 171:850–857. doi: 10.1164/rccm.200407-857OC. [DOI] [PubMed] [Google Scholar]
- 193.Reghunathan R, Jayapal M, Hsu LY, Chng HH, Tai D, Leung BP, Melendez AJ. 2005. Expression profile of immune response genes in patients with severe acute respiratory syndrome. BMC Immunol 6:2. doi: 10.1186/1471-2172-6-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Wiener-Kronish JP, Albertine KH, Matthay MA. 1991. Differential responses of the endothelial and epithelial barriers of the lung in sheep to Escherichia coli endotoxin. J Clin Invest 88:864–875. doi: 10.1172/JCI115388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Zhang Y, Li J, Zhan Y, Wu L, Yu X, Zhang W, Ye L, Xu S, Sun R, Wang Y, Lou J. 2004. Analysis of serum cytokines in patients with severe acute respiratory syndrome. Infect Immun 72:4410–4415. doi: 10.1128/IAI.72.8.4410-4415.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Chien JY, Hsueh PR, Cheng WC, Yu CJ, Yang PC. 2006. Temporal changes in cytokine/chemokine profiles and pulmonary involvement in severe acute respiratory syndrome. Respirology 11:715–722. doi: 10.1111/j.1440-1843.2006.00942.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Nicholls J, Dong XP, Jiang G, Peiris M. 2003. SARS: clinical virology and pathogenesis. Respirology 8(Suppl):S6–S8. doi: 10.1046/j.1440-1843.2003.00517.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Ooi GC, Daqing M. 2003. SARS: radiological features. Respirology 8(Suppl):S15–S19. doi: 10.1046/j.1440-1843.2003.00519.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Wong KT, Antonio GE, Hui DS, Lee N, Yuen EH, Wu A, Leung CB, Rainer TH, Cameron P, Chung SS, Sung JJ, Ahuja AT. 2003. Severe acute respiratory syndrome: radiographic appearances and pattern of progression in 138 patients. Radiology 228:401–406. doi: 10.1148/radiol.2282030593. [DOI] [PubMed] [Google Scholar]
- 200.Ajlan AM, Ahyad RA, Jamjoom LG, Alharthy A, Madani TA. 2014. Middle East respiratory syndrome coronavirus (MERS-CoV) infection: chest CT findings. AJR Am J Roentgenol 203:782–787. doi: 10.2214/AJR.14.13021. [DOI] [PubMed] [Google Scholar]
- 201.Chung M, Bernheim A, Mei X, Zhang N, Huang M, Zeng X, Cui J, Xu W, Yang Y, Fayad ZA, Jacobi A, Li K, Li S, Shan H. 2020. CT imaging features of 2019 novel coronavirus (2019-nCoV). Radiology 295:202–207. doi: 10.1148/radiol.2020200230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Colafrancesco S, Alessandri C, Conti F, Priori R. 2020. COVID-19 gone bad: a new character in the spectrum of the hyperferritinemic syndrome? Autoimmun Rev 19:102573. doi: 10.1016/j.autrev.2020.102573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Filipovich AH. 2008. Hemophagocytic lymphohistiocytosis and other hemophagocytic disorders. Immunol Allergy Clin North Am 28:293–313. doi: 10.1016/j.iac.2008.01.010. [DOI] [PubMed] [Google Scholar]
- 204.Abbas A, Raza M, Majid A, Khalid Y, Bin Waqar SH. 2018. Infection-associated hemophagocytic lymphohistiocytosis: an unusual clinical masquerader. Cureus 10:e2472. doi: 10.7759/cureus.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Kim WY, Montes-Mojarro IA, Fend F, Quintanilla-Martinez L. 2019. Epstein-Barr virus-associated T and NK-cell lymphoproliferative diseases. Front Pediatr 7:71. doi: 10.3389/fped.2019.00071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Daver N, McClain K, Allen CE, Parikh SA, Otrock Z, Rojas-Hernandez C, Blechacz B, Wang S, Minkov M, Jordan MB, La Rosée P, Kantarjian HM. 2017. A consensus review on malignancy-associated hemophagocytic lymphohistiocytosis in adults. Cancer 123:3229–3240. doi: 10.1002/cncr.30826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Lam CW, Chan MH, Wong CK. 2004. Severe acute respiratory syndrome: clinical and laboratory manifestations. Clin Biochem Rev 25:121–132. [PMC free article] [PubMed] [Google Scholar]
- 208.Sheng WH, Chiang BL, Chang SC, Ho HN, Wang JT, Chen YC, Hsiao CH, Hseuh PR, Chie WC, Yang PC. 2005. Clinical manifestations and inflammatory cytokine responses in patients with severe acute respiratory syndrome. J Formos Med Assoc 104:715–723. [PubMed] [Google Scholar]
- 209.Duan ZP, Chen Y, Zhang J, Zhao J, Lang ZW, Meng FK, Bao XL. 2003. Clinical characteristics and mechanism of liver injury in patients with severe acute respiratory syndrome. Zhonghua Gan Zang Bing Za Zhi 11:493–496. (In Chinese.) [PubMed] [Google Scholar]
- 210.Yu SY, Hu YW, Liu XY, Xiong W, Zhou ZT, Yuan ZH. 2005. Gene expression profiles in peripheral blood mononuclear cells of SARS patients. World J Gastroenterol 11:5037–5043. doi: 10.3748/wjg.v11.i32.5037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Alosaimi B, Hamed ME, Naeem A, Alsharef AA, AlQahtani SY, AlDosari KM, Alamri AA, Al-Eisa K, Khojah T, Assiri AM, Enani MA. 2020. MERS-CoV infection is associated with downregulation of genes encoding Th1 and Th2 cytokines/chemokines and elevated inflammatory innate immune response in the lower respiratory tract. Cytokine 126:154895. doi: 10.1016/j.cyto.2019.154895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Han H, Ma Q, Li C, Liu R, Zhao L, Wang W, Zhang P, Liu X, Gao G, Liu F, Jiang Y, Cheng X, Zhu C, Xia Y. 2020. Profiling serum cytokines in COVID-19 patients reveals IL-6 and IL-10 are disease severity predictors. Emerg Microbes Infect 9:1123–1130. doi: 10.1080/22221751.2020.1770129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.WHO. 2020. Coronavirus disease (COVID-19). Situation report–142. World Health Organization, Geneva, Switzerland. https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200610-covid-19-sitrep-142.pdf?sfvrsn=180898cd_6. Accessed 21 June 2020. [Google Scholar]