ABSTRACT
Despite advanced clinical treatments, mortality in patients with metastatic colorectal cancer (CRC) remains high. Three critical determinants in CRC progression include the epithelial proliferation checkpoints, epithelial‐to‐mesenchymal transition (EMT) and inflammatory cytokines in the tumour microenvironment. Genes involved in these three processes are regulated at the transcriptional and post-transcriptional level. Recent studies revealed previously unappreciated roles of non-coding ribonucleic acids (ncRNAs) in modulating the proliferation checkpoints, EMT, and inflammatory gene expression in CRC. In this review, we will discuss the mechanisms underlying the roles of ncRNAs in CRC as well as examine future perspectives in this field. Better understanding of ncRNA biology will provide novel targets for future therapeutic development.
KEYWORDS: Inflammation, non-coding RNA, lncRNA, miRNA, colorectal cancer, intestine, tumorigenesis, inflammatory cytokine
Introduction
Colorectal cancer (CRC) is the third most common cancer worldwide. Each year, there are over 100,000 newly diagnosed cases and greater than 50,000 related mortalities in the U.S. alone [1]. CRC is an adenocarcinoma originating from the epithelial cancer stem cells of the colon or rectum with pluripotency and self-renewal capabilities [2]. CRC initiation is characterized by the acquisition of genetic mutations in common signalling pathways, such as the wingless-type MMTV integration site (Wnt) and the transforming growth factor-beta (TGFβ) pathways, allowing for the bypass of cell cycle checkpoints [3–5]. Subsequent activation of the epithelial‐to‐mesenchymal transition (EMT) program is a critical step towards progression to invasive cancer and metastasis [6], which is a major cause of CRC-related mortality.
Recent high-throughput genomics efforts have identified a vast number of non-coding RNAs (ncRNAs) aberrantly expressed in CRC, fuelling a growing appreciation for their diverse roles in CRC initiation, growth and metastasis [7,8]. ncRNAs consist of two subgroups based on RNA length: the small non-coding RNAs (sncRNAs; less than 200 nucleotides) and the long-noncoding RNAs (lncRNAs; greater than 200 nucleotides). microRNAs (miRNAs) of the sncRNA family and diverse members of the lncRNA family are well studied in CRC and therefore form the major focus of this review.
General mechanisms underlying ncRNAs regulations of gene expression
sncRNAs and lncRNAs can form unique secondary and/or tertiary structures [9–11], enabling them to interact with diverse DNA, RNA, or protein partners to modulate processes at the levels of chromatin, transcription, translation and/or signalling transduction (Fig. 1A). miRNAs is the largest subset of sncRNAs known to be involved in CRC. miRNAs are ~22-nucleotides in length and are processed from the introns or exons of coding or non-coding transcripts [12]. By recruiting the RNA-induced silencing complex (RISC) to specific RNA targets through sequence complementarity, miRNAs control RNA degradation and/or protein translation [13]. miRNAs are counter-regulated by lncRNAs through three mechanisms (Fig. 1B). At the miRNA biogenesis step, lncRNAs can fine-tune miRNA processing and maturation [14]. LncRNAs can also act as ‘miRNA sponges’ [15]: by base complementarity, lncRNAs can bind to specific miRNAs and sequester them away from their canonical targets. Lastly, base pairing between lncRNA and mRNAs can physically limit miRNA access to their targets [16].
Figure 1.
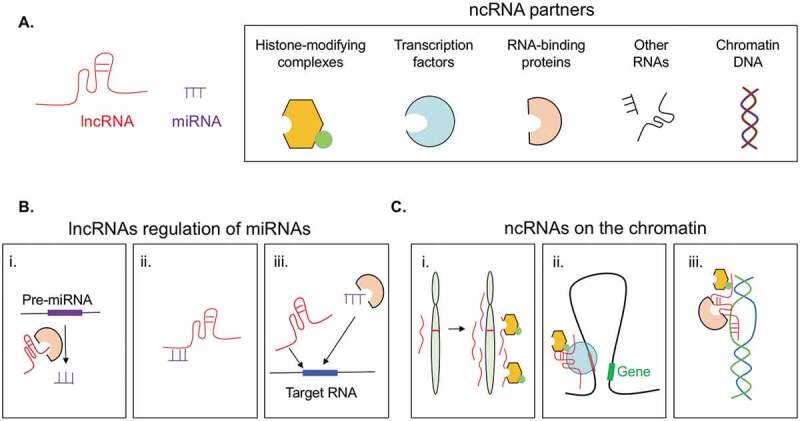
General principles and mechanisms underlying ncRNA functions. A. ncRNAs interact with diverse partners. B. LncRNAs counter regulate miRNAs by: i) modulating pre-miRNA processing, ii) direct sequestration, iii) competition for same target RNAs. C. LncRNAs regulate gene expression by i) modulating chromatin architecture of nearby genes in cis; ii) complexing with transcription factors or iii) forming RNA-DNA duplexes to regulate transcription of distal genomic loci.
In addition to controlling miRNA biogenesis and function, lncRNAs that are abundantly found in the nucleus can interact with chromatin DNA and regulate gene transcription (Fig. 1C). For instance, active transcription at ncRNA locus residing on regulatory elements such as enhancers and/or repressors can promote changes in the local chromatin architecture, resulting in activation or repression of nearby genes and/or the proper splicing of their transcripts [17]. Nuclear lncRNAs can also act in trans. By either base pairing with chromatin DNA to form RNA-DNA duplexes and/or tethering with DNA-binding transcription factors, lncRNAs can regulate chromatin accessibility and transcription of distal genes [18]. In the next section, we will discuss how many of these mechanisms underly ncRNA contribution to health and tumorigenesis in the intestine.
ncRNAs in epithelial proliferation checkpoints
Healthy intestinal epithelial cells (IECs) have a short life span of 3–5 days [19]. Mature IECs are replenished by newly differentiated cells derived from the transient amplifying cells residing at the base of intestinal crypts [19]. A main driver for epithelial turnover is Wnt signalling [20]. Wnt ligands binding to cell surface receptors promote the translocation of β-catenin into the nucleus and the transcriptional activation of the cell proliferation program [21]. Normally, the Wnt and β-catenin pathway is tightly regulated to prevent aberrant epithelial growth by the adenomatous polyposis coli (APC) protein complex. The APC complex targets β-catenin for proteasome-mediated destruction, putting a brake on epithelial growth [22]. In addition to the APC complex, tumour suppressor p53 also holds epithelial proliferation in check [23]. p53 activates the transcription of CDKN1A and PTEN to promote cell cycle arrest and facilitate apoptosis [24]. Loss of the APC and p53 checkpoints coupled with aberrant growth factor stimulation trigger tumorigenesis in humans and mice [25–28].
p53 is known to upregulate ncRNAs with potent anti-tumour activities, including miR-145, lincRNA-p21, and the growth arrest-specific transcript 5 (GAS5). miR-145 helps to dampen the expression of MYC, which encodes a one of the driver of epithelial proliferation [29]. LincRNA-p21 helps to shut down global gene transcription by recruiting the heterogeneous nuclear ribonucleoprotein K (hnRNP-K)-containing repressive complex onto chromatin DNA, halting CRC cell cycle progression [30]. When overexpressed, lincRNA-p21 attenuates the self-renewal capacity of CRC cancer stem cells by blocking Wnt/β-catenin signalling [31]. The p53-dependent lncRNA GAS5, encodes a cluster of small nucleolar RNAs (snoRNAs), is implicated in cell cycle arrest [32–34]. In response to growth factor stimulation, the insulin receptor substrate-1 (IRS-1) and AKT signalling triggers p53 ubiquitination and degradation, promoting epithelial proliferation. Under homoeostasis, miR-203a-3p and miR-126 negatively regulate IRS-1 and β-catenin transcripts to limit AKT-induced degradation of p53 and prevent aberrant proliferation [35,36].
ncRNAs in CRC growth and EMT
Tumour cells upregulate a unique set of lncRNAs with oncogenic activities to bypass the APC and p53 checkpoints, thereby driving cancer cell proliferation and EMT. At the transcription level, two well-studied examples include the nuclear lncRNAs DUXAP10 and CCAT1-L. LncRNA DUXAP10 is a transcriptional silencer. By forming a complex with the histone demethylase lysine-specific demethylase 1 (LSD1) complex, DUXAP10 shuts down the expression of p53 target genes, including CDKN1A and PTEN [37]. CCAT1-L is a 5200 nt lncRNA that is also highly upregulated in CRC [38]. It is transcribed 515 kb upstream of the MYC locus. CCAT1-L acts as a transcriptional regulator of MYC by binding to CTCF proteins and enhancing the interaction between the MYC promoter and its distal enhancers [38].
At the post-transcriptional level, CRCs hijack a diverse set of ncRNAs to evade cell cycle checkpoints. Several snoRNAs, ranging from 60 to 170 nucleotides in size, have been implicated in CRC (summarized in Table 1) [32,33,39–43]. Ectopic expression of SNORA42 strengthens CRC proliferation, invasion, and migration, while the inhibition of SNORA42 by CRISPR-Cas9 suppresses cell proliferation and invasion capacities [34]. The small nucleolar RNA host genes (SNHGs) encoding multiple snoRNAs are also involved in CRC. For instance, SNHG1 is a lncRNA encoding 8 snoRNAs, and is significantly upregulated in CRC [43]. SNHG1 sequesters miR-145/miR-154-5p [44,45] to promote MYC expression and cell cycle progression [40,41]. CRCs also overexpress miR-21 to target CDKN1A and PTEN transcripts for degradation [46].
Table 1.
ncRNAs involved in epithelial growth and transformation.
| Target Pathway | LncRNAs | miRNAs | snoRNAs |
|---|---|---|---|
| PI3K/AKT/MYC | CCAT [38] GHET1 [59] |
miR-203a-3p [60] miR-126 [61] miR-145 [29] |
SNORD126 [32] SNHG15 [42] |
| WNT/beta-Catenin | lincRNA-21 [31] CCAL [62] CTD903 [63] CASC11 [64] TINCR [65] |
miR-101 [66] miR-34 [67] miR-224 [68] miR-146a [69] miR-490-3p [70] miR-17-5p [71] |
SNHG1 [39,40] |
| p53/p21/PTEN | DUXAP10 [37] ZFAS1 [72] MEG3 [73] BANCR [74] lincRNA-p21 [30] |
miR-21 [75] | GAS5/SNORD44 [33] SNHG1 [41] |
| EMT |
AB073614 [76] TUG1 [77] |
miR-200s [78] | SNHG6 [43] |
EMT in CRCs is initiated by ZEB proteins [47,48]. ZEB1 transcripts are targeted for degradation by members of the miR-200 and miR-26a families in healthy epithelium [49,50]. miR-200 and miR-26a can be counteracted by lncRNAs, H19 and lncRNA-ATB acts as sponges for miR-200 to prevent ZEB1 transcript degradation and promote EMT and cancer progression [51–57]. And lncRNA SNHG6 counteracts miR-26a through similar mechanism to drive CRC invasion, migration and EMT [58]. Future studies will be needed to elucidate the exact molecular mechanisms underlying their contributions to CRC.
ncRNAs and inflammation
CRC is tightly associated with inflammation [79]. Patients with inflammatory bowel diseases (IBD) are at a higher risk of developing CRC [80]. Elevated immune-modulatory cytokines not only influence the function of immune populations infiltrated to the lesion, but also act directly on cancer cells to promote disease progression. Known CRC-related cytokines and their regulation by sncRNAs and lncRNAs are summarized in Table 2. The contributions of miRNAs to inflammatory cytokine expression have been extensively reviewed elsewhere and beyond the scope of this review [81]. In the next section, we will summarize lncRNA regulation of pro-inflammatory cytokines involved in CRC [82], including interleukin-6 (IL-6), tumour necrosis factor (TNFα), and interferon-gamma (IFNγ) (Fig. 2).
Table 2.
ncRNAs involved in inflammatory cytokine expression.
| Inflammatory cytokines | LncRNAs | miRNAs |
|---|---|---|
| IL-6 | Lethe [111] LincRNA-Cox2 [108] LincRNA-EPS [113] Lnc-IL7R [112] MARCKS [83] Mirt2 [110] NEAT1 [109] ROCK1 [83] |
miR-21 [84] miR-24 [85] miR-26a [86] miR-124 [87] miR-147 [88] |
| TNF | Lnc13 [120] THRIL [119] |
miR-24 [85] miR-124 [87] miR-147 [88] |
| IFN | LncRNA-CD244 [89] NeST [124] |
miR-29 [86] miR-155 [90] |
| IL-8 | NEAT1 [109] PANDA [91] |
miR10a [92] miR200 [93] miR203 [94] miR302 [95] |
| IL-1 | LSINCT5 [96] Mirt2 [110] |
miR-233 [97] |
Figure 2.
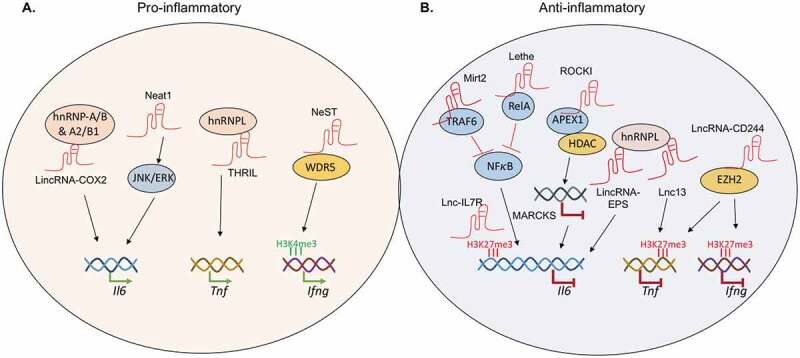
LncRNAs as positive and negative regulators of inflammation. A. Different lncRNAs recruit distinct partner proteins to regulate inflammatory cytokine expression. Il6 transcription is augmented by lincRNA-Cox2 and lncRNA NEAT1. TNF expression is upregulated by lncRNA THRIL. Transcription of the Ifng locus is regulated by lncRNA NeST. B. In humans, Il6 transcription is negatively regulated by Lnc-17R and ROCKI. In mice, Il6 transcription is negatively regulated by lncRNA Mirt2 and Lethe. Lnc13 interacts with hnRNPL to block the transcription of Tnf. LncRNA-CD244 recruits EZH2 to deposit repressive histone marks and shut down transcription at both the TNF and IFNG loci.
Increased expression of IL-6 is linked to advanced stages of CRC and decreased patient survival [98,99]. IL-6 activates the STAT3 signalling cascade [100] and turns on gene programs involved in CRC proliferation, migration, and angiogenesis [101–103]. In addition, IL-6 promotes the recruitment and expansion of immunosuppressive myeloid-derived suppressor cells (MDSCs) and inhibits the maturation of human dendritic cells in the tumour environment [104,105]. Neutralization of the IL-6 receptor as well as the genetic ablation of IL-6 and/or STAT3 reduces tumorigenesis in mouse models of CRC [102,106]. IL-6 expression is regulated by multiple lncRNAs. LincRNA-Cox2 and lncRNA NEAT1 promotes IL-6 expression via distinct mechanisms. LincRNA-Cox2 is induced by Toll-like receptor ligands and partners with the heterogeneous nuclear ribonucleoproteins hnRNP-A/B and hnRNP-A2/B1 to drive Il6 transcription in the nucleus [107,108]. LncRNA NEAT1, on the other hand, acts further upstream and potentiate IL-6 expression by promoting the activation of the JNK1/2 and ERK1/2 signalling cascades [109].
Other lncRNAs can limit IL-6 expression through negative feedback loops to prevent exacerbated inflammation. For example, the Toll-like receptor-induced lncRNA Mirt2 associates with TRAF6 in the cytoplasm and blocks its ubiquitination. Loss of TRAF6 ubiquitination diminishes the activation of NFκB and MAPK, putting a brake on Il6 transcription [110]. In the nucleus, three additional lncRNAs help to keep Il6 transcription in check. LncRNA Lethe acts as a decoy partner for NFκB and blocks its recruitment on the Il6 promoter [111]. Lnc-IL7R promotes the deposition of trimethylation on lysine 27 of histone H3 and sets up a repressive chromatin environment at the Il6 locus [112]. Lastly, LncRNA-EPS represses IL6 expression by partnering with hnRNPL to reduce chromatin accessibility [113].
Another CRC promoting inflammatory cytokine is TNFα [114]. TNFα signalling leading to the activation of NFκB drives CRC survival, proliferation, invasion, and metastasis [115]. Genetic ablation of the TNF receptor protects mice against chemical-induced colon tumour [114] and limits liver metastasis of transplanted CRC in mouse models [116]. Human IBD and CRC patients treated with anti-TNFα had less intestinal inflammation and decreased tumour burden [117,118]. TNFα expression is regulated by three lncRNAs. The TNFα and hnRNPL–related immunoregulatory lincRNA, THRIL, works together with hnRNPL to promote TNF transcription [119]. Lnc-13 and lncRNA-CD244 negatively regulate Tnf expression in mice and humans, respectively, through distinct mechanisms. Lnc-13 associates with hnRNPD to recruit the histone deacetylase, HDAC1, to remove the activation marks on the Tnf promoter [120]. In contrast, lncRNA-CD244 turns off TNF transcription by partnering with the enhancer of zeste homolog 2 (EZH2)-containing complex to deposit repressive tri-methylation marks on lysine 27 of histone H3 on the TNF locus.
Similar to IL-6 and TNFα, IFNγ also promotes CRC pathogenesis [121]. IFNγ signalling induces expression of immune checkpoint molecules, including PD-L1 [122], which promotes evasion from anti-tumour immune responses [123]. Transcription of the Ifng gene is regulated by lncRNA NeST, also known as Ifngas1 or Tmevpg1 [124]. The NeST locus is associated with an IBD susceptibility related SNP, rs7134599. Accordingly, elevation of NeST expression in ulcerative colitis patients positively correlates with enhanced IFNγ levels [125]. However, the link between rs7134599 and NeST to CRC has not been clearly demonstrated [126]. Mechanistically, NeST works together with the methyltransferase WDR5 to promote trimethylation on lysine 4 of histone H3 and transcriptional activation of the Ifng gene [124]. Together, lncRNAs and their associated transcription factors and epigenetic modifiers are critical regulators of inflammatory cytokine expression. Future studies will be needed to fully assess each of their contribution to CRC in vivo.
Emerging tools for studying ncRNAs
ncRNAs accomplishes a diverse range of biological functions by interacting with other RNAs, proteins, and chromatin DNA (Fig. 1). However, we have limited understanding of how these interactions contribute to ncRNA biology in the context of health and diseases. Emerging tools are now allowing researchers to tackle these challenges in the field. For instance, RNA–RNA interactions can be mapped using the MS2-tagged RNA affinity purification (MS2-TRAP) system in vitro as well as the cross-linking [127], ligation, and sequencing of hybrids (CLASH) assay in vivo [128]. Protein partners of ncRNAs can be elucidated using the comprehensive identification of RNA-binding proteins by mass spectrometry (ChIRP-MS) [129], click chemistry-assisted RNA-interactome capture (CARIC) [130], and the crosslinking and immunoprecipitation (iCLIP/eCLIP) assays [131,132]. ncRNA-DNA interactions on the chromatin can be revealed using the chromatin-associated RNA sequencing (ChAR-seq) [133] and the in situ global RNA interactions with DNA captured by deep sequencing (GRID-seq) approaches [134]. Furthermore, ncRNA secondary and tertiary structures are important determinants for the specificity and affinity of ncRNAs for their interaction partners. Future studies will need to better elucidate ncRNA structural information inside living cells with approaches such as the selective 2ʹ-hydroxyl acylation analysed by primer extension and sequencing (SHAPEseq) [135]. Together with cryogenic electron microscopy, researchers will soon be able to gain new lights on additional general principles underlying ncRNA biology.
Discussion
The human and mouse genome is estimated to have over 2,000 miRNAs [136], 50,000 lncRNAs [137], and 1,000 RNA-binding proteins [138]. However, only a handful has been characterized in the context of CRC. For instance, we know relatively little about the involvement of ncRNAs in the production of anti-inflammatory cytokines, such as IL-10 and TGFβ, which are known for negative regulating intestine inflammation [139,140]. In addition, there are several other families of ncRNAs with newly defined roles in CRC, such as the tRNA and tRNA-derivatives (reviewed in [141]). But the mechanistic details of their contribution remained to be elucidated. To close these knowledge gaps, high-throughput gain and loss of function screens, including the use of the latest CRISPR-Cas9 technologies [142], will be critical in future studies. Molecular insights can be revealed using a combined molecular, biochemical, genetic and genomic approach. Future studies should also evaluate whether the ncRNA mechanisms of action thus far are conserved or unique in different cell types under steady state and across different disease conditions. And, better understanding of how ncRNAs contribute to the crosstalk between tumour cells and their local environmental cues will facilitate the development of novel targets against inflammation-driven tumorigenesis.
Acknowledgments
We thank Juan Hernandez and Benjamin Cho for critical reading of our manuscript. S.M., T.L. and W.J.H. are partially funded by the Edward Mallinckrodt, Jr. Foundation, the Gabrielle’s Angel Foundation for Cancer Research, and the National Institutes of Health (NIGMS_1R01GM124494-01). The authors declare no competing financial interests.
Funding Statement
This work was supported by the Edward Mallinckrodt, Jr. Foundation;Gabrielle’s Angel Foundation for Cancer Research [105]; National Institute of General Medical Sciences (US) [1R01GM124494-01].
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
References
- [1].Siegel RL, Miller KD, Fedewa SA, et al. Colorectal cancer statistics, 2017. CA Cancer J Clin. 2017;67(3):177–193. [DOI] [PubMed] [Google Scholar]
- [2].Boman BM, Wicha MS.. Cancer stem cells: a step toward the cure. J Clin Oncol. 2008;26(17):2795–2799. [DOI] [PubMed] [Google Scholar]
- [3].Abdul Khalek FJ, Gallicano GI, Mishra L.. Colon cancer stem cells. Gastrointestinal Cancer Res: GCR. 2010;(Suppl 1):S16–S23. [PMC free article] [PubMed] [Google Scholar]
- [4].White BD, Chien AJ, Dawson DW. Dysregulation of Wnt/β-catenin signaling in gastrointestinal cancers. Gastroenterology. 2012;142(2):219–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Mishra L, Shetty K, Tang Y, et al. The role of TGF-beta and Wnt signaling in gastrointestinal stem cells and cancer. Oncogene. 2005;24(37):5775–5789. [DOI] [PubMed] [Google Scholar]
- [6].De Craene B, Berx G. Regulatory networks defining EMT during cancer initiation and progression. Nat Rev Cancer. 2013;13(2):97–110. [DOI] [PubMed] [Google Scholar]
- [7].Ragusa M, Barbagallo C, Statello L, et al. Non-coding landscapes of colorectal cancer. World J Gastroenterol. 2015;21(41):11709–11739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Xu MD, Qi P, Du X. Long non-coding RNAs in colorectal cancer: implications for pathogenesis and clinical application. Mod Pathol. 2014;27(10):1310–1320. [DOI] [PubMed] [Google Scholar]
- [9].Staple DW, Butcher SE. Pseudoknots: RNA structures with diverse functions. PLoS Biol. 2005;3(6):e213–e213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Qian X, Zhao J, Yeung PY, et al. Revealing lncRNA structures and interactions by sequencing-based approaches. Trends Biochem Sci. 2019;44(1):33–52. [DOI] [PubMed] [Google Scholar]
- [11].Tsai M-C, Manor O, Wan Y, et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329(5992):689–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 2014;15(8):509–524. [DOI] [PubMed] [Google Scholar]
- [13].Hausser J, Zavolan M. Identification and consequences of miRNA-target interactions–beyond repression of gene expression. Nat Rev Genet. 2014;15(9):599–612. [DOI] [PubMed] [Google Scholar]
- [14].Yu Y, Nangia-Makker P, Farhana L, et al. A novel mechanism of lncRNA and miRNA interaction: CCAT2 regulates miR-145 expression by suppressing its maturation process in colon cancer cells. Mol Cancer. 2017;16(1):155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Thomson DW, Dinger ME. Endogenous microRNA sponges: evidence and controversy. Nat Rev Genet. 2016;17(5):272–283. [DOI] [PubMed] [Google Scholar]
- [16].Faghihi MA, Zhang M, Huang J, et al. Evidence for natural antisense transcript-mediated inhibition of microRNA function. Genome Biol. 2010;11(5):R56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Engreitz JM, Haines JE, Perez EM, et al. Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature. 2016;539(7629):452–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Li Y, Syed J, Sugiyama H. RNA-DNA triplex formation by long noncoding RNAs. Cell Chem Biol. 2016;23(11):1325–1333. [DOI] [PubMed] [Google Scholar]
- [19].Williams JM, Duckworth CA, Burkitt MD, et al. Epithelial cell shedding and barrier function: a matter of life and death at the small intestinal villus tip. Vet Pathol. 2015;52(3):445–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].van der Flier LG, Clevers H. Stem cells, self-renewal, and differentiation in the intestinal epithelium. Annu Rev Physiol. 2009;71:241–260. [DOI] [PubMed] [Google Scholar]
- [21].Sansom OJ, KR Reed, AJ Hayes, et al. Loss of Apc in vivo immediately perturbs Wnt signaling, differentiation, and migration. Genes Dev. 2004;18(12):1385–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Kimelman D, Xu W. beta-catenin destruction complex: insights and questions from a structural perspective. Oncogene. 2006;25(57):7482–7491. [DOI] [PubMed] [Google Scholar]
- [23].Wawryk-Gawda E, Chylińska-Wrzos P, Lis-Sochocka M, et al. P53 protein in proliferation, repair and apoptosis of cells. Protoplasma. 2014;251(3):525–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Stambolic V, MacPherson D, Sas D, et al. Regulation of PTEN Transcription by p53. Mol Cell. 2001;8(2):317–325. [DOI] [PubMed] [Google Scholar]
- [25].Ashktorab H, Schäffer AA, Daremipouran M, et al. Distinct genetic alterations in colorectal cancer. PLoS One. 2010;5(1):e8879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Bruin SC, Klijn C, Liefers G-J, et al. Specific genomic aberrations in primary colorectal cancer are associated with liver metastases. BMC Cancer. 2010;10:662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Perri F, Pisconti S, Della Vittoria Scarpati G. P53 mutations and cancer: a tight linkage. Ann Transl Med. 2016;4(24):522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Yamada Y, Mori H. Multistep carcinogenesis of the colon in Apc(Min/+) mouse. Cancer Sci. 2007;98(1):6–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Sachdeva M, Zhu S, Wu F, et al. p53 represses c-Myc through induction of the tumor suppressor miR-145. Proc Natl Acad Sci U S A. 2009;106(9):3207–3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Huarte M, Guttman M, Feldser D, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142(3):409–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Wang J, Lei Z-J, Guo Y, et al. miRNA-regulated delivery of lincRNA-p21 suppresses beta-catenin signaling and tumorigenicity of colorectal cancer stem cells. Oncotarget. 2015;6(35):37852–37870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Fang X, Yang D, Luo H, et al. SNORD126 promotes HCC and CRC cell growth by activating the PI3K-AKT pathway through FGFR2. J Mol Cell Biol. 2017;9(3):243–255. [DOI] [PubMed] [Google Scholar]
- [33].Yuan S, Wu Y, Wang Y, et al. An oncolytic adenovirus expressing SNORD44 and GAS5 exhibits antitumor effect in colorectal cancer cells. Hum Gene Ther. 2017;28(8):690–700. [DOI] [PubMed] [Google Scholar]
- [34].Okugawa Y, Toiyama Y, Toden S, et al. Clinical significance of SNORA42 as an oncogene and a prognostic biomarker in colorectal cancer. Gut. 2017;66(1):107–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Xiao Z, Qu Z, Chen Z, et al. LncRNA HOTAIR is a prognostic biomarker for the proliferation and chemoresistance of colorectal cancer via MiR-203a-3p-mediated Wnt/ss-catenin signaling pathway. Cell Physiol Biochem. 2018;46(3):1275–1285. [DOI] [PubMed] [Google Scholar]
- [36].Zhou Y, Feng X, Liu Y-L, et al. Down-regulation of miR-126 is associated with colorectal cancer cells proliferation, migration and invasion by targeting IRS-1 via the AKT and ERK1/2 signaling pathways. PLoS One. 2013;8(11):e81203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Lian Y, Xu Y, Xiao C, et al. The pseudogene derived from long non-coding RNA DUXAP10 promotes colorectal cancer cell growth through epigenetically silencing of p21 and PTEN. Sci Rep. 2017;7(1):7312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Xiang JF, Yin Q-F, Chen T, et al. Human colorectal cancer-specific CCAT1-L lncRNA regulates long-range chromatin interactions at the MYC locus. Cell Res. 2014;24(5):513–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Yang H, Wang S, Kang Y-J, et al. Long non-coding RNA SNHG1 predicts a poor prognosis and promotes colon cancer tumorigenesis. Oncol Rep. 2018;40(1):261–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Zhu Y, Li B, Liu Z, et al. Up-regulation of lncRNA SNHG1 indicates poor prognosis and promotes cell proliferation and metastasis of colorectal cancer by activation of the Wnt/β-catenin signaling pathway. Oncotarget. 2017;8(67):111715–111727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Zhao Y, Qin Z-S, Feng Y, et al. Long non-coding RNA (lncRNA) small nucleolar RNA host gene 1 (SNHG1) promote cell proliferation in colorectal cancer by affecting P53. Eur Rev Med Pharmacol Sci. 2018;22(4):976–984. [DOI] [PubMed] [Google Scholar]
- [42].Saeinasab M, Bahrami AR, González J, et al. SNHG15 is a bifunctional MYC-regulated noncoding locus encoding a lncRNA that promotes cell proliferation, invasion and drug resistance in colorectal cancer by interacting with AIF. J Exp Clin Cancer Res. 2019;38(1):172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Yu C, Sun J, Leng X, et al. Long noncoding RNA SNHG6 functions as a competing endogenous RNA by sponging miR-181a-5p to regulate E2F5 expression in colorectal cancer. Cancer Manag Res. 2019;11:611–624. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [44].Xu M, Chen X, Lin K, et al. The long noncoding RNA SNHG1 regulates colorectal cancer cell growth through interactions with EZH2 and miR-154-5p. Mol Cancer. 2018;17(1):141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Tian T, Qiu R, Qiu X. SNHG1 promotes cell proliferation by acting as a sponge of miR-145 in colorectal cancer. Oncotarget. 2018;9(2):2128–2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Feng YH, Tsao CJ. Emerging role of microRNA-21 in cancer. Biomed Rep. 2016;5(4):395–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Serrano-Gomez SJ, Maziveyi M, Alahari SK. Regulation of epithelial-mesenchymal transition through epigenetic and post-translational modifications. Mol Cancer. 2016;15:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Krebs AM, Mitschke J, Lasierra Losada M, et al. The EMT-activator Zeb1 is a key factor for cell plasticity and promotes metastasis in pancreatic cancer. Nat Cell Biol. 2017;19(5):518–529. [DOI] [PubMed] [Google Scholar]
- [49].Davalos V, Moutinho C, Villanueva A, et al. Dynamic epigenetic regulation of the microRNA-200 family mediates epithelial and mesenchymal transitions in human tumorigenesis. Oncogene. 2011;31:2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Burk U, Schubert J, Wellner U, et al. A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Rep. 2008;9(6):582–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Yuan JH, Yang F, Wang F, et al. A long noncoding RNA activated by TGF-beta promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell. 2014;25(5):666–681. [DOI] [PubMed] [Google Scholar]
- [52].Cui H, Onyango P, Brandenburg S, et al. Loss of imprinting in colorectal cancer linked to hypomethylation of H19 and IGF2. Cancer Res. 2002;62(22):6442–6446. [PubMed] [Google Scholar]
- [53].Tsang WP, Ng EKO, Ng SSM, et al. Oncofetal H19-derived miR-675 regulates tumor suppressor RB in human colorectal cancer. Carcinogenesis. 2010;31(3):350–358. [DOI] [PubMed] [Google Scholar]
- [54].Ma C, Nong K, Zhu H, et al. H19 promotes pancreatic cancer metastasis by derepressing let-7’s suppression on its target HMGA2-mediated EMT. Tumour Biol. 2014;35(9):9163–9169. [DOI] [PubMed] [Google Scholar]
- [55].Kallen AN, Zhou X-B, Xu J, et al. The imprinted H19 lncRNA antagonizes let-7 microRNAs. Mol Cell. 2013;52(1):101–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Yue B, Qiu S, Zhao S, et al. LncRNA-ATB mediated E-cadherin repression promotes the progression of colon cancer and predicts poor prognosis. J Gastroenterol Hepatol. 2016;31(3):595–603. [DOI] [PubMed] [Google Scholar]
- [57].Li R-H, Chen M, Liu J, et al. Long noncoding RNA ATB promotes the epithelial-mesenchymal transition by upregulating the miR-200c/Twist1 axe and predicts poor prognosis in breast cancer. Cell Death Dis. 2018;9(12):1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Zhang X, Liu Z, Shu Q, et al. LncRNA SNHG6 functions as a ceRNA to regulate neuronal cell apoptosis by modulating miR-181c-5p/BIM signalling in ischaemic stroke. J Cell Mol Med. 2019;23(9):6120–6130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Yang F, Xue X, Zheng L, et al. Long non-coding RNA GHET1 promotes gastric carcinoma cell proliferation by increasing c-Myc mRNA stability. Febs J. 2014;281(3):802–813. [DOI] [PubMed] [Google Scholar]
- [60].Chen L, Gao H, Liang J, et al. miR-203a-3p promotes colorectal cancer proliferation and migration by targeting PDE4D. Am J Cancer Res. 2018;8(12):2387–2401. [PMC free article] [PubMed] [Google Scholar]
- [61].Yuan W, Guo Y-Q, Li X-Y, et al. MicroRNA-126 inhibits colon cancer cell proliferation and invasion by targeting the chemokine (C-X-C motif) receptor 4 and Ras homolog gene family, member A, signaling pathway. Oncotarget. 2016;7(37):60230–60244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Ma Y, Yang Y, Wang F, et al. Long non-coding RNA CCAL regulates colorectal cancer progression by activating Wnt/beta-catenin signalling pathway via suppression of activator protein 2alpha. Gut. 2016;65(9):1494–1504. [DOI] [PubMed] [Google Scholar]
- [63].Yuan Z, YU X, NI B, et al. Overexpression of long non-coding RNA-CTD903 inhibits colorectal cancer invasion and migration by repressing Wnt/beta-catenin signaling and predicts favorable prognosis. Int J Oncol. 2016;48(6):2675–2685. [DOI] [PubMed] [Google Scholar]
- [64].Zhang Z, Zhou C, Chang Y, et al. Long non-coding RNA CASC11 interacts with hnRNP-K and activates the WNT/beta-catenin pathway to promote growth and metastasis in colorectal cancer. Cancer Lett. 2016;376(1):62–73. [DOI] [PubMed] [Google Scholar]
- [65].Yu S, Wang D, Shao Y, et al. SP1-induced lncRNA TINCR overexpression contributes to colorectal cancer progression by sponging miR-7-5p. Aging (Albany NY). 2019;11(5):1389–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Strillacci A, Valerii MC, Sansone P, et al. Loss of miR-101 expression promotes Wnt/β-catenin signalling pathway activation and malignancy in colon cancer cells. J Pathol. 2013;229(3):379–389. [DOI] [PubMed] [Google Scholar]
- [67].Kim NH, Cha YH, Eun Kang S, et al. p53 regulates nuclear GSK-3 levels through miR-34-mediated Axin2 suppression in colorectal cancer cells. Cell Cycle. 2013;12(10):1578–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Li T, Lai Q, Wang S, et al. MicroRNA-224 sustains Wnt/beta-catenin signaling and promotes aggressive phenotype of colorectal cancer. J Exp Clin Cancer Res. 2016;35:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Hwang WL, Jiang J-K, Yang S-H, et al. MicroRNA-146a directs the symmetric division of Snail-dominant colorectal cancer stem cells. Nat Cell Biol. 2014;16(3):268–280. [DOI] [PubMed] [Google Scholar]
- [70].Zheng K, Zhou X, Yu J, et al. Epigenetic silencing of miR-490-3p promotes development of an aggressive colorectal cancer phenotype through activation of the Wnt/beta-catenin signaling pathway. Cancer Lett. 2016;376(1):178–187. [DOI] [PubMed] [Google Scholar]
- [71].Ma Y, Zhang P, Wang F, et al. Elevated oncofoetal miR-17-5p expression regulates colorectal cancer progression by repressing its target gene P130. Nat Commun. 2012;3:1291. [DOI] [PubMed] [Google Scholar]
- [72].Chen X, Zeng K, Xu M, et al. SP1-induced lncRNA-ZFAS1 contributes to colorectal cancer progression via the miR-150-5p/VEGFA axis. Cell Death Dis. 2018;9(10):982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Wang W, Xie Y, Chen F, et al. LncRNA MEG3 acts a biomarker and regulates cell functions by targeting ADAR1 in colorectal cancer. World J Gastroenterol. 2019;25(29):3972–3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Guo Q, ZHAO Y, CHEN J, et al. BRAF-activated long non-coding RNA contributes to colorectal cancer migration by inducing epithelial-mesenchymal transition. Oncol Lett. 2014;8(2):869–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Zheng P, Chen L, Yuan X, et al. Exosomal transfer of tumor-associated macrophage-derived miR-21 confers cisplatin resistance in gastric cancer cells. J Exp Clin Cancer Res. 2017;36(1):53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Xue J, Liao L, Yin F, et al. LncRNA AB073614 induces epithelial- mesenchymal transition of colorectal cancer cells via regulating the JAK/STAT3 pathway. Cancer Biomark. 2018;21(4):849–858. [DOI] [PubMed] [Google Scholar]
- [77].Qian W, Ren Z, Lu X. Knockdown of long non-coding RNA TUG1 suppresses nasopharyngeal carcinoma progression by inhibiting epithelial-mesenchymal transition (EMT) via the promotion of miR-384. Biochem Biophys Res Commun. 2019;509(1):56–63. [DOI] [PubMed] [Google Scholar]
- [78].Tian Y, Pan Q, Shang Y, et al. MicroRNA-200 (miR-200) cluster regulation by achaete scute-like 2 (Ascl2): impact on the epithelial-mesenchymal transition in colon cancer cells. J Biol Chem. 2014;289(52):36101–36115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Wang K, Karin M. Tumor-elicited inflammation and colorectal cancer. Adv Cancer Res. 2015;128:173–196. [DOI] [PubMed] [Google Scholar]
- [80].Jess T, Rungoe C, Peyrin-Biroulet L. Risk of colorectal cancer in patients with ulcerative colitis: a meta-analysis of population-based cohort studies. Clin Gastroenterol Hepatol. 2012;10(6):639–645. [DOI] [PubMed] [Google Scholar]
- [81].Tili E, Michaille -J-J, Piurowski V, et al. MicroRNAs in intestinal barrier function, inflammatory bowel disease and related cancers-their effects and therapeutic potentials. Curr Opin Pharmacol. 2017;37:142–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].West NR, McCuaig S, Franchini F, et al. Emerging cytokine networks in colorectal cancer. Nat Rev Immunol. 2015;15(10):615–629. [DOI] [PubMed] [Google Scholar]
- [83].Zhang Q, Chao T-C, Patil VS, et al. The long noncoding RNA ROCKI regulates inflammatory gene expression. Embo J. 2019;38(8):38:e100041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Feng J, Li A, Deng J, et al. miR-21 attenuates lipopolysaccharide-induced lipid accumulation and inflammatory response: potential role in cerebrovascular disease. Lipids Health Dis. 2014;13:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Yang J, Chen L, Ding J, et al. MicroRNA-24 inhibits high glucose-induced vascular smooth muscle cell proliferation and migration by targeting HMGB1. Gene. 2016;586(2):268–273. [DOI] [PubMed] [Google Scholar]
- [86].Zhang R, Tian A, Wang J, et al. miR26a modulates Th17/T reg balance in the EAE model of multiple sclerosis by targeting IL6. Neuromolecular Med. 2015;17(1):24–34. [DOI] [PubMed] [Google Scholar]
- [87].Xiao YT, Wang J, Lu W, et al. Downregulated expression of microRNA-124 in pediatric intestinal failure patients modulates macrophages activation by inhibiting STAT3 and AChE. Cell Death Dis. 2016;7(12):e2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Zheng CZ, Shu Y-B, Luo Y-L, et al. The role of miR-146a in modulating TRAF6-induced inflammation during lupus nephritis. Eur Rev Med Pharmacol Sci. 2017;21(5):1041–1048. [PubMed] [Google Scholar]
- [89].Wang Y, Zhong H, Xie X, et al. Long noncoding RNA derived from CD244 signaling epigenetically controls CD8+ T-cell immune responses in tuberculosis infection. Proc Natl Acad Sci U S A. 2015;112(29):E3883–E92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Trotta R, Chen L, Ciarlariello D, et al. miR-155 regulates IFN-gamma production in natural killer cells. Blood. 2012;119(15):3478–3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Peng C, Hu W, Weng X, et al. Over expression of long non-coding RNA PANDA promotes hepatocellular carcinoma by inhibiting senescence associated inflammatory factor IL8. Sci Rep. 2017;7(1):4186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Tangtanatakul P, Thammasate B, Jacquet A, et al. Transcriptomic profiling in human mesangial cells using patient-derived lupus autoantibodies identified miR-10a as a potential regulator of IL8. Sci Rep. 2017;7(1):14517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Chuang TD, Khorram O. miR-200c regulates IL8 expression by targeting IKBKB: a potential mediator of inflammation in leiomyoma pathogenesis. PLoS One. 2014;9(4):e95370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Qu JQ, Yi H-M, Ye X, et al. MiRNA-203 reduces nasopharyngeal carcinoma radioresistance by targeting IL8/AKT signaling. Mol Cancer Ther. 2015;14(11):2653–2664. [DOI] [PubMed] [Google Scholar]
- [95].Chen L, Min L, Wang X, et al. Loss of RACK1 promotes metastasis of gastric cancer by inducing a miR-302c/IL8 signaling loop. Cancer Res. 2015;75(18):3832–3841. [DOI] [PubMed] [Google Scholar]
- [96].Zhang X, SHA M, YAO Y, et al. Increased B-type-natriuretic peptide promotes myocardial cell apoptosis via the B-type-natriuretic peptide/long non-coding RNA LSINCT5/caspase-1/interleukin 1beta signaling pathway. Mol Med Rep. 2015;12(5):6761–6767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Haneklaus M, Gerlic M, Kurowska-Stolarska M, et al. Cutting edge: miR-223 and EBV miR-BART15 regulate the NLRP3 inflammasome and IL-1beta production. J Immunol. 2012;189(8):3795–3799. [DOI] [PubMed] [Google Scholar]
- [98].Knupfer H, Preiss R. Serum interleukin-6 levels in colorectal cancer patients–a summary of published results. Int J Colorectal Dis. 2010;25(2):135–140. [DOI] [PubMed] [Google Scholar]
- [99].Li X, Wang Y, Han C, et al. Colorectal cancer progression is associated with accumulation of Th17 lymphocytes in tumor tissues and increased serum levels of interleukin-6. Tohoku J Exp Med. 2014;233(3):175–182. [DOI] [PubMed] [Google Scholar]
- [100].Kishimoto T. Interleukin-6: from basic science to medicine–40 years in immunology. Annu Rev Immunol. 2005;23:1–21. [DOI] [PubMed] [Google Scholar]
- [101].Nagasaki T, Hara M, Nakanishi H, et al. Interleukin-6 released by colon cancer-associated fibroblasts is critical for tumour angiogenesis: anti-interleukin-6 receptor antibody suppressed angiogenesis and inhibited tumour-stroma interaction. Br J Cancer. 2014;110(2):469–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Grivennikov S, Karin E, Terzic J, et al. IL-6 and Stat3 are required for survival of intestinal epithelial cells and development of colitis-associated cancer. Cancer Cell. 2009;15(2):103–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Schmidt S, Schumacher N, Schwarz J, et al. ADAM17 is required for EGF-R-induced intestinal tumors via IL-6 trans-signaling. J Exp Med. 2018;215(4):1205–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Jiang M, Chen J, Zhang W, et al. Interleukin-6 trans-signaling pathway promotes immunosuppressive myeloid-derived suppressor cells via suppression of suppressor of cytokine signaling 3 in breast cancer. Front Immunol. 2017;8:1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Xu M, Zhao Z, Song J, et al. Interactions between interleukin-6 and myeloid-derived suppressor cells drive the chemoresistant phenotype of hepatocellular cancer. Exp Cell Res. 2017;351(2):142–149. [DOI] [PubMed] [Google Scholar]
- [106].Becker C, Fantini MC, Schramm C, et al. TGF-beta suppresses tumor progression in colon cancer by inhibition of IL-6 trans-signaling. Immunity. 2004;21(4):491–501. [DOI] [PubMed] [Google Scholar]
- [107].Carpenter S, Aiello D, Atianand MK, et al. A long noncoding RNA mediates both activation and repression of immune response genes. Science. 2013;341(6147):789–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Elling R, Robinson EK, Shapleigh B, et al. Genetic models reveal cis and trans immune-regulatory activities for lincRNA-Cox2. Cell Rep. 2018;25(6):1511–1524 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Zhang F, Wu L, Qian J, et al. Identification of the long noncoding RNA NEAT1 as a novel inflammatory regulator acting through MAPK pathway in human lupus. J Autoimmun. 2016;75:96–104. [DOI] [PubMed] [Google Scholar]
- [110].Du M, Yuan L, Tan X, et al. The LPS-inducible lncRNA Mirt2 is a negative regulator of inflammation. Nat Commun. 2017;8(1):2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Rapicavoli NA, Qu K, Zhang J, et al. A mammalian pseudogene lncRNA at the interface of inflammation and anti-inflammatory therapeutics. Elife. 2013;2:e00762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Cui H, Xie N, Tan Z, et al. The human long noncoding RNA lnc-IL7R regulates the inflammatory response. Eur J Immunol. 2014;44(7):2085–2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Atianand MK, Hu W, Satpathy AT, et al. A long noncoding RNA lincRNA-EPS acts as a transcriptional brake to restrain inflammation. Cell. 2016;165(7):1672–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Popivanova BK, Kitamura K, Wu Y, et al. Blocking TNF-alpha in mice reduces colorectal carcinogenesis associated with chronic colitis. J Clin Invest. 2008;118(2):560–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Aggarwal BB. Signalling pathways of the TNF superfamily: a double-edged sword. Nat Rev Immunol. 2003;3(9):745–756. [DOI] [PubMed] [Google Scholar]
- [116].Kitakata H, Nemoto-Sasaki Y, Takahashi Y, et al. Essential roles of tumor necrosis factor receptor p55 in liver metastasis of intrasplenic administration of colon 26 cells. Cancer Res. 2002;62(22):6682–6687. [PubMed] [Google Scholar]
- [117].Liu F, Ai F, Tian L, et al. Infliximab enhances the therapeutic effects of 5-fluorouracil resulting in tumor regression in colon cancer. Onco Targets Ther. 2016;9:5999–6008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Levin AD, Wildenberg ME, van den Brink GR. Mechanism of action of anti-TNF therapy in inflammatory bowel disease. J Crohns Colitis. 2016;10(8):989–997. [DOI] [PubMed] [Google Scholar]
- [119].Li Z, Chao T-C, Chang K-Y, et al. The long noncoding RNA THRIL regulates TNFalpha expression through its interaction with hnRNPL. Proc Natl Acad Sci U S A. 2014;111(3):1002–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Ray K. Coeliac disease: lnc13 and coeliac disease: a link to pathogenesis?. Nat Rev Gastroenterol Hepatol. 2016;13(6):314–315. [DOI] [PubMed] [Google Scholar]
- [121].Hanada T, Kobayashi T, Chinen T, et al. IFNgamma-dependent, spontaneous development of colorectal carcinomas in SOCS1-deficient mice. J Exp Med. 2006;203(6):1391–1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Mandai M, Hamanishi J, Abiko K, et al. Dual faces of IFN-gamma in cancer progression: a role of PD-L1 induction in the determination of Pro- and antitumor immunity. Clin Cancer Res. 2016;22(10):2329–2334. [DOI] [PubMed] [Google Scholar]
- [123].Valentini AM, Di Pinto F, Cariola F, et al. PD-L1 expression in colorectal cancer defines three subsets of tumor immune microenvironments. Oncotarget. 2018;9(9):8584–8596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Gomez JA, Wapinski O, Yang Y, et al. The NeST long ncRNA controls microbial susceptibility and epigenetic activation of the interferon-gamma locus. Cell. 2013;152(4):743–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Padua D, Mahurkar-Joshi S, Law IKM, et al. A long-noncoding RNA signature for ulcerative colitis identifies IFNG-AS1 as an enhancer of inflammation. Am J Physiol Gastrointest Liver Physiol. 2016;311(3):G446–G57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Mirza AH, Berthelsen CH, Seemann SE, et al. Transcriptomic landscape of lncRNAs in inflammatory bowel disease. Genome Med. 2015;7(1):39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Tsai BP, Wang X, Huang L, et al. Quantitative profiling of in vivo-assembled RNA-protein complexes using a novel integrated proteomic approach. Mol Cell Proteomics. 2011;10(4):M110 007385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Kudla G, Granneman S, Hahn D, et al. Cross-linking, ligation, and sequencing of hybrids reveals RNA-RNA interactions in yeast. Proc Natl Acad Sci U S A. 2011;108(24):10010–10015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Chu C, Chang HY. ChIRP-MS: RNA-Directed Proteomic Discovery. Methods Mol Biol. 2018;1861:37–45. [DOI] [PubMed] [Google Scholar]
- [130].Huang R, Han M, Meng L, et al. Capture and identification of RNA-binding proteins by using click chemistry-assisted RNA-interactome capture (CARIC) strategy. J Vis Exp. 2018;(140):e58580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Stork C, Zheng S. Genome-wide profiling of RNA-protein interactions using CLIP-Seq. Methods Mol Biol. 2016;1421:137–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Van Nostrand EL, Pratt GA, Shishkin AA, et al. Robust transcriptome-wide discovery of RNA-binding protein binding sites with enhanced CLIP (eCLIP). Nat Methods. 2016;13(6):508–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Bell JC, Jukam D, Teran NA, et al. Chromatin-associated RNA sequencing (ChAR-seq) maps genome-wide RNA-to-DNA contacts. Elife. 2018;7:pii:e27024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Li X, Zhou B, Chen L, et al. GRID-seq reveals the global RNA-chromatin interactome. Nat Biotechnol. 2017;35(10):940–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Mortimer SA, Trapnell C, Aviran S, et al. SHAPE-Seq: high-throughput RNA structure analysis. Curr Protoc Chem Biol. 2012;4(4):275–297. [DOI] [PubMed] [Google Scholar]
- [136].Friedlander MR, Lizano E, Houben AJ, et al. Evidence for the biogenesis of more than 1,000 novel human microRNAs. Genome Biol. 2014;15(4):R57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Derrien T, Johnson R, Bussotti G, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22(9):1775–1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Bryant CD, Yazdani N. RNA-binding proteins, neural development and the addictions. Genes Brain Behav. 2016;15(1):169–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Berg DJ, Davidson N, Kühn R, et al. Enterocolitis and colon cancer in interleukin-10-deficient mice are associated with aberrant cytokine production and CD4(+) TH1-like responses. J Clin Invest. 1996;98(4):1010–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Davidson NJ, Leach MW, Fort MM, et al. T helper cell 1-type CD4+ T cells, but not B cells, mediate colitis in interleukin 10-deficient mice. J Exp Med. 1996;184(1):241–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Zhu L, Ge J, Li T, et al. tRNA-derived fragments and tRNA halves: the new players in cancers. Cancer Lett. 2019;452:31–37. [DOI] [PubMed] [Google Scholar]
- [142].Esposito R, Bosch N, Lanzós A, et al. Hacking the cancer genome: profiling therapeutically actionable long non-coding RNAs using CRISPR-Cas9 screening. Cancer Cell. 2019;35(4):545–557. [DOI] [PubMed] [Google Scholar]


