Abstract
Mitochondria are the powerhouses of eukaryotic cells and the site of essential metabolic reactions. Complex I or NADH:ubiquinone oxidoreductase is the main entry site for electrons into the mitochondrial respiratory chain and constitutes the largest of the respiratory complexes. Its structure and composition vary across eukaryote species. However, high resolution structures are available only for one group of eukaryotes, opisthokonts. In plants, only biochemical studies were carried out, already hinting at the peculiar composition of complex I in the green lineage. Here, we report several cryo-electron microscopy structures of the plant mitochondrial complex I. We describe the structure and composition of the plant respiratory complex I, including the ancestral mitochondrial domain composed of the carbonic anhydrase. We show that the carbonic anhydrase is a heterotrimeric complex with only one conserved active site. This domain is crucial for the overall stability of complex I as well as a peculiar lipid complex composed of cardiolipin and phosphatidylinositols. Moreover, we also describe the structure of one of the plant-specific complex I assembly intermediates, lacking the whole PD module, in presence of the maturation factor GLDH. GLDH prevents the binding of the plant specific P1 protein, responsible for the linkage of the PP to the PD module.
Subject terms: Cryoelectron microscopy, Plant molecular biology, Cryoelectron microscopy
Electrons enter the mitochondrial respiratory chain via complex I. Here, the authors report high-resolution structures of mature plant complex I and one of its assembly intermediates, highlighting plant-specific features including an ancestral carbonic anhydrase domain.
Introduction
Complex I is the largest multimeric enzyme of the respiratory chain, composed of more than 40 protein subunits1. Fourteen are strictly conserved proteins subunits, vestige of complex I of bacterial origin2. The additional subunits, referred to as supernumerary subunits, were acquired during eukaryotes evolution3. Part of these additional subunits are conserved among other eukaryotes and play essential roles for the structure, function and the association with the other respiratory chain complexes, e.g., to assemble into respirasome in animals4. Recently, several high resolution 3D structures of the complete mitochondrial complex I of opisthokonts were determined by cryo-EM in mammalian species5–7 and in the aerobic yeast Yarrowia lipolytica8,9, revealing the organization of their additional specie-specific subunits. However, in plants, even though extensive biochemical characterization was conducted10,11, high-resolution structures of mitochondrial complex I are yet to be derived. Early negative staining studies11 revealed the presence of a large additional membrane attached domain, absent from animal and yeast species, hinting at the peculiar structure and composition of the plant complex I.
In order to obtain a high-resolution structure of the plant mitochondrial complex I and its additional subunits, we purified mitochondria from Brassica oleracea var. botrytis, a close relative to the model plant Arabidopsis (both belong to the group of Brassicaceae plants), as previously described12 (see “Methods” section). We recorded cryo-EM images of membrane complexes purified from sucrose gradient (see “Methods” section), corresponding to mitochondrial complex I. After particle sorting (see “Methods” section) we obtained cryo-EM reconstructions of two main complex I states: the full plant mitochondrial complex I, as well as reconstruction of a complex I assembly intermediate, without the PD module, and with the plant-specific assembly factor GLDH13 (Supplementary Fig. 1). Focused refinement and particle polishing were performed in RELION314 (see “Methods” section), allowing reconstruction of the full complex I at 3.7 Å resolution, focused classification on the membrane arm and PP module yielding 3.4 Å. The PD module being more flexible produced more scant densities reporting lower resolutions (Supplementary Figs. 1, 2). The cryo-EM maps allowed the determination of many well-resolved features and clear side-chain densities (Supplementary Fig. 3) that enabled modeling of the 45 proteins of the plant mitochondrial complex I (Supplementary Table 1), as well as several characteristic ligands that were directly identified from the density: the eight canonical FeS clusters of the matrix arm, the FMN of the 51 kDa subunit, the NADPH molecule in the 39 kDa subunit, three cardiolipins two phosphatidylinositol and one phosphatidylethanolamine (Supplementary Fig. 3). Ubiquinone was also clearly visible in the Q module (Supplementary Fig. 4).
Results and discussion
General description
The plant mitochondrial complex I has the classical open L-shape formed by the matrix and membrane arms (Fig. 1). In the matrix arm, electrons are transferred from NADH to ubiquinone along the FeS clusters with distances similar to what has been described in bacteria and opisthokonts mitochondrial complex I (Supplementary Fig. 5). The membrane part, composed of the proximal (PP) and distal (PD) modules relative to the matrix arm, where proton pumping takes place, is also conserved. Those highly conserved features are part of the 14 minimal protein subunits conserved in bacteria and other mitochondrial complex I1. The additional mitochondria specific proteins enhance the volume and mass of the complex I compared to bacteria. In plant, 25 proteins shared with aerobic yeast or mammals are present. Similarly to the previously obtained complex I structure5,7,9,15, the supernumerary subunits, specific to mitochondrial complex I, form a shell around the core subunits, adding nearly 400 kDa of proteins to the conserved subunits (Supplementary Fig. 5). These supernumerary subunits are mainly composed of α-helices and are largely interconnected with the core subunits. In the Q module of the matrix arm, our reconstructions allowed us to clearly visualize ubiquinone in the pocket formed by Nad1, PSST and Nad7 (Supplementary Fig. 4). Our results confirm the recently observed ubiquinone position described for Y. lipolytica8. During 3D classification of the mature complex, only one major conformation was observed. This would indicate that the plant complex I would be similar to the yeast complex I, where similar conformations are observed in both active and deactive states9, as opposed to mammalian complexes, where movement of the matrix arm relative to the membrane arm is observed between the states5.
Fig. 1. Overall structure of the plant mitochondrial complex I.
a Composite cryo-EM map of the complete plant complex I. The membrane part is displayed in blue shades, the matrix arm is displayed in green shades and the γ-carbonic anhydrase module is displayed in orange shades. Individual N, Q, PD, and PP modules are indicated. b The resulting atomic model, each individual 45 proteins are annotated and displayed with a different color. IMM stands for inner mitochondrial membrane, IMS inter-membrane space.
Carbonic anhydrase: an ancestral component of mitochondrial complex I
The main feature of the plant complex I is the presence of an additional globular matrix-exposed domain, bound to the membrane arm. It holds an overlapping position with the NDUFA10/42-kDa subunit found in mammalian complex I5. This domain is formed by a trimeric complex, the carbonic anhydrase (CA). CA are zinc-containing enzymes that catalyze the reversible hydration of CO2 to HCO3−, whose roles are usually carbon fixation and pH maintenance16. The carbonic anhydrase activity evolved independently several times during evolution, resulting in several CA enzyme families. The plant mitochondrial CA is part of the gamma-carbonic anhydrase (γCA) family, which was originally identified in the archaeon Methanosarcina thermophila17,18. Here, the overall structure of the γCA is conserved: it is a trimer composed of characteristic left-handed beta-helix monomers, forming triangular prisms (Fig. 2a). However, in contrast to the archaeal enzyme, the γCA present in plant complex I is an heterotrimer19. Indeed, in Arabidopsis five γCA genes were identified. Three have highly conserved catalytic domains and are termed γCA1-3, similar to the archaeal enzyme, while the two other show less homology to their prokaryotic homologs especially in the catalytic domain and were thus termed γCA-like proteins20,21. Our reconstruction shows that the γCA domain is formed by one copy of γCAL, built as γCAL1 representing the most proximal subunit to the membrane part and two copies of γCA. For the two γCA subunits, one was clearly identified as γCA1 but the scant density on the second, which could be explained by compositional heterogeneity, did not allow to clearly distinguish between γCA1 and γCA2, and was therefore built as γCA1 (Fig. 2a and Supplementary Fig. 6). Each subunits have long N and C-terminal extensions interconnecting the 3 subunits and contacting the membrane subunits of complex I (Fig. 2c). These extensions also serve as anchors for the γCA domain by interacting with the plant specific P2 protein N-terminal part which is anchored in the membrane. The C-terminal part of the canonical Nad6 protein also contacts the CA domain stabilizing it. γCA is an enzyme, where one histidine of one subunit and two histidine of the adjacent subunit contribute to coordinate a zinc atom. However, as γCAL proteins lack two of those three histidines, it impairs the formation of the zinc coordination with both γCA subunits. Hence the only conserved active site is at the interface of the two γCAs, which we confirmed by visualizing both the zinc and an additional density most likely corresponding to HCO3− (Fig. 2b and Supplementary Fig. 6). This strongly suggests that the γCA domain associated with complex I is indeed active, even if a role in HCO3− fixation and/or transport cannot be excluded, similarly to what is observed in cyanobacterial CcmM22. However, the function of complex I-associated γCA is not well understood. It was suggested to play a role in complex I assembly as γCA is found in early assembly intermediates of the complex and is essential for complex I formation23. Indeed, embryo development of γCA mutants is strongly delayed and seed development arrested before maturation. Moreover complemented mutants with inactive γCA variants is sufficient to complement the mutant phenotype, showing that carbonic anhydrase activity is not essential20. Recently it was shown that carbonic anhydrase activity is also present in photosynthetic cyanobacterial complex I, but is carried out by a different type of CA enzyme, positioned at the end of the PD module that would contribute to proton generation24. However, in plant mitochondrial complex I, the only conserved active site is positioned at the most distal position relative to the membrane. Nevertheless, one cannot exclude its contribution to proton generation, although its effect would be relatively modest.
Fig. 2. Hetero-trimeric γ-carbonic anhydrase of the plant mitochondrial complex I.
a–c Front and back view of the carbonic anhydrase showing individual subunits and contacting proteins. Active and inactive sites are shown in a. b Close up of the γCA1/γCA1 only conserved active site, showing the coordinate zinc by the three histidine residues between the two γCA1 subunits. Sequence alignment of γCA and γCAL highlight the catalytic histidines. Surrounding residues, Tyr207, Gln101, Arg86, are also shown and might be involved in HCO3−coordination. c Back view of the carbonic anhydrase, highlighting the N and C-ter extensions of the γCA and γCAL, as well as the proteins allowing to anchor the whole domain to the membrane arm. d The lipid block composed of one central cardiolipin and two phosphatidylinositol molecules is shown in a close-up view, with surrounding residues. The lipids are sandwiched between the carbonic anhydrase domain, the plant-specific protein P2, Nad2, and B14.5. Both inositol head groups contribute to coordinate the cardiolipin. CL stands for cardiolipin and PI for phosphatidylinositol.
Interestingly, the CA domain is in tight interaction with a peculiar lipid complex positioned below the latter and between the plant specific P2 and proteins B14.5b and Nad2, close to the Nad2–Nad4 junction (Fig. 2d). This lipid complex is composed of one cardiolipin coordinated by two phosphatidylinositol. Each inositol heads sandwich the glycerol linker of the cardiolipin and stabilize it. This lipid block was also observed in the assembly intermediate (see below) and appears to be very stable. Given the essential role of lipids for complex I activity, this lipid block might be crucial for the plant complex I efficiency and stability, which might be why CA mutants are so heavily affected8,25. Altogether, our analysis strongly suggest that the γCA is essential for the membrane part formation and stability, acting as an essential architectural factor that would remain even in mature complex.
Assembly intermediate of the plant complex I
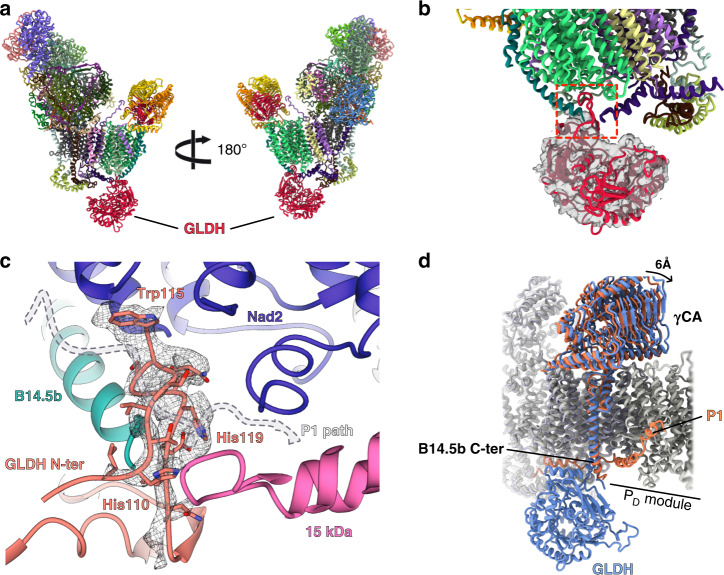
During 3D classification, two classes of complex I lacking the most distal proton pumping components of the membrane domain—Nad4 and Nad5 and their associated additional protein subunits—were found to naturally accumulate in our sample (Supplementary Fig. 1). One of the two classes presented an additional globular density on the intermembrane space exposed side of complex I. Such density wasn’t observed in full complex I classes. This class was thus attributed to an assembly intermediate of complex I (Fig. 3). In plants, such complex was already described to accumulate naturally. Indeed, extensive biochemical analyses identified complex I* as an assembly intermediate lacking the most distal membrane components and presenting an additional factor, GLDH (L-Galactono-1,4-lactone dehydrogenase). GLDH was identified in our mass spectrometry data (Supplementary Table 1), along with the other components of complex I. Likewise γCA, GLDH is an enzyme catalysing the last enzymatic step of the ascorbate biosynthetic pathway in plants26. It was never found in the full complex I, however, it was shown to associate with complex I intermediates, complex I* being the largest, as well as with smaller complex intermediates that were not observed here23. Moreover, in gldh knock-out mutant, complex I is undetectable, which hinted its role as a plant specific assembly factor13,27. Using focused classification and refinement, we were able to derive an intermediate-resolution reconstruction on the additional density corresponding to GLDH which was sufficient to attribute the density to the protein. Importantly, the complex I binding domain of GLDH is resolved at high resolution (3.8), thus validating our assignment (Fig. 3b–d). In this assembly intermediate, GLDH interacts with B14.5b—that is slightly displaced compared to the mature complex—and the 15 kDa subunit. Moreover the whole carbonic anhydrase is slightly shifted toward the missing PD module (Fig. 3c). We also successfully identified the N-terminal tail of GLDH bound to the maturing complex I, between B14.5b and 15 kDa preventing the plant specific protein P1 from docking to the PP module. P1 appears to functionally replace the NDUFB5 subunit, which is absent in plants23 (Supplementary Fig. 5). In the GLDH context the P1 path is blocked in the assembly intermediate due to B14.5b movement and N-ter tail of GLDH (Fig. 3). Thus, it appears that GLDH prevents the binding of P1 to the PP module, until its release, which will then allow association with the PD module. The clear mechanism of action of GLDH in the context of the assembly intermediate is however unclear. It was previously shown that GLDH can perform its original enzymatic activity, however this activity is not required from complex I formation, its functions are thus not linked13. Hence, in plants, GLDH might act as a stabilization factor of complex I assembly intermediates that would be released just before the final step of PD module docking to complex I*. It is unlikely that P1 alone could remove GLDH, thus this process would probably involve unknown additional trans-acting factor(s). Similarly, in yeast the assembly factor NDUFAF2 is released from the assembly intermediate by the joint action of three accessory subunits8.
Fig. 3. GLDH is a plant-specific assembly factor of mitochondrial complex I.
a Atomic model of the assembly intermediate, the whole PD module is missing and GLDH contacts the PP module on the intermembrane side. b GLDH is shown in cut-out and filtered density. The interaction point is highlighted and a zoomed view is shown in c. c Focused view of the interaction point of GLDH with the complex. The N-terminal part of GLDH was identified thanks to specific aromatic residues and is shown in its density. The N-ter part of GLDH, as well as the deviation of B14.5b block P1 path which is shown in dashed lines. d Overall movement in the assembly intermediate compared to the mature complex, B14.5b C-terminal part is deviated by 4 Å and the whole γCA domain is shifted by 6 Å toward the missing PD module, GLDH and absence of the P1 protein are also highlighted. Coral correspond to mature complex I and blue to assembly intermediate.
In conclusion, our high-resolution cryo-EM structures of the plant mitochondrial complex I, reveals plant-specific and ancestral features of this respiratory complex (Fig. 4). Indeed, γCA was identified as a complex I component in all Viridiplantae, including algae of the Chlorophyceae class11,28–31, as well as in Euglena gracilis, a photosynthetic protozoan related to trypanosomes32 and possibly Acanthamoeba castellanii, a protozoan ameboid33 and in non-photosynthetic organisms (e.g., Tetrahymena and Reclinomonas)33 (Fig. 4). Given the wide presence of the γCA in eukaryotes—members of the γCA are found in all eukaryotic lineages except opisthokonts—we present for the first time the structure of a mitochondrial complex I featuring this ancestral component. Moreover, we describe a yet unobserved plant-specific assembly intermediate, shedding light into the respiratory complex I maturation.
Fig. 4. Plant mitochondrial complex I specific subunits.
a Schematic representation of the five major eukaryote lineages. Organisms where γ-carbonic anhydrase is proposed to be part of mitochondrial complex I are listed next to their respective lineage. The red cross indicate the loss of mitochondrial γ-carbonic anhydrase in opisthokonta. b Comparison of mitochondrial complex I structures. Core subunits are shown in light gray and supernumary subunits in light blue. Plant specific proteins are highlighted in red-orange colors. Y. lipolytica (PDB:6RFR) and mammalian (PDB:6G2J) complex I do not have the γ-carbonic anhydrase domain.
Methods
Mitochondrial complex I purification
Cauliflower (Brassica oleracea var. botrytis) mitochondria were purified as previously described12. Quickly, fresh cauliflower inflorescence tissue was blended in extraction buffer containing 0.3 M mannitol, 30 mM sodium pyrophosphate (10.H2O), 0.5% BSA, 0.8% (w/v) polyvinylpyrrolidone-25, 2 mM beta-mercaptoethanol, 1 mM EDTA, 20 mM ascorbate and 5 mM cysteine, pH 7.5. Lysate was filtered and clarified by centrifugation at 1500 × g, 10 min at 4 °C. Supernatant was kept and centrifuged at 18,000 × g, 15 min at 4 °C. Organelle pellet was re-suspended in wash buffer (0.3 M mannitol and 10 mM phosphate buffer, 1 mM EDTA, pH 7.5) and the precedent centrifugations were repeated once. The resulting organelle pellet was re-suspended in wash buffer and loaded on a single-step 30% Percoll gradient (in wash buffer without EDTA) and run for 1h30 at 40,000 × g. Mitochondria were retrieved, washed two times and flash frozen in liquid nitrogen.
For complex I purification, mitochondria were re-suspended in Lysis buffer (20 mM HEPES-KOH, pH 7.6, 100 mM KCl, 1 mM DTT, 2% n-β-DDM, 1 mM EDTA, supplemented with proteases inhibitors (Complete)) incubated for 15 min in 4 °C. Lysate was clarified by centrifugation at 30,000 × g, 20 min at 4 °C. The supernatant was loaded on a 10–50% sucrose gradient buffer (20 mM HEPES-KOH, pH 7.6, 50 mM KCl, 1 mM DTT, 0.2% n-β-DDM, 1 mM EDTA, supplemented with proteases inhibitors (Complete)) and run for 16 h at 75,000 × g. The fraction corresponding to complex I was collected, pelleted and re-suspended in final resuspension buffer (same as sucrose gradient buffer without sucrose and only 0.1% n-β-DDM).
Grid preparation
Four microliter of the samples at a concentration of 1 µg/µl was applied onto Quantifoil R2/2 300-mesh holey carbon grid, coated with thin home-made continuous carbon film and glow-discharged (3 mA for 20 s). The sample was incubated on the grid for 30 s and then blotted with filter paper for 2.5 s in a temperature and humidity controlled Vitrobot Mark IV (T = 4 °C, humidity 100%, blot force 5) followed by vitrification in liquid ethane.
Single particle cryo-electron microscopy data collection
Data collection was performed on a Talos Arctica instrument (Thermofisher Company) at 200 kV using the SerialEM software for automated data acquisition. Data were collected at a nominal underfocus of −0.5 to −2.5 µm at a magnification of ×36,000 yielding a pixel size of 1.11 Å. Micrographs were recorded as movie stack on a K2 direct electron detector (GATAN Company), each movie stack were fractionated into 65 frames for a total exposure of 6.5 s corresponding to an electron dose of 45 ē/Å2.
Electron microscopy image processing
Drift and gain correction and dose weighting were performed using MotionCorr234. A dose weighted average image of the whole stack was used to determine the contrast transfer function with the software Gctf35. The following process has been achieved using RELION 3.014. Particles were picked using the general model of crYOLO 1.536. Output box files from crYOLO were imported in RELION and particles were extracted. After reference-free 2D classification, 650,844 particles were extracted with a box size of 360 pixels and binned threefold, resulting in a 120 pixels box size and used for 3D classification into 6 classes (Supplementary Fig. 1). CryoSPARC37 generated ab-initio cryo-EM map, low-pass filtered to 30 Å, was used as an initial reference for 3D classification. One subclass depicting high-resolution features have been selected for the mature complex I refinement with 65,018 particles. After Bayesian polishing a focused refinement has been performed using mask excluding the PD module yielding, respectively, 3.7 and 3.47 Å resolution. A second subclass containing the maturation factor GLDH composed of 155,644 particles was selected for further focused 3D classification, using a loose soft mask on the GLDH area. The 36,513 particles of GLDH enriched subclass were extracted for refinement and reached 3.8 Å. Determination of the local resolution of the final density map was performed using ResMap38.
Structure building and model refinement
The atomic model of the plant mitochondrial complex I was built into the high-resolution maps using Coot, Phenix, and Chimera. Atomic models from Y.lipolytica (6RFR) and M.musculus (6G2J) were used as starting points for protein identification and modelisation. Similarly to Waltz & Soufari 201912, as the genome of cauliflower is not sequenced, and the closest fully sequenced member of the family (Brassica oleracea subsp. oleracea) is poorly annotated, to facilitate comprehension and analysis we positioned Arabidopsis proteins in the cauliflower map. Still, protein sequence identities between members of the Brassicaceae family are higher than 90%, thus facilitating proteomics identification of cauliflower proteins. The online SWISS-MODEL service was used to generate initial models for bacterial and mitochondria conserved r-proteins. Models were then rigid body fitted to the density in Chimera39 and all subsequent modeling was done in Coot40. Extensions were built has polyalanine and mutated to the adequate sequences. A combination of regularization and real-space refine was performed in Coot for each proteins. The global atomic model was then subjected to real space refinement cycles using phenix.real_space_refine PHENIX41 function, during which protein secondary structure, Ramachandran and side chain rotamer restraints. Several rounds of refinement (manual in Coot and automated using the phenix.real_space_refine) were performed to obtain the final models, which were validated using the built-in validation tool of PHENIX, based on MolProbity. For the assembly intermediate, the model was built with the mature complex as template. Refinement and validation statistics were summarized in Supplementary Table 2.
Proteomic and statistical analyses of mitochondrial complex I composition
Mass spectrometry analyses of the complex I fractions were performed at the Strasbourg-Esplanade proteomic platform and performed as previously42. In brief, proteins were trypsin digested, mass spectrometry analyses and quantitative proteomics were carried out by nanoLC-ESI-MS/MS analysis on a QExactive+ (Thermo) mass spectrometer. Data were searched against the TAIR A. thaliana database with a target-decoy strategy (release TAIRv10, 27281 forward protein sequences), and home-made Brassica oleracea database extracted from UniProtKB (Swissprot+TrEMBL) including Brassica sub-taxonomy (release 2017_10, 59050 forward protein sequences). Proteins were validated respecting FDR < 1% (False Discovery Rate) and quantitative label-free analysis was performed through in-house bioinformatics pipelines.
Figure preparation
Figures featuring cryo-EM densities, as well as atomic models were visualized with UCSF ChimeraX43 and Chimera39.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Descriptions of Additional Supplementary Files
Acknowledgements
This work has benefitted from the facilities and expertize of the Biophysical and Structural Chemistry platform (BPCS) at IECB, CNRS UMS3033, Inserm US001, University of Bordeaux. We thank A. Bezault for assistance with the Talos Arctica electron microscope. We thank, J. Chicher and P. Hamman of the Strasbourg Espanade proteomic analysis for the proteomic analysis. This work was supported by a European Research Council Starting Grant (TransTryp ID:759120) and a Agence Nationale de la Recherche (ANR) grant [MITRA, ANR-16-CE11-0024-02] to Y.H.
Author contributions
F.W., H.S., and Y.H. designed and coordinated the experiments. F.W. purified the mitochondria and mitochondrial complex I. H.S. and C.P. prepared grids. H.S. acquired the cryo-EM data and processed the cryo-EM results with F.W. H.S. and F.W. built the atomic models. L.K. performed the mass-spectrometry experiments. F.W., H.S., and Y.H. interpreted the structure. F.W., H.S., C.P., and Y.H. wrote and edited the manuscript.
Data availability
The cryo-EM maps of plant mitochondrial complex I have been deposited at the Electron Microscopy Data Bank (EMDB): EMD-11614 for the mature complex, EMD-11513 for the focused reconstruction, EMD-11615 for the assembly intermediate. The corresponding atomic models been deposited in the Protein Data Bank (PDB) under the accession 7A23 for the mature complex and 7A24 for the assembly intermediate. Mass spectrometric data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD017847.
Competing interests
The authors declare no competing interests.
Footnotes
Peer review information Nature Communications thanks Friedrich Förster, Kutti Vinothkumar and Eduardo Zabaleta for their contribution to the peer review of this work. Peer review reports are available.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Florent Waltz, Email: florent.waltz@etu.unistra.fr.
Yaser Hashem, Email: yaser.hashem@inserm.fr.
Supplementary information
Supplementary information is available for this paper at 10.1038/s41467-020-18814-w.
References
- 1.Sazanov LA. A giant molecular proton pump: structure and mechanism of respiratory complex I. Nat. Rev. Mol. Cell Biol. 2015;16:375–388. doi: 10.1038/nrm3997. [DOI] [PubMed] [Google Scholar]
- 2.Baradaran R, Berrisford JM, Minhas GS, Sazanov LA. Crystal structure of the entire respiratory complex I. Nature. 2013;494:443–448. doi: 10.1038/nature11871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gray MW. Mitochondrial evolution. Cold Spring Harb. Perspect. Biol. 2012;4:a011403. doi: 10.1101/cshperspect.a011403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gu J, et al. The architecture of the mammalian respirasome. Nature. 2016;537:639–643. doi: 10.1038/nature19359. [DOI] [PubMed] [Google Scholar]
- 5.Agip A-NA, et al. Cryo-EM structures of complex I from mouse heart mitochondria in two biochemically defined states. Nat. Struct. Mol. Biol. 2018;25:548–556. doi: 10.1038/s41594-018-0073-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fiedorczuk K, et al. Atomic structure of the entire mammalian mitochondrial complex I. Nature. 2016;538:406–410. doi: 10.1038/nature19794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhu J, Vinothkumar KR, Hirst J. Structure of mammalian respiratory complex I. Nature. 2016;536:354–358. doi: 10.1038/nature19095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Parey K, et al. High-resolution cryo-EM structures of respiratory complex I: mechanism, assembly, and disease. Sci. Adv. 2019;5:eaax9484. doi: 10.1126/sciadv.aax9484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Parey K, et al. Cryo-EM structure of respiratory complex I at work. Elife. 2018;7:e39213. doi: 10.7554/eLife.39213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eubel H, Heinemeyer J, Braun H-P. Identification and characterization of respirasomes in potato mitochondria. Plant Physiol. 2004;134:1450–1459. doi: 10.1104/pp.103.038018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dudkina NV, Eubel H, Keegstra W, Boekema EJ, Braun H-P. Structure of a mitochondrial supercomplex formed by respiratory-chain complexes I and III. Proc. Natl Acad. Sci. USA. 2005;102:3225–3229. doi: 10.1073/pnas.0408870102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Waltz F, Soufari H, Bochler A, Giegé P, Hashem Y. Cryo-EM structure of the RNA-rich plant mitochondrial ribosome. Nat. Plants. 2020;6:377–383. doi: 10.1038/s41477-020-0631-5. [DOI] [PubMed] [Google Scholar]
- 13.Schimmeyer J, Bock R, Meyer EH. l-Galactono-1,4-lactone dehydrogenase is an assembly factor of the membrane arm of mitochondrial complex I in Arabidopsis. Plant Mol. Biol. 2016;90:117–126. doi: 10.1007/s11103-015-0400-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zivanov J, et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. Elife. 2018;7:e42166. doi: 10.7554/eLife.42166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Letts JA, Fiedorczuk K, Sazanov LA. The architecture of respiratory supercomplexes. Nature. 2016;537:644–648. doi: 10.1038/nature19774. [DOI] [PubMed] [Google Scholar]
- 16.Momayyezi, M., McKown, A. D., Bell, S. C. S. & Guy, R. D. Emerging roles for carbonic anhydrase in mesophyll conductance and photosynthesis. Plant J. tpj.14638 10.1111/tpj.14638 (2020). [DOI] [PubMed]
- 17.Tripp BC, Bell CB, Cruz F, Krebs C, Ferry JG. A role for iron in an ancient carbonic anhydrase. J. Biol. Chem. 2004;279:6683–6687. doi: 10.1074/jbc.M311648200. [DOI] [PubMed] [Google Scholar]
- 18.Alber BE, et al. Kinetic and spectroscopic characterization of the gamma-carbonic anhydrase from the methanoarchaeon methanosarcina thermophila. Biochemistry. 1999;38:13119–13128. doi: 10.1021/bi9828876. [DOI] [PubMed] [Google Scholar]
- 19.Parisi G, et al. Gamma carbonic anhydrases in plant mitochondria. Plant Mol. Biol. 2004;55:193–207. doi: 10.1007/s11103-004-0149-7. [DOI] [PubMed] [Google Scholar]
- 20.Fromm S, Senkler J, Zabaleta E, Peterhänsel C, Braun H-P. The carbonic anhydrase domain of plant mitochondrial complex I. Physiol. Plant. 2016;157:289–296. doi: 10.1111/ppl.12424. [DOI] [PubMed] [Google Scholar]
- 21.Perales M, et al. Gamma carbonic anhydrase like complex interact with plant mitochondrial complex I. Plant Mol. Biol. 2004;56:947–957. doi: 10.1007/s11103-004-6324-z. [DOI] [PubMed] [Google Scholar]
- 22.Cot SSW, So AKC, Espie GS. A multiprotein bicarbonate dehydration complex essential to carboxysome function in cyanobacteria. J. Bacteriol. 2008;190:936–945. doi: 10.1128/JB.01283-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ligas J, Pineau E, Bock R, Huynen MA, Meyer EH. The assembly pathway of complex I in Arabidopsis thaliana. Plant J. 2019;97:447–459. doi: 10.1111/tpj.14133. [DOI] [PubMed] [Google Scholar]
- 24.Schuller JM, et al. Redox-coupled proton pumping drives carbon concentration in the photosynthetic complex I. Nat. Commun. 2020;11:494. doi: 10.1038/s41467-020-14347-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jussupow A, Di Luca A, Kaila VRI. How cardiolipin modulates the dynamics of respiratory complex I. Sci. Adv. 2019;5:eaav1850. doi: 10.1126/sciadv.aav1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Millar AH, et al. Control of ascorbate synthesis by respiration and its implications for stress responses. Plant Physiol. 2003;133:443–447. doi: 10.1104/pp.103.028399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pineau B, Layoune O, Danon A, De Paepe R. l-Galactono-1,4-lactone dehydrogenase is required for the accumulation of plant respiratory complex I. J. Biol. Chem. 2008;283:32500–32505. doi: 10.1074/jbc.M805320200. [DOI] [PubMed] [Google Scholar]
- 28.Davies KM, Blum TB, Kühlbrandt W. Conserved in situ arrangement of complex I and III2 in mitochondrial respiratory chain supercomplexes of mammals, yeast, and plants. Proc. Natl Acad. Sci. USA. 2018;115:3024–3029. doi: 10.1073/pnas.1720702115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Subrahmanian N, Remacle C, Hamel PP. Plant mitochondrial Complex I composition and assembly: a review. Biochim. Biophys. Acta Bioenerg. 2016;1857:1001–1014. doi: 10.1016/j.bbabio.2016.01.009. [DOI] [PubMed] [Google Scholar]
- 30.Sunderhaus S, et al. Carbonic anhydrase subunits form a matrix-exposed domain attached to the membrane arm of mitochondrial complex I in plants. J. Biol. Chem. 2006;281:6482–6488. doi: 10.1074/jbc.M511542200. [DOI] [PubMed] [Google Scholar]
- 31.Klodmann J, Sunderhaus S, Nimtz M, Jansch L, Braun HP. Internal Architecture of Mitochondrial Complex I from Arabidopsis thaliana. Plant Cell. 2010;22:797–810. doi: 10.1105/tpc.109.073726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miranda-Astudillo HV, et al. The atypical subunit composition of respiratory complexes I and IV is associated with original extra structural domains in Euglena gracilis. Sci. Rep. 2018;8:9698. doi: 10.1038/s41598-018-28039-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gawryluk RMR, Gray MW. Evidence for an early evolutionary emergence of gamma-type carbonic anhydrases as components of mitochondrial respiratory complex I. BMC Evol. Biol. 2010;10:176. doi: 10.1186/1471-2148-10-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zheng SQ, et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods. 2017;14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang K. Gctf: Real-time CTF determination and correction. J. Struct. Biol. 2016;193:1–12. doi: 10.1016/j.jsb.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wagner T, et al. SPHIRE-crYOLO is a fast and accurate fully automated particle picker for cryo-EM. Commun. Biol. 2019;2:218. doi: 10.1038/s42003-019-0437-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Punjani A, Rubinstein JL, Fleet DJ, Brubaker M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods. 2017;14:290–296. doi: 10.1038/nmeth.4169. [DOI] [PubMed] [Google Scholar]
- 38.Kucukelbir A, Sigworth FJ, Tagare HD. Quantifying the local resolution of cryo-EM density maps. Nat. Methods. 2014;11:63–65. doi: 10.1038/nmeth.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pettersen EF, et al. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 40.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr. Sect. D. Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liebschner D, et al. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr. Sect. D. Struct. Biol. 2019;75:861–877. doi: 10.1107/S2059798319011471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Waltz F, et al. Small is big in Arabidopsis mitochondrial ribosome. Nat. Plants. 2019;5:106–117. doi: 10.1038/s41477-018-0339-y. [DOI] [PubMed] [Google Scholar]
- 43.Goddard TD, et al. UCSF ChimeraX: meeting modern challenges in visualization and analysis. Protein Sci. 2018;27:14–25. doi: 10.1002/pro.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Descriptions of Additional Supplementary Files
Data Availability Statement
The cryo-EM maps of plant mitochondrial complex I have been deposited at the Electron Microscopy Data Bank (EMDB): EMD-11614 for the mature complex, EMD-11513 for the focused reconstruction, EMD-11615 for the assembly intermediate. The corresponding atomic models been deposited in the Protein Data Bank (PDB) under the accession 7A23 for the mature complex and 7A24 for the assembly intermediate. Mass spectrometric data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD017847.