Significance
Alzheimer’s disease (AD) has complex pathological mechanisms. Our limited understanding of the molecular and cellular changes in AD hindered the identification of therapeutic targets. In this study, we profiled 169,496 nuclei from the prefrontal cortical samples of AD patients and healthy controls by single-nucleus RNA sequencing. The results indicated that there is a loss of neuroprotective glial cells in AD, which may contribute to impaired neurotransmitter recycling in astrocytes, demyelination in oligodendrocytes, and perturbed synaptic pruning in microglia. Moreover, the analysis revealed a role of antigen presentation by angiogenic endothelial cells in AD. Together, our findings offer important insights into the therapeutic potential of targeting glial- and endothelial-specific pathways to restore brain homeostasis in AD.
Keywords: synaptic signaling, synapse, neurodegenerative diseases, myelination, angiogenesis
Abstract
Alzheimer’s disease (AD) is the most common form of dementia but has no effective treatment. A comprehensive investigation of cell type-specific responses and cellular heterogeneity in AD is required to provide precise molecular and cellular targets for therapeutic development. Accordingly, we perform single-nucleus transcriptome analysis of 169,496 nuclei from the prefrontal cortical samples of AD patients and normal control (NC) subjects. Differential analysis shows that the cell type-specific transcriptomic changes in AD are associated with the disruption of biological processes including angiogenesis, immune activation, synaptic signaling, and myelination. Subcluster analysis reveals that compared to NC brains, AD brains contain fewer neuroprotective astrocytes and oligodendrocytes. Importantly, our findings show that a subpopulation of angiogenic endothelial cells is induced in the brain in patients with AD. These angiogenic endothelial cells exhibit increased expression of angiogenic growth factors and their receptors (i.e., EGFL7, FLT1, and VWF) and antigen-presentation machinery (i.e., B2M and HLA-E). This suggests that these endothelial cells contribute to angiogenesis and immune response in AD pathogenesis. Thus, our comprehensive molecular profiling of brain samples from patients with AD reveals previously unknown molecular changes as well as cellular targets that potentially underlie the functional dysregulation of endothelial cells, astrocytes, and oligodendrocytes in AD, providing important insights for therapeutic development.
The global prevalence of Alzheimer’s disease (AD), which is the most common form of dementia, is currently 24 million and is expected to double every 20 years (1). Its pathological hallmarks include the deposition of amyloid-beta peptides and neurofibrillary tangles as well as neuroinflammation (2). However, there is still no effective treatment for AD, which is in part because of the incomplete understanding of the molecular basis of cell type-specific responses during AD pathogenesis, including impaired synaptic functions (3–6), the loss of blood–brain barrier protection, and the loss of neurotrophic support (7–11), respectively.
Along AD progression, the disruption of the molecular pathways of specific cell types can contribute to their observed dysfunctions. Recent studies utilized single-nucleus transcriptome analysis to investigate the transcriptomic changes in AD brains (12–14) and have revealed molecular alterations at the single-cell level using readily available frozen brain tissues. In particular, such studies identified the dysregulated pathways in the most predominant neural cell types, such as neurons and oligodendrocytes, in AD. Although the data from these studies are valuable resources for researchers, some issues remain unresolved. First, the dysregulated pathways in other cell types, particularly endothelial cells, remain unknown. Second, because of cellular heterogeneity, it is unclear whether and how changes in cell subpopulations in AD contribute to the observed dysregulations. Therefore, it is essential to investigate the changes in cellular heterogeneity in AD in order to identify precise cellular targets for AD therapeutic development.
Accordingly, in order to comprehensively investigate the dysregulated molecular pathways in different cell types in AD and determine the cellular targets that contribute to the observed changes, we performed single-nucleus transcriptome analysis of the AD brain by profiling 169,496 nuclei from AD patients and healthy normal control (NC) subjects. Our unbiased transcriptome analysis showed that the AD-related, cell type-specific transcriptomic changes in endothelial cells, astrocytes, and oligodendrocytes are associated with the dysregulation of their respective functions, namely angiogenesis, synaptic signaling, and myelination. Subcluster analysis further identified that the induction of angiogenic endothelial cells and reduction of neuroprotective astrocytes and oligodendrocytes contribute to the transcriptomic changes in AD. In particular, the transcriptomic signature of angiogenic endothelial cells revealed their association with antigen presentation, suggesting that the dysregulated angiogenesis and antigen presentation in endothelial cells potentially contribute to AD pathogenesis. Hence, our comprehensive transcriptome profiling of brain tissues from AD patients provides important insights that could aid both prognostic evaluation and therapeutic targeting of endothelial- or glial-specific pathways in AD.
Results
Single-Nucleus Transcriptome Profiling of the Prefrontal Cortex in AD.
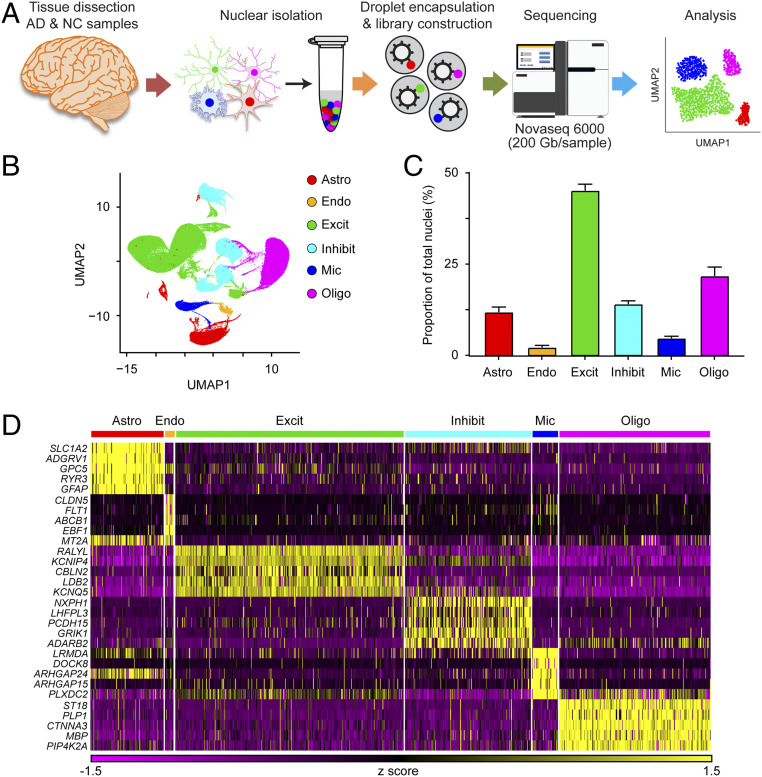
To investigate how the molecular and cellular profiles of brain tissues are altered in patients with AD compared to those in healthy NC subjects, we performed transcriptome analysis of 21 prefrontal cortex tissue samples from patients with AD (n = 12) and NC subjects (n = 9) at the single-cell level by single-nucleus RNA sequencing (snRNA-seq). A diagram of the experimental methodology is shown in Fig. 1A, and the sequencing characteristics of the samples are shown in SI Appendix, Table S1. We sampled 169,496 nuclei: 90,713 and 78,783 nuclei from AD and NC brain samples, respectively. Despite the variations in postmortem delay among samples, we showed that the sample quality—indicated by the mean numbers of transcripts and genes detected per nucleus—did not seem to be affected by the postmortem delay (SI Appendix, Fig. S1A). In addition, these parameters were comparable to those reported in a previous snRNA-seq study (15), confirming the quality of our samples prior to downstream analysis.
Fig. 1.
Single-nucleus transcriptome analysis of the prefrontal cortex in AD. (A) Single-nucleus transcriptome profiling workflow. (B–D) Unbiased identification of cell-type heterogeneity in the human prefrontal cortex. (B) UMAP plot showing the six major cell types isolated from prefrontal cortex. (C) Proportions of cell types among the 169,496 sampled nuclei. (D) Heatmap showing the top five most enriched genes for each cell type. All data are mean ± SEM. See also SI Appendix, Fig. S1 and Tables S1 and S2.
To establish a baseline profile of cell populations, we performed initial unbiased uniform manifold approximation and projection (UMAP) clustering on all 21 samples from both AD and NC subjects. This analysis yielded 43 unique cell clusters (SI Appendix, Fig. S1B), which we subsequently categorized into the following six major cell types according to their individual transcriptome profiles and previously reported cell-type markers: astrocytes (AQP4+, 11.9 ± 1.4% of total nuclei), endothelial cells (CLDN5+, 2.3 ± 0.5%), excitatory neurons (CAMK2A+, 45.2 ± 1.7%), inhibitory neurons (GAD1+, 14.1 ± 0.9%), microglia (C3+, 4.7 ± 0.6%), and oligodendrocytes (MBP+, 21.8 ± 2.5%) (Fig. 1 B and C and SI Appendix, Fig. S1 C and D). In addition to the previously reported cell-type markers, these six major cell types expressed the following unique signature genes, which can serve as novel cell-type markers: ADGRV1, GPC5, and RYR3 were expressed by astrocytes; ABCB1 and EBF1 by endothelial cells; CBLN2 and LDB2 by excitatory neurons; LHFPL3 and PCDH15 by inhibitory neurons; LRMDA and DOCK8 by microglia; and PLP1 and ST18 by oligodendrocytes (Fig. 1D and SI Appendix, Table S2). These results collectively and comprehensively reveal the cell-type heterogeneity in diseased and healthy brain tissues.
Cell Type-Specific Transcriptomic Changes Reveal Dysregulated Molecular Pathways in AD Brains.
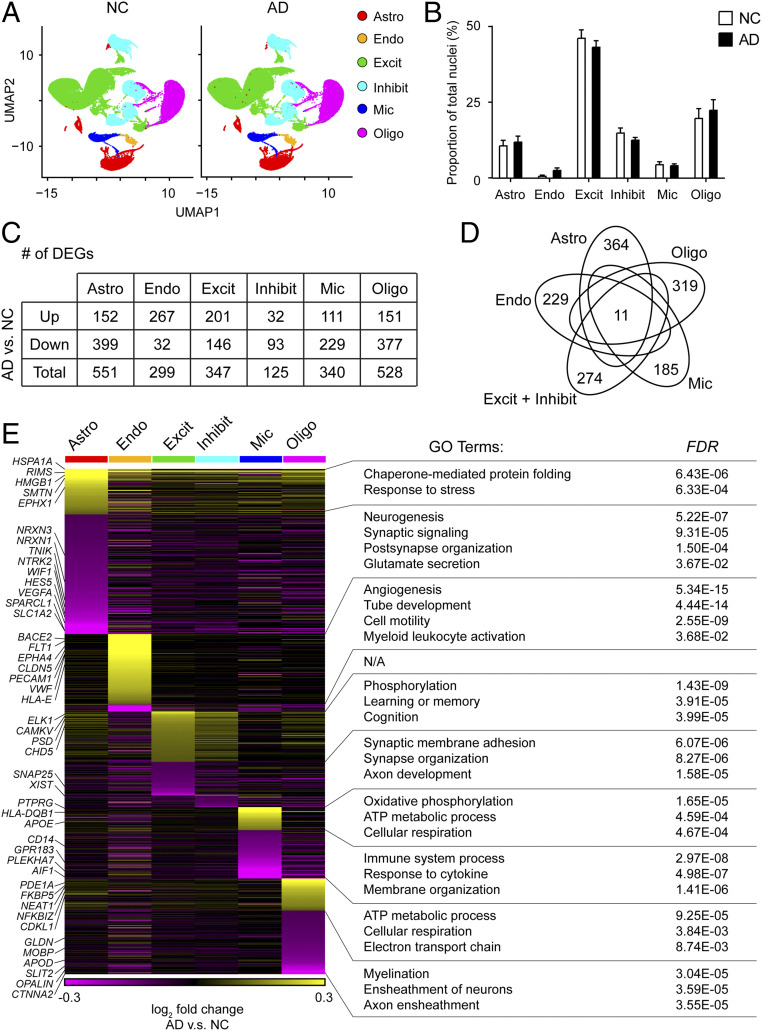
Following our initial cell-type characterization, we compared the proportions of different cell types between AD and NC brain samples. UMAP cluster analysis revealed that the proportions of astrocytes, excitatory neurons, inhibitory neurons, microglia, and oligodendrocytes were similar between the AD and NC brain samples. However, the proportion of endothelial cells was higher in the AD samples than the NC samples (3.0 ± 0.9% vs. 1.2 ± 0.3%, respectively; Fig. 2 A and B).
Fig. 2.
Dysregulated molecular pathways in AD according to cell type-specific transcriptomic changes. UMAP plots (A) and bar plot (B) showing the proportions of the six major cell types found in the AD and NC prefrontal cortical samples. (C and D) AD-associated transcriptomic changes were highly cell type-specific. (C) Numbers of DEGs between AD and NC samples within each cell type (adjusted P < 0.1, log2 fold change ≥ 0.1 or ≤ −0.1). Down: down-regulated; Up: up-regulated. (D) Venn diagram showing the 11 coregulated DEGs among all six cell types. Also shown are the numbers of DEGs specific to each cell type. (E) The cell type-specific transcriptomic changes in AD were associated with distinct molecular pathways. Heatmap showing the expression changes of DEGs in all six cell types in AD samples. Specific GO terms are listed at Right. Astro: astrocytes, Endo: endothelial cells, Excit: excitatory neurons, Inhibit: inhibitory neurons, Mic: microglia, and Oligo: oligodendrocytes. All data are mean ± SEM. See also SI Appendix, Figs. S2 and S4 and Table S3.
To examine the global transcriptomic changes in individual brain cell types in AD, we compared the individual cell-type transcriptome profiles between AD and NC samples. We identified the following 2,190 differentially expressed genes (DEGs) between the AD and NC brain samples: 551 in astrocytes, 299 in endothelial cells, 347 in excitatory neurons, 125 in inhibitory neurons, 340 in microglia, and 528 in oligodendrocytes (Fig. 2C and SI Appendix, Table S3). Among the 2,190 DEGs, only 11 were differentially expressed in all cell types (Fig. 2D), suggesting that most observed AD-associated transcriptomic changes are cell type-specific.
As changes in molecular phenotypes provide insights into the functional changes in individual cell types, we functionally annotated the DEGs associated with AD in individual cell types by performing pathway analysis (Fig. 2E and SI Appendix, Fig. S2). Our findings demonstrated that the DEGs in both excitatory and inhibitory neurons (e.g., SNAP25) were associated with synaptic organization and adhesion. Similarly, the down-regulated genes in astrocytes (e.g., HES5, NTRK2, SLC1A2, SPARCL1, and WIF1) were associated with synaptic signaling and glutamate secretion. Specifically, the expression levels of astrocytic neurexin genes (including NRXN1 and NRXN3), which regulate excitatory synaptogenesis (16, 17), were down-regulated in the AD samples.
Besides the transcriptomic changes in neurons and astrocytes, our pathway analysis also revealed that the DEGs in microglia (e.g., AIF1, CD14, GPR183, and PLEKHA7), endothelial cells (e.g., CLDN5, EPHA4, FLT1, PECAM1, and VWF), and oligodendrocytes (e.g., CTNNA2 and OPALIN) were associated with immune response, angiogenesis, and myelination, respectively (Fig. 2E). Of note, endothelial cells also exhibited dysregulation of immune response-related genes (e.g., HLA-E) in AD (Fig. 2E).
Together, our results reveal that the cell type-specific transcriptomic changes in AD are associated with four molecular pathways: angiogenesis in endothelial cells, immune response in endothelial cells and microglia, myelination in oligodendrocytes, and synaptic signaling in astrocytes and neurons (Fig. 2E).
Cross-Study Validation of the Transcriptomic Changes in AD Brains.
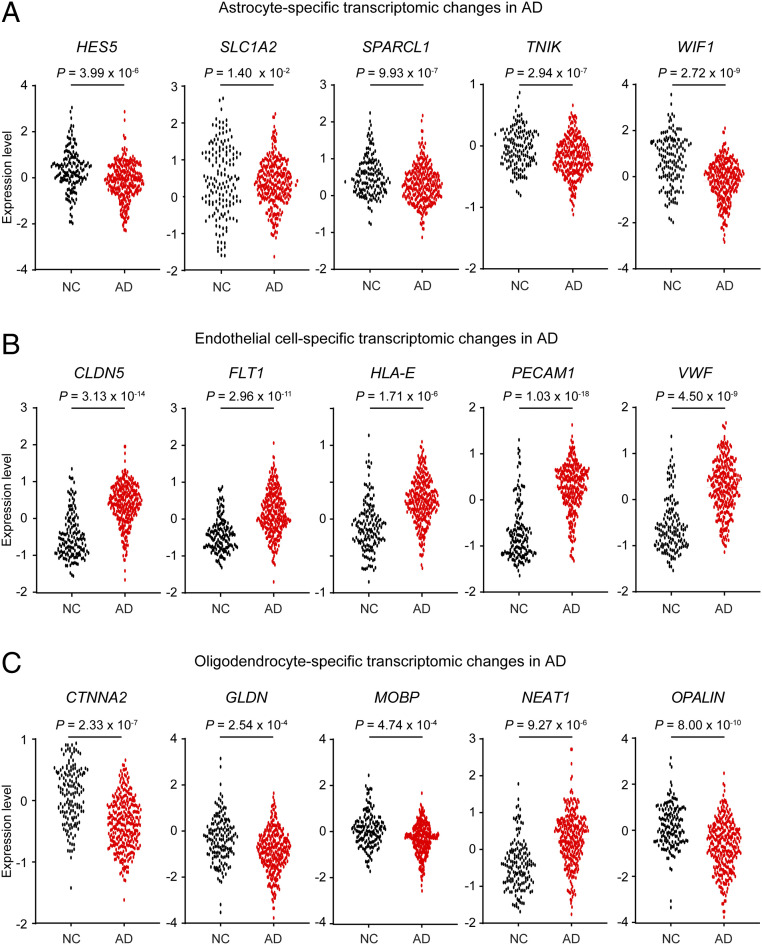
To validate the AD-associated transcriptomic changes detailed above, we compared our results with microarray data from large cohort studies that examined samples from the prefrontal cortex (AD: n = 310; NC: n = 157) or temporal cortex (AD: n = 106; NC: n = 135) (SI Appendix, Table S4) (18, 19). Among the DEGs identified in our snRNA-seq analysis, 1,113 and 764 genes were significantly differentially expressed in the microarray data from the prefrontal cortex and temporal cortex, respectively (adjusted P < 0.05) (SI Appendix, Table S4). In addition, 86.7% of the replicable DEGs in the prefrontal cortex microarray exhibited concordant changes in expression levels; these included changes specific to astrocytes (e.g., HES5, SLC1A2, SPARCL1, TNIK, and WIF1), endothelial cells (e.g., CLDN5, FLT1, HLA-E, PECAM1, and VWF), and oligodendrocytes (e.g., CTNNA2, GLDN, MOBP, NEAT1, and OPALIN) (Fig. 3 A–C). Stratification of the expression levels of these DEGs by sex showed that their expression changes were concordant in both sexes but to different degrees (SI Appendix, Fig. S3). These results validate the AD-associated transcriptomic changes identified in our snRNA-seq analysis.
Fig. 3.
Validation of the cell type-specific transcriptomic changes in AD based on data from a large cohort microarray study. Dot plots showing the expression levels of DEGs specific to astrocytes (A), endothelial cells (B), and oligodendrocytes (C) in the prefrontal cortex from patients with AD (n = 310) and NC (n = 157) samples. Each dot represents the expression level from an individual subject. Source: Narayanan et al. (18). See also SI Appendix, Fig. S3 and Table S4.
While comparison with bulk microarray data validated the transcriptomic changes observed in AD brains at the global transcriptome level, the bulk data were unable to distinguish cell type-specific changes. Therefore, we compared our findings with the snRNA-seq data reported by Mathys et al., who analyzed gene expression in the prefrontal cortex of patients with various degrees of AD pathology (SI Appendix, Fig. S4) (12). We found 273 cell type-specific DEGs in common between the two datasets, including 53 in astrocytes, 97 in excitatory neurons, 24 in inhibitory neurons, 13 in microglia, and 86 in oligodendrocytes (SI Appendix, Fig. S4 A, B, D, E, G, H, J, K, M, and N). More than 90% of the overlapping genes, especially the astrocyte-specific and neuron-specific DEGs, exhibited concordant changes in AD (SI Appendix, Fig. S4 B, E, and H). Moreover, pathway analysis showed that these overlapping DEGs were associated with transsynaptic signaling in astrocytes (SI Appendix, Fig. S4C), synaptic signaling in excitatory neurons (SI Appendix, Fig. S4F), mitochondrial functions in inhibitory neurons (SI Appendix, Fig. S4I), secretion in microglia (SI Appendix, Fig. S4L), and axonogenesis in oligodendrocytes (SI Appendix, Fig. S4O). Thus, these findings validate the cell type-specific dysregulated pathways identified in our study.
The Disrupted Subpopulation Heterogeneity of Astrocytes and Oligodendrocytes Contributes to Neuronal Dysfunction in AD.
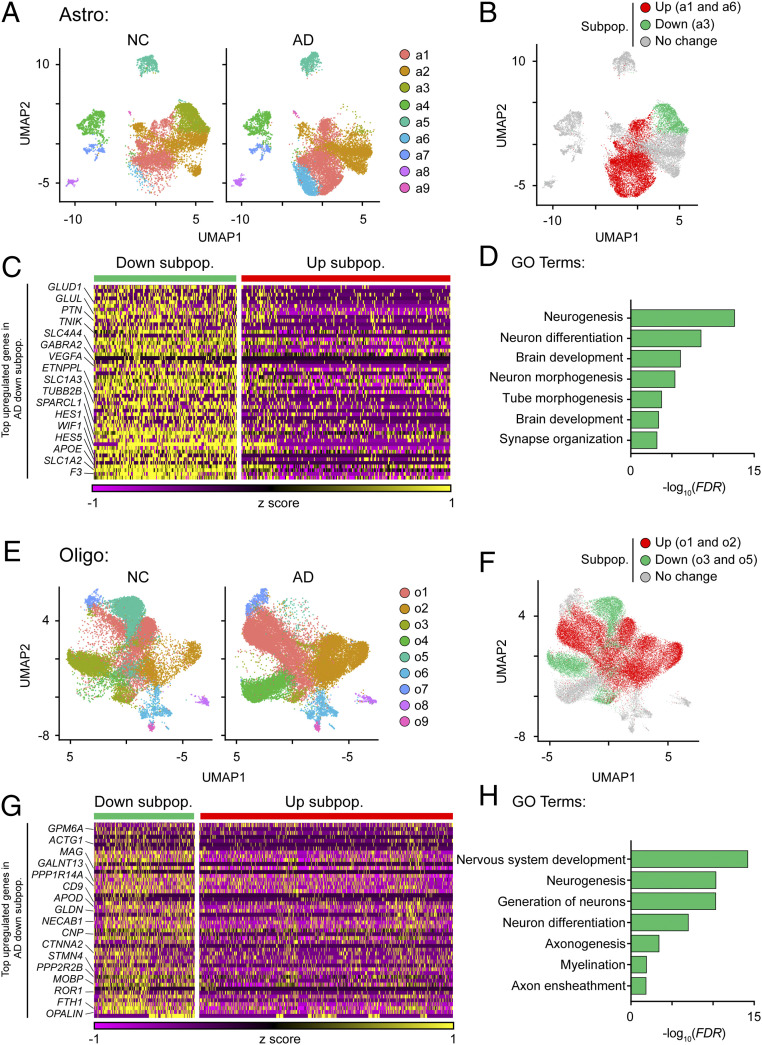
Next, we investigated the subpopulation heterogeneity within individual cell types in AD and NC samples. To this end, we performed subcluster analysis of individual cell types (except excitatory and inhibitory neurons, which exhibited only subtle changes in DEG expression levels). Subcluster analysis of astrocytes identified nine transcriptomically unique subpopulations (Fig. 4A). The relative proportions of subpopulations a2, a4, a5, a7, a8, and a9 were similar between the AD and NC samples (Fig. 4A and SI Appendix, Fig. S5A). However, compared to the NC samples, the relative proportions of a1 and a6 were 9.9% and 10.2% larger in the AD samples, respectively, while that of a3 was 23.5% smaller (Fig. 4A and SI Appendix, Fig. S5A). Only subpopulations a1, a3, and a6 expressed DEGs; specifically, subpopulations a1 and a6 were enriched with up-regulated signature genes, whereas subpopulation a3 was enriched in down-regulated signature genes (SI Appendix, Fig. S5B). Therefore, we categorized a1 and a6 as the “AD-up-regulated” subpopulations and a3 as the “AD-down-regulated” subpopulation (Fig. 4B).
Fig. 4.
The proportions of astrocytes and oligodendrocytes that regulate neuronal homeostasis are reduced in AD. (A) UMAP plots showing the distributions of astrocyte subpopulations (a1–a9) in AD and NC prefrontal cortical samples. (B–D) The astrocyte subpopulation associated with the maintenance of synaptic functions was reduced in AD. (B) UMAP plot showing the distribution of the AD-associated astrocyte subpopulations. (C) Heatmap showing the expression levels of the top enriched genes in the AD-down-regulated astrocyte subpopulation (adjusted P < 0.1, log2 fold change ≥ 0.1). Down subpop.: down-regulated subpopulation; Up subpop.: up-regulated subpopulation. (D) GO pathway analysis of the transcriptomic signature of the AD-down-regulated astrocyte subpopulation. (E) UMAP plots showing the distributions of oligodendrocyte subpopulations (o1–o9) in AD and NC prefrontal cortical samples. (F–H) Oligodendrocyte subpopulations associated with myelination were reduced in AD. (F) UMAP plot showing the distribution of the AD-associated oligodendrocyte subpopulations. (G) Top enriched genes in the AD-down-regulated oligodendrocyte subpopulations (adjusted P < 0.1, log2 fold change ≥ 0.1). Down subpop.: down-regulated subpopulation; Up subpop.: up-regulated subpopulation. (H) Pathway analysis of the transcriptomic signature of the AD-down-regulated oligodendrocyte subpopulations. See also SI Appendix, Figs. S5 and S6 and Table S5.
The transcriptome profile of the AD-down-regulated astrocyte subpopulation was characterized by enriched expression of genes associated with neurotransmitter metabolism, including SLC1A2 and GLUL (Fig. 4 C and D and SI Appendix, Fig. S5C and Table S5). Indeed, impaired recycling of neurotransmitters, especially glutamate, results in excitotoxicity and leads to neuronal death (6). The AD-up-regulated subpopulations exhibited enriched expression of stress response-associated genes including CRYAB (a heat shock protein), GFAP (a reactive astrocyte marker), and LINGO1 (a negative regulator of myelination) (SI Appendix, Fig. S5 D and E). Moreover, the expression of HMGB1, an alarmin that conveys injury signals to surrounding cells, was higher in the AD-up-regulated subpopulations than the AD-down-regulated subpopulation (SI Appendix, Fig. S5D) (20). Thus, our results show that in AD, (i) three specific astrocyte subpopulations contribute to the transcriptomic changes in astrocytes, (ii) the subpopulation of homeostatic astrocytes (a3), which are crucial for neurotransmitter recycling, is reduced, and (iii) alarmin-expressing astrocytes are induced (a1 and a6). These findings suggest that astrocytic dysfunction in AD contributes to the dysregulation of neurotransmitter recycling and exaggerated alarmin response.
Regarding oligodendrocytes, subcluster analysis identified nine subpopulations in NC and AD brains (Fig. 4E). Among these, only four subpopulations—o1, o2, o3, and o5—were enriched with genes that were differentially expressed in oligodendrocytes in AD (Fig. 4F and SI Appendix, Fig. S5 F and G). Pathway analysis revealed that the AD-down-regulated oligodendrocyte subpopulations o3 and o5 exhibited enriched expression of genes associated with myelination, including MAG, MOBP, and OPALIN (Fig. 4 G and H and SI Appendix, Fig. S5H and Table S5). Along the oligodendroglial lineage, myelinated mature oligodendrocytes exhibit enriched expression of MAG, MOBP, and OPALIN (21). Therefore, we concluded that the o3 and o5 subpopulations were mature myelinated oligodendrocytes. Notably, the transcriptome profiles of the AD-up-regulated oligodendrocyte subpopulations o1 and o2 were characterized by enriched expression of HSPA1A (a heat shock protein), NEAT1 (a nuclear noncoding RNA), and PDE1A (which encodes phosphodiesterase 1A) (SI Appendix, Fig. S5 I and J); this expression pattern resembles that of remyelinating oligodendrocytes in patients with multiple sclerosis (22). Thus, our results show that in AD, there is a decrease in the number of mature myelinated oligodendrocytes and an increase in the number of remyelinating oligodendrocytes.
Finally, subcluster analysis of microglia identified 13 subpopulations; only 3 of them—m1, m6, and m7—contributed to the transcriptomic changes in microglia in AD (SI Appendix, Fig. S6 A and B and Table S5). Notably, compared to the NC samples, the AD samples exhibited a smaller proportion of the m6 subpopulation, which expresses genes important for synaptic pruning (i.e., C1QA, C1QB, and C1QC, which encode complement component 1q) and cytokine response (i.e., IL4R and IL1RAP) (SI Appendix, Fig. S6 C and D). Given that aberrant complement signaling mediates nonselective synaptic pruning via phagocytosis (23), our results suggest that the loss of this typical microglial subpopulation can contribute to the imbalanced complement signaling and synaptic pruning in AD.
The Induction of Angiogenic Endothelial Cells Contributes to Immune Dysregulation in AD.
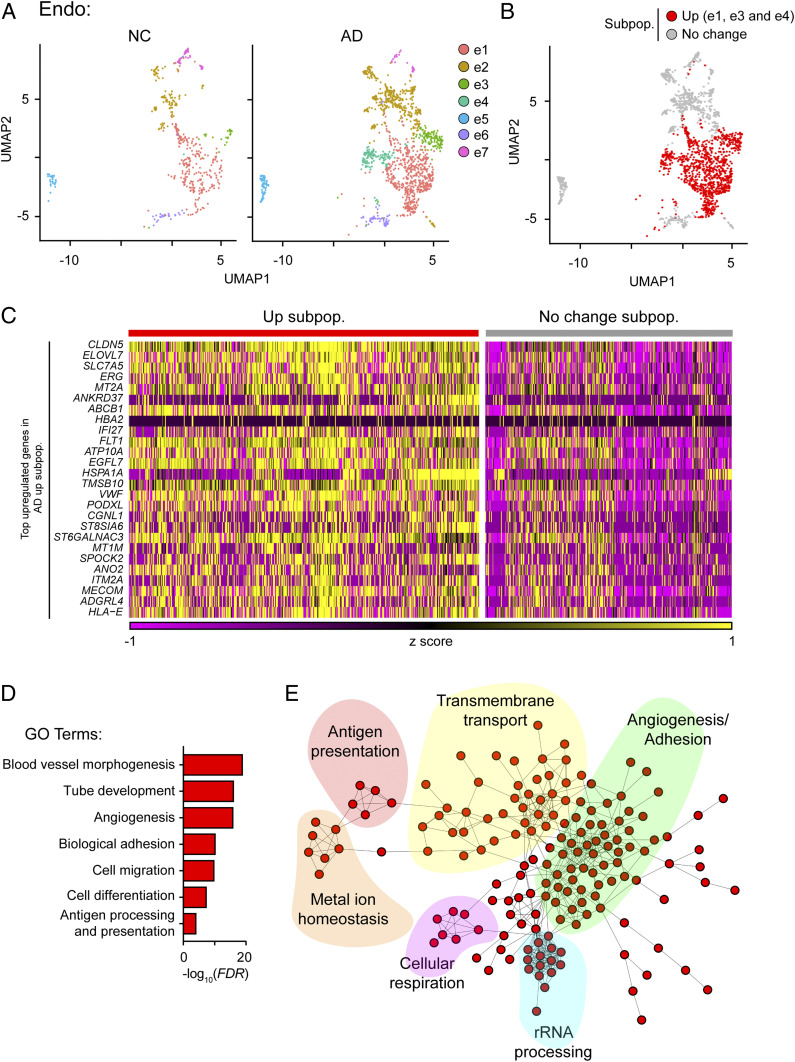
Our results demonstrate that the transcriptomic changes of endothelial cells in AD are associated with angiogenesis and immune response (Fig. 2E). Our subcluster analysis further revealed that only three of the seven identified endothelial cell subpopulations—e1, e3, and e4—contributed to the transcriptomic changes observed in endothelial cells in AD (Fig. 5A and SI Appendix, Fig. S7 A and B and Table S5). Of note, all three subpopulations were classified as AD-up-regulated subpopulations (Fig. 5B and SI Appendix, Fig. S7C). Transcriptome profiling showed that genes associated with angiogenesis (i.e., CLDN5, ERG, FLT1, and VWF) and antigen presentation—especially MHC-I (major histocompatibility complex class I) machinery (i.e., HLA-E)—were enriched in these three AD-up-regulated subpopulations (Fig. 5C and SI Appendix, Table S5). Pathway analysis and protein–protein interaction network analysis further revealed that the signature genes of these AD-up-regulated subpopulations form an interaction network associated with six major biological functions: angiogenesis/adhesion, transmembrane transport, antigen presentation, metal ion homeostasis, cellular respiration, and rRNA processing (Fig. 5 D and E).
Fig. 5.
An endothelial cell subpopulation associated with enhanced angiogenesis and antigen presentation is increased in AD. (A–C) Transcriptomically unique subpopulations of endothelial cells were present in AD samples. (A) UMAP plots showing the distributions of endothelial subpopulations (e1–e7) in AD and NC prefrontal cortical samples. (B) Distribution of AD-associated endothelial subpopulations. Red: AD-up-regulated subpopulations (i.e., e1, e3, and e4). (C) Expression levels of the top enriched genes in the AD-up-regulated subpopulations at the single-cell level (adjusted P < 0.1, log2 fold change ≥ 0.1). Up subpop.: up-regulated subpopulation. (D and E) The activated endothelial cells in AD were associated with angiogenesis and antigen presentation. GO analysis of the up-regulated genes in AD-up-regulated endothelial subpopulations (D), and STRING analysis (E) showing that the signature genes of activated endothelial cells form a protein–protein interaction network associated with six different functional pathways. See also SI Appendix, Fig. S7 and Table S5.
We subsequently investigated the potential mechanisms by which angiogenic endothelial cells are induced in AD by performing ingenuity pathway analysis using the signature genes in the previously identified AD-up-regulated endothelial subpopulations. The results revealed that several regulators of signature genes in the AD-up-regulated subpopulations, including IFNG, IRF7, and TCR, were associated with inflammatory response (SI Appendix, Fig. S7D). This suggests that the induction of angiogenic endothelial cells is regulated by a proinflammatory response. Moreover, analysis of bulk microarray data from mouse models of amyloid-beta deposition or Tau hyperphosphorylation also revealed transcriptome reprogramming similar to that observed in endothelial cells in patients with AD (SI Appendix, Fig. S7E), suggesting that the activation of endothelial cells in neurodegeneration is conserved between humans and mice.
Discussion
The identification of precise molecular and cellular targets for AD therapeutic development requires a comprehensive understanding of the cell type-specific responses and cellular heterogeneity in AD. Accordingly, our single-nucleus transcriptome analysis identified the four major molecular pathways that are dysregulated in specific neural cell types in patients with AD: impaired synaptic signaling in astrocytes and neurons, reduced myelination in oligodendrocytes, aberrant immune activation in microglia and endothelial cells, and enhanced angiogenesis in endothelial cells. Moreover, we showed that the dysregulation of these pathways in AD is due to changes in the subpopulation heterogeneity of respective cell types, including the reduced proportions of neuroprotective astrocytes and oligodendrocytes as well as the increased proportions of endothelial cells, which are associated with enhanced angiogenesis and antigen presentation. Together, our single-nucleus transcriptomic profiling highlights the potential roles of dysregulated angiogenesis in endothelial cells of AD patients.
The results of our single-nucleus transcriptomic profiling of AD brains are a useful resource for understanding the cellular dysregulation along AD progression. For example, we showed that the dysregulated pathways in neurons and oligodendrocytes are associated with synaptic signaling and myelination, respectively; these findings are similar to those in previous studies despite variations in study cohorts, brain regions, sample preparation, and sequencing protocols (12, 13, 23). In addition to these consistent changes, our findings provide insights into the cellular changes that occur in AD. For example, we found that the dysregulated pathways in endothelial cells are associated with angiogenesis and antigen presentation. However, such changes were not observed in previous snRNA-seq studies (12, 13), possibly due to a lower proportion of endothelial nuclei being sampled. Therefore, the addition of our dataset to the existing snRNA-seq datasets of AD brains enhances the comprehensiveness of cell type-specific transcriptomic changes in AD, especially in endothelial cells.
In addition to identifying dysregulated pathways involved in AD pathogenesis, our molecular characterization of the human AD brain supports the prevailing hypotheses based on studies of transgenic mouse models of AD. For example, previous studies using transgenic mouse models of AD have identified various mechanisms that lead to synaptic impairment in AD, including the accumulation of soluble amyloid-beta, Tau hyperphosphorylation, loss of VGF signaling, and glial state dysregulation (e.g., the induction of disease-associated microglia and disease-associated astrocytes) (9, 23–30). For astrocytes in particular, dysregulated communication with neurons can impair synaptic plasticity (31, 32). Indeed, we showed that the AD brain contains a reduced proportion of neuroprotective astrocytes, which are associated with glutamate recycling and synaptic signaling. This could lead to synaptic loss due to excitotoxicity (6, 33, 34) and reduced synaptogenesis due to the destabilization of nascent synapses (16, 17), which would result in a net loss of synapses during disease progression. Therefore, our findings suggest that the dysregulated pathways in astrocytes could contribute to synaptic impairment in AD.
Similarly, the loss of myelinated oligodendrocyte subpopulations suggests the disruption of oligodendrocyte maintenance in AD. Indeed, aging-related myelin loss is associated with cognitive impairment in AD (35). In animal models, demyelination can be triggered by the accumulation of amyloid-beta peptides, phosphorylated Tau, or proinflammatory cytokines (35–37). Consistent with recent findings (12, 13), the molecular changes observed in our study were associated with the demyelination process in AD. Interestingly, our results also show that oligodendrocytes adopt a remyelinating state in AD, suggesting a potential cell-intrinsic recovery mechanism. While proper remyelination can restore the impaired saltatory conduction in neurons (38), it remains unclear why remyelinating oligodendrocytes fail to overcome the demyelination observed in AD.
Our transcriptome profiling revealed that endothelial cells adopt an angiogenic state characterized by enhanced expression of genes involved in angiogenesis and antigen presentation. Endothelial cells are the primary component of the neurovascular system, which maintains blood–brain barrier integrity. Both patients with AD and AD animal models exhibit neurovascular system impairment including abnormalities in the number, diameter, and density of blood vessels, which lead to decreased brain perfusion and blood–brain barrier disruption (10, 39, 40). Compromised blood–brain barrier integrity permits the direct entry of neurotoxic thrombin and plasmin, leading to synapse loss (41, 42). Although it remains unclear how the dysregulation of endothelial cells leads to blood–brain barrier abnormalities in AD, our findings show that in AD, endothelial cells adopt an angiogenic state characterized by increased expression of angiogenic factors and receptors including ERG, FLT1, and VWF. Notably, increased expression of angiogenic genes in endothelial cells is associated with the blood vessel abnormalities observed in mouse models of neurodegeneration (40). Therefore, our study provides evidence of a potential link between endothelial angiogenesis and blood–brain barrier abnormalities in patients with AD.
Our findings also suggest a role of endothelial cells in antigen presentation, specifically that by MHC-I, in AD. MHC-I mostly presents endogenous proteins that originate in the cytoplasm, making it essential in the response to viral infection (43). Upon viral infection, endothelial MHC-I presents endogenous viral products and facilitates CD8+ T cell activation (44–46). Interestingly, several recent studies suggest that AD is associated with viral infection (47, 48). Therefore, the induction of MHC-I in endothelial cells in AD might reflect response to viral infection. Another possible role of this MHC-I induction is the triggering of CD8+ T cell activation and subsequent clonal expansion. Indeed, increased clonal expansion and CD8+ T cell infiltration in the brain are observed both in AD patients and during aging (48, 49). Once activated CD8+ T cells have infiltrated the brain, they can release IFN-γ to stimulate excessive microglial synaptic pruning or inhibit adult neural stem cell proliferation (44, 49), impairing neuronal homeostasis. Therefore, it is of great interest to investigate how MHC-I induction in endothelial cells contributes to AD pathogenesis.
In summary, our transcriptome profiling results constitute a useful resource for understanding the pathological roles of endothelial and glial cells in AD. Furthermore, our data can aid the identification of endothelial- and glial-specific molecular targets for the therapeutic restoration of neural homeostasis and the amelioration of the pathological progression of this devastating disease.
Methods
Selection Criteria for Brain Tissue Collection from AD Patients.
We obtained postmortem prefrontal cortex tissue samples (from the BA6, BA8, and BA9 domains) from patients with AD and NC subjects from the South West Dementia Brain Bank (SWDBB). The clinical diagnosis of AD was based on the Diagnostic and Statistical Manual of Mental Disorders-5 criteria for AD (SI Appendix, Table S1). For initial sample selection from the SWDBB, we excluded subjects with other neurodegenerative diseases, vascular diseases, an intoxicated state, infection, prions, inflammatory diseases, structural brain disorders, metabolic/nutritional diseases, trauma, delirium, genetic disorders (e.g., Down syndrome), or other systemic diseases (SI Appendix, Table S1). We categorized samples according to Braak stage (i.e., AD: ≥4 and NC: ≤2). For snRNA-seq library preparation, we subsequently selected subjects according to age and postmortem delay and ensured a balanced sex ratio. Thus, we included 21 subjects (mean age: AD, 74.6 y; NC, 85.4 y) including 8 males and 4 females with AD as well as 6 male and 3 female NC subjects. We did not intentionally include/exclude samples according to APOE genotype prior to snRNA-seq library preparation but confirmed APOE genotypes by TaqMan assay with a probe from Thermo Scientific (assay ID: C_3084793_20 and C_904973_10) following the manufacturer’s instructions.
Isolation of Nuclei from the Brain Tissues of AD Patients.
We isolated nuclei from frozen prefrontal cortex samples as previously described with minor modifications (50). Briefly, we placed frozen cortical tissues directly into a prechilled Dounce homogenizer with ice-cold homogenization buffer (0.25 M sucrose, 25 mM KCl, 5 mM MgCl2, 20 mM tricine-KOH [pH 7.8], 1 mM dithiothreitol, 0.15 mM spermine, 0.5 mM spermidine, protease inhibitors, 5 μg/mL actinomycin, 0.32% Nonidet P-40, and 0.04% bovine serum albumin). After 25 strokes with a loose pestle, we mixed the homogenate 1:1 in OptiPrep and centrifuged the solution at 10,000 × g for 20 min at 4 °C. We subsequently collected the separated nuclei, which were pelleted at the bottom of the centrifuge tube, washed them once to remove the OptiPrep, and resuspended them in Dulbecco's modified Eagle medium/F12 supplemented with 10% fetal bovine serum. We diluted the nuclei to 400 nuclei per microliter, ensuring this dilution by counting with a hemocytometer. We also assessed the purity of the single-nucleus suspensions by flow cytometry; this protocol routinely yielded high-purity, single-nucleus suspensions (i.e., >95% DAPI+ nuclei in all samples). All buffers and gradient solutions for nuclei extraction contained 60 U/mL RNAsin (Promega).
snRNA-seq Library Preparation.
We generated snRNA-seq libraries using the Chromium Single Cell 3′ Library Kit v3 (1000078; 10× Genomics) according to the manufacturer’s instructions. For snRNA-seq library construction, we used 40-μL diluted nucleus suspension (400 nuclei per µL) mixed with reverse-transcription reagent mix and loaded the sample into a chip for single-cell encapsulation. We then immediately incubated the encapsulated nuclei on a thermocycler to enable reverse transcription of the RNA and generate barcoded cDNA. We used this cDNA for library construction according to the manufacturer’s instructions. The concentrations of the final libraries were determined by Qubit (Thermo Fisher Scientific), and the fragment lengths were determined by a Fragment Analyzer (Advanced Analytical Technologies). We subjected libraries to paired-end sequencing on a NovaSeq 6000 system according to the manufacturer’s instructions (Novogene), and at least 200 GB of raw data were obtained per library.
snRNA-seq Analysis.
Preprocessing and quality control.
Given that snRNA-seq libraries capture reads from both unspliced pre-messenger RNAs (mRNAs) and mature mRNAs, we first generated a pre-mRNA reference genome according to the instructions provided by 10× Genomics. We subsequently aligned the demultiplexed FASTQ files from Novogene to the GRCh38 pre-mRNA reference genome using Cell Ranger (version 3.0.1) with the default settings (51). After performing the alignment, we used the default quality control settings in Cell Ranger as an initial quality control step. This initial quality control process retained only barcodes with unique molecular identifier counts within the top 10% of the 99th percentile of unique molecular identifier values among all barcodes and sorted them into cell-associated matrixes; the remaining barcodes were treated as background barcodes and were excluded. We subsequently used these cell-associated matrixes as input for the second round of quality control and downstream analysis in Seurat (version 3.0) (52).
For the second round of quality control, we controlled for the distributions of the numbers of genes, numbers of unique molecular identifier counts, and percentages of mitochondrial genes for each sample in Seurat. To exclude potential dead cells and cell debris from the dataset, we filtered out nuclei with ≤200 genes, ≥20,000 unique molecular identifiers, or ≥20% mitochondrial genes as described in several previous snRNA-seq studies (12, 13, 22, 53). The final filtered matrix contained 169,496 nuclei and 29,171 genes. During sample preparation, we targeted ∼16,000 nuclei per sample and found that while the number of nuclei that passed quality control varied among samples, the variation was independent of age, sex, postmortem delay, and pathology, and there was no batch effect (SI Appendix, Table S1). Although the postmortem delay varied among samples, we also found that the numbers of genes and detected transcripts were independent of postmortem delay variation (SI Appendix, Fig. S1A).
Cell-type identification by dimensionality reduction.
For integrative analysis, we followed the workflow described in the Seurat guided analysis. We first log-normalized the filtered matrixes and identified highly variable features for each sample using the FindVariableFeatures function with the parameters selection.method = vst, and nfeatures = 1000. To integrate all 21 samples, we identified features for anchoring the samples using the FindIntegrationAnchors function with the parameter dims = 1:20 and used the identified anchors to integrate the dataset using the IntegrateData function with the parameter dims = 1:20. We subsequently scaled the integrated matrix and performed linear dimensional reduction using the RunPCA function with the parameter npcs = 50. We visualized the P value distribution of each principal component using the JackStrawPlot function and opted to use the first 20 principal components for graph-based clustering.
We performed K-nearest neighbor clustering using the FindClusters function with the parameter resolution = 1 and UMAP clustering using the RunUMAP function with the parameter dims = 1:20, which initially yielded 43 cell clusters. We identified the DEGs in each cell cluster by the Wilcoxon rank-sum test using the FindAllMarkers function with the parameters logfc.threshold = 0.25 and test.use = wilcox. We then assigned a cell-type identity to each cell cluster according to the expression of known cell-type markers and identified additional cell type-specific marker genes by the Wilcoxon rank-sum test using the FindAllMarkers function with the parameters logfc.threshold = 0.25 and test.use = wilcox. For cell-type markers, the level of statistical significance was set at an adjusted P value <0.1. To further confirm the cell-type specificity of these markers, we compared our data with those of Jäkel et al. (22), which confirmed the expression patterns of these markers in the different cell types in our study.
Examination of cell type-specific transcriptomic changes.
To examine the cell type-specific transcriptomic changes in AD, we stratified samples according to our initial classification of AD and NC samples (based on Braak stage) and compared the transcriptome profiles of individual cell types between AD and NC samples by the Wilcoxon rank-sum test using the FindMarkers function with the parameters logfc.threshold = 0 and test.use = wilcox. The level of statistical significance for cell type-specific transcriptomic changes was set at an adjusted P < 0.1 and a log2 fold change ≥0.1 or ≤ −0.1.
Subcluster analysis.
For subcluster analysis, we first isolated individual cell types from the original Seurat dataset using the Subset function. We subsequently reclustered each cell type using an approach similar to that used for our initial cell type clustering. We performed K-nearest neighbor clustering using the FindClusters function with the parameters resolution = 0.2, 0.2, 0.3, and 0.2 for astrocytes, endothelial cells, microglia, and oligodendrocytes, respectively, as well as UMAP clustering using the RunUMAP function with the parameter dims = 1:20. To perform unbiased identification of the subpopulations of cells that contribute to AD-associated transcriptomic changes, we calculated the enrichment scores of AD-associated DEGs in each subpopulation by averaging the z-scores of all AD-associated DEGs. A subpopulation with an enrichment score > 0.5 was designated as an AD-associated subpopulation. We identified the transcriptomic signatures of AD-associated subpopulations by comparing up- and down-regulated subpopulations using the Wilcoxon rank-sum test with the FindMarkers function and the parameters logfc.threshold = 0 and test.use = wilcox. The level of statistical significance was set at an adjusted P < 0.1 and a log2 fold change ≥ 0.1 or ≤ −0.1.
Data Validation by Comparison with Previous Studies.
For data validation, we obtained datasets from published studies and compared them with our findings. We obtained DEG analysis results from a recent snRNA-seq study (12). We selected a list of cell type-specific DEGs between healthy and diseased samples with an adjusted P < 0.01 (two-sided Wilcoxon rank-sum test) and log2 fold change ≥ 0.25 or ≤ −0.25. However, the proportion of endothelial cells in the previous dataset was low, rendering it unsuitable for validating our findings in endothelial cells.
We also obtained the dataset from a study by Narayanan et al. (Gene Expression Omnibus [GEO] accession no. GSE33000), who performed bulk transcriptome microarray analysis of prefrontal cortical tissues in a large cohort (AD: n = 310; NC: n = 157) (18). We first filtered the samples according to disease status and kept only “Alzheimer’s disease” and “non-demented” samples for subsequent analysis. We removed genes that failed to be mapped to Entrez gene IDs. For genes mapped by multiple probes, we used the median value. To scale the variance, we performed log2 transformation and quantile normalization using the R limma package (54). For differential expression analysis, we fitted the gene expression profiles by linear regression, adjusting for age and sex. We used the empirical Bayes method provided in limma to calculate t-statistics and log-fold changes in differential expression. We adjusted the P values using the Benjamini–Hochberg procedure. We also used the same pipeline to process the microarray dataset of AD temporal cortical samples from Webster et al. (GEO accession no. GSE15222) (19).
We obtained transcriptome data for mouse models of amyloid-beta deposition and Tau hyperphosphorylation from MOUSEAC (55).
Data Visualization.
We visualized the data using Morpheus, Seurat’s DoHeatmap or DotPlot function, or Cytoscape (version 3.7.0) where appropriate. DEGs were functionally annotated according to Gene Ontology (GO) analysis (geneontology.org), ingenuity pathway analysis (QIAGEN), and STRING analysis (https://string-db.org).
Supplementary Material
Acknowledgments
We thank the SWDBB for providing brain tissues for this study. The SWDBB is part of the Brains for Dementia Research program, which is jointly funded by Alzheimer’s Research UK and the Alzheimer’s Society and is supported by Bristol Research into Alzheimer’s and Care of the Elderly and the Medical Research Council. We gratefully acknowledge Drs. Michael Greenberg (Harvard Medical School), William Renthal (Harvard Medical School), and Lisa Boxer (Harvard Medical School) for their expert advice on the snRNA sequencing protocol. We thank Drs. Yun Li (University of North Carolina, Chapel Hill), Tom Cheung (Hong Kong University of Science and Technology; HKUST) and Jiguang Wang (HKUST) for critical reading of the manuscript. We also thank Li Ouyang, Tiffany Mak, and Cara Kwong for their excellent technical assistance as well as other members of the N.Y.I. Laboratory for many helpful discussions. This study was supported in part by the Research Grants Council of Hong Kong (the Collaborative Research Fund [C6027-19GF], the Theme-Based Research Scheme [T13-607/12R], and the General Research Fund [HKUST16149616, HKUST16103017, HKUST16100418, and HKUST16102019]), National Key R&D Program of China Grants 2017YFE0190000 and 2018YFE0203600, the Areas of Excellence Scheme of the University Grants Committee AoE/M-604/16, the Innovation and Technology Commission Grant ITCPD/17-9, Guangdong Provincial Key S&T Program Grant 2018B030336001, and Shenzhen Knowledge Innovation Program Grants JCYJ20180507183642005 and JCYJ20170413173717055. S.-F.L. is a recipient of the Hong Kong PhD Fellowship Award.
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2008762117/-/DCSupplemental.
Data Availability.
Anonymized snRNA-seq sequencing data have been deposited in GEO (accession no. GSE157827). All study data are included in the article and supporting information.
References
- 1.Reitz C., Brayne C., Mayeux R., Epidemiology of Alzheimer disease. Nat. Rev. Neurol. 7, 137–152 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fu W. Y., Wang X., Ip N. Y., Targeting neuroinflammation as a therapeutic strategy for Alzheimer’s disease: Mechanisms, drug candidates, and new opportunities. ACS Chem. Neurosci. 10, 872–879 (2019). [DOI] [PubMed] [Google Scholar]
- 3.Snyder E. M. et al., Regulation of NMDA receptor trafficking by amyloid-β. Nat. Neurosci. 8, 1051–1058 (2005). [DOI] [PubMed] [Google Scholar]
- 4.Chen Y., Fu A. K. Y., Ip N. Y., Synaptic dysfunction in Alzheimer’s disease: Mechanisms and therapeutic strategies. Pharmacol. Ther. 195, 186–198 (2019). [DOI] [PubMed] [Google Scholar]
- 5.Scheff S. W., Price D. A., Schmitt F. A., Mufson E. J., Hippocampal synaptic loss in early Alzheimer’s disease and mild cognitive impairment. Neurobiol. Aging 27, 1372–1384 (2006). [DOI] [PubMed] [Google Scholar]
- 6.Hynd M. R., Scott H. L., Dodd P. R., Glutamate-mediated excitotoxicity and neurodegeneration in Alzheimer’s disease. Neurochem. Int. 45, 583–595 (2004). [DOI] [PubMed] [Google Scholar]
- 7.Parkhurst C. N. et al., Microglia promote learning-dependent synapse formation through brain-derived neurotrophic factor. Cell 155, 1596–1609 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Christopherson K. S. et al., Thrombospondins are astrocyte-secreted proteins that promote CNS synaptogenesis. Cell 120, 421–433 (2005). [DOI] [PubMed] [Google Scholar]
- 9.Liddelow S. A. et al., Neurotoxic reactive astrocytes are induced by activated microglia. Nature 541, 481–487 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zlokovic B. V., Neurovascular pathways to neurodegeneration in Alzheimer’s disease and other disorders. Nat. Rev. Neurosci. 12, 723–738 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao Z., Nelson A. R., Betsholtz C., Zlokovic B. V., Establishment and dysfunction of the blood-brain barrier. Cell 163, 1064–1078 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mathys H. et al., Single-cell transcriptomic analysis of Alzheimer’s disease. Nature 570, 332–337 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grubman A. et al., A single-cell atlas of entorhinal cortex from individuals with Alzheimer’s disease reveals cell-type-specific gene expression regulation. Nat. Neurosci. 22, 2087–2097 (2019). [DOI] [PubMed] [Google Scholar]
- 14.Wang X. et al., Deciphering cellular transcriptional alterations in Alzheimer’s disease brains. Mol. Neurodegener. 15, 38 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Habib N. et al., Massively parallel single-nucleus RNA-seq with DroNc-seq. Nat. Methods 14, 955–958 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stogsdill J. A. et al., Astrocytic neuroligins control astrocyte morphogenesis and synaptogenesis. Nature 551, 192–197 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Südhof T. C., Synaptic neurexin complexes: A molecular code for the logic of neural circuits. Cell 171, 745–769 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Narayanan M. et al., Common dysregulation network in the human prefrontal cortex underlies two neurodegenerative diseases. Mol. Syst. Biol. 10, 743 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Webster J. A. et al.; NACC-Neuropathology Group , Genetic control of human brain transcript expression in Alzheimer disease. Am. J. Hum. Genet. 84, 445–458 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harris H. E., Andersson U., Pisetsky D. S., HMGB1: A multifunctional alarmin driving autoimmune and inflammatory disease. Nat. Rev. Rheumatol. 8, 195–202 (2012). [DOI] [PubMed] [Google Scholar]
- 21.Marques S. et al., Oligodendrocyte heterogeneity in the mouse juvenile and adult central nervous system. Science 352, 1326–1329 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jäkel S. et al., Altered human oligodendrocyte heterogeneity in multiple sclerosis. Nature 566, 543–547 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hong S. et al., Complement and microglia mediate early synapse loss in Alzheimer mouse models. Science 352, 712–716 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sheng M., Sabatini B. L., Südhof T. C., Synapses and Alzheimer’s disease. Cold Spring Harb. Perspect. Biol. 4, 10 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tremblay M. È., Sierra A., Microglia in Health and Disease, (Springer, 2014). [Google Scholar]
- 26.El Gaamouch F. et al., VGF-derived peptide TLQP-21 modulates microglial function through C3aR1 signaling pathways and reduces neuropathology in 5xFAD mice. Mol. Neurodegener. 15, 4 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Butovsky O. et al., Identification of a unique TGF-β-dependent molecular and functional signature in microglia. Nat. Neurosci. 17, 131–143 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Habib N. et al., Disease-associated astrocytes in Alzheimer’s disease and aging. Nat. Neurosci. 23, 701–706 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Keren-Shaul H. et al., A unique microglia type Associated with restricting development of Alzheimer’s disease. Cell 169, 1276–1290.e17 (2017). [DOI] [PubMed] [Google Scholar]
- 30.Beckmann N. D. et al., Multiscale causal networks identify VGF as a key regulator of Alzheimer’s disease. Nat. Commun. 11, 3942 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hillen A. E. J., Burbach J. P. H., Hol E. M., Cell adhesion and matricellular support by astrocytes of the tripartite synapse. Prog. Neurobiol. 165-167, 66–86 (2018). [DOI] [PubMed] [Google Scholar]
- 32.Perea G., Navarrete M., Araque A., Tripartite synapses: Astrocytes process and control synaptic information. Trends Neurosci. 32, 421–431 (2009). [DOI] [PubMed] [Google Scholar]
- 33.Eid T., Tu N., Lee T. S. W., Lai J. C. K., Regulation of astrocyte glutamine synthetase in epilepsy. Neurochem. Int. 63, 670–681 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Petr G. T. et al., Conditional deletion of the glutamate transporter GLT-1 reveals that astrocytic GLT-1 protects against fatal epilepsy while neuronal GLT-1 contributes significantly to glutamate uptake into synaptosomes. J. Neurosci. 35, 5187–5201 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bartzokis G., Age-related myelin breakdown: A developmental model of cognitive decline and Alzheimer’s disease. Neurobiol. Aging 25, 5–18, author reply 49–62 (2004). [DOI] [PubMed] [Google Scholar]
- 36.Anderson J. M. et al., Abnormally phosphorylated tau is associated with neuronal and axonal loss in experimental autoimmune encephalomyelitis and multiple sclerosis. Brain 131, 1736–1748 (2008). [DOI] [PubMed] [Google Scholar]
- 37.Stadelmann C., Wegner C., Brück W., Inflammation, demyelination, and degeneration - recent insights from MS pathology. Biochim. Biophys. Acta 1812, 275–282 (2011). [DOI] [PubMed] [Google Scholar]
- 38.Franklin R. J. M., Ffrench-Constant C., Remyelination in the CNS: From biology to therapy. Nat. Rev. Neurosci. 9, 839–855 (2008). [DOI] [PubMed] [Google Scholar]
- 39.Sweeney M. D., Sagare A. P., Zlokovic B. V., Blood-brain barrier breakdown in Alzheimer disease and other neurodegenerative disorders. Nat. Rev. Neurol. 14, 133–150 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bennett R. E. et al., Tau induces blood vessel abnormalities and angiogenesis-related gene expression in P301L transgenic mice and human Alzheimer’s disease. Proc. Natl. Acad. Sci. U.S.A. 115, E1289–E1298 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mhatre M. et al., Thrombin, a mediator of neurotoxicity and memory impairment. Neurobiol. Aging 25, 783–793 (2004). [DOI] [PubMed] [Google Scholar]
- 42.Chen Z. L., Strickland S., Neuronal death in the hippocampus is promoted by plasmin-catalyzed degradation of laminin. Cell 91, 917–925 (1997). [DOI] [PubMed] [Google Scholar]
- 43.Neefjes J., Jongsma M. L. M., Paul P., Bakke O., Towards a systems understanding of MHC class I and MHC class II antigen presentation. Nat. Rev. Immunol. 11, 823–836 (2011). [DOI] [PubMed] [Google Scholar]
- 44.Di Liberto G. et al., Neurons under T Cell attack coordinate phagocyte-mediated synaptic stripping. Cell 175, 458–471.e19 (2018). [DOI] [PubMed] [Google Scholar]
- 45.Razakandrainibe R., Pelleau S., Grau G. E., Jambou R., Antigen presentation by endothelial cells: What role in the pathophysiology of malaria? Trends Parasitol. 28, 151–160 (2012). [DOI] [PubMed] [Google Scholar]
- 46.Pober J. S., Merola J., Liu R., Manes T. D., Antigen presentation by vascular cells. Front. Immunol. 8, 1907 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Eimer W. A. et al., Alzheimer’s disease-associated β-Amyloid is rapidly seeded by herpesviridae to protect against brain infection. Neuron 99, 56–63.e3 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gate D. et al., Clonally expanded CD8 T cells patrol the cerebrospinal fluid in Alzheimer’s disease. Nature 577, 399–404 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dulken B. W. et al., Single-cell analysis reveals T cell infiltration in old neurogenic niches. Nature 571, 205–210 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Renthal W. et al., Characterization of human mosaic Rett syndrome brain tissue by single-nucleus RNA sequencing. Nat. Neurosci. 21, 1670–1679 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zheng G. X. Y. et al., Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 8, 14049 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Butler A., Hoffman P., Smibert P., Papalexi E., Satija R., Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhou Y. et al., Human and mouse single-nucleus transcriptomics reveal TREM2-dependent and TREM2-independent cellular responses in Alzheimer’s disease. Nat. Med. 26, 131–142 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ritchie M. E. et al., Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Matarin M. et al., A genome-wide gene-expression analysis and database in transgenic mice during development of amyloid or tau pathology. Cell Rep. 10, 633–644 (2015). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Anonymized snRNA-seq sequencing data have been deposited in GEO (accession no. GSE157827). All study data are included in the article and supporting information.