Abstract
Background
The emergence and prevalence of plasmid-mediated colistin-resistant bacterial strains in recent years have raised great concerns in clinical medicine. It is urgently needed to develop a cheaper, faster, simpler, sensitive, and specific molecular detection method to identify and monitor the dissemination of the transferable resistant determinants.
Methods and Results
Herein, eight pairs of primers were designed to set up a multiplex PCR method for the rapid and efficient determination of reported mcr genes. This assay can give results within 85 min (35 min for amplification and 50 min for electrophoresis). We validated the feasibility of this assay by testing the presence of mcr genes in 60 colistin-resistant isolates.
Conclusion
Our multiplex PCR technique exhibits remarkable advantages in the light of clear identification, efficiency of amplification, as well as the time consuming for detection, and thus could be useful for the surveillance and epidemiological research of plasmid-mediated colistin resistance, particularly for the under-resourced laboratories.
Keywords: multiplex PCR, colistin resistance, mcr genes
Introduction
Colistin is a polymyxin antibiotic originating from the gram-positive organism Bacillus polymyxa. Due to its significant nephrotoxicity and neurotoxicity,1 the clinical application of colistin in the treatment of human bacterial infections has been restricted. The constantly increasing emergence and spread of multidrug-resistant bacteria is a serious threat to global health. The lack of effective antimicrobial agents in the treatment of diseases caused by resistant bacteria resulted in the reintroduction of colistin application in human infections. Unfortunately, colistin-resistant bacteria rapidly emerged owing to the improper use and abuse of colistin. Previously, resistance to colistin mostly involved chromosomal mutations.2 However, since the first description of plasmid-borne resistance gene mcr-1 by Liu et al in 2016, the acquired resistance mechanism of colistin has been placed on the center of focus.3 The prevalence of mobile colistin-resistant genes could be responsible for the rapid increase of colistin-resistant bacterial strains.3 In addition to mcr-1, nine other groups of mcr genes (mcr-2 to mcr-10) were described within the past 4 years, adding to the complexities of colistin resistance.4,5 The novel genes showed distinct similarity with mcr-1 in both nucleotide and amino acid sequences.6 Since mcr-6 has 86% nucleotide identity with mcr-2, it has been considered as an mcr-2 like gene.6 Most plasmid-mediated colistin-resistant strains carry a single mcr gene; however, co-presence of two mcr genes in a single isolate has been described recently.7–9 MCR family proteins are membrane-associated phosphoethanolamine (PEA) transferases which catalyze the addition of PEA moiety to the lipopolysaccharides (LPS)-lipid A, resulted in reduced affinity to colistin.10
To prevent the spread of colistin resistance, fast, cheap and efficient diagnostic approaches are desperately needed. Previously, several multiplex polymerase chain reaction (PCR) assays have been reported for the detection of mcr genes (mcr-1 to mcr-5).6,11–13 Apparently, this method needs to be updated due to the discovery of novel mcr genes (eg mcr-7, mcr-8, and mcr-9). Here, we developed a multiplex PCR method which allowed us to quickly and accurately detect currently reported mcr genes in a single mixture.
Materials and Methods
Primers and Multiplex PCR Reaction
The nucleotide sequences of eight mcr genes were retrieved from the NCBI database (https://www.ncbi.nlm.nih.gov/). Specific primers (Table 1) for the amplification of the eight mcr genes were designed by Primer3Plus (www.bioinformatics.nl). The compatibility of these primers for the multiplex PCR was verified by the in-silico PCR modeling feature at the UCSC Genome Bioinformatics website (http://genome.ucsc.edu). The ability of each primer pairs to bind to its variants was verified by in-silico analysis using FastPCR software (http://primerdigital.com/fastpcr.html). The multiplex PCR was performed in a 20 μL reaction mixture containing 10 μL of 2xFast Taq Master Mix (Novoprotein, Shanghai, China), 20 pmol of each primer for the amplification of the mcr-1 to mcr-5 genes, 10 pmol of each primer for the amplification of the mcr-7 to mcr-9 genes, and 20 ng of each template DNA. The multiplex PCR was performed with the following cycling conditions: denaturation at 94°C for 4 min, 20 cycles of amplification at 94°C for 5 seconds and 59°C for 15 seconds, and a final extension step at 72°C for 5 min. The resulting DNA products were analyzed by electrophoresis in a 2.0% agarose gel at 90 V for 50 min.
Table 1.
Primers Used for Multiplex PCR Detection of Mcr Genes
| Primer Name | Sequence (5ʹ-3ʹ) | Target Gene | Size (bp) |
|---|---|---|---|
| mcr-1_205F | TCCAAAATGCCCTACAGACC | mcr-1 | 205 |
| mcr-1_205R | GCCACCACAGGCAGTAAAAT | ||
| mcr-2_279F | CCTTTTGTGCTGATGGGTTT | mcr-2 | 279 |
| mcr-2_279R | ATTTTGGAGCATGGTGGTGT | ||
| mcr-3_347F | CTTGCTGAACCAATCCCATT | mcr-3 | 347 |
| mcr-3_347R | CCATCGTTCTCCTTCCAAAA | ||
| mcr-4_426F | GATCCGAAGCTGTGTTCTG | mcr-4 | 426 |
| mcr-4_426R | GCCAGCATTGGTACGCTAGT | ||
| mcr-5_522F | GGTTGGCCGAGAAGATAACA | mcr-5 | 522 |
| mcr-5_522R | ATGTTGCCAGAAGGTCCAAC | ||
| mcr-7_791F | GTCAGTTACGCCATGCTCAA | mcr-7 | 791 |
| mcr-7_791R | TTCTTGTCGCAGAACTGTGG | ||
| mcr-8_943F | AAACTGAACCCGGTACAACG | mcr-8 | 943 |
| mcr-8_943R | GCCATAGCACCTCAACACCT | ||
| mcr-9_635F | GCGGTTGTAAAGGCGTATGT | mcr-9 | 635 |
| mcr-9_635R | CAAATCGCGGTCAGGATTAT |
All of the mcr genes were synthesized by GenScript and cloned into pUC57 (Supplementary Table S1). To validate the specificity and efficiency of the primers and cycling conditions for the multiplex PCR assay, both isolated plasmid DNA of pUC57 harboring each mcr gene and the total DNA extracted from the liquid culture of E. coli Top10 containing the relevant plasmids by the boiling method were used as templates. In addition, different combinations of two or three mcr genes were applied to the multiplex PCR assay (Supplementary Table S2).
Detection of mcr Genes in Bacterial Isolates by the Multiplex PCR Assay
A total of 20 fecal samples of pigs were collected in 2019 from three different swine farms at Jilin province, China. Nineteen pet fecal samples (9 from cats and 10 from dogs) were collected at the Veterinary Teaching Hospital of Jilin University. Colistin-resistant strains were selectively isolated on MacConkey agar (Hopebio, Qingdao, China) containing 2 mg/L colistin (Sigma-Aldrich, St Louis, MO, USA). The resulting colonies were purified three times and characterized by 16S rRNA sequencing. The sources and species of the isolates were listed in Supplementary Table S3. Bacteria were cultured in 3 mL nutrient broth at 37°C for 16–24 hours. The total DNA was extracted by the boiling method. 0.5 μL of the total DNA was applied to the multiplex PCR assay described above to detect the presence of the currently reported mcr genes.
Results
Validation of the Multiplex PCR Method
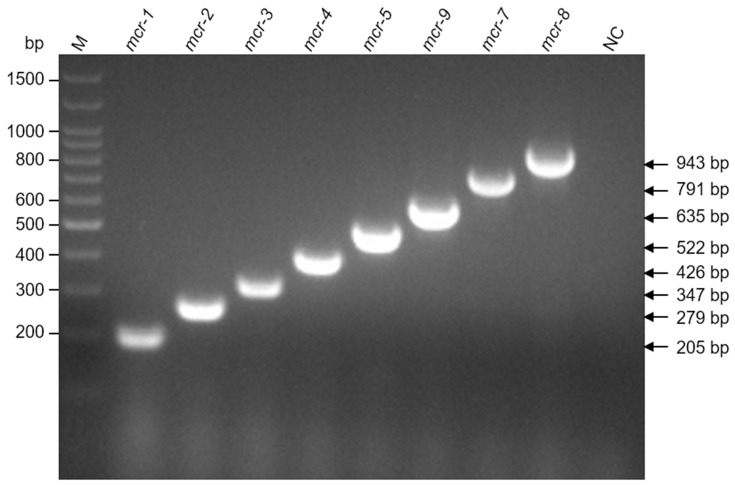
Eight pairs of primers were designed to amplify the internal fragments of mcr genes. The sizes of the amplicons were 205 bp (mcr-1), 279 bp (mcr-2), 347 bp (mcr-3), 426 bp (mcr-4), 522 bp (mcr-5), 791 bp (mcr-7), 943 bp (mcr-8), and 635 bp (mcr-9). As shown in Figure 1, when plasmid DNA of pUC57 carrying each mcr gene was used as template in the multiplex PCR mixture, unique and specific bands were visualized by agarose gel electrophoresis, suggesting the specificity of the primer sets. Additionally, we prepared the total DNA through boiling the broth cultures of E. coli strains harboring plasmid containing mcr genes. Similarly, amplification of the expected gene fragment was also observed when the total DNA was utilized as a template (Figure S1). The PCR products were recovered from the gel and the specific amplification was confirmed by sequencing. In order to test the detection limit of the method, different amounts of plasmid templates were applied to the reaction mixture. The detection limits for mcr-2, mcr-4, mcr-5, mcr-7, mcr-9 were less than 10 pg (Figure S2). Although at a lower efficiency, amplification of mcr-1, mcr-3 and mcr-8 could also be detected in the presence of 10 pg of plasmid templates (Figure S2). In addition, we tested the specificity of the method against other antibiotic resistance genes. No non-specific product or interference was detected when plasmid DNA harboring dim-1, spm-1, imp-1 or aim-1 was added to the multiplex PCR mixture (Figure S3). The PCR fragments were designed to be at least 60 bp difference in size, allowing us to easily differentiate the products. In addition, we utilized a 2% agarose gel and run for a longer time (50 min) to assure each band could be clearly distinguished. To accelerate the multiplex PCR detection process, we optimized the cycling conditions by reducing the denaturation time during cycling, annealing time and number of cycles. Since all the amplicons were less than 1000 bp in size, a final extension step was sufficient to allow extension of the PCR products. Apparently, omission of the extension step during cycling did not affect the amplification. Taken together, our method could allow the rapid identification of mcr genes with total detection time less than 85 min (amplification and electrophoresis).
Figure 1.
Multiplex PCR detection of mcr genes. Purified plasmid pUC57 carrying each mcr gene was used as templates. Agarose gel electrophoresis (2.0%) was used to separate multiplex PCR products. M indicates the molecular size marker (Trans100bp plus DNA ladder, TransGen Biotech, China). The size of each amplicon is indicated on the right side. NC, empty vector pUC57 was included as negative control.
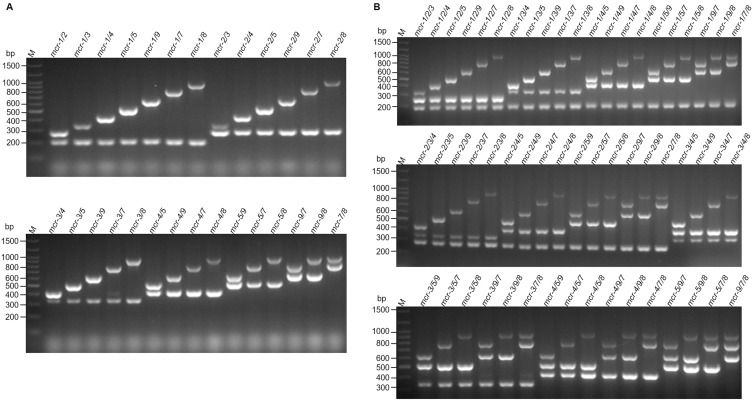
Since the co-occurrence of two mcr genes in a single isolate has been reported,7–9 we tested the ability of our multiplex PCR method to simultaneously detect mixed DNA templates in a single tube. As shown in Figure 2, the multiplex PCR system can effectively detect 28 combinations of two genes and 56 combinations of three genes. The band amplified for each gene was as expected.
Figure 2.
Multiplex PCR detection of two (A) or three (B) mcr genes. Different mixtures of double or triple plasmid DNA carrying a single mcr gene were used as templates. M indicates the molecular size marker.
Each of the mcr genes has multiple variants. We performed in-silico analysis to predict the ability of each primer pairs to bind to their variants. The results suggested that except for mcr-2.2 and mcr-3.25, all the mcr variants could specifically bind to the primers (Supplementary Table S4).
Application of the Multiplex PCR Method to Bacterial Isolates
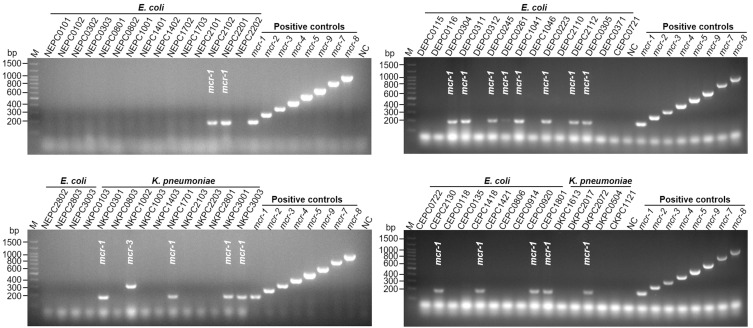
To test the feasibility of this method for clinical use, we isolated 60 colistin-resistant strains (43 isolates of E. coli and 17 isolates of Klebsiella pneumoniae) from fecal samples of pigs and pets by selective medium containing 2 mg/L of colistin (Supplementary Table S3). The total DNA of these strains was made by the boiling method from liquid cultures and added to the multiplex PCR mixture. Twenty out of the 60 isolates were determined to contain mcr genes (Figure 3). These included 19 strains positive for mcr-1 and 1 isolate positive for mcr-3 (Figure 3). Collectively, the results indicated the reliability of our method for the determination of mcr genes in bacterial isolates.
Figure 3.
Molecular detection of mcr genes in bacterial isolates by the multiplex PCR method. Colistin-resistant bacterial strains (43 isolates of E. coli and 17 isolates of K. pneumoniae) were tested for the presence of mcr genes. Total bacterial DNA was obtained by the boiling method and applied to the multiplex PCR assay. E. coli strain Top10 harboring the mcr-encoding plasmid was included as a positive control. NC, E. coli TOP10 with empty vector pUC57. M shows the molecular size marker.
Discussion
Colistin is one of the last-resort antimicrobial agents for the treatment of human infections caused by fatal and multidrug-resistant gram-negative bacteria. However, colistin resistance has increased significantly worldwide in the recent years, majorly attributed to the emergence and dissemination of plasmid-mediated resistance mechanisms.3 Since the first report of the plasmid-borne polymyxin resistance determinant mcr-1, nine groups of mcr genes with diverse similarities in nucleotide and amino acid sequences have been described.4
The development of efficient molecular diagnostics is urgently necessary to identify and constrain this fast-spreading menace. Recently, a series of molecular tests have been designed to detect MCR-producing strains, including microarray,14 loop-mediated isothermal amplification (LAMP),15 conventional PCR,3 multiplex PCR6,11–13 and real-time PCR.16,17 Considering the sensitivity, specificity, skill and equipment requirements, cost, as well as turnaround time, each detection approach has its own advantages and disadvantages. Apparently, most of the techniques only detect a single mcr gene. The microarray detection method and the real-time PCR method are sensitive, and capable of detecting multiple mcr genes, but their application is restricted due to the requirement of specific and expensive equipment. Previously, four multiplex PCR assays capable of screening for mcr-1 to mcr-5 with different sets of primers and cycling conditions were reported.6,11–13 This method represents an efficient and inexpensive way for the rapid determination of mcr-encoding bacterial isolates. However, novel groups of mcr genes were not included in these assays.6,11–13 Another multiplex PCR method aimed to amplify mcr-6 to mcr-9 was reported recently, which is a valuable supplement to the available mcr-1 to mcr-5 multiplex PCR assays.18 In this study, we developed a multiplex PCR technique that could detect the mcr genes in a single mixture. Apparently, our method shows a striking advantage in the time consuming for detection. Recently, mcr-10 was described as a novel group of mcr genes that has the highest similarity with mcr-9.5 In the future, it is necessary to include this gene for the development of a multiplex PCR method to detect all reported mcr genes.
Conclusion
Here we developed a multiplex PCR method that allowed efficient and rapid (less than 35 min for the amplification) detection of all described families of mcr gene (mcr-1, mcr-2, mcr-3, mcr-4, mcr-5, mcr-7, mcr-8 and mcr-9) within a single tube. We believe that this technique can be applied to periodic surveillance and epidemiological research in hospitals, farms, foods, and the environment.
Funding Statement
This work was supported by the Thousand Young Talents Program of the Chinese government (JZQ) and startup fund from Jilin University (JZQ).
Disclosure
The authors declare no conflicts of interest for this work.
References
- 1.Lim LM, Ly N, Anderson D, et al. Resurgence of colistin: a review of resistance, toxicity, pharmacodynamics, and dosing. Pharmacotherapy. 2010;30(12):1279–1291. doi: 10.1592/phco.30.12.1279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McPhee JB, Lewenza S, Hancock RE. Cationic antimicrobial peptides activate a two-component regulatory system, PmrA-PmrB, that regulates resistance to polymyxin B and cationic antimicrobial peptides in Pseudomonas aeruginosa. Mol Microbiol. 2003;50(1):205–217. doi: 10.1046/j.1365-2958.2003.03673.x [DOI] [PubMed] [Google Scholar]
- 3.Liu YY, Wang Y, Walsh TR, et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis. 2016;16(2):161–168. doi: 10.1016/S1473-3099(15)00424-7 [DOI] [PubMed] [Google Scholar]
- 4.Anyanwu MU, Jaja IF, Nwobi OC. Occurrence and characteristics of mobile colistin resistance (mcr) gene-containing isolates from the environment: a review. Int J Environ Res Public Health. 2020;17(3):1028. doi: 10.3390/ijerph17031028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang C, Feng Y, Liu L, Wei L, Kang M, Zong Z. Identification of novel mobile colistin resistance gene mcr-10. Emerg Microbes Infect. 2020;9(1):508–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lei S, Lv J, Gao S, Srinivas S, Feng Y. Developing an efficient multiplex PCR method to detect mcr-like genes. Sci China Life Sci. 2019;62(5):705–707. doi: 10.1007/s11427-019-9512-3 [DOI] [PubMed] [Google Scholar]
- 7.Touati M, Hadjadj L, Berrazeg M, Baron S, Rolain JM. Emergence of Escherichia coli harboring mcr-1 and mcr-3 gene in North West Algerian farmlands. J Glob Antimicrob Resist. 2019. [DOI] [PubMed] [Google Scholar]
- 8.Li R, Zhang P, Yang X, et al. Identification of a novel hybrid plasmid coproducing MCR-1 and MCR-3 variant from an Escherichia coli strain. J Antimicrob Chemother. 2019;74(6):1517–1520. doi: 10.1093/jac/dkz058 [DOI] [PubMed] [Google Scholar]
- 9.Creighton J, Anderson T, Howard J, Dyet K, Ren X, Freeman J. Co-occurrence of mcr-1 and mcr-3 genes in a single Escherichia coli in New Zealand. J Antimicrob Chemother. 2019;74(10):3113–3116. doi: 10.1093/jac/dkz311 [DOI] [PubMed] [Google Scholar]
- 10.Sun J, Zhang H, Liu YH, Feng Y. Towards understanding MCR-like colistin resistance. Trends Microbiol. 2018;26(9):794–808. doi: 10.1016/j.tim.2018.02.006 [DOI] [PubMed] [Google Scholar]
- 11.Rebelo AR, Bortolaia V, Kjeldgaard JS, et al. Multiplex PCR for detection of plasmid-mediated colistin resistance determinants, mcr-1, mcr-2, mcr-3, mcr-4 and mcr-5 for surveillance purposes. Eurosurveillance. 2018;23(6):29–39. doi: 10.2807/1560-7917.ES.2018.23.6.17-00672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lescat M, Poirel L, Nordmann P. Rapid multiplex polymerase chain reaction for detection of mcr-1 to mcr-5 genes. Diagn Microbiol Infect Dis. 2018;92(4):267–269. doi: 10.1016/j.diagmicrobio.2018.04.010 [DOI] [PubMed] [Google Scholar]
- 13.Jousset AB, Bernabeu S, Bonnin RA, et al. Development and validation of a multiplex polymerase chain reaction assay for detection of the five families of plasmid-encoded colistin resistance. Int J Antimicrob Agents. 2019;53(3):302–309. doi: 10.1016/j.ijantimicag.2018.10.022 [DOI] [PubMed] [Google Scholar]
- 14.Bernasconi OJ, Principe L, Tinguely R, et al. Evaluation of a new commercial microarray platform for the simultaneous detection of beta-lactamase and mcr-1 and mcr-2 genes in enterobacteriaceae. J Clin Microbiol. 2017;55(10):3138–3141. doi: 10.1128/JCM.01056-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Imirzalioglu C, Falgenhauer L, Schmiedel J, et al. Evaluation of a loop-mediated isothermal amplification-based assay for the rapid detection of plasmid-encoded colistin resistance gene mcr-1 in enterobacteriaceae isolates. Antimicrob Agents Chemother. 2017;61(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nijhuis RH, Veldman KT, Schelfaut J, et al. Detection of the plasmid-mediated colistin-resistance gene mcr-1 in clinical isolates and stool specimens obtained from hospitalized patients using a newly developed real-time PCR assay. J Antimicrob Chemother. 2016;71(8):2344–2346. [DOI] [PubMed] [Google Scholar]
- 17.Chabou S, Leangapichart T, Okdah L, Le Page S, Hadjadj L, Rolain JM. Real-time quantitative PCR assay with Taqman((R)) probe for rapid detection of MCR-1 plasmid-mediated colistin resistance. New Microbes New Infect. 2016;13:71–74. doi: 10.1016/j.nmni.2016.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Borowiak M, Baumann B, Fischer J, et al. Development of a novel mcr-6 to mcr-9 multiplex PCR and assessment of mcr-1 to mcr-9 occurrence in colistin-resistant salmonella enterica isolates from environment, feed, animals and food (2011–2018) in Germany. Front Microbiol. 2020;11:80. doi: 10.3389/fmicb.2020.00080 [DOI] [PMC free article] [PubMed] [Google Scholar]