Abstract
Association of platelet factor 4 (PF4) with heparin is a first step in formation of aggregates implicated in the development of heparin-induced thrombocytopenia (HIT), a potentially fatal immune disorder affecting 1–5% of patients receiving heparin. Despite being a critically important element in HIT etiology, relatively little is known about the specific molecular mechanism of PF4-heparin interactions. This work uses native mass spectrometry to investigate PF4 interactions with relatively short heparin chains (up to decasaccharides). The protein is shown to be remarkably unstable at physiological ionic strength in the absence of polyanions; only monomeric species are observed, and the extent of multiple charging of corresponding ions indicates a partial loss of conformational integrity. The tetramer signal remains at or below the detection threshold in the mass spectra until the solution’s ionic strength is elevated well above the physiological level, highlighting the destabilizing role played by electrostatic interactions vis-à-vis quaternary structure of this high-pI protein. The tetramer assembly is dramatically facilitated by relatively short polyanions (synthetic heparin-mimetic pentasaccharide), with the majority of the protein molecules existing in the tetrameric state even at physiological ionic strength. Each tetramer accommodates up to six pentasaccharides, with at least three such ligands required to guarantee the higher-order structure integrity. Similar results are obtained for PF4 association with longer and structurally heterogeneous heparin oligomers (decamers). These longer polyanions can also induce PF4 dimer assembly when bound to the protein in relatively low numbers, lending support to a model of PF4/heparin interaction in which the latter wraps around the protein, making contacts with multiple subunits. Taken together, these results provide a more nuanced picture of PF4-glycosaminoglycan interactions leading to complex formation. This work also advocates for a greater utilization of native mass spectrometry in elucidating molecular mechanisms underlying HIT, as well as other physiological processes driven by electrostatic interactions.
Significance
Platelet factor 4 (PF4)-heparin interactions remain a focal point of extensive research, with most studies aiming at identifying the hallmarks of PF4-heparin antigenicity. This puts a premium on characterizing all relevant species at the molecular level, which remains challenging because of the enormous structural diversity displayed by heparin and PF4-heparin complexes. Application of native mass spectrometry to characterize PF4 interactions with heparin oligomers provides a level of detail that was previously unattainable. It reveals a more nuanced picture of the interaction process, in which even acquisition of the native quaternary structure critically depends on the presence of polyanions. In addition to small PF4-polyanion complexes, mass spectrometry also reveals a range of transient species such as tetramers composed of partially unfolded units and heparin-stabilized dimeric PF4.
Introduction
Platelet factor 4 (PF4) is a small, high-isoelectric point protein (pI = 7.6) that was initially discovered as a prothrombotic factor in the blood of thrombocytopenic purpura patients (1). In addition to its prominent role in hemostasis and thrombosis (2), PF4 is intimately involved in a variety of other biological processes (3,4) via interactions with multiple physiological partners. One interaction that gained particular notoriety and remained a focal point of extensive research efforts in the past several decades is association of PF4 with a highly anionic glycosaminoglycan (GAG) heparin (5). This is a first step in the formation of antigenic aggregates that may lead to development of heparin-induced thrombocytopenia (HIT), a serious (and potentially fatal) immune disorder that affects 1–5% of patients receiving heparin as an anticoagulant (6). Despite PF4-heparin association being an important aspect in the etiology of HIT, relatively little is known about the specific molecular mechanism of this interaction. Even before the crystal structures of PF4 (showing the continuous ring of positive potential encircling the PF4 tetramer (7,8)) became available, suggestions were made that the heparin chain wraps around the tetramer to maximize electrostatic contacts (mostly with the lysine-rich C-termini of the constituent polypeptide chains) (9), a conjecture that was mostly confirmed by modeling the interaction using the coordinates of PF4 tetrameric structure (10). Later NMR studies expanded the repertoire of heparin-binding sites within PF4 to include several arginine, lysine, and histidine residues located outside of the C-terminal helix (11) and provided evidence that heparin may play an important role in fine-tuning the protein conformation and indeed control both PF4 folding and assembly of its quaternary structure (12).
On the other hand, the recently published crystal structure of PF4 complexed with a short synthetic heparin-mimetic pentasaccharide suggested relatively limited contacts between the highly anionic pentasaccharide and the PF4 tetramer, placing the number of highly anionic pentasaccharides bound to a single tetramer at two and finding only minimal conformational changes within PF4 resulting from this association (13). One of the two PF4-bound pentasaccharides was actually “shared” with another (symmetry-related) tetramer in the crystal structure, prompting a suggestion that this bidentate interaction is relevant vis-à-vis assembly of large immunogenic complexes. In a dramatic departure from the earlier models of PF4-heparin interactions, which invoked wrapping of a GAG chain around the tetramer (vide supra), the model emerging from the crystallographic data is built on the assumption that the initial PF4/heparin-binding event leads to the polyanion assuming a linear conformation, allowing it to associate with a second PF4 tetramer and eventually leading to formation of a large complex in which the proteins are tethered to a semirigid linear heparin chain (13).
Although extrapolation of the structure obtained with a short heparin-like oligosaccharide to longer GAG chains bears obvious dangers, until recently, crystallography remained the only technique capable of producing information on any relevant PF4-GAG oligomer assembly at the molecular level. Other biophysical techniques that have been used extensively in the recent past to characterize PF4-heparin complexes (reviewed in (14)) typically report only a single physicochemical characteristic of the protein/polyanion complexes averaged across the entire ensemble (e.g., average physical size) without providing sufficient details at the molecular level. The only exceptions are transmission electron microscopy and atomic force microscopy (AFM), which have been used to visualize large PF4-heparin complexes as well as their associations with antibodies (15,16). However, the resolution typically achieved in transmission electron microscopy and AFM measurements is not sufficient to deduce structural details of the substituents of the protein/polyanion complexes.
One aspect that makes characterization of PF4-heparin complexes (as well as heparin complexes with other proteins) particularly challenging is the structural heterogeneity of heparin (both intra- and interchain). One possible way of eliminating this problem takes advantage of the availability of highly homogeneous synthetic heparins (17). However, reducing the enormous complexity of intact heparin to a single structure raises the specter of inadvertent elimination of a subset of glycosaminoglycan chains exhibiting unique behavior toward the target protein. It is the heterogeneity of heparin that, until recently, limited the applications of native mass spectrometry (native MS; a potent biophysical tool frequently used to characterize biopolymer complexes (18)) in the field of protein-heparin interactions to small and relatively homogeneous systems (19,20). Recent advances in native MS, particularly the introduction of native LC/MS (21) and limited charge reduction in the gas phase (22) resulted in a dramatic expansion of the reach of this analytical technique in the field of highly heterogeneous biopolymers. In this work, we use native MS to evaluate PF4 interaction with relatively short heparin oligomers, including a homogeneous synthetic pentasaccharide (pS) and a structurally heterogeneous decasaccharide (dp10) derived from heparin. The results of this study provide clear evidence for the centrality of electrostatic interactions to PF4-heparin interactions, showing that the protein does act as a “heparin sponge” by readily accommodating up to six pentasaccharides on a single tetramer (as opposed to two pentasaccharides visualized by x-ray crystallography (13)). Intriguingly, native MS indicates that at least three short ligands are required for complete stabilization of the tetrameric structure, whereas lower ligand loads compromise either the quaternary structure of PF4 (causing its dissociation to monomers) or its ternary structure (resulting in partial unfolding of the polypeptide chains within the tetrameric assembly). Similar results are obtained for PF4 association with dp10. At the same time, native MS provides no evidence for bridging of PF4 tetramers by short oligoheparins. The study provides a more nuanced picture of PF4-glycosaminoglycan interactions, in which even the acquisition of the canonical quaternary structure by PF4 under physiologically relevant conditions is reliant on the availability of polyanionic segments (which are provided in vivo by relevant proteoglycans: chondroitin sulfate in the α-granules of platelets and heparan sulfate outside of the PF4 storage sites (23,24)).
Materials and Methods
Recombinant form of human PF4 was expressed using the previously published protocol (25) and stored in 1.8 M NaCl. The molecular weight of the polypeptide chain was measured by reversed-phase liquid chromatography/MS (isotopic mass 7891.26 Da, compared with the 7891.22 Da calculated for the oxidized form of the recombinant protein). The synthetic heparin-mimetic pentasaccharide with high affinity to antithrombin (fondaparinux) was purchased from Sigma-Aldrich (St. Louis, MO). Heparin dp10 produced by partial depolymerization of heparin was acquired from Iduron (Alderley Edge, UK). All other chemicals and solvents used in this work were of analytical grade or higher.
PF4 samples for MS analysis were prepared by dialyzing into 150 mM ammonium acetate (pH 6.9). Ionic strength was adjusted by adding additional ammonium acetate to the protein solution as needed. Native MS measurements were carried out using a Synapt G2-Si (Waters, Milford, MA) hybrid quadrupole/time-of-flight mass spectrometer equipped with an ion mobility analyzer and a nanospray source. The following ion optics parameters were typically used to ensure stability of noncovalent complexes in the gas phase: sampling cone voltage, 80 V; trap CE, 4 V; transfer CE, 0 V; and trap DC bias, 3 V. Ion selection before collision-induced dissociation and limited charge reduction was carried out by setting the quadrupole selection parameters (LM resolution) at 4. To trigger limited charge reduction, the trap wave height was set to 0.2 V, and the discharge current was optimized.
Molecular modeling of the full-length PF4 was carried out using Maestro modeling suite (release 2019-2; Schrödinger, New York, NY). The crystal structure of human PF4 (Protein Data Bank, PDB: 4R9Y (13)) was used as a template; the missing N-terminal peptides EAEED were appended to each monomer, followed by energy minimization of the engineered tetramer using the OPLS3e force field. The energy-minimized tetramers were subjected to a 6-ns molecular dynamics simulation at 300 K in explicit water and in the presence of 150 mM NaCl ions (Desmond).
Results and Discussion
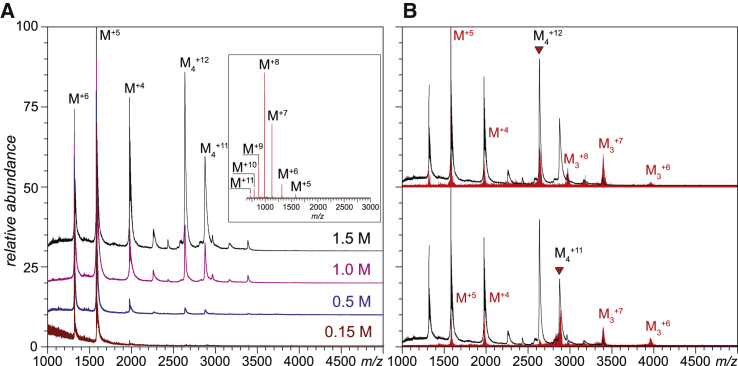
Assessment of the higher-order structure of PF4 at a physiological ionic strength (150 mM) and neutral pH (6.9) in the absence of GAGs carried out with native MS revealed the exclusively monomeric form of this protein (Fig. 1 A, bottom trace). The absence of any detectable signal of the tetrameric form of this protein might seem surprising because PF4 is frequently stated to have a stable tetrameric form at a physiological pH and ionic strength (5,26) without explicitly mentioning the critical importance of endogenous GAGs in stabilizing this form in vivo. However, the original work by Barber et al. noted that PF4 was released from platelets in the form of a complex with chondroitin sulfate proteoglycans (23). Separation of PF4 from its proteoglycan counterpart gave rise to a protein that could be readily dissolved at an acidic pH but remained insoluble under physiological conditions (27). All subsequent structural work with PF4 (e.g., x-ray crystallography studies) was carried out either under mildly acidic conditions or at an ionic strength that significantly exceeded the physiologically relevant levels (7,8). Elevation of the ionic strength of the PF4 solution gave rise to a progressively increasing ionic signal for the tetrameric form of this protein, although even at the highest ionic strength examined in these measurements (1.5 M), a prominent signal of the monomeric form of PF4 was still detected (Fig. 1 A). Although the instability of PF4 tetrameric assembly in the absence of polyanions is not surprising (given the highly localized cluster of positive charges on the protein surface (Fig. 2) that is likely to result in a significant repulsion among the monomeric components of the assembly, whose destabilizing effect is mitigated upon increasing the solution ionic strength because of a progressive decrease in Debye-Hückel length (28)), the prominence of PF4 monomers raises the question of whether these species retain a compact conformation with a native-like ternary structure.
Figure 1.
(A) shows mass spectra of 0.2 mg/mL aqueous solutions of PF4 acquired at pH 6.9 and different ionic strengths (the numbers in the right hand-side part of the panel indicate the NH4CH3CO2 concentrations in each protein solution); the inset shows a mass spectrum of PF4 acquired under denaturing conditions (3% formic acid, 50% acetonitrile by volume). (B) shows mass spectra of fragment ions produced by collision-induced dissociation of the two representative precursor ions (labeled with triangles) near the dissociation threshold (red traces). The black trace in each panel shows a reference mass spectrum of PF4 acquired in 1.5 M NH4CH3CO2, from which the precursor ions were selected. To see this figure in color, go online.
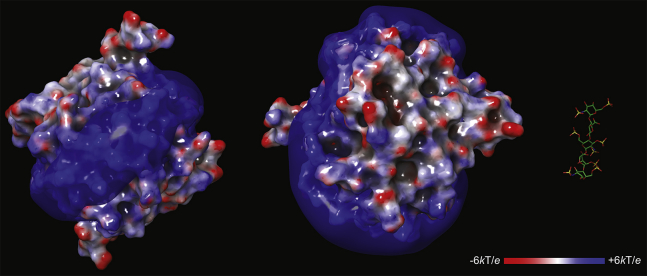
Figure 2.
A representative structure (two orthogonal views) of a PF4 tetramer composed of polypeptides with intact acidic N-termini. The structure is produced by molecular dynamics simulations (see the Experimental section for more detail) and colored according to the electrostatic potential. The semitransparent blue surface shows the +6 kT/e isopotential surface. The ball-and-stick model on the right represents the pentasaccharide structure (the same scale as the protein structure). To see this figure in color, go online.
Evaluation of the degree of compactness of monomeric polypeptide chains was carried out by analyzing protein ion charge state distributions in the mass spectra, a technique that is frequently used to assess the integrity of protein conformation in solution (29). In contrast to the broad distribution of charge states of PF4 polypeptide ions in the mass spectrum acquired under strongly denaturing conditions (inset in Fig. 1 A), charge distributions of PF4 monomer ions at neutral pH remain narrow (displaying only three charge states, +4 through +6) across the entire range of ionic strength examined in this work. The average charge density of these ions is also notably lower compared to those representing the fully denatured protein, indicating at least some degree of compactness. At the same time, it appears that the monomeric PF4 fails to maintain the native-like fold. Indeed, the gas-phase dissociation of the PF4 tetramers (in 1.5 M ammonium acetate) triggered by mild collisional activation proceeds through a classical asymmetric-charge-partitioning mechanism (30, 31, 32), with the low-charge-density trimers and complementary higher-charge-density monomers being the only dissociation products (Fig. 1 B). The low-mass, high-charge fragments are known to undergo the loss of the higher-order structure during their ejection from the noncovalent complexes (30, 31, 32), and because the extent of their multiple charging is very close to that exhibited by the PF4 monomer ions at neutral pH (Fig. 1 A), we conclude that the conformational integrity of PF4 monomers in solution is compromised.
A close examination of the evolution of the charge state distribution of the PF4 monomers as a function of the ionic strength reveals a small but detectable (and consistent) shift toward lower charge density at elevated ionic strength (Fig. 1 A). It might be tempting to ascribe this phenomenon to “tightening” of the monomer conformation in solution, leading to a lower average change accumulated by the protein ions upon transfer from solution to the gas phase because of a decrease of its solvent-accessible surface area (33). However, charge-state distributions of protein ions are known to be affected by the concentration of small electrolytes present in solution, with the elevated electrolyte concentrations leading to a moderate decrease in the extent of multiple charging of the protein ions without any noticeable conformational changes in solution (34). Although detection of small-scale conformational transitions based on the subtle changes in the ionic charge state distributions is possible (35), it requires that the electrolyte composition of the protein solution be kept nearly constant throughout the experiment. Obviously, this requirement cannot be met when the solution ionic strength changes 10-fold in the course of the measurements (as is the case in the studies reported in Fig. 1 A). Therefore, we conclude that the observed small-scale shift of the charge state distributions of PF4 monomer ions (the increase of the relative abundance of the M+4 ion in particular) is likely to reflect gas-phase processes in the ESI interface, rather than subtle conformational changes within the PF4 monomers in solution.
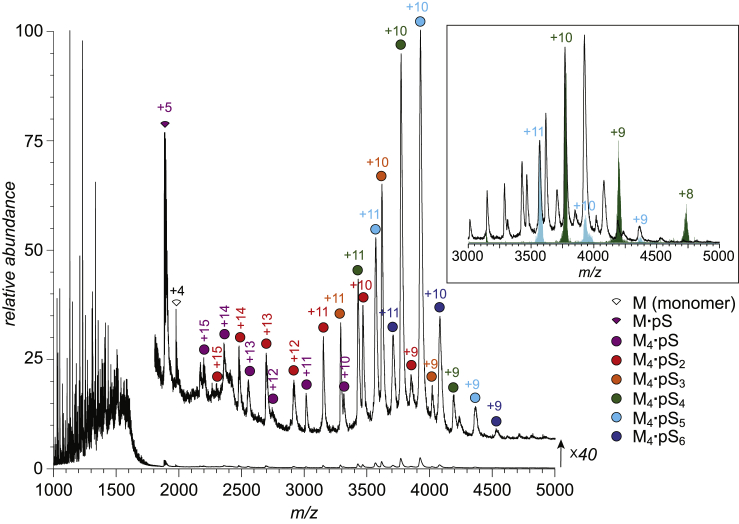
Addition of a nearly stoichiometric amount of the pS to the PF4 solution (on the PF4 monomer/pS molar ratio basis) results in a dramatic transformation of the appearance of the mass spectra acquired at the physiological ionic strength (Fig. 3). The high m/z region of the mass spectrum (>3000 u) becomes populated with abundant ionic signal. Although the large number of peaks present in the mass spectrum make the direct mass calculations challenging, grouping of multiple peaks into charge ladders corresponding to a single species can be readily made using limited charge reduction of the ionic species in the gas phase (22). Several examples of using the limited charge reduction for grouping ionic peaks into charge ladders and calculating masses of the corresponding species are shown in the inset in Fig. 3 (see Supporting Material for the entire data set). The majority of the ionic signals in the m/z range above 1700 u represent PF4 tetramers associated with pS polyanions in stoichiometric ratios ranging from one to six (the highest-abundance ions correspond to four and five polyanionic ligands bound to the protein tetramer, M4·pS4 and M4·pS5). The ligand load corresponding to the highest-abundance protein-polyanion complexes dramatically exceeds binding stoichiometry deduced from the x-ray crystallographic studies (in which only two pentasaccharides per PF4 tetramer could be visualized (13)). This discrepancy should not be surprising because the protein-glycosaminoglycan associations are dominated by ionic interactions, which frequently allow several binding modes (36,37). The resulting structural heterogeneity is known to present a significant challenge vis-à-vis crystallization of such complexes and/or producing interpretable electron density maps (36). On the other hand, native MS measurements are insensitive to the ligand localization on the protein surface as long as the assembly remains stable in the gas phase. Because neither ionic interactions nor hydrogen bonds within the protein-ligand complex are compromised in a solvent-free environment in which MS measurements are carried out (38), native MS provides reliable information on the stoichiometry of protein-glycosaminoglycan associations.
Figure 3.
A mass spectrum of an aqueous solution of a PF4-pS mixture (physiological pH and ionic strength). The inset shows examples of ionic peak grouping into charge ladders using limited charge reduction (only two examples are shown to avoid clutter: M4 × pS5, blue and M4 × pS4, green; the black trace in the inset shows a corresponding m/z region of the reference mass spectrum of the PF4/pS mixture). To see this figure in color, go online.
Another truly remarkable feature that becomes apparent upon close examination of the ionic signal corresponding to all PF4 tetramer-pS complexes is the bimodal charge state distribution of the ionic species with a low ligand load (M4·pS and M4·pS2). At the same time, ions representing complexes with higher ligand load (M4·pS3 through M4·pS6) exhibit narrow charge state distributions (+9 through +11) with lower values of the average charge, consistent with the notion of compact conformations. The bimodal charge state distributions of the M4·pS and M4·pS2 ions signal the existence of partially unfolded complexes (represented by charge states +12 through +15 alongside the compact species (+9 through +11 charge states)). This unique feature of the M4·pS and M4·pS2 ions indicates that the low number of short polyanionic ligands can promote association of the protein to the tetrameric form where at least some of the polypeptide chains fail to maintain native conformation. Furthermore, lower m/z range of the mass spectrum (<2000 u) reveals the presence of ionic signal representing PF4 monomers, including M·pS+5, in addition to the ligand-free monomer M+4 (Fig. 3). Thus, the entire complement of ionic species populating the mass spectrum of the PF4-pentasaccharide mixture is indicative of the pathway of the polyanion-assisted assembly of PF4 tetramers: the initial binding of the anionic ligand to the PF4 monomer (represented by the M·pS+5 ions) reduces the electrostatic repulsion between the polycationic polypeptides, allowing them to associate to tetramers even at physiological ionic strength (i.e., 150 mM). However, the higher-order structure of the resulting complexes in solution (represented in the mass spectrum shown in Fig. 3 by the higher-charge density M4·pSz+ and M4·pS2z+ ions, z = 12–15) is still significantly influenced by strong intramolecular electrostatic repulsion, preventing the polypeptide ions from maintaining highly compact (native) fold within such complexes. These repulsive forces are further mitigated by association with additional polyanionic ligands, giving rise to compact complexes in which all constituent polypeptide chains are natively folded (represented by M4·pSnz+ ions, where n = 3–6 and z = 9–11).
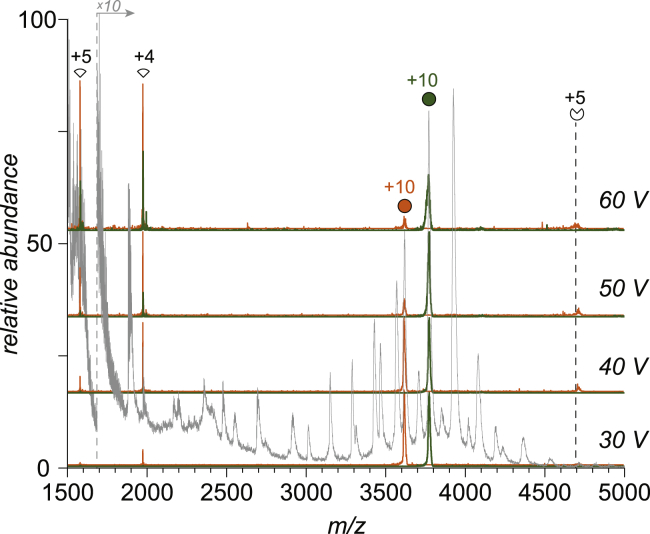
Native MS provides a unique way of evaluating the stabilizing role of polyanions beyond mitigation of electrostatic repulsion within the polycationic PF4 tetramer. Mass selection of PF4-pS complexes followed by their mild collisional activation in the gas phase reveals a strong dependence of the threshold collision energy (corresponding to the onset of complex dissociation) on the number of pS ligands within a set of ions having an identical total charge. This is illustrated in Fig. 4 using M4·pS310+ and M4·pS410+ ions as two representative examples (see Supporting Material for the entire data set). Dissociation of both ions gives rise to identical fragments (high-charge-density monomers and low-charge-density trimers, all of which appear to be ligand free). This suggests that the dissociation of the complex ions in the gas phase proceeds via shedding of the ligands, immediately followed by dissociation of the PF4 tetramer via the classical asymmetric-charge-partitioning route (ejection of highly charged monomer). It appears that the two steps are coupled because no signal was detected for a putative ionic species generated by either complete or partial loss of pS ligand(s) from the complex without compromising the tetrameric structure of the protein. Importantly, dissociation of the complex with the higher ligand load is significantly less effective (requiring both a higher threshold collision energy and exhibiting a higher fraction of surviving precursor ions at any collision energy tested in our work). Because both precursor ions were selected such that they have identical total charge (z = +10), the markedly increased stability of the ionic species representing complexes with higher ligand load indicates that the stabilizing action of pS extends beyond mere charge neutralization.
Figure 4.
Mass spectra of fragment ions generated upon collision-induced dissociation of M4 × pS310+ (orange) and M4 × pS410+ (olive) ions at different collisional energies as indicated on the graph. The gray trace shows a reference mass spectrum (MS1) of the PF4-pS mixture, from which the two precursor ions were selected. To see this figure in color, go online.
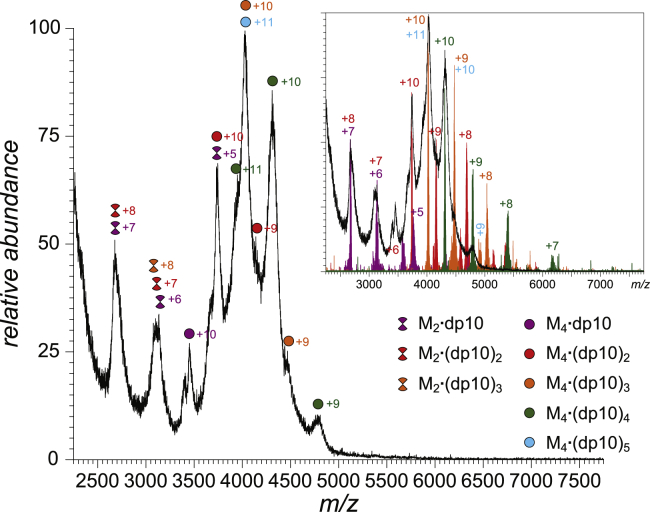
The complexity of the PF4-pS mass spectrum (Fig. 3) arises because of the presence of protein-polyanion complexes with different stoichiometries; however, most individual peaks appear to be well resolved. Replacing the structurally homogeneous synthetic pentasaccharide with a heparin decamer, produced by partial depolymerization of heparin (dp10), gives rise to a significantly more convoluted mass spectrum (Fig. 5) despite using identical sample preparation routines and MS measurement methods. None of the ionic peaks in this mass spectrum are resolved; the significant overlap of ionic signals is not surprising given the broad mass distribution of dp10 species covering a mass range spanning from 2200 to 3100 Da, with the most abundant ionic species populating the 2700–3000 Da range (see Fig. S3). Although the extreme heterogeneity of the dp10 species (caused primarily by variation in the levels of sulfation) does not allow any meaningful information to be extracted from the mass spectrum of the PF4-dp10 mixture with nearly equimolar polypeptide and polyanion concentrations, application of the limited charge reduction in the gas phase allows all ionic signals to be assigned (several examples are presented in the inset in Fig. 5). As was the case with PF4-pS interaction, the most abundant ionic signal in the PF4/dp10 mass spectrum represents PF4 tetramers complexed to multiple polyanion ligands (M4·dp10nz+ ions, where n = 2–4 and z = 9–10). The masses of these complexes are consistent with the notion of highly sulfated oligoheparin molecules bound to PF4 tetramers (the average sulfation level for such species is estimated to be 14–16 per decasaccharide chain, whereas the entire pool of dp10 species covers a significantly wider range, from six to 17 sulfate groups per decamer; see Fig. S3).
Figure 5.
A mass spectrum of an aqueous solution of a PF4-dp10 mixture (physiological pH and ionic strength). The inset shows examples of ionic peak grouping into charge ladders using limited charge reduction (the black trace in the inset shows a relevant m/z region of the reference mass spectrum of the PF4/dp10 mixture). To see this figure in color, go online.
The range of the ligand load exhibited by the PF4-dp10 complexes is somewhat diminished compared to that of the PF4-pS species (in which binding of up to six ligands to a single protein tetramer was observed, see Fig. 3). Interestingly, limited charge reduction measurements reveal a presence of a minor species M4·dp10511+ (its signal overlaps in the conventional mass spectrum with that of the abundant M4·dp10310+ ion; see inset in Fig. 5). The average sulfation level calculated for the dp10 species within the M4·dp105 complex is relatively modest, corresponding to only 10–12 sulfate groups per decasaccharide, indicating that ligand overcrowding is regulated by electrostatic repulsion. Binding preferences of proteins toward heparin oligomers with varying levels of sulfation are known to change depending on the binding stoichiometry (39), and in fact, oversulfation of synthetic heparin oligomers was shown to be detrimental vis-à-vis their affinity toward PF4 (17).
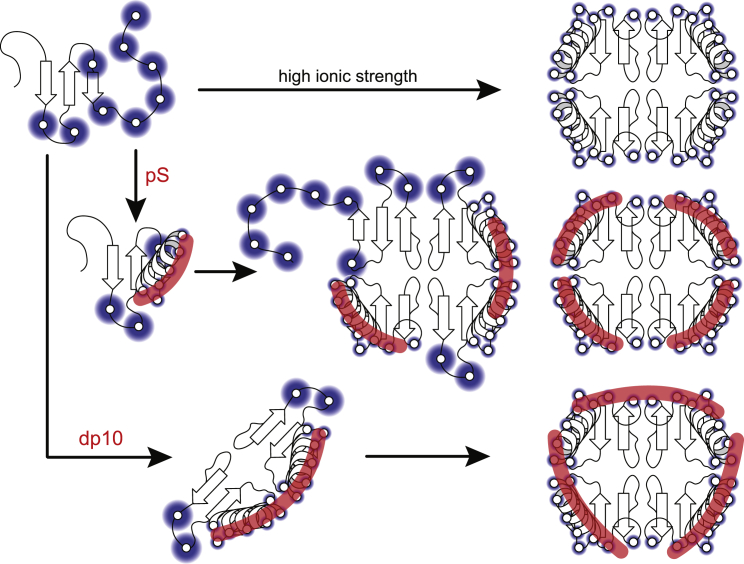
Another peculiar feature of the PF4-dp10 mass spectrum that was not observed for the PF4-pS system is the presence of PF4 dimers having one or two polyanionic ligands associated with them (M2·dp10kz+, k = 1–2, z = 6–8; see m/z range 2500–3500 in Fig. 5). The ability of heparin decamers to stabilize dimeric PF4 species is consistent with the earlier models of PF4-heparin interaction (9,10), in which the heparin chain “wraps around” the PF4 tetramer after the positive belt on its surface (somewhat reminiscent of the DNA wrapping around histones within nucleosomes). Because the decasaccharide length is not sufficient for encircling the entire tetramer, a single dp10 molecule associated with PF4 may not be able to stabilize the quaternary structure beyond the dimer level; stabilization of the entire tetramer requires involvement of a larger number of decameric polyanions (giving rise to M4 ⋅ dp10m, m = 2–4), as shown schematically in Fig. 6.
Figure 6.
Schematic representation of the influence of electrostatic interactions on the assembly of PF4 tetramers. At the top, high charge density within the lysine-rich C-terminus of the protein (as well as polycationic loops connecting β-strands) prevents the polypeptide chains from not only assembling to tetrameric structures but indeed maintaining stable ternary fold unless the ionic strength of the solvent is elevated above the physiological level to provide sufficient screening of the charges. In the middle, association of short polyanions (pS) with PF4 monomers at physiological ionic strength results in partial screening of positive charges, allowing the assembly process to proceed forward. The excessive (uncompensated) positive charge within the nascent tetramers prevents some of the polypeptides within the tetramer from acquiring a compact or native fold until additional polyanionic species are bound to the tetramer. At the bottom, association of longer polyanions (dp10) with PF4 monomers leads to formation of transient PF4 dimers, which proceed to form tetrameric species. To see this figure in color, go online.
Although the wrapping mode of oligoheparin interactions with PF4 supported by the MS data is in line with earlier models of PF4-heparin association (9,10), it contradicts the recently proposed model based on the crystal structure of a PF4-pS complex (13). The latter assumes that the initial heparin association with a PF4 tetramer results in “linearization” of the local structure of the GAG chain, maximizing its protein binding capacity. Recently, Whitelock et al. suggested that the linear configuration of GAG chain associated with a PF4 tetramer is likely to occur within the context of PF4-proteoglycan (serglycin or perlecan) complexes in which the close proximity of the GAG chains negatively affects their flexibility (24). In contrast, heparin chains are more flexible, which may allow them to maximize electrostatic contacts within the basic side chains on the PF4 surface by wrapping around the tetramer (Fig. 6), although the presence of a large number of heparin molecules within the so-called ultralarge complexes (15) may diminish this flexibility to minimize the unfavorable electrostatic interactions.
Another important distinction of the results of our work and the conclusions of the crystallographic study (13) relates to bivalency of short heparin oligomers vis-à-vis PF4 binding. The appearance of the PF4-pS crystal structure suggested that oligosaccharides as short as pS can bridge two PF4 tetramers (13), a conclusion that clearly contradicts the findings of this work. Indeed, native MS provides no evidence for the existence of two or more PF4 tetramers bridged by either pS or indeed dp10 chains (which are twice as long as pS and exhibit a high level of structural diversity because of the natural variation of the sulfation patterns). Should this bridging occur in solution, it would manifest itself in mass spectra via appearance of protein-oligoheparin complexes containing multiple PF4 tetramers (similar to heparin oligomer-bridged chemokines (39,40) or antithrombin (41)), none of which have been detected in this work. Likewise, no PF4 bridging by pS had been observed when AFM was used to visualize PF4 complexes with heparin preparations of various sizes (16). We also note that earlier examinations of the composition of PF4/heparin precipitate provided an estimate of the polysaccharide weight fraction as ∼16%, consistent with the notion of eight to 10 heparin disaccharide units per protein tetramer (42). This heparin/protein ratio corresponds to M4·pS4 and M4·(dp10)2 complexes, whose signals are very prominent in the mass spectra of PF4/pS and PF4/dp10 mixtures (Figs. 3 and 5). Although it might be tempting to propose that accumulation of such species could trigger formation of ultralarge complexes (following the implications made in (13)), it remains to be seen whether the small complexes studied in this work can be indeed converted to immunogenic ultralarge complexes, similar to the complexes formed by PF4 association with intact unfractionated heparin.
Conclusions
Because of their importance in medicine, PF4-heparin interactions remain a focal point of extensive research work. Most studies aim at identifying the hallmarks of PF4-heparin immunogenicity, which cannot be accomplished without the characterization of all relevant species at the molecular level. The latter task remains challenging because of the enormous structural diversity displayed by heparin and its complexes with PF4. An impressive arsenal of biophysical techniques that have been used to study PF4-heparin interactions provide important insights on various aspects of structure and behavior of PF4-heparin complexes (14), although no single technique has proved capable of exhaustive structural characterization of these elusive species. In recent years, MS has been gradually expanding its foothold in the field of GAG biology (43,44), including rapidly growing applications of MS to study protein binding to heparin and related polyanions (41,45,46). Application of native MS to characterization of PF4 interactions with relatively short (up to decasaccharide level) heparin oligomers described in this work provides a level of detail that was previously unattainable. Native MS provides strong evidence that even acquisition of the native quaternary structure by PF4 in solution under physiological conditions is impossible without assistance by polyanionic GAGs. Although this fact is frequently overlooked in modern literature, the critical importance of endogenous GAGs for PF4 assembly and storage has been previously demonstrated in animal models in which the lack of serglycin (the dominant platelet glycosaminoglycan) results in inability to store a range of α-granule-associated proteins, including PF4 (47). The dramatic facilitation of PF4 tetramer assembly, even by a relatively short polyanion (pS), highlights the importance of charge neutralization. Native MS reveals a number of pS-stabilized PF4 tetramers displaying a range of ligands accommodated on the protein surface. The charge-state-distribution analysis of ionic species representing these complexes indicates that only relatively high ligand load (at least three pS molecules per PF4 tetramer) guarantees structural integrity; a lower number of pS molecules fail to lock the entire protein in its native conformation. Longer and structurally heterogeneous heparin oligomers (decamers) are also very effective in stabilizing the tetrameric structure of PF4. When bound to the protein in small numbers, these heparin oligomers also give rise to PF4 dimers, lending support to the wrapping model of heparin-protein interactions. Although all PF4-heparin oligomer complexes characterized in this work are small complexes (no bridging of PF4 tetramers was observed), native MS will undoubtedly play an important role in elucidating the structure of significantly larger PF4-heparin systems, particularly those involved in etiology of HIT (48).
Author Contributions
C.N. planned and carried out the experimental work, interpreted the experimental data, and coedited the manuscript. Y.Y. carried out the experimental work, interpreted the experimental data, and coedited the manuscript. A.H. prepared the critical material for the experimental work (recombinant PF4). I.N. participated in planning the experimental work and coedited the manuscript. I.A.K. designed the study, planned the experimental work, interpreted the experimental data, and wrote the manuscript.
Acknowledgments
The authors are grateful to Dr. Cedric E. Bobst (University of Massachusetts-Amherst) for help with mass analyses of PF4 polypeptides.
This work was supported by a grant from the National Institutes of Health R01 GM112666. All measurements were carried out in the Mass Spectrometry Core Facility at University of Massachusetts-Amherst.
Editor: Alexandr Kornev.
Footnotes
Supporting Material can be found online at https://doi.org/10.1016/j.bpj.2020.04.012.
Supporting Material
References
- 1.Harrington W.J., Minnich V., Moore C.V. Demonstration of a thrombocytopenic factor in the blood of patients with thrombocytopenic purpura. J. Lab. Clin. Med. 1951;38:1–10. [PubMed] [Google Scholar]
- 2.Kowalska M.A., Rauova L., Poncz M. Role of the platelet chemokine platelet factor 4 (PF4) in hemostasis and thrombosis. Thromb. Res. 2010;125:292–296. doi: 10.1016/j.thromres.2009.11.023. [DOI] [PubMed] [Google Scholar]
- 3.Kasper B., Petersen F. Molecular pathways of platelet factor 4/CXCL4 signaling. Eur. J. Cell Biol. 2011;90:521–526. doi: 10.1016/j.ejcb.2010.12.002. [DOI] [PubMed] [Google Scholar]
- 4.Vandercappellen J., Van Damme J., Struyf S. The role of the CXC chemokines platelet factor-4 (CXCL4/PF-4) and its variant (CXCL4L1/PF-4var) in inflammation, angiogenesis and cancer. Cytokine Growth Factor Rev. 2011;22:1–18. doi: 10.1016/j.cytogfr.2010.10.011. [DOI] [PubMed] [Google Scholar]
- 5.Prechel M.M., Walenga J.M. Emphasis on the role of PF4 in the incidence, pathophysiology and treatment of heparin induced thrombocytopenia. Thromb. J. 2013;11:7. doi: 10.1186/1477-9560-11-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arepally G.M. Heparin-induced thrombocytopenia. Blood. 2017;129:2864–2872. doi: 10.1182/blood-2016-11-709873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang X., Chen L., Maione T.E. Crystal structure of recombinant human platelet factor 4. Biochemistry. 1994;33:8361–8366. doi: 10.1021/bi00193a025. [DOI] [PubMed] [Google Scholar]
- 8.St Charles R., Walz D.A., Edwards B.F. The three-dimensional structure of bovine platelet factor 4 at 3.0-A resolution. J. Biol. Chem. 1989;264:2092–2099. [PubMed] [Google Scholar]
- 9.Cowan S.W., Bakshi E.N., Isaacs N.W. Binding of heparin to human platelet factor 4. Biochem. J. 1986;234:485–488. doi: 10.1042/bj2340485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stuckey J.A., St Charles R., Edwards B.F. A model of the platelet factor 4 complex with heparin. Proteins. 1992;14:277–287. doi: 10.1002/prot.340140213. [DOI] [PubMed] [Google Scholar]
- 11.Mayo K.H., Ilyina E., Daly T.J. Heparin binding to platelet factor-4. An NMR and site-directed mutagenesis study: arginine residues are crucial for binding. Biochem. J. 1995;312:357–365. doi: 10.1042/bj3120357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mikhailov D., Young H.C., Mayo K.H. Heparin dodecasaccharide binding to platelet factor-4 and growth-related protein-alpha. Induction of a partially folded state and implications for heparin-induced thrombocytopenia. J. Biol. Chem. 1999;274:25317–25329. doi: 10.1074/jbc.274.36.25317. [DOI] [PubMed] [Google Scholar]
- 13.Cai Z., Yarovoi S.V., Greene M.I. Atomic description of the immune complex involved in heparin-induced thrombocytopenia. Nat. Commun. 2015;6:8277. doi: 10.1038/ncomms9277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Delcea M., Greinacher A. Biophysical tools to assess the interaction of PF4 with polyanions. Thromb. Haemost. 2016;116:783–791. doi: 10.1160/TH16-04-0258. [DOI] [PubMed] [Google Scholar]
- 15.Rauova L., Poncz M., Sachais B.S. Ultralarge complexes of PF4 and heparin are central to the pathogenesis of heparin-induced thrombocytopenia. Blood. 2005;105:131–138. doi: 10.1182/blood-2004-04-1544. [DOI] [PubMed] [Google Scholar]
- 16.Greinacher A., Gopinadhan M., Helm C.A. Close approximation of two platelet factor 4 tetramers by charge neutralization forms the antigens recognized by HIT antibodies. Arterioscler. Thromb. Vasc. Biol. 2006;26:2386–2393. doi: 10.1161/01.ATV.0000238350.89477.88. [DOI] [PubMed] [Google Scholar]
- 17.Nguyen T.H., Xu Y., Greinacher A. Characterization of the interaction between platelet factor 4 and homogeneous synthetic low molecular weight heparins. J Thromb Haemost. 2020;18:390–398. doi: 10.1111/jth.14657. Published online October 20, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tong W., Wang G. How can native mass spectrometry contribute to characterization of biomacromolecular higher-order structure and interactions? Methods. 2018;144:3–13. doi: 10.1016/j.ymeth.2018.04.025. [DOI] [PubMed] [Google Scholar]
- 19.Abzalimov R.R., Dubin P.L., Kaltashov I.A. Glycosaminoglycans as naturally occurring combinatorial libraries: developing a mass spectrometry-based strategy for characterization of anti-thrombin interaction with low molecular weight heparin and heparin oligomers. Anal. Chem. 2007;79:6055–6063. doi: 10.1021/ac0710432. [DOI] [PubMed] [Google Scholar]
- 20.Yu Y., Sweeney M.D., Leary J.A. Chemokine-glycosaminoglycan binding: specificity for CCR2 ligand binding to highly sulfated oligosaccharides using FTICR mass spectrometry. J. Biol. Chem. 2005;280:32200–32208. doi: 10.1074/jbc.M505738200. [DOI] [PubMed] [Google Scholar]
- 21.Kaltashov I.A., Pawlowski J.W., Lipatnikov A.N. LC/MS at the whole protein level: studies of biomolecular structure and interactions using native LC/MS and cross-path reactive chromatography (XP-RC) MS. Methods. 2018;144:14–26. doi: 10.1016/j.ymeth.2018.04.019. [DOI] [PubMed] [Google Scholar]
- 22.Abzalimov R.R., Kaltashov I.A. Electrospray ionization mass spectrometry of highly heterogeneous protein systems: protein ion charge state assignment via incomplete charge reduction. Anal. Chem. 2010;82:7523–7526. doi: 10.1021/ac101848z. [DOI] [PubMed] [Google Scholar]
- 23.Barber A.J., Käser-Glanzmann R., Lüscher E.F. Characterization of a chondroitin 4 -sulfate proteoglycan carrier for heparin neutralizing activity (platelet factor 4 ) released from human blood platelets. Biochim. Biophys. Acta. 1972;286:312–329. [PubMed] [Google Scholar]
- 24.Lord M.S., Cheng B., Whitelock J.M. Platelet factor 4 binds to vascular proteoglycans and controls both growth factor Activities and platelet activation. J. Biol. Chem. 2017;292:4054–4063. doi: 10.1074/jbc.M116.760660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huynh A., Arnold D.M., Nazy I. Development of a high-yield expression and purification system for platelet factor 4. Platelets. 2018;29:249–256. doi: 10.1080/09537104.2017.1378808. [DOI] [PubMed] [Google Scholar]
- 26.Bertini S., Fareed J., Naggi A. Characterization of PF4-heparin complexes by photon correlation spectroscopy and zeta potential. Clin. Appl. Thromb. Hemost. 2017;23:725–734. doi: 10.1177/1076029616685430. [DOI] [PubMed] [Google Scholar]
- 27.Moore S., Pepper D.S., Cash J.D. Platelet antiheparin activity. The isolation and characterisation of platelet factor 4 released from thrombin-aggregated washed human platelets and its dissociation into subunits and the isolation of membrane-bound antiheparin activity. Biochim. Biophys. Acta. 1975;379:370–384. [PubMed] [Google Scholar]
- 28.Lamm G., Pack G.R. Counterion condensation and shape within Poisson–Boltzmann theory. Biopolymers. 2010;93:619–639. doi: 10.1002/bip.21421. [DOI] [PubMed] [Google Scholar]
- 29.Kaltashov I.A., Abzalimov R.R. Do ionic charges in ESI MS provide useful information on macromolecular structure? J. Am. Soc. Mass Spectrom. 2008;19:1239–1246. doi: 10.1016/j.jasms.2008.05.018. [DOI] [PubMed] [Google Scholar]
- 30.Jurchen J.C., Williams E.R. Origin of asymmetric charge partitioning in the dissociation of gas-phase protein homodimers. J. Am. Chem. Soc. 2003;125:2817–2826. doi: 10.1021/ja0211508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Abzalimov R.R., Frimpong A.K., Kaltashov I.A. Gas-phase processes and measurements of macromolecular properties in solution: on the possibility of false positive and false negative signals of protein unfolding. Int. J. Mass Spectrom. 2006;253:207–216. [Google Scholar]
- 32.Sciuto S.V., Liu J., Konermann L. An electrostatic charge partitioning model for the dissociation of protein complexes in the gas phase. J. Am. Soc. Mass Spectrom. 2011;22:1679–1689. doi: 10.1007/s13361-011-0205-x. [DOI] [PubMed] [Google Scholar]
- 33.Kaltashov I.A., Mohimen A. Estimates of protein surface areas in solution by electrospray ionization mass spectrometry. Anal. Chem. 2005;77:5370–5379. doi: 10.1021/ac050511+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gumerov D.R., Dobo A., Kaltashov I.A. Protein-ion charge-state distributions in electrospray ionization mass spectrometry: distinguishing conformational contributions from masking effects. Eur. J. Mass Spectrom. (Chichester, Eng.) 2002;8:123–129. [Google Scholar]
- 35.Frimpong A.K., Abzalimov R.R., Kaltashov I.A. Gas-phase interference-free analysis of protein ion charge-state distributions: detection of small-scale conformational transitions accompanying pepsin inactivation. Anal. Chem. 2007;79:4154–4161. doi: 10.1021/ac0704098. [DOI] [PubMed] [Google Scholar]
- 36.Imberty A., Lortat-Jacob H., Pérez S. Structural view of glycosaminoglycan-protein interactions. Carbohydr. Res. 2007;342:430–439. doi: 10.1016/j.carres.2006.12.019. [DOI] [PubMed] [Google Scholar]
- 37.Seyrek E., Dubin P. Glycosaminoglycans as polyelectrolytes. Adv. Colloid Interface Sci. 2010;158:119–129. doi: 10.1016/j.cis.2010.03.001. [DOI] [PubMed] [Google Scholar]
- 38.Kaltashov I.A., Bobst C.E., Abzalimov R.R. Mass spectrometry-based methods to study protein architecture and dynamics. Protein Sci. 2013;22:530–544. doi: 10.1002/pro.2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Minsky B.B., Dubin P.L., Kaltashov I.A. Electrostatic forces as dominant interactions between proteins and polyanions: an ESI MS study of fibroblast growth factor binding to heparin oligomers. J. Am. Soc. Mass Spectrom. 2017;28:758–767. doi: 10.1007/s13361-017-1596-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao Y., Kaltashov I.A. Evaluation of top-down mass spectrometry and ion-mobility spectroscopy as a means of mapping protein-binding motifs within heparin chains. Analyst. 2020;145:3090–3099. doi: 10.1039/d0an00097c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhao Y., Abzalimov R.R., Kaltashov I.A. Interactions of intact unfractionated heparin with its client proteins can be probed directly using native electrospray ionization mass spectrometry. Anal. Chem. 2016;88:1711–1718. doi: 10.1021/acs.analchem.5b03792. [DOI] [PubMed] [Google Scholar]
- 42.Bock P.E., Luscombe M., Holbrook J.J. The multiple complexes formed by the interaction of platelet factor 4 with heparin. Biochem. J. 1980;191:769–776. doi: 10.1042/bj1910769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Staples G.O., Zaia J. Analysis of glycosaminoglycans using mass spectrometry. Curr. Proteomics. 2011;8:325–336. doi: 10.2174/157016411798220871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zaia J. Glycosaminoglycan glycomics using mass spectrometry. Mol. Cell. Proteomics. 2013;12:885–892. doi: 10.1074/mcp.R112.026294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zong C., Huang R., Boons G.-J. Integrated approach to identify heparan sulfate ligand requirements of Robo1. J. Am. Chem. Soc. 2016;138:13059–13067. doi: 10.1021/jacs.6b08161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li Z., Moniz H., Sharp J.S. High structural resolution hydroxyl radical protein footprinting reveals an extended Robo1-heparin binding interface. J. Biol. Chem. 2015;290:10729–10740. doi: 10.1074/jbc.M115.648410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Woulfe D.S., Lilliendahl J.K., Schick B.P. Serglycin proteoglycan deletion induces defects in platelet aggregation and thrombus formation in mice. Blood. 2008;111:3458–3467. doi: 10.1182/blood-2007-07-104703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Suvarna S., Espinasse B., Arepally G.M. Determinants of PF4/heparin immunogenicity. Blood. 2007;110:4253–4260. doi: 10.1182/blood-2007-08-105098. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.