Significance
Cockayne syndrome group B (CSB) belongs to the SWI2/SNF2 family chromatin remodelers and plays critical roles in DNA damage response, repair, and gene expression. Despite its broad roles in gene expression, it has remained elusive whether and how CSB functions as a chromatin remodeler, a transcription elongation factor, or both, in regulating RNA polymerase II (Pol II) transcription on chromatin. Here, we reveal that Rad26, the yeast CSB ortholog, is recruited to the upstream of Pol II as an ATP-dependent processivity factor that assists Pol II in overcoming downstream nucleosome barriers during transcription. This paper provides mechanistic insights into the roles of CSB in different aspects of transcription and DNA repair on chromatin, including release of transcription pausing and elongation.
Keywords: Cockayne syndrome, nucleosome bypass, chromatin remodeling, RNA polymerase II, transcription elongation
Abstract
While loss-of-function mutations in Cockayne syndrome group B protein (CSB) cause neurological diseases, this unique member of the SWI2/SNF2 family of chromatin remodelers has been broadly implicated in transcription elongation and transcription-coupled DNA damage repair, yet its mechanism remains largely elusive. Here, we use a reconstituted in vitro transcription system with purified polymerase II (Pol II) and Rad26, a yeast ortholog of CSB, to study the role of CSB in transcription elongation through nucleosome barriers. We show that CSB forms a stable complex with Pol II and acts as an ATP-dependent processivity factor that helps Pol II across a nucleosome barrier. This noncanonical mechanism is distinct from the canonical modes of chromatin remodelers that directly engage and remodel nucleosomes or transcription elongation factors that facilitate Pol II nucleosome bypass without hydrolyzing ATP. We propose a model where CSB facilitates gene expression by helping Pol II bypass chromatin obstacles while maintaining their structures.
During transcription, many cellular factors are required to facilitate transcriptional bypass of nucleosomes, inherent barriers to transcription initiation, and elongation (1, 2). These factors include ATP-dependent chromatin remodelers (3–8), histone-modifying enzymes (9, 10), histone chaperones (11–15), nucleosome-destabilizing factors (16), and transcription elongation factors (9, 17–20). Mechanistically, these factors can be divided into two classes: One functioning to reconfigure the chromatin template to reduce barriers to transcription, and another directly acting as part of the RNA Pol II complex to facilitate transcription elongation.
Chromatin remodelers, such as RSC (6, 12), Fun30 (8), and ISW2 (19), belong to the first class factors. They are able to couple ATP hydrolysis to the direct displacement or repositioning of nucleosomes to facilitate transcription elongation. Histone chaperones, including FACT (14, 15, 21) and NAP1 (12), bind histones to destabilize histone-DNA interactions thereby promoting transcription bypass of nucleosomes (21). Multiple histone-modifying enzymes weaken nucleosomal histone-DNA interactions by catalyzing specific histone modification events, such as H3K56 acetylation (9, 10). A common theme for these factors is that they enhance transcription elongation by acting directly on a chromatin template to remove or reduce the constraints (nucleosome barriers) imposed on an elongating Pol II.
In contrast to the protein factors that act directly on the nucleosome discussed above, most transcription elongation factors, including TFIIS, TFIIF, Spt4/5 (DSIF), and Elf1, stimulate transcription elongation by acting directly on Pol II (22). For example, TFIIS reactivates backtracked Pol II by cleaving RNA to facilitate the overall transcription bypass of the nucleosome (23, 24). TFIIF and Spt4/5 increase transcription passage of the nucleosome by decreasing the probability of pausing and shortening pause duration (2, 18, 20, 24, 25). A common feature of this class of factors is that their ability to enhance elongation involves a direct interaction with Pol II and does not require ATP hydrolysis.
The CSB is the product of a key disease gene as autosomal mutations in this gene have been linked to multisystem neurological disorders (26). CSB, a member of the SWI2/SNF2 ATPase family (27, 28), is conserved from yeast to humans, underscoring its functional importance. Indeed, CSB has been implicated in diverse cellular processes, such as DNA damage response and DNA repair (29–34), transcription (35–37), and chromatin maintenance (38). Although CSB is best known for its role in transcription-coupled nucleotide excision repair (TC-NER) (39), accumulating evidence suggests that CSB may act as a general Pol II elongation factor even in the absence of DNA damage (27, 36, 37, 40, 41), likely through its role in chromatin remodeling (35, 38, 41–43). Given its broad roles in transcriptional control, it has remained elusive whether and how CSB functions as a chromatin remodeler, a transcription elongation factor, or both. Resolving this will be key to our understanding of how a malfunctioning CSB leads to disease.
Most studies to date have focused on the functional consequences of CSB rather than its precise mechanism(s). Therefore, we sought to directly dissect how CSB works on chromatinized DNA by taking a reductionist approach and reconstituting transcription in vitro with biochemically defined components (44). Here, we present evidence that supports the role of CSB as an ATP-dependent Pol II-associated processivity factor that assists Pol II in overcoming nucleosome barriers during transcription elongation, suggesting that CSB acts as a general patroller of the genome to help stalled Pol II. This noncanonical mode of CSB function appears distinct from either canonical chromatin remodelers or transcription elongation factors (22, 45).
Results
CSB Facilitates Transcription Elongation on a Nucleosomal Template.
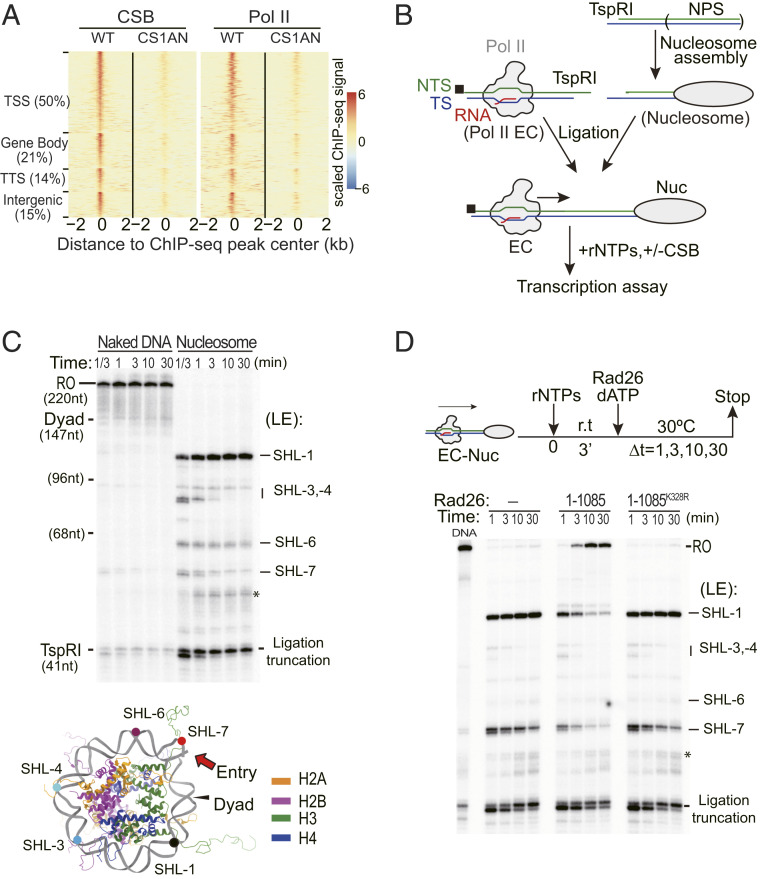
Previous results suggested a potential direct role for CSB in regulating Pol II transcription as an elongation factor in the absence of DNA damage (27, 36, 37, 40, 46). These functional data suggest that CSB may be recruited to specific genes or Pol II pausing sites during transcription. We, thus, reanalyzed an existing CSB chromatin immunoprecipitation-sequencing (ChIP-seq) data (40) to ask whether CSB shows specific colocalization with Pol II in the human genome. Indeed, we found that, although CSB is not present in all detectable Pol II pausing sites, it tightly colocalizes with Pol II in 1,154 established CSB binding sites (Fig. 1A and SI Appendix, Fig. S1 A and B), which is in good agreement with the original observation (40). These colocalization events occurred not only at TSSs (50%), but also in gene bodies ([GBs], 21%), near TTSs (14%) and within intergenic regions (15%), the latter of which likely correspond to active enhancers or some unannotated genes. Another related study also reported modest enrichment of human CSB peaks at TSS regions and enhancers (47). Interestingly, the authors found that the cells expressing a remodeling-defective CSB derivative have more significantly enriched CSB peaks at TSS regions (promoters and 5′ untranslated regions) than the cells expressing functional CSB proteins (47). One potential explanation for this enrichment is that recruitment of functional CSB to Pol II at TSS regions may utilize its remodeling activities to directly modulate Pol II transcription. Indeed, these previous studies further revealed that functional CSB (with chromatin remodeling activity) is important for directly regulating the expression of thousands of genes (such as NRG2, SYT9, and ZNFX-NC1) (40, 47), many of which are neuronal genes (40). Taken together, these observations suggest a direct role of CSB proteins in facilitating Pol II transcription.
Fig. 1.
An in vitro system to study the function of Rad26 in Pol II transcription elongation on a nucleosomal template. (A) CSB colocalizes with Pol II. Distribution of CSB and Pol II ChIP-seq signals within ±2-kb windows around CSB binding sites (n = 1,154) in wild type (WT) and CSB-deficient (CS1AN) cells. In each category, CSB binding sites are shown in descending order based on average CSB ChIP-seq signal intensity. Transcription start sites (TSSs) and transcription termination sites (TTSs). (B) Experimental setup to study the function of CSB in Pol II transcription on the nucleosome template. A Pol II EC segment and a nucleosome-containing DNA fragment were reconstituted individually and ligated by T4 ligase. A biotin label is shown as a black square. (C) The nucleosome is a strong barrier for Pol II transcription elongation. (Top) Mapping of Pol II pausing sites in the absence (Left) or presence (Right) of a nucleosome. A full gel mapping the pausing sites is shown in SI Appendix, Fig. S1. The nucleosomal arrest sites are indicated on the right. The asterisk indicates a transcript generated from unligated upstream template (see B). (Bottom) The locations of the pausing sites are shown in a structure of the nucleosome (PDB ID: 1KX5). For ease of viewing, most of the DNA beyond the dyad (i.e., away from the viewer) was removed from this representation. (D) Rad26 is able to promote Pol II bypass nucleosome barriers. (Top) Schematic of the transcription reaction. (Bottom) Mapping of pausing sites during Pol II transcription of a nucleosomal template in the absence of Rad26 (Left) or in the presence of either WT (Middle) or an ATPase-deficient mutant (Right) of Rad26. Arrest sites are indicated as in C. The ligation truncation corresponds to the transcripts from reconstituted ECs that did not ligate to the downstream nucleosome or naked DNA.
To investigate the function of CSB during Pol II transcription on chromatin and further explore the underlying mechanism, we reconstituted an in vitro transcription system on a nucleosomal template using purified components (17, 44, 48–50) and tested the effect of the yeast CSB ortholog, Rad26, on Pol II’s ability to bypass a nucleosome barrier during transcription elongation. In this system, we ligated a Pol II elongation complex (EC) to either a nucleosome-free or nucleosomal template and performed a transcription elongation assay in the presence or absence of Rad26 (Fig. 1B). In the absence of Rad26, we found that Pol II efficiently transcribed the nucleosome-free template and generated run-off (RO) full-length transcripts but paused at a series of superhelical locations (SHLs) within the nucleosome barrier leading up to SHL-1, immediately before the dyad (center of the nucleosomal DNA) where the strongest pausing took place and beyond which no transcription was observed (Fig. 1C and SI Appendix, Fig. S1C). This nucleosome-induced transcription pattern is fully consistent with previous biochemical studies (17, 44, 48–50) and corresponds well with in vivo Pol II pausing sites within GB nucleosomes (51).
We next asked how Rad26 might influence Pol II elongation on the nucleosome template by adding Rad26 to the reconstituted transcription system (see scheme on top of Fig. 1D). We found that Pol II efficiently bypassed the nucleosome barrier in the presence of Rad26, leading to a substantial increase in the RO product over the reaction period (Fig. 1D). Importantly, this Rad26-dependent Pol II bypass required ATP hydrolysis as addition of the ATPase inactive form (K328R) of Rad26 failed to stimulate nucleosome bypass (Fig. 1D). These data show that Rad26 helps elongating Pol II overcome a nucleosome barrier in an ATP-dependent manner.
Pol II-Dependent and Pol II-Independent Chromatin Remodeling Activities of Rad26.
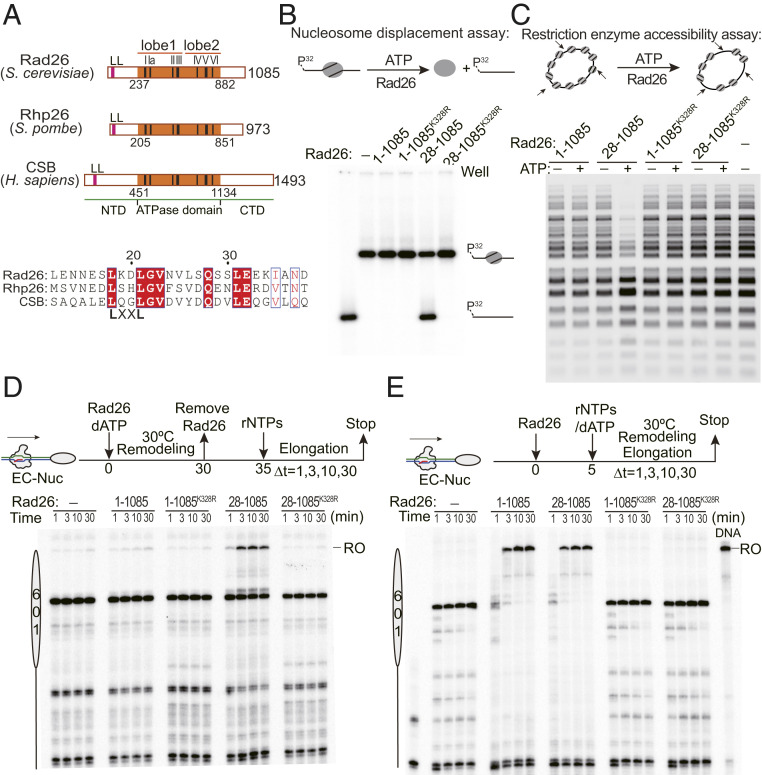
Canonical chromatin remodeling factors promote transcription elongation by directly evicting (6) or sliding nucleosome barriers away in an ATP-dependent manner (19), which is largely independent of Pol II action. These chromatin remodeling activities are often subjected to autoregulation from regions flanking the core ATPase domain or from other regulatory subunits (28). Our previous studies showed that the Schizosaccharomyces pombe CSB ortholog Rhp26 was able to robustly remodel nucleosomes in an ATP-dependent manner once the N-terminal leucine latch (LL) motif was removed (35, 42). This domain is conserved in the Saccharomyces cerevisiae CSB ortholog Rad26 (Fig. 2A). Using two different chromatin remodeling assays either with mononucleosomes (Fig. 2B) or with nucleosome arrays (Fig. 2C) as substrates, we found that full-length Rad26 was locked in an autorepressed state and was unable to remodel the substrates as was the case with Rhp26 (SI Appendix, Fig. S2). In contrast, deletion of the LL rendered Rad26 fully active in ATP-dependent chromatin remodeling (Fig. 2 B and C and SI Appendix, Fig. S2).
Fig. 2.
Rad26 promotes nucleosome bypass by Pol II in a Pol II-dependent manner. (A) Domain organization of CSB homologs. CSB contains a highly conserved ATPase core domain (orange) and a conserved inhibitory N-terminal LL motif (red). Seven hallmark helicase motifs are highlighted in black. Sequence alignment of the LL region is shown at the bottom. (B) Mononucleosome remodeling by full-length Rad26 and constitutively active Rad26 variant Rad26:28–1,085. (C) Remodeling by Rad26 on a nucleosome array. Deletion of the LL motif (Rad26:28–1,085) activates Rad26 in the two different assays. (D) Pol II nucleosome bypass assay where chromatin remodeling and transcription are allowed to take place sequentially. The workflow for experimental design is shown on top. Chromatin remodeling was performed in the absence of ribonucleoside 5′-triphosphates (rNTPs), thus, preventing transcription elongation, and, then, Rad26 was washed away, followed by the addition of rNTPs to allow transcription elongation. (E) Pol II nucleosome bypass assay where chromatin remodeling and transcription are allowed to take place at the same time. In this experimental setting (top). Position of the RO transcript is shown.
Since Rad26 can remodel chromatin in the absence of transcription, its ability to help Pol II overcome a nucleosome barrier may rely on one of two (nonmutually exclusive) mechanisms: 1) Rad26 could remodel the nucleosomal template as a canonical chromatin remodeler, independently of Pol II; or 2) Rad26 may function as a Pol II processivity factor that helps Pol II overcome the nucleosome barrier during transcription elongation. To differentiate between these two possibilities, we compared the activity of full-length Rad26 (Rad261−1,085, the autorepressed form) with that of the constitutively active Rad26 variant (Rad2628−1,085) using two different experimental schemes. In the first, we separated nucleosome remodeling and transcription by allowing them to happen sequentially (Fig. 2D). Rad2628−1,085 or Rad261−1,085 was added to the reactions under conditions that allow chromatin remodeling but not transcription to happen, and, then, Rad26 was removed before activation of Pol II transcription by the addition of rNTPs (Fig. 2D). This experimental setup allowed us to hold RNA Pol II in its original upstream location and test whether Rad26 could act as a canonical chromatin remodeler to remove the downstream nucleosome, independently of Pol II. Under these conditions, we only observed a full-length (i.e., RO) transcript in those reactions where the constitutively active form of Rad26 (Rad2628−1,085) was used; in the presence of full-length Rad26 (which is autoinhibited), Pol II was arrested within the nucleosome (Fig. 2D) as we had observed in transcription reactions performed in the absence of Rad26 (Figs. 1C and 2D). These results are consistent with Rad26’s ability to remove nucleosomes in a standard chromatin remodeling assay (Fig. 2 B and C), and with its acting, once activated, as a canonical remodeler that facilitates transcription by removing nucleosomal obstacles.
In the second experimental setup, we allowed chromatin remodeling and transcription elongation to happen at the same time, mimicking transcription on a physiological chromatinized template. In this scenario, Pol II is incubated with either Rad 26 or Rad2628−1,085 in the presence of rNTPs. In contrast to the previous experiment where remodeling would have taken place prior to transcription, we found that both the autorepressed full-length Rad26 and the activated variant Rad2628−1,085 efficiently facilitated the bypassing of a nucleosome barrier by Pol II (Fig. 2E). As before, this effect is dependent on Rad26’s ability to hydrolyze ATP as the ATPase mutant versions of both Rad26 constructs were inactive.
The main difference between the two assays is the fact that full-length Rad26 only promoted nucleosome bypass when transcription was allowed to take place at the same time as remodeling. A possible explanation for this difference is that full-length autoinhibited Rad26 must interact physically with a transcribing Pol II EC to become activated and remodel the nucleosome as a canonical chromatin remodeler. To test this, we asked whether Rad26 could remodel nucleosomes when an additional transcribing Pol II EC was added in trans. Rad26 was unable to remodel nucleosomes in this assay (SI Appendix, Fig. S3), suggesting that the presence of a Pol II EC is not sufficient to promote remodeling. We, thus, considered an alternative possibility: That Rad26 facilitates Pol II’s bypassing of a nucleosomal barrier by acting directly on Pol II as a processivity factor rather than as a canonical chromatin remodeler.
Rad26 Forms a Stable Complex with the Pol II Complex on Chromatinized DNA.
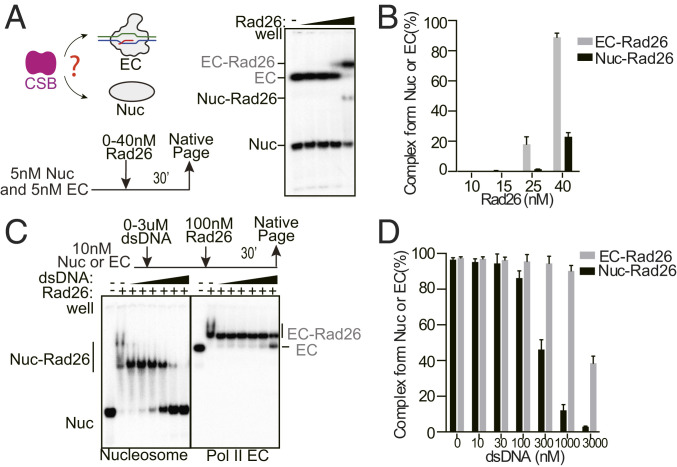
Previous studies have shown binding of Rad26 to both Pol II EC and nucleosomes (41, 42). Given those observations and the data presented above, we wondered whether full-length Rad26 formed a stable complex with Pol II EC in the presence of nucleosomes (Fig. 3A). To test this, we first set up a competition binding assay where we incubated full-length Rad26 with an equimolar mixture of Pol II EC and nucleosomes; Rad26 preferentially bound to the Pol II EC at a lower concentration (25 nM) and only started to bind to the nucleosome once the bulk of Pol II EC was in complex with Rad26 (89% for Pol II-Rad26 complex versus 23% for nucleosome-Rad26 complex at 40-nM Rad26) (Fig. 3 A and B). These results suggest that Rad26 preferentially binds Pol II EC over the nucleosome.
Fig. 3.
Rad26 preferentially binds to Pol II EC over nucleosomes. (A) Binding of Rad26 to an equimolar mixture of Pol II EC and the nucleosome. (B) Quantitation of the data in A. (C) Competition assay looking at the effect of adding dsDNA to the Rad26 nucleosome and Rad26-Pol II-EC complexes. The competitor DNA is a 25-bp dsDNA. The final concentration of the competitor DNA was 0, 10, 30, 100, 300, 1,000, and 3,000 nM. (D) Quantitation of the dsDNA competition assay.
We next compared the stabilities of the Rad26-Pol II EC and the Rad26-nucleosome complexes in a binding competition assay using a naked double-stranded DNA (dsDNA) as a competitor (Fig. 3C). We found that over 50% of the Rad26-nucleosome complex could be competed off in the presence of 300-nM dsDNA (∼30-fold excess over nucleosomal dsDNA), whereas a little Rad26-Pol II-EC complex could be displaced by dsDNA in the same concentration range (Fig. 3 C and D). Even at the highest concentration tested (∼300-fold excess), ∼50% of the Rad26-Pol II-DNA ternary complex remained (Fig. 3D). These data support the idea that full-length Rad26 is able to bind Pol II EC to form a stable complex, whereas its interaction with a nucleosome is weaker.
Upstream Duplex DNA Is Required for the Rad26-Mediated Transcription Bypass of a Nucleosome.
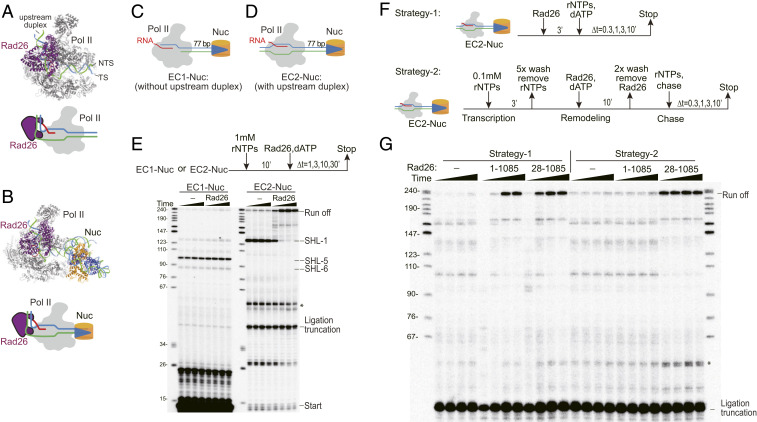
To further test the hypothesis that Rad26 facilitates Pol II’s bypassing of a nucleosomal barrier by acting directly on Pol II as a processivity factor rather than as a canonical chromatin remodeler, we sought to remove a feature that was required for the former activity but not for the latter. Current biochemical and structural studies suggest that Rad26 binds the upstream duplex DNA to promote Pol II forward translocation in an ATP-dependent manner (Fig. 4A) (41). We first modeled the Rad26-Pol II-nucleosome structure by combining the Rad26-Pol II-EC complex structure (41) with the Pol II-nucleosome structure (49) (Fig. 4B). There is no steric clash among Rad26, Pol II, and the nucleosome in our model with the nucleosome and Rad26, respectively, localizing at the downstream and upstream of the Pol II EC. This model then suggests that, upon ATP hydrolysis, Rad26 may move along the upstream template DNA to promote forward translocation of Pol II to overcome the nucleosome barrier in a fashion, similar to Rad26-mediated transcriptional bypass of repetitive DNA sequences or small lesions (41). By comparison, we also modeled Rad26 at the canonical binding site (SHL-2 position of the nucleosome) that is shared by most canonical chromatin remodelers (Position 2, SI Appendix, Fig. S4). If full-length Rad26 acts directly on Pol II as a processivity factor to promote transcriptional bypass of a nucleosome barrier, the upstream binding site (Position 1, SI Appendix, Fig. S4), which is 10–15 bp of dsDNA (41), would be required for its activity. On the other hand, removal of DNA upstream of Pol II should have no effect on remodeling of a downstream nucleosome if Rad26 were acting as a canonical remodeler.
Fig. 4.
Upstream duplex DNA of Pol II EC is required for Rad26-mediated transcription bypass of a nucleosome. (A) Structure of the Rad26-Pol II-EC complex (PDB ID:5VVR). Rad26, Pol II, template DNA, nontemplate DNA, and RNA are shown in purple, gray, cyan, green, and red, respectively. (B) A model of the Rad26-Pol II-nucleosome complex. H3 and H4 are shown in blue; H2A and H2B are shown in orange. (C and D) Schematics of two different Pol II EC-nucleosome complexes (EC1-Nuc and EC2-Nuc) used to study the functional importance of the upstream duplex DNA. EC1-Nuc contains no upstream duplex, whereas EC2-Nuc contains an intact upstream duplex. The length of duplex DNA between the Pol II active site and the nucleosome is 77 bp. (E) Rad26 was not able to promote Pol II elongation complex (EC1) bypass of a nucleosome barrier in the absence of an upstream DNA duplex but efficiently promotes Pol II EC (EC2) bypass of the nucleosome barrier in the presence of intact upstream DNA duplex. (F) Workflow for Pol II nucleosome bypass assay strategies to test whether Pol II EC can allosterically activate Rad26 remodeling activity in cis. In Strategy-1, Rad26 is loaded first with EC2-Nuc and is followed by incubation with rNTPs and 2′-deoxyadenosine 5′-triphosphate (dATP). In Strategy-2, sequential transcription-remodeling-chase experiments are performed. rNTPs are first added to allow Pol II to run into the nucleosome. rNTPs are then washed away, followed by the addition of Rad26 and dATP to allow chromatin remodeling in the absence of rNTPs, thus, preventing transcription elongation. Finally, Rad26 is washed away, followed by the addition of rNTPs to chase transcription. (G) Pol II EC cannot allosterically activate Rad26 to remodel downstream nucleosome in cis. Full-length Rad26 (1–1,085) and the constitutively active Rad26 variant Rad26 (28–1,085) were labeled at the top of the gel. Time points were 0.3, 1, 3, and 10 min, respectively.
We determined the importance of the upstream duplex DNA by comparing two different systems containing a Pol II EC with a downstream nucleosome barrier (EC-Nuc). In the first, the Pol II EC was generated by extension from a tailed scaffold (termed EC1-Nuc), whereas, in the second, the Pol II EC was generated by a full-bubble scaffold (termed EC2-Nuc) (schemes of Fig. 4 C and D). By design, EC2-Nuc but not EC1-Nuc would have an intact upstream duplex. As established earlier (50), the tailed template-generated Pol II EC mainly paused at SHL-6 and SHL-5 positions, and only a minor fraction of the EC was able to proceed further to be paused at the SHL-1 position in the absence of Rad26. Importantly, Rad26 failed to facilitate Pol II bypass across any of the nucleosome barriers in the EC1-Nuc system (Fig. 4E). In contrast, we found that Rad26 efficiently facilitated Pol II bypass of the nucleosome barrier in the EC2-Nuc system (Fig. 4E). These results show that the upstream duplex DNA is necessary for Rad26 to facilitate Pol II bypass of the nucleosome barrier, in agreement with our structure model.
Rad26-Pol II EC-Nucleosome Complex Cannot Allosterically Activate Rad26 to Remodel a Downstream Nucleosome.
One alternative possibility to explain the requirement of upstream duplex DNA is that this region is essential for the initial loading of full-length Rad26 onto the upstream DNA of the Pol II EC nucleosome. The Rad26-Pol II EC-nucleosome complex may allosterically activate Rad26 through some conformational rearrangement and allow Rad26 to remodel a downstream nucleosome directly as a canonical remodeler. To assess this possibility, we compared transcription results between a regular transcription/chromatin setting (strategy-1, control) and a sequential transcription-remodeling-chase setting (strategy-2) (Fig. 4 F and G). The strategy-1 setting is essentially the same as Fig. 2E: Rad26 is loaded first onto EC2-Nuc, followed by incubation with rNTPs and 2′-dATP. We obtained essentially the same results as in Fig. 2E: Rad26 promoted nucleosome bypass in this regular setting in a similar manner as the Rad26 variant Rad2628−1,085 (Figs. 2E and 4G, strategy-1 lanes). In contrast, in the sequential transcription-remodeling-chase setting (strategy-2), we first allowed Pol II to run into the nucleosome barrier by incubating the EC2-Nuc with rNTP. Then, rNTP was washed out from the system, and we incubated it with Rad26 and dATP to allow Rad26 loading and chromatin remodeling activity. In this case, Rad26 is able to be loaded on the Pol II EC nucleosome. If the Rad26-Pol II EC-nucleosome complex somehow allosterically activates Rad26 and allows Rad26 to remodel the downstream nucleosome as a canonical remodeler, we would expect the downstream nucleosome barrier to be removed by the allosterically activated Rad26 at this stage, leading to a full-length RO transcript after we chase the system with rNTP, even in the absence of Rad26. We used Rad26 variant Rad2628−1,085 with a constitutive remodeling activity as a positive control and demonstrated that the downstream nucleosome barrier can be removed by active Rad26 under this sequential transcription-remodeling-chase experiment condition, and we can obtain full-length RO transcripts (Fig. 4G, strategy-2, lanes 28–1,085). In sharp contrast, we found that Rad26 failed to support the full-length RO transcript under the same transcription-remodeling-chase condition (Fig. 4G, strategy-2, lanes 1–1,085). These results suggest that the Rad26-Pol II EC-nucleosome complex is unlikely to allosterically activate Rad26 to direct remodeling of the downstream nucleosome as a canonical remodeler under our experimental setting.
Rad26 Also Helps Pol II Overcome a Nonnucleosomal Barrier.
A unique requirement of Rad26’s ability to help Pol II bypass a nucleosome as compared to that of typical transcription elongation factors, such as TFIIS, TFIIF, Spt4/5, and Elf1 is ATP hydrolysis. We previously showed that upon binding to the upstream duplex DNA of the Pol II EC, Rad26 was able to actively translocate on the duplex DNA template in an ATP-dependent manner (41). This Rad26-mediated Pol II forward translocation would not only rescue a backtracked Pol II, but also help Pol II overcome nucleosome barriers or other types of barriers downstream. To test whether this may represent a general mechanism by which Rad26 facilitates the bypass of different types of barriers by Pol II EC, we engineered a strong Pol II translocation barrier with a noncovalent DNA binder, pyrrole-imidazole polyamide, and found that Rad26 but not Spt4/5 was able to help Pol II fully overcome the barrier (SI Appendix, Fig. S5). These results show that Rad26 is able to facilitate Pol II forward translocation to bypass transcription blockages in an ATP-dependent manner using a mechanism that is distinct from that used by canonical transcription elongation factors.
Rad26 Enhances Nucleosome Maintenance after Transcriptional Bypass.
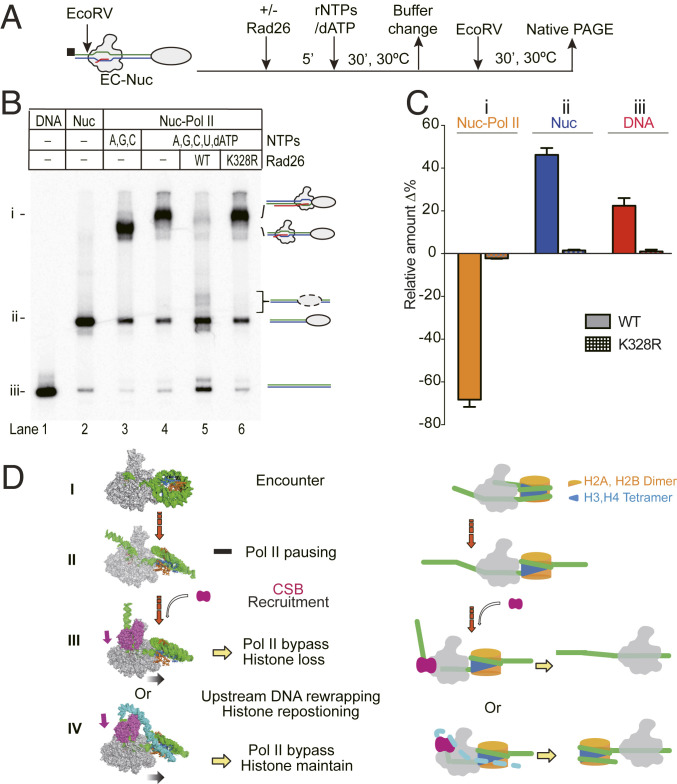
The survival of nucleosomes after transcription passage is functionally important to preserve the epigenome (52). To determine the fate of nucleosomes after Rad26-mediated transcriptional passage by Pol II, we analyzed the nucleosome template after the transcription reaction (schematically shown in Fig. 5A). The template was immobilized on beads and could be released by cleavage with EcoRV before or after transcription and analyzed by native page gel electrophoresis. In line with the transcription outcome (Fig. 1D), Pol II stalled at the nucleosome barrier in the absence of Rad26 (lane 4) or in the presence of the ATPase-dead Rad26K328R mutant (lane 6), both leading to the formation of slow-migrating bands that could be attributed to Pol II EC-nucleosome complexes (bands “i” in Fig. 5 B and C). Importantly, in the absence of Rad26, the nucleosome-paused Pol II EC is stable, and Pol II cannot dissociate from the template in 60 min (SI Appendix, Fig. S6). In sharp contrast, in the presence of full-length Rad26, the intensity of the Pol II EC-nucleosome complex (bands i) was dramatically decreased, indicating transcription passage of the nucleosome barrier in the presence of Rad26 (lane 5, Fig. 5 B and C). Strikingly, the intensity of the nucleosome bands greatly increased in those reactions (bands “ii,” lane 5, Fig. 5 B and C and SI Appendix, Fig. S7). These results suggest that a significant fraction of nucleosomes survived after Rad26-stimulated transcription bypass. Interestingly, we also observed a small portion of slower shifting bands that correspond to repositioned nucleosomes (moving upstream toward the center position of the DNA). Therefore, Rad26-mediated transcriptional bypass seems to preserve the integrity of the nucleosome. Based on this finding, coupled with structural modeling, we propose a model that could explain how Rad26 promotes nucleosome survival after transcriptional bypass (Fig. 5D and see Discussion).
Fig. 5.
Fate of the nucleosome after Rad26-mediated Pol II transcription bypass. (A) Experimental approach for analyzing the fate of nucleosomes after Rad26-mediated Pol II transcription bypass. The biotin label is shown as a black square. An EcoRV cleavage site was introduced in the upstream DNA to release the DNA from biotin beads. (B) Analysis of the released product by native polyacrylamide gel electrophoresis (PAGE). Three different species, the Pol II-Nuc complex, Nuc only, and DNA only, are labeled as i, ii, and iii, respectively, on the left. The mobilities of the released product are indicated on the right. In the presence of active Rad26, the majority of the arrested Pol II-nucleosome complex was converted into nucleosome bands. (C) Quantitation of the native PAGE assay. Lanes 4–6 from native gel were compared. The experiments were performed three times and shown as means with error bars. (D) Scheme of Rad26-mediated Pol II transcription bypass of a nucleosome. Pol II transcription encounters core nucleosome at SHL-6 (step I) and, then, proceeds to the major pausing site at SHL-1 (step II). In this step, an H2A-H2B dimer is exposed. Pausing of Pol II results in the recruitment of Rad26 which binds to the upstream of DNA of the elongation complex. Rad26 promotes the forward translocation of Pol II and helps Pol II bypass the nucleosome barrier (step III). Rad26 may also rewrap the upstream DNA and cover the exposed H2A-H2B to prevent the dissociation of the H2A-H2B dimer (step IV).
Discussion
Rad26 Is an ATP-Dependent Remodeling Factor that Recognizes Pol II as Its Substrate.
CSB was initially identified as a key factor in transcription-coupled repair (39). However, growing evidence indicates that CSB also plays important roles in transcription and transcriptional response to a variety of stresses (27, 36, 37, 40, 46, 53, 54). In this study, we provided biochemical evidence that Rad26, the budding yeast ortholog of CSB, facilitates nucleosome bypass by Pol II in a transcription-coupled and ATP-dependent manner.
Rad26 shares properties of both chromatin remodelers and transcription elongation factors. We present several lines of evidence that help elucidate the mechanism by which Rad26 promotes Pol II nucleosome bypass. First, we show that while full-length Rad26 can stimulate Pol II to go through a nucleosome barrier, the remodeling activity of full-length Rad26 is in an autorepressed state. Pol II-EC or the Pol II-EC-nucleosome complex cannot activate Rad26 remodeling activity on a downstream nucleosome either in cis (Fig. 4 F and G) or in trans (SI Appendix, Fig. S3). Second, we show that the Rad26-Pol II-EC complex is more stable than the Rad26-nucleosome complex in vitro, suggesting that Rad26 may preferentially bind Pol II EC over the nucleosome. Third, a structural model of the Rad26-Pol II nucleosome suggests that Rad26 associated with the upstream duplex would not be able to access the nucleosome in the same way as a canonical nucleosome remodeler would. Fourth, we showed that the duplex DNA upstream of Pol II EC is required to enable Rad26-dependent readthrough of the nucleosome barrier, while this region would not be expected to be needed for nucleosome remodeling activity by a canonical remodeler. Altogether, these results favor a noncanonical mechanism by which Rad26 stimulates the bypass of a nucleosome barrier by Pol II by acting as a Pol II processivity factor in an ATP-dependent manner.
Here, we propose that Rad26 can remodel a nucleosome in two different modes: canonical and noncanonical modes. Full-length Rad26 is in an autorepressed state and can be activated under certain conditions. Once activated, Rad26 can remodel nucleosomes in a canonical mode using ATP-dependent DNA translocation as is the case for the canonical remodelers in the Swi2/Snf2 family to which it belongs (35, 42). In addition to this canonical mode, here, we reveal that Rad26 has a noncanonical mode that is sufficient to promote Pol II transcription over nucleosomes. In this noncanonical mode, Rad26 recognizes Pol II EC as its substrate and stimulates Pol II to go through the nucleosome barrier, acting as a Pol II processivity factor. On the other hand, unlike other typical transcription elongation factors, Rad26 requires ATP hydrolysis to facilitate nucleosome bypass by Pol II. Therefore, Rad26 represents a specialized type of “dual mode” remodeling factor that is distinct from both “single mode” canonical chromatin remodelers and transcription elongation factors. It is also important to note that additional factors or stimuli may regulate the switch between two modes of Rad26, especially in the presence of DNA damage. Future structural studies on the Rad26-Pol II-EC-nucleosome complex will offer insights into Rad26-mediated transcriptional bypass of nucleosomes.
How Does Rad26 Maintain Nucleosome Integrity after Transcription?
Maintaining the integrity of nucleosomes during transcription elongation is crucial for epigenetic inheritance (55). In this study, we analyzed the fate of the nucleosome after the Rad26-mediated transcription passage (Fig. 5B). Our results indicate that the majority of nucleosomes are preserved after transcription bypass.
How does Rad26 promote nucleosome survival after transcription bypass? Previous biochemical (56) and atomic force microscopy (57) studies suggest that histones in front of an elongating Pol II may be transferred to the back via a “template looping” intermediate. A recent structural analysis of Pol II paused at the SHL-1 site postulates that the upstream exposed H2A-H2B dimer may be captured by a “foreign DNA” (49), thus, serving as a histone transfer intermediate for nucleosome survival after transcription bypass. Given that CSB has been proposed to undergo a conformational change as duplex DNA wraps around it (58) and induced a kink in an upstream DNA region upon binding to the Pol II EC (41), we speculate that Rad26 may facilitate nucleosome template looping in addition to the basal level of nucleosome template looping induced by Pol II (56). We propose that, upon Rad26 binding upstream of Pol II EC-nucleosome (Step III, Fig. 5D), Rad26 changes the conformation of the upstream DNA and promotes its association with the exposed H2A-H2B dimer thereby stabilizing the histone transfer intermediate during nucleosome bypass (Step IV, Fig. 5D). Our observation that only a portion of nucleosomes changed position after transcription passage is consistent with a previous study (23) especially with respect to the use of a strong nucleosome positioning sequence that not only enhances histone transfer, but also enhances their reassociation with DNA at the same location after transcription. A similar conclusion was reached in a previous atomic force microscopy analysis of transcriptional bypass of nucleosomes (57).
A Unified Mechanism for CSB in Transcription and TC-NER.
Our results also suggest a unified model where CSB uses the same mechanism for its roles in transcription elongation on a nucleosome template and in TC-NER (SI Appendix, Fig. S8). For CSB-dependent genes, its recruitment to Pol II is aided by binding to the upstream duplex DNA thereby promoting Pol II forward translocation on nucleosome templates. This function of CSB stimulates transcription elongation on the nucleosomal template. During DNA damage and repair, CSB may help distinguish between bulky DNA lesions from other forms of transcriptional arrest, thus, improving fidelity in recognizing DNA lesions. The Pol II-CSB complex would, then, initiate the TC-NER process (SI Appendix, Fig. S8). On the other hand, CSB may directly work on chromatin where, upon release of its autoinhibitory domain, it evicts histones as a canonical remodeler to facilitate other DNA repair processes, such as DBS repair (42, 59–61). Our findings have, thus, shed light on the multifaceted roles of CSB in different aspects of transcription and DNA repair on chromatin, including transcription pausing release and elongation (37, 40, 54).
Materials and Methods
Rhp26 WT and mutants were expressed in Escherichia coli strain Rosetta 2(DE3) (Novagen) and purified by an Ni-NTA agarose (Qiagen), Hi-Trap Heparin HP (GE Healthcare), Hi-Trap SP (GE Healthcare), and Superdex 200 10/300 GL columns (GE Healthcare). S. cerevisiae 12-subunit Pol II was purified by an IgG affinity column (GE Healthcare), followed by Hi-Trap Heparin (GE Healthcare) and Mono Q anion exchange chromatography columns (GE Healthcare). Mononucleosome remodeling assay and chromatin remodeling by restriction enzyme accessibility assays were performed as described (42). Detailed descriptions of protein purification, DNA sequences, chromatin-remodeling assay, transcription elongation assay, electrophoretic mobility-shift assay, and bioinformatics analysis are given in SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
We are grateful to Dr. Jianhua Fu for providing the Spt4/Spt5 protein. We thank Dr. Peter B. Dervan for providing pyrrole-imidazole (Py-Im) polyamides. This work was supported by Grants from the National Institutes of Health (R01 GM102362 and R01 GM092895 to D.W.; DK098808 to X.-D.F.; R01 GM092895 to A.E.L.). J.-Y.C. is supported by NIH K99 Grant (DK120952).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2013379117/-/DCSupplemental.
Data Availability.
All study data are included in the article and supporting information, and Protein Data Bank (PDB ID codes 1KX5 and 5VVR) (41, 62).
References
- 1.Li B., Carey M., Workman J. L., The role of chromatin during transcription. Cell 128, 707–719 (2007). [DOI] [PubMed] [Google Scholar]
- 2.Kujirai T., Kurumizaka H., Transcription through the nucleosome. Curr. Opin. Struct. Biol. 61, 42–49 (2020). [DOI] [PubMed] [Google Scholar]
- 3.Floer M. et al., A RSC/nucleosome complex determines chromatin architecture and facilitates activator binding. Cell 141, 407–418 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schwabish M. A., Struhl K., The Swi/Snf complex is important for histone eviction during transcriptional activation and RNA polymerase II elongation in vivo. Mol. Cell. Biol. 27, 6987–6995 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smolle M. et al., Chromatin remodelers Isw1 and Chd1 maintain chromatin structure during transcription by preventing histone exchange. Nat. Struct. Mol. Biol. 19, 884–892 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carey M., Li B., Workman J. L., RSC exploits histone acetylation to abrogate the nucleosomal block to RNA polymerase II elongation. Mol. Cell 24, 481–487 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Skene P. J., Hernandez A. E., Groudine M., Henikoff S., The nucleosomal barrier to promoter escape by RNA polymerase II is overcome by the chromatin remodeler Chd1. eLife 3, e02042 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee J. et al., Chromatin remodeller Fun30Fft3 induces nucleosome disassembly to facilitate RNA polymerase II elongation. Nat. Commun. 8, 14527 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guermah M., Palhan V. B., Tackett A. J., Chait B. T., Roeder R. G., Synergistic functions of SII and p300 in productive activator-dependent transcription of chromatin templates. Cell 125, 275–286 (2006). [DOI] [PubMed] [Google Scholar]
- 10.Kraus W. L., Kadonaga J. T., p300 and estrogen receptor cooperatively activate transcription via differential enhancement of initiation and reinitiation. Genes Dev. 12, 331–342 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Belotserkovskaya R. et al., FACT facilitates transcription-dependent nucleosome alteration. Science 301, 1090–1093 (2003). [DOI] [PubMed] [Google Scholar]
- 12.Kuryan B. G. et al., Histone density is maintained during transcription mediated by the chromatin remodeler RSC and histone chaperone NAP1 in vitro. Proc. Natl. Acad. Sci. U.S.A. 109, 1931–1936 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Angelov D. et al., Nucleolin is a histone chaperone with FACT-like activity and assists remodeling of nucleosomes. EMBO J. 25, 1669–1679 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang T. et al., The histone chaperone FACT modulates nucleosome structure by tethering its components. Life Sci. Alliance 1, e201800107 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu Y. et al., FACT caught in the act of manipulating the nucleosome. Nature 577, 426–431 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fei J. et al., NDF, a nucleosome-destabilizing factor that facilitates transcription through nucleosomes. Genes Dev. 32, 682–694 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kireeva M. L. et al., Nature of the nucleosomal barrier to RNA polymerase II. Mol. Cell 18, 97–108 (2005). [DOI] [PubMed] [Google Scholar]
- 18.Crickard J. B., Lee J., Lee T. H., Reese J. C., The elongation factor Spt4/5 regulates RNA polymerase II transcription through the nucleosome. Nucleic Acids Res. 45, 6362–6374 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gaykalova D. A. et al., A polar barrier to transcription can be circumvented by remodeler-induced nucleosome translocation. Nucleic Acids Res. 39, 3520–3528 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ehara H. et al., Structural insight into nucleosome transcription by RNA polymerase II with elongation factors. Science 363, 744–747 (2019). [DOI] [PubMed] [Google Scholar]
- 21.Hsieh F. K. et al., Histone chaperone FACT action during transcription through chromatin by RNA polymerase II. Proc. Natl. Acad. Sci. U.S.A. 110, 7654–7659 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sims R. J. 3rd, Belotserkovskaya R., Reinberg D., Elongation by RNA polymerase II: The short and long of it. Genes Dev. 18, 2437–2468 (2004). [DOI] [PubMed] [Google Scholar]
- 23.Bondarenko V. A. et al., Nucleosomes can form a polar barrier to transcript elongation by RNA polymerase II. Mol. Cell 24, 469–479 (2006). [DOI] [PubMed] [Google Scholar]
- 24.Ishibashi T. et al., Transcription factors IIS and IIF enhance transcription efficiency by differentially modifying RNA polymerase pausing dynamics. Proc. Natl. Acad. Sci. U.S.A. 111, 3419–3424 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luse D. S., Spangler L. C., Újvári A., Efficient and rapid nucleosome traversal by RNA polymerase II depends on a combination of transcript elongation factors. J. Biol. Chem. 286, 6040–6048 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karikkineth A. C., Scheibye-Knudsen M., Fivenson E., Croteau D. L., Bohr V. A., Cockayne syndrome: Clinical features, model systems and pathways. Ageing Res. Rev. 33, 3–17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Selby C. P., Sancar A., Human transcription-repair coupling factor CSB/ERCC6 is a DNA-stimulated ATPase but is not a helicase and does not disrupt the ternary transcription complex of stalled RNA polymerase II. J. Biol. Chem. 272, 1885–1890 (1997). [DOI] [PubMed] [Google Scholar]
- 28.Clapier C. R., Iwasa J., Cairns B. R., Peterson C. L., Mechanisms of action and regulation of ATP-dependent chromatin-remodelling complexes. Nat. Rev. Mol. Cell Biol. 18, 407–422 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hanawalt P. C., Spivak G., Transcription-coupled DNA repair: Two decades of progress and surprises. Nat. Rev. Mol. Cell Biol. 9, 958–970 (2008). [DOI] [PubMed] [Google Scholar]
- 30.Wei L. et al., DNA damage during the G0/G1 phase triggers RNA-templated, Cockayne syndrome B-dependent homologous recombination. Proc. Natl. Acad. Sci. U.S.A. 112, E3495–E3504 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Batenburg N. L., Thompson E. L., Hendrickson E. A., Zhu X. D., Cockayne syndrome group B protein regulates DNA double-strand break repair and checkpoint activation. EMBO J. 34, 1399–1416 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gregersen L. H., Svejstrup J. Q., The cellular response to transcription-blocking DNA damage. Trends Biochem. Sci. 43, 327–341 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lans H., Hoeijmakers J. H. J., Vermeulen W., Marteijn J. A., The DNA damage response to transcription stress. Nat. Rev. Mol. Cell Biol. 20, 766–784 (2019). [DOI] [PubMed] [Google Scholar]
- 34.Tufegdzic Vidakovic A. et al., Regulation of the RNAPII pool is integral to the DNA damage response. Cell 180, 1245–1261.e21 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang L. et al., Regulation of the Rhp26ERCC6/CSB chromatin remodeler by a novel conserved leucine latch motif. Proc. Natl. Acad. Sci. U.S.A. 111, 18566–18571 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee S. K., Yu S. L., Prakash L., Prakash S., Requirement for yeast RAD26, a homolog of the human CSB gene, in elongation by RNA polymerase II. Mol. Cell. Biol. 21, 8651–8656 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Malik S. et al., Rad26p, a transcription-coupled repair factor, is recruited to the site of DNA lesion in an elongating RNA polymerase II-dependent manner in vivo. Nucleic Acids Res. 38, 1461–1477 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Newman J. C., Bailey A. D., Weiner A. M., Cockayne syndrome group B protein (CSB) plays a general role in chromatin maintenance and remodeling. Proc. Natl. Acad. Sci. U.S.A. 103, 9613–9618 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Troelstra C. et al., ERCC6, a member of a subfamily of putative helicases, is involved in Cockayne’s syndrome and preferential repair of active genes. Cell 71, 939–953 (1992). [DOI] [PubMed] [Google Scholar]
- 40.Wang Y. et al., Dysregulation of gene expression as a cause of Cockayne syndrome neurological disease. Proc. Natl. Acad. Sci. U.S.A. 111, 14454–14459 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xu J. et al., Structural basis for the initiation of eukaryotic transcription-coupled DNA repair. Nature 551, 653–657 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang W. et al., Molecular basis of chromatin remodeling by Rhp26, a yeast CSB ortholog. Proc. Natl. Acad. Sci. U.S.A. 116, 6120–6129 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Citterio E. et al., ATP-dependent chromatin remodeling by the Cockayne syndrome B DNA repair-transcription-coupling factor. Mol. Cell. Biol. 20, 7643–7653 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kireeva M. L. et al., Nucleosome remodeling induced by RNA polymerase II: Loss of the H2A/H2B dimer during transcription. Mol. Cell 9, 541–552 (2002). [DOI] [PubMed] [Google Scholar]
- 45.Schier A. C., Taatjes D. J., Structure and mechanism of the RNA polymerase II transcription machinery. Genes Dev. 34, 465–488 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Le May N. et al., NER factors are recruited to active promoters and facilitate chromatin modification for transcription in the absence of exogenous genotoxic attack. Mol. Cell 38, 54–66 (2010). [DOI] [PubMed] [Google Scholar]
- 47.Lake R. J. et al., The sequence-specific transcription factor c-Jun targets Cockayne syndrome protein B to regulate transcription and chromatin structure. PLoS Genet. 10, e1004284 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ujvári A., Hsieh F. K., Luse S. W., Studitsky V. M., Luse D. S., Histone N-terminal tails interfere with nucleosome traversal by RNA polymerase II. J. Biol. Chem. 283, 32236–32243 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kujirai T. et al., Structural basis of the nucleosome transition during RNA polymerase II passage. Science 362, 595–598 (2018). [DOI] [PubMed] [Google Scholar]
- 50.Farnung L., Vos S. M., Cramer P., Structure of transcribing RNA polymerase II-nucleosome complex. Nat. Commun. 9, 5432 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weber C. M., Ramachandran S., Henikoff S., Nucleosomes are context-specific, H2A.Z-modulated barriers to RNA polymerase. Mol. Cell 53, 819–830 (2014). [DOI] [PubMed] [Google Scholar]
- 52.Teves S. S., Weber C. M., Henikoff S., Transcribing through the nucleosome. Trends Biochem. Sci. 39, 577–586 (2014). [DOI] [PubMed] [Google Scholar]
- 53.Vélez-Cruz R., Egly J. M., Cockayne syndrome group B (CSB) protein: At the crossroads of transcriptional networks. Mech. Ageing Dev. 134, 234–242 (2013). [DOI] [PubMed] [Google Scholar]
- 54.van den Heuvel D., et al. , A CSB-PAF1C axis restores processive transcription elongation after DNA damage repair. bioRxiv:10.1101/2020.01.04.894808 (5 January 2020). [DOI] [PMC free article] [PubMed]
- 55.Venkatesh S., Workman J. L., Histone exchange, chromatin structure and the regulation of transcription. Nat. Rev. Mol. Cell Biol. 16, 178–189 (2015). [DOI] [PubMed] [Google Scholar]
- 56.Kulaeva O. I. et al., Mechanism of chromatin remodeling and recovery during passage of RNA polymerase II. Nat. Struct. Mol. Biol. 16, 1272–1278 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bintu L. et al., The elongation rate of RNA polymerase determines the fate of transcribed nucleosomes. Nat. Struct. Mol. Biol. 18, 1394–1399 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Beerens N., Hoeijmakers J. H., Kanaar R., Vermeulen W., Wyman C., The CSB protein actively wraps DNA. J. Biol. Chem. 280, 4722–4729 (2005). [DOI] [PubMed] [Google Scholar]
- 59.Lake R. J., Geyko A., Hemashettar G., Zhao Y., Fan H. Y., UV-induced association of the CSB remodeling protein with chromatin requires ATP-dependent relief of N-terminal autorepression. Mol. Cell 37, 235–246 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cho I., Tsai P. F., Lake R. J., Basheer A., Fan H. Y., ATP-dependent chromatin remodeling by Cockayne syndrome protein B and NAP1-like histone chaperones is required for efficient transcription-coupled DNA repair. PLoS Genet. 9, e1003407 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Batenburg N. L. et al., ATM and CDK2 control chromatin remodeler CSB to inhibit RIF1 in DSB repair pathway choice. Nat. Commun. 8, 1921 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Davey C. A. et al., Solvent Mediated Interactions in the Structure of the Nucleosome Core Particle at 1.9 A Resolution. J. Mol. Biol. 319, 1097–1113 (2002). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and supporting information, and Protein Data Bank (PDB ID codes 1KX5 and 5VVR) (41, 62).