Abstract
The description of a so-called cytokine storm in patients with COVID-19 has prompted consideration of anti-cytokine therapies, particularly interleukin-6 antagonists. However, direct systematic comparisons of COVID-19 with other critical illnesses associated with elevated cytokine concentrations have not been reported. In this Rapid Review, we report the results of a systematic review and meta-analysis of COVID-19 studies published or posted as preprints between Nov 1, 2019, and April 14, 2020, in which interleukin-6 concentrations in patients with severe or critical disease were recorded. 25 COVID-19 studies (n=1245 patients) were ultimately included. Comparator groups included four trials each in sepsis (n=5320), cytokine release syndrome (n=72), and acute respiratory distress syndrome unrelated to COVID-19 (n=2767). In patients with severe or critical COVID-19, the pooled mean serum interleukin-6 concentration was 36·7 pg/mL (95% CI 21·6–62·3 pg/mL; I2=57·7%). Mean interleukin-6 concentrations were nearly 100 times higher in patients with cytokine release syndrome (3110·5 pg/mL, 632·3–15 302·9 pg/mL; p<0·0001), 27 times higher in patients with sepsis (983·6 pg/mL, 550·1–1758·4 pg/mL; p<0·0001), and 12 times higher in patients with acute respiratory distress syndrome unrelated to COVID-19 (460 pg/mL, 216·3–978·7 pg/mL; p<0·0001). Our findings question the role of a cytokine storm in COVID-19-induced organ dysfunction. Many questions remain about the immune features of COVID-19 and the potential role of anti-cytokine and immune-modulating treatments in patients with the disease.
Introduction
COVID-19 is a new and poorly understood disease, so much of our current understanding of organ dysfunction in severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection is extrapolated from other disorders with similar clinical features. Several studies have reported elevated serum concentrations of inflammatory cytokines, including interleukin (IL)-6, in severe COVID-19.1, 2 These observations have spurred comparisons with other syndromes of critical illness that are associated with elevated cytokines. Frequently invoked examples are acute respiratory distress syndrome (ARDS) and sepsis.3 Cytokine release syndrome in the setting of chimeric antigen receptor (CAR) T-cell therapy is another comparator of particular interest because it is a US Food and Drug Administration-approved indication for the drug tocilizumab.4 Tocilizumab is a humanised monoclonal antibody against the IL-6 receptor.5 On the basis of these comparisons, trials of anti-cytokine medications are ongoing in patients with COVID-19. Administration of these medications, including IL-6 antagonists, has become widespread while awaiting trial results.1 However, a systematic comparison of the inflammatory milieu in COVID-19-associated critical illness and these other disorders has not been done. Such a comparison might reveal important similarities and differences between these various syndromes and inform the successful application of immune-modulating therapy in COVID-19.
In this Rapid Review, we describe a rapid systematic review and meta-analysis of inflammatory cytokine and related biomarker concentrations in the COVID-19 literature. We compare the findings in patients with COVID-19 with those reported in landmark studies of patients with ARDS unrelated to COVID-19, sepsis, and CAR T cell-induced cytokine release syndrome. We discuss the implications for understanding of the pathobiology of each of these four syndromes, highlighting current uncertainties, and for future research and clinical practice.
Key messages.
-
•
Inflammatory cytokine elevations in patients with severe and critical COVID-19, including elevations of interleukin-6, are profoundly lower than those reported in patients with acute respiratory distress syndrome (ARDS) unrelated to COVID-19, sepsis, and chimeric antigen receptor (CAR) T cell-induced cytokine release syndrome
-
•
In contrast, several non-cytokine biomarkers, including D-dimer, C-reactive protein, and ferritin, are elevated to a similar or greater extent in patients with COVID-19 than in patients with these comparison disorders
-
•
As in other syndromes of critical illness, the role of inflammatory cytokine elevations in the pathobiology of COVID-19 remains unclear
-
•
The systemic inflammatory profile of COVID-19 is distinct from that of non-COVID-19 ARDS, sepsis, and CAR T cell-induced cytokine release syndrome; applying the descriptor cytokine storm to COVID-19 might be particularly problematic
-
•
Alternative models of organ dysfunction in COVID-19, such as endovasculitis, direct viral injury and lymphodepletion, or viral-induced immunosuppression, might be worth considering
Methods
Aims and overview
The aims of this Rapid Review were to synthesise and describe the reported pattern of inflammatory cytokines in COVID-19-induced respiratory failure and to compare this profile with those of other acute inflammatory syndromes. We did a rapid systematic review of the literature to describe inflammatory cytokine concentrations in severe and critical COVID-19. The primary response variable was plasma or serum IL-6. Additionally, concentrations of other cytokines, acute-phase reactants, and related biomarkers were recorded. Results were then compared with control populations identified from landmark trials of ARDS, sepsis, and cytokine release syndrome. A subanalysis distinguished results in severe versus critical COVID-19.
Search strategy and selection criteria
For COVID-19, we included original research studies that reported IL-6 concentrations for hospitalised patients with either severe or critical laboratory-confirmed COVID-19. Severe COVID-19 was identified by criteria of either WHO6 or the National Health Commission of China7 (appendix 1 p 8). We classified patients with critical COVID-19 as those who met the criteria of either WHO6 or the National Health Commission of China7 for COVID-19-induced ARDS (which align with the Berlin Definition of ARDS8), or who were admitted to an intensive care unit (ICU) and received invasive mechanical ventilation if severity was not otherwise specified. Case series including only deceased patients were also treated as having critical COVID-19. We excluded studies that had fewer than 20 participants, were not written in English, or for which measures of central tendency and distribution could not be obtained. A medical librarian (RP) designed and executed a comprehensive search strategy (appendix 1 p 2) of articles published between Nov 1, 2019, and April 14, 2020, in the Embase and MEDLINE databases. Additional searching was done in the medRxiv repository for relevant preprints. Titles and abstracts of results from all sources were screened by one reviewer (LR or DEL). The full text of relevant articles was then reviewed to identify studies for analysis.
For the comparator disorders, data were obtained from pre-specified landmark trials. For ARDS, we used data reported from the SAILS trial and from the pooled analysis of the ALVEOLI, ARMA, and FACCT trials.9, 10 For sepsis, data were obtained from the ACCESS, PROWESS, ProCESS, and GenIMS studies.11 For cytokine release syndrome, we obtained data from studies of a spectrum of haematological malignancies that were treated with CAR T-cell therapy.12, 13, 14, 15 We restricted the data to patients with cytokine release syndrome of grade 3 or higher, which generally involves organ dysfunction that prompts the administration of tocilizumab (grading criteria are given in appendix 1 p 9).
Study outcomes
The primary response variable was plasma or serum IL-6 concentration. Additional response variables of interest included tumour necrosis factor-α (TNFα), IL-8, IL-1β, IL-10, IL-2, IL-4, soluble IL-2 receptor (sIL-2R), interferon-γ (IFNγ), C-reactive protein (CRP), ferritin, D-dimer, procalcitonin, lactate dehydrogenase, erythrocyte sedimentation rate, albumin, total bilirubin, fibrinogen, lymphocyte count, lymphocyte percentage, and platelet count.
Accuracy of published data and inclusion of preprints
We adhered to a rapid (rather than traditional systematic) review methodology.16 We chose this approach in an attempt to balance systematic data collection with the need for rapid synthesis and reporting in the context of the pandemic. Given the rapid pace at which research related to COVID-19 is being disseminated, we were concerned about a relatively high frequency of corrections and errata. Accordingly, once all eligible studies were identified, one author (LR) reviewed each paper on the publisher's website to check whether any corrections had been issued.
The need for rapid dissemination of COVID-19 research had spurred many authors to post their studies as preprints, particularly on the website of medRxiv. We included these non-peer-reviewed preprints to capture a larger sample of observations during the brief time since COVID-19 has emerged. Given the descriptive nature of our research question (as opposed to a causal treatment effect), we reasoned that the risk of bias due to an absence of peer review was less concerning than potential biases due to the inclusion of only early manuscripts that had had time to navigate peer review.
This study did not operate in a prediction or causal inference framework, so we did not assess patient outcomes or their association with cytokine concentrations, to respect the limitations of descriptive modelling and inference; nor did we undertake a formal risk-of-bias assessment.17, 18
Data abstraction
One of two reviewers (DEL or LR) abstracted data from all studies that met inclusion criteria using a standardised data collection tool. It has been suggested that cytokine elevation is a late finding in COVID-19.4 To address this concern, we abstracted the peak IL-6 value for studies that reported multiple IL-6 concentrations. All included studies reported a measure of central tendency (mean or median) and dispersion (SD, standard error, IQR, or range) for IL-6 concentrations. For studies that did not report these statistics (eg, IL-6 reported as frequencies of categorical concentrations, or in a figure that was difficult to interpret), or that did not report them in the specific population of interest (eg, in an overall study population that included non-severe COVID-19), the data were requested from the corresponding authors. All of these authors were contacted at least three times over a 3-week period. If the corresponding author did not provide the data and the statistics could not be identified from the reporting format, the study was excluded. Measurement units were standardised across studies for comparison (more detail is provided in appendix 1 p 5).
Data analysis
Mean and SD were used when reported in selected studies. When median and IQR or range were reported, we adhered to Cochrane recommendations, estimating the mean using the method described by Wan and colleagues19 and the SD using the Cochrane handbook method.20 Because we expected many markers to display a beta distribution, we evaluated each response variable graphically before analysis and applied a log transformation if normality assumptions were violated. We included the analysis of the untransformed data as sensitivity analyses. Additionally, after data collection, we found that the central tendency was positively correlated with variance (ie, the homoscedasticity assumption was violated) for many biomarkers, including IL-6. Because the common practice of inverse-variance weighting would bias the estimates downwards in this situation,21 we instead weighted studies by the square root of the sample size in the primary analysis. However, we also did standard inverse-variance weighting as sensitivity analyses.
After computing study weights, pooled means were calculated from a generalised linear model with disorder as a class variable with the levels COVID-19, cytokine release syndrome, hypoinflammatory ARDS, hyperinflammatory ARDS, and sepsis, by estimating the least-squares means at each level. We constructed 95% CI and p values for the difference between COVID-19 and each of the other disorders using Dunnett's correction for multiple comparisons. To facilitate interpretation for variables that had been log-transformed, we back-transformed the results for reporting purposes. We calculated I 2 statistics to assess between-study heterogeneity within disorders.
For the secondary analyses, separate levels for severe and critical COVID-19 were included in the disorder variable. Unlike critical COVID-19, severe COVID-19 might have included a less severely ill population than that of non-COVID-19 ARDS. To address whether lower severity explained discrepancies in cytokine concentrations, we selected critical COVID-19 as the reference level for statistical hypothesis testing. As an additional sensitivity analysis, IL-6 concentrations were calculated among only COVID-19 studies that reported peak IL-6. Further details and rationale of the quantitative strategy are provided in appendix 1 (pp 6–7). The analyses were done in SAS University Edition. Figures were produced using GraphPad Prism 8. We provide the complete dataset used in these analyses in appendix 2. The study is registered with PROSPERO, CRD42020180350.
Results
Search results
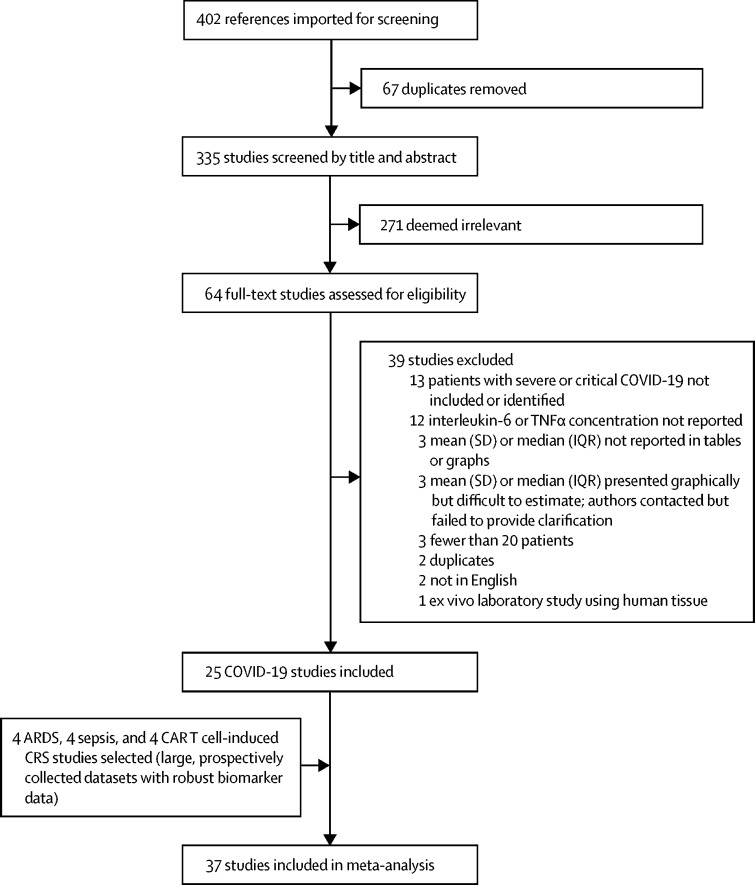
The COVID-19 search strategy returned 335 search results (figure 1 ). Abstract screening identified 64 studies for full-text review; 28 of these studies met the inclusion criteria. Six of these 28 studies did not report results in a format that easily facilitated data abstraction. All of these six reports indicated that data were available on request. Of the six corresponding authors whom we contacted, two provided data, one declined citing hospital data-sharing rules, and three did not answer after multiple contact attempts (appendix 1 p 10). Subsequently, we were able to estimate IL-6 concentrations from a figure in one of the four studies that did not provide requested data. Ultimately, 25 COVID-19 studies reflecting 1245 patients were included for analysis.2, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45 Of these, 15 studies (n=650 patients) either included only patients with severe COVID-19 or allowed data abstraction specifically within the subgroup of patients with severe disease, whereas ten studies (n=367 patients) included only patients with critical COVID-19 or allowed data abstraction specifically within the subgroup of patients with critical illness. COVID-19 study details are provided in appendix 1 (p 11).
Figure 1.
Selection of studies
ARDS=acute respiratory distress syndrome. CAR T cell-induced CRS=chimeric antigen receptor T cell-induced cytokine release syndrome.
Non-COVID-19 studies are summarised in appendix 1 (p 12). The four ARDS trials included 2767 patients.9, 10 Of these, 1899 had the hypoinflammatory phenotype and 868 had the hyperinflammatory phenotype. The four sepsis cohorts included 5320 patients.11 The four studies of cytokine release syndrome were smaller, with 72 patients included in the final analysis.12, 13, 14, 15
IL-6 in COVID-19 versus other disorders
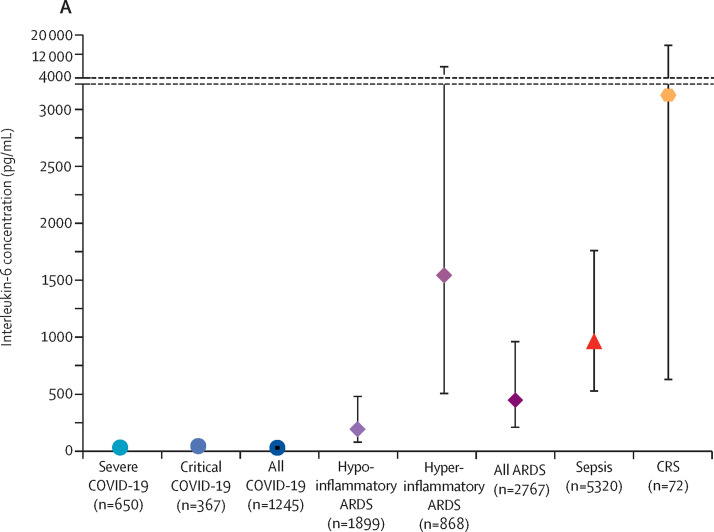
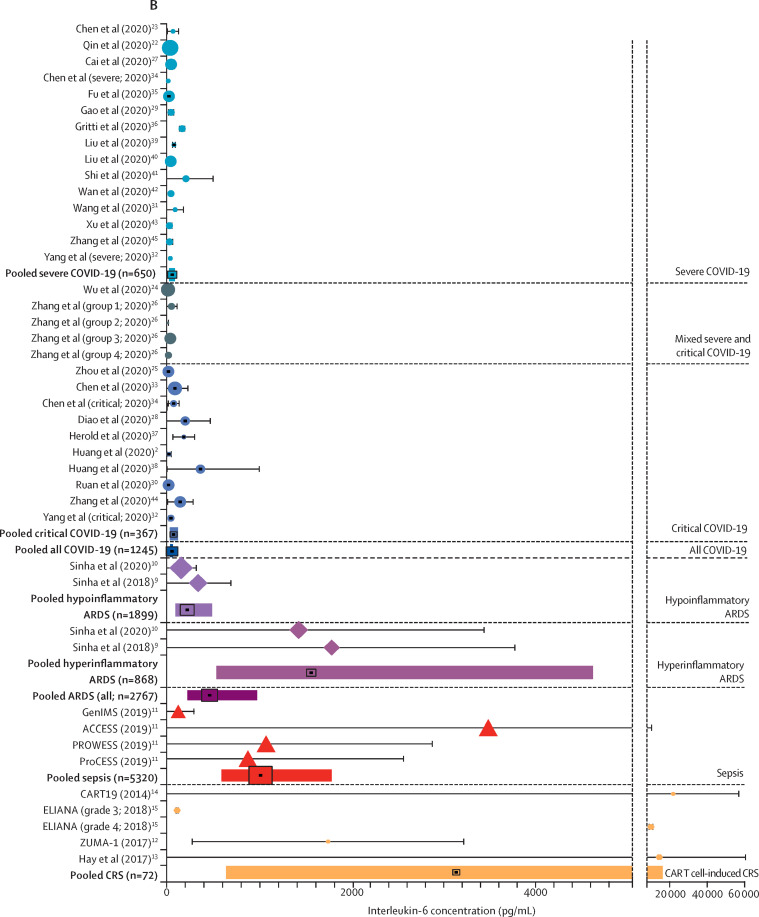
In the primary analysis, the estimated pooled mean for IL-6 concentrations in patients with COVID-19 was 36·7 pg/mL (95% CI 21·6–62·3 pg/mL; figure 2 , appendix 1 p 13). In contrast, the mean IL-6 serum concentration was 3110·5 pg/mL (632·3–15 302·9 pg/mL) in patients with CAR T cell-induced cytokine release syndrome, nearly 100 times higher than in patients with COVID-19 (difference 3074 pg/mL, 95% CI 325–26 735 pg/mL; p<0·0001). Similarly, the pooled mean IL-6 concentration was 1558·2 pg/mL (525·8–4617·6 pg/mL) in patients with hyperinflammatory ARDS (difference 1521·5 pg/mL, 324·7–26 735·0 pg/mL; p<0·0001) and 983·6 pg/mL (550·1–1758·4 pg/mL) in patients with sepsis (difference 947 pg/mL, 324–2648 pg/mL; p<0·0001). Even in patients with hypoinflammatory ARDS, the mean IL-6 concentration was 198·6 pg/mL (80·6–489·3 pg/mL), 5 times higher than the concentration in patients with COVID-19 (difference 162 pg/mL, 16–717 pg/mL; p=0·0085). Patients with ARDS unrelated to COVID-19 had significantly higher IL-6 concentrations than did patients with COVID-19 when analysed as a single disorder (mean 460·1 pg/mL, 216·3–978·7 pg/mL; difference 423·4 pg/mL, 106·9–1438·1 pg/mL; p<0·0001; appendix 1 p 14). In the sensitivity analysis using inverse-variance weighting, estimated means were lower than in the primary analysis for all groups, but between-group differences were similar to the primary analysis (appendix 1 p 13). In the sensitivity analyses in which IL-6 concentrations were not transformed, all mean estimates were higher and between-group differences were even larger than in the primary analysis, but the model fit appeared to be substantially worse (appendix 1 p 13).
Figure 2.
Interleukin-6 concentrations in patients with COVID-19 versus comparison disorders
(A) Pooled estimate for each disorder. Markers indicate point estimates and error bars indicate 95% CIs. (B) For individual studies, markers indicate study means and error bars indicate standard deviations. Markers are sized proportionately to the log weight of the study in the analysis. Pooled estimates are represented by the solid bars. The black marking in the centre of the bars indicates the point estimate for the disease. The width of the box is scaled according to the pooled number of participants, whereas the width of the bar indicates the 95% CI. ARDS=acute respiratory distress syndrome. CAR=chimeric antigen receptor. CRS=cytokine release syndrome.
IL-6 concentrations in patients with COVID-19 showed moderate heterogeneity (I 2=57·7%), with a range of 6·5–357·2 pg/mL, and 80·0% of the COVID-19 studies reported a mean IL-6 concentration lower than 100 pg/mL. Heterogeneity was lower for hyperinflammatory (I 2=0%) and hypoinflammatory ARDS (I 2=37·6%), but higher for cytokine release syndrome (I 2=77·0%) and sepsis (I 2=89·3%).
Additional inflammatory cytokines
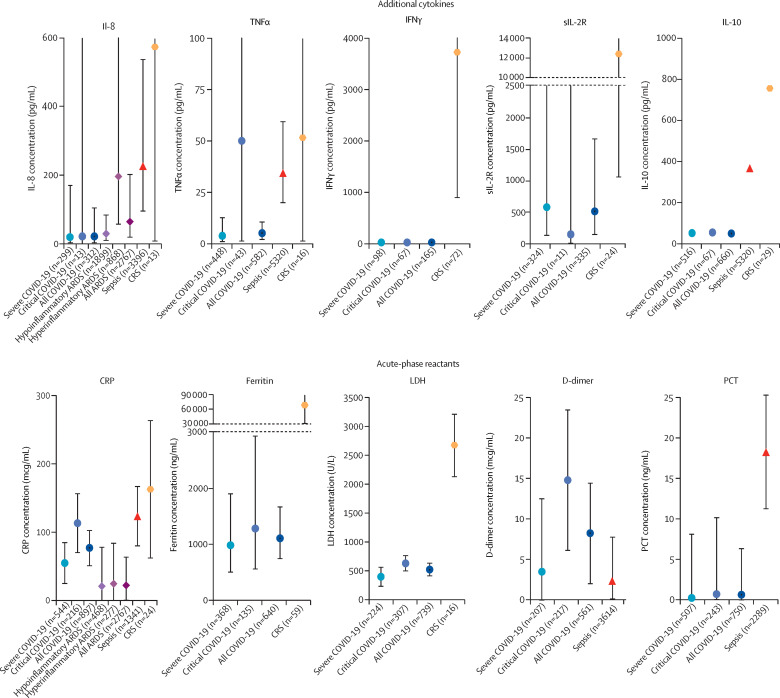
Most other cytokines were comparatively low in COVID-19 (figure 3 ; appendix 1 p 16). For example, the mean IL-8 concentration (neutrophil chemotactic factor) was 22 pg/mL (95% CI 5–108 pg/mL) in patients with COVID-19, compared with 228 pg/mL in patients with sepsis (difference 206 pg/mL, 95% CI 15–1371 pg/mL; p=0·021) and 196 pg/mL in patients with hyperinflammatory ARDS (difference 174 pg/mL, 5–1436 pg/mL; p=0·038). The mean IL-8 concentration in patients with cytokine release syndrome was 575 pg/mL; the difference between mean IL-8 concentration in patients with cytokine release syndrome versus COVID-19 was not statistically significant in the setting of a wide CI (difference 553 pg/mL, –47 to 47 502 pg/mL; p=0·11). However, the estimate for IL-8 concentration in patients with hypoinflammatory ARDS was 32 pg/mL, similar to that for COVID-19. TNFα concentrations were available for the four studies examining sepsis (n=5320) and one study examining cytokine release syndrome (n=16), and for ten COVID-19 studies (n=607 patients). Compared with a mean TNFα concentration of 5·0 pg/mL (2·3–10·7 pg/mL) in patients with COVID-19, mean concentration was 34·6 pg/mL (20·0–59·9 pg/mL) in patients with sepsis and 52·2 pg/mL (2·0–1390 pg/mL) in patients with cytokine release syndrome. All but one (92%) COVID-19 study had a mean TNFα concentration lower than 10 pg/mL. IFNγ concentrations were reported in seven COVID-19 studies (n=165) and sIL-2R concentrations were reported in three COVID-19 studies (n=335), and for two studies examining cytokine release syndrome (n=24). IFNγ concentration was not elevated in patients with COVID-19, with an average of 10·8 pg/mL, but was highly elevated in patients with cytokine release syndrome, averaging 3722·1 pg/mL (difference 3711 pg/mL, 624–21 838 pg/mL; p<0·0001). Mean sIL-2R was elevated in patients with COVID-19, but much less so than in patients with cytokine release syndrome (506 pg/mL vs 12 396 pg/mL; difference 11 890 pg/mL, 299–190 957 pg/mL; p=0·032). IL-2 and IL-4 concentrations were not available in any study of the comparison disorders, but IL-2 concentration was reported in nine COVID-19 studies and IL-4 concentration in ten COVID-19 studies. All of these COVID-19 studies reported these cytokines to be within normal physiological range.
Figure 3.
Additional cytokines and biomarkers in patients with COVID-19 versus comparison disorders
The figure shows pooled mean estimates for secondary analyses of inflammatory cytokines and markers. Markers indicate point estimates and error bars indicate 95% CIs. ARDS=acute respiratory distress syndrome. CRP=C-reactive protein. CRS=cytokine release syndrome. IFNγ=interferon-γ. IL=interleukin. LDH=lactate dehydrogenase. PCT=procalcitonin. sIL-2R=soluble interleukin-2 receptor. TNFα=tumour necrosis factor-α.
Other inflammatory and host-response markers
Acute-phase reactants were substantially elevated in patients with COVID-19 (figure 3; appendix 1 p 16). CRP concentrations were comparable in patients with COVID-19 and patients with sepsis, and higher in patients with cytokine release syndrome. D-dimer concentrations were available for COVID-19 and sepsis studies; these studies indicated that patients with COVID-19 had substantially higher D-dimer elevations than did patients with sepsis. Mean ferritin and lactate dehydrogenase concentrations were markedly higher in patients with cytokine release syndrome than in patients with COVID-19, but nonetheless highly elevated in patients with COVID-19. In contrast, procalcitonin concentrations were not elevated in patients with COVID-19 but were raised in sepsis. Absolute and relative lymphopenia were common in patients with COVID-19, but data were not available in the comparison groups. We report results for additional markers in appendix 1 (p 16).
Severe versus critical COVID-19 versus other disorders
The pooled mean IL-6 concentration in patients with critical COVID-19 was 55·3 pg/mL, and was not statistically greater than in patients with severe COVID-19 (mean 37·3 pg/mL; p=0·94; figure 2). This pooled mean IL-6 concentration in patients with critical COVID-19 was again significantly lower than in patients with all other non-COVID-19 comparator disorders. Sensitivity analysis using inverse-variance weighting showed the same result (appendix 1 p 17). Most of the within-COVID-19 heterogeneity in the primary analysis appeared to be driven by the group with critical COVID-19, in which the I 2 was 55·7%, compared with 1·1% in the group with severe COVID-19. The mean IL-6 concentration among studies of patients with critical COVID-19 ranged from 22·3 pg/mL to 136·8 pg/mL, with six of ten studies reporting mean IL-6 concentration lower than 100·0 pg/mL.
Other cytokine measures in the subgroup of patients with critical COVID-19 were similar to those observed in the primary analysis. In contrast, abnormalities of non-cytokine biomarkers appeared to be exaggerated in the group with critical versus severe COVID-19 (figure 3; appendix 1 p 18).
Peak IL-6 in COVID-19 versus other disorders
The time of IL-6 concentration measurement for all studies is shown in appendix 1 (p 19). Among COVID-19 studies reporting peak IL-6 concentration (six studies, n=245 patients), mean IL-6 concentration was 61·3 pg/mL in patients with COVID-19, significantly lower than in patients with sepsis, cytokine release syndrome, and hyperinflammatory ARDS (appendix 1 p 20). Results were similar when comparing peak IL-6 concentrations in patients with critical COVID-19 alone (mean 78·1 pg/mL) with those for the other disorders.
Discussion
In this Rapid Review of 25 studies reflecting 1245 patients with severe and critical COVID-19, plasma or serum IL-6 concentrations were at least an order of magnitude less than those reported in studies of patients with CAR T cell-induced cytokine release syndrome, sepsis, and non-COVID-19 ARDS. This finding was consistent across several sensitivity analyses. Most other cytokine concentrations also showed mild elevation in patients with COVID-19 as compared with the other disorders. In contrast, non-specific inflammatory markers appeared to be relatively comparable between COVID-19 and non-COVID-19 illnesses. These results build on a preliminary analysis in patients with COVID-19 versus patients with ARDS unrelated to COVID-19.46
Pathobiology of COVID-19
Our results suggest that the descriptor cytokine storm does not appropriately describe the milieu in COVID-19-induced organ dysfunction. Autopsy reports consistently note widespread dissemination of SARS-CoV-2 throughout diverse tissues.47 Lymphopenia is common, as we report here, and prognostic, as others have reported.1, 48 T lymphocytes are directly susceptible to SARS-CoV-2 infection,49 and are depleted in clinical COVID-19.22, 28 In this context, it is worth considering that the less pronounced cytokine elevations in COVID-19 could reflect a regulated, or even inadequate, inflammatory response to overwhelming viral infection. A predominantly hypoimmune state with subsequent (directly) virus-mediated tissue damage and dysregulated inflammation is consistent with both the apparent clinical and pathological abnormalities in COVID-19 and the high concentrations of circulating acute-phase reactants reported here (figure 4 ).3, 50
Figure 4.
Mechanistic comparison of inflammatory processes in patients with COVID-19 versus ARDS, sepsis, and CAR T cell-induced CRS
ARDS=acute respiratory distress syndrome. CAR T cell-induced CRS=chimeric antigen receptor T cell-induced cytokine release syndrome. CRP=C-reactive protein. DAMPs=damage-associated molecular patterns. IFN=interferon. IL=interleukin. LDH=lactate dehydrogenase. PAMPs=pathogen-associated molecular patterns. PCT=procalcitonin. ROS=reactive oxygen species. SARS-CoV-2=severe acute respiratory syndrome coronavirus 2. ssRNA=single-stranded RNA. vWF=von Willebrand factor. *Effector function measured by ex vivo functional assays.
In contrast to cytokine concentrations, similar or greater elevations of several acute-phase reactants and other biomarkers were found in patients with COVID-19. D-dimer concentrations were 5 times higher in patients with critical COVID-19 than in patients with sepsis, suggesting that the reported associations between D-dimer and severity in COVID-19 are a consistent and distinguishing signal. Although prediction inference is outside the scope of the present study, the ability of D-dimer and procalcitonin to discriminate COVID-19 from other infectious causes of respiratory distress might warrant further exploration.
Importantly, although the mortality benefit from dexamethasone treatment reported in patients with COVID-19 informs clinical practice,51 it is difficult to causally attribute this benefit to IL-6 suppression. Of the myriad effects of glucocorticoids relevant to critical illness (eg, inotropy, vasoconstriction in the more than 60% of critically ill patients with COVID-19 who require vasopressor support1), perhaps the most relevant is the ability of corticosteroids to suppress the late-onset fibrosis that leads to irreversible lung damage in ARDS.52 Notably, the large effect in the RECOVERY trial of dexamethasone was driven entirely by patients who were randomised more than 7 days after symptom onset.51
Pathobiology of ARDS
Because it arises from a range of precipitating causes, ARDS is associated with numerous pathobiological processes. Central to its pathogenesis is an acute inflammatory insult leading to pulmonary epithelial and endothelial injury. The extent to which these injuries are observed can depend on the site of insult. For example, circulating markers of epithelial injury are more elevated in patients with direct causes (eg, pneumonia, aspiration) than in those with indirect causes (eg, pancreatitis) of ARDS. Conversely, indirect causes are associated with higher concentrations of endothelial injury markers.53 Distinct hypoinflammatory and hyperinflammatory phenotypes of ARDS, which differ on the basis of systemic inflammatory profiles, have been robustly identified. The hyperinflammatory phenotype is associated with increased concentrations of IL-6, IL-8, and soluble TNF receptor 1, but lower concentrations of protein C.
In patients with COVID-19, the relative contributions of endothelial and epithelial injury remain unknown. Given that viral pneumonitis is a direct cause of lung injury, it might be anticipated that epithelial injury would be predominant. Numerous post-mortem studies in patients with COVID-19 ubiquitously identified diffuse alveolar damage in patients with severe disease.54, 55 Yet, these studies also describe severe endothelial damage and coagulopathic features in the pulmonary microvasculature.54, 55 These studies require cautious interpretation because they are subject to selection bias and the sample sizes are small. In an exploratory prospective study, the hyperinflammatory phenotype of ARDS was observed in 11–20% of patients with COVID-19 versus 35% of patients with non-COVID-19 ARDS.56 This finding substantiates the results of this meta-analysis, suggesting that circulating inflammatory responses are generally lower in patients with COVID-19 than in patients with hyperinflammatory ARDS.
Pathobiology of sepsis
Sepsis is defined as life-threatening organ dysfunction caused by a dysregulated host response to infection.57 The syndrome has diverse infectious causes and host substrates, which probably underlie the great heterogeneity in its manifestations. The precise biological events that precipitate transition from regulated to dysregulated host response remain unknown. The sine qua non of sepsis is organ dysfunction, often remote from the infectious source. Abnormalities include vasodilatory shock, ARDS, coagulopathy, and renal, hepatic, microcirculatory, and endocrine dysfunction. Immune dysfunction is another hallmark of sepsis, but conceptualising this dysfunction as hyperinflammation is probably too simplistic. While inflammatory cytokine concentrations are often exceptionally high, sepsis is also associated with immunosuppression marked by T cell exhaustion, neutrophil hyporesponsiveness to cytokine stimulation, and impaired innate cell phagocytosis and pathogen killing.58 Elevated cytokines coupled with impaired immune effector function is a pattern consistent with the peripheral resistance observed in multiple endocrine axes in sepsis. Therefore, whether inflammatory cytokine elevations in sepsis reflect a driver, a marker, or even an adaptive response to disease remains unknown.
Unanswered questions about the mechanistic role of cytokine elevations are shared between sepsis and COVID-19. However, despite much lower systemic cytokine concentrations, ex vivo stimulated blood mononuclear cells from patients with COVID-19 produced half as much TNFα and IFNγ as did cells from patients with sepsis and patients with critical illness without any infection.59 Therefore, immunosuppression might be even more pronounced in patients with COVID-19 than the paradoxical suppression frequently observed in sepsis.58, 59 Innate clearance capacity of microorganisms has not been investigated in patients with severe COVID-19 yet, and is probably a key question for future studies given the high risk of secondary infection among patients in ICUs.
Pathobiology of CAR T cell-induced cytokine release syndrome
Unlike sepsis and ARDS, CAR T cell-induced cytokine release syndrome has a well defined pathophysiology. After infusion, CAR T cells encounter cognate antigen, leading to activation, proliferation, and lysis of target cells with inflammatory cytokine release.60, 61, 62 CAR T-cell infusion is associated with fever, hypotension, coagulopathy and, in severe cases, multiorgan dysfunction that might include reversible neurotoxicity. In most patients, cytokine release syndrome develops shortly after infusion and resolves in the ensuing week with supportive care alone or in association with tocilizumab or corticosteroid treatment.60, 62, 63, 64 However, severe or prolonged cytokine release syndrome is associated with extraordinarily high serum concentrations of inflammatory cytokines, including IFNγ, IL-6, IL-10, IL-15, and TNF receptor p55, and chemokines, such as IL-8.60 Prompt resolution of fever and, often, hypotension after tocilizumab administration suggests that IL-6 contributes to the pathobiology of CAR T cell-induced cytokine release syndrome, although randomised evidence is lacking. Laboratory studies suggest that monocytes and macrophages are a major source of IL-6 after CAR T-cell therapy. Release of IL-1 appears to precede that of IL-6, so targeting of IL-1 signalling might mitigate or prevent cytokine release syndrome.65, 66 Ferritin concentrations rise substantially in patients with severe cytokine release syndrome, which might signify macrophage activation.
Elevations in ferritin, CRP, and cytokines such as IL-6 in patients with COVID-19 have spurred comparisons to CAR T cell-associated cytokine release syndrome. However, the comparatively low IL-6 concentrations and absence of substantial IFNγ elevations in patients with COVID-19 limit the analogy to cytokine release syndrome. Conversely, low IL-6 and high ferritin concentrations in patients with COVID-19 are in fact consistent with haemophagocytic lymphohistiocytosis or a macrophage activation syndrome.67 However, analogies are again limited by the absence of substantial IFNγ elevations in patients with COVID-19. Some researchers have proposed immunosubphenotypes of COVID-19, whereby some patients show so-called immunoparalysis and others a pattern similar to that of macrophage activation syndrome.68 Under this paradigm, the phenotype similar to macrophage activation syndrome represent a minority (<15%) of patients.68 Insufficient high-quality data on cytokine patterns in haemophagocytic lymphohistiocytosis or macrophage activation syndrome in a format conducive to analysis precluded the inclusion of these diagnoses in our analyses.
Implications for research and clinical practice
The results of our systematic review and meta-analysis raise concerns about the widespread off-label use of cytokine blockade in the treatment of COVID-19 before the results of randomised trials are available. Cytokine blockade has not been effective in patients with sepsis and ARDS, in whom inflammatory cytokine concentrations are far more elevated. IL-6 elevation might have a role in endothelial activation and precipitation of a pulmonary immune-mediated thrombosis, so ongoing trials might ultimately show that anti-cytokine treatment is beneficial in some patients with COVID-19.69 However, unencumbered use of these agents in the absence of randomised evidence seems premature. We note that the current Infectious Disease Society of America guidelines recommend against the use of tocilizumab in patients with COVID-19-associated ARDS outside the context of a clinical trial,70 but IL-6 and IL-1 antagonists have nevertheless been widely administered—to nearly 20% of patients with COVID-19 in ICUs in some studies.1
The intense focus on cytokine blockade has attracted substantial investment. Such focus might have incurred the opportunity cost of discouraging clinical exploration of other hypotheses, such as immunosupportive therapy. There are at least 20 trials of various IL-6 antagonists for COVID-19 registered with ClinicalTrials.gov. In contrast, there is a single unique trial for recombinant IL-7, which has been effective in previous randomised trials of severe viraemic illnesses. There are just four trials of interferons despite evidence that inhibition of IFN-1 signalling is an intrinsic mechanism of immune evasion by SARS-CoV-2. A single trial of all CTLA-4 and PD-1 or PD-L1 blockade agents is registered (NCT04335305), in which patients are randomised to pembrolizumab (a checkpoint inhibitor) and tocilizumab together versus standard of care.
Limitations of the study
Our systematic review and meta-analysis has important limitations. First, the criteria for severe COVID-19 probably select a population that is less acutely ill than the population of patients with non-COVID-19 ARDS, because not all patients were admitted to an ICU. However, many of the patients with severe COVID-19 meet the Berlin criteria for ARDS, all meet clinical criteria for sepsis, and all exhibit organ dysfunction consistent with cytokine release syndrome of grade 3 or higher.8, 57 Additionally, comparison of critical COVID-19 alone with the other, non-COVID-19 disorders yielded similar results. Second, in many instances, biomarker distributions violated many of the assumptions employed by common biostatistical modelling approaches, making the analyses complex. However, the differences between COVID-19 and comparator disorders are large enough to be visually obvious without formal statistical hypothesis testing, and various sensitivity analyses failed to impugn the stability of the primary analysis. Nevertheless, our complete dataset is available in appendix 2 to permit testing of alternative strategies. Third, our analysis considered only one IL-6 concentration per study. For COVID-19, we used the peak IL-6 concentration whenever studies reported multiple concentrations, whereas for sepsis and ARDS, we used the enrolment IL-6 concentration, which was generally recorded less than 24–48 h after presentation. Therefore, we might have underestimated differences in cytokine concentrations between these disorders. Even when comparing only peak IL-6 concentrations in patients with COVID-19 with those in the other disorders, IL-6 elevations were substantially lower in patients with COVID-19. Fourth, because tocilizumab is a receptor antagonist, its administration could have increased the measured concentrations in some of the studies examining cytokine release syndrome.13, 14, 15 The extent of treatment-induced IL-6 elevation appears to be disease variable, with small increases reported in rheumatoid arthritis (30 pg/mL) and large increases in Castleman disease (540 pg/mL) 14 days after administration.71 Tocilizumab probably affects IL-6 concentration measurement in cytokine release syndrome as well.72 However, reactive elevation in IL-6 concentration after tocilizumab alone is not expected to explain the marked differences in other cytokine concentrations between patients with cytokine release syndrome and patients with COVID-19. Fifth, whereas the studies examining COVID-19 and cytokine release syndrome are recent, those examining sepsis and ARDS extend over a longer time period, and secular changes in cytokine measurement procedures could have affected the results. Finally, reporting of cytokines other than IL-6 was variable, limiting our ability to do all designated secondary analyses.
Conclusions and future directions
Although cytokine concentrations are elevated in patients with severe and critical COVID-19, the degree of cytokinaemia is markedly less than that seen in other disorders associated with elevated cytokines. Given these findings, the descriptor cytokine storm is problematic and alternative mechanisms of COVID-19-induced organ dysfunction are worth considering. Various ongoing randomised trials will determine whether cytokine blockade (eg, treatments directed against IL-6 or IL-1) can improve outcomes in patients with severe and critical COVID-19. Conversely, immune-activating treatments (eg, interferons, IL-7, or checkpoint inhibition) merit investigation, but there are relatively few registered trials. More broadly, the immune features of COVID-19 remain largely unsettled. Deepening pathobiological understanding of severe SARS-CoV-2 infection and the host response it elicits must be prioritised. Deploying mechanistic studies nested within randomised trials offers an important avenue for basic scientists to explore the biology of COVID-19 within clinically relevant experimental systems, while also testing the efficacy and safety of potential therapeutics. Circulating markers might, ultimately, help to discriminate diagnoses and generate hypotheses. However, caution is needed in drawing inferences about the underlying processes that such markers reflect and their potential causal roles in disease. Even as new disease models and therapeutics gain traction, challenging assumptions is essential and scepticism is healthy.
Acknowledgments
Acknowledgments
We thank Yongwen Chen, Yingxia Liu, and Yang Yang for providing data used in this meta-analysis, and Christopher Seymour and Jason Kennedy for clarifications regarding the sepsis datasets used in this study.
Contributors
DEL conceived of the study, based on ideas from PS and CSC, and with input from LR, MDT, ML, and CSD. DEL, LR, and RP designed and executed the literature search. DEL and LR screened and reviewed the search results and abstracted the data. AVH and CJT provided key data. DEL designed and conducted the analysis with input from LR and MOH. All authors contributed to the interpretation of analysis. DEL, LR, and ML made the figures and tables. DEL and LR drafted the manuscript. All authors reviewed the manuscript and provided important intellectual content. DEL and LR take responsibility for the article as a whole.
Declaration of interests
MDT is supported in part by the US National Institute of General Medical Sciences, US National Institutes of Health (grant K08 GM 132794). CSC is supported in part by the US National Heart, Lung, and Blood Institute (R35 HL140026); she has received additional grant funding from Roche/Genentech (current) and Bayer (former), and consulting fees from Vasomune, Gen1e Life Sciences, and Quark Pharmaceuticals. CJT's institution, the Fred Hutchinson Cancer Research Center, has held equity in Juno Therapeutics; CJT has received research grants and personal fees for advisory board participation from Juno Therapeutics/Bristol Myers Squibb, during the conduct of the study. He declares fees and stock options in relation to a role on the scientific advisory boards of Precision Biosciences, Caribou Biosciences, Eureka Therapeutics, Myeloid Therapeutics, Century Therapeutics, and ArsenalBio; travel expenses and fees for participation on the advisory boards of Amgen, Novartis, Kite/Gilead, Humanigen, Aptevo, PACT Pharma, AstraZeneca, and Nektar Therapeutics; travel expenses from T-CURX; and research funding from AstraZeneca and Nektar Therapeutics, outside of the submitted work. CJT reports a patent (US patent number 10 653 756; issued May 19, 2020) for the “Identification of CD8+ T cells that are CD161hi and/or IL18Rαhi and have rapid drug efflux capacity for toxicity of CAR-T cells”, for which he receives royalties from the licensee, Juno Therapeutics; a patent pending for “Methods and compositions related to toxicity associated with cell therapy”; a patent pending for “Methods for the treatment of B cell malignancies using adoptive cell therapy”; and a patent pending for “Biomarkers and uses thereof for selecting pancreas immunotherapy intervention”. MOH is supported in part by the US National Heart Lung and Blood Institute (R00 HL 141678). ML discloses research funds from the French Ministry of Health, research support from Shingotec, lecture fees from Baxter and Fresenius, and consulting fees from Novartis. CSD is supported in part by the US National Institute of General Medical Sciences, US National Institutes of Health (R01 GM 121102). He has stock options with Enlivex Therapeutics, outside of the submitted work. CSD reports honoraria from Lippincott Williams & Wilkins (Scientific Editor Critical Care Medicine) and the New York University Department of Anesthesiology (visiting professor); non-financial support for participation in the Bernard-Wiggers Task Force (accommodation) and from the Society of Critical Care Medicine (meeting registration and accommodation); an honorarium, travel expenses, and accommodation from the International Society of Hematology and Thrombosis; and royalties from Elsevier for the textbook Evidence-Based Practice of Critical Care, outside of the submitted work. All other authors declare no competing interests.
Supplementary Materials
References
- 1.Cummings MJ, Baldwin MR, Abrams D. Epidemiology, clinical course, and outcomes of critically ill adults with COVID-19 in New York City: a prospective cohort study. Lancet. 2020;395:1763–1770. doi: 10.1016/S0140-6736(20)31189-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang C, Wang Y, Li X. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li H, Liu L, Zhang D. SARS-CoV-2 and viral sepsis: observations and hypotheses. Lancet. 2020;395:1517–1520. doi: 10.1016/S0140-6736(20)30920-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.England JT, Abdulla A, Biggs CM. Weathering the COVID-19 storm: lessons from hematologic cytokine syndromes. Blood Rev. 2020 doi: 10.1016/j.blre.2020.100707. published online May 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moore JB, June CH. Cytokine release syndrome in severe COVID-19. Science. 2020;368:473–474. doi: 10.1126/science.abb8925. [DOI] [PubMed] [Google Scholar]
- 6.WHO Clinical management of severe acute respiratory infection (SARI) when COVID-19 disease is suspected: interim guidance. https://apps.who.int/iris/handle/10665/331446
- 7.National Health Commission of China Diagnosis and treatment protocol for novel coronavirus pneumonia. https://www.chinadaily.com.cn/pdf/2020/1.Clinical.Protocols.for.the.Diagnosis.and.Treatment.of.COVID-19.V7.pdf
- 8.Ranieri VM, Rubenfeld GD, Thompson BT. Acute respiratory distress syndrome: the Berlin Definition. JAMA. 2012;307:2526–2533. doi: 10.1001/jama.2012.5669. [DOI] [PubMed] [Google Scholar]
- 9.Sinha P, Delucchi KL, Thompson BT, McAuley DF, Matthay MA, Calfee CS. Latent class analysis of ARDS subphenotypes: a secondary analysis of the statins for acutely injured lungs from sepsis (SAILS) study. Intensive Care Med. 2018;44:1859–1869. doi: 10.1007/s00134-018-5378-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sinha P, Delucchi KL, McAuley DF, O'Kane CM, Matthay MA, Calfee CS. Development and validation of parsimonious algorithms to classify acute respiratory distress syndrome phenotypes: a secondary analysis of randomised controlled trials. Lancet Respir Med. 2020;8:247–257. doi: 10.1016/S2213-2600(19)30369-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seymour CW, Kennedy JN, Wang S. Derivation, validation, and potential treatment implications of novel clinical phenotypes for sepsis. JAMA. 2019;321:2003–2017. doi: 10.1001/jama.2019.5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Neelapu SS, Locke FL, Bartlett NL. Axicabtagene ciloleucel CAR T-cell therapy in refractory large B-Cell lymphoma. N Engl J Med. 2017;377:2531–2544. doi: 10.1056/NEJMoa1707447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hay KA, Hanafi LA, Li D. Kinetics and biomarkers of severe cytokine release syndrome after CD19 chimeric antigen receptor-modified T-cell therapy. Blood. 2017;130:2295–2306. doi: 10.1182/blood-2017-06-793141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maude SL, Frey N, Shaw PA. Chimeric antigen receptor T cells for sustained remissions in leukemia. N Engl J Med. 2014;371:1507–1517. doi: 10.1056/NEJMoa1407222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maude SL, Laetsch TW, Buechner J. Tisagenlecleucel in children and young adults with B-Cell lymphoblastic leukemia. N Engl J Med. 2018;378:439–448. doi: 10.1056/NEJMoa1709866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tricco AC, Antony J, Zarin W. A scoping review of rapid review methods. BMC Med. 2015;13:224. doi: 10.1186/s12916-015-0465-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leisman DE, Harhay MO, Lederer DJ. Development and reporting of prediction models: guidance for authors from editors of respiratory, sleep, and critical care journals. Crit Care Med. 2020;48:623–633. doi: 10.1097/CCM.0000000000004246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lederer DJ, Bell SC, Branson RD. Control of confounding and reporting of results in causal inference studies. Guidance for authors from editors of respiratory, sleep, and critical care journals. Ann Am Thorac Soc. 2019;16:22–28. doi: 10.1513/AnnalsATS.201808-564PS. [DOI] [PubMed] [Google Scholar]
- 19.Wan X, Wang W, Liu J, Tong T. Estimating the sample mean and standard deviation from the sample size, median, range and/or interquartile range. BMC Med Res Methodol. 2014;14:135. doi: 10.1186/1471-2288-14-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Higgins JT, Chandler JJ, Cumpston M, Li T, Page M, Welch V. 2nd edn. John Wiley & Sons; Chichester: 2019. Cochrane handbook for systematic reviews of interventions. [Google Scholar]
- 21.Harrell FE. 1st edn. Springer-Verlag; New York, NY: 2001. Regression modeling strategies: with applications to linear models, logistic regression, and survival analysis. [Google Scholar]
- 22.Qin C, Zhou L, Hu Z. Dysregulation of immune response in patients with COVID-19 in Wuhan, China. Clin Infect Dis. 2020;71:762–786. doi: 10.1093/cid/ciaa248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen G, Wu D, Guo W. Clinical and immunological features of severe and moderate coronavirus disease 2019. J Clin Invest. 2020;130:2620–2629. doi: 10.1172/JCI137244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu C, Chen X, Cai Y. Risk factors associated with acute respiratory distress syndrome and death in patients with coronavirus disease 2019 pneumonia in Wuhan, China. JAMA Intern Med. 2020;180:934–943. doi: 10.1001/jamainternmed.2020.0994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou F, Yu T, Du R. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang B, Zhou X, Zhu C. Immune phenotyping based on the neutrophil-to-lymphocyte ratio and IgG level predicts disease severity and outcome for patients with COVID-19. Front Mol Biosci. 2020;7:157. doi: 10.3389/fmolb.2020.00157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cai Q, Huang D, Ou P. COVID-19 in a designated infectious diseases hospital outside Hubei Province, China. Allergy. 2020;75:1742–1752. doi: 10.1111/all.14309. [DOI] [PubMed] [Google Scholar]
- 28.Diao B, Wang C, Tan Y. Reduction and functional exhaustion of T cells in patients with coronavirus disease 2019 (COVID-19) Front Immunol. 2020;11:827. doi: 10.3389/fimmu.2020.00827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gao Y, Li T, Han M. Diagnostic utility of clinical laboratory data determinations for patients with the severe COVID-19. J Med Virol. 2020;92:791–796. doi: 10.1002/jmv.25770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ruan Q, Yang K, Wang W, Jiang L, Song J. Clinical predictors of mortality due to COVID-19 based on an analysis of data of 150 patients from Wuhan, China. Intensive Care Med. 2020;46:846–848. doi: 10.1007/s00134-020-05991-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Z, Yang B, Li Q, Wen L, Zhang R. Clinical Features of 69 Cases with coronavirus disease 2019 in Wuhan, China. Clin Infect Dis. 2020;71:769–777. doi: 10.1093/cid/ciaa272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang Y, Shen C, Li J. Plasma IP-10 and MCP-3 levels are highly associated with disease severity and predict the progression of COVID-19. J Allergy Clin Immunol. 2020;146:119. doi: 10.1016/j.jaci.2020.04.027. 27.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen J, Fan H, Zhang L. Retrospective Analysis of clinical features in 101 death cases with COVID-19. medRxiv. 2020 doi: 10.1101/2020.03.09.20033068. published online March 17. (preprint) [DOI] [Google Scholar]
- 34.Chen X, Zhao B, Qu Y. Detectable serum SARS-CoV-2 viral load (RNAaemia) is closely correlated with drastically elevated interleukin 6 (IL-6) level in critically ill COVID-19 patients. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa449. published online April 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fu S, Fu X, Song Y. Virologic and clinical characteristics for prognosis of severe COVID-19: a retrospective observational study in Wuhan, China. medRxiv. 2020 doi: 10.1101/2020.04.03.20051763. published online April 6. (preprint) [DOI] [Google Scholar]
- 36.Gritti G, Raimondi F, Ripamonti D. Use of siltuximab in patients with COVID-19 pneumonia requiring ventilatory support. medRxiv. 2020 doi: 10.1101/2020.04.01.20048561. published online April 3. (preprint) [DOI] [Google Scholar]
- 37.Herold T, Jurinovic V, Arnreich C. Elevated levels of IL-6 and CRP predict the need for mechanical ventilation in COVID-19. J Allergy Clin Immunol. 2020;146:128. doi: 10.1016/j.jaci.2020.05.008. 36.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang Y, Yang R, Xu Y, Gong P. Clinical characteristics of 36 non-survivors with COVID-19 in Wuhan, China. medRxiv. 2020 doi: 10.1101/2020.02.27.20029009. published online March 5. (preprint) [DOI] [Google Scholar]
- 39.Liu J, Li S, Liu J. Longitudinal characteristics of lymphocyte responses and cytokine profiles in the peripheral blood of SARS-CoV-2 infected patients. EBioMedicine. 2020;55 doi: 10.1016/j.ebiom.2020.102763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu T, Zhang J, Yang Y. The role of interleukin-6 in monitoring severe case of coronavirus disease 2019. EMBO Mol Med. 2020;12 doi: 10.15252/emmm.202012421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shi Y, Tan M, Chen X. Immunopathological characteristics of coronavirus disease 2019 cases in Guangzhou, China. Immunology. 2020;160:261–268. doi: 10.1111/imm.13223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wan S, Yi Q, Fan S. Characteristics of lymphocyte subsets and cytokines in peripheral blood of 123 hospitalized patients with 2019 novel coronavirus pneumonia (NCP) medRxiv. 2020 doi: 10.1101/2020.02.10.20021832. published online Feb 12. (preprint) [DOI] [Google Scholar]
- 43.Xu Y, Li Y-r, Zeng Q. Clinical characteristics of SARS-CoV-2 pneumonia compared to controls in Chinese Han population. medRxiv. 2020 doi: 10.1101/2020.03.08.20031658. published online March 10. (preprint) [DOI] [Google Scholar]
- 44.Zhang B, Zhou X, Qiu Y. Clinical characteristics of 82 death cases with COVID-19. PLoS One. 2020;15 doi: 10.1371/journal.pone.0235458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang H, Wang X, Fuz Z. Potential factors for prediction of disease severity of COVID-19 patients. medRxiv. 2020 doi: 10.1101/2020.03.20.20039818. published online March 23. (preprint) [DOI] [Google Scholar]
- 46.Sinha P, Matthay MA, Calfee CS. Is a “cytokine storm” relevant to COVID-19? JAMA Intern Med. 2020 doi: 10.1001/jamainternmed.2020.3313. https://doi.10.1001/jamainternmed.2020.3313 published online June 30. [DOI] [PubMed] [Google Scholar]
- 47.Puelles VG, Lütgehetmann M, Lindenmeyer MT. Multiorgan and renal tropism of SARS-CoV-2. N Engl J Med. 2020;383:590–592. doi: 10.1056/NEJMc2011400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tan L, Wang Q, Zhang D. Lymphopenia predicts disease severity of COVID-19: a descriptive and predictive study. Signal Transduct Target Ther. 2020;5:33. doi: 10.1038/s41392-020-0148-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang X, Xu W, Hu G. SARS-CoV-2 infects T lymphocytes through its spike protein-mediated membrane fusion. Cell Mol Immunol. 2020;17:894. doi: 10.1038/s41423-020-0498-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Leisman DE, Deutschman CS, Legrand M. Facing COVID-19 in the ICU: vascular dysfunction, thrombosis, and dysregulated inflammation. Intensive Care Med. 2020;46:1105–1108. doi: 10.1007/s00134-020-06059-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Horby P, Lim WS, Emberson JR. Dexamethasone in hospitalized patients with Covid-19—preliminary report. N Engl J Med. 2020 doi: 10.1056/NEJMoa2021436. https://doi.org.10.1056/NEJMoa2021436 published online July 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Matthay MA, Zemans RL, Zimmerman GA. Acute respiratory distress syndrome. Nat Rev Dis Primers. 2019;5:18. doi: 10.1038/s41572-019-0069-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Calfee CS, Janz DR, Bernard GR. Distinct molecular phenotypes of direct vs indirect ARDS in single-center and multicenter studies. Chest. 2015;147:1539–1548. doi: 10.1378/chest.14-2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ackermann M, Verleden SE, Kuehnel M. Pulmonary vascular endothelialitis, thrombosis, and angiogenesis in Covid-19. N Engl J Med. 2020;383:120–128. doi: 10.1056/NEJMoa2015432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schaller T, Hirschbühl K, Burkhardt K. Postmortem examination of patients with COVID-19. JAMA. 2020;323 doi: 10.1001/jama.2020.8907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sinha P, Calfee CS, Cherian S. Prevalence of phenotypes of acute respiratory distress syndrome in critically ill patients with COVID-19: a prospective observational study. Lancet Respir Med. 2020 doi: 10.1016/S2213-2600(20)30366-0. published online Aug 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Singer M, Deutschman CS, Seymour CW. The Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3) JAMA. 2016;315:801–810. doi: 10.1001/jama.2016.0287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hotchkiss RS, Monneret G, Payen D. Sepsis-induced immunosuppression: from cellular dysfunctions to immunotherapy. Nat Rev Immunol. 2013;13:862–874. doi: 10.1038/nri3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Remy KE, Mazer M, Striker DA. Severe immunosuppression and not a cytokine storm characterizes COVID-19 infections. JCI Insight. 2020;5 doi: 10.1172/jci.insight.140329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hay KA, Hanafi LA, Li D. Kinetics and biomarkers of severe cytokine release syndrome after CD19 chimeric antigen receptor-modified T-cell therapy. Blood. 2017;130:2295–2306. doi: 10.1182/blood-2017-06-793141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Brudno JN, Kochenderfer JN. Toxicities of chimeric antigen receptor T cells: recognition and management. Blood. 2016;127:3321–3330. doi: 10.1182/blood-2016-04-703751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lee DW, Gardner R, Porter DL. Current concepts in the diagnosis and management of cytokine release syndrome. Blood. 2014;124:188–195. doi: 10.1182/blood-2014-05-552729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Teachey DT, Lacey SF, Shaw PA. Identification of predictive biomarkers for cytokine release syndrome after chimeric antigen receptor t-cell therapy for acute lymphoblastic leukemia. Cancer Discov. 2016;6:664–679. doi: 10.1158/2159-8290.CD-16-0040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Park JH, Rivière I, Gonen M. Long-term follow-up of CD19 CAR therapy in acute lymphoblastic leukemia. N Engl J Med. 2018;378:449–459. doi: 10.1056/NEJMoa1709919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Giavridis T, van der Stegen SJC, Eyquem J, Hamieh M, Piersigilli A, Sadelain M. CAR T cell-induced cytokine release syndrome is mediated by macrophages and abated by IL-1 blockade. Nat Med. 2018;24:731–738. doi: 10.1038/s41591-018-0041-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Norelli M, Camisa B, Barbiera G. Monocyte-derived IL-1 and IL-6 are differentially required for cytokine-release syndrome and neurotoxicity due to CAR T cells. Nat Med. 2018;24:739–748. doi: 10.1038/s41591-018-0036-4. [DOI] [PubMed] [Google Scholar]
- 67.Ramos-Casals M, Brito-Zerón P, López-Guillermo A, Khamashta MA, Bosch X. Adult haemophagocytic syndrome. Lancet. 2014;383:1503–1516. doi: 10.1016/S0140-6736(13)61048-X. [DOI] [PubMed] [Google Scholar]
- 68.Blanco-Melo D, Nilsson-Payant BE, Liu WC. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell. 2020;181:1036. doi: 10.1016/j.cell.2020.04.026. 45.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.McGonagle D, O'Donnell JS, Sharif K, Emery P, Bridgewood C. Immune mechanisms of pulmonary intravascular coagulopathy in COVID-19 pneumonia. Lancet Rheumatol. 2020;2:e437–e445. doi: 10.1016/S2665-9913(20)30121-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bhimraj A, Morgan RL, Shumaker AH. Infectious Diseases Society of America guidelines on the treatment and management of patients with COVID-19. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa478. published online April 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nishimoto N, Terao K, Mima T, Nakahara H, Takagi N, Kakehi T. Mechanisms and pathologic significances in increase in serum interleukin-6 (IL-6) and soluble IL-6 receptor after administration of an anti-IL-6 receptor antibody, tocilizumab, in patients with rheumatoid arthritis and Castleman disease. Blood. 2008;112:3959–3964. doi: 10.1182/blood-2008-05-155846. [DOI] [PubMed] [Google Scholar]
- 72.Chen F, Teachey DT, Pequignot E. Measuring IL-6 and sIL-6R in serum from patients treated with tocilizumab and/or siltuximab following CAR T cell therapy. J Immunol Methods. 2016;434:1–8. doi: 10.1016/j.jim.2016.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.