Significance
Secondary organic aerosol (SOA) causes various adverse health outcomes, but identification of the toxic components remains infeasible for current methodology due to the existence of hundreds of unknown chemical components. In this study, we established a platform to tackle this challenge by combining atmospheric flow reactor, proteomics, reporter bioassays, and nontarget analysis. Short-lived unsaturated carbonyls were identified to be the components in naphthalene-derived SOA responsible for toxicity related to cysteine reactions.
Keywords: proteomics, cysteine alkylation, Nrf2-Keap1, atmospheric aging, toxicity dynamics
Abstract
Exposure to air pollution causes adverse health outcomes, but the toxicity mechanisms remain unclear. Here, we investigated the dynamic toxicities of naphthalene-derived secondary organic aerosol (NSOA) in a human bronchial epithelial cell line (BEAS-2B) and identified the chemical components responsible for toxicities. The chemical composition of NSOA was found to vary with six simulated atmospheric aging conditions (C1–C6), as characterized by high-resolution mass spectrometry and ion mobility mass spectrometry. Global proteome profiling reveals dynamic evolution in toxicity: Stronger proteome-wide impacts were detected in fresh NSOA, but the effects declined along with atmospheric aging. While Nrf2-regulated proteins (e.g., NQO1) were significantly up-regulated, the majority (78 to 97%) of proteins from inflammation and other pathways were down-regulated by NSOA exposure (e.g., Rho GTPases). This pattern is distinct from the reactive oxygen species (ROS)-mediated toxicity pathway, and an alternative cysteine reaction pathway was revealed by the decreased abundance of proteins (e.g., MT1X) prone to posttranslational thiol modification. This pathway was further validated by observing decreased Nrf2 response in reporter cells, after preincubating NSOA with cysteine. Ethynyl-naphthalene probe was employed to confirm the alkylation of cellular proteome thiols on the proteome-wide level by fresh NSOA via in-gel fluorescence imaging. Nontarget analysis identified several unsaturated carbonyls, including naphthoquinones and hydroxylated naphthoquinones, as the toxic components responsible for cysteine reactivity. Our study provides insights into the dynamic toxicities of NSOA during atmospheric aging and identifies short-lived unsaturated carbonyls as the predominant toxic components at the posttranslational level.
Exposure to ambient particulate matter (PM) has been associated with multiple adverse health outcomes (1–3). A major component of PM is secondary organic aerosol (SOA), composed of condensed-phase products formed from atmospheric oxidation of gas-phase precursors (4). SOA is made up of thousands of different organic compounds, many of which can be reactive toward proteins and lipids (5, 6). SOA composition is complex and dynamic upon atmospheric aging, and can therefore have highly variable toxicities (7, 8). Understanding the relationship between the chemical composition and the toxicity of SOA upon atmospheric aging is key to determining the health effects of SOA.
Naphthalene is the most abundant polycyclic aromatic hydrocarbon (PAH) and is emitted from wood combustion, traffic exhaust, and petroleum processing (9–11). Previous studies estimated that the contribution of PAHs to urban SOA formation is comparable to that of monocyclic aromatics, and, among PAHs, naphthalene has the greatest potential to form SOA (NSOA) under both high-NOx and low-NOx conditions (12–14). Chowdhury et al. (15, 16) reported a higher reactive oxygen species (ROS)-mediated toxicity of aged NSOA compared to fresh NSOA. Dynamic toxicities were also observed for other hydrocarbon-derived SOA upon atmospheric aging, with a higher DTT reactivity observed in fresh SOA (7). These studies highlight the role of chemical composition in determining the dynamic toxicities of SOA, and thus identification of the predominant toxic components is of great interest. Reactive intermediates like highly oxygenated molecules and environmentally persistent free radicals (EPFRs) in NSOA have been suggested to contribute toward the oxidative potential (17, 18), which can be further catalyzed by transition metal reactions such as the Fenton mechanism (19, 20). Selected compounds, for instance, naphthoquinone, were estimated to contribute 21% of the total measured oxidative potential of NSOA (21), and hydroperoxides may also be responsible for DTT reactivities based on correlation analysis (7). While these studies highlight the contributions of complicated NSOA components to ROS production, the toxicities for the majority of the chemical components in NSOA remain unidentified.
The complex nature of SOA suggests the potential for multiple toxicity pathways. While former studies are focused on ROS-mediated toxicities (21, 22), the well-known toxic components of NSOA, quinones, have been previously revealed to cause toxicities via alternative pathways, e.g., alkylating proteome thiols (23). The recent advancements of “omics” technologies provide an opportunity to investigate the toxicity at a molecular level (24). Exposure of human bronchial epithelial cell line (BEAS-2B) to SOA was observed to activate many toxicity pathways (e.g., DNA damage, p53 pathway) based on the transcriptome information from RNA-seq (25, 26). However, the expression levels of mRNA are poorly correlated to proteins (27), due to protein-specific posttranslational modifications (PTMs) and turnover rates. Investigating the toxicity by proteomics is therefore expected to directly provide additional posttranslation information. However, limited information is available on the toxicities of organic aerosol at the proteome level.
In this study, we aim to combine laboratory aerosol aging experiments with proteomics to investigate the dynamic chemical composition and toxicity of NSOA during atmospheric aging processes. Cellular thiol modifications caused by unsaturated carbonyls were identified by a combination of reporter bioassay and nontarget analysis as the major pathway for the dynamic toxicity of NSOA along with atmospheric aging.
Results and Discussion
Dynamic Changes of NSOA Chemical Components during Aging.
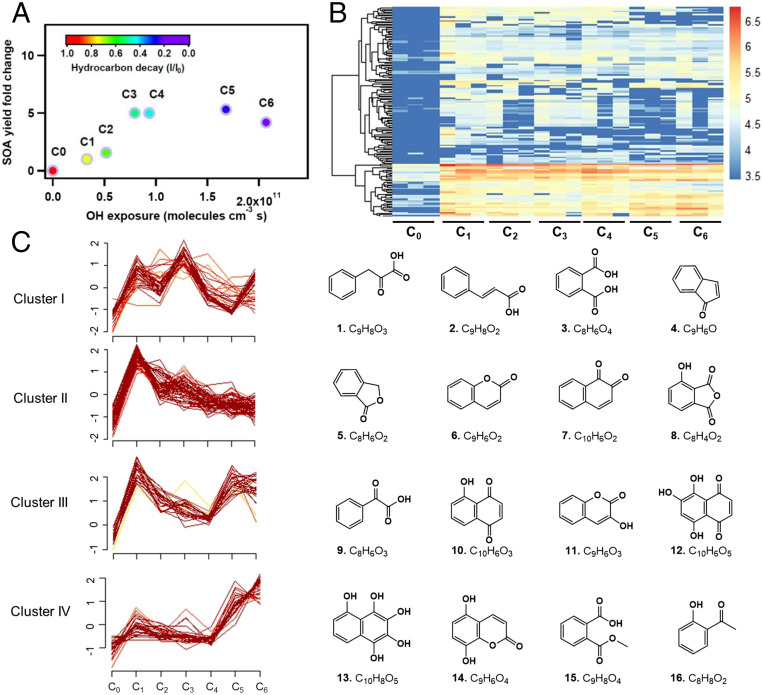
Fig. 1A illustrates the evolution of NSOA yields under various photooxidation conditions (C0–C6); see Materials and Methods for more experimental details. An increase in NSOA yield was observed at lower OH exposures (C0–C4), followed by a slight decrease (C5–C6) at higher OH exposures. A similar yield trend has been observed for isoprene, α-pinene (28), n-decane (29), and toluene (30) photooxidation at different OH exposure levels. This trend in SOA yield is a result of an overall transition from a functionalization-dominated to a fragmentation-dominated regime with increasing OH exposure (29, 31). A total of 156 photooxidation products were detected by liquid chromatography high-resolution mass spectrometry (LC-HRMS)-based nontarget analysis across six photooxidation conditions (Fig. 1B). Based on their abundances under the six photooxidation conditions, all 156 products were grouped into four clusters (Fig. 1C). Structures of 16 representative, high-abundant compounds were assigned by combining Orbitrap HRMS (elemental composition), ion mobility mass spectrometry (isomer assignment), and gas chromatography–mass spectrometry (GC-MS)-based National Institute of Standards and Technology (NIST) library matching (structure prediction) (details are shown in SI Appendix, Figs. S1–S4). The highly dynamic chemical compositions of NSOA along with atmospheric aging is consistent with previous studies, where the level of photooxidation affects the chemical composition, particulate mass, optical properties, phase state, and toxicity of SOA (7, 17, 32–34).
Fig. 1.
Dynamic changes in chemical composition of NSOA upon atmospheric aging. (A) Variation in NSOA yields under different OH exposure conditions. The color of each marker indicates the extent of hydrocarbon precursor decay, which was nearly complete in C6. (B) Hierarchical clustering of the 156 NSOA chemicals detected by LC-Orbitrap. Four major clusters were identified. The colors indicate log-transformed peak intensities. (C) Evolution of the four clusters under different OH exposure conditions. Molecular structures are shown for four representative chemicals detected in each cluster. The centralization of compounds is indicated from dark to light colors. The y axis represents the normalized abundances standardized to have a mean of zero and a SD of 1.
As shown in Fig. 1C, cluster I is mainly composed of carboxylic acids derived from naphthalene oxidation. Clusters II and III are compounds containing electrophilic functional groups, including quinolic compounds and other unsaturated carbonyls. Upon further oxidation, cluster II displays a monotonically decreasing trend along with photochemical aging (C1–C6), suggesting that these products are primary oxidation products and are consumed upon further atmospheric aging. While the trends of cluster I and cluster III are more complicated, a decreasing trend at C1–C4 was observed for cluster III. In contrast, cluster IV displays an increasing trend especially at C5–C6. Cluster IV are highly oxidized NSOA compounds with more oxygen atoms, which is likely a result of more extensive aging (12, 35, 36). The dynamic chemical composition accompanied by the dominance of highly oxidized products in NSOA under OH photochemical aging has been observed in previous studies (32, 37). This increase in oxygenation of NSOA along with the decrease in carbonyls detected by LC-HRMS is consistent with GC-MS results (SI Appendix, Figs. S3 and S4). Based on the GC-MS results matched to the NIST08 library, primary oxidation products (phenols, quinones) decreased with OH exposure, and are replaced by more highly oxygenated compounds. Phenols and quinones have been proposed to be toxic NSOA constituents due to their propensities to participate in redox cycling and catalyze the formation of radicals (38–40). The decrease in redox-active compounds is also consistent with the decrease in amount of ROS measured by electron paramagnetic resonance spectroscopy (EPR) and OP measured by the DTT assay, as shown in SI Appendix, Fig. S5. It should be noted that the EPR method applied in the current study is not only sensitive to radical species but can also serve as an indicator for H2O2, which can yield radicals via redox cycling and/or decomposition (18, 20, 41).
“Inverted U-Shaped” Proteome-Wide Effects of NSOA on BEAS-2B Cells.
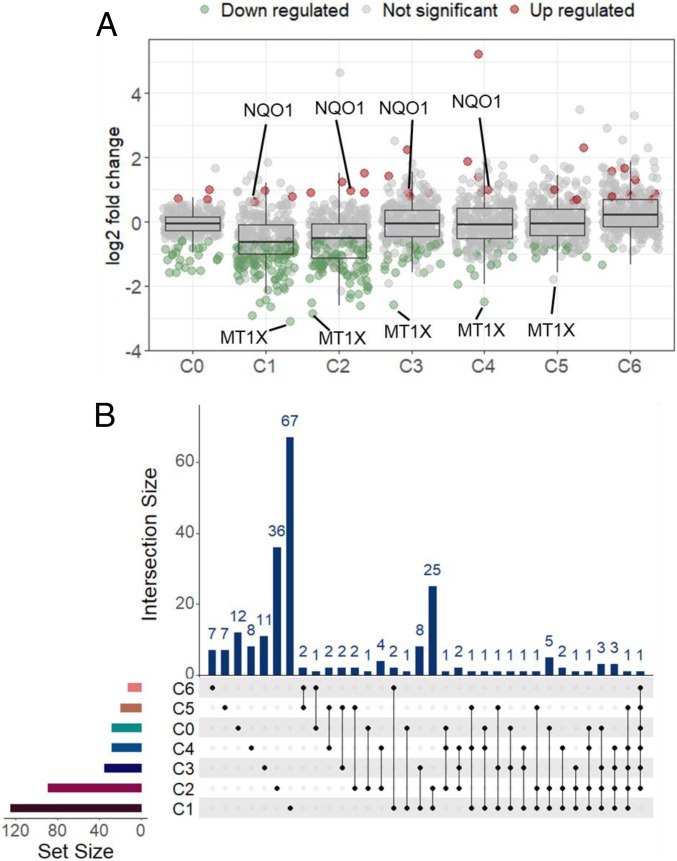
To investigate the impacts of dynamic chemical composition of NSOA on toxicities during aging, BEAS-2B cells were exposed to NSOA collected under the six photooxidation conditions. No cytotoxicity was observed up to 100 µM NSOA (SI Appendix, Fig. S6). Thus, 100 µM was selected as the concentration at which we investigated the subcytotoxicity impacts of NSOA on cellular proteome. A stable isotope dimethyl labeling-based proteomics method was employed (42, 43). In total, 1,212 proteins were detected with a false discovery rate of 1% (42). While few proteins were significantly changed (fold change [FC] > 1.5 and P < 0.05) in the C0 group (naphthalene without oxidation), greater impacts were observed in BEAS-2B cells exposed to C1 group by detecting 125 significantly altered proteins (Fig. 2A, the list of significantly changed proteins was shown in Dataset S1). This clearly shows an increase in proteome-wide impacts of NSOA on BEAS-2B cells, compared to naphthalene without photooxidation.
Fig. 2.
Distinct proteome profiles of BEAS-2B cells exposed to NSOA upon atmospheric aging. (A) Scatter plot and box plot showing the distribution of log2 expression fold change (FC) values of proteins detected in different exposure groups compared to vehicle control (0.1% DMSO); proteins with values of P < 0.05 are shown. FC > 1.5 was used as cutoff for significance in protein expression. Red, green, and gray dots indicate up-regulated proteins, down-regulated proteins, and not significantly changed proteins, respectively (B). UpSet plot showing overlap in significantly regulated proteins identified in different exposure groups. Each column of the top bar plot corresponds to an interaction that is containing the groups indicated by solid dots below. Total regulated proteins in different groups are represented by the left bar plot as “set size.”
Notably, the number of significantly altered proteins decreased with increasing OH exposure, and only 88, 34, 27, 19, and 12 proteins showed significant changes in C2–6 groups, respectively (Fig. 2A and SI Appendix, Fig. S7). Most of the impacted proteins observed in C1–6 treatments were overlapping, for example, 52 of the 88 proteins (59.1%) from C2 group were also significantly impacted in other treatment groups especially in C1 (25 proteins, Fig. 2B). These results demonstrate a common toxicity pathway of NSOA across aging, and, more importantly, fresh NSOA showed stronger proteome-wide impacts than aged NSOA. It should be noted that Chowdhury et al. (15, 16) reported stronger oxidative stress responses from aged NSOA, including the up-regulation of Nrf2 pathways in human lung cells. The difference between our study and their work may arise from the different extents of photooxidation. In their work, higher OH exposure concentrations were used for an aging timescale of 10 d, while we investigated the aging effects equivalent to 0 to 2 atmospheric processing days in this study. A higher oxidative potential in fresh SOA was also observed for other hydrocarbon precursors (e.g., isoprene, α-pinene, toluene), with declined toxicity observed during aging (7, 44).
NSOA Down-Regulated Cellular Proteome but Up-Regulated Nrf2 Pathway.
While the activation of oxidative stress and inflammatory pathways by SOA has been well documented, this study investigates the effects at the proteome level. Surprisingly, the expression for the majority of proteins was suppressed. For example, 121 of the 125 proteins (96.8%) significantly altered in the C1 treatment were down-regulated, while only 4 proteins (3.2%) were up-regulated (Fig. 2A and SI Appendix, Fig. S7). The predominance of down-regulated proteins is consistent across C1–5 treatments (78–97%), except for C6 treatment for which 10 of 12 proteins were up-regulated. This is different from the well-documented ROS-mediated toxicity mechanism by up-regulating gene expressions. For instance, 79.3% (23/29) of the genes in oxidative stress and inflammatory pathways were up-regulated in BEAS-2B cells exposed to isoprene-derived SOA, while only 20.7% (6/29) genes were down-regulated (45).
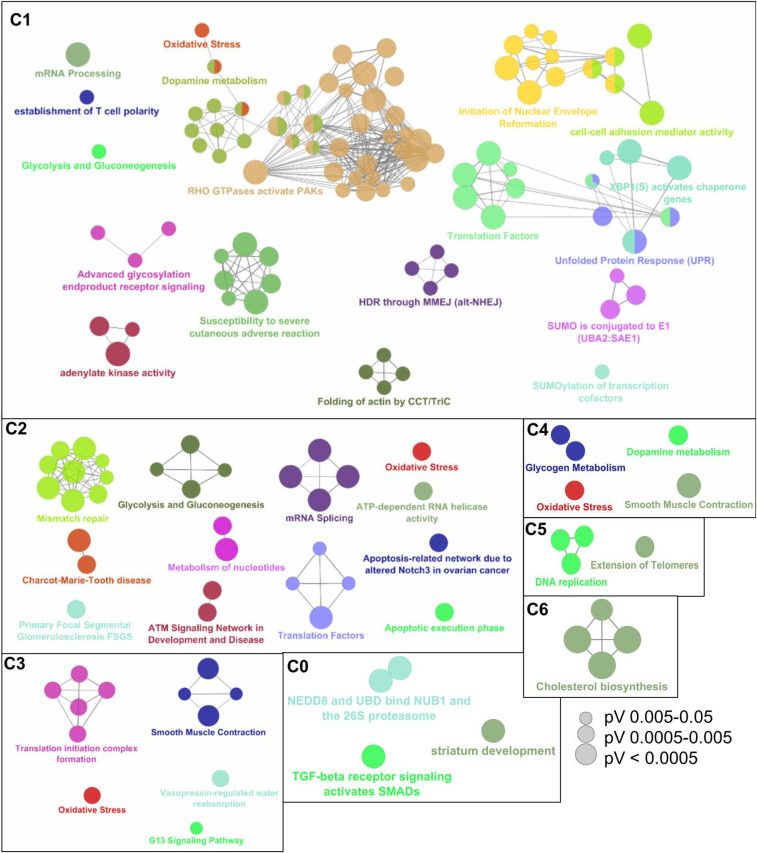
Pathway enrichment analysis showed that these down-regulated proteins were enriched in more than 10 pathways, especially Rho GTPases pathway in C1 treatment (Fig. 3). All three key members of Rho family of GTPases, including RhoA (0.63-fold, P = 0.022), Rac1 (0.60-fold, P = 0.016), and Cdc42 (0.39-fold, P = 0.018), were down-regulated in BEAS-2B exposed to C1 (SI Appendix, Fig. S8). Rho GTPases are key proteins regulating cell adhesion (46). Consistent with this, the expressions of proteins (e.g., JUP, TMOD3) in the “cell–cell adhesion mediator activity” pathway were also suppressed. Rho GTPases have been well documented to activate NF-κB–mediated inflammation, by inducing the degradation of IκB (47). Thus, the down-regulation of Rho GTPases observed in this study is expected to inactivate inflammation. NF-κB and cytokines (e.g., IL-6), the most common proteins/genes used to mark inflammation, were not detected due to their low abundances. However, several other inflammation-associated proteins, including HLA-B (0.55-fold, P = 0.037) and HLA-A (0.56-fold, P = 0.028), were significantly down-regulated in the C1 treatment, supporting the suppression of inflammation by NSOA.
Fig. 3.
Pathway enrichment analysis of proteins that were significantly altered by NSOA. The annotation network of significantly altered pathways was visualized by ClueGO, which is a Cytoscape plug-in. Each circle represents a biological term consisting of various related genes. Terms that pertain to the same pathway are displayed with the same color, and terms associated with two different pathways are shown in two-tone. The connectivity (edges) between the terms in functionally grouped network is derived from kappa score, which indicates the similarity of associated genes shared by different terms, and thicker edges indicate stronger similarity. The size of the circles positively correlates with statistical significance of the term enrichment. Only the name of the most significant term in each group is shown to reduce overlay.
In contrast to diverse down-regulated pathways, the few up-regulated proteins were specifically enriched in the “oxidative stress” pathway (Fig. 3), represented by NQO1 (1.90-fold in C1, P = 0.004) and TXNRD1 (1.95-fold in C1, P = 0.015) proteins (SI Appendix, Fig. S8). In particular, the up-regulation of NQO1 is consistently the most pronounced across C1–4 (Fig. 2A). NQO1 and TXNRD1 are both well known to be regulated by Nrf2–Keap1 protein complex (48), the major regulator of cytoprotective responses to oxidative stress. This demonstrates the activation of Nrf2 pathway in BEAS-2B cells by NSOA. The activation of Nrf2 pathway by SOA has been widely reported for other SOA systems in prior studies. For instance, up-regulation of TXNRD1 gene was observed in BEAS-2B cells exposed to isoprene-derived SOA (45).
Previous studies have investigated the oxidative potential of SOA by a variety of cellular and acellular assays (e.g., DTT) (49), among which NSOA consistently exhibits the highest oxidative potential (8, 22, 49). While these studies highlighted the health effects of NSOA mediated by the ROS pathway, our study applies proteomics to investigate the toxicity of NSOA. While both NSOA and ROS activate Nrf2 pathway, NSOA suppresses other pathways including inflammation, which is clearly different from well-documented ROS-mediated toxicities of other SOA systems followed by inflammation (50). The absence of inflammation for NSOA exposure was also observed in our previous in vivo study (51). This suggests the chemical composition of NSOA largely induced the proteome-wide impacts via a unique ROS-independent pathway, in addition to the ROS pathway, which has been well documented for many SOA systems.
Nrf2 Responses Were Caused by Posttranslational Cysteine Alkylation on Cellular Proteome.
To further understand the toxicity mechanisms of NSOA, metallothionein-1X (MT1X) called our attentions as its suppression is the most pronounced among all measured proteins across C1–5 (Fig. 2A). MT1X is another well-known antioxidant protein regulated by the Nrf2–Keap1 pathway (52). MT1X is a cysteine-rich metal-binding protein, with 11 cysteines located on α domain, and 9 cysteines on β domain (Fig. 4A). The peptide ([MDPNCSCSPVGSCACAGSCK]3+, m/z = 754.2909) (SI Appendix, Fig. S9) of β domain was consistently detected in samples across all six photooxidation conditions. Extraction of the raw chromatograms clearly showed the decrease in abundances of MT1X peptide in C1–5, but not in C0 or C6 treatments (Fig. 4B). After normalization by total proteins, the abundances of MT1X decreased by 0.16-, 0.14-, 0.17-, and 0.18-fold, in C1 (P = 0.001), C2 (P = 0.007), C3 (P = 0.0003), and C4 (P = 0.002) treatments, respectively (Fig. 4C). The down-regulation of MT1X is inconsistent with the up-regulation of two other Nrf2-regulated antioxidant proteins NQO1 and TXNRD1 as mentioned above, and hence should not be attributed to the process of transcription regulated by Nrf2. Multiple species of cysteines exist in cells, as free thiol, oxidized forms, or binding to metals or other electrophiles (Fig. 4D) (53). For example, ROS has been reported to cause the oxidation of cysteines to disulfate or sulfenic acid (54). Notably, most ROS oxidized cysteines can be reduced to free thiol by Tris(2-carboxyethyl)phosphine (TCEP) and then modified by iodoacetamide during proteomics sample preparation, except for irreversible sulfonic acid. To further evaluate whether the irreversible oxidation of cysteine to sulfonic acid is responsible for the decrease in abundances of MT1X, we used the MaxQuant to search the tryptic peptides with variable modifications of sulfonic acid (–SO3H). Only 13 peptides with sulfonic acid modification were detected, among which only 1 peptide (EVLRQALRCTNCK) from the RASSSF5 protein was detected at >1.5 times higher abundances in C1 treatment compared to control. This result further confirms the decrease in MT1X abundances should be not be attributed to ROS-mediated oxidations. Considering the high content of cysteines on MT1X, we hypothesize that the irreversible, posttranslational modifications by electrophiles (Fig. 4D) in NSOA could explain its decrease in abundances.
Fig. 4.
Posttranslational modification of MT1X by electrophiles in NSOA. (A) Sketch map and sequence of MT1X with 20 cysteines coordinated with metals. The sequence of MT1X is provided. Only the peptide of β-domain (labeled in red) was detected. (B) Chromatogram of isotopically labeled peptide of MT1X peptide across seven treatments. Compared to control, the abundances of MT1X are lower in C1–5 exposures. (C) FC of MT1X in different NSOA treatment groups (n = 3). **P < 0.01; ***P < 0.001. (D) Reduction, alkylation, and MS detection of cysteine species with different modifications. Note that most cysteine modifications via ROS (e.g., S–S, S–OH) are reversible and should be detected by proteomics. The irreversible oxidation of cysteine to sulfonic acid (–SO3H) are also detectable by proteomics searching with variable modifications. Modifications by electrophiles (i.e., S–R) are irreversible and nondetectable by proteomics.
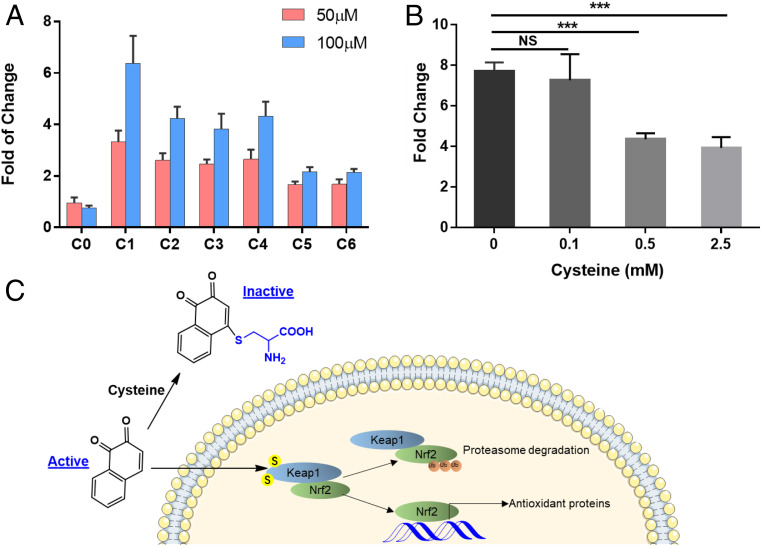
In order to confirm the electrophile-dependent toxicity pathway caused by NSOA, we employed a Nrf2 luciferase reporter cell line stably transfected with the pTA-Nrf2-luciferase reporter vector (55). In agreement with the expression patterns of Nrf2-regulated proteins NQO1 and TXNRD1, Nrf2 response was significantly induced by 6.4 ± 1.1-fold (P < 0.001) in the C1 treatment at 100 µM, but not in the C0 treatment (0.75 ± 0.10-fold) (Fig. 5A). The induction of Nrf2 response decreased along with aging, consistent with the inverted U-shaped curve as observed in proteomics experiments. The results verified Nrf2 reporter cell line as a high-throughput and quantitative way to monitor the toxicities of NSOA.
Fig. 5.
Activation of Nrf2 reporter cells by NSOA is supplemented by cysteine preincubation. (A) Induction of Nrf2 responses by NSOA in Nrf2 Luciferase reporter MCF7 stable cell line. Results are presented as fold relative to vehicle control group (n = 12). P values are <0.001 for all treatments. (B) The induction of Nrf2 responses by NSOA were supplemented by preincubation with free cysteine. Results are presented as fold relative to vehicle control group (n = 6). ***P < 0.001. (C) The schematic figure demonstrates that electrophilic NSOA chemicals (e.g., 1,2-naphthoquinone) are sequestered by free cysteine during preincubation, and hence reduce the activation of Nrf2 pathways inside the Nrf2 reporter cells.
To elucidate whether electrophilic reaction is responsible for the activation of Nrf2 pathway, NSOA was preincubated with free cysteine before it was dosed to Nrf2 luciferase reporter cells. The Nrf2 responses induced by NSOA were significantly suppressed by cysteine preincubation, and the induction was reduced from 7.7 ± 0.5- to 3.9 ± 0.5-fold (2.5 mM of cysteine) by 50.9% (P < 0.001, Fig. 5B). These results confirmed our hypothesis that electrophilic chemical species in NSOA are responsible for the activation of Nrf2 pathway, and these species are sequestered by free cysteine during preincubation (Fig. 6C). Electrophilic attack of cellular thiols has been reported for quinones (23) and endogenous electrophilic lipids via Michael addition (55). Interestingly, antiinflammatory effects of electrophilic lipids have been documented, which is attributed to the activation of Nrf2 pathway by covalently modifying the cysteines on Keap1 or NF-κB (55). This pattern is consistent with the impacts of NSOA on BEAS-2B cells as mentioned above. Thus, we conclude that electrophilic chemicals are responsible for the ROS-independent toxicities of NSOA, by attacking cysteine residues on cellular proteome at the posttranslation level.
Fig. 6.
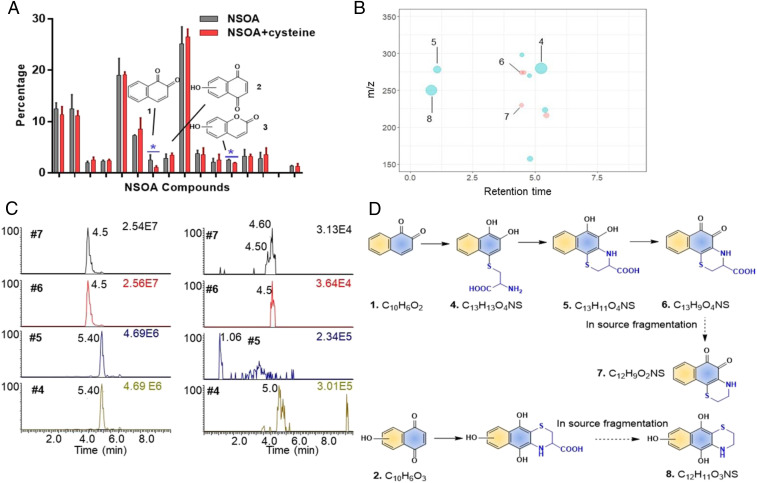
Nontargeted identification of unsaturated carbonyls as the cysteine reactive components. (A) Percentage of the 16 high abundant constituents of NSOA before and after incubation with cysteine. Only the abundances of chemical 1 and 3 were reduced significantly. (B) Cysteine adducts were detected after incubation with cysteine. The size of each dot is proportional to the peak intensity. Red represents peaks detected under ESI−, while blue represents peaks detected under ESI+. The x axis represents the retention time of cysteine adducts on C18 column in UPLC. (C) Authentic standard was used to confirm chemicals 4–7 as the cysteine adducts of 1,2-naphthoquinone (rt = 4.50 min for adduct 7) and 1,4-naphthoquinone (rt = 4.60 min for adduct 7). Left panel shows the chromatograms of cysteine adducts of 1,2-naphthequinone from authentic standards (10 µM). Right panel shows the chromatograms of cysteine adducts of 1,2-naphthequinone NSOA. Notice that adducts 4 and 5 were detected from authentic standards with the same m/z, due to the oxidation during ionization process under ESI+. (D) Proposed reaction pathways of unsaturated carbonyls 1 and 2 with cysteine.
To further confirm the posttranslational alkylation of cellular proteome by NSOA, we first tried to directly monitor the alkylated peptides by “dependent-peptide” search algorithm implemented in the MaxQuant. Only 32 peptides were detected with cysteine modifications, but only 5 of them were detected with the modification masses greater than 40 Da. As the dependent-peptide search algorithm is ineffective to identify low abundant alkylated proteins (56), we decided to employ chemical proteomics methods to monitor the alkylated proteins. To achieve this, a chemical probe of ethynyl-naphthalene SOA was used to covalently label the protein targets alkylated by NSOA on the proteome-wide level (see method in SI Appendix). In brief, the ethynyl-naphthalene probe was subjected to atmospheric oxidation to mimic NSOA, but with an additional ethynyl moiety. BEAS-2B cells were exposed to ethynyl-NSOA, and proteins alkylated by ethynyl-NSOA were further linked to fluorophore via copper-catalyzed click chemistry (57), followed by in-gel fluorescence imaging (workflow was described in SI Appendix, Fig. S10A). Hundreds of alkylated proteins were observed in BEAS-2B cells after exposure to ethynyl-NSOA (SI Appendix, Fig. S10B), which confirmed our hypothesis that numerous cellular proteins are alkylated by electrophilic NSOA compounds. Consistent with the Nrf2 results, more alkylated proteins were detected in cells exposed to C1 than those of C2–C6. To further confirm whether cysteine residuals are responsible for this alkylation, we employed 1,4-naphthoquinone, known to readily alkylate cellular thiols, to compete with ethynyl-NSOA (workflow was shown in SI Appendix, Fig. S11). The protein bands labeled by ethynyl-NSOA probes were abolished in a dose–response manner after preincubation with 1,4-naphthoquinone (SI Appendix, Fig. S11). The results validated cysteine residuals as the primary sites attacked by NSOA. Together with the MT1X, Nrf2-reporter assay, and ethynyl-NSOA probe, we confirmed that NSOA induced oxidative stress in BEAS-2B cells by alkylating cellular proteome. While protein thiols were revealed as the primary sites attacked by NSOA, previous studies have reported the reactions of electrophilic compounds with other amino acids, such as lysine and tyrosine (58). Future studies are warranted to investigate potential electrophilic attack on other amino acids by NSOA.
Nontarget Identification of Unsaturated Carbonyls as the Major Toxic Components.
In order to identify the constituents in NSOA that act as electrophiles to induce toxicities, we employed nontargeted analysis (59) with LC-HRMS to determine the cysteine-NSOA adducts. After incubation with cysteine, naphthoquinone 1 showed the most pronounced decrease by 57.6% (P = 0.007), while other higher abundant chemicals remain unchanged (Fig. 6A). Another chemical 3 with unsaturated carbonyl moiety also showed decrease in abundance, albeit with lesser extent (23.4%, P = 0.001). Nontarget analysis revealed the production of 11 new cysteine adducts from the incubation of NSOA with cysteine (Fig. 6B), among >20,000 background features. Elemental compositions and structures were successfully assigned to five of the cysteine adducts including two most abundant features 8 (peak intensity = 3.8 × 105) and 4 (peak intensity = 5.2 × 105). In agreement with the decrease in naphthoquinone 1, the most abundant feature 4 at retention time of 5.3 min and m/z = 280.0637 was predicted as the cysteine adduct of naphthoquinone 1 via Michael addition (C13H13O4NS, mass error = −0.38 ppm) (Fig. 6D). Similarly, features 5 and 6 were predicted as ring-closing products of feature 4, via reaction with primary amine on cysteine. Feature 7 was assigned as an in-source fragment of feature 6 during the electrospray ionization (ESI) process, considering their similar retention times and a loss of carboxylic acid group, which is commonly fragmented under ESI (60). While the cysteine adduct of chemical 3 was not detected, possibly due to its lower abundance, feature 8 was assigned as the cysteine adduct of another unsaturated carbonyl compound 2, hydroxylated naphthoquinone. The nontarget analysis results demonstrated that unsaturated carbonyls in NSOA are responsible for the cysteine reaction. This is consistent with the chemical properties of this class of compounds by reacting with thiol group on cellular proteome via Michael addition (61).
To further confirm the nontarget analysis results, we incubated the authentic standards of 1,2-naphthoquinone and 1,4-naphthoquinone with free cysteine. The products 4–7 were validated as the cysteine adducts of 1,2-naphthoquinone, as well as 1,4-naphthoquinone, by comparing the exact masses and retention times (Fig. 6C and SI Appendix, Fig. S12). The identification of unsaturated carbonyl compounds 1 (cluster II), 2 (cluster II), and 3 (cluster III) as major cysteine reactive chemicals, is also consistent with the inverted U-shape pattern that short-lived chemicals are responsible for the toxicities of NSOA.
We further employed glutathione (GSH), the physiologically relevant antioxidant, to identify thiol reactive chemicals from NSOA. The two chemicals 1 and 2 reactive toward free cysteines were also identified as the predominant GSH adducts (SI Appendix, Fig. S13). The identities of adducts 8–12 were confirmed as 1,4-naphthoquinone adducts by authentic standard, due to its higher reaction rates with GSH compared to 1,2- naphthoquinone (SI Appendix, Fig. S13B). Notably, the adducts of three other unsaturated carbonyls, 13, 14, and 15, were also identified, albeit with lower abundances. Together with the results from both cysteine and GSH incubation experiments, we conclude that short-lived unsaturated carbonyls are responsible for the proteome-wide impacts of NSOA, by forming Michael addition cysteine adducts with cellular proteome.
Conclusions
Characterizing the chemical components of SOA responsible for toxicities is a long-standing question. PAHs, in part, are regarded as a class of persistent environmental toxicants known for the carcinogenicity and mutagenicity (10, 62). Here, we focus on the aging of the most abundant PAH, naphthalene (14), and propose a robust strategy by combining proteomics, reporter cells, and nontarget analysis, to identify toxic components in SOA upon atmospheric aging. Our study reveals a unique toxicity pathway of NSOA via activating the Nrf2 pathway, while suppressing other pathways including inflammation, which is different from the ROS-mediated toxicities well studied for other SOA systems by activating Nrf2 and inflammation pathways. The detection of short-lived unsaturated carbonyl compounds as the toxic components in NSOA is significant at two different levels: The unique toxicity pathway of NSOA is partially due to its electron-rich aromatic rings to produce unsaturated carbonyls after reaction with OH radicals; these unsaturated carbonyls in NSOA are early-generation oxidation products but undergo oxidative decay on a timescale equivalent to 1 d of atmospheric photooxidation. As the lifetime of SOA is normally couple of days in urban area, the higher toxicities of fresh NSOA indicates potential stronger adverse health impacts in urban area with high population.
Up to 90% of PM organic constituents in urban areas are identified to be SOA (4). Such omics-based technique can be widely applied on a variety of SOA formed from different atmospheric aging conditions to study their detailed toxicological impacts upon inhalation so that it can provide evidence for mitigation of emission sources. One of the limitations for the current study is that only the Nrf2 pathway was confirmed to be caused by unsaturated carbonyls, but not for other suppressed pathways especially inflammation. Although the activation of Nrf2 and suppression of other pathways seems to be consistently described by the inverted U-shape, which indicates that the same compounds may be responsible for these toxicities. Another limitation is that the proteomic profile generated from in vitro exposure might be a simplified indicator of mechanism related to adverse health outcomes, while health impacts revealed by in vivo exposure to air pollutants should be further investigated in the future.
Materials and Methods
Chemicals and Reagents.
The reagents, solvents, and standards used are listed in SI Appendix.
NSOA Collection and Extraction.
To generate NSOA, purified air was passed through a glass container with naphthalene into a custom-built 10-L quartz tube for oxidation (63). O3 was produced by passing oxygen (99.6%; Linde) through an O3 generator (UVP) prior to the introduction into the flow tube. Inside the flow tube, gas-phase OH radicals were generated from UV photolysis of O3, which produced O1(D) that subsequently reacted with H2O vapor. The aerosol residence time within the flow tube was controlled to be around 300 s, with relative humidity (RH) and temperature in the flow tube monitored by an Omega HX94C RH/T transmitter. Blank experiments were conducted with naphthalene with the UV lights switched off. More details about the experimental setup can be found in our previous work (22).
The OH exposure, defined as the integrated product of OH radical concentration ([OH]) and residence time (t) inside the flow tube (64), was quantified by measuring the decay of gas-phase hexane using gas chromatography–flame ionization detector (GC-FID; SRI 8610C) equipped with a Tenax TA trap. By measuring the steady-state concentration of hexane before ([I]0) and after ([I]t) reaction with OH radical inside the flow tube, the OH exposure can be calculated as follows:
| [1] |
where is the second-order reaction rate constant between hexane and OH radicals (5.2 × 1012 cm3⋅molecule−1⋅s−1) (65). In this study, six different OH exposure conditions (3.3 × 1010 to 2.07 × 1011 molecules⋅cm−3⋅s) were achieved by varying the mixing ratios of O3 and water vapor. Based on an assumption of the averaged daily atmospheric OH radical concentration of 1.5 × 106 molecules⋅cm−3 (66), the range in equivalent ambient OH exposure was calculated to be 0 to 2 d. The mass concentration of SOA was derived by assuming a particle density of 1.5 g⋅cm−3 (12). SOA yields are expressed as the mass fraction of SOA formed relative to the mass of naphthalene reacted. Particles were collected on 47-mm quartz filters, and more details about online particle monitoring and filter extraction can be found in SI Appendix.
Characterizing NSOA Components by LC-Orbitrap, GC-MS, and Ion Mobility Spectrometry–Time-of-Flight–Mass Spectrometry.
To profile the chemical components of NSOA and predict corresponding structures, three mass spectrometric methods were combined (SI Appendix, Fig. S1). First, LC-Orbitrap high-resolution mass spectrometry (LC-HRMS) is used for nontargeted profiling of NSOA compounds. Exact masses of each individual component were determined by nontarget analysis (detailed are provided in SI Appendix), and corresponding elemental compositions were calculated within a mass tolerance of 3 ppm. All of the molecular formulas identified were categorized into four clusters by hierarchical clustering according to their timeseries as a function of the six atmospheric aging conditions. For each cluster, four components with high abundance were compared against GC-MS and ion mobility spectrometry–time-of-flight–mass spectrometry (IMS-TOF-MS) analysis to predict their molecular structures. Compounds were identified using GC-MS and the structures are confirmed by matching retention indices and EI mass spectra (NIST08). The structures are used to support the molecular formulas identified by LC-HRMS. Furthermore, collision cross-sections (CCS) were calculated from ion mobility measurements by IMS-TOF-MS (67) to further exclude isomers for structure prediction (SI Appendix, Fig. S2). Comparison of predicted CCS versus IMS-measured CCS can be found in SI Appendix, Fig. S4 and shows good agreement, further confirming the proposed structures. Details about the instrumentation are provided in SI Appendix.
BEAS-2B Cell Culture and Exposure.
BEAS-2B cells were seeded onto 10-cm Petri dishes and cultured with bronchial epithelial cell growth medium in humidified CO2 incubator at 37 °C. When the cells reached 70 to 80% confluence, growth medium was removed and replaced with 8 mL of exposure medium alongside 100 µM NSOA. The control group was treated with an equal amount of vehicle solvent (0.1% DMSO). After a 24-h exposure at 37 °C in the CO2 incubator, the exposure medium was removed by aspiration and 2 mL of cold phosphate-buffered saline (PBS) (Sigma) was used to wash the cells twice. The cells were collected with 1 mL of cold PBS on ice. After 5-min centrifugation at 3,000 × g, the supernatant was discarded and the cell pellet was stored in −80 °C for future experiments. In order to evaluate cytotoxicity upon NSOA exposure, the MTT Cell Viability Assay Kit (Biotium) was used to assess BEAS-2B cell viability according to the manufacturer’s protocol (details are provided in SI Appendix). Three biological replicates of each treatment were conducted by dosing NSOA extracts derived from the same OH exposure condition parallelly into different Petri dishes, for cytotoxicity and proteomics analysis.
Proteome Extraction and LC-MS/MS Analysis.
The proteome from BEAS-2B cells was extracted with 300 μL of lysis buffer (500 mM NaCl, 20 mM Hepes, 0.1% Nonidet P-40, protease inhibitor mixture, pH 7.4) according to previous studies (42). The details of protein extraction, digestion, and isotopic labeling are provided in SI Appendix.
For proteome analysis, samples were processed using the Q Exactive HF-X Hybrid Quadrupole-Orbitrap Mass Spectrometer (Thermo Fisher Scientific) equipped with an EASY-nLC 1200 nano-flow UHPLC (Thermo Fisher Scientific). Three microliters of each sample were loaded onto an in-house packed capillary column (inner diameter 75 μm × 10 cm) with luna C18 sorbent (3 μm, 100 Å; Phenomenex) for analytical separations of peptides. Details of LC gradients and mass spectrometry settings are provided in SI Appendix.
Exposure of Nrf2 Reporter Cell Line to NSOA.
In this study, a Nrf2 reporter cell line stably transfected with luciferase (Signosis) was used to assess the induction of Nrf2 pathway by NSOA. High-glucose Dulbecco’s modified Eagle’s medium (Sigma) supplemented with 10% (vol/vol) fetal bovine serum (FBS), 1% antibiotic antimycotic solution, and 75 mg/mL G418 was used as growth medium and the exposure medium had 0.1% (vol/vol) FBS (68). Details of cell culture and exposure are provided in SI Appendix.
To further investigate the mechanism of NSOA to induce Nrf2 responses, preincubating 100 μM NSOA from C1 condition with increasing concentrations of free cysteine (0.1 to 2.5 mM) at room temperature in dark for 4 h, was used to detect any alleviation of the Nrf2 responses. The mixture was then dosed to Nrf2 reporter cells for a 24-h incubation. Reduced Nrf2 responses toward NSOA was monitored as described in SI Appendix.
Nontargeted Identification of Thiol Reactive Chemicals.
Stock solution of C1 was diluted to a final concentration of 10 μM in 100 μL of HPLC water. One hundred micromolar cysteine or GSH was spiked into the solution, followed by incubation on the shaker at room temperature for 2 h (n = 3). For the control, no cysteine or GSH was added. Subsequently, the samples were centrifuged for 10 min at 14,000 × g and subjected to LC Orbitrap analysis. Nontarget analysis was conducted to determine the cysteine or GSH adducts, by comparing the features across treatments, control and cysteine/GSH solutions. Eleven features were detected in incubated samples with cysteines, significantly (P < 0.05) higher (FC > 10) than those in control and cysteine solutions. Elemental compositions and structures were assigned for 5 of the 11 detected cysteine adducts according to exact masses within a mass tolerance of 3 ppm. Twenty-five features were detected in C1 incubated with GSH, significantly (P < 0.05) higher (FC > 10) than those in control and GSH solutions. Elemental compositions and structures were assigned for 10 of the 25 detected GSH adducts according to exact masses within a mass tolerance of 3 ppm. Details about the instrumentation are provided in SI Appendix.
Statistical Analyses.
Statistical analyses were performed mainly via GraphPad Prism (v7.0.4; GraphPad Software) or R studio (v1.1.456; RStudio). Soft clustering was conducted with the Mfuzz R package (69). The peak intensities of NSOA chemicals were standardized with the default parameters for which mean value was set to 0 and the SD was set to 1. Soft clustering was implemented with four cluster centers (c = 4) and a fuzzification parameter of 1.2 (m = 1.2). The FASTA file of Homo sapiens was obtained from the UniProt database. MaxQuant algorithms (https://maxquant.org/; version 1.6.2.3) were utilized to process the proteomic raw files to identify and quantify the proteins. As for laboratory quantitative proteomics data analysis, considering the small number of replicates (n = 3), unadjusted P values (P < 0.05) and effect size cutoffs (FC > 1.5) were used to ascertain significantly changed proteins as widely employed for quantitative proteomics studies (70–76). More details about pathway enrichment analysis and nontarget analysis are provided in SI Appendix.
Supplementary Material
Acknowledgments
This research was supported by the National Sciences and Engineering Research Council (NSERC) Discovery Grant and Ontario Early Research Award. We acknowledge the support of instrumentation grants from the Canada Foundation for Innovation, the Ontario Research Fund, and the NSERC Research Tools and Instrument Grant.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. U.P. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2001378117/-/DCSupplemental.
Data Availability.
The raw mass spectrometry files of proteomics generated during this study have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the identifier PXD019348. The list of significantly changed proteins is provided in Dataset S1. Other relevant data have been included in the text and SI Appendix.
References
- 1.Burnett R. et al., Global estimates of mortality associated with long-term exposure to outdoor fine particulate matter. Proc. Natl. Acad. Sci. U.S.A. 115, 9592–9597 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cohen A. J. et al., Estimates and 25-year trends of the global burden of disease attributable to ambient air pollution: An analysis of data from the Global Burden of Diseases Study 2015. Lancet 389, 1907–1918 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dockery D. W. et al., An association between air pollution and mortality in six U.S. cities. N. Engl. J. Med. 329, 1753–1759 (1993). [DOI] [PubMed] [Google Scholar]
- 4.Hallquist M. et al., The formation, properties and impact of secondary organic aerosol: Current and emerging issues. Atmos. Chem. Phys. 9, 5155–5236 (2009). [Google Scholar]
- 5.Valavanidis A., Vlachogianni T., Fiotakis K., Loridas S., Pulmonary oxidative stress, inflammation and cancer: Respirable particulate matter, fibrous dusts and ozone as major causes of lung carcinogenesis through reactive oxygen species mechanisms. Int. J. Environ. Res. Public Health 10, 3886–3907 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saxena P., Hildemann L. M., Water-soluble organics in atmospheric particles: A critical review of the literature and application of thermodynamics to identify candidate compounds. J. Atmos. Chem. 24, 57–109 (1996). [Google Scholar]
- 7.Jiang H., Jang M., Dynamic oxidative potential of atmospheric organic aerosol under ambient sunlight. Environ. Sci. Technol. 52, 7496–7504 (2018). [DOI] [PubMed] [Google Scholar]
- 8.Tuet W. Y. et al., Chemical oxidative potential of secondary organic aerosol (SOA) generated from the photooxidation of biogenic and anthropogenic volatile organic compounds. Atmos. Chem. Phys. 17, 839–853 (2017). [Google Scholar]
- 9.Jia C., Batterman S., A critical review of naphthalene sources and exposures relevant to indoor and outdoor air. Int. J. Environ. Res. Public Health 7, 2903–2939 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim K. H., Jahan S. A., Kabir E., Brown R. J., A review of airborne polycyclic aromatic hydrocarbons (PAHs) and their human health effects. Environ. Int. 60, 71–80 (2013). [DOI] [PubMed] [Google Scholar]
- 11.Preuss R., Angerer J., Drexler H., Naphthalene—an environmental and occupational toxicant. Int. Arch. Occup. Environ. Health 76, 556–576 (2003). [DOI] [PubMed] [Google Scholar]
- 12.Chan A. W. H. et al., Secondary organic aerosol formation from photooxidation of naphthalene and alkylnaphthalenes: Implications for oxidation of intermediate volatility organic compounds (IVOCs). Atmos. Chem. Phys. 9, 3049–3060 (2009). [Google Scholar]
- 13.Zhou X. et al., Volatile organic compounds in a typical petrochemical industrialized valley city of northwest China based on high-resolution PTR-MS measurements: Characterization, sources and chemical effects. Sci. Total Environ. 671, 883–896 (2019). [DOI] [PubMed] [Google Scholar]
- 14.Huang G. et al., Potentially important contribution of gas-phase oxidation of naphthalene and methylnaphthalene to secondary organic aerosol during haze events in Beijing. Environ. Sci. Technol. 53, 1235–1244 (2019). [DOI] [PubMed] [Google Scholar]
- 15.Chowdhury P. H. et al., Exposure of lung epithelial cells to photochemically aged secondary organic aerosol shows increased toxic effects. Environ. Sci. Technol. Lett. 5, 424–430 (2018). [Google Scholar]
- 16.Chowdhury P. H. et al., Connecting the oxidative potential of secondary organic aerosols with reactive oxygen species in exposed lung cells. Environ. Sci. Technol. 53, 13949–13958 (2019). [DOI] [PubMed] [Google Scholar]
- 17.Tong H. et al., Radical formation by fine particulate matter associated with highly oxygenated molecules. Environ. Sci. Technol. 53, 12506–12518 (2019). [DOI] [PubMed] [Google Scholar]
- 18.Tong H. et al., Reactive oxygen species formed by secondary organic aerosols in water and surrogate lung fluid. Environ. Sci. Technol. 52, 11642–11651 (2018). [DOI] [PubMed] [Google Scholar]
- 19.Tong H. et al., Hydroxyl radicals from secondary organic aerosol decomposition in water. Atmos. Chem. Phys. 15, 1761–1771 (2016). [Google Scholar]
- 20.Yamazaki I., Piette L. H., EPR spin-trapping study on the oxidizing species formed in the reaction of the ferrous ion with hydrogen peroxide. J. Am. Chem. Soc. 113, 7588–7593 (1991). [Google Scholar]
- 21.McWhinney R., Zhou S., Abbatt J. P. D., Naphthalene SOA: Redox activity and naphthoquinone gas-particle partitioning. Atmos. Chem. Phys. 13, 9731–9744 (2013). [Google Scholar]
- 22.Wang S. et al., Relationship between chemical composition and oxidative potential of secondary organic aerosol from polycyclic aromatic hydrocarbons. Atmos. Chem. Phys. 18, 3987–4003 (2018). [Google Scholar]
- 23.Wang X. et al., Mechanism of arylating quinone toxicity involving Michael adduct formation and induction of endoplasmic reticulum stress. Proc. Natl. Acad. Sci. U.S.A. 103, 3604–3609 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martyniuk C. J., Are we closer to the vision? A proposed framework for incorporating omics into environmental assessments. Environ. Toxicol. Pharmacol. 59, 87–93 (2018). [DOI] [PubMed] [Google Scholar]
- 25.Lin Y. H. et al., Isoprene-derived secondary organic aerosol induces the expression of oxidative stress response genes in human lung cells. Environ. Sci. Technol. Lett. 3, 250–254 (2016). [Google Scholar]
- 26.Ahmed C. M. S. et al., Exposure to dimethyl selenide (DMSe)-derived secondary organic aerosol alters transcriptomic profiles in human airway epithelial cells. Environ. Sci. Technol. 53, 14660–14669 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu Y., Beyer A., Aebersold R., On the dependency of cellular protein levels on mRNA abundance. Cell 165, 535–550 (2016). [DOI] [PubMed] [Google Scholar]
- 28.Lambe A. T. et al., Effect of oxidant concentration, exposure time, and seed particles on secondary organic aerosol chemical composition and yield. Atmos. Chem. Phys. 15, 3063–3075 (2015). [Google Scholar]
- 29.Lambe A. T. et al., Transitions from functionalization to fragmentation reactions of laboratory secondary organic aerosol (SOA) generated from the OH oxidation of alkane precursors. Environ. Sci. Technol. 46, 5430–5437 (2012). [DOI] [PubMed] [Google Scholar]
- 30.Liu T. et al., Comparison of secondary organic aerosol formation from toluene on initially wet and dry ammonium sulfate particles at moderate relative humidity. Atmos. Chem. Phys. 18, 5677–5689 (2018). [Google Scholar]
- 31.Chacon-Madrid H. J., Donahue N. M., Fragmentation vs. functionalization: Chemical aging and organic aerosol formation. Atmos. Chem. Phys. 11, 10553–10563 (2011). [Google Scholar]
- 32.Lambe A. T. et al., Relationship between oxidation level and optical properties of secondary organic aerosol. Environ. Sci. Technol. 47, 6349–6357 (2013). [DOI] [PubMed] [Google Scholar]
- 33.Jimenez J. L. et al., Evolution of organic aerosols in the atmosphere. Science 326, 1525–1529 (2009). [DOI] [PubMed] [Google Scholar]
- 34.Kroll J. H., Lim C. Y., Kessler S. H., Wilson K. R., Heterogeneous oxidation of atmospheric organic aerosol: Kinetics of changes to the amount and oxidation state of particle-phase organic carbon. J. Phys. Chem. A 119, 10767–10783 (2015). [DOI] [PubMed] [Google Scholar]
- 35.Kautzman K. E. et al., Chemical composition of gas- and aerosol-phase products from the photooxidation of naphthalene. J. Phys. Chem. A 114, 913–934 (2010). [DOI] [PubMed] [Google Scholar]
- 36.Lambe A. T. et al., Characterization of aerosol photooxidation flow reactors: Heterogeneous oxidation, secondary organic aerosol formation and cloud condensation nuclei activity measurements. Atmos. Meas. Tech. 4, 445–461 (2011). [Google Scholar]
- 37.Riva M., Robinson E. S., Perraudin E., Donahue N. M., Villenave E., Photochemical aging of secondary organic aerosols generated from the photooxidation of polycyclic aromatic hydrocarbons in the gas-phase. Environ. Sci. Technol. 49, 5407–5416 (2015). [DOI] [PubMed] [Google Scholar]
- 38.Monks T. J., Hanzlik R. P., Cohen G. M., Ross D., Graham D. G., Quinone chemistry and toxicity. Toxicol. Appl. Pharmacol. 112, 2–16 (1992). [DOI] [PubMed] [Google Scholar]
- 39.Dellinger B. et al., Role of free radicals in the toxicity of airborne fine particulate matter. Chem. Res. Toxicol. 14, 1371–1377 (2001). [DOI] [PubMed] [Google Scholar]
- 40.O’Brien P. J., Molecular mechanisms of quinone cytotoxicity. Chem. Biol. Interact. 80, 1–41 (1991). [DOI] [PubMed] [Google Scholar]
- 41.Vejerano E. P., Rao G., Khachatryan L., Cormier S. A., Lomnicki S., Environmentally persistent free radicals: Insights on a new class of pollutants. Environ. Sci. Technol. 52, 2468–2481 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Peng H. et al., An unbiased chemical proteomics method identifies FabI as the primary target of 6-OH-BDE-47. Environ. Sci. Technol. 50, 11329–11336 (2016). [DOI] [PubMed] [Google Scholar]
- 43.Boersema P. J., Raijmakers R., Lemeer S., Mohammed S., Heck A. J., Multiplex peptide stable isotope dimethyl labeling for quantitative proteomics. Nat. Protoc. 4, 484–494 (2009). [DOI] [PubMed] [Google Scholar]
- 44.Liu Q. et al., Oxidative and toxicological evolution of engineered nanoparticles with atmospherically relevant coatings. Environ. Sci. Technol. 53, 3058–3066 (2019). [DOI] [PubMed] [Google Scholar]
- 45.Lin Y. H. et al., Gene expression profiling in human lung cells exposed to isoprene-derived secondary organic aerosol. Environ. Sci. Technol. 51, 8166–8175 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Warner H., Wilson B. J., Caswell P. T., Control of adhesion and protrusion in cell migration by Rho GTPases. Curr. Opin. Cell Biol. 56, 64–70 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tong L., Tergaonkar V., Rho protein GTPases and their interactions with NFκB: Crossroads of inflammation and matrix biology. Biosci. Rep. 34, 283–295 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tonelli C., Chio I. I. C., Tuveson D. A., Transcriptional regulation by Nrf2. Antioxid. Redox Signal. 29, 1727–1745 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bates J. T. et al., Review of acellular assays of ambient particulate matter oxidative potential: Methods and relationships with composition, sources, and health effects. Environ. Sci. Technol. 53, 4003–4019 (2019). [DOI] [PubMed] [Google Scholar]
- 50.Gaschen A. et al., Cellular responses after exposure of lung cell cultures to secondary organic aerosol particles. Environ. Sci. Technol. 44, 1424–1430 (2010). [DOI] [PubMed] [Google Scholar]
- 51.Ye J. et al., Development of a novel simulation reactor for chronic exposure to atmospheric particulate matter. Sci. Rep. 7, 42317 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Si M., Lang J., The roles of metallothioneins in carcinogenesis. J. Hematol. Oncol. 11, 107 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Paulsen C. E., Carroll K. S., Cysteine-mediated redox signaling: Chemistry, biology, and tools for discovery. Chem. Rev. 113, 4633–4679 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.van der Reest J., Lilla S., Zheng L., Zanivan S., Gottlieb E., Proteome-wide analysis of cysteine oxidation reveals metabolic sensitivity to redox stress. Nat. Commun. 9, 1581 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schopfer F. J., Cipollina C., Freeman B. A., Formation and signaling actions of electrophilic lipids. Chem. Rev. 111, 5997–6021 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tyanova S., Temu T., Cox J., The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 11, 2301–2319 (2016). [DOI] [PubMed] [Google Scholar]
- 57.Speers A. E., Cravatt B. F., Profiling enzyme activities in vivo using click chemistry methods. Chem. Biol. 11, 535–546 (2004). [DOI] [PubMed] [Google Scholar]
- 58.Shannon D. A., Weerapana E., Covalent protein modification: The current landscape of residue-specific electrophiles. Curr. Opin. Chem. Biol. 24, 18–26 (2015). [DOI] [PubMed] [Google Scholar]
- 59.Peng H. et al., Untargeted identification of organo-bromine compounds in lake sediments by ultrahigh-resolution mass spectrometry with the data-independent precursor isolation and characteristic fragment method. Anal. Chem. 87, 10237–10246 (2015). [DOI] [PubMed] [Google Scholar]
- 60.Xu Y. F., Lu W., Rabinowitz J. D., Avoiding misannotation of in-source fragmentation products as cellular metabolites in liquid chromatography-mass spectrometry-based metabolomics. Anal. Chem. 87, 2273–2281 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Khatik G. L., Kumar R., Chakraborti A. K., Catalyst-free conjugated addition of thiols to α,β-unsaturated carbonyl compounds in water. Org. Lett. 8, 2433–2436 (2006). [DOI] [PubMed] [Google Scholar]
- 62.Finlayson-Pitts B. J., Pitts J. N. Jr., Tropospheric air pollution: Ozone, airborne toxics, polycyclic aromatic hydrocarbons, and particles. Science 276, 1045–1052 (1997). [DOI] [PubMed] [Google Scholar]
- 63.Ye J., Gordon C. A., Chan A. W. H., Enhancement in secondary organic aerosol formation in the presence of preexisting organic particle. Environ. Sci. Technol. 50, 3572–3579 (2016). [DOI] [PubMed] [Google Scholar]
- 64.Smith J. et al., The heterogeneous reaction of hydroxyl radicals with sub-micron squalane particles: A model system for understanding the oxidative aging of ambient aerosols. Atmos. Chem. Phys. 9, 3209–3222 (2009). [Google Scholar]
- 65.Atkinson R., Arey J., Atmospheric degradation of volatile organic compounds. Chem. Rev. 103, 4605–4638 (2003). [DOI] [PubMed] [Google Scholar]
- 66.Mao J. et al., Airborne measurement of OH reactivity during INTEX-B. Atmos. Chem. Phys. 9, 163–173 (2009). [Google Scholar]
- 67.Zhang X. et al., A novel framework for molecular characterization of atmospherically relevant organic compounds based on collision cross section and mass-to-charge ratio. Atmos. Chem. Phys. 16, 12945–12959 (2016). [Google Scholar]
- 68.Sun J. et al., Identification of chemicals that cause oxidative stress in oil sands process-affected water. Environ. Sci. Technol. 51, 8773–8781 (2017). [DOI] [PubMed] [Google Scholar]
- 69.Kumar L., E Futschik M., Mfuzz: A software package for soft clustering of microarray data. Bioinformation 2, 5–7 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pascovici D., Handler D. C., Wu J. X., Haynes P. A., Multiple testing corrections in quantitative proteomics: A useful but blunt tool. Proteomics 16, 2448–2453 (2016). [DOI] [PubMed] [Google Scholar]
- 71.Di Bartolomeo F. et al., Absolute yeast mitochondrial proteome quantification reveals trade-off between biosynthesis and energy generation during diauxic shift. Proc. Natl. Acad. Sci. U.S.A. 117, 7524–7535 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Leung K. K. et al., Broad and thematic remodeling of the surfaceome and glycoproteome on isogenic cells transformed with driving proliferative oncogenes. Proc. Natl. Acad. Sci. U.S.A. 117, 7764–7775 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schmidt N. et al., An influenza virus-triggered SUMO switch orchestrates co-opted endogenous retroviruses to stimulate host antiviral immunity. Proc. Natl. Acad. Sci. U.S.A. 116, 17399–17408 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gao X., Yeom J., Groisman E. A., The expanded specificity and physiological role of a widespread N-degron recognin. Proc. Natl. Acad. Sci. U.S.A. 116, 18629–18637 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Reich S. et al., A multi-omics analysis reveals the unfolded protein response regulon and stress-induced resistance to folate-based antimetabolites. Nat. Commun. 11, 2936 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang C. et al., Integrated omics in Drosophila uncover a circadian kinome. Nat. Commun. 11, 2710 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw mass spectrometry files of proteomics generated during this study have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the identifier PXD019348. The list of significantly changed proteins is provided in Dataset S1. Other relevant data have been included in the text and SI Appendix.