Abstract
The ability to quantify protein concentrations and to measure protein interactions in vivo is key information needed for the understanding of complex processes inside cells, but the acquisition of such information from living cells is still demanding. Fluorescence-based methods like two-color fluorescence cross-correlation spectroscopy can provide this information, but measurement precision is hampered by various sources of errors caused by instrumental or optical limitations such as imperfect overlap of detection volumes or detector cross talk. Furthermore, the nature and properties of used fluorescent proteins or fluorescent dyes, such as labeling efficiency, fluorescent protein maturation, photostability, bleaching, and fluorescence brightness can have an impact. Here, we take advantage of previously published fluorescence lifetime correlation spectroscopy which relies on lifetime differences as a mean to discriminate fluorescent proteins with similar spectral properties and to use them for single-color fluorescence lifetime cross-correlation spectroscopy (sc-FLCCS). By using only one excitation and one detection wavelength, this setup avoids all sources of errors resulting from chromatic aberrations and detector cross talk. To establish sc-FLCCS, we first engineered and tested multiple green fluorescent protein (GFP)-like fluorescent proteins for their suitability. This identified a novel, to our knowledge, GFP variant termed short-lifetime monomeric GFP with the so-far shortest lifetime. Monte-Carlo simulations were employed to explore the suitability of different combinations of GFP variants. Two GFPs, Envy and short-lifetime monomeric GFP, were predicted to constitute the best performing couple for sc-FLCCS measurements. We demonstrated application of this GFP pair for measuring protein interactions between the proteasome and interacting proteins and for measuring protein interactions between three partners when combined with a red florescent protein. Together, our findings establish sc-FLCCS as a valid alternative for conventional dual-color fluorescence cross-correlation spectroscopy measurements.
Significance
The quantification of protein concentrations and protein-protein interactions in vivo is a crucial information needed for the understanding of complex processes inside cells. Determination of such information is, unfortunately, still challenging. A fluorescence-based method like fluorescence cross-correlation spectroscopy is the only method that provides this information in vivo and almost in the real time, however it suffers from limitations caused by experimental setup and biological origin of fluorescent proteins. We present single-color fluorescence lifetime cross-correlation spectroscopy as an alternative to fluorescence cross-correlation spectroscopy, which uses the information of fluorescence lifetime to overcome some of these limitations. We challenged the method and determined its advantages and limitations and demonstrated the applicability of the method on the proteins of yeast proteasome.
Introduction
The proteome of a cell is a complex mixture of millions of protein molecules of thousands of different species, all engaged in various types of interactions, from very transient ones to stable protein complexes. The fraction of an individual protein that is engaged in a functional interaction depends not only on the parameter that regulate the interaction, but also its own concentration and the concentration of its interaction partner(s) and of competing binding factors. Therefore, precise determination of protein in vivo concentrations, together with reliable measurements of association and dissociation constants, provides the necessary information to understand the dynamics of such a system. Methods such as immunoprecipitation or ex vivo studies using purified components (1,2) provide qualitative information about the biochemical properties of individual proteins and their interactions. However, they do not necessarily explain the behavior of proteins in the crowded cellular environment with its many constituents. Here, interactions with small molecules and regulatory activities can exert major influences on protein-protein interactions. To overcome this, a number of methods have been developed to study protein-protein interactions in the context of the complex environment of the cell, either using crude protein extracts from lysed cells (Biacore (3)), or using indirect in vivo strategies employing functionalized reporter molecules (such as the “two-hybrid” and “anchor away” techniques (4,5)). For a direct in vivo assessment of protein-protein interactions, fluorescence microscopy can be used to monitor the proximity of molecules using fluorescence resonance energy transfer (FRET) (6,7) or superresolution methods (8, 9, 10). For mobile and dynamic proteins, it is furthermore possible to estimate their interactions by quantification of comobility. One representative of these latter methods is dual-color fluorescence cross-correlation spectroscopy (dc-FCCS) (11,12). This method employs fluorescently labeled species of the molecules under investigation such as proteins tagged with different fluorescent protein reporters (13, 14, 15) or organic fluorescent dyes (16, 17, 18, 19), and it analyzes the fluorescence fluctuations that result from the movement of the labeled molecules in and out of a specified confocal detection volume. Statistical analysis of the fluctuations that result from one or several fluorescently labeled species is termed fluorescence correlation analysis and provides information about the concentration and diffusive behavior (i.e., the diffusion coefficient) of soluble proteins. When conducted for two protein species simultaneously, each labeled with a different fluorophore, dc-FCCS enables the quantification of the fraction of both species that exhibit codiffusion (20). In contrast to FRET, dc-FCCS provides reliable conclusions about the existence or the absence of a protein-protein interaction because the obtained information is independent on the steric arrangement of the fluorophore. A major drawback of fluorescence fluctuation measurements, however, are the many sources of uncertainty associated with the analysis of the data. These originate from constraints imposed by the small measurement volumes of diffraction limited high NA optical systems, the heterogenous optical properties of the cellular interior in which differences in the refractive indices of different cellular structures influence the shape of the detection volume, and a limited number of labeled species present in living cells. The situation is even further complicated by the in vivo properties of fluorescent proteins, e.g., slow maturation of their fluorophore or protein folding. Together with bleaching and the contribution of endogenous proteins, theses cause that not the entire population of the protein of interest is fluorescent. In addition, photophysical properties of the fluorophores, such as blinking or low quantum yield need to be considered (21).
Measurements via dc-FCCS make use of different fluorophores with their specific spectral characteristics. These measurements require different wavelengths for emission and detection of the signals to discriminate the fluorophores from each other. Such dual-color measurements are associated with additional errors that are caused by the different sizes of the detection volumes in each channel, and by light scattering inside the cells that might affect the shape and size of the detection volume in a wavelength-specific manner. This often yields an error that is influenced by the situation present in an individual cell and that is very difficult to correct for. In addition, dual-color measurements suffer from bleed-through of the emitted photons from one fluorophore into the detection channel of the other fluorophore. This bleed-through results in an aberrant cross-correlation signal in which the error associated with the bleed-through correction directly limits the sensitivity by which weak protein-protein interactions can be quantified. In a typical situation using endogenously expressed proteins, this limits the dynamic range for KD measurements to values below 500–1000 nM (22).
Therefore, it is difficult to obtain reliable in vivo estimates of protein concentration and protein-protein interactions from a dc-FCCS measurement conducted in an individual cell. To address these limitations, many measurements performed in a representative population of cells in combination with statistical analysis of the data are required to obtain reliable and reproducible estimates of the desired parameter (22).
To improve the reliability of individual FCCS measurements, it is desired to reduce the number of correction factors that are needed to analyze the fluorescence fluctuation data. To eliminate the volume-overlap problem, various single-excitation wavelength approaches has been reported (23, 24, 25, 26, 27, 28). These methods rely on the combination of fluorescence with light scattering (23), on the differences between the Stokes shifts of the fluorophores (24, 25, 26, 27), or on the two-photon excitation (28), and all of them require two detection channels.
Furthermore, cross-correlation artifacts caused by bleed-through of the emission of one fluorophore into the detection channel of the other can be avoided by pulsed interleaved excitation approach, in which the green and red fluorophores are sequentially excited by the different lasers and the photons are distinguished based on their arrival time with respect to the laser pulse (29).
Here, we now explore whether differences in the fluorescence lifetimes of different green fluorescent proteins (GFPs) can be used to eliminate the volume overlap and bleed-through problem in in vivo FCCS at the same time. For our analysis, we employ the principle of previously published fluorescence lifetime cross-correlation spectroscopy (FLCCS). FLCCS is a modification of FCCS, whereby a pulsed laser is used for fluorophore excitation and where each detected photon is weighted by a fluorescence lifetime, fluorophore-specific component based on its arrival time in relation to the excitation pulse. This enables to “filter” the photons in each detection channel (30, 31, 32) and to statistically eliminate photons that are not emitted from the investigated fluorophore. In principle, with FLCCS, it should also be possible to discriminate fluorophores with very similar spectral properties, provided that the lifetime histograms of their emitted photons are significantly different. With this, a single-color FLCCS (sc-FLCCS) setup could be used for fluorescence cross-correlation experiments, thereby eliminating the requirement for correction of two major sources of errors simultaneously: bleed-through and volume overlap. An extension to the FLCCS is filtered fluorescence correlation spectroscopy (fFCS), which additionally uses the species-specific anisotropy information (33); however, this approach is not implemented in our analysis.
Toward establishing sc-FLCCS, we evaluated first a broad range of GFP variants to identify suitable candidates with very short or very long fluorescence lifetimes. We use simulations, proof of principle in vivo measurements with synthetic constructs and real in vivo measurements with endogenously tagged proteins to challenge the method and to probe its limits. Our results indicate that sc-FLCCS is feasible and that it can be used to assess the interactions of multiple proteins in vivo.
Materials and Methods
Yeast strain construction and cell growth
All yeast strains were constructed using standard procedures as previously described (34) and validated by fluorescence intensity measurements, colony PCR, and, in some cases, by sequencing. Before FCS measurement, yeast cultures were grown in synthetic complete medium (2% glucose) at 30°C (230 rpm) over night until saturation, diluted to OD600 ∼0.1 and grown again to OD600 ∼0.5 (30°C, 230 rpm). Then, the cells were immobilized on the glass surface of the microscopy plates (Greiner Sensoplate; Greiner Bio-One, Kremsmünster, Austria) using Bio-conext (PSX1055; United Chemical Technologies, Levittown, PA) and concanavalin A (22) and covered by low-fluorescence medium (Synthetic Complete medium without riboflavin and folic acid). FCCS/FLCCS measurement was performed within the next 2–3 h by pointing the laser to the cytoplasm of budding cells.
Immunoblotting (Western blotting)
Whole-cell extracts were prepared using the sodium hydroxide and trichloroacetic acid method (35). Proteins were further separated by SDS-PAGE using a 12% polyacrylamide separating gel and transferred onto nitrocellulose membrane (XCell II Blot Module; Invitrogen, Carlsbad, CA). The membrane was incubated overnight with rabbit polyclonal primary anti-GFP antibodies (ab6556; Abcam, Cambridge UK). Peroxidase-conjugated goat anti-rabbit antibodies (111-035-003; Dianova, Hamburg, Germany) were used as secondary antibodies for detection. The visualization was performed on LAS-4000 imaging system (GE Healthcare Life Sciences, Chicago, IL).
Monte-Carlo simulations
As for Monte-Carlo simulations, we extended the work of Wohland et al. (36) by adding excited state lifetime information. The generated data were saved in TTTR data format, particularly in .pt3 files. Random positions of two types of particles (components), which differed in the excited state pattern and/or concentrations, were initially generated and then changed in each simulation step according to two-dimensional Brownian diffusion in a 6-μm square simulation box with periodic boundaries. The diffusion coefficient was fixed to the value of 2.25 μm2 s−1. The detection volume was approximated by a Gaussian profile with a beam waste radius of 300 nm. The simulations were run with 100 ns time steps, corresponding to 10 MHz repetition rate, with a sampling time of 32 ps per TCSPC (Time-Correlated Single Photon Counting) channel, each time trace was 180 s long. A TCSPC channel for every generated photon was chosen by random selection from a look-up table corresponding to component’s excited state pattern (exponential with different lifetimes, Gaussian with different peak positions, or experimental). The simulated molecular brightness was 100 kHz per molecule. To save simulation time, data with lower molecular brightness were generated from this 100-kHz data set by random deletion of individual events (photons). The nondelete probability corresponded to the ratio of the targeted and the original brightness for the given component.
Fluorescence cross-correlation spectroscopy
Fluorescence correlation spectroscopy (11,12) is based on the statistical analysis of the timescale intensity fluctuations I(t). Such dependence is described by the normalized auto- and cross-correlation functions GAC and GCC, which are defined as
| (1) |
whereτ is the time lag, angle brackets denote the averaging over all possible values of time t, and I1(t) and I2(t) are the intensity fluctuations in two different detection channels. In the case of Brownian motion in a three-dimensional Gaussian detection volume, assuming intersystem crossings and two species of molecules with distinct diffusion characteristics, the autocorrelation function can be described by the following equation:
| (2) |
where T and τ0 are the contribution and kinetics of intersystem crossing, and PN corresponds to the number of particles in the detection volume. τDa and τDb correspond to the average times of diffusing species a and b, for which the fluorescence molecules stay in the detection volume. A corresponds to the relative amplitude of the autocorrelation function with diffusion time τDa and is given by , where qi corresponds to the molecular brightness of the i-th species. The ω0 and ωz are the spatial parameters of the detection volume. Thus, the profile of the autocorrelation function bears the information about the concentration and diffusion properties. The concentration can be calculated as:
| (3) |
where PN1, PN2, and PNcc correspond to the number of particles of individual species and their complex, Veff is the effective confocal volume and NA is the Avogadro number. The interaction between two proteins of interest can be represented by the apparent dissociation constant which is defined as:
| (4) |
where c1, c2, and cCC are the concentrations of species in channel 1, 2, and their complex.
Fluorescence lifetime correlation spectroscopy
Fluorescence lifetime histograms are measured by time correlated single-photon counting, in which the time axis is divided into small parts (bins). The width of the bin depends on the time resolution. The mathematical expression for the overall fluorescence histogram Ij is:
| (5) |
where j corresponds to the bin number, and L corresponds to the number of decay components indexed by k. w(k) is the amplitude of the photon count contribution of the k-th species and pj(k) is the fluorescence decay pattern of k-th species alone, which is equal to its fluorescence lifetime histogram and is usually measured separately. In FLCCS (30, 31, 32,37) the normalized autocorrelation function is described by the following expression:
| (6) |
meaning that each photon is multiplied by a statistical filter fj, which corresponds to the fluorescence decay component k. The formula for the weighting factor fj(k)is:
| (7) |
where the dot, superscript T, and −1 denote matrix multiplication, transposition, and inversion, respectively. The is a L × L – dimensional diagonal matrix with diagonal elements corresponding to the fluorescence decay histogram , j = 1,…,L.
Microscope setup
FCCS/FLCCS were measured using a Nikon TiE body combining an inverted wide-field fluorescence microscope with a confocal MicroTime 200 unit (PicoQuant, Berlin, Germany) for time-resolved photon counting. Picosecond pulsed laser diode head (LDH-D-C-485; PicoQuant) with a 20 MHz repetition rate or a single-frequency continuous wave diode pumped laser (Cobolt Jive 561 nm; Cobolt, Solna, Sweden) were used for the excitation of green and red fluorophores. The collimated laser beam was coupled into an optical fiber for optical cleaning and then reflected using a beam splitter (zt 488/561rpc; AHF Analysentechnik, Tübingen, Germany) into the inverted microscope body (Nikon Eclipse Ti; Nikon Instruments Europe B.V., Amsterdam, the Netherlands). The sample was illuminated using a water immersion objective (Nikon, Plan Apo IR, 60×/NA 1.27; Nikon Instruments Europe B.V.) and the same objective was used for collection of the fluorescence light. Emitted light passed through the 50-μm pinhole, band-pass emission filters (ET525/50 m and ET632/60 m; Chroma Technology, Bellows Falls, VT), and was detected by a τ-SPAD single-photon avalanche photodiode (PicoQuant). Low laser intensities (<5 μW for 485 and 561 nm) were used to prevent photobleaching and pile-up effect (in case of 485-nm excitation). All data were measured at 19°C. The size of the detection volume was determined using calibration dyes Atto 488 (D = 390 μm2 s−1, T = 19°C) and Atto 565 (D = 390 μm2 s−1, T = 19°C). Volume overlap was determined using double-labeled in vitro FCCS standards for 488–543 nm (IBA life sciences, Göttingen, Germany).
Analysis pipeline
All experimental data was analyzed using a custom-made data analysis pipeline (Fig. S1) written in MATLAB (MathWorks, Natick, MA). For single-wavelength analysis using two different GFPs with different lifetime histograms, abbreviated as sc-FLCCS, we first measured fluorescence lifetime histograms of pure individual fluorescent proteins (FPs) (using protein fusions to the endogenous gene) and use them as reference patterns. Second, the reference patterns were loaded together with the overall lifetime histogram of the sample of interest and corresponding weighting filters were calculated according to the Eq. 7 (Document S1. Figs. S1–S8 and Table S1, Document S2. Article plus Supporting Material a). In the case of double-color excitation (excited by pulsed 485-nm laser and continuous 561-nm laser), abbreviated as dc-FLCCS for dual-color FLCCS, lifetime filtering was used to correct for the bleed-through of the photons that were excited by the pulsed 485-nm laser and detected in the red channel. The weighting filters were calculated from the fluorescence lifetime histograms (Eq. 7) detected in the green channel. The “positive” filter was used for filtering from the green detector and complementary “negative” filter was applied to the red detector (Fig. S2 b). Further mathematical operations (e.g., corrections and correlations) were common to both the sc-FLCCS and the dc-FLCCS.
Next, we corrected for photobleaching by dividing the overall intensity time trace into a set of shorter time intervals (3-s intervals in our case), which were correlated individually (Eq. 6; (38)). The final correlation curve was derived by averaging the multiple short-interval based correlation curves. Resulting auto- and cross-correlation curves were fitted with the model described in Eq. 2. Finally, the concentrations and apparent dissociation constants were calculated (Eqs. 3 and 4).
Results
Characterization of green fluorescence proteins
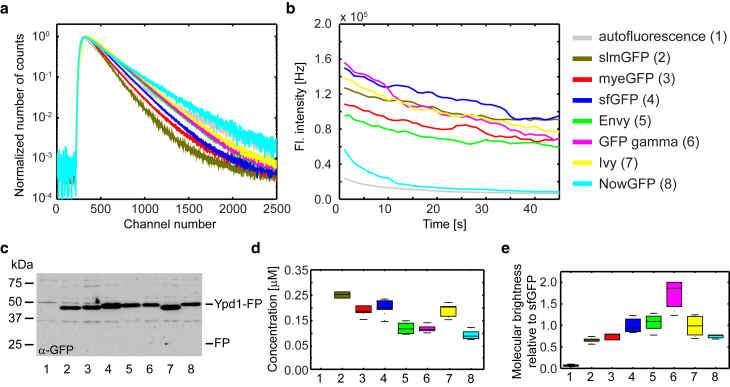
To establish sc-FLCCS, we first aimed to identify fluorescent proteins with optimal properties. To this end, there are many publications that report different GFPs; however, only for a few of them information about the fluorescence lifetime is available. To obtain this information, we selected 15 different GFP variants (Table 1): Envy and Ivy, NowGFP, GFPγ, monomeric yeast enhanced GFP (myeGFP), superfolder GFP with and without the F64L mutation for improved folding at 37°C (sfGFP) and circular permutations of sfGFP (cp3, cp7, and cp8), sfGFP without the superfolder mutation, mNeonGreen, Clover (with and without the F64L mutation for improved folding at 37°C), and slmGFP (a new GFP variant combining different mutations (39, 40, 41)). We furthermore used yeast-codon-optimized variants of the corresponding genes for their expression in yeast to test their fluorescence lifetimes and other properties (Table 1). This identified lifetimes in the range of 1.8–3.9 ns, with slmGFP exhibiting the shortest lifetime of all. Using cells without fluorescent protein expression, we also observed the weak cellular autofluorescence with a highly multiexponential lifetime in the range of 2.4 ns. This indicates multiple sources for this autofluorescence, in particular riboflavins, a common source of autofluorescence in yeast (42). Because of very low-fluorescence signal, we have neglected the contribution of the autofluorescence in the data analysis. For further analysis, we selected the three GFPs with the shortest lifetimes and the four GFPs with the longest lifetimes for further evaluation (Fig. 1 a; bold rows in Table 1).
Table 1.
Summary of Fluorescence Properties of Various Green Fluorescence Proteins
| Protein | Average Fluorescence Lifetime (ns) | Absmax (nm) | Emmax (nm) | Molecular Brightness (a.u.) | Fluorescence Stability (a.u.) | Maturation (min) |
|---|---|---|---|---|---|---|
| Autofluorescence | 2.40 ± 0.20 | |||||
| slmGFP | 1.80 ± 0.15 | 501 | 513 | 0.65 ± 0.07 | 0.97 ± 0.13 | 4a (59) |
| myeGFP (22) | 2.00 ± 0.18 | 501 | 513 | 0.76 ± 0.41 | 0.94 ± 0.19 | n.d. |
| sfGFP (60) | 2.50 ± 0.14 | 487 | 511 | 1.00 ± 0.18 | 0.90 ± 0.07 | 6 (61) |
| sfGFP_cp8 (62) | 2.52 ± 0.20 | 487 | 510 | n.d. | 0.77 ± 0.14 | n.d. |
| sfGFP_cp7 (62) | 2.72 ± 0.11 | 486 | 510 | n.d. | 0.87 ± 0.06 | n.d. |
| sfGFP_cp3 (62) | 2.78 ± 0.04 | 487 | 510 | n.d. | 0.84 ± 0.10 | n.d. |
| no-sfsfGFP (62) | 2.84 ± 0.06 | 489 | 511 | n.d. | 0.90 ± 0.08 | n.d. |
| mNeonGreen (63) | 2.90 ± 0.12 | 506 | 517 | 1.71 (63) | 0.80 ± 0.27 | <10 (63) |
| Clover (F64L) (62) | 3.00 ± 0.03 | 506 | 517 | n.d. | 0.67 ± 0.12 | n.d. |
| Clover (64) | 3.00 ± 0.09 | 505 | 517 | 1.56 (63) | 0.69 ± 0.37 | 30 (62) |
| sfGFP (L64F) (62) | 3.10 ± 0.01 | 487 | 511 | 0.78 ± 0.09 | n.d. | |
| Envy (65) | 3.23 ± 0.02 | 486 | 509 | 1.06 ± 0.21 | 0.92 ± 0.12 | n.d. |
| GFPγ (66) | 3.33 ± 0.08 | 493 | 509 | 1.71 ± 0.35 | 0.78 ± 0.25 | n.d. |
| Ivy (65) | 3.44 ± 0.06 | 500 | 515 | 0.98 ± 0.25 | 0.84 ± 0.12 | n.d. |
| NowGFP (10 s) (53) | 3.90 ± 0.09 | 492 | 502 | 0.79 ± 0.14 | 0.33 ± 0.13 | n.d. |
Proteins that were selected for further studies are indicated in bold. Average fluorescence lifetime corresponds the intensity weighted fluorescence lifetime. Fluorescence stability is defined as the percentage of fluorescence remaining after 20 s of illumination (corrected for the autofluorescence). Molecular brightness (as defined in the text) and fluorescence stability values correspond to the mean of five measurements per strain. Errors are expressed by standard deviations. Abs, absorption; a.u., arbitrary unit; Em, emission; n.d., not determined.
In contrast to the other maturation times that integrate protein folding and fluorophore maturation together, this value here only relates to fluorophore oxidation.
Figure 1.
Fluorescence properties of selected green fluorescence proteins. Different fluorescence proteins (color coding and numbering are consistent throughout the whole figure) were expressed in yeast as an endogenously expressed C-terminally tagged Ypd1 fusion protein. (a) Fluorescence lifetime histograms normalized to the maximum of each curve are shown. (b) Raw fluorescence intensity traces that were measured for 45 s are shown. Presented data in (a) and (b) correspond to average traces from five measurements. (c) Immunoblot and detection of the Ypd1-GFP fusion proteins using anti-GFP antibodies are shown. Please note that this does not allow direct comparison of protein levels because different GFP variants may cover different range of epitopes recognized by the polyclonal anti-GFP antibodies. No degradation products were detected, indicating that no free GFP is present inside the cells (62). (d) Absolute protein concentrations as determined by FCS are shown. (e) Fluorescence molecular brightness normalized to the mean of Ypd1-sfGFP. Data in (d) and (e) are the result of five measurements each. Boxes in the boxplot correspond to the first and third quartiles, the horizontal line represents the median. The whiskers show first quartile − 1.5 × interquartile range and third quartile + 1.5 × interquartile range, respectively (boxplot description is consistent throughout the whole manuscript).
To further characterize the selected GFP variants, we created yeast strains that endogenously expressed these proteins as C-terminal fusion to the cytoplasmic and nuclear localized yeast protein Ypd1. We then used these strains to compare different fluorescent proteins. To determine the photostability, we used a constant excitation intensity for all strains and acquired fluorescence intensity time traces for 45 s each. This revealed that NowGFP, the protein with the longest lifetime, was highly sensitive to photobleaching, whereas no major differences were detected between the other GFP variants (Fig. 1 b). Western blotting revealed that they were expressed to similar levels, indicating that none of the proteins affected the expression of the fusion protein in a major way (Fig. 1 c). For brightness comparison, we quantified the concentration of different Ypd1-GFP fusions using FCS and used these measurements (Fig. 1 d) to normalize the measured fluorescence intensities. This procedure gave us apparent molecular brightness (here, referred as molecular brightness) and it revealed that GFPγ is, by far, the brightest GFP variant, whereas all the others exhibited similar molecular brightness (Fig. 1 e; Table 1). It has to be noted that these values are only valid for the used system (excitation wavelength, 485 nm; major dichroic, zt 488/561 rpc; emission filter, ET525/50 m) because the different GFP variants exhibit different excitation and emission optima (Table 1).
Limits of sc-FLCCS examined by Monte-Carlo simulations
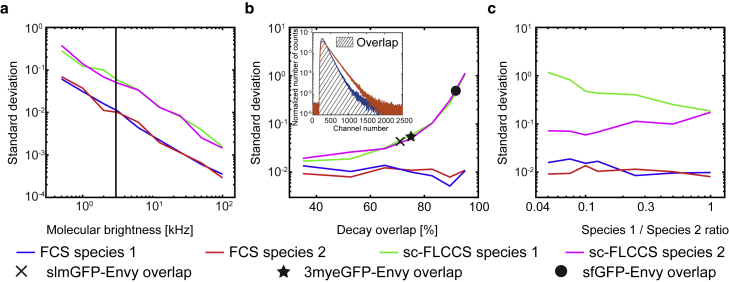
Molecular brightness, bleaching sensitivity, and the fluorescence lifetime histograms are the basic characteristics of fluorescence proteins that contribute to the detection sensitivity of protein-protein interactions in sc-FLCCS; however, systematical study of the impact of these parameters on the sc-FLCCS analysis is difficult to obtain experimentally. To explore how much each factor contributes to the overall performance of an individual fluorescent protein, we used Monte-Carlo simulations and conducted virtual sc-FLCCS experiments. Thereby, we simulated raw fluorescence photon events and generated simulated sc-FLCCS data. In contrast to real data, in simulated data, the origin of individual photons is known. The simulated data was then correlated using conventional FCS analysis, incorporating knowledge about the origin of the photons, or the data was correlated using sc-FLCCS approach, omitting the information about the origin of photons. The reference standard deviation, which allows comparison of signal-to-noise ratios between FCS and sc-FLCCS autocorrelation curves, was calculated from the values of the first 15 points of each autocorrelation curve. The examples of autocorrelation curves are shown in Fig. S3. We first used this strategy to test the impact of molecular brightness. As expected, increased molecular brightness of fluorescent species improves the quality of the correlation curves in both approaches, whereas lifetime filtering decreases the quality of the calculated correlation curves as indicated by an overall increased standard deviation for sc-FLCCS-derived curves (Fig. 2 a). Next, we tested how the overlap of fluorescence lifetime histograms between the two different green fluorescent proteins affects the quality of the correlation curves (Fig. 2 b). This revealed that the overlap has a significant impact, with a higher overlap worsening the quality of the correlation curves. To find out whether the impact of the lifetime filtering on the quality of the correlation curves does depend on the shape of the histograms, we tested two different series: one series based on exponential histograms (inspired by fluorescence lifetime filtering) and a second series based on Gaussian-distributed histograms (inspired by fluorescence spectral filtering (43)). This revealed no dependence on the shape of the histograms used for filtering (Fig. S4) and a sole dependence on the overlap of the area-normalized histograms.
Figure 2.
Quantification of the quality of resulting sc-FLCCS data using Monte-Carlo simulations. (a) Simulation of the impact of molecular brightness on the quality of autocorrelation curves – fluorescence lifetime histograms with 75% overlap is shown. (b) The quality of the autocorrelation curves depends on the extent of overlap of the fluorescence lifetime histograms. In the insert, schematic illustration of the overlap of area-normalized fluorescence lifetime histograms is shown. (c) Higher difference in the species abundance (and implicitly other fluorescence properties of the fluorophores) decreases the data quality for the low abundant species. Data in (b) and (c) are plotted for the molecular brightness of 3 kHz (indicated as a vertical line in (a)), which is a representative brightness of fluorescent proteins with our experimental conditions.
The quality of the correlation curves could depend on the relative abundance of the two fluorescent species. To address this, we varied the concentration of one species while keeping the concentration of the other constant (Fig. 2 c). In the case of standard FCS and assuming no spectral cross talk at all, the concentration does not affect the results. In contrast, sc-FLCCS showed a strong dependence on the relative abundance of species, with the best result for abundances of both species in the same concentration range. Together the analysis revealed that sc-FLCCS appears to be a valid alternative to dc-FCCS, but also that it has other intrinsic limitations imposed by the measurement principle.
Testing of selected FPs for sc-FLCCS analysis
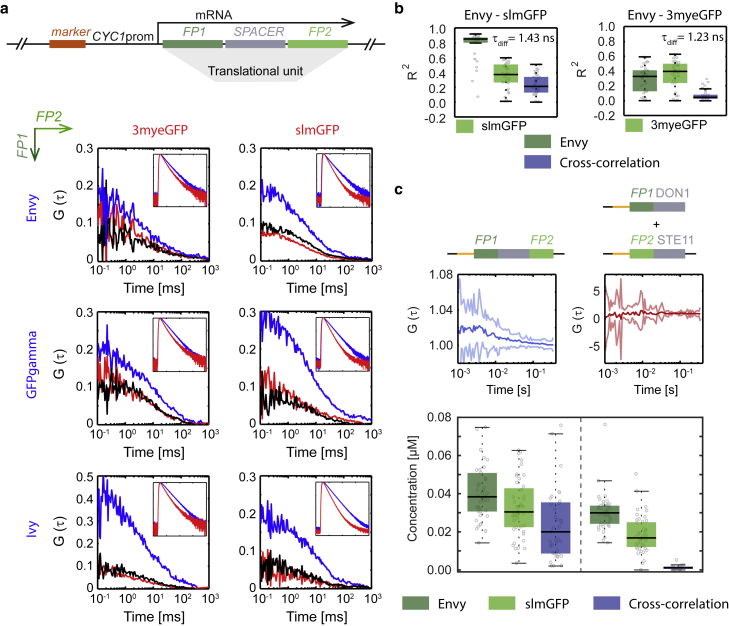
Our simulations demonstrate that the ability to separate correlation curves from different fluorophores with the same emission spectra depends on the difference in fluorescence lifetime histogram profiles, their molecular brightness, and, to some extent, also their relative concentrations. To establish sc-FLCCS in vivo, we chose the two GFP variants with the shortest lifetime (3myeGFP and slmGFP) and tested each of them in combination with the three GFP variants with the longest lifetimes (Envy, GFPγ, and Ivy). We constructed strains in which we expressed pairs of these GFP variants as N- and C-terminal fusion to the yeast protein Don1, which served as a spacer to keep the two fluorescent proteins with different lifetimes apart from each other (22). Next, we performed a sc-FLCCS measurement. For calculation of the fluorescence lifetime filters (see Materials and Methods), we used the strains that expressed individual Ypd1-FP fusion proteins (Fig. 1). This experiment revealed that the selected FP combinations provide data that is suitable for sc-FLCCS analysis (Fig. 3 a). As expected from our simulations (Fig. 2 b), the quality of the auto- and cross-correlation curves differs in terms of noise. Because both FPs are expressed as a part of the same translational unit, their concentrations and thus amplitudes of the autocorrelation functions should be the same. This is not the case in any of the tested FP combinations presumably because of the differences in fluorophore maturation time and photostability (Fig. 3 a; Table 1).
Figure 3.
Proof of principle. (a) Shown at the top is a schematic illustration of the tandem fluorescent protein fusion, with N- and C-terminally tagged spacer protein (Don1). Shown at the bottom, filtered auto- and cross-correlation functions were calculated for selected combinations of FPs. Fluorescence proteins in rows (blue lines) were attached N-terminally and proteins in columns C-terminally (red lines). Black lines correspond to the cross-correlation curves. Insets show corresponding fluorescence lifetime histograms (x axis is number of channels ranging from 0 to 2000 and y axis equate to normalized number of events ranging from 10−4 to 10°). (b) Comparison of the sc-FLCCS analysis for selected pairs of FPs as determined by R2-values. The higher the R2-value, the better the sc-FLCCS filtering and consecutive analysis. The difference in average fluorescence lifetimes between FPs (τdiff) is listed in the upper right corner of the corresponding box plot. (c) Comparison of the positive (left) and the negative (right) controls is shown. At the top is schematic illustrations of the strains. Orange lines denote weak CYC1 promoter. In the middle, cross-correlation curves, where solid dark line corresponds to the mean curve, light borders, denote standard deviations. At the bottom are the overall concentrations resulted from autocorrelation curves for each diffusing species and their complexes as determined by the sc-FLCCS analysis. Dashed line separates individual strains.
We further selected three combinations of FP pairs and tested them for sc-FLCCS application (Fig. 3 a; Fig. S5). The selected pairs were as follows: Envy and slmGFP (lifetime difference, 1.43 ns; histograms’ overlap, 71%), Envy and 3myeGFP (lifetime difference, 1.23 ns; histograms’ overlap, 75%), and Envy and sfGFP (lifetime difference, 0.73 ns; histograms’ overlap, 92%). The quality of the resulted auto- and cross-correlation curves was quantified using the R2-value of the fit (Fig. 3 b; Fig. S6 for Envy and sfGFP couple). The results correlate with the simulations, as shown by the black cross, black solid star, and black solid circle in Fig. 2. This confirms the result from the simulations that the difference in the fluorescence lifetime histograms of the two used fluorescent proteins is a major factor influencing sc-FLCCS measurements.
Because of the best resolution, the pair slmGFP and Envy was chosen for further studies. In addition to the tandem fusion of both proteins (Envy-Don1-slmGFP) we also designed a negative control in which both proteins are expressed independently (slmGFP-Don1 and Envy-Ste11) and no interaction was expected (22). We observed positive cross-correlation in the tandem fusion construct and no cross-correlation in the strain with independent expression units (Fig. 3 c). This validates the concept of sc-FLCCS.
In summary, the best FP candidates for lifetime filtering are slmGFP, as the partner with the short fluorescence lifetime, and Envy, as the long fluorescence lifetime partner, both with reasonable brightness and good photostability.
Challenging the limits of the sc-FLCCS analysis
FRET between two fluorophores depends on the spectral overlap, distance between the fluorophores, and their mutual orientation. Because the FPs in sc-FLCCS are spectrally highly similar and might be spatially relatively close to each other, we have to take into account the energy transfer. To test the impact of FRET, we prepared a construct in which FPs are in a tandem in one translational unit (Document S1. Figs. S1–S8 and Table S1, Document S2. Article plus Supporting Material a). This is an extreme case in which all the FPs are in the closest possible proximity, which is usually not the case for interacting tagged proteins. The fluorescence histograms were affected by the energy transfer, and correct lifetime filtering was not possible. Thus, we obtained zero cross-correlation (Fig. S7 b) and calculated concentrations were either overestimated (Envy) or underestimated (slmGFP; Document S1. Figs. S1–S8 and Table S1, Document S2. Article plus Supporting Material c; compare the values with Fig. 3 c). This indicates that sc-FLCCS is sensitive to FRET. Using appropriate controls, e.g., specimen in which only one of the components is tagged, is required to ensure that there is no FRET occurring.
We also tested performance of sc-FLCCS on the strains in which the expression levels of two tagged proteins significantly differ. We C-terminally tagged Ssk1 and Ypd1 (Document S1. Figs. S1–S8 and Table S1, Document S2. Article plus Supporting Material a), proteins of the high-osmolarity glycerol pathway, which have different expression levels as confirmed by the fluorescence lifetime histograms (Fig. S8 b) and FCS (Document S1. Figs. S1–S8 and Table S1, Document S2. Article plus Supporting Material c). Comparison of the single-tagged strains with the double-tagged strains shows that the concentration of higher abundant Ypd1 protein is similar; however, the concentration of low abundant Ssk1 proteins is underestimated in double-tagged strain (Document S1. Figs. S1–S8 and Table S1, Document S2. Article plus Supporting Material c). This is in full agreement with the simulations (Fig. 2 c). It has been reported that Ssk1 and Ypd1 proteins strongly interact (44), but we were not able to reproduce this interaction by the sc-FLCCS analysis of double-tagged strain. When the photons in the fluorescence lifetime histogram mostly correspond to just one of the species, proper calculation of the filters becomes difficult, which results in underestimation of the low abundant protein and no cross-correlation.
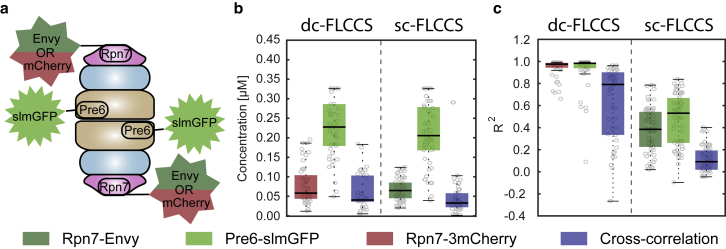
sc-FLCCS of proteasomal subunits
Next, we used sc-FLCCS to monitor the interaction between two proteins that are spatially well spaced so that FRET is unlikely to occur. We chose components of the proteasome, a large multisubunit protease that functions as the major proteolytic activity in the cytoplasm of the cell, with functions in protein degradation and regulation. The proteasome consists of different substructures, i.e., the core particle that contains the active sites of the protease, and the base and the lid complexes that can dynamically interact with the core particle and that regulate access of ubiquitylated substrate proteins to the core particle (Fig. 4 a; (45)). We constructed strains in which the core protein Pre6 was tagged with slmGFP and the lid protein Rpn7 was tagged with either Envy or 3mCherry. The distance between these proteins is ∼12 nm, well beyond the range of FRET. We performed fluorescence fluctuation measurement and compared the results from sc-FLCCS (single-color) and dc-FLCCS (dual-color) analysis (Fig. 4, b and c, dashed lines separate different strains). First, we determined the overall concentrations of individual tagged proteins and quantified the amount of those that form the proteasome (Fig. 4 b). We conclude that sc-FLCCS analysis of proteasomal proteins provide the same results as dc-FLCCS experiment. Considering R2 as a proxy for the quality of the data, we found that the sc-FLCCS data could be less well fitted compared to the dc-FLCCS data. This indicates that filtering of photons in sc-FLCCS, which contains statistical uncertainty of photon origin, leads to data that is noisier than the dc-FLCCS data, in which the origin of each photon is known. Nevertheless, the sc-FLCCS yielded valid data, demonstrating the applicability of the method (Fig. 4 c).
Figure 4.
Interplay between Pre6 and Rpn7 examined by sc-FLCCS analysis. (a) Schematic organization of the yeast proteasome is shown. Tagged proteins are indicated. Concentrations (b) and quality of the fit expressed by R2 values (c) are shown. Overall concentrations resulted from autocorrelation curves for each diffusing species and their complexes as determined by the sc-FLCCS analysis. Dashed lines separate individual single- or multiple-tagged strains.
Simultaneous monitoring of three proteasomal proteins using two channels
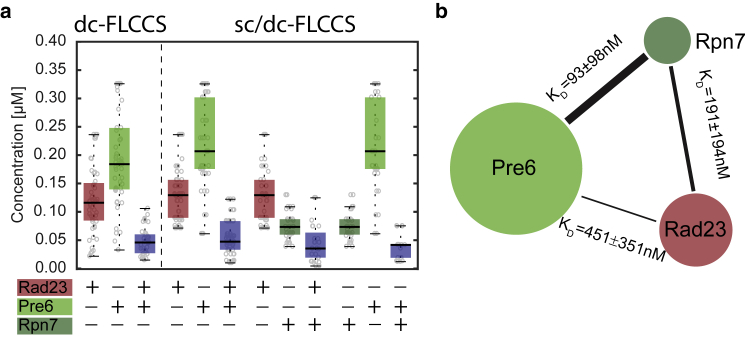
Measuring protein-protein interactions between three proteins inside the same cell is difficult. We decided to explore whether sc-FLCCS can be combined with dc-FLCCS. We chose the protein Rad23 as a third protein partner. Rad23 is a protein with an N-terminal ubiquitin like domain with several nuclear and cytoplasmic functions related to DNA damage repair and targeting of substrates to the proteasome (46, 47, 48). It has been reported that the E4 ubiquitin ligase Ufd2 and Rpn1, a component of the base part of the proteasome, compete for binding to Rad23. We used Rad23 tagged with 3mCherry, Pre6 with slmGFP and Rpn7 with Envy to perform a dual-color sc/dc-FLCCS experiment. For comparison, we used standard dc-FLCCS experiment in which only the interaction between Rad23 and Pre6 was monitored (Fig. 5 a). The concentrations of individual proteins were converted to the scheme and the strength of the interaction was quantified by the apparent dissociation constant (Fig. 5 b). These results confirmed that dual-color sc/dc-FLCCS enabled the detection of protein interactions between three different proteins of the proteasome using only one measurement in one strain.
Figure 5.
FLCCS analysis of a strain with three tagged proteasomal proteins. (a) Overall concentrations of individual proteins and concentrations resulted from cross-correlation are shown. Dashed line separates different strains. The analysis of triple-tagged strain required the combination of sc-FLCCS and dc-FLCCS (sc/dc-FLCCS). (b) Interaction map between Pre6, Rpn7, and Rad23 is shown. The size of the bubble illustrates the protein abundance, and the thickness of the line between bubbles refers to the strength of the interaction between corresponding proteins. The errors are represented by the standard deviations.
Discussion
So far, sc-FLCCS (30,37) was almost exclusively used for in vitro biophysical studies on model membrane systems (31,32,37,49). There are also reports on sc-FLCCS studies performed in in vivo (50, 51, 52), but up to date, it was not possible to perform fluorescence lifetime filtering on two different fluorescent proteins with matching spectral properties in vivo, either because of high similarity in their fluorescence lifetimes or because of insufficient photophysical properties of those proteins with respect to photostability and molecular brightness.
To use fluorescent lifetime filtering as a means to discriminate different fluorophores with the same spectral properties, we needed to identify GFP variants with optimal in vivo performance and extreme lifetimes, either very short or very long. From our experience, it is often not possible to derive these from published data because these values were often determined in vitro or using different organisms. This is exemplified for NowGFP, which was reported as highly stable variant in vitro and in vivo in Escherichia coli, Drosophila, and mammalian cells (53,54), but turned out to be highly photo-unstable in yeast, at least when imaged using our setup. This thus prompted us to evaluate several other green fluorescent proteins for sc-FLCCS applications ourselves.
We used Monte-Carlo simulations to explore the impact of different properties of fluorescence proteins on sc-FLCCS. We found that the critical parameter is indeed the difference in the fluorescence lifetime histograms of the used fluorescent proteins. A large overlap of the lifetime histogram means, in simple terms, that the uncertainty in photon assignment is too high for successful filtering. To compensate for this loss, optimal performance parameters for other properties of fluorescent proteins are needed to maximize the available overall “photon budget,” i.e., molecular brightness and photobleaching resistance. For further improvement, new fluorescent proteins with longer lifetimes would be needed. Alternatively, applications in which one of the GFPs is replaced by chemical dye labeling, e.g., using SNAP or HALO tags (55,56), should be possible in organisms in which such dye-labeling strategies are feasible (which is not easily the case in Saccharomyces cerevisiae). Chemical dyes exhibit a much broader range of physicochemical properties, including much longer lifetimes (57). Thereby, much more precise “photon assignment” can be reached, thus enabling “perfect” sc-FLCCS applications.
Our results demonstrate that sc-FLCCS analysis is an alternative and valid approach to the standard dual-color FCCS or dc-FLCCS methods. The advantage of the sc-FLCCS over standard dual-color methods is the reduction of two excitation wavelengths to only one while being able to resolve two spectrally similar fluorescence proteins. This is based on their differences in fluorescence lifetime histograms, but it also requires that these are determined for individual fluorescence proteins before the sc-FLCCS analysis. Moreover, the fluorescence lifetime does not depend on the protein abundance, excitation wavelength, filter configuration, and photobleaching, which is a definite advantage of this method.
A disadvantage of sc-FLCCS, however, apart from the “increased uncertainty due to filtering,” is the sensitivity of the method to FRET, which affects the lifetime histograms, impedes the analysis, and essentially makes the method useless in the case of two tightly interacting small proteins (Document S1. Figs. S1–S8 and Table S1, Document S2. Article plus Supporting Material c). However, the FRET efficiency decays with the sixth power of the distance, which makes sc-FLCCS suitable especially for large complexes. It is important to note that FRET does also affect dual-color FCCS. In the case of EGFP and mCherry, typical green and red FPs used in dual-color FCCS, which have highly overlapping emission (EGFP) and excitation spectra (mCherry), it causes an artificial underestimation (green channel) and overestimation (red channel) of the concentration of the individual fluorophores and it also affects the amount of interaction derived from cross-correlation. This fact is usually not considered when performing classical dual-color FCCS experiment in vivo. An additional disadvantage of sc-FLCCS over standard dc-FLCCS is the requirement that both concentrations shall be in the same range, at least for situations with two fluorescent proteins with a significant lifetime histogram overlap.
Nevertheless, in our proof of concept experiment, we have demonstrated that sc-FLCCS in combination with dc-FLCCS allows to distinguish and quantify the interaction of three proteins, measured simultaneously in one strain in one measurement using an instrument set up for two wavelengths only. Further improvements of sc-FLCCS toward four interaction partners is also thinkable, given the existence of new variations of red fluorescence proteins with prolonged fluorescence lifetime (e.g., mScarlet and its variants (58)). Combination of mScarlet (fluorescence lifetime 3.9 ns) and mCherry (fluorescence lifetime 1.5 ns) would be a perfect RFP-like couple for sc-FLCCS experiment in the red spectra. Thus, dual-color sc-FLCCS (sc in green + sc in red channels) could provide the information about four different proteins in just one measurement. In classical two-color dc-FLCCS experiment one would need pair wise measurements, which would require six strains to measure all interactions. Using dual-color sc-FLCCS decreases significantly the necessary measurement time when determining the interaction map of multiple proteins in vivo.
In summary, we have presented a possibility to use single-color FLCCS to analyze the interaction of two proteins in vivo and we have explored and discussed the advantages and disadvantages when compared to dc-FLCCS. Our in vivo measurements of important green fluorescent protein variants furthermore enables the selection of best couple of GFP-like fluorescence proteins not only for sc-FLCCS but also for potential applications such as fluorescence lifetime imaging, and we demonstrated the usefulness of the sc-FLCCS method in determination of the concentrations and intermolecular interactions in yeast cells.
Author Contributions
M.Š. and M.K. designed experiments. M.Š., K.H., and M.R. performed experiments and processed data. A.B. performed Monte-Carlo simulations. M.Š. and M.K. wrote the manuscript.
Acknowledgments
The authors thank to Dr. Alexander S. Mishin for providing the NowGFP-containing plasmids.
The authors acknowledge support from the German Research Foundation (Grant INST 35/1133-1 FUGG). M.Š. was supported by fellowships from Cellnetworks and the Alexander von Humboldt Foundation. M.Š. also thanks to the Czech Science Foundation (Grant 19-08304Y). A.B. acknowledges ERD Fund-Project No. CZ.02.1.01/0.0/0.0/16_013/0001775.
Editor: Samrat Mukhopadhyay.
Footnotes
Supporting Material can be found online at https://doi.org/10.1016/j.bpj.2020.06.039.
Contributor Information
Martin Štefl, Email: martin.stefl@jh-inst.cas.cz.
Michael Knop, Email: m.knop@zmbh.uni-heidelberg.de.
Supporting Material
References
- 1.Mazet F., Dunster J.L., Gibbins J.M. A high-density immunoblotting methodology for quantification of total protein levels and phosphorylation modifications. Sci. Rep. 2015;5:16995. doi: 10.1038/srep16995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Taylor S.C., Berkelman T., Hammond M. A defined methodology for reliable quantification of Western blot data. Mol. Biotechnol. 2013;55:217–226. doi: 10.1007/s12033-013-9672-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Leonard P., Hearty S., O’Kennedy R. Measuring protein-protein interactions using biacore. Methods Mol. Biol. 2017;1485:339–354. doi: 10.1007/978-1-4939-6412-3_17. [DOI] [PubMed] [Google Scholar]
- 4.Fields S., Song O. A novel genetic system to detect protein-protein interactions. Nature. 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 5.Haruki H., Nishikawa J., Laemmli U.K. The anchor-away technique: rapid, conditional establishment of yeast mutant phenotypes. Mol. Cell. 2008;31:925–932. doi: 10.1016/j.molcel.2008.07.020. [DOI] [PubMed] [Google Scholar]
- 6.Hannan L.A., Lisanti M.P., Edidin M. Correctly sorted molecules of a GPI-anchored protein are clustered and immobile when they arrive at the apical surface of MDCK cells. J. Cell Biol. 1993;120:353–358. doi: 10.1083/jcb.120.2.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Margineanu A., Chan J.J., French P.M.W. Screening for protein-protein interactions using Förster resonance energy transfer (FRET) and fluorescence lifetime imaging microscopy (FLIM) Sci. Rep. 2016;6:28186. doi: 10.1038/srep28186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hell S.W., Wichmann J. Breaking the diffraction resolution limit by stimulated emission: stimulated-emission-depletion fluorescence microscopy. Opt. Lett. 1994;19:780–782. doi: 10.1364/ol.19.000780. [DOI] [PubMed] [Google Scholar]
- 9.Betzig E., Patterson G.H., Hess H.F. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006;313:1642–1645. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- 10.Rust M.J., Bates M., Zhuang X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM) Nat. Methods. 2006;3:793–795. doi: 10.1038/nmeth929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Magde D., Elson E.L., Webb W.W. Fluorescence correlation spectroscopy. II. An experimental realization. Biopolymers. 1974;13:29–61. doi: 10.1002/bip.1974.360130103. [DOI] [PubMed] [Google Scholar]
- 12.Elson E.L., Magde D. Fluorescence correlation spectroscopy. I. Conceptual basis and theory. Biopolymers. 1974;13:1–27. doi: 10.1002/bip.1974.360130103. [DOI] [PubMed] [Google Scholar]
- 13.Rizzuto R., Brini M., Pozzan T. Chimeric green fluorescent protein as a tool for visualizing subcellular organelles in living cells. Curr. Biol. 1995;5:635–642. doi: 10.1016/s0960-9822(95)00128-x. [DOI] [PubMed] [Google Scholar]
- 14.Sheff M.A., Thorn K.S. Optimized cassettes for fluorescent protein tagging in Saccharomyces cerevisiae. Yeast. 2004;21:661–670. doi: 10.1002/yea.1130. [DOI] [PubMed] [Google Scholar]
- 15.Tanenbaum M.E., Gilbert L.A., Vale R.D. A protein-tagging system for signal amplification in gene expression and fluorescence imaging. Cell. 2014;159:635–646. doi: 10.1016/j.cell.2014.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keppler A., Gendreizig S., Johnsson K. A general method for the covalent labeling of fusion proteins with small molecules in vivo. Nat. Biotechnol. 2003;21:86–89. doi: 10.1038/nbt765. [DOI] [PubMed] [Google Scholar]
- 17.Gautier A., Juillerat A., Johnsson K. An engineered protein tag for multiprotein labeling in living cells. Chem. Biol. 2008;15:128–136. doi: 10.1016/j.chembiol.2008.01.007. [DOI] [PubMed] [Google Scholar]
- 18.Los G.V., Encell L.P., Wood K.V. HaloTag: a novel protein labeling technology for cell imaging and protein analysis. ACS Chem. Biol. 2008;3:373–382. doi: 10.1021/cb800025k. [DOI] [PubMed] [Google Scholar]
- 19.Stagge F., Mitronova G.Y., Jakobs S. SNAP-, CLIP- and Halo-tag labelling of budding yeast cells. PLoS One. 2013;8:e78745. doi: 10.1371/journal.pone.0078745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Medina M.Á., Schwille P. Fluorescence correlation spectroscopy for the detection and study of single molecules in biology. BioEssays. 2002;24:758–764. doi: 10.1002/bies.10118. [DOI] [PubMed] [Google Scholar]
- 21.Wachsmuth M., Conrad C., Ellenberg J. High-throughput fluorescence correlation spectroscopy enables analysis of proteome dynamics in living cells. Nat. Biotechnol. 2015;33:384–389. doi: 10.1038/nbt.3146. [DOI] [PubMed] [Google Scholar]
- 22.Maeder C.I., Hink M.A., Knop M. Spatial regulation of Fus3 MAP kinase activity through a reaction-diffusion mechanism in yeast pheromone signalling. Nat. Cell Biol. 2007;9:1319–1326. doi: 10.1038/ncb1652. [DOI] [PubMed] [Google Scholar]
- 23.Rika J., Binkert T. Direct measurement of a distinct correlation function by fluorescence cross correlation. Phys. Rev. A Gen. Phys. 1989;39:2646–2652. doi: 10.1103/physreva.39.2646. [DOI] [PubMed] [Google Scholar]
- 24.Hwang L.C., Wohland T. Dual-color fluorescence cross-correlation spectroscopy using single laser wavelength excitation. ChemPhysChem. 2004;5:549–551. doi: 10.1002/cphc.200301057. [DOI] [PubMed] [Google Scholar]
- 25.Hwang L.C., Wohland T. Single wavelength excitation fluorescence cross-correlation spectroscopy with spectrally similar fluorophores: resolution for binding studies. J. Chem. Phys. 2005;122:114708. doi: 10.1063/1.1862614. [DOI] [PubMed] [Google Scholar]
- 26.Kogure T., Karasawa S., Miyawaki A. A fluorescent variant of a protein from the stony coral Montipora facilitates dual-color single-laser fluorescence cross-correlation spectroscopy. Nat. Biotechnol. 2006;24:577–581. doi: 10.1038/nbt1207. [DOI] [PubMed] [Google Scholar]
- 27.Guan Y., Meurer M., Shah J.V. Live-cell multiphoton fluorescence correlation spectroscopy with an improved large Stokes shift fluorescent protein. Mol. Biol. Cell. 2015;26:2054–2066. doi: 10.1091/mbc.E14-10-1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim S.A., Heinze K.G., Schwille P. Two-photon cross-correlation analysis of intracellular reactions with variable stoichiometry. Biophys. J. 2005;88:4319–4336. doi: 10.1529/biophysj.104.055319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Müller B.K., Zaychikov E., Lamb D.C. Pulsed interleaved excitation. Biophys. J. 2005;89:3508–3522. doi: 10.1529/biophysj.105.064766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Böhmer M., Wahl M., Enderlein J. Time-resolved fluorescence correlation spectroscopy. Chem. Phys. Lett. 2002;353:439–445. [Google Scholar]
- 31.Kapusta P., Wahl M., Enderlein J. Fluorescence lifetime correlation spectroscopy. J. Fluoresc. 2007;17:43–48. doi: 10.1007/s10895-006-0145-1. [DOI] [PubMed] [Google Scholar]
- 32.Kapusta P., Macháň R., Hof M. Fluorescence lifetime correlation spectroscopy (FLCS): concepts, applications and outlook. Int. J. Mol. Sci. 2012;13:12890–12910. doi: 10.3390/ijms131012890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Felekyan S., Kalinin S., Seidel C.A.M. Filtered FCS: species auto- and cross-correlation functions highlight binding and dynamics in biomolecules. ChemPhysChem. 2012;13:1036–1053. doi: 10.1002/cphc.201100897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Janke C., Magiera M.M., Knop M. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast. 2004;21:947–962. doi: 10.1002/yea.1142. [DOI] [PubMed] [Google Scholar]
- 35.Knop M., Siegers K., Schiebel E. Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast. 1999;15:963–972. doi: 10.1002/(SICI)1097-0061(199907)15:10B<963::AID-YEA399>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 36.Wohland T., Rigler R., Vogel H. The standard deviation in fluorescence correlation spectroscopy. Biophys. J. 2001;80:2987–2999. doi: 10.1016/S0006-3495(01)76264-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Enderlein J., Gregor I. Using fluorescence lifetime for discriminating detector afterpulsing in fluorescence-correlation spectroscopy. Rev. Sci. Instrum. 2005;76:033102. [Google Scholar]
- 38.Delon A., Usson Y., Souchier C. Photobleaching, mobility, and compartmentalisation: inferences in fluorescence correlation spectroscopy. J. Fluoresc. 2004;14:255–267. doi: 10.1023/b:jofl.0000024557.73246.f9. [DOI] [PubMed] [Google Scholar]
- 39.Yoo T.H., Link A.J., Tirrell D.A. Evolution of a fluorinated green fluorescent protein. Proc. Natl. Acad. Sci. USA. 2007;104:13887–13890. doi: 10.1073/pnas.0701904104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Inouye S., Tsuji F.I. Aequorea green fluorescent protein. Expression of the gene and fluorescence characteristics of the recombinant protein. FEBS Lett. 1994;341:277–280. doi: 10.1016/0014-5793(94)80472-9. [DOI] [PubMed] [Google Scholar]
- 41.Zacharias D.A., Violin J.D., Tsien R.Y. Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science. 2002;296:913–916. doi: 10.1126/science.1068539. [DOI] [PubMed] [Google Scholar]
- 42.Maslanka R., Kwolek-Mirek M., Zadrag-Tecza R. Autofluorescence of yeast Saccharomyces cerevisiae cells caused by glucose metabolism products and its methodological implications. J. Microbiol. Methods. 2018;146:55–60. doi: 10.1016/j.mimet.2018.01.017. [DOI] [PubMed] [Google Scholar]
- 43.Benda A., Kapusta P., Gaus K. Fluorescence spectral correlation spectroscopy (FSCS) for probes with highly overlapping emission spectra. Opt. Express. 2014;22:2973–2988. doi: 10.1364/OE.22.002973. [DOI] [PubMed] [Google Scholar]
- 44.Posas F., Wurgler-Murphy S.M., Saito H. Yeast HOG1 MAP kinase cascade is regulated by a multistep phosphorelay mechanism in the SLN1-YPD1-SSK1 “two-component” osmosensor. Cell. 1996;86:865–875. doi: 10.1016/s0092-8674(00)80162-2. [DOI] [PubMed] [Google Scholar]
- 45.Tanaka K. The proteasome: overview of structure and functions. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2009;85:12–36. doi: 10.2183/pjab.85.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wade S.L., Auble D.T. The Rad23 ubiquitin receptor, the proteasome and functional specificity in transcriptional control. Transcription. 2010;1:22–26. doi: 10.4161/trns.1.1.12201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen L., Madura K. Rad23 promotes the targeting of proteolytic substrates to the proteasome. Mol. Cell. Biol. 2002;22:4902–4913. doi: 10.1128/MCB.22.13.4902-4913.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liang R.-Y., Chen L., Chuang S.-M. Rad23 interaction with the proteasome is regulated by phosphorylation of its ubiquitin-like (UbL) domain. J. Mol. Biol. 2014;426:4049–4060. doi: 10.1016/j.jmb.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Benda A., Fagul’ová V., Hof M. Fluorescence lifetime correlation spectroscopy combined with lifetime tuning: new perspectives in supported phospholipid bilayer research. Langmuir. 2006;22:9580–9585. doi: 10.1021/la061573d. [DOI] [PubMed] [Google Scholar]
- 50.Gartner M., Mütze J., Schwille P. Fluorescence lifetime correlation spectroscopy for precise concentration detection in vivo by background subtraction. Proc. SPIE. 2009;7368 73681V–73681V–7. [Google Scholar]
- 51.Chen J., Irudayaraj J. Fluorescence lifetime cross correlation spectroscopy resolves EGFR and antagonist interaction in live cells. Anal. Chem. 2010;82:6415–6421. doi: 10.1021/ac101236t. [DOI] [PubMed] [Google Scholar]
- 52.Padilla-Parra S., Audugé N., Tramier M. Dual-color fluorescence lifetime correlation spectroscopy to quantify protein-protein interactions in live cell. Microsc. Res. Tech. 2011;74:788–793. doi: 10.1002/jemt.21015. [DOI] [PubMed] [Google Scholar]
- 53.Sarkisyan K.S., Goryashchenko A.S., Mishin A.S. Green fluorescent protein with anionic tryptophan-based chromophore and long fluorescence lifetime. Biophys. J. 2015;109:380–389. doi: 10.1016/j.bpj.2015.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.George Abraham B., Sarkisyan K.S., Karp M. Fluorescent protein based FRET pairs with improved dynamic range for fluorescence lifetime measurements. PLoS One. 2015;10:e0134436. doi: 10.1371/journal.pone.0134436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Juillerat A., Gronemeyer T., Johnsson K. Directed evolution of O6-alkylguanine-DNA alkyltransferase for efficient labeling of fusion proteins with small molecules in vivo. Chem. Biol. 2003;10:313–317. doi: 10.1016/s1074-5521(03)00068-1. [DOI] [PubMed] [Google Scholar]
- 56.Los G.V., Wood K. The HaloTag: a novel technology for cell imaging and protein analysis. Methods Mol. Biol. 2007;356:195–208. doi: 10.1385/1-59745-217-3:195. [DOI] [PubMed] [Google Scholar]
- 57.Bosch P.J., Corrêa I.R., Jr., Subramaniam V. Evaluation of fluorophores to label SNAP-tag fused proteins for multicolor single-molecule tracking microscopy in live cells. Biophys. J. 2014;107:803–814. doi: 10.1016/j.bpj.2014.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bindels D.S., Haarbosch L., Gadella T.W.J., Jr. mScarlet: a bright monomeric red fluorescent protein for cellular imaging. Nat. Methods. 2017;14:53–56. doi: 10.1038/nmeth.4074. [DOI] [PubMed] [Google Scholar]
- 59.Iizuka R., Yamagishi-Shirasaki M., Funatsu T. Kinetic study of de novo chromophore maturation of fluorescent proteins. Anal. Biochem. 2011;414:173–178. doi: 10.1016/j.ab.2011.03.036. [DOI] [PubMed] [Google Scholar]
- 60.Pédelacq J.-D., Cabantous S., Waldo G.S. Engineering and characterization of a superfolder green fluorescent protein. Nat. Biotechnol. 2006;24:79–88. doi: 10.1038/nbt1172. [DOI] [PubMed] [Google Scholar]
- 61.Khmelinskii A., Keller P.J., Knop M. Tandem fluorescent protein timers for in vivo analysis of protein dynamics. Nat. Biotechnol. 2012;30:708–714. doi: 10.1038/nbt.2281. [DOI] [PubMed] [Google Scholar]
- 62.Khmelinskii A., Meurer M., Knop M. Incomplete proteasomal degradation of green fluorescent proteins in the context of tandem fluorescent protein timers. Mol. Biol. Cell. 2016;27:360–370. doi: 10.1091/mbc.E15-07-0525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shaner N.C., Lambert G.G., Wang J. A bright monomeric green fluorescent protein derived from Branchiostoma lanceolatum. Nat. Methods. 2013;10:407–409. doi: 10.1038/nmeth.2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lam A.J., St-Pierre F., Lin M.Z. Improving FRET dynamic range with bright green and red fluorescent proteins. Nat. Methods. 2012;9:1005–1012. doi: 10.1038/nmeth.2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Slubowski C.J., Funk A.D., Huang L.S. Plasmids for C-terminal tagging in Saccharomyces cerevisiae that contain improved GFP proteins, Envy and Ivy. Yeast. 2015;32:379–387. doi: 10.1002/yea.3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang C., Konopka J.B. A photostable green fluorescent protein variant for analysis of protein localization in Candida albicans. Eukaryot. Cell. 2010;9:224–226. doi: 10.1128/EC.00327-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.