Abstract
C57Bl6 (B6) mice devoid of glutathione peroxidases 1 and 2 (Gpx1/2‐DKO) develop ileitis after weaning. We previously showed germ‐free Gpx1/2‐DKO mice of mixed B6.129 background did not develop ileocolitis. Here, we examine the composition of the ileitis provoking microbiota in B6 Gpx1/2‐DKO mice. DNA was isolated from the ileum fecal stream and subjected to high‐throughput sequencing of the V3 and V4 regions of the 16S rRNA gene to determine the abundance of operational taxonomic units (OTUs). We analyzed the role of bacteria by comparing the microbiomes of the DKO and pathology‐free non‐DKO mice. Mice were treated with metronidazole, streptomycin, and vancomycin to alter pathology and correlate the OTU abundances with pathology levels. Principal component analysis based on Jaccard distance of abundance showed 3 distinct outcomes relative to the source Gpx1/2‐DKO microbiome. Association analyses of pathology and abundance of OTUs served to rule out 7–11 of 24 OTUs for involvement in the ileitis. Collections of OTUs were identified that appeared to be linked to ileitis in this animal model and would be classified as commensals. In Gpx1/2‐DKO mice, host oxidant generation from NOX1 and DUOX2 in response to commensals may compromise the ileum epithelial barrier, a role generally ascribed to oxidants generated from mitochondria, NOX2 and endoplasmic reticulum stress in response to presumptive pathogens in IBD. Elevated oxidant levels may contribute to epithelial cell shedding, which is strongly associated with progress toward inflammation in Gpx1/2‐DKO mice and predictive of relapse in IBD by allowing leakage of microbial components into the submucosa.
Keywords: IBD, ileitis, Lactobacillus, microbiome, mouse model, reactive oxygen species
Hydrogen peroxide (H2O2) and superoxide are generated by some bacterial genera and by the intestinal mucosa in response to bacteria. Gpx1/2‐double knockout mice are sensitive to intestinal H2O2 production, which causes crypt apoptosis and anoikis leading to inflammation. We investigate the normal ileum pathology inducing microbiota and use antibiotics to narrow down potential candidates. The principal candidate, Lactobacillus, stands out while not demonstrating unambiguous domination in provoking the pathology.

1. INTRODUCTION
Wild‐type, transgenic, and gene knockout (KO) rodent models of inflammatory bowel disease (IBD) most often demonstrate dependence on gut microbiota (Kiesler, Fuss, & Strober, 2015; Mizoguchi & Mizoguchi, 2010; Peloquin & Nguyen, 2013). This is in line with discoveries of bacterial alterations in IBD in humans (King & McCole, 2019; Sartor & Wu, 2017; Zuo & Ng, 2018). The question persists whether altered microbiomes cause IBD or are a result of IBD. Rodent models may provide some insight. Identities of provocative microbiota differ and will probably continue to differ model to model (Gkouskou, Deligianni, Tsatsanis, & Eliopoulos, 2014). Suspected pathogens and ordinary commensals might be acting as conditional pathogens depending on the genetic and environmental conditions contributing to each rodent model. Since the genetics of IBD is complex, as are the environments of sufferers, this might be the expectation for humans as well (Liu et al., 2015).
Oxidant generation from enzymes that use reduced nicotinamide adenine dinucleotide phosphate (NADPH oxidases), DUOX2 (H2O2), and NOX1 (superoxide) and from Lactobacillus participate in shaping the gut microbiota (Grasberger et al., 2015; Huang et al., 2016; Jones & Neish, 2017; Knaus, Hertzberger, Pircalabioru, Yousefi, & Branco Dos Santos, 2017; Lipinski et al., 2009; Matziouridou et al., 2018). NOX1‐dependent oxidant generation was observed in C57Bl6 (B6) ilea after oral gavage of Lactobacillus but not Escherichia coli (Jones et al., 2013). Lactobacillus thrives in an oxidant‐rich environment to the detriment of pathogens such as Citrobacter rodentium and Listeria monocytogenes (Pircalabioru et al., 2016). This is one basis for proposing the use of Lactobacillus sp. as probiotics in the treatment of inflammatory bowel disease (IBD) (Guandalini & Sansotta, 2019). Stimulation of crypt/gland localized NOX1 oxidant generation may be beneficial by sustaining proliferation and restitution (Alam et al., 2014; Coant et al., 2010; Jones et al., 2013; Kato et al., 2016; Singh, Hertzberger, & Knaus, 2018). Intestinal DUOX2 activation is downstream of NOD2 detection of cytosolic muramyl dipeptide, a component of bacteria cell walls (Lipinski et al., 2009). DUOX2 oxidant production may be in response to dysbiosis (Burgueno et al., 2019; Grasberger et al., 2015). Constraints on the expression of host NADPH oxidases and the presence of an antioxidant system normally prevent significant damage from occurring (Chu et al., 2017; Esworthy et al., 2014; Lee, Esworthy, Chu, Pfeifer, & Chu, 2006; Sommer & Backhed, 2015). However, typical oxidant‐producing Enterococcus faecalis caused more DNA damage in colon epithelial cells than a mutant strain with attenuated production after oral gavage of a bolus of the bacteria into antibiotic‐pretreated rats (Huycke, Abrams, & Moore, 2002). Oxidant generation in IBD is usually ascribed to ER stress in Paneth cells, NOX2 in macrophages and mitochondria, although DUOX2 mRNA levels are elevated up to 20 times in active Crohn's ileitis and ulcerative colitis (Hamm et al., 2010; Haberman et al., 2014; Li et al., 2010; Malhotra et al., 2008; MacFie et al., 2014; Yanai et al., 2015).
Glutathione peroxidases (GPX) 1 and 2 are antioxidant isoenzymes that reduce hydroperoxides to water or alcohols (Chu, Doroshow, & Esworthy, 1993). GPX1 is expressed in the luminal epithelium while GPX2 is concentrated in the crypts/glands (Esworthy, Swiderek, Ho, & Chu, 1998). Ileocolitis in homozygous Gpx1/2‐double knockout mice (DKO; Gpx1−/−Gpx2−/−) occurs spontaneously on 3 backgrounds B6, 129/Sv, and B6.129 (Esworthy et al., 2011; Esworthy, Smith, & Chu, 2010; Lee et al., 2006). Expression of one wild‐type Gpx1 or Gpx2 allele (non‐DKO mice; generated as littermates of DKO mice) suppresses the pathology, almost completely on the B6 background (Chu et al., 2017; Esworthy et al., 2014). In B6 DKO ilea, excessive crypt apoptosis and anoikis result in Paneth cell and crypt depletion from 27 to 35 days of age with infiltration of monocytes and neutrophils occurring around 27–28 days of age; pathology reaches its peak around 35 days of age (Chu et al., 2017; Chu, Esworthy, Shen, Gao, & Doroshow, 2019). Colon pathology begins soon after birth.
Studies were performed that link pathology in DKO ilea to DUOX2, NOX1, and microbiota. A germ‐free B6.129 Gpx1/2‐DKO colony did not exhibit pathology (Chu et al., 2004; Esworthy, Binder, Doroshow, & Chu, 2003). Ileocolitis in B6 DKO mice is driven by DUOX2 and NOX1 (Chu et al., 2017; Esworthy et al., 2014). This was shown in triple knockout lines (TKO), one in which the Nox1 gene was knocked out and the second in which the Duoxa locus (Duoxa1 and Duoxa2 maturation subunit genes) was modified to eliminate cell surface expression of DUOX1(barely expressed in the intestine) and DUOX2 and the ability to generate H2O2 (Grasberger et al., 2012). Lack of functional DUOX2 (Gpx1−/−Gpx2−/−Duoxa−/− TKO) eliminated crypt anoikis (exfoliation) and crypt abscesses. Excessive apoptosis remained resulting in partial loss of Paneth cells and crypts and slightly above background levels of macrophage numbers and monocyte infiltration (Chu et al., 2017). Gpx1−/−Gpx2−/−Nox1‐/Y or Nox1−/− (Nox1 gene on X‐chromosome) TKO mice had virtually no pathology (Esworthy et al., 2014).
Our hypothesis for linking microbiota to ileitis was based on the idea that Lactobacillus‐induced NOX1 oxidant generation produces damage on its own, which DUOX2 augments. We posited that antibiotic‐induced differences in Lactobacillus abundance in the ilea of B6 Gpx1/2‐DKO mice would be reflected in levels of pathology and E. coli abundance might not show a positive association with pathology since E. coli did not elicit NOX1 oxidant generation. To examine this, antibiotics were orally administered at the onset of ileum pathology to alter the evolution of the microbiota and possibly influence pathology levels. Three antibiotics were selected to be separately administered and produce distinct outcomes, impacting many operational taxonomic units (OTU; largely genus level). The primary analysis would be based on the association of abundance and pathology marker levels among the antibiotic‐treated and control sets. We were uncertain about the similarity of microbiotas between Gpx1/2‐DKO and non‐DKO mice (Gpx1+/−Gpx2−/− and Gpx1−/−Gpx2+/−). Since Gpx1/2‐DKO and non‐DKO mice were sibs or half‐sibs (cage mates), the differences might have been minor. Pathology in Gpx1/2‐DKO mice might alter the abundance of some OTUs from that of non‐DKO mice. This perspective was used to assess the antibiotic effects. Another viewpoint is that ileitis provoking OTUs might have some growth advantage in the pathological milieu over commensals as defined by abundances in wild‐type or other relatively normal mice (non‐DKO cage mates) and suppress the growth of more benign commensals. Thus, comparisons of OTUs with increased abundance in mice with ileitis compared to non‐DKO controls might indicate provoking candidates (Peloquin & Nguyen, 2013; Pircalabioru et al., 2016; Sartor & Wu, 2017). We also analyzed the results from this latter viewpoint.
2. MATERIALS AND METHODS
2.1. Mice
A genetically segregating breeding scheme (Gpx1−/−Gpx2−/− male x Gpx1+/−Gpx2−/− and Gpx1−/−Gpx2+/− females) provided homozygous Gpx1/2‐DKO mice and non‐DKO mice from most litters (Chu et al., 2017; Esworthy et al., 2014). Breeders were fed LabDiet 5062 (9% fat; LabDiet, St. Louis, MO). Pups were weaned at 21 days of age onto LabDiet 5061 (5% fat). The weight and condition of mice were recorded daily from 8 to 35 days of age. No morbidity was found. The study used homozygous Gpx1/2‐DKO and non‐DKO cage mates. Both males and females were included. The non‐DKO set was composed of 4 males (2 Gpx1+/Gpx2−/− and 2 Gpx1−/−Gpx2+/−) and 2 females (Gpx1+/Gpx2−/−). On the B6 background, there is no discernable difference in ileum histology between Gpx1+/1Gpx2−/−, Gpx1−/−Gpx2+/− mice and both genotypes closely resemble wild‐type mouse in histological features (Esworthy et al., 2014). Ileum histology of Gpx1/2‐DKO male and Gpx1/2‐DKO female mice is very similar, different only in the Paneth cells incidence marker.
2.2. Antibiotics
Metronidazole, streptomycin, and vancomycin were administered in drinking water; metronidazole concentration was 750 mg/L, streptomycin‐450 mg/L, and vancomycin‐50 mg/L (Ferreira et al., 2011; Sekirov et al., 2008; Wlodarska et al., 2011). Mice shun the metallic taste of metronidazole (Chu, Esworthy, Doroshow, & Shen, 2016). To overcome this, the water used to prepare antibiotics and the water for the control mice contained 0.4% of the sweetener, Splenda®. Mice were allowed free consumption from 22–35 days of age. The treated water was refreshed on the 7th day. At 35 days of age, the mice were euthanized by CO2 inhalation. Eighteen to 20 mice were in each group. Mice from multiple litters were used for each set (Splenda‐4 litters; Gpx1/2‐DKO and non‐DKO; metronidazole‐5 litters; vancomycin‐5 litters; streptomycin‐5 litters). The numbers of mice used were adequate to statistically distinguish sets of mice with intermediate pathology scores and marker values from DKO and non‐DKO or wild‐type mice in our previous studies (Chu et al., 2017; Esworthy et al., 2014). Eight Gpx1/2‐DKO mice with overall marker scores representing the average or median and 6 non‐DKO mice were selected for microbiome analysis. The number was based on examining comparable studies in B6 mice where the group sizes were generally 5–9 mice (Garidou et al., 2015; Gu et al., 2013; Jakobsson et al., 2015; Kar et al., 2017; Robertson et al., 2013; Tourret et al., 2017; Walk, Blum, Ewing, Weinstock, & Young, 2010).
2.3. Tissue sampling
The length of the small intestine (pylorus to ileocecal junction) was measured and the fecal stream of the ilea, 1–5 cm above the cecum, was expressed into sterile tubes and immediately frozen (−80°C). The length of the colon was measured. Ileum sections were fixed for histology (10% phosphate‐buffered formalin). Samples were processed for sectioning and stained with hematoxylin and eosin (H&E).
2.4. Histopathology
H&E histopathology was evaluated by enumerating crypt apoptosis, crypt exfoliation (anoikis), depletion of crypts and Paneth cells, and crypt abscesses, details in Figure 1 (Chu et al., 2017). The individual scoring was blinded to the identity of the slides.
FIGURE 1.
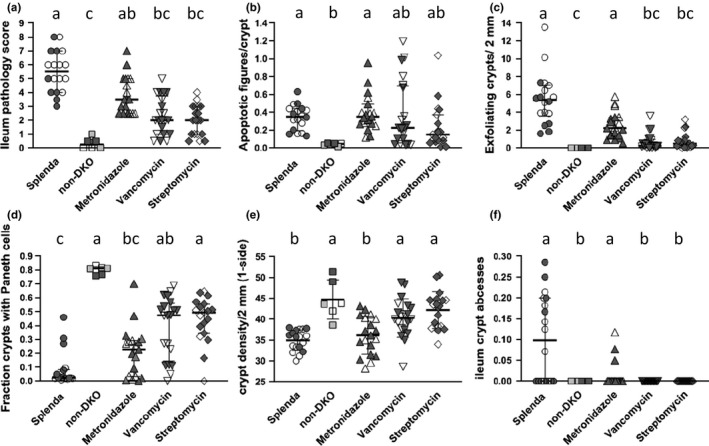
Ileum pathology scores and pathology marker levels in antibiotic‐treated DKO mice, Splenda‐treated DKO mice, and non‐DKO mice. Panel (A) is the overall score derived by adding values from appraisals of apoptosis (apoptosis figures/crypt >0.1 = 1; less = 0), Paneth cells (0.8–1 fraction crypts with Paneth cells = 0; 0–0.79 = 0.5; 0.34–0.59 = 1; 0.15–0.33 = 1.5; 0–0.14 = 2), crypt density (crypts/10× field one side; ≥39.1 = 0; 33.6–39 = 1; 27.8–33.5 = 2), inflammation foci/10× field both sides(0 = 0; 0.01–0.2 = 1; 0.21–0.5 = 2), and inflammation intensity (peak damage in section‐small abscesses = 1; large abscesses = 2; epithelial erosion/ulceration = 3; latter rarely found in B6); median and interquartile range. Panel B shows levels of crypt apoptosis, mean, and standard deviation. Panel (C) shows the fraction of crypts with anoikis (crypt exfoliation), median, and interquartile range. Panel (D) shows the fraction of crypts with Paneth cells, median and interquartile range. Panel (E) shows the number of crypt per 10× field, mean and standard deviation. Panel F shows the number of crypt abscesses per 10× field, median, and interquartile range. Data shown as median and interquartile range were evaluated by the Kruskal–Wallis test for multiple comparisons; data shown as mean and standard deviation were evaluated with 1‐way ANOVA with Tukey's multiple comparisons test. Common letters indicate no significant difference, otherwise a > b, etc. Open symbols for DKO sets (Splenda, metronidazole, vancomycin, and streptomycin) are males, dark symbols are females. For the non‐DKO mice, open symbols are Gpx1+/−Gpx2−/− males, dark symbols are Gpx1+/−Gpx2−/− females and half‐tone symbols are Gpx1−/−Gpx2+/− males
2.5. Isolation of DNA from ileum fecal stream, processing, and analysis of microbiome
DNA was isolated from the ileum fecal stream following the procedure described (Elson, Cong, Qi, Hershberg, & Targan, 2006; Esworthy et al., 2010). DNA was diluted to 0.1 μg/μl in TE (10 mM Tris, 1 mM EDTA, pH 8.0) and submitted to the Integrative Genomics Core City of Hope for processing and analysis. Amplicons of the V3‐V4 16S rRNA region were sequenced using the Illumina MiSeq platform. Duplicate 50‐μl PCRs were performed, each containing 50 ng of purified DNA, 0.2 mM dNTPs, 1.5 mM MgCl2, 1.25 U Platinum Taq DNA polymerase, 2.5 μl of 10× PCR buffer, and 0.5 μM of each primer designed to amplify the V3‐V4: F (5′‐CCTACACGACGCTCTTCCGATCTCCTACGGGNGGCWGCAG‐3′) and R (5′‐GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTGGACTACHVGGGTWTCTAAT‐3′). Cycling conditions were 94°C for 3 min, then 15 cycles of 94°C for 50 s, 51°C for 40 s, and 72°C for 1 min. 72°C for 5 min is used for the final elongation step. Amplicons were purified using the AxyPrep Mag PCR Clean‐up kit (Thermo Fisher Scientific). Up to 15 ng of PCR products were carried forward to library preparation using second‐round PCR. The Illumina primer PCR PE1.0 and index primers were used to allow the multiplexing of samples. Eight cycles of enrichment PCR were performed, and final libraries cleaned with AxyPrep Mag PCR Clean‐up kit. The library was quantified using ViiA™ 7 Real‐Time PCR System (Life Technologies) according to manufacturer's instructions and visualized for size validation on an Agilent 2100 Bioanalyzer (Agilent Technologies) using a high‐sensitivity DNA assay. The sequencing library pool was diluted to 4 nM until run on a MiSeq desktop sequencer (Illumina). 600 cycles chemistry (Illumina) was used according to the manufacturer's instructions to run the 6 pM library with 20% PhiX (Illumina).
Reads were merged using Mothur (v.1.33.3) make.contigs function and merged reads with any ambiguous bases were removed using screen.seqs function. Length selection of the merged reads was performed to retain merged reads with the length between 400 and 470 bp (range of expected V3 and V4 amplicon length), which were then classified against the SILVA ribosomal RNA gene database at the genus level using classify.seqs function with a confidence cutoff of 80%. Genera (or other OTUs) with abundance greater than 0.5% in at least one sample were included in the principal component analysis (PCA), which was performed based on Jaccard distance of abundance values. Differentially abundant genera or other OTUs in pairwise comparisons were identified requiring a fold‐change of 1.2 at limma‐voom FDR of 0.05. No samples submitted to the core were excluded from the analyses. Abundances and comparison sets (fold differences, p‐values, and FDR) are found in supporting data files (Esworthy, Chu, Shen, & Doroshow, 2020): https://doi.org/10.6084/m9.figshare.12167592
Evaluation of the OTUs as pathology candidates was based in part by identifying mice representing the extremes of pathology and analyzing OTUs between the groups for fold differences and statistical significance of the differences (pairwise t test and Bonferroni post‐test correction for multiple comparisons). This was followed up by ranking the results of the linear correlation of individual OTU abundance with the pathology scores and marker values from each of the DKO mice. This analysis used all results from Gpx1/2‐DKO mice in the Splenda control and antibiotic treatment groups. A third analysis was based on finding increased OTU abundances in Gpx1/2‐DKO versus non‐DKO mice (aggressive OTUs) or decreased in the abundance of OTUs in Gpx1/2‐DKO mice (protective OTUs). The evaluation of the statistical significance of differences for all analyses are presented in the supporting data: https://doi.org/10.6084/m9.figshare.12167592 [Excel file NONDKO vs. SPLENDACTRL and 13 other files; see Data Availability Statement] (Esworthy et al., 2020).
2.6. Statistical analysis
GraphPad Prism 7.01 was used for statistical analysis of pathology. Each data set was a check for a parametric distribution. Parametric sets were analyzed with a t test and nonparametric sets with the Mann–Whitney test with post‐tests for multiple samples. Post‐test adjustment for multiple samples was performed for all samples as described in the figure legends.
3. RESULTS
3.1. Moderately different microbiome in the non‐DKO cage mates
Pathology scores and small intestine lengths in the Splenda control Gpx1/2‐DKO and non‐DKO mice were consistent with prior observations (Figures 1 and 2). Splenda control Gpx1/2‐DKO ilea harbored microbiomes with some differences from non‐DKO cage mates (Figure 3). In DKO mice, significant decreases in abundance were found for Barnesiella, Desulfovibrio, and Porphyromonadaceae (11–27 fold; 0.005, FDR; Supporting Excel file: NONDKO‐vs‐SPLENDACTRL). Helicobacter had a significantly higher abundance in Gpx1/2‐DKO mice (9‐fold; 0.05, FDR) while Ureaplasma abundance was nearly 10‐fold higher, falling just short of significance (0.08, FDR). Eight OTUs are greater than 2‐fold more abundant in the Gpx1/2‐DKO mice compared to the non‐DKO mice and 2 other OTUs are more than 2‐fold less in the non‐DKO (not significant) (Figure 4d).
FIGURE 2.
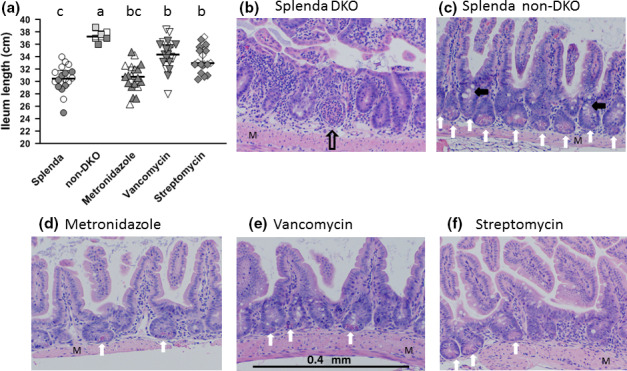
Impact of treatments or genotype on length of the small intestine (mean and standard deviation) panel A. Open symbols for DKO sets (Splenda, Metronidazole, vancomycin, and streptomycin) are males, dark symbols are females. For the non‐DKO mice, open symbols are Gpx1+/−Gpx2−/− males, dark symbols are Gpx1+/−Gpx2−/− females and half‐tone symbols are Gpx1−/−Gpx2+/− males. The values for non‐DKO mice resemble results from wild‐type mice. Statistics 1‐way ANOVA with Tukey's multiple comparisons test. Common letters indicate no significant difference, otherwise a > b, etc. Representative ileum histology (H&E) (panels B‐F). For histology, the original magnification was 10×. Non‐DKO mice (panel C) display histology nearly identical to wild‐type B6 with dense and orderly crypt/villus units (Esworthy et al., 2014). Most crypts have abundant Paneth (vertical white arrows) cells and goblet cells are visible (black arrowhead points to clusters). The control Splenda DKO mice (panel B) have disordered and distorted crypt/villus units that are less densely packed and lack Paneth and goblet cells. This particular image shows a probable crypt abscess (vertical open arrow). Panels D‐F show the generally beneficial impact of antibiotic treatment with some restoration of Paneth and goblet cells in better ordered crypt/villus units. These panels also show that some pathology remains, for example, less densely packed crypt/villus units in the metronidazole set (panel D)
FIGURE 3.
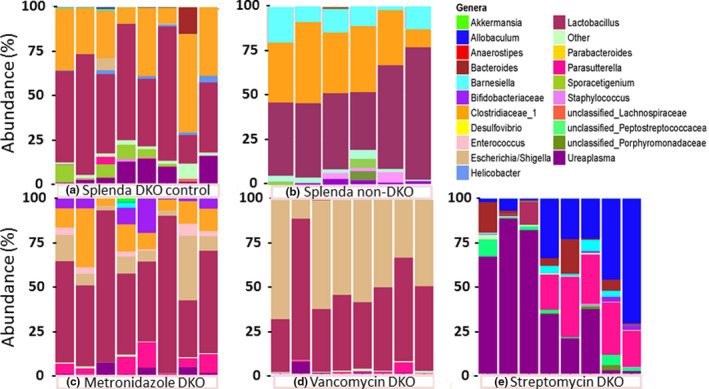
The microbiome of the Splenda control (panel a DKO; Splenda non‐DKO panel b) and antibiotic treated sets (panels c‐e; metronidazole, vancomycin, and streptomycin). Abundances of OTUs of each mouse are shown in a single column as a color spectrum portrayal generally at the genus level; exceptions noted for Clostridiaceae for which cluster 1 is shown and unclassified Lachnospiraceae, Peptostreptococcaceae, and Porphyromondaceae, which represent families. Bacteria OTU color codes are shown top to bottom in the same order as the color key—alphabetical). Table 1 provides the mean abundances for each OTU
FIGURE 4.
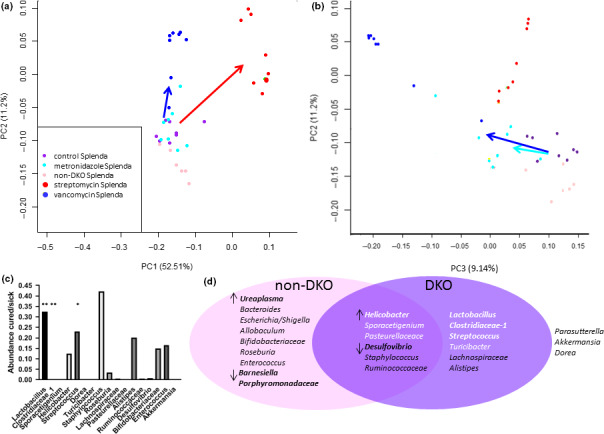
Results of 3 analyses for candidate provocative and beneficial OTUs. Principal component analysis of OTUs showing clustering of mice from Splenda DKO, non‐DKO, and antibiotic‐treated groups. This is based on Jaccard distances using abundance data. Panel a shows PC1 and PC2. Colored arrows indicate diverse microbiome groupings created by antibiotics relative to the Splenda DKO control; dark blue—vancomycin; red—streptomycin. Panel b has results for PC2 and PC3 and shows that non‐DKO mice remained grouped with the Splenda DKO control while the metronidazole group is slightly distanced (light blue arrow). Panel c represents the assessment provocative of OTUs (abundance in low pathology mice ÷sick mice <1). Statistical significance in pairwise t tests was found for an abundance of 3 OTUs (* and **). None passed adjustment for multiple samples. The best 2 OTU candidates are indicated by **. Panel d groups candidate OTUs based on contraction in DKO vs non‐DKO (↓) or expansion (↑) (statistically significant differences indicated by bold type), moderate to strong correlation with pathology markers (DKO purple oval stronger candidates in white letters; bold type for best candidates) or both (overlapping pink and purple ovals). The best candidates based on correlation are in bold white. The best consensus candidates are in whites letters and indicated by ↑ in the overlapping region. 3 OTUs found in Table 1 fall outside of both classifications. Data for results shown in panels c and d are found in Supporting Data Files
A core microbiota (less than 2‐fold difference; p > 0.09) was identified consisting of 8 OTUs accounting for 66.5% of the total in non‐DKO and 73% in DKO mice (Lactobacillus, Clostridiaceae‐1, Streptococcus, Lachnospiraceae, Turicibacter, Parasutterella, Dorea, and Alistipes). Principal component analysis (PCA), based on Jaccard distance of abundance values, places the Splenda control Gpx1/2‐DKO and non‐DKO sets as close neighbors, partially separated on the PC2 axis (Figure 4a,b).
3.2. Antibiotic effect on the ileum microbiome
Streptomycin depleted families Clostridiaceae, Enterobacteriaceae, Helicobacteraceae, Lactobacillaceae, and Pasteurellacease (Figure 3a,e). Clostridiaceae and Lactobacillaceae went from the 2 most abundant families (71% combined) to the minor presence (1.9%). The major opportunistic OTUs were Ureaplasma, Allobaculum, and Parasutterella. With metronidazole, Lactobacillus abundance was maintained while Clostridiaceae‐1 declined to one‐half (not significant; Supporting Excel file: METRO‐vs‐SPLENDACRTL). Of the rest of the most abundant genera, 4 declined in abundance while Escherichia/Shigella and Parasutterella increased in abundance each by 10‐fold. With vancomycin, Lactobacillus abundance was maintained while Clostridiaceace‐1 was down by 260‐fold. PCA showed partial to complete separation of the antibiotic treatment groups from DKO and non‐DKO mice (Figure 4a,b).
3.3. Antibiotic treatment effect on ileitis
A filter for assessing OTUs would be abundances in responsive and unresponsive Gpx1/2‐DKO mice. However, the penetrance of ileum pathology is 95%+; pathology scores were no less than 3 among 53 Gpx1/2‐DKO mice analyzed for this study and in prior work (Figure 1A) (Chu et al., 2019; Esworthy et al., 2014). Mice with scores of 3 exhibit elevated crypt apoptosis with reduction of Paneth cell incidence by half and/or crypt density less than the normal range. Non‐DKO mice had scores from 0 to 1 (Figure 1A). Antibiotic treatment expanded the range of pathology scores in the DKO mice from 0.5 to 8 (Figure 1A). Streptomycin and vancomycin had strong effects on the ileum pathology, while metronidazole had a moderate impact. Even though streptomycin treatment produces almost complete turnover of the microbiome, some pathology remained. Mice representing the extremes from DKO Splenda controls and antibiotic‐treated mice were identified and the OTU abundances compared. Six mice were deemed cured, having pathology scores of 0.5–1 (mean score 0.83 ± 0.26), comparable to non‐DKO mice. Ten mice were selected to represent the sick extreme, having pathology scores of 5 and above (mean score 5.9 ± 0.88). Seventeen OTUs were identified as possibly contributing to pathology by finding values less than 1 for—abundance in cured animals÷abundance in sick animals (Figure 4c). Second, we evaluated statistical significance for the difference in abundance for the respective OTUs (Supporting Excel file: Cured vs. Sick). Based on this, Lactobacillus, Clostridiaceae‐1, and Streptococcus stand out as candidate provocative OTUs (p‐value ≤0.026, pairwise comparisons). When corrected for multiple samples, the p‐value is 0.074 for Clostridiaceae‐1, 0.088 for Lactobacillus, and 0.44 for Streptococcus. Seven OTUs show suggestive signs as possible beneficial groups or opportunists by having abundance in cured animals÷abundance in sick animals greater than 1 (Ureaplasma, Bacteroides, Escherichia/Shigella, Parasutterella, Barnesiella, Allobaculum, and Porphyromonadaceae). The abundances of these OTUs are not significantly elevated in the cured mouse set after post‐test, multiple samples adjustment, although Bacteroides and Ureaplasma pass the pairwise t test (p ≤ 0.039).
To determine whether any individual OTU might be regarded as primarily responsible for pathology, we tested correlations between OTU abundances and pathology marker parameters, including final score, for all mice from the Gpx1/2‐DKO Splenda control and antibiotic sets. The best results were obtained with Lactobacillus, Clostridaceae–1, Pasteurellaceae, Streptococcus, and Helicobacter (Table 1a‐pathology column). The coefficients of 7 OTUs are listed with a minus sign indicating an inverse relationship between abundance and pathology; this set is identical to those identified as beneficial above. In many cases, the positive correlation reflects a uniform reduction in abundance in the presence of antibiotics (Helicobacter, e.g., Table 1a). Lactobacillus, Clostridaceae–1, and Streptococcus deviate from this pattern. Four OTUs show poor overall correlation with pathology markers, Dorea, Roseburia, Bifidobacteriaceae, and Enterococcus. Not all of the positive correlations look robust. Inspection of the correlation graphs reveals full variation in pathology markers values (i.e., pathology scores of 0.5–7) occur in mice with nearly zero abundance of Helicobacter, Pasteurellaceae, Sporacetigenium, and Turicibacter (Figure 5 and Figures S2‐S8). The low abundance of Lactobacillus and Clostridaceae–1 is more consistently associated with lower pathology for markers, while Streptococcus shows a mixed pattern. The Lactobacillus correlation with pathology score had a modest R‐squared value of 0.35. Clostridiaceae‐1 is in competition with Lactobacillus based on abundance and strength of correlation (R‐squared = 0.28 for pathology score). The R‐squared value is increased to 0.53 in the correlation of the combined abundances of Lactobacillus and Clostridiaceae‐1 to pathology scores (Figure 5 and Figures S2‐S7).
TABLE 1.
(a) Components of the Splenda control microbiome (bacteria OTUs represent genera, family, or cluster‐based on calls by the SILVA ribosomal RNA gene database) ranked by abundance (mean%; gray highlight)
| Bacteria | Abundance | Correlations | ||||
|---|---|---|---|---|---|---|
| Genus/family/cluster | Splenda | Non‐DKO | Metron | Vanco | Strep | Pathology |
| Lactobacillus | 44.6 | 39.54 | 55.97 | 43.4 | 1.77 | 0.506 |
| Clostridiaceae‐1 | 26.37 | 25.05 | 12.8 | 0.1 | 0.11 | 0.455 |
| Ureaplasma | 6.3 | 0.64 | 2.19 | 0.107 | 32.38 | −0.294 |
| Sporacetigenium | 3.76 | 0.68 | 0.1 | 0.1 | 0.1 | 0.343 |
| Bacteroides | 1.81 | 0.45 | 0.1 | 0.349 | 4.4 | −0.227 |
| Helicobacter | 1.22 | 0.14 | 0.11 | 0.11 | 0.17 | 0.378 |
| Escherichia/Shigella | 0.949 | 0.11 | 10.62 | 44.9 | 0.1 | −0.224 |
| Parasutterella | 0.656 | 0.46 | 6.12 | 1.23 | 11.73 | −0.266 |
| Streptococcus | 0.507 | 0.26 | 0.33 | 0.1 | 0.133 | 0.403 |
| Turicibacter | 0.477 | 0.32 | 0.2 | 0.1 | 0.1 | 0.308 |
| Dorea | 0.465 | 0.34 | 0.1 | 0.1 | 0.113 | 0.088 |
| Staphylococcus | 0.417 | 1.34 | 0.14 | 0.19 | 0.159 | 0.144 |
| Roseburia | 0.39 | 0.1 | 0.1 | 0.1 | 0.1 | 0.085 |
| Barnesiella | 0.33 | 8.33 | 0.348 | 0.1 | 1.4 | −0.178 |
| Lachnospiraceae | 0.266 | 0.27 | 0.1 | 0.1 | 0.104 | 0.183 |
| Pasteurellaceae | 0.26 | 0.1 | 0.1 | 0.1 | 0.1 | 0.435 |
| Alistipes | 0.21 | 0.28 | 0 | 0 | 0.001 | 0.174 |
| Ruminococcaceae | 0.19 | 0.44 | 0.1 | 0.1 | 0.1 | 0.139 |
| Allobaculum | 0.16 | 0.03 | 0.07 | 0 | 20.63 | −0.314 |
| Desulfovibrio | 0.11 | 1.31 | 0 | 0 | 0.0002 | 0.175 |
| Porphyromonadaceae | 0.11 | 1.17 | 0 | 0 | 0.58 | −0.299 |
| Bifidobacteriaceae | 0.11 | 0.016 | 5.86 | 0 | 0.79 | 0.099 |
| Enterococcus | 0.1 | 0.01 | 2.65 | 0.00065 | 0.044 | 0.044 |
| Akkermansia | 0 | 0.0003 | 0.25 | 0 | 0 | 0.122 |
|
Total abundance |
89.77 | 82.4 | 98.36 | 91.29 |
75.12 |
|
| Pathology parameters | |||||
|---|---|---|---|---|---|
| Pathology parameters | Splenda | Non‐DKO | Metron | Vanco | Strep |
| Score | 5.5 | 0.25 | 3.5 | 2 | 2 |
| Apoptosis | 0.34 | 0.04 | 0.39 | 0.38 | 0.24 |
| Exfoliation | 5.53 | 0 | 2.5 | 0.65 | 0.77 |
| Paneth cells | 0.09 | 0.8 | 0.21 | 0.4 | 0.44 |
| Crypt density | 35 | 44.7 | 36.2 | 40.4 | 42.2 |
| Abscesses | 0.1 | 0 | 0.01 | 0 | 0 |
Mean abundances for the non‐DKO mice and antibiotic‐treated mice (metron—metronidazole; vanco—vancomycin; and strep—streptomycin) are listed in adjacent columns. The last set of values (correlations) represent the average of the correlation coefficients for the pathology parameters listed in (b) for each mouse's pathology parameters versus the abundance of the OTUs for each mouse; all coefficients converted to positive values when larger OTU abundance is associated with greater pathology and converted to all negative values when larger OTU abundance is associated with less pathology. In (b), the median pathology score, the median level of crypt exfoliation, the median of the fraction of crypts with Paneth cells, the median value for crypt abscesses, and mean value for crypt apoptosis are listed.
FIGURE 5.
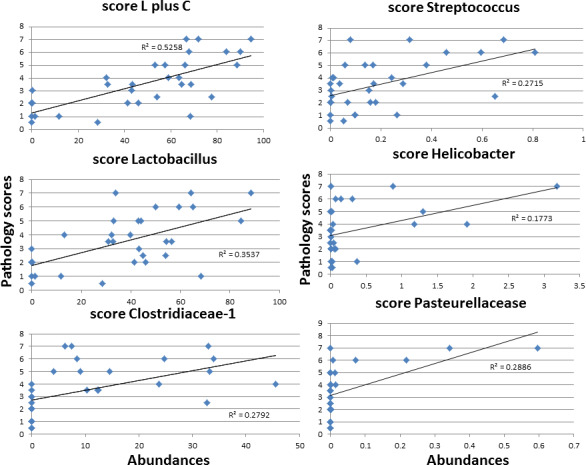
Correlation of OTU abundances with pathology scores using data from antibiotics and Splenda DKO sets. L plus C represents adding the abundances of Clostridiaceae‐1 and Lactobacillus and performing the correlation versus pathology score. This result was generated for comparison with the Clostridiaceae‐1 and Lactobacillus correlations. The remaining panels show some of the top candidates based on correlations across all markers (see Table 1 correlations pathology column)
4. DISCUSSION
We examined nine publications to determine whether the microbiomes found in the Gpx1/2‐DKO and non‐DKO ilea were typical of B6 mice (Garidou et al., 2015; Gu et al., 2013; Jakobsson et al., 2015; Kar et al., 2017; Mastrocola et al., 2018; Matziouridou et al., 2018; Robertson et al., 2013; Tourret et al., 2017; Walk et al., 2010). The reported microbiomes differed extensively, in some cases between replicate experiments within studies. One paper did not test for Clostridiaceae‐1 or a larger taxonomic unit containing the cluster and in others, Clostridia/Clostridium designation indicates clusters XIVa and IV. Lactobacillaceae or Lactobacillus abundances averaged 18% ± 21. Clostridia/Clostridium represented 5% ± 6 of reads with mice on 5001 and 5021 diets and a semi‐synthetic diet at the higher end of the range‐5%, 5%, 11%, and 18% (Clostridium sensu stricto, Clostridiaceae, Clostridia, and Clostridium) (Jakobsson et al., 2015; Mastrocola et al., 2018; Tourret et al., 2017; Walk et al., 2010). Clostridium sensu stricto corresponds to Clostridiaceae‐1 (Gupta & Gao, 2009). The bulk of reads among the sets consisted of Turicibacter (50%), unclassified‐Bacteriodales (38%), S24‐7 (2 reports; 63% and 55%), MIB (40%), Porphyromonadaceae (65%), Erysipelotrichaceace (2 reports; 22% and 50%), Lachnospiraceae (35%), and Allobaculum (22%). Our results may be one more example of the diversity among B6 ilea microbiomes, the high co‐abundance of Lactobacillus, and Clostridiaceae‐1 representing a unique combination. A link between facultative anaerobes like Lactobacillus and anaerobic Clostridia has been suggested to be due to the consumption of oxygen by Lactobacillus establishing anaerobic niches for Clostridia to thrive (Laukens, Brinkman, Raes, De Vos, & Vandenabeele, 2016). Not mentioned in reports on laboratory B6 mice but found in wild mice is Mycoplasma (Ureaplasma), found in the ilea of our mice (Suzuki & Nachman, 2016). Ureaplasma's status as a pathogen is unclear in humans (Combaz‐Sohnchen & Kuhn, 2017). Ureaplasma is a candidate as a provoking OTU based solely on the Gpx1/2‐DKO vs. non‐DKO comparison. Based on the other analyses Ureaplasma does not appear to be a pathogen in Gpx1/2‐DKO mice and is categorized as a potentially beneficial OTU, or as likely, an opportunistic OTU. Helicobacter was accidentally introduced into B6.129 Gpx1/2‐DKO mice soon after the line was established and transmitted to the B6 line (Esworthy et al., 2001). Helicobacter has long been noted as a possible pathogen in many rodent models of IBD (Peloquin & Nguyen, 2013). Helicobacter is the only OTU to show up as a tentative candidate in all 3 analytical approaches. However, it emerges as a strong candidate only in the Gpx1/2‐DKO vs. non‐DKO comparison with greater abundance in the Gpx1/2‐DKO.
A second question is whether the antibiotic treatments produced predictable effects on the microbiome. Our goal in using these 3 antibiotics was to achieve 3 distinct outcomes, which appeared likely based on results from prior studies using wild‐type B6 mice (Ferreira et al., 2011; Ju et al., 2017; Sekirov et al., 2008; Wlodarska et al., 2011). Once the composition of control DKO/non‐DKO microbiomes was determined, we were in a position to retrospectively examine whether the impact of the antibiotics would be expected. Metronidazole is supposed to target anaerobes. Thus, Clostridiaceae‐1 abundance should be down. In B6 colon, treatment with metronidazole at the dose used here for 4 days depleted Clostridium coccoides (cluster XIVa) and suppressed Bacteroidales allowing for increased abundance of Lactobacilli, Bifidobacteriaceae, and Enteroccaceae (Ju et al., 2017; Wlodarska et al., 2011). This is similar to what we found, although the impact on Clostridia was not nearly as strong in our samples. Vancomycin depleted Gram‐positive Clostridiaceae‐1, as expected. It did not affect Gram‐positive Lactobacillus, which could be anticipated due to similar findings in B6 colon at this concentration (Sekirov et al., 2008). The increase in abundance of the Gram‐negative Escherichia/Shigella OTU might have been expected from the loss of Clostridiaceae‐1 or other OTUs, although the overgrowth far exceeded that found in B6 colon at this concentration (Sekirov et al., 2008). The effect of streptomycin in the reference papers was to deplete C. coccoides and suppress Lactobacilli while leaving Bacteroidales unaffected. This is what we found in our samples. The only possible anomalies were the partial impact of metronidazole on Clostridiaceae‐1 abundance and the level of overgrowth by the Escherichia/Shigella OTU with a low concentration of vancomycin. The latter finding might be related to the ileum having higher oxygen content than colon favoring more overgrowth by facultative anaerobes like the Escherichia/Shigella OTU (Sommer & Backhed, 2016).
The Gpx1/2‐DKO and non‐DKO comparison can be used to examine the issue of whether pathology alters the microbiome. Ileum pathology was mild in the newly weaned B6 Gpx1/2‐DKO mice, confined to sporadic crypt exfoliation and sporadic crypt apoptosis that, in the majority of cases, was not over normal levels (Chu et al., 2017, 2019). Nox1 and TNF‐α mRNA levels were not elevated at this time. With that context, sibship of Gpx1/2‐DKO and non‐DKO mice and fostering by non‐DKO dams, we would expect they shared very similar microbiotas at weaning. The 2 sets of microbiomes are comparable when pathology reaches its high point (35 days and thereafter), although the non‐DKO microbiome is only marginally less different from the Splenda Gpx1/2‐DKO set than the metronidazole set based on PCA (non‐DKO separates in PC2; metronidazole in PC3). Four OTUs show up as having significantly altered abundances between Gpx1/2‐DKO and non‐DKO while 14 are altered between Gpx1/2‐DKO mice on Splenda and metronidazole‐treated mice (Esworthy et al., 2020). The possible impact of pathology in Gpx1/2‐DKO mice on the microbiota can be used as a reference for evaluating the effects of vancomycin and streptomycin.
Our experiment was designed so that antibiotics would have an opportunity to modify the microbiota before pathology significantly altered the architecture of the ileum with ensuing immune cell infiltration. Based on prior studies, there was a window from 22 to 26 days of age for the antibiotics to operate. Without antibiotics, pathology becomes more aggressive on and after 27 days of age (Chu et al., 2017, 2019). In our view, the early modification of the evolving microbiota would then influence subsequent pathology. The comparison of Gpx1/2‐DKO and non‐DKO microbiomes shows how pathology impacts the microbiota. The general effect of the antibiotics, vancomycin, and streptomycin was to significantly reduce pathology. Both produced a huge difference in the resulting microbiomes from the non‐DKO mice (Figure 4a,b). The antibiotic effect appears to dominate over any differences produced by differences in pathology levels. We seem to be largely observing the impact of microbiota on pathology.
The stratification of mice from the Splenda control and antibiotic Gpx1/2‐DKO sets into cured and sick divides the OTUs into provisional provocative and beneficial or opportunistic categories with statistical analysis for significance in either category. Bacteroides and Ureaplasma from the beneficial set pass statistical testing for differences in abundance in a pairwise t test and fail in after adjustment for multiple comparisons. The correlations with pathology markers for the beneficial set are generally weaker than in the provocative set (Table 1a). Lactobacillus, Clostridiaceae‐1, and Streptococcus were identified as candidate provocative OTUs in this analysis in pairwise testing. All fail to pass after adjustments for multiple samples with Lactobacillus and Clostridiaceae‐1 remaining as provisional candidates. Streptococcus is regarded as a probiotic (Koretz, 2018; Shiina et al., 2015). The literature on Clostridiaceae‐1 (C. perfringens excluded) is generally unfavorable to candidacy as a pathogen (Kanai, Mikami, & Hayashi, 2015; Lawson, Citron, Tyrrell, & Finegold, 2016; Peloquin & Nguyen, 2013; Peyrin‐Biroulet et al., 2012). While some species of Lactobacillus are used as probiotics, members of the genera exhibit properties that fit into our notion of a pathogen for Gpx1/2‐DKO mice by eliciting oxidant generation and/or generating oxidants (Jones et al., 2013; Knaus et al., 2017). Escherichia coli did not elicit ileum oxidant generation in earlier studies and this analysis, the associated OTU appeared to be eliminated as a provocative candidate. Some species of Streptococcus have a pyruvate oxidase activity and under aerobic conditions can generate H2O2 (Redanz et al., 2018). There is no information on whether Streptococcus elicits NOX1 oxidant generation by the host. The abundance is so low that its possible impact by these mechanisms would be minor compared to Lactobacillus.
Lactobacillus does not show a singular dominance in the correlation analysis with pathology markers and no other compelling candidates emerge. The reason for this may be that we are looking at genera and families rather than species. Alternatively, a superior correlation is found by using the combined abundance of Lactobacillus and Clostridiaceae‐1 and pathology markers. This improves the average of the correlation coefficients to 0.62 and more consistently associates high pathology maker values with high abundance. We examined this to investigate the idea that the response of Gpx1/2‐DKO mice may be to components of several pathology provoking OTUs. The residual pathology in the streptomycin set indicates at least a low‐level reaction to a different set of bacteria than found in the Gpx1/2‐DKO Splenda control (pathology scores of 3, 2, 2 and 2 for 4 of the last 5 mice in the streptomycin set, Figure 3e).
A second notion is that the displacement of Clostridiaceae‐1 by the Escherichia/Shigella OTU in the metronidazole and vancomycin sets might produce direct competition with Lactobacillus for microaerobic niches (Espey, 2013; Sommer & Backhed, 2016). Studies suggest that this would normally be an unlikely circumstance made possible by sustained antibiotic treatment. H2O2 and lactic acid production by Lactobacillus can provide an edge over E. coli for the population of mucosal sites (Gupta et al., 1998). The impact of high abundances of E. coli could be a reduction in oxidants both from bacteria and host (Jones et al., 2013). Adjusting for the presence of Escherichia/Shigella OTU as a factor diluting Lactobacillus improves the average of the correlation coefficients to 0.58 (EF supporting file Lactobacillus adjusted for E. coli dilution and Figure S10) (Esworthy et al., 2020). Catalase, present in many strains of E. coli, can be a significant sink for H2O2 (Rodriguez, Peiroten, Landete, Medina, & Arques, 2015). This would be another mechanism by which the Escherichia/Shigella OTU could counter Lactobacillus and host oxidant generation (Rodriguez et al., 2015). While 2 attempts to factor in the interaction between OTUs produce a better fit in correlations with pathology markers, neither yield a spectacularly enhanced outcome and employ opposed models (pathology enhancing interaction vs. counter pathological interaction) to yield similar improvements.
From the collective work on the Gpx1/2‐DKO mouse model (B6, 129/Sv and B6.129 backgrounds), we surmise that ileum and colon cell loss (the latter a major feature on the 129/Sv background) is driven by a subset of the microbiota primarily by stimulation of host oxidant generation (NOX1, DUOX2, and possibly ER stress in Paneth cells) (Chu et al., 2017, 2019). Inflammation is probably a result of leakage of microbial components into the submucosa since there was no indication of sterile inflammation in germ‐free B6.129 Gpx1/2‐DKO mice. In B6 and 129 Gpx1/2‐DKO mice, elevated lipopolysaccharides levels were detected in plasma indicating leakage through the epithelial barrier (Esworthy et al., 2011; Gao et al., 2010). Several mouse IBD models feature ileitis (Samp/YitFc, TNF∆ARE, Caspase‐8∆iec, Xbp‐1∆iec, NEMOtamIEC−KO, FADD∆iec, and N‐cadherin∆) (Cominelli, Arseneau, Rodriguez‐Palacios, & Pizarro, 2017). In common with most of these models is distress in or loss of Paneth cells due to aberrant autophagy and ER stress (Adolph et al., 2013). We found that the population of apoptosed and exfoliated cells while including Paneth cells was predominantly other cell types based on the general absence of lysozyme (Chu et al., 2017). Some of these ileitis models (Samp/YitFc, Caspase‐8∆iec, and FADD∆iec) exhibit sterile inflammation which can be augmented by the presence of microbes. In models with sterile inflammation, the Paneth cells may undergo necrosis or necroptosis, which are inherently inflammatory (Stolzer et al., 2020). Inflammatory signals may also derive from danger‐associated molecular patterns (DAMPs) generated from defective autophagy and unfolded proteins (unfolded protein response; UPR) in Paneth cells with sub‐lethal distress (Cadwell et al., 2008). Findings from genome‐wide association studies link genes in the UPR and autophagy pathways to Crohn's ileitis and Paneth cell defects (Cadwell, Stappenbeck, & Virgin, 2009). We see an outright loss of Paneth cells in Gpx1/2‐DKO mice, which has been proposed by others to be a possible consequence of ER stress (Kaser et al., 2008). ER stress increases the production of H2O2 as a by‐product of augmented protein folding activity (Delaunay‐Moisan & Appenzeller‐Herzog, 2015). Indications of ER stress were detected in tissues of B6 and 129 Gpx1/2‐DKO mice at very high levels of pathology and not at moderate levels of pathology (Esworthy et al., 2011; Gao et al., 2010). Since pathology in B6 Gpx1/2‐DKO mice shows a dependence on NOX1, we doubt that ER stress generates the ileitis, although when pathology is underway ER stress may contribute to the Paneth cell loss.
Pathology in the Xbp‐1∆iec and TNF∆ARE models is dependent on microbiota (Adolph et al., 2013; Schaubeck et al, 2016). In the Xbp‐1∆iec model (ER stress‐driven ileitis; B6; 129; FVB mixed background), the demonstration was limited to evaluation of germ‐free mice. As in the Xbp‐1∆iec model, we found nearly complete loss of Paneth cells and only partial loss of goblet cells (Chu et al., 2017; Kaser et al., 2008). In the TNF∆ARE model (TNFα over‐expression driven ileitis; B6/N background), the ilea showed a range of Paneth cell loss with the worst mice being similarly devoid of Paneth cells as Gpx1/2‐DKO mice. When the TNF∆ARE mice were treated with a combination of metronidazole and vancomycin for 4 weeks there was a partial but significant reduction in ileum pathology. That is what we would predict for a combined metronidazole and vancomycin effect on B6 Gpx1/2‐DKO mice. The cecum microbiome analysis showed a positive correlation between the abundance of unknown Clostriadales and ileitis scores (r = 0.48), while there was a strong negative correlation with Porphyromonadaceae (r = −0.79). We found a negative correlation for Porphyromonadaceae (r = −0.32). Unfortunately, we have no basis for comparing unknown Clostriadales to Clostridiaceae‐1. We are also comparing cecum to the ileum and the abundance of unknown Clostriadales was in the range of 1%–4% of total reads as opposed to 25% for Clostridiaceae in our samples. TNF∆ARE mice with low ileitis scores had microbiota compositions resembling wild‐type mice; the Splenda control Gpx1/2‐DKO mice closely resembled the non‐DKO controls. Loss of up to one‐half of the Paneth cells in B6 Gpx1/2‐DKO mice, largely through apoptosis, was associated with very mild, sporadic inflammation in Gpx1/2‐Duoxa‐TKO mice at 35 days at age and Gpx1/2‐DKO mice at 29 days at age (Chu et al., 2017). Macrophage numbers were elevated with few infiltrating monocytes and rare crypt abscesses. Further loss of Paneth cells was driven by anoikis/exfoliation in Gpx1/2‐DKO mice. This appeared at a later time than apoptosis and never attained high levels in Gpx1/2‐Duoxa‐TKO mice. While Paneth cell loss B6 Gpx1/2‐DKO mice is nearly complete by 32 days of age, we viewed this loss as a marker for the underlying processes of apoptosis and exfoliation and not a singular cause of inflammation. Lack of Paneth cells in B6 Gpx1/2‐DKO mice was not associated with a marked dysbiosis as gaged by the non‐DKO microbiome. However, increased susceptibility to the core microbiome could be due to the failure of anti‐bacterial defenses normally provided by Paneth cells. Exfoliation of any epithelial cell type can be inflammatory due to leakage of microbial components into the submucosa during this protracted process (Williams et al, 2015). After the treatment of IBD, high levels of exfoliation are prognostic of the potential for relapse (Kiesslich et al, 2012; Turcotte et al, 2012). In this way, we can link a process observed in Gpx1/2‐DKO mice to relapse in IBD and dependence on the microbiota.
CONFLICT OF INTERESTS
None declared. The content is solely the responsibility of the authors and does not necessarily represent the official views of NIH.
AUTHOR CONTRIBUTIONS
R. Steven Esworthy: Conceptualization (lead); formal analysis (lead); investigation (lead); methodology (lead); visualization (lead); writing – original draft (lead); writing – review & editing (lead). Fong‐Fong Chu: Funding acquisition (supporting); writing – review & editing (supporting). Binghui Shen: Funding acquisition (lead). James H. Doroshow: Funding acquisition (lead).
ETHICS STATEMENT
Studies were approved by the City of Hope IACUC (protocol #92008), which conforms to NIH and the American Association for Accreditation of Laboratory Animal Care (AAALAC) rules and standards.
ACKNOWLEDGMENTS
This study was supported by the National Cancer Institute [Contract No. HHSN261200800001E] [RO1CA085344 (BS)]; National Institutes of Health (NIH). COH core facilities services in this study included histology and immunohistochemistry performed by Solid tumor core, research histology service; housing and breeding mice by Animal Resource Center; construction of libraries, high‐throughput sequencing, and data analysis performed by the Integrative Genomics Core. All COH Cores used here are supported by NIH [P30CA33572]. NIH R01CA233664 (BS) supported the publication costs of this article.
Chu F‐F, Esworthy RS, Shen B, Doroshow JH. Role of the microbiota in ileitis of a mouse model of inflammatory bowel disease—Glutathione peroxide isoenzymes 1 and 2‐double knockout mice on a C57BL background. MicrobiologyOpen. 2020;9:e1107 10.1002/mbo3.1107
DATA AVAILABILITY STATEMENT
Supporting data sets generated and/or analyzed during the current study are available at figshare (Esworthy et al., 2020): https://doi.org/10.6084/m9.figshare.12167592; Excel files (EF) and supporting figures: (1) EF Abundances of OTUs; (2) EF NONDKO‐vs‐SPLENDACTRL‐differential‐abundance‐stats; (3) EF METRO‐vs‐SPLENDACTRL‐differential‐abundance‐stats; (4) EF STREP‐vs‐SPLENDACRTL‐differential‐abundance‐stats; (5) EF VANCO‐vs‐SPLENDACRTL‐ differential‐abundance‐stats; (6) EF Cured vs sick; (7) EF cured vs non‐DKO, non‐DKO vs metronidazole; (8) EF abundances correl abscesses; (9) EF abundances correl apoptosis; (10) EF abundances correl crypt density; (11) EF abundances correl exfoliation; (12) EF abundances correl Paneth cells; (13) EF abundances correl pathology scores; (14) EF Lactobacillus adjusted for E. coli dilution; (15) Figures S1‐S8—correlation graphs of top candidate provocative OTUs abundance with pathology markers; (16) Figure S9—pathology in colon; (17) Figure S10—correlation graphs Lactobacillus adjusted for E. coli dilution.
REFERENCES
- Adolph, T. E. , Tomczak, M. F. , Niederreiter, L. , Ko, H. J. , Bock, J. , Martinez‐Naves, E. , … Blumberg, R. S. (2013). Paneth cells as a site of origin for intestinal inflammation. Nature, 503, 272–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alam, A. , Leoni, G. , Wentworth, C. C. , Kwal, J. M. , Wu, H. , Ardita, C. S. , … Neish, A. S. (2014). Redox signaling regulates commensal‐mediated mucosal homeostasis and restitution and requires formyl peptide receptor 1. Mucosal Immunology, 7, 645–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgueno, J. F. , Fritsch, J. , Santander, A. M. , Brito, N. , Fernandez, I. , Pignac‐Kobinger, J. , … Abreu, M. T. (2019). Intestinal epithelial cells respond to chronic inflammation and dysbiosis by synthesizing H2O2 . Frontiers in Physiology, 10, 1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadwell, K. , Liu, J. Y. , Brown, S. L. , Miyoshi, H. , Loh, J. , Lennerz, J. K. , … Virgin, H. W. T. (2008). A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature, 456, 259–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadwell, K. , Stappenbeck, T. S. , & Virgin, H. W. (2009). Role of autophagy and autophagy genes in inflammatory bowel disease. Current Topics in Microbiology and Immunology, 335, 141–167. [DOI] [PubMed] [Google Scholar]
- Chu, F. F. , Doroshow, J. H. , & Esworthy, R. S. (1993). Expression, characterization, and tissue distribution of a new cellular selenium‐dependent glutathione peroxidase, GSHPx‐GI. The Journal of Biological Chemistry, 268, 2571–2576. [PubMed] [Google Scholar]
- Chu, F. F. , Esworthy, R. S. , Chu, P. G. , Longmate, J. A. , Huycke, M. M. , Wilczynski, S. , & Doroshow, J. H. (2004). Bacteria‐induced intestinal cancer in mice with disrupted Gpx1 and Gpx2 genes. Cancer Research, 64, 962–968. [DOI] [PubMed] [Google Scholar]
- Chu, F. F. , Esworthy, R. S. , Doroshow, J. H. , Grasberger, H. , Donko, A. , Leto, T. L. , … Shen, B. (2017). Deficiency in Duox2 activity alleviates ileitis in GPx1‐ and GPx2‐knockout mice without affecting apoptosis incidence in the crypt epithelium. Redox Biology, 11, 144–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu, F. F. , Esworthy, R. S. , Doroshow, J. H. , & Shen, B. (2016). NADPH oxidase‐1 deficiency offers little protection in Salmonella typhimurium‐induced typhlitis in mice. World Journal of Gastroenterology, 22, 10158–10165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu, F. F. , Esworthy, R. S. , Shen, B. , Gao, Q. , & Doroshow, J. H. (2019). Dexamethasone and Tofacitinib suppress NADPH oxidase expression and alleviate very‐early‐onset ileocolitis in mice deficient in GSH peroxidase 1 and 2. Life Sciences, 239, 116884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coant, N. , Ben Mkaddem, S. , Pedruzzi, E. , Guichard, C. , Treton, X. , Ducroc, R. , … Ogier‐Denis, E. (2010). NADPH oxidase 1 modulates WNT and NOTCH1 signaling to control the fate of proliferative progenitor cells in the colon. Molecular and Cellular Biology, 30, 2636–2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Combaz‐Sohnchen, N. , & Kuhn, A. (2017). A Systematic review of mycoplasma and ureaplasma in urogynaecology. Geburtshilfe und Frauenheilkunde, 77, 1299–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cominelli, F. , Arseneau, K. O. , Rodriguez‐Palacios, A. , & Pizarro, T. T. (2017). Uncovering pathogenic mechanisms of inflammatory bowel disease using mouse models of Crohn's disease‐like ileitis: What is the right model? Cellular and Molecular Gastroenterology and Hepatology, 4, 19–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaunay‐Moisan, A. , & Appenzeller‐Herzog, C. (2015). The antioxidant machinery of the endoplasmic reticulum: Protection and signaling. Free Radical Biology & Medicine, 83, 341–351. [DOI] [PubMed] [Google Scholar]
- Elson, C. O. , Cong, Y. , Qi, F. , Hershberg, R. M. , & Targan, S. R. (2006). Molecular approaches to the role of the microbiota in inflammatory bowel disease. Annals of the New York Academy of Sciences, 1072, 39–51. [DOI] [PubMed] [Google Scholar]
- Espey, M. G. (2013). Role of oxygen gradients in shaping redox relationships between the human intestine and its microbiota. Free Radical Biology & Medicine, 55, 130–140. [DOI] [PubMed] [Google Scholar]
- Esworthy, R. S. , Aranda, R. , Martin, M. G. , Doroshow, J. H. , Binder, S. W. , & Chu, F. F. (2001). Mice with combined disruption of Gpx1 and Gpx2 genes have colitis. American Journal of Physiology Gastrointestinal and Liver Physiology, 281, G848–G855. [DOI] [PubMed] [Google Scholar]
- Esworthy, R. S. , Binder, S. W. , Doroshow, J. H. , & Chu, F. F. (2003). Microflora trigger colitis in mice deficient in selenium‐dependent glutathione peroxidase and induce Gpx2 gene expression. Biological Chemistry, 384, 597–607. [DOI] [PubMed] [Google Scholar]
- Esworthy, R. S. , Kim, B. W. , Chow, J. , Shen, B. , Doroshow, J. H. , & Chu, F. F. (2014). Nox1 causes ileocolitis in mice deficient in glutathione peroxidase‐1 and ‐2. Free Radical Biology & Medicine, 68, 315–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esworthy, R. S. , Kim, B. W. , Larson, G. P. , Yip, M. L. , Smith, D. D. , Li, M. , & Chu, F. F. (2011). Colitis locus on chromosome 2 impacting the severity of early‐onset disease in mice deficient in GPX1 and GPX2. Inflammatory Bowel Diseases, 17, 1373–1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esworthy, R. S. , Smith, D. D. , & Chu, F. F. (2010). A strong impact of genetic background on gut microflora in mice. International Journal of Inflammation, 2010, 986046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esworthy, R. S. , Swiderek, K. M. , Ho, Y. S. , & Chu, F. F. (1998). Selenium‐dependent glutathione peroxidase‐GI is a major glutathione peroxidase activity in the mucosal epithelium of rodent intestine. Biochimica et Biophysica Acta, 1381, 213–226. [DOI] [PubMed] [Google Scholar]
- Esworthy, S. C. , Chu, C. , Shen, B. , & Doroshow, J. H. (2020). Supporting data‐ileum microbiome in 35 day‐old Gpx1/2‐DKO mice, antibiotic‐treated mice and sibling non‐DKO (Gpx1+/‐Gpx2‐/‐, Gpx1‐/‐Gpx2+/‐) mice. Figshare. 10.6084/m9.figshare.12167592 [DOI] [Google Scholar]
- Ferreira, R. B. , Gill, N. , Willing, B. P. , Antunes, L. C. , Russell, S. L. , Croxen, M. A. , & Finlay, B. B. (2011). The intestinal microbiota plays a role in Salmonella‐induced colitis independent of pathogen colonization. PLoS One, 6, e20338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, Q. , Esworthy, R. S. , Kim, B. W. , Synold, T. W. , Smith, D. D. , & Chu, F. F. (2010). Atherogenic diets exacerbate colitis in mice deficient in glutathione peroxidase. Inflammatory Bowel Diseases, 16, 2043–2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garidou, L. , Pomie, C. , Klopp, P. , Waget, A. , Charpentier, J. , Aloulou, M. , … Burcelin, R. (2015). The gut microbiota regulates intestinal CD4 T cells expressing RORgammat and controls metabolic disease. Cell Metabolism, 22, 100–112. [DOI] [PubMed] [Google Scholar]
- Gkouskou, K. K. , Deligianni, C. , Tsatsanis, C. , & Eliopoulos, A. G. (2014). The gut microbiota in mouse models of inflammatory bowel disease. Frontiers in Cellular and Infection Microbiology, 4, 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grasberger, H. , De Deken, X. , Mayo, O. B. , Raad, H. , Weiss, M. , Liao, X. H. , & Refetoff, S. (2012). Mice deficient in dual oxidase maturation factors are severely hypothyroid. Molecular Endocrinology, 26, 481–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grasberger, H. , Gao, J. , Nagao‐Kitamoto, H. , Kitamoto, S. , Zhang, M. , Kamada, N. , … Kao, J. Y. (2015). Increased expression of DUOX2 is an epithelial response to mucosal dysbiosis required for immune homeostasis in mouse intestine. Gastroenterology, 149, 1849–1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu, S. , Chen, D. , Zhang, J. N. , Lv, X. , Wang, K. , Duan, L. P. , … Wu, X. L. (2013). Bacterial community mapping of the mouse gastrointestinal tract. PLoS One, 8, e74957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guandalini, S. , & Sansotta, N. (2019). Probiotics in the treatment of inflammatory bowel disease. Advances in Experimental Medicine and Biology, 1125, 101–107. [DOI] [PubMed] [Google Scholar]
- Gupta, K. , Stapleton, A. E. , Hooton, T. M. , Roberts, P. L. , Fennell, C. L. , & Stamm, W. E. (1998). Inverse association of H2O2‐producing lactobacilli and vaginal Escherichia coli colonization in women with recurrent urinary tract infections. The Journal of Infectious Diseases, 178, 446–450. [DOI] [PubMed] [Google Scholar]
- Gupta, R. S. , & Gao, B. (2009). Phylogenomic analyses of clostridia and identification of novel protein signatures that are specific to the genus Clostridium sensu stricto (cluster I). International Journal of Systematic and Evolutionary Microbiology, 59, 285–294. [DOI] [PubMed] [Google Scholar]
- Haberman, Y. , Tickle, T. L. , Dexheimer, P. J. , Kim, M.‐O. , Tang, D. , Karns, R. , … Denson Lee A. (2014). Pediatric Crohn disease patients exhibit specific ileal transcriptome and microbiome signature. Journal of Clinical Investigation, 124(8), 3617–3633. 10.1172/jci75436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamm, C. M. , Reimers, M. A. , McCullough, C. K. , Gorbe, E. B. , Lu, J. , Gu, C. C. , … Hunt, S. R. (2010). NOD2 status and human ileal gene expression. Inflammatory Bowel Diseases, 16, 1649–1657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, C. , De Ravin, S. S. , Paul, A. R. , Heller, T. , Ho, N. , & Wu Datta, L. , … Consortium NIG (2016). Genetic risk for inflammatory bowel disease is a determinant of Crohn's disease development in chronic granulomatous disease. Inflammatory Bowel Diseases, 22, 2794–2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huycke, M. M. , Abrams, V. , & Moore, D. R. (2002). Enterococcus faecalis produces extracellular superoxide and hydrogen peroxide that damages colonic epithelial cell DNA. Carcinogenesis, 23, 529–536. [DOI] [PubMed] [Google Scholar]
- Jakobsson, H. E. , Rodriguez‐Pineiro, A. M. , Schutte, A. , Ermund, A. , Boysen, P. , Bemark, M. , … Johansson, M. E. (2015). The composition of the gut microbiota shapes the colon mucus barrier. EMBO Reports, 16, 164–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, R. M. , Luo, L. , Ardita, C. S. , Richardson, A. N. , Kwon, Y. M. , Mercante, J. W. , … Neish, A. S. (2013). Symbiotic lactobacilli stimulate gut epithelial proliferation via Nox‐mediated generation of reactive oxygen species. The EMBO Journal, 32, 3017–3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, R. M. , & Neish, A. S. (2017). Redox signaling mediated by the gut microbiota. Free Radical Biology & Medicine, 105, 41–47. [DOI] [PubMed] [Google Scholar]
- Ju, T. , Shoblak, Y. , Gao, Y. , Yang, K. , Fouhse, J. , Finlay, B. B. , … Willing, B. P. (2017). Initial gut microbial composition as a key factor driving host response to antibiotic treatment, as exemplified by the presence or absence of commensal Escherichia coli . Applied and Environmental Microbiology, 83(17), 1–15. 10.1128/AEM.01107-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai, T. , Mikami, Y. , & Hayashi, A. (2015). A breakthrough in probiotics: Clostridium butyricum regulates gut homeostasis and anti‐inflammatory response in inflammatory bowel disease. Journal of Gastroenterology, 50, 928–939. [DOI] [PubMed] [Google Scholar]
- Kar, S. K. , Jansman, A. J. M. , Benis, N. , Ramiro‐Garcia, J. , Schokker, D. , Kruijt, L. , … Wells, J. M. (2017). Dietary protein sources differentially affect microbiota, mTOR activity and transcription of mTOR signaling pathways in the small intestine. PLoS One, 12, e0188282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaser, A. , Lee, A. H. , Franke, A. , Glickman, J. N. , Zeissig, S. , Tilg, H. , … Blumberg, R. S. (2008). XBP1 links ER stress to intestinal inflammation and confers genetic risk for human inflammatory bowel disease. Cell, 134, 743–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato, M. , Marumo, M. , Nakayama, J. , Matsumoto, M. , Yabe‐Nishimura, C. , & Kamata, T. (2016). The ROS‐generating oxidase Nox1 is required for epithelial restitution following colitis. Experimental Animals, 65, 197–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiesler, P. , Fuss, I. J. , & Strober, W. (2015). Experimental models of inflammatory bowel diseases. Cellular and Molecular Gastroenterology and Hepatology, 1, 154–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiesslich, R. , Duckworth, C. A. , Moussata, D. , Gloeckner, A. , Lim, L. G. , Goetz, M. , … Watson, A. J. M. (2012). Local barrier dysfunction identified by confocal laser endomicroscopy predicts relapse in inflammatory bowel disease. Gut, 61(8), 1146–1153. 10.1136/gutjnl-2011-300695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King, S. J. , & McCole, D. F. (2019). Epithelial‐microbial diplomacy: Escalating border tensions drive inflammation in inflammatory bowel disease. Intestinal Research, 17, 177–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knaus, U. G. , Hertzberger, R. , Pircalabioru, G. G. , Yousefi, S. P. , & Branco Dos Santos, F. (2017). Pathogen control at the intestinal mucosa ‐ H2O2 to the rescue. Gut Microbes, 8, 67–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koretz, R. L. (2018). Probiotics in gastroenterology: How pro is the evidence in adults? The American Journal of Gastroenterology, 113, 1125–1136. [DOI] [PubMed] [Google Scholar]
- Laukens, D. , Brinkman, B. M. , Raes, J. , De Vos, M. , & Vandenabeele, P. (2016). Heterogeneity of the gut microbiome in mice: Guidelines for optimizing experimental design. FEMS Microbiology Reviews, 40, 117–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson, P. A. , Citron, D. M. , Tyrrell, K. L. , & Finegold, S. M. (2016). Reclassification of Clostridium difficile as Clostridioides difficile (Hall and O'Toole 1935) Prevot 1938. Anaerobe, 40, 95–99. [DOI] [PubMed] [Google Scholar]
- Lee, D. H. , Esworthy, R. S. , Chu, C. , Pfeifer, G. P. , & Chu, F. F. (2006). Mutation accumulation in the intestine and colon of mice deficient in two intracellular glutathione peroxidases. Cancer Research, 66, 9845–9851. [DOI] [PubMed] [Google Scholar]
- Li, G. , Scull, C. , Ozcan, L. , & Tabas, I. (2010). NADPH oxidase links endoplasmic reticulum stress, oxidative stress, and PKR activation to induce apoptosis. The Journal of Cell Biology, 191(6), 1113–1125. 10.1083/jcb.201006121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipinski, S. , Till, A. , Sina, C. , Arlt, A. , Grasberger, H. , Schreiber, S. , & Rosenstiel, P. (2009). DUOX2‐derived reactive oxygen species are effectors of NOD2‐mediated antibacterial responses. Journal of Cell Science, 122, 3522–3530. [DOI] [PubMed] [Google Scholar]
- Liu, J. Z. , van Sommeren, S. , Huang, H. , Ng, S. C. , Alberts, R. , Takahashi, A. , … Weersma, R. K. (2015). Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nature Genetics, 47, 979–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacFie, T. S. , Poulsom, R. , Parker, A. , Warnes, G. , Boitsova, T. , Nijhuis, A. , … Silver, A. (2014). DUOX2 and DUOXA2 form the predominant enzyme system capable of producing the reactive oxygen species H2O2 in active ulcerative colitis and are modulated by 5‐aminosalicylic acid. Inflammatory Bowel Diseases, 20(3), 514–524. 10.1097/01.mib.0000442012.45038.0e [DOI] [PubMed] [Google Scholar]
- Malhotra, J. D. , Miao, H. , Zhang, K. , Wolfson, A. , Pennathur, S. , Pipe S. W., & Kaufman, R. J. (2008). Antioxidants reduce endoplasmic reticulum stress and improve protein secretion. Proceedings of the National Academy of Sciences, 105(47), 18525–18530. 10.1073/pnas.0809677105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastrocola, R. , Ferrocino, I. , Liberto, E. , Chiazza, F. , Cento, A. S. , Collotta, D. , … Collino, M. (2018). Fructose liquid and solid formulations differently affect gut integrity, microbiota composition and related liver toxicity: A comparative in vivo study. The Journal of Nutritional Biochemistry, 55, 185–199. [DOI] [PubMed] [Google Scholar]
- Matziouridou, C. , Rocha, S. D. C. , Haabeth, O. A. , Rudi, K. , Carlsen, H. , & Kielland, A. (2018). iNOS‐ and NOX1‐dependent ROS production maintains bacterial homeostasis in the ileum of mice. Mucosal Immunology, 11, 774–784. [DOI] [PubMed] [Google Scholar]
- Mizoguchi, A. , & Mizoguchi, E. (2010). Animal models of IBD: Linkage to human disease. Current Opinion in Pharmacology, 10, 578–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peloquin, J. M. , & Nguyen, D. D. (2013). The microbiota and inflammatory bowel disease: Insights from animal models. Anaerobe, 24, 102–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peyrin‐Biroulet, L. , Billioud, V. , D'Haens, G. , Panaccione, R. , Feagan, B. , Panes, J. , … Sandborn, W. J. (2012). Development of the Paris definition of early Crohn's disease for disease‐modification trials: Results of an international expert opinion process. The American Journal of Gastroenterology, 107, 1770–1776. [DOI] [PubMed] [Google Scholar]
- Pircalabioru, G. , Aviello, G. , Kubica, M. , Zhdanov, A. , Paclet, M. H. , Brennan, L. , … Knaus, U. G. (2016). Defensive mutualism rescues NADPH oxidase inactivation in gut infection. Cell Host & Microbe, 19, 651–663. [DOI] [PubMed] [Google Scholar]
- Redanz, S. , Cheng, X. , Giacaman, R. A. , Pfeifer, C. S. , Merritt, J. , & Kreth, J. (2018). Live and let die: Hydrogen peroxide production by the commensal flora and its role in maintaining a symbiotic microbiome. Molecular Oral Microbiology, 33, 337–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson, S. J. , Zhou, J. Y. , Geddes, K. , Rubino, S. J. , Cho, J. H. , Girardin, S. E. , & Philpott, D. J. (2013). Nod1 and Nod2 signaling does not alter the composition of intestinal bacterial communities at homeostasis. Gut Microbes, 4, 222–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez, E. , Peiroten, A. , Landete, J. M. , Medina, M. , & Arques, J. L. (2015). Gut catalase‐positive bacteria cross‐protect adjacent bifidobacteria from oxidative stress. Microbes and Environments, 30, 270–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartor, R. B. , & Wu, G. D. (2017). Roles for intestinal bacteria, viruses, and fungi in pathogenesis of inflammatory bowel diseases and therapeutic approaches. Gastroenterology, 152, 327–339 e324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaubeck, M. , Clavel, T. , Calasan, J. , Lagkouvardos, I. , Haange, S. B. , Jehmlich, N. , Basic, M. , … Haller, D. (2016). Dysbiotic gut microbiota causes transmissible Crohn's disease‐like ileitis independent of failure in antimicrobial defence. Gut, 65(2), 225–237. 10.1136/gutjnl-2015-309333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekirov, I. , Tam, N. M. , Jogova, M. , Robertson, M. L. , Li, Y. , Lupp, C. , & Finlay, B. B. (2008). Antibiotic‐induced perturbations of the intestinal microbiota alter host susceptibility to enteric infection. Infection and Immunity, 76, 4726–4736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiina, T. , Shima, T. , Naitou, K. , Nakamori, H. , Sano, Y. , Horii, K. , … Shimizu, Y. (2015). Actions of probiotics on trinitrobenzenesulfonic acid‐induced colitis in rats. BioMed Research International, 2015, 528523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, A. K. , Hertzberger, R. Y. , & Knaus, U. G. (2018). Hydrogen peroxide production by lactobacilli promotes epithelial restitution during colitis. Redox Biology, 16, 11–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer, F. , & Backhed, F. (2015). The gut microbiota engages different signaling pathways to induce Duox2 expression in the ileum and colon epithelium. Mucosal Immunology, 8, 372–379. [DOI] [PubMed] [Google Scholar]
- Sommer, F. , & Backhed, F. (2016). Know your neighbor: Microbiota and host epithelial cells interact locally to control intestinal function and physiology. BioEssays: News and Reviews in Molecular, Cellular and Developmental Biology, 38, 455–464. [DOI] [PubMed] [Google Scholar]
- Stolzer, I. , Kaden‐Volynets, V. , Ruder, B. , Letizia, M. , Bittel, M. , Rausch, P. , … Gunther, C. (2020). Environmental microbial factors determine the pattern of inflammatory lesions in a murine model of Crohn's disease‐like inflammation. Inflammatory Bowel Diseases, 26, 66–79. [DOI] [PubMed] [Google Scholar]
- Suzuki, T. A. , & Nachman, M. W. (2016). Spatial heterogeneity of gut microbial composition along the gastrointestinal tract in natural populations of house mice. PLoS One, 11, e0163720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tourret, J. , Willing, B. P. , Dion, S. , MacPherson, J. , Denamur, E. , & Finlay, B. B. (2017). Immunosuppressive treatment alters secretion of ileal antimicrobial peptides and gut microbiota, and favors subsequent colonization by uropathogenic Escherichia coli . Transplantation, 101, 74–82. [DOI] [PubMed] [Google Scholar]
- Turcotte, J.‐F. , Wong, K. , Mah, S. J. , Dieleman, L. A. , Kao, D. , Kroeker, K. , … Liu, J. J. (2012). Increased epithelial gaps in the small intestine are predictive of hospitalization and surgery in patients with inflammatory bowel disease. Clinical and Translational Gastroenterology, 3(7), e19 10.1038/ctg.2012.13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walk, S. T. , Blum, A. M. , Ewing, S. A. , Weinstock, J. V. , & Young, V. B. (2010). Alteration of the murine gut microbiota during infection with the parasitic helminth Heligmosomoides polygyrus. Inflammatory Bowel Diseases, 16, 1841–1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, J. M. , Duckworth, C. A. , Burkitt, M. D. , Watson, A. J. M. , Campbell, B. J. , & Pritchard, D. M. (2015). Epithelial Cell Shedding and Barrier Function. Veterinary Pathology, 52(3), 445–455. 10.1177/0300985814559404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wlodarska, M. , Willing, B. , Keeney, K. M. , Menendez, A. , Bergstrom, K. S. , Gill, N. , … Finlay, B. B. (2011). Antibiotic treatment alters the colonic mucus layer and predisposes the host to exacerbated Citrobacter rodentium‐induced colitis. Infection and Immunity, 79, 1536–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanai, H. , Ben‐Shachar, S. , Baram, L. , Elad, H. , Gitstein, G. , Brazowski, E. , … Dotan, I. (2015). Gene expression alterations in ulcerative colitis patients after restorative proctocolectomy extend to the small bowel proximal to the pouch. Gut, 64(5), 756–764. 10.1136/gutjnl-2014-307387 [DOI] [PubMed] [Google Scholar]
- Zuo, T. , & Ng, S. C. (2018). The gut microbiota in the pathogenesis and therapeutics of inflammatory bowel disease. Frontiers in Microbiology, 9, 2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Supporting data sets generated and/or analyzed during the current study are available at figshare (Esworthy et al., 2020): https://doi.org/10.6084/m9.figshare.12167592; Excel files (EF) and supporting figures: (1) EF Abundances of OTUs; (2) EF NONDKO‐vs‐SPLENDACTRL‐differential‐abundance‐stats; (3) EF METRO‐vs‐SPLENDACTRL‐differential‐abundance‐stats; (4) EF STREP‐vs‐SPLENDACRTL‐differential‐abundance‐stats; (5) EF VANCO‐vs‐SPLENDACRTL‐ differential‐abundance‐stats; (6) EF Cured vs sick; (7) EF cured vs non‐DKO, non‐DKO vs metronidazole; (8) EF abundances correl abscesses; (9) EF abundances correl apoptosis; (10) EF abundances correl crypt density; (11) EF abundances correl exfoliation; (12) EF abundances correl Paneth cells; (13) EF abundances correl pathology scores; (14) EF Lactobacillus adjusted for E. coli dilution; (15) Figures S1‐S8—correlation graphs of top candidate provocative OTUs abundance with pathology markers; (16) Figure S9—pathology in colon; (17) Figure S10—correlation graphs Lactobacillus adjusted for E. coli dilution.


