ABSTRACT
RNA plays essential roles in not only translating nucleic acids into proteins, but also in gene regulation, environmental interactions and many human diseases. Nature uses over 150 chemical modifications to decorate RNA and diversify its functions. With the fast-growing RNA research in the burgeoning field of ‘epitranscriptome’, a term describes post-transcriptional RNA modifications that can dynamically change the transcriptome, it becomes clear that these modifications participate in modulating gene expression and controlling the cell fate, thereby igniting the new interests in RNA-based drug discovery. The dynamics of these RNA chemical modifications is orchestrated by coordinated actions of an array of writer, reader and eraser proteins. Deregulated expression of these RNA modifying proteins can lead to many human diseases including cancer. In this review, we highlight several critical modifications, namely m6A, m1A, m5C, inosine and pseudouridine, in both coding and non-coding RNAs. In parallel, we present a few other cancer-related tRNA and rRNA modifications. We further discuss their roles in cancer promotion or tumour suppression. Understanding the molecular mechanisms underlying the biogenesis and turnover of these RNA modifications will be of great significance in the design and development of novel anticancer drugs.
KEYWORDS: RNA modifications, epitranscriptome, cancer
Introduction
The past four decades have witnessed the growth of nucleic acid research, ranging from the discovery of new functions of RNAs as catalysts and regulators of numerous biochemical reactions to their conventional roles as carriers of genetic information, the adapters in protein synthesis and the structural scaffolds in subcellular organelles [1–5]. Accordingly, the essential roles that RNA can play as protein in the biological processes also led to the great interests in RNA-based drug discovery [6]. However, compared to proteins that contain 20 different amino acid residues, RNA only has four types of nucleobases. In order to achieve structural and functional diversity, nature uses a variety of chemical modifications to decorate RNAs in all the three primary domains of life [7]. Since the discovery of the first RNA modification almost 60 years ago in yeast, over 150 additional modifications have been identified in all types of RNA [8]. Great efforts have also been dedicated to the development of bioinformatic approaches (e.g. RNA Modification Database and MODOMICS [9]) to bridge the chemistry, the interacting enzymes and the biological effects of these modifications. Many of these modifications play critical roles in human diseases and biological processes such as embryonic stem cell differentiation, development, circadian rhythms, temperature adaptation, meiotic progression, and the regulation of RNA-RNA and RNA-protein binding interactions [10–16]. More interestingly, it is believed that these chemical modifications are the most evolutionarily conserved properties in RNAs, and some of the modified nucleobases are relics of the RNA World, where they may have enhanced the chemical diversity of RNA prior to protein [17].
Similar as epigenetic modifications at the DNA and protein levels, these posttranscriptional RNA modifications, also called ‘epitranscriptome’, can be dynamically and reversibly regulated by specific enzymes termed as ‘reader’ (translator), ‘writer’ (installer) and ‘eraser’ (demodifier), which represent a group of potential drug targets because of their ability to modulate RNA functions. Indeed, the linkages between several RNA modifying proteins to human diseases have been illustrated by the fast-growing applications of next-generation sequencing in genome-wide association studies (GWAS) [8]. With the most recent exciting progress focus on messenger RNA (mRNA), the core connection between DNA and protein, the significance of epitranscriptomics changes as new layers of gene regulation has been appreciated in other major RNA species, including transfer RNA (tRNA), which actually contains the most abundant and diverse modifications, ribosomal RNA (rRNA), small nuclear RNA (snRNA), microRNA (miRNA) and other non-coding RNAs responding to different physiological and environmental conditions. For example, methylation, the dominant RNA modification in mRNA as various forms like N6-methyladenosine (m6A), N1-methyladenosine (m1A), N7-methylguanidine (m7G), 5-methylcytidine (m5C) and 2ʹ-O-methylation (Nm), is widely present in all types of RNAs. Interestingly, even targeting the same type of methylation, the sets of methyltransferases and demethylase as its writers and erasers could be different in various types of RNA species [7]. These methylated moieties can modulate biological and pathological processes such as cell differentiation, stress response and tumorigenesis by providing diverse functions and dynamic regulation of RNA molecules. The research progress in this area has been extensively summarized [2,7,14,16,18–25]. In this review, we highlight several critical modifications, namely m6A, m1A, m5C, inosine and pseudouridine, which are relatively abundant and have been systematically detected, in both coding and non-coding RNA (ncRNA) contexts, and present their known roles in tumorigenesis. In addition, we summarize a few other cancer-related tRNA and rRNA modifications. The modifications are listed in Table 1 along with the types of cancer where it has been directly quantified or extrapolated from the expression level of writers or erasers enzymes. The chemical structures of modified nucleotides and the simplified catalytic pathways were depicted in Fig. 1. These modifications could play vital roles either as tumour-suppressive or tumour-promoting factors depending on the cellular and tumour types. As emerging new research areas, mapping new RNA modifications, studying the biological functions of RNA modification-related genes and understanding their pathogenic mechanisms will be of great significance in the design and development of novel anticancer drugs.
Table 1.
The known prominent RNA modifications associated with cancer types. Genes that have been analysed are shown in parentheses.
| RNA modifications | Cancer types (relevant genes) |
|---|---|
| m6A | Lung adenocarcinoma [57], AML [46], HER2 overexpressing subtypes breast cancer [26] and NANOG [53], t(11q23)/MLL rearranged, t(15;17)/PML-RARA, FLT3-ITD, and/or NPM1-mutated AMLs (ASB2 and RARA) [27] and GBM (FOXM1) [64]. |
| 2ʹ-O-methylation | Breast cancer [28,190], primary and metastatic prostate cancers [29] and squamous cell cervical carcinoma [30] |
| Pseudouridine (Ψ) | Leukaemia, lymphoma and multiple myeloma [113–115] |
| Inosine (A to I editing) | BLCA, BRCA, COAD, HNSC, LUAD, THCA, KICH [119,120], NSCLC (NEIL1 [122], AZIN1 [126], miR-381 [122]), SCLC (AZIN1) [119], HCC (AZIN1 [123], FLNB [128], GC (PODXL) [130], ESCC (FLNB) [124], ESCC (IGFBP7) [31], cervical cancer [121], CRC (RHOQ) [129], AML (PTPN6) [32], KIRP, KIC [119,120], breast cancer (Gabra3) [33], glioblastoma (GluR-B) [131], onco miR-21, miR-221, miR-222 [134], Glioma (miR-376a*) [135] and melanoma [34] (miR-455-5p) [136]. |
| 5mC/m5C | Circulating tumour cells in lung cancer [149] |
| m1A/m3C | PAAD [99], ESCA, COADREAD, LIHC, STAD [86], BLCA [83,84], LUAD [91], human prostate carcinoma [90], urothelial carcinomas [85], CRC (DLD-1), HCT116 [92], breast and ovarian cancer cells [87] |
AML: acute myeloid leukaemia; HER2: human epidermal growth factor receptor type 2; MLL: mixed lineage leukaemia; PML/RARA: promyelocytic leukaemia/retinoic acid receptor alpha; FLT3-ITD: Fms-related tyrosine kinase 3–internal tandem duplication; NPM1: nucleophosmin 1; RARA: retinoic acid receptor alpha; GBM: glioblastoma multiforme; BLCA: bladder urothelial carcinoma; BRCA: breast invasive carcinoma; COAD: colon adenocarcinoma; FOXM1: forkhead box protein M1; HNSC: head and neck squamous cell carcinoma; LUAD: lung adenocarcinoma; THCA: thyroid carcinoma; KICH: kidney chromophobe; NSCLC: non-small cell lung cancer; NEIL1: NEI-like protein 1; AZIN1: antizyme inhibitor 1; SCLC: small cell lung cancer; HCC: hepatocellular carcinoma; FlnB: Filamin B; GC: gastric cancer; PODXL: podocalyxin- like; ASB2: Ankyrin repeat and SOCS box containing 2; ESCC: oesophageal cell carcinoma; CRC: colorectal cancer; RHOQ: Ras homolog family member Q; PTPN6: protein tyrosine phosphatase non-receptor type 6; KIRP: kidney renal papillary cell carcinoma; Gabra3: Alpha-3 subunit of gamma-aminobutyric acid type A; GluR-B: glutamate R-B; IGFBP7: insulin-like growth factor-binding protein 7; PAAD: pancreatic adenocarcinoma; ESCA: oesophageal carcinoma; COADREAD: colorectal adenocarcinoma; LIHC: liver hepatocellular carcinoma; STAD: stomach adenocarcinoma.
Figure 1.
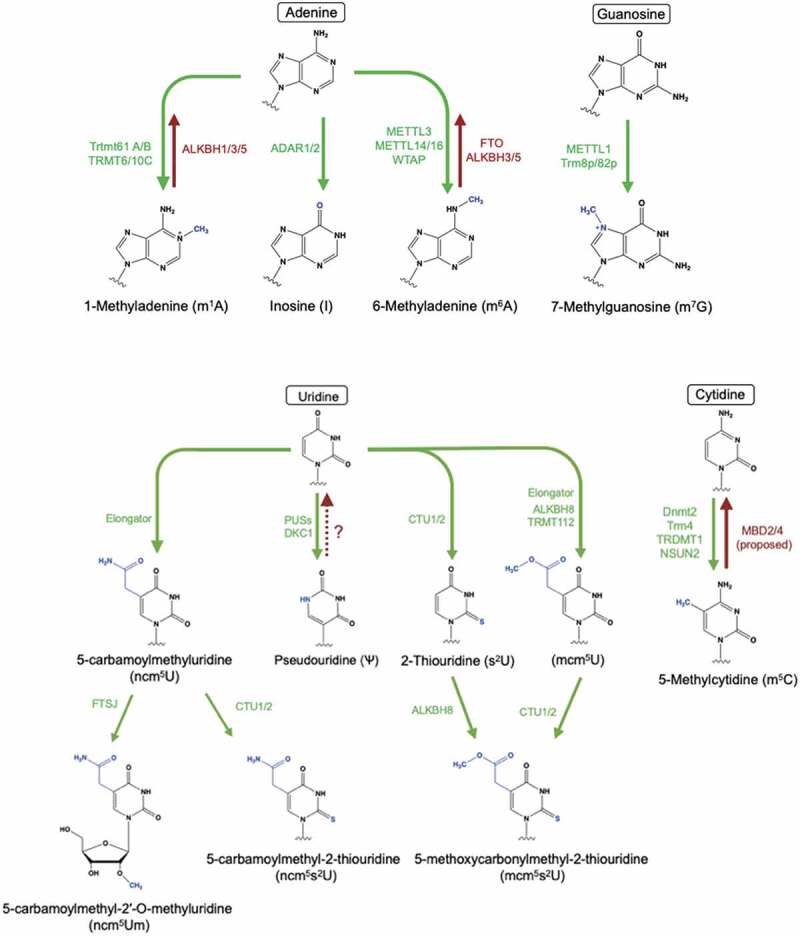
Chemical structures of RNA modifications on adenosine, cytosine and uridine. In green are the enzymes catalysing the reaction of the modification (writer) and in red are the putative enzymes removing the modification (erasers). The modification sites are coloured in blue. The abbreviations of the modified nucleosides are shown in the parenthesis.
N6-methyladenosine (m6A)
m6A in mRNA
Since our knowledge on structural mechanisms and functions of mRNA evolved from merely an adaptor molecule between DNA and protein to a gene regulator that can be modified and edited, manipulating mRNA has become a potential tool in developing novel therapeutics to treat a broad spectrum of diseases [35]. Among the various mRNA modifications, m6A is the most abundant form in eukaryotic cells, accounting for ~80% of all mRNA modifications, which is also represented by the extensive research in recent years. Using m6A specific antibodies to perform immunoprecipitation combined with high-throughput sequencing (MeRIP-seq), Meyer et al have shown that m6A was enriched in the three-prime untranslated region (3ʹ-UTR) and near stop codons [36]. Another study showed that m6A at the 5ʹ-UTR can promote cap-independent translation activities [37]. These lines of evidence suggested that site-specific modification could potentially affect the efficiency of translation and could possibly be an important regulator and even target for anticancer therapy. It has also been shown that the correct deposition of m6A in mRNA is essential for embryonic development and cell differentiation, which provides unique signalling in regulatory transcripts [13].
To further understand m6A modification in relation to cancer, one must first decipher the catalytic mediators of its writer, eraser and reader proteins. The reader proteins decode the message and signal downstream processes. There are many reader proteins have been identified for m6A, namely the YTH domain-containing proteins [38], eukaryotic initiation factor 3 (eIF3) [19], heterogeneous nuclear ribonucleoprotein (HNRNP) protein families [20] and insulin-like growth factor 2 mRNA-binding proteins (IGF2BP) which can recognize m6A in RNA and enhance mRNA stability and translation [39]. Two eraser proteins, fat mass and obesity-associated protein (FTO) and AlkB homolog 5 (ALKBH5), both of which are demethylase, have been demonstrated to effectively oxidize and demethylate the target m6A residues [40,41]. Liu et al. reported that m6A RNA methylation was catalysed by its writer protein complex consisting of human methyltransferase-like 14 (METTL14) and methyltransferase-like 3 (METTL3). The two proteins form a stable heterodimer, which functions in deposition of methyl on nuclear RNAs using SAM (S-adenosylmethionine) as cofactor [42]. Genetic inactivation or depletion of mouse and human METTL3 resulted in prolonged NANOG expression and delays embryonic stem cell (ESC) turnover from self-renewal, which in turns prevented the stem cell from differentiation into downstream lineages in the absence of m6A [43]. This indicates that m6A is required for stem cell signalling and regulation [10]. Another study has shown that in a mouse model, METTL3 knockout resulted in depletion of m6A in mRNA and subsequently led to early embryonic lethality [44]. This demonstrated the importance of m6A in embryo development and initiating the cell differentiation program. Another protein, Wilms’ tumour 1-associating protein (WTAP) also affects the methylation activities since in the absence of WTAP, the RNA-binding capability of METTL3 is strongly reduced, which suggested that WTAP is a regulatory subunit of the RNA m6A methyltransferase [45].
METTL3 mRNA and protein are found overexpressed in acute myeloid leukaemia (AML) cells compared to the healthy haematopoietic stem/progenitor cells (HSPCs) or other types of tumour cells [46]. It is also reported that METTL3 can control myeloid differentiation by conditionally depleting METTL3 in leukaemia cells, which resulted in cell differentiation and apoptosis and delayed leukaemia progression in recipient mice in vivo [46]. Mutations in METTL3 protein are known to be associated with poor haematopoietic proliferation and differentiation with the consequence leading to the accumulation of malignancy in myeloid cells [47]. Human hepatocellular carcinoma (HCC) is a dominant type of liver disease with low survival rate and therefore is considered to be one of the common cancer-related worldwide death [48]. METTL3 overexpression was also observed in human hepatocellular carcinoma (HCC). Knockdown of METTL3 in vitro showed that proliferation and colony formation of HCC cell are reduced. Moreover, knockout of METTL3 in vivo showed to suppress HCC tumorigenesis and lung metastasis. On the contrary, overexpression of METTL3 significantly promoted HCC growth [49]. Glioblastoma is the most common invasive malignant brain tumour diagnosed in the USA and associated with short life expectancy and poor prognosis [50]. A recent study showed that knockdown of METTL3 and METTL14 led to decreased level of m6A and resulted in self-renewal and tumorigenesis of glioblastoma stem cells [51]. Another report indicates that METTL3 is associated with breast cancer, in which METTL3 and oncogene hepatitis B X-interacting protein (HBXIP) are positively correlated in a way that HBXIP upregulates METTL3 and promotes the progression of breast cancer via inhibiting tumour suppressor miRNA let-7g [52]. Moreover, ALKBH5 can mediate m6A demethylation of NANOG in mRNA, leading to higher expression of NANOG mRNA and protein resulting in the breast cancer stem cell phenotype [53].
Another study also suggested that truncated membrane-associated guanylate kinase, WW and PDZ domain Containing 3 (MAGI3) mRNA lead to premature polyadenylation. This premature polyadenylation of MAGI3 mRNA is associated with low levels of m6A modification and can no longer function as tumour suppressor genes but turn into a non-functional gene in breast cancer [54]. Despite intensive research, breast cancer remains to be the top malignant tumour afflicting women and is responsible for high mortality rate and large number of deaths each year [50]. Likewise, cervical cancer is caused by human papillomavirus, and although it is a preventable disease, it is still the fourth most common cancer among women [55]. A myriad of evidence indicate that m6A is required for survival of health cells and reduced level of m6A in mRNA which is linked to the progression of human cervical cancer [56]. Non-small cell lung carcinoma (NSCLC) is the most common type of lung cancer accounts for about 85% of all cases, other 15% are small cell lung carcinoma [50]. METTL3 is also associated with NSCLC. It was demonstrated that the elevated level of METTL3 contributes to the tumorigenicity of lung cancer cells by enhancing the translation of oncogenic mRNA, such as epidermal growth factor receptor (RGFR) and protein coding gene Tafazzin (TAZ) [57]. Final remark to conclude this m6A modification in disease is that both METTL3 and METTL4 are overexpressed in human haematopoietic stem and progenitor cells (HSPCs), but are subsequently downregulated during the HSPC differentiation, suggesting that these two genes actually inhibit cell differentiation (Fig. 2) [56,57].
Figure 2.
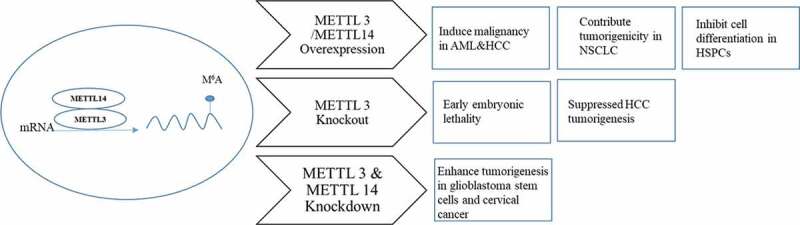
METTL3 is the main writer protein and works together with the substrate-recognizing subunit METTL14 to catalyse the methylation of m6A on mRNA. Overexpression of METTL3 was observed in acute myeloid leukaemia (AML), human hepatocellular carcinoma (HCC) and NSCLC (non-small cell lung carcinoma). Overexpression of both METTL3 and METTL4 was found in human haematopoietic stem and progenitor cells (HSPCs). Knockout of METTL3 in vivo causes early embryonic lethality and suppresses HCC tumorigenesis. By contrast, knockdown of both METTL3 and METTL 14 promoted tumorigenesis in brain and cervical cancers.
m6A in lncRNA
As previously noted, m6A is not only the most prevalent modification in mRNA but also is present in long-noncoding RNAs (lncRNAs). This modification can alter the secondary structure of lncRNA, and hence, affect many cellular processes such as splicing, transcription, translation and mRNA stability [58]. The writer methyltransferase includes the heterodimeric METTL3/METTL14 complex and WTAP. Two eraser demethylases are alkylated DNA repair protein B family named FTO and ALKBH5. Reader decoding proteins include YTH N6 methyladenosine RNA-binding protein 1–3 (YTHDF1-3) which belongs to the family of YT521-B homology domain [59]. METTL16 is a U6 small nuclear RNA (snRNA) methyltransferase which has the ability to regulate SAM synthetase and control SAM homoeostasis and promote intron retention [60]. METTL16 can also bind to the 3ʹ-terminal triple helix of metastasis associated with lncRNA of lung adenocarcinoma transcript 1 (MALAT1) [61]. In addition to lncRNA, microRNAs (miRNA), the small non-coding RNAs with about 18–25 nucleotides in length, have been demonstrated to play important roles in cell development, differentiation and the regulation of cell cycle in plants and animals by complementary base pairing with the 3ʹ-untranslated regions (3ʹUTR) of mRNA for cleavage or translational repression [62]. Methylated miRNAs are involved in apoptosis, proliferation, cell migration, angiogenesis and metastasis, indicating the broad impact of methylation on the process of oncogenic signalling pathways in cancer cells [63].
m6A in tRNA
ALKBH5 is regarded as a demethylase of m6A in transfer RNA (tRNA). It has been reported to play an oncogenic role in the development of glioblastoma (GBM) and breast cancer by affecting the self-renewal and proliferation of cancer stem cells [53,64,65]. The crystal structure of the N-terminal RNA-recognition motif (RRM) of ALKBH5 showed the recognition of hypermodification in tRNAs. In addition, demethylase from the same AlkB family, ALKBH3, was reported to form a complex with the activating signal cointegrator complex (ASCC) which is crucial for cancer cell proliferation [66]. A potential new mechanism by which ALKBH3 may promote tumour progression is associated with m6A demethylation in tRNA [67], although the activities of ALKBH3 that are most relevant in cancer are as yet unclear. Nevertheless, inhibiting ALKBH3 demethylase activity with small-molecule inhibitors shows high promises in preclinical cancer models [68–71].
N1- methyladenosine (m1A)
m1A in mRNA
N1 methylation on adenosine is another crucial posttranscriptional modification in RNA [72,73]. The addition of this methyl group is known to disrupt the base-pairing specificity, suggesting the regulatory functions of this modification in RNA [74]. The presence of m1A has been reported in mRNA, tRNA, rRNA and mitochondrial transcripts [75]. In mRNA, it predominantly appeared at the start codon upstream of the first splice site specifically enriched in the 5ʹ-UTR of mRNA and influence the translation [76]. In addition, it has the ability to stall reverse transcription and responds to stimuli in cellular stress environment [74]. The enzymes that recognize this modification site, ‘reader’ (YTHDF1, YTHDF2, YTHDF3, and YTHDC1) [73], and the ones regulate the level of m1A, the ‘writer’ (TRMT10C, Trmt61B, TRMT6/61A) and ‘eraser’ (ALKBH1, ALKBH3), have shown significant roles at the posttranscriptional stage of mRNA and ncRNAs [73,77–79]. It is worth mentioning that ALKBH1 and ALKBH3 were found as an eraser of m1A in single-stranded DNA and RNA [80–82].
The increased m1A level associated with hTrm6p/hTrm61p was found to promote urinary bladder cancer [83,84]. The loss of methylation by its demethylase, ALKBH3, led to progression, angiogenesis, and invasion of urothelial carcinomas by modulation through NADPH oxidase-2-reactive oxygen species (NOX-2-ROS) and the signals of the following complex: TNF-like weak inducer of apoptosis (TWEAK)/Fibroblast growth factor-inducible 14 (Fn14) and Vascular endothelial growth factor (VEGF) [85]. The recent report suggested that m1A regulates erb-b2 receptor tyrosine kinase 2 (ErbB2) and mechanistic target of rapamycin kinase (mTOR) pathways in gastrointestinal cancer (GI) [86]. In GI cancer cells, the level of the protein involved with modification m1A (writer: TRMT6, TRMT61A and TRT10C) (reader: ALKBH1/3) (eraser: YTHDF1-3 and YTHDC1) were observed to be mostly higher than the normal cell [86]. The demethylation of m1A has been shown to promote the breast and ovarian cancer cell invasiveness by stabilizing the mRNA of cytokine macrophage colony-stimulating factor (CSF-1) [87]. The longer the lifetime of CSF-1, the higher the activation of CSF-1R leading to promotion of metastasis. The expression of both ALKBH3 and CSF-1 is low in normal breast cell which agrees with the higer level of m1A on CFS-1 mRNA in normal breast cell in comparison to the cancer cells [87].
m1A in tRNA
Similar as the case of mRNA, the m1A modification in human mitochondria and cytoplasmic tRNA was catalysed by Trmt61B, TRMT6/61A and TRMT10C [78,79,88]; and the demethylation was catalysed by ALKBH1 and ALKBH3 [89]. The dynamic methylation of tRNA affects the cellular level of tRNAiMet and regulates translation initiation [89]. It has been reported that expression of ALKBH3 is elevated in pancreatic [90], lung [91] and urothelial cancers [85]. Also, ALKBH3 is suggested to promote the growth and progression of colorectal [92] and lung cancer cells [91]. ALKBH3 is a demethylase of m1A and m3C of tRNA in both HeLa and human embryonic kidney 293 cells, while not affecting the m7G, m1G and m5C levels [77]. The demethylation step leads to the progression of cancer. In vivo study showed that ALKBH3 can promote cancer cell proliferation, migration and invasion in addition to having the ability to regulate the growth of tumour xenografts. It was proposed that the demethylation of m1A leads to an increasing number of tRNA-derived small RNAs (tDRs) due to the higher susceptible of binding to angiogenin (ANG) cleavage (Fig. 3). The tRNA-derived fragment (tRFs) or tRNA-derived small RNAs (tDRs) are well conserved. They can strengthen the ribosome assembly and increase the translation rate and interactions with cytochrome C in order to prevent cell apoptosis [77]. In a non-small cell lung cancer model, tRF-Leu-CAG promoted cell proliferation and caused the G0/G1 cell cycle progression. The downregulation of tRF-Leu-CAG also repressed AURKA, indicating that tRF-Leu-CAG may be involved in regulating AURKA expression [93]. The high expression of tRF in a panel of cancer cell lines was also reported with strong relevance to cell proliferation [94].
Figure 3.
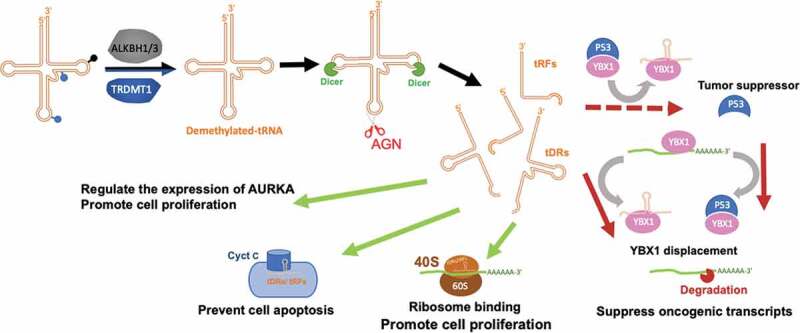
tRNA-derived small RNAs (tDRs) and tRNA-derived fragment (tRFs) affect multiple pathways to regulate gene expression and cell fate. The m6A, m1A and m3C on tRNA can be demethylated by ALKBH1 and ALKBH3 while TRDMT1 catalyzes 5mC demethylation. Demethylated-tRNA is prone to the formation of tDRs/tRFs through dicer and angiogenin (AGN) pathways. The tDRs promote cancer cell proliferation through three possible mechanisms (green arrows). On the contrary, the red arrow shows the modulation of invasion and metastatic lung colonization through tDRs-YBX1 binding, which tends to suppress oncogenic transcripts.
tsRNAs are tDRs that generated through the cleavage of the 3ʹ-end of pre-tRNA [95]. The dysregulation of tsRNA may exert oncogenic or tumour-suppressor functions in cancer [95,96]. Balatti et al. demonstrated that the overexpression of ts-46, ts-53 and ts-47 significantly reduced the clonal formation in lung cancer cells. They also showed the inhibitory effects in KRAS mutation cell lines and potential effects on the p53 pathway [96]. tRFs suppressed the invasion and metastatic lung colonization through the binding to Y-box binding protein1 (YBX1) protein, thereby resulting in the degradation of oncogenic transcripts (Fig. 3) [97]. YBX1 binding to P53 has been reported previously, which increased the DNA binding activity of p53, while simultaneously reduced the binding of YBX1 to the Y-box region of mRNA [98]. Together with the previous reports of tsRNA binding to YBX1 and releasing mRNA from YBX1-mRNA complex, tRFs may also be able to displace the YBX1 and release P53 from the P53-YBX1 complex with introducing another layer of gene regulation. The free tumour suppressor, P53, can bind to the YBX1 from YBX1-oncogenic transcripts, thus leading to a cancer-suppressive effect (dashed-red arrow in Fig. 3).
m1A in lncRNA
In addition to mRNA and tRNA, m1A modification is found in some lncRNAs. Although the exact roles of m1A still remain elusive and the writers of m1A are yet to be defined, the m1A modification level is positively correlated with protein production and the translational efficiency [76]. Two eraser proteins ALKBH1 and ALKBH3 are believed to account for the reversal of m1A modifications in lncRNAs [89]. High levels of ALKBH3 are found to be associated with human pancreatic cancer by supporting apoptotic resistance angiogenesis [99]. The readers are the same as m6A, including several YTH domain family: YTHDF1-3, and YTHDC1 [100].
Pseudouridine (ψ)
Pseudouridine (Ψ) is another RNA modification abundantly present in mRNA, tRNA, rRNA, snRNA and lncRNA. The single-base resolution mapping and precise quantification of this modification have been achieved recently. It was determined that the Ψ fractions in mRNA and lncRNA range from 30% to 84% in human cell lines [101]. The majority of ψ in mRNA functions as a regulator in response to environmental stress such as nutrient deprivation. It has also been shown that this modification plays a role in stabilizing RNA structure, altering translation initiation efficiency, ribosome pausing, RNA localization and RNA interference, thus providing an additional layer of control over gene expression [102]. The introduction of Ψ into eukaryotic RNA is mediated by the RNA-dependent H/ACA BOX snoRNA pseudouridine synthases (PUSs) or guide RNA-independent PUSs. A recent review by Penzo et al. has summarized the functional roles of pseudouridines in human pathologies [101,103].
The gene encoding the pseudouridine synthase dyskerin is DKC1, and the mutation of this gene could lead to the pseudouridylation defect and cause the X-linked Dyskeratosis Congenita (X-DC), a genetically uncommon and inherited disorder with mucocutaneous abnormalities and bone marrow failure in an X-lined autosomal dominant or recessive manner [104]. In addition to DKC1 mutations, it has also been shown that the autosomal form of X-DC is associated with the mutations in H/ACA-resembling domains in the RNA component of telomerase RNP, which are required for telomerase accumulation, stability, and 3ʹ-end processing [104–106].
Patients with X-DC have been reported to exhibit a higher risk for cancer development [107]. It has been hypothesized that a synergistic outcome of the impaired pseudouridylation on rRNA might account for the higher cancer susceptibility. As the studies in hypomorphic Dkc1-mutant mice suggested, the dysregulation of rRNA pseudouridylation indeed precedes cancer onset. The DKC1 mutation also results in the defect of the internal ribosome entry site (IRES), which is an RNA element allowing for translation initiation in a cap-independent manner, thus causing translational defect in some IRES-containing mRNAs. Similarly, ribosomes with the pseudouridine mutated rRNA show a much weaker binding affinity to the IRES elements [108]. Consequently, in hypomorphic DKC-1 mice, the translation of IRES-containing mRNAs, including the tumour suppressors p27 and p53, was perturbed, resulting in a higher incidence of cancer development in these mice [109–112]. Impaired translation of tumour suppressor mRNA might also be a key driving force of cancer in X-DC patients (Fig. 4B). Moreover, recent identification of widespread Ψ in mRNA introduces an additional layer of complexity and regulation of target RNA expressions [102]. Other than X-DC associated cancers, the downregulation of specific subsets of dyskerin-associated H/ACA snoRNAs has also been reported in haematological malignancies such as leukaemia, lymphoma and multiple myeloma [113–115], further strengthening the correlation between aberrant pseudouridylation and cancer.
Figure 4.
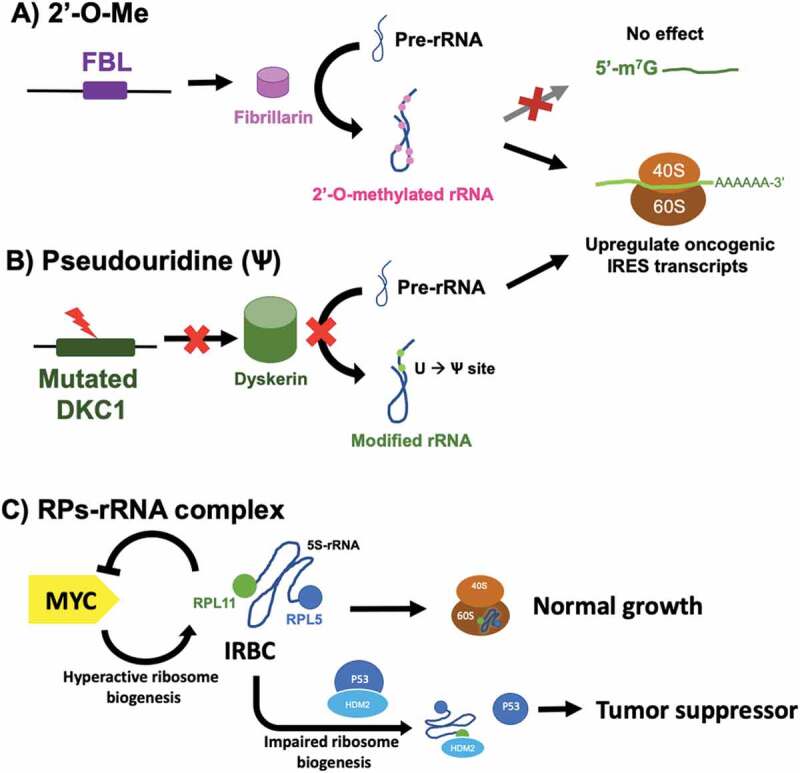
2ʹ-O-methylated rRNA showed direct correlation to cancer progression while pseudouridine modification was reported to have a negative correlation with cancer progression. (A) FBL regulates the methylation on rRNA leading to the upregulation of IRES-containing oncogenic transcripts, but has limited effects on m7G cap-dependent translation. (B) Pseudouridine on rRNA is regulated by DKC1. The knockdown DKC1 gene downregulates tumour suppressor proteins (P53 and P27) while upregulates the translation of VEGF. (C) The ribosomal protein complex with 5S-rRNA (IRBC) to regulate tumour proliferation by binding to HDM2 of the p53-HDM2 complex [220].
Inosine
Another common type of post-transcriptional modification is adenosine to inosine (A-to-I) RNA editing which is catalysed by the ADAR (adenosine deaminases acting on RNA) family of enzymes that include ADAR1-4 [116]. ADARs introduce inosine in both coding and non-coding RNAs to regulate transcription and translation.
Both hypo and hyper A-to-I editing occurred in the coding or noncoding RNA can affect fundamental cellular processes and thereby give rise to diverse diseases [117]. Most of the editing activities occur in the 3ʹUTR region, introns and other intergenic regions [118]. Studies focused on investigating the level of editing on non-coding region of mRNA such as introns of Arthrobacter luteus (Alu) and non-Alu mediated by ADAR 1 in tumour tissues in contrast to healthy tissues. The comparison was done by utilizing and analysing the database from The Cancer Genome Atlas (TCGA) project [119,120]. The result showed that high Alu editing levels in tumour tissues are a common feature among bladder urothelial carcinoma (BLCA), breast invasive carcinoma (BRCA), colon adenocarcinoma (COAD), head and neck squamous cell carcinoma (HNSC), lung adenocarcinoma (LUAD) and lastly, thyroid carcinoma (THCA) [117,121]. High ADAR1 expression was also noted in non-small cell lung cancer (NSCLC), causing the mutation on the NEI-like protein 1 (NEIL1) with single amino acid changed from lysine to arginine [122]. Encoding antizyme inhibitor 1 (AZIN1) was also upregulated by ADAR1 editing in hepatocellular carcinoma (HCC). ADAR1 not only increased the conversion of A to I but also caused serine to glycine substitution in AZIN1 proteins in promotion of tumorigenesis [117,123]. Other studies have reported similar aberrant editing activities by ADAR1 in oesophageal squamous cell carcinoma (ESCC) and breast cancer [124,125]. Recently, Hu et al. demonstrated that RNA editing of AZIN1 could promote malignant progression in non-small-cell lung cancers (NSCLC) [126]. Fritzell et al. reported similar trends in prostate, liver, chronic myoelogenous leukaemia (CML), colorectal and cervical cancers due to upregulation of ADAR1 within targeted mRNAs such as DHFR, AZIN1, BLCAP, RRUNE, thus promoting cancer progression [127]. Filamin B (FLNB) editing was shown to have an effect on both HCC and ESCC. ADAR1 hyperedited and ADAR 2 hypoedited the substitution of valine by methionine, attributing to HCC pathogenesis [128]. On the other hand, only ADAR1 hyperedited FLNB in ESCC [124].
Another editing event was reported on RhoQ gene with unidentified ADAR. The increased RhoQ mRNA editing is linked to colorectal cancer (CRC) and facilitates aggressiveness in metastasis [129]. High level of editing in miRNA also plays a role in cancer development. Overexpression of ADAR1 on miR381 was linked to NSCLC which caused enhancing of lung tumorigenesis [122]. Rather than hyperediting, kidney chromophobe (KICH) and kidney renal papillary cells both showed hypotherditing of ADAR 1 [120]. A low level of ADAR1 expression was also proposed to be linked with alpha-3 subunit of gamma-aminobutyric acid type A (Gabra3), which played a role in tumour progression in breast cancer. Gumireddy et al. reported that A-I mRNA editing was mainly found in non-invasive breast cancer which suggested that editing suppresses GABRA3-mediated Akt activation and breast cancer metastasis [120].
Hypoedited ADAR2 in podocalyxin-like (PODXL) was reported to be associated with gastric cancer. The editing caused a single amino acid change from histidine to arginine, which resulted in slower tumour growth and reduced invasive capability [130]. Hypoedited ADAR2 in GluR-B caused a glutamine-to-arginine substitution and led to malignant glioblastoma [131]. Insulin-like growth factor-binding protein 7 (IGFBP7), can be a target for ADAR2 mediated editing on the coding exon of position 284 from (AAG) lysine to (AIG) arginine with Inosine being read as Guanosine. This editing promoted apoptosis in ESCC. IGFBP 7 can potentially be a regulator of either promoting tumorigenesis in the case of under editing or cancer suppressor when over-edited [121]. In parallel to the mRNA hypoedition, low miRNA editing can also contribute to cancer development. Studies have suggested that ADAR2 editing is essential to suppress tumour growth in glioblastoma. This inhibition activity may be attributable to the regulation of onco-miRNA miR21, miR221 and miR222 [132,133]. ADAR2 served to reduce onco-miRNAs by editing miR21, miR221 and miR222 and thereby prevented them from maturing into onco-miRNA. The decreasing of microRNA editing activity mediated by ADAR2 can promote tumorigenesis in glioblastoma [134]. Moreover, Fritzell et al. reported that astrocytoma, glioblastoma, gastric, oesophageal and liver cancers all showed low editing activity of ADAR2, indicating that ADAR2 downregulation induced proto-oncogenic miRNA [127]. In human brain, microRNA-376a* can undergo A to I editing and its aberrant editing is associated with glioblasoma. Yukti et al. reported that attenuated A to I editing of miRNA-376a* could promote tumorigenesis and progression of cancer [135]. miRNA-455-4p can also be the target for ADAR1 editing. In the study conducted by Shoshan et al., it has been shown that under the increasing level of A to I caused by hypoediting of ADAR1 on miR-455-5p can inhibit the expression of tumour suppressor gene cytoplasmic polyadenylation element-binding protein (CPEB1), which leads to melanoma growth and metastasis [136]. Recent study on the level of ADAR3 in glioma cell reported that ADAR3 acted like a tumour suppressor and high level of ADAR3 can be predicted in lower-grade glioma (LGG) [137].
5-methylcytidine (m5c)
5-methylcytosine (m5C) has been identified in rRNA, tRNA and recently in mRNAs. It is particularly enriched in untranslated regions near Argonaute binding sites [138]. The methylation process was catalysed by the DNA methyltransferase homolog (Dnmt2) and the NOP2/Sun (NSUN 2 and 4) RNA methyltransferase family as m5C writers [139–141]. NSUN2 was reported to be targeted by critical transforming proteins such as c-MYC [140] and Aurora kinase B [142]. NSUN2 is reported to be upregulated in some tumour types by copy-number gains [143]. The overexpression of NSUN2 by DNA hypomethylation is associated with metastatic progression in human breast cancer [144]. However, there was also an observation of downregulation of NSUN2 in other classes of malignancies such as skin cancer, resulting in a reduction in protein translation rates and increase in the tumour-initiating population [145]. Mutations in NSUN2 can cause autosomal-recessive intellectual disability [146], and mutations in NSUN7 can cause sperm motility defects and infertility in male mice [147]. Evidence has also shown that m5C is actively involved in promoting mRNA export via m5C reader protein ALYREF, which acts as mRNA export adaptor in both in vitro and in vivo [148]. Although the detailed activities of these enzymes in tumorigenesis currently remain elusive and further investigation of the roles that m5C can play in cancer development is still required, the m5C levels have been increasingly recognized as a cancer marker. For example, increased RNA m5C levels could be detected in circulating tumour cells from lung cancer patients compared to those in whole blood cells [149].
In addition, it is also known that m5C in tRNA could regulate cancer progression through tRNA fragmentation process [150]. DNMT2 was found to be responsible for methylating cytidine at position 38 of tRNA specific for aspartate, which is similar to TRDMT1 activity [141]. Somatic cancer mutations were highly associated with the decreased methyltransferase activity of DNMT2 and the reduction of tRNA [151]. Similarly, NSUN3 is an RNA methyltransferase to regulate the level of 5-formylcytidine (f5C), a downstream analogue of m5C. The reduction of NSUN3 decreases the level of both m5C and f5C in tRNA, which is also linked with cancer [152].
Mcm5u34 in tRNA
The modification on tRNA and their related enzymes has long shown significance in the regulation of cancer pathogenesis [140,153,154]. In addition to the whole tRNA, the modified tRNA fragments or derivatives (tRFs/tsRNAs), which are cleaved by angiogenin, Dicer or RNase Z at different positions of the mature tRNA [94,155], also play crucial roles in RNA silencing, microenvironment monitoring and diseases like cancers [156]. The genome-wide tRNA profiling data revealed that the nuclear and mitochondrial encoded tRNAs were usually upregulated in pathogenic cells compared to healthy ones [157–160]. The disease-related tRNA modifications have been well summarized very recently [161]. We highlight here some discovery on cancer-associated wobble uridine modification, mcm5U34 in tRNA.
It is known that the modification at wobble position of tRNA affects the translational efficiency [162,163]. The 5-carbonylmethyluridine (cm5U) was methylated by ALKBH8 and tRNA methyltransferase 9-like (hTRM9L) to generate 5-methoxycarbonylmethyluridine (mcm5U) at wobble position [164]. In human breast cancer, the expression of U34-modifying enzymes, namely elongator complex protein 3 (Elp3) and cytoplasmic tRNA 2-thiolation protein 1 and 2 (Ctu1/2), which further catalysed the formation of mcm5s2-U34, promoted the translation of oncoprotein DEK and increased the translation of the oncogenic LEF-1 mRNA via binding to the LEF1-IRES sequence, leading to the invasion and metastasis of breast cancer cells [154]. Another study reported the high level of tRNA methyltransferase homolog 12 (TRMT12) in several breast cancer cell lines and tissues [153], although its molecular mechanisms were unclear. On 2016, it was reported that the methyltransferase ALKBH8, the Trm9 homologs in mammals, was highly expressed in bladder cancer and the absence of ALHBH8 promoted cell apoptosis due to the reduction of the anti-apoptotic protein surviving level [165,166].
In addition to sustaining the metastasis of breast and bladder cancers, U34-tRNA modifiers were shown to be the key regulators of the survival of malignant melanoma cells [154,166,167]. The elevated levels of ELP1, ELP3, CTU1 and CTU2 were observed in BRAFV600E cells which is the most common mutation among human melanoma patients and believed to be responsible for resistance to targeted therapy [161]. In addition, the development of BRAFV600E melanoma in a zebrafish model was compromised by the inactive ELP3 [167]. The depletion of ELP1, ELP3, ELP5 or ELP6 in melanoma cells contributed to the reduction of the migration and oncogenesis [168]. The high level of ELP3 was shown to link to the phosphorylation level of protein kinase B (AKT) in human hepatocellular carcinoma (HCC) cells [169].
The Elongator complex induced the radical SAM-dependent pathway for modification on position 5 of U34 with the involvement of Kti11/Dph3-Kti13 complex [170,171]. Kti11 (also known as Dph3) and Kti13 (also known as Ats1) were shown to influence in Elongator regulation process [172]. A hetero-dimer of Kti11 and Kti13 affected Elongator’s of U34 modification process by precipitating the Elongator subunits (Elp1, Elp2, Elp3, Elp5) [170,171,173–176]. Depletion of Kti11 diminished the U34 modification activity of Elongator while the loss of Kti13 reduced 20% of the tRNA modification [174,175,177].
Diphthamide is a posttranslational modification on histidine residue (His699 in yeast; His715 in humans) found on translation elongation factor 2 (EF2), which is an essential translation factor that mediates the translocation of the ribosome during elongation process. Diphthamide-EF2 was catalysed by a group of Dph family Dph1, Dph2, Dph4, Dph5, Dph6 and Dph7 including Kti11/Dph3 [178,179]. It has been found that Dph1 and Dph5 activity is associated with the proliferation of intestinal stem cell in Drosophila melanogaster [180]. Diphtheria toxin (DT) introduced cell death via inactivation of the translation factor through adenosine diphosphate (ADP)-ribosylation. It has been found that the loss of Kti11/Dph3 let to the cell resistance to DT suggesting that diphthamide-EF2 is the target of DT [170,174,178,179,181]. These data supported that diphthamide post-translation modification regulates cell growth and proliferation via stabilizing the reading frame and reducing the ribosomal errors [161,181–185]. In contrary, the methylation enzyme, hTRM9L, was reported to be down-regulated in breast, bladder, colorectal, cervix and testicular carcinomas and diminished in more aggressive SW620 and HCT116 colon carcinoma cell lines. Interestingly, the restored methylation dramatically suppressed tumour growth in vivo via LIN9 and HIF1-α-dependent mechanisms [164]. Therefore, the dysregulation of mcm5s2U34 modification seems to be regulated by many layers of protein-RNA complex chain interactions, which affect the level of modification on U34 of tRNA. The presence of modification on U34 may also be subjected to tRNA fragmentation formation leading to the cancer cell progression or inhibition of oncogene as showed in Fig. 3.
2′-O-Me modification in rRNA
An increasing body of evidence links the alteration of rRNA modification levels and the defect in components of the rRNA modification machinery to development, genetic diseases and cancer [186]. For example, as stated previously, pseudouridine modification is regulated by the pseudouridine synthase 1 (DKC1) gene, which played critical roles in dyskeratosis congenita (DC) and its associated cancers.
2′-O-Me modification in rRNA was reported to depend on the methylation level of rRNA. Ribosome expression is upregulated in most of cervical intraepithelial neoplasia (CIN) when compared to healthy tissue. The methylation level at cytosines in the CpG islands was significantly reduced in rDNA promoter region of CIN tissues together with the decondensation of rDNA chromatin. The methylation inhibition experiment by 5-aza-2ʹ-deoxycytidine (DAC) suggested the negative correlation between methylation level in rDNA promoter region and 45S-rDNA. The data indicated that the decreasing of methylation in rDNA promoter results in the development of human cervical cancer through an increasing of rRNA synthesis [187].
Studies showed the correlation of modulation function of FBL gene and 2′-O-Me patterns. Downregulation of FBL led to rRNA 2′-O-Me patterns, with a direct impact on ribosome function, in neurogenesis and stem cell differentiation [188,189]. In addition, maintaining the expression of FBL in stem cells prolonged their pluripotent state in mouse embryos [189]. In breast cancer cells, changes in FBL expression, which altered the level of 2′-O-Me in rRNA, affected translational accuracy and translational initiation efficiency of mRNAs containing internal ribosome entry site (IRES) elements [190–192]. Diminishing the rRNA 2ʹO-methyltransferase, fibrillarin, by FBL knocked down affected the ribosome biogenesis and global 2′-O-Me-rRNA in human cells [193]. The overexpression of FBL in tumours and cancer cells upregulated 2ʹ-O-Me modification [190]. Fibrillarin stimulates the cancer-promoting protein translation; IGF1R [194], c-Myc [195], FGF1/2 [196], and VEGFA [190,197]. Mutated p53 associated with tumorigenesis was reported to promote methylation level in rRNA and thus increased their translation fidelity and rate (Fig. 4A) [117].
On the contrary, the FBL knockdown resulted in the accumulation of p53, increasing IRES-driven de novo synthesis, possibly through the effect of UTR of the p53_mRNA [192]. Elimination of the methylation function affected translation from globin and GAPDH 5′UTR in the in vitro translation assay. However, 2′-O-Me does not significantly modulate the ability of ribosomes to initiate m7G-Cap-dependent translation. However, due to the dysregulation of FBL on the cancer cell growth and the interplay of p53, more studies are required to fully understand the mechanism of FBL modulation on the level of 2ʹ-O-Me and its effect on translational activity.
P53 is the key mediator [198] of the abolishing of cell differentiation and proliferation in mice liver caused by deletion of gene encoding 40S ribosomal protein S6 (RpS6 or eS6) [199]. During the impairment of 60S ribosome biogenesis, many precursor complexes newly synthesized 60S ribosomal protein L5 (RPL5, uL18), L11 (RPL11, uL5) and 5S rRNA. They target the E3 ubiquitin-protein ligase HDM2 (also known as MDM2 in mice) thereby preventing the ubiquitylation and degradation of p53 [200–205]. The free pre-ribosomal RPL5–RPL11–5S rRNA complex that binds to HDM2 has been termed ‘asimpaired ribosome biogenesis checkpoint’ (IRBC) complex [23]. The IRBC regulated p53–HCM2 interactions through stress-activated responses [206]. Among several ribosomal proteins (RPs), the studies showed that RPL5 and RPL11 are the main RPs that involved in stabilizing p53 in a mutually dependent manner (Fig. 4C) [204,207]. A crystal structure of HDM2-RPL11 complex revealed the binding site in the acidic region of HDM2 [208], which was hypothesized to mimic the 28S rRNA binding site for RPLL in the 60S subunit [23,208]. Additional analyses are needed to elucidating the HDM2-IRBC binding mechanism to facilitate the HDM2 inhibition therapeutic.
The IRBC was also reported to involve in hyperactive ribosome biogenesis. When MDM2 (HDM2) of Eμ-Myc mice failed to bind to IRBC because of the mutation on Mdm2C305F knock-in, the higher rate of lymphomagenesis than the expression in wild-type Mdm2 was observed [209]. A number of studies hypothesized that the upregulation of MYC translation would increase the number of RPL5 and RPL11 for HDM2-inhibition leading to an intrinsic tumour suppressor response [209–212]. It was also reported that RPL5 and RPL11 destabilize MYC-mRNA through the inhibition of MYC transcription (Fig. 4C) [213–216] which was supported by the observation of increasing the level of MYC protein on primary mouse embryonic fibroblasts with heterozygous deletion of Rpl11 [217]. The cellular senescence was induced by many stress signals including telomere shortening, oxidative stress and DNA damage, generally in a p53-dependent manner [218]. IRBC could activate cellular senescence as a barrier against tumour formation by preventing proliferation or by inducing immune-mediated clearance of pre-malignant cells [218] which supported that IRBC mediates tumour suppression [219].
lncRNAs with cancer
In the end, we think it is worthwhile highlighting the research progress of the three most studied cancer-related lncRNAs, which contain various of chemical modifications: the metastasis-associated lung adenocarcinoma transcript 1 (MALAT1), the Hox transcript antisense intergenic RNA (HOTAIR), and the X-inactive specific transcript (XIST). MALAT1 bears m6A and pseudouridine modifications and was reported to function as a regulator of metastasis to control cancer cell proliferation, migration and apoptosis in pancreatic, hepatic and ovarian cancers. HOTAIR which bears cytosine methylation can promote metastasis in gastric, colorectal, pancreatic, hepatic, breast and skin cancers. XIST has all three types of chemical modifications, including m6A, m5C and ψ, and can act either as oncogene or as a suppressor in leukaemia and colorectal cancers (Fig. 5) [59].
Figure 5.
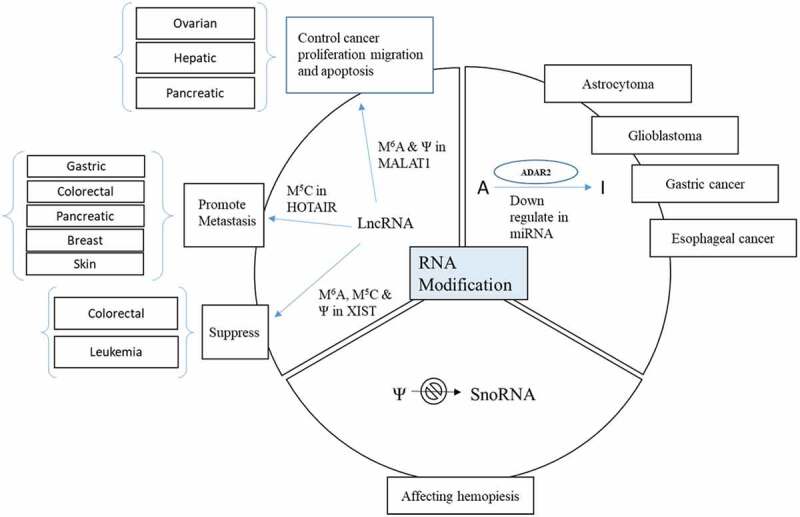
Examples of aberrant RNA modifications in cancer-related lncRNAs. M6A and Ψ in MALAT1 were reported to function as a regulator of metastasis to control cancer cell proliferation, migration and apoptosis in pancreatic, hepatic and ovarian cancers. M5C in HOTAIR can promote metastasis in gastric, colorectal, pancreatic, hepatic, breast and skin cancers.
Concluding remarks
As summarized above, epitranscriptomic levels and dynamic changes of RNA modifications may hold promise as new diagnostic biomarkers of clinical values. More importantly, the enzymes involved in regulating RNA metabolism may function as oncogenic regulators and serve as novel therapeutic targets. In the future, precision medicine based on epitranscriptomic signatures may be tailored to the diagnosis and treatment of specific tumour types within individual patients. However, the functionality of these chemical modifications in both coding and non-coding RNAs is not yet fully understood and still require collaborative endeavour to draw a clear connection between modifications in RNAs and cancer. Further improvements in sequencing sensitivity and functional validation methods are urgently needed to achieve this loft goal.
Acknowledgments
This work was supported by grants from NSF (CHE-1845486 MCB-1715234 to JS), Cancer Prevention and Research Institute of Texas (RP170660 to YZ), National Institute of Health grants (R01HL134780 to YH, R01HL146852 to YH, R01GM112003 to YZ), the Welch Foundation (BE-1913-20190330 to YZ), the American Cancer Society (RSG-18-043-01-LIB to YH; RSG-16-215-01-TBE to YZ). Ya Ying Zheng is currently supported by NIH training grant T32GM132066.
Funding Statement
This work was supported by the American Cancer Society [RSG-18-043-01-LIB]; American Cancer Society [RSG-16-215-01-TBE]; National Institutes of Health [R01HL134780, R01HL146852]; National Institutes of Health [R01GM112003]; National Science Foundation [1845486]; Welch Foundation [BE-1913-20190330].
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- [1].Jia G, Fu Y, He C.. Reversible RNA adenosine methylation in biological regulation. Trends Genet. 2013;29(2):108–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Noh JH, Kim KM, McClusky WG, et al. Cytoplasmic functions of long noncoding RNAs. Wiley Interdiscip Rev RNA. 2018;9(3):e1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Yin X, Liang C, Feng Y, et al. Research progress on synthetic scaffold in metabolic engineering - a review. Sheng Wu Gong Cheng Xue Bao. 2019;35(3):363–374. [DOI] [PubMed] [Google Scholar]
- [4].den Boon JA, Ahlquist P.. Organelle-like membrane compartmentalization of positive-strand RNA virus replication factories. Annu Rev Microbiol. 2010;64:241–256. [DOI] [PubMed] [Google Scholar]
- [5].Howard MJ, Liu X, Lim WH, et al. RNase P enzymes: divergent scaffolds for a conserved biological reaction. RNA Biol. 2013;10(6):909–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].!!! INVALID CITATION !!!
- [7].Lewis CJT, Pan T, Kalsotra A. RNA modifications and structures cooperate to guide RNA–protein interactions. Nat Rev Mol Cell Biol. 2017;18:202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Ontiveros RJ, Stoute J, Liu KF. The chemical diversity of RNA modifications. Biochem J. 2019;476(8):1227–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Boccaletto P, et al. MODOMICS: a database of RNA modification pathways. 2017 update. Nucleic Acids Res. 2018;46(D1):D303–D307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Batista PJ, Molinie B, Wang J, et al. m6A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem Cell. 2014;15(6):707–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Benegiamo G, Brown SA, Panda S. RNA dynamics in the control of circadian rhythm. Adv Exp Med Biol. 2016;907:107–122. [DOI] [PubMed] [Google Scholar]
- [12].Lorenz C, Lunse CE, Morl M. tRNA modifications: impact on structure and thermal adaptation. Biomolecules. 2017;7(2):E35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Roundtree IA, Evans ME, Pan T, et al. Dynamic RNA Modifications in Gene Expression Regulation. Cell. 2017;169(7):1187–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Frye M, Harada BT, Behm M, et al. RNA modifications modulate gene expression during development. Science. 2018;361(6409):1346–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Fazi F, Fatica A. Interplay between N (6)-methyladenosine (m(6)A) and non-coding RNAs in cell development and cancer. Front Cell Dev Biol. 2019;7:116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Zaccara S, Ries RJ, Jaffrey SR. Reading, writing and erasing mRNA methylation. Nat Rev Mol Cell Biol. 2019;20:608–624. [DOI] [PubMed] [Google Scholar]
- [17].Ma W. What does “the RNA world” mean to “the origin of life”? Life (Basel). 2017;7(4):E49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Esteller M, Pandolfi PP. The epitranscriptome of noncoding RNAs in cancer. Cancer Discov. 2017;7(4):359–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Meyer KD, Jaffrey SR. Rethinking m(6)A readers, writers, and erasers. Annu Rev Cell Dev Biol. 2017;33:319–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Zhao BS, Roundtree IA, He C. Post-transcriptional gene regulation by mRNA modifications. Nat Rev Mol Cell Biol. 2017;18(1):31–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Deng X, Su R, Feng X, et al. Role of N 6 -methyladenosine modification in cancer. Curr Opin Genet Dev. 2018;48:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Lian H, Wang Q-H, Zhu C-B, et al. Deciphering the epitranscriptome in cancer. Trends in Cancer. 2018;4(3):207–221. [DOI] [PubMed] [Google Scholar]
- [23].Pelletier J, Thomas G, Volarevic S. Ribosome biogenesis in cancer: new players and therapeutic avenues. Nat Rev Cancer. 2018;18(1):51–63. [DOI] [PubMed] [Google Scholar]
- [24].Tong J, Flavell RA, Li HB. RNA m(6)A modification and its function in diseases. Front Med. 2018;12(4):481–489. [DOI] [PubMed] [Google Scholar]
- [25].Delaunay S, Frye M. RNA modifications regulating cell fate in cancer. Nat Cell Biol. 2019;21(5):552–559. [DOI] [PubMed] [Google Scholar]
- [26].Tan A, Dang Y, Chen G, et al. Overexpression of the fat mass and obesity associated gene (FTO) in breast cancer and its clinical implications. Int J Clin Exp Pathol. 2015;8(10):13405–13410. [PMC free article] [PubMed] [Google Scholar]
- [27].Li Z, Weng H, Su R, et al. FTO Plays an oncogenic role in acute myeloid leukemia as a N 6 -methyladenosine RNA demethylase. Cancer Cell. 2017;31(1):127–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Appaiah HN, Goswami CP, Mina LA, et al. Persistent upregulation of U6: SNORD44small RNA ratio in the serum of breast cancer patients. Breast Cancer Res. 2011;13(5):R86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Koh CM, Gurel B, Sutcliffe S, et al. Alterations in nucleolar structure and gene expression programs in prostatic neoplasia are driven by the MYC oncogene. Am J Pathol. 2011;178(4):1824–1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Choi YW, Kim Y-W, Bae S-M, et al. Identification of differentially expressed genes using annealing control primer-based GeneFishing in human squamous cell cervical carcinoma. Clin Oncol (R Coll Radiol). 2007;19(5):308–318. [DOI] [PubMed] [Google Scholar]
- [31].Chen YB, et al. ADAR2 functions as a tumor suppressor via editing IGFBP7 in esophageal squamous cell carcinoma. Int J Oncol. 2017;50(2):622–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Beghini A, Ripamonti CB, Peterlongo P, et al. RNA hyperediting and alternative splicing of hematopoietic cell phosphatase (PTPN6) gene in acute myeloid leukemia. Hum Mol Genet. 2000;9(15):2297–2304. [DOI] [PubMed] [Google Scholar]
- [33].Gumireddy K, Li A, Kossenkov AV, et al. The mRNA-edited form of GABRA3 suppresses GABRA3-mediated Akt activation and breast cancer metastasis. Nat Commun. 2016;7:10715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Nemlich Y, Greenberg E, Ortenberg R, et al. MicroRNA-mediated loss of ADAR1 in metastatic melanoma promotes tumor growth. J Clin Invest. 2013;123(6):2703–2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Laina A, Gatsiou A, Georgiopoulos G, et al. RNA therapeutics in cardiovascular precision medicine. Front Physiol. 2018;9:953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Meyer KD, Saletore Y, Zumbo P, et al. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell. 2012;149(7):1635–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Meyer KD, Patil D, Zhou J, et al. 5ʹ UTR m(6)A promotes cap-independent translation. Cell. 2015;163(4):999–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Luo S, Tong L. Molecular basis for the recognition of methylated adenines in RNA by the eukaryotic YTH domain. Proc Natl Acad Sci U S A. 2014;111(38):13834–13839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Huang H, Weng H, Sun W, et al. Recognition of RNA N(6)-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat Cell Biol. 2018;20(3):285–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Jia G, Fu Y, Zhao X, et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol. 2011;7(12):885–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Zheng G, Dahl J, Niu Y, et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell. 2013;49(1):18–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Liu J, Yue Y, Han D, et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol. 2014;10(2):93–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Chambers I, Colby D, Robertson M, et al. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell. 2003;113(5):643–655. [DOI] [PubMed] [Google Scholar]
- [44].Geula S, Moshitch-Moshkovitz S, Dominissini D, et al. m6A mRNA methylation facilitates resolution of naïve pluripotency toward differentiation. Science. 2015;347(6225):1002–1006. [DOI] [PubMed] [Google Scholar]
- [45].Ping X-L, Sun B-F, Wang L, et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 2014;24(2):177–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Vu LP, Pickering BF, Cheng Y, et al. The N(6)-methyladenosine (m(6)A)-forming enzyme METTL3 controls myeloid differentiation of normal hematopoietic and leukemia cells. Nat Med. 2017;23(11):1369–1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].De Kouchkovsky I, Abdul-Hay M. Acute myeloid leukemia: a comprehensive review and 2016 update. Blood Cancer J. 2016;6(7):e441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Jemal A, Bray F, Center MM, et al. Global cancer statistics. CA Cancer J Clin. 2011;61(2):69–90. [DOI] [PubMed] [Google Scholar]
- [49].Chen M, Wei L, Law C-T, et al. RNA N6-methyladenosine methyltransferase-like 3 promotes liver cancer progression through YTHDF2-dependent posttranscriptional silencing of SOCS2. Hepatology. 2018;67(6):2254–2270. [DOI] [PubMed] [Google Scholar]
- [50].Liu J, Harada BT, He C. Regulation of gene expression by N(6)-methyladenosine in cancer. Trends Cell Biol. 2019;29(6):487–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Cui Q, Shi H, Ye P, et al. m 6 A RNA methylation regulates the self-renewal and tumorigenesis of glioblastoma stem cells. Cell Rep. 2017;18(11):2622–2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Cai X, Wang X, Cao C, et al. HBXIP-elevated methyltransferase METTL3 promotes the progression of breast cancer via inhibiting tumor suppressor let-7g. Cancer Lett. 2018;415:11–19. [DOI] [PubMed] [Google Scholar]
- [53].Zhang C, Samanta D, Lu H, et al. Hypoxia induces the breast cancer stem cell phenotype by HIF-dependent and ALKBH5-mediated m 6 A-demethylation of NANOG mRNA. Proc Natl Acad Sci U S A. 2016;113(14):E2047–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Ni TK, Elman JS, Jin DX, et al. Premature polyadenylation of MAGI3 is associated with diminished N(6)-methyladenosine in its large internal exon. Sci Rep. 2018;8(1):1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Lopez MS, ES Baker, Maza M, et al. Cervical cancer prevention and treatment in Latin America. J Surg Oncol. 2017;115(5):615–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Wang X, Li Z, Kong B, et al. Reduced m(6)A mRNA methylation is correlated with the progression of human cervical cancer. Oncotarget. 2017;8(58):98918–98930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Lin S, Choe J, Du P, et al. The m 6 A methyltransferase METTL3 promotes translation in human cancer cells. Mol Cell. 2016;62(3):335–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Zhou KI, et al. N(6)-methyladenosine modification in a long noncoding RNA hairpin predisposes its conformation to protein binding. J Mol Biol. 2016;428(5 Pt A):822–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Dinescu S, Ignat S, Lazar A, et al. Epitranscriptomic signatures in lncRNAs and their possible roles in cancer. Genes (Basel). 2019;10(1):52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Pendleton KE, Chen B, Liu K, et al. The U6 snRNA m 6 A methyltransferase METTL16 regulates SAM synthetase intron retention. Cell. 2017;169(5):824–835 e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Brown JA, CG Kinzig, SJ DeGregorio, et al. Methyltransferase-like protein 16 binds the 3ʹ-terminal triple helix of MALAT1 long noncoding RNA. Proc Natl Acad Sci U S A. 2016;113(49):14013–14018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. [DOI] [PubMed] [Google Scholar]
- [63].Loginov VI, Rykov SV, Fridman MV, et al. Methylation of miRNA genes and oncogenesis. Biochemistry (Mosc). 2015;80(2):145–162. [DOI] [PubMed] [Google Scholar]
- [64].Zhang S, Zhao BS, Zhou A, et al. m 6 A demethylase ALKBH5 maintains tumorigenicity of glioblastoma stem-like cells by sustaining FOXM1 expression and cell proliferation program. Cancer Cell. 2017;31(4):591–606 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Zhang C, Zhi WI, Lu H, et al. Hypoxia-inducible factors regulate pluripotency factor expression by ZNF217- and ALKBH5-mediated modulation of RNA methylation in breast cancer cells. Oncotarget. 2016;7(40):64527–64542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Thapar R, Bacolla A, Oyeniran C, et al. RNA modifications: reversal mechanisms and cancer. Biochemistry. 2019;58(5):312–329. [DOI] [PubMed] [Google Scholar]
- [67].Ueda Y, Ooshio I, Fusamae Y, et al. AlkB homolog 3-mediated tRNA demethylation promotes protein synthesis in cancer cells. Sci Rep. 2017;7:42271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Li Q, Huang Y, Liu X, et al. Rhein inhibits AlkB repair enzymes and sensitizes cells to methylated DNA damage. J Biol Chem. 2016;291(21):11083–11093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Nakao S, Mabuchi M, Shimizu T, et al. Design and synthesis of prostate cancer antigen-1 (PCA-1/ALKBH3) inhibitors as anti-prostate cancer drugs. Bioorg Med Chem Lett. 2014;24(4):1071–1074. [DOI] [PubMed] [Google Scholar]
- [70].Wang P, Wu J, Ma S, et al. Oncometabolite D-2-hydroxyglutarate inhibits ALKBH DNA repair enzymes and sensitizes IDH mutant cells to alkylating agents. Cell Rep. 2015;13(11):2353–2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Ueda M, Shimizu T, Mabuchi M, et al. Novel metabolically Stable PCA-1/ALKBH3 inhibitor has potent antiproliferative effects on DU145 cells In vivo. Anticancer Res. 2018;38(1):211–218. [DOI] [PubMed] [Google Scholar]
- [72].Dunn DB. The occurrence of 1-methyladenine in ribonucleic acid. Biochim Biophys Acta. 1961;46:198–200. [DOI] [PubMed] [Google Scholar]
- [73].Dai X, Wang T, Gonzalez G, et al. Identification of YTH domain-containing proteins as the readers for N1-methyladenosine in RNA. Anal Chem. 2018;90(11):6380–6384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Li X, Xiong X, Wang K, et al. Transcriptome-wide mapping reveals reversible and dynamic N1-methyladenosine methylome. Nat Chem Biol. 2016;12(5):311–316. [DOI] [PubMed] [Google Scholar]
- [75].Zhang C, Jia G. Reversible RNA Modification N(1)-methyladenosine (m(1)A) in mRNA and tRNA. Genomics Proteomics Bioinformatics. 2018;16(3):155–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Dominissini D, Nachtergaele S, Moshitch-Moshkovitz S, et al. The dynamic N(1)-methyladenosine methylome in eukaryotic messenger RNA. Nature. 2016;530(7591):441–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Chen Z, Qi M, Shen B, et al. Transfer RNA demethylase ALKBH3 promotes cancer progression via induction of tRNA-derived small RNAs. Nucleic Acids Res. 2019;47(5):2533–2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Chujo T, Suzuki T. Trmt61B is a methyltransferase responsible for 1-methyladenosine at position 58 of human mitochondrial tRNAs. RNA. 2012;18(12):2269–2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Safra M, Sas-Chen A, Nir R, et al. The m1A landscape on cytosolic and mitochondrial mRNA at single-base resolution. Nature. 2017;551(7679):251–255. [DOI] [PubMed] [Google Scholar]
- [80].Duncan T, Trewick SC, Koivisto P, et al. Reversal of DNA alkylation damage by two human dioxygenases. Proc Natl Acad Sci U S A. 2002;99(26):16660–16665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Aas PA, Otterlei M, Falnes PØ, et al. Human and bacterial oxidative demethylases repair alkylation damage in both RNA and DNA. Nature. 2003;421(6925):859–863. [DOI] [PubMed] [Google Scholar]
- [82].Trewick SC, Henshaw TF, Hausinger RP, et al. Oxidative demethylation by Escherichia coli AlkB directly reverts DNA base damage. Nature. 2002;419(6903):174–178. [DOI] [PubMed] [Google Scholar]
- [83].Shi L, Yang X-M, Tang -D-D, et al. Expression and significance of m1A transmethylase, hTrm6p/hTrm61p and its related gene hTrm6/hTrm61 in bladder urothelial carcinoma. Am J Cancer Res. 2015;5(7):2169–2179. [PMC free article] [PubMed] [Google Scholar]
- [84].Barraud P, Golinelli-Pimpaneau B, Atmanene C, et al. Crystal structure of thermus thermophilus tRNA m1A58 methyltransferase and biophysical characterization of its interaction with tRNA. J Mol Biol. 2008;377(2):535–550. [DOI] [PubMed] [Google Scholar]
- [85].Shimada K, Fujii T, Tsujikawa K, et al. ALKBH3 contributes to survival and angiogenesis of human urothelial carcinoma cells through NADPH oxidase and tweak/Fn14/VEGF signals. Clin Cancer Res. 2012;18(19):5247–5255. [DOI] [PubMed] [Google Scholar]
- [86].Zhao Y, Zhao Q, Kaboli PJ, et al. m1A regulated genes modulate PI3K/AKT/mTOR and ErbB pathways in gastrointestinal cancer. Transl Oncol. 2019;12(10):1323–1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Woo -H-H, Chambers SK. Human ALKBH3-induced m1A demethylation increases the CSF-1 mRNA stability in breast and ovarian cancer cells. Biochim Biophys Acta Gene Regul Mech. 2019;1862(1):35–46. [DOI] [PubMed] [Google Scholar]
- [88].Vilardo E, Rossmanith W. Molecular insights into HSD10 disease: impact of SDR5C1 mutations on the human mitochondrial RNase P complex. Nucleic Acids Res. 2015;43(13):6649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Liu F, Clark W, Luo G, et al. ALKBH1-mediated tRNA demethylation regulates translation. Cell. 2016;167(7):1897. [DOI] [PubMed] [Google Scholar]
- [90].Konishi N, Nakamura M, Ishida E, et al. High expression of a new marker PCA-1 in human prostate carcinoma. Clin Cancer Res. 2005;11(14):5090–5097. [DOI] [PubMed] [Google Scholar]
- [91].Tasaki M, Shimada K, Kimura H, et al. ALKBH3, a human AlkB homologue, contributes to cell survival in human non-small-cell lung cancer. Br J Cancer. 2011;104(4):700–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Luo J, Emanuele MJ, Li D, et al. A genome-wide RNAi screen identifies multiple synthetic lethal interactions with the Ras oncogene. Cell. 2009;137(5):835–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Shao Y, Sun Q, Liu X, et al. tRF-Leu-CAG promotes cell proliferation and cell cycle in non-small cell lung cancer. Chem Biol Drug Des. 2017;90(5):730–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Lee YS, Shibata Y, Malhotra A, et al. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs). Genes Dev. 2009;23(22):2639–2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Pekarsky Y, Balatti V, Palamarchuk A, et al. Dysregulation of a family of short noncoding RNAs, tsRNAs, in human cancer. Proc Natl Acad Sci U S A. 2016;113(18):5071–5076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Balatti V, Nigita G, Veneziano D, et al. tsRNA signatures in cancer. Proc Natl Acad Sci U S A. 2017;114(30):8071–8076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Goodarzi H, Liu X, Nguyen HB, et al. Endogenous tRNA-derived fragments suppress breast cancer progression via YBX1 displacement. Cell. 2015;161(4):790–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Okamoto T, Izumi H, Imamura T, et al. Direct interaction of p53 with the Y-box binding protein, YB-1: a mechanism for regulation of human gene expression. Oncogene. 2000;19(54):6194–6202. [DOI] [PubMed] [Google Scholar]
- [99].Yamato I, Sho M, Shimada K, et al. PCA-1/ALKBH3 contributes to pancreatic cancer by supporting apoptotic resistance and angiogenesis. Cancer Res. 2012;72(18):4829–4839. [DOI] [PubMed] [Google Scholar]
- [100].Dai D, Wang H, Zhu L, et al. N6-methyladenosine links RNA metabolism to cancer progression. Cell Death Dis. 2018;9(2):124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Zhang W, Eckwahl MJ, Zhou KI, et al. Sensitive and quantitative probing of pseudouridine modification in mRNA and long noncoding RNA. RNA. 2019;25(9):1218–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Carlile TM, Rojas-Duran MF, Zinshteyn B, et al. Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells. Nature. 2014;515(7525):143–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Penzo M, AN Guerrieri, Zacchini F, et al. RNA pseudouridylation in physiology and medicine: for better and for worse. Genes (Basel). 2017;8(11). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Mitchell JR, Wood E, Collins K. A telomerase component is defective in the human disease dyskeratosis congenita. Nature. 1999;402(6761):551–555. [DOI] [PubMed] [Google Scholar]
- [105].Jády BE, Bertrand E, Kiss T. Human telomerase RNA and box H/ACA scaRNAs share a common Cajal body-specific localization signal. J Cell Biol. 2004;164(5):647–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Mitchell JR, Cheng J, Collins K. A box H/ACA small nucleolar RNA-like domain at the human telomerase RNA 3ʹ end. Mol Cell Biol. 1999;19(1):567–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Penzo M, Casoli L, Ceccarelli C, et al. DKC1 gene mutations in human sporadic cancer. Histol Histopathol. 2013;28(3):365–372. [DOI] [PubMed] [Google Scholar]
- [108].Jack K, Bellodi C, Landry D, et al. rRNA pseudouridylation defects affect ribosomal ligand binding and translational fidelity from yeast to human cells. Mol Cell. 2011;44(4):660–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Montanaro L, Calienni M, Bertoni S, et al. Novel dyskerin-mediated mechanism of p53 inactivation through defective mRNA translation. Cancer Res. 2010;70(11):4767–4777. [DOI] [PubMed] [Google Scholar]
- [110].Bellodi C, Krasnykh O, Haynes N, et al. Loss of function of the tumor suppressor DKC1 perturbs p27 translation control and contributes to pituitary tumorigenesis. Cancer Res. 2010;70(14):6026–6035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Yoon A, Peng G, Brandenburg Y, et al. Impaired control of IRES-mediated translation in X-linked dyskeratosis congenita. Science. 2006;312(5775):902–906. [DOI] [PubMed] [Google Scholar]
- [112].Bellodi C, Kopmar N, Ruggero D. Deregulation of oncogene-induced senescence and p53 translational control in X-linked dyskeratosis congenita. Embo J. 2010;29(11):1865–1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Ronchetti D, Todoerti K, Tuana G, et al. The expression pattern of small nucleolar and small Cajal body-specific RNAs characterizes distinct molecular subtypes of multiple myeloma. Blood Cancer J. 2012;2:e96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Valleron W, Ysebaert L, Berquet L, et al. Small nucleolar RNA expression profiling identifies potential prognostic markers in peripheral T-cell lymphoma. Blood. 2012;120(19):3997–4005. [DOI] [PubMed] [Google Scholar]
- [115].Valleron W, Laprevotte E, Gautier E-F, et al. Specific small nucleolar RNA expression profiles in acute leukemia. Leukemia. 2012;26(9):2052–2060. [DOI] [PubMed] [Google Scholar]
- [116].Nishikura K. Functions and regulation of RNA editing by ADAR deaminases. Annu Rev Biochem. 2010;79:321–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Tusup M, Kundig T, Pascolo S. Epitranscriptomics of cancer. World J Clin Oncol. 2018;9(3):42–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Xu X, Wang Y, Liang H. The role of A-to-I RNA editing in cancer development. Curr Opin Genet Dev. 2018;48:51–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Han L, Diao L, Yu S, et al. The genomic landscape and clinical relevance of A-to-I RNA editing in human cancers. Cancer Cell. 2015;28(4):515–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Paz-Yaacov N, Bazak L, Buchumenski I, et al. Elevated RNA editing activity is a major contributor to transcriptomic diversity in tumors. Cell Rep. 2015;13(2):267–276. [DOI] [PubMed] [Google Scholar]
- [121].Chen Y, Wang H, Lin W, et al. ADAR1 overexpression is associated with cervical cancer progression and angiogenesis. Diagn Pathol. 2017;12(1):12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Anadon C, Guil S, Simó-Riudalbas L, et al. Erratum: gene amplification-associated overexpression of the RNA editing enzyme ADAR1 enhances human lung tumorigenesis. Oncogene. 2016;35(33):4422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Chen L, et al. Recoding RNA editing of AZIN1 predisposes to hepatocellular carcinoma. Nat Med. 2013;19(2):209–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Qin Y-R, Qiao -J-J, Chan THM, et al. Adenosine-to-inosine RNA editing mediated by ADARs in esophageal squamous cell carcinoma. Cancer Res. 2014;74(3):840–851. [DOI] [PubMed] [Google Scholar]
- [125].Fumagalli D, et al. Principles governing A-to-I RNA editing in the breast cancer transcriptome. Cell Rep. 2015;13(2):277–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Hu X, Chen J, Shi X, et al. RNA editing of AZIN1 induces the malignant progression of non-small-cell lung cancers. Tumour Biol. 2017;39(8):1010428317700001. [DOI] [PubMed] [Google Scholar]
- [127].Fritzell K, Xu L-D, Lagergren J, et al. ADARs and editing: the role of A-to-I RNA modification in cancer progression. Semin Cell Dev Biol. 2018;79:123–130. [DOI] [PubMed] [Google Scholar]
- [128].Chan TH, Lin CH, Qi L, et al. A disrupted RNA editing balance mediated by ADARs (Adenosine DeAminases that act on RNA) in human hepatocellular carcinoma. Gut. 2014;63(5):832–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Han S-W, Kim H-P, Shin J-Y, et al. RNA editing in RHOQ promotes invasion potential in colorectal cancer. J Exp Med. 2014;211(4):613–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Chan TH, Qamra A, Tan KT, et al. ADAR-mediated RNA editing predicts progression and prognosis of gastric cancer. Gastroenterology. 2016;151(4):637–650 e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Maas S, Patt S, Schrey M, et al. Underediting of glutamate receptor GluR-B mRNA in malignant gliomas. Proc Natl Acad Sci U S A. 2001;98(25):14687–14692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Galeano F, Rossetti C, Tomaselli S, et al. ADAR2-editing activity inhibits glioblastoma growth through the modulation of the CDC14B/Skp2/p21/p27 axis. Oncogene. 2013;32(8):998–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Karsy M, Arslan E, Moy F. Current progress on understanding microRNAs in glioblastoma multiforme. Genes Cancer. 2012;3(1):3–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Tomaselli S, Galeano F, Alon S, et al. Modulation of microRNA editing, expression and processing by ADAR2 deaminase in glioblastoma. Genome Biol. 2015;16:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Choudhury Y, Tay FC, Lam DH, et al. Attenuated adenosine-to-inosine editing of microRNA-376a* promotes invasiveness of glioblastoma cells. J Clin Invest. 2012;122(11):4059–4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Shoshan E, Mobley AK, Braeuer RR, et al. Reduced adenosine-to-inosine miR-455-5p editing promotes melanoma growth and metastasis. Nat Cell Biol. 2015;17(3):311–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Zhang Y, Wang K, Zhao Z, et al. ADAR3 expression is an independent prognostic factor in lower-grade diffuse gliomas and positively correlated with the editing level of GRIA2Q607R. Cancer Cell Int. 2018;18(1):196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Squires JE, et al. Widespread occurrence of 5-methylcytosine in human coding and non-coding RNA. Nucleic Acids Res. 2012;40(11):5023–5033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Metodiev MD, Spåhr H, Loguercio Polosa P, et al. NSUN4 is a dual function mitochondrial protein required for both methylation of 12S rRNA and coordination of mitoribosomal assembly. PLoS Genet. 2014;10(2):e1004110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Frye M, Watt FM. The RNA methyltransferase Misu (NSun2) mediates Myc-induced proliferation and is upregulated in tumors. Curr Biol. 2006;16(10):971–981. [DOI] [PubMed] [Google Scholar]
- [141].Goll MG, Kirpekar F, Maggert KA, et al. Methylation of tRNAAsp by the DNA methyltransferase homolog Dnmt2. Science. 2006;311(5759):395–398. [DOI] [PubMed] [Google Scholar]
- [142].Hussain S, Benavente SB, Nascimento E, et al. The nucleolar RNA methyltransferase Misu (NSun2) is required for mitotic spindle stability. J Cell Biol. 2009;186(1):27–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Frye M, Dragoni I, Chin S-F, et al. Genomic gain of 5p15 leads to over-expression of Misu (NSUN2) in breast cancer. Cancer Lett. 2010;289(1):71–80. [DOI] [PubMed] [Google Scholar]
- [144].Yi J, Gao R, Chen Y, et al. Overexpression of NSUN2 by DNA hypomethylation is associated with metastatic progression in human breast cancer. Oncotarget. 2017;8(13):20751–20765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145].Blanco S, Bandiera R, Popis M, et al. Stem cell function and stress response are controlled by protein synthesis. Nature. 2016;534(7607):335–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].Abbasi-Moheb L, Mertel S, Gonsior M, et al. Mutations in NSUN2 cause autosomal-recessive intellectual disability. Am J Hum Genet. 2012;90(5):847–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147].Harris T, Marquez B, Suarez S, et al. Sperm motility defects and infertility in male mice with a mutation in Nsun7, a member of the Sun domain-containing family of putative RNA methyltransferases. Biol Reprod. 2007;77(2):376–382. [DOI] [PubMed] [Google Scholar]
- [148].Yang X, Yang Y, Sun B-F, et al. 5-methylcytosine promotes mRNA export — NSUN2 as the methyltransferase and ALYREF as an m5C reader. Cell Res. 2017;27(5):606–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Huang W, Qi C-B, Lv S-W, et al. Determination of DNA and RNA methylation in circulating tumor cells by mass spectrometry. Anal Chem. 2016;88(2):1378–1384. [DOI] [PubMed] [Google Scholar]
- [150].Schaefer M, Pollex T, Hanna K, et al. RNA methylation by Dnmt2 protects transfer RNAs against stress-induced cleavage. Genes Dev. 2010;24(15):1590–1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [151].Elhardt W, Shanmugam R, Jurkowski TP, et al. Somatic cancer mutations in the DNMT2 tRNA methyltransferase alter its catalytic properties. Biochimie. 2015;112:66–72. [DOI] [PubMed] [Google Scholar]
- [152].Van Haute L, Dietmann S, Kremer L, et al. Deficient methylation and formylation of mt-tRNA(Met) wobble cytosine in a patient carrying mutations in NSUN3. Nat Commun. 2016;7:12039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153].Towns WL, Begley TJ. Transfer RNA methytransferases and their corresponding modifications in budding yeast and humans: activities, predications, and potential roles in human health. DNA Cell Biol. 2012;31(4):434–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Delaunay S, Rapino F, Tharun L, et al. Elp3 links tRNA modification to IRES-dependent translation of LEF1 to sustain metastasis in breast cancer. J Exp Med. 2016;213(11):2503–2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Ivanov P, Emara M, Villen J, et al. Angiogenin-induced tRNA fragments inhibit translation initiation. Mol Cell. 2011;43(4):613–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [156].Maute RL, Schneider C, Sumazin P, et al. tRNA-derived microRNA modulates proliferation and the DNA damage response and is down-regulated in B cell lymphoma. Proc Natl Acad Sci U S A. 2013;110(4):1404–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [157].Huang S-Q, Sun B, Xiong Z-P, et al. The dysregulation of tRNAs and tRNA derivatives in cancer. J Exp Clin Cancer Res. 2018;37(1):101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [158].Mahlab S, Tuller T, Linial M. Conservation of the relative tRNA composition in healthy and cancerous tissues. RNA. 2012;18(4):640–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [159].Goodarzi H, Nguyen HCB, Zhang S, et al. Modulated expression of specific tRNAs drives gene expression and cancer progression. Cell. 2016;165(6):1416–1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [160].Pavon-Eternod M, Gomes S, Geslain R, et al. tRNA over-expression in breast cancer and functional consequences. Nucleic Acids Res. 2009;37(21):7268–7280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [161].Hawer H, Hammermeister A, Ravichandran K, et al. Roles of elongator dependent tRNA modification pathways in neurodegeneration and cancer. Genes (Basel). 2018;10(1):19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [162].Novoa EM, Pavon-Eternod M, Pan T, et al. A role for tRNA modifications in genome structure and codon usage. Cell. 2012;149(1):202–213. [DOI] [PubMed] [Google Scholar]
- [163].Agris PF, Vendeix FAP, Graham WD. tRNA’s wobble decoding of the genome: 40 years of modification. J Mol Biol. 2007;366(1):1–13. [DOI] [PubMed] [Google Scholar]
- [164].Begley U, Sosa MS, Avivar‐Valderas A, et al. A human tRNA methyltransferase 9-like protein prevents tumour growth by regulating LIN9 and HIF1-alpha. EMBO Mol Med. 2013;5(3):366–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [165].Shimada K, Nakamura M, Anai S, et al. A novel human AlkB homologue, ALKBH8, contributes to human bladder cancer progression. Cancer Res. 2009;69(7):3157–3164. [DOI] [PubMed] [Google Scholar]
- [166].Ohshio I, Kawakami R, Tsukada Y, et al. ALKBH8 promotes bladder cancer growth and progression through regulating the expression of survivin. Biochem Biophys Res Commun. 2016;477(3):413–418. [DOI] [PubMed] [Google Scholar]
- [167].Rapino F, Delaunay S, Rambow F, et al. Codon-specific translation reprogramming promotes resistance to targeted therapy. Nature. 2018;558(7711):605–609. [DOI] [PubMed] [Google Scholar]
- [168].Close P, Gillard M, Ladang A, et al. DERP6 (ELP5) and C3ORF75 (ELP6) regulate tumorigenicity and migration of melanoma cells as subunits of Elongator. J Biol Chem. 2012;287(39):32535–32545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [169].Xu Y, Zhou W, Ji Y, et al. Elongator promotes the migration and invasion of hepatocellular carcinoma cell by the phosphorylation of AKT. Int J Biol Sci. 2018;14(5):518–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [170].Glatt S, Zabel R, Vonkova I, et al. Structure of the Kti11/Kti13 heterodimer and its double role in modifications of tRNA and eukaryotic elongation factor 2. Structure. 2015;23(1):149–160. [DOI] [PubMed] [Google Scholar]
- [171].Kolaj-Robin O, McEwen AG, Cavarelli J, et al. Structure of the Elongator cofactor complex Kti11/Kti13 provides insight into the role of Kti13 in Elongator-dependent tRNA modification. Febs J. 2015;282(5):819–833. [DOI] [PubMed] [Google Scholar]
- [172].Fichtner L, Schaffrath R. KTI11 and KTI13, Saccharomyces cerevisiae genes controlling sensitivity to G1 arrest induced by Kluyveromyces lactis zymocin. Mol Microbiol. 2002;44(3):865–875. [DOI] [PubMed] [Google Scholar]
- [173].Fichtner L, Jablonowski D, Schierhorn A, et al. Elongator’s toxin-target (TOT) function is nuclear localization sequence dependent and suppressed by post-translational modification. Mol Microbiol. 2003;49(5):1297–1307. [DOI] [PubMed] [Google Scholar]
- [174].Bar C, Zabel R, Liu S, et al. A versatile partner of eukaryotic protein complexes that is involved in multiple biological processes: kti11/Dph3. Mol Microbiol. 2008;69(5):1221–1233. [DOI] [PubMed] [Google Scholar]
- [175].Zabel R, Bär C, Mehlgarten C, et al. Yeast α-tubulin suppressor Ats1/Kti13 relates to the Elongator complex and interacts with Elongator partner protein Kti11. Mol Microbiol. 2008;69(1):175–187. [DOI] [PubMed] [Google Scholar]
- [176].Liu S, Leppla SH. Retroviral insertional mutagenesis identifies a small protein required for synthesis of diphthamide, the target of bacterial ADP-ribosylating toxins. Mol Cell. 2003;12(3):603–613. [DOI] [PubMed] [Google Scholar]
- [177].Huang B, Johansson MJ, Bystrom AS. An early step in wobble uridine tRNA modification requires the Elongator complex. RNA. 2005;11(4):424–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [178].Chen JY, Bodley JW, Livingston DM. Diphtheria toxin-resistant mutants of Saccharomyces cerevisiae. Mol Cell Biol. 1985;5(12):3357–3360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [179].Liu S, Milne GT, Kuremsky JG, et al. Identification of the proteins required for biosynthesis of diphthamide, the target of bacterial ADP-ribosylating toxins on translation elongation factor 2. Mol Cell Biol. 2004;24(21):9487–9497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [180].Obata F, Tsuda-Sakurai K, Yamazaki T, et al. Nutritional control of stem cell division through S-adenosylmethionine in drosophila intestine. Dev Cell. 2018;44(6):741–751 e3. [DOI] [PubMed] [Google Scholar]
- [181].Uthman S, Bär C, Scheidt V, et al. The amidation step of diphthamide biosynthesis in yeast requires DPH6, a gene identified through mining the DPH1-DPH5 interaction network. PLoS Genet. 2013;9(2):e1003334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [182].Ortiz PA, Ulloque R, GK Kihara, et al. Translation elongation factor 2 anticodon mimicry domain mutants affect fidelity and diphtheria toxin resistance. J Biol Chem. 2006;281(43):32639–32648. [DOI] [PubMed] [Google Scholar]
- [183].Abeyrathne PD, San Koh C, Grant T, et al. Ensemble cryo-EM uncovers inchworm-like translocation of a viral IRES through the ribosome. Elife. 2016;5:e14874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [184].Murray J, CG Savva, BS Shin, et al. Structural characterization of ribosome recruitment and translocation by type IV IRES. Elife. 2016;5:e13567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [185].Pellegrino S, Demeshkina N, Mancera-Martinez E, et al. Structural Insights into the Role of Diphthamide on Elongation Factor 2 in mRNA Reading-Frame Maintenance. J Mol Biol. 2018;430(17):2677–2687. [DOI] [PubMed] [Google Scholar]
- [186].Sloan KE, AS Warda, Sharma S, et al. Tuning the ribosome: the influence of rRNA modification on eukaryotic ribosome biogenesis and function. RNA Biol. 2017;14(9):1138–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [187].Zhou H, Wang Y, Lv Q, et al. Overexpression of ribosomal RNA in the development of human cervical cancer is associated with rDNA promoter hypomethylation. PLoS One. 2016;11(10):e0163340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [188].Recher G, Jouralet J, Brombin A, et al. Zebrafish midbrain slow-amplifying progenitors exhibit high levels of transcripts for nucleotide and ribosome biogenesis. Development. 2013;140(24):4860–4869. [DOI] [PubMed] [Google Scholar]
- [189].Watanabe-Susaki K, Takada H, Enomoto K, Miwata K, et al. Biosynthesis of ribosomal RNA in nucleoli regulates pluripotency and differentiation ability of pluripotent stem cells. Stem Cells. 2014;32(12):3099–3111. [DOI] [PubMed] [Google Scholar]
- [190].Marcel V, Ghayad S, Belin S, et al. p53 acts as a safeguard of translational control by regulating fibrillarin and rRNA methylation in cancer. Cancer Cell. 2013;24(3):318–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [191].Basu A, et al. Requirement of rRNA methylation for 80S ribosome assembly on a cohort of cellular internal ribosome entry sites. Mol Cell Biol. 2011;31(22):4482–4499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [192].Su H, Xu T, Ganapathy S, et al. Elevated snoRNA biogenesis is essential in breast cancer. Oncogene. 2014;33(11):1348–1358. [DOI] [PubMed] [Google Scholar]
- [193].Erales J, et al. Evidence for rRNA 2ʹ-O-methylation plasticity: control of intrinsic translational capabilities of human ribosomes. Proc Natl Acad Sci U S A. 2017;114(49):12934–12939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [194].Pollak MN, Schernhammer ES, Hankinson SE. Insulin-like growth factors and neoplasia. Nat Rev Cancer. 2004;4(7):505–518. [DOI] [PubMed] [Google Scholar]
- [195].Dang CV. MYC on the path to cancer. Cell. 2012;149(1):22–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [196].Billottet C, Elkhatib N, Thiery J-P, et al. Targets of fibroblast growth factor 1 (FGF-1) and FGF-2 signaling involved in the invasive and tumorigenic behavior of carcinoma cells. Mol Biol Cell. 2004;15(10):4725–4734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [197].Carmeliet P. VEGF as a key mediator of angiogenesis in cancer. Oncology. 2005;69(Suppl 3):4–10. [DOI] [PubMed] [Google Scholar]
- [198].Volarevic S, et al. Proliferation, but not growth, blocked by conditional deletion of 40S ribosomal protein S6. Science. 2000;288(5473):2045–2047. [DOI] [PubMed] [Google Scholar]
- [199].Turowski TW, Tollervey D. Cotranscriptional events in eukaryotic ribosome synthesis. Wiley Interdiscip Rev RNA. 2015;6(1):129–139. [DOI] [PubMed] [Google Scholar]
- [200].Zhang Y, Wolf GW, Bhat K, et al. Ribosomal protein L11 negatively regulates oncoprotein MDM2 and mediates a p53-dependent ribosomal-stress checkpoint pathway. Mol Cell Biol. 2003;23(23):8902–8912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [201].Bhat KP, Itahana K, Jin A, et al. Essential role of ribosomal protein L11 in mediating growth inhibition-induced p53 activation. Embo J. 2004;23(12):2402–2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [202].Dai MS, Lu H. Inhibition of MDM2-mediated p53 ubiquitination and degradation by ribosomal protein L5. J Biol Chem. 2004;279(43):44475–44482. [DOI] [PubMed] [Google Scholar]
- [203].Donati G, Peddigari S, Mercer C, et al. 5S ribosomal RNA is an essential component of a nascent ribosomal precursor complex that regulates the Hdm2-p53 checkpoint. Cell Rep. 2013;4(1):87–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [204].Bursac S, Brdovcak MC, Pfannkuchen M, et al. Mutual protection of ribosomal proteins L5 and L11 from degradation is essential for p53 activation upon ribosomal biogenesis stress. Proc Natl Acad Sci U S A. 2012;109(50):20467–20472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [205].Lohrum MAE, Ludwig RL, Kubbutat MHG, et al. Regulation of HDM2 activity by the ribosomal protein L11. Cancer Cell. 2003;3(6):577–587. [DOI] [PubMed] [Google Scholar]
- [206].Bode AM, Dong Z. Post-translational modification of p53 in tumorigenesis. Nat Rev Cancer. 2004;4(10):793–805. [DOI] [PubMed] [Google Scholar]
- [207].Fumagalli S, Ivanenkov VV, Teng T, et al. Suprainduction of p53 by disruption of 40S and 60S ribosome biogenesis leads to the activation of a novel G2/M checkpoint. Genes Dev. 2012;26(10):1028–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [208].Zheng J, Lang Y, Zhang Q, et al. Structure of human MDM2 complexed with RPL11 reveals the molecular basis of p53 activation. Genes Dev. 2015;29(14):1524–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [209].Macias E, Jin A, Deisenroth C, et al. An ARF-independent c-MYC-activated tumor suppression pathway mediated by ribosomal protein-Mdm2 Interaction. Cancer Cell. 2010;18(3):231–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [210].Nie Z, Hu G, Wei G, et al. c-Myc is a universal amplifier of expressed genes in lymphocytes and embryonic stem cells. Cell. 2012;151(1):68–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [211].Lin CY, Lovén J, Rahl P, et al. Transcriptional amplification in tumor cells with elevated c-Myc. Cell. 2012;151(1):56–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [212].Lowe SW, Cepero E, Evan G. Intrinsic tumour suppression. Nature. 2004;432(7015):307–315. [DOI] [PubMed] [Google Scholar]
- [213].Dai M-S, Arnold H, Sun -X-X, et al. Inhibition of c-Myc activity by ribosomal protein L11. Embo J. 2007;26(14):3332–3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [214].Dai M-S, Sun -X-X, Lu H. Ribosomal protein L11 associates with c-Myc at 5 S rRNA and tRNA genes and regulates their expression. J Biol Chem. 2010;285(17):12587–12594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [215].Challagundla KB, Sun -X-X, Zhang X, et al. Ribosomal protein L11 recruits miR-24/miRISC to repress c-Myc expression in response to ribosomal stress. Mol Cell Biol. 2011;31(19):4007–4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [216].Liao J-M, Zhou X, Gatignol A, et al. Ribosomal proteins L5 and L11 co-operatively inactivate c-Myc via RNA-induced silencing complex. Oncogene. 2014;33(41):4916–4923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [217].Morgado-Palacin L, et al. Partial loss of Rpl11 in adult mice recapitulates diamond-blackfan anemia and promotes lymphomagenesis. Cell Rep. 2015;13(4):712–722. [DOI] [PubMed] [Google Scholar]
- [218].Perez-Mancera PA, Young AR, Narita M. Inside and out: the activities of senescence in cancer. Nat Rev Cancer. 2014;14(8):547–558. [DOI] [PubMed] [Google Scholar]
- [219].Halazonetis TD, Gorgoulis VG, Bartek J. An oncogene-induced DNA damage model for cancer development. Science. 2008;319(5868):1352–1355. [DOI] [PubMed] [Google Scholar]
- [220].Patel S, Player MR. Small-molecule inhibitors of the p53-HDM2 interaction for the treatment of cancer. Expert Opin Investig Drugs. 2008;17(12):1865–1882. [DOI] [PubMed] [Google Scholar]


