Significance
The World Health Organization has declared antimicrobial resistance one of the greatest threats to human health. Understanding the molecular mechanisms by which bacteria resist the effects of antibiotic therapies is central to understanding antimicrobial resistance and to informing the development of new treatments that can circumvent these resistance mechanisms. This study reveals the molecular details of the mechanism of FusB-mediated resistance to fusidic acid, an important clinical treatment for Staphylococcus aureus infections, including methicillin-resistant Staphylococcus aureus. Here we elucidate an antibiotic resistance mechanism driven by an allosteric effect on the conformational flexibility of the drug target.
Keywords: antibiotic resistance, Elongation Factor G, NMR, FusB, conformational flexibility
Abstract
Antibiotic resistance in clinically important bacteria can be mediated by target protection mechanisms, whereby a protein binds to the drug target and protects it from the inhibitory effects of the antibiotic. The most prevalent source of clinical resistance to the antibiotic fusidic acid (FA) is expression of the FusB family of proteins that bind to the drug target (Elongation factor G [EF-G]) and promote dissociation of EF-G from FA-stalled ribosome complexes. FusB binding causes changes in both the structure and conformational flexibility of EF-G, but which of these changes drives FA resistance was not understood. We present here detailed characterization of changes in the conformational flexibility of EF-G in response to FusB binding and show that these changes are responsible for conferring FA resistance. Binding of FusB to EF-G causes a significant change in the dynamics of domain III of EF-GC3 that leads to an increase in a minor, more disordered state of EF-G domain III. This is sufficient to overcome the steric block of transmission of conformational changes within EF-G by which FA prevents release of EF-G from the ribosome. This study has identified an antibiotic resistance mechanism mediated by allosteric effects on the dynamics of the drug target.
Antibiotic resistance is an emerging public health crisis that threatens our ability to treat bacterial infections (1). To effectively tackle this problem, a comprehensive understanding of the mechanisms of resistance to these important drugs is crucial, especially because knowledge of these mechanisms in molecular detail could lead to strategies to overcome them and prolong the usefulness of current antibiotics. While many resistance mechanisms are broadly understood, there remain some notable gaps, particularly in our understanding of target protection mechanisms of resistance. In such cases, a resistance protein binds to the drug target, and through this protein–protein interaction protects the target protein from inhibition by the drug. Examples of target protection mechanisms that have been identified to date are the FusB-like proteins (2, 3) and the quinolone resistance protein Qnr (4), but an understanding of the mechanisms of resistance at the molecular level is lacking in both cases. However, the recent identification of ABC-F proteins as mediating resistance to a range of clinically important antibiotics through target protection (5) suggests that such mechanisms may be more clinically significant than previously understood.
Fusidic acid (FA) is one of few remaining antibiotics that can be administered orally to treat methicillin-resistant Staphylococcus aureus infections and is also used topically to treat skin and soft tissue infections, but resistance to this important antibiotic has risen significantly in recent years (6–8). The major clinical source of FA resistance is heterologous expression of the FusB family of proteins (8–11). While recent advances have provided insight into FusB-mediated FA resistance (3, 12, 13), questions remain regarding the underlying molecular mechanism of resistance. Here we elucidate the key mechanisms that allow FusB to confer FA resistance.
Elongation factor G (EF-G) is an essential translocase during protein synthesis, responsible for promoting translocation of transfer RNA (tRNA) from the A to the P site of the ribosome following peptide bond formation (14, 15). EF-G must then be released to allow further rounds of chain elongation to occur. FA binds to EF-G when it is bound to the ribosome during the translocation phase and prevents its release, stalling protein synthesis (16). The FusB family of proteins confer FA resistance by binding to EF-G and promoting dissociation of these stalled complexes via a target protection mechanism to allow protein synthesis to continue (2, 3, 12). We recently solved the solution structure of FusB bound to EF-GC3 (domains III to V of EF-G), showing that FusB binds to domain IV of EF-G and does not interact with the FA-binding site between domains II and III (12). Our structure showed conformational changes in both EF-GC3 and FusB upon forming the complex and also identified evidence of a change in the conformational flexibility of domain III in response to FusB binding. However, it was not possible to determine whether the FusB-induced structural changes or the change in conformational flexibility was responsible for conferring resistance to FA, leaving the key question of how FusB causes FA resistance unanswered.
In this study we have used NMR relaxation dispersion to characterize the differences in conformational flexibility in EF-GC3 in the apo- and FusB-bound forms to determine the effect of FusB binding on domain III of EF-G. By disrupting these dynamics, without perturbing FusB binding and the conformational changes in domains IV and V, we were able to separate the dynamic and structural effects of FusB binding on FA resistance to determine whether the conformational flexibility or changes in the domain orientations within EF-G are the most significant cause of FA resistance. Our data show that FusB binding increases the population of a minor state of EF-GC3 with a more disordered domain III. Preventing the formation of this increased minor state population in response to FusB binding prevents FusB from rescuing EF-G from FA-stalled ribosome complexes, showing that this change in domain III dynamics is key to FusB-mediated FA resistance.
Results
Binding to FusB Triggers a Change in Conformational Dynamics throughout Domain III of EF-GC3.
The 1H-15N amide NMR signals throughout domain III of EF-GC3 broaden upon binding to FusB, suggesting a change in the conformational flexibility of this domain upon FusB binding (12). It is remarkable that nearly all of the amide signals from a single domain become undetectable in the NMR spectrum. This may partly be a consequence of the conditions (pH 8.0 and 30 °C) used for our NMR experiments. If a state is involved with little or no protection from hydrogen exchange such as a partially or fully unfolded state, then the expectation would be that amide signals broaden beyond detection through increased hydrogen exchange. Therefore, to determine whether broadening of amide signals as a result of increased amide exchange is an important factor at the higher pH at which experiments are conducted, 1H-15N-TROSY-HSQC spectra were recorded over the pH range 6.6 to 8.5 (SI Appendix, Fig. S7). These spectra showed that only two additional resonances became visible at pH 6.6, suggesting that the majority of domain III peaks remain broadened at a lower pH. Due to the reduction in amide exchange rate at lower pHs, this lack of additional resonances suggests that the previously identified broadening of amide resonances from domain III (12) is not only due to amide exchange, but also likely reflects conformational exchange and that the combination of both processes may be responsible for the almost uniform disappearance of amide signals from domain III. Fortunately, the majority of 13C-1H ILVA methyl resonances remain visible in 1H-13C heteronuclear multiple quantum coherence (HMQC) spectra, while some resonances from domain III (I408, A426, A439, and L461) are also broadened beyond detection in these spectra in the FusB-bound state but present in the apo spectra. I450, while present in the HMQC spectrum, is also noticeably broadened in the FusB-bound state. This further supports the suggestion that conformational exchange causes considerable broadening in both amide and methyl groups. The higher quality and sensitivity of the methyl spectra allow this process to be more closely investigated.
To understand how the dynamics of EF-GC3 domain III change in response to FusB binding, we used 1H-13C ILVA methyl relaxation dispersion experiments (17) to compare dynamics within EF-GC3 in the presence and absence of FusB. These relaxation dispersion experiments are used to study residues that undergo chemical exchange on a μs-ms timescale between two states for which the NMR chemical shifts are different. Fitting of these data allows extraction of quantitative parameters for the rate of exchange between states, the populations of the two states, and the difference in chemical shift between them (|Δω|). This allows identification of which residues are involved in exchange processes and grouping of residues reporting on the same global exchange. To study dynamics in EF-GC3, methyl-TROSY 1H-13C multiple quantum Carr-Purcell-Meiboom-Gill (CPMG) relaxation dispersion was used (18), allowing identification of methyl groups involved in exchange.
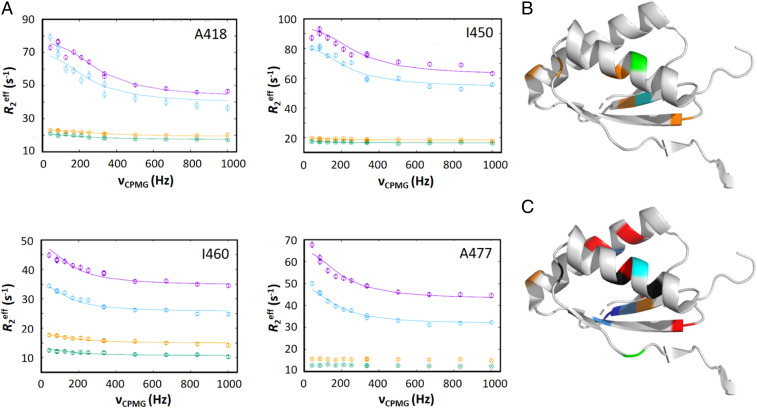
When bound to FusB, many residues within domain III of EF-GC3 showed relaxation dispersion profiles (Fig. 1 and SI Appendix, Fig. S1) that indicate these residues are undergoing exchange on the µs-ms timescale (V412, A418, L427, V448, I449, I450, I460, and A477). Several additional residues showed broadening or signs of dispersion indicating that these residues also undergo exchange, but the peaks became too broad to allow for accurate fitting of R2eff values or dispersion profiles (residues A426, A439, L430, and L461). Relaxation dispersion data were fitted to a standard two-state exchange model. Fitting of dispersion data for each affected residue within domain III independently showed a similar exchange rate and population of the minor state, suggesting a global effect throughout the domain in response to FusB binding. Therefore, fitting was repeated for all residues within domain III using a global exchange rate and population to reflect a single event. This identified an exchange process that occurred at a rate of 930 ± 90 s−1 with a minor state population of 4.7 ± 0.2%. Relaxation dispersion effects were also identified within domains IV and V (residues I600, I609, I619, V678, and A683, SI Appendix, Fig. S2).
Fig. 1.
Comparison of relaxation dispersion effects in domain III of EF-GC3 in the presence and absence of FusB. (A) Methyl relaxation dispersion curves measured for residues in domain III in EF-GC3:FusB at 950 MHz (magenta) and 750 MHz (blue) and in apo EF-GC3 at 950 MHz (orange) and 750 MHz (green), showing significant dispersion effects in the complex that are absent or greatly reduced in the unbound protein, as shown by a reduced difference in R2eff over the range of νCPMG field strengths. (B and C) Mapping of residues showing relaxation dispersion effects on the structure of domain III of EF-G in the apo state [Protein Data Bank (PDB) ID: 2XEX (24)] in the absence (B) and the presence (C) of FusB. Residues showing dispersion effects in each case are colored by the magnitude of the difference in chemical shift (blue: <0.2 ppm; light blue: 0.2 to 0.4 ppm; cyan: 0.4 to 0.6 ppm; green: 0.6 to 0.8 ppm; orange: >0.8 ppm; red: broadens and cannot be accurately fitted; dark gray: unassigned). Dispersion effects are spread more widely throughout domain III and involve more residues in the presence of FusB. All relaxation dispersion data were acquired at 30 °C with a relaxation delay of 24 ms. No structure of domain III in the complex state is currently available as NMR signal broadening prevented modeling of this domain in the complex structure (12). Errors in R2eff were determined through repeated measurement of two data points per experiment. Where error bars are not visible, they are within the limits of the data point. For each relaxation dispersion experiment, n = 13 at each field.
In apo EF-GC3, dispersion profiles similar to those seen in the presence of FusB were observed for residues I600, I609, I619, V678, and A683 in domains IV and V (SI Appendix, Fig. S2). This suggests that the dynamics seen for these residues in the FusB bound state are not caused by FusB binding. Therefore, these effects are not likely to influence FA resistance and are instead likely to be related to the conformational flexibility of EF-G that allows it to catalyze translocation of the ribosome. In contrast to the FusB bound form, few of the residues in domain III undergo dynamics on the µs-ms timescale in the apo state, and those that do (residues I408, A418, I449, and I460) have significantly reduced relaxation dispersion profiles compared to the FusB-bound state, as shown by significantly reduced differences in R2eff with changes in CPMG field strength (Fig. 1). Global fitting of dispersion profiles for residues in domain III in the apo state identified a similar exchange rate to the FusB-bound state (1,100 ± 120 s−1) but a reduced population of the minor state of 0.59 ± 0.03%. This suggests that binding of FusB is causing a change in the equilibrium of an exchange process that occurs within EF-GC3, rather than creating a new exchange process, increasing the population of a minor state that could have an effect on the ability of FA to inhibit release of EF-G from the ribosome.
To investigate the likely changes within domain III of EF-GC3 further, hydrogen exchange–mass spectrometry (HX-MS) was used to compare the apo and FusB-bound states of EF-GC3. These data showed that regions within domains IV and V show the greatest protection from exchange in the presence of FusB (Fig. 2 and SI Appendix, Fig. S3), confirming the role of these regions in FusB binding. In contrast, domain III shows widespread deprotection in response to FusB binding (i.e., residues throughout this domain become more solvent exposed and less hydrogen bonded). This occurs throughout the domain with the greatest effects seen within the α-helices and two of the β-strands (Fig. 2 and SI Appendix, Fig. S3). Most of the residues that show conformational exchange in the FusB-bound state, as determined by relaxation dispersion (Fig. 2C), fall within the most deprotected area of domain III or form interactions with residues in the deprotected regions. The widespread nature of the deprotection within domain III suggests that it may be less ordered in the FusB-bound state. Such disorder within domain III could promote FA resistance by preventing the steric block of transmission of conformational changes from domains I and II to domain III caused by FA binding to allow EF-G to take up the conformation necessary for ribosome release.
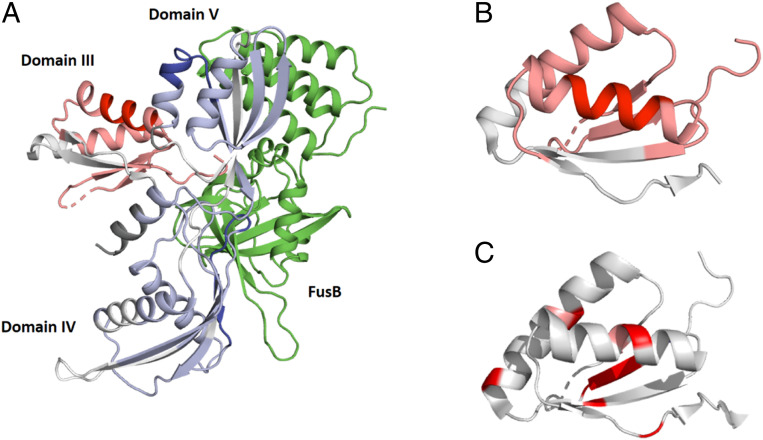
Fig. 2.
Comparison of hydrogen exchange mass spectrometry data in EF-GC3 in the apo- and FusB-bound states. (A) Regions of EF-GC3 that are protected (blue) or deprotected (red) from exchange in the presence of FusB are shown in the structure of the FusB:EF-GC3 complex (12) (PDB ID: 2MZW). The structure of domain III is modeled on the basis of the EF-G apo structure (24) (PDB ID: 2XEX) as no structure of this domain in the FusB-bound complex is available (12). Domains IV and V are protected from exchange upon FusB binding, particularly in the regions that contact FusB, while domain III becomes deprotected throughout the domain. FusB is shown in green. (B and C) Comparison of regions within domain III showing HX-MS deprotection. SI Appendix, Fig. S3 (B and C) Comparison of regions within domain III showing HX-MS deprotection (B) with residues showing relaxation dispersion effects upon FusB binding (C). (B) Dark red indicates greater levels of deprotection, as shown in SI Appendix, Fig. S3. (C) Residues colored red undergo dispersion in CPMG experiments in EF-GC3 bound to FusB. Both significant deprotection and relaxation dispersion occur in the center of the domain, suggesting that they reflect the same change. Data in B and C are shown on the structure of apo EF-G (24) (PDB ID: 2XEX) as no structure of this domain in the presence of FusB is available (12).
In light of the HX-MS data suggesting disorder in domain III in the minor state, we calculated amide exchange rates assuming domain III became unfolded in the FusB-bound state using the program SPHERE (https://protocol.fccc.edu/research/labs/roder/sphere/sphere.html) (19, 20). This predicted an average exchange rate of ∼160 s−1 at pH 8.0 and ∼5 s−1 at pH 6.6 for domain III residues. This suggests that at pH 8.0 amide NMR resonances from a disordered domain III are likely to be broadened while at pH 6.6 it is likely that some resonances would be visible. As amide resonances are not visible at pH 6.6, this suggests that solvent exchange in a disordered domain is not sufficient to explain the loss of amide signals. However, while not completely responsible for amide signal broadening, disorder in a minor state of domain III likely contributes to the widespread broadening of domain III resonances in addition to conformational exchange and could help to explain why so many signals from this domain are lost in the presence of FusB.
To determine whether the FusB-induced disordered domain III minor state is the result of domain III becoming unfolded in the presence of FusB, we compared the difference in chemical shift between the major and minor states extracted from the CPMG relaxation dispersion fitting (|Δω|, SI Appendix, Table S1) with the difference between observed chemical shifts and reference values for unfolded proteins (secondary shifts) (21) (SI Appendix, Fig. S4). While 1H |Δω| in the FusB-bound state correlated well with secondary shifts, the 13C |Δω|-to-secondary shift correlation is weak with large differences for some residues, suggesting that the minor state, while more disordered, does not consist of a fully unfolded domain III. Instead, it is more likely that domain III is becoming more flexible and potentially losing some secondary structure order without resembling a random coil.
Changes in the Dynamics of Domain III Are Key to FA Resistance.
Our previous structural model of the FusB:EF-GC3 complex identified both a change in the dynamics of domain III and also structural changes in domains IV and V in response to FusB binding (12), preventing an analysis of whether the changes in conformational flexibility are important in the mechanism of FA resistance. To determine whether the FusB-induced dynamics changes within domain III or the domain IV and V conformational changes are most important for FA resistance, mutations were introduced into domain III of EF-GC3 to disrupt the effects of FusB binding on domain III dynamics without disrupting the conformational changes in domains IV and V upon FusB binding. These mutations were H438P to restrain the movements of a flexible loop, H409K/M479E double mutant to introduce a salt bridge between β-strands within domain III, N470D to introduce a salt bridge between the end of an α-helix and a β-strand, and H438C/G451C double mutant to create a disulfide bond between β-strands that both show FusB-induced deprotection within domain III (SI Appendix, Fig. S5). For each of the 4 EF-GC3 variants, amide 1H-15N HSQC-TROSY and 1H-13C methyl HMQC spectra in the FusB-bound state overlaid well with the wild-type FusB-bound spectra with no evidence of apo EF-GC3 peaks. This showed that the mutations did not prevent FusB binding or the conformational changes that occur upon FusB binding (SI Appendix, Fig. S5). Analysis of amide chemical shift differences between wild-type and mutant proteins for EF-GC3 H438C/G451C, and EF-GC3 H438P (SI Appendix, Fig. S5) showed that chemical shift differences were small with a maximum of 0.26 ppm and were similar throughout the protein, suggesting that they are limited to experimental variation, with the exception of G446, G556, S586, and N661 in EF-GC3 H438P. However, G446 (the largest chemical shift difference) is close to the site of the introduced mutation and could reflect localized changes resulting from the introduced mutation while the remaining residues are spread throughout the protein. For 1H-15N TROSY-HSQC spectra of EF-GC3 H438C/G451C bound to FusB, in addition to the expected FusB-bound peaks, some additional peaks appear (SI Appendix, Fig. S5D). These resonances could be from residues within domain III that are broadened in response to FusB binding causing exchange on the µs-ms timescale (12) and could therefore suggest that the H438C/G451C mutation stabilizes the domain, disrupting the change in the dynamics of domain III induced by FusB binding, preventing broadening of these resonances.
ILVA methyl relaxation dispersion experiments were performed for each of these four variants in the FusB-bound state at 950 MHz to determine if the mutations caused any changes in the dynamics of the protein. As these data were acquired only at a single magnetic field strength, it is not possible to extract accurate parameters from fitting these data as discussed for the wild-type data at two field strengths. However, such data can be used for qualitative comparison to determine whether dynamics effects are different from those in the wild-type protein to identify those mutations that disrupt effects observed in the wild-type protein. For EF-GC3 H409K/M479E, residues showing dispersion profiles in the wild-type complex showed similar effects in the mutant complex, suggesting that this mutation did not affect the ability of FusB to induce changes in the dynamics of domain III (Fig. 3). Likewise, the N470D mutation showed little change in the dynamics of EF-GC3 compared with the wild-type protein. This suggests that the exchange process induced by FusB may not involve significant disruption of these interactions. In support of this, the H409K/M479E expected additional salt bridge involves a β-strand on the edge of domain III that shows the lowest levels of HX-MS deprotection in response to FusB binding, and this region of the protein may be less affected by FusB binding.
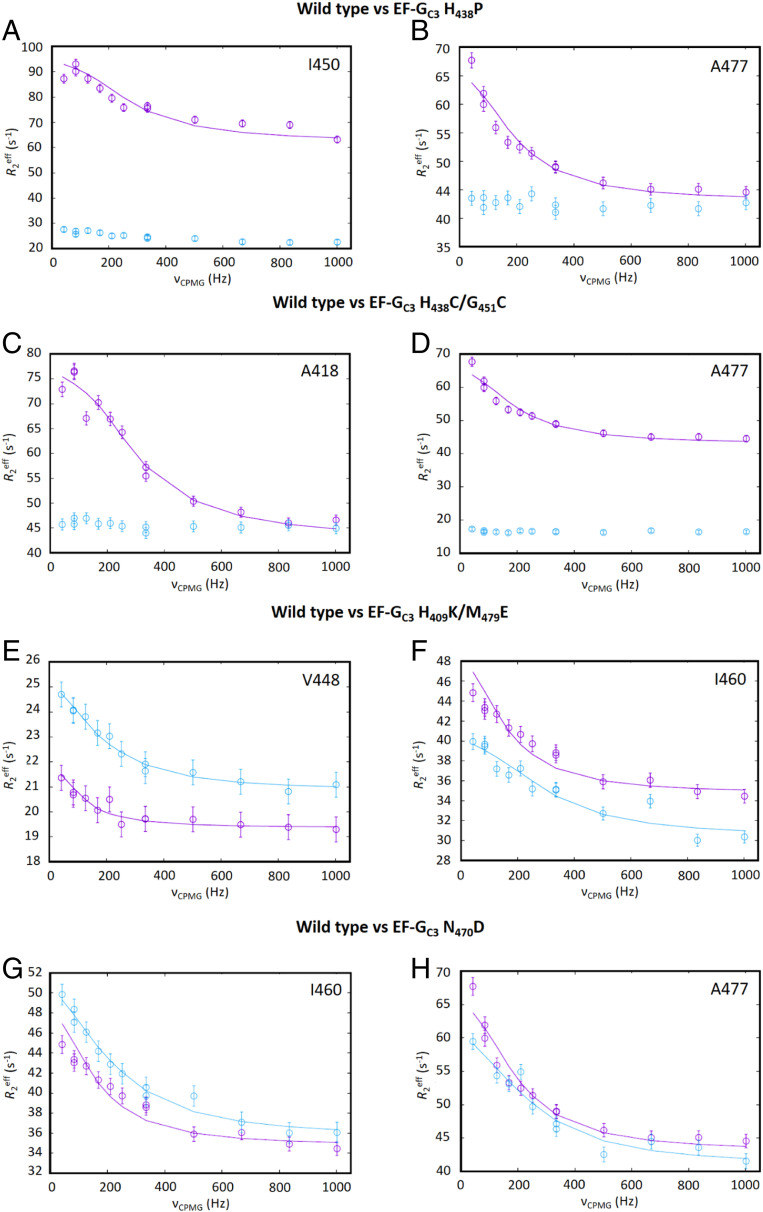
Fig. 3.
Comparison of the effects of mutations in EF-GC3 methyl relaxation dispersion. Comparison of methyl relaxation dispersion data for wild-type (cyan) and mutant (magenta) forms of EF-GC3 bound to FusB for (A and B) EF-GC3 H438P, (C and D) EF-GC3 H438C/G451C, (E and F) EF-GC3 H409K/M479E, and (G and H) EF-GC3 N470D showing EF-GC3 H409K/M479E and N470D have relaxation dispersion curves for residues in domain III that are similar to those in the wild-type protein while the remaining variants show little or no relaxation dispersion for these residues. Data were acquired at 950 MHz, 30 °C, with a relaxation delay of 24 ms. Errors in R2eff were determined through repeated measurement of two data points per experiment. For each experiment, n = 13.
For both EF-GC3 H438P and EF-GC3 H438C/G451C, ILVA methyl relaxation dispersion experiments in the presence of FusB showed marked differences from the wild-type complex. For both mutations, the residues in domain III that show large dispersion profiles in the wild-type complex showed no or significantly reduced dispersion profiles in the presence of FusB, suggesting that these mutations did indeed allow the dynamics in domain III to be restrained in the presence of FusB (Fig. 3). Comparison of relaxation dispersion curves for residues in domains IV and V showed that residues within these domains in the EF-GC3 H438C/G451C mutant gave similar dispersion to those of the wild type protein, suggesting the exchange that occurs in both apo- and FusB-bound states remains unchanged for this mutant (SI Appendix, Fig. S6). Although there is some suggestion of reduced dispersion curves in the EF-GC3 H438P mutant, the same residues show dispersion as those seen for wild-type EF-GC3, and it is possible that the differences could reflect changes that also occur in the apo state for this mutant. The fact that there are no significant differences between the FusB-bound NMR spectra for these proteins compared to the wild type shows that both mutations allow the separation of the conformational change upon FusB binding from the change in the dynamics of domain III, allowing these mutants to be used to determine which process is most important in conferring FA resistance.
As both mutations resulting in changes in the dynamics of EF-GC3 domain III upon FusB binding involve substitution of a histidine, 1H-13C HMQC and 1H-15N-TROSY-HSQC spectra of ILVA-labeled EF-GC3 bound to FusB were measured over a pH range of 6.6 to 8.5 to ensure that the relaxation dispersion profiles observed in the wild-type protein were not the result of histidine protonation/deprotonation. The HMQC spectra showed very little change in chemical shift with pH (SI Appendix, Fig. S7) over this range, suggesting that there is little effect of histidine titrations on the chemical shifts of these residues. The greatest effect of pH on chemical shift within domain III is for A477, which has changes in 1H and 13C chemical shifts of 0.084 and 0.138 ppm, respectively, suggesting that the titration of a sidechain in the region has an effect. However, plotting the chemical shift against pH suggests that this is responding to something with a pKa greater than 8.5 and so is unlikely to be the result of a histidine. Also, this effect is localized to A477, which is not close to the mutated H438, and it is unlikely that titration of this histidine could affect A477 but not residues closer to it. Furthermore, for the change in dynamics within domain III to be reflecting histidine titrations, the chemical shift changes would be expected to be much more widespread to resemble the extent of changes in relaxation dispersion. Therefore, it is unlikely that the change in relaxation dispersion data in the H438P and H438C/G451C mutants compared to the dispersion profiles observed in the wild-type protein reflects responses to histidine protonation/deprotonation. These results therefore reflect real changes in the interactions within domain III in response to FusB binding.
To assess the role of these mutations in FA resistance, a fluorescence-based assay to follow the buildup of stalled ribosome:EF-G:GDP complexes was employed (3). Escherichia coli ribosomes were mixed with EF-G and fluorescent BODIPY-FL-GDP, and increasing concentrations of FA were added. The formation of the ribosome:EF-G:GDP complex results in a large increase in the fluorescence of BODIPY-FL-GDP, and therefore the effect of increasing concentrations of FA in forming stalled complexes can be measured. Adding FusB in a 1:5 EF-G:FusB molar excess allows the determination of whether FusB can prevent the buildup of these stalled complexes. To ensure that the mutations in EF-G do not prevent ribosome binding or cause FA resistance, this assay was first performed in the absence of FusB. For EF-G H409K/M479E, H438P and H438C/G451C, similar fluorescence curves to those observed for wild-type EF-G were produced, showing that the proteins bind to the ribosome and can be inhibited by FA (Fig. 4). In order to produce the same magnitude of fluorescence change as the wild type, the concentration of EF-G H438P needed to be increased, suggesting that this mutation did reduce the overall affinity of EF-G for the ribosome. However, this was subsequently compensated for by an increased concentration of FusB to maintain the 1:5 molar ratio. For EF-G H438C/G451C, while a curve similar to that obtained with wild-type EF-G was produced, the total fluorescence change was slightly reduced after increasing the EF-G concentration compared to wild type, suggesting that this mutation also reduces the affinity of EF-G for the ribosome. However, the effects of FusB binding can still be assessed based on differences for this mutant in the presence and absence of FusB in the same 1:5 molar ratio.
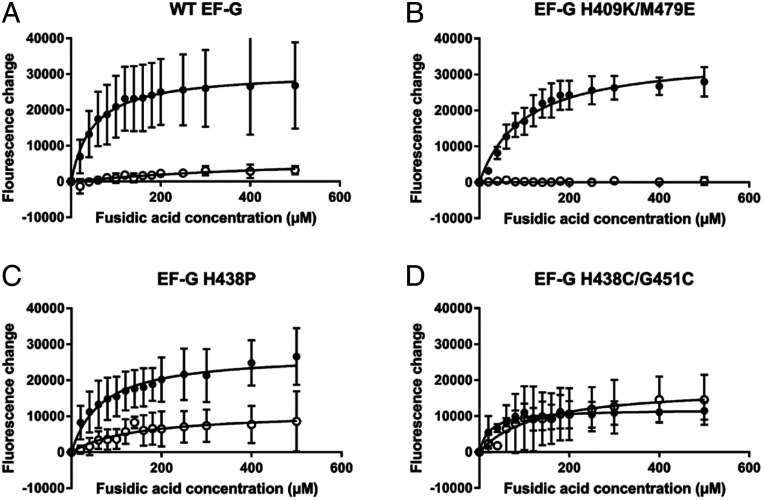
Fig. 4.
The impact of FusB upon accumulation of stalled ribosome:EF-G:GDP:FA complexes. Fluorescence of BODIPY-FL-GDP in response to increasing concentrations of FA in the absence (closed circles) and presence (open circles) of FusB for (A) wild-type EF-G, (B) EF-G H409K/M479E, (C) EF-G H438P, and (D) EF-G H438C/G451C. Data show that, for both wild-type and EF-G H409K/M479E, stalled complexes accumulate in the absence of FusB but little accumulation is observed in the presence of FusB, as FusB promotes dissociation of these complexes. In contrast, for both EF-G H438P and EF-G H438C/G451C, stalled complexes build up in the absence of FusB, similar to the wild-type protein, but both mutant proteins show an increase in fluorescence with increasing FA concentration in the presence of FusB. The large error bars for EF-G H438P mean that a significant difference from wild type cannot be shown with certainty, but the data suggest increased formation of stalled complexes in the presence of FusB. While not restored to levels in the absence of FusB for EF-G H438P, these data show that the H438P and H438C/G451C mutations at least partially restore FA sensitivity in the presence of FusB. This suggests that the changes in dynamics in response to FusB binding that are suppressed by these mutations are important in the mechanism of FusB-induced FA resistance. Each data point represents the mean value after three independent repeats with error bars representing the SD from the mean. For each experiment, n = 15.
In the presence of FusB, very little fluorescence change was observed for either the wild-type or EF-G H409K/M479E proteins (Fig. 4), showing that both can be prevented from forming the stalled complexes by FusB. However, for both EF-G H438P and EF-G H438C/G451C, there was a marked increase in fluorescence in response to increasing concentrations of FA even in the presence of FusB. This shows that both mutations impair the ability of FusB to rescue EF-G from forming stalled complexes. For EF-G H438C/G451C, the response in the presence of FusB is clearly different from the wild-type protein, but due to larger error bars, the response of EF-G H438P to FA in the presence of FusB cannot confidently be said to be significantly different from the wild-type protein. The increased fluorescence response, however, does suggest that the protein is more susceptible to FA than the wild type. For EF-G H438C/G451C, the response to FA addition in the presence of FusB closely resembled that in the absence of FusB, while in the case of EF-G H438P the profile of the curve was similar to that of the protein in the absence of FusB, but the maximum change in fluorescence was less. This shows that, while both mutations impair FusB-mediated FA resistance, the H438P mutation does not abolish it completely as the magnitude of the fluorescence increase does not return to levels seen in the absence of FusB. However, these effects show that the change in the dynamics of domain III in response to FusB binding are important in the mechanism of FusB-mediated FA resistance as the conformational change within domains IV and V is not sufficient to confer FA resistance. It is likely that the H438P mutation has impaired rather than abolished the changes within domain III, accounting for the inability of the mutation to completely abolish FA resistance.
Discussion
The FusB family of proteins comprises the major source of clinical resistance to FA (10), but, despite recent progress, questions remain over the molecular mechanism of resistance. Our previous work has shown that FusB binding causes both conformational changes in EF-GC3 and allosterically induced changes in the conformational flexibility of domain III (12). However, the relative importance of the conformational and dynamics changes in response to FusB binding and the nature of the changes in domain III dynamics could not be determined. In this work, we have characterized these changes in dynamics and determined their relative importance in the mechanism of resistance to FA.
Comparing relaxation dispersion data in the apo- and FusB-bound forms of EF-GC3, it is clear that there is a widespread change in the dispersion profiles within domain III in response to FusB binding. However, fitting the smaller dispersion profiles in the apo state suggests that FusB likely significantly increases the population of a minor state that occurs in apo EF-GC3, rather than instigating an exchange process that does not occur in the absence of FusB. The widespread deprotection observed by HX-MS, extending throughout both helices and the β-sheet, suggests that this minor state is likely to be a more disordered form of domain III, similar to the partially disordered nature of domain III in some crystal structures of EF-G (22, 23), rather than a defined minor state conformation, as reduction of amide hydrogen bonding within all secondary structure elements suggests destabilization of all of the structure of the domain. Differences in chemical shifts between major and minor states suggest that the minor state includes a more disordered, but not a fully unfolded, domain III. This agrees with the widespread resonance broadening in the NMR spectra and conformational exchange observed by relaxation dispersion, as a single conformational change would not be expected to cause effects throughout the domain.
EF-G undergoes several conformational changes throughout translocation that are central to its function. Comparison of structures of EF-G determined in the apo form (24, 25), nucleotide-bound form (23, 26), and resident on the ribosome pre- and post translocation (27–29) show that major structural rearrangements occur within EF-G throughout the translocation cycle, most of which involve domains I and II moving relative to domains III to V (30). Dissociation of EF-G from the ribosome following translocation requires conformational changes to be transmitted from domains I and II to domain IV to disrupt the contacts that this domain makes with the 30S subunit of the ribosome (29). FA inhibits dissociation of EF-G from the ribosome by binding to EF-G between domains II and III and apparently restricting this transmission of conformational change (27).
The increased population of a more disordered minor state observed in this study would cause EF-G, on average, to take up this minor state more often, and it is likely that this state is sufficient to allow release from the ribosome by preventing the steric block of conformational change in domains III to V conferred by FA (27). This increase in population could therefore be enough to allow release of EF-G from the stalled complexes to allow protein synthesis to continue. FusB-mediated resistance has been shown to confer FA resistance to around 16 µg/mL FA (31), a lower concentration than that which can be conferred by some mutations within EF-G itself (fusA-type resistance) [e.g., L461K > 256 µg/mL (32) for a mutation within the FA-binding site that is likely to reduce FA affinity]. If FusB acts to increase the population of a more disordered “ribosome-release” conformation of EF-G, then this would most likely have an upper limit to its ability to confer resistance. Too great an increase in the population of this state would itself inhibit protein synthesis by destabilizing the EF-G:ribosome binding interaction to the extent that EF-G could not function effectively in translocation, and the fact that the more disordered state remains a minor state would reflect that. This may well explain why resistance levels are more modest for FusB-type resistance than for mutations in EF-G expected to directly reduce the affinity for FA.
The increased buildup of FA-stalled complexes in the presence of FusB for those mutant EF-G proteins that do not allow FusB to confer the same change in the population of the minor state suggests that it is this change in the equilibrium between the native and the more disordered states of domain III, and not the conformational change in domains IV and V upon binding (12), that allows FusB to confer FA resistance. This model for the FusB-mediated resistance mechanism is consistent with previously published data that show that FusB can act as a release factor, increasing release of EF-G from the ribosome in the absence of FA (3) as increasing a more disordered ribosome-release state would necessarily cause increased dissociation of EF-G from the ribosome regardless of whether FA is present. The Kd of EF-G for FusB is in the nanomolar range (3), and EF-G binds to FusB in the absence of the ribosome so it is likely that a significant proportion of EF-G in the cell would be bound to FusB, thereby increasing release from the ribosome.
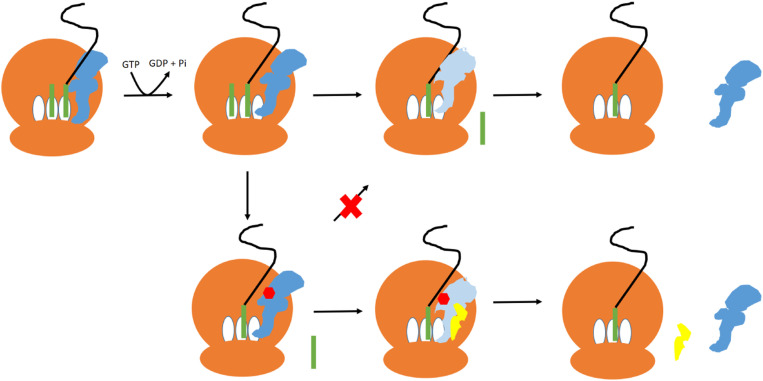
In light of these findings, we therefore propose that FusB-mediated FA resistance occurs through an allosterically induced change in the conformational flexibility of domain III of EF-G in response to FusB binding (Fig. 5). This increases the population of a more disordered state of domain III. During translocation, conformational changes within domain III are thought to cause movement of domains IV and V relative to domains I and II in response to conformational changes in the latter domains upon GTP hydrolysis (30). FA has been shown to prevent these changes by mimicking the GTP-bound state of domains I and II where they contact domain III (27). If FusB binding causes domain III to become more flexible, then this would allow movement of domains IV and V relative to domains I and II irrespective of conformational changes in these domains and so allow release of EF-G from the ribosome. In the presence of FA, this could well be enough to allow transient formation of the GDP-bound conformation and cause release of EF-G from the ribosome even in the presence of the inhibitory drug and therefore allow protein synthesis to continue. In the absence of FA, this same effect would lead to greater release of EF-G from the ribosome. Once released from the ribosome, EF-G has a much-reduced affinity for FA (14) so the drug would be expected to dissociate from EF-G, allowing protein synthesis to continue. This mechanism of FA resistance is consistent with all available data and previously published results (3, 12). As a small population of the minor state is present in the absence of FusB, this opens up the intriguing possibility that disorder in domain III plays a role in the normal release of EF-G from the ribosome where FusB is absent. However, further investigation will be needed to establish if this is the case.
Fig. 5.
The proposed mechanism of FusB-mediated FA resistance. In the absence of FA, EF-G (blue) binds to the ribosome (brown) and catalyzes translocation of the tRNA (green) from the A (acceptor) and P (peptidyl) to the P and E (exit) sites. GTP hydrolysis causes EF-G to undergo a conformational change (light blue), allowing EF-G to dissociate from the ribosome. In the presence of FA, FA (red) binds to EF-G following GTP hydrolysis and prevents the conformational change so that EF-G does not dissociate from the ribosome. FusB (yellow) binding to EF-G promotes an increased population of a minor state of EF-G with a more disordered domain III, allowing EF-G to undergo the conformational change required for release from the ribosome even while bound to FA.
Allosteric effects have been shown to play a significant role in only two resistance mechanisms to date: FusB-mediated FA resistance (12) and PBP2a-mediated methicillin resistance (33, 34), while a mechanism that requires changes in the dynamics of the drug target has not previously been reported to the best of our knowledge. However, given the importance of dynamics in the correct functioning of many proteins, it is likely that the mechanism outlined here represents a type of resistance mechanism that plays a role in resistance to other antibiotics.
Materials and Methods
Additional details are available in SI Appendix, Supplementary Materials and Methods.
Protein Overexpression and Purification.
FusB, EF-G, and EF-GC3 were expressed and purified as previously described (12). FusB and EF-G were produced with no specific labeling while EF-GC3 was produced with either no specific labeling, uniform 15N, 13C, and partial 2H labeling or ILVA methyl 13C 1H labeling on a uniformly 12C 2H 15N background as detailed in SI Appendix, Supplementary Materials and Methods.
NMR.
All NMR spectra were acquired using 200- to 250-µM protein samples in 20 mM Tris-HCl, 300 mM sodium chloride, 1 mM dithiothreitol (DTT), pH 8.0, except for samples containing the H438C/G451C mutations for which the DTT was omitted to allow for disulfide bond formation. Spectra were recorded on Bruker Avance spectrometers at field strengths of either 950 or 750 MHz, equipped with cryoprobes (TXO 5- mm probe or 3-mm TCI probe at 950 MHz and TCI 5-mm probe at 750 MHz). Spectra for pH titrations were acquired using 100 μM of protein on a Bruker Avance 600-MHz spectrometer equipped with a 5-mm QCI-P cryoprobe. For samples in the FusB-bound state, the protein under study was saturated with 1.5× nonisotopically enriched binding partner. Where appropriate, all experiments were performed using TROSY modifications (35) and deuterium decoupling. Data were processed in NMRPipe (36) before assignment and measurement of chemical shift perturbations by CCPN analysis (37). Peak intensity measurements were made using NMRView (38). Some data analysis was performed using NMRbox (39).
NMR Resonance Assignment.
ILVA methyl resonances for EF-GC3 in both the apo and complex states were assigned using CCH-TOCSY spectra measured at 750 MHz with reference to previously published backbone assignments (12). These assignments were supplemented with HMQC spectra of EF-GC3 harboring conservative mutations to remove selected Leu, Val, or Ala residues to identify residues that could not be otherwise assigned. Each of these samples contained one Ala-to-Gly, one Val-to-Ile, and one Leu-to-Ile mutation to allow assignments to be made based on loss of peaks from the methyl HMQC spectrum.
NMR–CPMG Relaxation Dispersion.
NMR methyl relaxation dispersion experiments were acquired at 30 °C at field strengths of both 950 and 750 MHz using EFGC3 with 1H-13C labeled Ile Cδ, Leu Cδ, Val Cγ, and Ala Cβ on a 2H-12C background in complex with unlabeled FusB. Experiments were conducted using the pulse program of Korzhnev et al. (18) with a T2 mixing time of 24 ms and CPMG field strengths of 41.8. 83.8 (repeated), 126.1, 168.6, 210.4, 251.9, 335.4 (repeated), 502.0, 668.7, 835.4, and 1002.0 Hz. Errors in peak intensities were estimated through the use of repeated CPMG field strengths. Methyl groups showing relaxation dispersion were identified as those with a difference in R2eff over the course of the νCPMG field range of at least 1 s−1. Methyl groups with overlapped peaks or those for which the systematic variation in R2eff was not less than the overall R2eff change were discounted. Methyl groups showing relaxation dispersion but for which peak height was reduced to the noise level for some data points were noted as showing dispersion but were not subsequently used for fitting as no accurate R2eff values could be obtained for those data points. Relaxation dispersion data were fitted to a standard two-state exchange model using CATIA (40), initially fitting data for each residue separately and then grouping data for residues showing similar kex and Pb values. A global fit to a single kex and Pb was then performed. For samples containing mutations to disrupt dynamics in EF-GC3, relaxation dispersion data were acquired at 950 MHz only.
Hydrogen Exchange–Mass Spectrometry.
HX-MS experiments were performed as previously outlined (41). Briefly, experiments were carried out using a HX robot (LEAP Technologies) interfaced to an Acquity HX manager and M-Class LC (Waters). Samples were prepared with either 8 µM of EF-GC3 or 8 µM of EF-GC3 and 10 µM FusB in 10 mM potassium phosphate buffer, pH 8.0 containing 300 mM NaCl. Thirty microliters of sample and 135 μL of deuterated buffer (10 mM potassium phosphate buffer, pH 8.0, 300 mM NaCl, 82% D2O) were mixed and incubated for 0.5, 2, 10, 30, or 60 min at 4 °C. For each condition and time point, measurements were performed in triplicate. The HX reaction was quenched by mixing 50 μL of the reaction with 100 μL of quench buffer (10 mM potassium phosphate, 2 M Gdn-HCl, pH 2.2). Fifty microliters of this sample was then injected into immobilized pepsin/aspergillopepsin columns (Affipro) (20 °C). A VanGuard Precolumn [Acquity UPLC BEH C18 (1.7 μm, 2.1 mm × 5 mm, Waters)] was used to trap peptides for 3 min, after which they were injected into a C18 column (75 μm × 150 mm, Waters). A 7-min gradient of 0 to 40% (vol/vol) acetonitrile (0.1% vol/vol formic acid) in H2O (0.3% vol/vol formic acid) at 40 μL • min−1 was used to separate the peptides. Separated peptides were infused into a Synapt G2Si mass spectrometer (Waters) operating in HDMSE mode. Peptides were separated by ion mobility before being subjected to fragmentation by CID (collision induced dissociation) (42). PLGS (v3.0.2) and DynamX (v3.0.0) (Waters) were used for data analysis. Peptides were filtered in DynamX using the following parameters: minimum intensity = 1,000; minimum products per amino acid = 0.3; maximum sequence length = 25; maximum parts-per-million error = 5; file threshold = 3. Deuterium uptake data for all time points are shown in SI Appendix, Table S2. Deuteros (43) was used to determine if increases/decreases in deuterium uptake between states were statistically significant and to visualize the data. The raw data have been made available in the ProteomeXchange Consortium via the PRIDE (44) partner repository with the dataset identifier PXD017491. A reporting summary (45) can be found in SI Appendix, Table S4.
FA Inhibition Assay.
To determine the effect of different mutations on the sensitivity of EF-G to FA and the ability of FusB to rescue EF-G from stalled complexes in the presence of FA, a fluorescence-based assay to measure the buildup of stalled complexes was used (3). All experiments were performed in triplicate. FA in the range 0 to 500 µM was added to a 50-µL solution containing 0.5 µM E. coli ribosomes (NEB), 0.05 µM BODIPY-FL-GDP (Invitrogen), and either 0.5 µM EF-G, 0.8 µM EF-G H409K/M479E, 2.5 µM EF-G H438P, or 5.0 µM EF-G H438C/G451C in 50 mM Tris–HCl, 70 mM NH4Cl, 30 mM KCl, 7 mM MgCl2, pH 7.5, in the presence and absence of FusB in a 5× molar excess with respect to EF-G. Fluorescence was measured with an excitation wavelength of 485 nm and an emission wavelength of 520 nm at 37 °C using a FLUOStar Omega plate reader.
Supplementary Material
Acknowledgments
We thank Prof. Flemming Hansen (University College London) for access to CATIA and processing scripts for NMR relaxation dispersion fitting and helpful discussions regarding its use. For access to the 950-MHz spectrometers, we acknowledge the Astbury Biostructure Laboratory BioNMR facility, which was funded by the University of Leeds and the Francis Crick Institute through provision of access to the Medical Research Council Biomedical NMR Centre. The Francis Crick Institute receives its core funding from Cancer Research UK (FC001029), the Medical Research Council (FC001029), and the Wellcome Trust (FC001029). This study made use of NMRbox: National Center for Biomolecular NMR Data Processing and Analysis. NMRBox is a Biomedical Technology Research Resource which is supported by NIH National Institute of General Medical Sciences Grant P41GM111135. This work was supported by a Royal Society Dorothy Hodgkin Fellowship (DH160089) for J.H.T. A.N.C. acknowledges support from a University Academic Fellowship from the University of Leeds.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2008577117/-/DCSupplemental.
Data Availability.
Assigned methyl chemical shifts for EF-GC3 in the apo- and FusB-bound states have been deposited in the Biological Magnetic Resonance Data Bank with the accession codes 50221 (46) and 50220 (47), respectively. HX-MS data have been deposited in the ProteomeXchange Consortium with the accession number PXD017491 (48). FA inhibition assay data are available in the Open Science Forum (OSF) (https://osf.io/uq4g7) (49). Raw relaxation dispersion data are available in the OSF (https://osf.io/95v4r/) (50). All other data are contained within the main text and SI Appendix.
References
- 1.O’Neill J., Antimicrobial resistance: Tackling a crisis for the health and wealth of nations. The reivew on antimicrobial resistance. http://amr-review.org/sites/default/files/AMR%20Review%20Paper%20-%20Tackling%20a%20crisis%20for%20the%20health%20and%20wealth%20of%20nations_1.pdf (2014). Accessed 20 November 2015.
- 2.O’Neill A. J., Chopra I., Molecular basis of fusB-mediated resistance to fusidic acid in Staphylococcus aureus. Mol. Microbiol. 59, 664–676 (2006). [DOI] [PubMed] [Google Scholar]
- 3.Cox G. et al., Ribosome clearance by FusB-type proteins mediates resistance to the antibiotic fusidic acid. Proc. Natl. Acad. Sci. U.S.A. 109, 2102–2107 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tran J. H., Jacoby G. A., Hooper D. C., Interaction of the plasmid-encoded quinolone resistance protein Qnr with Escherichia coli DNA gyrase. Antimicrob. Agents Chemother. 49, 118–125 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sharkey L. K. R., Edwards T. A., O’Neill A. J., ABC-F proteins mediate antibiotic resistance through ribosomal protection. MBio 7, e01975 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Howden B. P., Grayson M. L., Dumb and dumber–The potential waste of a useful antistaphylococcal agent: Emerging fusidic acid resistance in Staphylococcus aureus. Clin. Infect. Dis. 42, 394–400 (2006). [DOI] [PubMed] [Google Scholar]
- 7.Brown E. M., Thomas P., Fusidic acid resistance in Staphylococcus aureus isolates. Lancet 359, 803 (2002). [DOI] [PubMed] [Google Scholar]
- 8.McLaws F. B., Larsen A. R., Skov R. L., Chopra I., O’Neill A. J., Distribution of fusidic acid resistance determinants in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 55, 1173–1176 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McLaws F., Chopra I., O’Neill A. J., High prevalence of resistance to fusidic acid in clinical isolates of Staphylococcus epidermidis. J. Antimicrob. Chemother. 61, 1040–1043 (2008). [DOI] [PubMed] [Google Scholar]
- 10.Castanheira M., Watters A. A., Bell J. M., Turnidge J. D., Jones R. N., Fusidic acid resistance rates and prevalence of resistance mechanisms among Staphylococcus spp. isolated in North America and Australia, 2007-2008. Antimicrob. Agents Chemother. 54, 3614–3617 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Castanheira M., Watters A. A., Mendes R. E., Farrell D. J., Jones R. N., Occurrence and molecular characterization of fusidic acid resistance mechanisms among Staphylococcus spp. from European countries (2008). J. Antimicrob. Chemother. 65, 1353–1358 (2010). [DOI] [PubMed] [Google Scholar]
- 12.Tomlinson J. H., Thompson G. S., Kalverda A. P., Zhuravleva A., O’Neill A. J., A target-protection mechanism of antibiotic resistance at atomic resolution: Insights into FusB-type fusidic acid resistance. Sci. Rep. 6, 19524 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cox G., Edwards T. A., O’Neill A. J., Mutagenesis mapping of the protein-protein interaction underlying FusB-type fusidic acid resistance. Antimicrob. Agents Chemother. 57, 4640–4644 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bodley J. W., Zieve F. J., Lin L., Zieve S. T., Formation of the ribosome-G factor-GDP complex in the presence of fusidic acid. Biochem. Biophys. Res. Commun. 37, 437–443 (1969). [DOI] [PubMed] [Google Scholar]
- 15.Rodnina M. V., Savelsbergh A., Katunin V. I., Wintermeyer W., Hydrolysis of GTP by elongation factor G drives tRNA movement on the ribosome. Nature 385, 37–41 (1997). [DOI] [PubMed] [Google Scholar]
- 16.Tanaka N., Kinoshita T., Masukawa H., Mechanism of protein synthesis inhibition by fusidic acid and related antibiotics. Biochem. Biophys. Res. Commun. 30, 278–283 (1968). [DOI] [PubMed] [Google Scholar]
- 17.Otten R., Villali J., Kern D., Mulder F. A. A., Probing microsecond time scale dynamics in proteins by methyl (1)H Carr-Purcell-Meiboom-Gill relaxation dispersion NMR measurements. Application to activation of the signaling protein NtrC(r). J. Am. Chem. Soc. 132, 17004–17014 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Korzhnev D. M., Kloiber K., Kanelis V., Tugarinov V., Kay L. E., Probing slow dynamics in high molecular weight proteins by methyl-TROSY NMR spectroscopy: Application to a 723-residue enzyme. J. Am. Chem. Soc. 126, 3964–3973 (2004). [DOI] [PubMed] [Google Scholar]
- 19.Zhang Y.-Z., “Protine and peptide structure and interactions studied by hydrogen exchange and NMR,” PhD thesis, University of Pennsylvania, Philadelphia (1995).
- 20.Bai Y., Milne J. S., Mayne L., Englander S. W., Primary structure effects on peptide group hydrogen exchange. Proteins 17, 75–86 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kjaergaard M., Iešmantavičius V., Poulsen F. M., The interplay between transient α-helix formation and side chain rotamer distributions in disordered proteins probed by methyl chemical shifts. Protein Sci. 20, 2023–2034 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.al-Karadaghi S., Aevarsson A., Garber M., Zheltonosova J., Liljas A., The structure of elongation factor G in complex with GDP: Conformational flexibility and nucleotide exchange. Structure 4, 555–565 (1996). [DOI] [PubMed] [Google Scholar]
- 23.Hansson S., Singh R., Gudkov A. T., Liljas A., Logan D. T., Crystal structure of a mutant elongation factor G trapped with a GTP analogue. FEBS Lett. 579, 4492–4497 (2005). [DOI] [PubMed] [Google Scholar]
- 24.Chen Y., Koripella R. K., Sanyal S., Selmer M., Staphylococcus aureus elongation factor G: Structure and analysis of a target for fusidic acid. FEBS J. 277, 3789–3803 (2010). [DOI] [PubMed] [Google Scholar]
- 25.AEvarsson A. et al., Three-dimensional structure of the ribosomal translocase: Elongation factor G from Thermus thermophilus. EMBO J. 13, 3669–3677 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Czworkowski J., Wang J., Steitz T. A., Moore P. B., The crystal structure of elongation factor G complexed with GDP, at 2.7 A resolution. EMBO J. 13, 3661–3668 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gao Y.-G. et al., The structure of the ribosome with elongation factor G trapped in the posttranslocational state. Science 326, 694–699 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen Y., Feng S., Kumar V., Ero R., Gao Y.-G., Structure of EF-G-ribosome complex in a pretranslocation state. Nat. Struct. Mol. Biol. 20, 1077–1084 (2013). [DOI] [PubMed] [Google Scholar]
- 29.Zhou J., Lancaster L., Donohue J. P., Noller H. F., Crystal structures of EF-G-ribosome complexes trapped in intermediate states of translocation. Science 340, 1236086 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li W., Trabuco L. G., Schulten K., Frank J., Molecular dynamics of EF-G during translocation. Proteins 79, 1478–1486 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chopra I., Mechanisms of resistance to fusidic acid in Staphylococcus aureus. J. Gen. Microbiol. 96, 229–238 (1976). [DOI] [PubMed] [Google Scholar]
- 32.Besier S., Ludwig A., Brade V., Wichelhaus T. A., Molecular analysis of fusidic acid resistance in Staphylococcus aureus. Mol. Microbiol. 47, 463–469 (2003). [DOI] [PubMed] [Google Scholar]
- 33.Fishovitz J., Taghizadeh N., Fisher J. F., Chang M., Mobashery S., The Tipper–Strominger hypothesis and triggering of allostery in penicillin-binding protein 2a of methicillin-resistant Staphylococcus aureus (MRSA). J. Am. Chem. Soc. 137, 6500–6505 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fishovitz J. et al., Disruption of allosteric response as an unprecedented mechanism of resistance to antibiotics. J. Am. Chem. Soc. 136, 9814–9817 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pervushin K., Riek R., Wider G., Wüthrich K., Attenuated T2 relaxation by mutual cancellation of dipole-dipole coupling and chemical shift anisotropy indicates an avenue to NMR structures of very large biological macromolecules in solution. Proc. Natl. Acad. Sci. U.S.A. 94, 12366–12371 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Delaglio F. et al., NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293 (1995). [DOI] [PubMed] [Google Scholar]
- 37.Vranken W. F. et al., The CCPN data model for NMR spectroscopy: Development of a software pipeline. Proteins 59, 687–696 (2005). [DOI] [PubMed] [Google Scholar]
- 38.Schwarzinger S., Kroon G. J. A., Foss T. R., Wright P. E., Dyson H. J., Random coil chemical shifts in acidic 8 M urea: Implementation of random coil shift data in NMRView. J. Biomol. NMR 18, 43–48 (2000). [DOI] [PubMed] [Google Scholar]
- 39.Maciejewski M. W. et al., NMRbox: A resource for biomolecular NMR computation. Biophys. J. 112, 1529–1534 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hansen D. F., Vallurupalli P., Lundström P., Neudecker P., Kay L. E., Probing chemical shifts of invisible states of proteins with relaxation dispersion NMR spectroscopy: How well can we do? J. Am. Chem. Soc. 130, 2667–2675 (2008). [DOI] [PubMed] [Google Scholar]
- 41.Calabrese A. N. et al., Inter-domain dynamics in the chaperone SurA and multi-site binding to its outer membrane protein clients. Nat. Commun. 11, 2155 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cryar A., Groves K., Quaglia M., Online hydrogen-deuterium exchange traveling wave ion mobility mass spectrometry (HDX-IM-MS): A systematic evaluation. J. Am. Soc. Mass Spectrom. 28, 1192–1202 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lau A. M. C., Ahdash Z., Martens C., Politis A., Deuteros: Software for rapid analysis and visualization of data from differential hydrogen deuterium exchange-mass spectrometry. Bioinformatics 35, 3171–3173 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Perez-Riverol Y. et al., The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res. 47, D442–D450 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Masson G. R. et al., Recommendations for performing, interpreting and reporting hydrogen deuterium exchange mass spectrometry (HDX-MS) experiments. Nat. Methods 16, 595–602 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tomlinson J. H., Kalverda A. P., Staphylococcus aureus EF-GC3 methyl side chain chemical shifts. Biological Magnetic Resonance Data Bank. http://www.bmrb.wisc.edu/data_library/summary/index.php?bmrbId=50221. Deposited 1 April 2020.
- 47.Tomlinson J. H., Kalverda A. P., Staphylococcus aureus EF-GC3 methyl chemical shift assignments in complex with FusB. Biological Magnetic Resonance Data Bank. http://www.bmrb.wisc.edu/data_library/summary/index.php?bmrbId=50220. Deposited 1 April 2020.
- 48.Calabrese A. N., Tomlinson J. H., HX-MS data comparing EF-GC3 in the absence and presence of FusB. ProteomeXchange. http://proteomecentral.proteomexchange.org/cgi/GetDataset?ID=PXD017491. Deposited 20 February 2020.
- 49.Tomlinson J. H., Fusidic acid inhibition assay for EF-G mutants affecting FusB mediated resistance. Open Science Forum. https://osf.io/uq4g7. Deposited 17 September 2020.
- 50.Tomlinson J. H., Kalverda A. P., NMR relaxation dispersion data supporting PNAS paper “Fusidic acid resistance through changes in the dynamics of the drug target.” Open Science Forum. https://osf.io/95v4r/. Deposited 17 September 2020. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Assigned methyl chemical shifts for EF-GC3 in the apo- and FusB-bound states have been deposited in the Biological Magnetic Resonance Data Bank with the accession codes 50221 (46) and 50220 (47), respectively. HX-MS data have been deposited in the ProteomeXchange Consortium with the accession number PXD017491 (48). FA inhibition assay data are available in the Open Science Forum (OSF) (https://osf.io/uq4g7) (49). Raw relaxation dispersion data are available in the OSF (https://osf.io/95v4r/) (50). All other data are contained within the main text and SI Appendix.