A reliable pipeline to identify DNA methylation signatures in CSF ctDNA could serve as potential biomarkers for MB patients.
Abstract
Medulloblastoma (MB), the most common form of pediatric brain malignancy, has a low frequency of oncogenic mutations but pronouncedly abnormal DNA methylation changes. Epigenetic analysis of circulating cell-free tumor DNA (ctDNA) by liquid biopsy offers an approach for real-time monitoring of tumor status without tumor dissection. In this study, we identified 6598 differentially methylated CpGs in both MB tumors and the ctDNA isolated from matched cerebrospinal fluid (CSF) compared with normal cerebellum, which could be used to detect MB tumor occurrence and determine its subtype. Furthermore, DNA methylation changes in serial CSF samples could be used to monitor the treatment response and tumor recurrence. Integrating our data with large public datasets, we identified reliable MB DNA methylation signatures in ctDNA that have potential diagnostic and prognostic values. Our study sets the stage for exploiting epigenetic markers in CSF to improve the clinical management of patients with MB.
INTRODUCTION
Medulloblastoma (MB), the most common pediatric brain tumor, is considered an embryonal tumor of the cerebellum (1–6). Although the cellular origin of MB is unclear, it has been speculated that MB tumor cells arise from neuronal stem or progenitor cells during early life. On the basis of its molecular features, MB can be categorized into four subtypes: WNT (wingless) pathway–activated, SHH (Sonic hedgehog)pathway–activated, and the less well-characterized group 3 and group 4 (7–10). MB is well known for having very few genomic mutations compared with adult cancers and other pediatric cancers (6, 11). Recent studies have shown that different MB subtypes harbor distinct epigenetic signatures and undergo dynamic alterations in DNA methylation during tumor progression and clinical treatment (10). DNA hypomethylation is strongly associated with increased gene expression in MB, suggesting that DNA demethylation might play a pivotal role in MB pathogenesis (12). The Ten-eleven translocation family of dioxygenases plays a major role in DNA demethylation by catalyzing the oxidation of 5-methycytosine (5mC) to produce, in succession, 5-hydroxymethylation (5hmC), 5-formylcytosine, and 5-carboxycytosine (13). Among these oxidized 5mC derivatives, 5hmC is the predominant catalytic product and functions as an epigenetic mark to modulate chromatin accessibility and gene transcription (14). The diagnostic and prognostic value of 5hmC in circulating cell-free tumor DNA (ctDNA) or cancer tissues has been reported for several types of tumors (e.g., B cell lymphoma and colon cancer) (15, 16), but its value for pediatric brain tumors remains to be determined.
MB is currently diagnosed on the basis of clinical symptoms and radiographic findings, with final confirmation by histopathological examination. For group 3 and group 4 MBs, approximately 30% of patients show signs of metastasis at diagnosis (17). In addition to radiographic evaluation of detectable metastasis, lumbar puncture (LP) is often performed at diagnosis to complete the staging process, with the disseminated cases classified according to the Chang Staging System (i.e., M stage) (18). Routine magnetic resonance imagings (MRIs) are obtained during and after treatment to assess treatment response and to monitor recurrence. Repeated LPs are typically performed at the end of therapy and when clinically indicated. Recent advances in imaging-based diagnostic techniques have markedly improved the early detection and assessment of treatment outcomes (19). However, there remains a strong desire to have an independent method that can reliably monitor tumor treatment response and confidently detect early tumor recurrence, before anatomical or metabolic changes are evident by advanced imaging techniques [e.g., MRI, computed tomography, positron emission tomography, and single-photon emission computed tomography] (19, 20).
Liquid biopsy, which is based on the analysis of ctDNA, exosomes, or circulating tumor cells in plasma or other biological fluids, has emerged as a promising approach to aiding the timely detection, molecular profiling, and response monitoring of many solid tumors (21). However, because of the blood-brain barrier, plasma contains substantially lower amounts of ctDNA from brain tumors than from peripheral solid tumors. In contrast to plasma, cerebrospinal fluid (CSF) interacts more frequently with brain tumor cells in the central nervous system (CNS) and can serve as a substrate for liquid biopsy. It has been shown that the mutations in CSF ctDNA parallel the genetic alterations in the brain tumor and may serve as a reliable source to monitor brain tumor status (22–24). However, whether CSF ctDNA modifications faithfully reflect brain tumor epigenetic landscapes remains underexplored. In the current study, we obtained DNA methylomes and hydroxymethylomes using whole-genome bisulfite sequencing (WGBS) and anti–cytosine-5-methylenesulfonate (anti-CMS) immunoprecipitation sequencing (CMS-IP–seq) (25), respectively, for very small amounts of CSF ctDNA from pediatric patients with MB. We found a positive correlation between CSF samples and tumor tissues, suggesting that CSF ctDNA can be used as material to monitor changes in MB tumor DNA methylomes and hydroxymethylomes. Furthermore, by analyzing the DNA methylation dynamics in ctDNA purified from serial CSF samples in the same patients, the tumor-specific DNA methylation signatures occur before the positive cytology analysis, suggesting the high-sensitivity and potential high-clinic impact of our method to modulate the MB disease status. In addition, by correlating epigenetic changes in CSF ctDNA with the clinical outcomes for patients with MB, we identified DNA methylation markers of diagnostic and prognostic value detectable in CSF ctDNA to aid the management of MB.
RESULTS
DNA methylation and hydroxymethylation profiling of CSF ctDNA from pediatric patients with MB
To characterize the cell-free DNA in CSF, we performed the bioanalyzer analysis using the purified DNA from precentrifuged CSF samples (fig. S1A). We observed that the majority of DNA fragments ranged from 100 to 400 base pairs (bp) with the peak size at 200 bp, which is consistent with a previous report (26). This finding suggests that the majority of DNA from CSF in the analyzed samples is ctDNA. To profile the DNA methylation and hydroxymethylation patterns in MB ctDNA from CSF, we prepared sequencing libraries using these precentrifuged CSF samples with or without additional centrifugation after recovery from freezer (fig. S1, A and B, and table S1). After evaluating the conditions, we found that 200 μl of CSF without an additional centrifugation yielded sufficient ctDNA that could be used to generate high-quality libraries suitable for sequencing (fig. S1C and table S1). Compared to a previous method (27), our optimized protocol, which uses random priming to construct the bisulfite library, required much less CSF as input (0.2 ml versus 1 ml). However, a low-input volume combined with the small-fragment sizes of ctDNA can result in a low ratio of uniquely mapped reads and, consequently, high cost. We therefore developed LiBis (28), a method that can markedly increase the ratio of uniquely mapped reads for bisulfite sequencing of low-input DNA (fig. S1D). We then collected matched MB (SHH) tumor and CSF samples from patients 1, 2, and 3 and MB (WNT) CSF samples from patient 4 (Fig. 1A) for integrative epigenomic analyses. To test whether CSF can be used to monitor tumor status following treatment and to predict recurrence, we also collected a total of eight MB CSF samples from patients 2, 3, and 4 during and after treatment (Fig. 1A and table S1). The CSF collected from patients with hydrocephalus (n = 4) and the CSF samples collected from patients with acute lymphoblastic leukemia (n = 2) were used as surrogate nontumor CSF controls, and normal cerebellum tissues (n = 2) were used as tissue controls (Fig. 1A). Overall, we obtained an average of 600 million reads covering 13 million CpG sites at least three times by WGBS analysis, with bimodal distribution of the DNA methylation ratios and without M bias after trimming of the first 10 bp along the read (fig. S1, E and F, and table S1). The average DNA methylation level was near 0.6 to 0.8, which was consistent with a previous study (12) (fig. S1G). In parallel, we collected an average of 23 million reads to yield an average of 85,000 5hmC-enriched regions using the CMS-IP–seq method (table S1). The distribution patterns of ctDNA methylation and hydroxymethylation for metagenes were consistent with that of tumor tissues, as described in previous publications (14) (fig. S1H). In addition, we compared WGBS data of MB tissues obtained in this study with previously published WGBS data from MB tumors (12). We also observed a relatively high Pearson correlation coefficient (>0.5) between these two datasets regardless of MB subtypes (fig. S1I). Furthermore, 5hmC was enriched in common H3K27ac-enriched regions (9) among four subtypes in both ctDNA and MB tissues (fig. S1J), which was consistent with 5hmC distribution in other systems, including embryonic stem cells, T cells, and B cells (14). These data suggest that our optimized protocol yields DNA methylomes and hydroxymethylomes of high quality from CSF ctDNA.
Fig. 1. Cytosine modifications in CSF ctDNA are highly concordant with those detected in MB tumors in situ.
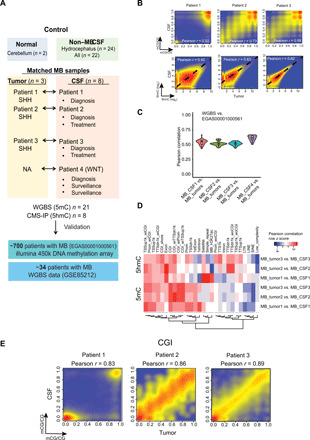
(A) Schematic of the experimental design. Normal cerebellum tissue (n = 2), non–MB CSF samples (n = 6, four patients with hydrocephalus without symptoms of other diseases and two patients with acute lymphoblastic leukemia without brain metastasis), matched MB tumor tissue, and CSF sample pairs from patients with MB SHH (patients 1, 2, and 3) and CSF samples from patient 4 (WNT) were used in this study. (B) Pearson correlation analysis of the DNA methylation status of common CpG sites (covered at least 10 times) and common hydroxymethylated regions shared between CSF ctDNA samples and their matched MB tumors. (C) Pearson correlation analysis of the DNA methylation status of the CpG sites that are common between indicated CSF ctDNA samples and published MB tumor samples (n = 34). The minimal coverage of selected CpG sites was 5×. (D) Pearson correlation analysis of the DNA methylation and hydroxymethylation at the indicated regulatory elements between CSF ctDNA and matched MB tumors. (E) Scatter plots showing the correlation of DNA methylation levels between CSF ctDNA and matched MB tumors within CpG island (CGI) regions (CpGs covered at least 10 times).
To further evaluate the data quality of WGBS data obtained in ctDNA in MB CSF, the Pearson correlation analysis shows that there is a relatively high Pearson correlation coefficient between matched MB CSF and tumor samples (average > 0.5), while the Pearson correlation between nontumor CSF samples and MB tumors are relatively low (average < 0.33) (Fig. 1B, top, and fig. S2, A and B). These findings suggest that CSF ctDNA can be used to faithfully elucidate DNA methylation profiles representative of MB tumors in situ. Similarly, the Pearson correlation of the DNA hydroxymethylation analysis between MB tumor and matched CSF ctDNA ranged from 0.4 to 0.8 (Fig. 1B, bottom). In parallel, to estimate the individual variations of these DNA epigenetic markers, we performed pairwise comparisons of the CSF ctDNA methylation and hydroxymethylation data among analyzed patients. We observed a significant positive correlation between the individuals, suggesting that DNA epigenetic markers might be relatively well conserved with low interpatient variation (fig. S2C).
Next, we compared DNA methylation and hydroxymethylation levels within the regulatory elements between MB tumor and matched MB CSF. We observed a relatively high Pearson correlation coefficient (average r = 0.7) between tumor and CSF within genic regions, including transcription start sites, exons, CpG islands (CGIs), and promoters (fig. S2D). A relatively low Pearson correlation coefficient (average r = 0.5) was found for repeat elements, including long interspersed nuclear elements (LINEs), short interspersed nuclear elements (SINEs), and long terminal repeats (LTRs) (Fig. 1D). Furthermore, the DNA methylation levels within CGIs were more strongly correlated between MB tumor tissue and MB CSF and were highly consistent among individuals (Fig. 1E, and fig. S2, E and F). These results suggest that DNA methylation status within the CGIs of CSF ctDNA could potentially serve as biomarkers to report the status of the original MB tumor.
DNA methylation and hydroxymethylation dynamics in MB CSF ctDNA
To evaluate whether differences in the DNA methylation and hydroxymethylation between MB and normal cerebellum tissue were recapitulated in CSF ctDNA, we identified differentially methylated regions (DMRs) or differentially hydroxymethylated regions (DHMRs) by comparing normal cerebellum data, MB tumor data, and MB CSF data (Fig. 2, A and B) using MOABS (29). Consistent with a previous report (12), MB tumors displayed a global decrease in DNA methylation but increased DNA methylation at CGIs (fig. S3A). Next, we selected the DMRs or DHMRs that we identified as shared (i.e., common) between the normal cerebellum and the MB tumor and between the normal cerebellum and the MB CSF. In total, we obtained 17,898 and 1777 common hyper- and hypo-DMRs, respectively, and 39,602 and 20,707 common hyper- and hypo-DHMRs, respectively (Fig. 2, A and B). The average Pearson correlation coefficient of DNA methylation of CpGs in DMRs and DHMRs was >0.6 between the MB CSF and the MB tumor (fig. S3B). The methylation differences between the MB CSF and the normal cerebellum and between the MB tumor and the normal cerebellum were highly consistent, with Pearson correlation coefficients varying from 0.96 to 0.98 within DMRs (Fig. 2C). The hydroxymethylation differences within DHMRs were also very consistent (Fig. 2D). These data strongly suggest that ctDNAs from MB tumors are present in CSF and can be used to faithfully mirror the DNA methylation status of MB tumors in situ.
Fig. 2. DNA modification changes in both MB tumors and MB CSF compared to normal cerebellum.
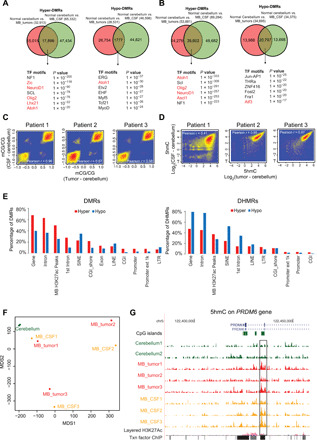
(A) Top: Venn diagrams of the number of hyper-DMRs (left) or hypo-DMRs (right) identified between cerebellum and MB tumor and between cerebellum and MB CSF. Bottom: Lists of transcription factor (TF) motifs that were enriched within shared hyper-DMRs (left) or hypo-DMRs (right). (B) The same analysis as described in (A) but for hydroxymethylation. (C and D) Scatter plots showing the correlation of differences in 5mC (C) or 5hmC (D) between the normal cerebellum and MB tumor and between cerebellum and MB CSF ctDNA. (E) The genome distributions of shared hyper-DMRs (red) and hypo-DMRs (blue) identified in (A) (left), shared hyper-DHMRs (red) and hypo-DHMRs (blue) identified in (B) (right). The y axes report the percentages of the DMRs or DHMRs relative to all DMRs or DHMRs, respectively. LINE, long interspersed nuclear element; SINE, short interspersed nuclear elements; LTR, long terminal repeat. (F) Multidimensional scaling (MDS) analysis of the 5hmC signals in the shared DHMRs identified in (B). (G) University of California, Santa Cruz (UCSC) genome browser view of 5hmC enrichment at the PRDM6 locus (chr5: 122,433,516 to 122,435,744) in cerebellum, MB tumor, and MB CSF ctDNA. The highlighted region exhibits increased 5hmC in both MB tumor and MB CSF samples.
To evaluate the function of the shared DMRs and DHMRs, we performed motif analysis and identified several neuronal function-associated transcription factor (TF) motifs that were enriched within DMRs and DHMRs, including Oligo2 (33, 38), Atoh1, and NeuroD1 (Fig. 2, A and B, bottom). Analysis using Genomic Regions Enrichment of Annotations Tool further revealed that these common DMRs and DHMRs were mainly enriched at genomic regions that are associated with genes important for cerebellar and CNS functions with particular enrichment in the MB tumor origin Purkinje cell–granule cell precursor cell signaling genes (fig. S3, C and D) (30, 31). Functional genome analysis further unveiled that, compared to hypo-DMRs, hyper-DMRs tend to occur more often at genic regions and distal regulatory regions marked by H3K27ac (Fig. 2E, left). In addition, the DHMRs displayed an enrichment pattern opposite that of the DMRs (Fig. 2E, right). For example, hypo-DHMRs were enriched to a greater degree than were hyper-DHMRs in genic regions (Fig. 2E). These data are consistent with the previously observed lower level of DNA methylation in repetitive regions and higher level of DNA methylation at genic regions in tumor cells.
It has been reported that DNA demethylation is closely associated with MB pathogenesis (12). 5hmC constitutes one of the most important intermediates during active DNA demethylation. We therefore performed multidimensional scaling (MDS) analysis using DHMRs between the normal cerebellum and MB tumors. Normal cerebellum tissues were clearly separated from MB tumor and MB CSF, whereas MB tumor and MB CSF samples were paired by person (Fig. 2F). Note that the tumor and CSF samples for patient 1 were closer to normal cerebellum tissue than to samples of other patients. We also noticed a relatively lower Pearson correlation coefficient of 5hmC analysis between CSF and tumor tissue in patient 1 compared with the two coefficients of other analyzed patients (Figs. 1B and 2B). It might be due to the clinical stage differences between these patients (table S2). However, our overall result indicates that 5hmC signatures obtained from CSF ctDNA can be used to detect MB tumor existence. For example, we observed a marked increase in 5hmC at the PRDM6 locus in both MB tumor and MB CSF compared with normal cerebellum (Fig. 2G). At this location, enhancer hijacking–driven activation of PRDM6 in MB has been reported previously (10). In summary, these analyses show that the identified DMRs and DHMRs are potentially associated with CNS function and that the dysregulation of DNA methylation and hydroxymethylation pathways might be culprits contribute to MB pathogenesis.
Next, we compared DNA methylation and hydroxymethylation differences within the shared DHMRs and DMRs, respectively. For brevity, we define characteristic DMRs (or DHMRs) as the DMRs (or DHMRs), common between normal cerebellum and MB tumor and between normal cerebellum and MB CSF. As sodium bisulfite–based DNA methylation analysis was unable to discriminate between 5hmC and 5mC (32), the identified DMRs might contain the changes of 5hmC. Moreover, since 5hmC is the catalytic product of 5mC, the identified DHMRs might also contain information regarding 5mC alterations. Therefore, we compared the DNA methylation within DHMRs and the genomic distribution of DMRs and DHMRs. We observed that more than half of the characteristic DHMRs (n = 16,017, 51.1% of all CpGs in hyper-DHMRs; n = 567, 56% of all CpGs in hypo-DHMRs) displayed less than 20% difference in DNA methylation ratios in both tumor and CSF compared to normal (fig. S4, A and B). This observation indicates that although the 5hmC signals in characteristic DHMRs are significantly different, more than half of the characteristic DHMRs are not characteristic DMRs, as they do not exhibit large WGBS signal differences in the tumor and CSF simultaneously. Detailed analysis of the WGBS data showed that the DNA methylation ratios of the CpGs in characteristic DHMRs were predominantly near 0.6 to 1.0 in the tumor tissue and CSF (fig. S4, C and D, dashed line). Only a small fraction of characteristic CpGs for hydroxymethylation, specifically, 27% for shared hyper-DHMRs (6.8% + 20.2%) and 10.1% for shared hypo-DHMRs (5.5% + 4.6%), displayed strong (>20%) DNA methylation changes in both tumors and CSF (fig. S4, A to D, solid line). Similarly, very few characteristic DMRs remained as characteristic DHMRs (figs. S4, E and F, and S5, A and B), mostly due to the sparsity of 5hmC. These findings suggest that characteristic DMRs and characteristic DHMRs mark both common and specific genomic regions and are complementary to each other. Both DNA methylation and hydroxymethylation analyses are required to yield a comprehensive picture of DNA modification dynamics in MB tumor and MB CSF.
Identifying the DNA methylation signatures of MB in CSF ctDNA
There were 40,056 hyper–differentially methylated cytosine (DMC) and 20,498 hypo-DMCs in the DMRs between the normal cerebellum and MB tumor. Among these 60,554 DMCs, 6598 CpG sites were differentially methylated in MB CSF in the same direction of change as in the MB tumor, that is, they paralleled each other, and thus formed shared DMRs between MB CSF and normal cerebellum and between MB tumor and normal cerebellum. We termed these 6598 sites as “MB CSF signature CpGs” (4253 hypermethylated and 2345 hypomethylated CpGs) in the context of DNA methylation of MB CSF (Fig. 3A). Since these three patients are SHH subtypes, MB CSF signature CpG defined that this way is an abbreviation for MB CSF (SHH) signature CpG. These CpGs are mostly scattered in the genome. When we merged adjacent CpGs if they are separated in less than 300 bp, these signature CpGs formed 705 regions with 2.35 CpGs per region, on average, and 4943 single CpGs. This result indicates that even under very strict criteria for MB CSF signature CpG selection, we observe that some signature CpGs share the same methylation dynamics while they are located in the same genomic locations. The functional genome annotation analysis showed that most of the 6598 CpGs were located in gene-rich regions. Around 50% of hypermethylated CpGs were located in MB tumor–specific H3K27ac-enriched regions (fig. S6A). Consistent with the analysis shown in Fig, 2A, the motifs of TFs that are important for neural function were enriched approximal within 100 bp of these CpGs [NeuroD1 (P = 1 × 10−28), Olig2 (P = 1 × 10−21), Atoh1 (P = 1 × 10−13), Oct6 (P = 1 × 10−5), and Pax6 (P = 1 × 10−5)]. These data suggested that abnormal DNA methylation is frequently observed in H3K27ac-marked enhancers that are approximal to potential neuron-specific TF binding sites in MB. Next, we measured the DNA methylation of ctDNA purified from nontumor individuals (patients with hydrocephalus) as controls to further validate our results. Since the ctDNA is not detected in the CSF of healthy individuals, we then used the CSF samples from patients with hydrocephalus as surrogate nontumor controls in our study. Among these 6598 MB CSF signature CpGs, by requiring minimum sequencing depth 5, we detected 2027 shared CpGs among all the analyzed samples, including two normal cerebellum tissues, four nontumor CSF samples, three MB tumor tissue samples, and three MB CSF samples (fig. S6B). We next performed MDS analysis using the DNA methylation status of these 2027 common DMCs. We observed distinct separation between normal cerebellum, nontumor CSF, and MB samples (tumor tissue and CSF), whereas we clustered the MB tumor and MB CSF together (Fig. 3B). To further confirm this data, we analyzed the DNA methylation levels of these 2027 common DMCs by integrating with published WGBS datasets in MB tumors. We observed that the DNA methylation level in these CpGs could not only distinguish between MB tumor (CSF and tumor) and nontumor (tissue and CSF) samples but also be able to separate the subtypes of MB tumor (fig. S6C). For example, the DNA methylation levels at NEUROD1, STARD13, and NCOR2 loci were significantly increased in MB tumor tissue and CSF samples compared with the levels in nontumor samples, including cerebellum and CSF samples from patients with hydrocephalus (Fig. 3C and fig. S6D). These data strongly suggest that the signature CpGs and their DNA methylation in MB CSF consistently reflect MB tumor signatures and can be used as a potential CSF biomarker to indicate the presence of MB.
Fig. 3. MB DNA methylation signatures in CSF ctDNA for cancer diagnosis.
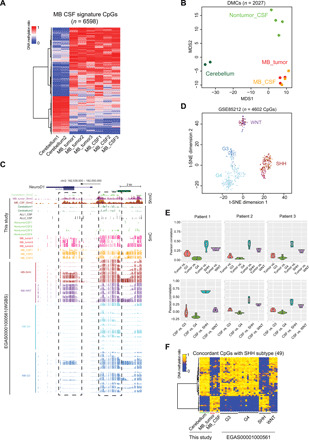
(A) Heatmap representation of DNA methylation levels at MB CSF signature CpGs (n = 6598). (B) MDS analysis of the DNA methylation levels at MB CSF signature CpGs detected in cerebellum (dark green), MB tumor (red), MB CSF (orange), and nontumor CSF (light green). (C) UCSC genome browser view of DNA methylation and hydroxymethylation levels at the NEUROD1 locus (chr2: 182,539,000 to 182,550,000) in the indicated samples. (D) t-distributed stochastic neighbor embedding (t-SNE) analysis of the DNA methylation levels of the common CpG sites between MB CSF signature CpGs and the published DNA methylation array data from approximately 600 patients with MB (8). WNT, purple; SHH, maroon; group 3 (G3), dark blue; group 4 (G4), light blue. (E) Pearson correlation analysis of DNA methylation status of the common CpGs between MB CSF signature CpGs and subtype-specific CpGs identified from public data (20) (n = 1047 CpGs). Top: The methylation status in MB tumor samples. Bottom: the methylation status in MB CSF samples. (F) Heatmap representation of the selected 49 subtype-specific CpGs (of 1047 CpG sites), which exhibited concordant DNA methylation status between the three SHH-subtype MB CSF and MB tumor samples (this study) and public MB tumors data.
To further test whether the signature CpGs could be used to characterize MB subtype, we analyzed the DNA methylation status of the MB CSF signature CpG sites using two public datasets. First, we studied the MB CSF signature CpG sites in the published Illumina HumanMethylation450 BeadChip data, which was obtained from approximately 600 MB patient tumor samples (8). Of the 6598 MB CSF signature CpGs, 4602 CpGs were covered by the array data. The DNA methylation status at these 4602 CpGs clearly identified the MB subtype of the 600 analyzed tumor samples (Fig. 3D). Second, we studied the MB CSF signature CpG sites in published WGBS data collected from pediatric patients with MB (12). As shown in fig. S6E, the four different MB subtypes displayed distinct DNA methylation patterns at the MB CSF signature CpG sites. To further obtain comprehensive MB subtype–specific DNA methylation signatures, we compared our results (Fig. 3A) with subtype-specific CpGs identified from published WGBS datasets (12) and identified 1047 MB subtype–specific CpG sites. By analyzing the DNA methylation levels within these sites in tumor and/or CSF samples collected from four patients with MB, we further confirmed that the DNA methylation status at these 1047 CpG sites could clearly reflect the tumor subtypes (Fig. 3E and fig. S6E). Next, we selected 49 CpGs (of the 1047 CpGs) displaying the most notable subtype-specific DNA methylation signatures in the SHH subtype. As shown in Fig. 3F, the DNA methylation levels at these 49 CpGs were consistent among the MB tumors and CSF in our own study, as well as with the published data from SHH MB tumors (12), but were markedly different from normal cerebellum and other MB tumor subtypes. Our results from the analysis with two large independent datasets converged to indicate that the DNA methylation patterns at MB CSF signature CpG sites can both reflect the presence of an MB tumor and be used to facilitate the identification of MB subtype, making it possible to determine the MB subtype from CSF collection at initial diagnosis and to further guide the treatment.
Using the DNA methylation signatures of MB in CSF ctDNA to probe the disease status
To further test whether the DNA methylation levels at the MB CSF signature CpG sites could be used to reflect the treatment response and tumor recurrence, we analyzed DNA methylation in CSF collected from two patients (2 and 3) at sequential time points (Fig. 4, A and B). For patient 2, CSF was collected at diagnosis, 10 months (during month 7 of treatment; Fig. 4A, top). We observed dynamic DNA methylation changes at the MB CSF signature CpG sites in CSF collected at different disease statuses (at diagnosis, during treatment, and before recurrence; Fig. 4A, bottom). Among 1106 of 6598 MB CSF signature CpGs, a total of 91 CpG sites (cluster 3) regained DNA methylation during treatment, suggesting that these CpGs can be used to assess treatment responses (Fig. 4A, bottom). We also observed that 88 CpG sites (cluster 4) and 227 CpG sites (clusters 5 and 10) showed a continuous increase and reduction in DNA methylation in the same patient regardless of treatment, suggesting treatment-independent epigenetic patterns associated with MB (Fig. 4A, bottom). For patient 3, CSF was collected at diagnosis, 6 months (start of treatment; Fig. 4B, top). Dynamic DNA methylation changes were also observed at the MB CSF signature CpG sites in CSF collected at different time points. Among 1336 of 6598 MB CSF signature CpGs, a total of 77 CpGs in cluster 6 showed an increase in DNA methylation levels after treatment, which indicates that these CpGs might be related to good treatment responses (Fig. 4B, bottom right). A total of 85 CpGs (cluster 14) exhibited decreased DNA methylation level (similar to normal cerebellum tissue) immediately after treatment. A total of 407 CpGs in clusters 4, 9, 10, and 11 maintained high DNA methylation level in all collected samples, which indicates that these CpGs might represent the malignant progress of tumors. We found that 61 overlapping CpGs, identified by comparing the 203 CpGs in clusters 2 and 5 in patient 3 and the 227 CpGs in clusters 5 and 10 in patient 2, displayed a continuous decrease in DNA methylation regardless of treatment (Fig. 4C). Ten CpGs were found to be overlapping between cluster 4 in patient 2 and clusters 4, 9, 10, and 11 in patient 3, which exhibit high DNA methylation levels in the MB group (tumors and CSFs; 0.8 to 1.0; Fig. 4, A and B) compared with that in normal cerebellums (<0.2). Considering that patients 2 and 3 received similar treatments within a comparable duration of time (table S2), these overlapping CpGs can be used as a confident CpG index to reflect the tumor status.
Fig. 4. Dynamic DNA methylation changes of MB CSF signature CpGs in CSF ctDNA along treatment.
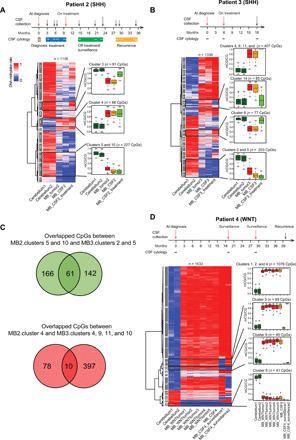
(A) Top: The timeline for serial CSF collections and CSF cytology results in patient 2. Arrows indicate the time points of CSF sample collection (red arrows, CSF samples used in this study). Bottom: Heatmap representation of DNA methylation status of MB CSF (SHH) signature CpGs in normal cerebellum tissues, MB tumor tissue, and CSF at diagnosis, during treatment. (B) Top: The timeline for serial CSF collections and CSF cytology results in patient 3. Bottom: Heatmap representation of DNA methylation levels of MB CSF (SHH) signature CpGs in normal cerebellum tissues, MB tumor tissue, and CSF at diagnosis and during treatment. (C) The Venn diagram representation of overlapped CpGs between clusters 5 and 10 (patient 2) and clusters 2 and 5 (patient 3) and between cluster 4 (patient 2) and clusters 4, 9, 10, and 11 (patient 3). (D) Top: The timeline for serial CSF collections and CSF cytology results in patient 4. Bottom: Heatmap representation of DNA methylation status of MB CSF (WNT) signature CpGs in normal cerebellum tissues, MB tumor tissue from patient 4, published MB (WNT) tumors, and CSF from patient 4 at diagnosis and after treatment.
In parallel, we also analyzed the DNA methylation in ctDNA purified from serial CSF samples obtained from patient 4 (WNT subtype). Consistent with our analysis, we observed a significant difference in DNA methylation between SHH and WNT tumor subtypes (fig. S7A). Since there is no matched MB tumor sample from patient 4, we compared public WGBS data from five WNT-subtype MB tumors (20) with the WGBS data from the CSF sample of patient 4 (collected at diagnosis; fig. S7B). Using the similar method for identification of MB CSF (SHH) signature CpGs, we identified 9373 MB CSF (WNT) signature CpGs that showed distinct DNA methylation levels in both WNT MB tumor and CSF compared with normal cerebellum (fig. S7C). A total of 146 CpGs were found to overlap between MB CSF (SHH) and MB CSF (WNT) signature CpGs, and the majority displayed higher DNA methylation compared with normal cerebellum (table S4 and fig. S7, D and E). We also measured the DNA methylation in ctDNA purified from serial CSF samples in patient 4 (at diagnosis, 19 months and 29 months after diagnosis without recurrence; Fig. 4D, top). Within MB CSF (WNT) signature CpGs (n = 9373), we observed 1632 CpGs exhibiting dynamic DNA methylation changes along the treatment (Fig. 4D, bottom). Specifically, DNA methylation levels of CpGs in cluster 9 dropped from 85 to 10% after treatment (both at 19th and 29th month after initial diagnosis), which was comparable to the level in normal cerebellum. For CpGs in cluster 3, their DNA methylation showed dynamic changes at 19th and 29th month (from 10 to 95%). For CpGs in clusters 1, 2, and 4, these CpGs exhibited high DNA methylation levels (>80%) in tumors, CSFs, and two surveillance samples, whereas CpGs in cluster 5 had a low DNA methylation level (<20%) (Fig. 4D, bottom). Notably, the cytology analysis results of most CSF samples in this study remained negative. However, we were able to detect and analyze DNA methylation in CSF ctDNA from these patients. Furthermore, we detected strong subtype-specific DNA methylation signatures at different disease stages in patients with MB, suggesting a high sensitivity and reliability of this method. These data clearly establish that the DNA methylation status of MB CSF signature CpG sites can be exploited to identify MB subtype and monitor disease progression (e.g., treatment response and recurrence).
Potential prognostic DNA methylation markers in CSF ctDNA for pediatric MB
To further test whether the DNA methylation of MB signature CpGs in CSF could be used as potential prognostic markers, we performed a univariate Cox proportional hazard analysis for the MB CSF (SHH) signature CpGs (Fig. 4A) and the information about overall survival (OS) from a previous study (8). We identified 224 probes that were significantly associated (P < 0.001) with the OS of patients with MB (table S3). For example, as shown in Fig. 5A, one CpG site (cg14582550; chr9: 97,786,878 to 97,786,879) located within the intron of C9orf3 displayed a marked increase in DNA methylation in MB samples (both tumor and CSF ctDNA) in our study, which was concordant with earlier reports (8, 12, 33) that used Illumina HumanMethylation450 BeadChip (Fig. 5, B and C). By associating the DNA methylation status at this CpG site with patient survival information, we found that patients with a high DNA methylation level (>0.8) at this CpG site showed significantly (P < 0.0001) lower survival rates compared with patients with a low DNA methylation level (≤0.8) at this site (Fig. 5D). We further examined the DNA methylation level of seven probes covering the gene body of C9orf3 (three probes up- and downstream of probe cg14582550, respectively) and observed consistent survival rates as using the single probe cg14582550 (fig. S7F). Next, we randomly selected 438 and 189 patients as training and validation data, respectively (8). By applying multivariate Cox regression analysis using MB signature CpGs on the training data, we selected the best linear model with the smallest root mean square error in the training set from models comprising all possible combinations of two to five CpG sites using a stepwise regression method. The best linear model is Y (survival score) = 2.343 + mCG/CG ratio of CpG#1 (cg27490391) * 3.528 + mCG/CG ratio of CpG#2 (cg27579805) * 1.598 + mCG/CG ratio of CpG#3(cg27638288) * (−0.771). The probe cg27490391 located on the gene body of LHFP, probe cg27579805 located at the 3′ untranslated region of TNRC6C, and probe cg27638288 located in nongenetic region. These three CpG sites displayed significantly increased DNA methylation in MB tumors compared with normal cerebellum (fig. S7G). We then calculated survival scores using this formula for each patient in the training dataset (n = 438). The patients could be clearly separated into low- and high-risk groups based on the medium survival score. The patients with high-survival scores had a significantly better outcomes than those with low-survival scores in both training and validation datasets (Fig. 5, E and F). In summary, our data clearly demonstrate that DNA methylation status at MB CSF signature CpG sites can be used as potential prognostic markers to predict the clinical outcomes of patients with MB.
Fig. 5. Exploring the prognostic value of MB DNA methylation signatures in CSF ctDNA for clinical outcome.
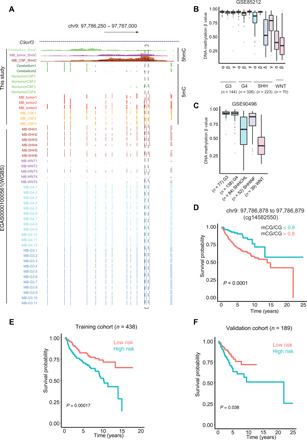
(A) UCSC genome browser view showing DNA methylation and hydroxymethylation levels of CpGs (chr9: 97,786,878 to 97,786,879) located within the intron of C9orf3 for normal cerebellum, nontumor CSF, MB CSF ctDNA, and MB tumors, including data from this study and 34 public WGBS datasets (12). (B and C) Box plots representing the DNA methylation levels at the single CpG site highlighted in Fig. 4A, using previously published DNA methylation array data [B (8) and C (41)]. (D) Kaplan-Meier survival curves of patients with MB separated according to a methylation ratio cutoff value of 80% at the single CpG site highlighted in Fig. 4A. (E and F) Kalan-Meier curves and log-rank tests were used to visualize and compare the OS between low-risk and high-risk groups in the training cohort (n = 438 patients) (E) and the validation cohort (n = 189) (F) using the methylation ratios at the three CpGs in the model.
DISCUSSION
The detection of ctDNA from the plasma of patients with cancer was reported previously; however, it is not suitable for patients with brain tumor because the ctDNA from the brain tumor might be blocked by blood-brain barrier (34). CSF is in constant and intimate contact with brain malignancies and has been reported to contain ctDNA (35). CSF might therefore serve as a better source than plasma for obtaining ctDNA for real-time monitoring of disease progression and response to treatment (23). Until now, most CSF ctDNA analyses have been geared toward the detection of cancer-associated mutations (22–24, 36). However, pediatric brain tumors have much lower frequencies of mutations compared to adult brain tumors (6), making the detection of tumor DNA via mutational analysis difficult. In contrast, alterations in epigenetic landscapes have been frequently observed in pediatric brain tumors (9, 12, 33, 37), making epigenetic markers a more ideal readout for diagnostic and prognostic purposes. We reason that epigenetic signatures inherent to the brain tumors in situ will be preserved in CSF ctDNA. Therefore, measuring the epigenetic signatures from CSF ctDNA could be a sensitive and accurate approach to monitoring brain tumor treatment response, progression, and relapse.
In this study, we profiled epigenetic markers, including DNA methylation and hydroxymethylation, in CSF ctDNA genome widely from pediatric patients with MB. We have shown that these two epigenetic markers can be detected in ctDNA extracted from MB CSF with an input volume as low as 200 μl. Since our CSF samples were immediately subjected to hard spins after collection, it is less likely that the purified DNA in CSF is from genomic DNA (gDNA) in circulating tumor or nontumor cells in CSF. Note that the DNA purified from MB tumor tissue and CSF displayed similar epigenetic signatures in the same patients, suggesting that DNA purified from CSF in patients with MB according to our protocols is mainly derived from MB tumor. However, during the analysis, we encountered low genomic coverage or relatively low bisulfite conversion efficiency in some samples. We only recovered 16 M CpGs in the MB CSF sample collected from patient 3 after treatment, although we sequenced ~1.5 billion total reads in this sample. During the analysis, there are only 116 million uniquely mapped reads after duplication removal, suggesting that most reads are from the same genomic regions and the original ctDNA might have relatively low genomic coverage. Both samples were collected from patients after chemotherapy treatment (table S2). It is possible that the chemotherapy might introduce DNA damage and perturb bisulfite conversion. Further analysis is needed to clarify this point. We have excluded these samples in this analysis due to the low data quality. On the basis of these analyses, not all the CSF samples are suitable for using 200 μl of CSF for DNA methylation and hydroxymethylation analysis. With samples that have low genomic coverage and possible DNA damage, it is most ideal to increase the input volume of CSF samples to avoid potential analysis bias. Regardless of these caveats, the ctDNA methylation and hydroxymethylation patterns match those of MB tumor tissues from the same patient, suggesting that CSF ctDNA can be used as material to monitor MB progression.
One point that needs to be noted is that the changes of DNA methylation level have been reported during aging (38). The MB samples used in this study had an age of <18 years old, and the nontumor CSF samples were from individuals with ages ranging from 32 to 66 years old (table S2). To test whether the DNA methylation changes within the MB signature CpGs were due to the age difference in Fig. 3B and fig. S6C, we monitored the DNA methylation level within these CpGs using published WGBS data from normal human brain tissues of ages ranging from 35 days to 55 years old (38). No differential DNA methylation was observed within these MB signature CpGs at different ages (fig. S7H), which rules out the possible influence of aging-associated DNA methylation changes at these CpG sites.
One limitation in our study is that the sample size is relatively small. In our analysis, we observed large intertumor heterogeneity based on DNA methylation analysis (Fig. 2F), suggesting that large sample size is preferred to obtain more reliable DNA methylation signatures. Nonetheless, even with the limited sample size, we were able to identify DNA methylation signature CpGs in CSF. We further validated our results using published datasets collected from more than 600 patients with MB, suggesting that our discovery could be widely applied to identify and classify patients with MB. In addition, the DNA methylation status at subsets of MB signature CpGs in CSF samples collected at different time points from the same patient reflected the tumor status during treatment and recurrence even in cytogenic negative samples, suggesting that our approach has superior sensitivity and high specificity. DNA methylation and hydroxymethylation at these CpG sites could be used as prognostic markers to stratify patients with MB into low- and high-risk groups. We did observe some variations in epigenetic markers in CSF ctDNA among patients with MB, which might be due to differences in sample collection times (e.g., before or after surgery and treatment) or the fact that the patients had MB belonging to different subgroups (22) or different stages. Further studies with a larger MB sample size could be used to refine the results from this study.
Overall, DNA methylation and hydroxymethylation signatures in CSF ctDNA can serve as valuable epigenetic markers to guide the clinical management of patients with MB. The epigenetic features detected in CSF ctDNA can be exploited for previously unidentified biomarker and prognostic marker development. Our method using extremely low input DNA to accurately profile DNA methylation and DNA hydroxymethylation genome widely in ctDNA represents a proof of concept for its use in other tumors beyond MB.
MATERIALS AND METHODS
Patient tumor tissue collection
Signed informed consent was obtained from the patients or their legal guardians before sample acquisition in accordance with an institutional review board–approved protocol. Freshly resected MB tumor specimens from three patients undergoing surgery at Texas Children’s Hospital were obtained for this study. Multiple CSF samples were collected during clinically indicated LPs. All samples were subjected to pathological diagnosis and were graded according to the World Health Organization system. Tumor tissues were snap-frozen in liquid nitrogen and preserved in a −80°C freezer. CSF was processed using a standardized protocol, then divided into aliquots, and stored immediately in a −80°C freezer.
MB tumor gDNA and CSF ctDNA purification
Freshly collected CSF samples were centrifuged at 1000g for 10 min before frozen in −80°C as described previously (36, 39). A total of 200 to 400 μl of CSF recovered from freezer was either directly subjected to centrifugation at 1000g at 4°C for 10 min before ctDNA purification or directly subjected to DNA purification without centrifugation. Cell-free DNA was purified from 200 to 400 μl of CSF using the QIAamp Circulating Nucleic Acid Kit (QIAGEN) according to the manufacturer’s instructions. For MB tumor tissues, as much as 20 mg of tumor tissue was snap-frozen with liquid nitrogen and ground. DNA was then isolated using the AllPrep DNA/RNA Mini Kit (QIAGEN) according to the manufacturer’s protocol. The concentration of the isolated DNA was measured by a Qubit 4 Fluorometer with the Qubit dsDNA (double-stranded DNA) High-Sensitivity Assay Kit (Thermo Fisher Scientific).
WGBS library preparation
WGBS analysis was used to measure the genome-wide DNA methylation profile. The ctDNA and tissue WGBS libraries were generated using the Pico Methyl-Seq Library Prep Kit (Zymo Research). Briefly, ctDNA was mixed with 0.1% unmethylated λ-bacteriophage DNA (w/w) (New England Biolabs), followed by sodium bisulfite conversion. The bisulfite-converted DNA was then annealed with random primers for initial amplification, followed by adaptor ligation and final amplification with Illumina TruSeq indices. Constructed libraries were run on a 2% agarose gel to assess size distribution, and the library concentration was measured by a Qubit 4 Fluorometer with the Qubit dsDNA High-Sensitivity Assay Kit. Normalized libraries were then pooled at an equimolar ratio and sequenced on a NextSeq 500 (Illumina) with a NextSeq 500/550 High Output Kit v2.5 (single-end reads, 75 cycles) according to the manufacturer’s protocols.
CMS-IP–seq library preparation
CMS-IP–seq was performed as described previously with some modifications. Briefly, purified MB tumor gDNA was sonicated to the 200- to 400-bp size range using an M220 Focused-ultrasonicator (Covaris). The CSF ctDNA was directly treated with sodium bisulfite without sonication. The sheared gDNA and purified ctDNA were treated with sodium bisulfite using the EZ DNA Methylation-Lightning Kit (Zymo Research) with the manufacturer’s protocols to convert 5hmC to CMS. Next, CMS-containing DNA fragments were immunoprecipitated using a CMS-specific antiserum conjugated to Protein A/G Dynabeads. Enriched DNA fragments were purified using conventional phenol/chloroform/isomyl-alcohol extraction and then amplified with random primers, followed by adaptor ligation and final amplification with Illumina TruSeq index using the Pico Methyl-Seq Library Prep Kit (Zymo Research). DNA library sizes and concentrations were verified by Bioanalyzer. The DNA libraries were then sequenced in NextSeq 500 (Illumina) with the NextSeq 500/550 High Output Kit v2.5 (single-end reads, 75 cycles) following the manufacturer’s protocols.
WGBS and CMS-IP–seq data analysis
For WGBS analysis, raw FASTQ files were mapped to the National Center for Biotechnology Information Human Reference Genome Build GRCh37 (hg19) using BSMAP (40) and LiBis (28). LiBis used a dynamic clipping method to rescue the unmapped reads. For each unmapped read, LiBis generated 40-bp (user defined) segments with a 5-bp (user defined) space (see workflow figure in https://github.com/Dangertrip/LiBis). One unmapped read became several 40-bp small fragments. Then, these small 40-bp fragments were remapped, and only the uniquely mapped reads were kept. Next, LiBis extended these uniquely mapped small fragments based on their mapping locations in the genome. The final step was to filter out the shorter extended fragments (<50 bp). The longer extended fragments (>50 bp) were counted as the highly confident rescued reads. The polymerase chain reaction duplicates were removed using Picard MarkDuplicates (http://broadinstitute.github.io/picard/). The MCALL module in MOABS was used to calculate the DNA methylation ratio and coverage for each CpG site. Differentially methylated CpGs and regions were identified using the MCOMP module considering variance among samples (--withVariance 1) in the MOABS software. Bisulfite conversion efficiencies were estimated using spike-in unmethylated λ-bacteriophage DNA. The output bedGraph files from MCALL include single-base resolution DNA methylation ratios, which were transformed to a bigWig file format. The bigWig files were uploaded to the University of California, Santa Cruz (UCSC) genome browser for visualization.
For CMS-IP–seq analysis, we used the CMS-IP software to detect DHMRs between normal cerebellum samples and MB tumor or CSF samples (https://github.com/lijinbio/cmsip). Raw CMS-IP–seq reads were aligned to hg19 using BSMAP (40). Uniquely mapped reads were retained for downstream analysis. Size factor estimation normalized the total WIG file sums of sample replicates. A CMS count table was tabulated for the 5hmC-enriched regions detected by MACS2. Adjusting for the estimated size factors, the normalized CMS count table was tested by the G test of goodness of fit. The G test examines whether the sums of counts fit the proportion of the numbers of replicates between two samples. CMS-IP calls the G.test() function in the R package RVAideMemoire. CMS-IP computes false discovery rate (FDR)–adjusted q values of the P values in G test using the Benjamini and Hochberg procedure. To reduce the loss of statistical power caused by the FDR adjustment, independent filtering is applied to rule out low-count regions, where the filtering criterion measures the average normalized CMS counts across sample replicates. DHMRs were identified by a q < 0.05. To facilitate the visualization of 5hmC signals, bigWig files for read coverage were generated from the aligned BAM files and visualized in the UCSC genome browser.
The smoothed scatterplots (R package: geneplotter) used the common CpGs or common 5hmC-enriched regions between two samples as input. Pearson correlations coefficients were calculated using the R cor function. The R package ggplot2 was used to plot violin plots and box plots. Heatmaps were plotted using the R package heatmap3 (www.rdocumentation.org/packages/heatmap3/versions/1.1.6/topics/heatmap3) by taking the CpGs shared among all samples as input. t-distributed stochastic neighbor embedding (t-SNE) analysis was performed using the R package Rtsne (https://github.com/jkrijthe/Rtsne).
Identification of prognostic CpG markers in CSF ctDNA for pediatric MB
We downloaded the DNA methylation 450k array data and clinical data from dataset GSE65362 (>600 patients with MB). We randomly selected 438 and 189 patients as the training dataset and the validation dataset, respectively. OS was defined as the time from the patient diagnosis to MB tumor–related death or last follow-up. To reduce noise, we first performed univariate Cox proportional hazard analysis in the training dataset to identify the CpGs significantly associated (P < 0.001) with OS. Among these CpGs, we only used those CpGs that were identified as candidate markers for prognostic evaluation both in DMC comparisons between normal cerebellum and MB tumors and DMC comparisons between normal cerebellum and MB CSF ctDNA. These candidate markers were then used in multivariate Cox regression analysis to construct linear models comprising all possible combinations of two to five markers. The best linear model was Y = 2.343 + mCG/CG ratio of cg27490391 * 3.528 + mCG/CG ratio of cg27579805 * 1.598 + mCG/CG ratio of cg27638288 * (−0.771). We use the above formula to estimate a survival score for each patient. We then separated patients into two groups, survival good and survival bad, based on the median values of the survival scores. Next, the R package survival was used to draw Kaplan-Meier survival curves. The log rank test was used to assess the difference in OS between the two groups.
Supplementary Material
Acknowledgments
Funding: We are grateful to J. Shen, the MD Anderson Cancer Center Next-Generation Sequencing Core at Smithville (CPRIT RP120348 and RP170002), and the Texas A&M Institute of Biosciences and Technology Epigenetics Core. This work was supported by grants from the Cancer Prevention and Research Institute of Texas (RR140053 to Y.H., RP170660 to Y.Z., and RP180131 to D.S.), the NIH (R01HL134780 and R01HL146852 to Y.H. and R01GM112003 to Y.Z.), the Welch Foundation (BE-1913-20190330 to Y.Z.), the American Cancer Society (RSG-18-043-01-LIB to Y.H. and RSG-16-215-01-TBE to Y.Z.), and by start-up funds from the Texas A&M University (to Y.H. and D.S.). Author contributions: X.-N.L., Y.H., and D.S. directed and oversaw the project. Jia Li and D.S. performed comprehensive bioinformatics analyses, including data quality control, publicly available data collection, integration analysis, and identification of the CpGs potentially associated with patient outcome. M.L. optimized CSF ctDNA sequencing library preparation and performed high-throughput sequencing. S.Z. and X.-N.L. collected and identified CSF and tumor samples. L.Y.B., Y.E., R.H.D., P.J.A.D., and D.W.P. provided intellectual input. Y.H. and Y.Z. wrote the manuscript, and all authors participated in discussion, data interpretation, and manuscript editing. Competing interests: J.L., S.B.Z., Y.H., X.-N.L., and D.S. are co-inventors on a pending patent application related to this work filed by Texas A&M University. The other authors declare no other competing interests. Data and materials availability: Raw and processed data are available at the Gene Expression Omnibus database under accession number GSE142241. All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/42/eabb5427/DC1
REFERENCES AND NOTES
- 1.Massimino M., Biassoni V., Gandola L., Garrè M. L., Gatta G., Giangaspero F., Poggi G., Rutkowski S., Childhood medulloblastoma. Crit. Rev. Oncol. Hematol. 105, 35–51 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Batora N. V., Sturm D., Jones D. T. W., Kool M., Pfister S. M., Northcott P. A., Transitioning from genotypes to epigenotypes: Why the time has come for medulloblastoma epigenomics. Neuroscience 264, 171–185 (2014). [DOI] [PubMed] [Google Scholar]
- 3.Dubuc A. M., Remke M., Korshunov A., Northcott P. A., Zhan S. H., Mendez-Lago M., Kool M., Jones D. T. W., Unterberger A., Morrissy A. S., Shih D., Peacock J., Ramaswamy V., Rolider A., Wang X., Witt H., Hielscher T., Hawkins C., Vibhakar R., Croul S., Rutka J. T., Weiss W. A., Jones S. J. M., Eberhart C. G., Marra M. A., Pfister S. M., Taylor M. D., Aberrant patterns of H3K4 and H3K27 histone lysine methylation occur across subgroups in medulloblastoma. Acta Neuropathol. 125, 373–384 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jones D. T. W., Northcott P. A., Kool M., Pfister S. M., The role of chromatin remodeling in medulloblastoma. Brain Pathol. 23, 193–199 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Remke M., Ramaswamy V., Taylor M. D., Medulloblastoma molecular dissection: The way toward targeted therapy. Curr. Opin. Oncol. 25, 674–681 (2013). [DOI] [PubMed] [Google Scholar]
- 6.Parsons D. W., Li M., Zhang X., Jones S., Leary R. J., Lin J. C.-H., Boca S. M., Carter H., Samayoa J., Bettegowda C., Gallia G. L., Jallo G. I., Binder Z. A., Nikolsky Y., Hartigan J., Smith D. R., Gerhard D. S., Fults D. W., Berg S. V., Berger M. S., Marie S. K. N., Shinjo S. M. O., Clara C., Phillips P. C., Minturn J. E., Biegel J. A., Judkins A. R., Resnick A. C., Storm P. B., Curran T., He Y., Rasheed B. A., Friedman H. S., Keir S. T., Lendon R. M., Northcott P. A., Taylor M. D., Burger P. C., Riggins G. J., Karchin R., Parmigiani G., Bigner D. D., Yan H., Papadopoulos N., Vogelstein B., Kinzler K. W., Velculescu V. E., The genetic landscape of the childhood cancer medulloblastoma. Science 331, 435–439 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schwalbe E. C., Lindsey J. C., Nakjang S., Crosier S., Smith A. J., Hicks D., Rafiee G., Hill R. M., Iliasova A., Stone T., Pizer B., Michalski A., Joshi A., Wharton S. B., Jacques T. S., Bailey S., Williamson D., Clifford S. C., Novel molecular subgroups for clinical classification and outcome prediction in childhood medulloblastoma: A cohort study. Lancet Oncol. 18, 958–971 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cavalli F. M. G., Remke M., Rampasek L., Peacock J., Shih D. J. H., Luu B., Garzia L., Torchia J., Nor C., Morrissy A. S., Agnihotri S., Thompson Y. Y., Kuzan-Fischer C. M., Farooq H., Isaev K., Daniels C., Cho B.-K., Kim S.-K., Wang K.-C., Lee J. Y., Grajkowska W. A., Perek-Polnik M., Vasiljevic A., Faure-Conter C., Jouvet A., Giannini C., Rao A. A. N., Li K. K. W., Ng H.-K., Eberhart C. G., Pollack I. F., Hamilton R. L., Gillespie G. Y., Olson J. M., Leary S., Weiss W. A., Lach B., Chambless L. B., Thompson R. C., Cooper M. K., Vibhakar R., Hauser P., van Veelen M.-L. C., Kros J. M., French P. J., Ra Y. S., Kumabe T., López-Aguilar E., Zitterbart K., Sterba J., Finocchiaro G., Massimino M., Van Meir E. G., Osuka S., Shofuda T., Klekner A., Zollo M., Leonard J. R., Rubin J. B., Jabado N., Albrecht S., Mora J., Van Meter T. E., Jung S., Moore A. S., Hallahan A. R., Chan J. A., Tirapelli D. P. C., Carlotti C. G., Fouladi M., Pimentel J., Faria C. C., Saad A. G., Massimi L., Liau L. M., Wheeler H., Nakamura H., Elbabaa S. K., Perezpeña-Diazconti M., de León F. C. P., Robinson S., Zapotocky M., Lassaletta A., Huang A., Hawkins C. E., Tabori U., Bouffet E., Bartels U., Dirks P. B., Rutka J. T., Bader G. D., Reimand J., Goldenberg A., Ramaswamy V., Taylor M. D., Intertumoral heterogeneity within medulloblastoma subgroups. Cancer Cell 31, 737–754.e6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin C. Y., Erkek S., Tong Y., Yin L., Federation A. J., Zapatka M., Haldipur P., Kawauchi D., Risch T., Warnatz H.-J., Worst B. C., Ju B., Orr B. A., Zeid R., Polaski D. R., Segura-Wang M., Waszak S. M., Jones D. T. W., Kool M., Hovestadt V., Buchhalter I., Sieber L., Johann P., Chavez L., Gröschel S., Ryzhova M., Korshunov A., Chen W., Chizhikov V. V., Millen K. J., Amstislavskiy V., Lehrach H., Yaspo M.-L., Eils R., Lichter P., Korbel J. O., Pfister S. M., Bradner J. E., Northcott P. A., Active medulloblastoma enhancers reveal subgroup-specific cellular origins. Nature 530, 57–62 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Northcott P. A., Buchhalter I., Morrissy A. S., Hovestadt V., Weischenfeldt J., Ehrenberger T., Gröbner S., Segura-Wang M., Zichner T., Rudneva V. A., Warnatz H.-J., Sidiropoulos N., Phillips A. H., Schumacher S., Kleinheinz K., Waszak S. M., Erkek S., Jones D. T. W., Worst B. C., Kool M., Zapatka M., Jäger N., Chavez L., Hutter B., Bieg M., Paramasivam N., Heinold M., Gu Z., Ishaque N., Jäger-Schmidt C., Imbusch C. D., Jugold A., Hübschmann D., Risch T., Amstislavskiy V., Gonzalez F. G. R., Weber U. D., Wolf S., Robinson G. W., Zhou X., Wu G., Finkelstein D., Liu Y., Cavalli F. M. G., Luu B., Ramaswamy V., Wu X., Koster J., Ryzhova M., Cho Y.-J., Pomeroy S. L., Herold-Mende C., Schuhmann M., Ebinger M., Liau L. M., Mora J., McLendon R. E., Jabado N., Kumabe T., Chuah E., Ma Y., Moore R. A., Mungall A. J., Mungall K. L., Thiessen N., Tse K., Wong T., Jones S. J. M., Witt O., Milde T., Deimling A. V., Capper D., Korshunov A., Yaspo M.-L., Kriwacki R., Gajjar A., Zhang J., Beroukhim R., Fraenkel E., Korbel J. O., Brors B., Schlesner M., Eils R., Marra M. A., Pfister S. M., Taylor M. D., Lichter P., The whole-genome landscape of medulloblastoma subtypes. Nature 547, 311–317 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morrissy A. S., Garzia L., Shih D. J. H., Zuyderduyn S., Huang X., Skowron P., Remke M., Cavalli F. M. G., Ramaswamy V., Lindsay P. E., Jelveh S., Donovan L. K., Wang X., Luu B., Zayne K., Li Y., Mayoh C., Thiessen N., Mercier E., Mungall K. L., Ma Y., Tse K., Zeng T., Shumansky K., Roth A. J. L., Shah S., Farooq H., Kijima N., Holgado B. L., Lee J. J. Y., Matan-Lithwick S., Liu J., Mack S. C., Manno A., Michealraj K. A., Nor C., Peacock J., Qin L., Reimand J., Rolider A., Thompson Y. Y., Wu X., Pugh T., Ally A., Bilenky M., Butterfield Y. S. N., Carlsen R., Cheng Y., Chuah E., Corbett R. D., Dhalla N., He A., Lee D., Li H. I., Long W., Mayo M., Plettner P., Qian J. Q., Schein J. E., Tam A., Wong T., Birol I., Zhao Y., Faria C. C., Pimentel J., Nunes S., Shalaby T., Grotzer M., Pollack I. F., Hamilton R. L., Li X.-N., Bendel A. E., Fults D. W., Walter A. W., Kumabe T., Tominaga T., Collins V. P., Cho Y.-J., Hoffman C., Lyden D., Wisoff J. H., Garvin J. H. Jr., Stearns D. S., Massimi L., Schüller U., Sterba J., Zitterbart K., Puget S., Ayrault O., Dunn S. E., Tirapelli D. P. C., Carlotti C. G., Wheeler H., Hallahan A. R., Ingram W., MacDonald T. J., Olson J. J., Van Meir E. G., Lee J.-Y., Wang K.-C., Kim S.-K., Cho B.-K., Pietsch T., Fleischhack G., Tippelt S., Ra Y. S., Bailey S., Lindsey J. C., Clifford S. C., Eberhart C. G., Cooper M. K., Packer R. J., Massimino M., Garre M. L., Bartels U., Tabori U., Hawkins C. E., Dirks P., Bouffet E., Rutka J. T., Wechsler-Reya R. J., Weiss W. A., Collier L. S., Dupuy A. J., Korshunov A., Jones D. T. W., Kool M., Northcott P. A., Pfister S. M., Largaespada D. A., Mungall A. J., Moore R. A., Jabado N., Bader G. D., Jones S. J. M., Malkin D., Marra M. A., Taylor M. D., Divergent clonal selection dominates medulloblastoma at recurrence. Nature 529, 351–357 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hovestadt V., Jones D. T. W., Picelli S., Wang W., Kool M., Northcott P. A., Sultan M., Stachurski K., Ryzhova M., Warnatz H.-J., Ralser M., Brun S., Bunt J., Jäger N., Kleinheinz K., Erkek S., Weber U. D., Bartholomae C. C., von Kalle C., Lawerenz C., Eils J., Koster J., Versteeg R., Milde T., Witt O., Schmidt S., Wolf S., Pietsch T., Rutkowski S., Scheurlen W., Taylor M. D., Brors B., Felsberg J., Reifenberger G., Borkhardt A., Lehrach H., Wechsler-Reya R. J., Eils R., Yaspo M.-L., Landgraf P., Korshunov A., Zapatka M., Radlwimmer B., Pfister S. M., Lichter P., Decoding the regulatory landscape of medulloblastoma using DNA methylation sequencing. Nature 510, 537–541 (2014). [DOI] [PubMed] [Google Scholar]
- 13.Huang Y., Rao A., Connections between TET proteins and aberrant DNA modification in cancer. Trends Genet. 30, 464–474 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li J., Wu X., Zhou Y., Lee M., Guo L., Han W., Mo W., Cao W.-M., Sun D., Xie R., Huang Y., Decoding the dynamic DNA methylation and hydroxymethylation landscapes in endodermal lineage intermediates during pancreatic differentiation of hESC. Nucleic Acids Res. 46, 2883–2900 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li W., Zhang X., Lu X., You L., Song Y., Luo Z., Zhang J., Nie J., Zheng W., Xu D., Wang Y., Dong Y., Yu S., Hong J., Shi J., Hao H., Luo F., Hua L., Wang P., Qian X., Yuan F., Wei L., Cui M., Zhang T., Liao Q., Dai M., Liu Z., Chen G., Meckel K., Adhikari S., Jia G., Bissonnette M. B., Zhang X., Zhao Y., Zhang W., He C., Liu J., 5-Hydroxymethylcytosine signatures in circulating cell-free DNA as diagnostic biomarkers for human cancers. Cell Res. 27, 1243–1257 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cai J., Chen L., Zhang Z., Zhang X., Lu X., Liu W., Shi G., Ge Y., Gao P., Yang Y., Ke A., Xiao L., Dong R., Zhu Y., Yang X., Wang J., Zhu T., Yang D., Huang X., Sui C., Qiu S., Shen F., Sun H., Zhou W., Zhou J., Nie J., Zeng C., Stroup E. K., Zhang X., Chiu B. C.-H., Lau W. Y., He C., Wang H., Zhang W., Fan J., Genome-wide mapping of 5-hydroxymethylcytosines in circulating cell-free DNA as a non-invasive approach for early detection of hepatocellular carcinoma. Gut 68, 2195–2205 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Srinivasan V. M., Ghali M. G. Z., North R. Y., Boghani Z., Hansen D., Lam S., Modern management of medulloblastoma: Molecular classification, outcomes, and the role of surgery. Surg. Neurol. Int. 7, S1135–S1141 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chang C. H., Housepian E. M., Herbert C. Jr., An operative staging system and a megavoltage radiotherapeutic technic for cerebellar medulloblastomas. Radiology 93, 1351–1359 (1969). [DOI] [PubMed] [Google Scholar]
- 19.Weintraub L., Miller T., Friedman I., Abbott R., Levy A. S., Misdiagnosing recurrent medulloblastoma: The danger of examination and imaging without histological confirmation. J. Neurosurg. Pediatr. 13, 33–37 (2014). [DOI] [PubMed] [Google Scholar]
- 20.Cristiano S., Leal A., Phallen J., Fiksel J., Adleff V., Bruhm D. C., Jensen S. Ø., Medina J. E., Hruban C., White J. R., Palsgrove D. N., Niknafs N., Anagnostou V., Forde P., Naidoo J., Marrone K., Brahmer J., Woodward B. D., Husain H., van Rooijen K. L., Ørntoft M.-B. W., Madsen A. H., van de Velde C. J. H., Verheij M., Cats A., Punt C. J. A., Vink G. R., van Grieken N. C. T., Koopman M., Fijneman R. J. A., Johansen J. S., Nielsen H. J., Meijer G. A., Andersen C. L., Scharpf R. B., Velculescu V. E., Genome-wide cell-free DNA fragmentation in patients with cancer. Nature 570, 385–389 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wan J. C. M., Massie C., Garcia-Corbacho J., Mouliere F., Brenton J. D., Caldas C., Pacey S., Baird R., Rosenfeld N., Liquid biopsies come of age: Towards implementation of circulating tumour DNA. Nat. Rev. Cancer 17, 223–238 (2017). [DOI] [PubMed] [Google Scholar]
- 22.Miller A. M., Shah R. H., Pentsova E. I., Pourmaleki M., Briggs S., Distefano N., Zheng Y., Skakodub A., Mehta S. A., Campos C., Hsieh W.-Y., Selcuklu S. D., Ling L., Meng F., Jing X., Samoila A., Bale T. A., Tsui D. W. Y., Grommes C., Viale A., Souweidane M. M., Tabar V., Brennan C. W., Reiner A. S., Rosenblum M., Panageas K. S., De Angelis L. M., Young R. J., Berger M. F., Mellinghoff I. K., Tracking tumour evolution in glioma through liquid biopsies of cerebrospinal fluid. Nature 565, 654–658 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.De Mattos-Arruda L., Mayor R., Ng C. K. Y., Weigelt B., Martínez-Ricarte F., Torrejon D., Oliveira M., Arias A., Raventos C., Tang J., Guerini-Rocco E., Martínez-Sáez E., Lois S., Marín O., de la Cruz X., Piscuoglio S., Towers R., Vivancos A., Peg V., y Cajal S. R., Carles J., Rodon J., González-Cao M., Tabernero J., Felip E., Sahuquillo J., Berger M. F., Cortes J., Reis-Filho J. S., Seoane J., Cerebrospinal fluid-derived circulating tumour DNA better represents the genomic alterations of brain tumours than plasma. Nat. Commun. 6, 8839 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pentsova E. I., Shah R. H., Tang J., Boire A., You D., Briggs S., Omuro A., Lin X., Fleisher M., Grommes C., Panageas K. S., Meng F., Selcuklu S. D., Ogilvie S., Distefano N., Shagabayeva L., Rosenblum M., De Angelis L. M., Viale A., Mellinghoff I. K., Berger M. F., Evaluating cancer of the central nervous system through next-generation sequencing of cerebrospinal fluid. J. Clin. Oncol. 34, 2404–2415 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang Y., Pastor W. A., Zepeda-Martínez J. A., Rao A., The anti-CMS technique for genome-wide mapping of 5-hydroxymethylcytosine. Nat. Protoc. 7, 1897–1908 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pan W., Gu W., Nagpal S., Gephart M. H., Quake S. R., Brain tumor mutations detected in cerebral spinal fluid. Clin. Chem. 61, 514–522 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seoane J., De Mattos-Arruda L., Le Rhun E., Bardelli A., Weller M., Cerebrospinal fluid cell-free tumour DNA as a liquid biopsy for primary brain tumours and central nervous system metastases. Ann. Oncol. 30, 211–218 (2019). [DOI] [PubMed] [Google Scholar]
- 28.Y. Yin, J. Li, J. Li, M. Lee, S. Zhao, L. Guo, J. Li, M. Zhang, Y. Huang, X.-N. Li, D. Sun, LiBis: An ultrasensitive alignment method for low-input bisulfite sequencing. BioRxiv 2020.05.14.096461 [Preprint]. 16 May 2020. 10.1101/2020.05.14.096461. [DOI]
- 29.Sun D., Xi Y., Rodriguez B., Park H. J., Tong P., Meong M., Goodell M. A., Li W., MOABS: Model based analysis of bisulfite sequencing data. Genome Biol. 15, R38 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang Z.-J., Ellis T., Markant S. L., Read T.-A., Kessler J. D., Bourboulas M., Schüller U., Machold R., Fishell G., Rowitch D. H., Wainwright B. J., Wechsler-Reya R. J., Medulloblastoma can be initiated by deletion of Patched in lineage-restricted progenitors or stem cells. Cancer Cell 14, 135–145 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schüller U., Heine V. M., Mao J., Kho A. T., Dillon A. K., Han Y.-G., Huillard E., Sun T., Ligon A. H., Qian Y., Ma Q., Alvarez-Buylla A., McMahon A. P., Rowitch D. H., Ligon K. L., Acquisition of granule neuron precursor identity is a critical determinant of progenitor cell competence to form Shh-induced medulloblastoma. Cancer Cell 14, 123–134 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang Y., Pastor W. A., Shen Y., Tahiliani M., Liu D. R., Rao A., The behaviour of 5-hydroxymethylcytosine in bisulfite sequencing. PLOS ONE 5, e8888 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pajtler K. W., Witt H., Sill M., Jones D. T. W., Hovestadt V., Kratochwil F., Wani K., Tatevossian R., Punchihewa C., Johann P., Reimand J., Warnatz H.-J., Ryzhova M., Mack S., Ramaswamy V., Capper D., Schweizer L., Sieber L., Wittmann A., Huang Z., van Sluis P., Volckmann R., Koster J., Versteeg R., Fults D., Toledano H., Avigad S., Hoffman L. M., Donson A. M., Foreman N., Hewer E., Zitterbart K., Gilbert M., Armstrong T. S., Gupta N., Allen J. C., Karajannis M. A., Zagzag D., Hasselblatt M., Kulozik A. E., Witt O., Collins V. P., von Hoff K., Rutkowski S., Pietsch T., Bader G., Yaspo M.-L., von Deimling A., Lichter P., Taylor M. D., Gilbertson R., Ellison D. W., Aldape K., Korshunov A., Kool M., Pfister S. M., Molecular classification of ependymal tumors across all CNS compartments, histopathological grades, and age groups. Cancer Cell 27, 728–743 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bettegowda C., Sausen M., Leary R. J., Kinde I., Wang Y., Agrawal N., Bartlett B. R., Wang H., Luber B., Alani R. M., Antonarakis E. S., Azad N. S., Bardelli A., Brem H., Cameron J. L., Lee C. C., Fecher L. A., Gallia G. L., Gibbs P., Le D., Giuntoli R. L., Goggins M., Hogarty M. D., Holdhoff M., Hong S.-M., Jiao Y., Juhl H. H., Kim J. J., Siravegna G., Laheru D. A., Lauricella C., Lim M., Lipson E. J., Marie S. K. N., Netto G. J., Oliner K. S., Olivi A., Olsson L., Riggins G. J., Sartore-Bianchi A., Schmidt K., Shih L.-M., Oba-Shinjo S. M., Siena S., Theodorescu D., Tie J., Harkins T. T., Veronese S., Wang T.-L., Weingart J. D., Wolfgang C. L., Wood L. D., Xing D., Hruban R. H., Wu J., Allen P. J., Schmidt C. M., Choti M. A., Velculescu V. E., Kinzler K. W., Vogelstein B., Papadopoulos N., Diaz L. A. Jr., Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci. Transl. Med. 6, 224ra24 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang Y., Springer S., Zhang M., McMahon K. W., Kinde I., Dobbyn L., Ptak J., Brem H., Chaichana K., Gallia G. L., Gokaslan Z. L., Groves M. L., Jallo G. I., Lim M., Olivi A., Quinones-Hinojosa A., Rigamonti D., Riggins G. J., Sciubba D. M., Weingart J. D., Wolinsky J.-P., Ye X., Oba-Shinjo S. M., Marie S. K. N., Holdhoff M., Agrawal N., Diaz L. A. Jr., Papadopoulos N., Kinzler K. W., Vogelstein B., Bettegowda C., Detection of tumor-derived DNA in cerebrospinal fluid of patients with primary tumors of the brain and spinal cord. Proc. Natl. Acad. Sci. U.S.A. 112, 9704–9709 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mouliere F., Mair R., Chandrananda D., Marass F., Smith C. G., Su J., Morris J., Watts C., Brindle K. M., Rosenfeld N., Detection of cell-free DNA fragmentation and copy number alterations in cerebrospinal fluid from glioma patients. EMBO Mol. Med. 10, e9323 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bender S., Tang Y., Lindroth A. M., Hovestadt V., Jones D. T. W., Kool M., Zapatka M., Northcott P. A., Sturm D., Wang W., Radlwimmer B., Højfeldt J. W., Truffaux N., Castel D., Schubert S., Ryzhova M., Seker-Cin H., Gronych J., Johann P. D., Stark S., Meyer J., Milde T., Schuhmann M., Ebinger M., Monoranu C.-M., Ponnuswami A., Chen S., Jones C., Witt O., Collins V. P., von Deimling A., Jabado N., Puget S., Grill J., Helin K., Korshunov A., Lichter P., Monje M., Plass C., Cho Y.-J., Pfister S. M., Reduced H3K27me3 and DNA hypomethylation are major drivers of gene expression in K27M mutant pediatric high-grade gliomas. Cancer Cell 24, 660–672 (2013). [DOI] [PubMed] [Google Scholar]
- 38.Lister R., Mukamel E. A., Nery J. R., Urich M., Puddifoot C. A., Johnson N. D., Lucero J., Huang Y., Dwork A. J., Schultz M. D., Yu M., Tonti-Filippini J., Heyn H., Hu S., Wu J. C., Rao A., Esteller M., He C., Haghighi F. G., Sejnowski T. J., Behrens M. M., Ecker J. R., Global epigenomic reconfiguration during mammalian brain development. Science 341, 1237905 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Connolly I. D., Li Y., Pan W., Johnson E., You L., Vogel H., Ratliff J., Gephart M. H., A pilot study on the use of cerebrospinal fluid cell-free DNA in intramedullary spinal ependymoma. J. Neurooncol 135, 29–36 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xi Y., Li W., BSMAP: Whole genome bisulfite sequence MAPping program. BMC Bioinformatics 10, 232 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Capper D., Jones D. T. W., Sill M., Hovestadt V., Schrimpf D., Sturm D., Koelsche C., Sahm F., Chavez L., Reuss D. E., Kratz A., Wefers A. K., Huang K., Pajtler K. W., Schweizer L., Stichel D., Olar A., Engel N. W., Lindenberg K., Harter P. N., Braczynski A. K., Plate K. H., Dohmen H., Garvalov B. K., Coras R., Hölsken A., Hewer E., Bewerunge-Hudler M., Schick M., Fischer R., Beschorner R., Schittenhelm J., Staszewski O., Wani K., Varlet P., Pages M., Temming P., Lohmann D., Selt F., Witt H., Milde T., Witt O., Aronica E., Giangaspero F., Rushing E., Scheurlen W., Geisenberger C., Rodriguez F. J., Becker A., Preusser M., Haberler C., Bjerkvig R., Cryan J., Farrell M., Deckert M., Hench J., Frank S., Serrano J., Kannan K., Tsirigos A., Brück W., Hofer S., Brehmer S., Seiz-Rosenhagen M., Hänggi D., Hans V., Rozsnoki S., Hansford J. R., Kohlhof P., Kristensen B. W., Lechner M., Lopes B., Mawrin C., Ketter R., Kulozik A., Khatib Z., Heppner F., Koch A., Jouvet A., Keohane C., Mühleisen H., Mueller W., Pohl U., Prinz M., Benner A., Zapatka M., Gottardo N. G., Driever P. H., Kramm C. M., Müller H. L., Rutkowski S., von Hoff K., Frühwald M. C., Gnekow A., Fleischhack G., Tippelt S., Calaminus G., Monoranu C.-M., Perry A., Jones C., Jacques T. S., Radlwimmer B., Gessi M., Pietsch T., Schramm J., Schackert G., Westphal M., Reifenberger G., Wesseling P., Weller M., Collins V. P., Blümcke I., Bendszus M., Debus J., Huang A., Jabado N., Northcott P. A., Paulus W., Gajjar A., Robinson G. W., Taylor M. D., Jaunmuktane Z., Ryzhova M., Platten M., Unterberg A., Wick W., Karajannis M. A., Mittelbronn M., Acker T., Hartmann C., Aldape K., Schüller U., Buslei R., Lichter P., Kool M., Herold-Mende C., Ellison D. W., Hasselblatt M., Snuderl M., Brandner S., Korshunov A., von Deimling A., Pfister S. M., DNA methylation-based classification of central nervous system tumours. Nature 555, 469–474 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/42/eabb5427/DC1


