Abstract
Objective
The coronavirus disease 2019 (COVID-19) pandemic has caused an exponential rise in death rates and hospitalizations. The aim of this study was to characterize the D614G substitution in the severe acute respiratory syndome coronavirus 2 (SARS-CoV-2) spike glycoprotein (S protein), which may affect viral infectivity.
Methods
The effect of D614G substitution on the structure and thermodynamic stability of the S protein was analyzed with use of DynaMut and SCooP. HDOCK and PRODIGY were used to model furin protease binding to the S protein RRAR cleavage site and calculate binding affinities. Molecular dynamics simulations were used to predict the S protein apo structure, the S protein–furin complex structure, and the free binding energy of the complex.
Results
The D614G substitution in the G clade of SARS-CoV-2 strains introduced structural mobility and decreased the thermal stability of the S protein (ΔΔG = −0.086 kcal mol−1). The substitution resulted in stronger binding affinity (Kd = 1.6 × 10−8) for furin, which may enhance S protein cleavage. The results were corroborated by molecular dynamics simulations demonstrating higher binding energy of furin and the S protein D614G mutant (−61.9 kcal mol−1 compared with −56.78 kcal mol−1 for wild-type S protein).
Conclusions
The D614G substitution in the G clade induced flexibility of the S protein, resulting in increased furin binding, which may enhance S protein cleavage and infiltration of host cells. Therefore, the SARS-CoV-2 D614G substitution may result in a more virulent strain.
Keywords: SARS-CoV-2, COVID-19, Furin, S protein, G clade, Interatomic binding, Thermodynamic stability, Molecular dynamics simulations
Introduction
Severe acute respiratory syndome coronavirus 2 (SARS-CoV-2), which was first detected in Wuhan (Hubei Province, China) in late 2019, causes upper respiratory tract infection resulting in severe pneumonia and bronchiolitis (Wang et al., 2020a, Zhu et al., 2020). The rapid rate of SARS-CoV-2 human-to-human transmission resulted in the virus spreading worldwide within months. Subsequently, the World Health Organization declared a coronavirus disease 2019 (COVID-19) pandemic. SARS-CoV-2 belongs to the lineage B family of betacoronaviruses (Chan et al., 2020) with a continuous nonsegmented 30-kb positive-sense linear single-stranded RNA genome (Vijgen et al., 2005). The SARS-CoV-2 genome encodes four structural proteins: glycosylated spike (S), envelope (E) (required to infiltrate host cells), M (membrane), and N (nucleocapsid) proteins. These structural proteins have 96% sequence homology with bat coronavirus RaTG13 (Coutard et al., 2020, Zhou et al., 2020).
The S protein of SARS-CoV-2 expressed by the ORF1b gene is trimeric in structure, where each monomer is 180 kDa in size and has two main subunits, S1 and S2 (Ou et al., 2020). SARS-CoV-2 utilizes the S protein to facilitate attachment to host cell surface membranes, whereby the S protein undergoes a significant structural rearrangement for the receptor-binding domain of the S1 subunit to interact with the host cell transmembrane protein angiotensin-converting enzyme 2 (ACE2) (Wan et al., 2020), consequently destabilizing the trimeric structure of the S protein to create a transition structure in which the S2 subunit is in a stable conformation. For the fusion process to occur, the S protein of coronaviruses must be cleaved by proteases, such as transmembrane protease serine 2 (TMPRSS2) or furin protease (Bertram et al., 2011). Unlike SARS-CoV and Middle East respiratory syndrome coronavirus, the SARS-CoV-2 S protein has a unique polybasic “RRAR” furin recognition site (Coutard et al., 2020). Furin is a convertase enzyme that cycles between the trans-Golgi network and the cell surface, where it recognizes the cleavage motif on protein precursors and converts them to functional proteins through cleavage (Braun and Sauter, 2019). Furin proteolysis occurs at a specific multibasic sequence (R-[X]-(R/K)-R), with arginine (R) being the most commonly favored residue at position 1 (Tian et al., 2011). The furin recognition site within the SARS-CoV-2 S protein is very similar to that of the highly virulent avian and human influenza viruses (Andersen et al., 2020).
Genome sequences of SARS-CoV-2 from various countries have been deposited in the GISAID database (Shu and McCauley, 2017). The sequencing of SARS-CoV-2 has shown amino acid substitution arising in various continents, called super clades (G, S, V, D, and I). Initially, the 23403A > G mutation (D614G) found in the G clade S protein was found in the European population. (Phan, 2020). Recently, Korber et al. (2020) showed that patients infected with SARS-CoV-2 D614G mutant had higher levels of viral RNA. Furthermore, D614G substitution demonstrated higher titers in pseudoviruses from in vitro assays, thus indicating that the D614G mutant is more infective than the wild type (WT). In addition, a structural analysis showed that the replacement of aspartate (D) by glycine (G) might alter the structure of the S protein, making it easy to cleave (Eaaswarkhanth et al., 2020). Thus, increasing S protein exposure to cleavage by furin may increase the virulence of SARS-CoV-2.
In this study, we further analyze the structure of the WT S protein (D614) and a mutant (D614G) to understand the stability of both states using an array of bioinformatics tools and molecular dynamics (MD) simulations. Also, we model the binding of furin to the S protein, where we measured the binding affinities of furin for the S protein (WT and mutant) to understand further the high virulence associated with the D614G substitution.
Results and discussion
S protein D614G substitution
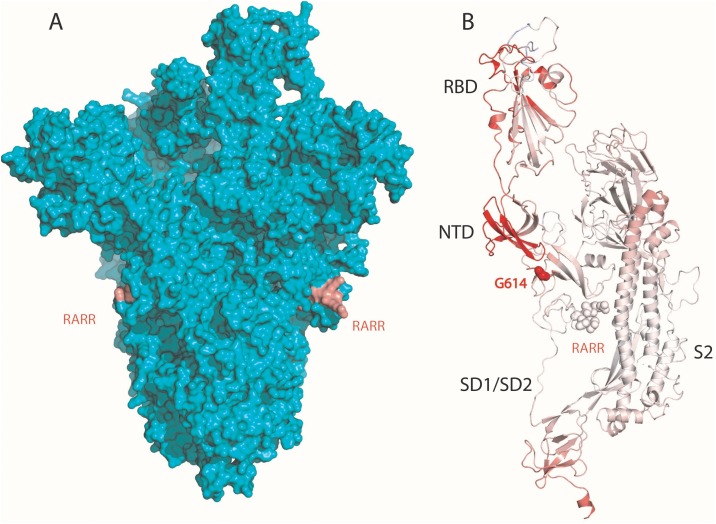
Genome sequences of SARS-CoV2 deposited in the GISAID database showed a D614G substitution on the S protein, which is the point of entry of the virus to the host cell, to be the most common in Europe and the world (Korber et al., 2020, Phan, 2020, Wang et al., 2020b). Therefore, understanding the structural changes in the mutated S protein is critical to understand its function and potential effects on the infectivity of SARS-CoV-2. Recently, Wrapp et al. (2020) solved a 3.5-Å-resolution cryogenic electron microscopy structure of the SARS-CoV-2 S protein (Protein Data Bank [PDB] ID 6VSB) (Figure 1 A). The missing loop regions of the S protein including the “RARR” furin cleavage site were modeled with a root-mean-square deviation (RMSD) of 0.25 Å between modeled and 6VSB structures. The D614G substitution is a change from a negatively charged aspartate (D) to glycine (G) in the S1 domain loop region of the S protein. As charged amino acids are important in stabilizing proteins through electrostatic interactions or hydrogen bonds (Zhou and Pang, 2018), this change may render the loop more flexible, resulting in the S protein “RARR” site being more accessible for furin cleavage.
Figure 1.
(A) Trimeric structure of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-S) spike glycoprotein (S protein) (Protein Data Bank ID 6VSB). (B) D614G mutant S protein monomer. The red region of the protein depicts the more flexible region of the protein due to the D614G substitution with decreased stability of ΔΔG = −0.086 kcal mol−1 and an increase in vibrational entropy (ΔΔSVib) of 0.137 kcal mol −1 K−1. NTD, NTD, N-terminal domain; RBD, receptor-binding domain.
Thermodynamic analysis of the D614G substitution
S protein stability analysis using the DynaMut webserver showed that the D614G substitution in the loop region resulted in a slightly more dynamic structure with ΔΔG of −0.086 kcal mol−1 and vibrational entropy (ΔΔS Vib) of 0.137 kcal mol−1 K−1 (Figure 1B). Protein loop regions possess greater motion dynamics (Yang et al., 2014), ranging from femtoseconds to seconds due to limited covalent interactions (Kamerzell and Middaugh, 2008). The increased entropy in the system resulting from substitutions exposes the region to surrounding moieties (Schrank et al., 2009), causing local unfolding, and so cleavage sites are readily attacked by proteases, such as furin. In addition, we compared temperature-dependent stability prediction curves for D614 S protein and G614 S protein. The folding free energy ΔG(T) was similar at 3.1 kcal mol−1 for both D614 S protein and G614 S protein, which indicates that the substitution did not affect the overall thermal stability of the protein (Aydin et al., 2014). Furthermore, there was no significant difference in melting temperature between the two structures (74.3 °C for D614 S protein and 74.2 °C for G614 S protein). However, the standard folding heat capacity was lower for D614 S protein (1.52 kcal mol−1 K−1) than for G614 S protein (1.56 kcal mol−1 K−1). A stable melting temperature was previously observed in the mutant S protein of SARS-CoV, where the difference in melting temperature between the WT and the mutant was small (Aydin et al., 2014). However, the substitutions in the study were in the S2 subunit of the S protein and not near the furin cleavage site. The standard folding enthalpy measured at the melting temperature was higher for G614 S protein at 62.6 kcal/mol compared with 60.9 kcal/mol for D614 S protein, indicating that G614 S protein is less stable at 25 °C (Spuergin et al., 1995). Substitutions can have either a stabilizing or a destabilizing effect on WT protein folding, whereby folding free energy measurements on RNA viral proteins with high substitution rates have demonstrated less stable protein structures (Wylie and Shakhnovich, 2011).
Interatomic interactions in D614G mutant S protein
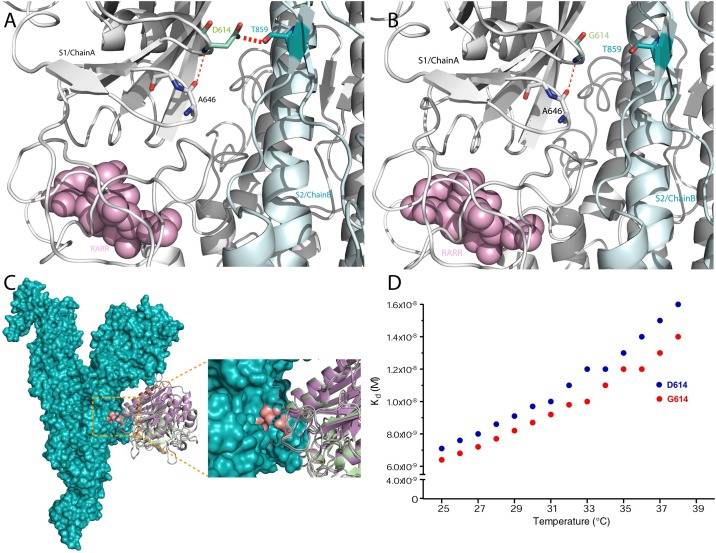
Substitutions can alter macromolecular interactions, such as hydrophobic and electrostatic interactions, van der Waals forces, and hydrogen-bonding networks (Jubb et al., 2017). Interatomic bonding analysis of the SARS-CoV-2 S protein revealed that D614 is close to T859 of the S2 domain of the adjacent monomer (chain B), whereby their side chains can form a short, 2.7-Å hydrogen bond (Figure 2 A). This hydrogen bond is lost in the D614G mutant since glycine lacks a hydrogen-bond donor or acceptor side chain (Figure 2B). Meanwhile, the hydrogen-bond strength is dependent on the distance and dihedral angle (Grzesiek et al., 2001); the 2.7-Å hydrogen bond between the donor (D614) and the acceptor (T859) is predicted to be important for the stability of the overall structure. Therefore, the loss of the hydrogen bond in the D614G mutant may have two consequences: (1) loss of hinging of the S2 subunit, which increases its flexibility in the transition state when interacting with the host cell receptor; and (2) increased flexibility, resulting in a more accessible furin cleavage site concomitant with increased infectivity of the D614G mutant.
Figure 2.
(A) Suggested hydrogen bonds (dashed red lines) of D614 (S1 domain chain A) with T859 (S2 domain chain B) and D614 and A646 of S1 domain chain A. (B) This hydrogen bond can be disrupted with the D614G substitution, altering the activity of the protein. (C) Modeled furin bound to the S protein RARR site. Furin bound to WT D614 S protein is depicted in green and bound to the D614G mutant is depicted in purple. (D) The dissociation constant Kd for furin bound to D614 S protein (blue) and G614 S protein (red).
Furin binding to D614 and G614 S proteins
As a result of thermodynamic analysis indicating that the S protein D614G substitution is less stable, we modeled the binding of furin (PDB ID 4Z2A) (Pearce et al., 2019) to the RRAR cleavage site of D614 S protein and G614 S protein (Figure 2C). To calculate the binding affinities of both structures, model 1 was chosen (Dudenhoeffer et al., 2019) from HDOCK analysis (Yan et al., 2020, Yan et al., 2017) of furin bound to D614 S protein (green) and G614 S protein (purple). Other proteases, such as TMPRSS2, elastase, and lysosomal cathepsins, bind to the S protein of coronaviruses (Matsuyama et al., 2010). However, arginine at position 1 (P1) and basic residues at P2 and P4 in the SARS-CoV-2 furin cleavage site are identical to the H5N1 hemagglutinin cleavage site, which may have contributed to the highly virulent virus outbreak of 1997 in Hong Kong (Claas et al., 1998).
Since the D614G substitution correlates with higher infectivity, we calculated the binding affinities (K d) of the furin bound to D614 S protein and G614 S protein structures at different temperatures using the PRODIGY server (Xue et al., 2016) (Figure 2D). Both D614 S protein and G614 S protein displayed a similar pattern in K d scores. There was an obvious shift in K d as the temperature increased from euthermic temperatures (≤36 °C) to hyperthermic temperatures (≥37 °C), whereby furin appeared to have stronger binding affinity for G614 S protein (K d = 1.4 × 10−8M at 38 °C) in comparison with D614 S protein (K d = 1.6 × 10−8M at 38 °C). It is expected that the loss of hydrogen bonds affects the stability of the S protein because such loss has been shown to substantially contribute to protein stability with an average of 1 kcal mol−1 for each hydrogen bond (Pace et al., 2014). This, in turn, increased the surface accessibility of the RARR cleavage site, inducing stronger binding affinity of the S protein variant at hyperthermic temperatures, which may explain the infectivity of the D614G mutant.
MD. simulations of D614 S protein and G614 S protein bound to furin
MD simulation is a technique for obtaining dynamic protein data at atomic spatial resolution. Therefore, to further characterize the structural stability and dynamic features of WT D614 apo-S protein and G614 apo-S protein and D614 S protein–furin and G614 S protein–furin complexes, we ran 100-ns simulations using the GROMACS package. The dynamic behavior of each system was monitored by our observing the RMSD and root-mean-square fluctuation (RMSF) trajectories of the Cα atoms. To further confirm the impact of the D614G substitution on the binding of furin, the total binding free energy was determined by the MMGBSA method (molecular mechanics, generalized Born model and solvent accessibility model).
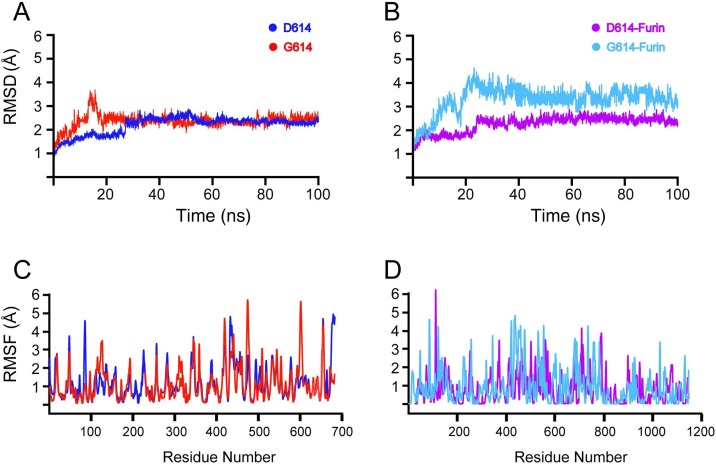
The RMSD of WT D614 apo-S protein and G614 apo-S protein (Figure 3 A) presented a stable structure with an average RMSD of 2.5 Å for most of the 100-ns simulation. The structure converged during the first 30 ns of the D614 S protein simulation with an RMSD increase from 1.25 to 2 Å. From 40 ns onward, the system reached equilibrium at 2.5 Å and this was maintained until the end of the simulation. The RMSD of the G614 apo-S protein structure increased from 1.5 to 3.5 Å in the initial 15 ns of the simulation. Subsequently, the RMSD of the G614 S protein structure decreased to 2.5 Å at 20 ns and remained uniform until the end of the simulation. This high convergence at the start of the simulation may have been the result of instability in the structure caused by the D614G substitution, resulting in a more flexible loop region. A more dynamic conformation may be a factor that contributed to the destabilization of the protein, making the region near the RARR furin cleavage site more accessible.
Figure 3.
Molecular dynamics simulations. Root-mean-square deviation (RMSD) plots for 100-ns simulations of D614 S protein and G614 S protein (A) and D614 S protein–furin and G614 S protein–furin complexes (B). Root-mean-square fluctuation (RMSF) plots of D614 S protein and G614 S protein (C) and D614 S protein–furin and G614 S protein–furin complexes (D).
The RMSDs demonstrate the stability and dynamics of WT D614 S protein–furin and G614 S protein–furin complexes, as shown in Figure 3B. The D614 S protein–furin complex demonstrated convergence from 1.25 to 2.5 Å in the initial 20 ns. Subsequently, the system did not show any convergence, and the average RMSD remained stable at 2.5 Å for the remainder of the simulation. The G614 S protein–furin complex demonstrated dynamic behavior, with an average RMSD that was higher than that of the WT D614 S protein–furin complex. The system exhibited a significant convergence from 0 to 15 ns as the RMSD increased from 1.5 to 3.5 Å. The complex continued to fluctuate between 3.5 and 4.5 Å until the end of the 100-ns simulation. RMSD results for the G614 S protein–furin complex signify a dynamic complex due to the effect of the substitution on the stability of the S protein and its binding to furin. The dynamic structure was probably due to the D614G substitution in the loop region, causing changes in conformational dynamics and ultimately affecting the interaction with furin.
The RMSF values from the 100-ns simulations of the WT D614 apo-S protein and G614 apo-S protein structures are depicted in Figure 3C and D. WT D614 S protein demonstrated relatively low residual flexibility in comparison with the G614 S protein structure. The G614 S protein mutant exhibited higher fluctuations, specifically in the loop region between residues 600 and 620. The D614G substitution can affect the internal dynamics of the protein, resulting in higher fluctuation than in the WT D614 S protein (Aier et al., 2016). Loop regions located far from or near the cleavage site play critical roles in dynamic and structural changes in enzyme–protein interactions. As a consequence, any conformational changes near the binding site can influence the enzyme cleavage process (Yang et al., 2014). Therefore, the higher RMSF demonstrated by the D614G mutant caused greater conformational mobility, which may expose the RARR cleavage site and result in a more favorable S protein–furin binding interaction through conformational selection or an induced-fit mechanism (Tsai et al., 2001, Yang et al., 2014).
Similarly, the G614 S protein–furin complex showed greater flexibility than the WT D614 S protein–furin complex (Figure 3D). The greater mobility observed close to the loop region between residues 600 and 620 could be the result of higher binding affinity of furin for the D614G mutant. The binding affinity of furin for G614 S protein was further corroborated by the binding free energy of −61.9 kcal mol−1 compared with −56.78 kcal mol−1 for furin bound to WT D614 S protein (Table 1 ). Furthermore, the WT D614 S protein–furin complex presented a van der Waals interaction of −105.34 kcal mol−1, whereas the G614 S protein–furin complex showed a van der Waals interaction of 103.67 kcal mol−1. These results indicate that furin favors binding to the G614 S protein RARR cleavage site over that of the WT D614 S protein. Therefore, the D614G mutant may result in more efficient cleavage of the S protein and subsequent interaction with ACE2, which may result in a more virulent strain.
Table 1.
The binding free energy of WT D614 S protein and D614G mutant S protein in complex with furin.
| Complex | vdW(kcal mol−1) | ELE (Å) | SASA (Å) | ΔG (kcal mol−1) |
|---|---|---|---|---|
| D614 S protein–furin | −105.34 | −559.47 | −11.97 | −56.78 |
| G614 S protein–furin | −103.67 | −456.58 | −11.65 | −61.90 |
ELE, ; SASA, ; vdW, van der Waals.
Summary
Although the D614G substitution did not affect the overall thermal stability of the S protein, the loop region of the D614G mutant showed a more dynamic structure compared with WT D614 S protein. Moreover, MD simulations of WT D614 S protein showed a profile similar to that for the D614G mutant, showing high convergence in the initial stages, which can be attributed to the substitution in the loop region. A dynamic loop region resulted from loss of the hydrogen bond between D614 (S1/chain A) and T859 (S2/chain B), where the loop was hinged to the α-helix of the corresponding residue. Such structural dynamics resulted in furin showing a higher binding affinity for the D614G mutant, which implies that increased infectivity may result from the D614G substitution. Hence, substitution near the binding interface can play a vital role in affecting the binding affinity of the SARS-CoV-2 S protein.
Conclusions
Our analysis showed that the D614G substitution disturbed the structural stability of the SARS-Cov-2 S protein, which consequently increased the binding affinity of furin for the S protein. This structural instability appears to be a more favorable state for furin cleavage. As a result, the D614G substitution may enhance viral entry to the host cell, causing higher infectivity. We used MD simulations to elucidate the effect of the substitution on the structure and stability of the S protein to formulate a hypothesis about potential factors involved in effects of the D614G mutant of the S protein. A larger-scale investigation will be critical for building a reliable model of these protein–protein interactions and their functional consequences.
Methods
Structural stability analysis
The structures of the S protein (PDB ID 6VSB) (Wrapp et al., 2020) and furin (PDB ID 4Z2A) (Pearce et al., 2019) were used as models for structural analysis. SWISS-MODEL (Waterhouse et al., 2018) was used to simulate the missing amino acids that were not visible in the cryogenic electron microscopy structure of the S protein. The DynaMut (Rodrigues et al., 2018) webserver was used to predict the effect of genetic variants on the stability and flexibility of D614G mutant S protein. The folding free energy, melting temperature, standard folding enthalpy measured at the melting temperature, and standard folding heat capacity were calculated by SCooP and online temperature-dependent stability prediction algorithms (Pucci et al., 2017).
Protein–protein docking
Protein–protein docking of WT (D614) and mutant (D614G) S protein (PDB ID 6VSB) with furin (PDB ID: 4Z2A) was done with the HDOCK server (Yan et al., 2020, Yan et al., 2017), which is based on a hybrid algorithm of template-based modeling and ab initio free docking. Residues 682, 683, 684, and 685 of the RRAR site were defined as binding site residues of chain A of the S protein structure (PDB ID 6VSB). Model 1, with the lowest docking energy score and the highest ligand RMSD, was selected to analyze binding energy scores (K d) using the PRODIGY server (Xue et al., 2016).
MD. simulations
An all-atom MD simulation was performed for all systems to estimate the dynamics of the apo and bound systems. For this purpose, we used GROMACS 5.1.2 (Abraham et al., 2015, Van Der Spoel et al., 2005) and the OPLS-AA force field (Kaminski et al., 2001) with the simple point charge water model to solvate the systems. The distance around the protein was kept at 20 Å because of the large size of the protein. To neutralize the pH, we added sodium counterions. Energy minimization of all systems was performed with 50,000 iterations. When the maximum force of 1000 kJ mol−1 nm−1 was applied, the steepest descent energy minimization was terminated. Following energy minimization, we equilibrated the systems with a constant temperature of 300 K, while the pressure was kept at 1 bar. For electrostatic contacts, we used particle mesh Ewald. The Berendsen thermostat was used as a temperature-coupling method (Bussi et al., 2007, Darden et al., 1993, Toukmaji et al., 2000). Finally, all systems were simulated for 100 ns in MD simulations, and the coordinates were saved at intervals of 2 ps.
Trajectories were analyzed after completion of the MD simulations to compare and observe the structural deviation between WT and mutant structures of the apo structures and complexes. The RMSD, which reflects the stability of the system, RMSF, which shows the flexibility of the protein, and the g_mmpbsa (MMGBSA) method for the binding free energy were used.
Ethical approval
Not required.
Conflict of interest
All authors have no conflict of interest to declare.
Author contributions
FA-M conceptualized the study. AM collected data and analyzed and proposed protein-protein interactions. EM calculated the protein kinetics. AM, EM, SKM, MA, JA, and FA-M wrote and edited the manuscript. All authors discussed the hypothesis, critically read and revised the manuscript, and gave final approval for publication.
Acknowledgment
This study was supported by the Kuwait Foundation for the Advancement of Sciences.
References
- Abraham M.J., Murtola T., Schulz R., Páll S., Smith J.C., Hess B., et al. GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1:19–25. [Google Scholar]
- Aier I., Varadwaj P.K., Raj U. Structural insights into conformational stability of both wild-type and mutant EZH2 receptor. Sci Rep. 2016;6 doi: 10.1038/srep34984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen K.G., Rambaut A., Lipkin W.I., Holmes E.C., Garry R.F. The proximal origin of SARS-CoV-2. Nat Med. 2020;26(4):450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aydin H., Al-Khooly D., Lee J.E. Influence of hydrophobic and electrostatic residues on SARS-coronavirus S2 protein stability: insights into mechanisms of general viral fusion and inhibitor design. Protein Sci. 2014;23(5):603–617. doi: 10.1002/pro.2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram S., Glowacka I., Muller M.A., Lavender H., Gnirss K., Nehlmeier I., et al. Cleavage and activation of the severe acute respiratory syndrome coronavirus spike protein by human airway trypsin-like protease. J Virol. 2011;85(24):13363–13372. doi: 10.1128/JVI.05300-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun E., Sauter D. Furin-mediated protein processing in infectious diseases and cancer. Clin Transl Immunol. 2019;8(8):e1073. doi: 10.1002/cti2.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bussi G., Donadio D., Parrinello M. Canonical sampling through velocity rescaling. J Chem Phys. 2007;126(1) doi: 10.1063/1.2408420. [DOI] [PubMed] [Google Scholar]
- Chan J.F., Kok K.H., Zhu Z., Chu H., To K.K., Yuan S., et al. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg Microbes Infect. 2020;9(1):221–236. doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claas E.C., Osterhaus A.D., van Beek R., De Jong J.C., Rimmelzwaan G.F., Senne D.A., et al. Human influenza A H5N1 virus related to a highly pathogenic avian influenza virus. Lancet. 1998;351(9101):472–477. doi: 10.1016/S0140-6736(97)11212-0. [DOI] [PubMed] [Google Scholar]
- Coutard B., Valle C., de Lamballerie X., Canard B., Seidah N.G., Decroly E. The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antiviral Res. 2020;176 doi: 10.1016/j.antiviral.2020.104742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darden T., York D., Pedersen L. Particle mesh Ewald: an N⋅log(N) method for Ewald sums in large systems. J Chem Phys. 1993;98(12):10089–10092. [Google Scholar]
- Dudenhoeffer B.R., Schneider H., Schweimer K., Knauer S.H. SuhB is an integral part of the ribosomal antitermination complex and interacts with NusA. Nucleic Acids Res. 2019;47(12):6504–6518. doi: 10.1093/nar/gkz442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaaswarkhanth M., Madhoun A.A., Al-Mulla F. Could the D614G substitution in the SARS-CoV-2 spike (S) protein be associated with higher COVID-19 mortality? Int J Infect Dis. 2020;96:459–460. doi: 10.1016/j.ijid.2020.05.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grzesiek S., Cordier F., Dingley A.J. Scalar couplings across hydrogen bonds. Methods Enzymol. 2001;338:111–133. doi: 10.1016/s0076-6879(02)38217-x. [DOI] [PubMed] [Google Scholar]
- Jubb H.C., Pandurangan A.P., Turner M.A., Ochoa-Montano B., Blundell T.L., Ascher D.B. Mutations at protein-protein interfaces: small changes over big surfaces have large impacts on human health. Prog Biophys Mol Biol. 2017;128:3–13. doi: 10.1016/j.pbiomolbio.2016.10.002. [DOI] [PubMed] [Google Scholar]
- Kamerzell T.J., Middaugh C.R. The complex inter-relationships between protein flexibility and stability. J Pharm Sci. 2008;97(9):3494–3517. doi: 10.1002/jps.21269. [DOI] [PubMed] [Google Scholar]
- Kaminski G.A., Friesner R.A., Tirado-Rives J., Jorgensen W.L. Evaluation and reparametrization of the OPLS-AA force field for proteins via comparison with accurate quantum chemical calculations on peptides. J Phys Chem B. 2001;105(28):6474–6487. [Google Scholar]
- Korber B., Fischer W.M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., et al. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182(4):812–827.e19. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuyama S., Nagata N., Shirato K., Kawase M., Takeda M., Taguchi F. Efficient activation of the severe acute respiratory syndrome coronavirus spike protein by the transmembrane protease TMPRSS2. J Virol. 2010;84(24):12658–12664. doi: 10.1128/JVI.01542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou X., Liu Y., Lei X., Li P., Mi D., Ren L., et al. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat Commun. 2020;11(1) doi: 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pace C.N., Fu H., Lee Fryar K., Landua J., Trevino S.R., Schell D., et al. Contribution of hydrogen bonds to protein stability. Protein Sci. 2014;23(5):652–661. doi: 10.1002/pro.2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce K.H., Overton L.K., Gampe R.T., Barrett G.B., Taylor J.D., McKee D.D., et al. BacMam production and crystal structure of nonglycosylated apo human furin at 1.89 Å resolution. Acta Crystallogr F Struct Biol Commun. 2019;75(Pt 4):239–245. doi: 10.1107/S2053230X19001419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan T. Genetic diversity and evolution of SARS-CoV-2. Infect Genet Evol. 2020;81 doi: 10.1016/j.meegid.2020.104260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pucci F., Kwasigroch J.M., Rooman M. SCooP: an accurate and fast predictor of protein stability curves as a function of temperature. Bioinformatics. 2017;33(21):3415–3422. doi: 10.1093/bioinformatics/btx417. [DOI] [PubMed] [Google Scholar]
- Rodrigues C.H., Pires D.E., Ascher D.B. DynaMut: predicting the impact of mutations on protein conformation, flexibility and stability. Nucleic Acids Res. 2018;46(W1):W350–5. doi: 10.1093/nar/gky300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrank T.P., Bolen D.W., Hilser V.J. Rational modulation of conformational fluctuations in adenylate kinase reveals a local unfolding mechanism for allostery and functional adaptation in proteins. Proc Natl Acad Sci U S A. 2009;106(40):16984–16989. doi: 10.1073/pnas.0906510106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu Y., McCauley J. GISAID: Global initiative on sharing all influenza data – from vision to reality. Euro Surveill. 2017;22(13):30494. doi: 10.2807/1560-7917.ES.2017.22.13.30494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spuergin P., Abele U., Schulz G.E. Stability, activity and structure of adenylate kinase mutants. Eur J Biochem. 1995;231(2):405–413. doi: 10.1111/j.1432-1033.1995.tb20713.x. [DOI] [PubMed] [Google Scholar]
- Tian S., Huang Q., Fang Y., Wu J. FurinDB: a database of 20-residue furin cleavage site motifs, substrates and their associated drugs. Int J Mol Sci. 2011;12(2):1060–1065. doi: 10.3390/ijms12021060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toukmaji A., Sagui C., Board J., Darden T. Efficient particle-mesh Ewald based approach to fixed and induced dipolar interactions. J Chem Phys. 2000;113(24):10913–10927. [Google Scholar]
- Tsai C.J., Ma B., Sham Y.Y., Kumar S., Nussinov R. Structured disorder and conformational selection. Proteins. 2001;44(4):418–427. doi: 10.1002/prot.1107. [DOI] [PubMed] [Google Scholar]
- Van Der Spoel D., Lindahl E., Hess B., Groenhof G., Mark A.E., Berendsen H.J. GROMACS: fast, flexible, and free. J Computat Chem. 2005;26(16):1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- Vijgen L., Keyaerts E., Moes E., Thoelen I., Wollants E., Lemey P., et al. Complete genomic sequence of human coronavirus OC43: molecular clock analysis suggests a relatively recent zoonotic coronavirus transmission event. J Virol. 2005;79(3):1595–1604. doi: 10.1128/JVI.79.3.1595-1604.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Y., Shang J., Graham R., Baric R.S., Li F. Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J Virol. 2020;94(7):e00127–20. doi: 10.1128/JVI.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Horby P.W., Hayden F.G., Gao G.F. A novel coronavirus outbreak of global health concern. Lancet. 2020;395(10223):470–473. doi: 10.1016/S0140-6736(20)30185-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., Hozumi Y., Yin C., Wei G.W. Decoding SARS-CoV-2 transmission and evolution and ramifications for COVID-19 diagnosis, vaccine, and medicine. J Chem Inf Model. 2020 doi: 10.1021/acs.jcim.0c00501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse A., Bertoni M., Bienert S., Studer G., Tauriello G., Gumienny R., et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46(W1):W296–303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367(6483):1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wylie C.S., Shakhnovich E.I. A biophysical protein folding model accounts for most mutational fitness effects in viruses. Proc Natl Acad Sci U S A. 2011;108(24):9916–9921. doi: 10.1073/pnas.1017572108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue L.C., Rodrigues J.P., Kastritis P.L., Bonvin A.M., Vangone A. PRODIGY: a web server for predicting the binding affinity of protein-protein complexes. Bioinformatics. 2016;32(23):3676–3678. doi: 10.1093/bioinformatics/btw514. [DOI] [PubMed] [Google Scholar]
- Yan Y., Tao H., He J., Huang S.Y. The HDOCK server for integrated protein-protein docking. Nat Protoc. 2020;15(5):1829–1852. doi: 10.1038/s41596-020-0312-x. [DOI] [PubMed] [Google Scholar]
- Yan Y., Zhang D., Zhou P., Li B., Huang S.Y. HDOCK: a web server for protein-protein and protein-DNA/RNA docking based on a hybrid strategy. Nucleic Acids Res. 2017;45(W1):W365–73. doi: 10.1093/nar/gkx407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L.Q., Sang P., Tao Y., Fu Y.X., Zhang K.Q., Xie Y.H., et al. Protein dynamics and motions in relation to their functions: several case studies and the underlying mechanisms. J Biomol Struct Dyn. 2014;32(3):372–393. doi: 10.1080/07391102.2013.770372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H.X., Pang X. Electrostatic interactions in protein structure, folding, binding, and condensation. Chem Rev. 2018;118(4):1691–1741. doi: 10.1021/acs.chemrev.7b00305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382(8):727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]