Abstract
Two new fungal genera and six species occurring on insects in the orders Orthoptera and Phasmatodea (superorder Orthopterida) were discovered that are distributed across three families in the Hypocreales. Sixty-seven sequences generated in this study were used in a multi-locus phylogenetic study comprising SSU, LSU, TEF, RPB1 and RPB2 together with the nuclear intergenic region (IGR). These new taxa are introduced as Metarhizium gryllidicola, M. phasmatodeae, Neotorrubiella chinghridicola, Ophiocordyceps kobayasii, O. krachonicola and Petchia siamensis. Petchia siamensis shows resemblance to Cordyceps mantidicola by infecting egg cases (ootheca) of praying mantis (Mantidae) and having obovoid perithecial heads but differs in the size of its perithecia and ascospore shape. Two new species in the Metarhizium cluster belonging to the M. anisopliae complex are described that differ from known species with respect to phialide size, conidia and host. Neotorrubiella chinghridicola resembles Torrubiella in the absence of a stipe and can be distinguished by the production of whole ascospores, which are not commonly found in Torrubiella (except in Torrubiella hemipterigena, which produces multiseptate, whole ascospores). Ophiocordyceps krachonicola is pathogenic to mole crickets and shows resemblance to O. nigrella, O. ravenelii and O. barnesii in having darkly pigmented stromata. Ophiocordyceps kobayasii occurs on small crickets, and is the phylogenetic sister species of taxa in the ‘sphecocephala’ clade.
Keywords: Clavicipitaceae, Cordycipitaceae, entomopathogenic fungi, new taxa, Ophiocordycipitaceae, taxonomy
INTRODUCTION
The majority of the entomopathogenic fungi belong to the order Hypocreales in the Ascomycota. A reclassification of cordycipioid fungi based on molecular phylogeny over a decade ago split Cordyceps s.lat. into three families: Cordycipitaceae, Ophiocordycipitaceae and Clavicipitaceae (Sung et al. 2007). However, many species still could not be confidently identified in the new classification system due to either a lack of ex-type cultures for molecular studies or specimens for morphological comparison, leaving these species without information relating to their taxonomy and phylogenetic position in Cordyceps s.lat. (Sung et al. 2007).
In a recent phylogenetic classification of the family Cordycipitaceae by Kepler et al. (2017), two new genera, Hevansia and Blackwellomyces, were described while nine genera were proposed to be protected including Akanthomyces, Ascopolyporus, Beauveria, Cordyceps, Gibellula, Hyperdermium, Simplicillium, and eight genera proposed to be rejected, including Evlachovaea, Granulomanus, Isaria, Lecanicillium, Microhilum, Phytocordyceps, Synsterigmatocystis, and Torrubiella, a sexually reproductive genus originally identified in Cordycipitaceae together with Cordyceps. Subsequently, a new genus Samsoniella was added to the family (Mongkolsamrit et al. 2018). The family is characterised by pallid to brightly coloured fleshy stromata possessing superficial to pseudo-immersed perithecia with filiform or bola-shaped whole ascospores or ascospores that disarticulate into part-spores.
The genus Ophiocordyceps belongs to Ophiocordycipitaceae, and is one of the most speciose genera in Cordyceps s.lat., with more than 260 species records (Index Fungorum continuously updated). The majority of the species possess darkly pigmented stromata with superficial to immersed perithecia that produce whole or non-disarticulating ascospores (Sung et al. 2007, Luangsa-ard et al. 2018). The type of Ophiocordyceps is O. blattae (Petch 1931). The asexual morphs linked with Ophiocordyceps are known as Hirsutella, Hymenostilbe, Paraisaria, Stilbella and Syngliocladium (Sung et al. 2007). Ophiocordycipitaceous fungi can be found on a broad range of substrates including insects of Coleoptera, Diptera, Hemiptera, Hymenoptera, Isoptera, Lepidoptera, Neuroptera, Odonata and Orthoptera species (Kobayasi 1941, Mains 1958, Kepler et al. 2013, Sanjuan et al. 2015), and even occur on the fungal genus Elaphomyces (Nees 1820).
The Clavicipitaceae is the most heterogeneous and diverse family in Hypocreales with species occurring on plants, insects and other invertebrates (Chaverri et al. 2005, Sung et al. 2007, Kepler et al. 2012, Luangsa-ard et al. 2017). It contains the genus Metarhizium that has a greenish appearance when sporulating on arthropod hosts or in culture. They can be pathogenic to plants (e.g., Claviceps, Balansia), to other fungi (e.g., Verticillium epiphytum, Tyrannicordyceps), and infect a broad range of insect orders (Spatafora et al. 2007). Metarhizium anisopliae is a generalist occurring on more than seven insect orders, whereas M. acridum is specific only to insects from the Acrididae family (Moon & Hue 2017), and plays an important role as controller of insect populations in nature (Mondal et al. 2016).
Out of the 31 insect orders, 20 are susceptible to infection by entomopathogenic fungi in all stages of the insects’ life cycle - eggs, larvae, pupae, nymphs and adults (Aráujo & Hughes 2016). The four major insect orders that are parasitized by entomopathogenic fungi are the Coleoptera, Lepidoptera, Hemiptera and Hymenoptera (Shrestha et al. 2016). In Thailand, entomopathogenic fungi have been found on various invertebrate hosts including c. 11 insect orders and spiders. They occur on Blattodea, Coleoptera, Diptera, Hemiptera, Hymenoptera, Isoptera, Lepidoptera, Mantodea, Neuroptera, Odonata, Orthoptera and the spider order Araneae. There have been many recent reports of fungi occurring in the superorder Orthopterida (Orthoptera and Phasmatodea) from the three families of entomopathogenic fungi, especially on adult grasshoppers, locusts and stick insects. Most of them were reported from the New World (Mains 1959), such as Beauveria acridophila, B. diapheromeriphila, B. locustiphila, Cordyceps grylli, C. gryllotalpae, C. monticola, C. parvula, C. trinidadensis, C. uleana, Metarhizium acridum, M. majus and M. robertsii, and Ophiocordyceps amazonica, but recent findings also report them from the Old World, including Beauveria loeiensis, B. gryllotalpidicola (Ariyawansa et al. 2015), B. bassiana, Cordyceps neogryllotalpae, C. mantidicola, Metarhizium acridum, M. anisopliae and M. majus.
The aims of this study were:
to establish a species list of entomopathogenic fungi parasitizing Orthopterida, which are rarely found in Thailand; and
to clarify the taxonomic and phylogenetic positions of these fungi by using partial DNA sequences of multiple genetic loci: small and large subunits of the ribosomal DNA (SSU, LSU), elongation factor 1-α (TEF), the largest and second largest subunits of RNA polymerase II (RPB1, RPB2) and the nuclear intergenic regions (IGR).
MATERIALS AND METHODS
Specimens and isolations
Collection trips were done regularly throughout the year but intensively during the rainy season from June to September from 2011–2016. The forest floor, leaf litter and the underside of leaves were carefully scanned for fungi growing on invertebrates, especially insects and spiders. Specimens were collected from Khao Yai National Park in Nakhon Ratchasima province, Khao Luang National Park in Nakhon Si Thammarat province, Khlong Nakha Wildlife Sanctuary in Ranong province, Ban Hua Thung Community Forest in Chiang Mai province and Ban Phao Thai Community Forest in Phitsanulok province. Fungal collection and isolation followed the protocols described in previous studies (Luangsa-ard et al. 2017, Mongkolsamrit et al. 2018).
Cultivation
Starter cultures were grown on Potato Dextrose Agar (PDA: potato 200 g, dextrose 20 g, agar 15 g) for 7 d at room temperature. Mycelial plugs were cut from an actively growing colony using 5-mm-diam cork borer and inoculated into three media used for comparative studies: PDA, Potato Sucrose Agar (PSA: potato 200 g, sucrose 20 g, calcium carbonate 5 g, agar 15 g) and quarter-strength Sabouraud dextrose agar with yeast extract (SDAY/4: dextrose 10 g, peptone 2.5 g, yeast extract 2.5 g, agar 15 g) (Bischoff et al. 2009). Ophiocordyceps were grown in the dark at 20 °C following Ban et al. (2015), while Metarhizium, Petchia and Neotorrubiella specimens were incubated at room temperature under daylight conditions. Morphological observations were recorded at 7, 14, 21 and 30 d depending on the sporulation of each species.
Morphology
Colours of fresh specimens and cultures on PDA, PSA and SDAY/4 after 7–30 d were characterised using the colour chart of Kornerup & Wanscher (1963), as well as the Online Auction Colour Chart (abbreviated ‘OAC’ herein) and the Naturalist’s Color Guide (Smithe 1975) due to the absence of some colour ranges in each colour chart. Photographs of fresh specimens were taken to document the colour of stromata and the hosts when possible (Nikon D5100). Fungal materials, such as the perithecia, asci, ascospores, synnemata, phialides and conidia were mounted in lactophenol cotton blue and measured using a compound microscope (Olympus SZ61). Measurements of important morphological characters such as length and width were made from 20–50 observations, and variability calculated using standard deviation (with absolute minima and maxima in brackets) and average +/– standard deviation values. Specimens were either air-dried or dried in an electric food dryer (50–55 °C) overnight and deposited in BIOTEC Bangkok Herbarium (BBH) for further study.
DNA isolation, PCR and sequencing
Genomic DNA was extracted from 5–10-d-old fungal cultures grown on PDA plates by a modified CTAB method (Doyle & Doyle 1987). The fungal mycelium was scraped out from the agar using a sterile spatula and lysed in 600 μL CTAB extraction buffer (1M Tris-HCl, 5M NaCl, 0.5M EDTA, CTAB and PVP-40). Mycelium was ground using a sterile pestle and incubated at 65 °C for 30 min. After incubation, 600 μL of Chloroform:Isoamyl Alcohol (24:1) was added and mixed by inverting the tube. The samples were centrifuged at 12 000 rpm for 15 min and the supernatant transferred to a new tube. To precipitate the DNA, 300 μL of ice-cold Isopropanol was added and placed in –20 °C for 1 h. Samples were centrifuged at 4 °C at 12 000 rpm for 20 min to precipitate the DNA. After centrifugation the DNA pellet was washed with 70 % ethanol and centrifuged at 12 000 rpm for 20 min. The DNA pellets were air dried and dissolved in 1X TE buffer and stored at –20 °C.
PCR was conducted in 25 mL reaction volumes consisting of 1× PCR buffer, 200 μM of each of the four dNTPs, 2.5 mM MgCl2, 1 U Taq DNA Polymerase, recombinant (Thermo Scientific, US), 0.5 μM of each primer and 50–100 ng DNA template. Sequences of the nuclear ribosomal small and large subunits (SSU and LSU), the largest and second largest subunits of RNA polymerase II (RPB1 and RPB2), elongation factor 1-α (TEF) and 5′ intron-rich region of elongation factor 1-α (5′TEF) were used for this analysis (White et al. 1990, Bischoff et al. 2009). In addition, to verify cryptic diversification within the M. anisopliae species complex, seven nuclear intergenic loci were sequenced as they have shown a good performance in separating species of the PARB and MGT clades (Kepler & Rehner 2013, Rehner & Kepler 2017). The PCR primers used to amplify the gene regions for this study were: NS1 and NS4 for SSU, LROR and LR7 for LSU (White et al. 1990), 983F and 2218R for TEF, CRPB1 and RPB1Cr for RPB1, fRPB2-5F2 and fRPB2-7cR for RPB2 (Castlebury et al. 2004), EF1T and EF2T for 5′TEF (Bischoff et al. 2009), BTIGS, MzFG543, MzFG546, MzIGS2, MzIGS3, MzIGS5, MzIGS7 for IGR (Kepler & Rehner 2013). Sequencing primers were the same as for amplification, and conditions as set in Sung et al. (2007).
Sequence alignment and phylogenetic analyses
Sequences from this study were assembled using BioEdit v. 7.2.3 (Hall 2004) and compared to sequences in GenBank via a BLAST search. Assembled sequences were aligned using MUSCLE (Edgar 2004) and manually refined. All alignments were analysed together with other fungi from previously published studies (Table 1). The dataset was analysed separately in each family within the order Hypocreales, using maximum parsimony (MP), bayesian inference (BI) and maximum likelihood (ML). The 5′TEF was analysed separately in a supplementary tree (see Appendix).
Table 1.
List of specimens and GenBank accession numbers of sequences used in this study. Bold accession numbers were generated from this study.
T= culture ex-type
1Abbreviations for collections: A.E.G = A.E. Glenn personal collection, USA; AFTOL = Assembling the Fungal Tree of Life, USA ; ARSEF = USDA-ARS Collection of Entomopathogenic Fungal Cultures, USA; ATCC = American Type Culture Collection, USA; BCC = BIOTEC Culture Collection, Thailand; CBS = Centraalbureau voor Schimmelcultures, The Netherlands; EFCC = Entomopathogenic Fungal Culture Collection, Korea; GJS = G.J. Samuels personal collection, USA; GZUH = Herbarium of Guizhou University, China; HMAS = Herbarium Mycologicum Academiae Sinicae, China; HUA = Herbarium Antioquia University, Colombia; IndGH = Indonesian isolate collected by USAID project; KEW = Royal Botanic Gardens, UK; KT = Kanoksri Tasanathai Collection, BIOTEC, Thailand; MCA = M. Catherine Aime personal collection, USA; MY = Mycology Laboratory in BIOTEC, Thailand; NBRC = NITE Biological Resource Center, Japan; NHJ = Nigel Hywel Jones Collection, BIOTEC, Thailand; OSC = Oregon State University Herbarium, USA; P.C. = Priscila Chaverri personal collection; QCNE = National Herbarium of Ecuador, Ecuador; RCEF = Research Center on Entomogenous Fungi, China; spat = Joseph W. Spatafora personal collection, USA; TBRC = Thailand Bioresource Research Center, Thailand; TNS = National Museum of Nature and Science, Tsukuba, Ibaraki, Japan.
A maximum parsimony analysis was conducted for SSU, LSU, TEF, RPB1, RPB2 and 5′TEF using parsimony via PAUP v. 4.0b10 (Swofford 2002). Heuristic MP bootstrap (MP BS) analyses (Felsenstein 1985) with TBR branch swapping option included 1 000 replicates, and 10 random addition replicates were performed to provide bootstrap support values.
Bayesian analysis was conducted using MrBayes v. 3.2.6 (Ronquist et al. 2012) to determine posterior probabilities (BI PP), and analysis of the nucleotide substitution model was determined by using MrModeltest v. 2.2 (Nylander 2004) in each family. MrBayes was run with four independent Markov chains Monte Carlo (MCMC) for 20 000 000 generations with tree and parameter sampling occurring every 100 generations.
Maximum likelihood analyses were run using a randomised accelerated maximum likelihood model in RAxML v. 8.2.10 on XSEDE platform on CIPRES Science Gateway Portal (Stamatakis 2014). Relative support of internal nodes was assessed by a rapid bootstrap with 1 000 replications (ML BS). Nodes were considered supported by bootstrap values greater than 70 %.
RESULTS
Fungal cultures
Growth rates on three kinds of media differed for each species. Neotorrubiella chinghridicola (30 d, 0.8–1.4 cm) and Ophiocordyceps kobayasii (21 d, 0.8–1.2 cm) grew slower among the fungi studied. In addition, the best growth rate and conidiation was observed using PSA medium for almost all species, except for Neotorrubiella. Hyphal growth and mycelium formation of Neotorrubiella chinghridicola was observed after prolonged cultivation (> 30 d). There was no significant difference between growth on PDA and SDAY/4 among the species studied.
Phylogenetic analyses
Sixty-seven sequences were generated from 15 specimens, which were obtained from Cordycipitaceae (10 sequences of two samples), Clavicipitaceae (35 sequences of eight samples) and Ophiocordycipitaceae (22 sequences of five samples). The six new species and two new genera were phylogenetically distinct from other known species previously reported from Orthopterida.
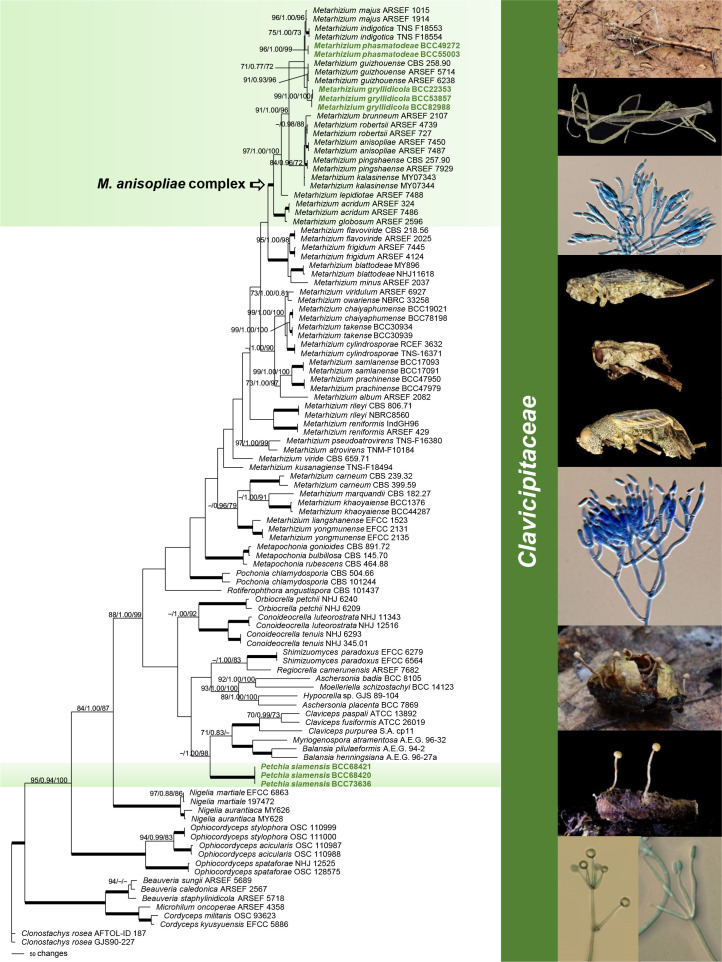
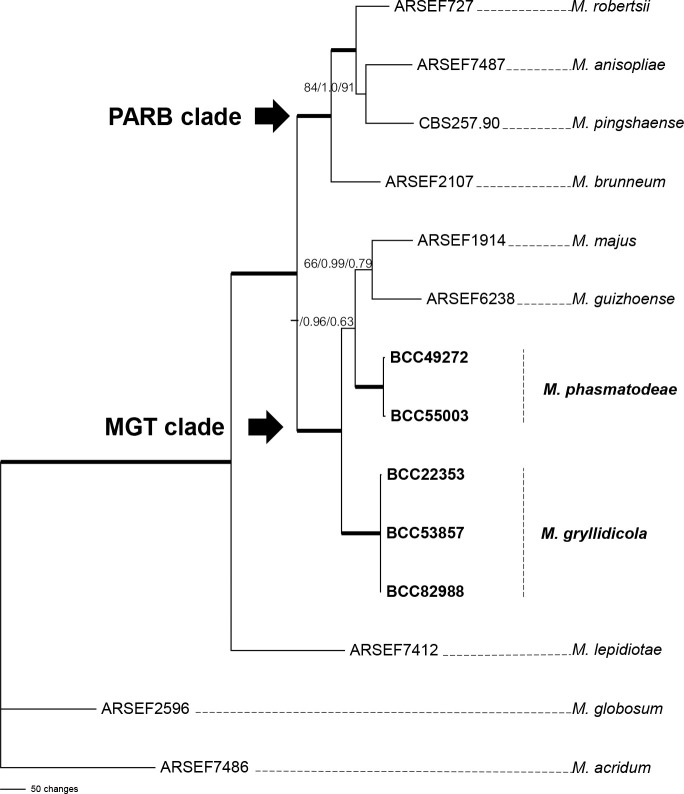
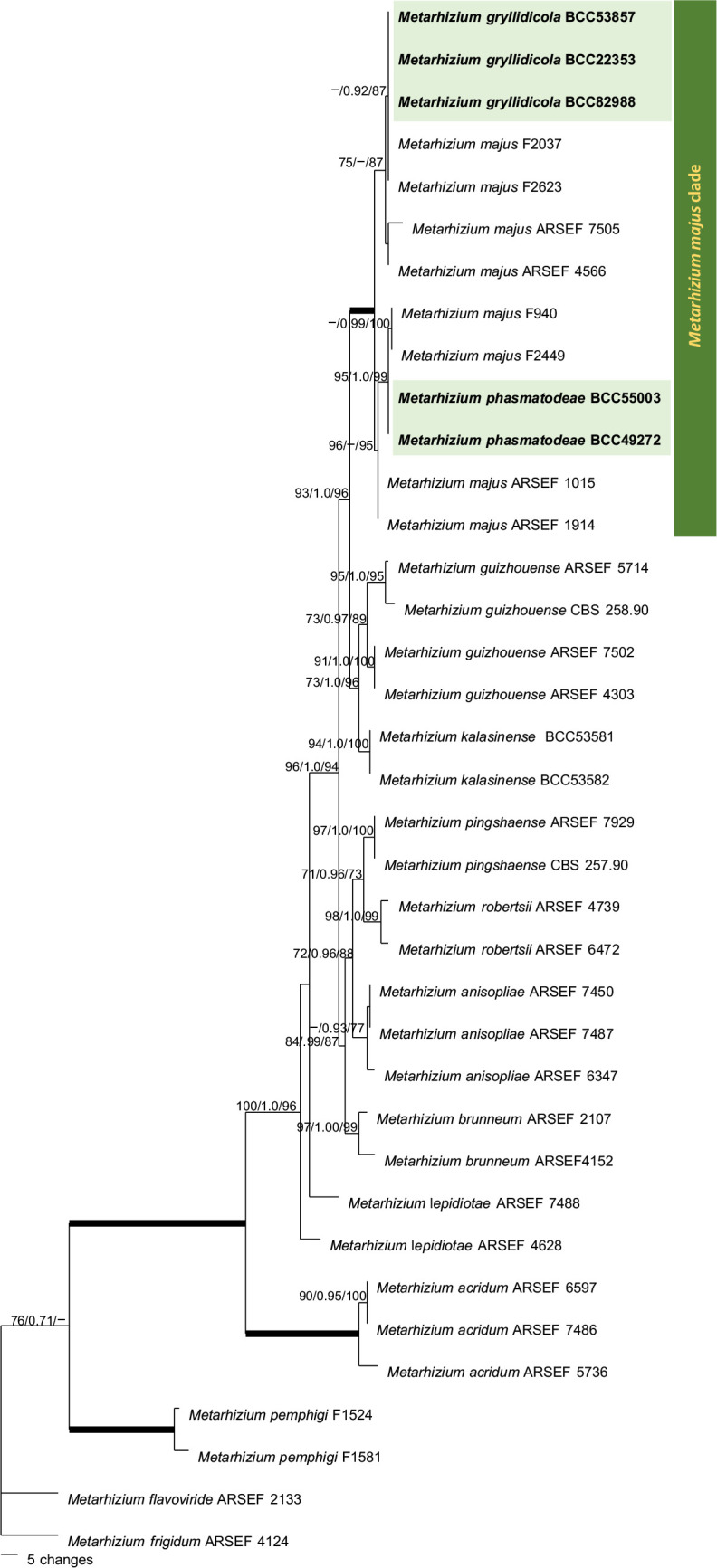
The first analysis of the family Clavicipitaceae (Fig. 1) was made to evaluate the preliminary identification of Metarhizium species complex and related genera selected from the dataset described by Luangsa-ard et al. 2017. Clonostachys rosea was designated as outgroup. The combined dataset of 107 taxa consisted of 4 292 bp (SSU 1 020 bp, LSU 872 bp, TEF 902 bp, RPB1 700 bp and RPB2 798 bp). Two species of Metarhizium, M. gryllidicola and M. phasmatodeae, clustered within the M. anisopliae species clade (Kepler & Rehner 2013, Kepler et al. 2014) are closely related to M. majus and M. guizhouense (MGT clade, Bischoff et al. 2009). One new genus, Petchia, which is resolved in these analyses, also nested within the Clavicipitaceae, closely related to the plant associated genera Balansia, Claviceps, Myriogenospora and Shimizuomyces, as well as the scale insect pathogens Hypocrella, Moelleriella and Regiocrella. It is separate from other known entomopathogenic genera. For this reason, C. mantidicola was transferred to Clavicipitaceae. This study also revealed that two isolates from stick insects, M. phasmatodeae (MP BS = 96 %, BI PP = 1.00, ML BS = 99 %), three isolates from crickets, Metarhizium gryllidicola (MP BS = 99 %, BI PP = 1.00, ML BS = 100 %) and three isolates occurring on egg cases (Ootheca) of praying mantis (Mantidae), Petchia (MP BS = 100 %, BI PP = 1.00, ML BS = 100 %) were strongly supported in Clavicipitaceae with high bootstrap values and posterior probabilities. Furthermore, in these analyses we evaluated the relationship within the M. anisopliae species complex by focusing on six nuclear intergenic loci (BTIGS, MzFG543, MzIGS2, MzIGS3, MzIGS5 and MzIGS7) of PARB and MGT clades, which includes M. pingshaense, M. anisopliae, M. robertsii, M. brunneum, M. majus and M. guizhouense. The result of this phylogenetic analysis (Fig. 2) showed that M. phasmatodeae (MP BS = 50 %, BI PP = 0.96, ML BS = 63 %) belongs to the MGT clade and is a sister taxon to M. gryllidicola which showed strong support as the basal species lineage to the MGT and M. phasmatodeae clade (MP BS = 100 %, BI PP = 1.00, ML BS = 100 %). Based on morphological and molecular studies, it is appropriate to describe a new genus and two new species in this family.
Fig. 1.
Phylogenetic reconstruction of the Clavicipitaceae obtained from the combined SSU, LSU, TEF, RPB1 and RPB2 sequences based on Maximum Parsimony, Bayesian analysis and RAxML. Number on the nodes are MP bootstrap/Bayesian posterior probability/ML bootstrap values above 70 %. Bold lines mean support for the three analyses were 100 %.
Fig. 2.
Phylogenetic reconstruction of the nuclear intergenic region of Metarhizium anisopliae species complex obtained from the combined BTIGS, MzFG543, MzIGS2, MzIGS3, MzIGS5 and MzIGS7 loci based on Maximum Parsimony, Bayesian analysis and RAxML. Number on the nodes are MP bootstrap/Bayesian posterior probability/ML bootstrap values above 50 %. Bold lines mean support for the three analyses were 100 %.
The second analysis of the family Cordycipitaceae (Fig. 3) was established to determine the taxonomic position of a new genus Neotorrubiella and a new species N. chinghridicola. The dataset used contained almost all genera belonging to this family (Kepler et al. 2017, Mongkolsamrit et al. 2018), and the analysis included sequences from related species occurring on Orthoptera: Beauveria acridophila, B. diapheromeriphila, B. locustiphila, B. loeiensis and B. gryllotalpidicola (Sanjuan et al. 2014, 2015, Ariyawansa et al. 2015) with other species of Torrubiella, especially T. wallacei (= Lecanicillium wallacei), which is one of the earliest diverging members of this family, and other known species. Sequences of Conoideocrella tenuis were used as outgroup. The alignment of 64 taxa is 4 390 bp (SSU 1 011 bp, LSU 870 bp, TEF 981 bp, RPB1 730 bp and RPB2 798 bp) long. The genus Neotorrubiella was strongly supported (MP BS = 100 %, BI PP = 1.00, ML BS = 100 %) in Cordycipitaceae as a separate genus from other genera in the phylogenetic reconstruction, closely related to the genera Hevansia and Gibellula. Therefore, a new species, Neotorrubiella chinghridicola, is proposed.
Fig. 3.
Phylogenetic reconstruction of the Cordycipitaceae obtained from the combined SSU, LSU, TEF, RPB1 and RPB2 sequences based on Maximum Parsimony, Bayesian analysis and RAxML. Number on the nodes are MP bootstrap/Bayesian posterior probability/ML bootstrap values above 70 %. Bold lines mean support for the three analyses were 100 %. Triangles represent taxa occurring on Orthoptera.
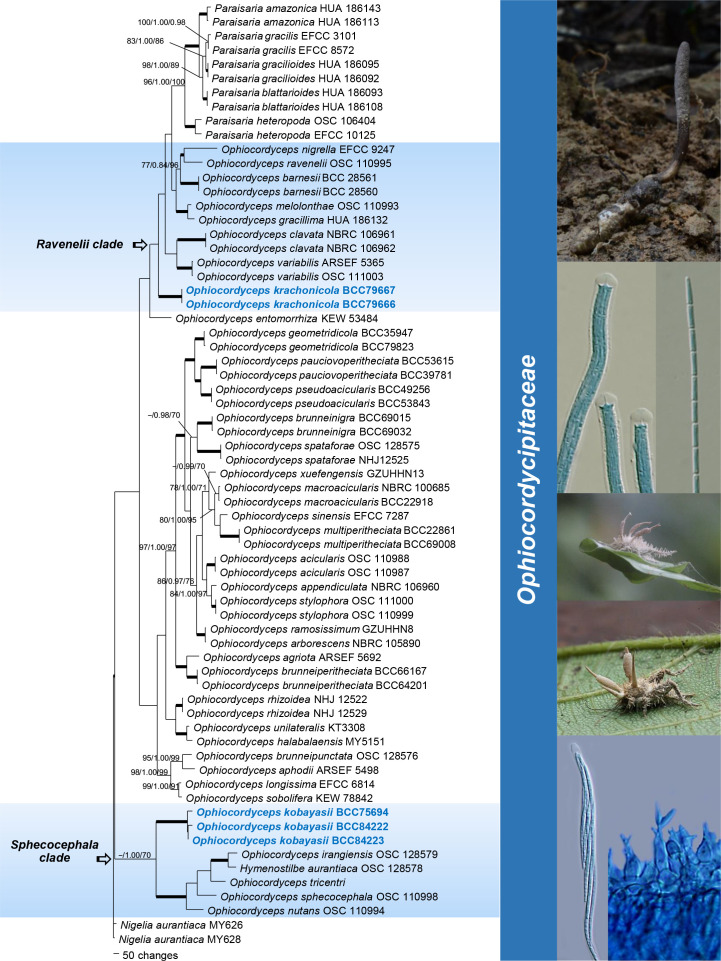
The analysis of the family Ophiocordycipitaceae (Fig. 4) was mainly targeted to elucidate the phylogenetic positions of putative new Ophiocordyceps species. The alignment consisted of 4 349 bp (SSU 1 012 bp, LSU 914 bp, TEF 904 bp, RPB1 725 bp and RPB2 794 bp) from 67 taxa including Nigelia aurantiaca as outgroup. The results of these analyses showed two well-supported clades in Ophiocordycipitaceae distinct from other known taxa. Two new species are identified: Ophiocordyceps kobayasii, which formed a sister clade to species producing a Hymenostilbe asexual morph in the sphecocephala clade, and O. krachonicola, which formed a separate clade basal to other species of Ophiocordyceps including O. variabilis, O. barnesii, O. clavata, O. melolonthae, O. gracillima, O. nigrella and O. ravenelii clades and Paraisaria (Mongkolsamrit et al. 2019), with credible bootstrap supports (MP BS = 100 %, BI PP = 1.00, ML BS = 100 %).
Fig. 4.
Phylogenetic reconstruction of the Ophiocordycipitaceae obtained from the combined SSU, LSU, TEF, RPB1 and RPB2 sequences based on Maximum Parsimony, Bayesian analysis and RAxML. Number on the nodes are MP bootstrap/Bayesian posterior probability/ML bootstrap values above 70 %. Bold lines mean support for the three analyses were 100 %.
TAXONOMY
Clavicipitaceae
Metarhizium gryllidicola Khons., Thanakitp. & Luangsa-ard, sp. nov. — MycoBank MB830169; Fig. 5
Fig. 5.
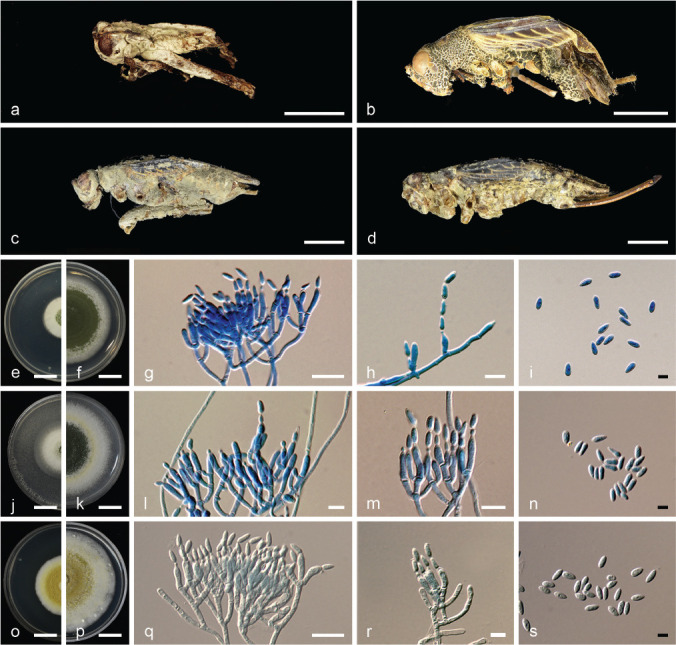
Metarhizium gryllidicola (BBH 44436, BCC82988). a–d. Fungus on adult cricket (Gryllidae) hosts; e–f. colony on PDA: (e) at 7 d (f) at 14 d; g–h. conidiophores bearing phialides and conidia; i. conidia; j–k. colony on PSA: (j) at 7 d (k) at 14 d; l–m. conidiophores bearing phialides and conidia; n. conidia; o–p. colony on SDAY/4: (o) at 14 d (p) at 21 d; q–r. conidiophores bearing phialides and conidia; s. conidia. — Scale bars: a, e–f, j–k, o–p = 10 mm; b–d = 5 mm; g, l, q = 10 μm; h–i, m–n, r–s = 5 μm.
Etymology. In reference to the family of the host, namely a cricket (Gryllidae).
Typus. Thailand, Nakhon Ratchasima Province, Khao Yai National Park, on adult cricket (Gryllidae), in leaf litter, 1 Nov. 2016, B. Sakolrak, D. Thanakitpipattana, N. Arnamnart, N. Kobmoo & R. Somnuk (holotype BBH 44436, ex-type culture BCC82988).
Sexual morph. Unknown.
Specimens found only on adult crickets (Gryllidae) in leaf litter on the forest floor. The host body is covered with greyish green (27C4, 29C6), pastel green (28A4) and dark green (28F8) conidia.
Culture characteristics — Colonies on PDA fast-growing, attaining 4.7 cm diam after 14 d at 25 °C, at first white with smooth and cottony mycelia. Conidiation starts in the middle of colony at 5 d, becoming lime green (No. 59), buff yellow (No. 53) and olive green (No. 46) with age. Colony reverse white-cream at the edges and sulphur yellow (No. 57) in the centre. Conidiophores arising from aerial mycelium, smooth, cylindrical. Phialides smooth-walled, cylindrical to clavate, without a distinct neck, (5–)6–9(–12) × 2–3 μm. Conidia smooth-walled, cylindrical, ovoid, (5–)5.7–6.9(–7) × (2–)2.2–2.9(–3) μm. Colonies on PSA fast-growing, attaining 5 cm diam after 14 d at 25 °C, at first white with smooth and cottony mycelia turning cream (No. 54). Conidiation starts in the middle of colony at 5 d, turning to yellow to olive green (No. 50), lime green (No. 59) and olive green (No. 46) with age. Colony reverse white-cream at the edges and straw yellow (No. 56) in the centre. Conidiophores arising from aerial mycelium, smooth, cylindrical. Phialides smooth-walled, cylindrical, clavate, without a distinct neck, (6–)7–10(–13) × 2 μm. Conidia smooth-walled, cylindrical, ovoid, obclavate, (5–)5.5–6.4(–7) × 2.5–3 μm. Colonies on SDAY/4 slow-growing, attaining 3.5 cm diam after 14 d at 25 °C, at first white with smooth and cottony mycelia becoming sulphur yellow (No. 157), spectrum yellow (No.55), straw yellow (No.56) and pale horn (No. 92) with age. Conidiation starts after 30 d, turning to olive yellow (No. 52) to citrine (No. 51) and olive green (No. 46). Colony reverse white-cream at the edges and straw yellow (No. 56) in the centre. Conidiophores arising from aerial mycelium smooth, cylindrical. Phialides smooth-walled, cylindrical, clavate, without a distinct neck, (6–)7–9.5(–11) × 2–3 μm. Conidia smooth-walled, cylindrical, ovoid, obclavate, (4–)5–6(–7) × 2–3 μm.
Additional materials examined. Thailand, Nakhon Ratchasima Province, Khao Yai National Park, on adult crickets (Gryllidae), in leaf litter, 11 July 2012, A. Khonsanit, K. Tasanathai, P. Srikitikulchai, S. Mongkolsamrit & W. Noisripoom (BBH 32733, BCC53857); ibid., 14 Aug. 2009, K. Tasanathai, P. Srikitikulchai & S. Mongkolsamrit (BBH 26529, BCC37915), (BBH 26533, BCC37918); ibid., 13 Sept. 2009, K. Tasanathai, P. Srikitikulchai & S. Mongkolsamrit (BBH 27261, BCC39045); ibid., 18 June 2008, B. Thongnuch, J.J. Luangsa-ard, K. Tasanathai, P. Srikitikulchai, R. Promharn & S. Mongkolsamrit (BBH 23876, BCC30917); ibid., 5 July 2006, B. Thongnuch, J.J. Luangsa-ard, K. Tasanathai, P. Srikitikulchai & S. Mongkolsamrit (BBH 18647, BCC22353).
Notes — Metarhizium gryllidicola is found only on insects in the family Gryllidae (Orthoptera). This species is in the M. anisopliae species complex, which is a sister species to M. majus, M. indigotica, M. guizhouense and M. taii, which is a later synonym of M. guizhouense (Bischoff et al. 2009) and M. phasmatodeae (Fig. 1, 2). It differs from these known species significantly in the size of its phialides and conidia. The conidial size of M. gryllidicola is the same as in M. phasmatodeae but smaller than in M. majus, the phialides of both species are shorter than those of M. majus and M. guizhouense. Metarhizium phasmatodeae can only be separated from M. gryllidicola in terms of host and genetic data (Table 2; Fig. 1, 2).
Table 2.
Morphological comparison of species (asexual morph).
| Species | Host | Colony colour | Phialides (μm) | Conidia (μm) | References |
|---|---|---|---|---|---|
| Metarhizium acridum | Orthoptera, Soil | Greyish yellow to greyish green | 4.5–13 × 2–4.5 | 5–7 × 2–4 | Bischoff et al. (2009) |
| Metarhizium anisopliae | Coleoptera, Hemiptera, Orthoptera | Greyish green | 8–11.5 × 2–3 | 5–7 × 2–3.5 | Bischoff et al. (2009) |
| Metarhizium brunneum | Coleoptera, Hemiptera, Soil | Pale-yellow to olive | 6–18 × 2–5 | 4.5–8 × 2–3.5 | Bischoff et al. (2009) |
| Metarhizium gryllidicola | Orthoptera | Pale horn to sulfur yellow | 6–11 × 2–3 | 4–7 × 2–3 | This study |
| Metarhizium guizhouense | Coleoptera, Diptera, Lepidoptera, Soil | – | 6–20 × 2–3.5 | 5–5.9 × 2–3.5 | Bischoff et al. (2009) |
| Metarhizium kalasinense | Coleoptera | Greenish olive and spectrum yellow | 8–12 × 2–3 | 6–8 × 2–3 | Luangsa-ard et al. (2017) |
| Metarhizium majus | Coleoptera, Lepidoptera, Soil | Olive to dark green | 9–23.5 × 2.5–4.5 | 8–14.5 × 2.5–5 | Bischoff et al. (2009) |
| Metarhizium phasmatodeae | Phasmatodea | Spectrum to sulfur-yellow and lime green | 5–11 × 2–3 | 5.5–8 × 2–3 | This study |
| Metarhizium pingshaense | Coleoptera, Isoptera | Olive | 7–17 × 2–3.5 | 4.5–8 × 2–3.5 | Bischoff et al. (2009) |
| Metarhizium robertsii | Coleoptera, Orthoptera, Soil | Olive to greyish green | 7–14.5 × 2–3.5 | 5–7.5 × 2–3.5 | Bischoff et al. (2009) |
Metarhizium phasmatodeae Khons., Thanakitp. & Luangsa-ard, sp. nov. — MycoBank MB830170; Fig. 6
Fig. 6.
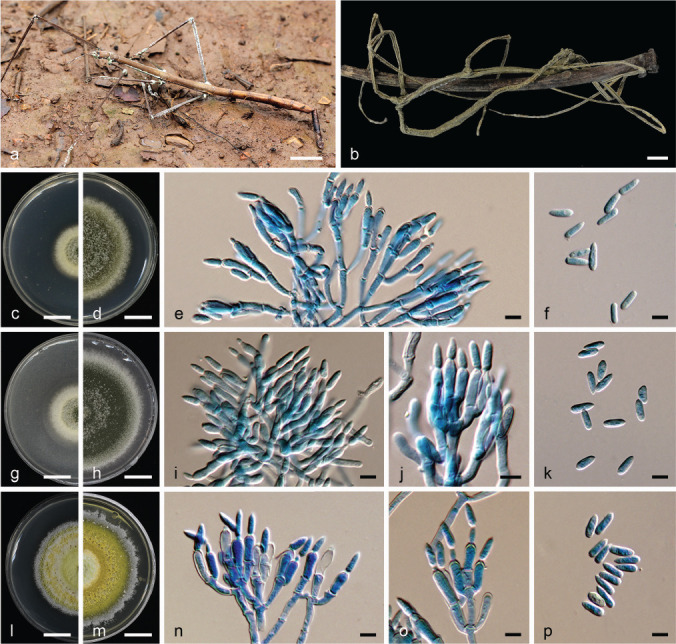
Metarhizium phasmatodeae (BBH 32532, BCC49272). a–b. Fungus on stick insect hosts; c–d. colony on PDA: (c) at 7 d (d) at 14 d; e. conidiophores bearing phialides and conidia; f. conidia; g–h. colony on PSA: (g) at 7 d (h) at 14 d; i–j. conidiophores bearing phialides and conidia; k. conidia; l–m. colony on SDAY/4: (l) at 14 d (m) at 21 d; n–o. conidiophores bearing phialides and conidia; p. conidia. — Scale bars: a–d, g–h, l–m = 10 mm; e–f, i–k, n–p =5 μm.
Etymology. In reference to the order of the insect host – Phasmatodea.
Typus. Thailand, Chiang Mai Province, Ban Hua Thung Community Forest, on stick insect (Phasmatodea), in leaf litter, 16 Aug. 2011, A. Khonsanit, J. Kumsao, K. Tasanathai, P. Srikitikulchai & S. Mongkolsamrit (holotype BBH 32532, ex-type culture BCC49272).
Sexual morph. Unknown.
Specimens found only on stick insects (Phasmatodea) in leaf litter on the forest floor. The host body is covered with green (28B6, 29C6) and deep green (27E8) conidia.
Culture characteristics — Colonies on PDA fast-growing, attaining 4 cm diam after 14 d at 25 °C, at first white with smooth and cottony mycelium turning to leaf-green (No. 146), greenish olive (No. 49) due to conidiation at the middle of colony after 5 d. Colony reverse clay (No. 123B) to cinnamon (No. 123B). Conidiophores arising from aerial mycelium, smooth, cylindrical. Phialides smooth-walled, cylindrical to clavate, without a distinct neck, (6–)7.5–10.5(–12) × 2–3 μm. Conidia smooth-walled, cylindrical to obclavate, (7–)7.5–9.5(–10) × 2–2.5(–3) μm. Colonies on PSA fast-growing, attaining 5.2 cm diam after 14 d at 25 °C, at first white with smooth and cottony mycelia, becoming yellowish to olive green (No. 50), lime green (No. 59) and greenish olive (No. 49) at 5 d due to the production of conidia. Colonies turn to light pink with age. Colony reverse beige (No. 219D). Conidiophores arising from aerial mycelium, smooth, cylindrical. Phialides smooth-walled, cylindrical to clavate, without a distinct neck, (7–)7.5–11(–12) × (2–)2.5–3 μm. Conidia smooth-walled, cylindrical to ovoid, (6–)7–8.5(–10) × 2–3 μm. Colonies on SDAY/4 moderate-growing, attaining 3.5 cm diam after 14 d at 25 °C, at first white, smooth, turning spectrum yellow (No. 55), sulphur yellow (No. 157), apple green (No. 61), citrine (No. 51), olive yellow (No. 52) and olive green (No. 46), the medium becoming lime green (No. 59). Conidiation starts after 16 d, turning to olive yellow (No. 52) to citrine (No. 51) and olive green (No. 46). Colony reverse olive green in the centre (No. 47-48) and olive grey (No. 42) at the edges. Conidiophores arising from aerial mycelium, smooth, cylindrical. Phialides smooth-walled, cylindrical, without a distinct neck, (5–)6.5–9.5(–11) × (2–)2.5–3 μm. Conidia smooth-walled, cylindrical, ovoid, obclavate (5.5–)6.5–7.5(–8) × 2–3 μm.
Additional materials examined. Thailand, Nong Khai Province, Amphoe Si Chiang Mai, on stick insect (Phasmatodea), in leaf litter, 30 Aug. 2009, K. Tasanathai, P. Srikitikulchai, T. Chohmee, N. Thanh & N. Toan (BBH 27078); Chiang Mai Province, Ban Hua Thung Community Forest, in leaf litter, 25 Oct. 2013, A. Khonsanit, D. Thanakitpipattana, K. Tasanathai, P. Srikitikulchai & W. Noisripoom (BBH 37785, BCC68409); Saraburi Province: Jedkod waterfall, in leaf litter, 27 Aug. 2012, A. Khonsanit, D. Thanakitpipattana, J.J. Luangsa-ard, S. Mongkolsamrit & W. Noisripoom (BBH 32525, BCC55003).
Notes — Metarhizium phasmatodeae is found only on stick insects (order Phasmatodea). It is a sister species of M. majus, M. indigotica, M. guizhouense, M. taii and M. gryllidicola. However, the conidial size of M. phasmatodeae is in the same range as in M. gryllidicola and could only be distinguished based on the host, but is smaller than M. majus, and the phialides of both aforementioned species are shorter than those of M. majus and M. guizhouense (Table 2).
Petchia Thanakitp., Mongkols. & Luangsa-ard, gen. nov. — MycoBank MB830167
Etymology. In honour of Tom Petch (1870–1948), renowned English mycologist and plant pathologist best remembered for his work on entomopathogenic fungi.
Type species. Petchia siamensis Thanakitp., Mongkols. & Luangsa-ard.
Asexual morph. Acremonium-like.
Stromata arising from the egg cases of praying mantis (Mantidae), multiple, mostly erect up to 2 cm high. Fertile part is at the terminal end of the stroma, pale cream (OAC900), globose, 1.5–1.8 cm diam. Perithecia narrowly ovoid, brown, immersed, with packed mycelium surrounding each perithecium. Asci cylindrical, 8-spored. Ascus cap rounded. Ascospores hyaline, filiform, multiseptate, whole. On PDA cultures hyphae branched, smooth, hyaline, sometimes fasciculate. Conidiogenous structures phialidic, conidiophores elongate, erect, non-septate, verticillately or irregularly branched, bearing phialides singly, or in whorls of one to five. Conidia ellipsoidal or reniform.
Habitat — Egg cases (Ootheca) of praying mantis (Mantidae) in leaf litter.
Distribution — Japan, China, Thailand.
Petchia siamensis Thanakitp., Mongkols. & Luangsa-ard, sp. nov. — MycoBank MB830168; Fig. 7
Fig. 7.
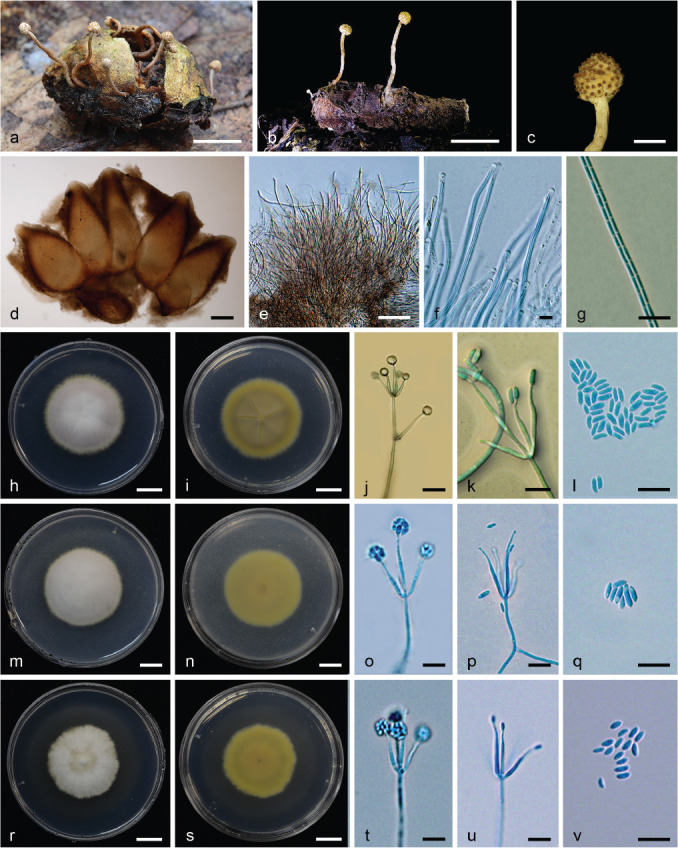
Petchia siamensis (BBH 39551, BCC73636). a–b. Fungus on egg case of praying mantis; c. ascomata; d. perithecia; e–f. asci; g. ascospores; h–i. colony on PDA at 21 d (h) obverse (i) reverse; j–k. phialides with conidia on PDA; l. conidia; m–n. colony on PSA at 21 d (m) obverse (n) reverse; o–p. phialides with conidia on PSA; q. conidia; r–s. colony on SDAY/4 at 21 d (r) obverse (s) reverse; t–u. phialides with conidia on SDAY/4; v. conidia. — Scale bars: a–b, h–i, m–n, r–s = 1 cm; c = 1 mm; d = 200 μm; e = 50 μm; f = 5 μm; g, j–l, o–q, t–v = 10 μm.
Etymology. Referring to ‘Siam’, old name for Thailand, where the species was collected.
Typus. Thailand, Nakhon Ratchasima Province, Khao Yai National Park, on ootheca of praying mantis (Mantidae), in leaf litter, 20 July 2014, K. Tasanathai, A. Khonsanit, W. Noisripoom & D. Thanakitpipattana (holotype BBH 39551, ex-type culture BCC73636).
Asexual morph. Acremonium-like.
Stromata erect, robust, multiple, arising from the egg case of insect host, pale cream (OAC900), cylindrical, up to 2 cm long, 0.8–1 cm wide. Fertile part occurs at the terminal end of stroma, globose, 1.5–1.8 mm diam. Perithecia pseudo-immersed, obpyriform, (600–)637–827(–870) × (320–)343–393(–400) μm. Asci cylindrical, 8-spored, up to 320 μm long, 2.5–3 μm wide. Ascus cap rounded, 2 × 2.5 μm. Ascospores whole, multiseptate, filiform, hyaline, septate, (200–)202–268(–300) × 1–1.5 μm.
Culture characteristics (from germinating ascospores) — Colonies on PDA moderate-growing, attaining c. 3 cm diam after 21 d at 20 °C. Colony purplish grey (OAC557), cottony, with high mycelial density. Colony reverse pale brown (OAC813). Vegetative hyphae smooth, septate, hyaline, c. 3 μm diam. Conidiogenous structures consisting of erect conidiophores arising from the vegetative hyphae and aerial hyphae. Conidiophores forming verticillate branches with phialides singly or in whorls of three to five. Phialides (10–)12–20(–28) × 1–2 μm, awl-shaped, acremonium-like. Conidia aggregated in slimy heads at the apex of phialides, hyaline, ellipsoidal or reniform, 1-celled, (3–)4–6 × 1–2 μm. Chlamydospores not observed. Colonies on PSA moderate-growing, attaining c. 3 cm diam after 21 d at 20 °C. Colony purplish grey (OAC557), cottony with high mycelial density. Colony reverse pale yellow (OAC813). Vegetative hyphae smooth, septate, hyaline, c. 3 μm diam. Conidiogenous structures consisting of erect conidiophores arising from the vegetative hyphae and aerial hyphae. Conidiophores forming verticillate branches with phialides singly or in whorls of three to five. Phialides 10–20(–25) × 1–2 μm, awl-shaped, acremonium-like. Conidia aggregated in slimy heads at the apex of phialides, hyaline, ellipsoidal 1-celled, (3–)4–5(–6) × 1–2 μm. Chlamydospores not observed. Colonies on SDAY/4 moderate-growing, attaining c. 2.5 cm diam after 21 d at 20 °C. Colony white with irregular edges, velvety. Colony reverse pale yellow (OAC813). Vegetative hyphae smooth, septate, hyaline, c. 3 μm diam. Conidiogenous structures consisting of erect conidiophores arising from the vegetative hyphae and aerial hyphae. Conidiophores forming verticillate branches with phialides singly or in whorls of three to five. Phialides 12–20(–35) × 1–2 μm, awl-shaped, acremonium-like. Conidia aggregated in slimy heads at the apex of phialides, hyaline, ellipsoidal or reniform, 1-celled, (3–)3.5–5(–6) × 1–2 μm. Chlamydospores not observed.
Additional materials examined. Thailand, Nakhon Ratchasima Province, Khao Yai National Park, on ootheca of praying mantis (Mantidae), in leaf litter, 2 June 2011, K. Tasanathai, P. Srikitikulchai, S. Mongkolsamrit, A. Khonsanit, K. Sansatchanon & W. Noisripoom (BBH 37736, BCC48062); Chiang Mai Province, Ban Hua Thung community forest, on ootheca of praying mantis (Mantidae), 25 Oct. 2013, K. Tasanathai, P. Srikitikulchai, A. Khonsanit, W. Noisripoom, D. Thanakitpipattana & S. Watcharapayungkit (BBH 38368, BCC68420), (BBH 37855, BCC68421), (BBH 37793, BCC68417); Ban Hua Thung community forest, on ootheca of praying mantis (Mantidae), 29 Oct. 2014, K. Tasanathai, A. Khonsanit, W. Noisripoom, D. Thanakitpipattana, P. Srikitikulchai & S. Wongkanoun (BBH 39627, BCC75724), (BBH 39660, BCC75750); Nakhon Ratchasima Province, Khao Yai National Park, on ootheca of praying mantis (Mantidae), in leaf litter, 20 July 2014, K. Tasanathai, A. Khonsanit, W. Noisripoom & D. Thanakitpipattana (BBH 39552, BCC73637).
Petchia mantidicola (Kobayasi & Shimizu) Thanakitp., Mongkols. & Luangsa-ard, comb. nov. — MycoBank MB833017
Basionym. Cordyceps mantidicola Kobayasi & Shimizu [as ‘mantidaecola’], Bull. Natl. Sci. Mus., Tokyo, Ser. B 9: 12. 1983.
Notes — Both P. siamensis and P. mantidicola occur on ootheca of praying mantis and produce cream terminal obovoid perithecial heads. However, in P. siamensis the perithecia are distinctly larger (600–870 × 320–400 μm) than those of P. mantidicola (400–450 × 200–250 μm), and the ascospores in P. siamensis are whole while they disarticulate into part-spores in P. mantidicola (Table 3).
Table 3.
Morphological comparison of species (sexual morph).
| Species | Host | Stromata (cm) | Fertile part (mm) | Perithecia (μm) | Asci (μm) | Ascospores (μm) | References |
|---|---|---|---|---|---|---|---|
| Beauveria locustiphila | Adults and nymphs of Acrididae | Claviform, 1–1.5 long | Clavate, 0.8–12 × 2–2.5 | Aggregated, immersed, 150 long | Cylindrical, clavate, 150–200 × 3–3.5 | Filiform, breaking into part-spores, 0.6–0.9 | Hennings (1904) |
| Beauveria loeiensis | Adult Orthoptera (Gryllacrididae) | Several, scattered, simple or branched, cylindrical to enlarged | – | Superficial, narrowly ovoid with acute apices, 650–710 × 280–320 | Cylindrical, 370–450 × 5 | Filiform, breaking into part-spores, 5–10 × 1 | Ariyawansa et al. (2015) |
| Cordyceps parvula | Orthoptera | Yellowish cream, scattered | – | Superficial, ovoid, 500–650 × 250–300 | Cylindrical, 400–500 × 5–6 | Filiform, 8–10 × 1 | Mains (1959) |
| Neotorrubiella chinghridicola | Adult Orthoptera (Gryllidae) | White mat of mycelium covering the host | – | Superficial, ovoid to obclavate, 220–280 × 130–170 | Cylindrical, 125–165 × 10–15 | Filiform, breaking into part-spores, 70–90 × 4–5 | This study |
| Ophiocordyceps amazonica | Adult Orthoptera (Romaleidae, Acrididae) | Simple, gregarious, 2–4.5 long | Subglobose, spherical, reddish brown, 2.5–5.5 | Immersed, ovoid-ellipsoid, 760–1100 × 220–400 | Cylindrical, 325–450 × 5 | 9–17 × 0.5–2 | Hennings (1904), Mains (1959), Sanjuan et al. (2015) |
| Ophiocordyceps kobayasii | Adult Gryllidae | Multiple, cylindrical, up to 0.8 long | Pale cream, densely packed in the middle of stromata, cylindrical to clavate, up to 3 long | Superficial, ovoid, 180–250 × 140–200 | Cylindrical, up to 125 × 10–15 | Whole ascospores, filiform, 45–72 × 2–2.5 | This study |
| Ophiocordyceps krachonicola | Nymph of Gryllotalpa orientalis (Gryllidae) | Solitary, cylindrical, 3.7–4.0 long | Cylindrical to clavate, 25 × 2–3 | Immersed, lanceolate, 460–580 × 180–300 | Cylindrical, 250–400 × 4–5 | Part spores, 4–10 × 1 | This study |
| Ophiocordyceps ravenelii | Beetle larvae (Coleoptera) | Clavate, single or occasionally two, 1.5–4.5 | Cylindrical, 2 × 0.8 | Immersed, narrowly flask-shaped or ovoid, 240–420 × 144–240 | Clavate-cylindrical, 170–220 × 8–10 | Whole ascospores, multiseptate, 102–164 × 2–3 | Berkeley (1856), Mains (1941) |
| Petchia mantidicola | Cocoon of Mantidae | Simple, usually solitary, 2.5–4 × 3–4 long | Spherical, pale rufous, 1.5–2.1 | 400–450 × 200–250 | Cylindrical | Breaking into part-spores, 3–5 × 1 | Kobayasi & Shimizu (1983) |
| Petchia mantidicola | Cocoon of Mantidae | Single or two caespitous stromata, 0.5–1.5 | 1.5–5 | 570–750 × 350–520 | Cylindrical, 180–200 × 1.8–2.3 | Breaking into part-spores, 3–5 × 1 | Liu et al. (1997) |
| Petchia siamensis | Ootheca of praying mantis | Multiple, cylindrical, up to 2 long | Globose, pale cream, 1.5–1.8 | Immersed, obpyriform, 600–870 × 320–400 | Cylindrical, up to 320 × 2.5–3 | Multiseptate, 200–300 × 1–1.5 | This study |
| Torrubiella aranicida | On spider | Dispersed or aggregate on subicula | – | Elongated conoid, 0.5–0.7 × 0.3–0.4 | Unitunicate, 350–400 × 4–6 | Filiform, not breaking into part-spores, 350–400 × 1–1.5 | Boudier (1885), Doi (1977) |
| Torrubiella gonylepticida | On a large spider | White film of mycelium | – | Superficial, ovoid to conoid, 300–400 long | – | – | Möller (1901), Petch (1937) |
Cordycipitaceae
Neotorrubiella Tasan., Thanakitp. & Luangsa-ard, gen. nov. — MycoBank MB830171
Etymology. Referring to the phenotypic similarity of the perithecial formation to Torrubiella.
Type species. Neotorrubiella chinghridicola Tasan., Thanakitp. & Luangsa-ard.
Asexual morph. Unknown.
Scant mycelium covering the host, flattened, scattered, white to cream. Perithecia superficial, pale cream, ovoid to obclavate arising all over the body and legs of the host. Asci cylindrical with thickened hyaline cap, 8-spored. Ascus cap rounded. Ascospores filiform, multiseptate, not breaking into part-spores. In culture the colonies on PDA, PSA, SDAY/4 are slow-growing, and do not produce conidiogenous structures, even after 1 mo.
Habitat — On Gryllidae, on underside of leaves.
Distribution — Thailand, known from Khlong Nakha Wildlife Sanctuary, Khao Luang and Khao Yai National Parks.
Notes — Neotorrubiella resembles the genus Torrubiella in its lack of a stipe, and the production of superficial perithecia directly on the body of the host. It differs from Torrubiella by its type of ascospores. Most species in Torrubiella have part-spores whereas Neotorrubiella produces whole ascospores. Species of Torrubiella occur mainly on spiders and hoppers while Neotorrubiella was only found on crickets.
Neotorrubiella chinghridicola Tasan., Thanakitp. & Luangsa-ard, sp. nov. — MycoBank MB830172; Fig. 8
Fig. 8.
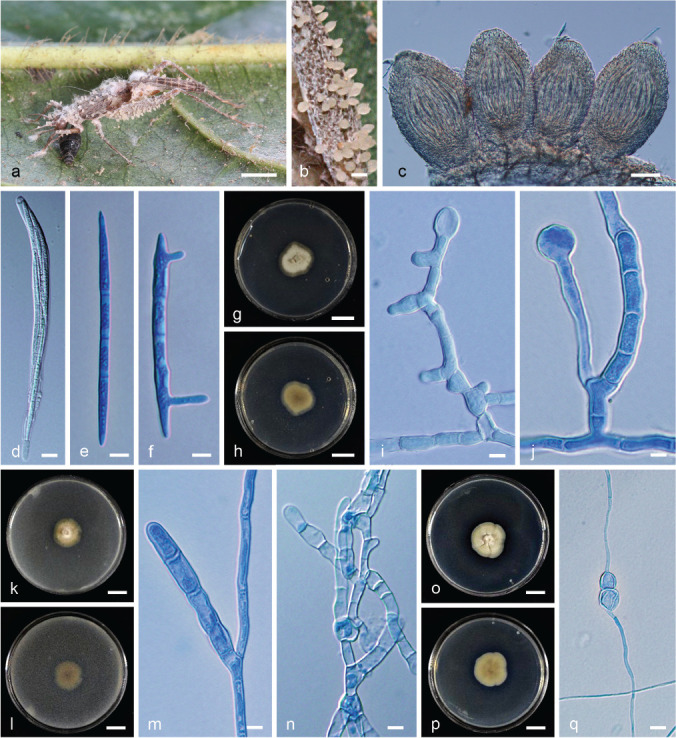
Neotorrubiella chinghridicola (BBH 41240, BCC80733). a. Fungus on Gryllidae host; b. superficial perithecia on host; c. perithecia; d. asci with ascospores; e. ascospore; f. germination of ascospore; g–h. colony on PDA at 30 d: (g) obverse (h) reverse; i–j. swollen hyphae produced on PDA; k–l. colony on PSA at 30 d: (k) obverse (l) reverse; m–n. swollen hyphae produced on PSA; o–p. colony on SDAY/4 at 30 d (o) obverse (p) reverse; q. swollen hyphae produced on SDAY/4. — Scale bars: a = 3 mm; b = 280 μm; c = 70 μm; d = 10 μm; e–f = 7 μm; g–h = 7 mm; i–j, m–n, q = 100 μm; k–l = 6 mm; o–p = 8 mm.
Etymology. Named after the host in Thai language, chringrid, meaning cricket.
Typus. Thailand, Nakhon Si Thammarat Province, Khao Luang National Park, on Gryllidae, on the underside of leaves, 26 Jan. 2016, K. Tasanathai, S. Mongkolsamrit, D. Thanakitpipattana, W. Noisripoom, R. Promharn, P. Srikitikulchai & S. Wongkanoun (holotype BBH 41240, ex-type culture BCC80733).
Asexual morph. Unknown.
Scant mycelium covering the host body, flattened, tomentose, white to cream. Perithecia superficial, pale cream (OAC900), ovoid to obclavate, (220–)226–261(–280) × (130–)145–167.5(–170) μm. Asci cylindrical with thickened ascus apex, 8-spored, (125–)134–162.5 × (9.5–)10–12.5(–15) μm. Ascus cap rounded. Ascospores filiform, multiseptate, not disarticulating into partspores, (68–)70–83.5(–87.5) × 4–5 μm.
Culture characteristics (from germinating ascospores) — Colonies on PDA slow-growing, attaining c. 1.2 cm diam after 30 d at 20 °C. Colony white cream (OAC795), cottony with high mycelial density, pigmented around the colony. Colony reverse cream yellow (OAC813). Colony on PDA did not produce any conidiogenous structures after 1 mo. Chlamydospores observed on PDA after 1 mo. Colonies on PSA slow-growing, attaining c. 0.8 cm diam after 30 d at 20 °C. Colony white cream (OAC816) with a brown clear zone at the colony edges. Colony reverse yellow brown (OAC799). Colony on PSA did not produce any conidiogenous structures after 1 mo. Chlamydospores observed after 1 mo. Colonies on SDAY/4 slow-growing, attaining c. 1.4 cm diam after 30 d at 20 °C. Colony cream (OAC815), cottony with high mycelial density. Colony reverse yellow cream (OAC814) with a brown clear zone at the colony edges. Similar to the colonies on PDA and PSA, it did not produce any conidiogenous structures after 1 mo. Chlamydospores observed after 1 mo.
Additional materials examined. Thailand, Ranong Province, Khlong Nakha Wildlife Sanctuary, on Gryllidae, on the underside of leaves, 11 Jan. 2006, K. Tasanathai, W. Chaygate, S. Mongkolsamrit, P. Srikitikulchai, B. Thongnuch, L. Hung & L. Yen (BBH 16541); Nakhon Ratchasima Province, Khao Yai National Park, on Gryllidae, on the underside of leaves, 14 Aug. 2009, K. Tasanathai, P. Srikitikulchai, S. Mongkolsamrit, T. Chohmee & R. Ridkaew (BBH 30241, BCC39684); Nakhon Ratchasima Province, Khao Yai National Park, on Gryllidae, on the underside of leaves, 6 Oct. 2010, K. Tasanathai, P. Srikitikulchai, A. Khonsanit, R. Somnuk, K. Sansatchanon & W. Noisripoom (BBH 30006, BCC46588).
Ophiocordycipitaceae
Ophiocordyceps kobayasii Mongkols., Thanakitp., Luangsa-ard & Hywel-Jones, sp. nov. — MycoBank MB830173; Fig. 9
Fig. 9.
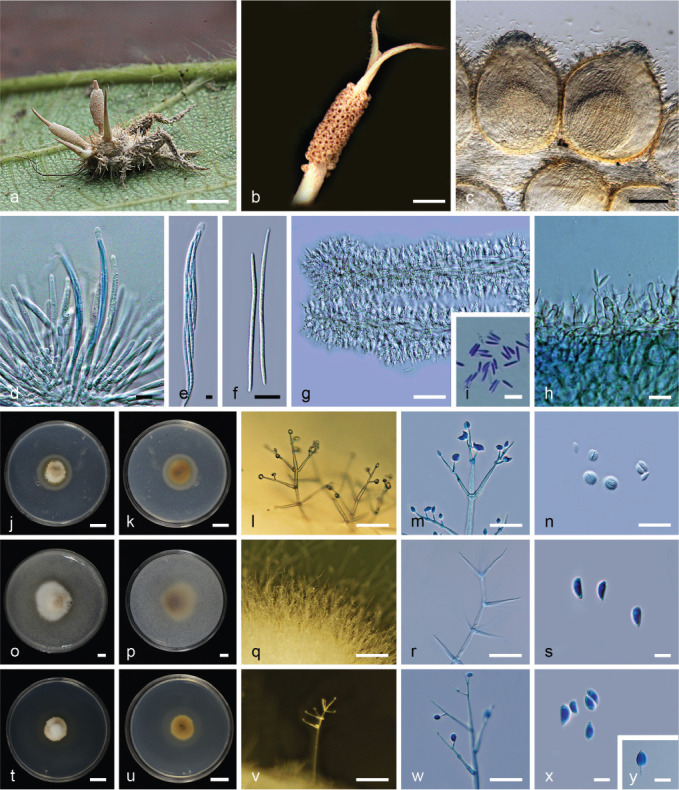
Ophiocordyceps kobayasii (BBH 39608, BCC75694). a. Fungus on Gryllidae host; b. perithecia on stroma; c. perithecia; d–e. asci; f. ascospores; g–i. phialides with conidia on synnema; j–k. colony on PDA at 21 d: (j) obverse (k) reverse; l–m. phialides with conidia on PDA; n. conidia; o–p. colony on PSA at 21 d: (o) obverse (p) reverse; q–r. phialides with conidia on PSA; s. conidia; t–u. colony on SDAY/4 at 21 d: (t) obverse (u) reverse; v–w. phialides with conidia on SDAY/4; x–y. conidia. — Scale bars: a, o–p = 5 mm; b = 1 mm; c = 80 μm; d, n, s, x–y = 5 μm; e–i, m = 10 μm; j–k, t–u = 1 cm; l, q–r, w = 20 μm; v = 50 μm.
Etymology. Named after Prof. Yosio Kobayasi (1907–1993), a Japanese mycologist best known for his work on Cordyceps and its allies.
Typus. Thailand, Nakhon Ratchasima Province, Khao Yai National Park, on Gryllidae, in leaf litter, 1 Oct. 2014, S. Mongkolsamrit, A. Khonsanit, W. Noisripoom, D. Thanakitpipattana & R. Somnuk (holotype BBH 39608, ex-type culture BCC75694).
Asexual morph. Hymenostilbe-like on the host and acremonium-like in culture.
Stromata multiple, arising from the head region and body of the insect host, pale cream (OAC900), cylindrical, up to 8 mm long. Densely packed perithecia occur in the middle of stromata, pale cream (OAC900), c. 3 mm long. Perithecia superficial, ovoid, (180–)190–230(–250) × (140–)150–175(–200) μm. Asci cylindrical, 8-spored, up to 125 μm long, 10–15 μm wide. Ascus cap rounded, 5 × 5 μm. Ascospores whole, filiform, hyaline, spirally arranged inside the ascus, aseptate, (45–)52–72(–90) × 2–2.5 μm. Asexual morph: Numerous synnemata growing from all over the host body. Occasionally hymenostilbe-like asexual morph is produced on the apex of the stroma and below the perithecial layer. Numerous synnemata arising from the body and legs of the host producing hymenostilbe-like asexual morph. The synnemata measured up to 1 cm long and c. 0.5 mm wide, powdery due to sporulation, pale cream (OAC900). Conidiophores arising from hyphae of synnemata, forming a hymenial layer. Phialides (6–)8–12(–15) × (3–)3.5–5 μm, hymenostilbe-like, clavate, cylindrical basal parts, bearing mini cylindrical projections, c. 1 × 0.5 μm. Conidia hyaline, smooth-walled, narrow fusoid, 1-celled, (5–)6–8 × 1–2 μm.
Culture characteristics (from germinating ascospores) — Colonies on PDA slow-growing, attaining c. 0.8 cm diam after 21 d at 20 °C. Colony pink greyish (OAC550), cottony with high mycelial density, brown pigment around the colony. Colony reverse pale brown (OAC638). Vegetative hyphae smooth, septate, hyaline, 2–3 μm diam. Conidiogenous structures consisting of erect conidiophores arising from the vegetative hyphae. Conidiophores consist of verticillate phialides, singly or in whorls of two to five. Phialides awl-shaped, acremonium-like, smooth-walled, (18–)26–43(–50) × 2–3.5(–5) μm, bearing numerous lateral necks along the main phialide, (2–)2.5–8(–10) × 1 μm. Conidia hyaline, ellipsoidal or reniform, 1-celled, (4–)4.5–6.5(–8) × (1.5–)2–2.5(–3) μm, aggregated in slimy heads (Fig. 9l). Chlamydospores not observed. Colonies on PSA slow-growing, attaining c. 1.2 cm diam after 21 d at 20 °C. Colony pinkish grey (OAC550), cottony with high mycelial density and brown pigment around the colony. Colony reverse pale brown (OAC638). Vegetative hyphae smooth, septate, hyaline, 2–3 μm diam. Conidiogenous structures consisting of erect conidiophores arising from the vegetative hyphae. Conidiophores consist of verticillate phialides, singly or in whorls of two to five. Phialides awl-shaped, acremonium-like, (25–)28–38(–42) × (2–)2.5–4 μm. Conidia hyaline, ellipsoidal or reniform, 1-celled, 5–6 × (2–)2.5–3.5(–4) μm, aggregated in slimy heads at the apex of phialides. Chlamydospores not observed. Colonies on SDAY/4 slow-growing, attaining c. 8 mm diam after 21 d at 20 °C. Colony pinkish grey (OAC550), cottony with high mycelial density. Colony reverse pale brown (OAC638). Vegetative hyphae smooth, septate, hyaline, 2–3 μm diam. Conidiogenous structures consisting of erect conidiophores arising from the vegetative hyphae. Conidiophores consist of verticillate phialides, singly or in whorls of two to five. Phialides awl-shaped, acremonium-like, (20–)28–44(–50) × (2–)2.5–4 μm, bearing numerous necks, (3–)4–6(–8) × 1 μm. Conidia hyaline, ellipsoidal or reniform, 1-celled, (5–)5.5–7 × 2–3 μm, aggregated in slimy heads. Chlamydospores not observed.
Additional materials examined. Thailand, Nakhon Ratchasima Province, Khao Yai National Park, on Gryllidae, on the underside of leaves, 8 Nov. 2012, S. Mongkolsamrit, A. Khonsanit, W. Noisripoom, P. Srikitikulchai & R. Somnuk (BBH 38353, BCC57812 and BCC58417), (BBH 35186); Saraburi Province, Khao Yai National Park, on the underside of leaves, 27 May 2015, K. Tasanathai, A. Khonsanit, W. Noisripoom, D. Thanakitpipattana, N. Kobmoo & J.J. Luangsa-ard (BBH 41178, BCC84222), (BBH 41180, BCC84223).
Ophiocordyceps krachonicola Tasan., Thanakitp. & Luangsa-ard, sp. nov. — MycoBank MB830174; Fig. 10
Fig. 10.
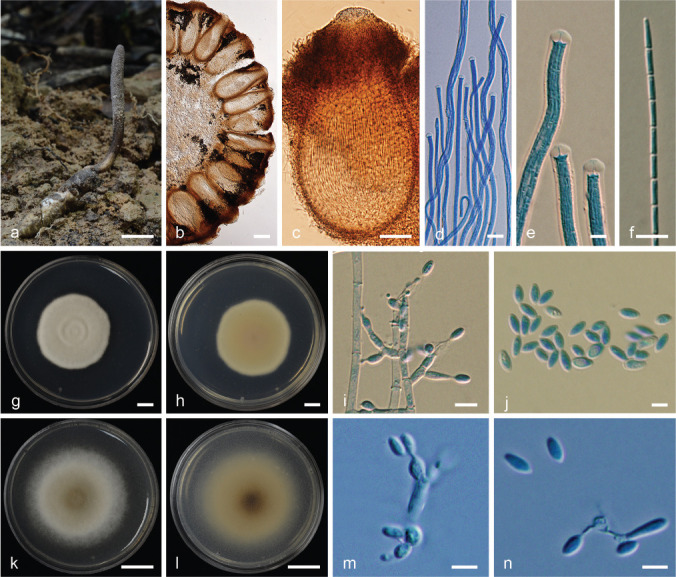
Ophiocordyceps krachonicola (BBH 40672, BCC79666). a. Stroma of fungus emerging from a mole cricket; b. cross section of stroma showing perithecia; c. perithecium; d. asci; e. part of asci with asci tip; f. part-spores; g–h. colony on PDA: (g) obverse (h) reverse; i. conidiogenous cells with conidia; j. conidia; k–l. colony on PSA: (k) obverse (l) reverse; m. conidiogenous cells with conidia; n. conidia. — Scale bars: a = 10 mm; b = 150 μm; c = 80 μm; d = 10 μm; e–f = 5 μm; g–h = 5 mm; i–j, m–n = 5 μm; k–l = 10 mm.
Etymology. Named after the host in Thai, ‘krachon’; meaning mole cricket.
Typus. Thailand, Phitsanulok Province, Ban Phao Thai Community Forest, on Gryllotalpa orientalis nymph (Gryllidae), buried in the ground, 10 Oct. 2015, S. Mongkolsamrit, D. Thanakitpipattana, W. Noisripoom & A. Khonsanit (holotype BBH 40672, ex-type culture BCC79666).
Asexual morph. Hirsutella-like.
Stroma solitary, cylindrical, arising from the head of cricket nymph buried in the ground, 37–40 × 1–1.5 mm. Fertile part cylindrical to clavate, pale grey (OAC907), ostiole dark grey (OAC901), up to 25 mm long, 2–3 mm wide. Perithecia completely immersed, ordinal in arrangement, ovoid to obclavate, (460–)474.5–539(–580) × (180–)193.5–258(–300) μm. Asci cylindrical with thickened ascus apex, 8-spored, (250–)286–377(–400) × 4–5 μm. Ascus cap rounded. Ascospores divided into 64 part-spores after maturity, (300–)351–416.5(–450) × 1 μm, part-spores cylindrical, (4–)4.4–7(–10) × 1 μm.
Culture characteristics (from germinating ascospores) — Colonies on PDA fast growing, attaining c. 2.2 cm diam after 21 d at 20 °C, white to cream with distinct margin. Phialides flask-shaped with distinct neck, (3–)3.5–6(–7) × 1.5–2 μm. Conidia not in chains, hyaline, 1-celled, fusiform, (3–)4–5.5(–7) × 2–2.5(–3) μm. Colonies on PSA fast growing, attaining c. 3 cm diam after 21 d at 20 °C, white to pale yellow (OAC858) with packed mycelium. Colony reverse pale brown (OAC638). Phialides flask-shaped, 3–5 × 2–3 μm. Conidia hyaline, 1-celled, fusiform, 4–6 × 2–3 μm. Colonies on SDAY/4 fast growing, attaining c. 3 cm diam after 21 d at 20 °C. Colony pinkish grey (OAC550) with cottony and high mycelial density. Colony reverse pale brown (OAC638). Colony on SDAY/4 did not produce any conidiogenous structures after 1 mo.
Additional materials examined. Thailand, Phitsanulok Province, Ban Phao Thai Community Forest, on Gryllotalpa orientalis nymph (Gryllidae), buried in the ground, 10 Oct. 2015, S. Mongkolsamrit, D. Thanakitpipattana, W. Noisripoom & A. Khonsanit (BBH 41219, BCC79667).
DISCUSSION
Clavicipitaceae
Two Metarhizium species that parasitize crickets and stick insects are nested in the core Metarhizium clade (Kepler et al. 2014, Luangsa-ard et al. 2017) within the Metarhizium anisopliae species complex (Kepler & Rehner 2013), and separated from Metarhizium acridum that was previously reported to be specific to Orthoptera (Fig. 1). Nishi & Sato (2017) classified similar specimens on stick insects (Phasmatodea) and crickets (Orthoptera) as Metarhizium majus by using DNA sequence data of the 5′TEF (see Appendix). However, our multi-gene phylogenetic analyses showed our samples on stick insects and on crickets to be distinct from M. majus. These results were confirmed using nuclear intergenic sequence markers, which showed better phylogenetic informativeness than other legacy genes (Kepler & Rehner 2013). Two new species are thus introduced as Metarhizium phasmatodeae on stick insects and Metarhizium gryllidicola on crickets. Morphologically, both new species are difficult to distinguish from other species in the M. anisopliae species complex (MGT clade) but can be circumscribed based on multi-loci and IGR phylogenetic analyses and host affinity. Recent genomic studies on species of Metarhizium has shown that generalists evolved from specialists via transitional species with intermediate host ranges (Hu et al. 2014, Zhang et al. 2019). Based on the phylogeny presented here, the two new species in the MGT clade (Fig. 2) exhibited host-specific interactions, as opposed to the intermediate host ranges reported for M. majus and M. guizhouense (Hu et al. 2014). However, as the discovery of these entomopathogens in the field are mostly of serendipitous nature, additional data are necessary to resolve these discrepancies to place the new taxa in their proper context.
Specimens on egg cases (ootheca) of praying mantis previously identified as Cordyceps mantidicola from Japan (Kobayasi & Shimizu 1983) look morphologically similar to Petchia siamensis (Fig. 7). Cordyceps mantidicola was reported occurring on cocoons of Mantidae (Kobayasi & Shimizu 1983, Liu et al. 1997). However, mantis do not produce cocoons but egg cases. Cordyceps mantidicola from Japan and China produces part-spores while Petchia siamensis produces whole ascospores. We believe the Chinese ‘C. mantidicola’ is different from the Thai and Japanese materials, but this can only be verified through molecular phylogenetic reconstruction using strains from Japan, China and Thailand. Multi-locus phylogenetic analyses clearly placed our new species in the Clavicipitaceae as a separate genus. We therefore proposed a new genus, Petchia, and transferred C. mantidicola (on ootheca of mantis from Japan) to this genus (Table 3).
Cordycipitaceae
Specimens producing superficial perithecia on crickets exhibited an outer appearance (lack of stipe and perithecial characters) similar to Torrubiella aranicida and T. gonylepticida occurring on spiders. However, our specimens showed a different type of ascospore morphology compared to the aforementioned species. It shares similarity with T. wallacei (Lecanicillium wallacei = Simplicillium wallacei) in producing whole ascospores, but differs in host as well as in phylogenetic position (Table 3). Many species of Torrubiella produce part-spores. In his monograph on Paecilomyces and related genera, Samson (1974) reported the asexual morph for both T. aranicida and T. gonylepticida as Cordyceps farinosa (= Paecilomyces farinosus = Isaria farinosa). From our molecular phylogenetic analyses, we clearly showed that our specimens were nested in Cordycipitaceae, but distinct from Cordyceps (Fig. 3). The production of the asexual morph was not observed in any of the three culture media used. We therefore described a new genus Neotorrubiella to accommodate these isolates.
Ophiocordycipitaceae
Known Ophiocordyceps species occurring on Orthoptera could be found on adult and nymph stages and have different morphologies from the newly described species as follows: O. amazonica was first reported on adult Orthoptera by Hennings (1904), producing solitary or single stroma to multiple or gregarious stromata, reddish brown and rounded cylindrical to compressed fertile heads (Sanjuan et al. 2015; Table 3). The stroma of O. krachonicola that parasitize Orthoptera larvae is solitary, simple, dark brown, and produces a hirsutella-like asexual morph, while O. kobayasii has multiple, pale cream, cylindrical stromata and also parasitizes adult orthopterans (small crickets). Our molecular phylogeny and host affiliations suggest that both Ophiocordyceps species represent new species from Thailand (Fig. 4).
Ecology
New species on Orthopterida (orders Orthoptera and Phasmatodea) were found buried in the ground, on the underside of leaves of dicotyledonous plants as well as lying loosely in leaf litter. These fungi occur on adults and nymphs, as well as on egg cases (ootheca) of praying mantis (Mantidae). All of the collections were derived from national parks and community forests where human disturbance is still low, with only a few individual collections. No specimens were found in agricultural ecosystems.
Acknowledgements
The authors are grateful to the Platform Technology Management Section, National Center for Genetic Engineering and Biotechnology (BIOTEC), Grant No. P19-50231 and CPMO Grant No. P15-51452 for their support of the biodiversity studies of invertebrate-pathogenic fungi in Thailand. We thank the Department of National Parks for their kind support and permission to collect fungi in the National Parks. We also thank Noppol Kobmoo for discussing the phylogenetic analyses, Paradorn Dokchan for the identification of insects, Philip Shaw and Amy Divinagracia for editing the manuscript. This study was supported by the National Science and Technology Development Agency (NSTDA). We are also grateful to the comments and suggestions of the two anonymous reviewers to help improve the manuscript.
Appendix
Phylogenetic reconstruction of Metarhizium using 5′TEF sequences based on Maximum Parsimony, Bayesian analysis and RAxML. Number on the nodes are MP bootstrap/Bayesian posterior probability/ML bootstrap values above 70 %. Bold lines mean support for the three analyses were 100 %.
REFERENCES
- Aráujo JPM, Hughes DP. 2016. Diversity of entomopathogenic fungi: Which groups conquered the insect body? Advances in Genetics 94: 1–39. [DOI] [PubMed] [Google Scholar]
- Ariyawansa HA, Hyde KD, Jayasiri SC, et al. 2015. Fungal diversity notes 111–252 – taxonomic and phylogenetic contributions to fungal taxa. Fungal Diversity 75: 27–274. [Google Scholar]
- Ban S, Sakane T, Nakagiri A. 2015. Three new species of Ophiocordyceps and overview of anamorph types in the genus and the family Ophiocordycipitaceae. Mycological Progress 14: 1–12. [Google Scholar]
- Berkeley MJ. 1856. On some entomogenous Sphaeria. Journal of the Proceedings of the Linnean Society. Botany 1(4): 159. [Google Scholar]
- Bischoff JF, Rehner SA, Humber RA. 2009. A multilocus phylogeny of the Metarhizium anisopliae lineage. Mycologia 101: 512–530. [DOI] [PubMed] [Google Scholar]
- Boudier JLÉ. 1885. Note sur un nouveau genre et quelques nouvelles espèces des Pyrenomycètes. Revue Mycologique Toulouse 7: 224–227. [Google Scholar]
- Castlebury LA, Rossman AY, Sung GH, et al. 2004. Multigene phylogeny reveals new lineage for Stachybotrys chartarum, the indoor air fungus. Mycological Research 108: 864–872. [DOI] [PubMed] [Google Scholar]
- Chaverri P, Bischoff JF, Evans HC, et al. 2005. Regiocrella, a new entomopathogenic genus with a pycnidial anamorph and its phylogenetic placement in the Clavicipitaceae. Mycologia 97: 1225–1237. [DOI] [PubMed] [Google Scholar]
- Doi Y. 1977. Two species of Torrubiella and their conidial states. Boletín de la Sociedad Argentina de Botánica 18: 110–114. [Google Scholar]
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemical Bulletin 19: 11–15. [Google Scholar]
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research 32: 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. 1985. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 39: 783–791. [DOI] [PubMed] [Google Scholar]
- Hall T. 2004. BioEdit. Version 6.0.7. Raleigh, North Carolina, Department of Microbiology, North Carolina State University; Available from: http://www.mbio.ncsu.edu/BioEdit/bioedit.html (accessed 16 Jan. 2019) [Google Scholar]
- Hennings P. 1904. Fungi amazonici II. a cl. Ernesto Ule collecti. Hedwigia 43: 246–249. [Google Scholar]
- Hu X, Xiao G, Zheng P, et al. 2014. Trajectory and genomic determinants of fungal-pathogen speciation and host adaptation. Proceedings of the National Academy of Sciences USA 111: 16796–16801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Index Fungorum. (continuously updated). Royal Botanic Gardens Kew. Available from http://www.indexfungorum.org (accessed Jan. 2019).
- Kepler RM, Ban S, Nakagiri A, et al. 2013. The phylogenetic placement of hypocrealean insect pathogens in the genus Polycephalomyces: An application of One Fungus One Name. Fungal Biology 117(9): 611–622. [DOI] [PubMed] [Google Scholar]
- Kepler RM, Humber RA, Bischoff JF, et al. 2014. Clarification of generic and species boundaries for Metarhizium and related fungi through multigene phylogenetics. Mycologia 106: 811–829. [DOI] [PubMed] [Google Scholar]
- Kepler RM, Luangsa-ard JJ, Hywel-Jones NL, et al. 2017. A phylogenetically-based nomenclature for Cordycipitaceae (Hypocreales). IMA Fungus 8: 335–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kepler RM, Rehner SA. 2013. Genome-assisted development of nuclear intergenic sequence markers for entomopathogenic fungi of the Metarhizium anisopliae species complex. Molecular Ecology Resources 13: 210–217. [DOI] [PubMed] [Google Scholar]
- Kepler RM, Sung GH, Harada Y, et al. 2012. Host jumping onto close relatives and across kingdoms by Tyrannicordyceps gen. nov. and Ustilaginoidea. American Journal of Botany 99(3): 552–561. [DOI] [PubMed] [Google Scholar]
- Kobayasi Y. 1941. The genus Cordyceps and its allies. Science Reports of the Tokyo Bunrika Daigaku 84: 53–260. [Google Scholar]
- Kobayasi Y, Shimizu D. 1983. Cordyceps species from Japan. 6. Bulletin of the National Science Museum Tokyo 9: 1–21. [Google Scholar]
- Kornerup A, Wanscher JH. 1963. Methuen handbook of colour. Methuen & Company Ltd., London. [Google Scholar]
- Liu ZY, Liang ZQ, Liu AY. 1997. Two uncommon species of Cordyceps collected in Guizhou province. Mycosystema 16: 97–101. [Google Scholar]
- Luangsa-ard JJ, Mongkolsamrit S, Thanakitpipattana D, et al. 2017. Clavicipitaceous entomopathogens: new species in Metarhizium and a new genus Nigelia. Mycological Progress 16: 369–391. [Google Scholar]
- Luangsa-ard JJ, Tasanathai K, Thanakitpipattana D, et al. 2018. Novel and interesting Ophiocordyceps spp. (Ophiocordycipitaceae, Hypocreales) with superficial perithecia from Thailand. Studies in Mycology 89: 125–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mains EB. 1941. Cordyceps stylophora and Cordyceps ravenelii. Mycologia 33: 611–617. [Google Scholar]
- Mains EB. 1958. North American entomogenous species of Cordyceps. Mycologia 50: 169–222. [Google Scholar]
- Mains EB. 1959. Cordyceps species. Bulletin of the Torrey Botanical Club 86: 46–58. [Google Scholar]
- Möller A. 1901. Botanische Mittheilungen aus den Tropen von Schimper AFW, Heft 9. Phycomyceten und Ascomyceten, untersuchungen aus Brasilien 9: 210. [Google Scholar]
- Mondal S, Baksi S, Koris A, et al. 2016. Journey of enzymes in entomopathogenic fungi. Pacific Science Review A: Natural Science and Engineering 18: 85–99. [Google Scholar]
- Mongkolsamrit S, Noisripoom W, Arnamnart N, et al. 2019. Resurrection of Paraisaria in the Ophiocordycipitaceae with three new species from Thailand. Mycological Progress 18: 1213–1230. [Google Scholar]
- Mongkolsamrit S, Noisripoom W, Thanakitpipattana D, et al. 2018. Disentangling cryptic species with isarioid morphs in Cordycipitaceae. Mycologia 110: 230–257. [DOI] [PubMed] [Google Scholar]
- Moon K, Hue SM. 2017. Mode of infection of Metarhizium spp. Fungus and their potential as biological control agents. Journal of Fungi 3: 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nees TFL. 1820. Synopsis generum plantarum mycetoidearum. Geschichte der merckwürdigsten Pilze 4: XLVII–CXII. [Google Scholar]
- Nishi O, Sato H. 2017. Species diversity of the entomopathogenic fungi Metarhizium anisopliae and M. flavoviride species complexes isolated from insects in Japan. Mycoscience 58: 472–479. [Google Scholar]
- Nylander JAA. 2004. MrModeltest 2.2: program distributed by the author. Evolutionary Biology Centre, Uppsala University. [Google Scholar]
- Petch T. 1931. Notes on entomogenous fungi. Transactions of the British Mycological Society 16: 55–75. [Google Scholar]
- Petch T. 1937. Notes on entomogenous fungi. Transactions of the British Mycological Society 21: 34–67. [Google Scholar]
- Rehner SA, Kepler RM. 2017. Species limits, phylogeography and reproductive mode in the Metarhizium anisopliae complex. Journal of Invertebrate Pathology 148: 60–66. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Teslenko M, Mark P, et al. 2012. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model selection across a large model space. Systematic Biology 61: 539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samson RA. 1974. Paecilomyces and some allied hyphomycetes. Studies in Mycology 6: 1–119. [Google Scholar]
- Sanjuan T, Franco-Molano AE, Kepler RM, et al. 2015. Five new species of entomopathogenic fungi from the Amazon and evolution of neotropical Ophiocordyceps. Fungal Biology 119: 901–916. [DOI] [PubMed] [Google Scholar]
- Sanjuan T, Tabima J, Restrepo S, et al. 2014. Entomopathogens of Amazonian stick insects and locusts are members of the Beauveria species complex (Cordyceps sensu stricto). Mycologia 106: 260–275. [DOI] [PubMed] [Google Scholar]
- Shrestha B, Tanaka E, Hyun MW, et al. 2016. Coleopteran and Lepidopteran hosts of the entomopathogenic genus Cordyceps sensu lato. The Journal of Mycology 2016: 1–14. doi: 10.1155/2016/7648219. [DOI] [Google Scholar]
- Smithe FB. 1975. Naturalist’s Color Guide. The American Museum of Natural History, New York. [Google Scholar]
- Spatafora JW, Sung GH, Sung JM, et al. 2007. Phylogenetic evidence for an animal pathogen origin for ergot and the grass endophytes. Molecular Ecology 16: 1701–1711. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30: 1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung GH, Hywel-Jones NL, Sung JM, et al. 2007. Phylogenetic classification of Cordyceps and the clavicipitaceous fungi. Studies in Mycology 57: 5–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford DL. 2002. PAUP*: phylogenetic analysis using parsimony (*and other methods), version 4.0b10. Sinauer Associates, Sunderland. [Google Scholar]
- White TJ, Bruns TD, Lee SB, et al. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, et al. (eds), PCR Protocols: a guide to methods and applications: 315–322. Academic Press, New York. [Google Scholar]
- Zhang Q, Chen X, Xu C, et al. 2019. Horizontal gene transfer allowed the emergence of broad host range entomopathogens. Proceedings of the National Academy of Sciences USA 116: 7982–7989. [DOI] [PMC free article] [PubMed] [Google Scholar]