Abstract
Pyruvate dehydrogenase (PDH) and 2‐oxoglutarate dehydrogenase (ODH) are critical enzymes in central carbon metabolism. In Corynebacterium glutamicum, an unusual hybrid complex consisting of CgE1p (thiamine diphosphate‐dependent pyruvate dehydrogenase, AceE), CgE2 (dihydrolipoamide acetyltransferase, AceF), CgE3 (dihydrolipoamide dehydrogenase, Lpd), and CgE1o (thiamine diphosphate‐dependent 2‐oxoglutarate dehydrogenase, OdhA) has been suggested. Here, we elucidated that the PDH‐ODH hybrid complex in C. glutamicum probably consists of six copies of CgE2 in its core, which is rather compact compared with PDH and ODH in other microorganisms that have twenty‐four copies of E2. We found that CgE2 formed a stable complex with CgE3 (CgE2‐E3 subcomplex) in vitro, hypothetically comprised of two CgE2 trimers and four CgE3 dimers. We also found that CgE1o exists mainly as a hexamer in solution and is ready to form an active ODH complex when mixed with the CgE2‐E3 subcomplex. Our in vitro reconstituted system showed CgE1p‐ and CgE1o‐dependent inhibition of ODH and PDH, respectively, actively supporting the formation of the hybrid complex, in which both CgE1p and CgE1o associate with a single CgE2‐E3. In gel filtration chromatography, all the subunits of CgODH were eluted in the same fraction, whereas CgE1p was eluted separately from CgE2‐E3, suggesting a weak association of CgE1p with CgE2 compared with that of CgE1o. This study revealed the unique molecular architecture of the hybrid complex from C. glutamicum and the compact‐sized complex would provide an advantage to determine the whole structure of the unusual hybrid complex.
This study shows the unique molecular architecture of the PDH‐ODH hybrid complex in Corynebacterium glutamicum, in which both CgE1p and CgE1o associate with the CgE2‐E3 subcomplex. The core‐forming CgE2‐E3 probably consists of six copies of CgE2, which is relatively compact compared with PDH and ODH in other microorganisms that have 24 copies of E2. The fragility of the CgPDH complex and the unusual multimerization of CgE1o are also presented.
1. INTRODUCTION
Pyruvate dehydrogenase (PDH) and 2‐oxoglutarate dehydrogenase (ODH) are critical enzymes in central carbon metabolism and generate vital high energy compounds acetyl‐CoA and succinyl‐CoA, respectively; both of which are major CoA derivatives in cell and serve as substrates for protein acetylation and succinylation (Komine‐Abe et al., 2017; Mizuno et al., 2016; Nagano‐Shoji et al., 2017). PDH and ODH are large enzyme complexes (∼10 MDa) composed of three subunits; E1, E2, and E3. E1 (EC 1.4.2.1) is involved in thiamine pyrophosphate‐dependent oxidative decarboxylation of 2‐oxoacid (pyruvate or 2‐oxoglutarate) with the concomitant transfer of the corresponding acyl group to a lipoyl group attached on a lysine residue of E2. E2 (EC 2.3.1.12) is a dihydrolipoyl acyltransferase that catalyzes the transfer of the acyl group to CoA. E3 (EC 1.8.1.4) is a dihydrolipoyl dehydrogenase that is responsible for FAD‐dependent reoxidation of dihydrolipoamide to lipoamide on E2 to generate NADH (Perham, 2000; Patel, Nemeria, Furey, & Jordan, 2014). The complexes are organized in both structural and functional aspects using E2 as the core. E2 protein consists of three different types of domains: one to three N‐terminal lipoyl domains (~80 amino acids each), a peripheral subunit‐binding domain (PSBD, ~45 amino acids), and a core‐forming C‐terminal catalytic acyltransferase domain (~250 amino acids). As demonstrated by the crystal structures of PDH and ODH from several organisms (Izard et al., 1999; Knapp et al., 1998; Mattevi et al., 1992, 1993; Wang et al., 2014), E2 trimer is the principal building block to be assembled into an octahedral (24‐mer) or icosahedral (60‐mer) symmetrical shape to form the inner core. Both E1 and E3 exist as a dimer and are noncovalently tethered to the PSBD of E2. In many organisms, including Escherichia coli, the E3 protein is shared by PDH and ODH, while specific E1 and E2 proteins are utilized for each enzyme complex.
PDH and ODH from the order Corynebacteriales including Corynebacterium glutamicum have several unique features. First, there is only a single gene encoding E2 in the genome, and the same E2, as well as E3, proteins are used by PDH and ODH. Second, there exists a variant protein of E1o with an extra E2 acyltransferase domain in the N‐terminal region, which corresponds to OdhA (CgE1o, NCgl1084) in C. glutamicum (Usuda et al., 1996) and Kgd or SucA in Mycobacterium (Tian et al. 2005; Wagner et al. 2019). The deletion of odhA resulted in the loss of ODH activity, and OdhA certainly functions as E1o (Hoffelder et al. 2010). Mycobacterium SucA functions as a multifunctional enzyme capable of reductive succinyl‐transfer to a lipoyl residue via the canonical dehydrogenase reaction (Wagner et al. 2011), nonreductive decarboxylation of α‐ketoglutarate (2‐oxoglutarate) to produce succinic semialdehyde (Tian et al., 2005; Wagner et al., 2011), and carboligation with glyoxylate to give 2‐hydroxy‐3‐oxoadipate (de Carvalho et al., 2010; Wagner et al., 2011). Third, a small regulatory protein (OdhI in C. glutamicum and GarA in Mycobacterium) interacts with the E1o variant to control the enzymatic activity (Niebisch et al., 2006; Nott et al., 2009). OdhI is composed of two domains: an N‐terminal domain, containing phosphorylation sites, and a C‐terminal forkhead‐associated (FHA) domain, recognizing phosphorylated serine/threonine. The FHA domain also establishes the binding to CgE1o (Krawczyk et al., 2010). When the relevant N‐terminal sites are phosphorylated, OdhI undergoes a conformational change through the interaction of the FHA domain with the phosphorylated sites, releasing ODH from the inhibition by OdhI (Barthe et al., 2009).
In a previous study that investigated the subunit organization of C. glutamicum, ODH, CgE2 (AceF, NCgl2126), and CgE3 (Lpd, NCgl0355) were copurified along with a tagged CgE1o as expected. Interestingly, CgE1p (AceE, NCgl2167) was also copurified with the ODH complex (Niebisch et al., 2006). Furthermore, when a tagged CgE1p was used as prey, all the components of the ODH complex were copurified (Niebisch et al., 2006). This was the first evidence to suggest the presence of a PDH‐ODH hybrid complex in C. glutamicum. Further enzymatic and genetic studies revealed that each E2 acyltransferase domain of CgE2 and CgE1o specifically catalyzes the transacetylase and transsuccinylase reaction, respectively, suggesting that the E2 catalytic domain of CgE2 and CgE1o is responsible for PDH and ODH activities (Hoffelder et al., 2010). It was also revealed that CgE1o and also CgE2 are required for the ODH activity (Hoffelder et al., 2010). Recent structural studies of Mycobacterium smegmatis SucA, which is the homolog of CgE1o, revealed that the E1o catalytic domain is involved in generating a ThDP‐bound decarboxylation intermediate derived from 2‐oxoglutarate. This decarboxylation intermediate undergoes conformational changes from the α‐carbanion to the enamine form and is reductively transferred to a lipoyl residue on E2 (Wagner et al., 2014; Wagner et al., 2019). All the pieces of evidence strongly support the presence of the PDH‐ODH hybrid complex. However, the molecular architecture, including the subunit stoichiometry and arrangement of the hybrid complex, remains to be elucidated. In the present study, we aimed to characterize the C. glutamicum PDH‐ODH complex in vivo and in vitro and show the unique molecular architecture of the hybrid complex.
2. MATERIALS AND METHODS
2.1. Bacterial strains and culture conditions
Corynebacterium glutamicum ATCC13869 was used as the wild‐type strain. All strains and plasmids used in this study are shown in Table 1. Corynebacterium glutamicum cells were grown in 25 ml of glutamate production medium (Mizuno et al., 2016) at 31.5°C in 500‐ml baffled flasks on a rotary shaker at 100 rpm (70 mm of the shaking width, Takasaki Kagaku, Japan). Tween 40 (1.5 g/l) was added after 3 h of cultivation to induce l‐glutamate production, and the same volume of distilled water was added for control (uninduced). Nine‐hour cultivated cells were used for the lysate preparation.
TABLE 1.
Bacterial strains and plasmids used in this study
| Description | Source or reference | |
|---|---|---|
| Corynebacterium glutamicum strain | ||
| ATCC13869 | Wild‐type strain | Laboratory stock |
| Plasmids | ||
| pAA62 | pET21a expressing C‐terminal His‐tagged CgE1o (OdhA) | Komine‐Abe et al. (2017) |
| pAA77 | pET21a expressing N‐terminal Strep‐tagged OdhI | Komine‐Abe et al. (2017) |
| pKH4 | pET21a expressing C‐terminal His‐tagged CgE1p (AceE) | This study |
| pKH13 | pRSFDuet expressing N‐terminal His‐ and Flag‐tagged CgE3 (Lpd) and CgE2 (AceF) | This study |
2.2. Ultracentrifuge analysis
Corynebacterium glutamicum cells were lysed in TESG15 buffer (50 mM TES‐NaOH [pH 7.6] and 15% glycerol) containing 1 mM PMSF, 1 mM DTT, 10 µg/ml DNase, and 10 µg/ml RNase at high pressure using EmulsiFlex‐B15 (Avestin Inc., Ottawa). After cell debris removal by centrifugation (7000 g, 10 min, 4°C), the cleared lysate containing 1.2 mg of protein was layered onto a 10%–30% or 15%–45% sucrose density gradient in 50 mM TES‐NaOH (pH 7.6). The gradient was centrifuged at 36,000 rpm (max. 230,000 g) in a HITACHI P40ST rotor for 17 h at 4°C. After centrifugation, the gradient was fractionated using a Piston Gradient Fractionator (BioComP Instruments, Fredericton) into 21 fractions of 0.5 ml and the bottom fraction. Escherichia coli DH5α cells were grown in 2 × YT medium until the OD660 reached 3.5 and processed using the same ultracentrifuge protocol. Aliquots (10 μl) were separated by SDS‐PAGE and subjected to Western blot analysis to detect each subunit of PDH and ODH. Denatured samples were prepared by boiling for 5 min in the presence of 1% SDS before ultracentrifugation.
2.3. Western blot analysis
Western blot analysis was performed as described previously (Mizuno et al., 2016). The rabbit polyclonal anti‐CgAceE and anti‐CgAceF antibodies were raised against the commercially synthesized NCgl2167 (904‐KFKLDDPTSVSVDPNAPEE‐922) and NCgl2126 (1‐MAFSVEMPELGESVT‐15) peptides, respectively (Sigma‐Genosys LP, Texas). Primary antibodies used are as follows: rabbit polyclonal anti‐CgAceE (1:10,000); rabbit polyclonal anti‐CgAceF (1:5000); rabbit polyclonal anti‐CgOdhA (1:10,000) (Kim et al., 2011); rabbit polyclonal anti‐EcAceE (1:10,000); rabbit polyclonal anti‐EcAceF (1:5000); rabbit polyclonal anti‐EcSucA (1:10,000); rabbit polyclonal anti‐EcSucB (1:10,000); rabbit polyclonal anti‐EcLpd (1:10,000) (provided by Shimada and Tanaka); rabbit polyclonal anti‐RplC from Bacillus subtilis (1:10,000) (provided by Kawamura); and rabbit anti‐lipoic acid antibody (Calbiochem, 437695) (1:2000).
2.4. Preparation of recombinant proteins
Recombinant CgE1o protein was expressed as a C‐terminal His‐tagged protein using the pET‐21a vector (pAA62) (Komine‐Abe et al., 2017). Recombinant CgE1p protein was expressed as a C‐terminal His‐ and Flag‐tagged protein using the pET‐21a vector (pKH4). Recombinant CgE2 and CgE3 proteins were co‐expressed using the pRSFDuet vector (pKH13). N‐terminal His‐ and Flag‐tagged CgE3 enabled copurification of CgE2 and CgE3 as a CgE2‐E3 subcomplex. Recombinant OdhI protein was expressed as an N‐terminal Strep‐tagged protein (pAA77) (Komine‐Abe et al., 2017).
All recombinant proteins were produced in Escherichia coli BL21‐CodonPlus (DE3) cells. Escherichia coli cells were grown in 2×YT medium at 37°C for 3 h and then cultivated at 25°C for 18 h in the presence of 0.5 mM IPTG to induce enzyme expression. The cells were harvested and lysed in TESG10N (100 mM TES‐NaOH [pH 7.6], 150 mM NaCl, and 10% [w/v] glycerol) containing 1 mM PMSF, 1 mM DTT, 10 µg/ml DNase, and 10 µg/ml RNase, followed by affinity purification using Ni‐NTA His Bind resin (Merck Millipore, Burlington). His‐tagged proteins were eluted with TESG10N containing 200 mM imidazole. The eluted proteins were dialyzed against the TESG10N buffer.
2.5. In vitro reconstitution of PDH and ODH
Dialyzed or gel filtration purified CgE2‐E3 proteins were mixed with CgE1p and/or CgE1o in TESG10N buffer and incubated on ice for 30 min. Then, the sample was used for enzyme assays or gel filtration chromatography.
2.6. Gel filtration chromatography
Gel filtration chromatography was performed using a HiLoad 26/600 Superdex 200 pg column (GE Healthcare, Chicago) or a Superose 6 10/300 GL column (GE Healthcare, Chicago) with TESG10N buffer. For a Superdex 200 column, 2 or 5 ml of sample was applied. Chromatography was performed at a flow rate of 1.0 ml per min for 600 min, and 5 ml fractions were collected. For a Superose 6 column, 0.2 ml of sample was applied. Chromatography was performed at a flow rate of 0.2 ml per min for 250 min, and 0.5 ml fractions were collected.
2.7. Enzyme assays
PDH and ODH activities were assayed as described previously (Komine‐Abe et al., 2017). Recombinant CgE2‐E3 and CgE1 proteins were mixed and kept on ice for 30 min. Aliquots (20–50 μl) of the mixture were incubated in a reaction mixture (1 ml) containing 100 mM TES‐NaOH (pH 7.6), 0.2 mM coenzyme A, 0.3 mM thiamine pyrophosphate, 3 mM L‐cysteine, 5 mM MgCl2, 1 mM oxidized form of 3‐acetylpyridine adenine dinucleotide (APAD+; Oriental Yeast Co., Ltd., Japan), and 6 mM pyruvate (for PDH) or 1 mM 2‐oxoglutarate (for ODH). Assays were performed at 30°C, and the initial rate of APAD+ reduction was monitored using spectrophotometry at 365 nm. Endogenous APAD+ reduction in the absence of the substrates was subtracted from the APAD+ reduction in the presence of the substrates. One unit of activity was defined as the reduction of 1 nmol of APAD+ (ε = 9.1 l μmol−1 cm−1) per min.
3. RESULTS
3.1. Detection of the PDH‐ODH hybrid complex in C. glutamicum lysates using ultracentrifuge analysis
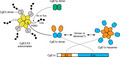
To investigate the molecular architectures of PDH and ODH in C. glutamicum lysates, we performed ultracentrifuge analysis using a sucrose density gradient. We first analyzed E. coli lysate samples, since the molecular architectures of the PDH and ODH complexes in E. coli are well characterized. Previous ultracentrifuge analysis showed that the E. coli PDH and ODH are large complexes with the molecular mass of 4.8 MDa and 2.4 MDa, respectively (Koike et al., 1960). Structural studies revealed that in both complexes, 24 copies of E2 subunit form an octahedral inner core, and up to 24 dimers of either E1 or E3 associate with the E2 core (Knapp et al., 1998; Murphy & Jensen, 2005; Reed et al., 1975). In our ultracentrifuge analysis of E. coli lysates using a 15%–45% sucrose density gradient (Figure 1a), the PDH subunit proteins AceE (E1p), AceF (E2p), and Lpd (E3) were detected in fractions 18‐19, representing a PDH complex. The three PDH subunits were also detected in fractions 8‐9. Previous studies reported that the PDH complex was present in multiple forms, a major 56‐60 S component (a full complex) as well as minor 86‐90 S and 20‐26 S (a partial complex) components (Danson et al., 1979; Koike et al., 1960). The PDH complexes in fractions 18‐19 and 8‐9 were assigned as the full and partial complexes of PDH, respectively. Meanwhile, SucA (E1o), SucB (E2o), and Lpd (E3) of the ODH complex were detected in fractions 15‐16 and 6‐7, which were likely the full and partial complexes of ODH, corresponding to major 37 S and minor 22 S components, according to a previous sedimentation analysis (Koike et al., 1960), respectively. We also detected the ribosomal large subunit protein L3 (RplC) in fractions 15‐17, representing ribosomes (an approx. molecular mass of 2.7 MDa). Thus, our centrifugation system could assign PDH, ODH, and ribosomes in fractions with the expected molecular sizes and confirmed that PDH and ODH are different complexes in E. coli. These results assured that our ultracentrifuge system was reliable to analyze the molecular architecture of large protein complexes like PDH and ODH.
FIGURE 1.
Detection of PDH and ODH in vivo. (a) Ultracentrifuge analysis of Escherichia coli lysates. The total lysate (TL) was sedimented using a 15%–45% sucrose density gradient. Aliquots (10 μl) of 21‐fractionated and the bottom (b) samples were separated using 8% or 10% SDS‐PAGE, followed by Western blot analysis to detect each subunit. Black bars indicate the positions of PDH, ODH, and ribosomes detected. PDHp and ODHp represent partial complexes of PDH and ODH, respectively. A representative data set of two biologically independent experiments is shown. (b) Ultracentrifuge analysis of Corynebacterium glutamicum lysates. The TL was sedimented using a 10%–30% sucrose density gradient. Uninduced, conditions without Tween 40 as an inducer; induced, Tween 40‐induced glutamate‐producing conditions; denatured, denatured samples of uninduced conditions. Detection of each subunit was performed as shown in (a). Black bars indicate the positions of Escherichia coli PDH, ODH, and ribosomes detected in the same conditions (see Figure A1). A representative data set of three biologically independent experiments is shown
We next analyzed C. glutamicum lysate samples prepared from cells grown in glutamate production medium without Tween 40 (as an inducer) using ultracentrifugation in a 10%–30% sucrose density gradient. CgE1p, CgE1o, and CgE2 proteins were detected in the “light” fraction 4 (and also 5‐6) (Figure 1b). CgE2 was migrated to the lighter fractions (2 and 3) in SDS‐treated denatured samples. We thus speculated that CgE1p, CgE1o, and CgE2 formed a PDH‐ODH hybrid complex which was eluted in fraction 4, although the fractions containing CgE3 could not be determined due to the unavailability of the specific antibody. The molecular mass of a hybrid PDH‐ODH complex could be much smaller than that of the E. coli PDH or ODH complex (Figure 1b, Figure A1). Interestingly, CgE1p and CgE1o migrated to higher molecular mass fractions (fraction 7 and more) which did not contain CgE2. This may suggest that the two E1 proteins interact with unknown proteins or are multimerized.
We also performed a similar analysis using lysates from cells grown in Tween 40‐triggered glutamate‐producing conditions and compared the sedimentation patterns of the PDH and ODH subunit proteins. The CgE2 was constantly detected in higher molecular mass fractions in glutamate‐producing conditions compared with nonproducing conditions (Figure 1b).
3.2. Reconstitution of the PDH and ODH activities in vitro
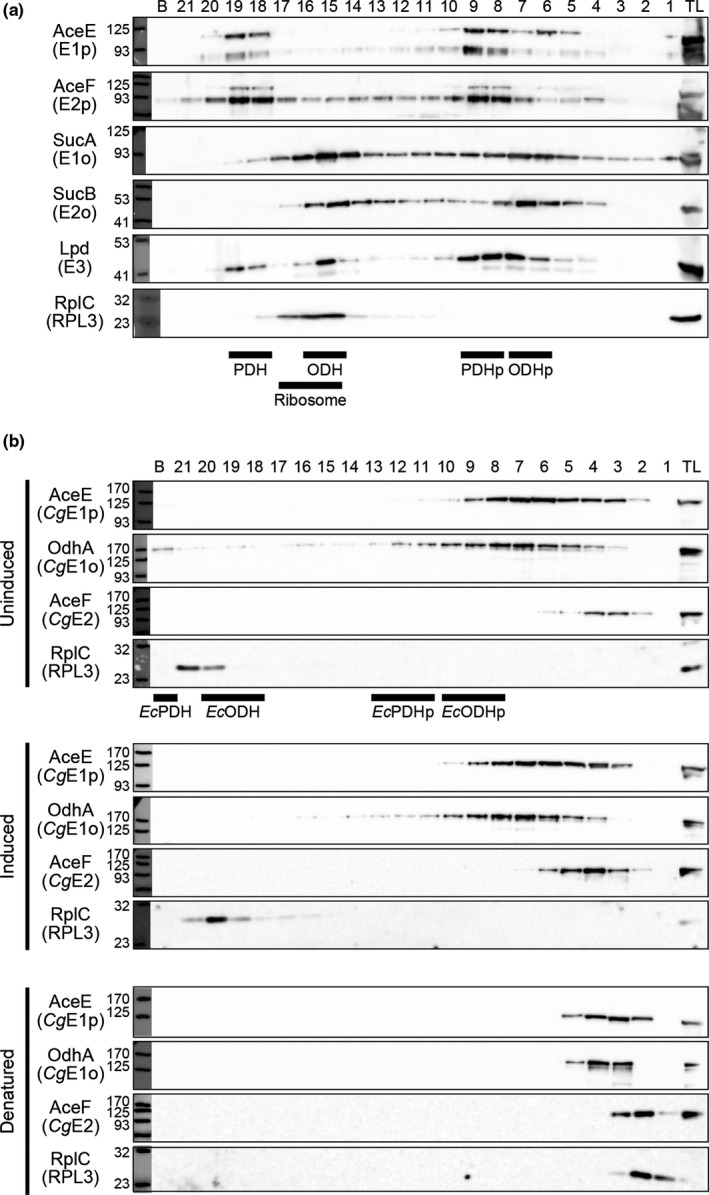
We tried to reconstitute PDH and ODH activities in vitro using subunit proteins prepared from E. coli. We successfully copurified CgE2 with CgE3 using the N‐terminal His‐tagging of CgE3 (Figure 2a). The interaction of CgE2 with CgE3 was retained in gel filtration chromatography (Figure 2d, Figure A2), suggesting the formation of an E2‐E3 subcomplex rather than a mixture of the two proteins. The recombinant CgE2 contained an attached lipoyl group, as detected using Western blot analysis with an anti‐lipoyl antibody (Figure 2b). Incubation of the affinity‐purified CgE2‐E3 with CgE1p and CgE1o resulted in PDH and ODH activities, respectively (Figure 2c). When both CgE1p and CgE1o were added to a reaction mixture, PDH and ODH activities were detected. These results indicated that PDH and ODH activities were successfully reconstituted in vitro.
FIGURE 2.
In vitro reconstitution of PDH and ODH activities. (a) Affinity purification of His‐tagged CgE1p, CgE1o, and CgE2‐E3 of Corynebacterium glutamicum. Affinity‐purified samples (1.5 μg) were separated using 10% SDS‐PAGE and detected using CBB staining. (b) Detection of lipoyl group attachment on CgE2. Affinity‐purified CgE2 (2 μg), separated using 10% SDS‐PAGE, was detected using Western blot analysis with anti‐lipoic acid (left) and anti‐CgE2 (right) antibodies. (c) CgE1p‐dependent PDH and CgE1o‐dependent ODH activities. Affinity‐purified CgE2‐E3 (110 μg) was mixed with CgE1p (70 μg), CgE1o (90 μg), or both and subjected to PDH (white columns) and ODH (black columns) activity assays. Data are shown as the mean and standard deviation of triplicate assays. A representative data set of three independent experiments is shown. (d) Gel filtration chromatogram of the reconstituted sample including CgE1p, CgE1o, and CgE2‐E3 using a Superdex 200 column (upper). Aliquots (5 μl) of fractionated samples (5 ml each) were analyzed using Western blotting to detect each subunit (lower). Samples of three peak fractions (a, b, and c) were subjected to PDH and ODH activity assays (right). Data are shown as the mean and standard deviation of triplicate assays
Next, we analyzed the reconstituted sample containing CgE1o, CgE1p, and CgE2‐E3 using gel filtration chromatography. Three peak fractions (a, b, and c) were observed in the chromatogram. Western blot analysis using specific antibodies revealed that the a fraction contained CgE1o, CgE2, and CgE3 proteins, the b fraction contained CgE1o and CgE3, and the c fraction majorly contained CgE1p (Figure 2d, left). ODH activity was detected in the a fraction, indicating that the peak a represents the ODH complex, in which CgE1o was associated with the CgE2‐E3 subcomplex. On the other hand, no PDH activity was detected in any peak fraction (Figure 2d, right). Also, when CgE2‐E3 was incubated only with CgE1p, CgE2‐E3 and CgE1p were eluted in separate fractions (Figure A2). These results suggested that the association of CgE1p with CgE2‐E3 was too weak to be retained during gel filtration.
3.3. Subunit stoichiometry of the CgE2‐E3 subcomplex and ODH complex
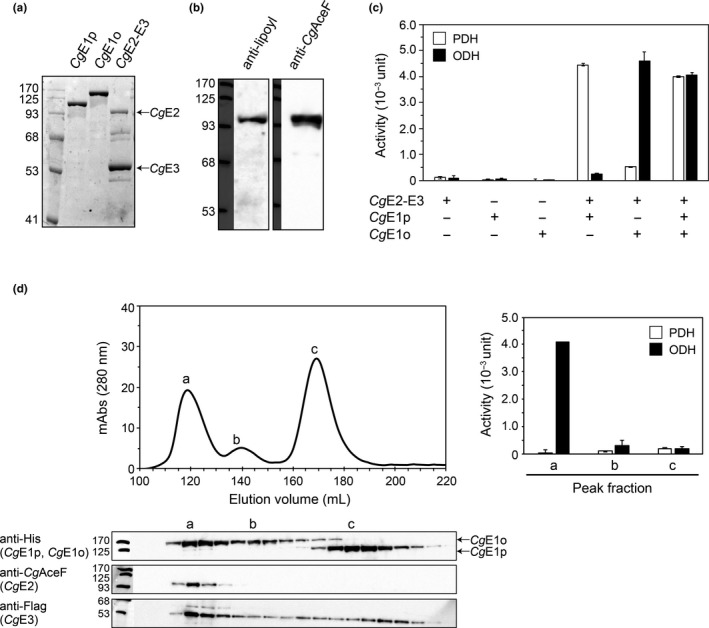
The subunit stoichiometry of CgE2‐E3 and ODH complexes was analyzed using gel filtration chromatography with molecular size standards. Although the CgE2‐E3 subcomplex was eluted outside the range of the size standards, the molecular mass of the subcomplex was estimated to be 780 kDa (Figure 3a). CgE2 and CgE3 proteins show 47% and 53% similarity with E. coli E2p and E3, respectively, and following relevant estimations were done according to previous research (Chandrasekhar et al., 2013; Wang et al., 2014). The subunit stoichiometry of the CgE2‐E3 subcomplex was estimated to be six copies of CgE2 (theoretical molecular mass of 70,905 Da) and eight copies of CgE3 (theoretical molecular mass of 50,652 Da), giving two E2 trimers and four E3 dimers (theoretically 831 kDa in total). CgE1o (theoretical molecular mass of 134,664 Da) was eluted in two separated peak fractions, corresponding to 750 kDa and 460 kDa for major and minor peaks, respectively (Figure 3a). The 750 kDa and 460 kDa multimers of CgE1o were estimated to be a hexamer (theoretically 808 kDa), and a tetramer (theoretically 539 kDa) or a trimer (theoretically 404 kDa). The fraction of 460 kDa (CgE1o460k) was collected and subjected to gel filtration chromatography, which provided two peaks of 750 kDa and 460 kDa again (Figure A3). Since both CgE1o‐containing fractions exhibited ODH activity when incubated with CgE2‐E3, they were not inactive aggregates but functional multimers (Figure 3b). Therefore, we assumed that CgE1o is present at equilibrium between the two multimer forms in solution.
FIGURE 3.
Estimation of molecular masses of the CgE1o component, the CgE2‐E3 subcomplex, and ODH complex. (a) Gel filtration chromatograms of the E1o (black line) and the E2‐E3 subcomplex (gray line) using a Superdex 200 column. The dotted line indicates a standard curve of molecular weight standards (thyroglobulin, 669 kDa; ferritin, 440 kDa; aldolase, 158 kDa; conalbumin, 75 kDa; and ovalbumin, 44 kDa). A representative data set of two independent experiments is shown. (b) Both CgE1o fractions of 750 kDa (CgE1o750k) and 460 kDa (CgE1o460k) provided ODH activity. CgE1o750k or CgE1o460k (20 μg) was mixed with E2‐E3 (15 μg) and subjected to enzyme assays. Data are shown as the mean and standard deviation of triplicate assays. (c) Gel filtration chromatograms of the ODH complex (a mixture of E1o with CgE2‐E3, broken line) using a Superose 6 column (upper). Chromatograms of individual CgE1o (black line) and CgE2‐E3 (gray line) are also shown. Aliquots (10 μl) of fractions (0.5 ml each) indicated were analyzed using 10% SDS‐PAGE, and the CBB staining gel image is also shown (lower)
Next, we tried to determine the molecular mass of the ODH complex. The mixture sample containing CgE1o and CgE2‐E3 provided a new peak fraction with a higher molecular mass than the individual CgE1o and CgE2‐E3 samples (Figure 3c). Since the new peak fraction contained CgE1o, CgE2, and CgE3 (Figure 3c) and exhibited ODH activity, the active ODH complex was indeed reconstituted in vitro. The exact molecular mass could not be estimated for the ODH complex because it ranged out of the molecular size standards. However, it was estimated to be approximately 940 kDa.
As described above, the full PDH complex, consisting of CgE1p, CgE2, and CgE3, was not detected in our gel filtration chromatography. The CgE1p (a theoretical molecular mass of 102,826 Da) was mainly eluted in a peak fraction with the estimated molecular size of 170 kDa, which likely represents a dimeric form (theoretically 206 kDa) in solution (Figure A2).
3.4. Kinetic analysis of in vitro reconstituted PDH and ODH
We performed kinetic analyses of the reconstituted PDH and ODH complexes. For this analysis, the affinity‐purified proteins were further purified using gel filtration chromatography to remove contaminated proteins (Figure A4). As a result of the enzyme assays using a mixture of CgE2‐E3 and CgE1p or CgE1o, shown in Figure 4, the apparent K m value of the reconstituted PDH complex for pyruvate was 3.5 ± 0.2 mM with a V max of 0.48 ± 0.01 nmol min−1 mg−1, and the apparent K m of the reconstituted ODH complex for 2‐oxoglutarate was 0.010 ± 0.001 mM with a V max of 0.48 ± 0.00 nmol min−1 mg−1. Our apparent K m value of the PDH complex for pyruvate was comparable to that reported previously using C. glutamicum lysates (1.7 mM), while our apparent K m of the ODH complex for 2‐oxoglutarate was one order smaller than that determined using lysates (0.13 mM) (Hoffelder et al., 2010).
FIGURE 4.
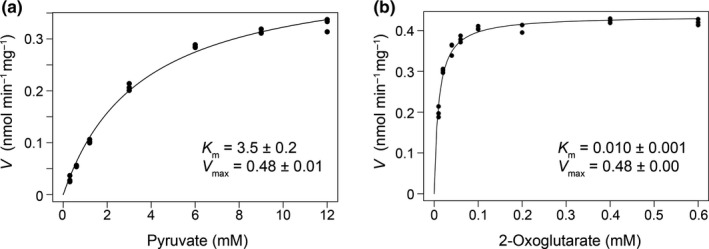
Kinetic analysis of PDH (a) and ODH (b) activities. A mixture of CgE1p (15 μg) or CgE1o (20 μg) with CgE2‐E3 (15 μg) was incubated on ice for 30 min for reconstitution and then subjected to enzyme assays. Dots show individual data of triplicate assays
3.5. CgE1p‐ and CgE1o‐dependent inhibition of ODH and PDH, respectively
CgE1p and CgE1o subunits associate with CgE2‐E3 in a complex. CgE1p (or CgE1o) may compete with CgE1o (or CgE1p) for binding to CgE2‐E3, which results in competitive inhibition of ODH and PDH via CgE1p and CgE1o, respectively. We examined the effect of CgE1o (or CgE1p) addition on PDH or ODH activity. We first examined the dose dependency of each CgE1 on the activity of the cognate enzyme. PDH activity was saturated when 15 μg (150 pmol) of CgE1p was added to a fixed amount (15 μg, 18 pmol) of CgE2‐E3 (Figure 5a). Meanwhile, ODH activity was not saturated even with 80 μg (600 pmol) of CgE1o in the reaction mixture containing the same amount of CgE2‐E3. Next, we added an increased amount of CgE1o to the above reconstituted PDH mixture. As the molar ratio of CgE1o to CgE1p increased, PDH activity decreased (Figure 5b, left, white columns). Similarly, ODH activity decreased as the molar ratio of CgE1p to CgE1o increased (Figure 5b, right, black columns). The inhibitory effect of CgE1o on PDH activity was stronger than that of CgE1p on ODH (Figure 5c). These results suggested that CgE1p and CgE1o competed with each other for CgE2‐E3 to form the PDH‐ODH hybrid complex.
FIGURE 5.
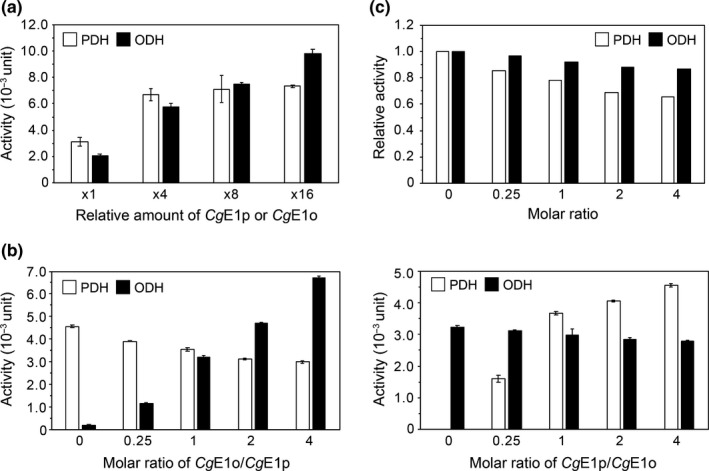
CgE1p‐dependent inhibition of ODH and CgE1o‐dependent inhibition of PDH. (a) Dose dependency of PDH and ODH activities. Various amounts (37, 150, 300, and 600 pmol) of CgE1p or CgE1o were mixed with a fixed amount of CgE2‐E3 (15 μg, 18 pmol) and subjected to enzyme assays. The CgE1p (3.8 μg, 37 pmol) and CgE1o (5.0 μg, 37 pmol) are defined as one amount. Data are shown as the mean and standard deviation of triplicate assays. (b) Competitive inhibition of PDH and ODH activities. PDH and ODH complexes were reconstituted by mixing CgE1p (15 μg, 150 pmol) and CgE1o (20 μg, 150 pmol) with CgE2‐E3 (15 μg, 18 pmol), separately; 37, 150, 300, and 600 pmol of CgE1o were added to the PDH complex (left), and 37, 150, 300, and 600 pmol of CgE1p were added to the ODH complex (right) and subjected to enzyme assays. The molar ratio of E1p and E1o is shown on the X‐axis. Data are shown as the mean and standard deviation of triplicate assays. A representative data set of two independent experiments is shown. (c) CgE1o‐dependent PDH inhibition (white columns) and CgE1p‐dependent ODH inhibition (black columns). The data were originated from Figure 5b and show the relative activities compared to the ones without the other E1 subunit
3.6. The effect of OdhI
OdhI inhibits ODH by binding to CgE1o (Krawczyk et al., 2010; Niebisch et al., 2006). However, it is unclear whether OdhI just associates with CgE1o in the ODH complex or elicits the dissociation of CgE1o from the full ODH complex to reduce the ODH activity. We examined the effect of OdhI on in vitro reconstituted ODH complex using gel filtration chromatography. It was observed that OdhI induced a peak shift in the gel filtration chromatogram and a substantial portion of OdhI coeluted with CgE1o as well as CgE2‐E3 (Figure 6). This result suggested that OdhI did not dissociate CgE1o from the ODH complex.
FIGURE 6.
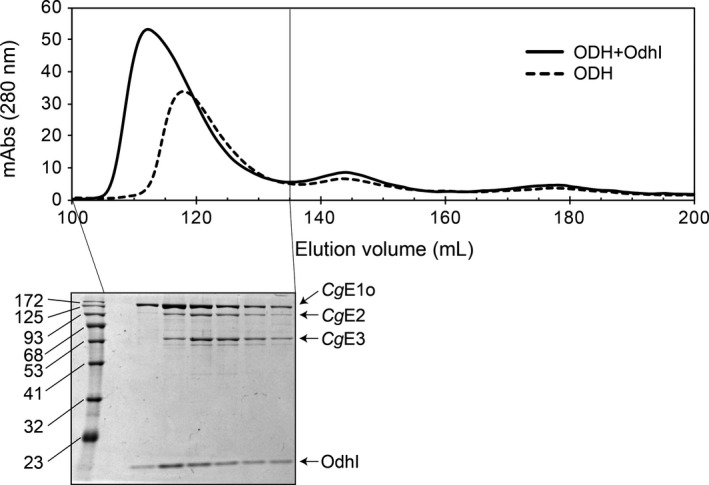
Effect of OdhI on the ODH complex. ODH complex was reconstituted by incubating CgE1o (4.0 mg) with CgE2‐E3 subcomplex (3.1 mg) on ice for 30 min. OdhI protein (4.6 mg) was added and further incubated for 30 min. The mixture was analyzed using gel filtration chromatography with a Superdex 200 column (upper). Aliquots (5 μl) of fractions (5 ml each) indicated were analyzed using 12% SDS‐PAGE, and the CBB staining gel image is also shown (lower)
4. DISCUSSION
In the present study, we performed a detailed molecular characterization of PDH and ODH from C. glutamicum in vivo and in vitro. Using ultracentrifuge analysis, we detected the PDH and ODH of C. glutamicum in the fractions much smaller than those of E. coli (Figure 1). It was shown that the catalytic domains of Azotobacter vinelandii and E. coli E2p, as well as E. coli E2o, are assembled in 24‐mer structures using gel filtration (Knapp et al., 1998; Schulze et al., 1991). On the other hand, CgE2 likely existed in a hexamer in the CgE2‐E3 subcomplex, which is one fourth of the octahedral PDH and ODH complexes, according to our gel filtration analysis (Figure 3). The in vivo evidence and in vitro evidence indicate that PDH and ODH of C. glutamicum are small and compact compared with those of other species.
A current schematic model of the CgPDH‐ODH hybrid complex is shown in Figure 7. The core‐forming CgE2‐E3 subcomplex (the predicted mass of 831 kDa) is hypothetically composed of two CgE2 trimers and four CgE3 dimers, in which six PSBD sites are available for E1 and E3 subunit binding. Four PSBD sites of CgE2 are occupied by the CgE3 dimer, and the remaining two sites are available for the CgE1 binding. Though we were not successful in detecting a PDH complex using gel filtration analysis, it seems reasonable that PDH activity was saturated with 150 pmol of CgE1p and 18 pmole of CgE2‐E3 (Figure 5a). Considering that 18 pmol of CgE2‐E3 contains 36 pmol of vacant PSBD sites, to which 72 pmol of CgE1p monomer (36 pmol of the dimer) can bind, our experimental data support our hypothesis on CgE2‐E3 stoichiometry. On the other hand, we successfully reconstituted the active ODH complex (Figure 2). Unlike EcE1p, EcE1o directly interacts with the E2 core, not the PSBD, via its N‐terminal extended region (Frank et al. 2007; Packman & Perham, 1986). The N‐terminal region of CgE1o has a structural similarity to the N‐terminal region of EcE1o, suggesting its key function in the association of CgE1o with the hybrid complex (Hoffelder et al., 2010; Niebisch et al., 2006). From the estimated molecular mass (940 kDa) of the ODH complex, it is likely speculated that one or two molecules of CgE1o (theoretical molecular mass of 135 kDa) associate with a single CgE2‐E3 subcomplex at most. CgE1o alone was present mainly in a hexamer in solution and equilibrium between multimer forms (Figure 3b). Mycobacterium smegmatis SucA (KGD), which has a high similarity to CgE1o, is reported to be a dimer in the crystal structure (Wagner et al., 2011, 2014). Taking the above observations into consideration, it can be speculated that CgE1o associates with the CgE2 core as a dimer to form the active ODH complex. The ODH activity was not saturated even when 600 pmol of CgE1o (300 pmol of CgE1o dimer) was added to 18 pmol of CgE2‐E3 (Figure 5a); an excess amount of CgE1o dimer were bound to the binding sites on the CgE2 core. We currently do not know the reason for the unsaturated ODH activity with the amount of CgE1o and need to study this further to understand the relationship between the molecular assembly and activity of CgODH.
FIGURE 7.
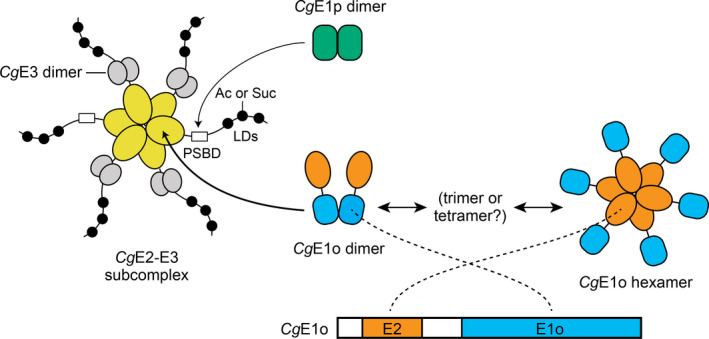
A current schematic model of the PDH‐ODH hybrid complex of Corynebacterium glutamicum. The core CgE2‐E3 subcomplex is hypothetically composed of two CgE2 (yellow) trimers and four CgE3 (gray) dimers. CgE1p (green) weakly associates with the PSBD of CgE2 to form the PDH complex. CgE1o stably exists in a hexamer and associates with the CgE2 core potentially as the dimer to form the ODH complex. The hexamer and dimer of CgE1o are putatively assembled via the E2 (orange) and E1o (blue) domains, respectively. CgE1p and CgE1o utilize lipoyl domains (LDs) on CgE2 for tethering the acetyl group (Ac) derived from pyruvate and the succinyl group (Suc) derived from 2‐oxoglutarate, respectively, which are then transferred to CoA
We observed CgE1o‐ and CgE1p‐dependent decrease in PDH and ODH activities, respectively (Figure 5b,c). This evidence is consistent with the formation of the PDH‐ODH hybrid complex, as suggested previously (Hoffelder et al., 2010; Niebisch et al., 2006), in which both CgE1p and CgE1o subunits associate with the single CgE2‐E3 subcomplex. As mentioned above, in E. coli, E1p and E1o utilize different sites (PSBD and the core, respectively) to bind to the cognate E2 (Packman & Perham, 1986; Frank et al., 2007; Arjunan et al., 2014), and CgE1p and CgE1o would not compete for binding to CgE2. Both CgE1p and CgE1o utilize lipoyl residues on CgE2 for tethering the acetyl and succinyl group derived from pyruvate and 2‐oxoglutarate, respectively. We thus speculated that CgE1p and CgE1o might compete for the availability of lipoyl residues on CgE2.
Since the active ODH complex was retained during gel filtration, but PDH was not (Figure 2, Figure A2), we assumed that the association of CgE1o with CgE2‐E3 was more stable than that of CgE1p with CgE2‐E3. Since the association of E1p with E2 is retained in gel filtration in other bacteria (Lessard et al., 1998; Schulze et al., 1993), CgPDH is likely more fragile compared to other PDHs. C. glutamicum PDH showed a K m value of 3.5 mM for pyruvate (Figure 4), which is comparable to the intracellular concentration (7.5 mM) of pyruvate reported in E. coli (Yang et al., 2001). Since the CgPDH complex seemed more fragile compared to CgODH, and PDH activity was more strongly affected by CgE1o than ODH activity was by CgE1p, the PDH activity might be altered in response to the pyruvate levels and the expression levels of CgE1o. Conversely, the K m value of CgODH for 2‐oxoglutarate (0.01 mM) was much lower than the intracellular concentration of 2‐oxoglutarate (0.44 mM) (Bennett et al., 2009) and the CgODH complex was stable. The ODH activity may be unperturbed by the 2‐oxoglutarate levels, and the availability of CgE1o and OdhI was perhaps critical to control the activity. The V max values of PDH and ODH were much lower than those previously reported using C. glutamicum lysates (148 and 134 nmol min−1 mg−1 for PDH and ODH, respectively) (Hoffelder et al., 2010). We assumed that it is because we used the NAD+ analog (APAD+) in enzyme assays, which, in some cases, give different specific activities from those with NAD+ (Kavanagh et al., 2007).
The regulation of PDH and ODH activities is a key event in glutamate overproduction in C. glutamicum. The OdhI‐dependent inhibition of ODH is well characterized as a critical determinant for glutamate production (Kim et al., 2010; Schultz et al., 2007), and the decrease in the PDH activity is observed in glutamate‐producing conditions (Hasegawa et al., 2008). Considering two enzyme activities on a single complex, the change in the subunit ratio of CgE1p and CgE1o would affect the balance of PDH and ODH activities. In this regard, it was intriguing that the shift of CgE2 toward higher molecular mass fractions was observed in glutamate‐producing conditions (Figure 1b). This may reflect the association of OdhI with the ODH (or hybrid) complex (Figure 6). Alternatively, the change in the molecular assembly and/or stoichiometry of the hybrid complex could occur, which would contribute to the control of PDH and ODH activities during glutamate overproduction.
We found that CgE1o alone formed multimers, mainly a hexamer, which were ready to form an active ODH complex (Figure 3). The in vitro multimerization of CgE1o agreed with the CgE1o detected in higher molecular mass fractions, independent of CgE2, in vivo (Figure 1b). The E1o component is usually a dimer (Frank et al., 2007; Wagner et al., 2011). However, considering that CgE1o has an extra E2‐catalytic domain capable of forming a trimer (Izard et al., 1999; Knapp et al., 1998; Mattevi et al., 1992, 1993; Wang et al., 2014), following possibilities for the detected hexamer were considered: three CgE1o dimers and two CgE1o trimers, and we prefer to consider the possibility of two CgE1o trimers assembled via the extra E2 domain (Figure 7). A structural study needs to be conducted to reveal the subunit assembly of the CgE1o hexamer. Meanwhile, CgE1p existed majorly as a dimer in vitro (Figure A2), and it also migrated to higher molecular mass fractions independent of CgE2 in vivo (Figure 1b). It might be speculated that CgE1p interacted with unknown proteins in vivo.
5. CONCLUSION
This study revealed the unique molecular architecture of C. glutamicum PDH and ODH. They can form compact‐sized PDH‐ODH hybrid complexes. At present, we are not sure whether all complexes were a hybrid form of PDH and ODH in vivo. Thus, we speculated that some might exist as independent PDH or ODH complexes. Our present study also revealed the fragility of the CgPDH complex and the unusual multimerization of CgE1o, which has not been reported. The compact size of the hybrid complex would provide an advantage to determine its whole structure and understand the molecular mechanism to balance the PDH and ODH activities, which should be critical to control the metabolic flux in C. glutamicum.
CONFLICT OF INTEREST
None declared.
AUTHOR CONTRIBUTIONS
Hirokazu Kinugawa: Formal analysis (equal); investigation (equal). Naoko Kondo: Formal analysis (equal); investigation (equal). Ayano Komine‐Abe: Formal analysis (supporting); investigation (supporting). Takeo Tomita: Methodology (equal). Makoto Nishiyama: Supervision (lead); writing – review & editing (equal). Saori Kosono: Conceptualization (lead); funding acquisition (lead); methodology (equal); project administration (lead); validation (lead); writing – original draft (lead); writing – review & editing (equal).
ETHICS STATEMENT
None required.
ACKNOWLEDGMENTS
We thank Takashi Hirasawa for providing the anti‐CgOdhA antibody. We thank Kan Tanaka for providing anti‐EcAceE, anti‐EcAceF, anti‐EcSucA, anti‐EcSucB, and anti‐EcLpd antibodies. We thank Fujio Kawamura for providing the anti‐BsRplC antibody. This work was partially supported by JSPS KAKENHI (grant no. 20K05803) and Noda Institute for Scientific Research.
Appendix A.
FIGURE A1.
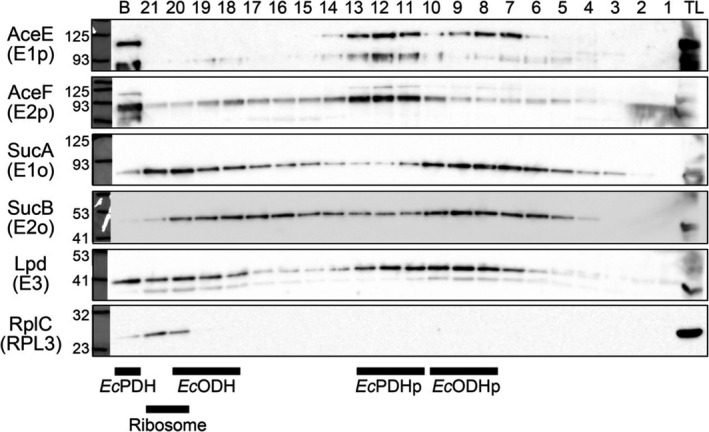
Ultracentrifuge analysis of Escherichia coli lysates using a 10%–30% sucrose density gradient. The total lysates (TL) of Escherichia coli were processed using the same ultracentrifuge protocol as Figure 1b. Aliquots (10 μl) of 21‐fractionated and the bottom (b) samples were separated using 8% or 10% SDS‐PAGE, followed by western blot analysis to detect each subunit. Black bars indicate the positions of EcPDH, EcODH and ribosome detected. EcPDHp and EcODHp represent partial complexes of EcPDH and EcODH, respectively. The positions of EcPDH, EcODH, EcPDHp, and EcODHp are indicated in Figure 1b
FIGURE A2.
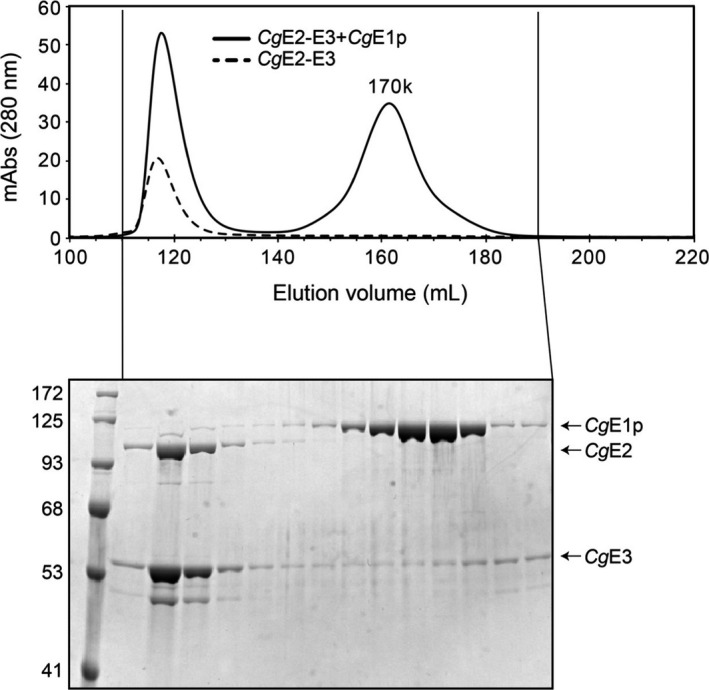
Gel filtration chromatography of a mixture sample of the CgE1p and the CgE2‐E3 subcomplex. Chromatograms of the mixture sample (solid line) and individual CgE2‐E3 (broken line) using a Superdex 200 column are shown (upper). Aliquots (5 μl) of fractions (5 ml each) were analyzed using 10% SDS‐PAGE and the CBB staining gel image is shown (lower). An estimated molecular mass of the peak fraction containing CgE1p was 170 kDa
FIGURE A3.
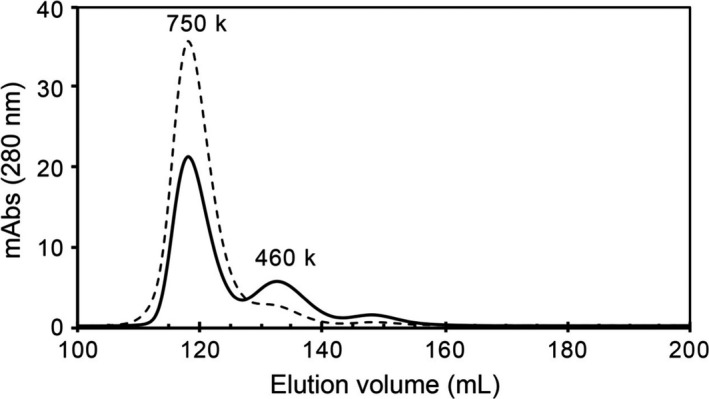
Gel filtration chromatography of the CgE1o460k fraction. The fraction containing CgE1o460k were collected and subjected to gel filtration chromatography, which provided two peaks of 750 kDa and 460 kDa again. The chromatography pattern of CgE1o in Figure 3a is shown with a broken line as a reference
FIGURE A4.
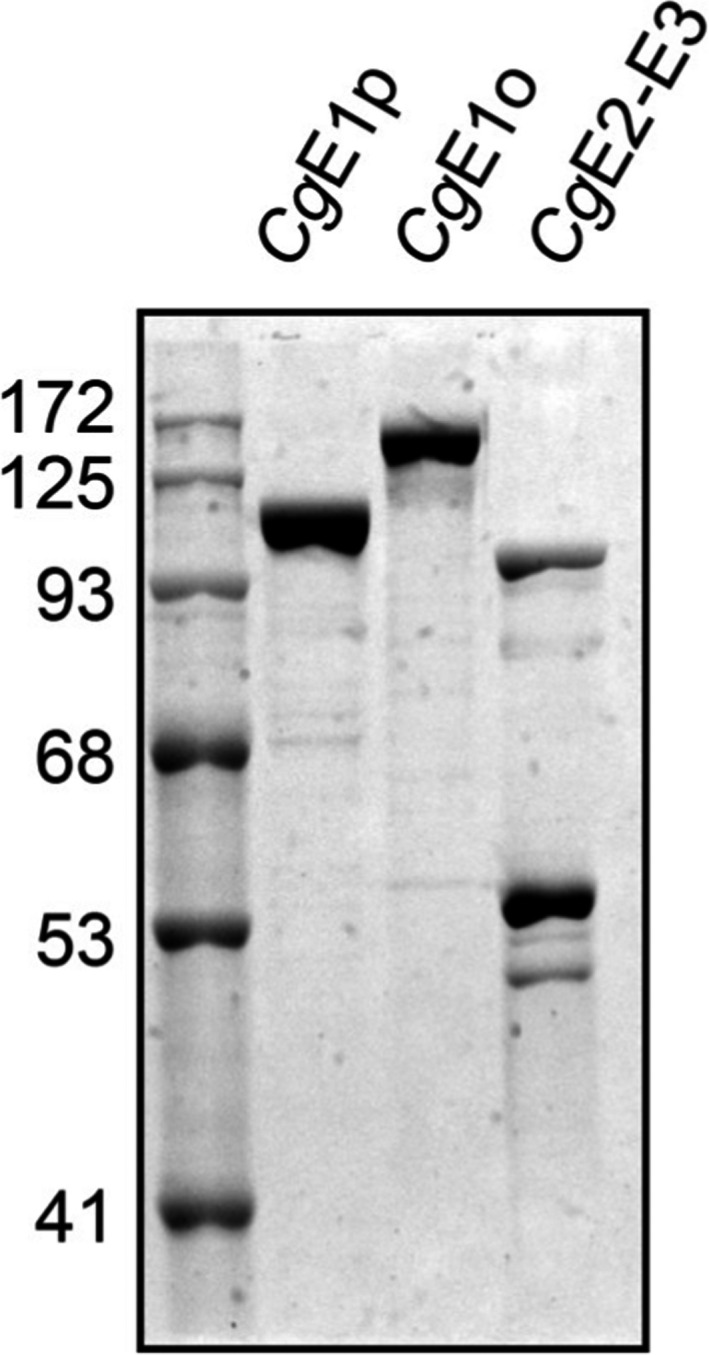
The purity of gel‐filtration purified His‐tagged CgE1p, CgE1o, and CgE2‐ E3 proteins. Purified samples (2 μg) were separated using 10% SDS‐PAGE and detected using CBB staining. These protein samples were used for kinetic analysis (Figure 4)
Kinugawa H, Kondo N, Komine‐Abe A, et al. In vitro reconstitution and characterization of pyruvate dehydrogenase and 2‐oxoglutarate dehydrogenase hybrid complex from Corynebacterium glutamicum . MicrobiologyOpen. 2020;9:e1113 10.1002/mbo3.1113
DATA AVAILABILITY STATEMENT
All data generated or analyzed during this study are included in this article.
References
- Arjunan, P. , Wang, J. , Nemeria, N. S. , Reynolds, S. , Brown, I. , Chandrasekhar, K. , … Furey, W. (2020). Novel binding motif and new flexibility revealed by structural analyses of a pyruvate dehydrogenase‐dihydrolipoyl acetyltransferase subcomplex from the Escherichia coli pyruvate dehydrogenase multienzyme complex. Journal of Biological Chemistry, 289, 30161–30176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barthe, P. , Roumestand, C. , Canova, M. J. , Kremer, L. , Hurard, C. , Molle, V. , & Cohen‐Gonsaud, M. (2009). Dynamic and structural characterization of a bacterial FHA protein reveals a new autoinhibition mechanism. Structure, 17, 568–578. [DOI] [PubMed] [Google Scholar]
- Bennett, B. D. , Kimball, E. H. , Gao, M. , Osterhout, R. , Van Dien, S. J. , & Rabinowitz, J. D. (2009). Absolute metabolite concentrations and implied enzyme active site occupancy in Escherichia coli . Nature Methods, 5, 593–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrasekhar, K. , Wang, J. , Arjunan, P. , Sax, M. , Park, Y.‐H. , Nemeria, N. S. , … Furey, W. (2013). Insight to the interaction of the dihydrolipoamide acetyltransferase (E2) core with the peripheral components in the Escherichia coli pyruvate dehydrogenase complex via multifaceted structural approaches. Journal of Biological Chemistry, 288, 15402–15417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danson, M. J. , Hale, G. , Johnson, P. , Perham, R. N. , Smith, J. , & Spragg, P. (1979). Molecular weight and symmetry of the pyruvate dehydrogenase multienzyme complex of Escherichia coli . Journal of Molecular Biology, 129, 603–617. [DOI] [PubMed] [Google Scholar]
- de Carvalho, L. P. S. , Zhao, H. , Dickinson, C. E. , Arango, N. M. , Lima, C. D. , Fischer, S. M. , … Rhee, K. Y. (2010). Activity‐based metabolomic profiling of enzymatic function: Identification of Rv1248c as a Mycobacterial 2‐hydroxy‐3-oxoadipate synthase. Chemistry & Biology, 17, 323–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank, R. A. W. , Price, A. J. , Northrop, F. D. , Perham, R. N. , & Luisi, B. F. (2007). Crystal structure of the E1 component of the Escherichia coli 2‐oxoglutarate dehydrogenase multienzyme complex. Journal of Molecular Biology, 368, 639–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa, T. , Hashimoto, K.‐I. , Kawasaki, H. , & Nakamatsu, T. (2008). Changes in enzyme activities at the pyruvate node in glutamate‐overproducing Corynebacterium glutamicum . Journal of Bioscience and Bioengineering, 105, 12–19. [DOI] [PubMed] [Google Scholar]
- Hoffelder, M. , Raasch, K. , van Ooyen, J. , & Eggeling, L. (2010). The E2 domain of OdhA of Corynebacterium glutamicum has succinyltransferase activity dependent on lipoyl residues of the acetyltransferase AceF. Journal of Bacteriology, 192, 5203–5211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izard, T. , Ævarsson, A. , Allen, M. D. , Westphal, A. H. , Perham, R. N. , Kok, A. D. , & Hol, W. G. J. (1999). Principles of quasi‐equivalence and Euclidean geometry govern the assembly of cubic and dodecahedral cores of pyruvate dehydrogenase complexes. Proceedings of the National Academy of Sciences of the United States of America, 96, 1240–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavanagh, K. L. , Elling, R. A. , & Wilson, D. K. (2007). Structure of Toxoplasma gondii LDH1: Active‐site differences from human lactate dehydrogenases and the structural basis for efficient APAD+ use. Biochemistry, 43, 879–889. [DOI] [PubMed] [Google Scholar]
- Kim, J. , Fukuda, H. , Hirasawa, T. , Nagahisa, K. , Nagai, K. , Wachi, M. , & Shimizu, H. (2010). Requirement of de novo synthesis of the OdhI protein in penicillin‐induced glutamate production by Corynebacterium glutamicum . Applied Microbiology and Biotechnology, 86, 911–920. [DOI] [PubMed] [Google Scholar]
- Kim, J. , Hirasawa, T. , Saito, M. , Furusawa, C. , & Shimizu, H. (2011). Investigation of phosphorylation status of OdhI protein during penicillin‐ and Tween 40‐triggered glutamate overproduction by Corynebacterium glutamicum . Applied Microbiology and Biotechnology, 91, 143–151. [DOI] [PubMed] [Google Scholar]
- Knapp, J. E. , Mitchell, D. T. , Yazdi, M. A. , Ernst, S. R. , Reed, L. J. , & Hackert, M. L. (1998). Crystal structure of the truncated cubic core component of the Escherichia coli 2‐oxoglutarate dehydrogenase multienzyme complex. Journal of Molecular Biology, 280, 655–668. [DOI] [PubMed] [Google Scholar]
- Koike, M. , Reed, L. J. , & Carroll, W. R. (1960). α‐Keto acid dehydrogenation complexes: I. Purification and properties of pyruvate and α‐ketoglutarate dehydrogenation complexes of Escherichia coli . Journal of Biological Chemistry, 235, 1924–1930. [PubMed] [Google Scholar]
- Komine‐Abe, A. , Nagano‐Shoji, M. , Kubo, S. , Kawasaki, H. , Yoshida, M. , Nishiyama, M. , & Kosono, S. (2017). Effect of lysine succinylation on the regulation of 2‐oxoglutarate dehydrogenase inhibitor, OdhI, involved in glutamate production in Corynebacterium glutamicum . Bioscience, Biotechnology, and Biochemistry, 81, 2130–2138. [DOI] [PubMed] [Google Scholar]
- Krawczyk, S. , Raasch, K. , Schultz, C. , Hoffelder, M. , Eggeling, L. , & Bott, M. (2010). The FHA domain of OdhI interacts with the carboxyterminal 2‐oxoglutarate dehydrogenase domain of OdhA in Corynebacterium glutamicum . FEBS Letters, 584, 1463–1468. [DOI] [PubMed] [Google Scholar]
- Lessard, I. A. D. , Domingo, G. J. , Borges, A. , & Perham, R. N. (1998). Expression of genes encoding the E2 and E3 components of the Bacillus stearothermophilus pyruvate dehydrogenase complex and the stoichiometry of subunit interaction in assembly in vitro . European Journal of Biochemistry, 258, 491–501. [DOI] [PubMed] [Google Scholar]
- Mattevi, A. , Obmolova, G. , Kalk, K. H. , Westphal, A. H. , de Kok, A. , & Hol, W. G. J. (1993). Refined crystal structure of the catalytic domain of dihydrolipoyl transacetylase (E2p) from Azotobacter vinelandii at 2.6 Å resolution. Journal of Molecular Biology, 230, 1183–1199. [DOI] [PubMed] [Google Scholar]
- Mattevi, A. , Obmolova, G. , Schulze, E. , Kalk, K. H. , Westphal, A. H. , Kok, A. D. , & Hol, W. G. J. (1992). Atomic structure of the cubic core of the pyruvate dehydrogenase multienzyme complex. Science, 255, 1544–1550. [DOI] [PubMed] [Google Scholar]
- Mizuno, Y. , Nagano‐Shoji, M. , Kubo, S. , Kawamura, Y. , Yoshida, A. , Kawasaki, H. , … Kosono, S. (2016). Altered acetylation and succinylation profiles in Corynebacterium glutamicumin response to conditions inducing glutamate overproduction. MicrobiologyOpen, 5, 152–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy, G. E. , & Jensen, G. J. (2005). Electron cryotomography of the E. coli pyruvate and 2‐oxoglutarate dehydrogenase complexes. Structure, 13, 1765–1773. [DOI] [PubMed] [Google Scholar]
- Nagano‐Shoji, M. , Hamamoto, Y. , Mizuno, Y. , Yamada, A. , Kikuchi, M. , Shirouzu, M. , … Kosono, S. (2017). Characterization of lysine acetylation of a phosphoenolpyruvate carboxylase involved in glutamate overproduction in Corynebacterium glutamicum . Molecular Microbiology, 104, 677–689. [DOI] [PubMed] [Google Scholar]
- Niebisch, A. , Kabus, A. , Schultz, C. , Weil, B. , & Bott, M. (2006). Corynebacterial protein kinase G controls 2‐oxoglutarate dehydrogenase activity via the phosphorylation status of the OdhI protein. Journal of Biological Chemistry, 281, 12300–12307. [DOI] [PubMed] [Google Scholar]
- Nott, T. J. , Kelly, G. , Stach, L. , Li, J. , Westcott, S. , Patel, D. , … Smerdon, S. J. (2009). An intramolecular switch regulates phosphoindependent FHA domain interactions in Mycobacterium tuberculosis . Science Signalling, 2, ra12. [DOI] [PubMed] [Google Scholar]
- Packman, L. C. , & Perham, R. N. (1986). Chain folding in the dihydrolipoyl acyltransferase components of the 2‐oxo-acid dehydrogenase complexes from Escherichia coli . FEBS Letters, 206, 193–198. [DOI] [PubMed] [Google Scholar]
- Patel, M. S. , Nemeria, N. S. , Furey, W. , & Jordan, F. (2014). The pyruvate dehydrogenase complexes: Structure‐based function and regulation. Journal of Biological Chemistry, 289, 16615–16623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perham, R. N. (2000). Swinging arms and swinging domains in multifunctional enzymes: Catalytic machines for multistep reactions. Annual Review of Biochemistry, 69, 961–1004. [DOI] [PubMed] [Google Scholar]
- Reed, L. J. , Pettit, F. H. , Eley, M. H. , Hamilton, L. , Collins, J. H. , & Oliver, R. M. (1975). Reconstitution of the Escherichia coli pyruvate dehydrogenase complex. Proceedings of the National Academy of Sciences of the United States of America, 72, 3068–3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz, C. , Niebisch, A. , Gebel, L. , & Bott, M. (2007). Glutamate production by Corynebacterium glutamicum: Dependence on the oxoglutarate dehydrogenase inhibitor protein OdhI and protein kinase PknG. Applied Microbiology and Biotechnology, 76, 691–700. [DOI] [PubMed] [Google Scholar]
- Schulze, E. , Westphal, A. H. , Hanemaaijer, R. , & de Kok, A. (1993). Structure/function relationships in the pyruvate dehydrogenase complex form Azotobacter vinelandii . European Journal of Biochemistry, 211, 591–599. [DOI] [PubMed] [Google Scholar]
- Schulze, E. , Westphal, A. H. , Obmolova, G. , Mattevi, A. , Hol, W. G. J. , & de Kok, A. (1991). The catalytic domain of the dihydrolipoyl transacetylase component of the pyruvate dehydrogenase complex from Azotobacter vinelandii and Escherichia coli . European Journal of Biochemistry, 201, 561–568. [DOI] [PubMed] [Google Scholar]
- Tian, J. , Bryk, R. , Itoh, M. , Suematsu, M. , & Nathan, C. (2005). Variant tricarboxylic acid cycle in Mycobacterium tuberculosis: Identification of α‐ketoglutarate decarboxylase. Proceedings of the National Academy of Sciences of the United States of America, 102, 10670–10675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usuda, Y. , Tujimoto, N. , Abe, C. , Asakura, Y. , Kimura, E. , Kawahara, Y. , … Matsui, H. (1996). Molecular cloning of the Corynebacterium glutamicum (“Brevibacterium lactofermenturn” AJ12036) odhA gene encoding a novel type of 2‐oxoglutarate dehydrogenase. Microbiology, 142, 3347–3354. [DOI] [PubMed] [Google Scholar]
- Wagner, T. , Barilone, N. , Alzari, P. M. , & Bellinzoni, M. (2014). A dual conformation of the post‐decarboxylation intermediate is associated with distinct enzyme states in mycobacterial KGD (α‐ketoglutarate decarboxylase). The Biochemical Journal, 457, 425–434. [DOI] [PubMed] [Google Scholar]
- Wagner, T. , Bellinzoni, M. , Wehenkel, A. , O'Hare, H. M. , & Alzari, P. M. (2011). Functional plasticity and allosteric regulation of α‐ketoglutarate decarboxylase in central Mycobacterial metabolism. Chemistry & Biology, 18, 1011–1020. [DOI] [PubMed] [Google Scholar]
- Wagner, T. , Boyko, A. , Alzari, P. M. , Bunik, V. I. , & Bellinzoni, M. (2019). Conformational transitions in the active site of mycobacterial 2‐oxoglutarate dehydrogenase upon binding phosphonate analogues of 2‐oxoglutarate: From a Michaelis‐like complex to ThDP adducts. Journal of Structural Biology, 208, 182–190. [DOI] [PubMed] [Google Scholar]
- Wang, J. , Nemeria, N. S. , Chandrasekhar, K. , Kumaran, S. , Arjunan, P. , Reynolds, S. , … Jordan, F. (2014). Structure and function of the catalytic domain of the dihydrolipoyl acetyltransferase component in Escherichia coli pyruvate dehydrogenase complex. Journal of Biological Chemistry, 289, 15215–15230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Y.‐T. , Bennett, G. N. , & San, K.‐Y. (2001). The effects of feed and intraclellular pyruvate levels on the redistribution of metabolic fluxes in Escherichia coli . Metabolic Engineering, 3, 115–123. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this article.


