Significance
Chromatin regulators and receptor tyrosine kinase (RTK) signaling proteins are both frequently mutated in bladder cancer. In this study, we sought to understand whether oncogenic signaling activation in the cytoplasm functionally interacted with loss-of-function mutations in chromatin regulators within the nucleus. The Cancer Genome Atlas cohort of muscle invasive bladder tumors revealed that loss-of-function mutations for the histone demethylase UTX (also known as KDM6A) significantly cooccurred with activating mutations in the RTK FGFR3. Here, we report that UTX loss and FGFR3 activation cooperate in bladder cells to reduce the expression of luminal genes, promoting a more basal, de-differentiated cellular state. These insights could prove useful in expanding therapeutic strategies for these tumors, which currently have limited options for effective treatment.
Keywords: bladder cancer, chromatin, receptor tyrosine kinase signaling
Abstract
Bladder cancer prognosis is closely linked to the underlying differentiation state of the tumor, ranging from the less aggressive and most-differentiated luminal tumors to the more aggressive and least-differentiated basal tumors. Sequencing of bladder cancer has revealed that loss-of-function mutations in chromatin regulators and mutations that activate receptor tyrosine kinase (RTK) signaling frequently occur in bladder cancer. However, little is known as to whether and how these two types of mutations functionally interact or cooperate to regulate tumor growth and differentiation state. Here, we focus on loss of the histone demethylase UTX (also known as KDM6A) and activation of the RTK FGFR3, two events that commonly cooccur in muscle invasive bladder tumors. We show that UTX loss and FGFR3 activation cooperate to disrupt the balance of luminal and basal gene expression in bladder cells. UTX localized to enhancers surrounding many genes that are important for luminal cell fate, and supported the transcription of these genes in a catalytic-independent manner. In contrast to UTX, FGFR3 activation was associated with lower expression of luminal genes in tumors and FGFR inhibition increased transcription of these same genes in cell culture models. This suggests an antagonistic relationship between UTX and FGFR3. In support of this model, UTX loss-of-function potentiated FGFR3-dependent transcriptional effects and the presence of UTX blocked an FGFR3-mediated increase in the colony formation of bladder cells. Taken together, our study reveals how mutations in UTX and FGFR3 converge to disrupt bladder differentiation programs that could serve as a therapeutic target.
The normal bladder urothelium is composed of three major cell types: Basal cells that contact the basement membrane and are believed to contain epithelial stem cells, intermediate cells, and differentiated superficial or umbrella cells that line the lumen (1). The gene-expression programs of these cell types are important for the molecular subtyping of bladder cancer. Urothelial carcinoma, by far the most common type of bladder cancer, can present as either superficial papillary (∼80% of urothelial carcinoma cases) or nonpapillary, high-grade invasive bladder cancer. Superficial papillary tumors are more benign and are often resected, but disease recurrence is common (2). Approximately 15% of these luminal papillary tumors eventually progress to invasive cancer and are characterized by the expression of luminal markers (e.g., KRT20, FOXA1, GATA3, Uroplakins), while the remaining nonpapillary invasive carcinomas express basal markers (e.g., KRT5, KRT14, CD44, TP63) and are typically the most aggressive subtype with limited therapy options currently available (2–5). Given these links between the differentiation state of a cancer cell and its prognosis, understanding the underlying mechanisms of maintaining cellular states and cell-state plasticity will provide valuable insights into bladder cancer and possible treatments.
Next-generation sequencing has allowed for extensive characterization of the mutational spectrum of bladder cancer. Bladder tumors have a relatively high mutational burden, underlying the importance of understanding the contexts in which certain alterations have functional roles in tumorigenesis (6). The Cancer Genome Atlas (TCGA) contains both sequencing and gene-expression data for over 400 muscle invasive bladder tumors, providing the opportunity to link transcriptional subtypes with mutational patterns (4). TCGA, in addition to many other sequencing studies, revealed that loss-of-function mutations in chromatin-modifying enzymes are common across all subtypes of bladder cancer, including those proteins known to posttranslationally modify histones (e.g., UTX/KDM6A, MLL3/KMT2C, MLL4/KMT2D, and EP300) and those that remodel nucleosomes (e.g., ARID1A) (7, 8). Included in this group is UTX (ubiquitously transcribed X chromosome tetratricopeptide repeat protein, also known as KDM6A), which is highly mutated in bladder cancer (26% of TCGA muscle invasive cohort and 22% of nonmuscle invasive cohort of 460 patients) while having a relatively lower mutation rate in TCGA studies of other tumor types (7–9). UTX is known to demethylate histone H3 at lysine 27 (H3K27) when it is trimethylated (10, 11), a repressive histone modification catalyzed by the EZH2 subunit of the polycomb repressive complex 2 (12). The catalytic activity of UTX is potentially relevant in bladder cancer as UTX-null cells were reported to be more sensitive to inhibition of the H3K27 methyltransferase EZH2 (13). Furthermore, a noncatalytic function for UTX at enhancers as part of MLL3- or MLL4-containing complexes has been described (14–16). MLL3- or MLL4-catalyzed monomethylation at H3K4, together with EP300/CBP-catalyzed H3K27 acetylation, mark active enhancer regions that promote transcription of specific genes (17–20). In the absence of UTX, activation of transcription by MLL3 or MLL4 complexes at enhancers is significantly reduced (14). While the function of UTX at enhancers has not been studied in bladder cancer, de-regulation of key enhancers upon UTX loss sensitizes pancreatic cancer cells to BET inhibitors and also restrains myeloid leukemogenesis (21, 22).
Sequencing of bladder cancer has also uncovered a high frequency of mutations that activate receptor tyrosine kinase (RTK) signaling pathways, including recurrent hotspot mutations in FGFR3, PIK3CA, RAS, and ERBB2 (4, 7, 8). The majority (71%) of all invasive bladder tumors from TCGA have at least one mutation contributing to the activation of RTK signaling (4). RTK signaling pathways impact cellular function in part by modulating transcriptional output, which integrates the regulation of transcription factors and the activity of chromatin-modifying enzymes (23). Despite the connection between signaling and transcription, the extent to which RTK activation and the loss of chromatin modifiers, such as UTX, functionally interact and potentially cooperate to promote tumorigenesis has not been extensively studied in bladder cancer. Here we investigate the tumor-suppressive functions of UTX in bladder cancer cells and find that UTX localizes to enhancers to regulate key bladder differentiation genes through a catalytic-independent mechanism. We identify RTK activation, specifically via FGFR3 (fibroblast growth factor receptor 3) hotspot mutations, as a context where UTX loss-of-function significantly cooccurs and has a functional role. The presence of UTX blunts transcriptional and phenotypic effects of aberrant FGFR3 activation, suggesting that the cooccurring UTX loss-of-function and FGFR3 mutations cooperate in bladder cancer to promote tumorigenesis.
Results
UTX Has a Catalytic-Independent Tumor-Suppressive Function while Localizing to Enhancers of Key Bladder Luminal Genes.
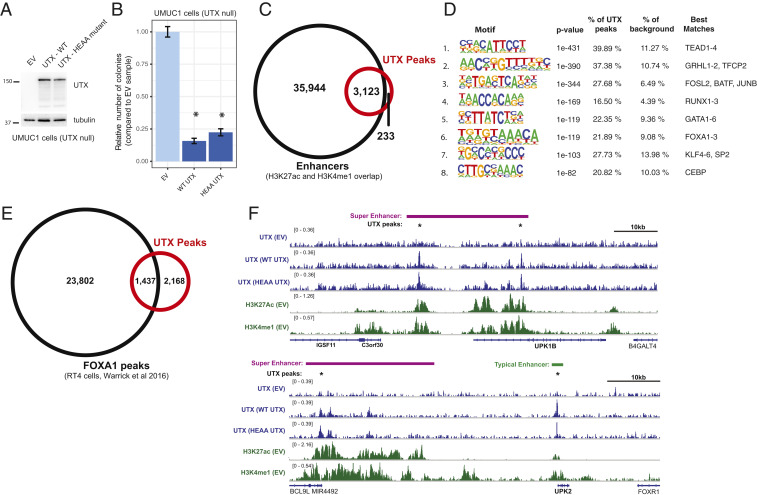
To study the role of UTX in bladder cancer cells, we generated mixed population, UTX-expressing UMUC1 cells (Fig. 1A). UMUC1 cells are a luminal bladder cancer cell line that harbors a truncating UTX mutation and has no detectable UTX protein by Western blot (24, 25). To address whether UTX catalytic function is important in bladder cancer, we also expressed a UTX transgene that carries two missense mutations in the demethylase domain, H1146A and E1148A (referred to as HEAA UTX), rendering it catalytically dead (26). Consistent with its role as a tumor suppressor, expression of wild-type or catalytically dead (HEAA) UTX reduced the ability of cells to form colonies in soft agar, suggesting that the demethylase activity is not required for this phenotype in UMUC1 cells (Fig. 1B).
Fig. 1.
UTX acts as a tumor suppressor in bladder cancer cells through a catalytic-independent mechanism and localizes to enhancers of luminal genes. (A) Western blot analysis of UMUC1 cells expressing wild type UTX or the catalytically dead HEAA mutant of UTX (both in pSIN-puro backbone). (B) Quantification of the number of colonies formed in agar by empty vector (EV) control versus UTX-expressing UMUC1 cells, normalized to the EV cells. Data from at least 12 wells from 3 separate experiments are shown, and *P < 0.05 by t test compared to EV cells. (C) Venn diagram depicting overlap of UTX ChIP-seq peaks (either in wild-type or HEAA UTX cells) with enhancers in UMUC1 cells. Enhancers are defined as genomic regions that contain both histone H3K27ac and H3K4me1 ChIP-seq peaks in either EV control, wild-type (WT) UTX, or HEAA UTX cells. (D) Sequence motifs that are enriched in UTX peaks. The motifs were generated by the Homer software package and the transcription factors whose motifs most closely resemble each identified motif is shown. (E) Venn diagram showing overlap between UTX ChIP-seq peaks (either in wild-type or HEAA UTX cells) in UMUC1 cells with FOXA1 ChIP-seq peaks in RT4 cells (25). (F) Genome browser (IGV) representation of ChIP-seq signal across select genomic regions that include key bladder differentiation genes (UPK1B, Upper, and UPK2, Lower). The antibody used for the ChIP experiment is shown on the left with the cell line indicated in parenthesis. UTX peaks are indicated with a star (*), super enhancers are depicted by purple bars, and all other enhancers with green bars.
Since UTX has known noncatalytic functions at transcriptional enhancers, we probed whether UTX regulates enhancers in UMUC1 cells by performing chromatin immunoprecipitation followed by sequencing (ChIP-seq) for UTX and several histone posttranslational modifications, including H3K27 acetylation (H3K27ac) and H3K4 monomethylation (H3K4me1), which together denote active enhancers, and H3K27 trimethylation (H3K27me3), which is the substrate for UTX catalytic activity. The UTX ChIP-seq signal from wild-type and HEAA UTX cells was very similar, suggesting negligible effect of catalytic function on the genomic localization of UTX (SI Appendix, Fig. S1A). This observation led us to use the combined set of UTX peaks for further analysis. Enhancer regions were operationally defined as any genomic location with overlapping H3K27ac and H3K4me1 peaks, and the vast majority of UTX peaks, or 93%, fell within these enhancers (Fig. 1C). We observed minor effects of UTX expression on the levels of either H3K27 acetylation or H3K4me1 around the entire UTX peak set (SI Appendix, Fig. S1B). Consistent with a noncatalytic role for UTX at these enhancers, H3K27me3 was also unaffected by the presence of UTX (SI Appendix, Fig. S1C). The primary known role for UTX at enhancers is as a critical component of MLL3- and MLL4-containing complexes. An analysis of all bladder cancers in the cBioPortal database revealed that MLL3 and MLL4 loss-of-function mutations commonly cooccur (P < 0.01, Fisher’s exact test), which is consistent with reports of their partially redundant functions. However, tumors that have mutations in both MLL3 and MLL4 rarely have a loss-of-function mutation for UTX (P < 0.01, Fisher’s exact test) (SI Appendix, Fig. S1D) (7, 8). This suggests that UTX, MLL3, and MLL4 likely have redundant functions and may act as tumor suppressors through overlapping mechanisms at specific enhancers. Consistent with this model and with previous studies showing that UTX is important for MLL3/4 complex assembly at enhancers (14), we found significant overlap between UTX and MLL4 ChIP-seq peaks (70% of MLL4 peaks, P < 0.01), and higher MLL4 signal within these regions of overlap in the UTX-expressing cells (SI Appendix, Fig. S1 E and F). The overall number of MLL4 peaks identified was also increased in UTX-expressing cells (empty vector [EV] cells: 123 peaks; wild-type UTX cells: 888 peaks; HEAA UTX cells: 1,116 peaks). These data demonstrate that UTX binds predominantly to enhancers in bladder cancer cells, at least partly in coordination with MLL3 and MLL4 complexes.
Enhancers can have wide-ranging effects on transcription and cellular phenotypes; thus, we sought to understand UTX function at these enhancer regions. Transcription factor motif analysis of UTX peak regions showed significant enrichment for the DNA motifs of factors associated with luminal bladder cells, including FOXA1, GATA3, and GHRL2 (Fig. 1D) (27). FOXA1 and GATA3 in particular have been shown to regulate differentiation pathways that promote the transition of a bladder cell from a basal to a luminal transcriptional state (25). We then compared our ChIP-seq data for UTX with a published dataset of FOXA1 ChIP-seq in RT4 bladder cancer cells (25). Given that both UMUC1 and RT4 cells have gene-expression patterns consistent with luminal tumors (25), overlap between these two sets of peaks would support a function for UTX at FOXA1-bound enhancers in luminal cells. We found that 40% (P < 0.01) of UTX peaks in UMUC1 cells overlap with FOXA1 peaks in RT4 cells (Fig. 1E). To further support a role for UTX in regulating the differentiation state of a bladder cell, we looked at UTX enrichment within genomic regions that contain a high concentration of enhancers, also known as super enhancers. These genomic elements drive the expression of genes that are important for establishing and maintaining cell identity during differentiation (28). Using our H3K27ac ChIP-seq data from UMUC1 cells, we identified super enhancers with the ROSE algorithm (28, 29). The majority (63.2%, P < 0.01) of all super enhancers in UTX-expressing UMUC1 cells contained at least one UTX peak, supporting the hypothesis that UTX is regulating genes involved in bladder cell identity (SI Appendix, Fig. S1G). As an example, the uroplakin genes UPK1B and UPK2, which are only expressed in differentiated luminal bladder cells, have proximal super enhancers and normal enhancers that overlap with UTX peaks (Fig. 1F). These data implicate UTX in the regulation of genes expressed in luminal bladder cells through a catalytic-independent mechanism at enhancers.
UTX Maintains Luminal Gene Expression in Bladder Cancer Cells.
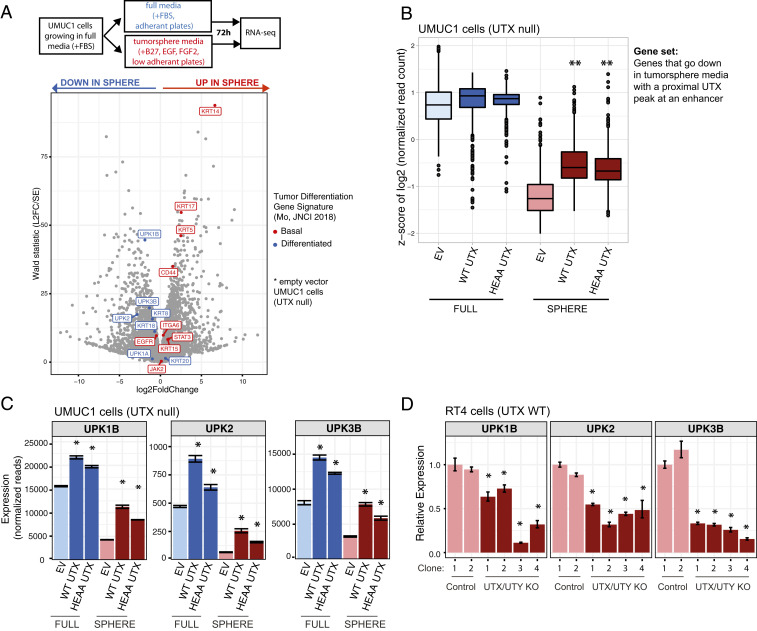
We then wanted to understand whether UTX occupancy at enhancers translated to changes in gene expression. To answer this question, we sought a cellular model that could mimic a transition between basal and luminal transcriptional states. Growing cells as tumorspheres in media containing B27 supplement, FGF2, and EGF has been used in other cancer types to enrich for a more stem-like cell population (30). Not all cell lines are amenable to tumorsphere growth conditions (31), so we tested this in UMUC1 cells by growing them in both tumorsphere and full media (contains fetal bovine serum), and then assaying gene expression using RNA-sequencing (RNA-seq) (Fig. 2A). The genes highlighted in the volcano plot are from a published tumor differentiation signature that was compiled with genes known to be biologically relevant in the bladder with the purpose of distinguishing between basal and luminal muscle invasive bladder tumors (32). We found that tumorsphere media caused an up-regulation of genes associated with basal bladder tumors, and a down-regulation of genes associated with luminal (or “differentiated”) tumors (Fig. 2A).
Fig. 2.
UTX hinders the transition to a more stem-like cellular state by regulating bladder differentiation genes. (A) Volcano plot showing gene-expression changes in EV control UMUC1 cells in full media conditions versus tumorsphere media conditions, as measured by RNA-seq. Cells were plated and grown in the indicated media for 72 h before harvest. Genes known to be expressed highly in basal bladder tumors are highlighted in red, while those expressed in luminal/differentiated tumors are highlighted in blue (32). (B) Plot showing the distribution of z-scores for UMUC1-derived cell lines in both regular and tumorsphere media conditions after filtering for genes that go down in tumorsphere conditions versus full media and have a UTX peak at a nearby enhancer. **P < 0.05 by Wilcoxon rank sum test compared to the EV-sphere condition. (C) Mean normalized read signal of select genes across three RNA-seq replicate samples, and adjusted *P < 0.05 as calculated by differential gene-expression analysis performed with DESeq2 compared to the EV control cell line. (D) qPCR analysis of RT4 cells with both UTX and UTY knocked out by CRISPR-Cas9. These cell lines are derived from single cell UTX and UTY knockout clones. qPCR was performed 24 h after plating. Mean expression was calculated from a representative experiment of three replicates and is relative to the first EV-infected cell line, called “Control-1,” and *P < 0.05 by t test compared to both Control-1 and Control-2 cell lines.
We then looked at how UTX impacted gene expression in this cellular system. Globally, there were minimal UTX-dependent transcriptional changes that required demethylase activity in either full media or tumorsphere media, consistent with our data showing a negligible role for UTX catalytic activity in colony formation of UMUC1 cells (SI Appendix, Fig. S2 A and B). When looking at all genes that had significantly reduced expression in tumorsphere media, and were annotated by an enhancer-overlapping UTX peak, we found that both wild-type and HEAA UTX collectively increased the expression of these genes in tumorsphere media (Fig. 2B) (33). To take a more focused approach, we examined how UTX alters the transcription of the genes from the tumor-differentiation signature that are highlighted in Fig. 2A. We found that in tumorsphere media wild-type and HEAA UTX increased the expression of the luminal markers from this signature that had appreciable levels of expression, while having minimal impact in full media (SI Appendix, Fig. S2C). Included among these luminal genes were the uroplakins UPK1B, UPK2, and UPK3B, which showed UTX-dependent increases in expression in both full and sphere media (Fig. 2C). In addition, UPK1B and UPK2 both had UTX-bound enhancers nearby (Fig. 1F), suggesting that the presence of UTX at these enhancers might be directly impacting gene expression. We also note that despite having a similar impact on colony formation compared to wild-type UTX, HEAA UTX-expressing cells did consistently show a slightly less pronounced transcriptional change, leaving open the possibility that catalytic function is playing a minor role for UTX at enhancers. We then studied the effect on uroplakin expression of knocking out the UTX gene, along with its closely related homolog UTY, in RT4 bladder cancer cells (16, 34). Sequential clonal knockouts of UTX and UTY using CRISPR-Cas9 showed a decrease in uroplakin expression (Fig. 2D and SI Appendix, Fig. S2 D and E). Taken together, these data suggest that UTX hinders the transcriptional transition of UMUC1 cells to a basal state, possibly through the regulation of enhancers proximal to key luminal genes.
FGFR3 Suppression of Luminal Gene Expression Correlates with Bladder Cancer Cell Viability.
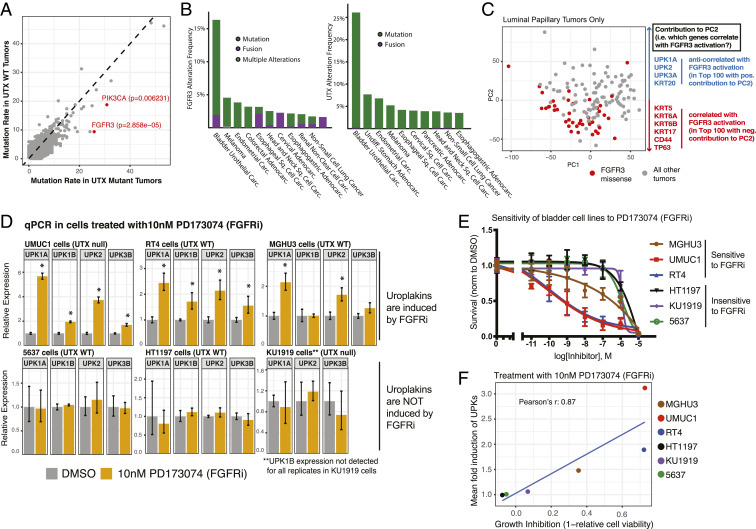
Next, we sought to understand specific contexts in which the regulation of differentiation genes by UTX might be functionally important. To address this question, we looked at TCGA data for muscle invasive bladder tumors, and found that activating mutations in two key RTK signaling proteins, FGFR3 and PIK3CA, significantly cooccur with UTX mutations (Fig. 3A, SI Appendix, Fig. S3A, and Dataset S1) (4). FGFR3 is an RTK that initiates the PI3K and MAPK signaling pathways, and PIK3CA (p110α) is the catalytic subunit of PI3K. In addition to hotspot mutations in FGFR3 and PIK3CA, the PI3K and MAPK signaling pathways are activated in bladder cancer through a variety of mechanisms (4, 7, 8). Furthermore, the tumorsphere media used in this study contains a number of molecules that activate RTKs. Treatment with the combination of MAPK and PI3K inhibitors prevented the reduction in uroplakin gene expression that is caused by the tumorsphere media, while treatment with either inhibitor alone resulted in a partial effect (SI Appendix, Fig. S3B) (35, 36). This suggests that RTKs and UTX promote opposing gene programs in bladder cancer: While RTK signaling promotes the shift to a more basal transcriptional state, UTX, in contrast, promotes luminal gene expression (Figs. 1 and 2). This observation raises the possibility that UTX loss creates a permissive transcriptional environment for FGFR3 and PIK3CA activation to impact cell state and promote tumorigenesis.
Fig. 3.
Loss-of-function UTX mutations cooccur with activating mutations of upstream kinases that regulate differentiation pathways. (A) Plot of mutation rate of proteins in either UTX mutant or UTX wild-type tumors present in TCGA muscle invasive bladder cancer data. Genes of interest, FGFR3 and PIK3CA, are highlighted in red. P values were obtained from the cBioPortal, and were calculated using a Fisher’s exact test. (B) Genetic alteration frequencies of FGFR3 (Left) and UTX (Right) in numerous TCGA studies are shown. Abbreviations: adenocarc. adenocarcinoma; carc., carcinoma; sq., squamous. (C) PCA of bladder cancer tumors in the luminal-papillary mRNA subtype of TCGA cohort based on RNA-seq gene-expression data. Tumors with an FGFR3 missense mutation are highlighted in red. The top 100 genes that are correlated and anticorrelated with PC2 were identified, and key differentiation genes on either end of PC2 in terms of correlation are shown at right. (D) qPCR analysis of a panel of bladder cancer cell lines treated with either DMSO or 10 nM of the FGFR inhibitor (FGFRi) PD173074 for 48 h. Treatments began 24 h after the cells were plated. Mean expression was calculated from a representative experiment of three replicates and is relative to the DMSO-treated cells, and *P < 0.05 by t test compared to DMSO-treated cells. (E) Cell viability of a panel of bladder cancer cell lines after treatment with increasing doses of PD173074. Cells were plated, treated the following day, and cell density was quantified after 96 h by CellTiter-Glo. These measurements were then normalized to the DMSO condition. At least two replicate experiments (each with three technical replicates) were performed for each cell line at the indicated doses. (F) Growth inhibition by PD173074 after 96 h of treatment is plotted against the mean induction of gene expression of UPK1A, UPK1B, UPK2, and UPK3B for the panel of bladder cancer cell lines. Note that UPK3A was not expressed in majority of cell lines so was excluded and UPK1B was excluded for the KU1919 calculation due to being on the edge of detection.
To further understand the role of RTK signaling in regulating luminal gene expression, we focused on FGFR3. Interestingly, FGFR3 has a high rate of mutation in bladder cancer compared to other tumor types, which is similar to UTX and suggests that it may have key functions that are specific to bladder cells, such as cell identity and differentiation (Fig. 3B). We first looked for evidence that FGFR3 regulates such genes in tumors using TCGA muscle invasive bladder cancer RNA-seq data. FGFR3 activating mutations are much more prevalent in the luminal-papillary mRNA subtype of bladder cancer as defined by TCGA (SI Appendix, Fig. S3C), and principal component analysis (PCA) of these 142 luminal-papillary tumors revealed that tumors with FGFR3 missense mutations cluster together (Fig. 3C). We then analyzed the contribution of all genes to the principal component (PC) by which FGFR3 mutant tumors segregate (PC2), focusing on genes related to bladder differentiation (Dataset S2). Using the tumor differentiation signature introduced in Fig. 2, the contribution of basal genes to PC2 was significantly negative, and therefore correlated with the direction of the FGFR3 mutant cluster, while the contribution of genes associated with luminal or more differentiated tumors was significantly positive, or anticorrelated with the FGFR3 mutant cluster (SI Appendix, Fig. S3D). Furthermore, when looking only at the top 100 genes on either end of PC2, many basal genes correlated with FGFR3 mutant tumors (e.g., KRT5, KRT6, TP63, CD44), while luminal genes were anticorrelated (e.g., UPK1A, UPK2, UPK3A, KRT20) (Fig. 3C and Dataset S2). This suggests that within the luminal-papillary cohort of tumors, FGFR3 mutant tumors skew toward a more basal, stem-like transcriptional profile.
We next aimed to validate these results in tissue culture models and measured uroplakin gene expression in a panel of six bladder cancer cell lines treated with two different FGFR inhibitors, PD173074 and BGJ398 (37, 38). These lines were selected to have a mix of luminal and basal gene expression signatures, UTX status, or have shown UTX-dependent phenotypes in prior publications (SI Appendix, Table S1) (24, 25, 39). Three of the six cell lines—specifically UMUC1, RT4, and MGHU3 cells—showed significant induction of gene expression in at least two uroplakin genes after treatment with either FGFR inhibitor, indicating that FGFR signaling is suppressing the expression of these luminal markers in these cells (Fig. 3D and SI Appendix, Fig. S3 E, Upper). However, treatment of 5637, HT1197, and KU1919 cells with either FGFR inhibitor did not result in the same increase in uroplakin expression (Fig. 3D and SI Appendix, Fig. S3 E, Lower), suggesting that FGFR suppression of these luminal genes is not a common feature among all bladder cancer cell lines. Ultimately, we were interested in whether the ability of FGFR signaling to reduce luminal gene expression is related to cell survival; therefore, we tested cell viability after FGFR inhibitor treatment. Among the six cell lines tested, increases in uroplakin expression upon treatment with both FGFR inhibitors strongly correlated with growth inhibition (Fig. 3 E and F and SI Appendix, Fig. S3 F and G). Sensitivity to FGFR inhibition also correlated with baseline uroplakin gene expression, demonstrating that luminal cell lines were more sensitive to these drugs, which is consistent with the fact that FGFR3 mutations are primarily found in luminal tumors (SI Appendix, Fig. S3 H and I). These data implicate FGFR signaling in suppressing the expression of uroplakins, and possibly connect this regulation of luminal genes to the role of FGFRs in bladder cell survival.
FGFR3-Driven Cellular Phenotypes Are Hindered by UTX.
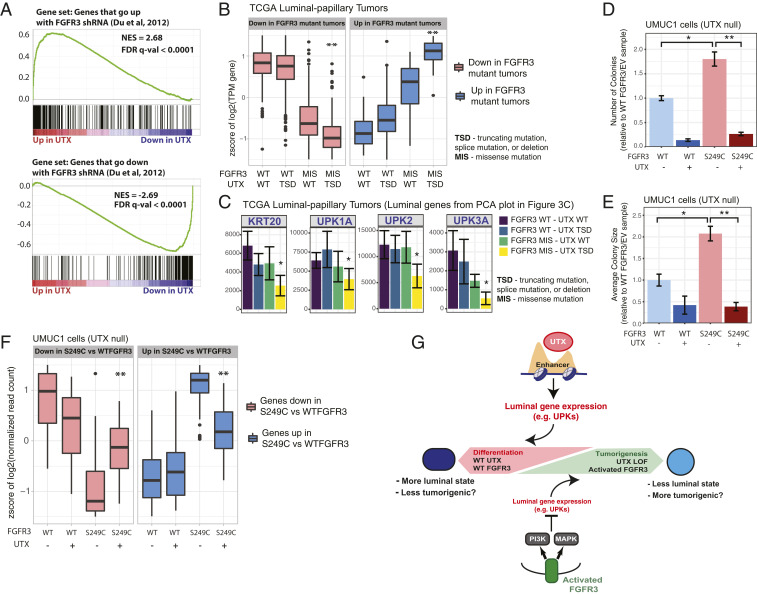
Thus far, our data point to opposing roles for UTX and FGFR3 in regulating the balance between luminal and basal gene expression in bladder cells. To test the hypothesis that there is an antagonistic transcriptional relationship between UTX and FGFR3, we compared UTX-dependent gene-expression changes in UMUC1 cells to a previously published gene-expression dataset from bladder cancer cells (RT112) with FGFR3 knocked down by shRNA (40). We found that the gene-expression changes after FGFR3 knockdown in RT112 cells were strongly correlated with the expression changes caused by the introduction of UTX into UMUC1 cells (Fig. 4A). These data indicate that FGFR3 and UTX have opposing effects on the transcription of a shared set of genes in the luminal RT112 and UMUC1 cells, respectively (25). The next question was whether UTX loss and FGFR3 activation cooperate to impact transcription. To address this possibility using TCGA muscle invasive bladder cancer data, we identified genes that were differentially expressed (both increased and decreased) in FGFR3-mutant luminal-papillary tumors compared to those that have no FGFR3 alterations, and then analyzed the expression of these genes after stratifying the tumors by both FGFR3 and UTX genotypes. Tumors that had MLL3 or MLL4 mutations were excluded from this analysis since these proteins are known to have a similar role at enhancers as UTX and are often mutated in bladder cancer (SI Appendix, Fig. S1D). Tumors with both an FGFR3 mutation and a UTX loss-of-function mutation had potentiated gene-expression changes in either direction (Fig. 4B). When we focused on the expression of key luminal genes that were identified in the PCA analysis (Fig. 3C), the same pattern is observed: The expression of these genes is lowest when both UTX is lost and FGFR3 is activated (Fig. 4C and SI Appendix, Fig. S4A). We also looked at the relationship between UTX loss and PI3K activation in TCGA tumors from the basal-squamous mRNA subtype. This analysis revealed a similar pattern where PIK3CA-dependent gene-expression changes were most significant in the context of UTX loss (SI Appendix, Fig. S4B). Taken together, these data suggest that UTX loss amplifies the gene-expression changes that are driven by the activation of upstream signaling, such as FGFR3 and PIK3CA.
Fig. 4.
UTX counteracts FGFR3 activation. (A) Gene set enrichment analysis showing how the top 200 genes that go up (Upper) or down (Lower) after shRNA knockdown of FGFR3 in RT112 bladder cells [from a published dataset (40)] are altered by wild-type UTX expression in the UMUC1 cells in full media when compared to EV cells. (B) Plot showing the distribution of z-scores for gene expression as measured by RNA-seq across FGFR3 and UTX genotypes for genes that go down (Left) or up (Right) in luminal-papillary (TCGA mRNA subtype) tumors with an FGFR3 missense mutation (MIS) compared to tumors with wild-type FGFR3 (log2 fold-change > 2 and P < 0.0005). “TSD” for UTX genotype indicates either a truncating, splice, or deletion mutation. Only tumors with wild-type MLL3 and MLL4 (KMT2C and KMT2D) were included in this analysis. **P < 0.05 by Wilcoxon rank sum test compared to the FGFR3 missense/UTX wild-type tumors for that group of genes. (C) Normalized RNA-seq read signal of select differentiation genes across the FGFR3 and UTX genotypes described in B within luminal-papillary (TCGA mRNA subtype) tumors. *P < 0.05 as calculated by differential gene-expression analysis performed with DESeq2. (D and E) Quantification of the number (D) and size (E) of colonies formed in agar by UMUC1 cells expressing FGFR3 and/or UTX. Data from at least 10 wells from two separate experiments are shown, and *P < 0.05 by t test for S249C FGFR3/EV cells compared to WT FGFR3/EV cells, while **P < 0.05 by t test for S249C FGFR3/EV cells compared to S249C FGFR3/UTX cells. (F) Plot showing the distribution of z-scores for gene expression as measured by RNA-seq from UMUC1 cells expressing FGFR3 and/or UTX for genes that go down (Left) or up (Right) in S249C FGFR3 versus wild-type FGFR3-expressing cells (log2 fold-change > 1.5 and P < 0.05). Cells were harvested 24 h after being plated. **P < 0.05 by Wilcoxon rank sum test compared to the FGFR3 S249C/UTX null cells for that group of genes. (G) Model for the antagonizing roles of UTX and FGFR3 in the regulation of differentiation genes and tumorigenesis.
To more directly test FGFR3 function in bladder cells, we expressed either wild-type or S249C FGFR3 (the most common activating mutation in bladder cancer) in UMUC1 cells (these transgenic cell lines are mixed population) (SI Appendix, Fig. S4C). The expression of S249C FGFR3 increased the number and size of colonies formed in soft agar and proliferation compared to wild-type FGFR3 expressing cells (Fig. 4 D and E and SI Appendix, Fig. S4D). Introduction of UTX (UMUC1 cells are UTX null) abolished the FGFR3-dependent increase in colony number and size, and partially reduced proliferation. Profiling by RNA-seq identified genes that were differentially expressed between S249C and wild-type FGFR3-expressing cells. Cells that coexpressed UTX showed blunted S249C FGFR3-dependent gene expression changes, and this pattern was true of many genes from the luminal gene signature used in Fig. 2A (Fig. 4F and SI Appendix, Fig. S4E). Furthermore, genes that go up in UTX-expressing cells and have a nearby UTX peak showed decreased expression in S249C FGFR3 cells compared to wild-type FGFR3 cells, which is again consistent with an antagonistic transcriptional relationship between the two proteins (SI Appendix, Fig. S4F). Altogether, these data support a model where UTX maintains a more luminal or differentiated state in a bladder tumor cell, and the loss of UTX in bladder tumors potentiates an FGFR3-dependent move toward a more basal and tumorigenic state cellular state (Fig. 4G).
Discussion
UTX function is commonly lost in bladder cancer, but the role of UTX in normal bladder cells, and the reasons why it is such a powerful tumor suppressor in this tissue, are not well understood. Our study shows that UTX and RTK signaling pathways have antagonistic roles in determining the differentiation state of a bladder cell. ChIP-seq experiments revealed that UTX localizes to enhancers near genes expressed in luminal cells of the bladder, and that these enhancers are enriched with motifs of transcription factors that determine the luminal cell fate. Using tumorspheres as a cellular model to enrich for a more stem/basal-like cell population, we found that UTX counteracted the reductions in gene expression caused by tumorsphere media. This included the expression of key luminal genes with nearby UTX-occupied enhancers, suggesting a functional role for UTX in regulating the balance between luminal and basal bladder cell state. Importantly, the phenotypes we have attributed to UTX—reduction in colony formation in soft agar and regulation of luminal cell identity genes—largely did not rely on the catalytic histone demethylase function of UTX. We did observe that the transcriptional effect of wild-type UTX was in general slightly stronger than HEAA UTX in UMUC1 cells, leaving open the possibility that some of the functions of UTX in bladder cells do rely on demethylation of H3K27. However, our data indicate that the bulk of UTX tumor-suppressive function does not require its catalytic activity. Our data are consistent with previous reports that UTX function at MLL3 or MLL4 complex-containing enhancers is independent of UTX demethylase activity (14). The particular vulnerability of bladder cells to perturbations in this complex is clear from the high rate of mutations in MLL3 and MLL4, in addition to UTX. Moreover, the patterns of these mutations also point to the noncatalytic, enhancer-localized UTX function as driving tumor suppression: MLL3 and MLL4 mutations commonly cooccur, while UTX is rarely mutated in these double MLL3/MLL4 mutant tumors (7, 8). These data align with a model where MLL3 and MLL4 have some redundant functions, while UTX alone is critical for complex function. While our data also showed that UTX and MLL4 colocalize in UMUC1 bladder cancer cells, further work is necessary to investigate functional overlap between UTX and the MLL3/MLL4 complex. It is also noteworthy that while the mutation rate of UTX is uniquely high in bladder cancer, MLL3 and MLL4 mutation rates are high in many different tumor types. This suggests a level of complexity for the functional role and regulation of MLL3/MLL4 complexes that goes beyond a simple model where UTX loss-of-function has the same effect on these complexes in all cell types.
The enhancers bound by UTX were enriched for motifs of transcription factors that direct bladder cells to the luminal cell fate, including FOXA1 and GATA3 (25, 27). While this reported connection between UTX and these transcription factors in bladder cells is unique, interactions between UTX and GATA3, and between MLL3 and FOXA1, have been described in breast cancer cells (41, 42). Furthermore, FOXA1 and GATA3 cooperate with peroxisome proliferator-activated receptor-γ (PPARγ) to drive luminal gene expression in bladder cells (25, 43, 44). This is particularly notable given a previously described role for MLL3/MLL4 and UTX complexes in PPARγ-mediated transcription during adipogenesis, where PPARγ is also known to have a fundamental role. Adipogenesis relies on MLL4 and is characterized by MLL4 and PPARγ genomic colocalization (45). Moreover, MLL3/MLL4 and UTX complexes bind to PTIP, a protein that is essential for PPARγ induction during adipogenesis (46, 47). Our data implicate UTX in these same transcriptional pathways in bladder cells, where they are known to drive luminal gene expression.
Like UTX expression, the inhibition of RTK signaling (PI3K and MAPK combined) counteracted the tumorsphere-induced shift to a more basal transcriptional state, suggesting that RTK activation could be contributing to tumorigenesis via de-differentiation of luminal bladder cells. We focused on FGFR3, which is often activated in tumors that have lost UTX function, and we found that FGFR inhibition increased luminal gene expression in a subset of bladder cancer cell lines. Interestingly, the extent to which luminal genes are induced by an FGFR inhibitor strongly correlated with growth inhibition after treatment with the same drug. We did not see a clear connection between UTX status and sensitivity to the FGFR inhibitors in our small cohort, but this is not necessarily expected given that UTX mutations are common across all subtypes of bladder cancer. Furthermore, the cell lines that were more luminal (as determined by baseline uroplakin expression) were also more sensitive to the FGFR inhibitor. If these data in cell lines translates to human cancer (i.e., luminal-papillary tumors are be more sensitive to FGFR regulation), it could provide insight into why FGFR3 activation primarily occurs in luminal-papillary tumors. These data also have therapeutic implications as they hint at a possible mechanism of action for FGFR inhibitors in bladder cancer, which have recently been approved to treat bladder cancer patients with FGFR mutations. Our data suggest that FGFR inhibition could be a form of differentiation therapy in bladder cancer. The strategy of using a drug to promote irreversible changes to the differentiation state of cancer cells, thereby stopping tumor growth, has been used in various tumor types with particular success in certain leukemias (48). Our data with PI3K and MAPK inhibitors in UMUC1 cells also suggest a broader role for RTK signaling in suppressing luminal gene expression, and perhaps PI3K and MAPK inhibitors would be effective at promoting differentiation of bladder cancer cells that have signaling activation caused by mutations that are not occurring in the FGFR3 gene.
Finally, we describe functional cooperation between UTX loss and FGFR3 activation both in tumors and in our cellular models. UTX expression prevented an FGFR3-dependent increase in colony formation, and blunted FGFR3-dependent gene-expression changes. It appears that UTX has a global effect on FGFR3-driven transcription, behaving like a rheostat to fine-tune the eventual transcriptional output of FGFR3 signaling, potentially in terms of both the amplitude and length of the signal. Upon loss of UTX, this may create a more plastic and aberrant chromatin environment that potentiates global transcriptional changes caused by FGFR3 signal activation, increasing the likelihood that this oncogene will promote tumorigenesis. Similar models have been described in the literature; for example, KRAS-induced transformation of lung cancer cells only occurred in the context of an altered epigenetic environment marked by an increase in DNA methylation that was caused by chronic exposure of cells to cigarette smoke (49). This model of UTX-mediated tumor suppression is particularly exciting because it can potentially be applied to the many other chromatin regulators that are commonly altered in cancer, such as MLL3, MLL4, and ARID1A, proteins for which the mechanism of tumor suppression has been difficult to definitively characterize. Perhaps this difficulty is due to the fact that their loss does not result in consistent gene-expression changes, but simply creates a more permissive genomic environment for tumorigenesis. Moreover, the transcriptional changes that drive cancer when these tumor suppressors are lost may not only depend on the cell type, but also specific oncogenic lesions that are present in that cell. This model could be particularly relevant for bladder cancer, where UTX and other chromatin regulators are mutated in the presence of many different RTK activation events. Understanding the precise mechanisms that underlie cooperativity between RTK activation and mutations in chromatin regulators could help to direct therapeutic strategies for specific mutation combinations. For example, our data would suggest that in the context of a UTX mutation, differentiation therapy could supplement FGFR3 inhibition, perhaps increasing potency or reducing the chance of resistance. However, in combination with other chromatin mutations, another mode of supplemental therapy could be necessary due to a different mechanism of transcriptional cooperation. Therefore, future work delving into these transcriptional relationships that exist between specific signaling and chromatin alterations could be very helpful in driving therapeutic decisions.
Materials and Methods
Additional materials and methods for the following are included in SI Appendix, SI Materials and Methods: qRT-PCR, ChIP for histone posttranslational modifications using fragmentation by sonication, ChIP for UTX and MLL4 using fragmentation with micrococcal nuclease, colony formation in soft agar, and cell proliferation.
Plasmids and Lentivirus Generation.
Full-length wild-type and HEAA mutant UTX in the pSIN-puromycin backbone with a C-terminal HA tag was a gift of Robert Roeder, The Rockefeller University, New York, NY, and Shu Ping Wang, Institute of Biomedical Sciences at Academia Sinica, Taiwan, China. Full-length UTX was also cloned into the pCDH-blasticidin backbone using Gibson assembly (New England Biolabs). Wild-type and S249C FGFR3 in the pBabe-puromycin gateway backbone, in addition to the empty vector, were gifts from Matthew Myerson, Harvard Medical School, Boston, MA (Addgene plasmid nos. 45711, 45713, 51070) (50, 51). To generate lentivirus, 293T cells were transfected with Lipofectamine 2000 (Life Technologies) with the above vectors along with helper plasmids (psPAX2 and pMD2.G) and supernatant was collected and filtered 48 h later for transduction.
Cell Culture Media and Drug Treatments.
Unless otherwise indicated, cell lines were grown in the following media supplemented with 10% fetal bovine serum (Sigma-Aldrich): UMUC1 (Sigma Aldrich), HT1197 (ATCC), and MGHU3 cells (gift of Alain Bergeron and Yves Fradet, Centre Hospitalier Universitaire de Quebec, Canada) were grown in Eagle’s Minimum Essential Medium (Corning, 10-009-CV), RT4 cells (ATCC) were grown in McCoy’s 5A Medium (Corning, 10-050-CV), and 5637 (ATCC) and KU1919 cells (DMSZ) were grown in RPMI Medium (Corning, 10-040-CV). For tumorsphere conditions, cells were washed once in empty media then plated in ultralow attachment plates (Corning, 3471) in serum-free media with 1× B27 supplement without vitamin A (Life Technoligies, 12587-010), human-FGF2 (bFGF) (20 ng/mL; Shenandoah Biotech), and human-EGF (20 ng/mL) (Shenandoah). The following small-molecule inhibitors were used: PD173074 (FGFRi – Selleck, S1264), GDC0941 (PI3Ki; gift of Ramon Parsons, Mount Sinai, New York, NY), and AZD6244/Selumetinib (MEK1/2i – Selleck, S1008). Doses and length of small-molecule drug treatment are described in the figure legend for each experiment.
Generation of CRISPR-Cas9 Edited Cell Lines.
Single-guide RNAs (sgRNAs) against UTX and UTY were cloned into px458 (Addgene 48138; a gift from F. Zhang, Massachusetts Institute of Technology, Cambridge, MA) (52). The UTX sgRNA targeted exon 2, and the following primers were used for cloning into the px458 vector: Forward: caccGAAATCTCACGAACCCAAAG; reverse: aaacCTTTGGGTTCGTGAGATTTC. A previously published UTY sgRNA was used that targets exon 1 (34), and the following primers were used for cloning on to the px458 vector: Forward: caccGTCTGTTAGCCTGACAGTCG; reverse: aaacCGACTGTCAGGCTAACAGAC. RT4 cells were transfected with the px458 vector with the specific UTX guides using Lipofectamine LTX (Life Technologies) and the cells were then sorted for GFP expression. Single-cell UTX knockout clones were obtained, and then two knockout clones and one control clone were used for subsequent UTY knockout and clonal growth. Two clones from each UTX/UTY pair, and control cells were collected resulting in four total UTX/UTY knockout cell lines, and two control cell lines.
Immunoblotting.
Cell pellets were lysed in Laemmli sample buffer (125 mM Tris pH 6.8, 4% SDS, 20% glycerol, 10% β-mercaptoethanol, 0.05% bromophenol-blue), and then were separated by SDS/PAGE, transferred to a PVDF membrane (Millipore, IPV00010), blocked with 5% nonfat milk in PBS containing 0.5% Tween-20 for at least 30 min at room temperature, probed with primary antibodies overnight at 4 °C, and detected with horseradish peroxidase-conjugated anti-rabbit or anti-mouse secondary antibodies (GE Healthcare). The following primary antibodies were used: anti-UTX (Cell Signaling, 33510), anti-UTY (Abcam, ab91236), anti-FGFR3 (Abcam, ab133644), anti–β-actin (Abcam, ab8224), and anti–β-tubulin (Sigma, T5201).
RNA-Seq.
RNA was isolated from cells using the RNeasy Mini Kit (Qiagen, 74104). Libraries were prepared using either the Illumina TruSeq protocol (UMUC1 cells in full and tumorsphere media introduced in Fig. 2) or the NEBNext Ultra II RNA Library Prep kit (UTX/FGFR3-expressing UMUC1 cells introduced in Fig. 4), and then sequenced on a NextSeq500 sequencer. Transcript abundance was computed from FASTQ files using Salmon and the GENCODE reference transcript sequences, transcript counts were imported into R with the tximport R Bioconductor package, and differential gene expression was determined with the DESeq2 R Bioconductor package (53–55). The data were visualized using either the ggplot2 R package for box/bar plots (56), or the pheatmap R package for heatmaps showing the expression changes of individual genes (57).
ChIP-Seq Analysis.
Low-quality reads from each FASTQ file were filtered using the ShortRead R Bioconductor package, and filtered FASTQ files were aligned to the hg19 reference genome obtained from the University of California, Santa Cruz using the Rsubread R Bioconductor package (58, 59). Bigwig files showing signal coverage over the genome were generated with the GenomicAlignments and rtracklayer R Bioconductor packages (60, 61). Peak calling was done with macs2 using default settings, with the exception of using the “–broad” flag for the histone H3K27ac, H3K27me3, and H3K4me1 ChIP-seq experiments (62). Enhancers were defined as H3K27ac peaks that directly overlap H3K4me1 peaks. For H3K27ac, two replicate ChIP-seq experiments were performed and the peaks that were present in both replicates were used to define enhancers. The UTX combined peak set used throughout this study was generated by first calling peaks separately in the wild-type and HEAA UTX cell lines, and then by finding the nonoverlapping union set of peaks. Overlaps between peak sets shown as Venn diagrams were determined using the ChIPpeakAnno R Bioconductor package with a maximum gap between peaks set to 1 kb (63). Hypergeometric statistical tests for overlap between two peak sets were also performed by the ChIPpeakAnno R Bioconductor package. Range-based heatmaps showing signal over genomic regions were generated using the profileplyr R Bioconductor package (64). “Super enhancer” regions were determined with the ROSE software (28, 29). Super enhancers present in both H3K27ac replicate ChIP-seq experiments were included in the analysis. Genic annotation of ChIP-seq peaks was accomplished with the GREAT algorithm (v3.0.0) (33). Any regions included in the ENCODE blacklisted regions of the genome were excluded from all region-specific analyses (65). ChIP-seq coverage tracks (Fig. 1F) were visualized using IGV (v2.3.91) (66).
Acquisition and Analysis of TCGA Data.
TCGA bladder cancer gene expression count data were retrieved using the TCGAbiolinks R Bioconductor package, and differential expression was determined using the DESeq2 R Bioconductor package (55, 67). Mutation data for each tumor and calculations of mutation cooccurrence and mutual exclusivity were downloaded directly from the cBioPortal for Cancer Genomics (7, 8).
Gene Set Enrichment Analysis.
The DESeq2 R Bioconductor package was used to generate a ranked list of genes based on the Wald statistic (log2foldchange/SE of counts) for the wild-type UTX-expressing UMUC1 cells versus the EV control cells in full media (68). The microarray data from Gene Expression Omnibus accession no. GSE41035 was retrieved and expression changes were determined using the affy and limma R Bioconductor packages (40, 69, 70). The average fold-change for each gene for all three FGFR3 shRNA replicates versus the control cells was calculated. Gene set enrichment analysis (v3.0) was run to look for enrichment of genes that go up or down by 1.5-fold after FGFR3 knockdown within the ranked list of UTX-dependent gene-expression changes in UMUC1 cells (68).
HOMER Analysis.
Enrichment of transcription factor motifs within the combined set of wild-type and HEAA UTX peaks in UMUC1 cells was measured using the “findMotifsGenome.pl” function from the HOMER suite of tools using the default background settings (v3.12) and the Homer de novo motif results are reported (71).
Statistical Analysis.
Experimental data are presented as means ± SEM unless stated otherwise. Statistical significance was calculated as indicated in the figure legends.
Supplementary Material
Acknowledgments
We thank members of the C.D.A. laboratory for their feedback regarding this project and manuscript; Leah Gates and Alexey Soshnev for input and illustrations that were helpful in creating the model in Fig. 4G; Dr. Robert Roeder (The Rockefeller University) and Dr. Shu Ping Wang (Institute of Biomedical Sciences at Academia Sinica, Taiwan) for providing reagents and helpful discussion; and The Rockefeller University Genomic Resource Center and Flow Cytometry Resource Center. This work was supported by the following funding from the NIH: R01GM040922 (to C.D.A.) and 5F32CA217068 (to D.B.) and by the St. Jude Epigenetics Collaborative Initiative (to C.D.A.).
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2008017117/-/DCSupplemental.
Data Availability.
The ChIP-seq and RNA-seq data have been deposited in the Gene Expression Omnibus database (accession no. GSE157091). R scripts used for analyses in the main figures are available at https://github.com/dougbarrows/PNAS_UTX_scripts.
References
- 1.Khandelwal P., Abraham S. N., Apodaca G., Cell biology and physiology of the uroepithelium. Am. J. Physiol. Renal Physiol. 297, F1477–F1501 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Czerniak B., Dinney C., McConkey D., Origins of bladder cancer. Annu. Rev. Pathol. 11, 149–174 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Choi W. et al., Identification of distinct basal and luminal subtypes of muscle-invasive bladder cancer with different sensitivities to frontline chemotherapy. Cancer Cell 25, 152–165 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Robertson A. G. et al.; TCGA Research Network , Comprehensive molecular characterization of muscle-invasive bladder cancer. Cell 171, 540–556.e25 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Damrauer J. S. et al., Intrinsic subtypes of high-grade bladder cancer reflect the hallmarks of breast cancer biology. Proc. Natl. Acad. Sci. U.S.A. 111, 3110–3115 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lawrence M. S. et al., Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 499, 214–218 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cerami E. et al., The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2, 401–404 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gao J. et al., Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 6, pl1 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hedegaard J. et al., Comprehensive transcriptional analysis of early-stage urothelial carcinoma. Cancer Cell 30, 27–42 (2016). [DOI] [PubMed] [Google Scholar]
- 10.Agger K. et al., UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature 449, 731–734 (2007). [DOI] [PubMed] [Google Scholar]
- 11.Hong S. et al., Identification of JmjC domain-containing UTX and JMJD3 as histone H3 lysine 27 demethylases. Proc. Natl. Acad. Sci. U.S.A. 104, 18439–18444 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cao R. et al., Role of histone H3 lysine 27 methylation in polycomb-group silencing. Science 298, 1039–1043 (2002). [DOI] [PubMed] [Google Scholar]
- 13.Ler L. D. et al., Loss of tumor suppressor KDM6A amplifies PRC2-regulated transcriptional repression in bladder cancer and can be targeted through inhibition of EZH2. Sci. Transl. Med. 9, eaai8312 (2017). [DOI] [PubMed] [Google Scholar]
- 14.Wang S. P. et al., A UTX-MLL4-p300 transcriptional regulatory network coordinately shapes active enhancer landscapes for eliciting transcription. Mol. Cell 67, 308–321.e6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shpargel K. B., Starmer J., Yee D., Pohlers M., Magnuson T., KDM6 demethylase independent loss of histone H3 lysine 27 trimethylation during early embryonic development. PLoS Genet. 10, e1004507 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shpargel K. B., Sengoku T., Yokoyama S., Magnuson T., UTX and UTY demonstrate histone demethylase-independent function in mouse embryonic development. PLoS Genet. 8, e1002964 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Creyghton M. P. et al., Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci. U.S.A. 107, 21931–21936 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heintzman N. D. et al., Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature 459, 108–112 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rada-Iglesias A. et al., A unique chromatin signature uncovers early developmental enhancers in humans. Nature 470, 279–283 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zentner G. E., Tesar P. J., Scacheri P. C., Epigenetic signatures distinguish multiple classes of enhancers with distinct cellular functions. Genome Res. 21, 1273–1283 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Andricovich J. et al., Loss of KDM6A activates super-enhancers to induce gender-specific squamous-like pancreatic cancer and confers sensitivity to BET inhibitors. Cancer Cell 33, 512–526.e8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gozdecka M. et al., UTX-mediated enhancer and chromatin remodeling suppresses myeloid leukemogenesis through noncatalytic inverse regulation of ETS and GATA programs. Nat. Genet. 50, 883–894 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu F., Wang L., Perna F., Nimer S. D., Beyond transcription factors: How oncogenic signalling reshapes the epigenetic landscape. Nat. Rev. Cancer 16, 359–372 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ghandi M. et al., Next-generation characterization of the cancer cell line encyclopedia. Nature 569, 503–508 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Warrick J. I. et al., FOXA1, GATA3 and PPARIγ cooperate to drive luminal subtype in bladder cancer: A molecular analysis of established human cell lines. Sci. Rep. 6, 38531 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang C. et al., UTX regulates mesoderm differentiation of embryonic stem cells independent of H3K27 demethylase activity. Proc. Natl. Acad. Sci. U.S.A. 109, 15324–15329 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fishwick C. et al., Heterarchy of transcription factors driving basal and luminal cell phenotypes in human urothelium. Cell Death Differ. 24, 809–818 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Whyte W. A. et al., Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 153, 307–319 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lovén J. et al., Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell 153, 320–334 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dontu G. et al., In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev. 17, 1253–1270 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Calvet C. Y., André F. M., Mir L. M., The culture of cancer cell lines as tumorspheres does not systematically result in cancer stem cell enrichment. PLoS One 9, e89644 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mo Q. et al., Prognostic power of a tumor differentiation gene signature for bladder urothelial carcinomas. J. Natl. Cancer Inst. 110, 448–459 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McLean C. Y. et al., GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 28, 495–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ahn J. et al., Target sequencing and CRISPR/Cas editing reveal simultaneous loss of UTX and UTY in urothelial bladder cancer. Oncotarget 7, 63252–63260 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Folkes A. J. et al., The identification of 2-(1H-indazol-4-yl)-6-(4-methanesulfonyl-piperazin-1-ylmethyl)-4-morpholin-4-yl-thieno[3,2-d]pyrimidine (GDC-0941) as a potent, selective, orally bioavailable inhibitor of class I PI3 kinase for the treatment of cancer. J. Med. Chem. 51, 5522–5532 (2008). [DOI] [PubMed] [Google Scholar]
- 36.Yeh T. C. et al., Biological characterization of ARRY-142886 (AZD6244), a potent, highly selective mitogen-activated protein kinase kinase 1/2 inhibitor. Clin. Cancer Res. 13, 1576–1583 (2007). [DOI] [PubMed] [Google Scholar]
- 37.Mohammadi M. et al., Crystal structure of an angiogenesis inhibitor bound to the FGF receptor tyrosine kinase domain. EMBO J. 17, 5896–5904 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guagnano V. et al., Discovery of 3-(2,6-Dichloro-3,5-dimethoxy-phenyl)-1-{6-[4-(4-ethyl- piperazin-1-yl)-phenylamino]-pyrimidin-4-yl}-1-methyl-urea (NVP-BGJ398), A potent and selective inhibitor of the fibroblast growth factor receptor family of receptor tyrosine kinase. J. Med. Chem. 54, 7066–7083 (2011). [DOI] [PubMed] [Google Scholar]
- 39.Nickerson M. L. et al., Concurrent alterations in TERT, KDM6A, and the BRCA pathway in bladder cancer. Clin. Cancer Res. 20, 4935–4948 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Du X. et al., FGFR3 stimulates stearoyl CoA desaturase 1 activity to promote bladder tumor growth. Cancer Res. 72, 5843–5855 (2012). [DOI] [PubMed] [Google Scholar]
- 41.Jozwik K. M., Chernukhin I., Serandour A. A., Nagarajan S., Carroll J. S., FOXA1 directs H3K4 monomethylation at enhancers via recruitment of the methyltransferase MLL3. Cell Rep. 17, 2715–2723 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu W. et al., GATA3 recruits UTX for gene transcriptional activation to suppress metastasis of breast cancer. Cell Death Dis. 10, 832 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Varley C. L. et al., Role of PPARgamma and EGFR signalling in the urothelial terminal differentiation programme. J. Cell Sci. 117, 2029–2036 (2004). [DOI] [PubMed] [Google Scholar]
- 44.Varley C. L., Stahlschmidt J., Smith B., Stower M., Southgate J., Activation of peroxisome proliferator-activated receptor-reverses squamous metaplasia and induces transitional differentiation in normal human urothelial cells. Am. J. Pathol. 164, 1789–1798 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee J. E. et al., H3K4 mono- and di-methyltransferase MLL4 is required for enhancer activation during cell differentiation. eLife 2, e01503 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cho Y. W. et al., PTIP associates with MLL3- and MLL4-containing histone H3 lysine 4 methyltransferase complex. J. Biol. Chem. 282, 20395–20406 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cho Y. W. et al., Histone methylation regulator PTIP is required for PPARgamma and C/EBPalpha expression and adipogenesis. Cell Metab. 10, 27–39 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.de Thé H., Differentiation therapy revisited. Nat. Rev. Cancer 18, 117–127 (2018). [DOI] [PubMed] [Google Scholar]
- 49.Vaz M. et al., Chronic cigarette smoke-induced epigenomic changes precede sensitization of bronchial epithelial cells to single-step transformation by KRAS mutations. Cancer Cell 32, 360–376.e6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Greulich H. et al., Functional analysis of receptor tyrosine kinase mutations in lung cancer identifies oncogenic extracellular domain mutations of ERBB2. Proc. Natl. Acad. Sci. U.S.A. 109, 14476–14481 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liao R. G. et al., Inhibitor-sensitive FGFR2 and FGFR3 mutations in lung squamous cell carcinoma. Cancer Res. 73, 5195–5205 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ran F. A. et al., Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8, 2281–2308 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Soneson C., Love M. I., Robinson M. D., Differential analyses for RNA-seq: Transcript-level estimates improve gene-level inferences. F1000 Res. 4, 1521 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Patro R., Duggal G., Love M. I., Irizarry R. A., Kingsford C., Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 14, 417–419 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wickham H., ggplot2: Elegant Graphics for Data Analysis, (Springer Verlag, 2016). [Google Scholar]
- 57.Kolde R., pheatmap: Pretty heatmaps. The Comprehensive R Archive Network (CRAN) (R package version 1.0.12, 2019).
- 58.Morgan M. et al., ShortRead: A bioconductor package for input, quality assessment and exploration of high-throughput sequence data. Bioinformatics 25, 2607–2608 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liao Y., Smyth G. K., Shi W., The R package Rsubread is easier, faster, cheaper and better for alignment and quantification of RNA sequencing reads. Nucleic Acids Res. 47, e47 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lawrence M. et al., Software for computing and annotating genomic ranges. PLOS Comput. Biol. 9, e1003118 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lawrence M., Gentleman R., Carey V., rtracklayer: An R package for interfacing with genome browsers. Bioinformatics 25, 1841–1842 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang Y. et al., Model-based analysis of ChIP-seq (MACS). Genome Biol. 9, R137 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhu L. J. et al., ChIPpeakAnno: A bioconductor package to annotate ChIP-seq and ChIP-chip data. BMC Bioinf. 11, 237 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Barrows D., Carroll T., profileplyr: Visualization and annotation of read signal over genomic ranges with profileplyr. Bioconductor (R package version 1.2.0, 2019).
- 65.Amemiya H. M., Kundaje A., Boyle A. P., The ENCODE blacklist: Identification of problematic regions of the genome. Sci. Rep. 9, 9354 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Robinson J. T. et al., Integrative genomics viewer. Nat. Biotechnol. 29, 24–26 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Colaprico A. et al., TCGAbiolinks: An R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res. 44, e71 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Subramanian A. et al., Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Nat. Acad. Sci. U.S.A. 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gautier L., Cope L., Bolstad B. M., Irizarry R. A., Affy—Analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 20, 307–315 (2004). [DOI] [PubMed] [Google Scholar]
- 70.Ritchie M. E. et al., Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Heinz S. et al., Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The ChIP-seq and RNA-seq data have been deposited in the Gene Expression Omnibus database (accession no. GSE157091). R scripts used for analyses in the main figures are available at https://github.com/dougbarrows/PNAS_UTX_scripts.