Novel coronavirus disease (COVID-19) pandemic is becoming a threat of global public health. Up to Aug 31, 2020, 182 countries and territories have reported a cumulative total of 24,854,140 COVID-19 cases; 838,924 of them have died (https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports). Recent data noticed that SARS-CoV-2 RNA test results of the recovered COVID-19 patients returned positive, termed “re-positive” phenomenon.1,2 However, the clinical characteristics and the immune biomarkers of “re-positive” COVID-19 patients are still far from being fully understood. We therefore conducted this retrospective study to clarify the clinical features and possible risk predictors of “re-positive” in discharged COVID-19 patients.
From Jan 17, 2020, to Mar 16, 2020, 503 COVID-19 patients were discharged from eight hospitals in Wenzhou City, Zhejiang Province, China. Patients were discharged if they met the criteria based on the National Health Commission of the People’s Republic of China, the diagnosis and treatment plan for the novel coronavirus disease (7th). We followed up 369 COVID-19 convalescent patients. The nasopharyngeal and cloacal swab samples were both detected for SARS-CoV-2 nucleic acid every week in one-month follow-up after discharge. Among them, 23 discharged patients identified positive results again. We collected and analyzed the demographic, clinical characteristics, and critical laboratory findings of 369 discharged COVID-19 patients on the first admission. The demo-graphics and clinical characteristics between COVID-19 patients with and without “re-positive” are presented in Table 1. Recent studies found that young COVID-19 patients are more likely to appear in the “re-positive” phenomenon.3 As shown in Table 1, there was no significant difference in age and gender distribution. There were no significant differences in initial clinical symptoms and laboratory findings at baseline between Re-positive and Non-re-positive group.
Table 1.
Characteristics at Baseline in COVID-19 Patients with and without Re-Positive Findings
| Variables | Re-Positive Group (N=23) | Non-Re-Positive Group (N=346) | p |
|---|---|---|---|
| Age (%) | 0.8123 | ||
| ≤45 | 9 (39.1) | 150 (44.0) | |
| >45 | 14 (60.9) | 191 (56.0) | |
| Sex (%) | 1 | ||
| F | 12 (52.2) | 171 (50.1) | |
| M | 11 (47.8) | 170 (49.9) | |
| Medical history | |||
| Hypertension - n (%) | 0.1063 | ||
| N | 15 (65.2) | 271 (81.4) | |
| Y | 8 (34.8) | 62 (18.6) | |
| Diabetes - n (%) | 0.7444 | ||
| N | 20 (87.0) | 304 (91.3) | |
| Y | 3 (13.0) | 29 (8.7) | |
| Coronary heart disease - n (%) | 0.4702 | ||
| N | 22 (95.7) | 331 (99.4) | |
| Y | 1 (4.3) | 2 (0.6) | |
| Chronic heart diseases - n (%) | 0.5786 | ||
| N | 21 (91.3) | 317 (95.2) | |
| Y | 2 (8.7) | 16 (4.8) | |
| Cerebrovascular disease - n (%) | 0.7473 | ||
| N | 22 (95.7) | 328 (98.8) | |
| Y | 1 (4.3) | 4 (1.2) | |
| Laboratory parameters on fist hospital admission | |||
| Procalcitonin, median [IQR], ng/mL | 0.08 [0.05, 0.25] | 0.10 [0.05, 0.25] | 0.5792 |
| Procalcitonin - n (%) | 1 | ||
| ≤0.046 | 4 (26.7) | 63 (24.8) | |
| >0.046 | 11 (73.3) | 191 (75.2) | |
| C-reactive protein, median [IQR], mg/L | 12.00 [3.42, 32.70] | 8.95 [3.51, 25.30] | 0.8316 |
| C-reactive protein - n (%) | 0.8704 | ||
| ≤8 | 10 (43.5) | 156 (47.6) | |
| >8 | 13 (56.5) | 172 (52.4) | |
| White-cell count, median [IQR], 10^9/L | 5.09 [4.20, 6.54] | 4.71 [3.80, 6.04] | 0.32 |
| White-cell count - n (%) | 0.8816 | ||
| [4, 10] | 16 (69.6) | 213 (65.3) | |
| <4 | 6 (26.1) | 101 (31.0) | |
| >10 | 1 (4.3) | 12 (3.7) | |
| Neutrophil count, median [IQR], 10^9/L | 3.20 [2.35, 4.10] | 2.90 [2.11, 4.03] | 0.32 |
| Neutrophil count - n (%) | 0.9241 | ||
| [1.8, 6.3] | 19 (82.6) | 258 (79.1) | |
| <1.8 | 3 (13.0) | 51 (15.6) | |
| >6.3 | 1 (4.3) | 17 (5.2) | |
| Lymphocyte count, median [IQR], 10^9/L | 1.33 [0.90, 1.53] | 1.30 [0.95, 1.58] | 0.9395 |
| Lymphocyte count - n (%) | NaN | ||
| [1.1, 3.2] | 15 (65.2) | 219 (67.2) | |
| <1.1 | 8 (34.8) | 107 (32.8) | |
| >3.2 | 0 (0.0) | 0 (0.0) | |
| Hemoglobin, median [IQR], g/L | 135.00 [125.00, 143.50] | 134.00 [124.00, 146.00] | 0.7933 |
| Hemoglobin - n (%) | 0.9017 | ||
| ≤110 | 1 (4.3) | 24 (7.4) | |
| >110 | 22 (95.7) | 302 (92.6) | |
| Platelet count, median [IQR], 10^9/L | 189.00 [149.00, 219.00] | 187.00 [153.00, 235.50] | 0.8365 |
| Platelet count - n (%) | 1 | ||
| ≤100 | 0 (0.0) | 7 (2.1) | |
| >100 | 23 (100.0) | 319 (97.9) | |
| Lymphocyte-to-white-cell ratio, median [IQR] | 0.26 [0.19, 0.30] | 0.28 [0.21, 0.35] | 0.3556 |
| Neutrophil-to-lymphocyte ratio, median [IQR] | 2.40 [2.04, 3.46] | 2.21 [1.56, 3.30] | 0.3545 |
| Platelet-to-lymphocyte ratio, median [IQR] | 145.03 [121.64, 195.21] | 150.53 [114.09, 205.04] | 0.9864 |
| Albumin, median [IQR], g/L | 42.20 [37.90, 43.70] | 41.00 [37.45, 43.50] | 0.4918 |
| Albumin - n (%) | 0.6428 | ||
| >40 | 12 (63.2) | 173 (54.9) | |
| ≤40 | 7 (36.8) | 142 (45.1) | |
| Globulin, median [IQR], g/L | 27.60 [26.50, 32.05] | 28.60 [25.55, 31.40] | 0.9201 |
| Globulin - n (%) | 0.6436 | ||
| [20, 40] | 19 (100.0) | 301 (95.6) | |
| <20 | 0 (0.0) | 7 (2.2) | |
| >40 | 0 (0.0) | 7 (2.2) | |
| Albumin-to-globulin ratio, median [IQR] | 1.42 [1.29, 1.57] | 1.44 [1.26, 1.62] | 0.6946 |
| Alanine aminotransferase, median [IQR], IU/L | 20.00 [16.00, 29.00] | 22.50 [14.00, 34.75] | 0.6563 |
| Alanine aminotransferase - n (%) | 0.7812 | ||
| ≤69 | 23 (100.0) | 315 (96.6) | |
| >69 | 0 (0.0) | 11 (3.4) | |
| Aspartate aminotransferase, median [IQR], IU/L | 23.00 [18.25, 28.25] | 24.00 [18.00, 34.00] | 0.4345 |
| Aspartate aminotransferase - n (%) | 0.6405 | ||
| ≤46 | 23 (100.0) | 311 (95.7) | |
| >46 | 0 (0.0) | 14 (4.3) | |
| Serum creatinine, median [IQR], μmol/L | 61.00 [55.00, 73.00] | 63.00 [54.00, 75.30] | 0.9513 |
| Creatine kinase, median [IQR], IU/L | 63.00 [46.50, 83.40] | 67.00 [48.70, 107.50] | 0.4705 |
| Creatine kinase - n (%) | 0.5435 | ||
| Abnormal∮ | 1 (5.0) | 35 (12.2) | |
| Normal∮ | 19 (95.0) | 252 (87.8) | |
| Lactate dehydrogenase, median [IQR], IU/L | 218.50 [189.50, 256.75] | 211.00 [174.00, 264.00] | 0.9627 |
| Lactate dehydrogenase - n (%) | 1 | ||
| ≤245 | 12 (66.7) | 187 (67.0) | |
| >245 | 6 (33.3) | 92 (33.0) | |
Notes: ∮Abnormal was defined when creatine kinase was greater than 174 IU/L for male and greater than 140 IU/L for female; normal was defined when creatine kinase was less than or equal to 174 IU/L for male and less than or equal to 140 IU/L for female. Data are presented as count (percentage) for categorical outcomes and as median [quartiles] for continuous outcomes. Groups comparison were using Chi-square test or Fisher’s exact test for categorical variables and Kruskal–Wallis rank sum test for quantitative variables. P-values of less than 0.05 was regarded as significant. P-values presented in this table have not been adjusted for multiplicity. No imputation was made for missing data. All statistical analyses were performed with R software (version 3.6.3; www.r-project.org).
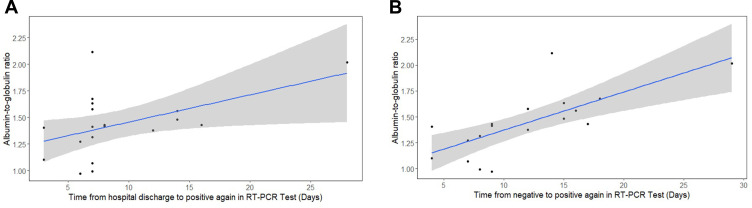
Subsequently, we did correlation analysis to demonstrate whether specific laboratory indicators are related to the critical period of these “re-positive” patients’ disease processes (Supplemental Table). These patients had experienced an average of 22 days (IQR: 16–29.5) of hospital stay, while 7 days (IQR: 7–13) of the duration from hospital discharge to positive again. Meanwhile, the mean duration of antivirus treatment was 12 days (IQR: 9–17). The average period from onset of symptoms to the end of antivirus treatment was 16 days (IQR: 11.25–22.75). Overall, there were no apparent differences in the two age groups. As shown in Figure 1A, correlation analysis indicated that there was a significant positive correlation existing between AGR at first admission and the period from hospital discharge to positive again in these 23 patients (r=0.49, p=0.0343). We also found that AGR at first admission was markedly positively correlated (r=0.72, p=0.00048) with the duration from last negative to positive again (Figure 1B).
Figure 1.
Correlation analysis for albumin/globulin ratio (AGR) and the period from hospital discharge to positive again (A) or the duration from last negative to positive again (B).
In summary, these 23 “re-positive” patients showed several features as a mild symptom, which is consistent with a previous study.3 However, there were no more cases in the lower age group in our study. This may be related to the sample size and age stratification of the present study. Hypertension, markedly common in older patients, maybe a host risk factor for severe SARS-CoV-2.4 AGR combines the nutritional as well as the inflammatory status in one measure, it may be a good indicator reflecting these two factors.5 It can be regarded as an index for predicting the prognosis of AECOPD and other inflammatory diseases.6 Recent studies indicated that a low AGR may respect a susceptible state to infection.5,6 We suggested that “re-positive” patients may not sufficiently apparent the virus. Our results indicated that AGR may potentially have a predictive effect in “re-positive” discharged COVID-19 patients. Further definitive largescale clinical studies are feasible and needed.
Acknowledgments
We acknowledge all health care workers from the eight hospitals who involved in the diagnosis and treatment of the COVID-19 patients in Wenzhou.
Funding Statement
This work was supported by: National Major Scientific and Technological Special Project of china for: Significant New Drugs Development(2020ZX09201002); Wenzhou Science and technology key problem program [ZY2020001]; The critical research and development program of Zhejiang Province [2019C03011]; The research was designed, conducted, analyzed, and interpreted by the authors entirely independently of the funding sources.
Ethics Approval and Consent to Participate
The study complied with the declaration of Helsinki and was approved by the Institutional Review Board (IRB) of Wenzhou Medical University (No.2020-004). Informed consent was obtained from all participants.
Disclosure
All authors report no potential conflicts of interest for this work.
References
- 1.Lan L, Xu D, Ye G, et al. Positive RT-PCR test results in patients recovered from COVID-19. JAMA. 2020;323(15):1502. doi: 10.1001/jama.2020.2783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li J, Long X, Fang X, et al. SARS-CoV-2 positivity in a discharged COVID-19 patient: a case report. Clin Microbiol Infect. 2020;26(8):1115–1117. doi: 10.1016/j.cmi.2020.04.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yuan J, Kou S, Liang Y, Zeng J, Pan Y, Liu L. PCR assays turned positive in 25 discharged COVID-19 patients. Clin Infect Dis. 2020. doi: 10.1093/cid/ciaa398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shi Y, Yu X, Zhao H, Wang H, Zhao R, Sheng J. Host susceptibility to severe COVID-19 and establishment of a host risk score: findings of 487 cases outside Wuhan. Crit Care. 2020;24(1):108. doi: 10.1186/s13054-020-2833-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xun Y, Yang Y, Yu X, Li C, Lu J, Wang S. A preoperative nomogram for sepsis in percutaneous nephrolithotomy treating solitary, unilateral and proximal ureteral stones. PeerJ. 2020;8:e9435. doi: 10.7717/peerj.9435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qin J, Qin Y, Wu Y, et al. Application of albumin/globulin ratio in elderly patients with acute exacerbation of chronic obstructive pulmonary disease. J Thorac Dis. 2018;10(8):4923–4930. doi: 10.21037/jtd.2018.07.47 [DOI] [PMC free article] [PubMed] [Google Scholar]