Abstract
Background
Multiple studies of depression indicated a significant role of gene-by-environment interactions; however, they are mainly limited to the examination of modulating effect of recent stressful life events. Other environmental factors occurring at different stages of ante- and postnatal development may affect the association between multiple genes and depression. The study aimed to analyze the main and haplotype-based effect of serotonergic system and HPA-axis gene polymorphisms on depression and to detect gene-by-environment interaction models explaining individual variance in depression in mentally healthy young adults from Russia.
Methods
Depression score was assessed using Beck Depression Inventory (BDI) in 623 healthy individuals (81% women; 17-25 years) of Caucasian origin (Russians, Tatars, Udmurts) from Russia. The main- and gene-based effects of 12 SNPs in SLC6A4 (5-HTTLPR, rs1042173), HTR2A (rs7322347), OXTR (rs7632287, rs2254298, rs13316193, rs53576, rs2228485, rs237911), AVPR1A (rs3803107, rs1042615), and AVPR1B (rs33911258) genes, and gene-by-environment interactions were tested with linear regression models (PLINK v.1.9) adjusted for multiple comparisons.
Results
We observed ethnicity-specific main effect of the AVPR1A rs3803107 (P = 0.003; PFDR = 0.047) and gene-based effect of the OXTR gene (Р = 0.005; Pperm = 0.034) on BDI-measured depression, and modifying effect of paternal care on OXTR rs53576 (P = 0.004; PFDR = 0.012) and birth order on OXTR rs237911 (P = 0.006; PFDR = 0.018) association with depression level.
Limitations
A hypothesis driven candidate gene approach, which examined a limited number of genetic variants in a moderately large sample, was used.
Conclusions
Our preliminary findings indicate that familial environment may play a permissive role modulating the manifestation of OXTR-based depression variance in mentally healthy subjects.
Keywords: Oxytocin, Arginine vasopressin, Serotonin, Depression, Family environment, GxE interaction, Behavioral neuroscience, Molecular neuroscience, Mental disorder, Genomics, Polymerase chain reaction, Human genetics, Biological psychiatry, Mental health, Psychological disorders
oxytocin; arginine vasopressin; serotonin; Depression; family environment; GxE interaction; Behavioral Neuroscience; Molecular Neuroscience; Mental Disorder; Genomics; Polymerase Chain Reaction; Human Genetics; Biological Psychiatry; Mental Health; Psychological Disorders.
1. Introduction
Major depression (MD) and depressive-like behavior represent important public health issues due to their high prevalence and incidence in the population (Mandelli and Serretti, 2013). Considering high economic costs of depression to society and a high frequency of depression-like behavior in non-clinical forms in population (10-15% of the general population experience a clinical depressive episode in their lifetime), the study of etiology of depression in population is of high relevance. According to family, twin, epidemiological, and molecular studies, depression is a multifactorial illness showing a highly complex genetic architecture with a large number of loci, each contributing a very small effect size to the phenotype (Gonda et al., 2018). Genetic factors account for ~30% in childhood increasing to ~40% in adolescence, while shared environmental influences decrease from 70% to 48% in the same period and individual environment accounts for a significant proportion of the association in adolescence only (~12%) in depression etiology (Hannigan et al., 2017).
During the past decades manifold biological factors and candidate genes have being examined in major depression. Although recent advances in this field such as consortium-based cohorts for GWAS of major depressive disorder (MDD) identified distinct SNPs at genome-wide significance level in case-control studies (Coleman et al., 2020; Howard et al., 2019; Mullins and Lewis, 2017), GWAS unconsidered gene-environment interaction (GxE) effects except for two research groups, which have applied a genome-wide by SLEs (stressful life events) interaction studies (GWEIS) to improve the percent of variance explained in depression liability in Scotland-UK cohort (Arnau-Soler et al., 2019), African Americans and Hispanics/Latinas (Dunn et al., 2016). However, together with recent SLEs other environmental factors occurring during antenatal and childhood periods of development were reported to modulate genetic liability to depression (Brummett et al., 2008; Kendler et al., 2018; Lahat et al., 2017; Lin and Tsai, 2019; Starr and Huang, 2019).
Candidate gene association studies represent another approach, which can deal with GxE interaction involving individual differences in manifold environmental factors. In particular, such studies have been mainly focused on the involvement of childhood adversity in later depression development. For instance, childhood trauma and maltreatment positively correlate with the strength of GxE interactions involving low-activity serotonergic system associated with low-expressing SLC6A4 and HTR2A gene variants (for review see (Lin and Tsai, 2019; Starr et al., 2019)). The genes belonging to hypothalamic-pituitary-adrenal (HPA) system (FKBP5, CRHR1, CRHBP) were also shown to significantly interact with traumatic life events, physical abuse and childhood maltreatment to affect depression (Lin and Tsai, 2019; Starr and Huang, 2019). However, other researchers failed to detect such effects (Culverhouse et al., 2018; Van der Auwera et al., 2017). Between study heterogeneity may be attributed to inherited epigenetic patterns (Jiang et al., 2019) and environmental factors other than childhood trauma, which cause induced differential health outcomes via epigenetic reprogramming (Jiang et al., 2019). Therefore, some scientists have focused on other psychosocial factors including peculiarities of child-parent relations, familial support, parenting behavior (Cao et al., 2018; Van Assche et al., 2016), and peer relationships with respect to depressive symptomatology; however, such attempts are scarce. One of recent large-scale studies evidence in the parent-offspring resemblance in MD and an additive effect of genetic factors and rearing experiences (Kendler et al., 2018). Functional studies suggested that parenting behaviors affected the neural correlates of emotion processing in children. Specifically, an interactive effect of parenting behaviors and HPA axis-related genes on amygdala activity and connectivity during emotion processing, and, in turn, on internalizing symptoms in children was reported (Pozzi et al., 2019). Together with parenting style, a significant effect of birth order (Easey et al., 2019), socioeconomic status (SES) (Brummett et al., 2008), season of birth (Kazantseva et al., 2015), gestational age (preterm birth) (Johns et al., 2019) and/or extremely low birth weight (ELBW, less than 1000 g) (Lahat et al., 2017; Mathewson et al., 2017) interacting with genetic factors on depressive symptoms was also established. Prenatal factors such as maternal nutrition (House et al., 2018), inflammatory response to infection, maternal depression (Nemoda and Szyf, 2017), maternal age at birth (Miller et al., 2019), and smoking (Kuja-Halkola et al., 2014) during gestation have been examined for their impact on offspring's behavioral problems under GxE interactions. These observations can be partially explained by differential methylation in genes involved in HPA-axis and immune functions, which may cause later mental health problems in children (Kantake et al., 2014; Nemoda and Szyf, 2017). In addition, a significant effect of addictive behavior (smoking, alcohol abuse/dependence) during adulthood on developing depression in a predictive framework based on interactions of multiple functional genetic variants has been reported (Schmitz et al., 2019; Tylee et al., 2018; Wong et al., 2012). In turn, increased exposure to smoking may significantly affect epigenetic changes in HPA-axis (Dogan et al., 2016) and serotonergic genes (Smolka et al., 2019).
Although the abovementioned research evidence in a significant role of family environment in stress and depression sensitivity, the GxE studies have been mainly focused on the role of the SLC6A4 and HPA-axis genes (NR3C1, NR3C2, CRHR1, FKBP5) in clinical manifestation of depression. Moreover, there is a controversy in GxE effect of the most examined 5-HTTLPR (SLC6A4) and SLEs (Coventry et al., 2010; Gonda et al., 2018; Mandelli and Serretti, 2013) thus indicating the possibility of moderating effect of other SNPs in stress-related depression liability (Kazantseva et al., 2008). Recently, several studies in the field of psychopathology have been focused on the analysis of rs1042173 located in the 3′-UTR regulatory region of the SLC6A4 (Resnick et al., 2015; Wang et al., 2018), since differential expression of the SLC6A4 gene caused by rs1042173 variants was reported (Seneviratne et al., 2009). A functional rationale to examine the role of another serotonergic system gene (HTR2A) in depressive traits involves the data on differential HTR2A level detected in neocortex of individuals with depression and suicidal behavior varying on childhood adversity (Underwood et al., 2018). The most widely studied genetic variant of the HTR2A gene (rs6311) have been implicated in depression development (Dressler et al., 2009; Gonda et al., 2018; Jokela et al., 2007). However, other genetic variants located in regulatory gene regions may be relevant to individual differences in depression. For instance, the main effect of rs7322347 in the HTR2A gene on increased aggression (Banlaki et al., 2015) and GxE effect of rs7322347×physical assault in childhood/adolescence on suicidal attempts (Ben-Efraim et al., 2013) were reported.
It should be noted that studies examining interaction between family environment and other HPA-axis genes (AVPR1A, AVPR1B) promoting individual sensitivity to stressors and, therefore, depression liability are absent to date. However, it is known that stress reaction causes enhanced cortisol release, which is regulated by the HPA axis and influenced by oxytocin (OXT) and arginine vasopressin (AVP) (Holmqvist Jämsen et al., 2017). Since activation of AVP and OXT receptors oppositely affects fear and anxiety-related behaviors, we can suggest that variations in AVPR1A and AVPR1B genes associated with their upregulation and lower OXTR gene expression may result in higher depressive-like behavior. To date multiple studies have focused on AVPR1A VNTR polymorphisms (RS1 and RS3) located in the 5′-flanking region and their relation to anxiety and social behavior (Procyshyn et al., 2017; Tansey et al., 2011; Yang et al., 2017). However, the studies on other regulatory regions (for instance, 3′-UTR) in the AVPR1A gene remain scarce. Namely, just few attempts to find relations between AVPR1A rs1042615 and rs3803107 and stress sensitivity were performed (Bernhard et al., 2016; Holmqvist Jämsen et al., 2017). Although studies dedicated to the involvement of AVPR1B SNPs in mood states (Gonzalez et al., 2019), emotional empathy and prosociality (Wu et al., 2015), MDD (Ben-Efraim et al., 2013; Szczepankiewicz et al., 2013) and personality traits (Kazantseva et al., 2014) were published, a pervasive modulating effect of environmental factors on AVPR1B gene variants has to be explored.
The presence of prominent sex-specific differences in depression (Labonté et al., 2017) dictates the analysis of genes encoding sex hormones including oxytocin receptor gene (OXTR) under GxE paradigm. Since oxytocin was shown to regulate different social behaviors including social recognition, affiliation and response to threat (Meyer-Lindenberg et al., 2011; Skuse and Gallagher, 2009) via regulation of HPA axis and attenuate amygdala's response to stress (Bernhard et al., 2016), the OXTR gene became explored with respect to depressive mood, stress reactivity, antisocial behavior, emotional loneliness (Inoue et al., 2010; Kang et al., 2017; Kawamura et al., 2010; LoParo et al., 2016; Lucht et al., 2009; Thompson et al., 2011). Congruent with a suggestion that variations in the OXTR gene may differentially affect oxytocin regulation moderated by the effect of family environment, several studies clarified the effect of gene-by-environment interactions based on the OXTR gene variants in clinical samples (Asherin et al., 2019; Choi et al., 2019; Elwood et al., 2019; Park et al., 2019; Parris et al., 2018). However, no findings of OXTR-by-environment effect on depressive-like behavior in non-clinical cohort were published to date.
Given that multiple common gene variants together with environmental factors are associated with clinical forms of depression, we sought to investigate the association of the serotonergic (SLC6A4, HTR2A) and HPA-axis genes (AVPR1A, AVPR1B, OXTR) with depression level in mentally healthy young adults from the Russian population. We aimed to analyze the main and haplotype-based effect of candidate gene polymorphisms on depression within the framework of stress-diathesis and plasticity gene models and to detect gene-by-environment interactions explaining individual variances in depression level. We hypothesized that: 1) sex and ethnicity modulate main/gene-based effect on depression; 2) environmental factors occurring within ante- and postnatal development may modulate the association between gene variants and depression score.
2. Materials and methods
2.1. Subjects
In total, 623 healthy individuals (81% women) from Russia comprised the sample. All participants were young adults (mean age ±SD: 19.53 ± 1.75 years, age range: 17-25 years), enrolled at the Universities in Russia. All participants were of Caucasian origin, from the Russian population (Slavic group of the Indo-European language family) (N = 225), Tatar population (Turkic group of the Altaic language family) (N = 141), Udmurt population (Finno-Ugric group of the Finn-Permian Branch of the Uralic language family) (N = 218) and individuals of mixed ethnicity (N = 39). Exclusion criteria were a self-reported individual or family history of any psychiatric disorder. Socio-demographic data including sex, ethnicity (by 3 generations), place of residence, birth order, number of children in the family (sibship size), family income, rearing in a full family (yes/no), maltreatment in childhood, bilingual rearing, the presence of severe chronic disease, smoking, weight at birth, mother age at birth were obtained from the participants. The study was approved by the Biological Ethics Committee at the Institute of Biochemistry and Genetics – Subdivision of the Ufa Federal Research Centre of Russian Academy of Sciences (Ufa, Russia), and written informed consent was obtained from all the participants after they were acquainted with all the procedures. All participants were informed about the voluntary and confidential nature of their participation. All procedures performed were in accordance with the Helsinki Declaration as revised 1989.
2.2. Psychological measures
Depression score was assessed using the Russian version of self-report Beck Depression Inventory (BDI) representing 21 multiple-choice questionnaire, which measures the severity of depression and consists of a cognitive-affective and somatic depression subscales. To assess the style of parental rearing, Parental Bonding Instrument (PBI) (Parker et al., 1979) was used, which consists of 25 items and estimates two bipolar scales (care and protection) with respect to maternal and paternal style of parenting.
Involved individuals were residents of the Republic of Bashkortostan (population = 4.072 million of citizens, 60.4% of urban status) and the Udmurt Republic (population = 1.517 million of citizens, 65% of urban status) of Russia. Place of childhood residence was determined as urban/rural. Urban status was given to individuals from medium and large metropolitan regions with a population between 60 000 and 1 100 000; while individuals from the localities with a population lower than 60 000 were recognized of rural status.
2.3. SNPs selection and genotyping
Genomic DNA was isolated from the whole blood using a standard phenol-chlorophorm technique. In total, 12 SNPs with a minor allele frequency above 5% (rs7632287, rs2254298, rs13316193, rs53576, rs2228485, rs237911 in the OXTR gene, rs3803107 and rs1042615 in the AVPR1A gene, rs33911258 in the AVPR1B gene, rs7322347 in the HTR2A gene, and 5-HTTLPR (rs4795541), rs1042173 in the SLC6A4 gene) were selected based on their relation to depression and anxiety-related behavior in previously published studies or due to their location in functional gene regions (5′-UTRs, 3′-UTRs) for their possible involvement in the regulation of gene expression. Six SNPs in the OXTR gene were genotyped for the better gene coverage to perform the haplotype-based association analysis to obtain higher statistical power. All SNPs demonstrated sufficient call rates and no deviation from Hardy-Weinberg equilibrium except for OXTR rs13316193 (P < 0.01) was detected.
Genotyping of examined SNPs was performed using a real-time PCR based on TaqMan technology using oligonucleotide probes with chemical modifications of locked nuclear acids (TaqMan-LNA) with subsequent fluorescent detection via a fluorescent resonance energy transfer approach (TestGen, Russia). DNA samples were amplified in a total volume of 10 μl with 20–50 ng of genomic DNA, Taq polymerase and Master Mix (TestGen, Russia) containing two DNA probes (to two alleles of the SNP) marked with different fluorescent labels and fluorescence absorbers. Alleles’ assignment was conducted via fluorescence end-point analysis using CFX96 Touch™ Real-Time PCR Detection System (BioRad, USA).
2.4. Statistical analysis
Genotype and allele frequencies of investigated SNPs as well as the Hardy-Weinberg equilibrium calculations were performed in a total sample (Table 1) using PLINK v.1.9 (Purcell et al., 2007). Haplotype blocks were delineated using the confidence interval method of Gabriel et al. (2002), and measures of linkage disequilibrium (LD, standardized D′) between markers were obtained using Haploview 4.2. Haplotypes with a frequency less than 1% were excluded from the further analysis.
Table 1.
The investigated SNPs.
| Gene | SNP | Chromosomal position, bp∗ | Location in gene | Minor allele/Major allele | Genotype frequency | PHWE | ||
|---|---|---|---|---|---|---|---|---|
| AVPR1A | rs3803107 | 63147054 | 3′-UTR | A/G | 0.025 | 0.280 | 0.695 | 0.775 |
| rs1042615 | 63150429 | exon 1 | A/G | 0.156 | 0.486 | 0.358 | 0.798 | |
| AVPR1B | rs33911258 | 206118034 | 5′-UTR | G/A | 0.020 | 0.293 | 0.687 | 0.201 |
| OXTR | rs7632287 | 8749760 | 3′-UTR | A/G | 0.041 | 0.318 | 0.642 | 0.901 |
| rs2254298 | 8760542 | intron 1 | A/G | 0.003 | 0.164 | 0.833 | 0.299 | |
| rs13316193 | 8761057 | intron 1 | C/T | 0.255 | 0.401 | 0.344 | <0.01 | |
| rs53576 | 8762685 | intron 1 | A/G | 0.213 | 0.498 | 0.290 | 1 | |
| rs2228485 | 8768017 | exon 1 | C/T | 0.042 | 0.283 | 0.674 | 0.149 | |
| rs237911 | 8768322 | 5′-UTR | G/A | 0.030 | 0.264 | 0.706 | 0.559 | |
| HTR2A | rs7322347 | 46835968 | intron 2 | A/T | 0.095 | 0.473 | 0.432 | 0.108 |
| SLC6A4 | 5-HTTLPR (rs4795541) | 30237299 | 5′-UTR | L/S | 0.235 | 0.490 | 0.275 | 0.635 |
| rs1042173 | 30197993 | 3′-UTR | T/G | 0.206 | 0.493 | 0.301 | 0.936 | |
according to NCBI36 genome build 36.3. PHWE – P-value for Hardy-Weinberg equilibrium test. UTR - untranslated region.
To test for the normality of distribution of the quantitative data (depression score, age, weight at birth and mother age at birth), Kolmogorov-Smirnov's test was used (SPSS v.23). Due to deviation of the depression score from normality, the Mann-Whitney U test and Kruskal–Wallis H test was used to estimate the influence of stress-related environmental factors, sex and ethnicity on depression (SPSS v.23). For independent categorical variables with a number of categories higher than two, a matrix of binary dummy variables was constructed with PLINK v.1.9, which were later used in the linear regression analysis. The main effects of the individual genotypes and haplotypes on depression were investigated using linear regression models adjusted for sex and ethnicity as covariates in a total sample followed by sex- and ethnicity-stratified analysis with PLINK v.1.9. The best statistical model (among additive, dominant, recessive, and dominance deviation from additivity ones) was selected based on Akaike information criterion (AIC). Multiple linear regression models controlled for sex and ethnicity were analyzed to estimate gene-by-environment interactions to test for the modulating effects of stress-related environmental factors and PBI scores on depression in healthy individuals. For the interaction effects, those with P-value less than 0.05 were considered for stratification analysis. Three gene-by-environment models were constructed: (1) Model 1 containing the main effect of SNP, sex and ethnicity, together with SNP-by-sex and SNP-by-ethnicity interaction terms; (2) Model 2 containing the main effect of SNP, sex, ethnicity and environmental factor together with SNP-by-environment interaction term; and (3) Model 3 containing the main effect of SNP, sex, ethnicity and environmental factor together with SNP-by-environment, SNP-by-sex and SNP-by-ethnicity interaction terms. For the models demonstrating an interaction effect of a specific SNP and environmental stress-related factor on depression score, we conducted stratification analysis to clarify the direction of the effect.
As multiple positive findings were expected, correction for multiple testing was performed via false discovery rate (FDR) procedure (Benjamini and Hochberg, 1995) for genotype-based effect, while permutation (10000) test was used for haplotype-based associations. Corrected P-values (PFDR, Pperm) are shown in the present study. Effect sizes were calculated for all statistical models. The effect sizes were reported as r2, which describes the proportion of variance in BDI-measured depression that is accounted for the differences in genotype or haplotype controlling for sex and ethnicity.
3. Results
3.1. Sample characteristics
Since depression score in our sample didn't coincide with the Gaussian distribution (P < 0.05), non-parametric tests (Mann-Whitney-Wilcoxon U test and Kruskal–Wallis H test) were used to estimate the association between stress-related and socio-demographic parameters and depression level in mentally healthy individuals. The descriptive statistics of the examined sample is shown in Table 2. Thus, sex (P = 0.019), ethnicity (Р = 0.001) together with such socio-demographic factors as birth order (Р = 0.017), sibship size (Р = 0.017), maltreatment in childhood (Р = 0.004), a presence of severe chronic disorders (Р = 0.033), maternal care (Р < 0.001) and protection (Р = 0.002), as well as paternal care (Р = 0.010) significantly affected depression score (Table 2). In particular, significantly higher depression level was more likely observed in women and/or in individuals who were the only child in a family and/or those reported childhood maltreatment or severe chronic disease. At the same time, a decreased mean depression score was significantly more prominent in third-born individuals. Congruent with previous research, participants with low maternal care and overprotection together with low paternal care scored significantly higher on depression compared to those, who reported the opposite style of parenting. A linear regression analysis conducted to estimate the influence of individual age, weight at birth (1500–4950 g) and maternal age at birth (16–44 years) on depression level revealed statistically significant effect of age (β = -0.408, Р = 0.010), whereas weight at birth (β < 0.001, Р = 0.863) and maternal age at birth (β = -0.508, Р = 0.309) failed to affect individual's variance in depression.
Table 2.
The sample structure according to examined socio-demographic parameters, mean depression score and Mann-Whitney U-test on depression score.
| Parameter | N (%) | Mean score ±SD | Mann-Whitney test (p)a |
|---|---|---|---|
| Sex | |||
| Men | 118 (18.90) | 7.23 ± 6.62 |
0.019 |
| Women |
505 (81.10) |
8.63 ± 6,98 |
|
| Ethnicity | |||
| Russians | 225 (36.11) | 8.78 ± 7.71 | 0.814 |
| Tatars | 141 (22.63) | 6.61 ± 5.60 | 0.001 |
| Udmurts | 218 (35.00) | 8.55 ± 6.36 | 0.134 |
| Others (mixed ethnicity) |
39 (6.26) |
11.54 ± 8.38 |
0.014 |
| Residence | |||
| Urban | 329 (52.81) | 8.28 ± 7.53 | 0.589 |
| Rural |
294 (47.19) |
8.03 ± 6.02 |
|
| Order of birth | |||
| 1 | 367 (58.91) | 8.32 ± 7.01 | 0.976 |
| 2 | 195 (31.30) | 8.69 ± 6.65 | 0.123 |
| >3 |
61 (9.79) |
6.50 ± 6.44 |
0.017 |
| Number of children in family | |||
| 1 | 130 (20.87) | 10.17 ± 8.20 | 0.017 |
| 2 | 302 (48.47) | 7.41 ± 6.01 | 0.116 |
| >3 |
191 (30.66) |
7.88 ± 6.69 |
0.685 |
| Family income | |||
| lower than average | 78 (12.52) | 9.38 ± 8.41 | 0.379 |
| average | 503 (80.74) | 7.99 ± 6.59 | 0.322 |
| higher than average |
42 (6.74) |
8.09 ± 5.64 |
0.689 |
| Rearing in full family | |||
| yes | 510 (81.86) | 8.18 ± 6.78 | 0.974 |
| no |
113 (18.14) |
8.37 ± 7.06 |
|
| Maltreatment | |||
| yes | 81 (13.00) | 10.58 ± 8.00 |
0.004 |
| no |
542 (87.00) |
7.70 ± 6.74 |
|
| Bilingual | |||
| yes | 326 (52.33) | 7.16 ± 5.61 | 0.498 |
| no |
297 (47.67) |
8.25 ± 7.48 |
|
| Chronic disease | |||
| yes | 172 (27.61) | 9.27 ± 7.68 |
0.033 |
| no |
451 (72.39) |
7.64 ± 6.64 |
|
| Smoking | |||
| yes | 42 (6.74) | 9.22 ± 8.37 | 0.796 |
| previously | 58 (9.31) | 10.39 ± 8.42 | 0.064 |
| no |
523 (83.95) |
7.93 ± 6.49 |
0.096 |
| Maternal care | |||
| high | 452 (72.55) | 7.26 ± 6.10 |
<0.001 |
| low |
171 (27.45) |
11.48 ± 9.01 |
|
| Maternal protection | |||
| high | 361 (57.95) | 9.39 ± 7.75 |
0.002 |
| low |
262 (42.05) |
7.06 ± 6.28 |
|
| Paternal care | |||
| high | 331 (53.13) | 7.41 ± 6.42 |
0.010 |
| low |
292 (46.87) |
9.60 ± 7.87 |
|
| Paternal protection | |||
| high | 282 (45.27) | 8.89 ± 8.23 | 0.854 |
| low | 341 (54.73) | 7.95 ± 6.15 | |
| Age (17–25 years) | 623 (100) | 19.53 ± 1.75 | 0.010b |
| Weight (1500–4950 g) | 623 (100) | 3381 ± 533 | 0.863b |
| Mother Age (16–44 years) | 623 (100) | 25.61 ± 5.43 | 0.309b |
Mann-Whitney U-test was performed for dummy variables (one variable vs others) in the case of a number of categorical variables higher than 2.
Linear regression analysis was performed for quantitative data instead of non-parametric Mann-Whitney test. SD - standard deviation. Statistically significant P-values are marked in bold.
3.2. Genotype-based association analysis
No statistically significant differences in allele and genotype frequencies distribution in all examined loci were detected between individuals of different ethnicity and sex (P > 0.05). Hence, the main effect estimate was carried out in both total sample and certain ethnic groups (Russians, Tatars, and Udmurts) and men and women separately. While testing for the main effects of the examined gene polymorphisms on BDI-measured depression, we detected associations between the rs3803107 A-allele of the AVPR1A gene and increased depression in the dominant model (AA + AG vs GG) in both total sample (β = 1,241; P = 0.046; PFDR = 0.644; r2 = 0.005), women (β = 1.468; P = 0.035; PFDR = 0.415; r2 = 0.008), and individuals of Russian ethnicity (β = 3.596; P = 0.003; PFDR = 0.047; r2 = 0.031) (Table 3). However, only the last association remained statistically significant after FDR-correction for multiple comparisons. We also detected a trend for sex-specific effect of 5-HTTLPR in the SLC6A4 gene and rs2228485 in the OXTR gene on depression level. Namely, men bearing 5-HTTLPR LL genotype under recessive model scored significantly higher on this trait compared to other genotype carriers (β = 3.798; P = 0.009; PFDR = 0.135; r2 = 0.044), while men (β = -2.684; P = 0.035; PFDR = 0.491; r2 = 0.036) or individuals of Tatar ethnic origin (β = -2.411; P = 0.019; PFDR = 0.139; r2 = 0.029) with rs2228485 С-allele demonstrated reduced depression level under dominant statistical model (Table 3). However, these associations became non-significant after FDR-correction.
Table 3.
Main effect of examined SNPs on depression score in different groups based on linear regression analysis∗.
| Gene | SNP (MiA/MaA) | Modela | Total (N = 623) |
Women (N = 504) |
Men (N = 117) |
Russians (N = 225) |
Udmurts (N = 218 |
Tatars (N = 141) |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| β | P-value | β | P-value | β | P-value | β | P-value | β | P-value | β | P-value | |||
| AVPR1A | rs3803107 A/G |
dominant | 1.241 | 0.046 | 1.468 | 0.035 | 0.763 | 0.570 | 3.596 | 0.003b | -0.059 | 0.948 | 0.080 | 0.941 |
| AVPR1B | rs1042615 A/G |
additive | -0.139 | 0.748 | 0.091 | 0.850 | -1.330 | 0.164 | -0.929 | 0.225 | 0.056 | 0.929 | -0.466 | 0.602 |
| rs33911258 G/A |
domdev | 0.711 | 0.238 | 0.634 | 0.294 | 0.832 | 0.532 | 0.041 | 0.971 | 1.456 | 0.426 | 2.101 | 0.047 | |
| OXTR | rs7632287 A/G |
additive | -0.013 | 0.980 | 0.199 | 0.728 | -0.544 | 0.588 | 0.030 | 0.973 | 0.027 | 0.981 | -1.177 | 0.180 |
| rs2254298 A/G |
additive | -0.808 | 0.273 | -1.016 | 0.226 | 0.290 | 0.850 | -1.192 | 0.378 | -0.855 | 0.530 | 0.016 | 0.989 | |
| rs53576 A/G |
additive | -0.053 | 0.894 | 0.189 | 0.675 | -0.875 | 0.305 | -0.141 | 0.853 | 0.411 | 0.522 | 0.950 | 0.146 | |
| rs2228485 C/T |
dominant | -0.679 | 0.258 | -0.081 | 0.905 | -2.684 | 0.035 | -0.834 | 0.444 | -0.243 | 0.800 | -2.411 | 0.019 | |
| rs237911 G/A |
domdev | 0.498 | 0.427 | 0.459 | 0.465 | -2.055 | 0.131 | 3.419 | 0.085 | 0.224 | 0.882 | -2.141 | 0.048 | |
| HTR2A | rs7322347 A/T |
additive | 0.741 | 0.094 | 0.779 | 0.105 | 0.588 | 0.602 | 0.257 | 0.746 | 0.995 | 0.199 | 0.475 | 0.550 |
| SLC6A4 | rs4795541 L/S |
recessive | 0.520 | 0.441 | -0.199 | 0.790 | 3.798 | 0.009 | 1.358 | 0.298 | 0.066 | 0.958 | -0.312 | 0.792 |
| rs1042173 T/ |
additive | -0.069 | 0.863 | -0.023 | 0.957 | 0.588 | 0.602 | -0.218 | 0.767 | 0.308 | 0.662 | -0.734 | 0.248 | |
Sex, ethnicity and age were included as covariates in linear regression models.
SNP model (additive, dominant, recessive, or dominance deviation from additivity) best explaining variance in depression score (based on AIC criterion) is reported. In the case of non-significant association between SNP and depression level in any examined group the values for additive model are shown.
PFDR = 0.047. domdev - dominance deviation from additivity model (A1A2 vs A1A1+A2A2). MiA/MaA – Minor allele/major allele. Statistically significant P-values after FDR correction are marked in bold.
3.3. Haplotype-based association analysis
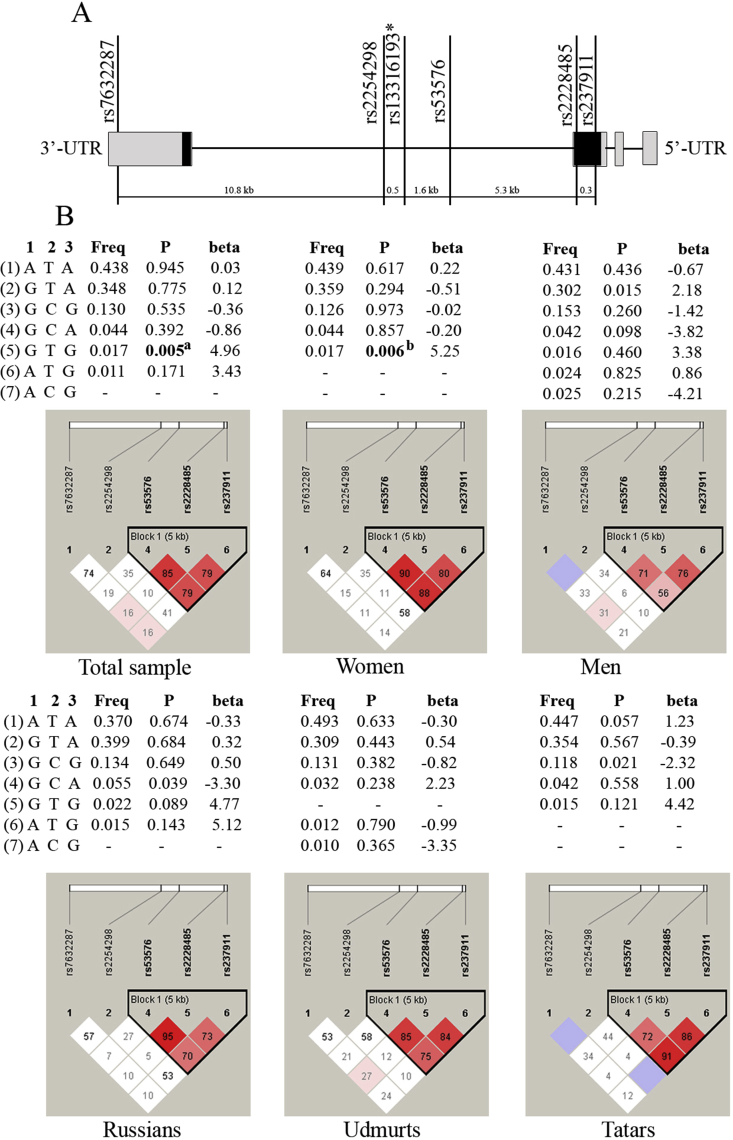
The analysis of pair-wise linkage disequilibrium revealed the presence of haplotype block in the AVPR1A gene (rs3803107, rs1042615) (D’ > 0.73) and in the OXTR gene (rs53576, rs2228485, rs237911) (D’ > 0.70), while no LD was detected between rs4795541 and rs1042173 in the SLC6A4 gene (D’ < 0.088) in all examined groups. Three haplotypes in the AVPR1A gene and eight haplotypes in the OXTR gene with haplotype frequencies above 1% have been observed (Table 4, Figure 1).
Table 4.
Haplotype frequencies in the AVPR1A gene (based on rs3803107 and rs1042615) and haplotype-based linear regression analysis in the examined groups.
| Group | N | Haplotype |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| G∗A |
A∗G |
G∗G |
||||||||
| Freq | β | P | Freq | β | P | Freq | β | P | ||
| Total | 623 | 0.394 | -0.21 | 0.628 | 0.163 | 1.23 | 0.034 | 0.439 | 0.55 | 0.206 |
| Women | 504 | 0.364 | -1.33 | 0.164 | 0.206 | 0.58 | 0.612 | 0.430 | 1.00 | 0.313 |
| Men | 117 | 0.400 | -0.002 | 0.996 | 0.153 | 1.52 | 0.023 | 0.441 | -0.88 | 0.067 |
| Russians | 225 | 0.393 | -1.07 | 0.162 | 0.127 | 2.90 | 0.015 | 0.472 | -0.25 | 0.743 |
| Udmurts | 218 | 0.401 | 0.09 | 0.881 | 0.219 | -0.002 | 0.998 | 0.379 | -0.09 | 0.879 |
| Tatars | 141 | 0.358 | -0.46 | 0.602 | 0.128 | 1.71 | 0.135 | 0.514 | -0.53 | 0.539 |
Statistically significant P-values before correction for multiple comparisons are marked in bold. Haplotypes with the frequencies less than 1% are not shown.
Figure 1.
The structure of the OXTR gene and association analysis of OXTR gene haplotypes (based on rs53576, rs2228485, rs237911) and depression level in healthy individuals. A. A schematic structure of the OXTR gene, examined SNPs and distance between them (kb). ∗rs13316193 was excluded from analysis due to deviation from the Hardy-Weinberg equilibrium. UTR – untranslated gene region. B. Haplotype frequencies of the OXTR gene in the examined groups (in total sample, women, men, individuals of Russian, Udmurt, and Tatar ethnic origin). 1 – rs53576, 2 – rs2228485, 3 – rs237911. Freq – haplotype frequency, P – P-value before correction for multiple comparisons. aR2 = 0.013, Pperm = 0.034, bR2 = 0.015, Pperm = 0.028. Statistically significant P-values after 10000 permutations are marked in bold. `Haplotypes with the frequencies less than 1% are not shown or marked with dashes. Constructed haplotype blocks of linked SNPs in the examined groups based on Lewontin's criterion (D′) are marked in triangles (Haploview v.4.2).
Haplotype analysis revealed association of OXTR GTG haplotype (based on rs53576, rs2228485, rs237911, respectively) and enhanced depression level in the total sample (β = 4.96; Р = 0.005; Pperm = 0.034; r2 = 0.013) and among women (β = 5.25; Р = 0.006; Pperm = 0.028; r2 = 0.015), which survived correction for multiple testing (Figure 1). At the same time, ethnicity-specific haplotype-based association of OXTR GCA haplotype in Russians (β = -3.30; Р = 0.039; Pperm = 0.187; r2 = 0.019) and GCG haplotype in Tatars (β = -2.32; Р = 0.021; Pperm = 0.106; r2 = 0.038) underachieved the level of statistical significance after FDR correction (Figure 1). In addition, a trend for carriers of AVPR1A AG haplotype (based on rs3803107 and rs1042615) to score higher on depression was observed in the total group (β = 1.23; Р = 0.034; Pperm = 0.084; r2 = 0.008) and among Russians (β = 0.127; Р = 0.015; Pperm = 0.059; r2 = 0.027).
3.4. Gene-environment interaction analysis
The analysis of gene-by-environment interactions was based on the inclusion of 16 different socio-demographic parameters and examined SNPs in linear regression models as main effects and interaction terms controlled for sex and ethnicity.
While examining gene-by-environment interactions with sex and ethnicity inclusion as covariates in linear regression models, we succeeded to detect a modifying effect of a sibship size (i.e. the number of children in a family) on the association of SLC6A4 rs1042173 and BDI-measured depression (Table 5) (Model 2, β = 2.89; Р = 0.017; r2 = 0.100). The inclusion of sex and ethnicity as interaction terms with rs1042173 weakened the rs1042173-by-sibship size interaction model (Model 3, β = 2.66; Р = 0.040; r2 = 0.116). Subsequent post hoc analyses revealed a trend for individuals bearing rs1042173 major GG genotype to score lower on depression while rearing in a large family (more than 3 children in a family) compared to those with less than two siblings, however, it was under the level of statistical significance (Z = -2.027; P = 0.043; PFDR = 0.129).
Table 5.
Significant multiple linear regression models (with GxE interaction) explaining variation in depression score controlled for sex and ethnicity.
| Items in linear regression model | Ref. group | Model 1 |
Model 2 |
Model 3 |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| β | T | P | β | T | P | β | T | P | ||
| rs1042173 (SLC6A4) | T-allele | -0.16 | -0.08 | 0.936 | -0.72 | -1.12 | 0.263 | 1.00 | 0.37 | 0.710 |
| Sex | Women | 1.33 | 1.12 | 0.263 | 2.23 | 2.4 | 0.015 | 2.20 | 1.46 | 0.143 |
| Ethnicity | Russians | -0.41 | -0.37 | 0.706 | -1.65 | -1.60 | 0.109 | -1.15 | -0.62 | 0.530 |
| Tatars | -1.79 | -1.56 | 0.119 | -4.07 | -4.20 | <0.001 | -1.85 | -1.11 | 0.266 | |
| rs1042173 x sex | T∗women | 0.11 | 0.10 | 0.915 | - | - | - | -0.13 | -0.10 | 0.916 |
| rs1042173 x ethnicity | Russians | 0.17 | 0.18 | 0.855 | - | - | - | -0.68 | -0.40 | 0.683 |
| Tatars | -0.71 | -0.68 | 0.491 | - | - | - | -2.57 | -1.66 | 0.097 | |
| Sibship size | ≥3 | - | - | - | -3.01 | -2.17 | 0.030 | -2.76 | -1.90 | 0.057 |
| rs1042173 x sibship size | T∗≥3 | - | - | - | 2.89 | 2.39 | 0.017 | 2.66 | 2.05 | 0.040 |
| rs53576 (OXTR) | A-allele | -2.68 | -1.42 | 0.154 | 4.33 | 2.52 | 0.012 | 2.27 | 0.79 | 0.431 |
| Sex | Women | 0.28 | 0.23 | 0.815 | 1.05 | 1.10 | 0.270 | 0.28 | 0.18 | 0.855 |
| Ethnicity | Russians | -0.71 | -0.66 | 0.508 | -0.11 | -0.12 | 0.904 | -1.27 | -0.86 | 0.392 |
| Tatars | -3.97 | -3.18 | 0.001 | -3.13 | -3.35 | <0.001 | 1.09 | 0.84 | 0.404 | |
| rs53576 x sex | A∗women | 1.17 | 1.18 | 0.234 | - | - | - | 0.72 | 0.57 | 0.571 |
| rs53576 x ethnicity | Russians | 0.35 | 0.37 | 0.708 | - | - | - | 1.21 | 0.91 | 0.362 |
| Tatars | 1.53 | 1.48 | 0.139 | - | - | - | 1.09 | 0.83 | 0.404 | |
| Paternal care | High | - | - | - | 0.29 | 0.23 | 0.810 | 0.25 | 0.20 | 0.841 |
| rs53576 x paternal care | A∗high | - | - | - | -2.52 | -2.38 | 0.017 | -2.49 | -2.33 | 0.020 |
| rs237911 (OXTR) | G-allele | -3.45 | -1.55 | 0.121 | 0.55 | 0.79 | 0.427 | -1.99 | -0.78 | 0.435 |
| Sex | Women | 0.63 | 0.73 | 0.466 | 1.25 | 1.60 | 0.108 | 0.56 | 0.60 | 0.547 |
| Ethnicity | Russians | -0.62 | -0.80 | 0.423 | -0.06 | -0.09 | 0.925 | 0.12 | 0.14 | 0.886 |
| Tatars | -2.27 | -1.59 | 0.112 | -2.67 | -3.48 | <0.001 | -1.88 | -2.18 | 0.030 | |
| rs237911 x sex | G∗women | 2.18 | 1.73 | 0.084 | - | - | - | 2.04 | 1.46 | 0.144 |
| rs237911 x ethnicity | Russians | 0.83 | 0.68 | 0.494 | - | - | - | -0.87 | -0.63 | 0.526 |
| Tatars | -2.27 | -1.59 | 0.112 | - | - | - | -2.78 | -1.87 | 0.062 | |
| OB | 2 | - | - | - | 1.59 | 2.02 | 0.043 | 1.59 | 2.03 | 0.042 |
| rs237911 x OB | G∗2 | - | - | - | -2.74 | -2.15 | 0.031 | -2.71 | -2.12 | 0.035 |
| rs33911258 (AVPR1В) | G-allele | 0.80 | 0.28 | 0.778 | 0.16 | 0.26 | 0.790 | 0.82 | 0.27 | 0.789 |
| Sex | Women | 1.58 | 1.77 | 0.075 | 1.59 | 2.09 | 0.036 | 1.78 | 1.89 | 0.058 |
| Ethnicity | Russians | -0.13 | -0.16 | 0.866 | -0.36 | -0.51 | 0.603 | -0.37 | -0.44 | 0.659 |
| Tatars | -2.74 | -3.16 | 0.002 | -2.26 | -3.01 | 0.002 | -2.80 | -3.18 | 0.001 | |
| rs33911258 x sex | G∗women | -0.41 | -0.28 | 0.777 | - | - | - | -0.62 | -0.39 | 0.692 |
| rs33911258 x ethnicity | Russians | -0.27 | -0.22 | 0.823 | - | - | - | 0.06 | 0.04 | 0.961 |
| Tatars | 1.04 | 0.71 | 0.477 | - | - | - | 1.87 | 1.21 | 0.225 | |
| Smoking | Previous | - | - | - | 0.48 | 0.40 | 0.689 | 0.47 | 0.38 | 0.701 |
| rs33911258∗smoking | G∗previous | - | - | - | 5.87 | 2.70 | 0.007 | 6.13 | 2.78 | 0.005 |
Additive effect of SNPs on depression score is shown while controlled for sex and ethnicity (Models 1, 2, 3). Smoking – previous smoking. OB – order of birth. Ref. group – reference group. Statistically significant P-values after FDR-correction for GxE interaction are shown in bold. Ethnicity was encoded as a set of dummy variables for the inclusion in the models. The number of children higher than three in the family is included in the model. Previous smoking is included in the model.
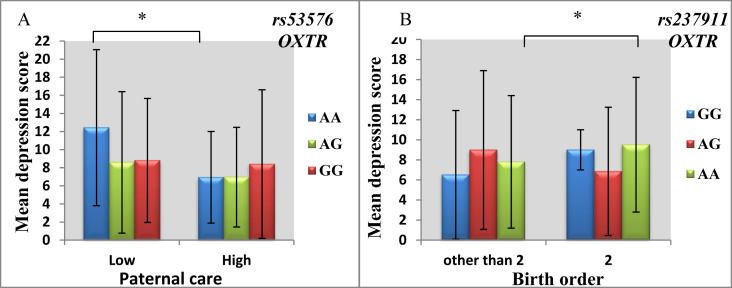
Next, we revealed a modifying effect of parenting style (namely, a degree of paternal care) on the association between the OXTR gene rs53576 and depression (Table 5) (Model 2, β = -2.52; Р = 0.017; r2 = 0.089). While including rs53576-by-sex and rs53576-by-ethnicity interaction terms in the model, the rs53576-by-paternal care interaction remained statistically significant (Model 3, β = -2.49; Р = 0.020; r2 = 0.104). The following stratified analysis revealed that carriers of rs53576 minor AA genotype demonstrated lower depression while rearing under the conditions of high paternal care compared to those, which reported low paternal interest in their rearing (Z = -2.898; P = 0.004; PFDR = 0.012) (Figure 2, a).
Figure 2.
Stratification analysis demonstrating a modulating effect of level of paternal care - on association of OXTR rs53576 (A), birth order - on association of OXTR rs237911 (B) gene variants and individual differences in depression under GxE paradigm while controlling for sex and ethnicity. The charts demonstrate medians and standard deviations. Statistically significant differences in depression score between stratified groups detected in non-parametric models are marked by brackets. ∗PFDR < 0.05.
Another GxE model explaining individual differences in depression included the OXTR rs237911-by-birth order interaction term (Table 5) (Model 2, β = -2.74; Р = 0.031; r2 = 0.049), which remained significant, however, attenuated after controlling for rs237911-by-sex and rs237911-by-ethnicity interaction (Model 3, β = -2.71; Р = 0.035; r2 = 0.073). Subsequent post-hoc analysis revealed that second-born individuals with rs237911 AA genotype scored significantly higher on depression compared to first- and third-borns (Z = -2.756; P = 0.006; PFDR = 0.018) (Figure 2, b).
Finally, an interaction effect of previous smoking and AVPR1B rs33911258 on depression was established (Table 5) (Model 2, β = 5.87; Р = 0.007; r2 = 0.057), while inclusion of rs33911258-by-sex and rs33911258-by-ethnicity facilitated GxE interaction model (Model 3, β = 6.13; Р = 0.005; r2 = 0.064). A stratified analysis by previous smoking demonstrated that rs33911258 G-allele carriers (GG + GA vs AA under the dominant statistical model), which reported previous smoking, tended to have higher depression level compared to non-smoking individuals (Z = -2.327; P = 0.020; PFDR = 0.060).
4. Discussion
In the present study, ethnicity-specific main effect of the AVPR1A rs3803107 and gene-based effect of the OXTR gene on BDI-measured depression, as well as a modulating effect of paternal care and birth order on gene-based association with depression has been observed. More specifically, an increased depression level was reported in individuals bearing OXTR rs53576 AA genotype while reported low level of paternal care and/or OXTR rs237911 AA genotype being the second-born individuals.
4.1. Serotonergic system genes (SLC6A4, HTR2A)
Given that changes in serotoninergic system functioning have been largely implicated in the etiology of depression and 5-HTT is a master regulator of the fine-tuning of 5-HT signaling, an extensive literature reported that attenuated SLC6A4 level caused by enhanced methylation of the SLC6A4 gene and associated with 5-HTTLPR S-allele (Bleys et al., 2018) was an elevated risk of depressive pathology (Dell’Osso et al., 2016). However, we failed to detect any association of the SLC6A4 gene polymorphisms (5-HTTLPR and rs1042173) with depression variance while considering either main effect or GxE interaction in healthy young adults. The most recent and methodologically different study, which have imputed and examined the effects of several highly explored VNTRs in a large GWAS dataset, also failed to confirm a significant effect of 5-HTTLPR in depression liability even at a liberal significance threshold (3.13 × 10−3 vs.5 × 10−8) (Border et al., 2019). Congruent with our results several researchers were unable to detect an interaction between 5-HTTLPR and stressful life events (SLEs) on increased depression risk in a community sample (Coventry et al., 2010) or MD patients (Gonda et al., 2018; Mandelli and Serretti, 2013). Such observation can be explained by recent conclusion that SLC6A4 gene expression is not only attributed to a classic assignment of S and L as low- and high-expressing alleles but is also affected by modulating polymorphisms such as rs25531, rs25532, intronic and 3′-UTR variations together with epigenetic regulatory mechanisms (Iurescia et al., 2016). Probably, a precise analysis of gene-based effect of the SLC6A4 gene has to be conducted in the study of depression etiology.
Recent findings reported the involvement of another gene belonging to the serotonergic system functioning (HTR2A, rs6311) in developing depressive symptoms while dealing with gene-by-environment interactions including smoking, chronic disease (Gonda et al., 2018) and urbal/rural residency (Dressler et al., 2009; Jokela et al., 2007). The rs7322347 examined in the present study moderated the effect of place of residency (rural/urban areas) (Mandelli and Serretti, 2013) on depression level and physical assault in childhood/adolescence on suicidal attempts (Ben-Efraim et al., 2013). Our findings indicate the absence of HTR2A rs7322347 effect on individual differences in depression level under the dialogue with environmental factors. Congruent with the present results, Mandelli and Serretti (2013) unobserved an interactive effect between lifetime SLEs or childhood trauma and HTR2A SNP on developing MD. It was suggested that the effect of stress exposure severity on depression might be mediated by partially different pathways and mechanisms of serotonergic functioning (Gonda et al., 2018).
4.2. Oxytocin receptor gene
In the present study we observed a haplotypic effect on depression with the OXTR GTG haplotype (based on rs53576, rs2228485, rs237911, respectively) carriers scoring higher on this trait, which was both true for the total sample and women group. The rationale behind the use of gene-based tests instead of analyzing the effects of SNPs individually is based on the suggestion that gene is a functional unit of the genome; thus, simultaneous analysis of gene SNPs increases the statistical power (Holmqvist Jämsen et al., 2017). Previously OXTR rs53576 has been associated with empathy and stress reactivity (Rodrigues et al., 2009), interaction with stress-protective effects such as social support (Chen et al., 2011), and responses to acute stressors (Moons et al., 2014). The attempts to find associations between rs2228485 (c.171C > T) and eating behavior (Kim et al., 2015), autism spectrum disorders (Kelemenova et al., 2010) and social recognition (Lucht et al., 2013) were undertaken. However, to date no findings depicting gene-based haplotypes including the same SNPs as in the present study were reported. Nevertheless, in methodologically similar study a specific haplotype in the OXTR gene (rs53576, rs2254298 and rs2228485) was associated with affectivity, emotional loneliness and cognitive functioning in mentally healthy individuals (Lucht et al., 2009). It should be noted that rs237911 has been unexamined for the association with psychological and psychiatric phenotypes before.
To date multiple efforts to unravel GxE interactions affecting depression-like behavior have been conducted, which demonstrated the interrelation of stress-related factors occurred at pre- and postnatal levels of development and OXTR gene polymorphisms. These studies reported a modulating effect of childhood adversity experienced by responders (Park et al., 2019) or by their mother in childhood (Elwood et al., 2019; Mileva-Seitz et al., 2013), maternal postnatal depression (Choi et al., 2019), and even their child's OXTR genotype (Asherin et al., 2019). Previously, an interaction between OXTR rs2254298 and the quality of the parental environment was reported to affect symptoms of depression and anxiety in female adolescents (Thompson et al., 2011). In the present study we also clarified the modulating effect of parenting style as GxE interaction between another SNP in the OXTR gene (rs53576) and paternal style of parenting. Interestingly, statistically significant differences in depression level were identified in individuals with a differential level of paternal care, which was demonstrated only in respondents bearing low-activity minor A-allele (AA + AG vs GG) of rs53576 (Parris et al., 2018). We suggest that modulating effect of enhanced level of paternal care on a reduced depression-like behavior is observed only under the conditions of OXTR deficit related to the presence of rs53576 A-allele. This effect was reported to be caused by a reduced methylation of exon 2 in the OXTR gene in the carriers of rs53576 GG genotype and, hence, elevated OXTR expression (Reiner et al., 2015). Together with allele-related association of OXTR gene expression, stressful life events are also known to result in differential OXTR methylation. Since an interaction between childhood maltreatment and OXTR methylation at multiple CpG sites predicted depressive symptoms later in ontogenesis (Misra et al., 2019; Smearman et al., 2016), it was suggested that OXTR methylation might moderate, rather than mediate, the association between childhood abuse and depression (Park et al., 2019). According to previous research consistent with a diathesis-stress model (Augustine et al., 2018), rs53576 A-allele was frequently associated with psychopathologies correlated with an increased sensitivity to stress (Saphire-Bernstein et al., 2011; Choi et al., 2019). Despite the suggestion that individuals homozygous for the G-allele may exhibit greater sensitivity to social experiences (Asherin et al., 2019), the present findings oppositely demonstrated that A-allele carriers appeared to be more sensitive to parenting style (presumably, to the level of paternal interest in their lives) compared to those with GG genotypes. Therefore, together with the “plasticity genes model” the data obtained allow to hypothesize that A-allele is a plasticity marker, contributing to a greater sensitivity to both positive and negative environmental influences. One of the possible mechanisms of the involvement of family environment in emotions processing is probably based on changes in neurobiological pathways (related to distinct gene variants) triggered by a specific family environment (Little et al., 2015; Pozzi et al., 2019).
Moreover, our findings indicate that the effect of rs237911 (с.-135C > T, located in the 5′-UTR of the OXTR gene) on depression score was modified by birth order controlling for sex and ethnicity. In particular, being the second-order of birth child in a family facilitated higher depression only among those with a major AA genotype of the rs237911 compared to first-born individuals. One of the probable explanations of this observation might be based on the “resource depletion theory”, which suggests that each subsequent child requires an amount of parental resources higher than they can provide (Härkönen, 2014). From another side, the middle position in a siblingship was shown to be related to specific unfavorable rearing patterns resulting in psychiatric prevalence (Richter et al., 1997). Accordingly, in the case of limited parental resources (i.e., if the individual is the middle sibling in a family), the differences in emotional processing would be associated with stress sensitivity depending on the presence of a certain rs237911 variant in the OXTR gene. As observed in the present study (mean depression score for the first-borns was 8.32 ± 7.00, for the second-borns was 8.69 ± 6.65, for the third-borns was 6.50 ± 6.44), in multiplex families birth order has V-shaped effects on several psychopathologies (for instance, ASD) with middle births being at high risk (Turner et al., 2011). From another side, our findings might be explained by differential methylation of depression-related genes caused by differences in birth order and several other predictors, which were shown to explain 75% of variably methylated regions (Teh et al., 2014). According to bioinformatics resources, rs237911 resides the region of intronic circular RNA (circRNA), which represents a class of small non-coding RNAs functioning as microRNA sponges, regulators of splicing and transcription, modifiers of parental gene expression, and interacting with RNA binding proteins (Qu et al., 2017). Moreover, due to circRNAs high expression in the brain, their role in early brain development and other brain-related processes (Zhuo et al., 2020) including depression-like behavior (Zhang et al., 2018) have been recently reported. In addition, birth order might have an indirect effect on depressive-like behavior via changes in intrauterine environment caused by previous delivery, which plays a key role in shaping offspring DNA methylation of genes related to “Nervous system development and function” pathway (Li et al., 2017).
The involvement of the OXTR gene in anxiety-related traits (BDI-measured depression) on the gene-based and SNP-by-environment interaction level is unsurprising, since oxytocin receptors might buffer stress reactivity associated with a reduced cortisol secretion (Cardoso et al., 2014). In turn, cortisol represents one of the primary stress hormones activating the body during a stress reaction and is regulated by the HPA axis together with oxytocin (Holmqvist Jämsen et al., 2017). It was previously suggested that the effect of OXTR SNPs on stress-related symptoms could be realized via either direct effects of SNPs on OXT function influencing cortisol levels or indirect effects of SNPs on social traits, which promote seeking of peer support during stress and hence could reduce stress symptoms and lower cortisol levels (Holmqvist Jämsen et al., 2017). In the present study there was an interaction between OXTR rs53576 and paternal care, as well as rs237911 and birth order, thus demonstrating the possibility of the influence of parental style and birth order on HPA axis reactivity with direct effect on OXTR functioning. Thus, the presence of high level of paternal care may attenuate HPA-axis activity caused by diminished OXTR level (rs53576 AA genotype) related to enhanced anxiety and stress (Landgraf and Neumann, 2004).
4.3. Arginine vasopressin receptor genes
Since arginine vasopressin receptor is a potential regulator of stress response, we succeed to demonstrate the main effect of AVPR1A rs3803107 on individual liability to increased depression-like behavior. Although a correlation between AVPR1A-related differences in circulating levels of AVP and emotional responses to an acute stressor was revealed (Moons et al., 2014), we failed to detect any moderating effect of stressful childhood conditions on AVPR1A-related association with depression. Recently it was predicted that miR-375 and miR-186 target regions were located at the rs3803107 locus and an increased AVPR1A mRNA was detected in the case of A-allele (GA/AA genotypes) compared to GG genotypes at rs3803107 (Zhang et al., 2019). In the present study we reported the association of rs3803107 A-allele and higher depression score in one ethnic group (Russians), which seems to be congruent with published data on association between lower AVPR1A level and reduced anxiety (Barrett et al., 2013) and the presence of rs3803107 GG genotype (Zhang et al., 2019). According to the literature, miR-375 is relevant to psychiatric conditions and psychological phenotypes (Bhinge et al., 2016; Dulcis et al., 2017), while chronic unpredictable stress can cause miR-375 increase thus affecting expression of relevant genes (Lotan et al., 2018). In turn, the role of miR-186 in synaptic scaling as a degree of stable neuronal activity affected by developmental processes was demonstrated (Silva et al., 2019). Therefore, future research on the connection between rs3803107 variations and depression liability via miR-375 and miR-186 binding appears to be promising. From the other side, observed association was ethnicity-specific, which can be explained by the existence of cross-cultural differences in depression and anxiety level (Zhao and Zhang, 2018). Previously we also demonstrated the involvement of another SNP located in 3′-UTR of the AVPR1A gene (rs11174811) in individual variation in personality traits in ethnicity-specific manner (Kazantseva et al., 2014). Moreover, the impact of maternal ethnicity on placental gene expression (HPA-axis genes, in particular) may mediate differences in depressive behavior (Capron et al., 2018). Although previous findings indicated the involvement of AVPR1B rs33911258 in manifestation of specific behavioral pattern (Self-transcendence measured with TCI-125) (Kazantseva et al., 2014), which resembles with a decreased depression, we failed to observe any impact of this SNP in depression level in the present study.
4.4. Strengths and limitations
The present study has a number of methodological strengths including homogeneity of the sample in respect to age and education. To our best knowledge, the present study is the first one to explore a modulating effect of multiple early postnatal factors occurring in childhood on genetic association of oxytocin and arginine-vasopressin receptor genes with individual liability to depression in a community sample of young adults. The majority of previously conducted studies sought to unravel genetic effect on depression under case-control paradigm, while the present study involved mentally healthy subjects based on the hypothesis of depression as a subtherapeutic continuum of affective disorders. Moreover, correction for multiple testing was used to decrease the possibility of false positive results under the false discovery rate (FDR) procedure (Simes procedure), since multiple independent hypotheses have been tested.
However, the present investigation study suffers from a number of limitations that should be reported. First, the study used a hypothesis driven candidate gene approach, which examined a limited number of genetic variants in a moderately large sample. Second, although we sought to conduct our study on the subjects of approximately the same age group (18-25 years) to protect against confounding by age, the findings may not generalize to younger or older ages. Third, we could not control for the type of parental transmission due to the absence of parental DNA samples; however, a type of allele inheritance (maternal/paternal) was described so far to affect liability to psychiatric phenotypes (Ben-Efraim et al., 2013). Although the analyses included multiple stress-related environmental factors occurring during childhood as modifiers between candidate genes and individual depression level, it is possible that we missed some other important environmental factors, such as maternal depression, recent and in utero stressful life events, social adversity.
5. Conclusion
This is a preliminary study, which indicates that specific interaction of alleles with environmental risk may be relevant to individual sensitivity in depression variance in non-clinical sample of healthy young adults. Congruent with the differential susceptibility model of depression, we demonstrated that distinct genetic context (based on OXTR rs53576, rs237911) modulated sensitivity to both positive and negative environmental influences thus resulting in plasticity-related individual differences in depression level in mentally healthy subjects. Future psychogenetic research of depression should seek to replicate and extend the present research by examining other stress-related environmental factors of ante- and postnatal development under gene-by-environment paradigm involving more biologically interacting pathways and to involve not only clinical forms of depression (for instance, MDD) but also population-based samples.
Declarations
Author contribution statement
A. Kazantseva: Conceived and designed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Yu. Davydova: Conceived and designed the experiments; Performed the experiments; Contributed reagents, materials, analysis tools or data.
R. Enikeeva, M. Lobaskova, S. Malykh: Conceived and designed the experiments; Contributed reagents, materials, analysis tools or data.
R. Mustafin: Performed the experiments; Contributed reagents, materials, analysis tools or data. .
Z. Takhirova: Analyzed and interpreted the data.
E. Khusnutdinova: Conceived and designed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data.
Funding statement
This work was supported by the state task of the Ministry of education of the Russian Federation (project no. AAAA-A16-116020350032-1), the Program of collections of biological resources of FASO of Russian Federation (project no. 007-030164/2) and partially supported by the grant of the Russian Foundation for Basic Research (project no. 17-29-02195_ofi_m) and by the grant of the Bashkortostan Republic to young scientists in 2020 (to Yu. Davydova, state contract no. 9GR).
Competing interest statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Acknowledgements
We are grateful to Dr. I. Gilyazova, Dr. N. Ekomasova, Dr. A. Nurgalieva, Prof. A. Karunas, and Dr. M. Dzhaubermezov for their assistance with samples collection.
Appendix ASupplementary data
The following is the supplementary data related to this article:
References
- Arnau-Soler A., Macdonald-Dunlop E., Adams M.J., Clarke T.-K., MacIntyre D.J., Milburn K., Navrady L., Generation Scotland, Major Depressive Disorder Working Group of the Psychiatric Genomics Consortium. Hayward C., McIntosh A.M., Thomson P.A. Genome-wide by environment interaction studies of depressive symptoms and psychosocial stress in UK Biobank and Generation Scotland. Transl. Psychiatry. 2019;9:14. doi: 10.1038/s41398-018-0360-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asherin R.M., Everhart K.D., Stophaeros S.L., Vogeli J.M., Fowler J., Phiel C.J., Kaplan P.S. Associations between maternal depression and mother and infant oxytocin receptor gene (OXTR_rs53576) polymorphisms. Dev. Psychobiol. 2019;62(4) doi: 10.1002/dev.21938. [DOI] [PubMed] [Google Scholar]
- Augustine M.E., Leerkes E.M., Smolen A., Calkins S.D. Relations between early maternal sensitivity and toddler self-regulation: exploring variation by oxytocin and dopamine D2 receptor genes. Dev. Psychobiol. 2018;60:789–804. doi: 10.1002/dev.21745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banlaki Z., Elek Z., Nanasi T., Szekely A., Nemoda Z., Sasvari-Szekely M., Ronai Z. Polymorphism in the serotonin receptor 2a (HTR2A) gene as possible predisposal factor for aggressive traits. PloS One. 2015;10 doi: 10.1371/journal.pone.0117792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett C.E., Keebaugh A.C., Ahern T.H., Bass C.E., Terwilliger E.F., Young L.J. Variation in vasopressin receptor (Avpr1a) expression creates diversity in behaviors related to monogamy in prairie voles. Horm. Behav. 2013;63:518–526. doi: 10.1016/j.yhbeh.2013.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Efraim Y.J., Wasserman D., Wasserman J., Sokolowski M. Family-based study of HTR2A in suicide attempts: observed gene, gene × environment and parent-of-origin associations. Mol. Psychiatr. 2013;18:758–766. doi: 10.1038/mp.2012.86. [DOI] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. Benjamini-1995.pdf. J. Roy. Stat. Soc. B. 1995 [Google Scholar]
- Bernhard R.M., Chaponis J., Siburian R., Gallagher P., Ransohoff K., Wikler D., Perlis R.H., Greene J.D. Variation in the oxytocin receptor gene (OXTR) is associated with differences in moral judgment. Soc. Cognit. Affect Neurosci. 2016;11:1872–1881. doi: 10.1093/scan/nsw103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhinge A., Namboori S.C., Bithell A., Soldati C., Buckley N.J., Stanton L.W. MiR-375 is essential for human Spinal motor neuron development and may Be involved in motor neuron degeneration. Stem Cell. 2016;34:124–134. doi: 10.1002/stem.2233. [DOI] [PubMed] [Google Scholar]
- Bleys D., Luyten P., Soenens B., Claes S. Gene-environment interactions between stress and 5-HTTLPR in depression: a meta-analytic update. J. Affect. Disord. 2018;226:339–345. doi: 10.1016/j.jad.2017.09.050. [DOI] [PubMed] [Google Scholar]
- Border R., Johnson E.C., Evans L.M., Smolen A., Berley N., Sullivan P.F., Keller M.C. No support for historical candidate gene or candidate gene-by-interaction hypotheses for major depression across multiple large samples. Am. J. Psychiatr. 2019;176:376–387. doi: 10.1176/appi.ajp.2018.18070881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brummett B.H., Boyle S.H., Siegler I.C., Kuhn C.M., Ashley-Koch A., Jonassaint C.R., Züchner S., Collins A., Williams R.B. Effects of environmental stress and gender on associations among symptoms of depression and the serotonin transporter gene linked polymorphic region (5-HTTLPR) Behav. Genet. 2008;38:34–43. doi: 10.1007/s10519-007-9172-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao C., Rijlaarsdam J., van der Voort A., Ji L., Zhang W., Bakermans-Kranenburg M.J. Associations between dopamine D2 receptor (DRD2) gene, maternal positive parenting and trajectories of depressive symptoms from early to mid-adolescence. J. Abnorm. Child Psychol. 2018;46:365–379. doi: 10.1007/s10802-017-0294-5. [DOI] [PubMed] [Google Scholar]
- Capron L.E., Ramchandani P.G., Glover V. Maternal prenatal stress and placental gene expression of NR3C1 and HSD11B2: the effects of maternal ethnicity. Psychoneuroendocrinology. 2018;87:166–172. doi: 10.1016/j.psyneuen.2017.10.019. [DOI] [PubMed] [Google Scholar]
- Cardoso C., Kingdon D., Ellenbogen M.A. A meta-analytic review of the impact of intranasal oxytocin administration on cortisol concentrations during laboratory tasks: moderation by method and mental health. Psychoneuroendocrinology. 2014;49:161–170. doi: 10.1016/j.psyneuen.2014.07.014. [DOI] [PubMed] [Google Scholar]
- Chen F.S., Kumsta R., von Dawans B., Monakhov M., Ebstein R.P., Heinrichs M. Common oxytocin receptor gene (OXTR) polymorphism and social support interact to reduce stress in humans. Proc. Natl. Acad. Sci. U.S.A. 2011;108:19937–19942. doi: 10.1073/pnas.1113079108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi D., Tsuchiya K.J., Takei N. Interaction effect of oxytocin receptor (OXTR) rs53576 genotype and maternal postpartum depression on child behavioural problems. Sci. Rep. 2019;9:7685. doi: 10.1038/s41598-019-44175-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman J.R.I., Gaspar H.A., Bryois J., Byrne E.M., Forstner A.J., Holmans P.A., de Leeuw C.A., Mattheisen M., McQuillin A., Whitehead Pavlides J.M., Pers T.H., Ripke S., Stahl E.A., Steinberg S., Trubetskoy V., Trzaskowski M., Wang Y., Abbott L., Abdellaoui A., Adams M.J., Adolfsson A.N., Agerbo E., Akil H., Albani D., Alliey-Rodriguez N., Als T.D., Andlauer T.F.M., Anjorin A., Antilla V., Van der Auwera S., Awasthi S., Bacanu S.A., Badner J.A., Bækvad-Hansen M., Barchas J.D., Bass N., Bauer M., Beekman A.T.F., Belliveau R., Bergen S.E., Bigdeli T.B., Binder E.B., Bøen E., Boks M., Boocock J., Budde M., Bunney W., Burmeister M., Buttenschøn H.N., Bybjerg-Grauholm J., Byerley W., Cai N., Casas M., Castelao E., Cerrato F., Cervantes P., Chambert K., Charney A.W., Chen D., Christensen J.H., Churchhouse C., St Clair D., Clarke T.K., Colodro-Conde L., Coryell W., Couvy-Duchesne B., Craig D.W., Crawford G.E., Cruceanu C., Czerski P.M., Dale A.M., Davies G., Deary I.J., Degenhardt F., Del-Favero J., DePaulo J.R., Derks E.M., Direk N., Djurovic S., Dobbyn A.L., Dolan C.V., Dumont A., Dunn E.C., Eley T.C., Elvsåshagen T., Escott-Price V., Fan C.C., Finucane H.K., Fischer S.B., Flickinger M., Foo J.C., Foroud T.M., Forty L., Frank J., Fraser C., Freimer N.B., Frisén L., Gade K., Gage D., Garnham J., Giambartolomei C., Goes F.S., Goldstein J., Gordon S.D., Gordon-Smith K., Green E.K., Green M.J., Greenwood T.A., Grove J., Guan W., Hall L.S., Hamshere M.L., Hansen C.S., Hansen T.F., Hautzinger M., Heilbronner U., van Hemert A.M., Herms S., Hickie I.B., Hipolito M., Hoffmann P., Holland D., Homuth G., Horn C., Hottenga J.J., Huckins L., Ising M., Jamain S., Jansen R., Johnson J.S., de Jong S., Jorgenson E., Juréus A., Kandaswamy R., Karlsson R., Kennedy J.L., Hassan Kiadeh F.F., Kittel-Schneider S., Knowles J.A., Kogevinas M., Kohane I.S., Koller A.C., Kraft J., Kretzschmar W.W., Krogh J., Kupka R., Kutalik Z., Lavebratt C., Lawrence J., Lawson W.B., Leber M., Lee P.H., Levy S.E., Li J.Z., Li Y., Lind P.A., Liu C., Olde Loohuis L.M., Maaser A., MacIntyre D.J., MacKinnon D.F., Mahon P.B., Maier W., Maier R.M., Marchini J., Martinsson L., Mbarek H., McCarroll S., McGrath P., McGuffin P., McInnis M.G., McKay J.D., Medeiros H., Medland S.E., Mehta D., Meng F., Middeldorp C.M., Mihailov E., Milaneschi Y., Milani L., Mirza S.S., Mondimore F.M., Montgomery G.W., Morris D.W., Mostafavi S., Mühleisen T.W., Mullins N., Nauck M., Ng B., Nguyen H., Nievergelt C.M., Nivard M.G., Nwulia E.A., Nyholt D.R., O’Donovan C., O’Reilly P.F., Ori A.P.S., Oruc L., Ösby U., Oskarsson H., Painter J.N., Parra J.G., Pedersen C.B., Pedersen M.G., Perry A., Peterson R.E., Pettersson E., Peyrot W.J., Pfennig A., Pistis G., Purcell S.M., Quiroz J.A., Qvist P., Regeer E.J., Reif A., Reinbold C.S., Rice J.P., Riley B.P., Rivas F., Rivera M., Roussos P., Ruderfer D.M., Ryu E., Sánchez-Mora C., Schatzberg A.F., Scheftner W.A., Schoevers R., Schork N.J., Schulte E.C., Shehktman T., Shen L., Shi J., Shilling P.D., Shyn S.I., Sigurdsson E., Slaney C., Smeland O.B., Smit J.H., Smith D.J., Sobell J.L., Spijker A.T., Steffens M., Strauss J.S., Streit F., Strohmaier J., Szelinger S., Tansey K.E., Teismann H., Teumer A., Thompson R.C., Thompson W., Thomson P.A., Thorgeirsson T.E., Traylor M., Treutlein J., Uitterlinden A.G., Umbricht D., Vedder H., Viktorin A., Visscher P.M., Wang W., Watson S.J., Webb B.T., Weickert C.S., Weickert T.W., Weinsheimer S.M., Wellmann J., Willemsen G., Witt S.H., Wu Y., Xi H.S., Xu W., Yang J., Young A.H., Zandi P., Zhang P., Zhang F., Zollner S., Adolfsson R., Agartz I., Alda M., Arolt V., Backlund L., Baune B.T., Bellivier F., Berger K., Berrettini W.H., Biernacka J.M., Blackwood D.H.R., Boehnke M., Boomsma D.I., Corvin A., Craddock N., Daly M.J., Dannlowski U., Domenici E., Domschke K., Esko T., Etain B., Frye M., Fullerton J.M., Gershon E.S., de Geus E.J.C., Gill M., Goes F., Grabe H.J., Grigoroiu-Serbanescu M., Hamilton S.P., Hauser J., Hayward C., Heath A.C., Hougaard D.M., Hultman C.M., Jones I., Jones L.A., Kahn R.S., Kendler K.S., Kirov G., Kloiber S., Landén M., Leboyer M., Lewis G., Li Q.S., Lissowska J., Lucae S., Madden P.A.F., Magnusson P.K., Martin N.G., Mayoral F., McElroy S.L., McIntosh A.M., McMahon F.J., Melle I., Metspalu A., Mitchell P.B., Morken G., Mors O., Mortensen P.B., Müller-Myhsok B., Myers R.M., Neale B.M., Nimgaonkar V., Nordentoft M., Nöthen M.M., O’Donovan M.C., Oedegaard K.J., Owen M.J., Paciga S.A., Pato C., Pato M.T., Pedersen N.L., Penninx B.W.J.H., Perlis R.H., Porteous D.J., Posthuma D., Potash J.B., Preisig M., Ramos-Quiroga J.A., Ribasés M., Rietschel M., Rouleau G.A., Schaefer C., Schalling M., Schofield P.R., Schulze T.G., Serretti A., Smoller J.W., Stefansson H., Stefansson K., Stordal E., Tiemeier H., Turecki G., Uher R., Vaaler A.E., Vieta E., Vincent J.B., Völzke H., Weissman M.M., Werge T., Andreassen O.A., Børglum A.D., Cichon S., Edenberg H.J., Di Florio A., Kelsoe J., Levinson D.F., Lewis C.M., Nurnberger J.I., Ophoff R.A., Scott L.J., Sklar P., Sullivan P.F., Wray N.R., Breen G. The genetics of the mood disorder spectrum: Genome-wide Association Analyses of More Than 185,000 Cases and 439,000 Controls. Biol. Psychiatry. 2020;88(2):168–184. doi: 10.1016/j.biopsych.2019.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coventry W.L., James M.R., Eaves L.J., Gordon S.D., Gillespie N.A., Ryan L., Heath A.C., Montgomery G.W., Martin N.G., Wray N.R. Do 5HTTLPR and stress interact in risk for depression and suicidality? Item response analyses of a large sample. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 2010;153B:757–765. doi: 10.1002/ajmg.b.31044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culverhouse R.C., Saccone N.L., Horton A.C., Ma Y., Anstey K.J., Banaschewski T., Burmeister M., Cohen-Woods S., Etain B., Fisher H.L., Goldman N., Guillaume S., Horwood J., Juhasz G., Lester K.J., Mandelli L., Middeldorp C.M., Olié E., Villafuerte S., Air T.M., Araya R., Bowes L., Burns R., Byrne E.M., Coffey C., Coventry W.L., Gawronski K.A.B., Glei D., Hatzimanolis A., Hottenga J.-J., Jaussent I., Jawahar C., Jennen-Steinmetz C., Kramer J.R., Lajnef M., Little K., Zu Schwabedissen H.M., Nauck M., Nederhof E., Petschner P., Peyrot W.J., Schwahn C., Sinnamon G., Stacey D., Tian Y., Toben C., Van der Auwera S., Wainwright N., Wang J.-C., Willemsen G., Anderson I.M., Arolt V., Åslund C., Bagdy G., Baune B.T., Bellivier F., Boomsma D.I., Courtet P., Dannlowski U., de Geus E.J.C., Deakin J.F.W., Easteal S., Eley T., Fergusson D.M., Goate A.M., Gonda X., Grabe H.J., Holzman C., Johnson E.O., Kennedy M., Laucht M., Martin N.G., Munafò M.R., Nilsson K.W., Oldehinkel A.J., Olsson C.A., Ormel J., Otte C., Patton G.C., Penninx B.W.J.H., Ritchie K., Sarchiapone M., Scheid J.M., Serretti A., Smit J.H., Stefanis N.C., Surtees P.G., Völzke H., Weinstein M., Whooley M., Nurnberger J.I., Breslau N., Bierut L.J. Collaborative meta-analysis finds no evidence of a strong interaction between stress and 5-HTTLPR genotype contributing to the development of depression. Mol. Psychiatr. 2018;23:133–142. doi: 10.1038/mp.2017.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dell’Osso L., Carmassi C., Mucci F., Marazziti D. Depression, serotonin and tryptophan. Curr. Pharmaceut. Des. 2016;22:949–954. doi: 10.2174/1381612822666151214104826. [DOI] [PubMed] [Google Scholar]
- Dogan M.V., Lei M.-K., Beach S.R.H., Brody G.H., Philibert R.A. Alcohol and tobacco consumption alter hypothalamic pituitary adrenal axis DNA methylation. Psychoneuroendocrinology. 2016;66:176–184. doi: 10.1016/j.psyneuen.2016.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dressler W.W., Balieiro M.C., Ribeiro R.P., Dos Santos J.E. Cultural consonance, a 5HT2A receptor polymorphism, and depressive symptoms: a longitudinal study of gene x culture interaction in urban Brazil. Am. J. Hum. Biol. 2009;21:91–97. doi: 10.1002/ajhb.20823. [DOI] [PubMed] [Google Scholar]
- Dulcis D., Lippi G., Stark C.J., Do L.H., Berg D.K., Spitzer N.C. Neurotransmitter Switching regulated by miRNAs controls changes in social preference. Neuron. 2017;95:1319–1333. doi: 10.1016/j.neuron.2017.08.023. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn E.C., Wiste A., Radmanesh F., Almli L.M., Gogarten S.M., Sofer T., Faul J.D., Kardia S.L.R., Smith J.A., Weir D.R., Zhao W., Soare T.W., Mirza S.S., Hek K., Tiemeier H., Goveas J.S., Sarto G.E., Snively B.M., Cornelis M., Koenen K.C., Kraft P., Purcell S., Ressler K.J., Rosand J., Wassertheil-Smoller S., Smoller J.W. GENOME-WIDE association study (GWAS) and genome-wide BY environment interaction study (GWEIS) OF depressive symptoms IN african AMERICAN and hispanic/latina women. Depress. Anxiety. 2016;33:265–280. doi: 10.1002/da.22484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Easey K.E., Mars B., Pearson R., Heron J., Gunnell D. Association of birth order with adolescent mental health and suicide attempts: a population-based longitudinal study. Eur. Child Adolesc. Psychiatr. 2019;28:1079–1086. doi: 10.1007/s00787-018-1266-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elwood J., Murray E., Bell A., Sinclair M., Kernohan W.G., Stockdale J. A systematic review investigating if genetic or epigenetic markers are associated with postnatal depression. J. Affect. Disord. 2019 doi: 10.1016/j.jad.2019.04.059. [DOI] [PubMed] [Google Scholar]
- Gabriel S.B., Schaffner S.F., Nguyen H., Moore J.M., Roy J., Blumenstiel B., Higgins J., DeFelice M., Lochner A., Faggart M., Liu-Cordero S.N., Rotimi C., Adeyemo A., Cooper R., Ward R., Lander E.S., Daly M.J., Altshuler D. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- Gonda X., Hullam G., Antal P., Eszlari N., Petschner P., Hökfelt T.G., Anderson I.M., Deakin J.F.W., Juhasz G., Bagdy G. Significance of risk polymorphisms for depression depends on stress exposure. Sci. Rep. 2018;8:3946. doi: 10.1038/s41598-018-22221-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez I., Polvillo R., Ruiz-Galdon M., Reyes-Engel A., Royo J.L. Dysmorphic contribution of neurotransmitter and neuroendocrine system polymorphisms to subtherapeutic mood states. Brain Behav. 2019;9 doi: 10.1002/brb3.1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannigan L.J., McAdams T.A., Eley T.C. Developmental change in the association between adolescent depressive symptoms and the home environment: results from a longitudinal, genetically informative investigation. JCPP (J. Child Psychol. Psychiatry) 2017;58:787–797. doi: 10.1111/jcpp.12689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Härkönen J. Birth order effects on educational attainment and educational transitions in West Germany. Eur. Socio Rev. 2014;30:166–179. [Google Scholar]
- Holmqvist Jämsen S., Johansson A., Westberg L., Santtila P., von der Pahlen B., Simberg S. Associations between vocal symptoms and genetic variants in the oxytocin receptor and arginine vasopressin 1A receptor gene. J. Speech Lang. Hear. Res. 2017;60:1843–1854. doi: 10.1044/2016_JSLHR-S-16-0059. [DOI] [PubMed] [Google Scholar]
- House J.S., Mendez M., Maguire R.L., Gonzalez-Nahm S., Huang Z., Daniels J., Murphy S.K., Fuemmeler B.F., Wright F.A., Hoyo C. Periconceptional maternal mediterranean diet is associated with favorable offspring behaviors and altered CpG methylation of imprinted genes. Front. cell Dev. Biol. 2018;6:107. doi: 10.3389/fcell.2018.00107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard D.M., Adams M.J., Clarke T.K., Hafferty J.D., Gibson J., Shirali M., Coleman J.R.I., Hagenaars S.P., Ward J., Wigmore E.M., Alloza C., Shen X., Barbu M.C., Xu E.Y., Whalley H.C., Marioni R.E., Porteous D.J., Davies G., Deary I.J., Hemani G., Berger K., Teismann H., Rawal R., Arolt V., Baune B.T., Dannlowski U., Domschke K., Tian C., Hinds D.A., Trzaskowski M., Byrne E.M., Ripke S., Smith D.J., Sullivan P.F., Wray N.R., Breen G., Lewis C.M., McIntosh A.M. Genome-wide meta-analysis of depression identifies 102 independent variants and highlights the importance of the prefrontal brain regions. Nat. Neurosci. 2019;22:343–352. doi: 10.1038/s41593-018-0326-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue H., Yamasue H., Tochigi M., Abe O., Liu X., Kawamura Y., Takei K., Suga M., Yamada H., Rogers M.A., Aoki S., Sasaki T., Kasai K. Association between the oxytocin receptor gene and amygdalar volume in healthy adults. Biol. Psychiatr. 2010;68:1066–1072. doi: 10.1016/j.biopsych.2010.07.019. [DOI] [PubMed] [Google Scholar]
- Iurescia S., Seripa D., Rinaldi M. Role of the 5-HTTLPR and SNP promoter polymorphisms on serotonin transporter gene expression: a closer look at genetic architecture and in vitro functional studies of common and uncommon allelic variants. Mol. Neurobiol. 2016;53:5510–5526. doi: 10.1007/s12035-015-9409-6. [DOI] [PubMed] [Google Scholar]
- Jiang S., Postovit L., Cattaneo A., Binder E.B., Aitchison K.J. Epigenetic modifications in stress response genes associated with childhood trauma. Front. Psychiatr. 2019;10:808. doi: 10.3389/fpsyt.2019.00808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johns C.B., Lacadie C., Vohr B., Ment L.R., Scheinost D. Amygdala functional connectivity is associated with social impairments in preterm born young adults. NeuroImage. Clin. 2019;21:101626. doi: 10.1016/j.nicl.2018.101626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jokela M., Lehtimäki T., Keltikangas-Järvinen L. The influence of urban/rural residency on depressive symptoms is moderated by the serotonin receptor 2A gene. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 2007;144B:918–922. doi: 10.1002/ajmg.b.30555. [DOI] [PubMed] [Google Scholar]
- Kang J.I., Kim H.W., Kim C.H., Hwang E.H., Kim S.J. Oxytocin receptor gene polymorphisms exert a modulating effect on the onset age in patients with obsessive-compulsive disorder. Psychoneuroendocrinology. 2017;86:45–52. doi: 10.1016/j.psyneuen.2017.09.011. [DOI] [PubMed] [Google Scholar]
- Kantake M., Yoshitake H., Ishikawa H., Araki Y., Shimizu T. Postnatal epigenetic modification of glucocorticoid receptor gene in preterm infants: a prospective cohort study. BMJ Open. 2014;4 doi: 10.1136/bmjopen-2014-005318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura Y., Liu X., Akiyama T., Shimada T., Otowa T., Sakai Y., Kakiuchi C., Umekage T., Sasaki T., Akiskal H.S. The association between oxytocin receptor gene (OXTR) polymorphisms and affective temperaments, as measured by TEMPS-A. J. Affect. Disord. 2010;127:31–37. doi: 10.1016/j.jad.2010.04.014. [DOI] [PubMed] [Google Scholar]
- Kazantseva A., Gaysina D., Kutlumbetova Y., Kanzafarova R., Malykh S., Lobaskova M., Khusnutdinova E. Brain derived neurotrophic factor gene (BDNF) and personality traits: the modifying effect of season of birth and sex. Prog. Neuro-Psychopharmacol. Biol. Psychiatry. 2015;56:58–65. doi: 10.1016/j.pnpbp.2014.08.001. [DOI] [PubMed] [Google Scholar]
- Kazantseva A.V., Gaysina D.A., Faskhutdinova G.G., Noskova T., Malykh S.B., Khusnutdinova E.K. Polymorphisms of the serotonin transporter gene (5-HTTLPR, A/G SNP in 5-HTTLPR, and STin2 VNTR) and their relation to personality traits in healthy individuals from Russia. Psychiatr. Genet. 2008;18:167–176. doi: 10.1097/YPG.0b013e328304deb8. [DOI] [PubMed] [Google Scholar]
- Kazantseva A.V., Kutlumbetova Y.Y., Malykh S.B., Lobaskova M.M., Khusnutdinova E.K. [Arginine-vasopressin receptor gene (AVPR1A, AVPR1B) polymorphisms and their relation to personality traits] Genetika. 2014;50:341–352. [PubMed] [Google Scholar]
- Kelemenova S., Schmidtova E., Ficek A., Celec P., Kubranska A., Ostatnikova D. Polymorphisms of candidate genes in Slovak autistic patients. Psychiatr. Genet. 2010;20:137–139. doi: 10.1097/YPG.0b013e32833a1eb3. [DOI] [PubMed] [Google Scholar]
- Kendler K.S., Ohlsson H., Sundquist K., Sundquist J. Sources of parent-offspring resemblance for major depression in a national Swedish extended adoption study. JAMA Psychiatry. 2018;75:194–200. doi: 10.1001/jamapsychiatry.2017.3828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.-R., Kim J.-H., Kim C.-H., Shin J.G., Treasure J. Association between the oxytocin receptor gene polymorphism (rs53576) and bulimia nervosa. Eur. Eat Disord. Rev. 2015;23:171–178. doi: 10.1002/erv.2354. [DOI] [PubMed] [Google Scholar]
- Kuja-Halkola R., D’Onofrio B.M., Larsson H., Lichtenstein P. Maternal smoking during pregnancy and adverse outcomes in offspring: genetic and environmental sources of covariance. Behav. Genet. 2014;44:456–467. doi: 10.1007/s10519-014-9668-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labonté B., Engmann O., Purushothaman I., Menard C., Wang J., Tan C., Scarpa J.R., Moy G., Loh Y.H.E., Cahill M., Lorsch Z.S., Hamilton P.J., Calipari E.S., Hodes G.E., Issler O., Kronman H., Pfau M., Obradovic A.L.J., Dong Y., Neve R.L., Russo S., Kazarskis A., Tamminga C., Mechawar N., Turecki G., Zhang B., Shen L., Nestler E.J. Sex-specific transcriptional signatures in human depression. Nat. Med. 2017;23:1102–1111. doi: 10.1038/nm.4386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahat A., van Lieshout R.J., Mathewson K.J., Mackillop J., Saigal S., Morrison K.M., Boyle M.H., Schmidt L.A. Extremely low birth weight babies grown up: gene-environment interaction predicts internalizing problems in the third and fourth decades of life. Dev. Psychopathol. 2017;29:837–843. doi: 10.1017/S0954579416000511. [DOI] [PubMed] [Google Scholar]
- Landgraf R., Neumann I.D. Vasopressin and oxytocin release within the brain: a dynamic concept of multiple and variable modes of neuropeptide communication. Front. Neuroendocrinol. 2004;25:150–176. doi: 10.1016/j.yfrne.2004.05.001. [DOI] [PubMed] [Google Scholar]
- Li S., Kim E., Wong E.M., Joo J.-H.E., Nguyen T.L., Stone J., Song Y.-M., Flander L.B., Saffery R., Giles G.G., Southey M.C., Sung J., Hopper J.L. Twin birth changes DNA methylation of subsequent siblings. Sci. Rep. 2017;7:8463. doi: 10.1038/s41598-017-08595-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin E., Tsai S.-J. Epigenetics and depression: an update. Psychiatry Investig. 2019;16:654–661. doi: 10.30773/pi.2019.07.17.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little K., Olsson C.A., Youssef G.J., Whittle S., Simmons J.G., Yücel M., Sheeber L.B., Foley D.L., Allen N.B. Linking the serotonin transporter gene, family environments, hippocampal volume and depression onset: a prospective imaging gene × environment analysis. J. Abnorm. Psychol. 2015;124:834–849. doi: 10.1037/abn0000101. [DOI] [PubMed] [Google Scholar]
- LoParo D., Johansson A., Walum H., Westberg L., Santtila P., Waldman I. Rigorous tests of gene-environment interactions in a lab study of the oxytocin receptor gene (OXTR), alcohol exposure, and aggression. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 2016;171:589–602. doi: 10.1002/ajmg.b.32359. [DOI] [PubMed] [Google Scholar]
- Lotan A., Lifschytz T., Wolf G., Keller S., Ben-Ari H., Tatarsky P., Pillar N., Oved K., Sharabany J., Merzel T.K., Matsumoto T., Yamawaki Y., Mernick B., Avidan E., Yamawaki S., Weller A., Shomron N., Lerer B. Differential effects of chronic stress in young-adult and old female mice: cognitive-behavioral manifestations and neurobiological correlates. Mol. Psychiatr. 2018;23:1432–1445. doi: 10.1038/mp.2017.237. [DOI] [PubMed] [Google Scholar]
- Lucht M.J., Barnow S., Sonnenfeld C., Rosenberger A., Grabe H.J., Schroeder W., Völzke H., Freyberger H.J., Herrmann F.H., Kroemer H., Rosskopf D. Associations between the oxytocin receptor gene (OXTR) and affect, loneliness and intelligence in normal subjects. Prog. Neuro-Psychopharmacol. Biol. Psychiatry. 2009;33:860–866. doi: 10.1016/j.pnpbp.2009.04.004. [DOI] [PubMed] [Google Scholar]
- Lucht M.J., Barnow S., Sonnenfeld C., Ulrich I., Grabe H.J., Schroeder W., Völzke H., Freyberger H.J., John U., Herrmann F.H., Kroemer H., Rosskopf D. Associations between the oxytocin receptor gene (OXTR) and “mind-reading” in humans--an exploratory study. Nord. J. Psychiatr. 2013;67:15–21. doi: 10.3109/08039488.2012.700731. [DOI] [PubMed] [Google Scholar]
- Mandelli L., Serretti A. Gene environment interaction studies in depression and suicidal behavior: an update. Neurosci. Biobehav. Rev. 2013;37(10):2375–2397. doi: 10.1016/j.neubiorev.2013.07.011. [DOI] [PubMed] [Google Scholar]
- Mathewson K.J., Chow C.H.T., Dobson K.G., Pope E.I., Schmidt L.A., Van Lieshout R.J. Mental health of extremely low birth weight survivors: a systematic review and meta-analysis. Psychol. Bull. 2017;143:347–383. doi: 10.1037/bul0000091. [DOI] [PubMed] [Google Scholar]
- Meyer-Lindenberg A., Domes G., Kirsch P., Heinrichs M. Oxytocin and vasopressin in the human brain: social neuropeptides for translational medicine. Nat. Rev. Neurosci. 2011 doi: 10.1038/nrn3044. [DOI] [PubMed] [Google Scholar]
- Mileva-Seitz V., Steiner M., Atkinson L., Meaney M.J., Levitan R., Kennedy J.L., Sokolowski M.B., Fleming A.S. Interaction between oxytocin genotypes and early experience predicts quality of mothering and postpartum mood. PloS One. 2013;8 doi: 10.1371/journal.pone.0061443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller M., Musser E.D., Young G.S., Olson B., Steiner R.D., Nigg J.T. Sibling recurrence risk and cross-aggregation of attention-deficit/hyperactivity disorder and autism spectrum disorder. JAMA Pediatr. 2019;173:147–152. doi: 10.1001/jamapediatrics.2018.4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra P., Liu S., Meng X. What DNA methylation modifications and/or genetic variations interact with childhood maltreatment in the development of depression: a systematic review. J. Affect. Disord. 2019;252:325–333. doi: 10.1016/j.jad.2019.04.049. [DOI] [PubMed] [Google Scholar]
- Moons W.G., Way B.M., Taylor S.E. Oxytocin and vasopressin receptor polymorphisms interact with circulating neuropeptides to predict human emotional reactions to stress. Emotion. 2014;14:562–572. doi: 10.1037/a0035503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullins N., Lewis C.M. Genetics of depression: progress at last. Curr. Psychiatr. Rep. 2017;19(8):43. doi: 10.1007/s11920-017-0803-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemoda Z., Szyf M. Epigenetic alterations and prenatal maternal depression. Birth Defects Res. 2017 doi: 10.1002/bdr2.1081. [DOI] [PubMed] [Google Scholar]
- Park C., Rosenblat J.D., Brietzke E., Pan Z., Lee Y., Cao B., Zuckerman H., Kalantarova A., McIntyre R.S. Stress, epigenetics and depression: a systematic review. Neurosci. Biobehav. Rev. 2019:139–152. doi: 10.1016/j.neubiorev.2019.04.010. [DOI] [PubMed] [Google Scholar]
- Parker G., Tupling H., Brown L.B. A parental bonding instrument. Br. J. Med. Psychol. 1979;52:1–10. [Google Scholar]
- Parris M.S., Grunebaum M.F., Galfalvy H.C., Andronikashvili A., Burke A.K., Yin H., Min E., Huang Y.-Y., Mann J.J. Attempted suicide and oxytocin-related gene polymorphisms. J. Affect. Disord. 2018;238:62–68. doi: 10.1016/j.jad.2018.05.022. [DOI] [PubMed] [Google Scholar]
- Pozzi E., Bousman C.A., Simmons J.G., Vijayakumar N., Schwartz O., Seal M., Yap M.B.H., Allen N.B., Whittle S.L. Interaction between hypothalamic-pituitary-adrenal axis genetic variation and maternal behavior in the prediction of amygdala connectivity in children. Neuroimage. 2019;197:493–501. doi: 10.1016/j.neuroimage.2019.05.013. [DOI] [PubMed] [Google Scholar]
- Procyshyn T.L., Hurd P.L., Crespi B.J. Association testing of vasopressin receptor 1a microsatellite polymorphisms in non-clinical autism spectrum phenotypes. Autism Res. 2017;10:750–756. doi: 10.1002/aur.1716. [DOI] [PubMed] [Google Scholar]
- Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J., Sham P.C. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu S., Zhong Y., Shang R., Zhang X., Song W., Kjems J., Li H. The emerging landscape of circular RNA in life processes. RNA Biol. 2017;14:992–999. doi: 10.1080/15476286.2016.1220473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiner I., Van IJzendoorn M.H., Bakermans-Kranenburg M.J., Bleich S., Beutel M., Frieling H. Methylation of the oxytocin receptor gene in clinically depressed patients compared to controls: the role of OXTR rs53576 genotype. J. Psychiatr. Res. 2015;65:9–15. doi: 10.1016/j.jpsychires.2015.03.012. [DOI] [PubMed] [Google Scholar]
- Resnick B., Klinedinst N.J., Yerges-Armstrong L., Choi E.Y., Dorsey S.G. The impact of genetics on physical resilience and Successful aging. J. Aging Health. 2015;27:1084–1104. doi: 10.1177/0898264315577586. [DOI] [PubMed] [Google Scholar]
- Richter J., Richter G., Eisemann M., Mau R. Sibship size, sibship position, parental rearing and psychopathological manifestations in adults: preliminary analysis. Psychopathology. 1997;30:155–162. doi: 10.1159/000285042. [DOI] [PubMed] [Google Scholar]
- Rodrigues S.M., Saslow L.R., Garcia N., John O.P., Keltner D. New doc 2017-11-30 (1) (3) Proc. Natl. Acad. Sci. U.S.A. 2009;106:21437–21441. doi: 10.1073/pnas.0909579106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saphire-Bernstein S., Way B.M., Kim H.S., Sherman D.K., Taylor S.E. Oxytocin receptor gene (OXTR) is related to psychological resources. Proc. Natl. Acad. Sci. U.S.A. 2011;108:15118–15122. doi: 10.1073/pnas.1113137108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz L.L., Gard A.M., Ware E.B. Examining sex differences in pleiotropic effects for depression and smoking using polygenic and gene-region aggregation techniques. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 2019;180:448–468. doi: 10.1002/ajmg.b.32748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seneviratne C., Huang W., Ait-Daoud N., Li M.D., Johnson B.A. Characterization of a functional polymorphism in the 3’ UTR of SLC6A4 and its association with drinking intensity. Alcohol Clin. Exp. Res. 2009;33:332–339. doi: 10.1111/j.1530-0277.2008.00837.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva M.M., Rodrigues B., Fernandes J., Santos S.D., Carreto L., Santos M.A.S., Pinheiro P., Carvalho A.L. MicroRNA-186-5p controls GluA2 surface expression and synaptic scaling in hippocampal neurons. Proc. Natl. Acad. Sci. U.S.A. 2019;116:5727–5736. doi: 10.1073/pnas.1900338116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skuse D.H., Gallagher L. Dopaminergic-neuropeptide interactions in the social brain. Trends Cognit. Sci. 2009 doi: 10.1016/j.tics.2008.09.007. [DOI] [PubMed] [Google Scholar]
- Smearman E.L., Almli L.M., Conneely K.N., Brody G.H., Sales J.M., Bradley B., Ressler K.J., Smith A.K. Oxytocin receptor genetic and epigenetic variations: association with child abuse and adult psychiatric symptoms. Child Dev. 2016;87:122–134. doi: 10.1111/cdev.12493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smolka M.N., Reimold M., Kobiella A., Reischl G., Rietschel M., Heinz A. Smoking moderates association of 5-HTTLPR and in vivo availability of serotonin transporters. Eur. Neuropsychopharmacol. 2019;29:171–178. doi: 10.1016/j.euroneuro.2018.08.509. [DOI] [PubMed] [Google Scholar]
- Starr L.R., Huang M. HPA-axis multilocus genetic variation moderates associations between environmental stress and depressive symptoms among adolescents. Dev. Psychopathol. 2019;31:1339–1352. doi: 10.1017/S0954579418000779. [DOI] [PubMed] [Google Scholar]
- Starr L.R., Vrshek-Schallhorn S., Stroud C.B. Serotonergic multilocus genetic variation moderates the association between major interpersonal stress and adolescent depressive symptoms: replication and candidate environment specification. J. Psychiatr. Res. 2019;117:55–61. doi: 10.1016/j.jpsychires.2019.06.020. [DOI] [PubMed] [Google Scholar]
- Szczepankiewicz A., Leszczyńska-Rodziewicz A., Pawlak J., Rajewska-Rager A., Wilkosc M., Zaremba D., Dmitrzak-Weglarz M., Skibinska M., Hauser J. Epistatic interaction between CRHR1 and AVPR1b variants as a predictor of major depressive disorder. Psychiatr. Genet. 2013;23:239–246. doi: 10.1097/YPG.0000000000000007. [DOI] [PubMed] [Google Scholar]
- Tansey K.E., Hill M.J., Cochrane L.E., Gill M., Anney R.J., Gallagher L. Functionality of promoter microsatellites of arginine vasopressin receptor 1A (AVPR1A): implications for autism. Mol. Autism. 2011;2:3. doi: 10.1186/2040-2392-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teh A.L., Pan H., Chen L., Ong M.-L., Dogra S., Wong J., MacIsaac J.L., Mah S.M., McEwen L.M., Saw S.-M., Godfrey K.M., Chong Y.-S., Kwek K., Kwoh C.-K., Soh S.-E., Chong M.F.F., Barton S., Karnani N., Cheong C.Y., Buschdorf J.P., Stünkel W., Kobor M.S., Meaney M.J., Gluckman P.D., Holbrook J.D. The effect of genotype and in utero environment on interindividual variation in neonate DNA methylomes. Genome Res. 2014;24:1064–1074. doi: 10.1101/gr.171439.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson R.J., Parker K.J., Hallmayer J.F., Waugh C.E., Gotlib I.H. Oxytocin receptor gene polymorphism (rs2254298) interacts with familial risk for psychopathology to predict symptoms of depression and anxiety in adolescent girls. Psychoneuroendocrinology. 2011;36:144–147. doi: 10.1016/j.psyneuen.2010.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner T., Pihur V., Chakravarti A. Quantifying and modeling birth order effects in autism. PloS One. 2011;6 doi: 10.1371/journal.pone.0026418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tylee D.S., Sun J., Hess J.L., Tahir M.A., Sharma E., Malik R., Worrall B.B., Levine A.J., Martinson J.J., Nejentsev S., Speed D., Fischer A., Mick E., Walker B.R., Crawford A., Grant S.F.A., Polychronakos C., Bradfield J.P., Sleiman P.M.A., Hakonarson H., Ellinghaus E., Elder J.T., Tsoi L.C., Trembath R.C., Barker J.N., Franke A., Dehghan A., 23 and Me Research Team, Inflammation Working Group of the CHARGE Consortium, METASTROKE Consortium of the International Stroke Genetics Consortium, Netherlands Twin Registry, neuroCHARGE Working Group, Obsessive Compulsive and Tourette Syndrome Working Group of the Psychiatric Genomics Consortium. Faraone S.V., Glatt S.J. Genetic correlations among psychiatric and immune-related phenotypes based on genome-wide association data. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 2018;177:641–657. doi: 10.1002/ajmg.b.32652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underwood M.D., Kassir S.A., Bakalian M.J., Galfalvy H., Dwork A.J., Mann J.J., Arango V. Serotonin receptors and suicide, major depression, alcohol use disorder and reported early life adversity. Transl. Psychiatry. 2018;8:279. doi: 10.1038/s41398-018-0309-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Assche E., Moons T., Van Leeuwen K., Colpin H., Verschueren K., Van Den Noortgate W., Goossens L., Claes S. Depressive symptoms in adolescence: the role of perceived parental support, psychological control, and proactive control in interaction with 5-HTTLPR. Eur. Psychiatr. 2016;35:55–63. doi: 10.1016/j.eurpsy.2016.01.2428. [DOI] [PubMed] [Google Scholar]
- Van der Auwera S., Peyrot W.J., Milaneschi Y., Hertel J., Baune B., Breen G., Byrne E., Dunn E.C., Fisher H., Homuth G., Levinson D., Lewis C., Mills N., Mullins N., Nauck M., Pistis G., Preisig M., Rietschel M., Ripke S., Sullivan P., Teumer A., Völzke H., Boomsma D.I., Wray N.R., Penninx B., Grabe H. Genome-wide gene-environment interaction in depression: a systematic evaluation of candidate genes. Am. J. Med. Genet. Part B Neuropsychiatr. Genet. 2017;177:40–49. doi: 10.1002/ajmg.b.32593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Li S., Li H., Jia C. Association of serotonergic pathway genes with smoking cessation in a Chinese rural male population. Addict. Behav. 2018;80:34–38. doi: 10.1016/j.addbeh.2018.01.001. [DOI] [PubMed] [Google Scholar]
- Wong M.-L., Dong C., Andreev V., Arcos-Burgos M., Licinio J. Prediction of susceptibility to major depression by a model of interactions of multiple functional genetic variants and environmental factors. Mol. Psychiatr. 2012;17:624–633. doi: 10.1038/mp.2012.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu N., Shang S., Su Y. The arginine vasopressin V1b receptor gene and prosociality: mediation role of emotional empathy. PsyCh J. 2015;4:160–165. doi: 10.1002/pchj.102. [DOI] [PubMed] [Google Scholar]
- Yang S.Y., Kim S.A., Hur G.M., Park M., Park J.-E., Yoo H.J. Replicative genetic association study between functional polymorphisms in AVPR1A and social behavior scales of autism spectrum disorder in the Korean population. Mol. Autism. 2017;8:44. doi: 10.1186/s13229-017-0161-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Liu J., Cheng P., Lv F. Correlation between miRNA target site polymorphisms in the 3’ UTR of AVPR1A and the risk of hypertension in the Chinese Han population. Biosci. Rep. 2019;39 doi: 10.1042/BSR20182232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Du L., Bai Y., Han B., He C., Gong L., Huang R., Shen L., Chao J., Liu P., Zhang Hongxing, Zhang Haisan, Gu L., Li J., Hu G., Xie C., Zhang Z., Yao H. CircDYM ameliorates depressive-like behavior by targeting miR-9 to regulate microglial activation via HSP90 ubiquitination. Mol. Psychiatr. 2018;9:1–6. doi: 10.1038/s41380-018-0285-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S., Zhang J. The association between depression, suicidal ideation and psychological Strains in college Students: a cross-national study. Cult. Med. Psychiatry. 2018;42:914–928. doi: 10.1007/s11013-018-9591-x. [DOI] [PubMed] [Google Scholar]
- Zhuo C.-J., Hou W.-H., Jiang D.-G., Tian H.-J., Wang L.-N., Jia F., Zhou C.-H., Zhu J.-J. Circular RNAs in early brain development and their influence and clinical significance in neuropsychiatric disorders. Neural Regen. Res. 2020;15:817–823. doi: 10.4103/1673-5374.268969. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.