Abstract
Background
Hepatitis E virus (HEV) genotype 3 has a worldwide distribution. The food-borne transmission of HEV associated with the consumption of products derived from domestic pig, wild boar has been reported in various countries. In this study the genetic diversity, evolutionary rates of HEV 3f, 3c among swine and wild boar in Italy were estimated.
Methods
Sampling was performed on a wild boar population living in an area located in Abruzzo region. The HEV RNA amplification was performed by real-time RT-PCR. Nested RT-PCR and sequencing of the ORF2 region were carried out by the Super Script III First-Strand Synthesis System. Sequencing of purified PCR products was carried out by the Genome Lab Dye Terminator Cycle Sequencing (DTCS) Quick Start Kit. The maximum likelihood trees were generated by using Phyml. The mean evolutionary rates and the dated trees were co-estimated by BEAST.
Results
The phylogenetic analysis showed that the HEV ORF2 isolates from Abruzzo region belonged to 3f subtype. The prevalent subtypes in Italy were those belonging to 3f and 3c. The estimated mean values of the HEV ORF2 capsid gene evolutionary rates were 1.915 × 10−2 substitutions/site/year (95% HPD: 1.64 × 10−3 – 3.97 × 10−2) and 2.81 × 10−2 substitutions/site/year (1.83 × 10−2 - 3.8 × 10−2) for 3f and 3c subtype datasets, respectively.
The HEV 3f dated back to 1985 (1960–2000), whereas the 3c subtype entered in Italy during the year 2006 (2005–2006). The majority of the HEV 3f sequences collected from swine didn't appear intermixed, except in two cases. The HEV 3c population circulating in Italy remained segregated without significant transfer to swine.
Conclusion
Our study provide insight into the evolution, circulation of HEV 3f and 3c in Italy.
Continued genomic surveillance of HEV in animal reservoir, as well as improving sanitary control measures are required.
Keywords: Bioinformatics, Microbiology, Epidemiology, Hepatitis E virus, Molecular evolution, Phylogeny, Animal reservoir, Genomic surveillance
Bioinformatics; Microbiology; Epidemiology; Hepatitis E virus; Molecular evolution; Phylogeny; Animal reservoir; Genomic surveillance.
1. Introduction
Hepatitis E virus (HEV) is a non-enveloped single stranded RNA virus, that belongs to the Hepeviridae family and is a major cause of viral hepatitis in humans worldwide (WHO, 2019). HEV has been classified into seven genotypes (Smith et al., 2014, 2016; Sridhar et al., 2017). In particular, genotypes 1 and 2 (Smith et al., 2014) infect only humans and are endemic in developing countries as Asia, Africa and Mexico and Central-South America, where are usually transmitted feco-orally by contaminated water; whereas genotypes 3 and 4 can infect humans and various animals, particularly pigs, wild boars, and other mammals (Smith et al., 2014; Pavio et al., 2017).
HEV genotype 3 has a worldwide distribution and most of the infections in humans are asymptomatic, self-limited, locally acquired.
The virus is spread worldwide and the most common subtypes in Europe are 3c, 3e and 3f (Adlhoch et al., 2016). The food-borne transmission of HEV associated with the consumption of liver, meat, sausages and offal products derived from domestic pig, wild boar, and deer has been reported in several studies conducted in various countries (Colson et al., 2010, 2012; Renou et al., 2014; Riveiro-Barciela et al., 2015; Tei et al., 2004; Yazaki et al., 2003).
In Italy, it was observed an active circulation of this virus probably related to a relatively high prevalence of HEV in domestic pigs and wild boar (Di Pasquale et al., 2019; De Sabato et al., 2019; Romanò et al., 2011; Martinelli et al., 2011; Martelli et al., 2008). A recent national study in blood donors reported a great variation in the anti-HEV prevalence among the different Italian regions, with values from 2.2% to 22.8% (Spada et al., 2018). High prevalence rates were observed in Abruzzo (22.8%) and in Sardinia (19.9%). In addition, overall rates between 10 and 15% were detected in several regions of Central Italy (Lazio, Umbria and Marche). Importantly, a prevalence of 31.6% has been detected at L'Aquila, the main city of Abruzzo region (Spada et al., 2018).
These high anti-HEV prevalences urged monitoring of food products that might be sources for HEV infection. Subtyping of the HEV virus through phylogenetic analysis, in association with the evolutionary rate estimate, can help identifying the viral evolution dynamics, possible origin, transmission and circulation.
The aim of the present study was to investigate the genetic diversity and circulation of HEV (subtype 3f and 3c) among swine and wild boar in Italy. The mean evolutionary rates and the date of origin of 3f and 3c subtypes clades, based on the HEV ORF2 partial gene sequences, were also estimated.
2. Materials and methods
2.1. Sample collection
Sampling was performed on a wild boar (Sus scrofa scrofa) population living in an area located in the province of L'Aquila, Abruzzo region. In agreement with the regional regulation, a demographic control program of wild boar was applied. During the slaughtering process, a liver sample was taken from each animal with a sterile lancet (used once and then discarded) and stored at -80 °C until processing. In 2017, liver samples from 106 animals were collected and tested for HEV RNA: twelve of them proved to be HEV RNA positive and an ORF2 sequence could finally be obtained from six of them by nested RT-PCR and sequencing (see below).
2.2. HEV RNA extraction and detection
HEV RNA extraction from liver samples and detection by real-time RT-PCR were carried out by the National Reference Laboratory for Foodborne Viruses at Istituto Superiore di Sanità as previously described (Di Pasquale et al., 2019). Briefly, 1 g of chopped tissue was digested by Trizol treatment as Szabo et al. (2015) have recently shown, and RNA was extracted from the recovered supernatant using the MiniMag platform (bioMerieux, Marcy-l'Étoile, France). HEV detection was carried out with an optimized real-time RT-PCR assay, using previously published primers/probe (Jothikumar et al., 2006; Garson et al., 2012) and thermal conditions (La Rosa et al., 2018).
2.3. Nested RT-PCR and sequencing
Nested RT-PCR and sequencing of ORF2 region of viral genome were carried out at the Istituto Superiore di Sanita by the National Reference Laboratory for Viral Hepatitis (NRL-VH). Extracted HEV RNA from real-time RT-PCR positive samples was reverse transcribed by the SuperScript III First-Strand Synthesis System for RT-PCR (Invitrogen) with random hexamers. Nested PCR was carried out with primer sequences made available by RIVM to European laboratories participating to the HEVnet database. The presence and size of amplification products were assessed by agarose gel electrophoresis. To control for PCR contamination, negative control sera were extracted and subjected to reverse transcription and nested PCR in each run, along with the sera to be assayed. Sequencing of purified PCR products was carried out by the GenomeLab Dye Terminator Cycle Sequencing (DTCS) Quick Start Kit and an automated DNA sequencer (Beckman Coulter, Inc., Fullerton, CA). The final length of genomic region obtained and used was 490 bp.
2.4. Phylogenetic datasets
The first dataset (490 bp) included six HEV ORF2 sequences obtained from wild boar liver samples at the National Institute of Health, Department of Infectious Diseases, Rome, Italy (collection year: 2017), together with n = 25 subtype specific reference sequences downloaded from NCBI database. The inclusion criteria for the 25 reference sequences were: i) all the reference sequences were downloaded from the National Centre for Biotechnology Information (NCBI) database; ii) all the reference sequences belonged to Hepatitis E virus; iii) all the reference sequences were already published in peer-reviewed journals; iv) no uncertainty about the subtype assignment. This dataset was used to determine the subtype of the six HEV isolates.
The second dataset (average length of the edited alignment: 368 bp) included a total of 198 HEV ORF2 sequences (genotype 3), 34 of which were HEV references with known subtype, 158 were HEV ORF2 sequences from swine and wild boar samples collected in Italy and downloaded from the NCBI database and six were the HEV ORF2 sequences from wild boar obtained at ISS (Istituto Superiore di Sanità). This dataset was used to extrapolate the most prevalent subtypes circulating in swine and wild boar in Italy.
The third dataset (average length of the edited alignment: 341 bp) included n = 38 HEV ORF2 subtype 3f sequences from swine and wild boar, circulating in Italy. The inclusion criteria for reference sequences in this dataset were: i) all the reference sequences were downloaded from the National Centre for Biotechnology Information (NCBI) database; ii) all the reference sequences belonged to Hepatitis E virus, genotype 3 - subtype f; iii) the references have been derived from swine or wild board collected in Italy.
This dataset has been used to estimate the mean evolutionary rate of 3f subtype, the date of tMRCA, to understand the HEV 3f circulation dynamics and possible transmission.
The fourth dataset (average length of the edited alignment: 322 bp) included n = 39 HEV ORF2 subtype 3c, from swine and wild boar circulating in Italy. The inclusion criteria for reference sequences in this dataset were: i) all the reference sequences were downloaded from the National Centre for Biotechnology Information (NCBI) database; ii) all the references belonged to Hepatitis E virus, genotype 3 - subtype c; ii) the references have been derived from swine or wild board collected in Italy. This dataset has been used to estimate the mean evolutionary rate of 3c subtype, the date of tMRCA, to understand the HEV 3c circulation dynamics and possible transmission.
2.5. Phylogenetic analysis
All sequences in the dataset were aligned and manually edited by using Bioedit (Hall, 1999). The best fitting evolutionary model for each dataset was estimated by using JModeltest (Posada, 2008; Darriba et al., 2012).
The phylogenetic signal in each dataset was investigated with the likelihood mapping method by analyzing groups of four randomly chosen sequences, called quartets through TREE-PUZZLE program and by using 10,000 random quartets (Strimmer and von Haeseler, 1997). A quartet has three possible un-rooted tree topologies. The likelihood of each topology is estimated with the maximum likelihood method and the three likelihoods are reported as a dot in an equilateral triangle (the likelihood map). The three corners represented fully resolved tree topologies (i.e., the presence of a treelike phylogenetic signal in the data), the center represents a star-like phylogeny, and the three areas on the sides that indicate a network-like phylogeny (i.e., the presence of recombination or conflicting phylogenetic signals) as previously reported (Montesano et al., 2016). A substantial star-like signal (i.e., a star-like outburst of multiple phylogenetic lineages) is indicated by > 33% dots falling within the central area, as confirmed by extensive simulation studies (Strimmer and von Haeseler, 1997). The maximum likelihood phylogenetic trees were generated on the first and second dataset by using Phyml v 3.0 (Guindon et al., 2010) with the GTR + I + G model of nucleotide substitution, previously estimated through JModeltest.
The statistical robustness and reliability of the branching order within the phylogenetic trees was confirmed by bootstrap analysis (bootstrap >75%).
2.6. Bayesian evolutionary rate estimate and dated trees
The mean evolutionary rate and the dated trees for the third and fourth data set were co-estimated using a Bayesian Monte Carlo Markov Chain (MCMC) approach under the GTR + I + G model and by using both a strict and an uncorrelated log-normal relaxed clock model by using BEAST (Drummond and Rambaut, 2007).
Three parametric demographic models of population growth (constant size, exponential, and expansion) and two non-parametric models (Bayesian skyline plot, BSP and the Smooth skyride plot Gaussian Markov Random Field – GMRF) were compared as coalescent priors.
The best fitting models were selected by means of a Bayes factor (BF, using marginal likelihoods) implemented in Beast (Drummond and Rambaut, 2007). In accordance with Kass and Raftery (1995), the strength of the evidence against H0 (null hypothesis) was evaluated as follows: 2lnBF <2 = no evidence; 2–6 = weak evidence; 6–10 = strong evidence; and >10 = very strong evidence. A negative 2lnBF indicates evidence in favor of H0. Only values ≥6 were considered significant.
The MCMC chains were run for at least 50 million generations and sampled every 5,000 steps. Convergence was assessed by estimating the effective sampling size (ESS) after a 10% burn-in, using Tracer software (http://tree.bio.ed.ac.uk/software/tracer/), and accepting ESS values of 250 or more. Uncertainty in the estimates was indicated by 95% highest posterior density (95% HPD) intervals.
The posterior probability has been used as statistical support for specific clades and clusters.
The trees were summarized by Tree Annotator, by choosing the tree with the maximum product of posterior probabilities (maximum clade credibility or MCC), after a 10% burn-in.
3. Results
3.1. Likelihood mapping
The percentages of dots falling in the central area of the likelihood map were 3%, 4.8%, 10.1% and 26% for the first, second, third and fourth data sets, respectively. As none of the datasets showed more than 33% of noise, they contained sufficient phylogenetic signal, meaning that sequence evolution followed a resolved tree topology.
3.2. Phylogenetic analysis
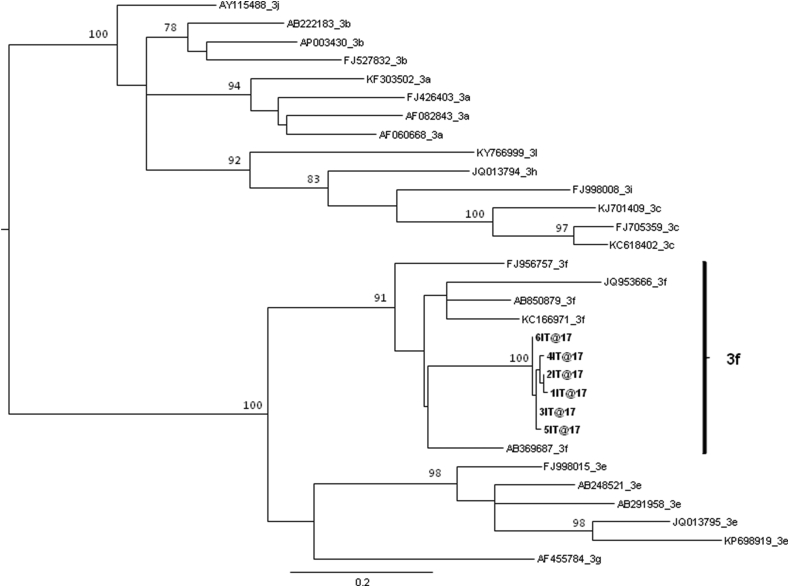
The maximum likelihood phylogenetic tree of the first dataset (Figure 1) showed that all the six HEV ORF 2 isolates from ISS belonged to 3f subtype. The phylogenetic relationships among the different clades were confirmed by bootstrap values >75 %.
Figure 1.
Maximum likelihood phylogenetic tree of HEV ORF2 gene, first dataset. Maximum likelihood phylogenetic tree of six HEV ORF2 sequences, derived from wild boar liver samples at Abruzzo Region, together with n = 25 subtype specific HEV reference sequences downloaded from the NCBI database. The tree was midpoint rooted. Branch lengths were estimated with the best fitting nucleotide substitution model (GTR + I + G) according to a hierarchical likelihood ratio test. Scale bar at the bottom represents 0.2 nucleotide substitutions per site. Values along the branches represents significant statistical support for the clusters subtending that branch (bootstrap support >75%). The clade representing HEV 3f subtype sequences was highlighted.
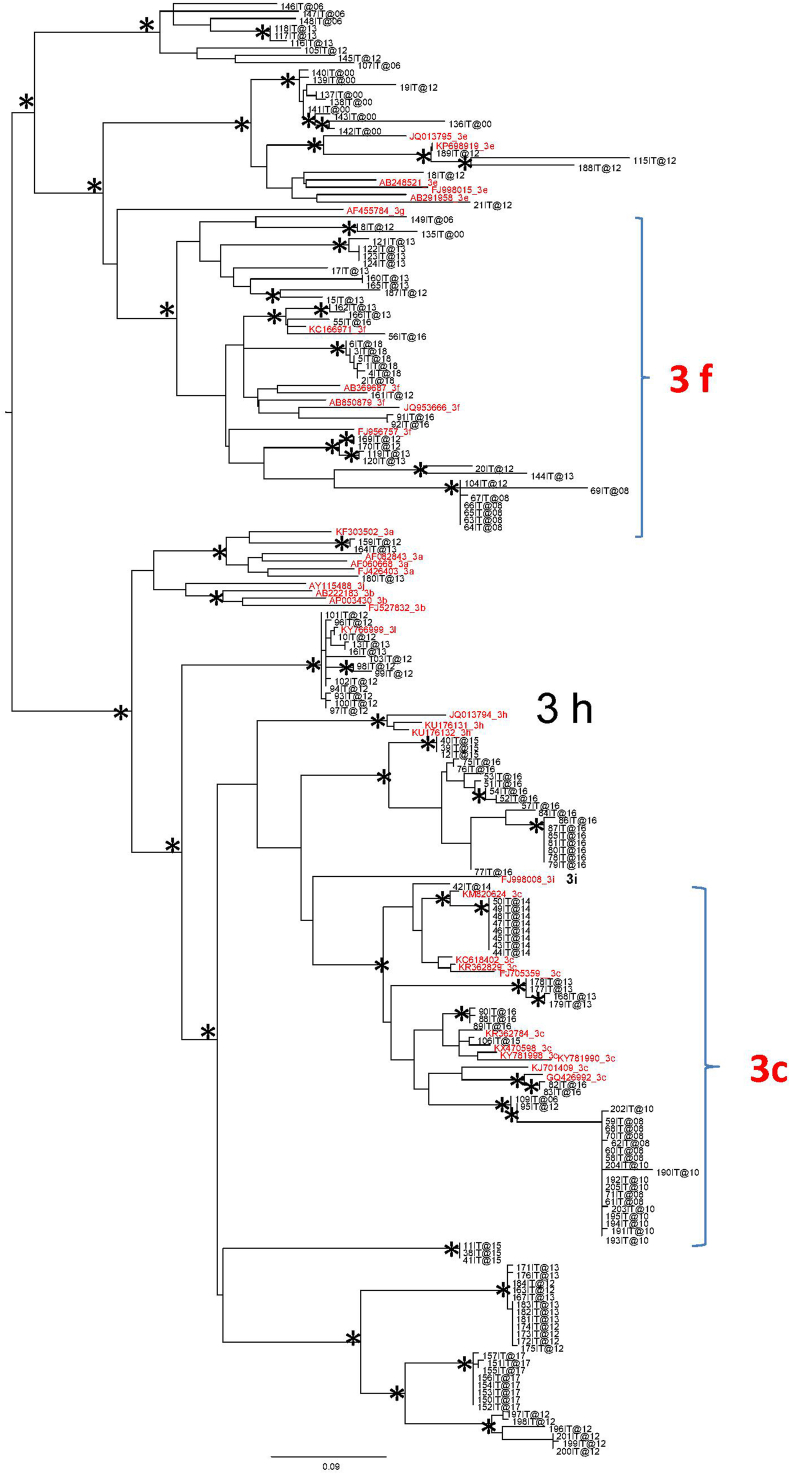
The maximum likelihood phylogenetic tree of the second dataset, provided in Supplementary Figure 1, clearly indicated that the most numerous clades were represented by the subtypes 3f and 3c (supported by bootstrap >75%). This analysis allowed to confirm and extrapolate HEV sequences from swine and wild boar, belonging to the most prevalent subtypes (3f and 3c) circulating in Italy and useful for the subsequent analysis.
3.3. Bayesian evolutionary rate estimate and dated trees
The Bayes Factor analysis showed that the third and fourth dataset fitted significantly better with the relaxed clock than with the strict clock (2 ln BF > 50 for the relaxed clock).
The BF analysis, under the relaxed clock, showed that expansion model was better than the other models (2 ln BF > 19) for the third dataset, while for the fourth dataset, under the relaxed clock, the BSP model was better than the other models (2lnBF >10).
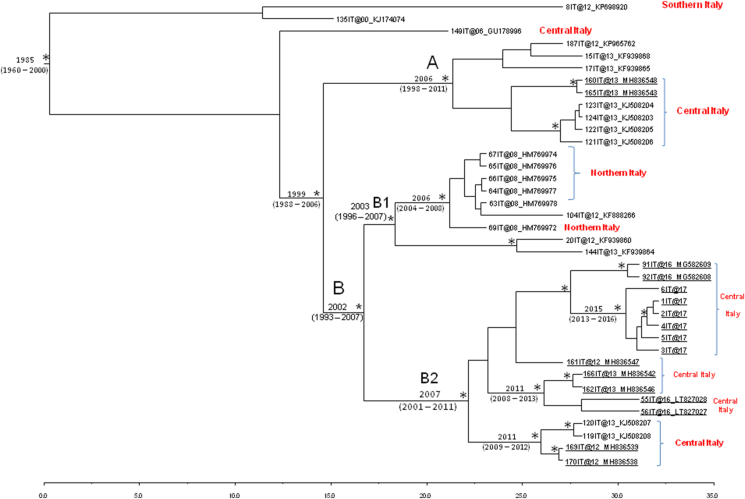
The estimated mean value of the HEV ORF2 capsid gene evolutionary rate estimated for 3f subtype dataset was 1.915 × 10−2 substitutions/site/year (95% HPD: 1.64 × 10−3 – 3.97 × 10−2). Figure 2 shows the Bayesian maximum clade credibility tree and the time to the most common recent ancestor (tMRCA) estimates performed on the third data set (HEV subtype 3f from swine and wild boar). The root of the tree dated back to the year 1985 (95% HPD: 1960–2000). A main statistically supported clade can be highlighted, that dated back to 1999 (95% HPD: 1988–2006).
Figure 2.
Maximum clade credibility (MCC) tree of HEV ORF2 subtype 3f sequences from swine and wild boar circulating in Italy, third dataset. The maximum clade credibility (MCC) tree of n = 38 HEV ORF2 subtype 3f sequences from swine and wild boar, circulating in Italy. The asterisks (∗) along the branches represent posterior probability (statistical supported values, pp > 0.90). The scale at the bottom of the tree represents time (years) before the last sampling time. Main clades were highlighted. The bold and underlining indicates sequences sampled from wild boar. The geographical origin of the clades or sequences, where available, was indicated as Northern, Central and Southern Italy.
Within the main clade, two clades (A and B) were found. Clade A, dated back to 2006 (95% HPD: 1998–2011), included a total of nine sequences, seven of them collected from swine and two from wild boar. The lower part of clade A included two HEV ORF 2 sequences from wild boar collected in Central Italy (Aquapendente, Viterbo province) and four HEV ORF 2 sequences from swine, also circulating in Central Italy.
Clade B, which dated to 2002 (95% HPD: 1993–2007), was splitted into two sub-clades, B1 and B2. The majority of sequences inside sub-clade B1, which originated in 2003 (95% HPD: 1996–2007), were collected from Northern Italy. The sub-clade B2, which dated back to 2007 (95% HPD: 2001–2011), included all HEV ORF2 sequences sampled in Central Italy. The six HEV – ISS isolates from wild boar (labeled as 1IT@17, 2IT@17, 3IT@17, 4IT@17, 5IT@17, 6IT@17) and sampled from L'Aquila (Abruzzo Region, Central Italy) appeared closely related together, in a supported cluster originating in 2015 (95% HPD: 2013–2016) but also related to two HEV sequences from wild boar collected from Viterbo (Lazio Region, Central Italy). Two further statistically supported clusters were highlighted inside B2. The first one included four HEV sequences collected from wild boar: two of them were collected from Montefiascone Lubriano (Viterbo province), the others were sampled in Umbria. The second one, dated to 2011 (95% HPD: 2009–2011), included two HEV sequences from swine (Central Italy) related to two HEV sequences from wild boar sampled at Bolsena (Viterbo province).
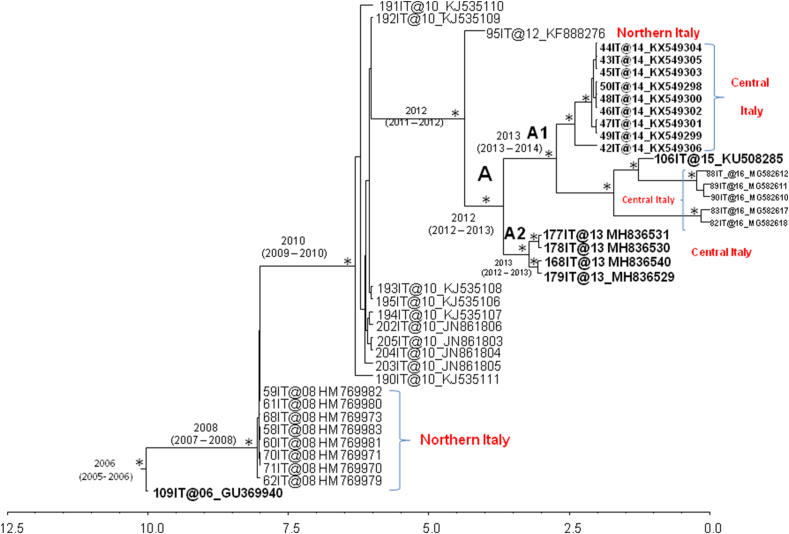
Figure 3 shows the dated tree of HEV sequences belonging to subtype 3c (fourth dataset). The estimated mean value of the HEV ORF 2 capsid gene subtype 3c evolutionary rate was 2.81 × 10−2 substitutions/site/year (1.83 × 10−2 - 3.8 × 10−2). The root of the tree dated back to the year 2006 (95% HPD: 2005–2006) and a main clade originating to the year 2008 (95% HPD: 2007–2008) was found. Inside the main clade, HEV sequences mainly segregated according to their geographical sampling locations, in fact HEV sequences sampled in Northern Italy were mainly found as out group (externally located).
Figure 3.
Maximum clade credibility (MCC) tree of HEV ORF2 subtype 3c sequences from swine and wild boar circulating in Italy, fourth dataset. The maximum clade credibility (MCC) tree of n = 39 HEV ORF2 subtype 3c, from swine and wild boar, circulating in Italy. The asterisks (∗) along the branches represent posterior probability (statistical supported values, pp > 0.90). The scale at the bottom of the tree represents time (years) before the last sampling time. Main clades were highlighted. The bold indicates sequences sampled from wild boar. The geographical origin of the clades or sequences, where available, was indicated as Northern and Central Italy.
An internal clade (A), dated to the year 2012 (95% HPD: 2012–2013), mostly included HEV sequences sampled from Central Italy. It splitted into two clusters, A1 and A2. Cluster A1 dated back to 2013 (95% HPD: 2013–2014) and included HEV wild boar sequences from Abruzzo and Lazio Regions; cluster A2, dated to 2013 (95% HPD: 2012–2013), included four sequences sampled from wild boar in the province of Viterbo (Montalto di Castro, Farnese and Tuscania).
4. Discussion
Hepatitis E is considered an emerging disease in industrialized countries, due to an increase in the autochthonous cases (Adlhoch et al., 2016; Aspinall et al., 2017). Studies conducted among the general population and blood donors in Europe found variable anti-HEV seroprevalence rates, with important variation even within the same country (Fogeda et al., 2012; Faber et al., 2012; Ijaz et al., 2009; Verhoef et al., 2012; Wenzel et al., 2014). In Italy, heterogeneous anti-HEV IgG prevalence rates of 1.0–4.3% and 0.7–9.1% were found in the general population (in the period between 1993 and 2011) and in blood donors (in the period between 1993 and 2013), respectively (Zanetti and Dawson, 1994; Vulcano et al., 2007; Masia et al., 2009). Those hetereogeneous values were most likely due to variable sensitivity of the different detection systems used in each study as well as to the geographical area from which the studied populations were enrolled. A recent study in 10,011 blood donors collected throughout Italy and centrally tested with a validated highly sensitive system showed an average national anti-HEV IgG sero-prevalence rate of 8.7% but with a great variation among the different Italian regions and some hyperendemic areas with prevalence rates approaching or even higher than 30% (Spada et al., 2018). This situation led to monitor the food products that might be sources for HEV infection. Swine and wild boars are considered important reservoirs for human infections. Recent studies performed in Italy focused the attention on the detection and genetic characterization of HEV in swine and wild boar, but none of them applied high resolution Bayesian phylogenetic analysis to study the evolutionary dynamics of specific HEV genotype 3 subtypes and the evolutionary rates.
In this study we analyzed the phylogenetic relationships of six HEV isolates from wild boar sampled in an area located in the province of L'Aquila (Abruzzo region, Central Italy), together with specific reference sequences to perform the subtyping. The province of L'Aquila, which covers part of the National Park of Abruzzo, Lazio and Molise is characterized by rich and varied vegetation forms together with a great animal biodiversity. We analyzed for the first time the circulation of HEV belonging to subtype 3f and 3c among swine and wild boar in Italy, together with the estimation of the mean evolutionary rates and dates of origin of these subtypes.
The maximum likelihood tree evidenced that the six HEV isolates from ISS, collected from wild boar in Abruzzo Region, were genotype 3 subtype f. Search for HEV genotype 3 swine and wild boar sequences collected in Italy in NCBI database and subsequent phylogenetic analysis revealed that the most prevalent subtypes were 3c and 3f, similarly to other European countries (Adlhoch et al., 2016). The estimated mean value of the HEV ORF 2 capsid gene evolutionary rate estimated for 3f subtype dataset was 1.915 × 10−2 substitutions/site/year (95% HPD: 1.64 × 10−3 – 3.97 × 10−2), which appeared comparable to those reported by other authors (Nakano et al., 2012; Zehender et al., 2014; Montesano et., 2016). Based on this temporal reconstruction, the HEV subtype 3f dated back to 1985 (1960–2000) and a main supported clade, including the majority of the sequences, has been identified. Looking at both the geographical distribution and the tree structure (Figure 2), it appears that two different groups of 3f strains circulated approximately in the same time period in Central Italy: the first one represented by the strains located in clade A (originating in 2006) and the second one represented by the strains in sub-clade B2 (originating in 2007). The sequences collected from Northern Italy segregated inside sub-clade B1, dated to 2003 (1996–2007). One sequence from Southern Italy appeared located outside the main clade. Overall, these data show a clear segregation according to the sampling location.
In our study, the majority of the HEV 3f sequences collected from swine didn't appear intermixed together with those isolated from wild boar, except in two cases: (a) in clade A two sequences from wild boar (160IT@13_MH836548 and 165IT@13_MH836543) appeared intermixed with those from swine; inside sub-clade B2, two sequences from swine (120IT@13_KJ508207 and 119IT@13_KJ508208) appeared to be genetically highly related to strains obtained from wild boar (169IT@12_MH836539 and 170IT@12_MH836538).
The swine become infected by direct contact with infected pig faeces or exposure to water/feed contaminated with HEV. The intermixing between swine and wild boar strains, found here, suggests that the transmission/circulation between swine and wild boar can potentially happen under specific circumstances, e.g. by direct contact, such as in case of open air fenced farms. The wild boar/pig interface and/or extensive farming practices in contact with the natural environment might play a role in the exposure of swine to the virus.
A mean evolutionary rate of 2.81 × 10−2 substitutions/site/year (1.83 × 10−2 - 3.8 × 10−2) has been estimated for subtype 3c dataset. According to this estimate, we can speculate that HEV 3c entered/circulated in Italy starting from the year 2006 (95% HPD: 2005–2006). The HEV 3c sequences collected from Northern and Central Italy appeared segregated in distinct clusters. Sequences from wild boar resulted separated respect to those sampled from swine, suggesting that HEV 3c exchange did not occur, i.e. wild boar HEV population remained segregated without significant transfer to swine.
An increase in consumption of pork between 1961–1981 was observed in high income countries, with parallel increase of production, until the first years of 2000s (Zehender et al., 2014). At the same time, farming in larger, fewer and totally confined farms than in the previous decades took over. In Italy, it was reported that from 1991 to 2001 the pork meat production was not self-sufficient and both pork meat and live animals were imported, mainly from The Netherlands (34%), Germany (19%), Denmark (17%) and France (14%) (Cozzi and Ragno, 2003). These factors led to a greater density of pigs per farm and, possibly, a general larger spread of HEV-3 infection. In order to prevent the spread of HEV infection in Italy, continued genomic surveillance of HEV in animal reservoir (i.e. swine and wild boar) as well as improving sanitary control measures are required.
Declarations
Author contribution statement
Alessandra Lo Presti: Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Roberto Bruni, Alessandro Grimaldi, Dario De Medici: Analyzed and interpreted the data; Wrote the paper.
Umbertina Villano, Cinzia Marcantonio, Michele Equestre, Elisabetta Suffredini, Simona Di Pasquale: Performed the experiments.
Massimo Ciuffetelli: Conceived and designed the experiments; Performed the experiments.
Anna Rita Ciccaglione: Conceived and designed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Funding statement
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Competing interest statement
The authors declare no conflict of interest.
Additional information
The six HEV sequences will be deposited in the NCBI Database after the acceptance of the manuscript.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
Supplementary Figure 1.
Maximum likelihood phylogenetic tree of HEV ORF2 gene, second dataset. Maximum likelihood phylogenetic tree of 198 HEV ORF2 sequences (genotype 3), 34 of which were references with known subtype, 158 were HEV ORF2 sequences from swine and wild boar samples from Italy and downloaded from the NCBI database, together with six HEV ORF2 gene sequences from wild boar obtained at ISS. The tree was midpoint rooted. Branch lengths were estimated with the best fitting nucleotide substitution model (GTR + I + G) according to a hierarchical likelihood ratio test. Scale bar at the bottom represents 0.09 nucleotide substitutions per site. The asterisks (∗) along the branches represents significant statistical support for the clusters subtending that branch (bootstrap support >75%). The main clades were highlighted. 3f and 3c subtype sequences have been highlighted with braces.
References
- Adlhoch C., Avellon A., Baylis S.A., Ciccaglione A.R., Couturier E., de Sousa R., Epstein J., Ethelberg S., Faber M., Feher A., Ijaz S., Lange H., Manďáková Z., Mellou K., Mozalevskis A., Rimhanen-Finne R., Rizzi V., Said B., Sundqvist L., Thornton L., Tosti M.E., van Pelt W., Aspinall E., Domanovic D., Severi E., Takkinen J., Dalton H.R. Hepatitis E virus: assessment of the epidemiological situation in humans in Europe, 2014/15. J. Clin. Virol. 2016;82:9–16. doi: 10.1016/j.jcv.2016.06.010. [DOI] [PubMed] [Google Scholar]
- Aspinall E.J., Couturier E., Faber M., Said B., Ijaz S., Tavoschi L., Takkinen J., Adlhoch C. Hepatitis E virus infection in europe: surveillance and descriptive epidemiology of confirmed cases, 2005 to 2015. Euro Surveill. 2017;22 doi: 10.2807/1560-7917.ES.2017.22.26.30561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colson P., Borentain P., Queyriaux B., Kaba M., Moal V., Gallian P., Heyries L., Raoult D., Gerolami R. Pigliver sausage as a source of hepatitis E virus transmission to humans. J. Infect. Dis. 2010;202:825–834. doi: 10.1086/655898. [DOI] [PubMed] [Google Scholar]
- Colson P., Romanet P., Moal V., Borentain P., Purgus R., Benezech A., Motte A., Gerolami R. Autochthonous infections with hepatitis E virus genotype 4, France. Emerg. Infect. Dis. 2012;18:1361–1364. doi: 10.3201/eid1808.111827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cozzi G., Ragno E. Meat production and market in Italy. Agric. Conspectus Sci. 2003;68(2):71–77. [Google Scholar]
- Darriba D., Taboada G.L., Doallo R., Posada D. jModelTest 2: more models, new heuristics and parallel computing. Nat. Methods. 2012;30:772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Sabato L., Amoroso M.G., Ianiro G., Esposito C., De Grossi L., Fusco G., Barone A., Martini E., Ostanello F., Di Bartolo I. Detection of hepatitis E virus in livers and muscle tissues of wild boars in Italy. Food. Environ. Virol. 2019;10 doi: 10.1007/s12560-019-09405-0. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Di Pasquale S., De Santis P., La Rosa G., Di Domenico K., Iaconelli M., Micarelli G., Martini E., Bilei S., De Medici D., Suffredini E. Quantification and genetic diversity of Hepatitis E virus in wild boar (Sus scrofa) hunted for domestic consumption in Central Italy. Food Microbiol. 2019;82:194–201. doi: 10.1016/j.fm.2019.02.005. Epub 2019 Feb 12. [DOI] [PubMed] [Google Scholar]
- Drummond A.J., Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faber M.S., Wenzel J.J., Jilg W., Thamm M., Höhle M., Stark K. Hepatitis E virus seroprevalence among adults, Germany. Emerg. Infect. Dis. 2012;18:1654–1657. doi: 10.3201/eid1810.111756. PMID: 23018055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogeda M., Avellón A., Echevarría J.M. Prevalence of specific antibody to hepatitis E virus in the general population of the community of Madrid, Spain. J. Med. Virol. 2012;84:71–74. doi: 10.1002/jmv.22270. PMID: 22095537. [DOI] [PubMed] [Google Scholar]
- Garson J.A., Ferns R.B., Grant P.R., Ijaz S., Nastouli E., Szypulska R., Tedder R.S. Minor groove binder modification of widely used TaqMan probe for hepatitis E virus reduces risk of false negative real-time PCR results. J. Virol. Methods. 2012;186:157–160. doi: 10.1016/j.jviromet.2012.07.027. [DOI] [PubMed] [Google Scholar]
- Guindon S., Dufayard J.F., Lefort V., Anisimova M., Hordijk W., Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- Hall T.A. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999;41:95–98. [Google Scholar]
- Ijaz S., Vyse A.J., Morgan D., Pebody R.G., Tedder R.S., Brown D. Indigenous hepatitis E virus infection in England: more common than it seems. J. Clin. Virol. 2009;44:272–276. doi: 10.1016/j.jcv.2009.01.005. PMID: 19217345. [DOI] [PubMed] [Google Scholar]
- Jothikumar N., Cromeans T.L., Robertson B.H., Meng X.J., Hill V.R. A broadly reactive one-step real-time RT-PCR assay for rapid and sensitive detection of hepatitis E virus. J. Virol. Methods. 2006;131:65–71. doi: 10.1016/j.jviromet.2005.07.004. [DOI] [PubMed] [Google Scholar]
- Kass R.E., Raftery A.E. Bayes factors. J. Am. Stat. Assoc. 1995;90:773–795. [Google Scholar]
- La Rosa G., Proroga Y.T.R., De Medici D., Capuano F., Iaconelli M., Della Libera S., Suffredini E. First detection of hepatitis E virus in shellfish and in seawater from production areas in southern Italy. Food. Environ. Virol. 2018;10:127–131. doi: 10.1007/s12560-017-9319-z. [DOI] [PubMed] [Google Scholar]
- Martelli F., Caprioli A., Zengarini M., Marata A., Fiegna C., Di Bartolo I., Ruggeri F.M., Delogu M., Ostanello F. Detection of hepatitis E virus (HEV) in a demographic managed wild boar (Sus scrofa scrofa) population in Italy. Vet. Microbiol. 2008;1(126):74–81. doi: 10.1016/j.vetmic.2007.07.004. [DOI] [PubMed] [Google Scholar]
- Martinelli N., Luppi A., Cordioli P., Lombardi G., Lavazza A. Prevalence of hepatitis E virus antibodies in pigs in Northern Italy. Infect. Ecol. Epidemiol. 2011;1 doi: 10.3402/iee.v1i0.7331. Epub 2011 Jul 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masia G., Orrù G., Liciardi M., Desogus G., Coppola R.C., Murru V., Argiolas M., Orrù G. Evidence of hepatitis E virus (HEV) infection in human and pigs in Sardinia, Italy. J. Prev. Med. Hyg. 2009;50:227–231. PMID: 20812518. [PubMed] [Google Scholar]
- Montesano C., Giovanetti M., Ciotti M., Cella E., Lo Presti A., Grifoni A., Zehender G., Angeletti S., Ciccozzi M. Hepatitis E virus circulation in Italy: phylogenetic and evolutionary analysis. Hepat. Mon. 2016;16 doi: 10.5812/hepatmon.31951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano T., Okano H., Kobayashi M., Ito K., Ohmori S., Nomura T., Kato H., Ayada M., Nakano Y., Akachi S., Sugimoto K., Fujita N., Shiraki K., Takei Y., Takahashi M., Okamoto H. Molecular epidemiology and genetic history of European type genotype 3 hepatitis E virus indigenized in the central region of Japan. Infect. Genet. Evol. 2012;12:1524–1534. doi: 10.1016/j.meegid.2012.06.002. [DOI] [PubMed] [Google Scholar]
- Pavio N., Doceul V., Bagdassarian E., Johne R. Recent knowledge on hepatitis E virus in Suidae reservoirs and transmission routes to human. Vet. Res. 2017;48:78. doi: 10.1186/s13567-017-0483-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posada D. jModelTest: phylogenetic model averaging. Mol. Biol. Evol. 2008;25:1253–1256. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- Renou C., Roque-Afonso A.M., Pavio N. Foodborne transmission of hepatitis E virus from raw pork liver sausage. France. Emerg. Infect. Dis. 2014;20:1945–1947. doi: 10.3201/eid2011.140791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riveiro-Barciela M., Mínguez B., Gironés R., Rodriguez-Frías F., Quer J., Buti M. Phylogenetic demonstration of hepatitis E infection transmitted by pork meat ingestion. J. Clin. Gastroenterol. 2015;49:165–168. doi: 10.1097/MCG.0000000000000113. [DOI] [PubMed] [Google Scholar]
- Romanò L., Paladini S., Tagliacarne C., Canuti M., Bianchi S., Zanetti A.R. Hepatitis E in Italy: a long-term prospective study. J. Hepatol. 2011;54:34–40. doi: 10.1016/j.jhep.2010.06.017. Epub 2010 Aug 20. [DOI] [PubMed] [Google Scholar]
- Smith D.B., Simmonds P., Jameel S., Emerson S.U., Harrison T.J., Meng X.J., Okamoto H., Van der Poel W.H., Purdy M.A., International Committee on Taxonomy of Viruses Hepeviridae Study Group Consensus proposals for classification of the family Hepeviridae. J. Gen. Virol. 2014;95:2223–2232. doi: 10.1099/vir.0.068429-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D.B., Simmonds P., Izopet J., Oliveira-Filho E.F., Ulrich R.G., Johne R., Koenig M., Jameel S., Harrison T.J., Meng X.J., Okamoto H., Van der Poel W.H.M., Purdy M.A. Proposed reference sequences for hepatitis E virus subtypes. J. Gen. Virol. 2016;97:537–542. doi: 10.1099/jgv.0.000393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spada E., Pupella S., Pisani G., Bruni R., Chionne P., Madonna E., Villano U., Simeoni M., Fabi S., Marano G., Marcantonio C., Pezzotti P., Ciccaglione A.R., Liumbruno G.M. A nationwide retrospective study on prevalence of hepatitis E virus infection in Italian blood donors. Blood. Transfus. 2018;16:413–421. doi: 10.2450/2018.0033-18. Epub 2018 May 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridhar S., Teng J.L.L., Chiu T.H., Lau S.K.P., Woo P.C.Y. Hepatitis E virus genotypes and evolution: emergence of camel hepatitis E variants. Int. J. Mol. Sci. 2017;18:E869. doi: 10.3390/ijms18040869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strimmer K., von Haeseler A. Likelihood-mapping: a simple method to visualize phylogenetic content of a sequence alignment. Proc. Natl. Acad. Sci. USA. 1997;94:6815–6819. doi: 10.1073/pnas.94.13.6815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo K., Trojnar E., Anheyer-Behmenburg H., Binder A., Schotte U., Ellerbroek L., Klein G., Johne R. Detection of hepatitis E virus RNA in raw sausages and liver sausages from retail in Germany using an optimized method. Int. J. Food Microbiol. 2015;23(215):149–156. doi: 10.1016/j.ijfoodmicro.2015.09.013. [DOI] [PubMed] [Google Scholar]
- Tei S., Kitajima N., Ohara S., Inoue Y., Miki M., Yamatani T., Yamabe H., Mishiro S., Kinoshita Y. Consumption of uncooked deer meat as a risk factor for hepatitis E virus infection: an age- and sex-matched case-control study. J. Med. Virol. 2004;74:67–70. doi: 10.1002/jmv.20147. [DOI] [PubMed] [Google Scholar]
- Verhoef L., Koopmans M., Duizer E., Bakker J., Reimerink J., Van Pelt W. Seroprevalence of hepatitis E antibodies and risk profile of HEV seropositivity in The Netherlands, 2006-2007. Epidemiol. Infect. 2012;140:1838–1847. doi: 10.1017/S0950268811002913. PMID: 22269886. [DOI] [PubMed] [Google Scholar]
- Vulcano A., Angelucci M., Candelori E., Martini V., Patti A.M., Mancini C., Santi A.L., Calvani A., Casagni L., Lamberti A. HEV prevalence in the general population and among workers at zoonotic risk in Latium Region. Ann Ig. 2007;19:181–186. PMID: 17658105. [PubMed] [Google Scholar]
- Wenzel J.J., Sichler M., Schemmerer M., Behrens G., Leitzmann M.F., Jilg W. Decline in hepatitis E virus antibody prevalence in southeastern Germany, 1996-2011. Hepatology. 2014;60:1180–1186. doi: 10.1002/hep.27244. PMID: 24912687. [DOI] [PubMed] [Google Scholar]
- WHO World Health organization - hepatitis E. 2019. https://www.who.int/news-room/fact-sheets/detail/hepatitis-E Available online:
- Yazaki Y., Mizuo H., Takahashi M., Nishizawa T., Sasaki N., Gotanda Y., Okamoto H. Sporadic acute or fulminant hepatitis E in Hokkaido, Japan, may be food-borne, as suggested by the presence of hepatitis E virus in pig liver as food. J. Gen. Virol. 2003;84:2351–2357. doi: 10.1099/vir.0.19242-0. [DOI] [PubMed] [Google Scholar]
- Zanetti A.R., Dawson G.J. Hepatitis type E in Italy: a seroepidemiological survey. J. Med. Virol. 1994;42:318–320. doi: 10.1002/jmv.1890420321. PMID: 8006645. [DOI] [PubMed] [Google Scholar]
- Zehender G., Ebranati E., Lai A., Luzzago C., Paladini S., Tagliacarne C., Galli C., Galli M., Ciccozzi M., Zanetti A.R., Romanò L. Phylogeography and phylodynamics of European genotype 3 hepatitis E virus. Infect. Genet. Evol. 2014;25:138–143. doi: 10.1016/j.meegid.2014.04.016. Epub 2014 Apr 28. [DOI] [PubMed] [Google Scholar]