Abstract
Different from typical LexA repressors in heterotrophic bacteria exerting SOS response by auto-cleavage, cyanobacterial LexAs, especially that of Synechocystis sp. PCC 6803 (S.6803), have been suggested be involved in regulation of a number of genes related to various cellular processes, rather than the typical SOS regulon. When and how cyanobacterial LexAs are triggered to regulate its target genes have remained unknown. In this study, we found the profound repressing effect of LexA on salt-stress inducible genes in S.6803. The repressing activity of LexA was likely to persist during salt stress and the salt response of these genes was mainly achieved by other regulators than LexA, suggesting that the physiological role of LexA is fine-tuning of gene expression in response to environmental changes. Although the amount and oligomeric state of LexA were unchanged upon salt stress, two-dimensional electrophoresis and liquid chromatography-tandem mass spectrometry analyses detected a change in posttranslational modification in a small fraction of LexA molecules, possibly dephosphorylation of Ser173, after 30 min upon the upshift in salt concentration. Activity of LexA in S.6803 may be under gradual control by posttranslational modification to fine-tune gene expression, which is contrasted with the digital switching-off regulation by auto-cleavage in heterotrophic bacteria.
Subject terms: Microbiology, Molecular biology, Plant sciences
Introduction
The LexA transcription factor is highly conserved among bacterial species and is well-characterized in Escherichia coli as the key regulator of the SOS response induced by DNA damage. Under non-stress conditions, LexA represses expression of the SOS regulon including genes involved in DNA repair, replication and cell division1–3. Upon SOS response, LexA interacts with RecA which is activated through binding to single-stranded DNAs formed at the sites of DNA damage. This interaction changes LexA to a cleavable conformation and auto-cleavage at the Ala84–Gly85 peptide bond is carried out by the catalytic dyad Ser119 and Lys156, leading to inactivation of LexA and the derepression of SOS regulon4–6.
LexA is conserved also in many cyanobacterial species, but the experimental data on model cyanobacterial species have been suggesting that regulatory roles of cyanobacterial LexAs are quite different from that of heterotrophic bacteria. Global transcriptome analyses of Synechocystis sp. PCC 6803 (S.6803) revealed that deletion of lexA positively or negatively affected expression levels of a number of genes related to various cellular processes but not genes involved in DNA repair7–9. Furthermore, the function of S.6803 LexA as a global regulator is suggested by the fact that LexA has been isolated as a binding factor to the upstream regions of various genes such as the hoxEFUYH operon encoding a bidirectional hydrogenase10,11, crhR encoding an RNA helicase12, sbtA encoding a sodium-dependent bicarbonate transporter13 and fab genes encoding fatty-acid biosynthetic enzymes14. Analyses of the LexA-overexpressing strain of Anabaena sp. PCC 7120 suggested that LexA is involved in the responses to various abiotic stresses such as oxidative stress, DNA damage, carbon limitation and heavy metal stress through regulation of diverse genes15.
When and how cyanobacterial LexAs are triggered to regulate its target genes have remained unknown so far. In Anabaena sp. PCC 7120, the auto-cleavage of LexA occurs neither at physiological pH even in the presence of the activated RecA in vitro nor under SOS-response inducing conditions in vivo, such as UV-B exposure and mitomycin C treatment16. In the case of S.6803, LexA lacks the Ala-Gly auto-cleavage site and the serine of the Ser-Lys catalytic dyad12. Oliveira and Lindblad17 reported that LexA in S.6803 exists in at least three forms with different isoelectric points. This may suggest posttranslational control of LexA activity but no information is available regarding the nature of the modification and the conditions under which modification pattern changes.
To clarify the physiological role of cyanobacterial LexA and the regulatory mechanism of its activity, here we focus on salt acclimation of S.6803, since our RNA-seq data revealed that expression levels of a set of genes related to accumulation of the osmoprotective compound glucosylglycerol (GG) were significantly affected in the lexA-disrupted mutant9. When exposed to salt-stress conditions, S.6803 starts to synthesize GG by a two-step reaction18. First GG-phosphate is synthesized from glycerol-3-phosphate and ADP-glucose by GG-phosphate synthase (GgpS) and then GG-phosphate is dephosphorylated by GG-phosphate phosphatase (GgpP) to yield GG. The ABC-type transporter for GG uptake composed of four subunits, GgtA, GgtB, GgtC and GgtD, is also induced upon the salt stress. The main function of this transporter is suggested to be the reuptake of GG leaked into the periplasm to avoid carbon loss19. In addition, the GG-degrading enzyme GghA encoded by a salt-inducible gene slr1670 was supposed to work for GG turnover in fluctuating salinities20. In the genome of S.6803, genes encoding these GG-related enzymes and transporters together with genes related to glycerol-3-phosphate metabolism (glp genes) are organized into four gene clusters and their expression is highly induced under non-stress conditions by disruption of lexA9.
In this study, the contribution of LexA to transcriptional regulation of GG-related gene clusters together with other salt stress-inducible genes was evaluated before and after salt stress. Although expression of many salt stress-inducible genes are directly or indirectly repressed by LexA under non-stress conditions, their salt response can be attained without LexA. We suggest that the physiological role of LexA in S.6803 is fine-tuning of gene expression in response to environmental changes. To achieve this, LexA activity is likely to be modulated through posttranslational modification instead of digital switching-off regulation employed by heterotrophic bacteria.
Results
Contribution of LexA to transcriptional regulation of four gene clusters involved in accumulation of glucosylglycerol
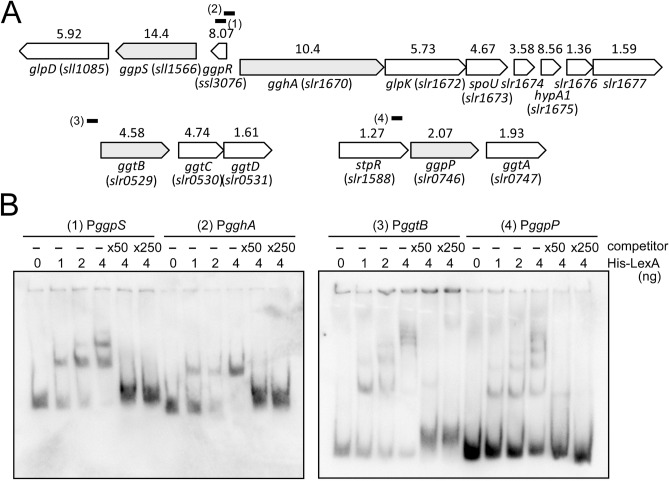
Figure 1A shows the genomic organization of four gene clusters containing GG biosynthesis- (ggp, glp) and uptake- (ggt) related genes in S.6803. The numerals above each gene indicate the induction ratio by disruption of lexA examined by RNA-seq analysis under non-stress conditions9. We performed DNA gel mobility shift assays to examine whether all of these four gene clusters were under the direct control of LexA. Approximately 200 bp of DNA fragments containing the upstream promoter region and the major transcription start site determined by differential RNA-seq (dRNA-seq) analysis21 were used as probes for the assay (short bold lines in Fig. 1A). The first gene cluster, ggpS–glpD (sll1566–sll1085), was reported to have its transcription start site within the upstream small ggpR gene encoding a negative regulator of ggpS 22. Thus, we employed a part of the ggpR coding sequence as the PggpS probe. Although ggpR was reported to show salt-independent constitutive expression22, high induction of ggpR transcripts by disruption of lexA (Fig. 1A) may imply that ggpR is also under the negative control by LexA independent of ggpS–glpD. The second gene cluster including gghA (slr1670) and glpK (slr1672) shares the promoter region with the divergently transcribed ggpS–glpD. The co-induction of gghA–glpK–spoU–slr1674–hypA1 by disruption of lexA (Fig. 1A) suggests polycistronic expression of these five genes. The third and fourth gene clusters are ggtBCD (slr0529–0531) and ggpP–ggtA (slr0746–0747). The upstream situated gene of ggpP–ggtA, stpR (slr1588) encoding a negative regulator of spsA23, was reported to be monocistronically transcribed based on the location of the transcription start sites21,23. Thus we used a part of the stpR coding sequence as the PggpP probe for DNA gel mobility shift assay. The results of DNA gel mobility shift assay are shown in Fig. 1B. His-LexA recombinant protein bound to the upstream region of all of these gene clusters. The addition of 50-fold and 250-fold excess amounts of the non-labeled specific promoter segments as a competitor abolished the formation of the shifted complex for each probe. These results suggest a mechanism for coordinated transcriptional regulation of GG-related genes by the specific binding of LexA.
Figure 1.
The genomic organization of four gene clusters containing glucosylglycerol biosynthesis- and uptake-related genes in S.6803 and binding of LexA to their upstream region. (A) Schematic view of the genomic organization. The first gene of each gene cluster is shown in gray. The numerals above each gene indicate the induction ratio by disruption of lexA examined by RNA-seq analysis under non-stress conditions9. Short bold bars indicate probes for DNA gel mobility shift assay. (B) DIG-labeled promoter segments of four gene clusters, PggpS, PgghA, PggtB and PggpP, were incubated with His-LexA added at indicated concentrations. 50-fold and 250-fold excess amounts of the non-labeled promoter segments were added as a competitor. Samples were separated on a 6% polyacrylamide gel.
In order to examine the role of LexA on salt-induced expression of GG-related genes, the wild-type (WT) strain and the lexA-disrupted (ΔlexA) mutant were transferred from non-stress to salt-stress conditions by the addition of NaCl. The growth of WT was not delayed even in the presence of 500 mM NaCl which has been generally used as salt stress conditions in S.6803 (Supplementary Fig. S1). The growth of ΔlexA was also not affected by NaCl but the mutant grew slower than WT as reported previously9.
Changes in transcript levels of GG-related gene clusters before and after the addition of 500 mM NaCl were detected by RNA gel blot analysis using ggpS, glpK, ggtB and ggpP probes, respectively (Fig. 2). As reported in the previous studies19,24, smearing of the bands was observed, which may be caused by both low stability of GG-related transcripts and existence of multiple transcription units within a gene cluster. Under non-stress conditions (0 h), expression of GG-related transcripts was tightly repressed in WT, whereas substantial expression was observed in the ΔlexA mutant, which is consistent with the previous RNA-seq results (Fig. 1A)9. Upon the addition of NaCl, GG-related transcripts were highly accumulated within 1 h. Although GG-related genes were derepressed in the ΔlexA mutant under non-stress conditions, further upregulation was clearly observed in response to salt stress. This observation suggests that induction of GG-related genes under salt-stress conditions is mainly governed by alternative, unidentified regulators.
Figure 2.
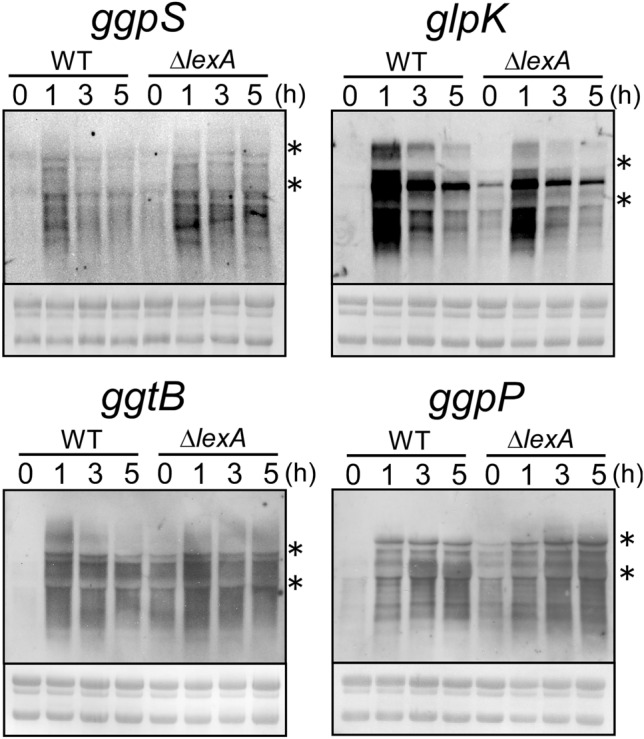
Changes in transcript levels of four GG-related gene clusters upon the addition of 500 mM NaCl in WT and ΔlexA strains. Total RNA was isolated from cells before and after the addition of NaCl at time points indicated. 5 μg of total RNA was applied to each lane for RNA gel blot analysis and transcripts were detected by using single-stranded riboprobes of ggpS, glpK, ggtB and ggpP. rRNA was visualized with methylene blue staining to show the equal RNA loading. The location of 23S and 16S rRNA was shown in each panel by asterisks.
The effect of the lack of repression by LexA on GgpS protein level was examined by immunoblot analysis (Fig. 3). The GgpS protein (57 kDa) was detected only after 3 h of the addition of NaCl in WT, whereas it was already accumulated under non-stress conditions and further increased after the addition of NaCl in the ΔlexA mutant. The amount of LexA (27 kDa) was almost unchanged before and after the addition of NaCl in WT (Fig. 3). Neither a decrease in the size of LexA nor the appearance of auto-cleavage products were detected.
Figure 3.
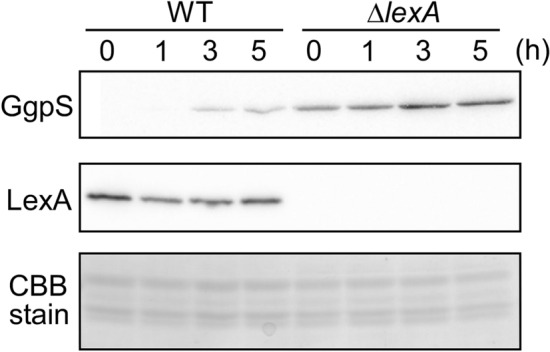
Changes in protein levels of GgpS and LexA upon the addition of 500 mM NaCl in WT and ΔlexA strains. Total proteins were isolated from cells before and after the addition of NaCl at time points indicated. 5 μg of total proteins were applied to each lane for immunoblot analysis and detected by specific antibodies. A part of Coomassie Brilliant Blue (CBB) stained SDS-PAGE gel was presented to show the equal protein loading.
Contribution of LexA to transcriptional regulation of salt-stress inducible genes
Previous DNA microarray studies revealed that expression of several hundreds of genes is induced or repressed under salt-stress conditions25,26. We wondered whether LexA is involved in regulation of not only GG-related genes but also other salt-stress responsive genes. To evaluate to what extent LexA contributes to salt-stress response, we listed 140 genes reported as NaCl-inducible in at least two of the previous reports25–28 in Supplementary Table S1 and added our previously generated RNA-seq data on induction ratio of each gene by disruption of lexA9. Among 140 NaCl-inducible genes, 63 genes (45%) were induced more than two-fold but no gene was repressed less than half under non-stress conditions by disruption of lexA. As for genes whose expression was repressed under salt stress25, the effect of disruption of lexA on their expression was quite limited (Supplementary Table S2), and the most affected gene among them, psaD, has been experimentally shown not to be a direct target of LexA9. Taken together, these data suggest the function of LexA as a general repressor of NaCl-inducible genes. Marin et al.26 categorized salt-stress inducible genes into four groups according to the time of maximum induction, 15 min, 30 min, 2 h and 6 h. As shown in Supplementary Table S3, highly induced genes by disruption of lexA were concentrated into a group with a maximum induction at 2 h, including GG-related genes such as ggpS, gghA and glpD.
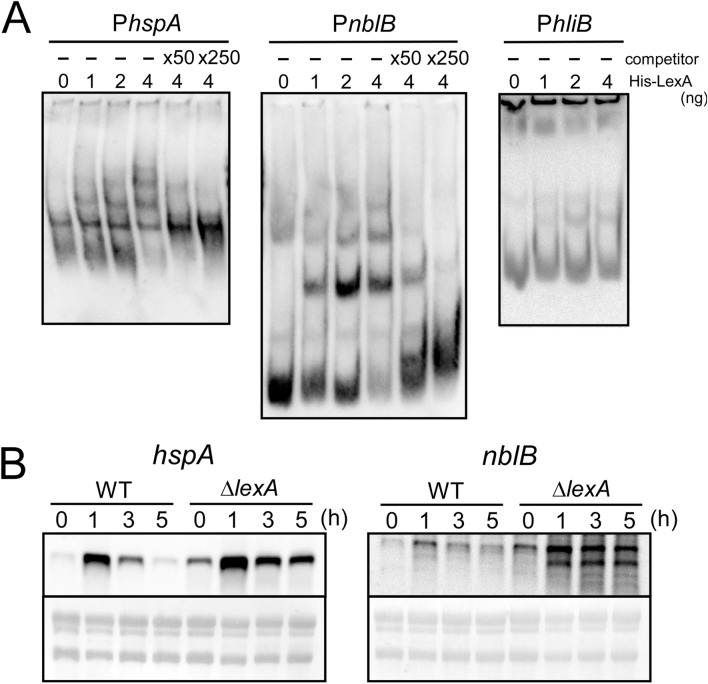
To test whether LexA directly regulates expression of other members than GG-related genes in this group, we chose hspA (sll1514) encoding a small heat-shock protein, hliB (ssr2595) encoding a high-light inducible protein and nblB (slr1687) encoding a protein involved in degradation of phycobilisome, and performed DNA gel mobility shift assays using the upstream intergenic region of these genes as DNA probe (Fig. 4A). Specific binding of His-LexA protein to the upstream region of hspA and nblB was observed, whereas band shift was not detected in the case of hliB. When examined by RNA gel blot analysis (Fig. 4B), the expression level of hspA and nblB under non-stress conditions (0 h) was clearly higher in the ΔlexA mutant than in WT. Upon the addition of 500 mM NaCl, the pattern of the salt response was similar between both strains but transcript levels were always higher in the mutant. The presumable degradation product of nblB was specifically detected in the mutant under salt stress conditions. These observations suggest that LexA binds to the upstream region of hspA and nblB and works as repressor irrespective of salt concentration.
Figure 4.
Regulation of salt-stress inducible genes by direct binding of LexA. (A) DNA gel mobility shift assay. DIG-labeled promoter segments of hspA, nblB and hliB were incubated with His-LexA added at indicated concentrations. 50-fold and 250-fold excess amounts of the non-labeled promoter segments were added as a competitor. Samples were separated on a 6% polyacrylamide gel. (B) RNA gel blot analysis. Total RNA was isolated from WT and ΔlexA cells before and after the addition of 500 mM NaCl at time points indicated. 5 μg of total RNA was applied to each lane and transcripts were detected by using single-stranded riboprobes of hspA and nblB. rRNA was visualized with methylene blue staining to show the equal RNA loading.
Next, we examined whether LexA is involved in downregulation of salt-stress inducible genes upon transfer from salt stress to non-stress conditions (Fig. 5). Incubation under salt stress conditions for 1 h resulted in accumulation of transcripts of ggpS, ggpP, hspA and nblB in both WT and the ΔlexA mutant. Upon the down-shift in salt concentration, ggpS and ggpP transcripts significantly decreased within 30 min in both strains and tended to gradually reaccumulate in the mutant. In the case of hspA and nblB, transcript levels were always higher in the mutant as observed in the up-shift experiment of salt concentration (Fig. 4B).
Figure 5.
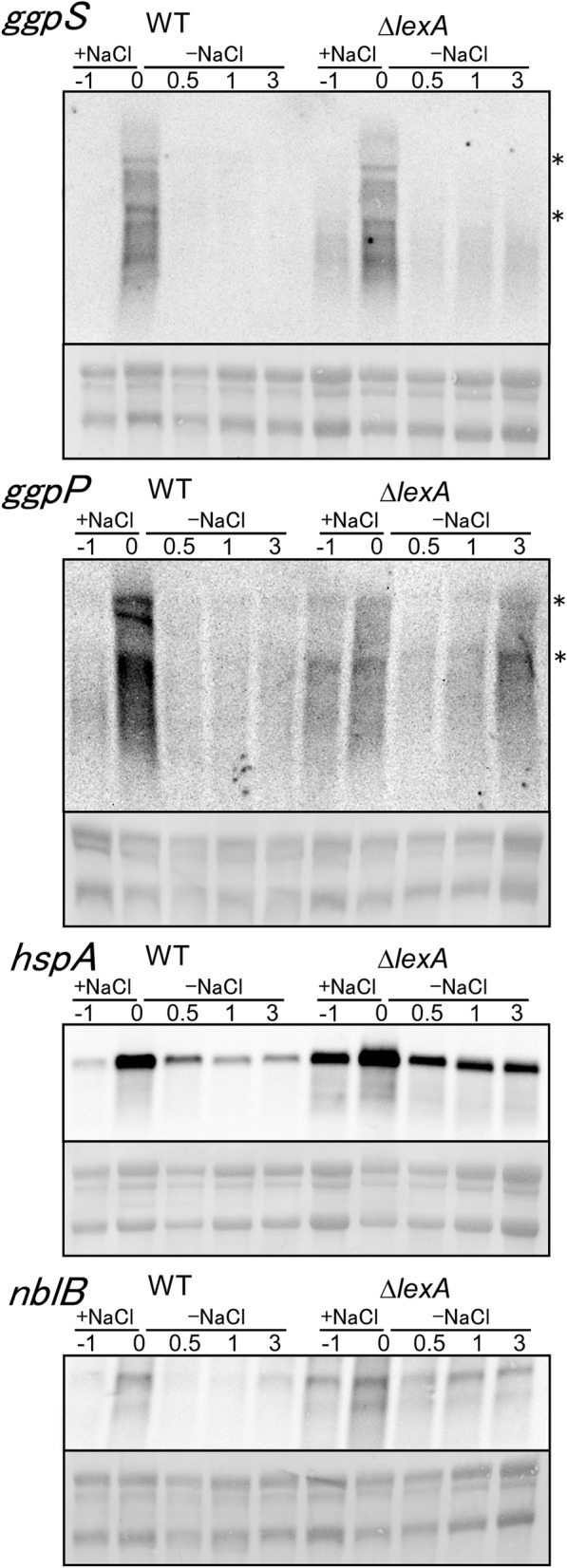
Changes in transcript levels upon the downshift in salt concentration in WT and ΔlexA strains. Total RNA was isolated from cells before (− 1 h) and after 1 h of incubation with 500 mM NaCl (0 h) and at time points indicated after transferred to the normal BG-11 medium (0.5, 1 and 3 h). 5 μg of total RNA was applied to each lane for RNA gel blot analysis and transcripts were detected by using single-stranded riboprobes of ggpS, ggpP, hspA and nblB. rRNA was visualized with methylene blue staining to show the equal RNA loading. The location of 23S and 16S rRNA was shown by asterisks.
Changes in posttranslational modification of LexA protein during salt stress
As shown in Fig. 3, the amount and size of the LexA protein did not change during salt stress. Furthermore, blue native PAGE (BN-PAGE) followed by immunodetection of LexA revealed that the oligomerization state of LexA did not change before and after salt treatment (Supplementary Fig. S2). The detected size was larger than the dimeric form of LexA (54 kDa), which may be due to slower mobility of LexA than the molecular weight marker in BN-PAGE. Since the presence of at least three forms of LexA with different isoelectric points (pIs), 5.6, 5.8 (theoretical pI of LexA) and 6.1 was reported17, we performed two-dimensional (2D) electrophoresis to detect changes in posttranslational modification of LexA after 0, 0.5, 1 and 3 h of incubation with NaCl. Immunodetection with anti-LexA antibody revealed three spots with different pIs, 5.6, 5.9 and 6.2 (Fig. 6A), which is consistent with the previous report. The spot with apparent pI of 5.6 disappeared after 30 min, but the modification state of the rest of LexA seemed unchanged during salt treatment (Fig. 6B).
Figure 6.
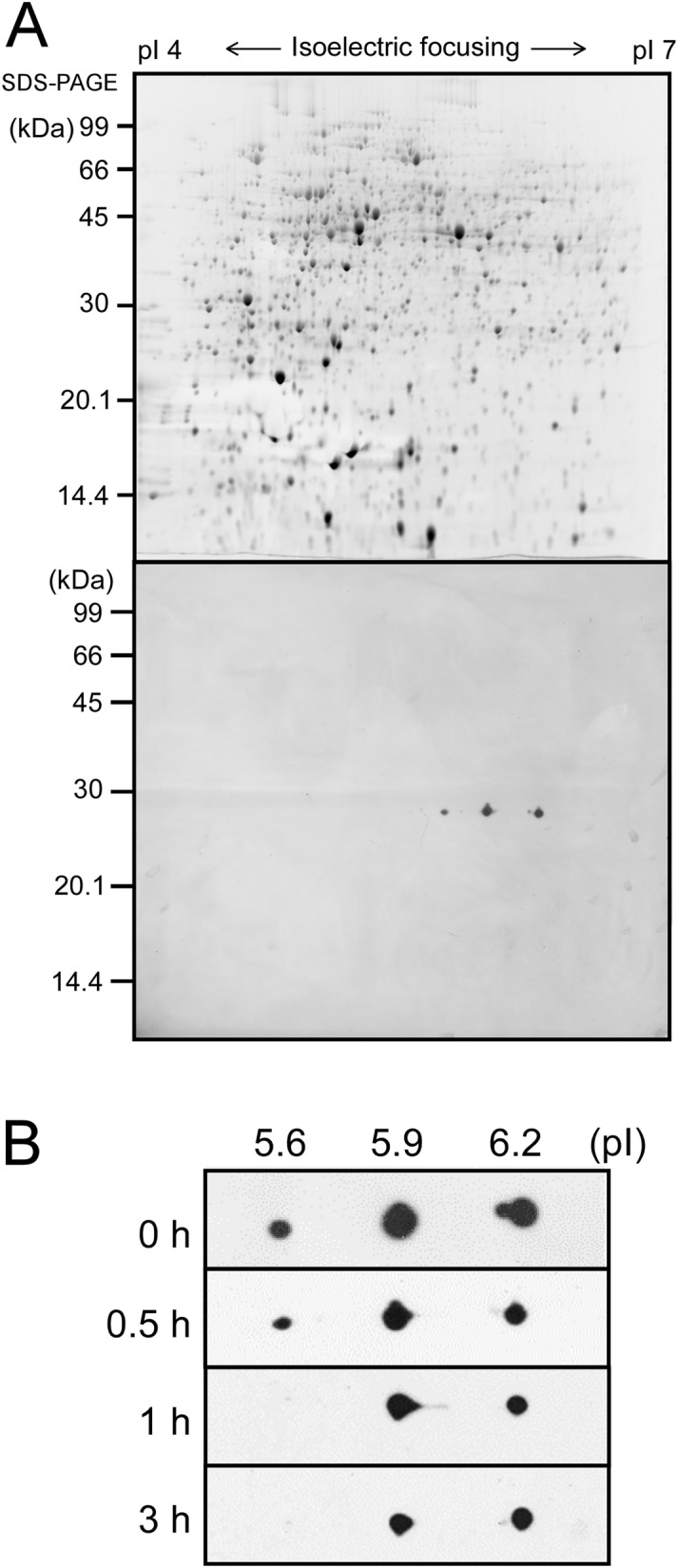
Changes in posttranslational modification of LexA upon the addition of 500 mM NaCl examined by 2D electrophoresis. (A) SYPRO Ruby-stained 2D gel with 270 μg of total protein from S.6803 grown under non-stress conditions (upper panel) and immunodetection of LexA using anti-LexA antibody (lower panel). At least three different forms of LexA with pIs of approximately 5.6, 5.9, and 6.2 were detected. (B) Changes in the spot pattern of LexA before and after the addition of NaCl at time points indicated.
To further test the possibility of regulation of LexA activity by posttranslational modification, we performed immunoprecipitation of LexA from S.6803 cell extracts after 0, 1 and 3 h of incubation with NaCl. After SDS-PAGE of the eluted fraction followed by silver staining (Supplementary Fig. S3), LexA bands were excised from the gel, digested with trypsin, and then analyzed with liquid chromatography-tandem mass spectrometry (LC–MS/MS). Obtained data suggested phosphorylation of several Ser, Thr and Tyr residues in LexA (Supplementary Table S4). In order to evaluate the change in the phosphorylation state of LexA in response to salt stress, the abundance of phosphopeptides was normalized by the abundance of LexA protein recovered from the excised gel and then the abundance ratio of 1 h/0 h and 3 h/0 h was calculated for each phosphopeptide (Supplementary Table S5). The most notable change was a decrease in the abundance ratio of the ASNNKGPGQELKASDVEIQGILMGVWR peptide to 0.26 after 1 h. The phosphorylation site in this peptide was identified as the second Ser (Supplementary Fig. S4), corresponding to Ser173 in LexA. However, the decrease in the phosphorylation rate upon salt stress could not be confirmed by the other peptides containing Ser173. The ASNNKGPGQELK and ASNNK peptides were not detected as phosphopeptides, probably because of low recovery by the reversed phase chromatography due to their low hydrophobicity. The VVLKASNNKGPGQELK phosphopeptide was detected but a change in the abundance ratio was not clearly observed probably due to its low abundance (Supplementary Table S5). The abundance of the phosphorylated form of the ASNNKGPGQELKASDVEIQGILMGVWR peptide (Supplementary Table S4, lines 50 and 51) was one order lower than that of the non-phosphorylated form (Supplementary Table S4, lines 52 and 54). Taken together, the results of LC–MS/MS analysis suggest that a small fraction of LexA molecules is phosphorylated at Ser173 under non-stress conditions and dephosphorylated within 1 h upon the addition of NaCl.
Discussion
DNA microarray analyses to examine the transcriptional response of S.6803 to salt stress revealed the induction of genes encoding proteins required for high salt acclimation as well as those encoding general stress response proteins25–28. By re-analysis of our previously generated RNA-seq data9, we found that disruption of lexA resulted in induction of about half of these salt-stress inducible genes more than two-fold under non-stress conditions (Supplementary Table S1). DNA gel mobility shift assay revealed that LexA directly regulates all of four gene clusters involved in GG accumulation, as well as general stress responsive genes, hspA and nblB (Figs. 1B, 4A). These results suggest that LexA works as a general repressor of salt-inducible genes. However, induction of these genes upon salt stress and repression upon transfer to normal growth conditions took place irrespective of the presence or absence of LexA (Figs. 2, 4B, 5). Changes in activity of transcription factors other than LexA and/or stability of transcripts may be mainly responsible for salt stress responses of these genes. Shoumskaya et al.28 reported that several two-component systems, namely the pairs of histidine kinase and response regulator, Hik33–Rre31, Hik10–Rre3, Hik16–Hik41–Rre17 and Hik34–Rre1, are involved in salt-induction of certain subsets of genes in response to various stresses indirectly caused by the addition of salt, such as generation of reactive oxygen species, redox imbalance and denatured proteins. In that study, hspA and nblB were suggested to be under the control of Rre1 and Rre31, respectively. There have been no reports of identification of regulators other than LexA involved in the coordinated regulation of all of the four GG-related gene clusters. As for ggpS, several studies suggest the existence of specific regulators. For example, GgpR encoded by ssl3076, a small ORF adjacent to ggpS, works as a repressor of ggpS22, the group 2 sigma factor SigB affects the timing of induction of ggpS upon salt stress24 and small RNA IsaR1 interacts with the 5′-UTR of ggpS to negatively affect its stability and translation29. Salt response of the other GG-related genes may also be attained by specific regulators not yet identified.
Based on the observation in this study, we propose that the contribution of LexA to salt acclimation is not induction of salt-stress responsive genes but optimization of energy distribution by repressing unnecessary transcription. In the ΔlexA mutant, the salt-stress responsive genes were derepressed under non-stress conditions (Figs. 2, 4B, Supplementary Table S1) and their induction levels upon salt stress were sometimes higher than WT (Figs. 2, 4B). Accumulation of GgpS protein under non-stress conditions was also observed in the mutant (Fig. 3). Considering that LexA is a global regulator involved in regulation of various cellular processes9, such uncontrolled expression of the LexA regulon must cause energy losses and delayed growth of the mutant both under normal and salt-stress conditions (Supplementary Fig. S1).
In heterotrophic bacteria such as E. coli, activity of LexA is controlled by digital switching-off mechanism through auto-cleavage in response to DNA damage. On the other hand, there have been no reports on physiological changes in the amount and oligomerization state as well as auto-cleavage in the case of cyanobacterial LexAs. In this study, we first expected switching-off of repressing activity of LexA upon salt stress. However, induction levels of some salt-responsive genes such as ggpS, hspA and nblB were always higher in the ΔlexA mutant than in WT (Figs. 2, 4B, 5), suggesting that repressing activity of LexA can persist during salt stress. We propose here that activity of LexA in S.6803 is not controlled in a digital ON–OFF manner in response to environmental changes, but there may exist gradual control through posttranslational modification.
We observed disappearance of the LexA spot with an apparent pI of 5.6 after 30 min of salt-treatment by 2D electrophoresis (Fig. 6B) and a decrease in the phosphorylation level of Ser173 within 1 h by immunoprecipitation followed by LC–MS/MS analysis (Supplementary Table S5). It is worth noting that both the pI 5.6 spot and phosphorylated Ser173 represent a minor fraction of the total LexA population. Upon salt acclimation, a large fraction of LexA may maintain the activity as a repressor but dephosphorylation of Ser173 after 30 min may cause decrease in the activity of some LexA molecules. This heterogeneity in LexA activity could explain the difference in the repressing effect of LexA among individual target genes (Figs. 2, 4B). Previously, we reported that the repressor activity of LexA on the fatty-acid biosynthetic genes, fabD, fabH and fabF, is enhanced under nitrogen-depleted conditions, and weakened under phosphate-depleted conditions14. Examination of changes in 2D spot pattern and phosphorylation level of Ser173 under different nutrient conditions will strengthen the view that LexA activity is modulated by posttranslational modification in S.6803.
Conclusions
We have revealed that LexA in S.6803 works as a general repressor of salt-inducible genes but is not the primary regulator of their salt response. Based on the observation, we hypothesize that the contribution of LexA to salt acclimation is optimization of energy distribution by repressing unnecessary transcription. In contrast to the digital switching-off regulation of LexA activity by auto-cleavage in heterotrophic bacteria, activity of LexA in S.6803 may be under more gradual control through posttranslational modification. It seems reasonable to employ such a regulatory mechanism assuming that physiological roles of LexA in S.6803 is fine-tuning of expression levels of genes related to various cellular processes in response to environmental changes. It is worth noting that Ser173 is not so much conserved, 26 out of 152 amino acid sequences of cyanobacterial LexAs according to our preliminary search (not shown). This may indicate the diversity in the function and regulatory mechanism of cyanobacterial LexAs. Based on the computational analysis of LexA binding sites across a large number of cyanobacterial genomes, Li et al.30 proposed the possibility that LexAs in most cyanobacteria can work as a regulator of SOS response. To obtain the comprehensive picture of the physiological role of cyanobacterial LexAs, we are now examining the distribution of the Ala-Gly auto-cleavage site, the Ser-Lys catalytic dyad, the putative phosphorylation site Ser173 and the recA gene with or without LexA binding sites throughout the phylogenetic tree of cyanobacteria.
Materials and methods
Strains and culture conditions
A glucose-tolerant non-motile strain (GT strain) of Synechocystis sp. PCC 6803 was grown at 32 °C in BG-11 medium containing 20 mM HEPES–NaOH, pH7.0, under continuous illumination at 20 μmol photons m−2 s–1 with bubbling of air. The lexA (sll1626)-disrupted mutant (ΔlexA) was generated by insertion of a kanamycin resistance cassette at nucleotide 1,319,123 of the coding region of lexA9 and grown under the same conditions as the wild-type cells, except that 20 μg mL–1 kanamycin was added to the medium. Cell density was estimated by measuring OD730 using a spectrophotometer (model UV-1800, Shimadzu). To perform salt stress experiments, WT and the ΔlexA cells grown in BG-11 medium to OD730 = 0.8 under non-stress conditions were diluted to OD730 = 0.3 and a solution of 3 M NaCl dissolved in BG-11 medium was added to 50 mL culture to give a final concentration of 500 mM. To perform downshift experiments of salt concentration, WT and the ΔlexA cells at OD730 = 0.3 incubated in BG-11 medium containing 500 mM NaCl for 1 h were harvested by centrifugation, washed with the normal BG-11, and resuspended with 50 mL of the same medium.
DNA gel mobility shift assay
Probes for DNA gel mobility shift assays were obtained by PCR amplification with primers shown in Supplementary Table S6 using genomic DNA as a template. The 3′ end of the DNA fragment for each probe was labeled with DIG-ddUTP using the terminal transferase method according to the manufacturer's instructions (DIG Gel Shift Kit, second generation; Roche). Overexpression and purification of the recombinant LexA protein with an N-terminal 6 × His-tag and DNA gel mobility shift assays by using a DIG Gel Shift Kit were performed as described in Kizawa et al.9.
RNA gel blot analysis
Isolation of total RNA by the hot phenol method and RNA gel blot analyses, using DIG RNA Labeling and Detection Kit (Roche), were performed as described previously31. Template DNA fragments for in vitro transcription to generate RNA probes were prepared by PCR using the primers shown in Supplementary Table S6. Original uncropped images of RNA Gel Blot Analysis were shown in Supplementary Fig. S5.
Immunoblot analysis
Total proteins were extracted from Synechocystis cells as described previously32 and separated by 15% (w/v) SDS-PAGE, followed by electroblotting onto PVDF membranes (Immobilon-P; Millipore). Immunodetection was done using a rabbit polyclonal antibody raised against the recombinant proteins in combination with a horseradish peroxidase-conjugated secondary antibody. Antibodies raised against GgpS were kindly provided by Prof. M. Hagemann (Universität Rostock). Original uncropped images of Immunoblot Analysis were shown in Supplementary Fig. S5.
BN-PAGE
Total proteins were extracted from Synechocystis cells as described previously32. Samples were added with 5 × BN-PAGE sample buffer, containing 240 mM Tris–HCl, pH8.0, 250 mM 6-amino caproic acid, 30% (v/v) glycerol, 0.57 M sucrose and 50 mg mL−1 Coomassie G-250. 10 μg protein per lane was applied to 5–20% native-PAGE gel (Kitasato Medical Service) and run according to the manufacturer’s instructions. After incubating the gels in resolubilization buffer containing 0.3% (w/v) Tris, 0.7% (w/v) Glycine and 0.2% (w/v) SDS, electroblotting and immunodetection of LexA were performed as described above.
2D gel electrophoresis
For 2D analysis, 350 mL of WT culture at OD730 = 0.5 was exposed to salt stress and harvested by centrifugation. The cell pellet was resuspended in 400 μL of 10 mM HEPES–NaOH buffer, pH7.0, containing 10 mM PMSF, supplemented with 10 mM NaF and 1 mM Na3VO433 and mixed with 3.6 g of zircon beads (diameter 0.1 mm, BioSpec Products) in a 2 mL tube. The cells were then disrupted with a Mini-Bead Beater (BioSpec Products) for three pulses of 50 s at 4 °C. After the cell debris was pelleted by centrifugation, the soluble protein fraction was obtained by ultracentrifugation at 100,000 g for 1 h at 4 °C, quantified by the Bio-Rad protein assay (Bio-Rad), and then concentrated by acetone precipitation. The dried pellet was resuspended with extraction buffer containing 8 M urea, 2 M thiourea, 1% (w/v) CHAPS, 19.4 mM dithiothreitol, and 0.5% Ampholytes (GE Healthcare) and protein concentration was adjusted to 15 mg mL−1. For two-dimensional analyses, 270 μg of total protein resuspended with extraction buffer was absorbed by 18-cm-long Immobiline DryStrip gels (GE Healthcare) with a pH range of 4 to 7 (linear), according to the manufacturer's instructions. The isoelectric-focusing step was carried out on CoolPhorestar IPG-IEF Type-PX system (Anatech), followed by 15% (w/v) SDS-PAGE in the second dimension. After completion of SDS-PAGE, total proteins on the gels were stained with SYPRO Ruby (Thermo Fisher Scientific) and images of the SYPRO Ruby-stained gels were acquired using FluoroPhorester 3000 (Anatech). After washing of the gels for 30 min twice with water and then incubating for 30 min in resolubilization buffer containing 0.3% (w/v) Tris, 0.7% (w/v) glycine and 0.2% (w/v) SDS, electroblotting and immunodetection of LexA were performed as described above.
Immunoprecipitation of LexA protein
For immunoprecipitation, 50 mL of WT culture at OD730 = 0.5 was exposed to salt stress and harvested by centrifugation. The cell pellet was resuspended in 500 μL of lysis buffer containing 50 mM HEPES–KOH, pH7.5, 140 mM NaCl, 1 mM EDTA, 1% Triton X-100 and 0.1% sodium deoxycholate, supplemented with Complete Mini EDTA-free protease inhibitor cocktail (Roche). Cells were added with 0.5 g of zircon beads (BioSpec Products), broken by vigorous vortexing, centrifuged at 14,000 g at 4 °C for 15 min and the supernatant was collected as the cell lysate for immunoprecipitation. After determination of the protein concentration using the Bio-Rad protein assay, the cell lysate containing 500 μg of protein was brought to a volume of 500 μL with cold lysis buffer, added with 20 μL of the pre-equilibrated Dynabeads Protein G (Thermo Fisher Scientific) and rotated for 1 h at 4 °C to avoid non-specific binding during the immunoprecipitation step. After removal of Dynabeads Protein G by magnetic separation, the cell lysate was added with 80 μg of anti-LexA antibody and rotated at 4 °C for overnight. Then 20 μL of the equilibrated Dynabeads Protein G were added and rotated at 4 °C for 1 h. After the binding reaction, Dynabeads Protein G was collected by magnetic separation, washed with 1.5 mL of lysis buffer by rotating at 4 °C for 5 min and again collected by magnetic separation. The washing steps were repeated sequentially using 1 mL of the following buffers: wash buffer1 (lysis buffer containing 500 mM NaCl), wash buffer 2 (10 mM Tris–HCl, pH8.0, 250 mM LiCl, 0.5% NP-40, 0.5% sodium deoxycholate, 1 mM EDTA) and TE (10 mM Tris–HCl, pH8.0, 1 mM EDTA). Elution was performed by incubation with 50 μL of elution buffer (1% SDS, 0.1 mM NaHCO3) at room temperature for 15 min with occasional vortexing. The supernatant was collected and the elution step was repeated once. Total 100 μL of eluates were subjected to SDS-PAGE and 27 kDa bands of LexA protein were excised after silver staining.
LC–MS/MS analysis
The silver-stained bands of LexA were de-stained and digested with a trypsin (TPCK-treated; Worthington Biochemical). The digestion mixture was separated on a nanoflow LC (Easy nLC; Thermo Fisher Scientific) using a nano-electrospray ionization spray column (NTCC analytical column; C18, φ75 μm × 100 mm, 3 μm; Nikkyo Technology) with a linear gradient of 0–80% buffer B (100% acetonitrile and 0.1% formic acid) in buffer A (0.1% formic acid) and a flow rate of 300 nL min−1 over 30 min, coupled on-line to a Q-Exactive mass spectrometer (Thermo Fisher Scientific) that was equipped with a nanospray ion source, based on the method described in Nagao et al.34. The mass spectrometer was operated in positive-ion mode, and MS and MS/MS spectra were acquired in a data dependent TOP 10 method. Protein were identified and quantified using Proteome Discoverer 2.2 (Thermo Fisher Scientific) with MASCOT program ver 2.6 (Matrix Science).
Supplementary information
Acknowledgements
We thank Prof. M. Hagemann (Universität Rostock) for helpful discussions and antibodies raised against GgpS. This work was supported by Japan Society for the Promotion of Science (JSPS) KAKENHI and the Cooperative Research Program of “Network Joint Research Center for Materials and Devices.” to Y.H.
Author contributions
K.T. and S.N. performed most of the experiments using cyanobacterial strains and recombinant proteins; A.K. made the lexA-disrupted mutant and performed RNA-seq analysis; T.S. and N.D. performed LC–MS/MS analysis; Y.H. conceived and supervised the project and wrote the article with contributions of all the authors.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-74534-7.
References
- 1.Fernández de Henestrosa AR, et al. Identification of additional genes belonging to the LexA regulon in Escherichia coli. Mol. Microbiol. 2000;35:1560–1572. doi: 10.1046/j.1365-2958.2000.01826.x. [DOI] [PubMed] [Google Scholar]
- 2.Courcelle J, Khodursky A, Peter B, Brown PO, Hanawalt PC. Comparative gene expression profiles following UV exposure in wild-type and SOS-deficient Escherichia coli. Genetics. 2001;158:41–64. doi: 10.1093/genetics/158.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wade JT, Reppas NB, Church GM, Struhl K. Genomic analysis of LexA binding reveals the permissive nature of the Escherichia coli genome and identifies unconventional target sites. Genes Dev. 2005;19:2619–2630. doi: 10.1101/gad.1355605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Horii T, et al. Regulation of SOS functions: Purification of E. coli LexA protein and determination of its specific site cleaved by the RecA protein. Cell. 1981;27:515–522. doi: 10.1016/0092-8674(81)90393-7. [DOI] [PubMed] [Google Scholar]
- 5.Slilaty SN, Little JW. Lysine-156 and serine-119 are required for LexA repressor cleavage: A possible mechanism. Proc. Natl. Acad. Sci. USA. 1987;84:3987–3991. doi: 10.1073/pnas.84.12.3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Luo Y, et al. Crystal structure of LexA: A conformational switch for regulation of self-cleavage. Cell. 2001;106:585–594. doi: 10.1016/S0092-8674(01)00479-2. [DOI] [PubMed] [Google Scholar]
- 7.Kamei, A. et al. Functional analysis of lexA-like gene, sll1626 in Synechocystis sp. PCC 6803 using DNA microarray. In PS2001 Proceedings: 12th International Congress on Photosynthesis S41-013 (CSIRO Publishing, 2001).
- 8.Domain F, Houot L, Chauvat F, Cassier-Chauvat C. Function and regulation of the cyanobacterial genes lexA, recA and ruvB: LexA is critical to the survival of cells facing inorganic carbon starvation. Mol. Microbiol. 2004;53:65–80. doi: 10.1111/j.1365-2958.2004.04100.x. [DOI] [PubMed] [Google Scholar]
- 9.Kizawa A, Kawahara A, Takimura Y, Nishiyama Y, Hihara Y. RNA-seq profiling reveals novel target genes of LexA in the cyanobacterium Synechocystis sp. PCC 6803. Front. Microbiol. 2016;7:193. doi: 10.3389/fmicb.2016.00193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gutekunst K, et al. LexA regulates the bidirectional hydrogenase in the cyanobacterium Synechocystis sp. PCC 6803 as a transcription activator. Mol. Microbiol. 2005;58:810–823. doi: 10.1111/j.1365-2958.2005.04867.x. [DOI] [PubMed] [Google Scholar]
- 11.Oliveira P, Lindblad P. LexA, a transcription regulator binding in the promoter region of the bidirectional hydrogenase in the cyanobacterium Synechocystis sp. PCC 6803. FEMS Microbiol. Lett. 2005;251:59–66. doi: 10.1016/j.femsle.2005.07.024. [DOI] [PubMed] [Google Scholar]
- 12.Patterson-Fortin LM, Colvin KR, Owttrim GW. A LexA-related protein regulates redox-sensitive expression of the cyanobacterial RNA helicase, crhR. Nucl. Acids Res. 2006;34:3446–3454. doi: 10.1093/nar/gkl426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lieman-Hurwitz J, et al. A cyanobacterial AbrB-like protein affects the apparent photosynthetic affinity for CO2 by modulating low-CO2-induced gene expression. Environ. Microbiol. 2009;11:927–936. doi: 10.1111/j.1462-2920.2008.01818.x. [DOI] [PubMed] [Google Scholar]
- 14.Kizawa A, et al. The LexA transcription factor regulates fatty acid biosynthetic genes in the cyanobacterium Synechocystis sp. PCC 6803. Plant J. 2017;92:189–198. doi: 10.1111/tpj.13644. [DOI] [PubMed] [Google Scholar]
- 15.Kumar A, Kirti A, Rajaram H. Regulation of multiple abiotic stress tolerance by LexA in the cyanobacterium Anabaena sp. strain PCC7120. Biochim. Biophys. Acta Gene Regul. Mech. 2018;1861:864–877. doi: 10.1016/j.bbagrm.2018.07.007. [DOI] [PubMed] [Google Scholar]
- 16.Kumar A, Kirti A, Rajaram H. LexA protein of cyanobacterium Anabaena sp. strain PCC 7120 exhibits in vitro pH-dependent and RecA-independent autoproteolytic activity. Int. J. Biochem. Cell Biol. 2015;59:84–93. doi: 10.1016/j.biocel.2014.12.003. [DOI] [PubMed] [Google Scholar]
- 17.Oliveira P, Lindblad P. Novel insights into the regulation of LexA in the cyanobacterium Synechocystis sp. strain PCC 6803. J. Bacteriol. 2011;193:3804–3814. doi: 10.1128/JB.00289-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hagemann M. Molecular biology of cyanobacterial salt acclimation. FEMS Microbiol. Rev. 2011;35:87–123. doi: 10.1111/j.1574-6976.2010.00234.x. [DOI] [PubMed] [Google Scholar]
- 19.Hagemann M, Richter S, Mikkat S. The ggtA gene encodes a subunit of the transport system for the osmoprotective compound glucosylglycerol in Synechocystis sp. strain PCC 6803. J. Bacteriol. 1997;179:714–720. doi: 10.1128/JB.179.3.714-720.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kirsch F, Pade N, Klähn S, Hess WR, Hagemann M. The glucosylglycerol-degrading enzyme GghA is involved in acclimation to fluctuating salinities by the cyanobacterium Synechocystis sp. strain PCC 6803. Microbiology. 2017;163:1319–1328. doi: 10.1099/mic.0.000518. [DOI] [PubMed] [Google Scholar]
- 21.Kopf M, et al. Comparative analysis of the primary transcriptome of Synechocystis sp. PCC 6803. DNA Res. 2014;21:527–539. doi: 10.1093/dnares/dsu018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klähn S, Höhne A, Simon E, Hagemann M. The gene ssl3076 encodes a protein mediating the salt-induced expression of ggpS for the biosynthesis of the compatible solute glucosylglycerol in Synechocystis sp. strain PCC 6803. J. Bacteriol. 2010;192:4403–4412. doi: 10.1128/JB.00481-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Song K, Hagemann M, Tan X, Lu X. The response regulator Slr1588 regulates spsA but is not crucial for salt acclimation of Synechocystis sp. PCC 6803. Front. Microbiol. 2017;8:1176. doi: 10.3389/fmicb.2017.01176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nikkinen HL, et al. The SigB σ factor regulates multiple salt acclimation responses of the cyanobacterium Synechocystis sp. PCC 6803. Plant Physiol. 2012;158:514–523. doi: 10.1104/pp.111.190058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kanesaki Y, Suzuki I, Allakhverdiev SI, Mikami K, Murata N. Salt stress and hyperosmotic stress regulate the expression of different sets of genes in Synechocystis sp. PCC 6803. Biochem. Biophys. Res. Commun. 2002;290:339–348. doi: 10.1006/bbrc.2001.6201. [DOI] [PubMed] [Google Scholar]
- 26.Marin K, et al. Gene expression profiling reflects physiological processes in salt acclimation of Synechocystis sp. strain PCC 6803. Plant Physiol. 2004;136:3290–3300. doi: 10.1104/pp.104.045047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marin K, et al. Identification of histidine kinases that act as sensors in the perception of salt stress in Synechocystis sp. PCC 6803. Proc. Natl. Acad. Sci. USA. 2003;100:9061–9066. doi: 10.1073/pnas.1532302100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shoumskaya MA, et al. Identical Hik–Rre systems are involved in perception and transduction of salt signals and hyperosmotic signals but regulate the expression of individual genes to different extents in Synechocystis. J. Biol. Chem. 2005;280:21531–21538. doi: 10.1074/jbc.M412174200. [DOI] [PubMed] [Google Scholar]
- 29.Rübsam H, et al. The iron-stress activated RNA 1 (IsaR1) coordinates osmotic acclimation and iron starvation responses in the cyanobacterium Synechocystis sp. PCC 6803. Environ. Microbiol. 2018;20:2757–2768. doi: 10.1111/1462-2920.14079. [DOI] [PubMed] [Google Scholar]
- 30.Li S, Xu M, Su Z. Computational analysis of LexA regulons in cyanobacteria. BMC Genomics. 2010;11:527. doi: 10.1186/1471-2164-11-527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Muramatsu M, Hihara Y. Transcriptional regulation of genes encoding subunits of photosystem I during acclimation to high-light conditions in Synechocystis sp. PCC 6803. Planta. 2003;216:446–453. doi: 10.1007/s00425-002-0859-5. [DOI] [PubMed] [Google Scholar]
- 32.Ishii A, Hihara Y. An AbrB-like transcriptional regulator, Sll0822, is essential for the activation of nitrogen-regulated genes in Synechocystis sp. PCC 6803. Plant Physiol. 2008;148:660–670. doi: 10.1104/pp.108.123505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mikkat S, Fulda S, Hagemann M. A 2D gel electrophoresis-based snapshot of the phosphoproteome in the cyanobacterium Synechocystis sp. strain PCC 6803. Microbiology. 2014;160:296–306. doi: 10.1099/mic.0.074443-0. [DOI] [PubMed] [Google Scholar]
- 34.Nagao R, et al. Structural basis for energy harvesting and dissipation in a diatom PSII–FCPII supercomplex. Nat. Plants. 2019;5:890–901. doi: 10.1038/s41477-019-0477-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.