Abstract
The multitude of varied, energy-dependent processes that exist in the cell necessitate a diverse array of macromolecular machines to maintain homeostasis, allow for growth, and facilitate reproduction. ATPases associated with various cellular activity are a set of protein assemblies that function as molecular motors to couple the energy of nucleoside triphosphate binding and hydrolysis to mechanical movement along a polymer lattice. A recent boom in structural insights into these motors has led to structural hypotheses on how these motors fulfill their function. However, in many cases, we lack direct kinetic measurements of the dynamic processes these motors undergo as they transition between observed structural states. Consequently, there is a need for improved techniques for testing the structural hypotheses in solution. Here, we apply transient-state fluorescence anisotropy and total fluorescence stopped-flow methods to the analysis of polypeptide translocation catalyzed by these ATPase motors. We specifically focus on the Hsp100-Clp protein system of ClpA, which is a well-studied, model ATPases associated with various cellular activity system that has both eukaryotic and archaea homologs. Using this system, we show that we can reproduce previously established kinetic parameters from the simultaneous analysis of fluorescence anisotropy and total fluorescence and overcome previous limitations of our previous approach. Specifically, for the first time, to our knowledge, we obtain quantitative interpretations of the translocation of polypeptide substrates longer than 100 aa.
Significance
Stopped-flow anisotropy time courses are uniquely difficult to quantitatively model because of their nonexponential behavior. Here, we have incorporated the n-step sequential mechanism to the method developed by Otto et al. to examine stopped-flow anisotropy time courses that report on polypeptide translocation catalyzed by a model of ATPases associated with various cellular activity translocase. This strategy uses the total fluorescence time course to account for the nonexponential nature of anisotropy time courses, allowing for the determination of kinetic parameters for translocation. The method overcomes previous limitations of using raw fluorescence alone and reveals further insights into enzyme-catalyzed polypeptide translocation. Moreover, the method will be broadly applicable to protein translocases that do not covalently modify the substrate they translocate.
Introduction
ATPases associated with various cellular activity (AAA+) is a large superfamily of protein translocases. Some of these translocases function by associating with a protease partner and act as both a motor and regulatory component. Examples include the eukaryotic 19S cap of the 26S proteasome (1), members of the bacterial Clp-Hsp100 (2) and HslU (3) families, and PAN-20S in archaea (4,5). The investigation of the mechanisms of translocation catalyzed by such systems has been accomplished through steady-state degradation of model substrates (6), like green fluorescent protein, and Förster resonance energy transfer using dyes located on the substrate and inside the protease (7,8). However, other translocases such as N-ethylmaleimide sensitive factor (9), katanin or spastin (10,11), VPS4 (12), ClpB-Hsp104 (13,14), and p97 (15) perform their functions without a proteolytic partner. In these examples, there are no covalent modifications of the substrates they translocate. This makes monitoring translocation catalyzed by these motors more challenging. Consequently, in many cases, it remains unclear if these motors fully translocate substrate through their axial channels or only partially translocate (16,17).
Recent advances in cryogenic electron microscopy (cryo-EM) techniques have led to a wealth of important new structural insights on AAA+ polypeptide translocases (18, 19, 20, 21, 22, 23, 24). With the new structures have come many structural hypotheses on the mechanism of polypeptide translocation catalyzed by AAA+ molecular motors. However, static structures are unable to reveal dynamic parameters such as step size (average distance traveled per turnover), coupling efficiency (ATP molecules hydrolyzed per step), elementary rate constants, and processivity. Despite this, the spacing of the polypeptide substrate’s contacts along the axial channel of these motors has been taken to indicate step size. Consequently, there is an urgent need for additional techniques that are able to test these structural hypotheses in solution.
We have previously developed a transient-state kinetics technique to investigate motors that do not associate with, or have been separated from, their proteolytic partners (25), specifically with the goal of determining elementary rate constants, step size, and processivity. This technique employed fluorescently modified synthetic polypeptide substrates (17,25,26). When these fluorescent substrates are bound to the motor proteins, there is a marked decrease in the observed fluorescence intensity. Upon translocation, there is a transition from a population of bound to unbound substrate as the enzyme and substrate dissociate. This results in an observed increase in fluorescence as a function of time. The difference between the bound and the unbound substrate fluorescence can arise from differences in the quantum yield of the fluorophore on each species, rotational artifacts of each species on the timescale of fluorescence (27), or a combination of the two. When using these techniques, we assume an all-or-none scenario in which the substrate is either in a bound or unbound fluorescent state.
The investigation of translocation catalyzed by ClpA on unstructured polypeptide substrates indicated that ClpA is translocating ∼14 aa between two rate-limiting steps (25). Given that the substrates used were 30–50 aa in length, only two to four steps were taken per polypeptide translocated. Although we have been able to determine a set of kinetic parameters describing the mechanism of translocation, some properties of translocation remain to be resolved. One important parameter is processivity, or the likelihood of the translocase to continue translocating versus dissociating from its lattice. ClpA has been reported as a highly processive motor that is able to translocate entire target substrates into its proteolytic partner ClpP for degradation (28, 29, 30). However, a quantitative estimate of processivity has not been reported. We were only able to report a lower limit of processivity at limiting ATP of p =∼0.88 (25). Longer polypeptide substrates will be required to quantitatively determine processivity at saturating ATP concentrations in which translocation is fast. However, longer substrates in our previously developed stopped-flow strategy did not yield interpretable signal changes (unpublished data).
In principle, anisotropy-based, stopped-flow techniques would be better suited for an examination of polypeptide translocation because it is sensitive to the presence of the motor on the lattice. That is to say, the polypeptide substrate being translocated is much smaller than hexameric ClpA (∼500,000 Da). Thus, a polypeptide substrate bound by ClpA should exhibit a larger anisotropy than substrate alone, and the anisotropy should decrease as ClpA dissociates from the peptide after complete translocation. In fact, we and others have reported stopped-flow anisotropy time courses that exhibit exactly that behavior for ClpA (7,25), ClpB (16), and Hsp104 (17). However, only a qualitative description of those time courses was reported. This is a consequence of anisotropy not following typical exponential behavior, and therefore, it cannot be described by sums of exponentials. Thus, the analysis is uniquely complex because anisotropy time courses are described by a ratio of sums of exponentials. Moreover, the ratios are scaled by both changes in anisotropy and quantum yield.
Here, we define a fluorescence anisotropy method for observing and modeling translocation of polypeptide substrates by protein translocases that do not covalently modify the polypeptide on which they translocate. These methods can account for unique intermediate species as well as distinguish the contribution of changes in quantum yield and rotational mobility to the overall signal. In this setup, translocation is monitored concurrently in two ways. First, it is monitored as total fluorescence, which is collected such that rotational artifacts are eliminated from the signal (31). As such, total fluorescence is defined by only changes in quantum yield. Second, translocation is monitored as fluorescence anisotropy that reports on the rotational mobility of the complex, which in turn, depends on the size of complex and is thereby sensitive to the presence of the motor on the lattice (31). Here, we report the simultaneous analysis of both total fluorescence and fluorescence anisotropy time courses using the techniques described by Otto et al. and our previous application of n-step sequential mechanisms (25,27,32,33). Although the simultaneous analysis of total fluorescence and anisotropy time courses has been done by others (27,34), to our knowledge, this is the first presentation of this approach to polypeptide translocation. With these techniques, we have been successful in both reproducing kinetic parameters from the analysis of the original polypeptides (25) and, to our knowledge, for the first time, gaining quantitative interpretations of the translocation of polypeptide substrates longer than 100 aa using stopped-flow anisotropy. Equally, the analysis of the time courses leads to a determination of the quantum yield and anisotropy of each species, which leads to additional insights into the molecular mechanisms of enzyme-catalyzed polypeptide translocation. With this technique in hand, we will be able to survey a large number of longer substrates both with and without structure.
Materials and Methods
Solutions
All solutions were prepared in double distilled water from a Siemens Water Technology Purelab Ultra Genetic System (Alpharetta, GA) with commercially available reagent grade chemicals. All experiments were carried out in buffer H300: 25 mM HEPES (pH 7.5) at 25°C, 300 mM NaCl, 10 mM MgCl2, 2 mM 2-mercaptoethanol, and 10% v/v glycerol.
Polypeptide substrates
SsrA polypeptides were synthesized and fluorescently labeled by CPC Scientific (Sunnyvale, CA) and were certified as >90% pure by liquid-chromatography-mass-spectrometry analysis. The peptides consist of the C-terminal SsrA tag flanked by truncations of the titin I27 domain. Each peptide has an N-terminal cysteine that has been labeled with a fluorescein-5-maleimide. αS1 casein truncations were constructed as detailed in our previously published work. Each substrate consists of a C-terminal truncation of αS1 casein that is labeled with fluorescein-5-maleimide on an N-terminal cysteine. These peptides have been previously shown to bind to ClpA through quantitative anisotropy titrations and were analyzed using circular dichroism spectroscopy to show that they lack any significant structure (35).
ClpA
ClpA was purified in a manner similar to that described in Veronese et al. (36). The only significant difference was the omission of the final Blue Sepharose Fast Flow column (Cytiva, Chicago, IL). The reported concentrations of ClpA were determined in H300 buffer using the molar extinction coefficient 31,000 (M monomer)−1 cm−1, and all concentrations are in monomer units.
Polypeptide translocation experiments
Polypeptide translocation experiments were carried out on an Applied Photophysics SX20 stopped-flow spectrometer (Leatherhead, UK). First, 4 μM ClpA was incubated with 300 μM ATPγS in H300 for 30 min at 25°C. Then, 200 nM fluorescent SsrA substrate was added and incubated for 15 min to achieve binding equilibrium. Equilibrium was assured by monitoring fluorescence change in a HORIBA Jobin Yvon Fluorolog-3 spectrofluorometer (Albany, NY) over 2 h. No change was observed after 15 min. Additionally, 6 mM ATP and 20 μM α-casein were incubated in H300 for 45 min at 25°C.
Raw fluorescence (RF) (no polarizers) stopped-flow experiments were collected in an L configuration using the longer 10-mm path length. Before each set of experimental acquisitions, the stopped-flow system is prepared by adjusting the photomultiplier tube (PMT) voltages. A single acquisition is taken, and the chemistry is allowed to react. Once the steady state of the final reaction conditions is met, the PMT voltages are autoadjusted by the instrument. The stopped-flow sample chamber, flow lines, and syringes are washed with buffer matching that of reaction conditions between each experiment. Each data set gathered represents the mean and standard deviation of at least four back-to-back acquisitions from the same reaction mixture.
Fluorescence anisotropy data for translocation were collected using a T-format (31). Before each set of experimental acquisitions, the stopped flow is prepared by adjusting the PMT voltages and determining the G factor. A single acquisition is taken, and the chemistry is allowed to react. Once the steady state of the final reaction conditions is met, the PMT voltages are set to 15%, and the G factor is obtained to a 10% level of precision as described in the instrument manual. The stopped-flow sample chamber, flow lines, and syringes are washed with buffer matching that of reaction conditions between each experiment. Each data set gathered represents the mean and standard deviation of at least four back-to-back acquisitions from the same reaction mixture.
Multistart Evolutionary Nonlinear Optimizer
All data analysis performed here was carried out in the custom-build MATLAB (The MathWorks, Natick, MA) toolbox MENOTR. Multistart Evolutionary Nonlinear Optimizer (MENOTR) is a hybrid multistart genetic and nonlinear least squares (NLLS) algorithm (N.W.S. and Z.M. Ingram, unpublished data). This hybrid fitting method utilizes not only the intrinsic strengths of both genetic and NLLS algorithms but also balances the strengths of each method to offset their corresponding limitations. The genetic algorithm is able to survey large and diverse error spaces to find candidate solutions. However, because of the lack of convergence, the genetic algorithm becomes computationally expensive and underperforms in the later stages of optimization (37,38). In contrast, the NLLS algorithm converges quickly on a local minimum but cannot guarantee a global minimum because of its inability to escape local minima or survey sufficient solution space (39, 40, 41). In this hybrid method, the genetic algorithm will quickly find improved initial guesses for the slower running NLLS algorithm and simultaneously assist in the escape from local minima. Then, the NLLS algorithm can refine the guesses to determine a “best-fit” set of solutions. The collaboration between the approaches yields an overall more robust and superior method of generating high-quality solutions to fitting problems compared with either algorithm alone. More information on this algorithm will be published in (Z.M. Ingram and N.W.S., unpublished data). MENOTR can use the model equations discussed in this manuscript to find a set of optimized parameters that best describe each data set.
Results
Application of raw fluorescence stopped-flow methods to examine polypeptide translocation catalyzed by ClpA
We have previously developed and reported a single-turnover, transient-state kinetics technique to examine the mechanism of polypeptide translocation catalyzed by ClpA, ClpAP, ClpB, and Hsp104 (16,17,25,26). The experimental design of the technique is schematized in Fig. 1, A and B. Briefly, in the context of ClpA, we premix ClpA with the slowly hydrolyzable ATP analog, ATPγS, and the polypeptide substrate that contains a fluorescein attached to a cysteine residue at the N-terminus and the 11-aa SsrA sequence at the C-terminus, a known binding sequence for ClpA (see Table 1). The ATPγS is included because ClpA requires nucleotide binding to form hexamers active in polypeptide substrate binding (42). This sample, which contains hexameric ClpA bound to a fluorescently modified polypeptide substrate, is loaded into syringe 1 of the stopped-flow system. Syringe 2 is loaded with a solution containing ATP and nonfluorescent polypeptide substrate. The nonfluorescent substrate is included at large excess over the prebound ClpA-substrate complex to serve as a trap for any ClpA that is free in solution or that dissociates from the polypeptide during or after translocation, thereby maintaining single-turnover conditions.
Figure 1.
Schematic outlining single-turnover translocation experiments in a SX20 stopped-flow spectrometer (Applied Photophysics). (A) Syringe 1, shown in blue, contains the preassembled ClpA/flu-polypeptide complex. This consists of 2 μM ClpA, 300 μM ATPγS, and 200 nM flu-polypeptide. Syringe 2, shown in red, contains 6 mM ATP to fuel translocation and 20 μM α-casein to serve as a trap for unbound ClpA, maintaining single-turnover conditions. The contents of the two syringes are rapidly mixed into the sample chamber, shown in purple, where the sample is excited, and emission is observed as detailed below. Upon mixing, the final concentration of each reagent in the sample chamber is half of its original in the premixing syringes. (B) Shown is the L-format setup for raw fluorescence, N.B. There are no polarizers, so both the excitation and emission are circularly polarized, we define this as raw fluorescence. The sample is excited with light at 494 nm. Fluorescence emission of the sample is collected 90° to the incident light through a 515-nm long-pass filter with a single photomultiplier tube (PMT) detector. (C) Shown is the T-format setup for fluorescence anisotropy. In this method, the fluorescein sample is excited with vertically polarized light at 494 nm. The fluorescence emission is measured at 90° to the incident light with two PMTs set 180° to one another. One PMT is fitted with a vertical polarizer, detecting emitted light parallel to excitation, and the other a horizontal polarizer that detects light perpendicular to excitation. Each PMT detector is fitted with a 515-nm long-pass filter to observe fluorescein fluorescence. To see this figure in color, go online.
Table 1.
Fluorescent Polypeptide Substrates
| Name | Length (aa) | Sequence or Source |
|---|---|---|
| SsrA 30-mer | 30 | Flu-CTKSAANLKVKELRSKKKLAAN DENYALAA |
| SsrA 40-mer | 40 | Flu-CTGEVSFQAANTKSAANLKVKEL RSKKKLAA |
| NDENYALAA | ||
| SsrA 50-mer | 50 | Flu-CLILHNKQLGMTGEVSFQAAN TKSAANLKVK |
| ELRSKKKLAANDENYALAA | ||
| αS1 casein 102-mer | 102 | C-terminal 102 aa of αS1 casein |
| αS1 casein 127-mer | 127 | C-terminal 127 aa of αS1 casein |
The contents of the two syringes are rapidly mixed, fluorescein is excited at 494 nm, and emission from fluorescein is monitored; see Fig. 2 A for two representative time courses. In this design, there are no polarizers used, so both the excitation light and the emission light are circularly polarized. We will refer to this as raw fluorescence. As we have previously reported and show in Fig. S1, A–C, fluorescein fluorescence is quenched when ClpA is bound relative to fluorescence of the free substrate (25). This signal change is likely due to interactions between the fluorophore and ClpA. However, in raw fluorescence, this signal change can also occur because of the changes in anisotropy upon ClpA binding to the polypeptide substrate. Nevertheless, the kinetic time courses shown in Fig. 2 A exhibit low fluorescence at time zero followed by an increase in fluorescence over time, indicating that ClpA is dissociating upon mixing with ATP and protein trap. These experiments are single turnover with respect to the polypeptide substrate, which means that ClpA cannot rebind the fluorescently modified substrate after the first round of translocation. Therefore, the kinetic time courses represent a single round of substrate translocation followed by dissociation. Consistent with our previous observations, the kinetic time courses shown in Fig. 2 A exhibit a constant fluorescence signal, or lag, between 0 and ∼2 s before the increase in fluorescence. The duration of the lag is observed to increase with increasing substrate length as seen in Fig. 2 A for representative kinetic time courses collected with two different substrate lengths of 30 and 50 aa. We have interpreted this increase in the lag with increasing substrate length to indicate that ClpA is taking more kinetic steps before dissociation on a longer substrate compared with a shorter substrate. This observation is consistent with translocation from the binding site at the C-terminus to the N-terminus.
Figure 2.
Raw fluorescence time courses of ClpA-catalyzed translocation of (A) SsrA and (B) αS1 casein substrates collected as described in Fig. 1. (A) Given are the single-turnover translocation time courses for SsrA 30-mer (red) and 50-mer (blue) shown in solid traces with best fits shown in black broken traces. Best fits were generated using MENOTR parameter optimization applied to Scheme 1 as described in the Materials and Methods. Analysis was performed globally across the 30-, 40-, and 50-mer SsrA peptides (40-mer data not shown). The translocation rate constant (kT) was found to be (1.5 ± 0.2) s−1, and the kinetic step size (m) was determined to be (14 ± 1) aa step−1. (B) Single-turnover translocation time courses for αS1 casein 102-mer (blue) and 127-mer (purple) are shown in solid traces. Parameter optimization using Scheme 1 failed to produce any viable best-fit results (data not shown). To see this figure in color, go online.
We developed this method using short synthetic polypeptides substrates ranging in length between 30 and 50 aa (see Table 1). Synthetic polypeptides longer than 50 aa are both difficult to synthesize and cost prohibitive. Thus, our attempts to examine substrates longer than 50 aa were performed with fluorescently labeled truncations of the protein αS1 casein, which were expressed recombinantly. This substrate was chosen for two reasons. First, αS1 casein is a natural substrate for ClpA. Second, αS1 casein is an intrinsically disordered protein. Although we are interested in examining translocation on folded proteins, we have shown that the short synthetic polypeptides are unfolded, and we have extensively characterized translocation on these substrates (35). Thus, for consistency, we are seeking to examine translocation on substrates with minimal structure.
To test translocation on substrates longer than 50 aa, we performed single-turnover, stopped-flow experiments with 2 μM ClpA, 300 μM ATPγS, and 200 nM fluorescent αS1 casein. The αS1 casein substrates are 102 and 127 aa in length (see Table 1). Syringe 2 is loaded with 6 mM ATP and 20 μM unlabeled α-casein to serve as a trap. Fig. 2 B shows representative time courses collected with the αS1 casein substrates. The observed signal does not show the same clear trend that was observed with the shorter synthetic substrates shown in Fig. 2 A.
There are multiple potential explanations for the signal observed with αS1 casein. For example, although casein is a natural substrate for ClpA, there is no clear binding site for ClpA. Therefore, we do not know if ClpA binds the N-terminus, C-terminus, or somewhere in between. Meaning, ClpA may bind close to or far away from the fluorescein dye at the N-terminus. It is tempting to conclude that ClpA must be interacting with the fluorescein because the fluorescence is enhanced at time zero and decays away as ClpA dissociates. However, because the time courses in Fig. 2 B represent raw fluorescence that is sensitive to changes in both quantum yield and anisotropy, it cannot be concluded that ClpA is interacting with the dye. Similarly, the signal change for the short synthetic polypeptide substrates shown in Fig. 2 A is also likely influenced by both direct interactions of the fluorescein with ClpA and changes in anisotropy. Moreover, recent cryo-EM structures with a polypeptide bound in the axial channel indicate that ∼25 aa of the substrate reside in the channel (24). Thus, for the polypeptides of length 30–50 aa, the signal likely has a larger contribution from direct interactions because one would expect ∼5–25 aa to be protruding out of the axial channel. In contrast, the α-casein substrates of 102 and 127 aa would have 77 and 102 aa protruding from the axial channel if ClpA binds at either the N- or C-terminus. If ClpA binds the C-terminus, then this would result in the fluorescein being far from ClpA; it is also possible that the polypeptide chain that protrudes out of ClpA could fold back and interact with ClpA. Alternatively, if ClpA binds to the N-terminus, then ClpA would directly interact with the fluorophore. For these reasons, it is difficult to conclude what causes the signal change upon binding when using raw fluorescence.
To overcome these unknowns and better interpret the signal changes, we sought to test translocation of the longer polypeptides using fluorescence anisotropy and total fluorescence stopped-flow methods. Anisotropy is sensitive to the size of the fluorescently modified species. Thus, fluorescently modified αS1 casein bound to hexameric ClpA (∼500 kDa) will exhibit a higher anisotropy value relative to free αS1 casein (∼12 kDa). We have previously shown this to be the case in steady-state anisotropy titrations and single-turnover translocation experiments (17,35,43). Thus, we hypothesized that an anisotropy time course collected with ClpA prebound to polypeptide substrate should exhibit a high anisotropy at time zero followed by a decrease toward an anisotropy value consistent with free substrate (see Fig. S2) as ClpA dissociates. Therefore, the anisotropy time course will reflect the residence time of ClpA on the polypeptide substrate regardless of where ClpA binds relative to the position of the fluorophore. Moreover, unlike raw fluorescence, total fluorescence reflects only changes in quantum yield because of the presence of the motor on the polypeptide chain and removes any signal change due to anisotropy changes.
Application of fluorescence anisotropy and total fluorescence stopped-flow methods to examine polypeptide translocation catalyzed by ClpA
To determine if fluorescence anisotropy and total fluorescence stopped flow is a viable method for examining polypeptide translocation catalyzed by ClpA, we first sought to determine if we could acquire the same kinetic parameters for our previously reported synthetic polypeptide substrates of 30–50 aa (see Table 1). Anisotropy stopped-flow experiments were carried out as schematized in Fig. 1 A, and the signal was monitored as schematized in Fig. 1 C. Briefly, the sample is excited with vertically polarized light at 494 nm. The emission path is set up in a T-format with two PMT detectors set 90° to the incident light and 180° to one another. One detector collects vertically polarized emitted light (), whereas the other collects horizontal emissions (I⊥). Each PMT is fitted with a 515-nm long-pass filter to block any excitation light.
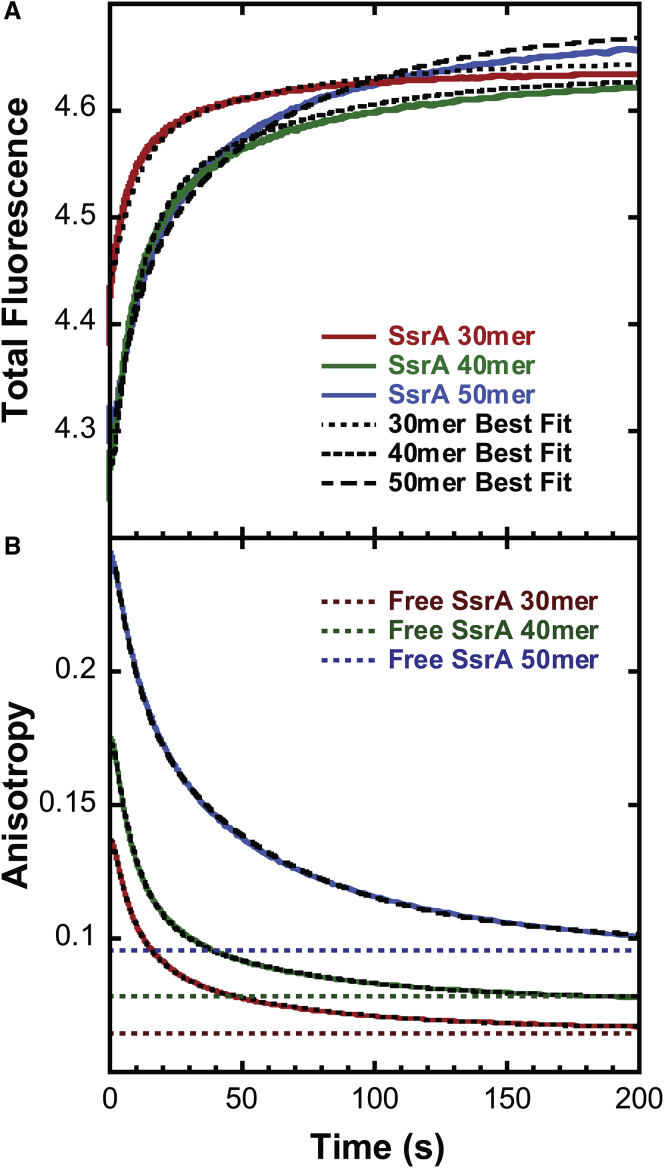
In this experiment, total fluorescence Eq. 1 and fluorescence anisotropy Eq. 2 are gathered simultaneously, meaning two time courses are collected for each acquisition. The time courses for ClpA-catalyzed translocation of SsrA 30-, 40-, and 50-mer are shown in Fig. 3, A and B for total fluorescence and anisotropy, respectively (plots of these time courses logarithmically spaced can be found in Fig. S7, A and B). In the total fluorescence time courses, we see an initially quenched fluorescence that remains constant for a short period followed by an increase in fluorescence to a final plateau. These data follow the same trends as described in the raw fluorescence time courses shown in Fig. 2 A and previously reported. As predicted, anisotropy starts at an initial high anisotropy value, and as shown in Fig. S2, the value is consistent with the anisotropy of the preassembled ClpA-peptide complex with no ATP. This observation indicates that no amplitude is lost in the deadtime of the instrument. The anisotropy is also constant for a short period followed by a decrease in signal eventually plateauing at an anisotropy value consistent with free peptide, in which the anisotropy of the free peptide is indicated for each peptide by the dashed line in Fig. 3 B. This observation indicates that all the ClpA dissociates from the polypeptide by the end of the time course.
Figure 3.
(A) Total fluorescence and (B) fluorescence anisotropy time courses of ClpA-catalyzed translocation of SsrA 30-, 40-, and 50-mer substrates (see Table 1). Solid traces show the representative time courses of 30-mer (red), 40-mer (green), and 50-mer (blue) that were collected at 3 mM [ATP] as described in Fig. 1. The anisotropy and total fluorescence data were subjected to MENOTR global analysis described in the Materials and Methods using Scheme 1. The resulting best fits are shown in the overlaid black broken traces. The optimized kinetic parameters for this fit are presented in Table 2 and below. Shown are the translocation rate constant kT = (1.8 ± 0.3) s−1, nonproductive rate constant kNP = (0.06 ± 0.02) s−1, slow step rate constant kC = (0.17 ± 0.05) s−1, and kinetic step size m = (14 ± 3) aa. The time courses were fitted globally across total fluorescence and fluorescence anisotropy for all three lengths. Steady-state anisotropy of free peptide is shown as dashed lines in (B). These anisotropy measurements of free peptide were collected in the stopped flow by filling syringe 1 with 200 nM peptide in H300 and rapid mixing it against H300 buffer in syringe 2. The anisotropy value reported in the figure represents the average signal collected over 50 s. All the steady-state measurements exhibited flat time courses over this period. The best-fit simulations better describe the anisotropy data compared with that of the total fluorescence. This is due to the relative error on the respective data with total fluorescence having an error, ±0.1, two orders of magnitude higher than that of anisotropy, ±0.001 (see Figs. S3 and S4). To see this figure in color, go online.
The question is, can we extract mechanistic information from the anisotropy time courses because the time courses cannot be described by a simple sum of exponentials (27)? As stated, when collecting fluorescence anisotropy experiments, two types of signals are simultaneously gathered: total fluorescence (TF) and fluorescence anisotropy (r). When exciting the sample with vertically polarized light, each signal is described by Eqs. 1 and 2, respectively.
| (1) |
| (2) |
Where and I⊥ are the measured fluorescence intensities detected parallel and perpendicular to the incident polarization (see Fig. 1 C). The G factor, G, is the ratio of the sensitivity of the two detectors. The G factor accounts for any differences in the optical paths of the two detectors.
To apply Eqs. 1 and 2 to the kinetic time courses collected for ClpA-catalyzed polypeptide translocation, we need these equations in a form that can be used to describe all the species that have unique fluorescence values (quantum yields) and unique anisotropy values as a function of time. Total fluorescence as a function of time is the summation of all the states, yi, multiplied by their respective quantum yields, qi, (see Eq. 3). Similarly, anisotropy is the summation of all the states multiplied by both their respective quantum yield and the anisotropy, ri. However, as can be seen in Eq. 2, anisotropy is normalized by total fluorescence. Therefore, an additional term in the time-dependent anisotropy function that accounts for this normalization is required. By dividing the anisotropy summation by the total fluorescence summation, the same normalization can be achieved in the time-dependent function (see Eq. 4).
| (3) |
| (4) |
Equations 3 and 4 are general and have been reported previously (27). However, to apply Eqs. 3 and 4 to a new system, the task at hand is to determine time-dependent functions, yi(t), for each species in the reaction. We have previously reported that Scheme 1 describes ClpA-catalyzed polypeptide translocation (25). From this scheme, we can construct a system of coupled differential equations that describe each species in the mechanism. It has been demonstrated that Laplace transforms can be applied to solve these systems of differential equations (33). The transformation of the system of differential equations into Laplace space results in a much simpler system of coupled algebraic equations, yi(s). This transformation not only simplifies the system but, more importantly, allows us to construct a model within Laplace space where the number of steps, n, is a fitting parameter of interest. This is essential because it is not initially known how many steps ClpA, or any translocating motor, is taking on a given substrate lattice (32,33). Once we acquire each yi(s), the task is to find the inverse Laplace transform to yield yi(t) as given by Eq. 5.
| (5) |
Scheme 1.
Reaction mechanism of ClpA-catalyzed translocation of polypeptide substrate.
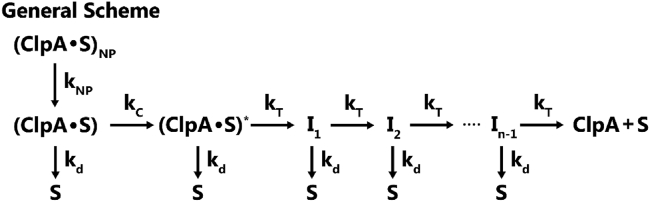
Scheme 1 depicts the reaction mechanism used to describe ClpA-catalyzed translocation of polypeptide substrate. Before mixing with ATP, ClpA is bound to fluorescent substrate in two forms: nonproductive ClpA complex (ClpA⋅S)NP and productive ClpA complex (ClpA⋅S). Upon mixing with ATP (ClpA⋅S)NP slowly isomerizes with rate constant kNP to form (ClpA⋅S). Because these two states are present before rapid mixing with ATP, we define the relative populations of the two states by the fraction of productively bound complexes, x, where x is the ratio of productive complex to total complex. Translocation begins and some number, n − 1, of intermediates, I, are formed with rate constant kT. The step size, m, of the motor is determined by relating the number of steps taken, n, to the length of the substrate being translocated, L. The model assumes homogenous stepping; however, the motor is translocating substrates with heterogenous sequences. Thus, the reported step size represents the average number of amino acids translocated between two rate-limiting steps. With the last translocation step, the ClpA-peptide complex will dissociate. Throughout the scheme it is possible for peptide, S, to dissociate from ClpA before the completion of translocation, these steps are described by the rate constant kd. Additionally, there is a slow conformational step to (ClpA⋅S)∗ described by the rate constant kC that repeats h number of times. We have previously shown that this is the minimal set of parameters required to describe the phenomenological features of the kinetic time courses (25,26,32,44). Applying Eq. 5 to Eqs. 3 and 4 yields Eqs. 6 and 7.
| (6) |
| (7) |
With a strategy for finding the time-dependent functions, yi(t), for each intermediate, the next step is to approximate the quantum yield values, qi, and the anisotropy values, ri. In our experimental design, the signal is sensitive to only fluorescently modified substrate. In the simplest case, only the bound and unbound peptides give unique quantum yields and anisotropy values, i.e., only two states exist. The time courses in Fig. 3, A and B indicate, at a minimum, that bound and free substrate have different anisotropy and total fluorescence values. Thus, the first level of analysis is to assume that the quantum yield and anisotropy of all the substrate-bound states are given by q1 and r1, respectively, and all free substrate is given by q2 and r2. To apply Eqs. 6 and 7 to the analysis of the time courses in Fig. 3, A and B, the summation is expanded and is given by Eqs. 8 and 9.
| (8) |
| (9) |
During analysis, MENOTR solves Eqs. 8 and 9 by numerically approximating the inverse Laplace transform of both equations. Note that for anisotropy, inverse Laplace transforms must be performed on the numerator and denominator separately before division. The transforms yield solutions in the time domain that represent the total fluorescence and anisotropy signals. For full expansions of the fitting functions, see Eqs. S1–S4.
The time courses from Fig. 3, A and B were subjected to global MENOTR analysis to determine a set of best-optimized kinetic parameters using Scheme 1 and the derived two-state model (Eqs. S1–S4). A description of the analysis can be found in the Materials and Methods. The resulting best fits of the data are plotted as broken black traces in Fig. 3, A and B. All the determined parameters (see Tables 2 and S1) agreed with those determined using raw fluorescence techniques, denoted below for each parameter as RF. The translocation rate constant was found to be kT = (1.8 ± 0.3) s−1 compared with kT(RF) = (1.5 ± 0.2) s−1, whereas the kinetic step size was found to be m = (14 ± 3) aa compared with m(RF) = (14 ± 1) aa. The slow pretranslocation transition rate constant was determined to be kNP = (0.06 ± 0.02) s−1 compared with kNP(RF) = (0.040 ± 0.003) s−1. The slow nontranslocation rate constant was found to be kC = (0.17 ± 0.05) s−1 compared with kC(RF) = (0.168 ± 0.008) s−1. We have previously shown that this is the minimal set of parameters required to describe the phenomenological features of the kinetic time courses and they are well determined (25). Thus, the only additional parameters that are required for this analysis are the quantum yield changes and anisotropy values, which represent scalar constants that are constrained by the magnitude of the signal change.
Table 2.
Kinetic Parameters Determined for ClpA-Catalyzed Translocation
| Technique: Substrate | kT (s−1) | kNP (s−1) | kC (s−1) | m (aa) |
|---|---|---|---|---|
| RF: SsrA substrates | 1.5 ± 0.2 | 0.040 ± 0.003 | 0.168 ± 0.008 | 14 ± 1 |
| TF and r: SsrA substrates | 1.8 ± 0.3 | 0.06 ± 0.02 | 0.17 ± 0.05 | 14 ± 3 |
| TF and r: αS1 casein substrates | 1.18 ± 0.03 | 0.014 ± 0.003 | 0.08 ± 0.02 | 11.9 ± 0.2 |
Errors associated with each parameter were determined through Monte Carlo analysis using 1000 simulations.
It can be seen in Fig. 3 A that the fit traces do not describe the total fluorescence time courses as well as the anisotropy time courses in Fig. 3 B. This is the consequence of the total fluorescence time courses exhibiting “drift” from acquisition to acquisition. Recall that the time courses represent the average of multiple acquisitions, in which each acquisition represents a time course collected with the same sample contained within the two syringes of the stopped flow (see Fig. 1 A). Although the shape of each time course from acquisition to acquisition is identical, its total fluorescence value is variable (see Fig. S3). In our previously published raw fluorescence results, we also observe this phenomenon. However, previously, the raw fluorescence time courses were offset so that they overlay each other. Because the shapes of the curves are all the same, this procedure of overlaying the curves does not influence the determination of the kinetic parameters. However, in the analysis of the total fluorescence time courses, we cannot offset the time courses because the absolute value of the fluorescence is required to analyze the anisotropy time courses, see Eqs. 3 and 4. Consequently, when we average multiple acquisitions, there is a standard deviation on each time point that accounts for the drift in the signal. Strikingly, the anisotropy time courses from acquisition to acquisition overlay nearly perfectly (see Fig. S3). Upon averaging the acquisitions, as expected because of drift, we see that there is a larger standard deviation on each total fluorescence time point compared with each anisotropy time point (see Fig. S4). Consequently, the anisotropy time points are weighted heavier in the global analysis than the total fluorescence time points because the analysis uses the standard deviation to weight each data point when finding the minimum of the fitting function. However, all the fit lines shown in Fig. 3, A and B fall within the standard deviation of the data points, and thus, we conclude that the combined fit of both total fluorescence and anisotropy is a good description of the data.
Fluorescence anisotropy and total fluorescence techniques applied to extended length polypeptide substrates
The analysis of the total fluorescence and the anisotropy time courses collected with synthetic polypeptide substrates yielded kinetic parameters consistent with our raw fluorescence time courses. Therefore, we next sought to test the anisotropy technique with the αS1 casein substrates. Recall, the αS1 casein substrates yielded time courses in raw fluorescence that we deemed uninterpretable, see Fig. 2 B. For these experiments, fluorescently modified αS1 casein 102-mer and 127-mer (see Table 1) were prebound to ClpA and loaded in syringe 1 of the stopped-flow spectrometer and, in syringe 2, was loaded with ATP and protein trap, as shown in Fig. 1 A. The contents of the two syringes were rapidly mixed and total fluorescence and anisotropy time courses were collected. The resultant average time courses are shown in Fig. 4, A and B (plots of these time courses logarithmically spaced can be found in Fig. S7, C and D).
Figure 4.
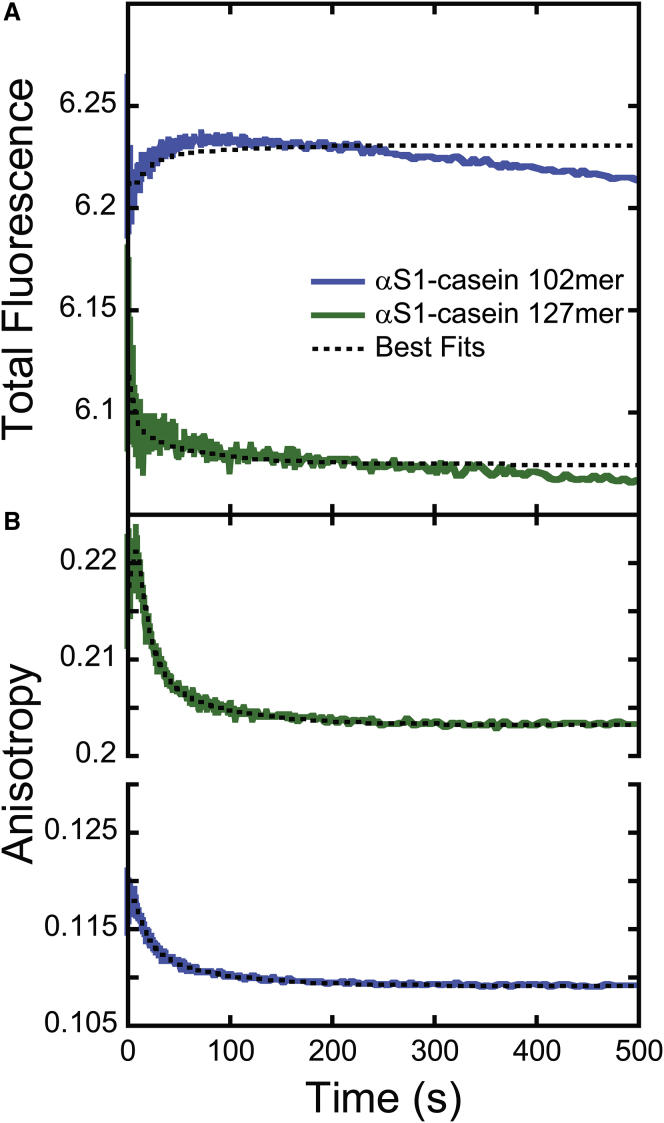
(A) Total fluorescence and (B) fluorescence anisotropy time courses of ClpA-catalyzed translocation of αS1 casein 102- and 127-mer (see Table 1). Solid traces show representative time courses of 102-mer (blue) and 127-mer (green) that were collected at 3 mM [ATP] as described in Fig. 1. The anisotropy and total fluorescence data were subjected to MENOTR global analysis described in the Materials and Methods using Scheme 1. The resulting best fits are shown in the overlaid black broken traces. The optimized parameters for this fit are provided in Table 2 and below. Shown are the translocation rate constant kT = (1.18 ± 0.03) s−1, dissociation rate constant kd was not detected, nonproductive rate constant kNP = (0.014 ± 0.003) s−1, slow step rate constant kC = (0.08 ± 0.02) s−1, and kinetic step size m = (11.9 ± 0.2) aa. The time courses were fitted globally across total fluorescence and fluorescence anisotropy and all substrate lengths. The best-fit simulations better describe the anisotropy data compared with that of the total fluorescence. This is due to the relative error on the respective data with total fluorescence having an error, ±0.1, two orders of magnitude higher than that of anisotropy, ±0.001 (see Figs. S5 and S6). To see this figure in color, go online.
The total fluorescence time course for 127-mer exhibits an initial lag followed by a clearly defined decrease, whereas anisotropy exhibited an initial rise followed by the expected decay. The trends in the 102-mer total fluorescence time courses are not the same as the 127-mer. The total fluorescence initially rises over the first 100 s and then decays back toward the initial total fluorescence value. On the other hand, the anisotropy time courses displayed an initial lag followed by the expected decay. It should also be noted that, in addition to being different from each other, the signals for the 102-mer and 127-mer are not the same as what we observe with the SsrA substrates.
To try and better understand the observed signal changes between bound and unbound peptide, steady-state fluorescent measurements of the αS1 casein and SsrA substrates alone as well as incubated with 15 μM ClpA and 300 μM ATPγS were collected in both raw fluorescence and total fluorescence modes (Fig. S1). The synthetic SsrA substrates (Fig. S1, A–C) all exhibit steady-state fluorescence signals that are consistent with the time courses collected here (Fig. 3 A) and our previous results. That is to say, unbound substrate has a higher fluorescence intensity compared with the quenched fluorescence of bound substrate and this is observed for both total fluorescence and raw fluorescence. In contrast to this, the αS1 casein steady-state fluorescence measurements showed an opposite effect upon binding to ClpA. Unbound substrate had lower fluorescent intensities than that of bound, i.e., fluorescein fluorescence is enhanced upon binding in both total fluorescence and raw fluorescence (Fig. S1, D and E).
We have previously reported steady-state anisotropy values of bound and unbound αS1 casein 127-mer and 102-mer. We found that 127-mer and 102-mer had steady-state anisotropy values of 0.11 and 0.08, respectively. When bound to another Hsp-Clp chaperone, ClpB, their steady-state anisotropy values increased to 0.23 and 0.18, respectively (43). With ClpA, we reported that 127-mer increased to an anisotropy value of ∼0.25 (35). We did not previously determine a value for 102-mer bound to ClpA; however, we expect to see an increase with this substrate as well. Based on the results of both the steady-state fluorescence and anisotropy for bound and unbound αS1 casein substrates, we predict that there will need to be at least two values of quantum yield and anisotropy used in the model to describe the data in Fig. 4.
Recall that the time courses collected with the synthetic polypeptide substrates were described assuming that all substrate-bound states had the same quantum yield and anisotropy value, and free substrate has a different quantum yield and anisotropy. We applied the same model used to describe ClpA translocation of the synthetic polypeptide substrates here with the αS1 casein substrates. However, the model did not adequately describe the time courses based both on visual inspection and poor chi-squared values (fits not shown). It is likely still the case that free αS1 casein substrate can be described by a single quantum yield and a single anisotropy value. Thus the differences in the observed signal compared with the SsrA substrates are likely due either to the prebound states ((ClpA⋅P)NP and (ClpA⋅P)) or the various translocation intermediates, Ii, exhibiting different quantum yields and anisotropies. To test this idea, we derived a model in which we assume that the initial prebound ClpA state exhibits a quantum yield, q1, and anisotropy, r1, whereas the summation of all the translocation intermediates exhibit a different quantum yield, q2, and anisotropy, r2. Finally, free peptide exhibits a quantum yield and anisotropy, q3 and r3, respectively. Other three-state and four-state models were tested, none of which produce statistically superior results than this simpler three-state model (results not shown). Applying this to Eqs. 6 and 7, we can expand the summations to Eqs. 10 and 11.
| (10) |
| (11) |
Three-state models were derived (see Eqs. S1 and S2) and used to perform global MENOTR analysis on the αS1 casein substrate data (see Tables 2 and S2). The resulting translocation rate constant was found to be kT = (1.18 ± 0.03) s−1, similar to what was determined for the synthetic substrates, kT = (1.8 ± 0.6) s−1. The kinetic step size found for the αS1 casein substrates, m = (11.9 ± 0.2) aa, was just under that of the synthetic substrates, m = (14 ± 3) aa. The other kinetic parameters determined for the αS1-casein substrates were also similar to the previous results, with kNP = (0.014 ± 0.003) s−1 and kC = (0.08 ± 0.02) s−1.
Discussion
Here, we have incorporated the n-step sequential mechanism into the method described by Otto et al. to simultaneously analyze anisotropy and total fluorescence time courses. This was done to examine the mechanism of single-turnover polypeptide translocation catalyzed by ClpA in the absence of its protease. Equally as important, we developed and report methods to quantitatively evaluate the resultant time courses to determine kinetic parameters that describe the translocation mechanism. With these techniques and methods, we show that we can overcome obstacles previously encountered in obtaining mechanistic information about the translocation of substrates greater than 100 aa in length. Moreover, we have shown that these techniques are suitable for monitoring the residence time of a translocase on its lattice independent of covalent modification of the lattice.
Monitoring ClpA-catalyzed translocation using fluorescence anisotropy and total fluorescence
Before applying florescence anisotropy to any novel investigations, we first sought to test whether this technique is sensitive to ClpA-catalyzed translocation. We have previously developed a single-turnover fluorescence stopped-flow technique sensitive to translocation catalyzed by ClpA (25). In that assay a monochromator is used to select the excitation wavelength and fluorescence emission is detected by a PMT containing a long-pass filter (see Fig. 1 B). Thus, the fluorescence signal detected in that technique represents the integrated area under the emission spectra for all wavelengths above the cutoff of the filter being used. Additionally, neither the excitation nor emission light pass through a polarizer, leaving all the light circularly polarized. We refer to this as “raw fluorescence” to distinguish it from the total fluorescence (Eq. 1) collected simultaneously with fluorescence anisotropy (Eq. 2), both of which are determined with polarized excitation and emission.
In our previous work, raw fluorescence time courses for the fluorescently modified polypeptides exhibited a substrate length-dependent lag followed by an increase to a final plateau. An increase in the extent of lag with increasing lattice length is evidence that ClpA is taking more kinetic steps as the lattice length is increased. This observation is consistent with directional translocation along the polypeptide chain (25). Thus, the analysis of those data allowed for the determination of several kinetic parameters describing the elementary steps of ClpA-catalyzed translocation.
Global analysis of time courses collected for both raw fluorescence and combined total fluorescence and fluorescence anisotropy produced kinetic parameters that agreed with one another. We showed here that the translocation rate constant, kT, and the kinetic step size, m, determined using the combined signals from anisotropy and total fluorescence were within error of those determined from raw fluorescence alone (25). This observation suggests that we are monitoring the same kinetic mechanism of polypeptide translocation in fluorescence anisotropy as previously observed using raw fluorescence. Thus, the application of these fluorescence and anisotropy-based methods to novel questions regarding ClpA and other potential translocases is appropriate and warranted.
Fluorescence anisotropy is normalized by total fluorescence. Thus, the anisotropy value of free peptide and ClpA bound peptide is a reproducible number. Consequently, fluorescence anisotropy measurements allow us to conclude if complete dissociation of ClpA from the polypeptide occurs or if some fraction of ClpA remains bound. We showed in Fig. S2 that ClpA-catalyzed translocation of all three synthetic polypeptide substrate lengths resulted in an anisotropy change consistent with complete dissociation of ClpA from its fluorescently modified substrates. Thus, we know that ClpA is not stalled on the lattice partially translocated or bound at the end of the substrate after complete translocation. This information is important in deciding on the mechanistic meanings behind the derived kinetic parameters obtained from analysis of these data. If some subpopulation of the motors are stalled on the lattice without dissociating, the schemes describing translocation would need to account for this phenomenon.
Changes in raw fluorescence can be due to both direct interactions with the fluorophore and changes in anisotropy upon binding. In contrast, total fluorescence removes any influence of anisotropy. Thus, the observation of a signal change in total fluorescence can be interpreted to indicate there are direct interactions with the fluorophore.
Recent cryo-EM structures of ClpA indicate that 25 aa of bound substrate are contained within the axial channel. Consequently, 5, 15, and 25 aa would protrude from the axial channel in the ClpA-peptide prebound complex for the 30-, 40-, and 50-aa substrates, respectively. Because ClpA binds to the 11-aa SsrA sequence at the C-terminus, this would result in the fluorescein on the N-terminus protruding outside of the axial channel. The observed quantum yield changes reported in Table S1 indicate that the signal changes between 4 and 9% for the three peptides. Thus, even though the fluorescein is outside of the axial channel, these quantum yield changes indicate the fluorescein does interact with ClpA in the bound complex because one would expect no quantum yield changes if there was not interaction at all. In contrast, the αS1 casein substrates exhibit a quantum yield change that is 10-fold smaller of 0.4 and 0.8% for the 102-mer and 127-mer, respectively (Table S2). Because 77 and 102 aa would be expected to protrude out of the axial channel in the ClpA-αS1-casein complex, the 10-fold reduction in quantum yield change compared with the short synthetic polypeptides indicates that the dye has less interaction with ClpA. Recall that αS1 casein does not contain a known binding motif for ClpA. However, if ClpA were binding to the N-terminus of the αS1 casein substrates, then ClpA would have direct interactions with the fluorescein, and the quantum yield change would be expected to be at least as large as that observed with the short polypeptides. In contrast, if ClpA is bound to the C-terminus of αS1 casein, we would expect the largest amount of distance between the fluorescein at the N-terminus and ClpA and, thus, the least amount of interaction. However, we do see a small quantum yield change between bound and unbound, which indicates some interaction. Thus, an analysis of the quantum yield changes reported in Tables S1 and S2 suggest that ClpA binds the C-terminus on the αS1 casein substrates, and some interaction between the fluorescein and ClpA must occur in the bound complex.
Examination of ClpA-catalyzed translocation of substrates greater than 100 aa in length
Processivity, P, represents the probability of a translocase taking another step forward along its lattice versus disassociating. Processivity can be quantitatively defined as the translocation rate constant, kT, divided by the summation of the translocation rate constant and the dissociation rate constant, kd (see Eq. 12 and Scheme 1) (45). Where P = 1 indicates that a motor completely translocates its lattice with every binding event, and P < 1 indicates the motor has some probability of dissociating before complete translocation of its lattice. Using this probability, it is possible to predict the average number of amino acids translocated, N, by a motor before dissociating from the polypeptide (see Eq. 13) (33). Recall that m is the kinetic step size of the motor, which represents the average number of aa translocated between two rate-limiting steps.
| (12) |
| (13) |
We have assumed that ClpA is a processive motor under saturating ATP concentrations because dissociation during translocation was not detected. However, a quantitative estimate of processivity was not possible (25,46). In those investigations, kd was only detectable at [ATP] below 300 μM, resulting in P = 0.876 at subsaturating [ATP] using Eq. 12. The average number of aa translocated per binding event was calculated to be N = ∼100 aa using Eq. 13 and p = 0.876. However, that estimate is twofold larger than the longest substrate used in the study. Thus, we conclude that this estimate of processivity represents a lower limit and a more precise estimate is needed.
Here, we have shown that fluorescence anisotropy and total fluorescence techniques can be used to collect time courses that can be quantitatively evaluated for substrates of lengths up to at least 127 aa. Strikingly, the results of the anisotropy translocation experiments performed on the fluorescently modified αS1 casein substrates, 102 and 127 aa in length, resulted in the determination of kinetic parameters that agreed with both the fluorescence anisotropy and raw fluorescence results for the shorter 30–50 aa substrates. However, the analysis of both the 102- and 127-aa αS1 casein substrates did not lead to a detectible kd precluding us from calculating a processivity based on Eq. 12. Thus, leading us to conclude that ClpA is translocating at least 127 aa per binding event at saturating [ATP] and updating our estimate of processivity to P > 0.893 using Eq. 13.
We have previously constructed unstructured polypeptide substrates as long as 177 aa for use in examining ClpA binding specificity (35). In that work, we found that at chain lengths greater than 127 aa, multiple ClpA hexamers can bind a single substrate. When using either raw fluorescence or the combined anisotropy and total fluorescence approach, it is imperative to have only one ClpA hexamer bound per polypeptide substrate. Consequently, when using fluorescence anisotropy and total fluorescence to examine substrates between 150 and 200 aa in length, we will have to perform these experiments under conditions that favor one-to-one binding of ClpA hexamer to substrate. Thus, there are still challenges to overcome to examine substrates long enough to report on processivity. On the other hand, we are now poised to begin examining folded proteins with specific ClpA binding sequences. In this case, the binding sequence is sufficiently short to accommodate only one ClpA hexamer and ClpA will not bind to the folded region. Thus, the combined total fluorescence and anisotropy technique will allow us to determine the impact of folded structures on the polypeptide translocation mechanism, which was also not possible with raw fluorescence.
In-solution methods for evaluating translocation mechanisms
Gates et al. recently published a review of 41 cryo-EM structures that revealed comparable spiral-staircase arrangements in multiple AAA+ motors (20, 21, 22,24,47, 48, 49, 50, 51, 52). From these structures, multiple structural hypotheses for AAA+-catalyzed translocation have been proposed. The hypotheses propose that translocation involves conformational shifts in motor protomers as a consequence of ATP binding and hydrolysis. These conformational changes create a rotating spiral-staircase arrangement in the motor that is suggested to support a hand-over-hand or ratchet-like mechanism for translocation. The mechanisms proposed in these studies are based on structural snapshots of motor proteins bound to model substrates in the presence of nucleotide.
The class I AAA+ motor proteins ClpB (23,50,53) and Hsp104 (22), and just recently ClpA as part of the ClpAP system, have been shown to adopt spiral-staircase architecture in both nucleotide binding domains. Multiple motor states determined in these studies are suggested to indicate successive hydrolysis events at domains 1 and 2 that catalyze a 2-aa translocation step per ATP hydrolysis cycle. This model is in contrast to the traditional power stroke model that has been established using in-solution biochemical assays (47). Additionally, the 2-aa step size is in direct contrast to the 5-aa step size for ClpAP determined by both transient-state kinetics and single-molecule optical tweezer experiments (26,54).
There are a limited number of tools that have the molecular level resolution to test the structural hypotheses being proposed by the new cryo-EM structures. Rapid mixing experiments that simultaneously monitor total fluorescence and anisotropy will aid in addressing questions raised by the wealth of new structural data. Identical to thermodynamic measurements, cryo-EM only reports on the final state of the system and are therefore path independent. Consequently, it is unknown where the observed states exist along the reaction pathway and if they represent reaction intermediates. Kinetic measurements are required to report on the path and to determine parameters like step-sizes, ATP coupling efficiencies, elementary rate constants, and processivity. Although these recent structural studies provide new hypotheses for AAA+ catalyzed translocation, there is a need to have path-dependent experiments that can test these hypotheses.
Methods for investigating protease-independent translocation
Many kinetic techniques applied to the investigation of AAA+ translocases utilize the degradation of substrate by proteolytic partners to monitor the reaction. These methods take advantage of proteolysis to monitor translocation by relating it to disappearance of substrate bands on an SDS-PAGE gel (55, 56, 57), loss of fluorescence as labeled lattice substrate is cleaved (6,58), or Förster resonance energy transfer signal as labeled lattice approaches a partner dye inside the protease (7,59). Although these coupled methods are excellent for monitoring AAA+ motors in the presence of their proteases, they assume that the mechanism of translocation is the same in both the presence and absence of the proteolytic partner. Additionally, there are many examples of AAA+ motors that do not interact with a proteolytic partner nor do they covalently modify the polypeptide chain on which they operate.
It has been shown that some AAA+ motors, including ClpA and ClpX (26,60), have translocation activity that is allosterically controlled by the presence of their proteases, ClpP. In the cases of ClpA and ClpX, the presence of ClpP has been shown to induce increases in rate and lead to changes in kinetic parameters like step size. Baytshtok et al. showed that ClpXP exhibited threefold higher unfoldase activity compared with ClpX and that ClpAP unfolds substrates sevenfold faster than ClpA (60). Additionally, Miller et al. showed that ClpAP translocates polypeptide with an almost twofold faster rate and translocates almost half as many aa per rate-limiting step compared with ClpA. To draw conclusions about these types of allosteric interactions between the partner and its chaperone, it is necessary to have methods that can measure the activity of the motor in the presence and absence of the protease.
AAA+ motors, such as N-ethylmaleimide sensitive factor, katanin, spastin, VPS4, ClpB, and Hsp104, do not natively associate with a partner protease. If these motors processively translocate a polypeptide through their axial channel, then no covalent modification of the polypeptide chain would occur. Consequently, determining if these motors are processive translocases has proven difficult to answer. One way this problem has been overcome was by a protein engineering strategy. For example, ClpB and Hsp104 have been mutated such that they include the helix-loop-helix IGL loop responsible for ClpA’s association with ClpP (55,56). As such, the corresponding variants BAP and HAP, named as either ClpB or Hsp104 that contains a ClpA-P-loop that contacts ClpP, are competent in forming a complex with ClpP. The idea being that if proteolytic degradation is observed in the presence of BAP or HAP then they must be threading the polypeptide chain through their axial channel and into the proteolytic chamber of ClpP the same way that ClpA and ClpX do naturally. In that work, they showed that when BAP-ClpP or HAP-ClpP provided polypeptide substrate and ATP, disappearance of substrate bands corresponding to α-casein on an SDS-PAGE gel was observed. They concluded that this is indicative of complete and processive threading of polypeptide substrate through the axial channel of both motors. It should be noted that only a single cleavage event is needed to observe this disappearance of substrate bands on a gel.
Recent transient-state kinetic analysis of ClpB and Hsp104 revealed both to be nonprocessive translocases in direct contrast to the BAP and HAP results (16,17). Both Hsp104WT and ClpBWT were shown to only take two kinetic steps on their substrates per binding event across all substrate lengths tested. In addition, Li et al. showed that the previous degradation assays performed on BAP discussed above (56) yielded substrate degradation in the absence of ATP. Thus, it is unclear whether any of the observed degradation in the BAP studies is the consequence of ATP-dependent translocation rather than ATP-independent degradation. Li et al. proposed that the ATP-independent degradation could be the consequence of dysregulation of ClpP activity (16,61). Similar dysregulation is seen in acyldepsipeptide (ADEPS) studies, in which ADEPS are small molecules that have been shown to bind to ClpP in a similar manner to the ClpA-P loops. Their binding induces opening of the ClpP axial pores that allows for unregulated degradation of substrate without a translocase (61).
We propose that the fluorescence anisotropy and total fluorescence techniques presented here overcome some of these types of problems. The fluorescence anisotropy methods do not rely on the presence of a partner protein and, thus, have the potential to be used in both protease-dependent and protease-independent investigations. Further, the only modification necessary to monitor fluorescence anisotropy is the addition of a fluorescent dye to the substrates being translocated. Thus, the motor remains unmodified, and wild-type actives of AAA+ motors that do not natively associate with a protease can be directly examined.
A robust method for investigating AAA+ translocases
Otto et al. showed that there is an absolute requirement to simultaneously analyze anisotropy with total fluorescence under conditions in which there is a change in total fluorescence (27). This is the consequence of the nonexponential behavior of the anisotropy time courses, as indicated by Eq. 4. They further illustrated that if anisotropy time courses were fitted without accounting for the change in total fluorescence, then the determined number of phases, rate constants, and amplitudes were all inaccurately determined. Finally, they also argued that kinetic experiments with combined total fluorescence and anisotropy data sets can provide more insight into the reaction mechanisms compared with either alone.
Here, we have incorporated the n-step sequential mechanism and numerical inverse Laplace transforms into the method described by Otto et al. By doing this, we have shown that the global analysis of total fluorescence and fluorescence anisotropy time courses represent a robust method for monitoring translocation catalyzed by AAA+ translocases and can yield additional information on the reaction mechanism. We have shown that we can determine the same kinetic parameters as previously determined using raw fluorescence alone and can access mechanistic information on substrates that were inaccessible by raw fluorescence alone.
It is important to note that we are not arguing that by simultaneously analyzing total fluorescence with anisotropy one will achieve improved fits or better constraints on the parameters. Rather, the simultaneous analysis is a requirement of the physics of the system (see Eqs. 3 and 4). The internal test as to whether one needs to simultaneously analyze total fluorescence and anisotropy together is not if a good fit of anisotropy alone can be achieved. Rather, the internal test is the detection of a change in total fluorescence in the reaction of interest. Only under conditions in which all qi are equal would it be appropriate to analyze the anisotropy time courses alone because, as can be seen in Eq. 4, all qi would factor out of the summations. However, as pointed out by Otto et al., from an analysis of anisotropy without knowledge of changes in total fluorescence, there is no way to know if the anisotropy time courses would have an influence of quantum yield changes, i.e., no internal test. This is because “good fits” will be achieved by applying Eq. 4 to anisotropy time courses without total fluorescence. However, Otto el al. showed that the error will simply show up as over or underestimates of the parameters and/or, worse, an apparent increase in the number of phases leading to an incorrect reaction mechanism. We have observed the same phenomena in our attempts to fit the anisotropy time courses in the absence of the total fluorescence time courses. What we observed was that a variety of good fits were achieved with vastly different parameters (data not shown). Moreover, if one did achieve identical fits of the anisotropy time courses both with and without the total fluorescence time courses, then this would not be evidence that this is a viable strategy that is generally applicable.
In contrast, fitting total fluorescence time courses in the absence of anisotropy, in some cases, will be equivalent to fitting raw fluorescence time courses. However, raw fluorescence and total fluorescence time courses will not always be the same. Here, we observed that the total fluorescence and raw fluorescence time courses for the αS1 casein substrates are clearly not the same. On the other hand, the raw fluorescence and total fluorescence time courses for the synthetic polypeptide substrates are similar. Therefore, comparing raw fluorescence with total fluorescence time courses serves as a test for whether or not rotational mobility is influencing the raw fluorescence time courses and to what magnitude. It would seem reasonable to recommend always using total fluorescence over raw fluorescence. However, this is not always practical. Collecting total fluorescence requires the use of polarizers in front of both the excitation light and emission light. Consequently, the observed intensity will be substantially reduced compared with the observed intensity in a raw fluorescence experiment. Thus, depending on the fluorophore of choice, concentration ranges, magnitude of signal change, etc., it may not be practical to choose total fluorescence over raw fluorescence. On the other hand, if it is practical to collect total fluorescence, then the recommendation from this work is to do so for two reasons. First, one can compare total and raw fluorescence to determine if there are influences of anisotropy on raw fluorescence. Second, upon collection of total fluorescence, one will simultaneously collect anisotropy, and the combined analysis of the two signals will allow for additional insights into the mechanism.
Although stopped-flow techniques involving fluorescence or anisotropy have been extensively used to observe biochemical reactions, few examples appear to exist in which the data have been examined by simultaneous analysis of anisotropy and total fluorescence. ClpA (7,25), ClpB-Hsp104 (16,17,51), p97 (62), 26S proteasome (59), Hsp70 or Hsp60 (63), and HslU (64) systems have all been examined using anisotropy-based techniques. However, all, including our previous work, have only supplied qualitative descriptions of the kinetic time courses, and none have attempted to couple fluorescence anisotropy to total fluorescence in the manner described in this manuscript. Thus, we hope that the strategies presented here will aid in gaining deeper insight into the mechanisms of polypeptide translocation catalyzed by AAA+ motors.
Author Contributions
N.W.S. wrote the manuscript and designed, performed, and analyzed experiments. A.L.L. wrote the manuscript and designed experiments. All authors read and approved final manuscript.
Acknowledgments
We thank Zachariah Ingram for his critical discussions of the manuscript and his partnership in designing the MENOTR fitting toolbox.
This work was supported by the National Science Foundation (grant MCB-1412624 to A.L.L.). Computational work done in data analysis was performed using the University of Alabama at Birmingham High Performance Computing Cheaha, which is supported in part by the National Science Foundation under grant no. OAC-1541310, the University of Alabama at Birmingham, and the Alabama Innovation Fund.
Editor: Doug Barrick.
Footnotes
Supporting Material can be found online at https://doi.org/10.1016/j.bpj.2020.08.018.
Supporting Material
References
- 1.Baumeister W., Lupas A. The proteasome. Curr. Opin. Struct. Biol. 1997;7:273–278. doi: 10.1016/s0959-440x(97)80036-x. [DOI] [PubMed] [Google Scholar]
- 2.Katayama-Fujimura Y., Gottesman S., Maurizi M.R. A multiple-component, ATP-dependent protease from Escherichia coli. J. Biol. Chem. 1987;262:4477–4485. [PubMed] [Google Scholar]
- 3.Kwon A.R., Trame C.B., McKay D.B. Kinetics of protein substrate degradation by HslUV. J. Struct. Biol. 2004;146:141–147. doi: 10.1016/j.jsb.2003.11.003. [DOI] [PubMed] [Google Scholar]
- 4.Sauer R.T., Baker T.A. AAA+ proteases: ATP-fueled machines of protein destruction. Annu. Rev. Biochem. 2011;80:587–612. doi: 10.1146/annurev-biochem-060408-172623. [DOI] [PubMed] [Google Scholar]
- 5.Horwitz A.A., Navon A., Goldberg A.L. ATP-induced structural transitions in PAN, the proteasome-regulatory ATPase complex in Archaea. J. Biol. Chem. 2007;282:22921–22929. doi: 10.1074/jbc.M702846200. [DOI] [PubMed] [Google Scholar]
- 6.Weber-Ban E.U., Reid B.G., Horwich A.L. Global unfolding of a substrate protein by the Hsp100 chaperone ClpA. Nature. 1999;401:90–93. doi: 10.1038/43481. [DOI] [PubMed] [Google Scholar]
- 7.Reid B.G., Fenton W.A., Weber-Ban E.U. ClpA mediates directional translocation of substrate proteins into the ClpP protease. Proc. Natl. Acad. Sci. USA. 2001;98:3768–3772. doi: 10.1073/pnas.071043698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kolygo K., Ranjan N., Weber-Ban E. Studying chaperone-proteases using a real-time approach based on FRET. J. Struct. Biol. 2009;168:267–277. doi: 10.1016/j.jsb.2009.07.003. [DOI] [PubMed] [Google Scholar]
- 9.Fleming K.G., Hohl T.M., Hanson P.I. A revised model for the oligomeric state of the N-ethylmaleimide-sensitive fusion protein, NSF. J. Biol. Chem. 1998;273:15675–15681. doi: 10.1074/jbc.273.25.15675. [DOI] [PubMed] [Google Scholar]
- 10.Hartman J.J., Mahr J., McNally F.J. Katanin, a microtubule-severing protein, is a novel AAA ATPase that targets to the centrosome using a WD40-containing subunit. Cell. 1998;93:277–287. doi: 10.1016/s0092-8674(00)81578-0. [DOI] [PubMed] [Google Scholar]
- 11.Eckert T., Le D.T.-V., Woehlke G. Spastin’s microtubule-binding properties and comparison to katanin. PLoS One. 2012;7:e50161. doi: 10.1371/journal.pone.0050161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Monroe N., Han H., Hill C.P. Structural basis of protein translocation by the Vps4-Vta1 AAA ATPase. eLife. 2017;6:e24487. doi: 10.7554/eLife.24487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mogk A., Kummer E., Bukau B. Cooperation of Hsp70 and Hsp100 chaperone machines in protein disaggregation. Front. Mol. Biosci. 2015;2:22. doi: 10.3389/fmolb.2015.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Doyle S.M., Wickner S. Hsp104 and ClpB: protein disaggregating machines. Trends Biochem. Sci. 2009;34:40–48. doi: 10.1016/j.tibs.2008.09.010. [DOI] [PubMed] [Google Scholar]
- 15.Dalal S., Rosser M.F.N., Hanson P.I. Distinct roles for the AAA ATPases NSF and p97 in the secretory pathway. Mol. Biol. Cell. 2004;15:637–648. doi: 10.1091/mbc.E03-02-0097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li T., Weaver C.L., Lucius A.L. Escherichia coli ClpB is a non-processive polypeptide translocase. Biochem. J. 2015;470:39–52. doi: 10.1042/BJ20141457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Durie C.L., Lin J., Lucius A.L. Hsp104 and potentiated variants can operate as distinct nonprocessive translocases. Biophys. J. 2019;116:1856–1872. doi: 10.1016/j.bpj.2019.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matyskiela M.E., Lander G.C., Martin A. Conformational switching of the 26S proteasome enables substrate degradation. Nat. Struct. Mol. Biol. 2013;20:781–788. doi: 10.1038/nsmb.2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Han H., Fulcher J.M., Hill C.P. Structure of Vps4 with circular peptides and implications for translocation of two polypeptide chains by AAA+ ATPases. eLife. 2019;8:e44071. doi: 10.7554/eLife.44071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sandate C.R., Szyk A., Roll-Mecak A. An allosteric network in spastin couples multiple activities required for microtubule severing. Nat. Struct. Mol. Biol. 2019;26:671–678. doi: 10.1038/s41594-019-0257-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.White K.I., Zhao M., Brunger A.T. Structural principles of SNARE complex recognition by the AAA+ protein NSF. eLife. 2018;7:e38888. doi: 10.7554/eLife.38888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gates S.N., Yokom A.L., Southworth D.R. Ratchet-like polypeptide translocation mechanism of the AAA+ disaggregase Hsp104. Science. 2017;357:273–279. doi: 10.1126/science.aan1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rizo A.N., Lin J., Southworth D.R. Structural basis for substrate gripping and translocation by the ClpB AAA+ disaggregase. Nat. Commun. 2019;10:2393. doi: 10.1038/s41467-019-10150-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lopez K.E., Rizo A.N., Southworth D.R. Conformational plasticity of the ClpAP AAA+ protease couples protein unfolding and proteolysis. Nat. Struct. Mol. Biol. 2020;27:406–416. doi: 10.1038/s41594-020-0409-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rajendar B., Lucius A.L. Molecular mechanism of polypeptide translocation catalyzed by the Escherichia coli ClpA protein translocase. J. Mol. Biol. 2010;399:665–679. doi: 10.1016/j.jmb.2010.03.061. [DOI] [PubMed] [Google Scholar]
- 26.Miller J.M., Lin J., Lucius A.L. E. coli ClpA catalyzed polypeptide translocation is allosterically controlled by the protease ClpP. J. Mol. Biol. 2013;425:2795–2812. doi: 10.1016/j.jmb.2013.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Otto M.R., Lillo M.P., Beechem J.M. Resolution of multiphasic reactions by the combination of fluorescence total-intensity and anisotropy stopped-flow kinetic experiments. Biophys. J. 1994;67:2511–2521. doi: 10.1016/S0006-3495(94)80741-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee C., Schwartz M.P., Matouschek A. ATP-dependent proteases degrade their substrates by processively unraveling them from the degradation signal. Mol. Cell. 2001;7:627–637. doi: 10.1016/s1097-2765(01)00209-x. [DOI] [PubMed] [Google Scholar]
- 29.Thompson M.W., Singh S.K., Maurizi M.R. Processive degradation of proteins by the ATP-dependent Clp protease from Escherichia coli. Requirement for the multiple array of active sites in ClpP but not ATP hydrolysis. J. Biol. Chem. 1994;269:18209–18215. [PubMed] [Google Scholar]
- 30.Hoskins J.R., Pak M., Wickner S. The role of the ClpA chaperone in proteolysis by ClpAP. Proc. Natl. Acad. Sci. USA. 1998;95:12135–12140. doi: 10.1073/pnas.95.21.12135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lakowicz J.R. Second Edition. Kluwer Academic/Plenum; New York: 1999. Principles of Fluorescence Spectroscopy. [Google Scholar]
- 32.Lucius A.L., Miller J.M., Rajendar B. Application of the sequential n-step kinetic mechanism to polypeptide translocases. Methods Enzymol. 2011;488:239–264. doi: 10.1016/B978-0-12-381268-1.00010-0. [DOI] [PubMed] [Google Scholar]
- 33.Lucius A.L., Maluf N.K., Lohman T.M. General methods for analysis of sequential “n-step” kinetic mechanisms: application to single turnover kinetics of helicase-catalyzed DNA unwinding. Biophys. J. 2003;85:2224–2239. doi: 10.1016/s0006-3495(03)74648-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Galletto R., Bujalowski W. Kinetics of the E. coli replication factor DnaC protein-nucleotide interactions. II. Fluorescence anisotropy and transient, dynamic quenching stopped-flow studies of the reaction intermediates. Biochemistry. 2002;41:8921–8934. doi: 10.1021/bi020127w. [DOI] [PubMed] [Google Scholar]
- 35.Li T., Lucius A.L. Examination of the polypeptide substrate specificity for Escherichia coli ClpA. Biochemistry. 2013;52:4941–4954. doi: 10.1021/bi400178q. [DOI] [PubMed] [Google Scholar]
- 36.Veronese P.K., Stafford R.P., Lucius A.L. The Escherichia coli ClpA molecular chaperone self-assembles into tetramers. Biochemistry. 2009;48:9221–9233. doi: 10.1021/bi900935q. [DOI] [PubMed] [Google Scholar]
- 37.Liu G.R., Han X., Lam K.Y. A combined genetic algorithm and nonlinear least squares method for material characterization using elastic waves. Comput. Methods Appl. Mech. Eng. 2002;191:1909–1921. [Google Scholar]
- 38.Liu Y., Tian P. A multi-start central force optimization for global optimization. Appl. Soft Comput. 2015;27:92–98. [Google Scholar]
- 39.Johnson M.L. Why, when, and how biochemists should use least squares. Anal. Biochem. 1992;206:215–225. doi: 10.1016/0003-2697(92)90356-c. [DOI] [PubMed] [Google Scholar]
- 40.Johnson M.L. Parameter correlations while curve fitting. Methods Enzymol. 2000;321:424–446. doi: 10.1016/s0076-6879(00)21207-x. [DOI] [PubMed] [Google Scholar]
- 41.Johnson M.L. Evaluation and propagation of confidence intervals in nonlinear, asymmetrical variance spaces. Analysis of ligand-binding data. Biophys. J. 1983;44:101–106. doi: 10.1016/S0006-3495(83)84281-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Veronese P.K., Rajendar B., Lucius A.L. Activity of E. coli ClpA bound by nucleoside diphosphates and triphosphates. J. Mol. Biol. 2011;409:333–347. doi: 10.1016/j.jmb.2011.02.018. [DOI] [PubMed] [Google Scholar]
- 43.Li T., Lin J., Lucius A.L. Examination of polypeptide substrate specificity for Escherichia coli ClpB. Proteins. 2015;83:117–134. doi: 10.1002/prot.24710. [DOI] [PubMed] [Google Scholar]
- 44.Miller J.M., Lucius A.L. ATPγS competes with ATP for binding at domain 1 but not domain 2 during ClpA catalyzed polypeptide translocation. Biophys. Chem. 2014;185:58–69. doi: 10.1016/j.bpc.2013.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McClure W.R., Chow Y. The kinetics and processivity of nucleic acid polymerases. Methods Enzymol. 1980;64:277–297. doi: 10.1016/s0076-6879(80)64013-0. [DOI] [PubMed] [Google Scholar]
- 46.Duran E.C., Weaver C.L., Lucius A.L. Comparative analysis of the structure and function of AAA+ motors ClpA, ClpB, and Hsp104: common threads and disparate functions. Front. Mol. Biosci. 2017;4:54. doi: 10.3389/fmolb.2017.00054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gates S.N., Martin A. Stairway to translocation: AAA+ motor structures reveal the mechanisms of ATP-dependent substrate translocation. Protein Sci. 2020;29:407–419. doi: 10.1002/pro.3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cooney I., Han H., Shen P.S. Structure of the Cdc48 segregase in the act of unfolding an authentic substrate. Science. 2019;365:502–505. doi: 10.1126/science.aax0486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Twomey E.C., Ji Z., Rapoport T.A. Substrate processing by the Cdc48 ATPase complex is initiated by ubiquitin unfolding. Science. 2019;365:eaax1033. doi: 10.1126/science.aax1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yu H., Lupoli T.J., Li H. ATP hydrolysis-coupled peptide translocation mechanism of Mycobacterium tuberculosis ClpB. Proc. Natl. Acad. Sci. USA. 2018;115:E9560–E9569. doi: 10.1073/pnas.1810648115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Deville C., Carroni M., Saibil H.R. Structural pathway of regulated substrate transfer and threading through an Hsp100 disaggregase. Sci. Adv. 2017;3:e1701726. doi: 10.1126/sciadv.1701726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ho C.M., Beck J.R., Zhou Z.H. Malaria parasite translocon structure and mechanism of effector export. Nature. 2018;561:70–75. doi: 10.1038/s41586-018-0469-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Deville C., Franke K., Saibil H.R. Two-step activation mechanism of the ClpB disaggregase for sequential substrate threading by the main ATPase motor. Cell Rep. 2019;27:3433–3446.e4. doi: 10.1016/j.celrep.2019.05.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Olivares A.O., Nager A.R., Baker T.A. Mechanochemical basis of protein degradation by a double-ring AAA+ machine. Nat. Struct. Mol. Biol. 2014;21:871–875. doi: 10.1038/nsmb.2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tessarz P., Mogk A., Bukau B. Substrate threading through the central pore of the Hsp104 chaperone as a common mechanism for protein disaggregation and prion propagation. Mol. Microbiol. 2008;68:87–97. doi: 10.1111/j.1365-2958.2008.06135.x. [DOI] [PubMed] [Google Scholar]
- 56.Weibezahn J., Tessarz P., Bukau B. Thermotolerance requires refolding of aggregated proteins by substrate translocation through the central pore of ClpB. Cell. 2004;119:653–665. doi: 10.1016/j.cell.2004.11.027. [DOI] [PubMed] [Google Scholar]
- 57.Gottesman S., Roche E., Sauer R.T. The ClpXP and ClpAP proteases degrade proteins with carboxy-terminal peptide tails added by the SsrA-tagging system. Genes Dev. 1998;12:1338–1347. doi: 10.1101/gad.12.9.1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Maglica Z., Kolygo K., Weber-Ban E. Optimal efficiency of ClpAP and ClpXP chaperone-proteases is achieved by architectural symmetry. Structure. 2009;17:508–516. doi: 10.1016/j.str.2009.02.014. [DOI] [PubMed] [Google Scholar]
- 59.Bhattacharyya S., Renn J.P., Matouschek A. An assay for 26S proteasome activity based on fluorescence anisotropy measurements of dye-labeled protein substrates. Anal. Biochem. 2016;509:50–59. doi: 10.1016/j.ab.2016.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Baytshtok V., Baker T.A., Sauer R.T. Assaying the kinetics of protein denaturation catalyzed by AAA+ unfolding machines and proteases. Proc. Natl. Acad. Sci. USA. 2015;112:5377–5382. doi: 10.1073/pnas.1505881112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Brötz-Oesterhelt H., Beyer D., Labischinski H. Dysregulation of bacterial proteolytic machinery by a new class of antibiotics. Nat. Med. 2005;11:1082–1087. doi: 10.1038/nm1306. [DOI] [PubMed] [Google Scholar]
- 62.Chou T.F., Bulfer S.L., Arkin M.R. Specific inhibition of p97/VCP ATPase and kinetic analysis demonstrate interaction between D1 and D2 ATPase domains. J. Mol. Biol. 2014;426:2886–2899. doi: 10.1016/j.jmb.2014.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bukau B., Horwich A.L. The Hsp70 and Hsp60 chaperone machines. Cell. 1998;92:351–366. doi: 10.1016/s0092-8674(00)80928-9. [DOI] [PubMed] [Google Scholar]
- 64.Burton R.E., Baker T.A., Sauer R.T. Nucleotide-dependent substrate recognition by the AAA+ HslUV protease. Nat. Struct. Mol. Biol. 2005;12:245–251. doi: 10.1038/nsmb898. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.