Abstract
Background
Body composition from computed tomography (CT) scans is associated with cancer outcomes including surgical complications, chemotoxicity, and survival. Most studies manually segment CT scans, but Automatic Body composition Analyser using Computed tomography image Segmentation (ABACS) software automatically segments muscle and adipose tissues to speed analysis. Here, we externally evaluate ABACS in an independent dataset.
Methods
Among patients with non‐metastatic colorectal (n = 3102) and breast (n = 2888) cancer diagnosed from 2005 to 2013 at Kaiser Permanente, expert raters annotated tissue areas at the third lumbar vertebra (L3). To compare ABACS segmentation results to manual analysis, we quantified the proportion of pixel‐level image overlap using Jaccard scores and agreement between methods using intra‐class correlation coefficients for continuous tissue areas. We examined performance overall and among subgroups defined by patient and imaging characteristics. To compare the strength of the mortality associations obtained from ABACS's segmentations to manual analysis, we computed Cox proportional hazards ratios (HRs) and 95% confidence intervals (95% CI) by tertile of tissue area.
Results
Mean ± SD age was 63 ± 11 years for colorectal cancer patients and 56 ± 12 for breast cancer patients. There was strong agreement between manual and automatic segmentations overall and within subgroups of age, sex, body mass index, and cancer stage: average Jaccard scores and intra‐class correlation coefficients exceeded 90% for all tissues. ABACS underestimated muscle and visceral and subcutaneous adipose tissue areas by 1–2% versus manual analysis: mean differences were small at −2.35, −1.97 and −2.38 cm2, respectively. ABACS's performance was lowest for the <2% of patients who were underweight or had anatomic abnormalities. ABACS and manual analysis produced similar associations with mortality; comparing the lowest to highest tertile of skeletal muscle from ABACS versus manual analysis, the HRs were 1.23 (95% CI: 1.00–1.52) versus 1.38 (95% CI: 1.11–1.70) for colorectal cancer patients and 1.30 (95% CI: 1.01–1.66) versus 1.29 (95% CI: 1.00–1.65) for breast cancer patients.
Conclusions
In the first study to externally evaluate a commercially available software to assess body composition, automated segmentation of muscle and adipose tissues using ABACS was similar to manual analysis and associated with mortality after non‐metastatic cancer. Automated methods will accelerate body composition research and, eventually, facilitate integration of body composition measures into clinical care.
Keywords: Body composition, Automation, Software, Adiposity, Muscle, Sarcopenia, Obesity, Cancer
Introduction
With the advent of computed tomography (CT) for diagnosis and surgical planning, secondary use of CT scans to study the relationship of body composition to clinical outcomes has grown exponentially. This is particularly true in oncology, where CT scans are standard‐of‐care for diagnosis and surveillance in many cancers, making secondary analysis highly feasible. CT measures of muscle and adipose tissue have been associated with surgical complications, chemotherapy toxicity, quality of life, recurrence, and survival across a variety of stages and types of cancer. 1 , 2 , 3 Thus, body composition data have the potential to improve risk stratification before surgery and chemotherapy as well as to personalize lifestyle interventions during cancer therapy and into the survivorship period. Segmentation of a single axial CT image at the third lumbar vertebra (L3) is a reference method for body composition assessment, 4 particularly in oncology research. 1 , 2 , 3
Despite its prognostic value, body composition is rarely assessed in cancer patients or used in clinical decision‐making. This may be due, in part, to a lack of time‐efficient, clinic‐friendly assessment tools that produce accurate muscle and adipose tissue quantifications. Analysis of CT images requires segmentation of different tissue areas by a trained rater with anatomical knowledge. Automated analysis of body composition has the potential to substantially reduce this workload and to accelerate research in body composition and chronic disease outcomes by leveraging the vast repositories of imaging data available within health systems.
Automated and semi‐automated software for analysis of body composition from medical imaging exists, 5 , 6 , 7 , 8 , 9 , 10 , 11 , 12 , 13 , 14 but most methods have not been externally evaluated in large, real‐world datasets. More typically, the performance of automated methods is reported only on the internal, training dataset or a small test dataset. Often, researchers manually correct the automated quantifications of muscle and adipose tissues, a semi‐automated process that likely over‐estimates the software's performance. ABACS (Automatic Body composition Analyzer using Computed tomography image Segmentation) is a commercially available software that automatically segments skeletal muscle and adipose tissue regions at L3 to estimate tissue areas and their mean radiodensities. Initially developed among 670 advanced cancer patients from a single clinical centre in Canada, ABACS had high accuracy in the test set of the derivation cohort. 15 However, ABACS has not been independently evaluated in a large, multi‐site, independent sample with CT scans collected across multiple hospitals and clinics.
The primary objective of this study is to evaluate the performance of ABACS for segmenting muscle and adipose tissues relative to manual analysis from CT scans in a large population of patients with non‐metastatic breast and colorectal cancer that was completely independent of the cohort used to develop the algorithm. As a secondary objective, to understand how the use of automated methods might change the observed association of body composition with clinical outcomes, we compared the magnitude of association with overall mortality after cancer diagnosis of muscle and adipose tissue estimates from ABACS to those from manual analysis.
Materials and methods
Study population
The patient population for this study was drawn from the Sarcopenia, Cancer And Near‐term Survival (SCANS) studies, which are completely distinct from the ABACS derivation cohort. Data collection and analysis methods for each of these studies have been described previously 16 , 17 but are summarized briefly here. The SCANS studies included all Kaiser Permanente Northern California (KPNC) health plan members diagnosed with non‐metastatic colorectal cancer (C‐SCANS) from 2005 to 2011 or with non‐metastatic, invasive breast cancer (B‐SCANS) from 2006 to 2013. Both studies found associations of body composition (assessed using manual analysis) with overall survival. 16 , 17 , 18 , 19 To be eligible, patients had to be aged 18 to 80 years at diagnosis, have no prior cancer history, and have an abdominal or pelvic CT scan available at diagnosis for analysis. CT scans came from a variety of clinical centres spread over KPNC's 21 hospitals and over 200 outpatient clinics and included contrast, non‐contrast, and PET‐CTs. Covariate data on patient (e.g. weight, height, sex, age, and race/ethnicity), tumour (site, stage, and subtype), and treatment characteristics (receipt of chemotherapy and/or radiation) were obtained by accessing the electronic medical record for prospectively collected clinical data. Clinical data were linked to the cancer registry and mortality files compiled from internal data, California state death data, and Social Security Administration data. The KPNC Institutional Review Board approved the study with a waiver of informed consent.
Image review
Prior to automated segmentation at L3, research assistants blinded to the results of both the manual and automated analysis qualitatively reviewed all the original, unlabelled Digital Imaging and Communications in Medicine (DICOM) images. Based on this blinded review, we excluded cases unsuitable for manual analysis due to metal implants (n = 2), abdominal skeletal muscle partially out of the image field (n = 7), severe anatomic abnormalities obstructing the abdominal muscle groups (fluid accumulation, diastasis recti, or hernia, n = 7), and severe photon starvation artefacts resulting from the patient's body pressing against the top and sides of the scanner (n = 205). Examples of these 221 exclusions, which were not included in the 5990 patient images analysed for this study, are shown in Supporting Information, Figure S1 . Still blinded to the results, we classified all remaining DICOM images according to characteristics commonly observed in real‐world clinical data for use in sensitivity analyses: notable streaking or graininess, object touching patient's trunk [limb or hardware (e.g. colostomy port and electrode)], less severe anatomic abnormalities (classified as emaciated body habitus, hernia, or fluid accumulation), skinfolds/pannus, or subcutaneous adipose tissue cut‐off (partially out of image field).
Manual segmentation
As part of the SCANS studies, two centrally trained researchers using SliceOmatic Software version 5.0 (TomoVision, Montreal, Quebec, Canada) selected a single slice at L3 and segmented the cross‐sectional area in centimetres squared (cm2) of each tissue area, distinguishing muscle and visceral from subcutaneous adipose tissues using anatomic knowledge and tissue‐specific Hounsfield Units (HU) ranges: −29 to 150 for skeletal muscle, −190 to −30 for subcutaneous and inter‐muscular adipose, and −150 to −50 for visceral adipose. Each CT scan was manually segmented following the Alberta protocol. 20 The first segmentation was used as the reference in this analysis. In inter‐rater reliability analysis in a subset of 50 scans, coefficients of variation (CV%) were 1.2%, 2.7%, 1.1%, and 9.0% for muscle and subcutaneous, visceral, and inter‐muscular adipose tissues between the two human raters, respectively.
Of note, a limitation of ABACS is that the algorithm for inter‐muscular adipose tissue segmentation is still in beta‐testing. By default, ABACS treats inter‐muscular adipose tissue (adipose tissue deposits within skeletal muscle) as subcutaneous adipose tissue. Thus, for comparability, we combined the manual labels for subcutaneous and inter‐muscular adipose tissue in this study; this combined tissue area is henceforth referred to as subcutaneous adipose tissue.
Automated segmentation
The development of the ABACS automated segmentation algorithm and its performance in the derivation cohort of 670 patients with advanced gastrointestinal, lung, and head and neck cancers has been previously described. 15 The algorithm behind ABACS follows a two‐step approach for the segmentation of muscle and adipose tissues from an input L3 CT slice. In the first step, a muscle region mask is determined using a template‐based segmentation methodology, wherein a binary template defining an initial shape of the muscle is deformed via non‐rigid registration to closely match the muscle region in the binarized version (obtained by thresholding within the muscle −29 to 150 HU range) of the input slice. The deformation process is guided by a statistical shape prior model that encodes a priori knowledge about the characteristic, cross‐sectional shape of the muscles in the L3 location. This aids the disambiguation of the muscle tissue from the neighbouring organs with overlapping HU ranges, leading to an accurate segmentation of the muscle region mask. The second step involves masking the input slice with the estimated muscle region segmentation and determining the final muscle, subcutaneous, and visceral adipose tissue regions of interest (ROIs) using the corresponding pre‐defined HU ranges for these tissues. Specifically, the pixels within the muscle region mask that have attenuation values ranging from −29 to 150 HU are used to obtain the muscle tissue ROI, whereas pixels not belonging to the muscle mask are used to determine the adipose tissue ROIs. The pixels lying ‘outside’ of the outer boundary of the muscle mask that have attenuation values in the −190 to −30 HU range comprise the subcutaneous adipose tissue ROI, while the pixels that are ‘inside’ the interior of the inner of the boundary of the muscle mask and have attenuation values in the −150 to −50 HU range are used to define the visceral adipose tissue ROI.
Of note, ABACS is not at present fully automated, because manual exclusion of aberrant images and manual identification of the L3 anatomic landmarks were conducted prior to image segmentation. Automated selection of the L3 is currently being beta‐tested. Variability in the L3 slice selection was not assessed as part of this evaluation.
The software is available commercially from Voronoi Health Analytics Inc., (Coquitlam, Canada, https://voronoihealthanalytics.com) and is integrated into the SliceOmatic (TomoVision, Magog, Canada, https://tomovision.com) software as a module.
Statistical analysis
We examined descriptive characteristics and ABACS performance overall and within subgroups defined by cancer site, age, body mass index (BMI), sex, stage, CT type (contrast versus non‐contrast or PET‐CT), and other common imaging characteristics. First, we computed Jaccard scores (also known as the Jaccard similarity coefficient or intersection over union score) for each tissue area, which quantify segmentation accuracy by measuring the pixel‐level overlap between the automated and the manual labels. Jaccard scores range from 0% (indicating no overlap) to 100% (indicating perfect overlap). To quantify the similarity between the automated and manual analysis, we computed the intra‐class correlations. To provide a visual means to evaluate a bias between the automated and manual analysis, we created Bland–Altman plots for each tissue area, reported the mean differences between the automated and manual analysis and constructed an agreement interval, within which 95% of these differences fall.
To determine whether patients were ranked equivalently with respect to muscularity or adiposity, we then categorized patients' tissue areas into tertiles and quintiles separately based on manual and then automated analysis and examined the kappa coefficients for agreement and percentage misclassification among these categories.
Finally, we compared the associations of muscle and subcutaneous and visceral adiposity with overall mortality after cancer diagnosis when each tissue area was categorized into tertiles based on the automated versus the manual segmentations. We fit Cox proportional hazards models separately by cancer site. Tertiles of muscle, subcutaneous, and visceral adipose tissues were included in the same models, which additionally adjusted for age, sex, race/ethnicity, stage and grade, receipt of chemotherapy and/or radiation, smoking status, and other relevant covariates (breast cancer models adjusted for hormone receptor and HER2 status and colorectal cancer models additionally adjust for tumour site, colon versus rectum).
In sensitivity analyses, we repeated analyses within categories of imaging characteristics that could influence ABACS performance, as described earlier: notable streaking or graininess, limb or hardware touching patient's trunk, abnormal anatomy, skinfolds/pannus, or subcutaneous adipose tissue cut‐off.
Results
Table 1 shows descriptive characteristics of the n = 5990 patients included in this study after exclusion of 221 images that were unsuitable for manual analysis. Mean (SD) age at diagnosis was 63 (11) years for patients with colorectal and 56 (12) years for patients with breast cancer; a majority of patients were overweight or obese 30% and 33%, respectively. In 32% of the images included in our analysis, blinded review of the original DICOM images found notable streaking or graininess (n = 249), skinfolds/pannus (n = 492), hardware (n = 87), a limb (n = 7) touching the patient's trunk, or abnormal anatomy (n = 31). Examples of these features are given in Figure 1 .
Table 1.
Characteristics of patients and computed tomography scans at diagnosis of non‐metastatic colorectal and breast cancer in an independent evaluation of ABACS software (n = 5990) a
| Characteristics | C‐SCANS (n = 3102) | B‐SCANS (n = 2888) | Combined (n = 5990) |
|---|---|---|---|
| Mean (SD) or N, % | |||
| Age at diagnosis, years | 62.6 (11.4) | 56 (11.8) | 59.4 (12.1) |
| Female, % | 1541, 49.7% | 2888, 100% | 4429, 73.9% |
| Stage, % | |||
| I | 935, 30.1% | 620, 21.5% | 1555, 26% |
| II | 973, 31.4% | 1320, 45.7% | 2293, 38.3% |
| III | 1194, 38.5% | 948, 32.8% | 2142, 35.8% |
| Body mass index, % | |||
| Underweight: <18.5 kg/m2 | 57, 1.8% | 45, 1.6% | 102, 1.7% |
| Normal weight: 18.5 to <25 kg/m2 | 984, 31.7% | 985, 34.1% | 1969, 32.9% |
| Overweight: 25 to <30 kg/m2 | 1141, 36.8% | 921, 31.9% | 2062, 34.4% |
| Class I obesity: 30 to <35 kg/m2 | 628, 20.2% | 563, 19.5% | 1191, 19.9% |
| Class II obesity: ≥35 kg/m2 | 292, 9.4% | 374, 13% | 666, 11.1% |
| Scan post‐surgery, % | 531, 17.1% | 1656, 57.3% | 2187, 36.5% |
| CT type, % | |||
| Contrast | 2989, 96.4% | 2162, 74.9% | 5151, 86% |
| Non‐contrast | 113, 3.6% | 142, 4.9% | 255, 4.3% |
| PET | 0, 0% | 584, 20.2% | 584, 20.2% |
| Common imaging characteristics, % | |||
| Notable streaking or graininess | 229, 7.4% | 20, 0.7% | 249, 4.1% |
| Skinfolds or pannus | 240, 7.7% | 252, 8.7% | 492, 8.2% |
| Subcutaneous adipose cut‐off | 736, 23.7% | 603, 20.9% | 1339, 22.4% |
| Limb touching trunk | 2, 0.1% | 5, 0.2% | 7, 0.1% |
| Hardware touching trunk | 61, 2% | 26, 0.9% | 87, 1.5% |
| Abnormal anatomy | 29, 0.9% | 2, 0.1% | 31, 0.5% |
| Manual analysis, cm2 | |||
| Skeletal muscle | 140.1 (37.7) | 114.3 (20) | 127.7 (33.1) |
| Visceral adipose | 152.8 (108.3) | 102.1 (77.5) | 128.3 (98) |
| Subcutaneous adipose | 216.8 (111.7) | 252.8 (120.9) | 234.2 (117.6) |
| ABACS analysis, cm2 | |||
| Skeletal muscle | 135.8 (36.5) | 114 (20.2) | 125.3 (31.7) |
| Visceral adipose | 151 (108.7) | 100 (77) | 126.4 (98.1) |
| Subcutaneous adipose | 213.8 (114) | 251.1 (123.9) | 231.8 (120.3) |
Inter‐muscular and subcutaneous adipose tissues are combined in this analysis.
Figure 1.
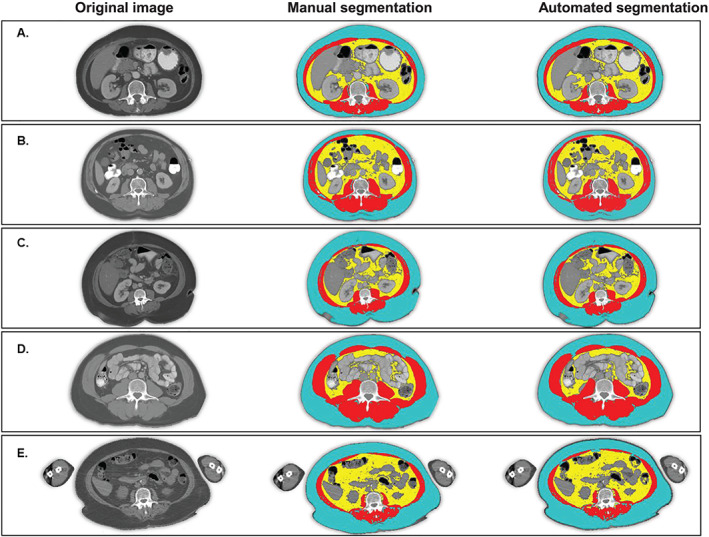
Manual and automated segmentation of body composition from CT produces similar results. Blue = subcutaneous adipose tissue; red = skeletal muscle tissue; yellow = visceral adipose tissue. (A) A normal weight patient with successful automated segmentation. Jaccard scores for muscle and visceral and subcutaneous adipose tissue are 95, 97, and 96, respectively. (B) An overweight patient with an electrode on the surface of the abdomen. Jaccard scores for muscle and visceral and subcutaneous adipose tissue are 97, 98, and 96, respectively. (C) A patient with class II obesity and skinfolds and pannus visible on the scan. Jaccard scores for muscle and visceral and subcutaneous adipose tissue are 96, 99, and 97, respectively. (D) A patient with class I obesity with some subcutaneous adipose tissue outside of the visual field. Jaccard scores for muscle and visceral and subcutaneous adipose tissue are 95, 95, and 98, respectively. (E) An overweight patient with a PET‐CT whose limbs are present in the visual field. Jaccard scores for muscle and visceral and subcutaneous adipose tissue are 93, 92, and 91, respectively.
Overall, there was strong agreement between ABACS and manual analysis. As shown in Table 2 and Figure 2 , average Jaccard scores (95% CI) were 91.5% (91.4, 91.7) for skeletal muscle and 91.5% (91.2, 91.9) for visceral and 93.9% (93.7, 94.0) for subcutaneous adipose tissue areas. Consistent with the high Jaccard scores, intra‐class correlation coefficients ranged from 0.96 to 0.99.
Table 2.
Agreement between ABACS segmentations of muscle and adipose tissue compared with manual analysis among non‐metastatic colorectal and breast cancer patients at Kaiser Permanente Northern California, overall and by body mass index category (n = 5990) a
| Colorectal cancer patients in C‐SCANS (n = 3102) | Breast cancer patients in B‐SCANS (n = 2888) | |||||
|---|---|---|---|---|---|---|
| Jaccard score b | Reliability coefficient c | Simple kappa across quintiles d | Jaccard score b | Reliability coefficient c | Simple kappa across quintiles d | |
| Overall results | Mean (95% confidence interval) | |||||
| Skeletal muscle | 92.5 (92.3, 92.7) | 0.98 (0.98, 0.98) | 0.85 (0.83, 0.87) | 90.5 (90.3, 90.7) | 0.96 (0.96, 0.96) | 0.72 (0.71, 0.74) |
| Visceral adipose | 93.1 (92.8, 93.5) | 0.98 (0.98, 0.98) | 0.96 (0.94, 0.97) | 89.8 (89.4, 90.3) | 0.97 (0.96, 0.97) | 0.94 (0.92, 0.96) |
| Subcutaneous adipose | 93.5 (93.3, 93.7) | 0.98 (0.98, 0.98) | 0.94 (0.92, 0.96) | 94.3 (94.1, 94.5) | 0.99 (0.98, 0.99) | 0.95 (0.93, 0.97) |
| Underweight: BMI <18.5 kg/m2 | N = 57 (1.8%) | N = 45 (1.6%) | ||||
| Skeletal muscle | 84.9 (82.8, 87.0) | 0.84 (0.75, 0.90) | 0.54 (0.41, 0.67) | 83.0 (80.4, 85.7) | 0.88 (0.80, 0.93) | 0.53 (0.38, 0.67) |
| Visceral adipose | 75.6 (70.5, 80.7) | 1.00 (1.00, 1.00) | 0.87 (0.74, 1.00) | 70.5 (64.0, 76.9) | 0.99 (0.98, 0.99) | 0.64 (0.49, 0.78) |
| Subcutaneous adipose | 86.3 (83.4, 89.2) | 1.00 (0.99, 1.00) | 0.91 (0.78, 1.04) | 87.8 (85.4, 90.2) | 0.98 (0.97, 0.99) | 0.89 (0.74, 1.03) |
| Normal weight: BMI 18.5 to <25 kg/m2 | N = 984 (31.7%) | N = 985 (34.1%) | ||||
| Skeletal muscle | 90.9 (90.6, 91.3) | 0.96 (0.95, 0.96) | 0.79 (0.76, 0.82) | 89.0 (88.6, 89.3) | 0.90 (0.89, 0.92) | 0.61 (0.58, 0.64) |
| Visceral adipose | 89.1 (88.4, 89.9) | 1.00 (1.00, 1.00) | 0.94 (0.91, 0.97) | 84.9 (84.1, 85.7) | 1.00 (1.00, 1.00) | 0.89 (0.86, 0.92) |
| Subcutaneous adipose | 92.5 (92.2, 92.9) | 0.99 (0.99, 1.00) | 0.89 (0.86, 0.92) | 93.3 (93.0, 93.6) | 0.99 (0.99, 0.99) | 0.89 (0.86, 0.92) |
| Overweight: BMI 25 to <30 kg/m2 | N = 1141 (36.8%) | N = 921 (31.9%) | ||||
| Skeletal muscle | 93.3 (93.1, 93.6) | 0.97 (0.97, 0.98) | 0.85 (0.82, 0.88) | 91.4 (91.1, 91.8) | 0.96 (0.95, 0.96) | 0.73 (0.7, 0.76) |
| Visceral adipose | 95.4 (94.9, 95.8) | 0.99 (0.99, 0.99) | 0.95 (0.92, 0.98) | 92.6 (92.1, 93.1) | 0.99 (0.99, 0.99) | 0.93 (0.90, 0.96) |
| Subcutaneous adipose | 93.9 (93.7, 94.2) | 0.97 (0.97, 0.98) | 0.92 (0.9, 0.95) | 95.0 (94.8, 95.2) | 0.99 (0.99, 0.99) | 0.89 (0.86, 0.92) |
| Obese: BMI ≥30 kg/m2 | N = 920 (29.7%) | N = 938 (32.5%) | ||||
| Skeletal muscle | 93.5 (93.2, 93.9) | 0.98 (0.98, 0.98) | 0.88 (0.84, 0.91) | 91.5 (91.1, 91.9) | 0.96 (0.96, 0.97) | 0.74 (0.71, 0.77) |
| Visceral adipose | 95.8 (95.1, 96.5) | 0.94 (0.94, 0.95) | 0.93 (0.90, 0.96) | 93.2 (92.4, 94.1) | 0.90 (0.88, 0.91) | 0.90 (0.87, 0.93) |
| Subcutaneous adipose | 94.4 (94.1, 94.8) | 0.94 (0.93, 0.94) | 0.91 (0.88, 0.94) | 94.9 (94.6, 95.3) | 0.95 (0.94, 0.95) | 0.92 (0.89, 0.96) |
Inter‐muscular and subcutaneous adipose tissues are combined in this analysis.
Jaccard scores measure pixel‐level image overlap between the automated segmentation by ABACS and the manual segmentation by trained raters.
Reliability coefficients, also known as intra‐class correlation coefficients, reflect not only degree of correlation between automated and manual segmentation but also agreement between the two measurements. Mathematically, reliability represents a ratio of true variance in the manual segmentations over true variance plus error variance (discrepancy between the manual and automated segmentations).
Cohen's kappa coefficient (κ) measures inter‐rater reliability across quintile categories and considers the potential of agreement occurring by chance.
Figure 2.
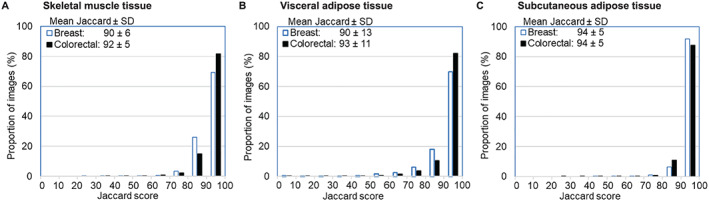
Distribution of Jaccard scores quantifying image overlap between ABACS automated and manual segmentation of body composition. Legends show the mean ± standard deviation (SD) Jaccard scores for each tissue area separately among breast and colorectal cancer patients. Jaccard scores measure pixel‐level overlap in image segmentation comparing ABACS to manual analysis by a trained rater with anatomic knowledge. Overall, the average Jaccard scores exceeded 90% for all tissues. Breast = 2888 non‐metastatic breast cancer patients in B‐SCANS study; colorectal = 3102 non‐metastatic colorectal cancer patients in C‐SCANS study. (A) Average muscle tissue Jaccard scores exceeded 90% for both breast and colorectal cancer patients. (B) Visceral adipose tissue Jaccard scores demonstrated a small number of total segmentation failures (scores <20). (C) Subcutaneous adipose tissue Jaccard scores combine subcutaneous and inter‐muscular adipose tissue.
As shown in the Bland–Altman plots in Figure 3 , on average, ABACS underestimated muscle and visceral and subcutaneous adipose areas relative to manual analysis by about 1% to 2% of total tissue area: mean differences (95% CI) were −2.35 (−2.52, −2.19), −1.97 (−2.51, −1.45), and −2.38 (−2.95, −1.80) cm2, respectively. In general, the degree of bias was correlated with the magnitude, with larger differences between automated and manual analysis for patients with larger tissue areas. The Bland–Altman limits of agreement (average difference between measurements) ± 2 × SD were broad at 10.65 to −15.35 for muscle, 39.55 to −43.44 for visceral, and 42.98 to −47.72 for subcutaneous adipose tissue. Despite differences in the absolute tissue area estimates, there was moderate to good agreement when tissue areas were categorized among quintiles based on the distribution of ABACS and compared with quintiles defined using manual analysis: among all patients, kappa coefficients (95% CIs) ranged from very good [0.80 (0.79, 0.81)] for skeletal muscle tissue to excellent [0.95 (0.94, 0.96)] for visceral adipose tissue. For skeletal muscle tissue, ABACS segmentations correctly classified all but 956 (16%) of patients into the same quintile as manual analysis. Of these misclassified patients, all but 41 were in an adjacent quintile. Kappa coefficients are shown separately by cancer site and subgroup among in Table 2 .
Figure 3.
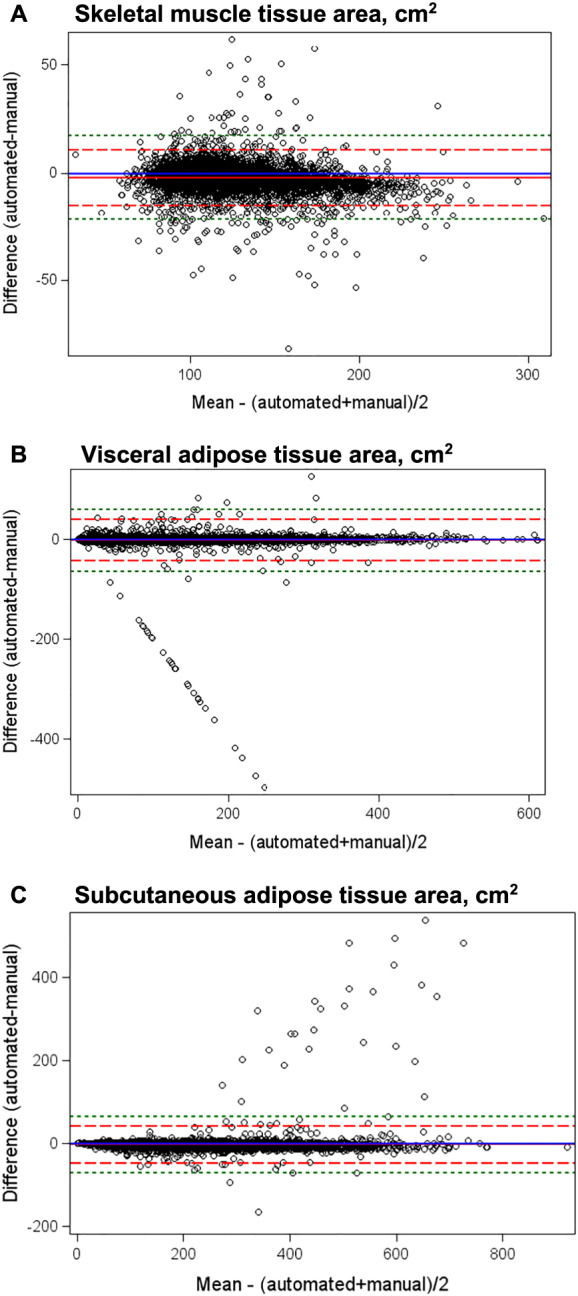
Bland–Altman plots of mean difference in skeletal muscle and visceral and subcutaneous adipose tissue areas between automated and manual analysis of computed tomography scans. The Bland–Altman plot is a graphical method for assessment of the magnitude of disagreement, both error and bias, between automated and manual segmentation of CT scans. The plot presents the difference versus the average of the automated and manual quantifications of body composition with reference lines at 0 (blue line indicating no difference between the manual and automated methods) and at ±2 standard deviations (SD, the dashed red lines) or ±3 SD (the dashed green lines) of the difference to aid in identification of outliers. Mean differences were −2.35 for muscle, −1.97 for visceral, and −2.38 for subcutaneous adipose tissue. Limits of agreement were broad, with ±2 SD limits of agreement at 10.65 to −15.35 for muscle, 39.55 to −43.44 for visceral, and 42.98 to −47.72 for subcutaneous adipose tissue and ±3 SD limits of agreement at 17.15 to −21.85 for muscle, 60.30 to −64.19 for visceral, and 65.65 to −70.39 for subcutaneous adipose tissue.
As shown in Table 3 , mortality associations were similar for ABACS versus manual analysis and confidence intervals overlapped. For example, the hazard ratios (95% CI) for death from any cause comparing the lowest versus highest tertile of skeletal muscle area from ABACS were weaker at 1.23 (1.00, 1.52) for patients with colorectal cancer and similar at 1.30 (1.01, 1.66) for patients with breast cancer, respectively, in contrast to 1.38 (1.11, 1.70) for patients with colorectal and 1.29 (1.00, 1.65) for those with breast cancer, respectively, when using manual segmentation. Of note, 10% of colorectal and 13% of breast cancer patients, respectively, were classified into a different tertile by ABACS versus manual analysis.
Table 3.
Association of muscle and adipose tissue tertiles with overall mortality using ABACS and manual analysis to assess body composition: C‐SCANS non‐metastatic colorectal patients and B‐SCANS breast cancer patients (n = 5990) a , b
| C‐SCANS (n = 3102) c | B‐SCANS (n = 2888) d | |||||||
|---|---|---|---|---|---|---|---|---|
| Events | ABACS | Events | Manual analysis | Events | ABACS | Events | Manual analysis | |
| HR (95% CI) | HR (95% CI) | HR (95% CI) | HR (95% CI) | |||||
| Skeletal muscle | ||||||||
| Tertile 1 | 339 | 1.23 (1.00, 1.52) | 342 | 1.38 (1.11, 1.70) | 212 | 1.30 (1.01, 1.66) | 207 | 1.29 (1.00, 1.65) |
| Tertile 2 | 250 | 1.01 (0.84, 1.23) | 260 | 1.16 (0.96, 1.41) | 159 | 0.95 (0.76, 1.19) | 172 | 1.01 (0.81, 1.27) |
| Tertile 3 (Ref.) | 222 | 1.0 (—) | 209 | 1.0 (—) | 176 | 1.0 (—) | 168 | 1.0 (—) |
| Continuous, per SD e | 811 | 0.86 (0.75, 0.98) | 811 | 0.74 (0.65, 0.86) | 547 | 0.89 (0.80, 0.99) | 547 | 0.89 (0.80, 1.00) |
| Subcutaneous adipose | ||||||||
| Tertile 1 (Ref.) | 293 | 1.0 (—) | 294 | 1.0 (—) | 174 | 1.0 (—) | 174 | 1.0 (—) |
| Tertile 2 | 263 | 0.96 (0.80, 1.15) | 262 | 0.93 (0.77, 1.11) | 174 | 1.04 (0.82, 1.32) | 176 | 1.01 (0.80, 1.28) |
| Tertile 3 | 255 | 0.98 (0.80, 1.21) | 255 | 0.97 (0.79, 1.19) | 199 | 1.28 (0.99, 1.67) | 197 | 1.24 (0.95, 1.62) |
| Continuous, per SD | 811 | 0.99 (0.91, 1.08) | 811 | 0.97 (0.88, 1.07) | 547 | 1.12 (1.01, 1.23) | 547 | 1.09 (0.98, 1.22) |
| Visceral adipose | ||||||||
| Tertile 1 (Ref.) | 283 | 1.0 (—) | 277 | 1.0 (—) | 156 | 1.0 (—) | 151 | 1.0 (—) |
| Tertile 2 | 242 | 0.78 (0.65, 0.93) | 246 | 0.82 (0.68, 0.99) | 174 | 0.84 (0.66, 1.06) | 180 | 0.92 (0.72, 1.17) |
| Tertile 3 | 286 | 0.93 (0.76, 1.14) | 288 | 0.99 (0.81, 1.22) | 217 | 0.98 (0.75, 1.28) | 216 | 1.03 (0.78, 1.36) |
| Continuous, per SD | 811 | 1.01 (0.92, 1.10) | 811 | 1.07 (0.97, 1.18) | 547 | 1.01 (0.91, 1.13) | 547 | 1.04 (0.93, 1.16) |
Tertiles are defined separately by cancer site (colorectal versus breast), sex (male versus female among colorectal cancer patients only), and data source (ABACS automated versus manual analysis).
Cox proportional hazards models mutually adjust muscle, subcutaneous and visceral adipose tissue tertiles, and additionally adjust for age, sex, race/ethnicity, stage and grade, receipt of chemotherapy and/or radiation, smoking status.
Models for overall survival after colorectal cancer additionally adjust for tumour site (colon versus rectum).
Models for overall survival after breast cancer additional adjust for hormone receptor and HER2 status.
SD = standard deviation units defined by dividing the individual patient's value for each tissue area by the population standard deviation for that tissue.
Sensitivity analyses revealed that sex, age, and cancer stage did not substantially impact accuracy of ABACS compared with manual segmentations (Supporting Information, Table S1 ) nor did imaging issues common in real‐world data. For example, the mean Jaccard scores were approximately 90% for all tissue areas in images both with and without subcutaneous adipose tissue cut‐off or pannus/skinfolds. However, there were some patient and imaging characteristics that reduced the accuracy of ABACS's segmentations. Most notably, among underweight patients BMI <18.5 kg/m2 (n = 108, <2% of the cohort), the mean Jaccard scores in all patients were moderate for muscle and visceral and subcutaneous adiposity at 83%, 70%, and 88%, respectively. Jaccard scores were also moderate in patients with gross anatomic abnormalities such as hernia, diastasis recti, or fluid accumulation (n = 31, <1% of the cohort) for muscle and visceral and subcutaneous adiposity at 81%, 71%, and 83%, respectively. Figure 4 displays representative examples of such cases.
Figure 4.
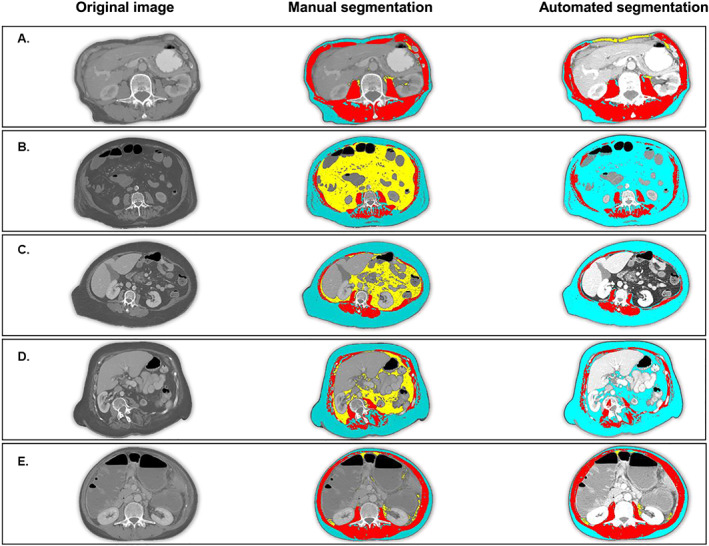
Cases of segmentation failure: Emaciated body habitus and anatomic abnormalities can cause segmentation failure because ABACS relies on the characteristic shape of the L3 muscle groups. Blue = subcutaneous adipose tissue; red = skeletal muscle tissue; yellow = visceral adipose tissue. In each case of segmentation failure, the abdominal muscle wall is atypical (not continuous or asymmetrical) and thus cannot be identified by the shape‐based prior upon which the ABACS algorithm relies. (A) Patient with emaciated body habitus, in which the thin subcutaneous adipose tissue layer and proximity of the organs to the abdominal muscle wall cause ABACS to mislabel visceral adipose tissue. Jaccard scores for muscle and visceral and subcutaneous adipose tissue are 75, 17, and 73, respectively. (B) Patient in whom muscle wasting (including substantial inter‐muscular adipose tissue) interrupt the continuity of the anterior abdominal wall, causing ABACS to fail to delineate subcutaneous from visceral adipose tissue. Jaccard scores for muscle and visceral and subcutaneous adipose tissue are 80, 0, and 39, respectively. (C) Proximity of organs and bulge in abdominal muscle wall cause visceral adipose tissue segmentation failure. Jaccard scores for muscle and visceral and subcutaneous adipose tissue are 76, 0, and 93, respectively. (D) Patient with scoliosis. Shape of abdominal muscles is not symmetrical and cannot be identified by shape‐based prior segmentation. Jaccard scores for muscle and visceral and subcutaneous adipose tissue are 63, 0, and 69, respectively. (E) Image lacks a continuous musculature in the anterior abdominal wall leading to misclassification of visceral adipose tissue. Jaccard scores for muscle and visceral and subcutaneous adipose tissue are 81, 54, and 81, respectively.
Finally, after completing the automated analysis using ABACS, we examined a random sample of images with total segmentation failures (Jaccard scores <20% for one or more tissue areas), which occurred in 38 patients (0.6% of the cohort). All were patients for whom ABACS segmented very little or no visceral adipose tissue (this is reflected in the visceral adipose tissue outliers visible as a linear pattern on the Bland–Altman plot in Figure 3 ). Two thirds of these patients were sarcopenic obese (muscle area < 40 cm2; BMI ≥ 30 kg/m2); only three were underweight (BMI < 18.5 kg/m2). Figure 4 displays representative examples of such cases. Of note, we also considered Jaccard scores <70% for one or more tissue areas to indicate poor segmentation, which occurred in 312 patients (5.2% of the cohort).
Discussion
We found that the ABACS automated software provides accurate segmentations of muscle and adipose tissues from CT scans among a large, community‐based cohort of nearly 6000 patients with non‐metastatic colorectal and breast cancer. We found good agreement between ABACS's segmentations and those obtained from manual analysis by trained raters overall and among subgroups defined by age, stage, sex, and other characteristics. Importantly, when muscle tissue was segmented using ABACS the magnitude of association with overall survival was similar to our prior studies using manual analysis for breast cancer patients but somewhat weaker for colorectal cancer patients, although confidence intervals overlapped. 16 , 17
Several limitations to accurate automatic segmentation were noted. As described previously, 15 ABACS's segmentation algorithm relies on the characteristic shape of the abdominal muscles to delineate subcutaneous from visceral adipose tissue. In cases where the integrity of the abdominal wall was compromised by anatomic abnormalities (e.g. severe muscle or subcutaneous adipose tissue wasting, organs pressed against the abdominal wall, severe fluid accumulation, diastasis recti, and/or extensive inter‐muscular adipose tissue infiltration), the software did not perform well. While such cases comprised <2% of our study population, tissue wasting can be common in other settings, such as advanced lung, head and neck, or pancreatic cancers. Researchers examining these patient populations should conduct additional, post‐segmentation screening of images to ensure data quality.
Various automatic methods have been developed for tissue segmentation from CT scans. 5 , 6 , 7 , 8 , 9 , 10 , 11 , 12 , 13 , 14 Many methods focused either on the segmentation of muscle 6 , 7 , 8 , 10 or adipose tissue 5 , 11 , 14 , 21 but not both. Most methods for muscle tissue segmentation only considered single muscles such as diaphragm, psoas major, and rectus abdominis, 6 , 7 , 8 as opposed to total L3 skeletal muscle tissue area. Most methods that segmented both muscle and adipose tissue were semi‐automated, requiring considerable manual corrections 13 , 22 or involved algorithms that need cohort‐specific parameter tuning via a heuristic approach. 9 These methods are akin to the semi‐automated automatic segmentation features such as region‐growing and thresholding available within popular DICOM viewers such as OsiriX (Primeo, Bernex, Switzerland, https://www.osirix‐viewer.com) and SliceOmatic. While the ABACS framework segments a single‐slice axial image into skeletal muscle, visceral adipose and subcutaneous adipose tissues in a fully automated manner once the L3 has been selected, users should be aware of the limitations of the software and visually inspect results to identify segmentation failures.
Recently, there have been efforts towards building fully automated pipelines using machine learning approaches like random forests 12 and deep learning techniques such as convolution neural networks 23 for obtaining the segmentation of both muscle and adipose tissues from a CT scan. However, these studies are not directly comparable because they have included smaller patient populations from a single clinical centre and/or reported the agreement of the automated and manual analysis based only on the derivation cohort, that is, agreement within the training dataset or on a test dataset that was generated from a small number of held‐out cases. Also, it should be noted that these methods are not available to researchers outside the institutions where they were developed. In addition to providing an efficient method to assess body composition automatically from clinically acquired CT scans, ABACS offers several advantages: it is commercially available and has already been used in oncology research. 15 , 24 , 25
There is accumulating evidence that body composition assessed from CT scans is associated with morbidity and mortality in cancer patients, often more strongly than BMI, which does not distinguish muscle from adipose tissue or describe adipose tissue distribution. 2 , 3 Thus, clinicians should consider that patients with similar BMI can have very different body composition when making treatment decisions and tailoring lifestyle guidance for cancer survivors. Automated methods such as ABACS accurately segment muscle and adipose tissue from single‐slice CT scans and can contribute to the growing evidence of the importance of body composition in cancer.
Limitations
Important limitations to the ABACS software and to this evaluation should be noted. First, at present, the software requires manual selection of the L3. While single‐slice L3 measures of body composition are commonly used, there may be utility to using larger regions for analysis, particularly for visceral adipose tissue where more stable measures may be obtained if multiple abdominal slices are analysed. Complete validation of this method in a given patient population would include evaluation of the impact of variability in patient positioning and L3 image slice selection, as well as how effective and consistent a research or clinical workflow is to detect images with poor image quality before applying ABACS. In addition, we have yet to evaluate ABACS's performance in quantifying longitudinal changes in body composition based on serial CT scans. Second, as noted, despite excellent performance in most cases, ABACS did not produce optimal segmentations in patients with poor contrast between muscle and adjacent tissue due to anatomic abnormalities or significant muscle or adipose tissue wasting. 15 It may therefore be prudent for researchers to pre‐screen DICOM images prior to analysis by either ABACS or manual analysis to ensure data quality. The criteria for pre‐screening images or conducting post hoc quality control will depend on the context in which the software is used for example, in large‐scale studies, the error introduced through the use of automated segmentation may be overcome with larger samples sizes Thus, an image quality control protocol for population‐based research might include manual checks on a subset of images to estimate the accuracy of ABACS in a specific patient population or a rapid visual review to identify and exclude total failures of segmentation. Meanwhile, for individual patient diagnostics in the clinical setting, more accurate algorithms must be developed.
Conclusions and future directions
This independent evaluation of a commercially available software found that automated segmentations of CT scans to assess body composition were similar to manual analysis and associated with mortality after non‐metastatic cancer. Future directions for this research that will enhance the utility of ABACS include the use of a deep learning approach for designing convolution neural networks to improve the segmentation algorithm and automate slice localization at all vertebral landmarks, building upon the recently published CNN‐based approaches for automatic L3 slice detection. 21 , 22 In the long‐term, rapid, accurate and cost‐effective methods for high‐throughput analysis of body composition across multiple anatomical areas is a pre‐requisite to using body composition data in clinical practice. In the immediate‐term, automation will accelerate body composition research.
Conflict of interest
Drs Popuri and Beg are co‐founders and actively direct Voronoi Health Analytics Incorporated, a Canadian corporation that sells commercial licences for the ABACS (Automated Body Composition Analyser using Computed tomography image Segmentation) software. Dr Prado declares honoraria and travel from Abbott Nutrition unrelated to the current work. Dr Meyerhardt received consulting fees from Taiho Pharmaceuticals, Ignyta, and COTA Healthcare, unrelated to current work.
Funding
Elizabeth M. Cespedes Feliciano was supported by National Cancer Institute grant K01CA226155; data collection was supported by National Cancer Institute grants R01CA175011 and R01CA184953.
Supporting information
Figure S1. Examples of scans excluded from analysis in blinded review of DICOM images
Table S1. Agreement between ABACS segmentations of muscle and adipose tissue compared to manual analysis among non‐metastatic colorectal and breast cancer patients
Acknowledgement
The authors of this manuscript certify that they comply with the ethical guidelines for authorship and publishing in the Journal of Cachexia, Sarcopenia and Muscle. 26
Cespedes Feliciano E. M., Popuri K., Cobzas D., Baracos V. E., Beg M. F., Khan A. D., Prado C. M., Xiao J., Liu V., Chen W. Y., Meyerhardt J., Albers K. B., and Caan B. J. (2020) Evaluation of automated computed tomography segmentation to assess body composition and mortality associations in cancer patients, Journal of Cachexia, Sarcopenia and Muscle, 11, 1258–1269, doi: 10.1002/jcsm.12573.
References
- 1. Shachar SS, Williams GR, Muss HB, Nishijima TF. Prognostic value of sarcopenia in adults with solid tumours: a meta‐analysis and systematic review. Eur J Cancer 2016;57:58–67. [DOI] [PubMed] [Google Scholar]
- 2. Prado CM, Cushen SJ, Orsso CE, et al. Sarcopenia and cachexia in the era of obesity: clinical and nutritional impact. Proc Nutr Soc 2016;75:188–198. [DOI] [PubMed] [Google Scholar]
- 3. Prado CM, Birdsell LA, Baracos VE. The emerging role of computerized tomography in assessing cancer cachexia. Curr Opin Support Palliat Care 2009;3:269–275. [DOI] [PubMed] [Google Scholar]
- 4. Heymsfield S, Ross R, Wang Z, Frager D. Imaging techniques of body composition: advantages of measurement and new uses. Emerging technologies for nutrition research: Potential for assessing military performance capability 1997;127–150. [Google Scholar]
- 5. Decazes P, Rouquette A, Chetrit A, Vera P, Gardin I. Automatic measurement of the total visceral adipose tissue from computed tomography images by using a multi‐atlas segmentation method. J Comput Assist Tomogr 2017;42(1):139–145. [DOI] [PubMed] [Google Scholar]
- 6. Kamiya N, Zhou X, Chen H, Muramatsu C, Hara T, Yokoyama R, et al. Automated segmentation of psoas major muscle in X‐ray CT images by use of a shape model: preliminary study. Radiol Phys Technol 2011;5:5–14. [DOI] [PubMed] [Google Scholar]
- 7. Kamiya N, Zhou X, Chen H , Muramatsu C , Hara T , Yokoyama R , Kanematsu M , Hoshi H , Fujita H Automated segmentation of recuts abdominis muscle using shape model in X‐ray CT images. In. 2011 Annual International Conference of the IEEE Engineering in Medicine and Biology Society: IEEE; 2011. [DOI] [PubMed]
- 8. Karami E, Wang Y, Gaede S, Lee T, Samani A. Anatomy‐based algorithm for automatic segmentation of human diaphragm in noncontrast computed tomography images. J Med Imag 2016;3:046004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kullberg J, Hedström A, Brandberg J, Strand R, Johansson L, Bergström G, et al. Automated analysis of liver fat, muscle and adipose tissue distribution from CT suitable for large‐scale studies. Sci Rep 2017;7:1–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lee H, Troschel FM, Tajmir S, Fuchs G, Mario J, Fintelmann FJ, et al. Pixel‐level deep segmentation: artificial intelligence quantifies muscle on computed tomography for body morphometric analysis. J Digit Imaging 2017;30:487–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lee SJ, Liu J, Yao J, Kanarek A, Summers RM, Pickhardt PJ. Fully automated segmentation and quantification of visceral and subcutaneous fat at abdominal CT: application to a longitudinal adult screening cohort. Br J Radiol 2018;9:20170968, 10.1259/bjr.20170968:20170968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Polan DF, Brady SL, Kaufman RA. Tissue segmentation of computed tomography images using a random forest algorithm: a feasibility study. Phys Med Biol 2016;61:6553–6569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Takahashi N, Sugimoto M, Psutka SP, Chen B, Moynagh MR, Carter RE. Validation study of a new semi‐automated software program for CT body composition analysis. Abdom Radiol 2017;42:2369–2375. [DOI] [PubMed] [Google Scholar]
- 14. Wang Y, Qiu Y, Thai T, Moore K, Liu H, Zheng B. A two‐step convolutional neural network based computer‐aided detection scheme for automatically segmenting adipose tissue volume depicting on CT images. Comput Methods Programs Biomed 2017;144:97–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Popuri K, Cobzas D, Esfandiari N, Baracos V, Jägersand M. Body composition assessment in axial CT images using FEM‐based automatic segmentation of skeletal muscle. IEEE Trans Med Imaging 2016;35:512–520. [DOI] [PubMed] [Google Scholar]
- 16. Caan BJ, Meyerhardt JA, Kroenke CH, Alexeeff S, Xiao J, Weltzien E, et al. Explaining the obesity paradox: the association between body composition and colorectal cancer survival (C‐SCANS study). Cancer Epidemiol Biomarkers Prev 2017;26:1008–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Caan BJ, Cespedes Feliciano EM, Prado CM, Alexeeff S, Kroenke CH, Bradshaw P, et al. Association of muscle and adiposity measured by computed tomography with survival in patients with nonmetastatic breast cancer. JAMA Oncol 2018;4:798–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bradshaw PT, Cespedes Feliciano EM, Prado CM, Alexeeff S, Albers KB, Chen WY, et al. Adipose tissue distribution and survival among women with nonmetastatic breast cancer. Obesity 2019;27:997–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Brown JC, Caan BJ, Prado CM, Cespedes Feliciano EM, Xiao J, Kroenke CH, et al. The association of abdominal adiposity with mortality in patients with stage i‐iii colorectal cancer. JNCI: J Natl Cancer Inst 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. TomoVision . sliceOmatic Alberta Protocol. http://www.tomovision.com/Sarcopenia_Help/index.htm (February 11, 2017; date last accessed).
- 21. Commandeur F, Goeller M, Betancur J, Cadet S, Doris M, Chen X, et al. Deep learning for quantification of epicardial and thoracic adipose tissue from non‐contrast CT. IEEE Trans Med Imaging 2018;37:1835–1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ozola‐Zālīte I, Mark EB, Gudauskas T, Lyadov V, Olesen SS, Drewes AM, et al. Reliability and validity of the new VikingSlice software for computed tomography body composition analysis. Eur J Clin Nutr 2018;73:54–61. [DOI] [PubMed] [Google Scholar]
- 23. Belharbi S, Chatelain C, Hérault R, Adam S, Thureau S, Chastan M, et al. Spotting L3 slice in CT scans using deep convolutional network and transfer learning. Comput Biol Med 2017;87:95–103. [DOI] [PubMed] [Google Scholar]
- 24. Williams GR, Deal AM, Muss HB, Weinberg MS, Sanoff HK, Guerard EJ, et al. Frailty and skeletal muscle in older adults with cancer. J Geriatr Oncol 2017;9:68–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Shachar SS, Deal AM, Weinberg M, Williams GR, Nyrop KA, Popuri K, et al. Body composition as a predictor of toxicity in patients receiving anthracycline and taxane‐based chemotherapy for early‐stage breast cancer. Clin Cancer Res 2017;23:3537–3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. von Haehling S, Morley JE, Coats AJS, Anker SD. Ethical guidelines for publishing in the Journal of Cachexia, Sarcopenia and Muscle: update 2019.J Cachexia Sarcopenia Muscle 2019;10:1143–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Examples of scans excluded from analysis in blinded review of DICOM images
Table S1. Agreement between ABACS segmentations of muscle and adipose tissue compared to manual analysis among non‐metastatic colorectal and breast cancer patients


