Abstract
We explored the effect of vincristine and prednisone on cellular and exosomal miR-181a expression in first time diagnosed leukemia and relapsed leukemia. Vincristine and prednisone induced apoptosis/pro-apoptotic genes in first time diagnosed leukemia, and suppressed the cellular and exosomal miR-181a expression. In contrast, vincristine and prednisone could not induce apoptosis/pro-apoptotic genes in relapsed leukemia, and could not change the expression of cellular or exosomal miR-181a. In conclusion, the non-suppressive nature of miR-181a in relapsed leukemia might contribute to the chemo-resistance and this suggests a potential role of miR-181a-inhibitor along with the chemotherapy in the treatment of relapsed leukemia.
Keywords: Acute lymphocytic leukemia; Exosomes; miR-181a, relapsed leukemia
Keywords: Abbreviations: Cell-miR-181a, cellular expression of miR-181a; Exo-miR-181a, exosomal expression of miR-181a
1. Introduction
Vincristine and prednisone are standard chemotherapeutic agents for the treatment of ALL (acute lymphoblastic leukemia) and other types of cancers. Vincristine induces apoptosis by blocking microtubule formation in the mitotic spindle [1] while prednisone is a catabolic steroid that binds to cytoplasmic receptors and inhibits DNA synthesis leading to apoptosis [2]. ALL is a cancer of the blood and bone marrow. It is one of the most common hematological cancers which impact both adults and children [3]. Epigenetic factors such as DNA methylation, gene regulation, and chromatin remodeling [4] play a major role in ALL development and progression, partially via changes in miRNA expression [5, 6]. There is evidence in B-precursor ALL, relapsed ALL differs from first time diagnosed ALL. Heterogeneity of relapsed ALL clones are distributed as; 8% same as diagnosis, 34% clonal evolution from diagnosis, 51% clonal evolution from a pre-leukemic clone, and 7% unrelated second leukemia [7]. There are several factors which contribute to the emergence of new sets of clones in relapsed ALL including a loss of heterozygosity (LOH), acquiring single nucleotide polymorphism (SNP), gene mutational status, changes in DNA promoter methylation and demethylation patterns, mRNA and miRNA expression profile [8], [9], [10]. Similarly, there is an evidence describing first time diagnosed AML (acute myeloid leukemia) is different from relapsed AML and these processes induce, single nucleotide polymorphism (SNP) patterns, mutational status, DNA methylation pattern of gene regulatory regions, mRNA (coding), and miRNA (non-coding) gene expression patterns [11]. This study aimed to explore how vincristine and prednisone regulate miR-181a expression in the first time diagnosed leukemia and relapsed leukemia. In short, miRNAs are endogenously expressed non-coding regulatory RNAs that act as gene silencers at the post-transcriptional level by inhibiting mRNA translation, inducing mRNA degradation, and destabilizing mRNA transcript through binding to the 5′ or 3′ untranslated regions (UTR) fragments [12], [13], [14]. Differential changes in miRNAs modulate biological processes such as cell differentiation, proliferation, apoptosis, and cell survival [15]. There are some reports which support that miR-181a specifically modulates cellular events at multiple levels such as cell proliferation, growth, survival, and even chemo-sensitivity in cancer [16, 17]. There is no report in the literature about the role of vincristine and prednisone on cellular and exosomal miR-181a regulation in ALL. Recently, we published that leukemia-derived exosomes induce leukemia cell proliferation via up-regulating miR-181a expression, and silencing of exosomal miR-181a reverses induced cell proliferation [18]. Briefly, exosomes are nanoparticles (30–150 nm) produced by cells. Exosomes originate from the cytoplasm of the cell by inward budding of multi-vesicular bodies [19]. Exosomes are composed of proteins, lipids, cytoplasmic mRNA, and most importantly non-coding micro-RNA [20, 21]. Exosomal cargo that is transferred as such is functionally active; enabling the target cells to reprogram and redefine their immunological and biological responses [22]. The whole idea of this study is to explore, novel anti-ALL molecules required to defeat chemotherapy, chemotherapy resistance, and reduce cytotoxicity and side effects. Either cocktails of miR mimics or anti-miR (miR inhibitor) may be an alternative for cancer therapy either along with chemotherapy regimen or alone.
2. Materials and methods
2.1. Cell lines and cell culture condition
First time diagnosed leukemia cell lines: JM1 (CRL-10,423™, ATCC), SUP-B15 (CRL-1929 ™, ATCC), NALM-6 (CRL-3273™, ATCC). Relapsed leukemia cell lines: REH (ACC-2, DSMZ), and NALM-16 (ACC-680, DSMZ). JM1 and SUP-B15 cells were expanded and cultured in IMDM (Iscove's Modified Dulbecco's Medium) supplemented with 0.05 mM β-mercaptoethanol, 10% FBS, and 1 × Penicillin-Streptomycin. SUP-B15 cells were cultured in IMDM (Iscove's Modified Dulbecco's Medium), supplemented with 1.5 g/L sodium bicarbonate, 0.05 mM β-mercaptoethanol, 20% fetal bovine serum (not heat-inactivated), and 1 × Penicillin-Streptomycin. NALM-16, REH, NALM-6 cells were cultured in RPMI-1640 supplemented with 10% FBS, and 1 × Penicillin-Streptomycin. All were incubated in a humidified incubator at 37 °C with 5% CO2.
2.2. Exosome depletion from the FBS
To avoid contamination by exogenous exosomes present in the fetal bovine serum (FBS) that is added to the culture medium, FBS was processed to make exosome-free FBS by ultracentrifugation [23]. Cell culture was carried out in a specific culture medium using exosome depleted FBS, where the purpose of the experiment was to isolate exosomes after treatment.
2.3. Cell viability MTS assay
Cells (JM1 and SUP-B15, NALM-6, REH, and NALM-16) were seeded in a 96-well culture plate (cell density: 105/100 µl) and treated with different doses of vincristine (0.1 to 4.0 µM) and prednisone (0.1 to 12.0 µM). Cells were treated for 24 and 48 h. Cell apoptosis/viability was measured by MTS assay (CellTiter 96® aqueous one solution, Promega). Each experiment was carried out in quadruplets; data represented here is the mean of each value (Mean ± S.D.).
2.4. Treatment of vincristine and prednisone on cell lines
Cell lines (JM1, Sup-B15, and NALM-6) were plated (2 × 106 cells /ml culture volume) in 24 well plates. Cells were treated with vincristine (4.0 µM) and prednisone (4.0 µM) for 24 h. After 24 h of exposure, cells were harvested and washed with cold PBS and centrifuged. Cell pellets were utilized for RNA extraction.
2.5. RNA extraction and cDNA preparation
Total RNA was extracted from cell pellets of SUP-B15, JM1, and NALM-6 by Trizol reagent (Invitrogen). The quality and quantity of RNA were evaluated by the NanoDrop ND1000 spectrophotometer. Between 1 to 5 µg of total RNA was used for cDNA synthesis, Oligo-dT primers (Invitrogen) and M-MLV reverse transcriptase (cat # 28,025–013, Invitrogen) were used for cDNA synthesis.
2.6. Gene expression by q-PCR
Primer sequences, probe numbers, and gene accession numbers from the universal probe library (UPL) of Roche Applied Science are described (Table 1). Gene amplification was carried out by q-PCR with cDNA as a template using the Eurogentec master mix. Fold change was calculated by comparing treated vs. untreated groups. Data were analyzed with RQ manager version 1.2.1 (Applied Biosystems, Foster City, CA). The data are expressed as fold change and GAPDH was used as a reference gene for fold change calculation.
Table 1.
List of human primers for q-PCR.
| SN | Gene name | Gene ID | Primers | UPL # |
|---|---|---|---|---|
| 1 | BAD | AF031523.1 | For: 5′-CGAGTTTGTGGACTCCTTTAAGA-3′ | 78 |
| Rev: 5′-CACCAGGACTGGAAGACTCG-3′ | ||||
| 2 | BAX | U19599.1 | For: 5‘-CAAGACCAGGGTGGTTGG-3′ | 55 |
| Rev: 5‘-CACTCCCGCCACAAAGAT-3′ | ||||
| 3 | GAPDH | NM_002046.3 | For: 5′-AGCCACATCGCTCAGACAC-3′ | 60 |
| Rev: 5′-GCCCAATACGACCAAATCC-3′ |
2.7. Primers for micro-RNA (miR) amplification
We purchased primers from Qiagen for miRNA amplification. Brief details of miR-181a-5p (cat # MS00008827, product number 218,300, Sanger accession MI0000289), which targets mature miRNA: MIMAT0000256–5′AACAUUCAACGCUGUCGGUGAGU. We chose housekeeping or endogenous miRNA, SNORD61 (cat# MS00033705, product# 218,300) for fold change calculation in the qPCR analysis.
2.8. Cell culture set up for exosome production
Each cell line was cultured in exo-free FBS/exosome depleted FBS cell culture medium when cells were expanded for the harvest of conditioned medium (CM) exosomes. Cells were plated (20 × 106 cells/plate/10 ml) in a 100 mm plate in an exo-free cell culture medium. After 48 h, the CM of each cell line was harvested and filtered through a 0.22-micron filter (Millipore). Filtered CM was used for exosome isolation by ultracentrifugation.
2.9. Exosome isolation and extraction of RNA from exosomes
Exosomes were isolated and purified by ultracentrifugation as described [23]. The quality of purified exosomes were confirmed by western blot (CD63 and calnexin expression) and size of the exosomes were confirmed by nanoparticle tracking analysis as shown [24]. Purified exosomes were dissolved into the Trizol reagent for exosomal RNA extraction. Purified exosomal RNA concentration and quality was determined by the NanoDrop (ND1000 spectrophotometer).
2.10. Cellular and exosomal miR-181a-5p expression by q-PCR
For cellular miR-181a-5p expression, cellular RNA was converted into cDNA as described. For exosomal miR-181a-5p expression, exosomal RNA converted into cDNA. Each condition of RNA was converted into cDNA by the miScript II RT kit (cat # 218,161, Qiagen). The miRNA expression was evaluated by qPCR using a miScript SYBR Green PCR Kit (cat # 218,073, Qiagen). Fold expression was calculated using Ct value and SNORD61 was used as endogenous control miR.
2.11. Statistical analysis
To compare the mean values between the two groups, the unpaired t-test was used. Statistical significance was defined as p<0.05. All results are represented as mean ± SD. Each experiment was performed three times in triplicates.
3. Results
3.1. Vincristine and prednisone induced cell death
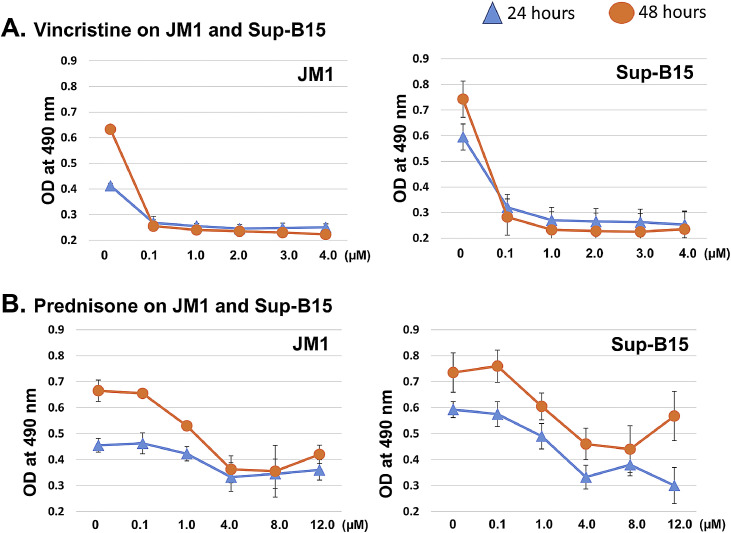
There is substantial evidence that vincristine and prednisone induce apoptosis in a variety of cell types. To evaluate the cytotoxic effects of vincristine and prednisone in our settings. JM1 and Sup-B15 cells were treated with vincristine (dose-ranging 0.1 to 4.0 µM) and prednisone (dose-ranging 0.1 to 12.0 µM) and incubated for 24 as well as 48 h. Cytotoxicity was measured by adding MTS reagent. We found that vincristine-induced cytotoxicity compared to vehicle control (Fig. 1A). Prednisone also showed significant cell death compared to untreated control (Fig. 1B). In both cell lines, cytotoxicity was similar at both time points (24 h and 48 h). For further experiments, we subsequently used 24 h of the incubation period for both vincristine and prednisone.
Fig. 1.
Vincristine and prednisone induce apoptosis in the first time diagnosed leukemia B cells. Cells (JM1 and SUP-B15) were seeded in a 96-well culture plate (cell density: 105/100 µl) and treated with different doses. A. Vincristine (0.1 to 4.0 µM) and B. Prednisone (0.1 to 12.0 µM) and cells were harvested at 24 and 48 h of post-treatment. Cell apoptosis was assessed by MTS assay. Each experiment was carried out in quadruplets; data represented here is the mean of each value (Mean ± S.D.).
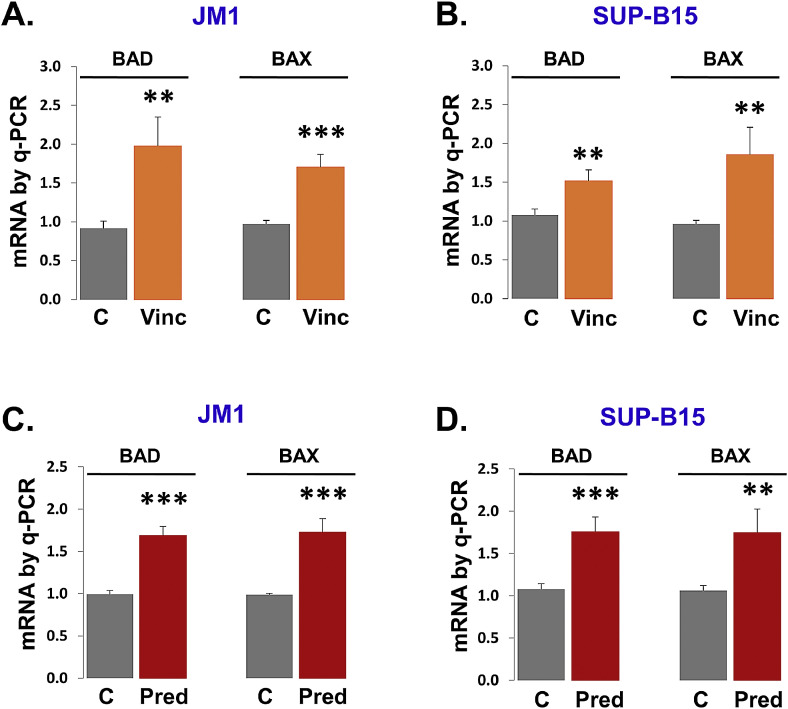
3.2. Apoptotic gene regulation by vincristine and prednisone
Since chemotherapeutic drugs such as vincristine and prednisone have been shown to induce apoptosis. Next, we explored pro-apoptotic genes such as BAD and BAX mRNA expression for further confirmation of apoptosis induction. Vincristine treatment-induced apoptosis with augmented expression of pro-apoptotic genes (BAD and BAX) mRNA in JM1 cells (Fig. 2A) and Sup-B15 cells (Fig. 2B). Prednisone exposure induced apoptosis by increasing expression of BAD and BAX mRNA in JM1 cells (Fig. 2C) and Sup-B15 cells (Fig. 2D). Both cell lines showed induced apoptosis upon treatment of vincristine and prednisone.
Fig. 2.
Vincristine and prednisone induce pro-apoptotic genes in the first time diagnosed leukemia B cells. JM1 (A) and Sup-B15 (B) cells were treated with vincristine (4.0 µM) for 24 h. The expression of BAD mRNA and BAX mRNA was analyzed was by q-PCR. Prednisone (4.0 µM) treatment was given to JM1 (C) and Sup-B15 (D) cells for 24 h. Expression of BAD mRNA and BAX mRNA was analyzed was by q-PCR and results were displayed in fold change. GAPDH gene was used as an endogenous control. Results were expressed in the fold. Data were analyzed from the untreated (control) vs. treated group. P-value *p<0.05, **p<0.01, ***p<0.001.
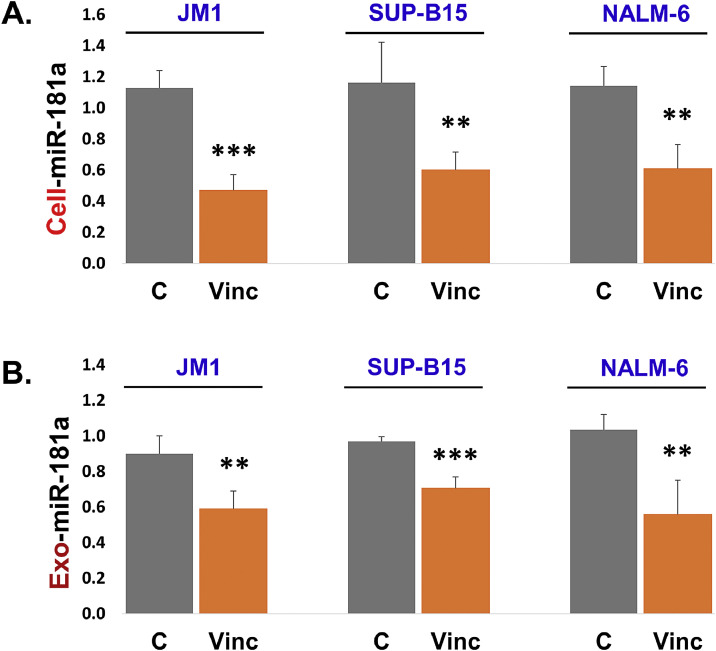
3.3. Vincristine suppresses cellular and exosomal miR-181a expression in first time diagnosed leukemia cells
We optimized the dose-response of vincristine, JM1, and Sup-B15 cells were exposed with vincristine in doses from 0.1 to 4.0 µM for 24 h. We found that vincristine suppressed the miR-181a expression most significantly at 4.0 µM in both cell lines (Supplemental Fig. S1 A and S1 B). Thus, we chose 4.0 µM vincristine dose and repeated in three leukemia cell lines (JM1, Sup-B15, and NALM-6). We found that cellular miR-181a expression was reduced by vincristine exposure compared to control in all three leukemia cell lines (Fig. 3A). Since miR-181a expression is down-regulated at the cellular level; we thought to explore miR-181a expression at the exosomal level as well. Cultures were set up for the purpose to isolate exosomes from the conditioned medium after vincristine treatment including the control sample. RNA was isolated from the exosomes (Exo-RNA). Exo-RNA was converted into cDNA using RT kit and miR-181a expression was assessed by q-PCR. We found that vincristine down-regulated miR-181a expression at the exosomal level as well compared to the control sample (Fig. 3B). Altogether, our data show that vincristine exposure suppresses miR-181a expression at both cellular and exosomal level.
Fig. 3.
Vincristine down-regulates cellular and exosomal miR-181a expression in the first time diagnosed leukemia B cells. JM1, Sup-B15, and NALM-6 cells were treated with vincristine (4.0 µM) for 24 h. A. Cellular miR-181a expression was quantified by qPCR, and results were displayed in fold change. B. Exosomal miR-181a expression was quantified by qPCR and results were displayed in fold change. SNORD61 was used as an endogenous control. (P-value *p<0.05, **p<0.01, ***p<0.001).
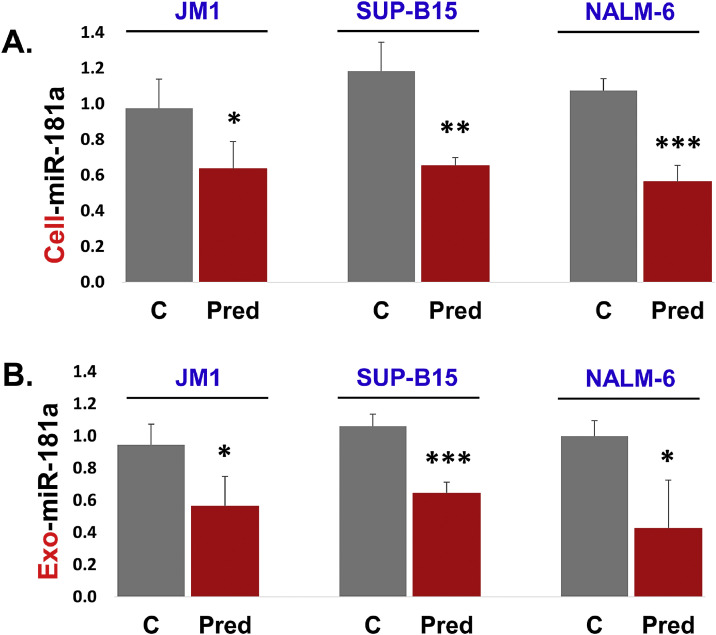
3.4. Prednisone down regulates cellular and exosomal miR-181a expression in first time diagnosed leukemia cells
We also tested the effect of prednisone on miR-181a expression. Initially, we optimized the dose-response of prednisone on JM1 and Sup-B15 cell lines. Both cell lines were exposed to prednisone (0.1 to 12.0 µM) for 24 h. Our qPCR data showed that prednisone exposure down-regulated miR-181a expression most significantly at 4.0 µM in both cell lines (Supplemental Fig. S1 C and S1 D). For further experiments, we chose 4.0 µM prednisone dose and repeated these experiments in three leukemia cell lines (JM1, Sup-B15, and NALM-6). We found that cellular miR-181a expression was reduced by prednisone compared to control cells in all three leukemia cell lines (Fig. 4A). We observed that miR-181a expression is down-regulated at the cellular level. Next, we thought to unravel miR-181a expression at the exosomal level as well. We found that prednisone down-regulated miR-181a expression in exosomes as well compared to controls (Fig. 4B). Altogether, our data show that the prednisone downregulates miR-181a expression at both cellular and exosomal level.
Fig. 4.
Prednisone down-regulates cellular and exosomal miR-181a expression in the first time diagnosed leukemia B cell lines. JM1, Sup-B15, and NALM-6 cells were treated with prednisone (4.0 µM) for 24 h. A. Cellular RNA was extracted and cellular miR-181a expression was quantified by qPCR, and results were displayed in fold change. B. Exosomal miR-181a expression was assessed by q-PCR and results were displayed in fold change. SNORD61 was used as an endogenous control. (P-value *p<0.05, **p<0.01, ***p<0.001).
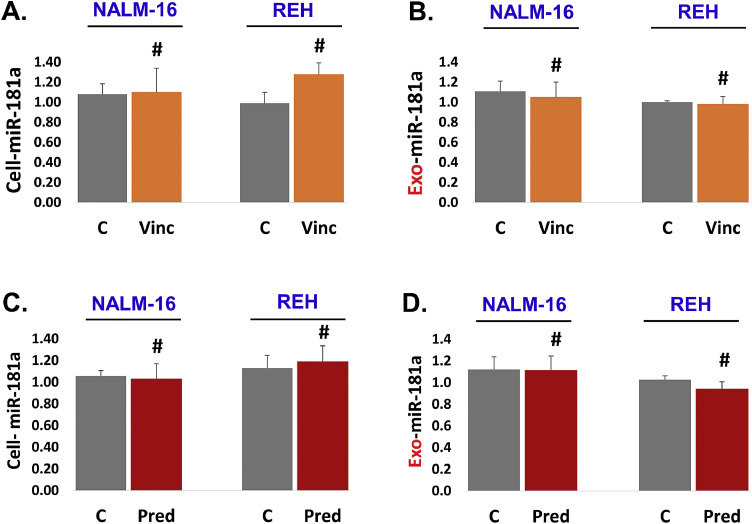
3.5. Vincristine and prednisone exposure did not change miR-181a expression in relapsed leukemia cells
There is a tremendous difference in the molecular nature of the first time diagnosed leukemia and relapsed leukemia. We chose to explore the effect of vincristine on miR-181a expression in relapsed leukemia cell lines (NALM-16 and REH). Our results showed that vincristine could not downregulate miR-181a expression in relapsed leukemia cells (Fig. 5A). We also checked miR-181a expression in exosomal RNA. We did not see any significant difference in miR-181a expression between the vincristine treated and the control group (Fig. 5B). Collectively, data shows that vincristine exposure did not change miR-181a expression either at the cellular or exosomal level in relapsed leukemia cell lines (NALM-16 and REH). This suggests that if an event is not taking place at the cellular level, it won't appear at the exosomal level.
Fig. 5.
Vincristine and prednisone exposure did not change miR-181a expression in relapsed leukemia cells. (A and B) Vincristine (4.0 µM) treatment was given to NALM-16 and REH cells for 24 h. A. Cellular miR-181a expression was measured by q-PCR. B. Exosomal miR-181a expression was assessed by q-PCR. (C and D) Prednisone (4.0 µM) exposure was given to NALM-16 and REH cells for 24 h. C. Cellular miR-181a expression was measured by q-PCR. D. Exosomal miR-181a expression was assessed by q-PCR. Results were displayed in fold change. SNORD61 was used as an endogenous control. (P-value, not significant # p >0.05).
Similarly, we chose to see the effect of prednisone on miR-181a expression in relapsed leukemia cell lines (NALM-16 and REH). Cell lines were treated with prednisone (4.0 µM) for 24 h. We found that prednisone could not downregulate miR-181a expression in relapsed leukemia cells (Fig. 5C). We also checked the level of miR-181a expression in exosomal RNA. We did not see any significant difference in miR-181a expression between prednisone treated and control group (Fig. 5D). Collectively, our data show that prednisone treatment did not change miR-181a expression either at the cellular or exosomal level in relapsed leukemia. This also confirms that if an event is not happening at a cellular level, it won't appear at the exosomal level either.
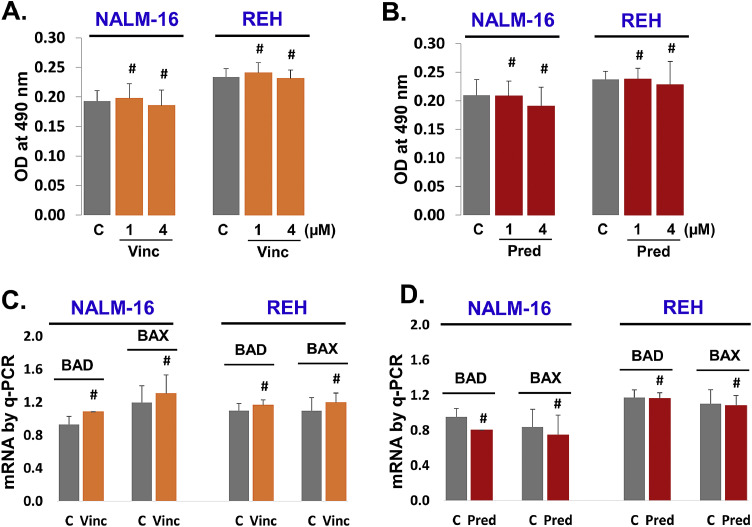
3.6. Vincristine and prednisone exposure neither induce apoptosis nor pro-apoptotic genes in relapsed leukemia cells
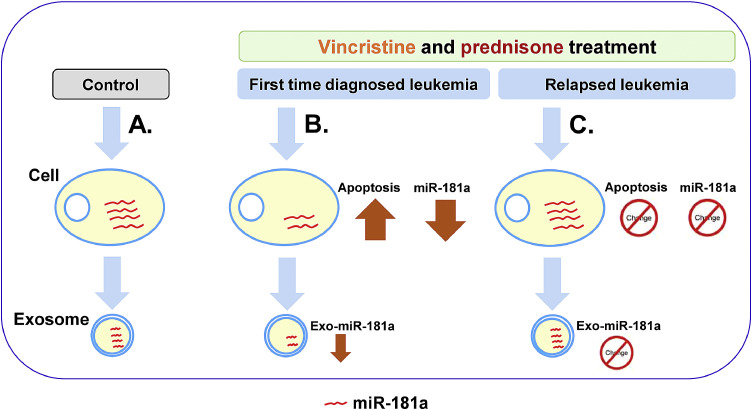
Vincristine and prednisone did not change the expression of miR-181a in relapsed leukemia. These findings intrigued us to explore the role of vincristine and prednisone on relapsed leukemia B cell apoptosis. Both NALM-16 and REH cells were exposed to vincristine (Fig. 6A) and prednisone ((Fig. 6B) for 24 h and apoptosis measured by MTS assay. We found that vincristine and prednisone could not induce significant apoptosis in relapsed leukemia cells compared to control groups. Further, we also looked for pro-apoptotic gene markers (BAD, and BAX mRNA) by q-PCR. Both NALM-16 and REH cells were treated with vincristine (Fig. 6C) and prednisone (Fig. 6D). We observed no significant change in BAD and BAX mRNA expression in response to vincristine or prednisone compared to control. Overall, these data confirm that vincristine and prednisone are not able to induce cell apoptosis in relapsed leukemia B cells. It appears that relapsed leukemia B cells are chemo-resistant with vincristine and prednisone. Schematically overall results summarized (Fig. 7). This diagram shows overall crux of the study.
Fig. 6.
Vincristine and prednisone exposure neither induce apoptosis nor pro-apoptotic genes (BAD and BAX) in relapsed leukemia cells. Relapsed leukemia cell lines (NALM-16 and REH) were seeded (105 cells/100 µl in 96 well plates) and exposed with vincristine and prednisone for 24 h and cell apoptosis was quantified by MTS assay. A. Cells were exposed with vincristine (1.0 and 4.0 µM). B. Cells were exposed with prednisone (1.0 and 4.0 µM). Pro-apoptotic genes by qPCR. Relapsed leukemia cells were plated (2 × 106 cells/1.0 ml) in 24 well culture plates and treated with vincristine and prednisone for 24 h. C. Cells were treated with vincristine (4.0 µM). D. Cells were treated with prednisone (4.0 µM). (P-value, not significant # p >0.05).
Fig. 7.
Schematically overview of the study. A. Cells were treated with vehicle control such as PBS or water. B. Exposure of vincristine and prednisone on first time diagnosed leukemia cells (JM1, and SUP-B15) resulted in induced apoptosis by downregulating cellular and exosomal miR-181a expression. C. Contrary, exposure of vincristine and prednisone on relapsed leukemia cells (NALM-16 and REH) resulted in no apoptosis induction and no change in cellular and exosomal miR-181a expression.
We observed in our study that both vincristine and prednisone are able to suppress miR-181a expression in the first time diagnosed leukemia cell lines (JM1, Sup-B15, and NALM-6) while no suppression of miR-181a was resulted in relapsed leukemia cell lines (NALM-16 and REH). One of the reviewers suggested to evaluate the base line expression of miR-181a in the first time diagnosed cell lines and the relapsed leukemia cell lines. We performed the experiments and found that relapsed leukemia cell lines have significant higher expression of miR-181a compared to the first time diagnosed cell lines (Supplemental Fig. S2).
4. Discussion
Despite several advantages, chemotherapy drugs have several levels of drawbacks and side effects, such as chemotherapy resistance and high non-specific toxicity [25, 26]. Here the idea is to develop an alternative or parallel therapy along with chemotherapy to reduce toxicity and side effects of chemotherapeutic drugs. One of the approaches is to understand how chemotherapy impairs and regulates miRNA expression and utilize this information to develop alternative therapy to cure cancers. This is well established and widely demonstrated that vincristine induces apoptosis in leukemia cells [27, 28] and prednisone induces apoptosis in leukemia [29, 30] and other types of cancers. In this study, we have explored the role of vincristine and prednisone on miR-181a expression in the first time diagnosed ALL and relapsed ALL. First, we checked, the apoptotic effect of the effect of vincristine and prednisone in JM1 and Sup-B15 cell lines. If these drugs induce apoptosis there must be the induction of pro-apoptotic genes. We found that vincristine and prednisone showed upregulated BAD and BAX mRNA (pro-apoptotic) expression and chemotherapeutic drugs (vincristine and prednisone) exposure induces cell apoptosis by enhancing pro-apoptotic genes, BID, BAX, caspase-3 induction, and downregulation of BCL-2 expression in ALL cells [31, 32].
In our previous study, we demonstrated that leukemia-derived exosomes induce leukemia cell proliferation via up-regulating miR-181a expression and silencing of exosomal miR-181a reverses induced cell proliferating [33]. That shows that miR-181a has a significant and pivotal role in cell proliferation and apoptosis pathway. However, we chose to expose first time diagnosed leukemia cells (JM1, Sup-B15, and NALM-6) with vincristine and prednisone and analyze cellular and exosomal miR-181a expression. We found that vincristine and prednisone suppress miR-181a expression at both cellular and exosomal level with consistency. Since exosomes are the fingerprint of the parent cells [20, 21], if miR-181a expression is downregulated at the cellular level then exosomes derived from the parent cells will also show a similar pattern of miR-181a downregulation. Overall, this study significantly established that vincristine and prednisone exposure suppresses cellular and exosomal miR-181a expression resulting in induced apoptosis in the first time diagnosed leukemia cells. Cell-derived components such as exosomes and extracellular matrix are one of the major players for the development of microenvironment in hematological malignancies. Some reports show that there is a better platform for cell culture coated with extracellular matrix-rich biomimetic substrate that can provide podocytes cell culture such as proliferation, differentiation and can be utilized for drug screening purpose [34], development of 3D culture can be employed in mimicking in vivo tissue system that can be exploited for the investigation of tumor biomarkers, drug screening, and understanding tumor progression and metastasis [35].
Relapsed leukemia is normally linked with high rates of treatment failure because of the induction of chemotherapy resistance. Augmented expression of miR-181a promotes chemo-resistance in leukemia and breast cancers [36, 37] including other micro-RNAs. Here we explored the effect of vincristine and prednisone on miR-181a expression in relapsed leukemia cells (NALM-16 and REH). We did not see downregulation of miR-181a expression in relapsed leukemia, neither at the cellular level nor at the exosomal level. This outcome is the opposite of the first time diagnosed leukemia cells, where vincristine and prednisone were able to suppress miR-181a expression. Our data explain to some extent that vincristine and prednisone exposure could not suppress miR-181a expression in relapsed leukemia cells and this non-suppressive nature of miR-181a making relapsed leukemia cells resistant to vincristine and prednisone. This suggests that miR-181a plays a role as chemo-resistance in relapsed leukemia. Although chemo-resistance induction by vincristine and prednisone is not a new avenue in cancer therapy, this was widely studied in the past by many investigators. Briefly, exposure with prednisone and dexamethasone showed relapsed childhood ALL cells are 5–30 fold more resistant than first time diagnosed ALL in terms of cytotoxicity [30]. The resistant nature of ALL cells can be converted into sensitized to prednisolone by silencing MCL1, which is demonstrated in BCP-ALL and T-ALL leukemic cells established as combination therapy [38]. Inhibition of glycolysis is also an explored pathway that modulates prednisolone resistance, a combination of 2-deoxyglucose (2-DG, a glycolysis inhibitor) with prednisone treatment can reverse prednisone induced resistant ALL cells into sensitive ALL cells a combination therapy for relapsed ALL treatment [39]. Vincristine and prednisone exposure did not induce cytotoxicity/apoptosis in relapsed ALL (NALM-16 and REH) in 24 h of exposure and also did not induce any change in pro-apoptotic genes such as BAD and BAX mRNA expression. Similarly, glucocorticoids (dexamethasone and prednisone) exposures neither induce apoptosis nor induce pro-apoptotic genes (BIM) pathway in REH cells while induced apoptosis and pro-apoptotic pathways in 697 cells (BCP-ALL cell line) [40]. Even in breast carcinoma, doxorubicin exposure induced cellular resistance (MCF-7/R), X-radiation exposure to MCF-7/R cells could not induce BAX, BAD protein expression [41].
Many factors contribute to the induction of cell resistance, not only microRNA but certain mRNAs/proteins are also a major factor for the augmentation of cell resistance in leukemia. It would be early to say miR-181a inhibitor combined with chemotherapy as a choice of leukemia treatment for relapsed leukemia. We speculate there is a need for thorough and broad investigation required to understand the entire spectrum of molecular change and induction of chemoresistance influenced by vincristine and prednisone. At the basal level, we found that relapsed leukemia cell lines have significantly higher expression of miR-181a compared to the first time diagnosed leukemia cell lines. Since relapsed leukemia cell lines are robust expressers of miR-181a, this could be one of the reasons why vincristine or prednisone could not suppress miR-181a expression in relapsed leukemia cells.
5. Conclusions
The chemotherapeutic drugs vincristine and prednisone induce apoptosis in general. Besides, vincristine and prednisone exposure downregulates miR-181a expression at cellular and exosomal level and induces apoptosis in first time diagnosed leukemia. Thus, it is tempting to speculate miR-181a plays important role in the vincristine and prednisone induced apoptosis in relapsed leukemia. To further support that, neither vincristine nor prednisone downregulates miR-181a expression nor apoptosis in the relapsed leukemia B cells. The Non-suppressive nature of miR-181a might be opening a new avenue to explore the chemo-resistant nature of relapsed leukemia B cells.
Authors contributions
S.H. performed the experiments, designed the experiments, analyzed the data, and wrote the manuscript. S.V. conceived the idea, designed the experiments, reviewed and edited the manuscript, and provided all required supports.
Competing interests
The authors declare no competing interests.
Acknowledgments
This work was supported by the Pediatric Oncology Fund at Staten Island University Hospital, Staten Island, New York, and The DiMartino Family Foundation and Island Auto group.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.lrr.2020.100221.
Appendix. Supplementary materials
References
- 1.Jordan M.A. Mechanism of action of antitumor drugs that interact with microtubules and tubulin, Current medicinal chemistry. Anti-Cancer Agents. 2002;2(1):1–17. doi: 10.2174/1568011023354290. [DOI] [PubMed] [Google Scholar]
- 2.Lanza L., Scudeletti M., Puppo F., Bosco O., Peirano L., Filaci G., Fecarotta E., Vidali G., Indiveri F. Prednisone increases apoptosis in in vitro activated human peripheral blood T lymphocytes. Clin. Exp. Immunol. 1996;103(3):482–490. doi: 10.1111/j.1365-2249.1996.tb08306.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pui C.H. Recent research advances in childhood acute lymphoblastic leukemia. J. Formos. Med. Assoc. 2010;109(11):777–787. doi: 10.1016/S0929-6646(10)60123-4. [DOI] [PubMed] [Google Scholar]
- 4.Payne K.J., Dovat S. Ikaros and tumor suppression in acute lymphoblastic leukemia. Crit. Rev. Oncog. 2011;16(1–2):3–12. doi: 10.1615/critrevoncog.v16.i1-2.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burke M.J., Bhatla T. Epigenetic modifications in pediatric acute lymphoblastic leukemia. Front. Pediatr. 2014;2:42. doi: 10.3389/fped.2014.00042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chatterton Z., Morenos L., Mechinaud F., Ashley D.M., Craig J.M., Sexton-Oates A., Halemba M.S., Parkinson-Bates M., Ng J., Morrison D., Carroll W.L., Saffery R., Wong N.C. Epigenetic deregulation in pediatric acute lymphoblastic leukemia. Epigenetics. 2014;9(3):459–467. doi: 10.4161/epi.27585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mullighan C.G. Molecular genetics of B-precursor acute lymphoblastic leukemia. J. Clin. Invest. 2012;122(10):3407–3415. doi: 10.1172/JCI61203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kunz J.B., Rausch T., Bandapalli O.R., Eilers J., Pechanska P., Schuessele S., Assenov Y., Stütz A.M., Kirschner-Schwabe R., Hof J., Eckert C., von Stackelberg A., Schrappe M., Stanulla M., Koehler R., Avigad S., Elitzur S., Handgretinger R., Benes V., Weischenfeldt J., Korbel J.O., Muckenthaler M.U., Kulozik A.E. Pediatric T-cell lymphoblastic leukemia evolves into relapse by clonal selection, acquisition of mutations and promoter hypomethylation. Haematologica. 2015;100(11):1442–1450. doi: 10.3324/haematol.2015.129692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mullighan C.G., Downing J.R. Genome-wide profiling of genetic alterations in acute lymphoblastic leukemia: recent insights and future directions. Leukemia. 2009;23(7):1209–1218. doi: 10.1038/leu.2009.18. [DOI] [PubMed] [Google Scholar]
- 10.Kirschner-Schwabe R., Lottaz C., Tödling J., Rhein P., Karawajew L., Eckert C., von Stackelberg A., Ungethüm U., Kostka D., Kulozik A.E., Ludwig W.D., Henze G., Spang R., Hagemeier C., Seeger K. Expression of late cell cycle genes and an increased proliferative capacity characterize very early relapse of childhood acute lymphoblastic leukemia. Clin. Cancer Res. 2006;12(15):4553–4561. doi: 10.1158/1078-0432.CCR-06-0235. [DOI] [PubMed] [Google Scholar]
- 11.Hackl H., Astanina K., Wieser R. Molecular and genetic alterations associated with therapy resistance and relapse of acute myeloid leukemia. J. Hematol. Oncol. 2017;10(1):51. doi: 10.1186/s13045-017-0416-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tuschl T., Zamore P.D., Lehmann R., Bartel D.P., Sharp P.A. Targeted mRNA degradation by double-stranded RNA in vitro. Genes Dev. 1999;13(24):3191–3197. doi: 10.1101/gad.13.24.3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 14.Morozova N., Zinovyev A., Nonne N., Pritchard L.L., Gorban A.N., Harel-Bellan A. Kinetic signatures of microRNA modes of action. RNA. 2012;18(9):1635–1655. doi: 10.1261/rna.032284.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garofalo M., Condorelli G.L., Croce C.M., Condorelli G. MicroRNAs as regulators of death receptors signaling. Cell Death Differ. 2010;17(2):200–208. doi: 10.1038/cdd.2009.105. [DOI] [PubMed] [Google Scholar]
- 16.Lin Y., Zhao J., Wang H., Cao J., Nie Y. miR-181a modulates proliferation, migration and autophagy in AGS gastric cancer cells and downregulates MTMR3. Mol. Med. Rep. 2017;15(5):2451–2456. doi: 10.3892/mmr.2017.6289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu Y., Wu J., Li S., Ma R., Cao H., Ji M., Jing C., Tang J. The function role of miR-181a in chemosensitivity to adriamycin by targeting Bcl-2 in low-invasive breast cancer cells. Cell. Physiol. Biochem. 2013;32(5):1225–1237. doi: 10.1159/000354521. [DOI] [PubMed] [Google Scholar]
- 18.Haque S., Vaiselbuh S.R. Silencing of exosomal miR-181a reverses pediatric acute lymphocytic leukemia cell proliferation. Pharmaceuticals (Basel) 2020;13(9) doi: 10.3390/ph13090241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yáñez-Mó M., Siljander P.R., Andreu Z., Zavec A.B., Borràs F.E., Buzas E.I., Buzas K., Casal E., Cappello F., Carvalho J., Colás E., Cordeiro-da Silva A., Fais S., Falcon-Perez J.M., Ghobrial I.M., Giebel B., Gimona M., Graner M., Gursel I., Gursel M., Heegaard N.H., Hendrix A., Kierulf P., Kokubun K., Kosanovic M., Kralj-Iglic V., Krämer-Albers E.M., Laitinen S., Lässer C., Lener T., Ligeti E., Linē A., Lipps G., Llorente A., Lötvall J., Manček-Keber M., Marcilla A., Mittelbrunn M., Nazarenko I., Nolte-’t Hoen E.N., Nyman T.A., O’Driscoll L., Olivan M., Oliveira C., É. Pállinger, Del Portillo H.A., Reventós J., Rigau M., Rohde E., Sammar M., Sánchez-Madrid F., Santarém N., Schallmoser K., Ostenfeld M.S., Stoorvogel W., Stukelj R., Van der Grein S.G., Vasconcelos M.H., Wauben M.H., De Wever O. Biological properties of extracellular vesicles and their physiological functions. J. Extracell. Vesicle. 2015;4:27066. doi: 10.3402/jev.v4.27066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thakur B.K., Zhang H., Becker A., Matei I., Huang Y., Costa-Silva B., Zheng Y., Hoshino A., Brazier H., Xiang J., Williams C., Rodriguez-Barrueco R., Silva J.M., Zhang W., Hearn S., Elemento O., Paknejad N., Manova-Todorova K., Welte K., Bromberg J., Peinado H., Lyden D. Double-stranded DNA in exosomes: a novel biomarker in cancer detection. Cell Res. 2014;24(6):766–769. doi: 10.1038/cr.2014.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Théry C., Zitvogel L., Amigorena S. Exosomes: composition, biogenesis and function, Nature reviews. Immunology. 2002;2(8):569–579. doi: 10.1038/nri855. [DOI] [PubMed] [Google Scholar]
- 22.Umezu T., Ohyashiki K., Kuroda M., Ohyashiki J.H. Leukemia cell to endothelial cell communication via exosomal miRNAs. Oncogene. 2013;32(22):2747–2755. doi: 10.1038/onc.2012.295. [DOI] [PubMed] [Google Scholar]
- 23.Théry C., Amigorena S., Raposo G., Clayton A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr. Protocol. Cell Biol. Chapter. 2006;3 doi: 10.1002/0471143030.cb0322s30. Unit 3.22. [DOI] [PubMed] [Google Scholar]
- 24.Haque S., Vaiselbuh S.R. Exosomes molecular diagnostics: direct conversion of exosomes into the cDNA for gene amplification by two-step polymerase chain reaction. J. Biol. Methods. 2018;5(3):e96. doi: 10.14440/jbm.2018.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schmiegelow K., Müller K., Mogensen S.S., Mogensen P.R., Wolthers B.O., Stoltze U.K., Tuckuviene R., Frandsen T. Non-infectious chemotherapy-associated acute toxicities during childhood acute lymphoblastic leukemia therapy. F1000Res. 2017;6:444. doi: 10.12688/f1000research.10768.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Caiado F., Maia-Silva D., Jardim C., Schmolka N., Carvalho T., Reforço C., Faria R., Kolundzija B., Simões A.E., Baubec T., Vakoc C.R., da Silva M.G., Manz M.G., Schumacher T.N., Norell H., Silva-Santos B. Lineage tracing of acute myeloid leukemia reveals the impact of hypomethylating agents on chemoresistance selection. Nat. Commun. 2019;10(1):4986. doi: 10.1038/s41467-019-12983-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chao M.W., Lai M.J., Liou J.P., Chang Y.L., Wang J.C., Pan S.L., Teng C.M. The synergic effect of vincristine and vorinostat in leukemia in vitro and in vivo. J. Hematol. Oncol. 2015;8:82. doi: 10.1186/s13045-015-0176-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Souza P.S., Vasconcelos F.C., De Souza Reis F.R., Nestal De Moraes G., Maia R.C. P-glycoprotein and survivin simultaneously regulate vincristine-induced apoptosis in chronic myeloid leukemia cells. Int. J. Oncol. 2011;39(4):925–933. doi: 10.3892/ijo.2011.1103. [DOI] [PubMed] [Google Scholar]
- 29.Bindreither D., Ecker S., Gschirr B., Kofler A., Kofler R., Rainer J. The synthetic glucocorticoids prednisolone and dexamethasone regulate the same genes in acute lymphoblastic leukemia cells. BMC Genomics. 2014;15(1):662. doi: 10.1186/1471-2164-15-662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ito C., Evans W.E., McNinch L., Coustan-Smith E., Mahmoud H., Pui C.H., Campana D. Comparative cytotoxicity of dexamethasone and prednisolone in childhood acute lymphoblastic leukemia. J. Clin. Oncol. 1996;14(8):2370–2376. doi: 10.1200/JCO.1996.14.8.2370. [DOI] [PubMed] [Google Scholar]
- 31.Ghasemi A., Khanzadeh T., Zadi Heydarabad M., Khorrami A., Jahanban Esfahlan A., Ghavipanjeh S., Gholipour Belverdi M., Darvishani Fikouhi S., Darbin A., Najafpour M., Azimi A. Evaluation of BAX and BCL-2 gene expression and apoptosis induction in acute lymphoblastic leukemia cell line CCRFCEM after high- dose prednisolone treatment. Asian Pacific J. Cancer Prevent. 2018;19(8):2319–2323. doi: 10.22034/APJCP.2018.19.8.2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Espinasse M.A., Pépin A., Virault-Rocroy P., Szely N., Chollet-Martin S., Pallardy M., Biola-Vidamment A. Glucocorticoid-Induced Leucine Zipper Is Expressed in Human Neutrophils and Promotes Apoptosis through Mcl-1 Down-Regulation. J. Innate Immun. 2016;8(1):81–96. doi: 10.1159/000439052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Haque S., Vaiselbuh S.R. Silencing of exosomal miR-181a reverses pediatric acute lymphocytic leukemia cell proliferation. Pharmaceuticals. 2020;13(9):241. doi: 10.3390/ph13090241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Satyam Abhigyan, Tsokos Maria G., Tresback Jason S., Zeugolis Dimitrios I., Tsokos G.C. Cell-derived extracellular matrix-rich biomimetic substrate supports podocyte proliferation, differentiation, and maintenance of native phenotype. Adv. Funct. Mater. 2020 doi: 10.1002/adfm.201908752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thippabhotla S., Zhong C., He M. 3D cell culture stimulates the secretion of in vivo like extracellular vesicles. Sci Rep. 2019;9(1):13012. doi: 10.1038/s41598-019-49671-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liao W., Liu X., Peng H., Xu L. The expression and functional study of miR-181a in pediatric acute lymphoblastic leukemia. Zhonghua Xue Ye Xue Za Zhi. 2015;36(1):53–57. doi: 10.3760/cma.j.issn.0253-2727.2015.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ouyang M., Li Y., Ye S., Ma J., Lu L., Lv W., Chang G., Li X., Li Q., Wang S., Wang W. MicroRNA profiling implies new markers of chemoresistance of triple-negative breast cancer. PLoS ONE. 2014;9(5):e96228. doi: 10.1371/journal.pone.0096228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ariës I.M., Hansen B.R., Koch T., van den Dungen R., Evans W.E., Pieters R., den Boer M.L. The synergism of MCL1 and glycolysis on pediatric acute lymphoblastic leukemia cell survival and prednisolone resistance. Haematologica. 2013;98(12):1905–1911. doi: 10.3324/haematol.2013.093823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hulleman E., Kazemier K.M., Holleman A., VanderWeele D.J., Rudin C.M., Broekhuis M.J., Evans W.E., Pieters R., Den Boer M.L. Inhibition of glycolysis modulates prednisolone resistance in acute lymphoblastic leukemia cells. Blood. 2009;113(9):2014–2021. doi: 10.1182/blood-2008-05-157842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sakurai N., Komada Y., Hanaki R., Morimoto M., Ito T., Nakato D., Hirayama M. Role of microRNAs in glucocorticoid-resistant B-cell precursor acute lymphoblastic leukemia. Oncol. Rep. 2019;42(2):708–716. doi: 10.3892/or.2019.7191. [DOI] [PubMed] [Google Scholar]
- 41.Chorna I.V., Datsyuk L.O., Stoika R.S. Expression of Bax, Bad and Bcl-2 proteins under x-radiation effect towards human breast carcinoma MCF-7 cells and their doxorubicin-resistant derivatives. Exp. Oncol. 2005;27(3):196–201. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.