Abstract
Partly due to the use of exhaustive-annotated data, deep networks have achieved impressive performance on medical image segmentation. Medical imaging data paired with noisy annotation are, however, ubiquitous, but little is known about the effect of noisy annotation on deep learning based medical image segmentation. We studied the effect of noisy annotation in the context of mandible segmentation from CT images. First, 202 images of head and neck cancer patients were collected from our clinical database, where the organs-at-risk were annotated by one of twelve planning dosimetrists. The mandibles were roughly annotated as the planning avoiding structure. Then, mandible labels were checked and corrected by a head and neck specialist to get the reference standard. At last, by varying the ratios of noisy labels in the training set, deep networks were trained and tested for mandible segmentation. The trained models were further tested on other two public datasets. Experimental results indicated that the network trained with noisy labels had worse segmentation than that trained with reference standard, and in general, fewer noisy labels led to better performance. When using 20% or less noisy cases for training, no significant difference was found on the segmentation results between the models trained by noisy or reference annotation. Cross-dataset validation results verified that the models trained with noisy data achieved competitive performance to that trained with reference standard. This study suggests that the involved network is robust to noisy annotation to some extent in mandible segmentation from CT images. It also highlights the importance of labeling quality in deep learning. In the future work, extra attention should be paid to how to utilize a small number of reference standard samples to improve the performance of deep learning with noisy annotation.
Keywords: deep learning, noisy annotation, radiation oncology, medical image segmentation
1. Introduction
Deep supervised networks have achieved impressive performance in medical image segmentation partly due to the use of high-quality exhaustive-annotated data (Liu et al 2017, Chen et al 2019, Hesamian et al 2019). However, in radiation oncology, it is hard or impossible to conduct sufficient high-quality image annotation. In addition to potential hurdles of funding acquisitions, time cost and patient privacy, accurate annotation of medical images always depends on scarce and expensive medical expertise (Greenspan et al 2016) and thereby, medical imaging data paired with noisy annotation is prevalent, particularly in radiation oncology.
Increasing attention has been paid to the issue of label noise in deeply supervised image classification (Hendrycks et al 2018, Tanaka et al 2018, Han et al 2019). These approaches to tackle the label noise could be generally categorized into two groups. One tends to analyze the label noise and to develop deep networks with noise-robust loss functions. Reed et al proposed a generic way to tackle inaccurate labels by augmenting the prediction objective function with a notion of perceptual consistency (Reed et al 2014). The consistency was defined as the confidence of predicted labels between different objective estimations computed from the same input data. Further, the authors introduced a convex combination of the known labels and predicted labels as the training target in self-learning. Patrini et al presented two procedures for loss function correction, and both the application domain and the network architecture were unknown (Patrini et al 2017). The computing cost is at most a matrix inversion and multiplication. Both procedures were proven to be robust to the noisy data, and importantly, the Hessian of the loss function was found independent from label noise for the ReLU networks. By generalizing the categorical cross entropy, Zhang and Sabuncu developed a theoretically grounded set of noise-robust loss functions (Zhang and Sabuncu 2018). These functions could be embedded into any deep networks to yield good performance in a wide range of noisy label scenarios. And notably, Luo et al designed a variance regularization term to penalize the Jacobian norm of a deep network on the whole training set (Luo et al 2019). Both theoretically deduced and experimental results showed that the regularization term can decrease the subspace dimensionality, improve the robustness, and generalize well to label noise. However, these approaches require prior knowledge or accurate estimation of the label noise distribution, which is not practical in real-world applications. The other group tends to figure out and to remove or correct noisy labels by using a small set of reference labels. Misra et al demonstrated that noisy labels from human-centric annotation are statistically dependent on the data, and thus, reference labels could be used to decouple this kind of human reporting bias and to improve image captioning performance (Misra et al 2016). Xiao et al introduced a general framework to train deep networks with a limited number of reference samples and massive noisy samples (Xiao et al 2015). The relationships among images, class labels, and label noises were quantified with a probabilistic graphical model, which was further integrated into an end-to-end deep learning system. Mirikharaji et al proposed a practical framework to learn from a limited number of clean samples in the training phase that assigned higher weights to pixels with gradient directions closer to those of reference data in a meta-learning approach (2019). This kind of approaches is feasible but significantly increases the computing complexity.
Many efforts have been made to tackle label noise in image classification, while little is known about the effect of annotation quality on object segmentation. Object segmentation can be viewed as pixel-wise image classification and requires high-quality exhaustive-annotated data for algorithm training. However, in radiation oncology, some organs-at-risk (OARs) may be roughly annotated due to the trade-off between time spent and radiation treatment planning quality. Such inconsistent and rough annotations can mislead the training of deep networks and result in ambiguous localization of anatomical structures. Thus, this study concerns the effect of annotation quality on medical image segmentation. It involves medical data annotated by dosimetrists in radiation treatment planning and differs from the aforementioned studies, which artificially generate noisy labels and do not reflect real-life scenarios. Further, two public datasets are used for cross-dataset validation. The primary purpose of this study is to investigate whether a deep network trained with noisy data can achieve comparative performance with that trained with reference standard. Specifically, the effect of different ratios of noisy cases in the training set is investigated in the context of deep learning based mandible segmentation from CT images.
2. Methods and materials
2.1. Data collection
Three datasets were analyzed. One is an in-house collection, named the UTSW dataset. It contained 202 images of head and neck (H&N) cancer patients (47 females and 155 males; mean age, 62 years). CT images and corresponding RT Structures were exported from Varian Eclipse Treatment Planning System (Eclipse v15.5, Varian Medical Systems). The isotropic in-plane voxel resolution was between 1.17 and 1.37 mm, and the slice thickness was 3.00 mm. The in-plane image size was [512, 512] and the slice number ranged between 127 and 264. All patient data are HIPPA-compliant de-identified and protected under an IRB for retrospective studies.
The second one is a public domain dataset from The Cancer Imaging Archive, named the TCIA dataset (Clark et al 2013, Nikolov et al 2018). It included 31 cases (4 females, 25 males and 2 unknown; mean age, 59 years), and to each case, 21 OARs were annotated by a single radiologist with a second arbitrating and compared with a ground truth from two further radiologists arbitrated by one of two independent oncology specialists. The in-plane voxel resolution was isotropic within the range of 0.94 and 1.27 mm, and the slice thickness was 2.50 mm. The in-plane image size was [512, 512] and the slice number was between 119 to 437.
The last one is the Public Domain Database for Computational Anatomy (PDDCA) dataset (Raudaschl et al 2017). As part of the challenge at the International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI) in 2015, the dataset was released for the segmentation of anatomical structures in the H&N region of CT images. In this study, 40 images (25 training images, 10 off-site testing images, and 5 on-site testing images) were collected, and to each image, 9 structures were manually re-contoured by experts for uniform quality and consistency (Santanam et al 2012). The isotropic in-plane voxel resolution was between 0.76 and 1.27 mm, and the slice thickness ranged in 1.25 and 3.00 mm. The in-plane image size was [512, 512] and the slice number was in the range of 76 to 360.
2.2. Mandible annotation and correction
Mandible, the largest bone in the human head, is one of the OARs in 3D-conformal radiotherapy or intensity-modulated radiotherapy for H&N cancers. The mandible regions are outlined in radiation treatment planning to avoid the potential development of jaw osteoradionecrosis. Anatomically, the mandible starts from the bottom chin area to the alveolar process and condyloid process, excluding the teeth.
To the UTSW dataset, 12 dosimetrists participated in image annotation. Among them, three dosimetrists each annotated more than 30 cases (31, 59, and 82 cases), and the others each annotated fewer than 10 cases. To address the issue of inaccurate annotation, the correction procedure was applied by a H&N radiation oncology specialist with 10+ year experience to get the reference standard. Given the reference annotation, figure 1 shows the distribution of voxel numbers in mandible regions, kept regions, deleted regions, and added regions after manual correction. Within the UTSW dataset, the mandible contained 14 09 ± 3236 voxels (≈ 57.89 ± 13.29 cm3); in the correction procedure, 95% voxels (13 398 ± 3474) were kept in the noisy labels; and to form the reference labels, 3887 ± 3254 and 699 ± 765 voxels are removed from and added to the noisy labels, respectively.
Figure 1.
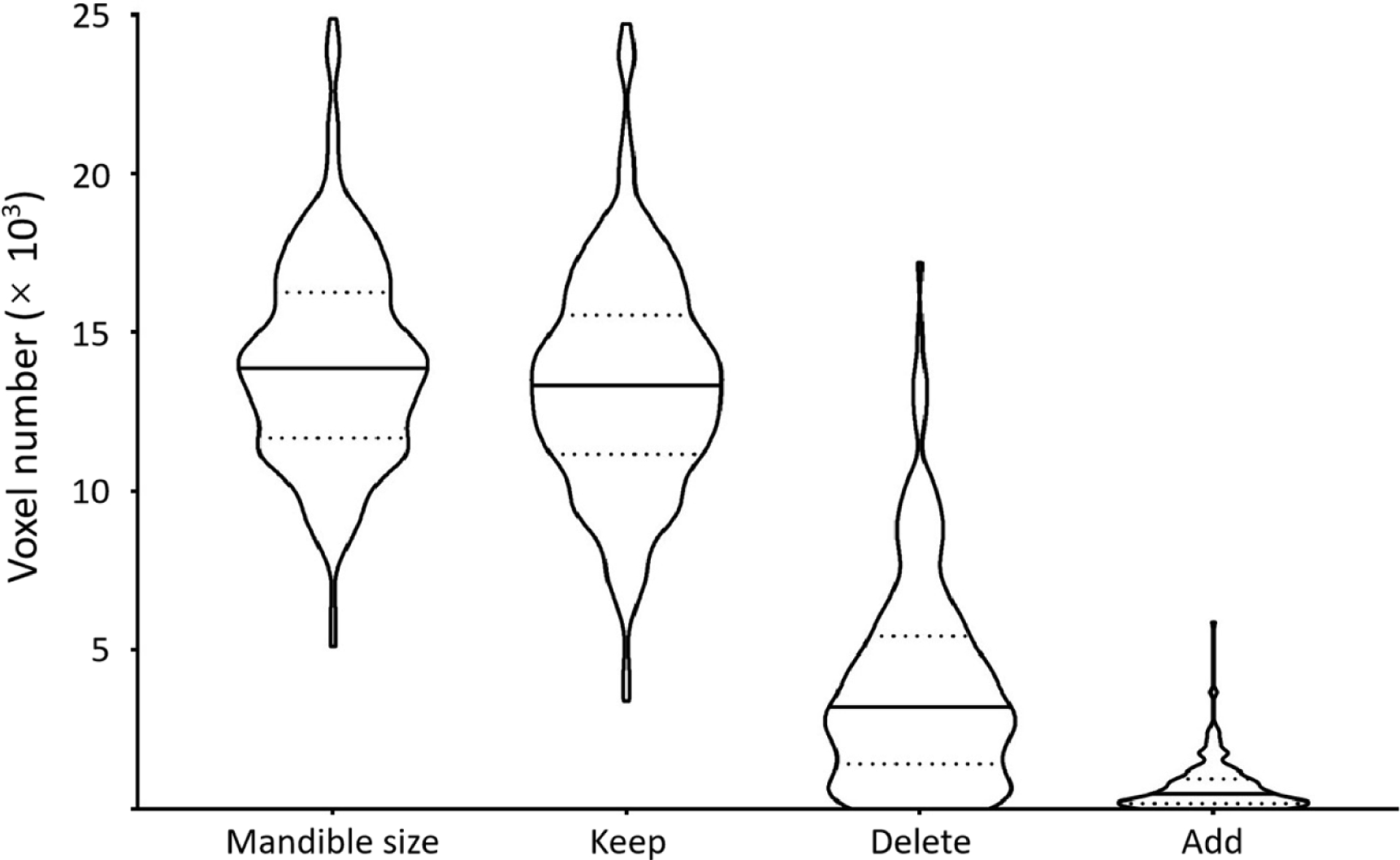
Voxel number analysis of the mandible regions. In each violin plot, the solid and dotted lines correspond to median and quartile values. It indicates the size of the mandible. Given the reference annotation, it further shows the distributions of the number of voxels kept in, deleted from, and added to the noisy labels in the correction procedure.
Figure 2 illustrates three representative examples of mandible contour before and after correction. In the figure, the top row shows the delineation of mandible regions for radiation treatment planning, and the bottom row shows the corresponding regions after correction. The red and green contours stand for the boundaries of mandible regions. In general, label noise of the mandible is from incomplete annotation ((a) vs (d)), different definitions of mandible regions ((b) with vs (e) without the teeth), and inaccurate contouring ((c) vs (f)). In addition to noisy annotation, it is observed that the major challenge in mandible segmentation comes from the correct exclusion of the teeth (b) and metal artifacts of dental implants in image acquisitions (c).
Figure 2.
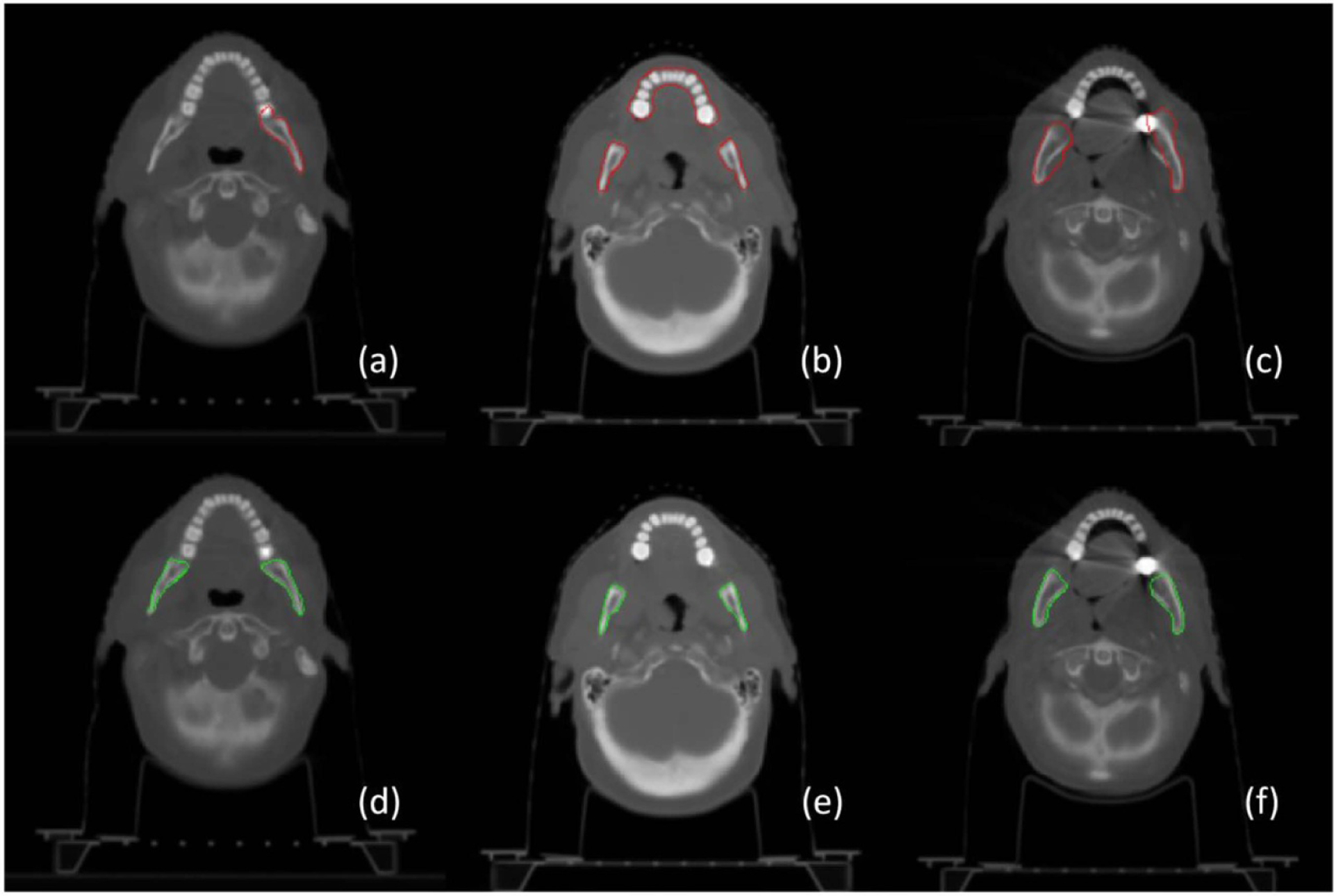
Perceived annotation difference via contour visualization. The top row shows rough contouring of the mandible in radiation treatment planning and the bottom row shows the corresponding mandible after label correction. It shows that the label noise mainly comes from incomplete annotation ((a) vs (d)), different definition of the mandible with the teeth (b) or without the teeth (e), and inaccurate delineation ((c) vs (f)).
2.3. Data preparation
CT images and corresponding label images of the UTSW, TCIA and PDDCA datasets were prepared in the same way as follows. First, to the label images, the mandible regions were assigned with value 1 and other regions were with 0. Then, CT images were transformed into RAS (right, anterior, superior) anatomical coordinate system, and tri-linear-interpolated to the same voxel resolution [1.17, 1.17, 3.00] mm3, and accordingly the label images. Third, one label image from the UTSW dataset was manually selected by the H&N radiation oncology specialist and the center of the mandible was set as the reference center in the image coordinate system. The centers of mandible regions of all other label images were translated to the reference center, and the CT images were translated accordingly. Finally, the centralized CT images and label images are cropped to the same matrix size of [256 256 128]. In addition, to highlight bone regions, the CT number (Hounsfield units) V of CT images was normalized by the mean μV>−100and standard deviation σV>−100following , where V > −100 denotes those pixels with CT number larger than −100, which is the threshold for typical fat tissue.
2.4. Experiment design
Table 1 shows the data splitting and experiment design. To the UTSW dataset, patient cases were divided into a training set (180 samples), a validation set (10 samples), and a test set (12 samples). For fair comparison, samples in both the validation and the test set were fixed. In addition, two open datasets, TCIA and PDDCA, were used for cross-dataset test after deep models were trained, validated and tested on the UTSW dataset.
Table 1.
Data splitting and experiment design.
| UTSW | TCIA | PDDCA | |||
|---|---|---|---|---|---|
| Train | Validation | Test | Test | Test | |
| Total scans(patients) | 180 (180) | 10(10) | 12(12) | 31(30) | 40 (40) |
To investigate the effect of noisy annotation on deep learning based mandible image segmentation, 8 designs were conducted on the UTSW dataset as shown in figure 3, where a rectangle denotes 18 (10%) samples, a rectangle with brown color indicates the CT images paired with noisy label images, and a rectangle with green color stands for the CT images paired with reference standard images. In each design, the percentages of noisy label cases were from 0% to 60% at 10% equal increment in addition to 100%, labeled alphabetically by (A) to (H). As for design B to G, each repeated six times of experiments by randomly splitting noisy cases for deep model training, validation and test, and results were reported on average. Thus, this study contained 8 designs and 38 times of experiments in total.
Figure 3.
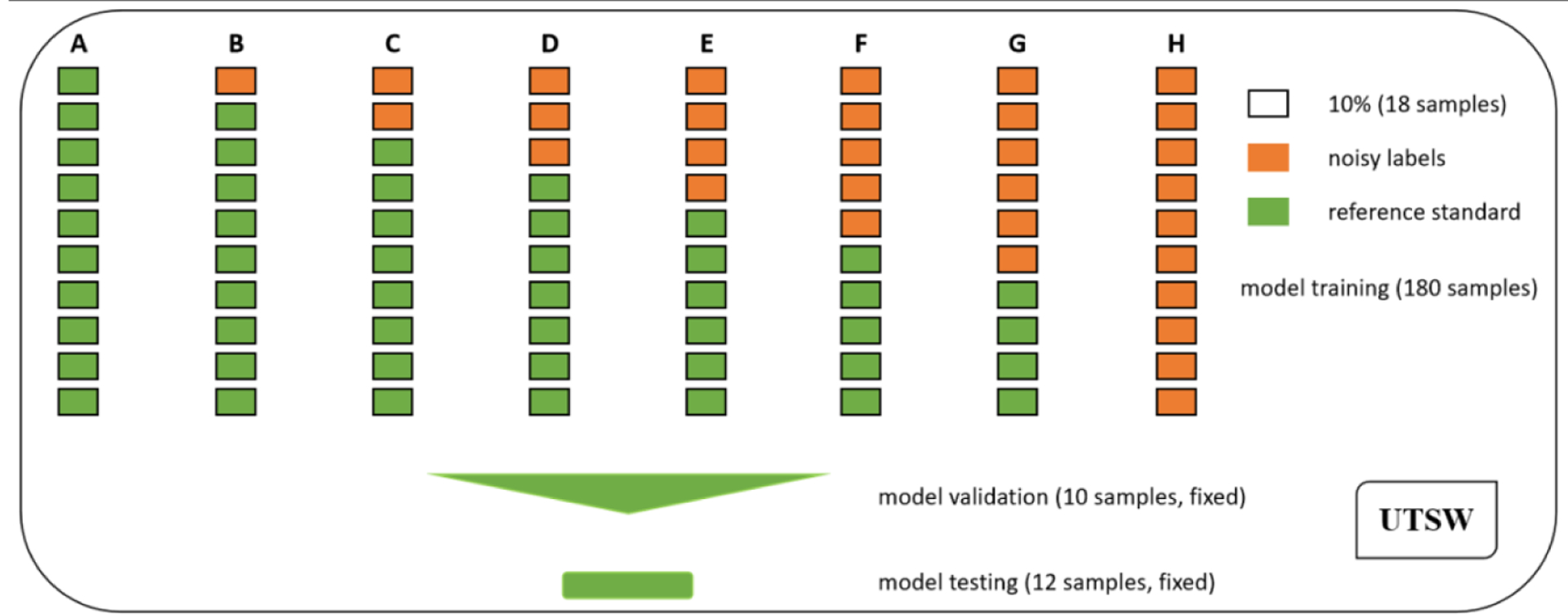
Experimental design with various ratios of noisy cases for mandible segmentation. Eight designs were conducted, and the ratios included 0% (A), 10% (B), 20% (C), 30% (D), 40% (E), 50% (F), 60% (G) and 100% (H). Moreover, each design of B to G was repeated six times based on random splitting of noisy samples for model training, validation and test.
2.5. A deep neural network and parameter settings
A compact deep network HighRes3DNet was used, which utilizes efficient and flexible elements, such as dilation convolution and residual connection, for volumetric image segmentation (Li et al 2017). It has been applied for brain parcellation and hyperintensity isolation (Li et al 2017, Kuijf et al 2019). In this study, parameters were set as follows. Both the input and output image size were [256, 256, 128], and the output images were in binary values. The Adam optimizer was used for hyper-parameter optimization (Kingma and Ba 2014). Binary cross entropy was set as the loss function, and ReLu as the activation function. The learning rate was 10−4, the number of iterations was 105, and the batch size is 1. For each CT image, 64 patches of size [96, 96, 96] were uniformly generated. Other parameters were set as default with random initialization. The model was validated per 103 iterations at the training stage, and 102 checkpoints were saved after the training. Neither fine-tuning nor data augmentation was used.
2.6. Model training, validation and selection
Figure 4 shows an example of model training (blue line) and validation (red circles) based on 100% noisy cases. In the training stage, the network inferred the data samples in the validation set per 103 iterations, and thus, 102 check points (red circles) of loss values were obtained after the training procedure. To select the best trained model, the validation losses were compared, and the check point with the least loss value indicated the model at this point was optimized. The selected model was used for follow-up mandible segmentation. In this example, the model validated at 9.6 × 104 iterations achieved the least loss value (8.0 × 10−5), and it was taken as well-trained.
Figure 4.
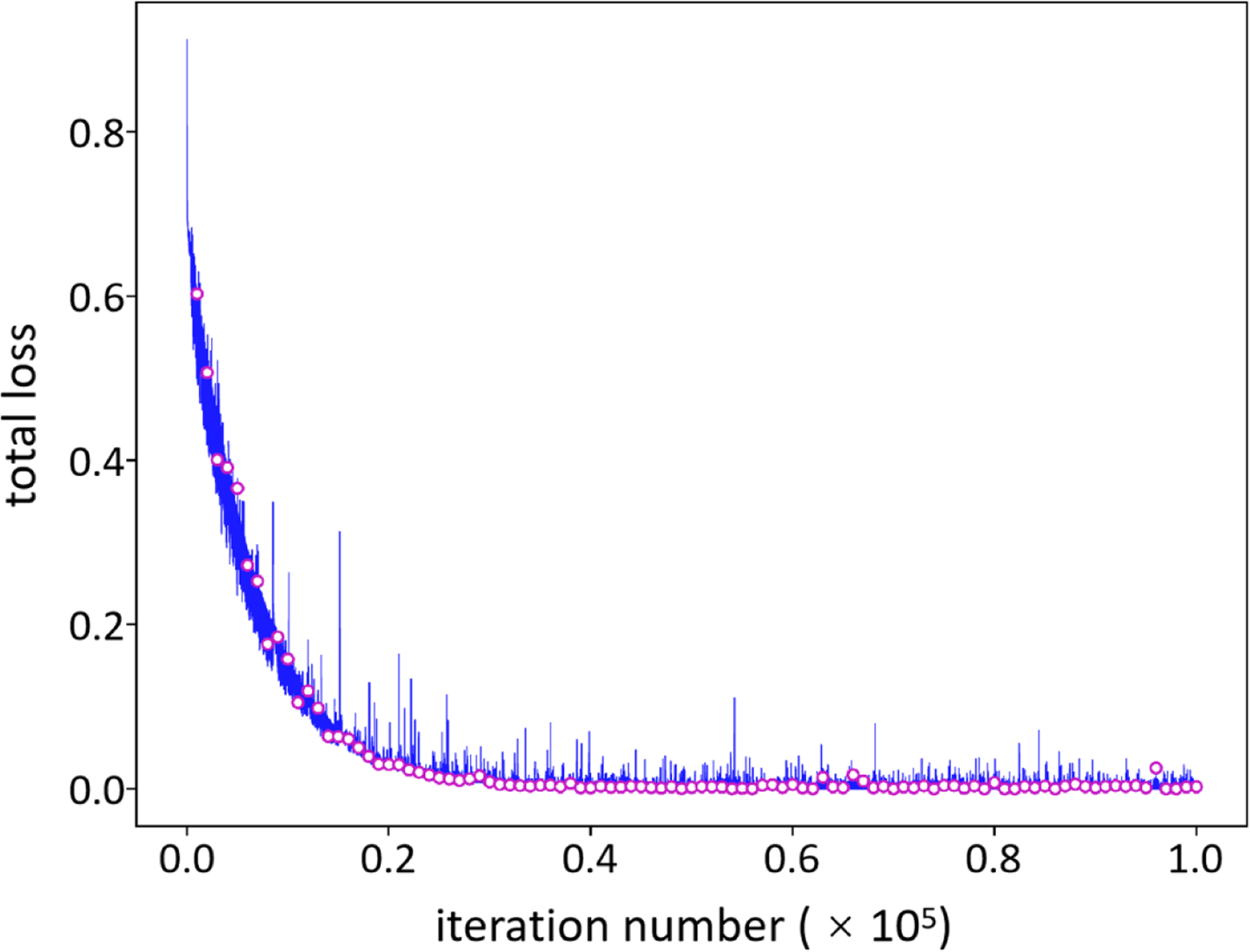
Model training, validation and selection. A model under training will be validated per 103 iterations which results in 102 check points (red circles). To select an optimal model, the validation loss is calculated and the check point with the least loss value is selected as the optimized model for follow-up mandible segmentation from CT images.
2.7. Performance evaluation
The segmentation performance was evaluated by using Dice similarity coefficient (DSC), percent volume error (PVE), 95% percentile Hausdorff distance (HD95) and contour mean distance (CMD) (Chen et al 2015, Taha and Hanbury 2015, Raudaschl et al 2017, Nikolov et al 2018). Given a reference annotation A, a segmented result B and their corresponding boundary point sets as X = {x1,x2,…,xm} and Y = {y1,y2,…,yn}, DSC is defined as and PVE is defined as where | · |stands for the number of voxels enclosed by contours. DSC values range in [0, 1] and larger values indicate better agreement between reference standard and segmentation results. PVE ranges from 0 to positive infinity, and 0 represents the best. Moreover, taking ||x − y||as the Euclidean distance of two points xand y, HD is defined as max(h(X,Y),h(Y,X))where and CMD is defined as max(d(X,Y),d(Y,X))where . Note that the operations of maxand minstand for getting the maximum and the minimum value, respectively. HD95 and CMD are reported since they are robust to outliers and noise on overall evaluation of segmentation quality, and a smaller value denotes a better performance.
2.8. Software and platform
Experiments were conducted on a 64-bit Windows 10 workstation with 8 Intel (R) Xeon (R) processors (3.60 GHz), 64.0 GB RAM, and a NVIDIA GeForce RTX 2080 Ti GPU card. HighRes3DNet was implemented in NiftyNet (Gibson et al 2018) (version 0.5.0, https://niftynet.io/), a deep learning toolbox dedicated for medical image analysis based on Tensorflow (version 1.13.2, https://www.tensorflow.org/).
3. Results
3.1. Annotation quality of dosimetrists
The mandible annotation of 12 dosimetrists showed inter-rater disagreement, such as the inclusion of teeth regions. Mandible is one of OARs delineated with less care, and the delineation is incomplete or over-segmented (figure 2). The initial annotation quality, however, was still acceptable since most voxels were kept in the correction procedure (figure 1). Given the reference standard, figure 5 shows the initial quality of mandible annotation (DSC, 0.86 ± 0.09; PVE, 0.34 ± 0.23; HD95, 7.84 ± 13.26 mm; and CMD, 1.96 ± 4.73 mm). It is found 20 cases with DSC < 0.75, 40 cases with PVE > 0.50, 6 cases with HD95 > 16.00 mm, and 8 cases with CMD > 4.00 mm.
Figure 5.
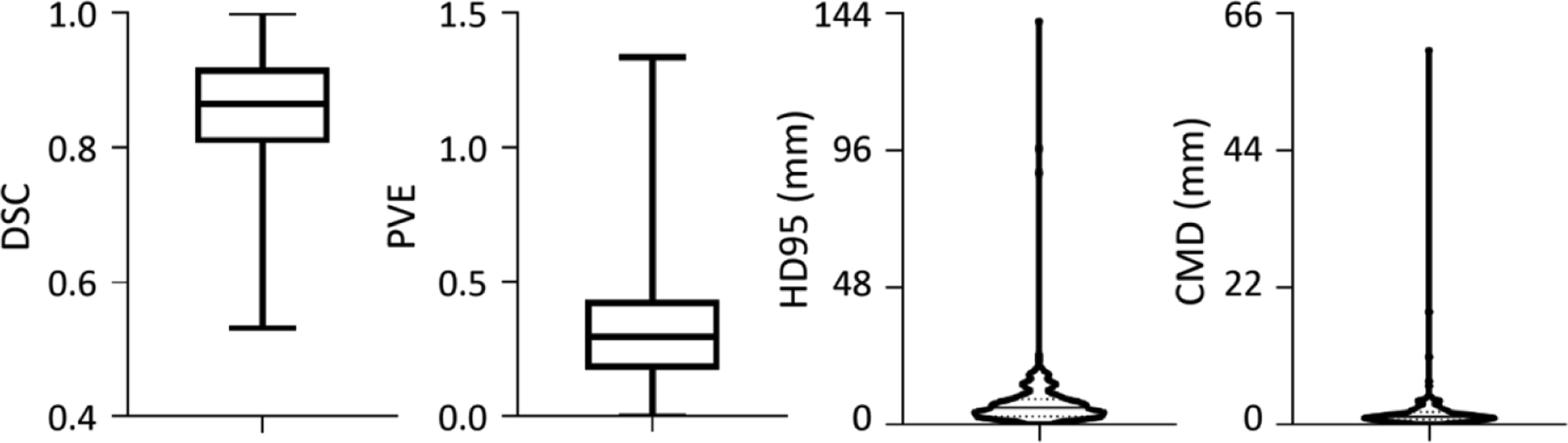
Initial quality of mandible annotation. Given the reference standard, the annotation quality is evaluated from DSC, PVE, HD95 and CMD metrics. The box and whiskers plots show the max, median and min values, and the violin plots shows the median and quartile.
3.2. Data quality for model training
Table 2 shows the data quality of the training set in each design. It is observed that when the number of noise cases increases, the data quality is degraded. On average, there is 0.15 decrease of the DSC and 0.34 increase of the PVE, and the distance of boundary points increase to 6.76 mm of the HD95 and to 1.82 mm of the CMD.
Table 2.
Annotation quality of the UTSW training set.
| DSC | PVE | HD95 (mm) | CMD (mm) | |
|---|---|---|---|---|
| A (0%) | 1.00 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 |
| B (10%) | 0.99 ± 0.05 | 0.03 ±0.12 | 0.48 ± 1.78 | 0.16 ±0.46 |
| C (20%) | 0.97 ± 0.07 | 0.07 ±0.18 | 1.38 ±4.65 | 0.31 ± 0.94 |
| D (30%) | 0.96 ± 0.08 | 0.11 ±0.21 | 2.29 ± 7.87 | 0.56 ± 2.38 |
| E (40%) | 0.94 ± 0.09 | 0.14 ±0.22 | 2.37 ± 9.70 | 0.80 ±3.15 |
| F (50%) | 0.93 ±0.10 | 0.17 ± 0.24 | 4.25 ± 10.54 | 0.94 ± 3.52 |
| G (60%) | 0.91 ±0.10 | 0.20 ± 0.25 | 5.54 ±11.37 | 1.17 ±3.76 |
| H (100%) | 0.85 ± 0.09 | 0.34 ± 0.24 | 6.76 ± 12.78 | 1.82 ± 4.48 |
3.3. Performance evaluation on the UTSW test set
The performance of selected models on the UTSW test set is shown in table 3 and the best result of each metric is boldfaced. Given the model (A) trained with the reference standard as the baseline, deep networks trained with noisy label images achieve slightly inferior results. When the percentage of noisy label images increases to 100%, each design still achieves competitive performance on the DSC (< 0.10 decrease), the PVE (< 0.20 increase), the HD95 (< 7.00 mm increase) and the CMD (< 1.70 mm increase). It should be highlighted that, when using 20% or 10% noisy cases for model training, no significant decrease of segmentation performance is found between the model trained with reference standard and the models trained with noisy cases on each metric (two sample t-test, p > 0.11).
Table 3.
Performance evaluation on the UTSW test set.
| DSC | PVE | HD95 (mm) | CMD (mm) | |
|---|---|---|---|---|
| A (0%) | 0.90 ± 0.07 | 0.21 ± 0.18 | 5.65 ± 7.38 | 1.46 ± 1.98 |
| B (1O%) | 0.89 ± 0.07 | 0.22 ±0.15 | 6.41 ± 7.02 | 1.64 ± 1.89 |
| C (20%) | 0.87 ± 0.09 | 0.25 ±0.15 | 7.14 ±6.58 | 1.82 ± 1.66 |
| D (30%) | 0.84 ±0.12 | 0.30 ± 0.22 | 9.65 ± 10.24 | 2.61 ± 2.74 |
| E (40%) | 0.85 ± 0.09 | 0.29 ±0.18 | 9.67 ±8.19 | 2.34 ± 2.09 |
| F (50%) | 0.87 ± 0.07 | 0.27 ±0.18 | 6.93 ± 6.32 | 1.89 ± 1.67 |
| G (60%) | 0.83 ±0.10 | 0.35 ± 0.20 | 12.53 ± 16.29 | 3.13 ±3.56 |
| H (100%) | 0.82 ± 0.07 | 0.40 ± 0.20 | 7.63 ± 5.08 | 1.91 ± 3.47 |
3.4. Performance of cross-dataset validation
The performance of cross-dataset validation test was summarized and the best result of each metric is boldfaced. Table 4 shows the segmentation results on the TCIA dataset. It indicates that the model (A) trained with reference annotation on the UTSW dataset achieves superior results with the highest DSC and the lowest PVE and HD95, and other models trained with noisy data obtain close results. Taking the model A as the baseline, some designs achieve rivaling performance on the DSC (design B, E, F, G and H with < 0.06 decrease), the PVE (design B, E, F, G and H with < 0.10 increase), the HD95 (design B, C, E, F and H with < 4.00 mm increase) and even better performance on the CMD (design B, C, F and H).
Table 4.
Performance of cross-dataset test on the TCIA dataset.
| DSC | PVE | HD95 (mm) | CMD (mm) | |
|---|---|---|---|---|
| A (0%) | 0.90 ± 0.02 | 0.20 ± 0.05 | 3.51 ± 1.42 | 2.02 ± 2.92 |
| B (10%) | 0.85 ± 0.05 | 0.28 ± 0.09 | 5.38 ±12.38 | 1.48 ± 1.44 |
| C (20%) | 0.82 ± 0.07 | 0.32 ±0.11 | 5.94 ± 3.24 | 1.80 ± 1.06 |
| D (30%) | 0.82 ±0.12 | 0.32 ±0.15 | 12.87 ± 40.00 | 2.99 ± 4.90 |
| E (40%) | 0.85 ± 0.08 | 0.28 ±0.11 | 7.22 ± 8.46 | 2.11 ± 1.91 |
| F (50%) | 0.86 ± 0.04 | 0.27 ± 0.08 | 7.16 ± 16.79 | 1.81 ± 2.04 |
| G (60%) | 0.85 ±0.10 | 0.28 ±0.14 | 11.30 ±23.63 | 2.66 ± 3.22 |
| H (100%) | 0.85 ± 0.06 | 0.28 ±0.10 | 4.70 ± 2.32 | 1.37 ± 0.60 |
Table 5 shows the results of cross-dataset evaluation on the PDDCA dataset. It is found that the model A trained with UTSW reference annotation achieves the best performance with the highest DSC and the lowest PVE, and the second best HD95 and CMD, and other models trained with noisy cases get competitive results. Notably, design F using 50% noisy cases of the training set achieves the second best DSC and PVE, the third best HD95, and the fourth best CMD.
Table 5.
Performance of cross-dataset test on the PDDCA dataset.
| DSC | PVE | HD95 (mm) | CMD (mm) | |
|---|---|---|---|---|
| A (0%) | 0.87 ± 0.04 | 0.28 ± 0.08 | 4.79 ± 1.69 | 1.78 ± 0.95 |
| B (1O%) | 0.82 ± 0.05 | 0.35 ± 0.08 | 4.63 ± 1.66 | 1.49 ± 0.50 |
| C (20%) | 0.80 ± 0.07 | 0.39 ±0.11 | 6.32 ± 5.28 | 2.02 ± 1.24 |
| D (30%) | 0.80 ±0.10 | 0.39 ±0.13 | 15.79 ±40.25 | 3.74 ± 7.38 |
| E (40%) | 0.83 ± 0.05 | 0.35 ± 0.08 | 7.22 ± 7.87 | 2.28 ± 1.72 |
| F (50%) | 0.84 ± 0.04 | 0.34 ± 0.09 | 5.92 ± 13.21 | 2.12 ±1.63 |
| G (60%) | 0.82 ± 0.09 | 0.35 ±0.12 | 10.36 ± 22.06 | 2.66 ± 2.94 |
| H (100%) | 0.82 ± 0.05 | 0.37 ± 0.08 | 21.70 ±64.88 | 4.74 ± 9.81 |
Figure 6 demonstrates the results of mandible segmentation from CT images. The models are trained and tested on the UTSW dataset, and further evaluated on the TCIA and the PDDCA dataset. From top to bottom rows are the metrics of DSC, PVE, HD95 and CMD, and from left to right columns are the results on the UTSW test set, and the TCIA and the PDDCA dataset. The left column indicates that the model A trained with the UTSW reference standard outperforms other models (design B to H) trained with noisy labels, meanwhile the model A achieves overall best results on cross-dataset evaluation. Moreover, from the first column, a general trend is observed on each metric values that increasing the number of noisy training samples leads to the decrease of segmentation performance on the UTSW dataset. Meanwhile, this decreasing trend holds but not strictly, in particular when the number of noisy cases reaches 40% and 50%, in the intra- and inter-dataset evaluation.
Figure 6.
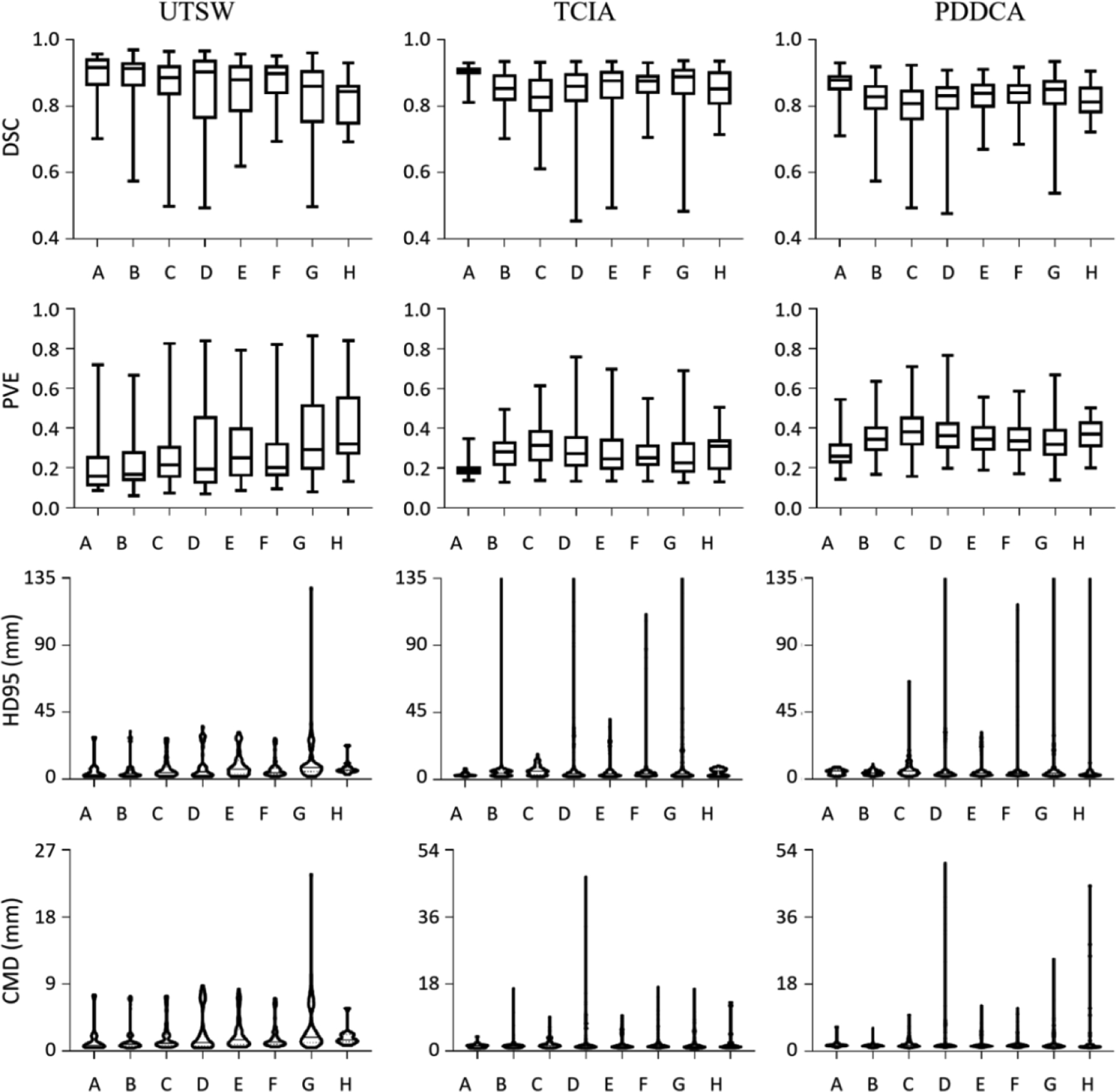
Experimental results on deep learning based mandible segmentation. The models are trained and tested on the UTSW dataset and further evaluated on two other datasets. The metrics of DSC, PVE, HD95 and CMD are shown from top to bottom rows, and the results on the dataset UTSW, TCIA and PDDCA are shown from left to right columns, correspondingly.
3.5. Performance of selected models on the training samples
The performance of selected models on the UTSW training set is additionally evaluated. The results are shown in tables 6 and 7, where the training labels and the reference standard, respectively, perform as the ground truth for quantitative assessment. Table 6 indicates that fewer noisy cases correspond to better model training. When the ratio of noisy cases increases to 100%, it causes up the most to 0.10 decrease on the DSC, 0.17 increase on the PVE, 7.83 mm increase on the HD95, and 2.08 mm increase on the CMD.
Table 6.
Evaluation of selected models on the UTSW training set by using the training labels as the ground truth to quantify the segmentation performance.
| DSC | PVE | HD95 (mm) | CMD (mm) | |
|---|---|---|---|---|
| A (0%) | 0.92 ± 0.05 | 0.16 ±0.09 | 4.00 ± 6.05 | 1.10 ± 1.08 |
| B (10%) | 0.90 ± 0.06 | 0.20 ±0.11 | 4.72 ±6.16 | 1.21 ± 0.99 |
| C (20%) | 0.86 ± 0.09 | 0.25 ±0.13 | 6.85 ± 8.44 | 1.67 ± 1.62 |
| D (30%) | 0.84 ±0.12 | 0.30 ± 0.20 | 11.18 ± 17.68 | 2.75 ± 3.61 |
| E (40%) | 0.84 ±0.10 | 0.32 ± 0.24 | 11.23 ± 15.35 | 2.73 ± 3.96 |
| F (50%) | 0.86 ± 0.08 | 0.28 ± 0.22 | 9.40 ± 17.32 | 2.35 ± 4.04 |
| G (60%) | 0.82 ±0.12 | 0.33 ± 0.25 | 11.83 ± 16.60 | 3.18 ±4.83 |
| H (100%) | 0.85 ± 0.09 | 0.29 ± 0.26 | 7.72 ± 12.31 | 2.36 ± 4.33 |
Table 7.
Evaluation of selected models on the UTSW training set when using the reference standard as the ground truth to quantify the performance metrics.
| DSC | PVE | HD95 (mm) | CMD (mm) | |
|---|---|---|---|---|
| A (0%) | 0.92 ± 0.05 | 0.16 ± 0.09 | 3.96 ± 6.08 | 1.03 ± 1.01 |
| B (10%) | 0.90 ± 0.06 | 0.18 ± 0.09 | 4.41 ±6.15 | 1.12 ±0.94 |
| C (20%) | 0.88 ± 0.08 | 0.23 ±0.11 | 6.59 ± 9.26 | 1.53 ± 1.68 |
| D (30%) | 0.85 ±0.12 | 0.28 ±0.18 | 10.74 ± 17.32 | 2.52 ± 3.02 |
| E (40%) | 0.85 ± 0.09 | 0.31 ±0.17 | 10.86 ± 13.57 | 2.45 ± 2.50 |
| F (50%) | 0.88 ± 0.06 | 0.25 ± 0.12 | 8.17 ± 15.36 | 1.84 ± 2.47 |
| G (60%) | 0.83 ±0.10 | 0.32 ± 0.16 | 11.65 ± 14.67 | 2.80 ± 3.34 |
| H (100%) | 0.82 ± 0.08 | 0.37 ±0.16 | 7.78 ± 8.73 | 1.89 ± 1.78 |
Table 7 indicates that increasing the number of noisy cases leads to worse segmentation results. When the ratio of noisy cases reaches 100%, the metric values get worse of 0.10, 0.21, 7.69 mm and 1.77 mm on the metrics DSC, PVE, HD95 and CMD on average, respectively.
It is worth noting that the difference is quite limited by comparing the metric values of each design in tables 6 and 7. The maximum difference is 0.03 on the metric DSC from design H, 0.08 on the metric PVE from design H, 1.23 mm on the metric HD95 from design F, and 0.47 mm on the metric CMD from design H.
3.6. Computation time
Based on the software and platform, it took about 0.89 s per iteration, 24.72 h to complete one training procedure, and 5.83 s to fulfill the test of one volume.
4. Discussion
This study concerns deep learning with noisy annotation and explores mandible segmentation from CT images for radiation treatment planning. It demonstrates the segmentation performance of deep network HighRes3DNet trained with different numbers of noisy cases, suggesting that the network is robust to noisy annotation to some extent. These trained models are further evaluated on two unseen datasets, indicating the models trained with noisy cases achieve competitive performance as the model trained with reference standard. The performance of mandible segmentation at the training stage was additionally compared by using noisy labels and reference annotation as the ground truth, and it shows that the entropy-based training loss is a good driver for the segmentation task regardless of the training data quality. It also suggests that the prediction model may improve with bootstrap ensemble approaches, since training with reference annotation results in most consistent high-quality performance.
4.1. The robustness of deep learning to noisy annotation
Experimental results indicate that the involved deep network is robust to noisy annotation to some extent. Based on noisy cases in design H (table 2), this study shows that the selected model was well-trained (table 7) and obtained good segmentation on the test set (table 3). In particular, there was no significant difference in segmentation metrics on the test set between the model trained with reference standard and the model trained with 10% or 20% noisy cases (table 3, two sample t-test, p > 0.11). A similar phenomenon has been observed in image classification. Van Horn et al claimed that if the training set is sufficiently large, a small label error rate of training data led to an acceptable small increase of prediction error in the test set (Van Horn et al 2015). Rolnick et al observed that deep learning was robust to label noise, and they figured out that a sufficient training set can accommodate a wide range of noise levels (Rolnick et al 2017). This kind of robustness to label noise can be explained from the capacity of deep networks on continual representation learning. Rolnick et al also pointed out that a deep architecture tends to learn the intrinsic pattern of objects instead of merely memorizing noise (Rolnick et al 2017). Arpit et al conducted close examinations at the memorization of deep networks with regard to the model capacity, generalization, and adversarial robustness (Arpit et al 2017). They showed that deep networks preferred to prioritize learning simple and general patterns before fitting the noise.
It should be noted that there is a general trend observed from each metric that more noisy training samples lead to worse segmentation results (tables 3–5), while some designs using more noisy cases still achieve competitive performance to those designs based on less noisy cases. For instance, design F using 50% noisy cases obtained slightly superior results on the DSC and the PVE metrics than design B that used 20% noisy cases for model training (tables 4 and 5). Two reasons could account for this finding. First, in each design, six times of random selection of noisy samples for model training are not sufficient, which is hard to reflect the intrinsic distribution of label noise. Second, the metrics DSC and PVE denote part of the segmentation quality, but not the overall accuracy. In spite of the slight randomness, the other metrics HD95 and CMD show that design F obtained worse performance than design B (tables 3–5). Thus, the general decreasing trend is reasonable and interpretable.
It should be also noted that while the performance degrades on the UTSW test set with more noisy labels (table 3), this is not that obvious with external datasets (tables 4 and 5), especially with the DSC metrics. As shown in tables 4 and 5, it is surprising that design C shows significantly worse DSC than design A and even worse than many other ones with more noisy samples. It might make sense with an external dataset, which has different data distribution and ground truth definition so that segmentation errors may show up randomly. The exact reason needs further investigation.
4.2. The importance of annotation quality in deep learning
The importance of labeling quality has been highlighted in previous studies, and this study further emphasizes this point. Given the annotation quality of dosimetrists (table 2), experimental results indicate that the selected models trained with 100% noisy cases (design H) achieved worse performance than the annotation quality of dosimetrists (table 3). Even in the training stage, the selected model obtained inferior results (table 7) than the initial annotation. On the contrary, when using reference standard for model training (design A), models reached superior performance on the training set (table 7) and the test set (table 3). The cross-validation test on the TCIA and the PDDCA dataset (tables 4 and 5) further verifies that a model trained with reference standard could generalize better on unseen datasets. Interestingly, when comparing each design on the UTSW training set (tables 6 and 7), the limited difference on segmentation performance implicitly suggests that when inconsistent labels exists in the training data, the deep network would try to get an average or balance model, which might be the reason why the trained models could achieve close metric values either using noisy labels or reference annotation as the ground truth.
A deep network is a computational model. It consists of multiple levels of abstraction for representation learning. Through an iterative process of forward pass and back propagation, a deep network attempts to learn an intricate structure from a large number of data samples, and its internal hyper-parameters are dynamically updated toward an optimal solution (Lecun et al 2015, Shen et al 2020). To deep supervised learning, any noisy annotation is bound to mislead the iterative process and subsequently, the learned structure may not be representative, and the determined parameters may not be optimal. Thus, to provide sufficient high-quality data samples is essential for deep learning based image segmentation.
4.3. Future work on deep learning with noisy annotation
In radiation oncology, it is possible to provide a small number of exhaustive-annotated samples and thus, how to utilize such limited samples to address the issue of deep learning with noisy annotation becomes an urgent problem.
Several studies for image classification have provided clues on this topic (Hendrycks et al 2018, Tanaka et al 2018, Han et al 2019, Tajbakhsh et al 2020). One way is to implicitly estimate the possibility of training samples with reference labels. Reed et al proposed a coherent bootstrapping model to evaluate the consistency between given labels and its predicted labels in the training stage, and both labels contributed to the resultant prediction in a convex combination (Reed et al 2014). Han et al trained two deep networks simultaneously, and the networks learned from each other and mutually exchanged probable reference labels to reduce the error flows (Han et al 2018). As such, each network could attenuate different types of labeling errors and lead to better performance. The other way is to explicitly guide the model training with reference labels. Given reference samples in the validation set, Ren et al designed an online meta-learning algorithm, which updated the weights of training examples using a gradient descent step on the current training example weights to minimize the loss on the validation set (Ren et al 2018). Mirikharaji et al developed an adaptive reweighting approach and commensurately treated both reference and noisy labels in the loss function (Mirikharaji et al 2019). They deployed a meta-learning approach to assign higher importance to pixels whose loss gradient direction was closer to those of reference data. Additionally, utilizing a noise-robust loss function could further improve the training effectiveness (Zhang and Sabuncu 2018).
The optimization of deep networks is content-aware and the purpose is to retrieve patterns shared by training samples (Lecun et al 2015). And thus, building multiple atlases for a specific application potentially improves the representation learning performance as reported in the literature. Ma et al utilized deep learning to localize the prostate region and to distinguish prostate pixels from the surround tissues (Ma et al 2017), and used similar atlases to refine the segmentation results. Zhu et al proposed a hybrid framework for the fusion of predicted hippocampus regions (Zhu et al 2020). The framework first used atlases to estimate the deformation of image labels, and then a fully convolutional network was designed to learn the relationship between pairs of image patches to correct the potential errors. Finally, both multi-atlas image segmentation and the fully convolutional network were used for label fusion. Vakalopoulou et al introduced a multi-network architecture to exploit domain knowledge (Vakalopoulou et al 2018). After co-aligning multiple anatomies through multi-metric non-rigid registration, each network performed CT image segmentation for interstitial lung disease in the atlas space. At last, segmentation results were fused in the source data space. Ding et al presented a deep learning based label fusion strategy (Ding et al 2019). It attempted to locally select a set of reliable atlases by deep learning, and finally, estimated labels were fused via plurality voting.
Deep learning with noisy annotation is challenging but clinically important (Rozario et al 2017, Min et al 2019, Sahiner et al 2019, Yang et al 2019). In this study, the deep network shows certain robustness to noisy annotation, while for further improvement, it should take advantage of reference samples either implicitly or explicitly to avoid the misleading of noisy annotation in the training stage. In general, multiple atlases provide diverse but representative contextures, and multi-atlas image segmentation provides good spatial consistency via deformable segmentation, both of which might contribute to the development of robust deep networks and may be generalizable to noisy annotation.
4.4. Limitations of the current study
There are several limitations in the current study. At first, the inter- and intra-reader variability of clinicians on mandible annotation were not explored. This study involved one specialist for mandible annotation as the reference standard. It may cause the model training biased toward that specific reference standard and thus, multiple clinicians should be involved in the preparation of reference annotation. Second, due to limited computing resources, the number of experiments in each design is not enough and more experiments should be conducted to represent the distribution of annotation noise. Third, the number of cases for model testing is small, and in the future studies, more patient cases should be collected. Then, the annotation quality can be directly stratified through metric values, such as DSC values, and the effect of noisy annotation on deep learning could be analyzed from other perspectives. Furthermore, other networks besides the one explored in this study, such as U-Net (Ronneberger et al 2015) and V-Net (Milletari et al 2016), or other techniques, such as data augmentation (Yu et al 2019) and the Dice loss function (Milletari et al 2016, Sudre et al 2017), may be applied and may make a great effect on the segmentation performance. Thus, in the future, massive experiments should be systematically conducted and the effect of noisy annotation on deep learning could be deeply understood.
5. Conclusions
This study concerns deep learning with noisy annotation in medical image segmentation. It shows that the involved deep network is robust to noisy annotation to some extent in mandible segmentation from CT images. In general, a deep network trained with noisy labels is inferior to that trained with reference annotation. Thus, how to maximize limited reference standard samples to improve the performance of deep learning with noisy annotation needs further investigation.
Acknowledgments
This work was partially supported by NIH R01 CA218402 and R01 CA235723.
References
- Arpit D, Jastrzębski S, Ballas N, Krueger D, Bengio E, Kanwal MS, Maharaj T, Fischer A, Courville A and Bengio Y 2017. A closer look at memorization in deep networks Proc. 34th Int. Conf. on Machine Learning pp 233–42 [Google Scholar]
- Chen H, Lu W, Chen M, Zhou L, Timmerman R, Tu D, Nedzi L, Wardak Z, Jiang S and Zhen X 2019. A recursive ensemble organ segmentation (REOS) framework: application in brain radiotherapy Phys. Med. Biol 64 025015. [DOI] [PubMed] [Google Scholar]
- Chen H, Zhen X, Gu X, Yan H, Cervino L, Xiao Y and Zhou L 2015. SPARSE: Seed Point Auto-Generation for Random Walks Segmentation Enhancement in medical inhomogeneous targets delineation of morphological MR and CT images J. Appl. Clin. Med. Phys 16 387–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark K et al. 2013. The Cancer Imaging Archive (TCIA): maintaining and operating a public information repository J. Digit. Imaging 26 1045–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Z, Han X and Niethammer M 2019. VoteNet: a deep learning label fusion method for multi-atlas segmentation Medical Image Computing and Computer Assisted Intervention – MICCAI 2019. Lecture Notes in Computer Science vol 11766, ed Shen D et al. (Berlin: Springer; ) pp 202–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson E, Li W, Sudre C, Fidon L, Shakir DI, Wang G, Eaton-Rosen Z, Gray R, Doel T and Hu Y 2018. NiftyNet: a deep-learning platform for medical imaging Comput. Methods Programs Biomed 158 113–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenspan H, Van Ginneken B and Summers RM 2016. Guest editorial deep learning in medical imaging: overview and future promise of an exciting new technique IEEE Trans. Med. Imaging 35 1153–9 [Google Scholar]
- Han B, Yao Q, Yu X, Niu G, Xu M, Hu W, Tsang I and Sugiyama M 2018. Co-teaching: robust training of deep neural networks with extremely noisy labels NIPS’18: Proc. 32nd Int. Conf. on Neural Information Processing Systems (Red Hook, NY: Curran Associates Inc; ) pp 8527–37 [Google Scholar]
- Han J, Luo P and Wang X 2019. Deep self-learning from noisy labels 2019 IEEE/CVF International Conference on Computer Vision (ICCV) (Piscataway, NJ: IEEE) pp 5138–47 [Google Scholar]
- Hendrycks D, Mazeika M, Wilson D and Gimpel K 2018. Using trusted data to train deep networks on labels corrupted by severe noise NIPS’18: Proc. 32nd Int. Conf. on Neural Information Processing Systems (Red Hook, NY: Curran Associates Inc; ) pp 10456–65 [Google Scholar]
- Hesamian MH, Jia W, He X and Kennedy P 2019. Deep learning techniques for medical image segmentation: achievements and challenges J. Digit. Imaging 32 582–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingma DP and Ba J 2014. Adam: a method for stochastic optimization (arXiv: 1412.6980) [Google Scholar]
- Kuijf HJ, Biesbroek JM, De Bresser J, Heinen R, Andermatt S, Bento M, Berseth M, Belyaev M, Cardoso MJ and Casamitjana A 2019. Standardized assessment of automatic segmentation of white matter hyperintensities and results of the WMH segmentation challenge IEEE Trans. Med. Imaging 38 2556–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecun Y, Bengio Y and Hinton G 2015. Deep learning Nature 521 436–44 [DOI] [PubMed] [Google Scholar]
- Li W, Wang G, Fidon L, Ourselin S, Cardoso MJ and Vercauteren T 2017. On the compactness, efficiency, and representation of 3D convolutional networks: brain parcellation as a pretext task Information Processing in Medical Imaging. IPMI 2017. Lecture Notes in Computer Science vol 10265, ed Niethammer M et al. (Berlin: Springer; ) pp 348–60 [Google Scholar]
- Liu Y, Stojadinovic S, Hrycushko B, Wardak Z, Lau S, Lu W, Yan Y, Jiang SB, Zhen X and Timmerman R 2017. A deep convolutional neural network-based automatic delineation strategy for multiple brain metastases stereotactic radiosurgery PLoS ONE 12 e0185844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y, Zhu J and Pfister T 2019. A simple yet effective baseline for robust deep learning with noisy labels (arXiv: 1909.09338) [Google Scholar]
- Ma L, Guo R, Zhang G, Tade F, Schuster DM, Nieh P, Master V and Fei B 2017. Automatic segmentation of the prostate on CT images using deep learning and multi-atlas fusion Proc. SPIE 10133 101332O. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milletari F, Navab N and Ahmadi SA 2016. V-net: fully convolutional neural networks for volumetric medical image segmentation 2016 Fourth Int. Conf. on 3D Vision (Piscataway, NJ: IEEE; ) pp 565–71 [Google Scholar]
- Min S, Chen X, Zha Z-J, Wu F and Zhang Y 2019. A two-stream mutual attention network for semi-supervised biomedical segmentation with noisy labels Proc. AAAI Conf. on Artificial Intelligence vol 33 (Palo Alto, CA: AAAI; ) pp 4578–85 [Google Scholar]
- Mirikharaji Z, Yan Y and Hamarneh G 2019. Learning to segment skin lesions from noisy annotations Domain Adaptation and Representation Transfer and Medical Image Learning with Less Labels and Imperfect Data. DART 2019, MIL3ID 2019. Lecture Notes in Computer Science vol 11795 ed Wang Q et al. (Berlin: Springer; ) pp 207–15 [Google Scholar]
- Misra I, Lawrence Zitnick C, Mitchell M and Girshick R 2016. Seeing through the human reporting bias: visual classifiers from noisy human-centric labels 2016 IEEE Conf. on Computer Vision and Pattern Recognition (CVPR) (Piscataway, NJ: IEEE; ) pp 2930–9 [Google Scholar]
- Nikolov S. et al. Deep learning to achieve clinically applicable segmentation of head and neck anatomy for radiotherapy (arXiv: 1809.04430) 2018 doi: 10.2196/26151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrini G, Rozza A, Krishna Menon A, Nock R and Qu L. 2017. Making deep neural networks robust to label noise: a loss correction approach 2017 IEEE Conf. on Computer Vision and Pattern Recognition (Piscataway, NJ: IEEE; ) pp 1944–52 [Google Scholar]
- Raudaschl PF et al. 2017. Evaluation of segmentation methods on head and neck CT: auto-segmentation challenge 2015 Med. Phys 44 2020–36 [DOI] [PubMed] [Google Scholar]
- Reed S, Lee H, Anguelov D, Szegedy C, Erhan D and Rabinovich A 2014. Training deep neural networks on noisy labels with bootstrapping (arXiv: 1412.6596) [Google Scholar]
- Ren M, Zeng W, Yang B and Urtasun R 2018. Learning to reweight examples for robust deep learning (arXiv: 1803.09050) [Google Scholar]
- Rolnick D, Veit A, Belongie S and Shavit N 2017. Deep learning is robust to massive label noise (arXiv: 1705.10694) [Google Scholar]
- Ronneberger O, Fischer P and Brox T 2015. U-net: convolutional networks for biomedical image segmentation Medical Image Computing and Computer-Assisted Intervention – MICCAI 2015. Lecture Notes in Computer Science vol 9351, ed Navab N, Hornegger J, Wells Wand Frangi A(Berlin: Springer; ) pp 234–41 [Google Scholar]
- Rozario T, Long T, Chen M, Lu W and Jiang S 2017. Towards automated patient data cleaning using deep learning: A feasibility study on the standardization of organ labeling (arXiv: 1801.00096) [Google Scholar]
- Sahiner B, Pezeshk A, Hadjiiski LM, Wang X, Drukker K, Cha KH, Summers RM and Giger ML 2019. Deep learning in medical imaging and radiation therapy Med. Phys 46 e1–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santanam L, Hurkmans C, Mutic S, van Vliet-vroegindeweij C, Brame S, Straube W, Galvin J, Tripuraneni P, Michalski J and Bosch W 2012. Standardizing naming conventions in radiation oncology Int. J. Radiat. Oncol. Biol. Phys 83 1344–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen C, Nguyen D, Zhou Z, Jiang SB, Dong B and Jia X 2020. An introduction to deep learning in medical physics: advantages, potential, and challenges Phys. Med. Biol 65 05TR01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudre CH, Li W, Vercauteren T, Ourselin S and Cardoso MJ 2017. Generalised Dice overlap as a deep learning loss function for highly unbalanced segmentations Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support. DLMIA 2017, ML-CDS 2017. Lecture Notes in Computer Science vol 10553, ed Cardoso M et al. (Berlin: Springer; ) pp 240–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taha AA and Hanbury A 2015. Metrics for evaluating 3D medical image segmentation: analysis, selection, and tool BMC Med. Imaging 15 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajbakhsh N, Jeyaseelan L, Li Q, Chiang JN, Wu Z and Ding X 2020. Embracing imperfect datasets: a review of deep learning solutions for medical image segmentation Med. Image. Anal 66 101693. [DOI] [PubMed] [Google Scholar]
- Tanaka D, Ikami D, Yamasaki T and Aizawa K 2018. Joint optimization framework for learning with noisy labels 2018 IEEE Conf. on Computer Vision and Pattern Recognition (Piscataway, NJ: IEEE; ) pp 5552–60 [Google Scholar]
- Vakalopoulou M, Chassagnon G, Bus N, Marini R, Zacharaki EI, Revel M-P and Paragios N 2018. AtlasNet: multi-atlas non-linear deep networks for medical image segmentation Medical Image Computing and Computer Assisted Intervention – MICCAI 2018. Lecture Notes in Computer Science vol 11073, ed Frangi A et al. (Berlin: Springer; ) pp 658–66 [Google Scholar]
- Van Horn G, Branson S, Farrell R, Haber S, Barry J, Ipeirotis P, Perona P and Belongie S 2015. Building a bird recognition app and large scale dataset with citizen scientists: the fine print in fine-grained dataset collection 2015 IEEE Conf. on Computer Vision and Pattern Recognition (Piscataway, NJ: IEEE; ) pp 595–604 [Google Scholar]
- Xiao T, Xia T, Yang Y, Huang C and Wang X 2015. Learning from massive noisy labeled data for image classification 2015Proc. IEEE Conf. on Computer Vision and Pattern Recognition (Piscataway, NJ: IEEE; ) pp 2691–9 [Google Scholar]
- Yang Q, Chao H, Nguyen D, Jiang S et al. 2019. A novel deep learning framework for standardizing the label of OARs in CT Artificial Intelligence in Radiation Therapy. AIRT 2019. Lecture Notes in Computer Science vol 11850 ed Nguyen D, Xing Land Jiang S(Berlin: Springer; ) pp 52–60 [Google Scholar]
- Yu S, Liu L, Wang Z, Dai G and Xie Y 2019. Transferring deep neural networks for the differentiation of mammographic breast lesions Sci. China Technol. Sci 62 441–7 [Google Scholar]
- Zhang Z and Sabuncu M 2018. Generalized cross entropy loss for training deep neural networks with noisy labels NIPS’18: Proc. 32nd Int. Conf. on Neural Information Processing Systems (Red Hook, NY: Curran Associates Inc.) pp 8778–88 [Google Scholar]
- Zhu H, Adeli E, Shi F and Shen D 2020. FCN based label correction for multi-atlas guided organ segmentation Neuroinformatics 18 319–31 [DOI] [PubMed] [Google Scholar]


