Abstract
Over the past decade, technical and methodological improvements in cryogenic electron microscopy (cryo-EM) single-particle analysis have enabled routine high-resolution structural analyses of biological macromolecules, resulting in a flood of new molecular insights into protracted biological questions. However, despite the tremendous progress and success of the field in recent years, opportunities for improvement remain in various aspects of the cryo-EM single-particle analysis workflow (e.g., sample preparation, image acquisition and processing, and structure validation). Here, we review recent advances that have contributed to the principal methods in cryo-EM and identify persisting challenges and bottlenecks that will require further methodological and hardware development.
Introduction
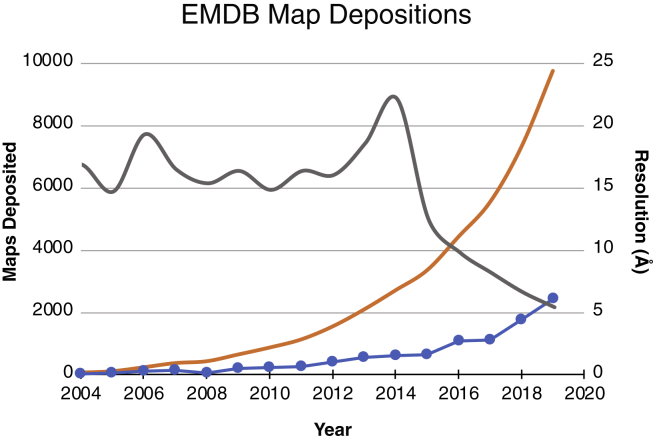
The transformative impact of cryogenic electron microscopy (cryo-EM) single-particle analysis (SPA) on the structural biology field has garnered considerable attention from the scientific community. The power of cryo-EM SPA lies in the preservation of macromolecules in frozen-hydrated states through rapid vitrification, which enables the visualization of unrestrained biological targets. These complexes are often observed in multiple conformations, providing insights to their dynamics and mechanisms of function. For this reason, cryo-EM SPA methodologies can be used to study a broad range of macromolecular assemblies that are typically too flexible or heterogeneous to be crystallized (1, 2, 3, 4). Many of the earlier limitations of cryo-EM (e.g., resolution, specimen size range, and low data throughput) have been diminished by technical and methodological advances established over the past decade, such as direct electron detectors (DEDs), modern transmission electron microscopes (TEMs) with stable optics, improved automated data collection software, and more sophisticated image processing algorithms. As a result, the annual number of cryo-EM SPA structures deposited to the Electron Microscopy Data Bank (EMDB) has been exponentially rising in conjunction with improvements to the median resolution, indicating that both the quantity and quality of new structures is increasing (Fig. 1). Indeed, cryo-EM SPA structures in the resolution range of 3 Å and better have now become near-routine, allowing for de novo model building of many macromolecules that have previously evaded structural study. Cryo-EM SPA has proven especially valuable for obtaining new insights to membrane proteins (5, 6, 7, 8, 9) and is also quickly establishing utility in studying small complexes (10, 11, 12). The breadth of this technique reflects the continuous co-evolution of TEM design and optics, specimen preparation, image acquisition and processing, and structure validation within the cryo-EM workflow. These developments, in addition to other recent breakthroughs, will be discussed in the following sections along with remaining challenges and promising new methodologies to look forward to on the horizon.
Figure 1.
Resolution and map deposition trends in single-particle cryo-EM. Shown are the annual (blue dotted curve) and cumulative (orange curve) map depositions into the Electron Microscopy Data Bank (EMDB). The median resolution of deposited maps per year is also shown (gray curve); future cryo-EM methodological and hardware developments, such as those discussed in this review, will likely improve the median resolution to 4 Å or better over the next decade. To see this figure in color, go online.
Sample preparation for cryo-EM
Cryo-EM analysis utilizes significantly lower quantities of sample than is typically required for x-ray crystallography or NMR studies, and specimens can be preserved through vitrification immediately after purification. Thus, cryo-EM analysis is particularly advantageous for proteins that are low yield and/or not stable over extended periods. However, the success of most cryo-EM SPA experiments begins (and, more importantly, ends) with the quality of the purified biological specimen. Ideally, samples for cryo-EM SPA should be biochemically stable in solution up to the point of vitrification, with minimal conformational and/or compositional heterogeneity. Confirming sample purity through size exclusion chromatography and sodium-dodecyl-sulfate-polyacrylamide-gel analysis is required, but not necessarily sufficient, for structural studies. Negative staining TEM can also be useful for visually assessing sample quality and homogeneity (though results from negative stain analysis do not necessarily translate to outcomes under cryogenic conditions (13)). Although satisfying purification requirements may entail extensive optimization (common strategies have been summarized elsewhere (13,14)), doing so is ultimately more time and cost effective than relying on brute force in silico methods to “purify” the target from contaminants through extensive classification or to identify subsets of useable data that will contribute to a meaningful reconstruction from a sample of subpar quality.
Once a sufficiently optimized sample has been obtained, it is rapidly cryogenically cooled in a thin layer of vitrified ice to yield a frozen-hydrated specimen (15), which enables the specimen to withstand the high vacuum of the TEM interior and mitigates the damaging effects of the high-energy electrons used for imaging. The standard sample vitrification process involves applying a small volume of purified sample onto an EM grid that has been covered with a holey film support that is made hydrophilic (usually by glow discharging or plasma cleaning). The sample on the grid is then blotted with filter paper, either manually or using a robotic instrument such as the Vitrobot (Thermo Fisher Scientific, Waltham, MA), Cryoplunge 3 (Gatan, Pleasanton, CA), or EM GP2 (Leica Microsystems, Wetzlar, Germany), to remove the majority of the sample, and then immediately plunged into a reservoir of liquid ethane or a liquid-ethane-propane mixture (16). The higher heat conductivity of liquid ethane and ethane-propane over liquid nitrogen enables the formation of vitreous (amorphous or glass-like) ice instead of crystalline ice, the latter of which can damage the biological specimen and reduce the image quality through electron diffraction.
Sample vitrification is a variable process because there are many factors at play that can have varying levels of impact on the quality and reproducibility of the vitrified specimen (e.g., sample stability, temperature, humidity, blotting time and force, grid substrate, and buffer components). Ultimately, the objective is to produce grids containing macromolecules that are distributed in random orientations and embedded in the thinnest possible layer of vitrified ice—ideally, ice that is only slightly thicker than the longest dimension of the targeted molecule (17,18). Because the atoms comprising biological specimens scatter electrons only slightly more than those in the surrounding buffer, ice thickness is inextricably tied to image contrast and signal-to-noise ratio (SNR), which in turn limits the available high-resolution information. Maximizing the image SNR is particularly critical for cryo-EM SPA studies of small complexes (19,20), which have fewer scattering atoms that contribute to signal.
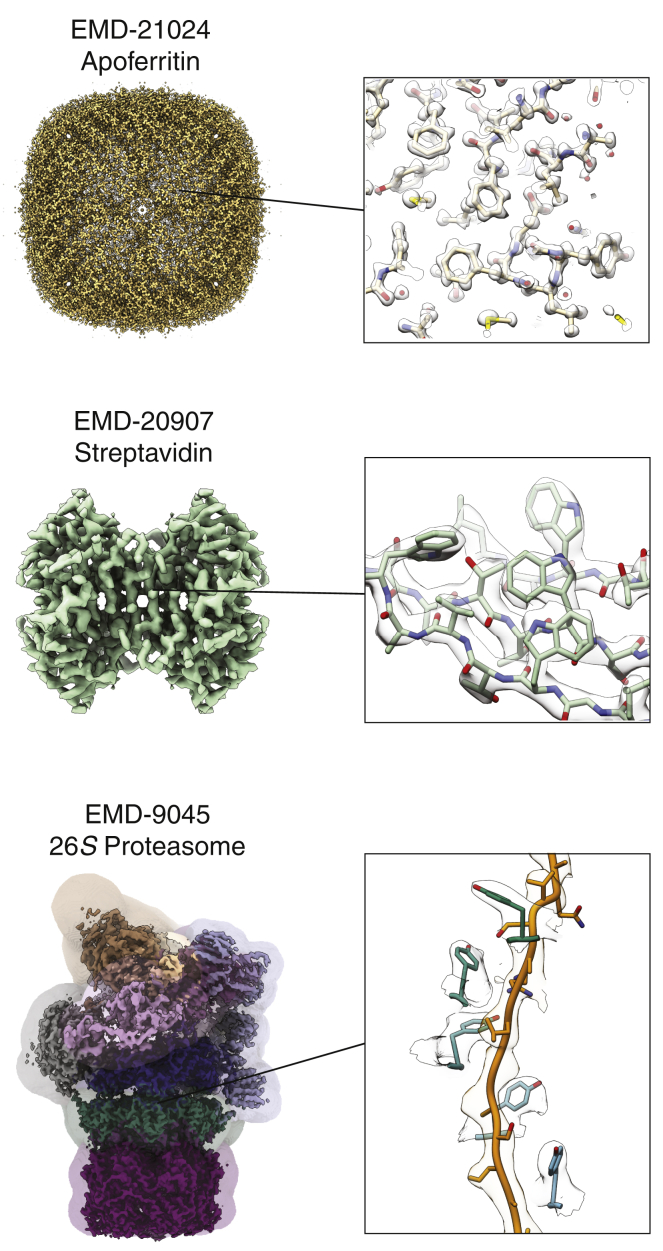
Achieving the ideal specimen grid is a nontrivial endeavor. Many oft-encountered challenges to specimen preparation are sample dependent, such as the proclivity of the macromolecule to distribute within the grid holes or dissociation of complexes. One major make-or-break factor is the unavoidable adsorption of the specimen to the hydrophobic air-water-interface (AWI) as a result of the multiple sample-AWI collisions that occur on a timescale shorter than that of the plunge-freezing process (21, 22, 23). Depending on the surface chemistry of the biological specimen, the resulting hydrophobic interactions at the AWI may promote partial to complete sample denaturation and/or the adoption of a single, preferential orientation relative to the AWI (21, 22, 23, 24). Although the latter issue may be overcome during data acquisition by employing a tilted collection strategy (25,26), a means to completely abolish sample adsorption to the AWI has yet to be developed. However, given the ubiquitously disastrous effects of the AWI on macromolecules, solutions are being eagerly pursued and will likely usher significant improvements to sample preparation. Several grid preparation strategies that mitigate the deleterious effects of the AWI have emerged in recent years, including the use of surfactants to block AWI interactions (27), as well as affinity grids (28) or monolayer supports (e.g., graphene and graphene oxide (29, 30, 31, 32, 33)) to sequester samples away from the AWI with additional benefits of increasing particle density and producing uniformly thin vitreous ice. Graphene monolayer supports have recently been used for high-resolution structure determination of the 52-kDa streptavidin complex (Fig. 2; (34)), demonstrating the utility of graphene for imaging small biological targets that normally adopt a preferred orientation at the AWI. Another promising solution lies in next-generation automated sample vitrification robots, such as Spotiton (35) (commercially Chameleon; TTP Labtech, Melbourn, UK), VitroJet (36) (CryoSol-World, Maastricht, the Netherlands), CryoWriter (37) (Nuonex, Basel, Switzerland), the time-resolved cryo-EM device (TED) (38), and Shake-it-off (39), which dispense small volumes of sample onto commercial or self-wicking (35,39) grids in a controlled process that can be monitored in real time to provide visual feedback on the quality of the vitrified specimen. These vitrification systems may overcome pitfalls from AWI interactions by shortening the sample dispense-to-plunge time (40). Furthermore, multiple inkjet sprayers can be used to mix samples directly onto the grid to enable time-resolved studies of complex assembly and/or short-lived ligand-induced conformational states, as was recently demonstrated for several biological systems (41). Additionally, such devices could be used to deposit more than one type of sample onto discrete areas of the same grid to increase throughput during specimen preparation and imaging over traditional blotting methods (35,42,43).
Figure 2.
Cryo-EM structures of biological macromolecules enabled by technical and methodological advances in SPA. Top: shown is mouse apoferritin determined to ∼1.2-Å resolution using a TEM equipped with a cold FEG and next-generation energy filter and direct detector. Middle: shown is the ∼2.6-Å resolution reconstruction of the 52-kDa streptavidin imaged over graphene monolayer support grids. Bottom: shown is the ∼4.2-Å resolution reconstruction of the substrate-engaged yeast 26S proteasome in the "4D" motor state. Focused classification and refinement strategies (colored by focusing area) were used to identify the distinct motor conformations and to generate composite reconstructions to facilitate model building for each state. The inset on the right shows density features of the substrate polypeptide encircled by a spiral-staircase arrangement of pore loop tyrosine residues within the central pore of the AAA+ motor. The EMDB accession code and full map (left) and density features with the corresponding atomic model docked in (right) are shown for each structure. To see this figure in color, go online.
TEMs and the costs versus benefits of keV
Modern TEMs have incorporated vast technological upgrades since the prototype was first developed in 1931 by Max Knoll and Ernst Ruska and now include improved vacuum systems, more stable specimen stages and automated grid loaders (autoloaders), constant-power optical lens systems, and—most importantly—more brilliant and coherent electrons from field emission guns (FEGs). However, the basic components and principles responsible for image formation in a TEM have remained unchanged. Within a TEM, electrons are emitted from an emission source under high vacuum and subsequently accelerated down the microscope column through a high potential difference. The theoretically achievable resolution of a TEM is limited by the wavelength of the accelerated electron. It therefore follows that TEMs operating at higher accelerating voltages are preferred for maximizing the achievable resolution (44). Indeed, three-condenser lens TEMs operating at 300 kV, such as the Thermo Fisher Scientific Titan Krios, and paired with a DED are considered the “industry standard” across the field for high-resolution cryo-EM because of the smaller inelastic scattering cross section, smaller defocus spread, minimized effects of specimen charging, better detector performance, and a flatter Ewald sphere associated with shorter electron wavelengths (44). The vast majority of cryo-EM SPA structures determined to date have been obtained using 300-kV TEMs. Furthermore, recent studies using prototype TEM hardware optimized to minimize the energy spread of the electron beam (e.g., using a cold FEG energy source and/or monochromator and energy filter) to greatly preserve information at high spatial frequencies have successfully generated cryo-EM densities that enable, for the first time using SPA approaches, visualization of atomic resolution features (Fig. 2; (45,46)). These exciting results demonstrate the potential of next-generation instruments for further extending resolution boundaries in the field.
However, the imaging benefits afforded by these instruments come with a high cost of purchase and maintenance that may be prohibitive to many institutions. In light of this, several groups have recently also demonstrated the utility of comparatively less expensive two-condenser lens TEMs operating at 200 kV, such as the Talos Arctica (47) and Glacios (48) (Thermo Fisher Scientific), for high-resolution structure determination. There are a few theoretical benefits that make imaging with lower-energy electrons worthwhile. Image contrast is higher at lower energies due to a greater elastic scattering cross-section and slightly larger contrast transfer function values at low spatial frequencies (22). Moreover, Peet et al. (49) showed that the “information coefficient” (the ratio between the elastic and inelastic scattering cross-sections) is greater at 200 keV than at 300 keV, therefore affording more information per unit of radiation damage. Interestingly, their study suggested that for the “average” cryo-EM SPA specimen (∼300 Å thick), the optimal electron energy is actually 100 keV. A preliminary demonstration of subnanometer-resolution reconstructions obtained at 100 keV has established a compelling rationale for the development of cheaper and more accessible lower kV TEMs for sample screening and routine imaging (50). Currently, the most pressing barriers to realizing this goal are the lack of a compatible detector due to the increased amount of backscattering at lower electron energies, as well as the substantially greater effects of chromatic aberration (Cc) resulting from increased energy spread. It will be of great interest to the structural biology community to examine the resolving capabilities at lower keVs once these hurdles are overcome. The “optimal” energy for cryo-EM is unquestionably a nuanced topic, and the choice of instrumentation ultimately depends on many factors, such as the nature of the biological specimen itself, the desired resolution, and—perhaps the biggest determining factor for many institutions—available resources.
Image acquisition
Cryo-EM data sets often consist of thousands of electron micrographs collected over the course of several days, which yield hundreds of thousands (and sometimes millions) of particle images. The number of particles required to produce a high-resolution reconstruction varies widely depending on the quality of the specimen (as discussed previously) as well as the conditions and/or instrumentation used for image acquisition, but generally more particle images correlate with increased resolution. Given the high cost of the operation of modern TEMs used for high-resolution data collection, as well as the dependence of resolution on particle number (51), it is prudent to maximize both the quality and quantity of data collected. For this reason, there has been a concerted push for automation in cryo-EM SPA image acquisition as well as processing over the past decades. Image acquisition software packages such as Leginon (52), SerialEM (53), and EPU (Thermo Fisher Scientific) interface with the TEM and enable automated navigation and exposure targeting in areas selected by the user. The rate of image acquisition can be accelerated by more than an order of magnitude by navigating across the grid through optical shifting of the beam (beam-image shift) instead of mechanically moving the stage, which requires an extended settling time after each movement. Historically, the faster beam-image shift strategy of data collection was avoided for high-resolution imaging because of the off-axis coma aberrations and phase shift errors introduced by beam tilt; however, it has recently been shown that these aberrations can be effectively minimized by a compensatory adjustment of the TEM deflection coils (54). Furthermore, it has been shown that these aberrations can be estimated and corrected for in silico (55,56). Additional image acquisition speedups are possible on three-condenser lens systems that allow for small nanoprobe beam sizes (∼600 nm in diameter or even smaller with the recent introduction of the “fringe-free illumination” scheme (57)), which enables targeting of multiple exposures around the perimeter of the same grid hole to further increase efficiency (1). Despite these advances, several major bottlenecks still remain during image acquisition. One of these is the time spent performing microscope alignments or adjustments during data collection, which could be reduced through automation or improvements to future TEMs. Additionally, a significant amount of user intervention and savvy is still required to identify and target the areas of the grid that will produce the best images. This limitation could be overcome by developing machine-learning algorithms to detect optimal acquisition areas (i.e., based off ice thickness measurements performed using an energy filter or aperture-limited scattering (18)) with minimal user intervention. Fully automated acquisition targeting in conjunction with on-the-fly image analysis (58, 59, 60) would considerably improve both the speed and quality of data collection.
A final critical factor in image acquisition lies in the use of DEDs, the development of which was the impetus for high-resolution cryo-EM SPA. Modern DEDs yield significantly improved image quality relative to previous charge-coupled devices by detecting charges generated directly from incident electrons, resulting in precise readouts of electron positions and higher detective quantum efficiency (61,62) (DQE, a measure of the SNR performance of the detector across spatial frequencies). More importantly, DEDs enable the collection of short video frames, which can then be aligned to each other to correct for the effects of image blurring from the beam-induced specimen motion that would otherwise dampen the high-resolution information (63,64). Dose weighting of individual frames to partially account for radiation damage can also be performed concurrently (65). Development of next-generation DEDs is well underway in the technical pursuit of the “perfect” detector, i.e., one with the DQE approaching 1 across all spatial frequencies, higher frame rates and readout times, and larger fields of view to maximize data collection efficiency. Looking ahead, it is likely that the rate-limiting bottleneck during imaging will alternate between hardware and software as technical advances in both areas continue to be achieved.
Image processing
The cryo-EM image processing workflow has become increasingly standardized over the past decade and begins with the computational correction of beam-induced specimen motion in the acquired images (motion correction) and filtering of image frames to account for accumulated radiation damage (dose-weighting). Biological specimens (“particles”) in the corrected micrographs are next identified using template- or gaussian-based algorithms, followed by two-dimensional (2D) rotational alignment, classification, and averaging of particle images to facilitate the initial culling of junk or low-quality particle picks. The selected particle subset is then subjected to iterative rounds of three-dimensional (3D) alignment and classification using a projection-matching approach to determine angular assignments for the particle images and to separate conformational and/or compositional heterogeneity. Angular assignments for these states are then refined separately to improve the resolution of the 3D reconstruction (66). Ancillary steps, such as finer correction of particle motion (particle “polishing” (58)) and aberration correction (55), can be employed to further improve map quality and resolution by restoring high spatial frequency information.
Macromolecules that are dynamic or contain continuously flexible components can be challenging to reconstruct because their projection images cannot be ascribed to a single structural state. In such cases, a focused classification and refinement approach may be useful, wherein focused 3D masks are applied to a specific region of interest on the complex to classify heterogeneity and/or to improve the alignment parameters within that region. Information outside of the region of interest may also be computationally removed altogether from the particle images themselves (“signal subtraction”) (67). Additionally, multibody refinement (68) and 3D variability analysis (69) approaches may be used to describe interdomain motions across the complex. The full continuum of conformational states adopted by a biological complex can also be described using manifold embedding, which defines the relative orientational and conformational relationships of all particles in a data set to generate a map of the free-energy landscape of the complex in solution (70). These strategies have enabled detailed structural studies of dynamic, multisubunit complexes (71, 72, 73, 74), including the substrate-engaged 26S proteasome in multiple states of substrate translocation (Fig. 2; (1)).
Although numerous image processing algorithms and software packages are available nowadays that can perform some or all of these steps (58, 59, 60), most of these programs are not fully automated and still require a significant amount of user intervention and judgement between steps, presenting yet another major bottleneck in cryo-EM SPA, particularly for inexperienced users. Furthermore, the wealth of available computational tools is somewhat of a double-edged sword; although it is to the user’s benefit to empirically test which combination of algorithms and parameters yields the best results for their particular data set, this approach is also very time consuming. Many popular software packages (such as RELION (58), cryoSPARC (60), Warp (59), and CisTEM (75), to name just a few) aim to overcome this by implementing guided and more user-friendly workflows and by utilizing GPU or high-end CPU processors to speed up computation time. However, there is still much room for improvement when it comes to the speed and capabilities of image processing algorithms. Many image reconstruction programs struggle with large data sets or particle image sizes and are less robust for challenging macromolecules (e.g., small complexes (12,76) or highly heterogeneous specimens). To assist with improving these programs, many in the community have uploaded raw data sets of challenging targets to the Electron Microscopy Public Image Archive (EMPIAR) (77) to serve as testbeds for algorithm development. Continuation of these practices, in addition to improvements to image SNR from next-generation detectors, will hopefully lead to more streamlined and robust image processing software in the future.
Resolution assessment and validation
The ultimate goal of most cryo-EM SPA studies is to produce an atomic model of a targeted biological specimen to gain residue-level insights into the molecular mechanisms associated with function. It is thus of utmost importance to verify that the experimental cryo-EM map and corresponding atomic model accurately represent the acquired data to prevent the formation of biological interpretations that exceed the structural details of the reconstruction. Structure validation is particularly critical for cryo-EM because the maps and atomic models are generated through independent processes (in contrast to x-ray crystallography, in which the maps and models are iteratively refined). Although validation methods and metrics for cryo-EM are not yet fully standardized (see below), analysis of the experimental map, model, and map-to-model fit are required for all cryo-EM publications and depositions.
The quality metric for cryo-EM maps is reported by the Fourier shell correlation (FSC) curve (78), which measures the agreement between two independently refined “half maps,” each containing a random half-subset of the data as a function of spatial resolution. The value at the FSC = 0.143 threshold is typically reported as the nominal resolution of the map (51), although numerous other metrics have been proposed (78,79). It is important to keep in mind, however, that the FSC is not a measure of resolution but rather of the global correlation between two halves of the data. For data sets of compositionally and conformationally homogeneous specimens that sample a wide range of orientations in ice, the FSC = 0.143 cutoff provides a sufficient estimation of the resolution of the reconstruction. However, even in these ideal cases, a single resolution value does not reflect the variability that is typically present in SPA reconstructions (80). Local resolution analysis can reveal map quality differences to provide insight into specific regions of structural heterogeneity (81,82). Stable regions, such as the core of the molecule, will typically be resolved to higher resolution than peripheral or flexible domains. It is also prudent to examine the FSC itself because the shape of the curve can inform on issues such as overfitting, sample heterogeneity, insufficient defocus spread, and duplicate particles, all of which manifest as distinctive pathologies in the FSC (79).
Given that most biological specimens contain one or more regions that preferentially adhere to the AWI, it is also important to consider that some cryo-EM reconstructions will not have isotropic directional resolution. Such reconstructions contain substantially less detailed structural information along the axes of the underpopulated orientations. In extreme cases in which preferential orientation dominates the data set, structural features along the axis orthogonal to the preferred view will appear distorted and stretched, yet the FSC may falsely report a high global resolution value. Thus, to more accurately assess resolution in cryo-EM maps, particularly those exhibiting strong preferential orientation, the 2D FSC curve can be extended into a 3D FSC to describe the overall resolution anisotropy of the resulting reconstruction (25,26).
The resulting atomic models must also be validated through various metrics that are commonplace in x-ray crystallography. Importantly, the correlation between the experimental map and the atomic model must also be assessed because a model with good statistics may not necessarily represent the data. Due to the inherent conformational heterogeneity of cryo-EM SPA structures, as well as the aspect that atomic resolution structures have not been obtained until recently, the field has yet to agree upon a standardized method for model validation in cryo-EM SPA; rather, multiple complementary validation programs are typically used to assess the quality of models built from cryo-EM data, including MolProbity (83), EMRinger (84), and CaBLAM (85). The agreement between the experimental data and map can also be assessed by calculating a map-to-model FSC, either globally or on the basis of local resolution (FSC-Q) (86). These statistics are collectively presented in published works as a supplementary table similar to what is prepared for models determined from x-ray crystal structures. Furthermore, given that the inherent resolution variation in cryo-EM maps cannot be sufficiently described using a single B-factor, multimodel convergence approaches have also been proposed for evaluating map and model quality (87). The rapid influx of cryo-EM structures nowadays necessitates more efforts toward standardizing model validation. In the interim, these validation metrics must be kept in mind, and appropriate caution taken, when interpreting published maps and models.
Concluding remarks
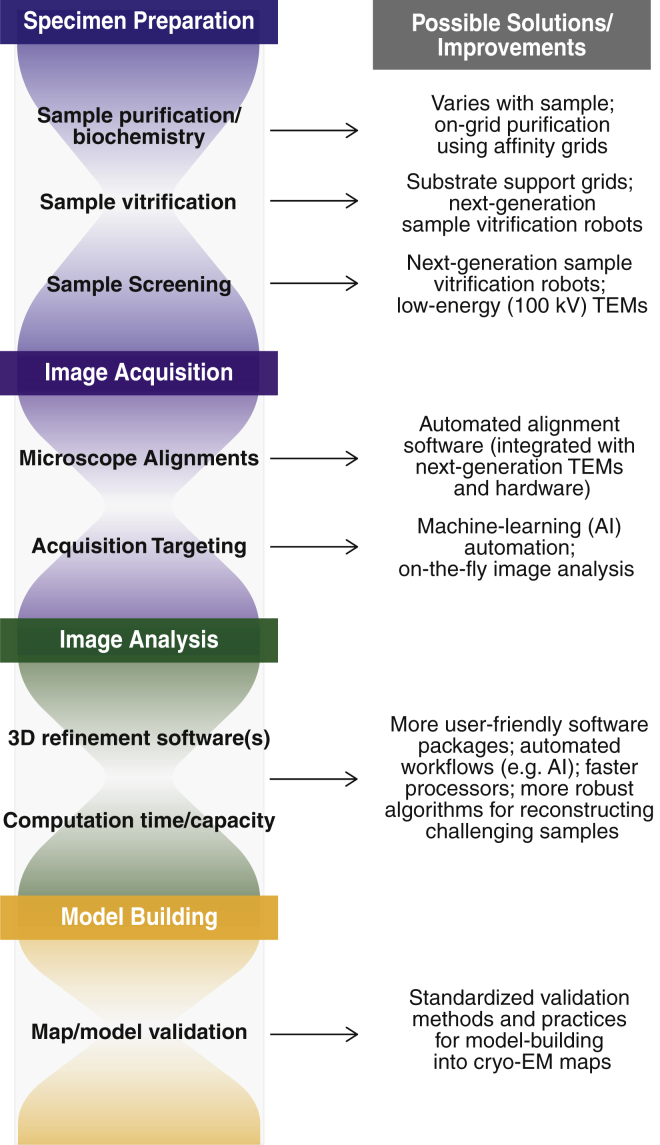
Salient technical breakthroughs in cryo-EM SPA over the past decade have enabled novel structural and mechanistic perspectives of a multitude of biological macromolecules. However, as the popularity of this technique increases, so too does the need for greater efficiency and accessibility. To this end, substantial strides are being made toward developing a more streamlined SPA workflow (Fig. 3). New sample vitrification robots can improve the reproducibility and quality of cryo-EM specimen grids by avoiding common pitfalls associated with conventional blotting methods and by providing a means to prescreen grid quality. Furthermore, the development of cheaper 100-kV instruments for routine sample screening may reduce the burden on higher-end TEMs. Another exciting prospect that has yet to be fully explored is the integration of machine-learning technology to guide or entirely replace steps that require significant user intervention, which would remove major bottlenecks in image acquisition and data processing (and could potentially be extended to other aspects of the workflow, such as specimen preparation). Collectively, these developments in conjunction with next-generation instruments and hardware will further expand the accessibility and utility of cryo-EM, particularly for high-throughput applications such as structure-based drug design, and it will be tremendously exciting to see how the field advances over the next decade.
Figure 3.
Current bottlenecks in the cryo-EM SPA workflow. Aspects of the workflow that impose significant limitations to efficiency, data quality, and/or reproducibility are indicated, along with potential future opportunities for reducing these bottlenecks. To see this figure in color, go online.
Acknowledgments
M.W. is supported as a National Science Foundation Graduate Student Research fellow. G.C.L. is supported by a young investigator award from Amgen and by National Institutes of Health grants DP2EB020402, R21AR072910, and R21AG061697.
Editor: Meyer Jackson.
References
- 1.de la Peña A.H., Goodall E.A., Martin A. Substrate-engaged 26 S proteasome structures reveal mechanisms for ATP-hydrolysis-driven translocation. Science. 2018;362:eaav0725. doi: 10.1126/science.aav0725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bai X.C., Yan C., Shi Y. An atomic structure of human γ-secretase. Nature. 2015;525:212–217. doi: 10.1038/nature14892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu Z., Gutierrez-Vargas C., Frank J. Structure and assembly model for the Trypanosoma cruzi 60S ribosomal subunit. Proc. Natl. Acad. Sci. USA. 2016;113:12174–12179. doi: 10.1073/pnas.1614594113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yan C., Wan R., Shi Y. Structure of a yeast activated spliceosome at 3.5 Å resolution. Science. 2016;353:904–911. doi: 10.1126/science.aag0291. [DOI] [PubMed] [Google Scholar]
- 5.Koehl A., Hu H., Kobilka B.K. Structure of the μ-opioid receptor-Gi protein complex. Nature. 2018;558:547–552. doi: 10.1038/s41586-018-0219-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liang Y.-L., Khoshouei M., Wootten D. Phase-plate cryo-EM structure of a biased agonist-bound human GLP-1 receptor-Gs complex. Nature. 2018;555:121–125. doi: 10.1038/nature25773. [DOI] [PubMed] [Google Scholar]
- 7.Kim J., Tan Y.Z., Mancia F. Structure and drug resistance of the Plasmodium falciparum transporter PfCRT. Nature. 2019;576:315–320. doi: 10.1038/s41586-019-1795-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saotome K., Teng B., Ward A.B. Structures of the otopetrin proton channels Otop1 and Otop3. Nat. Struct. Mol. Biol. 2019;26:518–525. doi: 10.1038/s41594-019-0235-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu X., Plotnikova O., Han S. Cryo-EM structures of the human glutamine transporter SLC1A5 (ASCT2) in the outward-facing conformation. eLife. 2019;8:e48120. doi: 10.7554/eLife.48120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Han B.G., Watson Z., Glaeser R.M. Monolayer-crystal streptavidin support films provide an internal standard of cryo-EM image quality. J. Struct. Biol. 2017;200:307–313. doi: 10.1016/j.jsb.2017.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang K., Li S., Chiu W. Cryo-EM structure of a 40 kDa SAM-IV riboswitch RNA at 3.7 Å resolution. Nat. Commun. 2019;10:5511. doi: 10.1038/s41467-019-13494-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Herzik M.A., Jr., Wu M., Lander G.C. High-resolution structure determination of sub-100 kDa complexes using conventional cryo-EM. Nat. Commun. 2019;10:1032. doi: 10.1038/s41467-019-08991-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lyumkis D. Challenges and opportunities in cryo-EM single-particle analysis. J. Biol. Chem. 2019;294:5181–5197. doi: 10.1074/jbc.REV118.005602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Renaud J.-P., Chari A., Wiesmann C. Cryo-EM in drug discovery: achievements, limitations and prospects. Nat. Rev. Drug Discov. 2018;17:471–492. doi: 10.1038/nrd.2018.77. [DOI] [PubMed] [Google Scholar]
- 15.Dubochet J., Adrian M., Schultz P. Cryo-electron microscopy of vitrified specimens. Q. Rev. Biophys. 1988;21:129–228. doi: 10.1017/s0033583500004297. [DOI] [PubMed] [Google Scholar]
- 16.Tivol W.F., Briegel A., Jensen G.J. An improved cryogen for plunge freezing. Microsc. Microanal. 2008;14:375–379. doi: 10.1017/S1431927608080781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim L.Y., Rice W.J., Carragher B. Benchmarking cryo-EM single particle analysis workflow. Front. Mol. Biosci. 2018;5:50. doi: 10.3389/fmolb.2018.00050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rice W.J., Cheng A., Potter C.S. Routine determination of ice thickness for cryo-EM grids. J. Struct. Biol. 2018;204:38–44. doi: 10.1016/j.jsb.2018.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Glaeser R.M. Limitations to significant information in biological electron microscopy as a result of radiation damage. J. Ultrastruct. Res. 1971;36:466–482. doi: 10.1016/s0022-5320(71)80118-1. [DOI] [PubMed] [Google Scholar]
- 20.Henderson R., McMullan G. Problems in obtaining perfect images by single-particle electron cryomicroscopy of biological structures in amorphous ice. Microscopy (Oxf.) 2013;62:43–50. doi: 10.1093/jmicro/dfs094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Glaeser R.M. Proteins, interfaces, and cryo-EM grids. Curr. Opin. Colloid Interface Sci. 2018;34:1–8. doi: 10.1016/j.cocis.2017.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Glaeser R.M. How good can single-particle cryo-EM become? What remains before it approaches its physical limits? Annu. Rev. Biophys. 2019;48:45–61. doi: 10.1146/annurev-biophys-070317-032828. [DOI] [PubMed] [Google Scholar]
- 23.Glaeser R.M., Han B.-G. Opinion: hazards faced by macromolecules when confined to thin aqueous films. Biophys. Rep. 2017;3:1–7. doi: 10.1007/s41048-016-0026-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Noble A.J., Dandey V.P., Carragher B. Routine single particle CryoEM sample and grid characterization by tomography. eLife. 2018;7:e34257. doi: 10.7554/eLife.34257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tan Y.Z., Baldwin P.R., Lyumkis D. Addressing preferred specimen orientation in single-particle cryo-EM through tilting. Nat. Methods. 2017;14:793–796. doi: 10.1038/nmeth.4347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Naydenova K., Russo C.J. Measuring the effects of particle orientation to improve the efficiency of electron cryomicroscopy. Nat. Commun. 2017;8:629. doi: 10.1038/s41467-017-00782-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen J., Noble A.J., Darst S.A. Eliminating effects of particle adsorption to the air/water interface in single-particle cryo-electron microscopy: bacterial RNA polymerase and CHAPSO. J Struct Biol X. 2019;1:100005. doi: 10.1016/j.yjsbx.2019.100005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kelly D.F., Dukovski D., Walz T. A practical guide to the use of monolayer purification and affinity grids. Methods Enzymol. 2010;481:83–107. doi: 10.1016/S0076-6879(10)81004-3. [DOI] [PubMed] [Google Scholar]
- 29.Han Y., Fan X., Yan N. High yield monolayer graphene grids for near-atomic resolution cryo-electron microscopy. bioRxiv. 2019 doi: 10.1101/827089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu N., Zhang J., Wang H.-W. Bioactive functionalized monolayer graphene for high-resolution cryo-electron microscopy. J. Am. Chem. Soc. 2019;141:4016–4025. doi: 10.1021/jacs.8b13038. [DOI] [PubMed] [Google Scholar]
- 31.Naydenova K., Peet M.J., Russo C.J. Multifunctional graphene supports for electron cryomicroscopy. Proc. Natl. Acad. Sci. USA. 2019;116:11718–11724. doi: 10.1073/pnas.1904766116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Palovcak E., Wang F., Cheng Y. A simple and robust procedure for preparing graphene-oxide cryo-EM grids. J. Struct. Biol. 2018;204:80–84. doi: 10.1016/j.jsb.2018.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pantelic R.S., Meyer J.C., Stahlberg H. The application of graphene as a sample support in transmission electron microscopy. Solid State Commun. 2012;152:1375–1382. [Google Scholar]
- 34.Han Y., Fan X., Yan N. High-yield monolayer graphene grids for near-atomic resolution cryoelectron microscopy. Proc. Natl. Acad. Sci. USA. 2020;117:1009–1014. doi: 10.1073/pnas.1919114117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dandey V.P., Wei H., Carragher B. Spotiton: new features and applications. J. Struct. Biol. 2018;202:161–169. doi: 10.1016/j.jsb.2018.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ravelli R.B.G., Nijpels F.J.T., Peters P.J. Cryo-EM structures from sub-nl volumes using pin-printing and jet vitrification. Nat. Commun. 2020;11:2563. doi: 10.1038/s41467-020-16392-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schmidli C., Rima L., Braun T. Miniaturized sample preparation for transmission electron microscopy. J. Vis. Exp. 2018;137:57310. doi: 10.3791/57310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kontziampasis D., Klebl D.P., White H.D. A cryo-EM grid preparation device for time-resolved structural studies. IUCrJ. 2019;6:1024–1031. doi: 10.1107/S2052252519011345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rubinstein J.L., Guo H., Kwok T. Shake-it-off: a simple ultrasonic cryo-EM specimen-preparation device. Acta Crystallogr. D Struct. Biol. 2019;75:1063–1070. doi: 10.1107/S2059798319014372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Klebl D.P., Gravett M.S.C., Muench S.P. Need for speed: examining protein behavior during CryoEM grid preparation at different timescales. Structure. 2020;S0969-2126:30282–30283. doi: 10.1016/j.str.2020.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dandey V.P., Budell W.C., Carragher B. Time-resolved cryo-EM using Spotiton. Nat. Methods. 2020;17:897–900. doi: 10.1038/s41592-020-0925-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ravelli R.B.G., Nijpels F.J.T., Peters P.J. Automated cryo-EM sample preparation by pin-printing and jet vitrification. bioRxiv. 2019 doi: 10.1101/651208. [DOI] [Google Scholar]
- 43.Jain T., Sheehan P., Potter C.S. Spotiton: a prototype for an integrated inkjet dispense and vitrification system for cryo-TEM. J. Struct. Biol. 2012;179:68–75. doi: 10.1016/j.jsb.2012.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Henderson R. The potential and limitations of neutrons, electrons and X-rays for atomic resolution microscopy of unstained biological molecules. Q. Rev. Biophys. 1995;28:171–193. doi: 10.1017/s003358350000305x. [DOI] [PubMed] [Google Scholar]
- 45.Nakane T., Kotecha A., Scheres S.H.W. Single-particle cryo-EM at atomic resolution. bioRxiv. 2020 doi: 10.1101/2020.05.22.110189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yip K.M., Fischer N., Stark H. Breaking the next Cryo-EM resolution barrier – atomic resolution determination of proteins! bioRxiv. 2020 doi: 10.1101/2020.05.21.106740. [DOI] [Google Scholar]
- 47.Wu M., Lander G.C., Herzik M.A., Jr. Sub-2 Angstrom resolution structure determination using single-particle cryo-EM at 200 keV. J Struct Biol X. 2020;4:100020. doi: 10.1016/j.yjsbx.2020.100020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hamdi F., Tüting C., Kastritis P.L. 2.7 Å cryo-EM structure of vitrified M. musculus H-chain apoferritin from 200 keV “screening microscope”. bioRxiv. 2019 doi: 10.1101/738724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Peet M.J., Henderson R., Russo C.J. The energy dependence of contrast and damage in electron cryomicroscopy of biological molecules. Ultramicroscopy. 2019;203:125–131. doi: 10.1016/j.ultramic.2019.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Naydenova K., McMullan G., Russo C.J. CryoEM at 100 keV: a demonstration and prospects. IUCrJ. 2019;6:1086–1098. doi: 10.1107/S2052252519012612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rosenthal P.B., Henderson R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J. Mol. Biol. 2003;333:721–745. doi: 10.1016/j.jmb.2003.07.013. [DOI] [PubMed] [Google Scholar]
- 52.Suloway C., Pulokas J., Carragher B. Automated molecular microscopy: the new Leginon system. J. Struct. Biol. 2005;151:41–60. doi: 10.1016/j.jsb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 53.Mastronarde D.N. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 2005;152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 54.Cheng A., Eng E.T., Carragher B. High resolution single particle cryo-electron microscopy using beam-image shift. J. Struct. Biol. 2018;204:270–275. doi: 10.1016/j.jsb.2018.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zivanov J., Nakane T., Scheres S.H.W. Estimation of high-order aberrations and anisotropic magnification from cryo-EM datasets in RELION-3.1. bioRxiv. 2019 doi: 10.1101/798066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cash J.N., Kearns S., Cianfrocco M.A. High-resolution cryo-EM using beam-image shift at 200 keV. bioRxiv. 2020 doi: 10.1101/2020.01.21.914507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Konings S., Kuijper M., Tiemeijer P. Advances in single particle analysis data acquisition. Microsc. Microanal. 2019;25:1012–1013. [Google Scholar]
- 58.Zivanov J., Nakane T., Scheres S.H.W. New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife. 2018;7:e42166. doi: 10.7554/eLife.42166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tegunov D., Cramer P. Real-time cryo-electron microscopy data preprocessing with Warp. Nat. Methods. 2019;16:1146–1152. doi: 10.1038/s41592-019-0580-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Punjani A., Rubinstein J.L., Brubaker M.A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods. 2017;14:290–296. doi: 10.1038/nmeth.4169. [DOI] [PubMed] [Google Scholar]
- 61.McMullan G., Faruqi A.R., Henderson R. Direct electron detectors. Methods Enzymol. 2016;579:1–17. doi: 10.1016/bs.mie.2016.05.056. [DOI] [PubMed] [Google Scholar]
- 62.Ruskin R.S., Yu Z., Grigorieff N. Quantitative characterization of electron detectors for transmission electron microscopy. J. Struct. Biol. 2013;184:385–393. doi: 10.1016/j.jsb.2013.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Brilot A.F., Chen J.Z., Grigorieff N. Beam-induced motion of vitrified specimen on holey carbon film. J. Struct. Biol. 2012;177:630–637. doi: 10.1016/j.jsb.2012.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li X., Mooney P., Cheng Y. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat. Methods. 2013;10:584–590. doi: 10.1038/nmeth.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zheng S.Q., Palovcak E., Agard D.A. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods. 2017;14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Penczek P.A. Image restoration in cryo-electron microscopy. Methods Enzymol. 2010;482:35–72. doi: 10.1016/S0076-6879(10)82002-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bai X.C., Rajendra E., Scheres S.H.W. Sampling the conformational space of the catalytic subunit of human γ-secretase. eLife. 2015;4:e11182. doi: 10.7554/eLife.11182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nakane T., Kimanius D., Scheres S.H. Characterisation of molecular motions in cryo-EM single-particle data by multi-body refinement in RELION. eLife. 2018;7:e36861.. doi: 10.7554/eLife.36861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Punjani A., Fleet D.J. 3D variability analysis: directly resolving continuous flexibility and discrete heterogeneity from single particle cryo-EM images. bioRxiv. 2020 doi: 10.1101/2020.04.08.032466. [DOI] [PubMed] [Google Scholar]
- 70.Frank J., Ourmazd A. Continuous changes in structure mapped by manifold embedding of single-particle data in cryo-EM. Methods. 2016;100:61–67. doi: 10.1016/j.ymeth.2016.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rogala K.B., Gu X., Sabatini D.M. Structural basis for the docking of mTORC1 on the lysosomal surface. Science. 2019;366:468–475. doi: 10.1126/science.aay0166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ramírez A.S., Kowal J., Locher K.P. Cryo-electron microscopy structures of human oligosaccharyltransferase complexes OST-A and OST-B. Science. 2019;366:1372–1375. doi: 10.1126/science.aaz3505. [DOI] [PubMed] [Google Scholar]
- 73.Chang L., Yang J., Barford D. Structure of the DOCK2-ELMO1 complex provides insights into regulation of the auto-inhibited state. Nat. Commun. 2020;11:3464. doi: 10.1038/s41467-020-17271-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Baretić D., Jenkyn-Bedford M., Yeeles J.T.P. Cryo-EM structure of the fork protection complex bound to CMG at a replication fork. Mol. Cell. 2020;78:926–940.e13. doi: 10.1016/j.molcel.2020.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Grant T., Rohou A., Grigorieff N. cisTEM, user-friendly software for single-particle image processing. eLife. 2018;7:e35383. doi: 10.7554/eLife.35383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fan X., Wang J., Wang H.-W. Single particle cryo-EM reconstruction of 52 kDa streptavidin at 3.2 Angstrom resolution. Nat. Commun. 2019;10:2386. doi: 10.1038/s41467-019-10368-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Iudin A., Korir P.K., Patwardhan A. EMPIAR: a public archive for raw electron microscopy image data. Nat. Methods. 2016;13:387–388. doi: 10.1038/nmeth.3806. [DOI] [PubMed] [Google Scholar]
- 78.van Heel M., Schatz M. Fourier shell correlation threshold criteria. J. Struct. Biol. 2005;151:250–262. doi: 10.1016/j.jsb.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 79.Penczek P.A. Resolution measures in molecular electron microscopy. Methods Enzymol. 2010;482:73–100. doi: 10.1016/S0076-6879(10)82003-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cardone G., Heymann J.B., Steven A.C. One number does not fit all: mapping local variations in resolution in cryo-EM reconstructions. J. Struct. Biol. 2013;184:226–236. doi: 10.1016/j.jsb.2013.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kucukelbir A., Sigworth F.J., Tagare H.D. Quantifying the local resolution of cryo-EM density maps. Nat. Methods. 2014;11:63–65. doi: 10.1038/nmeth.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Vilas J.L., Gómez-Blanco J., Sorzano C.O.S. MonoRes: automatic and accurate estimation of local resolution for electron microscopy maps. Structure. 2018;26:337–344.e4. doi: 10.1016/j.str.2017.12.018. [DOI] [PubMed] [Google Scholar]
- 83.Chen V.B., Arendall W.B., III, Richardson D.C. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Barad B.A., Echols N., Fraser J.S. EMRinger: side chain-directed model and map validation for 3D cryo-electron microscopy. Nat. Methods. 2015;12:943–946. doi: 10.1038/nmeth.3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Prisant M.G., Williams C.J., Richardson D.C. New tools in MolProbity validation: CaBLAM for CryoEM backbone, UnDowser to rethink “waters,” and NGL Viewer to recapture online 3D graphics. Protein Sci. 2020;29:315–329. doi: 10.1002/pro.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ramírez-Aportela E., Maluenda D., Sorzano C.O.S. FSC-Q: a cryoEM map-to-atomic model quality validation based on the local Fourier shell correlation. bioRxiv. 2020 doi: 10.1101/2020.05.12.069831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Herzik M.A., Jr., Fraser J.S., Lander G.C. A multi-model approach to assessing local and global cryo-EM map quality. Structure. 2019;27:344–358.e3. doi: 10.1016/j.str.2018.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]