Abstract
Coronaviruses are single stranded RNA viruses usually present in bats (reservoir hosts), and are generally lethal, highly transmissible, and pathogenic viruses causing sever morbidity and mortality rates in human. Several animals including civets, camels, etc. have been identified as intermediate hosts enabling effective recombination of these viruses to emerge as new virulent and pathogenic strains. Among the seven known human coronaviruses SARS-CoV, MERS-CoV, and SARS-CoV-2 (2019-nCoV) have evolved as severe pathogenic forms infecting the human respiratory tract. About 8096 cases and 774 deaths were reported worldwide with the SARS-CoV infection during year 2002; 2229 cases and 791 deaths were reported for the MERS-CoV that emerged during 2012. Recently ~ 33,849,737 cases and 1,012,742 deaths (data as on 30 Sep 2020) were reported from the recent evolver SARS-CoV-2 infection. Studies on epidemiology and pathogenicity have shown that the viral spread was potentially caused by the contact route especially through the droplets, aerosols, and contaminated fomites. Genomic studies have confirmed the role of the viral spike protein in virulence and pathogenicity. They target the respiratory tract of the human causing severe progressive pneumonia affecting other organs like central nervous system in case of SARS-CoV, severe renal failure in MERS-CoV, and multi-organ failure in SARS-CoV-2. Herein, with respect to current awareness and role of coronaviruses in global public health, we review the various factors involving the origin, evolution, and transmission including the genetic variations observed, epidemiology, and pathogenicity of the three potential coronaviruses variants SARS-CoV, MERS-CoV, and 2019-nCoV.
Electronic supplementary material
The online version of this article (10.1007/s13337-020-00632-9) contains supplementary material, which is available to authorized users.
Keywords: Coronavirus, SARS-CoV, MERS-CoV, SARS-CoV-2, COVID-19, Epidemiology
Introduction
Viral respiratory infections represent the major cause of morbidity and mortality in humans and animals worldwide since early 1930’s [15, 45, 96]. Almost about 200 antigenically different type of influenza viruses, rhino viruses, adenoviruses, coronaviruses, metapneumoviruses, orthopneumo viruses are known to cause respiratory illness in humans [45]. Among these, family of Coronaviridea is considered as one of the major viruses causing lethal respiratory infections [27]. They are crown shaped-enveloped-RNA viruses, widely spread, and are capable of infecting a wide range of hosts including human and other mammals [14, 88]. They are majorly divided into four sub-groups namely alpha, beta, gamma, and delta viruses of which Betacoronavirus genus are pathogenic to human [16, 34]. HCoV-229E, HCoV-OC43, HCoV-NL63, HCoV-HKU1, Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV), MERS-CoV (Middle East Respiratory Syndrome Coronavirus), and the recent SARS-CoV-2 (Severe Acute Respiratory Syndrome Coronavirus-2 otherwise known as COVID-19) are the seven types of Betacoronoviruse genus well-known for their respiratory illness caused in humans and are capable of co-evolving, which enhances their pathogenicity and the property of long term adaptation to human host [16, 99]. SARS-CoV, MERS-CoV, and SARS-CoV-2 are the potential members of the Betacoronoviridae family that are very well known for their pandemic outbreak of lethal respiratory infections in human whereas the other types are associated with mild respiratory illness [26]. Thus, the knowledge synthesis, new insights, and comprehensive information about these viruses is essential, in particular as consolidated and comprehensive information at one single source. With this motivation, the present review deals with the origin, spread, genetic variance, etc. of the deadly human respiratory pathogenic viruses SARS-CoV, MERS-CoV, and the SARS-CoV-2 (COVID-19), particularly in view of awareness, importance, and recent global pandemic.
Source, origin, and transmission of the pandemic coronaviruses causing severe acute respiratory syndrome
Corona virus is a fast-evolving virus as it has the ability to infect wide range of hosts by developing diverse genome variations and properties [16]. Generally, two different lineages of evolution of coronavirus are known, based on the phylogenic studies, namely the avian lineage and the mammalian lineage [99]. Avian lineage includes the gammacoronavirinae and deltacoronavirinae as commonly found pathogens of avian species whereas the mammalian lineage comprises the alphacoronavirinae and betacoronavirinae pathogens that infects the mammals like bats, murine, bovine, humans, etc., [92, 106]. Studies on the evolution and transmission of the coronavirus have always stated the importance of bats in the transmission of the virus over all geographical location [66]. The special features of bats and their huge species variation make them an important ancestral host of various viruses. Further, their unique behavior of multiple-species co-existence in a single colony, their long life span, and their long flying habits across geographical locations has helped in frequent evolution of recombinant viral pathogens and inter-species transmission [6, 35, 49, 117].
Balboni et al. [11] has reported that bats acted as the primary host for the evolution and spread of the first pandemic transmissible lethal respiratory SARS-CoV virus and civets as intermediate host [41]. SARS-CoV genome has shown 87–92% sequence homology to the SARS like coronavirus isolated from the bats through anal swabs and the one isolated from civets with a 29-nucleotide signature site in their genome. Whereas, the spike protein of the SARS-CoV was found to play an important role in differentiating it from the other SARS like CoV due to the presence of a highly variable S1 domain that attach to the host cell receptor namely ACE2 and the S2 domain (96% homology with SRAS like CoV) that fuses to the host cell membrane following a similar fusion mechanism as observed in the SARS like CoV viruses. Further, CoV virus isolated from the palm civets and raccoon dogs has shown 99.8% sequence homology with the human CoV viruses. CoVs isolated from the animals showed the presence of 29 nucleotide sequence upstream N gene in the C-terminal region, that was present in the human virus during initial phase of SARS-CoV infection and was not found in the viruses isolated from the later phase [16, 72, 85]. Similarly, [85] have also listed the animals that acted as intermediate host like red fox, cats, field rats, goose, ferret-badger, and wild boar.
MERS-CoV being the second most virulent virus of the genus betacoronaviridae, that caused severe to lethal respiratory infection in human was also found to be transmitted through bats, which acted as the ancestral host and camels as the possible intermediate host [6, 117]. Further, MERS-CoV was found to share genetic homology with many bat CoV species worldwide, which included the Egyptian tomb bats [5, 6, 43, 52], African bats [6, 25], Italian bats [66], and Chinese bats [56]. Similar to SARS-CoV, MERS-CoV also has two domains S1 and S2, of which S1 is the variant peptide that binds to the receptor dipeptidyl peptidase 4 (DPP4). S1 domain is located in the 240 residue of the C-terminal receptor binding domain and are composed of a core and external subdomain unit enabling their attachment, fusion, and entry in to the host cells [78]. Further, [67] has stated that MERS-CoV receptor binding domain was able to efficiently interact with Jamaican fruit bat DPP4 receptor and also stated that the MERS-CoV virus was able to replicate in the Jamaican fruit bat cell line.
Recently evolved SARS-CoV-2 (COVID-19), the seventh known coronavirus has shown similar properties of SARS-CoV with the presence of a highly variable S1 sub-unit of spike protein. It was also found that about five of the six residues of S1 domain differ in SARS-CoV-2 compared to the S1 domain of SARS-CoV virus [53, 101, 107, 116]. Further, the presence of polybasic cleavage site namely the three O-linked glycan [17] in the intersection of the S1 and S2 sub-domains also appear to play a vital role in the infective properties of the virus [100]. Although, the source, reservoir, and cause of transmission of SARS-CoV-2, however, is still not well understood, bat is strongly suspected as the ancestral host potentially including an intermediate animal host [4, 109].
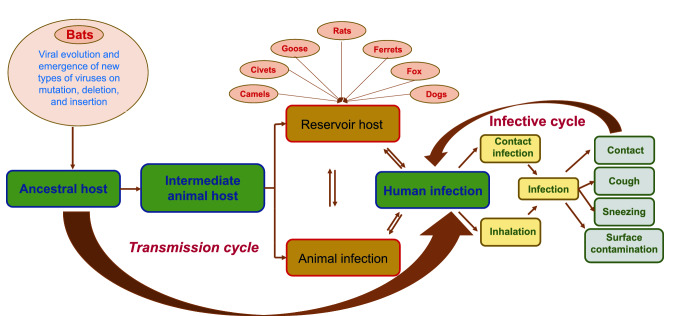
Thus, it is inferred that the variation in the S1 and S2 domains of the spike protein act as the major site of mutation, insertion, and deletion of genes during every transformation of the SARS-CoV-2 virus. This enables the evolution of the new infective virus forms that are capable of efficiently binding to the receptor binding domain, further enabling the effective invasion of viral particles into the human cells. Figure 1 provides an overview of the possible transmission cycle followed by the SARS-CoV, MERS-CoV, and SARS-CoV-2 in recent times. The general mode of transmission is initiated from the ancestral host, i.e. bat to other animals or directly to human. Infected animal develops symptoms or remain as reservoir host and transmit infection to human and other animals. Further, infected human spread infection to other human and animals through various modes that includes contact, surface infection, aerosolization of the virus particle during sneezing, coughing, etc., and sometime in the excretions [36, 38, 58, 61, 70, 75, 93, 105, 115].
Fig. 1.
Schematic representation of the origin, source, and transmission of the coronavirus
Genetic evolution of the three coronaviruses
Genetic evolution of the RNA viruses, especially the Coronaviridae family of the order Nidovirales with genome length of ~ 30 Kb long is generally rapid when compared to all other organisms [82]. This property of rapid genetic evolution helps these lethal virus types (SARS-CoV, MERS-CoV, and SARS-CoV-2) to jump the species barriers and cause severe morbidity and mortality to humankind. Generally, viral genomes encode the protein, which is pertinent to three functions, namely proteins for replication and transcription, structural proteins, and the proteins that enables the infectivity of the virus.
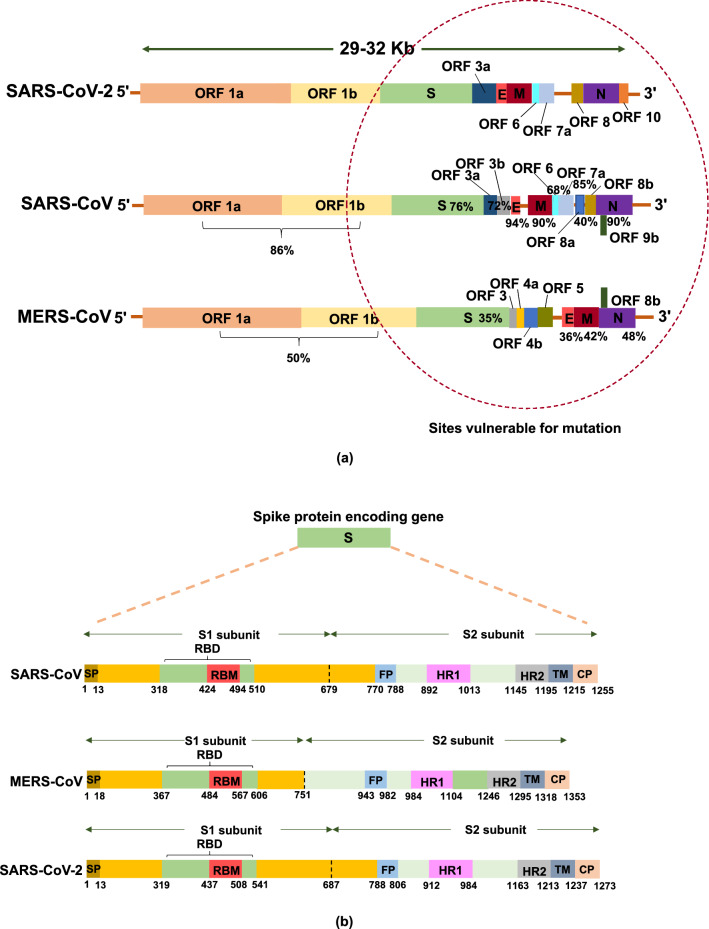
Viral genome contains open reading fragments (ORF) 1a and b (Fig. 2a), those are transcribed in to non-structural protein RNA-dependent-RNA-polymerase, which helps in the viral RNA replication and transcription, also called as R protein [64, 104]. These proteins help in the replication of the viral RNA and also in generating nested set of sg mRNAs (sub-genomic mRNA) coding for various structural and accessory proteins, and accounts for two thirds of the viral genome [79, 84]. The non-structural proteins encoded by ORF 1b (protein 12–16) is comparatively less expressed than the proteins encoded by ORF 1a (protein 1–11) [74]. Various non-structural proteins that play a vital role in the viral multiplication are as follows: protein 12 plays an important role in the viral RNA replication and transcription [32], protein 14, which is a 3′–5′ exonuclease with a clear proofreading activity [63], protein 15 with unclear endo-ribonuclease activity [44], protein 7 and 8, which acts as activating co-factors of polymerase, protein 10, acts as 2′ O-methyltransferase [30, 86, 114], protein 3, 4, and 6, are three transmembrane proteins that acts as anchoring protein of replicative transcription complex to the membrane [47], protein 13, a conserved region helps in unwinding the RNA from 5′–3′ direction [44, 51, 91], and protein 9, unique to coronavirus that protects the genome from degradation during replication [84, 62]. Downstream this are the S protein regions encoding the spike glycoproteins, M region coding the membrane proteins, E region coding for the envelop protein, and the N region encoding nucleocapsid. The ORFs scattered between these genes are the specific site of mutation, deletion and insertion that helps in the evolution of recombinant lethal virus types.
Fig. 2.
Schematic representation of the coronavirus RNA genome: a comparison of the SARS-CoV, MERS-CoV, and SARS-CoV-2 viral genome with the % sequence homology shared by SARS-CoV and MERS-CoV with the newly evolved SARS-CoV-2, b comparison of the spike protein (S) of SARS-CoV, MERS-CoV, and SARS-CoV-2-ORF (open reading frame), RBD (receptor-binding domain), NTD (N-terminal domain), CTD (C-terminal domain), FP (fusion peptide), and HR1-2 (heptad repeats 1–2)
Figure 2a shows the comparison of the genome structure and the genetic variance observed in the pandemic lethal viruses namely SARS-CoV, MERS-CoV, and SARS-CoV-2. The region marked with red dotted circle in Fig. 2a exhibits the region of the genome that are liable for mutation, deletion, and insertion enabling the evolution of the new viruses with improved virulence. From the available information, it is inferred that the SARS-CoV viruses have evolved from bat coronaviruses (Bat-CoVs) like the other human coronaviruses (HCoVs) as a zoonotic virus that has jumped the species barriers from bats to human involving civets as intermediate hosts [102]. It was found to share genetic homology with the other known HCoVs.
SARS-CoV virus was a genetically complex virus compared to the other earlier types of HCoV with a genome length of ~ 30 Kb. It was found to have specific functions that improved their virulence to the host cells. SARS-CoV possessed a large number of unique recombinant regions in the ORF 12 in the RNA-dependent-RNA-polymerase, ORF 9, ORF 10, and ORF 14 regions, which have resulted from a series of horizontal transmission and recombination. The 29-nucleotide deletion in the SARS-CoV in the ORF 8 region creating two segments namely ORF 8a and b has been widely discussed as the vital reason for the improved virulence in the SARS-CoV [72]. Initial isolates obtained from animals and human has shown the presence of single ORF 8 gene, whereas the isolates obtained during the middle and later phase of infection from human has shown only the presence of strains with 29-nucleotide deletion. This reveals the fact that the 29-nucleotide deletion enabled the strain to spread effectively from human to human [36, 50]. Further, the species transmission and adaptation to human host is said to be mainly achieved by the mutation in the S protein (Fig. 2b). The spike protein (S) contains a receptor binding domain that recognizes the human angiotensin-converting enzyme 2 (ACE2) as its receptor [55, 83]. A cascade of other proteins encoded by the genes present in the S region enables the viral invasion in to the host cell (Fig. 2b) which are as follows: cytoplasmic domain (CP), which helps in transport of the virions, fusion protein (FP), plays a vital role in the fusion of the virion to the cell membrane, heptad repeats (HP), helps in the structural modification of the spike proteins, receptor binding site (RBD), helps in recognition and binding to receptors, receptor binding motif (RBM), enhances attachment to the cell receptor, signal peptide (SP), and the transmembrane protein (TM), enabling transmission of the virion across the membrane [110]. Over expression of the protein 3b and 7a, the main protein, helps in the cell apoptosis and cell cycle arrest at the initial stage [89, 111, 112]. Moreover, the protein 3b and 6 play a vital role by functioning as interferon antagonists [76]. Furthermore, it was interesting to know that SARS-CoV lack the hemagglutinin-esterase protein, which acts as a major virulence factor in other HCoVs. Consequently, there is no observable space between R and S protein for the existence of gene encoding the protein hemagglutinin-esterase, perhaps the artifacts present in the neighboring region provides the evidence of large-scale recombination [48, 87]. This emphasizes the fact that on evolution, all the above stated factors have highly favored the viral attachment and invasion into the host cells enabling its pathogenicity.
The sixth variant of the coronavirus is the MERS-CoV, evolved from bats as the evolutionary host infecting human host by jumping species with Camels (especially Camelus dromedarius) as intermediate host [6, 117, 9, 24]. It has a single stranded RNA (ssRNA) genome of length ~ 29Kb encoding replicase polyprotein ORF 1a and b, structural proteins E, N, and M, surface glycoprotein S, and five non-structural protein coded by ORF 3, 4a, 4b, and 5 (Fig. 2a) [59]. Spike (S) protein of the MERS-CoV is cleaved into S1 and S2 domains (Fig. 2b), which helps in the membrane fusion by engaging the DPP4 receptors found in human cell surface associated with immune regulation, signal transduction, and apoptosis [78]. S1, a 240-residue receptor binding domain located in the C-terminal is composed of a core and external sub-domain that helps in the recognition of the DPP4 receptors [66]. Virion enters the host cells with the fusion enabled by the RBD region (Fig. 2b) with the cell receptors, which triggers a cascade of conformational changes leading to the formation of pre-hairpin intermediate of S2 protein. From this pre-hairpin structures the hydrophobic HR1 subunit extends and enables the fusion by inserting the fusion peptide into the host cell membrane. Then this intermediate refolds with HR2 forming six-helix bundle structures that pulls the host cell membrane much closer to the viral envelop enhancing the fusion [110, 77]. Further, [46] has reported that the isolates of MERS-CoV obtained from South Korean patients have shown the presence of 29-nucleotide inclusion and 12 amino acid variants compared to the genomes obtained from the other MERS-CoV strains. Similarly, [65] stated that though 99% nucleotide homology was shared between the 8 strains isolated from South Korea with Riyadh strain, about 13 variant nucleotides were observed in 24–27 nucleotides position across the genome namely 6 variations in ORF1ab gene, 5 in S gene, and 1 each in ORF 4a and ORF 5 evidencing the micro-evolution in the MERS-CoV during an out-break.
Recently evolved SARS-CoV-2 (COVID-19) is the seventh variant of the coronaviruses known to date. It has demonstrated high sequence similarity with SARS-CoV compared to MERS-CoV (Fig. 2a) [33]. Even though, there is only little information available about the genetic evolution of SARS-CoV-2, many studies are underway in understanding its genetic structure and evolution. From the available research reports, it is understood that the Spike (S) proteins (Fig. 2b) acts as the receptor binding domain and binds to the human ACE2 receptors much efficiently than the SARS-CoV. Wan et al. [101] have reported in their study on the S protein of SARS-CoV-2 that the six amino acids residues of the protein play a vital role in binding the virus to the ACE2 receptor and also in the host specificity. Further, they have also reported that five of the six amino acids differ in SARS-CoV-2 compared to SARS-CoV, this feature has enhanced its affinity and specificity to the ACE2 receptor of human with high receptor homology. The receptor binding domain of the SARS-CoV-2 has a core structure and receptor binding motif, which specifically recognizes the structure of the ACE2 receptor, stabilizes the two-virus binding hot-spots in the ACE2 receptors, and helps in binding the virion [55, 83]. Hence, these receptor binding motifs are highly variable among the different strains reported and are highly vulnerable for mutation deciding the host specificity and host range [109]. Shang et al. [83] has also suggested that the property of confirmation of the loops in human ACE2 binding ridge by receptor binding domain of SARS-CoV-2 is a significant difference observed in SARS-CoV-2 and SARS-CoV. Similarly, [4] suggested that the high specificity observed in the S protein of SARS-CoV-2 to human ACE2 is mainly based on the natural selection occurred in the human or human like ACE2 receptor. Structural changes observed in the human ACE2 binding ridge caused by the four amino acid residue motifs namely Gly-Val-Glu-Gly (residue position 482–485) has made the ridge more compact enabling better contact with the N-terminal helix of human ACE2 [109, 83]. The second important feature of the SARS-CoV-2 is the polybasic cleavage site located at the junction of the two subunits of the spike protein S1 and S2 [100], which are effectively cleaved by furin and other protease enzyme deciding the viral infectivity and the host range [68]. Furthermore, the unique feature observed is the 12-nucleotide insertion encoding proline probably plays an important role in the addition of three O-linked glycans that flank the polybasic cleavage site [17, 4]. Similar insertion of polybasic cleavage site in the hemagglutinin protein of the avian influenza virus has shown remarkable improvement in its pathogenicity [3]. Further, the possession of the O-linked glycan is suspected to act as a mucin-like domain, which might help in shielding the epitopes or the key virulent residues of the S protein of SARS-CoV-2 [10]. Additional detailed studies on the structure and genetic evolution of the recently evolved SARS-CoV-2 would enable us the better understanding of the viral virulence, transmission, and help us in finding a suitable vaccine.
Epidemiology and pathogenicity
HCoVs are known for their mild respiratory infections in human like common cold since 1960 and many other HCoVs have been identified till recent times. There were only two widely known HCoVs that were infecting human with mild respiratory symptoms namely HCoV-229E and HCoV-OC43 till the emergence of the SARS-CoV in 2003, that caused a severe acute respiratory syndrome with severe morbidity and mortality (Fig. 3a). Later, another variant of the coronavirus namely the MERS-CoV emerged for the first time in 2012 [31]. Further, the seventh variant of coronavirus has evolved recently during late 2019 that has emerged and caused widespread lethal upper respiratory disease taxing many lives worldwide (Fig. 3a). The pre-requisite of this genetic variance is the evolution, which is favored by the more flexible recombination property observed in the coronavirus. Genome of coronavirus exhibits several intrinsic point mutations in the order of about 1 × 10− 6 per site in the RNA-dependent-RNA-polymerase gene [57, 97] and possess a property of acquiring 100–1000 base-pair fragments from other coronaviruses during co-infection in single species [108]. These properties enable the virus to reemerge into a new type recombinant strain with improved virulence and wide range of host selection challenging the humankind.
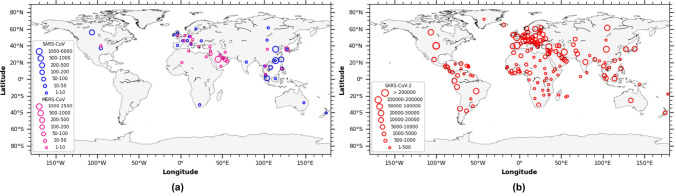
Fig. 3.
Global distribution based on the epidemiologic data of coronavirus: a Number of incidences observed globally for SARS-CoV and MERS-CoV outbreak, and b Number of incidences observed globally for SARS-CoV-2 outbreak until April 2020 (https://www.who.int/csr/sars/country/table2004_04_21/en/; https://www.who.int/csr/disease/coronavirus_infections/risk-assessment-august-2018.pdf; https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200420-sitrep-91-covid-19.pdf?sfvrsn=fcf0670b_4)
Symptoms observed in the patients infected with SARS-CoV include fever, myalgia, malaise, headache, and chills, which are generally followed by dry cough, dyspnea, and respiratory distress [87]. Initially the symptoms start with mild cold and fever and progress to Severe Acute Respiratory Illness (SARI) eventually causing multiple organ failure [35]. In acute cases the respiratory distress lasts for 5–7 days and ultimately leads to death. Other symptoms observed in patients with SARS-CoV are infection of gastrointestinal tract, kidney, liver, and brain. About 30–40% of the patients with gastrointestinal symptoms develop diarrhea and lymphopenia. Alveolar damage, hemophagocytosis, epithelial cell proliferation, and increased counts of macrophages are the symptoms developed in the lungs of the patients suffering SARS-CoV infection [87]. On the other hand, it invades the central nervous system through the olfactory route and cause severe neurological syndrome that could lead to encephalitis, other types of encephalopathy, and neuronal cell death [69, 94]. About 8096 cases with 20% of them suffering with SARI [95, 13] were reported worldwide covering more than 30 different countries with 774 (9%) deaths (Fig. 4a and Table S1) in 2003 outbreak [12, 21, 22]. It initially originated from the Foshan, Guangdong Province in southern China during November 2002 [20] and was found to spread to more than 30 countries including Asia, Europe, and North America in less than 5 months’ time (Fig. 3a). Being the place of origin, China reported about 5327 cases with 349 deaths followed by the Hong Kong with 1755 cases and 299 deaths. Following these countries, Taiwan and Canada reported 346 and 251 cases, and 37 and 43 deaths, respectively indicating the highest death rate in Canada compared to Taiwan (Fig. 3a and Table S1). Further, epidemiological studies have revealed that elderly people were found more susceptible to the infection with a mortality rate of about 50% and people working in food industries and people working as animal handlers were found to be the next vulnerable community to SARS-CoV [36]. Initial infection was observed among the people who were in contact with animals like civets in markets and later was found to spread causing severe infection in human population. The isolate obtained during 2002–2004 infection was found to have identical genome compared to the one isolated from the civets but with very high divergence improving its infectivity to human cells [38]. Rapid spread of the SARS-CoV was found to get transmitted from human to human through droplets, aerosols, and contaminated fomites [75]. A single infected individual could cause secondary infection to as many as 2.2–3.7 individual which is comparatively less to other known respiratory viral infection like Influenza etc. Whereas, countries with moderate to large infection (Hong Kong, Taiwan, and Canada) were found to experience super spreading events which enabled substantially large-scale transmission of the virus among human population (Fig. 3a) [87]. The recent studies have also highlighted the role and importance of international travel dynamics on the rapid spread of SARS-CoV-2 [37].
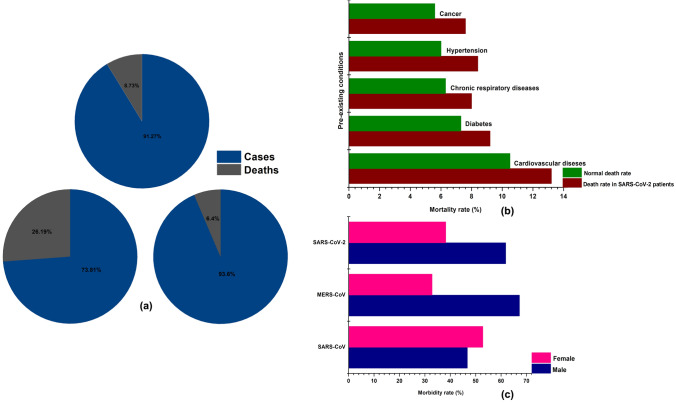
Fig. 4.
Epidemiological data of SARS-CoV, MERS-CoV, and SARS-CoV-2: a total number of cases and deaths observed during the outbreak, b death rates observed in individuals with SARS-CoV-2 infection and pre-existing comorbidities, c demographic information related to the morbidity ratio observed in male and female for SARS-CoV, MERS-CoV, and SARS-CoV-2 (https://www.worldometers.info/coronavirus/coronavirus-age-sex-demographics/)
MERS-CoV, sixth variant of coronavirus caused severe respiratory diseases leading to renal failure with a case fatality rate of about 35% (Fig. 4a) [66, 46]. It was first isolated from a 60 years old patient died out of severe respiratory illness during 2012 in Jeddah, Saudi Arabia [113]. Since then around 27 countries have reported MERS-CoV infection (Fig. 3a). Clinical symptoms of the viral infection include rapid progressive acute pneumonia leading to acute renal failure resulting in death which is a unique symptom of MERS-CoV compared to SARS-CoV [118]. The progressive stages of symptomatic pneumonia are respiratory distress, septic shock, consumptive coagulopathy, pericarditis, and renal failure which initially begin with fever, cough, chills, sore throat, myalgia, arthralgia, dyspnea leading to pneumonia [2, 7, 8]. Most of the cases end up with acute renal impairment and about one-third of them were found to suffer severe gastrointestinal symptoms including diarrhea and vomiting [28]. With the observed initial phase of infection confined to Middle East countries including Saudi Arabia, Qatar, Jordan, and United Arab Emirates, secondary spread through person to person contact was observed in countries like United Kingdom, Germany, France, Italy, and Tunisia (Fig. 3a) [87,2,8]. Total incidences reported worldwide was about 2229 cases (Table S1) with a mortality of 791 (35%) (Fig. 4a) which is relatively high compared to SARS-CoV with mortality rate of 9%. Middle East nation Saudi Arabia was found to report the highest number of cases of about 1854 followed by South Korea with 185 cases. Other countries with moderate infection were United Arab Emirates, Qatar, and Oman with 86, 19 and 11cases respectively. Whereas, the country wise death rate of the MERS-CoV is still not available (Table S1). Camels of Middle East and Africa played an imperative role in the evolution of the different recombinant variants of MERS-CoV as these animals were carrying multiple-lineage of the MERS-CoV in them resulting in the outbreak occurred during 2015 and 2018 in Middle East and Republic of Korea [1, 65, 80]. Further, it was estimated that it had an incubation period of 2–14 days involving an infection period of 11 days with a super spreading event rate of 83% [71].
SARS-CoV-2 (COVID-19), the recently evolved strain of the coronavirus during 2019, emerged in to a severe respiratory pathogen causing extensive morbidity and mortality globally covering about 210 countries and territories (Fig. 3b), was initially reported in Wuhan, Hubei province, China [116, 108]. As on 20th April 2020, it was found to cause 2,430,923 cases and 166,278 deaths (~ 7%) globally (Fig. 4a and Table S1). Though evolved in China, the incidences and deaths observed in China (82,747; 4632 respectively) are considerably low compared to other nations like United States of America (767,379; 40,750 respectively), Spain (200,210; 20,852 respectively), Italy (178,972; 23,660 respectively), France (152,894; 19,718 respectively), Germany (145,743; 4642 respectively), and United Kingdom (120,067; 16,060 respectively) with very high morbidity and mortality (Fig. 3b and Table S1) (Data for SARS-CoV-2 shown as on 20 April 2020). Whereas, high incidences of cases ranging from 10,000 to 100,000 was observed in many countries namely Saudi Arabia, Chile, South Korea, Japan, Israel, Sweden, Austria, Ireland, Peru, India, Portugal, Switzerland, Netherlands, Canada, Brazil, Belgium, Russia, Iran, and Turkey (Fig. 3b). Other countries have shown relatively moderate to low incidences of cases and deaths (Table S1). SARS-CoV-2 infected patients develop deadly pneumonia with associated symptoms including fever, cough, throat pain, shortness of breath [18, 19, 42, 54]. It spread from person to person mainly through contact with the secretions from the infected patients like droplets, aerosols, and also through the contaminated surfaces [98]. Further the infection is classified in to two based on the symptoms namely severe infection developing tachypnoea i.e. rapid breathing almost ≥ 30 breaths/min, oxygen saturation rate ≤ 93% (for patients at rest), and PiO2/FiO2 ratio ≤ 300 mm Hg, and chronic infection with the development of respiratory failure (ventilator dependent), septic shock, multi-organ dysfunction or failure [54]. Further, many patients developed severe liver and kidney impairment along with severe lung injury developing viral sepsis [54]. Also, it was found that about 71.4% of the patients at critical stage had developed intravascular coagulopathy i.e. almost ≥ 5 points according to the International Society on Thrombosis and Haemostasis criteria [90]. The exact reason for the intravascular coagulopathy is still not well understood and it is suspected that the over expression of the ACE2 receptors could directly attack the vascular endothelial cells leading to abnormal coagulation and sepsis [39]. Moreover, overexpression of ACE2 receptors in circulatory system could lead to blood hypotension as ACE2 plays a vital role in regulating blood pressure levels [103]. Furthermore, furin mediated pre-cleavage of the S protein plays an imperative role in the viral entry to the lung cells, especially through the proteolytic S protein activation by TMPRSS2 present in the epithelial and lung cells that hinder the expression of required levels of cathepsin L enabling the rapid invasion of lung cells by the virions leading to increased mortality levels [40, 73, 81]. Morbidity and mortality were observed to be high in elderly people of age ≥ 60 years and also with the people who already suffered pre-existing underlying conditions like cancer, hypertension, respiratory disease, diabetes, and cardiovascular diseases (Fig. 4b). Among the observed comorbid conditions cardiovascular disease associated SARS-CoV-2 infection has shown the highest mortality rate compared to the others (Fig. 4b). Further, demographic studies have shown that Male (61.8%) were mostly affected compared to Female (38.2%) similar to the scenario observed with MERS-CoV, but SARS-CoV sharing highest homology with SARS-CoV-2 was found to infect Female (52.8%) more than Male (46.7%) (Fig. 4c). All the above-mentioned factors could be the plausible reason for the improved virulence observed in the SARS-CoV-2 virus compared to the other two variants that caused similar respiratory syndrome. However, better insight on the mode of infectivity of the SARS-CoV in terms of its antigenicity, virulence, and spread could help in finding an appropriate vaccine and treatment measures to control the infection.
Concluding remarks
From the detailed review on the origin and evolution of the genetic variants, it is inferred that the bats acted as an important reservoir host and involved an intermediate animal host, enabling human infection of the HCoVs. Inter species mixing in the bats and animal hosts has played an imperative role in the evolution of new recombinant virulent strains that jumped the species barriers causing severe acute respiratory illness to humans with very high morbidity and mortality on global scale. Morbidity and mortality rates were found to increase with each evolution inferring the fact that genetic variations are more complex with each evolution improving the virulence properties of the virion. Prevalence and circulation of these viruses in animals pose a serious health risk to human population. Hence, it is highly essential to maintain a surveillance network to monitor the emergence of the potentially virulent viral strains. Further, it is necessary to understand the various factors involved in the epidemiological spread of the virus in the human population and proper precautionary measures must be laid down to prevent the spread of the infection.
Moreover, it is observed from the various studies worldwide that the spike (S) proteins and their subunits S1 and S2 play a vital role in the viral infection by enabling the viral binding and fusion to the host cell receptors. The subunits S1 and S2 are known to undergo strong genetic variations with each evolution. Though ACE2 is a target receptor of SARS-CoV and SARS-CoV-2, there was observable variance in the sequence homology of the gene encoding the spike protein. This genetic variation has enabled SARS-CoV-2 to bind efficiently to the receptor with firm attachment, improving the virulence compared to SARS-CoV causing very high morbidity and mortality challenging the humankind. Hence, many researchers worldwide have started targeting this region for the potential development of SARS-CoV-2 vaccine. Such advancement in the development of an appropriate vaccine could really solve similar problems that could be encountered in the future.
SARS-CoV-2 being the most potent member of the family has emerged as a great threat to human kind. It has affected a wide range of population belonging to different age group with elderly group of people being the more vulnerable group. Efforts have been made to understand the disease and the clinical features since the onset of the outbreak, the mechanism underlying the infection and huge spread, however, is still unclear. Several hypotheses have been discussed by various clinicians and pathologists globally to discover the mechanism of the SARS-CoV-2 pathogenicity based on the observations made on the autopsied dead individuals, and all such conclusions drawn so far remain either scientifically unvarified or inconclusive in the course of disease progression. At this moment it is important to bring the infection under control by developing an appropriate vaccine and applying treatment measures for controlling the disease spread.
Future research needs and measures to be followed
Future research must be focused on issues that could effectively control the spread and occurrence of disease. Bats are considered as the reservoir host enabling the genetic evolution of the virus, therefore monitoring and surveillance of the bats (reservoir host) and other animals (intermediate host) for the emergence of recombinant viral strain could be a possible solution to minimize the occurrence of the disease. Unprotected contacts with farm animals and wild animals should be minimized to control the spread of the disease. Once the disease occurs in a population, strict measures of isolation of the community from others should be implemented in order to control the further spread of the disease. Above all, healthy lifestyle and hygenic practices would enable us to totally eradicate the disease. Further, a considerable restriction and strong monitoring for the trade and tourism related travel worldwide would definitely help in controlling the spread of the disease across territories.
Better understanding of the viral target cells and receptors is highly essential for the establishment of the appropriate animal models unveiling the infective cycle adopted by the virion to infect the host cells. This would further help in understanding the immune response and adaptation of the immunomodulatory therapies for the effective treatment of the viral infection.
Most importantly, development of an appropriate vaccine could give us a promising result in controlling the morbidity and mortality worldwide. Several vaccines targeting the S proteins have been developed by the researchers and medical practitioners across the globe [29]. For examples, vaccine that block the receptor binding site, full length S protein vaccine, and the ones that blocks the fusion of the virion to the host cells. The efforts to put in these vaccines for effective use against the evolving coronavirus is yet to achieve any conclusive outcome. Currently practiced, plasma administration and broad-spectrum antiviral medications has been found to have promising effect in treating patients suffering from 2019-nCoV [23]. Moreover, administration of monoclonal antibodies could also be adopted for the effective control of the 2019-nCoV. Hence, researchers are aiming in the development of an effective and specific monoclonal antibodies for the treatment of the 2019-nCoV infection [60].
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
Authors acknowledge the financial support from Max Planck Partner Group on Bioaerosol Research at IITM. SK gratefully acknowledge Council of Scientific and Industrial Research (CSIR) for awarding the CSIR-Senior Research Associateship. SSG acknowledge the support and encouragement from the selected faculty colleagues from Dept. of Civil Engineering, IIT Madras. SK and SSG conceived the idea. SK wrote the manuscript with the inputs from SSG, RSV, and BS.
Funding
No funding was available/used for this study.
Availability of data and material
The data, codes, and post processed data used in this this study are available from the corresponding author upon reasonable request.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Ahmed H, Erica Billig R, Holly MB, et al. Middle east respiratory syndrome coronavirus, Saudi Arabia, 2017–2018. Emerg Infect Dis J. 2019;25:2149. doi: 10.3201/eid2511.190726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Al-Tawfiq JA, Hinedi K, Ghandour J, et al. Middle east respiratory syndrome coronavirus: a case-control study of hospitalized patients. Clin Infect Dis. 2014;59:160–5. doi: 10.1093/cid/ciu226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alexander DJ, Brown IH. History of highly pathogenic avian influenza. Rev Sci Tech. 2009;28:19–38. doi: 10.20506/rst.28.1.1856. [DOI] [PubMed] [Google Scholar]
- 4.Andersen KG, Rambaut A, Lipkin WI, Holmes EC, Garry RF. The proximal origin of SARS-CoV-2. Nat Med. 2020;26:450–2. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Annan A, Baldwin HJ, Corman VM, et al. Human betacoronavirus 2c EMC/2012-related viruses in bats, Ghana and Europe. Emerg Infect Dis. 2013;19:456–9. doi: 10.3201/eid1903.121503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Anthony SJ, Gilardi K, Menachery VD, et al. Further evidence for bats as the evolutionary source of middle east respiratory syndrome coronavirus. mBio. 2017;8:e00373–00317. doi: 10.1128/mBio.00373-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arabi YM, Arifi AA, Balkhy HH, et al. Clinical course and outcomes of critically Ill patients with middle east respiratory syndrome coronavirus onfection. Ann Intern Med. 2014;160:389–97. doi: 10.7326/M13-2486. [DOI] [PubMed] [Google Scholar]
- 8.Assiri A, Al-Tawfiq JA, Al-Rabeeah AA, et al. Epidemiological, demographic, and clinical characteristics of 47 cases of Middle East respiratory syndrome coronavirus disease from Saudi Arabia: a descriptive study. Lancet Infect Dis. 2013;13:752–61. doi: 10.1016/S1473-3099(13)70204-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Azhar EI, El-Kafrawy SA, Farraj SA, et al. Evidence for camel-to-human transmission of MERS coronavirus. N Engl J Med. 2014;370:2499–505. doi: 10.1056/NEJMoa1401505. [DOI] [PubMed] [Google Scholar]
- 10.Bagdonaite I, Wandall HH. Global aspects of viral glycosylation. Glycobiology. 2018;28:443–67. doi: 10.1093/glycob/cwy021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Balboni A, Battilani M, Prosperi S. The SARS-like coronaviruses: the role of bats and evolutionary relationships with SARS coronavirus. New Microbiol. 2012;35:1–16. [PubMed] [Google Scholar]
- 12.Braden CR, Dowell SF, Jernigan DB, Hughes JM. Progress in global surveillance and response capacity 10 years after severe acute respiratory syndrome. Emerg Infect Dis. 2013;19:864–9. doi: 10.3201/eid1906.130192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van den Brand JMA, Haagmans BL, van Riel D, Osterhaus ADME, Kuiken T. The pathology and pathogenesis of experimental severe acute respiratory syndrome and influenza in animal models. J Comp Pathol. 2014;151:83–112. doi: 10.1016/j.jcpa.2014.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cavanagh P. The artist as neuroscientist. Nature. 2005;434:301–7. doi: 10.1038/434301a. [DOI] [PubMed] [Google Scholar]
- 15.Cesario TC. Viruses associated with pneumonia in adults. Clin Infect Dis. 2012;55:107–13. doi: 10.1093/cid/cis297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chan PKS, Chan MCW. Tracing the SARS-coronavirus. J Thorac Dis. 2013;5:S118–S121. doi: 10.3978/j.issn.2072-1439.2013.06.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chan C-M, Woo PCY, Lau SKP, et al. Spike protein, S, of human coronavirus HKU1: role in viral life cycle and application in antibody detection. Exper Biol Med 2008;233:1527–36. doi: 10.3181/0806-RM-197. [DOI] [PubMed] [Google Scholar]
- 18.Chan JF-W, Yuan S, Kok K-H, et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020;395:514–23. doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen N, Zhou M, Dong X, et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507–13. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheng VCC, Lau SKP, Woo PCY, Yuen KY. Severe acute respiratory syndrome coronavirus as an agent of emerging and reemerging infection. Clin Microbiol Rev. 2007;20:660–94. doi: 10.1128/CMR.00023-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cherry JD. The chronology of the 2002–2003 SARS mini pandemic. Paediatr Respir Rev. 2004;5:262–9. doi: 10.1016/j.prrv.2004.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cherry JD, Krogstad P. SARS: the first pandemic of the 21st century. Pediatr Res. 2004;56:1–5. doi: 10.1203/01.PDR.0000129184.87042.FC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cohen J. New coronavirus threat galvanizes scientists. Science. 2020;367:492. doi: 10.1126/science.367.6477.492. [DOI] [PubMed] [Google Scholar]
- 24.Coleman CM, Frieman MB. Coronaviruses: important emerging human pathogens. J Virol. 2014;88:5209. doi: 10.1128/JVI.03488-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Corman VM, Ithete NL, Richards LR, et al. Rooting the phylogenetic tree of middle East respiratory syndrome coronavirus by characterization of a conspecific virus from an African bat. J Virol 2014;88:11297–303. doi: 10.1128/JVI.01498-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Desforges M, Le Coupanec A, Dubeau P, et al. Human coronaviruses and other respiratory viruses: underestimated opportunistic pathogens of the central nervous system? Viruses. 2020 doi: 10.3390/v12010014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Desforges M, Le Coupanec A, Stodola JK, Meessen-Pinard M, Talbot PJ. Human coronaviruses: viral and cellular factors involved in neuroinvasiveness and neuropathogenesis. Virus Res. 2014;194:145–58. doi: 10.1016/j.virusres.2014.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Drosten C, Seilmaier M, Corman VM, et al. Clinical features and virological analysis of a case of Middle East respiratory syndrome coronavirus infection. Lancet Infect Dis. 2013;13:745–51. doi: 10.1016/S1473-3099(13)70154-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Du L, He Y, Zhou Y, Liu S, Zheng B-J, Jiang S. The spike protein of SARS-CoV—a target for vaccine and therapeutic development. Nat Rev Microbiol. 2009;7:226–36. doi: 10.1038/nrmicro2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Egloff M-P, Ferron F, Campanacci V, et al. The severe acute respiratory syndrome-coronavirus replicative protein nsp9 is a single-stranded RNA-binding subunit unique in the RNA virus world. Proc Natl Acad Sci USA. 2004;101:3792. doi: 10.1073/pnas.0307877101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Farag EAB, Nour M, El Idrissi A, et al. Survey on implementation of one health approach for MERS-CoV preparedness and control in Gulf Cooperation Council and Middle East Countries. Emerg Infect Dis. 2019;25:e171702. doi: 10.3201/eid2503.171702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gorbalenya AE, Enjuanes L, Ziebuhr J, Snijder EJ, Nidovirales Evolving the largest RNA virus genome. Virus Res. 2006;117:17–37. doi: 10.1016/j.virusres.2006.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grifoni A, Sidney J, Zhang Y, Scheuermann RH, Peters B, Sette A. A sequence homology and bioinformatic approach can predict candidate targets for immune responses to SARS-CoV-2. Cell Host Microbe. 2020;27:671–80.e672. doi: 10.1016/j.chom.2020.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.de Groot RJ, Baker SC, Baric RS, et al. Commentary: middle east respiratory syndrome coronavirus (MERS-CoV)—announcement of the Coronavirus study group. J Virol. 2013;87:7790. doi: 10.1128/JVI.01244-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gu J, Gong E, Zhang B, et al. Multiple organ infection and the pathogenesis of SARS. J Exper Med 2005;202:415–24. doi: 10.1084/jem.20050828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guan Y, Zheng BJ, He YQ, et al. Isolation and characterization of viruses related to the SARS coronavirus from animals in Southern China. Science. 2003;302:276. doi: 10.1126/science.1087139. [DOI] [PubMed] [Google Scholar]
- 37.Gunthe SS, Patra SS. Impact of international travel dynamics on domestic spread of 2019-nCoV in India: origin-based risk assessment in importation of infected travelers. Glob Health. 2020;16:45. doi: 10.1186/s12992-020-00575-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guodong L, Qiuxia C, Jianguo X, et al. Laboratory diagnosis of four recent sporadic cases of community-acquired SARS, Guangdong Province, China. Emerg Infect Dis J. 2004;10:1774. doi: 10.3201/eid1010.040445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hamming I, Timens W, Bulthuis MLC, Lely AT, Navis GJ, van Goor H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J Pathol. 2004;203:631–7. doi: 10.1002/path.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hoffmann M, Kleine-Weber H, Schroeder S, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–80.e278. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hu B, Ge X, Wang L-F, Shi Z. Bat origin of human coronaviruses. Virol J 2015;12:221. doi: 10.1186/s12985-015-0422-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ithete NL, Stoffberg S, Corman VM, et al. Close relative of human Middle East respiratory syndrome coronavirus in bat, South Africa. Emerg Infect Dis. 2013;19:1697–9. doi: 10.3201/eid1910.130946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ivanov KA, Thiel V, Dobbe JC, van der Meer Y, Snijder EJ, Ziebuhr J. Multiple enzymatic activities associated with severe acute respiratory syndrome coronavirus helicase. J Virol 2004;78:5619–32. doi: 10.1128/JVI.78.11.5619-5632.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jartti T, Jartti L, Ruuskanen O, Söderlund-Venermo M. New respiratory viral infections. Curr Opin Pulm Med 2012;18:271–278. doi: 10.1097/MCP.0b013e328351f8d4. [DOI] [PubMed] [Google Scholar]
- 46.Kim Y-J, Cho Y-J, Kim D-W, et al. Complete genome sequence of middle east respiratory syndrome coronavirus KOR/KNIH/002_05_2015, isolated in South Korea. Genome Announc 2015;3:e00787-00715. doi: 10.1128/genomeA.00787-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Knoops K, Kikkert M, Worm SHEvd, et al. SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol. 2008;6:e226–6. doi: 10.1371/journal.pbio.0060226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Langereis MA, van Vliet ALW, Boot W, de Groot RJ. Attachment of mouse hepatitis virus to O-acetylated sialic acid Is mediated by hemagglutinin-esterase and not by the spike protein. J Virol. 2010;84:8970. doi: 10.1128/JVI.00566-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lau SKP, Chan JFW. Coronaviruses: emerging and re-emerging pathogens in humans and animals. Virol J 2015;12:209. doi: 10.1186/s12985-015-0432-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lau SKP, Woo PCY, Li KSM, et al. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc Natl Acad Sci USA. 2005;102:14040–5. doi: 10.1073/pnas.0506735102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee N-R, Kwon H-M, Park K, Oh S, Jeong Y-J, Kim D-E. Cooperative translocation enhances the unwinding of duplex DNA by SARS coronavirus helicase nsP13. Nucl Acids Res 2010;38:7626–36. doi: 10.1093/nar/gkq647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lelli D, Papetti A, Sabelli C, Rosti E, Moreno A, Boniotti MB. Detection of coronaviruses in bats of various species in Italy. Viruses. 2013;5:2679–2689. doi: 10.3390/v5112679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Letko M, Marzi A, Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat Microbiol 2020;5:562–9. doi: 10.1038/s41564-020-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li Q, Guan X, Wu P, et al. Early transmission dynamics in Wuhan, China, of novel coronavirus–infected pneumonia. N Engl J Med. 2020;382:1199–207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li W, Shi Z, Yu M, et al. Bats are natural reservoirs of SARS-like coronaviruses. Science. 2005;310:676. doi: 10.1126/science.1118391. [DOI] [PubMed] [Google Scholar]
- 56.Li Y, Zhiqiang W, Xianwen R, et al. MERS: related betacoronavirus in Vespertilio superans Bats, China. Emerg Infect Dis J. 2014;20:1260. doi: 10.3201/eid2007.140318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lu H, Zhao Y, Zhang J, et al. Date of origin of the SARS coronavirus strains. BMC Infect Dis. 2004;4:3. doi: 10.1186/1471-2334-4-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lun Z-R, Qu L-H. Animal-to-human SARS-associated coronavirus transmission? Emerg Infect Dis. 2004;10:959–9. doi: 10.3201/eid1005.040022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mackay IM, Arden KE. MERS coronavirus: diagnostics, epidemiology and transmission. Virol J. 2015;12:222–2. doi: 10.1186/s12985-015-0439-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Malik YS, Sircar S, Bhat S, et al. Emerging novel coronavirus (2019-nCoV)—current scenario, evolutionary perspective based on genome analysis and recent developments. Vet Q 2020;40:68–76. doi: 10.1080/01652176.2020.1727993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Martina BEE, Haagmans BL, Kuiken T, et al. Virology: SARS virus infection of cats and ferrets. Nature. 2003;425:915–5. doi: 10.1038/425915a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Miknis ZJ, Donaldson EF, Umland TC, Rimmer RA, Baric RS, Schultz LW. Severe acute respiratory syndrome coronavirus nsp9 dimerization is essential for efficient viral growth. J Virol 2009;83:3007–18. doi: 10.1128/JVI.01505-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Minskaia E, Hertzig T, Gorbalenya AE, et al. Discovery of an RNA virus 3′–5′ exoribonuclease that is critically involved in coronavirus RNA synthesis. Proc Natl Acad Sci USA. 2006;103:5108–13. doi: 10.1073/pnas.0508200103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mizutani T, Repass JF, Makino S. Nascent synthesis of leader sequence-containing subgenomic mRNAs in coronavirus genome-length replicative intermediate RNA. Virology. 2000;275:238–43. doi: 10.1006/viro.2000.0489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Moon-Woo S, So Yeon K, Victor Max C, et al. Microevolution of outbreak-associated middle east respiratory syndrome coronavirus, South Korea, 2015. Emerg Infect Dis J. 2016;22:327. doi: 10.3201/eid2202.151700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Moreno A, Lelli D, de Sabato L, et al. Detection and full genome characterization of two beta CoV viruses related to Middle East respiratory syndrome from bats in Italy. Virol J 2017;14:239. doi: 10.1186/s12985-017-0907-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Munster VJ, Adney DR, van Doremalen N, et al. Replication and shedding of MERS-CoV in Jamaican fruit bats (Artibeus jamaicensis) Sci Rep. 2016;6:21878. doi: 10.1038/srep21878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nao N, Yamagishi J, Miyamoto H, et al. Genetic predisposition to acquire a polybasic cleavage site for highly pathogenic avian influenza virus hemagglutinin. mBio. 2017;8:e02298-02216. doi: 10.1128/mBio.02298-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Netland J, Meyerholz DK, Moore S, Cassell M, Perlman S. Severe acute respiratory syndrome coronavirus infection causes neuronal death in the absence of encephalitis in mice transgenic for human ACE2. J Virol 2008;82:7264–75. doi: 10.1128/JVI.00737-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ng SKC. Possible role of an animal vector in the SARS outbreak at Amoy Gardens. Lancet. 2003;362:570–2. doi: 10.1016/S0140-6736(03)14121-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Oh M-D, Park WB, Park S-W, et al. Middle East respiratory syndrome: what we learned from the 2015 outbreak in the Republic of Korea. Korean J Intern Med. 2018;33:233–46. doi: 10.3904/kjim.2018.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Oostra M, de Haan CAM, Rottier PJM. The 29-nucleotide deletion present in human but not in animal severe acute respiratory syndrome coronaviruses disrupts the functional expression of open reading frame 8. J Virol. 2007;81:13876. doi: 10.1128/JVI.01631-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Park J-E, Li K, Barlan A, et al. Proteolytic processing of Middle East respiratory syndrome coronavirus spikes expands virus tropism. Proc Natl Acad Sci. 2016;113:12262. doi: 10.1073/pnas.1608147113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pasternak AO, Spaan WJM, Snijder EJ. Nidovirus transcription: how to make sense…? J Gen Virol 2006;87:1403–1421. doi: 10.1099/vir.0.81611-0. [DOI] [PubMed] [Google Scholar]
- 75.Peiris JSM, Guan Y, Yuen KY. Severe acute respiratory syndrome. Nat Med 2004;10:88–97. doi: 10.1038/nm1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pewe L, Zhou H, Netland J, et al. A severe acute respiratory syndrome-associated coronavirus-specific protein enhances virulence of an attenuated murine coronavirus. J Virol 2005;79:11335–42. doi: 10.1128/JVI.79.17.11335-11342.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Qian Z, Travanty EA, Oko L, et al. Innate immune response of human alveolar type II cells infected with severe acute respiratory syndrome-coronavirus. Am J Respir Cell Mol Biol. 2013;48:742–8. doi: 10.1165/rcmb.2012-0339OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Raj VS, Mou H, Smits SL, et al. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature. 2013;495:251–4. doi: 10.1038/nature12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rota PA, Oberste MS, Monroe SS, et al. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300:1394. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- 80.Sabir JSM, Lam TTY, Ahmed MMM, et al. Co-circulation of three camel coronavirus species and recombination of MERS-CoVs in Saudi Arabia. Science. 2016;351:81. doi: 10.1126/science.aac8608. [DOI] [PubMed] [Google Scholar]
- 81.Sakai K, Ami Y, Nakajima N, et al. TMPRSS2 independency for haemagglutinin cleavage in vivo differentiates influenza B virus from influenza A virus. Sci Rep 2016;6:29430–0. doi: 10.1038/srep29430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sevajol M. Insights into RNA synthesis, capping, and proofreading mechanisms of SARS-coronavirus. Virus Res 2014 doi: 10.1016/j.virusres.2014.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Shang J, Ye G, Shi K, et al. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020 doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Snijder EJ, Bredenbeek PJ, Dobbe JC, et al. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J Mol Biol 2003;331:991–1004. doi: 10.1016/s0022-2836(03)00865-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Song H-D, Tu C-C, Zhang G-W, et al. Cross-host evolution of severe acute respiratory syndrome coronavirus in palm civet and human. Proc Natl Acad Sci USA. 2005;102:2430. doi: 10.1073/pnas.0409608102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Su D, Lou Z, Sun F, et al. Dodecamer structure of severe acute respiratory syndrome coronavirus nonstructural protein nsp10. J Virol 2006;80:7902–8. doi: 10.1128/JVI.00483-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Su S, Wong G, Shi W, et al. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol. 2016;24:490–502. doi: 10.1016/j.tim.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Talbot HK, Griffin MR, Chen Q, Zhu Y, Williams JV, Edwards KM. Effectiveness of seasonal vaccine in preventing confirmed influenza-associated hospitalizations in community dwelling older adults. J Infect Dis 2011;203:500–8. doi: 10.1093/infdis/jiq076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tan Y-J, Fielding BC, Goh P-Y, et al. Overexpression of 7a, a protein specifically encoded by the severe acute respiratory syndrome coronavirus, induces apoptosis via a caspase-dependent pathway. J Virol 2004;78:14043–7. doi: 10.1128/JVI.78.24.14043-14047.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tang X, Wu C, Li X, Song Y, Yao X, Wu X, Duan Y, Zhang H, Wang Y, Qian Z, Cui J, Lu J. On the origin and continuing evolution of SARS-CoV-2. Nat Sci Rev. 2020;7(6):1012–1023. doi: 10.1093/nsr/nwaa036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tanner JA, Watt RM, Chai Y-B, et al. The severe acute respiratory syndrome (SARS) coronavirus NTPase/Helicase belongs to a distinct class of 5′ to 3′ viral helicases. J Biol Chem. 2003;278:39578–82. doi: 10.1074/jbc.C300328200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.To KKW, Hung IFN, Chan JFW, Yuen K-Y. From SARS coronavirus to novel animal and human coronaviruses. J Thorac Dis. 2013;5:S103–S108. doi: 10.3978/j.issn.2072-1439.2013.06.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tsang KW, Ho PL, Ooi GC, et al. A cluster of cases of severe acute respiratory syndrome in Hong Kong. N Engl J Med. 2003;348:1977–85. doi: 10.1056/NEJMoa030666. [DOI] [PubMed] [Google Scholar]
- 94.Tseng SS-Y. SARS, avian flu, bioterror: infection control awareness for the optometrist. Clin Exper Optom 2007;90:31–5. doi: 10.1111/j.1444-0938.2006.00086.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Vabret A, Dina J, Brison E, Brouard J, Freymuth F. Coronavirus humains (HCoV) Pathol Biol (Paris) 2009;57:149–60. doi: 10.1016/j.patbio.2008.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Vareille M, Kieninger E, Edwards MR, Regamey N. The airway epithelium: soldier in the fight against respiratory viruses. Clin Microbiol Rev. 2011;24:210. doi: 10.1128/CMR.00014-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Vega VB, Ruan Y, Liu J, et al. Mutational dynamics of the SARS coronavirus in cell culture and human populations isolated in 2003. BMC Infect Dis. 2004;4:32. doi: 10.1186/1471-2334-4-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Verity R, Okell LC, Dorigatti I, et al. Estimates of the severity of coronavirus disease 2019: a model-based analysis. Lancet Infect Dis. 2020 doi: 10.1016/S1473-3099(20)30243-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Vijaykrishna D, Smith GJD, Zhang JX, Peiris JSM, Chen H, Guan Y. Evolutionary insights into the ecology of coronaviruses. J Virol. 2007;81:4012. doi: 10.1128/JVI.02605-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Walls AC, Park Y-J, Tortorici MA, Wall A, McGuire AT, Veesler D. Structure, function and antigenicity of the SARS-CoV-2 spike glycoprotein. bioRxiv. 2020 doi: 10.1101/2020.02.19.956581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wan Y, Shang J, Graham R, Baric RS, Li F. Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J Virol. 2020;94:e00127-00120. doi: 10.1128/JVI.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wang L-F, Eaton BT. Bats, Civets and the emergence of SARS. Curr Top Microbiol Immunol. 2007;315:25. doi: 10.1007/978-3-540-70962-6_13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wang C, Horby PW, Hayden FG, Gao GF. A novel coronavirus outbreak of global health concern. Lancet. 2020;395:470–3. doi: 10.1016/S0140-6736(20)30185-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Weidmann M, Zanotto PMDA, Weber F, Spiegel M, Brodt HR, Hufert FT. High-efficiency detection of severe acute respiratory syndrome virus genetic material. J Clin Microbiol. 2004;42:2771. doi: 10.1128/JCM.42.6.2771-2773.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Weijun C, Minghua Y, Ling Y, et al. SARS-associated coronavirus transmitted from human to pig. Emerg Infect Dis J. 2005;11:446. doi: 10.3201/eid1103.040824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Woo PCY, Lau SKP, Huang Y, Yuen K-Y. Coronavirus diversity, phylogeny and interspecies jumping. Exper Biol Med 2009;234:1117–27. doi: 10.3181/0903-MR-94. [DOI] [PubMed] [Google Scholar]
- 107.Wrapp D, Wang N, Corbett KS, et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wu A, Peng Y, Huang B, et al. Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe. 2020;27:325–8. doi: 10.1016/j.chom.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wu K, Peng G, Wilken M, Geraghty RJ, Li F. Mechanisms of host receptor adaptation by severe acute respiratory syndrome coronavirus. J Biol Chem. 2012;287:8904–11. doi: 10.1074/jbc.M111.325803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Xu J, Jia W, Wang P, et al. Antibodies and vaccines against Middle East respiratory syndrome coronavirus. Emerg Microbes Infect 2019;8:841–56. doi: 10.1080/22221751.2019.1624482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Yuan X, Shan Y, Zhao Z, Chen J, Cong Y. G0/G1 arrest and apoptosis induced by SARS-CoV 3b protein in transfected cells. Virol J 2005;2:66. doi: 10.1186/1743-422X-2-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Yuan X, Wu J, Shan Y, et al. SARS coronavirus 7a protein blocks cell cycle progression at G0/G1 phase via the cyclin D3/pRb pathway. Virology. 2006;346:74–85. doi: 10.1016/j.virol.2005.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus ADME, Fouchier RAM. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med. 2012;367:1814–20. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 114.Zhai Y, Sun F, Li X, et al. Insights into SARS-CoV transcription and replication from the structure of the nsp7-nsp8 hexadecamer. Nat Struct Mol Biol. 2005;12:980–6. doi: 10.1038/nsmb999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zhong NS, Zheng BJ, Li YM, et al. Epidemiology and cause of severe acute respiratory syndrome (SARS) in Guangdong, People’s Republic of China, in February, 2003. Lancet (London, England) 2003;362:1353–1358. doi: 10.1016/s0140-6736(03)14630-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Zhou P, Yang X-L, Wang X-G, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–3. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ziad AM, Nischay M, Kevin JO, et al. Middle east respiratory syndrome coronavirus in bats, Saudi Arabia. Emerg Infect Dis J. 2013;19:1819. doi: 10.3201/eid1911.131172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Zumla A, Hui DS, Perlman S. Middle East respiratory syndrome. Lancet. 2015;386:995–1007. doi: 10.1016/S0140-6736(15)60454-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data, codes, and post processed data used in this this study are available from the corresponding author upon reasonable request.