Abstract
Upon acquiring two unique plasmids (pMT1 and pPCP1) and genome rearrangement during the evolution from Yersinia pseudotuberculosis, the plague causative agent Y. pestis is closely related to Y. pseudotuberculosis genetically but became highly virulent. We developed a pentaplex real‐time PCR assay that not only detects both Yersinia species but also differentiates Y. pestis strains regarding their plasmid profiles. The five targets used were Y. pestis‐specific ypo2088, caf1, and pst located on the chromosome, plasmids pMT1 and pPCP1, respectively; Y. pseudotuberculosis‐specific chromosomal gene opgG; and 18S ribosomal RNA gene as an internal control for flea DNA. All targets showed 100% specificity and high sensitivity with limits of detection ranging from 1 fg to 100 fg, with Y. pestis‐specific pst as the most sensitive target. Using the assay, Y. pestis strains were differentiated 100% by their known plasmid profiles. Testing Y. pestis and Y. pseudotuberculosis‐spiked flea DNA showed there is no interference from flea DNA on the amplification of targeted genes. Finally, we applied the assay for testing 102 fleas collected from prairie dog burrows where prairie dog die‐off was reported months before flea collection. All flea DNA was amplified by 18S rRNA; no Y. pseudotuberculosis was detected; one flea was positive for all Y. pestis‐specific targets, confirming local Y. pestis transmission. Our results indicated the assay is sensitive and specific for the detection and differentiation of Y. pestis and Y. pseudotuberculosis. The assay can be used in field investigations for the rapid identification of the plague causative agent.
Keywords: assay development, fleas, pentaplex real‐time PCR, Yersinia pestis, Yersinia pseudotuberculosis
Short abstract
We developed a pentaplex real‐time PCR assay including Yersinia pestis‐specific genes ypo2088, caf1, and pst located on the chromosome, as well as on plasmids pMT1 and pPCP1, respectively. Yersinia pseudotuberculosis‐specific chromosomal gene opgG and the 18S ribosomal RNA gene served as an internal control for flea DNA. All targets showed 100% specificity and high sensitivity with limits of detection ranging from 1 to 100 femtograms. Using the assay, Y. pestis was detected in field‐collected fleas and strains were differentiated 100% by their known plasmid profiles.
1. INTRODUCTION
Yersinia pestis, a Tier 1 select agent, is the bacterial causative pathogen of a plague that infects rodents and can be transmitted to other mammals, including humans through flea biting (Perry & Fetherston, 1997; Stenseth et al., 2008). Plague causes serious illness or death if not treated promptly, especially if it develops into the pneumonic plague.
The plague has had a big impact on human history through three devastating pandemics (Perry & Fetherston, 1997) causing millions of deaths, as well as by locally endemic occurrence which results in hundreds to thousands of annual cases worldwide. Currently, the plague is endemic in Asia, Africa, and the Americas (Bertherat, 2016). Multiple recent outbreaks, including that in China, Democratic Republic of Congo, Madagascar, Peru, and Uganda (Abedi et al., 2018; Andrianaivoarimanana et al., 2019; Randremanana et al., 2019; Respicio‐Kingry et al., 2016; Shi et al., 2018) warn us not to ignore this severe infectious disease.
Along with Y. pestis, Y. pseudotuberculosis, and Y. enterocolitica are the other two related species within the genus Yersinia that can cause diseases in both animals and humans (Perry & Fetherston, 1997). Nevertheless, symptoms of disease caused by enteropathogens Y. pseudotuberculosis and Y. enterocolitica are vastly different from the plague. Belonging to the same genus, these organisms, especially Y. pestis and Y. pseudotuberculosis, are genetically closely related (Moore & Brubaker, 1975). Previous studies have indicated that Y. pestis has diverged recently from Y. pseudotuberculosis through events of genome reduction and gene gain; thus, the high genomic similarity were observed between the two species (Achtman et al., 1999; Brubaker, 2004; Chain et al., 2004; Hu et al., 1998; Moore & Brubaker, 1975; Parkhill et al., 2001). In the genetic structure, all three species possess a single circular chromosome and a common virulence plasmid termed pCD1, or pIB, or pYV in Y. pestis, Y. pseudotuberculosis, and Y. enterocolitica, respectively (Cornelis & Wolf‐Watz, 1997; Iriarte & Cornelis, 1996; Perry & Fetherston, 1997). The most prominent genetic difference between Y. pestis and the enteropathogenic Yersinia species is the presence of pPCP1 and pMT1, two newly acquired plasmids in most but not all strains of Y. pestis (Ben‐Gurion & Shafferman, 1981; Ferber & Brubaker, 1981; Filippov, Oleinikov, Motin, Protsenko, & Smirnov, 1995; Filippov, Solodovnikov, & Protsenko, 1990; Hu et al., 1998; Portnoy & Falkow, 1981). These plasmids contain different genes that play important roles regarding pathogenesis. The plasmid pCD1 contains the highly conserved type three secretion system (T3SS) injectosome and effector proteins Yops (Rajanna et al., 2010); pPCP1 contains pla gene and pst gene that encode plasminogen activator (PLA) protease and the pesticin protein (Bearden, Fetherston, & Perry, 1997); pMT1 contains caf1 and ymt, encoding fraction 1 capsule antigen and murine toxin, respectively, as well as other putative virulence determinants (Hu et al., 1998).
Most Y. pestis strains isolated from humans, animals, or fleas contain all three typical plasmids. However, Y. pestis isolates lacking one or more plasmids may cause mild or even fatal disease, as previously been reported in South Africa and the United States (Beesley & Surgalla, 1970; Isaacson et al., 1973; Williams, Harrison, & Cavanaugh, 1975; Williams et al., 1978; Winter, Cherry, & Moody, 1960). Such variants have been reported infrequently. The natural occurrence of strains with atypical plasmid profiles might be greater than present data suggest.
The extreme pathogenicity of Y. pestis makes accurate and rapid detection of this bacterium a priority. Traditional methods of detecting Y. pestis include standard microbiological techniques (Norkina et al., 1994) and immunofluorescent staining (Feodorova & Devdariani, 2000). However, these methods take a long time and are relatively insensitive. In contrast, PCR‐based assays have the advantage of rapid detection with high sensitivity. Furthermore, PCR is often preferable with the select agent and biosafety concerns. PCR assays for Y. pestis detection are not uncommon. Several PCR assays have been developed (Campbell, Lowe, Walz, & Ezzell, 1993; Matero et al., 2009; Neubauer et al., 2000; Radnedge, Gamez‐Chin, McCready, Worsham, & Andersen, 2001; Riehm et al., 2011; Stewart, Satterfield, Cohen, O'Neill, & Robison, 2008; Tomaso et al., 2003; Woron et al., 2006). The pPCP1‐situated pla gene, known to be present in 150–200 copies per bacterium (Parkhill et al., 2001), has been widely used due to the very high sensitivity (Riehm et al., 2011; Stewart et al., 2008; Woron et al., 2006). Ideally, if we are dealing with typical Y. pestis strains that harbor all three plasmids, a pla‐based assay could be reasonable. However, a good portion (~25%) of Y. pestis strains existing in nature are deficient in one or more of the three plasmids (Filippov et al., 1990; Stewart et al., 2008) due to various host growth temperatures, as well as other unknown factors (Iqbal, Chambers, Goode, Valdes, & Brubaker, 2000). As such, a plasmid‐based assay may yield false‐negative results. On the other hand, a pla assay may cause false‐positive results because pla may also be present in other Enterobacteriaceae such as Escherichia coli and Citrobacter koseri (Armougom et al., 2016; Hänsch et al., 2015). Further, a homolog of pla has been reported in rodents of multiple species (Giles et al., 2016). In addition to plasmid‐based genes, chromosomal genes, such as entF3, were also applied for Y. pestis detection (Woron et al., 2006). The problem with using a chromosomal gene alone is it may involve non‐specific amplification due to the genetic relatedness of Y. pestis to the other Yersinia species. Amplification with a chromosomal gene may not indicate which species is present (Woron et al., 2006).
In the current study, we developed a pentaplex real‐time PCR assay that includes three Y. pestis‐specific targets, one Y. pseudotuberculosis‐specific target and one internal control for flea DNA since the assay is intended to test field‐collected fleas. The three Y. pestis‐specific targets in the assay are pPCP1‐situated pst, pMT1‐situated caf1, and chromosomal gene ypo2088. ypo2088 codes for a putative methyltransferase and is considered Y. pestis‐specific (Chain et al., 2004; Matero et al., 2009). The Y. pseudotuberculosis‐specific gene used in the assay is chromosomal gene opgG. Yersinia pestis lost this gene during its speciation from Y. pseudotuberculosis; hence, opgG can differentiate Y. pestis strains and Y. pseudotuberculosis strains (Quintard et al., 2015). For the internal control of flea DNA, the very conservative 18S ribosomal gene was used for a wide application for any flea species in general. The objective of this study was to allow accurate and rapid identification of all Y. pestis strains from field‐collected fleas; to discriminate Y. pestis from Y. pseudotuberculosis; and to differentiate Y. pestis strains in regard to their plasmid profiles.
2. EXPERIMENTAL PROCEDURES
2.1. Gene selection, primer, and probe design
Five genes were selected for developing the pentaplex real‐time assay: Yersinia pst, caf1, ypo2088, and opgG, and flea 18S rRNA. Sequences of primer and probe used in the assay were designed for the current study except for those for caf1 which were adapted from assays published (Liu et al., 2016) (Table 1). The TaqMan primers/probes design was conducted with the PrimerQuest program, Integrated DNA Technologies, Inc., Coralville, Iowa, https://www.idtdna.com/SciTools. Also, the pla gene of Y. pestis was tested using previously published primers and probe (Riehm et al., 2011) for comparison and validation purposes.
Table 1.
Description of the genes selected for the pentaplex real‐time PCR and the validation PCR, and the primers/probes sequences
| Gene/location | Detection | Primer/probe sequences (5′‐3′) | Product | Reference |
|---|---|---|---|---|
| pst/pPCP1 | Y. pestis strains with pPCP1 plasmid presence | Forward: GCGAAGCAAACAGGATTTATTG | 116 bp | This study |
| Reverse: GAGGTGCTGTTCTCACTTTATC | ||||
| Probe: FAM‐AGCCTCCTTCCCTCGAAGCATATAATACCC‐BHQ−1 | ||||
| caf1/pMT1 | Y. pestis strains with pMT1 plasmid presence | Forward: CCACTGCAACGGCAACTCTT | 71 bp | Liu et al. (2016) |
| Reverse: TGTAATTGGAGCGCCTTCCT | ||||
| Probe: QUAS705‐TTGAACCAGCCCGCATCACTCTTACA‐BHQ3 | ||||
| ypo2088/chromosome | All Y. pestis strains | Forward: TCGGCAACAGCTCAACACCT | 107 bp | This study |
| Reverse: ATGCATTGGACGGCATCACG | ||||
| Probe: CALRD610‐CGCCCTCGAATCGCTGGCCAACTGC‐BHQ2 | ||||
| opgG/chromosome | Y. pseudotuberculosis strains | Forward: ACGTGGGCGTGAATTCTCTCAA | 126 bp | This study |
| Reverse: GCCGTTGGGATCTCCACCAA | ||||
| Probe: QUAS670‐CCTGCGCCCAAGCGCGTGGG‐BHQ2 | ||||
| 18S rRNA | Internal control for flea DNA | CAGATACCGCCCTAGTTC TAA | 135 bp | This study |
| Reverse: GTTTCAGCTTTGCAACCATAC | ||||
| Probe: HEX‐TCATCGGAGGAACTTCGGCGGATC‐BHQ−1 | ||||
| pla/pPCP1 | Y. pestis strains with pPCP1 plasmid presence | Forward: Forward: GTAATAGGTTATAACCAGCGCTT | 232 bp | Rajanna et al. (2010) and Tomaso et al. (2003) |
| Reverse: AGACTTTGGCATTAGGTGTG | ||||
| Probe: HEX‐ATGCCATATATTGGACTTGCAGGCCAGT‐BHQ1 |
2.2. Template DNA
All Y. pestis and Y. pseudotuberculosis DNA templates used in the study were obtained from BEI Resources or the University of Texas Medical Branch (UTMB) unless otherwise stated (Table 2). Yersinia pestis: CO96‐3188 DNA was obtained from CDC Bacterial Diseases Branch. CO96‐3188 is a fully virulent (Pgm+, pMT1+, pCD1+, and pPCP1+) North American biovar Orientalis strain (Eisen et al., 2006; Engelthaler, Hinnebusch, Rittner, & Gage, 2000). For the other 16 Y. pestis strains, both pMT1 and pPCP1 were present in 11 strains; both pMT1 and pPCP1 were absent in one strain; pPCP1was absent in four more strains (Table 2). Yersinia pseudotuberculosis was represented by strain B15 and four other strains (Table 2). Two Y. enterocolitica strains were included for specificity checking (Table 2). Also, we included 12 non‐Yersinia species representing five Bartonella spp. (B. bovis, B. doshiae, B. elizabethae, B. henselae, and B. rochalimae), Rickettsia rickettsii, Anaplasma phagocytophilum, three Borrelia spp. (B. burgdorferi, B. miyamotoi, B. mayonii), Trypanosoma cruzi, and Leptospira interrogans for specificity testing (Table 3). The reason we included these species is that the transmission route of these bacteria is similar to the Yersinia species, either by fleas or other blood‐feeding vectors (Bai et al., 2017; Brook et al., 2015; Livengood, Hutchinson, Thirumalapura, & Tewari, 2020; Pawelczyk, Asman, & Solarz, 2019) or by consumption of contaminated water (Casanovas‐Massana, Pedra, Wunder, Begon, & Ko, 2018). All DNA from these non‐Yesinia species was obtained from CDC Bacterial Diseases Branch. DNA extracted from Xenopsylla cheopis fleas raised at the CDC colony were used as internal controls in the experiment. Finally, DNA extracted from alcohol‐preserved field‐collected fleas were tested to check whether 18S rRNA amplifies DNA of flea species other than X. cheopis, and if interference or inhibition would occur while applying the assay to field samples. These fleas were collected from guinea pigs (Cavia porcellus) in Peru and represent three species, including Pulex sp. (n = 6), Ctenocephalides felis (n = 5), and Tiamastus cavicola (n = 5). A previous study showed they were negative for Bartonella species (María et al., 2019).
Table 2.
Plasmid presence/absence in Yersinia pestis strains and PCR results in all strains of Y. pestis, Y. pesudotuberculosis, and Y. enterocolitica used to evaluate assay performance
| ID | Strain | Source | Plasmid presence | PCR | ||||
|---|---|---|---|---|---|---|---|---|
| pMT1 | pPCP1 | ypo2088 | caf1 | pst | opgG | |||
| Y. pestis | ||||||||
| NR‐4709 | KIM D23 | BEI | Yes | No | pos | pos | neg | neg |
| NR‐4713 | A12 D6 | BEI | Yes | No | pos | pos | neg | neg |
| NR‐4706 | KIM D2 | BEI | Yes | Yes | pos | pos | pos | neg |
| NR‐4717 | Yokohama D11 | BEI | Yes | Yes | pos | pos | pos | neg |
| NR‐4719 | Kimberley D13 | BEI | Yes | Yes | pos | pos | pos | neg |
| NR‐4727 | K25 D80 | BEI | Yes | Yes | pos | pos | pos | neg |
| NR‐4705 | KIM D19 | BEI | Yes | Yes | pos | pos | pos | neg |
| NR‐2645 | KIM10+ | BEI | Yes | No | pos | pos | neg | neg |
| NR‐4708 | KIM D22 | BEI | Yes | No | pos | pos | neg | neg |
| NR‐4714 | Kuma D7 | BEI | Yes | Yes | pos | pos | pos | neg |
| NR‐4716 | yokohama D10 | BEI | Yes | Yes | pos | pos | pos | neg |
| NR‐4718 | Kimberley D12 | BEI | Yes | Yes | pos | pos | pos | neg |
| NR‐4721 | K25 D72 | BEI | Yes | Yes | pos | pos | pos | neg |
| NR‐2718 | PB6 | BEI | No | No | pos | neg | neg | neg |
| NR‐2719 | Harbin 35 | BEI | Yes | Yes | pos | pos | pos | neg |
| NR‐2720 | Nepal 516 | BEI | Yes | Yes | pos | pos | pos | neg |
| Y. pseudotuberculosis | neg | neg | neg | pos | ||||
| NR‐4653 | YPIII(p+) | BEI | neg | neg | neg | pos | ||
| NR‐4651 | IP2775 | BEI | neg | neg | neg | pos | ||
| 4284 | 4284 | UTMB | neg | neg | neg | pos | ||
| 6904 | 6904 | UTMB | neg | neg | neg | pos | ||
| Y. enterocolitica | ||||||||
| NR‐3064 | Billups‐1803‐68 | BEI | neg | neg | neg | neg | ||
| NR‐3065 | WA | BEI | neg | neg | neg | neg | ||
Table 3.
PCR results on non‐Yersinia bacterial species for specificity testing of the pentaplex assay
| Bacterial species | YPO2088 | caf1 | pst | opgG | 18S rRNA |
|---|---|---|---|---|---|
| Anaplasma phagocytophilum | neg | neg | neg | neg | neg |
| Bartonella bovis | neg | neg | neg | neg | neg |
| Bartonella doshiae | neg | neg | neg | neg | neg |
| Bartonella elizabethae | neg | neg | neg | neg | neg |
| Bartonella henselae | neg | neg | neg | neg | neg |
| Bartonella rochalimae | neg | neg | neg | neg | neg |
| Borrelia bergdorferi | neg | neg | neg | neg | neg |
| Borrelia mayonii | neg | neg | neg | neg | neg |
| Borrelia miyamotoi | neg | neg | neg | neg | neg |
| Leptospira interrogans | neg | neg | neg | neg | neg |
| Rickettsia rickettsii | neg | neg | neg | neg | neg |
| Trypanosoma cruzi | neg | neg | neg | neg | neg |
2.3. Optimization of primer concentration
DNA of Y. pestis wild‐type strain CO96‐3188, Y. pseudotuberculosis strain B15 and Xenopsylla cheopis fleas raised at the CDC colony were used for the optimization of the primer concentration and identification of the limit of detection (LOD) of each target. Three DNA concentrations (100 pg/µl, 10 pg/µl, 1 pg/µl) of each template were used to test the targets independently with the corresponding primers at different concentrations, for example, DNA of Y. pestis strain CO96‐3188 for pst, caf1, and ypo2088; DNA of Y. pseudotuberculosis strain B15 for opgG; and DNA of Xenopsylla cheopis flea for 18S rRNA. The optimal primer concentrations were tested using a primer concentration matrix with final concentrations of 200 nM–1000 nM. PCR (25 µl) contained 12.5 µl of 2× PerfeCta MultiPlex qPCR SuperMix (Quanta Biosciences, Gaithersburg, MD), primers (forward and reverse), probes at a standard concentration of 200 nM, and 5 µl of DNA template. Real‐time PCR was performed on a CFX96 Real‐Time System (Bio‐Rad, Hercules, CA) with the following conditions: 95°C for 2 min followed by 45 cycles of 95°C for 15 s and 64°C for 60 s. Distilled water was always included as a negative control. A primer concentration that generates a sigmoidal shape of the amplification curve with a relatively lower Ct value compared to other primer concentration is considered an optimized concentration.
2.4. Sensitivity testing and specificity assessment
Sensitivity was estimated using the limit of detection (LOD). The LOD for each target was determined and verified by comparing the primer/probe in a single PCR and in a pentaplex PCR simultaneously using the optimized primer concentration by testing six replicates of each DNA template with the following dilutions: 2 pg, 1 pg, 0.5 pg, 0.2 pg, 0.1 pg, 50 fg, 20 fg, 10 fg, 5 fg, 2 fg, 1 fg, 0.5 fg, 0.2 fg, 0.1 fg, 0.05 fg, and 0.02 fg. The lowest dilution that was amplified in all six replicates in both single PCR and pentaplex PCR, with a threshold Ct value <40 in any of the reactions was considered the LOD. To validate the result, an identical set of real‐time PCR was performed using the widely used pla gene (Riehm et al., 2011).
The specificity of each primer/probe set was assessed by testing high concentration (500 pg) of nucleic acid from Y. pestis, Y. pseudotuberculosis, Y. enterocolitica strains, and other 12 non‐Yersinia microorganisms (Table 3) by running a pentaplex real‐time PCR using the optimized primer concentrations.
2.5. Evaluation of the pentaplex assay
To evaluate whether the pentaplex assay is applicable for real sample testing, sixteen fleas collected from guinea pigs in Peru and were preserved in alcohol before DNA extraction were first tested for the presence of flea DNA and any other DNA using the pentaplex PCR assay; then, the flea DNA was spiked with DNA of Y. pestis strain CO96‐3788 or Y. pseudotuberculosis strain B15. For comparison purposes, final DNA concentrations of Y. pestis strain or Y. pseudotuberculosis matched those used for sensitivity testing. Pentaplex PCR was performed to test the Y. pestis‐ and Y. pseudotuberculosis‐spiked flea DNA separately to check whether the flea DNA and alcohol would interfere or inhibit the amplification. Six replicates were tested for each concentration of each spiked DNA.
2.6. Differentiation of Yersinia species and elucidation of Y. pestis strains with different plasmid profiles
Genomic DNA of Y. pestis strains with different plasmid profiles, Y. pseudotuberculosis strains, and Y. enterocolitica strains were tested with all four sets of Yersinia primer/probe (pst, caf1, ypo2088, and opgG) with a correspondent optimized concentration of each primer in a multiplex PCR setting.
2.7. Application of the assay for testing field‐collected fleas
To evaluate the utility of the pentaplex real‐time PCR assay, we tested fleas (n = 102) collected from burrows in a prairie dog colony in Converse County, Wyoming in July 2019. A prairie dog die‐off was noticed in the area in February 2019 and plague activity was suspected. Collected fleas belonged to three species: Oropsylla hirsuta (n = 87), O. tuberculata (n = 14), and Thrassiis fotus (n = 1). The fleas were stored in 70% ethanol and kept at 4°C before testing. DNA extraction was prepared from individual fleas using a Kingfisher platform (Life Technologies, Inc., Grand Island, NY) following the instructions provided by the manufacturer.
3. RESULTS
3.1. Optimization of primer concentration
The amplification was grouped tightly by the concentration of DNA for each target (Figure 1). The higher the DNA concentration, the earlier the amplification started (lower Ct value). Primer concentrations had no significant differences in the amplification efficiencies with the change of Ct value <1. For example, for pst gene, the Ct value ranged 22.14–22.82 with primer concentrations of 200 nM–1000 nM when the DNA concentration was 100 pg. Based on our criteria (generating a sigmoidal shape of the amplification curve with lower Ct value), the optimized primer concentration is 200 nM for all Y. pestis‐associated targets (pst, caf1, and ypo2088) primers, 600 nM for opgG primers, and 500 nM for 18S rRNA primers (Table 4).
Figure 1.
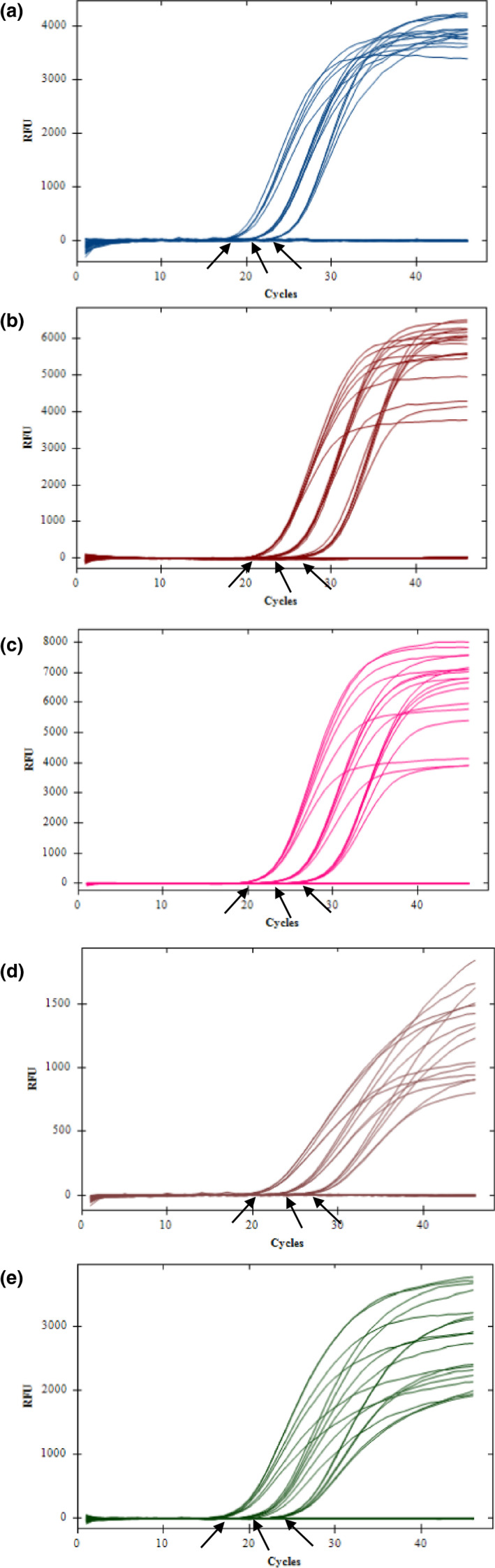
Amplification of DNA (100 pg/µl, 10 pg/µl, 1 pg/µl) with different primer concentrations ranging from 200 nM to 1000 nM. The amplification was grouped tightly by the concentration of DNA. The higher the DNA concentration, the earlier the amplification started (lower Ct value). Primer concentrations had no significant differences in the amplification efficiencies with a very small change of Ct value. The three arrows indicated the DNA concentration for each group was 100 pg/µl, 10 pg/µl, and 1 pg/µl from left to right. A. pst; B. caf1; C. ypo2088; D. opgG; E. 18S rRNA
Table 4.
Optimization of primer/probe concentration and identification of limit of detection (LOD) of each gene used in the pentaplex assay
| Gene | Primer concentration (nM) | Probe concentration (nM) | LOD (fg) |
|---|---|---|---|
| pst | 200 nM | 200 nM | 1 |
| caf1 | 200 nM | 200 nM | 50 |
| ypo2088 | 200 nM | 200 nM | 100 |
| opgG | 600 nM | 200 nM | 5 |
| 18S rRNA | 500 nM | 200 nM | 1 |
3.2. Sensitivity and specificity
All targets showed extremely high sensitivity (Figure 2). Among the three Y. pestis‐specific targets, pst was the most sensitive with a LOD of 1 fg per reaction, followed by caf1 (LOD 50 fg) and ypo2088 (LOD 100 fg) (Table 4). The Y. pseudotuberculosis‐specific opgG also showed very high sensitivity, with a LOD of 5 fg per reaction; the internal control target 18S rRNA for flea DNA had a LOD of 1 fg. Standard curves showed the Ct values were correlated to the DNA concentration for each target (Figure 2).
Figure 2.
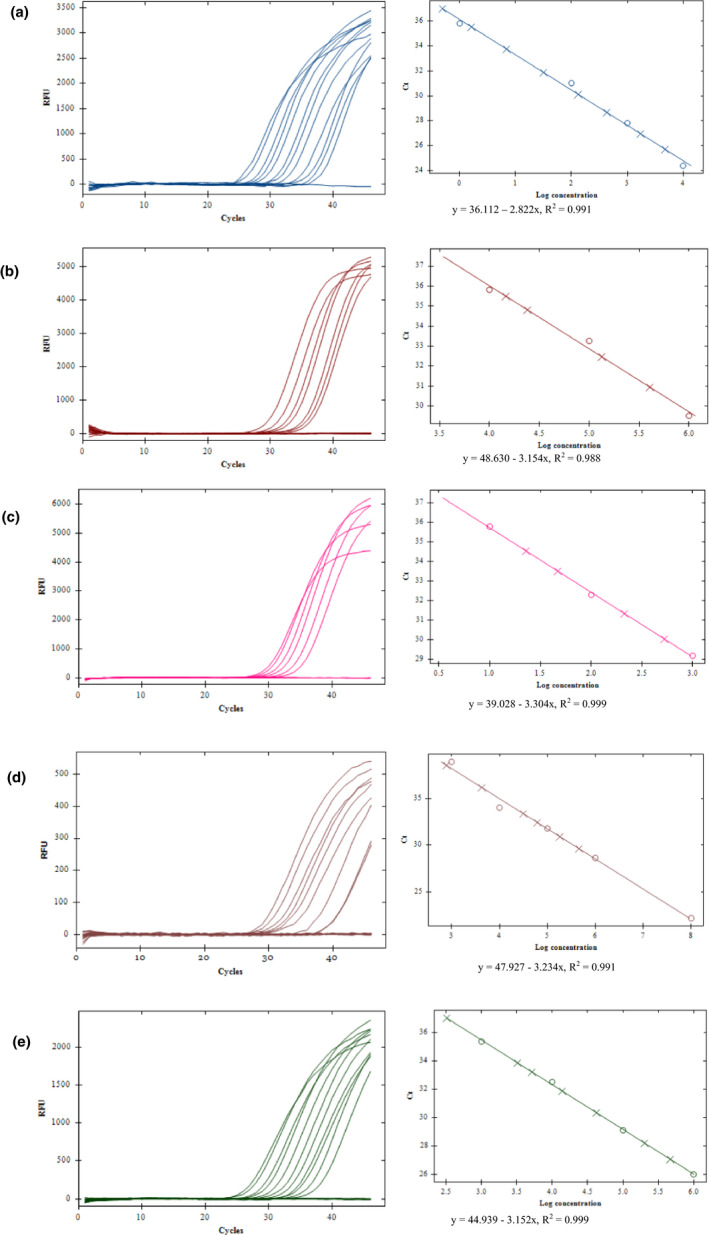
Sensitivity testing. Amplification (left panel) and standard curve (right panel) of each target with DNA dilutions 2 pg, 1 pg, 0.5 pg, 0.2 pg, 0.1 pg, 50 fg, 20 fg, 10 fg, 5 fg, 2 fg, 1 fg, 0.5 fg, 0.2 fg, 0.1 fg, 0.05 fg, and 0.02 fg. Ct values were correlated to the DNA concentration for each target. A. pst; B. caf1; C. ypo2088; D. opgG; E. 18S rRNA
To validate the above testing results, a PCR was performed using the widely used pla gene. The PCR detected 1 fg Y. pestis DNA as the LOD, which is identical to that of pst, suggesting that pst gene and pla gene are equivalent in terms of the sensitivity.
All primer/probe sets showed 100% specificity to the targets accordingly. DNA of Y. pestis strain CO96‐3188 was positive to pst, caf1, and ypo2088 but negative to opgG and 18S rRNA; DNA of Y. pseudotuberculosis strain B15 was positive to opgG but negative to all other genes; DNA of X. cheopis fleas was positive to 18S rRNA but negative to all other genes; no amplification to any of the genes was observed in Y. enterocolitica (Table 2) and any of the other 12 species of non‐Yersinia microorganisms (Table 3).
3.3. Differentiation between Y. pestis strains and other Yersinia species
Testing of all Yersinia DNA (16 Y. pestis, 4 Y. pseudotuberculosis, and 2 Y. enterocolitica) with the four primers/probe sets of Yersinia targets demonstrated that the two Y. enterocolitica were negative to all targets; the four Y. pseudotuberculosis were positive to Y. pseudotuberculosis‐specific opgG but to none of the Y. pestis‐specific genes; the 16 Y. pestis DNA samples were positive to different Y. pestis‐specific targets, which was associated with their plasmid profile (BEI resource): All strains were positive to the chromosomal gene ypo2088; the strain with pMT1 and pPCP1 absent was negative to both caf1 and pst; four strains with pPCP1 absence were negative to pst. None of the Y. pestis strains were positive to opgG (Table 2).
3.4. Pentaplex assay evaluation
Testing of 16 DNA of fleas collected from guinea pigs in Peru showed all fleas were amplified with 18S rRNA, with Ct values ranging 17.62–31.08 for the Pulex fleas; 20.54–34.33 for the Ctenocephalides felis fleas; and 21.13–31.05 for the Tiamastus cavicola fleas, showing that 18S rRNA worked equally for different flea species. All fleas were negative to ypo2088, pst, caf1, and opgG.
Testing of Y. pestis DNA‐spiked flea DNA showed that ypo2088, pst, caf1, and 18S rRNA were amplified, with LOD 1 fg, 50 fg, 100 fg for ypo2088, pst, caf1, respectively, all of which were identical to the LOD observed in the sensitive testing. All DNA in this group was negative to opgG.
Testing of Y. pseudotuberculosis DNA‐spiked flea DNA showed that opgG and 18S rRNA were amplified with LOD 5 fg for opg. The LOD was identical to that observed in the sensitive testing. All DNA in this group was negative to ypo2088, pst, and caf1.
3.5. Detection of Yersinia pestis in field‐collected fleas
By performing the pentaplex real‐time PCR, all 102 fleas collected from prairie dog burrows several months after a prairie dog die‐off was reported in Converse County, Wyoming in July 2019 were positive to 18S rRNA, with Ct value ranging 13.93–27.91. Among these, one Oropsylla tuberculata flea (Ct value 15.06 for 18S rRNA) was positive to all three Y. pestis genes, with Ct value 19.65, 21.08, and 21.74 for pst, caf1, and ypo2088, respectively (Figure 3). The other 101 fleas were negative to any of the three Y. pestis genes. No Y. pseudotuberculosis was detected in any fleas all of which was negative to opgG.
Figure 3.
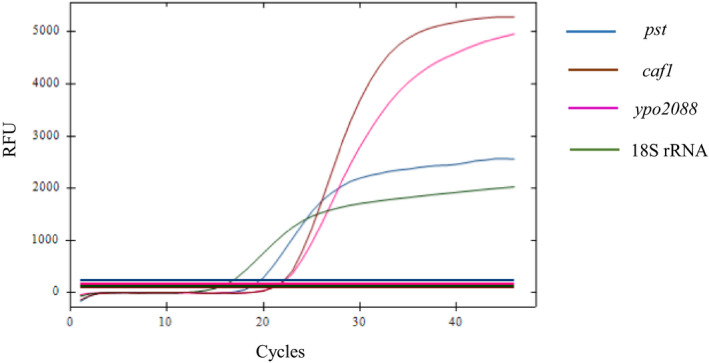
By performing the pentaplex real‐time PCR, one Oropsylla tuberculata flea collected from prairie dog burrows after a prairie dog die‐off in Wyoming in 2019 was positive to pst, caf1, ypo2088, and 18S rRNA, with Ct value 19.65, 21.08, 21.74, and 15.06, respectively, suggesting a local Y. pestis transmission
4. DISCUSSION
It is of supreme importance using analytical assays for Y. pestis which can show the presence of various targets located on the chromosome and plasmids. A pentaplex real‐time PCR assay developed in this study included three Y. pestis‐specific genes (pst, caf1, and ypo2088), one Y. pseudotuberculosis gene (opgG), and 18S rRNA as an internal control of flea DNA if the assay is to be applied to test fleas. Multiplex PCR assays for Y. pestis are not uncommon (Stewart et al., 2008; Tomaso et al., 2003; Woron et al., 2006) but, to our knowledge, this is the first to integrate five biologically meaningful targets.
Many assays have used the pla gene that is located on the pPCP1 plasmid for the detection of Y. pestis due to its high sensitivity with the presence of high copy numbers (150–200 copies) (Parkhill et al., 2001). Finding a homolog of pla in other bacteria or rodents (Armougom et al., 2016; Giles et al., 2016; Hänsch et al., 2015) has raised a concern of false identification, particularly when considering the frequency of flea‐feeding on rodent hosts. Instead of using pla, we used pst which is also located on pPCP1. Our results demonstrated pst is equally as sensitive as the pla gene with the same LOD (1 fg per reaction). Such results suggest that pst is a plausible diagnostic marker to replace pla. Like pla, the pMT1‐located caf1 is another commonly used target for Y. pestis detection in multiple studies (Norkina et al., 1994; Stewart et al., 2008; Tomaso et al., 2003; Woron et al., 2006). Similar to other reports, our results also showed that caf1 is highly sensitive, but somewhat less sensitive compared to pst. It is known that caf1 is present in only about two copies per bacterium (Parkhill et al., 2001), explaining the differences in sensitivity between the two genes.
We did not use any pCD1‐located genes in this study. This is because pCD1 is possessed by all currently recognized pathogenic Yersinia species. Detection based on pCD1‐located genes does not necessarily indicate the species of the presenting organism as previous studies have shown detection of Y. pestis, Y. enterocolitica, and Y. pseudotuberculosis using pCD1‐located lcrV or other targets (Stewart et al., 2008). Further, using a pCD1‐located gene may not be as efficient compared to pPCP1‐ and pMT1‐located genes because the pCD1 plasmid is absent in many more Y. pestis strains compared to pPCP1 and pMT1 plasmids in nature due to the intrinsic variability of pCD1 plasmids in Y. pestis (Stewart et al., 2008).
Unlike plasmids, chromosomal genes are not likely to be lost during laboratory cultivation or in nature; thus, chromosomal genes are more reliable targets than plasmid genes. Nevertheless, specific identification can be difficult if substantial genomic similarities are shared among multiple species. In the past, researchers have used the inv gene, the entF3 gene, the wzz gene, and the 16S rRNA gene in the identification of Y. pestis (Matero et al., 2009; Neubauer et al., 2000; Tomaso et al., 2003; Woron et al., 2006). However, non‐specific amplification has been observed with these genes. All of them not only amplify Y. pestis, but also Y. pseudotuberculosis. In our assay, we used ypo2088 and opgG, two chromosomal genes that are designed for Y. pestis and Y. pseudotuberculosis, respectively. Our results demonstrated that all Y. pestis strains were detected by ypo2088 and Y. pseudotuberculosis strains by opgG. Both ypo2088 and opgG showed high sensitivity (LOD 100 fg and 5 fg respectively) with 100% specificity to their targeted Yersinia species. Because opgG has been lost in Y. pestis, this makes it ideal for the differentiation of Y. pestis and Y. pseudotuberculosis. With a combined analysis of Y. pestis‐specific and Y. pseudotuberculosis‐specific assays, any detection will be elucidated by the exclusion of one from the other. No amplification was observed with Y. enterocolitica to either gene. Also, several non‐Yersinia microorganisms were tested using the assay for specificity assessment with no amplification observed in any of them. Our results demonstrated all primer/probe used in the assay was 100% specific.
Using the developed assay, we tested Y. pestis strains with known plasmid profiles (BEI Resource). All strains were successfully identified by pst and caf1 in accordance with the presence of the pPCP1 plasmid and pMT1 plasmid with 100% specificity.
Laboratory‐based PCR assays may struggle with different problems in reality, such as low concentration, and inhibition. To observe if that would be a case for our pentaplex assay, we spiked Y. pestis DNA and Y. pseudotuberculosis DNA into DNA of fleas that were collected from guinea pigs in Peru and preserved in alcohol before DNA extraction. The results showed the LOD of each primer/probe set on the Y. pestis‐ and Y. pseudotuberculosis‐spiked flea DNA was the same as that observed in the sensitive testing, suggesting no interference or inhibition from fleas or alcohol on the amplifications.
We applied the assay to test 102 fleas that were collected from prairie dog burrows where prairie dog die‐offs were reported 5 months before our field investigation. All fleas were successfully amplified with the 18S rRNA. One out of the 102 fleas was positive for Y. pestis by all three Y. pestis‐specific genes (pst, caf1, and ypo2088). The result confirmed the local transmission of Y. pestis in the prairie dog colony. Since the flea collection did not occur until 5 months afterward the prairie dog die‐off, the low number of positive samples may have suggested plague activity had diminished gradually after the epizootic peak. Flea numbers were noticeably lower compared to an earlier investigation conducted in the area (around 3 months after the die‐off). This indicated that most fleas may have died of starvation. The extremely low number of positive fleas was reasonably expected (Tripp, Gage, Montenieri, & Antolin, 2009). Not surprisingly, no Y. pseudotuberculosis was detected in any of these fleas.
In conclusion, the pentaplex real‐time PCR assay developed in this work is highly sensitive and 100% specific in the detection and differentiation of Y. pestis and Y. pseudotuberculosis. Further, the assay allows one for the elucidation of the presence/absence of Y. pestis pPCP1 plasmid and pMT1 plasmid in a particular strain, which can be applied to testing fleas and other field‐collected materials when the plague is suspected.
CONFLICT OF INTEREST
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions.
AUTHOR CONTRIBUTIONS
Ying Bai: Conceptualization (lead); data curation (lead); formal analysis (lead); investigation (lead); methodology (lead); project administration (equal); supervision (lead); validation (lead); visualization (lead); writing‐original draft (lead); writing‐review and editing (lead). Vladimir Motin: Conceptualization (lead); methodology (lead); writing‐review and editing (equal). Russell E. Enscore: Investigation (lead); writing‐review and editing (supporting). Lynn Osikowicz: Investigation (equal). Maria Rosales Rizzo: Investigation (equal). Andrias Hojgaard: Methodology (equal). Michael Kosoy: Conceptualization (equal); writing‐review and editing (equal). Rebecca J. Eisen: Writing‐review and editing (equal).
ETHICS STATEMENT
None required.
ACKNOWLEDGEMENTS
The following reagents were obtained through the NIH Biodefense and Emerging Infections Research Resources Repository, NIAID, NIH: Genomic DNA from Yersinia pestis, Strain KIM10+, NR‐2645; Genomic DNA from Yersinia pestis, Strain PB, NR‐2718; Genomic DNA from Yersinia pestis, Strain Harbin 35, NR‐2719; Genomic DNA from Yersinia pestis, Strain Nepal 516, NR‐2720; Genomic DNA from Yersinia pestis, Strain KIM Derivative 19 (D19), NR‐4705; Genomic DNA from Yersinia pestis, Strain KIM Derivative 2 (D2), NR‐4706; Genomic DNA from Yersinia pestis, Strain KIM Derivative 22 (D22), NR‐4708; Genomic DNA from Yersinia pestis, Strain KIM Derivative 22 (D22), NR‐4708; Genomic DNA from Yersinia pestis, Strain A12 Derivative 6 (D6), NR‐4713; Genomic DNA from Yersinia pestis, Strain Kuma Derivative 7 (D7), NR‐4714; Genomic DNA from Yersinia pestis, Strain yokohama Derivative 10 (D10), NR‐4716; Genomic DNA from Yersinia pestis, Strain yokohama Derivative 11 (D11), NR‐4717; Genomic DNA from Yersinia pestis, Strain Kimberley Derivative 12 (D12), NR‐4718; Genomic DNA from Yersinia pestis, Strain Kimberley Derivative 13 (D13), NR‐4719; Genomic DNA from Yersinia pestis, Strain K25 Derivative 72 (D72), NR‐4721; Genomic DNA from Yersinia pestis, Strain K25 Derivative 80 (D80), NR‐4727; ; Genomic DNA from Yersinia pseudotuberculosis, Strain IP2775, NR‐4651; Genomic DNA from Yersinia pseudotuberculosis, Strain YPIII(p+), NR‐4653; Genomic DNA from Yersinia enterocolita, Strain Billups‐1803‐68, NR‐3064; and Genomic DNA from Yersinia enterocolita, Strain Billups‐1803‐68, NR‐3064. We thank Scott Bearden from the Bacterial Disease Branch, Division of Vector‐Borne Diseases, Centers for Disease Control and Prevention for providing genomic DNA from Yersinia pestis, strain CO96‐3166 and genomic DNA from Yersinia pseudotuberculosis, strain B15. We thank Nicole Breuner, Erik Foster, Christina Parise, Sarah Maes, Aine Lehane, Christine Graham, Amy Fleshman, and Karen Boegler from the Bacterial Disease Branch, Division of Vector‐Borne Diseases, Centers for Disease Control and Prevention, and Daniel O’Leary, Wyoming Department of Health, for their participation in the field collection of fleas.
Bai Y, Motin V, Enscore RE, et al. Pentaplex real‐time PCR for differential detection of Yersinia pestis and Y. pseudotuberculosis and application for testing fleas collected during plague epizootics. MicrobiologyOpen. 2020;9:e1105 10.1002/mbo3.1105
DATA AVAILABILITY STATEMENT
All data generated or analyzed during this study are included in this published article.
REFERENCES
- Abedi, A. A. , Shako, J.‐C. , Gaudart, J. , Sudre, B. , Ilunga, B. K. , Shamamba, S. K. B. , Diatta, G. , … Piarroux, M. (2018). Ecologic features of plague outbreak areas, Democratic Republic of the Congo, 2004–2014. Emerging Infectious Diseases, 24, 210–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achtman, M. , Zurth, K. , Morelli, G. , Torrea, G. , Guiyoule, A. , & Carniel, E. (1999). Yersinia pestis, the cause of plague, is a recently emerged clone of Yersinia pseudotuberculosis . Proceedings of the National Academy of Sciences of the United States of America, 96, 14043–14048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrianaivoarimanana, V. , Piola, P. , Wagner, D. M. , Rakotomanana, F. , Maheriniaina, V. , Andrianalimanana, S. , … Rajerison, M. (2019). Trends of human plague, Madagascar, 1998–2016. Emerging Infectious Diseases, 25, 220–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armougom, F. , Bitam, I. , Croce, O. , Merhej, V. , Barassi, L. , Nguyen, T. T. , … Raoult, D. (2016). Genomic insights into a new Citrobacter koseri strain revealed gene exchanges with the virulence‐associated Yersinia pestis pPCP1 plasmid. Frontiers in Microbiology, 7, 340 10.3389/fmicb.2016.00340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai, Y. , Osikowicz, L. M. , Kosoy, M. Y. , Eisen, R. J. , Atiku, L. A. , Mpanga, J. T. , Gage, K. (2017). Comparison of zoonotic bacterial agents in fleas collected from small mammals or host‐seeking fleas from a Ugandan region where plague is endemic. mSphere, 2, e00402‐17 10.1128/mSphere.00402-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bearden, S. W. , Fetherston, J. D. , & Perry, R. (1997). Genetic organization of the yersiniabactin biosynthetic region and construction of avirulent mutants in Yersinia pestis . Infection and Immunity, 65, 1659–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beesley, E. D. , & Surgalla, M. J. (1970). Pesticinogeny: A characteristic useful for presumptive identification and isolation of Pasteurella pestis. Applied Microbiology, 19, 915–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben‐Gurion, R. , & Shafferman, A. (1981). Essential virulence determinants of different Yersinia species are carried on a common plasmid. Plasmid, 5, 183–187. [DOI] [PubMed] [Google Scholar]
- Bertherat, E. (2016). Plague around the world, 2010–2015. Weekly Epidemiological Record, 91, 89–104.26922822 [Google Scholar]
- Brook, C. E. , Bai, Y. , Dobson, A. P. , Osikowicz, L. M. , Ranaivoson, H. C. , Zhu, Q. , … Dittmar, K. (2015). Bartonella spp. in fruit bats and blood‐feeding Ectoparasites in Madagascar. PLOS Neglected Tropical Diseases, 9, e0003532 10.1371/journal.pntd.0003532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brubaker, R. R. (2004). The recent emergence of plague: A process of felonious evolution. Microbial Ecology, 47, 293–299. [DOI] [PubMed] [Google Scholar]
- Campbell, J. , Lowe, J. , Walz, S. , & Ezzell, J. (1993). Rapid and specific identification of Yersinia pestis by using a nested polymerase chain reaction procedure. Journal of Clinical Microbiology, 31, 758–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanovas‐Massana, A. , Pedra, G. G. , Wunder, E. A., Jr. , Diggle, P. J. , Begon, M. , & Ko, A. I. (2018). Quantification of leptospira interrogans survival in soil and water microcosms. Applied and Environment Microbiology, 84, e00507‐18 10.1128/AEM.00507-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chain, P. S. , Carniel, E. , Larimer, F. W. , Lamerdin, J. , Stoutland, P. O. , Regala, W. M. , … Garcia, E. (2004). Insights into the evolution of Yersinia pestis through whole‐genome comparison with Yersinia pseudotuberculosis. Proceedings of the National Academy of Sciences of the United States of America, 101, 13826–13831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornelis, G. R. , & Wolf‐Watz, H. (1997). The Yersinia Yop virulon: a bacterial system for subverting eukaryotic cells. Molecular Microbiology, 23, 861–867. [DOI] [PubMed] [Google Scholar]
- Eisen, R. J. , Bearden, S. W. , Wilder, A. P. , Montenieri, J. A. , Antolin, M. F. , & Gage, K. L. (2006). Early‐phase transmission of Yersinia pestis by unblocked fleas as a mechanism explaining rapidly spreading plague epizootics. Proceedings of the National Academy of Sciences of the United States of America, 103, 15380–15385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelthaler, D. M. , Hinnebusch, B. J. , Rittner, C. M. , & Gage, K. L. (2000). Quantitative competitive PCR as a technique for exploring flea‐Yersina pestis dynamics. American Journal of Tropical Medicine and Hygiene, 62, 552–560. [DOI] [PubMed] [Google Scholar]
- Feodorova, V. A. , & Devdariani, Z. L. (2000). Development, characterisation and diagnostic application of monoclonal antibodies against Yersinia pestis fibrinolysin and coagulase. Journal of Medical Microbiology, 49, 261–269. [DOI] [PubMed] [Google Scholar]
- Ferber, D. M. , & Brubaker, R. R. (1981). Plasmids in Yersinia pestis . Infection and Immunity, 31, 839–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippov, A. A. , Oleinikov, P. N. , Motin, V. L. , Protsenko, O. A. , & Smirnov, G. B. (1995). Sequencing of two Yersinia pestis IS elements, IS285 and IS100. Contributions to Microbiology and Immunology, 13, 306–309. [PubMed] [Google Scholar]
- Filippov, A. A. , Solodovnikov, N. S. , & Protsenko, O. A. (1990). Plasmid content in Yersinia pestis strains of different origin. FEMS Microbiology Letters, 67, 45–48. [DOI] [PubMed] [Google Scholar]
- Giles, T. A. , Greenwood, A. D. , Tsangaras, K. , Giles, T. C. , Barrow, P. A. , Hannant, D. , … Yon, L. (2016). Detection of a Yersinia pestis gene homologue in rodent samples. PeerJ., 4, e2216 10.7717/peerj.2216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hänsch, S. , Cilli, E. , Catalano, G. , Gruppioni, G. , Bianucci, R. , Stenseth, N. C. , … Pallen, M. J. (2015). The pla gene, encoding plasminogen activator, is not specific to Yersinia pestis . BMC Research Notes, 535, 1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, P. , Elliott, J. , McCready, P. , Skowronski, E. , Garnes, J. , Kobayashi, A. , … Garcia, E. (1998). Structural organization of virulence‐associated plasmids of Yersinia pestis . Journal of Bacteriology, 180, 5192–5202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iqbal, S. S. , Chambers, J. P. , Goode, M. T. , Valdes, J. J. , & Brubaker, R. R. (2000). Detection of Yersinia pestis by pesticin fluorogenic probe‐coupled PCR. Molecular and Cellular Probes, 14, 109–114. [DOI] [PubMed] [Google Scholar]
- Iriarte, M. , & Cornelis, G. R. (1996). Molecular determinants of Yersinia pathogenesis. Microbiologia (Madrid, Spain), 12, 267–280. [PubMed] [Google Scholar]
- Isaacson, M. , Levy, D. , Pienaar, B. J. , Bubb, H. D. , Louw, J. A. , & Genis, G. K. (1973). Unusual cases of human plague in Southern Africa. South African Medical Journal, 47, 2109–2113. [PubMed] [Google Scholar]
- Liu, J. , Ochieng, C. , Wiersma, S. , Ströher, U. , Towner, J. S. , Whitmer, S. , … Fields, B. (2016). Development of a TaqMan array card for acute‐febrile‐illness outbreak investigation and surveillance of emerging pathogens, including ebola virus. Journal of Clinical Microbiology, 54, 49–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livengood, J. , Hutchinson, M. L. , Thirumalapura, N. , & Tewari, D. (2020). Detection of Babesia, Borrelia, Anaplasma, and Rickettsia spp. in Adult Black‐Legged Ticks (Ixodes scapularis) from Pennsylvania, United States, with a Luminex Multiplex Bead Assay. Vector‐Borne and Zoonotic Diseases, 20, 406–411. 10.1089/vbz.2019.2551 [DOI] [PubMed] [Google Scholar]
- María, F. R. , Osikowicz, L. , Cáceres, A. G. , Luna‐Caipo, V. D. , Suarez‐Puyen, S. M. , Bai, Y. , & Kosoy, Y. (2019). Identification of Bartonella rochalimae in guinea pigs (Cavia porcellus) and fleas collected from rural peruvian households. American Journal of Tropical Medicine and Hygiene, 101, 1276–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matero, P. , Pasanen, T. , Laukkanen‐Ninios, R. , Tissari, P. , Tarkka, E. , Vaara, M. , & Skurnik, M. (2009). Real‐time multiplex PCR assay for detection of Yersinia pestis and Yersinia pseudotuberculosis . APMIS, 117, 34–44. [DOI] [PubMed] [Google Scholar]
- Moore, R. L. , & Brubaker, R. R. (1975). Hybridization of deoxyribonucleotide sequences of Yersinia enterocolitica and other selected members of Enterobacteriaceae . International Journal of Systematic Bacteriology, 25, 336–339. [Google Scholar]
- Neubauer, H. , Meyer, H. , Prior, J. , Aleksic, S. , Hensel, A. , & Splettstosser, W. (2000). A combination of different polymerase chain reaction (PCR) assays for the presumptive identification of Yersinia pestis . Journal of Veterinary Medicine, Series B, 47, 573–580. [DOI] [PubMed] [Google Scholar]
- Norkina, O. V. , Kulichenko, A. N. , Gintsburg, A. L. , Tuchkov, I. V. , Popov, Y. A. , Aksenov, M. U. , & Drosdov, I. G. (1994). Development of a diagnostic test for Yersinia pestis by the polymerase chain reaction. Journal of Applied Bacteriology, 76, 240–245. [DOI] [PubMed] [Google Scholar]
- Parkhill, J. , Wren, B. W. , Thomson, N. R. , Titball, R. W. , Holden, M. T. , Prentice, M. B. , … Barrell, B. G. (2001). Genome sequence of Yersinia pestis, the causative agent of plague. Nature, 413, 523–527. [DOI] [PubMed] [Google Scholar]
- Pawelczyk, O. , Asman, M. , & Solarz, K. (2019). The molecular detection of Anaplasma phagocytophilum and Rickettsia spp. in cat and dog fleas collected from companion animals. Folia Parasitologica, 66 10.14411/fp.2019.020 [DOI] [PubMed] [Google Scholar]
- Perry, R. D. , & Fetherston, J. D. (1997). Yersinia pestis – etiologic agent of plague. Clinical Microbiology Reviews, 10, 35–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portnoy, D. A. , & Falkow, S. (1981). Virulence‐associated plasmids from Yersinia enterocolitica and Yersinia pestis . Journal of Bacteriology, 148, 877–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintard, K. , Dewitte, A. , Reboul, A. , Madec, E. , Bontemps‐Gallo, S. , Dondeyne, J. , … Sebbane, F. (2015). Evaluation of the role of the opgGH operon in Yersinia pseudotuberculosis and its deletion during the emergence of Yersinia pestis . Infection and Immunity, 83, 3638–3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radnedge, L. , Gamez‐Chin, S. , McCready, P. M. , Worsham, P. L. , & Andersen, G. L. (2001). Identification of nucleotide sequences for the specific and rapid detection of Yersinia pestis . Applied and Environment Microbiology, 67, 3759–3762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajanna, C. , Revazishvili, T. , Rashid, M. H. , Chubinidze, S. , Bakanidze, L. , Tsanava, S. , … Sulakvelidze, A. (2010). Characterization of pPCP1 Plasmids in Yersinia pestis Strains Isolated from the Former Soviet Union. International Journal of Microbiology, 2010 10.1155/2010/760819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randremanana, R. , Andrianaivoarimanana, V. , Nikolai, B. , Ramasindrazana, B. , Paireau, J. , Ten Bosch, Q. A. , … Rajerison, M. (2019). Epidemiological characteristics of an urban plague epidemic in Madagascar, August‐November, 2017: An outbreak report. The Lancet Infectious Diseases, 19, 537–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Respicio‐Kingry, L. B. , Yockey, B. M. , Acayo, S. , Kaggwa, J. , Apangu, T. , Kugeler, K. J. , … Petersen, J. M. (2016). Two distinct Yersinia pestis populations causing plague among humans in the West Nile region of Uganda. PLoS Neglected Tropical Diseases, 10, e0004360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riehm, J. M. , Rahalison, L. , Scholz, H. C. , Thoma, B. , Pfeffer, M. , Razanakoto, L. M. , … Tomaso, H. (2011). Detection of Yersinia pestis using real‐time PCR in patients with suspected bubonic plague. Molecular and Cellular Probes, 25, 8–12. [DOI] [PubMed] [Google Scholar]
- Shi, L. , Yang, G. , Zhang, Z. , Xia, L. , Liang, Y. , Tan, H. , … Wang, P. (2018). Reemergence of human plague in Yunnan, China in 2016. PLoS One, 13, e0198067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenseth, N. C. , Atshabar, B. B. , Begon, M. , Belmain, S. R. , Bertherat, E. , Carniel, E. , … Rahalison, L. (2008). Plague: past, present, and future. PLoS Med, 5, e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart, A. , Satterfield, B. , Cohen, M. , O'Neill, K. , & Robison, R. (2008). A quadruplex real‐time PCR assay for the detection of Yersinia pestis and its plasmids. Journal of Medical Microbiology, 57, 324–331. 10.1099/jmm.0.47485-0 [DOI] [PubMed] [Google Scholar]
- Tomaso, H. , Reisinger, E. C. , Al, D. S. , Frangoulidis, D. , Rakin, A. , Landt, O. , & Neubauer, H. (2003). Rapid detection of Yersinia pestis with multiplex real‐time PCR assays using fluorescent hybridisation probes. FEMS Immunology and Medical Microbiology, 38, 117–126. [DOI] [PubMed] [Google Scholar]
- Tripp, D. W. , Gage, K. L. , Montenieri, J. A. , & Antolin, M. F. (2009). Flea abundance on black‐tailed prairie dogs (Cynomys ludovicianus) increases during plague epizootics. Vector‐Borne and Zoonotic Diseases, 9, 313–321. [DOI] [PubMed] [Google Scholar]
- Williams, J. E. , Harrison, D. N. , & Cavanaugh, D. C. (1975). Letter: Cryptic infection of rats with non‐encapsulated variants of Yersinia pestis . Transactions of the Royal Society of Tropical Medicine and Hygiene, 69, 171–172. [DOI] [PubMed] [Google Scholar]
- Williams, J. E. , Harrison, D. N. , Quan, T. J. , Mullins, J. L. , Barnes, A. M. , & Cavanaugh, D. C. (1978). Atypical plague bacilli isolated from rodents, fleas, and man. American Journal of Public Health, 68, 262–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter, C. C. , Cherry, W. B. , & Moody, M. D. (1960). An unusual strain of Pasteurella pestis isolated from a fatal human case of plague. Bulletin of the World Health Organization, 23, 408–409. [PMC free article] [PubMed] [Google Scholar]
- Woron, A. M. , Nazarian, E. J. , Egan, C. , McDonough, K. A. , Cirino, N. M. , Limberger, R. J. , & Musser, K. A. (2006). Development and evaluation of a 4‐target multiplex real‐time polymerase chain reaction assay for the detection and characterization of Yersinia pestis . Diagnostic Microbiology and Infectious Disease, 56, 261–268. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.


